Abstract
Tropical ecosystems hold an extremely diverse array of endophytic fungi, but their potential use still remains to be explored. In this study, we isolated an endophytic fungus from the leaves of Otoba gracilipes, a medicinal tree from a tropical rainforest in Colombia. Following isolation and cultivation, we evaluated its extracellular crude extract for antioxidant activity. Using traditional and molecular methods (ITS1, NL1 regions), the endophyte was identified as Fusarium oxysporum. Fresh spores from the fungal isolate were inoculated in liquid media (potato dextrose broth [PDB] and potato dextrose–yeast extract broth [PDYB]) and centrifuged for recovering extracellular polysaccharides from the exhausted medium after 30 days of cultivation. Crude extracts were recovered, purified, lyophilized, and evaluated for their ability to inactivate the free radical 2,2‐diphenyl‐1‐picrylhydrazyl (DPPH). The extracts obtained from PDB culture media had a 51.5% of scavenging effect on DPPH after 5 min of reaction compared with the extracts from PDBY (26.4%), which suggests a high antioxidant potential of these fungal extracts. Thus, our results suggest other fungi from tropical ecosystems should be explored as potential sources of novel enzymes and other metabolites with bioactivity.
Keywords: antioxidant activity, crude extract, endophytic fungi, Fusarium oxysporum, Otoba gracilipes, tropical forest
An endophytic fungus isolated from the leaves of Otoba gracilipes, a poorly explored medicinal plant from tropical rainforest in Colombia, was cultivated, and its extracellular crude extract was evaluated for antioxidant activity. Using traditional and molecular methods of characterization (ITS1, NL1 regions), the endophyte was identified as Fusarium oxysporum.

1. INTRODUCTION
Endophytic fungi are microorganisms that grow inside plant tissues without causing any adverse effects (Meenatchi, Ramesh, Bagyalakshmi, Shanmugaiah, & Rajendran, 2016). The identity and potential role of biologically active metabolites they produce as a result of their association with host plants have been investigated before (Bhardwaj, Sharma, & Agrawal, 2014; Meng et al., 2011; Schulz, Boyle, Draeger, Römmert, & Krohn, 2002). In particular, given that to colonize their host endophytes produce exoenzymes, they can be easily isolated, for they grow well in the host apoplastic washing fluid. Thus, previous studies have found that different endophytes produce secondary metabolites that mimic their host plant, with some showing a potential for pharmaceutical and medicinal applications (Hussain et al., 2014; Sharma, Pramanik, & Kumar, 2016; Strobel, 2003). For instance, several studies have shown that extracellular polysaccharides of endophytic fungi present antioxidant activity (Guo et al., 2013; Khiralla et al., 2015; Prihantini & Tachibana, 2017; Yadav, Yadav, & Yadav, 2014; Yan et al., 2016; Ye et al., 2013). On the other hand, endophytic microorganisms can be rich sources of novel enzymes and other metabolites with bioactivity as well (Hussain et al., 2014), although their potential function in biocatalysis has been poorly explored (Rodríguez, González, & Rodríguez, 2016).
Most antioxidants known today are industrially synthesized although being accounted for causing liver damage and carcinogenesis (Yuan, Zhang, Fan, & Yang, 2008). In contrast, natural‐derived antioxidants, like those produced by endophytes, have not been found to be harmful. In particular, due to a high biological diversity and biochemical evolution (Fernandes et al., 2009), endophytes have the ability of using several substrates, producing a wide array of secondary metabolites (Guanatilaka, 2006). These comprise a large but little explored proportion of fungal diversity (Perottoab et al., 2013; Yuanab, Chena, & Mac, 2011). For example, since the discovery of paclitaxel, a potent anticancer agent isolated from endophytic fungi such as Taxomyces andreanae and Pestalotia spp., endophytes have been recognized as potential new sources of anticancer, antimicrobial, and antimalarial bioactive metabolites, attracting much more attention from researchers (Cui, Guo, Ren, Zhang, & Wang, 2015). These metabolites include steroids, xanthines, phenols, isocoumarins, quinones, and terpenoids (Schulz et al., 2002), among others.
It is relevant to mention that the biotechnological use of endophyte metabolites for pharmaceutical or agrochemical products is still in the developmental stage. For example, rugulosin, a mycotoxin produced by a spruce endophyte, has been shown to be effective against pine worm (Miller, Mackenzie, Foto, Adams, & Findlay, 2002), but is still not commercially produced. In this study, we explored secondary metabolites produced by fungal endophytes of Otoba gracilipes (family Myristicaceae), a tropical medicinal tree not previously explored for potential bioactive metabolites (Bernal, García, & Quevedo, 2011). Since previous studies have shown that the leaves of other trees such as Quercus ilex and Nothapodytes foetida contain a high diversity and abundance of fungal endophytic strains (Fisher et al., 1994; Fhatima et al., 2013; Musavi & Balakrishnan, 2015), we predicted that leaves of O. gracilipes would contain a high diversity of endophytic fungi, with a high potential for producing secondary metabolites. For this purpose, we isolated, cultivated, and molecularly characterized a leaf endophyte of O. gracilipes. In addition, crude extracts composed mainly of polysaccharides were evaluated for antioxidant activity by a DPPH free radical test.
2. MATERIAL AND METHODS
2.1. Study Area and collection of plant material
Fresh and healthy leaves of two young trees of Otoba gracilipes were collected during the dry season in “La Carolina” (3°24'10.662"N, 76°36'52.774"W), Cali, Colombia, and a humid low montane forest at 1,600 m.a.s.l. The leaves were cut with a sterile scalpel and stored at 4°C in a sterile polyethylene bag until being used.
2.2. Isolation of the fungal strain
The isolated fungus was obtained following a modified method described by 1993). Leaves were surface‐sterilized by a thorough wash of sterile demineralized water followed by 70% ethanol for 1–2 min and 3% sodium hypochlorite for 15 min. Small pieces of plant tissue were then placed on potato dextrose agar (PDA, Merck®) medium pH 4.5 supplemented with antibiotic clindamycin (0.2 ml/100 ml) in Petri dishes and incubated at 28 ± 2°C until the fungus started to grow.
2.3. Crude extracts obtained from the endophytic fungal isolate grown on different culture media
Fresh fungal spores were suspended on a Tween‐80 sterile solution (0,01% v/v), from which 3 ml was inoculated on Erlenmeyer flasks containing 200 ml of potato dextrose broth (PDB) or potato dextrose broth and 0,5% yeast extract broth (PDYB). These were grown at a pH of 6 and incubated for 30 days at 29°C. After removal of mycelium by filtration, the supernatants were collected and concentrated (KNF® RC 900 vacuum rotary evaporator) at 40°C to a final volume of 20 ml.
The recovered fraction was added to two volumes of 96% (v/v) ethanol to precipitate the exopolysaccharides (EPS), which were recovered by centrifugation (1,440 g, 20 min) and resuspended in 25 ml of distilled water type I. This solution was then dialyzed (SnakeSkin® membrane of 3,500 molecular weight cutoff (MWCO)) against double‐distilled water for three days to obtain the crude extracellular polysaccharides and freeze‐dried (FDU‐1110.2110 EYELA® lyophilizer).
2.4. Phenotypic and molecular identification of the endophytic fungus
The isolated fungus was grown on Sabouraud dextrose agar (Merck®) and incubated during 5–7 days at 29°C. Fungal colonies were macroscopically characterized, and reproductive and vegetative structures were microscopically analyzed with lactophenol cotton blue (Merck®) and KOH (Merck®). Microscope slides were observed and photographed using a light microscope (Leica® DM500, coupled with a camera Leica® ICC50).
For molecular identification, fungal DNA was extracted using EZNA® Tissue DNA Kit (Omega Bio‐Tek), and total DNA was quantified using a NanoDrop Spectrophotometer ND‐1000 (NanoDrop, Wilmington, DE). Universal fungal primers were used to amplify the internal transcribed spacer (ITS1) ribosomal DNA region (forward 5'‐AGA GTT TGA TCH YTY AGA TGG‐3' and reverse 5'‐TTG TTA ACC ACG GYT CGA CTT‐3'), as well as the D1‐D2 region of the large‐subunit RNA gene (28S rDNA) using primers forward: D1/D2‐NL–GGTCCGTGTTTCAAGACGG and reverse: D1/D2‐NL1–GCATATCAATAAGCGGAGGAAAAG, by PCR and sequenced using automated dye termination sequencing. PCR amplification, PCR product purification, and sequencing of PCR products were the same for the ITS and D1‐D2 regions as follows. PCRs were run at 94°C for 1 min, and 35 cycles of 94°C: 30 s, specific primer Tm 60°C for 30 s, and 72°C for 5 min, using the Taq DNA polymerase (Thermo Fisher Scientific) in a Swift™ MiniPro Thermal Cycler (ESCO). PCR products were visualized in a 1% agarose gel and purified using Wizard SV Gel and PCR Clean‐Up System (Promega, USA) before being sequenced in an ABI prism 3500 sequencer (Applied Biosystems). DNA sequences were analyzed and compared with those available in GenBank via BLAST search (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Results were deposited in GenBank with accession number MN038127.1 (https://www.ncbi.nlm.nih.gov/nuccore/1679350429).
2.5. Antioxidant activity assay of fungal crude extracts
The antioxidant potential of EPS’ crude extracts was assessed by free radical scavenging using a DPPH assay following Prihantini and Tachibana (2017). Correspondingly, we used 1,1‐diphenyl‐2‐picrylhydrazyl (DPPH), a stable synthetic free radical widely used to evaluate the ability of compounds to act as free radical scavengers or radical hydrogen donors. To do this, crude extracts (2 mg/ml) were dissolved in a solution of water: methanol (3:1), and a volume of 50 μl was added to 100 µl of DPPH (100 μM in methanol) solution and mixed in a 96‐well plate at room temperature. The absorbance at 515 nm (A515) was measured spectrophotometrically (Synergy™ H1 Multi‐Mode Reader microplate) following a kinetic reaction of 30 min in quadruplicate every five minutes. Gallic acid (0.5 to 1.5 µM) was used as a positive control. The DPPH scavenging percentage (%) was calculated after García et al., 2012, as follows:
| (1) |
where A Blank is the absorbance of the blank (150 μl methanol without DPPH), A controlDPPH is the absorbance of the control (100 μl methanol solution of DPPH and 50 ml methanol), Asample is the absorbance of the sample and DPPH, and A sample blank is the absorbance of 100 µl methanol and 50 µl sample with no DPPH.
2.6. Statistical analyses
In order to estimate the DPPH scavenging percentage, we monitored the absorbance values of five minutes of kinetic activity. Given that the data followed a normal distribution, we ran an analysis of variance (ANOVA) and a Tukey (p ≤ 0.05) test to compare the difference in the bioactivity of the crude media extracts from the two fungal strain cultures. Statistical analyses were performed using the Minitab18 software (Minitab Inc., State College, PA, USA).
3. RESULTS AND DISCUSSION
Several studies have addressed endophytic fungal diversity and fungi–host interactions across different ecosystems (Bandra et al., 2001). However, fewer studies have explored the potential of endophytic fungi as secondary metabolite producers. According to Schulz and Boyle (2005), natural products continue to be an important source of new pharmaceutical products. Taking into account that six of the twenty drugs mostly prescribed in the United States are of fungal origin, and only 5% of the world's fungi have been described (Gloer, 1997; Hawksworth, 1991, 2001), there is an enormous potential for finding new pharmaceutical products in fungi. However, many studies have shown that the fungus–plant symbiosis is labile and varies from antagonistic, to neutral or mutualistic, depending on the stage of life of the host and the fungus, the genotype and abiotic and biotic conditions (Hamilton, Gundel, Helander, & Saikkonen, 2012). Hence, it is important to mention that the benefit that endophytic fungi provide in a diverse group of host plants has been mostly observed in nutrient‐poor environments or when plants are under stress such as drought, flood, or plant competition, or under herbivore and/or pathogen attack. Thus, in order to optimize the search for fungal endophyte secondary metabolites, it is necessary to consider that these metabolites may correspond to the fungal respective taxon and ecological niche and that only particular metabolic interactions can trigger the synthesis of secondary metabolites.
Tropical forests are known for holding a huge diversity of plants (Bernal, Gradstein, & Celis, 2016) and for having great structural complexity comprising different microclimates and both horizontal and vertical microhabitats (Gamboa‐Gaitán, 2006a,2006b). Correspondingly, there is a huge diversity and abundance of endophytic fungi (Gamboa‐Gaitán, Laureano, & Bayman, 2005; Gamboa‐Gaitán, 2013), and a high probability of finding metabolites with interesting pharmacological function.
3.1. Sample collection and isolation of endophytic fungi
A filamentous endophytic fungal strain was isolated from the leaves of Otoba gracilipes (Figure 1c) and grown in potato dextrose agar (PDA) (Figure 1a) and Sabouraud agar (Figure 1b).
Figure 1.

Fusarium oxysporum isolated from leaves of Otoba gracilipes in (a) PDA media, (b) Sabouraud dextrose agar media, and (c) Otoba gracilipes (https://fm-digital-assets.fieldmuseum.org/176/073/MYRI_otob_grac_col_1760717.jpg)
The isolate was coded as EBB‐ET‐01 (ecology, bioprocess, bioprospecting—tropical endophytes) and stored at −20°C in a glycerol solution (20%) in the strain collection of Universidad Icesi.
3.2. Phenotypic and molecular identification of the isolated endophytic fungus
After incubation on Sabouraud dextrose agar (SAB‐Merck®), we observed white‐pinkish (surface and reverse), cottony or woolly textured colonies of 5 cm in diameter, with no exudate. Septate, hyaline, and branching hyphae were microscopically (10× y 40×) observed, as well as hyaline, fusiform, curved, and septate macroconidia (Figure 2a), and ovoid to cylindrical microconidia (Figure 2a). Additionally, we found globular chlamydospores (Figure 2b) formed individually or in pairs. The identification of EBB‐ET‐01 based on morphological and microscopic features revealed the fungus was Fusarium sp.
Figure 2.
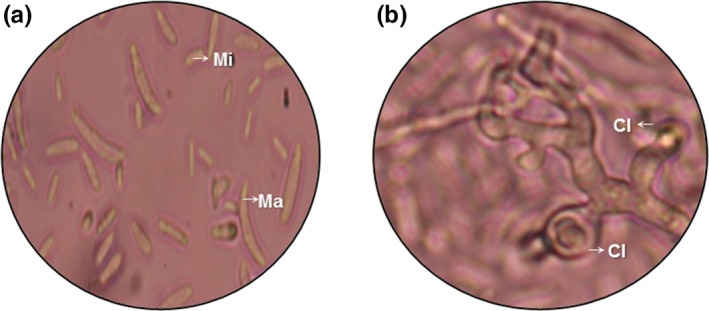
Conidia of the endophytic fungus Fusarium oxysporum (EBB‐ET‐01). (a) Macroconidia (Ma) and microconidia (Mi). (b) Chlamydospores (Cl)
Molecular identification was done using the amplified fungal sequences (ITS and Di‐D2 regions) as BLAST queries against the NCBI database in GenBank (Table 1), with a 99% match with related fungi. Sequence was deposited in GenBank with accession number MN038127.1 (https://www.ncbi.nlm.nih.gov/nuccore/1679350429).
Table 1.
Identification of fungal isolates from Otoba gracilipes by sequencing of the internal transcribed spacer 1 (ITS1) and the D1‐D2 region of rRNA gene compared with sequences listed in GenBank
| Region amplified by PCR from cultured fungus | GeneBank information | ||||||
|---|---|---|---|---|---|---|---|
| Sequences producing significant alignments | Max score | Total score | Query cover | E value | Ident | Accession N in GenBank | |
| ITS1 | Fusarium oxysporum isolate MC−26‐F internal transcribed spacer 1. | 962 | 962 | 100% | 0.0 | 99% | KU527806.1 |
| D1‐D2 | Fusarium oxysporum genomic DNA sequence contains 18S rRNA gene | 1,078 | 1,078 | 98% | 0.0 | 99% | LT746254.1 |
3.3. Antioxidant activity assay of fungal crude extracts
The DPPH free radical scavenging assay is widely used for testing antioxidant activity because it changes in coloration from intense violet to bright yellow when reacting with antioxidant compounds (García et al., 2012). Correspondingly, a higher percentage of scavenging corresponds to a higher antioxidant activity of the isolate being tested. In this study, the crude extract from the PDB culture had a scavenging of 51.5%, which was significantly higher than the PDYB culture (26.4%) (F 2 = 2,299.7; p < 0.001). Moreover, the fungal isolates from the PBD culture had the highest antioxidant activity because the DPPH free radical was inactivated in more than 50% after 5 min of reaction (Figure 3). These results suggest that PDB presumably induced a higher synthesis of secondary metabolites responsible for the observed bioactivity.
Figure 3.
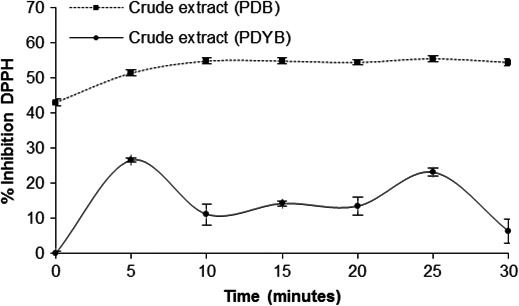
DPPH radical scavenging activity of crude extract obtained from Fusarium oxysporum strain EBB‐ET‐01 growing in potato dextrose broth (PDB) or potato dextrose broth and 0.5% yeast extract (PDYB) at pH 6.0 and 29°C for 30 days
On the other hand, it was impossible to compare the equivalent IC50 for gallic acid as DPPH radical inhibitor (0.85 µM) with the same values used for the crude extract, because these contained a no purified mixture of polysaccharides.
Recent studies have shown that endophytic fungi synthesize EPS involved in plant–endophyte interactions and that such biopolymers are characterized by structures that exhibit antioxidant activity (Chen et al., 2011; Liu et al., 2017; Orlandelli et al., 2017). In this study, an endophyte crude extract obtained from the leaves of Otoba gracilipes showed a 51.5% of antioxidant activity in 5 min. This is much higher than the 20% scavenging activity of DPPH reported by 2011) after 30 min of an EPS obtained from the culture of the strain Fusarium oxysporum Dzf17 and with a 10‐fold lower concentration (200 µg/ml). However, in Li et al., 2011, crude extracts were subjected to deproteination and decolorization processes. Using a different approach, 2017) isolated two endophytes (Aspergillus sp. y Fusarium sp.) from roots of a close relative of Otoba gracilipes, a Virola sp. tree, and found an EC50 of 17.4 µg/ml for antioxidant activity for the crude extracts of Aspergillus sp. Such activity was also measured on DPPH and found to be related to the presence of secondary metabolites such as flavonoids. However, no antioxidant activity was detected for crude extracts obtained from Fusarium sp. Thus, the high performance of culture media extracts of Fussarium oxysporium in this study suggests that the novel fungal strain we describe here has a greater potential for producing exopolysaccharides with antioxidant activity. Given that in our study the fungus was isolated from plant leaves and not from roots, it is possible that the production of secondary metabolites with antioxidant activity is tissue‐dependent. In addition, it is important to mention that exopolysaccharides with antioxidant activity were detected as secondary metabolites; therefore, the production of these compounds should be measured at different developmental stages of the fungal endophyte.
Considering that the antioxidant mechanisms of EPS are complex (Freitas, Torres, & Reis, 2016; Li et al., 2011), additional methods should be used in further investigations to re‐evaluate and compare the percentage of scavenging such as OH free radical, ABTS (Pan, Su, Cai, & Wei, 2017), and reducing powder. Simultaneously, studies on the chemical structure of EPS should be performed considering Fusarium oxysporum (EBB‐ET‐01) as a potential candidate for the production of bioactive compounds.
Finally, it is important to note that we found a high antioxidant activity of the crude extracts of only one endophytic fungus isolated from a single plant species (Otoba gracilipes) from a particular tropical forest in Colombia. Thus, there is still much to explore in the potentially hugely diverse array of fungal endophytes present across tropical ecosystems.
CONFLICT OF INTERESTS
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
N.H.C., A.F.D, and P.A.C conceived and designed experiments and contributed to the writing of the manuscript. P.A.P and A.Y.R conducted experiments.
ETHICS STATEMENT
Protocols and procedures employed in this investigation were reviewed and approved by the appropriate institutional review committee. Otoba gracilipes and Fusarium oxysporum were kept and handled in compliance with the guidelines of the “Autoridad Nacional de Licencias Ambientales (ANLA)‐Colombia” through permission “Permiso Marco de Recolección de Especímenes Silvestres de la Diversidad Biológica con fines de Investigación Científica No Comercial—Resolution 00,272, 14 March 2017.” For molecular identification of Fusarium oxysporum, The Universidad Icesi have the permission “Contrato de Acceso a Recursos Genéticos y sus Productos Derivados” from “Ministerio de ambiente y desarrollo sostenible, Colombia—Resolution 0,763, 9 May 2018.”
ACKNOWLEDGMENTS
This work was supported by Universidad ICESI. DNA sequencing was provided by Laboratorio de Medicina Genómica, Universidad ICESI. N.H.C., A.F.D., and P.A.C. were supported by Universidad ICESI. A.Y.R was supported by Universidad Pedagógica y Tecnológica de Colombia, UPTC. P.A.P participated as an undergraduate student. The sample collection of material of Otoba gracilipes and the access to the forest were performed with the "Departamento Administrativo de Gestión Medio Ambiente‐Dagma", Cali‐Colombia (Catalina Silva and Sandra Franco) autorizathion. The genetic resource access contract from ANLA was the number RGE244‐44#190.
Caicedo NH, Davalos AF, Puente PA, Rodríguez AY, Caicedo PA. Antioxidant activity of exo‐metabolites produced by Fusarium oxysporum: An endophytic fungus isolated from leaves of Otoba gracilipes . MicrobiologyOpen. 2019;8:e903 10.1002/mbo3.903
DATA AVAILABILITY STATEMENT
All data are provided fully in the Results section of this paper. The DNA sequence of ITS region from Fussarium oxysporum was submitted to NCBI with accession number MN038127.1 and is available at (https://www.ncbi.nlm.nih.gov/nuccore/1679350429).
REFERENCES
- Bandra, W. , Seneviratne, G. , & Kulasooroya, S. A. (2013). Infection among endophytic bacteria and fungi: effects and potential. J Biosci., 3(1), 645–650. [DOI] [PubMed] [Google Scholar]
- Bernal, H. Y. , García, M. H. , & Quevedo, S. F. (2011). Pautas para el conocimiento, conservación y uso sostenible de las plantas medicinales nativas en Colombia: Estrategia nacional para la conservación de plantas (p. 230). Bogotá: Instituto de Investigación de Recursos Biológicos Alexander von Humboldt. [Google Scholar]
- Bernal, R. , Gradstein, S. R. , & Celis, M. (Eds.) (2016). Catálogo de platas y líquenes de Colombia (p. 3060). Bogotá: Instituto de Ciencias Naturales, Universidad Nacional de Colombia. [Google Scholar]
- Bhardwaj, A. , Sharma, D. , & Agrawal, P. K. (2014). Isolation and characterization of endophytic fungi from spikes of Pinus roxburghii growing in Himalayan region. World Journal of Pharmaceutical Research, 3, 568–579. [Google Scholar]
- Chen, J. , Hu, K. X. , Hou, X. Q. , & Guo, S. X. (2011). Endophytic fungi assemblages from 10 Dendrobium medicinal plants (Orchidaceae). World J Microbiol Biotechnol., 27(5), 1009–1016. [Google Scholar]
- Cui, J. L. , Guo, T. T. , Ren, Z. X. , Zhang, N. L. , & Wang, M. L. (2015). Diversity and Antioxidant Activity of Culturable Endophytic Fungi from Alpine Plants of Rhodiola crenulata, R. angusta, and R. sachalinensis . PLoS ONE, 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fathima, S. , & Balakrishnan, R. M. (2013). Biodiversity, Antimicrobial Potential, and Phylogenetic Placement of an Endophytic Fusarium oxysporum NFX 06 Isolated from Nothapodytes foetida. Journal of Mycology, 10.1155/2013/172056. [DOI] [Google Scholar]
- Fernandes, M. , Silva, T. , Pfenning, L. , Costa, C. , Heinrich, T. , Alencar, S. , … Ikegaki, M. (2009). Biological activities of the fermentation extract of the endophytic fungus Alternaria alternata isolated from Coffea arabica L. Brazilian Journal of Pharmaceutical Sciences, 45, 677–685. 10.1590/S1984-82502009000400010 [DOI] [Google Scholar]
- Fisher, P. J. , Petrini, O. , Petrini, L. E. , & Sutton, B. C. (1994). Fungal endophytes from the leaves and twigs of Quercus ilex L. from England. Majorca and Switzerland. New Phytologist, 127, 133–137. [DOI] [PubMed] [Google Scholar]
- Freitas, F. , Torres, C. , & Reis, M. A. V. (2016). Engineering aspects of microbial exopolysaccharide production. Bioresource Technology, 245, 1674–1683. 10.1016/j.biortech.2017.05.092 [DOI] [PubMed] [Google Scholar]
- Gamboa‐Gaitán, M. A. (2006a). Colombian vanilla and its microbiota, I First report of Fusarium taxa from both wild and cultivated species. Acta Botanica Hungarica, 55, 239–245. [Google Scholar]
- Gamboa‐Gaitán, M. A. (2006b). Tropical Endophytic Fungi, current knowledge and perspectives. Acta Biológica Colombiana, 11, 3–20. [Google Scholar]
- Gamboa-Gaitan, M. (2013). COLOMBIAN VANILLA AND ITS MICROBIOTA, I First report of Fusarium taxa from both wild and cultivated species. Acta Botanica Hungarica, 55(3–4), 239–245. [Google Scholar]
- Gamboa‐Gaitán, M. A. , Laureano, S. , & Bayman, P. (2005). Endophytic Phomopsis Strains from Leaves of Guarea guidonia (Meliaceae). Caribbean Journal of Science, 41, 1–9. [Google Scholar]
- García, E. J. , Oldono, T. L. , Alencar, S. M. , Reis, A. , Loguercio, A. D. , & Grande, R. H. (2012). Antioxidant activity by DPPH assay of potential solutions to be applied on bleached teeth. Brazilian Dental Journal, 23, 22–27. 10.1590/S0103-64402012000100004 [DOI] [PubMed] [Google Scholar]
- Gloer, J. B. (1997). Applications of fungal ecology in the search for new bioactive natural products In Wicklow D. T., & Soderstrom B. E. (Eds.), The Mycota. Vol. IV. Environmental and microbial relationships (pp. 249–268). New York, NY: Springer Verlag. [Google Scholar]
- Guanatilaka, A. (2006). Natural products from plant‐associated microorganisms: Distribution, structural diversity, bioactivity, and implications of their occurrence. Journal of Natural Products, 69, 509–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, S. , Mao, W. , Li, Y. , Gu, Q. , Chen, Y. , Zhao, C. , … Liu, X. (2013). Preparation, structural characterization and antioxidant activity of an extracellular polysaccharide produced by the fungus Oidiodendron truncatum GW. Process Biochemistry, 48, 539–544. 10.1016/j.procbio.2013.01.014 [DOI] [Google Scholar]
- Hamilton, C. E. , Gundel, P. E. , Helander, M. , & Saikkonen, K. (2012). Endophytic mediation of reactive oxygen species and antioxidant activity in plants: A review. Fungal Diversity, 54, 1–10. 10.1007/s13225-012-0158-9 [DOI] [Google Scholar]
- Hawksworth, D. L. (1991). The fungal dimension of biodiversity: Magnitude, significance and conservation. Mycological Research, 95, 641–655. 10.1016/S0953-7562(09)80810-1 [DOI] [Google Scholar]
- Hawksworth, D. L. (2001). The magnitude of fungal diversity: The 1.5 million species estimate revisited. Mycological Research, 105, 1422–1431. 10.1017/S0953756201004725 [DOI] [Google Scholar]
- Hussain, H. , Kliche‐Spory, C. , Al‐Harrasi, A. , Al‐Rawahi, A. , Abbas, G. , Green, I. R. , … Shah, A. (2014). Antimicrobial constituents from three endophytic fungi. Asian Pacific Journal of Tropical Medicine, 7, S224–S227. [DOI] [PubMed] [Google Scholar]
- Katoch, M. , Singh, A. , Singh, G. , Wazir, P. , & Kumar, R. (2017). Phylogeny, antimicrobial, antioxidant and enzyme‐producing potential of fungal endophytes found in Viola odorata . Annals of Microbiology, 67, 529–540. 10.1007/s13213-017-1283-1 [DOI] [Google Scholar]
- Khiralla, A. , Mohamed, I. , Thomas, J. , Mignard, B. , Spina, R. , Yagi, S. , & Laurain, D. (2015). A pilot study of antioxidant potential of endophytic fungi from some Sudanese medicinal plants. Asian Pacific Journal of Tropical Medicine, 8, 701–704. 10.1016/j.apjtm.2015.07.032 [DOI] [PubMed] [Google Scholar]
- Li, P. , Luo, C. , Sun, W. , Lu, S. , Mou, Y. , Peng, Y. , & Zhou, L. (2011). In vitro antioxidant activities of polysaccharides from endophytic fungus Fusarium oxysporum Dzf17. African Journal of Microbiology Research, 5, 5990–5993. [Google Scholar]
- Liu, J. , Wang, X. , Pu, H. , Liu, S. , Kan, J. , & Jin, C. (2017). Recent advances in endophytic exopolysaccharides: Production, structural characterization, physiological role and biological activity. Carbohydrate Polymers, 157, 1113–1124. [DOI] [PubMed] [Google Scholar]
- Meenatchi, A. , Ramesh, V. , Bagyalakshmi, K. R. , Shanmugaiah, V. , & Rajendran, A. (2016). Diversity of endophytic fungi from the ornamental plant‐ Adenium obesum . Studies in Fungi, 1, 34–42. 10.5943/sif/1/1/3 [DOI] [Google Scholar]
- Meng, L. , Sun, P. , Tang, H. , Li, L. , Draeger, S. , Schulz, B. , … Yi, Y. (2011). Endophytic fungus Penicillium chrysogenum, a new source of hypocrellins. Biochemical Systematics and Ecology, 39, 163–165. 10.1016/j.bse.2011.02.003 [DOI] [Google Scholar]
- Miller, J. D. , Mackenzie, S. , Foto, M. , Adams, G. W. , & Findlay, J. A. (2002). Needles of white spruce inoculated with rugulosin producing endophytes contain rugulosin reducing spruce budworm growth rate. Mycological Research, 106, 471–479. 10.1017/S0953756202005671 [DOI] [Google Scholar]
- Orlandellia, R. C. , Corradi da Silva, M. L. , Dalberto, A. F. , Almeidaa, I. V. , Pimenta, V. , Prietoc, A. , … Alencar, J. (2017). β-(1→3,1→6)-d-glucans produced by Diaporthe sp. endophytes: Purification, chemical characterization and antiproliferative activity against MCF-7 and HepG2-C3A cells. International Journal of Biological Macromolecules, 94, 431–437. [DOI] [PubMed] [Google Scholar]
- Pan, F. , Su, T.‐J. , Cai, S.‐M. , & Wei, W. (2017). Fungal endophyte‐derived Fritillaria unibracteata var. wabuensis: Diversity, antioxidant capacities in vitro and relations to phenolic, flavonoid or saponin compounds. Scientific Reports, 7, 1–14. 10.1038/srep42008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perottoab, S. , Angelinic, P. , Bianciottob, V. , Bonfanteab, P. , Girlandaab, M. , Kulld, T. , … Seloss, M. A. (2013). Interactions of fungi with other organisms. Plant Biosystems‐An International Journal Dealing with All Aspects of Plant Biology, 147, 208–218. 10.1080/11263504.2012.753136 [DOI] [Google Scholar]
- Prihantini, A. I. , & Tachibana, S. (2017). Antioxidant compounds produced by Pseudocercospora sp. ESL 02, an endophytic fungus isolated from Elaeocarpus sylvestris . Asian Pacific Journal of Tropical Biomedicine, 7, 110–115. [Google Scholar]
- Rodríguez, P. , González, D. , & Rodríguez, S. (2016). Endophytic microorganisms: A source of potentially useful biocatalysts. Journal of Molecular Catalysis B: Enzymatic, 133, S569–S581. [Google Scholar]
- Schulz, B. , & Boyle, C. (2005). The endophytic continuum. Mycological Research, 109, 661–686. 10.1017/S095375620500273X [DOI] [PubMed] [Google Scholar]
- Schulz, B. , Boyle, C. , Draeger, S. , Römmert, A.‐K. , & Krohn, K. (2002). Endophytic fungi: A source of novel biologically active secondary metabolites. Mycological Research, 109, 996–1004. 10.1017/S0953756202006342 [DOI] [Google Scholar]
- Schulz, B. , Wanke, U. , Draeger, S. , & Aust, H.‐J. (1993). Endophytes from herbaceous plants and shrubs: Effectiveness of surface sterilization methods. Mycological Research, 97, 1447–1450. 10.1016/S0953-7562(09)80215-3 [DOI] [Google Scholar]
- Sharma, D. , Pramanik, A. , & Kumar, P. (2016). Evaluation of bioactive secondary metabolites from endophytic fungus Pestalotiopsis neglecta BAB‐5510 isolated from leaves of Cupressus torulosa D. Don. 3 Biotech, 6, 1–14. 10.1007/s13205-016-0518-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strobel, G. A. (2003). Endophytes as sources of bioactive products. Microbes and Infection, 5, 535–544. 10.1016/S1286-4579(03)00073-X [DOI] [PubMed] [Google Scholar]
- Yadav, M. , Yadav, A. , & Yadav, J. P. (2014). In vitro antioxidant activity and total phenolic content of endophytic fungi isolated from Eugenia jambolana Lam. Asian Pacific Journal of Tropical Medicine, 7, S256–S261. 10.1016/S1995-7645(14)60242-X [DOI] [PubMed] [Google Scholar]
- Yan, M. X. , Mao, W. J. , Liu, X. , Wang, S. Y. , Xia, Z. , Cao, S. J. , … Xian, H. L. (2016). Extracellular polysaccharide with novel structure and antioxidant property produced by the deep‐sea fungus Aspergillus versicolor N2bc. Carbohydrate Polymers, 147, 272–281. 10.1016/j.carbpol.2016.03.090 [DOI] [PubMed] [Google Scholar]
- Ye, Y. , Xiao, Y. U. , Ma, L. , Li, H. , Xie, Z. , Wang, M. , … Liu, J. (2013). Flavipin in Chaetomium globosum CDW7, an endophytic fungus from Ginkgo biloba, contributes to antioxidant activity. Applied Microbiology and Biotechnology, 97, 131–7139. 10.1007/s00253-013-5013-8 [DOI] [PubMed] [Google Scholar]
- Yuan, J. F. , Zhang, Z. Q. , Fan, Z. C. , & Yang, J. X. (2008). Antioxidant effects and cytotoxicity of three purified polysaccharides from Ligusticum chuanxiong Hort. Carbohydrate Polymers, 74, 822–827. 10.1016/j.carbpol.2008.04.040 [DOI] [Google Scholar]
- Yuanab, Z. , Chena, Y. , & Mac, X. (2011). Symbiotic fungi in roots of Artemisia annua with special reference to endophytic colonizers. Plant Biosystems‐An International Journal Dealing with All Aspects of Plant Biology, 145, 495–502. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are provided fully in the Results section of this paper. The DNA sequence of ITS region from Fussarium oxysporum was submitted to NCBI with accession number MN038127.1 and is available at (https://www.ncbi.nlm.nih.gov/nuccore/1679350429).


