Abstract
Microalgae are promising biocatalysts for applications in sustainable fuel, food, and chemical production. Here, we describe culture collection screening, down-selection, and development of a high-productivity, halophilic, thermotolerant microalga, Picochlorum renovo. This microalga displays a rapid growth rate and high diel biomass productivity (34 g m−2 day−1), with a composition well-suited for downstream processing. P. renovo exhibits broad salinity tolerance (growth at 107.5 g L−1 salinity) and thermotolerance (growth up to 40 °C), beneficial traits for outdoor cultivation. We report complete genome sequencing and analysis, and genetic tool development suitable for expression of transgenes inserted into the nuclear or chloroplast genomes. We further evaluate mechanisms of halotolerance via comparative transcriptomics, identifying novel genes differentially regulated in response to high salinity cultivation. These findings will enable basic science inquiries into control mechanisms governing Picochlorum biology and lay the foundation for development of a microalga with industrially relevant traits as a model photobiology platform.
Subject terms: Applied microbiology, Molecular engineering in plants, Green diesel, Transcriptomics
Lukas Dahlin et al. report the development of Picochlorum renovo, a high-productivity, halophilic, thermotolerant microalga. They report the nuclear and chloroplast genomes, and develop a system for inserting transgenes in both organelles.
Introduction
Microalgae are a source of renewable biomass and promising photosynthetic biocatalysts for the sustainable production of fuel and chemical intermediates1. Importantly, they are also valuable model systems for fundamental investigation of mechanistic photobiology2. These microbes possess a series of unique characteristics that make them well-suited for biotechnological applications, including year-round cultivation capacity in saline water on non-arable land, higher potential biofuel yields than terrestrial crops, and the ability to utilize CO2 as a sole carbon source3,4. Rising greenhouse gas emissions from anthropogenic sources has led to a resurgent interest in exploiting these organisms for concurrent CO2 capture and renewable biocommodity production5,6. However, at present, current model microalgal systems are not suitable for outdoor deployment, displaying low productivity under relevant environmental conditions (e.g., high light intensity, high temperature, seawater environments)7. Further, top-candidate deployment systems display low genetic throughput, often requiring weeks-to-months to generate and verify transgenic lines, which hinders fundamental mechanistic inquiry and metabolic engineering strategies in deployment-relevant microalgae7,8.
Since its first classification in 2004, the genus Picochlorum has been recognized for its distinct characteristics of broad thermotolerance, salinity tolerance, compact genome architecture, fast doubling time, and resilience to high light intensity6,9–13. An alga of the genus Picochlorum was recently shown to have the highest biomass productivity in a comparative analysis between a series of industrially relevant microalgae, underscoring this genera’s deployment potential6. However, to date, there are limited insights into Picochlorum halotolerance, biosynthetic capacity, biomass characterization, and genetic tractability, hindering its development as a fundamental platform and for biotechnological applications.
Here, we report the down-selection, characterization, and development of a novel alga of the genus Picochlorum, Picochlorum renovo sp. nov. We characterized the diel biomass productivity (34 g m−2 day−1) of this alga under simulated outdoor cultivation conditions, quantifying the protein, carbohydrate, and lipid content (20, 60, and 10% ash-free dry cell weight, respectively), thermotolerance (growth capacity up to 40 °C), and salinity tolerance (growth at 107.5 g L−1 salinity). Furthermore, we generated nuclear, chloroplast, and mitochondrial genome sequences and report comparative transcriptomic analyses under low- and high-salt conditions, enabling high-resolution genome annotation and providing novel insight into the mechanisms of halotolerance. Lastly, we developed a set of facile genetic tools that enable expression of multiple transgenes inserted into either the nuclear or chloroplast genomes. Combined, these data will enable fundamental insights into Picochlorum photobiology and inform targeted genetic engineering strategies to accelerate microalgal biotechnological applications in a deployment-relevant, emerging model microalga.
Results
Down-selection, physiology, and compositional analysis
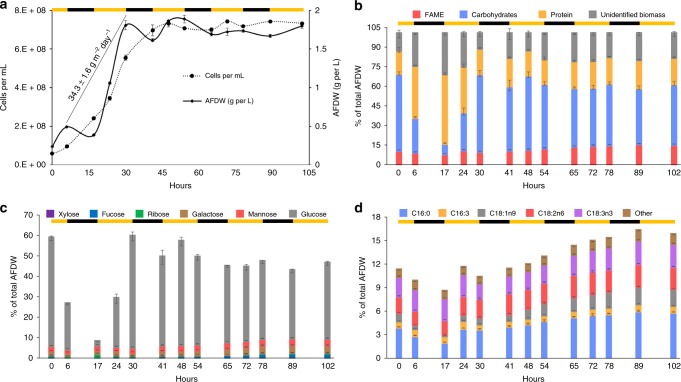
We screened a >300-strain microalgal culture collection, in order to identify halotolerant strains14,15. Over 100 unique halotolerant isolates were down-selected and screened under simulated (diurnal light and temperature cycling) summer growth conditions using a custom built photobioreactor, described in Dahlin et al.15. We identified one isolate that exhibited a noticeably faster growth rate relative to other isolates, including control strains Nannochloropsis oceanica (KA32) and Nannochloropsis salina (CCMP 1776), two top-candidate strains currently under evaluation for outdoor deployment (Fig. 1)15–17. This rapidly growing strain was down-selected for further analysis and development. Under batch growth this microalga displayed a diel biomass productivity of 34.3 g m−2 day−1, from hour 6 to 30, representative of one day and night of high-productivity growth (Fig. 2a). Dark period biomass loss was 0.25 g m−2 h−1 during the first 11-h dark period and 0.46 g m−2 h−1 in the second. Cell division occurs during both light and dark periods when grown under a diel cycle. Cessation of cell division and biomass accumulation occurs simultaneously. Nitrogen supplementation during stationary phase led to growth reinitiation (Supplementary Fig. 1a). We observed peak growth at 35 °C under continuous illumination, with growth capacity up to 40 °C (Supplementary Fig. 1b).
Fig. 1.
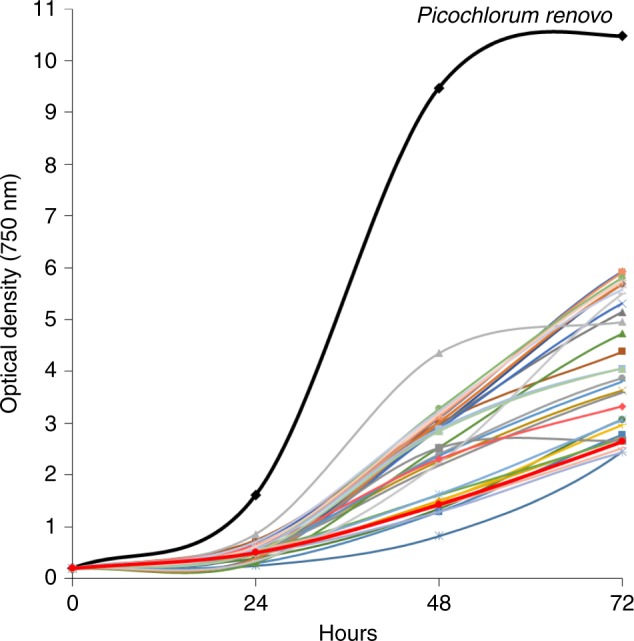
Representative culture collection growth screening data. The rapid growth and high optical density phenotype of P. renovo is highlighted in black. Nannochloropsis salina CCMP 1776 is bolded in red for reference
Fig. 2.
Overview of P. renovo productivity and associated biomass composition, as a function of time. Alternating black and yellow bars depict the light-dark growth cycle. a Growth curves as a function of ash-free dry weight (g per L) and cell density (cells per mL). Areal productivity is shown for hour 6 to 30, representing one light-dark cycle (day). b The biomass content of lipid (as FAMEs), carbohydrate, protein, and the fraction of biomass not identified. c Carbohydrate speciation via hydrolysis of biomass. d Fatty acid speciation via fatty acid methyl ester analysis, representative of the lipid fraction of the biomass. All data points are an average of n = 3 biological replicates; error bars depict the standard deviation of the replicates
Biomass composition varies as a function of growth phase, with fluctuations in carbohydrate and protein content observed throughout diel cycles (Fig. 2b). Notably there is a substantial decrease in glucose (derived from biomass hydrolysis) following inoculation into fresh media, with glucose declining from 52 to 1.4% of AFDW (ash-free dry weight) (Fig. 2c). Lipid content, as measured by fatty acid methyl esters, varied from 8.5 to 16.2%, with C16:0, C16:3, C18:1n9, C18:2n6, and C18:3n3 representing major lipid fractions (Fig. 2d). Thirty hours post-inoculation the cells enter stationary phase and have an ash-free composition of 10% FAME (fatty acid methyl ester), 20% protein, 59.5% carbohydrates (measured as hydrolyzed monomeric sugars), and 10.5% unidentified biomass components (Fig. 2b).
Genomic analysis and speciation
We conducted phylogenetic analysis of the isolate’s 18S rRNA, identifying high similarity (>99%) to numerous Picochlorum species, providing initial evidence for taxonomic classification. PacBio genome sequencing generated an assembled nuclear genome containing 29 contigs with 14.4 Mbps and 46.2% GC, similar to previously reported Picochlorum genomes12,13. In total, 8902 protein coding sequences were putatively annotated, with an average of 2.2 exons/1.2 introns per gene. The nuclear genome contains a homolog of the universally conserved meiosis associated gene, spo11–2 (E-value: 2E-22), and homologs of multiple meiotic and/or homologous recombination associated genes (with reported E-values), including four rad51 homologs (2E-107 to 4E-26), dmc1 (2E-121), pol2A (0), rfc1 (4E-34), polD1 (0), mre11 (2E-77), rad50 (0), rad54 (4E-134), mus81 (6E-15), msh4 (2E-10), msh5 (2E-50), rpa1 (3E-69), rpa2 (8E-25), and rpa3 (5E-16)18. Further evidence of genes putatively involved in meiosis is provided by the identification of oda2 (0) and bug22 (1E-72) homologs, which are flagella associated genes, implicated in gamete pairing prior to mating19–23. We also identified a putative chlorophyllide-a oxygenase (0), necessary for chlorophyll b production, and cell division was observed to occur by autosporulation (Supplementary Fig. 2), providing additional evidence for classification of this strain as a Picochlorum11. When compared to other available Picochlorum genomes, the novel Picochlorum isolate displayed 87–94% whole genome sequence identity (Supplementary Table 1). These data support that the isolate is a novel Picochlorum species, henceforth termed Picochlorum renovo sp. nov.24.
Chloroplast and mitochondria genome maps are presented in Supplementary Fig. 3. The 74 kb chloroplast genome displayed a non-canonical chloroplast genome architecture lacking an inverted repeat region, as noted by Krasovec et al. for the genus Picochlorum12. The 36 kb mitochondrial genome displayed a compact coding architecture, representing the highest mitochondrial coding density reported to date (1.05 CDS per kb) for the class Trebouxiophyceae, in line with the previously reported mitochondrial genome of Picochlorum costavermella12. Notably, genes encoding a homing endonuclease and protein of unknown function split the mitochondrial 23s rRNA, contributing, in part, to the higher coding density (Supplementary Fig. 3).
Transcriptome response to salinity
We observed broad halotolerance in P. renovo, as reported previously for this genus10,11, with cultivation capacity in minimal media salinity concentrations ranging from 8.75 to 107.5 g L−1 sea salts (Supplementary Fig. 1c, d). To better understand the genes involved in the salinity response, cultures were grown in 8.75 and 35 g L−1 salinity seawater and assessed via comparative transcriptomics. RNA from triplicate mid-log phase cultures was sequenced, and subsequent differential expression analysis was performed. In total, 3464 genes were differentially expressed at 35 g L−1 salinity (1934 down, 1530 up) at statistically significant values (q < 0.05), representing 39 percent of total coding sequences (Supplementary Data File 1). Gene ontology semantic analyses were used to deconvolute the large number of differentially expressed transcripts, implicating a subset of processes involved in the high-salt response, including previously reported genes governing proline metabolism (Supplementary Fig. 4)10. A series of previously unreported haloresponsive genes were also observed, including ppsA (E-value: 2E-54), ppsC (E-value: 2E-60), pks1 (E-value: 4E-30) and pks15 (E-value: 1E-28) (polyketide synthases), iput1 (inositol phosphorylceramide glucuronosyltransferase, E-value: 9E-75), cerk (ceramide kinase, E-value: 8E-34), rad54 (DNA repair and recombination protein, E-value: 2E-20), and dmc1 (disrupted meiotic cDNA 1, E-value 2E-121), discussed further below.
Nuclear and chloroplast engineering
We randomly integrated a linear PCR amplicon comprised of native promoter and terminator elements into the nuclear genome of P. renovo, directing transcription of 2A peptide-linked bleomycin resistance gene and the fluorescent reporter mcherry (Fig. 3a) via electroporation. mCherry was chosen as a reporter gene for nuclear expression based on prior reports of high signal to noise ratios in microalgae25.
Fig. 3.
Overview of P. renovo nuclear transformation. a Construct design showing genetic elements and primers used to generate DNA for electroporation (49 and 11) and subsequent PCR confirmation of transformants. b PCR verification of 12 clones utilizing primers shown in panel a. c Dot plot of fluorescent plate reader data of wild type and mCherry transformants, normalized to chlorophyll autofluorescence. Data is from three biological replicates. d Confocal microscopy images of wild type and transformant microalgae expressing mCherry. Green coloring represents chlorophyll autofluorescence, red coloring represents mCherry fluorescence, 10 µm scale bar
Per transformation, an average of 41 colonies were obtained, representing transformation efficiencies of 14 colonies per µg of DNA, and 9 × 10−8 colonies per electroporated cell. Seventy-five percent (9/12) of PCR screened transformants contained the entire transgene construct, while the remaining contained truncated versions (Fig. 3b). Transgene integration was also achieved via biolistics, however we observed approximately an order of magnitude lower transformation efficiency relative to electroporation. Positive transformants showed a two- to fourfold increase in mCherry fluorescence over wild type (Fig. 3c), and confocal microscopy confirmed mCherry fluorescence localized to the nucleus and cytoplasm in these cells (Fig. 3d). Additional promoter configurations, with and without their respective introns, were also evaluated, including elongation factor 1-alpha 2 (eef1A2) and photosystem I reaction center subunit II (psaD), both utilizing the eef1A2 terminator, which displayed comparable transformation efficiencies and mCherry fluorescence (Supplementary Data File 2). P. renovo is also sensitive to G418, which we have successfully used as a selection agent.
Figure 4a depicts the construct utilized for targeted engineering of the P. renovo chloroplast via biolistics. The native 16S ribosomal RNA promoter and 3′ UTR were utilized to direct transgene expression. The commonly utilized antibiotics spectinomycin and streptomycin failed to inhibit P. renovo growth, and the above utilized phleomycin (for nuclear transformation) was ineffective for isolation of viable chloroplast transformants. Therefore, we chose erythromycin for selection, following antibiotic sensitivity screening. Notably, there is 100% homology between the last 9 base pairs (anti-Shine-Dalgarno sequence) of the P. renovo 16S rRNA and the E. coli 16S rRNA26. Thus, a canonical E. coli ribosomal binding site (RBS, AGGAGGTTATAAAAA) was used to direct translation. The erythromycin resistance gene (ereB) was linked to the reporter super folder green fluorescent protein (sfGFP) in an operon27 for rapid identification of transgenic lines. When fully constructed with targeting homology arms, this plasmid readily yielded transformed microalgae using a conventional biolistic approach28–30.
Fig. 4.
Overview of P. renovo chloroplast transformation. a Construct design showing genetic elements utilized and homology directed integration into the chloroplast genome, along with the primers used for subsequent PCR confirmation of transformants. b PCR verification of 3 clones utilizing primers shown in panel a. c Dot plot of fluorescent plate reader data of wild type and sfGFP transformants, normalized to chlorophyll autofluorescence. Data is from three biological replicates. d Epifluorescent microscopy images of wild type and sfGFP transformant microalgae. Red coloring represents chlorophyll autofluorescence, green coloring represents sfGFP fluorescence, 10 µm scale bar
Transformants could be rapidly identified via the reporter gene by imaging of the bombarded plate in a gel imaging station with filter sets suitable for sfGFP detection. This procedure yielded an average (n = 3) of a single colony per transformation with efficiencies of 1.4 colonies per µg delivered DNA and 8 × 10−9 colonies per microalgal cell. Colonies positive for sfGFP were passaged on selective media and proper integration of the construct into the target region was verified via PCR using primers binding outside the homology region and within the transgene operon, depicted in Fig. 4a, b. 38–48-fold greater sfGFP fluorescence was observed over wild type when measured via fluorometry (Fig. 3c). Epifluorescent microscopy showed sfGFP fluorescence successfully localized to the chloroplast (Fig. 4d).
Discussion
P. renovo displayed a distinct rapid growth rate phenotype, in initial screening trials comparing over 100 unique isolates (Fig. 1). Peak growth rate at 35 °C (Supplementary Fig. 1b) and cultivation capacity up to ~3× seawater salinity (Supplementary Fig. 1d) indicates this strain is well-suited for outdoor cultivation in high temperature regions with saltwater access. These traits complement those of previously identified winter candidate deployment strains15,31, laying the foundation for crop rotation strategies17,32. Given the potential discordance between optical density and biomass density we further characterized P. renovo’s biomass productivity; the diel biomass productivity reported here exceeds the target productivity of 25 g m−2 day−1 reported by Davis, et al.33 for cost-competitive algal biofuels. Higher biomass productivities are likely achievable, given the suboptimal growth temperatures used in this study, which simulated outdoor cultivation in Mesa, Arizona. Future studies will evaluate outdoor productivity metrics to assess translatability of indoor metrics to outdoor systems in geographic regions better suited for high temperature cultivation.
Biomass analysis indicates the primary biomass hydrolysis product in P. renovo is glucose, which is a favorable feedstock for downstream biotechnological applications34. A drastic depletion of glucose was observed following inoculation into fresh media, similar to outdoor cultivation trends observed in other microalgal genera15 (Fig. 2c). Dark period biomass loss is characterized almost exclusively by a decrease in glucose. These phenomena putatively function as a mechanism to remobilize glucose as an energy source for cell division and cellular homeostasis35. Thus, as previously reported36, dark period biomass loss is an important parameter to consider when cultivating microalgae for biomass production. Under the conditions tested here, dark period biomass loss ranged from 0.25 to 0.46 g m−2 h−1. Combined, these data highlight the potential advantage of harvesting P. renovo biomass prior to dark period losses to maximize biomass and storage carbon yields.
Interestingly, cell division occurs during both the light and dark periods when grown under a diel cycle (Fig. 2a). This is contrary to many microalgae that synchronize cell division to occur at night; such is the case for the genera Chlamydomonas37, Nannochloropsis38, Chlorella, and Scenedesmus39. This continuous diurnal and nocturnal cell division, coupled with the compact genome(s), may partially explain the rapid doubling time and high biomass productivity of P. renovo, and represents an area warranting further research. Cell division and biomass accumulation cease concurrently, suggesting a non-photosynthetic state when nitrogen-deprived (Fig. 2a). This is notable as some microalgae will continue to accumulate biomass post-nitrogen deprivation, presenting another avenue for comparative analyses40. Importantly, addition of nitrogen following growth arrest resulted in reinitiated growth, implicating nitrogen deprivation as the key driver for entry into stationary phase under the conditions evaluated in this study (Supplementary Fig. 1a). Optimization of nitrogen levels and harvest point may lead to enhanced productivity and storage carbon content.
Comparative transcriptomic analyses identified a series of previously unreported, halo-responsive genes. dmc1, which is involved in homologous chromosome pairing during meiosis was one of the most highly upregulated transcripts at higher salinity41. rad54, encoding a putative DMC1-interacting protein known to function during homologous recombination, is concurrently upregulated42,43. The upregulation of these genes could be attributed to meiosis, or homologous recombination repair of double strand DNA breaks, due to increased double strand breaks at higher salinities44,45. The observation of differentially expressed genes putatively associated with meiosis and homologous recombination suggests P. renovo may participate in sexual mating, and is capable of DNA repair via nuclear homologous recombination, both powerful tools for genetic manipulation. Indeed, sexual mating has been leveraged for trait stacking in both microalgae and plants and presents a powerful approach for rapid development of production hosts. Though we acknowledge that gene homology is insufficient evidence to assert functionality, as reviewed by Fučíková et al., multiple morphological/cytological observations of syngamy and/or meiosis have been reported in the class Trebouxiophyceae and high conservation of meiotic genes is found within this class46–48.
Downregulation of genes encoding proteins putatively relating to lipid remodeling was observed under high-salt conditions, including pks1, pks15, ppsA, ppsC, iput1, and cerk. ppsA, ppsC, pks1, and pks15 are involved in the synthesis of diverse lipids and polyketides which have been implicated in cell wall permeability49. cerk is an enzyme that transfers a phosphate group to ceramide and is potentially acting in coordination with iput1 which transfers a glucuronic acid moiety to glycosyl inositol phosphorylceramides. Ceramides provide the lipid backbone for plant sphingolipids, and are primarily believed to be structural components of cellular membranes; however, ceramides have also been suggested to play a role in plant signaling50. The above data suggests that P. renovo is potentially using lipid remodeling to tune membrane permeability at differing salinities.
To facilitate P. renovo genetic and metabolic engineering, we developed tools enabling transgene expression in both the nucleus and chloroplast. Interestingly, only 9 of the 12 nuclear transgenic isolates screened showed insertion of the full transgene construct. Of the remaining three isolates, two were shown to have a truncated promoter or terminator, and one was shown to have an incomplete mCherry coding sequence, observed by the inability to generate a full-length coding sequence PCR product (Fig. 3b). It is not clear whether these truncated transgene constructs are the result of native P. renovo machinery cleaving the transgene construct or an incomplete PCR product integrating into the genome. Fluorescent plate reader analysis of the clones revealed increases in mCherry fluorescence over wild type for the 11 clones containing a full length mCherry coding sequence (Fig. 3b,c). As expected, the single clone without a full length mCherry coding sequence did not show an increase in mCherry fluorescence relative to wild type. The variation in mCherry fluorescence could be due to unique integration sites, or multiple integration events. mCherry fluorescence was primarily localized to the nucleus and cytoplasm of the transformant, with no observable chloroplast localization, as has been previously observed51. Preliminary analyses indicate stable nuclear transformation; mCherry fluorescence intensity remains constant following passaging on and off the selection marker.
Successful chloroplast transformation was phenotypically observed via high reporter expression, and epifluorescent microscopy confirmed successful localization of the sfGFP to the P. renovo chloroplast, evident by overlap with chlorophyll autofluorescence (Fig. 4c, d). The ability to confirm transgenic colonies via direct imaging of the high sfGFP signal will increase the throughput of control element screening, such as varied promoter strengths, in order to optimize metabolic engineering strategies. Additionally, the successful utilization of an E. coli RBS for operonic expression presents the potential for optimization of mRNA translation via the employ of established RBS prediction software26. Thus, these tools will be useful for biotechnological applications, such as overexpression of desired industrial enzymes28,52 or fine-tuned regulation of native and/or synthetic metabolic pathways for bioproduct formation53.
The transformation procedure presented herein is a facile protocol with relatively rapid turnaround time that can be completed in a few hours. Given the fast growth of this alga, transformant colonies can be generated in as few as 5 days, considerably faster than top-candidate deployment strains such as Nannochloropsis, wherein colonies need ~21 days of growth before verification analyses can be performed7. We have also provided the sequences of two additional nuclear promoters (elongation factor 1-alpha 2 and photosystem I reaction center subunit II, Supplementary Data File 2), which we have successfully utilized to generate transformants. These additional promoters could prove useful for expression of multiple transgenes from one nuclear targeting cassette.
The full biotechnological potential of microalgae has yet to be brought to bear at commercial scale, in part due to the lack of robust, high-productivity strains suitable for outdoor deployment. Further, microalgal genetics in non-model systems has proven to be a limiting factor in strain development and fundamental mechanistic probing of top-candidate deployment strains. Here, we characterize a novel high-productivity, halophilic, thermotolerant microalga, and report the development of genomic and genetic tools therein. In addition to possessing a series of traits suitable for outdoor deployment, this strain displays favorable characteristics for development as a model system, in part due to its compact genomic architecture and rapid genetic throughput. Combined, the above-described traits present a unique complement to extant model systems. Further genetic development of P. renovo will enable both fundamental and applied insights, including elucidation of key regulatory mechanisms governing rapid growth and halotolerance in microalgae, as well as strain engineering strategies targeting enhanced productivity and carbon partitioning in a deployment-relevant microalga.
Methods
Strain screening and characterization of algal growth
Microalgae were screened as previously reported15, under conditions representative of summer cultivation. Briefly, 100 mL microalgal cultures were sparged with 2% CO2 at 100 mL min−1. Temperature cycled from 21 to 32 °C while lighting cycled from 0 to 965 µmol m−2 s−1 (the maximum output of the utilized lights). This regime was designed to simulate the temperature and lighting diel cycles measured in outdoor raceway ponds located at the Arizona Center for Algae Technology and Innovation testbed site located in Mesa Arizona, during the time frame of 12 June to 21 July 2014. We utilized a modified f/2 medium for cultivation, termed NREL Minimal Medium (NM2), in seawater (Gulf of Maine, Bigelow Laboratory), the following were added to the indicated final concentrations followed by addition of 12 M HCl to attain pH 8.0: NH4Cl (5.0 × 10−3 M), NaH2PO4∙H2O (0.313 × 10−3 M), Na2SiO3∙9H2O (1.06 × 10−4 M), FeCl3∙6H2O (1.17 × 10−5 M), Na2EDTA∙2H2O (1.17 × 10−5 M), CuSO4∙5H2O (3.93 × 10−8 M), Na2MoO4∙2H2O (2.60 × 10−8 M), ZnSO4∙7H2O (7.65 × 10−8 M), CoCl2∙6H2O (4.20 × 10−8 M), MnCl2∙4H2O, (9.10 × 10−7 M), thiamine HCl (2.96 × 10−7 M), biotin (2.05 × 10−9 M), cyanocobalamin (3.69 × 10−10 M), Tris base (24.76 × 10−3 M). For genetic engineering, the concentration of seawater was diluted 4-fold with Milli-Q water (Millipore Corporation), ammonium bicarbonate was utilized in the place of ammonium chloride, and 1.5× vitamins (thiamine HCl, biotin, cyanocobalamin) were utilized. Agar (Bacto) plates were prepared by autoclaving 3% agar in Milli-Q water, followed by addition of an equal volume of sterile filtered NM2 (seawater diluted twofold) with 2× nutrients, trace metals, vitamins, and Tris buffer. Sterile filtered selection antibiotic was added as necessary to appropriate concentrations, defined below.
To obtain a more detailed analysis of P. renovo growth, the above conditions were utilized with a 120 mL culture volume. Mid-log phase seed culture was generated under the above diel conditions, and used to inoculate 36, 120 mL cultures at a starting optical density of 1.0, in biological triplicate. Inoculation occurred approximately halfway through the light cycle, and biomass samples were harvested at the initiation, mid-point, and end-point of the lighting cycle, as indicated in Fig. 2. Sterile water was added prior to samplings to account for evaporative loses. Cell counts were performed using an Improved Neubauer hemocytometer. To convert volumetric productivities to areal values, the cross-sectional area of the culture tubes (0.00459 m2) was employed.
Growth at varying salinities for Supplementary Fig. 1d were done in the same fashion as culture collection screening except salt levels were varied by addition of sea salts (Sigma S9883). 17.5 g L−1 salinity was achieved via addition of seawater to milli-Q water and higher salinities utilized addition of sea salts. One hundred milliliters of culture was harvested after 6 days of growth, utilizing the temperature and light cycling from the culture collection screening methods described above. Temperature optima data, represented in Supplementary Fig. 1b was generated by growing strains in NM2, with culture conditions of constant 400 µmol m−2 s−1 lighting, 2% constant CO2 sparging, and 100 mL volume. To determine growth rates, optical density (750 nm) measurements were taken daily, Supplementary Fig. 1b.
Compositional analysis
Compositional analysis was carried out as reported previously15, with the following modification; a Carbopac PA1 HPAEC column was utilized for sugar monomer (carbohydrate) analysis. Protein was quantified via CHN (carbon, hydrogen, and nitrogen) analysis, utilizing an Elementar VarioEL cube CHN analyzer according to the manufacture’s specifications. Briefly, a 5 mg sample is combusted at 950 °C, and subsequent gasses are carried via helium to reduction and adsorption tubes utilizing an intake pressure of 1200 psi and ultimately detected with a thermal conductivity detector. A nitrogen-to-protein conversion factor of 4.78 was used54.
Genome sequencing, assembly, and annotation
High molecular weight algal genomic DNA was extracted from cells imbedded in agarose, purified and concentrated using AMPure PB beads. The DNA was then fragmented using Covaris g-Tubes. Fragmented and purified DNA was processed for 20 kb SMRT bell library prep. The long insert libraries were size selected using a Blue Pippin instrument (Sage Sciences, Beverly, MA). The sequencing primer was annealed to the selected SMRT bell templates. The libraries were bound to DNA polymerase and loaded on the PacBio RSII for sequencing. Sequencing was completed using either C2/P4 or C3/P5 chemistry and 3-h movies. 8 SMRT cells of sequencing data were assembled with FALCON, version 0.2.255. The final assembly includes 29 contigs with an assembled genome size of 14.4 Mbp. Estimated fold coverage of the PacBio reads was 270×.
Genome annotation was performed using the BRAKER (v2) training and annotation pipeline56 utilizing the six sets of transcriptomic reads (described below in transcriptome response to salinity) to inform AUGUSTUS gene models57,58. Functional annotation of the 8902 genes was performed by InterProScan 559 and BLASTp searches against the UniProt60 protein blast database; reported E-values reflect this methodology. The P. renovo genome assembly and annotation is available for download at the Greenhouse Knowledgebase (greenhouse.lanl.gov).
Transcriptome response to salinity
In order to identify genes putatively conferring halotolerance, cells were cultivated under low- and high-salinity conditions, corresponding to 8.75 g L−1 and 35 g L−1 sea salts. Cells grow at approximately the same growth rate under these conditions (Supplementary Fig. 1c). Cells adapted to the appropriate salinity level were grown in NM2 medium, utilizing ammonium bicarbonate as a nitrogen source. Biological triplicate culture conditions were as follows: 33 °C, 400 µmol m−2 s−1 lighting, and 2% constant CO2 sparging in 100 mL volume. Seawater was employed as a source of salt, as this provides a more accurate proxy for halo-responsiveness compared to NaCl61. Seawater was diluted with distilled water to obtain appropriate salinity levels. The data from these methods are reflected in Supplementary Fig. 1c and salinity transcriptomics data. The above methods were done with the explicit goal of reducing culture shock, and subsequent global stress response, thus allowing a steady state comparison of RNA transcripts relating to salinity tolerance. RNA was obtained utilizing a QIAGEN RNeasy Plant Mini Kit following the manufacturer’s recommendations, cells were homogenized under liquid nitrogen using a mortar and pestle. Paired-end 150 bp Illumina read RNA seq data were received from Genewiz in the form of compressed fastq files. Samples were comprised of two conditions and three biological replicates of each condition, resulting in six total samples. Raw fastq reads were quality trimmed using HTStream62 and mapped via Salmon63 to the available genomic assembly. Coding regions were extracted from the full reference assembly prior to mapping. Read counts were formatted into a tab-separated file and migrated to R64 to perform differential expression using the edgeR65 package. Low-level transcripts were filtered and removed, and all libraries were normalized to each other. Transcript counts were fit to a generalized linear model and the Cox-Reid profile-adjusted likelihood method was used to estimate the dispersion of each transcript. Differential expression was performed by a quasi-likelihood test between each condition. Transcripts were determined as differentially expressed when the corrected p-value (also known as q-value, or False Discovery Rate) was less than or equal to 0.05 after a Benjamini-Hochberg correction for multiple hypothesis testing.
Whole genome alignments to other publicly available Picochlorum genomes were done as follows: six assemblies of different strains of Picochlorum sp. were compared using the nucmer utility in the large-scale alignment program MUMmer12,13,66,67. Maximal matches were found and total bases matching between the samples were summed and the percent identity was reported as the average identity among the maximal unique matches.
Gene ontology analysis was performed as follows: differentially expressed genes were assigned putative functions by extracting the FASTA sequence from the original list of genes and aligning the sequence against the available Chlamydomonas reinhardtii annotated assembly (version 5.5) via BLAST68. Protein identification numbers and putative annotations were then uploaded to the UniProt60 database and cross-referenced against the available gene ontology (GO) terms. GO terms were visualized on a semantic space scatterplot with the online software Revigo69.
Nuclear engineering
A nuclear integration cassette, as depicted in Fig. 3a, was synthesized and subcloned into the pUC19 plasmid backbone by Genewiz, Inc (South Plainfield, NJ). The selection marker, 2A peptide and mCherry were codon optimized to the P. renovo genome. The final linear PCR product (from primers LRD 49 and 11) for transformation was generated utilizing Q5 2× hot start master mix (NEB) and purified with a PureLink Quick PCR Purification kit (Invitrogen) following the manufacture’s protocol, modified to include a second wash step.
10 OD units (~475 × 106 cells) of early log phase cells per transformation were harvested and washed three times at room temperature in 375 mM D-Sorbitol (Sigma S6021). Washing utilized 2 mL Eppendorf tubes, 950 µL of 375 mM D-Sorbitol per wash, centrifuged at 8000 × g for 1 min. After washing, cells were resuspended in 100 µL of 375 mM D-Sorbitol; 3 µg of DNA at 850 ng per µL (concentrated on a vacuum centrifuge) was added to the cells and gently mixed. Cells and DNA were incubated for 3 min, transferred to an ice cold 2 mm gap electroporation cuvette (Bulldog Bio) and electroporated with a Gene Pulser Xcell (Bio-Rad) electroporator utilizing a set time constant and voltage protocol of 2200 volts with a 25 ms time constant. Immediately following the pulse, cells were transferred to 400 µL of media supernatant (from the above utilized cells) and incubated at room temperature for 15 min. Cells were then split equally between three selection plates (1.5% agarose) comprised of NM2 supplemented with 20 µg per mL of phleomycin (InvivoGen). Plates were placed in a Percival incubator with fluorescent lighting at 33 °C, 150 µmol m−2 s−1, and 1.5% CO2. Colonies were counted and passaged on selection after 5 days for further analysis. A table of all DNA fragments and PCR primers utilized in this study is supplied in the supplementary information (Supplementary Data File 2).
Chloroplast engineering
Homology arm sequences were PCR amplified from chloroplast genomic DNA using NEB Q5 Master Mix from New England Biolabs (Ipswich, MA). A promoter-RBS-sfGFP-RBS-ereB-3′ UTR cassette was synthesized by Genewiz, Inc (South Plainfield, NJ) as depicted Fig. 4a. The chloroplast integration cassette was assembled into a pUC19 backbone using 2× Gibson Assembly Mix from New England Biolabs, following the manufacturer’s protocol. Complete vector sequences were confirmed by Sanger sequence analysis (Genewiz, South Plainfield, NJ).
Biolistic transformation was employed to deliver DNA into the chloroplast, as reported previously28,70. Ten micrograms of plasmid DNA (QIAprep spin miniprep kit QIAGEN) was precipitated onto 550 nm gold sphere nanoparticles (Seashell Inc.) under constant vortexing. Ten microliters of plasmid DNA (1 µg/µL) was added to 60 µL of gold particles (50 mg per mL), followed by dropwise addition of 50 µL of 2.5 M CaCl2 and 20 µL of 0.1 M spermidine (Sigma S0266-1G). This was vortexed for 5 min, incubated for 1 min at room temperature, briefly centrifuged, and washed with 140 µL of isopropanol. Following removal of wash supernatant, the gold particles were resuspended in 60 µL of isopropanol and gently sonicated in a bath sonicator to resuspend the pellet. To assay loading efficiency of the DNA onto the gold, a 9 µL aliquot was taken, washed in 9 µL of water and assayed for DNA concentration utilizing a NanoDrop 2000 spectrophotometer.
To transform P. renovo, an overnight culture was grown to early log phase in NM2, concentrated to 2.5 OD units in 170 µL, and spread evenly onto a 100 × 15 mm NM2 agar plate supplemented with 800 µg per mL erythromycin (Sigma E5389-5G). A Biolistic PDS-1000/He Particle Delivery System (metal case version) (Bio-Rad) was used for bombardment, which was accomplished by drying 9 µL of the above DNA loaded gold particles onto the macrocarrier (fast, low humidity drying was accomplished by placing the loaded macrocarrier into the bombardment chamber and pulling vacuum71), and bombarding cells 5 cm below the macrocarrier with a 1100 psi rupture disk. After bombardment, plates were placed into the same growth conditions described above. Following 7 days of growth, the plates were imaged with a FluorChemQ gel imaging station (Protein Simple) with 475/35 and 573/35 nm respective excitation and emission filters, which allowed direct imaging of sfGFP positive colonies.
To assess construct integration into the genomes of P. renovo, genomic DNA was extracted from cells passaged on the selection marker utilizing a MasterPureTM Yeast DNA Purification kit (Lucigen). PCR was performed utilizing Q5 Hot Start High-Fidelity polymerase (New England Biolabs) according to the manufacturer’s recommendations. A table of the utilized primers is provided in the supplementary information (Supplementary Data File 2). Images of uncropped gels (depicted in Figs. 3 and 4) are provided in Supplementary Fig. 5.
Fluorescent plate reader analysis
Colonies were restreaked onto fresh agar plates supplemented with the appropriate selection marker (phleomycin 20 µg per mL and erythromycin 800 µg per mL for the nucleus and chloroplast, respectively) and grown in triplicate in 3 mL of growth media (no selection marker) in standard glass cell culture tubes mixed daily via vortexing. Cultures were grown in the above described Percival incubator conditions (33 °C, 150 µmol m−2 s−1, 1.5% CO2). Early log phase cells were analyzed for mCherry and sfGFP fluorescence utilizing 200 µL of cell culture in a black 96 well plate and a FLUOstar Omega plate reader v. 5.11 R3 (BMG Labtech). To quantify mCherry a 584 nm excitation filter and 620/10 nm emission filter were utilized with gain set to 2500; to quantify sfGFP a 485/12 nm excitation and 520 nm emission filter set was used with gain set to 1200. Data was normalized to chlorophyll content, which was determined by using a 485/12 nm excitation and 680/10 emission filters, with gain set to 1500. Data is represented as a fold increase over the wild type alga.
Statistics and reproducibility
All experiments in this study utilized triplicate biological replicates. Error is represented by the sample standard deviation of the replicates, unless otherwise noted. All source data for main text figures and charts is provided in Supplementary Data File 3.
Microscopy
Mid-log phase chloroplast sfGFP transformants and wild type were imaged with a Nikon Eclipse 80i microscope, equipped with a Nikon Intensilight C-HGFI mercury lamp light source, a Nikon Plan Apo VC 100× objective lens, and a Nikon DS-QiMc camera. NIS-Elements BR 4.30.01 software was utilized for imaging chlorophyll and sfGFP (31017 – Chlorophyll Bandpass Emission and 41017 – Endow GFP/EGFP Bandpass, both from CHORMA®). Imaging of wild type and transgenic lines employed equivalent exposure time and gain settings. ImageJ was used for post imaging analysis.
Nuclear mCherry transformants and wild type were imaged with a Nikon C1si confocal microscope, equipped with EZ-C1 3.60 software. Chlorophyll was imaged with a 650 LP filter. mCherry was imaged with a 590/50 filter. Both chlorophyll and mCherry were excited with a 561.4 nm laser. Laser intensity, pin hole size, pixel dwell time, and gain were set using an mCherry clone. Equivalent settings were utilized for imaging wild type cells. ImageJ was used for post imaging analysis.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary items
Acknowledgements
The authors would like to thank Dr. Brian Vogler for insightful discussions regarding design and delivery of nuclear DNA constructs into eukaryotic microalgae, and Andy Politis, Bonnie Panczak, and Brittany Thornton for assistance in compositional analyses. This research was supported by the Department of Energy, Office of Energy Efficiency and Renewable Energy (EERE) under Agreements No. 22000 and DE-NL0029949. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the United States Government or any agency thereof. Neither the United States Government nor any agency thereof, nor any of their employees, makes any warranty, expressed or implied, or assumes any legal liability or responsibility for the accuracy, completeness, or usefulness of any information, apparatus, product, or process disclosed, or represents that its use would not infringe privately owned rights.
Author contributions
L.D., M.P., and M.G. designed and carried out initial strain screening to identify P. renovo. L.D. designed and carried out detailed characterization of P. renovo growth, S.V. performed compositional analysis. Genome sequencing and annotation was done by Y.K., B.H., S.S., and A.G. Salinity transcriptomics were designed and evaluated by L.D. and M.G., A.G. performed gene ontology and differential expression analysis, along with whole genome alignments. Nuclear and chloroplast constructs were designed and evaluated by L.D., C.H., J.L., M.P. and M.G.
Data availability
The algal strain (Picochlorum renovo), DNA elements, and raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher. The genome for Picochlorum renovo is publicly available at https://greenhouse.lanl.gov/greenhouse/organisms. Genomic sequence data can also be accessed at NCBI, Project Number PRJNA558990. The raw data supporting the conclusions of salinity transcriptomics is available from the Sequence Read Archive, under Project Number PRJNA553204.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s42003-019-0620-2.
References
- 1.Sheehan, J., Dunahay, T., Benemann, J. & Roessler, P. Look back at the U. S. Department of Energy’s Aquatic Species Program: Biodiesel from Algae; Close-Out Report. (1978).
- 2.Harris EH. Chlamydomonas as a model organism. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:363–406. doi: 10.1146/annurev.arplant.52.1.363. [DOI] [PubMed] [Google Scholar]
- 3.Wijffels RH, Barbosa MJ. An outlook on microalgal biofuels. Science. 2010;329:796–799. doi: 10.1126/science.1189003. [DOI] [PubMed] [Google Scholar]
- 4.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 5.Ajjawi I, et al. Lipid production in Nannochloropsis gaditana is doubled by decreasing expression of a single transcriptional regulator. Nat. Biotechnol. 2017;35:647–652. doi: 10.1038/nbt.3865. [DOI] [PubMed] [Google Scholar]
- 6.Weissman Joseph C., Likhogrud Maria, Thomas Dylan C., Fang Wei, Karns Devin A.J., Chung Jeffrey W., Nielsen Robert, Posewitz Matthew C. High-light selection produces a fast-growing Picochlorum celeri. Algal Research. 2018;36:17–28. doi: 10.1016/j.algal.2018.09.024. [DOI] [Google Scholar]
- 7.Kilian O, Benemann CSE, Niyogi KK, Vick B. High-efficiency homologous recombination in the oil-producing alga Nannochloropsis sp. Proc. Natl Acad. Sci. USA. 2011;108:21265–21269. doi: 10.1073/pnas.1105861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Radakovits R, et al. Draft genome sequence and genetic transformation of the oleaginous alga Nannochloropsis gaditana. Nat. Commun. 2012;3:686. doi: 10.1038/ncomms1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Foflonker F, et al. Genome of the halotolerant green alga P icochlorum sp. reveals strategies for thriving under fluctuating environmental conditions. Environ. Microbiol. 2015;17:412–426. doi: 10.1111/1462-2920.12541. [DOI] [PubMed] [Google Scholar]
- 10.Foflonker F, et al. The unexpected extremophile: tolerance to fluctuating salinity in the green alga Picochlorum. Algal Res. 2016;16:465–472. doi: 10.1016/j.algal.2016.04.003. [DOI] [Google Scholar]
- 11.Henley WJ, et al. Phylogenetic analysis of the ‘Nannochloris-like’ algae and diagnoses of Picochlorum oklahomensis gen. et sp. nov. (Trebouxiophyceae, Chlorophyta) Phycologia. 2004;43:641–652. doi: 10.2216/i0031-8884-43-6-641.1. [DOI] [Google Scholar]
- 12.Krasovec Marc, Vancaester Emmelien, Rombauts Stephane, Bucchini François, Yau Sheree, Hemon Claire, Lebredonchel Hugo, Grimsley Nigel, Moreau Hervé, Sanchez-Brosseau Sophie, Vandepoele Klaas, Piganeau Gwenael. Genome Analyses of the Microalga Picochlorum Provide Insights into the Evolution of Thermotolerance in the Green Lineage. Genome Biology and Evolution. 2018;10(9):2347–2365. doi: 10.1093/gbe/evy167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Foflonker F, Mollegard D, Ong M, Yoon HS, Bhattacharya D. Genomic analysis of Picochlorum species reveals how microalgae may adapt to variable environments. Mol. Biol. Evol. 2018;35:2702–2711. doi: 10.1093/molbev/msy167. [DOI] [PubMed] [Google Scholar]
- 14.Elliott LG, et al. Establishment of a bioenergy-focused microalgal culture collection. Algal Res. 2012;1:102–113. doi: 10.1016/j.algal.2012.05.002. [DOI] [Google Scholar]
- 15.Dahlin LR, et al. Down-selection and outdoor evaluation of novel, halotolerant algal strains for winter cultivation. Front. Plant Sci. 2018;9:1513. doi: 10.3389/fpls.2018.01513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McGowen J, et al. The Algae Testbed Public-Private Partnership (ATP3) framework; establishment of a national network of testbed sites to support sustainable algae production. Algal Res. 2017;25:168–177. doi: 10.1016/j.algal.2017.05.017. [DOI] [Google Scholar]
- 17.Lammers PJ, et al. Review of the cultivation program within the National Alliance for Advanced Biofuels and Bioproducts. Algal Res. 2017;22:166–186. doi: 10.1016/j.algal.2016.11.021. [DOI] [Google Scholar]
- 18.Wang, Y. & Copenhaver, G. P. Meiotic recombination: mixing it up in plants. 10.1146/annurev-arplant-042817 (2018). [DOI] [PubMed]
- 19.Kamiya R. Functional diversity of axonemal dyneins as studied in Chlamydomonas mutants. Int. Rev. Cytol. 2002;219:115–155. doi: 10.1016/S0074-7696(02)19012-7. [DOI] [PubMed] [Google Scholar]
- 20.Meng D, Cao M, Oda T, Pan J. The conserved ciliary protein Bug22 controls planar beating of Chlamydomonas flagella. J. Cell Sci. 2014;127:281–287. doi: 10.1242/jcs.140723. [DOI] [PubMed] [Google Scholar]
- 21.Silflow CD, Lefebvre PA. Assembly and motility of eukaryotic cilia and flagella. lessons from Chlamydomonas reinhardtii. Plant Physiol. 2001;127:1500–1507. doi: 10.1104/pp.010807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pan J, Snell WJ. Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 2000;3:596–602. doi: 10.1016/S1369-5274(00)00146-6. [DOI] [PubMed] [Google Scholar]
- 23.Frenkel J, Vyverman W, Pohnert G. Pheromone signaling during sexual reproduction in algae. Plant J. 2014;79:632–644. doi: 10.1111/tpj.12496. [DOI] [PubMed] [Google Scholar]
- 24.Chung, M., Munro, J. B., Tettelin, H. & Dunning Hotopp, J. C. Using core genome alignments to assign bacterial species. mSystems3, e00236–18 (2018). [DOI] [PMC free article] [PubMed]
- 25.Rasala BA, et al. Expanding the spectral palette of fluorescent proteins for the green microalga Chlamydomonas reinhardtii. Plant J. 2013;74:545–556. doi: 10.1111/tpj.12165. [DOI] [PubMed] [Google Scholar]
- 26.Tian T, Salis HM. A predictive biophysical model of translational coupling to coordinate and control protein expression in bacterial operons. Nucleic Acids Res. 2015;43:7137–7151. doi: 10.1093/nar/gkv635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quesada-Vargas Tania, Ruiz Oscar N., Daniell Henry. Characterization of Heterologous Multigene Operons in Transgenic Chloroplasts. Transcription, Processing, and Translation. Plant Physiology. 2005;138(3):1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Georgianna DR, et al. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta. Algal Res. 2013;2:2–9. doi: 10.1016/j.algal.2012.10.004. [DOI] [Google Scholar]
- 29.Randolph-Anderson, B. et al. Sub-micron gold particles are superior to larger particles for efficient Biolistic® transformation of organelles and some cell types. https://www.bio-rad.com/webroot/web/pdf/lsr/literature/Bulletin_2015.pdf US/EG Bulletin 2015. Bio-rad Literature On-Line (2015).
- 30.Bio-Rad Laboratories, M. B. G. Biolistic® PDS-1000/He Particle Delivery System.
- 31.Řezanka T, Nedbalová L, Lukavský J, Střížek A, Sigler K. Pilot cultivation of the green alga Monoraphidium sp. producing a high content of polyunsaturated fatty acids in a low-temperature environment. Algal Res. 2017;22:160–165. doi: 10.1016/j.algal.2016.12.017. [DOI] [Google Scholar]
- 32.White Rebecca L., Ryan Rebecca A. Long-Term Cultivation of Algae in Open-Raceway Ponds: Lessons from the Field. Industrial Biotechnology. 2015;11(4):213–220. doi: 10.1089/ind.2015.0006. [DOI] [Google Scholar]
- 33.Davis, R. et al. Process Design and Economics for the Production of Algal Biomass: Algal Biomass Production in Open Pond Systems and Processing Through Dewatering for Downstream Conversion. (2016).
- 34.Dong Tao, Knoshaug Eric P., Davis Ryan, Laurens Lieve M.L., Van Wychen Stefanie, Pienkos Philip T., Nagle Nick. Combined algal processing: A novel integrated biorefinery process to produce algal biofuels and bioproducts. Algal Research. 2016;19:316–323. doi: 10.1016/j.algal.2015.12.021. [DOI] [Google Scholar]
- 35.Graf, A., Schlereth, A., Stitt, M. & Smith, A. M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Natl Acad. Sci. USA107, 9458–9463 (2010). [DOI] [PMC free article] [PubMed]
- 36.Edmundson SJ, Huesemann MH. The dark side of algae cultivation: Characterizing night biomass loss in three photosynthetic algae, Chlorella sorokiniana, Nannochloropsis salina and Picochlorum sp. Algal Res. 2015;12:470–476. doi: 10.1016/j.algal.2015.10.012. [DOI] [Google Scholar]
- 37.Cross FR, Umen JG. The Chlamydomonas cell cycle. Plant J. 2015;82:370–392. doi: 10.1111/tpj.12795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poliner E, et al. Transcriptional coordination of physiological responses in Nannochloropsis oceanica CCMP1779 under light/dark cycles. Plant J. 2015;83:1097–1113. doi: 10.1111/tpj.12944. [DOI] [PubMed] [Google Scholar]
- 39.Bišová K, Zachleder V. Cell-cycle regulation in green algae dividing by multiple fission. J. Exp. Bot. 2014;65:2585–2602. doi: 10.1093/jxb/ert466. [DOI] [PubMed] [Google Scholar]
- 40.Breuer G, Lamers PP, Martens DE, Draaisma RB, Wijffels RH. The impact of nitrogen starvation on the dynamics of triacylglycerol accumulation in nine microalgae strains. Bioresour. Technol. 2012;124:217–226. doi: 10.1016/j.biortech.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 41.Nara T, et al. Isolation of a LIM15/DMC1 homolog from the basidiomycete Coprinus cinereus and its expression in relation to meiotic chromosome pairing. Mol. Gen. Genet. MGG. 1999;262:781–789. doi: 10.1007/s004380051141. [DOI] [PubMed] [Google Scholar]
- 42.Mazin AV, Mazina OM, Bugreev DV, Rossi MJ. Rad54, the motor of homologous recombination. DNA Repair (Amst.). 2010;9:286–302. doi: 10.1016/j.dnarep.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Heyer W-D, Li X, Rolfsmeier M, Zhang X-P. Rad54: the Swiss Army knife of homologous recombination? Nucleic Acids Res. 2006;34:4115–4125. doi: 10.1093/nar/gkl481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dmitrieva NI, Bulavin DV, Burg MB. High NaCl causes Mre11 to leave the nucleus, disrupting DNA damage signaling and repair. Am. J. Physiol. Physiol. 2003;285:F266–F274. doi: 10.1152/ajprenal.00060.2003. [DOI] [PubMed] [Google Scholar]
- 45.Kültz D, Chakravarty D. Hyperosmolality in the form of elevated NaCl but not urea causes DNA damage in murine kidney cells. Proc. Natl. Acad. Sci. U. S. A. 2001;98:1999–2004. doi: 10.1073/pnas.98.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fučíková K, Pažoutová M, Rindi F. Meiotic genes and sexual reproduction in the green algal class Trebouxiophyceae (Chlorophyta) J. Phycol. 2015;51:419–430. doi: 10.1111/jpy.12293. [DOI] [PubMed] [Google Scholar]
- 47.Rasala BA, Chao S-S, Pier M, Barrera DJ, Mayfield SP. Enhanced genetic tools for engineering multigene traits into green algae. PLoS ONE. 2014;9:e94028. doi: 10.1371/journal.pone.0094028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Russell, G. E. Plant Breeding for Pest and Disease Resistance: Studies in the Agricultural and Food Sciences. (Butterworth & Co, 1978).
- 49.Camacho LR, et al. Analysis of the phthiocerol dimycocerosate locus of Mycobacterium tuberculosis. Evidence that this lipid is involved in the cell wall permeability barrier. J. Biol. Chem. 2001;276:19845–19854. doi: 10.1074/jbc.M100662200. [DOI] [PubMed] [Google Scholar]
- 50.Hou Q, Ufer G, Bartels D. Lipid signalling in plant responses to abiotic stress. Plant. Cell Environ. 2016;39:1029–1048. doi: 10.1111/pce.12666. [DOI] [PubMed] [Google Scholar]
- 51.Plucinak TM, et al. Improved and versatile viral 2A platforms for dependable and inducible high-level expression of dicistronic nuclear genes in Chlamydomonas reinhardtii. Plant J. 2015;82:717–729. doi: 10.1111/tpj.12844. [DOI] [PubMed] [Google Scholar]
- 52.Rasala BA, et al. Robust Expression and Secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE. 2012;7:e43349. doi: 10.1371/journal.pone.0043349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galarza JI, Gimpel JA, Rojas V, Arredondo-Vega BO, Henríquez V. Over-accumulation of astaxanthin in Haematococcus pluvialis through chloroplast genetic engineering. Algal Res. 2018;31:291–297. doi: 10.1016/j.algal.2018.02.024. [DOI] [Google Scholar]
- 54.Laurens L. M. L., Olstad J. L., Templeton D. W. Methods in Molecular Biology. Totowa, NJ: Humana Press; 2018. Total Protein Content Determination of Microalgal Biomass by Elemental Nitrogen Analysis and a Dedicated Nitrogen-to-Protein Conversion Factor. [DOI] [PubMed] [Google Scholar]
- 55.FALCON Assembler — FALCON 0.5 documentation. https://pb-falcon.readthedocs.io/en/latest/. (Accessed 25 April 2019).
- 56.Hoff, K. J., Lange, S., Lomsadze, A., Borodovsky, M. & Stanke, M. BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMark-ET and AUGUSTUS. Bioinformatics32, 767–769 (2016). [DOI] [PMC free article] [PubMed]
- 57.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 58.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinforma. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jones P, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.UniProt Consortium, T. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2018;46:2699. doi: 10.1093/nar/gky092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Osterhout WJV. Extreme toxicity of sodium chloride and its prevention by other salts. J. Biol. Chem. 1906;1:363–369. [Google Scholar]
- 62.HTStream. https://github.com/ibest/HTStream.
- 63.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.RFFSC R Development Core Team. R: a language and environment for statistical computing. 409 (2011).
- 65.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Delcher AL, et al. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gonzalez-Esquer, C. R., Twary, S. N., Hovde, B. T. & Starkenburg, S. R. Nuclear, chloroplast, and mitochondrial genome sequences of the prospective microalgal biofuel strain Picochlorum soloecismus. Genome Announc. 6, e01498-17 (2018). [DOI] [PMC free article] [PubMed]
- 68.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 69.Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pena, L. et al. Transgenic Plants Methods and Protocols. 10.13387/j.cnki.nmld.2013.02.001 (2005).
- 71.Smith, F. D., Harpending, P. R. & Sanford, J. C. Biolistic transformation of prokaryotes: factors that affect biolistic transformation of very small cells. J. General Microbiol.138, 239–248 (2019). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary items
Data Availability Statement
The algal strain (Picochlorum renovo), DNA elements, and raw data supporting the conclusions of this manuscript will be made available by the authors, without undue reservation, to any qualified researcher. The genome for Picochlorum renovo is publicly available at https://greenhouse.lanl.gov/greenhouse/organisms. Genomic sequence data can also be accessed at NCBI, Project Number PRJNA558990. The raw data supporting the conclusions of salinity transcriptomics is available from the Sequence Read Archive, under Project Number PRJNA553204.