Summary
FTO, a mRNA N6-methyladenosine (m6A) demethylase, was reported to promote leukemogenesis. Using structure-based rational design, we have developed two promising FTO inhibitors, namely FB23 and FB23-2, which directly bind to FTO and selectively inhibit FTO’s m6A demethylase activity. Mimicking FTO depletion, FB23-2 dramatically suppresses proliferation and promotes the differentiation/apoptosis of human acute myeloid leukemia (AML) cell line cells and primary blast AML cells in vitro. Moreover, FB23-2 significantly inhibits the progression of human AML cell lines and primary cells in xenotransplanted mice. Collectively, our data suggest that FTO is a druggable target and that targeting FTO by small-molecule inhibitors holds potential to treat AML.
Introduction
RNA epitranscriptomics represents a recently identified layer of regulation of genetic information (He, 2010; Roundtree et al., 2017a). N6-methyladenosine (m6A), discovered in the mid-1970s (Wei et al., 1975; Wei et al., 1976), is the most abundant internal modification in eukaryotic mRNA and noncoding RNAs (Esteller and Pandolfi, 2017). The discovery of the fat-mass and obesity-associated protein (FTO) as a m6A demethylase corroborates the m6A modification as a dynamic process (Jia et al., 2011). The m6A is enriched near stop codons, 3’ untranslated regions (Dominissini et al., 2012; Meyer et al., 2012), as well as coding regions (Rana and Tuck, 1990), which is critical for the regulation of mRNA stability (Huang et al., 2018; Wang et al., 2014a; Zhao et al., 2014), splicing (Xiao et al., 2016; Zhao et al., 2014), transport (Tuck, 1992), translation (Meyer et al., 2015; Wang et al., 2015b), primary microRNA processing (Alarcon et al., 2015), and protein-RNA interactions (Liu et al., 2015). Precise regulation of the m6A modification on RNAs plays important roles in various biological and pathological processes (Fustin et al., 2013; Geula et al., 2015; Lichinchi et al., 2016; Wang et al., 2014b; Xiang et al., 2017; Zheng et al., 2013; Zhou et al., 2015). The m6A levels depend on the functional interplay among several proteins. METTL3 and METTL14, the m6A methyltransferases, form a heterodimer with the support of cofactors to induce m6A methylation (Bokar et al., 1997; Feng et al., 2014; Knuckles et al., 2018; Ping et al., 2014; Wang et al., 2014b). The two demethylases FTO and ALKBH5, which belong to the Fe2+- and 2-oxoglutarate (2OG)-dependent AlkB dioxygenase family, predominantly catalyze m6A demethylation (Fu et al., 2013; Jia et al., 2011; Zheng et al., 2013), although FTO was initially identified as a demethylase of N3-methylthymidine and N3-methyluridine in vitro (Gerken et al., 2007; Jia et al., 2008). N6,2’-O-dimethyladenosine (m6Am) was recently discovered to be another substrate of FTO (Mauer et al., 2017). Importantly, m6Am is part of the 5’ cap. The m6A reader proteins, YTHDF1/2/3, YTHDC1/2, and IGF2BP1/2/3, preferentially recognize m6A in mRNA (Dominissini et al., 2012; Hsu et al., 2017; Huang et al., 2018; Li et al., 2017a; Roundtree et al., 2017b; Shi et al., 2017; Wang et al., 2014a; Xiao et al., 2016).
The discovery of small-molecule inhibitors of FTO enabled temporal intervention of mRNA methylation. Rhein, an FTO inhibitor, globally increases the cellular m6A on mRNA (Chen et al., 2012). The structural complexes of rhein bound to either FTO (Aik et al., 2013) or AlkB (Li et al., 2016) revealed that the molecule uses different mechanisms to inhibit AlkB enzymes (Rose et al., 2011). Meclofenamic acid (MA), an anti-inflammatory drug, was repositioned to increase m6A in cells through the inhibition of FTO rather than ALKBH5 (Huang et al., 2015). Inspired by the structural similarity to MA, fluorescein was shown to simultaneously inhibit and label FTO (Wang et al., 2015a). Selective inhibition of FTO was also achieved by a compound that contains the tethered nucleotide mimic with the 2OG-binding component (Toh et al., 2015). Recent developments of FTO inhibitors revealed nonconserved small-molecule binding sites in the FTO protein (He et al., 2015; Padariya and Kalathiya, 2016). In addition, a high-throughput screening was developed on the basis of FTO-demethylating fluorescent RNA aptamers (Svensen and Jaffrey, 2016). Cellular activity of these inhibitors is still limited, however.
The critical impact of the dysfunction of FTO demethylation has been associated with human diseases, especially in cancer (Deng et al., 2018a; Deng et al., 2018b). Therefore, several studies have been dedicated to the application of FTO inhibitors in a variety of m6A-related diseases. The ethyl ester of MA has been shown to suppress glioblastoma stem cell (GSC)-initiated tumorigenesis (Cui et al., 2017). Rhein or MA could be uniquely positioned to either prevent or override tyrosine kinase inhibitor resistance by inhibiting FTO demethylation on a subset of mRNA (Yan et al., 2018). Recently, R-enantiomer of 2-hydroxyglutarate (R-2HG) was demonstrated to be active for anti-proliferation of AML cells, mainly by targeting FTO (Su et al., 2018). Moreover, FTO inhibitors were demonstrated to change the distribution of adipose tissue and bone mineral density (McMurray et al., 2015), show anticonvulsant activity in a mouse model (Zheng et al., 2014), and decrease local translation of axonal GAP-43 mRNA (Yu et al., 2018). The action modes of these FTO inhibitors have not been adequately understood, however, and none seem clinically applicable due to their relatively poor activity and/or selectivity. Thus, there is an unmet need to develop selective and more effective inhibitors of FTO demethylation for therapeutic applications.
Results
Development of a selective and potent FTO inhibitor (FB23)
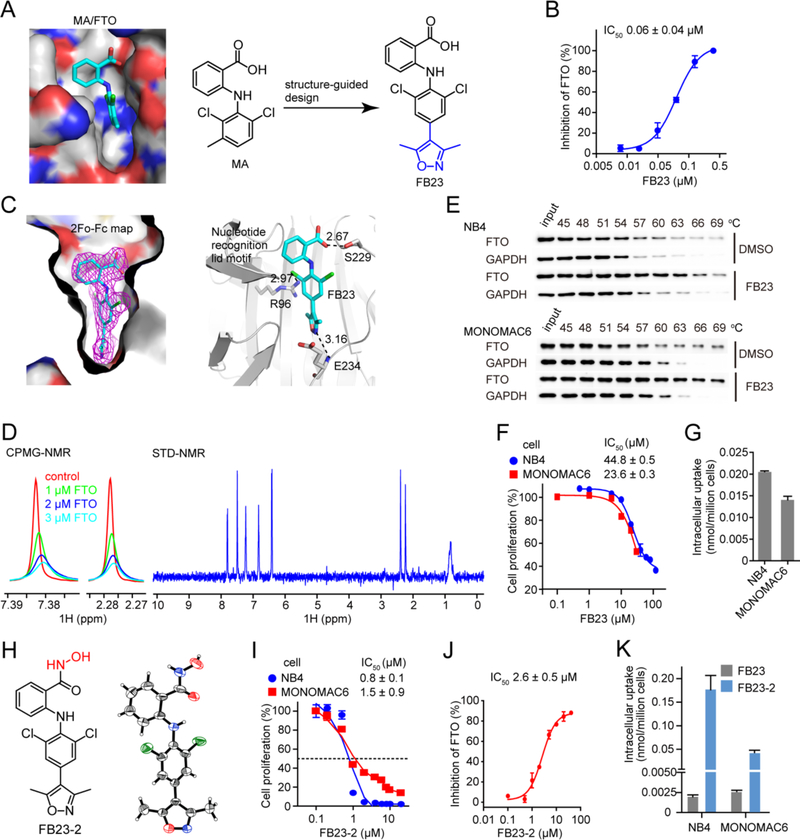
Previously, we identified MA as an inhibitor of FTO demethylation over ALKBH5 (Huang et al., 2015). The structural complex of FTO/MA clearly elucidated the principles underlying MA’s selectivity, facilitating a structure-guided design for potent FTO inhibitors. To this end, we applied the following two principles: i) to keep the benzyl carboxylic acid as the key element of MA contributing selectivity for FTO over ALKBH5, and ii) to extend the dichloride-substituted benzene to a deeper pocket that could be fully occupied by a bulky ligand (Figures 1A and S1A). Synthesis of the inhibitors involves introducing a 5-membered heterocyclic ring to MA through cross-coupling chemistry (Figure S1B). Among them, FB23 is much more potent than MA in inhibiting FTO-mediated demethylation, with an IC50 of 0.06 μM (Figure 1B) and thus a 140-fold increase over that of MA (Huang et al., 2015).
Figure 1. Design and characteristic profiling of FTO inhibitors.
(A) Structure-guided design of inhibitor FB23. The MA binding pocket in FTO is shown, and MA is colored in cyan.
(B) Effect of FB23 on FTO demethylation of m6A in RNA in vitro using HPLC quantification.
(C) Structural complex of FTO bound with FB23. 2Fo-Fc density map contoured to 1.0 sigma was shown in magenta. Dark dotted lines indicate hydrogen bonding, and the distance in Å is labelled.
(D) NMR measurement of FB23 interacting with FTO. The CPMG-NMR spectra are recorded for FB23 without FTO (red), and with FTO at 1.0 μM (green), 2.0 μM (blue), and 3.0 μM (cyan), respectively. The STD-NMR spectrum for FB23 is recorded with 5 μM FTO.
(E) Representative western blots for the effects of 50 μM FB23 on thermal stabilization of FTO protein. CETSA was assayed in cell lysates. The results are derived from three biological replicates.
(F) Effect of FB23 treatment of 72 hr on proliferation of AML cells.
(G) Determination of cellular uptake of FB23 by LC-MS/MS quantitation. AML cells were treated with 10 μM FB23 for 24 hr.
(H) Structure of FB23-2. Its absolute configuration was determined by X-ray.
(I) Effect of FB23-2 treatment of 72 hr on proliferation of AML cells.
(J) Inhibition of FB23-2 on FTO demethylation of m6A in RNA in vitro using HPLC quantification.
(K) Determination of cellular uptake of FB23-2 by LC-MS/MS quantitation. FB23, the hydrolysate of FB23-2 was also detected. AML cells were treated with 10 μM FB23-2 for 24 hr.
Error bars, mean ± SD, n = 3. See also Figure S1 and Table S1.
To validate the direct binding of FB23 to FTO, we established co-crystal structure of FB23 bound with the FTO protein. The crystal structure was solved by molecular replacement and refined to 2.20 Å resolution (Table S1). The superimposition of structural complexes of FTO bound with dm3T ligand or inhibitor revealed no gross differences in overall protein folding (Figure S1C). The 2Fo-Fc density map contoured to 1.0 sigma (Figure 1C), and the simulated annealing Fo-Fc OMIT density map contoured to 3.0 sigma (Figure S1C), demonstrating that FB23 showed an extraordinary shape complementary with the substrate-binding site, occupying the entire binding pocket. Similar to interactions observed in the FTO/MA complex, the phenyl ring in FB23 bearing carboxyl acid substituent forms hydrophobic interactions with the nucleotide recognition lid, thereby ruling out nonspecific binding to either RNA demethylase ALKBH5 or DNA repair enzymes ALKBH2 and ALKBH3. Hydrogen bonding occurs between the carboxyl group in FB23 and the side chain from the Ser229 residue of FTO directly. In FB23 one chlorine atom directly contacts the guanidinium group in Arg96 of FTO. In addition, extra hydrogen bonding was observed between nitrogen or oxygen in the extended heterocyclic ring of FB23 and the amide backbone of Glu234 of FTO, which likely allows the inhibitor FB23 to show enhanced inhibitory activity on FTO compared to MA. Collectively, the FTO/FB23 structure revealed that FB23 possesses specificity for and improved inhibition of FTO.
We further investigated the interaction between FTO and FB23. Dose-dependent attenuation of signals was observed in Carr-Purcell-Meiboom-Gill (CPMG) Nuclear Magnetic Resonance (NMR) titrations (Figures 1D and S1D), and positive saturation transfer difference (STD) signals were also detected (Figure 1D), which indicates that FTO interferes with the state of FB23. We also performed a Cellular Thermal Shift Assay (CETSA) to further validate their interactions in cellular conditions (Martinez Molina et al., 2013). As expected, the presence of FB23 induced an obvious thermal shift of the FTO protein in NB4 and MONOMAC6 AML cells (Figure 1E). Thus, the NMR titration and CETSA assays further demonstrate that FB23 is a direct FTO inhibitor.
FB23 exhibits moderate anti-proliferation effects and its derivative (FB23-2) shows significantly improved activity
We next sought to examine the anti-proliferative effect of FB23 on AML cells. However, FB23 only moderately inhibited the proliferation of NB4 and MONOMAC6 cells, with an IC50 of 44.8 μM and 23.6 μM, respectively (Figure 1F). As detected by LC-MS/MS analysis, we found that the intracellular concentration of FB23 is a mere 0.02 nmol/million in NB4 cells and 0.015 nmol/million in MONOMAC6 cells (Figure 1G). Thus, the limited inhibitory effect of FB23 on AML cell proliferation is likely due to the low cellular uptake of FB23.
The structure of the FTO/FB23 complex suggests that the optimization on the carboxylic acid of FB23 would not disturb the affinity and specificity for FTO. To improve the permeability of FB23, we synthesized derivatives of the benzyl carboxylic acid on the basis of the bioisosterism principle. The benzohydroxamic acid, termed as FB23-2 (Figures 1H and S1B), displays significantly improved anti-proliferative activity on NB4 and MONOMAC6 cells with an IC50 of 0.8 – 1.5 μM (Figure 1I), and maintains inhibitory activity on FTO demethylation in vitro (Figure 1J). To establish the absolute configuration, we determined the X-ray crystal structure of FB23-2, which unambiguously shows an intramolecular hydrogen bond between the amino hydrogen and the carbonyl of hydroxamic acid (Figure 1H, right panel). In addition, we analyzed the relative configuration of FB23-2 in solution using the Nuclear Overhauser Effect (NOE), which is a transfer of nuclear spin polarization through space, rather than chemical bonds. The strong NOE correlation between H-1 and H-10 in the NOESY spectrum also supports the intramolecular hydrogen bonding (Figure S1E). With this evidence in hand, the docking of FB23-2 to FTO resulted in an excellent fit of FB23-2 in a position of perfect overlap to the crystallographically determined binding mode of inhibitor FB23 bound to FTO (Figure S1F). Next, we detected the cellular uptake of FB23-2 by LC-MS/MS quantitation (Figure 1K). Of note, FB23-2 was detected around 0.05–0.2 nmol/million cells in MONOMAC6 and NB4 cells, which is several folds higher than the cellular uptake of FB23 (see Figure 1G). Meanwhile, FB23 was also detected in tiny amounts in the FB23-2 treated AML cells, which is likely a hydrolysis product of FB23-2. The increased intracellular concentration of FB23-2 likely contributes to its improved anti-proliferation effect in AML cells.
FB23-2 increases RNA methylation in a panel of AML cells
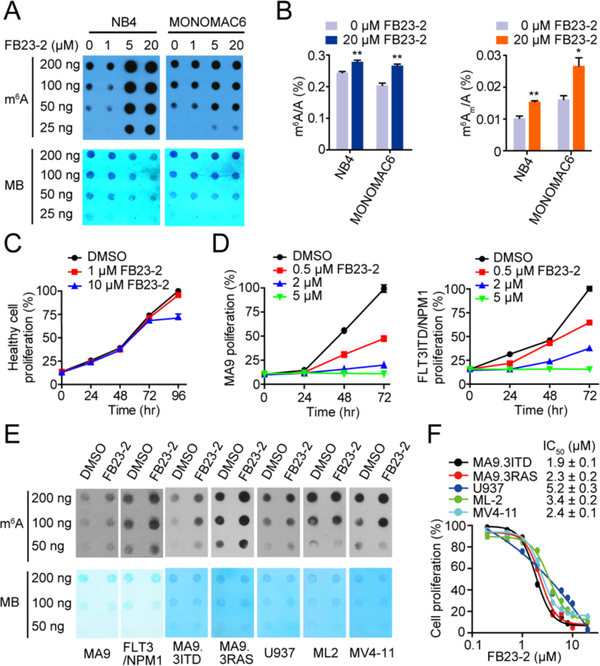
We checked the changes of m6A on RNA when AML cells were treated by the FTO inhibitor. The treatment of NB4 and MONOMAC6 cells with FB23-2 resulted in a substantial increase of m6A abundance in transcriptomes as detected by m6A dot blot assay (Figure 2A). LC-MS/MS quantitation further confirmed the increase of cellular m6A in mRNA of AML cells after exposure to FB23-2 (Figure 2B). The m6Am is another substrate of FTO, and we observed similar increases of m6Am abundance in AML cells exposed to FB23-2 (Figure 2B). The overall level of m6Am is much lower than that of m6A, however. Since recent reports independently showed that FTO minimally affected the expression levels of mRNA starting with m6Am (Akichika et al., 2019; Sun et al., 2019; Wei et al., 2018), it is likely that m6A, rather than m6Am, is the main substrate of FTO in AML cells (Su et al., 2018).
Figure 2. FB23-2 displays anti-proliferation effect via upregulating global m6A levels.
(A) Determination of m6A abundance in mRNA in NB4 and MONOMAC6 cells upon FB23-2 treatment for 72 hr via dot blot assay. MB (Methylene Blue) represents loading control of RNA samples. The results are derived from two biological replicates.
(B) Quantitation of the percentage of m6A/A and m6Am/A ratios in mRNA by LC-MS/MS in NB4 and MONOMAC6 cells treated with 20 μM FB23-2 for 72 hr.
(C) Effect of FB23-2 on proliferation of human normal BM cells isolated from a healthy donor.
(D) Effect of FB23-2 on proliferation in MA9 and FLT3ITD/NPM1 mouse BM cells.
(E) Determination of m6A abundance in MA9 and FLT3/NPM1 primary cells isolated from AML mice and in the five human AML cell lines upon 5 μM FB23-2 treatment for 72 hr by dot blot assay.
(F) Effect of FB23-2 treatment of 96 hr on proliferation of a panel of AML cell lines with different genetic backgrounds and molecular mutations.
*, p < 0.05; **, p < 0.01; unpaired Student’s t-test. Error bar, mean ± SD, n = 3.
FB23-2 minimally altered proliferation of human normal bone marrow (BM) cells isolated from a healthy donor (Figure 2C). Consistent with the effect of FTO KD on the proliferation of MLL-AF9 (MA9) and FLT3ITD/NPM1 AML murine cells (Li et al., 2017b), FB23-2 significantly suppressed the proliferation of BM cells from these two models in a dose-dependent manner (Figure 2D), accompanied with increased m6A abundance (Figure 2E). Moreover, we determined the anti-leukemia effects of FB23-2 in a panel of additional AML cell lines with different genetic backgrounds, including MA9.3ITD (with MLL-AF9 and FLT3ITD mutation), MA9.3RAS (with MLL-AF9 and NRAS mutations), U937 (with t(10;11) translocation), ML-2 (with t(6;11) translocation), and MV4-11 (with t(4;11) translocation). As expected, FB23-2 efficiently inhibited the proliferation of these AML cell lines with IC50 ranging from 1.9 μM to 5.2 μM, and increased m6A abundance in these cell lines as well (Figures 2E and 2F). Together, these results demonstrate that FB23-2 exhibits FTO inhibition and anti-leukemia effects broadly.
FB23-2 displays a high selectivity toward FTO
We performed the selectivity profile of FTO inhibitors. Similar to MA, neither FB23 nor FB23-2 inhibits ALKBH5 demethylation in vitro (Figure S2A). As shown by Drug Affinity Responsive Target Stability (DARTS) assay (Lomenick et al., 2009), FB23-2 could not directly bind to ALKBH5 in AML cell lysates (Figure S2B), and showed only marginal effects on the transcription level and protein stability of ALKBH5 (Figures S2C and S2D). In addition, we checked the inhibitory effects of FTO inhibitors on epigenetic targets involved in AML and/or other cancers, including the Histone deacetylases (HDAC), Disruptor of Telomere Silencing 1-like (DOTL1), Bromodomain-containing “reader” proteins (BRD), Lysine-specific demethylase 1 (LSD1), and Jumonji domain-containing histone demethylases (Shortt et al., 2017). FB23 and FB23-2 slightly attenuated the activities of these targets in vitro, while the positive control inhibitors display significant activities (Table S2). Similarly, 20 μM FB23-2 minimally inhibited TET1 protein in vitro (Figure S2E) and did not alter the abundance of 5mC or 5hmC in AML cells (Figure S2F). The major histone methylations in NB4 cells were unaltered by FB23-2 (Figure S2G). In addition, we performed a much broader enzymatic specificity test of FB23-2. The inhibitory effect of 10 μM FB23-2 on activities of 405 human kinases was mapped onto the kinome phylogenetic tree (Figure S2H). Kinases with more than 40% of inhibition were further evaluated. Six kinases were inhibited by FB23-2 with IC50 around 3.0 – 13.4 μM. The inhibitory efficiency of FB23-2 on these kinases is much lower than that of well-established kinase inhibitors. FB23-2 also barely inhibited the oncogenic proteases (Table S3). Although MA primary inhibits both COX-1 and COX-2 to different extents (Vane et al., 1998), neither FB23 nor FB23-2 was observed to significantly inhibit cyclooxygenases even at 50 μM (Figure S2I). Taken together, these results indicate that our inhibitors display a high enzymatic selectivity for FTO.
FB23-2 exhibits FTO-dependent anti-proliferation activity and promotes myeloid differentiation and apoptosis
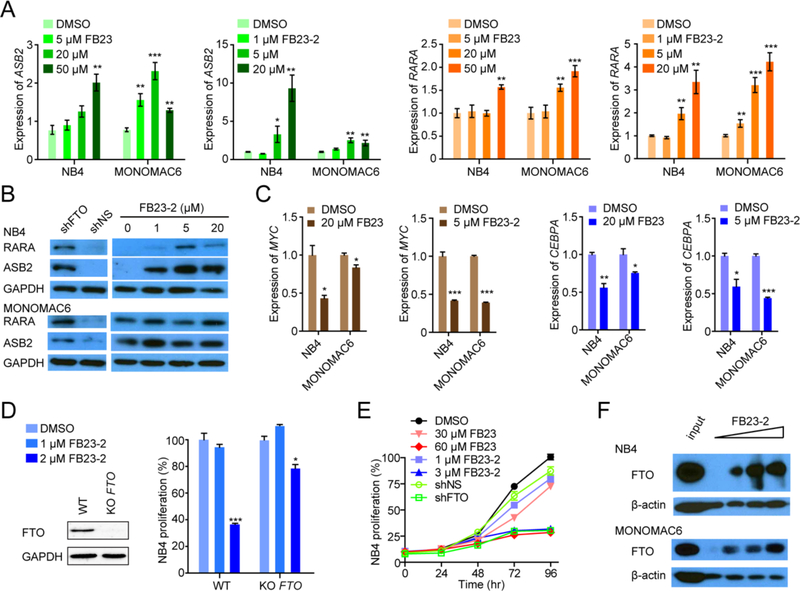
In line with the negative regulation of FTO on ASB2 and RARA expression in AML cells (Li et al., 2017b), FB23 and FB23-2 treatment significantly increased their abundance at the mRNA and protein levels in NB4 and MONOMAC6 cells (Figures 3A and 3B). FTO positively regulates MYC and CEBPA in an m6A modification-dependent manner (Su et al., 2018). Similar with shRNA-induced FTO KD, FB23 or FB23-2 indeed inhibited MYC and CEBPA expression in both NB4 and MONOMAC6 cells (Figure 3C).
Figure 3. Regulatory pathway and target engagement of FTO inhibitors.
(A) Effects of FB23 and FB23-2 treatment of 72 hr on ASB2 and RARA mRNA expression in NB4 and MONOMAC6 AML cells by RT-qPCR.
(B) Effects of FTO KD (shFTO) and FB23-2 treatment of 72 hr on RARA and ASB2 abundance in AML cells by western blot. shNS, the control shRNA. The results are derived from two biological replicates.
(C) Effects of FB23 and FB23-2 treatment of 72 hr on MYC and CEBPA mRNA expression in AML cells by RT-qPCR.
(D) Effect of FB23-2 treatment of 72 hr on proliferation of FTO KO NB4 cells. FTO abundance was measured by western blot. The percentage of each stable cell line treated with FB23-2 was normalized to that treated with DMSO.
(E) Effects of FTO inhibitors and shFTO on proliferation of NB4 cells.
(F) Representative DARTS results for FTO levels by western blot. AML cell lysates with 50 μM, 200 μM, and 500 μM FB23-2 were incubated for 1 hr at room temperature before pronase digestion. The results are derived from three biological replicates.
*, p < 0.05; **, p < 0.01; ***, p < 0.001; unpaired Student’s t-test. Error bar, mean ± SD, n = 3. See also Figure S2 and Table S2–S3.
In order to further determine whether the inhibitory effect of FB23-2 on AML cell proliferation relies on FTO, we generated stable FTO KO NB4 AML cells using CRISPR-Cas9. FB23-2 dramatically suppressed proliferation of AML cells but exhibited a much milder effect on AML cells with stable FTO KO (Figure 3D), suggesting that the inhibitory effect of FB23-2 on the proliferation of AML cells depends on the suppression of an activated FTO signaling. Consistently, we found that the FTO KD and 60 μM FB23 or 3 μM FB23-2 had comparable effects in NB4 cells (Figure 3E). To verify the direct interaction between FB23-2 and FTO, we performed a DARTS assay. As expected, the FTO protein becomes protease-resistant in the presence of FB23-2 (Figure 3F), indicating that FB23-2 indeed binds to FTO in cell lysates.
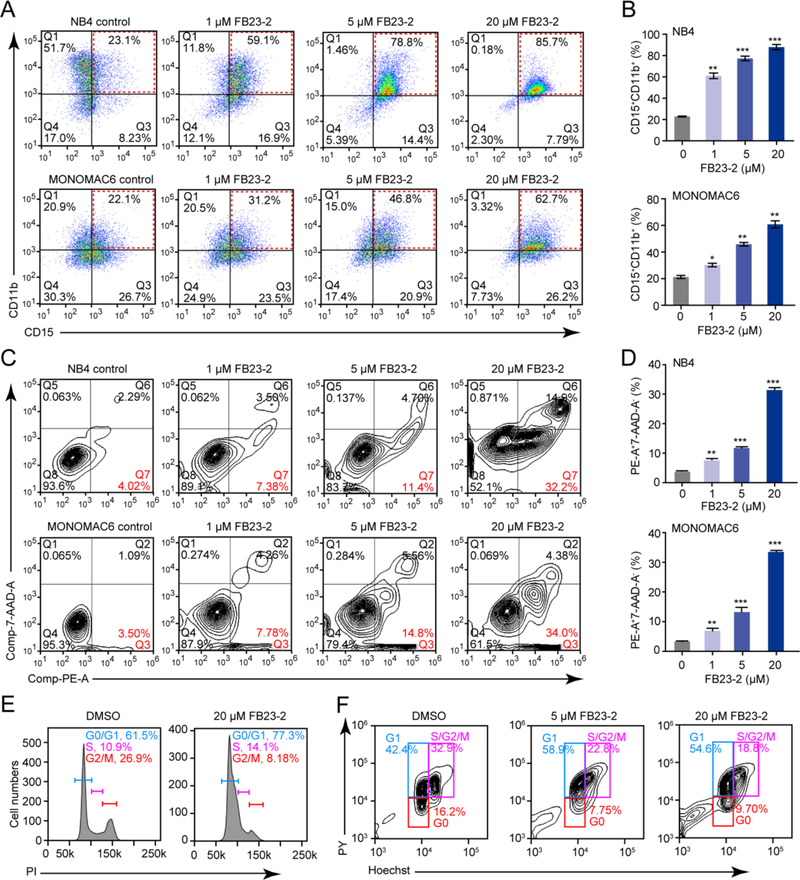
We further characterized the effects of FB23-2 on AML cells. In line with the inhibitory effects of FTO on myeloid differentiation and apoptosis in AML cells (Li et al., 2017b), FB23-2 substantially accelerated all-trans retinoic acid (ATRA)-induced myeloid differentiation in NB4 and MONOMAC6 cells in a dose-dependent manner (Figures 4A and 4B). Furthermore, FB23-2 induced apoptosis (Figures 4C and 4D) and cell cycle arrest at G1 stage in AML cells (Figures 4E and 4F). Collectively, these results suggest that FB23-2 exhibits FTO-dependent activity in AML cells.
Figure 4. The impact of FB23-2 on AML cell differentiation, apoptosis, and cycle arrest in vitro.
(A and B) The effect of FB23-2 on ATRA-induced myeloid differentiation in AML cells was analysed by FACS (A) and the percentage of CD15+CD11b+ cells was quantified (B). NB4 cells were treated with 200 nM ATRA, while MONOMAC6 cells were treated with 1 μM ATRA for 48 hr.
(C and D) The effect of FB23-2 on cell apoptosis was analysed by FACS (C) and the percentage of cells positive for Annexin V and 7-AAD staining was quantified (D). NB4 cells were treated for 48 hr, while MONOMAC6 cells for 72 hr.
(E and F) Determination of the effect of FB23-2 on cell cycle arrest by FACS based on PI staining (E) or Hoechst/Pyronin Y staining (F) in MONOMAC6 cells after 24 hr of treatment.
*, p < 0.05; **, p < 0.01; ***, p < 0.001; unpaired Student’s t-test. Error bars, mean ± SD, n = 3.
FB23 and FB23-2 target similar signaling pathways to FTO KD in AML cells
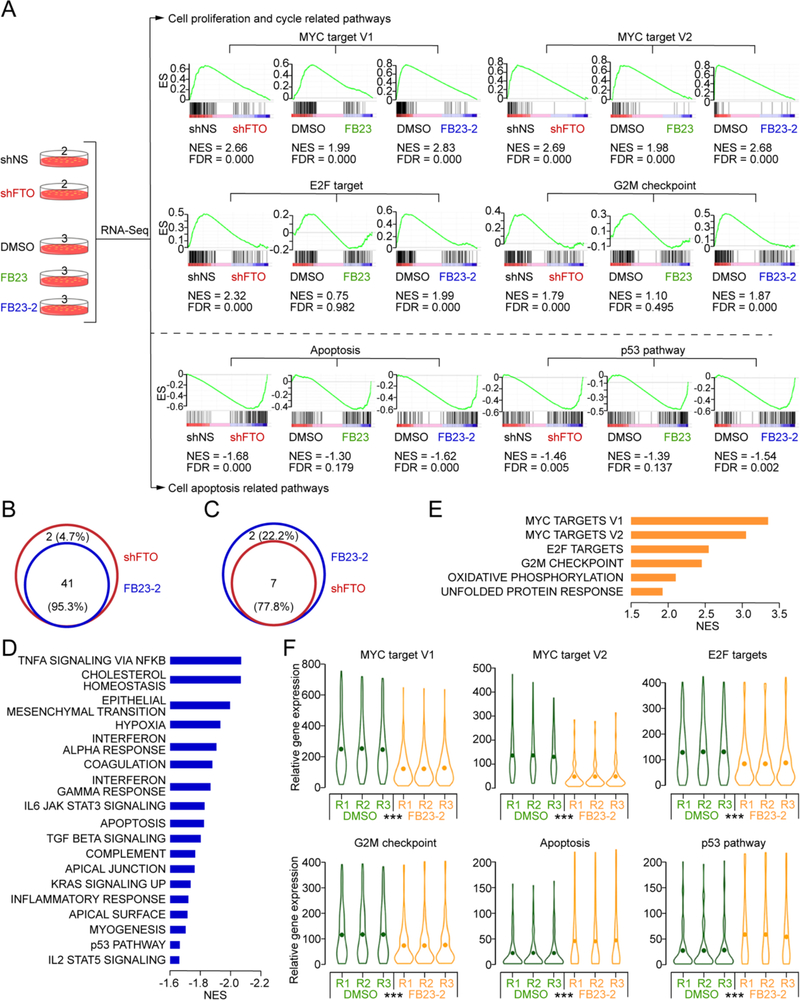
To investigate which genes and signaling pathways are responsible for the anti-leukemic function of FTO inhibitors, we performed transcriptome-wide RNA-sequencing (RNA-seq) analysis of FTO KD, FB23 treated, or FB23-2 treated NB4 AML cells as well as control cells. Via independent analysis of three different comparisons, we found that FTO KD, FB23 treatment, and FB23-2 treatment all caused the significant suppression of MYC targets, E2F targets, and G2M checkpoint signal cascades, which may contribute to the inhibitory effects of FTO inhibitors and FTO KD on cell cycle and proliferation (Figures 5A and S3A–S3D). In addition, all three treatments consistently activated apoptosis and p53 pathways (Figure 5A). Global gene set enrichment analysis (GSEA) indicated that FTO KD and FB23 or FB23-2 treatment display similar effects on regulating a set of functionally important signaling pathways (Figures S3E–S3G). Notably, the vast majority of pathways (41 out of 43, 95.3%) increased by FTO KD could also be enriched by FB23-2 (Figures 5B and S3H); similarly, the majority of signaling pathways suppressed by FB23-2 are also inhibited by FTO KD (Figures 5C and S3H). These results strongly suggest that FTO inhibitors, especially FB23-2, had the same effect on critical signalling pathways that control cell cycle, cell proliferation, and cell survival in AML cells as FTO KD.
Figure 5. Transcriptome-wide RNA-seq assays to identify potential targets of FTO inhibitors in AML cells.
(A) Transcriptome strategy of RNA-seq conducted on NB4 cells exposed to 5 μM inhibitor for 48 hr. shFTO and shNS groups contain two biological replicates; FB23, FB23-2, and DMSO groups contain three replicates. Gene set enrichment analysis (GSEA) was used to analyse the signaling pathways enrichment in different groups. Normalized enrichment score (NES) indicated the analysis results across gene sets. False discovery rate (FDR) presented if a set was significantly enriched. ES, enrichment score.
(B) Venn diagram of the shared pathways among the increased signaling pathways in FTO KD and FB23-2 treated NB4 cells.
(C) Venn diagram of the shared pathways among the decreased signaling pathways in FTO KD and FB23-2 treated NB4 cells.
(D and E) The core enriched signaling pathways, including increased (D) and decreased (E), in FTO inhibited (shFTO + FB23 + FB23-2) cells compared to control (shNS + DMSO). The NES values of the pathways with p < 0.001 are presented.
(F) Violin plots showing the relative abundance of genes involved in the MYC pathway, G2M checkpoint, E2F targets, apoptosis, and p53 pathway in DMSO and FB23-2 treated NB4 cells. ***, p < 0.001; paired t-test.
Furthermore, we considered shFTO, FB23, and FB23-2 as a single group and re-analyzed the sequencing data between this group and the control group (including shNS group and DMSO group). Consistently, FTO KD and inhibition stimulated apoptosis and p53 pathway (Figures 5D and S3I, and Table S4); meanwhile, MYC targets, G2M checkpoint, and E2F targets were repressed (Figures 5E and S3I, and Table S4). FB23-2 treatment dramatically down-regulated genes enriched in MYC target V1, MYC target V2, E2F targets, and the G2M checkpoint signatures; meanwhile, genes enriched in apoptosis and the p53 pathway signatures were downregulated in AML cells (Figure 5F).
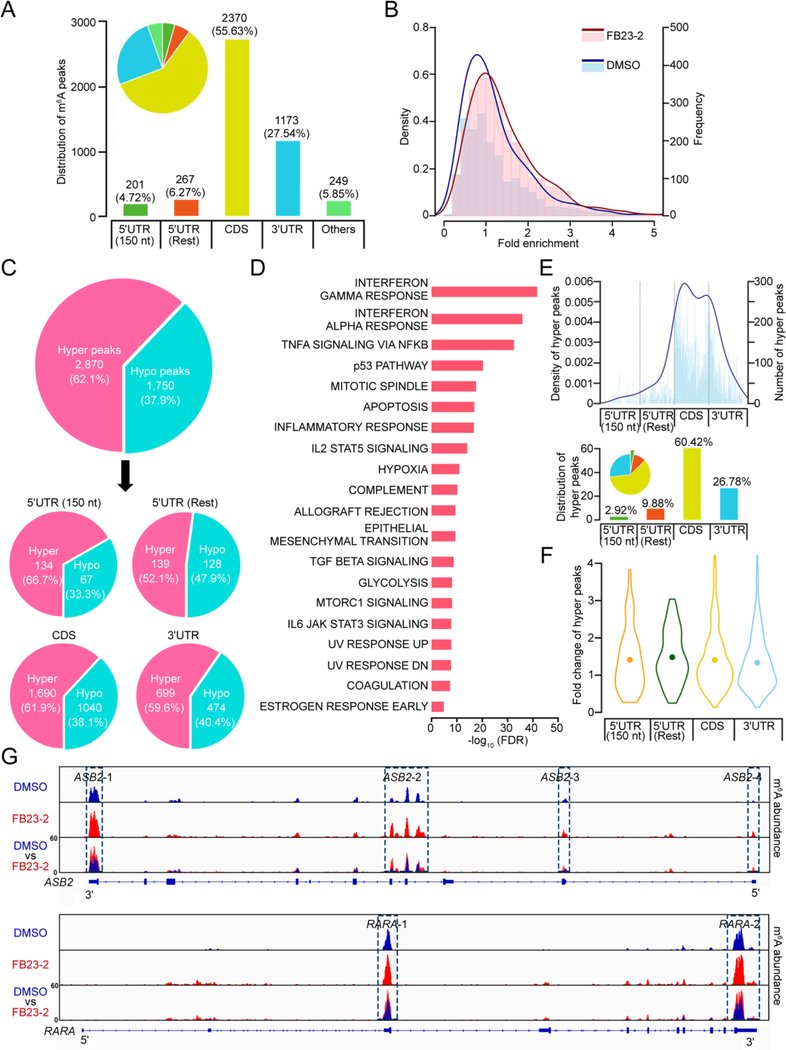
To determine the effects of FB23-2 on global m6A modification and to confirm its inhibition on FTO enzyme, we carried out m6A-seq with mRNA samples isolated from MONOMAC6 AML cells upon DMSO or FB23-2 treatment. The data showed that the vast majority of m6A peaks are distributed in the CDS (55.63%) and 3’UTR (27.54%) regions (Figure 6A). Consistent with m6A dot blot and LC-MS/MS quantitation assays (see Figures 2A and 2B), FB23-2 treatment induced an obvious increase of m6A peaks (Figure 6B). Indeed, amongst the m6A peaks identified from both DMSO and FB23-2 treated groups, the majority (62.1%) are increased upon FB23-2 treatment (Figure 6C). Interestingly, we found that genes with hyper peaks are also enriched in the p53 pathway and apoptosis-signaling pathway (Figures 6D, S4A, and S4B), suggesting that FB23-2 activates p53 and apoptosis pathways (see Figure 5) likely through an m6A-depedent mechanism. We also determined the potential effect of FB23-2 on cap m6Am modification. We observed that a mere 2.92% of hyper peaks were located in the 5’ end of 5’UTR (covering the 5’ cap m6Am site) and we did not observe any further significant fold changes of these hyper peaks in MONOMAC6 AML cells (Figures 6E and 6F), which is consistent with reported observations in AML cells with FTO KD or R-2HG treatment (Su et al., 2018). Such data suggests that FTO-mediated demethylation of m6Am may play a minimal role in the FB23-2-induced inhibition of cell proliferation and activation of apoptosis in AML cells.
Figure 6. Transcriptome-wide m6A-seq assays to confirm the effects of FB23-2 in AML cells.
(A) Distribution of m6A peaks in different regions of mRNA as detected in m6A-seq assays conducted on MONOMAC6 cells upon treatment with DMSO or 5 μM FB23-2 for 72 hr. 5’UTR (150 nt) represents the first 150 nt of 5’ end of 5’UTR, while 5’UTR (Rest) represents the remaining regions of 5’ end of 5’UTR.
(B) The density (line) and frequency (histogram) of m6A peaks from m6A-seq assays conducted in FB23-2 treated (red) and DMSO treated (blue) MONOMAC6 cells.
(C) Distribution of FB23-2-increased m6A peaks (termed hyper peaks) and FB23-2-decreased m6A peaks (termed hypo peaks) from m6A-seq assays conducted in FB23-2 and DMSO treated MONOMAC6 cells. The peaks which were significantly (p < 0.001) altered in FB23-2 group compared with DMSO group are presented.
(D) GSEA analysis of the genes with increased m6A abundance upon FB23-2 treatment identified by m6A-seq in MONOMAC6 cells.
(E) The adjusted density (line, top) and distribution (histogram, bottom) of hyper peaks from (C) across different mRNA regions.
(F) Fold changes of hyper peaks from (C) in different regions of mRNAs.
(G) The m6A abundance in ASB2 and RARA transcripts. The m6A peaks were called by exomePeak.
See also Figure S4.
Finally, we analyzed the m6A modification on RARA and ASB2 mRNAs upon FB23-2 treatment in MONOMAC6 cells and observed an obvious increase of m6A abundance (Figures 6G, S4C, and S4D). This data suggests that the FB23-2/FTO axis-induced upregulation of RARA and ASB2 and downregulation of MYC and CEBPA might depend on an increased abundance of m6A on mRNA. The effects of FB23-2 inhibition of FTO on regulation of its target mRNAs likely relies on m6A modification rather than m6Am as none of them belong to m6Am-initiated mRNAs (Mauer et al., 2017). Indeed, FTO mainly affects the expression levels of mRNA containing internal m6A rather than mRNA starting with m6Am (Akichika et al., 2019). These results also support our hypothesis that regulation of m6A acts as the major effector of FB23-2 in AML cells.
FB23-2 is safe in mice and displays a favorable pharmacokinetic profile
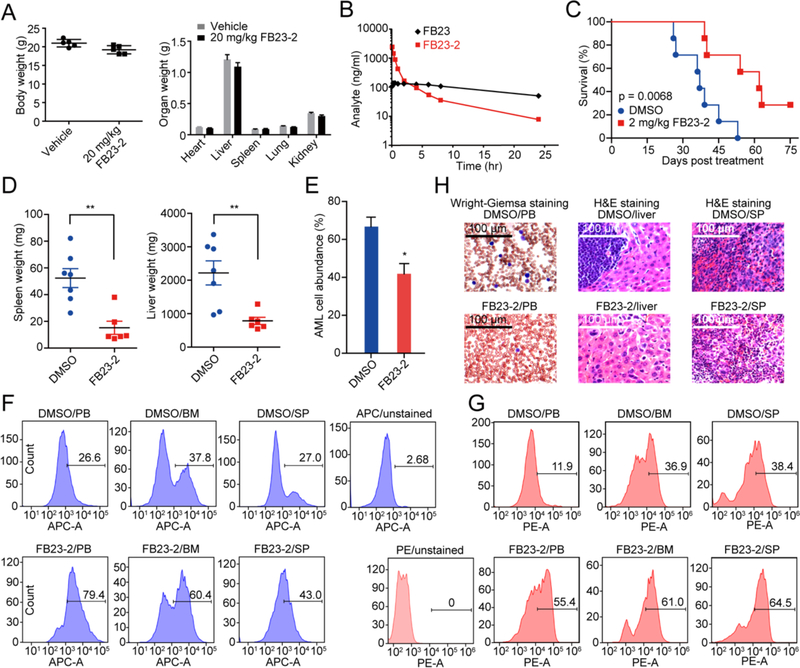
To determine if FB23-2 is safe for in vivo treatment, we examined the toxic effects of multi-doses of FB23-2 in BALB/c mice over a two-week time frame. The BALB/c mice (n = 5) were treated by way of intraperitoneal injection (i.p.) daily with 10, 20, 40, and 80 mg/kg FB23-2 respectively, for 14 days. Under a dosing scheme of 20 mg/kg FB23-2, we observed no evidence of body weight loss (Figures 7A and S5A); nor was any physical damage observed on different organs (Figures 7A, S5A, and S5B). Blood was collected, and further hematology and plasma biochemistry analysis showed that no significant difference was observed in hematopoiesis among the vehicle control and the 20 mg/kg inhibitor-treated mice (Tables S5 and S6). These data indicate that FB23-2 in a dosage of 20 mg/kg is safe for exploring in vivo efficacy.
Figure 7. FB23-2 delays leukemogenesis in vivo.
(A) The weight of body and organs of female BALB/c mice (n = 5) treated with vehicle or 20 mg/kg FB23-2 daily for 14 days. The weight was recorded at day 15.
(B) Pharmacokinetics of FB23-2. The concentration of FB23-2 and FB23 in the serum was quantitated by LC-MS/MS after i.p. administration of 3 mg/kg to rat.
(C) Kaplan-Meier survival curves of MONOMAC6 xeno-transplanted mice (n = 7 for each group) after vehicle or FB23-2 treatment. The p value was calculated with the log-rank test.
(D) The weight of spleen and liver of mice from (C) at the endpoint.
(E) The percentage of human AML cells in the PB of mice from (C) at the endpoint was analysed by FACS.
(F-G) FACS analysis of the distribution of human AML cells in PB, BM, and spleen (SP) of mice from (C) at the endpoint and stained with anti-human CD15 (F) and anti-human CD11b (G).
(H) Staining of PB, liver, and SP of mice from (C) at the endpoint.
*, p < 0.05; **, p < 0.01; unpaired Student’s t-test. Error bars, mean ± SEM. See also Figure S5 and Tables S5–S7.
Next, a single dose of 3 mg/kg FB23-2 was i.p. administrated to Sprague Dawley (SD) rats for the pharmacokinetic profile (Figure 7B and Table S7). The Cmax and Tmax value of FB23-2 were 2421.3 ± 90.9 ng/ml and 0.08 hr, respectively. FB23-2 elimination half-life, T1/2 was 6.7 ± 1.3 hr, and the AUC0–24 was 2184 ± 152 hr × ng/ml. Meanwhile, FB23 was also detected, with Cmax and Tmax as 142.5 ± 26.1 and 0.4 ± 0.1 hr, respectively. The metabolic stability of FB23-2 in the SD rat liver microsome was also determined, with an estimated T1/2 of 128 min, and an intrinsic clearance of 19.7 ml/min/kg. Lastly, we measured the degree of protein binding by FB23-2. Nearly 100% FB23-2 inhibitor was bound to plasma proteins. In summary, FB23-2 displayed a favorable pharmacokinetic profile for in vivo study.
FB23-2 suppresses leukemia progression and improves the survival of leukemic mice
We next assessed the therapeutic effects of FB23-2 in vivo with a xeno-transplantation leukemic model. NOD/LtSz-scid IL2RG-SGM3 (NSGS) mice (Wunderlich et al., 2010) were xeno-transplanted with MONOMAC6 AML cells, and 10 days post xeno-transplantation, FB23-2 (2 mg/kg) or vehicle control was intraperitoneally injected into the individual mice daily for 10 days. Notably, FB23-2 injection substantially delayed the onset of full-blown leukemic symptoms and significantly prolonged survival by almost doubling the median survival (Figure 7C). Compared with the vehicle, FTO inhibitor treatment suppressed leukemia malignancy, including reduced splenomegaly and hepatomegaly (Figure 7D). FACS analysis confirmed that FB23-2 injection suppressed the abundance of human AML cells in the recipient mice (Figures 7E and S5C). To further interpret the effect of FB23-2 on differentiation of AML cells in vivo, we collected peripheral blood (PB), BM, and spleen samples of FB23-2- and vehicle control- treated xenograft mice and stained them with anti-human CD15 and anti-human CD11b. As determined by FACS, FB23-2 treatment promoted AML cell differentiation in vivo (Figures 7F and 7G). Wright-Giemsa staining of PB smears revealed that leukemic blasts from FTO inhibitor-treated AML mice were inhibited and partially differentiated; consistently, H&E staining of spleen and liver also showed less AML cell dissemination in FB23-2-treated mice (Figure 7H). Taken together, our data suggests that pharmacological inhibition of FTO by FB23-2 substantially suppresses leukemia progression and prolongs survival.
FB23-2 exhibits therapeutic efficacy in treating a patient-derived xeno-transplantation (PDX) AML mouse model
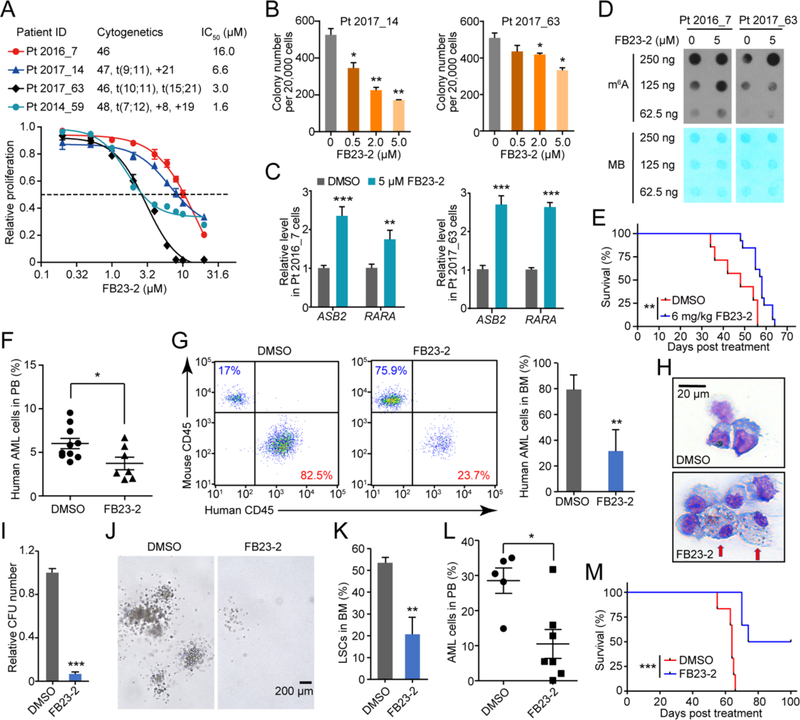
We assessed the therapeutic potential of FB23-2 in treating human primary AML cells. Four AML patients with diverse cytogenetics were tested (Table S8). FB23-2 suppressed proliferation of all four sets of primary AML cells, with IC50 values ranging from 1.6 μM to 16 μM (Figure 8A). FB23-2 also induced cell apoptosis (Figure S6A), decreased colony-forming unit (CFU) capacity (Figure 8B), and accelerated ATRA-mediated myeloid differentiation (Figure S6B) of these primary AML cells. Furthermore, FB23-2 treatment also upregulated the expression of both ASB2 and RARA (Figure 8C), two direct targets of FTO, and elevated global mRNA m6A abundance (Figure 8D), thus supporting our conclusion that FB23-2 displays therapeutic effects via directly targeting FTO signaling in patient-derived primary AML cells.
Figure 8. Therapeutic efficacy of FB23-2 in PDX mouse model.
(A) Effect of FB23-2 treatment of 96 hr on proliferation of primary BM cells collected from four leukemic patients. The proliferation is relative to vehicle group.
(B) Effect of FB23-2 on colony formation in AML patient cells for 12 days.
(C) Expression of ASB2 and RARA in control and 5 μM FB23-2 treated primary BM cells for 72 hr detected by RT-qPCR.
(D) Determination of m6A abundance in the poly(A)+ RNA samples upon FB23-2 treatment of 72 hr in primary BM cells via dot blot assay. The results are derived from two biological replicates.
(E) Kaplan-Meier analysis of patient AML cell-bearing NSGS mice with i.p. administration of DMSO (n = 7) or FB23-2 (n = 13). The BM sample from patient Pt 2017_63 was used for transplantation. The graph starts from the first day after transplantation.
(F and G) FACS analysis of the percentage of patient AML (human CD45+) cells in PB collected from PDX mice one day after the 17-day full treatment of DMSO (n = 10) or FB23-2 (n = 7) (F), and in BM collected when the mice became moribund (n = 3 – 4) (G).
(H) Wright-Giemsa staining to show the effect of FB23-2 on differentiation of human leukemia cells from primary PDX mice BM when the mice became moribund (n = 3 – 4). Arrows indicated differentiated cells.
(I-J) Relative CFUs (I) and morphology of colonies (J) of patient AML cells from primary PDX mice (n = 3 – 4).
(K) FACS analysis of the percentage of LSCs (human CD34+CD38− cells) in BM from primary PDX mice when they became moribund (n = 3 – 4).
(L) FACS analysis of the percentage of human AML cells (human CD45+) in PB isolated from the secondarily transplanted PDX mice 8 weeks post transplantation, which were reconstituted with the same number of leukemia cells from primary PDX mice treated with DMSO (n = 5) or FB23-2 (n = 7).
(M) Kaplan–Meier analysis of the secondary recipient mice transplanted with AML cells collected from primary PDX mice treated at indicated (n = 6).
*, p < 0.05; **, p < 0.01; ***, p < 0.001; unpaired Student’s t-test. Error bars, mean ± SEM; if not indicated, n = 3. See also Figure S6 and Table S8.
Lastly, we tested the in vivo therapeutic efficacy of FB23-2 in a PDX AML mouse model. Primary AML cells were xeno-transplanted into sublethally irradiated NSGS mice. We monitored the engraftment of AML leukemia cells in vivo by FACS analysis of the percentage of donor AML cells in PB in recipient mice. When recipient mice had 3–5% donor-derived AML cells, the recipient mice were treated with FB23-2 or DMSO for 17 days. The disease latency of FB23-2-treated mice (median survival time of 58 days) was significantly prolonged compared with that of control mice (median survival time of 48 days) (Figure 8E). Furthermore, FACS analysis of engrafted AML cells in recipient mice revealed a significantly reduced proportion of AML blast cells in PB (Figure 8F) and BM (Figure 8G) upon FB23-2 treatment. Consistent with our findings that FB23-2 induced differentiation of AML cell lines in vitro, we found that more differentiated myeloid cells with an increased ratio of cytoplasm/nucleus were present in FB23-2-treated mice (Figure 8H). The leukemia cells from FB23-2-treated PDX mice gave rise to significantly fewer CFUs with markedly reduced sizes of colonies than the leukemia cells from the DMSO-treated PDX mice did (Figures 8I and 8J), thus suggesting that the leukemia malignancy of FB23-2 treated AML cells was significantly impaired. Notably, not only were the bulk AML cells affected, but also leukemia stem cells (LSCs, defined by CD34+CD38−) were significantly eliminated by FB23-2 in vivo in the treated mice (Figure 8K).
To further evaluate the number of functional LSCs in primary PDX mice, we performed a secondary transplantation. The secondary recipients of AML cells from primary DMSO-treated PDX mice (control) had markedly higher engraftment compared to the secondary recipient mice with AML cells from primary FB23-2 treated PDX mice (Figure 8L). All of the control, secondary PDX mice died within 66 days while 50% of the secondary PDX mice with FB23-2-treated AML cells still survived after 100 days (Figure 8M), thus suggesting that the number of functional LSCs that are able to regenerate leukemia in vivo in secondary recipients was significantly reduced after FB23-2 treatment in the primary recipient mice. Taken together, our data indicate that FB23-2 induced differentiation of AML cells significantly reduced the number of functional primary AML LSCs in vivo.
Discussion
Epitranscriptomics is a rapidly evolving field in biology. Emerging evidence suggests that the deregulation of m6A modification on RNA contributes to leukemogenesis. METTL3 and METTL14, the m6A methyltransferases, have been reported to control and/or maintain myeloid leukemia, highlighting their oncogenic roles in leukemogenesis (Barbieri et al., 2017; Vu et al., 2017; Weng et al., 2018). In addition, the m6A demethylase FTO has been found to play an oncogenic role in a subset of AMLs (Li et al., 2017b). Also, by suppression of FTO activity, R-2HG exhibits significant anti-tumor effects in AML (Su et al., 2018). These results suggest that FTO inhibitors alone or in combination with standard chemotherapeutic agents represent possible effective therapeutics for the treatment of AML, especially those with high FTO expression. However, none of the known FTO inhibitors seems to be clinically applicable due to the limited activities. Therefore, it is urgently needed to develop highly effective FTO inhibitors, which can be used as tools to study the biological roles of FTO in normal and malignant development as well as in various biological processes involving m6A modification.
Few inhibitors for regulation of RNA methylation have been characterized, which exists in sharp contrast to factors of DNA and histone epigenetics. Here we report that through structure-based rational designs, we have successfully developed more effective small-molecule inhibitors of FTO. The MA-derived inhibitor FB23 displays significantly improved inhibitory activity on FTO demethylation of m6A-RNA in vitro. Next, we optimized the physicochemical property of FB23, thus leading to the identification of FB23-2 with a significantly improved ability to hinder the proliferation of a panel of AML cell lines, and also inhibits primary AML LSCs in PDX mice, thus suggesting that FTO might serve as a potential molecular target in LSCs in order to inhibit leukemogenesis. The discovery of FB23-2 and its anti-proliferative effects on AML would increase the current intense interest in RNA methylation, especially with regard to the pharmacology.
Importantly, we tend to show our inhibitors target FTO and impair its demethylation, and by targeting FTO our inhibitor causes a significant biological impact. We validated that the effects of FTO inhibitors on AML are linked to certain downstream targets, e.g., MYC, CEBPA, RARA, and ASB2 RNA transcripts. It remains unknown whether FB23-2 impairs FTO’s binding to target transcripts in cells, however. The target engagement of current inhibitors needs further explorations with a more depth, which could show the potential for these inhibitors to help propel the field of epitranscriptomics forward.
In summary, we provide here a proof-of-concept that small-molecule targeting of oncogenic FTO demethylase may be an effective therapeutic strategy for the treatment of AML. Our study demonstrates the feasibility of attenuated FTO demethylation for the induction of differentiation of AML cells. This effect is likely achieved through specifically regulating expression of critical genes and signalling pathways as a result of elevated m6A levels in mRNA transcripts of these genes that are induced by FTO inhibitors. As FTO-mediated demethylation has also been linked to a variety of cancer types, our findings may have a broad impact on cancer therapy by targeting epitranscriptomic RNA methylation.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Cai-Guang Yang (yangcg@simm.ac.cn).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Leukemic patients sample sections
The leukemia patient samples were obtained according to a protocol approved by the institutional review board of the Cincinnati Children’s Hospital Medical Center, and written informed consent was obtained from all patients for the use of their samples. Patients’ mutation profiles are showed in Table S8.
Cell cultures
Leukemia cells NB4, U937, MV4-11, and ML-2 were cultured in RPMI1640 supplemented with 10% fetal bovine serum (FBS) (GIBCO). MONOMAC6 (ACC-124) cells were cultured in RPMI1640 with 10% FBS, antibiotics, 10 mM HEPES, 2 mM L-Glutamine (25030081, ThermoFisher), non-essential amino acids (11140050, ThermoFisher), 1 mM sodium pyruvate (11360070, ThermoFisher), and 10 μg/ml human insulin (12585014, ThermoFisher). MA9.3ITD (MLL-AF9 plus FLT3-ITD) and MA9.3RAS (MLL-AF9 plus NRasG12D) were established by Dr. James C. Mulloy (Wunderlich et al., 2013), and cultured in IMDM supplemented with 20% FBS. 293T cells were cultured in DEME with 10% FBS, antibiotics, and 10 mM HEPES, and used for lentivirus package. The leukemic patient’s samples (Pt 2016_7, Pt 2017_14, Pt 2017_63, and Pt 2014_59) were maintained in IMDM medium supplemented with 20% FBS, antibiotics, and 10 ng/ml recombinant human SCF, TPO, FLT3-3L, IL-3, and IL-6 (PeproTech) cytokines. The mouse bone marrow cells (FLT3ITD/NPM1 and MA9) were kept in RPMI1640 supplemented with 10% FBS, antibiotics, 100 ng/ml recombinant mouse SCF, 10 ng/ml mouse IL-3 and IL-6 (PeproTech).
Care and maintenance of animals
The BALB/c mice and rat were obtained from Shanghai SLAC Laboratory Animals Co. Ltd. and Shanghai Sippr-BK Laboratory Animal Co. Ltd, respectively, and the NSGS mice were bred in the University of Cincinnati Vontz Center animal core facility, the University of Illinois at Chicago and the University of Florida animal facility. The Institutional Animal Care and Use Committee (IACUC) of the Shanghai Institute of Materia Medica, the University of Cincinnati, the University of Illinois at Chicago, and the University of Florida approved all animal procedures and conformed to all regulatory standards.
Animal model
The NSGS mice were bred and subjected to the xeno-transplantation model. For the AML mouse model, 0.2 × 106 MONOMAC6 cells were directly transplanted into NSGS mice via tail vein. After 10 days, FB23-2 (2 mg/kg/day) and DMSO vehicle were intraperitoneally injected into the mice for a continuous 10 days. The mice were euthanized by CO2 inhalation if they exhibited classical AML symptoms including hunched posture, paralysis, and reduced body weight. Meanwhile, the PB, spleen, and liver samples were collected for further analysis.
PDX models were generated by injecting primary BM cells from AML patient Pt 2017_63 (2 X 106 per mouse) into the tail veins of 6- to 8-week-old sublethally irradiated (2.5 Gy) NSGS mice (NOD.Cg-PrkdcscidIl2rgtm1WjlTg(CMV-IL3, CSF2, KITLG)1Eav/MloySzJ), which were purchased from The Jackson Laboratory. When recipient mice had 3–5% donor-derived AML cells in PB, 6 mg/kg/day FB23-2 was delivered by i.p. for 17 days, vehicle DMSO was administrated as control. Mice were weighed daily during treatment and doses were recalculated to make sure the mice received a consistent dose of 6 mg/kg/day. One day after the 17-day full treatment, mice were randomly picked up, and then PB cells were collected and analyzed for the engraftment of leukemia cells by FACS using anti-human-CD45 and anti-mouse-CD45 (eBioscience). When the mice became moribund, BM cells were collected and analyzed for the engraftment of leukemia cells by FACS using anti-human-CD45 and anti-mouse-CD45 (eBioscience). In addition, the LSCs population was determined as the human CD34+CD38− (eBioscience) population. For second transplantation, the patient AML cells, collected from the spleen of primary NSGS mice which were transplanted with BM cells from AML patients and received FB23-2 or DMSO treatment, were transplanted into NSGS mice irradiated at 2.5 Gy. 8 weeks post transplantation, PB cells were collected for FACS analysis using anti-human-CD45 and anti-mouse-CD45 and the mice were continued to monitor for survival.
METHOD DETAILS
Protein expression and purification
The N-terminal 31 residues truncated FTO gene was subcloned into pET28a vector, encoding a His-tag fusion FTO protein. The expression and purification of FTO were modified from previously reported methods (Huang et al., 2015). For crystallization, 5 mM EDTA was added to the gel filtration buffer in order to remove the endogenous binding ions. FTOΔN31 fractions were concentrated to 20 mg/ml and stored in an EDTA-free buffer at −80 °C.
The N-terminal 66 residues truncated alkbh5 gene was cloned into pGEX-6p-1 vector. The protein was purified by Nickle-affinity chromatography, followed by a step of PreScission protease digestion to remove both His- and GST-tags. Digested mixtures were loaded onto the cation exchange column (Mono S 5/50 GL, GE Healthcare) and eluted with a buffer of 20 mM Tris-HCl (pH 8.0), 500 mM NaCl. Fractions were collected and checked by 12% SDS-PAGE. ALKBH5ΔN66 was stored at −80 °C for further bioassays.
HPLC-based assay of the inhibition of m6A demethylation in RNA
In vitro ssRNA demethylations were performed with some modifications on the reported assay (Huang et al., 2015). The reactions, containing 0.25 μM FTOΔN31 or 3 μM ALKBH5ΔN66, 5 μM 15-mer ssRNA (5′-AUUGUCA(m6A)CAGCAGC-3′), 300 μΜ 2OG, 280 μΜ (NH4)2Fe(SO4)2, 2 mM L-ascorbic acid, and inhibitors at required concentrations in 50 mM Tris-HCl (pH 7.5 – 8.0), were incubated at 25 °C for 30 min. The reactions were terminated by heating for 5 min at 90 °C, and then the mixtures were subjected to digestion by nuclease P1 (N8630, Sigma) and alkaline phosphatase (P5931, Sigma). The IC50 values were quantitated based on the inhibitory percentages of m6A demethylation in the presence of inhibitors at indicated concentrations, using nonlinear regression, dose-response fit on GraphPad Prism 5.0™. All reactions were performed in triplicate.
Crystallization and structure determination of FTO/FB23 complex
Crystallizations were conducted with hanging-drop vapor-diffusion method at 18 °C. 8 mg/ml of FTOΔN31 protein was incubated with 5-folds FB23 and mixed with a reservoir solution containing 100 mM sodium citrate (pH 5.4), 11.5% (w/v) polyethylene glycol 3350, and 8% isopropanol. The crystals were cryo-protected using extra 20% (v/v) glycerol. Diffraction data were collected on the BL18U1 and BL17U1 beamline at the Shanghai Synchrotron Research Facility (SSRF). All X-ray data were processed using HKL2000 programs (Otwinowski and Minor, 1997), and converted to structure factors within the CCP4 program (Collaborative Computational Project, 1994). The structure was solved by molecular replacement in Phaser using the structure of FTO/MA complex (PDB code 4QKN) as the searching model. The model of structural complex FTO/FB23 was computationally refined with the program REFMAC5.
Nuclear Magnetic Resonance (NMR) titration
Phosphate buffer (20 mM sodium phosphate (pH 7.4), 100 mM NaCl, 5% DMSO) was used for NMR data acquisition on a Bruker Avance III-600 MHz spectrometer equipped with a cryogenically cooled probe (Bruker biospin, Germany) at 25 °C. Experimental samples contained 200 μM FB23 and FTO protein at 0 μM, 1 μM, 2 μM, and 3 μM, respectively.
Cellular thermal shift assay (CETSA)
CETSA was conducted according to the protocol as previously described (Martinez Molina et al., 2013). NB4 and MONOMAC6 cells were collected and lysed in 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, and 2 mM DTT. 50 μM FB23 or DMSO was added to the supernatant and incubated at 25 °C for 25 min. After denaturing at various temperatures for 5 min, samples were centrifuged, and the supernatants were analyzed by western blot. All experiments were performed in triplicate.
Cell proliferation assays
5,000 cells/well NB4, FTO KO NB4, and MONOMAC6 AML cells were seeded and treated with DMSO or FTO inhibitors for 72 hr. The cell proliferations were determined with CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (G4100, Promega) according to the manufacturer’s instructions. 10,000 cells/well human AML cells (MA9.3ITD, MA9.3RAS, U937, ML2, and MV4-11) and four primary cells from AML patients were seeded and subjected to FTO inhibitor treatment for 96 hr as indicated. 10,000 cells/well MA9 and FLT3/NPM1 primary cells isolated from AML mice and 5,000 cells/well shNS and shFTO NB4 cells were seeded and treated with FTO inhibitors for 24 hr, 48 hr, 72 hr, and 96 hr for proliferation determination.
Quantitation of FB23 and FB23-2 in AML cells
NB4 and MONOMAC6 cells were treated with 10 μM FB23 or FB23-2 for 24 hr, respectively. Viable cells were distinguished with 0.1% trypan blue, counted and then harvested with PBS by several washings. Cells were diluted into 100 μl with 50% (v/v) water/methanol and followed by several shock freeze-thaw cycles. The supernatants were collected for analysis. The Ultimate 3000 system coupled with a TSQ Quantiva mass spectrometer (Thermo, USA) was applied to determine the cellular concentration of compound FB23 and FB23-2. Analytes were separated on a XSELECT™ HSS T3 column (100 mm × 3.0 mm, 2.5 μm; Waters, USA). The mobile phases used for elution were (A) 0.1% (v/v) formic acid/water and (B) 0.1% (v/v) formic acid/acetonitrile. The mass spectrometer was operated in the negative MRM mode. Parent-to-product transitions were m/z 375.1→339.1, 375.1→298.1 for FB23, and m/z 390.3→318.0, 390.3→289.9 for FB23-2, respectively.
m6A dot blot assay
NB4 and MONOMAC6 cells were treated with DMSO or FB23-2 at varying concentrations for 72 hr, while MA9 and FLT3/NPM1 primary cells isolated from AML mice, five human AML cell (MA9.3ITD, MA9.3RAS, U937, ML2, and MV4-11), and human primary AML cells were treated with DMSO or 5 μM FB23-2 for 72 hr for dot blot assay. Total RNA was separated with miRNeasy Mini Kit (217004, Qiagen), and poly (A)+ RNA was further enriched with PolyATract mRNA isolation System IV (Z5310, Promega) in accordance with the manufacturer’s instructions. The RNA samples were diluted in RNA binding buffer, denatured at 65 °C for 5 min. Then one volume of 20 x SSC buffer was added into the RNA samples before dotted onto the Amersham Hybond-N+ membrane (RPN119B, GE Healthcare) with Bio-Dot Apparatus (#170-6545, Bio-Rad). The RNA samples were cross-linked onto the membrane via UV irradiation. The membrane was stained with 0.02% methylene blue (MB) as loading control. After UV crosslinking and MB staining, the membrane was washed with PBST, blocked with 5% nonfat dry milk for 1 hr at room temperature and incubated with m6A antibody (202003, Synaptic Systems, 1 : 2000) at 4 °C overnight. Finally, the membrane was then incubated with the HRP-conjugated goat anti-rabbit IgG (sc-2030, Santa Cruz Biotechnology) and developed with Amersham ECL Prime Western Blotting Detection Reagent (RPN2232, GE Healthcare).
LC-MS/MS quantitation of m6A and m6Am in AML cells
NB4 and MONOMAC6 cells were cultured with DMSO or 20 μM FB23-2 for 72 hr. mRNA was isolated in line with the dot blot assay, followed by the removal of contaminated rRNA with RiboMinus Transcriptome Isolation Kit (A1083708, Invitrogen). 300 ng mRNA was decapped with 5 units RppH (M0356S, NEB) with the thermopol buffer (B9004S, NEB), then the products were digested by nuclease P1 (N8630, Sigma) for 1 hr at 42 °C. Subsequently, 1 unit of alkaline phosphatase (P5931, Sigma) and NH4HCO3 (100 mM) were added and incubated for another 1 hr at 37 °C. The Ultimate 3000 system coupled with a TSQ Quantiva mass spectrometer (Thermo, USA) was applied to quantitate the cellular levels of A, m6A, and m6Am. Samples were centrifuged and loaded onto a XSELECT™ CSH™ C18 column (100 mm × 3.0 mm, 2.5 μm; Waters, USA) and eluted by the gradient methanol. The parent-to-product transitions for A, m6A, and m6Am were 268.1/136.1, 282.1/150.1, and 296.2/150.1, respectively.
Lentivirus production and infection
Lentivirus-induced FTO KD was conducted in NB4 and MONOMAC6 cells as reported previously with some modifications (Li et al., 2017b). Briefly, 0.5 μg pMD2.G (#12259, addgene), 0.3 μg pMDLg/pRRE (#60488, addgene) and 0.7 μg pRSV-Rev (#12253, addgene), and 1.5 μg of pLKO.1-shFTO or pLKO.1-shNS were co-transfected into 293T cells in 60 mm cell-culture dishes with Effectene transfection reagent (301427, Qiagen) to package lentivirus. The lentivirus particles were harvested for 48 hr and 72 hr post transfection, directly pipetted into NB4 and MONOMAC6 cells with existence of 4 μg/ml polybrene (H9268, Sigma-Aldrich). Then the AML cells including lentivirus were subjected into spinoculation at 32 °C, 1,200 rpm for 90 min. Finally, 1 μg/ml puromycin (P8833, Sigma-Aldrich) was added into the cultured AML cells 48 hr post spinoculation to select positive infected-cells.
CRISPR-Cas9-based genome editing
The CRISPR–Cas9 system used in this study was constructed by the Genoarray Company (Suzhou, China). Briefly, a genome-wide CRISPR sgRNA library with 6 sgRNAs was designed to construct the Cas9 expression plasmids (pWSLV-cas02-FTO-KO). NB4 AML cells stably expressing Cas9 endonuclease were constructed with lentiviral infection of the sgRNA libraries. FTO KO cells were sorted by the FACS after infection for 48 hr and continually cultured for PCR amplifications from genomic DNA and sequencing.
Real-time Quantitative PCR (RT-qPCR)
NB4, MONOMAC6, and human primary AML cells were treated with vehicle control or FB23-2 at indicated concentrations for 72 hr. Total RNA isolated with the TRIZOL reagent was subjected to reverse transcription using the PrimeScript™ RT reagent Kit (RR047Q, Takara). RT-qPCR reactions were performed with the THUNDERBIRDSYBR qPCR Mix (QPS201, TOYOBO) and primers listed in Table S9. Gene expression was calculated using the comparative ΔΔCT method with the GAPDH for normalization.
Immunoblotting (Western blot)
NB4 and MONOMAC6 cells were harvested after FB23-2 treatment for 72 hr and lysed with the RIPA Lysis buffer solution at 4 °C. Supernatants were centrifuged and the protein concentration was quantified with BCA Protein Assay (23225, ThermoFisher). An equal amount of whole-cell lysate from each sample was loaded to the SDS-PAGE. The proteins were transferred onto nitrocellulose membrane (Millipore, USA) and blocked with 5% skim milk and incubated with antibodies against FTO, ALKBH5, RARA, ASB2, GAPDH etc. HRP conjugated Goat Anti-mouse (sc-2055, Santa Cruz Biotechnology) or Anti-rabbit IgG (sc-2030, Santa Cruz Biotechnology) was used as secondary antibody.
Drug affinity responsive target stability (DARTS) assay
DARTS was run in AML cells following the previously published protocol with some modifications (Lomenick et al., 2009). Briefly, 6 × 107 NB4 or MONOMAC6 cells were collected, washed with chilled PBS and lysed in M-PER buffer (78501, ThermoFisher) supplemented with protease inhibitor cocktail (78430, ThermoFisher) and phosphatase inhibitor cocktail (78420, ThermoFisher). The cell pellets were lysed on ice for 10 min and then subjected to centrifuge at 18,000 × g for 10 min at 4 °C. The supernatant was transferred into TNC buffer (50 mM Tris-HCl (pH 8.0), 50 mM NaCl and 10 mM CaCl2) and calibrated with BCA assay for protein concentration. Then the cell lysates were incubated with 50 μM, 200 μM, and 500 μM FB23-2 as well as DMSO for 1 hr at room temperature and subsequently digested with Pronase as indicated (10165921001, Roche) for 30 min. The reaction was quenched by addition of protease inhibitor cocktail (78429, ThermoFisher Scientific) and the sample was chilled immediately on ice. Finally, the cell lysates were subjected to western blot to determine the abundance of FTO or ALKBH5.
Inhibition of FTO inhibitors on epigenetic enzymes in vitro
The activity of TET1 enzyme in vitro was performed by following the reported assay with some modifications (Blaschke et al., 2013). Recombinant TET1 protein was commercially purchased (31417, Active motif). 2.5 μM dsDNA oligos (Sequence: 5’-CAGTAACTGTGGTC/5mC/GGTAACTGACTTGCA-3’) were incubated with 250 ng TET1 protein in 50 μl buffer containing 50 mM HEPES (pH 8.0), 50 μM Fe(NH4)2(SO4)2, 100 μM 2OG, 2 mM L-ascorbic acid, 100 μM ATP, 3 mM DTT and varying concentrations of FB23-2 or NOG at 37 °C for 2 hr. The reaction mixture was concentrated to 10 μl, followed by adding 0.1N NaOH, and then denatured at 99 °C for 5 min. After neutralized by adding 0.1 volume of 6.6 M ammonium acetate, 1 μl sample was spotted on Amersham Hybond-N+ membrane and crosslinked under UV irradiation. Anti-5hmC antibody (1:5000, Active Motif) was applied to detect the product of TET1 oxidation.
The in vitro enzyme activities of HDACs were determined by a protease-coupled assay (Lobera et al., 2013). Varying concentrations of inhibitors were incubated with full-length recombinant HDAC1 or fragment of HDAC9 (BPS Biosciences) at room temperature, and the reactions were initiated by adding Ac-peptide-AMC substrates. After incubation for 1 hr, reactions were terminated by adding a stop solution containing trypsin. Fluorescent AMC released from substrate was measured using filter sets of an excitation wavelength at 355 nm and an emission wavelength at 460 nm.
The inhibitory assays on histone demethylase LSD1, JMJD2A, JMJD3, JARID1A, and FBXL11 were performed using AlphaLISA technology according to the manufacturer’s constructions (PerkinElmer). Briefly, inhibitors were pre-incubated with enzymes at room temperature for 15 min. For LSD1, reactions were initiated by adding the H3 derived peptide solution; for JMJD2A, JMJD3, FBXL11, and JARID1A, reactions were initiated by adding mixtures of the biotinylated H3 derived peptide, L-ascorbic acid, 2OG, and (NH4)2Fe(SO4)2; for BRD4(1,2), the binding assay was initiated by addition of the biotinylated H4 derived acetylated peptide. All reactions were incubated for 60 min at room temperature and terminated by addition of the acceptor and donor beads mix (PerkinElmer). After incubated for 1 hr at room temperature, the AlphaScreen signals were read on EnSpire (PerkinElmer) in Alpha Mode.
To determine IC50 value for BRD4(1,2) inhibition, the HTRF-based biochemical binding assay was performed. Briefly, different concentrations of FB23-2 were transferred to the 384-well plate by Echo® 550 liquid handler (Labcyte, USA), and then pre-incubated with BRD4(1,2) (BPS Biosciences) for 15 min at room temperature, which was followed by addition of the biotinylated H4 derived acetylated peptide to each well. Then the mixture of anti GST-Donor Antibody and Streptavidin-Acceptor were added to each well, and the assay plate was covered with a sealer to be incubated at room temperature for 1 hr. Read fluorescence emission at wavelengths of 665 nm and 620 nm on EnVision (PerkinElmer). The signal was treated as HTRF Ratio = Acceptor signal (665 nm)/Donor Signal (620 nm).
The radioactivity inhibition assay of DOT1L was conducted with minor modification (Daigle et al., 2011). Human DOT1L protein (Reaction Biology Corp.) and compounds were firstly incubated for 15 min at room temperature. The substrate mixtures of oligonuclesomes and adenosyl-L-methionine S-[methyl-3H] (3H-SAM, PerkinElmer) were added to initiate the reaction. The whole reaction lasted for 120 min at room temperature and was stopped by adding cold SAM. Then the reaction system was transferred to MultiScreen plate (Merck Millipore), which was incubated with 0.5% PEI for 15 min in advance. Methylated oligonuclesomes were bind to the filter and were washed by ddH2O for three times via vacuum. Finally, scintillation liquids were added for signals read on MicroBeta (PerkinElmer).
Effect of FB23-2 on 5mC and 5hmC in AML cells
NB4 and MONOMAC6 AML cells were seeded and treated with varying concentrations of FB23-2. Genomic DNA was separated with a DNeasy Blood and Tissue Kit (69506, QIAGEN) and the levels of 5hmC and 5mC were determined with dot blot assay. Anti-5hmC antibody (39769, Active Motif) and anti-5mC antibody (39649, Active Motif) was used as the primary antibody.
Effect of FB23-2 on histone methylations in NB4 cells
NB4 AML cells were incubated with two dosages of FB23-2, and the known histone demethylase inhibitors JIB-04 (HY-13953, MCE) (Wang et al., 2013), Ciclopirox (T1482, Topscience) (Yang et al., 2017), and IOX-1 (T6545, Topscience) (Schiller et al., 2014) for 48 hr, respectively. Viable cells were counted with 0.1% typan blue and denatured with 1× SDS-PAGE loading buffer at 95 °C for 30 min. Samples were loaded for SDS-PAGE and further western blot procedures. Primary antibodies used were listed in Key Recourses Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| 5-Hydroxymethylcytosine (5hmC) antibody | Active Motif | Cat#39769; RRID: AB_10013602 |
| 5-Methylcytosine (5mC) antibody (mAb) | Active Motif | Cat#39649; RRID: AB_2687950 |
| Anti-ALKBH5 antibody | Abcam | Cat#ab195377; RRID: AB_1267666 |
| Anti-ASB2 antibody | Abcam | Cat#ab13710; RRID: AB_300589 |
| Anti-CEBP Alpha antibody | Abcam | Cat#ab15048; RRID: AB_2077890 |
| Anti-c-MYC antibody [Y69] | Abcam | Cat#ab32072; RRID: AB_731658 |
| Anti-FTO antibody [EPR6895] | Abcam | Cat#ab124892; RRID: AB_10972698 |
| Anti-RARA antibody | BioLegend | Cat#616801; RRID: AB_2253600 |
| Anti-Histone H3 | Proteintech | Cat#17168–1-AP; RRID: AB_2716755 |
| Anti-Histone H4 | Cell Signaling Technology | Cat#13919S |
| Anti-H3K4me1 | Abcam | Cat#ab176877; RRID: AB_2637011 |
| Anti-H3K4me2 | Active Motif | Cat#39914; RRID: AB_2614976 |
| Anti-H3K4me3 | Cell Signaling Technology | Cat#9751T; RRID: AB_2616028 |
| Anti-H3K9me2 | Abcam | Cat#ab1220; RRID: AB_449854 |
| Anti-H3K9me3 | Cell Signaling Technology | Cat#13969T |
| Anti-H3K27me2 | Abcam | Cat#ab24684; RRID: AB_448222 |
| Anti-H3K27me3 | Active Motif | Cat#39157; RRID: AB_2636821 |
| Anti-H3K36me2 | Active Motif | Cat#39892; RRID: AB_2793383 |
| Anti-H3K36me3 | Cell Signaling Technology | Cat#4909T; RRID: AB_1950412 |
| Anti-H3K79me1 | Active Motif | Cat#39922; RRID: AB_2793394 |
| Anti-H3K79me2 | Active Motif | Cat#39924; RRID: AB_2793395 |
| Anti-H3K79me3 | Abcam | Cat#ab2621; RRID: AB_303215 |
| Anti-H3R17me2a | Active Motif | Cat#39710; RRID: AB_2793315 |
| Anti-H4K20me1 | Abcam | Cat#ab177188 |
| Anti-H4K20me2 | Active Motif | Cat#39540; RRID: AB_2793248 |
| Anti-H4K20me3 | Active Motif | Cat#39672; RRID: AB_2650526 |
| Donkey anti-rabbit IgG-HRP | Santa Cruz | Cat#sc-2077; RRID: AB_631745 |
| GAPDH antibody (0411) | Santa Cruz | Cat#sc-47724; RRID: AB_627678 |
| Goat anti-mouse IgG-HRP | Santa Cruz | Cat#sc-2055; RRID: AB_631738 |
| m6A (N6-methyladenosine) antibody | Synaptic Systems | Cat#202003; RRID: AB_2279214 |
| Biological Samples | ||
| Leukemic patients sample sections | Cincinnati Children’s Hospital Medical Center | N/A |
| Chemical, Peptides, and Recombinant protein | ||
| ATRA | Sigma-Aldrich | Cat#223018 |
| Ciclopirox | Topscience | Cat#T1482 |
| FBS | GIBCO | Cat#10099–141 |
| Giemsa Stain | EMD chemicals | Cat#620G-75 |
| IOX-1 | Topscience | Cat#T6545 |
| JIB-04 | MCE | Cat#HY-13953 |
| May-Grunwald Stain | Sigma-Aldrich | Cat#MG-500 |
| Polybrene | Sigma-Aldrich | Cat#H9268 |
| Pronase from Streptomyces griseus | Roche | Cat#10165921001 |
| Puromycin dihydrochloride | Sigma-Aldrich | Cat#P8833 |
| Recombinant Human Flt3-Ligand | PeproTech | Cat#300–19 |
| Recombinant Human IL-3 | PeproTech | Cat#200–03 |
| Recombinant Human IL-6 | PeproTech | Cat#200–06 |
| Recombinant Human SCF | PeproTech | Cat#300–07 |
| Recombinant Human TPO | PeproTech | Cat#300–18 |
| Recombinant Mouse IL-3 | PeproTech | Cat#213–03 |
| Recombinant Mouse SCF | PeproTech | Cat#250–03 |
| RppH | NEB | Cat#M0356S |
| TET1 | Active Motif | Cat#31417 |
| Critical Commercial Assays | ||
| CellTiter 96 Non-Radioactive Cell Proliferation Assay (MTT) | Promega | Cat#G4100 |
| COX Fluorescent Inhibitor Screening Assay Kit | Cayman | Cat#700100 |
| Epigenetic Screening | Shanghai ChemPartner Co., Ltd | CPB-P16–20033 |
| FITC Annexin V Apoptosis Detection Kit I | BD Biosciences | Cat#556547 |
| IC50Profiler™ | Eurofins Pharma Discovery Services UK Limited | UK022–0004210 |
| KinaseProfiler™ | Eurofins Pharma Discovery Services UK Limited | UK022–0004141 |
| miRNeasy Mini Kit | QIAGEN | Cat#217004 |
| PE Annexin V Apoptosis Detection Kit I | BD Biosciences | Cat#559763 |
| PolyATract mRNA isolation System IV | Promega | Cat#Z5310 |
| ProteaseProfiler™ | Eurofins Panlabs Discovery Services Taiwan Limited | TW04–0002669 |
| RiboMinus Transcriptome Isolation Kit | Invitrogen | Cat#A1083708 |
| Deposit Data | ||
| m6A-seq | This paper | GEO: GSE103496 |
| RNA-seq | This paper | GEO: GSE103494, GSE103495 |
| Structural complex of FTO/FB23 | This paper | PDB: 6AKW |
| X-ray structure of FB23-2 | This paper | CCDC: 1558332 |
| Experimental Models: Cell line | ||
| 293T | ATCC | CRL-3216 |
| FLT3ITD/NPM1 | Isolated from BM of AML mouse induced by FLT3ITD/NPM1 | N/A |
| MA9 | Isolated from BM of AML mouse induced by MA9 oncogene | N/A |
| MA9.3ITD | A gift from James C. Mulloy lab | N/A |
| MA9.3RAS | A gift from James C. Mulloy lab | N/A |
| ML-2 | DSMZ | ACC-15 |
| MONOMAC6 | DSMZ | ACC-124 |
| MV4–11 | ATCC | CRL-9591 |
| NB4 | DSMZ | ACC-207 |
| U937 | ATCC | CRL-1593.2 |
| Experimental Models: Organisms/Strains | ||
| BALB/c mouse | Shanghai SLAC Laboratory Animals Co. Ltd. | N/A |
| NSGS mouse | The Jackson Laboratory | Stock#013062 |
| SD rat | Shanghai SLAC Laboratory Animals Co. Ltd. | N/A |
| Oligonucleotides | ||
| 5′-AUUGUCA(m6A)CAGCAGC-3′ | TAKARA, China | N/A |
| 5’-CAGTAACTGTGGTC(5mC)GGTAACTGACTTGCA-3’ | Sangon Biotech, China | N/A |
| See Table S9 for primers | Generay, China | N/A |
| Recombinant DNA | ||
| pLKO.1-shFTO | This paper | N/A |
| pLKO.1-shNS | This paper | N/A |
| pWSLV-cas02-FTO-KO | This paper | N/A |
Kinases and proteases profiling
The kinases and proteases profiling were conducted by Eurofins Pharma Discovery Services. Inhibitory kinases profiling was conducted against a panel of 405 kinases in the presence of FB23-2 at 1 μM and 10 μM, respectively. Taken the Met (h) kinase as an example of kinases profiling, Met (h) was incubated with 8 mM MOPS (pH 7.0), 0.2 mM EDTA, 250 μM KKKGQEEEYVFIE, 1 mM sodium orthovanadate, 5 mM sodium-6-glycerophosphate, 10 mM Mg(OAc)2, and [γ−33P]-ATP (specific activity and concentration as required). The reaction was initiated by the addition of the Mg(OAc)2/ATP mix. After incubation for 40 min at room temperature, the reaction was terminated by the addition of phosphoric acid to a final concentration of 0.5%. 10 μl of the reaction was then spotted onto a P30 filtermat and washed four times for 4 min in 0.425% phosphoric acid and once in methanol prior to drying and scintillation counting. The results were calculated with an equation, Inhibition (%) = (Max-Signal)/(Max-Min) × 100%. The reactions without enzyme but all other components served as Min, and the reactions with DMSO served as Max. The percentage inhibitions of 405 kinases by 10 μM were mapped on to the kinome phylogenetic tree (Illustration reproduced courtesy of Cell Signaling Technology, Inc. (www.cellsignal.com)). Each group had two repeats. For IC50 determination, the inhibitory percentage of FB23-2 at varying concentrations was obtained, and the IC50 value for each tested kinase was calculated with nonlinear regression analysis using equation in GraphPad Prism 5.
Taken the Caspase2 as an example of inhibitory proteases profiling, 1 μM or 10 μM FB23-2 was pre-incubated with human recombinant Caspase2 in a reaction buffer of 50 mM HEPES, pH 7.4, 100 mM NaCl, 0.1% CHAPS, 1 mM EDTA, 10% Glycerol, 10 mM DTT at 37 °C for 15 min, followed by adding the 25 μM substrate Z-VDVAD-AFC. After incubation for 1 hr, signals of AFC were quantified with spectrofluorimetric method and the DMSO group was treated as 100%. Each group had two repeats.
Effect of FTO inhibitors on COX-1 and COX-2 enzymes
The inhibitions of COX1 and COX2 enzymes by FTO inhibitors FB23 and FB23-2 were evaluated using the COX Fluorescent Inhibitor Screening Assay Kit (700100, Cayman) following the manufacturer’s protocols. Briefly, COX-1 and COX-2 were incubated with test compounds at room temperature for 5 min, respectively, then 10 μl ADHP (10-acetyl-3,7-dihydroxyphenoxazine) was added to the sample and background wells (without COX enzymes). Reactions were initiated by quickly adding 10 μl of Arachidonic Acid and incubated for 2 min at room temperature. An excitation wavelength of 535 nm and an emission wavelength of 595 nm were used to obtain the signals.
FACS analysis
NB4 and MONOMAC6 AML cells were induced to myeloid differentiation for 48 hr by ATRA (223018, Sigma-Aldrich), 200 nM for NB4 and 1000 nM for MONOMAC6, coupled with the existence of FB23-2 or DMSO. The induced AML cells were collected, washed with chilled phosphate buffered saline (PBS) and stained with PE-conjugated anti-CD11b (101208, BioLegend) and APC-conjugated anti-CD14 (17-0149-41, eBioscience) at 4 °C for 30 min before subjected to FACS analysis. Similarly, AML patient primary cells were induced to myeloid differentiation for 96 hr by ATRA at indicated concentration with existence of FB23-2 or not and stained with PE-conjugated anti-CD11b and APC-conjugated anti-CD15 (17-0158-41, eBioscience).
PE Annexin V Apoptosis Detection Kit I and FITC Annexin V Apoptosis Detection Kit I (BD Biosciences) were used to determine the effects of FTO inhibitor on cell apoptosis following the manufacturer’s instructions. Briefly, NB4 and MONOMAC6 AML cells were treated with 1 μM, 5 μM, and 20 μM FB23-2 or DMSO for 48 hr (for NB4) and 72 hr (for MONOMAC6), washed with cold PBS, resuspended in 1 × binding buffer, and stained with PE Annexin V and 7-AAD. Bone marrow cells from AML patients were treated with FB23-2 as indicated for 48 hr before staining with FITC Annexin V and PI. Finally, the apoptosis in each group was assessed by FACS. To investigate the effects of FB23-2 on AML cells abundance in AML mouse model, the PB samples were collected at end point, lysed with ammonium chloride solution (07850, Stemcell technologies), washed with cold PBS, blocked nonspecific binding with affinity-purified anti–mouse CD16/32 (eBioscience), stained with PE-conjugated anti-human CD33 (12-0338-42, ThermoFisher scientific) for 30 min at 4 °C and finally subjected to FACS analysis.
MONOMAC6 cells were treated with varying concentrations of FB23-2 for 24 hr before cell cycle analysis. For PI staining, cells were resuspended and incubated in Krishan’s reagent (0.05 mg/ml PI, 0.1% trisodium citrate, 0.02 mg/ml ribonuclease A, 0.3% NP-40) at 37°C for 30 min, and then subjected to FACS analysis. For Hoechst/Pyronin Y staining, cells were suspended in cell culture medium, incubated at 37°C for 45 min with 10 ug/ml Hoechst 33342, and then further incubated at 37 °C for 15 min with Pyronin Y before FACS analysis.
RNA-seq and m6A-seq assays and data analysis
For RNA-seq assay, total RNA samples were extracted from NB4 cells upon FTO KD and inhibitors treatment for 48 hr with miRNeasy mini kit (217004, Qiagen) and poly (A)+ mRNAs were purified with NEBNext Poly(A) mRNA Magnetic Isolution Module. Libraries were constructed with PrepX mRNA library kit (WaferGen) and sequenced to 50 bp using TruSeq SBS kit on Illumina Hiseq system. Differential gene expression was analyzed following standard Illumina sequence analysis pipeline. Gene Set Enrichment Analysis (GSEA) (Subramanian et al., 2005) was used to analyze the enriched signaling pathways.
m6A-seq assays were carried on according to the published protocol (Dominissini et al., 2013) with minor modifications. Poly(A)+ RNA was enriched from total RNA samples isolated from DMSO and FB23-2 treated MONOMAC6 AML cells for 72 hr and randomly fragmented with RNA fragmentation reagents (Ambion). Specific m6A antibody (202003, Synaptic Systems) was applied for immunoprecipitation (IP). The m6A IP samples from both groups were subjected into next-generation sequencing on the Illumina Hiseq 2500. exomePeak (Meng et al., 2014) with default setting was used to m6A peak calling and Integrative Genomics Viewer (IGV) (Robinson et al., 2011) was used to visualize the distribution and abundance of m6A peaks on each mRNA transcript.
Toxicity study
6 to 8-week old BALB/c mice weighed 20 ± 2 g were purchased from Shanghai SLAC Laboratory Animals Co. Ltd. (Shanghai, China). A veterinary health check was performed to select healthy BALB/c mice. Mice were randomly grouped and treated daily with either vehicle control or FB23-2 intraperitoneally for 14 days. The mice were housed five per ventilated polysulfone cage and maintained under constant temperature (18 – 26 °C), humidity (30 – 70%) and lighting conditions (12 hr light and 12 hr dark). After 14 days, animals were euthanized. Then PB samples were collected for complete blood content analysis and plasma biochemical analysis (Shanghai Meixuan Biological Science & Technology LTD, China). The vital organs (heart, kidney, lung, liver, and spleen) were collected and weighed.
Pharmacokinetics
Inhibitor FB23-2 was formulated in DMSO at 3 mg/ml. SD rat (male, 7 – 8 weeks old, n = 3) were treated intraperitoneally with 1 ml/kg formulated compound. Blood samples were collected by retro-orbital bleeding at 5 min, 0.25, 0.5, 1, 2, 4, 6, 8, and 24 hr after the intraperitoneal administration. Blood was collected into EDTA-containing tubes and plasma was obtained by centrifugation at 2,000 g for 5 min. FB23-2 and its hydrolysis metabolite FB23 concentrations in plasma were quantitated by LC-MS/MS method. Noncompartmental analysis with Phoenix 1.4 (Pharsight, USA) was used for all analytical measurements. Area under the concentration-time curve (AUC) was calculated using trapezoidal method. AUC0−∞ = AUC0-t + Ct/ke, ke is elimination rate constant. Elimination half-life (T1/2) = 0.693/ke, mean residence time (MRT) = AUMC/AUC.
Quantitation of FB23-2 in plasma
Calibration curve concentrations ranged from 1.00 to 500 ng/ml for FB23-2 and FB23. 50 μl of rat plasma was precipitated by adding 150 μl acetonitrile immediately and vortexed to stabilize FB23-2 at each sample collection. 50 μl of study sample supernatant, 25 μl internal standard solution (probenecid and estrone-3-sulfate: 400/100 nmol/l), and 50.0 μl of 5 mM ammonium acetate solution (containing 0.1% formic acid) were added to a 1.5 ml polypropylene tube, then vortexed and centrifuged at 11,000 × g for 10 min, the supernatant was injected to LC-MS/MS. A LC-30AD liquid chromatographic system (Shimadzu, Kyoto, Japan) coupled to a Triple Quad 5500 mass spectrometer (AB Sciex, Concord, ON, Canada) was used for acquiring LC-MS/MS data. Analytes were separated on an Eclipse Plus C18 column (100 mm × 4.6 mm I.D., 3.5 μm; Agilent, USA). The mobile phases used for isocratic elution were 25% (A) 5 mM ammonium acetate-formic acid (100/0.1, v/v) and 75% (B) acetonitrile. The flow rate was 0.6 ml/min. The mass spectrometer was operated in the negative MRM mode. The parent-to-product transitions were m/z 390.2→318.0 for FB23-2, m/z 283.9→239.9 for probenecid (internal standard of FB23-2), m/z 375.2→298.2 for FB23, m/z 349.2→269.2 for estrone-3-sulfate (internal standard of FB23). The collision energy was set at −16, −30, −28, and −43 eV. The dwell time for each transition was set at 100 ms.
Microsomal stability assay
The assay was conducted as the previously reported (Di et al., 2006). Briefly, 3 μM FB23-2 was incubated with 0.5 mg/ml rat liver microsomal protein at 37 °C in the presence of 1 mM NADPH cofactor. After incubation for 0, 5, 15, 30 and 60 min, respectively, cold acetonitrile was added to terminate the reactions. The solution was centrifuged, and the supernatants were analyzed using LC–MS/MS method similar with the quantitation of FB23 and FB23-2 in AML cells. The parent-to-product transition of probenecid, internal standard of FB23-2, was m/z 283.9→239.9. The formula for calculation was T1/2 = −0.693/k, and the inherent clearance rate CLint = (0.693/in vitro T1/2) × (incubation volume/mg of microsomal protein) × (mg of microsomal protein/gram of liver) × (gram of liver/kg body weight). Each time-point group includes two repeats.
Plasma protein binding
The plasma protein binding assay was conducted with the Rapid Equilibrium Dialysis (RED) (Wang and Williams, 2013). 3 μM FB23-2 was added to rat plasma, which were vortexed well before placing 100 μl in the red chamber of the RED device (Thermo, USA). 300 μl of PBS (pH 7.4) was added to the corresponding white chamber, the base plate was covered with a gas-permeable membrane, and then incubated in a CO2 incubator (MCO-18AC, Panasonic, Japan) set at 250 rpm and 37 °C for 4 hr. Samples collected from both chambers were analyzed with LC-MS/MS to determine the compound concentrations (C0 - initial concentration, Cf - ultrafiltrate and Cp - plasma). Percentage of protein binding (PPB) using the RED device was calculated with the formula, PPB (%) = (1- Cf /Cp) × 100%. The experiments were performed in triplicate.
H&E staining
The vital organs from BALB/c mice for toxicity studies and spleen or liver samples from xeno-transplanted AML mice were fixed in 4% formaldehyde solution, which were subsequently processed and embedded in paraffin wax, and serially sectioned at 4 – 7 μm thickness. Each section of rat tissues was mounted on a glass slide, then observed, photographed, and analyzed using hematoxylin and eosin (H&E) staining.
Wright-Giemsa staining
1 × 105 leukemia cells isolated from xenograft mice BM when the mice became moribund were suspended in 100 μl PBS and performed with cytospin at 500 rpm for 5 min. After air dry, the slides were placed in May-Grunwald Stain (Sigma) and washed with PBS for 5 min, respectively, followed by incubation in Giemsa Stain (EMD) solution (diluted 1:20 with ddH2O) for another 20 min. Then the slides were rinsed gently with ddH2O and dried for evaluations in conventional light-field microscopy using an optical microscope (CKX53, Olympus, Japan).
Colony formation with primary cells isolated from AML patients
Human methylcellulose complete media (HSC003, R&D systems) was selected to determine the function of FB23-2 on colony forming capacity in AML patient cells according to the manufacturer’s instruction. Briefly, 20,000 primary cells, with or without FB23-2 treatment, were suspended in 200 μl suspension buffer and seeded in 35 mm culture dishes with 1.5 ml methylceulose media. The cells were cultured under humid atmosphere in incubator for 12 days and the colony numbers were determined.
Synthesis of FTO inhibitors
As shown in Supplementary Figure S1B, starting with 2-iodobenzoic acid 1, copper-catalysed Ullman-coupling reaction between 1 and 4-bromo-2,6-dichloroaniline 2 provided 2-((4-bromo-2,6-dichlorophenyl)amino) benzoic acid 3. The ethyl 2-((4-bromo-2,6-dichlorophenyl)amino)benzoate 4 was obtained by acid-prompted esterification. Suzuki-coupling reaction of 4 and (3,5-dimethylisoxazol-4-yl)boronic acid 5 catalysed by Pd(dppf)Cl2 yielded ethyl 2-((2,6-dichloro-4-(3,5-dimethylisoxazol-4-yl)phenyl)amino) benzoate 6. Hydrolysis of 6 under alkaline conditions gave 2-((2,6-dichloro-4-(3,5-dimethylisoxazol-4-yl) phenyl) amino)benzoic acid FB23. Condensation of acid chloride of FB23 with hydroxylamine yielded the target compound FB23-2.
General methods
All solvents and reagents were purchased from commercial sources and used as received. 1H NMR and 13C NMR spectra were recorded with a Varian-MERCURY Plus-400 NMR spectrometer at room temperature. NMR spectra were calibrated to the solvent signals of DMSO-d6 or CDCl3. Chemical shifts are reported in ppm (δ scale) as referenced to TMS and coupling constant (J) values are reported in hertz (Hz). Data are presented as follows: chemical shift, multiplicity (s = singlet, d = doublet, dd = doublet of doublet, t = triplet, q = quartet, m = multiplet, br = broad), coupling constant in Hz, and integration. Low-resolution electrospray ionization mass spectrometry (LRESI) was recorded on a Finnigan LCQ/DECA spectrometer, and high-resolution electrospray ionization mass spectrometry (HRESI) was recorded on a Micromass Ultra Q-TOF spectrometer. Flash and column chromatography were performed using silica gel (230 – 400 mesh). Analytical TLC was performed on silica gel plates and visualized under ultraviolet light (254 nm).
Synthesis of 2-((4-bromo-2,6-dichlorophenyl)amino)benzoic acid (3)
The mixture of 2-iodobenzoic acid 1 (30 g, 120 mmol), 4-bromo-2,6-dichloroaniline 2 (24 g, 100 mmol), and N-methylmorpholine (15.2 g, 150 mmol) in DMF (500 ml) was stirred at 150 °C for 24 hr in the presence of anhydrous Cu(OAc)2 (9 g, 5 mmol) under N2 atmosphere. After the consumption of 2-iodobenzoic acid 1 monitored by TLC, 500 ml water was added to the reaction system, adjusting pH to 3.0 using 2.0 M diluted HCl. The precipitates were collected by filtration and purified by silica chromatography to provide product 3 (9.8 g, 27%). 1H NMR (400 MHz, DMSO-d6) δ 13.20 (s, 1H), 9.47 (s, 1H), 7.92 – 8.00 (2H), 7.90 (m, 1H), 7.33 (t, J = 7.8 Hz, 1H), 6.81 (t, J = 7.2 Hz, 1H), 6.25 (dd, J = 7.2, 2.8 Hz, 1H). 13C NMR (126 MHz, DMSO-d6) δ 170.43, 147.09, 137.49, 135.09, 134.91, 134.67, 132.01, 119.49, 118.21, 113.60, 112.53; LRESI m/z (% relative abundance) 360.0 ([M-H]−, 100).
Synthesis of ethyl 2-((4-bromo-2,6-dichlorophenyl)amino) benzoate (4)
To an ice-cooled solution of compound 3 (3.6 g, 10 mmol) in ethanol (200 ml) was added concentrated H2SO4 (20 ml) slowly. The reaction mixture was refluxed for 72 hr. The reaction mixture was condensed via vacuum. The residue was re-dissolved in water, adjusted pH to 10.0 using saturated Na2CO3 solution and extracted with ethyl acetate (50 ml × 3). The combined organic phase was washed with brine (10 ml × 3), dried over anhydrous Na2SO4. Then the filtrate was evaporated and purified through silica chromatography, giving compound 4 as a white solid (3.1 g, 80% yield). 1H NMR (400 MHz, CDCl3) δ 9.40 (s, 1H), 8.04 (d, J = 8.0 Hz, 1H), 7.60 – 7.77 (2H), 7.36 – 7.25 (m, 1H), 6.82 (t, J = 7.6 Hz, 1H), 6.36 (m, 1H), 4.42 (q, J = 7.2 Hz, 2H), 1.45 (t, J = 7.8 Hz, 3H). 13C NMR (126 MHz, DMSO-d6) δ 168.13, 146.87, 137.51, 135.24, 134.83, 132.05, 131.52, 119.62, 118.38, 113.88, 111.95, 61.22, 14.59. LREI m/z (% relative abundance) 389 ([M]+, 100).
Synthesis of ethyl 2-((2,6-dichloro-4-(3,5-dimethylisoxazol-4-yl)phenyl)amino) benzoate (6)
A mixture of compound 4 (3.1 g, 8 mmol), (3,5-dimethylisoxazol-4-yl)boronic acid 5 (1.36 g, 9.6 mmol), anhydrous K2CO3 (1.68 g, 12 mmol), and Pd(dppf)Cl2 (584 mg, 0.8 mmol) in co-solvent of 1,4-dioxane and water (60 ml, 4:1/v:v) was stirred at 100 °C for 24 hr under N2 atmosphere. The reaction mixture was evaporated and subjected directly to silica chromatography, providing compound 6 as a white solid (1.98 g, 61% yield). 1H NMR (500 MHz, CDCl3) δ 9.49 (s, 1H), 8.05 (dd, J = 8.0, 1.5 Hz, 1H), 7.37 – 7.33 (m, 1H), 7.32 (s, 2H), 6.86 – 6.81 (m, 1H), 6.44 (d, J = 7.5 Hz, 1H), 4.43 (q, J = 7.0 Hz, 2H), 2.49 (s, 3H), 2.34 (s, 3H), 1.46 (t, J = 7.0 Hz, 3H); 13C NMR (151 MHz, CDCl3) δ 168.52, 165.96, 158.21, 146.80, 134.76, 134.07, 133.78, 131.40, 129.70, 129.08, 117.80, 114.34, 113.87, 112.29, 60.79, 14.30, 11.71, 10.82; LREI m/z (% relative abundance) 404 ([M]+, 100).
Synthesis of 2-((2,6-dichloro-4-(3,5-dimethylisoxazol-4-yl) phenyl)amino)benzoic acid (FB23)
To an ice-cooled solution of compound 6 (1.78 g, 4.4 mmol) in THF (36 ml) and EtOH (72 ml) was added a solution of NaOH (880 mg, 22 mmol) in H2O (9 ml) slowly. Then the reaction mixture was stirred at 45 °C overnight. After the consumption of compound 6, the mixture was concentrated via vacuum to afford a suspension. The suspension was acidified to pH 3.0 by 2.0 M HCl, filtered and the cake was washed with water (20 ml), and dried to give compound FB23 as a white solid (1.6 g, 96% yield). 1H NMR (400 MHz, DMSO-d6) δ 13.21 (s, 1H), 9.62 (s, 1H), 7.92 (d, J = 8.0 Hz, 1H), 7.68 (s, 2H), 7.37 (t, J = 8.0 Hz, 1H), 6.82 (t, J = 7.6 Hz, 1H), 6.35 (d, J = 8.0 Hz, 1H), 2.47 (s, 3H), 2.29 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 170.51, 166.73, 158.56, 147.33, 134.60, 134.41, 133.98, 131.98, 130.60, 129.77, 118.13, 114.18, 113.78, 112.52, 11.87, 10.81; LRESI m/z (% relative abundance) 375.1 ([M-H]−, 100); HRESI m/z calcd for C18H13Cl2N2O3 [M-H]− 375.0303, found 375.0315.
Synthesis of 2-((2,6-dichloro-4-(3,5-dimethylisoxazol-4-yl) phenyl)amino)-N-hydroxybenzamide (FB23-2)
A solution of compound FB23 (760 mg, 2.0 mmol) in SOCl2 (1.60 ml, 22 mmol) with catalytic amount of anhydrous DMF (two drops) was stirred at 65 °C for 2 hr. The mixture was concentrated and dried via vacuum to give acid chloride intermediate as a yellow solid, which was used without further purification. To a solution of hydroxylamine (660 mg, 10 mmol) and TEA (400 mg, 4 mmol) in anhydrous THF (30 ml) was added dropwise a solution of acid chloride intermediate of FB23 (2 mmol) in anhydrous THF (20 ml) at 0 °C. The resulting mixture was stirred at 25 °C for 1 hr. The mixture was evaporated via vacuum to afford a crude residue. The residue was purified by prep-HPLC (Phenomenex Synergi Max-RP column, 250 × 50 mm × 10 μm) to give FB23-2 as an off-white solid (218 mg, 28% yield). 1H NMR (600 MHz, DMSO-d6) δ 11.32 (s, 1H), 9.53 (s, 1H), 9.18 (s, 1H), 7.62 (s, 2H), 7.51 (d, J = 7.2 Hz, 1H), 7.26 (t, J = 7.8 Hz, 1H), 6.79 (t, J = 7.2 Hz, 1H), 6.34 (d, J = 8.4 Hz, 1H), 2.44 (s, 3H), 2.26 (s, 3H); 13C NMR (126 MHz, DMSO-d6) δ 166.90, 166.63, 158.58, 144.71, 134.86, 132.59, 132.16, 129.78, 129.52, 128.27, 118.59, 116.08, 114.29, 114.20, 11.87, 10.81; LRESI m/z (% relative abundance) 390.1 ([M-H]−, 100); HRESI m/z calcd for C18H14Cl2N3O3 [M-H]− 390.0418, found 390.0420.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data were analyzed with GraphPad Prism 5 and were presented as mean ± SEM or mean ± SD as indicated. Two-tailed Student’s t-test was used to compare means between groups as indicated and p < 0.05 was considered significant. For dot blot and western blot results, representative figures from two or three biological replicates were shown.
DATA AND SOFTWARE AVAILABILITY
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB, www.pdb.org) under accession ID code 6AKW for the structure of FTO/FB23. The crystal structure of compound FB23-2 has been deposited in the Cambridge Crystallography Data Centre under deposition number CCDC 1558332. The RNA-seq and m6A-seq data have been deposited in Gene expression Ominibus (GEO) repository with the accession numbers GSE103494, GSE103495, and GSE103496.
Supplementary Material
Table S4. List of differentially regulated core-enriched genes, Related to Figure 5.
Significance.
As the most abundant internal mRNA modification, m6A impacts various biological processes. As a major m6A demethylase, FTO is overexpressed in certain subtypes of AMLs and promotes leukemogenesis. Thus, the development of effective inhibitors to target FTO’s aberrant m6A demethylase activity is urgently needed for leukemia therapy. Here we report two selective FTO inhibitors that efficiently reverse/suppress FTO-mediated aberrant epitranscriptome in AML cells and significantly inhibit AML progression in vivo. Our studies provide the proof-of-concept evidence demonstrating that small-molecule inhibitors targeting oncogenic FTO represent a promising targeted therapeutic strategy for the effective treatment of AML. Moreover, given the overexpression of FTO in various cancers, our work may have a broad impact on cancer therapy by targeting the FTO-mediated epitranscriptome.
Acknowledgments
We thank all staff for data collection support from BL17U1 and BL18U1 beamline at Shanghai Synchrotron Radiation Facility. This work was supported in part by the National Natural Science Foundation of China (21725801 to C.-G.Y., 21807103 to Z.D., and 91753000 to H.J.), the National Key R&D Program of China (2016YFA0501500 to C.-G.Y.), the Science and Technology Commission Shanghai Municipality (18YF1428500 to Y.H. and 18431907100 to H.J.), China Postdoctoral Science Foundation (2018M640434 to Y.H., 2017M621570 to Z.D., and 2018M632187 to T.Z.), the Chinese Academy of Sciences (XDA12020359 to C.-G.Y.), and the National Institutes of Health RO1 Grants (CA214965, CA211614, and CA178454 to C.J., HL131444, and DK107615 to Z.Q.). J.C. is a Leukemia & Lymphoma Society (LLS) Scholar. We thank Dr. Chuan He at the University of Chicago for constructive comments, and S.F. Reichard, MA for contributing editing.
Footnotes
Declaration of Interests
Y.H., R.S., Z.D., J.C., and C.-G.Y. are named inventors of pending patent applications (CN201810172219.8 and PCT/CN2018/077789, to the Chinese Patent Office) related to the work described.
References
- Aik W, Demetriades M, Hamdan MK, Bagg EA, Yeoh KK, Lejeune C, Zhang Z, McDonough MA, and Schofield CJ (2013). Structural basis for inhibition of the fat mass and obesity associated protein (FTO). J. Med. Chem 56, 3680–3688. [DOI] [PubMed] [Google Scholar]
- Akichika S, Hirano S, Shichino Y, Suzuki T, Nishimasu H, Ishitani R, Sugita A, Hirose Y, Iwasaki S, Nureki O, and Suzuki T (2019). Cap-specific terminal N (6)-methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 363. [DOI] [PubMed] [Google Scholar]
- Alarcon CR, Lee H, Goodarzi H, Halberg N, and Tavazoie SF (2015). N6-methyladenosine marks primary microRNAs for processing. Nature 519, 482–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri I, Tzelepis K, Pandolfini L, Shi J, Millan-Zambrano G, Robson SC, Aspris D, Migliori V, Bannister AJ, Han N, et al. (2017). Promoter-bound METTL3 maintains myeloid leukaemia by m(6)A-dependent translation control. Nature 552, 126–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaschke K, Ebata KT, Karimi MM, Zepeda-Martínez JA, Goyal P, Mahapatra S, Tam A, Laird DJ, Hirst M, Rao A, et al. (2013). Vitamin C induces Tet-dependent DNA demethylation and a blastocyst-like state in ES cells. Nature 500, 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokar JA, Shambaugh ME, Polayes D, Matera AG, and Rottman FM (1997). Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6-adenosine)-methyltransferase. RNA 3, 1233–1247. [PMC free article] [PubMed] [Google Scholar]
- Chen B, Ye F, Yu L, Jia G, Huang X, Zhang X, Peng S, Chen K, Wang M, Gong S, et al. (2012). Development of cell-active N6-methyladenosine RNA demethylase FTO inhibitor. J. Am. Chem. Soc 134, 17963–17971. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project, N. (1994). The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cui Q, Shi H, Ye P, Li L, Qu Q, Sun G, Sun G, Lu Z, Huang Y, Yang CG, et al. (2017). m6A RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 18, 2622–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle SR, Olhava EJ, Therkelsen CA, Majer CR, Sneeringer CJ, Song J, Johnston LD, Scott MP, Smith JJ, Xiao Y, et al. (2011). Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell 20, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Su R, Feng X, Wei M, and Chen J (2018a). Role of N(6)-methyladenosine modification in cancer. Curr. Opin. Genet. Dev 48, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Su R, Weng H, Huang H, Li Z, and Chen J (2018b). RNA N(6)-methyladenosine modification in cancers: current status and perspectives. Cell Res. 28, 507–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di L, Kerns EH, Li SQ, and Petusky SL (2006). High throughput microsomal stability assay for insoluble compounds. Int. J. Pharm 317, 54–60. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Salmon-Divon M, Amariglio N, and Rechavi G (2013). Transcriptome-wide mapping of N(6)-methyladenosine by m(6)A-seq based on immunocapturing and massively parallel sequencing. Nat. Protoc 8, 176–189. [DOI] [PubMed] [Google Scholar]
- Dominissini D, Moshitch-Moshkovitz S, Schwartz S, Salmon-Divon M, Ungar L, Osenberg S, Cesarkas K, Jacob-Hirsch J, Amariglio N, Kupiec M, et al. (2012). Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 485, 201–206. [DOI] [PubMed] [Google Scholar]
- Esteller M, and Pandolfi PP (2017). The Epitranscriptome of Noncoding RNAs in Cancer. Cancer Discov. 7, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng C, Liu Y, Wang G, Deng Z, Zhang Q, Wu W, Tong Y, Cheng C, and Chen Z (2014). Crystal structures of the human RNA demethylase Alkbh5 reveal basis for substrate recognition. J. Biol. Chem 289, 11571–11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Jia G, Pang X, Wang RN, Wang X, Li CJ, Smemo S, Dai Q, Bailey KA, Nobrega MA, et al. (2013). FTO-mediated formation of N6-hydroxymethyladenosine and N6-formyladenosine in mammalian RNA. Nat. Commun 4, 1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fustin JM, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, Isagawa T, Morioka MS, Kakeya H, Manabe I, and Okamura H (2013). RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155, 793–806. [DOI] [PubMed] [Google Scholar]
- Gerken T, Girard CA, Tung YC, Webby CJ, Saudek V, Hewitson KS, Yeo GS, McDonough MA, Cunliffe S, McNeill LA, et al. (2007). The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 318, 1469–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geula S, Moshitch-Moshkovitz S, Dominissini D, Mansour AA, Kol N, Salmon-Divon M, Hershkovitz V, Peer E, Mor N, Manor YS, et al. (2015). m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science 347, 1002–1006. [DOI] [PubMed] [Google Scholar]
- He C (2010). Grand challenge commentary: RNA epigenetics? Nat. Chem. Biol 6, 863–865. [DOI] [PubMed] [Google Scholar]
- He W, Zhou B, Liu W, Zhang M, Shen Z, Han Z, Jiang Q, Yang Q, Song C, Wang R, et al. (2015). Identification of A Novel Small-Molecule Binding Site of the Fat Mass and Obesity Associated Protein (FTO). J. Med. Chem 58, 7341–7348. [DOI] [PubMed] [Google Scholar]
- Hsu PJ, Zhu Y, Ma H, Guo Y, Shi X, Liu Y, Qi M, Lu Z, Shi H, Wang J, et al. (2017). Ythdc2 is an N(6)-methyladenosine binding protein that regulates mammalian spermatogenesis. Cell Res. 27, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Weng H, Sun W, Qin X, Shi H, Wu H, Zhao BS, Mesquita A, Liu C, Yuan CL, et al. (2018). Recognition of RNA N(6)-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nat. Cell. Biol 20, 285–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yan J, Li Q, Li J, Gong S, Zhou H, Gan J, Jiang H, Jia GF, Luo C, and Yang CG (2015). Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 43, 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Fu Y, Zhao X, Dai Q, Zheng G, Yang Y, Yi C, Lindahl T, Pan T, Yang YG, and He C (2011). N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol 7, 885–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia G, Yang CG, Yang S, Jian X, Yi C, Zhou Z, and He C (2008). Oxidative demethylation of 3-methylthymine and 3-methyluracil in single-stranded DNA and RNA by mouse and human FTO. FEBS Lett. 582, 3313–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knuckles P, Lence T, Haussmann IU, Jacob D, Kreim N, Carl SH, Masiello I, Hares T, Villasenor R, Hess D, et al. (2018). Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m(6)A machinery component Wtap/Fl(2)d. Genes Dev. 32, 415–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Chen YS, Ping XL, Yang X, Xiao W, Yang Y, Sun HY, Zhu Q, Baidya P, Wang X, et al. (2017a). Cytoplasmic m(6)A reader YTHDF3 promotes mRNA translation. Cell Res. 27, 444–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Huang Y, Liu X, Gan J, Chen H, and Yang CG (2016). Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage. J. Biol. Chem 291, 11083–11093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Weng H, Su R, Weng X, Zuo Z, Li C, Huang H, Nachtergaele S, Dong L, Hu C, et al. (2017b). FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell 31, 127–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichinchi G, Zhao BS, Wu Y, Lu Z, Qin Y, He C, and Rana TM (2016). Dynamics of Human and Viral RNA Methylation during Zika Virus Infection. Cell Host Microbe 20, 666–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Dai Q, Zheng G, He C, Parisien M, and Pan T (2015). N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 518, 560–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobera M, Madauss KP, Pohlhaus DT, Wright QG, Trocha M, Schmidt DR, Baloglu E, Trump RP, Head MS, Hofmann GA, et al. (2013). Selective class IIa histone deacetylase inhibition via a nonchelating zinc-binding group. Nat. Chem. Biol 9, 319–325. [DOI] [PubMed] [Google Scholar]
- Lomenick B, Hao R, Jonai N, Chin RM, Aghajan M, Warburton S, Wang J, Wu RP, Gomez F, Loo JA, et al. (2009). Target identification using drug affinity responsive target stability (DARTS). Proc. Natl. Acad. Sci. U S A 106, 21984–21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez Molina D, Jafari R, Ignatushchenko M, Seki T, Larsson EA, Dan C, Sreekumar L, Cao Y, and Nordlund P (2013). Monitoring drug target engagement in cells and tissues using the cellular thermal shift assay. Science 341, 84–87. [DOI] [PubMed] [Google Scholar]
- Mauer J, Luo X, Blanjoie A, Jiao X, Grozhik AV, Patil DP, Linder B, Pickering BF, Vasseur JJ, Chen Q, et al. (2017). Reversible methylation of m6Am in the 5’ cap controls mRNA stability. Nature 541, 371–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray F, Demetriades M, Aik W, Merkestein M, Kramer H, Andrew DS, Scudamore CL, Hough TA, Wells S, Ashcroft FM, et al. (2015). Pharmacological inhibition of FTO. PLoS One 10, e0121829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Lu Z, Liu H, Zhang L, Zhang S, Chen Y, Rao MK, and Huang Y (2014). A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods 69, 274–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, and Jaffrey SR (2015). 5’ UTR m(6)A Promotes Cap-Independent Translation. Cell 163, 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer KD, Saletore Y, Zumbo P, Elemento O, Mason CE, and Jaffrey SR (2012). Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 149, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Padariya M, and Kalathiya U (2016). Structure-based design and evaluation of novel N-phenyl-1H-indol-2-amine derivatives for fat mass and obesity-associated (FTO) protein inhibition. Comput. Biol. Chem 64, 414–425. [DOI] [PubMed] [Google Scholar]
- Ping XL, Sun BF, Wang L, Xiao W, Yang X, Wang WJ, Adhikari S, Shi Y, Lv Y, Chen YS, et al. (2014). Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 24, 177–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana AP, and Tuck MT (1990). Analysis and in vitro localization of internal methylated adenine residues in dihydrofolate reductase mRNA. Nucleic Acids Res. 18, 4803–4808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose NR, McDonough MA, King ON, Kawamura A, and Schofield CJ (2011). Inhibition of 2-oxoglutarate dependent oxygenases. Chem. Soc. Rev 40, 4364–4397. [DOI] [PubMed] [Google Scholar]
- Roundtree IA, Evans ME, Pan T, and He C (2017a). Dynamic RNA Modifications in Gene Expression Regulation. Cell 169, 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roundtree IA, Luo G-Z, Zhang Z, Wang X, Zhou T, Cui Y, Sha J, Huang X, Guerrero L, Xie P, et al. (2017b). YTHDC1 mediates nuclear export of N6-methyladenosine methylated mRNAs. eLife 6, e31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller R, Scozzafava G, Tumber A, Wickens JR, Bush JT, Rai G, Lejeune C, Choi H, Yeh TL, Chan MC, et al. (2014). A cell-permeable ester derivative of the JmjC histone demethylase inhibitor IOX1. ChemMedChem 9, 566–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Wang X, Lu Z, Zhao BS, Ma H, Hsu PJ, Liu C, and He C (2017). YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 27, 315–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortt J, Ott CJ, Johnstone RW, and Bradner JE (2017). A chemical probe toolbox for dissecting the cancer epigenome. Nat. Rev. Cancer 17, 160–183. [DOI] [PubMed] [Google Scholar]
- Su R, Dong L, Li C, Nachtergaele S, Wunderlich M, Qing Y, Deng X, Wang Y, Weng X, Hu C, et al. (2018). R-2HG Exhibits Anti-tumor Activity by Targeting FTO/m(6)A/MYC/CEBPA Signaling. Cell 172, 90–105 e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, and Mesirov JP (2005). Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Zhang M, Li K, Bai D, and Yi C (2019). Cap-specific, terminal N(6)-methylation by a mammalian m(6)Am methyltransferase. Cell Res. 29, 80–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svensen N, and Jaffrey SR (2016). Fluorescent RNA Aptamers as a Tool to Study RNA-Modifying Enzymes. Cell Chem. Biol 23, 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toh JDW, Sun L, Lau LZM, Tan J, Low JJA, Tang CWQ, Cheong EJY, Tan MJH, Chen Y, Hong W, et al. (2015). A strategy based on nucleotide specificity leads to a subfamily-selective and cell-active inhibitor of N6-methyladenosine demethylase FTO. Chem. Sci 6, 112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuck MT (1992). The formation of internal 6-methyladenine residues in eucaryotic messenger RNA. Int. J. Biochem 24, 379–386. [DOI] [PubMed] [Google Scholar]
- Vane JR, Bakhle YS, and Botting RM (1998). Cyclooxygenases 1 and 2. Annu. Rev. Pharmacol. Toxicol 38, 97–120. [DOI] [PubMed] [Google Scholar]
- Vu LP, Pickering BF, Cheng Y, Zaccara S, Nguyen D, Minuesa G, Chou T, Chow A, Saletore Y, MacKay M, et al. (2017). The N(6)-methyladenosine (m(6)A)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med 23, 1369–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, and Williams NS (2013). A mass balance approach for calculation of recovery and binding enables the use of ultrafiltration as a rapid method for measurement of plasma protein binding for even highly lipophilic compounds. J. Pharm. Biomed. Anal 75, 112–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Chang J, Varghese D, Dellinger M, Kumar S, Best AM, Ruiz J, Bruick R, Peña-Llopis S, Xu J, et al. (2013). A small molecule modulates Jumonji histone demethylase activity and selectively inhibits cancer growth. Nat. Commun 4, 2035–2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Hong T, Huang Y, Su H, Wu F, Chen Y, Wei L, Huang W, Hua X, Xia Y, et al. (2015a). Fluorescein Derivatives as Bifunctional Molecules for the Simultaneous Inhibiting and Labeling of FTO Protein. J. Am. Chem. Soc 137, 13736–13739. [DOI] [PubMed] [Google Scholar]
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, Fu Y, Parisien M, Dai Q, Jia G, et al. (2014a). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 505, 117–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, Weng X, Chen K, Shi H, and He C (2015b). N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 161, 1388–1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li Y, Toth JI, Petroski MD, Zhang Z, and Zhao JC (2014b). N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat. Cell. Biol 16, 191–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1975). Methylated nucleotides block 5’ terminus of HeLa cell messenger RNA. Cell 4, 379–386. [DOI] [PubMed] [Google Scholar]
- Wei CM, Gershowitz A, and Moss B (1976). 5’-Terminal and internal methylated nucleotide sequences in HeLa cell mRNA. Biochemistry 15, 397–401. [DOI] [PubMed] [Google Scholar]
- Wei J, Liu F, Lu Z, Fei Q, Ai Y, He PC, Shi H, Cui X, Su R, Klungland A, et al. (2018). Differential m(6)A, m(6)Am, and m(1)A Demethylation Mediated by FTO in the Cell Nucleus and Cytoplasm. Mol. Cell 71, 973–985 e975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng H, Huang H, Wu H, Qin X, Zhao BS, Dong L, Shi H, Skibbe J, Shen C, Hu C, et al. (2018). METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m(6)A Modification. Cell Stem Cell 22, 191–205 e199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wunderlich M, Chou FS, Link KA, Mizukawa B, Perry RL, Carroll M, and Mulloy JC (2010). AML xenograft efficiency is significantly improved in NOD/SCID-IL2RG mice constitutively expressing human SCF, GM-CSF and IL-3. Leukemia 24, 1785–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Laurent B, Hsu CH, Nachtergaele S, Lu Z, Sheng W, Xu C, Chen H, Ouyang J, Wang S, et al. (2017). RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature 543, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, et al. (2016). Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol. Cell 61, 507–519. [DOI] [PubMed] [Google Scholar]
- Yan F, Al-Kali A, Zhang Z, Liu J, Pang J, Zhao N, He C, Litzow MR, and Liu S (2018). A dynamic N(6)-methyladenosine methylome regulates intrinsic and acquired resistance to tyrosine kinase inhibitors. Cell Res. 28, 1062–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Milasta S, Hu D, AlTahan AM, Interiano RB, Zhou J, Davidson J, Low J, Lin W, Bao J, et al. (2017). Targeting Histone Demethylases in MYC-Driven Neuroblastomas with Ciclopirox. Cancer Res. 77, 4626–4638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Chen M, Huang H, Zhu J, Song H, Zhu J, Park J, and Ji SJ (2018). Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 46, 1412–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Yang Y, Sun BF, Shi Y, Yang X, Xiao W, Hao YJ, Ping XL, Chen YS, Wang WJ, et al. (2014). FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 24, 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Cox T, Tribbey L, Wang GZ, Iacoban P, Booher ME, Gabriel GJ, Zhou L, Bae N, Rowles J, et al. (2014). Synthesis of a FTO inhibitor with anticonvulsant activity. ACS Chem. Neurosci. 5, 658–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Dahl JA, Niu Y, Fedorcsak P, Huang CM, Li CJ, Vagbo CB, Shi Y, Wang WL, Song SH, et al. (2013). ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 49, 18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Wan J, Gao X, Zhang X, Jaffrey SR, and Qian SB (2015). Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature 526, 591–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. List of differentially regulated core-enriched genes, Related to Figure 5.
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB, www.pdb.org) under accession ID code 6AKW for the structure of FTO/FB23. The crystal structure of compound FB23-2 has been deposited in the Cambridge Crystallography Data Centre under deposition number CCDC 1558332. The RNA-seq and m6A-seq data have been deposited in Gene expression Ominibus (GEO) repository with the accession numbers GSE103494, GSE103495, and GSE103496.