Abstract
Purpose
Unlike conventional Magnetic Resonance Spectroscopy (MRS), which only measures metabolite concentrations, multiparametric MRS also quantifies their longitudinal (T1) and transverse (T2) relaxation times, as well as the radiofrequency transmitter inhomogeneity (B1+). To test whether knowledge of these additional parameters can improve the clinical utility of brain MRS, we compare the conventional and multiparametric approaches in terms of expected classification accuracy in differentiating controls from patients with neurological disorders.
Methods
A literature review was conducted to compile metabolic concentrations and relaxation times in a wide range of neuropathologies and regions of interest. Simulations were performed to construct receiver operating characteristic curves and compute the associated areas (AUC) to examine the sensitivity and specificity of MRS for detecting each pathology in each region. Classification accuracy was assessed using metabolite concentrations corrected using population-averages for T1, T2 and B1+ (conventional MRS); using metabolite concentrations corrected using per-subject values (multiparametric MRS); and using an optimal linear multiparametric estimator comprised of the metabolites’ concentrations and relaxation constants (multiparametric MRS). Additional simulations were conducted to find the minimal intra-subject precision needed for each parameter.
Results
Compared to conventional MRS, multiparametric approaches yielded AUC improvements for almost all neuropathologies and regions of interest. The median AUC increased by 0.14 over the entire dataset, and by 0.24 over the ten instances with the largest individual increases.
Conclusions
Multiparametric MRS can substantially improve the clinical utility of MRS in diagnosing and assessing brain pathology, motivating the design and use of novel multiparametric sequences.
Keywords: Quantitative MRI, multiparametric MRI, MRS, single-voxel spectroscopy, qMRI
1. Introduction
In recent years, there has been substantial interest in transforming the field of magnetic resonance into quantitative and multiparametric, capable of producing simultaneous estimates of multiple tissue parameters, such as the proton density, longitudinal and transverse relaxation coefficients (T1, T2), and hardware radiofrequency transmitter imperfections (B1+) within clinical timeframes. Several approaches to this end have been suggested, ranging from explicit model based estimations (1–3), to simulated echo modulation curves (4,5), to magnetic resonance fingerprinting (6–8). These schemes are almost universally multiparametric: they simultaneously uncover all, or most parameters. Since T1 and T2 are known to change in disease, these additional data can be used to enhance clinical assessment and diagnosis. Similar interest has been shown recently in the field of Magnetic Resonance Spectroscopy (MRS) (9,10), where knowledge of metabolites’ relaxation constants and transmitter inhomogeneity serves two purposes:
Metabolites’ T1s and T2s probe microscopic changes in specific cellular environments. An example of this is n-acetyl-aspartate (NAA), which is mostly found in neurons and is an established biomarker of neuronal health (11). While NAA’s concentrations reflect neuronal metabolism, NAA’s T1 and T2 independently reflect the equally-important neuronal microenvironment.
In the context of MRS, a second, unique use of multiparametric data arises: The acquired metabolite concentrations are “weighted” by T1 and T2. Knowledge of each metabolite’s T1 and T2 constants, as well as of B1+, is required to properly correct for this weighting (12). Due to the long acquisition times conventionally required for their accurate per-subject determination – using, e.g., inversion recovery and multi-echo sequences – these are not acquired on a per-subject basis in clinical settings. As an approximation, tabulated values of metabolites’ T1s and T2s, representing population averages, are conventionally employed. However, the very use of tabulated values does not account for the large inter-subject and regional variability in T1 and T2, and introduces unwanted errors into the correction process. This holds even when using advanced sequences which minimize the contribution of B1+, such as LASER (13), semiLASER (14,15) or STRESS (16,17).
Herein, we examine by how much multiparametric spectroscopic data – yielding not only metabolite concentrations, but also their subject-specific T1s, T2s, and B1+ – improves the detection and assessment of several well-known neuropathologies using single-voxel MRS. To carry out this analysis, we treat the tissue’s metabolic parameters as binary classifiers which can be used to distinguish between two states of tissue: healthy and pathological. We then evaluate the utility of these classifiers using the concept of receiver operating characteristic (ROC) curves and, in particular, the area under the curve (AUC). The AUC ranges from 0.5 (useless; equivalent to a coin toss) to 1.0 (perfect classifier), and is used ubiquitously in assessing the performance of binary classifiers (18,19). Increases to the AUC translate to improved accuracy in clinical practice, or to smaller sample sizes when using MRS as an endpoint in a clinical trial. We compare the AUC of three binary classifiers in order of expected increasing accuracy:
C1. Metabolite concentrations, as obtained using conventional, non-multiparametric MRS, after correcting for signal weighting using population average values of T1 and T2, and assuming B1+=1.
C2. Metabolite concentrations, corrected using subject-specific values of T1 and T2, as well as knowledge of B1+, as acquired using multiparametric MRS.
C3. An optimal linear classifier comprised of corrected metabolite levels (same as (C2)), as well as subjects’ T1 and T2, which themselves change in pathology.
All multiparametric schemes will inevitably exhibit some amount of intra-subject variability. Its effects will also be examined and used to derive guidelines regarding what constitutes “acceptable” variability which should be demanded of multiparametric sequences. These comparisons will serve to assess the importance of high quality multiparametric quantitative acquisitions to MRS, as well as to potentially motivate the development of methodologies capable of acquiring such data within clinical scan times.
Theory
A Simple Model for Quantification in MRS
To estimate the gains to the AUC obtained by fully knowing each subject’s individual T1, T2 and B1+, one must assume a model for their effect on the signal. This will depend on the sequence employed. For a simple semiLASER-like model, whereby only the first pulse introduces B1+ weighting and the pulse spacing is sufficiently close so as to neglect any alterations to T1 weighting beyond the simple pulse-acquire scheme, the MRS signal becomes weighted according to:
| (1) |
where Cmet, are the metabolite’s concentration and relaxation constants, , B1− the receiver sensitivity and FA the flip angle. The MRS signal is conventionally corrected for relaxation by dividing by a factor of the form:
| (2) |
Conventionally, the values of are taken from literature data, where available. Since most clinical protocols do not quantify transmitter inhomogeneities, B1+,c=1 is often assumed. For absolute quantification, the acquired signal is divided by a second reference signal, often water, exhibiting the same behavior as shown in Eq. (1) and requiring its own correction factor, fref, with its own , but the same B1+,c This is then multiplied by some assumed concentration of the reference signal, , such that the final estimated quantity is:
| (3) |
For a perfectly accurate multiparametric MRS sequence, the values of are B1+,c are assumed known, leading to fmet=fref
Construction of Optimal Linear Multiparametric Classifiers
Given a collection of M normally distributed uncorrelated binary classifiers X1, X2, …, XM, , form X=(X1,X2,…,XM), their multivariate joint distribution. Assume X~N(μh, ∑h) in healthy tissue and X~N(μp, ∑p) in pathology, with μ = (μ1,μ2,…,μM) and ∑ the diagonal covariance matrix, . The linear combination Y = a1X1+ a2X2 +…+aMXM = aTX is then normally distributed with Y~N(aTμh,aT∑ha) in healthy tissue and Y~N(aTμp,aT∑pa) in pathology. The AUC of the combined classifier is (20):
| (4) |
where Φ is the cumulative normal distribution function. The coefficients a1,a2,…,aM can be chosen in a way which maximizes Eq. (4):
| (5) |
Using these coefficients, the AUC of the combined classifier Y becomes (20):
| (6) |
In this manner, several classifiers, such as concentrations, T1 and T2 can be optimally combined to improve their sensitivity and specificity.
Methods
Literature Search
A literature search was conducted on metabolite concentrations as well as metabolite and water relaxation times and their standard deviations (SDs) in brain pathologies. The following criteria were applied: adult populations (>18 years of age); only proton MRS studies; only NAA, choline (Cho) and creatine (Cr), due to the almost complete lack of concentration and relaxation data of other metabolites; for the concentrations, only millimolar (mM) values showing statistically significant changes in pathology (p<0.05) and obtained by absolute quantification (i.e. studies reporting institutional units were not included); for the relaxation data, only studies done at field strength of 1.5T or higher. A data entry (disease/region) was created for each instance of reported metabolic or water relaxation times in disease. Even though no published relaxation times could be found for two regions in mild cognitive impairment (MCI), they were nevertheless included to match the regions listed for the closely related Alzheimer’s Disease (AD). When multiple relaxation values for a particular tissue type were found within a single study, an averaged mean±SD was recorded.
Quantitative MRS data for many pathologies and regions is often partial. Where pathological values were missing for either T1 or T2, we assumed them to remain unchanged in pathology and substituted for them with values from healthy controls from a similar region (or at least tissue type).
Comparison of Conventional and Multiparametric MRS
A single-voxel acquisition was assumed with the following parameters: TE=40 ms, to minimize T2 weighting; FA=90°, to minimize B1+ weighting; and TR=1500 ms, approximately equal to 1.2 T1, to maximize sensitivity (21). Using these parameters, and the assumed signal model of Eq. (1), the AUC was calculated for each of the literature-reviewed cases for (C1)-(C3) listed in the Introduction. Each metabolite was considered independently.
To assess the AUC for (C1), Monte Carlo simulations were employed to randomly generate values of , Cmet, Cref and B1+ for a large number of controls and patients (Nsub=10,000). We’ve made the simplifying assumptions that tissue parameters are normally distributed: and , and uncorrelated. This is physiologically reasonable: the concentration of a metabolite (C) should not be correlated to its microenvironment (T1, T2) or to hardware related artifacts (B1+). Water was used as the reference signal. Since water concentrations are rarely quantified, we assumed them to be stable, fully known, and equal to some arbitrary constant. Patients’ and controls’ means and variances of the concentrations and relaxation times for both the metabolite of interest and water were taken from the results of our literature review. B1+ is pathology independent and therefore we assumed that . At clinical (3T) field strengths, a ±10% variation in B1+ is common (22), and thus σB = 0.1. The values of B1+, concentrations and relaxation times were fed into Eq. (3) to generate absolutely-quantified metabolite concentrations, , for each control and patient. To calculate the AUC, a classification threshold, TH, was fixed; in pathologies where metabolic concentrations decline, values of were classified as controls, while were classified as patients (and vice versa). The threshold TH was varied from 0 mM to 20 mM in steps of 0.05 mM, and the fraction of false positives and true positives was calculated as a function of TH. An ROC curve was constructed by plotting the true positive fraction as a function of the false positive fraction, and the AUC was calculated via numerical integration. To ensure accuracy, three simulations were run consecutively, and cases where the resulting AUCs disagreed by more than 0.5% were repeated by increasing Nsub by 5,000, until the target 0.5% accuracy was obtained.
For (C2), the same Monte Carlo simulations for (C1) were repeated, but assuming the exact values of T1, T2 and B1+ were known for each subject for both the metabolite being quantified and the reference water signal. For (C3), an optimal linear classifier Λ = ηCC + η1T1 + η2T2 was formed, with coefficients calculated using Eq. (5). Its AUC was calculated using Eq. (6).
We note that several sources reported mean±SD with coefficients of variation (CV=SD/mean) larger than 30%. These cases are problematic to model as Gaussian distributions since the probability of generating a meaningless negative value over a population of Nsubj=10,000 samples is non-negligible. Thus, we further employed a “cutoff” value in our simulations: quantities which dipped below 70% of the mean value were discarded and re-generated. This has the effect of slightly shifting the mean and standard deviation of the generated (non-Gaussian) distribution, although for the vast majority of cases these shifts are small. We marked (but did not omit) cases with substantial skews, where reported CVs exceed 30% for at least one of the variables in either the patient or control population.
Assessing the Effect of Each Spin Parameter on Classification Accuracy
The AUC was also calculated for three additional cases: (V1) The case where B1+ was assumed known on a per-subject basis, e.g., using an auxiliary B1 mapping sequence; (V2) An ultra-long TR sequence (TR/TE=10000/40 ms), minimizing T1 and some B1+ weighting; and (V3) An ultra-short TE sequence (TR/TE=1500/0.1 ms), minimizing T2 weighting. The same Monte Carlo pipeline used for (C1) was employed for each of the cases (V1)-(V3), using population-averaged values for T1 and T2. The purpose of these additional comparisons was to assess the relative impact of each variable (T1, T2, B1+) on the classification accuracy, and shed light on whether some variables are more important to quantify than others.
Assessing the Effect of Sequence Precision on Multiparametric MRS
MRS data is noisy, and thus any multiparametric MRS technique will suffer from some degree of error, which will result in intra-subject variability. At some point, this variability might become so significant so as to introduce greater uncertainty than the one it was intended to correct. To assess the point at which this happens, our simulations for case (C2) and (C3) were repeated while including a non-zero intra-subject coefficient of variation (CVs) for each quantity (T1, T2, C, B1+) and recalculating the AUCs. The CVs for each parameter were varied from 0% up to 25% while keeping all other CVs at zero. Note that these simulations quantify the effect of intra-subject variability, in contrast to cases (V1)-(V3) considered above, which quantify the effect of inter-subject variability.
For recalculating (C2), the Monte-Carlo simulations were repeated, and additional variability was added to each subject’s parameter. For example, for T1, it was assumed the method yielded , where is that subject’s true T1 value. All other steps, including generation of the ROC curves and numerical evaluation of their associated AUCs, remained the same. For recalculating (C3), the optimal combination coefficients (Eq. (5)) of the metabolite’s concentrations, T1 and T2 were calculated as before according to the population means and SDs obtained from the literature search. The intra-subject variability was added to each variable’s variance – e.g., , reasonably assuming lack of correlation between intra- and inter-subject sources of variability – and the AUC of the linear combination was then calculated using Eq. (4) directly.
Results
Comparison of Conventional and Multiparametric MRS
Table 1 lists the results of the conducted literature review of concentrations and relaxation times in neurological disorders. A total of 18 regions (including abnormal-appearing tissue and tumors) within 7 neuropathologies were identified. One hundred sixty-two instances of published metabolite and water relaxation times were found in disease. Most were measured at 1.5T and 3T, with single cases at 2.0T and 4T. The remaining 129 relaxation time entries in Table 1 were replaced with values reported for healthy controls.
Table 1.
Literature review on the variations of T1, T2 and metabolite concentrations in seven neurological disorders, displayed as mean±standard deviation (SD), with numbers in parentheses indicating values for matched controls. Not all studies report sample standard deviations; where missing, they are marked by an asterisk (*) and are assumed as ±10%. Where T1 or T2 values in pathology were missing from the literature, only values in healthy subjects from the same region or tissue type are given (in parenthesis). If region-specific healthy subject GM values were not available, (g) and (o) indicate that they were substituted with a global GM, or other regional GM, respectively. For multiple sclerosis (MS) lesions the healthy values shown are from WM, while for glioma and meningioma they are from GM. Hippocampal pathology can be lateralized, therefore, we specify whether the values were reported for the average (a), left (l), or right (r) hippocampus. “N/A” under “Concentrations” refers to lack of reports on statistically significant concentration changes between patients and controls, measured in millimolar. Other abbreviations: AD=Alzheimer’s Disease, ALS=amyotrophic lateral sclerosis, CG=cingulate gyrus, HIV=human immunodeficiency virus, MCI=mild cognitive impairment, NAGM=normal-appearing gray matter, NAWM=normal-appearing white matter, PCG=posterior cingulate gyrus.
| Disorder | Region | Metab. | Concentration (mM) | 1.5 T | 3.0 T | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Metabolite | Water | Metabolite | Water | ||||||||
| T1 | T2 | T1 | T2 | T1 | T2 | T1 | T2 | ||||
| AD | NAWM | NAA | 9.6±1.2 (10.8±1.2) (47) 9.4±1.6 (10.6±1.2) (47) 7.6±2.0 (9.7±0.8) (48) 8.3±1.2 (9.2±0.8) (49) |
1250±125* (1160±116*) (50) 1500±700 (1500±300) (48) |
469±262 (272±58) (48) | (656±16) (51) | 101±7 (100±8) (52) 87±9 (87±10) (53) 68±3 (67±2) (54) 70±1 (66±3) (55) |
(1560±60) (56) (1350±268) (38) |
(301±18) (57) (295±65) (38) |
(832±44) (58) | (80±3) (58) |
| Cho | 1.6±0.3 (1.8±0.2) (47) 1.7±0.2 (1.8±0.3) (47) |
980±98* (1030±103*) (50) 1400±600 (1400±500) (48) |
382±91 (341±90) (48) | (1210±130) (56) (1080±134) (38) |
(222±17) (57) (187±45) (38) |
||||||
| Cr | N/A | 1150±115* (1080±108*) (50) 1700±300 (1600±400) (48) |
201±38 (190±45) (48) | (1400±60) (56) (1240±223) (38) |
(178±9) (57) (156±45) (38) |
||||||
| PCG | NAA | 8.9±1.4 (9.8±1.0) (47,59) 8.7±1.3 (9.8±1.1) (60) 8.3±0.8* (9.3±0.9*) (50) 9.4±1.2 (10.5±1.6) (49) |
1110±111* (1150±115*) (50) | 456±40 (402±147) (50) | (1188±69) (51) o | (86±9*) (61) | (1470±80) (56) (1470±134) (38) |
254±36 (235±24) (62) | (1356±174) (58) | 61±11 (64±29) (62) | |
| Cho | N/A | 1300±130* (1250±125*) (50) | 418±42 (386±19) (50) | (1250±220) (56) (1300±134) (38) |
190±43 (176±37) (62) | ||||||
| Cr | 6.3±0.8 (6.9±0.8) (60) | 1330±133* (1120±112*) (50) | 201±28 (186±42) (50) | (1330±130) (56) (1460±157) (38) |
151±21 (141±16) (62) | ||||||
| Region | Metab. | Concentdation (mM) | 1.5 T | 2.0 T | |||||||
| Metabolite | Water | Metabolite | Water | ||||||||
| T1 | T2 | T1 | T2 | T1 | T2 | T1 | T2 | ||||
| Hippo-campus | NAA | 6.8±1.1 (8.5±1.0) (47) r 6.8±1.1 (8.7±1.0) (47) l 8.6±0.7 (9.8±1.2) (63) l 6.6±0.7 (7.6±0.8) (64) a 6.9±1.2 (8.4±1.3) (60) l 7.0±1.2 (8.3±1.4) (60) r 7.8±0.3 (9.3±0.3) (65) l 7.7±0.2 (9.1±0.2) (65) r 11.7±2.4 (13.3±3.2) (66) |
(1100±300) (67) r | (222±121) (67) r | (1300±174) (68) a | 73±8 (81±5) (69) r 130±15 (98±5) (52) a 99±10 (94±7) (53) a 98±10 (95±8) (70) a 79±7 (78±6) (71) a 98±7 (90±3) (72) a |
(1408±253) (73) a | (283±62) (73) a | N/A | 93±10 (85±6) (63) l | |
| Cho | 1.7±0.3 (1.9±0.3) (60) l 1.7±0.4 (1.9±0.3) (47) r 1.6±0.3 (1.9±0.3) (47) l |
(1600±800) (67) r | (359±300) (67) r | (1438±365) (73) a | (334±106) (73) a | ||||||
| Cr | 5.6±0.8 (6.1±0.5) (64) a 6.6±1.2 (7.1±1.0) (47) r 6.5±1.0 (7.1±1.1) (47) l |
(2100±1600) (67) r | (221±77) (67) r | (1554±317) (73) a | (191±43) (73) a | ||||||
| ALS | Primary motor cortex | NAA | 9.1±0.7 (10±0.6) (74) 12±1.3 (13.9±0.9) (75) |
1420±142* (1460±146*) (76) | 315±31 (317±32) (76) | (1188±69) (51) o | (78±8*) (61) o | (1460±220) (56) | (247±13) (57) | (1331±57) (58) g | (110±9) (58) g |
| Cho | 1.3±0.2 (1.1±0.1) (77) | 1230±123* (1440±144*) (76) | 297±23 (307±21) (76) | (1470±200) (56) | (222±15) (57) | ||||||
| Cr | 6.8±0.8 (7.4±0.8) (75) | 1280±128* (1250±125*) (76) | 210±21 (206±17) (76) | (1380±160) (56) | (162±16) (57) | ||||||
| Brain-stem | NAA | N/A | N/A | N/A | N/A | N/A | N/A | 272±10 (351±58) (78) | (1331±57) (58) g | (94±9) (79) | |
| Cho | N/A | N/A | N/A | N/A | 223±55 (245±50) (78) | ||||||
| Cr | N/A | N/A | N/A | N/A | 132±17 (184±42) (78) | ||||||
| HIV | NAWM | NAA | 9.6±0.3 (10.5±0.1) (80) | 2264±1206 (1734±1052) (81) | 414±129 (292±118) (81) | 896±62 (869±43) (82) |
75±6 (71±5) (82) | (1560±60) (56) (1350±268) (38) |
(301±18) (57) (295±65) (38) |
(832±44) (58) | (80±3) (58) |
| Cho | 1.9±1.2 (1.6±0.2) (83) 1.8±0.05 (1.6±0.04) (84) |
1477±404 (1597±952) (81) | 405±254 (334±195) (81) | (1210±130) (56) (1080±134) (38) |
(222±17) (57) (187±45) (38) |
||||||
| Cr | 6.3±0.13 (5.8±0.16) (84) 9.1±3.0 (7.0±2.6) (85) |
1647±647 (2184±424) (81) | 331±157 (307±122) (81) | (1400±60) (56) (1240±223) (38) |
(178±9) (57) (156±45) (38) |
||||||
| Ab-normal WM | NAA | N/A | 2061±730 (1734±1052) (81) | 492±199 (292±118) (81) | 957±114 (869±43) (82) |
77±3 (71±5) (82) | N/A | N/A | N/A | N/A | |
| Cho | N/A | 1367±815 (1597±952) (81) | 289±168 (334±195) (81) | N/A | N/A | ||||||
| Cr | N/A | 1764±543 (2184±424) (81) | 285±63 (307±122) (81) | N/A | N/A | ||||||
| MCI | NAWM | NAA | 9.7±1.4 (10.8±1.2) (47) 9.5±1.3 (10.6±1.2) (47) |
(1190±90) (56) (1340±280) (86) (1276±132) (87) |
(450±45*) (88) (336±46) (87) |
(656±16) (51) | (77±8*) (61) g | (1560±60) (56) (1350±268) (38) |
(301±18) (57) (295±65) (38) |
(832±44) (58) | (80±3) (58) |
| Cho | 1.6±0.3 (1.8±0.2) (47) | (1040±120) (56) (1480±280) (86) (1091±132) (87) |
(330±33*) (88) (352±52) (87) |
(1210±130) (56) (1080±134) (38) |
(222±17) (57) (187±45) (38) |
||||||
| Cr | 4.8±0.7 (5.2±0.6) (47) | (1150±80) (56) (1240±160) (86) (1363±137) (87) |
(240±24) (88) (219±29) (87) |
(1400±60) (56) (1240±223) (38) |
(178±9) (57) (156±45) (38) |
||||||
| PCG | NAA | 9.0±1.5 (9.8±1.0) (47,59) | (1270±50) (56) (1290±250) (86) |
(330±33*) (88) (388±11) (86) |
(1188±69) (51) o | (86±9*) (61) | (1470±80) (56) (1470±134) (38) |
254±34 (235±24) (62) | (1356±174) (58) | 63±10 (64±29) (62) | |
| Cho | N/A | (1150±120) (56) (1390±380) (86) |
(380±38) (88) (395±55) (86) |
(1250±220) (56) (1300±134) (38) |
183±43 (176±37) (62) | ||||||
| Cr | N/A | (1240±60) (56) (1290±190) (86) |
(250±25*) (88) (207±4) (86) |
(1330±130) (56) (1460±157) (38) |
147±16 (141±16) (62) | ||||||
| Hippo-campus | NAA | 7.6±1.1 (8.5±1.0) (47) r 7.5±0.9 (8.7±1.0) (47) l |
(1270±50) (56) o (1290±250) (86) o |
(330±33*) (88) o (388±11) (86) o |
(1300±174) (68) a | (86±2) (89) a | (1470±80) (56) o (1470±134) (38) o |
(324±29) (90) l | (1331±57) (58) g | (85±3) (91) a | |
| Cho | N/A | (1150±120) (56) o (1390±380) (86) o |
(380±38) (88) o (395±55) (86) o |
(1250±220) (56) o (1300±134) (38) o |
(312±65) (90) l | ||||||
| Cr | 6.6±1.0 (7.1±1.1) (47) l 5.6±0.6 (6.1±0.5) (64) a |
(1240±60) (56) o (1290±190) (86) o |
(250±25*) (88) o (207±4) (86) o |
(1330±130) (56) o (1460±157) (38) o |
(180±20) (90) l | ||||||
| MS | NAWM | NAA | 9.7±0.6 (10.2±0.7) (92) 7.8±0.6 (8.4±0.6) (93) 8.3±0.8* (8.8±0.9*) (94) 8.5±0.6 (9.2±0.7) (95) 7.7±1.4 (8.4±1.2) (40) |
(1190±90) (56) (1340±280) (86) (1276±132) (87) |
434±43* (441±44*) (96) | 767±25 (746±23) (97) 664±49 (670±51) (98) 749±75* (677±68*) (99) 669±29 (630±23) (100) 687±85 (642±90) (101) |
91±10 (85±7) (102) 76±7 (72±7) (103) |
1680±100 (1860±200) (104) | 250±50 (230±50) (104) 296±48 (395±41) (105,106) 269±19 (296±28) (107) |
1190±400 (830±200) (104) 975±53 (891±23) (108) 926±34 (923±40) (109) 895±41 (855±15) (110) 836±30 (821±31) (111) 888±39 (871±99) (112) |
57±0.5 (56±0.5) (113) 65±2 (62±1) (114) 102±9 (99±8) (112) 113±11 (106±4) (115) |
| Cho | 1.6±0.1 (1.4±0.1) (116) | (1040±120) (56) (1480±280) (86) (1091±132) (87) |
355±36* (296±30*) (96) | 1140±100 (1320±200) (104) | 200±40 (180±30) (104) 222±41 (256±19) (105,106) 204±34 (221±26) (107) |
||||||
| Cr | 5.5±0.5 (5.1±0.3) (92) 5.3±0.4 (4.9±0.2) (116) 5.1±0.6 (4.3±0.7) (117) |
(1150±80) (56) (1240±160) (86) (1363±137) (87) |
237±24* (244±24*) (96) | 1590±200 (1740±100) (104) | 140±10 (140±10) (104) 166±27 (184±16) (105,106) 174±35 (168±21) (107) |
||||||
| NAGM (cortex) | NAA | 7.4±0.7 (8.3±1.0) (93) 8.2±0.8* (9.0±0.9*) (94) 8.5±0.5 (9.3±1.0) (95) |
(1270±50) (56) (1290±250) (86) |
(330±33*) (88) (388±11) (86) |
1383±94 (1306±49) (97) 1075±93 (1041±97) (101) |
(78±8*) (61) | (1470±80) (56) (1470±134) (38) |
243±50 (274±18) (105,106) | 1429±23 (1410±17) (110) 1382±55 (1369±66) (111) |
64±0.5 (62±0.5) (113) 124±5 (115±3) (118) 126±9 (124±6) (115) |
|
| Cho | 1.0±0.1* (1.2±0.12*) (94) | (1150±120) (56) (1390±380) (86) |
(380±38) (88) (395±55) (86) |
(1250±220) (56) (1300±134) (38) |
228±54 (289±46) (105,106) | ||||||
| Cr | N/A |
(1240±60) (56) (1290±190) (86) |
(250±25*) (88) (207±4) (86) |
(1330±130) (56) (1460±157) (38) |
157±23 (166±30) (105,106) | ||||||
| Thala-mus | NAA | 8.1±1.0 (8.9±0.3) (119) 9.8±2.2 (11.4±1.6) (120) |
(1410±90) (56) (1400±140*) (88) |
(340±34*) (88) | 969±39 (963±38) (97) 959±66 (926±54) (101) 942±94* (927±93*) (121) |
(76±8*) (61) | (1570±80) (56) | 237±31 (280±39) (105,106) | 1086±70 (1075±57) (108) 1156±57 (1160±48) (111) |
61±1 (56±0.5) (113) | |
| Cho | N/A | (1100±170) (56) (1200±120*) (88) |
(320±32*) (88) | (1380±220) (56) | 224±31 (246±39) (105,106) | ||||||
| Cr | N/A |
(1270±110) (56) (1750±175*) (88) |
(200±20*) (88) | (1450±160) (56) | 152±11 (158±16) (105,106) | ||||||
| Lesions | NAA | 8.0±0.7 (8.7±0.7) (122) 10.8±2.0 (13.4±1.7) (104) 6.8±2.0 (9.0±0.9) (123) |
969±250 (1124±233) (124) | 411±41* (441±44*) (96) |
1071±18 (739±8) (124) 929±93* (677±68*) (99) |
129±31 (85±7) (102) 94±7.5 (63±15) (125) |
(1560±60) (56) (1350±268) (38) |
264±30 (296±28) (107) 305±81 (395±41) (105,106) |
1353±185 (891±23) (108) 1225±131 (923±40) (109) 1363±147 (821±31) (111) |
(80±3) (58) | |
| Cho | 1.5±0.2 (1.3±0.2) (122) | 896±276 (1103±193) (124) | 394±40* (296±30*) (96) | (1210±130) (56) (1080±134) (38) |
187±30 (221±26) (107) 219±45 (256±19) (105,106) |
||||||
| Cr | N/A | 1103±333 (1287±295) (124) |
253±25* (244±24*) (96) | (1400±60) (56) (1240±223) (38) |
154±20 (168±21) (107) 176±39 (256±19) (105,106) |
||||||
| Schizo-phrenia | NAWM | NAA | 9.2±0.7 (9.6±0.7) (126) 8.7±0.9 (10.5±0.6) (127,128) 5.6±1.0 (6.6±1.0) (129) |
(1190±90) (56) (1340±280) (86) (1276±132) (87) |
317±20 (309±18) (127,128) | 684±78 (658±37) (68) | 77±12 (60±7) (127,128) 55±3 (53±2) (68) 73±4 (72±3) (130) 84±8*(82±8*) (131) |
(1560±60) (56) (1350±268) (38) |
317±32 (339±27) (126) 190±23 (210±24) (132) |
(832±44) (58) | 68±3 (69±3) (126) 66±5 (61±2) (132) |
| Cho | N/A | (1040±120) (56) (1480±280) (86) (1091±132) (87) |
(330±33*) (88) (352±52) (87) |
(1210±130) (56) (1080±134) (38) |
220±28 (221±20) (126) | ||||||
| Cr | N/A | (1150±80) (56) (1240±160) (86) (1363±137) (87) |
(240±24) (88) (219±29) (87) |
(1400±60) (56) (1240±223) (38) |
168±19 (174±14) (126) | ||||||
| Hippo-campus | NAA | 11.0±0.8 (11.6±1.1) (133) l | (1270±50) (56) o (1290±250) (86) o |
(330±33*) (88) o (388±11) (86) o |
1360±207 (1300±174) (68) a | 65±5 (61±4) (68) a | (1470±80) (56) o (1470±134) (38) o |
(324±29) (90) l | (1331±57) (58) g | (85±3) (91) a | |
| Cho | N/A | (1150±120) (56) o (1390±380) (86) o |
(380±38) (88) o (395±55) (86) o |
(1250±220) (56) o (1300±134) (38) o |
(312±65) (90) l | ||||||
| Cr | N/A | (1240±60) (56) o (1290±190) (86) o |
(250±25*) (88) o (207±4) (86) o |
(1330±130) (56) o (1460±157) (38) o |
(180±20) (90) l | ||||||
| Region | Metab. | Concentration (mM) | 4.0 T | ||||||||
| Metabolite | Water | ||||||||||
| T1 | T2 | T1 | T2 | ||||||||
| CG | NAA | 10.2±2.8 (13.1±2.0) (134) 10.8±0.8 (10.2±0.9) (135) |
(1267±141) (136) | 222±39 (240±43) (137) | (1724±51) (138) | (63±6) (138) | |||||
| Cho | N/A | (1111±136) (136) | 208±25 (238±43) (137) | ||||||||
| Cr | N/A | (1487±146) (136) | 142±23 (146±22) (137) | ||||||||
| Tumors | Glioma | NAA | 4.6±3.2 (7.9±1.1) (139) 6.6±2.2 (12.5±2.1) (140) 2.6±1.7 (11±0.4) (141) 7.4±6.3 (12.4±3.8) (142) 2.4±0.8 (8.4±1.5) (143) |
1060±230 (1440±140) (144) | 113±11* (312±31*) (145) 208±46 (369±23) (141) 225±23 (338±23) (144) 226±169 (413±212) (142) |
(1188±69) (51) | 173±20 (96±13) (142) 156±16* (100±10*) (145) 175±41 (90±9) (141) 116±6 (64±15) (125) |
1330±390 (1525±35) (139) 1380±210 (1380±130) (140) |
224±55 (249±53) (140) 256±41 (291±36) (143) |
(1331±57) (58) g | 145±30 (81±6) (143) |
| Cho | 4.0±1.6 (1.1±0.1) (139) 2.6±0.6 (2.1±0.1) (141) 1.6±0.7 (1.3±0.5) (142) 2.6±1.0 (1.5±0.2) (143) |
1140±210 (1240±140) (144) | 403±40* (318±32*) (145) 293±42 (266±18) (141) 219±26 (311±15) (144) 410±161 (378±223) (142) |
1160±200 (1210±60) (139) 1000±210 (1060±110) (140) |
204±30 (169±27) (140) 285±55 (240±6) (143) |
||||||
| Cr | 6.3±2.3 (6.1±0.1) (139) 5.8±1.1 (8.4±1.5) (140) 4.3±0.6 (7.7±0.4) (141) 4.5±1.3 (7.4±1.9) (142) 5.0±2.2 (6.3±1.4) (143) |
1450±320 (1580±150) (144) | 250±25* (192±19*) (145) 175±24 (205±10) (141) 211±32 (225±26) (144) 216±100 (229±80) (142) |
1840±300 (1690±85) (139) 1370±200 (1380±130) (140) |
148±12 (139±16) (140) 168±18 (149±15) (143) |
||||||
| Menin-gioma | NAA | N/A | N/A | N/A | (1188±69) (51) | 97±17 (96±13) (142) 107±11* (100±10*) (145) |
N/A | N/A | (1331±57) (58) g | (110±9) (58) g | |
| Cho | 1.9±0.7 (1.3±0.5) (142) | (1150±120) (56) (1390±380) (86) |
459±46* (318±32*) (145) 955±1039 (378±223) (142) |
(1250±220) (56) (1300±134) (38) |
(207±36) (38) (265±29) (57) |
||||||
| Cr | 3.1±1.5 (7.4±1.9) (142) | (1240±60) (56) (1290±190) (86) |
168±17* (192±19*) (145) 262±180 (229±80) (142) |
(1330±130) (56) (1460±157) (38) |
(152±16) (38) (161±10) (57) |
||||||
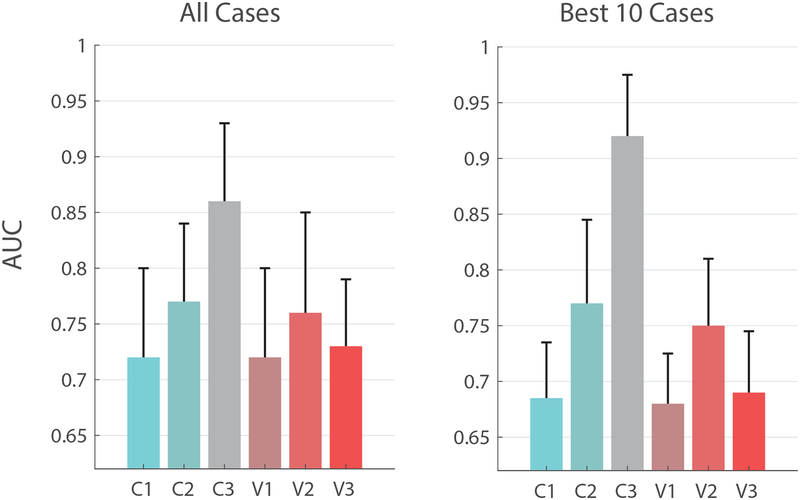
Table 2 lists the AUCs computed for each case: (C1)-(C3) and (V1)-(V3), using the averaged data from each pathology, metabolite and region. Included were only those cases for which there were documented differences in metabolic concentration, and at least one metabolite relaxation time was reported in the disease, region and field strength (n=37). As expected, multiparametric MRS improves classification for concentrations alone ((C2) vs. (C1)) and offers further improvements when relaxation values are incorporated into the predication ((C2) vs (C3)). The median ± median absolute deviation (MAD) taken over all pathologies and regions yields (Fig. 1): AUCC1=0.72±0.07, AUCC2=0.78±0.07, AUCC3=0.86±0.07. While the increases are substantial, the global statistics confound the underlying improvement observed in several particular cases: confining the calculation to the 10 most substantial improvements yields AUCC1=0.68±0.05, AUCC2=0.77±0.07, AUCC3=0.92±0.05. These cases, listed in Table 2 in bold, consist of five applications in MS, two in AD, two in human immunodeficiency virus (HIV) infection and one in amyotrophic lateral sclerosis (ALS).
Table 2.
Area under the receiver operating characteristic curve (AUC) for the cases in Table 1 for which there were documented differences in metabolic concentration, and for which at least one metabolite relaxation time was reported in the particular disease, region and field strength. For each such case, the average±SD is noted for concentrations and relaxation times of both metabolites and reference water values. In case of multiple values in Table 1, an averaged mean±SD is shown: the averaged mean was calculated by averaging the means; and the average SD was calculated by averaging their variances and taking the square root. Relaxation times exhibiting a CV of at least 30% are marked with a dagger (†). The AUC is calculated for conventional MRS concentrations (C1), multiparametric MRS concentrations (C2), and multiparametric estimation combining concentrations and relaxation times (C3). The top ten cases with the largest AUC improvements expected with multiparametric MRS (AUCC3 – AUCC1) are shown in bold. Also shown are AUCs for conventional MRS, using per-subject knowledge of B1+ (V1), very long TRs with negligible T1 weighting (V2) and ultrashort TEs with negligible T2 weighting (V3). Cases (V1–V3) exhibit AUCs that lie between those of (C1) and (C2).
| Disorder | Region | Metab. | Concentration (mM) | Field strength | T1 (metabolites) | T2 (metabolites) | T1 (water) | T2 (water) | C1 | C2 | C3 | V1 | V2 | V3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | NAWM | NAA | 8.7±1.5 (10.1±1.0) | 1.5 | 1375±503† (1330±227) | 469±262† (272±58) | 656±16 (656±16) | 82±6 (80±7) | 0.71 | 0.78 | 0.86 | 0.71 | 0.78 | 0.7 |
| Cho | 1.6±0.3 (1.8±0.3) | 1190±430† (1215±361)† | 382±91 (341±90) | 0.62 | 0.68 | 0.72 | 0.63 | 0.66 | 0.63 | |||||
| PCG | NAA | 8.8±1.2 (9.9±1.2) | 1.5 | 1110±111 (1150±115) | 456±40 (402±147)† | 1188±69 (1188±69) | 86±9 (86±9) | 0.67 | 0.74 | 0.78 | 0.67 | 0.68 | 0.72 | |
| 3.0 | 1470±110 (1470±110) | 254±36 (235±24) | 1356±174 (1356±174) | 61±11 (64±29)† | 0.64 | 0.74 | 0.78 | 0.64 | 0.65 | 0.7 | ||||
| Cr | 6.3±0.8 (6.9±0.8) | 1.5 | 1330±133 (1120±112) | 201±28 (186±42) | 1188±69 (1188±69) | 86±9 (86±9) | 0.64 | 0.7 | 0.91 | 0.64 | 0.67 | 0.67 | ||
| 3.0 | 1395±144 (1395±144) | 151±21 (141±16) | 1356±174 (1356±174) | 61±11 (64±29)† | 0.62 | 0.7 | 0.74 | 0.61 | 0.61 | 0.65 | ||||
| ALS | Primary motor cortex | NAA | 10.6±1.0 (11.9±0.8) | 1.5 | 1420±142 (1460±146) | 315±31 (317±32) | 1188±69 (1188±69) | 78±8 (78±8) | 0.75 | 0.84 | 0.85 | 0.76 | 0.79 | 0.78 |
| Cho | 1.3±0.2 (1.1±0.1) | 1230±123 (1440±144) | 297±23 (307±21) | 0.76 | 0.81 | 0.93 | 0.77 | 0.79 | 0.78 | |||||
| Cr | 6.8±0.8 (7.4±0.8) | 1280±128 (1250±125) | 210±21 (206±17) | 0.66 | 0.7 | 0.72 | 0.66 | 0.67 | 0.68 | |||||
| HIV | NAWM | NAA | 9.6±0.3 (10.5±0.1) | 1.5 | 2264±1206† (1734±1052)† | 414±129† (292±118)† | 896±62 (869±43) | 75±6 (71±5) | 0.55 | 1 | 1 | 0.53 | 0.76 | 0.55 |
| Cho | 1.9±0.8 (1.6±0.1) | 1477±404 (1597±952)† | 405±254† (334±195)† | 0.58 | 0.65 | 0.67 | 0.57 | 0.63 | 0.58 | |||||
| Cr | 7.7±2.1 (6.4±1.8) | 1647±647† (2184±424) | 331±157† (307±122)† | 0.63 | 0.68 | 0.8 | 0.64 | 0.66 | 0.64 | |||||
| MCI | PCG | NAA | 9.0±1.5 (9.8±1.0) | 3.0 | 1470±110 (1470±110) | 254±34 (235±24) | 1356±174 (1356±174) | 63±10 (64±29)† | 0.62 | 0.67 | 0.74 | 0.61 | 0.62 | 0.65 |
| MS | NAWM | NAA | 8.4±0.9 (9.0±0.8) | 1.5 | 1269±186 (1269±186) | 434±43 (441±44) | 707±58 (673±57) | 84±9 (79±7) | 0.64 | 0.69 | 0.7 | 0.63 | 0.66 | 0.64 |
| 3.0 | 1680±100 (1860±200) | 272±42 (307±41) | 963±182 (862±103) | 84±7 (81±5) | 0.64 | 0.69 | 0.87 | 0.64 | 0.67 | 0.64 | ||||
| Cho | 1.6±0.1 (1.4±0.1) | 1.5 | 1204±192 (1204±192) | 355±36 (296±30) | 707±58 (673±57) | 84±9 (79±7) | 0.78 | 0.92 | 0.97 | 0.78 | 0.87 | 0.8 | ||
| 3.0 | 1140±100 (1320±200) | 209±39 (219±25) | 963±182 (862±103) | 84±7 (81±5) | 0.76 | 0.92 | 0.95 | 0.77 | 0.86 | 0.79 | ||||
| Cr | 5.3±0.5 (4.8±0.5) | 1.5 | 1251±130 (1251±130) | 237±24 (244±24) | 707±58 (673±57) | 84±9 (79±7) | 0.7 | 0.76 | 0.77 | 0.71 | 0.72 | 0.72 | ||
| 3.0 | 1590±200 (1740±100) | 160±26 (164±16) | 963±182 (862±103) | 84±7 (81±5) | 0.68 | 0.76 | 0.84 | 0.68 | 0.73 | 0.7 | ||||
| NAGM (cortex) | NAA | 8.0±0.7 (8.9±1.0) | 3.0 | 1470±110 (1470±110) | 243±50 (274±18) | 1406±42 (1390±48) | 105±6 (100±4) | 0.74 | 0.77 | 0.83 | 0.75 | 0.77 | 0.74 | |
| Cho | 1.0±0.1 (1.2±0.1) | 1275±182 (1275±182) | 228±54 (289±46) | 0.85 | 0.92 | 0.95 | 0.85 | 0.9 | 0.85 | |||||
| Thala-mus | NAA | 8.9±1.7 (10.2±1.2) | 1570±80 (1570±80) | 237±31 (280±39) | 1121±64 (1118±53) | 61±1 (56±1) | 0.72 | 0.73 | 0.86 | 0.72 | 0.72 | 0.72 | ||
| Lesions | NAA | 8.5±1.7 (10.4±1.2) | 1.5 | 969±250 (1124±233) | 411±41 (441±44) | 1000±67 (708±48) | 112±23 (74±12) | 0.75 | 0.82 | 0.87 | 0.75 | 0.78 | 0.77 | |
| 3.0 | 1455±194 (1455±194) | 285±61 (346±35) | 1314±156 (878±32) | 80±3 (80±3) | 0.79 | 0.82 | 0.9 | 0.79 | 0.82 | 0.78 | ||||
| Cho | 1.5±0.2 (1.3±0.2) | 1.5 | 896±276† (1103±193) | 394±39 (296±30) | 1000±67 (708±48) | 112±23 (74±12) | 0.68 | 0.76 | 0.99 | 0.68 | 0.72 | 0.7 | ||
| 3.0 | 1145±132 (1145±132) | 203±38 (239±23) | 1314±156 (878±32) | 80±3 (80±3) | 0.71 | 0.76 | 0.86 | 0.72 | 0.74 | 0.73 | ||||
| Schizo-phrenia | NAWM |
NAA | 7.8±0.9 (8.9±0.8) | 1.5 | 1269±186 (1269±186) | 317±20 (309±18) | 684±78 (658±37) | 72±8 (67±6) | 0.73 | 0.82 | 0.83 | 0.73 | 0.78 | 0.75 |
| 3.0 | 1455±194 (1455±194) | 254±28 (275±26) | 832±44 (832±44) | 67±4 (65±3) | 0.75 | 0.82 | 0.86 | 0.75 | 0.8 | 0.75 | ||||
| CG | NAA | 10.5±2.1 (11.6±1.6) | 4.0 | 1267±141 (1267±141) | 222±39 (240±43) | 1724±51 (1724±51) | 63±6 (63±6) | 0.64 | 0.66 | 0.7 | 0.64 | 0.65 | 0.65 | |
| Tumors | Glioma | NAA | 4.7±3.4 (10.4±2.1) | 1.5 | 1060±230 (1440±140) | 193±89† (358±108)† | 1188±69 (1188±69) | 155±24 (88±12) | 0.91 | 0.92 | 0.99 | 0.91 | 0.92 | 0.91 |
| 3.0 | 1355±313 (1453±95) | 240±49 (270±45) | 1331±57 (1331±57) | 145±30 (81±6) | 0.9 | 0.92 | 0.94 | 0.9 | 0.92 | 0.91 | ||||
| Cho | 2.7±1.1 (1.5±0.3) | 1.5 | 1140±210 (1240±140) | 318±87 (318±113)† | 1188±69 (1188±69) | 155±24 (88±12) | 0.84 | 0.85 | 0.87 | 0.84 | 0.85 | 0.85 | ||
| 3.0 | 1080±205 (1135±89) | 245±44 (205±20) | 1331±57 (1331±57) | 145±30 (81±6) | 0.84 | 0.85 | 0.91 | 0.85 | 0.85 | 0.84 | ||||
| Cr | 5.2±1.6 (7.2±1.3) | 1.5 | 1450±320 (1580±150) | 213±55 (213±43) | 1188±69 (1188±69) | 155±24 (88±12) | 0.8 | 0.83 | 0.85 | 0.8 | 0.82 | 0.8 | ||
| 3.0 | 1605±255 (1535±110) | 158±15 (144±16) | 1331±57 (1331±57) | 145±30 (81±6) | 0.79 | 0.83 | 0.88 | 0.8 | 0.81 | 0.81 | ||||
| Menin-gioma | Cho | 1.9±0.7 (1.3±0.5) | 1.5 | 1270±282 (1270±282) | 707±735† (348±159)† | 1188±69 (1188±69) | 102±14 (98±12) | 0.73 | 0.76 | 0.8 | 0.74 | 0.74 | 0.75 | |
| Cr | 3.1±1.5 (7.4±1.9) | 1265±141 (1265±141) | 215±128† (211±58) | 0.95 | 0.96 | 0.96 | 0.95 | 0.96 | 0.96 |
Fig 1.
The benefits of multiparametric MRS. Left: median AUCs, taken over all pathologies and regions (Table 2), for conventional spectroscopy (C1), multiparametric MRS concentrations (C2) and optimal linear classifiers (C3), as well as the effect of knowing B1+ (V1), ultra-long TR (V2) and ultra-short TE (V3). Error bars represent median absolute deviations (MADs). Right: Median±MAD AUCs using the ten cases from Table 2 which show the most marked improvement to AUC (compared between (C1) and (C3)).
Assessing the Effect of Each Spin Parameters on Classification Accuracy
When considering the relative impact of each sequence parameter on the classification of pathology using only metabolite concentrations (cases V1–V3, Table 2), we find that the median±MAD AUCs, taken over all cases, are: AUCV1=0.72±0.08, AUCV2=0.76±0.09 and AUCV3=0.73±0.06 (Fig. 1). These numbers all fall between AUCC1, where typical parameters TR/TE=1500/40 ms are used, and AUCC2, which completely removes all T1, T2 and B1+ weighting from the concentrations. Our results imply that long TRs (V2) are the most beneficial in removing unwanted signal weighting. Shortening TE (V3) has less of a dominant effect on quantification, providing some motivation for ultrashort-TE sequences such as STEAM (23), SPECIAL (24) or STRESS (16,17), but not a particularly strong one; however, this statement only applies to singlets and does not take into account the improvements to quantification conferred by ultrashort echo times, such as minimal J-coupling dephasing. Our calculations also do not take into account the effects ultrashort TEs and ultralong TRs have on the signal-to-noise ratio and, commensurately, on the accuracy of quantifying metabolite concentrations. Finally, B1+ quantification (V1) seems to have a negligible effect on the overall accuracy of MRS, making the addition of B1+ mapping sequences to an imaging protocol of secondary importance. These results hold also when we confine ourselves to the ten cases which show the greatest improvement, as before, for which AUCV1=0.69±0.04, AUCV2=0.74±0.06, AUCV3=0.7±0.06 (Fig. 1).
Assessing the Effect of Sequence Precision on Multiparametric MRS
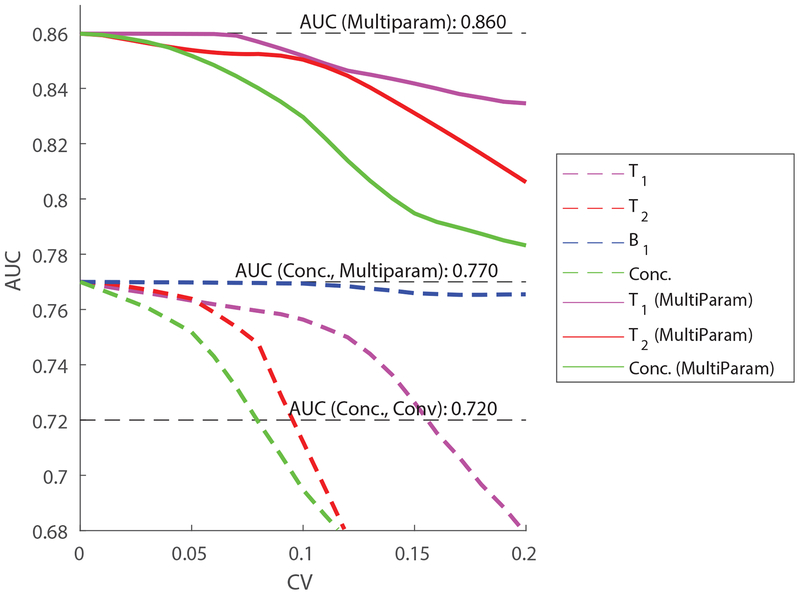
Cases (C2) and (C3) above assume each subject’s relaxation values are perfectly known. The introduction of real-world intra-subject variability will unavoidably degrade those benefits, which will be reflected in a commensurate decline in the AUC. Fig. 2 shows how the AUC varies as a function of the intra-subject variability of each measured quantity. The curves represent the median taken over all cases in Table 2. The four dashed curves show how intra-subject variability in T1 (magenta), T2 (red), B1+ (blue) and concentrations (green) affect the usability of metabolite concentrations alone (case C2). The three solid curves show how intra-subject variability in T1 (magenta), T2 (red) and concentrations (green) affect the performance of the optimal linear multiparametric classifier (case C3). Note that B1+ does not directly affect the linear multiparametric classifier, but can indirectly affect the quantification of T1, T2 and the concentrations themselves. The exact way in which this might occur will depend on the specific workings of the multiparametric MRS sequence and hence are not explicitly modeled in our simulations.
Fig. 2.
The effect of precision (intra-subject variability) on multiparametric MRS. The AUC is plotted as a function of the multiparametric MRS’s method coefficient of variation (CV) for each of the variables: T1, T2 and B1+, for both cases (C2) (using only the concentrations corrected using per-subject metabolite concentrations) and (C3) (using a full multiparametric classifier). Dashed lines correspond to (C2) while solid lines correspond to (C3). Note that, for multiparametric classification, only errors in the concentrations and relaxation parameters (but not B1+) are modeled.
The results reveal several features of interest. When considering concentrations alone, the most significant source of error comes from imprecise estimation of the concentrations themselves. A CV of 5–10% is sufficient to completely undermine any gains made by multiparametric MRS. Lack of precision in quantifying relaxation times has a somewhat less pronounced, but still important effect on quantitative MRS, with a precision of approximately 10% in T2 and 15% in T1 sufficient to reduce the AUC to that of conventional spectroscopy. Finally, variability in B1+ has a weak effect on the quantification of concentrations, with a CV of even 20% leading to a decline of the AUC from 0.77 to 0.76. This is in accordance with our simulations above, which have shown that knowledge of B1+ improves the AUC of conventional MRS only marginally.
Interestingly, uncertainty in T2 has the most pronounced deleterious effect on the AUC. This finding does not invalidate our previous result that ultralong TR sequences (V2) increase the AUC more than ultrashort TE sequences (V3), since these two comparisons evaluate the effects of intra-subject and inter-subject variabilities, respectively. In practice, T1 exhibits greater inter-subject variability compared to T2, as evident in Table 2. An additional reason for this discrepancy could also be that ultralong TRs minimize both T1 and B1+ weighting, confounding two effects.
Similar trends to the AUC are observed when one considers linear optimal classifiers. Here, however, multiparametric MRS retains its edge even for fairly large intra-subject CVs of up to, and even above 20%. Intra-subject CVs of several percent are unavoidable for any method. These results serve to motivate the importance of quantitative multiparametric MRS for realistic sequences and signal-to-noise ratios.
Discussion
Making Sense of the AUC: What do the Increases Mean?
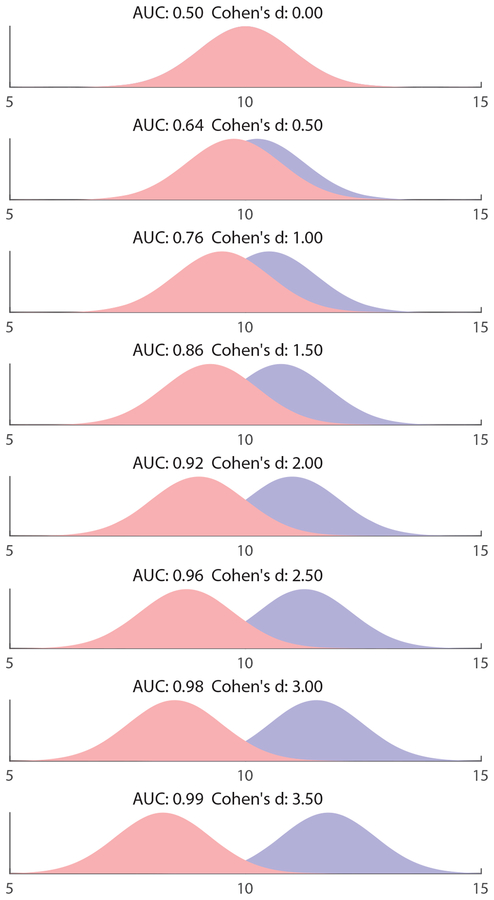
The AUC compactly describes the ROC curves of a binary classifier. It is used ubiquitously in the assessment of binary classifiers in fields ranging from medicine to machine learning and geology. It can be shown that, if a certain quantity increases in pathology, its corresponding AUC equals the probability that a randomly chosen pathological specimen has a higher value of that quantity compared to a randomly chosen healthy specimen (19). Several works have linked the AUC to several other widely used performance metrics. Swets et al. have shown that, for normally distributed variables, the AUC is related to Cohen’s d, which is used to assess effect sizes, via , where z(x) is the normal z-distribution (25). Figure 3 plots several equal-variance normal distributions with varying means, and their related AUCs and Cohen’s d. It provides a visual aid for interpreting the AUC.
Fig. 3.
Visual interpretation of the AUC. Shown are two normal distributions of unit standard deviation, which represent some quantity which varies between patients and controls. As the distance between the distributions increases, so do the AUC and Cohen’s d, both variables used to describe effect size.
Several caveats of the AUC should be mentioned. First, there are cases in which a high AUC is not necessarily desired. Rather, high sensitivity – that is, a low rate of false negatives – is more appropriate, since the implications of not treating a patient are sometimes direr than those of mistakenly treating a healthy individual; unfortunately, there is no simple, closed-form correspondence between sensitivity and the AUC, and it is not straightforward to relate an increase in one to an increase in the other. The optimal linear combination of several weak classifiers with poor AUCs may in practice result in smaller gains than predicted, due to the inherent difficulties and large sample sizes required to estimate the AUC precisely (26). Finally, while the AUC remains widely used, several criticisms have been leveled against its practical usefulness; for example, it takes into account regions of the ROC curves which are not usually used, and disregards the goodness-of-fit of the model being considered. Interested readers are referred to the literature for a comprehensive discussion (27,28).
Where is Multiparametric MRS Most Effective?
We set out to calculate the AUC for multiple neuropathologies, regions and metabolites. In all cases, multiparametric MRS had higher classification accuracy than conventional MRS, as revealed by Table 2, although the improvements to the AUC exhibit a large range. We note several cases where multiparametric MRS could make a substantial contribution. The largest number of improvements were observed in MS, with most for normal-appearing white matter (NAWM), a hallmark region for MS pathology, often studied with MRS in clinical trials (29–32). Overall in MS there were 5 instances showing AUCC3>0.9, which also include gray matter and lesions. Two of the largest AUC increases were in AD, which has a staggering incidence, but suffers from a lack of MR biomarkers. Marked AUC improvements were also seen in HIV infection and ALS, with the former recording the largest single increase across our study (0.55 for NAA in NAWM).
Although multiparametric MRS showed the smallest AUC increases in tumors, the technique can still have an impact in neuro-oncology. Due to the large concentration differences between healthy and diseased tissue, currently the most common clinical use of brain MRS is in tumor assessment. This is reflected in Table 2, which shows high AUC values for conventional MRS (AUCC1>0.8 for 7 out of 8 instances). Further increases due to multiparametric MRS, although projected to be modest, would improve the diagnostic accuracy of a technique already in clinical use.
We note that while our literature search was confined to adult neurological disorders, reports of altered concentrations and metabolic relaxation times in children with autism (33,34) suggest a role for multiparametric MRS in the pediatric population. Finally, it is important to note that our results show the potential benefit of multiparametric MRS not only by way of AUC, but also by providing additional, hitherto-unexplored disease biomarkers of the molecular environment within the intracellular space.
The Practical Challenges of Multiparametric MRS
The current work purposely avoided assuming a particular multiparametric approach, since research into such sequences is preliminary at best, and since the implementation details can have a substantial impact on the performance and limitations of each method. However, for the sake of completeness, we mention here some of the potential challenges we foresee for future approaches, alongside possible solutions.
Existing multiparametric MRS acquisitions often entail varying sequence parameters (TE, TR, FA) to encode the relaxation parameters into the acquired signal (9,10). While optimal for relaxation measurements, these approaches can detract from the SNR per unit time and, consequently, impair quantification of metabolite concentrations. As argued in the literature, quantification accuracy improves marginally beyond a certain SNR threshold (35). For voxel sizes of several cm3, the major singlets (NAA, Cho, Cr) have a high SNR and their quantification would be only slightly impaired even if some SNR is lost. For lower concentration metabolites, this should be more carefully considered and weighted against the potential gains of multiparametric data. For example, the detection of 2HG in gliomas (36) could outweigh the information offered by its relaxation values, and it might very well be that, for that particular application, a sequence aimed at maximizing SNR could outperform a multiparametric alternative.
There are cases in which multiparametric approaches could also benefit SNR. For example, clinical protocols might resort to long TRs to minimize the confounding effects of T1-weighting, leading to sub-optimal SNR per unit time (21). A multiparametric approach would most likely use shorter TRs in order to encode T1 relaxation into the signal. Depending on their specific implementation details, such acquisitions might improve the SNR per unit time compared to the alternative long TR protocol, while also providing an estimate of the metabolites’ relaxation values.
No single multiparametric protocol could be optimized for all metabolites, given the spread of T1s and T2s found in-vivo. Even if confined to a specific metabolite, the inter-subject and regional variability relaxation times makes it impossible to simultaneously address all pathologies, subjects and regions. However, given that most in-vivo relaxation values are confined to a range of several hundred milliseconds, it is reasonable to expect that, even if sub-optimal, many solutions are expected to function reasonably well for a wide range of pathologies and metabolites. This is not unlike the problem of inversion recovery, where inversion times are fixed in advance and yet used to successfully fit the T1 values of multiple metabolites (37–39).
The current study was confined to NAA, Cho and Cr, the major singlets, due to lack of literature data on relaxation times of coupled metabolites in pathology. Many coupled metabolites have shown clinical promise, such as myo-inositol in AD (40,41) or glutamate in traumatic brain injury (42), and preliminary reports have shown the feasibility of multiparametric approaches in measuring such coupled metabolites’ relaxation times (10). However, the variable TE of existing multiparametric implementations could lead to unwanted scalar coupling evolution, making the quantification of some of the metabolites more difficult compared to a sequence with one optimal, fixed TE. It is also possible that the converse holds: since specific metabolites are often easier to quantify at specific TEs – such as the Glx pseudo-singlet at around 2.35 ppm at TE=80 ms (43), the 1.9 ppm resonance of 2HG at TE=110 ms (44), or the 2.25 ppm resonance of 2HG at TE=97 ms (45,46) – it might turn out that acquiring data at multiple TEs could increase the fitting reliability of MRS data using advanced algorithms, much like a 2D resolved experiment. The extension of existing multiparametric approaches to include J-coupled, weaker metabolites should keep in mind these challenges.
Limitations of Current Work
The work reported herein provides a quantitative impetus to multiparametric MRS. Several assumptions made and caveats encountered should be kept in mind when evaluating our conclusions, beyond the drawbacks of using the AUC as an evaluation metric that were raised above.
Relaxation times were not found for many pathologies. While in Table 2 we only compile those cases where at least one metabolic relaxation time was reported in disease, in many cases the remaining metabolic and water relaxation times for a particular AUC calculation were substituted with values from healthy controls. This assumes that those T1 or T2 do not change in pathology (likely untrue), which underestimates the improvement to the AUC that comes from forming linear multiparametric estimators (case C3). However, using control values retains the natural inter-subject variability of T1 and T2; this variability impairs the absolute quantification of metabolites’ concentrations, and is corrected by multiparametric MRS (case C2). Thus, knowledge of the missing relaxation times would probably lead to even more pronounced improvements in the median AUC, although it is impossible to estimate the magnitude of these improvements. Furthermore, multiparametric MRS cannot be used where a particular peak is missing. Such is the case for NAA in meningiomas (Table 1).
Our conclusions have some degree of publication bias, for two reasons: (i) some metabolites, pathologies and regions are more widely studied and (ii) we have only included concentration results from statistically significant findings. Reason (i) is likely behind the fact that most improvements were found for NAWM, a relatively simple region for performing MRS relaxation time measurements. The corollary explains that many regions which are key in certain diseases are missing from this report, e.g. the hippocampus in AD, MCI and schizophrenia. Therefore, it is very likely that there are other instances where multiparametric MRS significantly outperforms conventional MRS, even beyond what is reported in Table 2. Reason (ii) can be illustrated with an example for NAWM in MS: as shown in Table 1 there are five statistically significant reports of NAA changes, substantially more than the single report found for Cho. As shown in Table 2, a multiparametric classifier based on Cho’s concentrations and relaxation times offers a better AUC (0.95 at 3T) than an estimator based on NAA (0.87 at 3T). However, Table 1 also demonstrates the large degree of inter-study variability in MRS, evidenced by the spread of effect sizes observed for NAA’s concentrations in MS. It is possible that the single statistically significant result reported for Cho happens to represent a deviation, or a special characteristic of the particular cohort considered in that study. Thus, the exact AUCs in Table 2 should not be treated as clinical recommendations, but rather as general guidelines, indicating which metabolites could be potentially interesting to target in future studies.
Typical CVs within both patients and controls were between 10%−20%, although some pathologies, such as brain tumors, exhibit a very large variability in reported concentrations and relaxation times. Such a large spread of values might be attributed to artifacts typically found in spectra from cancer patients, such as the presence of intense lipid peaks. However, in the absence of any additional specific information or access to patient data, it is impossible to apply any consistent exclusion criteria. We have chosen to include these published cases and mark them appropriately in Table 2. We have assumed throughout that all variables are normally distributed. This is clearly at odds with several cases in Table 1 which exhibit CVs larger than ≈20%−25%, which lead to non-physical negative values, which were dealt with by setting a lower cutoff value for all simulated quantities. Cases for which this happens are marked in Table 2. However, the relative fraction of these cases is fairly small and has negligible effect on the overall AUC. For example, even with a CV of 30%, a distribution of randomly chosen numbers from X~N(X0,(CV⋅X0)2 will only have approximately 1% of all values which will dip below the X0−70%⋅X0 lower threshold set in our Monte Carlo simulations.
It is impossible to ascertain what part of the reported variability (the SDs in Table 1) originates from true biological variation in the population, and what originates from methodological errors and noisy or corrupted data. We have treated all variability as inter-subject variability, and have also run additional simulations to quantify the effect of additional intra-subject variability.
The values for the concentrations and relaxation times for the same disorder and region often originate from different publications and, hence, from different patient cohorts. The values used to calculate a single AUC, therefore, may be mismatched in terms of disease stage and patient characteristics related to study entry criteria, medication use, etc. It is not possible to correct for these discrepancies, given the lack of studies which simultaneously quantify metabolic concentrations, as well as metabolite and water relaxation times. Our results should therefore be considered an approximation of the expected AUC improvements, and may not match the results obtained in practice.
Even when all are taken into account, these drawbacks do not invalidate the reported improvements. MRS relaxation times and concentrations often exhibit inter-scan CVs of approximately 10–20%, which are on the same order as the differences observed in most pathologies. This is clearly observed in many of the entries of Tables 1 and 2. Thus, the median AUC behavior, summarized in Fig. 1 and reported in the Results, captures the essence of the expected benefits of multiparametric MRS.
Conclusions
Our results show that, compared to conventional MRS, multiparametric MRS confers benefits to metabolite quantification, by removing T1, T2 and B1+ weighting from the MRS signal and by integrating additional metabolite-specific biomarkers (T1, T2). Quantitatively, we have shown that by doing so, the median AUC, calculated for a wide range of brain pathologies, increases from 0.72 to 0.78. Furthermore, forming multiparametric classifiers, consisting not only of metabolite concentrations but also of their relaxation times, provides an additional and substantial boost to the power of MRS, increasing the AUC again to a median of 0.86. When considering the ten cases showcasing the greatest improvement, the median AUC increases from 0.68 to 0.92. Our results demonstrate tangible and substantial potential clinical benefits of quantitative, multiparametric MRS sequences and data processing pipelines, and serve to motivate their pursuit.
Sponsors, Grants & Funding
The research reported in this publication was supported by the National Institute of Neurological Disorders and Stroke of the National Institutes of Health (NIH) under award R21NS112853. Assaf Tal acknowledges the support of the Monroy-Marks Career Development Fund, the Minerva Foundation and the historic generosity of the Harold Perlman Family. Ivan Kirov acknowledges the support of NIH through award R01NS097494, through pilot grant funding from the Alzheimer’s Disease Center at NYU Langone (NIH award P30AG008051), as well as the support of the Center for Advanced Imaging Innovation and Research (CAI2R, www.cai2r.net) through NIH award P41EB017183. The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
References
- 1.Wang G, El-Sharkawy AM, Bottomley PA. Minimum acquisition methods for simultaneously imaging T(1), T(2), and proton density with B(1) correction and no spin-echoes. J Magn Reson 2014;242:243–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Warntjes JB, Dahlqvist O, Lundberg P. Novel method for rapid, simultaneous T1, T2*, and proton density quantification. Magnetic resonance in medicine 2007;57(3):528–537. [DOI] [PubMed] [Google Scholar]
- 3.Voigt T, Nehrke K, Doessel O, Katscher U. T1 corrected B1 mapping using multi-TR gradient echo sequences. Magnetic resonance in medicine 2010;64(3):725–733. [DOI] [PubMed] [Google Scholar]
- 4.Ben-Eliezer N, Sodickson DK, Block KT. Rapid and accurate T2 mapping from multi-spin-echo data using Bloch-simulation-based reconstruction. Magnetic resonance in medicine 2015;73(2):809–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shepherd TM, Kirov II, Charlson E, Bruno M, Babb J, Sodickson DK, Ben-Eliezer N. New rapid, accurate T2 quantification detects pathology in normal-appearing brain regions of relapsing-remitting MS patients. NeuroImage Clinical 2017;14:363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen O, Zhu B, Rosen MS. MR fingerprinting Deep RecOnstruction NEtwork (DRONE). Magnetic resonance in medicine 2018;80(3):885–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mazor G, Weizman L, Tal A, Eldar YC. Low-rank magnetic resonance fingerprinting. Medical physics 2018;45(9):4066–4084. [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, Griswold MA. Magnetic resonance fingerprinting. Nature 2013;495(7440):187–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kulpanovich A, Tal A. The application of magnetic resonance fingerprinting to single voxel proton spectroscopy. NMR in biomedicine 2018;31(11):e4001. [DOI] [PubMed] [Google Scholar]
- 10.An L, Li S, Shen J. Simultaneous determination of metabolite concentrations, T1 and T2 relaxation times. Magnetic resonance in medicine 2017;78(6):2072–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moffett JR, Ross B, Arun P, Madhavarao CN, Namboodiri AM. N-Acetylaspartate in the CNS: from neurodiagnostics to neurobiology. Prog Neurobiol 2007;81(2):89–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jansen JF, Backes WH, Nicolay K, Kooi ME. 1H MR spectroscopy of the brain: absolute quantification of metabolites. Radiology 2006;240(2):318–332. [DOI] [PubMed] [Google Scholar]
- 13.Garwood M, DelaBarre L. The return of the frequency sweep: designing adiabatic pulses for contemporary NMR. J Magn Reson 2001;153(2):155–177. [DOI] [PubMed] [Google Scholar]
- 14.Scheenen TW, Heerschap A, Klomp DW. Towards 1H-MRSI of the human brain at 7T with slice-selective adiabatic refocusing pulses. MAGMA 2008;21(1–2):95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jhaveri K, Guo L, DeVito T. Feasibility of in-vivo semi-LASER renal magnetic resonance spectroscopy (MRS): Pilot study in healthy volunteers. Magn Reson Imaging 2017;40:12–16. [DOI] [PubMed] [Google Scholar]
- 16.Volovyk O, Tal A. Application of phase rotation to STRESS localization scheme at 3 T. Magnetic resonance in medicine 2018;79(5):2481–2490. [DOI] [PubMed] [Google Scholar]
- 17.Tal A, Gonen O. Spectroscopic localization by simultaneous acquisition of the double-spin and stimulated echoes. Magnetic resonance in medicine 2015;73(1):31–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mason SJ, Graham NE. Areas beneath the relative operating characteristics (ROC) and relative operating levels (ROL) curves: Statistical significance and interpretation. Q J Roy Meteor Soc 2002;128(584):2145–2166. [Google Scholar]
- 19.van Erkel AR, Pattynama PM. Receiver operating characteristic (ROC) analysis: basic principles and applications in radiology. European journal of radiology 1998;27(2):88–94. [DOI] [PubMed] [Google Scholar]
- 20.Kang L, Liu A, Tian L. Linear combination methods to improve diagnostic/prognostic accuracy on future observations. Stat Methods Med Res 2016;25(4):1359–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goelman G, Liu S, Hess D, Gonen O. Optimizing the efficiency of high-field multivoxel spectroscopic imaging by multiplexing in space and time. Magnetic resonance in medicine 2006;56(1):34–40. [DOI] [PubMed] [Google Scholar]
- 22.Pohmann R, Speck O, Scheffler K. Signal-to-noise ratio and MR tissue parameters in human brain imaging at 3, 7, and 9.4 tesla using current receive coil arrays. Magnetic resonance in medicine 2016;75(2):801–809. [DOI] [PubMed] [Google Scholar]
- 23.Knight-Scott J, Shanbhag DD, Dunham SA. A phase rotation scheme for achieving very short echo times with localized stimulated echo spectroscopy. Magn Reson Imaging 2005;23(8):871–876. [DOI] [PubMed] [Google Scholar]
- 24.Fuchs A, Luttje M, Boesiger P, Henning A. SPECIAL semi-LASER with lipid artifact compensation for 1H MRS at 7 T. Magnetic resonance in medicine 2013;69(3):603–612. [DOI] [PubMed] [Google Scholar]
- 25.Swets JA. Indices of discrimination or diagnostic accuracy: their ROCs and implied models. Psychol Bull 1986;99(1):100–117. [PubMed] [Google Scholar]
- 26.Yan L, Tian L, Liu S. Combining large number of weak biomarkers based on AUC. Stat Med 2015;34(29):3811–3830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo JM, Jimenez-Valverde A, Real R. AUC: a misleading measure of the performance of predictive distribution models. Global Ecol Biogeogr 2008;17(2):145–151. [Google Scholar]
- 28.Peterson AT, Papes M, Soberon J. Rethinking receiver operating characteristic analysis applications in ecological niche modeling. Ecol Model 2008;213(1):63–72. [Google Scholar]
- 29.Narayanan S, De Stefano N, Francis GS, Arnaoutelis R, Caramanos Z, Collins DL, Pelletier D, Arnason BGW, Antel JP, Arnold DL. Axonal metabolic recovery in multiple sclerosis patients treated with interferon beta-1b. J Neurol 2001;248(11):979–986. [DOI] [PubMed] [Google Scholar]
- 30.Khan O, Shen Y, Bao F, Caon C, Tselis A, Latif Z, Zak I. Long-term study of brain 1H-MRS study in multiple sclerosis: effect of glatiramer acetate therapy on axonal metabolic function and feasibility of long-Term H-MRS monitoring in multiple sclerosis. J Neuroimaging 2008;18(3):314–319. [DOI] [PubMed] [Google Scholar]
- 31.Arnold DL, Narayanan S, Antel S. Neuroprotection with glatiramer acetate: evidence from the PreCISe trial. J Neurol 2013;260(7):1901–1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Filippi M, Rocca MA, Pagani E, De Stefano N, Jeffery D, Kappos L, Montalban X, Boyko AN, Comi G, Group AS. Placebo-controlled trial of oral laquinimod in multiple sclerosis: MRI evidence of an effect on brain tissue damage. J Neurol Neurosurg Psychiatry 2014;85(8):851–858. [DOI] [PubMed] [Google Scholar]
- 33.Friedman SD, Shaw DW, Artru AA, Richards TL, Gardner J, Dawson G, Posse S, Dager SR. Regional brain chemical alterations in young children with autism spectrum disorder. Neurology 2003;60(1):100–107. [DOI] [PubMed] [Google Scholar]
- 34.Friedman SD, Shaw DW, Artru AA, Dawson G, Petropoulos H, Dager SR. Gray and white matter brain chemistry in young children with autism. Arch Gen Psychiatry 2006;63(7):786–794. [DOI] [PubMed] [Google Scholar]
- 35.Bartha R Effect of signal-to-noise ratio and spectral linewidth on metabolite quantification at 4 T. NMR in biomedicine 2007;20(5):512–521. [DOI] [PubMed] [Google Scholar]
- 36.Emir UE, Larkin SJ, de Pennington N, Voets N, Plaha P, Stacey R, Al-Qahtani K, McCullagh J, Schofield CJ, Clare S, Jezzard P, Cadoux-Hudson T, Ansorge O. Noninvasive Quantification of 2-Hydroxyglutarate in Human Gliomas with IDH1 and IDH2 Mutations. Cancer Res 2016;76(1):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xin L, Schaller B, Mlynarik V, Lu H, Gruetter R. Proton T1 relaxation times of metabolites in human occipital white and gray matter at 7 T. Magnetic resonance in medicine 2013;69(4):931–936. [DOI] [PubMed] [Google Scholar]
- 38.Mlynarik V, Gruber S, Moser E. Proton T (1) and T (2) relaxation times of human brain metabolites at 3 Tesla. NMR in biomedicine 2001;14(5):325–331. [DOI] [PubMed] [Google Scholar]
- 39.Brief EE, Whittall KP, Li DK, MacKay A. Proton T1 relaxation times of cerebral metabolites differ within and between regions of normal human brain. NMR in biomedicine 2003;16(8):503–509. [DOI] [PubMed] [Google Scholar]
- 40.Llufriu S, Kornak J, Ratiney H, Oh J, Brenneman D, Cree BA, Sampat M, Hauser SL, Nelson SJ, Pelletier D. Magnetic resonance spectroscopy markers of disease progression in multiple sclerosis. JAMA Neurol 2014;71(7):840–847. [DOI] [PubMed] [Google Scholar]
- 41.Zhu X, Schuff N, Kornak J, Soher B, Yaffe K, Kramer JH, Ezekiel F, Miller BL, Jagust WJ, Weiner MW. Effects of Alzheimer disease on fronto-parietal brain N-acetyl aspartate and myo-inositol using magnetic resonance spectroscopic imaging. Alzheimer Dis Assoc Disord 2006;20(2):77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shutter L, Tong KA, Holshouser BA. Proton MRS in acute traumatic brain injury: role for glutamate/glutamine and choline for outcome prediction. J Neurotrauma 2004;21(12):1693–1705. [DOI] [PubMed] [Google Scholar]
- 43.Schubert F, Gallinat J, Seifert F, Rinneberg H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. Neuroimage 2004;21(4):1762–1771. [DOI] [PubMed] [Google Scholar]
- 44.Berrington A, Voets NL, Plaha P, Larkin SJ, McCullagh J, Stacey R, Yildirim M, Schofield CJ, Jezzard P, Cadoux-Hudson T, Ansorge O, Emir UE. Improved localisation for 2-hydroxyglutarate detection at 3T using long-TE semi-LASER. Tomography 2016;2(2):94–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi C, Ganji SK, DeBerardinis RJ, Hatanpaa KJ, Rakheja D, Kovacs Z, Yang XL, Mashimo T, Raisanen JM, Marin-Valencia I, Pascual JM, Madden CJ, Mickey BE, Malloy CR, Bachoo RM, Maher EA. 2-hydroxyglutarate detection by magnetic resonance spectroscopy in IDH-mutated patients with gliomas. Nat Med 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi C, Ganji S, Hulsey K, Madan A, Kovacs Z, Dimitrov I, Zhang S, Pichumani K, Mendelsohn D, Mickey B, Malloy C, Bachoo R, Deberardinis R, Maher E. A comparative study of short- and long-TE (1)H MRS at 3 T for in vivo detection of 2-hydroxyglutarate in brain tumors. NMR in biomedicine 2013;26(10):1242–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Watanabe T, Shiino A, Akiguchi I. Absolute quantification in proton magnetic resonance spectroscopy is useful to differentiate amnesic mild cognitive impairment from Alzheimer’s disease and healthy aging. Dement Geriatr Cogn Disord 2010;30(1):71–77. [DOI] [PubMed] [Google Scholar]
- 48.Christiansen P, Schlosser A, Henriksen O. Reduced N-acetylaspartate content in the frontal part of the brain in patients with probable Alzheimer’s disease. Magn Reson Imaging 1995;13(3):457–462. [DOI] [PubMed] [Google Scholar]
- 49.Schuff N, Amend DL, Meyerhoff DJ, Tanabe JL, Norman D, Fein G, Weiner MW. Alzheimer disease: quantitative H-1 MR spectroscopic imaging of frontoparietal brain. Radiology 1998;207(1):91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moats RA, Ernst T, Shonk TK, Ross BD. Abnormal cerebral metabolite concentrations in patients with probable Alzheimer disease. MagnResonMed 1994;32:110–115. [DOI] [PubMed] [Google Scholar]
- 51.Rooney WD, Johnson G, Li X, Cohen ER, Kim SG, Ugurbil K, Springer CS Jr., Magnetic field and tissue dependencies of human brain longitudinal 1H2O relaxation in vivo. Magn Reson Med 2007;57(2):308–318. [DOI] [PubMed] [Google Scholar]
- 52.Kirsch SJ, Jacobs RW, Butcher LL, Beatty J. Prolongation of magnetic resonance T2 time in hippocampus of human patients marks the presence and severity of Alzheimer’s disease. Neurosci Lett 1992;134(2):187–190. [DOI] [PubMed] [Google Scholar]
- 53.Laakso MP, Partanen K, Soininen H, Lehtovirta M, Hallikainen M, Hanninen T, Helkala EL, Vainio P, Riekkinen PJ Sr., MR T2 relaxometry in Alzheimer’s disease and age-associated memory impairment. Neurobiol Aging 1996;17(4):535–540. [DOI] [PubMed] [Google Scholar]
- 54.Bartzokis G, Sultzer D, Lu PH, Nuechterlein KH, Mintz J, Cummings JL. Heterogeneous age-related breakdown of white matter structural integrity: implications for cortical “disconnection” in aging and Alzheimer’s disease. Neurobiol Aging 2004;25(7):843–851. [DOI] [PubMed] [Google Scholar]
- 55.Bartzokis G, Sultzer D, Mintz J, Holt LE, Marx P, Phelan CK, Marder SR. In vivo evaluation of brain iron in Alzheimer’s disease and normal subjects using MRI. Biol Psychiatry 1994;35(7):480–487. [DOI] [PubMed] [Google Scholar]
- 56.Ethofer T, Mader I, Seeger U, Helms G, Erb M, Grodd W, Ludolph A, Klose U. Comparison of longitudinal metabolite relaxation times in different regions of the human brain at 1.5 and 3 Tesla. Magnetic resonance in medicine 2003;50(6):1296–1301. [DOI] [PubMed] [Google Scholar]
- 57.Traber F, Block W, Lamerichs R, Gieseke J, Schild HH. 1H metabolite relaxation times at 3.0 tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J Magn Reson Imaging 2004;19(5):537–545. [DOI] [PubMed] [Google Scholar]
- 58.Wansapura JP, Holland SK, Sunn RS, Ball WS. NMR relaxation times in the human brain at 3.0 Tesla. J Magn Reson Imag 1999;9:531–538. [DOI] [PubMed] [Google Scholar]
- 59.Watanabe T, Shiino A, Akiguchi I. Hippocampal metabolites and memory performances in patients with amnestic mild cognitive impairment and Alzheimer’s disease. Neurobiol Learn Mem 2012;97(3):289–293. [DOI] [PubMed] [Google Scholar]
- 60.Shiino A, Watanabe T, Shirakashi Y, Kotani E, Yoshimura M, Morikawa S, Inubushi T, Akiguchi I. The profile of hippocampal metabolites differs between Alzheimer’s disease and subcortical ischemic vascular dementia, as measured by proton magnetic resonance spectroscopy. J Cereb Blood Flow Metab 2012;32(5):805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whittall KP, MacKay AL, Graeb DA, Nugent RA, Li DK, Paty DW. In vivo measurement of T2 distributions and water contents in normal human brain. Magn Reson Med 1997;37(1):34–43. [DOI] [PubMed] [Google Scholar]
- 62.Dumoulin MC, Zimmerman EA, Hurd R, Hancu I. Increased brain metabolite T2 relaxation times in patients with Alzheimer’s disease. 2005; Miami. [Google Scholar]
- 63.Dixon RM, Bradley KM, Budge MM, Styles P, Smith AD. Longitudinal quantitative proton magnetic resonance spectroscopy of the hippocampus in Alzheimer’s disease. Brain 2002;125(Pt 10):2332–2341. [DOI] [PubMed] [Google Scholar]
- 64.Foy CM, Daly EM, Glover A, O’Gorman R, Simmons A, Murphy DG, Lovestone S. Hippocampal proton MR spectroscopy in early Alzheimer’s disease and mild cognitive impairment. Brain Topogr 2011;24(3–4):316–322. [DOI] [PubMed] [Google Scholar]
- 65.Schuff N, Amend D, Ezekiel F, Steinman SK, Tanabe J, Norman D, Jagust W, Kramer JH, Mastrianni JA, Fein G, Weiner MW. Changes of hippocampal N-acetyl aspartate and volume in Alzheimer’s disease. A proton MR spectroscopic imaging and MRI study. Neurology 1997;49(6):1513–1521. [DOI] [PubMed] [Google Scholar]
- 66.Jessen F, Gur O, Block W, Ende G, Frolich L, Hammen T, Wiltfang J, Kucinski T, Jahn H, Heun R, Maier W, Kolsch H, Kornhuber J, Traber F. A multicenter (1)H-MRS study of the medial temporal lobe in AD and MCI. Neurology 2009;72(20):1735–1740. [DOI] [PubMed] [Google Scholar]
- 67.Christiansen P, Toft P, Larsson HB, Stubgaard M, Henriksen O. The concentration of N-acetyl aspartate, creatine + phosphocreatine, and choline in different parts of the brain in adulthood and senium. Magn Reson Imaging 1993;11(6):799–806. [DOI] [PubMed] [Google Scholar]
- 68.Williamson P, Pelz D, Merskey H, Morrison S, Karlik S, Drost D, Carr T, Conlon P. Frontal, temporal, and striatal proton relaxation times in schizophrenic patients and normal comparison subjects. Am J Psychiatry 1992;149(4):549–551. [DOI] [PubMed] [Google Scholar]
- 69.Haley AP, Knight-Scott J, Fuchs KL, Simnad VI, Manning CA. Shortening of hippocampal spin-spin relaxation time in probable Alzheimer’s disease: a 1H magnetic resonance spectroscopy study. Neurosci Lett 2004;362(3):167–170. [DOI] [PubMed] [Google Scholar]
- 70.Pitkanen A, Laakso M, Kalviainen R, Partanen K, Vainio P, Lehtovirta M, Riekkinen P, Soininen H. Severity of hippocampal atrophy correlates with the prolongation of MRI T2 relaxation time in temporal lobe epilepsy but not in Alzheimer’s disease. Neurology 1996;46(6):1724–1730. [DOI] [PubMed] [Google Scholar]
- 71.Campeau NG, Petersen RC, Felmlee JP, O’Brien PC, Jack CR Jr., Hippocampal transverse relaxation times in patients with Alzheimer disease. Radiology 1997;205(1):197–201. [DOI] [PubMed] [Google Scholar]
- 72.Wang H, Yuan H, Shu L, Xie J, Zhang D. Prolongation of T(2) relaxation times of hippocampus and amygdala in Alzheimer’s disease. Neurosci Lett 2004;363(2):150–153. [DOI] [PubMed] [Google Scholar]
- 73.Choi CG, Frahm J. Localized proton MRS of the human hippocampus: metabolite concentrations and relaxation times. Magn Reson Med 1999;41(1):204–207. [DOI] [PubMed] [Google Scholar]
- 74.Gredal O, Rosenbaum S, Topp S, Karlsborg M, Strange P, Werdelin L. Quantification of brain metabolites in amyotrophic lateral sclerosis by localized proton magnetic resonance spectroscopy. Neurology 1997;48(4):878–881. [DOI] [PubMed] [Google Scholar]
- 75.Pohl C, Block W, Karitzky J, Traber F, Schmidt S, Grothe C, Lamerichs R, Schild H, Klockgether T. Proton magnetic resonance spectroscopy of the motor cortex in 70 patients with amyotrophic lateral sclerosis. Arch Neurol 2001;58(5):729–735. [DOI] [PubMed] [Google Scholar]
- 76.Block W, Karitzky J, Traber F, Pohl C, Keller E, Mundegar RR, Lamerichs R, Rink H, Ries F, Schild HH, Jerusalem F. Proton magnetic resonance spectroscopy of the primary motor cortex in patients with motor neuron disease: subgroup analysis and follow-up measurements. Arch Neurol 1998;55(7):931–936. [DOI] [PubMed] [Google Scholar]
- 77.Bowen BC, Pattany PM, Bradley WG, Murdoch JB, Rotta F, Younis AA, Duncan RC, Quencer RM. MR imaging and localized proton spectroscopy of the precentral gyrus in amyotrophic lateral sclerosis. AJNR Am J Neuroradiol 2000;21(4):647–658. [PMC free article] [PubMed] [Google Scholar]
- 78.Hanstock CC, Cwik VA, Martin WR. Reduction in metabolite transverse relaxation times in amyotrophic lateral sclerosis. J Neurol Sci 2002;198(1–2):37–41. [DOI] [PubMed] [Google Scholar]
- 79.Kumar R, Delshad S, Woo MA, Macey PM, Harper RM. Age-related regional brain T2-relaxation changes in healthy adults. J Magn Reson Imaging 2012;35(2):300–308. [DOI] [PubMed] [Google Scholar]
- 80.Lentz MR, Kim WK, Lee V, Bazner S, Halpern EF, Venna N, Williams K, Rosenberg ES, Gonzalez RG. Changes in MRS neuronal markers and T cell phenotypes observed during early HIV infection. Neurology 2009;72(17):1465–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wilkinson ID, Paley M, Chong WK, Sweeney BJ, Shepherd JK, Kendall BE, Hall-Craggs MA, Harrison MJ. Proton spectroscopy in HIV infection: relaxation times of cerebral metabolites. Magn Reson Imaging 1994;12(6):951–957. [DOI] [PubMed] [Google Scholar]
- 82.Wilkinson ID, Paley MN, Hall-Craggs MA, Chinn RJ, Chong WK, Sweeney BJ, Kendall BE, Miller RF, Newman SP, Harrison MJ. Cerebral magnetic resonance relaxometry in HIV infection. Magn Reson Imaging 1996;14(4):365–372. [DOI] [PubMed] [Google Scholar]
- 83.Chang L, Ernst T, Leonido-Yee M, Walot I, Singer E. Cerebral metabolite abnormalities correlate with clinical severity of HIV-1 cognitive motor complex. Neurology 1999;52(1):100–108. [DOI] [PubMed] [Google Scholar]
- 84.Chang L, Ernst T, Witt MD, Ames N, Gaiefsky M, Miller E. Relationships among brain metabolites, cognitive function, and viral loads in antiretroviral-naive HIV patients. Neuroimage 2002;17(3):1638–1648. [DOI] [PubMed] [Google Scholar]
- 85.Bairwa D, Kumar V, Vyas S, Das BK, Srivastava AK, Pandey RM, Sharma SK, Jagannathan NR, Sinha S. Case control study: magnetic resonance spectroscopy of brain in HIV infected patients. BMC Neurol 2016;16:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kreis R, Ernst T, Ross BD. Absolute quantitation of water and metabolites in the human Brain. II. Metabolite Concentrations. J Magn Reson Series B 1993;102:9–19. [Google Scholar]
- 87.Rutgers DR, van der Grond J. Relaxation times of choline, creatine and N-acetyl aspartate in human cerebral white matter at 1.5 T. NMR Biomed 2002;15(3):215–221. [DOI] [PubMed] [Google Scholar]
- 88.Frahm J, Bruhn H, Gyngell ML, Merboldt KD, Hanicke W, Sauter R. Localized proton NMR spectroscopy in different regions of the human brain in vivo. Relaxation times and concentrations of cerebral metabolites. Magnetic resonance in medicine 1989;11(1):47–63. [DOI] [PubMed] [Google Scholar]
- 89.Duncan JS, Bartlett P, Barker GJ. Technique for measuring hippocampal T2 relaxation time. AJNR Am J Neuroradiol 1996;17(10):1805–1810. [PMC free article] [PubMed] [Google Scholar]
- 90.Allaili N, Valabregue R, Auerbach EJ, Guillemot V, Yahia-Cherif L, Bardinet E, Jabourian M, Fossati P, Lehericy S, Marjanska M. Single-voxel (1)H spectroscopy in the human hippocampus at 3 T using the LASER sequence: characterization of neurochemical profile and reproducibility. NMR in biomedicine 2015;28(10):1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luo Z, Zhuang X, Kumar D, Wu X, Yue C, Han C, Lv J. The correlation of hippocampal T2-mapping with neuropsychology test in patients with Alzheimer’s disease. PLoS One 2013;8(9):e76203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hattingen E, Magerkurth J, Pilatus U, Hubers A, Wahl M, Ziemann U. Combined (1)H and (31)P spectroscopy provides new insights into the pathobiochemistry of brain damage in multiple sclerosis. NMR in biomedicine 2011;24(5):536–546. [DOI] [PubMed] [Google Scholar]
- 93.Kapeller P, McLean MA, Griffin CM, Chard D, Parker GJ, Barker GJ, Thompson AJ, Miller DH. Preliminary evidence for neuronal damage in cortical grey matter and normal appearing white matter in short duration relapsing-remitting multiple sclerosis: a quantitative MR spectroscopic imaging study. J Neurol 2001;248(2):131–138. [DOI] [PubMed] [Google Scholar]
- 94.Chard DT, Griffin CM, McLean MA, Kapeller P, Kapoor R, Thompson AJ, Miller DH. Brain metabolite changes in cortical grey and normal-appearing white matter in clinically early relapsing-remitting multiple sclerosis. Brain 2002;125(Pt 10):2342–2352. [DOI] [PubMed] [Google Scholar]
- 95.Tiberio M, Chard DT, Altmann DR, Davies G, Griffin CM, McLean MA, Rashid W, Sastre-Garriga J, Thompson AJ, Miller DH. Metabolite changes in early relapsing-remitting multiple sclerosis. A two year follow-up study. J Neurol 2006;253(2):224–230. [DOI] [PubMed] [Google Scholar]
- 96.van Walderveen MA, Barkhof F, Pouwels PJ, van Schijndel RA, Polman CH, Castelijns JA. Neuronal damage in T1-hypointense multiple sclerosis lesions demonstrated in vivo using proton magnetic resonance spectroscopy. Ann Neurol 1999;46(1):79–87. [DOI] [PubMed] [Google Scholar]
- 97.Vrenken H, Geurts JJ, Knol DL, van Dijk LN, Dattola V, Jasperse B, van Schijndel RA, Polman CH, Castelijns JA, Barkhof F, Pouwels PJ. Whole-brain T1 mapping in multiple sclerosis: global changes of normal-appearing gray and white matter. Radiology 2006;240(3):811–820. [DOI] [PubMed] [Google Scholar]
- 98.Ropele S, Strasser-Fuchs S, Augustin M, Stollberger R, Enzinger C, Hartung HP, Fazekas F. A comparison of magnetization transfer ratio, magnetization transfer rate, and the native relaxation time of water protons related to relapsing-remitting multiple sclerosis. AJNR Am J Neuroradiol 2000;21(10):1885–1891. [PMC free article] [PubMed] [Google Scholar]
- 99.Griffin CM, Parker GJ, Barker GJ, Thompson AJ, Miller DH. MTR and T1 provide complementary information in MS NAWM, but not in lesions. Mult Scler 2000;6(5):327–331. [DOI] [PubMed] [Google Scholar]
- 100.Davies GR, Hadjiprocopis A, Altmann DR, Chard DT, Griffin CM, Rashid W, Parker GJ, Tofts PS, Kapoor R, Thompson AJ, Miller DH. Normal-appearing grey and white matter T1 abnormality in early relapsing-remitting multiple sclerosis: a longitudinal study. Mult Scler 2007;13(2):169–177. [DOI] [PubMed] [Google Scholar]
- 101.Griffin CM, Chard DT, Parker GJ, Barker GJ, Thompson AJ, Miller DH. The relationship between lesion and normal appearing brain tissue abnormalities in early relapsing remitting multiple sclerosis. J Neurol 2002;249(2):193–199. [DOI] [PubMed] [Google Scholar]
- 102.Papanikolaou N, Papadaki E, Karampekios S, Spilioti M, Maris T, Prassopoulos P, Gourtsoyiannis N. T2 relaxation time analysis in patients with multiple sclerosis: correlation with magnetization transfer ratio. Eur Radiol 2004;14(1):115–122. [DOI] [PubMed] [Google Scholar]
- 103.Whittall KP, MacKay AL, Li DK, Vavasour IM, Jones CK, Paty DW. Normal-appearing white matter in multiple sclerosis has heterogeneous, diffusely prolonged T(2). Magn Reson Med 2002;47(2):403–408. [DOI] [PubMed] [Google Scholar]
- 104.Srinivasan R, Sailasuta N, Hurd R, Nelson S, Pelletier D. Evidence of elevated glutamate in multiple sclerosis using magnetic resonance spectroscopy at 3 T. Brain 2005;128(Pt 5):1016–1025. [DOI] [PubMed] [Google Scholar]
- 105.Kirov II, Liu S, Fleysher R, Fleysher L, Babb JS, Herbert J, Gonen O. Brain metabolite proton T2 mapping at 3.0 T in relapsing-remitting multiple sclerosis. Radiology 2010;254(3):858–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kirov II, Fleysher L, Fleysher R, Patil V, Liu S, Gonen O. Age dependence of regional proton metabolites T2 relaxation times in the human brain at 3 T. Magn Reson Med 2008;60(4):790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schubert F, Seifert F, Elster C, Link A, Walzel M, Mientus S, Haas J, Rinneberg H. Serial 1H-MRS in relapsing-remitting multiple sclerosis: effects of interferon-beta therapy on absolute metabolite concentrations. MAGMA 2002;14(3):213–222. [DOI] [PubMed] [Google Scholar]
- 108.Parry A, Clare S, Jenkinson M, Smith S, Palace J, Matthews PM. White matter and lesion T1 relaxation times increase in parallel and correlate with disability in multiple sclerosis. J Neurol 2002;249(9):1279–1286. [DOI] [PubMed] [Google Scholar]
- 109.Jurcoane A, Wagner M, Schmidt C, Mayer C, Gracien RM, Hirschmann M, Deichmann R, Volz S, Ziemann U, Hattingen E. Within-lesion differences in quantitative MRI parameters predict contrast enhancement in multiple sclerosis. J Magn Reson Imaging 2013;38(6):1454–1461. [DOI] [PubMed] [Google Scholar]
- 110.Gracien RM, Jurcoane A, Wagner M, Reitz SC, Mayer C, Volz S, Hof SM, Fleischer V, Droby A, Steinmetz H, Groppa S, Hattingen E, Deichmann R, Klein JC. Multimodal quantitative MRI assessment of cortical damage in relapsing-remitting multiple sclerosis. J Magn Reson Imaging 2016;44(6):1600–1607. [DOI] [PubMed] [Google Scholar]
- 111.Steenwijk MD, Vrenken H, Jonkman LE, Daams M, Geurts JJ, Barkhof F, Pouwels PJ. High-resolution T1-relaxation time mapping displays subtle, clinically relevant, gray matter damage in long-standing multiple sclerosis. Mult Scler 2016;22(10):1279–1288. [DOI] [PubMed] [Google Scholar]
- 112.Reitz SC, Hof SM, Fleischer V, Brodski A, Groger A, Gracien RM, Droby A, Steinmetz H, Ziemann U, Zipp F, Deichmann R, Klein JC. Multi-parametric quantitative MRI of normal appearing white matter in multiple sclerosis, and the effect of disease activity on T2. Brain Imaging Behav 2017;11(3):744–753. [DOI] [PubMed] [Google Scholar]
- 113.Shepherd TM, Kirov II, Charlson E, Bruno M, Babb J, Sodickson DK, Ben-Eliezer N. New rapid, accurate T2 quantification detects pathology in normal-appearing brain regions of relapsing-remitting MS patients. Neuroimage Clin 2017;14:363–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Neema M, Goldberg-Zimring D, Guss ZD, Healy BC, Guttmann CR, Houtchens MK, Weiner HL, Horsfield MA, Hackney DB, Alsop DC, Bakshi R. 3 T MRI relaxometry detects T2 prolongation in the cerebral normal-appearing white matter in multiple sclerosis. Neuroimage 2009;46(3):633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Hasan KM, Walimuni IS, Abid H, Wolinsky JS, Narayana PA. Multi-modal quantitative MRI investigation of brain tissue neurodegeneration in multiple sclerosis. J Magn Reson Imaging 2012;35(6):1300–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kirov II, Tal A, Babb JS, Herbert J, Gonen O. Serial proton MR spectroscopy of gray and white matter in relapsing-remitting MS. Neurology 2013;80(1):39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Vrenken H, Barkhof F, Uitdehaag BM, Castelijns JA, Polman CH, Pouwels PJ. MR spectroscopic evidence for glial increase but not for neuro-axonal damage in MS normal-appearing white matter. Magn Reson Med 2005;53(2):256–266. [DOI] [PubMed] [Google Scholar]
- 118.Gracien RM, Reitz SC, Hof SM, Fleischer V, Zimmermann H, Droby A, Steinmetz H, Zipp F, Deichmann R, Klein JC. Assessment of cortical damage in early multiple sclerosis with quantitative T2 relaxometry. NMR Biomed 2016;29(4):444–450. [DOI] [PubMed] [Google Scholar]
- 119.Geurts JJ, Reuling IE, Vrenken H, Uitdehaag BM, Polman CH, Castelijns JA, Barkhof F, Pouwels PJ. MR spectroscopic evidence for thalamic and hippocampal, but not cortical, damage in multiple sclerosis. Magnetic resonance in medicine 2006;55(3):478–483. [DOI] [PubMed] [Google Scholar]
- 120.Wylezinska M, Cifelli A, Jezzard P, Palace J, Alecci M, Matthews PM. Thalamic neurodegeneration in relapsing-remitting multiple sclerosis. Neurology 2003;60(12):1949–1954. [DOI] [PubMed] [Google Scholar]
- 121.Niepel G, Tench Ch R, Morgan PS, Evangelou N, Auer DP, Constantinescu CS. Deep gray matter and fatigue in MS: a T1 relaxation time study. J Neurol 2006;253(7):896–902. [DOI] [PubMed] [Google Scholar]
- 122.Kapeller P, Brex PA, Chard D, Dalton C, Griffin CM, McLean MA, Parker GJ, Thompson AJ, Miller DH. Quantitative 1H MRS imaging 14 years after presenting with a clinically isolated syndrome suggestive of multiple sclerosis. Mult Scler 2002;8(3):207–210. [DOI] [PubMed] [Google Scholar]
- 123.Kapeller P, Ropele S, Enzinger C, Lahousen T, Strasser-Fuchs S, Schmidt R, Fazekas F. Discrimination of white matter lesions and multiple sclerosis plaques by short echo quantitative 1H-magnetic resonance spectroscopy. J Neurol 2005;252(10):1229–1234. [DOI] [PubMed] [Google Scholar]
- 124.Brief EE, Vavasour IM, Laule C, Li DK, Mackay AL. Proton MRS of large multiple sclerosis lesions reveals subtle changes in metabolite T(1) and area. NMR Biomed 2010;23(9):1033–1037. [DOI] [PubMed] [Google Scholar]
- 125.Darwin RH, Drayer BP, Riederer SJ, Wang HZ, MacFall JR. T2 estimates in healthy and diseased brain tissue: a comparison using various MR pulse sequences. Radiology 1986;160(2):375–381. [DOI] [PubMed] [Google Scholar]
- 126.Tunc-Skarka N, Weber-Fahr W, Hoerst M, Meyer-Lindenberg A, Zink M, Ende G. MR spectroscopic evaluation of N-acetylaspartate’s T2 relaxation time and concentration corroborates white matter abnormalities in schizophrenia. Neuroimage 2009;48(3):525–531. [DOI] [PubMed] [Google Scholar]
- 127.Aydin K, Ucok A, Guler J. Altered metabolic integrity of corpus callosum among individuals at ultra high risk of schizophrenia and first-episode patients. Biol Psychiatry 2008;64(9):750–757. [DOI] [PubMed] [Google Scholar]
- 128.Aydin K, Ucok A, Cakir S. Quantitative proton MR spectroscopy findings in the corpus callosum of patients with schizophrenia suggest callosal disconnection. AJNR Am J Neuroradiol 2007;28(10):1968–1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chang L, Friedman J, Ernst T, Zhong K, Tsopelas ND, Davis K. Brain metabolite abnormalities in the white matter of elderly schizophrenic subjects: implication for glial dysfunction. Biol Psychiatry 2007;62(12):1396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Supprian T, Hofmann E, Warmuth-Metz M, Franzek E, Becker T. MRI T2 relaxation times of brain regions in schizophrenic patients and control subjects. Psychiatry Res 1997;75(3):173–182. [DOI] [PubMed] [Google Scholar]
- 131.Pfefferbaum A, Adalsteinsson E, Spielman D, Sullivan EV, Lim KO. In vivo spectroscopic quantification of the N-acetyl moiety, creatine, and choline from large volumes of brain gray and white matter: effects of normal aging. Magn Reson Med 1999;41(2):276–284. [DOI] [PubMed] [Google Scholar]
- 132.Du F, Cooper A, Cohen BM, Renshaw PF, Ongur D. Water and metabolite transverse T2 relaxation time abnormalities in the white matter in schizophrenia. Schizophrenia research 2012;137(1–3):241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Klar AA, Ballmaier M, Leopold K, Hake I, Schaefer M, Bruhl R, Schubert F, Gallinat J. Interaction of hippocampal volume and N-acetylaspartate concentration deficits in schizophrenia: a combined MRI and 1H-MRS study. Neuroimage 2010;53(1):51–57. [DOI] [PubMed] [Google Scholar]
- 134.Bustillo JR, Rowland LM, Mullins P, Jung R, Chen H, Qualls C, Hammond R, Brooks WM, Lauriello J. 1H-MRS at 4 tesla in minimally treated early schizophrenia. Mol Psychiatry 2010;15(6):629–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jessen F, Fingerhut N, Sprinkart AM, Kuhn KU, Petrovsky N, Maier W, Schild HH, Block W, Wagner M, Traber F. N-acetylaspartylglutamate (NAAG) and N-acetylaspartate (NAA) in patients with schizophrenia. Schizophr Bull 2013;39(1):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hetherington HP, Mason GF, Pan JW, Ponder SL, Vaughan JT, Twieg DB, Pohost GM. Evaluation of cerebral gray and white matter metabolite differences by spectroscopic imaging at 4.1T. Magn Reson Med 1994;32(5):565–571. [DOI] [PubMed] [Google Scholar]
- 137.Ongur D, Prescot AP, Jensen JE, Rouse ED, Cohen BM, Renshaw PF, Olson DP. T2 relaxation time abnormalities in bipolar disorder and schizophrenia. Magnetic resonance in medicine 2010;63(1):1–8. [DOI] [PubMed] [Google Scholar]
- 138.Jezzard P, Duewell S, Balaban RS. MR relaxation times in human brain: measurement at 4 T. Radiology 1996;199(3):773–779. [DOI] [PubMed] [Google Scholar]
- 139.Landheer K, Sahgal A, Myrehaug S, Chen AP, Cunningham CH, Graham SJ. A rapid inversion technique for the measurement of longitudinal relaxation times of brain metabolites: application to lactate in high-grade gliomas at 3 T. NMR Biomed 2016;29(10):1381–1390. [DOI] [PubMed] [Google Scholar]
- 140.Li Y, Srinivasan R, Ratiney H, Lu Y, Chang SM, Nelson SJ. Comparison of T(1) and T(2) metabolite relaxation times in glioma and normal brain at 3T. J Magn Reson Imaging 2008;28(2):342–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Isobe T, Matsumura A, Anno I, Yoshizawa T, Nagatomo Y, Itai Y, Nose T. Quantification of cerebral metabolites in glioma patients with proton MR spectroscopy using T2 relaxation time correction. Magn Reson Imaging 2002;20(4):343–349. [DOI] [PubMed] [Google Scholar]
- 142.Usenius JP, Kauppinen RA, Vainio PA, Hernesniemi JA, Vapalahti MP, Paljarvi LA, Soimakallio S. Quantitative metabolite patterns of human brain tumors: detection by 1H NMR spectroscopy in vivo and in vitro. J Comput Assist Tomogr 1994;18(5):705–713. [DOI] [PubMed] [Google Scholar]
- 143.Madan A, Ganji SK, An Z, Choe KS, Pinho MC, Bachoo RM, Maher EM, Choi C. Proton T2 measurement and quantification of lactate in brain tumors by MRS at 3 Tesla in vivo. Magn Reson Med 2015;73(6):2094–2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sijens PE, Oudkerk M. 1H chemical shift imaging characterization of human brain tumor and edema. Eur Radiol 2002;12(8):2056–2061. [DOI] [PubMed] [Google Scholar]
- 145.Manton DJ, Lowry M, Blackband SJ, Horsman A. Determination of proton metabolite concentrations and relaxation parameters in normal human brain and intracranial tumours. NMR Biomed 1995;8(3):104–112. [DOI] [PubMed] [Google Scholar]