A set of AP2-like genes regulate rice axillary meristem determination and evolved during the African and Asian domestications. These genes are new targets for crop improvement.
Keywords: AP2/EREBP-like genes, development, domestication, inflorescence, rice
Abstract
Rice yield is influenced by inflorescence size and architecture, and inflorescences from domesticated rice accessions produce more branches and grains. Neither the molecular control of branching nor the developmental differences between wild and domesticated rice accessions are fully understood. We surveyed phenotypes related to branching, size, and grain yield across 91 wild and domesticated African and Asian accessions. Characteristics related to axillary meristem identity were the main phenotypic differences between inflorescences from wild and domesticated accessions. We used whole transcriptome sequencing in developing inflorescences to measure gene expression before and after the transition from branching axillary meristems to determinate spikelet meristems. We identified a core set of genes associated with axillary meristem identity in Asian and African rice, and another set associated with phenotypic variability between wild and domesticated accessions. AP2/EREBP-like genes were enriched in both sets, suggesting that they are key factors in inflorescence branching and rice domestication. Our work has identified new candidates in the molecular control of inflorescence development and grain yield, and provides a detailed description of the effects of domestication on phenotype and gene expression.
Introduction
Rice produces grains on complex raceme inflorescences called panicles, which consist of a series of branches of different orders. The rachis is the main axis, and primary, secondary, and tertiary branches form higher-order axes (see Supplementary Fig. S1 at JXB online). Grain yield in rice is linked to inflorescence branching, because the number of spikelets produced on the higher-order branches determines the number of grains per panicle (Doebley et al., 2006; Xing and Zhang, 2010; Olsen and Wendel, 2013). Inflorescence size and architecture are key targets in selective breeding for improved rice grain yield and quality (Doust, 2007; Wang and Li, 2011), and these characteristics are different between domesticated and wild accessions.
Inflorescence branching and the number of grains per inflorescence vary between clades, species, and accessions of rice in the genus Oryza (Yamaki et al., 2010; Taguchi-Shiobara et al., 2011). Asian and African clades diverged around 2 million years ago and were domesticated independently (Zhu and Ge, 2005). Domesticated Asian rice, Oryza sativa, diverged 10 000 years ago via a complex domestication process from the wild Asian rice species, Oryza rufipogon. The O. sativa species complex involves a network of subspecies, including O. sativa ssp. indica and O. sativa ssp. japonica (Huang et al., 2012; Choi et al., 2017; Stein et al., 2018). Domesticated African rice, Oryza glaberrima, has a simpler history. It was domesticated from its wild relative, Oryza barthii, 3000 years ago, without subsequent introgression (Vaughan et al., 2008; Cubry et al., 2018). In both African and Asian clades, domesticated species produce inflorescences that are more complex and ramified than those of their wild relatives (Linares, 2002; Yamaki et al., 2010; Ta et al., 2017).
Panicle structure is established early after floral transition. The vegetative shoot meristem develops into a reproductive rachis meristem, which produces axillary meristems until its abortion. The axillary meristems on the rachis all develop as primary branches, which themselves produce a variable number of axillary meristems. Axillary meristems on the primary branches can differentiate into secondary branches, some of which may also produce tertiary branch meristems. Axillary meristems on the primary branches can also differentiate directly into spikelet meristems. Finally, all axillary meristems and terminal meristems differentiate gradually from the top to the base of the panicle into spikelet meristems and then florets. Each rice grain is produced from a single spikelet (reviewed by Itoh et al., 2005).
The complexity of panicle branching and the number of grains are determined by two basic developmental outcomes: the number of axillary meristems produced along all panicle axes; and the rate of meristem fate transition, which determines whether an axillary meristem grows into a higher-order branch or differentiates into a spikelet and gives rise to a grain. The maturation rate and identity of reproductive and axillary meristems are controlled by transcriptional regulators, some of which have been identified in O. sativa (Xing and Zhang, 2010; Wang and Li, 2011). Genes including G1 LIKE PROTEIN 5 (TAWAWA1/TAW1), LAX PANICLE 1 (LAX1), and LAX PANICLE 2 (LAX2) control the branching phase via establishment and activity of indeterminate meristems (Komatsu et al., 2001; Tabuchi et al., 2011; Yoshida et al., 2013). Genes such as SUPERNUMERARY BRACT GENE (SNB), FRIZZY PANICLE (FZP), and LEAFY HULL STERILE 1 (LHS1) regulate the transition from indeterminate meristems to determinate spikelet meristems and the subsequent transition from spikelet to floret meristem (Jeon et al., 2000; Komatsu et al., 2001; Agrawal et al., 2005; Chen et al., 2006; Lee et al., 2007; Khanday et al., 2013). Regulatory changes in some genes, including FZP, WEALTHY FARMER’S PANICLE (IDEAL PLANT ARCHITECTURE 1, WFP/IPA1/SPL14), GRAIN SIZE 2 (G22/GRF4), and GRF6, are also associated with modified panicle phenotype (Jiao et al., 2010; Miura et al., 2010; Bai et al., 2016; Duan et al., 2015; Huang et al., 2018).
Despite advances in understanding molecular mechanisms that regulate panicle branching in rice (Furutani et al., 2006; Harrop et al., 2016), and characterization of individual genes associated with variation in panicle branching (Bai et al., 2012; Ikeda et al., 2013), the transcriptional control of axillary meristem identity that underlies the phenotypic diversity of branching across rice species is not fully understood. The independent domestications of African and Asian rice in the genus Oryza provide a comparative context to study the evolution of agronomic traits such as panicle architecture, the mechanisms underlying parallel evolution of phenotype, and the basic molecular control of inflorescence branching. Here, we phenotyped panicles from domesticated and wild accessions of Asian and African rice, and used whole-transcriptome sequencing (RNAseq) to reveal gene expression patterns associated with the diversity of inflorescence architecture. Our analysis reveals an association between expression of AP2/EREBP-like genes, panicle architecture and domestication.
Materials and methods
Plant material and growth conditions
Panicle morphological traits were measured in 91 accessions of O. rufipogon, O. sativa, O. glaberrima, and O. barthii, grown in Cali, Colombia and Montpellier, France (Supplementary Table S1). At panicle maturity, we collected the three main panicles from three plants per accession, per replicate (i.e. 18 panicles per accession). We used four accessions for expression analysis: O. sativa ssp. indica IR64, O. rufipogon W1654, O. glaberrima Tog5681, and O. barthii B88. These accessions were grown in a greenhouse in Montpellier, France, in June 2014, under long day conditions (14 h light–10 h dark). After 6–8 weeks they were transferred to short day conditions (11 h light–13 h dark) to induce flowering. To confirm panicle phenotypes in the growth conditions used for RNAseq, we evaluated panicle traits for nine panicles from each accession, which were grown in the greenhouse under the same growth conditions. The crl5 and smos1-3 mutants (Kitomi et al., 2011; Aya et al., 2014) were grown in a greenhouse in Montpellier, France, in October 2017 under short day conditions (11 h light–13 h dark). At least 18 panicles were used for panicle phenotyping. All greenhouse plants were grown at 28 °C with 80% relative humidity. For phenotyping analyses, each panicle was spread out and fixed on white paper using adhesive tape. Panicles were photographed and the images were used for panicle structure and seed number analysis with P-TRAP software (AL-Tam et al., 2013).
Tissue collection and RNA sequencing
For expression analysis, we collected 15 immature panicles each from at least 10 plants per accession, per stage, collected from 4 to 15 d after floral induction (i.e. transfer to short day conditions) for each biological replicate. For sample collection, leaves surrounding the young panicle were removed by hand and the reproductive tissue was cut with a sharp blade under a Stemi 508 (Zeiss, Germany) stereo microscope to identify developmental stage. The reproductive tissues were immediately frozen in liquid nitrogen, and total RNA including small RNA was extracted using the RNeasy Plant Mini kit with RLT and RWT buffers (Qiagen, Germany). DNase treatments were performed using the RNase-free DNase set (Qiagen, Germany). RNA integrity numbers of the extracted RNA, measured using a 2100 Bioanalyzer (Agilent, USA), were between 8.6 and 10. Stage specificity was validated with quantitative real-time RT-PCR (qPCR) using stage-specific marker genes (Supplementary Table S2); 400 ng of total RNA was used for each sample for RNAseq library preparation with the TruSeq Stranded Total RNA with Ribo-Zero Plant kit (Illumina, USA). After quantification with a 2100 Bioanalyzer, 125-base paired-end reads were generated on a HiSeq 2500 (Illumina) by the GeT platform (Toulouse, France).
qPCR
cDNA was synthesized from 1 μg of DNase-treated total RNA using the SuperScript III First-Strand Synthesis System (Invitrogen, USA). A Biomark HD Microfluidic Dynamic Array (Fluidigm, USA) was used for large-scale qPCR. A 96×96 Dynamic Array Integrated Fluidic Circuit (Fluidigm, USA) was loaded with cDNA and primer combinations after 15 cycles of specific target amplification and exonuclease I treatment. A fast cycling protocol with EvaGreen dye (Bio-Rad Laboratories, USA) was used for amplification. Three biological replicates were performed for each sample. Data were normalized using four genes (LOC_Os06g11170, LOC_Os06g48970, LOC_Os01g16970, and LOC_Os03g61680). Gene expression relative to the normalization factors was estimated using the 2-∆∆CT method without a calibrator sample (Livak and Schmittgen, 2001). Primer sequences are listed in Supplementary Table S2.
Data analysis
We trimmed reads and removed adaptors with cutadapt (Martin, 2011), before mapping to the MSU v7 annotation of the Oryza sativa ssp. japonica cv. Nipponbare reference genome (Ouyang et al., 2007) using STAR in two-pass mode (Dobin et al., 2013). To generate per-library gene expression cutoffs, we used the 95th percentile of reads that mapped to intergenic regions of the genome, as described previously (Harrop et al., 2016). We used DESeq2 (Love et al., 2014) for differential expression analysis of genes that passed the cutoff. We used annotations from PlnTFDB v3.0 (Pérez-Rodríguez et al., 2010) and PlantTFDB v4.0 (Jin et al., 2017) to analyse expression of transcription factors. Soft clustering of transcription factor genes was performed with Mfuzz (Kumar and Futschik, 2007), and enrichment of transcription factor family genes was tested with the GSEA method using the FGSEA package (Subramanian et al., 2005; Sergushichev, 2016, Preprint).
Reproducibility and data availability
Raw sequence data are hosted at the National Center for Biotechnology Information Sequence Read Archive (NCBI SRA) under accession PRJNA518559. The code we used to analyse the RNAseq data and panicle phenotype data is hosted at https://github.com/tomharrop/5acc, and the code for qPCR analysis is at https://github.com/othomantegazza/5acc-qpcr. We used snakemake (Köster and Rahmann, 2012) to arrange analysis steps into workflows and monitor dependencies, and Singularity (Kurtzer et al., 2017) to capture the computing environment. The final results and all intermediate steps can be exactly reproduced from the raw data with a single command using snakemake and Singularity. The source for this manuscript is hosted at https://github.com/tomharrop/ird-5acc-paper.
Results
Parallel changes in panicle architecture between wild and domesticated accessions
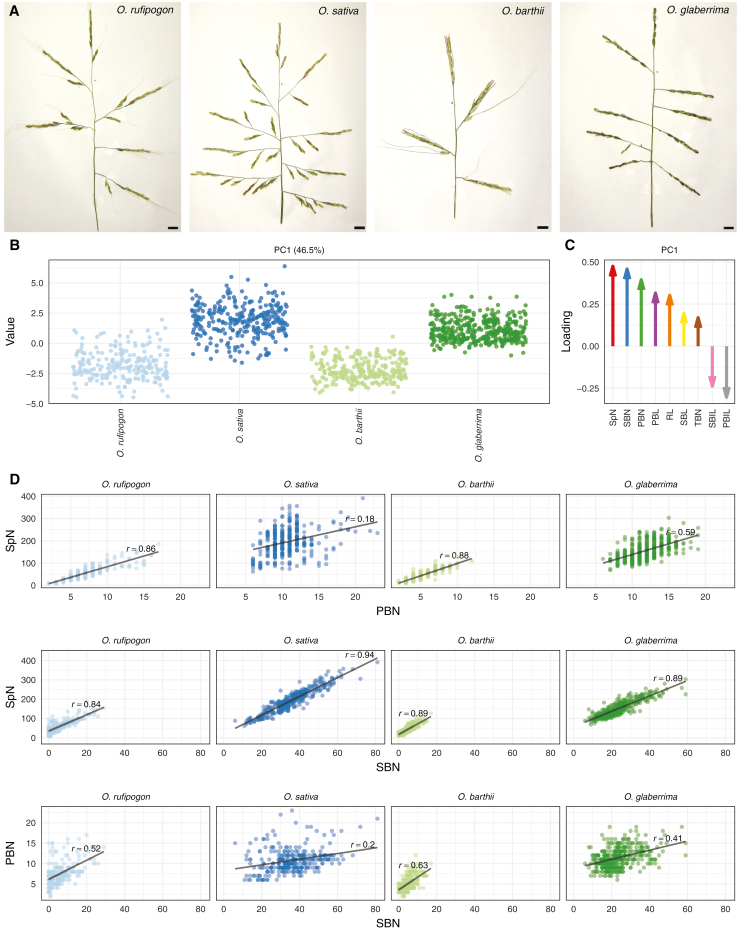
To measure the diversity of panicle architecture, we phenotyped 91 rice accessions (Supplementary Table S1), including wild Asian rice (Oryza rufipogon), domesticated Asian rice (Oryza sativa), wild African rice (Oryza barthii), and domesticated African rice (Oryza glaberrima), using P-TRAP image analysis software for automated measurement of traits (Fig. 1A; Supplementary Fig. S1; Supplementary Tables S1, S3; AL-Tam et al., 2013). Principal components analysis (PCA) of the phenotyping data identified a major coordinate (PC1) that accounts for 46.5% of variability (Fig. 1B). PC1 separates domesticated and wild accessions, but not Asian and African accessions, and is the only component that separates panicles from different accessions (Supplementary Fig. S2). Spikelet number, secondary branch number, and primary branch number have the highest loadings on PC1, whereas length traits have lower absolute loading on PC1 (Fig. 1C). For all species, spikelet number correlates more with secondary branch number than it does with primary branch number. Primary branch number correlates with spikelet number more in wild than in domesticated species, but this correlation is weaker in Asian species than in African species. Primary and secondary branch numbers do not correlate, suggesting they are controlled by different genetic mechanisms (Fig. 1D). Our phenotypic analysis indicates similar changes in panicle architecture between wild and domesticated accessions in the independent African and Asian domestication processes, which presumably result from parallel, artificial selection on panicle architecture. Spikelet number, secondary branch number and primary branch number are the main contributors to these differences in panicle architecture, and these phenotypes are all related to axillary meristem formation and fate transition (Teo et al., 2014; Zhang and Yuan, 2014).
Fig. 1.
Panicle complexity of 91 rice accessions. The main component of variablity in panicle phenotypes splits accessions by domestication status, and is related to spikelet number, secondary branch number, and primary branch number. (A) We measured traits using spread panicles from O. rufipogon, O. sativa, O. barthii, and O. glaberrima. (B) The first principal component (PC1) in the panicle phenotype data accounts for 46.5% of variability and separates wild and domesticated accessions independently of continent. (C) Spikelet number (SpN), secondary branch number (SBN), and primary branch number (PBN) have the highest loadings on PC1. (D) Correlation between the main panicle traits that contribute to panicle architecture diversity. Primary branch number and spikelet number correlate in wild species. Secondary branch number and spikelet number correlate more in cultivated species than in wild species. Primary and secondary branch numbers do not correlate. PBIL, primary branch internode length; PBL, primary branch length; RL, rachis length; SBIL, secondary branch internode length; SBL, secondary branch length; TBN, tertiary branch number.
Measurement of gene expression in developing panicles
We investigated gene expression changes associated with the diversity of panicle architecture and differences between the Asian and African domestication processes via RNAseq. We used a single accession each of domesticated Asian rice (O. sativa ssp. indica IR64) and its wild relative (O. rufipogon W1654), and domesticated African rice (O. glaberrima Tog5681) and its wild relative (O. barthii B88). Based on the extensive phenotyping described above, the chosen accessions are consistent with species-wide patterns of panicle architecture (Supplementary Fig. S3). To measure whole-transcriptome gene expression in these accessions, we collected immature panicles from each accession at four developmental stages: rachis meristem (RM); indeterminate meristem (IM), including panicles displaying primary branch initiation, elongation of primary branches, and axillary meristem initiation; determinate meristem (DM), including panicles wherein axillary meristems had transitioned into early spikelet differentiation; and floret meristem (FM), with early differentiation of floral organs (Supplementary Fig. S4A). We first confirmed staging of the panicles by extracting RNA from pooled immature panicles at each stage and measuring expression of markers of panicle development by quantitative real-time RT-PCR (qPCR) (Supplementary Fig. S4B). Because branching complexity is related to branch meristem establishment and meristem fate transition (Kyozuka et al., 2014), and secondary branch number and spikelet number contribute to differences between wild and domesticated accessions (Fig. 1), we chose the IM and DM stages for RNAseq. cDNA libraries for sequencing were constructed from rRNA-depleted RNA samples from three biological replicates at both stages for all four accessions. Using the O. sativa ssp. japonica cv. Nipponbare reference genome to map reads (Ouyang et al., 2007), we obtained an average of more than 20 million uniquely mapped reads within exons for each accession, including African rice species (Supplementary Table S4). Our analysis was limited to transcripts from the four studied accessions that have homologs in the reference annotation that are similar enough for reads to map unambiguously. To allow for differences in mapping between accessions, we compared genes using between-stage read count differences within accessions, rather than read count differences between accessions. Pairwise distances between samples, calculated on the number of reads per gene from all detected genes, grouped samples first by stage, then by accession, and then by continent (Supplementary Fig. S5). We did not observe grouping by domestication status, suggesting that transcriptome-wide changes during domestication are subtle compared with differences between species.
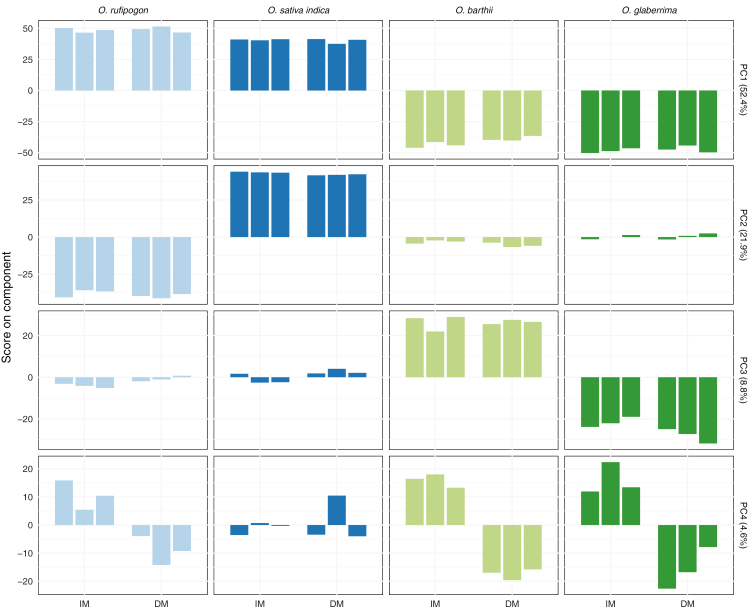
We used PCA on transformed raw counts to investigate general patterns of variation in the transcriptomes (Fig. 2). PC1 separates African and Asian accessions, and PC2 and PC3 separate wild and domesticated accessions in Asian and African samples, respectively. PC1–PC3 may relate to species-specific differences unrelated to panicle architecture, or mapping biases introduced by mapping all libraries against the O. sativa ssp. japonica reference. While the first three PCs separate different combinations of accessions, PC4 separates developmental stages across all four accessions, although separation is weaker in O. sativa ssp. indica. This suggests a conserved mechanism that controls the transition from indeterminate to determinate phase of axillary meristem development in all accessions.
Fig. 2.
Principal components analysis of transformed read counts for each library. Principal component 4 (PC4) separates RNAseq samples by developmental stage, and explains 4.6% of total variability. The first three components explain 83.1% of variability, and separate RNAseq samples by species. Bars show single samples (three replicates per accession per stage).
AP2/EREBP-like transcription factors are core regulators of panicle branching
To identify the core set of genes involved in axillary meristem determination in all four accessions, we used differential expression (DE) tests to find genes that were up- or down-regulated between stages across all accessions. Positive log2-fold change values (L2FCs) indicate higher expression in DM than in IM. There were 153 genes that had at least 1.5-fold DE between stages in all species at a false-discovery rate of 0.1, including 88 genes up-regulated in DM samples and 65 genes down-regulated in DM samples (Supplementary Table S5). There was an enrichment of transcription factor (TF) genes in the list of 153 differentially expressed genes (37 TF genes; P=6.0×10−12, hypergeometric test), including LHS1, LAX1, PANICLE PHYTOMER2 (PAP2), and MOSAIC FLORAL ORGANS 1 (MFO1), which regulate inflorescence architecture or meristem fate transition in rice (Komatsu et al., 2001; Ohmori et al., 2009; Kobayashi et al., 2010, 2012; Khanday et al., 2013). This indicates that RNAseq of developing panicles at the IM and DM stage identifies genes that control branching, and suggests that transcription factors are prominent among these genes.
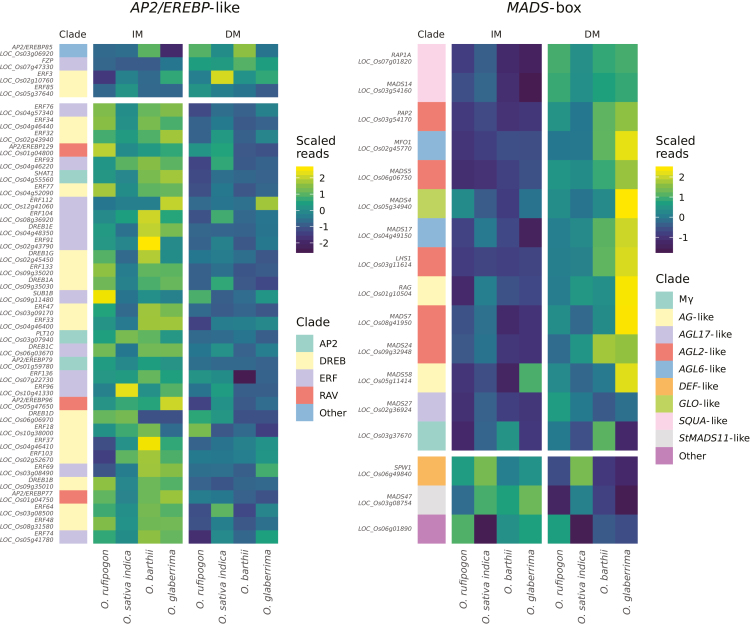
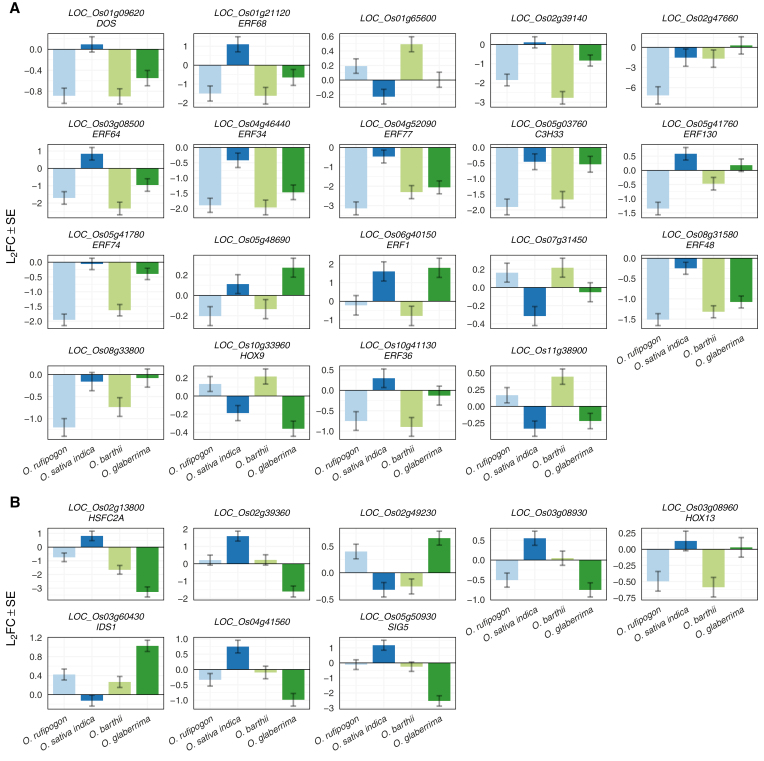
We used gene set enrichment analysis (GSEA; Subramanian et al., 2005; Sergushichev, 2016, Preprint) on genes ranked by L2FC to test for family-level enrichment among transcription factors. AP2/EREBP-like genes and MADS-box genes were both enriched (Padj=3.5×10−5 and Padj=4.0×10−4, respectively, GSEA permutation t-test; Supplementary Table S6). Correspondingly, the list of 153 differentially expressed genes included 10 MADS-box genes and seven AP2/EREBP-like genes (Supplementary Table S5). Most differentially expressed MADS-box genes are more highly expressed at the DM stage, whereas most differentially expressed AP2/EREBP-like genes are more highly expressed at the IM stage. The majority of AP2/EREBP-like genes that have higher expression at the IM stage are from the ERF and DREB clades (Fig. 3). The DE results are consistent with the role of transcriptional regulation in panicle branching, and highlight a set of candidate core regulators of axillary meristem determination and branching that are conserved between rice species. The pattern of expression of AP2/EREBP-like genes may indicate a role in the promotion of indeterminate axillary meristem identity or suppression of the transition from axillary meristem to spikelet meristem. MADS-box genes may have an inverse role as promoters of determinate meristem. Co-regulation of members of TF families, sometimes at the clade level, highlights the redundant or overlapping functions of TF families in meristem establishment and fate transition.
Fig. 3.
AP2/EREBP-like and MADS-box transcription factors change expression between IM and DM. For each family, we plotted genes that were in the top 10% of all genes by absolute L2FC between IM and DM, without filtering on adjusted P-value. Genes in the upper panels had higher expression at the DM stage, whilst genes in the lower panels had higher expression at the IM stage. Most AP2/EREBP-like genes that pass the 10% cutoff were more highly expressed in the IM. AP2/EREBP-like genes that were more highly expressed in IM mainly belong to the ERF and DREB clades. In contrast, most MADS-box genes that pass the cutoff were more highly expressed at the DM stage. Clades for AP2/EREBP-like genes are from the Plant Transcription Factor Database v4.0 (Jin et al., 2017) and Sharoni et al. (2011). MADS-box clades were manually tabulated from Arora et al. (2007).
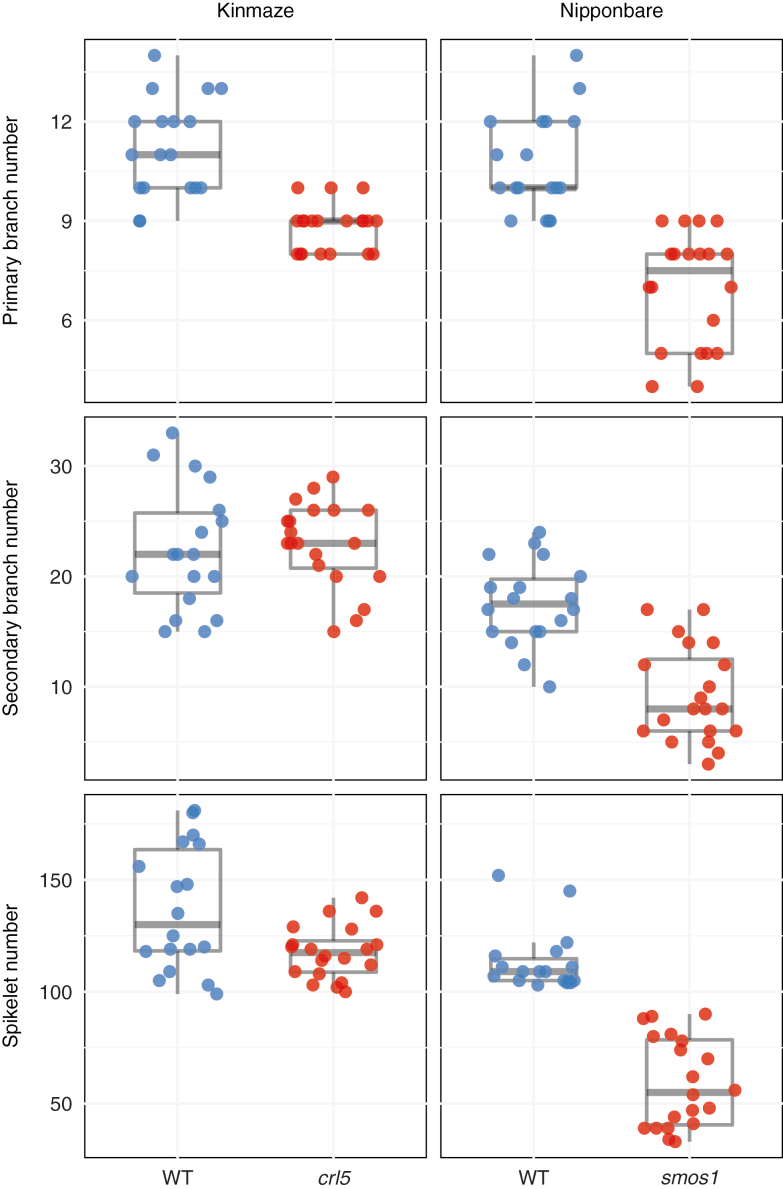
To test the role of AP2/EREBP-like genes in the control of panicle architecture, we phenotyped panicles from two loss-of-function mutants. The crown rootless5 (crl5) mutant of the AP2-like gene PLETHORA 8 (PLT8; Kitomi et al., 2011) produced panicles with a shorter rachis with fewer primary branches (Fig. 4; Supplementary Table S7), consistent with a peak of PLT8 expression in rachis meristem tissues from O. sativa ssp. japonica (Supplementary Fig. S6; Harrop et al., 2016). Panicles from the small organ size1 (smos1) mutant of ERF142 (Aya et al., 2014) have a reduced number of primary and secondary branches, and fewer spikelets (Fig. 4; Supplementary Table S7). ERF142 expression is highest in primary branch and elongating primary branch meristem tissues in O. sativa ssp. japonica (Supplementary Fig. S6; Harrop et al., 2016). Although neither gene was differentially expressed in our RNAseq dataset, these phenotypes support the involvement of AP2/EREBP-like genes in control of panicle architecture.
Fig. 4.
Mutants in two AP2/EREBP-like genes, PLT8 and ERF142, have defects in panicle architecture compared with their background accessions. The crl5 mutant of PLT8 (LOC_Os07g03250) produced fewer primary branches and spikelets, and the smos1-3 mutant of ERF142 (LOC_Os05g32270) produced fewer primary branches, secondary branches, and spikelets.
AP2/EREBP-like gene expression is associated with panicle diversity and domestication
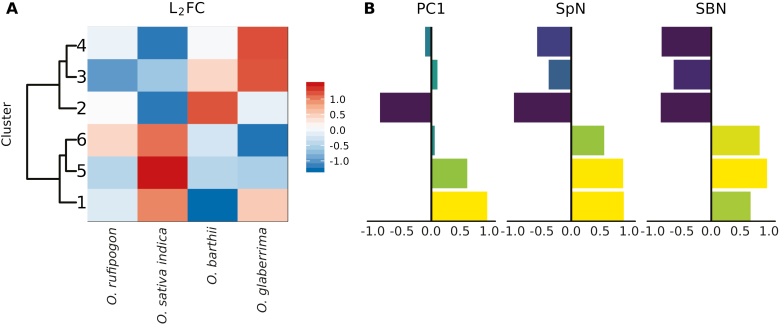
To identify common patterns of expression of transcriptional regulators related to variation in panicle phenotypes, we used soft clustering of scaled L2FCs between IM and DM on the subset of annotated TF genes that were detected in our RNAseq dataset. We recovered six clusters comprising a total of 119 genes (Fig. 5; Supplementary Table S8). To determine which clusters were related to domestication, we calculated correlations between the mean L2FC value of genes in each cluster and PC1 in the phenotyping data, and between mean L2FC and the number of secondary branches and spikelets from repeat panicle phenotyping for the accessions used for RNAseq (Figs 1, 5B; Supplementary Fig. S7).
Fig. 5.
Gene expression clusters correlate with the main component of diversity of panicle architecture (PC1) and the number of secondary branches (SBN) and spikelets (SpN). Clusters contained 19–31 genes each (Supplementary Table S8). (A) Mean, scaled log2-fold change (L2FC) of genes by cluster and accession. (B) Pearson correlation with PC1, SBN, and SpN. PC1 is the main principal component that separates panicles from domesticated and wild accessions of rice (Fig. 1). Correlations with SBN and SpN are based on repeat panicle phenotyping for the accessions used for RNAseq in greenhouse conditions (Supplementary Fig. S8; Supplementary Table S10).
Clusters 3, 4, and 6 correlated with spikelet number (SpN) and secondary branch number (SBN), but not with PC1, meaning that the L2FC of genes in those clusters does not correlate with the phenotypic differences between wild and domesticated accessions. Clusters of genes with high L2FC in O. sativa ssp. indica have a positive correlation with SBN and SpN, whereas clusters of genes with low L2FC in O. sativa ssp. indica have a negative correlation with SBN and SpN. Cluster 4 had an enrichment of HB genes (9 out of 31 genes; Padj=2.5×10−4). It also contained three MIKCC-type MADS-box genes (LHS1, MFO1, and MADS14), which promote spikelet meristem determination (Jeon et al., 2000; Ohmori et al., 2009; Kobayashi et al., 2012), and three AP2/EREBP-like genes including OsINDETERMINATE SPIKELET 1 (OsIDS1), which also controls inflorescence architecture (Chuck et al., 2008; Lee and An, 2012). L2FCs of genes in this cluster are low in O. sativa ssp. indica, high in O. glaberrima and intermediate in the two wild accessions. Although these genes may be involved in regulation of panicle complexity, their expression did not appear to have changed in parallel in the two domestications. L2FCs of genes in clusters 3 and 6 change between accessions from the two continents. In cluster 3, L2FCs are higher in African species than in Asian species, meaning that the genes are more highly expressed in DM stages in African species. Genes in cluster 6 have the opposite pattern, with higher L2FCs in Asian species compared with African species. Cluster 3 contained LAX1 and ABERRANT PANICLE ORGANIZATION 2 (FLO-LFY HOMOLOG OF RICE, AP02/RFL), which are involved in axillary meristem establishment and outgrowth and promotion of indeterminate meristematic activity in rice, respectively (Komatsu et al., 2001; Ikeda-Kawakatsu et al., 2012). Their higher expression at the DM stage in panicles from both wild and domesticated African accessions could be associated with a reduced number of spikelets.
Clusters 1, 2, and 5 correlated with the main principal component (PC1) in the phenotyping data, which separates wild and domesticated species independently of continent. Clusters 1 and 5 are also positively correlated with spikelet number and secondary branch number, whereas cluster 2 is negatively correlated. The correlation of L2FC patterns with PC1 suggests that genes in these clusters may be associated with changes in panicle architecture between wild and domesticated species. Cluster 1 and cluster 5 both had a positive correlation with PC1. L2FCs are higher in domesticated accessions for genes in cluster 1, meaning that they are more highly expressed at the DM stage in domesticated accessions. Genes in cluster 2 have lower L2FCs in domesticated accessions, meaning that they are more highly expressed at the DM stage in wild accessions. Cluster 2 also had the strongest negative correlation with PC1, and low L2FCs in O. sativa ssp. indica. The lower L2FCs in domesticated accessions could implicate cluster 2 genes in promotion of determinate meristem fate, because their lower expression at the DM stage may result in more activity of indeterminate axillary meristems. Eight GROWTH-REGULATING FACTOR1 (GRF) family genes, which are involved in the regulation of cell proliferation (Kim and Tsukaya, 2015), were detected in our dataset, and three of them were present in cluster 2 (Padj=7.6×10−3, hypergeometric test). In contrast to cluster 2, most of the genes in cluster 5 have L2FCs close to zero in O. sativa ssp. indica, and negative L2FCs in the other accessions (Supplementary Fig. S8), meaning that the expression of these genes decreases at the DM stage in all accessions except O. sativa ssp. indica. The lack of repression of cluster 5 genes and to a lesser extent cluster 1 genes at the DM stage in O. sativa ssp. indica could result in more branching via the promotion of indeterminate axillary meristem identity. Cluster 5, which had the highest mean L2FC in O. sativa ssp. indica, had an enrichment of AP2/EREBP-like genes (6 out of 17 genes; Padj=7.6×10−3, hypergeometric test), and cluster 1 also contains 3 AP2/EREBP-like genes (Supplementary Table S8). We used qPCR to confirm these patterns in all four stages of each accession for all AP2/EREBP-like genes in cluster 5 (Supplementary Fig. S9). The enrichment of AP2/EREBP-like genes in cluster 5 and the presence of three AP2/EREBP-like genes in cluster 1 suggest that their pattern of expression is associated with differences in panicle architecture across wild and domesticated accessions.
To find TF genes associated with parallel changes in panicle architecture during domestication, we tested the stage×domestication interaction for O. rufipogon, O. sativa ssp. indica, O. barthii and O. glaberrima at a false discovery rate of 0.1 (Supplementary Table S9). We detected 19 genes with a stage×domestication interaction, including nine AP2/EREBP-like genes (P=4.4×10−7, hypergeometric test; Fig. 6A). These genes are putative targets of parallel selection on panicle architecture that occurred during domestication. AP2/EREBP-like genes were also prominent when we tested the stage×accession interaction separately for each domestication (12 out of 85 genes in Asian accessions; 8 out of 50 genes in African accessions; Supplementary Table S9). Consistent with its presence in cluster 4, OsIDS1 was also differentially expressed in both Asian and African domestications, although the direction of change was different (Fig. 6B). Genes with this pattern of expression in the four accessions may have also been targets of selection on panicle architecture, but evolved divergently.
Fig. 6.
Parallel and divergent evolution of gene expression associated with domestication. (A) Expression of genes with a stage×domestication interaction when all four accessions were tested together. We used this test to identify genes where the change in L2FC between indeterminate (IM) and determinate (DM) stages changed in the same direction in both African and Asian domesticated accessions. (B) Expression of genes with a stage×domestication interaction when tested separately for African and Asian accessions. These genes have divergent changes between wild and domesticated accessions. The genes plotted in (B) had an interaction in both of the separate tests, but not in the single test used to identify the genes in (A).
The prominence of AP2/EREBP-like genes among putative core regulators of branching in all four Oryza species, and among genes associated with differences between wild and cultivated accessions, suggest that they were key targets of artificial selection for improvement in panicle architecture, and were involved in changes to the regulatory network controlling branching that occurred during domestication.
Discussion
The purpose of this study was to identify genetic factors underlying the diversity of panicle architecture, which influences grain number in rice. Our transcriptomic comparison of panicles at indeterminate and determinate stages of axillary meristem development revealed a core set of transcription factors associated with axillary meristem phase transition in wild and domesticated African and Asian rice (Fig. 2). Some of these transcription factors, including AP2/EREBP-like and MIKCC-type MADS-box genes, appear to be co-regulated at the family or clade level (Fig 3). Our phenotypic survey of 91 accessions showed that characteristics related to axillary meristem formation and fate transition are the main factors separating wild and domesticated rice (Fig. 1). Three clusters of gene expression correlated with the major component of phenotypic variability between wild and domesticated accessions, containing enrichments of GRF and AP2/EREBP-like transcription factor genes (Fig. 5). We observed a correlation between expression of AP2/EREBP-like transcription factors, domestication status, and derived phenotypes, suggesting that expression of these genes has changed as a result of artificial selection during domestication. As well as basic insights into molecular control of branching in rice, this work provides an overview of the outcome of the domestication process at phenotypic and whole-transcriptome levels.
A set of AP2/EREBP-like genes decrease in expression over the course of wheat spike development (Li et al., 2018), and the molecular function of some individual AP2/EREBP-like genes has been reported in relation to inflorescence or root development. FZP represses the formation of axillary meristem and induces transition from spikelet to floral meristem (Komatsu et al., 2001). Along with the AP2/EREBP-like gene FZP, AP2/EREBP85, ERF3, and ERF85 were more highly expressed at the DM stage across all four accessions, consistent with a role in the regulation of axillary meristem identity (Fig. 3). Conversely, most of the AP2/EREBP-like genes that change expression between stages had lower expression in DM than in IM, including mainly ERF and DREB clade genes and three RAV-like genes. In Arabidopsis, RAV orthologs repress flowering genes (Matías-Hernández et al., 2014), and ERF3 interacts with the HB gene WOX11 to promote crown root development (Zhao et al., 2015). Our results suggest additional roles for these genes in promoting reproductive axillary meristem initiation. This family- and clade-level view of their expression suggests that AP2/EREBP-like play a central role in the regulation of phase transition from indeterminate to determinate state.
AP2/EREBP-like genes are also associated with domestication. The AP2-like wheat domestication gene Q regulates inflorescence and glume shape and spike length, and has variation in binding sites for microRNA miR172 between wild and domesticated species (Simons et al., 2006; Debernardi et al., 2017; Greenwood et al., 2017). Recently, characterization of a quantitative trait locus associated with grain yield and panicle branching and variation in the promoter region showed that FZP regulates panicle architecture and is associated with rice domestication (Bai et al., 2017; Fujishiro et al., 2018; Huang et al., 2018). Our analysis indicates a modified expression pattern of some AP2/EREBP-like genes between wild and domesticated rice accessions. One of the clusters of genes with expression patterns that correlate to the main phenotypic differences between wild and domesticated accessions contained an enrichment of AP2/EREBP-like genes. In addition, nine out of the 19 genes with a stage×domestication interaction across the entire dataset were AP2/EREBP-like genes (Fig. 6A). Eight of these nine genes have lower expression in panicles at the DM stage, and may be involved in promotion of indeterminate axillary meristem and/or suppression of the transition from axillary meristem to spikelet meristem. Among these genes, ERF130 (MULTI-FLORET SPIKELET1, MFS1) regulates the timing of the transition of spikelet meristems to terminal spikelets and positively regulates the expression of OsIDS1 and SNB (Ren et al., 2013). Taken with the roles of AP2 mutants in modification of rice panicle architecture (this study; Komatsu et al., 2001; Lee and An, 2012), our results implicate several AP2/EREBP-like genes as putative targets of artificial selection during rice domestication resulting in parallel evolution of expression.
OsIDS1 controls panicle branching in rice, and positively regulates branch meristem identity by repressing genes that specify spikelet identity (Lee and An, 2012). We found a stage×domestication interaction in expression of OsIDS1, but the change was divergent between African and Asian accessions (Fig. 6B). This suggests that even if genes are involved in similar regulatory networks, their expression may diverge during selection. Parallel morphological evolution during Asian and African rice domestication is sometimes associated with different genomic modifications (Furuta et al., 2015; Cubry et al., 2018; Hu et al., 2018; Lv et al., 2018), illustrating that the interaction between genetic variation and the diverse selection pressures associated with domestication can result in diverse genomic outcomes.
Our interspecific analysis showed that parallel domestication of African and Asian rice resulted in similar modifications in panicle architecture, with changes in branch number contributing to derived phenotypes more than branch length traits (Fig. 1B, C). Secondary branch number correlated with spikelet number in all species, but primary branch number only correlates with spikelet number in wild species (Fig. 1D). Yield improvements seem to have occurred mainly through increased ramification of branching, rather than, for example, elongation of the primary axis. This suggests that increased production of secondary branches from axillary meristems rather than direct differentiation into spikelets was important in both domestications.
The molecular control of phase transition from indeterminate to determinate axillary meristems is not fully understood in rice panicle development. Microarray analysis of developing panicles has identified a small set of differentially expressed genes, enriched for TFs (Furutani et al., 2006). In microdissected meristem tissues, gene expression changes gradually during transition in axillary meristem identity (Harrop et al., 2016), similar to the gradual meristem maturation during tomato inflorescence development (Park et al., 2012). We also observed a small set of genes that were differentially expressed between IM and DM consistently in all accessions (153 of 25 229 tested), with an enrichment of TFs (37 of 153 genes). More genes were upregulated in DM than down-regulated, as in wheat spike development (Wang et al., 2017). These results suggest that the gene regulatory network controlling phase transition from IM to DM is controlled by a small subset of core, conserved genes in Asian and African rice.
Because of redundant and overlapping activities of TFs, gene regulatory networks are robust to perturbations in single genes, allowing them to produce a stable transcriptional output in variable cellular and environmental conditions (Gitter et al., 2009; Dai et al., 2009; Wu and Lai, 2015). AP2/EREBP-like and MADS-box genes were enriched among differentially expressed genes. Expression of AGL6-like, AP1/FUL-like, and four of the five SEPALLATA-like MADS-box genes all increased between indeterminate and determinate stage, as expected given their roles in spikelet determination and inflorescence development (Yoshida and Nagato, 2011; Zhang and Yuan, 2014). This suggests family-level or clade-level co-regulation of TFs in rice panicle development. Our observation that some genes appear to be co-expressed at the family or clade level may indiciate partial functional redundancy between groups of homologous TFs in the rice genome that act in the gene regulatory network controlling axillary meristem determination.
This work has revealed the core set of genes that are associated with the determination of axillary meristem identity across Asian and African rice accessions. We have also discovered candidate targets of artificial selection on panicle phenotype during domestication. In particular, the interspecific expression pattern of AP2/EREBP-like genes suggests that they regulate axillary meristem determination and have evolved during domestication. Along with efforts to characterize the molecular function of the candidate genes we have identified and to measure their expression in other accessions, further investigation of the gene regulatory network controlling meristem identity in developing panicles would continue to drive an understanding of the process of inflorescence development in rice and its relationship to grain yield.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Spread mature rice panicle.
Fig. S2. Principal component analysis (PCA) of panicle phenotyping data showing components 1–4.
Fig. S3. The accessions used for RNAseq are consistent with species-wide patterns of panicle architecture.
Fig. S4. Early stages of rice panicle development used for gene expression analysis.
Fig. S5. Heatmap of pairwise distances between RNAseq samples.
Fig. S6. Expression of AP2/EREBP-like genes in O. sativa spp. japonica cv. Nipponbare meristems (data from Harrop et al., 2016).
Fig. S7. Phenotyping of the five Oryza accessions used for RNAseq.
Fig. S8. Most genes in cluster 5 have negative L2FCs between IM and DM in O. rufipogon, O. barthii, and O. glaberrima, but L2FCs in O. sativa spp. indica are closer to zero.
Fig. S9. Expression analysis along early panicle development of AP2/EREBP-like genes present in cluster 5.
Table S1. Rice accessions used in this study.
Table S2. Sequences of primers used.
Table S3. Quantification of panicle traits in 91 accessions from wild and domesticated Asian and African rice species.
Table S4. Read and mapping statistics for all RNAseq samples.
Table S5. Differential expression test results between stages across all species; we used an arbitrary differential expression threshold of 1.5-fold change in expression and adjusted P-value (false discovery rate) less than 0.1.
Table S6. Transcription factor family enrichment by L2FC.
Table S7. Quantification of panicle traits from crl5 and smos1 mutants.
Table S8. Clustered genes.
Table S9. Differential expression test results for the stage×accession interaction in Asian and African accessions.
Table S10. Detailed quantification of panicle traits from rice accessions used for sequencing analysis.
Funding
This research was funded by Agropolis Foundation through the Investissements d’avenir programme (ANR-10-LABX-0001-01), Fondazione Cariplo (EVOREPRICE 1201-004), and the CGIAR Research Program on Rice. We also received support from the Excellence Scholarship Program of the Embassy of France in Vietnam (LAM).
Author contributions
HA and SJ designed the research with input from TWRH and OM; HA, TWRH, OM, KB, and AML performed the research; TWRH, OM, HA, and ML performed data analysis, collection, and interpretation; TWRH, SJ, and HA wrote the manuscript.
Acknowledgements
Dr Ko Hirano and Prof. Yoshiaki Inukai from Nagoya University, Japan, kindly provided smos1-3 and crl5 mutant seeds, respectively. We thank Lady Johanna Arbelaez Rivera (CIAT, Colombia) for plant care and Céline Cardi and Hélène Vignes (Grand plateau technique régional de génotypage, CIRAD, Montpellier) for support with high-throughput qPCR and cDNA library preparation. Sophie Chéron and Harold Chrestin assisted with plant care and phenotyping, and Christine Tranchant-Dubreuil, Axel Verdier, and the IRD bioinformatics platform assisted with technical support in Montpellier. We also thank James Tregear for feedback on the manuscript and Alain Ghesquière for discussion on the project.
References
- Agrawal GK, Abe K, Yamazaki M, Miyao A, Hirochika H. 2005. Conservation of the E-function for floral organ identity in rice revealed by the analysis of tissue culture-induced loss-of-function mutants of the OsMADS1 gene. Plant Molecular Biology 59, 125–135. [DOI] [PubMed] [Google Scholar]
- Al-Tam F, Adam H, Anjos Ad, Lorieux M, Larmande P, Ghesquière A, Jouannic S, Shahbazkia HR. 2013. P-TRAP: a panicle trait phenotyping tool. BMC Plant Biology 13, 122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S. 2007. MADS-box gene family in rice: genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8, 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. 2014. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant & Cell Physiology 55, 897–912. [DOI] [PubMed] [Google Scholar]
- Bai X, Huang Y, Hu Y, Liu H, Zhang B, Smaczniak C, Hu G, Han Z, Xing Y. 2017. Duplication of an upstream silencer of FZP increases grain yield in rice. Nature Plants 3, 885–893. [DOI] [PubMed] [Google Scholar]
- Bai X, Huang Y, Mao D, Wen M, Zhang L, Xing Y. 2016. Regulatory role of FZP in the determination of panicle branching and spikelet formation in rice. Scientific Reports 6, 19022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Wu B, Xing Y. 2012. Yield-related QTLs and their applications in rice genetic improvement. Journal of Integrative Plant Biology 54, 300–311. [DOI] [PubMed] [Google Scholar]
- Chen ZX, Wu JG, Ding WN, Chen HM, Wu P, Shi CH. 2006. Morphogenesis and molecular basis on Naked Seed Rice, a novel homeotic mutation of OsMADS1 regulating transcript level of AP3 homologue in rice. Planta 223, 882–890. [DOI] [PubMed] [Google Scholar]
- Choi JY, Platts AE, Fuller DQ, Hsing YI, Wing RA, Purugganan MD. 2017. The rice paradox: multiple origins but single domestication in Asian rice. Molecular Biology and Evolution 34, 969–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuck G, Meeley R, Hake S. 2008. Floral meristem initiation and meristem cell fate are regulated by the maize AP2 genes ids1 and sid1. Development 135, 3013–3019. [DOI] [PubMed] [Google Scholar]
- Cubry P, Tranchant-Dubreuil C, Thuillet AC, et al. 2018. The rise and fall of African rice cultivation revealed by analysis of 246 new genomes. Current Biology 28, 2274–2282.e6. [DOI] [PubMed] [Google Scholar]
- Dai Z, Dai X, Xiang Q, Feng J. 2009. Robustness of transcriptional regulatory program influences gene expression variability. BMC Genomics 10, 573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debernardi JM, Lin H, Chuck G, Faris JD, Dubcovsky J. 2017. microRNA172 plays a crucial role in wheat spike morphogenesis and grain threshability. Development 144, 1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley JF, Gaut BS, Smith BD. 2006. The molecular genetics of crop domestication. Cell 127, 1309–1321. [DOI] [PubMed] [Google Scholar]
- Doust A. 2007. Architectural evolution and its implications for domestication in grasses. Annals of Botany 100, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan P, Ni S, Wang J, Zhang B, Xu R, Wang Y, Chen H, Zhu X, Li Y. 2015. Regulation of OsGRF4 by OsmiR396 controls grain size and yield in rice. Nature Plants 2, 15203. [DOI] [PubMed] [Google Scholar]
- Fujishiro Y, Agata A, Ota S, Ishihara R, Takeda Y, Kunishima T, Ikeda M, Kyozuka J, Hobo T, Kitano H. 2018. Comprehensive panicle phenotyping reveals that qSrn7/FZP influences higher-order branching. Scientific Reports 8, 12511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta T, Komeda N, Asano K, et al. 2015. Convergent loss of awn in two cultivated rice species Oryza sativa and Oryza glaberrima is caused by mutations in different loci. G3 5, 2267–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani I, Sukegawa S, Kyozuka J. 2006. Genome-wide analysis of spatial and temporal gene expression in rice panicle development. The Plant Journal 46, 503–511. [DOI] [PubMed] [Google Scholar]
- Gitter A, Siegfried Z, Klutstein M, Fornes O, Oliva B, Simon I, Bar-Joseph Z. 2009. Backup in gene regulatory networks explains differences between binding and knockout results. Molecular Systems Biology 5, 276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JR, Finnegan EJ, Watanabe N, Trevaskis B, Swain SM. 2017. New alleles of the wheat domestication gene Q reveal multiple roles in growth and reproductive development. Development 144, 1959–1965. [DOI] [PubMed] [Google Scholar]
- Harrop TW, Ud Din I, Gregis V, Osnato M, Jouannic S, Adam H, Kater MM. 2016. Gene expression profiling of reproductive meristem types in early rice inflorescences by laser microdissection. The Plant Journal 86, 75–88. [DOI] [PubMed] [Google Scholar]
- Hu M, Lv S, Wu W, et al. 2018. The domestication of plant architecture in African rice. The Plant Journal 94, 661–669. [DOI] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, et al. 2012. A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Zhao S, Fu Y, et al. 2018. Variation in the regulatory region of FZP causes increases in secondary inflorescence branching and grain yield in rice domestication. The Plant Journal 96, 716–733. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Miura K, Aya K, Kitano H, Matsuoka M. 2013. Genes offering the potential for designing yield-related traits in rice. Current Opinion in Plant Biology 16, 213–220. [DOI] [PubMed] [Google Scholar]
- Ikeda-Kawakatsu K, Maekawa M, Izawa T, Itoh J, Nagato Y. 2012. ABERRANT PANICLE ORGANIZATION 2/RFL, the rice ortholog of Arabidopsis LEAFY, suppresses the transition from inflorescence meristem to floral meristem through interaction with APO1. The Plant Journal 69, 168–180. [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. 2005. Rice plant development: from zygote to spikelet. Plant & Cell Physiology 46, 23–47. [DOI] [PubMed] [Google Scholar]
- Jeon JS, Jang S, Lee S, et al. 2000. Leafy Hull Sterile1 is a homeotic mutation in a rice MADS box gene affecting rice flower development. The Plant Cell 12, 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Wang Y, Xue D, et al. 2010. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nature Genetics 42, 541–544. [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. 2017. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research 45, D1040–D1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanday I, Yadav SR, Vijayraghavan U. 2013. Rice LHS1/OsMADS1 controls floret meristem specification by coordinated regulation of transcription factors and hormone signaling pathways. Plant Physiology 161, 1970–1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Tsukaya H. 2015. Regulation of plant growth and development by the GROWTH-REGULATING FACTOR and GRF-INTERACTING FACTOR duo. Journal of Experimental Botany 66, 6093–6107. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. 2011. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. The Plant Journal 67, 472–484. [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Maekawa M, Miyao A, Hirochika H, Kyozuka J. 2010. PANICLE PHYTOMER2 (PAP2), encoding a SEPALLATA subfamily MADS-box protein, positively controls spikelet meristem identity in rice. Plant & Cell Physiology 51, 47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K, Yasuno N, Sato Y, Yoda M, Yamazaki R, Kimizu M, Yoshida H, Nagamura Y, Kyozuka J. 2012. Inflorescence meristem identity in rice is specified by overlapping functions of three AP1/FUL-like MADS box genes and PAP2, a SEPALLATA MADS box gene. The Plant Cell 24, 1848–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M, Maekawa M, Shimamoto K, Kyozuka J. 2001. The LAX1 and FRIZZY PANICLE 2 genes determine the inflorescence architecture of rice by controlling rachis-branch and spikelet development. Developmental Biology 231, 364–373. [DOI] [PubMed] [Google Scholar]
- Köster J, Rahmann S. 2012. Snakemake—a scalable bioinformatics workflow engine. Bioinformatics 28, 2520–2522. [DOI] [PubMed] [Google Scholar]
- Kumar L, E Futschik M. 2007. Mfuzz: a software package for soft clustering of microarray data. Bioinformation 2, 5–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtzer GM, Sochat V, Bauer MW. 2017. Singularity: Scientific containers for mobility of compute. PLoS ONE 12, e0177459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyozuka J, Tokunaga H, Yoshida A. 2014. Control of grass inflorescence form by the fine-tuning of meristem phase change. Current Opinion in Plant Biology 17, 110–115. [DOI] [PubMed] [Google Scholar]
- Lee DY, An G. 2012. Two AP2 family genes, SUPERNUMERARY BRACT (SNB) and OsINDETERMINATE SPIKELET 1 (OsIDS1), synergistically control inflorescence architecture and floral meristem establishment in rice. The Plant Journal 69, 445–461. [DOI] [PubMed] [Google Scholar]
- Lee DY, Lee J, Moon S, Park SY, An G. 2007. The rice heterochronic gene SUPERNUMERARY BRACT regulates the transition from spikelet meristem to floral meristem. The Plant Journal 49, 64–78. [DOI] [PubMed] [Google Scholar]
- Li Y, Fu X, Zhao M, Zhang W, Li B, An D, Li J, Zhang A, Liu R, Liu X. 2018. A genome-wide view of transcriptome dynamics during early spike development in bread wheat. Scientific Reports 8, 15338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares OF. 2002. African rice (Oryza glaberrima): History and future potential. Proceedings of the National Academy of Sciences, USA 99, 16360: –16365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, Wu W, Wang M, et al. 2018. Genetic control of seed shattering during African rice domestication. Nature Plants 4, 331–337. [DOI] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17, 10–12. [Google Scholar]
- Matías-Hernández L, Aguilar-Jaramillo AE, Marín-González E, Suárez-López P, Pelaz S. 2014. RAV genes: regulation of floral induction and beyond. Annals of Botany 114, 1459–1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Ikeda M, Matsubara A, Song XJ, Ito M, Asano K, Matsuoka M, Kitano H, Ashikari M. 2010. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nature Genetics 42, 545–549. [DOI] [PubMed] [Google Scholar]
- Ohmori S, Kimizu M, Sugita M, Miyao A, Hirochika H, Uchida E, Nagato Y, Yoshida H. 2009. MOSAIC FLORAL ORGANS1, an AGL6-like MADS box gene, regulates floral organ identity and meristem fate in rice. The Plant Cell 21, 3008–3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Wendel JF. 2013. Crop plants as models for understanding plant adaptation and diversification. Frontiers in Plant Science 4, 290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang S, Zhu W, Hamilton J, et al. 2007. The TIGR rice genome annotation resource: improvements and new features. Nucleic Acids Research 35, D883–D887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Jiang K, Schatz MC, Lippman ZB. 2012. Rate of meristem maturation determines inflorescence architecture in tomato. Proceedings of the National Academy of Sciences, USA 109, 639–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Rodríguez P, Riaño-Pachón DM, Corrêa LG, Rensing SA, Kersten B, Mueller-Roeber B. 2010. PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Research 38, D822–D827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Li Y, Zhao F, et al. 2013. MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiology 162, 872–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergushichev A. 2016. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv 060012. [Preprint]. [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S. 2011. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant & Cell Physiology 52, 344–360. [DOI] [PubMed] [Google Scholar]
- Simons KJ, Fellers JP, Trick HN, Zhang Z, Tai YS, Gill BS, Faris JD. 2006. Molecular characterization of the major wheat domestication gene Q. Genetics 172, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, et al. 2018. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nature Genetics 50, 285–296. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK et al. 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences, USA 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ta KN, Adam H, Staedler YM, Schönenberger J, Harrop T, Tregear J, Do NV, Gantet P, Ghesquière A, Jouannic S. 2017. Differences in meristem size and expression of branching genes are associated with variation in panicle phenotype in wild and domesticated African rice. EvoDevo 8, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi H, Zhang Y, Hattori S, et al. 2011. LAX PANICLE2 of rice encodes a novel nuclear protein and regulates the formation of axillary meristems. The Plant Cell 23, 3276–3287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi-Shiobara F, Kojima Y, Ebitani T, Yano M, Ebana K. 2011. Variation in domesticated rice inflorescence architecture revealed by principal component analysis and quantitative trait locus analysis. Breeding Science 61, 52–60. [Google Scholar]
- Teo ZW, Song S, Wang YQ, Liu J, Yu H. 2014. New insights into the regulation of inflorescence architecture. Trends in Plant Science 19, 158–165. [DOI] [PubMed] [Google Scholar]
- Vaughan DA, Lu B-R, Tomooka N. 2008. The evolving story of rice evolution. Plant Science 174, 394–408. [Google Scholar]
- Wang Y, Li J. 2011. Branching in rice. Current Opinion in Plant Biology 14, 94–99. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yu H, Tian C, Sajjad M, Gao C, Tong Y, Wang X, Jiao Y. 2017. Transcriptome association identifies regulators of wheat spike architecture. Plant Physiology 175, 746–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu WS, Lai FJ. 2015. Functional redundancy of transcription factors explains why most binding targets of a transcription factor are not affected when the transcription factor is knocked out. BMC Systems Biology 9 Suppl 6, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Zhang Q. 2010. Genetic and molecular bases of rice yield. Annual Review of Plant Biology 61, 421–442. [DOI] [PubMed] [Google Scholar]
- Yamaki S, Miyabayashi T, Eiguchi M, Kitano H, Nonomura K-I, Kurata N. 2010. Diversity of panicle branching patterns in wild relatives of rice. Breeding Science 60, 586–596. [Google Scholar]
- Yoshida A, Sasao M, Yasuno N et al. 2013. TAWAWA1, a regulator of rice inflorescence architecture, functions through the suppression of meristem phase transition. Proceedings of the National Academy of Sciences, USA 110, 767–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Nagato Y. 2011. Flower development in rice. Journal of Experimental Botany 62, 4719–4730. [DOI] [PubMed] [Google Scholar]
- Zhang D, Yuan Z. 2014. Molecular control of grass inflorescence development. Annual Review of Plant Biology 65, 553–578. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Cheng S, Song Y, Huang Y, Zhou S, Liu X, Zhou DX. 2015. The interaction between rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. The Plant Cell 27, 2469–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Q, Ge S. 2005. Phylogenetic relationships among A-genome species of the genus Oryza revealed by intron sequences of four nuclear genes. New Phytologist 167, 249–265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.