Abstract
The mycobacterial cell envelope is crucial to host–pathogen interactions as a barrier against antibiotics and the host immune response. In addition, cell envelope lipids are mycobacterial virulence factors. Cell envelope lipid biosynthesis is the target of a number of frontline tuberculosis treatments and has been the focus of much research. However, the transport mechanisms by which these lipids reach the mycomembrane remain poorly understood. Many envelope lipids are exported from the cytoplasm to the periplasmic space via the mycobacterial membrane protein large (MmpL) family of proteins. In other bacteria, lipoproteins can contribute to outer membrane biogenesis through direct binding of substrates and/or protein–protein associations with extracytoplasmic biosynthetic enzymes. In this report, we investigate whether the lipoprotein LpqN plays a similar role in mycobacteria. Using a genetic two-hybrid approach, we demonstrate that LpqN interacts with periplasmic loop domains of the MmpL3 and MmpL11 transporters that export mycolic acid–containing cell envelope lipids. We observe that LpqN also interacts with secreted cell envelope biosynthetic enzymes such as Ag85A via pulldown assays. The X-ray crystal structures of LpqN and LpqN bound to dodecyl-trehalose suggest that LpqN directly binds trehalose monomycolate, the MmpL3 and Ag85A substrate. Finally, we observe altered lipid profiles of the ΔlpqN mutant during biofilm maturation, pointing toward a possible physiological role for the protein. The results of this study suggest that LpqN may act as a membrane fusion protein, connecting MmpL transporters with periplasmic proteins, and provide general insight into the role of lipoproteins in Mycobacterium tuberculosis cell envelope biogenesis.
Keywords: Mycobacterium tuberculosis, lipoprotein, crystal structure, membrane transport, glycolipid, lipid transport, trehalose, cell envelope, periplasm, virulence
Introduction
Mycobacterium tuberculosis (Mtb)2 is the causative agent of the human disease tuberculosis (TB). TB is one of the most devastating human diseases, and it remains a major public health concern. In 2017, there were ∼10 million new cases and 1.3 million deaths caused by TB (1). The cell envelope of Mtb is a waxy, lipid-rich structure that acts as a robust physical barrier to host antimicrobial defenses and antibiotics. This intrinsic resistance is largely conferred by mycolic acids, extremely long chain (C60–90) α-alkyl-β-hydroxy fatty acids that are major constituents of the mycobacterial cell envelope (2). Mycolic acids and their derivatives form an outer layer analogous to the outer membrane of Gram-negative bacteria, termed the mycomembrane. The mycomembrane consists of an inner leaflet of mycolylated arabinogalactan anchored to peptidoglycan (the mAGP complex) and an outer leaflet of predominantly mycolylated free lipids (3, 4). In addition to acting as a passive physical barrier, certain cell envelope components can directly interfere with host immune responses (5, 6). Therefore, the Mtb cell envelope plays a crucial role at the host–pathogen interface to promote bacterial survival.
The biogenesis of the mycobacterial cell envelope is complex and incompletely described. One class of proteins involved in transport of cell envelope components is the mycobacterial membrane protein large (MmpL) family. The MmpLs are polytopic membrane proteins that mediate transport of large hydrophobic substrates from the cytoplasm of the bacterium to the periplasmic space, where they are subsequently incorporated into the cell envelope (7–9). The MmpLs are considered members of the resistance–nodulation–cell division (RND) superfamily of membrane transporters that typically couple the proton motive force to export of their substrates (10, 11). A subset of MmpLs (MmpL1/2/4/5) are accompanied in the genome by genes encoding mycobacterial membrane protein small (MmpS) proteins (12) that are predicted to contain one or two transmembrane segments and are likely involved in substrate transport by their cognate MmpL protein.
The substrates of a number of MmpL transporters have been identified. Of particular interest are those MmpL proteins that transport mycolic acid–containing lipids. Most importantly, MmpL3 (Rv0206c) transports trehalose monomycolate (TMM) into the periplasmic space (13). There, mycolic acid transfer catalyzed by members of the antigen 85 (Ag85) enzyme complex generates the free lipid trehalose dimycolate (TDM) and arabinogalactan-anchored mycolic acids, which together form the mycomembrane (13–15). MmpL3 is therefore an essential Mtb protein, because of the central role of mycolic acids in mycobacterial physiology (16, 17). Another important MmpL transporter is MmpL11 (Rv0202c), which we showed exports species-specific mycolic acid–containing storage lipids such as the mycolate wax ester and long-chain triacylglycerols in Mtb and meromycolyl diacylglycerol in Mycobacterium smegmatis (18, 19). These lipids are important for Mtb persistence, because MmpL11-deficient Mtb exhibit attenuated survival during in vitro and in vivo models of infection and in an in vitro model of dormancy (10, 19).
Many of the insights into MmpL transporter function have been gleaned from comparison with other protein members of the larger RND transporter superfamily. RND superfamily proteins are primarily associated with antibiotic resistance and stress response in Gram-negative bacteria (reviewed in Ref. 20). They typically function in concert with a membrane fusion protein (MFP) and an outer membrane factor to form a tripartite export apparatus that extrudes substrates through the periplasm directly to the extracellular environment (21–25). Although Mtb exhibits pseudo–Gram-negative membrane organization, it remains to be determined whether the MmpLs interact with periplasmic and outer membrane proteins to form a similar tripartite pump that would enable efficient translocation and proper localization of their substrates. Some secreted lipoproteins direct the proper localization of virulence-associated lipids within the cell envelope, suggesting that these proteins may be functioning as MFPs in Mtb. Notably, translocation of phthiocerol dimycocerosate (PDIM) to the mycomembrane depends on the lipoprotein LppX, presumably through direct binding of PDIM by LppX (26). Similarly, the lipoprotein LprG binds both the virulence-associated glycolipid lipoarabinomannan and triacylglycerides and is required for translocation of these substrates to the cell envelope (27). Furthermore, LprG interacts with the Ag85A mycolyltransferase, implicating this lipoprotein in the transmycolylation processes that generate the mycomembrane (28). The MmpS proteins are also candidate MFPs, given that MmpS4/5 interact with their cognate MmpL4/5 partners and are required for transport of their siderophore substrates (29). However, only a minority of MmpL transporters have cognate MmpS proteins (MmpS1/2/4/5), and MmpL-MmpS protein–protein interactions have not been demonstrated for MmpS1/2. This raises the possibility that there are unidentified Mtb proteins that function as quasi-MFPs to assist in delivering MmpL-transported lipids through the periplasm to their cell envelope destinations.
We sought to identify proteins that might function as MFPs with MmpL3 and MmpL11. Using genetic methods, we identified the secreted lipoprotein LpqN (Rv0583c) as a periplasmic interacting partner of both MmpL3 and MmpL11. Biochemical and genetic studies suggested that LpqN may also associate with members of the Ag85 enzyme complex and plays a role in cell envelope lipid changes during biofilm maturation. These results suggest that LpqN may function as an MFP of MmpL3/11. This model is supported by the crystal structures of apo- and lipid-bound LpqN reported herein.
Results
LpqN interacts with D2 loops of mycolate lipid transporters MmpL3 and MmpL11
MmpL3 and MmpL11 are structurally similar and categorized into the hydrophobe/amphiphile efflux 3 (HAE3) subfamily of the larger RND transporter superfamily (29, 30). They possess two periplasmic loop domains, termed D1 and D2, as well as a cytosolic C-terminal domain (31). The D1, D2, and C-terminal domains are therefore the most likely regions of interaction between MmpL11 and biosynthetic enzymes or other transport machinery. To identify periplasmic proteins that interact with MmpL11TB, we used the mycobacterial protein fragment complementation (M-PFC) system (32). In this two-hybrid system, the “bait” and “prey” are expressed independently as fusion proteins with two fragments of the murine dihydrofolate reductase (DHFR [F1,2] and DHFR [F3]). Functional reconstitution by two interacting mycobacterial proteins when expressed in M. smegmatis results in trimethoprim resistance. Integrative plasmids were constructed containing D1 and D2 domains in-frame with the DHFR [F3] fragment and then transformed into M. smegmatis to generate a bait strain. An H37Rv Mtb prey library of over 106 independent clones was generated in pUAB300. Clones encoding putative proteins that interact with MmpL11TB were obtained by transforming the library constructs into the M. smegmatis bait strain and selecting for trimethoprim resistance. Ultimately, no positive clones were identified using the D1 domain. Using the D2 construct, we isolated two independent clones that contained a DHFR [F1,2]–LpqN fusion. LpqN was the only positive clone identified using the MmpL11 D2 domain as bait. This suggests that LpqN (Rv0583c) interacts with the periplasmic D2 domain of MmpL11TB.
MmpL11 and MmpL3 are topologically similar conserved transporters of mycolic acid–containing lipids. Therefore, we theorized that LpqN might interact with the periplasmic domains of MmpL3 as well. To address this and validate our initial M-PFC screen, we assessed the interactions between the MmpL3 and MmpL11 periplasmic D1 and D2 loops with LpqN using a directly cloned (nonlibrary) LpqN prey construct, which contained the soluble domain of LpqN fused to DHFR [F1,2]. We found that the D2, but not the D1, loops of both MmpL3 and MmpL11 conferred trimethoprim resistance when co-expressed with LpqN (Table 1). These results suggest that LpqN interacts with the D2 periplasmic regions of both MmpL3 and MmpL11.
Table 1.
M-PFC assessed via trimethoprim minimal inhibitory concentrations of MmpL3/11TB D1 and D2 domain–LpqN interactions
MIC, minimal inhibitory concentration.
| Insert in pUAB200 | Insert in pUAB300 | Trim MIC (μg/ml) |
|---|---|---|
| — | Rv2763 (dfr) positive control | >200 |
| — | LpqN | <6.25 |
| MmpL3 D1 | — | <6.25 |
| MmpL3 D1 | LpqN | <6.25 |
| MmpL11 D1 | — | <6.25 |
| MmpL11 D1 | LpqN | <6.25 |
| MmpL3 D2 | — | <6.25 |
| MmpL3 D2 | LpqN | 100 |
| MmpL11 D2 | — | <6.25 |
| MmpL11 D2 | LpqN | 50 |
To verify the interaction between LpqN and MmpL11 in vivo, we heterologously expressed affinity-tagged Mtb protein in M. smegmatis. Although both proteins were successfully expressed, we were unfortunately unable to co-purify LpqN with MmpL11 in the absence or presence of cross-linking (Fig. S1).
LpqN interacts with cell envelope lipid biosynthetic enzymes
We hypothesized that LpqN might act as an adaptor protein in the periplasm to facilitate interactions between MmpL proteins and periplasmic proteins. To identify secreted interacting partners of LpqN, we performed a pulldown purification of affinity-tagged LpqN incubated with Mtb culture filtrate proteins (CFPs) in the presence/absence of formaldehyde as a protein cross-linking reagent. Interacting proteins were subsequently identified via MS and categorized as weak or strong LpqN-interacting partners based on whether or not protein cross-linking was required for co-purification (Table 2). As a control, we analyzed the background binding of CFP to the metal affinity resin. Intriguingly, secreted cell envelope biosynthetic enzymes, such as the mycolyltransferase Ag85A and the mycocerosic acid synthase Mas, were enriched in the LpqN co-purified samples relative to background. Ag85B also copurified with LpqN, although to a lesser degree. Ag85 and Mas enzymes are responsible for the biosynthesis of the cell envelope lipids TDM and PDIM, respectively. These data suggest that LpqN interacts with periplasmic cell envelope biosynthetic enzymes.
Table 2.
Interacting partners of LpqN in the culture filtrate
| Protein | CFP (background)a | CFP + LpqN (strong interactions) | Cross-linked (weak interactions) |
|---|---|---|---|
| Rv0583c | lipoprotein lpqN | 54 | 1862 | 1774 |
| Rv2220 | glutamine synthetase glnA1 | 15 | 54 | 121 |
| Rv3804c | fibronectin-binding protein antigen fbpA, Ag85A | 18 | 35 | 100 |
| Rv1908c | catalase-peroxidase-peroxynitritase T katG | 22 | 46 | 83 |
| Rv2780 | secreted l-alanine dehydrogenase ald | 11 | 32 | 76 |
| Rv1475c | aconitate hydratase A acn | 3 | 25 | 60 |
| Rv1980c | immunogenic protein mpt64 | 16 | 21 | 20 |
| Rv0350 | chaperone protein dnaK | 50 | 60 | 51 |
| Rv0896 | citrate synthase I gltA2 | 3 | 2 | 57 |
| Rv006c | isocitrate dehydrogenase icd2 | 0 | 0 | 33 |
| Rv3248c | adenosylhomocysteinase sahH | 4 | 4 | 49 |
| Rv0440 | 60-kDa chaperonin groEL2 | 9.818 | 15.6 | 18.947 |
| Rv1098c | fumarate hydratase fumC | 8 | 8 | 20 |
| Rv0211 | phosphoenolpyruvate carboxykinase pckA | 4 | 6 | 26 |
| Rv2940c | mycocerosic acid synthase mas | 0 | 3 | 27 |
| Rv0363c | fructose-bisphosphate aldolase fba | 2 | 11 | 23 |
| Rv2467 | aminopeptidase N pepN | 2 | 5 | 23 |
| Rv3418 | 10-kDa chaperonin groS | 7 | 4 | 22 |
| Rv2030c | uncharacterized protein | 1 | 8 | 21 |
| Rv0462 | dihydrolipoamide dehydrogenase lpd | 0 | 2 | 20 |
| Rv1017c | ribose-phosphate pyrophosphokinase prsA | 7 | 13 | 20 |
| Rv1074c | acetyl-CoA acetyltransferase fadA3 | 0 | 3 | 20 |
| Rv1886c | secreted fibronectin-binding protein fbpB, Ag85B | 0 | 3 | 19 |
| Rv1093 | serine hydroxymethyltransferase 1 glyA1 | 3 | 6 | 19 |
| Rv2244 | meromycolate extension acyl carrier protein acpM | 14 | 13 | 18 |
| Rv2031c | α-crystallin hspX | 1 | 4 | 17 |
| Rv0315 | probable β-1,3-glucanase precursor | 0 | 0 | 12 |
| Rv2146c | N-acetyltransferase eis | 0 | 4 | 11 |
| Rv1837c | malate synthase G glcB | 0 | 0 | 7 |
| Rv0934 | periplasmic phosphate-binding lipoprotein pstS1 | 0 | 1 | 6 |
| Rv0379 | protein transport protein secE2 | 2 | 0 | 5 |
| Rv1860 | alanine and proline rich secreted protein apa | 2 | 0 | 5 |
| Rv1392 | S-adenosylmethionine synthetase metK | 0 | 0 | 5 |
| Rv0164 | conserved hypothetical protein TB18.5 | 0 | 0 | 4 |
| Rv1448c | transaldolase tal | 0 | 0 | 4 |
| Rv1449c | transketolase tkt | 0 | 0 | 4 |
| Rv3628 | inorganic pyrophosphatase ppa | 0 | 0 | 4 |
a|Cell values are the summed spectral counts of identified peptides corresponding to the indicated protein, from three biological replicate experiments.
The possible interaction between Ag85A and LpqN was interesting, given the key role that the Ag85 complex plays in the biogenesis of the mycobacterial cell envelope. Furthermore, the primary substrate of the Ag85 enzymes is TMM, which is exported by MmpL3. Attempts to demonstrate a direct interaction between LpqN and Ag85A using a heterologous dual-expression M. smegmatis co-immunopurification strategy were unsuccessful (Fig. S2). However, there is precedent for interactions between lipoproteins and members of the Ag85 complex, as evidenced by the recently reported LprG–Ag85A interaction (28). LprG (Rv1411c) binds phosphatidylinositol-containing glycolipids rather than mycolic acids (33). Nevertheless, LprG appears to contribute to mycolylation of cell wall analogues (28). We performed head-to-head M-PFC assays to determine whether the MmpL3 or MmpL11 D2 domains are capable of interacting with other LpqN family members (LpqT, Mtc28) or LprG. LpqT and Mtc28 also interact with the MmpL11 D2 domain, but not the MmpL3 D2 domain, via M-PFC (Table S1). The structurally distinct LprG protein also interacted with the MmpL11, but not MmpL3, D2 domain via M-PFC. These results suggest the possibility that multiple lipoproteins act as adaptors for the MmpL transporters.
LpqN contributes to Mtb biofilm lipid composition
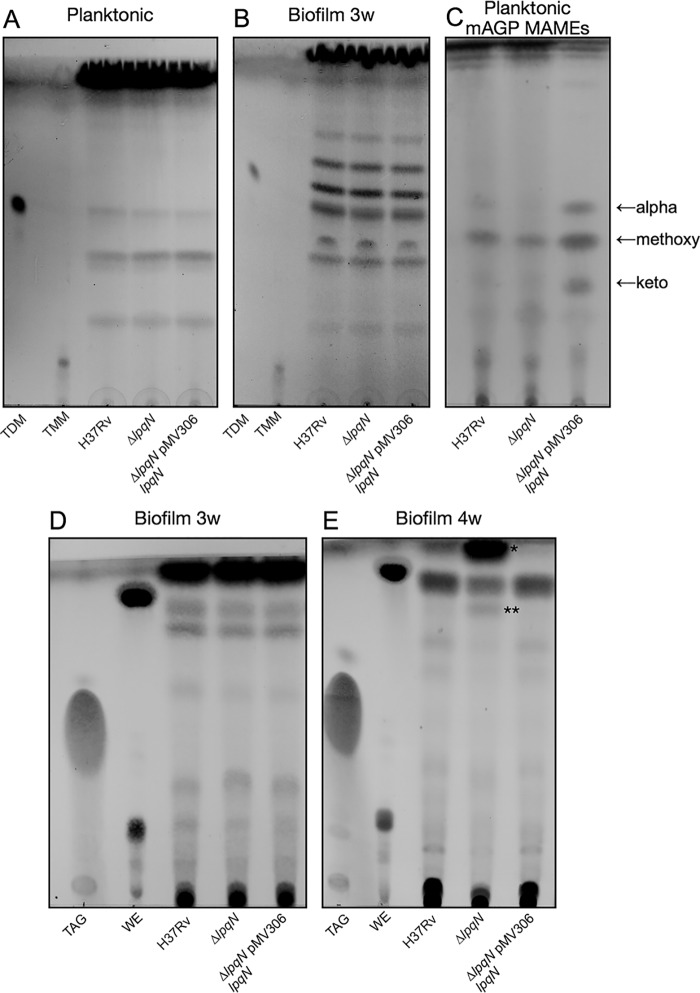
MmpL3 transports TMM, which is essential for replication, whereas MmpL11 transports cell envelope lipids that are important for biofilm formation. To investigate a possible role for LpqN in mycobacterial cell envelope biogenesis, we generated an lpqN-deletion mutant in H37Rv Mtb (ΔlpqN) via allelic exchange (Fig. S3, A and B). There was not a growth defect in either 7H9 or Sauton's medium (Fig. S3, C and D). We then characterized Mtb WT, ΔlpqN, and complemented strain cell envelope lipids from bacteria grown planktonically and in biofilms by TLC. We did not observe any differences in TMM and TDM levels between the WT, mutant, or complemented mutant (Mtb ΔlpqN::lpqN) lipid profiles using the chloroform:methanol:ammonium hydroxide (80:20:2) solvent system (Fig. 1, A and B). We also analyzed the mycolic acid methyl esters of mAGP, which did not differ significantly between strains (Fig. 1C). Consistent with these results, we also did not see significant differences in the LC/MS profiles among the lipid extracts, in particular, of TMM and TDM (Figs. S4 and S5). This was not surprising because the ΔlpqN mutant did not possess a growth defect.
Figure 1.
LpqN contributes to biofilm lipid composition in late biofilm cultures. Mtb was grown in either complete 7H9 medium or Sauton's medium lacking Tween for planktonic and biofilm cultures, respectively. Mtb cell envelope surface lipids were harvested by hexanes extraction from planktonic culture (A) or from biofilms harvested at 3 (B and D) or 4 (E) weeks and analyzed via TLC. mAGP was isolated from planktonic Mtb cell walls and MAMEs generated and visualized by TLC (C). The solvent systems used for TLC were 80:20:2, v/v/v, CHCl3:MeOH:NH4OH (A and B); 99:5, v/v, hexanes:ethyl acetate (C); and 99:1, v/v, toluene:acetone (D and E). TDM, TMM, triacylglycerol (TAG), and wax ester (WE) standards are indicated (Sigma). All TLCs were developed by charring in 10% molybdophosphoric acid. In E, * and ** indicate species that accumulate in the lpqN mutant.
Our analyses of Mtb biofilms over time indicate that the lipid profiles shift as the biofilm matures. Although there were no differences between WT and ΔlpqN mutant lipids at 3 weeks (Fig. 1D), we noted the relative accumulation of an apolar lipid in the lpqN mutant samples that ran with a similar Rf to the wax ester standard in 4 weeks (Fig. 1E, asterisk). We speculated that the apolar lipid (Fig. 1E, asterisk) in the mutant was a wax ester. However, mass spectrometric analysis of the extract of this apolar TLC spot did not yield mass spectra that directly identified any known mycobacterial lipids. These results countered our assumption that the spot is wax ester and are consistent with HPLC/MS results that showed no difference in wax ester between WT and LpqN (Fig. S4E). The ΔlpqN mutant biofilm also appeared to accumulate a less-apolar band that was similarly recalcitrant to identification (Fig. 1E, asterisks). Together, these biofilm lipid profiles suggest that LpqN may contribute to cell envelope lipid turnover or remodeling during biofilm development.
The LpqN crystal structure suggests that LpqN may directly bind lipids
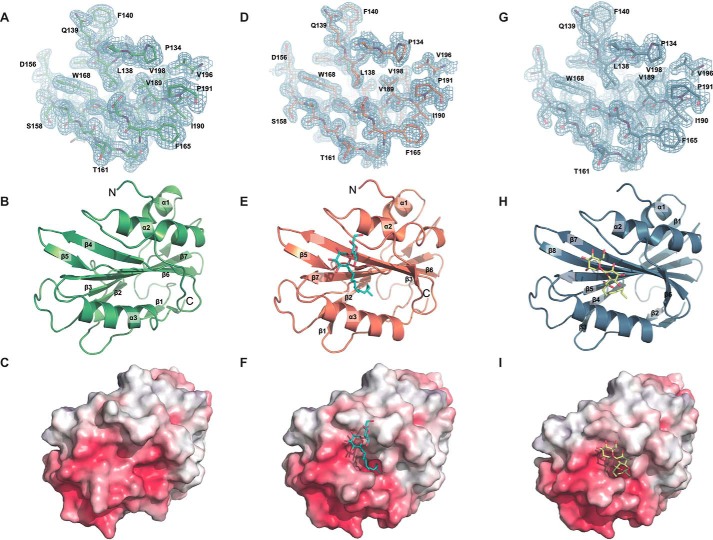
To gain additional insight into LpqN function, the crystal structure of apo-LpqN was determined at a resolution of 1.65 Å using single anomalous dispersion (Table 3 and Fig. 2). Because LpqN might play a role in cell envelope biogenesis, we also determined structures of LpqN in complexes with dodecyl trehalose (6LT) and trehalose 6-decanoate (T6D) to resolutions of 1.37 and 1.74 Å, respectively. We resolved these two ligand-bound LpqN structures by using molecular replacement and utilizing the apo-LpqN structure as a search model (Table 3 and Fig. 2). Both 6LT and T6D are water-soluble compounds that are structurally similar to the insoluble lipid molecule TMM in that they contain a two-ring system of the trehalose moiety. Therefore, 6LT and T6D are ideal compounds to mimic TMM with respect to the interaction with LpqN. The crystals of apo-LpqN, LpqN–6LT, and LpqN–T6D took the space groups C121, I4, and I4, respectively. A single molecule of LpqN was found in the asymmetric unit of each crystal. The molecule of LpqN consists of three α-helices and seven β-strands. These helices and strands are designated numerically from the N to C termini: α1 (61–68), β1 (84–86), β2 (104–108), β3 (120–124), α2 (134–147), β4 (170–176), β5 (179–182), β6 (184–192), β7 (195–206), and α3 (210–222). Based on the orientation of apo-LpqN within the asymmetric unit, the dimensions of this protein can be measured to be 45 Å × 40 Å × 35 Å. Accordingly, β4, β5, β6, β7, and α3 are involved in forming a shallow cavity, presumably constituting a ligand binding site. The interior wall of this cavity consists mainly of hydrophobic and polar residues including Leu-87, Ile-121, Val-183, Ala-184, Leu-201, Ala-203, and Met-214.
Table 3.
Data collection, phasing, and structural refinement statistics
| apo-LpqN (6E5D) | LpqN–6LT (6E5F) | LpqN–T6D (6MNA) | SeMet–LpqN | |
|---|---|---|---|---|
| Wavelength (Å) | 0.9792 | 0.9792 | 0.9792 | 0.9792 |
| Resolution (Å) | 48.11–1.65 (1.71–1.65) | 56.57–1.37 (1.41–1.37) | 56.25–1.74 (1.85–1.74) | 48.59–2.20 (2.32–2.20) |
| Unit cell parameters | ||||
| a, b, c (Å) | 99.3, 55.5, 44.6 | 80.0, 80.0, 59.2 | 79.5, 79.5, 60.0 | 99.0, 56.4, 44.5 |
| α, β, γ (°) | 90.0, 103.9, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 104.8, 90.0 |
| Space group | C121 | I4 | I4 | C121 |
| Total no. of reflections | 187,144 | 334,562 | 337,062 | 155,435 |
| Total no. of unique reflections | 27,327 | 39,223 | 19,157 | 12,099 |
| Wilson B factor (Å2) | 24.2 | 20.73 | 27.5 | 42.6 |
| Completeness (%) | 96.1 (87.3) | 99.7 (98.0) | 99.3 (98.5) | 99.6 (98.2) |
| Multiplicity | 6.9 (5.3) | 8.4 (3.2) | 17.6 (17.6) | 12.8 (13.3) |
| CC1/2 (%) | 99.9 (95.6) | 99.9 (64.5) | 99.9 (64.3) | 99.8 (98.2) |
| I/σ(I) | 19.0 (2.9) | 18.2 (1.9) | 11.1 (1.5) | 15.3 (5.1) |
| Phasing | ||||
| No. of sites | 2 | |||
| Figure of merit | 0.305 | |||
| Refinement | ||||
| Rwork (%)a | 18.57 | 17.8 | 18.45 | |
| Rfree (%)b | 20.85 | 20.35 | 21.75 | |
| No. of. atoms | ||||
| Protein | 2654 | 2661 | 2559 | |
| Water | 140 | 107 | 101 | |
| Ligand | 81 | 74 | ||
| B factors (Å2) | ||||
| Protein | 35.17 | 30.96 | 34.88 | |
| Water | 41.72 | 37.98 | 40.13 | |
| Ligand | 70.05 | 69.63 | ||
| RMSDs | ||||
| Bond lengths (Å) | 0.002 | 0.004 | 0.012 | |
| Bond angle (°) | 0.545 | 0.764 | 1.242 | |
| Ramachandran plot (%) | ||||
| Favored regions | 96.84 | 98.14 | 100 | |
| Allowed regions | 3.2 | 1.8 | 0 | |
| Outliers | 0 | 0 | 0 |
a|Rwork = Σhkl ||Fobs − Fcalc||/Σhkl|Fobs|.
b|Rfree was calculated as for Rwork but using 5% of the data that were excluded from the refinement calculation.
Figure 2.
Structures of the M. tuberculosis LpqN protein. A, representative section of electron density of apo-LpqN. The solvent-flattened electron density (50–1.65 Å) is contoured at 1.0 σ and superimposed with the final refined model (green, carbon; red, oxygen; blue, nitrogen). B, ribbon diagram of a protomer of LpqN. The secondary structural elements of LpqN are colored green. C, surface representation of the electrostatic surface potentials of LpqN colored by charge (red, negative −15 kT/e; blue, positive +15 kT/e). D, representative section of electron density of LpqN–6LT. The solvent-flattened electron density (50–1.37 Å) is contoured at 1.0 σ and superimposed with the final refined model (orange, carbon; red, oxygen; blue, nitrogen). E, ribbon diagram of a protomer of LpqN–6LT. The secondary structural elements of LpqN are colored orange. The bound 6LT is in cyan. F, surface representation of the electrostatic surface potentials of LpqN–6LT colored by charge (red, negative −15 kT/e; blue, positive +15 kT/e). The bound 6LT is in cyan. A specific deep binding pocket, which was not observed in apo-LpqN, was found to create on the LpqN–6LT surface. G, representative section of electron density of LpqN–T6D. The solvent-flattened electron density (50–1.74 Å) is contoured at 1.0 σ and superimposed with the final refined model (sky blue, carbon; red, oxygen; blue, nitrogen). H, ribbon diagram of a protomer of LpqN–T6D. The secondary structural elements of LpqN are colored sky blue. The bound T6D is in yellow. I, surface representation of the electrostatic surface potentials of LpqN–T6D colored by charge (red, negative −15 kT/e; blue, positive +15 kT/e). The bound T6D is in yellow. A specific deep binding pocket, which was not observed in apo-LpqN, was found to create on the LpqN–T6D surface.
Superimposition of the structures of apo-LpqN and LpqN–6LT results in an overall root-mean-square deviation (RMSD) of 0.8 Å, suggesting that these two structures are similar in conformation. However, detailed inspection indicates that the two structures are quite different. The main difference between the two conformations is the shift in location of α3 and the rearrangement of the secondary structures of several random loops located at the 6LT-binding site. These loops, including residues 93–96, 158–160, and 167–170, convert into β strands after binding 6LT. These changes result in a larger internal cavity for LpqN–6LT compared with that of apo-LpqN. In addition, a specific deep binding pocket, which was not observed in apo-LpqN, was found in LpqN–6LT to coordinate with the binding of the carbon chain of the 6LT ligand. Because the secondary structure of LpqN–6LT is very different, we reassigned its structural features to facilitate the description of this ligand-bound protein. The secondary structure assignments of 6LT-bound LpqN from the N to C termini are: α1 (61–67), β1 (93–96), β2 (104–109), α2 (134–147), β3 (158–160), β4 (167–176), β5 (179–185), β6 (187–192), β7 (195–206), and α3 (210–223).
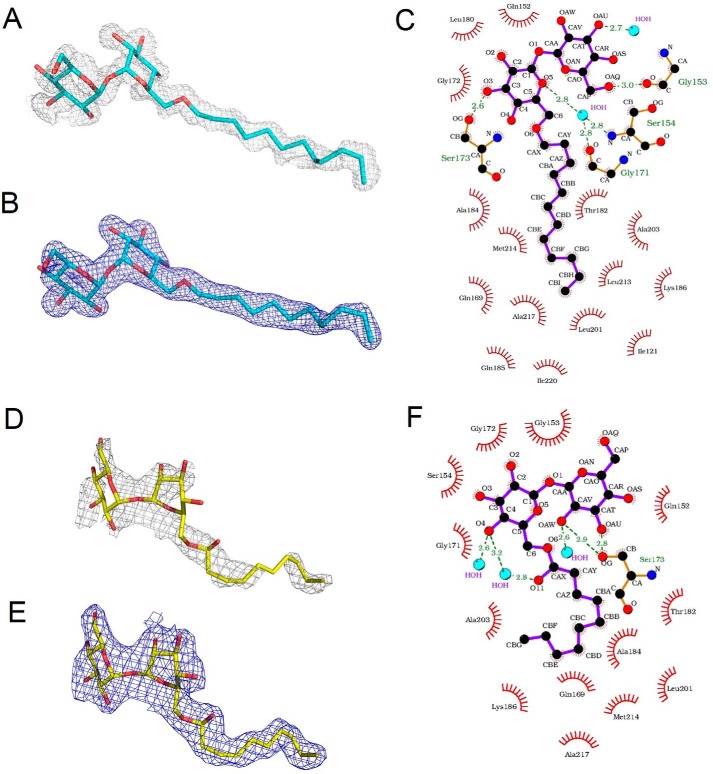
In the structure of LpqN–6LT, the 6LT molecule is bound within the cavity surrounded by β4, β5, β7, and α3. The dodecyl tail of 6LT is completely buried inside the LpqN protein, leaving its two sugar rings of the trehalose head group partially exposed to solvent. Within 6 Å of the dodecyl group of 6LT, 12 amino acids contact and secure the hydrocarbon tail (Fig. 3). Although the trehalose moiety of 6LT only partially contacts the LpqN protein, the binding of this two-ring head group is extensive. Ser-173 of LpqN forms a hydrogen bond with the trehalose head group to anchor this ligand. Gly-153 also donates its backbone oxygen to make another hydrogen bond with the trehalose moiety. Ser-154 and Gly-171 also participate in binding this head group. In addition, Gln-152 is 3.7 Å from the trehalose head group and interacts with the ligand via electrostatic interaction.
Figure 3.
The LpqN ligand-binding site. A, the Fo − Fc simulated annealing electron density map of the bound 6LT within the LpqN protein. The bound 6LT is shown as a stick model (cyan, carbon; red, oxygen). The simulated annealing Fo − Fc electron density map is contoured at 3.0 σ (gray mesh). B, the 2Fo − Fc electron density map of the bound 6LT within the LpqN protein. The bound 6LT is shown as a stick model (cyan, carbon; red, oxygen). The 2Fo − Fc electron density map is contoured at 1.0 σ (blue mesh). C, schematic representation of the LpqN and 6LT interactions. Amino acid residues within 3.5 Å from the bound 6LT are included. Dotted lines depict the hydrogen bonds. The hydrogen-bonded distances are also indicated in this figure. D, the Fo − Fc simulated annealing electron density map of the bound T6D within the LpqN protein. The bound T6D is shown as a stick model (yellow, carbon; red, oxygen). The simulated annealing Fo − Fc electron density map is contoured at 3.0 σ (gray mesh). E, the 2Fo − Fc electron density map of the bound T6D within the LpqN protein. The bound T6D is shown as a stick model (yellow, carbon; red, oxygen). The 2Fo − Fc electron density map is contoured at 1.0 σ (blue mesh). F, schematic representation of the LpqN and T6D interactions. Amino acid residues within 3.5 Å from the bound T6D are included. Dotted lines depict the hydrogen bonds. The hydrogen-bonded distances are also indicated in this figure.
The structure of LpqN–T6D resembles that of LpqN–6LT. Superimposition of these two structures leads to an RMSD of 0.1 Å, suggesting that these two structures are almost identical. Again, the main difference between the conformation of these two structures is the switch of local secondary structures from random loops to β strands. For example, residues 70–74, 84–86, 120–129, 160–162, 165–167, 185–187, and 225–227 are found to form flexible loops in the structure of LpqN–6LT. These residues are all incorporated into different β strands. Based on these structures, it appears that the LpqN protein is able to easily switch its secondary structures to accommodate for substrate binding. As a result, the LpqN–6LT forms three α-helices and nine β-strands: α1 (61–67), β1 (70–74), β2 (84–86), β3 (93–96), β4 (104–109), β5 (120–129), α2 (134–147), β6 (158–162), β7 (165–176), β8 (179–192), β9 (195–206), α3 (210–223), and β10 (225–227).
Like the binding of 6LT, the decanoate tail of T6D is completely buried in the binding cavity formed by LpqN. Within 6 Å of this hydrocarbon chain, at least eight amino acids are actively involved in anchoring the bound T6D. The trehalose head group of T6D is partially exposed to solvent and is surrounded by six amino acids. Ser-173 also participates by forming two hydrogen bonds with the sugar-ring system of T6D to secure the ligand (Fig. S4). The crystal structures of LpqN indeed support the idea that this lipoprotein is capable of binding cell envelope lipids, such as TMM.
Discussion
Using genetic methods, we demonstrated that LpqN interacts with the periplasmic D2 loop domains of the mycolate transporters MmpL3 and MmpL11. As such, LpqN may act as an MFP to facilitate substrate transport by MmpL3 and MmpL11. The outcome of this interaction is likely proper maturation of lipid substrates and/or the correct localization of these lipids within the cell envelope. We hypothesize that LpqN accomplishes these functions through direct binding of MmpL-transported lipids, as our structural data suggest.
Several observations suggest that there is substantial redundancy in this system. The cell envelope lipid profile of planktonic and 3-week biofilm-grown Mtb ΔlpqN mutant did not differ from the WT, implying that the absence of LpqN does not affect the cell envelope lipid composition during these growth phases. Moreover, Mtb ΔlpqN does not exhibit attenuated growth in planktonic culture (Fig. S1C). Because mycolic acid biosynthesis and MmpL3 are essential, we would expect that perturbing mycolic acid incorporation within the cell envelope would impact Mtb growth or viability. Given that lpqN-deficient Mtb grows at a normal rate and has a normal cell envelope lipid profile, LpqN is unlikely to be the sole facilitator of periplasmic mycolic acid transport in Mtb. In support of this, we note that the protein family to which LpqN belongs (PF10738) contains other secreted Mtb proteins such as the lipoprotein LpqT (Rv1016c) and the secreted proline-rich Mtc28 (Rv0040c) (31).
Lipid binding may be a shared feature of the LpqN protein family. The published structure of Mtc28 exhibits structural homology to eukaryotic lipid-binding proteins such as CERT and STARD13, as assessed by the DALI server (34–37). Both LpqT and Mtc28, as well as the unrelated lipoprotein LprG (Rv1411c), interact with the MmpL11 D2 domain, but not the MmpL3 D2 domain, via M-PFC (Table S1). Whether LpqT and Mtc28 are capable of binding lipids is unknown, but LprG binds phosphatidylinositol-containing glycolipids and interacts with Ag85A (28, 33). Thus, there are presumably multiple interconnected and redundant pathways of cell envelope biogenesis operating in the periplasm that are mediated by lipoproteins and other secreted proteins. Some periplasmic lipid mediators, like LpqN, appear capable of interacting with both MmpL3 and MmpL11. Other potential mediators, like LpqT, Mtc28, and LprG, appear to preferentially interact with MmpL11. Although MmpL3 and MmpL11 both export mycolic acid–containing lipids, MmpL3 is essential and therefore more crucial to cell envelope physiology. Because lpqN-deficient Mtb is physiologically unimpaired, there are likely additional MmpL3-interacting periplasmic lipid mediators that remain to be identified. The recent characterization of the MmpL3-associated protein TtfA in M. smegmatis supports the hypothesis that MmpLs function in concert with accessory proteins to transport their lipid substrates (38). LpqN and LpqN family proteins are therefore likely redundant periplasmic mediators of lipid transport.
Our work raises a number of questions regarding the function of LpqN. An M-PFC screen performed in M. smegmatis initially identified LpqN as an interacting partner of MmpL11. We also used this system to show LpqN interaction with MmpL3. However, we were unable to demonstrate a direct biochemical interaction between LpqN and MmpL11 when heterologously co-expressed in M. smegmatis, suggesting a weak interaction (Fig. S1). We also cannot exclude the possibility that the LpqN–MmpL3/11 interaction is indirect and that some putative mycobacterial adaptor protein or molecule is required for LpqN to associate with these transporters. LpqN also appeared to preferentially interact with Ag85A when incubated with purified Mtb culture filtrate proteins. Similarly, we could not successfully co-purify these two proteins when heterologously co-expressed in M. smegmatis (Fig. S2). Again, this may reflect an indirect interaction with a species specific adaptor protein. The observation that Mtb ΔlpqN biofilms appear to mature differently than WT Mtb biofilms potentially suggests temporal modulation of LpqN function and is the focus of ongoing investigation.
Recent work demonstrated that LpqN contributes to Mtb virulence in a manner apart from its role in cell envelope biogenesis. LpqN appears to interact with the host ubiquitin ligase Cbl that down-regulates antibacterial immunity and potentiates Mtb growth during ex vivo macrophage infection (39). Although this capacity of LpqN to function as a virulence factor is ostensibly unrelated to its apparent role in cell envelope biogenesis, this finding underscores the critical importance of the mycobacterial cell envelope and its components to pathogenesis.
Experimental procedures
Bacterial strains and growth conditions
The Mtb WT strain H37Rv was obtained from the ATCC. Mycobacterial strains were routinely maintained in Middlebrook 7H9 liquid medium (Difco) with 0.05% Tween 80 or on Middlebrook 7H10 agar (Difco), both supplemented with albumin dextrose salts (ADSs) containing 8.1 mg/ml NaCl, 50 mg/ml BSA, and 20 mg/ml dextrose. Glycerol was added to liquid 7H9 to a final concentration of 0.5%. Kanamycin (25 μg/ml) and hygromycin (50 μg/ml) were used to maintain bacterial selection when required.
The Mtb ΔlpqN mutant was created via allelic exchange. Upstream and downstream regions of lpqN homology were amplified by PCR using ΔlpqN 5′/3′ Forward/Reverse primers (Table S2) and cloned to flank the hygromycin resistance gene in pYUB854-rpsL (40). The resulting linear hygromycin product flanked by regions of lpqN homology was amplified by PCR, purified, concentrated, and used to transform electrocompetent H37Rv/pJV53 (41). Transformants were selected on 7H10 agar containing ADS and hygromycin (75 μg/ml). Individual colonies were transferred to 7H10 agar containing hygromycin (50 μg/ml). Deletion of lpqN was confirmed via PCR using flanking primers. Loss of the pJV53 recombineering plasmid was accomplished through iterative culturing on 7H10 agar in the absence of kanamycin and confirmed by susceptibility to kanamycin (25 μg/ml). For complementation, lpqN + ∼1000-bp upstream sequence was amplified via PCR with lpqN −865/+1637 primers and cloned into the integrative vector pMV306 (42). The resulting complementation plasmid was transformed into the Mtb ΔlpqN mutant and transformants selected on 7H10 agar containing ADS and kanamycin (25 μg/ml) and confirmed via PCR (Fig. S1, A and B).
Mycobacterial biofilms were grown in Sauton's medium containing 0.5 g/liter K2HPO4, 0.5g/liter MgSO4, 4.0 g/liter l-asparagine, 0.05 g/liter ferric ammonium citrate, 4.76% glycerol, and 1.0 mg/liter ZnSO4, with a final pH of 7.0. Mtb biofilms were inoculated to A600 = 0.05 in Sauton's medium and incubated at 37 °C in 5% CO2 in tightly sealed polystyrene bottles. At 2 weeks, the lids were loosened to permit gas exchange.
Lipid isolation and analyses
Mtb lipids were harvested by collecting and centrifuging planktonic or biofilm cultures in 50-ml conical vials, washing once with PBS, and then moving to glass test tubes containing 3-mm glass beads. Surface lipids were extracted by shaking with hexanes (∼5 ml) for 2 min, followed by pelleting (1000 × g/10 min). Supernatants were removed into fresh glass tubes and treated with HPLC-grade CHCl3:MeOH (2:1, v/v) prior to removal from biosafety containment. For total lipid extraction, bacterial pellets were autoclaved prior to removal from biosafety containment, then extracted with ∼5 ml of CHCl3:MeOH (2:1, v/v) and 1 ml of HPLC-grade H2O by shaking and bath sonication. The organic layer was separated by centrifugation (1000 × g/10 min), transferred to preweighed vial, and dried under inert N2 gas. mAGP was isolated as previously described (43). Mycolic acid methyl esters (MAMEs) were obtained by incubating mAGP in 15% tetrabutylammonium hydroxide (overnight, 100 °C) followed by iodomethane (4 h, room temperature) and extraction with dichloromethane. Extracted lipids were resuspended in CHCl3 or dichloromethane (MAMEs) and loaded onto aluminum-backed silica plates (EMD Millipore) for TLC analysis. Solvent systems used were chloroform:methanol:ammonium hydroxide (80:20:2, v/v/v), hexanes:ethyl acetate (99:5, v/v, three developments), and toluene:acetone (99:1, v/v) (Fisher). Lipids were visualized via spraying with 12-molybdophosphoric acid (Alfa Aesar; 10%, w/v in EtOH) and charring. The extracted lipids were also subjected to LC/MS analysis using Agilent 6550A QTOF system in conjunction with Agilent 1290 HPLC. HPLC separation and MS data acquisition were performed as described previously (52).
Mycobacterial protein fragment complementation assay
The M-PFC screen was previously described and relies on the reconstitution of two domains of the DHFR protein, which confers resistance to trimethoprim (32). The MmpL3 and MmpL11 D1 and D2 periplasmic domains were amplified by PCR, and the products were subcloned into pGEM-T Easy (Promega). Once confirmed by sequencing, the pUAB200 integrative plasmids were constructed such that the D1 and D2 domains were in-frame with the DHFR [F3] fragment. The pUAB200 was then transformed into M. smegmatis mc2155 generating bait strains. An H37Rv Mtb prey library of over 106 independent clones was generated in pUAB300. Briefly, H37Rv genomic DNA was partially digested with HpaI, and fragments between 0.5 and 2 kb were cloned into the ClaI site of pUAB300. Library DNA was generated by a MaxiPrep (Qiagen). To screen the library, electrocompetent bait strains were transformed with the Mtb library DNA and plated to Kan25/Hyg50/Trim50 plates. Screens were considered successful if they resulted in 106 kanRhygR transformants. Clones that grew on trimethoprim were screened by PCR to eliminate those that grew because of the presence of pUAB300 plasmids containing the Mtb dfrA (Rv2763c) gene. Sequencing of the prey Mtb genomic library plasmid identified the genes fused to DHFR [F1,2]. The MmpL11 D2–LpqN interaction was confirmed by direct cloning of LpqN lacking its signal sequence into pUAB300 to generate in frame fusions with the DHFR [F1,2] fragment. The MmpL D1 and D2–LpqN interactions were measured by determining the trimethoprim minimal inhibitory concentration. Similarly, LpqT, LprG, and Mtc28 were cloned without their signal sequences into pUAB300 to assess their interaction with MmpL3/11 D2 domains. A pUAB300 plasmid containing the Mtb dfrA (Rv2763c) gene was used as a positive control. Strains were serially diluted onto 7H11 agar containing 2-fold dilutions of trimethoprim from 6.25 to 200 μg/ml.
Protein purification
The Escherichia coli LpqN expression construct was created by amplifying lpqN lacking its signal sequence via PCR with primers lpqN.61/687 followed by subcloning into pGEM-T Easy. Upon sequence confirmation, the product was cloned into pET-15b (Novagen) in frame with an N-terminal six-histidine tag. This construct was transformed into E. coli BL21λDE3, and protein production was induced at A600 = ∼0.5 with 0.4 mm IPTG (∼1 h/37 °C). Bacteria were harvested via centrifugation and lysed via sonication. Protein was affinity-purified over HisPur Cobalt Superflow agarose resin (Thermo) and confirmed by Western blotting analysis.
LpqN–CFP pulldowns
Recombinantly expressed LpqN and CFPs (BEI Resources, NR-14825, lot no. 610560541) (500 μg each) were incubated overnight at 4 °C in buffer A (300 mm NaCl, 50 mm NaPO4). Cross-linking was performed using 1% formaldehyde for 30 min at room temperature, followed by quenching with 225 mm glycine for 5 min. Cross-linked samples were buffer-exchanged back into buffer A prior to LpqN purification on HisPur Cobalt Superflow agarose resin (Thermo). LpqN from the non–cross-linked sample was similarly purified. Background CFP binding was assessed by flowing 500 μg of CFP alone over the cobalt resin. All resin-immobilized samples were washed (twice in buffer A and once in buffer A + 10 mm imidazole) and eluted in buffer A + 200 mm imidazole. LpqN-interacting proteins and CFP background were identified by MS at the Oregon Health & Science University Proteomics Shared Resource.
Protein mass spectrometry
0.5 ml of each eluate was concentrated and buffer-exchanged with Amicon Ultra 3-kDa molecular mass cutoff filter. Samples were solubilized in 0.05% ProteaseMAX, reduced with DTT, alkylated with iodoacetamide, digested with trypsin overnight, and dried. Each sample was dissolved in 20 μl of 5% formic acid, and 20 μl/sample was injected into Orbitrap Fusion. Sample digests were loaded onto an Acclaim PepMap 0.1 × 20 mm NanoViper C18 peptide trap (Thermo Scientific) for 5 min at a 5 μl/min flow rate in a 2% acetonitrile, 0.1% formic acid mobile phase, and peptides separated using a PepMap RSLC C18, 2-μm particle, 75-μm × 25-cm EasySpray column (Thermo Scientific) using a 7.5–30% acetonitrile gradient over 60 min in mobile phase containing 0.1% formic acid and a 300 nl/min flow rate using a Dionex NCS-3500RS UltiMate RSLC nano UPLC system. Tandem MS data were collected using an Orbitrap Fusion Tribrid mass spectrometer configured with an EasySpray NanoSource (Thermo Scientific). Survey scans were performed in the Orbitrap mass analyzer at 120,000 resolution, and data-dependent MS2 scans were performed in the linear ion trap using higher-energy collisional dissociation following isolation with the instrument's quadrupole. Comet (version 2015.01, revision 1) was used to search MS2 Spectra against an April 2017 version of the uniprot_Mycobacterium.tuberculosis_h37rv_both.fasta (8352 entries) FASTA protein database, with concatenated sequence-reversed entries to estimate error thresholds and 179 common contaminant sequences and their reversed forms (44). The database processing was performed with Python scripts available at https://github.com/pwilmart/fasta_utilities.git.3 Comet searches for all samples were performed with trypsin enzyme specificity. The average parent ion mass tolerance was 2.5 Da. Monoisotopic fragment ion mass tolerance was 1.005 Da. A static modification of +57.02146 Da was added to all cysteine residues. A variable modification of +15.99491 Da on methionine residues was also allowed.
LpqN expression and purification for crystallization studies
Briefly, the LpqN protein containing a His6 tag at the C terminus was overproduced in E. coli BL21(DE3) cells possessing pET15bΩlpqN. The cells were grown in 2 liters of LB medium with 100 μg/ml ampicillin at 37 °C. When the A600 reached 0.5, the culture was treated with 0.2 mm IPTG to induce LpqN expression, and the cells were harvested within 3 h. The collected bacterial cells were suspended in 100 ml of ice-cold buffer containing 20 mm Na-HEPES (pH 7.2), 200 mm NaCl, 10 mm MgCl2, and 0.2 mg DNase I (Sigma–Aldrich). The cells were then lysed with a French pressure cell. Cell debris was removed by centrifugation for 45 min at 4 °C and 20,000 rpm. The crude lysate was filtered through a 0.2-μm membrane and was loaded onto a 5-ml Hi-Trap Ni2+-chelating column (GE Healthcare Biosciences, Pittsburgh, PA) pre-equilibrated with 20 mm Na-HEPES (pH 7.2) and 200 mm NaCl. To remove unbound proteins and impurities, the column was first washed with six column volumes of buffer containing 50 mm imidazole, 250 mm NaCl, and 20 mm Na-HEPES (pH 7.2). The LpqN protein was then eluted with four column volumes of buffer containing 300 mm imidazole, 250 mm NaCl, and 20 mm Na-HEPES (pH 7.2). The purity of the protein was judged using 12.5% SDS-PAGE stained with Coomassie Brilliant Blue. The purified protein was extensively dialyzed against buffer containing 20 mm Na-HEPES (pH 7.5), and concentrated to 15 mg/ml.
For the His6 selenomethionyl (SeMet)–substituted LpqN protein expression, a 1-ml LB broth overnight culture containing E. coli BL21(DE3)/pET15bΩlpqN cells was transferred into 20 ml of LB broth containing 100 μg/ml ampicillin and grown at 37 °C. When the A600 value reached 1.2, the cells were harvested by centrifugation at 3000 × g/min for 10 min and then washed two times with 10 ml of M9 minimal salt solution. The cells were resuspended in 20 ml of M9 medium and then transferred into a 2-liter prewarmed M9 solution containing 100 μg/ml ampicillin. The cell culture was incubated at 37 °C with shaking. When the A600 reached 0.4, 100 mg/liter of lysine, phenylalanine, and threonine; 50 mg/liter isoleucine, leucine, and valine; and 60 mg/liter of l-selenomethionine were added. The culture was continued to incubate at 37 °C with shaking for 15 min. Protein expression was the induced with 0.2 mm IPTG, and the cells were harvested within 3 h after induction. The procedures for purifying His6 SeMet– LpqN were identical to those of native LpqN.
Crystallization of LpqN
All crystals of the His6 LpqN protein were obtained using hanging-drop vapor diffusion. The LpqN crystals were grown at room temperature in 24-well plates with the following procedures. A 20-μl protein solution containing 15 mg/ml LpqN protein in 20 mm Na-HEPES (pH 7.5) was mixed with a 2 μl of reservoir solution containing 0.1 m sodium citrate (pH 6.0) and 1.9 m (NH4)2SO4. The resultant mixture was equilibrated against 500 μl of the reservoir solution. Crystals grew to a full size in the drops within 2 weeks. Cryoprotection was achieved by raising the glycerol concentration stepwise to 25% with a 5% increment in each step.
Crystals of SeMet– LpqN were prepared using similar procedures. The reservoir solution for crystallizing SeMet– LpqN is the same as that for native LpqN. These SeMet crystals grew to a full size in the drops within 2 weeks. Cryoprotection was achieved by raising the glycerol concentration stepwise to 25% with a 5% increment in each step.
The LpqN–6LT or LpqN–T6D crystals were prepare by incubating the purified LpqN protein (15 mg/ml) with 0.1% 6LT or 0.1% T6D for 2 h at 4 °C before crystallization. Crystals of LpqN–6LT and LpqN–T6D were then prepared using similar procedures as above. Their crystallization conditions are identical to each other. The reservoir solution for these complex crystals is the same as that for apo-LpqN (0.1 m sodium citrate, pH 6.0, and 1.9 m (NH4)2SO4). Both crystals of LpqN–6LT and LpqN–T6D grew to a full size in the drops within 2 weeks. Cryoprotection was achieved by raising the glycerol concentration stepwise to 25% with a 5% increment in each step.
Data collection, structural determination, and refinement
All diffraction data were collected at 100 K at Beamline 24ID-C located at the Advanced Photon Source, using a Pilatus 6M detector (Dectris Ltd.). Diffraction data were processed using DENZO and scaled using SCALEPACK (45). Crystals of LpqN, LpqN–6LT, LpqN–T6D, and SeMet– LpqN belong to space groups C121, I4, I4, and C121, respectively (Table 3).
The LpqN protein contains two methionines (excluding the N-terminal methionine). Within the asymmetric unit of SeMet– LpqN, these two selenium sites were identified using SHELXC and SHELXD (46) as implemented in the HKL2MAP package (47). Single anomalous dispersion was employed to obtain experimental phases using the program Autosol (48) in PHENIX (49). The resulting phases were then subjected to density modification and NCS averaging using the program PARROT (50) using the native structure factor amplitudes. The SeMet sites were also used to trace the molecule by anomalous difference Fourier maps, where we could ascertain the proper registry of SeMet residues. The initial model of LpqN was constructed manually using program Coot (51). Then the model was refined using PHENIX, leaving 5% of reflections in the Free-R set. Iterations of refinement were performed using PHENIX. Model buildings were done using Coot, which led to the current model (Table 3).
The crystal structures of LpqN–6LT and LpqN–T6D were determined by molecular replacement, utilizing the final structure of LpqN as a search model. The procedures for structural refinement and model building were the same as those of LpqN (Table 3).
LpqN–MmpL11/Ag85A interaction
HA-tagged LpqN was amplified by PCR using lpqN −570.F/+HA.R primers and the resulting product cloned into the pOLYG plasmid to generate pOLYG lpqN HA where LpqN is expressed from its native promoter. MmpL11 was amplified using mmpL11 TAP.F/R primers and ligated in-frame with the tandem affinity-tagged (TAP; His6 and FLAG tag) tag into pOLYG-TAP plasmid to generate pOLYG mmpL11-TAP plasmid. MmpL11 expression in the pOLYG mmpL11 TAP is driven by its native promoter. The lpqN-HA fragment was ligated into the pOLYGmmpL11–TAP plasmid to generate the pOLYGlpqN–HA/mmpL11–TAP co-expression plasmid. HIS-tagged Ag85A was amplified using Ag85A.F/R primers and ligated into the pOLYG plasmid via NEBuilder (New England Biolabs) to generate pOLYGag85A-HIS. The lpqN HA fragment was ligated into the pOLYGag85a-HIS plasmid to generate the pOLYGlpqN-HA/ag85A-HIS co-expression plasmid, as above. To test the LpqN/MmpL11 interaction, M. smegmatis mc2155 was transformed with either pOLYGlpqN-HA or pOLYGlpqN-HA/mmpL11-TAP, and grown in 7H9 liquid medium, as above. Protein cross-linking was induced by adding formaldehyde to a final concentration of 1% for 30 min at room temperature. All samples were quenched with 2.5 m glycine, washed with 1× PBS, and lysed in urea lysis buffer (8 m urea, 300 mm NaCl, 0.5% Nonidet P-40, 50 mm NaH2PO4, 50 mm Tris, Roche Complete protease inhibitor, pH 7.0) via sonication. The lysates were incubated on buffer-equilibrated HisPur Cobalt Superflow agarose resin (Thermo Scientific), washed extensively, and eluted in urea lysis buffer adjusted to 300 mm imidazole. Resin flow through and elutions were separated via SDS-PAGE (10%) and analyzed by Western blotting (1:2500 mouse monoclonal α-FLAG/α-HA; Thermo Scientific). To test the LpqN/Ag85A interaction, M. smegmatis was transformed with pOLYGag85a–His or pOLYGlpqN–HA/ag85A–HIS and cross-linked as above. Washed cell pellets were lysed in 1× PBS + Roche Complete protease inhibitor via sonication and incubated on Pierce HA epitope tag antibody agarose conjugate (Thermo Scientific). Resin-bound proteins were extensively washed in lysis buffer, eluted in 50 mm NaOH, and immediately quenched with 1.0 m Tris, pH 8.5. Lysates, resin flow-through, and elutions were separated via SDS-PAGE (15%) and analyzed by Western blotting (1:1000 rabbit polyclonal α-HIS/α-HA; Thermo Scientific).
Author contributions
G. C. M. conceptualization; G. C. M., H. S., J. L. D., F. F. H., M. R., C.-C. S., E. W. Y., and G. E. P. investigation; G. C. M. writing-original draft; G. C. M., F. F. H., E. W. Y., and G. E. P. writing-review and editing; F. F. H., M. R., C.-C. S., E. W. Y., and G. E. P. formal analysis; E. W. Y. and G. E. P. supervision; G. E. P. funding acquisition; G. E. P. project administration.
Supplementary Material
Acknowledgments
Protein mass spectrometric analysis was performed by the Oregon Health & Science University Proteomics Shared Resource with partial support from National Institutes of Health Core Grants P30EY010572, P30CA069533, and S10OD012246. Lipid mass spectrometric analysis was performed by the Washington University Biomedical Mass Spectrometry Research Resource, which is supported by National Institutes of Health Grants P41-GM103422, P60-DK-20579, and P30-DK56341. This work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). The Pilatus 6M detector on 24-ID-C beam line is funded by a NIH-ORIP HEI grant (S10 RR029205). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357.
This work was supported by NIAID, National Institutes of Health Grants R21 AI113074 (to G. E. P.), R01 AI123148 (to G. E. P. and E. W. Y.), and by Grant T32 AI007472 (to G. C. M.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Tables S1 and S2 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 6E5D, 6E5F, and 6MNA) have been deposited in the Protein Data Bank (http://wwpdb.org/).
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- Mtb
- M. tuberculosis
- TB
- tuberculosis
- M-PFC
- mycobacterial protein fragment complementation
- RMSD
- root-mean-square deviation
- RND
- resistance–nodulation–cell division
- TMM
- trehalose monomycolate
- TDM
- trehalose dimycolate
- MFP
- membrane fusion protein
- PDIM
- phthiocerol dimycocerosate
- DHFR
- dihydrofolate reductase
- CFP
- culture filtrate protein
- 6LT
- dodecyl trehalose
- T6D
- trehalose 6-decanoate
- ADS
- albumin dextrose salt
- MAME
- mycolic acid methyl ester
- IPTG
- isopropyl-β-d-thiogalactopyranoside
- SeMet
- selenomethionyl.
References
- 1. Global Tuberculosis Report (2018) World Health Organization, Geneva, Switzerland [Google Scholar]
- 2. Barry C. E. 3rd, Lee R. E., Mdluli K., Sampson A. E., Schroeder B. G., Slayden R. A., and Yuan Y. (1998) Mycolic acids: structure, biosynthesis and physiological functions. Prog. Lipid Res. 37, 143–179 10.1016/S0163-7827(98)00008-3 [DOI] [PubMed] [Google Scholar]
- 3. Chiaradia L., Lefebvre C., Parra J., Marcoux J., Burlet-Schiltz O., Etienne G., Tropis M., and Daffé M. (2017) Dissecting the mycobacterial cell envelope and defining the composition of the native mycomembrane. Sci. Rep. 7, 12807 10.1038/s41598-017-12718-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marrakchi H., Lanéelle M.-A., and Daffé M. (2014) Mycolic acids: structures, biosynthesis, and beyond. Chem. Biol. 21, 67–85 10.1016/j.chembiol.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 5. Blanc L., Gilleron M., Prandi J., Song O.-R., Jang M.-S., Gicquel B., Drocourt D., Neyrolles O., Brodin P., Tiraby G., Vercellone A., and Nigou J. (2017) Mycobacterium tuberculosis inhibits human innate immune responses via the production of TLR2 antagonist glycolipids. Proc. Natl. Acad. Sci. U.S.A. 114, 11205–11210 10.1073/pnas.1707840114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cambier C. J., Takaki K. K., Larson R. P., Hernandez R. E., Tobin D. M., Urdahl K. B., Cosma C. L., and Ramakrishnan L. (2014) Mycobacteria manipulate macrophage recruitment through coordinated use of membrane lipids. Nature 505, 218–222 10.1038/nature12799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chalut C. (2016) MmpL transporter-mediated export of cell-wall associated lipids and siderophores in mycobacteria. Tuberculosis 100, 32–45 10.1016/j.tube.2016.06.004 [DOI] [PubMed] [Google Scholar]
- 8. Viljoen A., Dubois V., Girard-Misguich F., Blaise M., Herrmann J.-L., and Kremer L. (2017) The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol. Microbiol. 104, 889–904 10.1111/mmi.13675 [DOI] [PubMed] [Google Scholar]
- 9. Melly G., and Purdy G. E. (2019) MmpL proteins in physiology and pathogenesis of M. tuberculosis. Microorganisms 7, E70 10.3390/microorganisms7030070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Domenech P., Reed M. B., and Barry C. E. 3rd (2005) Contribution of the Mycobacterium tuberculosis MmpL protein family to virulence and drug resistance. Infect. Immun. 73, 3492–3501 10.1128/IAI.73.6.3492-3501.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ruggerone P., Murakami S., Pos K. M., and Vargiu A. V. (2013) RND efflux pumps: structural information translated into function and inhibition mechanisms. Curr. Top. Med. Chem. 13, 3079–3100 10.2174/15680266113136660220 [DOI] [PubMed] [Google Scholar]
- 12. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E. 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 10.1038/31159 [DOI] [PubMed] [Google Scholar]
- 13. Grzegorzewicz A. E., Pham H., Gundi V. A. K. B., Scherman M. S., North E. J., Hess T., Jones V., Gruppo V., Born S. E., Korduláková J., Chavadi S. S., Morisseau C., Lenaerts A. J., Lee R. E., McNeil M. R., and Jackson M. (2012) Inhibition of mycolic acid transport across the Mycobacterium tuberculosis plasma membrane. Nat. Chem. Biol. 8, 334–341 10.1038/nchembio.794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Backus K. M., Dolan M. A., Barry C. S., Joe M., McPhie P., Boshoff H. I., Lowary T. L., Davis B. G., and Barry C. E. 3rd (2014) The three Mycobacterium tuberculosis antigen 85 isoforms have unique substrates and activities determined by non-active site regions. J. Biol. Chem. 289, 25041–25053 10.1074/jbc.M114.581579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Belisle J. T., Vissa V. D., Sievert T., Takayama K., Brennan P. J., and Besra G. S. (1997) Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276, 1420–1422 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- 16. Degiacomi G., Benjak A., Madacki J., Boldrin F., Provvedi R., Palù G., Kordulakova J., Cole S. T., and Manganelli R. (2017) Essentiality of mmpL3 and impact of its silencing on Mycobacterium tuberculosis gene expression. Sci. Rep. 7, 43495 10.1038/srep43495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li W., Obregón-Henao A., Wallach J. B., North E. J., Lee R. E., Gonzalez-Juarrero M., Schnappinger D., and Jackson M. (2016) Therapeutic potential of the Mycobacterium tuberculosis mycolic acid transporter, MmpL3. Antimicrob. Agents Chemother. 60, 5198–5207 10.1128/AAC.00826-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pacheco S. A., Hsu F. F., Powers K. M., and Purdy G. E. (2013) MmpL11 protein transports mycolic acid–containing lipids to the mycobacterial cell wall and contributes to biofilm formation in Mycobacterium smegmatis. J. Biol. Chem. 288, 24213–24222 10.1074/jbc.M113.473371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wright C. C., Hsu F. F., Arnett E., Dunaj J. L., Davidson P. M., Pacheco S. A., Harriff M. J., Lewinsohn D. M., Schlesinger L. S., and Purdy G. E. (2017) The Mycobacterium tuberculosis MmpL11 cell wall lipid transporter is important for biofilm formation, intracellular growth and non-replicating persistence. Infect. Immun. 10.1128/IAI.00131-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Delmar J. A., Su C. C., and Yu E. W. (2014) Bacterial multidrug efflux transporters. Annu. Rev. Biophys. 43, 93–117 10.1146/annurev-biophys-051013-022855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Okusu H., Ma D., and Nikaido H. (1996) AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178, 306–308 10.1128/jb.178.1.306-308.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ma D., Cook D. N., Alberti M., Pon N. G., Nikaido H., and Hearst J. E. (1993) Molecular cloning and characterization of acrA and acrE genes of Escherichia coli. J. Bacteriol. 175, 6299–6313 10.1128/jb.175.19.6299-6313.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dinh T., Paulsen I. T., and Saier M. H. (1994) A family of extracytoplasmic proteins that allow transport of large molecules across the outer membranes of gram-negative bacteria. J. Bacteriol. 176, 3825–3831 10.1128/jb.176.13.3825-3831.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Paulsen I. T., Park J. H., Choi P. S., and Saier M. H. (1997) A family of Gram-negative bacterial outer membrane factors that function in the export of proteins, carbohydrates, drugs and heavy metals from Gram-negative bacteria. FEMS Microbiol. Lett. 156, 1–8 10.1016/S0378-1097(97)00379-0 [DOI] [PubMed] [Google Scholar]
- 25. Zgurskaya H. I., and Nikaido H. (1999) Bypassing the periplasm: reconstitution of the AcrAB multidrug efflux pump of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 96, 7190–7195 10.1073/pnas.96.13.7190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Sulzenbacher G., Canaan S., Bordat Y., Neyrolles O., Stadthagen G., Roig-Zamboni V., Rauzier J., Maurin D., Laval F., Daffé M., Cambillau C., Gicquel B., Bourne Y., and Jackson M. (2006) LppX is a lipoprotein required for the translocation of phthiocerol dimycocerosates to the surface of Mycobacterium tuberculosis. EMBO J. 25, 1436–1444 10.1038/sj.emboj.7601048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martinot A. J., Farrow M., Bai L., Layre E., Cheng T.-Y., Tsai J. H., Iqbal J., Annand J. W., Sullivan Z. A., Hussain M. M., Sacchettini J., Moody D. B., Seeliger J. C., and Rubin E. J. (2016) Mycobacterial metabolic syndrome: LprG and Rv1410 regulate triacylglyceride levels, growth rate and virulence in Mycobacterium tuberculosis. PLoS Pathog. 12, e1005351–26 10.1371/journal.ppat.1005351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Touchette M. H., Van Vlack E. R., Bai L., Kim J., Cognetta A. B. 3rd, Previti M. L., Backus K. M., Martin D. W., Cravatt B. F., and Seeliger J. C. (2017) A screen for protein–protein interactions in live mycobacteria reveals a functional link between the virulence-associated lipid transporter LprG and the mycolyltransferase antigen 85A. ACS Infect. Dis. 3, 336–348 10.1021/acsinfecdis.6b00179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sandhu P., and Akhter Y. (2017) Siderophore transport by MmpL5-MmpS5 protein complex in Mycobacterium tuberculosis. J. Inorg. Biochem. 170, 75–84 10.1016/j.jinorgbio.2017.02.013 [DOI] [PubMed] [Google Scholar]
- 30. Tseng T. T., Gratwick K. S., Kollman J., Park D., Nies D. H., Goffeau A., and Saier M. H. (1999) The RND permease superfamily: an ancient, ubiquitous and diverse family that includes human disease and development proteins. J. Mol. Microbiol. Biotechnol. 1, 107–125 [PubMed] [Google Scholar]
- 31. Chim N., Torres R., Liu Y., Capri J., Batot G., Whitelegge J. P., and Goulding C. W. (2015) The structure and interactions of periplasmic domains of crucial MmpL membrane proteins from Mycobacterium tuberculosis. Chem. Biol. 22, 1098–1107 10.1016/j.chembiol.2015.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Singh A., Mai D., Kumar A., and Steyn A. J. (2006) Dissecting virulence pathways of Mycobacterium tuberculosis through protein–protein association. Proc. Natl. Acad. Sci. U.S.A. 103, 11346–11351 10.1073/pnas.0602817103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Drage M. G., Tsai H.-C., Pecora N. D., Cheng T.-Y., Arida A. R., Shukla S., Rojas R. E., Seshadri C., Moody D. B., Boom W. H., Sacchettini J. C., and Harding C. V. (2010) Mycobacterium tuberculosis lipoprotein LprG (Rv1411c) binds triacylated glycolipid agonists of Toll-like receptor 2. Nature Struct. Mol. Biol. 17, 1088–1095 10.1038/nsmb.1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holm L., and Rosenström P. (2010) Dali server: conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kundu P., Biswas R., Mukherjee S., Reinhard L., Dutta A., Mueller-Dieckmann J., Weiss M. S., Pal N. K., and Das A. K. (2016) Structure-based epitope mapping of Mycobacterium tuberculosis secretary antigen MTC28. J. Biol. Chem. 291, 13943–13954 10.1074/jbc.M116.726422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kudo N., Kumagai K., Tomishige N., Yamaji T., Wakatsuki S., Nishijima M., Hanada K., and Kato R. (2008) Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide. Proc. Natl. Acad. Sci. U.S.A. 105, 488–493 10.1073/pnas.0709191105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Thorsell A.-G., Lee W. H., Persson C., Siponen M. I., Nilsson M., Busam R. D., Kotenyova T., Schüler H., and Lehtiö L. (2011) Comparative structural analysis of lipid binding START domains. PLoS One 6, e19521 10.1371/journal.pone.0019521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Fay A., Czudnochowski N., Rock J. M., Johnson J. R., Krogan N. J., Rosenberg O., and Glickman M. S. (2019) Two accessory proteins govern MmpL3 mycolic acid transport in mycobacteria. mBio. 10, e00850–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Penn B. H., Netter Z., Johnson J. R., Von Dollen J., Jang G. M., Johnson T., Ohol Y. M., Maher C., Bell S. L., Geiger K., Golovkine G., Du X., Choi A., Parry T., Mohapatra B. C., et al. (2018) An Mtb-human protein–protein interaction map identifies a switch between host antiviral and antibacterial responses. Mol. Cell 71, 637–648.e5 10.1016/j.molcel.2018.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bardarov S., Bardarov S. Jr, Pavelka M. S. Jr, Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., and Jacobs W. R. Jr. (2002) Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017 10.1099/00221287-148-10-3007 [DOI] [PubMed] [Google Scholar]
- 41. van Kessel J. C., and Hatfull G. F. (2007) Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 10.1038/nmeth996 [DOI] [PubMed] [Google Scholar]
- 42. Stover C. K., de la Cruz V. F., Fuerst T. R., Burlein J. E., Benson L. A., Bennett L. T., Bansal G. P., Young J. F., Lee M. H., and Hatfull G. F. (1991) New use of BCG for recombinant vaccines. Nature 351, 456–460 10.1038/351456a0 [DOI] [PubMed] [Google Scholar]
- 43. Daffe M., Brennan P. J., and McNeil M. (1990) Predominant structural features of the cell wall arabinogalactan of Mycobacterium tuberculosis as revealed through characterization of oligoglycosyl alditol fragments by gas chromatography/mass spectrometry and by 1H and 13C NMR analyses. J. Biol. Chem. 265, 6734–6743 [PubMed] [Google Scholar]
- 44. Wilmarth P. A., Riviere M. A., and David L. L. (2009) Techniques for accurate protein identification in shotgun proteomic studies of human, mouse, bovine, and chicken lenses. J. Ocul. Biol. Dis. Infor. 2, 223–234 10.1007/s12177-009-9042-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Otwinowski Z., and Minor W. (1997) [20] Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 46. Schneider T. R., and Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr. 58, 1772–1779 10.1107/s0907444902011678 [DOI] [PubMed] [Google Scholar]
- 47. Pape T., and Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Cryst. 37, 843–844 10.1107/s0021889804018047 [DOI] [Google Scholar]
- 48. Terwilliger T. C., Adams P. D., Read R. J., McCoy A. J., Moriarty N. W., Grosse-Kunstleve R. W., Afonine P. V., Zwart P. H., and Hung L. W. (2009) Decision-making in structure solution using Bayesian estimates of map quality: the PHENIX AutoSol wizard. Acta Crystallogr D Biol Crystallogr. 65, 582–601 10.1107/S0907444909012098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Adams P. D., Grosse-Kunstleve R. W., Hung L.-W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 58, 1948–1954 10.1107/s0907444902016657 [DOI] [PubMed] [Google Scholar]
- 50. Cowtan K. (2010) Recent developments in classical density modification. Acta Crystallogr D Biol Crystallogr. 66, 470–478 10.1107/S090744490903947X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 52. Howard N. C., Marin N. D., Ahmed M., Rosa B. A., Martin J., Bambouskova M., Sergushichev A., Loginicheva E., Kurepina N., Rangel-Moreno J., Chen L., Kreiswirth B. N., Klein R. S., Balada-Llasat J. M., Torrelles J. B., et al. (2018) Mycobacterium tuberculosis carrying a rifampicin drug resistance mutation reprograms macrophage metabolism through cell wall lipid changes. Nat. Microbiol. 3, 1099–1108 10.1038/s41564-018-0245-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.