Key Points
Question
What are the molecular features of conjunctival melanoma?
Findings
In this case series of 8 patients with conjunctival melanoma examined, 1 had BRAF/V600E mutation, 3 had NRAS/Q61R mutation, 3 had NF1 mutations, and 1 had triple-wild type; mutation burden ranged from 1.1 to 15.6 mutations per megabase. A patient with an unresectable tumor and BRAF/V600E mutation was treated with combined systemic BRAF and MEK inhibitors, and after 3 months of therapy, conjunctival melanoma responded and the residual tumor was excised.
Meaning
Conjunctival melanoma appears to show some distinctive molecular features and its mutational profile may be worth evaluating in patients with advanced disease where neoadjuvant/adjuvant therapy may be considered.
Abstract
Importance
Greater understanding of molecular features of conjunctival melanoma (CM) may improve its clinical management.
Objective
To evaluate molecular features of CM and application of this information into clinical care.
Design, Setting, and Participants
In a prospective case series of CM with integrative exome and transcriptome analysis, 8 patients at an academic ocular oncology setting were evaluated. The study was conducted from November 2015 to March 2018.
Interventions/Exposures
Integrative exome and transcriptome analysis of CMs and clinical management of a patient’s care by using this information.
Main Outcomes and Measures
Molecular characterization of CM and its potential clinical application.
Results
In the 8 patients (4 men) included in analysis, 4 subgroups of CM were observed, including the BRAF V600E mutation in 1 tumor, NRAS Q61R mutation in 3 tumors, NF1 mutations (Q1188X, R440X, or M1215K+ S15fs) in 3 tumors, and triple-wild type (triple-WT) in 1 tumor. The triple-WT case had CCND1 amplification and mutation in the CIC gene (Q1508X). Five tumors, including the triple-WT, also harbored mutations in MAPK genes. In addition to the genes linked to mitogen-activated protein kinase and phosphoinositol 3-kinase pathways, those involved in cell cycle and/or survival, ubiquitin-mediated protein degradation, and chromatin remodeling/epigenetic regulation (ATRX being the most frequently mutated: noted in 5 tumors) may play an important role. Other frequently mutated genes included PREX2 (n = 3), APOB (n = 4), and RYR1/2 (n = 4), although their relevance remains to be determined. The mutation burden ranged from 1.1 to 15.6 mutations per megabase (Mut/Mb) and was 3.3 Mut/Mb or less in 3 tumors and more than 10 Mut/Mb in 2 tumors. A patient with a large tumor and BRAF V600E mutation was treated with combined systemic BRAF (dabrafenib) and MEK (trametinib) inhibitors. After 3 months of therapy, her CM responded substantially and the residual tumor was removed by local surgical excision.
Conclusions and Relevance
The NRAS Q61R and NF1 mutations were more common than the BRAF V600E mutation in this series. Although small tumors (where incisional biopsy is not indicated) are treated with surgical excision regardless of mutational profile, in large tumors carrying the BRAF V600E mutation, neoadjuvant therapy with combined systemic BRAF and MEK inhibitors followed by local excision may be used as an alternative to exenteration. Integrative omics analysis of CM may be informative and guide clinical management and treatment in selected cases.
This case series examines the use of integrative exome and transcriptome analysis of genes and mutations in 8 patients with conjunctival melanoma.
Introduction
Molecular features of conjunctival melanoma (CM) are not well defined. Previous studies reported BRAF (OMIM 164757) mutations in 14% to 60%, NRAS (OMIM 164790) mutations in 18% to 20%, NF1 (OMIM 613113) mutations in 20% to 33%, and TERT (OMIM 187270) promoter mutations in 20% to 41% of CMs.1,2,3,4,5,6 In the Translational Pathology Department, University of Michigan, integrative exome and transcriptome analysis is performed, in which a panel of more than 1700 cancer-related genes are captured for parallel sequencing of tumor and germline DNA.7 To our knowledge, this is currently the largest available panel of cancer-related genes. Herein, we used this panel to evaluate the molecular features of 8 CMs and applied this information for globe-saving management in 1 patient with extensive CM that would otherwise be exenterated. Although the use of systemic BRAF/MEK inhibitors in management of recurrent or metastatic CM has been reported in a few studies,8,9,10 to our knowledge, this is the first report of the use of combined systemic BRAF and MEK inhibitors as neoadjuvant therapy to regress primary CM so that it can be excised locally.
Methods
Integrative exome and transcriptome analysis was performed prospectively using standard protocols described previously.7 OncoSeq test is a Clinical Laboratory Improvement Amendments-certified test developed by Michigan Oncology Sequencing Center, designed to use next-generation sequencing technology to identify nonsynonymous somatic point mutations in a panel of more than 1700 genes with suggestive links to cancer by comparing tumor against normal samples obtained from the same patient.7 The test also evaluates somatic insertions and deletions (indels), copy number variations, gene fusions, and outlier gene expression. Flash-frozen tumor tissue collected during surgical excision and matched normal sample (blood, sputum, cheek swab) collected at the same time were used for integrative analysis. Hematoxylin-eosin–stained tissue sections were reviewed by a pathologist to identify areas with highest tumor content to be used for sequencing.
The study was conducted from November 2015 to March 2018 and was approved by the University of Michigan Institutional Review Board and adhered to the tenets of the Declaration of Helsinki.11 Written informed consent was obtained from all participants from whom clinical data, features, and samples were collected. Participants did not receive financial compensation.
Results
Clinical features and findings of 8 patients (4 men) with CM are presented in the eTable in the Supplement. Molecular characteristics of their CMs are presented in the Table.
Table. Summary of OncoSeq Results in Conjunctival Melanoma Samples.
| Patient No./Sex | Tumor Content, % | Mutation Burden (Mut/Mb), UV Signature | Copy No. Variations | Somatic Point Mutationsa | Somatic Indels | Outlier Gene Expressionb,c | Potential Therapies |
|---|---|---|---|---|---|---|---|
| 1/M | 30 | 7.8, UV signature | 1 Copy loss (TP53, NF1, CBL) | TP53 p.V197G, NF1 p.Q1188X, CBL p.P417L, ATRX p.R2131X; on ChrX, MAPK15 p.G347R, RYR2 p.R2267C, APOB p.R3291C | NA | NA | Dual SFK and RTK inhibitors |
| 2/M | 87 | 6.9, UV signature | Amplification (RAF1), homozygous deletion (CDKN2A/2B, HLA-A) | NRAS p.Q61R, ROS1 p.E1642K, FGFR2 p.P666S, ATM p.G2765C, RYR1 p.D1905N, APOB p.D3240N, PREX2 p.R140K | Frameshift insertion (ATRX, CREBBP) | RAF1 | Pan-RAF or MEK inhibitor |
| 3/F | 78 | 2.2 | Amplification (CCND1, MED29) | CIC p.Q1508X, MAPK4 p.E453K | NA | CCND1, MED29 | CDK inhibitors? |
| 4/F | 85 | 15.6, UV signature | Amplification (CCND1, AIP, BTG3, FADD, SUV420H1, KDM2A) | NF1 p.R440X, APOB p.Q2173X, RYR1 p.H4754Y, RYR2 p.FR331X, ATRX p.T154S, p.M56I, MAPK8 p.R150Q, PREX2 p.R117C, PDGFRB p.A619T, HDAC9 p.E692D | Frameshift insertion (MEF2B) | CCND1, AIP, BTG3, FADD, SUV420H1, KDM2A | MEK/mTOR inhibitor, CCND1 inhibitor |
| 5/F | 86 | 3.3 | Aneuploid (no focal amplification or deletion) | BRAF p.V600E, FBXW7 splice acceptor; copy neutral LOH, PIK3CB p.D1067A, PREX2 p.G393E | NA | CDK2, ETV5, MET, PAX3 | BRAF, PIK3CB inhibitors |
| 6/M | 74 | 14.4, UV signature | Aneuploid with gain or loss of several large chromosome regions, MAPK1 amplification (no focal deletions) | NRAS p.Q61R, TP53 p.I232T, ATRX splice acceptor, MAP3K5 p.G467E, PIK3CG p.D521N, PPP6C p.P186S, HDAC9 p.G167E, SETD5 p.H121Y | Disruptive in-frame deletion (SETD2) | CDK2, SOX10, ETV5 | |
| 7/F | 38 | 1.1 | Aneuploid, amplification (RYR2, BTG2) | NRAS p.Q61R, EMSY p.Q1059X, IGF1 p.G194E | NA | CDK2, SOX10, PAX3 | |
| 8/M | 76 | 9.4, UV signature | Genome-wide polyploidy (no focal gains/losses) | NF1 p.M1215K, CBL p.P417L, ALK splice acceptor, ERBB4 p.R168W, p.S173F, MAPK4 p.A546V, APOB p.H4422Y, HDAC4 p.T660S, SETD1A p.S349L | Frameshift insertion (NF1), Frameshift deletion (ATRX, PIK3CA) | TNFRSF14, SOX10, TBX2 | MEK inhibitor, dual SFK and RTK inhibitors |
Abbreviations: Mut/Mb, mutations per megabase; NA, not applicable.
Total number of mutations identified in each tumor ranged from 3 to more than 300; the most relevant ones are included in the Table. Additional genes that appeared to be frequently mutated in our series (eg, PREX2, APOB, RYR1/2 genes) are also included, although their relevance remains to be determined.
No notable driver gene fusions were detected in our series.
Melanoma markers (eg, MLANA, MITF, TYR) were expressed at high levels in our samples (not shown in the Table).
Somatic tumor mutation burden (TMB) ranged from 1.1 to 15.6 mutations per megabase (Mut/Mb) and was 3.3 Mut/Mb or lower in 3 cases and more than 10 Mut/Mb in 2 cases. The UV light–related mutational signature (C>T transitions at dipyrimidine sites accounted for >60% or CC>TT mutations >5% of TMB) was detected in 5 cases.
Focal or complex structural rearrangements were detected in all tumors (eFigure in the Supplement), while somatic indels were observed in 4 tumors. Somatic point mutation and indel analysis revealed 4 subgroups, including BRAF V600E mutation in 1 patient, NRAS Q61R mutation in 3 patients, NF1 mutations (Q1188X, R440X, or M1215K+S15fs) in 3 patients, and triple-wild type in 1 patient. RAF1 (OMIM 164760) amplification was observed in 1 tumor with NRAS mutation and PIK3CA/B/G (OMIM 171834/602925/601232) mutations were detected in 3 tumors. The BRAF-mutated tumor also showed FBXW7 (OMIM 606278) mutation. Of 3 NF1-mutated tumors, 1 had CCND1 (OMIM 168461) amplification and 2 harbored CBL (OMIM 165360) mutations. Triple-WT tumor showed CCND1 amplification and CIC (612082) mutation (Q1508X). Five tumors, including the triple-WT, also harbored mutations in MAPK genes.
Somatic ATRX (300032) mutations were detected in 5 tumors with either NF1 (n = 3) or NRAS (n = 2) mutations. Of 5 ATRX-mutated tumors, 4 also showed mutations in genes involved in histone modification (SETD and/or HDAC genes or CREBBP [OMIM 600140]). Other frequently mutated genes were PREX2 (OMIM 612139) (n = 3), APOB (OMIM 107730) n = 4), and RYR1/2 (OMIM 180901/180902) (n = 4), although their relevance remains to be determined. Moreover, 4 CMs showed outlier expression of CDK2 (OMIM 116953) and/or SOX10 (OMIM 602229).
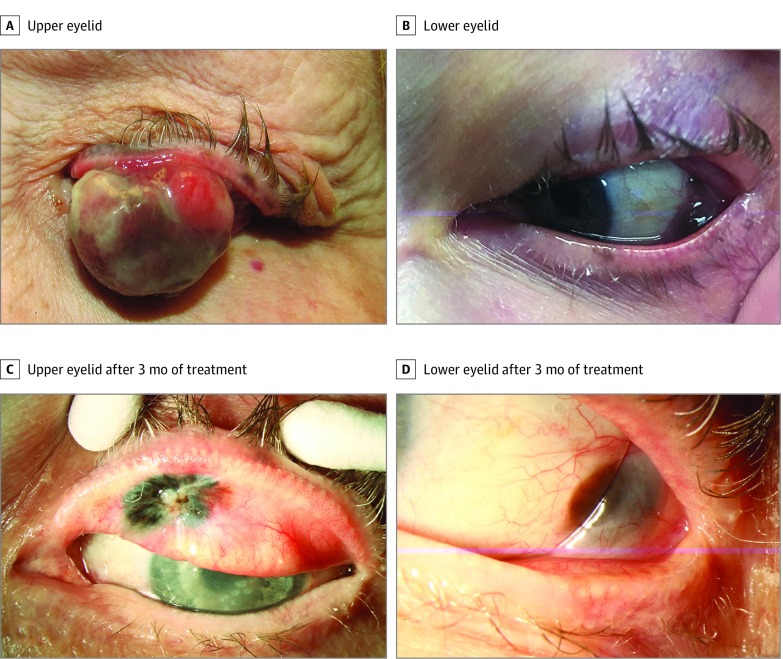
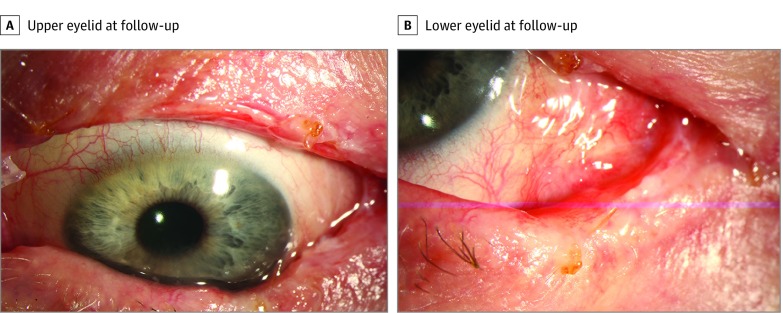
The genetic information was used for management of a woman in her 70s who presented with a multinodular, extensive CM of the left eye (case 5; eTable in the Supplement; Figure 1A and B). Her tumor was found to harbor a BRAF V600E mutation. Given the extent of disease, an incisional biopsy was done for diagnosis prior to planned exenteration. However, given the mutational profile and following discussions with the medical oncologist and patient, systemic BRAF inhibitor (dabrafenib) and MEK inhibitor (trametinib) were started. After 3 months of therapy, the CM regressed substantially (Figure 1C and D) and she underwent surgical excision of residual tumor and sentinel lymph node biopsy, with negative results. During follow-up, she showed no local recurrence (Figure 2), but after 1 year she developed metastasis.
Figure 1. Multinodular Extensive Conjunctival Melanoma (CM) of the Left Eye .
A and B, The patient presented with a CM that involved the upper and lower eyelids and bulbar conjunctiva (eTable in the Supplement, case 5). Given the extent of the disease, an incisional biopsy was done for diagnosis prior to planned exenteration. The tumor was found to harbor the BRAF V600E mutation and, following discussions with the medical oncologist and patient, neoadjuvant therapy with systemic BRAF and MEK inhibitors was started. C and D, After 3 months of therapy, the conjunctival melanoma regressed.
Figure 2. Regressed/Residual Conjunctival Melanoma Removed by Surgical Excision .
Surgery was performed on the patient (case 5) shown in Figure 1. During follow-up, the patient showed no local recurrence on the upper eyelid (A) and on the lower eyelid and bulbar conjunctiva (B); however, after 1 year, she developed metastasis.
Discussion
UV mutational signature was observed in 5 of 8 CMs, suggesting the role of UV light exposure in the pathogenesis of a subgroup of CMs.
Most melanomas have been shown to harbor mutations affecting the mitogen-activated protein kinase (MAPK) and phosphoinositol 3-kinase pathways.12,13 The Cancer Genome Atlas project has classified 318 cutaneous melanomas into 4 subgroups as BRAF mutated, RAS mutated, NF1 mutated, and triple-WT.12 Of these cutaneous melanomas, 52% had BRAF mutation, 28% harbored NRAS mutation, 14% had NF1 mutation, and 14% lacked BRAF, RAS, and NF1 mutations (triple-WT).12 Patients who lacked BRAF, RAS, and NF1 somatic mutations had KIT (OMIM 164920) mutations and focal amplifications and complex structural arrangements.12 Similarly, in our CM study, we observed 4 main subgroups (BRAF mutated, NRAS mutated, NF1 mutated, and triple-WT), which represent 4 different mechanisms that can activate the MAPK pathway. In our study, the triple-WT tumor harbored CCND1 amplification coupled with CIC mutation, which appears to be a novel finding. CIC encodes a transcriptional repressor whose deficiency has been linked to MAPK pathway activation. We also noted that most CMs in our series (n = 5), including the triple-WT, harbored mutations in MAPK genes.
Scholz et al1 reported NF1 mutations in 33% of CMs, followed by mutations in BRAF (25%) and RAS genes (NRAS and KRAS [OMIM 190070]; 19%). Similarly in our series, NF1 and NRAS mutations were more common than BRAF mutations, although only 8 cases were evaluated. In the BRAF-mutated tumor, we also detected a mutation in FBXW7, which is a tumor-suppressor gene involved in ubiquitin-mediated oncoprotein degradation. Similar to a recent small whole-exome sequencing study,5 we also observed CBL (n = 2) and ATRX (n = 5) mutations in CM. ATRX belongs to the switch/sucrose nonfermentable (SWI/SNF) family of chromatin remodelers and its protein product plays a role in DNA methylation and alternative telomere lengthening. Other frequently mutated genes (eg, PREX2, APOB, RYR1/2) noted in our study warrant further investigation.
Molecular and genetic information obtained in this study was used for clinical management in 1 patient with a BRAF-mutated large CM who wanted to avoid exenteration. The patient was given systemic BRAF and MEK inhibitors as neoadjuvant therapy to regress her extensive primary tumor followed by local excision, enabling a globe-preserving therapy. Although novel, this approach needs to be further studied for its long-term effects.
Immune checkpoint inhibitors are used to treat recurrent or metastatic conjunctival or orbital melanomas.14,15 Goodman et al16 divided melanomas and non–small cell lung cancers into 3 groups based on TMB and immunotherapy response (low, 1-5 Mut/Mb; intermediate, 6-19 Mut/Mb; and high, >20 Mut/Mb) and showed that high TMB may be a biomarker for response to PD-1/PD-L1 inhibitor treatment. In our series, 3 tumors had TMB 3.3 Mut/Mb or fewer while only 2 had more than 10 Mut/Mb.
Limitations
This case series has limitations. CM is a rare cancer and the main limitation of this case series is the small sample size. Therefore, our study findings cannot be generalized until confirmed in larger series and studies.
Conclusions
Four main subgroups of somatic mutations were noted in this study: BRAF, NRAS, and NF1 mutations and triple-WT. In this series, NF1 and NRAS mutations were the most common, although with only 8 patients, our findings should be considered provisional until confirmed in large studies. In addition to MAPK and phosphoinositol 3-kinase pathways-related genes, those involved in cell cycle/survival, ubiquitin-mediated protein degradation, chromatin remodeling, and epigenetic regulation may also play important role. Overall, CM appears to show some similar but also several distinctive features compared with cutaneous melanoma. Moreover, the tumor mutational profile may be worth evaluating in patients with advanced and/or unresectable disease and high-risk histopathologic findings where neoadjuvant or adjuvant therapy may be considered.
eTable. Clinical Features, Management, and Prognosis of Conjunctival Melanoma Patients
eFigure. Copy Number Plots of Conjunctival Melanomas
References
- 1.Scholz SL, Cosgarea I, Süsskind D, et al. . NF1 mutations in conjunctival melanoma. Br J Cancer. 2018;118(9):1243-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Griewank KG, Westekemper H, Murali R, et al. . Conjunctival melanomas harbor BRAF and NRAS mutations and copy number changes similar to cutaneous and mucosal melanomas. Clin Cancer Res. 2013;19(12):3143-3152. [DOI] [PubMed] [Google Scholar]
- 3.Gear H, Williams H, Kemp EG, Roberts F. BRAF mutations in conjunctival melanoma. Invest Ophthalmol Vis Sci. 2004;45(8):2484-2488. [DOI] [PubMed] [Google Scholar]
- 4.Spendlove HE, Damato BE, Humphreys J, Barker KT, Hiscott PS, Houlston RS. BRAF mutations are detectable in conjunctival but not uveal melanomas. Melanoma Res. 2004;14(6):449-452. [DOI] [PubMed] [Google Scholar]
- 5.Swaminathan SS, Field MG, Sant D, et al. . Molecular characteristics of conjunctival melanoma using whole-exome sequencing. JAMA Ophthalmol. 2017;135(12):1434-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Griewank KG, Murali R, Schilling B, et al. . TERT promoter mutations in ocular melanoma distinguish between conjunctival and uveal tumours. Br J Cancer. 2013;109(2):497-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robinson DR, Wu YM, Lonigro RJ, et al. . Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dagi Glass LR, Lawrence DP, Jakobiec FA, Freitag SK. Conjunctival melanoma responsive to combined systemic BRAF/MEK inhibitors. Ophthalmic Plast Reconstr Surg. 2017;33(5):e114-e116. [DOI] [PubMed] [Google Scholar]
- 9.Maleka A, Åström G, Byström P, Ullenhag GJ. A case report of a patient with metastatic ocular melanoma who experienced a response to treatment with the BRAF inhibitor vemurafenib. BMC Cancer. 2016;16:634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mor JM, Heindl LM. Systemic BRAF/MEK inhibitors as a potential treatment option in metastatic conjunctival melanoma. Ocul Oncol Pathol. 2017;3(2):133-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310(20):2191-2194. [DOI] [PubMed] [Google Scholar]
- 12.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hayward NK, Wilmott JS, Waddell N, et al. . Whole-genome landscapes of major melanoma subtypes. Nature. 2017;545(7653):175-180. [DOI] [PubMed] [Google Scholar]
- 14.Kini A, Fu R, Compton C, Miller DM, Ramasubramanian A. Pembrolizumab for recurrent conjunctival melanoma. JAMA Ophthalmol. 2017;135(8):891-892. [DOI] [PubMed] [Google Scholar]
- 15.Pinto Torres S, André T, Gouveia E, Costa L, Passos MJ. Systemic treatment of metastatic conjunctival melanoma. Case Rep Oncol Med. 2017;2017:4623964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goodman AM, Kato S, Bazhenova L, et al. . Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16(11):2598-2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Clinical Features, Management, and Prognosis of Conjunctival Melanoma Patients
eFigure. Copy Number Plots of Conjunctival Melanomas