Abstract
Aging research has experienced a burst of scientific efforts in the last decades as the growing ratio of elderly people has begun to pose an increased burden on the healthcare and pension systems of developed countries. Although many breakthroughs have been reported in understanding the cellular mechanisms of aging, the intrinsic and extrinsic factors that contribute to senescence on higher biological levels are still barely understood. The dog, Canis familiaris, has already served as a valuable model of human physiology and disease. The possible role the dog could play in aging research is still an open question, although utilization of dogs may hold great promises as they naturally develop age-related cognitive decline, with behavioral and histological characteristics very similar to those of humans. In this regard, family dogs may possess unmatched potentials as models for investigations on the complex interactions between environmental, behavioral, and genetic factors that determine the course of aging. In this review, we summarize the known genetic pathways in aging and their relevance in dogs, putting emphasis on the yet barely described nature of certain aging pathways in canines. Reasons for highlighting the dog as a future aging and gerontology model are also discussed, ranging from its unique evolutionary path shared with humans, its social skills, and the fact that family dogs live together with their owners, and are being exposed to the same environmental effects.
Keywords: hallmarks of aging, animal aging models, family dogs, aging genetics, dementia research
Introduction
Dogs (Canis familiaris) are special in the animal kingdom in many aspects. Being the oldest domesticated species, they accompany humans for approximately 15,000–100,000 years (estimates depend on the different approaches used to study their origin; see Vila, 1997; Larson et al., 2012; Thalmann et al., 2013; Frantz et al., 2016; Botigué et al., 2017). Consequently, they have adapted to the special social environment of human communities in a way unmatched by any other species and thus earned their rightful title as man’s best friend. They have also gained diverse functionality throughout the millennia, which resulted in a phenotypic variability unrivalled by any other species. Recently, the species has also been promoted to be a model of human physiology and disease. The sequencing of the dog genome (Lindblad-Toh et al., 2005) and the development of high-resolution genotyping arrays to support genome wide association studies (GWAS) (Hayward et al., 2016) were important steps to open up new perspectives of genetic investigations in dogs. Recently, a growing number of full genome sequences from various dog breeds have also been added to the genetic toolkit of canine researchers (Dreger et al., 2016a; Dreger et al., 2016b; Kim et al., 2018). The importance of these improvements is clearly visible through the expanding list of reported disease associated polymorphisms in dogs. Examples like the famous case of the narcolepsy causing mutation in the canine Hypocretin Receptor 2 gene (Lin et al., 1999), which turned the focus of researchers to its human homolog’s role in narcolepsy, have demonstrated how canine genetics can benefit humans.
Genetic analyses have also helped to shed light on the origin and evolution of dogs and on the divergence of breeds (vonHoldt et al., 2010; Axelsson et al., 2013; Skoglund et al., 2015; Frantz et al., 2016; Freedman et al., 2016; Wang et al., 2016) and have revealed numerous genetic variants responsible for their phenotypic variability. For example, several genes have been shown to affect the body size variability of dogs (Sutter et al., 2007; Hoopes et al., 2012; Rimbault et al., 2013; Plassais et al., 2017), which is unmatched by any other mammalian species. Importantly, dogs also show marked differences in their expected lifespan in connection with body mass. On average, giant sized breeds (above 50 kg) have an expected lifespan of 6–8 years, while small sized breeds (below 10 kg) can live up to 14–16 years (Jimenez, 2016).
This wide range of expected lifespans, together with other aspects, has made dogs promising as model organisms for aging research (Gilmore and Greer, 2015; Creevy et al., 2016; Kaeberlein et al., 2016; Mazzatenta et al., 2017; Hoffman et al., 2018). In this regard, family dogs living as animal companions with their owners could be even more relevant than laboratory dogs (Kaeberlein, 2016). Although laboratory dogs had been traditionally used for a wide range of investigations, including aging research (Cotman and Head, 2008), they have some major limitations. For example, they experience a less complex nutritional and environmental history during their life course than family dogs do, which could lead to major deviances in their base level behavioral parameters. Also, they usually represent only a few breeds. These aspects clearly reduce the power of laboratory dogs to correspond with highly variable natural populations, especially when we consider the range of different aging phenotypes. Actually, describing and characterizing human aging phenotypes is a main goal of researchers, as such variability leads to fundamental differences in individual courses of aging. Living a long life with poor health can negatively affect the welfare of both the elderly and their surroundings. Age-related dementia can especially make a major impact in this regard, rendering patients unable to live an independent life. Therefore, lifespan and healthspan are considered partly independent attributes of human aging, leading to the distinction between healthy aging and pathological aging. Furthermore, in respect to cognitive decline, which can hinder welfare even if no other diseases are present, some authors suggested to discriminate successful aging as a subtype of healthy aging, which is characterized by maintained ability to live an autonomous life until death (Rowe and Kahn, 1987; Rowe and Kahn, 2015). In this regard, family dogs clearly surpass laboratory dogs as models, because they are more abundant, are more variable, and are much more likely to reach an old age and to encounter various aging courses. Although the detailed phenotypic categorization of aging in dogs will require further efforts, definitions for frailty, for example, have already been proposed in the species (Hua et al., 2016).
Nevertheless, it still remains a question how exactly dogs, especially family dogs, can fit in the puzzle of aging genetics among many already well-established experimental models. Despite the huge progress in understanding the genetic basis of morphological variability of dogs, still very little is known about the functional relevance of canine homologs of conserved longevity genes. Currently, this may stand as an obstacle in the way of effectively utilizing dogs as aging models. As family dogs can provide unique insights into many aspects of human aging, the current lack of detailed information about the canine genetic pathways of aging should be overcome by future research approaches. In this review, we provide an overview of the evolutionary conserved biological mechanisms that contribute to aging, following the classification system proposed by López-Otín et al., 2013, and we summarize current knowledge about these pathways in dogs. We also briefly discuss the benefits and limitations of family dogs in aging research and propose possible future directions for canine aging genetic studies.
Dogs as Model Animals in Aging Research
A plethora of different species has been involved in aging studies to unravel the genetic factors behind this complex biological process. Due to their short lifespan and easy handling under laboratory conditions, the yeast (Saccharomyces cerevisiae), the nematode worm Caenorhabditis elegans, the fruit fly (Drosophila melanogaster), and rodents (mice: Mus musculus and rats: Rattus norvegicus) all became important contributors to the discovery of longevity affecting genes. Recently, the turquoise killifish (Nothobranchius furzeri) has also been added to this palette (Hu and Brunet, 2018). The applicability of various genetic approaches (e.g., induced mutagenesis, RNA interference, gene trapping) in these organisms allowed researchers to specifically target genes for further investigations or to efficiently search for phenotype–genotype associations in mutants.
As most of the revealed pathways turned out to be highly conserved, findings made on model organisms seemed translatable to humans in most cases. However, human aging has characteristics, like the occurrence of age-related dementia, which do not have counterparts in many model organisms. Although this limitation has been overcome by different techniques to induce neurodegenerative processes in the central nervous system (CNS) of model animals, the findings of such studies may not be easily implemented in humans (Jucker, 2010; Puzzo et al., 2015; De Felice and Munoz, 2016). In addition, the interaction between genetic factors and environmental conditions can also vary in humans, meaning that certain variants can have beneficial effects in one context and adverse effects in another (Ukraintseva et al., 2016).
Therefore, studies on human populations are inevitable to understand human aging in its full complexity. Apparently, in this case the genetic toolkit is reduced to associative approaches. With the advent of the genomic era, this has become less of a problem, and several genome wide association studies (GWAS) have actually reported lifespan affecting loci in humans (Deelen et al., 2011; Nebel et al., 2011; Sebastiani et al., 2012; Sebastiani and Perls, 2012; Beekman et al., 2013). However, longitudinal studies and testing the effects of anti-aging interventions are still more challenging in humans than in short-lived animals.
When all of these considerations are taken into account, the dog may rise as an excellent midline solution for the limitations of simple organisms and for the challenges human studies hold (Waters, 2011). Here are some examples, why:
Family dogs, on average, age about six to seven times faster than humans. The mean lifespan of companion dogs (purebred and crossbred together) from Europe and Japan were shown to be 12 and 13.7 years, respectively (O’Neill et al., 2013; Inoue et al., 2018), while the mean lifespan of European humans is 77.2, according to a UN report (Anon, 2019) and is around 83 years for Japanese people (Tokudome et al., 2016). Therefore, follow-up studies are much easier in the case of dogs, and have already been performed by several research groups to measure immunological, neuropathological, and metabolomic changes related to canine aging (Su et al., 2005; Greeley et al., 2006; Cotman and Head, 2008).
The fact that the mean lifespan of dogs can range from 5.5 to 14.5 years (Michell, 1999; O’Neill et al., 2013; Jimenez, 2016), depending on body size and breed, suggests that dogs, sharing their lives with humans, gained considerable advantages from this alliance by doubling their mean expected lifespan compared to wild wolves (Mech, 2006). This artificially enhances the proportion of individuals with age-related pathologies, which often show strong correspondences with human diseases, and thus can provide opportunities for translational studies.
Dogs are prone to develop human-like neurodegenerative disorders and are susceptible to age-related cognitive abnormalities. Almost one third of 11–12-year-old dogs and 70% of 15–16-year-old dogs were reported to show cognitive disturbances with symptoms corresponding to human senile dementia: spatial disorientation, social behavior disorders (e.g., problems with recognizing family members), repetitive (stereotype) behavior, apathy, increased irritability, sleep–wake cycle disruption, incontinence, and reduced ability to accomplish tasks (Neilson et al., 2001). Together, these symptoms constitute a typical, age-related, progressive pathological decline in dogs’ mental abilities, which is usually referred to as “Canine Cognitive Dysfunction Syndrome” (CCD) (Cummings et al., 1996; Landsberg et al., 2012). To this day, a vast amount of literature has accumulated about CCD (Szabó et al., 2016; Chapagain et al., 2018), yet there is weak knowledge about the genetic factors influencing it. Importantly, cognitive decline in dogs was associated with β-amyloid accumulation in the prefrontal cortex, noradrenergic neuron loss in the locus coeruleus (Insua et al., 2010), and, lately, with the formation of tau tangles (Schmidt et al., 2015; Smolek et al., 2016), which can all be seen in humans in early stages of neurodegenerative diseases.
Dogs also correspond very well to humans in several metabolic and physiological features, some of which are consequences of domestication (Axelsson et al., 2013). These features have already been thoroughly described in laboratory dogs, as traditional test animals of the pharmacological industry. Therefore, the intestinal absorption profiles of many drugs and supplements are actually known to be very similar in dogs and humans (Roudebush et al., 2005).
Several studies from the last two decades (for a review on the history of dog behavioral research, see Feuerbacher and Wynne, 2011) have supported the notion that dogs possess cognitive abilities that are similar to human social skills in communication and learning (Topál et al., 2009; Bensky et al., 2013; Miklósi, 2014). Also, they have a prolonged postnatal period with high sensitivity for human contact and usually live in a close proximity with people, which makes them able to easily interpret many human actions (Miklósi and Kubinyi, 2016). Therefore, dogs can participate in special experimental protocols, which would not be possible with less trainable and sociable species.
Dogs share more ancestral genomic sequence with humans than rodents do (Lindblad-Toh et al., 2005), and linkage disequilibrium regions can be extensive within dog breeds, making it easier to pinpoint phenotype–marker associations, which can be later narrowed down by interbreed investigations. This provides particular prospects for GWAS (Boyko, 2011; Vaysse et al., 2011; Schoenebeck and Ostrander, 2014; Hayward et al., 2016).
Family dogs are plentiful and easily available at very little cost, so large datasets can be collected via the help of dog owners and veterinarians under citizen science approaches (Hecht and Rice, 2015; Stewart et al., 2015).
These points suggest that family dogs can become valuable models to study complex human traits like aging. However, researchers have to face some obstacles and limitations as well, which have to be addressed properly.
One of these limitations is the still deficient knowledge about the exact functions that conserved genetic pathways play in canine aging. On the one hand, this may seem to be a minor question, as all fundamental cellular senescence mechanisms were reported to be conserved. On the other hand, divergences may occur in each species regarding some of these mechanisms, as for example, both the telomere biology of flies and the somatic telomerase expression of mice were reported to show marked differences from humans (Kipling and Cooke, 1990; Levis et al., 1993; Prowse and Greidert, 1995). Furthermore, the genes and their functions linked to human age-related neurodegeneration may be fundamentally different from their homologs found in model organisms, or even missing from other species (Bitar and Barry, 2017). Consequently, in an ideal setting, each genetic pathway should be evaluated in each species intended for translational studies before further efforts are put into costly and time-consuming investigations.
The variable living environment of family dogs has been discussed as a potential advantage over laboratory dogs; however, it also brings serious challenges to optimal study design. Integrative and cooperative approaches based on large datasets could help to overcome this limitation. Large-scale retrospective studies, which were based on veterinary databases, have already led to important findings regarding the differences in life expectancies between various breeds (Proschowsky et al., 2003; O’Neill et al., 2013; Inoue et al., 2018), or between lean and obese dogs (Salt et al., 2018). In this regard, citizen science approaches can have promising prospects in family dog research (Hecht and Rice, 2015; Stewart et al., 2015), as it was already indicated by a few examples (Hecht and Rice, 2015; Stewart et al., 2015; Ilska et al., 2017). Also, if studies need to involve pathological, histological, and molecular data about dogs that suffered from CCD, citizen science approaches must be expanded to involve a wider range of professionals, including veterinarians. Such interdisciplinary studies may become especially important in cases where family dogs are used as preclinical models to test the anti-aging effects of drugs (Kaeberlein et al., 2016).
Until now few studies involved family dogs in cellular and molecular level investigations, as this may require invasive methods or even the sacrifice of animals. Apparently, such approaches are not applicable in the case of family dogs, which, however, represent the valuable genetic and behavioral variability of the species. These issues were encountered by researchers, who aimed to study brains of non-laboratory dogs, and found it difficult to collect both behavioral and molecular data (which required medically advised euthanasia of the animals) from the same individuals within the time-frame of the study (Ghi et al., 2009). In this regard, the establishment and long-term maintenance of databanks and biobanks that collect behavioral, lifestyle, medical data, and biological materials from family dogs, by providing the opportunity for owners to donate their dogs’ bodies for research purposes under appropriate ethical considerations, would be advantageous for canine genomics and aging research.
Similarly to invasive methods, genetic manipulations may seem less applicable in dogs than in experimental model organisms. Nevertheless, some groups have already applied targeted genetic manipulations in laboratory dogs to create better models of certain medical conditions (Zou et al., 2015). More importantly, therapeutic applications of gene editing have recently been applied on pet dogs suffering from Duchenne muscular atrophy, with promising results (Amoasii et al., 2018). Hence, it is likely that this line of canine genetics and medical research will continue to unfold its potentials.
Currently, methods, by which cognitive aging can be effectively assessed in dogs, are limited. Effective phenotypical categorization of canine age-related pathologies, including CCD, will be crucial for studies, which intend to assess the effects of anti-aging interventions on dogs.
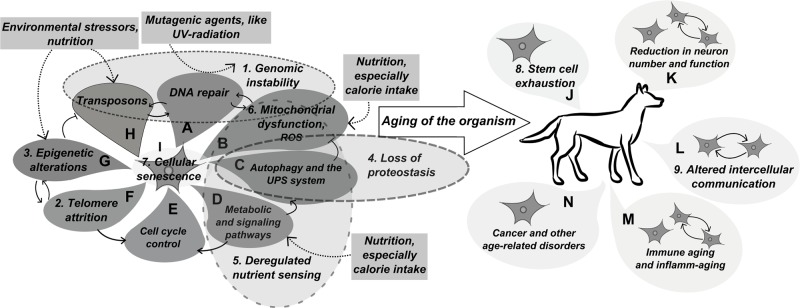
The Hallmarks of Aging in Dogs
Most aging-related genes are components of essential metabolic and signaling pathways ( Figure 1 ), like the ones regulating autophagic activity. Other genes make contribution to cellular processes that affect genomic integrity, either in a protective role (DNA repair mechanisms) or in a destructive manner (oxidative stress, transposons). Some genes may affect aging in a somewhat programmed manner, either through epigenetic modulations or by the altered maintenance of telomeres. Because of this large number of involved genetic—and environmental—factors, establishment of a conceptual framework that can systematically comprise all of them would be a first step to provide better insight into the aging process in its entirety. In this regard, recently nine main factors have been designated as fundamental hallmarks that contribute to the aging of animals, with a main focus on mammals (López-Otín et al., 2013). Each hallmark had to meet three criteria: they must affect longevity and healthspan either in a negative or positive manner and have to show age-related changes in measurable parameters. Thus, the following phenomena were defined as main contributors to mammalian aging ( Figure 1 ): 1) genomic instability; 2) telomere attrition; 3) epigenetic alterations; 4) disruption of proteostasis; 5) deregulation of nutrient sensing; 6) mitochondrial dysfunction; 7) cellular senescence; 8) stem cell exhaustion; and 9) altered intercellular communication. Although some of these could not perfectly fit all of the criteria, they still make an effective framework to work with. Providing a systematic overview of the genetic pathways involved in the aging of dogs is also of high relevance, as it can help defining directions in canine aging research to support the progression of the species into an effective translational model. For example, in the case of the apolipoprotein E (APOE) gene, which has polymorphisms strongly associated with average lifespan and Alzheimer’s risk in humans (Nebel et al., 2011; Broer et al., 2015), the translational relevance of the canine homolog is debatable, because APOE’s sequence was reported to have low conservation between the two species (Sarasa et al., 2010). Nevertheless, the function of the expressed protein may still be conserved. As APOE variants are major risk factors of human dementia, clarifying this question would be an important step to ensure clinical translatability of canine CCD research. In general, exploring more details about genetic pathways and gene variants involved in canine aging and age-related pathologies should be a major consideration of researchers who utilize dogs in aging research.
Figure 1.
Main mechanisms of aging.The figure depicts the mechanism that underlie cellular aging and, consequently, aging of a multicellular organism. The main mechanisms defined as hallmarks of aging by López-Otín et al. in 2013 are numbered according to their order in the main text. (A) The DNA repair machinery provides the first line of defence against mutagenic agents and also interacts with mobile DNA elements through repairing double strand breaks induced by transposons. Impaired function of this machinery can be a main contributor to Genomic instability (1). (B) Reactive oxygen species produced by mitochondrial respiration represent an inner source for DNA lesions, thus interact with the DNA repair machinery: proper functionality of DNA repair enzymes is required to protect cells from oxidative damage, however elevation in the level of oxidative stress may overburden the repair machinery. Age-related Mitochondrial dysfunction (6) can lead to increased oxidative and apoptotic burden. In addition, increased oxidative stress can also affect proteostasis. (C) Macroautophagy is a protective mechanism against malfunctioning mitochondria that might cause oxidative stress, while together with other clearance mechanisms it also functions to remove misfolded proteins and protein aggregates. It also functions as a mechanism for programmed cell death (indicated with dashed line between autophagy and cell cycle control). Together with the ubiquitin-proteasome system (not indicated separately on the figure) its malfunction may lead to Loss of proteostasis (4) in cells. (D) The activity of the autophagy machinery is strictly regulated by different signalling pathways, many of which function in metabolite sensing and cell growth control. Dysruption in these signaling pathways is summarized as Deregeulated nutrient sensing (5) by López-Otín et al (2013). (E) Cell cycle control is a main determinant of Cellular senescence (7). Also, on a multicellular level, dysregulation of cell cycle control may decrease lifespan by initiating tumour formation. (F) In many species telomeres function as a measuring mechanisms to limit the number of potential cell cycles. When telomere length reaches a critical shortness, it will activate cell cycle control mechanisms to render the cell into a senescent state. The telomerase enzyme was also shown to interact with global genomic chromatin maintenance. In general, Telomere attrition (2) is a main hallmark of aging in most mammalian species. (G) Epigenetic regulation involves two main mechanisms, the methylation of CpG islands and modification of the chromatin structure through histone proteins. Chromatin structure at telomeres is important for telomere maintenance and the repression of retroelements by CpG methylation may prevent DNA damage caused by transposon mobilisation. Epigenetic alterations (3) are also linked to aging in both humans and model organisms. (H) Derepression of mobile DNA elements, primarily of retroelements in mammalian genomes, may result in an increased frequency of double strand breaks and insertion mutagenesis leading to increased Genomic instability (1). (I) Altogether, the molecular mechanisms of aging eventually result in Cellular senescence (7). (J) Cellular senescence will lead to reduced function of tissues in a multicellular organism. Depletion of tissue renewing stem cells (8) is also a main hallmark of organismal aging and is a result of cellular senescence and increased activation / differentiation of dormant stem cells in certain tissues. (K) Loss of stem cells and cellular senescene will lead to functional decline in the central nervous system. (L) Altered intercellular communication (9) is also considered a main hallmark of aging in multicellular organisms, as coordinated activity of cells is essential for proper tissue functionality. (M) The immune system can be affected considerably by altered intercellular communication, and this could also lead to increased inflammation in the body, called inflamm-aging. The elevated levels of inflammation may also result from the increasing number of senescent / apoptotic cells. (N) Together with intracellular changes, mainly loss of genomic integrity, disrupted proteostasis and signalling pathways, the imbalanced function of the immune system will contribute to the occurrence of age-related diseases.
Genomic Instability
As organisms age, various forms of damage may accumulate in their genomes, leading to mutations, chromosomal rearrangements, and aneuploidy (Faggioli et al., 2012; Forsberg et al., 2012; Moskalev et al., 2013). Increased mutational burden in somatic cells eventually hinders cellular function and leads to terminal cellular senescence or apoptosis. In cases when cells escape death/senescence inducing processes, malignant transformations can occur as a consequence of genomic damage. Therefore, various protective mechanisms have evolved to prevent or correct DNA damage. Genomic instability arises when the occurrence of deleterious events exceeds the capacity of the DNA damage response system. DNA damaging agents can originate from various extrinsic or intrinsic sources. Intrinsic factors involve oxidative damage, telomere attrition, and transposon insertions.
The DNA Repair Machinery
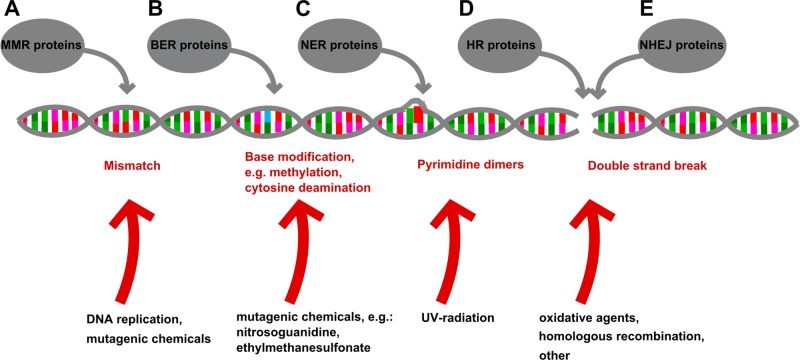
The DNA repair machinery involves divergent pathways, each aimed to correct certain forms of DNA damage ( Figure 2 ). These protective mechanisms have been in the focus of cancer and aging research for a long time (Zimmermann, 1971; Lombard et al., 2005; Cho and Suh, 2014). Defects in DNA repair genes, like the Bloom syndrome RecQ like helicase (BLM) and the Werner syndrome RecQ like helicase (WRN), can lead to severe illnesses, called progeria syndromes in humans, which are characterized by premature aging and other symptoms, including cognitive disabilities and a higher rate of tumorigenesis (Ellis et al., 1995; Yu et al., 1996; Martin, 2005; Arora et al., 2014). Mutations in other DNA repair genes were also reported to increase cancer risk (Jeggo et al., 2016) and thus lead to a reduction in expected lifespan. More importantly, polymorphisms in several genes of the DNA damage response machinery have been actually linked to longevity in humans (Cho and Suh, 2014). Intriguingly, no canine progeria syndrome has been documented in scientific literature. On the other hand, several studies that investigated various forms of canine cancer revealed alterations in the DNA repair machinery, which corresponded to findings in human cancers. For example, a reduced DNA damage response capacity was observed in lymphomas of Golden retriever dogs (Thamm et al., 2013), and a lower expression of the ATM serine/threonine kinase (ATM) gene was found in canine mammary tumors (Raposo-Ferreira et al., 2016). Genetic variations in the breast cancer 1 (BRCA1) and tumor protein p53 (TP53) genes and the MTAP-CDKN2A locus were also linked to various forms of cancer in dogs (Kirpensteijn et al., 2008; Rivera et al., 2009; Shearin et al., 2012). While these findings clearly promote the dog as a natural model of human cancers, it is still unclear how exactly variations in DNA repair capacity contribute to the expected lifespan of dogs. A more detailed discussion of DNA repair in dogs can be found in the review of Grosse et al. (2014).
Figure 2.
The protective roles of the DNA repair machinery. The DNA repair machinery counteracts the effects of variable DNA damaging processes. In healthy cells, the repair machinery can balance these deleterious effects (represented by red arrows); however, in the case of increased mutagenic burden (e.g., exposure to UV radiation), or when members of the repair machinery are not functioning properly, the balance can be lost and a growing number of DNA lesions may cause the cells to die or turn malignant. (A) The function of the Mismatch Repair system (MMR) is coupled to DNA replication where mismatching base pairs can be formed spontaneously and are being identified and repaired by MMR proteins. (B) The Base Excision Repair (BER) system can detect damaged/chemically modified bases in the DNA helix, and remove them, resulting in an apurinated site, which will induce endonucleases to cut back the DNA strand. The single strand break is repaired by a DNA polymerase based on the sequence of the complementary strand, and the newly synthesized sequence is ligated to the original DNA strand by a ligase enzyme. (C) Mutations that disrupt the normal topology of the DNA double helix, like the UV-light induced formation of pyrimidine dimers, are corrected by the nucleotide excision repair (NER) system. This machinery recognizes aberrant DNA structure caused by chemically modified nucleotides and removes these nucleotides resulting in a single strand break, which will be filled in by DNA polymerase and ligase enzymes. (D, E) The most destructive form of DNA damage is double strand break (DSB), which could trigger an immediate apoptotic response if it fails to be repaired. Two distinct mechanisms are used by cells to repair DSB: one is homologous recombination (HR) and the other is non-homologous end joining (NHEJ). HR is a fundamental process also linked to meiosis in eukaryotic cells, and it provides a possibility to recover the damaged DNA strand in full length, by using a homologous DNA helix (e.g., the sister chromatid) as template. In contrast, NHEJ may link ends of double stranded DNA together randomly, which could lead to loss of sequences around the breakpoint. All types of DNA repair are indispensable for normal cellular and organismal function.
Nuclear Architecture
Genomic instability may also rise from altered nuclear architecture. Good examples are the Hutchinson-Gilford and the Néstor-Guillermo progeria syndromes, which were linked to mutations in lamin genes responsible for formation of the nuclear lamina (De Sandre-Giovannoli et al., 2003; Eriksson et al., 2003; Cabanillas et al., 2011). In addition, both the accumulation of progerin, which is an aberrant form of lamin A, and the reduced expression of lamin B1 were linked to aging (Freund et al., 2012; Golubtsova et al., 2016; Hilton et al., 2017), and lamins were also shown to regulate DNA damage response (Gonzalez-Suarez et al., 2009). Thus, the canine homologs of lamins could be promising targets in aging research. So far, a few studies have investigated them, mainly in regard to their possible role in hereditary diseases, like dilated cardiomyopathy of Doberman pinschers and Newfoundland dogs (Wiersma et al., 2007; Meurs et al., 2008) and elbow dysplasia in Bernese Mountain Dogs (Pfahler and Distl, 2012); however, only in the latter case an association was reported between disease occurrence and the lamin B1 gene.
Oxidative Damage
Oxidative damage in cells mainly results from chemical interactions between cellular constituents and reactive oxygen species (ROS), which chemically act as free radicals, characterized by a high oxidative activity. These agents target macromolecules, such as DNA, lipids (Rubbo et al., 1994), and proteins (Stadtman and Levine, 2006), and thus may make a huge impact on cellular function. The sources of ROS are many: mitochondrial respiration (Liu et al., 2002; Murphy, 2009), ionizing radiation (Riley, 1994), and the activity of specific enzymes, such as the NADPH oxidase (Babior, 2004) and dual oxidase (DUOX) (Edens et al., 2001), are the main examples.
The amount of oxidative DNA lesions has been well documented to increase with age in different species. For example, in rats, Fraga et al. (1990) reported the age-related accumulation of 8-hydroxy-2-deoxyguanosine, which is a typical product of DNA oxidation. Furthermore, several studies confirmed that aged dogs show elevated levels of oxidative damage in their brains, indicated by the accumulation of carbonyl groups (Head et al., 2002; Skoumalova et al., 2003), lipofuscin (Rofina et al., 2004), 4-hydroxynonenal (Papaioannou et al., 2001; Rofina et al., 2004; Hwang et al., 2008), and malondialdehyde (Head et al., 2002) in neural tissue. In addition, as the reduced expression of antioxidant enzymes may also contribute to the increased oxidative burden in cells (Kiatipattanasakul et al., 1996), their role in neural aging and neurodegeneration should also be considered. In humans, for example, a mutation in superoxide dismutase 1 (SOD1), which is a main antioxidant enzyme in cells, has been linked to amyotrophic lateral sclerosis (ALS) (Orrell, 2000). Importantly, a mutation in the canine homolog of SOD1 was also linked to an ALS like neurodegenerative process, called degenerative myelopathy (DM) (Awano et al., 2009).
Surprisingly, ROS have also been indicated as important and evolutionary conserved signaling molecules, which function in pathways that respond to availability of nutrients, changes in environmental oxygen levels, and exercise (Schieber and Chandel, 2014; Merry and Ristow, 2016). Hydrogen peroxide (H2O2), for example, plays an important role as signal transducer in the MAPK and Nf-Kβ pathways (Allen and Tresini, 2000) and also serves as activator of peroxiredoxins (Wood et al., 2003), which are crucial for maintaining redox balance of cells. Nitric oxide (NO) has long been indicated to play various physiological roles, with emphasis on the immune and cardiovascular systems (Lundberg et al., 2008). Thus, maintaining optimal levels of oxidative stress in cells could actually be more important for healthy aging than maximizing the neutralization of ROS by antioxidants. In accordance with this, some studies reported controversial effects of oxidative stress in aging and metabolic parameters. For example, it was demonstrated by Ristow et al. (2009) in a human study that supplementation with high doses of extrinsic antioxidants ameliorated the beneficial effects of exercise in volunteers. Furthermore, elevated levels of ROS were reported to either increase lifespan in yeast and worms (Doonan et al., 2008; Van Raamsdonk and Hekimi, 2009; Mesquita et al., 2010) while having no effect on mortality in mice (Van Remmen et al., 2003; Zhang et al., 2009). Also, the lifespan extension of worms promoted by reduced glucose availability was found to be accompanied by elevated levels of ROS in cells (Schulz et al., 2007). Such findings led to the reconsideration of the role oxidative stress plays in cellular senescence and resulted in a more refined view (López-Otín et al., 2013; Shadel and Horvath, 2015). In this regard, it is a question yet to be addressed, how lifelong antioxidant supplementation, often provided by high-quality commercial foods, may affect the healthspan of dogs.
Transposable Elements
The mobilization of endogenous transposable elements, called transposons, has recently gained attention as an intrinsic contributor to cellular senescence (Gorbunova et al., 2014). Transposons are present in the genomes of all organisms, from bacteria to mammals, and possess the ability to change their position in or between chromosomes. They can be categorized into two groups. Retrotransposons move by a replicative “copy and paste” mechanism, increasing in numbers in their host genome ( Figure 3 ), while DNA transposons mainly follow a “cut and paste” mechanism, leaving only a short footprint behind (Wicker et al., 2007; Mandal and Kazazian, 2008). Since the human genome project revealed that around 55% of the human genome is composed of remains of transposable elements, mainly of retrotransposons, an increased attention has been paid to their role in genome evolution (Kazazian, 2004), especially in the formation of gene regulatory networks (Sundaram et al., 2014). While all DNA transposons have lost their mobility in the course of human evolution (Pace and Feschotte, 2007), several retroelements found in our genome are still active and can cause insertional mutations (Hancks and Kazazian, 2016). Human retroelements can be categorized into three groups: the LTR (Long Terminal Repeat) elements, the LINE (Long Interspersed Nuclear Element) transposons, and the SINE (Short Interspersed Nuclear Element) transposons (Mandal and Kazazian, 2008). Similar types of retroelements can be found in the dog genome; however, transposon-derived sequences make up only 34% of it (Lindblad-Toh et al., 2005). Importantly, active LINE and SINE elements are present in both species.
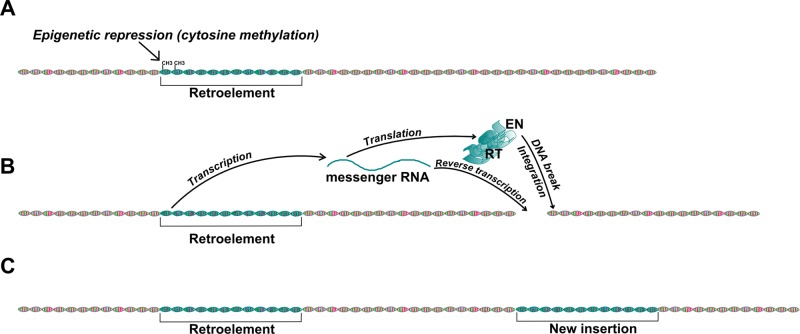
Figure 3.
Mobilization of retroelements in the genome. This picture shows the basic mechanism of retrotransposon mobilization. (A) Normally, activity of functional retroelements, like LINE-1, is repressed in somatic cells by methylation of CpG islands in their promoter regions. (B) Demethylation of the transposon promoter may result in transcriptional activation. The transcribed mRNA encodes the proteins necessary for retrotransposition, the Integrase (Int) and Reverse Transcriptase (RT), and also serves as template for reverse transcription. The reverse transcribed transposon DNA will be integrated into the genome by the Int protein, which first induces a double strand break. (C) The retroelement has copied itself into a new genomic region.
Active retroelements have been found responsible for several hereditary diseases in dogs by causing insertional mutations. For example, a LINE-1 (L1) insertion in the gene of Factor IX was shown to segregate with mild hemophilia in German Wirehaired Pointers (Brooks et al., 2003), while a similar insertion in the dystrophin gene leads to Duchenne-like muscular dystrophy in Pembroke Welsh Corgi dogs (Smith et al., 2011). SINE elements were also shown to cause several inherited diseases, like recessive centronuclear myopathy in Labrador Retrievers (Tiret et al., 2005) and early canine retinal degeneration, which was linked to the serine/threonine kinase 38 like (STK38L) gene in Norwegian Elkhound–Beagle outcrosses by linkage mapping (Goldstein et al., 2010). A form of progressive retinal atrophy (PRA) in Tibetan Spaniels and Tibetan Terriers was also associated with a SINE insertion, but in the family with sequence similarity 161 member A (FAM161A) gene (Downs et al., 2014). Bandera’s neonatal ataxia in Coton de Tulear dogs was shown to be caused by the disruption of the glutamate metabotropic receptor 1 (GRM1) gene by recent retrotransposon mobilization, as the insertion was not found in other breeds (Zeng et al., 2011). Interestingly, several examples of non-disease causing insertional mutations are known, which alter morphology (Parker et al., 2009; Marchant et al., 2017) or coat color (Clark et al., 2006; Dreger and Schmutz, 2011) and thus have become selection criteria in many breeds. Beyond these examples, where the integration event can be revealed by a phenotypic effect or disease, the mobilization of retroelements seems common in dogs, as analyses of individual dog genomes showed that approximately half of annotated dog genes contain a SINEC_Cf type insertion in their introns (Wang and Kirkness, 2005). This high activity of retrotransposons in the lineage of domestic dogs can be explained by intense selection pressures that resulted from domestication, breeding strategies, and changing environment (Capy et al., 2000; Chénais et al., 2012). This hypothesis was actually supported by the findings of Koch et al. (2016), who compared the methylation patterns of wolf and dog genomes and found that almost half of the sites potentially relevant in domestication contained a LINE or SINE insertion.
Beyond the germ-line mutations discussed so far, a vast body of evidence indicates that retroelements can mobilize in somatic cells (Collier and Largaespada, 2007; Hunter et al., 2015) although this is strictly controlled by specific non-coding small RNAs and epigenetic regulation, including hypermethylation and transcriptional repression (Levin and Moran, 2011; Pizarro and Cristofari, 2016). As the general hypomethylation of the genome has long been documented to be an attribute of aging (Wilson and Jones, 1983; Singhal et al., 1987), more and more researchers have suggested a main role for somatic transposon mobilization in cellular senescence (Murray, 1990; Sturm et al., 2015; Pal and Tyler, 2016). Importantly, this hypothesis was also supported by experimental findings. For example, the artificial downregulation of the yeast Ty1 element resulted in lower levels of age-related chromosome rearrangements in aged cells (Maxwell et al., 2011). Also, many of the 118 L1 subfamilies of mice showed an elevated expression with age (De Cecco et al., 2013). In humans the hypomethylation of LINE-1 and Alu (a SINE element abundant in the human genome) elements has been linked to cancer susceptibility (Zhu et al., 2011; Luo et al., 2014).
Because the activation of transposable elements can be induced by various environmental stressors as well (Hunter et al., 2015), including heavy metal toxicity (Morales et al., 2015), certain genotoxic agents (Stribinskis and Ramos, 2006), and even nutrition (Waterland and Jirtle, 2003), they represent another possible intracellular interface between the living environment/lifestyle and aging. In this regard, family dogs, which share their environment with their owners, can be valuable models to study how retrotransposons may contribute to aging and mortality under various circumstances. The age-related activity of retroelements has not yet been specifically assessed in dogs. However, in a study that investigated the elevated blood levels of SINE sequences in dogs with mammary tumors, it was shown that tumor-affected dogs above 10 years of age had higher levels of circulating SINE elements than younger dogs with tumors (Gelaleti et al., 2014).
Telomere Attrition and Aging
In most eukaryotic cells, shortening of the protective sequences at chromosome ends ( Figure 4A ), called telomeres, occurs with each DNA replication. Therefore, telomere shortening has been proposed as a key mechanism of cellular senescence and it also suggested the existence of an aging program in cells (Shampay and Blackburn, 1988; Harley et al., 1990; Hastie et al., 1990). This so called replicative aging limits the number of cell cycles a cell can go through before reaching its Hayflick limit and entering a senescent state (Hayflick, 1976). Furthermore, a recent study supported the conserved role of this aging program on the level of whole organisms by reporting clear correlations between the rate of telomere shortening and the lifespan of different mammalian and bird species (Whittemore et al., 2019). Importantly, telomere shortening is a characteristic only of somatic cells, while in germ line cells, telomere sequences are constantly restored by telomerase enzymes. The limited proliferative potential of somatic cells may seem disadvantageous for an individual, yet it may increase fitness by limiting the growth of malignant cells. In line with this, recent studies have suggested a trade-off between telomere length and cancer occurrence (Zhang et al., 2015; Stone et al., 2016). On the other hand, loss of telomeres can result in end-to-end chromosomal fusions, which might also lead to tumorigenesis (Hastie et al., 1990). These findings indicated that fine-tuning of telomere dynamics in somatic cells might be crucial for healthy aging, at the cost of reducing the maximal lifespan. In fact, polymorphisms in genes associated with telomerase function were shown to be linked with expected lifespan and disease predisposition in human populations (Atzmon et al., 2010; Soerensen et al., 2012b; Codd et al., 2013). Telomere dynamics may also play an influential role in neurodegeneration, as patients with Alzheimer’s showed shorter average telomere length than healthy controls (Forero et al., 2016). Therefore, understanding the links between telomeres and age-related changes on the cellular level, which can lead to pathological processes, is a main goal in aging research. However, in some animals, including common laboratory models, telomerase biology does not entirely correspond to that described in humans. The laboratory mouse, for instance, was shown to exhibit a high variability of telomere length and telomerase expression in adult tissues (Kipling and Cooke, 1990; Prowse and Greidert, 1995; Greenberg et al., 1998; Martín-Rivera et al., 1998), indicating a lesser role of constant telomere attrition as a programmed aging inducer. More importantly, D. melanogaster was shown to possess a fundamentally different telomere structure than found in other animals, as chromosome ends of fruit flies are capped by transposon derived sequences (Levis et al., 1993). These facts clearly limit the applicability of these species as models of human telomere function (Wright and Shay, 2000; Smogorzewska and De Lange, 2002). Nevertheless, longevity of mice was still shown to be positively affected by gene therapy induced telomerase expression (de Jesus et al., 2012).
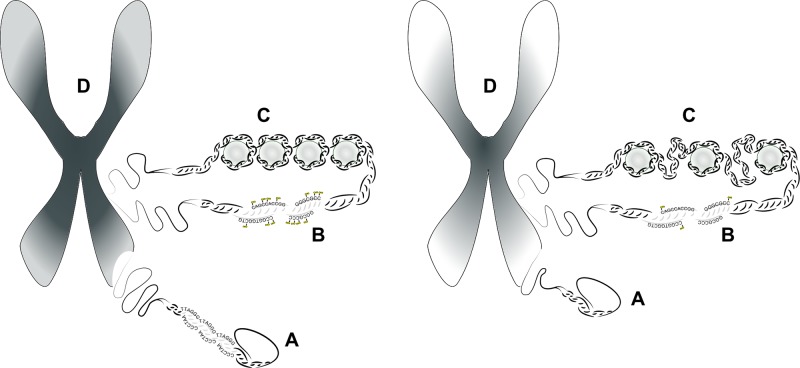
Figure 4.
The role of telomeres and epigenetics in chromosomal integrity and aging. The figure illustrates how shortening of telomeres and changes in the epigenetic pattern affect the overall structure of chromosomes. (A) Chromosome ends are protected by repetitive sequences called telomeres in most eukaryotic organisms. This telomere sequence, consisting of TTAGGG repeats, shortens with each DNA replication, which eventually triggers cellular senescence. (B) Chromatin changes occur on the first level of DNA packaging, when the DNA double strand is coiled up on nucleosomes. Tight coiling on nucleosomes results in a heterochromatic state, when the DNA double helix is not accessible to many proteins, including the transcription machinery. In contrast, reduction in the number of nucleosomes leads to a less coiled and less dense state rendering the DNA more open to transcription. (C) DNA methylation at CpG islands cause chemical changes directly in the DNA double helix. Cytosine methylation is usually linked to silencing of transcription. Methylation also interacts with chromatin structure: increased CpG methylation is usually linked to heterochromatic state. (D) Changes in chromosomal structure during aging is characterized by a decrease of heterochromatic regions (symbolized by darker color) and an increase of euchromatic regions (symbolized by lighter colors).
Contrary to mice, dogs were reported to have low or no telomerase expression in normal somatic tissues, a pattern similar to that in humans (Nasir et al., 2001). It was also reported by Yazawa et al. (1999) Yazawa et al. (2001). Furthermore, tumors in dogs often showed high levels of telomerase expression, similarly to human malignancies (Vonderheide et al., 1999; Lamb et al., 2015). Although very little is known about the molecular mechanisms regulating telomere maintenance and cell cycle arrest in dogs, such findings indicate that dogs may also share basic telomere biology with humans. Importantly, telomere length was shown to be variable across different dog breeds and was in correlation with expected lifespan (Fick et al., 2012). Also, telomere length in individual dogs was found to decrease with age (Nasir et al., 2001), similarly as described in humans (Harley et al., 1990; Hastie et al., 1990; Lindsey et al., 1991).
Epigenetic Alterations
Epigenetics refers to mechanisms that modulate gene expression by determining how the transcription apparatus can access different sections of the genomic DNA. The condensation procedure, which literally packs the DNA double helix into a dense structure, called chromatin, is one of the main mechanisms to provide epigenetic regulatory potentials ( Figure 4C ). The structure of chromatin is determined by histone proteins, which constitute the basic building blocks for DNA condensation, the nucleosomes. The more densely packed heterochromatic state renders the DNA inaccessible for RNA polymerases and thus inhibits gene expression, while genes positioned in euchromatic sites are open to transcription. However, not all of these genes can be transcribed, even if appropriate activating factors are present, as another epigenetic mechanism, the methylation of cytosines at specific GC-rich sites (called CpG islands), may block transcription ( Figure 4B ). This process has an important role in cellular differentiation and probably also acts as genomic “memory,” storing information about the fate of individual cells (Bird, 2002; Halley-Stott and Gurdon, 2013). Abnormal somatic alterations in DNA methylation have been linked to various diseases, including schizophrenia (Hannon et al., 2015; Wockner et al., 2015). Furthermore, changes in chromatin structure and methylation pattern are often found in cancer (Daniel and Tollefsbol, 2015), where the disruption of cellular identity and concurrent dedifferentiation is a common phenomenon.
Interestingly, the genomic methylation pattern is erased and rewritten during spermato- and oogenesis and after fertilization in mammals (Geiman and Muegge, 2009; Seisenberger et al., 2012; Smith et al., 2012). The exact role these mechanisms play in aging, however, is still unknown.
In general, systemic changes in the ratio of heterochromatic and euchromatic regions ( Figure 4D ) and a global hypomethylation of the genome have been shown to accompany aging (Gentilini et al., 2013; Pal and Tyler, 2016). When focusing on specific genomic regions, however, both hypomethylation and hypermethylation should be taken into account (Maegawa et al., 2010). Actually, senescence-related changes in the DNA methylation profile may include both the activation of pro-aging genes and the repression of anti-aging genes, as in the case of WRN and LMNA (Fraga and Esteller, 2007). The remodeling of chromatin structure, induced by methylation and acetylation of certain histone protein residues, also shows complex age-related patterns (Fraga and Esteller, 2007; Han and Brunet, 2012). Importantly, both chromatin dynamics and DNA methylation were shown to interact with other age-related genetic pathways, like telomere-length control (Blasco, 2007). In turn, the telomerase enzyme was found to affect chromatin structure and DNA repair mechanisms (Masutomi et al., 2005). In addition, the epigenetic pattern is regulated by many factors other than developmental status, like stress, exercise, and diet (Daniel and Tollefsbol, 2015), which therefore can also affect aging through altering the expression of certain genes.
Although age associated changes in chromatin structure and DNA methylation patterns have been reported in several model animals, there can be major differences between species. For example, epigenetic regulation in C. elegans seems to be limited to chromatin remodeling by histone modifications, as m5C DNA methylation pattern does not exist in this organism (Bird, 2002), limiting its utilization as a model to study epigenetic changes in aging. Nevertheless, the histone demethylase UTX-1 was shown to regulate aging in worms (Jin et al., 2011).
In dogs, an increasing body of evidence has suggested epigenetic regulation behind species and breed-specific traits (Koch et al., 2016; Banlaki et al., 2017; Cimarelli et al., 2017). Importantly, a recent study demonstrated that changes in methylation status in DNA regions, which were homologous to regions with known age-sensitive methylation patterns in humans, were in strong correlation with chronological age in dogs and wolves (Thompson et al., 2017). This finding supported the applicability of the dog as a model of age-related epigenetic changes, while it also provided a molecular approach to determine the biological age of individual canines.
Regulation of Epigenetic Pattern
The regulation and maintenance of the epigenetic pattern are coordinated by various enzymes, which act downstream of metabolic and signaling pathways. Altered functions of these enzymes were shown to have a major impact on health and aging. Most importantly, sirtuin genes were among the first shown to affect longevity in yeast (Kaeberlein et al., 1999), C. elegans (Tissenbaum and Guarente, 2001), Drosophila (Rogina and Helfand, 2004), and mice (Calvanese et al., 2009). Sirtuins exert various enzymatic functions, including histone deacetylation, and thus play a key role in the maintenance of chromatin structure (Longo and Kennedy, 2006; Fraga and Esteller, 2007). They also interact with many signaling and metabolic pathways, and regulate oxidative metabolism, stress response, autophagy, and the maintenance of telomeres (Jia et al., 2012; Kim et al., 2012). In mammals, seven sirtuins are known with divergent functions (Guarente, 2011) and at least three of them—SIRT1, SIRT3, and SIRT6—have been implicated to modulate aging. Importantly, polymorphisms in sirtuin genes have been actually linked to human longevity (Kim et al., 2012; Albani et al., 2014).
Sirtuins and other histone-modifying enzymes, together with DNA methyltransferases, have been barely studied in dogs so far. However, as the sequence and function of sirtuin genes show a highly conserved nature (Greiss and Gartner, 2009; Gaur et al., 2017), they are likely to play similar roles in the aging of dogs as in other species. In fact, altered expression of sirtuin genes, mainly that of SIRT1, have been implicated in canine tumors (Marfe et al., 2012), similarly as in humans (Brooks and Gu, 2009). Several sirtuin-targeting drugs have been proposed as promising pharmacological interventions to fight disease and aging (Dai et al., 2018); therefore, they are likely to be utilized in the future as anti-aging therapeutics and may be applied in dogs as well. In this regard, some compounds that interact with histone-regulating enzymes have already been tested in dogs for various reasons. For example, the histone deacetylase inhibitors AR-42 and panobiostat showed promising results in dog cell line models of prostate cancer and B-cell lymphoma, respectively (Elshafae et al., 2017; Dias et al., 2018). More importantly, resveratrol, which has sirtuin-activating effect (Gertz et al., 2012), was reported to positively affect the immune function of healthy pet dogs (Mathew et al., 2018) and it also effectively inhibited the growth of canine hemangiosarcoma in vitro (Alderete et al., 2017). Actually, resveratrol is one of the most comprehensively studied naturally occurring compounds with suggested beneficial effects on health and aging. It was shown to activate SIRT1 and improve mitochondrial function in mice (Lagouge et al., 2006) and to reverse age-related cognitive decline in learning and memory in rats (Gocmez et al., 2016). However, its longevity benefits are still dubious. For example, it only increased the relative survival of mice when the animals received a high calorie diet (Baur et al., 2006). Longitudinal follow-up studies on family dogs may help to clarify this question, as these animals represent a naturally variable population regarding diet and genetic background.
Age-Related Changes in Gene Expression
Alterations in the epigenetic pattern, together with the availability of transcription factors and activation of signaling pathways, can influence the whole expressed mRNA content (the transcriptome) in cells. Not surprisingly, altered gene expression patterns were shown to correlate with aging in mice, humans, and dogs (Lee et al., 1999; Lee et al., 2000; Lu et al., 2004; Zahn et al., 2007; Swanson et al., 2009). Comparisons between species-specific expression profiles have already been implicated as powerful tools to identify evolutionary conserved regulatory pathways (de Magalhães et al., 2009). In this regard, further gene expression data from dogs, especially from individuals with CCD, may also help researchers to pinpoint the shared molecular pathways of human and canine neurodegeneration.
The expression of some microRNAs (miRNAs), which are small non-coding RNAs with important regulatory functions, was also shown to correlate with aging in humans and mice (Somel et al., 2010; Drummond et al., 2011; Inukai et al., 2012; Zhang et al., 2012). Furthermore, age-associated miRNAs—named as gero-miRNAs—were identified in various organisms and were shown to target mRNAs associated with longevity pathways (Gonskikh and Polacek, 2017). Thus, characterization of gero-miRNAs would be a crucial step in dog aging research to further support the role of the dog as a translational model of human aging. Efforts have already been made to provide a detailed annotation of canine miRNAs (Penso-Dolfin et al., 2016), including the establishment of a miRNA tissue atlas in Beagle dogs (Koenig et al., 2016).
Disruption of Proteostasis
Proteins represent the key functional components of cells. The totality of all protein types expressed simultaneously in a cell is called the proteome. Proteome integrity is indispensable for the optimal functionality of cells; therefore, several mechanisms have evolved to maintain its homeostasis—called proteostasis. Impairments in proteostasis can lead to cellular senescence and even severe diseases, called proteinopathies, which mainly affect the CNS and are caused by the excessive accumulation and aggregation of misfolded proteins (Pievani et al., 2014). Loss of proteostasis is hypothesized to be a general attribute of aging cells across different taxa (Koga et al., 2011). For example, it was reported to be an early sign of aging in worms (Ben-Zvi et al., 2009) and to be a characteristic change during both premature and normal aging in mice (Wilson et al., 2015).
Proteostasis is maintained by the orchestrated function of mechanisms, which provide protein quality control, support the folding of synthesized proteins, protect them from various stressors, and eventually remove aberrant or senescent proteins from the cell. The folding and stability of proteins are mainly supervised by so called chaperone proteins, while the efficient removal of unnecessary, damaged, or senescent proteins is handled by two machineries: the ubiquitin-proteasome system (UPS) and the autophagy-lysosome pathway.
Chaperones and Protein Quality Control
Chaperone proteins play an important role in the post-translational maturation of nascent proteins by facilitating their folding. They also function as protectors of mature proteins under various stressful conditions, by helping to maintain their natural conformation and by preventing aggregation. Actually, the first identified chaperones were named heat shock proteins (Hsp), because their expression was induced by elevated temperatures. Importantly, many of these stress responsive chaperones were reported to show reduced expression with aging (Calderwood et al., 2009), and genetic manipulations that affected the expression of certain heat shock proteins resulted in altered aging phenotypes in model organisms. Overexpression or upregulation of Hsp-s was shown to extend lifespan, together with providing increased stress resistance, in both worms and flies (Walker and Lithgow, 2003; Morrow et al., 2004; Chiang et al., 2012), while reduced chaperone function caused accelerated aging in mice (Min et al., 2008).
In humans, chaperones, together with other proteostasis maintenance mechanisms, were suggested to play important roles in neurodegenerative diseases (Morimoto, 2008). This role, however, may not be entirely protective, as some Hsp-s were actually indicated to augment propagation of malformed proteins in proteinopathies (Dickey et al., 2007; Luo et al., 2007).
In dogs, the few studies that investigated chaperone proteins in relation to aging reported similar age-related changes as in humans. For example, blood levels of the Hsp70 chaperone were shown to decrease with age in dogs (Alexander et al., 2018), similarly to what had been previously reported in humans (Deguchi et al., 1988). Interestingly, a research group that investigated the hippocampi of donated pet dogs from various breeds (Ghi et al., 2009) reported an age-related increase in Hsp90 levels. This finding could indicate both a compensatory response to the accumulation of damaged proteins and a more direct link between Hsp90 and age-related neural decline in dogs, similarly as it was suggested in humans, where Hsp90 was implicated as a factor that may actually drive spreading of taupathy (Dickey et al., 2007; Luo et al., 2007). Based on these possible similarities between canine and human chaperone functions in the brain, dogs can be suitable to test various pharmacological interventions and small molecular chaperones (Calamini et al., 2012), which modify or complement chaperone activity to support proteostasis and reduce neurodegenerative pathologies. Such interventions have already been successfully tested in rodents (Gehrig et al., 2012).
The Ubiquitin-Proteasome System
The UPS is responsible for the selective removal of misfolded and senescent proteins in cells. Mutations in genes that encode subunits of the proteasome and proteins responsible for proteasomal targeting can lead to accumulation of aberrant proteins, and have been actually linked to several types of neurodegenerative diseases, including Alzheimer’s disease and Parkinson’s disease (Zheng et al., 2016). Especially in the case of AD, causative links were described between disturbances in the UPS and progression of the disease (de Vrij et al., 2004; Liu et al., 2014).
Importantly, the UPS was also linked to longevity in model organisms. In flies, loss-of-function mutations in the ubiquitin activating enzyme E1 were shown to reduce lifespan and cause disturbances in motor function (Liu and Pfleger, 2013); meanwhile, extended lifespan in worms was associated with increased expression of a proteasome subunit (Vilchez et al., 2012).
In dogs, an increased density of ubiquitinated bodies was reported to be present in the brains of aged individuals (Ferrer et al., 1993; Borràs et al., 1999), and further signs of impaired proteostasis were also indicated (reviewed by Romanucci and Della Salda, 2015). The same group that reported age-related increase in the Hsp90 chaperone in dog brains also found incongruent changes in the abundance of various proteasomal proteins, suggesting complex impairments and compensatory mechanisms in the regulation of the UPS in aged dogs (Ghi et al., 2009).
Interestingly, a homozygous lethal mutation in the proteasome β2 subunit was reported as the possible causative variant behind the unique harlequin coat color of Great Dane dogs (Clark et al., 2011). Further studies may shed light on the possible health or longevity effects of this mutation.
Importantly, as proteasome activation by pharmacological agents has been proposed as a promising approach to delay aging and the development of age-related diseases (Chondrogianni et al., 2015), dogs may provide an appropriate large animal model for pre-clinical testing of these interventions, especially in the case of brain pathologies.
Autophagy
While the UPS eliminates individual proteins or small aggregates tagged by ubiquitin, the autophagic machinery is capable of targeting greater amounts of cellular content for lysosomal degradation, including mitochondria and large protein aggregates. In fact, autophagy is a fundamental mechanism in eukaryotic cells and was often found indispensable for the ontogenesis of multicellular organisms, including the embryonic development of mice (Cecconi and Levine, 2008) and the metamorphosis of flies (Juhász et al., 2003). Three main types of autophagy have been described in the literature. In the case of macroautophagy, targeted cytoplasmic constituents get isolated by double membrane vesicles, called autophagosomes. These then fuse with lysosomes, leading to the degradation of their content into small molecular components ( Figure 5 ), which can be recycled thereafter (Klionsky, 2005). The other two types, microautophagy and chaperone-mediated autophagy, utilize different targeting mechanisms and may be less capable for bulk degradation of intracellular content. Therefore, the name autophagy usually refers to macroautophagy in the literature. In general, all types of autophagy play a crucial role in cellular metabolism (Rabinowitz and White, 2010); pathogen resistance (Levine, 2005; Deretic, 2006); inflammation (Levine et al., 2011); and cleansing of macromolecular debris, like protein aggregates seen in Alzheimer’s disease (Rubinsztein, 2006; Mizushima et al., 2008; Nixon and Yang, 2011) and in programmed cell death (Tsujimoto and Shimizu, 2005; Ouyang et al., 2012). Impairments in autophagy were linked to several disease phenotypes in model organisms, as well as in dogs (listed below) and humans (Levine and Kroemer, 2008). Importantly, reduced autophagic activity in the adult brain was shown to promote neurodegeneration in mice (Hara et al., 2006; Komatsu et al., 2006).
Figure 5.
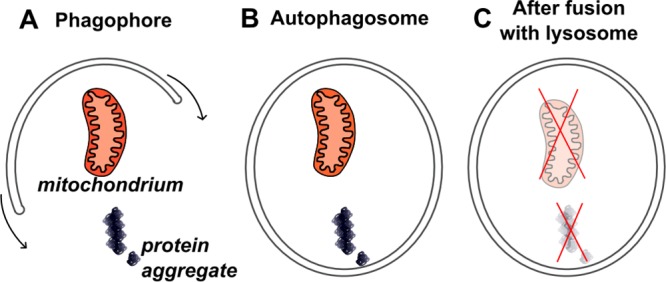
Macroautophagy. This figure depicts macroautophagy, which is the only mechanism in cells able to remove aberrant mitochondria and large protein aggregates. (A) First, formation of a double membrane structure, the phagophore, is initiated around the target. (B) The expansion of the double membrane structure around the target will eventually form a vesicle, called autophagosome. (C) When the autophagosome fuses with a lysosome, degradation of the autophagosome’s content can take place and resulting molecular compounds can be recycled thereafter.
Based on these findings, it is not surprising that autophagy has been proposed as a major factor in aging regulation. In C. elegans, loss-of-function mutations in autophagy genes shortened lifespan, while disruption of signaling pathways that downregulate autophagy led to a significant increase in expected lifespan (Hars et al., 2007; Tóth et al., 2008). Similar findings were reported from yeast, flies, and mice (Juhász et al., 2007; Simonsen et al., 2008; Eisenberg et al., 2009; Pyo et al., 2013), although with a less pronounced lifespan extension in the latter. Importantly, the longevity effect of caloric restriction (CR)—which is discussed in the Supplementary Section “Beyond Genetics”—was shown to be dependent on the proper functioning of autophagy (Jia and Levine, 2007). Chaperon-mediated autophagy was also reported to directly affect cellular senescence through the selective elimination of soluble proteins (Cuervo and Dice, 2000; Massey et al., 2006; Zhang and Cuervo, 2008). In the livers of aged mice, but not of young animals, impaired function of chaperone-mediated autophagy resulted in increased loss of proteostasis (Schneider et al., 2015).
Surprisingly, cohort studies have reported little or no association between autophagy linked genes and longevity in humans, implicating that the effects of mutations, which alter autophagic activity, are less pronounced, or that such mutations are not common in people. Nevertheless, the role autophagy has in neurodegenerative processes is indisputable in humans (Jiang and Mizushima, 2014). For example, mutations in the WD repeat domain 45 (WDR45) gene, which functions in formation of the double membrane structures (“phagophores”), were shown to cause static encephalopathy of childhood with neurodegeneration in adulthood (SENDA). Also, both Alzheimer’s and Parkinson’s diseases were characterized by accumulation of autophagic vacuoles, indicating a disruption in their turnover (Nixon et al., 2005; Nixon and Yang, 2011). Consonantly, loss-of-function mutations in the Parkinson’s disease associated protein DJ-1 gene were linked to reduced basal levels of autophagy (Krebiehl et al., 2010). On the other hand, enhanced levels of autophagy have been linked to neuron loss in ALS (Sasaki, 2011; Chen et al., 2012), marking it as a possible driver of neurodegeneration in this case. Such controversial findings may result from the complex roles autophagy plays in cellular homeostasis, stress resistance, and also in programmed cell death (White and DiPaola, 2009; Tung et al., 2012), calling for further research to clarify its contribution to different types of neurodegeneration. In this regard, the dog could serve as a model more closely related to human physiology than rodents. Some canine hereditary diseases have already been linked to mutations in autophagy genes and many of these diseases have human homologs. For example, a polymorphism in the Ras-Related Protein Rab-24 (RAB24) gene, a member of the RAS oncogene family, which encodes a protein necessary for autophagosome trafficking, was found responsible for juvenile onset ataxia in some breeds (Agler et al., 2014). A missense mutation in the autophagy related 4D cysteine peptidase (ATG4D) gene was linked to vacuolar storage deficiency and neurodegeneration in Lagotto Romagnolo dogs (Kyöstilä et al., 2015). A study investigating juvenile onset neuroaxonal dystrophy in Spanish Water Dogs identified a non-synonymous mutation in the tectonin beta-propeller repeat containing 2 (TECPR2) gene, which had been linked to autophagosome formation (Hahn et al., 2015). A very similar type of neuroaxonal dystrophy exists in humans; hence, this finding could have actually suggested a possible genetic background to look for in affected people.
DM is another example of a naturally occurring neurodegenerative disease in dogs and shows a high degree of similarity to human ALS. Both DM and ALS have been linked to mutations in the ROS neutralizing SOD1 gene, suggesting a shared genetic and metabolic background. Importantly, the possible contribution of autophagy to motor neuron loss was reported to be controversial both in DM (Ogawa et al., 2015) and in ALS (Chen et al., 2012). Autophagy also has a similarly controversial role in muscular atrophy in humans and dogs (Sandri, 2010; Pagano et al., 2015). Altogether, these findings indicate many homologies between dogs and humans regarding the regulation of autophagy in aging and disease.
Deregulation of Nutrient Sensing
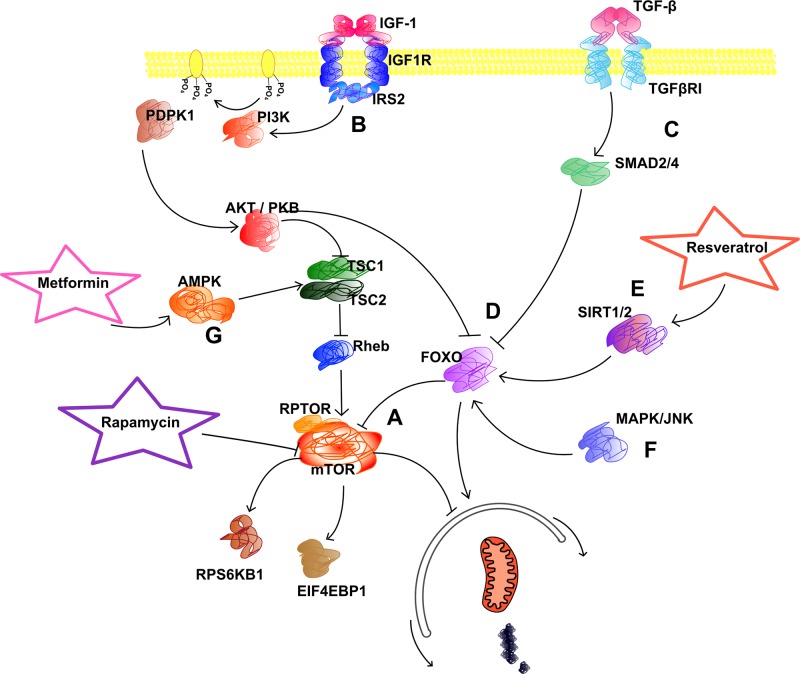
Cellular metabolism, protein synthesis, and autophagy are strictly regulated by various signaling pathways ( Figure 6 ) (Martindale and Holbrook, 2002; He and Klionsky, 2009). Most of these have evolved to synchronize cell growth and metabolism with nutrient availability; hence, they are often referred to as nutrient sensing pathways. Many of them converge on the target of rapamycin (TOR) kinase ( Figure 6A ), a main factor in determining rates of protein turnover and metabolism (Wullschleger et al., 2006). In mammals, the TOR kinase may function in different complexes, named mTORC1 or mTORC2, depending on its protein partners. The mTORC1 complex, which includes the regulatory associated protein of MTOR (RPTOR) protein, corresponds to the invertebrate TOR complex in its regulatory interactions, while mTORC2 controls other intracellular processes. Knock-down of TOR expression by RNA interference was shown to increase lifespan of C. elegans by threefold (Vellai et al., 2003). Later, similar effects of inhibiting TOR or its homologs were reported in S. cerevisiae (Kaeberlein et al., 2005), D. melanogaster (Kapahi et al., 2004), and laboratory mice (Wu et al., 2013), emphasizing its conserved role in the aging process (Kapahi et al., 2010).
Figure 6.
Signaling pathways. This figure illustrates some of the many signaling pathways that have been connected to aging. Activating interactions are shown with arrows, while inhibiting interactions are represented by bar headed lines. (A) Almost all of the age-related signaling pathways converge on the metabolic signal integrator mTORC1 complex, which includes the mTOR kinase together with RPTOR and other proteins. mTORC1 integrates stimuli to fine-tune metabolic processes, protein synthesis, cell growth, and autophagy. Downstream targets of mTORC1 include ribosomal proteins and translation initiation factors, like RPS6KB1 and EIF4EBP1, as well as ULK1, which is an activator of autophagy. As its name indicates, mTOR is the main target of rapamycin, which inhibits its function. (B) The IGF1 signaling is considered to be the main modulator that links autophagy to aging. Upregulation of this pathway leads to repression of autophagy and activation of protein synthesis by mTOR. This pathway includes many proteins, most of which have kinase activity. The PI3K enzymes transmit the signal from the IGF1 receptor by phosphorylating phosphatidylinositol molecules in the membrane, which then activate PDPK1. From here, the signal is forwarded to AKT (also known as PKB) by phosphorylation. AKT then inhibits the function of the TSC1 and TSC2 proteins, and consequently releases RHEB from inhibition. RHEB directly binds and activates the mTORC1 complex. (C) Another signaling pathway, which acts parallel to IGF1, is the TGF-β signalization. It is implicated in cellular growth control and also in tumorigenesis and inhibits autophagy. SMAD proteins transduce the TGF-β signal to downstream targets. An important target of SMAD2/4 is the FOXO gene family. (D) FOXO transcription factors have an evolutionary conserved function in aging regulation and integrate several pathways to upregulate autophagy and inhibit mTORC1. (E) Sirtuins (SIRT1/2) act contrary to the TGF-β pathway as they upregulate FOXO and thus autophagy. Resveratrol and caloric restriction exert their anti-aging effect through the activation of sirtuins. (F) The MAPK proteins were also shown to play a role in aging by regulating FOXO. They serve as important early responsive elements of different cellular stimuli and also play a role in apoptotic cell death induction in the case of UV-light damage. (G) AMPK integrates metabolite sensing information and acts contrary to the IGF1 pathway: activation of AMPK leads to down-regulation of mTOR and activation of autophagy. AMPK is the main target of metformin. mTOR, mechanistic target of rapamycin; RPTOR, regulatory associated protein of MTOR; RPS6KB1, ribosomal protein S6 kinase, 70kD, polypeptide 1; EIF4EBP1, eukaryotic translation initiation factor 4E binding protein 1; ULK1, unc-51 like autophagy activating kinase 1; IGF1, insulin-like growth factor 1; PIK3, phosphatidylinositol-4,5-bisphosphate 3-kinase; PDPK1, 3-phosphoinositide dependent protein kinase 1; AKT, AKT serine/threonine kinase 1; TSC1, tuberous sclerosis 1; TSC2, tuberous sclerosis 2; RHEB, Ras homolog, mTORC1 binding protein b; complex 1; TGF-β, transforming growth factor β; SMAD, MAD, mothers against decapentaplegic; FOXO, forkhead box O; SIRT, sirtuin; MAPK, mitogen-activated protein kinase; AMPK, adenosine monophosphate kinase.
Importantly, the function of mTOR can be efficiently inhibited by rapamycin, which is an already approved immunosuppressant in human medicine, and therefore has been proposed as a promising anti-aging compound to be used in humans. However, it was reported to cause severe side effects in medical dosages (Hartford and Ratain, 2007). Therefore, optimal dosages, which do not cause undesirable syndromes, yet still exert longevity promoting effects should be carefully determined in preclinical studies. Actually, pharmaceutical studies have already been initiated to investigate the effects of rapamycin on the lifespan of dogs (Kaeberlein et al., 2016; Urfer et al., 2017).
One of the main signaling pathways that regulate TOR activity is the insulin and IGF1 signaling (IIS) pathway ( Figure 6B ). It was first linked to aging when strains of C. elegans with doubled lifespan revealed a mutation in daf-2, the worm homologue of the IGF1 receptor (IGF1R) gene (Kenyon et al., 1993). Later, a threefold elongation in the non-replicative lifespan of S. cerevisiae was also linked to two genes that functioned in the glucose sensing pathway (Thevelein and de Winde, 1999). In flies and mice with hypomorphic alleles of IGF1R homologs, a significant increase in lifespan was observed together with characteristic pleiotropic effects (Tatar et al., 2001; Holzenberger et al., 2003). However, the longevity effect was less pronounced in mice, pointing at the possibility that the relative contribution of IIS to aging regulation may differ in various taxa.
Importantly, comparisons between centenarian and younger human cohorts also showed associations between expected lifespan and serum IGF1 levels or genetic polymorphisms in related genes (Barbieri et al., 2003; Van Heemst et al., 2005). Furthermore, functional variants in IGF1R were shown to be enriched in centenarians (Suh et al., 2008). Polymorphisms in other genes of the IIS pathway were also linked to longevity in GWAS, although not without contradictions (Soerensen et al., 2012a). In fact, some studies reported a decrease in GH and IGF1 plasma levels during normal aging (Breese et al., 1991; Sonntag et al., 1997). In this regard, it was hypothesized that the reduction in IGF1 levels may actually serve as a first line compensatory mechanism when age-related damage starts to accumulate in cells (Schumacher et al., 2008; López-Otín et al., 2013). Although low basic IIS signaling may delay aging, the overcompensation resulting from continuously accumulating damage in aged individuals may lead to insufficiencies in IGF1 signaling, and this can cause further decline. This effect may be particularly relevant in the case of neural aging, because the brain has special metabolic properties and a high need for optimal glucose levels. In support of this hypothesis, some age-related neurodegenerative states were linked to low IGF1 levels (Sonntag et al., 2000; Sonntag et al., 2005; Moloney et al., 2010). In addition, lower plasma IGF1 levels were shown to impair vascular maintenance in the brain (Sonntag et al., 1997). In this regard, dogs, which are more similar to humans in brain physiology and function than rodents, also seem promising to further unfold the relationship between IGF1 signaling and healthy aging.
Importantly, functional mutations in the IGF1 and IGF1R genes have already been linked to body size variability in dogs (Sutter et al., 2007; Hoopes et al., 2012; Rimbault et al., 2013), similarly to humans and laboratory animals (Liu et al., 1993; Ishida et al., 1998; Perry and Dominy, 2009). The notion that small dog breeds usually live longer than large breeds (Galis et al., 2007; Greer et al., 2007; Kraus et al., 2013) also hints at the potential role of the IIS pathway in canine lifespan determination. In fact, the genomic region harboring the IGF1 locus was linked to size and lifespan across different breeds (Jones et al., 2008) and serum IGF1 levels were shown to correlate with age and obesity (Greer et al., 2011) in individual dogs.
Some other pathways linked to body size in dogs (Rimbault et al., 2013; Schoenebeck and Ostrander, 2014) are also known to regulate TOR and autophagy. The SMAD family member 2 (SMAD2) gene, which functions in the transforming growth factor beta (TGF-β) pathway ( Figure 6C ) (Vellai, 2009), was associated with body size (Rimbault et al., 2013) and was previously found to be in linkage with mortality of dog breeds (Jones et al., 2008). The growth hormone receptor (GHR) and growth hormone (GH) genes, which also modulate dogs’ body size, were shown to affect longevity in humans (Soerensen et al., 2012a; van der Spoel et al., 2016), and in mice (Bartke et al., 2001; Flurkey et al., 2001; Kinney et al., 2001; Amador-Noguez et al., 2004).
Both IIS and TGF-β signaling have several targets beyond TOR, and many of them were implicated in aging. For example, the forkhead box O (FOXO) transcription factors are targeted by both IGF1 and TGF-β signaling ( Figure 6D ) and were shown to have an important role in tumor suppression (Greer and Brunet, 2005) and age-related diseases (Hesp et al., 2015). The worm homologue of the mammalian FOXO genes, daf-16, was one of the first genes linked to extreme longevity in C. elegans, as it was found necessary for the longevity effect observed in daf-2 deficient worms (Kenyon et al., 1993; Larsen et al., 1995; Lin et al., 1997; Ruvkun et al., 1997). Out of the four mammalian orthologues, FOXO3a has been associated with aging in human cohort studies (Willcox et al., 2008; Anselmi et al., 2009; Soerensen et al., 2010). FOXO1a SNPs were also reported to affect longevity, however, in a gender-specific manner (Li et al., 2009). In addition, FOXO3 was shown to regulate autophagy in skeletal muscle and plays a role in muscular atrophy (Mammucari et al., 2007). Despite the emphasized role of FOXOs in disease and aging, the canine homologues have not yet been studied in detail.
Sirtuins ( Figure 6E ) and the 5’ AMP-activated protein kinase (AMPK) also play an important role in nutrient sensing. Sirtuins function as nicotinamide adenine dinucleotide (NAD) dependent protein deacetylases, sensing the levels of NAD in cells. Decreased NAD levels were shown to reduce their activity, and thus NAD replacement therapies have been suggested as possible anti-aging interventions (Chini et al., 2017). AMPK detects the levels of adenosine monophosphate (AMP) in cells, and it is able to counteract IGF1 signaling by inhibiting TOR (Inoki et al., 2003; Gwinn et al., 2008) and activating FOXO3a (Sanchez et al., 2012). It was also suggested to play a major neuroprotective role by activating the UNC-51 Like 1 (ULK1) kinase (Kim et al., 2011), a key autophagy inducing protein (Wong et al., 2013).
Both sirtuins and AMPK are targets of several proposed anti-aging drugs. For example, the anti-diabetic drugs, metformin and acarbose, were both shown to exert an anti-aging effect in model animals (Anisimov et al., 2011; Cabreiro et al., 2013; Martin-Montalvo et al., 2013; Harrison et al., 2014) and to modulate various aging-related intracellular processes, including the activation of AMPK (Cho et al., 2015; Lu et al., 2015; Chan et al., 2016). Importantly, metformin was actually reported to decrease mortality of diabetic people in comparison to patients treated with other drugs in a retrospective large-scale study (Bannister et al., 2014). Consequently, metformin has been suggested as a promising candidate for anti-aging interventions (Barzilai et al., 2016). It may also be easily applied in family dogs to test its longevity enhancing potential, because it was already shown to have relatively mild side effects in dogs (Heller, 2007).
However, it is important to note that all of these drugs, including rapamycin, may exert pleiotropic effects in organisms through various cellular signaling and regulatory mechanisms. For example, metformin was implicated to modify the composition of the gut microbiome in diabetic patients (Forslund et al., 2015), which in turn can indirectly affect aging and neural function (see below in the section Microbiome). Such pleiotropic effects should be thoroughly considered in humans and family dogs, as both are exposed to variable environmental stimuli, have diverse genetic background, and may use other medications, which can alter the mechanisms of actions of anti-aging compounds through complex interactions.
A fairly recently emerged possible regulator of aging that interacts with IGF1 signaling and FOXO activity is the klotho hormone, which was first identified in mice as a longevity factor (Kuro-o et al., 1997; Kurosu et al., 2005). The klotho (KL) gene represents an example of longevity genes that are missing from invertebrate models, but show functional polymorphisms associated with human longevity (Arking et al., 2002).
Mitochondrial Dysfunction
Nutrient sensing pathways converge on the regulation of mitochondrial activity, as these organelles are the main sources of energy (in the form of adenosine triphosphate, ATP) in eukaryotic cells under normal circumstances, when enough oxygen is present. The availability of nutrients determines the rate of mitochondrial respiration, which, however, generates not only ATP but also chemical by-products, including ROS ( Figure 7A ). The oxidative burden created by mitochondria may be especially high in neurons, which solely depend on aerobic mitochondrial respiration as energy source. In accordance with this, associations between chronological age and a higher ROS production rate of mitochondria in brain cells were demonstrated in rodents (Navarro and Boveris, 2004; Petrosillo et al., 2008), humans (Mecocci et al., 1993), and also in dogs (Head et al., 2009). Oxidative DNA lesions could also occur in the mitochondrial genome and consequently modify gene expression and optimal function of mitochondria, possibly leading to a positive feedback mechanism in the generation of extensive levels of ROS. In fact, age-related changes in mitochondrial gene expression profiles were reported in mice (Manczak et al., 2005) and the accumulation of mitochondrial DNA mutations, mainly deletions, in certain brain regions was linked to impaired mitochondrial respiration in humans (Corral-Debrinski et al., 1992; Kraytsberg et al., 2006). Importantly, mitochondrial dysfunction is considered to be a main driver of the pathophysiology of neurodegenerative diseases, including Alzheimer’s disease, Parkinson’s disease, and ALS (Lin and Beal, 2006). For example, gain-of-function mutations in the Leucine-rich repeat kinase 2 (LRRK2) gene causing autosomal dominant form of PD (Di Fonzo et al., 2005) were shown to result in hampered mitochondrial function (Mortiboys et al., 2010). Although the generation of malfunctioning mitochondria may be counterbalanced by elevated levels of mitophagy, a form of macroautophagy that is able to degrade mitochondria, it is not yet clear in the literature how mitophagic activity is generally changed in affected cells of PD patients (Chu, 2019).
Figure 7.
Mitochondria and oxidative stress. (A) Mitochondria represent the main source for reactive oxygen species (ROS) within eukaryotic cells as the oxidative respiration processes take place in the inner membrane of mitochondria, utilizing a special electron transport chain. Increased respiration rate due to metabolic changes and reduced antioxidant accessibility may also increase generation of ROS, which can damage the mitochondrial genome as well (indicated by thick arrow). (B) Accumulation of mutations in the mitochondrial genome may lead to malfunction in the electron transport chain in aberrant mitochondria that consequently produce an elevated rate of ROS. Removal of aberrant mitochondria is a key process for maintaining oxidative balance in cells.
The role of mitochondrial dysfunction and increased oxidative burden in neural aging was also investigated in dogs. In general, dog brains were shown to accumulate oxidative damage with age (for more details, see the section Genomic Instability). In a study published by Head et al., 2009, mitochondrial ROS production and complex I driven respiration rate showed significant alterations between old and young laboratory Beagle dogs from the same colony, with aged dogs having higher ROS production and lower respiration rate. The same study reported that a diet enriched with antioxidants and mitochondrial cofactors improved mitochondrial respiration rate and reduced ROS production in aged dogs, and also had a positive effect on their cognitive performance. In another study, a ketosis-inducing diet was shown to modify mitochondrial function and, to some extent, reduce amyloid-β deposition in dog brains (Studzinski et al., 2008). Interestingly, Christie et al. reported in 2009 that short-term supplementation with lipoic acid (LA), which is both an important mitochondrial cofactor and a powerful antioxidant, did not improve cognitive function of aged Beagle dogs, contrary to their previous findings where LA had been used together with other antioxidants (Cotman et al., 2002; Milgram et al., 2002a; Milgram et al., 2002b). Furthermore, another mitochondrial cofactor, acetyl-L-carnitine (ALCAR), when supplemented in itself, was shown to decrease cognitive performance (Christie et al., 2009). In a paper, Snigdha et al., 2016 who also failed to replicate findings about the beneficial effects of antioxidant enriched diet on the cognition of aged Beagle dogs, suggested that these controversies could have resulted from differences in the baseline nutrition of dogs.
Nevertheless, such findings could also result from a more complicated interaction between ROS and aging, as it was suggested by several authors, who re-evaluated the classical theories about the connection between mitochondria, ROS, and general longevity, based on the increasing body of experimental evidences (Hekimi et al., 2011; López-Otín et al., 2013). Accordingly, the ambiguous results listed above could result from altered mitochondrial homeostasis (mithormesis) in cells and not solely from impaired mitochondrial respiration and ROS overproduction. The concept of mithormesis suggests that mild mitochondrial stressors may actually benefit cellular health and longevity (López-Otín et al., 2013). Such minor stress, which, for example, can be induced by pharmaceutical agents, can boost mitochondrial turnover and activate defensive mechanisms (Haigis and Yankner, 2010). As it was shown that malfunctioning mitochondria can also directly affect aging by other mechanisms than increased oxidative stress (Trifunovic et al., 2004; Vermulst et al., 2008; Edgar et al., 2009; Hiona et al., 2010), for example by inducing apoptosis (Kroemer et al., 2007) ( Figure 7B ), elevated mitochondrial turnover can actually protect cells from these deleterious effects. Since both resveratrol and metformin have been hypothesized to be mild mitochondrial toxins, they may also exert their anti-aging effect at least partly by inducing mithormesis (López-Otín et al., 2013).
Altogether, mitochondria-targeting interventions may require mindful considerations, especially in populations with high genetic variability. Genetic variants in mitochondrial genomes are known to cause disorders in humans (Koopman et al., 2012), and they may also substantially alter the capability of cells to cope with mitochondrial poisons (Finsterer and Frank, 2016). This means that interactions between mitochondrial genotypes and specific chemical compounds should also be considered in anti-aging intervention studies. In this regard, dogs can again become ideal models with good translatability. Several mitochondrial diseases are known in dogs, which have human homologs, such as the sensory ataxic neuropathy found in Golden Retriever dogs (Baranowska et al., 2009) or the familial dilated cardiomyopathy in Doberman Pinschers (Meurs et al., 2012). As several promising anti-aging drugs are likely to be tested in dogs in preclinical studies, looking into their effects on mitochondrial function and testing their possible interactions with mitochondrial genotypes can be highly relevant for humans.
Cellular Senescence
All the mechanisms discussed so far act congruently to modulate cellular metabolism, growth, proliferation, and, eventually, senescence, which is characterized by a permanent cell cycle arrest and stereotyped phenotypic changes (Campisi and d’Adda di Fagagna, 2007; Collado et al., 2007). Specific mechanisms serve as effectors of senescence in response to unrepaired DNA damage, mitochondrial malfunction, and other forms of excessive stress. Most famously, telomere attrition represents a somewhat genetically programmed route to cellular senescence as it was implied from experiments with human fibroblasts (Harley et al., 1990; Allsopp et al., 1992). However, the fact that cultured murine cells also reached senescence despite their telomerase positivity suggested that other mechanisms, like increased oxidative damage in cultured conditions, should have contributed to their limited proliferative potential (Sherr and DePinho, 2000).
Among other examples, the activation of the p16INK4a/Rb and the p14ARF/p53 signaling pathways is able to induce cellular senescence in response to time-dependent changes, including the accumulation of DNA damage (d’Adda di Fagagna, 2008). The expression of the INK4/ARF locus, which encodes the p16INK4, p15INK4, and p14ARF proteins in humans, was reported to correlate with chronological age in various tissues of rodents and humans (Krishnamurthy et al., 2004; Krishnamurthy et al., 2006; Ressler et al., 2006; Liu et al., 2009b), nominating it as an ideal biomarker of aging. Importantly, the INK4/ARF locus can directly affect healthspan and longevity, as it was shown experimentally in mice (Matheu et al., 2009). Furthermore, simultaneous overexpression of INK4/ARF and p53 caused extended lifespan in mice, accompanied by cancer resistance and reduced neural decline (Matheu et al., 2007; Carrasco-Garcia et al., 2015). The human INK4/ARF locus was also found to be the most strongly associated locus with several age-related pathologies in a meta-analysis of GWAS (Jeck et al., 2012), and it was associated with longevity in a smaller cohort study (Emanuele et al., 2010). Interestingly, the elevated expression and activity of p53 in itself did not increase longevity in experimental studies (García-Cao et al., 2002; Mendrysa et al., 2006). Actually, certain hyperactive variants of p53 were shown to reduce lifespan in mice, although with simultaneously prompting increased cancer resistance (Tyner et al., 2002; Dumble et al., 2007).
In this regard, it is important to note that the p14/p53 pathway, together with p16/pRb, are fundamental tumor suppressor mechanisms; therefore, they unquestionably contribute to healthy aging by forcing potentially malignant cells into a senescent state or into programmed cell death (Hickman et al., 2002).
This anti-tumor effect of induced cellular senescence can explain the contradictory findings regarding the role of p53 and other tumor suppressor mechanisms in aging (Rodier and Campisi, 2011), as a possible trade-off exists between longevity and cancer occurrence (Matheu et al., 2008).
So far, no studies have investigated the canine homologs of the INK4/ARF locus and p53 in relation to aging. However, and not surprisingly, their role and regulation in tumorigenesis showed high similarities between dogs and humans (Rowell et al., 2011; Lutful Kabir et al., 2013), suggesting that their roles in aging, especially in healthy aging, are also conserved in dogs.
Accumulation of Senescent Cells in Tissues
The time-dependent upregulation of senescence inducing mechanisms means that the ratio of aged cells may gradually increase in the tissues of older individuals. Indeed, a marked elevation of senescent cell numbers was reported in old mice (Wang et al., 2009), although not in all tissues. Importantly, this accumulation process can result from both the increased generation of senescent cells and a decreased activity of macrophages that are able to eliminate aged or apoptotic cells from tissues. As the activity of the innate immune system was shown to decrease with age (Plowden et al., 2004; Mahbub et al., 2011), it is likely that reduced phagocytic capacity also contributes to organismal aging through the disrupted elimination of senescent cells (López-Otín et al., 2013). Furthermore, senescent cells were shown to produce inflammatory signals and create a special inflammatory microenvironment around themselves (Kuilman et al., 2010; Rodier and Campisi, 2011), which may directly contribute to tissue aging by creating a positive feedback loop and inducing cellular senescence in neighboring cells (Nelson et al., 2012). This so-called “bystander” effect was actually shown to modulate the number of senescent cells in vivo in mice (da Silva et al., 2019).
Little is known about the accumulation of senescent cells in canine tissues, although this phenomenon is also likely to show fundamental similarities with other mammalian species. As there is a growing interest toward pharmacological approaches to deplete senescent cells in tissues by specific apoptosis inducing agents (senolytic drugs) (Kirkland et al., 2017), dogs may eventually be involved in testing these types of anti-aging interventions.
Stem Cell Exhaustion
Tissue renewal depends on the abundance and replicative capacity of tissue-specific stem cells, which can replace cells lost by terminal senescence or apoptosis. Thus, the age-related increase in cellular senescence may also result in elevated stem cell activation and differentiation, eventually causing the depletion of stem cell pools. In fact, early exhaustion of stem cells in certain tissues was shown to accelerate aging in flies and mice (Cheng et al., 2000; Kippin et al., 2005; Rera et al., 2011). Furthermore, it was shown that increased basic fibroblast growth factor (FGF2) signaling in muscle tissues of aged mice accelerated depletion of stem cells by forcing them to leave quiescent state (Chakkalakal et al., 2012).
Importantly, senescence may also directly affect stem cells, depriving them from the ability to replicate and differentiate even if they are still present in tissues. For example, hematopoietic stem cells (HSCs) were reported to have reduced replicative capacity in both aged mice and humans, mainly because of accumulating DNA damage (de Haan and Lazare, 2018). This reduction can explain the old age anemia of elderly people (Patel, 2008). Importantly, similar forms of age-associated changes in blood parameters, including anemia, were reported in dogs (Strasser et al., 1993; Radakovich et al., 2017).
Stem cell quiescence and activation is regulated by many of the already discussed aging pathways, including p53 and IIS (Liu et al., 2009a; Xian et al., 2012). Thus, pharmacological interventions that act on these could also affect stem cell dynamics. In this regard, the inhibition of mTOR was shown to have beneficial effects on aging by promoting cellular rejuvenation (Castilho et al., 2009; Chen et al., 2009; Yilmaz et al., 2012). Furthermore, pharmacological inhibitors of the Cell Division Cycle 42 (CDC42) protein, which is an inducer of HSC senescence, were shown to promote rejuvenation of HSC pools in mice (Florian et al., 2012).
Besides pharmacological interventions, stem cell therapy has also been suggested as a possible anti-aging intervention, with highlighted promises to treat certain forms of neurodegeneration (Lindvall et al., 2004; Trounson and DeWitt, 2016). In this regard, stem cell therapy trials conducted on dogs affected with CCD or other forms of neurodegeneration could represent a crucial step before progressing to human trials. In the case of the Golden Retriever model for Duchenne muscular dystrophy, successful stem cell-based interventions had actually preceded human clinical trials (Pelatti et al., 2016). Other instances of dog stem cell therapy trials were discussed by Hoffman and Dow (2016).
Altered Intercellular Communication
In the course of evolution, several mechanisms have evolved to establish efficient communication between cells in multicellular organisms. Intercellular communication types include paracrine, endocrine, and neurocrine signaling, and all of these can be involved in the aging process. Especially hormones and other endocrine signal transducers can have a main role in systemic aging regulation. Actually, GH and the insulin/IGF1 signaling pathway belong to these main systemic regulators, most of which are supervised by the hypothalamic–pituitary–adrenal (HPA) and –thyroid axes. Endocrine signaling also involves hormones synthetized by the digestive system and reproductive glands. Furthermore, small molecules produced by gut bacteria can also have systemic effects on the host organism (Donia and Fischbach, 2015).
Neuroendocrine Signaling
The CNS mainly functions as conductor, coordinating various processes of the organism according to intrinsic and extrinsic stimuli. Signals provided by the CNS—together with the digestive system—can affect every part of the body. In this regard, “neural aging” has recently gained more focus as a central mechanism, which could impact the systemic aging of the whole organism (Weir and Mair, 2016). In support of this theory, both neuronal and intestinal genetic manipulations, which reduced mitochondrial electron transport chain function, were shown to extend lifespan in C. elegans, while similar manipulations in other tissues had no longevity effect (Durieux et al., 2011).
Importantly, several signaling pathways have been hypothesized to play fundamental roles in both neural senescence and systemic aging. For example, IGF1, together with the brain-derived neurotrophic factor (BDNF) and serotonin, were shown to affect brain aging and modulate metabolic changes linked to caloric restriction across the body (Mattson et al., 2004). The hypothalamus also has major implications in aging. For example, reproductive aging was shown to be controlled by the gonadotropin releasing hormone (GnRH), which is produced by special cells in the hypothalamus (Yin and Gore, 2006). Age-related reduction in GnRH levels, in response to activation of inflammatory pathways, was suggested to aggravate frailty and neurodegeneration in the elderly (Zhang et al., 2013). Altogether, age-related changes in the hypothalamus and, consequently, in HPA regulation seem to play a central role in the systemic regulation of aging (Deuschle et al., 1997; Kim and Choe, 2019). The activity of the hypothalamus and the HPA axis was reported to show similar general attributes in dogs as in humans and age-related changes in the HPA axis were already assessed in dogs (Reul et al., 1991; Rothuizen et al., 1991). However, further studies will be needed to investigate the function of GnRH and other hormones in canine aging.
Parabiosis Experiments and Systemic Factors of Aging
Most molecular effectors of systemic aging are excreted into the blood, by which they can reach every part of the body. This mediatory function of the blood was proven by parabiosis experiments in rodents, when the artificial connection of the circulatory systems of old and young animals resulted in beneficial effects on the cognitive performance of aged individuals (Katsimpardi et al., 2014; Villeda et al., 2014). Several of the possible effector molecules behind this phenomenon have been revealed since (Loffredo et al., 2013; Demontis et al., 2014; Elabd et al., 2014). Interestingly, some of the systemic factors present in human umbilical cord plasma were shown to beneficially influence brain aging when applied experimentally in mice (Castellano et al., 2017), indicating conserved functions for these molecules. It is important to note, however, that other blood-borne factors were shown to actually promote aging. For example, the β2 microglobulin was reported to negatively affect cognitive performance and regenerative potentials in aged mice (Smith et al., 2015). Although parabiosis is not really applicable in humans and in family dogs, the identified systemic factors seem promising as effectors or targets for anti-aging interventions in both species and may be introduced to preclinical studies conducted on dogs.
Extracellular Vesicles
In addition to hormones and metabolites, extracellular vesicles released by cells into the blood, called exosomes and ectosomes, have emerged as important transducers of various cellular signals (Meldolesi, 2018) and their content, including miRNAs, may provide diagnostic and prognostic measures for many diseases, including AD (Cheng et al., 2014; Cheng et al., 2015; Thind and Wilson, 2016; Van Giau and An, 2016). Consequently, exosomes may also modulate aging and neurodegeneration (Cheng et al., 2015). In support of this, it was recently demonstrated by Zhang et al. (2017) that the stem cells of the hypothalamus could affect the speed of aging by exosomal miRNAs secreted into the cerebrospinal fluid in mice.
Exosome research in dogs have been limited until recently. However, blood miRNA levels—which were hypothesized to be mainly found in exosomes—were reported to correlate with disease phenotypes in canine Duchenne muscular dystrophy (Mizuno et al., 2011). Similarly, miRNA content in circulating exosomes was shown to correlate with progression of secondary heart failure in cases of myxomatous mitral valve disease in dogs (Yang et al., 2017). Direct links were suggested between alterations in urinary exosome formation, miRNA content, and occurrence of kidney disease in dogs by Ichii et al., 2017. Furthermore, a recent study reported exosome derived miRNAs as biomarkers for canine mammary tumors (Fish et al., 2018). Altogether, investigations about the connections between exosome content and aging or age-related pathologies in dogs may lead to the identification of diagnostic markers with potential translational prospects into human studies.
Immunaging and Inflamm-Aging
Together with the CNS, the immune system has a main systemic regulatory function in the organism. Most immune cells synthesize various signaling molecules that act either in a paracrine or endocrine manner and can also provide defense against various pathogens. Macrophages, which are part of the innate immune system, can wander throughout the body and have important roles in tissue homeostasis by cleaning cellular debris and pathogens.
The human immune system is known to experience a general age-related decline in its function and in the abundance of some cell types, although the exact details of reported changes may vary between studies (Pawelec, 2018). In general, reduced numbers of naïve CD8+ T cells and moderately elevated numbers of memory T cells were found to be linked to aging. Importantly, bone marrow derived macrophages were also shown to lose phagocytic capacity with aging (Kim et al., 2017; Li et al., 2017), which can contribute to the accumulation of senescent cells in tissues.
In addition, it has long been hypothesized that systemic age-related changes in certain immune components linked to inflammation will lead to a so-called inflamm-aging phenomenon. Importantly, the exact interactions between immunosenescence and inflamm-aging have not yet been clarified (Fulop et al., 2018); hence, further studies using systems biology approaches may shed light on the detailed mechanisms that underlie them (Ostan et al., 2017).
Both immunosenescence and inflamm-aging were proposed as contributors to aging and age-related pathologies in dogs (Day, 2010). Large-scale hematologic and serum phenotyping studies done in various breeds (Faldyna et al., 2001; Lawrence et al., 2013; Chang et al., 2016) showed that several of the assessed blood parameters correlated with chronological age. Regarding the immune system, T and B lymphocytes were mainly affected in most cases; however, the directions of these changes were contradictory (Faldyna et al., 2001; Massimino et al., 2003; HogenEsch et al., 2004; Reis et al., 2005; Greeley et al., 2006). However, a recent study reported that changes in naïve and memory T cell numbers in old dogs were similar to those previously described in most human studies (Withers et al., 2018). Importantly, it was shown that lifelong calorie restriction positively affected lymphocyte numbers in aged dogs (Greeley et al., 2006). Taken together, the dog may become one of the most applicable model animals to study immunosenescence and inflamm-aging, and to test interventions that could attenuate the deterioration of the immune system.
Microbiome
Recent findings suggest that both systemic metabolism and immune function can be modulated by bacteria inhabiting the gut, termed gut microbiome (Tremaroli and Bäckhed, 2012). The microbiome can interact with the host organism through various chemical signals, and some of these may directly affect the function of distant organs, like the brain (Sharon et al., 2016). Therefore, the microbiome may fundamentally affect health and disease, and possibly aging (Zapata and Quagliarello, 2015). This was supported by findings that reported consistent changes in the composition of the microbiome in elderly people and centenarians (Biagi et al., 2010; Biagi et al., 2017). Although these correlations do not necessarily indicate causative links (Saraswati and Sitaraman, 2015), the theory of microbial modulation of aging has been gaining more and more scientific interest. Importantly, experimental evidences from rodents have already shown that the microbiome can affect the progression of neurodegeneration (Sampson et al., 2016). Furthermore, probiotics and prebiotics, which can beneficially alter the composition of the microbiome, were reported to positively influence the aging of the gut and systemic inflammation in people (Patel et al., 2014; Vaiserman et al., 2017).
Currently, not much is known about age-related changes in the canine gut microbiome; however, there is growing resear ch interest in this field. Future findings may have direct implications to humans as well, because the composition of the canine microbiome was shown to be more similar to humans than that of mice and pigs (Coelho et al., 2018) and actual correlations between the microbiomes of dogs and people living in the same household were also reported (Misic et al., 2015). Because dogs age faster than humans, they can be ideal models to test the potential aging effects of prebiotics and probiotics in longitudinal follow-up studies. Importantly, some probiotics used in humans were already suggested to promote health in dogs (Grześkowiak et al., 2015), and this can facilitate their adaptation for systemic anti-aging intervention trials.
It is important to note that other microbial niches on the human body may also affect aging and disease, as it was implicated when oral microbiota and inflammation were linked to the progression of Alzheimer’s disease (Pritchard et al., 2017). As periodontitis is also a serious health issue in aged dogs (Albuquerque et al., 2012), this link between oral microbiome and neurodegeneration undoubtedly requires further focus in canine aging research and veterinary medicine and may also benefit humans by translational studies (An et al., 2018).
Conclusions and Perspectives
Considering the remarkably complex nature of biological processes that underlie aging, it is not surprising that finding a biological model for aging that would unify all relevant aspects is challenging. Family dogs have been proposed as ideal models to complement findings from other model organisms ( Figure 8 ); however, the still limited knowledge about the exact genetic and regulatory mechanisms that underlie their aging may restrict their applicability in translational studies. Although searching for links between genetic variants and aging phenotypes would be more challenging than in the case of other phenotypic parameters, which are easier to measure, such approaches seem indispensable to gain insight into the main genetic mechanisms that modulate aging variability in dogs. Actually, there have been some efforts along these lines (Jones et al., 2008); however, interbreed comparisons have a limited potential to reveal the exact variants responsible for longevity differences between individuals. Future studies should aim at intrabreed approaches, for which both genetic and aging related data should be available from the same animals. Furthermore, gene expression mapping could be a novel approach to pinpoint at pathways that show changes between young and old dogs or between dogs with short and long lifespan. Although it could be challenging to obtain good quality tissues from a large number of family dogs with known lifespan and other parameters, biobanks created following human examples may help to overcome this limitation in the long term.
Figure 8.
Model organisms of aging. The figure illustrates common aging model organisms, including small animal models and large aninmal models used to study various aspetcs of aging. (A–C) Yeast (Saccharomyces cerevisiae) and the invertebrates Caenorhabditis elegans and Drosophila melanogaster are ideal to experimentally study the basic, conserved mechanisms of cellular – and organismal – aging. On the other hand, they show less biological complexity than vertebrates in many aspects, and they do not naturally develop neurodegeneration. (D–E) Vertebrate small animal models, like the turqoise killifish (Nothobranchius furzeri) and rodents (Mus musculus and Rattus norvegicus) are ideal to study the biological mechanisms that may be absent in invertebrates, and they can still be rather easily used in experimental studies, including genetic manipulations. However, they typically do not develop age-related neurodegeneration, and may lack many aspects of the complex social and environmental influencers of human aging. (F–G) Dogs show similarities to humans in their physiology and they tend to naturally develop age-related cognitive decline. Laboratory dogs (F) are traditional large animal models in pharmacology reserach. However, the same way as other laboratory models, they do not represent the natural genetic and environmental variability typical for human populations. Family dogs, (G) on the other hand, live in the same environment as humans do, and show a special population genetic stratification, with the presence of genetically isolated, diverse populations (breeds). (H) Primates are the closest related to humans, thus they may seem to be the most appropriate animals to study human aging. However, primates are not suited for large-scale sudies for many reasons, including ethical and financial ones. Although they tend to develop human-like age-related neurodegeneration, they still lack the genetic and environmental complexity (both in the laboratory and in their natural habitats), which may influence human aging phenotypes in human populations. (I) Human aging shows many unique attributes, including a high prevalence of neurodegeneration. Age-related neurodegeneration is hard to study in most animals, and translational experiments have had many limitations so far. Brain aging may be fundamentally affected by non-genetic factors, including diet, exercise and social environment, which seem challenging to be modelled under laboratory conditions to reflect the natural circumstances of human populations.
As recent findings have increased the palette of possible anti-aging interventions, making almost all of the nine hallmarks of aging (López-Otín et al., 2013) targetable by drugs, a growing interest for preclinical testing of these compounds is expected. Consequently, the dog may gain more and more attention as a preclinical model species. Because family dogs are exposed to almost the same background effects, which can modify the outcome of interventions, as are people, they might even become inevitable to provide a suitable model to assess the effects of anti-aging interventions on natural populations.
It is important to emphasize that characterizing the aging process of dogs and establishing effective interventions within the species may benefit humans not only by clarifying scientific questions but also by making it possible to increase the healthy lifespan of companion and service animals. Owning a guide dog or service dog can lead to great improvements in the quality of life of disabled people. Also, service dogs may facilitate human–human interactions and contribute to the socio-emotional well-being of their owners. Caron-Lormier et al. (2016) reported that most guide dogs were retired due to age-related diseases or simply old age, after an average of 8.5 years of service. Increasing the lifespan and healthspan of working dogs could be emotionally beneficial for their owners, and also could be financially advantageous for societies, as the training of these animals is time consuming and expensive. Furthermore, providing average family dogs an elongated healthspan may also benefit their owners. Several studies have reported a positive correlation between dog walking, physical activity, and health variables in owners, although results were often controversial, suggesting the need for further research on this topic (Brown and Rhodes, 2006; Lentino et al., 2012; Christian et al., 2018). In some cases, improvements were most pronounced in older cohorts (Thorpe et al., 2006; Toohey et al., 2013; Garcia et al., 2015; Curl et al., 2016). Experiences from animal-assisted therapy also suggested that animal–human interactions may help the elderly to experience a successful aging course (Baun and Johnson, 2010). Therefore, providing a long and healthy life for companion animals may benefit the health and welfare of their owners as well.
Taken together, strong scientific evidence suggests that utilizing dogs as models of human aging and anti-aging interventions may hold prospects unattainable by other model organisms, if the complex interactions between genetics and environmental factors are taken into consideration. Thus, canine studies on aging may bring forward results that can eventually benefit the elderly as well as their pets.
Author Contributions
SS collected literature and wrote manuscript parts regarding genetic pathways in aging and in dogs. EK collected literature and wrote manuscript parts regarding dogs as models in general, environmental, and cognitive factors. Both authors worked on reviewing the final text. EK provided funding.
Funding
This project has received funding from the European Research Council (ERC) under the European Unions Horizon 2020 research and innovation program (Grant Agreement No. 680040), the János Bolyai Research Scholarship of the Hungarian Academy of Sciences, the Bolyai+ ÚNKP-18-4 New National Excellence Program of the Ministry of Human Capacities, and the Hungarian Brain Research Program 2017-1.2.1-NKP-2017-00002.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.00948/full#supplementary-material
References
- Agler C., Nielsen D. M., Urkasemsin G., Singleton A., Tonomura N., Sigurdsson S., et al. (2014). Canine hereditary ataxia in old English sheepdogs and Gordon Setters is associated with a defect in the autophagy gene encoding RAB24. PLoS Genet. 10, e1003991. 10.1371/journal.pgen.1003991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albani D., Ateri E., Mazzuco S., Ghilardi A., Rodilossi S., Biella G., et al. (2014). Modulation of human longevity by SIRT3 single nucleotide polymorphisms in the prospective study “Treviso Longeva (TRELONG)”. Age (Omaha) 36, 469–478. 10.1007/s11357-013-9559-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albuquerque C., Morinha F., Requicha J., Martins T., Dias I., Guedes-Pinto H., et al. (2012). Canine periodontitis: the dog as an important model for periodontal studies. Vet. J. 191, 299–305. 10.1016/j.tvjl.2011.08.017 [DOI] [PubMed] [Google Scholar]
- Alderete K. S., Carlson A., Grant M., Sharkey L., Zordoky B. (2017). The anti-cancer effects of resveratrol on canine hemangiosarcoma cells. FASEB J. 31, 996, 15,–996. 15. 10.1096/fasebj.31.1_supplement.996.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J. E., Colyer A., Haydock R. M., Hayek M. G., Park J. (2018). Understanding how dogs age: longitudinal analysis of markers of inflammation, immune function, and oxidative stress. J. Gerontol. Ser. A 73, 720–728. 10.1093/gerona/glx182 [DOI] [PubMed] [Google Scholar]
- Allen R., Tresini M. (2000). Oxidative stress and gene regulation. Free Radic. Biol. Med. 28, 463–499. 10.1016/S0891-5849(99)00242-7 [DOI] [PubMed] [Google Scholar]
- Allsopp R. C., Vaziri H., Patterson C., Goldstein S., Younglai E. V., Futcher A. B., et al. (1992). Telomere length predicts replicative capacity of human fibroblasts. Proc. Natl. Acad. Sci. 89, 10114–10118. 10.1073/pnas.89.21.10114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amador-Noguez D., Yagi K., Venable S., Darlington G. (2004). Gene expression profile of long-lived Ames dwarf mice and little mice. Aging Cell 3, 423–441. 10.1111/j.1474-9728.2004.00125.x [DOI] [PubMed] [Google Scholar]
- Amoasii L., Hildyard J. C. W., Li H., Sanchez-Ortiz E., Mireault A., Caballero D., et al. (2018). Gene editing restores dystrophin expression in a canine model of Duchenne muscular dystrophy. Science 362, 86–91. 10.1126/science.aau1549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. Y., Darveau R., Kaeberlein M. (2018). Oral health in geroscience: animal models and the aging oral cavity. GeroScience 40, 1–10. 10.1007/s11357-017-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimov V. N., Berstein L. M., Popovich I. G., Zabezhinski M. A., Egormin P. A., Piskunova T. S., et al. (2011). If started early in life, metformin treatment increases life span and postpones tumors in female SHR mice. Aging (Albany NY) 3, 148–157. 10.18632/aging.100273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anon (2019). [ebook] Available at: (http://www.un.org/en/development/desa/population/publications/pdf/ageing/WPA2017_Highlights.pdf) (Accessed 19 Mar. 2019).
- Anselmi C. V., Malovini A., Roncarati R., Novelli V., Villa F., Condorelli G., et al. (2009). Association of the FOXO3A locus with extreme longevity in a southern Italian centenarian study. Rejuvenation Res. 12, 95–104. 10.1089/rej.2008.0827 [DOI] [PubMed] [Google Scholar]
- Arking D. E., Krebsova A., Macek M., Macek M., Arking A., Mian I. S., et al. (2002). Association of human aging with a functional variant of klotho. Proc. Natl. Acad. Sci. U. S. A. 99, 856–861. 10.1073/pnas.022484299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora H., Chacon A. H., Choudhary S., McLeod M. P., Meshkov L., Nouri K., et al. (2014). Bloom syndrome. Int. J. Dermatol. 53, 798–802. 10.1111/ijd.12408 [DOI] [PubMed] [Google Scholar]
- Atzmon G., Cho M., Cawthon R. M., Budagov T., Katz M., Yang X., et al. (2010). Genetic variation in human telomerase is associated with telomere length in Ashkenazi centenarians. Proc. Natl. Acad. Sci. 107, 1710 LP–1717. 10.1073/pnas.0906191106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awano T., Johnson G. S., Wade C. M., Katz M. L., Johnson G. C., Taylor J. F., et al. (2009). Genome-wide association analysis reveals a SOD1 mutation in canine degenerative myelopathy that resembles amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U. S. A. 106, 2794–2799. 10.1073/pnas.0812297106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsson E., Ratnakumar A., Arendt M.-L., Maqbool K., Webster M. T., Perloski M., et al. (2013). The genomic signature of dog domestication reveals adaptation to a starch-rich diet. Nature 495, 360–364. 10.1038/nature11837 [DOI] [PubMed] [Google Scholar]
- Babior B. M. (2004). NADPH oxidase. Curr. Opin. Immunol. 16, 42–47. 10.1016/j.coi.2003.12.001 [DOI] [PubMed] [Google Scholar]
- Banlaki Z., Cimarelli G., Viranyi Z., Kubinyi E., Sasvari-Szekely M., Ronai Z. (2017). DNA methylation patterns of behavior-related gene promoter regions dissect the gray wolf from domestic dog breeds. Mol. Genet. Genomics 292, 685–697. 10.1007/s00438-017-1305-5 [DOI] [PubMed] [Google Scholar]
- Bannister C. A., Holden S. E., Jenkins-Jones S., Morgan C. L., Halcox J. P., Schernthaner G., et al. (2014). Can people with type 2 diabetes live longer than those without? A comparison of mortality in people initiated with metformin or sulphonylurea monotherapy and matched, non-diabetic controls. Diabetes Obes. Metab. 16, 1165–1173. 10.1111/dom.12354 [DOI] [PubMed] [Google Scholar]
- Baranowska I., Jäderlund K. H., Nennesmo I., Holmqvist E., Heidrich N., Larsson N.-G., et al. (2009). Sensory ataxic neuropathy in golden retriever dogs is caused by a deletion in the mitochondrial tRNATyr gene. PLoS Genet. 5, e1000499. 10.1371/journal.pgen.1000499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri M., Bonafè M., Franceschi C., Paolisso G. (2003). Insulin/IGF-I-signaling pathway: an evolutionarily conserved mechanism of longevity from yeast to humans. Am. J. Physiol. - Endocrinol. Metab. 285. 10.1152/ajpendo.00296.2003 [DOI] [PubMed]
- Bartke A., Brown-Borg H., Mattison J., Kinney B., Hauck S., Wright C. (2001). Prolonged longevity of hypopituitary dwarf mice. Exp. Gerontol. 36, 21–28. 10.1016/S0531-5565(00)00205-9 [DOI] [PubMed] [Google Scholar]
- Barzilai N., Crandall J. P., Kritchevsky S. B., Espeland M. A. (2016). Metformin as a tool to target aging. Cell Metab. 23, 1060–1065. 10.1016/j.cmet.2016.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baun M., Johnson R. (2010). Human/animal interaction and successful aging. Handb. Anim. Ther. 283–299. 10.1016/B978-0-12-381453-1.10015-7 [DOI]
- Baur J. A., Pearson K. J., Price N. L., Jamieson H. A., Lerin C., Kalra A., et al. (2006). Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444, 337–342. 10.1038/nature05354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman M., Blanché H., Perola M., Hervonen A., Bezrukov V., Sikora E., et al. (2013). Genome-wide linkage analysis for human longevity: genetics of healthy aging study. Aging Cell 12, 184–193. 10.1111/acel.12039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Zvi A., Miller E. A., Morimoto R. I. (2009). Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proc. Natl. Acad. Sci. 106, 14914–14919. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensky M. K., Gosling S. D., Sinn D. L. (2013). The world from a dog’s point of view: a review and synthesis of dog cognition research. Adv. Study Behav. 45, 209–406. 10.1016/B978-0-12-407186-5.00005-7 [DOI] [Google Scholar]
- Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., et al. (2010). S AND CENTENARIANS. PLoS One 5, e10667. 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E., Rampelli S., Turroni S., Quercia S., Candela M., Brigidi P. (2017). The gut microbiota of centenarians: signatures of longevity in the gut microbiota profile. Mech. Ageing Dev. 165, 180–184. 10.1016/j.mad.2016.12.013 [DOI] [PubMed] [Google Scholar]
- Bird A. (2002). DNA methylation patterns and epigenetic memory. Genes Dev. 16, 6–21. 10.1101/gad.947102 [DOI] [PubMed] [Google Scholar]
- Bitar M. A., Barry G. (2017). Multiple innovations in genetic and epigenetic mechanisms cooperate to underpin human brain evolution. Mol. Biol. Evol. 35, 263–268. 10.1093/molbev/msx303 [DOI] [PubMed] [Google Scholar]
- Blasco M. A. (2007). The epigenetic regulation of mammalian telomeres. Nat. Rev. Genet. 8, 299–309. 10.1038/nrg2047 [DOI] [PubMed] [Google Scholar]
- Borràs D., Ferrer I., Pumarola M. (1999). Age-related changes in the brain of the dog. Vet. Pathol. 36, 202–211. 10.1354/vp.36-3-202 [DOI] [PubMed] [Google Scholar]
- Botigué L. R., Song S., Scheu A., Gopalan S., Pendleton A. L., Oetjens M., Bobo D. (2017). Ancient European dog genomes reveal continuity since the early Neolithic. Nat. Commun. 8, 16082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyko A. R. (2011). The domestic dog: man’s best friend in the genomic era. Genome Biol. 12, 216. 10.1186/gb-2011-12-2-216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breese C. R., Ingram R. L., Sonntag W. E. (1991). Influence of age and long-term dietary restriction on plasma insulin-like growth factor-1 (IGF-1), IGF-1 gene expression, and IGF-1 binding proteins. J. Gerontol. 46, B180–B187. 10.1093/geronj/46.5.B180 [DOI] [PubMed] [Google Scholar]
- Broer L., Buchman A. S., Deelen J., Evans D. S., Faul J. D., Lunetta K. L., et al. (2015). GWAS of longevity in CHARGE consortium confirms APOE and FOXO3 candidacy. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 70, 110–118. 10.1093/gerona/glu166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks C. L., Gu W. (2009). How does SIRT1 affect metabolism, senescence and cancer? Nat. Rev. Cancer 9, 123–128. 10.1038/nrc2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks M. B., Gu W., Barnas J. L., Ray J., Ray K. (2003). A line 1 insertion in the factor IX gene segregates with mild hemophilia B in dogs. Mamm. Genome 14, 788–795. 10.1007/s00335-003-2290-z [DOI] [PubMed] [Google Scholar]
- Brown S. G., Rhodes R. E. (2006). Relationships among dog ownership and leisure-time walking in western Canadian adults. Am. J. Prev. Med. 30, 131–136. 10.1016/j.amepre.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Cabanillas R., Cadiñanos J., Villameytide J. A. F., Pérez M., Longo J., Richard J. M., et al. (2011). Néstor-Guillermo progeria syndrome: a novel premature aging condition with early onset and chronic development caused by BANF1 mutations. Am. J. Med. Genet. Part A 155, 2617–2625. 10.1002/ajmg.a.34249 [DOI] [PubMed] [Google Scholar]
- Cabreiro F., Au C., Leung K.-Y., Vergara-Irigaray N., Cochemé H. M., Noori T., et al. (2013). Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell 153, 228–239. 10.1016/j.cell.2013.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamini B., Silva M. C., Madoux F., Hutt D. M., Khanna S., Chalfant M. A., et al. (2012). Small-molecule proteostasis regulators for protein conformational diseases. Nat. Chem. Biol. 8, 185–196. 10.1038/nchembio.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood S. K., Murshid A., Prince T. (2009). The shock of aging: molecular chaperones and the heat shock response in longevity and aging–a mini-review. Gerontology 55, 550–558. 10.1159/000225957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvanese V., Lara E., Kahn A., Fraga M. F. (2009). The role of epigenetics in aging and age-related diseases. Ageing Res. Rev. 8, 268–276. 10.1016/j.arr.2009.03.004 [DOI] [PubMed] [Google Scholar]
- Campisi J., d’Adda di Fagagna F. (2007). Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740. 10.1038/nrm2233 [DOI] [PubMed] [Google Scholar]
- Capy P., Gasperi G., Biémont C., Bazin C. (2000). Stress and transposable elements: co-evolution or useful parasites? Heredity (Edinb.) 85, 101–106. 10.1046/j.1365-2540.2000.00751.x [DOI] [PubMed] [Google Scholar]
- Caron-Lormier G., England G. C. W., Green M. J., Asher L. (2016). Using the incidence and impact of health conditions in guide dogs to investigate healthy ageing in working dogs. Vet. J. 207, 124–130. 10.1016/j.tvjl.2015.10.046 [DOI] [PubMed] [Google Scholar]
- Carrasco-Garcia E., Arrizabalaga O., Serrano M., Lovell-Badge R., Matheu A. (2015). Increased gene dosage of Ink4/Arf and p53 delays age-associated central nervous system functional decline. Aging Cell 14, 710–714. 10.1111/acel.12343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano J. M., Mosher K. I., Abbey R. J., McBride A. A., James M. L., Berdnik D., et al. (2017). Human umbilical cord plasma proteins revitalize hippocampal function in aged mice. Nature 544, 488–492. 10.1038/nature22067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho R. M., Squarize C. H., Chodosh L. A., Williams B. O., Gutkind J. S. (2009). mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell 5, 279–289. 10.1016/j.stem.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecconi F., Levine B. (2008). The role of autophagy in mammalian development: cell makeover rather than cell death. Dev. Cell 15, 344–357. 10.1016/j.devcel.2008.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakkalakal J. V., Jones K. M., Basson M. A., Brack A. S. (2012). The aged niche disrupts muscle stem cell quiescence. Nature 490, 355–360. 10.1038/nature11438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K.-C., Yu M.-H., Lin M.-C., Huang C.-N., Chung D.-J., Lee Y.-J., et al. (2016). Pleiotropic effects of acarbose on atherosclerosis development in rabbits are mediated via upregulating AMPK signals. Sci. Rep. 6, 38642. 10.1038/srep38642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y.-M., Hadox E., Szladovits B., Garden O. A. (2016). Serum biochemical phenotypes in the domestic dog. PLoS One 11, e0149650. 10.1371/journal.pone.0149650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapagain D., Range F., Huber L., Virányi Z. (2018). Cognitive aging in dogs. Gerontology 64, 165–171. 10.1159/000481621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C., Liu Y., Liu Y., Zheng P. (2009). mTOR regulation and therapeutic rejuvenation of aging hematopoietic stem cells. Sci. Signal. 2, ra75. 10.1126/scisignal.2000559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhang X., Song L., Le W. (2012). Autophagy dysregulation in amyotrophic lateral sclerosis. Brain Pathol. 22, 110–116. 10.1111/j.1750-3639.2011.00546.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chénais B., Caruso A., Hiard S., Casse N. (2012). The impact of transposable elements on eukaryotic genomes: from genome size increase to genetic adaptation to stressful environments. Gene 509, 7–15. 10.1016/j.gene.2012.07.042 [DOI] [PubMed] [Google Scholar]
- Cheng L., Doecke J. D., Sharples R. A., Villemagne V. L., Fowler C. J., Rembach A., et al. (2015). Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol. Psychiatry 20, 1188–1196. 10.1038/mp.2014.127 [DOI] [PubMed] [Google Scholar]
- Cheng L., Sharples R. A., Scicluna B. J., Hill A. F. (2014). Exosomes provide a protective and enriched source of miRNA for biomarker profiling compared to intracellular and cell-free blood. J. Extracell. Vesicles 3, 23743. 10.3402/jev.v3.23743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng T., Rodrigues N., Shen H., Yang Y., Dombkowski D., Sykes M., et al. (2000). Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808. 10.1126/science.287.5459.1804 [DOI] [PubMed] [Google Scholar]
- Chiang W.-C., Ching T.-T., Lee H. C., Mousigian C., Hsu A.-L. (2012). HSF-1 regulators DDL-1/2 link insulin-like signaling to heat-shock responses and modulation of longevity. Cell 148, 322–334. 10.1016/j.cell.2011.12.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini C. C. S., Tarragó M. G., Chini E. N. (2017). NAD and the aging process: role in life, death and everything in between. Mol. Cell. Endocrinol. 455, 62–74. 10.1016/j.mce.2016.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho K., Chung J. Y., Cho S. K., Shin H.-W., Jang I.-J., Park J.-W., et al. (2015). Antihyperglycemic mechanism of metformin occurs via the AMPK/LXRα/POMC pathway. Sci. Rep. 5, 8145. 10.1038/srep08145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho M., Suh Y. (2014). Genome maintenance and human longevity. Curr. Opin. Genet. Dev. 26, 105–115. 10.1016/j.gde.2014.07.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrogianni N., Voutetakis K., Kapetanou M., Delitsikou V., Papaevgeniou N., Sakellari M., et al. (2015). Proteasome activation: an innovative promising approach for delaying aging and retarding age-related diseases. Ageing Res. Rev. 23, 37–55. 10.1016/j.arr.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Christian H., Bauman A., Epping J. N., Levine G. N., McCormack G., Rhodes R. E., et al. (2018). Encouraging dog walking for health promotion and disease prevention. Am. J. Lifestyle Med 12, 233–243. 10.1177/1559827616643686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie L.-A., Opii W. O., Head E., Araujo J. A., de Rivera C., Milgram N. W., et al. (2009). Short-term supplementation with acetyl-l-carnitine and lipoic acid alters plasma protein carbonyl levels but does not improve cognition in aged beagles. Exp. Gerontol. 44, 752–759. 10.1016/j.exger.2009.08.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C. T. (2019). Multiple pathways for mitophagy: a neurodegenerative conundrum for Parkinson’s disease. Neurosci. Lett. 697, 66–71. 10.1016/j.neulet.2018.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimarelli G., Virányi Z., Turcsán B., Rónai Z., Sasvári-Székely M., Bánlaki Z. (2017). Social behavior of pet dogs is associated with peripheral OXTR methylation. Front. Psychol. 8, 549. 10.3389/fpsyg.2017.00549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark L. A., Tsai K. L., Starr A. N., Nowend K. L., Murphy K. E. (2011). A missense mutation in the 20S proteasome β2 subunit of Great Danes having harlequin coat patterning. Genomics 97, 244–248. 10.1016/j.ygeno.2011.01.003 [DOI] [PubMed] [Google Scholar]
- Clark L. A., Wahl J. M., Rees C. A., Murphy K. E. (2006). Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc. Natl. Acad. Sci. U. S. A. 103, 1376–1381. 10.1073/pnas.0506940103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd V., Nelson C. P., Albrecht E., Mangino M., Deelen J., Buxton J. L., et al. (2013). Identification of seven loci affecting mean telomere length and their association with disease. Nat. Genet. 45, 422–427. 10.1038/ng.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho L. P., Kultima J. R., Costea P. I., Fournier C., Pan Y., Czarnecki-Maulden G., et al. (2018). Similarity of the dog and human gut microbiomes in gene content and response to diet. Microbiome 6, 72. 10.1186/s40168-018-0450-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado M., Blasco M. A., Serrano M. (2007). Cellular senescence in cancer and aging. Cell 130, 223–233. 10.1016/j.cell.2007.07.003 [DOI] [PubMed] [Google Scholar]
- Collier L. S., Largaespada D. A. (2007). Transposable elements and the dynamic somatic genome. Genome Biol. 8, S5. 10.1186/gb-2007-8-s1-s5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corral-Debrinski M., Horton T., Lott M. T., Shoffner J. M., Flint Beal M., Wallace D. C. (1992). Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat. Genet. 2, 324–329. 10.1038/ng1292-324 [DOI] [PubMed] [Google Scholar]
- Cotman C. W., Head E. (2008). The canine (dog) model of human aging and disease: dietary, environmental and immunotherapy approaches. J. Alzheimer’s Dis. 15, 685–707. 10.3233/JAD-2008-15413 [DOI] [PubMed] [Google Scholar]
- Cotman C. W., Head E., Muggenburg B. A., Zicker S., Milgram N. W. (2002). Brain aging in the canine: a diet enriched in antioxidants reduces cognitive dysfunction. Neurobiol. Aging 23, 809–818. 10.1016/S0197-4580(02)00073-8 [DOI] [PubMed] [Google Scholar]
- Creevy K. E., Austad S. N., Hoffman J. M., O’Neill D. G., Promislow D. E. L. (2016). The companion dog as a model for the longevity dividend. Cold Spring Harb. Perspect. Med. 6, a026633. 10.1101/cshperspect.a026633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuervo A. M., Dice J. F. (2000). Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275, 31505–31513. 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- Cummings B. J., Head E., Ruehl W., Milgram N. W., Cotman C. W. (1996). The canine as an animal model of human aging and dementia. Neurobiol. Aging 17, 259–268. 10.1016/0197-4580(95)02060-8 [DOI] [PubMed] [Google Scholar]
- Curl A. L., Bibbo J., Johnson R. A. (2016). Dog walking, the human–animal bond and older adults’ physical health. Gerontologist 35, gnw051. 10.1093/geront/gnw051 [DOI] [PubMed] [Google Scholar]
- d’Adda di Fagagna F. (2008). Living on a break: cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522. 10.1038/nrc2440 [DOI] [PubMed] [Google Scholar]
- da Silva P. F. L., Ogrodnik M., Kucheryavenko O., Glibert J., Miwa S., Cameron K., et al. (2019). The bystander effect contributes to the accumulation of senescent cells in vivo. Aging Cell 18, e12848. 10.1111/acel.12848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai H., Sinclair D. A., Ellis J. L., Steegborn C. (2018). Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 188, 140–154. 10.1016/j.pharmthera.2018.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M., Tollefsbol T. O. (2015). Epigenetic linkage of aging, cancer and nutrition. J. Exp. Biol. 218, 59–70. 10.1242/jeb.107110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day M. J. (2010). Ageing, immunosenescence and inflammageing in the dog and cat. J. Comp. Pathol. 142, S60–S69. 10.1016/j.jcpa.2009.10.011 [DOI] [PubMed] [Google Scholar]
- De Cecco M., Criscione S. W., Peterson A. L., Neretti N., Sedivy J. M., Kreiling J. A. (2013). Transposable elements become active and mobile in the genomes of aging mammalian somatic tissues. Aging (Albany NY) 5, 867–883. 10.18632/aging.100621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Felice F. G., Munoz D. P. (2016). Opportunities and challenges in developing relevant animal models for Alzheimer’s disease. Ageing Res. Rev. 26, 112–114. 10.1016/j.arr.2016.01.006 [DOI] [PubMed] [Google Scholar]
- de Haan G., Lazare S. S. (2018). Aging of hematopoietic stem cells. Blood 131, 479–487. 10.1182/blood-2017-06-746412 [DOI] [PubMed] [Google Scholar]
- de Jesus B. B., Vera E., Schneeberger K., Tejera A. M., Ayuso E., Bosch F., et al. (2012). Telomerase gene therapy in adult and old mice delays aging and increases longevity without increasing cancer. EMBO Mol. Med. 4, 691–704. 10.1002/emmm.201200245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Magalhães J. P., Curado J., Church G. M. (2009). Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881. 10.1093/bioinformatics/btp073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sandre-Giovannoli A., Bernard R., Cau P., Navarro C., Amiel J., Boccaccio I., et al. (2003). Lamin a truncation in Hutchinson-Gilford progeria. Science 300, 2055. 10.1126/science.1084125 [DOI] [PubMed] [Google Scholar]
- de Vrij F. M. S., Fischer D. F., van Leeuwen F. W., Hol E. M. (2004). Protein quality control in Alzheimer’s disease by the ubiquitin proteasome system. Prog. Neurobiol. 74, 249–270. 10.1016/j.pneurobio.2004.10.001 [DOI] [PubMed] [Google Scholar]
- Deelen J., Beekman M., Uh H.-W., Helmer Q., Kuningas M., Christiansen L., et al. (2011). Genome-wide association study identifies a single major locus contributing to survival into old age; the APOE locus revisited. Aging Cell 10, 686–698. 10.1111/j.1474-9726.2011.00705.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguchi Y., Negoro S., Kishimoto S. (1988). Age-related changes of heat shock protein gene transcription in human peripheral blood mononuclear cells. Biochem. Biophys. Res. Commun. 157, 580–584. 10.1016/S0006-291X(88)80289-4 [DOI] [PubMed] [Google Scholar]
- Demontis F., Patel V. K., Swindell W. R., Perrimon N. (2014). Intertissue control of the nucleolus via a myokine-dependent longevity pathway. Cell Rep. 7, 1481–1494. 10.1016/j.celrep.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V. (2006). Autophagy as an immune defense mechanism. Curr. Opin. Immunol. 18, 375–382. 10.1016/j.coi.2006.05.019 [DOI] [PubMed] [Google Scholar]
- Deuschle M., Gotthardt U., Schweiger U., Weber B., Körner A., Schmider J., et al. (1997). With aging in humans the activity of the hypothalamus–pituitary–adrenal system increases and its diurnal amplitude flattens. Life Sci. 61, 2239–2246. 10.1016/S0024-3205(97)00926-0 [DOI] [PubMed] [Google Scholar]
- Di Fonzo A., Rohé C. F., Ferreira J., Chien H. F., Vacca L., Stocchi F., et al. (2005). A frequent LRRK2 gene mutation associated with autosomal dominant Parkinson’s disease. Lancet 365, 412–415. 10.1016/S0140-6736(05)17829-5 [DOI] [PubMed] [Google Scholar]
- Dias J. N. R., Aguiar S. I., Pereira D. M., André A. S., Gano L., Correia J. D. G., et al. (2018). The histone deacetylase inhibitor panobinostat is a potent antitumor agent in canine diffuse large B-cell lymphoma. Oncotarget 9, 28586–28598. 10.18632/oncotarget.25580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickey C. A., Kamal A., Lundgren K., Klosak N., Bailey R. M., Dunmore J., et al. (2007). The high-affinity HSP90-CHIP complex recognizes and selectively degrades phosphorylated tau client proteins. J. Clin. Invest. 117, 648–658. 10.1172/JCI29715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia M. S., Fischbach M. A. (2015). Small molecules from the human microbiota. Science (80-.). 349, 1254766–1254766. 10.1126/science.1254766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan R., McElwee J. J., Matthijssens F., Walker G. A., Houthoofd K., Back P., et al. (2008). Against the oxidative damage theory of aging: superoxide dismutases protect against oxidative stress but have little or no effect on life span in Caenorhabditis elegans. Genes Dev. 22, 3236–3241. 10.1101/gad.504808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs L. M., Mellersh C. S., Staden R., Sivachenko A., Cibulskis K. (2014). An intronic SINE insertion in FAM161A that causes exon-skipping is associated with progressive retinal atrophy in Tibetan spaniels and Tibetan terriers. PLoS One 9, e93990. 10.1371/journal.pone.0093990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger D. L., Davis B. W., Cocco R., Sechi S., Di Cerbo A., Parker H. G., et al. (2016. a). Commonalities in development of pure breeds and population isolates revealed in the genome of the Sardinian Fonni’s dog. Genetics 204, 737–755. 10.1534/genetics.116.192427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger D. L., Rimbault M., Davis B. W., Bhatnagar A., Parker H. G., Ostrander E. A. (2016. b). Whole-genome sequence, SNP chips and pedigree structure: building demographic profiles in domestic dog breeds to optimize genetic-trait mapping. Dis. Model. Mech. 9, 1445–1460. 10.1242/dmm.027037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreger D. L., Schmutz S. M. (2011). A SINE insertion causes the black-and-tan and saddle tan phenotypes in domestic dogs. J. Hered. 102, S11–S18. 10.1093/jhered/esr042 [DOI] [PubMed] [Google Scholar]
- Drummond M. J., McCarthy J. J., Sinha M., Spratt H. M., Volpi E., Esser K. A., et al. (2011). Aging and microRNA expression in human skeletal muscle: a microarray and bioinformatics analysis. Physiol. Genomics 43, 595–603. 10.1152/physiolgenomics.00148.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumble M., Moore L., Chambers S. M., Geiger H., Van Zant G., Goodell M. A., et al. (2007). The impact of altered p53 dosage on hematopoietic stem cell dynamics during aging. Blood 109, 1736–1742. 10.1182/blood-2006-03-010413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durieux J., Wolff S., Dillin A. (2011). The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. 10.1016/j.cell.2010.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edens W. A., Sharling L., Cheng G., Shapira R., Kinkade J. M., Lee T., et al. (2001). Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 154, 879–91. 10.1083/jcb.200103132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar D., Shabalina I., Camara Y., Wredenberg A., Calvaruso M. A., Nijtmans L., et al. (2009). Random point mutations with major effects on protein-coding genes are the driving force behind premature aging in mtDNA mutator mice. Cell Metab. 10, 131–138. 10.1016/j.cmet.2009.06.010 [DOI] [PubMed] [Google Scholar]
- Eisenberg T., Knauer H., Schauer A., Büttner S., Ruckenstuhl C., Carmona-Gutierrez D., et al. (2009). Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 11, 1305–1314. 10.1038/ncb1975 [DOI] [PubMed] [Google Scholar]
- Elabd C., Cousin W., Upadhyayula P., Chen R. Y., Chooljian M. S., Li J., et al. (2014). Oxytocin is an age-specific circulating hormone that is necessary for muscle maintenance and regeneration. Nat. Commun. 5, 4082. 10.1038/ncomms5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis N. A., Groden J., Ye T.-Z., Straughen J., Lennon D. J., Ciocci S., et al. (1995). The Bloom’s syndrome gene product is homologous to RecQ helicases. Cell 83, 655–666. 10.1016/0092-8674(95)90105-1 [DOI] [PubMed] [Google Scholar]
- Elshafae S. M., Kohart N. A., Altstadt L. A., Dirksen W. P., Rosol T. J. (2017). The effect of a histone deacetylase inhibitor (AR-42) on canine prostate cancer growth and metastasis. Prostate 77, 776–793. 10.1002/pros.23318 [DOI] [PubMed] [Google Scholar]
- Emanuele E., Fontana J. M., Minoretti P., Geroldi D. (2010). Preliminary evidence of a genetic association between chromosome 9p21.3 and human longevity. Rejuvenation Res. 13, 23–26. 10.1089/rej.2009.0970 [DOI] [PubMed] [Google Scholar]
- Eriksson M., Brown W. T., Gordon L. B., Glynn M. W., Singer J., Scott L., et al. (2003). Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423, 293–298. 10.1038/nature01629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faggioli F., Wang T., Vijg J., Montagna C. (2012). Chromosome-specific accumulation of aneuploidy in the aging mouse brain. Hum. Mol. Genet. 21, 5246–5253. 10.1093/hmg/dds375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faldyna M., Levá L., Knötigová P., Toman M. (2001). Lymphocyte subsets in peripheral blood of dogs—a flow cytometric study. Vet. Immunol. Immunopathol. 82, 23–37. 10.1016/S0165-2427(01)00337-3 [DOI] [PubMed] [Google Scholar]
- Ferrer I., Pumarola M., Rivera R., Zújar M. J., Cruz-Sánchez F., Vidal A. (1993). Primary central white matter degeneration in old dogs. Acta Neuropathol. 86, 172–175. 10.1007/BF00334884 [DOI] [PubMed] [Google Scholar]
- Feuerbacher E. N., Wynne C. D. L. (2011). A history of dogs as subjects in North American experimental psychological research. Comp. Cogn. Behav. Rev. 6, 46–71. 10.3819/ccbr.2011.60001 [DOI] [Google Scholar]
- Fick L. J., Fick G. H., Li Z., Cao E., Bao B., Heffelfinger D., et al. (2012). Telomere length correlates with life span of dog breeds. Cell Rep. 2, 1530–1536. 10.1016/j.celrep.2012.11.021 [DOI] [PubMed] [Google Scholar]
- Finsterer J., Frank M. (2016). Propofol is mitochondrion-toxic and may unmask a mitochondrial disorder. J. Child Neurol. 31, 1489–1494. 10.1177/0883073816661458 [DOI] [PubMed] [Google Scholar]
- Fish E. J., Irizarry K. J., DeInnocentes P., Ellis C. J., Prasad N., Moss A. G., et al. (2018). Malignant canine mammary epithelial cells shed exosomes containing differentially expressed microRNA that regulate oncogenic networks. BMC Cancer 18, 832. 10.1186/s12885-018-4750-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian M. C., Dörr K., Niebel A., Daria D., Schrezenmeier H., Rojewski M., et al. (2012). Cdc42 activity regulates hematopoietic stem cell aging and rejuvenation. Cell Stem Cell 10, 520–530. 10.1016/j.stem.2012.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flurkey K., Papaconstantinou J., Miller R. A., Harrison D. E. (2001). Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc. Natl. Acad. Sci. 98, 6736–6741. 10.1073/pnas.111158898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forero D. A., González-Giraldo Y., López-Quintero C., Castro-Vega L. J., Barreto G. E., Perry G. (2016). Meta-analysis of telomere length in Alzheimer’s disease. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 71, 1069–1073. 10.1093/gerona/glw053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg L. A., Rasi C., Razzaghian H. R., Pakalapati G., Waite L., Thilbeault K. S., et al. (2012). Age-related somatic structural changes in the nuclear genome of human blood cells. Am. J. Hum. Genet. 90, 217–228. 10.1016/j.ajhg.2011.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forslund K., Hildebrand F., Nielsen T., Falony G., Le Chatelier E., Sunagawa S., et al. (2015). Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266. 10.1038/nature15766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga C. G., Shigenaga M. K., Park J. W., Degan P., Ames B. N. (1990). Oxidative damage to DNA during aging: 8-hydroxy-2’-deoxyguanosine in rat organ DNA and urine. Proc. Natl. Acad. Sci. U. S. A. 87, 4533–4537. 10.1073/pnas.87.12.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraga M. F., Esteller M. (2007). Epigenetics and aging: the targets and the marks. Trends Genet. 23, 413–418. 10.1016/j.tig.2007.05.008 [DOI] [PubMed] [Google Scholar]
- Frantz L. A. F., Mullin V. E., Pionnier-Capitan M., Lebrasseur O., Ollivier M., Perri A., et al. (2016). Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science (80-.). 352, 1228–1231. 10.1126/science.aaf3161 [DOI] [PubMed] [Google Scholar]
- Freedman A. H., Lohmueller K. E., Wayne R. K. (2016). Evolutionary history, selective sweeps, and deleterious variation in the dog. Annu. Rev. Ecol. Evol. Syst. 47, 73–96. 10.1146/annurev-ecolsys-121415-032155 [DOI] [Google Scholar]
- Freund A., Laberge R.-M., Demaria M., Campisi J. (2012). Lamin B1 loss is a senescence-associated biomarker. Mol. Biol. Cell 23, 2066–2075. 10.1091/mbc.e11-10-0884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulop T., Larbi A., Dupuis G., Le Page A., Frost E. H., Cohen A. A., et al. (2018). Immunosenescence and inflamm-aging as two sides of the same coin: friends or foes? Front. Immunol. 8, 1960. 10.3389/fimmu.2017.01960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galis F., Van Der Sluijs I., Van Dooren T. J. M., Metz J. A. J., Nussbaumer M. (2007). Do large dogs die young? J. Exp. Zool. Part B Mol. Dev. Evol. 308B, 119–126. 10.1002/jez.b.21116 [DOI] [PubMed] [Google Scholar]
- García-Cao I., García-Cao M., Martín-Caballero J., Criado L. M., Klatt P., Flores J. M., et al. (2002). Super p53mice exhibit enhanced DNA damage response, are tumor resistant and age normallyc. EMBO J. 21, 6225–6235. 10.1093/emboj/cdf595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D. O., Wertheim B. C., Manson J. E., Chlebowski R. T., Volpe S. L., Howard B. V., et al. (2015). Relationships between dog ownership and physical activity in postmenopausal women. Prev. Med. (Baltim) 70, 33–38. 10.1016/j.ypmed.2014.10.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur U., Tu J., Li D., Gao Y., Lian T., Sun B., et al. (2017). Molecular evolutionary patterns of NAD+/Sirtuin aging signaling pathway across taxa. PLoS One 12, e0182306. 10.1371/journal.pone.0182306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrig S. M., van der Poel C., Sayer T. A., Schertzer J. D., Henstridge D. C., Church J. E., et al. (2012). Hsp72 preserves muscle function and slows progression of severe muscular dystrophy. Nature 484, 394–398. 10.1038/nature10980 [DOI] [PubMed] [Google Scholar]
- Geiman T. M., Muegge K. (2009). DNA methylation in early development. Mol. Reprod. Dev. 77, n/a–n/a. 10.1002/mrd.21118 [DOI] [PubMed] [Google Scholar]
- Gelaleti G. B., Granzotto A., Leonel C., Jardim B. V., Moschetta M. G., Carareto C. M. A., et al. (2014). Short interspersed CAN SINE elements as prognostic markers in canine mammary neoplasia. Oncol. Rep. 31, 435–441. 10.3892/or.2013.2827 [DOI] [PubMed] [Google Scholar]
- Gentilini D., Mari D., Castaldi D., Remondini D., Ogliari G., Ostan R., et al. (2013). Role of epigenetics in human aging and longevity: genome-wide DNA methylation profile in centenarians and centenarians? Offspring. Age (Omaha) 35, 1961–1973. 10.1007/s11357-012-9463-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertz M., Nguyen G. T. T., Fischer F., Suenkel B., Schlicker C., Fränzel B., et al. (2012). A molecular mechanism for direct sirtuin activation by resveratrol. PLoS One 7, e49761. 10.1371/journal.pone.0049761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghi P., Di Brisco F., Dallorto D., Osella M. C., Orsetti M. (2009). Age-related modifications of egr1 expression and ubiquitin-proteasome components in pet dog hippocampus. Mech. Ageing Dev. 130, 320–327. 10.1016/j.mad.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Gilmore K. M., Greer K. A. (2015). Why is the dog an ideal model for aging research? Exp. Gerontol. 71, 14–20. 10.1016/j.exger.2015.08.008 [DOI] [PubMed] [Google Scholar]
- Gocmez S. S., Gacar N., Utkan T., Gacar G., Scarpace P. J., Tumer N. (2016). Protective effects of resveratrol on aging-induced cognitive impairment in rats. Neurobiol. Learn. Mem. 131, 131–136. 10.1016/j.nlm.2016.03.022 [DOI] [PubMed] [Google Scholar]
- Goldstein O., Kukekova A. V., Aguirre G. D., Acland G. M. (2010). Exonic SINE insertion in STK38L causes canine early retinal degeneration (erd). Genomics 96, 362–368. 10.1016/j.ygeno.2010.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubtsova N. N., Filippov F. N., Gunin A. G. (2016). Lamin B1 and lamin B2 in human skin in the process of aging. Adv. Gerontol. 6, 275–281. 10.1134/S2079057016040068 [DOI] [PubMed] [Google Scholar]
- Gonskikh Y., Polacek N. (2017). Alterations of the translation apparatus during aging and stress response. Mech. Ageing Dev. 168, 30–36. 10.1016/j.mad.2017.04.003 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Suarez I., Redwood A. B., Gonzalo S. (2009). Loss of A-type lamins and genomic instability. Cell Cycle 8, 3860–3865. 10.4161/cc.8.23.10092 [DOI] [PubMed] [Google Scholar]
- Gorbunova V., Boeke J. D., Helfand S. L., Sedivy J. M. (2014). Sleeping dogs of the genome. Science 346, 1187–1188. 10.1126/science.aaa3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greeley E. H., Spitznagel E., Lawler D. F., Kealy R. D., Segre M. (2006). Modulation of canine immunosenescence by life-long caloric restriction. Vet. Immunol. Immunopathol. 111, 287–299. 10.1016/j.vetimm.2006.02.002 [DOI] [PubMed] [Google Scholar]
- Greenberg R. A., Allsopp R. C., Chin L., Morin G. B., DePinho R. A. (1998). Expression of mouse telomerase reverse transcriptase during development, differentiation and proliferation. Oncogene 16, 1723–1730. 10.1038/sj.onc.1201933 [DOI] [PubMed] [Google Scholar]
- Greer E. L., Brunet A. (2005). FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 24, 7410–7425. 10.1038/sj.onc.1209086 [DOI] [PubMed] [Google Scholar]
- Greer K. A., Canterberry S. C., Murphy K. E. (2007). Statistical analysis regarding the effects of height and weight on life span of the domestic dog. Res. Vet. Sci. 82, 208–214. 10.1016/j.rvsc.2006.06.005 [DOI] [PubMed] [Google Scholar]
- Greer K. A., Hughes L. M., Masternak M. M. (2011). Connecting serum IGF-1, body size, and age in the domestic dog. Age (Omaha) 33, 475–483. 10.1007/s11357-010-9182-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiss S., Gartner A. (2009). Sirtuin/Sir2 phylogeny, evolutionary considerations and structural conservation. Mol. Cells 28, 407–415. 10.1007/s10059-009-0169-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse N., Van Loon B., Bley C. R. (2014). DNA damage response and DNA repair—dog as a model? BMC Cancer 14, 1–8. 10.1186/1471-2407-14-203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grześkowiak Ł., Endo A., Beasley S., Salminen S. (2015). Microbiota and probiotics in canine and feline welfare. Anaerobe 34, 14–23. 10.1016/j.anaerobe.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. (2011). Sirtuins, aging, and metabolism. Cold Spring Harb. Symp. Quant. Biol. 76, 81–90. 10.1101/sqb.2011.76.010629 [DOI] [PubMed] [Google Scholar]
- Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., et al. (2008). AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol. Cell 30, 214–226. 10.1016/j.molcel.2008.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn K., Rohdin C., Jagannathan V., Wohlsein P., Baumgärtner W., Seehusen F., et al. (2015). TECPR2 associated neuroaxonal dystrophy in Spanish water dogs. PLoS One 10, 1–18. 10.1371/journal.pone.0141824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haigis M. C., Yankner B. A. (2010). The aging stress response. Mol. Cell 40, 333–344. 10.1016/j.molcel.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halley-Stott R. P., Gurdon J. B. (2013). Epigenetic memory in the context of nuclear reprogramming and cancer. Brief. Funct. Genomics 12, 164–173. 10.1093/bfgp/elt011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S., Brunet A. (2012). Histone methylation makes its mark on longevity. Trends Cell Biol. 22, 42–49. 10.1016/j.tcb.2011.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks D. C., Kazazian H. H. (2016). Roles for retrotransposon insertions in human disease. Mob. DNA 7, 9. 10.1186/s13100-016-0065-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon E., Spiers H., Viana J., Pidsley R., Burrage J., Murphy T. M., et al. (2015). Methylation QTLs in the developing brain and their enrichment in schizophrenia risk loci. Nat. Neurosci. 19, 48–54. 10.1038/nn.4182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., et al. (2006). Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Harley C. B., Futcher B. A., Greider C. W. (1990). Telomeres shorten during ageing of human fibroblasts. Nature 345, 458–460. 10.1038/345458a0 [DOI] [PubMed] [Google Scholar]
- Harrison D. E., Strong R., Allison D. B., Ames B. N., Astle C. M., Atamna H., et al. (2014). Acarbose, 17-α-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging Cell 13, 273–282. 10.1111/acel.12170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hars E. S., Qi H., Jin S. V., Cai L., Hu C., Liu L. F. (2007). Autophagy regulates ageing in C. elegans. Autophagy 3, 93–95. 10.4161/auto.3636 [DOI] [PubMed] [Google Scholar]
- Hartford C. M., Ratain M. J. (2007). Rapamycin: something old, something new, sometimes borrowed and now renewed. Clin. Pharmacol. Ther. 82, 381–388. 10.1038/sj.clpt.6100317 [DOI] [PubMed] [Google Scholar]
- Hastie N. D., Dempster M., Dunlop M. G., Thompson A. M., Green D. K., Allshire R. C. (1990). Telomere reduction in human colorectal carcinoma and with ageing. Nature 346, 866–868. 10.1038/346866a0 [DOI] [PubMed] [Google Scholar]
- Hayflick L. (1976). The cell biology of Human aging. N. Engl. J. Med. 295, 1302–1308. 10.1056/NEJM197612022952308 [DOI] [PubMed] [Google Scholar]
- Hayward J. J., Castelhano M. G., Oliveira K. C., Corey E., Balkman C., Baxter T. L., et al. (2016). Complex disease and phenotype mapping in the domestic dog. Nat. Commun. 7, 10460. 10.1038/ncomms10460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He C., Klionsky D. J. (2009). Regulation mechanisms and signaling pathways of autophagy. Annu. Rev. Genet. 43, 67–93. 10.1146/annurev-genet-102808-114910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Head E., Liu J., Hagen T. M., Muggenburg B. A., Milgram N. W., Ames B. N., et al. (2002). Oxidative damage increases with age in a canine model of human brain aging. J. Neurochem. 82, 375–381. 10.1046/j.1471-4159.2002.00969.x [DOI] [PubMed] [Google Scholar]
- Head E., Nukala V. N., Fenoglio K. A., Muggenburg B. A., Cotman C. W., Sullivan P. G. (2009). Effects of age, dietary, and behavioral enrichment on brain mitochondria in a canine model of human aging. Exp. Neurol. 220, 171–176. 10.1016/j.expneurol.2009.08.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht J., Rice E. S. (2015). Citizen science: a new direction in canine behavior research. Behav. Processes 110, 125–132. 10.1016/j.beproc.2014.10.014 [DOI] [PubMed] [Google Scholar]
- Hekimi S., Lapointe J., Wen Y. (2011). Taking a “good” look at free radicals in the aging process. Trends Cell Biol. 21, 569–576. 10.1016/j.tcb.2011.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller J. B. (2007). Metformin overdose in dogs and cats. Vet. Med. 231–234.
- Hesp K., Smant G., Kammenga J. E. (2015). Caenorhabditis elegans DAF-16/FOXO transcription factor and its mammalian homologs associate with age-related disease. Exp. Gerontol. 72, 1–7. 10.1016/J.EXGER.2015.09.006 [DOI] [PubMed] [Google Scholar]
- Hickman E. S., Moroni M. C., Helin K. (2002). The role of p53 and pRB in apoptosis and cancer. Curr. Opin. Genet. Dev. 12, 60–66. 10.1016/S0959-437X(01)00265-9 [DOI] [PubMed] [Google Scholar]
- Hilton B. A., Liu J., Cartwright B. M., Liu Y., Breitman M., Wang Y., et al. (2017). Progerin sequestration of PCNA promotes replication fork collapse and mislocalization of XPA in laminopathy-related progeroid syndromes. FASEB J. 31, 3882–3893. 10.1096/fj.201700014R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiona A., Sanz A., Kujoth G. C., Pamplona R., Seo A. Y., Hofer T., et al. (2010). Mitochondrial DNA mutations induce mitochondrial dysfunction, apoptosis and sarcopenia in skeletal muscle of mitochondrial DNA mutator mice. PLoS One 5, e11468. 10.1371/journal.pone.0011468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman A. M., Dow S. W. (2016). Concise review: stem cell trials using companion animal disease models. Stem Cells 34, 1709–1729. 10.1002/stem.2377 [DOI] [PubMed] [Google Scholar]
- Hoffman J. M., Creevy K. E., Franks A., O’Neill D. G., Promislow D. E. L. (2018). The companion dog as a model for human aging and mortality. Aging Cell 17, e12737. 10.1111/acel.12737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- HogenEsch H., Thompson S., Dunham A., Ceddia M., Hayek M. (2004). Effect of age on immune parameters and the immune response of dogs to vaccines: a cross-sectional study. Vet. Immunol. Immunopathol. 97, 77–85. 10.1016/j.vetimm.2003.08.010 [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont J., Ducos B., Leneuve P., Géloën A., Even P. C., et al. (2003). IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421, 182–187. 10.1038/nature01298 [DOI] [PubMed] [Google Scholar]
- Hoopes B. C., Rimbault M., Liebers D., Ostrander E. A., Sutter N. B. (2012). The insulin-like growth factor 1 receptor (IGF1R) contributes to reduced size in dogs. Mamm. Genome 23, 780–790. 10.1007/s00335-012-9417-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu C.-K., Brunet A. (2018). The African turquoise killifish: a research organism to study vertebrate aging and diapause. Aging Cell 17, e12757. 10.1111/acel.12757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J., Hoummady S., Muller C., Pouchelon J.-L., Blondot M., Gilbert C., et al. (2016). Assessment of frailty in aged dogs. Am. J. Vet. Res. 77, 1357–1365. 10.2460/ajvr.77.12.1357 [DOI] [PubMed] [Google Scholar]
- Hunter R. G., Gagnidze K., McEwen B. S., Pfaff D. W. (2015). Stress and the dynamic genome: steroids, epigenetics, and the transposome. Proc. Natl. Acad. Sci. U. S. A. 112, 6828–6833. 10.1073/pnas.1411260111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang I. K., Yoon Y. S., Yoo K.-Y., Li H., Choi J. H., KIM D. W., et al. (2008). Differences in lipid peroxidation and Cu,Zn-superoxide dismutase in the hippocampal CA1 region between adult and aged dogs. J. Vet. Med. Sci. 70, 273–277. 10.1292/jvms.70.273 [DOI] [PubMed] [Google Scholar]
- Ichii O., Ohta H., Horino T., Nakamura T., Hosotani M., Mizoguchi T., et al. (2017). Urinary exosome-derived microRNAs reflecting the changes of renal function and histopathology in dogs. Sci. Rep. 7, 40340. 10.1038/srep40340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilska J., Haskell M. J., Blott S. C., Sánchez-Molano E., Polgar Z., Lofgren S. E., et al. (2017). Genetic characterization of dog personality traits. Genetics 206, 1101–1111. 10.1534/genetics.116.192674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K., Zhu T., Guan K.-L. (2003). TSC2 mediates cellular energy response to control cell growth and survival. Cell 115, 577–590. 10.1016/S0092-8674(03)00929-2 [DOI] [PubMed] [Google Scholar]
- Inoue M., Kwan N. C. L., Sugiura K. (2018). Estimating the life expectancy of companion dogs in Japan using pet cemetery data. J. Vet. Med. Sci. 80, 1153–1158. 10.1292/jvms.17-0384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insua D., Suárez M.-L., Santamarina G., Sarasa M., Pesini P. (2010). Dogs with canine counterpart of Alzheimer’s disease lose noradrenergic neurons. Neurobiol. Aging 31, 625–635. 10.1016/j.neurobiolaging.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Inukai S., de Lencastre A., Turner M., Slack F. (2012). Novel MicroRNAs differentially expressed during aging in the mouse brain. PLoS One 7, e40028. 10.1371/journal.pone.0040028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishida T., Suzuki J., Duangchan P., Settheetham-Ishida W. (1998). Preliminary report on the short stature of Southeast Asian forest dwellers, the Manni, in southern Thailand: lack of an adolescent spurt in plasma IGF-I concentration. Southeast Asian J. Trop. Med. Public Heal. 29, 62–65. [PubMed] [Google Scholar]
- Jeck W. R., Siebold A. P., Sharpless N. E. (2012). Review: a meta-analysis of GWAS and age-associated diseases. Aging Cell 11, 727–731. 10.1111/j.1474-9726.2012.00871.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeggo P. A., Pearl L. H., Carr A. M. (2016). DNA repair, genome stability and cancer: a historical perspective. Nat. Rev. Cancer 16, 35–42. 10.1038/nrc.2015.4 [DOI] [PubMed] [Google Scholar]
- Jia G., Su L., Singhal S., Liu X. (2012). Emerging roles of SIRT6 on telomere maintenance, DNA repair, metabolism and mammalian aging. Mol. Cell. Biochem. 364, 345–350. 10.1007/s11010-012-1236-8 [DOI] [PubMed] [Google Scholar]
- Jia K., Levine B. (2007). Autophagy is required for dietary restriction-mediated life span extension in C. elegans. Autophagy 3, 597–599. 10.4161/auto.4989 [DOI] [PubMed] [Google Scholar]
- Jiang P., Mizushima N. (2014). Autophagy and human diseases. Cell Res. 24, 69–79. 10.1038/cr.2013.161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez A. G. (2016). Physiological underpinnings in life-history trade-offs in man’s most popular selection experiment: the dog. J. Comp. Physiol. B 186, 813–827. 10.1007/s00360-016-1002-4 [DOI] [PubMed] [Google Scholar]
- Jin C., Li J., Green C. D., Yu X., Tang X., Han D., et al. (2011). Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 14, 161–172. 10.1016/j.cmet.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Jones P., Chase K., Martin A., Davern P., Ostrander E. A., Lark K. G. (2008). Single-nucleotide-polymorphism-based association mapping of dog stereotypes. Genetics 179, 1033–1044. 10.1534/genetics.108.087866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jucker M. (2010). The benefits and limitations of animal models for translational research in neurodegenerative diseases. Nat. Med. 16, 1210–1214. 10.1038/nm.2224 [DOI] [PubMed] [Google Scholar]
- Juhász G., Csikós G., Sinka R., Erdélyi M., Sass M. (2003). The Drosophila homolog of Aut1 is essential for autophagy and development. FEBS Lett. 543, 154–158. 10.1016/S0014-5793(03)00431-9 [DOI] [PubMed] [Google Scholar]
- Juhász G., Erdi B., Sass M., Neufeld T. P. (2007). Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 21, 3061–3066. 10.1101/gad.1600707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M. (2016). The biology of aging: citizen scientists and their pets as a bridge between research on model organisms and human subjects. Vet. Pathol. 53, 291–298. 10.1177/0300985815591082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Creevy K. E., Promislow D. E. L. (2016). The dog aging project: translational geroscience in companion animals. Mamm. Genome 27, 279–288. 10.1007/s00335-016-9638-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., McVey M., Guarente L. (1999). The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 13, 2570–2580. 10.1101/gad.13.19.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M., Powers R. W., Steffen K. K., Westman E. A., Hu D., Dang N., et al. (2005). Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–6. 10.1126/science.1115535 [DOI] [PubMed] [Google Scholar]
- Kapahi P., Chen D., Rogers A. N., Katewa S. D., Li P. W.-L., Thomas E. L., et al. (2010). With TOR, less is more: a key role for the conserved nutrient-sensing TOR pathway in aging. Cell Metab. 11, 453–465. 10.1016/j.cmet.2010.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi P., Zid B. M., Harper T., Koslover D., Sapin V., Benzer S. (2004). Regulation of lifespan in drosophila by modulation of genes in the TOR signaling pathway. Curr. Biol. 14, 885–890. 10.1016/J.CUB.2004.03.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsimpardi L., Litterman N. K., Schein P. A., Miller C. M., Loffredo F. S., Wojtkiewicz G. R., et al. (2014). Vascular and neurogenic rejuvenation of the aging mouse brain by young systemic factors. Science (80-.). 344, 630–634. 10.1126/science.1251141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian H. H. (2004). Mobile elements: drivers of genome evolution. Science 303 (5664), 1626–1632. 10.1126/science.1089670 [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang J., Gensch E., Rudner A., Tabtiang R. (1993). A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464. 10.1038/366461a0 [DOI] [PubMed] [Google Scholar]
- Kiatipattanasakul W., Nakamura S., Hossain M. M., Nakayama H., Uchino T., Shumiya S., et al. (1996). Apoptosis in the aged dog brain. Acta Neuropathol. 92, 242–248. 10.1007/s004010050514 [DOI] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., Guan K.-L. (2011). AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13, 132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Williams F. J., Dreger D. L., Plassais J., Davis B. W., Parker H. G., et al. (2018). Genetic selection of athletic success in sport-hunting dogs. Proc. Natl. Acad. Sci. U. S. A. 115, E7212–E7221. 10.1073/pnas.1800455115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Choe H. K. (2019). Role of hypothalamus in aging and its underlying cellular mechanisms. Mech. Ageing Dev. 177, 74–79. 10.1016/j.mad.2018.04.008 [DOI] [PubMed] [Google Scholar]
- Kim O.-H., Kim H., Kang J., Yang D., Kang Y.-H., Lee D. H., et al. (2017). Impaired phagocytosis of apoptotic cells causes accumulation of bone marrow-derived macrophages in aged mice. BMB Rep. 50, 43–48. 10.5483/BMBRep.2017.50.1.167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Bi X., Czarny-Ratajczak M., Dai J., Welsh D. A., Myers L., et al. (2012). Telomere maintenance genes SIRT1 and XRCC6 impact age-related decline in telomere length but only SIRT1 is associated with human longevity. Biogerontology 13, 119–131. 10.1007/s10522-011-9360-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney B. A., Meliska C. J., Steger R. W., Bartke A. (2001). Evidence that Ames dwarf mice age differently from their normal siblings in behavioral and learning and memory parameters. Horm. Behav. 39, 277–284. 10.1006/hbeh.2001.1654 [DOI] [PubMed] [Google Scholar]
- Kipling D., Cooke H. J. (1990). Hypervariable ultra-long telomeres in mice. Nature 347, 400–402. 10.1038/347400a0 [DOI] [PubMed] [Google Scholar]
- Kippin T. E., Martens D. J., van der Kooy D. (2005). p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 19, 756–767. 10.1101/gad.1272305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkland J. L., Tchkonia T., Zhu Y., Niedernhofer L. J., Robbins P. D. (2017). The clinical potential of senolytic drugs. J. Am. Geriatr. Soc. 65, 2297–2301. 10.1111/jgs.14969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirpensteijn J., Kik M., Teske E., Rutteman G. R. (2008). TP53 gene mutations in canine osteosarcoma. Vet. Surg. 37, 454–460. 10.1111/j.1532-950X.2008.00407.x [DOI] [PubMed] [Google Scholar]
- Klionsky D. J. (2005). Autophagy. Curr. Biol. 15, R282–R283. 10.1016/j.cub.2005.04.013 [DOI] [PubMed] [Google Scholar]
- Koch I. J., Clark M. M., Thompson M. J., Deere-Machemer K. A., Wang J., Duarte L., et al. (2016). The concerted impact of domestication and transposon insertions on methylation patterns between dogs and grey wolves. Mol. Ecol. 25, 1838–1855. 10.1111/mec.13480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig E. M., Fisher C., Bernard H., Wolenski F. S., Gerrein J., Carsillo M., et al. (2016). The beagle dog MicroRNA tissue atlas: identifying translatable biomarkers of organ toxicity. BMC Genomics 17, 649. 10.1186/s12864-016-2958-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H., Kaushik S., Cuervo A. M. (2011). Protein homeostasis and aging: the importance of exquisite quality control. Ageing Res. Rev. 10, 205–215. 10.1016/j.arr.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., et al. (2006). Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Koopman W. J. H., Willems P. H. G. M., Smeitink J. A. M. (2012). Monogenic mitochondrial disorders. N. Engl. J. Med. 366, 1132–1141. 10.1056/NEJMra1012478 [DOI] [PubMed] [Google Scholar]
- Kraus C., Pavard S., Promislow D. E. L. (2013). The size–life span trade-off decomposed : why large dogs die young. Am. Nat. 181, 492–505. 10.1086/669665 [DOI] [PubMed] [Google Scholar]
- Kraytsberg Y., Kudryavtseva E., McKee A. C., Geula C., Kowall N. W., Khrapko K. (2006). Mitochondrial DNA deletions are abundant and cause functional impairment in aged human substantia nigra neurons. Nat. Genet. 38, 518–520. 10.1038/ng1778 [DOI] [PubMed] [Google Scholar]
- Krebiehl G., Ruckerbauer S., Burbulla L. F., Kieper N., Maurer B., Waak J., et al. (2010). Reduced basal autophagy and impaired mitochondrial dynamics due to loss of Parkinson’s disease-associated protein DJ-1. PLoS One 5, e9367. 10.1371/journal.pone.0009367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J., Ramsey M. R., Ligon K. L., Torrice C., Koh A., Bonner Weir S., et al. (2006). p16INK4a induces an age-dependent decline in islet regenerative potential. Nature 443, 453–457. 10.1038/nature05092 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy J., Torrice C., Ramsey M. R., Kovalev G. I., Al-Regaiey K., Su L., et al. (2004). Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 114, 1299–1307. 10.1172/JCI22475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G., Galluzzi L., Brenner C. (2007). Mitochondrial membrane permeabilization in cell death. Physiol. Rev. 87, 99–163. 10.1152/physrev.00013.2006 [DOI] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Mooi W. J., Peeper D. S. (2010). The essence of senescence. Genes Dev. 24, 2463–2479. 10.1101/gad.1971610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuro-o M., Nabeshima Y., Matsumura Y., Aizawa H., Kawaguchi H., Suga T., et al. (1997). Mutation of the mouse klotho gene leads to a syndrome resembling ageing. Nature 390, 45–51. 10.1038/36285 [DOI] [PubMed] [Google Scholar]
- Kurosu H., Yamamoto M., Clark J. D., Pastor J. V., Nandi A., Gurnani P., et al. (2005). Suppression of aging in mice by the hormone Klotho. Science (80-.). 309, 1829–1833. 10.1126/science.1112766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyöstilä K., Syrjä P., Jagannathan V., Chandrasekar G., Jokinen T. S., Seppälä E. H., et al. (2015). A missense change in the ATG4D gene links aberrant autophagy to a neurodegenerative vacuolar storage disease. PLoS Genet. 11, 1–22. 10.1371/journal.pgen.1005169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., et al. (2006). Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127, 1109–1122. 10.1016/j.cell.2006.11.013 [DOI] [PubMed] [Google Scholar]
- Lamb R., Ozsvari B., Bonuccelli G., Smith D. L., Pestell R. G., Martinez-Outschoorn U. E., et al. (2015). Dissecting tumor metabolic heterogeneity: telomerase and large cell size metabolically define a sub-population of stem-like, mitochondrial-rich, cancer cells. Oncotarget 6, 21892–21905. 10.18632/oncotarget.5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landsberg G. M., Nichol J., Araujo J. A. (2012). Cognitive dysfunction syndrome. a disease of canine and feline brain aging. Vet. Clin. North Am. - Small Anim. Pract. 42, 749–768. 10.1016/j.cvsm.2012.04.003 [DOI] [PubMed] [Google Scholar]
- Larsen P. L., Albert P. S., Riddle D. L. (1995). Genes that regulate both development and longevity in Caenorhabditis elegans. Genetics 139, 1567–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson G., Karlsson E. K., Perri A., Webster M. T., Ho S. Y. W., Peters J., et al. (2012). Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc. Natl. Acad. Sci. 109, 8878–8883. 10.1073/pnas.1203005109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence J., Chang Y.-M. R., Szladovits B., Davison L. J., Garden O. A. (2013). Breed-specific hematological phenotypes in the dog: a natural resource for the genetic dissection of hematological parameters in a mammalian species. PLoS One 8, e81288. 10.1371/journal.pone.0081288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.-K., Klopp R. G., Weindruch R., Prolla T. A. (1999). Gene expression profile of aging and its retardation by caloric restriction. Science (80-.). 285, 1390–1393. 10.1126/science.285.5432.1390 [DOI] [PubMed] [Google Scholar]
- Lee C.-K., Weindruch R., Prolla T. A. (2000). Gene-expression profile of the ageing brain in mice. Nat. Genet. 25, 294–297. 10.1038/77046 [DOI] [PubMed] [Google Scholar]
- Lentino C., Visek A. J., McDonnell K., DiPietro L. (2012). Dog walking is associated with a favorable risk profile independent of a moderate to high volume of physical activity. J. Phys. Act. Heal. 9, 414–420. 10.1123/jpah.9.3.414 [DOI] [PubMed] [Google Scholar]
- Levin H. L., Moran J. V. (2011). Dynamic interactions between transposable elements and their hosts. Nat. Rev. Genet. 12, 615–627. 10.1038/nrg3030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B. (2005). Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell 120, 159–162. 10.1016/j.cell.2005.01.005 [DOI] [PubMed] [Google Scholar]
- Levine B., Kroemer G. (2008). Autophagy in the pathogenesis of disease. Cell 132, 27–42. 10.1016/j.cell.2007.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine B., Mizushima N., Virgin H. W. (2011). Autophagy in immunity and inflammation. Nature 469, 323–335. 10.1038/nature09782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R. W., Ganesan R., Houtchens K., Tolar L. A., Sheen F. (1993). Transposons in place of telomeric repeats at a Drosophila telomere. Cell 75, 1083–1093. 10.1016/0092-8674(93)90318-K [DOI] [PubMed] [Google Scholar]
- Li Y., Wang W.-J., Cao H., Lu J., Wu C., Hu F.-Y., et al. (2009). Genetic association of FOXO1A and FOXO3A with longevity trait in Han Chinese populations. Hum. Mol. Genet. 18, 4897–4904. 10.1093/hmg/ddp459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jiao Y., Fan E. K., Scott M. J., Li Y., Li S., et al. (2017). Aging-impaired filamentous actin polymerization signaling reduces alveolar macrophage phagocytosis of bacteria. J. Immunol. 199, 3176–3186. 10.4049/jimmunol.1700140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K., Dorman J. B., Rodan A., Kenyon C. (1997). daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278 (5341), 1319–1322. 10.1126/science.278.5341.1319 [DOI] [PubMed] [Google Scholar]
- Lin L., Faraco J., Li R., Kadotani H., Rogers W., Lin X., et al. (1999). The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell 98, 365–376. 10.1016/S0092-8674(00)81965-0 [DOI] [PubMed] [Google Scholar]
- Lin M. T., Beal M. F. (2006). Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443, 787–795. 10.1038/nature05292 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C. M., Mikkelsen T. S., Karlsson E. K., Jaffe D. B., Kamal M., et al. (2005). Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819. 10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Lindsey J., McGill N. I., Lindsey L. A., Green D. K., Cooke H. J. (1991). In vivo loss of telomeric repeats with age in humans. Mutat. Res. 256, 45–48. 10.1016/0921-8734(91)90032-7 [DOI] [PubMed] [Google Scholar]
- Lindvall O., Kokaia Z., Martinez-Serrano A. (2004). Stem cell therapy for human neurodegenerative disorders–how to make it work. Nat. Med. 10, S42–S50. 10.1038/nm1064 [DOI] [PubMed] [Google Scholar]
- Liu H.-Y., Pfleger C. M. (2013). Mutation in E1, the ubiquitin activating enzyme, reduces Drosophila lifespan and results in motor impairment. PLoS One 8, e32835. 10.1371/journal.pone.0032835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.-P., Baker J., Perkins A. S., Robertson E. J., Efstratiadis A. (1993). Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r). Cell 75, 59–72. 10.1016/S0092-8674(05)80084-4 [DOI] [PubMed] [Google Scholar]
- Liu Y., Elf S. E., Miyata Y., Sashida G., Liu Y., Huang G., et al. (2009. a). p53 regulates hematopoietic stem cell quiescence. Cell Stem Cell 4, 37–48. 10.1016/j.stem.2008.11.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Fiskum G., Schubert D. (2002). Generation of reactive oxygen species by the mitochondrial electron transport chain. J. Neurochem. 80, 780–787. 10.1046/j.0022-3042.2002.00744.x [DOI] [PubMed] [Google Scholar]
- Liu Y., Hettinger C. L., Zhang D., Rezvani K., Wang X., Wang H. (2014). The proteasome function reporter GFPu accumulates in young brains of the APPswe/PS1dE9 Alzheimer’s disease mouse model. Cell. Mol. Neurobiol. 34, 315–322. 10.1007/s10571-013-0022-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Sanoff H. K., Cho H., Burd C. E., Torrice C., Ibrahim J. G., et al. (2009. b). Expression of p16 INK4a in peripheral blood T-cells is a biomarker of human aging. Aging Cell 8, 439–448. 10.1111/j.1474-9726.2009.00489.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo F. S., Steinhauser M. L., Jay S. M., Gannon J., Pancoast J. R., Yalamanchi P., et al. (2013). Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell 153, 828–839. 10.1016/j.cell.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard D. B., Chua K. F., Mostoslavsky R., Franco S., Gostissa M., Alt F. W. (2005). DNA repair, genome stability, and aging. Cell 120, 497–512. 10.1016/j.cell.2005.01.028 [DOI] [PubMed] [Google Scholar]
- Longo V. D., Kennedy B. K. (2006). Sirtuins in aging and age-related disease. Cell 126, 257–268. 10.1016/j.cell.2006.07.002 [DOI] [PubMed] [Google Scholar]
- López-Otín C., Blasco M. A., Partridge L., Serrano M., Kroemer G. (2013). The hallmarks of aging. Cell 153, 1194–1217. 10.1016/j.cell.2013.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J., Shi J., Li M., Gui B., Fu R., Yao G., et al. (2015). Activation of AMPK by metformin inhibits TGF-β-induced collagen production in mouse renal fibroblasts. Life Sci. 127, 59–65. 10.1016/j.lfs.2015.01.042 [DOI] [PubMed] [Google Scholar]
- Lu T., Pan Y., Kao S.-Y., Li C., Kohane I., Chan J., et al. (2004). Gene regulation and DNA damage in the ageing human brain. Nature 429, 883–891. 10.1038/nature02661 [DOI] [PubMed] [Google Scholar]
- Lundberg J. O., Weitzberg E., Gladwin M. T. (2008). The nitrate–nitrite–nitric oxide pathway in physiology and therapeutics. Nat. Rev. Drug Discov. 7, 156–167. 10.1038/nrd2466 [DOI] [PubMed] [Google Scholar]
- Luo W., Dou F., Rodina A., Chip S., Kim J., Zhao Q., et al. (2007). Roles of heat-shock protein 90 in maintaining and facilitating the neurodegenerative phenotype in tauopathies. Proc. Natl. Acad. Sci. 104, 9511–9516. 10.1073/pnas.0701055104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Lu X., Xie H. (2014). Dynamic Alu methylation during normal development, aging, and tumorigenesis. Biomed Res. Int. 2014, 784706. 10.1155/2014/784706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutful Kabir F. M., Agarwal P., DeInnocentes P., Zaman J., Bird A. C., Bird R. C. (2013). Novel frameshift mutation in the p16/INK4A tumor suppressor gene in canine breast cancer alters expression from the p16/INK4A/p14ARF locus. J. Cell. Biochem. 114, 56–66. 10.1002/jcb.24300 [DOI] [PubMed] [Google Scholar]
- Maegawa S., Hinkal G., Kim H. S., Shen L., Zhang L., Zhang J., et al. (2010). Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20, 332–340. 10.1101/gr.096826.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahbub S., Brubaker A. L., Kovacs J. (2011). Aging of the innate immune system: an update. Curr. Immunol. Rev. 7, 104–115, E. 10.2174/157339511794474181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P., et al. (2007). FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 6, 458–471. 10.1016/j.cmet.2007.11.001 [DOI] [PubMed] [Google Scholar]
- Manczak M., Jung Y., Park B. S., Partovi D., Reddy P. H. (2005). Time-course of mitochondrial gene expressions in mice brains: implications for mitochondrial dysfunction, oxidative damage, and cytochrome c in aging. J. Neurochem. 92, 494–504. 10.1111/j.1471-4159.2004.02884.x [DOI] [PubMed] [Google Scholar]
- Mandal P. K., Kazazian H. H. (2008). SnapShot: vertebrate transposons. Cell 135, 192–192.e1. 10.1016/j.cell.2008.09.028 [DOI] [PubMed] [Google Scholar]
- Marchant T. W., Johnson E. J., McTeir L., Johnson C. I., Gow A., Liuti T., et al. (2017). Canine brachycephaly is associated with a retrotransposon-mediated missplicing of SMOC2. Curr. Biol. 27, 1573–1584.e6. 10.1016/j.cub.2017.04.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfe G., De Martino L., Tafani M., Irno-Consalvo M., Pasolini M. P., Navas L., et al. (2012). A multicancer-like syndrome in a dog characterized by p53 and cell cycle-checkpoint kinase 2 (CHK2) mutations and Sirtuin gene (SIRT1) down-regulation. Res. Vet. Sci. 93, 240–245. 10.1016/j.rvsc.2011.07.030 [DOI] [PubMed] [Google Scholar]
- Martin-Montalvo A., Mercken E. M., Mitchell S. J., Palacios H. H., Mote P. L., Scheibye-Knudsen M., et al. (2013). Metformin improves healthspan and lifespan in mice. Nat. Commun. 4, 2192. 10.1038/ncomms3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Rivera L., Herrera E., Albar J. P., Blasco M. A. (1998). Expression of mouse telomerase catalytic subunit in embryos and adult tissues. Proc. Natl. Acad. Sci. 95 (18), 10471–10476. 10.1073/pnas.95.18.10471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G. M. (2005). Genetic modulation of senescent phenotypes in Homo sapiens. Cell 120, 523–532. 10.1016/j.cell.2005.01.031 [DOI] [PubMed] [Google Scholar]
- Martindale J. L., Holbrook N. J. (2002). Cellular response to oxidative stress: signaling for suicide and survival. J. Cell. Physiol. 192, 1–15. 10.1002/jcp.10119 [DOI] [PubMed] [Google Scholar]
- Massey A. C., Zhang C., Cuervo A. M. (2006). Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 73, 205–235. 10.1016/S0070-2153(05)73007-6 [DOI] [PubMed] [Google Scholar]
- Massimino S., Kearns R. J., Loos K. M., Burr J., Park J. S., Chew B., et al. (2003). Effects of age and dietary β-carotene on immunological variables in dogs. J. Vet. Intern. Med. 17, 835–842. 10.1111/j.1939-1676.2003.tb02523.x [DOI] [PubMed] [Google Scholar]
- Masutomi K., Possemato R., Wong J. M. Y., Currier J. L., Tothova Z., Manola J. B., et al. (2005). The telomerase reverse transcriptase regulates chromatin state and DNA damage responses. Proc. Natl. Acad. Sci. U. S. A. 102, 8222–8227. 10.1073/pnas.0503095102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matheu A., Maraver A., Collado M., Garcia-Cao I., Cañamero M., Borras C., et al. (2009). Anti-aging activity of the Ink4/Arf locus. Aging Cell 8, 152–161. 10.1111/j.1474-9726.2009.00458.x [DOI] [PubMed] [Google Scholar]
- Matheu A., Maraver A., Klatt P., Flores I., Garcia-Cao I., Borras C., et al. (2007). Delayed ageing through damage protection by the Arf/p53 pathway. Nature 448, 375–379. 10.1038/nature05949 [DOI] [PubMed] [Google Scholar]
- Matheu A., Maraver A., Serrano M. (2008). The Arf/p53 pathway in cancer and aging. Cancer Res. 68, 6031–6034. 10.1158/0008-5472.CAN-07-6851 [DOI] [PubMed] [Google Scholar]
- Mathew L. M., Woode R. A., Axiak-Bechtel S. M., Amorim J. R., DeClue A. E. (2018). Resveratrol administration increases phagocytosis, decreases oxidative burst, and promotes pro-inflammatory cytokine production in healthy dogs. Vet. Immunol. Immunopathol. 203, 21–29. 10.1016/j.vetimm.2018.07.013 [DOI] [PubMed] [Google Scholar]
- Mattson M. P., Maudsley S., Martin B. (2004). A neural signaling triumvirate that influences ageing and age-related disease: insulin/IGF-1, BDNF and serotonin. Ageing Res. Rev. 3, 445–464. 10.1016/j.arr.2004.08.001 [DOI] [PubMed] [Google Scholar]
- Maxwell P. H., Burhans W. C., Curcio M. J. (2011). Retrotransposition is associated with genome instability during chronological aging. Proc. Natl. Acad. Sci. U. S. A. 108 (51), 20376–20381. 10.1073/pnas.1100271108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzatenta A., Carluccio A., Robbe D., Giulio C. Di (2017). The companion dog as a unique translational model for aging. Semin. Cell Dev. Biol. 70, 141–153. 10.1016/J.SEMCDB.2017.08.024 [DOI] [PubMed] [Google Scholar]
- Mech L. D. (2006). Estimated age structure of wolves in Northeastern Minnesota. J. Wildl. Manage. 70, 1481–1483. 10.2193/0022-541X(2006)70[1481:EASOWI]2.0.CO;2 [DOI] [Google Scholar]
- Mecocci P., MacGarvey U., Kaufman A. E., Koontz D., Shoffner J. M., Wallace D. C., et al. (1993). Oxidative damage to mitochondrial DNA shows marked age-dependent increases in human brain. Ann. Neurol. 34, 609–616. 10.1002/ana.410340416 [DOI] [PubMed] [Google Scholar]
- Meldolesi J. (2018). Exosomes and ectosomes in intercellular communication. Curr. Biol. 28, R435–R444. 10.1016/j.cub.2018.01.059 [DOI] [PubMed] [Google Scholar]
- Mendrysa S. M., O’Leary K. A., McElwee M. K., Michalowski J., Eisenman R. N., Powell D. A., et al. (2006). Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 20, 16–21. 10.1101/gad.1378506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merry T. L., Ristow M. (2016). Do antioxidant supplements interfere with skeletal muscle adaptation to exercise training? J. Physiol. 594, 5135–5147. 10.1113/JP270654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesquita A., Weinberger M., Silva A., Sampaio-Marques B., Almeida B., Leão C., et al. (2010). Caloric restriction or catalase inactivation extends yeast chronological lifespan by inducing H2O2 and superoxide dismutase activity. Proc. Natl. Acad. Sci. 107, 15123–15128. 10.1073/pnas.1004432107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurs K. M., Hendrix K. P., Norgard M. M. (2008). Molecular evaluation of five cardiac genes in Doberman Pinschers with dilated cardiomyopathy. Am. J. Vet. Res. 69, 1050–1053. 10.2460/ajvr.69.8.1050 [DOI] [PubMed] [Google Scholar]
- Meurs K. M., Lahmers S., Keene B. W., White S. N., Oyama M. A., Mauceli E., et al. (2012). A splice site mutation in a gene encoding for PDK4, a mitochondrial protein, is associated with the development of dilated cardiomyopathy in the Doberman pinscher. Hum. Genet. 131, 1319–1325. 10.1007/s00439-012-1158-2 [DOI] [PubMed] [Google Scholar]
- Michell A. R. (1999). Longevity of British breeds of dog and its relationships with sex, size, cardiovascular variables and disease. Vet. Rec. 145, 625–629. 10.1136/vr.145.22.625 [DOI] [PubMed] [Google Scholar]
- Miklósi Á. (2014). Dog behaviour, evolution, and cognition. (Oxford: Oxford University Press; ). 10.1093/acprof:oso/9780199646661.001.0001 [DOI] [Google Scholar]
- Miklósi Á., Kubinyi E. (2016). Current trends in canine problem-solving and cognition. Curr. Dir. Psychol. Sci. 25, 300–306. 10.1177/0963721416666061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milgram N. W., Head E., Muggenburg B., Holowachuk D., Murphey H., Estrada J., et al. (2002. a). Landmark discrimination learning in the dog: effects of age, an antioxidant fortified food, and cognitive strategy. Neurosci. Biobehav. Rev. 26, 679–695. 10.1016/S0149-7634(02)00039-8 [DOI] [PubMed] [Google Scholar]
- Milgram N. W., Zicker S. C., Head E., Muggenburg B. A., Murphey H., Ikeda-Douglas C. J., et al. (2002. b). Dietary enrichment counteracts age-associated cognitive dysfunction in canines. Neurobiol. Aging 23, 737–745. 10.1016/S0197-4580(02)00020-9 [DOI] [PubMed] [Google Scholar]
- Min J.-N., Whaley R. A., Sharpless N. E., Lockyer P., Portbury A. L., Patterson C. (2008). CHIP deficiency decreases longevity, with accelerated aging phenotypes accompanied by altered protein quality control. Mol. Cell. Biol. 28, 4018–4025. 10.1128/MCB.00296-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misic A. M., Davis M. F., Tyldsley A. S., Hodkinson B. P., Tolomeo P., Hu B., et al. (2015). The shared microbiota of humans and companion animals as evaluated from Staphylococcus carriage sites. Microbiome 3, 2. 10.1186/s40168-014-0052-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno H., Nakamura A., Aoki Y., Ito N., Kishi S., Yamamoto K., et al. (2011). Identification of muscle-specific MicroRNAs in serum of muscular dystrophy animal models: promising novel blood-based markers for muscular dystrophy. PLoS One 6, e18388. 10.1371/journal.pone.0018388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Levine B., Cuervo A. M., Klionsky D. J. (2008). Autophagy fights disease through cellular self-digestion. Nature 451, 1069–1075. 10.1038/nature06639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney A. M., Griffin R. J., Timmons S., O’Connor R., Ravid R., O’Neill C. (2010). Defects in IGF-1 receptor, insulin receptor and IRS-1/2 in Alzheimer’s disease indicate possible resistance to IGF-1 and insulin signalling. Neurobiol. Aging 31, 224–243. 10.1016/j.neurobiolaging.2008.04.002 [DOI] [PubMed] [Google Scholar]
- Morales M. E., Servant G., Ade C., Roy-Engel A. M. (2015). Altering genomic integrity: heavy metal exposure promotes transposable element-mediated damage. Biol. Trace Elem. Res. 166, 24–33. 10.1007/s12011-015-0298-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimoto R. I. (2008). Proteotoxic stress and inducible chaperone networks in neurodegenerative disease and aging. Genes Dev. 22, 1427–1438. 10.1101/gad.1657108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G., Samson M., Michaud S., Tanguay R. M. (2004). Overexpression of the small mitochondrial Hsp22 extends Drosophila life span and increases resistance to oxidative stress. FASEB J. 18, 598–599. 10.1096/fj.03-0860fje [DOI] [PubMed] [Google Scholar]
- Mortiboys H., Johansen K. K., Aasly J. O., Bandmann O. (2010). Mitochondrial impairment in patients with Parkinson disease with the G2019S mutation in LRRK2. Neurology 75, 2017–2020. 10.1212/WNL.0b013e3181ff9685 [DOI] [PubMed] [Google Scholar]
- Moskalev A. A., Shaposhnikov M. V., Plyusnina E. N., Zhavoronkov A., Budovsky A., Yanai H., et al. (2013). The role of DNA damage and repair in aging through the prism of Koch-like criteria. Ageing Res. Rev. 12, 661–684. 10.1016/j.arr.2012.02.001 [DOI] [PubMed] [Google Scholar]
- Murphy M. P. (2009). How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13. 10.1042/BJ20081386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray V. (1990). Are transposons a cause of ageing? Mutat. Res. 237, 59–63. 10.1016/0921-8734(90)90011-F [DOI] [PubMed] [Google Scholar]
- Nasir L., Devlin P., Mckevitt T., Rutteman G., Argyle D. J. (2001). Telomere lengths and telomerase activity in dog tissues: a potential model system to study human telomere and telomerase biology. Neoplasia 3, 351–359. 10.1038/sj.neo.7900173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A., Boveris A. (2004). Rat brain and liver mitochondria develop oxidative stress and lose enzymatic activities on aging. Am. J. Physiol. Integr. Comp. Physiol. 287, R1244–R1249. 10.1152/ajpregu.00226.2004 [DOI] [PubMed] [Google Scholar]
- Nebel A., Kleindorp R., Caliebe A., Nothnagel M., Blanché H., Junge O., et al. (2011). A genome-wide association study confirms APOE as the major gene influencing survival in long-lived individuals. Mech. Ageing Dev. 132, 324–330. 10.1016/j.mad.2011.06.008 [DOI] [PubMed] [Google Scholar]
- Neilson J. C., Hart B. L., Cliff K. D., Ruehl W. W. (2001). Prevalence of behavioral changes associated with age-related cognitive impairment in dogs. J. Am. Vet. Med. Assoc. 218, 1787–1791. 10.2460/javma.2001.218.1787 [DOI] [PubMed] [Google Scholar]
- Nelson G., Wordsworth J., Wang C., Jurk D., Lawless C., Martin-Ruiz C., et al. (2012). A senescent cell bystander effect: senescence-induced senescence. Aging Cell 11, 345–349. 10.1111/j.1474-9726.2012.00795.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nixon R. A., Wegiel J., Kumar A., Yu W. H., Peterhoff C., Cataldo A., et al. (2005). Extensive involvement of autophagy in Alzheimer disease: an immuno-electron microscopy study. J. Neuropathol. Exp. Neurol. 64, 113–122. 10.1093/jnen/64.2.113 [DOI] [PubMed] [Google Scholar]
- Nixon R. A., Yang D.-S. (2011). Autophagy failure in Alzheimer’s disease—locating the primary defect. Neurobiol. Dis. 43, 38–45. 10.1016/j.nbd.2011.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill D. G., Church D. B., Mcgreevy P. D., Thomson P. C., Brodbelt D. C. (2013). Longevity and mortality of owned dogs in England. Vet. J. 198, 638–643. 10.1016/j.tvjl.2013.09.020 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Uchida K., Yamato O., Mizukami K., Chambers J. K., Nakayama H. (2015). Expression of autophagy-related proteins in the spinal cord of Pembroke Welsh Corgi dogs with canine degenerative myelopathy. Vet. Pathol. 52, 1099–1107. 10.1177/0300985815570070 [DOI] [PubMed] [Google Scholar]
- Orrell R. W. (2000). Amyotrophic lateral sclerosis: copper/zinc superoxide dismutase (SOD1) gene mutations. Neuromuscul. Disord. 10, 63–68. 10.1016/S0960-8966(99)00071-1 [DOI] [PubMed] [Google Scholar]
- Ostan R., Borelli V., Castellani G. (2017). Inflammaging and human longevity in the omics era. Mech. Ageing Dev. 165, 129–138. 10.1016/j.mad.2016.12.008 [DOI] [PubMed] [Google Scholar]
- Ouyang L., Shi Z., Zhao S., Wang F.-T., Zhou T.-T., Liu B., et al. (2012). Programmed cell death pathways in cancer: a review of apoptosis, autophagy and programmed necrosis. Cell Prolif. 45, 487–498. 10.1111/j.1365-2184.2012.00845.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace J. K., Feschotte C. (2007). The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 17, 422–432. 10.1101/gr.5826307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagano T. B., Wojcik S., Costagliola A., De Biase D., Iovino S., Iovane V., et al. (2015). Age related skeletal muscle atrophy and upregulation of autophagy in dogs. Vet. J. 206, 54–60. 10.1016/j.tvjl.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Pal S., Tyler J. K. (2016). Epigenetics and aging. Sci. Adv. 2, 1–20. 10.1126/sciadv.1600584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papaioannou N., Tooten P. C. J., van Ederen A. M., Bohl J. R. E., Rofina J., Tsangaris T., et al. (2001). Immunohistochemical investigation of the brain of aged dogs. I. Detection of neurofibrillary tangles and of 4-hydroxynonenal protein, an oxidative damage product, in senile plaques. Amyloid 8, 11–21. 10.3109/13506120108993810 [DOI] [PubMed] [Google Scholar]
- Parker H. G., VonHoldt B. M., Quignon P., Margulies E. H., Shao S., Mosher D. S., et al. (2009). An expressed fgf4 retrogene is associated with breed-defining Chondrodysplasia in domestic dogs. Science (80-). 325, 995–998. 10.1126/SCIENCE.1173275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker H. G., Dreger D. L., Rimbault M., Davis B. W., Mullen A. B., Carpintero-Ramirez G., Ostrander E. A. (2017). Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Reports 19 (4), 697–708. 10.1016/j.celrep.2017.03.079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel P. J., Singh S. K., Panaich S., Cardozo L. (2014). The aging gut and the role of prebiotics, probiotics, and synbiotics: A review. J. Clin. Gerontol. Geriatr. 5, 3–6. 10.1016/j.jcgg.2013.08.003 [DOI] [Google Scholar]
- Patel K. V. (2008). Epidemiology of anemia in older adults. Semin. Hematol. 45, 210–217. 10.1053/j.seminhematol.2008.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawelec G. (2018). Age and immunity: what is “immunosenescence”? Exp. Gerontol. 105, 4–9. 10.1016/j.exger.2017.10.024 [DOI] [PubMed] [Google Scholar]
- Pelatti M. V., Gomes J. P. A., Vieira N. M. S., Cangussu E., Landini V., Andrade T., et al. (2016). Transplantation of human adipose mesenchymal stem cells in non-immunosuppressed GRMD dogs is a safe procedure. Stem Cell Rev. Reports 12, 448–453. 10.1007/s12015-016-9659-3 [DOI] [PubMed] [Google Scholar]
- Penso-Dolfin L., Swofford R., Johnson J., Alföldi J., Lindblad-Toh K., Swarbreck D., et al. (2016). An improved microRNA annotation of the canine genome. PLoS One 11, e0153453. 10.1371/journal.pone.0153453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry G. H., Dominy N. J. (2009). Evolution of the human pygmy phenotype. Trends Ecol. Evol. 24, 218–225. 10.1016/j.tree.2008.11.008 [DOI] [PubMed] [Google Scholar]
- Petrosillo G., Matera M., Casanova G., Ruggiero F., Paradies G. (2008). Mitochondrial dysfunction in rat brain with aging. Involvement of complex I, reactive oxygen species and cardiolipin. Neurochem. Int. 53, 126–131. 10.1016/j.neuint.2008.07.001 [DOI] [PubMed] [Google Scholar]
- Pfahler S., Distl O. (2012). Identification of quantitative trait loci (QTL) for canine hip dysplasia and canine elbow dysplasia in Bernese Mountain dogs. PLoS One 7, e49782. 10.1371/journal.pone.0049782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pievani M., Filippini N., van den Heuvel M. P., Cappa S. F., Frisoni G. B. (2014). Brain connectivity in neurodegenerative diseases—from phenotype to proteinopathy. Nat. Rev. Neurol. 10, 620–633. 10.1038/nrneurol.2014.178 [DOI] [PubMed] [Google Scholar]
- Pizarro J. G., Cristofari G. (2016). Post-transcriptional control of LINE-1 retrotransposition by cellular host factors in somatic cells. Front. Cell Dev. Biol. 4, 14. 10.3389/fcell.2016.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassais J., Rimbault M., Williams F. J., Davis B. W., Schoenebeck J. J., Ostrander E. A. (2017). Analysis of large versus small dogs reveals three genes on the canine X chromosome associated with body weight, muscling and back fat thickness. PLOS Genet. 13, e1006661. 10.1371/journal.pgen.1006661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowden J., Renshaw-Hoelscher M., Engleman C., Katz J., Sambhara S. (2004). Innate immunity in aging: impact on macrophage function. Aging Cell 3, 161–167. 10.1111/j.1474-9728.2004.00102.x [DOI] [PubMed] [Google Scholar]
- Pritchard A. B., Crean S., Olsen I., Singhrao S. K. (2017). Periodontitis, microbiomes and their role in Alzheimer’s disease. Front. Aging Neurosci. 9, 336. 10.3389/fnagi.2017.00336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proschowsky H. F., Rugbjerg H., Ersbøll A. K. (2003). Mortality of purebred and mixed-breed dogs in Denmark. Prev. Vet. Med. 58, 63–74. 10.1016/S0167-5877(03)00010-2 [DOI] [PubMed] [Google Scholar]
- Prowse K. R., Greidert C. W. (1995). Developmental and tissue-specific regulation of mouse telomerase and telomere length. Cell Biol. 92, 4818–4822. 10.1073/pnas.92.11.4818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puzzo D., Gulisano W., Palmeri A., Arancio O. (2015). Rodent models for Alzheimer’s disease drug discovery. Expert Opin. Drug Discov. 10, 703–711. 10.1517/17460441.2015.1041913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyo J.-O., Yoo S.-M., Ahn H.-H., Nah J., Hong S.-H., Kam T.-I., et al. (2013). Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat. Commun. 4, 2300. 10.1038/ncomms3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz J. D., White E. (2010). Autophagy and metabolism. Science 330, 1344–1348. 10.1126/science.1193497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radakovich L. B., Pannone S. C., Truelove M. P., Olver C. S., Santangelo K. S. (2017). Hematology and biochemistry of aging-evidence of “anemia of the elderly” in old dogs. Vet. Clin. Pathol. 46, 34–45. 10.1111/vcp.12459 [DOI] [PubMed] [Google Scholar]
- Raposo-Ferreira T. M. M., Bueno R. C., Terra E. M., Avante M. L., Tinucci-Costa M., Carvalho M., et al. (2016). Downregulation of ATM gene and protein expression in canine mammary tumors. Vet. Pathol. 53, 1154–1159. 10.1177/0300985816643367 [DOI] [PubMed] [Google Scholar]
- Reis A. B., Carneiro C. M., Carvalho M., das G., Teixeira-Carvalho A., Giunchetti R. C., et al. et al. (2005). Establishment of a microplate assay for flow cytometric assessment and it is use for the evaluation of age-related phenotypic changes in canine whole blood leukocytes. Vet. Immunol. Immunopathol. 103, 173–185. 10.1016/j.vetimm.2004.08.014 [DOI] [PubMed] [Google Scholar]
- Rera M., Bahadorani S., Cho J., Koehler C. L., Ulgherait M., Hur J. H., et al. (2011). Modulation of longevity and tissue homeostasis by the Drosophila PGC-1 homolog. Cell Metab. 14, 623–634. 10.1016/j.cmet.2011.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler S., Bartkova J., Niederegger H., Bartek J., Scharffetter-Kochanek K., Jansen-Durr P., et al. (2006). p16 INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell 5, 379–389. 10.1111/j.1474-9726.2006.00231.x [DOI] [PubMed] [Google Scholar]
- Reul J. M. H. M., Rothuizen J., de Kloet E. R. (1991). Age-related changes in the dog hypothalamic–pituitary–adrenocortical system: neuroendocrine activity and corticosteroid receptors. J. Steroid Biochem. Mol. Biol. 40, 63–69. 10.1016/0960-0760(91)90168-5 [DOI] [PubMed] [Google Scholar]
- Riley P. A. (1994). Free radicals in biology: oxidative stress and the effects of ionizing radiation. Int. J. Radiat. Biol. 65, 27–33. 10.1080/09553009414550041 [DOI] [PubMed] [Google Scholar]
- Rimbault M., Beale H. C., Schoenebeck J. J., Hoopes B. C., Allen J. J., Kilroy-Glynn P., et al. (2013). Derived variants at six genes explain nearly half of size reduction in dog breeds. Genome Res. 23, 1985–1995. 10.1101/GR.157339.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ristow M., Zarse K., Oberbach A., Klöting N., Birringer M., Kiehntopf M., et al. (2009). Antioxidants prevent health-promoting effects of physical exercise in humans. Proc. Natl. Acad. Sci. U. S. A. 106, 8665–8670. 10.1073/pnas.0903485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera P., Melin M., Biagi T., Fall T., Häggström J., Lindblad-Toh K., et al. (2009). Mammary tumor development in dogs is associated with BRCA1 and BRCA2. Cancer Res. 69, 8770–8774. 10.1158/0008-5472.CAN-09-1725 [DOI] [PubMed] [Google Scholar]
- Rodier F., Campisi J. (2011). Four faces of cellular senescence. J. Cell Biol. 192, 547–556. 10.1083/jcb.201009094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rofina J. E., Singh K., Skoumalova-Vesela A., van Ederen A. M., van Asten A. J., Wilhelm J., et al. (2004). Histochemical accumulation of oxidative damage products is associated with Alzheimer-like pathology in the canine. Amyloid 11, 90–100. 10.1080/13506120412331285779 [DOI] [PubMed] [Google Scholar]
- Rogina B., Helfand S. L. (2004). Sir2 mediates longevity in the fly through a pathway related to calorie restriction. PNAS 101, 15998–16003. 10.1073/pnas.0404184101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanucci M., Della Salda L. (2015). Oxidative stress and protein quality control systems in the aged canine brain as a model for human neurodegenerative disorders. Oxid. Med. Cell. Longev. 2015, 1–8. 10.1155/2015/940131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothuizen J., Reul J. M., Rijnberk A., Mol J. A., de Kloet E. R. (1991). Aging and the hypothalamus–pituitary–adrenocortical axis, with special reference to the dog. Acta Endocrinol. (Copenh) 125 Suppl 1, 73–76. [PubMed] [Google Scholar]
- Roudebush P., Zicker S. C., Cotman C. W., Milgram N. W., Muggenburg B. A., Head E. (2005). Nutritional management of brain aging in dogs. J. Am. Vet. Med. Assoc. 227, 722–728. 10.2460/javma.2005.227.722 [DOI] [PubMed] [Google Scholar]
- Rowe J., Kahn R. (1987). Human aging: usual and successful. Science (80-.). 237, 143–149. 10.1126/science.3299702 [DOI] [PubMed] [Google Scholar]
- Rowe J. W., Kahn R. L. (2015). Successful aging 2.0: conceptual expansions for the 21st century. Journals Gerontol. Ser. B Psychol. Sci. Soc. Sci. 70, 593–596. 10.1093/geronb/gbv025 [DOI] [PubMed] [Google Scholar]
- Rowell J. L., McCarthy D. O., Alvarez C. E. (2011). Dog models of naturally occurring cancer. Trends Mol. Med. 17, 380–388. 10.1016/j.molmed.2011.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbo H., Radi R., Trujillo M., Telleri R., Kalyanaraman B., Barnes S., et al. (1994). Nitric oxide regulation of superoxide and peroxynitrite-dependent lipid peroxidation. Formation of novel nitrogen-containing oxidized lipid derivatives*. J. Biol. Chem. 269, 26066–26075. [PubMed] [Google Scholar]
- Rubinsztein D. C. (2006). The roles of intracellular protein-degradation pathways in neurodegeneration. Nature 443, 780–786. 10.1038/nature05291 [DOI] [PubMed] [Google Scholar]
- Ruvkun G., Ogg S., Paradis S., Gottlieb S., Patterson G. I., Lee L., et al. (1997). The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature 389, 994–999. 10.1038/40194 [DOI] [PubMed] [Google Scholar]
- Salt C., Morris P. J., Wilson D., Lund E. M., German A. J. (2018). Association between life span and body condition in neutered client-owned dogs. J. Vet. Intern. Med. 33, jvim.15367. 10.1111/jvim.15367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson T. R., Debelius J. W., Thron T., Janssen S., Shastri G. G., Ilhan Z. E., et al. (2016). Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell 167, 1469–1480.e12. 10.1016/j.cell.2016.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A. M., Csibi A., Raibon A., Cornille K., Gay S., Bernardi H., et al. (2012). AMPK promotes skeletal muscle autophagy through activation of forkhead FoxO3a and interaction with Ulk1. J. Cell Biochem. 113, 695–710. 10.1002/jcb.23399 [DOI] [PubMed] [Google Scholar]
- Sandri M. (2010). Autophagy in skeletal muscle. FEBS Lett. 584, 1411–1416. 10.1016/j.febslet.2010.01.056 [DOI] [PubMed] [Google Scholar]
- Sarasa L., Gallego C., Monleón I., Olvera A., Canudas J., Montañés M., et al. (2010). Cloning, sequencing and expression in the dog of the main amyloid precursor protein isoforms and some of the enzymes related with their processing. Neuroscience 171, 1091–1101. 10.1016/j.neuroscience.2010.09.042 [DOI] [PubMed] [Google Scholar]
- Saraswati S., Sitaraman R. (2015). Aging and the human gut microbiota—from correlation to causality. Front. Microbiol. 5, 764. 10.3389/fmicb.2014.00764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S. (2011). Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J. Neuropathol. Exp. Neurol. 70, 349–359. 10.1097/NEN.0b013e3182160690 [DOI] [PubMed] [Google Scholar]
- Schieber M., Chandel N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. 10.1016/j.cub.2014.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt F., Boltze J., Jäger C., Hofmann S., Willems N., Seeger J., et al. (2015). Detection and quantification of β-amyloid, pyroglutamyl Aβ, and tau in aged canines. J. Neuropathol. Exp. Neurol. 74, 912–923. 10.1097/NEN.0000000000000230 [DOI] [PubMed] [Google Scholar]
- Schneider J. L., Villarroya J., Diaz-Carretero A., Patel B., Urbanska A. M., Thi M. M., et al. (2015). Loss of hepatic chaperone-mediated autophagy accelerates proteostasis failure in aging. Aging Cell 14, 249–264. 10.1111/acel.12310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoenebeck J. J., Ostrander E. A. (2014). Insights into morphology and disease from the dog genome project. Annu. Rev. Cell Dev. Biol. 30, 535–560. 10.1146/annurev-cellbio-100913-012927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T. J., Zarse K., Voigt A., Urban N., Birringer M., Ristow M. (2007). Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 6, 280–293. 10.1016/j.cmet.2007.08.011 [DOI] [PubMed] [Google Scholar]
- Schumacher B., van der Pluijm I., Moorhouse M. J., Kosteas T., Robinson A. R., Suh Y., et al. (2008). Delayed and accelerated aging share common longevity assurance mechanisms. PLoS Genet. 4, e1000161. 10.1371/journal.pgen.1000161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P., Perls T. T. (2012). The genetics of extreme longevity : lessons from the New England Centenarian study. Front. Genet. 3, 1–7. 10.3389/fgene.2012.00277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastiani P., Solovieff N., DeWan A. T., Walsh K. M., Puca A., Hartley S. W., et al. (2012). Genetic signatures of exceptional longevity in humans. PLoS One 7, e29848. 10.1371/journal.pone.0029848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seisenberger S., Andrews S., Krueger F., Arand J., Walter J., Santos F., et al. (2012). The Dynamics of genome-wide DNA methylation reprogramming in mouse primordial germ cells. Mol. Cell 48, 849–862. 10.1016/j.molcel.2012.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel G. S., Horvath T. L. (2015). Mitochondrial ROS signaling in organismal homeostasis. Cell 163, 560–569. 10.1016/j.cell.2015.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shampay J., Blackburn E. H. (1988). Generation of telomere-length heterogeneity in Saccharomyces cerevisiae. Genetics 85, 534–538. 10.1073/pnas.85.2.534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharon G., Sampson T. R., Geschwind D. H., Mazmanian S. K. (2016). The central nervous system and the gut microbiome. Cell 167, 915–932. 10.1016/j.cell.2016.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearin A. L., Hedan B., Cadieu E., Erich S. A., Schmidt E. V., Faden D. L., et al. (2012). The MTAP-CDKN2A locus confers susceptibility to a naturally occurring canine cancer. Cancer Epidemiol. Biomarkers Prev. 21, 1019–1027. 10.1158/1055-9965.EPI-12-0190-T [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherr C. J., DePinho R. A. (2000). Cellular senescence: mitotic clock or culture shock? Cell 102, 407–410. 10.1016/S0092-8674(00)00046-5 [DOI] [PubMed] [Google Scholar]
- Simonsen A., Cumming R. C., Brech A., Isakson P., Schubert D. R., Finley K. D. (2008). Promoting basal levels of autophagy in the nervous system enhances longevity and oxidant resistance in adult Drosophila. Autophagy 4, 176–184. 10.4161/auto.5269 [DOI] [PubMed] [Google Scholar]
- Singhal R. P., Mays-Hoopes L. L., Eichhorn G. L. (1987). DNA methylation in aging of mice. Mech. Ageing Dev. 41, 199–210. 10.1016/0047-6374(87)90040-6 [DOI] [PubMed] [Google Scholar]
- Skoglund P., Ersmark E., Palkopoulou E., Dalén L. (2015). Ancient wolf genome reveals an early divergence of domestic dog ancestors and admixture into high-latitude breeds. Curr. Biol. 25 (11), 1515–1519. 10.1016/j.cub.2015.04.019 [DOI] [PubMed] [Google Scholar]
- Skoumalova A., Rofina J., Schwippelova Z., Gruys E., Wilhelm J. (2003). The role of free radicals in canine counterpart of senile dementia of the Alzheimer type. Exp. Gerontol. 38, 711–719. 10.1016/S0531-5565(03)00071-8 [DOI] [PubMed] [Google Scholar]
- Smith B. F., Yue Y., Woods P. R., Kornegay J. N., Shin J.-H., Williams R. R., et al. (2011). An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab. Investig. 91, 216–231. 10.1038/labinvest.2010.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith L. K., He Y., Park J.-S., Bieri G., Snethlage C. E., Lin K., et al. (2015). β2-microglobulin is a systemic pro-aging factor that impairs cognitive function and neurogenesis. Nat. Med. 21, 932–937. 10.1038/nm.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Z. D., Chan M. M., Mikkelsen T. S., Gu H., Gnirke A., Regev A., et al. (2012). A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 484, 339–344. 10.1038/nature10960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., De Lange T. (2002). Different telomere damage signaling pathways in human and mouse cells. EMBO J. 21, 4338–4348. 10.1093/emboj/cdf433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolek T., Madari A., Farbakova J., Kandrac O., Jadhav S., Cente M., et al. (2016). Tau hyperphosphorylation in synaptosomes and neuroinflammation are associated with canine cognitive impairment. J. Comp. Neurol. 524, 874–895. 10.1002/cne.23877 [DOI] [PubMed] [Google Scholar]
- Snigdha S., de Rivera C., Milgram N. W., Cotman C. W. (2016). Effect of mitochondrial cofactors and antioxidants supplementation on cognition in the aged canine. Neurobiol. Aging 37, 171–178. 10.1016/j.neurobiolaging.2015.09.015 [DOI] [PubMed] [Google Scholar]
- Soerensen M., Dato S., Christensen K., McGue M., Stevnsner T., Bohr V. A., et al. (2010). Replication of an association of variation in the FOXO3A gene with human longevity using both case-control and longitudinal data. Aging Cell 9, 1010–1017. 10.1111/j.1474-9726.2010.00627.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M., Dato S., Tan Q., Thinggaard M., Kleindorp R., Beekman M., et al. (2012. a). Human longevity and variation in GH/IGF-1/insulin signaling, DNA damage signaling and repair and pro/antioxidant pathway genes: Cross sectional and longitudinal studies. Exp. Gerontol. 47, 379–387. 10.1016/j.exger.2012.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soerensen M., Thinggaard M., Nygaard M., Dato S., Tan Q., Hjelmborg J., et al. (2012. b). Genetic variation in TERT and TERC and human leukocyte telomere length and longevity: a cross-sectional and longitudinal analysis. Aging Cell 11, 223–227. 10.1111/j.1474-9726.2011.00775.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somel M., Guo S., Fu N., Yan Z., Hu H. Y., Xu Y., et al. (2010). MicroRNA, mRNA, and protein expression link development and aging in human and macaque brain. Genome Res. 20, 1207–1218. 10.1101/gr.106849.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag W. E., Lynch C. D., Cooney P. T., Hutchins P. M. (1997). Decreases in cerebral microvasculature with age are associated with the decline in growth hormone and insulin-like growth factor 1*. Endocrinology 138, 3515–3520. 10.1210/endo.138.8.5330 [DOI] [PubMed] [Google Scholar]
- Sonntag W. E., Lynch C., Thornton P., Khan A., Bennet S., Ingram R. (2000). The effects of growth hormone and IGF-1 deficiency on cerebrovascular and brain ageing. J. Anat. 197, 575–585. 10.1017/S002187829900713X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonntag W. E., Ramsey M., Carter C. S. (2005). Growth hormone and insulin-like growth factor-1 (IGF-1) and their influence on cognitive aging. Ageing Res. Rev. 4, 195–212. 10.1016/j.arr.2005.02.001 [DOI] [PubMed] [Google Scholar]
- Stadtman E. R., Levine R. L. (2006). Protein Oxidation. Ann. N. Y. Acad. Sci. 899, 191–208. 10.1111/j.1749-6632.2000.tb06187.x [DOI] [PubMed] [Google Scholar]
- Stewart L., MacLean E. L., Ivy D., Woods V., Cohen E., Rodriguez K., et al. (2015). Citizen science as a new tool in dog cognition research. PLoS One 10, e0135176. 10.1371/journal.pone.0135176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone R. C., Horvath K., Kark J. D., Susser E., Tishkoff S. A., Aviv A. (2016). Telomere length and the cancer–atherosclerosis trade-off. PLOS Genet. 12, e1006144. 10.1371/journal.pgen.1006144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser A., Niedermüller H., Hofecker G., Laber G. (1993). The effect of aging on laboratory values in dogs. J. Vet. Med. Ser. A 40, 720–730. 10.1111/j.1439-0442.1993.tb00689.x [DOI] [PubMed] [Google Scholar]
- Stribinskis V., Ramos K. S. (2006). Activation of human long interspersed nuclear element 1 retrotransposition by benzo(a)pyrene, an ubiquitous environmental carcinogen. Cancer Res. 66, 2616–2620. 10.1158/0008-5472.CAN-05-3478 [DOI] [PubMed] [Google Scholar]
- Studzinski C. M., MacKay W. A., Beckett T. L., Henderson S. T., Murphy M. P., Sullivan P. G., et al. (2008). Induction of ketosis may improve mitochondrial function and decrease steady-state amyloid-β precursor protein (APP) levels in the aged dog. Brain Res. 1226, 209–217. 10.1016/j.brainres.2008.06.005 [DOI] [PubMed] [Google Scholar]
- Sturm Á., Ivics Z., Vellai T. (2015). The mechanism of ageing: primary role of transposable elements in genome disintegration. Cell. Mol. Life Sci. 72, 1839–1847. 10.1007/s00018-015-1896-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su M.-Y., Tapp P. D., Vu L., Chen Y.-F., Chu Y., Muggenburg B., et al. (2005). A longitudinal study of brain morphometrics using serial magnetic resonance imaging analysis in a canine model of aging. Prog. Neuropsychopharmacol. Biol. Psychiatry 29, 389–397. 10.1016/j.pnpbp.2004.12.005 [DOI] [PubMed] [Google Scholar]
- Suh Y., Atzmon G., Cho M.-O., Hwang D., Liu B., Leahy D. J., et al. (2008). Functionally significant insulin-like growth factor I receptor mutations in centenarians. Proc. Natl. Acad. Sci. U. S. A. 105, 3438–3442. 10.1073/pnas.0705467105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., et al. (2014). Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24, 1963–1976. 10.1101/gr.168872.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N. B., Bustamante C. D., Chase K., Gray M. M., Zhao K., Zhu L., et al. (2007). A single IGF1 allele is a major determinant of small size in dogs. Science 316, 112–115. 10.1126/science.1137045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson K. S., Vester B. M., Apanavicius C. J., Kirby N. A., Schook L. B. (2009). Implications of age and diet on canine cerebral cortex transcription. Neurobiol. Aging 30, 1314–1326. 10.1016/j.neurobiolaging.2007.10.017 [DOI] [PubMed] [Google Scholar]
- Swindell W. R., Masternak M. M., Kopchick J. J., Conover C. A., Bartke A., Miller R. A. (2009). Endocrine regulation of heat shock protein mRNA levels in long-lived dwarf mice. Mech. Ageing Dev. 130, 393–400. 10.1016/j.mad.2009.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabó D., Gee N. R., Miklósi Á. (2016). Natural or pathologic? Discrepancies in the study of behavioral and cognitive signs in aging family dogs. J. Vet. Behav. 11, 86–98. 10.1016/j.jveb.2015.08.003 [DOI] [Google Scholar]
- Tatar M., Kopelman A., Epstein D., Tu M.-P., Yin C.-M., Garofalo R. S. (2001). A mutant Drosophila insulin receptor homolog that extends life-span and impairs neuroendocrine function. Science 292, 107–110. 10.1126/science.1057987 [DOI] [PubMed] [Google Scholar]
- Thalmann O., Shapiro B., Cui P., Schuenemann V. J., Sawyer S. K., Greenfield D. L., et al. (2013). Complete mitochondrial genomes of ancient canids suggest a European origin of domestic dogs. Science (80-.). 342, 871–874. 10.1126/science.1243650 [DOI] [PubMed] [Google Scholar]
- Thamm D. H., Grunerud K. K., Rose B. J., Vail D. M., Bailey S. M. (2013). DNA repair deficiency as a susceptibility marker for spontaneous lymphoma in Golden Retriever dogs: a case-control study. PLoS One 8, 1–8. 10.1371/journal.pone.0069192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevelein J. M., de Winde J. H. (1999). Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol. Microbiol. 33, 904–918. 10.1046/j.1365-2958.1999.01538.x [DOI] [PubMed] [Google Scholar]
- Thind A., Wilson C. (2016). Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 5, 31292. 10.3402/jev.v5.31292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. J., Holdt B., Horvath S., Pellegrini M. (2017). An epigenetic aging clock for dogs and wolves. Aging (Albany NY) 9, 1055–1068. 10.18632/aging.101211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorpe R. J., Simonsick E. M., Brach J. S., Ayonayon H., Satterfield S., Harris T. B., et al. (2006). Dog ownership, walking behavior, and maintained mobility in late life. J. Am. Geriatr. Soc. 54, 1419–1424. 10.1111/j.1532-5415.2006.00856.x [DOI] [PubMed] [Google Scholar]
- Tiret L., Kessler J., Pele M., Tiret L., Kessler J., Blot S., et al. (2005). SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum. Mol. Genet. 14, 1417–1427. 10.1093/hmg/ddi151 [DOI] [PubMed] [Google Scholar]
- Tissenbaum H. A., Guarente L. (2001). Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410, 227–230. 10.1038/35065638 [DOI] [PubMed] [Google Scholar]
- Tokudome S., Hashimoto S., Igata A. (2016). Life expectancy and healthy life expectancy of Japan: the fastest graying society in the world. BMC Res. Notes 9, 482. 10.1186/s13104-016-2281-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toohey A. M., McCormack G. R., Doyle-Baker P. K., Adams C. L., Rock M. J. (2013). Dog-walking and sense of community in neighborhoods: implications for promoting regular physical activity in adults 50 years and older. Health Place 22, 75–81. 10.1016/j.healthplace.2013.03.007 [DOI] [PubMed] [Google Scholar]
- Topál J., Miklósi Á., Gácsi M., Dóka A., Pongrácz P., Kubinyi E., et al. (2009). Chapter 3: The dog as a model for understanding human social behavior. Adv. Study Behav. 39, 71–116. 10.1016/S0065-3454(09)39003-8 [DOI] [Google Scholar]
- Tóth M. L., Sigmond T., Borsos É., Barna J., Erdélyi P., Takács-Vellai K., et al. (2008). Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy 4, 330–338. 10.4161/auto.5618 [DOI] [PubMed] [Google Scholar]
- Tremaroli V., Bäckhed F. (2012). Functional interactions between the gut microbiota and host metabolism. Nature 489, 242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- Trifunovic A., Wredenberg A., Falkenberg M., Spelbrink J. N., Rovio A. T., Bruder C. E., et al. (2004). Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429, 417–423. 10.1038/nature02517 [DOI] [PubMed] [Google Scholar]
- Trounson A., DeWitt N. D. (2016). Pluripotent stem cells progressing to the clinic. Nat. Rev. Mol. Cell Biol. 17, 194–200. 10.1038/nrm.2016.10 [DOI] [PubMed] [Google Scholar]
- Tsujimoto Y., Shimizu S. (2005). Another way to die: autophagic programmed cell death. Cell Death Differ. 12, 1528–1534. 10.1038/sj.cdd.4401777 [DOI] [PubMed] [Google Scholar]
- Tung Y.-T., Wang B.-J., Hu M.-K., Hsu W.-M., Lee H., Huang W.-P., et al. (2012). Autophagy: A double-edged sword in Alzheimer’s disease. J. Biosci. 37, 157–165. 10.1007/s12038-011-9176-0 [DOI] [PubMed] [Google Scholar]
- Tyner S. D., Venkatachalam S., Choi J., Jones S., Ghebranious N., Igelmann H., et al. (2002). p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53. 10.1038/415045a [DOI] [PubMed] [Google Scholar]
- Ukraintseva S., Yashin A., Arbeev K., Kulminski A., Akushevich I., Wu D., et al. (2016). Puzzling role of genetic risk factors in human longevity: “risk alleles” as pro-longevity variants. Biogerontology 17, 109–127. 10.1007/s10522-015-9600-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer S. R., Kaeberlein T. L., Mailheau S., Bergman P. J., Creevy K. E., Promislow D. E. L., et al. (2017). A randomized controlled trial to establish effects of short-term rapamycin treatment in 24 middle-aged companion dogs. GeroScience 39, 117–127. 10.1007/s11357-017-9972-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiserman A. M., Koliada A. K., Marotta F. (2017). Gut microbiota: a player in aging and a target for anti-aging intervention. Ageing Res. Rev. 35, 36–45. 10.1016/j.arr.2017.01.001 [DOI] [PubMed] [Google Scholar]
- van der Spoel E., Jansen S. W., Akintola A. A., Ballieux B. E., Cobbaert C. M., Slagboom P. E., et al. (2016). Growth hormone secretion is diminished and tightly controlled in humans enriched for familial longevity. Aging Cell 15, 1126–1131. 10.1111/acel.12519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Giau V., An S. S. A. (2016). Emergence of exosomal miRNAs as a diagnostic biomarker for Alzheimer’s disease. J. Neurol. Sci. 360, 141–152. 10.1016/j.jns.2015.12.005 [DOI] [PubMed] [Google Scholar]
- Van Heemst D., Beekman M., Mooijaart S. P., Heijmans B. T., Brandt B. W., Zwaan B. J., et al. (2005). Reduced insulin/IGF-1 signalling and human longevity. Aging Cell 4, 79–85. 10.1111/j.1474-9728.2005.00148.x [DOI] [PubMed] [Google Scholar]
- Van Raamsdonk J. M., Hekimi S. (2009). Deletion of the mitochondrial superoxide dismutase sod-2 extends lifespan in Caenorhabditis elegans. PLoS Genet. 5, e1000361. 10.1371/journal.pgen.1000361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Remmen H., Ikeno Y., Hamilton M., Pahlavani M., Wolf N., Thorpe S. R., et al. (2003). Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics 16, 29–37. 10.1152/physiolgenomics.00122.2003 [DOI] [PubMed] [Google Scholar]
- Vaysse A., Ratnakumar A., Derrien T., Axelsson E., Rosengren Pielberg G., Sigurdsson S., et al. (2011). Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 7, e1002316. 10.1371/journal.pgen.1002316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vellai T. (2009). Autophagy genes and ageing. Cell Death Differ. 16, 94–102. 10.1038/cdd.2008.126 [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai K., Zhang Y., Kovacs A. L., Orosz L., Müller F. (2003). Influence of TOR kinase on lifespan in C. elegans. Nature 426, 620–620. 10.1038/426620a [DOI] [PubMed] [Google Scholar]
- Vermulst M., Wanagat J., Kujoth G. C., Bielas J. H., Rabinovitch P. S., Prolla T. A., et al. (2008). DNA deletions and clonal mutations drive premature aging in mitochondrial mutator mice. Nat. Genet. 40, 392–394. 10.1038/ng.95 [DOI] [PubMed] [Google Scholar]
- Vila C. (1997). Multiple and ancient origins of the domestic dog. Science 276, 1687–1689. 10.1126/science.276.5319.1687 [DOI] [PubMed] [Google Scholar]
- Vilchez D., Morantte I., Liu Z., Douglas P. M., Merkwirth C., Rodrigues A. P. C., et al. (2012). RPN-6 determines C. elegans longevity under proteotoxic stress conditions. Nature 489, 263–268. 10.1038/nature11315 [DOI] [PubMed] [Google Scholar]
- Villeda S. A., Plambeck K. E., Middeldorp J., Castellano J. M., Mosher K. I., Luo J., et al. (2014). Young blood reverses age-related impairments in cognitive function and synaptic plasticity in mice. Nat. Med. 20, 659–663. 10.1038/nm.3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonderheide R. H., Hahn W. C., Schultze J. L., Nadler L. M. (1999). The telomerase catalytic subunit is a widely expressed tumor-associated antigen recognized by cytotoxic T lymphocytes. Immunity 10, 673–679. 10.1016/S1074-7613(00)80066-7 [DOI] [PubMed] [Google Scholar]
- vonHoldt B. M., Pollinger J. P., Lohmueller K. E., Han E., Parker H. G., Quignon P., et al. (2010). Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464, 898–902. 10.1038/nature08837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker G. A., Lithgow G. J. (2003). Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2, 131–139. 10.1046/j.1474-9728.2003.00045.x [DOI] [PubMed] [Google Scholar]
- Wang C., Jurk D., Maddick M., Nelson G., Martin-Ruiz C., Von Zglinicki T. (2009). DNA damage response and cellular senescence in tissues of aging mice. Aging Cell 8, 311–323. 10.1111/j.1474-9726.2009.00481.x [DOI] [PubMed] [Google Scholar]
- Wang G.-D., Zhai W., Yang H.-C., Wang L., Zhong L., Liu Y.-H., et al. (2016). Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 26, 21–33. 10.1038/cr.2015.147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Kirkness E. F. (2005). Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 15, 1798–1808. 10.1101/gr.3765505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterland R. A., Jirtle R. L. (2003). Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Mol. Cell. Biol. 23, 5293–5300. 10.1128/MCB.23.15.5293-5300.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters D. J. (2011). Aging research 2011: exploring the pet dog paradigm. ILAR J. 52, 97–105. 10.1093/ilar.52.1.97 [DOI] [PubMed] [Google Scholar]
- Weir H. J., Mair W. B. (2016). SnapShot: neuronal regulation of aging. Cell 166, 784–784.e1. 10.1016/j.cell.2016.07.022 [DOI] [PubMed] [Google Scholar]
- White E., DiPaola R. S. (2009). The double-edged sword of autophagy modulation in cancer. Clin. Cancer Res. 15, 5308–5316. 10.1158/1078-0432.CCR-07-5023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore K., Vera E., Martínez-Nevado E., Sanpera C., Blasco M. A. (2019). Telomere shortening rate predicts species life span. Proc. Natl. Acad. Sci. 116, 15122–15127. 10.1073/pnas.1902452116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T., Sabot F., Hua-Van A., Bennetzen J. L., Capy P., Chalhoub B., et al. (2007). A unified classification system for eukaryotic transposable elements. Nat. Rev. Genet. 8, 973–982. 10.1038/nrg2165 [DOI] [PubMed] [Google Scholar]
- Wiersma A. C., Stabej P., Leegwater P. A. J., Van Oost B. A., Ollier W. E., Dukes-McEwan J. (2007). Evaluation of 15 candidate genes for dilated cardiomyopathy in the Newfoundland dog. J. Hered. 99, 73–80. 10.1093/jhered/esm090 [DOI] [PubMed] [Google Scholar]
- Willcox B. J., Donlon T. A., He Q., Chen R., Grove J. S., Yano K., et al. (2008). FOXO3A genotype is strongly associated with human longevity. Proc. Natl. Acad. Sci. U. S. A. 105, 13987–13992. 10.1073/pnas.0801030105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C. H., Shalini S., Filipovska A., Richman T. R., Davies S., Martin S. D., et al. (2015). Age-related proteostasis and metabolic alterations in Caspase-2-deficient mice. Cell Death Dis. 6, e1615–e1615. 10.1038/cddis.2014.567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson V. L., Jones P. A. (1983). DNA methylation decreases in aging but not in immortal cells. Science 220, 1055–1057. 10.1126/science.6844925 [DOI] [PubMed] [Google Scholar]
- Withers S. S., Moore P. F., Chang H., Choi J. W., McSorley S. J., Kent M. S., et al. (2018). Multi-color flow cytometry for evaluating age-related changes in memory lymphocyte subsets in dogs. Dev. Comp. Immunol. 87, 64–74. 10.1016/j.dci.2018.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wockner L. F., Morris C. P., Noble E. P., Lawford B. R., Whitehall V. L. J., Young R. M., et al. (2015). Brain-specific epigenetic markers of schizophrenia. Transl. Psychiatry 5, e680. 10.1038/tp.2015.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.-M., Puente C., Ganley I. G., Jiang X. (2013). The ULK1 complex. Autophagy 9, 124–137. 10.4161/auto.23323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood Z. A., Poole L. B., Karplus P. A. (2003). Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science (80-.) 300, 650 LP–65 653. 10.1126/science.1080405 [DOI] [PubMed] [Google Scholar]
- Wright W. E., Shay J. W. (2000). Telomere dynamics in cancer progression and prevention: fundamental differences in human and mouse telomere biology. Nat. Med. 6, 849–851. 10.1038/78592 [DOI] [PubMed] [Google Scholar]
- Wu J. J., Liu J., Chen E. B., Wang J. J., Cao L., Narayan N., et al. (2013). Increased mammalian lifespan and a segmental and tissue-specific slowing of aging after genetic reduction of mTOR expression. Cell Rep. 4, 913–920. 10.1016/J.CELREP.2013.07.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M. N. (2006). TOR signaling in growth and metabolism. Cell 124, 471–484. 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]
- Xian L., Wu X., Pang L., Lou M., Rosen C. J., Qiu T., et al. (2012). Matrix IGF-1 maintains bone mass by activation of mTOR in mesenchymal stem cells. Nat. Med. 18, 1095–1101. 10.1038/nm.2793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang V. K., Loughran K. A., Meola D. M., Juhr C. M., Thane K. E., Davis A. M., et al. (2017). Circulating exosome microRNA associated with heart failure secondary to myxomatous mitral valve disease in a naturally occurring canine model. J. Extracell. Vesicles 6, 1350088. 10.1080/20013078.2017.1350088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazawa M., Okuda M., Setoguchi A., Iwabuchi S., Nishimura R., Sasaki N., et al. (2001). Telomere length and telomerase activity in canine mammary gland tumors. Am. J. Vet. Res. 62, 1539–1543. 10.2460/ajvr.2001.62.1539 [DOI] [PubMed] [Google Scholar]
- Yazawa M., Okuda M., Setoguchi A., Nishimura R., Sasaki N., Hasegawa A., et al. (1999). Measurement of telomerase activity in dog tumors. J. Vet. Med. Sci. 61, 1125–1129. 10.1292/jvms.61.1125 [DOI] [PubMed] [Google Scholar]
- Yilmaz Ö. H., Katajisto P., Lamming D. W., Gültekin Y., Bauer-Rowe K. E., Sengupta S., et al. (2012). mTORC1 in the Paneth cell niche couples intestinal stem-cell function to calorie intake. Nature 486, 490–495. 10.1038/nature11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W., Gore A. C. (2006). Neuroendocrine control of reproductive aging: roles of GnRH neurons. Reproduction 131, 403–414. 10.1530/rep.1.00617 [DOI] [PubMed] [Google Scholar]
- Yu C. E., Oshima J., Fu Y. H., Wijsman E. M., Hisama F., Alisch R., et al. (1996). Positional cloning of the Werner’s syndrome gene. Science 272, 258–62. 10.1126/science.272.5259.258 [DOI] [PubMed] [Google Scholar]
- Zahn J. M., Poosala S., Owen A. B., Ingram D. K., Lustig A., Carter A., et al. (2007). AGEMAP: a gene expression database for aging in mice. PLoS Genet. 3, e201. 10.1371/journal.pgen.0030201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapata H. J., Quagliarello V. J. (2015). The microbiota and microbiome in aging: potential implications in health and age-related diseases. J. Am. Geriatr. Soc. 63, 776–781. 10.1111/jgs.13310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng R., Farias F. H. G., Johnson G. S., McKay S. D., Schnabel R. D., Decker J. E., et al. (2011). A truncated retrotransposon disrupts the GRM1 coding sequence in Coton de Tulear dogs with Bandera’s neonatal ataxia. J. Vet. Intern. Med. 25, 267–272. 10.1111/j.1939-1676.2010.0666.x [DOI] [PubMed] [Google Scholar]
- Zhang C., Cuervo A. M. (2008). Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14, 959–965. 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Doherty J. A., Burgess S., Hung R. J., Lindström S., Kraft P., et al. (2015). Genetic determinants of telomere length and risk of common cancers: a Mendelian randomization study. Hum. Mol. Genet. 24, 5356–5366. 10.1093/hmg/ddv252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Li J., Purkayastha S., Tang Y., Zhang H., Yin Y., et al. (2013). Hypothalamic programming of systemic ageing involving IKK-β, NF-κB and GnRH. Nature 497, 211–216. 10.1038/nature12143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Azhar G., Wei J. Y. (2012). The expression of microRNA and microRNA clusters in the aging heart. PLoS One 7, e34688. 10.1371/journal.pone.0034688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ikeno Y., Qi W., Chaudhuri A., Li Y., Bokov A., et al. (2009). Mice deficient in both Mn superoxide dismutase and glutathione peroxidase-1 have increased oxidative damage and a greater incidence of pathology but no reduction in longevity. Journals Gerontol. Ser. A Biol. Sci. Med. Sci. 64A, 1212–1220. 10.1093/gerona/glp132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Kim M. S., Jia B., Yan J., Zuniga-Hertz J. P., Han C., et al. (2017). Hypothalamic stem cells control ageing speed partly through exosomal miRNAs. Nature 548, 52–57. 10.1038/nature23282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Q., Huang T., Zhang L., Zhou Y., Luo H., Xu H., et al. (2016). Dysregulation of ubiquitin-proteasome system in neurodegenerative diseases. Front. Aging Neurosci. 8, 303. 10.3389/fnagi.2016.00303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z.-Z., Sparrow D., Hou L., Tarantini L., Bollati V., Litonjua A. A., et al. (2011). Repetitive element hypomethylation in blood leukocyte DNA and cancer incidence, prevalence, and mortality in elderly individuals: the Normative Aging Study. Cancer Causes Control 22, 437–447. 10.1007/s10552-010-9715-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann F. K. (1971). Genetic aspects of carcinogenesis. Biochem. Pharmacol. 20, 985–995. 10.1016/0006-2952(71)90322-4 [DOI] [PubMed] [Google Scholar]
- Zou Q., Wang X., Liu Y., Ouyang Z., Long H., Wei S., et al. (2015). Generation of gene-target dogs using CRISPR/Cas9 system. J. Mol. Cell Biol. 7, 580–583. 10.1093/jmcb/mjv061 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.