The identification of NTRC partners in vivo confirms the interaction of NTRC and 2-Cys PRX in Arabidopsis plastids and provides further evidence for a role for NTRC in chloroplast redox regulation.
Keywords: Chloroplast, NTRC, peroxiredoxin, proteomics, redox regulation, TAP-Tag
Abstract
Redox regulation in heterotrophic organisms relies on NADPH, thioredoxins (TRXs), and an NADPH-dependent TRX reductase (NTR). In contrast, chloroplasts harbor two redox systems, one that uses photoreduced ferredoxin (Fd), an Fd-dependent TRX reductase (FTR), and TRXs, which links redox regulation to light, and NTRC, which allows the use of NADPH for redox regulation. It has been shown that NTRC-dependent regulation of 2-Cys peroxiredoxin (PRX) is critical for optimal function of the photosynthetic apparatus. Thus, the objective of the present study was the analysis of the interaction of NTRC and 2-Cys PRX in vivo and the identification of proteins interacting with them with the aim of identifying chloroplast processes regulated by this redox system. To assess this objective, we generated Arabidopsis thaliana plants expressing either an NTRC–tandem affinity purification (TAP)-Tag or a green fluorescent protein (GFP)–TAP-Tag, which served as a negative control. The presence of 2-Cys PRX and NTRC in complexes isolated from NTRC–TAP-Tag-expressing plants confirmed the interaction of these proteins in vivo. The identification of proteins co-purified in these complexes by MS revealed the relevance of the NTRC–2-Cys PRX system in the redox regulation of multiple chloroplast processes. The interaction of NTRC with selected targets was confirmed in vivo by bimolecular fluorescence complementation (BiFC) assays.
Introduction
Chloroplasts are equipped with a large set of thioredoxins (TRXs), small polypeptides with a conserved active site formed by two cysteines that regulate the activity of target enzymes via the reduction of specific disulfide groups (Meyer et al., 2012; Geigenberger et al., 2017). A classical scheme for the regulation of photosynthesis emerged by way of the TRX-dependent reductive activation of biosynthetic enzymes, such as those of the Calvin–Benson cycle (Michelet et al., 2013). During the day, these enzymes are maintained in a reduced and active state by the action of TRXs, which in turn are reduced via photosynthetically reduced ferredoxin (Fd) and an Fd-dependent TRX reductase (FTR) (Schürmann and Buchanan, 2008). Different methodologies developed to trap proteins interacting with TRXs together with the advance in proteomics has brought an impressive increase in the number of putative TRX targets, thus extending the processes under TRX-dependent redox regulation beyond the Calvin–Benson cycle in plant chloroplasts, green algae, and cyanobacteria (Motohashi et al., 2001; Balmer et al., 2003; Lindahl and Florencio, 2003; Lindahl and Kieselbach, 2009; Montrichard et al., 2009).
The notion of chloroplast redox regulation as a light-dependent process, which uses reducing power provided by photosynthetically reduced Fd, was modified by the discovery of a chloroplast-localized NADPH-dependent TRX reductase, termed NTRC, which has a joint TRX domain at the C-terminus (Serrato et al., 2004). It was later shown that NTRC is able to conjugate both NTR and TRX activities to efficiently reduce 2-Cys PRXs (Moon et al., 2006; Perez-Ruiz et al., 2006; Alkhalfioui et al., 2007; Perez-Ruiz and Cejudo, 2009), which led to the proposal of an antioxidant function for NTRC. Indeed, it was shown that the hydrogen peroxide-scavenging activity of the NTRC/2-Cys PRX system has a protective effect on Mg-protoporphyrin monomethyl ester cyclase, an enzyme of the chlorophyll biosynthesis pathway (Stenbaek et al., 2008). However, further analyses have shown the participation of NTRC in redox regulation of chloroplast processes previously shown to be regulated by TRXs such as the biosynthesis of tetrapyrroles (Richter et al., 2013; Pérez-Ruiz et al., 2014) and starch (Michalska et al., 2009; Lepistö et al., 2013).
Therefore, it is now well established that chloroplast redox regulation relies on two redox pathways, the light-dependent Fd–FTR–TRXs system and NTRC, which can act independently of light since NADPH is also produced from sugars by the oxidative pentose phosphate pathway (Spínola et al., 2008; Cejudo et al., 2012). The use of Arabidopsis mutants combining the deficiency of NTRC and different types of TRXs (Thormählen et al., 2015; Da et al., 2017; Ojeda et al., 2017) or NTRC and FTR (Yoshida and Hisabori, 2016) led to the proposal that the two chloroplast redox systems act cooperatively via the regulation of common targets, a notion further supported by the finding that NTRC interacts with TRXs and TRX-regulated proteins (Nikkanen et al., 2016). However, our group has recently reported that decreased levels of 2-Cys PRXs exert a suppressor effect on the ntrc phenotype and, based on these results, we have proposed that the redox balance of 2-Cys PRXs, which is regulated by NTRC, plays an essential role in maintaining chloroplast redox homeostasis (Perez-Ruiz et al., 2017). The finding of a close functional relationship between NTRC and 2-Cys PRXs implies that both proteins may interact; however, albeit there is extensive evidence showing the interaction of the two proteins in vitro (Perez-Ruiz et al., 2006; Bernal-Bayard et al., 2012, 2014), less is known about this interaction in vivo.
The characterization of Arabidopsis mutants devoid of NTRC has revealed the participation of this enzyme in photosynthetic performance (Carrillo et al., 2016; Naranjo et al., 2016), carbon fixation (Perez-Ruiz et al., 2006), starch metabolism (Michalska et al., 2009; Lepistö et al., 2013), and the biosynthesis of tetrapyrroles (Richter et al., 2013; Pérez-Ruiz et al., 2014). Moreover, the activity of NTRC as an efficient reductant of 2-Cys PRXs suggested its participation in the mechanism of chloroplast antioxidant defense (Moon et al., 2006; Perez-Ruiz et al., 2006) affecting the response to abiotic (Chae et al., 2013) and biotic stress (Ishiga et al., 2012, 2016). Altogether, these data indicate that NTRC participates in the regulation of a wide variety of processes, yet approaches to trap NTRC targets based on the formation of mixed disulfide led to the identification of a surprisingly low number of targets from either cyanobacteria (Mihara et al., 2016) or plant chloroplasts (Yoshida and Hisabori, 2016). Here, we have addressed the issue of identifying in vivo partners of NTRC by using the tandem affinity purification (TAP)-Tag technology in Arabidopsis. Our data show the in vivo interaction of NTRC and 2-Cys PRXs; moreover, the identification of proteins present in complexes containing NTRC and 2-Cys PRX by MS reveals the central function of this redox system in redox regulation of multiple chloroplast processes.
Materials and methods
Plant material and growth conditions
Arabidopsis thaliana (ecotype Columbia) plants were grown in soil in culture chambers under short-day (8 h light/16 h darkness) or long-day (16 h light/8 h darkness) conditions at 22 °C during the light and 20 °C during darkness.
Generation of C-TAPa constructs and plant transformation
NTRC full-length cDNA was amplified from total RNA isolated from Arabidopsis leaves using oligonucleotides containing the attB sequence and cloned into the pDONR221 plasmid (GATEWAY; Invitrogen). PCR was performed using two pairs of oligonucleotides (Supplementary Table S1 at JXB online), one for gene-specific amplification (attB1-for-AtNTRC/attB2-rev-AtNTRC, without a stop codon) and the second to include the whole attB sequences (attB1-adapter-for-AtNTRC/attB2-adapter-rev-AtNTRC). For control plants, the transit peptide of Arabidopsis NTRC (75 N-terminal residues) was fused to the green fluorescent protein (GFP) coding sequence. NTRC transit peptide was amplified from pUNI-AtNTRC using oligonucleotides indicated in Supplementary Table S1. This cDNA was digested with XhoI/AgeI and cloned into pEGFP-1. The resulting ptNTRC–eGFP fusion was amplified using oligonucleotides attB1-for-AtNTRC/attB2-rev-GFP and attB1-adapter-for-AtNTRC/attB2-adapter-rev-AtNTRC (Supplementary Table S1). In both cases, NTRC and ptNTRC–eGFP, the resulting PCR products were recombined with pDONR221 using the BP clonase reaction and all constructs were confirmed by DNA sequencing. Both constructs were cloned in pCTPAa using the LR clonase reaction, and the resulting plasmids were transformed into Agrobacterium, and were used for Arabidopsis transformation by floral dipping (Clough and Bent, 1998).
The analysis of the recombinant protein levels in the different transgenic lines was performed with plants grown under long-day conditions. Leaf extracts (30 µg of protein) of the different independent lines were fractionated by SDS–PAGE (10% acrylamide), immunoblotted, and probed with either anti-myc or anti-GFP antibodies, which were purchased from Agrisera (Sweden), or with the anti-NTRC antibody, which was raised in our laboratory, as previously reported (Serrato et al., 2004).
Protein extraction and double affinity chromatography
Prior to extraction, leaves dissected from 65-day-old short-day-grown plants were vacuum-infiltrated with 1% (v/v) formaldehyde in phosphate-buffered saline (PBS) for 30 min and, after washing with 300 mM glycine, vacuum-infiltrated with 300 mM glycine. Finally, the tissue was washed twice with PBS and frozen in liquid nitrogen. Protein extracts were prepared with 50 mM Tris–HCl pH 7.5, containing 150 mM NaCl, 0.1% (w/v) NP-40, and 10% (v/v) glycerol. For each line, five samples, 18 g FW each, were resuspended in 40 ml of extraction buffer containing protease inhibitors [1 mM phenylmethylsulfonyl fluoride (PMSF) and Sigma protein inhibitor cocktail] and the total extract obtained from these samples was divided into six samples that were processed independently thereafter. Samples were filtered through a nylon membrane and centrifuged at 200 g for 3 min at 4 °C. Aliquots (1.2 ml) of IgG beads (IgG Sepharose 6 fast flow, GE Healthcare) were washed with 10 ml of 0.1 M glycine pH 2.7 to remove unbound IgG. Beads were subsequently washed twice with 10 ml of extraction buffer, and once with 10 ml of extraction buffer containing protease inhibitors. Then, protein extracts were incubated with beads for 4 h at 4 °C with gentle shaking. The solution was centrifuged at 200 g for 3 min to discard unbound proteins, and unspecifically bound proteins were removed by three washes with extraction buffer without protease inhibitors. Bound proteins were released by overnight incubation with 5 ml of extraction buffer containing 10 μl of 3C Protease (PreScission, GE-Healthcare) and 1 mM E-64 at 4 °C with gentle shaking. Released proteins were collected by centrifugation and recovery of the supernatant. Beads were washed once more with 5 ml of extraction buffer and both supernatants were mixed for subsequent steps. The IgG eluate was then added to Ni-NTA agarose (Invitrogen) and incubated with gentle shaking for 4 h at 4 °C. Beads were allowed to settle, transferred to a polypropylene column, and washed with 30 ml of extraction buffer. Proteins were then eluted with buffer supplemented with 20 mM (fraction E1) and 500 mM (fractions E2–E5) imidazole. Aliquots of the fractions collected from the six different samples processed during the purification procedure were mixed, concentrated, analyzed by SDS–PAGE (10% acrylamide), and immunoblotted with anti-myc antibodies. For detection of 2-Cys PRXs, blots were probed with an anti-2-Cys PRX antibody that was raised by immunization of rabbits with the purified His-tagged protein from rice.
For LC-MS/MS analysis, proteins were concentrated in Amicon Ultra 3K columns (Millipore) followed by trichloroacetic acid (TCA)/acetone precipitation. Pellets were dissolved in 50 mM ammonium bicarbonate (digestion buffer) and proteins were reduced with 5 mM DTT (final concentration) at 95 °C for 5 min. Alkylation buffer, 10 mM iodoacetamide, final concentration, was added and samples were incubated in the dark at room temperature for 20 min. An aliquot (1 μl) of activated TPCK-treated trypsin (New England Biolabs) was added to the samples and incubated at 37 °C for 3 h. Then, an additional 1 μl aliquot of activated trypsin (0.1 μg μl−1 concentration) was added to the samples, which were incubated at 30 °C overnight. Samples were precipitated with TCA/acetone and the supernatant was mixed with sample buffer to reach a final concentration of 0.5% trifluoroacetic acid (TFA), 0.05% formic acid. Samples were cleaned by reverse-phase chromatography in C18 spin columns (Pierce). The peptides were eluted from the column with 70% acetonitrile, 0.1% formic acid, and samples were vacuum dried. The experiment was repeated three times, and the corresponding samples were analysed by MS/MS independently.
Label-free LC-MS/MS analysis
Peptides were separated by reverse phase chromatography using an Eksigent™ nanoLC ultra 2D+ nano pump fitted with a column from Eksigent™ (ChromXP nanoLC column 75 µm id×15 cm, ChromXP C18 3 µm 120 Å). Samples were first loaded for desalting and concentration during 5 min into a 0.5 cm length 350 μm ID pre-column packed with the same chemistry as the separating column (ChromXP nanoLC Trap column 350 µm id×0.5 mm, ChromXP C183 µm 120 Å). The mobile phases were 100% water, 0.1% formic acid (buffer A) and 100% acetonitrile, 0.1% formic acid (buffer B). Column gradient was developed in a 70 min two-step gradient from 5% to 40% B buffer in 40 min and 40% to 60% B buffer in 5 min. The column was equilibrated in 95% B buffer for 5 min and 5% B buffer for 15 min. Peptides eluted from the column were analyzed using an ABSciex 5600 TripleTOF™ plus system. Information-dependent data was acquired upon a survey scan performed in a mass range from 350 m/z up to 1250 m/z using a scan time of 250 ms. The 25 most intense peaks on every survey were selected for fragmentation. Minimum accumulation time for MS/MS was set to 50 ms, giving a total cycle time of 1550 ms. Product ions were scanned in a mass range from 100 m/z up to 1500 m/z and excluded for further fragmentation during 15 s. After MS/MS analysis, data files were processed using ProteinPilot™ 4.5 software from ABSciex, which uses the algorithm Paragon™ for database search and Progroup™ for data grouping (Shilov et al., 2007), and searched against a specific A. thaliana Uniprot database. The false discovery rate was determined using a non-lineal fitting method (Tang et al., 2008) and displayed results were those reporting a confidence of 99% in the global false discovery rate analysis. Data were analyzed using two technical replicates from each sample obtained in an independent purification experiment. The peak list was generated in PeakView™ 1.1.1 Software from ABSciex, using the combined database search results generated in ProteinPilot™ 4.5 software. The peak list matrix generated was exported to MarkerView™ 1.2.1 software for principal component analysis (PCA) (Ivosev et al., 2008). Sample comparison was performed using the first two components that explained a total of 78.1% of the variance between samples. Sample dispersion was measured using a t-test, and proteins with extreme t-values were chosen as candidates for validation.
Generation of constructs for bimolecular fluorescence complementation (BiFC) assays
To generate the constructs for BiFC assays, full-length cDNAs of 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase (DAHPS: TAIR At1g22410), glutamine synthetase 2 (GS2: TAIR At5g35630), and chloroplast heat shock protein 70-1 (CPHSP70-1: TAIR At4g24280) were amplified using pairs of oligonucleotides containing attB recombination sites, without stop codons in the reverse primers, to allow C-terminal translational fusions with yellow fluorescent protein (YFP) N- and C- terminal fragments (Supplementary Table S2). Once amplified, a second set of primers (attB1-adapter-for-AtNTRC/attB2-adapter-rev-AtNTRC) was used to include the whole attB sequences. PCR products were cloned into pDNOR221 by a BP clonase reaction. After sequencing, the cDNAs were cloned into pSPYCE-35S_GW and pSPYNE-35S_GW gateway-compatible vectors, carrying the C- and N-terminal fragments of YFP, respectively, by a LR clonase reaction. Agrobacterium tumefaciens (GV3101) cells were transformed with the resulting plasmids. Transformed cells were grown in liquid YEB and co-infiltrated, together with an Agrobacterium strain carrying the p19 gene-silencing suppressor plasmid, into Nicotiana benthamiana leaves. To this end, overnight grown cultures of Agrobacterium carrying the YFP construct moieties were co-infiltrated with p19 suppressor (Silhavy et al., 2002). Leaves of 4-week-old plants were infiltrated as previously described (Raynaud et al., 2016) and analyzed after 3 d in a growth chamber using a Leica TCS SP2 confocal laser-scanning microscope (Leica Microsystems). For the detection of yellow fluorescence, cells were excited at 488 nm with an argon laser and fluorescence emission was detected at 510–575 nm. Chlorophyll fluorescence was detected, after excitation at 633 nm with a HeNe laser, at 650–700 nm.
Results
Generation of Arabidopsis plants expressing NTRC–TAPa-Tag or GFP–TAPa-Tag fusion proteins
To gain more insight into the function of NTRC in chloroplast redox regulation, we have performed the identification of in vivo targets of the enzyme making use of the TAPa-Tag technology, which has been successfully used for multiprotein complex isolation from Arabidopsis (Rubio et al., 2005). With that purpose, the full-length cDNA encoding NTRC from Arabidopsis, including the putative transit peptide to drive plastid-specific localization, was cloned into the pC-TAPa vector (Rubio et al., 2005), according to the scheme depicted in Supplementary Fig. S1. This construct allowed the expression of NTRC tagged at the C-terminus with nine copies of the myc epitope (9×myc), a six histidine tag (6×His), the sequence for the 3C protease cleavage site (3C), and two copies of the protein A IgG-binding domain (2×IgG-BD). Expression was driven by two copies of the Cauliflower mosaic virus (CaMV) 35S promoter and a Tobacco mosaic virus (TMV) U1Ω translational enhancer (2×35S::TMVU1Ω). As negative control, the NTRC cDNA was replaced by the cDNA encoding GFP, which was C-terminally fused to the predicted transit peptide of NTRC (Supplementary Fig. S1).
Arabidopsis wild-type (WT) plants were transformed with both NTRC–TAPa-Tag and GFP–TAPa-Tag constructs, and transgenic lines with a high expression of NTRC–TAPa-Tag, line #5-24, or GFP–TAPa-Tag, line #3-21-6, as determined by western blotting probed with the anti-myc antibody (Supplementary Fig. S2A), were selected for further analysis. The anti-NTRC antibody confirmed the presence of both the tagged NTRC and the endogenous NTRC in the transgenic lines, while extracts from untransformed WT plants showed exclusively the band corresponding to the endogenous NTRC, which was absent in the ntrc mutant (Supplementary Fig. S2B). Similarly, line #3-21-6 showed a high expression of the tagged GFP as revealed by western blotting probed with the anti-GFP antibody (Supplementary Fig. S2B). Because NTRC is a plastid-localized enzyme, we first tested that the predicted NTRC transit peptide effectively targeted the fusion proteins to these organelles. Confocal microscopy analyses of the line expressing the GFP–TAPa-Tag showed fluorescence of the tagged GFP, hence confirming correct folding of the expressed protein (Supplementary Fig. S3A–D). Moreover, the localization of the tagged protein to chloroplasts (Supplementary Fig. S3A–D) confirmed the functionality of the predicted transit peptide of NTRC. Chloroplast localization of the tagged NTRC was tested by fractionation analysis (Supplementary Fig. S3E). Chloroplasts isolated from leaf extracts of WT and transgenic lines expressing the NTRC–TAP-Tag and GFP–TAP-Tag were further fractionated into stromal and thylakoid. Western blot analysis of these fractions showed the presence of the endogenous and tagged NTRC in the chloroplast stroma and associated with the thylakoid (Supplementary Fig. S3E), in agreement with previous results (Serrato et al., 2004). Leaf extracts resulting from chloroplast purification, hence free of chloroplasts, were analyzed to test the absence of endogenous and tagged NTRC in these extracts (Supplementary Fig. S3E). These results confirm the plastid localization of the tagged proteins in the transgenic lines and allow the presence of these proteins in other cell compartments to be ruled out. Finally, to test the functionality of the expressed tagged NTRC, the NTRC–TAPa-Tag was expressed in the ntrc mutant background (Supplementary Fig. S4). Independent transgenic lines #19 and #66 expressing the tagged NTRC (Supplementary Fig. S4A, B) showed a better growth rate than the ntrc mutant, as indicated by the rosette fresh weight (Supplementary Fig. S4C); in contrast, no recovery of the growth rate was observed in control transgenic lines #4-9 and #4-23 expressing the tagged GFP in the ntrc background (Supplementary Fig. S4C). The complementation of the ntrc phenotype by expression of the tagged NTRC indicates the functionality of the tagged enzyme; however, the recovery of the growth phenotype is only partial, thus suggesting that the tagged NTRC shows lower activity than the endogenous NTRC.
In vivo interaction of NTRC and 2-Cys PRXs
NTRC-containing protein complexes were purified from leaf extracts of plants expressing the NTRC–TAPa-Tag (line #5-24) according to the protocol depicted in Fig. 1, and the purification procedure was followed with the anti-myc antibody (Fig. 2A). In parallel, protein complexes were purified from the GFP–TAPa-Tag line, which was used as negative control (Fig. 2B). Complexes bound to IgG beads were eluted by cleavage with the 3C protease, which generated the corresponding tagged protein with the expected lower molecular mass due to the loss of the IgG-binding domain (Figs 1, 2A, B). The second purification step was performed by Ni2+-nitrilotriacetic acid (NTA) chromatography taking advantage of the 6×His tag. After extensive washing, protein complexes were eluted with buffer containing imidazole at concentrations of 20 mM (E1 fraction) or 500 mM (E2–E5 fractions) (Fig. 2A, B).
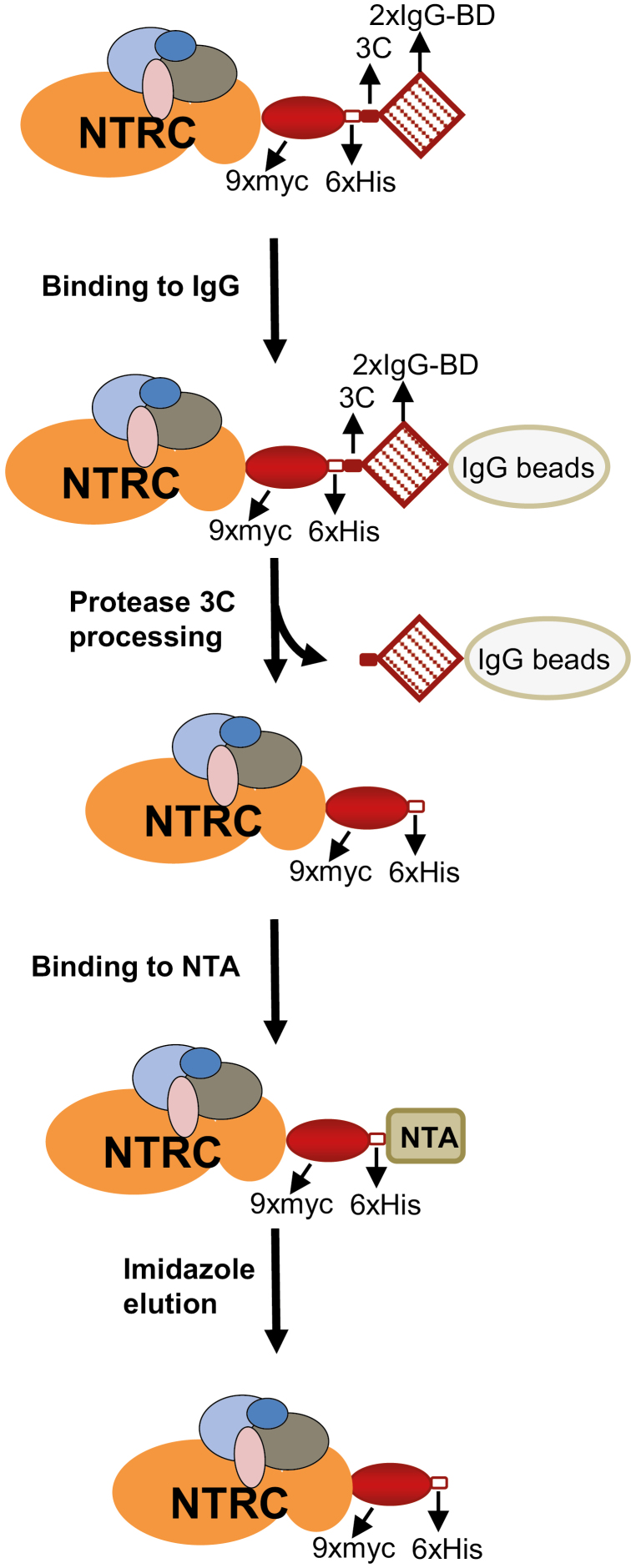
Fig. 1.
Procedure for the isolation of NTRC-containing protein complexes. NTRC-containing protein complexes were purified from Arabidopsis plants expressing NTRC tagged at the C-terminus with nine copies of the myc epitope (9×myc), six histidine residues (6×His), a 3C protease cleavage site (3C), and two copies of the protein A IgG-binding domain (2×IgG-BD). Leaf protein extracts were incubated with IgG beads as a first purification step; after removal of unspecifically bound proteins by washing, bound complexes were released by rhinovirus 3C protease-mediated processing. The released fraction was subjected to a second step of affinity chromatography with Ni-NTA beads, and NTRC-containing complexes were eluted with imidazole.
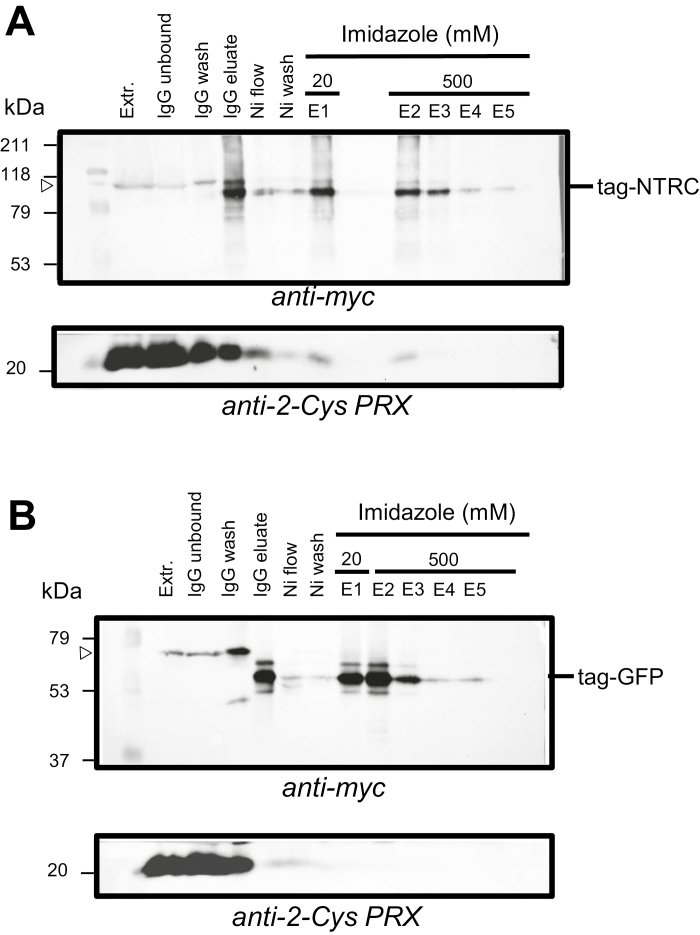
Fig. 2.
Purification of cTAPa–NTRC and cTAPa–GFP complexes by double affinity chromatography. NTRC- (A) or GFP- (B) containing complexes were purified from crude leaf extracts of the corresponding transgenic Arabidopsis plants. Purification steps were followed by western blot of the fractions. Proteins were fractionated by SDS–PAGE (10% acrylamide) and electrotransferred to nitrocellulose filters, which were probed with the anti-myc antibody. Fractions correspond to crude protein extract (Extr.), proteins unbound to IgG beads after 4 h of incubation (IgG unbound); proteins released by washing of the IgG beads (IgG wash), proteins released by 3C protease (IgG eluate); Ni-bead flow through (Ni flow), Ni-bead wash (Ni wash), and fractions eluted with 20 mM (E1) or 500 mM imidazole (E2–E5). The molecular mass in kDa of protein markers (M) is indicated on the left. Tag-NTRC, tagged NTRC; tag-GFP, tagged GFP.
The first goal of this work was to establish the in vivo interaction of NTRC and 2-Cys PRX. Thus, fractions from the purification steps were analyzed by western blotting probed with the anti-2-Cys PRX antibody. After the second purification step from the NTA column, 2-Cys PRX was detected in fractions eluted with 20 mM (E1) or 500 mM (E2) imidazole from plants expressing the NTRC-tagged protein, but not from control plants expressing the GFP-tagged protein (Fig. 3A). These results show the presence of this protein specifically in complexes containing the tagged NTRC. In fact, despite the reducing conditions, 2-Cys PRX was detected in the eluted fractions in monomeric and dimeric forms (Fig. 3A). Both endogenous and tagged NTRC were detected in imidazole fractions from plants expressing the NTRC-tag (line #5-24) but not from control plants (line #3-21-6) (Fig. 3B), showing the interaction of NTRC with itself, in agreement with previous data showing that the catalytically active form of NTRC is a dimer (Perez-Ruiz and Cejudo, 2009). Moreover, a significant amount of the enzyme was detected in aggregated form (Fig. 3B), which is in line with the tendency of purified NTRC to aggregate in vitro (Pérez-Ruiz et al., 2009; Wulff et al., 2011).
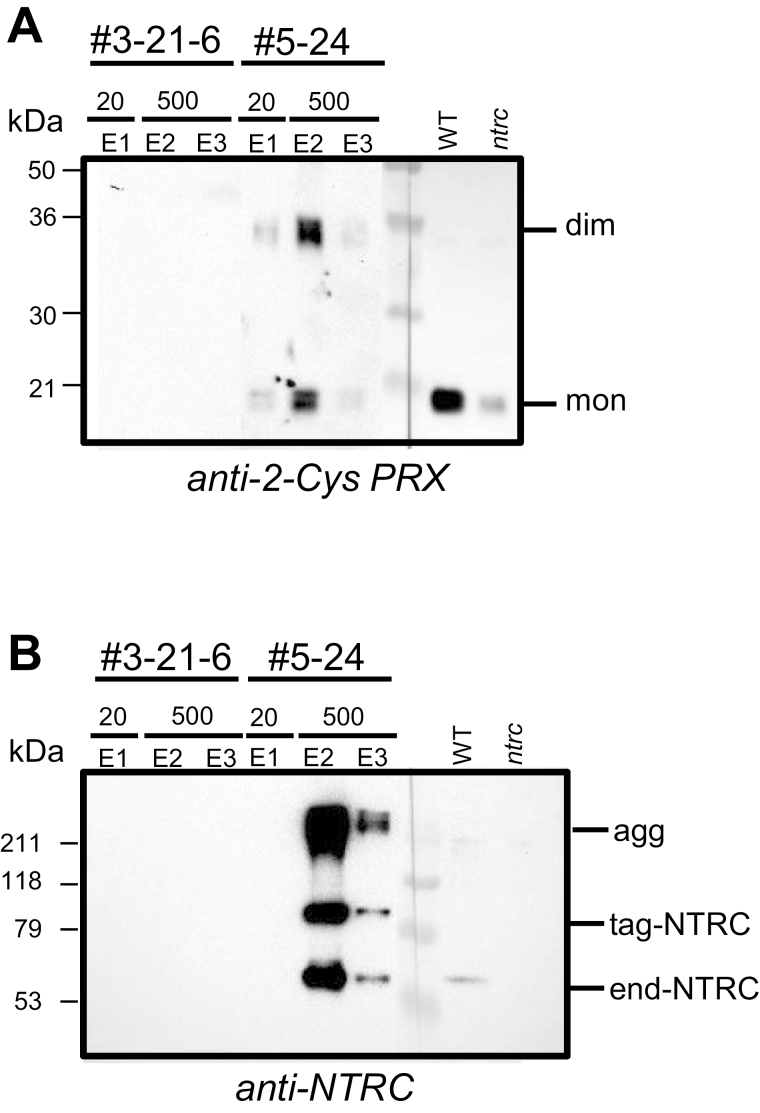
Fig. 3.
NTRC and 2-Cys PRXs are present in the same fractions. Western blot analysis of fractions eluted with imidazole from GFP- (line #3-21-6) and NTRC- (line #5-24) expressing plants probed with the anti-2-Cys PRX (A) or the anti-NTRC antibody (B). As reference, leaf crude extracts (30 µg of protein) from WT and ntrc plants were loaded and, after blotting, the filter was excised in two parts at the molecular markers line to allow a more extended exposure of a part of the filter. The molecular mass in kDa of protein markers (M) is indicated on the left. end-NTRC, endogenous NTRC; tag-NTRC, tagged NTRC; agg, aggregated; mon, monomer; dim, dimer.
NTRC-interacting proteins identify chloroplast processes potentially regulated by the NTRC/2-Cys PRX redox system
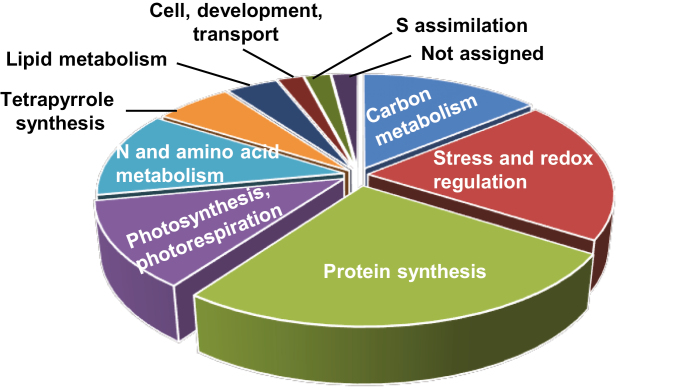
Having established the in vivo interaction of NTRC and 2-Cys PRXs, we looked at the possibility of identifying potential processes under the regulation of this redox system via the identification of additional proteins present in the NTRC-containing complexes. To that end, three independent complex purification experiments were performed and proteins were identified by LC-MS/MS. Only proteins detected in at least two of the experiments that were at least 2-fold more abundant in NTRC–TAPa-Tag eluates than in GFP–TAPa-Tag control plants were considered (Table 1). Of the 50 proteins identified that fulfilled these conditions, most of them (45) show the expected chloroplast localization, though some (14) also show localization in other cell compartments (Table 1). The four proteins with expected localization in cell compartments other than the chloroplast (Table 1) were considered unspecific. The chloroplast-localized proteins, except one that could not be assigned, were classified into nine biological functions (Table 1; Fig. 4).
Table 1.
Classification of identified NTRC-interacting proteins in A. thaliana leaves
| Entry Uniprot KB | TAIR | Protein name | Localization | No. of experiments | Max. ratio | No. of Cys residues | Trx target |
|---|---|---|---|---|---|---|---|
| Photosynthesis and photorespiration | |||||||
| Q9LMQ2 | At1g15820 | LHCB6, Chlorophyll A-B binding protein | C | 2 | 2.91 | 0 | x |
| P27521 | At3g47470 | CAB4, Chlorophyll a-b binding protein 4 | C | 2 | 24.24 | 1 (0) | |
| Q9SY97 | At1g61520 | LHCA3, PSI type III chlorophyll a/b-binding protein | C | 2 | 2.3 | 0 | |
| P56767 | AtCg00340 | D1, PSI P700 chlorophyll a apoprotein PsaB | C | 2 | 12.89 | 2 (2) | |
| P16972 | At1g60950 | Ferredoxin-2 | C | 2 | 2.7 | 5 (5) | x |
| Q8L3U4 | At5g36700 | PGLP-1, Phosphoglycolate phosphatase 1, PGLP-1 | C, Cyt | 2 | 6.97 | 8 (5) | |
| Carbon metabolism: Calvin cycle, starch, glycolysis, OPP, and TCA | |||||||
| O03042 | AtCg00490 | RBCL, Ribulose bisphosphate carboxylase large chain | C | 2 | 2.37 | 9 (9) | x |
| F4KA76 | At5g38410 | RBCS3B, Ribulose bisphosphate carboxylase small chain 3B | C | 2 | 9.53 | 5 (5) | x |
| P10795 | At1g67090 | RBCS1A, Ribulose bisphosphate carboxylase small chain 1A | C | 2 | 2.46 | 0 | x |
| Q9LD57 | At3g12780 | PGK1, Phosphoglycerate kinase 1 | C, Cyt, M, N | 3 | 4.64 | 2 (2) | x |
| Q9LZS3 | At5g03650 | SBE2.2, Starch-branching enzyme 2-2 | C | 2 | 2.1 | 7 (7) | x |
| Q93Z53 | At1g32440 | PKP3, Plastidial pyruvate kinase 3 | C | 2 | 2.8 | 6 (5) | x |
| F4K874 | At5g14740 | BETA CA2, Beta carbonic anhydrase 2 | C, Cyt | 2 | 3.69 | 9 (6) | x |
| Lipid metabolism | |||||||
| P56765 | AtCg00500 | ACCD, Acetyl-CoA carboxylase carboxyltransferase beta subunit | C | 2 | 9.85 | 12 (12) | x |
| Q38882 | At3g15730 | PLDALPHA1, Phospholipase D alpha 1 | C, Mit, N, M, V, Cyt | 2 | 2.69 | 8 (8) | |
| N and amino acid metabolism | |||||||
| Q43127 | At5g35630 | GS2, Glutamine synthetase 2 | C, Mit, M | 2 | 2.6 | 7 (6) | x |
| Q9LPR4 | At1g18500 | IPMS1, 2-Isopropylmalate synthase 1 | C | 3 | 2.4 | 9 (9) | |
| Q9SK84 | At1g22410 | Class-II DAHP synthase-like protein | C | 2 | 6.45 | 7 (7) | x |
| Q9FVP6 | At1g48860 | EPSPS, 5-Enolpyruvylshikimate-3-phosphate synthase | C | 2 | 14.77 | 10 (9) | |
| D7MUW5 | At5g54810 | TRPB, Tryptophan synthase beta-subunit | C, M | 2 | 5.89 | 5 (5) | x |
| Q9LU63 | At5g51110 | PDL1, PCD/DCoH-like protein (4-alpha-hydroxytetrahydrobiopterin dehydratase activity) | C | 3 | 4.6 | 5 (4) | |
| S assimilation | |||||||
| Q9LIK9 | At3g22890 | APS1, ATP sulfurylase 1 | C, M | 3 | 2.98 | 1 (1) | |
| Tetrapyrrole synthesis | |||||||
| Q9SFH9 | At1g69740 | HEMB1, Delta-aminolevulinic acid dehydratase 1 | C, Cyt | 2 | 2.12 | 8 (6) | |
| P16127 | At4g18480 | CHL1I, Magnesium-chelatase subunit ChlI-1 | C, Cyt | 2 | 2.71 | 5 (4) | x |
| P21218 | At4g27440 | PORB, NADPH-protochlorophyllide oxidoreductase B | C | 2 | 3.41 | 4 (4) | |
| Stress and redox regulation | |||||||
| Q9STW6 | At4g24280 | CPHSP70-1, Chloroplast heat shock protein 70-1 | C | 2 | 3.58 | 2 (2) | x |
| Q9SLJ2 | At1g54410 | HIRD11, Dehydrin 11 kDa | C, Cyt | 2 | 40.76 | 0 | |
| O22229 | At2g41680 | NTRC, NADPH-dependent thioredoxin reductase 3 | C | 3 | 26.16 | 7 (7) | |
| Q8LE52 | At5g16710 | DHAR3, Dehydroascorbate reductase | C | 2 | 4.43 | 4 (3) | x |
| Q949U7 | At3g52960 | PRXIIE, Peroxiredoxin-IIE | C | 2 | 9.57 | 2 (2) | x |
| Q8LEA5 | At5g06290 | 2-CYS PRXB, 2-cys peroxiredoxin B | C | 3 | 12.73 | 3 (2) | x |
| Q96291 | At3g11630 | 2-CYS PRXA, 2-cys peroxiredoxin A | C | 3 | 8.25 | 2 (2) | x |
| F4HUL6 | At1g20620 | CAT3, Catalase 3 | C, Mit, M, N, V | 2 | 3.52 | 7 (7?) | |
| Q9ZQ80 | At2g03440 | NRP1, Nodulin-related protein 1 | 2 | 2.79 | 0 | ||
| Q9C5D0 | At4g34120 | CBS domain-containing protein CBSX2 | C | 3 | 18.13 | 0 | |
| Protein synthesis and ribosomal structure | |||||||
| P56799 | AtCg00380 | RPS4, 30S ribosomal protein S4 | C | 2 | 3.12 | 2 (2) | |
| P56801 | AtCg00770 | RPS8, 30S ribosomal protein S8 | C | 2 | 2.72 | 1 (1) | |
| P16180 | At1g79850 | RPS17, 30S ribosomal protein small subunit protein 17 | C | 2 | 7.34 | 1 (1) | |
| P56807 | AtCg00650 | RPS18, 30S ribosomal protein S18 | C | 2 | 4.13 | 0 | |
| Q94K97 | At5g24490 | Putative 30S ribosomal protein | C | 3 | 47.77 | 5 (3) | x |
| Q9M385 | At3g54210 | RPL17, 50S ribosomal protein L17 | C | 2 | 3.62 | 0 | |
| Q8RXX5 | At5g47190 | 50S ribosomal protein L19-2 | C, M | 2 | 3.41 | 0 | |
| P92959 | At5g54600 | RPL24, 50S ribosomal protein L24 | C | 3 | 3.91 | 2 (1) | |
| P56796 | AtCg00640 | RPL33, 50S ribosomal protein L33 | C | 2 | 27.58 | 4 (2) | |
| P41377 | At1g54270 | EIF4A-2, Eukaryotic initiation factor 4A-2 | Cyt, M, V | 3 | 17.67 | 6 | |
| Q8GUN2 | At3g56490 | HINT1, His triad family protein (Adenylylsulfatase HINT1) | P, M | 2 | 108.11 | 1 | |
| Q9M0Y8 | At4g04910 | NSF, N-ethylmaleimide sensitive factor (Vesicle-fusing ATPase) | G, M, V | 2 | 9.84 | 9 | |
| Q84WV1 | At5g26360 | T-complex protein 1 subunit gamma, TCP-1/ cpn60 chaperonin family protein | Cyt | 2 | 42.26 | 10 | |
| Cell development and transport | |||||||
| F4J3Q8 | At3g10350 | GET3B, Guided entry of tail-anchored proteins 3B (P-loop containing nucleoside triphosphate hydrolases superfamily protein) | C, M | 2 | 8.83 | 5 (4) | |
| Not assigned | |||||||
| Q94K48 | At3g62530 | ARM repeat superfamily protein, Armadillo/ beta-catenin-like repeat-containing protein | C, Mit, N | 2 | 4.34 | 3 (2) |
Entry Uniprot KB, Uniprot accession number; TAIR, gene name; Localization, C- chloroplast, Cyt- cytosol, G- Golgi, M- membrane, Mit- mitochondria, V- vacuole, P- peroxisome; No. of experiments, number of experiments in which the protein was identified; Max. ratio, relative level of protein in NTRC complexes compared with GFP samples; No. of Cys residues, total number of Cys residues in the protein; shown in parentheses is the number of Cys residues in the mature protein; Trx target, identified as a Trx target in previous studies.
Fig. 4.
Biological function of proteins identified in NTRC-containing complexes. Chloroplast proteins identified in the NTRC-containing complexes, isolated by the double chromatography approach, were classified according to their biological function using the Map Man tool (Thimm et al., 2004).
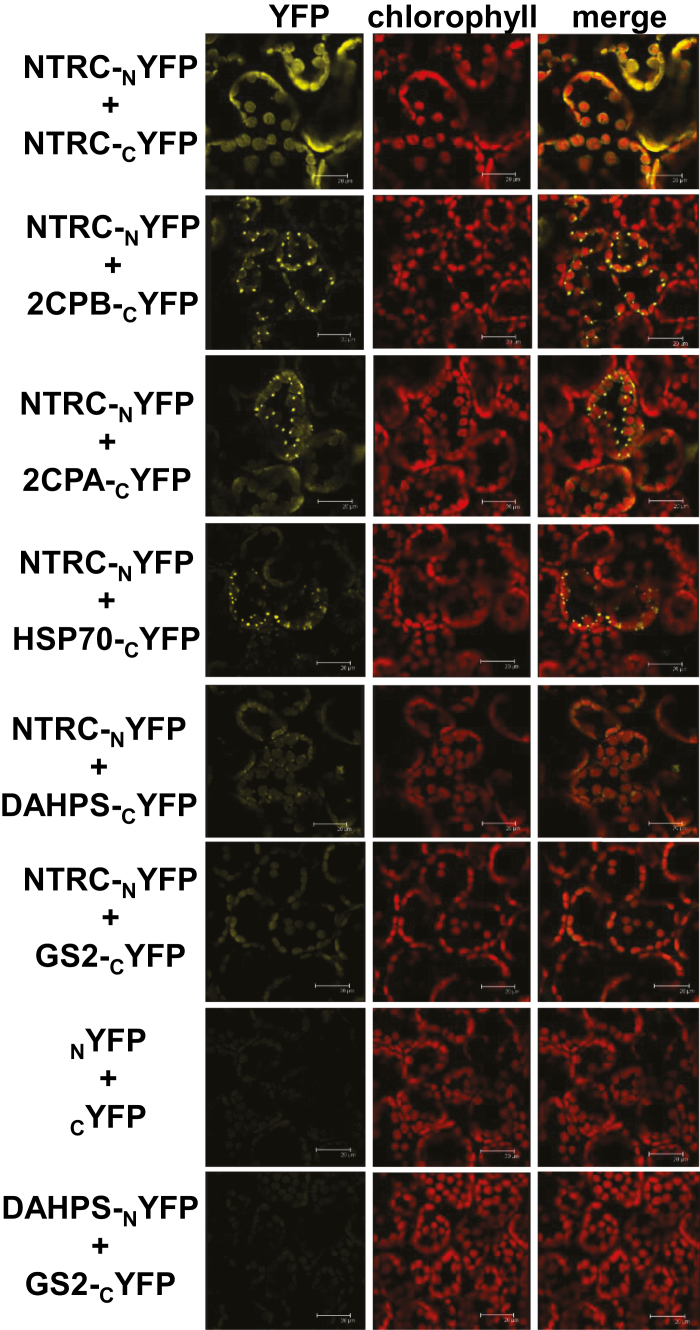
We then selected targets from different categories to test their interaction with NTRC by BiFC analysis after agro-infiltration in leaves of N. benthamiana plants. To this end, we chose 2-Cys PRXs A and B, to confirm the in vivo interaction with NTRC, and extended the analysis to the chaperone CPHSP70. In addition, DAHPS and GS2 were chosen as enzymes putatively redox regulated and involved in nitrogen and amino acid metabolism, the redox regulation of which is poorly known. A first set of experiments was performed in which NTRC was fused to the N-terminus of YFP and the tested protein to the C-terminus of YFP (Fig. 5; Supplementary Fig. S5). To rule out position effects, the opposite orientation, namely NTRC fused to the C-terminus and the tested interacting protein to the N-terminus of YFP, was also analyzed (Supplementary Fig. S6). Finally, a set of samples is shown at lower magnification (Supplementary Fig. S7), so that yellow fluorescence indicating interaction could be distinguished from background signal from chlorophyll fluorescence, thus serving as internal negative control of the BiFC approach. These analyses confirmed the interaction of NTRC with itself and with both 2-Cys PRXs A and B, in these cases displaying the formation of speckles (Fig. 5). Similarly, BiFC assays showed the interaction of NTRC with CPHSP70-1, also with formation of speckles (Fig. 5; Supplementary Figs S5, S6) and with DAHPS and GS2, the signal with these enzymes being less intense (Fig. 5; Supplementary Figs S5, S6), though well above the background level shown by negative controls obtained with the empty YFP vectors (Fig. 5) and also above the background observed in lower magnification images (Supplementary Fig. S7). As an additional negative control, the co-expression of fusion proteins of DAHPS and GS2, proteins that are not known to interact, produced background signal similar to that of the empty vectors (Fig. 5).
Fig. 5.
BiFC analysis of the in vivo interaction of NTRC with selected partners. The interaction of NTRC, fused to the N-terminus of YFP, with selected targets, fused to the C-terminus of YFP, was analyzed by confocal microscopy of mesophyll cells of Nicotiana benthamiana leaves agro-infiltrated with the indicated constructs. Images were acquired 3 d after infiltration. Red, chlorophyll autofluorescence; yellow, YFP fluorescence. Bars correspond to 20 μm. NTRC interaction with itself served as a positive control. Negative controls correspond to the signal obtained with the empty vectors. The interaction of DAHPS with GS2 fused to the N- and C-terminus of YFP, respectively, is also shown as a negative control.
Discussion
Recent reports have uncovered the key function of the redox state of 2-Cys PRXs for the light-dependent reductive activation of chloroplast biosynthetic enzymes (Perez-Ruiz et al., 2017) and for the oxidative inactivation of these enzymes in the dark (Ojeda et al., 2018; Vaseghi et al., 2018; Yoshida et al., 2018). Since NTRC is the most efficient reductant of 2-Cys PRXs (Kirchsteiger et al., 2009; Pulido et al., 2010), these findings imply that both proteins may have a tight interaction in vivo. In support of this notion, different reports, based on in vitro analyses, have shown the interaction between NTRC and 2-Cys PRXs (Pérez-Ruiz et al., 2006; Pérez-Ruiz and Cejudo, 2009; Bernal-Bayard et al., 2012; Yoshida and Hisabori, 2016); however, no evidence of their interaction in vivo has been reported so far. To address this issue, we have used the TAP-Tag approach, which has been previously used to identify in vivo interacting proteins in plants (Rubio et al., 2005). Both confocal microscopy analysis (Fig. S3A–D) and western blotting of extracts of isolated chloroplasts (Supplementary Fig. S3E) confirmed the correct localization of the expressed tagged proteins in the chloroplast. A relevant issue of this approach is to establish the functionality of the tagged NTRC, which was addressed by expressing the tagged enzyme in the ntrc mutant. As shown in Supplementary Fig. S4, these transgenic lines show a partial recovery of the mutant phenotype, thereby suggesting that the tagged NTRC shows a significant level of functionality although it is less active than the endogenous enzyme.
This approach clearly showed the in vivo interaction of NTRC and 2-Cys PRXs in Arabidopsis chloroplasts, a notion further supported by the presence of both proteins in the purified protein complexes, as shown by western blot analysis (Fig. 3A, B) and identification by MS (Table 1). Furthermore, BiFC analysis confirmed this interaction in chloroplasts of N. benthamiana (Fig. 5). Intriguingly, while the NTRC–NTRC interaction produced a homogeneous signal, suggesting an even distribution of the enzyme in the chloroplast stroma (Fig. 5), the pattern of interaction of NTRC with either 2-Cys PRX A or B showed the presence of speckles (Fig. 5), suggesting the formation of protein aggregates. The interaction of NTRC with CPHSP70-1 and, though with weaker signal, with DAHPS also showed the formation of speckles (Fig. 5; Supplementary Figs S5, S6). It is well known that 2-Cys PRXs show a tendency to form aggregates (König et al., 2002; Kirchsteiger et al., 2009) and that this has a great effect on the function of these enzymes since the aggregated form lacks peroxidase activity but shows chaperone activity (Dietz, 2011). Thus, the formation of speckles could reflect the formation of protein aggregates; however, it could also indicate a particular suborganellar localization.
Beside the confirmation of the in vivo interaction of NTRC and 2-Cys PRXs, the purification of protein complexes containing the tagged NTRC gave us the opportunity of identifying other proteins present in the complexes to gain insight on the chloroplast processes putatively modulated by this redox system. The number of partners in each category suggests a relevant role for the NTRC/2-Cys PRX redox system in protein synthesis, response to stress and redox regulation, and carbon metabolism, the system also being important for photosynthesis and photorespiration, nitrogen and amino acid metabolism, tetrapyrrole biosynthesis, and lipid metabolism (Fig. 4). Some of these categories are in agreement with functions already established for NTRC; in addition, NTRC partners identified in this study suggest biological functions, such as protein synthesis or lipid metabolism, among others (Table 1), not previously recognized as NTRC regulated. The significance of the putative partners in each of the biological categories is discussed below.
Stress and redox regulation
As expected, NTRC and 2-Cys PRXs A and B, the proteins used to validate the purification methodology (Figs. 2, 3), were among the proteins identified in this category (Table 1). The presence of PRX IIE suggests the functional relationship of NTRC with other chloroplast-localized PRXs, while the presence of dehydroascorbate reductase, DHAR3, indicates the link of the PRX-dependent antioxidant system with ascorbate metabolism. It should be noted that DHAR3 was identified as a 2-Cys PRX partner (Cerveau et al., 2016) and, thus, the identification of this partner in the protein complexes might be due to its interaction with 2-Cys PRX rather than with NTRC. Although catalase CAT3 was also identified (Table 1) and this protein has predicted chloroplast localization, the presence of catalases in this organelle has not been demonstrated. The identification of stress-responsive proteins such as CPHSP70-1 (Table 1), which was confirmed by BiFC analysis (Fig. 5; Supplementary Fig. S5), might be related to the temperature-sensitive phenotype of the Arabidopsis NTRC knockout mutant (Chae et al., 2013). In this regard, it has been shown that the yeast thiol-specific antioxidant (Tsa1) 2-Cys PRX interacts with HSP70 and that overoxidation of Tsa1 is required for the recruitment of HSP70 to misfolded proteins (Hanzén et al., 2016). Of note, TRXs were not identified as NTRC partners, in line with the inefficient activity of NTRC as a TRX reductant (Bohrer et al., 2012).
Protein synthesis and ribosomal structure
Most of the proteins identified in this category are components of the large and small subunits of chloroplast ribosomes. Redox regulation of chloroplast translation was previously established (Trebitsh and Danon, 2001), and was confirmed by the large number of components of the translation machinery so far identified as TRX targets (Montrichard et al., 2009). Moreover, the identification of ribosome components among the NTRC-containing complexes might be due to their interaction with 2-Cys PRX, which was identified as a ribosome-associated antioxidant in yeast (Trotter et al., 2008). This finding suggests a potential participation of the NTRC/2-Cys PRX system in antioxidant defense of chloroplast translation, which deserves future attention.
Carbon metabolism
The function of NTRC in photosynthetic carbon assimilation is supported by the lower rate of carbon fixation of the ntrc mutant (Perez-Ruiz et al., 2006), which might be related to the finding of large and small subunits of Rubisco as NTRC partners (Table 1). Carbonic anhydrase (CA) was identified as a putative TRX target in previous studies (Balmer et al., 2003; Lee et al., 2004). The activity of a thylakoid CA associated with PSII allows CO2 flux to the chloroplast stroma and the efficient operation of Rubisco (Igamberdiev and Roussel, 2012; Igamberdiev, 2015). Although it has been shown that the activity of the oxidized form of the enzyme is restored by reducing agents, redox regulation of the plant enzyme has not been demonstrated (Balmer et al., 2003). The relevance of CA in providing carbon dioxide for Rubisco could be related to the reduced carbon assimilation of the ntrc mutant.
An interesting NTRC partner in this category is pyruvate kinase 3, PKP3. An Arabidopsis mutant deficient in pyruvate kinase 1 and 2 shows severe alteration in seed fatty acid biosynthesis and seed germination (Baud et al., 2007). Thus, NTRC might participate in the redox regulation of this glycolytic enzyme, which affects fatty acid biosynthesis (Andre et al., 2007). Another NTRC partner in this class, phosphoglycerate kinase (PGK), has been shown to be redox regulated in Synechocystis and Phaeodactylum tricornutum, but not in land plants (Morisse et al., 2014).
It is known that the Arabidopsis ntrc mutant presents decreased starch content and impaired redox regulation of ADP-glucose pyrophosphorylase (AGPase) (Michalska et al., 2009; Lepistö et al., 2013). However, AGPase was identified only in one of the experiments in this study and, thus, was not included as an NTRC partner. In addition, the identification of starch branching enzyme 2 as an NTRC partner may extend the possible NTRC-dependent redox regulation of starch metabolism.
Photosynthesis and photorespiration
Although NTRC-deficient plants show decreased efficiency of photosynthesis (Thormählen et al., 2015; Carrillo et al., 2016; Naranjo et al., 2016), the identification as NTRC partners of proteins involved in photochemical reactions such as PsaB and subunits of the PSI and PSII light-harvesting complexes (Table 1) is somewhat surprising because of the membrane localization of these proteins. Moreover, the lack of Cys in the mature forms of most of these partners (Table 1) suggests that these proteins are not redox regulated. Although the high NPQ shown by the ntrc mutant at low light intensities led to the finding of impaired redox regulation of the γ subunit of ATP synthase in the mutant (Carrillo et al., 2016; Naranjo et al., 2016), this subunit was not found among the identified partners. Thus, the mechanistic basis of the participation of NTRC in the regulation of photosynthetic performance and energy production remains uncertain. An interesting partner of NTRC in this category is Fd, which is the source of electrons for the FTR–TRX redox system and for Fd-NADP reductase (FNR) to generate NADPH, the electron donor of NTRC. Thus, Fd might be a regulatory link between the NTRC with its own source of reducing power and the FTR–TRX redox system.
Phosphoglycolate phosphatase (PGPL) has an important role in the regulation of the levels of 2-phosphoglycolate (2PG) generated by the oxygenase activity of Rubisco. Although redox regulation of PGPL has not been reported, reversible oxidation of three reactive Cys residues was recently described for the highly conserved mammalian enzyme (Seifried et al., 2016). Interestingly, this enzyme has also been identified as a 2-Cys PRX partner in previous studies (Cerveau et al., 2016).
Nitrogen and amino acid metabolism
The partners identified in this category suggest the participation of NTRC in ammonia assimilation via glutamine synthetase and the biosynthesis of amino acids, remarkably of aromatic amino acids such as Trp. The identification of DAHPS, which catalyzes the first step of the shikimate pathway, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), and Trp synthase is in line with the altered levels of these amino acids in the ntrc mutant (Lepistö et al., 2009; Thormählen et al., 2015) and, most importantly, of the lower level of auxin (Lepistö et al., 2009), the synthesis of which derives from Trp (Kasahara, 2016). Isopropylmalate synthase (IPMS), which catalyzes the first committed step of Leu biosynthesis (de Kraker et al., 2007), was also identified as an NTRC-interacting protein. Although no redox regulation of this enzyme has been reported, ntrc mutant plants show altered levels of Leu (Lepistö et al., 2009; Thormählen et al., 2015).
Tetrapyrrole biosynthesis
The pathway of tetrapyrrole biosynthesis is the source of chlorophylls, hence being essential for chloroplast function (Tanaka et al., 2011). There is evidence in support of the redox regulation of chlorophyll biosynthesis (Brzezowski et al., 2015) and, thus, the identification of enzymes of this pathway as NTRC partners is in line with the decreased level of chlorophyll in NTRC-deficient plants. Among the identified partners (Table 1), the I subunit of Mg-chelatase (CHLI) was previously shown to be regulated by NTRC (Pérez-Ruiz et al., 2014). Though HEMB1 contains several Cys residues, no evidence of redox regulation of this enzyme has been reported. In contrast, GluTR and MgP methyltransferase, enzymes reported as regulated by NTRC (Richter et al., 2013), were not identified in this study.
Other biological functions
Additional biological functions, such as lipid metabolism, were represented by a low number of partners (Table 1). Although the participation of NTRC in redox regulation of lipid metabolism has not been analyzed, the identification of subunits of acetyl-CoA carboxylase (ACCase) as partners of NTRC (Table 1) suggests the participation of the enzyme in the redox regulation of fatty acid synthesis, which would thus be coordinated with the regulation of the synthesis of sugars and amino acids. There is evidence showing that ACCase is a TRX-regulated enzyme (Sasaki et al., 1997). Finally, the identification of phospholipase Dα, which is involved in abscisic acid (ABA) signaling (Wang, 2002), suggests that NTRC participates in this signaling pathway. The ntrc mutant is hypersensitive to salt and drought stress (Serrato et al., 2004; Perez-Ruiz et al., 2006) and shows increased stomatal transpiration (Lepistö et al., 2009), which suggests a possible alteration of ABA signaling in these plants.
Concluding remarks
The key role of NTRC and 2-Cys PRXs in chloroplast redox regulation is based on genetic analyses (Perez-Ruiz et al., 2017). However, the knowledge of the targets of NTRC is still scarce and, thus, the molecular basis of the effect of these enzymes on chloroplast redox regulation is poorly understood. Approaches to identify TRX targets by affinity chromatography (Montrichard et al., 2009) use cell extracts and are based on the formation of mixed disulfide, and hence are inefficient for identifying proteins that form part of multienzyme complexes in vivo. In this study we have overcome these limitations using the TAP-Tag methodology with double tagged NTRC as bait. Both western blot and MS analyses showed the presence of 2-Cys PRX among the NTRC-containing complexes, thus indicating the in vivo interaction of these proteins, which was further confirmed by BiFC analysis.
The identification of additional partners in the NTRC-containing complexes points to a relevant role for this enzyme in the redox regulation of processes such as carbon assimilation, tetrapyrrole and amino acid biosynthesis, and photosynthesis, confirming previous results (Kirchsteiger et al., 2009; Michalska et al., 2009; Lepistö et al., 2013; Richter et al., 2013; Pérez-Ruiz et al., 2014; Carrillo et al., 2016; Naranjo et al., 2016). In addition, the identification of components of ribosomes and lipid metabolism suggest previously unknown additional roles for NTRC, which deserve further analyses. It should be noted that the presence in NTRC-containing complexes does not necessarily mean a redox interchange of these proteins with NTRC since these complexes are expected to contain proteins that have non-redox interaction with NTRC as well as proteins interacting with partners such as 2-Cys PRX. In this regard, some of the proteins identified here, including DHA3, PORB, RbcL, and PGLP, were also identified as 2-Cys PRX partners (Cerveau et al., 2016). This would also explain the presence of putative partners without Cys residues.
Finally, the absence of plastidial TRXs as NTRC partners was somewhat surprising. It should be mentioned that although TRXs f1 and m were identified in some of the experiments performed in this study, these proteins were not considered since they were not detected in at least in two of them. In any case, the interaction of NTRC with plastid TRXs is still controversial; while BiFC assays showed the interaction of NTRC with TRXs x and y1, but not with TRX z (Nikkanen et al., 2016), in vitro analyses showed high affinity of NTRC in its interaction with TRX z but not with TRXs x and y (Yoshida and Hisabori, 2016).
Supplementary data
Supplementary data are available at JXB online.
Table S1. Oligonucleotides used for the generation of C-TAPa constructs.
Table S2. Oligonucleotides used for the generation of SPYCE and SPYNE constructs.
Fig. S1. Scheme of the pC-NTRC–TAPa-tag and pC-GFP–TAPa-tag expression cassettes.
Fig. S2. Selection of transgenic lines with a high expression of TAPa-tagged NTRC or GFP.
Fig. S3. Subcellular localization of NTRC and GFP in transgenic Arabidopsis plants expressing pC-GFP–TAPa-tag.
Fig. S4. Phenotype of transgenic plants expressing TAPa-TAG–NTRC or TAPa-TAG–GFP in the ntrc mutant background.
Fig. S5. BiFC analysis of the in vivo interaction of NTRC with selected partners.
Fig. S6. BiFC analysis of the in vivo interaction of NTRC with selected partners.
Fig. S7. BiFC analysis of the in vivo interaction of NTRC with selected partners.
Supplementary Material
Acknowledgements
This work was supported by a European Regional Development Fund-cofinanced grant (BIO2017-85195-C2-1-P) from the Spanish Ministry of Innovation and Competiveness (MINECO), Spain. TAPa-Tag vectors used in this work were provided by Dr V. Rubio (Centro Nacional de Biotecnología, Madrid, Spain). The technical assistance of Alicia Orea (Microscopy Service, Instituto de Bioquímica Vegetal y Fotosíntesis, Sevilla, Spain) is gratefully acknowledged. This article is dedicated to Cristina Spínola, in memoriam
Glossary
Abbreviations
- BiFC
bimolecular fluorescence complementation
- Fd
ferredoxin
- FTR
ferredoxin thioredoxin reductase
- NTRC
NADPH thioredoxin reductase C
- PRX
peroxiredoxin
- TAP
tandem affinity purification
- TRX
thioredoxin
References
- Alkhalfioui F, Renard M, Montrichard F. 2007. Unique properties of NADP-thioredoxin reductase C in legumes. Journal of Experimental Botany 58, 969–978. [DOI] [PubMed] [Google Scholar]
- Andre C, Froehlich JE, Moll MR, Benning C. 2007. A heteromeric plastidic pyruvate kinase complex involved in seed oil biosynthesis in Arabidopsis. The Plant Cell 19, 2006–2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB. 2003. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proceedings of the National Academy of Sciences, USA 100, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Wuillème S, Dubreucq B, de Almeida A, Vuagnat C, Lepiniec L, Miquel M, Rochat C. 2007. Function of plastidial pyruvate kinases in seeds of Arabidopsis thaliana. The Plant Journal 52, 405–419. [DOI] [PubMed] [Google Scholar]
- Bernal-Bayard P, Hervás M, Cejudo FJ, Navarro JA. 2012. Electron transfer pathways and dynamics of chloroplast NADPH-dependent thioredoxin reductase C (NTRC). Journal of Biological Chemistry 287, 33865–33872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernal-Bayard P, Ojeda V, Hervás M, Cejudo FJ, Navarro JA, Velázquez-Campoy A, Pérez-Ruiz JM. 2014. Molecular recognition in the interaction of chloroplast 2-Cys peroxiredoxin with NADPH-thioredoxin reductase C (NTRC) and thioredoxin x. FEBS Letters 588, 4342–4347. [DOI] [PubMed] [Google Scholar]
- Bohrer AS, Massot V, Innocenti G, Reichheld JP, Issakidis-Bourguet E, Vanacker H. 2012. New insights into the reduction systems of plastidial thioredoxins point out the unique properties of thioredoxin z from Arabidopsis. Journal of Experimental Botany 63, 6315–6323. [DOI] [PubMed] [Google Scholar]
- Brzezowski P, Richter AS, Grimm B. 2015. Regulation and function of tetrapyrrole biosynthesis in plants and algae. Biochimica et Biophysica Acta 1847, 968–985. [DOI] [PubMed] [Google Scholar]
- Carrillo LR, Froehlich JE, Cruz JA, Savage LJ, Kramer DM. 2016. Multi-level regulation of the chloroplast ATP synthase: the chloroplast NADPH thioredoxin reductase C (NTRC) is required for redox modulation specifically under low irradiance. The Plant Journal 87, 654–663. [DOI] [PubMed] [Google Scholar]
- Cejudo FJ, Ferrández J, Cano B, Puerto-Galán L, Guinea M. 2012. The function of the NADPH thioredoxin reductase C–2-Cys peroxiredoxin system in plastid redox regulation and signalling. FEBS Letters 586, 2974–2980. [DOI] [PubMed] [Google Scholar]
- Cerveau D, Kraut A, Stotz HU, Mueller MJ, Couté Y, Rey P. 2016. Characterization of the Arabidopsis thaliana 2-Cys peroxiredoxin interactome. Plant Science 252, 30–41. [DOI] [PubMed] [Google Scholar]
- Chae HB, Moon JC, Shin MR, et al. . 2013. Thioredoxin reductase type C (NTRC) orchestrates enhanced thermotolerance to Arabidopsis by its redox-dependent holdase chaperone function. Molecular Plant 6, 323–336. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Da Q, Wang P, Wang M, Sun T, Jin H, Liu B, Wang J, Grimm B, Wang HB. 2017. Thioredoxin and NADPH-dependent thioredoxin reductase C regulation of tetrapyrrole biosynthesis. Plant Physiology 175, 652–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Kraker JW, Luck K, Textor S, Tokuhisa JG, Gershenzon J. 2007. Two Arabidopsis genes (IPMS1 and IPMS2) encode isopropylmalate synthase, the branchpoint step in the biosynthesis of leucine. Plant Physiology 143, 970–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz KJ. 2011. Peroxiredoxins in plants and cyanobacteria. Antioxidants & Redox Signaling 15, 1129–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigenberger P, Thormählen I, Daloso DM, Fernie AR. 2017. The unprecedented versatility of the plant thioredoxin system. Trends in Plant Science 22, 249–262. [DOI] [PubMed] [Google Scholar]
- Hanzén S, Vielfort K, Yang J, et al. . 2016. Lifespan control by redox-dependent recruitment of chaperones to misfolded proteins. Cell 166, 140–151. [DOI] [PubMed] [Google Scholar]
- Igamberdiev AU. 2015. Control of Rubisco function via homeostatic equilibration of CO2 supply. Frontiers in Plant Science 6, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igamberdiev AU, Roussel MR. 2012. Feedforward non-Michaelis–Menten mechanism for CO2 uptake by Rubisco: contribution of carbonic anhydrases and photorespiration to optimization of photosynthetic carbon assimilation. Bio Systems 107, 158–166. [DOI] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Ikeda Y, Matsuura T, Mysore KS. 2016. NADPH-dependent thioredoxin reductase C plays a role in nonhost disease resistance against Pseudomonas syringae pathogens by regulating chloroplast-generated reactive oxygen species. PeerJ 4, e1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Wangdi T, Mysore KS, Uppalapati SR. 2012. NTRC and chloroplast-generated reactive oxygen species regulate Pseudomonas syringae pv. tomato disease development in tomato and Arabidopsis. Molecular Plant-Microbe Interactions 25, 294–306. [DOI] [PubMed] [Google Scholar]
- Ivosev G, Burton L, Bonner R. 2008. Dimensionality reduction and visualization in principal component analysis. Analytical Chemistry 80, 4933–4944. [DOI] [PubMed] [Google Scholar]
- Kasahara H. 2016. Current aspects of auxin biosynthesis in plants. Bioscience, Biotechnology, and Biochemistry 80, 34–42. [DOI] [PubMed] [Google Scholar]
- Kirchsteiger K, Pulido P, González M, Cejudo FJ. 2009. NADPH thioredoxin reductase C controls the redox status of chloroplast 2-Cys peroxiredoxins in Arabidopsis thaliana. Molecular Plant 2, 298–307. [DOI] [PubMed] [Google Scholar]
- König J, Baier M, Horling F, Kahmann U, Harris G, Schürmann P, Dietz KJ. 2002. The plant-specific function of 2-Cys peroxiredoxin-mediated detoxification of peroxides in the redox-hierarchy of photosynthetic electron flux. Proceedings of the National Academy of Sciences, USA 99, 5738–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Lee J, Kim Y, Bae D, Kang KY, Yoon SC, Lim D. 2004. Defining the plant disulfide proteome. Electrophoresis 25, 532–541. [DOI] [PubMed] [Google Scholar]
- Lepistö A, Kangasjärvi S, Luomala EM, Brader G, Sipari N, Keränen M, Keinänen M, Rintamäki E. 2009. Chloroplast NADPH-thioredoxin reductase interacts with photoperiodic development in Arabidopsis. Plant Physiology 149, 1261–1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepistö A, Pakula E, Toivola J, Krieger-Liszkay A, Vignols F, Rintamäki E. 2013. Deletion of chloroplast NADPH-dependent thioredoxin reductase results in inability to regulate starch synthesis and causes stunted growth under short-day photoperiods. Journal of Experimental Botany 64, 3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Florencio FJ. 2003. Thioredoxin-linked processes in cyanobacteria are as numerous as in chloroplasts, but targets are different. Proceedings of the National Academy of Sciences, USA 100, 16107–16112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl M, Kieselbach T. 2009. Disulphide proteomes and interactions with thioredoxin on the track towards understanding redox regulation in chloroplasts and cyanobacteria. Journal of Proteomics 72, 416–438. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Belin C, Delorme-Hinoux V, Reichheld JP, Riondet C. 2012. Thioredoxin and glutaredoxin systems in plants: molecular mechanisms, crosstalks, and functional significance. Antioxidants & Redox Signaling 17, 1124–1160. [DOI] [PubMed] [Google Scholar]
- Michalska J, Zauber H, Buchanan BB, Cejudo FJ, Geigenberger P. 2009. NTRC links built-in thioredoxin to light and sucrose in regulating starch synthesis in chloroplasts and amyloplasts. Proceedings of the National Academy of Sciences, USA 106, 9908–9913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Morisse S, et al. . 2013. Redox regulation of the Calvin–Benson cycle: something old, something new. Frontiers in Plant Science 4, 470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mihara S, Yoshida K, Higo A, Hisabori T. 2016. Functional significance of NADPH-thioredoxin reductase C in the antioxidant defense system of cyanobacterium Anabaena sp. PCC 7120. Plant & Cell Physiology 58, 86–94. [DOI] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. 2009. Thioredoxin targets in plants: the first 30 years. Journal of Proteomics 72, 452–474. [DOI] [PubMed] [Google Scholar]
- Moon JC, Jang HH, Chae HB, et al. . 2006. The C-type Arabidopsis thioredoxin reductase ANTR-C acts as an electron donor to 2-Cys peroxiredoxins in chloroplasts. Biochemical and Biophysical Research Communications 348, 478–484. [DOI] [PubMed] [Google Scholar]
- Morisse S, Michelet L, Bedhomme M, Marchand CH, Calvaresi M, Trost P, Fermani S, Zaffagnini M, Lemaire SD. 2014. Thioredoxin-dependent redox regulation of chloroplastic phosphoglycerate kinase from Chlamydomonas reinhardtii. Journal of Biological Chemistry 289, 30012–30024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. 2001. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proceedings of the National Academy of Sciences, USA 98, 11224–11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naranjo B, Mignée C, Krieger-Liszkay A, Hornero-Méndez D, Gallardo-Guerrero L, Cejudo FJ, Lindahl M. 2016. The chloroplast NADPH thioredoxin reductase C, NTRC, controls non-photochemical quenching of light energy and photosynthetic electron transport in Arabidopsis. Plant, Cell & Environment 39, 804–822. [DOI] [PubMed] [Google Scholar]
- Nikkanen L, Toivola J, Rintamäki E. 2016. Crosstalk between chloroplast thioredoxin systems in regulation of photosynthesis. Plant, Cell & Environment 39, 1691–1705. [DOI] [PubMed] [Google Scholar]
- Ojeda V, Pérez-Ruiz JM, Cejudo FJ. 2018. 2-Cys peroxiredoxins participate in the oxidation of chloroplast enzymes in the dark. Molecular Plant 11, 1377–1388. [DOI] [PubMed] [Google Scholar]
- Ojeda V, Pérez-Ruiz JM, González M, Nájera VA, Sahrawy M, Serrato AJ, Geigenberger P, Cejudo FJ. 2017. NADPH thioredoxin reductase C and thioredoxins act concertedly in seedling development. Plant Physiology 174, 1436–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Cejudo FJ. 2009. A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Letters 583, 1399–1402. [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, González M, Spínola MC, Sandalio LM, Cejudo FJ. 2009. The quaternary structure of NADPH thioredoxin reductase C is redox-sensitive. Molecular Plant 2, 457–467. [DOI] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Guinea M, Puerto-Galán L, Cejudo FJ. 2014. NADPH thioredoxin reductase C is involved in redox regulation of the Mg-chelatase I subunit in Arabidopsis thaliana chloroplasts. Molecular Plant 7, 1252–1255. [DOI] [PubMed] [Google Scholar]
- Perez-Ruiz JM, Naranjo B, Ojeda V, Guinea M, Cejudo FJ. 2017. NTRC-dependent redox balance of 2-Cys peroxiredoxins is needed for optimal function of the photosynthetic apparatus. Proceedings of the National Academy of Sciences, USA 114, 12069–12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Ruiz JM, Spínola MC, Kirchsteiger K, Moreno J, Sahrawy M, Cejudo FJ. 2006. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. The Plant Cell 18, 2356–2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido P, Spínola MC, Kirchsteiger K, Guinea M,Pascual MB, Sahrawy M, Sandalio LM, Dietz KJ, González M, Cejudo FJ. 2010. Functional analysis of the pathways for 2-Cys peroxiredoxin reduction in Arabidopsis thaliana chloroplasts. Journal of Experimental Botany 61, 4043–4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raynaud S, Ragel P, Rojas T, Mérida Á. 2016. The N-terminal part of Arabidopsis thaliana starch synthase 4 determines the localization and activity of the enzyme. Journal of Biological Chemistry 291, 10759–10771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter AS, Peter E, Rothbart M, Schlicke H, Toivola J, Rintamäki E, Grimm B. 2013. Posttranslational influence of NADPH-dependent thioredoxin reductase C on enzymes in tetrapyrrole synthesis. Plant Physiology 162, 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio V, Shen Y, Saijo Y, Liu Y, Gusmaroli G, Dinesh-Kumar SP, Deng XW. 2005. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. The Plant Journal 41, 767–778. [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Kozaki A, Hatano M. 1997. Link between light and fatty acid synthesis: thioredoxin-linked reductive activation of plastidic acetyl-CoA carboxylase. Proceedings of the National Academy of Sciences, USA 94, 11096–11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann P, Buchanan BB. 2008. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidants & Redox Signaling 10, 1235–1274. [DOI] [PubMed] [Google Scholar]
- Seifried A, Bergeron A, Boivin B, Gohla A. 2016. Reversible oxidation controls the activity and oligomeric state of the mammalian phosphoglycolate phosphatase AUM. Free Radical Biology & Medicine 97, 75–84. [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Pérez-Ruiz JM, Spínola MC, Cejudo FJ. 2004. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. Journal of Biological Chemistry 279, 43821–43827. [DOI] [PubMed] [Google Scholar]
- Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Molecular & Cellular Proteomics 6, 1638–1655. [DOI] [PubMed] [Google Scholar]
- Silhavy D, Molnár A, Lucioli A, Szittya G, Hornyik C, Tavazza M, Burgyán J. 2002. A viral protein suppresses RNA silencing and binds silencing-generated, 21- to 25-nucleotide double-stranded RNAs. The EMBO Journal 21, 3070–3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spínola MC, Pérez-Ruiz JM, Pulido P, Kirchsteiger K, Guinea M, González M, Cejudo FJ. 2008. NTRC new ways of using NADPH in the chloroplast. Physiologia Plantarum 133, 516–524. [DOI] [PubMed] [Google Scholar]
- Stenbaek A, Hansson A, Wulff RP, Hansson M, Dietz KJ, Jensen PE. 2008. NADPH-dependent thioredoxin reductase and 2-Cys peroxiredoxins are needed for the protection of Mg-protoporphyrin monomethyl ester cyclase. FEBS Letters 582, 2773–2778. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Kobayashi K, Masuda T. 2011. Tetrapyrrole metabolism in Arabidopsis thaliana. The Arabidopsis Book 9, e0145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WH, Shilov IV, Seymour SL. 2008. Nonlinear fitting method for determining local false discovery rates from decoy database searches. Journal of Proteome Research 7, 3661–3667. [DOI] [PubMed] [Google Scholar]
- Thimm O, Bläsing O, Gibon Y, Nagel A, Meyer S, Krüger P, Selbig J, Müller LA, Rhee SY, Stitt M. 2004. MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. The Plant Journal 37, 914–939. [DOI] [PubMed] [Google Scholar]
- Thormählen I, Meitzel T, Groysman J, Öchsner AB, von Roepenack-Lahaye E, Naranjo B, Cejudo FJ, Geigenberger P. 2015. Thioredoxin f1 and NADPH-dependent thioredoxin reductase C have overlapping functions in regulating photosynthetic metabolism and plant growth in response to varying light conditions. Plant Physiology 169, 1766–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trebitsh T, Danon A. 2001. Translation of chloroplast psbA mRNA is regulated by signals initiated by both photosystems II and I. Proceedings of the National Academy of Sciences, USA 98, 12289–12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotter EW, Rand JD, Vickerstaff J, Grant CM. 2008. The yeast Tsa1 peroxiredoxin is a ribosome-associated antioxidant. The Biochemical Journal 412, 73–80. [DOI] [PubMed] [Google Scholar]
- Vaseghi MJ, Chibani K, Telman W, Liebthal MF, Gerken M, Schnitzer H, Mueller SM, Dietz KJ. 2018. The chloroplast 2-cysteine peroxiredoxin functions as thioredoxin oxidase in redox regulation of chloroplast metabolism. eLife 7, e38194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X. 2002. Phospholipase D in hormonal and stress signaling. Current Opinion in Plant Biology 5, 408–414. [DOI] [PubMed] [Google Scholar]
- Wulff RP, Lundqvist J, Rutsdottir G, Hansson A, Stenbaek A, Elmlund D, Elmlund H, Jensen PE, Hansson M. 2011. The activity of barley NADPH-dependent thioredoxin reductase C is independent of the oligomeric state of the protein: tetrameric structure determined by cryo-electron microscopy. Biochemistry 50, 3713–3723. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Hara A, Sugiura K, Fukaya Y, Hisabori T. 2018. Thioredoxin-like2/2-Cys peroxiredoxin redox cascade supports oxidative thiol modulation in chloroplasts. Proceedings of the National Academy of Sciences, USA 115, E8296–E8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Hisabori T. 2016. Two distinct redox cascades cooperatively regulate chloroplast functions and sustain plant viability. Proceedings of the National Academy of Sciences, USA 113, E3967–E3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.