Forest communities with more distantly related trees have higher productivity, which is regulated by soil fungal pathogens.
Abstract
The relationship between plant diversity and productivity and the mechanisms underpinning that relationship remain poorly resolved in species-rich forests. We combined extensive field observations and experimental manipulations in a subtropical forest to test how species richness (SR) and phylogenetic diversity (PD) interact with putative root-associated pathogens and how these interactions mediate diversity-productivity relationships. We show that (i) both SR and PD were positively correlated with biomass for both adult trees and seedlings across multiple spatial scales, but productivity was best predicted by PD; (ii) significant positive relationships between PD and productivity were observed in nonsterile soil only; and (iii) root fungal diversity was positively correlated with plant PD and SR, while the relative abundance of putative pathogens was negatively related to plant PD. Our findings highlight the key role of soil pathogenic fungi in tree diversity-productivity relationships and suggest that increasing PD may counteract negative effects of plant-soil feedback.
INTRODUCTION
Phylogenetic diversity (PD) is increasingly regarded as a key indicator of functionally important aspects of diversity (1). Recent findings show that PD can predict ecosystem functioning (2–5), and phylogenetic-based biodiversity indices are stronger predictors of plant productivity in communities than species richness (SR) alone (6). However, despite progress in describing biodiversity–ecosystem function (BEF) relationships [e.g., (7)], the mechanisms that underpin the relationship remain uncertain. It is increasingly recognized that soil microbes can mediate plant interactions and thus may contribute to biodiversity-productivity relationships (8–11), but the link between soil pathogen diversity and PD of plant communities is unclear.
Soil-borne pathogens mediate host-specific and density-dependent negative plant-soil feedbacks in forests (9, 12, 13) and can have strong negative effects on plant growth when communities have low species diversity and high abundance of particular plant hosts. Therefore, soil-borne pathogens can strongly limit per capita biomass and community-level productivity through density-dependent regulation in low diversity mixtures but have less negative impacts in high diversity assemblages (10, 11). This can create a positive diversity-productivity relationship even in the absence of niche complementarity and sampling effects (10). The host ranges of pathogens are phylogenetically constrained, so the probability of infecting two different plant species decreases with greater phylogenetic distance between them (13–15). Congruent phylogenies between tree hosts and their fungal associates also suggest that closely related pathogens tend to infect closely related host species (16), and disease pressure on a host in a local community is explained by the abundance of all species in the community weighted by their phylogenetic distance to the host (15). Still, we do not know how this phylogenetic signal in plant-microbe interactions may apply to plant community productivity and the relationship between PD and ecosystem function (PDEF).
Previous studies of PDEF relationships provide limited evidence because they have either been reanalyses of experiments that were originally designed to examine BEF relationships of grasslands (6, 17) or simplified laboratory experiments attempting to simulate microbial communities (2, 18). Generating experimental evidence from forest ecosystems is hampered by the long life span of trees, large number of species, and high structural complexity of forest communities [but see (7, 19)]. Moreover, although tropical and subtropical regions harbor the greatest biodiversity, lower diversity temperate regions dominate studies in the BEF and PDEF literature, possibly biasing our understanding of community productivity response to the loss of diversity (20).
Here, we report observational and experimental data on (i) the relationship between plant PD and productivity and on (ii) the role of soil microbes, particularly fungal pathogens, in driving PDEF relationships in a subtropical forest. We tested for a PDEF relationship in seedling data from 1200 1-m2 quadrats and in adult trees from nonoverlapping quadrats of increasing area within a 50-ha plot. To examine the effects of soil microbes as a mechanism driving PDEF relationships, we conducted two controlled experiments. In the first experiment, we created independent gradients of both plant SR and PD, and we tested whether soil microbes affected the relationship between seedling diversity and productivity using soil sterilization. We also used molecular tools to measure how pathogen diversity and abundance are related to plant SR and PD. In the second experiment, we compared the effects of soil microbes associated with closely or distantly related trees on PDEF relationships. We predicted that because closer relatives are more likely to share pathogens, pots with fewer host species and lower PD should show lower fungal SR and greater pathogen relative abundance; hence, the negative effects of soil fungi on productivity should be strongest for tree communities comprising closely related species.
RESULTS
Biodiversity-productivity relationships in nature
We found that plant biomass production increased as a function of both SR and PD of trees at local scales (Fig. 1 and table S1). PD explained tree biomass better than SR, with lower P and Akaike information criterion (AIC) values at all scales from 10 m by 10 m to 50 m by 50 m (table S1). We observed the lowest AIC values when SR and PD were both included in the models, indicating that PD explained additional variation beyond that explained by SR (table S1). Together, this extensive observational evidence suggests that PD provided stronger explanatory power than SR of tree biomass in nature.
Fig. 1. Results of regression models predicting biomass production in response to SR or Faith’s PD of tree communities.
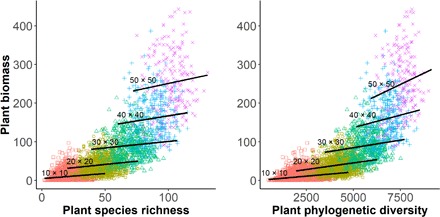
Nonoverlapping quadrats of the 50-ha permanent plot were analyzed at five different spatial scales (10 m by 10 m, 20 m by 20 m, 30 m by 30 m, 40 m by 40 m, and 50 m by 50 m). Statistics associated with the regressions are given in table S1.
We found even stronger relationships between productivity and diversity at the seedling stage. In the 2017 census of 1200 1 m by 1 m quadrats, seedling total height, a measure of total productivity, was strongly and positively correlated with both SR and PD of seedlings (P < 0.001; Fig. 2). When SR and PD were included together in the model, PD provided a clear additional explanation beyond that provided by SR (table S2).
Fig. 2. The relationship between diversity among neighboring tree seedlings and productivity at the seedling stage.
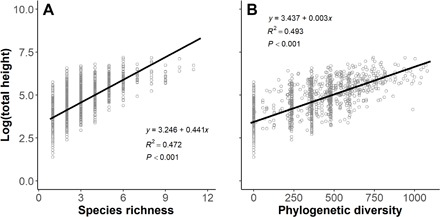
The total height of seedlings in each 1-m2 seedling quadrat was significantly related to (A) plant SR and (B) plant PD in the 2017 census.
BEF and PDEF relationships in controlled experiments
In the experiment to test how soil microorganisms altered the relationship between productivity and SR and PD of seedling communities (experiment 1; Fig. 3), mean biomass significantly increased with increasing SR and PD for species assemblages for both field soil and sterilized soil treatments (Fig. 4 and table S3). Although sterilized soil pots were recolonized by some generalist pathogens, we confirmed that field soil pots contained significantly higher pathogen richness and relative abundance than sterilized soil pots (fig. S1), and fungal pathogens in field soil were compositionally distinct from sterilized soil [permutational analysis of variance (PERMANOVA): df = 1, F = 1.8875, and P = 0.036]. In live field soil, variation in biomass production was better explained by PD than by SR (AIC = 964 and 981, respectively; table S3). When both SR and PD were included in the same models, PD had a strong influence in the fitted model in live field soil but not in sterilized soil pots (table S3). We found that PD was the most important determinant for seedling biomass, followed by SR and soil treatment, which had diminishing but still important effects (fig. S2). The regression slope for pot mean biomass as a function of PD was significantly greater in live field soil than in sterilized soil (P = 0.049; Fig. 4B), and a similar but nonsignificant effect was also found for the relationships between seeding biomass and SR (P = 0.350; Fig. 4A), indicating that soil microbes promoted the BEF and PDEF relationships for seedling communities and sterilizing the soil flattened the diversity-productivity relationship. We partitioned net biodiversity effects into complementarity and selection effects and found that compared to sterilized soil pots, field soil inoculation significantly and strongly promoted complementary effects among tree seedlings, while the selection effects remained small and showed no difference between the field and sterilized soil (fig. S3).
Fig. 3. Phylogenetic relationships of focal tree species and experimental design for the shade-house experiments.
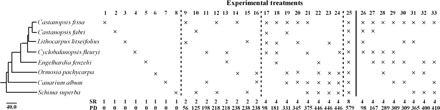
The phylogeny was built with the online software Phylomatic. In the first experiment (treatments 1 to 25), each pot included eight seedlings representing one of four SR levels: monocultures (SR1), treatments 1 to 8; two species (SR2), treatments 9 to16; four species (SR4), treatments 17 to 24; and eight species (SR8), treatment 25. In the second experiment (treatments 26 to 33), we transplanted four seedlings into each pot with a fixed SR of four, with Castanopsis fissa included as a shared species in all treatments. Treatments varied in PD from 98 to 410 in units of branch length.
Fig. 4. The relationship between plant biomass production and seedling diversity in pots in shade-house experiment 1.
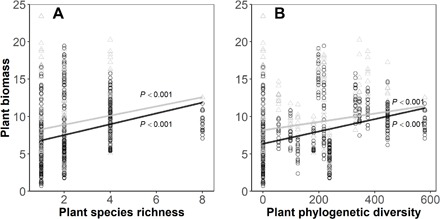
(A) SR and (B) PD. Lines are estimated from linear regressions, with black symbols and lines showing results of pots containing field soil and gray for pots containing initially sterilized soil. Statistics for lines are given in table S3.
Total richness of fungal operational taxonomic units (OTUs) per pot was not related to either plant SR or PD (Fig. 5, A and B). However, the richness of pathogenic fungi increased with increasing SR and PD (Fig. 5, C and D), while relative abundance of pathogens (as a proportion of all fungi in the sample) exhibited a significant downward trend (Fig. 5, E and F). Because we used the exact same field soil inocula at the beginning of seedling transplanting, these results indicated that more diverse plant assemblages could support a more diverse assemblage of fungal pathogens. However, pathogen relative abundance was sensitive to the relative density of any particular plant host or close relatives; lower diversity treatments had higher densities of individual plant species and thus a greater opportunity for pathogen enhancement in the soil, leading to greater pathogen read number in low SR or low PD treatments. This pattern is congruent with a dilution effect of high plant diversity on pathogenic fungi. Fungal OTU diversity, pathogen richness, and pathogen relative abundance were lower in the sterile than in the live treatment and were not significantly correlated with seedling diversity in the sterilized soil pots (fig. S4).
Fig. 5. Variation in total fungal richness, richness of pathogenic fungal OTUs, and read abundance of pathogenic fungi as functions of plant SR and PD in each field soil–treated pot in shade-house experiment 1.
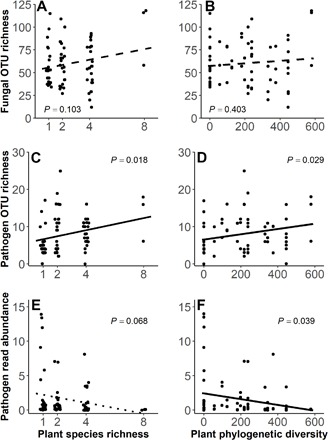
(A and B) OTU richness was calculated on the basis of internal transcribed spacer (ITS) ribosomal DNA sequencing data in each pot. (C and D) Pathogen richness was calculated as number of fungal OTUs per pot identified as pathotrophs. (E and F) Pathogen read abundance was the percentage of the total number of fungal reads from each pot that were identified as pathogenic fungal OTUs. Each dot represents the results for an individual pot; points on the left (A, C, and E) are positioned to include a small amount of random variation along the x axis when necessary to avoid overplotting.
In the experiment to test how soil microbial community composition affects PDEF relationships of tree seedlings (experiment 2; Fig. 3), we observed significantly positive relationships between mean plant biomass and PD when seedlings were inoculated with soil microbes but no effect in sterilized soil (Fig. 6 and table S4). While total fungal richness and the richness of pathogenic fungal OTUs were not correlated with plant PD in all treatments, pathogen read abundance was significantly and negatively related to PD in field soil from Castanopsis fissa and the four-species field soil but not in sterilized soil (fig. S5). The PDEF relationship did not differ when microbial inoculum was collected under adults of each of the four species in that pot or only under adults of the common species C. fissa (Fig. 6A). We found a similar relationship when analysis included only the biomass of C. fissa in each pot (Fig. 6B). However, the PDEF relationships were not significant in any of the three soil treatments when the biomass of C. fissa was excluded (Fig. 6C), showing that there was a phylogenetic signal in the effects of soil microbes on C. fissa; the effect of microbes decreased when seedlings of C. fissa were mixed with more distantly related species.
Fig. 6. The relationship between PD and total plant biomass production with different soil biota treatments in shade-house experiment 2.
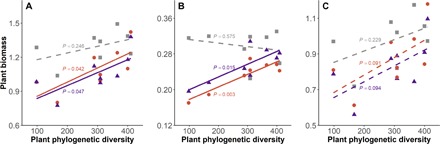
Seedling standardized mean biomass were calculated for (A) all four species, (B) only C. fissa, and (C) all species excluding C. fissa. Solid lines indicate significant (P < 0.05) linear regressions. Colored symbols indicate different soil biota treatments: purple triangles, combined field soil collected under adults of all four species in that pot; orange circles, field soil from beneath C. fissa; gray squares, sterilized soil. Statistics for lines are given in table S4.
DISCUSSION
We found notable evidence that tree productivity of seedlings and adult trees was strongly explained by the PD of communities. The positive effect of PD on productivity was seen for both adult trees and seedlings across multiple scales, suggesting that the patterns were not driven by patchiness in soil properties. Moreover, similar patterns were seen in synthesized communities in shade-house experiments where soil properties were held constant but the composition of microbial communities was manipulated. Therefore, our observational and experimental results together provide compelling evidence that soil fungi likely influence the relationship between tree PD and productivity in subtropical forest. We found a positive relationship between PD and productivity of seedlings in shade-house experiment 2 but only in nonsterile soil (Fig. 6); in shade-house experiment 1, we did see an effect of PD in the sterilized soil. Together, the results indicate that plants in communities of closely related species (low PD) produced less biomass when grown in live soil compared to sterile soil. There are at least two reasons to explain the different responses in the sterilized soil treatments in the two experiments. First, experiment 1 contained manipulations of both SR and PD gradients, while experiment 2 only comprised a PD treatment. Our results showed that SR per se promoted the complementarity effect among tree species (figs. S2 and S3), and this effect may not have been generated through interactions with plant pathogens but instead may be caused by competition for light and soil nutrients, which can happen in both live and sterilized soil. Second, although sterilized soil pots had significantly fewer pathogens than in live soil (fig. S1), some limited recolonization by pathogens could have promoted the PDEF relationship in these pots. These findings provide direct evidence of a strong phylogenetic signal between communities of plants and putative pathogenic fungi and that soil fungi might be an important driver of PDEF relationships in forests. Plant-soil feedbacks are prevalent in ecosystems (21), and previous work has shown that plants often perform differently in soil from beneath conspecific and heterospecific species (12, 22, 23). In tropical forest, seedling growth is reduced in soils from conspecific adults, indicating that soil microorganisms induce overall negative plant-soil feedback (12). Our findings provide further mechanistic insight by showing that soil microorganisms can regulate the positive relationships between productivity and both species and PD.
Recent work suggests that the ability of plants to form symbioses with either arbuscular or ectomycorrhizal fungi explains the direction of feedback in temperate trees (22). Our experiments used mixtures of plants that form either arbuscular (three species) or ectomycorrhizal (five species) symbioses, broadly reflecting patterns of coexistence and community structure in the Heishiding forest, as well as in species-rich Southeast Asian tropical forests. While our experiments were not explicitly designed to tease apart the effects of these host types, the analysis indicated that there were no systematic differences between plants that form different types of mycorrhiza. Regardless of host type, we found that mixtures of soil representing diverse plant communities (in terms of either SR or PD) significantly dampened positive effects of community PD on productivity compared to soil from beneath monocultures (Fig. 6).
We also found that the diversity of root-associated fungi and fungal pathogens was positively related to plant diversity and the relative abundance of fungal pathogens decreased with increasing seedling diversity. In a previous study, we sequenced field soil supporting different adult densities in the study site and found that greater pathogen frequency was significantly associated with reduced seedling performance for almost all focal species (24). Similar results were also reported in recent studies [e.g., (25)], which confirmed that relative and absolute pathogen DNA quantities in seedling roots were significantly correlated with the proportion of roots infected and that infection reduced seedling growth. Hence, the accumulation of plant pathogens signified by higher relative abundance of pathogenic OTU reads led to more negative effects on seedling performance at low SR and PD (Figs. 4 and 6). Thus, as greater plant diversity drives greater diversity in associated fungal communities, the ability of pathogens to proliferate in seedling roots appears to diminish; fungal diversity itself may therefore be a key mechanism in reducing pathogenicity of soil inocula. This supports recent findings that the richness of both fungal mutualists and pathogens can regulate the strength of plant-soil feedbacks (26) and adds to the broader view that soil biodiversity is a key driver of ecosystem multifunctionality (27). The mechanism behind fungal diversity–plant diversity relationships is likely multidimensional and may reflect competition among fungi for niche space in soil and roots [e.g., (28)], direct competition between individual fungal taxa (29), and host preference, especially of pathogens (30). We detected a significant trend that the proportional abundance of OTU reads identified as pathogens decreased with increasing plant PD (Fig. 5F), which was consistent with our expectation. However, read abundance of a group of taxa may not accurately reflect active fungal biomass, as potentially systematic biases inherent in sample processing, amplification, and sequencing may alter read abundance and create uncertainty in the use of quantitative metrics in metabarcoding datasets (31, 32). Our measure of fungal pathogen abundance is necessarily approximate, but it is the best currently available, and we have no reason to expect systematic variation in amplification of pathogenic fungi across treatments that would change the overall pattern. We also acknowledge that while FUNGuild is useful for fungal functional group assignment [e.g., (26)], many OTUs were placed into the unassigned group, as the FUNGuild database is currently incomplete (33). In addition, because many plant pathogens are facultative saprotrophs, fungal taxa assigned as pathogens may act differently on the specific host species used in our study. All these factors may influence our appraisal of fungal communities and their effects on host performance. Nevertheless, given the same initial soil biota inoculum, the significant relationships between plant community diversity and both fungal richness and pathogen abundance in roots (Fig. 5) indicate that plant diversity had nonrandom dilution effects on root pathogenic infection. This can explain the different diversity-productivity relationships between nonsterilized and sterilized soil (Fig. 4).
Our findings inform the debate on how biodiversity affects productivity of tree species. At the local scale, SR has been shown to be positively associated with tree productivity (34), and large-scale manipulations in subtropical forest have demonstrated positive effects of SR and PD on productivity in the first 8 years of tree establishment (7). Closely related species often have similar functional traits, niches, and ecological interactions; hence, PD is more likely than SR to describe the functional trait space represented by a community and therefore more likely to explain how community function is affected through different mechanisms (6, 17). Few studies have teased apart the mechanisms by which biodiversity drives productivity in trees, and some have found results that contrast with those in our study. For example, Grossman et al. (19) found that productivity of trees was best explained by community-weighted mean functional trait values rather than PD. Their findings did not detect a PDEF relationship driven by soil-borne pathogens, which may be due to the relatively low seedling density (6.9 seedlings per m2) compared to the density used in our experiments, which was more than 100 seedlings per m2. In addition, their experimental design did not include inoculation with pathogens and instead included only mycorrhizal fungal inocula that may mask pathogen effects on productivity. Further studies will require examining and measuring specific traits that could mediate pathogen effects to integrate the roles of functional traits and PD in regulating community productivity.
Our findings provide insight into the mechanisms underpinning relationships between PD and community productivity, namely, that the effects of soil-borne pathogens on interspecific interactions between plants have a strong phylogenetic signal. The finding that PD of trees interacts with soil fungal pathogens to affect plant productivity has implications for restoration of subtropical forests and suggests that maximum productivity should be achieved when phylogenetically diverse seedling communities are established.
MATERIALS AND METHODS
Study site
The study was located at the Heishiding Nature Reserve (111°53′E, 23°27′N) in south China, which consists of approximately 4200 ha of subtropical evergreen broad-leaved forest with an altitude range from 150 to 927 m. This site has a subtropical moist monsoon climate, and the Tropic of Cancer runs through its center. Mean monthly high temperatures range from 10.6°C in January to 28.4°C in July, and mean precipitation is 1744 mm, with 79% of annual rainfall occurring between April and September. The reserve contains at least 245 tree species belonging to 160 genera and 71 families. The dominant trees in this region are species in the Fagaceae and Lauraceae.
Relationships between PD and SR and biomass of trees in nature
To estimate how the PD of adult tree communities was correlated with aboveground productivity in nature, we used data from a 50-ha permanent plot at the field site established in 2012–2013. We tagged all adult trees and saplings with diameters at breast height (DBH) ≥ 1 cm and identified each to species, mapped their locations, and measured DBH and height. A total of 161,377 individuals were recorded, belonging to 214 species, 130 genera, and 61 families.
We divided the 50-ha permanent plot into nonoverlapping square quadrats of five spatial scales: 10 m by 10 m, 20 m by 20 m, 30 m by 30 m, 40 m by 40 m, and 50 m by 50 m. At each scale, we calculated the aboveground biomass based on DBH, using the equation , which is the best proxy for the aboveground biomass in the study area (35). We also calculated tree SR and Faith’s PD (sum of phylogenetic distance among species in a community) (36), which incorporate relative abundances and phylogenetic distances at each spatial scale. The angiosperm supertree structure and divergent age data were acquired from (37), which were based on APG IV (38). We then used a Phylomatic program and the BLADJ algorithm of the Phylocom version 4.2 software package to obtain an ultrametric tree with branch lengths scaled to divergent time. The measurements of diversity were calculated using R packages “picante” and “vegan” (39). We compared simple linear models of aboveground biomass against community diversity at each spatial scale, using AIC to detect whether PD was the stronger predictor for plot biomass.
To investigate the effect of PD of neighboring seedlings on growth and biomass at the seedling stage, in spring 2008, we demarcated 1200 1 m by 1 m quadrats, which were regularly spaced within six 1-ha permanent plots at the field site (13). In these quadrats, we surveyed seedlings of all woody plants (DBH < 1 cm) every spring from 2009 to 2017. At each census, we tagged all seedlings, determined their species, and measured their heights. In total, over the 9 years, we recorded 17,824 individuals belonging to 130 species, 82 genera, and 48 families. We summed seedling heights in each of the 1200 quadrats as a proxy of seedling community biomass and calculated corresponding seedling SR and Faith’s PD. We used linear models to test whether the diversity indices were correlated with seedling biomass in the 2017 census.
Experiments to test mechanisms underpinning biodiversity-productivity relationships
We conducted two independent but related microcosm experiments to evaluate BEF and PDEF relationships in seedlings and to investigate how soil microbes affect these relationships. For these experiments, we selected eight common tree species in the study area that had a wide range of phylogenetic relatedness (Fig. 3, left), including the ectomycorrhizal-forming species C. fissa (Fagaceae), Castanopsis fabri (Fagaceae), Lithocarpus litseifolius (Fagaceae), Cyclobalanopsis fleuryi (Fagaceae), and Engelhardia fenzelii (Juglandaceae) and the arbuscular mycorrhizal–forming species Ormosia pachycarpa (Fabaceae), Canarium album (Burseraceae), and Schima superba (Theaceae). Focal species with the two main mycorrhizal types were both selected, as recent evidence suggested that different types of mycorrhizal associations could affect the overall strength of plant-soil feedbacks (22).
We collected seeds and fruits for all focal species throughout the study site during winter 2013. Surface-sterilized seeds were kept at 4°C until February 2014 and then germinated in wet sterilized sand. There were sufficient newly germinated seedlings for all eight species after 6 to 8 weeks, and a total number of 8880 seedlings were included in the experiments (Fig. 3; see detailed designs below). For each treatment, the seedlings were randomly selected and transplanted into plastic pots (21 cm in diameter and 25 cm tall) containing background soil substrate. The soil was collected from a common understory at the study site, sterilized with 25 kilogray of γ radiation, and thoroughly mixed to eliminate nutrient differences among treatments. One week after transplanting, we replaced seedlings that were dead or growing poorly because of transplant injuries. We then added different field soil inocula to the rhizosphere of the seedlings (described below), filling about one-sixth of the total volume of each pot, which was then covered with 1-cm sterilized background soil.
Shade-house experiment 1
Experiment 1 tested how soil microorganisms altered the relationship between productivity and SR and PD of seedling communities. We collected 4-kg live (nonsterile) soil beneath three randomly selected adult trees of each of the eight focal species. We thoroughly mixed all soil from the 24 trees together (8 species × 3 adult trees) to ensure that the potential host-specific soil biota of each focal species was included, and different diversity treatments received exactly the same inoculum at the beginning. Two soil treatments were created: Half the field soil was placed directly in the pots to inoculate focal species with soil biota, and the other half was first sterilized as described previously and served as a sterile control. We transplanted eight seedlings into each pot at four levels of SR (Fig. 3): (i) monoculture, treatments 1 to 8; (ii) two species, treatments 9 to 16; (ii) four species, treatments 17 to 24; and (iv) eight species, treatment 25. We also selected eight different species combinations for the two-species and the four-species treatments, respectively, to set up a series of PD gradients (Fig. 3). The experimental units comprised 15 blocks (replicates), each block containing all 25 species combinations (n = 750 pots = 25 species treatments × 2 soil treatments × 15 replicates).
Shade-house experiment 2
Experiment 2 investigated how soil microbial community composition affects PDEF relationships of tree seedlings. In this experiment, we transplanted four seedlings into each pot with one seedling for each focal species (treatments 26 to 33 in Fig. 3). Each of the eight treatments contained four species and always included C. fissa based on seedling availability and phylogenetic relatedness among focal species, which resulted in considerable variation in PD among the communities. We established three different soil inoculum treatments: (i) soil from beneath C. fissa, (ii) a mixture of soil from the rhizosphere of the four focal species in the pot, and (iii) sterilized soil. For each of the eight species, we collected live soil beneath three randomly selected adult trees and then thoroughly mixed them, as above. For the four-species mixture, we mixed equal amounts of soil from the same four species corresponding to the different species combinations (treatments 26 to 33 in Fig. 3). This pooling approach was done to generate microbial communities associated with phylogenetically distinct tree species, on which PDEF relationships could be tested. It was not used to ascribe causation to plant-soil feedbacks, where pooling samples has been criticized (40). We also thoroughly mixed soil of all eight species, which was then sterilized and used for the sterilized soil treatment. This experiment comprised 30 blocks (8 species combinations × 3 soil treatments × 30 replicates = 720 pots).
For both shade-house experiments, we randomly placed the pots within each block and separated all blocks by a distance of 0.5 m in a shade-house located at the field site, with temperature and humidity conditions similar to the forest understory. We regularly watered the seedlings and monitored seedling survival weekly for 12 months and then harvested all plants. Shoots and roots of each seedling were oven-dried separately at 60°C for 48 hours to determine biomass.
Fungal community composition and DNA sequencing
To assess the fungal community composition associated with root samples of different experimental treatments for both experiments, we randomly chose three pots for each treatment, collected three fine root subsamples (that included ectomycorrhizas in those species that form them), and mixed them together to create a composite sample of 5-g fresh weight. We surface-sterilized root samples for 1 min in 1% NaOCl and 1 min in 70% alcohol, then rinsed twice in sterile Ringer’s solution (0.9% NaCl, 0.042% KCl, 0.048% CaCl2, and 0.02% NaHCO3), and performed a final rinse with distilled water. We extracted total genomic DNA directly from each root sample according to a standard procedure (E.Z.N.A. Tissue DNA Kit; Omega Bio-tek, Norcross, GA, USA). The nuclear ribosomal internal transcribed spacer region [ITS ribosomal DNA gene] was amplified by polymerase chain reaction (PCR) using the fungal primer set of ITS1F (CTTGGTCATTTAGAGGAAGTAA) and ITS2 (GCTGCGTTCTTCATCGATGC) (41). PCR was performed with standard protocols (see Supplementary Methods). We sequenced DNA samples using the Illumina MiSeq platform (Illumina, San Diego, CA, USA). Raw FASTQ files were demultiplexed, quality-filtered by Trimmomatic, and merged by FLASH with the following criteria: (i) The reads were truncated at any site receiving an average quality score <20 over a 50–base pair (bp) sliding window. (ii) Primers were exactly matched allowing two nucleotide mismatching, and reads containing ambiguous bases were removed. (iii) Sequences with overlap longer than 10 bp were merged according to their overlap sequence. OTUs were clustered with 97% similarity cutoff using UPARSE (version 7.1; http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each sequence was analyzed by RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the UNITE ITS sequences database (42), with an expected e value of <10−3 and a minimum identity of 97%. We assigned each identified fungal species to putative pathogenic or nonpathogenic fungi using the FUNGuild algorithm (33), following the approach used in several recent studies [e.g., (26, 43, 44)]. Of the total identified reads, 2.7% was assigned to plant pathogens, with 52.5% of the pathogenic OTUs at the “highly probable” confidence level and the others in the “probable” confidence ranking.
Statistical analysis
In shade-house experiment 1, we used linear regressions to test the relationships between mean plant biomass per pot and SR or PD for the different soil treatments. Data from the field soil and sterilized soil were analyzed separately; experimental block was treated as a random effect, and SR, PD, and their interaction were fixed effects in the models. We conducted PERMANOVA to test whether fungal pathogens in field soil were compositionally distinct from sterilized soil. We then fitted linear models with the whole dataset and included plant diversity, soil treatments, and their interaction as independent variables to test whether the different soil treatments (live versus sterile soil) significantly affect the slope of the response of productivity to plant richness and PD. We used the additive partitioning method of Loreau and Hector (45) to partition net biodiversity effects into complementarity and selection effects. We also quantified the relative importance of SR, PD, soil treatments, and their interactions using the R package “relaimpo” (46).
In experiment 2, we calculated standardized mean biomass of each species and tested how pot mean biomass correlated with PD of each pot. To investigate how root-associated fungal communities were affected by tree species treatments, we calculated, for each pot, the total richness of all fungal OTUs, the richness of pathogenic OTUs, and the proportion of the total reads of plant pathogenic fungal OTUs as a relative measure of the abundance of pathogenic fungi. We then assessed how each varied with SR or PD using simple linear regressions. All calculations and analyses were performed using R for Windows 3.4.0 (39).
Supplementary Material
Acknowledgments
We are grateful to S. Liu and W. Ye for assistance in the field. Funding: This research was funded by the National Key Research and Development Program of China (project no. 2017YFA0605100) and the National Natural Science Foundation of China (NSFC grants 31870403, 31500334, and 31770466). D.J. receives partial support from the N8 AgriFood programme and funding from the Natural Environment Research Council (no. NE/R004986/1). Author contributions: X.L., M.L., and S.Y. designed the study. M.L., X.L., Y.Z., and S.L. conducted the experiments. M.L. performed statistical analyses, with substantial input from I.M.P., G.S.G., and D.J. on data interpretation. M.L. wrote the first draft of the manuscript, and all authors contributed to revisions. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Shade-house experimental data are available at Figshare (https://doi.org/10.6084/m9.figshare.8863592). Field census data are available upon reasonable request from the ForestGEO data portal at http://ctfs.si.edu/datarequest/. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaax5088/DC1
Supplementary Methods
Fig. S1. Soil inoculum and sterile effects on community structure of plant pathogens in the shade-house experiments.
Fig. S2. The relative importance of plant richness, PD, soil microbes, and their interaction for plant productivity in shade-house experiment 1.
Fig. S3. Partition of complementarity and selection effects of plant diversity on seedling biomass in shade-house experiment 1.
Fig. S4. Variation in diversity of total and pathogenic fungal OTUs as functions of plant diversity in the sterilized soil of shade-house experiment 1.
Fig. S5. Regression coefficients of read abundance of pathogenic fungi (top) total fungal richness (middle), and richness of pathogenic fungal OTUs (bottom) as functions of plant PD in different soil biota treatments in shade-house experiment 2.
Table S1. Results of linear models predicting the biomass production to SR and Faith’s PD of tree communities in test quadrats at five spatial scales of the 50-ha plot, as shown in Fig. 2.
Table S2. Results of linear models predicting the total height of seedlings in each 1-m2 seedling quadrat as a function of SR or Faith’s PD among neighboring tree seedlings at the seedling stage, as shown in Fig. 2.
Table S3. Results of linear models predicting pot biomass production to SR and Faith’s PD in shade-house experiment 1, as shown in Fig. 4.
Table S4. Results of linear models of the relationship between PD and total plant biomass production in shade-house experiment 2, as shown in Fig. 6.
REFERENCES AND NOTES
- 1.Crozier R. H., Preserving the information content of species: Genetic diversity, phylogeny and conservation worth. Annu. Rev. Ecol. Syst. 28, 243–268 (1997). [Google Scholar]
- 2.Maherali H., Klironomos J. N., Influence of phylogeny on fungal community assembly and ecosystem functioning. Science 316, 1746–1748 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Cadotte M. W., Cavender-Bares J., Tilman D., Oakley T. H., Using phylogenetic, functional and trait diversity to understand patterns of plant community productivity. PLOS ONE 4, e5695 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mouquet N., Devictor V., Meynard C. N., Munoz F., Bersier L. F., Chave J., Couteron P., Dalecky A., Fontaine C., Gravel D., Hardy O. J., Jabot F., Lavergne S., Leibold M., Mouillot D., Münkemüller T., Pavoine S., Prinzing A., Rodrigues A. S., Rohr R. P., Thébault E., Thuiller W., Ecophylogenetics: Advances and perspectives. Biol. Rev. Camb. Philos. Soc. 87, 769–785 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Srivastava D. S., Cadotte M. W., MacDonald A. A. M., Marushia R. G., Mirotchnick N., Phylogenetic diversity and the functioning of ecosystems. Ecol. Lett. 15, 637–648 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Cadotte M. W., Cardinale B. J., Oakley T. H., Evolutionary history and the effect of biodiversity on plant productivity. Proc. Natl. Acad. Sci. U.S.A. 105, 17012–17017 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Chen Y., Castro-Izaguirre N., Baruffol M., Brezzi M., Lang A., Li Y., Härdtle W., von Oheimb G., Yang X., Liu X., Pei K., Both S., Yang B., Eichenberg D., Assmann T., Bauhus J., Behrens T., Buscot F., Chen X.-Y., Chesters D., Ding B.-Y., Durka W., Erfmeier A., Fang J., Fischer M., Guo L.-D., Guo D., Gutknecht J. L. M., He J.-S., He C.-L., Hector A., Hönig L., Hu R.-Y., Klein A.-M., Kühn P., Liang Y., Li S., Michalski S., Scherer-Lorenzen M., Schmidt K., Scholten T., Schuldt A., Shi X., Tan M.-Z., Tang Z., Trogisch S., Wang Z., Welk E., Wirth C., Wubet T., Xiang W., Yu M., Yu X.-D., Zhang J., Zhang S., Zhang N., Zhou H.-Z., Zhu C.-D., Zhu L., Bruelheide H., Ma K., Niklaus P. A., Schmid B., Impacts of species richness on productivity in a large-scale subtropical forest experiment. Science 362, 80–83 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Klironomos J. N., Feedback with soil biota contributes to plant rarity and invasiveness in communities. Nature 417, 67–70 (2002). [DOI] [PubMed] [Google Scholar]
- 9.van der Heijden M. G. A., Bardgett R. D., van Straalen N. M., The unseen majority: Soil microbes as drivers of plant diversity and productivity in terrestrial ecosystems. Ecol. Lett. 11, 296–310 (2008). [DOI] [PubMed] [Google Scholar]
- 10.Maron J. L., Marler M., Klironomos J. N., Cleveland C. C., Soil fungal pathogens and the relationship between plant diversity and productivity. Ecol. Lett. 14, 36–41 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Schnitzer S. A., Klironomos J. N., HilleRisLambers J., Kinkel L. L., Reich P. B., Xiao K., Rillig M. C., Sikes B. A., Callaway R. M., Mangan S. A., van Nes E. H., Scheffer M., Soil microbes drive the classic plant diversity–productivity pattern. Ecology 92, 296–303 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Mangan S. A., Schnitzer S. A., Herre E. A., Mack K. M. L., Valencia M. C., Sanchez E. I., Bever J. D., Negative plant–soil feedback predicts tree-species relative abundance in a tropical forest. Nature 466, 752–755 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Liu X., Liang M., Etienne R. S., Wang Y., Staehelin C., Yu S., Experimental evidence for a phylogenetic Janzen-Connell effect in a subtropical forest. Ecol. Lett. 15, 111–118 (2012). [DOI] [PubMed] [Google Scholar]
- 14.Gilbert G. S., Webb C. O., Phylogenetic signal in plant pathogen–host range. Proc. Natl. Acad. Sci. U.S.A. 104, 4979–4983 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parker I. M., Saunders M., Bontrager M., Weitz A. P., Hendricks R., Magarey R., Suiter K., Gilbert G. S., Phylogenetic structure and host abundance drive disease pressure in communities. Nature 520, 542–544 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Liu X., Liang M., Etienne R. S., Gilbert G. S., Yu S., Phylogenetic congruence between subtropical trees and their associated fungi. Ecol. Evol. 6, 8412–8422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flynn D. F. B., Mirotchnick N., Palmer M. J. M. I., Naeem S., Functional and phylogenetic diversity as predictors of biodiversity–ecosystem–function relationships. Ecology 92, 1573–1581 (2011). [DOI] [PubMed] [Google Scholar]
- 18.Venail P. A., Vines M. J., Phylogenetic distance and species richness interactively affect the productivity of bacterial communities. Ecology 94, 2529–2536 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Grossman J. J., Cavender-Bares J., Hobbie S. E., Reich P. B., Montgomery R. A., Species richness and traits predict overyielding in stem growth in an early-successional tree diversity experiment. Ecology 98, 2601–2614 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Clarke D. A., York P. H., Rasheed M. A., Northfield T. D., Does biodiversity-ecosystem function literature neglect tropical ecosystems? Trends Ecol. Evol. 32, 320–323 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Lekberg Y., Bever J. D., Bunn R. A., Callaway R. M., Hart M. M., Kivlin S. N., Klironomos J., Larkin B. G., Maron J. L., Reinhart K. O., Remke M., van der Putten W. H., Relative importance of competition and plant–soil feedback, their synergy, context dependency and implications for coexistence. Ecol. Lett. 21, 1268–1281 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Bennett J. A., Maherali H., Reinhart K. O., Lekberg Y., Hart M., Klironomos J., Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science 355, 181–184 (2017). [DOI] [PubMed] [Google Scholar]
- 23.Teste F. P., Kardol P., Turner B. L., Wardle D. A., Zemunik G., Renton M., Laliberté E., Plant-soil feedback and the maintenance of diversity in Mediterranean-climate shrublands. Science 355, 173–176 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Liang M., Liu X., Gilbert G. S., Zheng Y., Luo S., Huang F., Yu S., Adult trees cause density-dependent mortality in conspecific seedlings by regulating the frequency of pathogenic soil fungi. Ecol. Lett. 19, 1448–1456 (2016b). [DOI] [PubMed] [Google Scholar]
- 25.Moein S., Mazzola M., Spies C. F. J., McLeod A., Evaluating different approaches for the quantification of oomycete apple replant pathogens, and their relationship with seedling growth reductions. Eur. J. Plant Pathol. 154, 243–257 (2019). [Google Scholar]
- 26.Semchenko M., Leff J. W., Lozano Y. M., Saar S., Davison J., Wilkinson A., Jackson B. G., Pritchard W. J., De Long J. R., Oakley S., Mason K. E., Ostle N. J., Baggs E. M., Johnson D., Fierer N., Bardgett R. D., Fungal diversity regulates plant-soil feedbacks in temperate grassland. Sci. Adv. 4, eaau4578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Delgado-Baquerizo M., Maestre F. T., Reich P. B., Jeffries T. C., Gaitan J. J., Encinar D., Berdugo M., Campbell C. D., Singh B. K., Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hutchings M. J., John E. A., Wijesinghe D. K., Toward understanding the consequences of soil heterogeneity for plant populations and communities. Ecology 84, 2322–2334 (2003). [Google Scholar]
- 29.Hazard C., Johnson D., Does genotypic and species diversity of mycorrhizal plants and fungi affect ecosystem function? New Phytol. 220, 1122–1128 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Sarmiento C., Zalamea P. C., Dalling J. W., Davis A. S., Stump S. M., U’Ren J. M., Arnold A. E., Soilborne fungi have host affinity and host-specific effects on seed germination and survival in a lowland tropical forest. Proc. Natl. Acad. Sci. U.S.A. 114, 11458–11463 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amend A. S., Seifert K. A., Bruns T. D., Quantifying microbial communities with 454 pyrosequencing: Does read abundance count? Mol. Ecol. 19, 5555–5565 (2010). [DOI] [PubMed] [Google Scholar]
- 32.Nichols R. V., Vollmers C., Newsom L. A., Wang Y., Heintzman P. D., Leighton M., Green R. E., Shapiro B., Minimizing polymerase biases in metabarcoding. Mol. Ecol. Resour. 18, 927–939 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Nguyen N. H., Song Z., Bates S. T., Branco S., Tedersoo L., Menke J., Schilling J. S., Kennedy P. G., FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016). [Google Scholar]
- 34.Liang J., Crowther T. W., Picard N., Wiser S., Zhou M., Alberti G., Schulze E.-D., McGuire A. D., Bozzato F., Pretzsch H., de-Miguel S., Paquette A., Hérault B., Scherer-Lorenzen M., Barrett C. B., Glick H. B., Hengeveld G. M., Nabuurs G.-J., Pfautsch S., Viana H., Vibrans A. C., Ammer C., Schall P., Verbyla D., Tchebakova N., Fischer M., Watson J. V., Chen H. Y. H., Lei X., Schelhaas M.-J., Lu H., Gianelle D., Parfenova E. I., Salas C., Lee E., Lee B., Kim H. S., Bruelheide H., Coomes D. A., Piotto D., Sunderland T., Schmid B., Gourlet-Fleury S., Sonké B., Tavani R., Zhu J., Brandl S., Vayreda J., Kitahara F., Searle E. B., Neldner V. J., Ngugi M. R., Baraloto C., Frizzera L., Bałazy R., Oleksyn J., Zawiła-Niedźwiecki T., Bouriaud O., Bussotti F., Finér L., Jaroszewicz B., Jucker T., Valladares F., Jagodzinski A. M., Peri P. L., Gonmadje C., Marthy W., O’Brien T., Martin E. H., Marshall A. R., Rovero F., Bitariho R., Niklaus P. A., Alvarez-Loayza P., Chamuya N., Valencia R., Mortier F., Wortel V., Engone-Obiang N. L., Ferreira L. V., Odeke D. E., Vasquez R. M., Lewis S. L., Reich P. B., Positive biodiversity-productivity relationship predominant in global forests. Science 354, aaf8957 (2016). [DOI] [PubMed] [Google Scholar]
- 35.Liu W., Yu S., Wang Y., Lian J., Comparison of the biomass estimation methods for the forest at Heishiding nature reserve Acta Sci. Nat. Univ. Sunyatseni 41, 80,– 84 (2002). [Google Scholar]
- 36.Faith D. P., Conservation evaluation and phylogenetic diversity. Biol. Conserv. 61, 1–10 (1992). [Google Scholar]
- 37.Gastauer M., Meira-Neto J. A. A., Updated angiosperm family tree for analyzing phylogenetic diversity and community structure. Acta Bot. Brasilica. 31, 191–198 (2017). [Google Scholar]
- 38.The Angiosperm Phylogeny Group, Chase M. W., Christenhusz M. J. M., Fay M. F., Byng J. W., Judd W. S., Soltis D. E., Mabberley D. J., Sennikov A. N., Soltis P. S., Stevens P. F., An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 181, 1–20 (2016). [Google Scholar]
- 39.R Core Team, “R: A language and environment for statistical computing” R Foundation for Statistical Computing, 2017; www.R-project.org/.
- 40.Rinella M. J., Reinhart K. O., Toward more robust plant-soil feedback research. Ecology 99, 550–556 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Epp L. S., Boessenkool S., Bellemain E. P., Haile J., Esposito A., Riza T., Erseus C., Gusarov V. I., Edwards M. E., Johnsen A., Stenøien H. K., New environmental metabarcodes for analysing soil DNA: Potential for studying past and present ecosystems. Mol. Ecol. 21, 1821–1833 (2012). [DOI] [PubMed] [Google Scholar]
- 42.Abarenkov K., Nilsson R. H., Larsson K. H., Alexander I. J., Eberhardt U., Erland S., Høiland K., Kjøller R., Larsson E., Pennanen T., Sen R., The UNITE database for molecular identification of fungi—Recent updates and future perspectives. New Phytol. 186, 281–285 (2010). [DOI] [PubMed] [Google Scholar]
- 43.Cline L. C., Hobbie S. E., Madritch M. D., Buyarski C. R., Tilman D., Cavender-Bares J. M., Resource availability underlies the plant-fungal diversity relationship in a grassland ecosystem. Ecology 99, 204–216 (2018). [DOI] [PubMed] [Google Scholar]
- 44.Leff J. W., Bardgett R. D., Wilkinson A., Jackson B. G., Pritchard W. J., Long J. R., Oakley S., Mason K. E., Ostle N. J., Johnson D., Baggs E. M., Predicting the structure of soil communities from plant community taxonomy, phylogeny, and traits. ISME J. 12, 1794–1805 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loreau M., Hector A., Partitioning selection and complementarity in biodiversity experiments. Nature 412, 72–76 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Groemping U., Relative importance for linear regression in R: The package relaimpo. J. Stat. Softw. 17, 1–27 (2006). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/10/eaax5088/DC1
Supplementary Methods
Fig. S1. Soil inoculum and sterile effects on community structure of plant pathogens in the shade-house experiments.
Fig. S2. The relative importance of plant richness, PD, soil microbes, and their interaction for plant productivity in shade-house experiment 1.
Fig. S3. Partition of complementarity and selection effects of plant diversity on seedling biomass in shade-house experiment 1.
Fig. S4. Variation in diversity of total and pathogenic fungal OTUs as functions of plant diversity in the sterilized soil of shade-house experiment 1.
Fig. S5. Regression coefficients of read abundance of pathogenic fungi (top) total fungal richness (middle), and richness of pathogenic fungal OTUs (bottom) as functions of plant PD in different soil biota treatments in shade-house experiment 2.
Table S1. Results of linear models predicting the biomass production to SR and Faith’s PD of tree communities in test quadrats at five spatial scales of the 50-ha plot, as shown in Fig. 2.
Table S2. Results of linear models predicting the total height of seedlings in each 1-m2 seedling quadrat as a function of SR or Faith’s PD among neighboring tree seedlings at the seedling stage, as shown in Fig. 2.
Table S3. Results of linear models predicting pot biomass production to SR and Faith’s PD in shade-house experiment 1, as shown in Fig. 4.
Table S4. Results of linear models of the relationship between PD and total plant biomass production in shade-house experiment 2, as shown in Fig. 6.


