Abstract
The human gut microbiota, which underpins nutrition and systemic health, is compositionally sensitive to the availability of complex carbohydrates in the diet. The Bacteroidetes comprise a dominant phylum in the human gut microbiota whose members thrive on dietary and endogenous glycans by employing a diversity of highly specific, multi-gene polysaccharide utilization loci (PUL), which encode a variety of carbohydrases, transporters, and sensor/regulators. PULs invariably also encode surface glycan-binding proteins (SGBPs) that play a central role in saccharide capture at the outer membrane. Here, we present combined biophysical, structural, and in vivo characterization of the two SGBPs encoded by the Bacteroides ovatus mixed-linkage β-glucan utilization locus (MLGUL), thereby elucidating their key roles in the metabolism of this ubiquitous dietary cereal polysaccharide. In particular, molecular insight gained through several crystallographic complexes of SGBP-A and SGBP-B with oligosaccharides reveals that unique shape complementarity of binding platforms underpins specificity for the kinked MLG backbone vis-à-vis linear β-glucans. Reverse-genetic analysis revealed that both the presence and binding ability of the SusD homolog BoSGBPMLG-A are essential for growth on MLG, whereas the divergent, multi-domain BoSGBPMLG-B is dispensable but may assist in oligosaccharide scavenging from the environment. The synthesis of these data illuminates the critical role SGBPs play in concert with other MLGUL components, reveals new structure–function relationships among SGBPs, and provides fundamental knowledge to inform future (meta)genomic, biochemical, and microbiological analyses of the human gut microbiota.
Electronic supplementary material
The online version of this article (10.1007/s00018-019-03115-3) contains supplementary material, which is available to authorized users.
Keywords: Microbiota, Microbiome, Dietary fiber, Bacteroidetes, Beta-glucan, Cereal
Introduction
Trillions of microbial (mainly bacterial) cells make the human gut microbiota one of the most complex and dynamic ecosystems on the planet [1]. This microbial community has far-reaching influences on diverse aspects of human physiology and health [2] including, but not limited to, links to obesity [3, 4], asthma [5], allergies [6], and cancer [7]. The abundance of individual members of the human gut microbiota is driven by our diet, especially the influx of complex polysaccharides into the large intestine [8–12]. Indeed, regular ingestion of plant polysaccharides is integral to maintaining a healthy balance of microbes in our lower gastrointestinal tract [13–15].
The human genome is remarkably bereft of genes encoding carbohydrate-active enzymes (CAZymes) targeting dietary glycans. As such, we are critically dependent on members of the human gut microbiota to metabolize otherwise indigestible “dietary fiber” [15, 16]. Of the dominant bacterial phyla that comprise the microbiota, the Bacteroidetes in particular possess an enormous arsenal of CAZyme genes [17], which are co-localized along with genes encoding cognate surface glycan-binding proteins (SGBPs), TonB-dependent transporters (TBDTs), and transcriptional regulators into polysaccharide utilization loci (PULs) [18]. The gene products of a single PUL work in concert to sense, bind, cleave, and import a particular complex polysaccharide. Reflecting the large natural diversity of complex glycans encountered by the human gut microbiota, Bacteroidetes possess a plethora of PULs (e.g., 88 and 112 PULs in Bacteroides thetaiotaomicron and B. ovatus, respectively [19]), each of which is specifically upregulated by a target polysaccharide [19]. The abundance and broad distribution of PULs underscores their importance to human gut microbiota metabolism and, consequently, human nutrition and health.
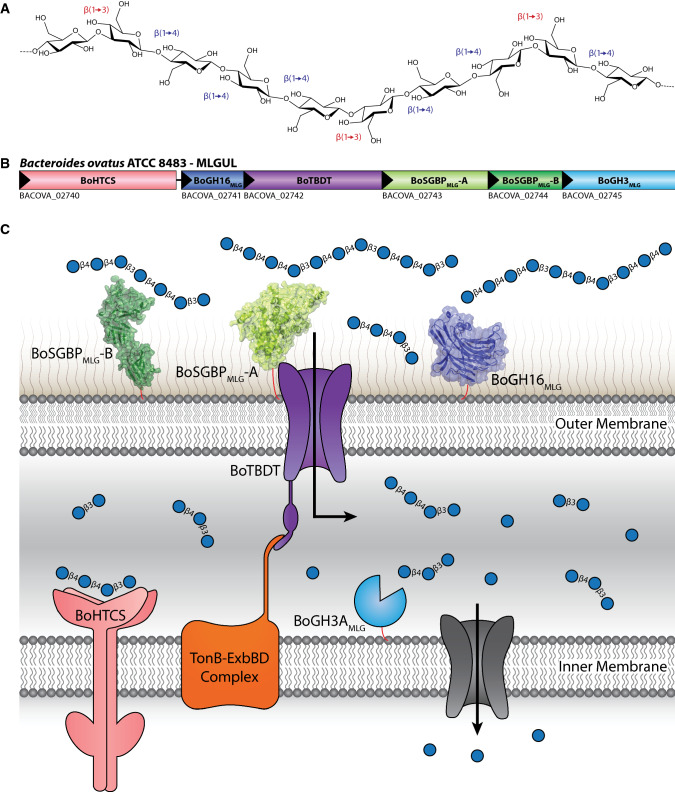
Mixed-linkage β-glucans (β(1,3)/β(1,4)-glucans, MLGs) are abundant in cereal grains such as oats and barley (Fig. 1a), and comprise a key group of human dietary glycans with recognized healthful effects. For example, the benefits of cereal-derived MLGs in ameliorating hypertension, diabetes mellitus, cardiovascular diseases, and cholesterol levels have been reported [15, 20–22]. Although the mechanisms behind these health benefits are not fully understood, the prebiotic activity of MLGs [23], a direct result of fermentability by the human gut microbiota [24], is likely to be a major factor.
Fig. 1.
Mixed-linkage glucan utilization locus (MLGUL). a Chemical structure of MLG targeted by this PUL, consisting of a linear glucan chain of β(1,4)-linked cellotriosyl and cellotetraosyl units separated by single β(1,3) bonds. b Genetic organization of the B. ovatus MLGUL. c Model of MLGUL machinery at the cell envelope showing the concerted mechanism of MLG utilization. Protein products are colored analogously to the gene locus. HTCS, hybrid two-component system sensor/regulator; GH, glycoside hydrolase, with family number indicated; TBDT, TonB-depdendent transporter (SusC homolog); SGBP, cell surface glycan-binding protein (SGBP-A is a SusD homolog, SGBP-B is highly sequence divergent); BACOVA_n, Bacteroides ovatus ATCC 8483 gene locus tag
We recently identified an MLG utilization locus (MLGUL) in B. ovatus (Fig. 1b), syntenic homologs of which enable select Bacteroides species in the human gut microbiota to metabolize this cereal glycan, and biochemically characterized its cognate glycoside hydrolases (GHs). Structural enzymology detailed the high specificity of the outer membrane GH16 endo-β-glucanase for MLG and supported a concerted model in which polysaccharide cleavage at the cell surface, oligosaccharide transport via a SusC-like TBDT, and periplasmic hydrolysis by GH3 exo-β-glucosidase enable complete MLG saccharification to glucose (Fig. 1c). As part of this study, we also demonstrated that MLGUL are essentially ubiquitous in human gut metagenomes [25].
This model also predicts the involvement of two cell surface glycan-binding proteins (SGBPs) encoded by the MLGUL (Fig. 1b) in the initial capture of the polysaccharide at the cell surface as a prelude to backbone hydrolysis, and/or facilitating product transport through the TBDT (Fig. 1c). Indeed, previous studies have outlined these roles for SGBPs in the archetypal starch utilization system (Sus) and other PULs [26–31], and SusC/SusD (TBDT/SGBP-A) homologs are known to form an intimate structural association in the membrane [32]. However, the molecular structures, substrate specificities, and individual contributions to MLG utilization are currently unknown for BoSGBPMLG-A (a SusD homolog) and BoSGBPMLG-B (a sequence-divergent “SusE-positioned” gene product) from the MLGUL. Here, we present the comprehensive biochemical, crystallographic, and reverse-genetic characterization of these two SGBPs to reveal the distinct roles they play in MLG metabolism by B. ovatus in the context of the human gut microbiota.
Results
MLGUL SGBP are highly specific for mixed-linkage β(1,3)/β(1,4)-glucans
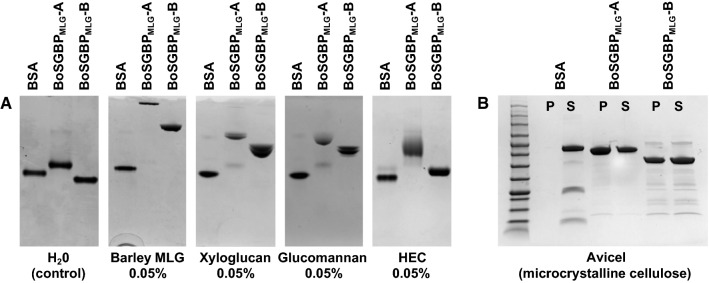
Recombinant BoSGBPMLG-A and BoSGBPMLG-B were produced in E. coli from constructs designed to exclude the predicted signal peptide and N-terminal lipidation site (Cys-1 of the mature protein; Fig S1). Carbohydrate binding was first screened qualitatively against a library of soluble polysaccharides (Fig. S2) by affinity gel electrophoresis (AGE). As suggested by their context in the MLGUL, the migration of both BoSGBPMLG-A and BoSGBPMLG-B was strongly retarded in a gel containing barley mixed-linkage β-glucan (bMLG, Fig. 2a). Qualitatively weaker interactions with tamarind xyloglucan (XyG), konjac glucomannan, and hydroxyethylcellulose were also observed (Fig. 2a), all of which contain stretches of β(1,4)-linked backbone glucosyl residues (Fig. S2). No binding was observed to a range of other polysaccharides, including diverse β(1,3)-glucans, mannans, xylans, dextran, ulvan, and anionic carboxymethylcellulose (Fig. S3). Isothermal titration calorimetry (ITC) confirmed the specific binding of both BoSGBPMLG-A and BoSGBPMLG-B to bMLG (Ka (4.41 ± 0.65) × 105 M−1 and (1.04 ± 0.1) × 104 M−1, respectively (Fig. S4, Table 1). In comparison, the affinity of BoSGBPMLG-A for xyloglucan was two orders of magnitude lower, while BoSGBPMLG-B binding to xyloglucan was too weak to be quantified (Fig. S4, Table 1).
Fig. 2.
BoSGBPMLG-A and BoSGBPMLG-B binding analyses. a Affinity electrophoresis gels against soluble polysaccharides that were bound by BoSGBPMLG-A and BoSGBPMLG-B with BSA as a non-binding control in each gel. b SDS-PAGE gel of pull-down assay against insoluble polysaccharide bound by BoSGBPMLG-A and BoSGBPMLG-B with BSA as a non-binding control
Table 1.
Summary of thermodynamic parameters for wild-type BoSGBPMLG-A and BoSGBPMLG-B obtained by isothermal titration calorimetry at 25 °C
| Carbohydrate | Ka (M−1) | ΔG (kcal mol−1) | ΔH (kcal mol−1) | TΔS (kcal mol−1) | n | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| BoSGBPMLG-A | BoSGBPMLG-B | BoSGBPMLG-A | BoSGBPMLG-B | BoSGBPMLG-A | BoSGBPMLG-B | BoSGBPMLG-A | BoSGBPMLG-B | BoSGBPMLG-A | BoSGBPMLG-B | |
| bMLGa | (4.41 ± 0.65) × 105 | (1.04 ± 0.1) × 104 | − 7.7 | − 5.5 | − 45.3 ± 1.6 | − 14.5 ± 1.6 | − 37.6 | − 9 | 1.11 ± 0.03 | 0.79 ± 0.07 |
| XyGb | (7.19 ± 1.9) × 103 | Weakc | − 5.2 | Weak | − 5.9 ± 1.8 | Weak | − 0.7 | Weak | 1.69 ± 0.53 | Weak |
| Cellotetraose | NBc | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| Cellopentaose | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| Cellohexaose | (3.39 ± 0.03) × 103 | (3.1 ± 0.06) × 103 | − 4.8 | − 4.8 | − 11.4 ± 0.05 | − 6.3 ± 0.09 | − 6.6 | − 1.5 | 1 | 1 |
| G3G4G4G | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| G4G4G3G | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| G4G3G4G | NB | NB | NB | NB | NB | NB | NB | NB | NB | NB |
| G4G3G4G4G3G | (1.44 ± 0.04) × 104 | (3.16 ± 0.19) × 103 | − 5.7 | − 2.4 | − 11.8 ± 0.3 | − 7.0 ± 0.4 | − 6.1 | − 2.2 | 1.49 ± 0.04 | 1 |
| G4G4G3G4G4G3G | (4.51 ± 0.34) × 104 | (4.69 ± 0.13) × 103 | − 6.3 | − 5 | − 11.9 ± 0.4 | − 13.9 ± 0.2 | − 5.6 | − 8.9 | 1.19 ± 0.05 | 1 |
| G4G4G3G4G4G3G | (4.07 ± 0.08) × 103 | − 4.9 | − 29 ± 0.4 | − 24.1 | 0.5 | |||||
Corresponding thermograms are shown in Figs. S4–S6. All parameters were allowed to vary independently during data fitting, with the exception of BoSGBPMLG-A and BoSGBPMLG-B binding cellohexaose, and BoSGBPMLG-B binding the MLGOs G4G3G4G4G3G and G4G4G3G4G4G3G, for which n was fixed at 1 or 0.5 as indicated
NB no binding observed
aKa values were calculated on a molar basis from MLG polysaccharide concentrations in g/L by assuming a hexasaccharide binding motif, based on crystal complex structures with MLGOs
bKa values were calculated on a molar basis from XyG polysaccharide concentrations in g/L by assuming a Glc8-backbone (XyGO dimer) oligosaccharide binding motif
cWeak: Ka < 500 M−1
Following the observed weak binding toward the soluble cellulose derivative hydroxyethyl cellulose (Fig. 2a), pull-down assays were conducted to assess binding to insoluble cellulose (β(1,4)-glucan) and β(1,4)-mannan. Both BoSGBPMLG-A and BoSGBPMLG-B indeed bind crystalline cellulose (Avicel, Fig. 2b), whereas neither bound insoluble β(1,4)-mannan (Fig. S2B). To quantify binding affinities to crystalline cellulose, green fluorescent protein (GFP) fusion proteins were produced to conduct depletion binding-isotherm experiments. GFP-BoSGBPMLG-A and GFP-BoSGBPMLG-B bound Avicel with lower affinity (Ka (2.04 ± 0.54) × 104 M−1 and (8.52 ± 1.50) × 103 M−1, respectively) compared to bMLG, but with higher affinity than xyloglucan (Fig. S7, Table 1).
The GH16 endo-β-glucanase of the MLGUL (BoGH16MLG, Fig. 1) specifically cleaves β(1,4) bonds of glucosyl residues to which a β(1,3)-linked glucosyl residue is attached, producing G4G3G and G4G4G3G as the limit-hydrolysis products from MLG [25]. As such, these would be the smallest mixed-linkage gluco-oligosaccharides (MLGOs) that could be encountered by the SGBPs. ITC analysis, however, showed that neither BoSGBPMLG-A nor BoSGBPMLG-B bind these mixed-linkage trisaccharide and tetrasaccharide products (Fig. S5, Table 1). Hence, longer MLGOs were obtained by controlled hydrolysis of oat MLG with BoGH16MLG, followed by fractionation with size exclusion chromatography. The mixed-linkage hexasaccharide G4G3G4G4G3G (MLG6) and heptasaccharide G4G4G3G4G4G3G (MLG7) were bound by BoSGBPMLG-A and BoSGBPMLG-B with respective affinities ca. one order of magnitude less than those for the full-length polysaccharide, with marginally stronger binding observed for the heptasaccharide for both SGBPs. (Fig. S5, Table 1).
Analogous to results for short MLGOs, neither SGBP was able to bind the all-β(1,4)-linked cellotetraose and cellopentaose (Fig. S5, Table 1). Reflecting the observed binding to insoluble cellulose and the apparent requirement for longer oligosaccharides, both SGBPs quantifiably bound cellohexaose (Fig. S6, Table 1). Notably, the affinity of BoSGBPMLG-A to cellohexaose was one order of magnitude weaker (Ka 103 M−1) than to the MLG hexa- and heptasaccharides (Ka 104 M−1), while the binding of BoSGBPMLG-B to cellohexaose and these MLGOs was comparably weak (Ka 103 M−1, Table 1).
SGBP crystallography illuminates the molecular basis of MLG specificity
To reveal the molecular basis of the substrate specificity of the SGBPs in the context of their contribution to MLG utilization by B. ovatus, we solved the three-dimensional structure of these proteins in several unliganded and oligosaccharide-complexed forms by X-ray crystallography (Table 2).
Table 2.
Crystallographic data and refinement statistics table
| BoSGBPMLG-A (6E60) | BoSGBPMLG-A cellohexaose (6DMF) | BoSGBPMLG-A MLG7 (6E61) | BoSGBPMLG-B cellohexaose (6E57) | BoSGBPMLG-B MLG7 (6E9B) | |
|---|---|---|---|---|---|
| Data collection | |||||
| Beamline | SSRL 9-2 | APS 21-ID-G | SSRL 9-2 | APS 21-ID-F | CLS 08B1-1 |
| Wavelength (λ) | 0.979 | 0.979 | 0.979 | 0.979 | 0.979 |
| Resolution range (Å) | 37.25–1.50 (1.59–1.50) | 49.60–2.40 (2.49–2.40) | 39.65–2.51 (2.66–2.51) | 78.34–2.71 (2.81–2.71) | 34.33–3.15 (3.34–3.15) |
| Space group | P 21 21 21 | P 61 | P 21 21 21 | P 21 21 21 | P 21 21 21 |
| Unit cell | |||||
| a, b, c (Å) | 47.351 89.484 121.104 | 228.845 228.845 246.517 | 86.977 93.137 155.589 | 156.438 243.652 76.059 | 155.621 241.102 74.884 |
| α, β, γ (°) | 90 90 90 | 90 90 120 | 90 90 90 | 90 90 90 | 90 90 90 |
| Total reflections | 363,678 (57,305) | 2,422,857 (206,443) | 279,444 (34,379) | 447,089 (44,223) | 371,772 (58,717) |
| Unique reflections | 81,232 (12,744) | 283,613 (28,197) | 43,053 (6405) | 78,048 (7714) | 50,094 (7894) |
| Redundancy | 4.5 (4.5) | 8.5 (7.3) | 6.5 (5.4) | 5.7 (5.7) | 7.4 (7.4) |
| Completeness (%) | 97.6 (95.6) | 99.94 (99.52) | 98.6 (92.3) | 97.74 (98.32) | 99.8 (99.7) |
| I/σI | 9.84 (1.49) | 8.40 (1.11) | 14.61 (3.51) | 10.44 (2.42) | 15.6 (1.73) |
| Wilson B-factor (Å2) | 14.5 | 34.87 | 28.7 | 50.08 | 90.3 |
| R-meas | 0.119 (0.975) | 0.2396 (2.007) | 0.12 (0.462) | 0.1353 (0.914) | 0.136 (1.277) |
| CC (1/2) | 0.997 (0.605) | 0.996 (0.58) | 0.997 (0.887) | 0.992 (0.7) | 0.999 (0.651) |
| Molecules in AU | 1 | 10 | 2 | 4 | 4 |
| Refinement | |||||
| R-work | 0.1578 (0.309) | 0.1850 (0.2850) | 0.1727 (0.255) | 0.1943 (0.2975) | 0.221 (0.2724) |
| R-free | 0.1839 (0.310) | 0.2399 (0.3387) | 0.2228 (0.310) | 0.2382 (0.3568) | 0.269 (0.3236) |
| Number of non-hydrogen atoms | |||||
| All | 4987 | 42441 | 8765 | 12333 | 11352 |
| Macromolecules | 4374 | 40685 | 8181 | 11933 | 11161 |
| Ligands | 16 | 812 | 164 | 260 | 123 |
| Water | 597 | 944 | 420 | 140 | 68 |
| RMS deviations | |||||
| Bond lengths (Å) | 0.012 | 0.01 | 0.005 | 0.009 | 0.01 |
| Bond angles (°) | 1.581 | 1.24 | 0.958 | 1.27 | 1.31 |
| Ramachandran statistics | |||||
| Favored (%) | 98 | 96 | 97 | 95 | 87 |
| Allowed (%) | 2 | 3.8 | 3 | 4.4 | 11 |
| Outliers (%) | 0 | 0.2 | 0 | 0.6 | 2 |
| Average B-factor (Å2) | |||||
| All | 15.12 | 40.4 | 28.61 | 58.7 | 100.4 |
| Macromolecules | 14.88 | 40.2 | 29.86 | 58.6 | 100.5 |
| Carbohydrate ligands | 50.05 | 30.36 | 74.15 | 74.2 | |
| Solvent | 25.69 | 37.1 | 25.4 | 43.8 | 60.48 |
BoSGBPMLG-A is a canonical SusD homolog
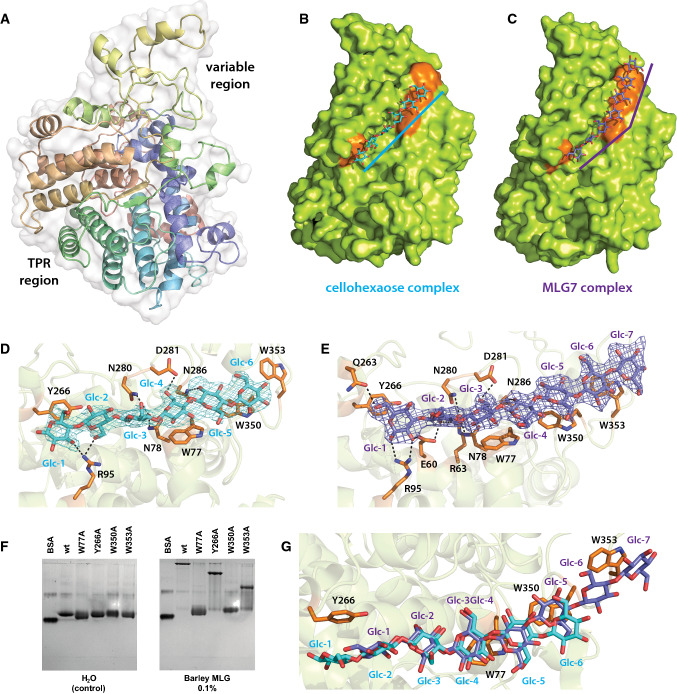
The crystal structure of unliganded BoSGBPMLG-A (1.50 Å, Rwork = 15.8%, Rfree = 18.4%; Table 2) revealed a single globular domain with a canonical “SusD-like” fold [27], which is dominated by α helices (Fig. 3a). As in the amylose-binding SusD, a series of these α helices are organized into four tetratricopeptide repeat (TPR) units that form a super-helical fold along the convex surface of the protein. The concave side of the TPR motif cradles the remainder of the polypeptide chain comprising many loops and short α helices, where the ligand binding site is found. This region is variable among SusD homologs, thus providing a tunable platform optimized to discriminate cognate substrates [26], e.g., starch [27], xyloglucan [29], mucin O-glycan [33], sialic acid [34], laminarin/pustulan [35], and chitin [36].
Fig. 3.
BoSGBPMLG-A crystal structure. a Overall structure of BoSGBPMLG-A in cartoon representation with the polypeptide color ramped from blue to red (from N- to C-termini) and the transparent surface shown in white. b Surface representation of the cellohexaose complex with ligand shown in cyan and the aromatic residues that form the binding platform shown in orange. Ten molecules are found in the asymmetric unit, each displaying no significant protein structural differences from the others; chain E is shown as a representative. c Surface representation of the MLG7 complex with ligand shown in slate and the aromatic residues that form the binding platform shown in orange. Two molecules are found in the asymmetric unit, each displaying no significant structural difference from the other; chain A is shown as a representative. d Close-up of the binding site of the cellohexaose complex with interacting residues shown as opaque orange sticks. Cellohexaose is colored cyan and potential hydrogen bonding interactions are shown as black dashed lines (within 3.5 Å of the ligand). Omit map for the ligand (generated by Privateer [79]) is shown contoured to 3σ. e Close-up of the binding site of the MLG7 complex with interacting residues shown as opaque orange sticks. MLG7 is colored slate and potential hydrogen bonding interactions are shown as black dashed lines (within 3.5 Å of the ligand). Omit map for the ligand is shown contoured to 3σ. f Affinity gel electrophoresis of binding platform site-directed mutants. g Overlay of cellohexaose and MLG7 bound to the binding platform; cellohexaose is shown in cyan and MLG7 in slate
Co-crystallization of BoSGBPMLG-A with cellohexaose (2.40 Å, Rwork = 18.5%, Rfree = 24.0%; Table 2) and the MLG heptasaccharide G4G4G3G4G4G3G (BoSGBPMLG-A MLG7: 2.51 Å, Rwork = 17.3%, Rfree = 22.3%; Table 2) clearly revealed this platform as the substrate-binding site. Superposition of the two complexes with the unliganded structure reveals that there are no major changes in global conformation or sidechain positioning upon substrate binding (Fig. S8). This is similar to the SGBP-A homolog of the xyloglucan utilization locus (XyGUL) [23], but contrary to SusD, in which two loops undergo a large conformational change to enable a tyrosine sidechain to stack against the α-glucan ligand [21]. Four surface-exposed aromatic residues (Y266, W77, W350, and W353) of BoSGBPMLG-A are arranged in a linear fashion to constitute a long (ca. 36 Å) binding platform (Fig. 3b–e).
In the BoSGBPMLG-A:cellohexaose complex, electron density was observed for all six glucose residues in nine of ten protein molecules in the asymmetric unit, with five glucose residues observed in the remaining molecule. All glucose residues were in the lowest energy 4C1 (chair) conformation (Fig. 3b, d; Table S2). The reducing end glucose, Glc-1, displays an aromatic stacking interaction [37, 38] with Y266 of the binding platform, Glc-4 is positioned over W77, and the non-reducing end glucose, Glc-6, stacks against W350 (Fig. 3b, d). In the BoSGBPMLG-A:G4G4G3G4G4G3G (MLG7) complex, all seven glucosyl residues could be convincingly modeled into the electron density in the 4C1 conformation in both molecules in the asymmetric unit (Fig. 3c, e; Table S2), with the oligosaccharide chain oriented in the same direction. In contrast to cellohexaose, G4G4G3G4G4G3G spans the entire length of the binding platform with the sugar rings of Glc-1, Glc-3, Glc-5, and Glc-7 stacking against the aromatic side chains of Y266, W77, W350, and W353, respectively (Fig. 3c, e). A limited number of potential hydrogen bonding interactions were observed with both oligosaccharide ligands. These hydrogen bonds are confined to one-half of the binding platform flanked by Y266 and W77; no additional hydrogen bonding interactions are observed to either ligand in the vicinity of W350 and W353 (Fig. 3d, e). Additionally, most heteroatom interatomic distances are greater than 3.0 Å, suggesting that these hydrogen bonds are moderate/weak and mostly electrostatic [39].
The importance of each aromatic residue comprising the binding platform is underscored by AGE analysis of single site-directed mutants (W77A, Y266A, W350A, and W353A). The two central tryptophan residues are critical as both W77A and W350A variants independently fail to bind bMLG and other β(1,4)-glucosyl-containing polysaccharides (Fig. 3f, S9). The two flanking residues, Y266 and W350, though not as critical as the central tryptophan residues, also contribute to binding MLG as evidenced from the diminished binding relative to wild type when either is replaced with an alanine. Although AGE analysis as performed here is only semi-quantitative, the data suggest that W350 may have a greater contribution to binding than Y266 (Fig. 3f).
BoSGBPMLG-B is a novel extended, multimodular MLG-binding protein
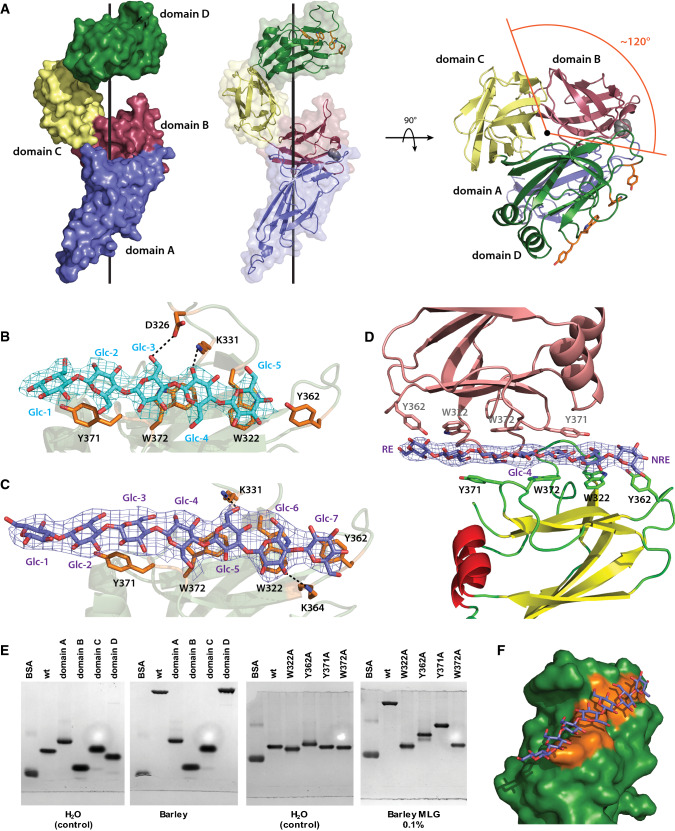
Distinct from BoSGBPMLG-A, crystal structures of BoSGBPMLG-B in complex with cellohexaose (2.71Å, Rwork = 19.4%, Rfree = 23.8%; Table 2) and G4G4G3G4G4G3G (BoSGBPMLG-B MLG7: 3.15 Å, Rwork = 22.1%, Rfree = 26.9%, residues 12–399; Table 2) reveal a multimodular architecture comprising four discrete immunoglobulin (Ig)-like domains in an extended arrangement: domain A (residues 22–133), domain B (residues 134–219), domain C (residues 220–309), and domain D (residues 310–420) (Fig. 4a). Such multi-domain architecture is typical of SGBPs, which are usually encoded directly downstream of the corresponding SGBP-A (SusD homolog) in PULs [18, 28–31], yet this architecture is difficult to predict due to very low sequence similarity among these proteins. Likewise, there is little tertiary structural homology among these proteins, such that a Dali search [40, 41] of the Protein Data Bank using the full-length structure or individual domains failed to return any matches with other SGBPs [28–30]; results were limited to unrelated proteins with Z-scores less than 11.
Fig. 4.
BoSGBPMLG-B crystal structure. a Overall structure of BoSGBPMLG-B in surface/cartoon representation with each domain colored differently: domain A—blue, domain B—raspberry, domain C—pale yellow, domain D—forest. A side view and a top view are shown with the black line representing the imaginary axis around which the domains wrap. The single interdomain proline is shown as gray spheres and the aromatic sidechains comprising the binding platform are shown as orange sticks. b Close-up of the binding site of the cellohexaose complex with interacting residues shown as opaque orange sticks. Cellohexaose is colored cyan and potential hydrogen bonding interactions are shown as black dashed lines (within 3.5 Å of the ligand). Omit map for the ligand (generated by Privateer [79]) is shown contoured to 3σ. Of the four molecules in the asymmetric unit, data from chain A is shown a representative. c Close-up of the binding site of the MLG7 complex with interacting residues shown as opaque orange sticks. MLG7 is colored slate and potential hydrogen bonding interactions are shown as black dashed lines (within 3.5 Å of the ligand). Omit map for the ligand is shown contoured to 3σ. Of the four molecules in the asymmetric unit, data from chain D are shown as representative. d A single MLG7 ligand being shared between two BoSGBPMLG-B molecules belonging to neighboring asymmetric units. The bottom molecule is colored according to secondary structure (yellow β-strands, red α-helices, and green loops), the top molecule from a different asymmetric unit is colored salmon, and the ligand is colored slate. Omit map for the ligand is shown contoured to 3σ. RE reducing end, NRE non-reducing end. An analogous orientation was observed for the cellohexaose complex (not shown, PDB ID 6E57). e Affinity gel electrophoresis of individual BoSGBPMLG-B domains and binding platform site-directed mutants. f Surface representation of the binding platform of domain D in complex with MLG7. Aromatic sidechains comprising the binding platform are colored orange and MLG7 is colored slate
The number of domains in these SGBPs is variable [28–30], ranging from three in SusE [28] to six in the SGBP-B from the heparin/heparan-sulfate PUL [30]. The four domains of BoSGBPMLG-B are arranged in a right-handed helical configuration in which the domains are rotated 120° around the central axis (three domains per helical turn such that domains A and D overlap when looking down the axis; Fig. 4a). Whereas the presence of a single proline residue in each interdomain linker is a feature of the previously solved SGBP-B structures [28–30], in BoSGBPMLG-B a proline residue was found only in the linker after domain A (Fig. 4a), which may suggest reduced conformational rigidity vis-à-vis related SGBPs [29]. However, the identical domain arrangement of all four molecules in the asymmetric unit (for both cellohexaose and MLG7 complexes, Table S2) could be evidence of a lack of conformational flexibility.
In the structure of BoSGBPMLG-B co-crystallized with cellohexaose, electron density was observed for five of the expected six glucosyl residues, all in the favored 4C1 conformation (Fig. 4b, Table S2). Oligosaccharide binding was mediated by the following aromatic stacking interactions: Glc-1 (reducing end) and Glc-2 with Y371, Glc-3, and Glc-4 with W373, and Glc-5 (non-reducing end) with W322. In the structure of BoSGBPMLG-B in complex with G4G4G3G4G4G3G (MLG7), electron density was observed for all seven glucosyl residues in the 4C1 conformation (Fig. 4c, Table S2). Reducing end Glc-1 only makes contact with a residue from a molecule in a neighboring asymmetric unit (vide infra), Glc-2 and Glc-3 exhibit stacking interaction with Y371, Glc-4 and Glc-5 with W372, Glc-6 with W322, and Glc-7 with Y362. As for BoSGBPMLG-A, very few hydrogen bonding interactions were observed with either oligosaccharide (Fig. 4b, c).
Notably, in the BoSGBPMLG-B cellohexaose and G4G4G3G4G4G3G (MLG7) complexes (Table 2), two out of the four molecules in the asymmetric unit each bound to a ligand that is shared with a molecule from a neighboring asymmetric unit, such that each oligosaccharide is sandwiched between two binding platforms presented on the C-terminal domain D (Fig. 4d, S10). In the other two molecules in the asymmetric unit that are not involved in the ligand-sharing crystal contact, the density of the ligand was very poor and no sugars were modeled. The molecules that share a common ligand are symmetry related through a twofold rotation operation about an axis orthogonal to the length of the oligosaccharide ligand, straight through the central Glc-4 of G4G4G3G4G4G3G (Fig. 4d). As such, the binding platforms are oriented in opposite directions to either side of the oligosaccharide chain, which suggests a degree of plasticity in MLG recognition.
As suggested by the complexed structures, domain D provides the only substrate-binding site in BoSGBPMLG-B. AGE analysis of the four domains produced independently demonstrated that indeed only domain D binds bMLG (Fig. 4e, S9). This feature is similar to the XyGUL SGBP-B and the heparin/heparan-sulfate PUL SGBP, although in the case of the latter, the binding platform spans two of the distal C-terminal domains, D5 and D6 [29, 30]. In contrast, the archetypal SusE and SusF SGBPs contain two and three starch-binding sites, respectively, on individual domains and thus represent the only SGBPs known to possess multiple binding sites [28].
The β-sandwich fold of domain D comprises two sheets of four and three antiparallel β-strands, with two of the connecting loops incorporating α-helices. The substrate-binding site is located above the smaller, top β-sheet, although residues important for binding are all borne on loops connecting the β-strands (Fig. 4d). The aromatic sidechains of Y371, W372, W322, and Y362 constitute a ca. 28 Å, flat binding platform (Fig. 4f). These aromatic residues are critical for substrate binding, as demonstrated by AGE analysis: Mutation of either of the central tryptophans (W322A and W372A) completely abrogates binding, whereas mutation of the tyrosines at the ends of the platform (Y362A and Y371A) severely diminishes binding to bMLG, xyloglucan, and hydroxyethylcellulose (Fig. 4e, S9).
SGBP-A and SGBP-B have distinct functions in vivo
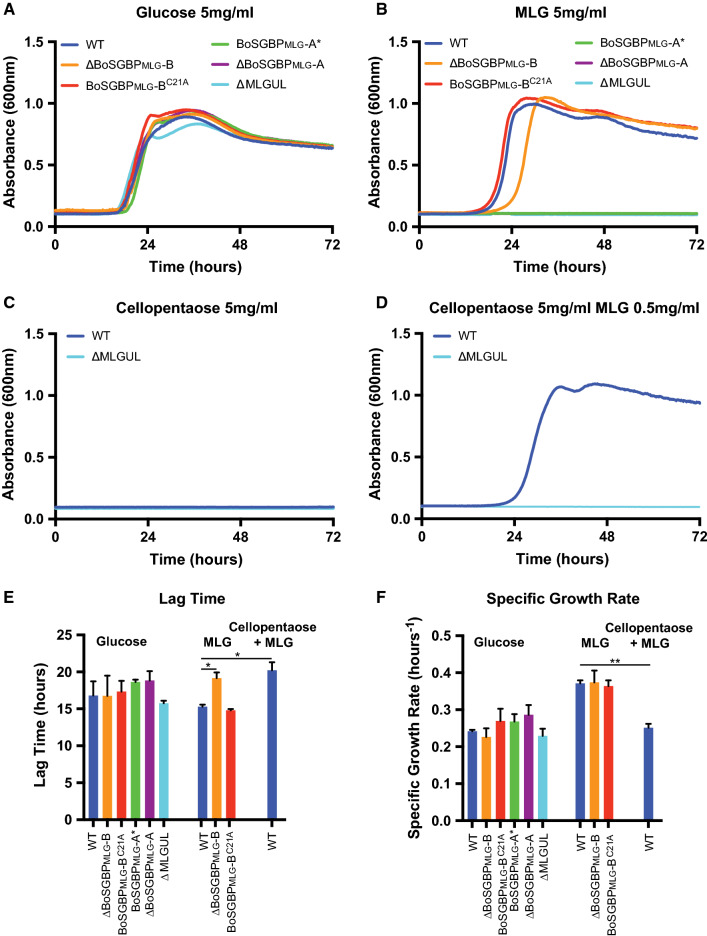
With a firm understanding of substrate specificity and tertiary structures of BoSGBPMLG-A and BoSGBPMLG-B, we then sought to determine how polysaccharide binding at the cell surface contributes to the growth of B. ovatus on MLG. Strains with in-frame deletions and binding site-deficient alleles of both genes were generated by performing allelic exchange in a ∆tdk strain of B. ovatus [27], which is subsequently referred to as “wild type.” To monitor anaerobic growth, cells were first cultured overnight in minimal medium (MM) containing glucose, then back-diluted 1:200 into parallel cultures containing glucose or glucan substrate. Wild-type B. ovatus (∆tdk) grows on high viscosity MLG, while cells in which the complete MLGUL (Fig. 1) was deleted (∆MLGUL) do not, which confirmed that the MLGUL is essential and solely responsible for growth on this β-glucan (Fig. 5a, b) [25].
Fig. 5.
MLGUL surface glycan-binding proteins facilitate growth on MLG and cellopentaose. Average growth curve of B. ovatus MLGUL strains on a 5 mg/mL glucose, b 5 mg/mL high viscosity MLG, c 5 mg/mL cellopentaose, d 5 mg/mL cellopentaose with 0.5 mg/mL MLG. e Lag time from the growth curves in a, b, and d. f Specific growth rates were calculated at O.D.600 = 0.5 for growth curves in a, b, and d. Bars denoted with a ‘*’ have a p < 0.05 and that with a ‘**’ has a p < 0.005. Statistically significant differences were determined using the two-tailed unpaired Student’s t test
The BoSGBPMLG-A single-gene knock-out (∆SGBPMLG-A) likewise cannot grow on MLG. This finding is consistent with the essential role of the SGBP-A (SusD) homologs for the uptake of starch via the Sus of B. thetaiotaomicron and the uptake of xyloglucan via the XyGUL of B. ovatus [29, 42]. In both the Sus and the XyGUL, an allele encoding a glycan-binding-deficient version of the native protein (SusD* and SGBPXyG-A*, respectively) restored growth on the cognate substrate [29, 42]. However, when the binding-deficient BoSGBPMLG-A* allele (a W77A/W350A double mutant) was exchanged into the ΔBoSGBPMLG-A background to restore the MLGUL genetic structure, growth was not observed on MLG (Fig. 5a, b). These data indicate that glycan-binding activity is essential to BoSGBPMLG-A function for growth on MLG, and furthermore suggests the importance to glycan import that is distinct from homologs in the Sus and XyGUL.
While BoSGBPMLG-A must be both present and functional for growth on MLG, BoSGBPMLG-B is not required. Deletion of BoSGBPMLG-B results in a longer lag before entering exponential phase on MLG compared to wild type, but does not affect the specific growth rate (Fig. 5a, b, e, f). In light of this result and based on previous experience, we were concerned that the BoSGBPMLG-B gene deletion might affect transcription of the MLGUL. Our previous work on the Sus of B. thetaiotaomicron indicated that deletion of the gene encoding the SGBP SusF decreases the transcription of the downstream susG, which encodes the essential, cell surface amylase. We therefore created a Cys21-Ala mutant to mis-traffic BoSGBPMLG-B to the periplasm by removing the N-terminal lipidation site of the mature protein [28, 43], while retaining the overall MLGUL genetic structure. Indeed, cells expressing BoSGBPMLG-BC21A grow similarly to wild-type cells on MLG, suggesting that the lag seen for ΔBoSGBPMLG-B is likely due to a negative impact on transcription of other MLGUL genes. Correspondingly, qPCR on B. ovatus ΔBoSGBPMLG-B and BoSGBPMLG-BC21A strains grown to mid-exponential phase on MLG revealed a 1000-fold decrease in the TBDT transcript in the knock-out versus the wild-type and BoSGBPMLG-BC21A strains (Fig. S11). The magnitude of this defect was surprising because all cells were harvested at the same O.D.600 (after the lag growth defect of the knock-out had passed), but clearly indicates the BoSGBPMLG-B deletion influences overall transcript stability (Fig. 1b, S11). Moreover, this observation also suggests that sustained upregulation of this PUL is not required to support wild-type level growth rates on MLG, as long as sufficient amounts of MLGUL components eventually populate the cell surface and periplasm (Fig. 5f).
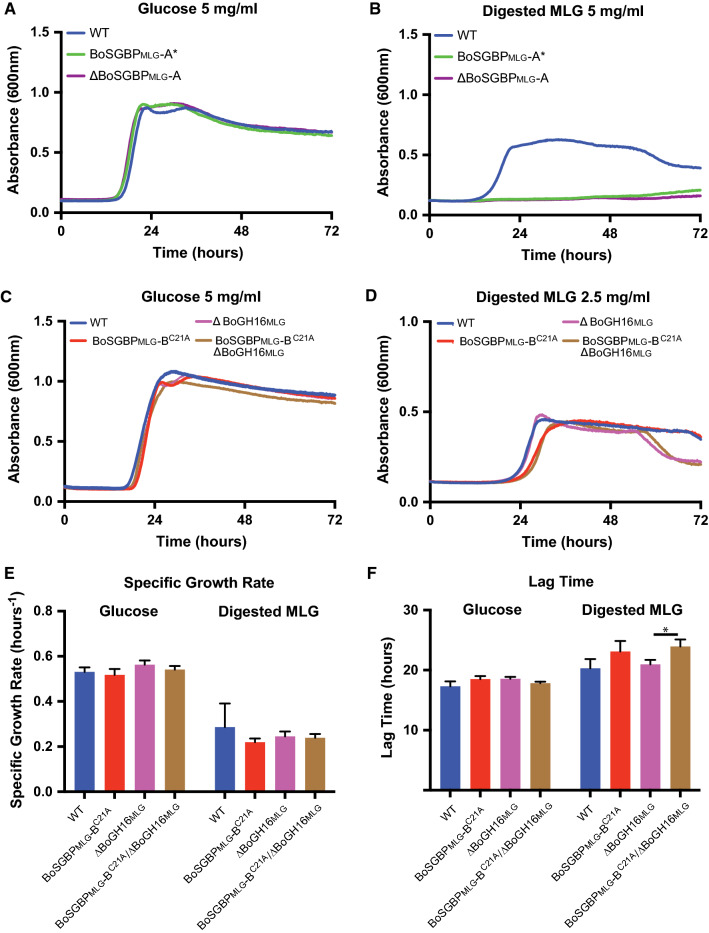
A key question regarding the MLGUL and related Sus-like systems is the size of the transported oligosaccharide. Our biochemical data demonstrate that the BoSGBPMLG-A and BoSGBPMLG-B proteins preferentially bind oligosaccharides with degree of polymerization ≥ 6 and, in particular, do not bind the limit-digest products of BoGH16MLG (Fig. S4, S5; Table 1). To mimic the likely MLG fragments that are transported by a putative TBDT/BoSGBPMLG-A(/BoSGBPMLG-B) complex, MLG was partially digested by recombinant BoGH16MLG in vitro to generate a profile of oligosaccharides that span a broad range of lengths (Fig. S13). Control experiments revealed that although a B. ovatus ΔGH16 mutant cannot grow on the native MLG polysaccharide, it does grow similarly to wild type on this mixture, thereby demonstrating competent uptake through the TBDT/BoSGBPMLG-A complex (Fig. 6, S14). Underscoring the essential role of the BoSGBPMLG-A in capturing longer oligosaccharides, the ΔBoSGBPMLG-A and BoSGBPMLG-A* mutants cannot grow on the MLG digest (Fig. 6a, b). This phenotype is identical to that observed on native MLG polysaccharide (Fig. 5). Interestingly, the mistrafficked BoSGBPMLG-BC21A mutant displays a longer lag time compared to the wild type, regardless of the presence of BoGH16MLG, although specific growth rates were similar (Fig. 6d–f).
Fig. 6.
MLGUL surface glycan-binding proteins capture BoGH16MLG-digested MLG. Average growth curve of B. ovatus MLGUL strains on a, c 5 mg/mL glucose, b, d 5 mg/mL digested MLG. e Lag time and from the growth curves in c and d. f Specific growth rates were calculated at O.D.600 = 0.25 for growth curves in c and d. Bars denoted with a ‘*’ have a p < 0.05. Statistically significant differences were determined using the two-tailed unpaired Student’s t test
Bacteroides ovatus is unable to grow on insoluble cellulose as a sole carbon source [19]. However, because the MLGUL-encoded SGBPs have weak affinity for cello-oligosaccharides, we wanted to explore the contribution of these proteins to growth. Wild-type cells fail to grow on cellopentaose as the sole carbon source (Fig. 5c), yet the addition of 0.5 mg/mL MLG to upregulate MLGUL [19] enables growth on cellopentaose to a cell density greater than that achievable on 0.5 mg/ml MLG alone (Fig. 5d, S12). These results are concordant with the observation that the HTCS of the MLGUL (Fig. 1) is not activated by cello-oligosaccharides [19]. The addition of 0.5 mg/mL MLG does not support growth on cellopentaose in the ΔMLGUL strain, demonstrating that growth on cello-oligosaccharides is indeed dependent on MLGUL expression (Fig. 5d). Approximately, 10% of the MLG structure is composed of longer regions (d.p. 5–9) of β(1,4)-glucosyl residues lacking β(1,3) kinks [44, 45] and the ability to utilize cellopentaose via the MLGUL undoubtedly is a consequence of the known catalytic promiscuity of the GH3 exo-β-glucosidase for both β(1,3)/β(1,4)-mixed-linkage gluco-oligosaccharides and all-β(1,4)-linked gluco-oligosaccharides [25].
Discussion
SGBPs are an important class of PUL components that effect target glycan capture at the outer membrane surface for backbone cleavage and import (Fig. 1). All PULs encode at least one SGBP: a SusD homolog, SGBP-A, which forms a functional complex with a partner TBDT [29, 32, 42]. Indeed, the tandem TBDT/SGBP-A (susC/susD homolog) pair is a signature feature that can be used to identify PULs in sequenced genomes [46]. As exemplified by the BoSGBPMLG-A structures reported here (Fig. 3), SGBP-A homologs are single-domain globular proteins built upon prominent tetratricopeptide repeats (TPRs), as first revealed by the structure of SusD from the starch utilization system [27, 29, 33, 35, 36]. Recent crystallography of two SGBP-A/TBDT homologs demonstrated that SGBPs-A associate closely with the extracellular side of the TBDTs, forming a “pedal bin” lid [32].
Although this structural arrangement might suggest a role in recognizing and directing cognate substrates into the transporter [32], previous studies on the Sus and XyGUL using non-binding SGBP-A* mutants in vivo have failed to demonstrate that substrate binding is a prerequisite for growth [29, 42]. In these systems, the expression of additional SGBPs (i.e., SusE, BoSGBPXyG-B) have been shown to supplement the loss of substrate binding by SusD and BoSGBPXyG-A deletion, respectively [29, 42, 47]. Deletion of the corresponding SGBP-A in these systems does cripple growth on the cognate substrates, consistent with the critical structural role in complex formation with the TBDT. Here, we showed that either complete removal of the BoSGBPMLG-A from the cell surface (by gene knock-out, ∆SGBP-A) or abrogation of its ability to bind substrate (by removing crucial amino acid sidechains, SGBP-A*) prevents MLG utilization by B. ovatus (Fig. 5). Thus, the present study is, to our knowledge, the first to demonstrate directly an essential role of substrate binding by an SGBP-A (SusD) homolog, which extends beyond the obvious requirement for its physical presence at the cell surface [29, 32, 42].
Additional SGBP(s) can also be encoded proximal to SGBP-A homologs in PULs (usually immediately downstream), although these proteins generally have such low sequence similarity and tertiary structural homology that they cannot be confidently identified as SGBPs by bioinformatic approaches. Of the functionally characterized representatives, the archetypal Sus system contains two such SGBPs (SusE and SusF) [28], whereas the XyGUL [29], heparin/heparan-sulfate PUL [30], xylan PULs (PUL-XylL and PUL-XylS) [31], and the MLGUL studied here contain only one additional SGBP (denoted “SGBP-B” or “SusE-positioned” in the literature). As exemplified by the structures of BoSGBPMLG-B (Fig. 4), these proteins generally comprise a multi-domain “beads-on-a-string” arrangement that presents a critical carbohydrate-binding site on the C-terminal domain most distal to the N-terminal membrane-anchoring lipid.
Vis-à-vis BoSGBPMLG-A, the role of the BoSGBPMLG-B is less easily deduced in the context of the MLG utilization system encoded by the MLGUL. BoSGBPMLG-B is dispensable for growth on MLG and is not able to compensate for the BoSGBPMLG-A* mutant (Fig. 5). Although it is clearly highly specific for MLG, the affinity of BoSGBPMLG-B for the cognate polysaccharide of the MLGUL is ca. tenfold lower than that of BoSGBPMLG-A. This comparatively weaker binding is consistent with XyGUL SGBPs, but in the case of the Sus SGBPs, SusD (SGBP-A) and SusE (SGBP-B) have comparable affinities toward α-cyclodextrin while SusF (SGBP-C) has lower affinity [27–29]. In vivo, SGBP-B-like proteins have been shown to play multiple roles during glycan capture. For example, the SGBP SusE influences the lengths of malto-oligosaccharide that can be taken up by the TBDT of the starch utilization system, SusC [47], while both SusE and SusF appear to offset the diffusion barrier at the cell surface established by the capsular polysaccharide, thereby aiding in starch capture [42].
The slight, but reproducible increase in growth lag on pre-digested MLG (Fig. 6) suggests that BoSGBPMLG-B functions in the capture of medium-length MLGOs, possibly including those in the extracellular environment that are not generated proximal to the cell surface by the BoGH16MLG. This could allow B. ovatus to access MLGOs liberated from neighboring species. Indeed, work studying cooperative growth between Bacteroides species and other members of the microbiota on xylans, inulin, and dietary pectins has demonstrated that members of this genus benefit from the uptake of oligosaccharides released by neighbors [48, 49]. These studies have primarily focused on the roles of glycosidases in the communal breakdown of polysaccharides; however, the potential contributions of glycan-binding proteins in these processes remain uncharacterized.
Our structural studies of BoSGBPMLG-B revealed an additional aspect of MLG recognition that suggests a more complex interplay of molecular interactions at the cell surface than previously observed for this class of proteins. Specifically, the observation of bi-directional ligand recognition (Fig. 4d) in crystallo implies the possibility of avidity effect through dual binding of one MLG chain from the opposite faces of the polysaccharide. This binding mode may be biologically relevant: when the symmetry operation bisecting the ligand is applied to the entire BoSGBPMLG-B molecule, both N-termini (through which the protein is anchored to the outer cell membrane via a canonical Cys lipidation site [50]) are orientated in the same direction (Fig. S10). BoSGBPMLG-B does not dimerize on its own in solution, as determined by size exclusion chromatography (data not shown), and there is minimal direct contact between the two symmetry-related protein molecules in crystallo. Unfortunately, ITC analysis was unable to resolve the binding stoichiometry within the limits of the current data (Table 1). However, we also note that no ligand density is observed in protein molecules that do not participate in this sandwich interaction.
Together, biochemical, structural, and microbiological data for BoSGBPMLG-A and BoSGBPMLG-B suggest that the external recognition machinery for MLG acquisition is tailored for the capture of longer oligo/polysaccharides, rather than the limit-digest products (tri- and tetrasaccharides) generated by BoGH16 of the MLGUL [25]. Other PUL-encoded SGBP systems similarly target longer partial digest saccharides: XyGUL SGBPs display stronger affinity toward xyloglucan oligosaccharides with a backbone of eight Glc residues or more, versus limit-digest oligosaccharides with Glc4 backbones [29]; xylan PUL SGBPs bind xylo-oligosaccharides with affinities that increase with chain length [31]; and the substrate-binding affinity of the heparin/heparan-sulfate PUL SGBP-B likewise increases with oligosaccharide degree of polymerization [30]. Preferential targeting and transport of longer oligosaccharides pre- and post-hydrolysis by the vanguard endo-glycanase encoded by a PUL is anticipated to be an effective strategy for Bacteroides species to rapidly acquire multiple glucose equivalents with minimal loss in a competitive environment.
The present study also provides specific molecular insight into the selectivity of the SGBPs of the MLGUL for mixed-linkage β(1,3)/β(1,4)-glucans from cereal grains over all-β(1,4)-glucans, i.e., cellulose and cello-oligosaccharides. Notably, B. ovatus does not grow on crystalline cellulose [19] and exhibits a significant growth lag on soluble cello-oligosaccharides ([19] and Fig. 5e). Comparison of the MLGO and cello-oligosaccharide complex structures of both BoSGBPMLG-A and BoSGBPMLG-B (Figs. 3, 4) reveals that shape complementarity of the glycan with the binding platforms rationalizes the primary specificity for MLG (Fig. 2; Table 1).
On one hand, the faces of the aromatic platform residues are angled with respect to each other in both SGBPs (Figs. 3d, e, g, 4b, c, f), which complements the twisted conformation that β-glucans natively adopt in solution [51]. In comparison, canonical cellulose-binding modules (“type A” CBMs) generally present aromatic sidechains in a flat, co-linear arrangement that matches the planar cellulose crystal [52]. On the other hand, the non-linear topology of the binding surface of both SGBPs is complementary to the intrinsically bent shape of the MLG chain. In the case of BoSGBPMLG-A, this enables optimal interaction with all four aromatic sidechains of the binding platform, which is then unavailable to the strictly linear β(1,4)-linked cello-oligosaccharides (Fig. 3b, c). In the case of BoSGBPMLG-B, an inherent curvature of the binding surface likewise complements the twist of the MLG chain. (Figure 4f). Similar conformation-dependent specificity was observed for the archetypal SGBP-A homolog, SusD, which bound cyclodextrins with higher affinity than linear malto-oligosaccharides, due to an arced binding platform evolved to fit the natural helical conformation of amylose [27]. Thus, although both BoSGBPMLG-A and BoSGBPMLG-B broadly recognize polysaccharides containing contiguous β(1,4)-linked glucosyl residues, they do so with significantly lower affinity than for MLG (Figs. 2, S6; Table 1). We anticipate that future structural studies may reveal contrary specificity determinants in SGBPs that predominantly bind cellulose [53–55].
Although BoSGBPMLG-A and BoSGBPMLG-B exhibit some degree of off-target affinity vis-à-vis the cognate substrate of the MLGUL, this is unlikely to be evolutionarily disadvantageous in the context of the human gut where a diverse array of dietary glycans is present. Indeed, as one component of the plant cell wall, MLG is generally found associated with cellulose and other hemicelluloses [56], such that non-specific binding may improve bacterial adhesion to insoluble particles. Additionally, in the context of the gut ecosystem, the ability to scavenge the products of possible para-crystalline cellulose degradation may allow B. ovatus to compete for an additional, privileged nutrient niche, as many gut species lack the ability to utilize cello-oligosaccharides larger than cellobiose [19, 57, 58].
Conclusion
Effective manipulation of the gut microbiota for therapeutic purposes—a topic of growing recent interest—will be significantly informed by a holistic understanding of the metabolism of the microbiota, including the mechanisms of complex dietary polysaccharide utilization which fuel this ecosystem [12, 15, 23, 59]. The present study reveals the essential roles that two SGBPs play in cereal β-glucan utilization by working in concert with the glycoside hydrolases and TBDT of the MLGUL from the human symbiont B. ovatus. On one hand, this study contributes directly to the currently limited but growing pool of structure–function relationships among SGBPs. On the other, the identification of syntenic MLGUL in other members of the human gut microbiota indicates that this data may be extrapolated more broadly in metagenomic analyses [25, 43, 60]. Expanding our knowledge of the interplay of SGBP and other PUL components is a critical step toward developing novel strategies to manipulate microbial communities [61–63] in the human gut and beyond.
Methods
Substrates and polysaccharides
Polysaccharides Barley beta-glucan (high viscosity), yeast beta-glucan, curdlan, tamarind xyloglucan, konjac glucomannan, carob galactomannan, wheat arabinoxylan, beechwood xylan were purchased from Megazyme International (Bray, Ireland). Laminarin (from Laminaria digitata) was purchased from Sigma Aldrich (St. Louis, MO, USA). Carboxymethyl cellulose was purchased from Acros Organics (Morris Plains, NJ, USA). Hydroxyethyl cellulose was purchased from Amresco (Solon, OH, USA). Xanthan gum was purchased from Spectrum (New Brunswick, NJ, USA). Ulvan (from Ulva sp.) was purchased from Elicityl (Crolles, France).
Oligosaccharides Cellobiose (G4G) was purchased from Acros Organics. Cellotriose (G4G4G), cellotetraose (G4G4G4G), cellopentaose (G4G4G4G4G), cellohexaose (G4G4G4G4G4G), laminaribiose (G3G), laminaritriose (G3G3G), laminaritetraose (G3G3G3G), laminaripentaose (G3G3G3G3G), mixed-linkage glucotriose A (G3G4G), mixed-linkage glucotriose B (G4G3G), mixed-linkage glucotetraose A (G3G4G4G), mixed-linkage glucotetraose B (G4G4G3G), and mixed-linkage glucotetraose C (G4G3G4G) were purchased from Megazyme. Gentiobiose (G6G) was purchased from Carbosynth (Compton, UK). MLG partial digest mixture, mixed-linkage hexasaccharide (MLG6), and mixed-linkage heptasaccharide (MLG7) were produced in-house as described by McGregor et al. [64] using BoGH16MLG [25] in 50 mM sodium phosphate, pH 7.0.
Cloning, expression, and purification of recombinant proteins
Gene sequences were obtained from B. ovatus ATCC 8483 genome available on the Integrated Microbial Genomes database from the Joint Genome Institute. PCR primers were synthesized by Integrated DNA technologies.
Cloning Open reading frames encoding BACOVA_02743 and BACOVA_02744 were amplified by PCR using Q5 high fidelity polymerase (NEB) with appropriate primers (Table S1) and genomic B. ovatus DNA as template. All primers were designed to amplify constructs truncated to exclude predicted signal peptides (prediction by SignalP 4.1 [65, 66]) and N-terminal lipidation cysteine residues (prediction by LipoP 1.0 [67]). NdeI and XhoI restriction sites were included in the forward and reverse primers for subsequent digestion (all restriction enzymes from NEB) and ligation (T4 ligase from Thermo Scientific) into the pET28 vector. Both constructs were designed to harbor an N-terminal his6-tag fusion in the translated recombinant peptide. The gene encoding sfGFP was fused to their N-termini by restriction enzyme-based cloning using a BamHI site between the sfGFP and BACOVA_02743 or BACOVA_02744, and NheI and XhoI to the corresponding sites on the pET28 vector. Site-directed mutant constructs were generated using the QuikChange II site-directed mutagenesis kit (Agilent) according to the manufacturer’s instructions. Oligonucleotides used in this study are listed in Table S1. Successful generation of clones and mutants were verified by Sanger sequencing (Genewiz).
Expression Plasmids harboring the gene of interest were transformed into chemically competent E. coli BL21 (DE3) and cultured in lysogeny broth (LB) containing 50 μg/mL kanamycin. Cells were grown on a large scale at 37 °C until mid-logarithmic growth phase was reached (O.D.600 = 0.4–0.6) at which point expression was induced by addition of isopropyl β-d-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and temperature was lowered to 16 °C. Induction of recombinant protein production continued overnight, after which the cells were collected by centrifugation at 4000g for 20 min.
Purification The harvested cell pellet was resuspended in binding buffer (20 mM sodium phosphate pH 7.4, 500 mM sodium chloride, 20 mM imidazole) and lysed using a Sonic Dismembrator F550 Ultrasonic Homogenizer (Fisher Scientific). Cell debris was pelleted by centrifugation at 15,000 rpm for 45 min and the supernatant was loaded onto a 2 mL HisTrap IMAC FF nickel–nitrilotriacetic acid column (GE Healthcare), a nickel-based matrix, using a BioLogic FPLC system (BioRad). After washing with 10 column volumes of binding buffer, his6-tagged protein was eluted using a linear gradient of 0–100% elution buffer (20 mM sodium phosphate pH 7.4, 500 mM sodium chloride, 500 mM imidazole) over 10 column volumes. Fractions were monitored by A280 and eluted protein fractions were pooled and buffer exchanged into 50 mM sodium phosphate pH 7.0 using Vivaspin centrifugal filters (GE Healthcare). After concentrating, aliquots were flash frozen in liquid nitrogen and stored at − 80 °C. Protein purity was determined by SDS-PAGE analysis and mass was confirmed by intact protein mass spectrometry on a Waters Xevo Q–TOF with nanoACQUITY UPLC system (data not shown), as described previously [68]. Protein concentrations were determined by spectrophotometry on an Epoch Microplate Spectrophotometer (BioTek) using the following molar extinction coefficients: 106690 M−1cm−1 for BACOVA_02743, and 51340 M−1cm−1 for BACOVA_02744. Typical yields were around 50 mg for BoSGBPMLG-A and 40 mg for BoSGBPMLG-B from 1 L of LB culture.
Selenomethionine protein production and purification For selenomethionine-substituted BoSGBPMLG-B the pET28-BoSGBPMLG-B plasmid was transformed into Rosetta (DE3) pLysS E. coli and plated onto LB supplemented with kanamycin (50 µg/mL) and chloramphenicol (20 µg/mL). After 16 h of growth at 37 °C, colonies were harvested from the plates and used to inoculate 100 mL of M9 minimal media supplemented with kanamycin (30 µg/mL) and chloramphenicol (20 µg/mL) and grown at 37 °C for 16 h. This overnight culture was used to inoculate a 2 L baffled flask containing 1 L of Molecular Dimensions SelenoMet Premade Medium supplemented with 50 mL of the recommended sterile nutrient mix, chloramphenicol, and kanamycin. Cultures were grown at 37 °C to an O.D.600 ≈ 0.45 before adjusting the temperature to 20 °C, and supplementing each flask with 100 mg each of l-lysine, l-threonine, l-phenylalanine, and 50 mg each of l-leucine, l-isoleucine, l-valine, and l-selenomethionine [69]. After 20 additional minutes of growth, the cells were induced with 0.5 mM IPTG, and cultures were grown for an additional 48 h. For the purification of selenomethionine-substituted protein, cells were thawed and lysed via sonication in His-Buffer (25 mM NaH2PO4, 500 mM NaCl, 20 mM imidazole pH 7.5) and purified via immobilized nickel affinity chromatography (His-Trap, GE Healthcare) using a gradient of 20–300 mM imidazole, according to the manufacturer’s instructions. The fractions were inspected for purity via SDS-PAGE, then pooled and dialyzed against 20 mM HEPES and 100 mM NaCl (pH 7.0) and concentrated using Vivaspin 15 (10,000 MWCO) centrifugal concentrators (Vivaproducts, Inc.).
Affinity gel electrophoresis
Qualitative assessment of binding was carried out on the following soluble substrates: barley β-glucan, laminarin, yeast β-glucan, curdlan, xyloglucan, glucomannan, galactomannan, xylan, arabinoxylan, xanthan gum, dextran, carboxymethylcellulose, and hydroxyethylcellulose. Native polyacrylamide gels consisting of 10% (w/v) acrylamide in 40 mM Tris–HCl at pH 8.8 were prepared. The final concentration of 0.05% or 0.1% (w/v) substrate (or water for control) was added to the gel solution prior to polymerization. 5 μg of BoSGBPMLG-A and BoSGBPMLG-B, along with bovine serum albumin (BSA) as a non-interacting negative control, were loaded on the gels and subjected to electrophoresis under non-denaturing conditions at 100 V for 3 h at room temperature. Proteins were visualized by staining with Coomassie Brilliant Blue.
Isothermal titration calorimetry
All isothermal titration calorimetry (ITC) experiments were performed using a MicroCal VP-ITC titration calorimeter calibrated to 25 °C. All titrations were performed in 50 mM sodium phosphate, pH 7.0, with the exception of BoSGBPMLG-B with bMLG, which was performed in 10 mM HEPES pH 7.0. Proteins (20–100 µM) were placed in the sample cell and a first injection of 2 µL was performed followed by 24 subsequent injections of 10 µL of 2.5–4.0 mg/mL polysaccharide or 1–2 mM oligosaccharides (see Figs. S3–S5 for exact compositions of each protein–ligand pair). The solution was stirred at 280 rpm and the resulting heat of reaction was recorded. Data were analyzed using the OriginPro graphing software. Ka values were calculated on a molar basis from MLG polysaccharide concentrations in g/L by assuming a hexasaccharide binding motif, based on crystal complex structures with MLGOs. Ka values were calculated on a molar basis from XyG polysaccharide concentrations in g/L by assuming a Glc8-backbone (XyGO dimer) oligosaccharide binding motif.
Insoluble polysaccharide binding assay
Qualitative assessment of binding to insoluble polysaccharides, cellulose, and mannan was carried out by a pull-down assay. 10 mg of substrate and 100 μg of protein were mixed in 200 μL of 50 mM sodium phosphate, pH 7.0. After incubation at 4 °C for 4 h with end-over-end rotation, the samples were centrifuged at 16,000g for 5 min. The supernatant containing unbound protein was collected, and the pellet was washed three times with 200 μL of 50 mM sodium phosphate, pH 7.0. Bound protein was released from the substrate by resuspending the pellet in 200 μL of 1× SDS running buffer and heating to 80 °C for 10 min. Eluted bound protein was collected by centrifugation and both fractions were subjected to SDS-PAGE. BSA was used as a non-binding negative control.
Quantitative assessment of binding to cellulose was conducted by depletion isotherm. 10 mg/mL Avicel, 5.9–140.4 μg of GFP_BoSBGP-A or 4.1–196.8 μg of GFP_BoSGBPMLG- B, and 0.1 mg/mL BSA were mixed in 1 mL of 50 mM sodium phosphate pH 7.0. After incubation at 4 °C for 4 h with end-over-end rotation, the samples were centrifuged at 16,000g for 5 min and the supernatant was collected. Concentration of unbound GFP-fused protein was determined by fluorescence using an Infinite M1000 Pro multifunction plate reader (Tecan Ltd.) with an excitation filter of 485 nm and an emission filter of 510 nm. Fluorescence measurement of the same concentration range of GFP-fused protein in the absence of Avicel was determined to construct a standard curve of total protein and unbound protein was subtracted to determine the concentration of bound protein. OriginPro graphing software was used to fit the isotherms to the equation [PC] = [FP][PC]max/(Kd + [FP]), where PC represents the concentration of protein bound to Avicel and FP represents the concentration of free protein in the supernatant.
X-ray crystallography
Selenomethionine-substituted BoSGBPMLG-B protein crystals with cellohexaose were obtained directly from the SaltRx crystallization screen (Hampton Research) via hanging drop vapor diffusion at room temperature. The protein (21.1 mg/ml with 10 mM cellohexaose) was mixed 1:1 with a crystallization solution comprising 1.8 M ammonium phosphate monobasic and 0.1 M sodium acetate trihydrate, pH 4.6. The selenomethionine-substituted BoSGBPMLG-B crystals were flash frozen in a cryoprotectant comprising mother liquor supplemented with 20% ethylene glycol and X-ray data were collected at the Life Sciences Collaborative Access Team (LSCAT) beamline 21-ID-F of the Advanced Photon Source (APS) at Argonne National Laboratory. X-ray data were processed in HKL2000 [70] and scaled with Scalepack [70]. Phasing via the anomalous selenium signal was performed in AutoSol [71] from the Phenix package [72], followed by partial refinement in phenix.refine [73] for use with the native protein data collected for BoSGBPMLG-B.
Crystals of BoSGBPMLG-A with cellohexaose were obtained directly from Crystal Strategy Screen I (Molecular dimensions) via hanging drop vapor diffusion at room temperature. The protein (14.9 mg/mL with 10 mM cellohexaose) was mixed 1:1 with the crystallization solution comprising 0.2 M MgCl2, 0.1 M Tris acetate, pH 8.5, 25% PEG 2000 monomethylether. Native crystals of BoSGBPMLG-B were obtained directly from JCSG Plus screen (Molecular dimensions) via hanging drop vapor diffusion at room temperature. BoSGBPMLG-B (20.1 mg/mL with 10 mM cellohexaose) was mixed 1:1 with the crystallization solution comprising 0.8 M NaH2PO4, 0.8 M KH2PO4, and 0.1 M sodium HEPES, pH 7.5. All crystals were flash frozen in a cryoprotectant comprising mother liquor supplemented with 20% ethylene glycol and X-ray data were collected at the LSCAT beamline 21-ID-G of the APS at Argonne National Laboratory. X-ray data for BoSGBPMLG-A were processed in HKL2000 and scaled with Scalepack, while the X-ray data for BoSGBPMLG-B were processed and scaled in DIALS/Xia2 [74, 75]. Molecular replacement was performed in Phaser [76] from the Phenix package using the homologous SGBP-A structure 6DK2 as the search model for BoSGBPMLG-A with cellohexaose, and the partially refined selenomethionine-substituted model of BoSGBPMLG-B was used with the native data collected for BoSGBPMLG-B with cellohexaose. The native BoSGBPMLG-A and BoSGBPMLG-B structures with cellohexaose were refined in Refmac5 [77], with alternate rounds of manual model building in Coot [78]. Validation of the carbohydrates was performed with Privateer [79], and validation of the model fit with Phenix.validate [72].
Initial sitting drop crystal screens for unliganded BoSGBPMLG-A at 21.7 mg/mL, BoSGBPMLG-A at 20.3 mg/mL with 6.6 mM MLG7, and BoSGBPMLG-B at 20.4 mg/mL with 6.3 mM MLG7 were set up using a Phoenix robot (Art Robbin). Crystals of unliganded BoSGBPMLG-A were obtained at room temperature in the JSCS + screen (Qiagen) condition H1: 0.2 M MgCl2, 0.1 M Bis–Tris, pH 5.5, 25% (w/v) PEG 3350. Crystals of BoSGBPMLG-A co-crystallized with MLG7 were obtained at room temperature in the Classics II screen (Qiagen) condition G12: 0.2 M MgCl2, 0.1 M HEPES pH 7.5, 25% (w/v) PEG 3350. Crystals of BoSGBPMLG-B co-crystallized with MLG7 were obtained at room temperature in the JCSG + screen (Qiagen) condition A6: 0.2 M Li2SO4, 0.1 M phosphate-citrate, pH 4.2, 20% (w/v) PEG 1000. For all three initial hits, crystals were readily reproduced by hand in larger hanging drops by screening around the condition varying the buffer pH in one dimension and PEG concentration in the other. The crystals obtained from these optimizations were used for data collection by flash freezing in cryoprotectant comprising mother liquor supplemented with 25% ethylene glycol. X-ray data for unliganded BoSGBPMLG-A and MLG7-bound BoSGBPMLG-A were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline 9-2. X-ray data for MLG7-bound BoSGBPMLG-B at the Canadian Macromolecular Crystallography Facility (CMCF) beamline 08B1-1 at the Canadian Light Source (CLS). All three data sets were indexed and integrated using XDS [80]. BoSGBPMLG-A structures (unliganded and MLG7-bound) were determined by molecular replacement in Phaser from the CCP4i2 package [81, 82] using the cellohexaose-bound BoSGBPMLG-A structure (with the ligand and waters removed) as the search model, and refined in Refmac5 with alternate rounds of manual model building in Coot. MLG7-bound BoSGBPMLG-B structure was determined by molecular replacement in Molrep [83] using the cellohexaose-bound BoSGBPMLG-B (with the ligand and waters removed) structure as the search model, and refined in Buster [84] with alternate rounds of manual model building in Coot.
B. ovatus genetics and anaerobic growth study
Bacterial strains and culture conditions For these experiments and to generate all of the mutant MLGUL strains used in these experiments, the B. ovatus ATCC-8483 Δtdk (ΔBACOVA_03071) strain was employed to facilitate allelic exchange, as previously described [27, 28]. For clarity we refer to the Δtdk strain as wild type, as this parent strain retains a wild-type MLGUL. Mutations were generated using the counter-selectable allelic exchange vector pExchange-tdk as previously described [27]. Oligonucleotides used in this study are listed in Table S1.
Bacteroides ovatus was cultured in a 37 °C Coy anaerobic chamber (5% H2/10% CO2/85% N2) from freezer stocks into tryptone–yeast extract–glucose (TYG) medium and grown for 24 h, to an O.D.600 ~ 1.0. The following day cells were back-diluted 1:100 into Bacteroides minimal media (MM) including 5 mg ml−1 glucose (Sigma) as noted and grown overnight (16 h). For kinetic growth experiments in a plate reader, MM–glucose grown cells were back-diluted 1:200 into MM with the experimental carbohydrate, and in parallel to MM with glucose. Thus, both glucose controls and experimental MLG and oligosaccharide grown cultures were started at the same initial O.D.600 in the plate reader. Kinetic growth experiments were performed at 37 °C in 96-well plates and O.D.600 were recorded every 10–30 min. All plate reader growth experiments were performed in three replicates and the averages are reported in each figure. However, all biological experiments were repeated at least twice to verify consistent growth phenotypes from day to day.
Quantitative RT-PCR (qRT-PCR)B. ovatus strains were cultured in 5 mL of MM containing 5 mg/mL glucose or MLG. Duplicate bacterial cultures were arrested at mid-log phase (O.D.600 ∼ 0.8) with RNAprotect (Qiagen), then stored at − 80 °C overnight, before purification with RNeasy kit (Qiagen). RNA purity was assessed spectrophotometrically, and 1 μg of RNA was used immediately for reverse transcription (QuantiTect Reverse Transcription kit, Qiagen). RT-qPCR was performed in a 96-well plate on a LightCycler 480 System (Roche) with FastStart Essential DNA Green Master (Roche) using the standard primer. Reactions were carried out in 10 μL, consisting of 5 μL of SYBR Green mix, 20 ng of cDNA, and 1 μM (MLGUL TBDT gene Bacova_02742) or 0.125 μM (16S ribosomal RNA) primer mix. Reaction conditions were 95 °C for 600 s, followed by 45 cycles of 95 °C for 10 s, 55 °C for 10 s, 72 °C for 10 s. Cq values (cycle at which an amplification signal is first detected) were calculated using a LightCycler 480 SW 1.5. Data were normalized to 16S rRNA transcript levels, and a change in expression level was calculated as a fold change compared with MM–glucose cultures.
Accession numbers
All coordinates and structure factors have been deposited in the Protein Data Bank (PDB) with accession codes as follows: 6E60 (BoSGBPMLG-A), 6DMF (BoSGBPMLG-A cellohexaose complex), 6E61 (BoSGBPMLG-A MLG7 complex), 6E57 (BoSGBPMLG-B cellohexaose complex), 6E9B (BoSGBPMLG-B MLG7 complex).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
We thank Associate Professor Russ Algar (Dept. Chemistry, University of British Columbia) for use of a fluorescence microplate reader. We thank Associate Professor Eric Martens and his laboratory for the use of a microplate reader in the anaerobic chamber and qPCR thermal cycler. We thank Prof. Charles Haynes (Michael Smith Laboratories, UBC) for access to ITC equipment and invaluable technical advice. We thank the Canadian Macromolecular Crystallography Facility for access to beamline 08B1-1 at the Canadian Light Source, which is supported by the Canada Foundation for Innovation, Natural Sciences and Engineering Research Council of Canada, the University of Saskatchewan, the Government of Saskatchewan, Western Economic Diversification Canada, the National Research Council Canada, and the Canadian Institutes of Health Research. We thank the Life Sciences Collaborative Access Team for access to beamline 21-ID-F and 21-ID-G at the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory (Argonne, IL, USA) under Contract No. DE-AC02-06CH11357. We thank the Stanford Synchrotron Radiation Lightsource at the SLAC National Accelerator Laboratory (Menlo Park, CA, USA) for access to beamline 9-2, the use of which is supported by the U.S. DOE, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Author contributions
KT cloned, expressed and purified recombinant SGBPs, GFP-fusions, and site-directed mutants, conducted AGE and pull-down depletion isotherm analyses, produced and purified MLG partial digest oligosaccharides, solved crystal structures of BoSGBPMLG-A, BoSGBPMLG-A_MLG7 and BoSGBPMLG-B_MLG7 with BRG, and co-wrote the article. MHF conducted reverse genetics and growth phenotype analyses, and co-wrote the article. BRG solved crystal structures of BoSGBPMLG-A, BoSGBPMLG-A_MLG7 and BoSGBPMLG-B_MLG7 with KT. GD conducted ITC experiments. MS expressed, purified and crystallized SeMet BoSGBPMLG-B_cellohexaose. CMEB crystallized BoSGBPMLG-A_cellohexaose and native BoSGBPMLG-B_cellohexaose. ALC assisted with ITC data collection and analysis. FVP, NMK, and HB designed and directed research, and co-wrote the article with input from all authors.
Funding
Work in the Brumer group was supported by operating grants from the Canadian Institutes of Health Research (MOP-137134 and MOP-142472) and infrastructure support from the Canadian Foundation for Innovation (Project #30663) and the British Columbia Knowledge Development Fund. Work in the Koropatkin group was funded by the National Institutes of Health (NIH R01 GM118475). Work in the van Petegem group was supported by the Canadian Institutes of Health Research (MOP-119404). K.T. was partially supported by a four-year doctoral fellowship from the University of British Columbia. M.H.F. was partially supported by a predoctoral fellowship from the Cellular Biotechnology Training Program (T32GM008353).
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Kazune Tamura and Matthew H. Foley contributed equally to this work.
Contributor Information
Nicole M. Koropatkin, Email: nkoropat@umich.edu
Harry Brumer, Email: brumer@msl.ubc.ca.
References
- 1.Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body. Plos Biol. 2016 doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas S, Izard J, Walsh E, Batich K, Chongsathidkiet P, Clarke G, Sela DA, Muller AJ, Mullin JM, Albert K, Gilligan JP, DiGuilio K, Dilbarova R, Alexander W, Prendergast GC. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Can Res. 2017;77(8):1783–1812. doi: 10.1158/0008-5472.can-16-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- 4.Ridaura VK, Faith JJ, Rey FE, Cheng JY, Duncan AE, Kau AL, Griffin NW, Lombard V, Henrissat B, Bain JR, Muehlbauer MJ, Ilkayeva O, Semenkovich CF, Funai K, Hayashi DK, Lyle BJ, Martini MC, Ursell LK, Clemente JC, Van Treuren W, Walters WA, Knight R, Newgard CB, Heath AC, Gordon JI. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341(6150):1079. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arrieta MC, Stiemsma LT, Dimitriu PA, Thorson L, Russell S, Yurist-Doutsch S, Kuzeljevic B, Gold MJ, Britton HM, Lefebvre DL, Subbarao P, Mandhane P, Becker A, McNagny KM, Sears MR, Kollmann T, Mohn WW, Turvey SE, Finlay BB, Investigators CS. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci Transl Med. 2015;7(307):14. doi: 10.1126/scitranslmed.aab2271. [DOI] [PubMed] [Google Scholar]
- 6.Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015;17(5):592–602. doi: 10.1016/j.chom.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. doi: 10.1038/nrc3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, Harris HMB, Coakley M, Lakshminarayanan B, O’Sullivan O, Fitzgerald GF, Deane J, O’Connor M, Harnedy N, O’Connor K, O’Mahony D, van Sinderen D, Wallace M, Brennan L, Stanton C, Marchesi JR, Fitzgerald AP, Shanahan F, Hill C, Ross RP, O’Toole PW. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488(7410):178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 9.Fujimura KE, Slusher NA, Cabana MD, Lynch SV. Role of the gut microbiota in defining human health. Expert Rev Anti Infect Ther. 2010;8(4):435–454. doi: 10.1586/eri.10.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10(5):323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorham JB, Kang S, Williams BA, Grant LJ, McSweeney CS, Gidley MJ, Mikkelsen D. Addition of arabinoxylan and mixed linkage glucans in porcine diets affects the large intestinal bacterial populations. Eur J Nutr. 2017;56(6):2193–2206. doi: 10.1007/s00394-016-1263-4. [DOI] [PubMed] [Google Scholar]
- 13.Desai MS, Seekatz AM, Koropatkin NM, Kamada N, Hickey CA, Wolter M, Pudlo NA, Kitamoto S, Terrapon N, Muller A, Young VB, Henrissat B, Wilmes P, Stappenbeck TS, Nunez G, Martens EC. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell. 2016;167(5):1339–1353. doi: 10.1016/j.cell.2016.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sonnenburg ED, Sonnenburg JL. Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab. 2014;20(5):779–786. doi: 10.1016/j.cmet.2014.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams BA, Grant LJ, Gidley MJ, Mikkelsen D. Gut fermentation of dietary fibres: physico-chemistry of plant cell walls and implications for health. Int J Mol Sci. 2017 doi: 10.3390/ijms18102203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol. 2013;11(7):497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- 17.Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. doi: 10.1038/nature13178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grondin JM, Tamura K, Dejean G, Abbott DW, Brumer H. Polysaccharide utilization loci: fueling microbial communities. J Bacteriol. 2017;199(15):1–15. doi: 10.1128/jb.00860-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN, Gordon JI. Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol. 2011;9(12):1–16. doi: 10.1371/journal.pbio.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Othman RA, Moghadasian MH, Jones PJH. Cholesterol-lowering effects of oat beta-glucan. Nutr Rev. 2011;69(6):299–309. doi: 10.1111/j.1753-4887.2011.00401.x. [DOI] [PubMed] [Google Scholar]
- 21.El Khoury D, Cuda C, Luhovyy BL, Anderson GH. Beta glucan: health benefits in obesity and metabolic syndrome. J Nutr Metab. 2012;2012:851362. doi: 10.1155/2012/851362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gunness P, Michiels J, Vanhaecke L, De Smet S, Kravchuk O, Van de Meene A, Gidley MJ. Reduction in circulating bile acid and restricted diffusion across the intestinal epithelium are associated with a decrease in blood cholesterol in the presence of oat β-glucan. FASEB J. 2016;30(12):4227–4238. doi: 10.1096/fj.201600465R. [DOI] [PubMed] [Google Scholar]
- 23.Fehlbaum S, Prudence K, Kieboom J, Heerikhuisen M, van den Broek T, Schuren FHJ, Steinert RE, Raederstorff D. In vitro fermentation of selected prebiotics and their effects on the composition and activity of the adult gut microbiota. Int J Mol Sci. 2018 doi: 10.3390/ijms19103097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsson U, Johansson M, Nilsson A, Björck I, Nyman M. Dietary supplementation with beta-glucan enriched oat bran increases faecal concentration of carboxylic acids in healthy subjects. Eur J Clin Nutr. 2008;62(8):978–984. doi: 10.1038/sj.ejcn.1602816. [DOI] [PubMed] [Google Scholar]
- 25.Tamura K, Hemsworth GR, DeJean G, Rogers TE, Pudlo NA, Urs K, Jain N, Davies GJ, Martens EC, Brumer H. Molecular mechanism by which prominent human gut Bacteroidetes utilize mixed-linkage beta-glucans. Major health-promoting cereal polysaccharides. Cell Rep. 2017;21(2):417–430. doi: 10.1016/j.celrep.2017.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bolam DN, Koropatkin NM. Glycan recognition by the bacteroidetes Sus-like systems. Curr Opin Struct Biol. 2012;22(5):563–569. doi: 10.1016/j.sbi.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 27.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16(7):1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM, Martens EC. Multidomain carbohydrate-binding proteins involved in bacteroides thetaiotaomicron starch metabolism. J Biol Chem. 2012;287(41):34614–34625. doi: 10.1074/jbc.M112.397380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H, Koropatkin NM. Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. Mbio. 2016;7(2):15. doi: 10.1128/mBio.02134-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cartmell A, Lowe EC, Basle A, Firbank SJ, Ndeh DA, Murray H, Terrapon N, Lombard V, Henrissat B, Turnbull JE, Czjzek M, Gilbert HJ, Bolam DN. How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc Natl Acad Sci USA. 2017;114(27):7037–7042. doi: 10.1073/pnas.1704367114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng HJ, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ, Bolam DN. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:15. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Basle A, Firbank SJ, Zheng HJ, Robinson CV, Winterhalter M, Kleinekathofer U, Bolam DN, van den Berg B. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature. 2017;541(7637):407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koropatkin N, Martens EC, Gordon JI, Smith TJ. Structure of a SusD homologue, BT1043, involved in mucin O-glycan utilization in a prominent human gut symbiont. Biochemistry. 2009;48(7):1532–1542. doi: 10.1021/bi801942a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phansopa C, Roy S, Rafferty JB, Douglas CWI, Pandhal J, Wright PC, Kelly DJ, Stafford GP. Structural and functional characterization of NanU, a novel high-affinity sialic acid-inducible binding protein of oral and gut-dwelling Bacteroidetes species. Biochem J. 2014;458:499–511. doi: 10.1042/bj20131415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mystkowska AA, Robb C, Vidal-Melgosa S, Vanni C, Fernandez-Guerra A, Hohne M, Hehemann JH. Molecular recognition of the beta-glucans laminarin and pustulan by a SusD-like glycan-binding protein of a marine Bacteroidetes. FEBS J. 2018;285(23):4465–4481. doi: 10.1111/febs.14674. [DOI] [PubMed] [Google Scholar]
- 36.Larsbrink J, Zhu Y, Kharade SS, Kwiatkowski KJ, Eijsink VGH, Koropatkin NM, McBride MJ, Pope PB. A polysaccharide utilization locus from Flavobacterium johnsoniae enables conversion of recalcitrant chitin. Biotechnol Biofuels. 2016;9:16. doi: 10.1186/s13068-016-0674-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hudson KL, Bartlett GJ, Diehl RC, Agirre J, Gallagher T, Kiessling LL, Woolfson DN. Carbohydrate–aromatic interactions in proteins. J Am Chem Soc. 2015;137(48):15152–15160. doi: 10.1021/jacs.5b08424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asensio JL, Ardá A, Cañada FJ, Jiménez-Barbero J. Carbohydrate–aromatic interactions. Acc Chem Res. 2013;46(4):946–954. doi: 10.1021/ar300024d. [DOI] [PubMed] [Google Scholar]
- 39.Jeffrey GA. An introduction to hydrogen bonding: topics in physical chemistry. New York: Oxford University Press; 1997. [Google Scholar]
- 40.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L, Laakso LM. Dali server update. Nucleic Acids Res. 2016;44(W1):W351–W355. doi: 10.1093/nar/gkw357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM, Martens EC. Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. MBIO. 2014;5(5):1–12. doi: 10.1128/mBio.01441-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC, Brumer H. A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature. 2014;506(7489):498–502. doi: 10.1038/nature12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Barsanti L, Passarelli V, Evangelista V, Frassanito AM, Gualtieri P. Chemistry, physico-chemistry and applications linked to biological activities of beta-glucans. Nat Prod Rep. 2011;28(3):457–466. doi: 10.1039/c0np00018c. [DOI] [PubMed] [Google Scholar]
- 45.Wood PJ, Weisz J, Blackwell BA. Structural studies of (1-3), (1-4)-beta-d-glucans by C(13)-nuclear magnetic-resonance spectroscopy and by rapid analysis of cellulose-like regions using high-performance anion-exchange chromatography of oligosaccharides released by lichenase. Cereal Chem. 1994;71(3):301–307. [Google Scholar]
- 46.Terrapon N, Lombard V, Drula E, Lapebie P, Al-Masaudi S, Gilbert HJ, Henrissat B. PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 2018;46(D1):D677–D683. doi: 10.1093/nar/gkx1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Foley MH, Martens EC, Koropatkin NM. SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron. Mol Microbiol. 2018;108(5):551–566. doi: 10.1111/mmi.13949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rakoff-Nahoum S, Coyne MJ, Comstock LE. An ecological network of polysaccharide utilization among human intestinal symbionts. Curr Biol. 2014;24(1):40–49. doi: 10.1016/j.cub.2013.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luis AS, Briggs J, Zhang XY, Farnell B, Ndeh D, Labourel A, Basle A, Cartmell A, Terrapon N, Stott K, Lowe EC, McLean R, Shearer K, Schuckel J, Venditto I, Ralet MC, Henrissat B, Martens EC, Mosimann SC, Abbott DW, Gilbert HJ. Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat Microbiol. 2018;3(2):210–219. doi: 10.1038/s41564-017-0079-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. Signal peptidases. Chem Rev. 2002;102(12):4549–4579. doi: 10.1021/cr010166y. [DOI] [PubMed] [Google Scholar]
- 51.Lazaridou A, Biliaderis CG, Micha-Screttas M, Steele BR. A comparative study on structure-function relations of mixed-linkage (1 - > 3), (1 - > 4) linear beta-d-glucans. Food Hydrocolloids. 2004;18(5):837–855. doi: 10.1016/j.foodhyd.2004.01.002. [DOI] [Google Scholar]
- 52.Gilbert HJ, Knox JP, Boraston AB. Advances in understanding the molecular basis of plant cell wall polysaccharide recognition by carbohydrate-binding modules. Curr Opin Struct Biol. 2013;23(5):669–677. doi: 10.1016/j.sbi.2013.05.005. [DOI] [PubMed] [Google Scholar]
- 53.Mackenzie AK, Pope PB, Pedersen HL, Gupta R, Morrison M, Willats WG, Eijsink VG. Two SusD-like proteins encoded within a polysaccharide utilization locus of an uncultured ruminant Bacteroidetes phylotype bind strongly to cellulose. Appl Environ Microbiol. 2012;78(16):5935–5937. doi: 10.1128/AEM.01164-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mackenzie AK, Naas AE, Kracun SK, Schückel J, Fangel JU, Agger JW, Willats WG, Eijsink VG, Pope PB. A polysaccharide utilization locus from an uncultured Bacteroidetes phylotype suggests ecological adaptation and substrate versatility. Appl Environ Microbiol. 2015;81(1):187–195. doi: 10.1128/AEM.02858-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Naas AE, Mackenzie AK, Mravec J, Schückel J, Willats WG, Eijsink VG, Pope PB. Do rumen Bacteroidetes utilize an alternative mechanism for cellulose degradation? MBio. 2014;5(4):e01401–e01414. doi: 10.1128/mBio.01401-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kiemle SN, Zhang X, Esker AR, Toriz G, Gatenholm P, Cosgrove DJ. Role of (1,3)(1,4)-beta-glucan in cell walls: interaction with cellulose. Biomacromol. 2014;15(5):1727–1736. doi: 10.1021/bm5001247. [DOI] [PubMed] [Google Scholar]
- 57.McNulty NP, Wu M, Erickson AR, Pan CL, Erickson BK, Martens EC, Pudlo NA, Muegge BD, Henrissat B, Hettich RL, Gordon JI. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013;11(8):20. doi: 10.1371/journal.pbio.1001637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cann I, Bernardi RC, Mackie RI. Cellulose degradation in the human gut: Ruminococcus champanellensis expands the cellulosome paradigm. Environ Microbiol. 2016;18(2):307–310. doi: 10.1111/1462-2920.13152. [DOI] [PubMed] [Google Scholar]
- 59.Haskey N, Gibson DL. An examination of diet for the maintenance of remission in inflammatory bowel disease. Nutrients. 2017 doi: 10.3390/nu9030259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Armstrong Z, Mewis K, Liu F, Morgan-Lang C, Scofield M, Durno E, Chen HM, Mehr K, Withers SG, Hallam SJ. Metagenomics reveals functional synergy and novel polysaccharide utilization loci in the Castor canadensis fecal microbiome. ISME J. 2018;12(11):2757–2769. doi: 10.1038/s41396-018-0215-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shepherd ES, DeLoache WC, Pruss KM, Whitaker WR, Sonnenburg JL. An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature. 2018;557(7705):434–438. doi: 10.1038/s41586-018-0092-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Joglekar P, Sonnenburg ED, Higginbottom SK, Earle KA, Morland C, Shapiro-Ward S, Bolam DN, Sonnenburg JL. Genetic variation of the SusC/SusD homologs from a polysaccharide utilization locus underlies divergent fructan specificities and functional adaptation in Bacteroides thetaiotaomicron strains. mSphere. 2018;3(3):5. doi: 10.1128/mspheredirect.00185-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrar MD, Whitehead TR, Lan J, Dilger P, Thorpe R, Holland KT, Carding SR. Engineering of the gut commensal bacterium Bacteroides ovatus to produce and secrete biologically active murine interleukin-2 in response to xylan. J Appl Microbiol. 2005;98(5):1191–1197. doi: 10.1111/j.1365-2672.2005.02565.x. [DOI] [PubMed] [Google Scholar]
- 64.McGregor N, Morar M, Fenger TH, Stogios P, Lenfant N, Yin V, Xu XH, Evdokimova E, Cui H, Henrissat B, Savchenko A, Brumer H. Structure-function analysis of a mixed-linkage beta-glucanase/xyloglucanase from the key ruminal Bacteroidetes Prevotella bryantii B(1)4. J Biol Chem. 2016;291(3):1175–1197. doi: 10.1074/jbc.M115.691659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nature Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 66.Nielsen H. Predicting secretory proteins with signalP. Methods Mol Biol. 2017;1611:59–73. doi: 10.1007/978-1-4939-7015-5_6. [DOI] [PubMed] [Google Scholar]
- 67.Juncker AS, Willenbrock H, Von Heijne G, Brunak S, Nielsen H, Krogh A. Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 2003;12(8):1652–1662. doi: 10.1110/ps.0303703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sundqvist G, Stenvall M, Berglund H, Ottosson J, Brumer H. A general, robust method for the quality control of intact proteins using LC–ESI–MS. J Chromatogr B Anal Technol Biomed Life Sci. 2007;852(1–2):188–194. doi: 10.1016/j.jchromb.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 69.Doublié S. Production of selenomethionyl proteins in prokaryotic and eukaryotic expression systems. Methods Mol Biol. 2007;363:91–108. doi: 10.1007/978-1-59745-209-0_5. [DOI] [PubMed] [Google Scholar]
- 70.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol Macromol Crystallogr Pt A. 1997;276:307–326. doi: 10.1016/s0076-6879(97)76066-x. [DOI] [PubMed] [Google Scholar]
- 71.Terwilliger TC, Adams PD, Read RJ, McCoy AJ, Moriarty NW, Grosse-Kunstleve RW, Afonine PV, Zwart PH, Hung LW. Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr Sect D Biol Crystallogr. 2009;65:582–601. doi: 10.1107/s0907444909012098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/s0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr Sect D Struct Biol. 2012;68:352–367. doi: 10.1107/s0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Winter G, Waterman DG, Parkhurst JM, Brewster AS, Gildea RJ, Gerstel M, Fuentes-Montero L, Vollmar M, Michels-Clark T, Young ID, Sauter NK, Evans G. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr Sect D Struct Biol. 2018;74:85–97. doi: 10.1107/s2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Winter G. xia2: an expert system for macromolecular crystallography data reduction. J Appl Crystallogr. 2010;43:186–190. doi: 10.1107/s0021889809045701. [DOI] [Google Scholar]
- 76.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/s0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/s0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/s0907444904019158. [DOI] [PubMed] [Google Scholar]
- 79.Agirre J, Iglesias-Fernandez J, Rovira C, Davies GJ, Wilson KS, Cowtan KD. Privateer: software for the conformational validation of carbohydrate structures. Nat Struct Mol Biol. 2015;22(11):833–834. doi: 10.1038/nsmb.3115. [DOI] [PubMed] [Google Scholar]
- 80.Kabsch W. XDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/s0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, McNicholas SJ, Murshudov GN, Pannu NS, Potterton EA, Powell HR, Read RJ, Vagin A, Wilson KS. Overview of the CCP4 suite and current developments. Acta Crystallogr Sect D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/s0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Potterton L, Agirre J, Ballard C, Cowtan K, Dodson E, Evans PR, Jenkins HT, Keegan R, Krissinel E, Stevenson K, Lebedev A, McNicholas SJ, Nicholls RA, Noble M, Pannu NS, Roth C, Sheldrick G, Skubak P, Turkenburg J, Uski V, von Delft F, Waterman D, Wilson K, Winn M, Wojdyr M. CCP4i2: the new graphical user interface to the CCP4 program suite. Acta Crystallogr Sect D Struct Biol. 2018;74:68–84. doi: 10.1107/s2059798317016035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:22–25. doi: 10.1107/s0907444909042589. [DOI] [PubMed] [Google Scholar]
- 84.Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr Sect D Struct Biol. 2004;60:2210–2221. doi: 10.1107/s0907444904016427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.