Abstract
In the past decade, more cancer researchers have begun to understand the significance of cancer prevention, which has prompted a shift in the increasing body of scientific literature. An area of fascination and great potential is the human microbiome. Recent studies suggest that the gut microbiota has significant roles in an individual’s ability to avoid cancer, with considerable focus on the gut microbiome and colorectal cancer. That in mind, racial disparities with regard to colorectal cancer treatment and prevention are generally understudied despite higher incidence and mortality rates among Non-Hispanic Blacks compared to other racial and ethnic groups in the United States. A comprehension of ethnic differences with relation to colorectal cancer, dietary habits and the microbiome is a meritorious area of investigation. This review highlights literature that identifies and bridges the gap in understanding the role of the human microbiome in racial disparities across colorectal cancer. Herein, we explore the differences in the gut microbiota, common short chain fatty acids produced in abundance by microbes, and their association with racial differences in cancer acquisition.
Keywords: Diet, Epigenetics, Microbiome, Disparities, Colorectal, Colon, Cancer
Core tip: In this paper, we summarize the literature in relation to the gut microbiome and colorectal cancer. We provide unique perspectives and identify new areas of interest that will progress the field with relation to colorectal cancer disparities. This is significant because the comprehension of the microbiome is quickly becoming paramount for personalized medicine and combating disease progression.
INTRODUCTION
It is safe to classify cancer as the disease of the 21st century. Ongoing investigations within the scientific community seek to identify novel approaches to remedy this debilitating disease, and much progress has been made in recent decades. From the discovery of immunologic drugs, to technological advancements in laser therapy, the wide array of research being done to resolve the ever-complicated ailment known as cancer continues to grow as researchers set out to find an effective treatment or cure[1-6]. Both cancer and chemo prevention are areas that have grown considerably as of late[7-10].The interest in these fields continues to thrive partially because of the high cost of cancer remediation in addition to the numerous side effects associated with chemotherapy drugs on the market[11-13]. In an effort to promote and conserve quality of life, in particular for African American and low income individuals, efforts to alter one’s environment and increase the avoidance of carcinogenesis is a much needed area of investigation.
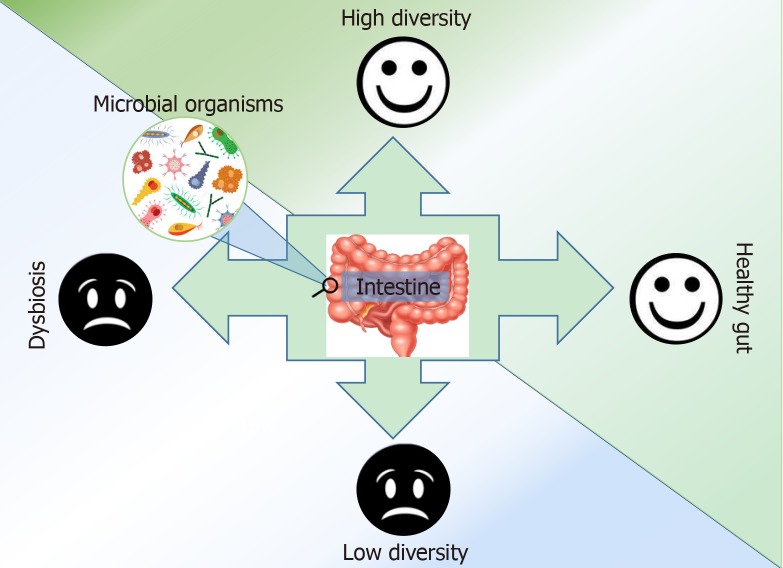
The microbiome is important in human health and disease, and while the microbiome is extremely complex, can change over time, and varies from person to person, we can still gain pertinent information that will help treat and cure disease. Researchers have undertaken the task of understanding the roles of the microbiome. It is common knowledge that a vast majority of commensal microbes have symbiotic relationships with their host organisms; we now seek to understand those relationships and the information we can gain about disease. Several studies have shown variance in microbial abundance and diversity among healthy and non-healthy individuals[10,14] (Figure 1). According to recent investigations, there are differences and trends of association in the gut microbiota based on race and ethnicity, sex, geography, diet, and other such demographics[15-17]. As a result of a study conducted by Chen et al[18], we now know that there are over 400 genes that can be used to distinguish the specific microbiomes of Asian, European, and American populations. Interestingly, Chen et al[19] at the Mayo Clinic in Rochester Minnesota report significant associations in microbial β-diversity according to body mass index, race, gender, and alcohol consumption. This study provides supporting evidence for the influence of environmental factors that affect the microbiome.
Figure 1.
Gut health and the influence of gut microbes: Microbial diversity is associated with better health outcomes. A greater abundance of varied microbial species can be influenced by environmental and lifestyle factors. This diversity can be associated with better health outcomes. The above graphic summarizes the relationship between microbes human health.
With respect to cancer and the roles of the microbes indigenous to the human gut, the need for further investigation and application is apparent. While we know there is an association with the microbiota and human health, we do not fully understand the significance of that association. It is believed that microbial transplantation could have some benefit in cancer prevention and or remediation[20,21]. If this is the case, trials need to be launched to assess this. We know that the consumption of certain foods can result in epigenetic modulation of oncogenes and tumor suppressors[22-25]. We also understand that metabolic processes are instrumental in the utilization of foods for nutrient absorption and energy production[26,27]. The question that arises from acknowledging the role microbes have in human health and disease becomes one of understanding how much of the metabolizing of nutrients is dependent on commensal microbes, and how these microbes drive epigenetic processes in the human organism. To begin unraveling these mysteries, we must first have a clear understanding of the typical microbial profile of a diseased and healthy individual. We (the scientific community) must also take on the daunting task of determining the functionality of the by-products of the microbes identified through our efforts.
Understanding the complexities of the microbiome gives rise to a better understanding of the complexities associated with the individuality of disease development and progression among different races and ethnicities. Diseases like cancer are difficult to appropriately address in part due to differences in chemotherapy effectiveness and adherence. Non-Hispanic Whites (NHW) tend to have overall better health outcomes when compared to Non-Hispanic Blacks (NHB) and other racial minorities in the United States[28-30]. A full comprehension of the differences among these groups at the microbiome level in concert with epigenetic and genetic differences will help decrease the health disparity in these populations.
RACE AND COLORECTAL CANCER IN THE UNITED STATES
Using incidence data from the Surveillance, Epidemiology and End Results Program, cancer registries and National Center for Health Statistics, incidence rate of colorectal cancer (CRC) was 49.2 per 100000 in NHB, higher than in NHW (40.2 per 100000) and Asian/Pacific Islander (API, 32.2 per 100000). Similarly, mortality rates from the disease using data from 2010 to 2014 were higher among NHB (20.5 per 100000) compared to NHW (14.6 per 100000) and APIs (10.3 per 100000). Among adults younger than 55 years, though mortality rates declined among blacks in the period 1970 (8.1 per 100000) to 2014 (to 6.1 per 100000), the rates were higher than for whites (3.6/100000 in 1970 to 4.1/100000 in 2014)[31]. Several factors could explain higher CRC rates in NHB. Blacks are usually diagnosed at more advanced stages of the disease and hence have higher mortality rates[32]. Socioeconomic disparities account for a significant proportion of the difference in CRC incidence between racial/ethnic groups, and these socioeconomic differences have been attributed to the higher prevalence of obesity, unhealthy diet and smoking[11,33]. Mortality differentials between racial groups can also be explained by inequalities in screening rates and access[34], and healthcare access and follow up care following abnormal findings on screening[35-37].
RACIAL DIFFERENCES IN DIET IN THE UNITED STATES
Several studies have reported a higher prevalence of unhealthy food intake among NHB compared to other racial groups. Dunford et al[38] examined trends in energy intake among United States adults aged 19 years and above from 1977 to 2012 using multiple national survey data. They found that NHB had a higher prevalence of snacking energy intake and salty snacks in the period reviewed. Using the Healthy Eating Index (HEI), Nowlin et al[39] analyzed NHANES data of United States adults aged 20 years and older between 2007 and 2012 and showed that among non-diabetics, NHB had significantly lower HEI scores compared to NHW. The authors also reported that the racial disparities were related to age, gender smoking status and time spent in the United States.
Another approach to assessing dietary quality was employed by Rehm et al[40]. The authors computed a composite variable to assess overall diet quality using 7 NHANES cycle from 1999 to 2012. They found that the estimated percentage of non-Hispanic white adults with a poor diet significantly declined (53.9% to 42.8%), while the corresponding figures were 64.7% to 57.7% among NHB and 66% to 58.9% among Mexican American adults. Furthermore, for specific food items, total vegetables, whole grains, unprocessed red meat, and milk consumption remained higher over time among non-Hispanic white adults compared to non-Hispanic black or Mexican American adults. Another noteworthy study conducted by Wang et al[41] used a composite index of dietary intake. The authors investigated trends in dietary quality among adults from 1999 to 2010 using the NHANES dataset and found only minimal improvements in dietary quality over time among NHB. Additionally, in each survey cycle, they found a significantly lower mean Alternate Healthy Eating Index-2010 among NHB compared to Mexican Americans and NHW, though the difference between NHW and NHB disappeared after adjusting for socioeconomic factors such as income and education. The authors suggested that the observed differences between NHW and NHB were likely explained by socioeconomic variables. Among children and adolescents, using the NHANES data over a twenty-year period (1988 to 2008), Kant et al[42] found a greater increase in the number of eating occasions, the amount of energy from all beverages and non-nutritive beverages in NHB compared to other racial groups. Taken together, these studies indicate the poor dietary quality of NHB compared to other racial groups, and that could explain part of the racial disparities in different cancers, especially those cancers where type of diet is an established risk factor.
THE INTERPLAY BETWEEN DIET, THE MICROBIOME, AND CANCER
Cancer is an extremely complex and life altering disease which brings to rise the urgency to unravel its complexities. Focus areas in the cancer research community encompass a broad range of topics, and there has been a mass effort to minimize cancer’s impact on quality of life and longevity. There are many approaches that can be taken; however, our interests lie in chemo adherence and prevention through modifiable factors. Knowledge about the disease and its causation associated with environmental exposures will help promote awareness and aversion to certain lifestyles that lead to mutations or epigenetic aberrancies, and cancer development. Since epigenetics is a field that focuses on changes in gene expression with no change in the underlying coding sequence[24], the regulation of epigenetic abnormalities is thought to have strong potential in the reversing cancer. One such Food and Drug Administration approved epigenetic therapy currently on the market treats cutaneous T cell lymphoma. Suberanilohydroxamic acid (SAHA), known commercially as Vorinostat, is a well-documented histone deacetylase (HDAC) inhibitor[43].
Researchers have suspected that lifestyle factors contribute to human health and disease for quite some time now. It was in 1964 that the surgeon general, Luther Terry and team, published the report that smoking had a direct impact on cancer development[44]. While this spurred changes in the regulations on tobacco, we believe that this report, and reports like it, placed a higher importance on environmental factors that lead to cancer. Takeshi Hirayama published one of the earlier documents on the impact of nutrition on cancer in 1979. Hirayama placed emphasis on the idea that lifestyle improvement (better diet, more exercise and smoking cessation) could be an effective community cancer plan[45]. Since then, many developments have been made with respect to cancer development and the environment over the decades. Fast-forwarding to 2003, Margaret Mason published a review paper that focused on the idea that diet could potentially modulate molecular signaling in cancer[8]; and to date, several studies have provided further evidence of this[7,9,23,24,46].
We now know with certainty that the foods we consume can have a direct impact on the state of human health and disease[39,47,48]. Several studies have begun to explore how these foods modulate gene expression of oncogenes and tumor suppressors[7,24,25,43]. Two recent papers detail how certain natural and dietary compounds promote cell death in breast cancer cell lines[7,25]. They demonstrate that sulforaphane, an isothiocyanate abundantly in cruciferous vegetables, and withaferin A a steroidal lactone from the Indian winter cherry, are efficient at downregulating epigenetic enzymes that are typically over expressed in breast cancer[25]. Further, this group also found that the prior-mentioned compounds increase the expression of tumor suppressor gene p21 through transcriptional activation at the promoter region through the up-regulation of a histone methylation marker associated with gene activation[7]. With respect to the epigenetic impact of nutritive compounds, other studies have found metabolic links among microbial organisms in the gut and choline consumption. Choline is a vital nutrient that acts as a source for the methyl groups needed for metabolic processes in humans. Apparently, choline utilizing bacteria can compete with the host for this nutrient, thereby impacting global DNA methylation patterns in mice[27]. In 2010, Moestue et al[49] noted that in breast cancer models, there were elevated levels of choline metabolites, and more recently it was found that while elevated choline metabolites were present in resistant breast cancers, chemotherapy induced an even greater increase in these metabolites[50]. Further study may reveal that choline-consuming bacteria like, Escherichia coli 536, are capable of altering epigenetic aberrancies (i.e., hyper/hypo-methylation and tumor-suppressor silencing and oncogene activation respectively) that lead to cancer.
Interestingly, other studies have also reported anticancer effects at the gene regulatory level with respect to immune responsiveness. Recently, Rubio-Patino et al[51] published a study on the effects of a low-protein diet on inositol requiring enzyme 1α (IRE1α)-dependent anticancer immunosurvellience. IRE1α is partially responsible for the activation of the endoribonuclease domain that catalyzes splicing of X-box binding protein 1. In addition to this function, IRE1α RNAse activity has roles in IRE1-dependent decay of mRNA, rRNA, and microRNAs which in turn can lead to modulation of adaptive immunity[52]. Mediterranean, vegetable based, Japanese diets, and others all show promise at decreasing cancer risk and mortality[46] . Soldati et al[46] detail in their 2018 paper, that diet affects cancer progression through either systemic or local effects within the tumor microenvironment. Higher glucose levels are known to have an impact on immune activity, which in turn destabilized immune functionality. That being said, regulatory T cell function can be regulated by the metabolism, and the metabolism is regulated by patterns of dietary intake and physical activity[53].
The gut microbiome also has key roles in cancer risk reduction. In this modern era of research and medicine, the comprehension of the microbiome has expanded considerably. Causal mechanisms for cancer have been identified in microbes[54]. Some of these mechanisms are immunologic, and others appear to be epigenetic[55,56]. Iida et al[57] published one such immunologic study that details the influence of the microbiome in disease progression in 2013. They found that antibiotics actually decreased the efficacy of the tested immunotherapy leading to the conclusion that commensal bacteria have an effect on chemo adherence by modulating immunologic factors in the tumor microenvironment. More recently, studies have indicated that immune checkpoint inhibitors are regulated by gut microbes[58], with crosstalk occurring between intestinal epithelial and lymphatic cells. Interestingly, regulatory T cells appear to be inducible by intestinal microbial organisms of the clostridium genus which may suggest a therapeutic approach to immune response[59].
Maryann Kwa and co-authors published an article a couple years ago that suggests the bacterial species housed in the human intestine regulate estrogen metabolism. Interestingly, gut microbiota may affect the risk of acquiring estrogen-receptor-positive breast cancer due to certain microbes being capable of metabolizing estrogens in what they coin as the estrobolome[60]. In addition to this unique perspective, other leading minds have discovered that the bacterial organism Clostridium perfringens enterotoxin has the ability to suppress claudin-4 (a regulator of proliferation and cancer cell metastasis) protein expression, kill gastric cancer cells in-vitro, and inhibit tumor growth in mice xenografts[61]. Other studies have suggested that microbes can be used to biosynthesize nanoparticles designed to target and treat various forms of cancer[62]. Considering the broader implications of microbial composition and its utilization in cancer elimination, the field is subject to grow considerably.
Routine exercise and proper nutrition have proven to be effective in reducing the risk of carcinogenic related fatality in cancer patients. Individuals who consume more plant matter have overall better health outcomes according to a number of epidemiological studies. Clear patterns in dietary intake and cancer risk can be observed among different cultures, races, and ethnicities, which have led to the generation of hypotheses centered on the idea that racial and cultural differences are involved in dietary habits. Researchers have found that there are several variables apart from genetic factors that have been associated with increased cancer mortality in minority populations, including socioeconomic status, and the availability of healthy food options. That being said, it is important to recognize the significance these two modifiable factors have in cancer prevention in relation to the microbiome. Monda et al[26] state that exercise is capable of enhancing the amount of positive microbial organisms resulting in a richer diversity. According to another report, harmful changes with associated polychlorinated biphenyls, pollutants in air and water, are decreased in the gut microbiome of exercising mice[63]. This provides a bit of insight on lifestyle and environmental factors that regulate the abundance or lack of commensal bacteria in host organisms and how they impact health. It would be interesting to conduct an in depth study to determine if microbial diversity is regulated by diet and if microbial abundances attributable to certain diets are in turn responsible for regulation of carcinogenic processes. One study discusses fermentable carbs, or non-digestible carbohydrates that undergo fermentation by microbial organisms to produce short chain fatty acids (SCFAs) that can be utilized by the host[64]. It would be meritorious to observe the impact of SCFAs produced in this manner in cell metabolism. We may yet find that fermentable carbohydrates are capable of reducing the persistent activation of aerobic glycolysis in cancer.
CANCER RISK AND PREVENTION AMONG RACES AND THE INFLUENCE OF THE MICROBIOME
Several studies have indicated that microbial composition is strongly linked to dietary intake and differences among racial and ethnic groups unassociated with diet. We know that there are a number of events responsible for the progression of disease. As research progresses, the scientific community has found there to be trends and correlations with microbial composition, regulation of inflammatory processes, and immune response with microbes playing a significant role; at the very least serving as bio-indicators of some sort. It remains unknown as to whether or not these organisms and their by-products aid in the direct regulation of epigenetic mechanisms that help regulate the human organism’s biological processes; however, it is plausible. With respect to African American susceptibility to cancer and a greater overall disease mortality, we may find a difference in the abundance of microbes in NHB when compared to racial groups with better disease outcomes. Pilot studies conducted by researchers confirm differences in microbial diversity in healthy individuals by race[18,65]. Alternatively, we may find that gut equilibrium conducive to individual survival differs from person to person.
Findley et al[14] published a comprehensive review in 2016 that noted the importance of not assuming race and ethnicity to be causal factors in microbial diversity. We recognize that there are numerous cultural differences that could potentially lead to the differences reported in the studies conducted to date. It remains that NHB and persons of low SES have a number of stressors and lifestyle factors that could directly influence the microbiome[66]. Despite these cautions (avoiding assumptions), one 2015 study shows, that Western diet consumed by individuals of African lineage can be damaging and creates an environment less capable of seamlessly carrying out biological processes (i.e., cellular division and differentiation)[67]. With respect to choline, introduced in the previous section, O’Keefe et al[67] found that Africans on their original diet had a lower level of choline in their fecal water as compared to NHB on the Western diet suggesting the greater microbial diversity reported in the African’s profile results in a greater metabolism of choline. In addition, epigenetic control of the expression or lack of expression of certain genes that are vital in the regulation of tumor suppression and oncogenesis is thought to be linked the gut flora. Histone modifying enzymes are sensitive to microbial metabolites, and they have been shown to mediate phenotype through programing gene expression thereby regulating cellular functionality[68]. This leads to the belief that NHB cancer susceptibility can somehow be linked to gut microbial composition, their metabolites, and their influence on the epigenetic regulation of processes that control oncogenes and tumor suppressors. Another paper that begins to establish the link between health disparities and the gut microbiome details key differences in the abundance or lack of certain SCFAs in the stool of African Americans, Caucasians and Asians[69].
We recognize the weakness in some of these studies stems from the fact that these data are self-reported questionnaires, which limit the accuracy of the similarities/ differences of diet and environment. However, it is intriguing that preliminary results show significant differences in NHB SCFA production, fecal pH, and microbial patterns compared with other races[69]. It can also be noted that there is a clear difference in the microbial profiles of healthy and non-healthy individuals[70]. Interestingly, patients appear to be more susceptible to mortality from a disease when they have lower microbial diversity[71]. Since efficiency of certain immunotherapies for cancer treatment is thought to be influenced by the gut flora[56], it is becoming increasingly apparent how significant the interaction between the microbiome and cancer prevention and therapy is.
SHORT CHAIN FATTY ACIDS
There are a number by-products and metabolites resultant from the consumption of food products. The abundance and variety of these by-products, much like the abundance and variety of certain microbes, is determined by environmental and lifestyle factors (i.e., diet and exercise). The main products absorbed as a result of digestion are long chain fatty acids, simple sugars like glucose, and monoacylglycerol, a class of glycerides[72]. In addition to these products are SCFAs, which are major nutrients resultant of bacterial fermentation. We briefly touched base on these fatty acids in the previous sections. The most studied SCFAs that have shown anti-cancer potential are acetate (much of which may be primarily derived from diet), butyrate and propionate, with acetate being the most abundant SCFA in the colon and propionate and butyrate being found at very low amounts in diseased individuals[73]. Interestingly, Krautkramer et al[74] conducted a study that demonstrated how the Western diet in mice limits SCFA production of microbes and negatively impacts the host’s chromatin. They further display SCFA supplementation to be sufficient in mimicking epigenetic phenotypes resultant of gut microbiota, thereby demonstrating the importance of these microbial metabolites in host homeostasis[74]. Here on we will discuss the significance of these metabolites in their regulation of common cancers.
Acetate
Acetate is a SCFA found in abundance in the human colon. Over the years, this fatty acid has been reported to impact tumor cells as nutrient[75,76] and as an inducer of apoptosis[77-81]. Despite the contradictory reports, there is considerable evidence supporting acetate’s importance in human physiological processes. Acetate is mandatory for histone acetylation[82,83], an epigenetic mechanism known for its roles regulating chromatin accessibility. Most notably, acetate is partially responsible for the appropriate binding of transcription factors for the activation of tumor suppressor genes. Conversely, inappropriate histone acetylation can allow for the activation of oncogenes. This subsection will delve into the roles of acetate in common cancers that affect the United Sates.
Colon cancer/CRC is one of the Western world’s most common cancers in men and women with combined total of approximately 140250 new cases per year according to the American Cancer Society. There has been a considerable amount of research conducted over the years suggesting the significant roles of the microbiome in this particular cancer. Since acetate has been reported to be found in abundance in the colon and to be used in cancer cell metabolism, the regulation of this particular SCFA is of extreme intrigue within the scientific community. A number of studies have sought to determine the effect of acetate on CRC cells in-vitro. One in particular shows that this SCFA inhibits cell growth, decreases cell viability and induces apoptosis in two CRC cell lines[80]. Alternatively, in brain cancer, acetate competes with glucose for the generation of TCA cycle intermediates despite glucose being more abundant[84,85]. An interesting review from 2014 highlights the importance of acetate in fueling cancer cells[86]. Interestingly, this does not appear to be the case in CRC[77,80,87]. One paper that attempts to unravel the role of acetate in CRC cell death demonstrates the ability for this SCFA to increase the expression of MCT1 and MCT4, two plasma membrane lactate transporters and CD147, a multipurpose transmembrane protein associated with inflammation and tumor invasion[87]. The comprehension of the mechanisms involved in acetate’s ability to influence CRC shows much therapeutic potential and promise for further scientific investigation.
Butyrate
A highly sighted review article published in 2011 by Berni Canani et al[22] highlights the numerous roles of butyrate (microbiome dependent) in modulating human health and disease. They detail how this particular SCFA has “potent regulatory effects on gene expression”. Butyrate has been shown to regulate inflammation by inhibiting NFkB[88], it can improve memory function in Alzheimer’s disease mouse models[89], and it has been shown to negatively impact cancer cell progression across a number of tumor types[23,90-93]. Butyrate also has significant roles in providing energy for colonocytes so that they may multiply and divide appropriately and avoid self-digestion. Interestingly, it accounts for approximately 70% of total colon cell energy utilization[94], but only comprises about 15% of the SCFA found in the colonic lumen[22]. There is considerable evidence in support of higher levels of butyrate being associated with better health outcomes, lower pH, and diets rich in fiber. Though butyrate has been shown to promote the growth and proliferation of colon cells, it is capable of inducing apoptosis in mutated cancer cells[91,92,95]. This SCFA has been studied quite a bit as an anticancer agent. A quick search of butyrate and cancer on PubMed will produce thousands of results with relevant material.
Several studies have shown butyrate to be an anti-cancerous agent resultant of bacterial fermentation of non-digestible carbohydrates[96]. Microbial organisms take up fibers that the host organism cannot digest in the colon and produce butyrate as one of three primary SCFAs. In 2017, Li et al[97] published an interesting study on butyrate in CRC cell lines. They report that this SCFA is capable of decreasing cancer cell migratory ability by inhibiting Akt/ERK signaling with dependency on HDAC3[97]. It is our opinion that the study could have been strengthened had the investigators chose to observe the effects of butyrate on HDAC3 gene and protein expression. There is quite a bit of evidence in support of butyrate’s ability to inhibit HDAC activity across a number of cancer types[98-100]. A team recently published an article demonstrating butyrate’s ability to act synergistically with the toxic chemo drug irinotecan[90].
The mechanisms by which butyrate has been demonstrated to promote CRC cell senescence are varied. A number of reports reveal a complex network of oncogenes and tumor suppressors that have been modulated by this fatty acid. One study found that neuropilin-1, a receptor for vascular endothelial growth factor and a key player in apoptotic signaling, is down regulated by butyrate[101]. Other studies show butyrate to regulate CRC cell apoptosis through its impact on the pro-apoptotic pathway by up-regulating BAK and activating caspase-3[92]. There have been a number of studies that attribute butyrate’s apoptotic ability to its regulation of epigenetic genes and microRNAs. In 2015, Hu et al[102] found that miRNA-92a, known for its role in promoting cancer cell proliferation, was inhibited by butyrate. It can be inferred that the inhibition of this oncogenic miRNA is in part due to butyrate’s HDAC inhibitory abilities as they show similar results when comparing to the HDACi SAHA[102]. The fact that a naturally occurring compound can have such a beneficial impact is a significant discovery. Findings like these help usher in new discoveries that help improve the quality of life in cancer patients.
Propionate
Propionate is an important SCFA produced from the dicarboxylic acid pathway or the acrylate pathway and a substrate for hepatic gluconeogenesis[103]. It appears to play a key role in cholesterol metabolism principally as an inhibitory agent and potentially has a cholesterol lowering function[47,104]. Propionate is also known for its antifungal and antibacterial effects, anti-inflammatory and anti-carcinogenic properties[105]. Although butyrate has been consistently shown to be the most potent of the SCFA, propionate also demonstrated anticancer activity in many studies[9,106]. In addition, propionate enhances the anti-proliferative function of butyrate due to its inhibition of cell growth[107]. Concerning histone acetylation, Kiefer et al[108] showed that butyrate and propionate, but not acetate significantly enhanced histone acetylation in colon cancer cells after 24 h incubation. Furthermore, the additive effects of butyrate and propionate when combined in a mixture also amplified histone acetylation[108].
There is compelling evidence of the importance of SCFA including propionate in the cancer prevention and the potential of these chemicals in treatment. A study by Emenaker et al[106] treated fresh surgical colon cancer cells with SCFA and found all SCFA inhibited primary invasive human colon cancer invasion and significantly altered protein expression levels of established cancer genes: p53, Bax, Bcl2 and PCNA. Giardina et al[109] using HT-29 CRC cell lines showed that propionate and butyrate, but not acetate, could alter the metabolism of reactive oxygen species and increase cellular peroxide generation. This ability to produce peroxide enables the induction of apoptosis in CRC cell lines.
There is evidence that the anti-carcinogenic effects of propionate could differ from those of other SCFA. For example, Tedelind et al[105] compared the anti-inflammatory properties of butyrate, propionate and acetate and found that propionate and butyrate were equally effective in suppressing NF-KB reporter activity, immune related gene expression and cytokine release in vitro. The authors concluded the SCFA could be useful for treating inflammatory bowel disease. Another study using proteomic and cellomic analysis methods[95] showed that propionate had different mechanisms of action on cellular proteins compared to other SCFA such as butyrate and valerate. Specifically, propionate had less pronounced effects on keratins and intermediate filaments, and on b-tubulin isotypes expression and microtubules compared to butyrate and acetate. Concerning colitis associated CRC, propionate as part of a SCFAs mix prevented development of tumors and attenuated the colonic inflammation in a mouse model of colitis-associated CRC[110], and holds promise in the management of colitis associated CRC.
RACIAL DIFFERENCES IN THE PROFILE OF GUT MICROBIOTA AND SCFA
The bacterial composition of the gut and SCFA content has been shown to differ between racial/ethnic groups. Hester et al[69] compared bacteria and SCFA in the stools of a small pilot sample of 20 apparently healthy participants (five each were NHW, NHB, Hispanics and American Indians). The authors found lower acetate, butyrate and total SCFA content and a higher pH in NHB compared to the other racial groups. In addition, levels of Firmicutes bacteria were higher in NHB compared to Whites and Hispanics, and lower levels of Lachnospiraceae, known to be involved in the production of butyrate. Moreover, the ratio of Firmicutes compared to Bacteriodes, that has been associated with obesity was also higher among blacks and could partly explain the links between obesity and colon cancer.
Two recent studies have reported that Bacteroides were more commonly found among NHB. Farhana et al[15] showed that Bacteroides were more abundant in colonic effluents of AAs compared to NHWs, especially Fusobacterium nucleatuma and Enterobacter species, whereas Akkermansia muciniphilia and Bifidobacterium were higher in NHW. The authors also showed that AA had decreased microbial diversity compared to NHW. Bacteroides were also shown to be more common among AA compared to NHW in a study of stress and microbiota among healthy individuals[66]. Higher stool counts of Bacteroides were also found in NHB in an older study that compared dietary habits and fecal microbiota of NHB and NHW[111]. In addition, NHB reported lower dietary calcium, magnesium and Vitamins A, C, D, and E.
A recent study compared colonic biopsies of healthy mucosa of CRC cases and tumor free controls and found a greater abundance of sulfidogenic bacteria among NHB compared to NHWs among cases and controls[65]. Additionally, Bilophilia wadsworthia (evidenced to be promoted by saturated fats from Western diets low in fiber[112,113]) and Pyramidobacter spp were significantly higher among AA cases compared to controls[65]. The study also showed a higher consumption of meat, fat and protein intake among NHB. David et al[113] also report that diet has the ability to alter the human gut microbiome. With regard to obesity, a known risk factor for various types of cancer, microbiota of the gut were shown to be instrumental in the regulation of diet induced obesity in lymphotoxin deficient mice[112]. This shows that this particular molecule key in gut immunity, is capable of modulating commensal responses enabling diet induced obesity, and we have a better understanding of the pathways involved in microbiota induced weight gain[112]. Comparison of the gut microbiota between populations with wide variations in dietary composition could shed light on the potential mechanisms involved in the role of diet and gut microbiota in carcinogenesis. A small study by Ou et al[114] studied the difference in gut microbiota between native Africans (NA) and NHB and showed that SFCA, total bacteria, microbial genes encoding for methanogenesis and hydrogen sulphide production were higher among NA while secondary bile acid was higher among NHB in fecal samples. The findings point to a higher saccharolytic fermentation and lower proteolytic fermentation among NA compared to NHB. Concerning specific bacteria, Prevotella was more common in NA while Bacteriodes were more abundant in NHB. The authors ascribed the observed differences in stool samples to NHB eating more dietary meat and fat and less complex carbohydrate and fiber. The same authors also showed in another study[115] comparing a small sample of high colon cancer risk in NA, NHB and NHW, that the levels of experimentally carcinogenic secondary bile acids (lithocholic and deoxycholic acids) was higher among NHB and NHW compared to NA. A related study[116] also showed that NA harbored a more diverse population of, methanogenic Archaea compared to NHB and NHW. The populations of other hydrogenotropic bacteria such as sulphate reducing bacteria was also more distinct among NA. Another study involving NA[117] examined total colonic evacuants for SCFA, vitamins, nitrogen and minerals and found total SCFA and butyrate were higher, but calcium, iron and zinc were lower among NA compared to NHB and NHW, supporting the mediatory role of these chemicals in the effects of microbiota in colon cancer development.
NA have been shown to have a propensity for methanogenic rather than sulfidogenic disposal of hydrogen generated from microbial fermentation in the human colon. In a South African study, NA excreted more methane in their breath compared to whites in the same country[118]. Furthermore, stool samples of European ancestry populations exhibited more of sulfate reduction activity. The differences observed in the bacterial composition of stool samples of NA and NHB is due to the dietary difference with the Africans consuming more coarse grains and vegetables.
Apart from intestinal microbiome, some other studies have examined the role of the oral microbiome in CRC risk using a multi-ethnic sample. For example, Li et al[16] showed unique differences in the patterns of saliva microbiome between individuals living in Alaska, Germany and Africa. In another study, Yang et al[17] using a nested case control study showed that Treponema denticola and Prevotella intermedia were associated with increased CRC risk among a sample made of predominantly EA and AA. Even though this study showed similar associations in AA and EA, they were more significant among AA, although AA were about three times more than EA in the sample. The authors however alluded to the fact that differences in the oral microbiome by race could explain the difference in association between the racial groups.
DISCUSSION AND CONCLUSION
We have reviewed the differences between race/ethnic groups concerning dietary habits, how different diets affect the gut’s environment, and the potential role of these differences in explaining disparities in CRC incidence in the United States and between native African populations. Our review reveals that unique opportunities exist in taking advantage of the racial differences in diet, other diet-related CRC risk factors such as obesity and diabetes, gut microbiome, and the genetic architecture of CRC for a greater understanding of the complexity of CRC etiology and carcinogenesis. Much of the literature we cite includes studies that show that environment, diet, and lifestyle can alter the microbiome and are associated with cancer risk, but whether these co-associations are linked is still unknown.
Several studies revealed that there is a direct link between dietary intake and the microbial profile of an individual, and this ultimately influences the risk of several diseases including cancer. A study we mention earlier in this review corroborates the link between dietary intake and microbiota. The authors switched regular diets between South African blacks and AA during a two-week period. Food exchanges of low fat, high fiber for AA and high fat low fiber for NA resulted in increased saccharolytic fermentation and suppressed secondary bile acid synthesis in AA[67]. Studies also consistently showed that AA consume less healthy diets and this could explain the higher rates of CRC among this population. Foods from an unbalanced diet deficient in fiber and high in meat, such as those consumed by AA, promotes proteolytic rather than saccharolytic fermentation and this results in branched chain SCFAs such as isobutyrate, isovaleric and 2-methylbutyroic acid. Moreover, proteolytic products including nitrogenous metabolites such as hipurate and ammonia have been shown to increase inflammation and carcinogenesis[48].
Evidence from studies demonstrating direct links between dietary habits, the gut microbiome and CRC risk indicates there is an urgent need for multiethnic studies of gut microbiome and SCFA and CRC. Moreover, such studies need to incorporate dietary intake, blood levels of nutrients and their metabolites, genes involved in the metabolism of various diets, and epigenetic factors. In addition, greater insights into colorectal etiology could be gained by exploring the large differences in diet, and several other environmental factors between native African populations and African Americans living in the United States. Recent estimates revealed greater than 10-fold difference in incidence rates of CRC between the United States and most of sub-Saharan Africa[119,120]. Multipronged approaches (such as integrative molecular epidemiology) to study the associations between dietary habits, profile of the gut microbiome and CRC across populations could provide great insights into the factors responsible for the widely varying CRC incidence rates across populations. Finally, most of the studies reviewed had small sample sizes that precluded adequate power to investigate CRC subtypes. Large sample multiethnic cohorts will afford greater understanding of CRC subtypes especially given the clear differences in the etiology, prognosis and approaches to the management of these subtypes.
ACKNOWLEDGEMENTS
We would like to thank Dr. Jack Gilbert for his willingness to converse to discuss the field of microbiology, and Dr. Eugene Chang for his critical review of this manuscript; We would also like to acknowledge the University of Chicago Institute for Translational Medicine TL-1 program.
Footnotes
Conflict-of-interest statement: The authors have no conflicts of interest to declare.
Open-Access: This article is an open-access article which was selected by an in-house editor and fully peer-reviewed by external reviewers. It is distributed in accordance with the Creative Commons Attribution Non Commercial (CC BY-NC 4.0) license, which permits others to distribute, remix, adapt, build upon this work non-commercially, and license their derivative works on different terms, provided the original work is properly cited and the use is non-commercial. See: http://creativecommons.org/licenses/by-nc/4.0/
Manuscript source: Unsolicited manuscript
Peer-review started: March 14, 2019
First decision: June 3, 2019
Article in press: July 26, 2019
Specialty type: Oncology
Country of origin: United States
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cao ZF, Jeong KY S-Editor: Yan JP L-Editor: A E-Editor: Wu YXJ
Contributor Information
Kendra J Royston, Division of Hematology Oncology, University of Chicago, Chicago, IL 60637, United States. kendraroyston@gmail.com.
Babatunde Adedokun, Center for Clinical Cancer Genetics and Global Health Department of Medicine, University of Chicago, Chicago, IL 60637, United States.
Olufunmilayo I Olopade, Division of Hematology Oncology, University of Chicago, Chicago, IL 60637, United States.
References
- 1.Breda E, Catarino R, Monteiro E. Transoral laser microsurgery as standard approach to hypopharyngeal cancer. Survival analysis in a hospital based population. Acta Otorrinolaringol Esp. 2018;69:1–7. doi: 10.1016/j.otorri.2016.11.009. [DOI] [PubMed] [Google Scholar]
- 2.Mothes AR, Runnebaum M, Runnebaum IB. Ablative dual-phase Erbium:YAG laser treatment of atrophy-related vaginal symptoms in post-menopausal breast cancer survivors omitting hormonal treatment. J Cancer Res Clin Oncol. 2018;144:955–960. doi: 10.1007/s00432-018-2614-8. [DOI] [PubMed] [Google Scholar]
- 3.Zeinizade E, Tabei M, Shakeri-Zadeh A, Ghaznavi H, Attaran N, Komeili A, Ghalandari B, Maleki S, Kamrava SK. Selective apoptosis induction in cancer cells using folate-conjugated gold nanoparticles and controlling the laser irradiation conditions. Artif Cells Nanomed Biotechnol. 2018;46:1026–1038. doi: 10.1080/21691401.2018.1443116. [DOI] [PubMed] [Google Scholar]
- 4.Fridman WH. From Cancer Immune Surveillance to Cancer Immunoediting: Birth of Modern Immuno-Oncology. J Immunol. 2018;201:825–826. doi: 10.4049/jimmunol.1800827. [DOI] [PubMed] [Google Scholar]
- 5.Ljunggren HG, Jonsson R, Höglund P. Seminal immunologic discoveries with direct clinical implications: The 2018 Nobel Prize in Physiology or Medicine honours discoveries in cancer immunotherapy. Scand J Immunol. 2018;88:e12731. doi: 10.1111/sji.12731. [DOI] [PubMed] [Google Scholar]
- 6.Ou W, Thapa RK, Jiang L, Soe ZC, Gautam M, Chang JH, Jeong JH, Ku SK, Choi HG, Yong CS, Kim JO. Regulatory T cell-targeted hybrid nanoparticles combined with immuno-checkpoint blockage for cancer immunotherapy. J Control Release. 2018;281:84–96. doi: 10.1016/j.jconrel.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 7.Royston KJ, Paul B, Nozell S, Rajbhandari R, Tollefsbol TO. Withaferin A and sulforaphane regulate breast cancer cell cycle progression through epigenetic mechanisms. Exp Cell Res. 2018;368:67–74. doi: 10.1016/j.yexcr.2018.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manson MM. Cancer prevention -- the potential for diet to modulate molecular signalling. Trends Mol Med. 2003;9:11–18. doi: 10.1016/s1471-4914(02)00002-3. [DOI] [PubMed] [Google Scholar]
- 9.Scharlau D, Borowicki A, Habermann N, Hofmann T, Klenow S, Miene C, Munjal U, Stein K, Glei M. Mechanisms of primary cancer prevention by butyrate and other products formed during gut flora-mediated fermentation of dietary fibre. Mutat Res. 2009;682:39–53. doi: 10.1016/j.mrrev.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 10.Vogtmann E, Goedert JJ. Epidemiologic studies of the human microbiome and cancer. Br J Cancer. 2016;114:237–242. doi: 10.1038/bjc.2015.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Enewold L, Horner MJ, Shriver CD, Zhu K. Socioeconomic disparities in colorectal cancer mortality in the United States, 1990-2007. J Community Health. 2014;39:760–766. doi: 10.1007/s10900-014-9824-z. [DOI] [PubMed] [Google Scholar]
- 12.Given CW, Given BA, Bradley CJ, Krauss JC, Sikorskii A, Vachon E. Dynamic Assessment of Value During High-Cost Cancer Treatment: A Response to American Society of Clinical Oncology and European Society of Medical Oncology. J Oncol Pract. 2016;12:1215–1218. doi: 10.1200/JOP.2016.012401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou M, Ng KF. Cancer Treatment In China: How Are Policy And Practice In Tier 1 Versus Tier 2/3 Cities Impacting Patient Access To High-Cost Therapies. Value Health. 2014;17:A739. doi: 10.1016/j.jval.2014.08.131. [DOI] [PubMed] [Google Scholar]
- 14.Findley K, Williams DR, Grice EA, Bonham VL. Health Disparities and the Microbiome. Trends Microbiol. 2016;24:847–850. doi: 10.1016/j.tim.2016.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farhana L, Antaki F, Murshed F, Mahmud H, Judd SL, Nangia-Makker P, Levi E, Yu Y, Majumdar AP. Gut microbiome profiling and colorectal cancer in African Americans and Caucasian Americans. World J Gastrointest Pathophysiol. 2018;9:47–58. doi: 10.4291/wjgp.v9.i2.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Quinque D, Horz HP, Li M, Rzhetskaya M, Raff JA, Hayes MG, Stoneking M. Comparative analysis of the human saliva microbiome from different climate zones: Alaska, Germany, and Africa. BMC Microbiol. 2014;14:316. doi: 10.1186/s12866-014-0316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, Long J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int J Cancer. 2019;144:2381–2389. doi: 10.1002/ijc.31941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen L, Zhang YH, Huang T, Cai YD. Gene expression profiling gut microbiota in different races of humans. Sci Rep. 2016;6:23075. doi: 10.1038/srep23075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J, Ryu E, Hathcock M, Ballman K, Chia N, Olson JE, Nelson H. Impact of demographics on human gut microbial diversity in a US Midwest population. PeerJ. 2016;4:e1514. doi: 10.7717/peerj.1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khoruts A, Weingarden AR. Emergence of fecal microbiota transplantation as an approach to repair disrupted microbial gut ecology. Immunol Lett. 2014;162:77–81. doi: 10.1016/j.imlet.2014.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen D, Wu J, Jin D, Wang B, Cao H. Fecal microbiota transplantation in cancer management: Current status and perspectives. Int J Cancer. 2018 doi: 10.1002/ijc.32003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berni Canani R, Di Costanzo M, Leone L. The epigenetic effects of butyrate: Potential therapeutic implications for clinical practice. Clin Epigenetics. 2012;4:4. doi: 10.1186/1868-7083-4-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bishop KS, Xu H, Marlow G. Epigenetic Regulation of Gene Expression Induced by Butyrate in Colorectal Cancer: Involvement of MicroRNA. Genet Epigenet. 2017;9:1179237X17729900. doi: 10.1177/1179237X17729900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Royston KJ, Tollefsbol TO. The Epigenetic Impact of Cruciferous Vegetables on Cancer Prevention. Curr Pharmacol Rep. 2015;1:46–51. doi: 10.1007/s40495-014-0003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Royston KJ, Udayakumar N, Lewis K, Tollefsbol TO. A Novel Combination of Withaferin A and Sulforaphane Inhibits Epigenetic Machinery, Cellular Viability and Induces Apoptosis of Breast Cancer Cells. Int J Mol Sci. 2017;18:pii: E1092. doi: 10.3390/ijms18051092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monda V, Villano I, Messina A, Valenzano A, Esposito T, Moscatelli F, Viggiano A, Cibelli G, Chieffi S, Monda M, Messina G. Exercise Modifies the Gut Microbiota with Positive Health Effects. Oxid Med Cell Longev. 2017;2017:3831972. doi: 10.1155/2017/3831972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano KA, Martinez-Del Campo A, Kasahara K, Chittim CL, Vivas EI, Amador-Noguez D, Balskus EP, Rey FE. Metabolic, Epigenetic, and Transgenerational Effects of Gut Bacterial Choline Consumption. Cell Host Microbe. 2017;22:279–290.e7. doi: 10.1016/j.chom.2017.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malat J, Mayorga-Gallo S, Williams DR. The effects of whiteness on the health of whites in the USA. Soc Sci Med. 2018;199:148–156. doi: 10.1016/j.socscimed.2017.06.034. [DOI] [PubMed] [Google Scholar]
- 29.O'Keefe EB, Meltzer JP, Bethea TN. Health disparities and cancer: Racial disparities in cancer mortality in the United States, 2000-2010. Front Public Health. 2015;3:51. doi: 10.3389/fpubh.2015.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yedjou CG, Tchounwou PB, Payton M, Miele L, Fonseca DD, Lowe L, Alo RA. Assessing the Racial and Ethnic Disparities in Breast Cancer Mortality in the United States. Int J Environ Res Public Health. 2017;14:pii: E486. doi: 10.3390/ijerph14050486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A, Jemal A. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67:177–193. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 32.Tawk R, Abner A, Ashford A, Brown CP. Differences in Colorectal Cancer Outcomes by Race and Insurance. Int J Environ Res Public Health. 2015;13:ijerph13010048. doi: 10.3390/ijerph13010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doubeni CA, Major JM, Laiyemo AO, Schootman M, Zauber AG, Hollenbeck AR, Sinha R, Allison J. Contribution of behavioral risk factors and obesity to socioeconomic differences in colorectal cancer incidence. J Natl Cancer Inst. 2012;104:1353–1362. doi: 10.1093/jnci/djs346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lansdorp-Vogelaar I, Kuntz KM, Knudsen AB, van Ballegooijen M, Zauber AG, Jemal A. Contribution of screening and survival differences to racial disparities in colorectal cancer rates. Cancer Epidemiol Biomarkers Prev. 2012;21:728–736. doi: 10.1158/1055-9965.EPI-12-0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coughlin SS, Blumenthal DS, Seay SJ, Smith SA. Toward the Elimination of Colorectal Cancer Disparities Among African Americans. J Racial Ethn Health Disparities. 2016;3:555–564. doi: 10.1007/s40615-015-0174-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tammana VS, Laiyemo AO. Colorectal cancer disparities: Issues, controversies and solutions. World J Gastroenterol. 2014;20:869–876. doi: 10.3748/wjg.v20.i4.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Laiyemo AO, Doubeni C, Pinsky PF, Doria-Rose VP, Bresalier R, Lamerato LE, Crawford ED, Kvale P, Fouad M, Hickey T, Riley T, Weissfeld J, Schoen RE, Marcus PM, Prorok PC, Berg CD. Race and colorectal cancer disparities: health-care utilization vs different cancer susceptibilities. J Natl Cancer Inst. 2010;102:538–546. doi: 10.1093/jnci/djq068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dunford EK, Popkin BM. Disparities in Snacking Trends in US Adults over a 35 Year Period from 1977 to 2012. Nutrients. 2017;9:pii: E809. doi: 10.3390/nu9080809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowlin SY, Cleland CM, Vadiveloo M, D'Eramo Melkus G, Parekh N, Hagan H. Explaining Racial/Ethnic Dietary Patterns in Relation to Type 2 Diabetes: An Analysis of NHANES 2007-2012. Ethn Dis. 2016;26:529–536. doi: 10.18865/ed.26.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rehm CD, Peñalvo JL, Afshin A, Mozaffarian D. Dietary Intake Among US Adults, 1999-2012. JAMA. 2016;315:2542–2553. doi: 10.1001/jama.2016.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang DD, Leung CW, Li Y, Ding EL, Chiuve SE, Hu FB, Willett WC. Trends in dietary quality among adults in the United States, 1999 through 2010. JAMA Intern Med. 2014;174:1587–1595. doi: 10.1001/jamainternmed.2014.3422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kant AK, Graubard BI. 20-Year trends in dietary and meal behaviors were similar in U.S. children and adolescents of different race/ethnicity. J Nutr. 2011;141:1880–1888. doi: 10.3945/jn.111.144915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lewis KA, Jordan HR, Tollefsbol TO. Effects of SAHA and EGCG on Growth Potentiation of Triple-Negative Breast Cancer Cells. Cancers (Basel) 2018;11:pii: E23. doi: 10.3390/cancers11010023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clement KW. Smoking and Health. J Natl Med Assoc. 1964;56:201. [PMC free article] [PubMed] [Google Scholar]
- 45.Hirayama T. Epidemiology of prostate cancer with special reference to the role of diet. Natl Cancer Inst Monogr. 1979;(53):149–155. [PubMed] [Google Scholar]
- 46.Soldati L, Di Renzo L, Jirillo E, Ascierto PA, Marincola FM, De Lorenzo A. The influence of diet on anti-cancer immune responsiveness. J Transl Med. 2018;16:75. doi: 10.1186/s12967-018-1448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Havenaar R. Intestinal health functions of colonic microbial metabolites: A review. Benef Microbes. 2011;2:103–114. doi: 10.3920/BM2011.0003. [DOI] [PubMed] [Google Scholar]
- 48.Russell WR, Gratz SW, Duncan SH, Holtrop G, Ince J, Scobbie L, Duncan G, Johnstone AM, Lobley GE, Wallace RJ, Duthie GG, Flint HJ. High-protein, reduced-carbohydrate weight-loss diets promote metabolite profiles likely to be detrimental to colonic health. Am J Clin Nutr. 2011;93:1062–1072. doi: 10.3945/ajcn.110.002188. [DOI] [PubMed] [Google Scholar]
- 49.Moestue SA, Borgan E, Huuse EM, Lindholm EM, Sitter B, Børresen-Dale AL, Engebraaten O, Maelandsmo GM, Gribbestad IS. Distinct choline metabolic profiles are associated with differences in gene expression for basal-like and luminal-like breast cancer xenograft models. BMC Cancer. 2010;10:433. doi: 10.1186/1471-2407-10-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Asten JJ, Vettukattil R, Buckle T, Rottenberg S, van Leeuwen F, Bathen TF, Heerschap A. Increased levels of choline metabolites are an early marker of docetaxel treatment response in BRCA1-mutated mouse mammary tumors: An assessment by ex vivo proton magnetic resonance spectroscopy. J Transl Med. 2015;13:114. doi: 10.1186/s12967-015-0458-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rubio-Patiño C, Bossowski JP, De Donatis GM, Mondragón L, Villa E, Aira LE, Chiche J, Mhaidly R, Lebeaupin C, Marchetti S, Voutetakis K, Chatziioannou A, Castelli FA, Lamourette P, Chu-Van E, Fenaille F, Avril T, Passeron T, Patterson JB, Verhoeyen E, Bailly-Maitre B, Chevet E, Ricci JE. Low-Protein Diet Induces IRE1α-Dependent Anticancer Immunosurveillance. Cell Metab. 2018;27:828–842.e7. doi: 10.1016/j.cmet.2018.02.009. [DOI] [PubMed] [Google Scholar]
- 52.Maurel M, Chevet E, Tavernier J, Gerlo S. Getting RIDD of RNA: IRE1 in cell fate regulation. Trends Biochem Sci. 2014;39:245–254. doi: 10.1016/j.tibs.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 53.De Rosa V, Di Rella F, Di Giacomo A, Matarese G. Regulatory T cells as suppressors of anti-tumor immunity: Role of metabolism. Cytokine Growth Factor Rev. 2017;35:15–25. doi: 10.1016/j.cytogfr.2017.04.001. [DOI] [PubMed] [Google Scholar]
- 54.Boccellato F, Meyer TF. Bacteria Moving into Focus of Human Cancer. Cell Host Microbe. 2015;17:728–730. doi: 10.1016/j.chom.2015.05.016. [DOI] [PubMed] [Google Scholar]
- 55.Yang T, Owen JL, Lightfoot YL, Kladde MP, Mohamadzadeh M. Microbiota impact on the epigenetic regulation of colorectal cancer. Trends Mol Med. 2013;19:714–725. doi: 10.1016/j.molmed.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gopalakrishnan V, Helmink BA, Spencer CN, Reuben A, Wargo JA. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018;33:570–580. doi: 10.1016/j.ccell.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iida N, Dzutsev A, Stewart CA, Smith L, Bouladoux N, Weingarten RA, Molina DA, Salcedo R, Back T, Cramer S, Dai RM, Kiu H, Cardone M, Naik S, Patri AK, Wang E, Marincola FM, Frank KM, Belkaid Y, Trinchieri G, Goldszmid RS. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science. 2013;342:967–970. doi: 10.1126/science.1240527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yi M, Yu S, Qin S, Liu Q, Xu H, Zhao W, Chu Q, Wu K. Gut microbiome modulates efficacy of immune checkpoint inhibitors. J Hematol Oncol. 2018;11:47. doi: 10.1186/s13045-018-0592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ivanov II, Honda K. Intestinal commensal microbes as immune modulators. Cell Host Microbe. 2012;12:496–508. doi: 10.1016/j.chom.2012.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwa M, Plottel CS, Blaser MJ, Adams S. The Intestinal Microbiome and Estrogen Receptor-Positive Female Breast Cancer. J Natl Cancer Inst. 2016:108. doi: 10.1093/jnci/djw029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang ZY, Kang X, Chen H, Wang M, Guan WX. Effect of Clostridium perfringens enterotoxin on gastric cancer cells SGC7901 which highly expressed claudin-4 protein. World J Gastrointest Oncol. 2017;9:153–159. doi: 10.4251/wjgo.v9.i4.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li X, Xu H, Chen ZS, Chen G. Biosynthesis of Nanoparticles by Microorganisms and Their Applications. J Nanomater. 2011;2011:270974. [Google Scholar]
- 63.Choi JJ, Eum SY, Rampersaud E, Daunert S, Abreu MT, Toborek M. Exercise attenuates PCB-induced changes in the mouse gut microbiome. Environ Health Perspect. 2013;121:725–730. doi: 10.1289/ehp.1306534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brooks L, Viardot A, Tsakmaki A, Stolarczyk E, Howard JK, Cani PD, Everard A, Sleeth ML, Psichas A, Anastasovskaj J, Bell JD, Bell-Anderson K, Mackay CR, Ghatei MA, Bloom SR, Frost G, Bewick GA. Fermentable carbohydrate stimulates FFAR2-dependent colonic PYY cell expansion to increase satiety. Mol Metab. 2016;6:48–60. doi: 10.1016/j.molmet.2016.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yazici C, Wolf PG, Kim H, Cross TL, Vermillion K, Carroll T, Augustus GJ, Mutlu E, Tussing-Humphreys L, Braunschweig C, Xicola RM, Jung B, Llor X, Ellis NA, Gaskins HR. Race-dependent association of sulfidogenic bacteria with colorectal cancer. Gut. 2017;66:1983–1994. doi: 10.1136/gutjnl-2016-313321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carson TL, Wang F, Cui X, Jackson BE, Van Der Pol WJ, Lefkowitz EJ, Morrow C, Baskin ML. Associations Between Race, Perceived Psychological Stress, and the Gut Microbiota in a Sample of Generally Healthy Black and White Women: A Pilot Study on the Role of Race and Perceived Psychological Stress. Psychosom Med. 2018;80:640–648. doi: 10.1097/PSY.0000000000000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.O'Keefe SJ, Li JV, Lahti L, Ou J, Carbonero F, Mohammed K, Posma JM, Kinross J, Wahl E, Ruder E, Vipperla K, Naidoo V, Mtshali L, Tims S, Puylaert PG, DeLany J, Krasinskas A, Benefiel AC, Kaseb HO, Newton K, Nicholson JK, de Vos WM, Gaskins HR, Zoetendal EG. Fat, fibre and cancer risk in African Americans and rural Africans. Nat Commun. 2015;6:6342. doi: 10.1038/ncomms7342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seidel C, Schnekenburger M, Dicato M, Diederich M. Histone deacetylase modulators provided by Mother Nature. Genes Nutr. 2012;7:357–367. doi: 10.1007/s12263-012-0283-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hester CM, Jala VR, Langille MG, Umar S, Greiner KA, Haribabu B. Fecal microbes, short chain fatty acids, and colorectal cancer across racial/ethnic groups. World J Gastroenterol. 2015;21:2759–2769. doi: 10.3748/wjg.v21.i9.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8:51. doi: 10.1186/s13073-016-0307-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taur Y, Jenq RR, Perales MA, Littmann ER, Morjaria S, Ling L, No D, Gobourne A, Viale A, Dahi PB, Ponce DM, Barker JN, Giralt S, van den Brink M, Pamer EG. The effects of intestinal tract bacterial diversity on mortality following allogeneic hematopoietic stem cell transplantation. Blood. 2014;124:1174–1182. doi: 10.1182/blood-2014-02-554725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ratnayake WM, Galli C. Fat and fatty acid terminology, methods of analysis and fat digestion and metabolism: A background review paper. Ann Nutr Metab. 2009;55:8–43. doi: 10.1159/000228994. [DOI] [PubMed] [Google Scholar]
- 73.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front Microbiol. 2016;7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Krautkramer KA, Kreznar JH, Romano KA, Vivas EI, Barrett-Wilt GA, Rabaglia ME, Keller MP, Attie AD, Rey FE, Denu JM. Diet-Microbiota Interactions Mediate Global Epigenetic Programming in Multiple Host Tissues. Mol Cell. 2016;64:982–992. doi: 10.1016/j.molcel.2016.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lakhter AJ, Hamilton J, Konger RL, Brustovetsky N, Broxmeyer HE, Naidu SR. Glucose-independent Acetate Metabolism Promotes Melanoma Cell Survival and Tumor Growth. J Biol Chem. 2016;291:21869–21879. doi: 10.1074/jbc.M115.712166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, Goodwin LM, Smethurst E, Mason S, Blyth K, McGarry L, James D, Shanks E, Kalna G, Saunders RE, Jiang M, Howell M, Lassailly F, Thin MZ, Spencer-Dene B, Stamp G, van den Broek NJ, Mackay G, Bulusu V, Kamphorst JJ, Tardito S, Strachan D, Harris AL, Aboagye EO, Critchlow SE, Wakelam MJ, Schulze A, Gottlieb E. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell. 2015;27:57–71. doi: 10.1016/j.ccell.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chia CF, Chen SC, Chen CS, Shih CM, Lee HM, Wu CH. Thallium acetate induces C6 glioma cell apoptosis. Ann N Y Acad Sci. 2005;1042:523–530. doi: 10.1196/annals.1338.064. [DOI] [PubMed] [Google Scholar]
- 78.Franchini A, Marchesini E, Ottaviani E. Corticosterone 21-acetate in vivo induces acute stress in chicken thymus: Cell proliferation, apoptosis and cytokine responses. Histol Histopathol. 2004;19:693–699. doi: 10.14670/HH-19.693. [DOI] [PubMed] [Google Scholar]
- 79.Maioral MF, Stefanes NM, Bigolin Á, Zatelli GA, Philippus AC, de Barcellos Falkenberg M, Santos-Silva MC. MICONIDINE acetate, a new selective and cytotoxic compound with synergic potential, induces cell cycle arrest and apoptosis in leukemia cells. Invest New Drugs. 2018 doi: 10.1007/s10637-018-0694-6. [DOI] [PubMed] [Google Scholar]
- 80.Marques C, Oliveira CS, Alves S, Chaves SR, Coutinho OP, Côrte-Real M, Preto A. Acetate-induced apoptosis in colorectal carcinoma cells involves lysosomal membrane permeabilization and cathepsin D release. Cell Death Dis. 2013;4:e507. doi: 10.1038/cddis.2013.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh RP, Agrawal P, Yim D, Agarwal C, Agarwal R. Acacetin inhibits cell growth and cell cycle progression, and induces apoptosis in human prostate cancer cells: Structure-activity relationship with linarin and linarin acetate. Carcinogenesis. 2005;26:845–854. doi: 10.1093/carcin/bgi014. [DOI] [PubMed] [Google Scholar]
- 82.Bolduc JF, Hany L, Barat C, Ouellet M, Tremblay MJ. Epigenetic Metabolite Acetate Inhibits Class I/II Histone Deacetylases, Promotes Histone Acetylation, and Increases HIV-1 Integration in CD4+ T Cells. J Virol. 2017;91:28539453. doi: 10.1128/JVI.01943-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Bulusu V, Tumanov S, Michalopoulou E, van den Broek NJ, MacKay G, Nixon C, Dhayade S, Schug ZT, Vande Voorde J, Blyth K, Gottlieb E, Vazquez A, Kamphorst JJ. Acetate Recapturing by Nuclear Acetyl-CoA Synthetase 2 Prevents Loss of Histone Acetylation during Oxygen and Serum Limitation. Cell Rep. 2017;18:647–658. doi: 10.1016/j.celrep.2016.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, Petersen KF, Kibbey RG, Goodman AL, Shulman GI. Acetate mediates a microbiome-brain-β-cell axis to promote metabolic syndrome. Nature. 2016;534:213–217. doi: 10.1038/nature18309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mashimo T, Pichumani K, Vemireddy V, Hatanpaa KJ, Singh DK, Sirasanagandla S, Nannepaga S, Piccirillo SG, Kovacs Z, Foong C, Huang Z, Barnett S, Mickey BE, DeBerardinis RJ, Tu BP, Maher EA, Bachoo RM. Acetate is a bioenergetic substrate for human glioblastoma and brain metastases. Cell. 2014;159:1603–1614. doi: 10.1016/j.cell.2014.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lyssiotis CA, Cantley LC. Acetate fuels the cancer engine. Cell. 2014;159:1492–1494. doi: 10.1016/j.cell.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 87.Ferro S, Azevedo-Silva J, Casal M, Côrte-Real M, Baltazar F, Preto A. Characterization of acetate transport in colorectal cancer cells and potential therapeutic implications. Oncotarget. 2016;7:70639–70653. doi: 10.18632/oncotarget.12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Segain JP, Raingeard de la Blétière D, Bourreille A, Leray V, Gervois N, Rosales C, Ferrier L, Bonnet C, Blottière HM, Galmiche JP. Butyrate inhibits inflammatory responses through NFkappaB inhibition: Implications for Crohn's disease. Gut. 2000;47:397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Govindarajan N, Agis-Balboa RC, Walter J, Sananbenesi F, Fischer A. Sodium butyrate improves memory function in an Alzheimer's disease mouse model when administered at an advanced stage of disease progression. J Alzheimers Dis. 2011;26:187–197. doi: 10.3233/JAD-2011-110080. [DOI] [PubMed] [Google Scholar]
- 90.Encarnação JC, Pires AS, Amaral RA, Gonçalves TJ, Laranjo M, Casalta-Lopes JE, Gonçalves AC, Sarmento-Ribeiro AB, Abrantes AM, Botelho MF. Butyrate, a dietary fiber derivative that improves irinotecan effect in colon cancer cells. J Nutr Biochem. 2018;56:183–192. doi: 10.1016/j.jnutbio.2018.02.018. [DOI] [PubMed] [Google Scholar]
- 91.Mu D, Gao Z, Guo H, Zhou G, Sun B. Sodium butyrate induces growth inhibition and apoptosis in human prostate cancer DU145 cells by up-regulation of the expression of annexin A1. PLoS One. 2013;8:e74922. doi: 10.1371/journal.pone.0074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ruemmele FM, Dionne S, Qureshi I, Sarma DS, Levy E, Seidman EG. Butyrate mediates Caco-2 cell apoptosis via up-regulation of pro-apoptotic BAK and inducing caspase-3 mediated cleavage of poly-(ADP-ribose) polymerase (PARP) Cell Death Differ. 1999;6:729–735. doi: 10.1038/sj.cdd.4400545. [DOI] [PubMed] [Google Scholar]
- 93.Salimi V, Shahsavari Z, Safizadeh B, Hosseini A, Khademian N, Tavakoli-Yaraki M. Sodium butyrate promotes apoptosis in breast cancer cells through reactive oxygen species (ROS) formation and mitochondrial impairment. Lipids Health Dis. 2017;16:208. doi: 10.1186/s12944-017-0593-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gonçalves P, Martel F. Regulation of colonic epithelial butyrate transport: Focus on colorectal cancer. Porto Biomed J. 2016;1:83–91. doi: 10.1016/j.pbj.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kilner J, Waby JS, Chowdry J, Khan AQ, Noirel J, Wright PC, Corfe BM, Evans CA. A proteomic analysis of differential cellular responses to the short-chain fatty acids butyrate, valerate and propionate in colon epithelial cancer cells. Mol Biosyst. 2012;8:1146–1156. doi: 10.1039/c1mb05219e. [DOI] [PubMed] [Google Scholar]
- 96.Slavin J. Fiber and prebiotics: Mechanisms and health benefits. Nutrients. 2013;5:1417–1435. doi: 10.3390/nu5041417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li Q, Ding C, Meng T, Lu W, Liu W, Hao H, Cao L. Butyrate suppresses motility of colorectal cancer cells via deactivating Akt/ERK signaling in histone deacetylase dependent manner. J Pharmacol Sci. 2017;135:148–155. doi: 10.1016/j.jphs.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 98.Chou AH, Chen SY, Yeh TH, Weng YH, Wang HL. HDAC inhibitor sodium butyrate reverses transcriptional downregulation and ameliorates ataxic symptoms in a transgenic mouse model of SCA3. Neurobiol Dis. 2011;41:481–488. doi: 10.1016/j.nbd.2010.10.019. [DOI] [PubMed] [Google Scholar]
- 99.Kim HJ, Leeds P, Chuang DM. The HDAC inhibitor, sodium butyrate, stimulates neurogenesis in the ischemic brain. J Neurochem. 2009;110:1226–1240. doi: 10.1111/j.1471-4159.2009.06212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mathew OP, Ranganna K, Yatsu FM. Butyrate, an HDAC inhibitor, stimulates interplay between different posttranslational modifications of histone H3 and differently alters G1-specific cell cycle proteins in vascular smooth muscle cells. Biomed Pharmacother. 2010;64:733–740. doi: 10.1016/j.biopha.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu DC, Waby JS, Chirakkal H, Staton CA, Corfe BM. Butyrate suppresses expression of neuropilin I in colorectal cell lines through inhibition of Sp1 transactivation. Mol Cancer. 2010;9:276. doi: 10.1186/1476-4598-9-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hu S, Liu L, Chang EB, Wang JY, Raufman JP. Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer. 2015;14:180. doi: 10.1186/s12943-015-0450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol. 2017;19:29–41. doi: 10.1111/1462-2920.13589. [DOI] [PubMed] [Google Scholar]
- 104.Venter CS, Vorster HH, Cummings JH. Effects of dietary propionate on carbohydrate and lipid metabolism in healthy volunteers. Am J Gastroenterol. 1990;85:549–553. [PubMed] [Google Scholar]
- 105.Tedelind S, Westberg F, Kjerrulf M, Vidal A. Anti-inflammatory properties of the short-chain fatty acids acetate and propionate: A study with relevance to inflammatory bowel disease. World J Gastroenterol. 2007;13:2826–2832. doi: 10.3748/wjg.v13.i20.2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Emenaker NJ, Basson MD. Short chain fatty acids differentially modulate cellular phenotype and c-myc protein levels in primary human nonmalignant and malignant colonocytes. Dig Dis Sci. 2001;46:96–105. doi: 10.1023/a:1005661809250. [DOI] [PubMed] [Google Scholar]
- 107.Beyer-Sehlmeyer G, Glei M, Hartmann E, Hughes R, Persin C, Böhm V, Rowland I, Schubert R, Jahreis G, Pool-Zobel BL. Butyrate is only one of several growth inhibitors produced during gut flora-mediated fermentation of dietary fibre sources. Br J Nutr. 2003;90:1057–1070. doi: 10.1079/bjn20031003. [DOI] [PubMed] [Google Scholar]
- 108.Kiefer J, Beyer-Sehlmeyer G, Pool-Zobel BL. Mixtures of SCFA, composed according to physiologically available concentrations in the gut lumen, modulate histone acetylation in human HT29 colon cancer cells. Br J Nutr. 2006;96:803–810. doi: 10.1017/bjn20061948. [DOI] [PubMed] [Google Scholar]
- 109.Giardina C, Inan MS. Nonsteroidal anti-inflammatory drugs, short-chain fatty acids, and reactive oxygen metabolism in human colorectal cancer cells. Biochim Biophys Acta. 1998;1401:277–288. doi: 10.1016/s0167-4889(97)00140-7. [DOI] [PubMed] [Google Scholar]
- 110.Tian Y, Xu Q, Sun L, Ye Y, Ji G. Short-chain fatty acids administration is protective in colitis-associated colorectal cancer development. J Nutr Biochem. 2018;57:103–109. doi: 10.1016/j.jnutbio.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 111.Mai V, McCrary QM, Sinha R, Glei M. Associations between dietary habits and body mass index with gut microbiota composition and fecal water genotoxicity: An observational study in African American and Caucasian American volunteers. Nutr J. 2009;8:49. doi: 10.1186/1475-2891-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Upadhyay V, Poroyko V, Kim TJ, Devkota S, Fu S, Liu D, Tumanov AV, Koroleva EP, Deng L, Nagler C, Chang EB, Tang H, Fu YX. Lymphotoxin regulates commensal responses to enable diet-induced obesity. Nat Immunol. 2012;13:947–953. doi: 10.1038/ni.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ, Turnbaugh PJ. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ou J, Carbonero F, Zoetendal EG, DeLany JP, Wang M, Newton K, Gaskins HR, O'Keefe SJ. Diet, microbiota, and microbial metabolites in colon cancer risk in rural Africans and African Americans. Am J Clin Nutr. 2013;98:111–120. doi: 10.3945/ajcn.112.056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ou J, DeLany JP, Zhang M, Sharma S, O'Keefe SJ. Association between low colonic short-chain fatty acids and high bile acids in high colon cancer risk populations. Nutr Cancer. 2012;64:34–40. doi: 10.1080/01635581.2012.630164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Nava GM, Carbonero F, Ou J, Benefiel AC, O'Keefe SJ, Gaskins HR. Hydrogenotrophic microbiota distinguish native Africans from African and European Americans. Environ Microbiol Rep. 2012;4:307–315. doi: 10.1111/j.1758-2229.2012.00334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.O'Keefe SJ, Ou J, Aufreiter S, O'Connor D, Sharma S, Sepulveda J, Fukuwatari T, Shibata K, Mawhinney T. Products of the colonic microbiota mediate the effects of diet on colon cancer risk. J Nutr. 2009;139:2044–2048. doi: 10.3945/jn.109.104380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.O'Keefe SJ, Kidd M, Espitalier-Noel G, Owira P. Rarity of colon cancer in Africans is associated with low animal product consumption, not fiber. Am J Gastroenterol. 1999;94:1373–1380. doi: 10.1111/j.1572-0241.1999.01089.x. [DOI] [PubMed] [Google Scholar]
- 119.Jemal A, Center MM, DeSantis C, Ward EM. Global patterns of cancer incidence and mortality rates and trends. Cancer Epidemiol Biomarkers Prev. 2010;19:1893–1907. doi: 10.1158/1055-9965.EPI-10-0437. [DOI] [PubMed] [Google Scholar]
- 120.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912. [DOI] [PubMed] [Google Scholar]