Abstract
Background
Human metapneumovirus (hMPV) causes respiratory tract infection in influenza‐like illness. The role of hMPV infections in all age groups in Thailand has not yet been investigated. Thus, the objective of this study was to determine prevalence of hMPV infection in all age groups in Thailand during 2011.
Methods
A total of 1,184 nasopharyngeal washes were collected from hospitalized patients and sent to the Department of Microbiology, Siriraj Hospital, for influenza A virus detection. Real‐time polymerase chain reaction (PCR) was used to detect hMPV infection. Partially, F gene from hMPV positive samples were sequenced and used for genotyping by phylogenetic tree analysis.
Results
The prevalence of hMPV for all age groups was 6.3%. The highest prevalence of hMPV infection was in children aged <2 years. Of 71 hMPV‐positive patients, three (4.2%) were coinfected with respiratory syncytial virus (RSV), two with rhinovirus (2.8%), one with coronavirus (1.4%), and one with RSV and adenovirus (1.4%). Phylogenetic analysis of F gene revealed that 96.8% of hMPV detected was subgenotype B1, 1.6% was sublineage A2a, and 1.6% was A2b. Genetic variation of F gene was much conserved.
Conclusion
We demonstrated the prevalence of hMPV subgenotype B1 circulating in Thailand during 2011.
Keywords: human metapnuemovirus, prevalence, influenza‐like illness, Thailand
INTRODUCTION
During 2009, there was an epidemic of influenza pdm2009 virus in the United States and Mexico. It spread through many countries, including Thailand. After the first case of influenza pdm2009 in Thailand was reported, the virus spread rapidly throughout the country. The symptoms of influenza pdm2009 were fever, cough, sore throat, and myalgia. During the epidemic, there were some patients who had similar clinical symptoms but were not infected by the influenza pdm2009 virus. Thus, influenza‐like illness (ILI) was used to describe a clinical syndrome that may be attributed to influenza and others respiratory viruses. ILI is caused by many viruses, including influenza virus, adenoviruses, respiratory syncytial virus (RSV), enteroviruses, human metapneumovirus (hMPV), and parainfluenza viruses 1. Hombrouck et al. reported that during the epidemic of influenza pdm2009 virus in Belgium, ILI in children aged <5 years was caused by influenza viruses (25%), RSV (19%), rhinovirus (17%), hMPV (9%), and parainfluenza viruses (7%; 2). When patients were infected by these viruses, there were some overlapping symptoms that rendered clinical diagnosis unreliable.
In 2001, Van den Hoogen et al. identified hMPV from acute respiratory infection in children in the Netherlands 3. hMPV was classified in the family Paramyxoviridae, subfamily Paramyxovirinae, and genus Metapneumovirus. It is an enveloped virus with negative sense, single‐stranded RNA of ≈13 kb. The primary target for hMPV are young children, immunocompromised hosts, and patients who have underlying conditions 4, 5, 6, 7, 8, 9. The symptoms of hMPV infection in young children vary from mild upper to severe lower respiratory tract disease. hMPV can trigger asthma in adults and young children 8.
The circulation of hMPV is worldwide, and seasonal distribution is predominantly in late winter to early spring in temperate climates and late spring to summer in tropical regions 10. The spreading of hMPV varies between populations and in timing. Sometimes the same strain may spread in different locations and at different times. The spread is still complicated and unclear.
Based on nucleotide sequence analysis of different hMPV F genes, hMPV can be classified into two genotypes, A and B. Both genotypes can be classified into subgenotypes A1, A2a, A2b, B1, and B2 11, 12, 13. The clinical differences between the genotypes are still unclear, but genotype A is responsible for the most severe disease 14.
Vaccine for hMPV is underdeveloped. Most vaccine targets are fragments of the F and G genes. Thus, genetic diversity of these genes is important for vaccine development. Therefore, this study analyzed F gene sequence and genotype in hMPV from patients with ILI who were negative for influenza A virus in Thailand.
MATERIALS AND METHODS
Sample Collection and Screening
A total 1,184 nasopharyngeal washes from patients presenting with ILI during May to September 2011 were collected in viral transport medium and sent to the Department of Microbiology, Siriraj Hospital, for influenza pdm2009 testing. NucliSens easyMAG (bioMérieux, Marcy l'Etoile, France) was used to extract viral RNA from 200 μl nasopharyngeal secretion. The extracted RNA was eluted with 80 μl elution buffer and examined for pandemic influenza virus by ProFast+ (Genprobe, Bedford, MA) and ProFlu+ (Genprobe Bedford, MA). All 1,134 samples that were negative for influenza A virus RNA were examined for hMPV using the commercial PrimerDesign™ genesig Kit for HMPV (Genesig, Southampton, UK).
RT‐PCR and Sequencing
Positive samples for hMPV by real‐time polymerase chain reaction (PCR) were used to amplify partial F gene by RT‐PCR (where RT is reverse‐transcription) followed by nested PCR. The primers used to amplify F gene were Fm1–18 (5′‐CATCTTGAATTCATGTCTTGGAAAGTGGTG‐3′) and Fm1620–1601 (5′‐CGACTGAAGCTTCTAATTATGTGGTATGAAGC‐3′; 11) for RT‐PCR, and Fm475 (5′‐GCCACTGCAGTGAGAGAGC‐3′) and Fm1620–1601 (5′‐CGACTGAAGCTTCTAATTATGTGGTATGAAGC‐3′) for nested PCR. RT‐PCR was done in One‐step RT‐PCR (Bioline, London) with the following conditions: reverse transcription at 45°C for 20 min; 95°C for 1 min, 35 cycles of 94°C for 1 min, 46°C for 1 min, 72°C for 2 min, with final extension at 72°C for 10 min. For F gene seminested PCR, 1 U Platinum Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) and 2 μl PCR products were used. The conditions for seminested F gene PCR were as follows: denaturation at 94°C for 2 min, 35 cycles of 94°C for 1 min, 56°C for 1 min, 68°C for 1 min, and final extension at 68°C for 10 min. The seminested PCR yielded amplicons of 1,145 bp. All positive PCR products were purified using HiYield Gel/PCR Fragment Extraction kit (RBC, Taipei, Taiwan). Purified PCR products were sequenced directly using primer Fm475 (5′‐GCCACTGCAGTGAGAGAGC‐3′) and Fm1620–1601 (5′‐CGACTGAAGCTTCTAATTATGTGGTATGAAGC‐3′). Sequencing reactions were carried out using ABI PRISM BigDye Terminator v 3.1 Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, CA). The products were processed by capillary electrophoresis using ABI 3130 DNA Analyzer (Applied Biosystems), and analyzed using DNA Sequencing Analysis (Applied Biosystems) and DNABaser (Heracle BioSoft S.R.L., Pitesti, Romania).
Nucleotide Sequence Analysis and Accession Numbers
A neighbor‐joining tree was constructed using Mega software (version 5.0) with 1,000 bootstrap replicates. The nucleotide and deduced amino acid sequences of the partial F gene were compared with those of the hMPV strains available from the GenBank database. The 71 partial sequences of the F gene were deposited in GenBank under accession numbers JQ181560‐JQ181584 and JQ745049‐JQ745094.
Statistical Analysis
The significance of the difference in rates was tested using the Pearson χ2 test and likelihood ratio. Analyses were performed using SPSS version 13.0 software (SPSS, Inc., Chicago, IL).
RESULTS
Patient Characteristics
The study was carried out in 1,184 patients with clinically suspected respiratory infections between May and September 2011. All samples were tested for influenza A detection by real‐time PCR. The result showed 1,134 cases were negative for influenza A viruses and used for hMPV study. These 1,134 patients aged from 8 days to 104 years (mean, 31.9 years; and median, 13 years). Four hundred fifty‐five (40.1%) of the 1,134 samples were from patients aged <5 years, 335 (29.6%) from adults aged 21–60 years, and 344 (30.3%) from patients aged >60 years. There were 588 (51.8%) male and 546 (48.2%) female patients.
Prevalence of hMPV
hMPVs were found in 6.3% (71/1,134) during May–September 2011, which correlated with rainfall and high humidity in Thailand. From the period of study, hMPV starts increasing in July and peaks in September (11.90%) as shown in Figure 1. Patients aged <5 years were most frequently affected by hMPV infection (35.2%). This was followed by those aged >60 years (31%). The youngest hMPV‐positive patient was 1‐month‐old, and the oldest was 104 years. Single hMPV infection was found in 65 specimens (91.5%). Among hMPV‐positive patients, six (8.5%) were coinfected with another respiratory virus: three with RSV (4.2%), two with rhinovirus (2.8%), one with RSV and adenovirus (1.4%), and one with coronavirus (1.4%). The age of patients coinfected with hMPV and RSV was <1 year. Among the 588 samples from male patients, 21 (3.6%) were hMPV positive; and among the 546 samples from female patients, 50 (9.2%) were hMPV positive. The ratio of female to male hMPV‐positive patients was 2.38:1 (P < 0.05). Age and sex distribution of the samples and patients positive for hMPV is presented in Table 1.
Figure 1.
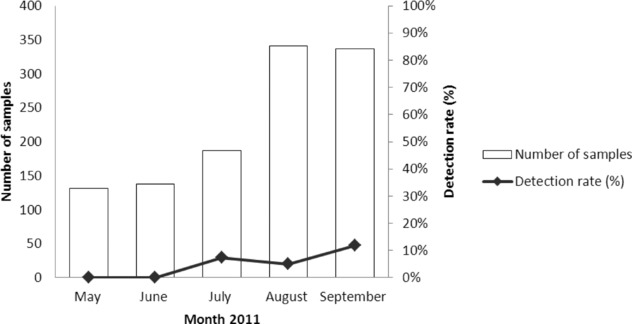
Distribution of hMPV with acute respiratory tract infection, May–September 2011.
Table 1.
Characteristics of hMPV‐Positive Patients
| Characteristics | No. of negative influenza A by real‐time PCR (N = 1,134) | No. of positive hMPV patients by real‐time PCR (N = 71) |
|---|---|---|
| Age | ||
| 0–5 years | 40.1% (455/1,134) | 35.2% (25/71) |
| 6–20 years | 13.5% (153/1,134) | 9.9% (7/71) |
| 21–60 years | 16.1% (182/1,134) | 23.9% (17/71) |
| >60 years | 30.3% (344/1,134) | 31% (22/71) |
| Mean | 31.9 | 33.54 |
| Median | 13 | 27 |
| Sex | ||
| Male | 51.8% (588/1,134) | 29.6% (21/71) |
| Female | 48.2% (546/1,134) | 70.4% (50/71) |
Phylogenetic Analysis of Partial F Gene Sequence
Partial F gene sequences were obtained from 62 of the 71 hMPV‐positive specimens. The phylogenetic analysis was constructed by comparing the four main genetic lineages of reference hMPV and avian pneumovirus C as an outgroup (Table 1, Table 2 and Fig. 2). Fifty‐four (96.8%) of the 62 viruses belonged to subgenotype B1, one (1.6%) belonged to sublineage A2a, and one (1.6%) belonged to sublineage A2b.
Table 2.
Reference Strain of hMPV for Construction of F Gene Phylogenetic Tree
| Strain | Origin | GenBank accession number | Lineage |
|---|---|---|---|
| TN96‐12 | Tennessee, USA | JN184399 | A1 |
| JPS03‐180 | Japan | AY530092.1 | A1 |
| NL/00/1 | Netherlands | AF371337 | A1 |
| Can00‐16 | Canada | AY14530.1 | A2a |
| Jpn03‐1 | Japan | AB503857 | A2a |
| NL/00/17 | Netherlands | AY355324 | A2a |
| Can97‐83 | Canada | NC004148 | A2a |
| JPS03‐240 | Japan | AY530095 | A2b |
| BJ1887 | China | DQ843659.1 | A2b |
| gZ01 | China | GQ153651 | A2b |
| NL/1/99 | Netherlands | AY304361 | B1 |
| TN89‐1‐13 | Tennessee, USA | EU857569.1 | B1 |
| TN93‐3‐2 | Tennessee, USA | EU857589.1 | B2 |
| NL/94/01 | Netherlands | FJ168778.1 | B2 |
| BJ1816 | China | DQ843658.1 | B2 |
| Can98‐75 | Canada | AY297748.1 | B2 |
| Avian metapneumovirus | Colorado | AY579780 | Avian metapneumovirus |
Figure 2.

Phylogenetic tree of partial F gene of hMPV was constructed by neighbor‐joining method with 1,000 replicate bootstraps. Strains are indicated by •, isolated during 2011; and ▴, isolated during 2010.
To observe circulation of hMPV during 2010, nine positive specimens from hMPV patients during 2010 were selected randomly. Partial F genes were amplified and sequenced. A phylogenetic tree was constructed that included specimens from 2010 for comparison (Fig. 2). The genotypic analysis found that all nine specimens during 2010 belonged to subgenotype B1.
Variation of hMPV F Gene
The nucleotide sequence analysis indicated that the identity of partial F gene in subgenotype B1, sublineage A2a, and sublineage A2b were 98.01%, 98.6%, and 97.6%, respectively. To identify lineage specificity, amino acid substitution in the fusion open reading frame at position 260–407 (location related to strain NL/00/01) was analyzed (Table 3). Genotype A could be distinguished from genotype B by amino acid substitution at V286, K296, Q312, K348, and N404. However, subgenotype A1 could not be distinguished from the sublineage A2a and A2b by amino acid substitution (Fig. 3). Cysteine residues were conserved at position 282, 300, 325, 334, and 349. One sample showed amino acid substitution at position 156 from cysteine to tryptophan (data not showed).
Table 3.
Lineage‐Specific Amino Acid Substitution in Fusion Open Reading Frame of F Gene
| Lineage | A1 | A2a | A2b | B1 | B2 |
|---|---|---|---|---|---|
| aa 286a | V | V | V | I | I |
| aa 296a | K | K | K | N | D |
| aa 312a | Q | Q | Q | K | K |
| aa 348a | K | K | K | R | R |
| aa 404a | N | N | N | P | P |
Number of amino acid residue related to amino acid strain NL/00/01, GenBank accession number AF371337.
Single letters refer to amino acid (aa): D, Aspartate; I, Isoleucine; K, Lysine; N, Aspargine; P, Proline; Q,Glutamine; R, Arginine; V, Valine
Figure 3.
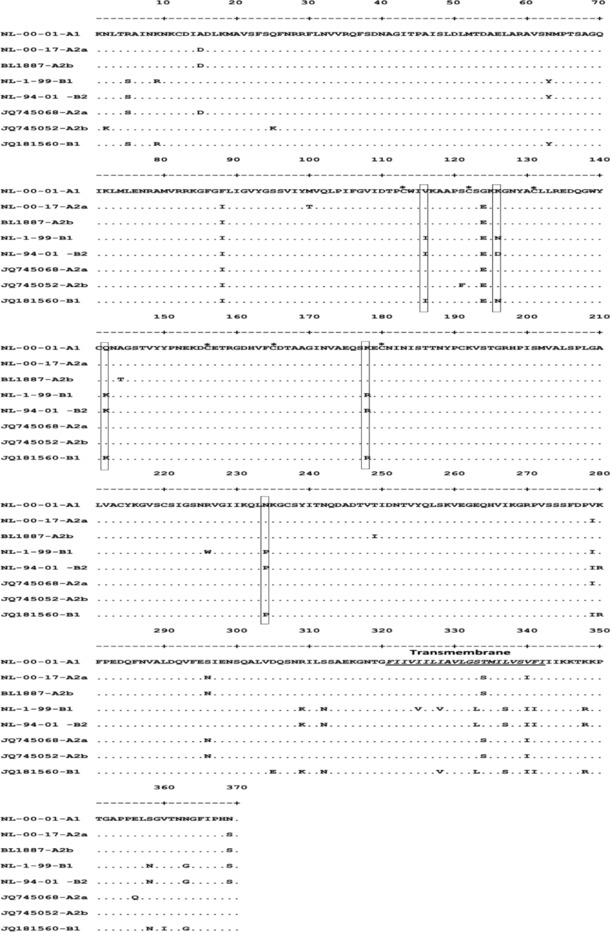
Comparison of amino acid sequence of fusion protein of reference strains (NL‐00‐01‐A1, NL‐00‐17‐A2a, BL1887‐A2b, NL‐1‐99‐B1, and NL‐94‐01‐B2) and each subgenotype isolated (JQ745068‐A2a, JQ745052‐A2b, and JQ181560‐B1). Box represents amino acid substitution for genotype distinct. Asterisk represents cysteine residue.
DISCUSSION
Acute respiratory infection can be caused by many viruses. For case management, it is important to differentiate infections caused by influenza viruses from other respiratory viruses. ILI is used worldwide to identify suspected cases of influenza. In Thailand, during a study of the influenza pdm2009 outbreak, it was found that 28% of cases were caused by influenza A virus, 24% by RSV, 2% by parainfluenza type I, 5% by hMPV, 3% by influenza B virus, 3% by parainfluenza type 3, and 1.5% by parainfluenza type 2 15.
In the present study, we detected hMPV in patients with ILI during 2011. A total of 1,184 specimens were sent to the laboratory for detection of influenza A virus. One thousand one hundred thirty‐four specimens were negative for influenza A virus by real‐time PCR and were used for hMPV detection. The prevalence of hMPV in the population was 6.26% (71/1,134) compared with 1.7% reported from Cambodia 16, 7.5% from the Netherlands 17, 14.8% from Canada 18, and 6.8% from China 19. A similar prevalence has been reported previously in Thailand 20. hMPV can be found in July and peaked in September, which is the rainy season in Thailand. This is in agreement with previous reports during the rainy season in tropical countries 16, 21. However, some countries such as Singapore can detect hMPV all year without any obvious peak 22. These results suggest that hMPV has different epidemiological patterns in different countries. Our analysis of age distribution indicated that 35.2% (25/71) of cases of hMPV infection were in children aged <5 years, and the highest infection rate (23.9% [17/71]) was observed in those aged <2 years, which is consistent with previous data 23, 24, 25. In this study, hMPV was more common in female (70.4%), which is similar to previous report 19. But some reports have shown that hMPV infection was more common in male than females 26. Some host factors may influence hMPV infection.
Many studies have reported coinfections with hMPV and others respiratory viruses, especially human RSV 27, 28, 29. We found that the rate of hMPV coinfection was 8.5%, and the most frequently detected viruses were RSV and rhinovirus, which is similar to previous reports 16, 19, 30. RSV was found to be the most frequent virus involved in coinfection with hMPV, which may be because it has a similar seasonal distribution 31. We did not find any severe illness caused by hMPV and RSV coinfection in the present study (data not showed).
F and G proteins, two major transmembrane glycoproteins, are important in stimulating protective immune responses and genotyping. There are several reports of genotypic data for hMPV in Southeast Asia 19, 20, 22, 32. To investigate hMPV genotype distribution, we amplified partially the F gene (1,300 bp) and constructed a phylogenetic tree. hMPV subgenotype B1 (96.8%) predominated during the time of this study, followed by sublineages A2a and A2b. The circulation of hMPV can vary among locations and annually, which is similar to other human respiratory viruses 33. We selected samples during 2010 to represent hMPV genotype circulation during that period. Most hMPV in 2010 were subgenotype B1 (Fig. 2). These data suggest that B1 was found predominantly in 2010–2011, whereas A2a and A2b cocirculated in 2011. We did not find subgenotype A1, which is similar to the studies of Loo et al. (2007) and Arnott et al. (2013; 16, 22). Our data differed from those in a 2008 study that found that genotype A was the predominant strain in Bangkok 34. In another study in Cambodia, subgenotypes B2, B1, and A2b cocirculated, while subgenotype B2 was predominant in 2008 16.
Amino acid sequence analysis demonstrated five substitutions in the fusion domains (positions 286, 296, 312, 348, and 404) and one substitution near the fusion domain (position 233), which was similar to the studies of Van den Hoogen et al. (2004) and Wang et al. (2008; 11, 35). One sample (JQ745075) had tryptophan substitution for cysteine conserved position 156 (related to NL/00/01 position), which has not been found previously. The effect of amino acid change might involve structure of F gene which affect neutralizing antibody response. The amino acid change position 156 should be studied further. Variation in the fusion domain allows us to differentiate genotype A from genotype B but not subgenotype. Many factors influence circulation of hMPV, such as antigenic variability, viral infectivity, and immunity. Our study reveals that genetic variability of F gene is less heterogeneous; thus, the preexisting immune response can control viral infection. Further investigation of A2 genotype is needed because Vicente et al. (2006) have found that genotype A is more pathogenic than genotype B, which results in greater clinical severity in children 14. There are still discrepant reports about the relationship between the subtype of hMPV and disease severity, and further study should be performed.
In conclusion, this study confirmed that hMPV subgenotype B1, sublineage A2a and A2b cocirculated in Thailand during 2010–2011. The subgenotype B1 predominated during circulation. The genetic variability of F gene from hMPV subgenotype B1 showed that it was highly conserved over time.
ACKNOWLEDGMENT
This research was supported by Faculty of Medicine Siriraj Hospital, Mahidol University.
Ethical approval. This study was approved by Siriraj Institutional Review Board (COA 652/2011).
REFERENCES
- 1. Laguna‐Torres VA, Gómez J, Ocaña V, et al. Influenza‐like illness sentinel surveillance in Peru. PloS One 2009;4(7):e6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hombrouck A, Sabbe M, Van Casteren V, et al. Viral aetiology of influenza‐like illness in Belgium during the influenza A(H1N1)2009 pandemic. Eur J Clin Microbiol Infec Dis 2012;31(6):999–1007. [DOI] [PubMed] [Google Scholar]
- 3. Van den Hoogen BG, de Jong JC, Groen J, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med 2001;7:719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Englund JA, Boeckh M, Kuypers J, et al. Brief communication: Fatal human metapneumovirus infection in stem‐cell transplant recipients. Ann Intern Med 2006;144(5):344–349. [DOI] [PubMed] [Google Scholar]
- 5. Larcher C, Geltner C, Fischer H, Nachbaur, D , Muller LC, Huemer HP. Human metapneumovirus infection in lung transplant recipients: Clinical presentation and epidemiology. J Heart Lung Transplant 2005;24(11):1891–1901. [DOI] [PubMed] [Google Scholar]
- 6. Madhi SA, Ludewick H, Abed Y, Klugman KP, Boivin G. Human metapneumovirus‐associated lower respiratory tract infections among hospitalized human immunodeficiency virus type 1 (HIV‐1)‐infected and HIV‐1‐uninfected African infants. Clin Infect Dis 2003;37(12):1705–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vicente D, Montes M, Cilla G, Perez‐Trallero E. Human metapneu‐ movirus and chronic obstructive pulmonary disease. Emerg Infect Dis 2004;10(7):1338–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams JV, Crowe JE Jr, Enriquez R, et al. Human metapneumovirus infection plays an etiologic role in acute asthma exacerbations requiring hospitalization in adults. J Infect Dis 2005a;192(7):1149–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Williams JV, Martino R, Rabella N, et al. A prospective study comparing human metapneumovirus with other respiratory viruses in adults with hematologic malignancies and respiratory tract infections. J Infect Dis 2005b;192(6):1061–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kroll JL, Weinberg A. Human metapneumoviruses. Semin Respir Crit Care Med 2011;32:447–453. [DOI] [PubMed] [Google Scholar]
- 11. Van den Hoogen BG, Herfst S, Sprong L, et al. Antigenic and genetic variability of human metapneumoviruses. Emerg Inf Dis 2004;10:658–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Huck B, Scharf G, Neumann‐Haefelin D, Puppe W, Weigl J, Falcone V. Novel human metapneumovirus sublineage. Emerg Infect Dis 2006;12:147–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ishiguro N, Ebihara T, Endo R, et al. High genetic diversity of the attachment (G) protein of human metapneumovirus. J Clin Microbiol 2004;42:3406–3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Vicente D, Montes M, Cilla G, Perez‐Yarza EG, Perez‐Trallero E. Differences in clinical severity between genotype A and genotype B humanmetapneumovirus infection in children. Clin Infect Dis 2006;42:e111–e113. [DOI] [PubMed] [Google Scholar]
- 15. Phumethum S. Viral pathogens of influenza‐like illness and clinical presentation in Thai children during the 2009 pandemic influenza A (H1N1). J Prapokklao Hosp Clin Med Educ Cent 2012;29:43–46. [Google Scholar]
- 16. Arnott A, Vong S, Sek M, et al. Genetic variability of human metapneumovirus amongst an all ages population in Cambodia between 2007 and 2009. Infect Genet Evol 2013;15:43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jartti T, Van den Hoogen B, Garofalo RP, Osterhaus AD, Ruuskanen O. Metapneumovirus and acute wheezing in children. Lancet 2002;360(9343):1393–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bastien N, Ward D, Van Caeseele P, et al. Human metapneumovirus infection in the Canadian population. J Clin Microbiol 2003;41(10):4642–4646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao NG, Xie ZP, Zhang B, et al. Prevalence and clinical and molecular characterisation of human metapneumovirus in children with acute respiratory infection in China. Pediatr Infect Dis J 2010;29:131–134. [DOI] [PubMed] [Google Scholar]
- 20. Samransamruajkit R, Thanasugarn W, Prapphal N, Theamboonlers A, Poovorawan, Y . Human metapneumovirus in infants and young children in Thailand with lower respiratory tract infections; molecular characteristics and clinical presentations. J Infect 2006;52:254–263. [DOI] [PubMed] [Google Scholar]
- 21. Teeratukulpisarn J, Ekalaksananan T, Pientong C, Limwattananon C. Human metapneumovirus and respiratory syncytial virus detection in young children with acute bronchiolitis. Asian Pac J Allergy Immunol 2007;25(2–3):139–145. [PubMed] [Google Scholar]
- 22. Loo LH, Tan BH, Ng LM, Tee NW, Lin RT, Sugrue RJ. Human metapneumovirus in children, Singapore. Emerg Infect Dis 2007;13:1396–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Freymouth F, Vabret A, Legrand L, et al. Presence of the new human metapneumovirus in French children with bronchiolitis. Pediatr Infect Dis J 2003;22:92–94. [DOI] [PubMed] [Google Scholar]
- 24. Esper F, Boucher D, Weibel C, et al. Human metapneumovirus infection in the United States: Clinical manifestations associated with a newly emerging respiratory infection in children. Pediatrics 2003;111:1407–1410. [DOI] [PubMed] [Google Scholar]
- 25. Van den Hoogen BG, van Doornum GJ, Fockens JC, et al. Prevalence and clinical symptoms of human metapneumovirus infection in hospitalized patients. J Infect Dis 2003;188:1571–1577. [DOI] [PubMed] [Google Scholar]
- 26. Mahalingam S, Schwarze J, Zaid A, et al. Perspective on the host response to human metapneumovirus infection: What can we learn from respiratory syncytial virus infections? Microbes Infect 2006;8:285–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Foulongne V, Guyon G, Rodière M, Segondy M. Human metapneumovirus infection in young children hospitalized with respiratory tract disease. Pediatr Infect Dis J 2006;25(4):354–359. [DOI] [PubMed] [Google Scholar]
- 28. Chung JY, Han TH, Kim SW, Hwang ES. Genotype variability of human metapneumovirus, South Korea. J Med Virol 2008;80(5):902–905. [DOI] [PubMed] [Google Scholar]
- 29. Kim HR, Cho AR, Lee MK, Yun SW, Kim TH. Genotype variability and clinical features of human metapneumovirus isolated from Korean children, 2007 to 2010. J Mol Diagn 2012;14(1):61–64. [DOI] [PubMed] [Google Scholar]
- 30. Ljubin‐Sternak S, Santak M, Cepin‐Bogović J, et al. Detection of genetic lineages of human metapneumovirus in Croatia during the winter season 2005/2006. J Med Virol 2008;80(7):1282–1287. [DOI] [PubMed] [Google Scholar]
- 31. Paranhos‐Baccalà G, Komurian‐Pradel F, Richard N, Vernet G, Lina B, Floret D. Mixed respiratory virus infections. J Clin Virol 2008;43(4):407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pavlin, JA , Hickey AC, Ulbrandt N, et al. Human metapneumovirus reinfection among children in Thailand determined by ELISA using purified soluble fusion protein. J Infect Dis 2008;198:836–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peret TC, Hall CB, Schnabel KC, Golub JA, Anderson LJ. Circulation patterns of genetically distinct group A and B strain of human respiratory syncytial virus in a community. J Gen Virol 1998;79(Pt 9):2221–2229. [DOI] [PubMed] [Google Scholar]
- 34. Gaunt ER, Jansen RR, Poovorawan Y, Templeton KE, Toms GL, Simmonds P. Molecular epidemiology and evolution of human respiratory syncytial virus and human metapneumovirus. PLoS One 2011;6(3):e17427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang HC, Huang SW, Wang SW, et al. Co‐circulating genetically divergent A2 human metapneumovirus strains among children in southern Taiwan. Arch Virol 2008;153(12):2207–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]


