Abstract
Background
microRNAs play a critical role in many biological processes such as cell proliferation and maturation, apoptosis, regulation of chronic inflammation and development of cancer.
Methods
In this study is described a protocol for the isolation of RNA from serum and subsequent determination of miRNA expression levels using TaqMan‐based MGB Real‐Time PCR detection. RNA was extracted using two different isolation methods including available kits RNAzol and a modified RNAzol protocol. In all cases, RNA was eluted in RNase free H2O, kept frozen until analysis and the presence of contaminants assessed by NanoDrop spectrophotometry.
Results
Higher RNA quantity was observed in RNAzol (378.8 ng/μl) vs RNAzol modified protocol (226.5 ng/μl) and a better performance in terms of RNA extraction yield and purity. Subsequently, measurements of endogenous miRNAs (RNU43), cellular miRNAs (mir155 and mir146a) and EBV miRNAs (mirBART2‐5p, mirBART15 and mirBART22) were performed by RT‐qPCR.
Conclusion
In contrast to the findings in terms of purity and quantity, the amplifiable RNA was more abundant using RNAzol modified protocol compared to not modified protocol.
Keywords: extraction, MGB probe, microRNA, real‐time PCR, retrotrascription, serum, stem‐loop primers
INTRODUCTION
The term microRNA (miRNA) comprises a family of highly conserved small noncoding RNAs (about 22 nt), which regulate gene expression at posttranscriptional level. Discovered in 1993, these endogenous noncoding transcripts represent approximately 1–2% known genes in eukaryotes and negatively regulate gene expression by repression or degradation of target mRNAs 1, 2. miRNAs play a critical role in many biological processes such as cell proliferation and maturation, apoptosis, regulation of chronic inflammation, and cancer development 3. Furthermore, these molecules play critical roles in different human diseases, including cancers, neurodegenerative disorders, and autoimmune diseases 4, 5, 6. The extremely small size of miRNAs renders most conventional biological amplifications tools less effective, taking in consideration that small RNAs could be less efficiently precipitated in alcohol solutions during the purification process. Furthermore, the close similarities among family members of miRNAs have introduced many challenges for developing miRNA‐specific detection assays, inducing deep differences in miRNAs’ detection and quantification approaches 7, 8.
miRNAs have been considered promising candidates for the next generation of diagnostic biomarkers thanks to the strong correlation between miRNA expression patterns and disease status, demonstrated not only in animal models but also in patients 9. While disease‐associated miRNA signatures have been largely identified in solid tissues obtained by invasive procedures, recent proof‐of‐concept studies suggest that serum miRNAs are reliable indicators of disease status 10, asserting their potential value in early and noninvasive diagnosis. Importantly, miRNAs are remarkably stable in serum for extended periods of time because of two mechanisms: (1) formation of a ribonucleoprotein complex with Argonaute proteins and (2) incorporation into exosomes 11.
Next‐generation sequencing initiated and expanded the identification of several new, rare or tissue‐specific small RNAs in an unprecedented manner. In recent years, hundreds of new small RNA variants have been identified by next‐generation sequencing 12. However, one important limitation of the current RNA sequencing approaches for studying small RNAs is the complexity of sample preparation that makes it difficult to generate quantitative data across large sample numbers 13. For this reason, to generate quantitative data through several sample sets of newly identified or well‐known small RNAs, Northern‐blot or quantitative PCR remains important technique in these studies.
Stem‐loop RT‐qPCR was developed for specific and efficient quantification of small RNAs and became a widely used technique primarily for canonical, well‐characterized small RNAs 14. An advantage of the method is that it enables the specific detection of the mature, processed miRNA molecules even from nanograms of total RNA. This technique, widely used for detecting well‐annotated miRNA molecules from humans or common model organisms, includes two steps: small RNA specific stem‐loop primer (SLP) based reverse transcription (RT) and quantification of RT products using conventional TaqMan assay with a small RNA specific TaqMan probe and forward primer 14.
In this study is described a protocol development for the isolation of RNA from serum and subsequent determination of miRNA expression levels using TaqMan Minor Groove Binder (MGB) based real‐time PCR detection with a total turnaround time of approximately 8 h.
MATERIALS AND METHODS
Study Population
Whole‐blood samples were previously collected, during a randomized, double‐blind controlled trial, in red‐top Vacutainer tubes (Becton–Dickinson, Rutherford, NJ) from 12 patients diagnosed with PID (Primary Immunodeficiency Disease) and from 20 healthy blood donors. Blood samples were left at room temperature for 30 min to allow complete coagulation. Coagulated samples were then spun at 1500 g for 15 min at 4°C to separate serum. The serum was transferred to a new CryoTube (Nunc, Rochester, NY) with care not to disturb the buffy coat. Serum samples were immediately frozen at −80°C until RNA extraction.
RNA Extraction Methods
Two different commercial kits/reagents were employed, following manufacturer's instructions, to test which was most suitable for obtaining RNA from human sera samples: RNAzol‐RT (Cat nu R4533, Sigma‐Aldrich, St. Louis) and RNAzol protocol modified by Taylor et al. 15. In both cases, a starting volume of 400 μL of thawed serum or freshly isolated serum was used.
RNA Quality and Integrity Evaluation
RNA purity and concentration were evaluated by spectrophotometry using NanoDrop ND‐2000 (Thermo Fisher Scientific, Wilmington, DE). 260/230 and 260/280 absorbance ratios were used to assess the presence of contaminants: peptides, phenols, aromatic compounds, or carbohydrates and proteins.
Reverse Transcription
miRNAs RT (starting from 500 ng of total RNA) was carried out with Gene Amp RNA PCR kit (Life Technologies, TX) including some modifications: 50 U of MMLV RT, 1 mM dNTPs, 5 mM MgCl2, 1 U RNase Inhibitor, 1× PCR Buffer II, and 0.5 μg of specific SLP (Table 1). Primer design was obtained using Primer express 3.0 (Life Technologies, Carlsbad, CA, USA). The reaction was performed with an initial incubation at 16°C for 30 min and a following step at 42°C for 1 h. In order to terminate the RT step, a final incubation at 99°C for 5 min occurred.
Table 1.
Primers and Probes Utilized in Stem‐Loop RT Real‐Time PCR
| Target | RNU43 |
|---|---|
| Sequence | GAACUUAUUGACGGGCGGACAGAAACUGUGUGCUGAUUGUCACGUUCUGAUU |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAATCAG |
| Forward | TGACGGGCGGACAGAAA |
| Probe MGB fam | TGTGTGCTGATTGTCA |
| Target | Mir155 |
| Sequence | UUAAUGCUAAUCGUGAUAGGGGU |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCACCCCT |
| Forward | CGCAGTTAATGCTAATCGTGATA |
| Probe MGB fam | GGGGTGGCTCTGG |
| Target | Mir146a |
| Sequence | UGAGAACUGAAUUCCAUGGGUU |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCAACCCA |
| Forward | CGCAGTGAGAACTGAATTCCAT |
| Probe MGB fam | GGGTTGGCTCTGG |
| Target | MirBART2‐5p |
| Sequence | UAUUUUCUGCAUUCGCCCUUGC |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCGCAAGG |
| Forward | CGCAGTATTTTCTGCATTCGC |
| Probe MGB fam | CCTTGCGGCTCTGG |
| Target | MirBART15 |
| Sequence | GUCAGUGGUUUUGUUUCCUUGA |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCTCAAGG |
| Forward | CGCAGGTCAGTGGTTTTGTT |
| Probe MGB fam | TCCTTGAGGCTCTGG |
| Target | mirBART22 |
| Sequence | UUACAAAGUCAUGGUCUAGUAGU |
| SLP | GGCTCTGGTGCAGGGTCCGAGGTATTCGCACCAGAGCCACTACT |
| Forward primer | CGCAGTTTTTTACAAAGTCATGG |
| Probe MGB fam | TCTAGTAGTGGCTCTGG |
| Universal reverse primer | TGCAGGGTCCGAGGTATTC |
miRNA Expression Levels Measurement by Real‐Time MGB PCR
After the RT step, an asymmetric PCR using 300 nM of specific forward primer (Table 1), 0.1 U of GoTaq Hot Start polymerase (Promega, Bergamo, Italy), 4 μl of 5× Colorless GoTaq Flexi Buffer, and 2 μl of cDNA, obtaining a final volume of 20 μl, was performed. The thermal profile used was as follows: 95°C for 2 min, 30 cycles of 94°C for 15 sec, 55°C for 30 sec, and 72°C for 20 sec.
Forward primer and MGB probe were specific for each assay and designed by the software Primer express 3.0. Five microliters of enriched cDNA denominated ccDNA were added to 35 μL of reaction mix containing 800 nM of forward primer, 1,000 nM of reverse primer, 200 nM of MGB probe, and 1× TaqMan Universal PCR Master Mix (P/N: 4324018, Life Technologies) in a final volume of 40 μl. The amplifications were performed on ABI 7500 Real Time PCR system in a 96‐well plate at 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min. Each sample was run in triplicate.
Statistical Analysis
Student t‐test P‐values and SDs were calculated by GraphPAD Prism5 (GraphPad Software, La Jolla, CA).
RESULTS
RNA was extracted using two different isolation methods including available kits RNAzol and a modified RNAzol protocol by Taylor et al. 15. In all cases, RNA was eluted in RNase‐free H2O and kept frozen until analysis. The presence of contaminants was assessed by NanoDrop spectrophotometry. A 1.6‐fold higher absorbance at 260 nm, implying a higher RNA quantity isolated, was observed in RNAzol extracted RNA (378.8 ± 28.45 ng/μl) compared to RNAzol modified protocol (226.5 ± 13.34 ng/μl, Table 2). This difference was statistically significant with a P‐value <0.0001.
Table 2.
Quality, Quantity, and Amplificability of Total RNA Obtained With RNAzol and RNAzol Modified Protocols
| Method | Target | RNA amount (ng/μl) | RNA yield (μg/ml serum) | OD 260/280 | C t real‐time PCR |
|---|---|---|---|---|---|
| RNAzol miRNA specific extraction | RNU43 | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 12.3 ± 3 |
| Mir155 | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 25.06 ± 3.76 | |
| Mir146a | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 18.42 ± 1.27 | |
| MirBART2‐5p | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 15.18 ± 2.7 | |
| MirBART15 | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 14.7 ± 1.18 | |
| MirBART22 | 378.8 ± 28.45 | 0.95 | 1.41 ± 0.03 | 18.1 ± 2.4 | |
| RNAzol modified by Taylor et al. | RNU43 | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 9.07 ± 2.18 |
| Mir155 | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 18.25 ± 3.2 | |
| Mir146a | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 13.64 ± 1.48 | |
| MirBART2‐5p | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 11.1 ± 1.65 | |
| MirBART15 | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 10.38 ± 0.88 | |
| MirBART22 | 226.5 ± 13.34 | 0.57 | 1.07 ± 0.01 | 13.29 ± 1.65 |
OD 260/280, absorbance ratio at 260 and 280 nm wavelength.
By using 260/280 nm absorbance ratio, RNA purity was assessed. The different extraction methods showed poor 260/280 ratios (Table 2). Moreover, RNAzol protocol seemed to be more performant, as the mean OD ratio 260/280 nm were 1.41 ± 0.03 and 1.07 ± 0.01 for RNAzol and RNA modified protocol, respectively (Fig. 1), showing a statistically significant difference with a P‐value <0.0001.
Figure 1.
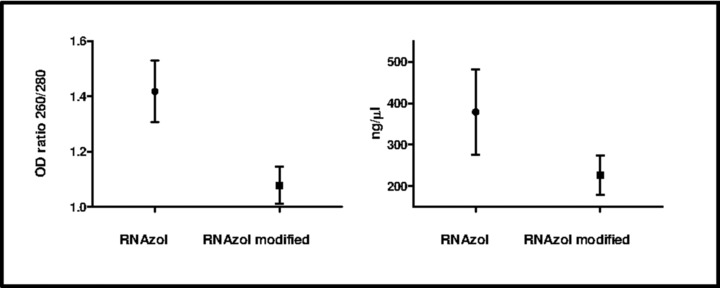
Quality and quantity of total RNA obtained with RNAzol and RANzol modified protocols.
On the basis of RNA amount, yield, and purity, the RNAzol protocol specific for miRNA extraction showed the best performance results among the tests used. RNA isolation was performed in triplicate and followed by triple RT‐qPCR measurements of endogenous miRNAs (RNA43), cellular miRNAs (mir155 and mir146a), and EBV miRNAs (mirBART2‐5p, mirBART15, and mirBART22). In contrast to the previous findings, in terms of RNA purity and quantity, the amplifiable RNA was approximately 10‐fold higher using RNAzol modified protocol (Table 2), showing statistical differences for each target tested (mir146a, P < 0.0001; mir155, P = 0.0004; mirBART2‐5p, P = 0.0007; mirBART15, P < 0.0001; and mirBART22, P < 0.0001).
The impact of SLP concentration in cDNA synthesis of miRNAs and subsequent amplification of cDNA was also evaluated in this study (Fig. 2). The RT reaction using 0.5 μm of SLP showed very low C t values between 9.95 and 14, with a mean value of 11.636 and 1.2 SDs, in the subsequent amplification reaction (Fig. 2), while, using respectively 0.1 or 0.01 μm of SLP, C ts between 16.42 and 21.92, with a mean value of 19.42 and 1.55 SDs, and between 26 and 32.17, with a mean value of 29.77 and 1.73 SDs, were obtained.
Figure 2.
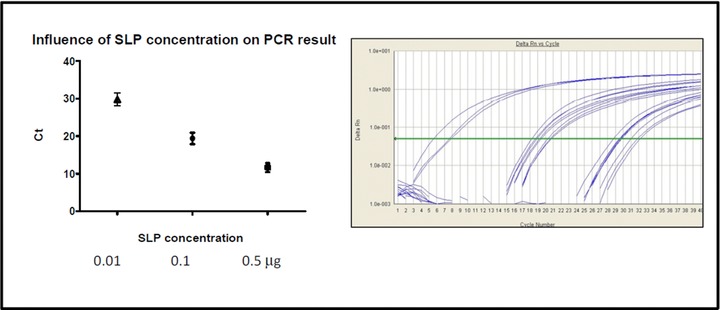
Effect of the concentration of SLP on the real‐time PCR results. The results of the experiment are shown as scatter plot (left) and real‐time amplification plot (right).
DISCUSSION
Our results indicate that the RNAzol modified by Taylor et al. 15 RNA isolation method outperforms other methods we tested, at least for what concerned isolation of small RNAs from serum. This finding, furthermore, should be viewed in light of the relative easiness of execution of the RNAzol modified protocol method. Ease of use and short processing time are certainly appropriate considerations when results are not compromised. However, additional optimization of existing methods is encouraged, since repeated extractions and temperature during isolation 16, different carriers and spike‐ins, and elution volumes are among the parameters that could be optimized for specific RNA isolation kits.
To inactivate serum RNases, RNase‐inhibitor denaturing reagents are routinely used. However, if RNase‐inhibitor denaturing reagents are not removed properly from the RNA isolated, those could inhibit enzymatic reactions such as RT or polymerase chain reaction (PCR), thereby compromising the sensitivity of downstream quantification assays. Li and Kowdley suggested the employment of a filtering agent to remove residual denaturing agents 17. However, our data showed that RNA isolation by RNAzol modified protocol was better in terms of purity compared to RNA isolation by RNAzol, confirmed by RNA isolated and amplifiable without use of filtering agent 17. As miRNA abundance in serum is significantly lower than that in solid tissues, low RNA yield would result in failure of detection of miRNA signatures present in low abundance. Among the two kits we tested, the RNAzol kit yielded the highest RNA concentration (Table 2). In addition, the relative abundance of six circulating miRNAs (mir146a, mir155, RNU43, mirBART2‐5, mirBART15, and mirBART22), recently found as potential biomarkers in diagnosis of cancer, autoimmune and inflammatory disease 3, 18 or cancer, and CAEBV (chronic active Epstein‐Barr virus infection) 19, 20, was measured by a novel real‐time PCR assay. The results obtained indicated that RNAzol modified protocol was able to recover more endogenous serum miRNA than RNAzol protocol (Table 2). Total RNA extracted was also measured using NanoDrop spectrophotometer in order to evaluate individual variations. Among 32 human serum samples, the average quantity of recovered total RNA from 400 μl of serum was 378.86 ± 28.45 ng/μl (mean ± SD) with RNAzol and 226.53 ± 13.34 ng/μl with RNAzol modified protocol. This quantity was significantly higher than that described in previous studies 9, wherein RNA recovery was reported to be approximately 120 ng of total RNA yield from 200 μl serum 21 or similar 17. These results indicated that the total RNA abundance in serum samples was highly variable among individuals. Notably, total RNA yield and quality between RNAzol and RNAzol modified protocols was significantly different (P < 0.01). Comparing real‐time PCR assay results, and in particular C ts obtained amplifying reverse‐transcribed serum miRNAs by RNAzol and RNAzol modified protocols, a statistical difference was observed among the RNA isolation protocols (P < 0.01) and, in detail, RNAzol modified protocol showed a better amplification performance. The C t differences between isolation replicates, about half a cycle or less, were lower compared to other publications on the same subject that reported a statistical significant differential expression between miRNA 9. Taking in consideration that in these publications twofold higher miRNA expression was attested, it is clear that accurate and precise isolation and amplification techniques could be important to avoid apparent differences declared in miRNAs expression, especially in studies with low number of samples. Taken together, our results indicate that serum miRNA isolated by RNAzol modified protocol is suitable for real‐time PCR‐based miRNA profiling experiments. Some papers reinforced the idea that the best way for miRNA detection and quantification is PCR and its variants with Locked Nucleic Acids or other annealing loop strategies 22, 23, 24, 25. In this article, a stem‐loop RT real‐time PCR described by Chen et al. 14 using MGB fluorescent probe was assessed. There are few different stem‐loop RT real‐time PCR methods available and companies that support them. Direct stem‐loop RT real‐time PCR allows total RNA (or <40 nt RNA) to be directly reverse transcribed and subsequently amplified by PCR. On the contrary, indirect RT‐PCR performs an initial polyadenilation of the miRNA of interest before reverse transcription and PCR amplification (e.g., NCode miRNA RT‐qPCR System from Invitrogen). The reasons behind the choice of using direct RT‐qPCR with the stem‐loop RT primer were the following: compared to the nondirect approach, (1) there is less handling of RNA and therefore less contamination or RNA degradation risk, (2) one less enzymatic step (each enzymatic step can introduce variation or reduce RNA recovery), (3) the stem‐loop method discriminates between mature and pre‐ or pri‐miRNA by 100‐fold, and (4) the stem‐loop method discriminates between single base mismatches by 100‐fold 14. The most critical parameter taken in consideration was the refolding of the stem loop into its stable structure. When this primer is subjected to high temperatures, indeed, some melting may occur. In its incompletely folded state, the reverse transcription of nonspecific cDNA may occur. If the miRNA of interest is rare, nonspecific cDNA could exceed the specific cDNA and potentially contribute to nonspecific PCR amplification. Given the combined specificities of the forward primer and probe, nonspecific PCR amplification should not contribute to the fluorescent signal. In this study, the impact of SLP concentration in cDNA synthesis of miRNAs and subsequent amplification of cDNA was also evaluated (Fig. 2. An SLP concentration equal to 0.5 μm resulted in a more efficient amplification compared to 0.1 or 0.01 μm concentrations (Fig. 2).
Circulating miRNA signatures can be identified in either serum or plasma 24; however, because serum is free of anticoagulants such as heparin, a potent inhibitor of downstream PCR, it delivers more consistent measurements 26. Because of this, the use of archived plasma samples is discouraged for biomarker identification. Also important to note when processing patient blood, serum samples may be less affected by hemolysis than plasma samples. A recent study suggested that significant variation in miRNA expression in plasma may be contributed by blood cell specific and platelet‐specific miRNAs from hemolysis 27. Improper handling of plasma samples could increase the risk of hemolysis and dilute the quality of downstream data analysis. By contrast, the standard serum isolation procedure is relatively more consistent, as whole‐blood samples are set to coagulate naturally. This study has several limitations, which may be helpful to review as they indicate opportunities for advances in this field. A limited number of endogenous miRNAs in this study (mir146a, mir155, mirBART2‐5, mirBART15, and mirBART22) were measured and recovery may differ from one method to another on an miRNA‐by‐miRNA basis, similar to the differences reported by Kim et al. 28, in their retraction letter. This could be due to GC content, length, or other features. Assuming availability of appropriate resources, an miRNome‐wide comparison of isolation methods might be highly useful. Also, an evaluation whether the differences observed were due to inhibitors, differential RNA recovery, or both was not performed.
Many RNA isolation kits are available, along with many method modifications, and only a small portion was explored. We encourage other researchers to join us in comparing methods and working toward standardization as well as improvement of existing practices. Thus, data comparison of serum samples collected at various research centers would be more feasible. In conclusion, we established a protocol to isolate miRNA from 400 μl of human serum. This protocol was able to extract high‐quality miRNAs with enough yield for high‐throughput real‐time PCR profiling experiments that could be as useful alternative to custom‐designed assays, allowing cost reduction when performing a large number of specific assays.
CONFLICT OF INTEREST
The authors state that there are no conflicts of interest.
ACKNOWLEDGMENTS
We are grateful for collaboration with the participating hospitals “Città della salute e della scienza di Torino” and the staff. The authors thank the members of the Department of Public Health and Pediatrics for interesting discussions, and Director L. Cordero di Montezemolo. This project was supported by research funded by the university (ex 60%) in the year 2013.
Grant sponsor: University of Turin
REFERENCES
- 1. Duttagupta R, DiRienzo S, Jiang R, et al. Genome‐wide maps of circulating miRNA biomarkers for ulcerative colitis. PLoS One 2012;7:e31241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Duttagupta R, Jiang R, Gollub J, Getts RC, Jones KW. Impact of cellular miRNAs on circulating mirna biomarker signatures. PLoS One 2011;6:e20769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Iborra M, Bernuzzi F, Invernizzi P, Danese S. MicroRNAs in autoimmunity and inflammatory bowel disease: Crucial regulators in immune response. Autoimmun Rev 2012;11:305–314. [DOI] [PubMed] [Google Scholar]
- 4. Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med 2010;16:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tili E, Michaille JJ, Costinean S, Croce CM. MicroRNAs, the immune system and rheumatic disease. Nat Clin Pract Rheumatol 2008;4:534–541. [DOI] [PubMed] [Google Scholar]
- 6. Du L, Pertsemlidis A. Cancer and neurodegenerative disorders: Pathogenic convergence through microRNA regulation. J Mol Cell Biol 2011;3:176–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang Z, Yang B. MicroRNA Expression Detection Methods. Springer; 2010. [Google Scholar]
- 8. McAlexander M A, Phillips MJ, Witwer KW. Comparison of methods for miRNA extraction from plasma and quantitative recovery of RNA from cerebrospinal fluid. Front Genet 2013;4:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li Y, Li J, Belisle S, Baskin CR, Tumpey TM, Katze MG. Differential microRNA expression and virulence of avian, 1918 reassortant, and reconstructed 1918 influenza A viruses. Virology 2011;421:105–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moussay E, Wang K, Cho JH, et al. MicroRNA as biomarkers and regulators in B‐cell chronic lymphocytic leukemia. Proc Natl Acad Sci USA 2011;108:6573–6578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA 2011;108:5003–5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Witten D, Tibshirani R, Gu SG, Fire A, Lui WO. Ultra‐high throughput sequencing‐based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol 2010;8:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ozsolak F, Milos PM. RNA sequencing: Advances, challenges and opportunities. Nat Rev Genet 2011;12:87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen C, Ridzon DA, Broomer AJ, et al. Real‐time quantification of microRNAs by stem‐loop RT‐PCR. Nucleic Acids Res 2005;33:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Taylor CT, Satoor SN, Ranjan AK, Pereira E, Cotta MV, Joglekar MV. A protocol for measurement of noncoding RNA in human serum. Exp Diabetes Res 2012. doi: 10.1155/2012/168368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Burgos KL, Javaherian A, Bomprezzi R, et al. Identification of extracellular miRNA in human cerebrospinal fluid by next‐generation sequencing. RNA 2013;19:712–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li Y, Kowdley KV. Method for microRNA isolation from clinical serum samples. Anal Biochem 2012;431:69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kopp KL, Ralfkiaer U, Gjerdrum LMR, et al. STAT5‐mediated expression of oncogenic mir‐155 in cutaneous T‐cell lymphoma. Cell Cycle 2013;12:1939–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kawano Y, Iwaka S, Kawada J, et al. Plasma viral microRNA profiles reveal potential biomarkers for chronic active Epstein‐Barr virus infection. J Infect Dis 2013;208:771–779. [DOI] [PubMed] [Google Scholar]
- 20. Lopes LF, Ruiz Miyazawa KW, de Almeida ER, et al. Epstein‐Barr virus (EBV) micrornas: Involvement in cancer pathogenesis and immunopathology. Int Rev Immunol 2013;32:271–281. [DOI] [PubMed] [Google Scholar]
- 21. Qi P, Cheng SQ, Wang H, Li N, Chen YF, Gao CF. Serum microRNAs as biomarkers for hepatocellular carcinoma in Chinese patients with chronic hepatitis B virus infection. Plos One 2011;6:e28486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pradervand S, Weber J, Lemoine F, et al. Concordance among digital gene expression, microarrays, and qPCR when measuring differential expression of microRNAs. Biotechniques 2010;48:219–222. [DOI] [PubMed] [Google Scholar]
- 23. Shi R, Chiang VL. Facile means for quantifying microRNA expression by real‐time PCR. Biotechniques 2005;39:519–525. doi 10.2144/000112010. [DOI] [PubMed] [Google Scholar]
- 24. Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: A new potential biomarker for cancer diagnosis and prognosis. Cancer Sci 2010;101:2087–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Czimmerer Z, Hulvely J, Simandi Z, et al. A versatile method to design stem‐loop primer‐based quantitative PCR assays for detecting small regulatory RNA molecules. PLoS One 2013;8(1):e55168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yokota M, Tatsumi N, Nathalang O, Yamada T, Tsuda I. Effect of heparin on polymerase chain reaction for blood white cells. J Clin Lab Anal 1999;13:133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pritchard CC, Kroh E, Wood B, et al. Blood cell origin of circulating microRNAs: A cautionary note for cancer biomarker studies. Cancer Prev Res 2011;5:492–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim YK, Yeo J, Ha M, Kim B, Kim VN. Retraction notice to: Cell adhesion dependent control of microRNA decay. Mol. Cell. 2012;43:1005–1014. [DOI] [PubMed] [Google Scholar]


