Summary
Despite improved methods for MHC affinity prediction, the vast majority of computationally predicted tumor neoantigens are not immunogenic experimentally, indicating that high-quality neoantigens are beyond current algorithms to discern. To enrich for neoantigens with the greatest likelihood of immunogenicity, we developed an analytic method to parse neoantigen quality through rational biological criteria across five clinical datasets for 318 cancer patients. We explored four quality metrics, including analysis of dissimilarity to the non-mutated proteome that was predictive of peptide immunogenicity. In patient tumors, neoantigens with high dissimilarity were unique, enriched for hydrophobic sequences, and correlated with survival after PD-1 checkpoint therapy in patients with non-small cell lung cancer independent of predicted MHC affinity. We incorporated our neoantigen quality analysis methodology into an open source tool, antigen.garnish, to predict immunogenic peptides from bulk computationally predicted neoantigens for which the immunogenic “hit rate” is currently low.
Graphical Abstract

eTOC:
Richman et al. identify dissimilarity to the non-mutated proteome as a predictor of peptide immunogenicity. With this metric implemented in neoantigen quality analysis software, peptide dissimilarity identifies high-quality neoantigens that correlate with survival in clinical datasets.
Introduction
Tumor mutational burden (TMB), defined by non-synonymous single amino acid mutations, correlates with clinical response to immune checkpoint blockade (Cristescu et al., 2018; Samstein et al., 2019; Yarchoan et al., 2017). Although tumor-specific neoantigens derived from these somatic mutations are thought to be the target of antitumor T cell responses mediated by immunotherapy (Carreno et al., 2015; Le et al., 2017; Ott et al., 2017; Tran et al., 2016), in practice, TMB has been a far more commonly used predictor of response than neoantigen burden (Lee et al., 2018; Topalian et al., 2016). To date, direct analysis of neoantigens adds little improvement to prediction of outcomes by TMB, and in only a small number of tumor types. Computational algorithms have been developed for neoantigen identification, but the vast majority of computationally predicted neoantigens are not immunogenic in vivo (González et al., 2018; Sarkizova and Hacohen, 2017; Topalian et al., 2016). Moreover, gene expression signatures of T cell tumor infiltration also correlate with response to checkpoint therapy, but TMB and such signatures themselves are weakly correlated (Cristescu et al., 2018; Spranger et al., 2016). Thus, a gap in understanding hinders the design of personalized immunotherapy that relies on selection of neoantigens from dozens or hundreds a particular tumor putatively expresses.
On the other hand, the prediction and rank order of peptide-MHC binding affinities has become increasingly accurate, and strong MHC affinity is dominant among selection criteria for neoantigen-targeted therapies (Carreno et al., 2015; Keskin et al., 2019; Ott et al., 2017). Other properties that predict peptide immunogenicity, and thus neoantigen quality, have recently emerged. One such quality metric, differential agretopicity index (DAI), is defined as the ratio of MHC affinity of the mutant peptide to MHC affinity of the non-mutated counterpart (Duan et al., 2014; Sercarz et al., 1993). In an analysis of 6,324 patients across 27 cancer types, we found that high DAI neoantigens correlated with patient survival (Rech et al., 2018). Another proposed metric of neoantigen quality involves comparison to known immunogenic peptides, and when combined with DAI and variant clonality, this approach stratified survival in patients with pancreatic ductal adenocarcinoma, melanoma, and non-small-cell lung cancer (NSCLC) (Balachandran et al., 2017; Łuksza et al., 2017). Overall, these studies suggest that criteria in addition to absolute MHC affinity may dictate the likelihood of a predicted neoantigen to drive an antitumor T cell response.
Here, to identify neoantigens with the greatest antitumor potential, we performed ensemble MHC affinity prediction and neoantigen quality analysis on 318 patient samples from five clinical trial datasets. We examined DAI and similarity to known immunogenic peptides, and also investigated the use of dissimilarity to the non-mutated (reference) proteome to discriminate high quality predicted neoantigens. Our quality assessment yielded non-overlapping classes of neoantigens that predict immunogenicity and response to immune checkpoint blockade. We incorporated our approach into an open source tool for neoantigen prediction and quality analysis, antigen.garnish, which is described herein.
Results
Ensemble Approach Improves Outlier MHC Affinity Prediction
antigen.garnish parses input variants to generate all possible neopeptide sequences. After filtering against wild-type sequences, peptides were then further analyzed for quality based on proteome-wide DAI, similarity to known immunogenic epitopes in IEDB (Balachandran et al., 2017; Łuksza et al., 2017; Vita et al., 2015), and dissimilarity to the non-mutated proteome (Figure 1A and Figure S1). Allelic fraction or variant clonality can be incorporated such that the final output of antigen.garnish is a prioritized table of predicted neoantigens. We first compared the ensemble prediction approach used by antigen.garnish with each individual component algorithm using a publicly available database of 167,112 measured peptide-MHC affinities (Kim et al., 2014). Across peptide lengths in the dataset, the accuracy of ensemble prediction was comparable to the highest performing single prediction algorithms measured by Spearman’s rank correlation coefficient (Figure 1B). The antigen.garnish ensemble prediction yielded fewer outlier predictions, demonstrated by the lowest interquartile range for the ratio of predicted MHC-binding affinity to measured values (p < 2.2e-16 for all comparisons to the ensemble method) (Figure 1C). Furthermore, for peptides in the top decile of variance between tools, the ensemble method had the lowest prediction error (p < 1e-9 for all comparisons to the ensemble method) (Figure 1D). These results demonstrate that the antigen.garnish ensemble method maintains accuracy, consistent with single prediction algorithms, and increases precision of affinity predictions.
Figure 1: antigen.garnish Workflow and Validation of Ensemble Neoantigen Prediction Method.
(A) Overview of antigen.garnish workflow. Blue: input data; orange: functions performed by antigen.garnish; red: output data. Dashed lines indicate optional steps.
(B) Bootstrapped Spearman’s rank correlation coefficients for predictions compared to measured affinities in the Kim et al. (2014) dataset for each peptide length. “a.g_ensemble” indicates the antigen.garnish ensemble method.
(C) Ratio of predicted to measured affinity for 9mers in the Kim et al. (2014) dataset. Interquartile range (IQR) for the entire distribution is indicated by the horizontal bracket. The black vertical line indicates the median. A value of 1 indicates perfect prediction. “***” indicates p < 0.001 for the antigen.garnish (a.g) ensemble method compared to all other methods using the post-hoc pairwise Wilcoxon rank sum test with the Bonferroni correction, as determined by bootstrap analysis. The extremes of the distribution outside the axis limits are not shown.
(D) Absolute value of error in predicted affinity as a percentage of measured affinity for peptides in the top decile of variance between tools in the Kim et al. (2014) dataset. “***” indicates p < 0.001 for the antigen.garnish (a.g) ensemble method compared to all other methods using the post-hoc pairwise Wilcoxon rank sum test with the Bonferroni correction.
Dissimilarity to the Non-mutated Proteome Enriches for Immunogenic Peptides
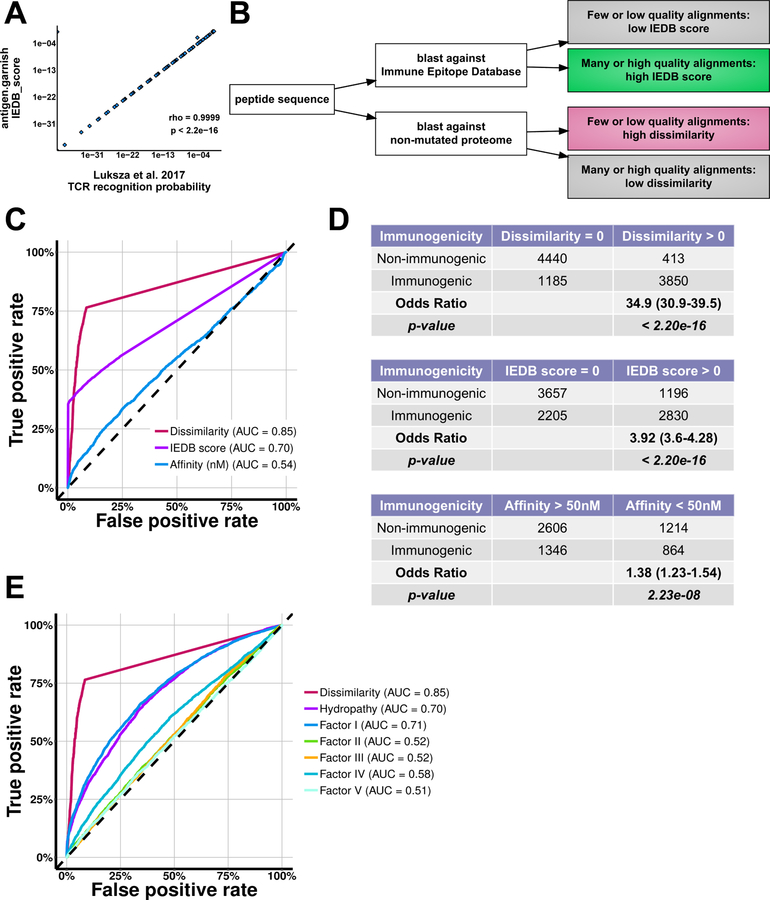
We hypothesized that peptide sequences with high dissimilarity to the non-mutated proteome would be less subject to self-tolerance and therefore more immunogenic. We defined dissimilarity as low sequence alignment of a mutant peptide and its sub-peptides to the non-mutated (reference) proteome. To assess dissimilarity to the non-mutated proteome, we reimplemented the methodology used by Łuksza et al. (2017) to model T cell receptor (TCR) binding energies for homologous sequences (Figure 2A–B). We first assessed if IEDB score, dissimilarity, and predicted affinity could classify immunogenic peptides. Dissimilarity and IEDB scores were calculated for a database of mass spectrometry-confirmed MHC binding peptides with experimentally determined T cell immunogenicity (Chowell et al., 2015). Both IEDB score and dissimilarity discriminated immunogenic peptides in these data (IEDB score: AUC = 0.70; dissimilarity: AUC = 0.85) (Figure 2C–D and S2A). In contrast, ensemble MHC affinity and MHC affinity calculated with individual prediction tools were weak classifiers (AUC = 0.54).
Figure 2: Non-mutated Proteome Dissimilarity Enriches for Immunogenic Peptides.
(A) Correlation between antigen.garnish “IEDB score” computed in the R statistical programming language and “TCR recognition probability” computed using provided Python source code from the complete input data from Łuksza et al. (2017). Spearman’s rho and associated p-value are shown.
(B) Schematic demonstrating the Immune Epitope Database (IEDB) score and dissimilarity metrics.
(C) Receiver-operator characteristic curve for immunogenicity in the Chowell et al. (2015) dataset of peptides with mass spectrometry-confirmed MHC binding. Dissimilarity and IEDB score were computed for all 9,888 unique entries and affinity was determined using the antigen.garnish ensemble method for the 6,050 entries with 4-digit MHC alleles. “AUC” denotes area-under-the-curve.
(D) Contingency tables for non-mutated proteome dissimilarity, IEDB score, and MHC affinity analysis applied to the dataset from Chowell et al. (2015). Odds ratios with confidence intervals and Fisher’s Exact test p-values are shown.
(E) Receiver-operator characteristic curve for classifying immunogenicity in the Chowell et al. (2015) dataset for dissimilarity, mean Kyte-Doolittle Hydropathy, and mean values for the five Atchley et al. (2005) factors. “AUC” denotes area-under-the-curve.
Peptide amino acid properties such as hydropathy can impact TCR interaction and immunogenicity (Chowell et al., 2015). We therefore next evaluated to what extent hydropathy (Kyte and Doolittle, 1982) and Atchley factors, which quantify biochemical properties of amino acids (Atchley et al., 2005), could predict immunogenicity of peptides. Both mean Kyte-Doolittle hydropathy and Atchley factor I, which reflects polarity, showed predictive potential for immunogenicity (AUC = 0.70 and 0.71), yet dissimilarity outperformed both (AUC = 0.85) (Figure 2E). These results suggest that dissimilarity captures distinct peptide properties beyond known biochemical metrics of amino acid sequence.
High Dissimilarity Identifies Molecularly Distinct Neoantigens
We then compared high dissimilarity to other metrics of immunogenicity using published data from five clinical trials of immune checkpoint blockade (Hellmann et al., 2018; Nathanson et al., 2017; Riaz et al., 2017; Rizvi et al., 2015; Snyder et al., 2014; Van Allen et al., 2015). We evaluated: (i) classically defined neoantigens (CDNs, defined by MHC affinity less than 50nM); alternatively defined neoantigens (ADNs, DAI greater than 10) (Rech et al., 2018); (iii) IEDB high neoantigens (IEDB score > 0.9) (Łuksza et al., 2017); and (iv) high dissimilarity neoantigens (dissimilarity > 0.75). We used the accepted minimum binding threshold of 500nM for all neoantigens except CDNs, which are defined by the 50nM threshold. We chose strict cutoffs for IEDB score and dissimilarity based on natural breaks in the distribution of scores (Figure 3A and S2B–C).
Figure 3: Dissimilarity to the Non-mutated Proteome Enriches for Unique Hydrophobic Neoantigens.
(A) Distribution of dissimilarity values for all neoantigens from: Hellmann et al. (2018), Riaz et al. (2017), Rizvi et al. (2015), Snyder et al. (2014), Van Allen et al. (2015). The grey region indicates “high dissimilarity” neoantigens (dissimilarity > 0.75).
(B) Classification of predicted neoantigens from an example patient as classically defined neoantigens (CDNs), alternatively defined neoantigens (ADNs), Immune Epitope Database-homology (IEDB) high neoantigens, and high dissimilarity neoantigens (see methods).
(C) Alignments to the non-mutated proteome for all predicted neoantigens. The median number of alignments by alignment length are shown. Vertical error bars indicate 95% confidence intervals. Global Kruskal-Wallis hypothesis test rejected the null hypothesis for all positions. “***” indicates adjusted p < 0.001 for comparison of high dissimilarity neoantigens to all other groups at each alignment length using post-hoc pairwise Wilcoxon rank sum tests with Bonferroni correction.
(D) Sequence logo analysis of neoantigens predicted to bind to HLA-A*02. All 9mer neoantigens exclusive to a single classification from HLA-A*02 patients were used to calculate sequence consensus. Letter height is proportional to prevalence of the indicated amino acid at that position.
(E) Median Kyte-Doolittle hydropathy at each amino acid position for all neoantigens and control non-binding peptides (predicted MHC affinity 1000–5000nM, “Non-binders”). Positive hydropathy index reflects an enrichment of hydrophobic amino acids. Vertical error bars indicate 95% confidence intervals. Global Kruskal-Wallis hypothesis test rejected the null hypothesis for all positions. “***” indicates adjusted p < 0.001 for comparison of high dissimilarity neoantigens to all other groups at each position using post-hoc pairwise Wilcoxon rank sum tests with Bonferonni correction.
(F) Venn diagram showing overlap between predicted neoantigen classes as a percent of all MHC binders for all neoantigens in the combined 318 patients.
For a sample patient with a tumor mutational burden of 290, a total of 728 peptides were predicted to bind patient MHC, and binding peptides were then stratified by quality criteria (Figure 3B). Across all 318 patient samples, antigen.garnish predicted 291,376 MHC binding putative neoantigens. Of these, 16.9% were CDNs, 10.7% were ADNs, 10.2% were IEDB high neoantigens, and 1.2% were high dissimilarity neoantigens. High dissimilarity neoantigens showed the fewest number of alignments to the self-proteome, as expected (Figure 3C and S3A). Predicted neoantigens meeting each quality criteria showed sequence conservation at the anchor positions 2 and 9 (Figure 3D). Unexpectedly, high dissimilarity neoantigens also showed sequence conservation at all other peptide positions, with a preference for non-polar, hydrophobic amino acids at non-anchor positions 3 to 8 compared to other neoantigen groups (Figure 3D and S3B–D). Increased prevalence of hydrophobic amino acids in high dissimilarity neoantigens from all HLA alleles was reflected by a greater median Kyte-Doolitle hydropathy index at all positions (Figure 3E). Overall, high dissimilarity neoantigens were rare and showed little overlap with other classes (Figure 3F and S3E).
High Dissimilarity Neoantigens Correlate with TMB and Progression-Free Survival after Immune Checkpoint Blockade
To understand the potential clinical relevance of neoantigen quality criteria, we determined the correlation between neoantigen classes, TMB, and progression-free survival (PFS). All neoantigen classes correlated with TMB, as expected (Spearman’s rho = 0.8525 to 0.9488) (Figure 4A). Consistent with Hellmann et al., (2018), we found that median TMB was predictive of PFS (Figure 4B–C). This was true in each NSCLC dataset analyzed separately and after normalizing to the median for each metric and combining datasets (Hellmann: HR = 2.83, 1.29 – 6.18; Rizvi: HR = 5.89, 2.1 – 16.51; Combined: HR = 3.61, 1.97 – 6.61). Notably, unfiltered neoantigens (“all MHC binders”, < 500 nM) did not correlate with PFS in any dataset.
Figure 4: Predicted Neoantigen Classes Correlate with Tumor Mutational Burden and Progression-free Survival.
(A) Correlation of tumor mutational burden with all MHC binders, classically defined neoantigens (CDNs), alternatively defined neoantigens (ADNs), Immune Epitope Database-homology (IEDB) high neoantigens, and high non-mutated proteome dissimilarity neoantigens. The black line shows a linear regression fit, with the 95% confidence interval in grey. Spearman’s rho and associated p-value are shown.
(B–C) Heatmaps of hazard ratios and adjusted logrank test p-values (FDR) for the Cox proportional hazard model for progression-free survival. Patients are stratified at the median for each metric. “Combined NSCLC” is all patients from the Rizvi et al. (2015) and Hellmann et al. (2018) datasets. Comparisons with FDR > 0.05 are shown as empty white tiles.
We then assessed whether the addition of quality metrics could improve the predictive ability of neoantigens for PFS after immune checkpoint blockade. CDNs were the least reliable predictors, only reaching statistical significance in the combined dataset (HR = 2.12, 1.20 – 3.74) (Figure 4B–C). ADNs and IEDB high neoantigens were more predictive, reaching statistical significance in the Rizvi et al. (2015) and combined datasets (ADNs - Rizvi: HR = 2.70, 1.12 – 6.51; Combined: HR = 2.19, 1.24 – 3.88; IEDB high - Rizvi: HR = 2.60, 1.08 – 6.27, Combined: HR = 2.28, 1.28 – 4.04). High dissimilarity neoantigens were predictive of PFS in the Hellmann et al. (2018) and combined datasets (Hellmann: HR = 3.37, 1.36 – 8.34; Combined: HR = 2.73, 1.45 – 5.15). Furthermore, the abundance of all dissimilar neopeptides – independent of MHC binding affinity – predicted PFS in each of the three datasets we evaluated (Hellmann: HR = 3.29, 1.50 – 7.22; Rizvi: HR = 3.40, 1.37 – 8.47, Combined: HR = 3.24, 1.79 – 5.85).
Discussion
Our studies highlight neoantigen quality as a predictor of peptide immunogenicity and clinical outcomes. We validated this approach using peptide-MHC affinity datasets, peptide immunogenicity datasets, and scrutiny of tumor exome variants from five clinical trials of immune checkpoint blockade. Our use of dissimilarity to the non-mutated proteome identified a subset of neoantigens with distinct hydrophobic properties, high likelihood of immunogenicity, and correlation with PFS in patients receiving PD-1 checkpoint blockade. Importantly, the abundance of high dissimilarity neopeptides correlates with PFS independently of predicted MHC affinity. These insights advance us closer to a refined understanding of what is, and what is not, “self” in the tumor genome and offer more precise methods for selection of neoantigens for cancer immunotherapy.
Neoantigens with high dissimilarity had greater prevalence of hydrophobic amino acid residues. Enrichment for immunogenicity by dissimilarity may therefore be understood in light of highly immunogenic intracellular pathogens, such as viruses, which are well-known to have lower GC content that favors hydrophobic amino acids and preferentially triggers TCR recognition (Chowell et al., 2015). Furthermore, at the level of antigen processing machinery, another rate limiting step, exposed hydrophobic residues significantly enhance proteasomal degradation and MHC presentation, suggesting that high dissimilarity neoantigens may be preferentially processed compared to other peptides (Seong and Matzinger, 2004). Notably, analysis of T cell repertoires in human cancer has found that tumor infiltrating lymphocytes are enriched for specificities that recognize hydrophobic epitopes, suggesting that hydrophobic epitopes, such as high dissimilarity neoantigens, may influence the composition of the tumor-immune interface (Li et al., 2016). We found several examples of immunogenic dissimilar neoantigens in mismatch-repair deficient tumors, pancreatic ductal adenocarcinoma, diffuse intrinsic pontine glioma, and vaccine induced responses in glioblastoma multiforme (analyses available online, see Methods) (Balachandran et al., 2017; Chheda et al., 2018; Keskin et al., 2019; Le et al., 2017).
Correlations between responses to immune checkpoint blockade and neoantigens are less reliable than correlations between response and TMB, perhaps because of interaction with additional tumor properties such as T cell gene expression profile (Cristescu et al., 2018). Our results suggest that correlation between clinical benefit and neoantigens could be improved through neoantigen quality criteria. High dissimilarity neoantigens and other quality metrics correlated with survival after PD-1 blockade, while neoantigens defined by MHC binding affinity at two thresholds correlated poorly. The extreme rarity of bona fide immunogenic neoantigens among those predicted has been a barrier to the design of neoantigen vaccines, which have prioritized MHC affinity (Carreno et al., 2015; Keskin et al., 2019; Ott et al., 2017). Because application of these quality criteria logarithmically narrows the scale of prioritized neoantigens from a tumor, our data suggest that the success of vaccines may be improved by integrating neoantigen quality into an immunogenicity classifier to sort the immunogenic “needles” from the bulk peptide “haystack”. In future work, a more sophisticated model could reveal new determinants of immunogenicity for MHC class I and even less well characterized class II antigens (Dhanda et al., 2018).
We developed antigen quality analysis software, antigen.garnish, as an open source tool to prioritize immunogenic neoantigens beyond predicted MHC affinity alone for clinical use. It is written in R to facilitate integration with Bioconductor and other toolsets for bioinformatics (Huber et al., 2015). antigen.garnish is human and murine input compatible and production pipeline ready: transparent, reproducible, fully documented, covered by unit tests, and employs continuous integration. Software is available for download at https://github.com/immune-health/antigen.garnish.
STAR Methods
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Robert H. Vonderheide (rhv@upenn.edu). This study did not generate new unique reagents.
Method Details
antigen.garnish Workflow
antigen.garnish is implemented in the R statistical programming language. Computationally intensive functions are internally parallelized using the ‘mclapply’ function (parallel R package). The main function, ‘garnish_affinity’ performs ensemble MHC affinity prediction and neoantigen quality analysis. Upstream preprocessing functions handle different forms of input and downstream output functions provide sample-level summary tables and plots. Dependencies include the ncbi-BLAST+ command-line tool (Camacho et al., 2009), netMHCI/II and netMHCI/IIpan suite of tools (Andreatta and Nielsen, 2016; Jensen et al., 2018; Nielsen and Andreatta, 2016), MHCnuggets (Bhattacharya et al., 2017), MHCflurry (O’Donnell et al., 2018), and multiple R packages available from the Comprehensive R Archive Network (http://cran.r-project.org) and Bioconductor (Huber et al., 2015). A list of dependencies and references is provided in the Key Resources Table. Full functionality of antigen.garnish requires Linux. A single script installs and configures antigen.garnish. Unit tests are implemented using the ‘testthat’ package for R.
KEY RESOURCES TABLE
Somatic Variant Input Parsing
Input to the main prediction function, ‘garnish_affinity’ can be in the form of VCFs, peptide sequences, cDNA transcripts, or Ensemble transcript IDs and HGVS-style cDNA annotations. For input VCFs, HGVS notation for SNV and indel variants annotated by SnpEff (Cingolani et al., 2012) are parsed for downstream analysis by the ‘garnish_variants’ function. Variants are imported using the vcfR R package (Knaus and Grünwald, 2017) and filtered for SnpEff errors and warnings. For fusion variants, output from JAFFA, an RNA-level fusion prediction algorithm, are parsed by the ‘garnish_jaffa’ function (Davidson et al., 2015). Only high- or medium-confidence fusions that span an exon-exon junction across two genes are kept for neoantigen prediction.
Mutant Sequence Prediction
For annotated input VCFs, gene symbols or Ensembl gene IDs are used to retrieve Ensembl transcript IDs for each variant using the GRCh38 and GRCm38 genome builds, Ensemble release 91 (retrieved on June 20th, 2018 via the biomaRt R package (Durinck et al., 2009)). The HGVS annotation for each variant is used to substitute the mutation and the mutant protein is then translated using the Biostrings R package.
Peptide Generation and Filtering
All 8–15mers are generated from input protein sequences using a sliding window method over the amino acid position of the missense mutation or single amino acid indel, similar to methods previously described (Hundal et al., 2016). For frameshift mutations, the sliding window is shifted and peptide generation is repeated for each sequential residue until reaching a predicted stop site. Fusion sequences are processed using the last amino acid of the N-terminal fusion gene as the anchor for the sliding window. Sequences are checked to ensure that all peptides generated contain at minimum one amino acid from the C-terminal fusion protein.
All peptides derived from input sequences are filtered against the non-mutated (reference) proteome for perfect matches (GRCh38 and GRCm38 Ensemble release 90, retrieved September 27th, 2017). This filter can be disabled to perform antigen quality analysis on wild-type peptide sequences.
Ensemble Prediction Method
MHC affinity prediction performance varies across peptide binding algorithms, especially for MHC alleles and peptide lengths sparsely represented in training data (Bhattacharya et al., 2017). Therefore, to decrease the number of outlier predictions and take advantage of diverse learning approaches for estimating peptide affinity, we integrated MHC prediction models from the NetMHCI/II, netMHCI/IIpan, MHCflurry, and MHCnuggets (GRU and LSTM trained) algorithms (Andreatta and Nielsen, 2016; Bhattacharya et al., 2017; Jensen et al., 2018; Nielsen and Andreatta, 2016; O’Donnell et al., 2018) to produce a single ensemble affinity score derived from all models that support the peptide-MHC pairing. Affinities are averaged to generate the ensemble score. antigen.garnish returns both the ensemble value and individual algorithm prediction affinities from each tool.
Proteome-wide Minimum Differential Agretopicity Calculation
Differential agretopicity calculates the difference in MHC binding affinity for the mutant peptide and the wild-type sequence from which it is derived (Duan et al., 2014; Rech et al., 2018). antigen.garnish expands upon this to determine a proteome-wide minimum differential agretopicity to account for the possibility that a missense mutation results in a peptide that matches elsewhere in the non-mutated proteome. To calculate this value, input sequences are aligned against the non-mutated proteome by BLAST and the closest ungapped alignment is retrieved using the Smith-Waterman algorithm with the BLOSUM62 substitution matrix (Biostrings R package). For every mutant peptide, the proteome-wide peptide with the highest alignment score is then passed to the prediction algorithms to calculate the proteome-wide minimum differential agretopicity. In the case of multiple alignments with the same score, the match with strongest MHC binding affinity is used. ADNs were defined as MHC binders with a proteome-wide DAI > 10 as done previously (Rech et al., 2018).
Immune Epitope Database (IEDB) Homology Analysis
The “TCR recognition probability” component of the immune fitness model of Łuksza et al. (2017) was re-implemented from the original Python source code into the R language. BLASTp and the Biostrings R package were used to perform sequence alignment. BLAST parameters were the same as those used in the original study, permitting multi-residue mismatches (expected with multi-amino acid variants such as fusions, frameshifts, and retained introns) but with high cost to gapped alignments (Camacho et al., 2009). The two model parameters affecting the slope (k) and horizontal displacement (a) were maintained as originally published, ~4.87 and 26 respectively. The resulting value reported by antigen.garnish, IEDB score, demonstrated 1:1 correlation to values produced using the original Python method (Figure 2A). Values range from 0–1, with 1 indicating greater homology to the IEDB database. The cutoff for IEDB high neoantigens was set to IEDB score > 0.9 based on a natural break in the discrete distribution of values from all neoantigens in the five clinical datasets with ensemble MHC affinity score < 500nM (Figure S2B). At this strict clinical cutoff, IEDB score was predictive of immunogenicity in the Chowell et al. (2015) validation dataset (OR = 10.9, 9.59 – 12.5, p < 2.2e-16) (Figure S2C).
Dissimilarity from the Non-mutated Proteome
Mutant peptides were aligned against a database constructed from the non-mutated (reference) proteome (GRCh38 for humans and GRCm38 for mice, Ensemble release 90, retrieved September 27th, 2017) using methods identical to those for IEDB score. Smith-Waterman alignments for each homologous sequence identified by BLAST were then passed to the partition function to substitute for TCR binding energies (Łuksza et al., 2017) which then generated a dissimilarity value for each peptide. The partition function parameter k, the slope of the sigmoidal curve, was kept the same as the value used by Łuksza et al. (2017). The partition function parameter a, which modulates the horizontal displacement of the sigmoidal curve, was set to 32. This value was determined using the method of Łuksza et al. by calculating the average alignment score for all mutant peptide alignments in four datasets (Hellmann et al., 2018; Rizvi et al., 2015; Snyder et al., 2014; Van Allen et al., 2015). This increase in the mean alignment score is expected due to the greater average alignment of self-peptides to the self-proteome compared to alignment of self-peptides to IEDB entries. No additional parameter tuning was explored and no parameter optimization was used. No information from the Chowell et al. (2015) dataset was used to create or optimize our model. The performance of the dissimilarity metric was assessed using Chowell et al. (2015) as a validation set.
The dissimilarity metric ranges from 0–1, with 1 indicating poor alignment to the non-mutated proteome, corresponding to higher dissimilarity. For example, a mutant peptide that poorly aligns to the non-mutated proteome has high dissimilarity. In contrast, a mutant peptide that contains sub-peptides with many non-mutated proteome alignments has low dissimilarity, regardless of whether the sub-peptides contain the mutant amino acid(s). The cutoff for high dissimilarity neoantigens was set to dissimilarity metric > 0.75 based on a natural break in the distribution of values from all neoantigens in the five clinical datasets (Figure 3A) with ensemble MHC affinity score < 500nM. We hypothesized that this restrictive threshold would have greater potential clinical utility by generating a list of the most likely immunogenic peptides, and by better reflecting the oligoclonal nature of anti-tumor T cell responses. At this strict cutoff, dissimilarity was predictive of immunogenicity in the Chowell et al. (2015) dataset (OR = 14.1, 10.3 – 19.6, p < 2.2e-16) (Figure S2C).
Data analysis:
Ensemble Prediction Method Validation
The MHC-peptide affinity data were retrieved from the IEDB website (http://tools.iedb.org/static/main/binding_data_2013.zip, accessed July 2nd, 2018). Only peptide measurements from murine and human MHC alleles with lengths supported by antigen.garnish (8 to 15 amino acids) were included in the analysis. Spearman’s rank correlation coefficient was computed for 2,000 bootstraps of 100 randomly sampled predictions per iteration from each tool. Hypothesis testing for interquartile range of prediction error was performed by bootstrap analysis of 2,000 iterations of 1,000 random peptide-MHC pairs. Interquartile range was chosen because it is robust to outliers. High variance peptide-MHC were selected by computing the variance in MHC affinity prediction for each peptide across all individual tools and taking the top decile. Prediction error was calculated as the absolute value of the ratio of predicted to measured affinity minus one.
Immune Checkpoint Blockade Response Dataset Curation
Survival data and MHC haplotypes for 318 patients were obtained from published databases (Hellmann et al., 2018; Nathanson et al., 2017; Riaz et al., 2017; Rizvi et al., 2015; Van Allen et al., 2015). Variants for the Riaz et al. (2017), Rizvi et al. (2015), Snyder et al. (2014), and Van Allen et al. (2015) studies were assembled from published supplementary tables. Variant calls from Hellman et al. (2018) were retrieved in VCF format from the European Variation Archive (https://www.ebi.ac.uk/eva/), and variants with an alternate allele count of less than 15 were filtered out, consistent with the original methods.
Tumor mutational burden was determined as the total number of non-synonymous variants using SnpEff annotations, with the exception of the Riaz et al. (2017) dataset, for which TMB was determined using the published variant table. Default antigen.garnish settings were used for neoantigen prediction, including removal of any peptides with identical non-mutated proteome matches.
Quantification and Statistical Analysis
Non-mutated Proteome Alignment Analysis
All predicted neoantigens exclusive to a single classification were aligned to a database constructed from the non-mutated proteome using BLASTp. BLASTp parameters were the same as those published by Łuksza et al. (2017). Ungapped alignments meeting the (E)-value cutoff for each neoantigen were retrieved. Alignment lengths greater than or equal to 6 were found to be the shortest alignments that frequently met the BLAST algorithm (E)-value threshold, therefore a length of 6 was set as a minimum.
Sequence Logo Analysis
All predicted 9mer neoantigens exclusive to a single classification from patients with HLA-A*02, HLA-A*68, HLA-B*07, or HLA-C*12 were included in the analysis. The gglogo R package was used to generate the sequence logo diagrams and a bitscore was generated for each amino acid position.
Hydropathy Analysis
Kyte-Doolittle hydropathy indices (Kyte and Doolittle, 1982) were used to calculate hydropathy for each position for all 9-mer peptides that were exclusive to a single classification. Bootstrapped subsampling was performed to account for differences in the total number of neoantigens for each classification. The median hydropathy was calculated for 2,000 samples of 298 neoantigens from each group. Hypothesis testing was performed on the bootstrapped median hydropathy values.
Peptide Immunogenicity Analysis
Non-mutated proteome dissimilarity and IEDB scores were calculated for all peptides using the Chowell et al. (2015) peptide dataset. MHC affinity was predicted with antigen.garnish for entries that provided four-digit HLA types (6,050 out of of 9,888 peptides). Hypothesis testing for the 2 × 2 contingency tables was performed using Fisher’s Exact test and a p-value and odds ratio confidence interval were calculated using 2,000 bootstraps as implemented in the ‘fisher.test’ R stats package function. Receiver-operator characteristic curves and AUC for MHC affinity, IEDB score, dissimilarity, mean peptide Kyte-Doolittle hydropathy, and mean values across the peptide for each of the five Atchley factors were computed using the “pROC” package (Robin et al., 2011). Contingency tables for both the strict cutoffs (to maximize clinical utility) and permissive cutoffs (to increase sensitivity) are shown (Figure 2D and Figure S2C).
Survival Analysis
Patients were stratified into high and low groups by the median value for each metric of interest, as previously described (Hellmann et al., 2018). PFS was used for Rizvi et al. (2015) and Hellmann et al. (2018). PFS was chosen instead of overall survival because it is less likely to be confounded by additional therapies (Sobrero et al., 2008; Van Cutsem et al., 2009). Hazard ratios (low:high) and log-rank p-values for survival analysis were calculated using the Cox proportional hazards model, as implemented in the survival R package ‘coxph’ function (Therneau and Grambsch, 2000). In the heatmap representations of false discovery rate, multiple comparisons were Benjamini-Hochberg corrected by passing the log-rank test p-values within each column to the ‘p.adjust’ function from the stats R package. For analysis of combined datasets, survival was recalculated relative to the median survival within each dataset, and TMB and neoantigen metrics were normalized to the median within each dataset to account for differences in variant calling methodologies before combination. CDNs, ADNs, IEDB high, and high dissimilarity neoantigens were classified as indicated in the methods above. The total number of dissimilar neopeptides (“all dissimilar neopeptides”) were determined by generating all possible 9-amino acid long neopeptides from input variants and computing the total number of neopeptides with dissimilarity > 0.75.
Software benchmarking
Benchmarking was performed using the “microbenchmark” R package using an Amazon Web Services T3.2xLarge instance (8-core Intel Xeon Platinum 8000 series, 32GB RAM). One hundred bootstraps of 10 randomly selected input variants from the Hellmann et al. (2018) dataset were passed to the ìgarnish_affinityè function with and without including the ìedb_scorè and ìgarnish_dissimilarityè functions. Ensemble MHC affinity prediction was performed for the HLA-A*02:01 allele. The number of neopeptides analyzed per second were computed for each condition.
Statistical Analysis and Data Visualization
The following R packages were used for statistical analysis and data visualization: data.table (data exploration), stats (statistical tests), ggplot2 (graphics), pheatmap (heatmaps), VennDiagram (Venn diagrams), DiagrammeR (flowcharts), gglogo (sequence logo diagrams), pROC (receiver-operator characteristics).
Supplementary Material
Highlights:
High dissimilarity to self identifies a unique set of predicted neoantigens
Dissimilarity to the self-proteome predicts peptide immunogenicity
Dissimilarity and other metrics of neoantigen quality predict clinical outcomes
antigen.garnish is an open-source R package for neoantigen quality analysis
Acknowledgements
This work was supported by NIH grants R01 CA229803 and P30 CA016520 (to R.H.V.) and the Parker Institute for Cancer Immunotherapy (to R.H.V. and A.J.R.). We gratefully acknowledge Drs. David Balli, Katelyn T. Byrne, Beatriz M. Carreno, Gerald P. Linette, and Alexander P. Morrison for helpful discussions.
R.H.V. reports having received consulting fees or honoraria from Apexigen, AstraZeneca, Celgene, Genentech, Janssen, Lilly, Medimmune, Merck and Verastem; he has received research funding from Apexigen, FibroGen, Inovio, Janssen, and Lilly.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
L.P.R. and A.J.R. declare no competing interests.
Data and Code Availability
Input data and source code for data analysis and figure generation are available at https://github.com/leeprichman/Richman_2019_Cell_Systems (DOI: 10.5281/zenodo.3353687). Releases, open source code, and one-line installation instructions for antigen.garnish are available at https://github.com/immune-health/antigen.garnish (DOI: 10.5281/zenodo.3358290).
References
- Andreatta M, and Nielsen M (2016). Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 32, 511–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atchley WR, Zhao J, Fernandes AD, and Drüke T (2005). Solving the protein sequence metric problem. Proc. Natl. Acad. Sci. U.S.A 102, 6395–6400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, et al. (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551, 512–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya R, Sivakumar A, Tokheim C, Guthrie VB, Anagnostou V, Velculescu VE, and Karchin R (2017). Evaluation of machine learning methods to predict peptide binding to MHC Class I proteins. BioRxiv 154757.
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie W-R, Hildebrand WH, Mardis ER, et al. (2015). A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348, 803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chheda ZS, Kohanbash G, Okada K, Jahan N, Sidney J, Pecoraro M, Yang X, Carrera DA, Downey KM, Shrivastav S, et al. (2018). Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. J. Exp. Med 215, 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowell D, Krishna S, Becker PD, Cocita C, Shu J, Tan X, Greenberg PD, Klavinskis LS, Blattman JN, and Anderson KS (2015). TCR contact residue hydrophobicity is a hallmark of immunogenic CD8+ T cell epitopes. Proc. Natl. Acad. Sci. U.S.A 112, E1754–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, Land SJ, Lu X, and Ruden DM (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, Sher X, Liu XQ, Lu H, Nebozhyn M, et al. (2018). Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 362, eaar3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson NM, Majewski IJ, and Oshlack A (2015). JAFFA: High sensitivity transcriptome-focused fusion gene detection. Genome Medicine 7, 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhanda SK, Karosiene E, Edwards L, Grifoni A, Paul S, Andreatta M, Weiskopf D, Sidney J, Nielsen M, Peters B, et al. (2018). Predicting HLA CD4 Immunogenicity in Human Populations. Front Immunol 9, 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan F, Duitama J, Al Seesi S, Ayres CM, Corcelli SA, Pawashe AP, Blanchard T, McMahon D, Sidney J, Sette A, et al. (2014). Genomic and bioinformatic profiling of mutational neoepitopes reveals new rules to predict anticancer immunogenicity. J. Exp. Med 211, 2231–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durinck S, Spellman PT, Birney E, and Huber W (2009). Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nature Protocols 4, 1184–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González S, Volkova N, Beer P, and Gerstung M (2018). Immuno-oncology from the perspective of somatic evolution. Semin. Cancer Biol 52, 75–85. [DOI] [PubMed] [Google Scholar]
- Hellmann MD, Nathanson T, Rizvi H, Creelan BC, Sanchez-Vega F, Ahuja A, Ni A, Novik JB, Mangarin LMB, Abu-Akeel M, et al. (2018). Genomic features of response to combination immunotherapy in patients with advanced non-small-cell lung cancer. Cancer Cell 33, 843–852.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. (2015). Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, and Griffith M (2016). pVAC-Seq: A genome-guided in silico approach to identifying tumor neoantigens. Genome Med 8, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, and Nielsen M (2018). Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keskin DB, Anandappa AJ, Sun J, Tirosh I, Mathewson ND, Li S, Oliveira G, Giobbie-Hurder A, Felt K, Gjini E, et al. (2019). Neoantigen vaccine generates intratumoral T cell responses in phase Ib glioblastoma trial. Nature 565, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Sidney J, Buus S, Sette A, Nielsen M, and Peters B (2014). Dataset size and composition impact the reliability of performance benchmarks for peptide-MHC binding predictions. BMC Bioinformatics 15, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus BJ, and Grünwald NJ (2017). vcfR: a package to manipulate and visualize variant call format data in R. Molecular Ecology Resources 17, 44–53. [DOI] [PubMed] [Google Scholar]
- Kyte J, and Doolittle RF (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Le DT, Durham JN, Smith KN, Wang H, Bartlett BR, Aulakh LK, Lu S, Kemberling H, Wilt C, Luber BS, et al. (2017). Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science 357, 409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C-H, Yelensky R, Jooss K, and Chan TA (2018). Update on tumor neoantigens and their utility: why it is good to be different. Trends Immunol 39, 536–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Li T, Pignon J-C, Wang B, Wang J, Shukla SA, Dou R, Chen Q, Hodi FS, Choueiri TK, et al. (2016). Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat. Genet 48, 725–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łuksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, et al. (2017). A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551, 517–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson T, Ahuja A, Rubinsteyn A, Aksoy BA, Hellmann MD, Miao D, Van Allen E, Merghoub T, Wolchok JD, Snyder A, et al. (2017). Somatic mutations and neoepitope homology in melanomas treated with CTLA-4 blockade. Cancer Immunol Res 5, 84–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen M, and Andreatta M (2016). NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Medicine 8, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell TJ, Rubinsteyn A, Bonsack M, Riemer AB, Laserson U, and Hammerbacher J (2018). MHCflurry: Open-source Class I MHC binding affinity prediction. Cell Systems 7, 129–132.e4. [DOI] [PubMed] [Google Scholar]
- Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, et al. (2017). An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547, 217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rech AJ, Balli D, Mantero A, Ishwaran H, Nathanson KL, Stanger BZ, and Vonderheide RH (2018). Tumor immunity and survival as a function of alternative neopeptides in human cancer. Cancer Immunol Res 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, et al. (2017). Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171, 934–949.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. (2015). Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348, 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez J-C, and Müller M (2011). pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 12, 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samstein RM, Lee C-H, Shoushtari AN, Hellmann MD, Shen R, Janjigian YY, Barron DA, Zehir A, Jordan EJ, Omuro A, et al. (2019). Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet 51, 202–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkizova S, and Hacohen N (2017). How T cells spot tumour cells. Nature 551, 444–446. [DOI] [PubMed] [Google Scholar]
- Seong S-Y, and Matzinger P (2004). Hydrophobicity: an ancient damage-associated molecular pattern that initiates innate immune responses. Nat. Rev. Immunol 4, 469–478. [DOI] [PubMed] [Google Scholar]
- Sercarz EE, Lehmann PV, Ametani A, Benichou G, Miller A, and Moudgil K (1993). Dominance and crypticity of T cell antigenic determinants. Annu. Rev. Immunol 11, 729–766. [DOI] [PubMed] [Google Scholar]
- Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, et al. (2014). Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med 371, 2189–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobrero AF, Maurel J, Fehrenbacher L, Scheithauer W, Abubakr YA, Lutz MP, Vega-Villegas ME, Eng C, Steinhauer EU, Prausova J, et al. (2008). EPIC: phase III trial of cetuximab plus irinotecan after fluoropyrimidine and oxaliplatin failure in patients with metastatic colorectal cancer. J. Clin. Oncol 26, 2311–2319. [DOI] [PubMed] [Google Scholar]
- Spranger S, Luke JJ, Bao R, Zha Y, Hernandez KM, Li Y, Gajewski AP, Andrade J, and Gajewski TF (2016). Density of immunogenic antigens does not explain the presence or absence of the T-cell-inflamed tumor microenvironment in melanoma. Proc. Natl. Acad. Sci. U.S.A 113, E7759–E7768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Therneau TM, and Grambsch PM (2000). Modeling survival data: extending the Cox model (New York: Springer; ). [Google Scholar]
- Topalian SL, Taube JM, Anders RA, and Pardoll DM (2016). Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat. Rev. Cancer 16, 275–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E, Robbins PF, Lu Y-C, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, et al. (2016). T-Cell transfer therapy targeting mutant KRAS in cancer. N. Engl. J. Med 375, 2255–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, et al. (2015). Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350, 207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Cutsem E, Köhne C-H, Hitre E, Zaluski J, Chang Chien C-R, Makhson A, D’Haens G, Pintér T, Lim R, Bodoky G, et al. (2009). Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N. Engl. J. Med 360, 1408–1417. [DOI] [PubMed] [Google Scholar]
- Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, et al. (2015). The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43, D405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarchoan M, Johnson BA, Lutz ER, Laheru DA, and Jaffee EM (2017). Targeting neoantigens to augment antitumour immunity. Nat. Rev. Cancer 17, 209–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.