Abstract
Recent studies of the tumor genome seek to identify cancer pathways as groups of genes in which mutations are epistatic with one another or, specifically, “mutually exclusive.” Here, we show that most mutations are mutually exclusive not due to pathway structure but to interactions with disease subtype and tumor mutation load. In particular, many cancer driver genes are mutated preferentially in tumors with few mutations overall, causing mutations in these cancer genes to appear mutually exclusive with numerous others. Researchers should view current epistasis maps with caution until we better understand the multiple cause-and-effect relationships among factors such as tumor subtype, positive selection for mutations, and gross tumor characteristics including mutational signatures and load.
Cancer genomes are the products of evolutionary trajectories, in which each new somatic mutation is selected only if it confers a fitness advantage over the existing genetic landscape (Nowell, 1976). Consequently, whether a specific driver mutation arises may depend on the presence of complementary mutations that act synergistically with that particular driver in carcinogenesis. Inversely, a driver mutation is less likely to occur in the presence of an earlier mutation that has common or redundant functional effects in the same molecular pathway. Such dependencies reflect a type of genetic epistasis, as the multiple genes in a pathway contribute non-additively to tumor fitness as a phenotype (Figure 1A).
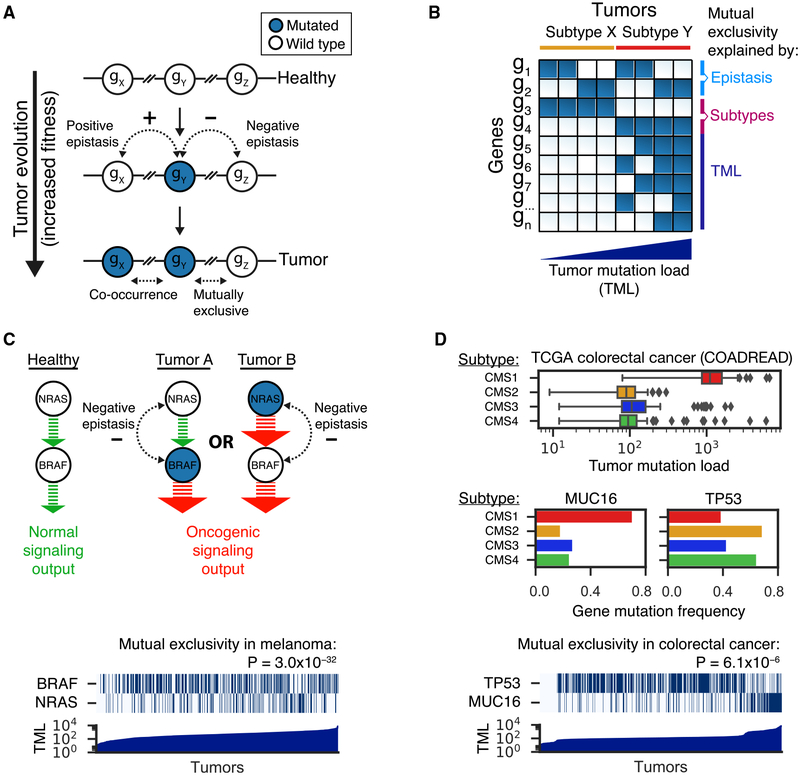
Figure 1. Genetic Epistasis, Tumor Subtypes, and Mutation Load Drive Patterns of Mutual Exclusivity in Cancer Genomes.
(A) Tumor evolution is shown as a stepwise process in which random mutations are sequentially selected if they confer a fitness advantage over the existing genomic landscape. Combinations of mutations in genes that show positive epistasis (gX – gY) result in a surprisingly strong fitness increase that exceeds the additive effects of the individual mutations. Hence, such mutations are more likely to occur together within the same tumor (co-occurrence). Combinations of mutations in genes showing negative epistasis (gY – gZ) result in a surprisingly weak fitness increase that is less than expected from their individual effects. This situation may arise (1) if two mutations deregulate the same molecular pathway and a single mutation is sufficient for a tumor to develop or (2) if the mutations are lethal in combination (synthetic lethality). Therefore, mutations that show negative epistasis are only rarely observed together within the same tumor (mutually exclusive). Blue and white circles represent mutated and wild-type genes, respectively.
(B) Binary matrix of tumors (columns) by genes (rows), in which blue and white squares represent mutated and wild-type states, respectively. Tumors are ranked by subtype and tumor mutation load. Gene mutation patterns are driven by (negative) epistasis (g{1,2}), tumor subtypes (g{3,4}), and tumor mutation load (TML; g{5,6,7,.,n}, with n = total number of genes). Multiple gene pairs show mutual exclusivity, whereas this pattern reflects epistasis only in the case of gene pair g1-g2. The mutual exclusivity between mutations in gene pair g3-g4 is driven by differences in the frequencies of these mutations in subtypes X and Y. As subtype X has relatively low tumor mutation load, the enrichment of g3 mutations within this subtype results in an anti-correlation between the gene mutation pattern of g3 and tumor mutation load. However, most genes (g{5,6,7,.,n}) show the opposite behavior and become more frequently mutated as tumor mutation load increases. This phenomenon drives many cases of mutual exclusivity between mutations in g3 and g{5,6,7,.,n}.
(C) Negative epistasis in cancer signaling pathways can result in patterns of mutual exclusivity, as exemplified by NRAS and BRAF in the TCGA skin cutaneous melanoma cohort. These two melanoma driver genes are successive signaling nodes within the RTK-RAS-RAF-MAPK pathway. Mutations in either of the two genes amplify the signaling output to a level sufficient for tumor progression. Once the first mutation is present, a second mutation does not further increase fitness (less than additive effect – negative epistasis) and is hence not positively selected. Analogous to gene pair g1-g2 in (B), these mutations follow an OR function and are strongly mutually exclusive across the full range of tumor mutation loads (bottom; p value by DISCOVER test). In the heatmap, blues and whites) and represent mutated and wild-type genes, respectively. Tumors were ranked on tumor mutation load (TML).
(D) The interplay between colorectal cancer subtypes (consensus molecular subtypes, CMS tumor mutation load drives strong mutual exclusivity between the genes TP53 and MUC16. CMS1 tumors comprise those with microsatellite instability and hypermutation in comparison to other subtypes (top; boxes, whiskers, and dots indicate quartiles, 1.5 interquartile ranges, and outliers outside these ranges, respectively). MUC16 follows the background mutation rate and is therefore very frequently mutated in CMS1 tumors (middle left). In contrast, mutations in TP53 are relatively infrequent in CMS1 tumors (middle right). This leads to strong mutual exclusivity between mutations in TP53 and MUC16 (bottom; p value by DISCOVER test). Thus, analogous to the many cases of mutual exclusivity between g3 and g{5,6,7,.,n}) in (B), the case of TP53-MUC16 can be explained by subtypes and tumor mutation load. In the heatmap, blues and whites represent mutated and wild-type genes, respectively. Tumors were ranked on tumor mutation load (TML).
Guided by this premise, numerous studies have searched cancer genomes for patterns of mutations that might indicate genetic epistasis and thus the structure of pathways that drive cancer (Babur et al., 2015; Canisius et al., 2016; Ciriello et al., 2012; Hua et al., 2016; Jerby-Arnon et al., 2014; Kim et al., 2015, 2017; Leiserson et al., 2013, 2015a, 2015b, 2016; Lu et al., 2015; Miller et al., 2011; Mina et al., 2017; Szczurek and Beerenwinkel, 2014; Vandin et al., 2012; Wappett et al., 2016; Zhao et al., 2012, 2017). Large-scale tumor-sequencing efforts, including many studies of The Cancer Genome Atlas (TCGA) Research Network (Brennan et al., 2013; Cancer Genome Atlas Network, 2012a, 2012b; Cancer Genome Atlas Research Network, 2008, 2012, 2013, 2014b, 2014c, 2014d, 2017; Ciriello et al., 2015; Ding et al., 2018; Fish-bein et al., 2017; Ge et al., 2018; Kandoth et al., 2013; Ley et al., 2013; Robertson et al., 2017; Sanchez-Vega et al., 2018; Schaub et al., 2018; Seiler et al., 2018) and others (Bolouri et al., 2018; Ma et al., 2018; Reddy et al., 2017), have also conducted epistasis testing as integral analyses. Generally, these studies look for “mutual exclusivity” among gene mutations, i.e., sets of genes in which joint mutation (more than one gene mutated per tumor) is less common than would be expected by random chance (Figure 1B, g1, g2). For example, activating mutations in NRAS or BRAF show strong mutual exclusivity in cutaneous melanoma, consistent with their functional roles as successive signaling nodes within the mitogen-activated protein kinase signaling pathway (Figure 1C).
Genetic epistasis within cancer pathways is not the only explanation for mutual exclusivity, however. To the extent that different tumor subtypes are characterized by different driver mutations, mutual exclusivity can simply reflect differences in driver mutation frequencies between subtypes (Figure 1B, g3, g4). For example, GATA3 mutations are frequent in the luminal A and B subtypes of breast cancer, creating mutual exclusivity and thus apparent epistasis with TP53, which is preferentially mutated in the basal-like and HER2-enriched subtypes (Cancer Genome Atlas Network, 2012b).
We considered that tumor subtype might especially complicate mutual-exclusivity-based epistasis detection when sub-types are linked to tumor mutation load, i.e., the genome-wide number of somatic mutations observed in a tumor. Namely, mutations that drive tumor subtypes with low mutation load might be mutually exclusive—and thus appear epistatic—with the vast majority of other genes (Figure 1B, g3, g{5,6,7,.,n}). For example, mutation load is associated with the well-defined consensus molecular subtypes (CMSs) of colorectal cancer and is particularly high in CMS1, which is characterized by hypermutation and loss of microsatellite stability (Figure 1D, top) (Guinney et al., 2015). Through an increased background mutation rate, most genes have higher mutation frequency in a high-mutation-load subtype like CMS1, which is especially clear for long genes such as MUC16 (Figure 1D, middle left). Contrary to this general trend, however, mutations in the cancer gene TP53 are less frequent in CMS1 than in other subtypes (Figure 1D, middle right). Therefore, TP53 mutations exhibit strong mutual exclusivity with mutations in MUC16 (Figure 1D, bottom). This mutual exclusivity is unlikely to be a functional epistatic interaction as the vast majority of mutations in MUC16 are thought to be passenger events (Lawrence et al., 2013; Martincorena et al., 2017). Moreover, TP53 mutations might exhibit strong mutual exclusivity with many other genes like MUC16, whose mutation frequencies steadily increase with tumor mutation load. Other examples of this phenomenon are provided by EGFR, an oncogene that is frequently altered in lung adenocarcinomas of nonsmoking patients, which as a class has low tumor mutation load (Cancer Genome Atlas Research Network, 2014c), and CDH1, which is most often mutated in genomically stable gastric cancer (Cancer Genome Atlas Research Network, 2014a). If such anecdotes were widespread, one would expect that many mutually exclusive gene mutations might arise from an artifact of subtypes and tumor mutation load rather than pathway structure and epistasis.
How Entangled Are Genetic Dependencies with Mutation Load?
To determine the extent of the mutation load artifact, we analyzed somatic mutation data from eight tumor types, employing 13 cohorts from TCGA and the Memorial Sloan Kettering Cancer Center (MSKCC). Focusing on a cancer gene panel of 341 established oncogenes and tumor suppressor genes (Table S1), we used logistic regression to calculate a standardized score of association between the mutation likelihood of each gene and the tumor mutation load (STAR Methods). This score, henceforth called the “mutation load association” (MLA), takes on a value of 0 in cases of no association with mutation load, whereas negative or positive values reflect higher mutation frequencies in tumors with low or high mutation loads, respectively.
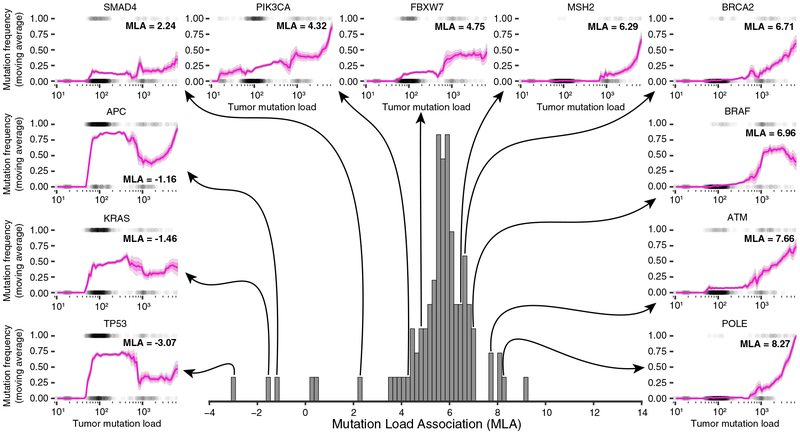
We first analyzed results for the TCGA colorectal adenocarcinoma cohort (COADREAD, 559 tumors). Over all genes within the cancer gene panel, we observed a strong positive association between gene mutation frequency and mutation load (median MLA = 5.74; Figure 2, center histogram). However, this positive association was not universally the case for specific cancer genes (MLA range −3.07 to +9.10). For TP53, the MLA was strongly negative (MLA = −3.07), consistent with previous reports (Cancer Genome Atlas Network, 2012a; Ciriello et al., 2013). We found that the established colorectal cancer drivers KRAS and APC were also more likely to be altered in tumors with low mutation load, with MLAs of −1.46 and −1.16, respectively. As a function of tumor mutation load, the mutation frequencies of these genes followed a complex pattern, in contrast to the generally monotonic functions followed by genes with strongly positive MLAs (e.g., ATM and POLE in Figure 2). We observed a similar MLA spectrum for all other tumor cohorts analyzed (Figure S1A), with the specific MLA of each gene dependent on cancer type (Figures S1B and S1C). Thus, there exists a broad spectrum of tumor-type-specific associations between gene mutation frequency and tumor mutation load, ranging from strongly negative to strongly positive.
Figure 2. Broad Spectrum of Associations between Gene Mutation Frequency and Tumor Mutation Load.
The center histogram shows the distribution of mutation load associations (MLAs) with each cancer gene on the TCGA colorectal adenocarcinoma cohort (COADREAD, 559 tumors). Each MLA represents the association between a gene’s mutation frequency and the mutation load across tumor samples. In the periphery, genes across the whole range of MLAs are explored by plotting their mutation frequencies (y axis) within a moving window of the tumor mutation load (x axis, bin size 0.5 on a log10 scale). Standard error (SE) and 0.5 × SE are plotted in light and dark magenta, respectively. Tumors are plotted as gray dots (1, mutated; 0, not mutated).
Broadening the analysis to encompass all genes in the human genome, we found that the 25 genes with lowest MLA were enriched for known cancer genes, in all tumor types tested (odds ratios 5.4–49.6, p = 1.0 × 10−2 to 7.9 × 10−12; Figure S1D). Conversely, although some known cancer genes had high MLA (e.g., ATM and BRAF; Figure 2), as a set, high-MLA genes were not enriched for cancer genes (Figure S1D). Thus, genes with low MLA tend to be known cancer genes.
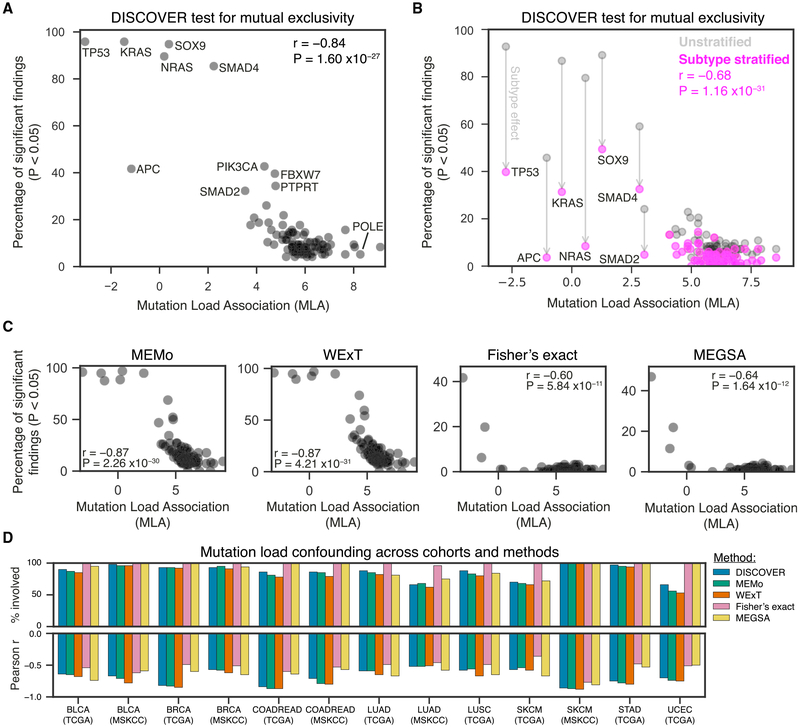
Next, we explored whether mutual exclusivity is more likely detected for genes with low MLA. We considered five methods: DISCOVER (Canisius et al., 2016), MEMo (Ciriello et al., 2012), WExT (Leiserson et al., 2016), Fisher’s exact test, and MEGSA (Hua et al., 2016). Strikingly, for all methods we found strong negative correlations between the MLA of a gene and the number of statistically significant findings (Pearson r range −0.60 to −0.87; Figures 3A–3C). This correlation, which we call “mutation load confounding,” implies that the more negative a gene’s MLA, the higher the number of other genes that show mutual exclusivity with that particular gene. For example, of 96 tested gene-gene combinations involving TP53, the gene having the most negative MLA, nearly all were reported by DISCOVER, MEMo, and WExT to be mutually exclusive (92, 95, and 95, respectively). Being the more conservative tests for mutual exclusivity (Canisius et al., 2016), Fisher’s exact and MEGSA detected fewer mutual exclusivities in general and, hence, also for TP53 (40 and 48, respectively). Together, the 15 genes with lowest MLA were responsible for 78% or more of the significant interactions reported by a method: DISCOVER: 622 of 724 (86%); MEMo: 769 of 948 (81%); WExT: 819 of 1053 (78%); Fisher’s exact test: 65 of 65 (100%); MEGSA: 85 of 85 (100%). This pervasive mutation load confounding generalized to all tumor cohorts tested (Figures 3D and S2–S6) and held for more stringent significance thresholds (data not shown) as well as when both mutations and copy-number alterations were considered (Figure S7).
Figure 3. A Gene’s Association to Tumor Mutation Load Predicts the Number of Significant Findings in Mutual Exclusivity Tests.
(A) Scatterplot of the percentage of the full set of pairwise interactions of a gene that had a p value below 0.05 according to the DISCOVER mutual-exclusivity-detection method, plotted against the gene’s MLA. Results were obtained on the TCGA COADREAD cohort (559 tumors).
(B) As in (A), with pink and gray dots corresponding to results obtained by a subtype-stratified and unstratified analysis, respectively. Results were obtained on the 498 tumors of the TCGA COADREAD cohort for which consensus molecular subtype (CMS) classification was available.
(C) Scatterplots of the percentage of the full set of pairwise interactions of a gene that had a p value below 0.05 according to the MEMo, WExT, Fisher’s exact test, and MEGSA mutual-exclusivity tests, respectively, plotted against the gene’s MLA. Results were obtained on the TCGA COADREAD cohort (559 tumors).
(D) Bar plots showing mutation load confounding across all 13 tumor cohorts analyzed, for the DISCOVER, MEMo, WExT, Fisher’s exact, and MEGSA mutual-exclusivity tests. Positive bars indicate the percentage of statistically significant interactions that involved at least 1 of the 15 genes with lowest MLA within that cohort (% involved). Negative bars indicate the Pearson correlation between the percentage of the full set of pairwise interactions of a gene that had a p value below 0.05 and MLA (as in A–C).
BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; LUAD, lung adenocarcinoma; SCKM, skin cutaneous melanoma; COADREAD, colorectal adenocarcinoma; STAD, stomach adenocarcinoma; LUSC, lung squamous cell carcinoma; UCEC, uterine corpus endometrial carcinoma; TCGA, The Cancer Genome Atlas; MSKCC, Memorial Sloan Kettering Cancer Center.
We next investigated whether mutation load confounding could be reduced by correcting for tumor subtype. Using colorectal cancer as a case study, we first stratified tumors into their predefined CMSs and then ran DISCOVER to identify mutually exclusive gene mutations corrected for subtype (STAR Methods). This correction greatly reduced the number of gene pairs reported to show mutual exclusivity, especially for pairs that included genes with low MLA (Figure 3B). In total, 58% of all significant findings could be attributed to CMS structure, with some mutation load confounding still present in the stratified analysis (Pearson r = –0.68, p = 1.2 × 10−12). Thus, tumor subtypes contribute to widespread patterns of mutual exclusivity in a manner that partially, but not completely, explains mutation load confounding.
Interactions between Gene Mutations, Tumor Subtype, and Mutational Processes
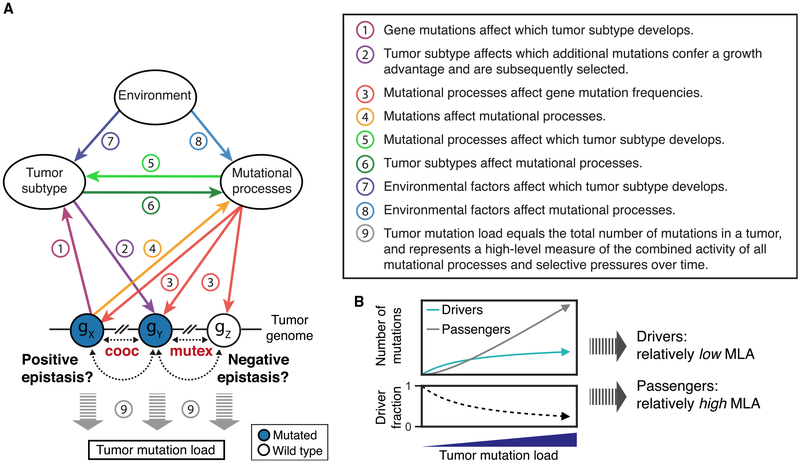
Collectively, the above observations suggest that patterns of mutual exclusivity and co-occurrence of mutations in tumor genomes are reflections of an underlying complex web of interacting factors. Although these factors can include selective pressures on gene function, leading to genetic epistasis, this selective pressure is apparently less frequent, and for some genes perhaps weaker, than confounding influences such as tumor sub-type, mutational processes and load, and environment. Although translating correlations among a set of factors to specific cause-and-effect relationships should be attempted with caution, some possible scenarios are as follows, all of which are supported by specific anecdotes from the cancer literature (Figure 4A).
Figure 4. Conceptual Model of Direct Epistasis and Indirect Paths to Mutual Exclusivity.
(A) The complex interplay between gene mutation frequencies, tumor subtypes, mutational processes, and environment. All of these factors (except environment) can be causally affected by every other factor, leading to a complex system in which both direct gene-gene interactions (epistasis) and indirect causal paths may drive patterns of mutual exclusivity (mutex) or co-occurrence (cooc) between mutations.
(B) The total number of driver mutations per tumor generally increases with tumor mutation load, but with ever decreasing rates (Martincorena et al., 2017). Therefore, driver mutations are less strongly positively associated with tumor mutation load (shallow slope in top; low MLA) compared to passenger mutations (steep slope in top; high MLA). This relatively low MLA may lead to epistasis-independent patterns of mutual exclusivity involving driver genes.
Interactions between Tumor Subtype and Specific Gene Mutations.
Specific mutations can determine which tumor subtype develops (e.g., germline BRCA1 mutations lead to basal-like breast cancer [Mavaddat et al., 2012; Figure 4A, 1]), and inversely, the biological characteristics of a tumor subtype can affect which driver mutations confer a functional advantage and are subsequently selected (e.g., immunogenic CMS1 colorectal cancers lose MHC-I antigen presentation through mutation of B2M [Cancer Genome Atlas Network, 2012a; Kloor et al., 2007; Figures 4A, 2]).
Interactions between Mutational Processes and Specific Gene Mutations.
Active mutational processes determine overall gene mutation frequencies as well as which DNA sequence motifs and genes are most likely to be mutated (Alexandrov et al., 2013) (e.g., mismatch repair deficiency broadly increases gene mutation frequencies, especially in genes with microsatellites [Maruvka et al., 2017; Figure 4A, 3]. Conversely, certain mutational processes can be activated through mutation of specific genes (e.g., BRCA1/2 mutations lead to homologous recombination deficiency and impaired repair of double-strand DNA breaks [Roy et al., 2011; Figure 4A, 4]).
Interactions between Mutational Processes and Tumor Subtypes. Mutational processes can spark the development of a specific tumor subtype (e.g., mismatch repair deficiency leads to CMS1 colorectal cancer [Guinney et al., 2015; Figure 4A, 5]), and in turn, the biology of a tumor sub-type can result in the activation of mutational processes (e.g., HER2-enriched breast cancers have consistently high APOBEC3B expression and show extensive APOBEC cytidine deaminase mutagenesis [Roberts et al., 2013; Figure 4A, 6]).
Environmental Effects on Tumor Subtypes and Mutational Processes.
Environmental factors can underlie the development of certain tumor subtypes (e.g., Epstein-Barr virus drives EBV+ gastric cancer [Cancer Genome Atlas Research Network, 2014a; Figure 4A, 7]) or determine the activity of specific mutational processes (e.g., ultraviolet light exposure leads to DNA damage through C>T transitions [Alexandrov et al., 2013; Figure 4A, 8]).
Within this web of interrelated factors, indirect causal paths can drive misleading patterns of mutual exclusivity in the absence of epistasis. For example, mutations in CDH1 lead to genomically stable (diffuse) gastric cancer (Figure 4A, 1), a sub-type with relatively inactive mutational processes (Figure 4A, 6) (Cancer Genome Atlas Research Network, 2014a; Hansford et al., 2015). The resulting low mutation frequency of other genes in CDH1-mutated tumors (Figure 4A, 3) then leads to mutual exclusivity between CDH1 and these genes.
The result that genes with low MLA tend to be cancer driver genes can be explained in two ways. First, whereas some driver mutations can be associated with tumor subtypes with low tumor mutation load (low MLA), passenger mutations tend to follow the background mutation rate of the tumor and will therefore become more likely as tumor mutation load increases (high MLA). Second, it has been shown that the total number of driver mutations per tumor generally increases with tumor mutation load, but with ever decreasing rates (Martincorena et al., 2017). Thus, as tumor mutation load increases, driver mutations represent an ever-smaller proportion of the total mutation load of the tumor. Although this fact alone is insufficient to result in a negative MLA for cancer genes, it will result in their general shift toward independence from tumor mutation load relative to genes that are predominantly impacted by passenger mutations (MLA = 0; Figure 4B). This logic implies that prioritizing the genes with lowest MLA in a tumor cohort should provide a route to identifying new cancer genes, although we have not pursued that idea here.
Disentangling the Web
Given these complexities, how might we strive to better understand the true epistatic effects of cancer mutations and chart their pathway relationships? First, functional experiments are needed to better understand the cause-and-effect interactions between specific mutations and global mutational processes, as recently explored using DNA-repair-deficient organoids (Drost et al., 2017). Second, statistical models are needed that adjust for mutation load confounding and its underlying factors. While controlling for established cancer subtypes provided a partial adjustment (Figure 3B), a complete solution is likely to require background models of co-mutation that consider the complex, non-linear relationships (Figures 2 and 4A) between gene mutation frequencies and tumor subtypes, mutational processes, and positive selection for mutations, while preventing overfitting. Third, statistical and experimental approaches should be used in a complementary manner, by rigorously validating epistatic relationships arising in genome analysis by, for example, directed perturbations via CRISPR and RNAi (Han et al., 2017; Horlbeck et al., 2018; McDonald et al., 2017; Rauscher et al., 2018; Shen et al., 2017) coupled to appropriate functional readouts. Conversely, statistical models of epistasis within cancer patient genomes are not limited by the artificial context inherent to cell lines and other model systems, and they could therefore be used to confirm the biological and clinical relevance of epistatic relationships identified in experimental models. As these studies progress, they enable the creation of ever-more-precise maps of epistasis in human cancers and, in turn, open up new therapeutic avenues based on these genetic dependencies.
As cancers are driven by combinations of mutations, understanding the epistatic interactions among these mutations will be critical to predict which phenotypes and vulnerabilities are to be expected based on a patient’s cancer genome. Although we are still far away from solving this complex combinatorial problem, we hope that our observations provide a foundation for future studies toward this goal. In the meantime, current epistatic maps of the cancer genome should be interpreted with caution.
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank Michael Kramer, Justin Huang, Jianzhu Ma, Rachel Marty, Philipp Jaeger, Hannah Carter, and members of the Ideker, Carter, Wessels, and Voest laboratories for scientific discussion. We would like to thank Anton Berns for critical reading of the manuscript. We would like to acknowledge the TCGA Research Network and the Memorial Sloan Kettering Cancer Center for providing the data used in the analyses. This work was supported by grants to T.I. from the US National Institutes of Health (U54CA209891 and R01HG009979), to L.F.A.W. from the Netherlands Organization for Scientific Research Gravitation Program (http://cancergenomics.nl/), and to E.E.V. from the Dutch Cancer Society (NKI 2016–1/10014).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.cell.2019.05.005.
DECLARATION OF INTERESTS
T.I. is cofounder of Data4Cure and has an equity interest. T.I. has an equity interest in Ideaya BioSciences. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
REFERENCES
- Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, et al. ; Australian Pancreatic Cancer Genome Initiative; ICGC Breast Cancer Consortium; ICGC MMML-Seq Consortium; ICGC PedBrain (2013). Signatures of mutational processes in human cancer. Nature 500, 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babur Ö , Gönen M, Aksoy BA, Schultz N, Ciriello G, Sander C, and Demir E (2015). Systematic identification of cancer driving signaling pathways based on mutual exclusivity of genomic alterations. Genome Biol 16, 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolouri H, Farrar JE, Triche T Jr., Ries RE, Lim EL, Alonzo TA, Ma Y, Moore R, Mungall AJ, Marra MA, et al. (2018). The molecular landscape of pediatric acute myeloid leukemia reveals recurrent structural alterations and age-specific mutational interactions. Nat. Med 24, 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, Zheng S, Chakravarty D, Sanborn JZ, Berman SH, et al. ; TCGA Research Network (2013). The somatic genomic landscape of glioblastoma. Cell 155, 462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012a). Comprehensive molecular characterization of human colon and rectal cancer. Nature 487, 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network (2012b). Comprehensive molecular portraits of human breast tumours. Nature 490, 61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2008). Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2012). Comprehensive genomic characterization of squamous cell lung cancers. Nature 489, 519–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2013). Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature 499, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014a). Comprehensive molecular characterization of gastric adenocarcinoma. Nature 513, 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014b). Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 507, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014c). Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network (2014d). Integrated genomic characterization of papillary thyroid carcinoma. Cell 159, 676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network, et al. (2017). Integrated genomic and molecular characterization of cervical cancer. Nature 543, 378–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canisius S, Martens JW, and Wessels LF (2016). A novel independence test for somatic alterations in cancer shows that biology drives mutual exclusivity but chance explains most co-occurrence. Genome Biol 17, 261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Cerami E, Sander C, and Schultz N (2012). Mutual exclusivity analysis identifies oncogenic network modules. Genome Res 22, 398–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, and Sander C (2013). Emerging landscape of oncogenic signatures across human cancers. Nat. Genet 45, 1127–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciriello G, Gatza ML, Beck AH, Wilkerson MD, Rhie SK, Pastore A, Zhang H, McLellan M, Yau C, Kandoth C, et al. ; TCGA Research Network (2015). Comprehensive Molecular Portraits of Invasive Lobular Breast Cancer. Cell 163, 506–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding L, Bailey MH, Porta-Pardo E, Thorsson V, Colaprico A, Bertrand D, Gibbs DL, Weerasinghe A, Huang KL, Tokheim C, et al. (2018). Perspective on Oncogenic Processes at the End of the Beginning of Cancer Genomics. Cell 173, 305–320.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drost J, van Boxtel R, Blokzijl F, Mizutani T, Sasaki N, Sasselli V, de Ligt J, Behjati S, Grolleman JE, van Wezel T, et al. (2017). Use of CRISPR-modified human stem cell organoids to study the origin of mutational signatures in cancer. Science 358, 234–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishbein L, Leshchiner I, Walter V, Danilova L, Robertson AG, Johnson AR, Lichtenberg TM, Murray BA, Ghayee HK, Else T, et al. ; Cancer Genome Atlas Research Network (2017). Comprehensive Molecular Characterization of Pheochromocytoma and Paraganglioma. Cancer Cell 31, 181–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z, Leighton JS, Wang Y, Peng X, Chen Z, Chen H, Sun Y, Yao F, Li J, Zhang H, et al. (2018). Integrated Genomic Analysis of the Ubiquitin Pathway across Cancer Types. Cell Rep 23, 213–226.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guinney J, Dienstmann R, Wang X, de Reyniès A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. (2015). The consensus molecular subtypes of colorectal cancer. Nat. Med 21, 1350–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Jeng EE, Hess GT, Morgens DW, Li A, and Bassik MC (2017). Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat. Biotechnol 35, 463–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansford S, Kaurah P, Li-Chang H, Woo M, Senz J, Pinheiro H, Schrader KA, Schaeffer DF, Shumansky K, Zogopoulos G, et al. (2015). Hereditary Diffuse Gastric Cancer Syndrome: CDH1 Mutations and Beyond. JAMA Oncol 1, 23–32. [DOI] [PubMed] [Google Scholar]
- Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR, et al. (2018). Mapping the Genetic Landscape of Human Cells. Cell 174, 953–967.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua X, Hyland PL, Huang J, Song L, Zhu B, Caporaso NE, Landi MT, Chatterjee N, and Shi J (2016). MEGSA: A Powerful and Flexible Framework for Analyzing Mutual Exclusivity of Tumor Mutations. Am. J. Hum. Genet 98, 442–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L, Pfetzer N, Waldman YY, McGarry L, James D, Shanks E, Seashore-Ludlow B, Weinstock A, Geiger T, Clemons PA, et al. (2014). Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 158, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, Benz CC, et al. ; Cancer Genome Atlas Research Network (2013). Integrated genomic characterization of endometrial carcinoma. Nature 497, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YA, Cho DY, Dao P, and Przytycka TM (2015). MEMCover: integrated analysis of mutual exclusivity and functional network reveals dysregulated pathways across multiple cancer types. Bioinformatics 31, i284–i292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YA, Madan S, and Przytycka TM (2017). WeSME: uncovering mutual exclusivity of cancer drivers and beyond. Bioinformatics 33, 814–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloor M, Michel S, Buckowitz B, Rüschoff J, Büttner R, Holinski-Feder E, Dippold W, Wagner R, Tariverdian M, Benner A, et al. (2007). Beta2-microglobulin mutations in microsatellite unstable colorectal tumors. Int. J. Cancer 121, 454–458. [DOI] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Siva-chenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. (2013). Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Blokh D, Sharan R, and Raphael BJ (2013). Simultaneous identification of multiple driver pathways in cancer. PLoS Comput. Biol 9, e1003054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Vandin F, Wu HT, Dobson JR, Eldridge JV, Thomas JL, Papoutsaki A, Kim Y, Niu B, McLellan M, et al. (2015a). Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat. Genet 47, 106–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Wu HT, Vandin F, and Raphael BJ (2015b). CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol 16, 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiserson MD, Reyna MA, and Raphael BJ (2016). A weighted exact test for mutually exclusive mutations in cancer. Bioinformatics 32, i736–i745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley TJ, Miller C, Ding L, Raphael BJ, Mungall AJ, Robertson A, Hoadley K, Triche TJ Jr., Laird PW, Baty JD, et al. ; Cancer Genome Atlas Research Network (2013). Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N. Engl. J. Med 368, 2059–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Megchelenbrink W, Notebaart RA, and Huynen MA (2015). Predicting human genetic interactions from cancer genome evolution. PLoS ONE 10, e0125795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Liu Y, Liu Y, Alexandrov LB, Edmonson MN, Gawad C, Zhou X, Li Y, Rusch MC, Easton J, et al. (2018). Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, Davies H, Stratton MR, and Campbell PJ (2017). Universal Patterns of Selection in Cancer and Somatic Tissues. Cell 171, 1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruvka YE, Mouw KW, Karlic R, Parasuraman P, Kamburov A, Polak P, Haradhvala NJ, Hess JM, Rheinbay E, Brody Y, et al. (2017). Analysis of somatic microsatellite indels identifies driver events in human tumors. Nat. Biotechnol 35, 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mavaddat N, Barrowdale D, Andrulis IL, Domchek SM, Eccles D, Nevanlinna H, Ramus SJ, Spurdle A, Robson M, Sherman M, et al. ; HEBON; EMBRACE; GEMO Study Collaborators; kConFab Investigators; SWEBRCA Collaborators; Consortium of Investigators of Modifiers of BRCA1/2 (2012). Pathology of breast and ovarian cancers among BRCA1 and BRCA2 mutation carriers: results from the Consortium of Investigators of Modifiers of BRCA1/2 (CIMBA). Cancer Epidemiol. Biomarkers Prev 21, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald ER 3rd, de Weck A, Schlabach MR, Billy E, Mavrakis KJ, Hoffman GR, Belur D, Castelletti D, Frias E, Gampa K, et al. (2017). Project DRIVE: A Compendium of Cancer Dependencies and Synthetic Lethal Relationships Uncovered by Large-Scale, Deep RNAi Screening. Cell 170, 577–592.e10. [DOI] [PubMed] [Google Scholar]
- Miller CA, Settle SH, Sulman EP, Aldape KD, and Milosavljevic A (2011). Discovering functional modules by identifying recurrent and mutually exclusive mutational patterns in tumors. BMC Med. Genomics 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mina M, Raynaud F, Tavernari D, Battistello E, Sungalee S, Saghafinia S, Laessle T, Sanchez-Vega F, Schultz N, Oricchio E, et al. (2017). Conditional Selection of Genomic Alterations Dictates Cancer Evolution and Oncogenic Dependencies. Cancer Cell 32, 155–168.e6. [DOI] [PubMed] [Google Scholar]
- Nowell PC (1976). The clonal evolution of tumor cell populations. Science 194, 23–28. [DOI] [PubMed] [Google Scholar]
- Rauscher B, Heigwer F, Henkel L, Hielscher T, Voloshanenko O, and Boutros M (2018). Toward an integrated map of genetic interactions in cancer cells. Mol. Syst. Biol 14, e7656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, Leppa S, Pasanen A, Meriranta L, Karjalainen-Lindsberg ML, et al. (2017). Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 171, 481–494.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SA, Lawrence MS, Klimczak LJ, Grimm SA, Fargo D, Stojanov P, Kiezun A, Kryukov GV, Carter SL, Saksena G, et al. (2013). An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet 45, 970–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson AG, Shih J, Yau C, Gibb EA, Oba J, Mungall KL, Hess JM, Uzunangelov V, Walter V, Danilova L, et al. (2017). Integrative Analysis Identifies Four Molecular and Clinical Subsets in Uveal Melanoma. Cancer Cell 32, 204–220.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Chun J, and Powell SN (2011). BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat. Rev. Cancer 12, 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. (2018). Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 173, 321–337.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaub FX, Dhankani V, Berger AC, Trivedi M, Richardson AB, Shaw R, Zhao W, Zhang X, Ventura A, Liu Y, et al. (2018). Pan-cancer Alterations of the MYC Oncogene and Its Proximal Network across the Cancer Genome Atlas. Cell Syst 6, 282–300.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, Smith PG, Cancer Genome Atlas Research Network, Buonamici S, and Yu L (2018). Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell Rep 23, 282–296.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen JP, Zhao D, Sasik R, Luebeck J, Birmingham A, Bojorquez-Gomez A, Licon K, Klepper K, Pekin D, Beckett AN, et al. (2017). Combinatorial CRISPR-Cas9 screens for de novo mapping of genetic interactions. Nat. Methods 14, 573–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczurek E, and Beerenwinkel N (2014). Modeling mutual exclusivity of cancer mutations. PLoS Comput. Biol 10, e1003503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandin F, Upfal E, and Raphael BJ (2012). De novo discovery of mutated driver pathways in cancer. Genome Res 22, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wappett M, Dulak A, Yang ZR, Al-Watban A, Bradford JR, and Dry JR (2016). Multi-omic measurement of mutually exclusive loss-of-function enriches for candidate synthetic lethal gene pairs. BMC Genomics 17, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang S, Wu LY, and Zhang XS (2012). Efficient methods for identifying mutated driver pathways in cancer. Bioinformatics 28, 2940–2947. [DOI] [PubMed] [Google Scholar]
- Zhao D, Lu X, Wang G, Lan Z, Liao W, Li J, Liang X, Chen JR, Shah S, Shang X, et al. (2017). Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer. Nature 542, 484–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.