Abstract
Introduction:
Due to the relatively low mutation rate and high frequency of copy number variation, finding actionable genetic drivers of high grade serous carcinoma (HGSC) is a challenging task. Furthermore, emerging studies show that genetic alterations are frequently poorly represented at the protein level adding a layer of complexity. With improvements in large-scale proteomic technologies, proteomics studies have the potential to provide robust analysis of the pathways driving high HGSC behavior.
Areas covered:
This review summarizes recent large-scale proteomics findings across adequately sized ovarian cancer sample sets. Key words combined with ‘ovarian cancer’ including 'proteomics', 'proteogenomic', 'reverse-phase protein array', 'mass spectrometry', and 'adaptive response', were used to search PubMed.
Expert opinion:
Proteomics analysis of HGSC as well as their adaptive responses to therapy can uncover new therapeutic liabilities, which can reduce the emergence of drug resistance and potentially improve patient outcomes. There is a pressing need to better understand how the genomic and epigenomic heterogeneity intrinsic to ovarian cancer is reflected at the protein level and how this information could be used to improve patient outcomes.
Keywords: Ovarian cancer, proteomics, reverse phase protein array, mass spectrometry, adaptive responses, targeted therapy
1. Introduction
High grade serous ovarian carcinoma (HGSC) is the most common form of ovarian cancer and is amongst the deadliest of all women’s cancer. The main reason behind this poor outcome is diagnosis of the majority of patients at an advanced stage. Although most HGSC respond to initial platinum-based chemotherapy, development of chemo-resistance and recurrence is frequent. Unfortunately, there are few effective treatment options for patients with chemo-resistant recurrent disease, showing the urgent need to improve therapeutic approaches available for the treatment of HGSC patients [1]. In the past decade, several groups have investigated the development and progression of HGSC. Perhaps the most striking new concept is that most if not all HGSC arise from the fallopian tube. The initial events in the process appears to be acquisition of aberrations in p53 function followed by mutation in p53. Aberrant cells arising from the fallopian tube colonize the ovary and subsequently the remainder of the peritoneal cavity [2]. These studies have led to a better understanding of the molecular origins of the disease and uncovered alterations in several therapeutically relevant pathways. By integrating data from mutations, copy number alterations and gene expression levels from patients with established ovarian cancer, genetic studies have found that several pathways are altered at high frequency in HGSC [3]. For example, in addition to the universal mutation in tp53, the RB1, PI3K/RAS and NOTCH pathways are deregulated in 67%, 45% and 22% of cases, respectively, while homologous recombination (HR) DNA repair pathways is defective in about 50% of the cases [3, 4]. Responsiveness to conventional chemotherapy based of a platin and a taxane derivative is associated with HR aberrations [5]. Indeed, HR deficiency and RB aberrations are associated with exceptional responders that have an improved outcome likely due to therapeutic sensitivity [5]. In contrast, amplification and overexpression of cyclin E, which is essentially mutually exclusive with BRCA1/2 aberrations and HR deficiency, represents a therapy-resistant population with a particular need for new approaches [4]. While these pathway alterations clearly represent therapeutics opportunities and challenges, the identification of patients who would benefit from a specific drug or drug combination remains a challenge as is the identification of therapeutic approaches that would capitalize on the suite of genomic aberrations present in individual ovarian cancer patients. Indeed, although the broad range of genomic alterations that are observed in HGSC converge into a limited set of functional events identifying reliable genomic biomarkers that could be tested routinely in the clinical environment for personalized treatment approaches remains elusive. It is now clear that the underlying biology of HGSC is not fully captured by genomic analysis and therefore the acquisition of additional layers of information is necessary. Further, analysis of genomic aberrations fails to capture the complexity of the tumor ecosystem that includes tumor cell intrinsic effects as well as the interactions with stroma, immune system and other cellular and extracellular matrix components. Because proteins are the main effectors of cell functions, many groups have started integrating large-scale proteomics with genetic studies, so called proteogenomic analysis [6, 7]. Indeed, proteomics can add valuable information and potentially facilitate the identification of targetable pathways involved in HGSC development and progression. In this review, we describe the advances in integration of proteomics analysis into ovarian cancer research.
2. Ovarian cancer proteogenomic landscape
2.1. Genomics and transcriptomics
Compared to most other cancer types, HGSC display a relatively low mutation rate associated with a high rate of copy number variation (CNV) [3, 8]. TP53 represents the most consistently mutated gene with mutations found in nearly 100% of HGSC. BRCA1 and BRCA2 germline and somatic mutations are found in 17% and 3% of cases, respectively [3, 9]. Less frequent mutations can also be observed in other genes such as RB1, NF1, FAT3, CSMD3, GABRA6 and CDK12 [3]. In rare cases, mutations were also found in oncogenes such as BRAF, PIK3CA, KRAS and NRAS, revealing the possible role of the PI3K-AKT and RAS-MAPK pathways in the development and progression of the disease [3]. Since HGSC tumors present with a large number of CNV of variable location and size, the identification of major drivers has been challenging. Earlier studies had implicated multiple drivers in the 3q26 amplicon, which is a common low level amplicon in HGSC, including PIK3CA, PRKCi, EVI1/MECOM, SNON and a series of microRNAs [10-15]. TCGA studies identified recurrent regional aberrations that comprises eight amplified regions and 22 deletions that occur in more than 50% of the tumors [3]. Analysis of these sequences revealed that CCNE1, MYC, MECOM, ZMYND8, IRF2BP2, ID4, PAX8 and TERT are frequently amplified, while deletion regions frequently cover genes such as PTEN, RB1, NF1 and CREBBP [3]. However, whether these aberrations represent driving events in the CNV and whether the CNVs themselves are drivers, at all, has not been definitively determined.
At the transcriptional level, HGSC can be separated into four molecular subtypes based on expression cluster analysis: the immunoreactive group expresses high level of T-cell chemokine ligands and receptors, the differentiated cluster is associated with high expression of MUC1, MUC16 and SLP1, the proliferative cluster has high expression of proliferative markers and the mesenchymal group has high expression of genes associated with epithelial mesenchymal transition (EMT) or with stromal cells [3, 16]. Finally, by integrating mutation, CNV and transcriptomic data, several groups have developed signatures of homologous recombination repair deficiency (HRD) and found that HRD is present in about 50% of HGSC [17-21]. This finding represents a major therapeutic opportunity, since HRD tumors are particularly sensitive to platin-based chemotherapy and poly (ADP-ribose) polymerase (PARP) inhibitors.
2.2. DNA-to-RNA-to-protein discrepancy
There is no doubt that genomic analyses have improved our understanding of HGSC. However, these studies have had only modest success at identifying new therapeutic liabilities that could improve patient care. Indeed, although most genomic alterations are executed at the protein level, a discordance between DNA alterations, RNA and protein expression has been recorded in different systems, including ovarian cancer. Furthermore, post-translational modifications including formation of functional protein complexes allow an additional level of modification of protein function that cannot be adequately captured by genomic analysis [22]. Since proteins are the main effectors of the cells, many groups have demonstrated that proteomics has the potential to provide better coverage of biological system alterations. Studies have shown that although most autosomal gene duplications are propagated to the protein level, 23-33% of genes located in CNV are not detected at the protein level, indicating the presence of post-transcription mechanisms that help attenuate the impact of CNV [23]. For example, proteins that are part of complexes can be unstable when other proteins of that same complex are absent, attenuating the effect of gene amplification [23]. In the case of HGSC, studying the influence of CNV on cellular processes has been challenging, since many of the CNV are unique to a limited set of tumors and cover very broad regions of the chromosome. Interestingly, by taking proteins into consideration in the analysis of CNV, Zhang et al. revealed the convergence of multiple common CNV into a limited number of protein networks [24]. Indeed, pathway analysis of proteins associated with common CNV revealed an enrichment of proteins associated with cell invasion and migration as well as proteins that are related to immune functions [24]. In addition to the discrepancy between DNA alterations and expression of the protein product, a similar observation occurs at the transcriptional level, where gene expression levels frequently correlate only weakly with protein amounts. Indeed, we and others have demonstrated that the correlation between protein and RNA has an r value of about 0.5 [25-27]. This represents a mixture of proteins with high and low correlations. The relatively low correlation is the consequence of post-transcriptional or post-translational modification as well as effects of miRNA and lncRNA on RNA stability and translation [28]. Interestingly, Zhang et al. [24] analyzed the correlation between mRNA and protein expression in HGSC and found that highly expressed and stable structural proteins tends to have a weaker correlation with mRNA expression than less stable proteins that are known to be transcriptionally regulated in response to environmental changes or stress. Conversely, mRNA with lower stability shows a relatively poor correlation with protein expression, which can be particularly problematic when studying transient molecular events. Another proteogenomic study performed by Johansson et al. [29] in breast tumors demonstrated that about 30% of mRNA transcripts did not correlate with protein expression. Interestingly, this discrepancy was not associated with the half-life of the protein or mRNA. The proteins that had the least correlation with their mRNA counterpart were proteins that are known to be sensitive to ubiquitination, signaling and proliferation related proteins as well as structural ribosomal and mitochondrial proteins [29]. Taken together, these studies demonstrate that assessment of protein expression and post-translational modifications offer an additional layer of functional heterogeneity beyond genomic aberrations and reinforce the need to consider the tumor proteome when studying the consequences of genomic alterations. Furthermore, these studies indicate the importance of analyzing the proteome in determining mechanisms underlying tumor initiation and progression as well as identification of effective therapies to counter heterogeneity and biomarkers to select patients likely to benefit from particular therapies.
2.3. Proteomics
RPPA has been extensively used in ovarian cancer studies. This method consists of a high throughput dot blot approach where protein lysates are printed on a slide and probed with high quality validated antibodies that allows quantification of up to 500 total and post-translationally modified proteins in up to 1,000 samples simultaneously [27, 30-32]. This technology can be used to study cellular processes such as apoptosis, migration, proliferation and DNA damage response. It is also particularly efficient in measuring major signaling pathway activity, including RAS-MAPK, PI3K-AKT, TSC-mTOR and RTK signaling [25, 33]. This antibody-based assay has been proven in cases where high quality antibodies are available to be more sensitive than other proteomics approaches, such as mass spectrometry (MS), particularly for detection and quantification of post-translational modifications [27]. However, it is highly dependent on the quality and validation of antibodies for the technology [24, 32]. Also, the coverage of multiple phosphorylation sites and splice variants remains challenging with RPPA. However, where high quality antibodies are available to phosphorylation sites of functional relevance, RPPA can be more sensitive than mass spectrometry and can detect a number of regulatory phosphorylation sites that are not detected by mass spectrometry due to low prevalence or their presence in a site that is not well resolved by mass spectrometry [27]. Although RPPA does not cover the complete proteome, the diversity of proteins covered by the assay allowed multiple groups to create signatures that can predict patient outcome. For example, the protein-driven index of ovarian cancer (PROVAR) was developed based on RPPA data analysis of HGSC and aimed at predicting time to recurrence [16]. This nine protein signature is an independent predictor of both overall (OS) and progression free survival (PFS). These include five proteins associated with a longer PFS (AR, Bid, HSP70, and phosphorylated TAZ and EGFR) and four proteins associated with a shorter PFS (EEF2, STAT5alpha, and phosphorylated PKCalpha and MEK1). Interestingly, although the signature seemed to be related to EGFR pathway activity, applying a hierarchical unsupervised clustering of the samples showed that the protein signature was dispersed amongst four main protein clusters that define ovarian cancer, indicating that this signature covers more than a single pathway. In another RPPA-based study, Carey et al. showed that the TGF-beta signaling pathway is an important predictor of chemoresistance in advanced HGSC. In their study, they associated RPPA data with normalization of CA125 values after the third course of chemotherapy and found that several proteins involved in the TGF-beta signaling pathways were increased in the group that retained higher CA125 values. CA125 normalization is defined as a reduction of CA125 to less than 35U/L (normal levels) after chemotherapy treatment in patients that have elevated CA125 levels at diagnosis [34, 35] and has been shown to be associated to survival in HGSC patients [35]. In addition, an unsupervised clustering of the RPPA data revealed two major groups of HGSC. Group 1 was associated to a worst outcome and displayed high levels of cyclin D1, cyclin E2, stromal markers and phosphorylated AKT. The second group had better PFS and was associated with high expression of Cyclin B and E1, ER and phosphorylated ER, Rb, mTOR and c-Myc proteins [30]. Several of the proteins are markers of cell cycle progression and the improved PFS may be due to sensitivity of this group to platin and taxol based chemotherapy. Finally, RPPA data has also been shown to be useful in improving proteogenomic study quality, more specifically in cases involving surgically resected tumors. Indeed, Mertins et al. analyzed proteins that are altered during ischemia [27]. Their study highlighted a list of proteins and pathways, primarily post-translational modifications, that are altered due to cold ischemia due to a delay between freezing and collection of a tissue sample. Their findings showed that the main pathways affected by cold ischemia are the MAPK stress-response, apoptosis and transcriptional regulation [27].
Many groups have investigated the proteome of ovarian tumors in order to define a better molecular classification and identify new therapeutic targets. For example, the Clinical Proteomic Tumor Analysis Consortium (CPTAC) used MS approaches to study the proteome and phosphoproteome of ovarian tumors. In the CPTAC study, the proteome and phosphoproteome of ovarian tumors was quantified using iTRAQ labelling and LC-MS/MS (Box1). Protein abundance analysis from 169 tumors highlighted the presence of five distinct HGSC tumor groups that display biological differences [24]. Four of those groups were consistent with the mesenchymal, proliferative, immunoreactive and differentiated subtypes that were previously identified at the transcriptional level by TCGA ovarian cancer study [3]. The fifth group was smaller and displayed higher expression of microenvironment markers, which could represent an artifact of low tumor content samples [24]. Based on proteome analysis, the CPTAC group developed protein signatures of chromosomal instability (CIN) and HRD. The CIN signature was composed of 128 proteins, with chromatin remodeling proteins CHD4 and CHD5 having the strongest association with CIN. The HRD signature was composed of 30 proteins that could differentiate HR deficient and HR proficient patients. Interestingly, several of these proteins are involved in histone modification, such as HDAC1, RBBP4, RBBP7, EP300 and HUS1 [24, 36]. By analyzing phosphopeptide abundance, the authors found 15 signaling pathways that are associated with a shorter survival of HGSC patients, with many of them being therapeutically tractable. These pathways included the RhoA, PDGFR, integrin-like kinase, Notch, HER2/Neu, Rac1, CxCR4, Thrombin, IL-12 and Thrombaxane pathways [24]. Interestingly, phosphoproteins quantification allowed a higher statistical significance for correlations with outcome when compared to transcriptomic or proteome analysis consistent with the need to assess protein function which is reflected by phosphorylation in comparison to protein amount to predict cellular behavior [24]. The CPTAC analysis failed to detect the presence of the majority sequence changes encoded by DNA aberrations at the protein level. This may be due to mutation mediated mRNA decay, protein instability particularly for truncated proteins or a low level of coverage at the single amino acid level precluding detection of aberrations. Nevertheless, this further emphasizes the challenges in predicting changes in protein function by analysis of the cancer genome. The database developed by the CPTAC effort provides an incredible resource for exploration by others. For example, Yu et al. [37] used machine learning models to reanalyze the CPTAC data and found a protein signature associated to chemotherapy response in HGSC patients. In their 24 protein signature, 14 proteins were overrepresented in the chemoresistant group, while 10 protein were expressed at higher levels in chemosensitive tumors. Pathway analysis revealed that most of the proteins associated with chemosensitivity are involved in oxidative phosphorylation, RNA transport or mineral absorption pathways, display Ran GTPase binding function, and are involved in the cellular response to zinc or in ATP synthesis and metabolism [37]. CPTAC data from both ovary and breast cancer studies, was further used to demonstrate that RNA editing results in the production of aberrant proteins that can be detected by MS contributing to proteomic diversity in ovarian cancers [38].
Box 1: Mass spectrometry.
There are three major approaches to quantify proteins using mass spectrometry (MS). The first one is shotgun/bottom-up proteomics where complex protein mixtures are enzymatically digested and the resulting peptides are separated using liquid chromatography (LC) before injection into a tandem mass spectrometer (MS/MS). Peptides sequences are then aligned against a database and proteins are identified. In this approach, the peptides can be labeled using isotopic or isobaric tags such as SILAC and iTRAQ, which allows a more accurate quantification [86, 87]. This data-dependent approach allows identification and quantification of hundreds to thousands of proteins. However, one of the main issues with this approach is that detection of specific peptides depends on the abundance and size of the proteins. Therefore, low abundance proteins are usually underrepresented, unless higher abundance proteins are depleted prior to the experiment or their peptides excluded from the MS/MS measurement [86]. This is particularly problematic when mutations or post-translational modifications that affect only a single amino acid in the sequence are sought. The second MS approach is targeted proteomics and involves a pre-defined list of peptides that will be accurately measured through selected reaction monitoring (SRM), parallel reaction monitoring (PRM), multiple reaction monitoring (MRM) or related approaches. After enzymatic digestion of the proteins and separation of the resulting peptides through LC, quantification of the peptides can be performed through the addition of internal standard composed of stable isotope-labeled (SIL) synthetic peptides representing the analytes of interest. Prior knowledge of the protein of interest sequence allows the selection of unique amino acid sequences that will be monitored during the experimental run and used to quantify the protein. These methods are particularly useful for experiments that aim at comparing protein abundance across samples, conditions or time points and are frequently used in the clinical environment because of their high reproducibility. The other advantage of this method is that low abundance peptides can be tracked and quantified accurately [7, 86-88]. SWATH-MS and related approaches represent the third approach and is complementary to traditional shotgun and targeted proteomics approaches. Label-free data-independent acquisition method allows precise quantification of all peptides found within a specific preselected m/z range [89]. Briefly, this method generate spectra of all precursor ions within a pre-defined rention time (RT) and m/z range, which can then be analyzed using spectral libraries. Spectral libraries are generated using shotgun proteomics on any sample of interest and contains information on precursors, fragment ions and RT. In theory, this approach allows the quantification of any peptides, independent of their abundance. However, the accuracy of this approach is highly dependent on the quality of the spectral library that is being use and the ability to achieve the deconvolution of the data [89, 90]. Although MS is incredibly robust and reproducible, there is one major issue related to this technology, which is the sensitivity. MS can accurately measure the relative expression of proteins and determine mutations. However, low protein coverage frequently occurs because of peptide nature. For example, too long or too short tryptic peptides or highly hydrophobic or hydrophilic peptides will not be measured accurately. For those reasons, many mutations are difficult to identify and some proteins might be impossible to quantify by MS [7, 24]. In these cases, antibody-based technologies such as western blotting or RPPA can provide important complementary information.
Multiparameter mass cytometry (CyTOF) has been used to characterize 37 candidate proteins in 800 000 single cells from 17 HGSC tumors providing a novel spatial and functional map of the proteome in ovarian cancer [39] . Briefly, the approaches involved a panel of 37 stable metal isotope labeled antibodies targeting intracellular and membrane proteins. Gonzalez et al. identified multiple cancer cell subsets that were not obvious from the bulk analysis done by RPPA or MS [39]. Importantly, the relative cell frequency of dominant cell types ranged from 16% to 83% per tumor, revealing a high level of heterogeneity. Not surprisingly, a higher level of protein heterogeneity was associated with a higher risk of a relapse albeit in a small sample set that requires confirmation in larger sets [39]. Expression of E-cadherin and vimentin as well as CD24, CD13, CD10, C73, CD61, CD491, CD90, CD44, CD133, endoglin and ROR1 was used to investigate potential stem cell phenotypes and cellular plasticity [39]. The antibody panel also identified three different clusters that were consistent with dysregulated cell cycle [39]. Taken together, this study demonstrated that HGSC display high level of protein heterogeneity at the single cell level that needs to be taken into account when developing and implementing new therapies.
3. Proteomics and precision medicine
3.1. Adaptive response to therapy
Although characterizing ovarian tumors prior to treatment helps to identify potential subtypes and molecular aberrations, one of the main challenges encountered in the treatment of HGSC is the development of drug resistance. There are three major classes of drug resistance: inherent, adaptive and acquired. Inherent resistance is intrinsic and is characterized by tumors not responding to a specific therapy or progressing through initial therapy. This is usually the result of pre-existing genetic alterations and tumor heterogeneity that can potentially be identified prior to treatment with proteogenomics approaches [40]. Adaptive and acquired resistance are tightly connected and both develop in response to therapy. Biological systems are used by normal cells to adapt to stress and to respond to stress rapidly primarily through non-genomic mechanisms. The adaptive response is a cellular state that is often not permanent and is the result of the cells rewiring networks to enable survival during stress. The underlying mechanisms have been co-opted by tumor cells to escape therapeutic stress. Adaptive responses involve mechanisms such as epigenetic changes and protein network rewiring through post-translational modification [41, 42]. This phenotypic plasticity takes place within hours or days after treatment initiation and allows the cells to survive until the stress is removed or until new genetic alterations occurs or are selected, leading to acquired resistance [41, 43]. In contrast to the adaptive response, acquired resistance is permanent, usually takes months to appear and is generally intractable therapeutically. Thus, determining adaptive responses to therapy early during the course of treatment represents an invaluable window of opportunity to prevent the development of acquired resistance. Our group and others have shown that proteomics is a robust approach to study adaptive responses of ovarian cancer.
Although targeted therapies have had considerable success in the clinical setting for multiple cancer types, emerging studies have shown that cancer cells can acquire resistance to many of these drugs within a relatively short period [33, 44]. Thus, single-agent targeted therapies are unlikely to be sufficient to provide complete or prolonged therapeutic responses in complex epithelial cancers such as ovarian cancer. Interestingly, a window of opportunity trial with HGSC patients that were treated for a short period with a PARP inhibitor indicated that adaptive responses to therapy can be detected by proteomics approaches early during the course of treatment [33]. More importantly, this study and many others have shown that targeting the adaptive response with a second drug can improve the efficacy, depth and duration of treatment as well as increase the spectrum of patients that could benefit the treatment [44-53]. For that reason, a better understanding of adaptive responses to therapy might help overcome acquisition of drug resistance and improve outcome of patients.
3.2. Chemotherapy
The standard treatment of HGSC consists of a tumor debulking surgery followed by chemotherapy treatment that is usually comprised of a combination of carboplatin and paclitaxel. In a subset of patients with advanced disease, neoadjuvant therapy is performed prior to surgery in order to reduce the tumor burden. In those cases, the neoadjuvant treatment usually consists of a combination of taxane/platin or liposomal doxorubicin/carboplatin [54]. Although most HGSC patients respond to primary therapy, resistance development and recurrence is observed in the majority of cases. Interestingly, many groups have described adaptive responses to chemotherapy and inhibition of these pathways led to increased treatment efficacy. For example, Lee et al. demonstrated that a transient expression of CXCR4 in ovarian cancer cell lines treated with cisplatin, doxorubicin and paclitaxel allows a cell subpopulation to enter dormancy until the treatment is stopped. Moreover, they showed that CXCR4 antagonists have synergistic effects in the killing of the cancer cells when combined with chemotherapeutic agents [55]. In another study, Choi et al. used RPPA analysis to identify protein alterations that can be detected early during paclitaxel treatment. They found that S6 phosphorylation is strongly upregulated in response to paclitaxel. Interestingly, S6 is a downstream target of the mTOR pathway, which has been described as a pro-survival mechanism in many biological systems [45]. Furthermore, the inhibition of S6 phosphorylation using PI3K pathway inhibitor drugs such as BX795 or CCT128930, in combination with paclitaxel, decreased viability of ovarian cancer cells [45]. Finally, other mechanism of adaptive response to chemotherapy were reported by other groups such as overexpression of PGC1a, a protein involved in mitochondrial biogenesis [56], as well as TRAP1. TRAP1 is proposed to contribute to the adaptive response to cisplatin through increased oxidative phosphorylation, leading to the secretion of cytokines and epithelial-to-mesenchymal transition (reviewed in [57]).
3.3. VEGF/VEGFR inhibitors
Angiogenesis is an important mediator of ovarian cancer pathogenesis, stimulating tumor growth and progression. The vascular endothelial growth factor (VEGF) and its receptor VEGFR are expressed by ovarian cancer cells and inhibitors of these proteins such as bevacizumab, an anti-VEGF monoclonal antibody, are being used as maintenance therapy for patients that were previously treated with cisplatin. Several clinical studies have shown that angiogenesis inhibition can increase progression free survival of ovarian cancer patients (Reviewed in [58]). However, although angiogenesis inhibitors increase the PFS of HGSC patients, most patients rapidly develop resistance to the drug. Several adaptive responses to VEGF have been observed, such as increased activity of the pro-survival pathways PI3K-AKT and P70S6K, as well as increased angiopoietin-2 signaling, which improves endothelial cells functions and survival through the recruitment of pericytes [59, 60]. Combining VEGF inhibition with inhibitors of these pathways has been shown to increase sensitivity of the tumors to VEGR inhibitors [59, 60]. Increased expression of the protein CSF1R, which is expressed in tumor-associated macrophages, can also mediate adaptive resistance to anti-VEGF. CSF1R controls the growth and differentiation of macrophages and AC708 that inhibits CSF1R signaling restores sensitivity to anti-VEGF antibodies in a mouse model [61]. With the entry of CSF1R inhibitors into clinical trials, there is a potential to translate these studies to clinical trials in ovarian cancer in the near future.
3.4. Bromodomain inhibitors
Although epigenetic modulators are yet to be approved by the FDA for the treatment of HGSC, several studies have shown that bromodomain proteins such as BRD4 contribute to the growth and survival of ovarian cancer cells. BRD4 inhibitors such as JQ1 have been shown to be effective in reducing ovarian cancer cell growth both in vitro and in vivo . However, as it is the case for many other targeted therapies, resistance development occurs rapidly. An interesting study performed by Kurimchak et al. showed that BET inhibitor JQ1 induces kinome reprogramming in ovarian cancer cells, which eventually gives rise to drug resistance. By using multiplexed inhibitor beads and quantitative mass spectrometry approaches (MIB/MS), they have demonstrated that this adaptive response involves the upregulation of RTK activity as well as increased downstream signaling through the PI3K-AKT and MAPK pathways [62]. More importantly, a number of different RTK were induced in a subset of ovarian cancer cells, indicating that the adaptive response depends on cell specific intrinsic RTK programs. Finally, combining JQ1 with inhibitors of these specific adaptive responses demonstrated an increase sensitivity to JQ1.
3.5. PARP inhibitors
PARP is an important mediator of both base excision repair and alternative-non homologous end joining. It is responsible for sensing DNA damage and it PARylates and activates several DNA repair proteins. Inhibition of PARP enzymatic activity leads to the accumulation of SSB that eventually transform into DSB (reviewed in [63-65]). More recently, the role of PARP in replication fork protection has been elucidated with PARPi inhibitors inducing replication stress and DNA damage [47]. Because of the extensive DNA damage induced by PARP inhibitors, cells rely on HR to accurately repair DSBs. In the case of HR defective tumors, such as tumors with BRCA mutations, PARP inhibitors create chromosomal instability that leads to synthetic lethality [63-66]. Interestingly, several PARP inhibitors have shown an additional lethal activity by trapping PARP on DNA [67-69]. The DNA replication machinery crashes into trapped PARP molecules, leading to the replication fork collapse and additional DSBs. Although PARP inhibitors improve the PFS of HGSC patients in the maintenance setting, they have only a modest effect on OS. While a subset of patients show a long-term benefit, there is rapid development of PARP inhibitor resistance in most patients. Fortunately, emerging studies suggests that PARP-based combination therapies are more effective than PARP monotherapy. Furthermore, targeting adaptive responses to PARP inhibitors demonstrates synergistic activity in many models including early studies in patients. Several adaptive responses to PARP inhibitors have been identified using proteomics. First, one of the most conserved adaptive responses is the activation of the G2-M DNA damage checkpoint. Indeed, several groups including ours, have demonstrated that a combination of PARP and G2-M DNA damage checkpoint inhibitors has synergistic activity in reducing cancer cell growth [33, 47]. A second adaptive response that has been demonstrated is the activation of pro-survival signaling pathways such as the RAS-MAPK and PI3K pathways [33, 44, 50, 70]. Interestingly, in addition to their pro-survival function, using an inhibitor specific to one of these pathways seems to reduce the ability of the cancer cells to repair DNA damage through the HR pathway [44, 50, 71]. Finally, other proteins such as BRD4 and FOXM1 were shown to be involved in the activation of the HR DNA repair pathway following PARP inhibitor treatment. Inhibition of FOXM1 or BRD4 can trigger an HRD state and increase the efficacy of PARP inhibitors [46, 53].
4. Expert opinion
For the past several decades, there has been limited progress made in overall survial of HGSC patients although there has been an improvement in 5 year survival. Large-scale genomics technologies have been successfully used to study various cancer types, uncovering new biomarkers and therapeutic targets, resulting in improved patient outcomes. Unfortunately, because of its unique genetic features, genomics approaches have not been sufficient to capture the complexity of HGSC or to provide a suite of actionable targets [3, 8]. Our group and others have demonstrated that proteomics can add valuable information to genomics data and help better understand ovarian cancer pathophysiology. Indeed, proteogenomic studies can help discriminate between silent and non-silent genomic alterations, identify the functional events that result from genetic, epigenetic or post-translational alteration, as well as identify neoantigens that could be therapeutically relevant [29]. Although some genetic features can predict response to specific therapeutic agents, proteomics can be used to measure and validate the activity of therapeutically tractable pathways that are not always represented or interpretable at the genomic level [25, 29-33, 37, 42, 44, 53, 72-76]. Indeed, cancer cells exist in multiple transient states that can be triggered by stimuli such as environmental or therapeutic stresses. These states are mostly regulated through post-translational modifications and epigenetic events [41, 42, 77-80]. We have demonstrated that studying post-translational modifications in cancer cells that have been treated with a therapeutic agent for a short period of time can inform not only on the tumor response to the drug but also on possible drug combinations that could effectively kill the cancer cells [33, 44, 47]. Indeed, a protein network rewiring rapidly occurs during therapeutic stress as a result of the cancer cells trying to survive until the stress is removed or until the cell develops genetic alterations that would render it resistant to the stress. Targeting these adaptive responses can be a very effective means to kill cancer cells and overcome the development of resistance [33, 44-53]. Unfortunately, ovarian cancer proteomics studies have been limited in both number and scope and we strongly believe that the field would benefit from large-scale proteomics studies that would better characterize the proteome and phosphoproteome of untreated and treated tumors. Having a better understanding of the proteome could lead to clinically relevant proteomic test that would allow more efficient personalized treatment and improved patient outcomes.
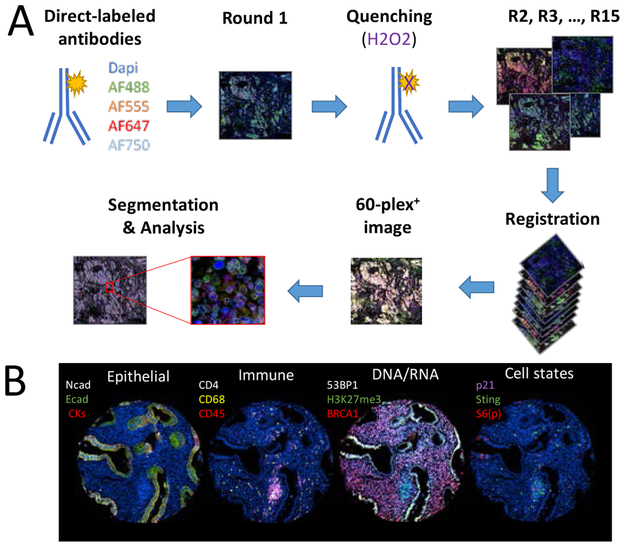
Although there has been massive improvement in proteomics technologies, sensitivity, specificity or coverage are still limiting factors particularly when attempts are made to capture heterogeneity at the single cell level. While MS has the potential to cover a broad range of proteins and is not dependent on the quality of commercially available antibodies, sensitivity as well as sample input remains a challenge. On the other hand, a number of antibody-based assays such as RPPA and scanning mass cytometry, multiplex IHC and cyclic immunofluorescence can be more sensitive in analyzing post-translational events, but are limited by the availability and quality of antibodies. Another limitation of the current proteomics approaches remains in being able to analyze a large number of proteins at the single cell level, while retaining the spatial organization of the tissue. It is clear that HGSC are heterogeneous tumors and bulk analysis of the proteome might provide insufficient information. There is limited information on how HGSC genetic intratumoral heterogeneity is reflected at the protein level and how this affects responses to targeted therapies. In a recent study published by our group, we have demonstrated that adaptive responses of HGSC to PARP inhibitors show very limited interlesion heterogeneity, suggesting that protein heterogeneity might not be as complex as the genomic heterogeneity [33]. Many genomic alterations converge into a limited number of functional events and single cell proteomics could help solve these questions. In the past few years, several new technologies have been developed, such as mass cytometry based approaches, multiplex IHC and cyclic immunofluorescence. For example, cyclic immunofluorescence (Figure 1) is an attractive way of studying tumor heterogeneity as well as monitoring the tumor microenvironment including immune populations. In this assay, antibodies are conjugated with fluorescent molecules and are applied on a tissue sections in cycles of four antibodies. Between each cycle, an image of the staining is acquired and then the signal is quenched. By using different bioinformatics tools, the different images are then registered into a single multiplex image that allow the quantification of more than 60 proteins markers at the single cell level [81]. Although the deconvolution of the data is still a challenge, these technologies have been successfully used for other cancer types, such as breast and prostate cancer and have helped characterize interaction between cancer cells and their environment and immune cells, as well as cell states. Applying these technologies to ovarian cancer research would clearly help understand how ovarian cancer interact with their environment, how heterogeneous the tumors really are and how different lesions can be treated efficiently.
Figure 1. The use of cyclic-immunofluorescence for spatially oriented single cell proteomics.
(A)Primary antibodies are directly conjugated to fluorescent molecules. During the first round, fours antibodies conjugate with different Alexa-Fluor molecules are incubated on the tissue sample. Dapi is added to stain the cell nucleus and an image is acquired using a slide scanner. The signal is then quenched with hydrogen peroxide solution. Staining is then repeated for multiple rounds. The images are aligned during registration, which allows the creation of a multiplex image. Segmentation is then performed in order to analyze the expression and localization of each markers. (B) Example of a multiplex image acquired through cyclic immunofluorescence. Each panel represent a single region of a fallopian tube sample, with the fallopian tube being the source of ovarian cancer. In the first panel on the left, epithelial markers (N-cadherin, E-cadherin and a mix of cytokeratin-5, −7, −18 and −19) are being used to identify epithelial cells. In the second panel, immune cells are identified (CD68, CD4 and CD45). In the third panel, DNA and RNA related proteins (BRCA1, H3K27me3 and 53BP1) are stained. In the last panel on the right, cell states are identified (phosphorylated S6_ser235/ser236, p21 and STING).
Overall, proteomics is still an emerging field that will benefit from new technology development and improvement in the ability to obtain and interpret data. Current proteomics technologies are in most cases much more expensive than genomics and transcriptomic technologies and are not as evolved as these approaches. Unfortunately, there is no equivalent of polymerase chain reaction that can amplify protein signals quantitatively and efficiently. Increasing the availability of high quality antibodies as well as improving depth of coverage and the need for large input material in mass spectrometry is essential in order to improve our ability to study the proteome of cancer cells particularly rare cell populations and small tissue samples available from human patients. In addition, there is an urgent need for spatially oriented single cell proteomics that will have a major impact on our understanding of the tumoral heterogeneity and how to target heterogeneity in patients. Indeed, the ability to perform unbiased deep protein analysis on single cells while capturing not only total proteins but rare events such as mutations, posttranslational modifications or splicing variants remains the holy grail for proteomic analysis. There is also a need to be able to deal with challenging tissues such as formalin fixed paraffin embeded samples that are the mainstay in the pathology laboratory. Rendering these technologies more affordable and improving our ability to deconvolute the data will be essential in order to increase the scalability of the approach. In the time being, combining proteomics with genomics approaches provides an incredible opportunity to maximize the information that can be extracted from a tumor [6, 7, 40].
There are major challenges remaining in the analysis and interpretation of proteomic data. The incredible diversity of proteins based on millions of variants generated by splicing variations, RNA editing and post translational modifications as well as mutations and fusion genes in cancer results in a limited coverage of each specific protein form. This results in challenges in quantification and in some cases full identification of the proteome. Furthermore, proteins are labile undergoing degradation and post translational modification after collection of samples. Together, these challenges can lead to both false positive and false negative identification of signals associated with specific events such as early diagnosis or prediction of patient outcomes.
Although we now have a better understanding of the protein networks involved in HGSC tumor progression and therapy response, we still know little about how to use this information to improve outcomes for ovarian cancer patients. Given the fact that HGSC presents with a high level of intra-tumoral and inter-lesion heterogeneity at the genetic level, it will be important to determine how much of this heterogeneity is being translated to the protein level. Many single-cell spatially oriented proteomics technologies have been developed in the past few years, including multiplexed ion beam imaging by time-of-flight, multiplex immunohistochemistry and cyclic-immunofluorescence [81-84]. These technologies could be particularly helpful in better characterizing cancer cell populations as well as their interaction with their microenvironment and immune cells [85]. Furthermore, more studies will be required to determine if adaptive responses to therapy are homogenous amongst cancer cells from the tumor or if different cell populations display different adaptive responses. This information will be necessary in order to better implement new personalized treatment approaches where patients would be given a drug based on specific vulnerabilities of their tumors.
Article highlights.
HGSC presents with low mutation rate and high frequency of copy number variation, which makes it difficult to identify reliable therapeutics targets.
Discrepancy between DNA, RNA and Protein levels has been observed in ovarian cancer likely due to post-transcriptional or post-translational modification as well as effects of miRNA and lncRNA on RNA stability and translation.
Proteomics studies using reverse phase protein arrays have allowed the identification of proteins associated to patient outcome, chemoresistance and progression of ovarian cancer.
The Clinical Proteomic Tumor Analysis Consortium CPTAC group used mass spectrometry to subclassify ovarian cancer and identify signature of chromosomal instability (CIN) and homologous recombination defect (HRD)
Proteomics can uncover how ovarian cancer cells and the tumor ecosystem adapts to therapy and has the potential to improve patient outcomes by development and implementation of effective drug combinations.
Acknowledgments
Funding
G.B. Mills is supported by a kind gift from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Ovarian Cancer Research Foundation, The Breast Cancer Research Foundation, The Komen Foundation SAC110052, and U.S. Department of Health and Human Services, National Institutes of Health, National Cancer Institute grants: CA217685, CA217842, and CA098258. M. Labrie is supported by the Ovarian Cancer Research Alliance and and Ruth and Steve Anderson, in honor of Shae Anderson Gerlinger.
Footnotes
Declaration of interest
G.B. Mills is an advisory board member for AstraZeneca, ImmunoMET, Ionis, Nuevolution, PDX bio, Signalchem, Symphogen, and Tarveda, holds stock options for Catena Pharmaceuticals, ImmunoMet, SignalChem, Spindle Top Ventures and Tarveda, travel support from Chrysallis Bio, and has licensed technology to Nanostring and Myriad Genetics. L. Campbell is a consultant for Quantitative Imaging System.
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewer disclosures
Peer reviewers on this manuscript have no relevant financial or other relationships to disclose.
References
Papers of special note have been highlighted as:
* of interest
** of considerable interest
- 1.Jayson GC, Kohn EC, Kitchener HC and Ledermann JA, Ovarian cancer. Lancet. 384(9951): p. 1376–88. (2014) [DOI] [PubMed] [Google Scholar]
- 2.Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. , High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun. 8(1): p. 1093 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cancer Genome Atlas Research, N., Integrated genomic analyses of ovarian carcinoma. Nature. 474(7353): p. 609–15. (2011)** This research article describes the genomics of ovarian cancer patient.
- 4.Berger AC, Korkut A, Kanchi RS, Hegde AM, Lenoir W, Liu W, et al. , A Comprehensive Pan-Cancer Molecular Study of Gynecologic and Breast Cancers. Cancer Cell. 33(4): p. 690–705 e9. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng G and Mills GB, Surviving Ovarian Cancer: An Affair between Defective DNA Repair and RB1. Clin Cancer Res. 24(3): p. 508–510. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dimitrakopoulos L, Prassas I, Diamandis EP and Charames GS, Onco-proteogenomics: Multi-omics level data integration for accurate phenotype prediction. Crit Rev Clin Lab Sci. 54(6): p. 414–432. (2017) [DOI] [PubMed] [Google Scholar]
- 7.Dimitrakopoulos L, Prassas I, Diamandis EP, Nesvizhskii A, Kislinger T, Jaffe J, et al. , Proteogenomics: Opportunities and Caveats. Clin Chem. 62(4): p. 551–7. (2016)** Review on proteogenomics technologies.
- 8.Yarchoan M, Albacker LA, Hopkins AC, Montesion M, Murugesan K, Vithayathil TT, et al. , PD-L1 expression and tumor mutational burden are independent biomarkers in most cancers. JCI Insight. 4(6). (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hennessy BT, Timms KM, Carey MS, Gutin A, Meyer LA, Flake DD 2nd, et al. , Somatic mutations in BRCA1 and BRCA2 could expand the number of patients that benefit from poly (ADP ribose) polymerase inhibitors in ovarian cancer. J Clin Oncol. 28(22): p. 3570–6. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki S, Moore DH 2nd, Ginzinger DG, Godfrey TE, Barclay J, Powell B, et al. , An approach to analysis of large-scale correlations between genome changes and clinical endpoints in ovarian cancer. Cancer Res. 60(19): p. 5382–5. (2000) [PubMed] [Google Scholar]
- 11.Shayesteh L, Lu Y, Kuo WL, Baldocchi R, Godfrey T, Collins C, et al. , PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet. 21(1): p. 99–102. (1999) [DOI] [PubMed] [Google Scholar]
- 12.Eder AM, Sui X, Rosen DG, Nolden LK, Cheng KW, Lahad JP, et al. , Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci U S A. 102(35): p. 12519–24. (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nanjundan M, Nakayama Y, Cheng KW, Lahad J, Liu J, Lu K, et al. , Amplification of MDS1/EVI1 and EVI1, located in the 3q26.2 amplicon, is associated with favorable patient prognosis in ovarian cancer. Cancer Res. 67(7): p. 3074–84. (2007) [DOI] [PubMed] [Google Scholar]
- 14.Chaluvally-Raghavan P, Jeong KJ, Pradeep S, Silva AM, Yu S, Liu W, et al. , Direct Upregulation of STAT3 by MicroRNA-551b-3p Deregulates Growth and Metastasis of Ovarian Cancer. Cell Rep. 15(7): p. 1493–1504. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nanjundan M, Cheng KW, Zhang F, Lahad J, Kuo WL, Schmandt R, et al. , Overexpression of SnoN/SkiL, amplified at the 3q26.2 locus, in ovarian cancers: a role in ovarian pathogenesis. Mol Oncol. 2(2): p. 164–81. (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang JY, Yoshihara K, Tanaka K, Hatae M, Masuzaki H, Itamochi H, et al. , Predicting time to ovarian carcinoma recurrence using protein markers. J Clin Invest. 123(9): p. 3740–50. (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chao A, Lai CH, Wang TH, Jung SM, Lee YS, Chang WY, et al. , Genomic scar signatures associated with homologous recombination deficiency predict adverse clinical outcomes in patients with ovarian clear cell carcinoma. J Mol Med (Berl). 96(6): p. 527–536. (2018) [DOI] [PubMed] [Google Scholar]
- 18.Ashley CW, Da Cruz Paula A, Kumar R, Mandelker D, Pei X, Riaz N, et al. , Analysis of mutational signatures in primary and metastatic endometrial cancer reveals distinct patterns of DNA repair defects and shifts during tumor progression. Gynecol Oncol. 152(1): p. 11–19. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peng G, Chun-Jen Lin C, Mo W, Dai H, Park YY, Kim SM, et al. , Genome-wide transcriptome profiling of homologous recombination DNA repair. Nat Commun. 5: p. 3361 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Knijnenburg TA, Wang L, Zimmermann MT, Chambwe N, Gao GF, Cherniack AD, et al. , Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 23(1): p. 239–254 e6. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abkevich V, Timms KM, Hennessy BT, Potter J, Carey MS, Meyer LA, et al. , Patterns of genomic loss of heterozygosity predict homologous recombination repair defects in epithelial ovarian cancer. Br J Cancer. 107(10): p. 1776–82. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vogel C and Marcotte EM, Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 13(4): p. 227–32. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goncalves E, Fragoulis A, Garcia-Alonso L, Cramer T, Saez-Rodriguez J and Beltrao P, Widespread Post-transcriptional Attenuation of Genomic Copy-Number Variation in Cancer. Cell Syst. 5(4): p. 386–398 e4. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. , Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell. 166(3): p. 755–765. (2016)** Describes the protein and genetic landscape of HGSC.
- 25.Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, et al. , A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nat Commun. 5: p. 3887 (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao W, Li J, Akbani R, Liang H and Mills GB, Credentialing Individual Samples for Proteogenomic Analysis. Mol Cell Proteomics. 17(8): p. 1515–1530. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mertins P, Yang F, Liu T, Mani DR, Petyuk VA, Gillette MA, et al. , Ischemia in tumors induces early and sustained phosphorylation changes in stress kinase pathways but does not affect global protein levels. Mol Cell Proteomics. 13(7): p. 1690–704. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanhaei S, Nikpour P, Ghaedi K, Rabiee F, Homayouni Moghadam F and Nasr-Esfahani MH, RNA/Protein Discordant Expression of Fndc5 in Central Nervous System Is Likely to Be Mediated Through microRNAs. DNA Cell Biol. 37(4): p. 373–380. (2018) [DOI] [PubMed] [Google Scholar]
- 29.Johansson HJ, Socciarelli F, Vacanti NM, Haugen MH, Zhu Y, Siavelis I, et al. , Breast cancer quantitative proteome and proteogenomic landscape. Nat Commun. 10(1): p. 1600 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Carey MS, Agarwal R, Gilks B, Swenerton K, Kalloger S, Santos J, et al. , Functional proteomic analysis of advanced serous ovarian cancer using reverse phase protein array: TGF-beta pathway signaling indicates response to primary chemotherapy. Clin Cancer Res. 16(10): p. 2852–60. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Akbani R, Becker KF, Carragher N, Goldstein T, de Koning L, Korf U, et al. , Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report: the RPPA (Reverse Phase Protein Array) society. Mol Cell Proteomics. 13(7): p. 1625–43. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Ling S, Hegde AM, Byers LA, Coombes K, Mills GB, et al. , Using reverse-phase protein arrays as pharmacodynamic assays for functional proteomics, biomarker discovery, and drug development in cancer. Semin Oncol. 43(4): p. 476–83. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrie M, Kim TB, Ju Z, Lee S, Zhao W, Fang Y, et al. , Adaptive responses in a PARP inhibitor window of opportunity trial illustrate limited functional interlesional heterogeneity and potential combination therapy options. Oncotarget. 10(37): p. 3533–3546. (2019)* Describes the adaptive response of ovarian cancer cells to PARP inhibitor.
- 34.Le T, Faught W, Hopkins L and Fung-Kee-Fung M, Importance of CA125 normalization during neoadjuvant chemotherapy followed by planned delayed surgical debulking in patients with epithelial ovarian cancer. J Obstet Gynaecol Can. 30(8): p. 665–670. (2008) [DOI] [PubMed] [Google Scholar]
- 35.Skaznik-Wikiel ME, Sukumvanich P, Beriwal S, Zorn KK, Kelley JL, Richard SD, et al. , Possible use of CA-125 level normalization after the third chemotherapy cycle in deciding on chemotherapy regimen in patients with epithelial ovarian cancer: brief report. Int J Gynecol Cancer. 21(6): p. 1013–7. (2011) [DOI] [PubMed] [Google Scholar]
- 36.Cai RL, Yan-Neale Y, Cueto MA, Xu H and Cohen D, HDAC1, a histone deacetylase, forms a complex with Hus1 and Rad9, two G2/M checkpoint Rad proteins. J Biol Chem. 275(36): p. 27909–16. (2000) [DOI] [PubMed] [Google Scholar]
- 37.Yu KH, Levine DA, Zhang H, Chan DW, Zhang Z and Snyder M, Predicting Ovarian Cancer Patients' Clinical Response to Platinum-Based Chemotherapy by Their Tumor Proteomic Signatures. J Proteome Res. 15(8): p. 2455–65. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peng X, Xu X, Wang Y, Hawke DH, Yu S, Han L, et al. , A-to-I RNA Editing Contributes to Proteomic Diversity in Cancer. Cancer Cell. 33(5): p. 817–828 e7. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gonzalez VD, Samusik N, Chen TJ, Savig ES, Aghaeepour N, Quigley DA, et al. , Commonly Occurring Cell Subsets in High-Grade Serous Ovarian Tumors Identified by Single-Cell Mass Cytometry. Cell Rep. 22(7): p. 1875–1888. (2018)* Single cells proteomics analysis of ovarian cancer tumors.
- 40.McGranahan N and Swanton C, Clonal Heterogeneity and Tumor Evolution: Past, Present, and the Future. Cell. 168(4): p. 613–628. (2017) [DOI] [PubMed] [Google Scholar]
- 41.Hammerlindl H and Schaider H, Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J Cell Commun Signal. 12(1): p. 133–141. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emmons MF, Faiao-Flores F and Smalley KSM, The role of phenotypic plasticity in the escape of cancer cells from targeted therapy. Biochem Pharmacol. 122: p. 1–9. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holohan C, Van Schaeybroeck S, Longley DB and Johnston PG, Cancer drug resistance: an evolving paradigm. Nat Rev Cancer. 13(10): p. 714–26. (2013) [DOI] [PubMed] [Google Scholar]
- 44.Sun C, Fang Y, Yin J, Chen J, Ju Z, Zhang D, et al. , Rational combination therapy with PARP and MEK inhibitors capitalizes on therapeutic liabilities in RAS mutant cancers. Sci Transl Med. 9(392). (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi JI, Park SH, Lee HJ, Lee DW and Lee HN, Inhibition of Phospho-S6 Kinase, a Protein Involved in the Compensatory Adaptive Response, Increases the Efficacy of Paclitaxel in Reducing the Viability of Matrix-Attached Ovarian Cancer Cells. PLoS One. 11(5): p. e0155052 (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang P, Madden JA, Neums L, Moulder RK, Forrest ML and Chien J, Olaparib-induced Adaptive Response Is Disrupted by FOXM1 Targeting that Enhances Sensitivity to PARP Inhibition. Mol Cancer Res. 16(6): p. 961–973. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fang Y, McGrail DJ, Sun C, Labrie M, Chen X, Zhang D, et al. , Sequential Therapy with PARP and WEE1 Inhibitors Minimizes Toxicity while Maintaining Efficacy. Cancer Cell. 35(6): p. 851–867 e7. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hew KE, Miller PC, El-Ashry D, Sun J, Besser AH, Ince TA, et al. , MAPK Activation Predicts Poor Outcome and the MEK Inhibitor, Selumetinib, Reverses Antiestrogen Resistance in ER-Positive High-Grade Serous Ovarian Cancer. Clin Cancer Res. 22(4): p. 935–47. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iavarone C, Zervantonakis IK, Selfors LM, Palakurthi S, Liu JF, Drapkin R, et al. , Combined MEK and BCL-2/XL Inhibition Is Effective in High-Grade Serous Ovarian Cancer Patient-Derived Xenograft Models and BIM Levels Are Predictive of Responsiveness. Mol Cancer Ther. 18(3): p. 642–655. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Konstantinopoulos PA, Barry WT, Birrer M, Westin SN, Cadoo KA, Shapiro GI, et al. , Olaparib and alpha-specific PI3K inhibitor alpelisib for patients with epithelial ovarian cancer: a dose-escalation and dose-expansion phase 1b trial. Lancet Oncol. 20(4): p. 570–580. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu JF, Tolaney SM, Birrer M, Fleming GF, Buss MK, Dahlberg SE, et al. , A Phase 1 trial of the poly(ADP-ribose) polymerase inhibitor olaparib (AZD2281) in combination with the anti-angiogenic cediranib (AZD2171) in recurrent epithelial ovarian or triple-negative breast cancer. Eur J Cancer. 49(14): p. 2972–8. (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matulonis UA, Wulf GM, Barry WT, Birrer M, Westin SN, Farooq S, et al. , Phase I dose escalation study of the PI3kinase pathway inhibitor BKM120 and the oral poly (ADP ribose) polymerase (PARP) inhibitor olaparib for the treatment of high-grade serous ovarian and breast cancer. Ann Oncol. 28(3): p. 512–518. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun C, Yin J, Fang Y, Chen J, Jeong KJ, Chen X, et al. , BRD4 Inhibition Is Synthetic Lethal with PARP Inhibitors through the Induction of Homologous Recombination Deficiency. Cancer Cell. 33(3): p. 401–416 e8. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stewart C, Ralyea C and Lockwood S, Ovarian Cancer: An Integrated Review. Semin Oncol Nurs. 35(2): p. 151–156. (2019) [DOI] [PubMed] [Google Scholar]
- 55.Lee HH, Bellat V and Law B, Chemotherapy induces adaptive drug resistance and metastatic potentials via phenotypic CXCR4-expressing cell state transition in ovarian cancer. PLoS One. 12(2): p. e0171044 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shen L, Sun B, Sheng J, Yu S, Li Y, Xu H, et al. , PGC1alpha promotes cisplatin resistance in human ovarian carcinoma cells through upregulation of mitochondrial biogenesis. Int J Oncol. 53(1): p. 404–416. (2018) [DOI] [PubMed] [Google Scholar]
- 57.Amoroso MR, Matassa DS, Agliarulo I, Avolio R, Maddalena F, Condelli V, et al. , Stress-Adaptive Response in Ovarian Cancer Drug Resistance: Role of TRAP1 in Oxidative Metabolism-Driven Inflammation. Adv Protein Chem Struct Biol. 108: p. 163–198. (2017) [DOI] [PubMed] [Google Scholar]
- 58.Reinthaller A, Antiangiogenic therapies in ovarian cancer. Memo. 9(3): p. 139–143. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Previs RA, Armaiz-Pena GN, Ivan C, Dalton HJ, Rupaimoole R, Hansen JM, et al. , Role of YAP1 as a Marker of Sensitivity to Dual AKT and P70S6K Inhibition in Ovarian and Uterine Malignancies. J Natl Cancer Inst. 109(7). (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang X, Shen F, Hu W, Coleman RL and Sood AK, New ways to successfully target tumor vasculature in ovarian cancer. Curr Opin Obstet Gynecol. 27(1): p. 58–65. (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lyons YA, Pradeep S, Wu SY, Haemmerle M, Hansen JM, Wagner MJ, et al. , Macrophage depletion through colony stimulating factor 1 receptor pathway blockade overcomes adaptive resistance to anti-VEGF therapy. Oncotarget. 8(57): p. 96496–96505. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurimchak AM, Shelton C, Duncan KE, Johnson KJ, Brown J, O'Brien S, et al. , Resistance to BET Bromodomain Inhibitors Is Mediated by Kinome Reprogramming in Ovarian Cancer. Cell Rep. 16(5): p. 1273–1286. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ohmoto A and Yachida S, Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther. 10: p. 5195–5208. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lord CJ and Ashworth A, PARP inhibitors: Synthetic lethality in the clinic. Science. 355(6330): p. 1152–1158. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilie PG, Tang C, Mills GB and Yap TA, State-of-the-art strategies for targeting the DNA damage response in cancer. Nat Rev Clin Oncol. 16(2): p. 81–104. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vanderstichele A, Busschaert P, Olbrecht S, Lambrechts D and Vergote I, Genomic signatures as predictive biomarkers of homologous recombination deficiency in ovarian cancer. Eur J Cancer. 86: p. 5–14. (2017) [DOI] [PubMed] [Google Scholar]
- 67.Murai J, Huang SY, Das BB, Renaud A, Zhang Y, Doroshow JH, et al. , Trapping of PARP1 and PARP2 by Clinical PARP Inhibitors. Cancer Res. 72(21): p. 5588–99. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shen Y, Aoyagi-Scharber M and Wang B, Trapping Poly(ADP-Ribose) Polymerase. J Pharmacol Exp Ther. 353(3): p. 446–57. (2015) [DOI] [PubMed] [Google Scholar]
- 69.Murai J, Huang SY, Renaud A, Zhang Y, Ji J, Takeda S, et al. , Stereospecific PARP trapping by BMN 673 and comparison with olaparib and rucaparib. Mol Cancer Ther. 13(2): p. 433–43. (2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ibrahim YH, Garcia-Garcia C, Serra V, He L, Torres-Lockhart K, Prat A, et al. , PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2(11): p. 1036–47. (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vena F, Jia R, Esfandiari A, Garcia-Gomez JJ, Rodriguez-Justo M, Ma J, et al. , MEK inhibition leads to BRCA2 downregulation and sensitization to DNA damaging agents in pancreas and ovarian cancer models. Oncotarget. 9(14): p. 11592–11603. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen M-J, Li J, Akbani R, Wang Y, Lu Y, Mills GB, et al. , TCPA v6.0: An Integrative Platform for Pan-cancer Analysis of Functional Proteomic Data Mol Cell Proteomics In press. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen MM, Li J, Wang Y, Akbani R, Lu Y, Mills GB, et al. , TCPA v3.0: An Integrative Platform to Explore the Pan-cancer Analysis of Functional Proteomic Data. Mol Cell Proteomics. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grote T, Siwak DR, Fritsche HA, Joy C, Mills GB, Simeone D, et al. , Validation of reverse phase protein array for practical screening of potential biomarkers in serum and plasma: accurate detection of CA19-9 levels in pancreatic cancer. Proteomics. 8(15): p. 3051–60. (2008) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee J, Geiss GK, Demirkan G, Vellano CP, Filanoski B, Lu Y, et al. , Implementation of a Multiplex and Quantitative Proteomics Platform for Assessing Protein Lysates Using DNA-Barcoded Antibodies. Mol Cell Proteomics. 17(6): p. 1245–1258. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li J, Lu Y, Akbani R, Ju Z, Roebuck PL, Liu W, et al. , TCPA: a resource for cancer functional proteomics data. Nat Methods. 10(11): p. 1046–7. (2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cohen AA, Geva-Zatorsky N, Eden E, Frenkel-Morgenstern M, Issaeva I, Sigal A, et al. , Dynamic proteomics of individual cancer cells in response to a drug. Science. 322(5907): p. 1511–6. (2008) [DOI] [PubMed] [Google Scholar]
- 78.Kalluri R and Weinberg RA, The basics of epithelial-mesenchymal transition. J Clin Invest. 119(6): p. 1420–8. (2009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gascoigne KE and Taylor SS, Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell. 14(2): p. 111–22. (2008) [DOI] [PubMed] [Google Scholar]
- 80.Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, et al. , A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 141(1): p. 69–80. (2010) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin JR, Fallahi-Sichani M, Chen JY and Sorger PK, Cyclic Immunofluorescence (CycIF), A Highly Multiplexed Method for Single-cell Imaging. Curr Protoc Chem Biol. 8(4): p. 251–264. (2016) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tsujikawa T, Kumar S, Borkar RN, Azimi V, Thibault G, Chang YH, et al. , Quantitative Multiplex Immunohistochemistry Reveals Myeloid-Inflamed Tumor-Immune Complexity Associated with Poor Prognosis. Cell Rep. 19(1): p. 203–217. (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Keren L, Bosse M, Marquez D, Angoshtari R, Jain S, Varma S, et al. , A Structured Tumor-Immune Microenvironment in Triple Negative Breast Cancer Revealed by Multiplexed Ion Beam Imaging. Cell. 174(6): p. 1373–1387 e19. (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lin JR, Fallahi-Sichani M and Sorger PK, Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method. Nat Commun. 6: p. 8390 (2015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eng JG, Luoh S, Gray J, Chang Y and Chin K, Cyclic Multiplexed-Immunofluorescence (cmIF), a Highly Multiplexed Method for Single-Cell Analysis, in Biomarkers for Immunotherapy of Cancer. Methods Molecular Biology. 2055 (2019) [DOI] [PubMed] [Google Scholar]
- 86.Faria SS, Morris CF, Silva AR, Fonseca MP, Forget P, Castro MS, et al. , A Timely Shift from Shotgun to Targeted Proteomics and How It Can Be Groundbreaking for Cancer Research. Front Oncol. 7: p. 13 (2017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang B, Whiteaker JR, Hoofnagle AN, Baird GS, Rodland KD and Paulovich AG, Clinical potential of mass spectrometry-based proteogenomics. Nat Rev Clin Oncol. 16(4): p. 256–268. (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chang CY, Picotti P, Huttenhain R, Heinzelmann-Schwarz V, Jovanovic M, Aebersold R, et al. , Protein significance analysis in selected reaction monitoring (SRM) measurements. Mol Cell Proteomics. 11(4): p. M111 014662 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gillet LC, Navarro P, Tate S, Rost H, Selevsek N, Reiter L, et al. , Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 11(6): p. O111 016717 (2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC and Aebersold R, Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol Syst Biol. 14(8): p. e8126 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]