Abstract
Tooth morphogenesis involves dynamic changes in shape and size as it proceeds through the bud, cap, and bell stages. This process requires exact regulation of cell proliferation and differentiation. Smad7, a general antagonist against transforming growth factor–β (TGF-β) signaling, is necessary for maintaining homeostasis and proper functionality in many organs. While TGF-β signaling is widely involved in tooth morphogenesis, the precise role of Smad7 in tooth development remains unknown. In this study, we showed that Smad7 is expressed in the developing mouse molars with a high level in the dental epithelium but a moderate to weak level in the dental mesenchyme. Smad7 deficiency led to a profound decrease in tooth size primarily due to a severely compromised cell proliferation capability in the dental epithelium. Consistent with the tooth shrinkage phenotype, RNA sequencing (RNA-seq) analysis revealed that Smad7 ablation downregulated genes referred to epithelial cell proliferation and cell cycle G1/S phase transition, whereas the upregulated genes were involved in responding to TGF-β signaling and cell cycle arrest. Among these genes, the expression of Cdkn1a (encoding p21), a negative cell proliferation regulator, was remarkably elevated in parallel with the diminution of Ccnd1 encoding the crucial cell cycle regulator cyclin D1 in the dental epithelium. Meanwhile, the expression level of p-Smad2/3 was ectopically elevated in the developing tooth germ of Smad7 null mice, indicating the hyperactivation of the canonical TGF-β signaling. These effects were reversed by addition of TGF-β signaling inhibitor in cell cultures of Smad7−/− molar tooth germs, with rescued expression of cyclin D1 and cell proliferation rate. In sum, our studies demonstrate that Smad7 functions primarily as a positive regulator of cell proliferation via inhibition of the canonical TGF-β signaling during dental epithelium development and highlight a crucial role for Smad7 in regulating tooth size.
Keywords: tooth size, cell proliferation, tooth morphogenesis, negative modulator, TGF-β signaling, dental epithelium
Introduction
Tooth morphogenesis, including growth and patterning, is a complex process based on epithelial-mesenchymal interactions and requires exact temporal and spatial regulation of cell proliferation and differentiation. Many signaling molecules are expressed in the dental epithelium and mesenchyme and are involved in the regulation of growth and eventually tooth size. For example, inactivation of Shh in the dental epithelium results in a cap stage tooth rudiment with a severely reduced tooth size (Dassule et al. 2000). Fgf10, a dental mesenchymal factor, stimulates cell proliferation in the dental epithelium during tooth development (Jernvall and Thesleff 2000; Kettunen et al. 2000). Moreover, our previous study has shown that Wnt5a-deficient mice exhibit smaller teeth as a result of the reduced levels of cell proliferation in both dental epithelium and mesenchyme (Lin et al. 2011). However, the regulatory mechanism of tooth growth remains elusive.
Transforming growth factor–β (TGF-β) signaling plays a key role in tooth development, with a close association with odontoblast maturation and root development (Oka et al. 2007; Li et al. 2017). In addition, TGF-β signaling participates in cell proliferation and apoptosis as a bifunctional regulator during tooth development (Zhao et al. 2008). In the canonical TGF-β signaling pathway, TGF-β ligand binds to the TGF-β type II receptor (TGF-βRII) and then type I receptor (TGF-βRI), and the latter further phosphorylates intracellular Smad protein (Smad2/3). Phosphorylated Smad2/3 (p-Smad2/3) forms a complex with the common Smad (Smad4) and translocates into the nucleus, resulting in transcriptional regulation of target genes (Derynck and Zhang 2003).
As a general antagonist against the TGF-β superfamily, Smad7 prevents the activation of Smad2/3 by competing for binding to TGF-βRI (Sapkota et al. 2006). Moreover, Smad7-induced degradation of TGF-βRI after recruitment of E3 ubiquitin ligases also contributes to the inhibition of TGF-β signaling (Yu et al. 2017). It was reported previously that attenuation of Smad7 in early embryonic tooth germs by antisense oligonucleotide treatment in organ culture inhibited tooth development with increased apoptotic activity in the enamel organ epithelium (Ito et al. 2001). On the other hand, overexpression of Smad7 in the dental epithelium led to furrowed and duplicated tooth formation, resulting from interrupted ameloblast layers and dentin organization (Klopcic et al. 2007). Collectively, while these studies have implicated the functional importance of Smad7 in tooth morphogenesis, the precise role of Smad7 in tooth development remains elusive.
In this study, we investigated the function of Smad7 during tooth morphogenesis using a loss-of-function mouse model. We showed that Smad7 deficiency led to reduced tooth size with attenuation of the cell proliferation rate, especially in the dental epithelium. The abnormally elevated/activated canonical TGF-β signaling in the absence of Smad7 upregulated the level of p21, which was further responsible for inhibition of cyclin D1 expression. Our findings demonstrate that Smad7 orchestrates a network to regulate tooth growth by modulating canonical TGF-β signaling activity during tooth development.
Materials and Methods
Generation of Transgenic Mice
The conditional Smad7 floxed (JAX#017008; The Jackson Laboratory) and Meox2Cre/+ (JAX#003755; The Jackson Laboratory) mouse lines have been described previously (Tallquist and Soriano 2000; Kleiter et al. 2010). Smad7+/− mice were generated by mating Smad7F/F with Meox2Cre/+ mice, followed by segregation of the Meox2Cre/+ allele. The animal studies were approved by the Institutional Animal Care and Use Committee of Tulane University.
Histological Analysis and Immunostaining
Embryonic heads harvested from timed pregnant female mice or newborn pups were fixed in 4% paraformaldehyde (PFA) at 4°C overnight and then decalcified in 10% ethylenediaminetetraacetic acid (EDTA) (pH 7.4) for 2 to 5 d depending on the age of samples. For cryosections, decalcified samples were dehydrated in 30% sucrose phosphate-buffered saline (PBS) solution overnight at 4°C, embedded in optimal cutting temperature compound (Tissue-Plus; Fisher Healthcare), and frozen by liquid nitrogen for solidification. Embedded samples were cryosectioned at 8 µm and subjected to immunofluorescent staining as described previously (Ye et al. 2015; Xu et al. 2018). Information for antibodies used in this study is provided in the Appendix.
RNA Sequencing and Quantitative Reverse Transcription Polymerase Chain Reaction Analyses
For RNA sequencing (RNA-seq), the first lower molar germs were dissected out from E15.5 Smad7−/− and control mice (Smad7+/−) and pooled (3 groups for each genotype), respectively, and subjected to RNA extraction (RNeasy Mini Kit, cat. 74134; Qiagen). Complementary DNA (cDNA) library preparation and sequencing were performed at the Translational Research Center of Tulane University. A total of 260 million 150-bp pair-end reads were obtained on Illumina NextSeq 550 equipment for 3 pairs of samples. Reads were aligned to mm10 using HISAT2 (Pertea et al. 2016). For each library, raw counts for each annotated gene were obtained using the featureCounts software from the Subread package. Differentially expressed genes were identified using DESeq2 (Love et al. 2014) and presented by selecting transcripts that displayed significant changes (P < 0.05) after Benjamini and Hochberg correction. For visualization, aligned reads were uploaded to Integrated Genome Viewer (Yu et al. 2012). A 2-way hierarchical clustering heat map using Euclidean distance and average linkage showed 2 distinct groups of genes from Smad7−/− and Smad7+/− control mice. The raw data have been deposited with Gene Expression Omnibus (GEO) database with accession number GSE130841.
For quantitative reverse transcription polymerase chain reaction (RT-qPCR), the first mandibular molar germs were isolated from E15.5 Smad7−/− and Smad7+/− mice (n = 5 for each genotype) and subjected to RNA extraction. The RNAs were subsequently reversely transcribed into cDNAs. SYBR green and gene-specific primers (Appendix Table) were used and transcript levels were examined by a 7500 Fast Real-Time PCR System (Applied Biosystems). Statistical difference of the RT-qPCR data was analyzed by Student’s t test, and results were presented as mean ± standard deviation. P < 0.05 was considered significant.
Cell Culture, Western Blotting, and CCK8 Assay
For cell culture, the first mandibular molars were isolated and pooled from E15.5 and P0 Smad7−/− and Smad7+/− mice, respectively. To obtain single-cell suspension, isolated tooth germs were subjected to digestion by 4 mg/mL dispase at 37°C for 20 min and then exposed to PBS containing 2.5% pancreatin and 0.5% trypsin at 37°C for another 10 min. Cells were then cultured in α–Modified Eagle’s Medium (α-MEM; HyClone) supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1% penicillin/streptomycin (10,000 U/mL) (Thermo Fisher Scientific), with medium change every 2 d until harvest. Tooth germ cells from either E15.5 or P0 Smad7−/− and control mice were harvested 5 d after treatment with 10 ng/mL SB431542 (Sigma-Aldrich) or mock treated with an equal volume of DMSO as control. For immunoblotting and CCK8 assay, see the Appendix.
Results
Smad7 Is Expressed Primarily in the Dental Epithelium in Developing Embryonic Molars
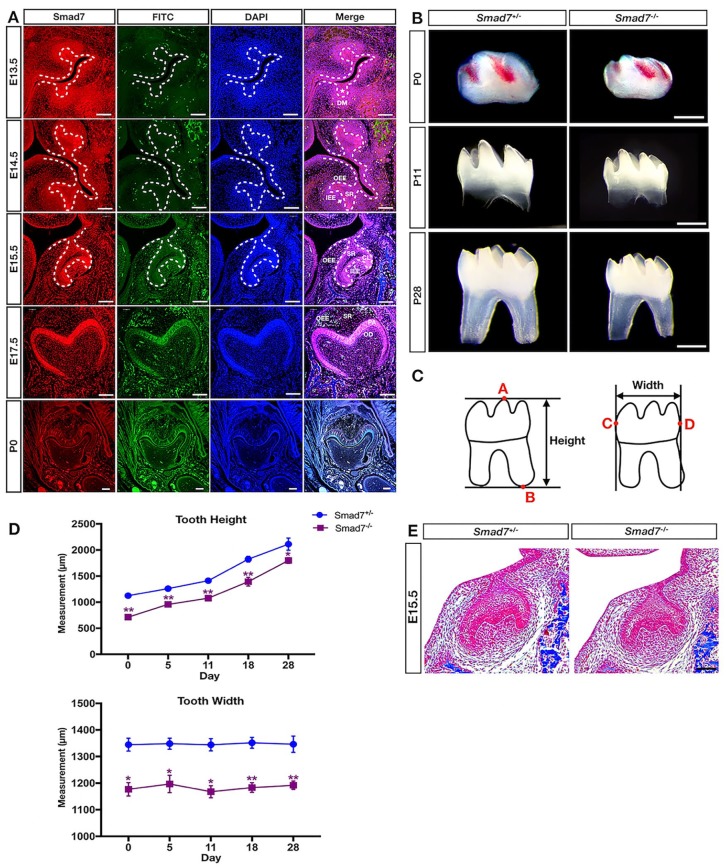
To investigate the role of Smad7 in tooth development, we started with examining the spatial and temporal expression of Smad7 by immunofluorescent staining in the developing embryonic molars (Fig. 1A). At E13.5 (the bud stage), strong Smad7 staining was detected in the dental epithelium (DE), while moderate to weak staining for Smad7 was seen in the dental mesenchyme (DM). As tooth morphogenesis advanced to the cap stage (E14.5), moderate to intense staining was found in the inner enamel epithelium (IEE), while weak signals were detected in the stellate reticulum (SR), outer enamel epithelium (OEE), primary enamel knot (PEK), and cervical loop (CL). At E15.5 (the late cap stage), intense staining was present in DM, IEE, OEE, and SR. At the bell stage (E17.5), strong staining was found in the preodontoblasts and preameloblasts. In contrast, Smad7 expression was downregulated and became barely detectable at postnatal day 0 (P0).
Figure 1.
Smad7 deficiency leads to reduced tooth size. (A) Immunofluorescent staining of Smad7 during tooth morphogenesis at the bud stage (E13.5), cap stage (E14.5), early bell stage (E15.5), late bell stage (E17.5), and postnatal stage (P0). (B) Representative images of control and Smad7−/− lower first molar at P0, P11, and P28 show unaltered dental patterning with reduced size in the mutants. (C) Schematic representation of the methods used for measuring the molar size, including height and width. Point A: the tip of the highest cusp of the molar; point B: the bottom of the molar root; point C: the most convex point on the mesial surface of the tooth crown; point D: the most convex point on the distal surface of the tooth crown. (D) Measurements of tooth size of the control and Smad7−/− first mandibular molars at different postnatal time points (n = 4 for each time point). Statistical analysis was performed using Student’s t test. *P < 0.05. **P < 0.01. (E) Representative histology from the control and Smad7−/− first mandibular molars at E15.5 shows discernably reduced size in the mutant. CL, cervical loop; DE, dental epithelium (marked by asterisk); DM, dental mesenchyme; IEE, inner enamel epithelium; OEE, outer enamel epithelium; PEK, primary enamel knot (indicated by arrow); SR, stellate reticulum. Scale bars: 500 µm (B); 100 µm (A and E).
Smad7-Deficient Mice Exhibit Reduced Size of the First Mandibular Molar
Since Smad7 is expressed in the both dental epithelium and mesenchyme, we subsequently inactivated Smad7 in each tissue component using Smad7 floxed mice compounded with Wnt1-Cre or K14-Cre allele. However, we did not find any obvious tooth phenotype in Wnt1-Cre;Smad7F/F mice but observed a recognizable reduction only in tooth height in the K14-Cre;Smad7F/F molar at weaned age (Appendix Fig. 1). We then decided to assess teeth from Smad7 null mice (Smad7−/−), which survived to adulthood as reported previously (Xu et al. 2003; Tojo et al. 2012). Surprisingly, we found significantly reduced size of the molar teeth in Smad7 mutants at various ages, beginning as early as P0 (Fig. 1B–D), despite that crown and root formation and patterning, tooth eruption, as well as differentiation status assessed by histology, appeared comparable to controls (Fig. 1B and data not shown). Smad7 apparently exerts its effects during the embryonic stage, consistent with its strong expression in the developing molar before birth (Fig. 1A). Indeed, histological examination identified discernible reduced size of the molar at E15.5 in the mutants (Fig. 1E). It is worth noting that skeletal preparations and visual inspection revealed identical size of the heads and incisors from mutants and controls at P0, indicating a specific function of Smad7 in molar development (Appendix Figs. 2 and 3).
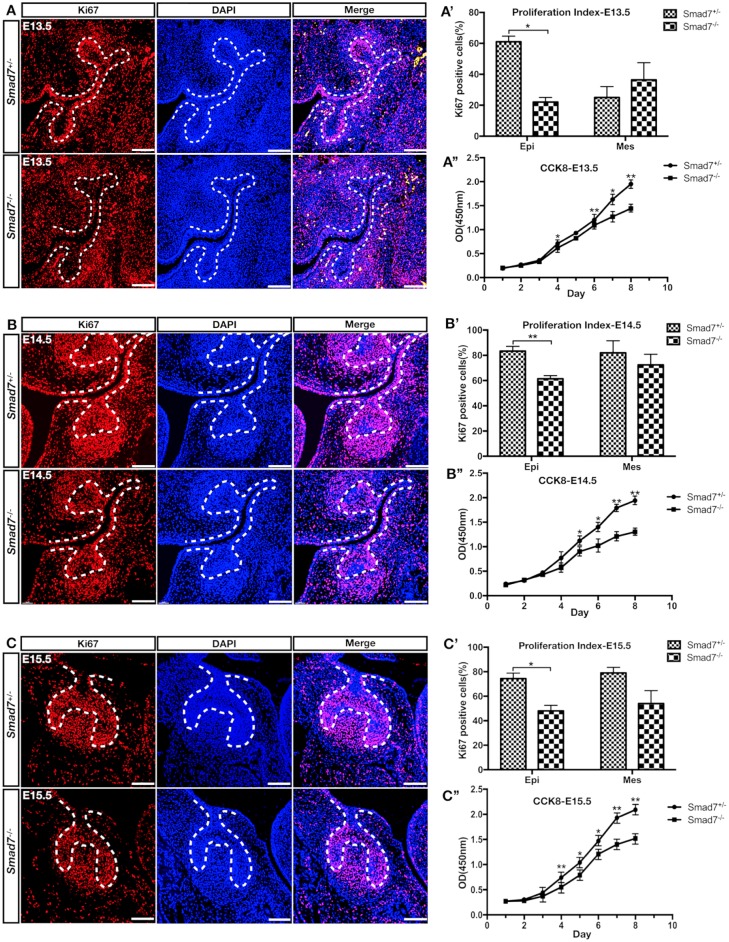
Disruption of Smad7 Causes Decreased Cell Proliferation Primarily in the Dental Epithelium
Cell proliferation and apoptosis are 2 critical factors contributing to the size of an organ or tissue. To explore if Smad7 could act on cell proliferation, we performed a cell proliferation assay using Ki-67 antibody from E13.5 to E15.5, stages before and after the size phenotype became recognizable (Fig. 2A–C). As shown in Figure 2A′–C′, the ratio of Ki-67+ cells in the Smad7−/− dental epithelium was statistically significantly reduced compared to control group throughout the stages examined (P < 0.05). In contrast, the molar mesenchyme did not show much difference in terms of the ratio of Ki-67+ cells (no statistical significance). Moreover, in vitro CCK8 assay demonstrated that the cell proliferation rate in the Smad7−/− molar was much lower compared to that of control from day 4 to day 8 at all timepoints (Fig. 2A′′–C′′). However, cell apoptosis assay at E14.5 and E15.5 using cleaved caspase-3 antibody for immunostaining and Annexin V-FITC/PI double staining for flow cytometry gave rise to comparable results between mutants and controls (Appendix Fig. 4). These results indicate that the loss of Smad7 primarily compromises the proliferative capability of the dental epithelial cells, representing a cellular defect responsible for the small tooth phenotype.
Figure 2.
Inactivation of Smad7 leads to compromised cell proliferation in the developing molar. (A–C) Immunofluorescent staining of Ki-67 on sections of control and Smad7−/− molar germs at E13.5 (A), E14.5 (B), and E15.5 (C). (A′–C′) Quantification of Ki-67–positive cells in the dental epithelium and mesenchyme of control and Smad7−/− tooth germs at E13.5 (A′), E14.5 (B′), and E15.5 (C′). (A′′–C′′) Growth curves of control and Smad7−/− tooth germ cells plotted from CCK8 assays at E13.5 (A′′), E14.5 (B′′), and E15.5 (C′′). Statistical analysis was performed using Student’s t test. *P < 0.05. **P < 0.01. Scale bars: 100 µm.
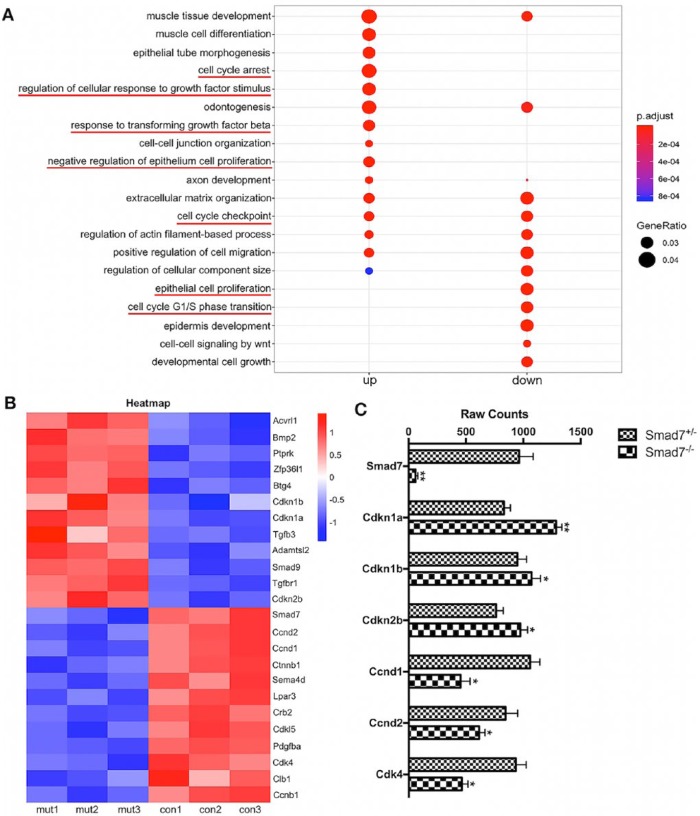
Inhibition of Smad7 Activates TGF-β Signaling and Orchestrates a Transcriptional Regulation of Cell Proliferation
Smad7 is known to regulate both Smad-dependent and Smad-independent TGF-β signaling pathways but is also extensively involved in crosstalks with other signaling pathways. To investigate the underlying molecular mechanism, we conducted RNA-seq analysis on E15.5 molars from Smad7 mutant and control mice. Gene ontology (GO) analysis demonstrated that in Smad7−/− tooth, the upregulated genes were primarily involved in cellular response to transforming growth factor stimulus and cell cycle arrest, while the downregulated genes were mainly referred to epithelium cell proliferation and cell cycle G1/S transition (Fig. 3A). In particular, the augmented signaling pathways were mostly related to canonical and noncanonical TGF-β signaling pathways, including JNK, MAPK, and ERK cascades, where Smad7 functions as an inhibitor. Among the differentially expressed genes, we identified several genes that play crucial roles in cell proliferation (i.e., Ccnd1, Ccnd2, Cdkn1a, Cdk4, and Ccnb1) and the regulation of TGF-β signaling pathways (i.e., Acvrl1, Tgfb3, Tgfbr1, Smad7, and Adamtsl2), as exemplified in Figures 3B, C.
Figure 3.
RNA sequencing (RNA-seq) analysis identifies the gene expression profile regulated by Smad7. (A) Gene ontology (GO) analysis shows that genes that were upregulated and downregulated, respectively, in the Smad7−/− first mandibular molar at E15.5. (B) Heatmap shows z scores (interpreted as a measure of SD away from the mean) for some selected genes from E15.5 RNA-seq data. The color scheme is based on the z scores, with upregulation in red, downregulation in blue, and undetermined directionality in white. (C) Raw counts from RNA-seq illustrate the magnificently decreased expression levels of Smad7 and 6 selected genes, including Cdkn1a, Cdkn1b, Cdkn2b, Ccnd1, Ccnd2, and Cdk4, that are critical for cell proliferation. Statistical analysis was performed using Student’s t test. *P < 0.05. **P < 0.01. This figure is available in color online.
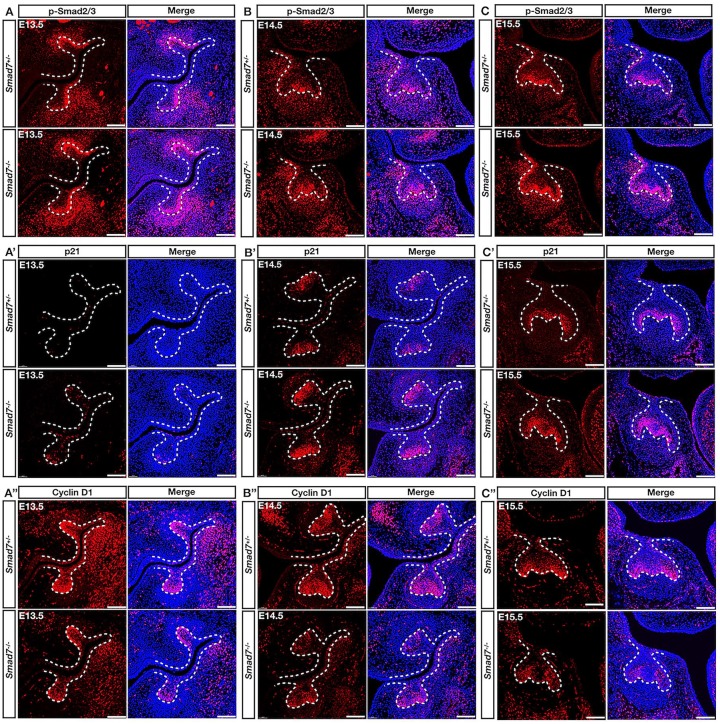
Because the RNA-seq analysis assayed a heterogeneous population, we further validated the altered expression of interesting candidate genes in the first mandibular molar by immunostaining. At E13.5, the level of p-Smad2/3 was increased in both the dental epithelium and mesenchyme in Smad7−/− mice compared to controls, suggesting the enhanced canonical TGF-β signaling activity (Fig. 4A–C). Interestingly, in the same region of the dental epithelium, we observed an ectopic activation of p21 that negates cell proliferation by linking DNA damage to cell cycle arrest (Fig. 4A′–C′). Meanwhile, the expression of cyclin D1, a positive regulator contributing to the cell cycle transition, was remarkably reduced in the dental epithelium (Fig. 4A′′–C′′). At E14.5 and E15.5, the boosted expression of p-Smad2/3 and p21 in parallel with the dramatically decreased cyclin D1 gradually led to identifiable small tooth size. Taken together, these results demonstrate that Smad7 fine-tunes the canonical TGF-β signaling to regulate a cell proliferation-related network primarily in the developing dental epithelium.
Figure 4.
Hyperactivation of canonical transforming growth factor–β (TGF-β) signaling and altered expression of p21 and cyclin D1 in the Smad7−/− dental epithelium. (A–C) Immunostaining shows the enhanced activity of p-Smad2/3 in the dental epithelium of Smad7−/− molar at E13.5 (A), E14.5 (B), and E15.5 (C). (A′–C′) Immunostaining of p21 demonstrates dramatically enhanced expression level in the dental epithelium of Smad7−/− tooth at all stages examined, which colocalizes with the enhanced p-smad2/3 activity. (A′′–C′′) Immunostaining shows significantly decreased expression level of cyclin D1 in the dental epithelium at all stages examined in Smad7−/− tooth, overlapping with hyperactivated sites of p-smad2/3 and the enhanced expression level of p21. Scale bars: 100 µm.
TGF-β Signaling Attenuates Cell Proliferation via Activation of p21 Expression and Suppression of Cyclin D1 Expression in the Dental Epithelium
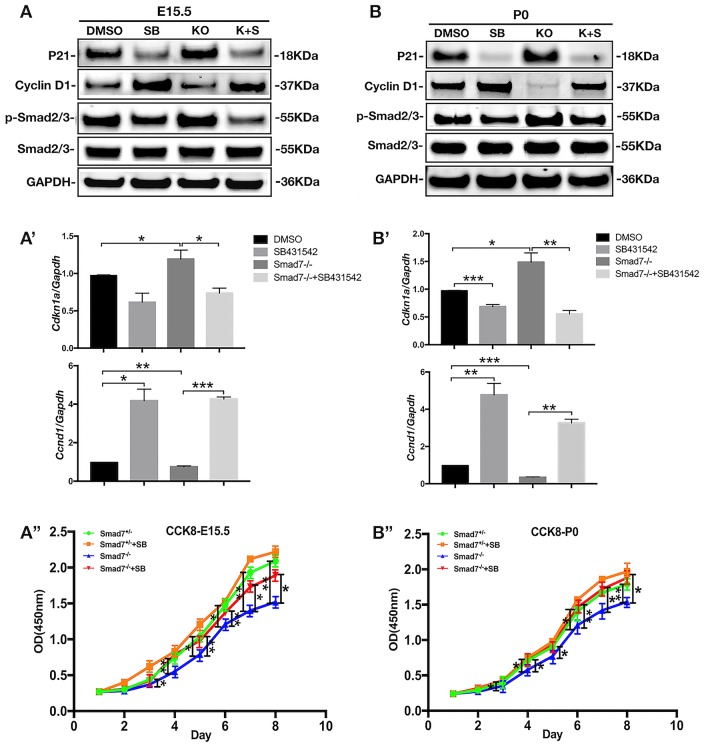
We next set to determine whether the enhanced canonical TGF-β signaling activity in the Smad7−/− molar epithelium was responsible for the reduced cell proliferation rate. We cultured cells suspended from molar germs isolated from E15.5 or P0 Smad7−/− and control mice in the presence and absence of SB431542, a potent antagonist of TGF-βRI (Koo et al. 2015). After 5 d in culture, Western blot and RT-qPCR analyses showed an augmented level of both p-Smad2/3 and p21 in Smad7−/− cells in the absence of SB431542, as compared to controls (Fig. 5A, 5B, 5A′, 5B′). On the other hand, cyclin D1 expression was pronouncedly decreased simultaneously. Notably, in the presence of SB431542, Smad7−/− cells exhibited a reverse behavior, showing reduced expression levels of p-Smad2/3 and p21, along with significantly elevated expression of cyclin D1. The results of CCK8 assay were in line with the Western blot and RT-qPCR analyses, showing increased growth of Smad7−/− dental cells in the presence of SB431542 (Fig. 5A′′, 5B′′). These results provide strong evidence for a specific function of Smad7 in regulating cell proliferation in the developing dental epithelium by modulating the activity of TGF-β signaling that in turn controls the expression of cell proliferation regulators, including p21 and cyclin D1. However, the possible contribution of Smad7 in the dental mesenchyme to the regulation of cell proliferation cannot be ruled out.
Figure 5.
Smad7 ablation compromises cell proliferation via the enhanced canonical transforming growth factor–β (TGF-β) signaling. (A, B) Western blot analyses show the levels of p21, cyclin D1, and p-Smad2/3 in control and Smad7−/− tooth germ cells harvested at E15.5 (A) and at P0 (B) after 5 d in cell culture in the presence or absence of SB431542. (A′, B′) Quantitative reverse transcription polymerase chain reaction analyses of Cdkn1a and Ccnd1 expression in control and Smad7−/− tooth germ cells harvested at E15.5 (A′) and at P0 (B′) after 5 d in cell culture in the presence or absence of SB431542. (A′′, B′′) Growth curves, plotted from CCK8 assays, of control and Smad7−/− tooth germ cells harvested at E15.5 (A′′) and at P0 (B′′) in the presence or absence of SB431542. Statistical analysis was performed using Student’s t test. *P < 0.05. **P < 0.01. ***P < 0.001.
Discussion
As a key event during tooth morphogenesis, cell proliferation is regulated by various genes and signaling molecules. Although many lines of evidence have supported that Smad7 is necessary for maintaining homeostasis and proper functionality in many organs and is also implicated in tooth development, the precise in vivo function of Smad7 in tooth development remained unknown. In this study, we have presented direct evidence that Smad7 plays a critical role in molar development by modulating the canonical TGF-β signaling activity to regulate cell proliferation. We found that size reduction occurs initially in the embryonic stage, consistent with restricted Smad7 expression in the developing tooth before birth. Despite Smad7 expression in the both dental epithelium and mesenchyme, the impaired cell proliferation was found primarily in the dental epithelium, which leads to reduced tooth size. Since tissue-specific inactivation of Smad7 in the dental epithelium did not produce the similar small tooth phenotype as seen in Smad7 null mice, Smad7 expression in the dental mesenchyme apparently also contributes to the regulation of tooth growth. Taking advantage of genome-wide gene expression profiling technology, we further identified several molecules critical to cell proliferation regulation, including p21 and cyclin D1, whose expression was dramatically altered along with enhanced canonical TGF-β signaling activity in tooth germs lacking Smad7. These altered levels of p21 and cyclin D1 expression as well as impaired cell proliferation could be rescued in cultured Smad7−/− dental cells by inhibition of canonical TGF-β signaling activity, pinpointing to the precise function of Smad7 in modulating TGF-β signaling during embryonic tooth development.
TGF-β signaling controls multiple fundamental aspects of cellular behavior and biological process, including odontogenesis. Loss of TGF-βRI (ALK5) resulted in delayed tooth initiation and development with increased apoptosis in the dental epithelium (Zhao et al. 2008). Furthermore, abrogation of TGF-βRII and Smad2 function using antisense ODN increased the tooth size and advanced the stage of tooth formation during mandibular morphogenesis, owing to increased cell proliferation of enamel organ (Chai et al. 1999; Ito et al. 2001). In addition, Smad2 overexpression reduced the level of the oral epithelium proliferation (Alotaibi et al. 2014). Known as an inhibitor of TGF-β superfamily, Smad7 can antagonize the canonical TGF-β signaling by interfering with the recruitment of Smad2/3 and induce degradation of ALK5 (Lebrun et al. 1999). More important, as demonstrated in our study, the spatial and temporal distribution (strong in the dental epithelium and relatively weak in the mesenchyme) of Smad7 matches precisely with the distribution of TGF-β ligand and its cognate receptors during early tooth development (Li and Pan 2017; Kahata et al. 2018). This overlapped expression strengthens a negative feedback regulatory role of Smad7 in TGF-β signaling during tooth development.
Cytostatic effect is one of the well-defined functions of TGF-β signaling. It has been proven that TGF-β–induced growth arrest occurs in the mid-late G1 phase and mainly through 2 interconnected processes (Matsuura et al. 2004). The first one comes from the repression of expression of certain growth-promoting transcription factors like c-Myc, Id1, Id2, and Id3, which leads to growth inhibition (Kang et al. 2003; Siegel et al. 2003; Morikawa et al. 2016). As presented in our study, enhanced TGF-β signaling activity in the absence of Smad7 resulted in compromised proliferation capability of the developing tooth germ. Especially, cyclin D1, an essential regulator of the G1-S transition in response to growth factor, was remarkably downregulated in the Smad7−/− molar epithelium. There was an overlap in the region of decreased cyclin D1 and increased p-Smad2/3 in the IEE. In line with the in vivo observations, in vitro assay also showed that the attenuated expression of cyclin D1 in Smad7−/− dental cells could be rescued after suppression of TGF-β signaling. Enhanced canonical TGF-β signaling is thus responsible for the inhibition of cyclin D1 expression, which in turn leads to a reduced level of cell proliferation. Cyclin D1 is known to regulate cell cycle via binding to cyclin-dependent kinase (CDK) to form the regulatory complex. TGF-β1 has been shown to inhibit CDK expression and therefore restrain cyclin D1 in intestinal epithelial cells (Ko et al. 1995). However, in chondrocytes, TGF-β signaling stimulates cyclin D1 expression through activation of β-catenin signaling (Li et al. 2006). Under certain circumstances, cyclin D1 can be manipulated as a downstream target of p21, a cell cycle negative regulator (Sandor et al. 2000; Ungefroren et al. 2011; Dai et al. 2017). Nevertheless, the underlying molecular mechanism of how cyclin D1 expression is regulated by TGF-β signaling remains obscure and warrants future investigation.
Induction of expression of CDK inhibitors is another significant mechanism of the TGF-β signaling-induced cytostatic effect. As a potent CDK inhibitor, p21 can be induced by activation of TGF-β signaling and leads to cell cycle arrest and antiproliferation (Murray 2004; Zhang et al. 2017). In the developing tooth, p21 has been shown to be a critical regulator involved in enamel knot formation (Jernvall et al. 1998; Kwon et al. 2015). In our study, the increased level of p21 expression apparently did not affect tooth patterning, as evidenced by the unaffected number of tooth cusp, cusp shape, and normal tooth morphogenesis. Nevertheless, while the upregulated p21 does not affect tooth patterning, its inhibitory role in cell proliferation represents a major factor contributing to the reduced rate of cell proliferation and ultimately to the small tooth phenotype.
In sum, our results presented here demonstrate a novel role for Smad7 in the regulation of cell proliferation in the dental epithelium by mediating the expression of p21 and cyclin D1 via the modulation of canonical TGF-β signaling. Given the fact that the TGF-β superfamily, including TGF-β and BMP subfamilies, plays multiple roles in tooth development, our study highlights the critical function of a negative modulator in organogenesis.
Author Contributions
Z. Liu, T. Chen, contributed to conception, design, data acquisition, and analysis, drafted the manuscript; D. Bai, W. Tian, Y. Chen, contributed to conception, design, and data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034519872487 for Smad7 Regulates Dental Epithelial Proliferation during Tooth Development by Z. Liu, T. Chen, D. Bai, W. Tian and Y. Chen in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
This work was supported by a National Institutes of Health grant (R01 DE024152) to Y. Chen, a grant (2017YFA 0104800) from National Key Research and Development Program of China to W. Tian, and a grant (81870804) from the Natural Science Foundation of China to D. Bai. Both Z. Liu and T. Chen were supported in part by a fellowship from the China Scholarship Council.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Alotaibi MK, Kitase Y, Shuler CF. 2014. Smad2 overexpression reduces the proliferation of the junctional epithelium. J Dent Res. 93(9):898–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Zhao J, Mogharei A, Xu B, Bringas P, Jr, Shuler C, Warburton D. 1999. Inhibition of transforming growth factor-beta type II receptor signaling accelerates tooth formation in mouse first branchial arch explants. Mech Dev. 86(1–2):63–74. [DOI] [PubMed] [Google Scholar]
- Dai M, Al-Odaini AA, Fils-Aime N, Villatoro MA, Guo J, Arakelian A, Rabbani SA, Ali S, Lebrun JJ. 2017. Erratum to: cyclin D1 cooperates with p21 to regulate TGFbeta-mediated breast cancer cell migration and tumor local invasion. Breast Cancer Res. 19(1):43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 127(22):4775–4785. [DOI] [PubMed] [Google Scholar]
- Derynck R, Zhang YE. 2003. Smad-dependent and smad-independent pathways in TGF-beta family signalling. Nature. 425(6958):577–584. [DOI] [PubMed] [Google Scholar]
- Ito Y, Zhao J, Mogharei A, Shuler CF, Weinstein M, Deng C, Chai Y. 2001. Antagonistic effects of Smad2 versus Smad7 are sensitive to their expression level during tooth development. J Biol Chem. 276(47):44163–44172. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. 1998. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 125(2):161–169. [DOI] [PubMed] [Google Scholar]
- Jernvall J, Thesleff I. 2000. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 92(1):19–29. [DOI] [PubMed] [Google Scholar]
- Kahata K, Dadras MS, Moustakas A. 2018. TGF-beta family signaling in epithelial differentiation and epithelial-mesenchymal transition. Cold Spring Harb Perspect Biol. 10(1). pii: a022194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y, Chen CR, Massague J. 2003. A self-enabling TGFbeta response coupled to stress signaling: Smad engages stress response factor ATF3 for Id1 repression in epithelial cells. Mol Cell. 11(4):915–926. [DOI] [PubMed] [Google Scholar]
- Kettunen P, Laurikkala J, Itaranta P, Vainio S, Itoh N, Thesleff I. 2000. Associations of FGF-3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 219(3):322–332. [DOI] [PubMed] [Google Scholar]
- Kleiter I, Song J, Lukas D, Hasan M, Neumann B, Croxford AL, Pedre X, Hovelmeyer N, Yogev N, Mildner A, et al. 2010. Smad7 in T cells drives T helper 1 responses in multiple sclerosis and experimental autoimmune encephalomyelitis. Brain. 133(Pt 4):1067–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopcic B, Maass T, Meyer E, Lehr HA, Metzger D, Chambon P, Mann A, Blessing M. 2007. TGF-beta superfamily signaling is essential for tooth and hair morphogenesis and differentiation. Eur J Cell Biol. 86(11–12):781–799. [DOI] [PubMed] [Google Scholar]
- Ko TC, Sheng HM, Reisman D, Thompson EA, Beauchamp RD. 1995. Transforming growth factor-beta 1 inhibits cyclin D1 expression in intestinal epithelial cells. Oncogene. 10(1):177–184. [PubMed] [Google Scholar]
- Koo BH, Kim Y, Cho YJ, Kim DS. 2015. Distinct roles of transforming growth factor-beta signaling and transforming growth factor-beta receptor inhibitor SB431542 in the regulation of p21 expression. Eur J Pharmacol. 764:413–423. [DOI] [PubMed] [Google Scholar]
- Kwon HJ, Park EK, Jia S, Liu H, Lan Y, Jiang R. 2015. Deletion of Osr2 partially rescues tooth development in Runx2 mutant mice. J Dent Res. 94(8):1113–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun JJ, Takabe K, Chen Y, Vale W. 1999. Roles of pathway-specific and inhibitory Smads in activin receptor signaling. Mol Endocrinol. 13(1):15–23. [DOI] [PubMed] [Google Scholar]
- Li J, Parada C, Chai Y. 2017. Cellular and molecular mechanisms of tooth root development. Development. 144(3):374–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Pan Y. 2017. Differential expression of transforming growth factor-beta1, connective tissue growth factor, phosphorylated-SMAD2/3 and phosphorylated-ERK1/2 during mouse tooth development. J Mol Histol. 48(5–6):347–355. [DOI] [PubMed] [Google Scholar]
- Li TF, Chen D, Wu Q, Chen M, Sheu TJ, Schwarz EM, Drissi H, Zuscik M, O’Keefe RJ. 2006. Transforming growth factor-beta stimulates cyclin d1 expression through activation of beta-catenin signaling in chondrocytes. J Biol Chem. 281(30):21296–21304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Li L, Liu C, Liu H, He F, Yan F, Zhang Y, Chen Y. 2011. Wnt5a regulates growth, patterning, and odontoblast differentiation of developing mouse tooth. Dev Dyn. 240(2):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura I, Denissova NG, Wang G, He D, Long J, Liu F. 2004. Cyclin-dependent kinases regulate the antiproliferative function of Smads. Nature. 430(6996):226–231. [DOI] [PubMed] [Google Scholar]
- Morikawa M, Derynck R, Miyazono K. 2016. TGF-beta and the TGF-beta family: context-dependent roles in cell and tissue physiology. Cold Spring Harb Perspect Biol. 8(5). pii: a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW. 2004. Recycling the cell cycle: cyclins revisited. Cell. 116(2):221–234. [DOI] [PubMed] [Google Scholar]
- Oka S, Oka K, Xu X, Sasaki T, Bringas P, Jr, Chai Y. 2007. Cell autonomous requirement for TGF-beta signaling during odontoblast differentiation and dentin matrix formation. Mech Dev. 124(6):409–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. 2016. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 11(9):1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor V, Senderowicz A, Mertins S, Sackett D, Sausville E, Blagosklonny MV, Bates SE. 2000. P21-dependent g(1)arrest with downregulation of cyclin d1 and upregulation of cyclin E by the histone deacetylase inhibitor fr901228. Br J Cancer. 83(6):817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota G, Knockaert M, Alarcon C, Montalvo E, Brivanlou AH, Massague J. 2006. Dephosphorylation of the linker regions of smad1 and Smad2/3 by small C-terminal domain phosphatases has distinct outcomes for bone morphogenetic protein and transforming growth factor-beta pathways. J Biol Chem. 281(52):40412–40419. [DOI] [PubMed] [Google Scholar]
- Siegel PM, Shu W, Massague J. 2003. Mad upregulation and Id2 repression accompany transforming growth factor (TGF)-beta-mediated epithelial cell growth suppression. J Biol Chem. 278(37):35444–35450. [DOI] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. 2000. Epiblast-restricted cre expression in more mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 26(2):113–115. [DOI] [PubMed] [Google Scholar]
- Tojo M, Takebe A, Takahashi S, Tanaka K, Imamura T, Miyazono K, Chiba T. 2012. Smad7-deficient mice show growth retardation with reduced viability. J Biochem. 151(6):621–631. [DOI] [PubMed] [Google Scholar]
- Ungefroren H, Groth S, Sebens S, Lehnert H, Gieseler F, Fandrich F. 2011. Differential roles of Smad2 and Smad3 in the regulation of TGF-beta1-mediated growth inhibition and cell migration in pancreatic ductal adenocarcinoma cells: control by Rac1. Mol Cancer. 10:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Huang Z, Wang W, Tan X, Li H, Zhang Y, Tian W, Hu T, Chen YP. 2018. FGF8 signaling alters the osteogenic cell fate in the hard palate. J Dent Res. 97(5):589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Jeong L, Han J, Ito Y, Bringas P, Jr, Chai Y. 2003. Developmental expression of Smad1-7 suggests critical function of TGF-beta/BMP signaling in regulating epithelial-mesenchymal interaction during tooth morphogenesis. Int J Dev Biol. 47(1):31–39. [PubMed] [Google Scholar]
- Ye W, Wang J, Song Y, Yu D, Sun C, Liu C, Chen F, Zhang Y, Wang F, Harvey RP, et al. 2015. A common Shox2-Nkx2-5 antagonistic mechanism primes the pacemaker cell fate in the pulmonary vein myocardium and sinoatrial node. Development. 142(14):2521–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang LG, Han Y, He QY. 2012. Clusterprofiler: an R package for comparing biological themes among gene clusters. OMICS. 16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Gu S, Li W, Sun C, Chen F, Xiao M, Wang L, Xu D, Li Y, Ding C, et al. 2017. Smad7 enables STAT3 activation and promotes pluripotency independent of TGF-beta signaling. Proc Natl Acad Sci USA. 114(38):10113–10118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Alexander PB, Wang XF. 2017. TGF-beta family signaling in the control of cell proliferation and survival. Cold Spring Harb Perspect Biol. 9(4). pii: a022145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Oka K, Bringas P, Kaartinen V, Chai Y. 2008. TGF-beta type I receptor Alk5 regulates tooth initiation and mandible patterning in a type II receptor-independent manner. Dev Biol. 320(1):19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034519872487 for Smad7 Regulates Dental Epithelial Proliferation during Tooth Development by Z. Liu, T. Chen, D. Bai, W. Tian and Y. Chen in Journal of Dental Research