Abstract
Background
Polymerase chain reaction‐restriction fragment length polymorphism (PCR‐RFLP) is a common and mature method of detecting the single nucleotide polymorphism (SNP). But, for the polymorphism site rs3863242 of telomeric repeat binding factor 1(TERF1) gene, there is no appropriate restriction enzyme to recognize it, which limits the research between the variants of rs3863242 and human diseases.
Methods
The reverse primer was designed based on turning the 3rd base T into the mismatch base G. After PCR amplification, a new restriction enzyme site was introduced into the TERF1 gene amplification products. Two hundred forty samples from Chinese Han individuals were genotyped to evaluate this method.
Results
A new restriction enzyme site for Cvi QI was introduced into the PCR products. The genotype frequencies of 240 samples from Chinese Han individuals were 4.17% for A/A, 29.58% for A/G, 66.25% for G/G respectively. The allele frequencies were 18.96% for A and 81.04% for G respectively. The genotyping results of PCR products were consistent with the gene sequencing result.
Conclusions
We developed a simple, direct and economical technique for analyzing the polymorphism of TERF1 rs3863242. It may be applied to the colony screening of other SNPs, mutation‐screening of tumor‐related gene or mutations in some specific genes on a large scale, in the future.
Keywords: PCR‐RFLP, rs3863242, Single nucleotide polymorphism, TERF1
Introduction
Telomere is a DNA‐protein complexes located in the terminus of eukaryotic chromosome, comprising with repetitive DNA sequence and shelterin.1 And the shelterin is composed by six subunit protein complex, including TERF1, TERF2, POT1, TPP1, TIN2, and Rap1.2 TERF1 (Telomeric repeat binding factor 1), one important telomeric DNA‐binding protein of the shelterin,3 plays crucial roles in telomere protection,4 the resolution of sister telomeres and the alteration of telomere length.5, 6, 7 Related studies suggested that the TERF1 gene polymorphism is implicated in some diseases, such as: osteosarcoma,8 aplastic anemia9 and so forth.10 And the polymorphism of rs3863242 in TERF1 showed the association with T2D risk.11
The genotyping of SNP could be detected by about four types of approaches: primer extension, ligation amplification, hybridization, and enzyme cleavage. The primer extension is based on common primer extension and could detect multiple SNPs, which needs the expensive instrument and apparatus for discrimination, such as: matrix assisted laser desorption/ionization time‐of‐flight mass spectrometry (MALDI‐TOF MS). The specificity of ligase enzymes is required in the ligation amplification to achieve genotyping. The method of hybridization is according to the differences in thermal stability of double‐stranded DNA to distinguish genotyping,12 which also relies on the expensive instrument and apparatus or GeneChip® array technology.13 The approach of enzyme cleavage, for instance: PCR‐Restriction Fragment Length Polymorphism (PCR‐RFLP) is used to detect the genotypes commonly.14
Although the PCR‐RFLP is a simple, direct and economical technology to detect polymorphisms, it is not perfect. For the SNP site rs3863242, there is no sites recognized by restriction enzymes available now, thus it could not be detected by PCR‐RFLP. In order to circumvent this problem, we introduced a new restriction site into TERF1 amplification products by mismatching primer and verified the possibility of colony screening polymorphism with this technique on a large scale in the Han nationality. The technique is based on distinguishing restriction sites which are artificially created by PCR site‐directed mutagenesis of allele specific site.15, 16
The “TERF1 rs3863242” information was searched from NCBI official website (http://www.ncbi.nlm.nih.gov). The sequence analysis by NEB cutter program (http://nc2.neb.com/NEBcutter2/) showed that no restriction enzymes of New England Biolabs can distinguish the single nucleotide polymorphism site (rs3863242) in the TERF1 gene. Therefore, in order to detect the TERF1 rs38636242 genotypes, the restriction enzyme site of CviQI was designed by mismatching primer.
A pair of primers was designed with the primer premier 5.0 software (PREMIER Biosoft, Palo Alto, CA, USA). And the PCR primer sequences for TERF1 rs3863242 were as below: forward primer 5′‐AATGCTTAGTTTCTTAAAGACGGAAGAC‐3′ and reverse primer 5′‐CAGTAATAATAATGACCCCTAAAAATGGTA‐3′. The bold italic letter with underline was the mismatched base which replaced natural base T. The mechanism that created CviQI restriction enzyme site by mismatching primer was demonstrated in Figure 1A. The corresponding amplification sequence was as follows: AATGCTTAGTTTCTTAAAGACGGAAGACttctctaaacatttcttagatttttaaaagattttacatttatcRTACCATTTTTAGGGGTCATTATTATTACTG.
Figure 1.
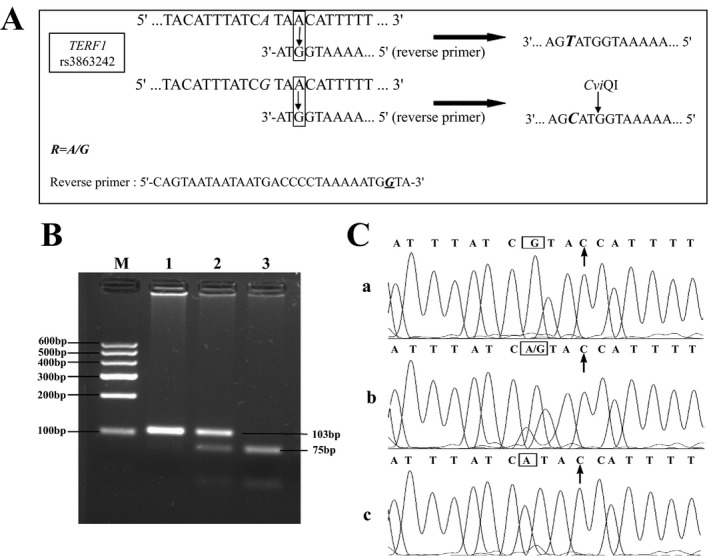
(A) The mechanism of creating Cvi QI restriction enzyme site by mismatching primer. The polymorphism bases R was shown with the letter “A” and “G” in bold italic. The way of mismatching bases were shown in frames. The PCR product was shown on the right. The recognition site GTAT with wave line could not be cut by restriction enzyme Cvi QI, but GTAC could. The G base in bold italic with underline represented the mismatching base of reverse primer. (B) PCR‐RFLP for TERF1 rs3863242 polymorphism by 3% agarose gel. After digestion with Cvi QI, the PCR products with homozygous A/A yielded one uncut band (103 bp), homozygous G/G yielded two bands of 75 bp and 28 bp, and the heterozygous A/G allelic variations yielded three bands of 103 bp, 75 bp and 28 bp. The length of the digestion products were shown on the right of the figure. Genotype analysis (from lane 1 to lane 3), Lane1: TERF1‐A/A; Lane2: TERF1‐A/G; Lane3: TERF1‐G/G. Lane M: molecular weight markers. (C) DNA sequences of the TERF1 PCR products. The C base with vertical arrows corresponds to the mismatched base of reverse primer. The polymorphic site (A/G) was marked with frames. The G base in frame of C1: corresponds to the genotype “G/G”; The A and G bases in frame of C2: correspond to the genotype “A/G” and the A base in frame of C3: corresponds to the genotype “A/A”.
Genomic DNA was extracted from leukocytes in human peripheral blood using genomic DNA isolation kit (Bio Teke Corporation, Beijing, China). And then, the PCR was performed in a final volume of 15 μL, containing 100 ng of genomic DNA, 0.3 μmol/L each primer (Sangon Biotech, Shanghai, China), 2×PCR Mix (Lifefeng Biotech, Shanghai, China) and adjusted with double‐distilled water to 15 μL/reaction. The PCR thermal cycles: lid temperature 96°C; Pre‐degeneration at 95°C for 5 minutes; 35 cycles of 30 seconds at 95°C, 30 seconds at 53.1°C and 30 seconds at 72°C for 30 seconds each, postcycle at 72°C for 5 minutes and at 4°C ended. The PCR products were separated by electrophoresis on the 3% agarose gel stained with EB (ethidium bromide) for 30 minutes, and then investigated the results under the “Gel Doc 2000” UV transilluminator (Bio‐Rad, California, CA, USA).
Enzyme digestion was conducted in a 20 μL final volume and consisted of 5 U of CviQI enzyme (New England Biolab, Cambridge, UK) and 10 μL of PCR product. The reaction was performed overnight at 37°C and the digested products were separated by electrophoresis on the 3% agarose gel containing EB for 40 min, and then observed under the UV transilluminator.
Based on the created restriction enzyme site theory, the TERF1 gene polymorphism could be identified directly. As shown in Figure 1B, the PCR products were separated by electrophoresis on the 3% agarose gel. Apparently, the PCR amplified the fragment of 103 bp. After incubation at 37°C with CviQI, the PCR products with homozygous A/A was not cut and yielded one band (103 bp), homozygous G/G was cleaved into two bands of 75 bp and 28 bp, and the heterozygous A/G allelic variations yielded three bands of 103 bp, 75 bp, and 28 bp.
The polymorphism of TERF1 rs3863242 in 240 Chinese Han individuals were genotyped by the above method. The genotype frequencies of homozygous A/A, G/G or heterozygous A/G allelic variations were 4.17%, 66.25%, and 29.58%. The allele frequencies were 18.96% for A and 81.04% for G. The Genotype distribution for the TERF1 polymorphism conformed to the Hardy–Weinberg equilibrium expectation (χ2 = 0.333, P > .05). To verify the accuracy of our method, we compared the result of enzyme digestion with DNA sequencing (Sangon Biotech, Shanghai, China). The results of DNA sequencing were shown in the Figure 1C. Three figures (from 1 to 3) represent the genotype of TERF1‐G/G, TERF1‐A/G, and TERF1‐A/A respectively. The genotyping results of PCR products proved to be consistent completely with the method of gene sequencing.
Polymerase chain reaction‐restriction fragment length polymorphism, as a rapid and sensitive method has been applied in detecting the gene polymorphism since 1988.17 However, there are still some limitations. For the SNP rs3863242 in TERF1 locus, there are no specific recognition sites for restriction enzymes. In this study, a new PCR‐RFLP assay was created by mismatching the primer to detect the SNP rs3863242 in TERF1 locus. As a result, the DNA amplification carried out successfully though without a perfect primer matched, and the polymorphisms of TERF1 were genotyped accurately.
In conclusion, we developed a precise, direct and economical technique for analyzing the polymorphism of TERF1 rs3863242 accurately. It could be applied to the colony screening of other SNPs, mutation‐screening of tumor‐related gene or mutations in some specific genes et al., in the future.
Compliance with ethical standards
All authors have declared that they have no potential conflicts of interest about this article. The research work was approved by Institutional Ethics Committee of Zhengzhou University. All participants provided written informed consent.
Acknowledgments
The work was supported by the National Natural Science Foundation of China (NSFC81001239), the International Cooperation Projects of Henan Province (152102410007) and Technologies R & D Program of Zhengzhou in Henan (121PPTGG503‐2).
Wang P, Yang Y, Wang S, et al. Detecting the polymorphism of TERF1 gene by an improved PCR‐RFLP method. J Clin Lab Anal. 2018;32:e22171 10.1002/jcla.22171
References
- 1. Blackburn EH, Epel ES, Lin J. Human telomere biology: a contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350:1193–1198. [DOI] [PubMed] [Google Scholar]
- 2. Schmutz I, de Lange T. Shelterin. Current Biol. 2016;26:R397–R399. [DOI] [PubMed] [Google Scholar]
- 3. Lewis Karen A, Wuttke Deborah S. Telomerase and telomere‐associated proteins: structural insights into mechanism and evolution. Structure. 2012;20:28–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ohishi T, Muramatsu Y, Yoshida H, Seimiya H. TRF1 ensures the centromeric function of Aurora‐B and proper chromosome segregation. Mol Cell Biol. 2014;34:2464–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Iwano T, Tachibana M, Reth M, Shinkai Y. Importance of TRF1 for functional telomere structure. J Biol Chem. 2003;279:1442–1448. [DOI] [PubMed] [Google Scholar]
- 6. Canudas S, Houghtaling BR, Kim JY, Dynek JN, Chang WG, Smith S. Protein requirements for sister telomere association in human cells. EMBO J. 2007;26:4867–4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. McKerlie M, Zhu X‐D. Cyclin B‐dependent kinase 1 regulates human TRF1 to modulate the resolution of sister telomeres. Nat Commun. 2011;2:371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mirabello L, Richards EG, Duong LM, et al. Telomere length and variation in telomere biology genes in individuals with osteosarcoma. Int J Mol Epidemiol Genet. 2011;2:19–29. [PMC free article] [PubMed] [Google Scholar]
- 9. Savage SA, Calado RT, Xin ZT, Ly H, Young NS, Chanock SJ. Genetic variation in telomeric repeat binding factors 1 and 2 in aplastic anemia. Exp Hematol. 2006;34:664–671. [DOI] [PubMed] [Google Scholar]
- 10. Brossard M, Fang S, Vaysse A, et al. Integrated pathway and epistasis analysis reveals interactive effect of genetic variants at TERF1 and AFAP1L2 loci on melanoma risk. Int J Cancer. 2015;137:1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zee RY, Ridker PM, Chasman DI. Genetic variants of 11 telomere‐pathway gene loci and the risk of incident type 2 diabetes mellitus: the Women's Genome Health Study. Atherosclerosis. 2011;218:144–146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gibson NJ. The use of real‐time PCR methods in DNA sequence variation analysis. Clin Chim Acta. 2006;363:32–47. [DOI] [PubMed] [Google Scholar]
- 13. Kim S, Misra A. SNP genotyping: technologies and biomedical applications. Annu Rev Biomed Eng. 2007;9:289–320. [DOI] [PubMed] [Google Scholar]
- 14. Kucukkal TG, Yang Y, Chapman SC, Cao W, Alexov E. Computational and experimental approaches to reveal the effects of single nucleotide polymorphisms with respect to disease diagnostics. Int J Mol Sci. 2014;15:9670–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Feng X, Wang S, Duan X, et al. An improved PCR‐RFLP assay for the detection of a polymorphism rs2289487 of PLIN1 gene. J Clin Lab Anal. 2016;30:986–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Haliassos A, Chomel JC, Tesson L, et al. Modification of enzymatically amplified DNA for the detection of point mutations. Nucleic Acids Res. 1989;17:3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Deng GR. A sensitive non‐radioactive PCR‐RFLP analysis for detecting point mutations at 12th codon of oncogene c‐Ha‐ras in DNAs of gastric cancer. Nucleic Acids Res. 1988;16:6231. [DOI] [PMC free article] [PubMed] [Google Scholar]


