Abstract
Lanthanide (Ln)-dependent methanol dehydrogenases (MDHs) have been recently shown to be widespread in methylotrophic bacteria. Along with the core MDH protein, XoxF, these systems comprise two other proteins, XoxG (a c-type cytochrome) and XoxJ (a periplasmic binding protein of unknown function), about which little is known. Here, we biochemically and structurally characterize these proteins from the methyltroph, Methylobacterium extorquens AM1. In contrast to results obtained via an artificial assay system, assays of XoxFs metallated with LaIII, CeIII, and NdIII using their physiological electron acceptor, XoxG, display Ln-independent activities, but the Km for XoxG markedly increases from La to Nd. This result suggests that XoxG’s redox properties are tuned specifically for lighter Lns in XoxF, an interpretation supported by the unusually low reduction potential of XoxG (+172 mV). The x-ray crystal structure of XoxG provides a structural basis for this reduction potential and insight into the XoxG-XoxF interaction. Finally, the x-ray crystal structure of XoxJ reveals a large hydrophobic cleft and suggests a role in activation of XoxF. These studies enrich our understanding of the underlying chemical principles that enable the activity of XoxF with multiple lanthanides in vitro and in vivo.
Keywords: cytochrome c, lanthanides, methanol dehydrogenase, rare earths, reduction potential
Table of contents
Methylotrophic bacteria can grow on light, but not heavy, lanthanides by using a lanthanide-dependent methanol dehydrogenase, but the chemical basis for this preference is incompletely understood. Through characterization of this enzyme’s partner proteins, we show that the redox properties of the system are specifically tuned for utilization of light lanthanides.
INTRODUCTION
Methanol dehydrogenases (MDHs) are abundant periplasmic enzymes in methylotrophic bacteria, catalyzing the oxidation of methanol to formaldehyde and enabling use of methane and/or methanol as these organisms’ sole carbon source.[1] The active site of the canonical MDH, MxaFI, contains a CaII ion coordinated to a pyrroloquinoline quinone (PQQ) cofactor.[1b, 2] The CaII ion facilitates activation of the PQQ cofactor for the formal hydride transfer reaction from methanol to PQQ.[1b, 3] The electrons extracted from methanol by this cofactor are transferred to the respiratory complex via a relay of two c-type cytochromes: a cytochrome cL (MxaG) interacts directly with the MDH and transfers electrons to cytochrome cH (Scheme 1A).[1b, 4] Recently, it was discovered that many methylotrophs also encode MDHs[5] and other PQQ-dependent alcohol dehydrogenases[6] that utilize a lanthanide ion (LnIII) rather than calcium in their active sites. The Ln-dependent MDH, XoxF, contains an analogous active site, except with the PQQ cofactor coordinated to a LnIII ion and an additional carboxylate ligand to the metal ion.[5b, 7] Presumably, the greater Lewis acidity of LnIII ions over that of CaII is advantageous for increasing the reduction potential of PQQ to facilitate this reaction.[6d, 8]
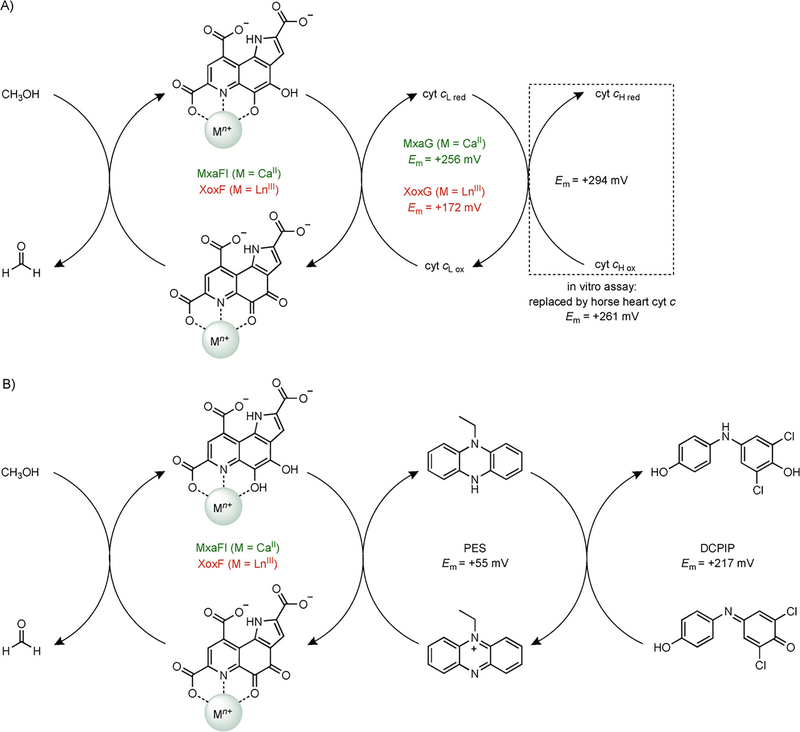
Scheme 1.
In vivo and in vitro oxidizing systems required for MDH activity. (A) Physiological pathway for electron flow from methanol to cytochrome cH in Ca- and Ln-dependent MDHs, and the protein-linked assay system for MDH activity measurements. Electron transfer from XoxG to cytochrome cH is assumed based on analogy to MxaG but has not yet been demonstrated in vitro. The in vitro assay monitors cytochrome c reduction at 550 nm. The M. extorquens XoxG reduction potential (Em) is determined in the present work. (B) Dye-linked assay system for MDH activity measurements, adapted from ref. [8d]. The assay monitors DCPIP reduction at 600 nm.
An unusual feature of the characterized Ln-dependent enzymes, relative to most other metalloenzymes, is that multiple metal ions are able to support their activity. Accordingly, there is significant interest in understanding the chemical underpinnings of the preference that these enzymes, as well as the bacteria that encode them, exhibit for the more abundant, lighter Lns – especially La, Ce, Pr, and Nd – over the subsequent less abundant, heavier Lns, Sm-Lu. The systems for which the most information is currently available are Methylacidiphilum fumariolicum SolV and Methylobacterium extorquens AM1. The first detailed biochemical and structural characterization of a Ln-dependent MDH was the XoxF from M. fumariolicum,[5b] an organism that requires Lns for growth, with La-Nd preferred, although Sm, Eu, and Gd can also support weak growth. Further studies[8d, 9] demonstrated that this XoxF had maximal activity in the standard artificial dye-linked assay (Scheme 1B) when metallated with Pr and Nd. Activity was ~30% lower with La and fell off quickly beyond Nd. This biphasic behavior was attributed to competition between the Lewis acidity of the LnIII ion, increasing across the series and therefore enhancing reactivity of the PQQ cofactor, with other, opposing factors. These other factors may include coordination number preferences and substrate binding affinity but are poorly understood.[8d]
For M. extorquens, the subject of the present study, it was shown[10] that only La-Nd, and Sm to a lesser extent, supported Ln-dependent growth on methanol in the absence of the Ca-MDH. The same series of Lns selectively activated the xox promoter. At the time, it was unclear whether this effect was due to sub-optimal cellular uptake of heavy Lns, diminished activity of XoxF with heavy Lns, or both. Recently, we demonstrated, using a genetically encoded fluorescent sensor for Lns,[11] that only light Lns, La-Nd, are efficiently imported into the bacterial cytosol, a process likely facilitated by a secreted metallophore.[11c] Therefore, it appears that on multiple levels – including, at least, regulation,[10] uptake,[11c, 12] and trafficking,[11a] – M. extorquens is fine-tuned for utilization of solely light Lns.
An understudied aspect of Ln-dependent alcohol dehydrogenases is the involvement of partner proteins in their activity inside the cell. PQQ-dependent alcohol dehydrogenases such as XoxF and MxaFI are found in operons[6d, 13] along with genes encoding the cytochrome cL (XoxG and MxaG, respectively) and a protein of unknown function (XoxJ and MxaJ, respectively). MxaJ was recently characterized as a member of the periplasmic binding protein (PBP) family – proteins typically associated with membrane-bound transport systems for small molecules – but its substrate is unknown.[14] Deletion of mxaJ disrupts activation of the Ca-MDH, suggesting a role for MxaJ in this process, which is not well understood in any MDH system.[15] By extension, we presume that XoxJ plays an analogous role in Ln-MDH activation. MxaG is the electron acceptor[4c] for MxaFI.[4a, 4b, 16] Recently, Chistoserdova and co-workers[17] reported the first purification of a XoxG protein, from Methylomonas sp. LW13, and showed that it has an absorption spectrum consistent with bound heme and that it can act as the electron acceptor for La-XoxF, but no detailed biochemical or kinetic characterization was performed.
We set out to provide a physiological context for understanding the activity of M. extorquens XoxF metallated with different Lns, by characterizing the other proteins encoded by the xox operon. We focused on the effect of LnIII coordination on the midpoint reduction potential (Em) of the PQQ cofactor in XoxF. This property is expected to increase as a function of LnIII Lewis acidity, via enhanced withdrawal of electron density from the reactive C5 carbonyl of the cofactor. Lewis acidity increases left to right across the period due to decreasing ionic radius,[18] but we presume that XoxG’s reduction potential is relatively fixed. Therefore, after a certain point in the Ln series, XoxG may become a progressively less efficient oxidant for the reduced LnIII-PQQ cofactor, providing a possible chemical rationale for the unsuitability of Lns beyond Nd in vivo. To test this hypothesis, we characterized XoxG and its interaction with XoxF. We show that the Vmax parameters of XoxFs metallated with La, Ce, and Nd are similar when assayed with XoxG. However, the Km for XoxG increases sharply from La-XoxF to Nd-XoxF. Biochemical characterization of XoxG reveals a surprisingly low Em for a c-type cytochrome, and structural characterization shows that, compared to MxaG, the heme binding site is significantly more solvent exposed. These findings may account for the observed redox properties and modulation of Km. We also complete the characterization of the xox operon by reporting the first biochemical and structural studies of a XoxJ protein, and we discuss potential functions in activation of XoxF, about which little is currently known. These studies yield insight into the selectivity of Ln-utilizing bacteria for light Lns and emphasize the importance of understanding enzymatic systems in the context of their physiological partners.
RESULTS
Kinetic characterization of XoxF metallated with La, Ce, and Nd
XoxF was purified from its native levels as described, by ammonium sulfate precipitation, cation exchange chromatography, and size exclusion chromatography (SEC),[11a] from M. extorquens AM1 cultures grown in the presence of 1 μM LaCl3, CeCl3, or NdCl3 (Figure S1). The UV-visible spectra of these proteins are shown in Figure S2, with the characteristic absorption band at ~360 nm and a shoulder at ~410 nm indicative of the metal-bound PQQ cofactor.[1b, 5b, 6a, 6b, 11a, 15b, 19] The protein yields, metal contents, and A280nm/A359nm ratios for each XoxF were similar (Table S1), indicating similar cofactor loading.
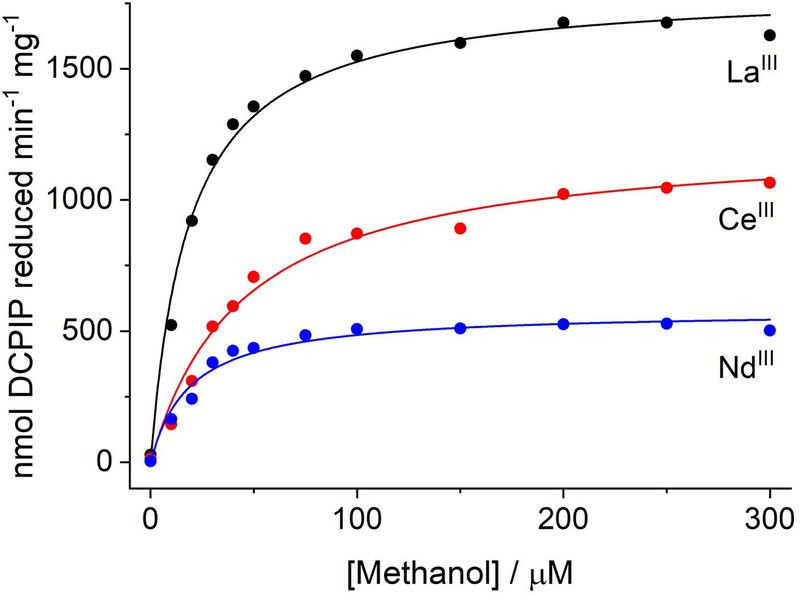
The most commonly used assay for MDH activity[20] spectrophotometrically monitors the reduction of 2,6-dichlorophenolindophenol (DCPIP, Em = +217 mV at pH 7.0, 25 °C[21]) by the MDH, with either phenazine ethosulfate (PES, Em = +55 mV) or phenazine methosulfate (PMS, Em = +80 mV) as mediator (Scheme 1B).[21] Using this dye-linked assay system at pH 7.0, we characterized the La-, Ce-, and Nd-XoxFs (Figure 1, Table 1). Our results are similar to a previous report of M. extorquens AM1 XoxF overexpressed and metallated with La and Nd, [19b] with Nd-XoxF exhibiting ~50% the maximum velocity (Vmax) as La-XoxF, although this observation correlated with lower metal content of the Nd-enzyme. (Unfortunately, these activities cannot be compared with those obtained for Ca-MDH using the dye-linked assay, as that assay must be carried out at pH 9 and with ammonia to accelerate the reaction.[4a]) We did not observe a significant difference in Km values for methanol for the three enzymes, but the higher kcat of La-XoxF resulted in a larger kcat/Km value. The findings from both studies on the M. extorquens enzyme are distinct from those in M. fumariolicum XoxF using the same assay. In the latter system, Pr and Nd enable highest activity, ~50% higher than that with La. However, these samples were grown with EuIII and rely on in vitro metal substitution by assaying in the presence of micromolar levels of added LnIII.[8d] Consequently, the differences between M. extorquens and M. fumariolicum XoxF activities may reflect distinct preferences for Lns or differences in extent of protein metalation. Additionally, the physiological insights into activity differences of different Ln-XoxFs that can be obtained with this artificial assay system are limited, motivating our efforts to measure XoxF activity in the presence of its physiological electron acceptor, XoxG.
Figure 1.
Representative Michaelis-Menten curves for La-XoxF (black), Ce-XoxF (red), and Nd-XoxF (blue), assayed with methanol using the dye-linked activity assay.
Table 1.
Kinetic parameters for M. extorquens La-, Ce-, and Nd-XoxF, determined using the dye-linked activity assay at 30 °C. Mean ± SD for 3 (La, Ce) or 2 (Nd) independent experiments.
| La-XoxF | Ce-XoxF | Nd-XoxF | |
|---|---|---|---|
| Vmax (nmol min−1 mg−1) | 1860 ± 110 | 1290 ± 100 | 560 ± 30 |
| Km, methanol (μM) | 22 ± 3 | 49 ± 11 | 18 ± 5 |
| kcat (s−1) | 4.0 ± 0.2 | 2.8 ± 0.2 | 1.2 ± 0.1 |
| kcat/Km (μM−1 s−1) | 0.19 ± 0.03 | 0.057 ± 0.014 | 0.067 ± 0.019 |
XoxG exhibits an unusually low reduction potential
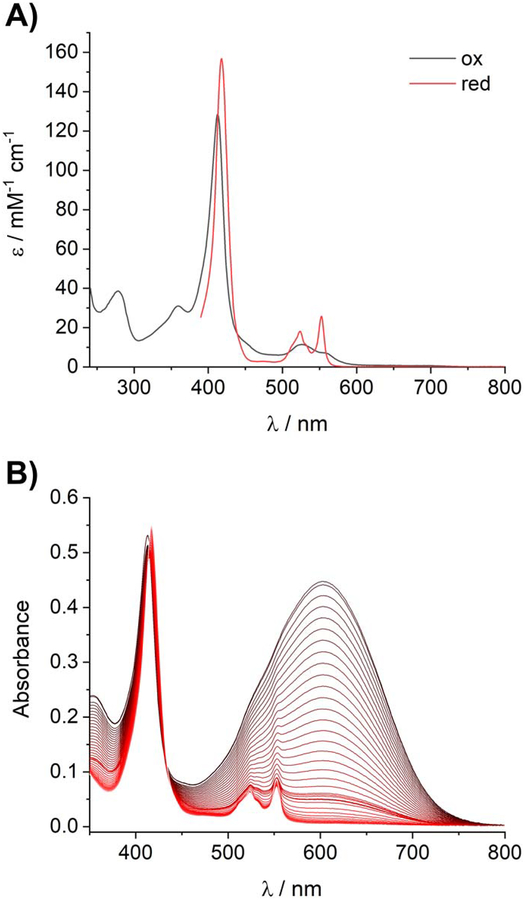
We heterologously expressed XoxG in E. coli in the presence of a plasmid encoding cytochrome c maturation proteins[22] and purified the protein from the periplasmic fraction (Figure S3). The N-terminal residue of XoxG was Gln27, following cleavage of the periplasmic signal peptide (Figure S4). As expected, XoxG purified with bound heme and displayed UV-visible absorption features characteristic of a c-type cytochrome (Figure 2A), including a weak band at 695 nm indicative of methionine coordination to the ferric heme[4b, 23] (Figure S5). The Em of XoxG at pH 7.0 was determined by spectrophotometric titration according to the method of Massey[24] using DCPIP as the reference dye and the xanthine/xanthine oxidase system as the source of reducing equivalents.[25] The Em of XoxG was measured to be 172 ± 1 mV (Figure 2B, Figure S6). This reduction potential is significantly lower than that for the corresponding cytochrome cL, MxaG, of the M. extorquens Ca-MDH, +256 mV.[4b] The measured value for XoxG is also relatively low for a class I c-type cytochrome with His/Met ligation (typically +200 to +370 mV) as well as for other cytochromes cL.[4c, 26] This result was not anticipated, as we had initially expected that the greater Lewis acidity of LnIIIs in the MDH active site versus CaII – and therefore, putatively a higher MDH Em – would necessitate a higher potential for the cytochrome cL. To our knowledge, the reduction potential of Ca-MDH or Ln-MDHs has not been reported, but the value for free PQQ is +90 mV.[27]
Figure 2.
Spectroscopic and redox characterization of XoxG. (A) UV-visible absorption spectra of oxidized XoxG and dithionite-reduced XoxG. (B) Spectrophotometric determination of the reduction potential of XoxG. A solution of oxidized XoxG and DCPIP was reduced by xanthine/xanthine oxidase over ~1 h (black to red lines: spectra acquired every 1 min, oxidized to reduced). Sharp features associated with reduced XoxG in the 520–560 range do not appear until late in the titration, showing that Em of XoxG is significantly lower than that of DCPIP. Data analysis is shown in Figure S6.
Kinetic characterization of Ln-XoxFs with XoxG as electron acceptor
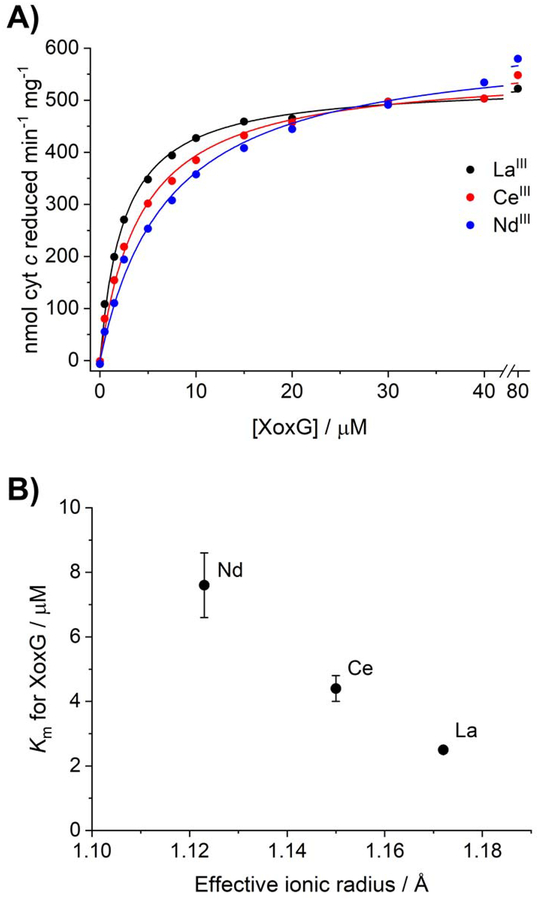
We hypothesized that the reduction potential of XoxG may be specifically optimized for transfer of electrons from PQQ, bound to lighter LnIIIs, to the cytochrome. To understand the efficiency of this step in the reaction, we evaluated kcat and Km for La-, Ce-, and Nd-XoxF proteins using XoxG as electron acceptor. While the analogous experiment using MxaG and the Ca-MDH (which can be carried out at pH 7, unlike the dye-linked assays) has been reported,[4a, 20] there is no report of such a full characterization with XoxG and Ln-XoxFs to our knowledge (while this manuscript was under review, biochemical characterization of the M. fumariolicum cytochrome cL was reported,[28] see Discussion). In this assay (Scheme 1A), reduction of the cytochrome cL is coupled to reduction of horse heart cytochrome c (Em = +261 mV[25]), which itself is unable to directly interact with the MDH. The results of this assay (Figure 3A, Table 2) were distinct from those obtained with the Ln-XoxFs using the dye-linked system. Whereas the Vmax was not significantly different for all three Ln-XoxFs, the Km for XoxG increased three-fold from La to Nd (Figure 3B, decreasing ionic radius being a proxy for increasing Lewis acidity). Both the Vmax and Km for XoxG of La-XoxF are comparable to those of MxaFI assayed with the MxaG/cytochrome c system (800 nmol min−1 mg−1 and 3 μM, respectively).[4a] With the exception of Nd, the overall Vmax values were generally lower for XoxFs assayed with XoxG than with the dye-linked system. Together, these data suggest that the dye-linked and XoxG-linked activity assays are measuring different aspects of XoxF turnover, and that a step involving XoxG (bypassed by the dye-linked assay) is likely rate-limiting for methanol reduction in vitro and presumably also inside the cell. Therefore, the XoxG Km is important for understanding the Ln-dependence of XoxF activity in vivo. We propose that the increasing Kms for XoxG reflect elevated reduction potentials of the LnIII-PQQ cofactor as Lewis acidity of the lanthanide ion increases from LaIII to NdIII (see Discussion).
Figure 3.
Km, but not Vmax, values of XoxF proteins assayed with XoxG are Ln-dependent. (A) Representative Michaelis-Menten plots of La-XoxF (black), Ce-XoxF (red), and Nd-XoxF (blue), with 20 mM methanol as substrate and assayed in the presence of XoxG and horse heart cytochrome c. (B) Km of XoxF for XoxG versus ionic radius[29] of LnIII.
Table 2.
Kinetic parameters for M. extorquens La-, Ce-, and Nd-XoxF, determined using the XoxG/cyt c-linked activity assay at 30 °C (mean ± SD for 3 independent experiments).
| La-XoxF | Ce-XoxF | Nd-XoxF | |
|---|---|---|---|
| Vmax (nmol min−1 mg−1) | 570 ± 30 | 560 ± 10 | 610 ± 20 |
| Km, XoxG (μM) | 2.5 ± 0.1 | 4.4 ± 0.4 | 7.6 ± 1.0 |
| kcat (s−1) | 1.2 ± 0.1 | 1.2 ± 0.1 | 1.3 ± 0.1 |
| kcat/Km (μM−1 s−1) | 0.49 ± 0.03 | 0.28 ± 0.03 | 0.17 ± 0.02 |
Structural characterization of XoxG
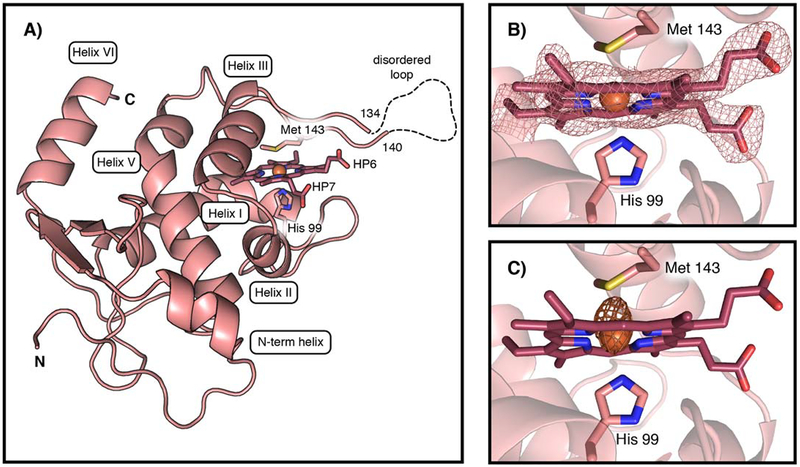
These biochemical observations led us to structurally characterize XoxG, in an effort to understand XoxG’s unusual redox properties and their impact on physiological methanol oxidation. The x-ray crystal structure of XoxG, solved to 2.71 Å resolution, reveals structural features[26, 30] conserved across class I cytochromes c (Figure 4A, Table S2 for data collection and refinement statistics). The heme c moiety, which is covalently attached to the protein through two thioether bonds to C95 and C98 via the signature CXXCH motif for heme attachment, is enclosed in a hydrophobic pocket formed by three core α-helices (helices I, III, and V). This binding motif leaves one of the heme edges open to solvent (Figure S7 for structure comparisons with other cytochromes c).[16, 31] Typical of most class I c-type cytochromes,[26] the FeIII is axially ligated by a His residue contributed by helix I (H99) and a Met residue from the loop between helices III and V (M143) (Figure 4B,C). Unlike most other class I cytochromes c,[16] XoxG lacks helix IV (Figure 4A), and this region is instead a 19-residue loop, the end of which is partially disordered (residues 135–139 were not modeled). MxaG also possesses a loop in place of helix IV, although it is significantly shorter (13 residues) and fully modeled in the reported x-ray structure (Figure S8). The function of the extended loop in XoxG is unknown, but it may facilitate interaction with its MDH binding partner (vide infra).
Figure 4.
The x-ray structure of the c-type cytochrome XoxG. (A) A ribbon diagram of M. extorquens XoxG with selected amino acid ligands and heme cofactor shown in stick format. The FeIII ion is shown as an orange sphere. A 5-residue disordered loop region (dashed line) is located near the Met ligand. HP6 and HP7 designate the two heme propionate substituents. The heme cofactor is buried in a hydrophobic core composed of α helices (I, III, and V) conserved in class I cytochromes c. (B) The XoxG heme cofactor is ligated by Met 143 and His 99. A 2Fo-Fc electron density map (pink mesh) is shown contoured to 1.5σ around the cofactor. (C) An iron anomalous difference Fourier map (orange mesh) is shown contoured to 5σ.
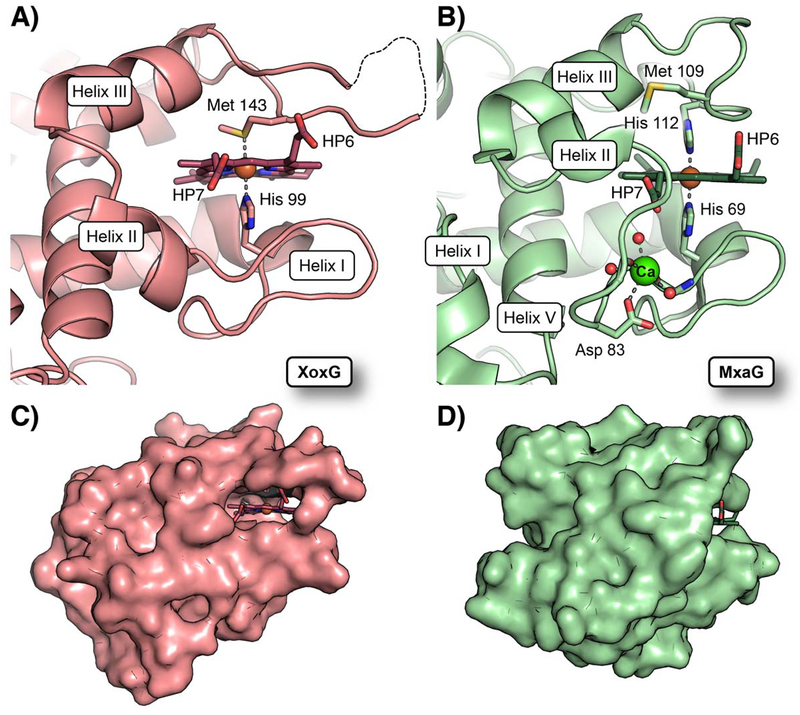
Two of the most important factors affecting reduction potentials of heme proteins are the electrostatic environment and solvent accessibility of the heme.[32] To understand the unusually low Em of XoxG, we compared its structure to that of M. extorquens MxaG (Figure 5) and other c-type cytochromes (Figure S7), all of which possess more typical Em values ~+260 mV. The other structures utilized in the comparison include MxaG (PDB ID: 2C8S), [16] Rhodothermus marinus cytochrome c (PDB ID: 3CP5),[31a] and Saccharomyces cerevisiae mitochondrial cytochrome c (PDB ID: 1YCC).[31b] The latter protein has been used to define core structural features in monoheme cytochromes, and the other two candidates were selected based on their structural/functional similarity to XoxG. The Met face of the XoxG heme is enclosed by hydrophobic residues, and the heme propionates (HP6 and HP7) in XoxG are engaged in hydrogen bonding interactions with Arg118 and Lys132, respectively (Figure 5A). These features are common to other cytochromes, but equivalent basic residues are not present in MxaG. Instead, a CaII ion is bound in the vicinity of HP7 and coordinated by an Asp residue, two backbone carbonyls, and three solvent molecules. One of the coordinated waters resides within hydrogen-bonding distance of HP7 (Figure 5B). The low degree of charge compensation at this site may elevate the heme reduction potential in MxaG. The coordination of CaII also has another important effect in MxaG: it partially disrupts helix II, bringing a connecting loop in front of the heme. This structural change largely blocks solvent access to the heme propionate face (Figure 5D). By contrast, in XoxG, helix II is positioned to open the heme propionates to the bulk (Figure 5C). In other cytochromes c, helix II is in the same position as in XoxG (Figure S7), but these proteins instead use helix IV to block this face, which constitutes a different strategy to achieve limitation of solvent access to the heme. We propose that the combination of a distinct lack of helix IV in XoxG with lack of the CaII binding site in helix II give rise to a uniquely solvent-exposed heme. This structural environment would be expected to decrease the Em of the heme cofactor based on previous studies of the effect of solvent exposure on heme reduction potentials.[32–33]
Figure 5. Comparison of heme solvent exposure in XoxG and MxaG.
(A) A ribbon diagram of the XoxG structure near the heme cofactor. The heme is highly solvent exposed because helix II is more extended and it resides further away from the heme propionate face. Heme propionate substituents are designated HP6 and HP7. (B) A ribbon diagram of MxaG (PDB ID: 2C8S) in the vicinity of the heme reveals that Helix II unwinds to coordinate a CaII (green sphere) via the side chain of Asp 83, backbone carbonyls of Tyr 85 and Gly 80, and three water molecules (red spheres). This structural change positions a loop in front of the inner propionate ligand. An FeIII ion (orange sphere) is bound to His 69 and His 112 in the crystal structure, but the latter ligand is replaced by Met 109 in solution. A surface representation of XoxG (C) and MxaG (D) shows that both heme propionate (HP) groups are exposed in XoxG, whereas the inner propionate of MxaG is buried. Increased access to solvent in XoxG should render the heme more easily reduced.
Finally, in order to explore potential interaction modes for XoxF and XoxG, a search of the PDB for structurally similar proteins using the Dali server[34] yielded a structure of a PQQ-dependent alcohol dehydrogenase natively fused to its c-type cytochrome (PDB code: 1YIQ[35]) as one of the top hits (z-score = 8.6). The crystal structure of M. extorquens XoxF has not been reported, but a homology model of this protein was generated[36] using the crystal structure of M. fumariolicum XoxF (PDB code: 4MAE)[5b], with which the M. extorquens enzyme shares 53% sequence identity, as a template. Alignment of XoxG with the cytochrome c domain and the XoxF homology model with the dehydrogenase domain suggests a plausible XoxF binding interface on XoxG. In the model, the loops between helices I and II and between III and V in XoxG are positioned to interact with XoxF (Figure S9). If the HP6/7 face of XoxG remains open to solvent in the complex with XoxF, as suggested by the model, the reduction potential of isolated XoxG measured in this work may reasonably approximate the potential of the cytochrome in complex with XoxF.
Biochemical and structural characterization of XoxJ
The third conserved gene in xox operons is xoxJ. The function of XoxJ is unknown, as is that of the analogous protein in the Ca-MDH system, MxaJ. However, MxaJ and XoxJ are predicted to be members of the periplasmic binding protein (PBP) family. An x-ray crystal structure of MxaJ[14] confirms that it adopts the characteristic architecture of this protein family, with two domains surrounding a ligand binding site at the domain interface. Various ligands have been proposed for these proteins, including the substrate and product of the MDH,[6d] methanol and formaldehyde; unlike for most bacterial PBPs, however, a membrane-bound ATP-binding cassette (ABC) transporter system is not encoded nearby mxaJ or xoxJ in the genome. Alternatively, it has been noted that MxaJ occasionally co-purifies with the Ca-MDH,[37] and it has been proposed to be part of the active MDH complex (albeit one not necessary for activity in vitro), for example in facilitating electron transfer from PQQ to MxaG.[14] However, deletion of mxaJ in M. extorquens leads to the absence of detectable levels of the two subunits of the Ca-MDH, MxaF and MxaI, despite the presence of MxaG, suggesting a more critical role of MxaJ in the Ca-MDH system.[15d] Deletion of mxaJ in Paracoccus denitrificans leads to lower than wild-type levels of MxaFI but no methanol oxidation activity,[15a] suggesting that MxaJ is involved in activation of Ca-MDH. We reasoned that XoxJ would be similarly essential in the Ln-MDH, and we hypothesized it was involved in PQQ insertion into apo-XoxF. PQQ is synthesized in the cytosol,[38] but the proteins that use it are periplasmic.[13] It is unknown how PQQ gets to the periplasm, but the reactivity of PQQ with nucleophiles at its C5 position[39] would likely necessitate a chaperoned process, as is the case for other reactive enzyme cofactors such as heme[40] and iron-sulfur clusters.[41]
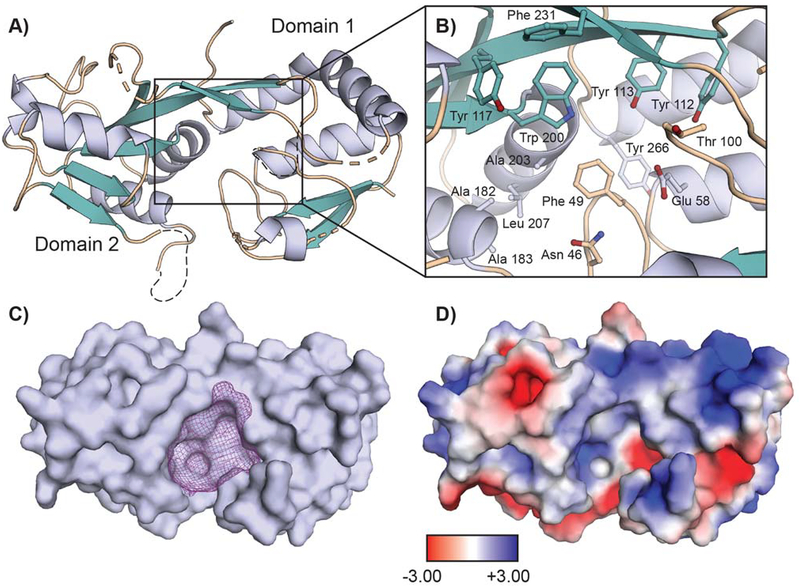
To investigate potential roles of XoxJ in the Ln-MDH system, we expressed XoxJ in E. coli and purified it from the periplasmic fraction using anion exchange, hydrophobic interaction, and size exclusion chromatographies (Figure S10). In order to inform our biochemical studies, we pursued crystallographic characterization of the protein. At the time at which we solved the structure (before the MxaJ structure was reported[14]), no protein in the Protein Data Bank was able to serve as a suitable molecular replacement model, so initial phase information was obtained from a SeMet-XoxJ crystal by single-wavelength anomalous diffraction methods, followed by solution of the wild-type XoxJ structure by molecular replacement, to 2.27 Å resolution (Table S3 for data collection and refinement statistics). The resulting model (Figure 6A) reveals the characteristic di-domain fold of a periplasmic binding protein, surrounding a putative substrate-binding cavity (Figure 6B). Overall, the fold of XoxJ is very similar to that of MxaJ, with which it shares 29% sequence identity. Secondary-structure matching superposition of the two proteins yields a root mean square deviation of 1.45 Å over 215 atoms.[42] As in MxaJ, domain 1 of XoxJ (Figure 6A) is missing three out of five β strands when compared with typical PBPs.[14] In place of these missing strands is a relatively unstructured region between Gln80 (after β2) and Tyr112 (the start of the domain-spanning strand, β3). Of these 32 residues, 14 are not visible in our structure (residues 81–82, 89–93, 101–107) due to disorder. The analogous region in MxaJ also adopts a long loop structure, although it is ordered in the crystallographic model. XoxJ additionally possesses a disordered loop (residues 170–176) on domain 2. In both structures, these two loops are located on the periphery of the putative substrate-binding cleft between the two domains, a large pocket of ~1750 Å3 in XoxJ (Figure 6C).[43] In MxaJ, this cleft contains six Tyr residues and six acidic residues. In XoxJ, four of these aromatic residues (Phe 49 and Tyr 112, 113, and 117) and one acidic residue (Glu 58) are retained, but many of the residues that are negatively charged in MxaJ have been replaced by hydrophobic or polar but uncharged residues (Figure 6B).
Figure 6.
A 2.27 Å resolution crystal structure of M. extorquens XoxJ. (A) Overall structure of XoxJ, shown as a ribbon diagram and colored by secondary structure. XoxJ consists of two globular domains linked by a rigid β-strand. Missing loops are denoted as dashed lines. (B) Selected residues that line the surface of the central cavity are shown in stick format. (C) Surface representation of XoxJ, highlighting the central cavity. The ~1750 Å3 cavity is displayed as purple mesh and was calculated using a 10 Å probe radius in HOLLOW.[43a] (D) A map of electrostatic surface potential of XoxJ (-3 kT/e to +3 kT/e), calculated by the APBS plugin in PYMOL and shown in the same orientation as viewed in panel C.
The large number of hydrophobic residues in the putative ligand-binding region of XoxJ is a shared feature of the active site of MDHs. In the latter protein, the PQQ cofactor is stacked between a conserved Trp residue and an unusual disulfide bond that forms between adjacent Cys residues (Figure S9).[1b, 5b, 7, 44] Given the presumed necessity of a PQQ chaperone, we considered PQQ as a potential ligand for XoxJ, which also contains a Trp side chain in the central cavity (Trp 200, Figure 6B). In order to test this model, we followed PQQ binding to XoxJ via titrations monitoring Trp fluorescence. XoxJ contains only three Trp residues, and only Trp 200 is in the putative ligand-binding cavity. Quenching of XoxJ fluorescence by PQQ was observed, whereas similar experiments carried out in a W200F variant of XoxJ showed negligible change in fluorescence (Figure S11), suggesting that PQQ binds to XoxJ near Trp 200. However, attempts to assess PQQ binding directly by isothermal titration calorimetry were inconclusive, and soaks of PQQ into XoxJ crystals were also unsuccessful. Therefore, we conclude that the fluorescence quenching may have resulted from non-specific interaction between PQQ and the hydrophobic pocket of XoxJ. In the Discussion, we consider alternative hypotheses for XoxJ’s role in the XoxF system.
DISCUSSION
In this manuscript, we have reported the first detailed characterization of XoxF activity with its physiological electron acceptor, XoxG. These studies reveal that the redox properties of XoxG are tuned to favor the lighter Lns in XoxF, with catalytic efficiencies markedly decreasing even over the range of Lns tolerated by M. extorquens for growth. Structural characterization of XoxG suggests a potential rationale for these observations. Here we focus on the ramifications of these data for understanding the physiological activities of XoxF systems. We also discuss the insights that our structural and biochemical characterization of XoxJ, the third and most enigmatic member of the xox operon, offers into the function of this protein and of MxaJ in the Ca-MDH.
Physiological relevance of XoxG’s redox properties
Characterization of XoxG allows for a synthesis of the activity data obtained with Ca- and Ln-MDHs using the dye- and protein-linked activity assays. Activity measurements of the Ca-MDHs using the dye-linked assay must be performed at high pH and with ammonia to facilitate activation of the C5 carbonyl of PQQ for hydride transfer, whereas MxaG-linked assays can be carried out at pH 7 without activator.[4a] This observation suggests that the coupling reagents in the dye-linked assay, which possess lower reduction potentials than MxaG (Scheme 1A), cannot provide sufficient driving force for the Ca-MDH reaction at neutral pH. By contrast, both dye-linked and XoxG-linked assays of Ln-MDHs can be carried out at pH 7.[6a, 10–11, 17] Together, these results suggest either that the reduction potential of the LnIII-PQQ cofactor in XoxFs is lower than that of the CaII-PQQ cofactor in MxaFI, or that the Ln-MDH reaction requires less driving force to be provided by the electron acceptor. The former is unlikely based on consideration of the Lewis acidity of the coordinating metal ions. Therefore, we propose that a chemical rationale for XoxG’s diminished Em, by 80 mV, relative to that of MxaG, is that the stronger Lewis acidity of the LnIII cofactor[8a, 8c] makes methanol oxidation more favorable in the Ln-MDH versus in the Ca-MDH, and therefore requires a lesser driving force to be provided via electron extraction from the metal-bound-PQQ cofactor by the cytochrome cL. Based on our crystallographic characterization of XoxG viewed in the context of MxaG and other cytochromes c, the structural rationale for this lower potential may be linked to greater solvent accessibility of the heme pocket in XoxG (Figure 5, Figure S7).
We can also consider potential physiological rationales for XoxG’s redox properties. The XoxG-linked assay results suggest that the Km for XoxG may be an important determinant of activity in vivo. The marked increase in Km across the Ln series is unlikely to be a result of significant Ln-dependent structural differences in XoxF that would interfere with XoxF-XoxG interaction (and therefore affect Km), as the crystal structures of Ce- vs. Eu-substituted XoxFs from M. fumariolicum are nearly identical.[9] Therefore, we propose that this increase in Km reflects a higher reduction potential of the LnIII-PQQ cofactor with more Lewis-acidic Lns, resulting in less efficient electron transfer and a higher concentration of XoxG needed for maximal activity. The observation that the Vmax values for each of the Ln-XoxFs assayed with XoxG are not significantly different may suggest that XoxG association or dissociation, rather than a chemical step, may be rate-limiting overall for MDH inside the cell. This explanation would also account for the lower Vmax values measured in XoxG- versus dye-linked assays. Notably, the Vmax for Nd-XoxF assayed with the dye-linked system coincides with that measured using XoxG as electron acceptor. A possible explanation is that with Nd as cofactor, the chemical steps catalyzed by the enzyme are no longer faster than the physical protein-protein interaction step(s) involving XoxG. Beyond Nd, the relationship between Km and ionic radius in Figure 3B would predict a Km of 10 μM for Sm-XoxF. In addition to poor uptake of Sm (and heavier Lns) by M. extorquens AM1,[11c] and while the periplasmic concentrations of XoxF and XoxG are not known, the Kms of XoxFs metallated with Lns beyond Sm may be too high to achieve sufficient methanol reduction rates in vivo at the physiological levels of XoxG. This idea is a different conclusion than would be drawn from considering the dye-linked assay results alone, which would suggest that unsuitability of later Lns for catalysis in vivo might be primarily related to a kcat effect. These findings underscore the importance of examination of MDH activities with their physiological electron acceptors.
XoxFs and XoxF-like proteins have been phylogenetically classified into five primary clades,[45] with M. extorquens XoxF being in the XoxF5 clade. Another XoxF5 enzyme from Methylomonas sp. LW13[6e] has also been characterized, but only with a La cofactor. This organism, like M. extorquens, cannot use Lns beyond Sm. Chistoserdova and co-workers have noted that phylogenetically distinct XoxF/XoxG pairs exist – for example, that xoxGs from other clades cannot complement in vivo the deletion of the xoxG associated with the XoxF5 enzyme in Methylomonas.[6e] Combining this observation with the conclusions of our study, we hypothesize that the reduction potentials of XoxGs are also tuned to the different subsets of Lns present in their associated XoxFs. Different organisms appear to acquire and use different sets of Lns based on metal availability in their environmental niches. Our findings suggest that XoxG reduction potential modulation may be another component of this adaptation. As one example, whereas M. fumariolicum XoxF is tolerant of Lns up to Gd in vivo,[5b] M. extorquens cannot operate efficiently on Lns beyond Nd. It is possible that one aspect of these selectivity differences is dictated by differences in the cellular uptake machinery:[10, 11c, 12] M. fumariolicum is cultured at pH 2.7[5b], where all Lns are far more soluble than in culture conditions for M. extorquens (pH 6.75). As a result, secretion of a chelator highly selective for La-Nd uptake, as our studies in M. extorquens indicated,[11c] may be unnecessary for M. fumariolicum. However, our hypothesis linking XoxG reduction potential and Ln-XoxF Km for XoxG predicts that the reduction potential of M. fumariolicum XoxG would be higher than that of M. extorquens XoxG, to allow for utilization of heavier Lns that may be bioavailable for this organism. Indeed, while our manuscript was under review, the former reduction potential was reported to be ~+240 mV,[28] supporting our proposal. The intersection of Ln uptake mechanisms and their selectivity, an organism’s environmental niche, and redox properties of XoxF and XoxG pairs will be an intriguing area of study.
Finally, from a practical biochemical perspective, our results also demonstrate that the increasing reduction potentials of the Ln-PQQ cofactor in XoxF across the Ln series may complicate interpretation of activity assays performed using the dye-linked system. Like XoxG, the dyes used (in typical experiments, 1 mM PES and 100 μM DCPIP) also operate at fixed potential (Em = +55 and +217 mV, respectively). Although these dyes are present in substantial excess, it is possible that prior observations of decreasing activity in M. fumariolicum in Ln-substituted XoxFs between Nd and Lu[8d] may reflect, in part, reduced efficiency of electron transfer to the dye as XoxF reduction potential increases, rather than intrinsically lower substrate turnover. Ln substitution in XoxF likely impacts multiple aspects of the reaction, some in opposing ways – Lewis acidity, ionic radius, and coordination number preferences of the metal ion, substrate binding and product release, and other chemical steps have all been discussed.[8d] However, the relative contributions of these factors to the observed rate constants are not well understood at present. Therefore, we suggest that interpretation of in vitro activity measurements over the full range of Lns, while intriguing, should be treated with caution until the redox properties of the Ln-XoxF systems are more thoroughly characterized.
Potential functions of XoxJ
In this work we have also characterized XoxJ for the first time. Our initial hypothesis was that this protein, unusual in that it is a periplasmic binding protein that is not obviously associated with an ABC transporter system, might be a PQQ chaperone for XoxF. Our biochemical data did not support this function, but the XoxJ structure reveals a large hydrophobic cavity surrounded by disordered loops, perhaps optimized to bind a large, hydrophobic protein or peptide target. The MxaJ crystal structure contains a similar feature,[14] suggesting analogous but distinct protein substrates for these two proteins (e.g., XoxF and MxaF, respectively). The authors of the MxaJ structural study speculated that MxaJ interacts with mature MxaFI to facilitate electron transfer from PQQ to MxaG, although we believe that genetic knockout experiments lend stronger support to the proposal that MxaJ is essential for MDH activation, not turnover.[15a, 15d] As XoxJ’s role in the Ln-MDH is likely analogous to that of MxaJ in the Ca-MDH, we propose instead that XoxJ’s role in XoxF activation is to bind a hydrophobic portion of a partially folded apo-XoxF to aid in cofactor insertion.
Crystal structures show that the MDH active site is buried with a narrow channel as the sole route for substrate access and product egress.[1b] In mature MDH, it is not obvious how metal and PQQ could be inserted into the apoprotein without a significant conformational change. Holo-XoxF is a dimer, but treatment of the enzyme with EGTA leads to partial irreversible inactivation, dissociation of PQQ, and reversion to a monomer (Figure S12). These observations indicate that cofactor removal disrupts the dimer interface, implying that the reverse reaction, cofactor insertion, should be accompanied by an ordering of this region of the protein and dimerization. Based on conserved structures of MDHs in general,[5b, 7, 44] and a homology model of M. extorquens XoxF derived from the x-ray structure of M. fumariolicum XoxF more specifically,[5b] we speculate that the C-terminus of the protein could undergo the most significant conformational change upon holoenzyme assembly. This region forms a helical domain that clamps over the active site, near the PQQ cofactor, in the homology model template structure (Figure S13). Overlay of this C-terminal region with the XoxJ cavity (Figure S14) shows that the XoxF domain is approximately the correct size and shape to bind in the XoxJ cavity. We are currently pursuing further studies to characterize the postulated XoxJ-XoxF interaction and test our hypothesis.
Conclusion
Biology’s preference for lighter lanthanides to oxidize methanol, while perhaps necessitated by elemental abundance considerations, is nevertheless an intriguing chemical problem on multiple levels – from selective metal ion recognition and incorporation into XoxF, to chemical ramifications of utilization of a range of LnIIIs in one enzyme. The availability of the entire LnIII series and YIII for in vitro analysis offers a unique opportunity to dissect these many intersecting factors in a single enzyme system. Our work provides the first insights into the implications for cofactor electron transfer, necessary for enzyme activity in vivo, that are associated with substitution of Lns in XoxFs. Not only has Nature developed sophisticated (and yet to be fully elucidated) mechanisms for selective uptake[11c] and regulation[6c, 10, 46] of light Lns – it has carefully tuned redox properties of the MDH and its electron acceptor, XoxG, to prefer these metals over the later Lns. Furthermore, our characterization of the third and least understood member of the xox operon, XoxJ, provides a path forward to unraveling the mechanism of activation of MDHs with PQQ and metal ions. An understanding of this mechanism would facilitate studies of Ln substitution in XoxF, providing a deeper understanding of how and why methylotrophs have harnessed Ln chemistry for one of their most crucial reactions.
EXPERIMETAL SECTION
General considerations.
Chemical reagents were obtained from Sigma-Aldrich, unless noted otherwise, at the highest purity available. Rare earth element salts were at a minimum purity of 99.9% rare earth metal content. Primers and gBlocks were ordered from Integrated DNA Technologies (IDT). E. coli strains [5alpha and BL21(DE3)] for cloning and recombinant protein expression, as well as cloning reagents (restriction enzymes, Q5 DNA polymerase, OneTaq DNA polymerase, T4 DNA ligase, and KLD Enzyme Mix) were obtained from New England Biolabs. PCR cleanup and miniprep kits were from Omega Bio-tek, and gel extractions used the Zymoclean gel DNA recovery kit from Zymo Research. M. extorquens AM1 (ATCC 14718, NCIB 9133) was purchased from the American Type Culture Collection. pEC86 was a kind gift of Prof. Ming Tien, Penn State University (Table S6). Protein gel electrophoresis was carried out using Life Tech 16% Tris-glycine gels and a mini gel apparatus. Automated protein chromatography was carried out on a GE Healthcare Biosciences Akta Pure fast protein liquid chromatography (FPLC) system. UV-visible absorption spectra were obtained on an Agilent Cary 60 UV-visible spectrophotometer using a quartz cuvette (Starna Cells). Well plate analyses were carried out using a BioTek Synergy H1 microplate reader. Fluorescence titrations were carried out on a Cary Eclipse fluorescence spectrophotometer using a quartz micro fluorometer cuvette (10 mm pathlength) with a septum and screw cap (Starna Cells). Inductively coupled plasma mass spectrometry (ICP-MS) analysis was carried out on a Thermo Fisher Scientific X Series 2 ICP-MS instrument at the Penn State Institutes of Energy and the Environment, Laboratory for Isotopes and Metals in the Environment. Further detailed methods are described in the Supporting Information.
Growth and purification of M. extorquens Ln-XoxFs.
XoxF was purified from its native levels in M. extorquens AM1 cells grown in the presence of 1 μM LaCl3, CeCl3, or NdCl3, as described,[11a] with three exceptions. First, cells were lysed by sonication (20 cycles of 20 s on, 40 s off, 50% amplitude) using a QSonica Q500 sonicator. Second, it was determined that XoxF activity elutes from the HiPrep SP-FF column in two peaks, at 70–80 mM and 100–120 mM NaCl, respectively, with the second peak having higher activity. This latter peak was pooled and carried forward in the purification. Third, the HiLoad Superdex 16/600 200pg column was eluted using 20 mM MOPS, 100 mM KCl, pH 7.0.
Dye-linked activity assays.
MDH activity assays on purified protein using the artificial, dye-linked system were carried out using a modification of the reported method[11a] in order to accommodate detection using a plate reader. An assay mix (98 μL) containing 100 mM Tris, pH 9.0, 15 mM NH4Cl, 1 mM PES (MP Biomedicals), 0.1 mM DCPIP, and 67 nM XoxF (dimer concentration) in a Greiner Bio-One 96-well half area plate, was incubated in the plate reader at 30 °C for 30 min while monitoring the A600nm. This step was important to equilibrate the assay mixture and to remove co-purifying background activity (in the absence of added substrate) typical of MDHs.[20] The assay was initiated by addition of 2 μL of 0–15 mM methanol (final concentration: 0–300 μM), and DCPIP reduction was monitored via the decrease in A600nm for 5 minutes, with reads every 10 s. The rate of DCPIP reduction was calculated from the initial slope of the line (up to 40 s), using a pathlength of 0.63 cm for 100 μL in the well, from which XoxF specific activity was determined. Activity assays during purification steps were performed using the same assay on a UV-visible spectrophotometer with 500 μL volume, initiated with 20 mM methanol.
Expression and purification of XoxG.
XoxG was expressed in the presence of pEC86, a plasmid encoding cytochrome c maturation factors.[22] Chemically competent E. coli BL21(DE3) cells were co-transformed with pET24a-XoxG (Table S6) and pEC86 (CmR) and plated on LB-agar plates containing 50 μg/mL kanamycin (Km) and 30 μg/mL chloramphenicol (Cm) (these antibiotic concentrations in all media), and grown at 30 °C. A single colony was used to inoculate 2 mL TB-Km/Cm, which was grown overnight at 30 °C with 200 rpm shaking. This colony was used to inoculate a 100 mL culture at 100× dilution, which was grown under the same conditions. This culture in turn was used to inoculate 2 × 1.5 L TB-Km/Cm at 100× dilution, which was grown under the same conditions to OD600 = 0.6 (~8–9 h), at which point isopropyl β-D-1-thiogalactopyranoside (IPTG) was added to 1 μM and protein expression was induced overnight (~15 h). Cells were harvested by centrifugation at 7000 × g, 7 min, at 4 ℃, yielding ~8 g cell paste per L culture. The cell pellets were used immediately to harvest the periplasmic fraction containing XoxG.
The cell paste was resuspended by pipetting and stirring at 40 mL/g in 30 mM Tris, 20% (w/v) sucrose, 1 mM EDTA, pH 7.4, at room temperature (20 °C). The suspension was stirred at 150 rpm at room temperature for 10 min and then centrifuged for 20 min at 8000 × g, 4 °C. The supernatant was decanted and the pellet was resuspended in 20 mL/g (original wet weight) ice-cold 5 mM MgSO4. The suspension was stirred for 10 min at 4 °C and then centrifuged for 10 min at 8000 × g, 4 °C. The supernatant (the periplasmic extract) was decanted and 1/20th (0.05) volume of 1 M Tris, pH 7.4 was added with stirring. This solution was passed through a 20 mL (2.5 × 4 cm) DEAE Sepharose Fast Flow column pre-equilibrated with 50 mM Tris, pH 7.4, and the column was washed with 2 column volumes (CV) of the same buffer; the light red flowthrough contained XoxG. The flowthrough and wash were pooled and ammonium sulfate was added to 35% saturation (194 g/L)[47] with stirring, and the solution was loaded onto a 20 mL (2.5 × 4 cm) phenyl sepharose FF column pre-equilibrated in 50 mM Tris, 35% ammonium sulfate, pH 7.4. The column was washed with 5 CV of the same buffer and eluted with 50 mM Tris, 20% ammonium sulfate, pH 7.4. The resulting XoxG-containing fractions (as judged by A418nm) were pooled and concentrated by centrifugal filtration (Amicon Ultra 10 kDa MWCO) to <2 mL and loaded to a HiLoad Superdex 75pg 16/600 column with 20 mM MOPS, 100 mM KCl, pH 7.0 (Buffer A) for polishing and buffer exchange. The purification yielded 6 mg per L culture. The absorption spectra and extinction coefficients of the oxidized and reduced forms of XoxG were determined as described in the Supporting Information. Maximum absorptions for oxidized XoxG were ε413nm = 129 mM−1cm−1 and ε527nm = 11.5 mM−1cm−1, and for the reduced form were ε418nm = 157 mM−1cm−1 and ε553nm = 25.8 mM−1cm−1.
Determination of the reduction potential of XoxG.
The reduction potential of the heme cofactor of XoxG was determined spectrophotometrically as described by Raven and co-workers[25] based on a method developed by Massey,[24] using 2,6-dichlorophenolindophenol (DCPIP, Em = +217 mV at 25 °C, pH 7.0[21]) as a standard. The UV-visible spectrum of DCPIPox in the 350–800 nm range was determined in Buffer A, using ε600 = 20.6 mM−1 cm−1, pH 7.0,[48] and the spectrum of DCPIPred was determined by addition of a solution of sodium dithionite. In a total volume of 500 μL, Buffer A, 300 μM xanthine (from 30 mM solution in 0.5 M NaOH), 5 mM D-glucose, 25 μg glucose oxidase (from an 8 mg/mL stock in Buffer A), and 2.5 μg bovine liver catalase were mixed in a capped cuvette. The solution was incubated for 5 min with periodic inversion to achieve anaerobic conditions. XoxG (final concentration of 4 μM, from a stock of 250 μM) and dye (final concentration 20–25 μM from a 2.5 mM stock) were added and mixed, and an initial spectrum at 350–800 nm was acquired. Reduction was initiated by addition of xanthine oxidase (final concentration: 20 nM) and inversion, and spectra were acquired every minute for ~40–60 min (4800 nm/min scan rate), until spectra were constant for >5 min.
The initial concentrations of protein and dye were determined by manual fitting of the initial spectrum with spectra of DCPIP and XoxG. For each time point, the concentrations of DCPIPox and DCPIPred were calculated from the A600 value (DCPIPred and XoxG contribute negligibly to the A600 value and can be ignored); [DCPIPred] was assumed to be equal to the difference between the initial [DCPIPox] and [DCPIPox] at each time point. The concentration of reduced XoxG was calculated by subtraction of the contribution of DCPIPox from the A418nm of each timepoint, subtraction of the initial A418 contribution of XoxGox, and division by ε418red-ε418ox = 56 mM−1 cm−1. From this value and the total concentration of XoxG, the concentration of XoxGox was calculated. Em was determined by plotting y-axis = 12.5×ln([DCPIPox]/[DCPIPred]) vs. x-axis = 25×ln([XoxGox]/[XoxGred]). The plot has a slope of ~1 and a y-intercept of Em,protein – Em,dye. Points at the beginning and end of the experiment were not at equilibrium and were ignored in the analysis.
XoxF activity assay using XoxG as electron acceptor.
The protocol was adapted from Day and Anthony,[20] initially developed to assay MxaFI using MxaG as electron acceptor. The final assay (100 μL final volume) contained 50 μM cytochrome c from equine heart, 20 mM methanol, 67 nM XoxF, and 0–80 μM XoxG, in 20 mM MOPS, pH 7.0. All assay components except for XoxG were added to the plate (Greiner Bio-One 96-well half area) and equilibrated in the plate reader 30 °C for 20 min, during which time A550nm did not change significantly. The assay was then initiated by addition of 5 μL XoxG at a range of concentrations, and the increase in A550nm associated with cytochrome c reduction was monitored every 10 s for 10 min. The rate of reduction was calculated from the average slope over the first 2 min of the assay, from which the specific activity of XoxF was calculated.
Expression and purification of XoxJ.
Chemically competent E. coli BL21(DE3) cells were transformed with pET24a-XoxJ and plated on LB-agar plates containing 50 μg/mL kanamycin (Km) and grown at 37 °C. A single colony was used to inoculate 100 mL of LB (50 μg/mL Km in all growth media), which was grown for ~16 h at 37 °C with shaking at 220 rpm. This culture was used to inoculate three 2 L cultures (in 6 L flasks) at 100× dilution, and the cultures were grown at 37 °C with shaking at 200 rpm. At OD600nm ~ 0.5, IPTG was added to a final concentration of 0.2 mM; after 2 h further incubation, the cells were pelleted by centrifugation for 7 min at 7000 × g, 4 °C, yielding ~3 g cell paste per L culture. The cell pellets were used immediately to harvest the periplasmic fraction.
Periplasmic extraction was carried out as described for XoxG. The extract was buffered by addition of 1/20th (0.05) volume of 1 M Tris, pH 7.4, with stirring, and solid NaCl was added to a final concentration of 50 mM. The solution was applied to a DEAE Sepharose Fast Flow column (2.5 × 4 cm, 20 mL) pre-equilibrated in 50 mM Tris, 50 mM NaCl, pH 7.4 (Buffer B) and washed with 5 CV of Buffer B. XoxJ did not bind appreciably to this column, and the flowthrough and wash were pooled. Ammonium sulfate was added slowly with stirring to 35% saturation (194 g/L at 4 °C). The solution was then loaded to a Phenyl Sepharose 6 Fast Flow (low substitution) column (2.5 × 2 cm, 10 mL) pre-equilibrated in 50 mM Tris, 5% glycerol, pH 7.4, containing 35% sat. (NH4)2SO4 (Buffer C). The column was washed with 15 CV of Buffer C, and eluted with 10 CV 50 mM Tris, 5% glycerol, pH 7.4 (Buffer D). The eluted protein was concentrated to ~5 mL using an Amicon Ultra 10-kDa MWCO centrifugal filter device.
After addition of solid (NH4)2SO4 to ~35% saturation, XoxJ was further purified by FPLC using a HiPrep Phenyl FF (low sub) 16/10 column (GE Healthcare) with a 5 mL capillary loop. The column was pre-equilibrated with Buffer C containing 1 mM EDTA and, following sample injection, was washed at 2 mL/min with 4 CV of the same buffer, followed by elution with a 200 mL linear gradient of 0–100% Buffer D containing 1 mM EDTA at 2 mL/min, with 4 mL fractions collected in peak fractionation mode (10 mAU threshold). XoxJ-containing fractions, as judged by the major peak at 280 nm eluting at 25–15% sat. (NH4)2SO4, were concentrated by centrifugal filtration (Amicon Ultra 10 kDa MWCO) to <2 mL. Buffer was exchanged into 20 mM MOPS, 100 mM NaCl, pH 7.0 (Buffer E), and a minor contaminant at ~15 kDa was removed, by size-exclusion chromatography on a HiLoad 16/600 Superdex 200 pg column. After column equilibration, the protein sample was loaded using a 2 mL capillary loop, washed with 4 mL Buffer E, and eluted with 1 CV Buffer E at 0.75 mL/min, with 2 mL fractions collected. XoxJ-containing fractions were pooled and concentrated by centrifugal filtration. The concentration of XoxJ was determined using ε280nm = 34.0 mM−1cm−1,[49] and the purification yielded ~3 mg/L culture.
General crystallographic methods.
All crystallographic datasets were collected at the Berkeley Center for Structural Biology at the Advanced Light Source (Lawrence Berkeley National Laboratory) and the National Institute of General Medical Science and National Cancer Institute Collaborative Access Team (GM/CA-CAT) beamline at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL). Diffraction images were processed with the HKL2000 package.[50] Initial auto-built models were further modified and refined using Coot[51] and Refmac5,[52] respectively. Final refinements were carried out in Phenix.[53] Structures were validated and analyzed for Ramachandran outliers with the Molprobity server.[54] Figures were prepared with the PyMOL molecular graphics software package (Schrödinger, LLC). Details of the crystallization methods for XoxG, SeMet-XoxJ, and XoxJ are provided in the Supporting Information.
Supplementary Material
Acknowledgements
We thank T. N. Laremore and J. A. Fecko for assistance with mass spectrometry experiments, as well as Matthew Gonzales and David Welsch for ICP-MS analysis. Portions of this work were performed at the Advanced Photon Source (APS), a US Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract DE-AC02–06CH11357, for use of its resources. The General Medical Sciences and National Cancer Institute Collaborative Access Team at the APS was funded in whole or in part with Federal funds, National Cancer Institute Grant ACB-12002 and National Institute of General Medical Sciences (NIGMS) Grant AGM-12006. The Eiger 16M detector was funded by NIH Office of Research Infrastructure Programs, High-End Instrumentation Grant 1S10OD012289-01A1. Use of the Life Sciences Collaborative Access Team Sector 21 was supported by the Michigan Economic Development Corporation and Michigan Technology Tri-Corridor Grant 085P1000817. We also acknowledge the Berkeley Center for Structural Biology at the Advanced Light Source (ALS), supported in part by the NIH, NIGMS, and the Howard Hughes Medical Institute. The ALS is a DOE Office of Science User Facility under Contract DE-AC02–05CH11231. J.A.C. acknowledges the Penn State Department of Chemistry, the Huck Institutes for the Life Sciences, and a Louis Martarano Career Development Professorship for funding. We also acknowledge NIH grant GM119707 (to A.K.B.).
Biography

Joseph Cotruvo, Jr. earned his Ph.D. in Biological Chemistry from MIT under the guidance of Prof. JoAnne Stubbe, where he identified the novel dimanganese-tyrosyl radical cofactor in class Ib ribonucleotide reductase and elucidated its mechanism of biosynthesis. As a Jane Coffin Childs postdoctoral fellow in the lab of Prof. Chris Chang at the University of California Berkeley, he used biochemistry and chemical biology to probe how copper modulates important cellular signaling pathways. Joey joined the Department of Chemistry at the Pennsylvania State University in 2016, where he is Assistant Professor and Louis Martarano Career Development Professor. His lab applies biochemical and chemical biology approaches to study selective metal ion recognition in biological systems.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Supporting information and the ORCIDs for the authors of this article can be found at ***: supplementary experimental procedures, structural data, additional tables, figures, and references.
Accession Codes:
The atomic coordinates and structure factors of XoxG and XoxJ have been deposited into the Protein Data Bank as entries 6ONQ and 6ONP, respectively.
References
- [1].a) Anthony C, in Enzyme-Catalyzed Electron and Radical Transfer: Subcellular Biochemistry (Eds.: Holzenburg A, Scrutton NS), Springer US, Boston, MA, 2000, pp. 73–117; [Google Scholar]; b) Anthony C, Williams P, Biochim. Biophys. Acta 2003, 1647, 18–23; [DOI] [PubMed] [Google Scholar]; c Chistoserdova L, Kalyuzhnaya MG, Lidstrom ME, Annu. Rev. Microbiol 2009, 63, 477–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ghosh M, Anthony C, Harlos K, Goodwin MG, Blake CCF, Structure 1995, 3, 177–187. [DOI] [PubMed] [Google Scholar]
- [3].Zhang X, Reddy SY, Bruice TC, Proc. Natl. Acad. Sci. USA 2007, 104, 745–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].a) Cox JM, Day DJ, Anthony C, Biochim. Biophys. Acta 1992, 1119, 97–106; [DOI] [PubMed] [Google Scholar]; b) O’Keeffe DT, Anthony C, Biochem. J 1980, 192, 411–419; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Anthony C, Biochim. Biophys. Acta 1992, 1099, 1–15. [PubMed] [Google Scholar]
- [5].a) Nakagawa T, Mitsui R, Tani A, Sasa K, Tashiro S, Iwama T, Hayakawa T, Kawai K, PLoS ONE 2012, 7, e50480; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Pol A, Barends TRM, Dietl A, Khadem AF, Eygensteyn J, Jetten MSM, Op den Camp HJM, Environ. Microbiol 2014, 16, 255–264. [DOI] [PubMed] [Google Scholar]
- [6].a) Good NM, Vu HN, Surlano CJ, Subuyuj GA, Skovran E, Martinez-Gomez NC, J. Bacteriol 2016, 198, 3109–3118; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Wehrmann M, Billard P, Martin-Meriadec A, Zegeye A, Klebensberger J, mBio 2017, 8, e00570–00517; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Wehrmann M, Berthelot C, Billard P, Klebensberger J, mSphere 2018, 3, e00376–00318; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Keltjens JT, Pol A, Reimann J, Op den Camp HJM, Appl. Microbiol. Biotechnol 2014, 98, 6163–6183; [DOI] [PubMed] [Google Scholar]; e) Huang J, Yu Z, Chistoserdova L, Front. Microbiol 2018, 9, 10.3389/fmicb.2018.01366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Deng YW, Ro SY, Rosenzweig AC, J. Biol. Inorg. Chem 2018, 23, 1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].a) Bogart JA, Lewis AJ, Schelter EJ, Chemistry 2015, 21, 1743–1748; [DOI] [PubMed] [Google Scholar]; b) Martinez-Gomez NC, Vu HN, Skovran E, Inorg. Chem 2016, 55, 10083–10089; [DOI] [PubMed] [Google Scholar]; c) Prejanò M, Marino T, Russo N, Chem. Eur. J 2017, 23, 8652–8657; [DOI] [PubMed] [Google Scholar]; d) Lumpe H, Pol A, Op den Camp HJM, Daumann LJ, Dalton Trans. 2018, 47, 10463–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jahn B, Pol A, Lumpe H, Barends TRM, Dietl A, Hogendoorn C, Op den Camp HJM, Daumann LJ, ChemBioChem 2018, 19, 1147–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Vu HN, Subuyuj GA, Vijayakumar S, Good NM, Martinez-Gomez NC, Skovran E, J. Bacteriol. 2016, 198, 1250–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].a) Cotruvo JA Jr., Featherston ER, Mattocks JA, Ho JV, Laremore TN, J. Am. Chem. Soc 2018, 140, 15056–15061; [DOI] [PubMed] [Google Scholar]; b) Cook EC, Featherston ER, Showalter SA, Cotruvo JA Jr., Biochemistry 2019, 58, 120–125; [DOI] [PubMed] [Google Scholar]; c) Mattocks JA, Ho JV, Cotruvo JA Jr., J. Am. Chem. Soc 2019, 141, 2857–2861. [DOI] [PubMed] [Google Scholar]
- [12].Ochsner AM, Hemmerle L, Vonderach T, Nüssli R, Bortfeld-Miller M, Hattendorf B, Vorholt JA, Mol. Microbiol 2019, 10.1111/mmi.14208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Goodwin PM, Anthony C, Adv. Microb. Physiol 1998, 40, 1–80. [DOI] [PubMed] [Google Scholar]
- [14].Choi JM, Cao T-P, Kim SW, Lee KH, Lee SH, Proteins 2017, 85, 1379–1386.28295618 [Google Scholar]
- [15].a) Van Spanning RJ, Wansell CW, De Boer T, Hazelaar MJ, Anazawa H, Harms N, Oltmann LF, Stouthamer AH, J. Bacteriol 1991, 173, 6948–6961; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Richardson IW, Anthony C, Biochem. J 1992, 287, 709–715; [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Morris CJ, Kim YM, Perkins KE, Lidstrom ME, J. Bacteriol 1995, 177, 6825–6831; [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Amaratunga K, Goodwin PM, O’connor CD, Anthony C, FEMS Microbiol. Lett 1997, 146, 31–38. [DOI] [PubMed] [Google Scholar]
- [16].Williams P, Coates L, Mohammed F, Gill R, Erskine P, Bourgeois D, Wood SP, Anthony C, Cooper JB, J. Mol. Biol 2006, 357, 151–162. [DOI] [PubMed] [Google Scholar]
- [17].Zheng Y, Huang J, Zhao F, Chistoserdova L, mBio 2018, 9, e02430–02417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].a) Cotton SA, Raithby PR, Coord. Chem. Rev 2017, 340, 220–231; [Google Scholar]; b) Cheisson T, Schelter EJ, Science 2019, 363, 489–493. [DOI] [PubMed] [Google Scholar]
- [19].a) Wu ML, Wessels HJCT, Pol A, Op den Camp HJM, Jetten MSM, van Niftrik L, Keltjens JT, Appl. Environ. Microbiol 2015, 81, 1442–1451; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Good NM, Moore RS, Suriano CJ, Martinez-Gomez NC, Sci. Rep 2019, 9, 4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Day DJ, Anthony C, Meth. Enzymol 1990, 188, 210–216. [DOI] [PubMed] [Google Scholar]
- [21].Clark WM, Oxidation-Reduction Potentials of Organic Systems, The Williams and Wilkins Company, Baltimore, 1960. [Google Scholar]
- [22].Londer YY, in Heterologous Gene Expression in E.coli: Methods and Protocols (Eds.: Evans JTC, Xu M-Q), Humana Press, Totowa, NJ, 2011, pp. 123–150. [Google Scholar]
- [23].Afolabi PR, Mohammed F, Amaratunga K, Majekodunmi O, Dales L, Gill R, Thompson D, Cooper B, Wood P, Goodwin M, Anthony C, Biochemistry 2001, 40, 9799–9809. [DOI] [PubMed] [Google Scholar]
- [24].Massey V, in Flavins and Flavoproteins 1990 (Eds.: Curtis B, Roncho S, Zanetti G), Walter de Gruyter & Co., Berlin, 1991, pp. 59–66. [Google Scholar]
- [25].Efimov I, Parkin G, Millett ES, Glenday J, Chan CK, Weedon H, Randhawa H, Basran J, Raven EL, FEBS Lett. 2014, 588, 701–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu J, Chakraborty S, Hosseinzadeh P, Yu Y, Tian S, Petrik I, Bhagi A, Lu Y, Chem. Rev 2014, 114, 4366–4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Frank J, Dijkstra M, Duine JA, Balny C, Eur. J. Biochem 1988, 174, 331–338. [DOI] [PubMed] [Google Scholar]
- [28].a) Versantvoort W, Pol A, Daumann LJ, Larrabee JA, Strayer AH, Jetten MSM, van Niftrik L, Reimann J, Op den Camp HJM, Biochim. Biophys. Acta - Proteins and Proteomics 2019, 1867, 595–603; [DOI] [PubMed] [Google Scholar]; b) Kalimuthu P, Daumann LJ, Pol A, Op den Camp HJM, Bernhardt PV, Chem. Eur. J 2019, doi: 10.1002/chem.201900525. [DOI] [PubMed] [Google Scholar]
- [29].Shannon RD, Acta Cryst. A 1976, 32, 751–767. [Google Scholar]
- [30].Bertini I, Cavallaro G, Rosato A, Chem. Rev 2006, 106, 90–115. [DOI] [PubMed] [Google Scholar]
- [31].a) Stelter M, Melo AMP, Pereira MM, Gomes CM, Hreggvidsson GO, Hjorleifsdottir S, Saraiva LM, Teixeira M, Archer M, Biochemistry 2008, 47, 11953–11963; [DOI] [PubMed] [Google Scholar]; b) Brayer GD, Louie GV, J. Mol. Biol 1990, 214, 527–555. [DOI] [PubMed] [Google Scholar]
- [32].Banci L, Bertini I, Luchinat C, Turano P, in Biological Inorganic Chemistry: Structure and Reactivity (Eds.: Bertini I, Gray HB, Stiefel EI, Valentine JS), University Science Books, Sausalito, CA, 2007, pp. 229–261. [Google Scholar]
- [33].Tezcan FA, Winkler JR, Gray HB, J. Am. Chem. Soc 1998, 120, 13383–13388. [Google Scholar]
- [34].Holm L, Laakso LM, Nucl. Acids Res 2016, 44 (W1), W351–W355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Toyama H, Chen ZW, Fukumoto M, Adachi O, Matsushita K, Mathews FS, J. Mol. Biol 2005, 352, 91–104. [DOI] [PubMed] [Google Scholar]
- [36].Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T, Nucl. Acids Res 2018, 46, W296–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Matsushita K, Takahashi K, Adachi O, Biochemistry 1993, 32, 5576–5582. [DOI] [PubMed] [Google Scholar]
- [38].a) Klinman JP, Bonnot F, Chem. Rev 2014, 114, 4343–4365; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Koehn EM, Latham JA, Armand T, Evans RL, Tu X, Wilmot CM, Iavarone AT, Klinman JP, J. Am. Chem. Soc 2019, 141, 4398–4405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Dekker RH, Duine JA, Frank J, Verweil PEJ, Westerling J, Eur. J. Biochem 1982, 125, 69–73. [DOI] [PubMed] [Google Scholar]
- [40].Sweeny EA, Singh AB, Chakravarti R, Martinez-Guzman O, Saini A, Haque MM, Garee G, Dans PD, Hannibal L, Reddi AR, Stuehr DJ, J. Biol. Chem 2018, 293, 14557–14568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Stehling O, Wilbrecht C, Lill R, Biochimie 2014, 100, 61–77. [DOI] [PubMed] [Google Scholar]
- [42].Krissinel E, Henrick K, Acta Crystallogr., Sect. D: Biol. Crystallogr 2004, 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- [43].a) Ho BK, Gruswitz F, BMC Struct. Biol 2008, 8, 49; [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Voss NR, Gerstein M, Nucl. Acids Res 2010, 38, W555–W562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Williams PA, Coates L, Mohammed F, Gill R, Erskine PT, Coker A, Wood SP, Anthony C, Cooper JB, Acta. Cryst. D 2005, 61, 75–79. [DOI] [PubMed] [Google Scholar]
- [45].Chistoserdova L, Environ. Microbiol 2011, 13, 2603–2622. [DOI] [PubMed] [Google Scholar]
- [46].Chu F, Beck DAC, Lidstrom ME, PeerJ 2016, 4, e2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Englard S, Seifter S, Meth. Enzymol 1990, 182, 287–300. [DOI] [PubMed] [Google Scholar]
- [48].Armstrong JD, Biochim. Biophys. Acta 1964, 86, 194–197. [DOI] [PubMed] [Google Scholar]
- [49].Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A, in The Proteomics Protocols Handbook (Ed.: Walker JM), Humana Press, Totowa, NJ, 2005, pp. 571–607. [Google Scholar]
- [50].Otwinowski Z, Minor W, Methods Enzymol. 1997, 276, 307–326. [DOI] [PubMed] [Google Scholar]
- [51].Emsley P, Cowtan K, Acta Crystallogr., Sect. D: Biol. Crystallogr 2004, 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- [52].Murshudov GN, Vagin AA, Dodson EJ, Acta Crystallogr., Sect. D: Biol. Crystallogr 1997, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- [53].Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echoo ls N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH, Acta Cryst. 2010, D66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC, Acta Crystallogr., Sect. D: Biol. Crystallogr 2010, 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.