Abstract
Transcriptome data can facilitate the interpretation of the effects of rare genetic variants. Here, we introduce ANalysis of Expression VAriation (ANEVA) to quantify genetic variation in gene dosage from allelic expression (AE) data in a population. Application of ANEVA to the Genotype-Tissues Expression (GTEx) data showed that this variance estimate is robust and correlated with selective constraint in a gene. Using these variance estimates in a Dosage Outlier Test (ANEVA-DOT) applied to AE data from 70 Mendelian muscular disease patients showed accuracy in detecting genes with pathogenic variants in previously resolved cases, and lead to one confirmed and several potential new diagnoses. Using our reference estimates from GTEx data, ANEVA-DOT can be incorporated in rare disease diagnostic pipelines to utilize RNA-seq data more effectively.
One Sentence Summary:
New statistical framework for modeling allelic expression characterizes genetic regulatory variation in populations and informs diagnosis in rare disease patients
Background
Large reference databases of human exomes and genomes have enabled the characterization of genomic variation in human populations (1–3). These data have been used to summarize genic intolerance to damaging variants, where depletion of gene disrupting variants (e.g. stop gain variants) indicates deleterious fitness consequences (1, 4, 5). Such analyses are essential for prioritizing rare and de novo coding variants that can underlie Mendelian disease and provide a genetic diagnosis for 25–50% of the patients (6, 7). However, despite advances in DNA sequencing, the search for rare disease-causing variants outside the coding sequence has been hindered by the difficulty of interpreting rare regulatory variants and identifying their target genes.
Integration of genome and transcriptome sequencing data has provided improved diagnosis via better detection of rare variants with functional effects (6, 8–10). However, the often laborious analysis is further complicated by the transcriptome being affected by the environment, disease state, and technical variation. This has made it challenging to quantify when an effect is genetic and beyond the normal population range. Thus, most analyses have been limited to only a small fraction of variants that induce clear alterations in the transcriptome, such as total loss of expression and splice defects.
One promising data type is the allelic expression (AE) which measures the relative expression of the paternal and maternal haplotype of a gene in an individual. Departure from equal AE, allelic imbalance, is largely unaffected by environmental and technical factors with a reported heritability of 85% (11), and therefore, has a unique sensitivity to capture cis-acting genetic effects including those induced by rare variants (6, 12–14) (15). However, a quantitative framework for interpreting this unique data type to identify rare pathogenic variants has been lacking.
Here, we quantify the effects of genetic regulatory variation in populations using a mechanistic model of cis-regulatory variation. Specifically, for each gene we estimate VG, the expected variance in the dosage that is due to inter-individual genetic differences within a population. Next, we use VG as a reference to identify genes affected by potentially pathogenic regulatory variants in patients.
Results
A generative model for population allelic expression data and the ANEVA method
Cis-regulatory variant effect sizes can be quantified with allelic fold change (aFC; (16)). AFC has an analytical link to gene dosage, which would allow calculation of VG if all regulatory variants were known (Eq. 1–7). In practice, we can use AE data to estimate the overall distribution of regulatory effects on a gene without having to identify these variants explicitly. Across individuals, AE data represents a series of comparisons between net expression effects of all variants on two random haplotypes at a time. A major complication for applications of AE data is that within a population, it has diverse patterns depending on the properties of regulatory variants present and the SNP used to measure the allelic expression (aeSNP; Fig. 1A–D; (14, 15)). We derive a generative model for population AE data under a realistic scenario where a gene is regulated by several regulatory variants of which only some are identifiable. Under this assumption, population AE data is described by a constrained mixture of Binomial-Logit-Normal (BLN) probability distribution functions (Eq. 8–19). We fit this model to population AE data (Eq. 20–28, Fig. 1E–H) and use the maximum likelihood parameters to estimate VG indirectly (Eq. 29–30). We refer to this method as ANalysis of Expression Variation (ANEVA). Simulations show that the inferred VG is accurate (R2 =0.92, Fig. S1). Thus, ANEVA allows one to derive biologically interpretable estimates of genetic variation in gene expression within a population from AE read count data.
Figure 1: Cis-regulatory variation, allelic expression, and ANEVA.
A–D) Examples of allelic expression across individuals (dots) for four genes with a single aeSNP each. In (A) similar haplotype expression levels for the gene indicate little cis-regulatory variation. In (B–D) there is relatively more variation. In (C–D) there are distinct clusters driven by different haplotype combinations of a common, strong regulatory variant and the aeSNP, with strong linkage disequilibrium in (D). These examples illustrate the challenge of consistently modeling the underlying regulatory variants. E) Schematic representation of ANEVA, which uses a generative model of population AE data and a mechanistic model of cis-regulatory variation to estimates the magnitude of genetic variation in expression for each gene. F–H) A generative model of population AE data, represented mechanistically (F), in population AE data (G), and as Bayesian plate diagram (H; Eq. 20–22). AE data is modeled with one distinctly strong regulatory bi-allelic variant. If present, this variant is specified by its effect size, SH,L, and its LD with the aeSNP. Residual cis-regulatory variation is modeled as an infinite-allelic regulatory variant summarized by variance term . Allelic expressions er and ea are measured at a heterozygous aeSNP with reference (R) and alternative (A) alleles, and s*r,a is the aeSNP reference allele alignment bias. Haplotypes h1 and h2, basal expression level eb, and N cis-regulatory variant sites v1… vn, are components of our complete formal model of cis-regulatory variation.
ANEVA estimates from AE data are consistent with eQTL data and heritability of gene expression
We applied ANEVA to 10,361 RNA-seq samples from 48 tissues and 620 individuals with whole genome sequencing (WGS) data from the GTEx v7 data (17, 18). Overall, we estimated VG at a median of 43,219 autosomal aeSNPs per tissue. Gene-level VG was derived as a weighted harmonic mean of SNP-level estimates for a median of 4,962 genes per tissue, and a total of 14,084 genes (Fig. 2A–B, Table S1). First, we ensured that our AE-derived estimates of VG were consistent with what is expected from eQTL data (median corr. SDG = 0.73; Figs. 2C, S2, Table S2). Next, we benchmarked ANEVA estimates against gene expression cis-heritability (h2). For GTEx whole blood, we calculated the ratio of AE and eQTL-derived VG to the total variance of gene expression (VT). These ANEVA-based h2 estimates were consistent and comparable with those from standard methods and larger data sets, confirming that VG measures the genetic variation in gene expression (Figs. 2D, S3). Since AE-based ANEVA VG estimates are better applicable to AE-based outlier detection, we used these estimates for all subsequent analyses (Fig. S14, (19)).
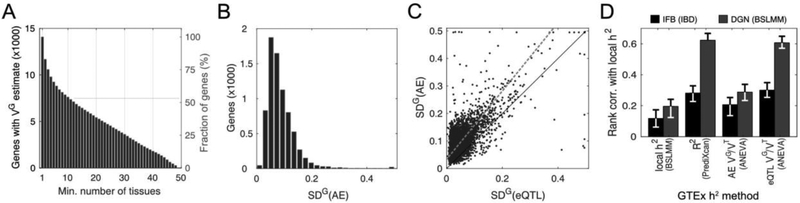
Figure 2: Estimates of genetic regulatory variation in GTEx.
A) Number of genes with VG estimates across 1 to 49 GTEX tissues; B–C) Distribution of SDG, , for 7556 genes in GTEx subcutaneous adipose (B), and its comparison to eQTL data (C; corr.=0.71). The red line is Deming regression fit (Fig. S2). SDG is capped at 0.5 for visualization. D) Benchmarking of ANEVA by gene expression heritability (h2) estimates. GTEx h2 was calculated by the linear mixed model based BSLMM, PrediXcan R2, and ANEVA (19). These were compared to two larger cohorts: BLSMM h2 from the DGN cohort (n=922; (20)), and local identity-by-descent (IBD) based h2 from the IFB cohort (n=722; (21)).
Genetically driven variation in gene expression across tissues, populations and gene sets
Next, we analyzed how VG varies between tissues and populations. The estimates were well correlated between tissues (median corr. SDG =0.57; Fig. 3A). For a given gene, VG tends to be smaller in tissues where the gene is more highly expressed (Wilcoxon signed rank test P<10–300; Fig. 3B). Since this was not an artifact of differences in read depth (Fig. S4), it suggests that there is an increased dosage sensitivity and a higher selective constraint in tissues where the gene has a more pronounced functional role (see Fig. S5 for an example). To analyze population differences in VG, we used ANEVA on AE data from three European and one African subpopulation from GEUVADIS data (22). We found a high correlation between estimates from all subpopulations (corr. range: [0.75, 0.83]; Fig. S6). This suggests that the total amount of genetic dosage variation is not highly variable between populations, and approaches that aggregate genetic effects at the gene level may have better applicability across populations than analyses of individual variants.
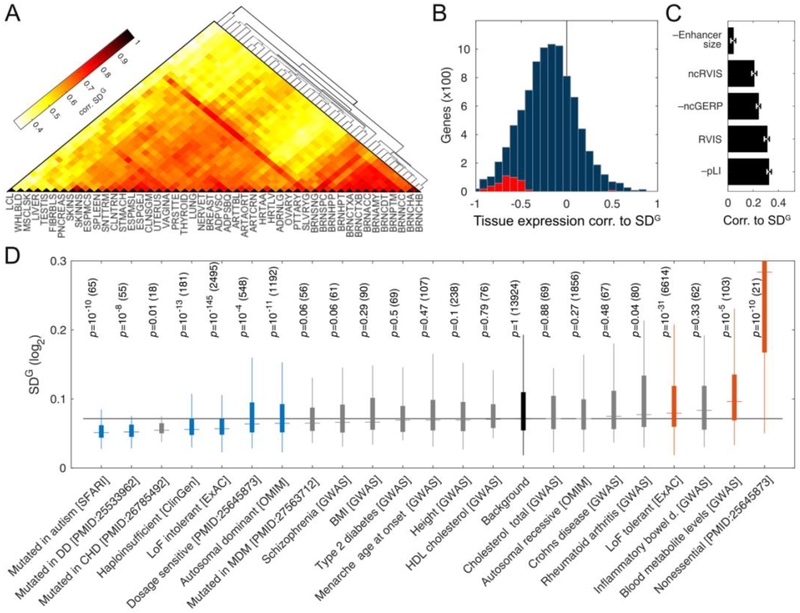
Figure 3: Biological sources of regulatory variation between genes.
A) Correlation of genetic regulatory variation across GTEx tissues (see Table S1 for tissue names). B) Rank correlation between median expression in a tissue and VG for 9,158 genes with VG estimates in at least five tissues. The distribution is shifted (median rank corr. −0.20). Significant genes are shown in red (5% FDR). C) Rank correlation of with enhancer size, coding constraint (RVIS, pLI), and noncoding constraint (ncRVIS) and conservation (ncGERP) in UTRs and promoters. D) for different gene sets (DD: Developmental disorder, CHD: Congenital heart disease, MDM: Congenital Muscular dystrophies and myopathies; Table S3), with nominal p-values from ranksum test compared to the background of all genes (p-value ≤ 0.01 highlighted), with the number of genes in parentheses. Boxes span the middle 50% values, and the whiskers span ±1.5 IQR from first and the third quartile.
To characterize differences in the amount of genetic regulatory variation between genes, we correlated VG to statistics of gene regulation and constraint. For each gene, we calculated a weighted harmonic mean of VG across tissues (; Table S1). Gene enhancer size had a minimal correlation to (Fig. 3C; (23)), suggesting that the size of the mutational target, a proxy for the background mutation rate plays a minor role. Genes with high purifying selection for coding gene disrupting variants, or noncoding variants in the promoter or UTR regions, were depleted of genetic regulatory variation (Fig. 3C), as previously observed by eQTL analysis (1). Rare disease genes had lower , while loss of function tolerant genes had higher (Fig. 3D), showing that dosage sensitivity is captured by both exome and regulatory variation analysis. Genes identified by genome wide association studies (GWAS) showed little deviation from the background, but schizophrenia genes having the lowest VG, and blood metabolite genes the highest suggests a link to genetic architecture of these traits. Altogether, the amount of genetic regulation variation measured as VG can complement previous coding and regulatory variation analyses of selective constraint on genes and traits.
Genetically driven variation in gene expression and dosage outlier testing from AE data
In addition to these biological insights, VG has a direct practical application in identifying population outliers that may be pathogenic. To this end, we developed ANEVA Dosage Outlier Test (ANEVA-DOT) to identify genes likely affected by a heterozygous genetic variant with an unusually strong effect on gene dosage. Using VG for each gene, ANEVA-DOT tests against the null hypothesis that the observed allelic imbalance in an individual is consistent with dosage variation in the general population (Fig. 4A) while accounting for a number of additional technical and biological sources of variation (Eq. 31–42). We used extensive simulations to ensure that the test is well calibrated (Fig. S7). ANEVA-DOT is implemented in R, and it runs in a few seconds per sample (24).
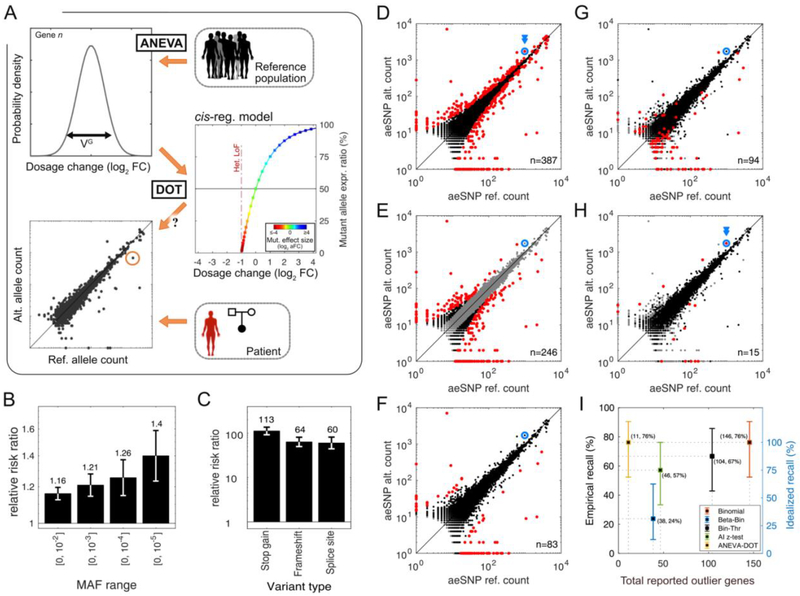
Figure 4: Regulatory outliers detected with the ANEVA Dosage Outlier Test.
A) Illustration of the ANEVA-DOT method. For each gene, the null distribution of allelic imbalance is estimated using the VG and the model of cis-regulatory genetic effect. Allelic counts in a test individual are compared to this null, accounting for sampling noise, sequencing noise, reference bias, and the variant haplotype. B–C) Enrichment of all rare variants in ANEVA-DOT genes as a function of allele frequency (B) and for putative gene-disrupting variants (MAF<1%; C). D–H) An example of AE data for all genes from one previously diagnosed muscle dystrophy patient (N13). The disease gene is shown in blue. Outlier genes identified by different tests (5% FDR) are marked in red: binomial (n=387; D), binomial with a 15% allelic imbalance threshold (Bin-Thr, n=246; E), beta-binomial (Beta-Bin, n=83; F), excess allelic imbalance against GTEx data via z-test (AI z-test, n=94; G), and ANEVA-DOT (n=15; H). Genes marked in grey are excluded from each test. I) Fraction of true causal genes identified in previously diagnosed patients (recall) and its 95% bootstrap confidence intervals versus the number of outliers reported. Empirical recall (left) is calculated using all cases where imbalanced AE would be expected (n=21), while idealized recall (right) excludes five cases in which detecting the gene from AE data is impossible (e.g. when the causal gene is not expressed; Fig. S12).
We first tested ANEVA-DOT in the general population of 466 skeletal muscle samples from GTEx. Each sample had a median of 3,390 genes tested and 10 genes identified as outliers at 5% FDR (hereafter ANEVA-DOT genes; 90% range: [3, 22]). An average of 56% of the genes previously implicated in neuromuscular disorders (6, 25), and up to 46% of the highly expressed genes were testable per individual (Fig. S8). As a quality filter, 113 out of 5848 tested genes that appeared as outliers in >1% of the individuals were excluded from further analysis (Fig. S8D–F, Table S4). After this step, a median of 4.5 ANEVA-DOT genes were retained per individual (90% range: [1, 14]; Fig. 4B). ANEVA-DOT genes were highly enriched for rare heterozygous variants in a 10kb window upstream of the TSS and in the gene body (Fig. 4B). This enrichment was particularly pronounced for rare putative gene disrupting variants that are expected to have a strong effect on gene expression levels via nonsense-mediated decay (Fig. 4B–C). This confirms that ANEVA-DOT captures rare genetic effects on gene dosage.
Next, we evaluated how sensitive ANEVA-DOT is to differences in the reference population where VG is calculated. First, using the GEUVADIS data (22), we looked for ANEVA-DOT genes in 86 European (GBR) individuals using VG estimates derived from two European (FIN and TSI), and one African (YRI) populations. The three reference populations performed similarly with an average of 74% (69%–78%) of ANEVA-DOT genes identified using one confirmed by another (Fig. S9), suggesting that the lack of full concordance is likely driven by noise and threshold effects. However, larger sample sizes will be needed for a comprehensive evaluation of the population effects._Next, we checked if ANEVA-DOT genes in GTEx skeletal muscle could be identified by analyzing other accessible tissues of these individuals. The detection rate varied from 23.3% in fibroblast to 12.3% in whole blood, which indicates that ANEVA-DOT can capture some outlier effects also from proxy tissues (Fig. S10).
ANEVA-DOT accurately identifies disease genes in AE data from rare disease patients
To test ANEVA-DOT’s performance in the diagnosis of rare disease patients, we applied it to AE data from 70 rare Mendelian muscle dystrophy and myopathy (MDM) patients using VG reference from GTEx skeletal muscle (Figs. S11–S17, Table S5). Out of the 65 patients with high quality data, 32 have a previous diagnosis, of which 21 are expected to lead to allelic imbalance (6). These cases were used as positive controls to benchmark ANEVA-DOT against previous tests of allelic imbalance: binomial and beta-binomial tests, binomial test with an allelic imbalance threshold, and a naive population-aware test of excess allelic imbalance against GTEx data via z-test (Fig. 4D–H, Fig. S12). ANEVA-DOT identified a median of 11 outlier genes per individual (out of a median of 2190 tested), substantially less than other tests, (Fig. 4H). This small number of outliers always included the previously diagnosed gene when there was a detectable allelic imbalance present (76%; Figs. S11–S12), typically (69%) among the top-five most significant genes (Table S5). ANEVA-DOT’s high recall and precision outperformed all the other tests by a substantial margin (Figs. 4I, S12–S14, (19)).
In the 33 patients without a genetic diagnosis from previous WES and/or WGS or RNA-seq analysis (6), we found a median of nine ANEVA-DOT genes per sample (in total 349 genes), which included at least one neuromuscular disease gene (6, 25) in 12 patients (in total 17 genes; Figs. S15–S16). One of these potential new diagnoses from ANEVA-DOT was confirmed: Patient N10, with limb-girdle muscular dystrophy-like phenotype, had 13 ANEVA-DOT genes, with the one known Mendelian muscle disease gene, DES being the most significant. Further RNA-sequencing and RT-PCR analysis identified a pseudo-exon insertion caused by a variant creating an intronic splice site. This had been missed by the prior gene panel, WES, WGS and RNA-seq analysis due to challenging in silico interpretation of intronic variants and the relatively low number of RNA-seq reads. The variant is in trans with a pathogenic missense variant that had not been identified as a diagnosis due to the lack of a second variant (Fig. S18). Additionally, ANEVADOT identified strong candidates in six cases and possible candidates in 11 others (19). By design, ANEVADOT does not rely on identifying which variant underlies the dosage outlier effect, but genetic analysis can be applied after prioritizing genes by ANEVADOT. This is currently mostly limited to gene or splice disrupting variants due to their easier annotation compared to rare regulatory variant candidates that may also exist. Overall, we expect up to 10.5 of the 17 known MDM, and 18.8 of all 349 identified ANEVADOT genes in the 33 undiagnosed patients to be true disrupted causative genes (19).
Discussion
In this study, we introduce a method, ANalysis of Expression VAriation (ANEVA), and its extension ANEVA Dosage Outlier Test (ANEVA-DOT) to quantify genetic variation in gene dosage in the general population, and to identify genes where a patient appears to carry a heterozygous variant with an unusually strong effect on gene expression. This enables individual transcriptome comparison to previously generated reference data without the caveats of technical and reverse causation noise in total gene expression analysis.
The ANEVA framework uses biologically interpretable units of gene dosage, allowing interpretation of regulatory and coding gene disrupting variants on the same scale. Furthermore, the statistical methods introduced here for modeling allelic expression data are applicable to other uses of this data type.
ANEVA-DOT is a fast and powerful approach for finding genes with likely disease effects, with the small numbers of outliers making further manual curation feasible in a clinical setting without compromising on sensitivity. The use of VG estimates from GTEx as a shared reference for ANEVA-DOT analysis of patients is analogous to use of coding constraint metrics for prioritization of pathogenic coding variants. ANEVA-DOT outlier genes can be further prioritized by candidate gene lists and by tools that are currently used in exome sequencing follow-up (1, 2, 5, 26, 27). Since ANEVA-DOT captures transcriptome outcomes of genetic effects without having to identify rare regulatory variants themselves, this method is particularly advantageous for rare genetic effects from poorly defined regulatory elements, but it will also detect, for example, variants triggering transcript decay. However, identifying the specific variants underlying ANEVA-DOT outliers is still challenging despite existing variant prioritization approaches, especially for noncoding regions (28–30).
Despite these advantages, our methods have several limitations. The main caveat is that AE data is sparse, and VG estimates may be lacking or noisy for genes with few common coding variants due to small size or high coding constraint, or low expression levels. These issues will, however, improve with increasingly large RNA-seq data sets. ANEVA-DOT is only applicable to about half of expressed genes per individual that have an aeSNP. Finally, allelic imbalance is not informative of recessive effects without family analysis. Thus, similarly to other genetic diagnosis tools, ANEVA-DOT should be used in conjunction with other methods to capture different types of rare variants underlying disease. We envision that in clinical genetics, when practically feasible, transcriptome data will become a powerful additional layer of data for interpreting the genome and its disease-contributing variants.
Supplementary Material
Acknowledgements
We would like to thank the GTEx consortium, in particular, F. Aguet, and N. Ferraro, and S. Montgomery.
Funding: This work was funded by following NIH grants: UL1TR002550 (P.M.), UL1TR001114 (P.M.), K99HG009916 (S.E.C.), R01MH106842 (T.L., P.H.), UM1HG008901 (T.L., J.E.), R01GM122924 (T.L.). R01MH107666 (H.K.I. and H.E.W.), P30DK020595 (H.K.I.), R15HG009569 (H.E.W.), UM1 HG008900 (D.G.M.), and intramural funds from the NIH (C.G.B., A.R.F., Pa.M., S.D.).
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Competing interests: D.G.M. is a founder with equity, and T.L. is an advisor in Goldfinch Bio. S.E.C. is a co-founder, chief technology officer, and stock owner in Variant Bio. T.L. is an advisor with equity in Variant Bio. H.K.I. has received honoraria from GSK and AbbVie. The other authors have no competing interests.
Data and materials availability: Software packages for the presented methods are available online: BLN distribution functions (31), ANEVA (32), ANEVA-DOT (24). Outlier summary statistics for all GTEx tissues are available in (33). The GTEx v7 data is available in dbGap (phs000424.v7.p2), and the MDM cohort data is available in dbGap phs000655. Gene level AE data and ANEVA-DOT results in MDM cohort is given in supplementary Data S1.
References
- 1.Lek M et al. , Analysis of protein-coding genetic variation in 60,706 humans. Nature. 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karczewski KJ et al. , Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. BioRxiv (2019), doi: 10.1101/531210. [DOI] [Google Scholar]
- 3.1000 Genomes Project Consortium et al. , A global reference for human genetic variation. Nature. 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Havrilla JM, Pedersen BS, Layer RM, Quinlan AR, A map of constrained coding regions in the human genome. Nat. Genet 51, 88–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB, Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet 9, e1003709 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cummings BB et al. , Improving genetic diagnosis in Mendelian disease with transcriptome sequencing. Sci. Transl. Med 9 (2017), doi: 10.1126/scitranslmed.aal5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farnaes L et al. , Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med 3, 10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kremer LS et al. , Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat. Commun 8, 15824 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gonorazky HD et al. , Expanding the boundaries of RNA sequencing as a diagnostic tool for rare mendelian disease. Am. J. Hum. Genet 104, 466–483 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fresard L et al. , Identification of rare-disease genes using blood transcriptome sequencing and large control cohorts. Nat. Med 25, 911–919 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buil A et al. , Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat. Genet 47, 88–91 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li X et al. , The impact of rare variation on gene expression across tissues. Nature. 550, 239–243 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Goldstein DB, Enhancer redundancy predicts gene pathogenicity and informs complex disease gene discovery. BioRxiv (2018), doi: 10.1101/459123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKean DM et al. , Loss of RNA expression and allele-specific expression associated with congenital heart disease. Nat. Commun 7, 12824 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glassberg EC, Gao Z, Harpak A, Lan X, Pritchard JK, Evidence for weak selective constraint on human gene expression. Genetics. 211, 757–772 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mohammadi P, Castel SE, Brown AA, Lappalainen T, Quantifying the regulatory effect size of cis-acting genetic variation using allelic fold change. Genome Res 27, 1872–1884 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GTEx Consortium, The Genotype-Tissue Expression (GTEx) project. Nat. Genet 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.GTEx Consortium et al. , Genetic effects on gene expression across human tissues. Nature. 550, 204–213 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.See supplementary materials on Science Online.
- 20.Battle A et al. , Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res 24, 14–24 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Price AL et al. , Single-tissue and cross-tissue heritability of gene expression via identity-by-descent in related or unrelated individuals. PLoS Genet 7, e1001317 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lappalainen T et al. , Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 501, 506–511 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fishilevich S et al. , GeneHancer: genome-wide integration of enhancers and target genes in GeneCards. Database (Oxford). 2017 (2017), doi: 10.1093/database/bax028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammadi P, Sousa C, ANEVA-DOT software. (2019), url: 10.5281/zenodo.3406690. [DOI]
- 25.Kaplan J-C, Hamroun D, The 2016 version of the gene table of monogenic neuromuscular disorders (nuclear genome). Neuromuscul. Disord 25, 991–1020 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Birgmeier J et al. , AMELIE accelerates Mendelian patient diagnosis directly from the primary literature. BioRxiv (2017), doi: 10.1101/171322. [DOI] [Google Scholar]
- 27.Deelen P et al. , Improving the diagnostic yield of exome-sequencing by predicting gene-phenotype associations using large-scale gene expression analysis. Nat. Commun 10, 2837 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M, CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.di Iulio J et al. , The human noncoding genome defined by genetic diversity. Nat. Genet 50, 333–337 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Anderson D, Lassmann T, A phenotype centric benchmark of variant prioritisation tools. NPJ Genom. Med 3, 5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mohammadi P, Hoffman P, R package for BLN distribution functions. (2019), url: 10.5281/zenodo.3406692. [DOI]
- 32.Mohammadi P, ANEVA software. (2019), url: 10.5281/zenodo.3406688. [DOI]
- 33.Mohammadi P, PejLab/Datasets: GTEx v7 Vg and freq. estimates. (2019), url: 10.5281/zenodo.3406717. [DOI]
- 34.Castel SE, Levy-Moonshine A, Mohammadi P, Banks E, Lappalainen T, Tools and best practices for data processing in allelic expression analysis. Genome Biol 16, 195 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anders S, Huber W, Differential expression analysis for sequence count data. Genome Biol 11, R106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wheeler HE et al. , Survey of the Heritability and Sparse Architecture of Gene Expression Traits across Human Tissues. PLoS Genet 12, e1006423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gamazon ER et al. , A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet 47, 1091–1098 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Petrovski S et al. , The intolerance of regulatory sequence to genetic variation predicts gene dosage sensitivity. PLoS Genet 11, e1005492 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zarrei M, MacDonald JR, Merico D, Scherer SW, A copy number variation map of the human genome. Nat. Rev. Genet 16, 172–183 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Homsy J et al. , De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 350, 1262–1266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deciphering Developmental Disorders Study, Large-scale discovery of novel genetic causes of developmental disorders. Nature. 519, 223–228 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Castel SE, Mohammadi P, Chung WK, Shen Y, Lappalainen T, Rare variant phasing and haplotypic expression from RNA sequencing with phASER. Nat. Commun 7, 12817 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van de Geijn B, McVicker G, Gilad Y, Pritchard JK, WASP: allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods 12, 1061–1063 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baran Y et al. , The landscape of genomic imprinting across diverse adult human tissues. Genome Res 25, 927–936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McLaren W et al. , The ensembl variant effect predictor. Genome Biol 17, 122 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorvaldsdottir H, Robinson JT, Mesirov JP, Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinformatics 14, 178–192 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gusev A et al. , Integrative approaches for large-scale transcriptome-wide association studies. Nat. Genet 48, 245–252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.