Abstract
Introduction:
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) has continued development and standardization of methodologies for biomarkers and has provided an increased depth and breadth of data available to qualified researchers. This review summarizes the over 400 publications using ADNI data during 2014 and 2015.
Methods:
We used standard searches to find publications using ADNI data.
Results:
(1) Structural and functional changes, including subtle changes to hippocampal shape and texture, atrophy in areas outside of hippocampus, and disruption to functional networks, are detectable in presymptomatic subjects before hippocampal atrophy; (2) In subjects with abnormal b-amyloid deposition (Aβ+), biomarkers become abnormal in the order predicted by the amyloid cascade hypothesis; (3) Cognitive decline is more closely linked to tau than Aβ deposition; (4) Cerebrovascular risk factors may interact with Aβ to increase white-matter (WM) abnormalities which may accelerate Alzheimer’s disease (AD) progression in conjunction with tau abnormalities; (5) Different patterns of atrophy are associated with impairment of memory and executive function and may underlie psychiatric symptoms; (6) Structural, functional, and metabolic network connectivities are disrupted as AD progresses. Models of prion-like spreading of Aβ pathology along WM tracts predict known patterns of cortical Aβ deposition and declines in glucose metabolism; (7) New AD risk and protective gene loci have been identified using biologically informed approaches; (8) Cognitively normal and mild cognitive impairment (MCI) subjects are heterogeneous and include groups typified not only by “classic” AD pathology but also by normal biomarkers, accelerated decline, and suspected non-Alzheimer’s pathology; (9) Selection of subjects at risk of imminent decline on the basis of one or more pathologies improves the power of clinical trials; (10) Sensitivity of cognitive outcome measures to early changes in cognition has been improved and surrogate outcome measures using longitudinal structural magnetic resonance imaging may further reduce clinical trial cost and duration; (11) Advances in machine learning techniques such as neural networks have improved diagnostic and prognostic accuracy especially in challenges involving MCI subjects; and (12) Network connectivity measures and genetic variants show promise in multimodal classification and some classifiers using single modalities are rivaling multimodal classifiers.
Discussion:
Taken together, these studies fundamentally deepen our understanding of AD progression and its underlying genetic basis, which in turn informs and improves clinical trial design.
Keywords: Alzheimer’s disease, Mild cognitive impairment, Amyloid, Tau, Biomarker, Disease progression
1. Introduction
The Alzheimer’s Disease Neuroimaging Initiative (ADNI) began in 2005 as a naturalistic longitudinal study to develop and validate biomarkers for subject selection and as surrogate outcome measures in clinical trials of Alzheimer’s disease (AD)-modifying therapies. The initial 5-year study, known as ADNI-1, enrolled 800 participants from 56 study sites in the United States and Canada, in the three groups: normal cognitive aging (CN), mild cognitive impairment (MCI), and early Alzheimer’s disease (AD) [1]. This was extended by a Grand Opportunities grant (ADNI-GO) in 2009, and a competitive renewal of ADNI-1 (ADNI-2) in 2011 [1] with each successive grant enrolling earlier stage patients and incorporating newly developed techniques. A further competitive renewal, ADNI-3, extends the study for another 5 years from 2016 to 2021 [2].
ADNI is structured as a public-private partnership overseen by the Private Partner Scientific Board comprising representatives of private, for-profit entities, and nonprofit organizations which facilitates precompetitive collaboration [3]. ADNI has been described as an exemplar of how these partnerships can impact both clinical and basic science research [4]. First, it has systematically optimized biomarkers for clinical trials through validation, and reproducibility studies, statistical analysis, and the avoidance of bias [5]. Second, ADNI has been an unmitigated success from the standpoint of generating new knowledge about the underlying physiopathology and genetic contributions to AD [6]. These advances have been largely predicated on the development of standardized protocols for use in multiple centers, the emphasis of the initiative on studying multiple modalities, and a policy of open data sharing [7]. ADNI’s approach has proved so successful that its framework has provided inspiration for similar consortia around the world. These include worldwide studies modeled on ADNI [8], as well as initiatives focused on biomarker discovery for diseases such as multiple sclerosis and Parkinson’s disease [1], and Down’s syndrome [9].
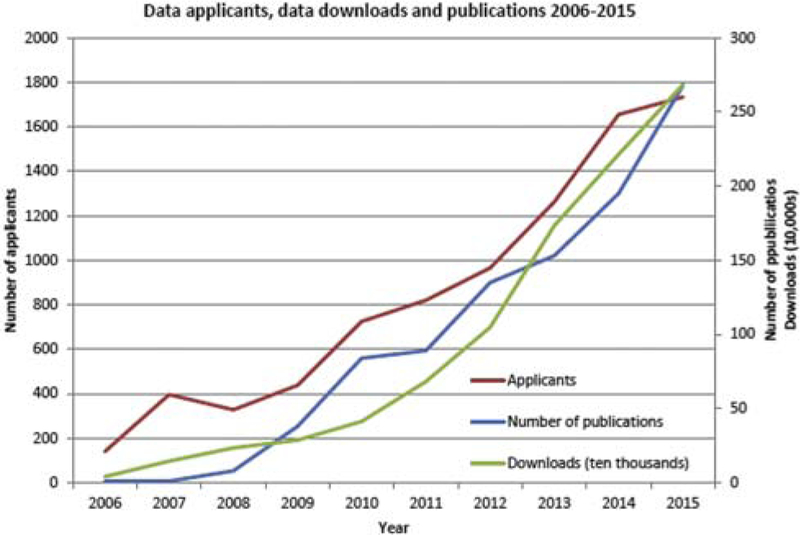
The impact of ADNI’s policy of open data sharing cannot be overemphasized [10]. All data generated by the eight ADNI cores [1] are deposited in the Laboratory of Neuro Imaging (LONI) at the University of Southern California, an informatics infrastructure which, after quality-control procedures, disseminates ADNI data to a continually growing number of investigators in the wider scientific community [11]. LONI has received nearly 1800 applications for data from scientists from multiple disciplines ranging from neuroscience to radiology to genetics to computer science. These investigators have downloaded over 7 million neuroimages and clinical data sets from the ADNI repository [11] resulting in the burgeoning number of scientific studies published using ADNI data over the past decade (Fig. 1).
Fig. 1.
Applications for use of ADNI data, download activity, and the number of ADNI publications per year, 2006–2015. Abbreviation: ADNI, Alzheimer’s Disease Neuroimaging Initiative.
The purpose of this review was to provide a comprehensive overview of the advances in the field of dementia from all studies published (to the best of our knowledge) in 2014 and 2015 using ADNI data (approximately 400). We hope that this will allow investigators to determine what analyses have already been done on ADNI data to help prevent duplication and to identify which questions remain to be answered. Previous successive reviews compiled summaries of publications using ADNI data until the end of 2011 [12], mid-2012 [13], and the end of 2013 [1]. The complete list of ADNI studies may be found at http://adni-info.org/Scientists/ADNIScientistsHome/ADNIPublications.html.
The review is structured in a thematic manner in three parts to reflect evolving views of AD progression. The first part outlines primarily technical advances made by the ADNI Clinical, Magnetic Resonance Imaging (MRI), Positron Emission Tomography (PET), Biomarker, and Genetics cores that do not pertain directly to disease progression or the improvement of clinical trials. The second part takes a holistic approach to the discussion of disease progression, incorporating advances from many ADNI cores. This section addresses in particular (1) the extension of the disease continuum to include the preclinical stage; (2) the importance of Aβ positivity at even preclinical stages; (3) the concept of the disease disrupting structural, functional, and metabolic connectivity in the brain; and (4) the role of white-matter (WM) disease in alternative pathways to dementia. The final part of the study discusses the application of these advances in our knowledge of disease progression to the improvement of clinical trials for AD preventive or modifying therapies, the ultimate goal of ADNI.
2. Cognitive and clinical aspects of Alzheimer’s disease
As cognition lies at the heart of AD, so too does cognitive characterization of the ADNI cohort. This is central to the development of all other biomarkers and to the improvement of clinical trial efficiency. With the focus of disease intervention shifting to the presymptomatic phase, there has been substantial effort in adapting cognitive tests to improve their sensitivity at early disease stages. Recognition that CN and MCI groups selected on the basis of cognition in fact represent a heterogeneous mix of pathologies has spurred studies to identify the basis of that heterogeneity and ultimately increase the power of clinical trials by selecting cohorts with defined pathology. Other studies have identified cognitive measures capable of predicting future decline. Beyond improving clinical trial efficiency, studies have investigated the associations between neuropsychiatric symptoms, or clinical risk factors in AD, and imaging and fluid biomarkers.
During ADNI-2, ADNI’s Clinical Core, led by Dr. Paul Aisen, focused on characterizing the trajectory of subjects in the early stages of disease. This required the development of cognitive and functional measures able to detect the first of subtle cognitive changes [14]. A subjective memory concern (SMC) group with self-reported memory problems was enrolled in ADNI-2 to facilitate investigation of the very earliest cognitive changes. This group was selected using a quantitative approach based on 12 episodic memory items from the self-rating form of the 20 item Cognitive Change Index [15–17]. The Clinical Core then used the self and informant versions of the Everyday Cognition instrument to study the relationship to AD biomarker measures [18]. Across all groups, a total of 1182 subjects were enrolled and followed during ADNI-2 (Table 1), many of whom will be followed in the next phase of the study which will incorporate computerized cognitive assessments.
Table 1.
Clinical and cognitive characteristics of ADNI-2 subjects
| N | CN, N = 314 | SMC, N = 107 | EMCI, N = 300 | LMCI, N = 311 | AD, N = 150 | Combined, N = 1182 | P-value | |
|---|---|---|---|---|---|---|---|---|
| Age | 1182 | 74.1 (5.8) | 72.2 (5.6) | 71.2 (7.4) | 73.1 (7.4) | 74.7 (8.2) | 73.0 (7.0) | <.00l1 |
| Sex: female | 1182 | 158 (50%) | 62 (58%) | 133 (44%) | 127 (41%) | 62 (41%) | 542 (46%) | .0092 |
| Education (years) | 1182 | 16.5 (2.6) | 16.8 (2.5) | 16.0 (2.7) | 16.1 (2.8) | 15.8 (2.7) | 16.2 (2.7) | .0071 |
| Marital | 1182 | |||||||
| Married | 226 (72%) | 70 (65%) | 227 (76%) | 242 (78%) | 130 (87%) | 895 (76%) | <.0012 | |
| Widowed | 42 (13%) | 15 (14%) | 21 (7%) | 38 (12%) | 14 (9%) | 130 (11%) | ||
| Divorced | 33 (11%) | 12 (11%) | 34 (11%) | 26 (8%) | 5 (3%) | 110 (9%) | ||
| Never married | 13 (4%) | 10 (9%) | 14 (5%) | 3 (1%) | 1 (1%) | 41 (3%) | ||
| Unknown | 0 (0%) | 0 (0%) | 4 (1%) | 2 (1%) | 0 (0%) | 6 (1%) | ||
| Ethnicity | 1182 | |||||||
| Not Hisp/Latino | 300 (96%) | 103 (96%) | 284 (95%) | 304 (98%) | 141 (94%) | 1132 (96%) | ||
| Hisp/Latino | 13 (4%) | 2 (2%) | 15 (5%) | 7 (2%) | 8 (5%) | 45 (4%) | ||
| Unknown | 1 (0%) | 2 (2%) | 1 (0%) | 0 (0%) | 1 (1%) | 5 (9%) | .142 | |
| Race | 1182 | |||||||
| Am Indian/Alaskan | 1 (0%) | 0 (0%) | 1 (0%) | 0 (0%) | 0 (0%) | 2 (0%) | ||
| Asian | 7 (2%) | 0 (0%) | 4 (1%) | 5 (2%) | 5 (3%) | 21 (2%) | .0492 | |
| Hawaiian/Other PI | 0 (0%) | 0 (0%) | 1 (0%) | 1 (0%) | 0 (0%) | 2 (0%) | ||
| Black | 21 (7%) | 3 (3%) | 5 (2%) | 10 (3%) | 6 (4%) | 45 (4%) | ||
| White | 283 (90%) | 101 (94%) | 279 (93%) | 294 (95%) | 137 (91%) | 1094 (93%) | ||
| More than one | 2 (1%) | 3 (3%) | 7 (2%) | 1 (0%) | 2 (1%) | 15 (1%) | ||
| Unknown | 0 (0%) | 0 (0%) | 3 (1%) | 0 (0%) | 0 (0%) | 3 (0%) | ||
| CDR-SB | 1182 | 0.030 (0.126) | 0.075 (0.179) | 1.286 (0.757) | 1.630 (0.910) | 4.507 (1.696) | 1.341 (1.622) | <.00l1 |
| ADAS-11 | 1178 | 5.8 (3.0) | 5.6 (2.7) | 7.9 (3.5) | 11.1 (4.6) | 20.8 (7.1) | 9.6 (6.4) | <.00l1 |
| MMSE | 1182 | 29.1 (1.2) | 29.0 (1.2) | 28.4 (1.6) | 27.5 (1.8) | 23.1 (2.1) | 27.7 (2.4) | <.00l1 |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; CN, cognitively normal; SMC, subjective memory concern; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment; AD, Alzheimer’s disease; CDR-SB, Clinical Dementia Rating–Sum of Boxes; ADAS-11, Alzheimer’s Disease Assessment Scale, 11 point; MMSE, Mini–Mental State Examination; SD, standard deviation.
NOTE. Mean (SD) for continuous variables. N is the number of nonmissing values. Tests used:
F test;
Pearson test.
2.1. Psychometric analysis of cognitive tests
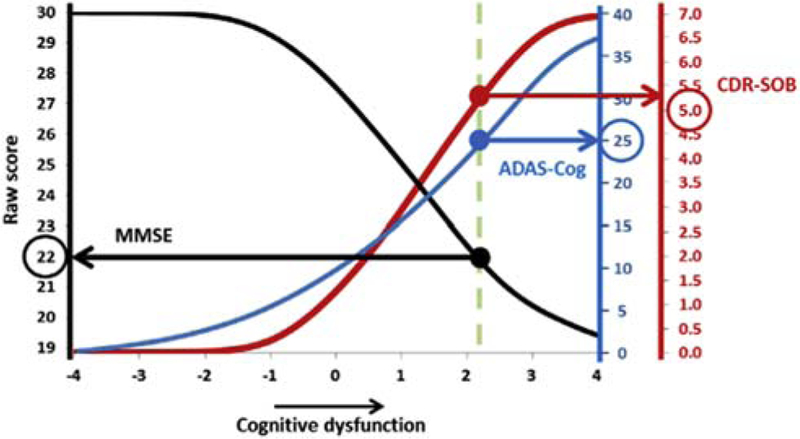
Psychometric analysis was applied to cognitive tests to improve their reliability, to allow the comparison of different cognitive tests, and to increase understanding of the cognitive processes underlying each test. The comparison of studies of cognitive decline is often hampered by the use of multiple cognitive tests. Gross et al. [19] derived summary factors based on the strength of association between cognitive change on a particular test and changes in cortical thickness and hippocampal volume biomarkers. These represented the average rate of cognitive decline and allowed the direct comparison of longitudinal decline on different cognitive tests. Trzepacz et al. provided a conversion table permitting translation of scores between the Montréal Cognitive Assessment and Mini–Mental State Examination (MMSE) [20]. Balsis et al. [21] determined the correspondence between scores on the Alzheimer’s Disease Assessment Scale–cognitive (ADAS-cog), MMSE, and Clinical Dementia Rating–Sum of Boxes (CDR-SB) measures in the entire ADNI cohort (Fig. 2). Their analysis suggested that ADAS-cog and CDR-SB were more precise than MMSE at measuring the severity of cognitive dysfunction. The reliability of ADAS-cog scores for measuring change in more cognitively intact subjects between baseline and 1 year was improved by reweighting the scale subtests for AD subjects, but not MCI subjects, although overall reliability remained low (0.39–0.61 for MCI subjects and 0.53–0.64 for AD subjects) [22].
Fig. 2.
The correspondence among three measures of cognitive dysfunction in Alzheimer’s disease. Latent AD–related cognitive dysfunction was calculated using Item Response Theory methodologies estimated from ADAS-cog, MMSE, and CDR-SOB. Abbreviations: AD, Alzheimer’s disease; ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive subscale; CDR-SOB, Clinical Dementia Rating–Sum of Boxes; MMSE, Mini–Mental State Examination. Reproduced with permission from [21].
Application of a psychometric model to the free recall task of ADAS-cog suggested that impaired patients have deficits in both long-term memory encoding, and short-term memory retrieval, in addition to poorer transfer into long-term memory of items successfully retrieved from short-term memory, and poorer retention of items encoded into long-term memory after long delays [23]. Their immediate recall of encoded words and long-term memory were unaffected. Using a psychometric dual retrieval model, Brainerd et al. [24] found that differences in reconstructive retrieval, rather than recollective retrieval in the delayed recall component of the Rey Auditory Verbal Learning Test (RAVLT) distinguished MCI from AD subjects, and predicted conversion of MCI to AD more accurately than APOE ε4 status, supporting the idea that declines in nonrecollective processes characterize memory loss in AD. A comparison of methods for quantifying how quickly a list of words is learned in a verbal learning test concluded that simple slope calculations, while less highly correlated with structural brain changes, offered ease of calculation advantages over regression-based methods [25]. Finally, psychometric analysis showed that the RALVT 30-minute delayed recall score was the best predictor of Aβ pathology with an accuracy equal to the best imaging biomarker, regional [18F]-fluorodeoxyglucose (FDG) PET measures (area under receiver operating curve [AUC] = 0.67 for both) [26] and that the addition of imaging biomarkers did not significantly improve either predictor. Overall, cognitive tests were more predictive of Aβ status in APOE4 ‒ subjects.
2.2. Associations between cognitive measures and AD biomarkers
Several studies investigated the underlying neural correlates of cognitive measures. In early-stage risk groups (CN to early MCI [EMCI]), Aβ was highly associated with APOE genotype, whereas EMCI subjects characterized by subtle memory performance changes were associated with decreases on structural MRI and metabolism on PET [16]. Episodic memory decline in MCI patients was associated with hippocampal atrophy and basal forebrain degeneration in Aβ+ subjects [27], and the association was mediated by hypometabolism in domain-specific cortical networks. Cognitive impairment in MCI subjects in the absence of hippocampal volume loss was accounted for by changes in hippocampal texture [28]. In Aβ− MCI subjects, episodic memory decline was correlated with hypometabolism in multiple regions outside the temporoparietal areas associated with memory deficits in Aβ+ MCI subjects [29]. Attentional control was associated with basal forebrain degeneration in MCI subjects [27]. Impairment of daily function was associated with greater hypometabolism in middle frontal and orbitofrontal regions [30] and temporal atrophy [31]. Worsening impairment of instrumental activities of daily living was associated with baseline middle frontal and posterior cingulate hypometabolism [30] and predicted by baseline parietal and temporal atrophy [31].
2.3. Associations between neuropsychiatric symptoms and AD biomarkers
Most patients with dementia suffer from neuropsychiatric symptoms (NPSs) such as anxiety, depression, apathy, and psychosis. MCI subjects differed in their trajectories of NPSs, with one group characterized by an initial low NPS burden that remained stable, a second group by an initial moderate NPS burden that worsened, and a final group with an initial high NPS burden that decreased over 2 years [32]. The group with worsening symptoms had the most rapid declines in cognition and function and had a 1.74 fold chance of being diagnosed with AD than the stable group.
Anxiety and irritability are common NPSs endorsed by cognitively impaired subjects. Anxiety was correlated with greater Aβ deposition [33]. Anxiety severity was correlated with increased rate of progression from MCI to AD above and beyond the effects of depression, memory loss, or atrophy and predicted greater rate of entorhinal cortex atrophy [34]. Therefore, anxiety may accelerate cognitive decline by affecting the entorhinal cortex and influencing Aβ deposition. Irritability was correlated with Aβ deposition in parietal regions in AD subjects [33].
Approximately 40% of AD subjects suffer from depressive symptoms. Depression may either be a risk factor for developing dementia, a symptom of dementia, or an early reaction to cognitive loss. MCI converters (MCIc) with depressive symptoms had earlier ages of progression [35] and those with chronic depressive symptoms had a 60% shorter progression time to AD than subjects without this history of depression [36]. These symptoms may exert their effect via modulation of Aβ load, tau pathology, brain structure, and/or metabolism. Amnestic MCI patients with a lifetime history of major depression had higher Aβ deposition in the frontal cortex than controls [37]. Current depressive symptoms in Aβ+ MCI subjects were associated with a higher Aβ load in the frontal, temporal, and insular cortices and with hypometabolism in the frontal cortices compared with nondepressed controls [38]. Depressed AD subjects had a greater correlation between levels of total tau (t-tau) and cortical thickness in the precuneus and parahippocampal cortex [39]. MCI subjects with depressive symptoms who converted to AD within 2 years had greater left hippocampal volume loss compared with converters without depressive symptoms [35]. Chronic depressive symptomatology was associated with accelerated cortical atrophy in the frontal lobe and anterior cingulate [36], and AD subjects with depressive symptoms had greater cortical thinning in the left parietal and temporal regions [39]. Finally, CN, but not MCI or AD, subjects with subsyndromal depression had greater frontal WM lesion volume and smaller orbitofrontal cortical volumes than controls [40].
Psychosis in the form of symptoms, such as delusions, physical aggression, and hallucinations, has long been recognized in AD and is now known to be an independent predictor of more rapid cognitive decline. Substantial evidence suggests that AD with psychosis is a distinct variant of AD with neuropathological specificity and localization [41]. Current psychosis in AD subjects was associated with reduced orbitofrontal brain metabolism, and functional decline, and decline on the MMSE [42], and the onset of psychosis in MCI or AD subjects was most significantly associated with increased atrophy in the lateral frontal lobe [43]. Delusional MCI and AD subjects had greater atrophy in the right frontotemporal regions compared with those without delusions [44]. Hallucinations and apathy also appear to be associated with both changes in brain structure and in metabolism. MCI patients with apathy had decreased metabolism in the posterior cingulate cortex, a landmark region for hypometabolism in AD, compared to subjects without apathy [45]. Greater temporal and parietal atrophy at baseline in CN, MCI, and AD subjects was associated with worsening apathy and hallucinations over 3 years [46]. These studies support the idea that psychosis predominantly affects frontal brain regions, with concomitant reductions in regional glucose metabolism, resulting in an acceleration of cognitive decline.
2.4. Other clinical risk or protective factors
A variety of other clinical factors have been associated with accelerated cognitive decline or in the preservation of cognition. Sleep breathing abnormalities such as snoring or complete airway obstruction have a high prevalence in the elderly and may be associated with cognitive impairment. Subjects with sleep-disordered breathing had an earlier age of progression from both CN to MCI, and MCI to AD than subjects without sleep-disordered breathing, but treatment with continuous positive airway pressure almost completely offset this effect and delayed MCI onset [47]. Epidemiological studies have suggested a link between dietary supplementation with fish oil, cancer history, and educational attainment, among other factors, and preservation of cognition. The use of fish oil supplements, containing omega-3 polyunsaturated fatty acids, in CN APOE4− subjects, was associated with preserved cognition, lower cortical gray-matter (GM) and hippocampal atrophy, and lower ventricular expansion, suggesting that fish oil supplementation may influence cognition by inhibiting brain morphology changes [48]. A history of cancer was associated with a later onset age of AD. Across the ADNI cohort, the number of previous cancer incidences incrementally increased the age of AD onset, from 81.7 to 84.3 to 85.7 years for subjects with zero, one, and two previous cancer incidences, respectively, and patients with a history of cancer showed regional atrophy in the frontal gyrus compared to patients with no cancer history [49]. These results suggest that alternative mechanisms to the metabolic survival theory (positing the metabolic survival of GM in these patients due to the presence of cancer cells that do not undergo apoptosis) may account for the delay in the onset of dementia in cancer survivors. The protective effect of educational attainment on cognition has led to the concept of cognitive reserve although the mechanisms underlying cognitive reserve remain uncertain. Higher education and larger hippocampal volume were significantly associated in AD but not CN or MCI subjects [50]. However, large intracranial volume, a measure of brain reserve, increased the risk of progression to AD and increased the rate of cognitive decline and brain atrophy in male MCI APOE4 noncarriers [51].
Falls are common in cognitively impaired elderly and can have serious consequences—around 13% of falls on the ADNI cohort are reported to be either serious or severe adverse events [52]. Alzheimer’s medication use was associated with hazard of fall after adjusting for age and Beers list medications use, suggesting that clinicians need to be aware of this additional risk when managing medications for AD patients [52]. In addition, the use of medications with high anticholinergic activity in CN subjects was associated with decreases on structural MRI and metabolism on FDG PET, in addition to the known risk of these drugs for cognitive decline [53].
2.5. Conclusions
Recent publications of predominantly clinical and cognitive analyses have steadily expanded our knowledge in numerous areas. Psychometric analyses have resulted in methods of correlating cognitive tests, characterized the nature of memory loss in AD, and even shown that cognitive measures can predict Aβ pathology. Neuropsychiatric symptoms in MCI subjects were associated with more rapid decline and generally found to accelerate atrophy, increase Aβ burden, and decrease metabolism. Psychosis symptoms appeared to mainly affect frontal brain regions. The neuroprotective effects of education, cancer history, and fish oil suggested by epidemiological approaches were supported by studies investigating their association with brain morphology and APOE4 status. Finally, changes on neuroimaging biomarkers in CN subjects were used to demonstrate the adverse effects of anti-cholinergic medications.
3. Magnetic resonance imaging
The ADNI MRI Core has played a central role in the development of biomarkers for clinical trials. Jack et al. [54] reviewed the contributions of the MRI Core over the course of ADNI-1, ADNI-GO, and ADNI-2. Major accomplishments of this Core include the development of standardized protocols for use across different scanner platforms, quality-control methodologies, and algorithms to measure longitudinal change for use as potential outcome measures in clinical trials. The MRI Core has been instrumental in standardizing imaging approaches for clinical trials in addition to structural MRI that reflect both technological development and evolving views of disease progression. These include sequences to image cerebrovascular disease (Fluid Attenuation Inversion Recovery [FLAIR]) and cerebral microbleeds (T2* gradient echo), and functional measures such as perfusion MRI (arterial spin labeling), diffusion MRI (diffusion tensor imaging [DTI]), and task-free functional MRI (TF-MRI, resting-state fMRI). Functional measures were introduced primarily because of their potential to detect early disease-related changes occurring before the atrophic changes detected by structural MRI. Jack et al. [54] present an excellent review of studies pertaining to the MRI Core over the course of ADNI-2.
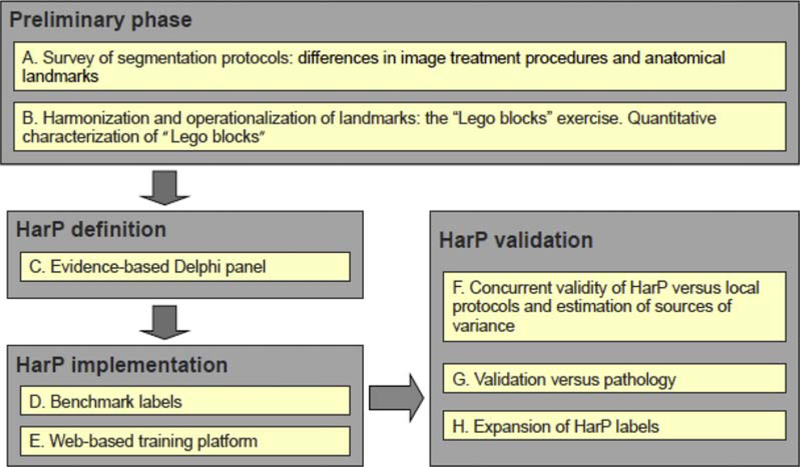
In 2008, ADNI entered into collaboration with the European Alzheimer’s Disease Consortium to develop a harmonized protocol for manual hippocampal segmentation. The goal of this international endeavor was to create a standard definition for hippocampal boundaries and standard data sets to facilitate use of hippocampal volumetry in clinical trials. This project had many phases and succeeded through the concerted effort of many scientists. A summary of how the harmonized segmentation protocol (HarP) was established is presented in Fig. 3.
Fig. 3.
Steps followed to develop the European Alzheimer’s Disease Consortium–ADNI harmonized protocol for manual hippocampal segmentation (HarP). Reproduced with permission from [55].
In the preliminary phase, the most reliable standard orientation for hippocampal volumetry was determined to be perpendicular to the anterior-posterior commissure [56]. Landmark variability across 12 manual segmentation protocols was reduced to four discrete and measurable segmentation units: the alveus/fimbria, whole hippocampal tail, and medial border of the body [57]. An international Delphi panel agreed on the inclusion of these segmentation units, finding that this definition captured 100% of hippocampal tissue and all of AD-related atrophy, and had good intrarater and interrater reliability estimates [58]. Two steps in the implementation of the HarP were the provision of benchmark labels which produced high intraclass and interclass correlation coefficients and could be used for training human tracers [59], and the development of a platform for training and qualifying new tracers to perform manual segmentation using the HarP [60]. Further validation for the protocol came from comparison of hippocampi segmented by tracers following local protocols and then segmented following the HarP [55]. Local protocols were in low agreement compared to the high measurement stability and good reproducibility within and among human tracers using the HarP. Pathological validation revealed that hippocampal volume was significantly correlated to Braak and Braak staging, tau, Aβ burden, and neuronal count and that hippocampal subfields were associated with Aβ, tau, and neuronal count [61]. A set of reference hippocampal labels in the HarP is publicly available on the Web for training and qualification of human tracers and automated algorithms [55,62].
Although manual segmentation is currently considered the gold standard approach to determining hippocampal morphology, the method is time consuming and dependent on the experience of tracers. Therefore, there is much interest in developing automated methods that can successfully segment this highly variable structure in clinical settings as well as for large brain imaging initiatives such as the Enhancing Neuro Imaging Genetics through Meta Analysis (ENIGMA) consortium. The subregion segmentation module in the FreeSurfer software package had high test-retest reliability and trans-platform reliability in 11 of the 12 human hippocampal subregions [63]. FreeSurfer had superior reproducibility of hippocampal volume change over 1 year to manual segmentation after removal of initially visible incorrect automated segmentation [64]. A large number of novel methods for robust and fully automated hippocampal segmentation have been developed and tested on ADNI data. These are summarized in Table 2.
Table 2.
Methods for automatic hippocampal segmentation
| Challenge | Approach and results | Reference |
|---|---|---|
| Hippocampal segmentation | ||
| Multi-atlas segmentation | Presents a unified algorithm, Hippocampal Unified Multi-Atlas Networks (HUMAN), that combines the accuracy of multi-atlas approaches with artificial neural network classification. The algorithm was robust and accurate compared to manual segmentation. | [65] |
| Proposes a learning-based atlas selection method that learns the relationship between each pair of atlases and target images. The method improved segmentation accuracy over other widely used multi-atlas segmentation methods. | [66] | |
| Present a novel segmentation method that uses a manifold learning technique to obtain spatially local weights for atlas label fusion. The weights depend on all pairwise similarities of the population. Segmentation using this method was highly correlated with manual segmentation. | [67] | |
| Propose a graphical approach to labeling using a Markov random field formulation which constructs graphs connecting atlases and the target image. This unified framework allows more efficient label propagation. The method was robust and accurate. | [68] | |
| Present an algorithm Multiple Automatically Generated Templates (MAGeT-Brain) which propagates atlas segmentation is to template library. These are then propagated to each target image and fused using a label fusion method. The method was compared with existing methods including FIRST and FreeSurfer. MAGeT-Brain achieved a higher Dice’s Similarity Coefficient with manual segmentation volumes than produced by FreeSurfer and FIRST. | [69] | |
| Use of hippocampal shape information | Uses spectral Laplace-Beltrami wavelets to obtain high-resolution hippocampal shaped deformations. This resulted in a sensitivity of 96% and a specificity of 90% in the classification of AD versus NC using hippocampal shapes. | [70] |
| Present a method using linear registration of brain images to a standard template, feature extraction, and voxel classification using a random Forest algorithm to determine whether voxels belong to the hippocampus or not. Outperformed FreeSurfer. | [71] | |
| Constructed a high-resolution atlas from manually segmented hippocampal substructures which included manual annotations for neighboring structures. The atlas, released as part of FreeSurfer (version 6.0), outperforms the atlas and FreeSurfer version 5.3. | [72] | |
| Combined that the use of FreeServer, FIRST, and SPHARM software packages to create an atlas by mapping interpolated subfields automation onto the average surface. Atlas has good reproducibility using ADNI data. | [73] | |
| Automated hippocampal segmentation for clinical use | Uses a fully automated multi-atlas segmentation. Found a high Dice Similarity Coefficient with manual segmentation. Suggests that NeuroReader could have clinical applications. | [74] |
| Present a fully automated pipeline using an affine registration step and classification of voxels using a Random Forest classifier. Classification was performed slice by slice along each of three orthogonal directions and achieved comparable results to manual segmentation. | [75] | |
| Present a fully automated pipeline which is atlas based and uses Statistical Parametric Mapping (SPM) software. The automated pipeline was computationally inexpensive, accurate, and is freely available as an SPM8 toolbox. | [76] | |
| Development of a robust hippocampal atrophy biomarker | Describe development of a longitudinal hippocampal atrophy biomarker which is not confounded by factors such as acquisition noise or artifacts, and physiological variations. Biomarker detects hippocampal atrophy due to disease and not to other factors such as long-term aging. In combination with baseline volumes, the method was highly accurate in discriminating patient groups. | [77] |
| Patch label fusion | Proposed a novel patch-based labels fusion method that combines the two approaches via matrix completion. The method results in more accurate segmentation than either with the reconstruction-based or the classification-based approaches. | [78] |
| Present a novel patch-based label fusion framework that uses an optimized PatchMatch Label Fusion (OPAL) strategy. OPAL produced a segmentation accuracy highly correlated with manual segmentation. | [79] | |
| Introduce three new label fusion contributions: (1) the feature representation for each image patch encodes local information; (2) each atlas image patch is further partitioned into partial image patches; (3) label fusion is improved with a hierarchical approach. The improvements proposed resulted in improved accuracy of segmentation. | [80] |
Abbreviations: AD, Alzheimer’s disease; NC, normal cognition; ADNI, Alzheimer’s Disease Neuroimaging Initiative.
The ADNI set of MRI images has been instrumental in the development of new methodologies for improving imaging at multiple stages and in the assessment of existing methodologies. These run the gamut from image acquisition at different field strengths [81–85] to brain extraction strategies [86,87] to improvements in registration and segmentation [88–96] to approaches for measuring longitudinal change [97–107] to cortical thickness estimation [108,109] to better morphometry techniques [110–117]. Many of these studies have been detailed by Jack et al. [54] and are summarized in Table 3.
Table 3.
Approaches for the improvement of MRI methodology
| Challenge | Approach and results | Reference |
|---|---|---|
| Image acquisition | ||
| Evaluate measurement properties of the Brain Atrophy and Lesion Index (BALI) [118] at 1.5-T and 3-TMRI. | Compared T1- and T2-weighted imaging at different field strengths for their ability to correctly diagnose patient groups. Found that 1.5-T BALI scores were similar to those obtained using 3-T images. | [81] |
| Compare 1.5-T and 3-T MRI for automated hippocampal segmentation | Compared the ability of baseline MRI data of patients scanned at both 1.5 T and 3 T to make a clinical diagnosis based on hippocampal radial distance. Found that both field strengths yielded comparable hippocampal atrophy patterns but that 3 T had a superior signal-to-noise ratio and ability to detect atrophy. | [82] |
| Effect of 1.5-T versus 3-T field strengths and image registration strategy on VBM | Tested different diffeomorphic spatial registration strategies over two field strengths for their ability to detect AD-related atrophy. Registration strategy affected the estimation of AD-related atrophy, whereas field strengths affected assessment of brain anatomy in the cerebellum, the precentral gyrus, and the thalamus bilaterally. | [83] |
| Change in 3.0-T MRI image acquisition scheme between ADNI-1 and subsequent grants | Used voxel-based morphometry to compare 3.0-T Tl-weighted volumes obtained in ADNI-1 and ADNI-2. The protocol used in ADNI-2 resulted in increased gray matter and localized decreases in white matter compared to ADNI-1 images and the total volumes of gray matter, white matter, and cerebrospinal fluid also differed. These results should be considered when comparing images obtained during these two protocols using VBM. | [84] |
| Analyzing the effect of geometric distortions on different scanner/protocol combinations. | Used the ADNI phantom to measure MRI image distortion. Found that the size of distortion field varied between scanners and protocols but that corrections applied reduced distortion to 1 mm or less. | [85] |
| Preprocessing | ||
| Different approaches for the normalization of regional volumes by intracranial volume (ICV) may influence the relationship between hippocampus and cognition. | Tested the effect of the three methods: raw volumes, volume to ICV fractions, or regional volumes, on the relationship between hippocampal volume and cognition. Found that the three approaches did not alter this association but had small effects on the prediction of diagnostic status. | [86] |
| Robust and accurate automatic brain extraction across diverse subject groups. | Proposed a method combining an atlas-based approach and a deformable surface-based approach guided by prior information on local intensities and specific populations. Found that the method was accurate across all disease states and across human lifespan and performed favorably compared to existing protocols. | [87] |
| Registration and segmentation | ||
| Selection of the most discriminative features for deformable image registration. | Proposed an image registration framework that uses deep learning to discover morphological patterns in image patches. Achieved more accurate registration results compared to state-of-the-art methods. | [88] |
| Development of a brain parcellation tool based on multi-atlas algorithms that is robust for many different imaging protocols | Used a multiple atlas, likelihood fusion algorithm to test parcellation of the entire brain using six protocols across different manufacturers and field strengths. Found that there was little effect of different protocols on the variability of brain volumes. | [89] |
| Optimal selection of the regularization parameter | Presents a nonregression approach for the selection of the regularization parameter based on the Variational-Bayesian cycle. Found this is more computationally efficient than other methods of noise reduction. | [90] |
| Presents a novel method based on full Bayesian inference on a probabilistic registration model, for inferring spatially varying regularization in nonlinear registration. The proposed model is data driven and its spatially adaptive prior provides better localization of regional volume change. | [91] | |
| Test-retest reliability of automated segmentation methods | Used FreeSurfer to process intrasession and day-to-day scans of subjects. Found that intersession variability exceeded intrasession variability for some regions | [92] |
| Faster image registration | Used three approaches to accelerate the image registration package elastix: (1) parallelization on the CPU; (2) parallelization on the GPU; and (3) improvements of the B-spline transformation model. Reported an acceleration factor of 4 to 5 fold and that the accelerated version had similar classification accuracies to the original version. | [93] |
| Accurate partial volume estimation in tissue labeling | Proposed a fast algorithm based on a Bayesian maximum a posteriori formulation. Algorithm enhanced diagnostic accuracy in ADNI standardized data set. | [94] |
| Automated segmentation of other regions | Proposed a novel automated method for the segmentation of the human brainstem into midbrain and pons called Landmark-based Automatic Brainstem Segmentation (LABS) which uses a revised landmark-based approach integrated with a thresholding method. LABS correlated highly with manual segmentation. | [95] |
| Present a novel segmentation algorithm for measuring change in MTL volume. Baseline MTL volume is defined as an atlas image and mapped onto the corresponding follow-up image to measure volume change. The automated approach measured significant differences between clinical groups, unlike existing FreeSurfer software. | [96] | |
| Longitudinal scans | ||
| Develop scoring and training methods for BALI for the accurate quantitative and of whole-brain structural changes | Trained raters using a step-by-step BALI process. New raters achieved >90% accuracy following 2 weeks of training and achieved both high interrater and intrarater correlation coefficients. Suggests that BALI is a robust method for assessing the whole-brain health in MCI and AD patients. | [97] |
| Use of accelerated versus unaccelerated scans in serial MRI to detect longitudinal change | Used symmetric diffeomorphic image normalization (SyN) to normalize serial scans obtained using TBM. Found that groupwise discrimination and sample size estimates were comparable using accelerated and unaccelerated scans but that the two protocols resulted in differences in TBM-Syn. | [98] |
| Compared the impact of nonaccelerated versus accelerated scans on brain atrophy using the means normalized boundary shift interval (KN-BSI) and deformation-based morphometry. Found differences in measured atrophy rates using scanners from different vendors but little difference between nonaccelerated and accelerated baseline scans and follow-up scans. | [99] | |
| Used morphometry to compare numerical summaries of accelerated versus nonaccelerated scans across patient groups over 6- and 12-month scanned intervals. Scan acceleration had minimal effects on the TBM-derived atrophy measures. | [100] | |
| Prediction of a brain image at a particular time point given minimal longitudinal data | Presented an algorithm for the simultaneous registration of N longitudinal image pairs. Information from each pier is used to constrain the registration of each other pair. The use of a groupwise consistency prior can predict an image act and third time point not included in the registration step. | [101] |
| Presented an algorithm to incorporate information from the entire patient group to predict longitudinal change, as they share similar spatial distributions of volume change. Use longitudinal registration was a groupwise coupling prior and found it able to estimate change robustly. | [102] | |
| Proposed a method for supplementing the lack of longitudinal information for an individual patient with cross-sectional data from the population. Used a probabilistic model based on James Stein estimators to improve geodesic estimation. Method allowed prediction of brain changes of images over time. | [103] | |
| Accounting for spatially inhomogeneous longitudinal data | Proposed a method based on the Sandwich Estimator to account for within-subject correlation in longitudinal data. Found that the method was flexible and fit within-and between-subject effects on the single model in an unbalanced longitudinal data set. | [104] |
| Measuring longitudinal gray-matter volume change in the default mode network | Proposed use of a volume standardized with global gray-matter volume. Method detected significant differences in longitudinal gray matter in the default mode network across patient groups. | [105] |
| Daily changes in brain volume resulting from physiological fluctuations may impact ability of imaging to detect longitudinal changes in brain volume. | Used statistical modeling of MRI images, measuring the brain parenchymal fraction to account for variations in head size. Found a statistically significant time of day effect on brain parenchymal fraction. Suggests that an acquisition time bias should be accounted for in brain volumetric studies. | [106] |
| Improvement of the boundary shift interval for measuring longitudinal change in brain volume | Proposed an extension to the boundary shift interval which uses probabilistic segmentations and then estimates a nonbinary exclusive or a region of interest to better capture patterns of brain atrophy. | [107] |
| Cortical thickness estimation | ||
| Cortical thickness estimation | Presented an algorithm driven by the graph spectrum and heat kernel theory to estimate cortical thickness. Successfully detected statistical differences between patient classes. | [108] |
| Tested the ability of voxel-based morphometry (VBM) to measure cortical thickness. Found that the VBM was less sensitive to cortical atrophy as it was biased to medial temporal lobe atrophy and that FreeSurfer was more sensitive to cortical thinning. | [109] | |
| Morphometry | ||
| Cortical pattern analysis | Proposed a multi-resolution approach which prescribes shape descriptors that characterize the signal at each mesh vertex. Method showed increased sensitivity and statistical power to detect group-level differences. | [110] |
| Ventricular morphology analysis | Presented a novel system for ventricular morphometry based on the hyperbolic Ricci flow method and tensor morphometry statistics. The TBM statistics enhanced surface shape analysis and the method revealed shape differences close to the temporal lobe and posterior cingulate. Correlations were detected between ventricular morphometry, neuropsychological measures, and metabolism. | [111] |
| Representation of overall brain morphology | Proposed a novel approach to deformation-based morphometry, regional flux analysis, based on the scheme halts decomposition of deformations parameterized by stationary velocity fields. The framework unifies voxel-based and regional approaches and had good power to discover shapes deformations both cross-sectionally and longitudinally. | [112] |
| Introduced BrainPrint, a fully automated framework which generates a compact representation of brain morphology by capturing shape information from both cortical and subcortical structures. Method was efficient and discriminative. | [113] | |
| Presented a mass univariate framework that uses longitudinal VBM data and Bayesian inference to analyze whole-brain structural changes over time. The probabilistic model detects individual and group trajectories of disease progression. | [114] | |
| Measuring patterns of brain morphological changes in populations. | Proposed a data-driven probabilistic unsupervised framework that automatically segments heterogeneous set of images using an atlas-based method and clusters images into homogeneous subgroups. It constructs separate probabilistic atlases for each cluster. Found that combining segmentation and atlas construction led to improved segmentation accuracy and clusters generated coincided with clinical subgroups. | [115] |
| Identification of shape deformation patterns | Developed a data-driven global analysis of brain anatomy using kernel partial least squares and a regression model to quantify shape changes that explain variations in clinical neuropsychological measures. Method identified similar patterns in AD to predefined ROIs as well as other new patterns of deformation. | [116] |
| Presented a framework for intrinsic comparison of surface metric structures and curvatures based on a Riemannian framework. Framework was able to efficiently detect boundaries between functionally and structurally distinct regions. | [117] |
Abbreviations: MRI, magnetic resonance imaging; AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; MTL, medial temporal lobe; ROI, regions of interest.
Finally, the ADNI MRI data set has been used in investigations of other neurological conditions. The ADNI control set was used in the comparison of NeuroQuant, an automated measure of brain volume in patients with traumatic brain injury, with manual interpretation of scans [119], and in the investigation of systematic differences in corpus callosum for morphology and periventricular nodular heterotopia [120].
4. Positron emission tomography
The ADNI PET Core, under the leadership of Dr. William J. Jagust, has collected longitudinal data on glucose metabolism, reflecting changes in neuronal metabolism, from FDG PET and on Aβ deposition from the uptake of radiotracers, 11C-Pittsburgh compound (PiB) tracer in ADNI-1, and subsequently the 18F-labeled florbetapir [121].
4.1. FDG PET
The ADNI PET Core has been responsible for the development of standardized procedures for FDG PET [121]. The importance of standardized procedures for FDG PET was underscored by a Cochrane systematic review of studies, including three from ADNI, that used baseline FDG PET measures to predict future MCI to AD progression [122]. The meta-analysis found considerable variability in specificity values, thought to be due to the lack of standardization and a lack of a threshold value for abnormality, and the review concluded that these obstacles precluded the use of FDG PET as a diagnostic modality in clinical practice. The PET Core has addressed these issues by developing image registration strategies to improve the consistency of qualitative values extracted from the scans, improving the quality-control process, and producing standardized sets of preprocessed images available for download from LONI. The PET Core discontinued conducting FDG PET scans on ADNI participants in 2014, having amassed a considerable library of longitudinal scans for future analysis [121].
4.2. Amyloid PET
The importance of Aβ status is underscored by its inclusion in the revised diagnostic criteria for AD [123] and by its use in the selection of asymptomatic subjects likely to progress for therapeutic clinical trials. The ADNI PET Core has worked on methodological quality assurance and control, as well as the standardization of Aβ PET images [121]. Collection of longitudinal data has enabled the examination of rates of Aβ accumulation and its effect on cognitive decline [121]. Longitudinal changes in cortical florbetapir standardized uptake value ratios (SUVRs) were more accurately measured by the use of subcortical WM reference regions compared with the cerebellum or pons [124]. These reference regions also increased the power to detect longitudinal increases in fibrillar Aβ and significant associations between Aβ increases and clinical decline over 24 months and improved the evaluation of Aβ-modifying treatment effects in Aβ+ subjects and CN APOE4 carriers [125].
Are the different Aβ ligands themselves equivalent in terms of detecting Aβ? Chiotis et al. [126] compared 11C PiB and florbetapir PET imaging and found similar mean regional uptake patterns and strong correlations between regions of interest across patient groups. Landau et al. [127] found that cortical retention between three radiotracers, 11C PiB, florbetapir, and 18F-flutemetamol were highly correlated. Although flutemetamol had higher WM retention and florbetapir had lower WM retention compared with PiB, the threshold values for Aβ positivity were consistent when the values were converted using PiB values as an intermediary. Thus, comparison of results obtained using different radiotracers appears to be valid, a conclusion supported by the comparable results using all three measures obtained by Nosheny et al. [128] in their investigation of the effect of Aβ positivity on hippocampal atrophy. A focus of the next phase of ADNI will be the development of the Centiloid scale for the direct comparison of amyloid tracers [2].
4.3. Tau PET imaging
The accumulation of Aβ plaques is only minimally associated with cognitive decline, which appears to be more closely associated with neurofibrillary tangles (NFTs) formed by tau amyloid fibrils [129]. Insoluble fibrillar species of tau assemble into intraneuronal inclusions known as NFTs as well as neuropil threads in neuronal processes, which represent >80% of tau pathology in AD compared with NFTs [130]. Notably, NFTs and neuropil threads in AD brains display all the features of amyloids [131]. Soluble tau detected in cerebrospinal fluid (CSF) is a putative indicator of neuronal damage as indicated by increases in CSF tau after traumatic brain injury [132]. New developments in tracer technology have led to the development of PET ligands that track tau fibrillary amyloid accumulation. The inclusion of this imaging in ADNI-3 [2] will likely help to unfold the contribution of this pathological event to the disease process [133].
5. CSF and blood biomarkers
The study of CSF and blood biomarkers in ADNI is led by the Penn Biomarker Core and overseen by Drs. Leslie M. Shaw and John Q. Trojanowski. A detailed account of progress throughout ADNI-2 and future plans is given by Kang et al. [129]. The primary goals of the Biomarker Core have been to develop CSF and plasma biomarkers signatures to identify AD subjects, CN to MCI progressors, and MCI to AD progressors, and to establish the longitudinal trajectories of CSF and plasma biomarkers. Multimodal data analyses in collaboration with other ADNI Cores have established the temporal order of changes in clinical measures, imaging data, and CSF biomarkers and allowed a greater understanding of AD pathophysiology. A secondary goal has been to develop biomarkers to detect common copathologies such as Lewy bodies, vascular disease, TDP-43 inclusions, and hippocampal sclerosis. Potential novel biomarkers have been identified from genome-wide association studies (GWAS) in collaboration with the Genetics Core and using proteomics approaches. These biomarkers may be able to not only detect AD pathology but also indicate the presence of mixed pathology. Through January 2016, the Biomarker Core received and processed a total of 10,279 biofluids (CSF, plasma, and serum) and prepared and stored 161,301 aliquots for use by qualified investigators. They provided a total of 2635 plasma, 1051 serum, and 3622 CSF blinded aliquot samples to 15 investigators whose request for samples was reviewed and approved during ADNI-2 by the Resource Allocation Review Committee. To track the longitudinal progression of all biomarkers, 265 ADNI subjects have provided series of three or more CSF samples over as much as 8 years. A total of 1248 subjects have provided at least three longitudinal samples of serum and plasma. These have proven invaluable for the study of CSF Aβ42, t-tau, and p-tau181 trajectories in individual subjects [129] and will help establish the trajectories of new biomarkers such as YKL-40, Vilip-1, total and phosphorylated α-SYN (Ser129-α-SYN), as well as neurogranin.
5.1. Established CSF biomarkers
5.1.1. Methodological improvements
The Biomarker Core has improved standard methodologies that allow the accurate comparison of CSF biomarker results across multiple centers. They have developed fully automated analyses of Aβ42, t-tau, and p-tau181 using the accuracy- and precision-based Roche Elecsys immunoassay platform, which improves on the AlzBio3 immunoassay platform. To circumvent difficulties associated with standardizing immunoassays across multiple centers (differences in antibodies, matrix problems, the lack of a CSF-based standard reference material), they have validated a mass spectrometry (MS) assay for Aβ42, Aβ40, and Aβ38 using a 2D-UPLC/MS-MS platform [129,134], calibrated with a surrogate calibrator matrix prepared from artificial CSF plus 4 mg/mL bovine serum albumin. This assay had equivalent diagnostic utility to the AlzBio3 immunoassay in quantifying CSF Aβ42 differences between controls and AD subjects (sensitivity = 92.7%, specificity = 84.5%) [134]. This reference method was developed as part of an international effort to develop reference methodology for Aβ42 [135]. It is expected that this will strongly support ongoing efforts to obtain harmonization across methods and platforms used worldwide for this essential CSF AD biomarker [136].
Another methodological improvement was described by Vidoni et al. [137] who demonstrated that the use of 24 bore atraumatic needles reduced the incidence of postlumbar puncture headache.
5.1.2. Comparison of CSF and PET measures of Aβ
CSF Aβ42 and amyloid PET measures are often assumed to be equivalent, but this may not be a valid assumption because the structure and biophysical properties of Aβ fibrils (measured by amyloid PET) and soluble Aβ (measured by CSF Aβ42) differ [138]. Some CN and stable MCI subjects have abnormally low CSF Aβ measures but no evidence of Aβ amyloid deposits by PET measures, suggesting that low CSF Aβ42 may not always be indicative of the accumulation of PET-detectable fibrillary Aβ deposits or alternatively that CSF Aβ42 becomes abnormal before amyloid PET [139]. However, pathologically low CSF levels of Aβ42 were strongly associated with AD diagnosis and cortical Aβ accumulation independent of APOE genotype, suggesting that abnormally low CSF levels of Aβ42 reflect cortical Aβ deposition and not the APOE genotype [140].
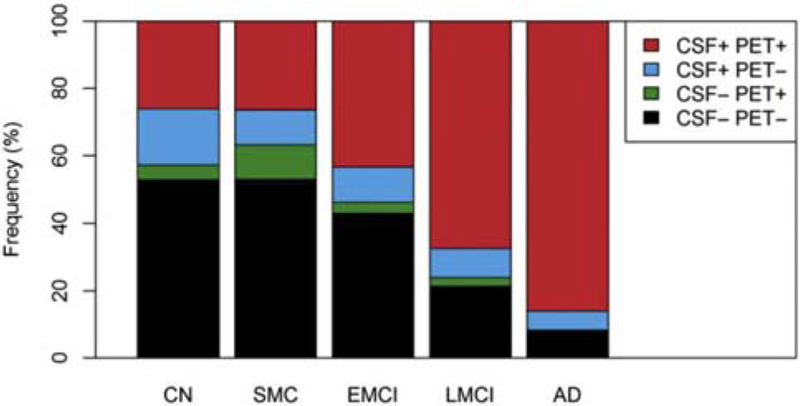
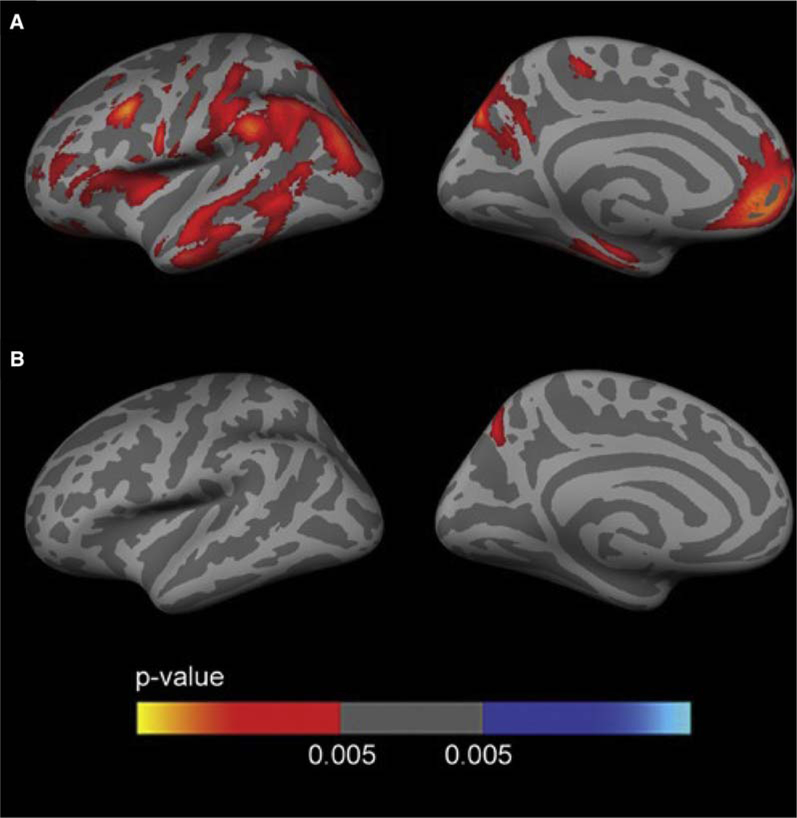
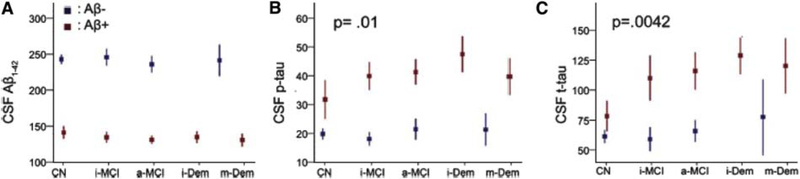
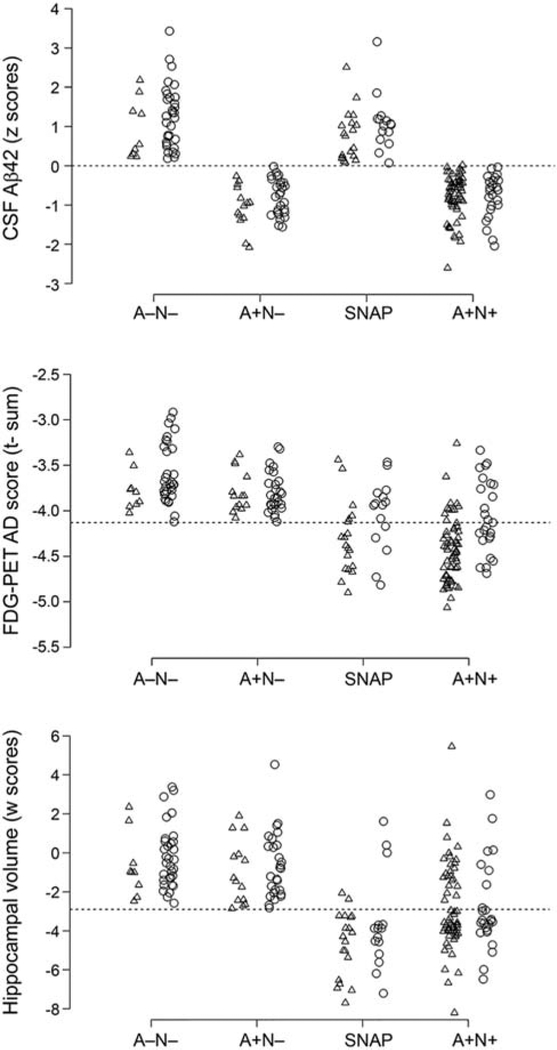
However, the two measures appeared comparable in the classification of MCI converters or AD subjects [139] compared to CN subjects, although florbetapir PET had a greater specificity than CSF Aβ42 for the latter classification. These measures provided partly different and independent information according to a further study by the same group [141]. CSF Aβ42 was more strongly related to APOE4 genotype, whereas PET Aβ was more strongly associated with levels of CSF tau and ADAS-cog scores. Furthermore, discordance between the two measures was most common in CN and SMC subjects (Fig. 4) and concordance between the two was more common in late-stage AD. The results suggest that CSF Aβ42 is a more sensitive marker of very early emergence of Aβ pathologies and that PET Aβ may better reflect later stage AD progression.
Fig. 4.
Frequencies of different CSF and PET Aβ profiles in different diagnostic groups. Subjects were dichotomized by CSF Aβ42 or florbetapir PET and classified as concordant negative (CSF− PET−), discordant (CSF+ PET− and CSF− PET+), and concordant positive (CSF+ PET−). Abbreviations: AD, Alzheimer’s disease; CN, cognitively normal; CSF, cerebrospinal fluid; EMCI, early MCI; LMCI, late MCI; MCI, mild cognitive impairment; PET, positron emission tomography; SMC, subjective memory concern. Reproduced with permission from [141].
5.1.3. Amyloid pathway
Aβ is generated by the sequential action of b-site amyloid precursor protein (APP) cleaving enzyme 1 (BACE1), the target of γ-secretase inhibitors, and β-secretase on the trans-membrane Aβ precursor protein. This process also generates a soluble N-terminal fragment, s-APPβ, reported to be correlated with BACE-1 activity [142]. Independent studies found no differences in either activity of BACE1 [142–144] or concentrations of s-APPβ [142,143] in CSF across all patient groups. Although these measures cannot be used to differentiate between healthy elderly and AD individuals, the results suggest that CSF levels of s-APPβ may be used as a surrogate for BACE-1 activity in clinical trials of β-secretase inhibitors.
5.2. Other CSF and blood analytes
Beyond established CSF biomarkers, Aβ, and p-tau181, there has been much interest in alternative CSF and blood analytes that are in some way associated with the disease process. Some have been already identified as AD risk factors or as being linked to common copathologies. Other novel analytes have been identified using Rules Based Medicine immunoassay technology and may have diagnostic or prognostic utility.
5.2.1. Associations between known AD risk factors in CSF and blood and AD pathology
Abnormally high blood homocysteine is a major cardiovascular risk factor as well as a risk factor for AD and was previously shown to be associated with lower regional WM and GM volumes in ADNI [145]. High levels of plasma homocysteine were associated with a cortical signature of reduced GM thicknesses, volumes, and surface areas in memory networks and the default mode network (DMN) [146], both of which are susceptible to Aβ deposition, metabolic disruption, and atrophy, and consistently implicated in AD. As elevated homocysteine is easily treatable, this cortical biomarker signature may have utility in assessing interventions for lowering homocysteine.
Low plasma levels of the obesity-related hormone leptin have been linked to AD pathogenesis. Conversely, high levels of leptin may function in a protective manner by regulating levels of Aβ in neurons through the inhibition of β-secretase activity and by modulating tau kinases to reduce tau phosphorylation [147]. Johnston et al. [148] reported that leptin levels in plasma reflected those in CSF, and that women had two-fold higher plasma levels of leptin than men in all groups. In women but not men, leptin levels were associated with body mass index. Seventy percent of MCI subjects, of whom half were APOE ε4 allele carriers, had lower plasma leptin than CN subjects. Given that reduced plasma leptin levels have been established in MCI and AD subjects, Maioli et al. [147] examined brain leptin levels across AD progression, including cellular localization of leptin and leptin receptors in the hippocampus and frontal cortex. Although they found no differences in CSF leptin levels across all diagnostic groups, they found that leptin localization in the brain was altered. In AD subjects, distribution shifted to the hippocampus where leptin translocated to become more abundant in reactive astrocytes and less so in neurons. A similar translocation was observed in Aβ+ and APOE ε4+ mice, suggesting an impairment of leptin signaling in AD in the presence of constant levels of the hormone. Animal models of Aβ accumulation indicated that changes in leptin signaling occurred before a downregulation of leptin receptors. The authors suggest a mechanism in which APOE ε4 allele, in conjunction with Aβ accumulation, transiently enhances leptin signaling leading to a leptinresistant state over time and subsequent decrease in cognition. Changes in leptin signaling are likely behind the observation that increased body mass index in the middle age is a risk factor for AD [149].
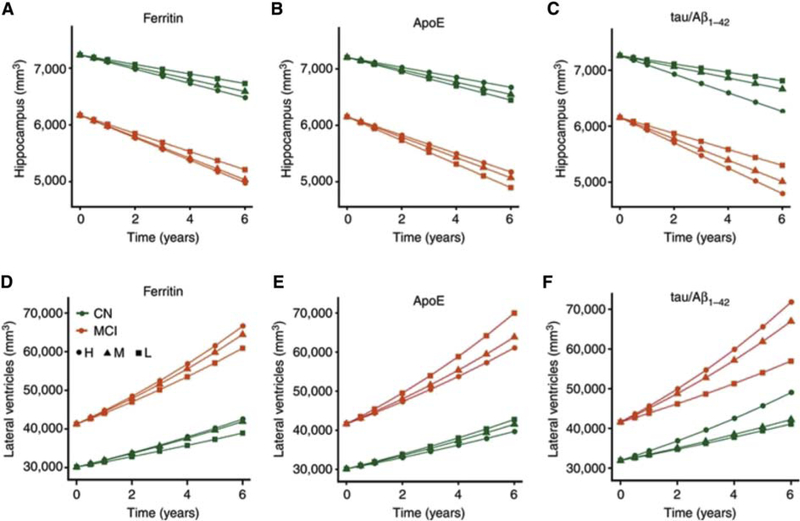
How do the gene products of major AD risk alleles exert their effect on the brain? Several groups have used multi-modal ADNI data to gain insight into this critical question, bridging the gap between genetics and pathophysiology. Three isoforms of the APOE gene product, apolipoprotein E (ApoE), corresponding to the ε2, ε3, and ε4 alleles, are found in CSF and blood. Plasma ApoE was mildly correlated with CSF ApoE, but not with longitudinal changes in cognition or atrophy [150], and was associated with left hippocampal volume in APOE4+ MCI subjects [151]. The APOE ε4 allele may increase neurodegeneration via a mechanism involving brain iron levels. CSF ferritin levels, reflecting cortical iron levels, were strongly associated with CSF ApoE levels in AD subjects and were elevated by the APOE ε4 allele [152]. They were also associated with ADAS-cog scores and greater rates of hippocampal atrophy and ventricular expansion (Fig. 5) and predicted MCI to AD progression [152]. Interestingly, ferritin affected cognitive performance to a similar degree as ApoE and p-tau/Aβ42. However, the analytes differed in the level of their effect over time; the effect of ferritin was constant, whereas the effect of both ApoE and p-tau/Aβ42 increased with disease severity resulting in a greater decrease in cognitive performance over time. ApoE may raise the baseline iron load of the brain and so lower the threshold for iron-mediated neuronal loss.
Fig. 5.
CSF ferritin independently predicts brain structural changes. (A–C) Longitudinal hippocampal volumetric changes based on tertiles of CSF: (A) ferritin, (B) ApoE, (C) tau/Aβ42. (D–F) Longitudinal volumetric ventricular changes based on tertiles of CSF: (D) ferritin, (E) ApoE, (F) tau/Aβ42. Abbreviations: CN, cognitively normal; CSF, cerebrospinal fluid; H, highest tertile; M, middle tertile; MCI, mild cognitive impairment. Reproduced with permission from [152].
Although Apo E4 appears to underlie neurodegeneration, Apo E2 or Apo E3 may exert a neuroprotective effect. In carriers of the APOE ε2 and ε3 alleles but not the ε4 allele, increased levels of CSF ApoE were associated with higher p-tau, an indicator of neuronal damage, whereas decreased baseline levels of ApoE were associated with worse longitudinal cognitive decline, MCI progression, and atrophy rate independent of CSF p-tau/Aβ42 ratio [150]. Therefore, Apo E2 or Apo E3 may be increased in CSF in response to neuronal injury and protect against neurodegeneration by decreasing neuronal damage independent of tau and Aβ deposition.
CLU, the gene for clusterin, has been identified and confirmed as an AD genetic risk factor [153–155]. Deming et al. [156] searched for additional SNPs associated with clusterin levels and used gene ontology analyses to identify molecular mechanisms by which CLU may exert its action. They found that CSF, but not plasma, levels of clusterin were significantly higher in AD subjects and were correlated with both p-tau/Aβ42 ratio and CSF ApoE, suggesting that clusterin and ApoE may interact to influence Aβ deposition. Clusterin may contribute to immune system changes observed in AD or disrupt healing after neurodegeneration [156].
5.2.2. Associations between novel CSF and blood analytes and AD pathology
Mattsson et al. [157] selected a panel of 70 CSF proteins involved in biological functions such as Aβ metabolism, microglia activity, and synaptic/neuronal function that may be altered in the early stages of AD. At baseline, several proteins in addition to Aβ and tau were mildly associated with atrophy over 4 years in specific regions: apolipoprotein D, α−1-micro-globulin, apolipoprotein CIII, apolipoprotein H, and interleukin 6 with the middle temporal cortex; apolipoprotein CIII, apolipoprotein D, α−1-microglobulin, apolipoprotein H, and interleukin 16 with the inferior temporal cortex. Several proteins (ferritin, S100b, apolipoprotein CIII, apolipoprotein H, and hepatocyte growth factor) were associated with atrophy rates that interacted with Aβ, suggesting that they act via an Aβ-dependent mechanism, but others appear to act via an Aβ-independent mechanism. The proteins identified, if replicated, may represent novel prognostic biomarkers of AD-related atrophy in CN subjects.
Synaptic dysfunction occurs in the early stages of AD and is followed by neurodegeneration. Several studies have concentrated on finding novel CSF analytes as potential biomarkers for this process. Levels of a postsynaptic protein neurogranin appear to reflect synaptic degeneration. CSF neurogranin levels were significantly higher in AD subjects and MCI converters than in CN subjects [158]. Moreover, neurogranin levels predicted decreased MMSE and increased ADAS-cog scores and were correlated with longitudinal reductions in cortical glucose metabolism and hippocampal volume. Neurogranin levels were significantly increased in Aβ+ subjects, and elevated levels were detected even in non-symptomatic subjects, suggesting that it may be an independent novel biomarker for synaptic pathology. Paterson et al. [159] identified several analytes using a CSF multiplex analyte panel that influence the rate of neurodegeneration in Aβ+ subjects. Low trefoil factor 3 was strongly associated with higher rates of whole-brain atrophy, ventricular expansion, and hippocampal atrophy. High cystatin predicted higher whole-brain atrophy and hippocampal atrophy rates, and low vascular endothelial growth factor (VEGF) and chromogranin Awere associated with higher whole-brain atrophy rate. After adjustment for baseline volume, p-tau, age, sex, APOE4 status, and diagnosis, trefoil factor 3 was still associated with increased hippocampal atrophy rate and may be a valuable biomarker for decline in AD.
AD is characterized by degradation of WM tracts which progressively disconnect cortical and subcortical regions. Neurofilaments (NFs) are structural components of the neural cytoskeleton and are abundant in large caliber myelinated axons of the WM tracts. Zetterberg et al. [160] investigated the associations of its low molecular weight subunit, NFL, in CSF with diagnostic status, cognitive decline, and WM change. CSF NFL was elevated in AD subjects compared to stable MCI and CN subjects, and correlated with accelerated cognitive decline (MMSE and ADAS-cog), WM change, and increased whole-brain atrophy, ventricular expansion, and hippocampal atrophy in MCI subjects. These results suggest that NFL may be a useful biomarker for tracking axonal degeneration, where NFs are most abundant, and this is associated with the degradation of WM tracts in cognitive decline.
VEGF has been implicated as a neuroprotective factor in AD, possibly neutralizing the damaging effects of the AD pathological cascade through improvements in vascular survival. Hohman et al. [161] found that higher levels of VEGF were associated with larger baseline hippocampal volume, lower hippocampal atrophy over time, lower episodic memory decline, and lower executive function decline over time. The neuroprotective effect of VEGF, which was greatest in MCI subjects, appeared to be enhanced in the presence of AD CSF biomarkers. In Aβ+ subjects, higher VEGF levels were associated with better memory performance, and in tau1 subjects, higher VEGF levels were associated with lower hippocampal atrophy, and decline in memory and executive function. These results suggest that angiogenic factors may be important in individuals with biomarker abnormalities consistent with early AD. The mechanism by which VEGF reduces risk of neurodegeneration is unknown. Leung et al. [162] searched for CSF analytes associated with Aβ positivity and also identified fatty acid binding protein and VEGF, along with less strongly associated analytes including resistin, CD40 antigen, prolactin, lipoprotein A, and hepatocyte growth factor. These analytes are involved in different aspects of AD pathophysiology, including inflammatory response, lipid metabolism, atherosclerosis, and insulin resistance, and are potential biomarkers that require future validation in independent cohorts.
Another factor reported to be protective against AD, possibly through its modulation of Aβ accumulation, is brain-derived neurotrophic factor (BDNF), a widely distributed neurotrophin with multiple functions including neuronal differentiation, regulation of synaptic function, and modulation of hippocampal long-term potentiation, learning, and memory formation. Hwang et al. [163] found a significant negative association between BDNF protein levels in plasma and brain Aβ burden, measured by PiB PET, in the lateral temporal, inferior parietal, and inferior frontal, anterior and posterior cingulate, and orbital frontal regions. In a separate study [164], the Val66Met polymorphism which is associated with lower BDNF secretion in vitro was not associated with hippocampal volume or memory. Therefore, plasma BDNF levels may act as a biomarker of Aβ pathology but not of volumetric or cognitive changes in the brain. Analytes identified in these studies are summarized in Table 4.
Table 4.
Novel CSF blood biomarkers
| Analyte | CSF or plasma | Association with AD pathology | Notes | Reference |
|---|---|---|---|---|
| α-1-microglobulin | CSF | Regional atrophy | [157] | |
| Apolipoprotein CIII | CSF | Regional atrophy | [157] | |
| Apolipoprotein D | CSF | Regional atrophy | [157] | |
| Brain-derived neurotrophic factor | Plasma | Aβ | Neuroprotective | [165] |
| Brain-derived neurotrophic factor | Plasma | NOT HV, M | Val66Met polymorphism | [164] |
| CD40 antigen | CSF | Aβ | [162] | |
| Chromogranin A | CSF | WBA | Aβ dependent | [159] |
| Chromogranin A | CSF | MCI to AD | [166] | |
| Cystatin | CSF | WBA, HA | Aβ dependent | [159] |
| Factor H | CSF | CD, VE | [167] | |
| Fatty acid binding protein | CSF | Aβ | [162] | |
| Fatty acid binding protein | CSF | CN to MCI | [168] | |
| Fatty acid binding protein | CSF | MCI to AD | [166] | |
| Ferritin | CSF | Regional atrophy | Aβ independent | [157] |
| Hepatocyte growth factor | CSF | Regional atrophy | Aβ independent | [157] |
| Hepatocyte growth factor | CSF | Aβ | [162] | |
| Interleukin 16 | CSF | Regional atrophy | [157] | |
| Interleukin 6 | CSF | Regional atrophy | [157] | |
| Lipoprotein A | CSF | Aβ | [162] | |
| Neurofilament light | CSF | CS, CD, WMC WBA, C, HA | [160] | |
| Neurogranin | CSF | CS, C, Met, HV | Aβ dependent | [158] |
| Prolactin | CSF | Aβ | [162] | |
| Resistin | CSF | Aβ | [162] | |
| S100β | CSF | Regional atrophy | Aβ independent | [157] |
| Trefoil factor 3 | CSF | WBA, HA, VE | Aβ dependent | [159] |
| Vascular endothelial growth factor | CSF | WBA | Aβ dependent | [159] |
| Vascular endothelial growth factor | CSF | HV, HA, CD | Neuroprotective, effect enhanced in Aβ+ | [161] |
| Vascular endothelial growth factor | CSF | Aβ | [162] |
Abbreviations: CSF, cerebrospinal fluid; AD, Alzheimer’s disease; HV, hippocampal volume; M, memory; WBA, whole brain atrophy; HA, hippocampal atrophy; CD, cognitive decline; VE, ventricular expansion; CN, cognitively normal; MCI, mild cognitive impairment; CS, clinical status; WMC, white-matter change; Met, metabolism; C, cognition.
5.2.3. CSF and plasma analytes for the detection of copathologies
There is a growing awareness that copathologies such as Lewy bodies, vascular disease, TDP43 inclusions, and hippocampal sclerosis are common and may explain the variability in AD progression [169]. The development of biomarkers to predict coincident pathologies that are frequently observed in clinically diagnosed AD patients would be of great clinical utility and may improve the diagnostic and prognostic abilities of established CSF biomarkers alone [129]. One of the most common comorbidities in AD, present in 10%–40% of AD patients, is the presence of Lewy bodies, composed of α-synuclein (α-syn). Symptoms of dementia with Lewy bodies include hallucinations, visual spatial impairment, and executive dysfunction but it is not known how reduced CSF α-syn relates to AD symptoms. Mackin et al. [170] reported that 20% of AD, 13% of MCI, and 8% of CN subjects reported hallucinations and that CSF α-syn was reduced in these AD subjects. Lower CSF α-syn was associated with decreases in memory and language, as well as executive function, suggesting that Lewy bodies may be related to overall cognitive decline in addition to the characteristic hallucinations of dementia with Lewy bodies.
A non-AD pathway to dementia was outlined by Alcolea et al. [144]. They reported that preclinical subjects as well as subjects with suspected non-Alzheimer’s pathology (SNAP) had higher levels of YKL40, a marker of neuroinflammation, than Aβ+ subjects early in disease progression. This suggests that neuroinflammation can emerge through a non–Aβ-related pathway and that it is also detectable in CSF in preclinical stages in non-Aβ degenerative disorders. The correlation between YLK40 levels and age regardless of APOE status suggests that low-grade inflammatory processes are present in the brain even in the absence of Aβ. These findings suggested that CSF YKL40 levels increase with aging, preclinical AD, and SNAP and correlate closely with markers of neurodegeneration.
5.3. Conclusions
Considerable progress has been made in 2014 and 2015 by the ADNI Biomarker Core in improving methodologies for measuring established CSF biomarkers and establishing relationships between different measures of Aβ. The ADNI Biomarker Core has also emphasized the need to detect copathologies, such as α-syn amyloid deposits in Lewy bodies which were associated with symptoms specific to AD in addition to those characterizing dementia with Lewy bodies. SNAP may be associated with YLK40, a marker of neuroinflammation. Cerebrovascular risk factors homocysteine and body mass index, as reflected in CSF leptin levels, have been linked to aspects of AD pathology. A number of studies have addressed the mechanism of action of the APOE ε4 allele via its CSF and blood ApoE protein. ApoE may modulate iron levels leading to oxidative damage–induced neurodegeneration or act in conjunction with Aβ to enhance leptin signaling. Confirmed AD genetic risk factor, CLU, may exert its effect via immune response. Another focus has been the use of proteomic approaches to identify primarily CSF analytes associated with atrophy, and cortical Aβ load, WM degradation, and cognitive decline that capture distinct information from those identified by traditional CSF biomarkers. These include fatty acid binding protein, apolipoproteins AII, CIII, D, and H, interleukins 6 and 16, ferritin, and chromogranin A. Conversely, several studies point to VEGF and BDNF as being neuroprotective.
6. Studies of genetic associations
Studies of ADNI genetics data have been instrumental in deepening our understanding of AD pathophysiology by bridging the gap between the genetic underpinnings and biochemical mechanisms of the disease. The Genetics Core, under the aegis of Dr. Andrew Saykin, has collected blood samples at every patient visit, extracted both DNA and RNA, and performed APOE, TOMM40, and genome-wide array genotyping, whole-exome sequencing, and whole-genome sequencing (the latter generously supported by the Brin-Wojcicki Foundation and the Alzheimer’s Association [171]). The rich ADNI longitudinal data set contains many biomarkers that can serve as quantitative endophenotypes for genetic association studies, increasing the power to detect biologically meaningful associations. In accordance with ADNI policy, all genetics data, like other data, have been made available to the wider scientific community, resulting in well over 300 publications by 2014 that analyzed ADNI data sets. This section describes the work of the Genetics Core and the approximately 80 publications arising from ADNI genetics data in 2014 and 2015.
6.1. Reviews
Saykin et al. [171] outlined the progress and plans of the ADNI Genetics Core and systematically reviewed 106 articles published between 2009 and 2012. These investigations ran the gamut from large meta-analytic case-control GWAS to GWAS of quantitative endophenotypes, to studies which select genes from a particular pathway thought to be involved in AD, to those pinpointing the effects of a particular polymorphism or gene. GWAS conducted by a multistudy consortia have identified and replicated approximately 20 genes in addition to APOE that confer risk or have protective roles in AD [172]. A number of these (APOE, BIN1, CLU, ABCA7, CR1, PICALM, MS4A6A, CD33, MS4A4E, and CD2AP) have been identified or confirmed using ADNI data. In addition, many novel candidate risk and protective loci cut have been identified using ADNI genetics data and quantitative imaging, cognitive, and fluid phenotypes (Fig. 6) [172]. However, these loci account for only a portion of disease heritability. As AD is a multigenic disorder influenced by environmental factors and epistasis, the Genetics Core has emphasized a Systems Biology perspective in which candidate genes identified in GWAS or from likely mechanistic biological pathways are studied for their associations with mRNA and other phenotypes. It is hoped that this integrative functional genomics approach will capture the biological complexity at multiple levels including genomic, transcriptomic, proteomic, and metabolomic (Fig. 7).
Fig. 6.
Word cloud of genes names reported in articles using ADNI genetic data. The color and size of the gene name corresponds to the number of abstracts mentioning the gene. Abbreviation: ADNI, Alzheimer’s Disease Neuroimaging Initiative. Reproduced with permission from [172].
Fig. 7.
Converging “multi-omics” in ADNI. This figure illustrates the landscape of multiple “-omics” domains relevant to AD and how they contribute to an integrated Systems Biology approach to discovering the underlying genetic architecture of AD. *Data from ADNI-1. **Data from ADNI-GO/2. Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative. Reproduced with permission from [171].
ADNI genetic studies also hold promise for improving clinical trial design and identifying therapeutic targets. Stratification and subject selection, currently limited to APOE, could likely be improved by the incorporation of combinatorial sets of genes that have a biologically relevant underpinning such as a target pathway. In a similar way, an improved understanding of pathways and mechanisms of association between AD risk genes in disease will provide therapeutic targets [171]. In the future, the ADNI Genetics Core will collect samples for the production of induced pluripotent stem cells. This will allow development of in vitro models that can bridge the gap between animal models and clinical development and that can improve phenotypic characterization of disease heterogeneity.
ADNI genetics data have contributed to a number of meta-analytic studies, most notably the ENIGMA consortium, a global collaborative network of over 500 scientists involved in neuroimaging genetics [173,174]. Using neuroimaging data from nearly 13,000 subjects, ENIGMA is studying 12 major brain diseases including schizophrenia, bipolar illness, and major depression and has identified a number of genetic loci that affect brain volumes, and how they may act to increase the risk of these diseases.
6.2. Genetic association studies
Two fundamentally different approaches have been used to characterize associations between genetic loci and phenotypes using ADNI data. Genotype approaches select candidate SNPs, genes, or pathways from a priori knowledge of biological function and test for their association with selected imaging or fluid biomarkers. Although this approach cannot discover new genes, it can examine the effect of interactions between loci and phenotypic variation. The main alternative approach is to begin with a phenotype and search the genome for significantly associated loci. This approach can discover new genes but is often hampered by problems of ultrahigh dimensionality and multiple comparisons and ignores spatial information in imaging data and correlations between genetic markers due to linkage disequilibrium and epistatic effects. A popular hybrid approach is to select candidate loci and then apply further biologically informed enrichment methodologies. Genetic association studies from 2014 and 2015 are summarized in Table 5; confirmed and novel risk or protective loci identified from ADNI studies during this period are detailed in Table 6.
Table 5.
Studies of genetic associations
| Category | # Of papers | Example paper | ||
|---|---|---|---|---|
| Genotype | APOE alone | 6 [175–180] |
Sampedro et al. 2015, Oncotarget | Female carriers of the APOE ε4 allele had widespread hypometabolism and cortical thinning compared with female noncarriers, whereas male carriers had a higher risk of cerebral microbleeds, suggesting a differential sex bias of the APOE ε4 allele. |
| Candidate SNPs and genes | 16 [181–195] |
Lazaris et al. 2015, Neurology Genetics | SNPs in BIN1, CD2AP, and CR1 modulated the association between plasma ApoE and Aβ load, independent of APOE4 status. | |
| Candidate pathways | 1 [196] |
Biffi et al. 2014, Neurobiol Aging | Variants in oxidative phosphorylation genes, summarized as a genetic risk score, were associated with clinical status, and hippocampal and entorhinal cortex volumes, suggesting an overlap of the genetic structure of AD and stroke. | |
| Epistatic interactions | 3 [197–199] |
Hohman et al. 2014, Neurobiol Aging | Found epistatic interactions between GSK3β (tau protein kinase 1) and Aβ-related genes such as APP. Combined interactions explained up to 1.5% of the variance in Aβ deposition. | |
| Phenotype | Case-control | 12 [200–211] |
Escott-Price et al. 2014, PLoS One | A novel gene-wide statistical approach in a mega meta-analysis of genome-wide data sets identified two novel loci, TP53INP1 and IGHV1 associated with AD, and found evidence of a gene-wide association for loci previously identified by SNP analysis. |
| Structural imaging | 6 [212–217] |
Xu et al. 2014, PLoS One | Used longitudinal structural MRI data as phenotypes in a GWAS and identified a larger number of SNP-phenotypes associations than from baseline data. APOE and TOMM40 were top hits; this is a novel SNP that was identified in APOE. | |
| Aβ deposition (PET, CSF) | 6 [218–223] |
Ramanan et al. 2015, Brain | Used longitudinal florbetapir PET data in a GWAS and identified an intronic SNP in IL1RAP, rs12053868, significant in the absence of APOE ε4 allele. The minor allele was associated with greater cortical Aβ burden, atrophy, rate of MCI to AD progression, and cognitive decline. IL1RAP is involved in microglial activation. | |
| Fluid (CSF, blood) | 3 [224,225] |
Kauwe et al. 2014, PLoS Genet | GWAS of panel of CSF analytes involved in a range of imported AD processes such as endocytosis, cholesterol metabolism, inflammatory and immune response, identified associations with proteins involved in Aβ processing or proinflammatory signaling. | |
| Neuropsychological assessments | 2 [226,227] |
Sherva et al. 2014, Alz Dem | Used longitudinal changes in ADAS-cog to identified variants in the SPON1 gene whose minor alleles were associated with more rapid cognitive decline. Spondin 1 inhibits APP cleavage by BACE. | |
| Other studies | 3 [228–230] |
Chen et al. 2015, Nat Commun | ADNI one of six cohorts identifying polygenic architecture of human cortex. Variability of cortex surface area explained by additive effects of genome-wide SNPs with those in evolutionary conserved areas contributing more to medial and temporal cortices. |
Abbreviations: APOE, apolipoprotein E; SNP, small nucleotide polymorphism; AD, Alzheimer’s disease; APP, Aβ precursor protein; PET, positron emission tomography; CSF, cerebrospinal fluid; GWAS, genome-wide association study; MCI, mild cognitive impairment; ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive subscale; BACE, beta-secretase 1.
Table 6.
Confirmed and novel risk and protective loci identified in ADNI studies 2014 to 2015
| Gene | SNP | Phenotype | Proposed biological function | Reference |
|---|---|---|---|---|
| APOE ε4 | Hippocampal atrophy rate | Modulates Aβ accumulation and clearance | [175,176] | |
| Left hippocampal deformation | [177] | |||
| Limbic region atrophy rate | [176] | |||
| Ventricular expansion and morphology | [178] | |||
| Females: hypometabolism, cortical thinning | [179] | |||
| Males: cerebral cortex microbleeds | ||||
| APOE* | Minor allele | Disease status | [204] | |
| APOE-BCHE | rs509208 minor G allele | Cortical Aβ load | Butyrylcholinesterase disrupts synaptic functioning | [218] |
| APOC1 | Top SNPs | CSF Aβ42 | Synaptic transmission, transmission of nerve impulses, and trait class | [219] |
| BIN1* | Plasma ApoE | Interacts with APOE | [181] | |
| CBLB | Minor allele | Clinical status | [204] | |
| CD2AP* | Plasma ApoE | Interacts with APOE | [181] | |
| CETP | rs5882G allele | Decreased white-matter integrity | Cholesteryl ester transfer protein may increase AD risk through the degradation of WM | [192] |
| CFH | Not associated with AD in Chinese cohort | [187] | ||
| CR1* | Plasma ApoE | Interacts with APOE | [181] | |
| CR1 | +CVD risk factors, reduced hippocampal volume | Increase of neurodegeneration in presence of CVD | [189] | |
| DAT1* | rs6347C allele | Decreased cognitive performance, greater ventricular expansion, dementia risk | Regulates neurotransmission by modulating dopamine receptors | [194] |
| EPHA1 | rs11771145A allele | Lower hippocampal atrophy, greater metabolism | [183] | |
| EPHA1 + CVD risk factors | Reduced hippocampal volume | Increase of neurodegeneration in presence of CVD | [189] | |
| GRIN2B | Disease status | Glutamate signaling | [201] | |
| HMGCR | rs3846662G allele | Delayed age of AD onset, reduced risk of AD | [193] | |
| MMP1 | Minor allele | Clinical status | [204] | |
| NAPRT1 | Minor allele | Clinical status | [204] | |
| SORL1 | rs2070045G allele | Increased CSF tau, hippocampal atrophy | May increase neurodegeneration | [186] |
| TOMM40 | Minor allele | Clinical status | [204] | |
| TOMM40 | Top SNPs | CSF Aβ42 | Synaptic transmission, transmission of nerve impulses, and trait class | [219] |
| TREM2 | Minor allele | Clinical status | [204] | |
| TREML2 | p.R47H | Increased risk of AD | [184] | |
| CLU-MS4A4E* | Gene interaction decreased AD incidence | [188] | ||
| RYR3-CACNA1C | Minor alleles | Increased cortical Aβ load | Regulation of intracellular calcium homoeostasis | [197] |
| GSK3β-APP | rs334543 minor allele-3SNPs | Increased cortical Aβ load | Tau kinases interact with APP to increase deposition | [199] |
| C9-IER6 | Increased cortical Aβ load | Neuroinfiammation may enhance Aβ deposition | [198] | |
| UNC5C | Hippocampal and precuneal atrophy | Unc-5 Netrin Receptor C—triggers apoptosis | [182] | |
| NME8 | rs2718058G allele | Higher CDR-SB scores, lower atrophy higher metabolism | May inhibit neurodegeneration | [185] |
| OPRD1 | rs678849 | Regional brain volume differences, CSF biomarkers | Delta opioid receptor may promote the processing of APP | [231] |
| NAV2 | Episodic memory scores | Neuron navigator 2, neurite outgrowth, and cell migration | [191] | |
| TP53INP1 | Disease status | Proapoptotic tumor suppressor | [200] | |
| IGHV1 | Disease status | Unknown | ||
| ZNF827* | CSF Aβ42 | Transcription factor | [203] | |
| ZNF628* | Minor allele | Disease status | Transcription factor | [204] |
| KDM2B* | p-tau181 | [203] | ||
| NANP* | Regional atrophy | |||
| OXPHOS (oxidative phosphorylation genes) | Disease status, CN to MCI progression, hippocampal volume, entorhinal cortex volume | Overlap of AD and stroke genes influence disease trajectory | [196] | |
| AEN | Disease status | Signaling network involved cholesterol metabolism | [202] | |
| ADAMTS12 | ||||
| PSMA5 | ||||
| FXN | ||||
| NTM | ||||
| LARP1 | ||||
| WDTC1 | ||||
| SEMA7A VK0RC1L1 | ||||
| COL5A3 | ||||
| H0MER2 | Disease status, right thalamus volume | Transcription factor linked to APP | [212] | |
| PML | Hippocampal shape, insular cortices | [214] | ||
| STOML1 | Hippocampal shape, insular cortices | |||
| SYNJ2-PI4KA | Inferior lateral ventricle atrophy | Inositol pathway | [216] | |
| REST* | rs3796529 minor T allele | Less hippocampal atrophy, greater medial temporal lobe metabolism | Negative transcriptional regulator of hippocampal neurogenesis | |
| CTXN | Plasma Aβ42 and Aβ40 | Cortexin modulates Aβ42 secretion | [220] | |
| ILRAP* | rs12053868 minor G allele | Longitudinal Aβ accumulation | Proinflammatory cytokine, activation of microglia | [221] |
| SULG2* | CSF Aβ42, cognitive decline | |||
| FRA10AC1* | Fragile site | CSF Aβ42, cognitive decline | [222] | |
| POT1 | rs4728029 | CSF p-tau181, ventricular expansion | Neuroinflammatory mechanism related to interleukin 6 receptor | [224] |
| Genes mediated by miR-33 |
CSF p-tau181 | Disruption of cellular cholesterol homoeostasis | ||
| SPON1 | Minor alleles | More rapid decline on ADAS-cog | Spondin 1 inhibits cleavage of APP by BACE | [226] |
| EXOC4, GABRG3, VAT1L | More rapid decline on ADAS-cog | Neuronal maintenance, neurotransmission, calcium signaling | ||
| CAMK4, CYCS, NCS1, CACNA1G | More rapid decline on ADAS-cog | Homoeostasis | ||
| RNASE13 (+PTK2B, PICALM, MS4A2, and APP from pathway analysis) | Cognitive resilience | Neuron loss, presynaptic membrane, and postsynaptic density | [227] |
Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; SNP, small nucleotide polymorphism; BCHE, butyrylcholinesterase; CSF, cerebrospinal fluid; AD, Alzheimer’s disease; WM, white matter; CVD, cardiovascular disease; APP, Aβ precursor protein; CDR-SB, Clinical Dementia Rating–Sum of Boxes; CN, cognitively normal; MCI, mild cognitive impairment; ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive subscale; BACE, beta-secretase.
Independent of APOE status. Novel loci are in bold, and blue shaded boxes represent protective alleles.
6.2.1. Genotype
6.2.1.1. APOE
The APOE ε4 allele is the single locus that confers the greatest risk for sporadic late-onset AD. Carriers of this allele have greater hippocampal atrophy rates than noncarriers [175,176], significant morphological deformation of the left hippocampus [177], accelerated rates of atrophy in limbic and cortical areas, particularly the amygdala [176], faster ventricular expansion, and regional patterns of ventricle morphology [178]. Although a family history of AD was associated with greater regional longitudinal atrophy rates in MCI participants, these were accounted for by APOE4 genotype [232]. These studies suggest that there is a genotype-specific network of brain regions associated with the APOE ε4 allele that undergoes faster atrophy and morphological changes. The effect of this allele is stronger in women than in men. Sampedro et al. [179] found that female APOE4 carriers had widespread brain hypometabolism and cortical thinning in several regional clusters compared to female noncarriers, whereas male APOE4 carriers differed only slightly from male noncarriers (Fig. 8). However, male APOE4 carriers had a higher risk of cerebral cortex microbleeds then the male noncarriers, suggesting that there is a differential sex bias of the APOE4 allele on diverse aspects of disease pathology.
Fig. 8.
Sex-stratified FDG analyses. Analysis between APOE4 carriers and APOE4 noncarriers (P <.005 uncorrected) in (A) females and (B) males, covaried for age and years of education across the lateral and medial views of the cerebral cortex. As shown, female APOE4 carriers showed widespread clusters of decreased metabolism with respect to female APOE4 noncarriers (A), whereas male APOE4 carriers only showed an isolated cluster of decreased metabolism (P <.005) in the precuneus with respect male noncarriers (B). Abbreviations: FDG, [18F]-fluorodeoxyglucose; APOE4, apolipoprotein E ε4 allele. Reproduced with permission from [179].
6.2.1.2. Candidate SNPs and genes
6.2.1.2.1. SNPs in confirmed risk alleles
Candidate SNPs for association studies are chosen because of their location in confirmed AD risk alleles or in genes of suspected importance on the basis of biological function. SNPs in BIN1 (rs744373), CD2AP (rs9349407), and CR1 (rs3818361) (AD risk variants implicated in Aβ deposition) modulated the association between plasma ApoE levels and cortical Aβ load in different directions (Fig. 9), independent of APOE4 carrier status, suggesting that these genes interact with APOE to modulate Aβ accumulation and clearance [181]. Genetic variants in the recently confirmed AD risk factor, UNC5C, were associated with the atrophy rate of the left hippocampus and right precuneus [182]. The protective A allele of rs11771145 in EPHA1 prevented hippocampal atrophy in MCI subjects and was associated with lower atrophy and greater metabolism in the temporal gyri in AD subjects [183]. Whole-exome sequencing identified a coding missense variant, p.S144G, in TREML2 responsible for the previously noted protective effect against AD [184]. This functional variant was associated with a reduced risk of AD, whereas a missense variant in TREM2, p.R47H had the opposite effect, independent of p.S144G. Cell-based analyses implicated the involvement of these variants in the modulation of microglial activation, possibly influencing Aβ clearance. Minor allele G carriers of rs2718058 in a recently confirmed preventive locus adjacent to NME8 had higher CDR-SB scores, lower occipital gyrus atrophy, greater metabolic rate and ventricular expansion, and a lower rate of hippocampal atrophy than carriers of the major A allele, suggesting that the minor allele may confer its protective effect by inhibiting neurodegeneration [185]. The G allele in rs2070045 within SORL1 was associated with increased CSF tau and hippocampal atrophy, and a haplotype in this region was associated with higher tau and p-tau181, suggesting that SORL1 may increase AD risk by increasing neurodegeneration [186]. Finally, top risk allele, CFH, but not risk alleles CR1, CD33, CLU, and TREML2, were strongly associated with AD in a Chinese cohort [187], suggesting that the genetics of AD may differ among diverse populations.
Fig. 9.
Significance maps displaying the associations between cortical Aβ binding (Pittsburgh compound B) and plasma ApoE protein. Plasma apoE levels were associated with Pittsburgh compound B SUVR in the pooled sample in all brain regions apart from the sensorimotor and entorhinal cortex (top panel). Plasma apoE levels were associated with Pittsburgh compound B SUVR in BIN1 rs 744373 minor allele carriers (second panel) and in CD2AP rs 9349407 and CR1 rs 38118361 minor allele noncarriers (third and fourth panel, respectively). Abbreviation: SUVR, standardized uptake value ratio. Reproduced with permission from [181].
The gene-gene interaction between CLU and MS4A4E was associated with APOE4− status and may have a possible dominant effect [188]. Lack of these risk alleles was estimated to decrease AD incidence by 8%. CR1 and EPHA1 interacted with cardiovascular disease risk factors to reduce hippocampal volume [189]. Cardiovascular risk dominated the genetic risk of these loci in terms of interaction effect such that at low genetic risk, high cardiovascular risk factors had a more detrimental effect (Fig. 10). These findings indicate that CR1 and EPHA1 may contribute to the etiology of late-onset neurodegeneration in the presence of cardiovascular disease.
Fig. 10.
Effect of interactions between CR1 or EPHA1 and cardiovascular disease risk factors on hippocampal volume. The estimated interaction effect on hippocampal volume for both risk genes is dominated by high cardiovascular disease risk. High genetic risk appears to reduce the interaction effect in the presence of high cardiovascular disease risk, suggesting that cardiovascular disease risk factors are more detrimental under low genetic risk. Abbreviations: CVD, cardiovascular risk; G, genetic risk. Reproduced with permission from [189].
6.2.1.2.2. SNPs in genes targeted for biological function
Several studies have targeted genes with biological functions implicated in AD. Rare variants of PLD3 are confirmed AD susceptibility loci; phospholipase D3 modulates APP processing. However, a common variant in the same gene, rs10407447, was associated with regional metabolism and lateral ventricular volume in CN and MCI subjects [190]. A study of SNPs found in genes preferentially expressed in the hippocampus identified a novel locus, NAV2 (neuron navigator 2), associated with episodic memory scores [191]. Neuron navigator 2 is involved in neurite outgrowth and cell migration.
Abnormal cholesterol levels increase the risk of AD and are influenced by several common genetic variants. Increased G allele dosage of rs5882 (p.I405V) in CETP (cholesterylester transfer protein) was associated with measures of WM integrity (lower fractional anisotropy and higher radial and mean diffusivities) in older individuals, suggesting that high cholesterol may increase AD risk through the degradation of WM integrity [192]. 3-hydroxy-3-methylglutaryl-CoA reductase plays a central role in the production of cholesterol and is the target of statins which reduce the risk of sporadic AD by as much as 70% in midlife. AD carriers of the G allele of rs3846662 in HMGCR had both a delayed age of onset and a reduced risk of AD [193]. The effect was greatest in women. In MCI subjects, the G allele reduced progression to AD over 3 years, even in APOE4+ subjects, providing further support for the protective role of this allele.
Altered dopamine transmission affects many aspects of brain function, including the formation of Aβ, and contributes to cognitive impairment. Dopamine transporter protein, encoded by the DAT1 gene, regulates neurotransmission by modulating dopamine receptors. The minor C allele of rs6347 in DAT1 was associated with poorer cognitive performance, greater ventricular expansion, and greater dementia risk, independent of APOE genotype [194]. Although the mechanism of action of this polymorphism is unknown, dopamine neurotransmission may be a useful target for anti-dementia drugs. Delta opioid receptors promote the processing of APP and are implicated in various psychiatric and neurological disorders. A common variant (rs678849) in the OPRD1 gene was associated with regional brain volume differences in healthy elderly and may predict levels of CSF biomarkers [231].
6.2.1.3. Candidate pathways and epistatic interactions
The top 10 AD susceptibility genes explain only 35% of the variability in disease risk [233]. It has become apparent that studies of single loci are not sufficient to unravel the complex genetic etiology of the disease. It is thought that the “missing heritability” may come from the association of multiple variants in genes interacting in functional pathways. Variation in genes involved in oxidative phosphorylation (OXPHOS), summarized by an OXPHOS genetic risk score, is associated with an increased risk of stroke. Stroke increases the risk of AD, and the OXPHOS genetic risk score was associated with AD clinical status, CN to MCI progression, and volumes of hippocampus and entorhinal cortex in CN and MCI, but not AD subjects [196]. This suggests that the genetic structure of AD and stroke overlap and that OXPHOS variants exert an early influence on disease trajectory.
Epistatic interactions may also account for some of the risk variability of AD. Koran et al. [197] investigated genes from AD pathways, including Aβ processing, apoptosis, calcium homoeostasis, free radical production, and mitochondrial dysfunction, defined by the Kyoto Encyclopedia of Genes and Genomes database. An interaction between minor alleles in RYR3 (ryanodine receptor 3) and CACNA1C (αC subunit of voltage-dependent L-type calcium channels) was associated with a higher Aβ load and explained 6% of Aβ load variance. Both genes encode proteins involved in the regulation of intracellular calcium homoeostasis. As ryanodine receptor–driven calcium release has been associated with increased Aβ levels, and Aβ modulates the function of L-type calcium channels, the authors suggest that interaction between these two variants may increase intracellular calcium levels by disrupting calcium homoeostasis and lead to increased Aβ production and deposition. Aβ accumulation was also associated with two SNP-SNP interactions between C9 (complement 9) and ILR6 (interleukin 6r), suggesting that neuroinflammation may exert its effect on AD by enhancing Aβ deposition [198].
According to the amyloid hypothesis, Aβ deposition precedes tau pathology. However, it is possible that the two pathologies arise independently due to upstream genetic interactions and that genes that confer risk for tau pathology also confer risk for Aβ pathology via complex epistatic relationships. Hohman et al. [199] searched for epistatic interactions between genes for tau kinases and genes involved in Aβ deposition associated with Aβ load. They found three interactions between rs334543 in GSK3β and different Aβ genes. A minor allele that interacted with APP was related to high levels of Aβ deposition. The combined interactions explained between 1.2% and 1.5% of the Aβ deposition variance, suggesting that Aβ burden may be increased by a combination of GSK3β and APP-related genes.
In a genome-wide search of all possible SNP-SNP interactions that affect regional brain volumes, one interaction between an SNP in a region encoding two transcription factors (rs1345203) and an intergenic SNP (rs1213205) explained 1.9% of the variance in temporal lobe volume [195]. The biological relevance of this interaction has yet to be determined.
6.2.2. Phenotype
A great strength of ADNI is its wealth of quantitative phenotypes that can be leveraged to identify novel susceptibility loci in GWAS, and more recently, gene-based GWAS.
6.2.2.1. Case-control studies
Complex patterns of the association may not be reflected solely in single SNPs. A mega meta-analysis of genome-wide data sets, including ADNI’s, as part of the International Genomics of Alzheimer’s Disease Consortium, identified two novel loci, TP53INP1 and IGHV1 [200]. The first locus encodes a proapoptotic tumor suppressor of interest due to the inverse association between cancer and AD, whereas the function of the latter locus is unknown. A similar analysis evaluated single gene associations in a network context, then use gene ontology and pathway enrichment methods to identify biologically plausible interactions [201]. Genes involved in the glutamatergic synapse, including GRIN2B, appeared to be overrepresented in AD subjects. Glutamate signaling regulates many biological processes such as learning, memory, and synaptic plasticity, and glutaminergic neurons located in the hippocampus and other brain areas are affected by Aβ plaques and tau tangles. Top genes identified by the computation of a multimarker genetic score associated with disease status (AEN, ADAMTS12, PSMA5, FXN, NTM, LARP1, WDTC1, SEMA7A, VKORC1L1, and COL5A3) were integrated into a hypothetical signaling network incorporating a priori protein-protein interaction data, which highlighted their function in cholesterol metabolism [202].
Genetic subtypes of AD may have specific molecular mechanisms of pathogenesis. A GWAS in APOE4 noncarriers identified three novel loci located in three haplotype blocks, ZNF827 (zinc finger protein 827), KDM2B (lysinespecific demethylase 2B), and NANP (N-acetylneuraminic acid phosphatase), which were associated with CSF Aβ42, p-tau181, and brain atrophy, respectively [203].
Some of the missing heritability of complex diseases may be accounted for by rare variants; low-frequency allele variants have a higher probability of functional significance. A gene-based analysis of alleles with frequencies lower than 3% identified one novel association between ZNF628 (zinc finger protein 628) and AD that reached genome-wide significance after adjustment for APOE4 [204]. The study also identified minor alleles in APOE, TOMM40, MMP1, TREM2, CBLB, and NAPRT1 that may contribute to AD heritability. The zinc finger proteins identified in these studies suggest that transcriptional regulation may play an important role in pathogenesis.
6.2.2.2. Structural imaging phenotypes
The use of structural MRI quantitative phenotypes in identifying genetic variants associated with AD has continued to be a powerful strategy. The development of machine learning approaches has facilitated previously challenging tasks such as the selection of imaging features with power to detect genetic associations. A polygenic approach using a penalized regression method to select 109 brain wide regional measures identified a novel marker, HOMER2, associated with right thalamus volume and with AD clinical status [212]. Homer2 is a transcription factor which may be linked to APP processing or the trafficking of APP to the cell surface. A GWAS of GM density data [205] jointly considered the effects of all polymorphisms, how they fit into biologically defined pathways, and the functional relationships of 20 genes involved in SNP-SNP interactions. Enrichment analysis identified visual perception, DNA repair and replication, and olfactory pathways. A statistically significant synergistic interaction between two SNPs in an olfactory gene cluster suggested that this pathway may be involved in AD. Using a subset of imaging features able to discriminate between CN and AD subjects as quantitative phenotypes, genetic variation in a relatively small number of genes (ESR1, BIN1, LDLR, SORCS1, APP, LRAT, and TF) was found to be associated with ventricular enlargement, hippocampal atrophy, and cortical atrophy, suggesting that these loci could have potential in diagnostic classification [213]. Another study selected neuroimaging biomarkers associated with disease state on the basis of global shape analysis and found differential associations between SNPs and regional volumes at different disease stages [214]. Notably, in MCI subjects, SNPs in PML (promyelocytic leukemia) and STOML1 (stomatin-like 1) were associated with shapes of the hippocampi and insular cortices.
Although most GWAS analyses focus only on baseline phenotypes, two studies used ADNI’s longitudinal neuroimaging data that, by capturing rates of change, have greater statistical power to detect genetic associations. Longitudinal structural MRI data identified a much larger number of SNP-phenotype associations than cross-sectional data [215]. A second study used longitudinal change in ventricular volume as a quantitative phenotype for a pathway-based gene-gene interaction analysis [216] and identified a novel interaction between SYNJ2 (synaptojanin 2) and PI4KA (phosphatidylinositol-4 kinase) that was associated with inferior lateral ventricle atrophy. Both proteins are involved the synthesis of phosphatidylinositol, and the authors suggest that these genes might modulate its synthesis leading to deficits in neuroprotective mechanisms.
A whole-exome sequencing study by Nho et al. [217] using DNA from ADNI-1 and an extreme phenotype approach identified a functional exonic single nucleotide variant that was associated with a slower rate of hippocampal atrophy in MCI subjects. The minor T allele of the missense variant rs3796529 in REST, a negative transcriptional regulator of adult hippocampal neurogenesis [206], conferred a protective effect on hippocampal loss in MCI and AD subjects. A subsequent study showed that carriers of this minor allele had greater medial temporal lobe metabolism compared to noncarriers, independent of APOE4 status [207] and that it was associated with reduced AD risk and did not confer susceptibility to AD [208]. However, a meta-analytic study using the ENIGMA cohort did not find that the variant conferred a significant effect on six subcortical regions including the hippocampus [209]. Further investigation is required to determine the relative enrichment of the variant in diagnostic groups and whether it protects against hippocampal atrophy in CN subjects.
Most ADNI studies are concerned with late-onset AD. However, the cohort contains a small number of subjects aged 55 to 65 years who have a relatively early onset form of the disease and who are characterized by much lower frequencies of amnestic MCI [234]. Early onset AD is most commonly autosomal dominant, caused by mutations in the APP, PS1, and PS2 genes [235], but some subjects lack these risk alleles. Moon et al. [210] used the LONI pipeline [211] to identify 20 neuroimaging shape and atrophy changes specific to MCI subjects aged 55 to 65 years, for use as GWAS quantitative phenotypes. These included not only hippocampal volumes but precuneus atrophy and shape changes, consistent with previous studies of early onset AD. A set of 15 SNPs best able to discriminate between early-onset MCI and early-onset AD subjects was associated with the volumetric and shape changes. The most significant loci, located in the genes for Janus kinase and microtubule interacting protein-1 and neuropilin 1, differed from those identified in the late onset form of the disease.
6.2.2.3. Measures of Aβ deposition
GWAS using either amyloid imaging data or fluid levels of Aβ have furthered our understanding of the genetic basis of Aβ deposition. Using florbetapir data, the minor G allele of rs509208 located upstream of butyrylcholinesterase (BCHE) and the APOE loci together accounted for 15% of the variance in baseline cortical Aβ load (Fig. 11) [218]. BCHE is a biologically plausible gene; decreased activity of butyrylcholinesterase, which is enriched in Aβ plaques and the target of cholinesterase inhibitors, increases acetylcholine levels and disrupt synaptic functioning eventually leading to neurodegeneration. A gene-set enrichment analysis performed on top SNPs identified in a GWAS of CSF levels of Aβ42, including those located in TOMM40 and near APOC1, revealed two clusters associated with synaptic transmission, transmission of nerve impulses, and trait class, and identified eight novel polymorphisms [219]. A subsequent gene-gene interaction network analysis identified GRIN2A as interacting with the most AD genes, followed by APOC1 and TOMM40 (Fig. 12). A GWAS of plasma concentrations of Aβ40 and Aβ42 identified 18 suggestive loci, the most strongly associated of which was CTXN3 (cortexin 3) [220]. Pathway enrichment analysis identified a variety of canonical pathways including some directly associated with Aβ peptide properties. Cortexin was found to modulate Aβ42 secretion.
Fig. 11.
The effects of APOE ε4 and rs509208 (BCHE) on cortical Aβ levels. The APOE ε4 allele and the minor allele (G) of rs509208 of BCHE exerted independent and additive effects on cortical Aβ burden. Bars represent mean cortical Aβ levels ± standard errors. Abbreviations: BCHE, butyrylcholinesterase; SUVR, standardized uptake value ratio. Reproduced with permission from [218].
Fig. 12.
Genome-wide CSF Aβ42 associations. GeneMANIA networks showing the interaction results of (A) associated genes and (B) highly associated genes. (C) Novel polymorphisms identified in study. Abbreviation: CSF, cerebrospinal fluid. Reproduced with permission from [219].
Although Aβ deposition is strongly linked to APOE4 and nearby genes in linkage disequilibrium (TOMM40, APOC1), Aβ deposition is still observed in APOE4 noncarriers. A GWAS of longitudinal Aβ accumulation in these subjects [221] reported that the minor G allele of an intronic SNP (rs12053868) in interleukin 1 receptor accessory protein (IL1RAP) accounted for 7.1% of the phenotypic variance (Fig. 13). Deep sequencing of IL1RAP identified additional rare variants associated with the rate of change in Aβ burden. Gene- and pathway-based GWAS identified pathways related to cell adhesion and immune response, consistent with the role of IL1RAP as a proinflammatory cytokine involved in the activation of microglia. The minor G allele was associated with higher rates of atrophy in the temporal cortex, a higher rate of MCI to AD progression, and faster cognitive decline. Other novel loci identified in GWAS of CSFAβ42 levels in APOE4− subjects are SULG2 (GTP-specific β-subunit, succinyl-CoA ligase), which accounted for 10.7% of the variance in Aβ42 levels in these subjects [222], and variants in the FRA10AC1 fragile site and in the intergenic 15q21 locus [223]. FRA10AC1 encodes a nuclear phosphoprotein of unknown function.
Fig. 13.
Association and the effect of ILRAP rs12053868-G on longitudinal change in cortical Aβ PET burden. The minor G allele of rs12053868 in ILRAP was associated with higher rates of amyloid accumulation compared to the major A allele. Mean annualized percent change and global cortical 18F-florbetapir SUVR ± standard error. Abbreviation: SUVR, standardized uptake value ratio. Reproduced with permission from [221].
6.2.2.4. Other CSF and blood phenotypes
A GWAS of CSF tau levels identified rs4728029 in protection of telomeres 1 (POT1), which modified the relationship between p-tau181 and ventricular expansion [224]. This novel locus explained 2.6% of the variance in p-tau181 and modified the relationship between p-tau181 and both ventricular dilation and memory performance. As this SNP was related to levels of interleukin 6 receptor, it may exert its effect via a neuroinflammatory mechanism. This GWAS also enriched targeted genes mediated by miR-33, which regulates lipid metabolism genes and disrupts cellular cholesterol homoeostasis.
A complementary approach to GWAS of CSF Aβ42 and tau [225] instead used CSF analytes involved in the processes such as endocytosis, cholesterol metabolism, and inflammatory and immune responses that are recognized to play important roles in AD pathogenesis beyond Aβ and tau pathology. This study identified five genetic associations between CSF proteins and angiotensin-converting enzyme, chemokine (C-C motif) ligands 2 and 4, interleukin 6 receptor, and matrix metalloproteinase 3. All identified proteins are involved in Aβ processing or proinflammatory signaling.
6.2.2.5. Neuropsychological assessments
In addition to neuroimaging and fluid biomarker measures, neuropsychological assessments offer another route to understanding AD genetics. Variants in the spondin 1 (SPON1) gene whose minor alleles were associated with more rapid progression were identified using longitudinal changes in ADAS-cog as a phenotype for a GWAS [226]. Spondin 1 inhibits cleavage of APP by BACE. Other significant associations were reported in genes involved in neuronal maintenance and neurotransmission, and calcium signaling (EXOC4, GABRG3, VAT1L), and homoeostasis (CAMK4, CYCS, NCS1, CACNA1G). Mukherjee et al. [236] investigated the genetic basis of cognitive resilience, observed in patients whose cognitive function is better than predicted by neuroimaging and fluid biomarkers. The top hit in their gene-based GWAS was RNASE13, and subsequent pathway analysis identified pathways involved in neuron loss, presynaptic membrane, and postsynaptic density. These included genes associated with AD such as PTK2B, PICALM, MS4A2, and APP.
6.2.3. Other association studies
Imaging genetic studies using ADNI data have not only focused on AD. Two meta-analytic GWAS identified variants influencing human subcortical structures [228] and cortical surface area [229]. Polymorphisms in the dopamine D2 receptor gene, which increases genetic risk for addictive disorders, altered regional brain volumes in areas implicated in addiction [230].
6.3. Methods
The biological insights that we have gained from these genetic association studies, particularly those using neuroimaging data, would not have been possible without a foundation of statistical methodologies. These have progressed from univariate analysis in standard GWAS to a variety of multivariate regression approaches, as well as ways to detect epistatic interactions or to discover associations at the gene or pathway level. Univariate analysis comparing single SNPs with single traits suffers from problems of high dimensionality and type-I errors and may not reveal significant associations without meta-analytic approaches. ADNI studies have assessed the extent of these problems [237,238], improved the computational efficiency of mass univariate analyses [239–242], and developed methods for the selection of the most informative SNPs or quantitative features to improve power to detect associations [243–247]. Two studies have developed summary measures representing associations between selected SNPs and traits of interest [202,248]. Nho et al. [249] developed specific methodologies for the analysis of whole-genome sequencing data, and several groups have developed methodologies for identifying and visualizing genetic interactions [189,205,250–253]. Finally, two groups described methods to select discriminative SNPs to improve diagnostic classification or prediction of future decline [254,255]. The studies are summarized in Table 7.
Table 7.
Approaches for the improvement of genetic studies
| Challenge | Approach and results | Reference |
|---|---|---|
| Avoiding biased interference when using secondary phenotypes | Tested whether a standard analysis of secondary phenotypes encountered problems such as type-I errors and reduced power for association testing. Although the analysis was generally valid, authors recommend caution when analyzing these types of data. | [237] |
| Improving computational efficiency of mass univariate analyses | Presented a functional mixed-effects modeling framework to jointly analyze high-dimensional imaging data with genetic markers and clinical covariants. Tested associations of candidate genes (CR1, CD2AP, PICALM) with MRI brain regions. Detected regional clusters of voxels associated with candidate genes and different patient groups. Method was computationally efficient. | [239] |
| Presented a fast voxelwise genome-wide association analysis framework able to search for sparse signals while controlling for family-wise error rate. When tested on ADNI data with 708 subjects, 193,275 voxels, and 501,584 SNPs, the total processing time was 203,645 seconds for a single CPU, a substantial improvement over traditional methods. | [240] | |
| Presented modifications to mass univariate analyses by using dimensionality reduction techniques on both MRI imaging data and genomics data, and a new multiple testing adjustment method. Experiments suggest procedure has more power to detect associations. | [241] | |
| Presented a method based on a generalization of the partial least squares correspondence analysis which can simultaneously analyze behavioral and genetic data. | [242] | |
| Selecting most informative SNPs or quantitative features | Presented method that uses tree-guided sparse learning to learn the most informative SNPs and MRI measures and that models the a priori hierarchical grouping structure among SNPs. Experiments suggest method can identify informative SNPs. | [243] |
| Adopted a generalized estimation equations methodology to test the association between single SNPs and multiple quantitated traits. Found the method was general and flexible when tested on ADNI data using seven MRI-derived multivariate traits. Outperformed principal component analysis or canonical correlation analysis for dimensionality reduction. | [244] | |
| Presented a sparse projection regression modeling framework. Incorporates two novel heritability ratios to simultaneously perform dimensionality reduction, response selection, estimation, and testing. | [245] | |
| Evaluated several sparse canonical correlation analysis methods that can reveal complex multi-SNP, multiquantitative trait associations. Suggest that the estimation of covariate structure is limited in these methods. | [246] | |
| Tested three Bayesian network supervised learning methods on a whole-genome sequencing data to identify causal AD SNPs and the gene-SNP interactions. Reported that Markoff blanket-based methods outperformed both naïve Bayes and tree-augmented naïve Bayes methods in selecting SNPs strongly associated with AD from top-ranked susceptibility genes. | [247] | |
| Developing a summary measure of associations between multiple SNPs and traits of interest | Adapted the Rasch model to compute a multimarker genetic summary score which accounts for statistical issues such as inflated false-positive rates, linkage disequilibrium. Genetic summary score can then be used for association analysis. | [202] |
| Developed a summary score based on an asymptotically normal and consistent estimate of the parameter vector to be tested and its covariance matrix. The derived score vector extended several score-based tests to mixed-effects models. | [248] | |
| Accurately calling multisample variants for whole-genome sequencing | Compared two multisample variant-calling methods for the detection of small nucleotide variants and short on solutions and deletions using a whole-genome sequencing data from ADNI subjects. Found that the JOINT method, which first calls variants individually and then genotypes the variant files for all samples, outperformed the second method, REDUCE. | [249] |
| Identifying interaction effects | Combined kernel machine regression and kernel distance covariance to identify associated genetic markers with multidimensional phenotypes. Identified SNPs in FLJ16124 that exhibit pairwise interaction effects correlated with volumetric changes. | [250] |
| Proposed a general kernel machine-based method to jointly detecting genetic and nongenetic variables and their interactions. Framework consists of a genetic kernel to capture epistasis, and a nongenetic kernel which can model the joint effects of multiple variables. | [189] | |
| Proposed a new Bayesian generalized low-rank regression model to characterize the association between genetic variants and brain imaging phenotypes, while accounting the impact of other covariance. Tested using 20 most significant SNPs from ADNI and identified loci associated with brain regions. Method was more computationally efficient and less noisy because it reduced the number of parameters to be sampled and tested. | [251] | |
| Presented a method to consider joint effects for polymorphisms in a biologically defined pathway. Determines SNP-SNP interactions using a quantitated multifactor dimensionality reduction technique, infers functional relationships between selected genes, and uses gene-set enrichment analysis to determine whether genes and functional network occur more frequently than expected by chance and biological pathways defined by gene ontology. | [205] [252] | |
| Proposed a versatile likelihood ratio test to detect mean and variance heterogeneity present in loci due to biological disruption, gene by gene or gene by environment interactions or linkage disequilibrium. | ||
| Visualizing genetic interaction networks | Used 3D printing to visualize a statistical epistasis network of 34 significant SNPs. Suggest that a 3D physical model may make interpretation of data easier than from a digital representation. | [253] |
| Imputing common APOE SNPs missing from genome-wide genotyping arrays. | Compared directly genotyped SNPs versus SNPs imputed via reference panel compiled by the Thousands Genome Project. Reported that the imputation method is highly accurate. | [238] |
| Learning predictive models or progression profiles | Applied three feature selection methods (multiple kernel learning, high-order graph matching-based feature selection, sparse multimodal learning) to classification challenges using multidimensional imaging genomics data and biochemical markers. Found that higher order graph matching-based feature selection gave best results. | [254] |
| Used the principal component analysis to select most important SNPs associated with clinical diagnosis and used these data along with hippocampal surface information to predict MCI to AD progression. | [255] |
Abbreviations: MRI, magnetic resonance imaging; ADNI, Alzheimer’s Disease Neuroimaging Initiative; SNP, small nucleotide polymorphism; AD, Alzheimer’s disease; MCI, mild cognitive impairment.
6.4. Conclusions
Much of the heritability of AD remains elusive, reflecting both the polygenic nature of the disease and the influence of environmental factors. The ADNI Genetics Core adopted a Systems Biology perspective and approaches that integrate multiple “omics” to characterize biological complexity on multiple levels [171]. ADNI genetics studies from 2014 and 2015 were notable for moving beyond simple GWAS using one-SNP-simple phenotype univariate analyses to an expanded array of increasingly sophisticated biologically informed approaches, such as pathway enrichment, network analysis, and gene set enrichment, aimed at bridging the gap between genetic information and biochemical disease mechanisms. The concomitant development of a wide range of statistical methodologies supported these efforts (Table 7). An excellent example of the power of these integrative approaches to place genetic results in biologically plausible context can be seen in the hypothetical signaling network constructed by Wang et al. [202] from their application of a Rasch genetic multimarker model to ADNI case-control GWAS data (Fig. 14).
Fig. 14.
Hypothetical signaling network integrating top genes identified through Rasch analysis. A Rasch model was applied to the genes of ADNI GWAS data and supports APOE as a major susceptibility gene for AD, and functionally links other top genes (AEN, ADANTS12, PSMA5, FXN, NTRN, LARP1, WDTC1, SEMA7A, VKORC1L1, COL5A3) to AD. A hypothetical signaling network was generated from a pathway analysis of these genes based on known proteinprotein, functional, and phenomenological interactions. Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; ARC, activity-regulated cytoskeleton-associated protein; EEF2K, eukaryotic elongation factor-2 kinase, activated by GRM5 receptor, regulates global protein synthesis; HDAC3, histone deacetylase; MDM2, negative modulator of TP53 tumor suppression gene; PLXNC1, plexin C1 receptor for semaphorins; PTK2: FAK, kinase implicated in integrin signaling; FYN, src family tyrosine kinase, downstream target of GRM5 receptor; RPTOR, regulatory protein associated with MTORC1 complex. Reproduced with permission from [202].
Genetic association studies begin either with a genotype, such as an SNP, gene, or a sets of genes in a pathway of interest, or with a phenotype (Table 5). Several GWAS notably capitalized on longitudinal phenotypes from ADNI’s rich data set to increase the power to capture significant associations [215,216,221]. Strategies for finding the “missing heritability” of the disease included searching for epistatic interactions, examining the associations of low-frequency variants, and extending analysis from a target gene to genes in the same biological pathway. The APOE ε4 allele was associated with atrophy and shape changes in the hippocampus and other regions [176–178] and was found to have differential effects in males and females [179]. Studies of APOE ε4 noncarriers identified loci responsible for disease phenotypes such as Aβ deposition in these subjects [188,222,223] (Table 6). Novel protective and risk loci associated with atrophy, metabolism, Aβ load, CSF biomarkers, and cognitive decline and with early onset AD in the absence of established autosomal-dominant genes were identified (Table 6). Risk genes lay in pathways involved in APP processing and Aβ clearance, apoptosis, cholesterol metabolism, neurotransmission, immune and inflammatory responses, microglial activation, and other cellular processes. Interestingly, stroke and AD risk genes appeared to overlap, implicating WM disease in AD pathology. Identified protective genes were involved in the inhibition of neurodegeneration and cholesterol metabolism, and cancer. Finally, ADNI genetics data have contributed to understanding other diseases with the inclusion of the cohort in various meta-analyses [228–230] and in the ENIGMA consortium [173].
7. Disease progression
A better understanding of the AD pathological pathway is central to ADNI’s goal of developing biomarkers for the improvement of clinical trials. AD is pathologically characterized by amyloid plaques composed largely of fibrillar forms of Aβ, and neurofibrillary tangles, composed of hyperphosphorylated tau (p-tau181) that is associated with synapse loss and neurodegeneration. The amyloid cascade hypothesis [256] has dominated research over the last two decades, to the point that diagnostic criteria for AD now include Aβ abnormalities. The amyloid hypothesis states that incorrect processing of the APP to form Aβ, together with an imbalance in the clearance of Aβ accumulation, triggers a cascade of events: the formation of Aβ plaques, the accumulation of fibrillar tau in cells as NFTs and neuropil threads, neuronal death and synaptic dysfunction, disruption of glucose metabolism, atrophy, and eventual cognitive decline. However, although cortical hypometabolism is largely linked to global amyloid burden, regional amyloid plaque deposition has little or no association with regional hypometabolism [257]. Jack et al. [258] proposed a hypothetical model describing the order in which biomarkers become abnormal during disease progression which largely follows this cascade of events (Fig. 15). The Biostatistics Core has been instrumental characterizing longitudinal trajectories of biomarkers through their biostatistical analyses that integrate data across the breadth of ADNI studies [259]. Numerous studies of ADNI data have supported this model and have been described in a previous review [1].
Fig. 15.
Schematic outlining of the current understanding of the hypothetical timeline for the onset and progression of AD neurodegeneration and cognitive impairments. Age is indicated at the bottom, whereas the green, blue, and red bars indicate the time at which preventive, disease-modifying, and symptomatic interventions, respectively, are likely to be most effective. Within the aqua bar, milestones are shown in the pathobiology of AD that culminate in death and autopsy confirmation of AD. The proposed ADNI model of the temporal ordering of biomarkers of AD pathology relative to stages in the clinical onset and progression of AD is shown in the insert at the upper right based on Jack et al. [258], whereas the insert at the left illustrates the defining plaque and tangle pathologies of AD and common comorbid pathologies including Lewy body pathology (SYN), TDP-43, and hippocampal sclerosis. In the insert on the right, clinical disease is on the horizontal axis and it is divided into three stages: CN, MCI, and dementia. The vertical axis indicates the range from normal to abnormal for each of the biomarkers and the measures of memory and functional impairments. Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CN, cognitively normal; CSF, cerebrospinal fluid; FDG, [18F]-fluorodeoxyglucose; MCI, mild cognitive impairment; MRI, magnetic resonance imaging; PET, positron emission tomography. Reproduced with permission from [129].
However, mounting evidence suggests that AD progression is a far more complicated tale. At autopsy, a substantial number of subjects have copathologies such as Lewy bodies, hippocampal sclerosis, and transactive response DNA binding protein 43 kDa (TDP-43) inclusions [260] (Fig. 15). The ADNI Biostatistics Core concluded that although Aβ positivity accelerates the progression from MCI to AD and subsequent cognitive decline, there is substantial evidence for alternative pathways to this end point [259]. The purpose of this section, therefore, is to outline evidence for the amyloid cascade hypothesis, primarily gained from studies of Aβ+ subjects, and for other possible pathways to dementia.
7.1. Disease progression in Aβ-positive subjects
In an increasingly complex view of AD progression, the pathway from presymptomatic CN Aβ− to CN Aβ+ to predementia MCI Aβ+ to AD Aβ+ remains the backbone of the process. Moreover, a patient may transition from a negative to a positive Aβ status at any time (Fig. 16). In dissecting out different pathways of disease progression, a number of studies have dichotomized subjects on the basis of Aβ positivity, defined either on the basis of a CSF Aβ42 levels, or from cortical Aβ load determined by amyloid PET. In this way, investigators have begun to tease out, even in those with normal cognition, the associations between Aβ deposition, and other factors in the disease such as metabolism, atrophy, APOE4 status, cerebral blood flow, and WM architecture. These studies have profound implications for our understanding of AD progression.
Fig. 16.
Model for Aβ status in disease progression. Illustrating the view of progression from presymptomatic Aβ negative to presymptomatic Aβ positive to MCI Aβ positive to AD Aβ positive as the primary pathway to AD, with the switch to Aβ positivity also occurring anywhere in the progression. A small percentage of clinically diagnosed AD patients lack Aβ pathology at autopsy. Abbreviations: AD, Alzheimer’s disease; MCI, mild cognitive impairment.
7.1.1. Aβ and the ordering of biomarkers
Aβ status clearly affects disease progression. A cross-sectional study [261] found that levels of CSF t-tau and p-tau181 only became abnormal in Aβ+ subjects and that trajectories of the CSF biomarkers across disease stages were distinct in the dichotomized groups (Fig. 17). Young et al. [262] developed an event-based model which used distributions of biomarker values rather than predetermined biomarker cut points to determine the sequence in which AD biomarkers become abnormal. In Aβ+ or APOE4 participants, the first CSF biomarker to become abnormal was Aβ42, followed by t-tau and p-tau181, the order predicted by the Jack model. In Aβ+ subjects, several studies [261–264] reported an ordering of biomarkers consistent with the Jack model: levels of CSF Aβ42 becoming abnormal in the preclinical stage, tau reaching maximum abnormality in the MCI stage, and imaging and cognitive markers beginning to decline in the asymptomatic stage but accelerating with advanced clinical stage [261,264]. These studies favor the concept of abnormal Aβ deposition as a prequel for tauopathy in AD.
Fig. 17.
Differential trajectories of CSF biomarkers in Aβ+ and Aβ− subjects across disease progression. The means (±2 standard error of the mean) in ng/mL of (A) CSFAβ42; (B) CSF p-tau; (C) CSF p-tau181 are shown. In Aβ− groups, the levels of all three CSF biomarkers did not significantly increase across disease stage, whereas in Aβ+ subjects, CSF p-tau and t-tau, but not Aβ, increased across disease stages. Abbreviations: a-MCI, (advanced) mild cognitive impairment; Aβ+, subjects with abnormal brain Aβ; Aβ−, subjects without abnormal brain Aβ; CN, cognitively normal; CSF, cerebrospinal fluid; i-Dem, (incipient) dementia; iMCI, (incipient) mild cognitive impairment; m-Dem, (mild) dementia. Reproduced with permission from [261].
7.1.2. Aβ and brain atrophy
Several studies support a link between Aβ positivity and increased regional atrophy in the brains of CN subjects. Hippocampal atrophy, a nonspecific characteristic of AD, accelerates throughout the disease process but also occurs in normal aging. Nosheny et al. [128] found that Aβ positivity contributed to, but did not entirely account for, hippocampal atrophy rate in CN participants (Fig. 18). Furthermore, atrophy preceding that in medial temporal regions has been detected by several groups, specifically in the precuneus [265]; the right supramarginal/inferior parietal gyrus [261]; and the posterior cingulate, amygdala, putamen, precuneus, and brainstem (before the Aβ42 cut point of 192 ng/mL insula) [266] (Fig. 19).
Fig. 18.
Hippocampal atrophy rate in Aβ+ and Aβ− CN and MCI subjects. The percentage of hippocampal atrophy rate attributable to Aβ status was calculated from the difference in hippocampal atrophy rate between Aβ+ and Aβ− subgroups. Aβ was measured using florbetapir PET. *P < .01, **P < .001, and ***P < .0001. Abbreviations: NC, normal cognition; MCI, mild cognitive impairment; PET, positron emission tomography. Reproduced with permission from [128].
Fig. 19.
Regions of Aβ-related atrophy ordered by acceleration and stabilization points. Regions, including the insula, posterior cingulate, amygdala, putamen, and precuneus, show early signs of atrophy before the hippocampus and entorhinal cortex. Parietal regions appear to have a shorter transition compared to temporal lobe regions with respect to Aβ. Red, yellow, and black dots represent significant (P <.05), marginally significant (.10 > P <.05), and nonsignificant (P >.10) acceleration or deceleration, respectively. Scale is pg/mL CSF Aβ42. Abbreviation: CSF, cerebrospinal fluid. Reproduced with permission from [266].
Cholinergic neurons in the basal forebrain regulate the supply of acetylcholine to areas of the temporal cortex and to the amygdala thereby influencing memory and attention, and their degeneration is regarded as a key event in AD pathogenesis. Aβ burden has been associated with basal fore-brain degeneration independent of APOE4 status in emergent Aβ+ asymptomatic subjects [267–269]. Moreover, basal forebrain volume classified preclinical patients as Aβ+ or Aβ− more accurately than hippocampal volume [269]. In MCI subjects, basal forebrain degeneration was more associated with impaired memory and attentional control, whereas hippocampal atrophy was more associated with memory deficits. This association was mediated by hypometabolism in domain-specific cortical networks and was not affected by Aβ status [27]. These studies support cholinergic basal forebrain neurodegeneration and the concomitant deposition of Aβ as early events in AD.
Two studies place glucose hypometabolism as an intermediary event between Aβ positivity and atrophy in accordance with the Jack model for the temporal ordering of biomarkers [258]. Kljajevic et al. [133] examined baseline FDG PET and structural MRI scans of Aβ+ CN, EMCI, and late MCI (LMCI) subjects compared with Aβ− CN elderly subjects. Glucose hypometabolism originated in the posterior parietotemporal regions before atrophy which originated in medial temporal regions (Fig. 20). Araque Caballero et al. [265] also reported that atrophy originated in the medial temporal lobe in MCI Aβ+ subjects and, furthermore, that this pattern of atrophy correlated with patterns of hypometabolism, suggesting that by the time the first cognitive symptoms arise, both these kinds of neurodegeneration are spatially associated. Although Kljajevic et al. [133] posit that abnormal Aβ deposition may affect synaptic activity leading to subsequent neuronal loss and hypometabolism, the results from Dowling et al. [270] suggest that this is an earlier event in pathogenesis and therefore more weakly related to changes in hypometabolism and subsequent changes than intraneuronal neurofibrillary degeneration.
Fig. 20.
Hypometabolism originates earlier than atrophy in Aβ+ subjects. CN Aβ+ subjects displayed significant hypometabolism in medial parietal and bilateral parietal temporal regions compared to Aβ− subjects, whereas there was no difference in GM volume between these two groups, indicating that hypometabolism precedes atrophy in Aβ+ subjects (A) hypometabolism (red) and (B) atrophy (blue). Abbreviations: Aβ+, subjects with abnormal brain Aβ deposition; Aβ−, subjects without abnormal brain Aβ deposition; CN, cognitively normal; EMCI, early mild cognitive impairment; GM, gray matter; LMCI, late mild cognitive impairment. Reproduced with permission from [133].
7.1.3. Aβ and cognitive deficits
The relationship between Aβ positivity and cognition may be more complex than the causal sequence of pathological events implied by the Jack model in which altered levels of CSF peptides exert a neurotoxic effect which impairs cell function and leads to decreased glucose metabolism, ultimately resulting in cognitive decline. First, Aβ positivity may affect cognition at a far earlier stage than previously thought. Susanto et al. [261] reported that CN Aβ+ subjects had impairments in executive functioning/processing speed which was accompanied by atrophy at the right supra marginal/inferior parietal gyrus. Mattsson et al. [271] reported that CN Aβ+ subjects had lower memory scores and smaller GM volumes in several regions including the hippocampus. Both studies reported that MCI Aβ+ subjects had widespread atrophy and impairments to multiple cognitive domains.
Second, Aβ positivity may exert its effect on cognition via multiple pathways. Mattsson et al. [271] used a mediation analysis to investigate the role of atrophy and hypometabolism in mediating the effect of Aβ on episodic memory in MCI patients. Hippocampal atrophy mediated approximately 25% of this association, and approximately 40% was mediated by a combination of hippocampal atrophy and hypometabolism (Fig. 21). Interestingly, the fact that 60% of the association between Aβ and memory was not mediated by hypometabolism and/or atrophy suggests that other mechanisms must exist. A study by Byun et al. [272] supports this idea. In subjects with significantly lower CSF Aβ42 levels compared to controls, they identified four subtypes of AD characterized by different patterns of regional brain atrophy and rates of progression: primarily hippocampal atrophy (19%), primarily cortical atrophy (17.7%), both hippocampal and cortical atrophy (41.1%), and neither region (10.4%). They found that subtypes characterized by primarily hippocampal atrophy exhibited more severe impairment of the memory domain (ADNI-Mem), whereas executive function (ADNI-EF) was more impaired in subtypes with predominantly cortical atrophy (Fig. 22). The finding that heterogeneous atrophy patterns exist with different rates of progression in the presence of Aβ neuropathology was also reported by Mattsson et al. [273] who found that a subset of CN participants with normal baseline levels of CSF Aβ42 that decreased over time, or with reduced baseline levels of CSF Aβ42, had increased frontal and parietal cortical atrophy but no accelerated temporal atrophy longitudinally. Byun et al. [272] postulated that these subtypes correlate with distribution of neurofibrillary tangles, suggesting a closer relationship between tau neuropathology and cognition than Aβ neuropathology and cognition, a hypothesis supported by two further studies. First, Fortea et al. [274] used correlation analysis to determine that p-tau-dependent cortical thinning was found only in Aβ+ subjects and not in Aβ− subjects. In the absence of abnormal p-tau, abnormal CSF Aβ42 was related to cortical thickening. The authors posited a two-phase phenomenon in which there is initial cortical thickening as Aβ levels become abnormal followed by cortical thinning as p-tau reaches pathological levels. Second, Dowling et al. [270] used a mediation analysis to examine changes in brain glucose metabolism, longitudinal changes in global cognition, their association over time, and the impact of baseline CSF measures on these associations. Glucose hypometabolism in all AD-associated brain regions mediated the relationship between CSF indicators of neuronal damage (t-tau, p-tau181, and their ratios), and cognitive decline. In contrast, only hypometabolism in the middle inferior temporal gyrus mediated the effect of baseline levels of Aβ42 on cognition. Overall, there is general (but not universal) agreement that initial development of Aβ plaques accelerates development of tau tangles which leads to synapse loss and neurodegeneration. However, TF-fMRI activity is disrupted in CN elders who are Aβ+ [275], suggesting that Aβ plaques alone may affect brain function before significant accumulation of tau tangles. Hopefully, the use of both amyloid and tau PET in future studies including ADNI-3 will shed more light on this matter.
Fig. 21.
Hippocampal volume and hypometabolism mediate the effect of Aβ on longitudinal change in Logical Memory Delayed Recall. Path analysis showing how hippocampal volume and angular FDG PET mediate the effect Aβ of on longitudinal change in Logical Memory delayed recall. (A) The direct effects of Aβ on memory; (B) hippocampal volume mediating the effects of Aβ on memory; (C) angular FDG PET mediating the effects of Aβ on memory; and (D) the combination of hippocampal volume and FDG PET mediating effects of Aβ on memory. The figure includes the following standardized regression coefficients: a, the effects of Aβ on hippocampal volume or FDG PET; b, the effects of hippocampal volume or FDG PET on the memory when adjusting for Aβ; c, the direct association between Aβ and memory (without adjusting for hippocampal volume or FDG PET); c′, the association between Aβ and memory when adjusting for hippocampal volume and/or FDG PET; and c-c′, the mediated effect on memory (with % mediation). *P < .05. Abbreviations: FDG, [18F]-fluorodeoxyglucose; PET, positron emission tomography. Reproduced with permission from [271].
Fig. 22.
Longitudinal changes in cognition in subtypes of Aβ+ cognitively normal subjects. Identified subtypes of cognitively normal subjects consisted of those with predominantly hippocampal atrophy, predominantly cortical atrophy, hippocampal and cortical atrophy combined, or neither type of atrophy. Baseline, 1-year, and 2-year follow-up data on (A) MMSE indicating global cognition, (B) ADNI-Mem indicating memory function, and (C) ADNI-EF indicating executive function are plotted, with means and standard errors. Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; BI, both impaired; BS, both spared; CA, cortical atrophy only; EF, executive function; HA, hippocampal atrophy only; Mem, memory domain; MMSE, Mini–Mental State Examination. Reproduced with permission from [272].
Disease progression in subjects dichotomized by APOE4 status appears to be similar to that in subjects dichotomized by Aβ status: the two groups have the same defined sequence of CSF biomarkers [262] as well as faster trajectories of CSF t-tau, and p-tau [276], and faster hippocampal atrophy [128]. But the APOE4 allele may actually modulate disease progression in Aβ+ subjects. Susanto et al. [261] reported that MCI and AD carriers of this allele had more severe atrophy of the medial temporal lobe and worse memory impairment but higher executive functioning/processing speed than noncarriers, suggesting that APOE4 modulates trajectories of both cognition and atrophy.
7.1.4. Aβ and cerebral blood flow
Mattsson et al. [277] determined the association of Aβ with variations in cerebral blood flow, a measurement of brain activity, across the cognitive spectrum. With all subjects, brain Aβ was associated with reduced cerebral blood flow in temporoparietal regions, but with increased cerebral flow in the posterior cingulate suggesting a compensatory mechanism for Aβ neurotoxicity in the latter region. Dichotomization of subjects by Aβ status revealed reduced cerebral blood flow is in several regions in Aβ+ compared to Aβ− participants. Aβ load was more associated with reduced cerebral blood flow than atrophy in CN subjects but the opposite was true in LMCI or AD patients. In keeping with the Jack model of disease progression [258], these results suggest Aβ is more associated with functional and synaptic loss leading to reduced cerebral blood flow early in disease progression and more associated with GM loss leading to atrophy later in disease progression.
7.2. Alternative pathways to dementia?
Beckett et al. [259] concluded that although Aβ positivity accelerates the progression from MCI to AD and subsequent cognitive decline, there is substantial evidence for alternative pathways to this end point. In their data-driven model of biomarker changes in AD, Young et al. [262] observed that in the pooled samples, t-tau was the first biomarker to become abnormal, followed by p-tau181 and Aβ42; only in Aβ+ or APOE4+ subjects was Aβ42 first biomarker to become abnormal, as predicted by the Jack et al. model [258]. Furthermore, trajectories of CSF biomarkers across disease stages in Aβ− subjects were distinct from Aβ+ subjects and did not feature the stereotypical decreased Aβ42 and increased p-tau181 and t-tau [261]. These results imply that a substantial proportion of ADNI subjects have atypical disease progression.
7.2.1. Heterogeneity of diagnostic groups
7.2.1.1. Subtle cognitive impairment
The appearance of subtle cognitive impairment (SCI), defined using 5th percentile cutoffs of composite memory and/or executive function scores, follows stereotypical sequential changes in Aβ and neurodegeneration biomarkers. A newly identified group, accounting for 5% of CN subjects, had SCI without neuronal injury in the presence or absence of abnormal Aβ [278]. Toledo et al. [279] reported that 27.6% of ADNI healthy controls had SCI and further defined three categories within this group—memory, executive, and multidomain—that differed in biomarker profiles and in the rate of progression to MCI and AD. The multidomain group, characterized by having the fastest progression to MCI, the most abnormal levels of Aβ42, atrophy, and greatest posterior cingulate FDG PET hypometabolism, was most related to AD. The memory group, characterized by slightly abnormal Aβ42 and increased atrophy, had a slower progression to MCI and the executive group had the slowest progression to MCI.
7.2.1.2. MCI
Several studies identified MCI subtypes using cluster analysis of neuropsychological data [280–282]. An amnestic subtype characterized by isolated memory impairment and abnormal CSF biomarkers, representing “typical” AD, was identified in all studies (summarized in Table 8). However, this cluster comprised only 25.7% to 58.6% of MCI subjects. The remaining MCI subjects were clustered into a number of different groups, most commonly a dysexecutive/mixed group typically characterized by significant deficits predominantly in executive function, elevated p-tau181, and the fastest progression to AD (12.4%–33%, identified in 4/5 studies), and a normal group, comparable to CN controls (31.3% to 41.3%, identified in 3/5 studies). The normal group had the lowest rate of progression to AD (10.7% at follow-up), and a significant rate of regression to normalcy (9.2% at follow-up), suggesting that ADNI MCI criteria may have a high rate of false-positive diagnostic errors leading to misclassification of subjects [280]. These criteria include a subjective memory concern reported by either the subject or a study partner, a single memory score (delayed recall of Story A from the Wechsler Logical Memory II test), and a global CDR score of 0.5 that may not capture variability in cognitive phenotype. Subjective memory impairment was overestimated by MCI subjects in the normal cluster but underestimated by MCI subjects in the amnestic group, suggesting that subjective memory concerns are not reliable in making an MCI diagnosis [282]. The false-positive diagnostic errors are also consistent with reports of high reversion rates of MCI subjects to cognitively normal [281].
Table 8.
Cluster-derived MCI subtypes from ADNI studies
| Cluster-derived MCI subtypes (% of MCI group) | |||||
|---|---|---|---|---|---|
| Edmonds et al. [282] | Edmonds et al. [281] | Peter et al. [283] | Bondi et al. [280], Actuarial† | Bondi et al. [280], ADNI criteria | |
| Amnestic (typical AD) | 25.7 | 34.9 | 32.0 | 58.6 | 56 |
| Dysexecutive/mixed | 33.0* | 12.4 | - | 12.9 | 19.7 |
| Normal | 41.3 | 34.2 | - | - | 31.3 |
| Language/naming impaired | - | 18.5 | 17 | 21 | - |
| Visuospatial impaired | - | - | 42 | - | - |
| Focal intrusions | - | - | 9 | - | - |
Abbreviations: MCI, mild cognitive impairment; ADNI, Alzheimer’s Disease Neuroimaging Initiative; AD, Alzheimer’s disease.
Included impairments in naming.
Classified 401 subjects as MCI (compared to 846 using conventional ADNI criteria).
The observation that MCI reverters differed little from CN subjects in levels of CSF biomarkers, APOE4 status, hippocampal volume, hypometabolism, and other measures but differed substantially from other MCI subjects supports this idea [284]. Bondi et al. [280] reported that MCI subjects classified using an alternative actuarial method (which diagnosed fewer ADNI subjects with MCI) did not contain the cluster-derived normal group. Interestingly, 3/5 studies identified a group with language or naming impairments (17%–21%), and one study identified a large cluster (42%) characterized by visuospatial impairments and a small cluster (9%) characterized by a focal intrusions (recalled items which were not part of the list) [283]. The language impairment group was characterized by fast progression to AD and a high frequency of APOE4 [281,283]. Language deficits may reflect neuronal loss in the left hemisphere. Pravata et al. [285] found greater atrophy in the Brodmann area 20 of the left fusiform gyrus 12 months before progression, and in the left hemisphere GM 12 months after progression in MCI converters with language impairment compared to those without.
Cluster analysis of MCI subjects on the basis of baseline MRI, CSF, and serum biomarkers identified four clusters with distinct biomarker patterns [286]. Like studies based on neuropsychological data, one group (44%) appeared to have “typical AD” characteristics (and a small group appeared to be pre-AD), and another was similar to normal controls (14%). However, the latter group also had pronounced memory deficits and smaller hippocampal volume, although they rarely converted to AD, and the authors suggest that the stable group may be undergoing a non-AD process such as hippocampal sclerosis. The final group (37%), characterized by severe ventricular expansion, hippocampal atrophy, and progression to AD but near normal levels of tau and Aβ, also appeared to be on a different path to dementia. Although further investigation is required to refine MCI subtypes, these studies imply that multiple pathological pathways underlie the substantial heterogeneity of this group.
7.2.1.3. Cortical atrophy
Patterns of atrophy differ in subjects stratified by Aβ status (Section 7.2.2) but also differ across patient groups. Ventricular expansion reflects cortical atrophy in regions associated with early AD. Madsen et al. [287] reported that 2-year ventricular expansion was associated with baseline cortical volume and thickness in combined patient groups and with thinning in areas of temporal, frontal, and parietal cortices affected by AD in MCI participants. Distinct patterns of cortical atrophy were present in three neuropathologically defined subtypes of AD [163]. Compared with AD subjects with a diffuse pattern of cortical thinning, AD subjects with a pattern of medial temporal thinning had more glucose hypometabolism in hippocampus and bilateral frontal cortices, and worse memory performance, and AD subjects with thinning in predominantly parietal regions were younger, had more glucose hypometabolism in parietal and occipital cortices, and showed Aβ accumulation in most regions [163]. No differences in CSF Aβ42 or tau levels were seen in any groups. Cortical atrophy patterns may reflect differing underlying pathologies.
7.2.1.4. Significant memory concerns
Current MCI diagnostic criteria include a significant memory concern (SMC) from either the patient, clinician, or someone close to the patient in addition to quantitative evidence of cognitive impairment with relative preservation of functional abilities [288]. ADNI-2 enrolled an additional cohort of subjects who were clinically evaluated as CN but who had SMCs in the interest of capturing the earliest cognitive decline. By most measures—cognitive performance, hippocampal volume, Aβ deposition, and metabolism—these subjects were indistinguishable from normal controls [259]. However, one study reported increased frontal atrophy in this group [279], and a cluster analysis of both SMC and CN subjects revealed three distinct groups (Fig. 23) [259]. The first cluster had normal levels of Aβ, normal metabolism, a low frequency of the APOE ε4 allele, but evidence of hippocampal atrophy. The second cluster appeared normal by all measures, and the third cluster aligned with biomarkers of “typical AD,” having abnormal Aβ, a high-frequency of the APOE ε4 allele, and slight hippocampal atrophy. Two studies examined SMCs in MCI subjects, one reporting that these concerns were only related to verbal episodic learning performance and not to neuroimaging biomarkers, executive functioning, language, or other cognitive domains [289], and the other suggesting that these concerns were only weakly associated with objective functioning [282].
Fig. 23.
Heterogeneity of the subjective memory concern cohort. Cluster analysis identified three distinct subgroups in both the normal cognition (NC) and subjective memory concern (SMC) groups. The first subgroup (1) had elevated brain amyloid, decreased CSF Aβ, and substantially reduced hippocampal volume; the second subgroup (2) was similar to group 1 but with less hippocampal atrophy and was thought to correspond to the Jack sequence for early signs of AD, and the third group (3) was normal by all measures. Abbreviations: AD, Alzheimer’s disease; CSF, cerebrospinal fluid; ICV, intracranial volume; SUVR, standardized uptake value ratio. Reproduced with permission from [259].
7.2.2. SNAP
Cluster analyses consistently identify a group characterized by signs of neuronal injury such as elevated p-tau181 or atrophy and accelerated progression to AD. This cluster is consistent with a group, comprising around a quarter of CN and MCI subjects, that typically has neurodegeneration in the absence of Aβ pathology and low frequencies of the APOE ε4 allele, recently been termed suspected non-Alzheimer’s pathology (SNAP) [290,291] (Fig. 24, Table 3). Over 7 years, more than half (12/19) of MCI progressors with SNAP developed AD; 5/19 progressed to a frontotemporal dementia, and 2 to Lewy body dementia [291]. It is possible that SNAP subjects who progressed to AD were misdiagnosed initially as being Aβ− due to limitations in the CSF Aβ42 assay or to Aβ levels being close to the cut point for amyloid positivity used (Amyloid positivity is defined either on the basis of a CSF Aβ42 level of greater than 192 pg/mL [292] or from cortical Aβ load determined by amyloid PET using the radiotracers Pittsburgh compound B or florbetapir [293]. Aβ+ subjects have levels of amyloid above either or both of these threshold values, and Aβ− subjects have levels of amyloid below the threshold values).
Fig. 24.
Biomarker abnormality in A− N−, A+ N−, SNAP, and A+ N+ MCI patient groups, disaggregated by progressive cognitive deterioration. MCI subjects were grouped on the basis of absence or presence of abnormal levels of amyloid and neurodegeneration. SNAP subjects were neurodegeneration positive but amyloid negative. All four groups significantly differed in CSF Aβ42 concentrations, hypometabolism on FDG PET, and hippocampal volume. SNAP subjects were characterized by more severe hippocampal atrophy than other groups in the absence of abnormal amyloid. Triangles denote progressors, whereas circles denote nonprogressors. Abbreviations: AD, Alzheimer’s disease; A− N−, amyloid negative neurodegeneration negative; A+ N−, amyloid positive neurodegeneration negative; A+ N+, amyloid positive neurodegeneration positive; CSF, cerebrospinal fluid; FDG, [18F]-fluorodeoxyglucose; MCI, mild cognitive impairment; PET, positron emission tomography; SNAP, suspected non-Alzheimer’s pathology. Reproduced with permission from [291].
Cognitive deterioration in SNAP subjects appeared more related to neuronal damage than to Aβ pathology. Medial temporal tau pathology may underlie SNAP and is referred to as primary age-related tauopathy (PART). It is possible that SNAP represents an aging process separate from AD [290]. The inclusion of tau PET imaging in ADNI-3 may help to resolve the involvement of PART in SNAP subjects. SNAP subjects also had higher levels of YKL40, a marker of neuroinflammation, than Aβ+ presymptomatic subjects [144], supporting the idea. Although the pathophysiology of this group is not yet known, it may involve Aβ unrelated pathologies such as hippocampal sclerosis, argyrophilic grain disease, Lewy body disease, or frontotemporal degeneration, with a variety of different pathologies found at autopsy. Caroli et al. [291] proposed a further subdivision of SNAP subjects into a subgroup with severe cortical damage and no hippocampal atrophy with underlying frontotemporal degeneration, and a second subgroup with preserved cortical metabolism but hippocampal atrophy with underlying hippocampal sclerosis or argyrophilic grain disease.
7.2.3. The role of WM disease
One source of biological heterogeneity in AD may be small-vessel cerebrovascular disease [1,54]. WM lesions are highly prevalent in AD and may represent microvascular ischemic and/or demyelinating changes linked to cerebrovascular disease [294]. Recognition of the contribution of cerebrovascular disease to AD is reflected in NIA-AA criteria for AD that include a diagnosis of possible AD dementia in circumstances where patients have an etiologically mixed presentation including a severe burden of white-matter hyperintensities (WMHs), a widely accepted measure of small-vessel cerebrovascular disease [294]. However, as WM microstructural and cerebrovascular disruptions are observed in both elderly and demented subjects, the threshold separating WM lesion burden in these two patient groups has yet to be determined and the specific WM disease contribution to AD remains unclear. Is WM disease a factor that increases AD risk and clinical severity independent of the prevailing pathogenic models, or does it promote AD neurodegenerative changes? ADNI publications over the last 2 years have begun to address these issues and to integrate the role of WM disease in AD into current models of disease progression.
Although subjects with hemispheric infarctions at baseline are excluded from ADNI, subject assessment includes several neuroimaging biomarkers for small-vessel pathology. T2-weighted or FLAIR MRI sequences are used to detect WMHs, and cerebral microbleeds are visualized using a T2* gradient echo MRI sequence [54]. The extent of cerebrovascular disease in the ADNI cohort was investigated by Ramirez et al. [295] who reported that the ADNI-1 sample had a significantly lower WMH burden relative to other elderly and dementia cohort studies. Despite their conclusion that ADNI-1 sample can be considered a relatively pure cohort with little to no vascular burden compared with community populations, ADNI studies investigating the role of WM disease suggest that a low burden may still have a significant impact on disease progression.
7.2.3.1. Vascular disease and risk factors are associated with cognitive decline and worsening clinical outlook
Numerous measures reflecting vascular disease have been associated with worsening clinical outlook. Greater baseline WMH volume was associated with lower processing speed [296] and rapid cognitive decline (.6 points/year MMSE) [297]. A vascular index score (summarizing past or present hypertension, hyperlipidemia, diabetes, myocardial infarction, atrial fibrillation, smoking, and stroke) was associated with greater memory impairment [296], and metabolic syndrome (obesity, hyperglycemia, hyperlipidemia, and hypertension) increased the risk of AD and was associated with a faster decline in WM volume [298]. Higher homocysteine levels were associated with lower processing speed [296]; higher cholesterol was associated with a greater rate of increase in global cognition and memory impairment [296]; hyperglycemia was associated with cognitive decline, whole-brain volume decline, and rate of progression to AD [299]; past or present hypertension predicted WMH volumes in CN subjects [300]; and a history of cigarette smoking (associated with oxidative stress and small hippocampal volume [301]) was associated with lower performance on cognitive tests [302]. An alternative marker of cerebrovascular dysfunction, physiological fluctuations in white matter (PFWM), was associated with glucose metabolism and composite memory but not ventricular or hippocampal volume, executive function, or CSF biomarkers [303]. Peripheral insulin resistance was differentially associated with either hypometabolism or hypermetabolism in different areas depending on whether subjects progressed to AD or not, suggesting that it may increase AD risk by affecting glucose metabolism [304]. Vascular burden may be evident at very early stages of cognitive decline as several associations were found in CN as well as MCI subjects [296,298,300,305].
Considerable evidence suggests that cardiovascular risk factors may accelerate neurodegeneration and subsequent cognitive decline via Aβ independent pathways. In CN subjects, Aβ deposition was more associated with APOE4 positivity than with a history of cigarette smoking, although APOE4+ smokers had the lowest glucose metabolism and poorest learning and memory scores of all groups, suggesting that cerebrovascular disease may worsen the effects of the APOE ε4 allele (Fig. 25) [302]. Similarly, stroke risk, comprising cardiovascular factors such as hypertension and cigarette smoking, was most related to hippocampal volume and memory performance in the absence of Aβ pathology (Fig. 26) [306]. Finally, the association between type-II diabetes mellitus and regional-reduced cortical thickness was modulated by levels of p-tau181, but not by cortical Aβ load [307].
Fig. 25.
Effect of interaction of smoking history and APOE ε4 genotype on amyloid level and hypometabolism. ADNI participants were grouped on the basis of smoking history and APOE4 status. Smoking status interacted with APOE ε4 carrier status such that APOE4 smokers had higher levels of amyloid and worse hypometabolism than other groups. (A) Florbetapir retention level across groups. Higher values indicate greater Aβ level. Levels above the horizontal line indicate Aβ positivity. Mean ± standard error of the mean. (B) Composite glucose uptake level across groups. Higher values indicate greater glucose metabolism. Mean ± standard error of the mean. Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; FDG, [18F]-fluorodeoxyglucose; SUVR, standardized uptake value ratio. Reproduced with permission from [302].
Fig. 26.
The effect of stroke risk on hippocampal volume and memory performance in Aβ-positive and Aβ-negative subjects. Stroke risk, assessed by the Framingham Stroke Risk Profile, was associated with decreased baseline hippocampal volume and decreased memory performance in both Aβ+ and Aβ− subjects. Worst performance on both measurements was observed in subjects with both abnormal amyloid and high stroke risk. Error bars represent 95% confidence intervals. Abbreviations: ADNI-MEM, Alzheimer’s Disease Neuroimaging Initiative–memory domain; CSF, cerebrospinal fluid; ICV, intracranial volume. Reproduced with permission from [306].
Vascular disease may act synergistically with neurodegeneration to accelerate cognitive decline. Tosto et al. [297] reported that low baseline entorhinal cortex volume, a marker of neurodegeneration due to AD, and high baseline WMH volume independently predicted rapid cognitive decline of a similar magnitude. Moreover, individuals with high entorhinal cortex volume and low WMH burden had a significantly lowered risk of rapid cognitive decline (Fig. 27).
Fig. 27.
Cumulative survival of individuals based on their high entorhinal cortex volume (ECV) and level of white-matter hyperintensities (WMHs). ADNI MCI subjects were dichotomized according to the median split of their ECV and levels of WMH. Individuals with high ECV and low WMH had low likelihood of rapid decline, whereas subjects with low ECV and low WMH or low ECV and high WMH appear to progress most rapidly. Solid line indicates high ECV, low WMH; dashed line indicates high ECV, high WMH; dotted line indicates low ECV, low WMH; dash-dotted line indicates low ECV, high WMH. Abbreviations: ADNI, Alzheimer’s Disease Neuroimaging Initiative; MCI, mild cognitive impairment. Reproduced with permission from [297].
7.2.3.2. WM disease and morphological changes in disease progression
Two distinct factors in AD subjects were identified in a factor analysis study of hippocampal volume, AD signature cortical thickness, ventricular volume, total WM volume, and volume of WM changes [308]. The first was associated with diffusivity, total volume of WM changes, and ventricular expansion. A second factor was more strongly related to MMSE and cortical thickness changes typical of AD and was associated with worse parahippocampal WM microstructure. These results imply that there are two sets of independently covarying degenerative changes: age-related vascular changes that are associated with ventricular expansion and volume of WM lesions, and WM changes in the parahippocampal gyrus that are associated with hippocampal atrophy and the classical patterns of AD neurodegeneration. WM burden may be more than a simple comorbidity due to vascular disease and may play a specific role in the latter pathway that is not considered in traditional models. Further evidence to support this comes from a study of changes in the quality of WM signal abnormalities. Accelerated before and after MCI progression to AD and echoed the acceleration of hippocampal atrophy at this time [309].
7.2.3.3. How do WM abnormalities interact with Aβ and tau deposition?
Clearly, cerebral WM disease influences disease trajectory. Do WMHs act independently of or interactively with CSF biomarkers? WM changes in the elderly have been linked to cognitive deficits but the relationship between the cerebral Aβ deposition and WM microstructure is not well understood. Wolf et al. [310] investigated the relationship between Aβ deposition and WM microstructure in CN subjects using DTI. They found that the relationship between Aβ deposition and DTI metrics of WM integrity (fractional anisotropy, mean diffusivity, radial diffusivity, and axial diffusivity) was not linear. In fact, at lower levels of Aβ burden, increasing Aβ load was associated with increases in fractional anisotropy and decreases in mean diffusivity and radial diffusivity, suggesting an improvement in WM integrity. At higher Aβ burden, increases in Aβ load were associated with DTI measures indicating the opposite. At low Aβ burden, compensatory mechanisms may act to preserve cognitive functioning, but these may be overcome by higher Aβ burden leading to damage to WM structure, which may, in turn, initiate cognitive decline.
In some instances, Aβ load may be correlated with WM lesions. Past or present hypertensive CN subjects had greater WMH volumes at a given burden of Aβ (Fig. 28) [300]. Although the extracellular Aβ deposition measured by FLAIR MRI in this instance is not directly correlated with cerebral Aβ angiopathy, these results support the idea that Aβ accumulation within blood vessels could exacerbate cerebrovascular injury processes that contribute to WMHs. Aβ load in the medial prefrontal cortex and posterior cingulate cortex was correlated with WM lesion load in CN subjects in periventricular and frontal regions, suggesting a link between microvascular damage and Aβ pathology at the early stages of disease [311]. The location of cerebral microbleeds was differentially associated with CSF Aβ and p-tau181 levels. Lobar microbleeds at the cortico-subcortical junction, which reflect Aβ angiopathy, and not microbleeds in deep GM, which reflect subject hypertension, were associated with abnormal levels of CSF Aβ and p-tau181 [312]. Furthermore, subjects with cortical microbleeds were more likely to carry the APOE4 allele, suggesting that WM damage at this location is associated with typical AD pathology.
Fig. 28.
The effect of hypertension and Aβ status on estimated trends of white-matter hyperintensities (WMHs) volume as a function of age. WMH volumes were predicted for the population average intracranial volume by age, exposure to elevated blood pressure, and CSF Aβ42 burden. High blood pressure increases WMH over time in both Aβ+ and Aβ− subjects, but the greatest effect in Aβ+ subjects. Abbreviation: CSF, cerebrospinal fluid. Reproduced with permission from [300].
Do WMH levels modify the effect of tau in neurodegeneration? Subjects with low baseline t-tau and higher frontal and parietal WMH volumes had greater entorhinal cortex atrophy than subjects with lower baseline t-tau and lower regional WMH volumes [313]. In addition, elevated WMH, particularly in the parietal lobes, predicted MCI to AD progression, and risk of progression increased in subjects with high levels of tau [313]. Thus, WMH burden may act as the “second hit” that is required, in addition to abnormal levels of CSF biomarkers, to produce neurodegenerative and cognitive changes associated with AD (Fig. 29).
Fig. 29.
Conceptual model linking white-matter hyperintensities to clinical progression of AD. Regional white-matter hyperintensities (WMHs) affect tau directly (a), affect regional atrophy and clinical progression directly (b), and modify the effect of tau on disease progression (c). The interaction between regional WMHs and Aβ has yet to be elucidated. Abbreviation: AD, Alzheimer’s disease. Reproduced with permission from [313].
Taken together, these studies offer an intriguing glimpse into how the effect of WM disease may be integrated into the traditional model for AD progression. Vascular risk factors may enhance cognitive decline by interacting with Aβ in the early disease stages to increase WM abnormalities. Regional WM abnormalities may then act as a “second hit” to augment the effects of tau abnormalities and neurodegeneration, thus accelerating the remaining disease process. The fact that these findings are from the ADNI cohort which has been characterized as having far lower burden of WM disease than other community samples [295] suggests that the contribution of WM abnormalities to dementia in the wider population may be even more profound.
7.3. AD as a disconnection syndrome
Several investigators have suggested that AD is a disconnection syndrome, based on the progressive synaptic and neural degeneration across the continuum of the disease [314,315]. Considerable evidence exists that specific structural and functional brain networks are increasingly disturbed, hypothetically due to neuronal injury caused by the abnormal deposition of Aβ and/or tau [275,316,317]. The different underlying biological substrates of these structural and functional networks, namely WM fiber tract networks, cortical thickness networks, and resting-state functional networks, can be mapped by applying graph theoretical analyses to different imaging modalities in the burgeoning field of “connectomics” [318]. Graph theory uses “nodes” to designate brain regions thought of as hubs, and “edges” to represent the connections between them to construct a topological map of the connectome and to generate measures (strength, weighted local efficiency, weighted clustering coefficient, and characteristic path length) that describe the organization of the network [319,320]. The inclusion of diffusion MRI for studying WM tract geometry, and TF-fMRI for mapping functional networks, in the ADNI-2 protocol has produced a set of longitudinal data across the disease spectrum that is central to number of studies targeted at understanding the role of brain network disruptions in AD.
7.3.1. Structural connectivity
Diffusion MRI can be used to study WM fiber integrity and microstructure based on measures of local water diffusion such as fractional anisotropy (FA), mean diffusivity (MD), axial diffusivity (DA), and radial diffusivity (DR) which respectively reflect the degree of myelination, cell death and edema, axonal injury and demyelination, and loss of oligodendrocytes and reactive astrocytosis [321]. The trajectory and structural connectivity of axonal fibers can be inferred using tractography based on DTI data. Studies of the ADNI cohort have found both global and local changes to WM tracts during disease progression. Sun et al. [322] found increased global DA and increased DR in prodromal patients, and a further increase in these measures, along with decreased global FA, in demented subjects, consistent with widespread WM damage. Reduced WM fiber integrity may be associated with GM atrophy. Deterioration in WM integrity may decrease GM volume, particularly in the hippocampus. Simultaneous changes in GM volume and FA values were reported in AD subjects compared to CN subjects primarily in the temporal lobe/hippocampuscingulum, frontal/cingulate gyrus–corpus callosum, and temporal/occipital/parietal lobe–corpus callosum/corona radiata [323]. However, there was no correlation between MMSE score and diffusion indices in regions known to affect memory [324] implying that memory loss is an event considerably downstream of network deterioration. Changes in WM structure have been observed in regions of the brain involved in known mechanisms of disease pathology—WM tracts known to be connected to areas of AD pathology appear most prone to deterioration. MCI subjects had increased DR and DA in the external capsules of the lateral cholinergic pathway, a finding consistent with the degeneration of cholinergic neurons of the basal forebrain as a distinct neuropathological feature of AD [322]. AD and MCI subjects had increased total diffusivity and DR and decreased FA in the optic nerves and optic tract, consistent with common visual deficits observed in AD despite relative lack of damage to the visual cortex, and suggesting that the visual pathway from eye to brain may be damaged [325]. The number of nerves fibers of limbic system touching the hippocampus, thalamus, and amygdala decreased from CN to EMCI to LMCI, and this metric was able to distinguish between EMCI and LMCI subjects (Fig. 30) [326]. The cingulum angular WM bundles, connected to the hippocampus, showed progressive deterioration in MCI and AD subjects, and their integrity was associated with hippocampal volume [321]. However, controlling for hippocampal volume did not remove all group differences, suggesting that WM damage additionally contributes to AD through an alternative mechanism.
Fig. 30.
Visualization of neuronal fibers touching limbic system ROIs in typical early MCI (left) and late MCI (right) patients. Abbreviation: MCI, mild cognitive impairment. Reproduced with permission from [326].
AD also appears to affect the topology of the structural connectome. This is characterized by small-world properties such as a high mean clustering coefficient reflecting the concentration of highly connected brain regions (“hubs”) and low characteristic path length representing the WM fibers connecting them [320]. Significant group differences in these metrics were reported by Prescott et al. [327] indicating that the AD structural connectome undergoes progressive deterioration with disease progression. Another attribute of the structural connectome, described by the rich club coefficient, is that its hubs are more interconnected than predicted by chance and play a central role in network communications [320]. Daianu et al. [328] used whole-brain tractography to reconstruct structural brain connectivity networks and to map connections between cortical regions. In MCI and AD subjects, there was a progressive disruption of global measures of network integrity (nodal degree, clustering coefficient, characteristic path length, and efficiency) but a relative preservation of the rich club coefficient. Thus, network disruptions were primarily in peripheral areas of low connectivity and left the highly connected rich club core, consisting of hubs in the superior frontal, insular, posterior cingulate, precuneus, and superior parietal region, relatively undisturbed (Fig. 31). Higher baseline characteristic path length and lower baseline mean clustering coefficient values were correlated with greater volumetric changes over 6 months in a preliminary study of MCI subjects [329]. Decreased clustering in the right pars opercularis, left superior parietal node, and left pericalcarine node was significantly associated with patterns of volumetric changes. Lower betweenness centrality (a measure of the number of short communication paths a node participates in) in the right temporal lobe was associated with greater atrophy, suggesting that this hub facilitates integration between anatomically unconnected regions. These studies suggest that the degree of integration across distributed brain regions and locally within regions decreases with disease progression as the small-world architecture of the brain is disturbed.
Fig. 31.
Average brain networks showing common connections at 90% of healthy controls (CN), MCI, and AD participants at k = 20 nodal degree threshold. Although individual connections (red edges) erode with disease progression, centrally positioned hubs (light blue nodes) are preserved in diagnostic groups. These hubs are in the superior frontal (SF), insula (I), posterior cingulate (PC), precuneus (P), and superior parietal cingulate regions (SP). Abbreviations: AD, Alzheimer’s disease; MCI, mild cognitive impairment. Reproduced with permission from [328].
Both APOE4 and Aβ positivity appear to influence the structure of the connectome. The number of APOE ε4 alleles was negatively correlated with the number of WM fibers touching the hippocampus, thalamus, and amygdala [326]. The shortest path length was increased and global efficiency decreased in preclinical Aβ+ compared to Aβ− participants, with no concomitant differences in either hippocampal volume or metabolism [330]. These WM changes were specific to network structure and were not explained by changes in global WM integrity. The degree of Aβ burden was more strongly associated with changes in graph theoretical measures than with diagnostic group and affected large-scale structural networks more severely in CN subjects than in MCI or AD subjects [327]. The decreasing effect of accumulating Aβ burden with time may be because there are fewer viable connections to degenerate [327]. Both studies suggest that damage to the structural connectome may occur very early in pathophysiological development of AD, perhaps closely following Aβ deposition.
7.3.2. Functional connectivity
In addition to structural connectivity, the human brain possesses functional connectivity which reflects signaling and communication events that unfold within the underlying structural network [320]. Functional networks are derived from statistical descriptions of time series data reflecting changes in blood flow measured as a blood-oxygen-level dependent (BOLD) signal on TF-fMRI. They differ from structural networks in that they are transitory, modulated by task, and link many structurally unconnected node pairs. Major functional hubs with high connectivity to other regions are located in the ventral and dorsal precuneus, posterior and anterior cingulate gyrus, ventromedial frontal cortex, and inferior parietal regions, and these have significant overlap with the DMN, which consists of an anatomically defined set of hubs and subsystems located in the cingulate cortex [320]. As the DMN is implicated in the process of encoding new memories, and AD impairs memory function, AD pathology may specifically target this functional network [331]. Increased WM lesion load was correlated with decreased functional connectivity of the DMN, and reduced neuronal activity in the temporal cortex [311].
Changes in functional connectivity appear to occur early in the disease process and can distinguish between EMCI and LMCI subjects. The magnitude of these changes in the posterior cingulate cortex, precuneus, right lingual gyrus, thalamus, and right parahippocampal gyrus increased across patient groups and was related to cognitive performance (MMSE) and the Geriatric Depression Scale [332]. Decreased functional connectivity in the thalamo–hippocampus, thalamo–temporal, thalamo–visual, and thalamo–DMN networks was observed between EMCI and LMCI subjects [333], highlighting the importance of the thalamus, a crucial brain area believed to coordinate communication (e.g., memory, attention, and perception). Interestingly, there were areas of increased functional connectivity between the thalamus and left fusiform gyrus, right middle occipital gyrus, left and right precuneus, right middle temporal gyrus, and left inferior temporal gyrus in amnestic MCI subjects, suggesting that they may be able to use additional brain resources to compensate for loss of cognitive function.
APOE4 may modulate brain functional connectivity in early stages of the disease. APOE4 noncarriers had the strongest functional connectivity, whereas EMCI subjects who were APOE4 carriers had the greatest dysfunction [334]. The pattern of functional connectivity disruptions in EMCI subjects overlapped to a large degree with that of APOE4 subjects. However, only the EMCI group, and not the APOE4 group, had decreased connectivity in the prefrontal cortex areas, suggesting a more multidimensional pathology in EMCI than is accounted for by the presence of an APOE ε4 allele alone [334].
Grothe et al. [335] used multimodal imaging data to assess the relationships between seven previously defined major functional connectivity networks (the DMN, frontoparietal-control network, dorsal- and ventral attention networks, limbic network, visual network, and somatomotor networks [Fig. 32]) and Aβ deposition, hypometabolism, and GM atrophy. Aβ deposition was widely distributed across the cerebral cortex, with the highest deposition in the DMN and frontoparietal-control network. This pattern did not change significantly across subject groups, consistent with Aβ deposition being an early event in AD progression. Atrophy occurred primarily in the anterior limbic network, followed by the DMN. The pattern of hypometabolism was a mixture of both Aβ- and atrophy-related profiles. The distribution of atrophy and hypometabolism increased with disease progression (Fig. 33). These results suggest that despite the high vulnerability of the DMN for changes in imaging abnormalities, distinct pathologic markers of AD have differential network specificities targeting different neuronal networks.
Fig. 32.
Overview of intrinsic connectivity networks. The figure shows standardized maps of seven intrinsic conductivity networks projected on the cortical surface and a midsagittal section of the reference template. This map estimates the functional conductivity architecture of the human cerebral cortex based on resting state functional conductivity projected on the cortical surface and a midsagittal section of the reference template. Abbreviations: blue, limbic network; cyan, somatomotor network; green, dorsal attention network; pink, ventral attention network; purple, visual network; red, default mode network; yellow, frontoparietal-control network. Reproduced with permission from [335].
Fig. 33.
Severity of AD-related imaging abnormalities within intrinsic connectivity networks. Plots depict means and 95% confidence intervals of averaged Z scores of Aβ deposition (top), hypometabolism (middle), and gray-matter atrophy (bottom) within the distinct intrinsic connectivity networks for each AD stage. The widespread distribution of amyloid deposition across the cerebral cortex appeared similar in all patient groups with highest amyloid load in the DMN and FPN. Hypometabolism was most pronounced in the AD group and occurred across most ICNs except the VIS and SMN. Likewise, atrophy was most pronounced in the AD group, which displayed a different relative pattern of atrophy severity across ICNs with atrophy most pronounced in the LIN followed by the DMN and relative sparing of the FPN. Abbreviations: blue, limbic network (LIN); cyan, somatomotor network (SMN); green, dorsal attention network (DAN); pink, ventral attention network (VAN); purple, visual network (VIS); red, default mode network (DMN); yellow, frontal parietal control network (FTN); AD, Alzheimer’s disease; CN, cognitively normal; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment. Reproduced with permission from [335].
A study by Jones et al. [336] replicated many of the aforementioned results and offered a model describing how microscale proteinopathy affects macroscale brain networks to ultimately result in clinical symptomatology. Their multi-modal study tracked the evolution of connectivity changes within and between the ventral, posterior, anterior ventral, and anterior dorsal subsystems of the DMN (Fig. 34) across the course of the disease. They found both increased and decreased connectivity in different regions, consistent with a previous study [332]. Connections within the posterior and ventral subsystems decreased linearly, whereas connections between posterior and ventral, and posterior and anterior dorsal subsystems, increased linearly across disease course. Decreasing posterior subsystem connectivity was marginally associated with elevated Aβ levels but not with hippocampal volume, whereas increased connectivity between the posterior and ventral DMN subsystems was associated with elevated Aβ levels and decreased hippocampal volume. Like McKenna et al. [334], they found that APOE4 positivity had specific effects on network connectivity. In CN Aβ− subjects, APOE4 was associated with lower posterior DMN connectivity only, suggesting that failure of this network may be the earliest critical event in pathophysiology, preceding Aβ deposition. Finally, the decrease in connectivity in the medial temporal lobe–ventral DMN was associated with decreased memory performance.
Fig. 34.
Subsystems of the default mode network. Nodes within the default mode network segregate into distinct subsystems. Abbreviations: aMPFC, anterior or medial prefrontal cortex; blue: dorsal medial prefrontal cortex system; dMPFC, dorsal medial prefrontal cortex; green, medial temporal lobe memory system; yellow, midline core regions; PCC, posterior cingulate cortex; Rsp, retrosplenial cingulate. Reproduced with permission from [336].
The results support a model (Fig. 35) in which pathophysiology, possibly instigated by the APOE ε4 allele, originates in the posterior subsystem of the DMN. This results in a transient increase in connectivity between the posterior DMN networks and other systems that are associated with Aβ deposition and hippocampal atrophy. The increased connectivity may indicate high processing burden or inefficient synaptic communications and requires high metabolism which may trigger a cascade of downstream of molecular events associated with AD. The processing burden may then proliferate to downstream networks. Highly connected regions in the brain may be particularly vulnerable to Aβ deposition because of their increased synaptic activity, according to the “nodal stress” hypothesis [335,337]. Indeed, the authors postulated that the shifting of processing burden may lead to APP processing changes and Aβ deposition. Overloading of specific networks may also exacerbate preexisting primary age-related tauopathy in the medial temporal lobe and thereby accelerate tau-mediated neurodegeneration.
Fig. 35.
Schematic of the proposed cascading network failure model of Alzheimer’s disease. Phase 0: The posterior DMN (pDMN) serves as the central hub processing and integrating association cortices and is highly metabolically active. Independently, the medial temporal lobe (MTL) has accumulated age-related damage from neocortical processing of a different kind contributing to primary age-related tauopathy (PART) in these regions. Phase 1: Declining posterior DMN transfers information-processing duties to the neocortical regions including the ventral DMN and/or the anterior dorsal DMN. Aberrant betweenneocortical network synaptic activity leads to dysregulated amyloid precursor protein (APP) processing promoting Aβ plaque formation in neocortical layers. Phase 2: Given that the hippocampus is continually processing information from the same regions, noise in these cortical systems is propagated down to the hippocampus. This increased burden on the hippocampus accelerates the preexisting PART. Phase 3: Neurodegeneration expands to adjacent systems. This creates a detrimental positive feedback loop because degeneration lowers the noise-handling capacity of the system leading to further degeneration. MCI phase: Posterior brain regions supporting memory succumb to the degenerative feedback loop as hippocampal regions increases processing. Later, the frontal brain regions begin to bear the high connectivity burden. Early Alzheimer’s disease phase: The high frontal connectivity firmly establishes the neurodegenerative feedback loop in these systems before declining as Alzheimer’s disease progresses. Abbreviations: DMN, default mode network; MCI, mild cognitive impairment. Reproduced with permission from [336].
7.3.3. Structural covariance networks
A complementary approach to diffusion-based and TF-fMRI studies compares patterns of structural covariance of GM volume within selected structural correlative networks. Aβ+ AD subjects had decreased structural association in the medial temporal lobe subsystem of the DMN (Fig. 35), with specific decreases between the entorhinal cortex and the medial prefrontal and dorsolateral prefrontal cortices, and in the midline core DMN subsystem [338]. The results suggest that these early structural disruptions between the heteromodal association cortices and the entorhinal cortex may isolate the hippocampus leading to memory loss. Concomitant increased connectivity was observed in other areas such as the salience and executive control networks [338], consistent with the Jones et al. model [336]. Both the salience and executive control networks affect frontal regions of the brain and are likely affected in AD rather than in earlier stages of the disease. The results support the concept of AD as a disconnection syndrome targeting specific large-scale brain networks.
7.3.4. Metabolic connectivity
Metabolic connectivity and the topological organization of metabolic brain networks can be surmised by the analysis of metabolic covariance between node regions, using FDG PET, which, like TF-fMRI BOLD measurements, reflects the brain’s metabolism. APOE4+ CN, MCI, and AD subjects had decreased metabolism in the parahippocampal gyrus and increased metabolism in the medial frontal gyrus and inferior frontal gyrus compared to APOE4− subjects [339]. The metabolic networks of both groups had small-world properties, but APOE4+ subjects had lower clustering coefficients and significant decreases in nodal centrality, a measure of the number of connections maintained by each hub, in six hub brain regions. APOE4+ subjects also had abnormally increased local short distance and decreased long-distance interregional correlations. The results suggest that APOE ε4 allele carriers have a less optimal metabolic network than noncarriers. A second study reported that the MCI group had higher clustering coefficients, shorter characteristic path length, and lower betweenness centrality than control subjects, supporting the idea that the small-world characteristics of hub nodes are affected during disease progression [340]. Hub nodes targeted by AD progression were identified in the left anterior cingulum, right superior parietal, left fusiform, right inferior temporal gyrus, and right cuneus.
Hypometabolism in the posterior cingulate cortex is an early marker of AD. Teipel et al. [341] hypothesized that decreased metabolism in the posterior cingulate cortex is related to disrupted input from connected regions, such as the hippocampus, that undergo early atrophy. In CN and EMCI subjects, they found that posterior cingulate cortex hypometabolism was exclusively associated with remote hippocampal atrophy. In LMCI subjects, it was associated with both hippocampal atrophy and local atrophy within the posterior cingulate cortex, as well as with Aβ load. In AD subjects, posterior cingulate cortex hypometabolism was solely associated with local atrophy. These results may result from a diaschisis-like mechanism in which loss of function of the posterior cingulate cortex is caused by connection to the damaged hippocampus and are consistent with a progressive disconnection of the posterior cingulate cortex from subcortical brain regions.
7.3.5. Models for the spread and propagation of Aβ and tau along brain networks
Recent neuropathological evidence supports a “prion-like” spreading mechanism for AD pathology in which misfolded pathological proteins trigger the misfolding of adjacent same species proteins and cascade along neuronal pathways via trans-synaptic or trans-neuronal spread [342,343]. The initial misfolding of these proteins and their subsequent propagation along structural brain networks is postulated to result in hypometabolism, atrophy, and ultimately cognitive decline. Two ADNI studies have developed or tested models based on this idea. Raj et al. [344] found that their earlier model describing a prion-like diffusive progression along the fiber pathways defined by the brain connectivity network [345] accurately predicted future patterns of atrophy and hypometabolism based on baseline volume and metabolism. These patterns were in agreement with the stereotypical progression of atrophy from temporal to parietal to frontal regions. They also found that Aβ positivity predicted a faster rate of progression. Iturria et al. [346] developed a model based on similar assumptions for the spread of Aβ pathology, namely that the spread of Aβ as an infectious agent is constrained by the brain’s connectional architecture. Their stochastic epidemic spreading model also incorporated the brain’s clearance response to misfolded proteins and reproduced the characteristic patterns of Ab deposition across the disease process (Fig. 36). They found that the Aβ clearance rate was inversely correlated both with worsening clinical diagnosis and increasing number of APOE ε4 alleles and that it partially accounted for variance in CSF t-tau and p-tau181 levels, supporting the idea of an interrelated pathway between Aβ pathophysiology and tauopathy. In addition, the model identified the posterior and anterior cingulate cortices as the most likely starting seed regions for the propagation process. The negative relationship between regional Aβ deposition patterns and effective anatomical distances from these regions suggested that the propagation of misfolded proteins from outbreak regions is modulated by the topology of the WM tracts in the brain.
Fig. 36.
Characteristic regional Aβ deposition patterns in healthy and pathological brains. An epidemic spreading model that predicts propagation/deposition of Aβ reproduces the characteristic Aβ deposition patterns in the ADNI cohort. (A) PET-based mean regional Aβ deposition probabilities in cognitively normal healthy controls (HC), early MCI (EMCI), late MCI (LMCI), and AD groups. Nodes correspond to 78 regions covering all the brains gray matter, with node sizes proportional to the associated Aβ burden. The progressive expansion of Aβ deposition starts mainly from the DMN regions and expands to the rest of the brain. (B) Correspondence between the estimated and PET-based mean regional Aβ deposition probabilities for the different clinical groups. Abbreviations: AD, Alzheimer’s disease; ADNI, Alzheimer’s Disease Neuroimaging Initiative; DMN, default mode network; MCI, mild cognitive impairment; PET, positron emission tomography. Reproduced with permission from [346].
7.4. Conclusions
ADNI studies in 2014 and 2015 have provided ample evidence to support not only the traditional view of an Aβ deposition-instigated cascade of events leading to dementia but also the complexity of disease progression which may involve copathologies or alternative pathways with as yet unidentified underlying pathology.
Cluster analysis of both MCI and CN subjects consistently identify a subtype with attributes of “typical” AD: abnormal levels of CSF biomarkers, hippocampal atrophy, regional hypometabolism, and impaired memory [279,281,283,305]. This group appears to largely overlap with Aβ-positive subjects, who have been identified in ADNI-2 by amyloid imaging and extensively characterized and who appear to follow “typical” AD progression in which biomarkers become abnormal in the sequence defined by Jack et al. [258]. The APOE ε4 allele may act to modulate this trajectory [128,261,262,276]. Novel findings indicated that atrophy may begin in the presymptomatic phase in areas outside the hippocampus such as the basal forebrain [267,268], precuneus [265,266], parietal gyrus [261], and posterior cingulate [266]. This is an agreement with observations from connectomics experiments which view AD as a disconnection syndrome with the earliest disruptions to metabolic connectivity occurring in the posterior cingulate cortex [328,332]. The model of Jones et al. [336] of a cascading network failure which begins in the posterior subsystem of the DMN, and which propagates connectivity changes along the WM tracts of the structural connectome, provides a provocative framework for functionally linking the many observations of “typical” AD progression. The observation that the APOE ε4 allele may instigate the process or modulate in some way the structural and functional connectome [310,326,327,334,336] also is consistent with studies of Aβ+ subjects. Models of a prion-like mechanism [344,346] by which misfolded Aβ induces further pathological misfolding along structural brain networks predict patterns of Aβ deposition, hypometabolism, and atrophy and provide a mechanistic basis for network failure. The association of WM changes caused by cerebrovascular disease with abnormal Aβ and with markers of neurodegeneration may also be viewed in the light of these models. The association of Aβ positivity with increased cerebral blood flow in the posterior cingulate [277], and with improvements in global measures of WM integrity at low Aβ burden [310], may be evidence of the initial compensatory changes in connectivity predicted by the model of Jones et al. [336]. Aβ pathology and microvascular damage may be linked at the early stages of disease [311], and WM changes in the parahippocampal gyrus were associated with hallmarks of AD decline [308]. Degradation of specific, disease-related regions of the structural connectome by cerebrovascular disease may exacerbate disconnection, consistent with the theory that WM damage causes a “second hit” in disease progression. However, WM changes do not always lead to AD; the total volume of WM changes was found to be more related to age-related vascular changes [308].
Most MCI and CN subjects were not identified by cluster analysis as following “typical” AD progression. In fact, a substantial proportion were normal by most measures [280–282,286], suggesting that there is a high rate of false diagnostic positives in MCI subjects. Various other subtypes were identified, chief among them a group typified by deficits in executive function and abnormal tau, but having normal Aβ [280,281]. These subjects may be related to Aβ−, neurodegeneration positive SNAP subjects, who were found to progress to frontotemporal dementia and Lewy body disease in addition to AD [291]. Conversely, a subtype of Aβ+ patients had primarily cortical atrophy and deficits in executive function which correlated with the distribution of neurofibrillary tangles [157,272]. These individuals likely had atypical forms of AD characterized by predominantly isocortical tau pathology. Finally, most of the associations between Aβ and memory were mediated by neither hypometabolism nor atrophy, further implicating p-tau in the process. The tale of tau has yet to be told; the inclusion of tau PET imaging in ADNI-3 promises to shed light on this conundrum.
The studies have had a profound impact on how we consider the disease process, which, in turn, has many implications for the development of AD preventive treatments and clinical trial design. Many questions remain to be answered: what is the underlying pathology of the Aβ− subtypes? How does abnormal Aβ mechanistically prompt tau propagation? How does tau propagate in the absence of Aβ? How does WM damage in different regions affect structural, functional, and metabolic connectivity? What is a genetic contribution to these different pathways? The answers will undoubtedly lie in the consideration and integration of results from multiple approaches. The breadth and depth of ADNI positions it ideally at the cusp of discovery.
8. Improvement of clinical trials
The overall goal of ADNI is to validate biomarkers for AD clinical trials. A large body of recent ADNI publications describes improved approaches for diagnostic classification, the prediction of future decline and the selection of trial participants likely to decline, and development of outcome measures sensitive to early changes wrought by the disease. To some extent, progress in this area dovetails with advances in our understanding of disease progression; recognition of the heterogeneity and pathophysiology underlying MCI and now even CN participants, and of the wide variety of clinical trajectories, informs many aspects of clinical trial design. As trials move to presymptomatic cohorts, biomarkers will be crucial in identifying those with preclinical AD. Furthermore, biomarkers may provide more sensitive and specific markers of progression. Certainly, this is one of the major hopes for tau PET [347]. Knowledge of how sources of disease heterogeneity influence biomarker changes will enable the selection and monitoring of subjects most likely to benefit from a targeted therapy. These advances improve statistical power to detect a slowing of clinical decline in predementia populations, a major challenge which has plagued AD clinical trials to date.
8.1. Diagnosis and prediction
8.1.1. Use of established modalities
8.1.1.1. Multimodal classifiers
Clinical trial design fundamentally depends on the ability to accurately diagnose the clinical group to which a subject belongs and to predict their likelihood of measurable progression within the time frame of the trial. As no single modality has been shown to be effective in all classification or prediction challenges, researchers have continued to focus on the use of multiple modalities. Optimum combinations of modalities differ between challenges. Although MRI and PET (FDG and florbetapir) were roughly equivalent in their ability to discriminate between AD and CN subjects and between AD and MCI subjects, changes in cortical thickness outperformed other measures for EMCI and CN subjects and may be an early indicator of neurodegeneration [348]. A systematic appraisal of the diagnostic and prognostic abilities of hippocampal volume and CSF biomarkers, alone and in combination, in both APOE4 carriers and non-carriers found that their multimodal classifier outperformed either single modality [349]. Aβ42 levels contributed to discriminating between CN and MCI or AD subjects but not between MCI and AD subjects. Conversely, p-tau181, a marker of neurodegeneration, played a role in discriminating between AD and MCI or CN subjects but not between MCI and CN subjects [349].
8.1.1.2. Prediction of CN to MCI progression
Biomarkers that predict cognitive decline in CN subjects also reflect disease progression. Based on the hypothetical AD model [258], successive preclinical AD stages have been proposed comprising firstly Aβ deposition (stage 1), followed by evidence of neuronal injury biomarkers (stage 2), and finally subtle cognitive impairment (stage 3) [278]. Consistent with the stages, Aβ42, but not p-tau181, predicted CN to MCI, but not MCI to AD progression [350]. A cut point of <220 pg/mL Aβ42, well above the established cut point for Aβ positivity of 192 pg/mL [292], was the best CSF predictor of decline on ADAS-cog in CN subjects, although t-tau and p-tau181 alone and their ratios with Aβ42 at modified cut points were also good predictors [350]. A combination of all CSF biomarkers using the modified cut points predicted progression to MCI with an accuracy of 65% [350]. In another study, volumetric changes and t-tau/Aβ42 were associated with a higher risk of progression in CN subjects, and lower baseline memory measures were the strongest predictors of progression [278]. It is important to note that although neuronal injury is included as a diagnostic criterion in AD, different markers of neuronal injury (hippocampal volume, regional glucose metabolism, levels of CSF p-tau181) were not found to be equivalent [278,351].
8.1.1.3. Prediction of MCI to AD progression
Predictors of MCI progression to AD also reflect the biomarker changes occurring at that stage of disease progression. Combinations of regional volumetric, glucose metabolism, and cortical deposition measures predicted progression with accuracies of 72% [352] and 76% [353]. Neuropsychological measures alone (clock drawing + AVLT) [354], or in combination with hippocampal volume [355], reached similar accuracies. A point-based tool with a range of 0 to 9 which included scores from the Functional Activities Questionnaire (FAQ), ADAS-cog and Clock-drawing tests, middle temporal cortical thinning, and hippocampal atrophy also had good predictive accuracy: 91% of MCI subjects with 7 to 9 points converted to AD within 3 years [356]. The primary drivers of prediction in the Disease State Index [357] were MRI features; additional neuropsychological and CSF markers improved accuracy only slightly [358]. A comparison of effect sizes of neuropsychological, MRI, FDG PET, and CSF measures for cognitive decline over 4 years in MCI nonconverters and converters (Fig. 37) [354] illustrates the relative importance of these measures, particularly atrophy in temporal regions, functional and memory impairment, and changes in glucose metabolism compared to CSF biomarkers in this transition.
Fig. 37.
Patterns of decline of the different classes of markers. The greatest effect sizes for MCI converters were for functional measures (Functional Activities Questionnaire [FAQ]) and for cognitive measures such as the ADAS-cog. Effect sizes for volumetric and CSF biomarker measures were much smaller. Panel 1: effect sizes for the difference in cognitive and functional measures between baseline and each one of the follow-ups from months 12–48: (A) MCI converters, (B) stable MCI. Panel 2: effect sizes in MRI morphometry, FDG PET HCI, and CSF biomarkers between baseline and months 12–36 follow-ups: (C) MCI converters, (D) stable MCI. Abbreviations: ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive; CSF, cerebrospinal fluid; FDG, [18F]-fluorodeoxyglucose; HCI, hyperbolic convergence index; MCI, mild cognitive impairment; MRI, magnetic resonance imaging; PET, positron emission tomography. Reproduced with permission from [354].
8.1.1.4. The effect of APOE4 status on the prediction of progression
Inclusion of APOE4 status substantially improved the prediction of MCI progression. In APOE4+ subjects, a hippocampal volume classifier had an accuracy of 76% [349], and a classifier based on FDG PETand volumetric MRI measures had an accuracy of 86.8% [359]. In APOE4 subjects, the best predictor of progression was a CSF classifier including p-tau181 that achieved 78% accuracy [349], suggesting that APOE4 plays a role in modulating disease trajectory, and may account for a portion of the observed disease heterogeneity.
8.1.1.5. Cognitive and clinical factors in prediction and diagnosis
Although multimodal classifiers currently predict future decline with the best overall accuracy, the use of cognitive or clinical factors that can be determined at a clinical visit can reduce cost, avoid invasive lumbar puncture procedures, and save time. A brief clinical index which included being female, several neuropsychiatric symptoms, and measures of cognition and functional dependence classified subjects into low (14% converted over 3 years), moderate (51% converted), and high-risk groups (91% converted) and had a Harrell’s c-statistic of 0.71, a predictive power similar to other commonly used prediction indexes such as the Framingham cardiovascular risk indicator (Fig. 38) [360]. Specific cognitive tests may be effective predictors. Clock drawing and RAVLT trial 5 were equally predictive of MCI to AD progression (AUC 0.78) as other single cognitive, FDG PET, structural MRI, and CSF biomarkers [354]. Two items on the FAQ (paying attention and understanding TV program, paying bills/balancing checkbook) predicted greater hazard of progression from CN to MCI. Similarly, the Everyday Cognition scale item “keeping mail and papers organized” predicted CN to MCI progression with hazard ratio of 2.27 [361]. These studies support the idea that simple questions regarding function can predict progression to MCI in CN elderly.
Fig. 38.
Observed versus predicted progression from amnestic MCI to AD over 3 years by Brief Clinical Index point score. The solid line shows the proportion of subjects predicted to progress from amnestic MCI to AD over 3 years as a function of the Brief Clinical Index point score. The dotted line shows the actual proportions that progressed at each point score value based on 3-year Kaplan-Meier estimates. The vertical bars showed the number of individuals at each point score value (right vertical axis). Abbreviations: AD, Alzheimer’s disease; MCI, mild cognitive impairment. Reproduced with permission from [360].
8.1.1.6. MRI biomarkers in prediction and diagnosis
MRI, although more costly than cognitive tests, is noninvasive and accessible in most settings. Automatic MR imaging analysis may assist a clinician in making an initial diagnosis. An automatic medial temporal lobe atrophy measurement predicted progression from SMC to MCI, and from MCI to AD with sufficient accuracy for clinical utility (AUCs of 0.904 and 0.810, respectively) [362]. Two studies have investigated use of the established Visual Rating Scale which obviates the need for research-oriented volumetric segmentation methods such as FreeSurfer. The entorhinal cortex visual rating system outperformed other visual rating systems against FreeSurfer for the AD versus CN classification (AUCs of 0.87 and 0.86, for entorhinal cortex and Free-Surfer, respectively) and for the MCI versus CN classification (AUCs of 0.73 and 0.75, for entorhinal cortex and FreeSurfer, respectively) [363]. Both visual rating systems and volumetric measures were good predictors of MCI to AD progression, with the entorhinal cortex visual rating system outperforming both the hippocampal visual rating system and volumetric analysis of the entorhinal cortex [363]. The visual rating scale for medial temporal atrophy outperformed equivalent visual rating scales of global cortical atrophy and posterior atrophy, achieving AUCs of 0.838 and 0.624 for the classification of AD versus CN, and MCIc versus MCInc, respectively, when optimized cutoffs for specific age ranges were used [364]. Visual rating scales may also have clinical utility in detecting sources of disease heterogeneity. A visual rating scale, the frontal subscale of the global cortical atrophy scale, reliably detected frontal atrophy and reductions in cortical volume and thickness that characterize the executive subtype of AD [365].
Further gains in the predictive ability of MRI, particularly in CN subjects, have come from new approaches which can identify changes in subtle structural features that occur before hippocampal atrophy. For example, hippocampal texture in the absence of hippocampal volume loss accounted for cognitive impairment in MCI subjects [28]. A hippocampal grading score which detected subtle patterns of neurodegeneration predicted progression to AD with an accuracy of 72.5% 7 years in advance of this transition [366]. Other features with prognostic potential include changes in the hippocampal subfield and presubiculum volumes [367], shape changes and atrophy rate of the corpus callosum [368], greater WM signal intensity ratio [369], and signal and texture-related features [370]. Data-driven feature selection outperformed region of interest (ROI)-based approaches in the prediction of MCI progression [371], suggesting that the subtle relationships between different brain areas detected by methods such as recursive feature elimination are highly effective predictors (Table 9).
Table 9.
Summary of classification and prediction methods
| Classification challenge | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Modality | Feature selection/dimensionality reduction | Classifier | N | AD versus CN | MCI versus CN | MCI versus AD | MCIc versus MCInc | Other | Unique approach | Reference | |||||
| MRI | Multi-atlas | SVM | 459 | ACC | 91.6 | ACC | 72.4 | Subjects registered to multiple atlases, features selected from each atlas, then jointly selected | [372] | ||||||
| SEN | 88.6 | SEN | 72.1 | ||||||||||||
| SPE | 93.9 | SPE | 72.6 | ||||||||||||
| AUC | 0.87 | AUC | 0.67 | ||||||||||||
| MRI | Multi-atlas | SVM | 459 | ACC | 90.7 | ACC | 73.7 | Joint learning of optimum representation of features from multiple atlases and classifier using maximum margin approach | [373] | ||||||
| SEN | 87.6 | SEN | 76.4 | ||||||||||||
| SPE | 93.0 | SPE | 70.8 | ||||||||||||
| MRI | Multitemplate | SVM | 459 | ACC | 93.3 | ACC | 80.9 | Inherent structure-based multiview learning (ISML) clusters subjects within a specific class into subclasses based on features learned using multiple templates | [374] | ||||||
| SEN | 92.8 | SEN | 86.0 | ||||||||||||
| SPE | 95.7 | SPE | 78.4 | ||||||||||||
| MRI | Independent component analysis | SVM | 818 | ACC | 86.4 | ACC | 70.2 | ACC | 69.8 | Uses novel ICA-based feature extraction method and linear SVM for classification | [375] | ||||
| SEN | 88.3 | SEN | 72.9 | SEN | 73.4 | ||||||||||
| SPE | 84.0 | SPE | 67.5 | SPE | 66.2 | ||||||||||
| MRI | Spatially weighted principal component analysis | SVM | 390 | Average misclassification %: 0.216 | Incorporates spatial structure by using both global and local spatial weights for feature selection to improve on PCA alone | [376] | |||||||||
| MRI | Circular harmonic function + principal component analysis | SVM | NA | ACC | 83.8 | ACC | 69.5 | ACC | 62.1 | Uses circular harmonic functions to extract local features from the hippocampus and posterior cingulate cortex and PCA for dimensionality reduction | [377] | ||||
| SEN | 88.2 | SEN | 74.8 | SEN | 75.1 | ||||||||||
| SPE | 79.1 | SPE | 62.5 | SPE | 49.0 | ||||||||||
| MRI | Gaussian process models | 415 | AUC | 0.94 | AUC | 0.91 | Generates quantitative maps of z-scores for WM, GM, and CSF abnormalities for clinical decision support | [378] | |||||||
| MCIc versus CN | |||||||||||||||
| MRI | Manifold learning: relevant variable selection (learned ROIs) | MR1 disease state score | 363 | ACC | 61 | Novel MRI disease state score (MRI DSS). Also used to predict MMSE scores and achieved separation between CN, EMCI, MCInc, MCIc, and AD groups | [379] | ||||||||
| ADNI-GO | SEN | 50 | |||||||||||||
| SPE | 72 | ||||||||||||||
| EMCI versus CN | |||||||||||||||
| SVM | 511 | ACC | 86 | ACC | 71 | ACC | 82 | ||||||||
| ADNI | SEN | 86 | SEN | 75 | SEN | 81 | |||||||||
| SPE | 85 | SPE | 67 | SPE | 83 | ||||||||||
| MCIc versus CN | |||||||||||||||
| MRI | Manifold learning: self-organizing map | SVM | 818 | ACC | 90 | ACC | 84 | Uses self-organizing map to select maximally discriminative ROIs and computes relative importance measure for each ROI | [380] | ||||||
| SEN | 87 | SEN | 82 | ||||||||||||
| SPE | 92 | SPE | 87 | ||||||||||||
| AUC | 0.92 | AUC | 0.84 | ||||||||||||
| MRI | Manifold learning: local binary map + custom masks | SVM | 363 | ACC | 82.8 | ACC | 61.5 | Uses local binary maps to directly select disease-specific patterns on a voxelwise basis without nonrigid registration. Also uses seven knowledge-based ROIs | [381] | ||||||
| SEN | 80.4 | SEN | 61.5 | ||||||||||||
| SPE | 82.7 | SPE | 63.5 | ||||||||||||
| AUC | 0.87 | AUC | 0.64 | ||||||||||||
| MRI | Multivariate regression: elastic net | Low density separation | 834 | AUC | 0.73 | Used a semisupervised learning method, low density separation, which uses unlabeled data to improve prediction of progression over supervised methods | [382] | ||||||||
| MRI | Multivariate regression: Ordinal regression | ORCHID score: probabilistic (not binary) | 564 | ACC | 91 | ACC | 70 | Models all clinical groups simultaneously as a continuum of disease, generates a summary score, ORCHID (Ordinal Regression Characteristic Index of Dementia) | [383] | ||||||
| AUC | 0.95 | AUC | 0.75 | ||||||||||||
| MRI | Multivariate regression: knockout strategy | Random Forests | 245 | SEN | 98.0 | Iterative multivariate knockout algorithm that uses Random Forests to construct set of relevant features | [384] | ||||||||
| MRI | Tree-structured sparse learning | SVM | 830 | ACC | 90.2 | ACC | 70.7 | ACC | 87.2 | Uses new tree construction method to cluster similar discriminative voxels, then a tree-structured sparse learning step and SVM classifier | [385] | ||||
| SEN | 85.3 | SEN | 56.2 | SEN | 80.1 | ||||||||||
| SPE | 94.3 | SPE | 80.9 | SPE | 92.2 | ||||||||||
| MCIc versus CN | |||||||||||||||
| MRI | FreeSurfer to select ROIs, cortical thickness measures | v-one class SVM | 814 | ACC | 84.3 | ACC | 54.4 | Focuses on detection of outliers who have abnormal brain patterns in a cognitively normal group. Trained on CN. Produces a brain abnormality index. | [386] | ||||||
| Labeled as outliers. In control group, ACC = 67.5% | |||||||||||||||
| MRI | Recursive feature elimination significance (p) maps from SVM weights | 509 | AUC | 0.92 | AUC | 0.74 | AUC | 0.69 | AUC | 0.61 | Uses p-maps of SVM weights as selection features in a wrapper method | [387] | |||
| MRI | Recursive feature elimination—SVM | 370 | ACC | 95.1 | Wrapper method based on recursive feature elimination and SVM selected GM and WM ROIs for classification with no need for dimensionality reduction step | [388] | |||||||||
| MRI (VBM) | Probability distribution function based | SVM | 260 | ACC | 89.7 | Extracts statistical patterns at multiple levels including use of probability distribution function of VOIs to represent statistical patterns | [389] | ||||||||
| SEN | 87.7 | ||||||||||||||
| SPE | 91.6 | ||||||||||||||
| AUC | 0.95 | ||||||||||||||
| MRI | Hierarchical fusion of multilevel classifiers | 652 | ACC | 92.0 | ACC | 85.3 | Divides classification of high dimensional features into multiple low-dimensional classification problems and integrates features hierarchically | [390] | |||||||
| SEN | 90.9 | SEN | 82.3 | ||||||||||||
| SPE | 93.0 | SPE | 88.2 | ||||||||||||
| MRI | Predefined ROIs and routine feature ranking (Pearson Correlation Coefficient) | Distance informed metric learning | 321 | ACC | 83.1 | ACC | 71.6 | Semisupervised metric learning framework that uses pairwise constraints that specify the relative distance between a pair of patients, according to their classification group | [391] | ||||||
| SEN | 75.0 | SEN | 77.6 | ||||||||||||
| SPE | 91.2 | SPE | 65.6 | ||||||||||||
| MRI | Graph-based multiple instance learning | SVM | 834 | ACC | 89.2 | ACC | 69.3 | Extracts local intensity patches as features and uses a novel graph-based multiple instance learning approach to assign disease labels to patches | [392] | ||||||
| SEN | 85.1 | SEN | 66.7 | ||||||||||||
| SPE | 92.6 | SPE | 71.2 | ||||||||||||
| MRI | t-tests of predefined cortical thickness measures | C4.5 decision tree | 364 | SEN | 80.0 | Encodes selected features into an ontology and uses C4.5 algorithm for clinical decision support | [393] | ||||||||
| SPE | 80.0 | ||||||||||||||
| MRI | Pseudo-Zernike moments | Neural network | 500 | ACC | 97.3 | ACC | 95.6 | ACC | 94.9 | Uses Pseudo-Zernike moments (30) for feature selection and neural network scaled conjugate gradient back propagation algorithm as classifier | [394] | ||||
| SEN | 96.6 | SEN | 95.9 | SEN | 94.2 | ||||||||||
| SPE | 97.8 | SPE | 95.3 | SPE | 95.6 | ||||||||||
| fMRI | Sparse inverse covariance estimation matrix + Kernel-based PCA | SVM | 82 | ACC | 73.2 | Uses a sparse inverse covariance estimation technique to model brain connectivity | [395] | ||||||||
| Note: EMCI | |||||||||||||||
| fMRI | Sparse learning using dictionary learning algorithm | SVM | 210 | ACC | 94.1 | ACC | 92.0 | ACC | 92.3 | Functional features are represented in a weighted dictionary matrix | [396] | ||||
| Note: SMC | |||||||||||||||
| FDG PET | Spatially weighted principal component regression | 196 | Average misclassification 0.117% | Spatially weighted selection of both individual features and neighboring patterns | [397] | ||||||||||
| FDG PET | Voxel-based longitudinal | SVM | 233 | ACC | 92.6 | ACC | 70.2 | Extension of [398] using whole-brain voxel approach for longitudinal analysis of FDG PET | [399] | ||||||
| FDG PET | Multi-channel pattern analysis + 3D Gabor wavelets feature extraction | SVM | 369 | MAP | 56.3 | MAP | 76.2 | Multi-channel approach integrates multiple patterns of hypometabolism from different patient groups using different analysis tools | [400] | ||||||
| FDG PET | Gaussian mixture model | SVM | 84 | ACC | 89.1 | ACC | 63.2 | ACC | 80.2 | Semi-data-driven approach to feature selection defines clusters of ROIs from an NC image and uses these to extract features from scans of MCI and AD patients | [401] | ||||
| SEN | 92.0 | SEN | 65.0 | SEN | 80.0 | ||||||||||
| SPE | 86.0 | SPE | 61.0 | SPE | 80.0 | ||||||||||
| FDG PET | Optimization of preprocessing steps for SPM8 | t-sum score | 108 | ACC | 68.0 | Modified preprocessing steps using motion correction, custom FDG template, different regions for intensity scaling and varied smoothing. | [402] | ||||||||
| SEN | 70.0 | ||||||||||||||
| SPE | 68.0 | ||||||||||||||
| AUC | 0.83 | ||||||||||||||
| MRI | Recursive feature elimination | Random Forests | 575 | ACC | 89.6 | SEN | 78.0 | Compares efficacy of Random Forest classifiers in several datasets including ADNI. RF model outperformed SVM, best model combined volumetric and cortical thickness measures with APOE | [403] | ||||||
| APOE4 demographics | SEN | 90.7 | Note: 12-month progression | ||||||||||||
| SPE | 82.9 | ||||||||||||||
| MRI | Regularized logistic regression | Low density separation (SVM) | 825 | AUC | 0.90 | Uses semisupervised learning to construct an aggregate biomarker by leading a separate MRI biomarker first and subsequently combining it with age and cognitive measures | [404] | ||||||||
| Cognitive | |||||||||||||||
| MRI | Multivariate regression: LASSO based + novel loss function | SVM | 202 | ACC | 95.9 | ACC | 82.0 | ACC | 72.6 | Uses a novel loss function combined with a group lasso method for joint feature selection. Multimodal classification outperformed single modalities | [405] | ||||
| PET | SEN | 95.7 | SEN | 98.0 | SEN | 48.5 | |||||||||
| CSF | SPE | 98.6 | SPE | 60.1 | SPE | 94.4 | |||||||||
| AUC | 0.99 | AUC | 87.0 | AUC | 0.79 | ||||||||||
| MRI | Multivariate regression: fused LASSO + LDDMM | SVM | 103 | SEN | 84.0 | Comparison of spatial regularization techniques for detecting longitudinal hippocampal shape changes in MCI progression | [406] | ||||||||
| HC shape longitudinal | SPE | 48.0 | |||||||||||||
| NPV | 0.93 | ||||||||||||||
| PPV | 0.27 | ||||||||||||||
| MRI | Multivariate regression: weak hierarchical LASSO | Random Forest | 293 | ACC | 74.8 | Uses hierarchical constraints and sparsity regularization to capture underlying interactions between biosignatures | [407] | ||||||||
| APOE4 | SEN | 66.7 | |||||||||||||
| Cognitive | SPE | 81.4 | |||||||||||||
| Demographics | |||||||||||||||
| MRI | Multitask learning:: K means clustering + l 2,1 regression | SVM | 202 | ACC | 95.1 | ACC | 79.5 | Uses a novel clustering approach to discover mulitpeak data distributions. These are accounted for in a subsequent a multitask learning step in a l 2,1 regression framework | [408] | ||||||
| FDG PET | SEN | 94.0 | SEN | 88.9 | |||||||||||
| SPE | 96.3 | SPE | 62.0 | ||||||||||||
| AUC | 0.96 | AUC | 0.78 | ||||||||||||
| MRI | Multitask learning: K means clustering + l 2,1 regression | SVM | 202 | ACC | 94.3 | ACC | 80.1 | ACC | 74.6 | Similar approach to [408]. NOTE: MCI versus AD classification uses ONLY MRI + PET data | [409] | ||||
| FDG PET | SEN | 94.0 | SEN | 86.8 | SEN | 46.7 | |||||||||
| CSF | SPE | 94.3 | SPE | 67.3 | SPE | 89.0 | |||||||||
| AUC | 0.96 | AUC | 0.82 | AUC | 0.72 | ||||||||||
| MRI | Multitask learning: relational function + l 2,1 regression | SVM | 202 | ACC | 95.7 | ACC | 79.9 | ACC | 72.4 | Introduces a function to conserve information about feature-feature relations, response-response relations, and sample-sample relations while jointly solving both classification and prediction of continuous variables | [410] | ||||
| FDG PET | SEN | 96.6 | SEN | 97.0 | SEN | 49.1 | |||||||||
| SPE | 98.2 | SPE | 59.2 | SPE | 94.6 | ||||||||||
| AUC | 0.98 | AUC | 0.85 | AUC | 0.83 | ||||||||||
| MRI | Multitask learning: label-aligned regularization | SVM | 202 | ACC | 95.6 | ACC | 80.3 | ACC | 69.8 | Uses a label-aligned regularization term in multitask learning step to use relationships across subjects, as well as across modalities in the feature selection step | [411] | ||||
| FDG PET | SEN | 95.1 | SEN | 85.0 | SEN | 66.7 | |||||||||
| SPE | 96.5 | SPE | 70.8 | SPE | 71.4 | ||||||||||
| AUC | 0.97 | AUC | 0.81 | AUC | 0.69 | ||||||||||
| MRI | Multitask learning: graph-guided + latent | 199 | ACC | 93.0 | ACC | 80.0 | Uses new latent group LASSO penalty combined with an undirected graph approach to select correlated features that can jointly predict class label and clinical scores | [412] | |||||||
| FDG PET | LASSO | ||||||||||||||
| CSF | group penalty | ||||||||||||||
| MRI, demographics, clinical, APOE4, PICALM | Statistical learning embedded in SVM-based multiple kernel framework | 213 | ACC | 91.0 | Longitudinal, multimodal data are leveraged by an embedded novel statistical learning approach in a multikernel framework. | [413] | |||||||||
| SEN | 95.0 | ||||||||||||||
| SPE | 80.0 | ||||||||||||||
| MRI | Domain transfer feature and sample selection | SVM | 202 | ACC | 76.5 | Uses both target domain (MCI) and auxiliary domains (CN and AD) to extract features from imaging and CSF modalities in domain transfer method. Fuses selected features with a domain transfer SVM | [414] | ||||||||
| FDG PET | SEN | 81.2 | |||||||||||||
| CSF | SPE | 71.9 | |||||||||||||
| AUC | 0.84 | ||||||||||||||
| MRI | Multimodal manifold-regularized transfer learning | 202 | ACC | 80.1 | Uses auxiliary domain information (as in [414]). Integrates kernel-based maximum mean discrepancy criterion and a manifold regularization function into single learning algorithm for both feature selection and classification | [415] | |||||||||
| FDG PET | SEN | 85.3 | |||||||||||||
| CSF | SPE | 73.3 | |||||||||||||
| AUC | 0.85 | ||||||||||||||
| MRI | Multimodal canonical correlation analysis | 103 | ACC | 95.1 | Used canonical correlation analysis to fuse data in multimodal classification as it preserves intermodal relationships. | [416] | |||||||||
| FDG PET | |||||||||||||||
| CSF | |||||||||||||||
| MRI | Deep architecture + multitask learning | SVM | 202 | ACC | 95.1 | ACC | 80.193.9 | ACC | 74.1 | Uses novel deep learning architecture that discards uninformative features in a hierarchical manner during multitask learning | [417] | ||||
| PET | SEN | 92 | SEN | 53.7 | SEN | 50.5 | |||||||||
| CSF | SPE | 98.3 | SPE | SPE | 92.7 | ||||||||||
| MRI/PET/CSF | MRI/PET | MRI/PET | |||||||||||||
| MRI | Deep learning with Deep Boltzman Machine | Hierarchical SVM | 398 | ACC | 95.4 | ACC | 85.7 | ACC | 75.9 | Uses deep learning with a Boltzman Machine to find hierarchical feature representation from MRI features and then fuses these with complementary information from PET | [418] | ||||
| PET | SEN | 94.7 | SEN | 95.4 | SEN | 48 | |||||||||
| SPE | 95.2 | SPE | 65.9 | SPE | 95.2 | ||||||||||
| AUC | 0.99 | AUC | 0.88 | AUC | 0.75 | ||||||||||
| MRI | PCA/LASSO + multitask deep learning with dropout technique | SVM | 202 | ACC | 91.4 | ACC | 77.4 | ACC | 57.4 | Uses dropout technique as method of regularization for deep learning with small data | [419] | ||||
| PET | |||||||||||||||
| CSF | |||||||||||||||
| Demographics | |||||||||||||||
| MRI | Independent component analysis | Discriminant classification analysis | 320 | ACC | 94.3 | ACC | 83.3 | ACC | 84.1 | ACC | 80.0 | Derived GM covariates patterns using independent component analysis and used for diagnostic classification in combination with cognitive performance | [304] | ||
| Demographics | SEN | 94.9 | SEN | 76.7 | SEN | 86.1 | SEN | 78.3 | |||||||
| SPE | 94.0 | SPE | 89.1 | SPE | 73.0 | SPE | 81.5 | ||||||||
| MRI | Stacked auto-encoder + multitask deep learning with zero-mask strategy | Softmax logistic regression | 331 | ACC | 91.4 | ACC | 82.1 | Uses stacked autoencoders to extract high-level features and a zero-masking strategy to extract synergy between modalities | [420] | ||||||
| PET | SEN | 92.3 | SEN | 60.0 | |||||||||||
| SPE | 90.4 | SPE | 92.3 | ||||||||||||
Abbreviations: AD, Alzheimer’s disease; CN, cognitively normal; MCI, mild cognitive impairment; MCIc, mild cognitive impairment converters (progressive MCI); MCInc, mild cognitive impairment non-converters (stable MCI); MRI, magnetic resonance imaging; SVM, support vector machine; ACC, accuracy; SEN, sensitivity; SPE, specificity; AUC, area under the receiver operating curve; ICA, independent component analysis; PCA, principal component analysis; WM, white matter; GM, gray matter; CSF, cerebrospinal fluid; ADNI, Alzheimer’s Disease Neuroimaging Initiative; EMCI, early mild cognitive impairment; ROI, regions of interest; VBM, voxel-based morphometry; fMRI, functional magnetic resonance imaging; SMC, subjective memory concern; FDG PET, 18F-flurodeoxyglucose positron emission tomography; MAP, mean average precision; APOE4, apolipoprotein ε4 allele; PET, positron emission tomography; HC, hippocampus; LASSO, least absolute shrinkage and selection operator; LDDMM, large deformations via diffeomorphisms.
8.1.2. Novel CSF and blood biomarkers
8.1.2.1. Novel CSF biomarkers
Proteomic approaches have identified numerous potential novel AD biomarkers in both CSF and blood. Potential AD biomarkers can be identified using a multiplex panel of selected CSF analytes that reflect the brain proteome. The p-tau181/Aβ42 ratio, a strong predictor of progression from CN to MCI over a 3- to 4-year period, was used as a continuous variable and endophenotype for AD to identify novel CSF analytes from a proteomic panel [168]. The most significantly associated analyte was heart fatty acid binding protein (FABP), which was comparable to p-tau181/Aβ42 in its ability to predict progression from CN to impaired cognition over 4 years. Other significant predictors were macrophage migration inhibitory factor and VEGF. A panel of 24 CSF analytes outperformed a combination of MRI and CSF measures in predicting MCI progression to AD (accuracy 94.1% vs. 76.5%) and included FABP, chromogranin A, matrix metalloproteinase 2, and pancreatic polypeptide as the strongest predictors [166]. The panel also discriminated between MCI and CN subjects with an accuracy of 91.5%, a sensitivity of 87.7%, and a specificity of 94.3%. Markers of inflammation, complement 3 and factor H, were associated with increasing ADAS-cog scores and lateral ventricular volume over time, suggesting they may have prognostic, but not diagnostic, utility [167].
MS is an alternative proteomics technology to multiplex immunoassay panels that can increase the speed, sensitivity, and quantitative precision of biomolecule analysis limited to a subset of analytes. The Foundation for the National Institutes of Health Biomarkers Consortium identified CSF-based AD biomarkers using a Multiple Reaction Monitoring MS panel of analytes selected for their relevance to AD and from previous multiplex immunoassay findings. The most significant peptides able to differentiate between MCIc and MCInc subjects were the blood proteins, hemoglobin A, hemoglobin B, and superoxide dismutase, consistent with the hypothesis that the blood brain barrier is leaky in AD. FABP was the top peptide for discriminating between MCI or AD and CN subjects, consistent with findings from immunoassay approaches. The top peptides predicting MCI to AD progression were putative markers of synaptic loss and neuronal injury/degeneration, neuronal pentraxin 2, VEGF, and secretogranin-2 (AUC 0.79).
8.1.2.2. Blood-based biomarkers
Because a blood-based test for AD is less costly and less invasive than a CSF-based test, there has been considerable effort in identifying blood-based diagnostic and prognostic AD biomarkers. Although CSF proteins are generally thought to reflect the brain proteome, the relationship between proteins in plasma and CSF is not clear. The cell bound fraction of Aβ42 was associated with left hippocampal and left entorhinal cortex volumes, suggesting that blood Aβ42 at least partially reflects Aβ production and/or clearance in the brain [421]. A blood biomarker–based signature comprising chemokine ligand 13, immunoglobulin M-1, interleukin 17, pancreatic polypeptide, and vascular cell adhesion protein-1 predicted neocortical Aβ burden with a sensitivity of 79% and a specificity of 76% [422]. These two studies suggest that Aβ accumulation in the blood is associated with neocortical Aβ burden and characteristic AD atrophy and that blood-based assessment of Aβ burden could have clinical utility.
Changes in plasma levels of bone morphogenetic protein 6, selectin, matrix metalloproteinase 10, and neuronal cell adhesion molecule were correlated with atrophy in temporal lobe structures, the posterior cingulate cortex, and thalamus, and diagnosed AD with a sensitivity of 93% and a specificity of 92% [423]. A panel of plasma proteins including interleukin 16, thyroxine-binding globulin, and peptide YY were predictive of MCI progression (accuracy 62.5%) [423]. Redox-reactive antiphospholipid autoantibodies may be increased early in disease due to neurodegenerative changes in cell membranes, and APOE4 may act by increasing redox-reactive Fe during disease progression [152]. Elevated levels of redox-reactive autoantibodies against phosphatidylserine, phosphatidylethanolamine, and phosphatidylcholine were elevated in MCI compared to CN subjects, and a panel of these autoantibodies discriminated between MCI and CN with a sensitivity of 80% and a specificity of 83.3% [424].
In conclusion, at the time of this review, no established validated blood test for AD exists. However, there is a pressing need for a test with relatively good sensitivity (even if lacking some specificity) to identify subjects at high risk for AD, who would then undergo further testing such as Aβ PET for diagnosis. There is a great deal of work going on in this field, and the availability of ADNI plasma samples hopefully will provide material for further investigation.
8.1.2.3. Combinations of CSF and blood biomarkers
A combination of both CSF and blood analytes may have even greater predictive power than either fluid alone. Lehallier et al. [425] examined multiple groups of variables—clinical, genetic, volumetric, cognitive, and CSF and plasma analytes—to identify a biomarker signature that could predict the time to progression from MCI to AD 1 to 6 years before clinical diagnosis. Variables from different categories were differentially associated and appeared to carry complementary information about the disease (Fig. 39). For example, CSF and plasma “communicomes” containing proteins involved in intracellular communication were highly associated with each other but not with established CSF biomarkers, whereas genetic factors (APOE4, TOMM40) were predominantly associated with AD CSF biomarkers. Predictive models for each progression time point in 6- or 12-month intervals identified six variables as top predictors in most models: CDR-SB, CSF tumor necrosis factor–related apoptosis-inducing ligand receptor 3 (TRAIL-R3), plasma apolipoprotein A-II (ApoAII), and CSF fibroblast growth factor 4 (FGF-4). Most analytes had the highest accuracies in predicting progression within 2 to 3 years. A final model consisting of two plasma (ApoAII, cortisol), and four CSF (FGF-4, FABP-H, calcitonin, and TRAIL-R3) analytes predicted MCI progression within 3 years with a sensitivity of 88% and a specificity of 70%. A comparison of this model with other groups of variables is shown in Fig. 40 and supports the idea that novel plasma and CSF biomarkers may outperform modalities traditionally used for prediction such as MRI volumes, cognitive scores, or AD CSF biomarkers and APOE status.
Fig. 39.
Associations between 249 variables shown by a circular visualization of correlation plot. Data from 928 patients with mild cognitive impairment were used to produce a network visualization of the complex relationships between and within variables in ADNI. Lines represent the Spearman rank correlation coefficient |(r)| = .3 between two variables. 1—sex, 2—years education, 3—age, 4—MMSE, 5—ADAS total score, 6—ADAS modified, 7—CDR composite score, 8—CDR-SOB composite score, 9—FAQ, 10—GDS, 11—Hachinski Ischemic Scale score, 12—NIQ total score, 13—brain volume, 14—intracranial volume, 15—ventricular volume, 16—hippocampal volume, 17—inferior lateral ventricular volume, 18—middle temporal volume, 19—inferior temporal volume, 20—fusiform cortical volume, 21—entorhinal cortex volume, 22—APOE4 carrier, 23—no. of APOE4 alleles, 24—TOMM40 polyT allele one, 25—TOMM40 polyT allele 2, 26—8-Iso-PGF, 27—8,12-iso-IPFa, 28—CSF white blood cell count, 29—CSF red blood cell count, 30—CSF total protein concentration, 31—CSF glucose level, 32—total plasma homocysteine level, 33—plasma Aβ40 level, 34—plasma Aβ42 level, 35—plasma Aβ40:AB42 ratio, 36—CSF Aβ42 level, 37—CSF t-tau level, 39—CSF Aβ42:t-tau ratio, 40—CSF Aβ42-p-tau ratio, 41—CSF p-tau:t-tau ratio, 42 to 115—74 CSF analytes measured by multiplex assay, 116 to 249—hundred and 34 plasma analytes measured by multiplex assay. Abbreviations: ADAS, Alzheimer’s Disease Assessment Scale; ADNI, Alzheimer’s Disease Neuroimaging Initiative; CDR-SOB, Clinical Dementia Rating–Sum of Boxes; CSF, cerebrospinal fluid; FAQ, Functional Activities Questionnaire; MMSE, Mini–Mental State Examination. Reproduced with permission from [425].
Fig. 40.
Prediction of progression from mild cognitive impairment to Alzheimer’s disease within 3 years. Markers in plasma or CSF predicted progression with a relatively high sensitivity compared to standard AD CSF biomarkers, regional MRI volumes, or cognitive scores. Prediction includes seven models combining different subsets of variables. Correct classification rate of the top 20 variables was estimated on the test data set after 1000-fold resampling of the learning and test data sets. Sex and age were included in all models. APOE4 indicates apolipoprotein ε4. Abbreviations: AD, Alzheimer’s disease; CSF, cerebrospinal fluid; MRI, magnetic resonance imaging. Reproduced with permission from [425].
8.1.3. Arterial spin labeling
Hypoperfusion of brain tissue, detected by arterial spin labeling MRI, precedes atrophy and may therefore be a useful biomarker for early changes in the disease process. Cerebral blood flow was marginally outperformed by structural MRI (AUCs of 0.87 compared to 0.89) in the classification of AD versus CN subjects [426], but the technique may have more utility in classifications such as MCI versus CN or in predicting cognitive decline in subjects with normal cognition.
8.1.4. Network connectivity measures
The concept of AD as a disconnection syndrome posits that some of the earliest changes in disease progression are disruptions to network connectivity. Network connectivity measures, which have been shown to differ between patient groups [321–323,329,333], may therefore have prognostic and diagnostic utility.
8.1.4.1. Structural connectivity measures
A variety of structural connectivity measures have been tested for their diagnostic or prognostic abilities. These include connectivity measures of WM fiber pathways [427], the tensor–based Fiber Assignment by Continuous Tracking algorithm [428], a new tractographic feature, average tract length [429], and a novel flow-based measure of brain connectivity [430]. The results were variable with accuracies ranging from 71.25% [428] to 97% [429] for the AD versus CN challenge, and from 57.3% [428] to 59.2% [430] for the MCI versus CN challenge. One difficulty in using structural connectivity measures is the high dimensionality of WM tracts. Nir et al. [431] used a novel maximum density pathway algorithm to find the shortest path between probabilistic ROIs and made a compact representation of WM tracts from whole-brain tractography. They used their low-dimensionality representation for classification, achieving accuracies of 84.9% and 79% for the AD versus CN, and MCI versus CN challenges, respectively.
8.1.4.2. Functional connectivity measures
Progressive deterioration of functional connectivity appears to be characteristic in AD and is reflected in changes to the network small-world properties and other measures. MCI subjects had lower clustering coefficients and longer characteristic path length than CN subjects in one study, and alterations in these properties in the DMN discriminated between EMCI and CN with an accuracy of 79.7% [432]. A topological metric, compression flow, derived from network centrality criteria, outperformed individual small-world metrics alone. It monotonically followed impairment progression in each patient group, and discriminated between all patient groups (CN, EMCI, LMCI, and AD) [433]. Other studies developed a novel multifractal feature [434] and created a compact representation of the brain network [395] for classification purposes. Machine learning techniques were used for dimensionality reduction and the selection of the most discriminative regions.
8.1.4.3. Cortical atrophy networks
Cortical atrophy networks which capture the spread of at rophy have been used for classification in several studies. Surface connectivity maps describing the center of each cortical region affected, the individual volume loss of these regions, and how they move apart were used to define functional regions of the brain [435]. This approach outperformed whole-brain or hippocampal volume in predicting the progression of MCI subjects and achieved AUCs of 0.88, 0.78, and 0.77 for AD versus CN, MCI versus CN, and MCI versus AD classifications, respectively. Instead of solely finding connectivity between areas of cortical atrophy, Friedman [436] developed a new approach, Directed Progression Networks (DPNets), based on the idea of a prionlike spread of the AD pathology along axonal tracts in a characteristic and systematic pattern. DPNets found the direction of connections between network hubs to capture the temporal progression of brain disease and outperformed undirected networks in the classification of AD versus CN subjects, achieving an AUC of 0.87. Interestingly, the network generated by this method did not have the small-world properties observed in almost all other brain networks probably because this network reflects the spread of disorder in the brain, and not computation.
8.1.4.4. Future directions
The use of connectivity measures in classification and prediction is still in its infancy. A study which combined metabolic and structural connectivity patterns by constructing networks from both FDG PET and structural MRI data was able to discriminate between AD and CN, MCI and CN, and MCI and AD subjects with AUCs of 0.96, 0.91, and 0.88, respectively [437]. The results for classifications involving MCI compare favorably with other state-of-the-art multimodal approaches, hinting at the potential power of multimodal network connectivity approaches. Methodological improvements will aid in the development of this exciting new area. Technical issues were found to influence interpretation of graph theory measures [438] and may be overcome by the standardization of connectomics methods. One study improved classification sensitivity and specificity by including a more modern group-wise DTI registration step [439] and likely represents just the initial phase of improvements in this area.
8.1.5. Single nucleotide polymorphisms
For the first time, ADNI studies demonstrated that diagnostic classification and the prediction of future clinical decline could be improved by the addition of selected SNPs to other modalities. One hundred eighty-nine SNPs selected for their discriminative ability improved classification accuracy by 5% and 3% for the MCI versus CN, and AD versus MCI classifications, respectively [254]. Top hits included CTNNA3 and PON2, but not APOE. SNPs did not improve the AD versus CN classification in combination with any other modality. The addition of 101 top SNPs from a GWAS to MRI features such as hippocampal volumetric and surface morphology data improved prediction of MCI to AD progression over 48 months, achieving an AUC of 0.95 [255]. The SNP markers selected for prognosis differed from those associated with increased disease risk, suggesting that genes underlying the MCI to AD progression differ from those that increase disease risk. This is an exciting advance and holds much promise for the incorporation of similar data to improve stratification and subject selection.
8.2. Diagnosis and prediction methods
A plethora of novel machine learning approaches to diagnosis and prediction have been developed and tested using ADNI data in 2014 and 2015, leading to an overall improvement in classification and prediction accuracy. The use of standardized ADNI data sets has, in some cases, facilitated comparison of results. Deep learning algorithms that use multiple processing layers to model high-level data abstractions have proved exceptionally successful in selecting and combining optimal features from multiple modalities. In combination with some form of dimensionality reduction and a classifier such as a support vector machine or treebased algorithm, either as sequential steps or as an embedded wrapper algorithm, deep learning has improved the effectiveness of MRI as a single modality to the point where it rivals multimodal approaches for classification accuracy. Several studies reported MRI-based methods that could discriminate between AD and CN subjects with accuracies between 92% and 97% [374,387,388,394] or predict MCI progression to AD with accuracies exceeding 80% [374]. Notably, Gorji et al. [394] used pseudo-Zernike moments, powerful shape descriptors used in image recognition, to characterize global and local pattern shapes and select discriminative features that were used by a supervised neural network for classification. Their results were comparable with other state-of-theart methods for the AD versus CN classification but substantially outperformed other methods, including multimodal approaches, for the more challenging problems of MCI versus CN (sensitivity 95.9%, specificity 95.3%, and accuracy of 95.6%), and AD versus MCI (sensitivity 94.2%, specificity 95.6%, and accuracy 94.9%) classification. This publication illustrates the vast potential of neural networks in tackling these classification problems, and further rapid improvement in this area is likely concomitant with the burgeoning development of this area of machine learning.
However, the best classifiers generally still incorporated multimodal information; deep learning approaches improved feature selection, in part by preserving intermodal relationships. The best multimodal classifiers for the AD versus CN, and MCI versus CN challenges reached accuracies of >95% [405,408,410,411,416–418] and >82% [304,405,417,420], respectively. An approach which incorporated longitudinal multimodal data, and which notably included PICALM status, predicted progression of MCI subjects to AD with an accuracy of 91% [413]. It is beyond the purview of this review to detail all of these efforts; they are instead summarized in Table 10. Instead of binary classification, one study used machine learning of structural MR images of a normative population to define a brain abnormality index [386], and others tackled multi-class classifications such as AD versus MCI versus CN (Table 11). Finally, a number of publications focused on addressing technical problems such as incomplete or imbalanced data in multimodal classification or problems with imperfect reference tests (Table 12).
Table 10.
Prediction of continuous variables
| Classification challenge | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Modality | Feature selection/dimensionality reduction | Classifier | N | ADAS-cog | MMSE | RALVT | Unique approach | Reference | |
| MRI Cortical surface | Multivariate regression: l 2,1 norm | SVM | 718 | RMSE | 0.7663 ± 0.0375 | 0.8325 ± 0.0399 | 0.9167 ± 0.0471 | Propose sparse multitask learning model Group-Sparse Multi-task Regression and Feature Selection (G-SMuRFS) to select cortical surface markers | [440] |
| CC | 0.6438 ± 0.0258 | 0.5277 ± 0.0539 | 0.3985 ± 0.0533 | ||||||
| MRI | Multitask learning: relational function + l 2,1 regression | SVM | 202 | CC(AD vs. MCI) | 0.680 | 0.682 | Introduces a function to conserve information about feature-feature relations, response-response relations, and sample-sample relations while jointly solving both classification and prediction of continuous variables | [410] | |
| PET | CC (MCI vs. CN) | 0.520 | 0.508 | ||||||
| CC (MCIc vs. MCInc) | 0.591 | 0.622 | |||||||
| MRI | Canonical correlation analysis + novel sparse multitask learning | SVM | 202 | CC | 0.740 ±0.18 | 0.675 ± 0.23 | Uses canonical correlation analysis to determine correlations between features of different modalities | [441] | |
| FDG PET | |||||||||
| Note: for AD vs. MCI vs. CN | |||||||||
| MRI | Canonical correlation analysis + novel sparse multitask learning | SVM | 202 | RMSE | 4.201 ± 0.82 | 2.110 ± 0.41 | Similar to [441] | [442] | |
| FDG PET | CC | 0.719 ± 0.81 | 0.655 ± 0.31 | Note: for AD vs. MCI vs. CN | |||||
| MRI | Multitask learning: graph-guided + latent | 199 | CC | 0.740 ± 0.002 | 0.745 ± 0.002 | Uses new latent group LASSO penalty combined with an undirected graph approach to select correlated features that can jointly predict class label and clinical scores. | [412] | ||
| FDG PET | LASSO group penalty | ||||||||
| CSF | |||||||||
| MRI | Multivariate regression: sparse model | Sparse Bayesian | 393 | CC | 0.767 | 0.758 | Models cognitive scores as nonlinear functions of neuroimaging variables; models correlations between regression coefficients. Derives sparse Bayesian learning algorithm for learning. | [443] | |
| MRI (TBM) | Multivariate TBM + Convex Fused Sparse Group LASSO | Prediction of continuous variables: ADAS-cog | 616 | Prediction of ADAS-cog scores (RMSE) | Uses Convex Fused Sparse Group LASSO for multitask learning using multivariate TBM data to predict ADAS-cog scores at 6, 12, 24, 36, and 48 months. | [444] | |||||
| HC shape, Clinical, APOE4, baseline MMSE | Months | 6 | 12 | 24 | 36 | 48 | |||||
| w/o mTBM | 5.259 ± 0.87 | 5.653 ± 1.14. 7 | 5.532 ± 1.03 | 4.777 ± 0.83 | 4.367 ± 1.18 | ||||||
| With mTBM | 4.534 ± 0.88 | 4.989 ± 1.13 | 4.885 ± 1.09 | 4.055 ± 1.02 | 3.164 ± 1.09 | ||||||
Abbreviations: ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive subscale; MMSE, Mini–Mental State Examination; RAVLT, Rey’s Auditory Verbal Learning Test; MRI, magnetic resonance imaging; SVM, support vector machine; RMSE, root mean square error; CC, correlation coefficient; PET, positron emission tomography (FDG and Aβ); AD, Alzheimer’s disease; MCIc, mild cognitive impairment converters (progressive MCI); MCInc, mild cognitive impairment nonconverters (stable MCI); FDG PET, 18F-flurodeoxyglucose positron emission tomography; MCI, mild cognitive impairment; CSF, cerebrospinal fluid; CN, cognitively normal; TBM, tensor-based morphometry; HC, hippocampus; APOE4, apolipoprotein ε4 allele; mTBM, multivariate tensor-based morphometry; LASSO, least absolute shrinkage and selection operator.
Table 11.
Multiclass classification
| Modality | Feature selection/dimensionality reduction | Classifier | N | Cross-validated | Classification challenge | Unique approach | Reference | |||
|---|---|---|---|---|---|---|---|---|---|---|
| MRI | Canonical correlation analysis + novel sparse multitask learning | SVM | 202 | Y | Multiclass classification | Uses canonical correlation analysis to determine correlations between features of different modalities. Applies to multiclass classification problem. | [441] | |||
| PET | AD versus MCI versus CN | AD versus MCIc versus MCInc versus NC | ||||||||
| ACC | 71.9 | ACC | 61.9 | |||||||
| MRI | Canonical correlation analysis + novel sparse multitask learning | SVM | 202 | Y | ACC | 68.6 | ACC | 56.2 | Similar to [441]. | [442] |
| PET | ||||||||||
| MRI | Linear discriminant analysis + locality preserving projection | SVM | 202 | Y | ACC | 76.4 | ACC | 61.1 | Combines two subspace learning techniques, linear discriminant analysis and locality preserving projection for feature selection and applied to multiclass and binary classification problems. | [445] |
| PET | ||||||||||
| MRI | Deep architecture + multitask learning | SVM | 202 | Y | ACC | 62.5 | ACC | 52.5 | Uses novel deep learning architecture that discards uninformative features in a hierarchical manner during multitask learning. | [417] |
| FDG PET | ||||||||||
| CSF | ||||||||||
Abbreviations: MRI, magnetic resonance imaging; PET, positron emission tomography; SVM, support vector machine; AD, Alzheimer’s disease; MCI, mild cognitive impairment; CN, cognitively normal; MCIc, mild cognitive impairment converters (progressive MCI); MCInc, mild cognitive impairment nonconverters (stable MCI); NC, normal cognition; ACC, accuracy; CSF, cerebrospinal fluid; FDG PET, 18F-flurodeoxyglucose positron emission tomography.
Table 12.
Methods that address technical problems in classification and prediction
| Problem | Approach | Results and conclusions | Reference |
|---|---|---|---|
| Variance in validation strategies can make comparison of the performance of supervised classification algorithms difficult. | Use of an empirical estimator that performs validation on two disjoint sets and comparison with different variance estimators in an AD versus CN classification experiment. | Proposed estimator is constant with sample size and is unbiased with regard to training set size. Recommend against the use of leave one out cross-validation because of its high variance. | [446] |
| Estimation of the sensitivity of a biomarker to the early diseased stage based on its performance on the fully diseased stage. | Use of empirical likelihood-based (ELB) confidence intervals. | ELB method is more robust than parametric methods and more accurate than nonparametric methods of confidence interval estimation. | [447] |
| Lack of a method for selecting diagnostic cut points in a multistage disease that is not dependent on correct classification rates. | Developed new measure, maximum absolute determinant for diseases with k stages, which selects cut points using all available classification data. | When applied to ADNI biomarker data, the proposed method more accurately selected cut points for the early diseased stage than existing methods. | [448] |
| Patterns of atrophy in normal aging can confound multivariate models of atrophy in AD. | Compared two age correction approaches on AD versus CN classification and prediction of MCI to AD progression: (1) using age as a covariate in MRI-derived measures; (2) linear detrending of age-related changes based on CN measures. | Both models improved classification and prediction accuracy. Analysis of incorrectly classified subjects suggested that the influence of cognitive impairment, APOE genotype, and gender is partially masked by age effects. | [449] |
| Errors in reference test for AD biomarkers (clinical diagnosis or Aβ PET in the absence of a gold standard) cause bias in their diagnostic accuracy. | Uses Bayesian method to determine diagnostic accuracy of AD biomarkers taking imperfectness of reference test into account. | Proposed methodology improved estimates of exact diagnostic values of three CSF biomarkers in the AD versus CN classification. | [450] |
| Imperfect reference test can lead to bias in accuracy of a combination of diagnostic biomarkers. | Uses Bayesian method to select combination of biomarkers that maximizes the AUC while taking imperfectness of reference test into account. | Proposed methodology improved estimates of AUCs of AD biomarkers over traditional logistic regression model. | [451] |
| Incomplete/imbalanced data biases estimation of diagnostic accuracy of AD biomarkers. | Novel approach uses augmented weighted estimator for covariate-specific time-dependent receiver operator curves using information from subjects with incomplete data. | Proposed estimator corrected bias and improved efficiency of classification in incomplete data sets over other estimators. | [452] |
| Incomplete/imbalanced data in multimodal classification. | Extensive and systematic analysis of effectiveness of combinations of sampling techniques (undersampling, oversampling, and a combination), six common feature selection algorithms, and Random Forest and SVM classifiers on AD/MCIc versus AD and MCI versus CN classification problems. | K-Medoids undersampling technique gave best results on imbalanced data sets. Sparse logistic regression with stability selection was best feature selection technique. | [453] |
| Novel approach based on collection of feature values into a large incomplete matrix, and subsequent matrix shrinkage and completion using a multitask learning algorithm. | Improved classification accuracy over two recent methods for accounting for missing data (including Incomplete Multi-Source Feature Learning [454]). | [455] | |
| Novel approach, Multi-Task Linear Programming Discriminant analysis which decomposes the classification problem into different classification tasks, adaptively chooses different feature subsets for each task, then solves them jointly. | Improved classification accuracy of MCIc versus MCInc classification over Incomplete Multi-Source Feature Learning [454]. | [456] | |
| Novel approach (3D-CNN) based on a convolutional neural network that can estimate missing data in an output modality (PET images) using data from an input modality (MR images). | 3D-CNN approach used on incomplete data sets achieved similar classification accuracies to using complete data sets in AD versus CN, MCI versus CN, and MCIc versus MCInc tasks and outperformed two commonly used missing data estimation methods. | [457] | |
| Conventional false discovery rate procedures for voxel level multiple testing ignore correlations between neighboring voxels. | Novel approach extends the local significance index procedure with a Markov random field model to consider spatial correlations along neighboring voxels. | When method was applied to ADNI FDG PET data, it outperformed other false discovery rate procedures. | [458] |
Abbreviations: AD, Alzheimer’s disease; CN, cognitively normal; ADNI, Alzheimer’s Disease Neuroimaging Initiative; MCI, mild cognitive impairment; MRI, magnetic resonance imaging; CSF, cerebrospinal fluid; AUC, area under the receiver operating characteristic; SVM, support vector machine; MCIc, mild cognitive impairment converters (progressive MCI); MCInc, mild cognitive impairment nonconverters (stable MCI); MR, magnetic resonance; PET, positron emission tomography.
8.3. Subject selection and outcome measures
With the transition of AD preventive and treatment clinical trials to preclinical and presymptomatic populations, the selection of subjects likely to decline in the near future, and the selection of clinical end points able to detect a treatment effect, are of paramount importance. Inclusion criteria include Aβ positivity, tau pathology, hippocampal atrophy, or combinations of these and genetic markers and are fundamentally guided by the need to decrease heterogeneity in the trial population. Both prognostic enrichment (selecting patients likely to progress) and predictive enrichment (selecting patients likely to respond to the therapy being trialed) need to be considered. Cognitive end points must be sensitive to early cognitive changes such as delayed recall, function, and attention, and imaging end points must also detect early changes. Once again, selection of inclusion criteria and outcome measures are informed by knowledge of disease progression. Understanding how covariates influence disease progression, even in an enriched MCI population, aids in selecting populations likely to progress.
A model predicting cognitive decline on the CDR-SB in MCI subjects found that the rate of disease progression was influenced by current CDR-SB score, baseline levels of CSF biomarkers, and baseline Delayed Logical Memory and Trails A scores [459]. Subjects selected for either low or high p-tau181/Aβ42 progressed at different rates, and the model identified a subpopulation with low likelihood of disease progression based on this ratio (Fig. 41). A different model found that baseline severity measured by CDR-SB, baseline hippocampal volume, and APOE4 predicted shorter times the clinical worsening [460] and estimated N80s of around 550 CN subjects using either low hippocampal volume or APOE4 positivity as an enrichment strategy for a 2-year trial. However, baseline values of biomarkers may not be sufficiently informative to predict clinical decline. Longitudinal biomarker progressions predicted substantially more variability in cognitive decline than baseline measures; ventricular expansion explained 40% of variability compared to a mere 8.7% of variability explained by baseline ventricular volume in MCI subjects [461]. Therefore, the use of longitudinal biomarker progressions for enrichment could improve the statistical power of clinical trials.
Fig. 41.
Nonlinear disease progression model capturing longitudinal Clinical Dementia Rating–Sum of Boxes (CDR-SB) scores. Visual predictive check simulations suggest that the model describes longitudinal progression of CDR-SB scores in both late MCI and mild AD subjects. Stratification using p-tau181/Aβ42 ratio reveals a lack of disease progression in biomarker negative subgroups. The upper, middle, and lower profiles indicated by the open circles represent the 95th, 50th, and 5th percentiles of the observed data, respectively. The upper, middle, and lower curves indicated by the lines are the median model–based predictions for the 95th, 50th, and 5th percentiles, respectively. The shaded areas are the 90% confidence intervals of the corresponding percentiles of the simulations based on the model. Abbreviations: AD, Alzheimer’s disease; LMCI, late MCI; MCI, mild cognitive impairment. Reproduced with permission from [459].
8.3.1. Enrichment biomarkers
Aβ positivity is currently used as an inclusion criterion for CN participants in the anti-Aβ A4 secondary prevention trial which aims to slow cognitive decline in CN elders using solanezumab [462]. Improvement of quantitative accuracy of amyloid PET scans by refinement of acquisition protocols and tracer administration, subject management, and image quality control and processing could reduce sample sizes for trials of intervention effects [463]. Prediction of Aβ positivity by florbetapir PETwas improved by the determination of a cutoff value of 1.24 based on three regions of interest (posterior cingulate cortex, precuneus, medial frontal cortex) which achieved an accuracy of 98.5% [464]. Visual inspection of florbetapir PET images was as effective as SUVR in determining Aβ positivity [465]. Alternative automated methods such as Syngo-PET Aβ Plaque [466], and a method based on a two-point correlation metric, wS2 [467], were comparable to SUVR and offered some advantages such as the ability to detect subtle changes in patterns of Aβ deposition [468].
Hippocampal volume is the first imaging biomarker to be qualified by the European Medicines Agency to select amnestic MCI subjects at risk of imminent rapid clinical deterioration for clinical trials [469]. As hippocampal volume is an established biomarker of AD, it can help to differentiate between the approximately two thirds of subjects who have MCI attributable to AD and the approximately one third of MCI subjects on a different disease pathways not necessarily attributable to Aβ pathology. Yu et al. [470] describes the systematic operationalization of hippocampal volume as an enrichment biomarker by defining specific cut points of hippocampal volume, comparing different commonly used algorithms for hippocampal volume quantification (FreeSurfer, HMAPS, LEAP, NeuroQuant), and considering three cognitive end points, MMSE, ADAS-cog, and CDR-SB. Hippocampal volume, calculated by any of the quantification methods, reduced sample sizes for all cognitive outcomes by 40% to 60%, with the greatest reduction in sample sizes at a cut point of 10% of normal volume. Trial costs were estimated to be reduced by 30% to 40% over a range of hippocampal volume cut points, although little effect on trial duration was predicted, regardless of cognitive end point (Fig. 42). Designed to guide clinical trial design, this analysis illustrates the trade-off between statistical and practical considerations.
Fig. 42.
Implications of hippocampal volume–based enrichment for clinical trials of amnestic MCI subjects. Estimates of trial costs and total duration for scenarios in which patients are enriched or not enriched with hippocampal volume are given for different outcome measures. (A–C) Trial cost and (D–F) trial execution time, as a function of cut point for (A and D) MMSE, (B and E) ADAS-cog, and (C and F) CDR-SB. Results are expressed as fractions of the unenriched scenario and are shown for four different hippocampal volume computational algorithms. Variance due to test-retest variability is shown as the shaded area for one of the four algorithms (LEAP). Abbreviations: ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive; CDR-SB, Clinical Dementia Rating–Sum of Boxes; MCI, mild cognitive impairment; MMSE, Mini–Mental State Examination. Reproduced with permission from [470].
Deep learning algorithms may also have great potential for selecting inclusion criteria that increase statistical power and reduce sample sizes in clinical trials. Ithapu et al. [471] described a new method of enrichment using inclusion criteria derived from FDG PET, florbetapir PET, and structural MRI features learned using their novel randomized denoising auto-encoder marker. Using CDR-SB scores as an outcome measure, this method reduced MCI sample sizes at least fivefold compared with no enrichment (N80s: 281 vs. 1586) and improved on enrichment with hippocampal volume alone (N80: 389) for a 2-year trial, suggesting that the learning algorithm can capture data from different modalities most representative of changes during disease progression.
8.3.2. Outcome measures
As clinical trials move to presymptomatic subjects, the development of outcome measures which can detect a subtle treatment effect is vital. Some investigations have focused on developing a clinical measure sensitive to very early cognitive changes; others have developed candidate biomarkers as potential surrogate outcome measures. A composite score developed from existing clinical end points and consisting of Word Recall, Delayed Word Recall, Orientation, 13-item CDR-SB, and FAQ had greater sensitivity to detect change in MCI subjects enriched with APOE4, hippocampal volume, and CSF Aβ42 than either CDR-SB alone or ADAS-cog [472]. The primary outcome measure in the A4 study is the Alzheimer’s Disease Cooperative Study Preclinical Alzheimer’s Cognitive Composite, comprising Total Recall, Delayed Recall, Digit Symbol Substitution, and MMSE [473]. Aβ+ CN participants had greater decline in this measure than Aβ− participants on this measure across different cohorts. With a cohort size of 500, it was predicted that the A4 study would be able to detect treatment benefit of about 0.5 units of the measure at 80% power over 3 years [473], suggesting that the measure is sufficiently sensitive and specific to detect Aβ-related decline in this trial population.
The quest for a surrogate outcome measure with greater power to detect decline than cognitive measures has progressed with several systematic studies comparing the efficacy of various biomarkers. Imaging biomarkers, especially MRI, have been of particular interest as longitudinal changes in them can be precise and reproducible. Caroli et al. [474] compared the power and performance of MRI and FDG PET biomarkers, and ADAS-cog, and found that for a 2-year trial with yearly biomarker assessment, a measure of the longitudinal whole-brain atrophy, KN-BSI [475], had the greatest power, with N80s of 48 and 85 for MCI subjects selected for Aβ positivity, or hippocampal atrophy, respectively. In comparison, they calculated N80s of approximately 1000 using ADAS-cog as an outcome measure with either enrichment strategy. Longer trials, but not more frequent biomarker assessments, increased power. Although measures of atrophy such as KN-BSI are well established, they may be outperformed by machine learning algorithms that are optimized for power to detect brain change. Gutman et al. [476] extended a previous data-driven feature selection approach [477] to maps of whole-brain volume change derived from TBM measures and combined this with ventricular surface measures to generate one summary atrophy score. This outperformed either measure alone and statistically defined ROIs in 1- or 2-year trials in both AD and MCI subjects, with calculated N80s of 31 for AD, and 56 for MCI subjects for a 2-year trial.
A systematic comparison of selection criteria and cognitive end points in CN elderly [478] found that a combination of the composite measure comprising the 11-point ADAS-cog, Delayed Recall on AVLT, and Trails B, and subject selection with three or four abnormal biomarkers (from APOE4, Aβ+, tau+, or hippocampal atrophy+) reached 79% power with N = 1500 participants per arm over a 3-year trial, assuming a 25% effect size of treatment (Fig. 43). Trial durations of longer than 4 years were required to achieve greater than 80% power using this combination of enrichment criteria and cognitive outcome measure. Recruiting subjects with multiple pathologies increased the power to detect change by 15% to 20%, but this strategy would also increase costs and limit the pool of eligible participants. A structural MRI surrogate outcome measure may improve the feasibility of clinical trials in CN subjects by lowering costs and increasing the power to detect disease progression and may be especially suited to trials of therapies that aim to repair brain tissue rather than clear Aβ. Hua et al. [479] investigated the utility of longitudinal TBM biomarkers for tracking brain changes, comparing the power of single numerical summary of brain atrophy scores reflecting disease-related change in the temporal lobe (temporal-ROI) [480], statistically defined ROIs, or over the whole brain over different trial lengths, and with different subject groups, enrichment biomarkers, and scan intervals. The whole-brain measure and statistically defined ROI measure produced N80s of 127 and 241, respectively, for CN subjects, 141 and 314, respectively, for EMCI subjects, and 72 and 162, respectively for LMCI subjects for a 2-year trial using a 12-month scan interval. Both measures are trainingbased approaches which select regions with the most consistent and detectable change. Enrichment using APOE4 or Aβ further reduced sample sizes to around 100 subjects regardless of diagnostic group (Fig. 44). Of 10 AD risk alleles tested as enrichment criteria, only APOE4 produced any gains in statistical power. Both statistically defined ROIs and the whole-brain measure based on linear discriminant analysis are training approaches which differ in training sample size and region selected; statistically defined ROI requires a small training set and selects regions such as the temporal lobe which are likely to be affected in AD but has low statistical power, whereas the whole-brain approach requires a larger training set and may not select patterns of brain change germane to the treatment but has higher performance and statistical power. Trade-offs between end-point measures and selection biomarkers will ultimately represent a balance of cost and power and will reflect the drug mechanism.
Fig. 43.
Ability of cognitive end points to detect change in cognitively normal subjects selected for multiple pathologies. Composite cognitive tests were more able to capture decline in cognitively normal (CN) subjects over 7 years than any measure of a single cognitive domain or ADAS-cog alone. Enrichment with three or more pathologies optimally enhanced this effect. Groups with 0, 1, 2, or 3+ pathologies (APOE4, Aβ+, tau+, or hippocampal atrophy1) plotted for each standardized cognitive measure with 7 years of follow-up. Composite #1: ADAS-11, Trails B, and Logical Memory II. Composite #2: ADAS-11, Trails B, and dALVT. Abbreviations: ADAS, Alzheimer’s Disease Assessment Scale; ADAS-cog, Alzheimer’s Disease Assessment Scale–cognitive; dAVLT, delayed Rey Auditory Verbal Learning Test. Reproduced with permission from [478].
Fig. 44.
The effect of clinical trial enrichment using APOE4 status or brain Aβ load. After screening participants for APOE4 status or brain Aβ load, sample size requirements are around 100 subjects for a 2-year trial. Sample size estimates (n80s) after trial enrichment using APOE4 status (A), brain Aβ load at screening (B), or both combined (C). Abbreviations: AD, Alzheimer’s disease; CN, cognitively normal; EMCI, early mild cognitive impairment; LMCI, late mild cognitive impairment. Reproduced with permission from [479].
8.4. Other improvements to clinical trials
The use of a placebo arm in addition to the treatment arm in clinical trials raises ethical issues. The Placebo Group Simulation Approach [481] models the placebo group based on baseline data of the treatment group and forecasts the distribution of quantified outcomes in MCI subjects. A comparison of empirically observed and simulated data suggested that the model approximated placebo data well. Of the baseline variables, cognitive performance predicted the trajectory of cognitive decline most accurately.
8.5. Conclusions
The overarching theme of publications focused on improving clinical trials from 2014 and 2015 has been a shift to optimizing enrichment strategies and developing outcome measures suited to trials in CN cohorts. Knowledge of disease progression has guided this development; the effectiveness of established biomarkers at diagnostic and prognostic challenges reflects their position in the temporal ordering of biomarkers as described by the first model of the pathological cascade [482]. The prediction of imminent decline in CN participants may require adjustment of established cut points of CSF biomarkers, the use of subtle changes to MRI features instead of atrophy measures, or measures of cognitive domains first affected in the disease. Multimodal approaches remain the most effective at prediction and classification, and the addition of novel modalities such as selected SNPs or network connectivity measures may improve accuracy even further. Panels of CSF and/or blood biomarkers now rival established multimodal biomarkers for diagnostic and prognostic utility. A substantial number of machine learning approaches have been tested using ADNI data and illustrate the potential power of neural networks in selecting highly discriminative features in single modalities, and in detecting optimum intermodal relationships using multiple modalities. The greatest gains in accuracy have been made in the more difficult classification (MCI vs. CN, AD vs. MCI) and prediction (CN to MCI, MCI to AD) challenges. Systematic studies of enrichment biomarkers, outcome measures, and combinations of the two have provided critical guidance to inform clinical trial design. Enrichment strategies aim to reduce the heterogeneity of trial cohorts and select populations both at risk of imminent decline and likely to respond to the trial treatment. Some studies refined cognitive measures as outcome measures, whereas others developed surrogate outcome measures using MRI approaches. Combinations of subject selection using multiple inclusion criteria and MRI longitudinal measures as surrogate outcomes had the greatest power to detect treatment effect and the most feasible sample sizes for early intervention trials in CN subjects.
9. Summary
Studies using ADNI data have produced an unprecedented body of research in 2014 and 2015. What then are the fundamental conclusions from this work? If you have read this entire review, hearty congratulations on your perseverance; if not we offer the following summary. ADNI studies have been supported by the continued development and standardization of methodologies, sample collection and distribution, and curating and sharing of data by the various ADNI Cores. The longitudinal data extending now over a decade and the breadth of modalities represent a unique resource to the scientific community that has been used in a myriad of ways to expand and deepen our knowledge of AD progression and to inform AD clinical trial design.
Studies of subjects selected for Aβ positivity have largely confirmed the order in which biomarkers become abnormal during disease progression and have suggested that APOE modulates disease progression through Aβ clearance mechanisms. However, there appears to be variability in this relatively well-defined process. For example, Aβ+ subjects with predominantly cortical atrophy had impaired executive function, whereas those with predominantly hippocampal atrophy had impaired memory function. Cluster analysis studies of CN and MCI subjects also consistently identified an executive function-impaired subtype, in addition to the more typical memory–impaired amnestic subtype, and differences in underlying atrophy patterns may result in to diverse neuropsychiatric symptoms observed in AD. These subtypes may correspond to differing distributions of neurofibrillary tangles. Multiple lines of evidence suggest that deposition of abnormal Aβ alone does not result in memory impairments typical of AD. Instead, Aβ deposition may be a prequel for the development of neurofibrillary tangles which may be more strongly associated with cognition. The precise role of tau fibrillar Aβ is unknown, and the introduction of tau PET imaging in ADNI-3 is expected to shed light on the issue. Regional WM abnormalities, increased by vascular risk factors acting in conjunction with Aβ, may additionally augment the effects of tau abnormalities, accelerating the AD disease process.
Two diverse approaches attempted to integrate knowledge from disparate facets of AD research. The concept of AD as a disconnection syndrome was enabled by the inclusion of diffusion imaging and TF-fMRI in the ADNI-2 protocol to track perturbations to structural, functional, and metabolic, and cortical atrophy networks during AD progression. Network changes were observed in presymptom atic subjects and support the earliest pathological changes occurring in the posterior cingulate cortex, consistent with MRI studies of CN subjects in which initial atrophy was observed in areas outside the hippocampus, such as the precuneus. Models for the spread of pathology based on a prionlike misfolding of Aβ along WM tracts predicted patterns of cortical Aβ deposition, glucose hypometabolism, and atrophy. Likewise, a Systems Biology approach integrating genomics, transcriptomics, proteomics, and metabolomics has begun to capture the biological complexity of AD engendered in part by its polygenic nature. Biologically informed strategies have allowed insights into how genetic underpinnings of AD are associated with biochemical mechanisms. The search for the “missing heritability” of AD has identified novel risk alleles and highlighted the involvement of processes such as inflammation, synaptic function, cell migration, cholesterol metabolism, and calcium signaling.
CN and MCI groups have proven to be highly heterogeneous; a sizeable portion of MCI subjects was virtually indistinguishable from CN subjects in all regards, and conversely, many CN subjects harbored pathology predictive of future clinical decline and were more aligned with the MCI group. This likely reflects the fact that MCI, even amnestic MCI, can be associated with a variety of pathologies, one of which is AD. It also implies that pathological events have already occurred in a portion of presymptomatic subjects. A group of MCI subjects with neurodegeneration in the absence of abnormal Aβ deposition appeared to be on a different pathway to dementia. The development of biomarkers of common comorbidities observed at autopsy such as Lewy bodies (α-SYN) and hippocampal sclerosis (TDP-43) will help future researchers to untangle the complex and seemingly intertwined trajectories to dementia.
Technological advances in machine learning such as the use of neural networks for feature selection have substantially improved the ability to discriminate between CN and MCI subjects, and MCI and AD subjects, and the ability to predict cognitive deterioration in CN and MCI subjects. Multimodal approaches to classification and prediction of future decline remain powerful, especially with the addition of new network connectivity measures or SNP data. However, the application of improved machine learning approaches to single modalities such as structural MRI has achieved accuracies rivaling state-of-the-art multimodal methods, and a cost-effective, widely available, and noninvasive approaches such as this may ultimately prove preferable for diagnosis.
Improvement of clinical trials of AD therapies, the overarching goal of the ADNI study, has been guided by a deeper understanding of AD progression and subject group heterogeneity. As AD therapies began to target presymptomatic patients, the choice of inclusion criteria that select subjects at risk of imminent decline, and outcome measures that are sufficiently sensitive to detect treatment effect within a feasible time frame is critical. Systematic studies have examined combinations of subject selection criteria such as APOE4 status, Aβ positivity, and hippocampal volume, together with cognitive and imaging outcome measures. Subject selection on the basis of a single pathology improves trial power; selection on the basis of multiple pathologies results in further gains in power. The sensitivity of cognitive end points to detect a subtle treatment effect in early phases of the disease has improved. However, surrogate outcome biomarkers such as longitudinal imaging have been predicted to significantly reduce sample sizes, trial duration, and trial cost and thus improve the feasibility of trials in presymptomatic subjects.
The remarkable advances described in ADNI papers over the course of 2 years have fundamentally altered our view of AD progression and offered significant guidance for clinical trial design. As the pace of discovery accelerates with the accumulation and sharing of an increasingly diverse longitudinal data set, ADNI will undoubtedly make even greater contributions to the field in the coming years.
RESEARCH IN CONTEXT.
Systematic review: The authors identified publications using data from the Alzheimer’s Disease Neuroimaging Initiative from their submission to the Data and Publications Committee, by traditional sources (PubMed and Google Scholar) and by personal communication with authors.
Interpretation: Substantial progress was made in 2014 and 2015 in improving biomarkers for clinical trials of Alzheimer’s disease (AD) therapies and the understanding of AD progression. Subtle structural and functional changes occur in presymptomatic subjects. Subjects positive for abnormal β-amyloid deposition progress according to the amyloid cascade hypothesis; other groups of mixed pathology may have different trajectories. Models of prion-like spreading β-amyloid pathology along white-matter tracts predict aspects of disease progression and emphasize the importance of structural, functional, and metabolic connectivity in AD. A Systems Biology approach identified novel risk gene loci. Diagnostic and prognostic accuracies improved using deep learning strategies.
Future directions: This knowledge will inform and improve clinical trial design.
Acknowledgments
This work was supported by NIH grant 5U01AG024904–10 funded by the National Institute on Aging to Dr. Michael Weiner. Dr. Beckett receives research funding from NIH/NCI (P30CA093373, RO1CA159447, R01CA115483, R01CA199668, R01CA199725), NIH/NIA (P30AG010129, P30AG043097, U01AG024904, R01AG047827), the California Department of Justice (14–6100), and the National Institute of Justice (2014-R2-CX-0012). Dr. Cairns receives research funding from the NIH/NIA (P50 AG005681 and P01 AG003991). Dr. Jack receives research funding from the NIH (R01-AG011378, RO1-AG041851, U01-AG06786, U01-AG024904, R01-AG37551, R01-AG043392, R01-NS092625) and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation. Dr. Harvey receives research funding from NIH/NIA (P30AG010129, R01AG047827, R01AG048252), NIH/NINDS (U54NS079202), NIH/NICHD (U54HD079125), DOD (W81XWH-12-2-0012, W81XWH-13-1-0259, W81XWH-14-1-0462). Dr. Green receives research funding from NIH (U01-HG006500, U19-HD077671, R01-HG005092, R01-AG047866, U01-HG008685, U41-HG006834), the Broad Institute and the Department of Defense. Dr. Jack receives research funding from the NIH (R01-AG011378, RO1-AG041851, U01-AG06786, U01-AG024904, R01-AG043392, R01-NS092625, U19-AG010483, UF1-AG032438, U01-AG042791, R01-NS089757, R01-AG049704) and the Alexander Family Alzheimer’s Disease Research Professorship of the Mayo Foundation. Dr. Saykin receives research funding from the NIH/NIA (R01AG19771, P30AG10133, R44AG049540 and U01AG024904), NIH/LM (R01LM011360) and NIH/NCI (R01CA129769). Dr. Shaw receives research funding from NIH (R01 MH 098260; R01 AG 046171; 1RFAG 051550); MJFox Foundation for PD Research (BioFIND). Dr. Petersen receives research funding from NIH (P50 AG016574, U01 AG006786, R01 AG011378, R01 AG041581, U01 AG024904). Work performed at LONI and by Dr. Toga was supported by NIH 5U01AG024904, P41EB015922 (Toga) and 1U54EB020406 (Toga). Dr. Trojanowski receives funding from AG-010124 and NS-053488 while he may accrue revenue in the future on patents submitted by the University of Pennsylvania wherein he is a coinventor and he received revenue from the sale of Avid to Eli Lily as a coinventor on imaging-related patents submitted by the University of Pennsylvania. He receives research support from the NIH, GSK, Janssen, Biogen, and several nonprofits.
Michael W. Weiner has served on the scientific advisory boards for Lilly, Araclon and Institut Catala de Neurociencies Aplicades, Gulf War Veterans Illnesses Advisory Committee, VACO, Biogen Idec, and Pfizer; has served as a consultant for Astra Zeneca, Araclon, Medivation/Pfizer, Ipsen, TauRx Therapeutics LTD, Bayer Healthcare, Biogen Idec, Exonhit Therapeutics, SA, Servier, Synarc, Pfizer, and Janssen; has received funding for travel from NeuroVigil, Inc., CHRU-Hopital Roger Sale-ngro, Siemens, AstraZeneca, Geneva University Hospitals, Lilly, University of California, San Diego–ADNI, Paris University, Institut Catala de Neurociencies Aplicades, University of New Mexico School of Medicine, Ipsen, CTAD (Clinical Trials on Alzheimer’s Disease), Pfizer, AD PD meeting, Paul Sabatier University, Novartis, Tohoku University; has served on the editorial advisory boards for Alzheimer’s & Dementia and MRI; has received honoraria from NeuroVigil, Inc., Insitut Catala de Neurociencies Aplicades, PMDA/Japanese Ministry of Health, Labour, and Welfare, and Tohoku University; has received commercial research support from Merck and Avid; has received government research support from DOD and VA; has stock options in Synarc and Elan; and declares the following organizations as contributors to the Foundation for NIH and thus to the NIA funded Alzheimer’s Disease Neuroimaging Initiative: Abbott, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Anonymous Foundation, AstraZeneca, Bayer Healthcare, BioClinica, Inc. (ADNI 2), Bristol-Myers Squibb, Cure Alzheimer’s Fund, Eisai, Elan, Gene Network Sciences, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson & Johnson, Eli Lilly & Company, Medpace, Merck, Novartis, Pfizer Inc., Roche, Schering Plough, Synarc, and Wyeth. Dr. Aisen has served as a consultant to the following companies: NeuroPhage, Eisai, Bristol-Myers Squibb, Eli Lilly, Merck, Roche, Genentech, Abbott, Pfizer, Novartis, AstraZeneca, Janssen, Ichor, Lundbeck, Biogen, iPerian, Probiodrug, Anavex, Abbvie, Janssen, Cohbar, aTyr, Axovant, and CogRx. Dr. Green receives compensation for speaking or consulting from AIA, Helix, Illumina, Invitae, Prudential, and Roche. Dr. Jack has provided consulting services for Eli Lily and owns stock in Johnson and Johnson. Dr. Jagust consults for Genentech Inc., Banner Alzheimer Institute, Bioclinica, and Novartis. Dr. Petersen consults for Roche Inc., Merck Inc., Genetech Inc., and Biogen Inc. Dr. Saykin receives support for a collaborative research grant from Eli Lilly and is MPI of an NIH SBIR grant with Arkley Biotek. Avid Radiopharmaceuticals provides precursor for tau PET imaging research at Indiana University. Dr. Saykin also receives editorial office support as a editor-in-chief of Brain Imaging and Behavior, published by Springer. Dr. Shaw is a consultant for Eli Lilly; Novartis; Roche.
References
- [1].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Cedarbaum J, et al. 2014 Update of the Alzheimer’s Disease Neuro-imaging Initiative: A review of papers published since its inception. Alzheimers Dement 2015;11:e1–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Weiner MW, Veitch D, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. and the Alzheimer’s Disease Neuroimaging Initiative. The Alzheimer’s Disease Neuroimaging Initiative 3: continued innovation for clinical trial improvement. Alzheimers Dement 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Liu E, Luthman J, Cedarbaum JM, Schmidt ME, Cole PE, Hendrix J, et al. Perspective: The Alzheimer’s Disease Neuroimaging Initiative and the role and contributions of the Private Partner Scientific Board (PPSB). Alzheimers Dement 2015;11:840–9. [DOI] [PubMed] [Google Scholar]
- [4].Jones-Davis DM, Buckholtz N. The impact of the Alzheimer’s Disease Neuroimaging Initiative 2: What role do public-private partnerships have in pushing the boundaries of clinical and basic science research on Alzheimer’s disease? Alzheimers Dement 2015; 11:860–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Anderson DC, Kodukula K. Biomarkers in pharmacology and drug discovery. Biochem Pharmacol 2014;87:172–88. [DOI] [PubMed] [Google Scholar]
- [6].Weiner MW, Veitch DP. Introduction to special issue: Overview of Alzheimer’s Disease Neuroimaging Initiative. Alzheimers Dement 2015;11:730–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Villemagne VL, Kim SY, Rowe CC, Iwatsubo T. Imago Mundi, Imago AD, Imago ADNI. Alzheimers Res Ther 2014;6:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Hendrix JA, Finger B, Weiner MW, Frisoni GB, Iwatsubo T, Rowe CC, et al. The Worldwide Alzheimer’s Disease Neuroimaging Initiative: An update. Alzheimers Dement 2015;11:850–9. [DOI] [PubMed] [Google Scholar]
- [9].Hartley D, Blumenthal T, Carrillo M, DiPaolo G, Esralew L, Gardiner K, et al. Down syndrome and Alzheimer’s disease: Common pathways, common goals. Alzheimers Dement 2015;11:700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Thies WH. Alzheimer’s Disease Neuroimaging Initiative: A decade of progress in Alzheimer’s disease. Alzheimers Dement 2015; 11:727–9. [DOI] [PubMed] [Google Scholar]
- [11].Toga AW, Crawford KL. The Alzheimer’s Disease Neuroimaging Initiative informatics core: A decade in review. Alzheimers Dement 2015;11:832–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement 2012; 8:S1–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, et al. The Alzheimer’s Disease Neuroimaging Initiative: a review of papers published since its inception. Alzheimers Dement 2013; 9:e111–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Aisen PS, Petersen RC, Donohue M, Weiner MW. Alzheimer’s Disease Neuroimaging Initiative 2 Clinical Core: Progress and plans. Alzheimers Dement 2015;11:734–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rattanabannakit C, Risacher SL, Gao S, Lane KA, Brown SA, McDonald BC, et al. The Cognitive Change Index as a Measure of Self and Informant Perception of Cognitive Decline: Relation to Neuropsychological Tests. J Alzheimers Dis 2016;51:1145–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Risacher SL, Kim S, Nho K, Foroud T, Shen L, Petersen RC, et al. APOE effect on Alzheimer’s disease biomarkers in older adults with significant memory concern. Alzheimers Dement 2015; 11:1417–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saykin AJ, Wishart HA, Rabin LA, Santulli RB, Flashman LA, West JD, et al. Older adults with cognitive complaints show brain atrophy similar to that of amnestic MCI. Neurology 2006;67:834–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rueda AD, Lau KM, Saito N, Harvey D, Risacher SL, Aisen PS, et al. Self-rated and informant-rated everyday function in comparison to objective markers of Alzheimer’s disease. Alzheimers Dement 2015;11:1080–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gross AL, Sherva R, Mukherjee S, Newhouse S, Kauwe JS, Munsie LM, et al. Calibrating longitudinal cognition in Alzheimer’s disease across diverse test batteries and datasets. Neuroepidemiology 2014;43:194–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Trzepacz PT, Hochstetler H, Wang S, Walker B, Saykin AJ. Alzheimer’s Disease Neuroimaging I. Relationship between the Montreal Cognitive Assessment and Mini-mental State Examination for assessment of mild cognitive impairment in older adults. BMC Geriatr 2015;15:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Balsis S, Benge JF, Lowe DA, Geraci L, Doody RS. How Do Scores on the ADAS-Cog, MMSE, and CDR-SOB Correspond? Clin Neuropsychol 2015;29:1002–9. [DOI] [PubMed] [Google Scholar]
- [22].Grochowalski JH, Liu Y, Siedlecki KL. Examining the reliability of ADAS-Cog change scores. Neuropsychol Dev Cogn B Aging Neuropsychol Cogn 2015;28:1–17. [DOI] [PubMed] [Google Scholar]
- [23].Alexander GE, Satalich TA, Shankle WR, Batchelder WH. A cognitive psychometric model for the psychodiagnostic assessment of memory-related deficits. Psychol Assess 2016;28:279–93. [DOI] [PubMed] [Google Scholar]
- [24].Brainerd CJ, Reyna VF, Gomes CF, Kenney AE, Gross CJ, Taub ES, et al. Dual-retrieval models and neurocognitive impairment. J Exp Psychol Learn Mem Cogn 2014;40:41–65. [DOI] [PubMed] [Google Scholar]
- [25].Gifford KA, Phillips JS, Samuels LR, Lane EM, Bell SP, Liu D, et al. Associations between Verbal Learning Slope and Neuroimaging Markers across the Cognitive Aging Spectrum. J Int Neuropsychol Soc 2015;21:455–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kandel BM, Avants BB, Gee JC, Arnold SE, Wolk DA. Neuropsychological Testing Predicts Cerebrospinal Fluid Amyloid-beta in Mild Cognitive Impairment. J Alzheimers Dis 2015;46:901–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Grothe MJ, Heinsen H, Amaro E Jr, Grinberg LT, Teipel SJ Alzheimer’s Disease Neuroimaging Initiative. Cognitive Correlates of Basal Forebrain Atrophy and Associated Cortical Hypometabolism in Mild Cognitive Impairment. Cereb Cortex 2016;26:2411–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Sorensen L, Igel C, Liv Hansen N, Osler M, Lauritzen M, Rostrup E, et al. Early detection of Alzheimer’s disease using MRI hippocampal texture. Hum Brain Mapp 2016;37:1148–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Seo EH, Choo IH Alzheimer’s Disease Neuroimaging Initiative. Amyloid-independent functional neural correlates of episodic memory in amnestic mild cognitive impairment. Eur J Nucl Med Mol Imaging 2016;43:1088–95. [DOI] [PubMed] [Google Scholar]
- [30].Roy K, Pepin LC, Philiossaint M, Lorius N, Becker JA, Locascio JJ, et al. Regional fluorodeoxyglucose metabolism and instrumental activities of daily living across the Alzheimer’s disease spectrum. J Alzheimers Dis 2014;42:291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Marshall GA, Lorius N, Locascio JJ, Hyman BT, Rentz DM, Johnson KA, et al. Regional cortical thinning and cerebrospinal biomarkers predict worsening daily functioning across the Alzheimer’s disease spectrum. J Alzheimers Dis 2014;41:719–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].David ND, Lin F, Porsteinsson AP. Trajectories of Neuropsychiatric Symptoms and Cognitive Decline in Mild Cognitive Impairment. Am J Geriatr Psychiatry 2016;24:70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Bensamoun D, Guignard R, Furst AJ, Derreumaux A, Manera V, Darcourt J, et al. Associations between Neuropsychiatric Symptoms and Cerebral Amyloid Deposition in Cognitively Impaired Elderly People. J Alzheimers Dis 2015;49:387–98. [DOI] [PubMed] [Google Scholar]
- [34].Mah L, Binns MA, Steffens DC. Anxiety symptoms in amnestic mild cognitive impairment are associated with medial temporal atrophy and predict conversion to Alzheimer disease. Am J Geriatr Psychiatry 2015;23:466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Chung JK, Plitman E, Nakajima S, Chakravarty MM, Caravaggio F, Takeuchi H, et al. Depressive Symptoms and Small Hippocampal Volume Accelerate the Progression to Dementia from Mild Cognitive Impairment. J Alzheimers Dis 2015;49:743–54. [DOI] [PubMed] [Google Scholar]
- [36].Sacuiu S, Insel PS, Mueller S, Tosun D, Mattsson N, Jack CR Jr, et al. Chronic Depressive Symptomatology in Mild Cognitive Impairment Is Associated with Frontal Atrophy Rate which Hastens Conversion to Alzheimer Dementia. Am J Geriatr Psychiatry 2016;24:126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chung JK, Plitman E, Nakajima S, Chakravarty MM, Caravaggio F, Gerretsen P, et al. Cortical Amyloid beta Deposition and Current Depressive Symptoms in Alzheimer Disease and Mild Cognitive Impairment. J Geriatr Psychiatry Neurol 2016;29:149–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Brendel M, Pogarell O, Xiong G, Delker A, Bartenstein P, Rominger A. Depressive symptoms accelerate cognitive decline in amyloid-positive MCI patients. Eur J Nucl Med Mol Imaging 2015;42:716–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lebedeva A, Westman E, Lebedev AV, Li X, Winblad B, Simmons A, et al. Structural brain changes associated with depressive symptoms in the elderly with Alzheimer’s disease. J Neurol Neurosurg Psychiatry 2014;85:930–5. [DOI] [PubMed] [Google Scholar]
- [40].Lee JY, Park S, Mackin S, Ewers M, Chui H, Jagust W, et al. Differences in prefrontal, limbic, and white matter lesion volumes according to cognitive status in elderly patients with first-onset subsyndromal depression. PLoS One 2014;9:e87747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Koppel J, Greenwald BS. Optimal treatment of Alzheimer’s disease psychosis: challenges and solutions. Neuropsychiatr Dis Treat 2014;10:2253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Koppel J, Sunday S, Goldberg TE, Davies P, Christen E, Greenwald BS. Psychosis in Alzheimer’s disease is associated with frontal metabolic impairment and accelerated decline in working memory: findings from the Alzheimer’s Disease Neuroimaging Initiative. Am J Geriatr Psychiatry 2014;22:698–707. [DOI] [PubMed] [Google Scholar]
- [43].Rafii MS, Taylor CS, Kim HT, Desikan RS, Fleisher AS, Katibian D, et al. Neuropsychiatric symptoms and regional neocortical atrophy in mild cognitive impairment and Alzheimer’s disease. Am J Alzheimers Dis Other Demen 2014;29:159–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Ting WK, Fischer CE, Millikin CP, Ismail Z, Chow TW, Schweizer TA. Grey matter atrophy in mild cognitive impairment/early Alzheimer disease associated with delusions: a voxel-based morphometry study. Curr Alzheimer Res 2015;12:165–72. [DOI] [PubMed] [Google Scholar]
- [45].Delrieu J, Desmidt T, Camus V, Sourdet S, Boutoleau-Bretonniere C, Mullin E, et al. Apathy as a feature of prodromal Alzheimer’s disease: an FDG-PET ADNI study. Int J Geriatr Psychiatry 2015;30:470–7. [DOI] [PubMed] [Google Scholar]
- [46].Donovan NJ, Wadsworth LP, Lorius N, Locascio JJ, Rentz DM, Johnson KA, et al. Regional cortical thinning predicts worsening apathy and hallucinations across the Alzheimer disease spectrum. Am J Geriatr Psychiatry 2014;22:1168–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Osorio RS, Gumb T, Pirraglia E, Varga AW, Lu SE, Lim J, et al. Sleep-disordered breathing advances cognitive decline in the elderly. Neurology 2015;84:1964–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Daiello LA, Gongvatana A, Dunsiger S, Cohen RA, Ott BR. Association of fish oil supplement use with preservation of brain volume and cognitive function. Alzheimers Dement 2015;11:226–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Nudelman KN, Risacher SL, West JD, McDonald BC, Gao S, Saykin AJ. Association of cancer history with Alzheimer’s disease onset and structural brain changes. Front Physiol 2014;5:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Shpanskaya KS, Choudhury KR, Hostage C Jr, Murphy KR, Petrella JR, Doraiswamy PM. Educational attainment and hippocampal atrophy in the Alzheimer’s disease neuroimaging initiative cohort. J Neuroradiol 2014;41:350–7. [DOI] [PubMed] [Google Scholar]
- [51].An H, Son SJ, Cho S, Cho EY, Choi B, Kim SY. Large intracranial volume accelerates conversion to dementia in males and APOE4 non-carriers with mild cognitive impairment. Int Psychogeriatr 2016;28:769–78. [DOI] [PubMed] [Google Scholar]
- [52].Epstein NU, Guo R, Farlow MR, Singh JP, Fisher M. Medication for Alzheimer’s disease and associated fall hazard: a retrospective cohort study from the Alzheimer’s Disease Neuroimaging Initiative. Drugs Aging 2014;31:125–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Risacher SL, McDonald BC, Tallman EF, West JD, Farlow MR, Unverzagt FW, et al. Association Between Anticholinergic Medication Use and Cognition, Brain Metabolism, and Brain Atrophy in Cognitively Normal Older Adults. JAMA Neurol 2016; 76:721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Jack CR Jr, Barnes J, Bernstein MA, Borowski BJ, Brewer J, Clegg S, et al. Magnetic resonance imaging in Alzheimer’s Disease Neuroimaging Initiative 2. Alzheimers Dement 2015;11:740–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Frisoni GB, Jack CR. HarP: The EADC-ADNI Harmonized Protocol for manual hippocampal segmentation. A standard of reference from a global working group. Alzheimers Dement 2015;11:107–10. [DOI] [PubMed] [Google Scholar]
- [56].Boccardi M, Bocchetta M, Apostolova LG, Preboske G, Robitaille N, Pasqualetti P, et al. Establishing magnetic resonance images orientation for the EADC-ADNI manual hippocampal segmentation protocol. J Neuroimaging 2014;24:509–14. [DOI] [PubMed] [Google Scholar]
- [57].Boccardi M, Bocchetta M, Ganzola R, Robitaille N, Redolfi A, Duchesne S, et al. Operationalizing protocol differences for EADC-ADNI manual hippocampal segmentation. Alzheimers Dement 2015;11:184–94. [DOI] [PubMed] [Google Scholar]
- [58].Boccardi M, Bocchetta M, Apostolova LG, Barnes J, Bartzokis G, Corbetta G, et al. Delphi definition of the EADC-ADNI Harmonized Protocol for hippocampal segmentation on magnetic resonance. Alzheimers Dement 2015;11:126–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Bocchetta M, Boccardi M, Ganzola R, Apostolova LG, Preboske G, Wolf D, et al. Harmonized benchmark labels of the hippocampus on magnetic resonance: The EADC-ADNI project. Alzheimers Dement 2015;11:151–160.e5. [DOI] [PubMed] [Google Scholar]
- [60].Duchesne S, Valdivia F, Robitaille N, Mouiha A, Valdivia FA, Bocchetta M, et al. Manual segmentation qualification platform for the EADC-ADNI harmonized protocol for hippocampal segmentation project. Alzheimers Dement 2015;11:161–74. [DOI] [PubMed] [Google Scholar]
- [61].Apostolova LG, Zarow C, Biado K, Hurtz S, Boccardi M, Somme J, et al. Relationship between hippocampal atrophy and neuropathology markers: a 7T MRI validation study of the EADC-ADNI Harmonized Hippocampal Segmentation Protocol. Alzheimers Dement 2015; 11:139–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Boccardi M, Bocchetta M, Morency FC, Collins DL, Nishikawa M, Ganzola R, et al. Training labels for hippocampal segmentation based on the EADC-ADNI harmonized hippocampal protocol. Alzheimers Dement 2015;11:175–83. [DOI] [PubMed] [Google Scholar]
- [63].Whelan CD, Hibar DP, van Velzen LS, Zannas AS, Carrillo-Roa T, McMahon K, et al. Heritability and reliability of automatically segmented human hippocampal formation subregions. Neuroimage 2016;128:125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mulder ER, de Jong RA, Knol DL, van Schijndel RA, Cover KS, Visser PJ, et al. Hippocampal volume change measurement: quantitative assessment of the reproducibility of expert manual outlining and the automated methods FreeSurfer and FIRST. Neuroimage 2014;92:169–81. [DOI] [PubMed] [Google Scholar]
- [65].Amoroso N, Errico R, Bruno S, Chincarini A, Garuccio E, Sensi F, et al. Hippocampal unified multi-atlas network (HUMAN): protocol and scale validation of a novel segmentation tool. Phys Med Biol 2015;60:8851. [DOI] [PubMed] [Google Scholar]
- [66].Sanroma G, Wu G, Gao Y, Shen D. Learning-Based Atlas Selection for Multiple-Atlas Segmentation. Conf Comput Vis Pattern Recognit Workshops 2014;2014:3111–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li XW, Li QL, Li SY, Li DY. Local manifold learning for multiatlas segmentation: application to hippocampal segmentation in healthy population and Alzheimer’s disease. CNS Neurosci Ther 2015; 21:826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Koch LM, Rajchl M, Tong T, Passerat-Palmbach J, Aljabar P, Rueckert D. Multi-atlas Segmentation as a Graph Labelling Problem: Application to Partially Annotated Atlas Data. Inf Process Med Imaging 2015;24:221–32. [DOI] [PubMed] [Google Scholar]
- [69].Pipitone J, Park MT, Winterburn J, Lett TA, Lerch JP, Pruessner JC, et al. Multi-atlas segmentation of the whole hippocampus and subfields using multiple automatically generated templates. Neuroimage 2014;101:494–512. [DOI] [PubMed] [Google Scholar]
- [70].Tan M, Qiu A. Spectral Laplace-Beltrami wavelets with applications in medical images. IEEE Trans Med Imaging 2015;34:1005–17. [DOI] [PubMed] [Google Scholar]
- [71].Tangaro S, Amoroso N, Boccardi M, Bruno S, Chincarini A, Ferraro G, et al. Automated voxel-by-voxel tissue classification for hippocampal segmentation: methods and validation. Phys Med 2014;30:878–87. [DOI] [PubMed] [Google Scholar]
- [72].Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, et al. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: Application to adaptive segmentation of in vivo MRI. Neuroimage 2015;115:117–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Cong S, Rizkalla M, Du EY, West J, Risacher S, Saykin A, et al. Building a Surface Atlas of Hippocampal Subfields from MRI Scans using FreeSurfer, FIRST and SPHARM. Conf Proc (Midwest Symp Circuits Syst) 2014;2014:813–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ahdidan J, Raji CA, DeYoe EA, Mathis J, Noe KO, Rimestad J, et al. Quantitative Neuroimaging Software for Clinical Assessment of Hippocampal Volumes on MR Imaging. J Alzheimers Dis 2015; 49:723–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Inglese P, Amoroso N, Boccardi M, Bocchetta M, Bruno S, Chincarini A, et al. Multiple RF classifier for the hippocampus segmentation: Method and validation on EADC-ADNI Harmonized Hippocampal Protocol. Phys Med 2015;31:1085–91. [DOI] [PubMed] [Google Scholar]
- [76].Suppa P, Hampel H, Spies L, Fiebach JB, Dubois B, Buchert R, et al. Fully Automated Atlas-Based Hippocampus Volumetry for Clinical Routine: Validation in Subjects with Mild Cognitive Impairment from the ADNI Cohort. J Alzheimers Dis 2015;46:199–209. [DOI] [PubMed] [Google Scholar]
- [77].Chincarini A, Sensi F, Rei L, Gemme G, Squarcia S, Longo R, et al. Integrating longitudinal information in hippocampal volume measurements for the early detection of Alzheimer’s disease. Neuroimage 2016;125:834–47. [DOI] [PubMed] [Google Scholar]
- [78].Sanroma G, Wu G, Gao Y, Thung KH, Guo Y, Shen D. A transversal approach for patch-based label fusion via matrix completion. Med Image Anal 2015;24:135–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Giraud R, Ta VT, Papadakis N, Manjon JV, Collins DL, Coupe P. An Optimized PatchMatch for multi-scale and multi-feature label fusion. Neuroimage 2016;124:770–82. [DOI] [PubMed] [Google Scholar]
- [80].Wu G, Kim M, Sanroma G, Wang Q, Munsell BC, Shen D. Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. Neuroimage 2015;106:34–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Guo H, Song X, Vandorpe R, Zhang Y, Chen W, Zhang N, et al. Evaluation of common structural brain changes in aging and Alzheimer disease with the use of an MRI-based brain atrophy and lesion index: a comparison between T1WI and T2WI at 1.5T and 3T. AJNR Am J Neuroradiol 2014;35:504–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chow N, Hwang KS, Hurtz S, Green AE, Somme JH, Thompson PM, et al. Comparing 3Tand 1.5T MRI for mapping hippocampal atrophy in the Alzheimer’s Disease Neuroimaging Initiative. AJNR Am J Neuroradiol 2015;36:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Marchewka A, Kherif F, Krueger G, Grabowska A, Frackowiak R, Draganski B. Influence of magnetic field strength and image registration strategy on voxel-based morphometry in a study of Alzheimer’s disease. Hum Brain Mapp 2014;35:1865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Brunton S, Gunasinghe C, Jones N, Kempton MJ, Westman E, Simmons A. A voxel-based morphometry comparison of the 3.0T ADNI-1 and ADNI-2 volumetric MRI protocols. Int J Geriatr Psychiatry 2015;30:531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Healy A, Fredriksson J, Goodenough D, Mallozzi R, Olafsdottir H, Kristbjornsson A. SU-D-303–04: A Survey of MRI Distortion Measurements. Med Phys 2015;42:3215. [Google Scholar]
- [86].Voevodskaya O, Simmons A, Nordenskjold R, Kullberg J, Ahlstrom H, Lind L, et al. The effects of intracranial volume adjustment approaches on multiple regional MRI volumes in healthy aging and Alzheimer’s disease. Front Aging Neurosci 2014;6:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Wang Y, Nie J, Yap PT, Li G, Shi F, Geng X, et al. Knowledge-guided robust MRI brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PLoS One 2014; 9:e77810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wu G, Kim M, Wang Q, Munsell BC, Shen D. Scalable High-Performance Image Registration Framework by Unsupervised Deep Feature Representations Learning. IEEE Trans Biomed Eng 2016;63:1505–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Liang Z, He X, Ceritoglu C, Tang X, Li Y, Kutten KS, et al. Evaluation of Cross-Protocol Stability of a Fully Automated Brain Multi-Atlas Parcellation Tool. PLoS One 2015;10:e0133533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Osadebey M, Bouguila N, Arnold D. Optimal selection of regularization parameter in total variation method for reducing noise in magnetic resonance images of the brain. Biomed Eng Lett 2014;4:80–92. [Google Scholar]
- [91].Simpson IJ, Cardoso MJ, Modat M, Cash DM, Woolrich MW, Andersson JL, et al. Probabilistic non-linear registration with spatially adaptive regularisation. Med Image Anal 2015;26:203–16. [DOI] [PubMed] [Google Scholar]
- [92].Maclaren J, Han Z, Vos SB, Fischbein N, Bammer R. Reliability of brain volume measurements: a test-retest dataset. Sci Data 2014; 1:140037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Shamonin DP, Bron EE, Lelieveldt BP, Smits M, Klein S, Staring M. Fast parallel image registration on CPU and GPU for diagnostic classification of Alzheimer’s disease. Front Neuroinform 2013;7:50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Roche A, Forbes F. Partial volume estimation in brain MRI revisited. Med Image Comput Comput Assist Interv 2014;17:771–8. [DOI] [PubMed] [Google Scholar]
- [95].Nigro S, Cerasa A, Zito G, Perrotta P, Chiaravalloti F, Donzuso G, et al. Fully automated segmentation of the pons and midbrain using human T1 MR brain images. PLoS One 2014;9:e85618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kazemifar S, Drozd JJ, Rajakumar N, Borrie MJ, Bartha R. Automated algorithm to measure changes in medial temporal lobe volume in Alzheimer disease. J Neurosci Methods 2014;227:35–46. [DOI] [PubMed] [Google Scholar]
- [97].Guo H, Song X, Schmidt MH, Vandorpe R, Yang Z, LeBlanc E, et al. Evaluation of whole brain health in aging and Alzheimer’s disease: a standard procedure for scoring an MRI-based brain atrophy and lesion index. J Alzheimers Dis 2014;42:691–703. [DOI] [PubMed] [Google Scholar]
- [98].Vemuri P, Senjem ML, Gunter JL, Lundt ES, Tosakulwong N, Weigand SD, et al. Accelerated vs. unaccelerated serial MRI based TBM-SyN measurements for clinical trials in Alzheimer’s disease. Neuroimage 2015;113:61–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Leung KK, Malone IM, Ourselin S, Gunter JL, Bernstein MA, Thompson PM, et al. Effects of changing from non-accelerated to accelerated MRI for follow-up in brain atrophy measurement. Neuroimage 2015;107:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ching CR, Hua X, Hibar DP, Ward CP, Gunter JL, Bernstein MA, et al. Does MRI scan acceleration affect power to track brain change? Neurobiol Aging 2015;36:S167–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Fleishman GM, Gutman BA, Fletcher PT, Thompson PM. Simultaneous Longitudinal Registration with Group-Wise Similarity Prior. Inf Process Med Imaging 2015;24:746–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Fleishman GM, Gutman BA, Fletcher PT, Thompson P. A Transformation Similarity Constraint for Groupwise Nonlinear Registration in Longitudinal Neuro Imaging Studies. Proceedings of SPIE–the International Society for Optical Engineering. 2015;9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Fleishman GM, Fletcher PT, Gutman BA, Prasad G, Wu Y, Thompson PM. Geodesic Refinement Using James-Stein Estimators. Mathematical Foundations of Computational Anatomy. 60. [Google Scholar]
- [104].Guillaume B, Hua X, Thompson PM, Waldorp L, Nichols TE. Fast and accurate modelling of longitudinal and repeated measures neuro-imaging data. Neuroimage 2014;94:287–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Goto M, Abe O, Aoki S, Hayashi N, Ohtsu H, Takao H, et al. Longitudinal gray-matter volume change in the default-mode network: utility of volume standardized with global gray-matter volume for Alzheimer’s disease: a preliminary study. Radiol Phys Technol 2015;8:64–72. [DOI] [PubMed] [Google Scholar]
- [106].Nakamura K, Brown RA, Narayanan S, Collins DL, Arnold DL. Diurnal fluctuations in brain volume: Statistical analyses of MRI from large populations. Neuroimage 2015;118:126–32. [DOI] [PubMed] [Google Scholar]
- [107].Prados F, Cardoso MJ, Leung KK, Cash DM, Modat M, Fox NC, et al. Measuring brain atrophy with a generalized formulation of the boundary shift integral. Neurobiol Aging 2015;36:S81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Wang G, Zhang X, Su Q, Shi J, Caselli RJ, Wang Y. A novel cortical thickness estimation method based on volumetric Laplace-Beltrami operator and heat kernel. Med Image Anal 2015;22:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Diaz-de-Grenu LZ, Acosta-Cabronero J, Chong YF, Pereira JM, Sajjadi SA, Williams GB, et al. A brief history of voxel-based grey matter analysis in Alzheimer’s disease. J Alzheimers Dis 2014; 38:647–59. [DOI] [PubMed] [Google Scholar]
- [110].Kim WH, Singh V, Chung MK, Hinrichs C, Pachauri D, Okonkwo OC, et al. Multi-resolutional shape features via non-Euclidean wavelets: applications to statistical analysis of cortical thickness. Neuroimage 2014;93 Pt 1:107–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Shi J, Stonnington CM, Thompson PM, Chen K, Gutman B, Reschke C, et al. Studying ventricular abnormalities in mild cognitive impairment with hyperbolic Ricci flow and tensor-based morphometry. Neuroimage 2015;104:1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Lorenzi M, Ayache N, Pennec X. Regional flux analysis for discovering and quantifying anatomical changes: An application to the brain morphometry in Alzheimer’s disease. Neuroimage 2015; 115:224–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wachinger C, Golland P, Kremen W, Fischl B, Reuter M. BrainPrint: A discriminative characterization of brain morphology. Neuroimage 2015;109:232–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Ziegler G, Penny WD, Ridgway GR, Ourselin S, Friston KJ Alzheimer’s Disease Neuroimaging Initiative. Estimating anatomical trajectories with Bayesian mixed-effects modeling. Neuroimage 2015;121:51–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Ribbens A, Hermans J, Maes F, Vandermeulen D, Suetens P. Unsupervised segmentation, clustering, and groupwise registration of heterogeneous populations of brain MR images. IEEE Trans Med Imaging 2014;33:201–24. [DOI] [PubMed] [Google Scholar]
- [116].Singh N, Fletcher PT, Preston JS, King RD, Marron JS, Weiner MW, et al. Quantifying anatomical shape variations in neurological disorders. Med Image Anal 2014;18:616–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Gutman B, Fletcher T, Cardoso MJ, Fleishman G, Lorenzi M, Thompson P, et al. A Riemannian Framework for Intrinsic Comparison of Closed Genus-Zero Shapes. Inf Process Med Imaging 2015; 9123:205–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Zhang N, Song X, Zhang Y, Chen W, D’Arcy RC, Darvesh S, et al. An MRI brain atrophy and lesion index to assess the progression of structural changes in Alzheimer’s disease, mild cognitive impairment, and normal aging: a follow-up study. J Alzheimers Dis 2011;26:359–67. [DOI] [PubMed] [Google Scholar]
- [119].Ross DE, Ochs AL, DeSmit ME, Seabaugh JM, Havranek MD. Man Versus Machine Part 2: Comparison of Radiologists’ Interpretations and NeuroQuant Measures of Brain Asymmetry and Progressive Atrophy in Patients With Traumatic Brain Injury. J Neuropsychiatry Clin Neurosci 2015;27:147–52. [DOI] [PubMed] [Google Scholar]
- [120].Pardoe HR, Mandelstam SA, Hiess RK, Kuzniecky RI, Jackson GD. Quantitative assessment of corpus callosum morphology in periventricular nodular heterotopia. Epilepsy Res 2015;109:40–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Jagust WJ, Landau SM, Koeppe RA, Reiman EM, Chen K, Mathis CA, et al. The Alzheimer’s Disease Neuroimaging Initiative 2 PET Core: 2015. Alzheimers Dement 2015;11:757–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Smailagic N, Vacante M, Hyde C, Martin S, Ukoumunne O, Sachpekidis C. (18)F-FDG PET for the early diagnosis of Alzheimer’s disease dementia and other dementias in people with mild cognitive impairment (MCI). Cochrane Database Syst Rev 2015; 1:Cd010632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sperling RA, Aisen PS, Beckett LA, Bennett DA, Craft S, Fagan AM, et al. Toward defining the preclinical stages of Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7:280–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Landau SM, Fero A, Baker SL, Koeppe R, Mintun M, Chen K, et al. Measurement of longitudinal beta-amyloid change with 18F-florbetapir PET and standardized uptake value ratios. J Nucl Med 2015; 56:567–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Chen K, Roontiva A, Thiyyagura P, Lee W, Liu X, Ayutyanont N, et al. Improved power for characterizing longitudinal amyloid-beta PET changes and evaluating amyloid-modifying treatments with a cerebral white matter reference region. J Nucl Med 2015;56:560–6. [DOI] [PubMed] [Google Scholar]
- [126].Chiotis K, Carter SF, Farid K, Savitcheva I, Nordberg A. Amyloid PET in European and North American cohorts; and exploring age as a limit to clinical use of amyloid imaging. Eur J Nucl Med Mol Imaging 2015;42:1492–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Landau SM, Thomas BA, Thurfjell L, Schmidt M, Margolin R, Mintun M, et al. Amyloid PET imaging in Alzheimer’s disease: a comparison of three radiotracers. Eur J Nucl Med Mol Imaging 2014;41:1398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Nosheny RL, Insel PS, Truran D, Schuff N, Jack CR Jr, Aisen PS, et al. Variables associated with hippocampal atrophy rate in normal aging and mild cognitive impairment. Neurobiol Aging 2015; 36:273–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kang JH, Korecka M, Figurski MJ, Toledo JB, Blennow K, Zetterberg H, et al. The Alzheimer’s Disease Neuroimaging Initiative 2 Biomarker Core: A review of progress and plans. Alzheimers Dement 2015;11:772–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Mitchell TW, Nissanov J, Han LY, Mufson EJ, Schneider JA, Cochran EJ, et al. Novel method to quantify neuropil threads in brains from elders with or without cognitive impairment. J Histochem Cytochem 2000;48:1627–38. [DOI] [PubMed] [Google Scholar]
- [131].Schmidt ML, Schuck T, Sheridan S, Kung MP, Kung H, Zhuang ZP, et al. The fluorescent Congo red derivative, (trans, trans)-1-bromo-2,5-bis-(3-hydroxycarbonyl-4-hydroxy)styrylbenzene (BSB), labels diverse beta-pleated sheet structures in postmortem human neurodegenerative disease brains. Am J Pathol 2001;159:937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Zetterberg H, Smith DH, Blennow K. Biomarkers of mild traumatic brain injury in cerebrospinal fluid and blood. Nat Rev Neurol 2013; 9:201–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Kljajevic V, Grothe MJ, Ewers M, Teipel S. Distinct pattern of hypometabolism and atrophy in preclinical and predementia Alzheimer’s disease. Neurobiol Aging 2014;35:1973–81. [DOI] [PubMed] [Google Scholar]
- [134].Korecka M, Waligorska T, Figurski M, Toledo JB, Arnold SE, Grossman M, et al. Qualification of a surrogate matrix-based absolute quantification method for amyloid-beta(4)(2) in human cerebrospinal fluid using 2D UPLC-tandem mass spectrometry. J Alzheimers Dis 2014;41:441–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Kuhlmann J, Andreasson U, Pannee J, Bjerke M, Portelius E, Leinenbach A, et al. CSF Abeta - an excellent but complicated Alzheimer’s biomarker - a route to standardisation. Clin Chim Acta 2016. [DOI] [PubMed] [Google Scholar]
- [136].Bjerke M, Andreasson U, Kuhlmann J, Portelius E, Pannee J, Lewczuk P, et al. Assessing the commutability of reference material formats for the harmonization of amyloid beta measurements. Clin Chem Lab Med 2016;54:1177–91. [DOI] [PubMed] [Google Scholar]
- [137].Vidoni ED, Morris JK, Raider K, Burns JM. Reducing post-lumbar puncture headaches with small bore atraumatic needles. J Clin Neurosci 2014;21:536–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Tycko R. Amyloid polymorphism: structural basis and neurobiological relevance. Neuron 2015;86:632–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Mattsson N, Insel PS, Landau S, Jagust W, Donohue M, Shaw LM, et al. Diagnostic accuracy of CSF AB42 and florbetapir PET for Alzheimer’s disease. Ann Clin Transl Neurol 2014;1:534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Lautner R, Palmqvist S, Mattsson N, Andreasson U, Wallin A, Palsson E, et al. Apolipoprotein E genotype and the diagnostic accuracy of cerebrospinal fluid biomarkers for Alzheimer disease. JAMA Psychiatry 2014;71:1183–91. [DOI] [PubMed] [Google Scholar]
- [141].Mattsson N, Insel PS, Donohue M, Landau S, Jagust WJ, Shaw LM, et al. Independent information from cerebrospinal fluid amyloid-beta and florbetapir imaging in Alzheimer’s disease. Brain 2015; 138:772–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Perneczky R, Alexopoulos P Alzheimer’s Disease Neuroimaging Initiative. Cerebrospinal fluid BACE1 activity and markers of amyloid precursor protein metabolism and axonal degeneration in Alzheimer’s disease. Alzheimers Dement 2014;10:S425–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Savage MJ, Holder DJ, Wu G, Kaplow J, Siuciak JA, Potter WZ. Soluble BACE-1 Activity and sAbetaPPbeta Concentrations in Alzheimer’s Disease and Age-Matched Healthy Control Cerebrospinal Fluid from the Alzheimer’s Disease Neuroimaging Initiative-1 Baseline Cohort. J Alzheimers Dis 2015;46:431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Alcolea D, Martinez-Lage P, Sanchez-Juan P, Olazaran J, Antunez C, Izagirre A, et al. Amyloid precursor protein metabolism and inflammation markers in preclinical Alzheimer disease. Neurology 2015; 85:626–33. [DOI] [PubMed] [Google Scholar]
- [145].Rajagopalan P, Hua X, Toga AW, Jack CR Jr, Weiner MW, Thompson PM. Homocysteine effects on brain volumes mapped in 732 elderly individuals. Neuroreport 2011;22:391–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Madsen SK, Rajagopalan P, Joshi SH, Toga AW, Thompson PM. Higher homocysteine associated with thinner cortical gray matter in 803 participants from the Alzheimer’s Disease Neuroimaging Initiative. Neurobiol Aging 2015;36:S203–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Maioli S, Lodeiro M, Merino-Serrais P, Falahati F, Khan W, Puerta E, et al. Alterations in brain leptin signalling in spite of unchanged CSF leptin levels in Alzheimer’s disease. Aging Cell 2015;14:122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Johnston JM, Hu WT, Fardo DW, Greco SJ, Perry G, Montine TJ, et al. Low plasma leptin in cognitively impaired ADNI subjects: gender differences and diagnostic and therapeutic potential. Curr Alzheimer Res 2014;11:165–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cronk BB, Johnson DK, Burns JM. Body Mass Index and Cognitive Decline in Mild Cognitive Impairment. Alzheimer Dis Assoc Disord 2010;24:126–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Toledo JB, Da X, Weiner MW, Wolk DA, Xie SX, Arnold SE, et al. CSF Apo-E levels associate with cognitive decline and MRI changes. Acta Neuropathol 2014;127:621–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Teng E, Chow N, Hwang KS, Thompson PM, Gylys KH, Cole GM, et al. Low plasma ApoE levels are associated with smaller hippocampal size in the Alzheimer’s disease neuroimaging initiative cohort. Dement Geriatr Cogn Disord 2015;39:154–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Ayton S, Faux NG, Bush AI. Ferritin levels in the cerebrospinal fluid predict Alzheimer’s disease outcomes and are regulated by APOE. Nat Commun 2015;6:6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 2009;41:1088–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat Genet 2009; 41:1094–9. [DOI] [PubMed] [Google Scholar]
- [155].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet 2013; 45:1452–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Deming Y, Xia J, Cai Y, Lord J, Holmans P, Bertelsen S, et al. A potential endophenotype for Alzheimer’s disease: cerebrospinal fluid clusterin. Neurobiol Aging 2016;37:208.e1–e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Mattsson N, Insel P, Nosheny R, Trojanowski JQ, Shaw LM, Jack CR Jr, et al. Effects of cerebrospinal fluid proteins on brain atrophy rates in cognitively healthy older adults. Neurobiol Aging 2014;35:614–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Portelius E, Zetterberg H, Skillback T, Tornqvist U, Andreasson U, Trojanowski JQ, et al. Cerebrospinal fluid neurogranin: relation to cognition and neurodegeneration in Alzheimer’s disease. Brain 2015;138:3373–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Paterson RW, Bartlett JW, Blennow K, Fox NC, Alzheimer’s Disease Neuroimaging Initiative, Shaw LM, et al. Cerebrospinal fluid markers including trefoil factor 3 are associated with neurodegeneration in amyloid-positive individuals. Transl Psychiatry 2014; 4:e419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Zetterberg H, Skillback T, Mattsson N, Trojanowski JQ, Portelius E, Shaw LM, et al. Association of Cerebrospinal Fluid Neurofilament Light Concentration With Alzheimer Disease Progression. JAMA Neurol 2016;73:60–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Hohman TJ, Bell SP, Jefferson AL. The role of vascular endothelial growth factor in neurodegeneration and cognitive decline: exploring interactions with biomarkers of Alzheimer disease. JAMA Neurol 2015;72:520–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Leung YY, Toledo JB, Nefedov A, Polikar R, Raghavan N, Xie SX, et al. Identifying amyloid pathology-related cerebrospinal fluid biomarkers for Alzheimer’s disease in a multicohort study. Alzheimers Dement 2015;1:339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Hwang J, Kim CM, Jeon S, Lee JM, Hong YJ, Roh JH, et al. Prediction of Alzheimer’s disease pathophysiology based on cortical thickness patterns. Alzheimers Dement 2015;2:58–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Kim A, Fagan AM, Goate AM, Benzinger TL, Morris JC, Head D. Lack of an association of BDNF Val66Met polymorphism and plasma BDNF with hippocampal volume and memory. Cogn Affect Behav Neurosci 2015;15:625–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Hwang KS, Lazaris AS, Eastman JA, Teng E, Thompson PM, Gylys KH, et al. Plasma BDNF levels associate with Pittsburgh compound B binding in the brain. Alzheimers Dement 2015;1:187–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Khan W, Aguilar C, Kiddle SJ, Doyle O, Thambisetty M, Muehlboeck S, et al. A Subset of Cerebrospinal Fluid Proteins from a Multi-Analyte Panel Associated with Brain Atrophy, Disease Classification and Prediction in Alzheimer’s Disease. PLoS One 2015;10:e0134368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Toledo JB, Korff A, Shaw LM, Trojanowski JQ, Zhang J. Low levels of cerebrospinal fluid complement 3 and factor H predict faster cognitive decline in mild cognitive impairment. Alzheimers Res Ther 2014;6:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Harari O, Cruchaga C, Kauwe JS, Ainscough BJ, Bales K, Pickering EH, et al. Phosphorylated tau-Abeta42 ratio as a continuous trait for biomarker discovery for early-stage Alzheimer’s disease in multiplex immunoassay panels of cerebrospinal fluid. Biol Psychiatry 2014;75:723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Nelson PT, Abner EL, Schmitt FA, Kryscio RJ, Jicha GA, Smith CD, et al. Modeling the association between 43 different clinical and pathological variables and the severity of cognitive impairment in a large autopsy cohort of elderly persons. Brain Pathol 2010;20:66–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Mackin RS, Insel P, Zhang J, Mohlenhoff B, Galasko D, Weiner M, et al. Cerebrospinal fluid alpha-synuclein and Lewy body-like symptoms in normal controls, mild cognitive impairment, and Alzheimer’s disease. J Alzheimers Dis 2015;43:1007–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, et al. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement 2015;11:792–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Shen L, Thompson PM, Potkin SG, Bertram L, Farrer LA, Foroud TM, et al. Genetic analysis of quantitative phenotypes in AD and MCI: imaging, cognition and biomarkers. Brain Imaging Behav 2014;8:183–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Thompson PM, Andreassen OA, Arias-Vasquez A, Bearden CE, Boedhoe PS, Brouwer RM, et al. ENIGMA and the individual: Predicting factors that affect the brain in 35 countries worldwide. Neuroimage 2017;145:389–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav 2014;8:153–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Manning EN, Barnes J, Cash DM, Bartlett JW, Leung KK, Ourselin S, et al. APOE epsilon4 is associated with disproportionate progressive hippocampal atrophy in AD. PLoS One 2014;9:e97608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Hostage CA, Choudhury KR, Murali Doraiswamy P, Petrella JR. Mapping the effect of the apolipoprotein E genotype on 4-year atrophy rates in an Alzheimer disease-related brain network. Radiology 2014;271:211–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Shi J, Lepore N, Gutman BA, Thompson PM, Baxter LC, Caselli RJ, et al. Genetic influence of apolipoprotein E4 genotype on hippocampal morphometry: An N = 725 surface-based Alzheimer’s disease neuroimaging initiative study. Hum Brain Mapp 2014;35:3903–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Roussotte FF, Gutman BA, Madsen SK, Colby JB, Narr KL, Thompson PM. Apolipoprotein E epsilon 4 allele is associated with ventricular expansion rate and surface morphology in dementia and normal aging. Neurobiol Aging 2014;35:1309–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Sampedro F, Vilaplana E, de Leon MJ, Alcolea D, Pegueroles J, Montal V, et al. APOE-by-sex interactions on brain structure and metabolism in healthy elderly controls. Oncotarget 2015; 6:26663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Cacciottolo M, Christensen A, Moser A, Liu J, Pike CJ, Smith C, et al. The APOE4 allele shows opposite sex bias in microbleeds and Alzheimer’s disease of humans and mice. Neurobiol Aging 2016;37:47–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Lazaris A, Hwang KS, Goukasian N, Ramirez LM, Eastman J, Blanken AE, et al. Alzheimer risk genes modulate the relationship between plasma apoE and cortical PiB binding. Neurol Genet 2015;1:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Sun JH, Wang HF, Zhu XC, Yu WJ, Tan CC, Jiang T, et al. The Impact of UNC5C Genetic Variations on Neuroimaging in Alzheimer’s Disease. Mol Neurobiol 2016;53:6759–67. [DOI] [PubMed] [Google Scholar]
- [183].Wang HF, Tan L, Hao XK, Jiang T, Tan MS, Liu Y, et al. Effect of EPHA1 genetic variation on cerebrospinal fluid and neuroimaging biomarkers in healthy, mild cognitive impairment and Alzheimer’s disease cohorts. J Alzheimers Dis 2015;44:115–23. [DOI] [PubMed] [Google Scholar]
- [184].Benitez BA, Jin SC, Guerreiro R, Graham R, Lord J, Harold D, et al. Missense variant in TREML2 protects against Alzheimer’s disease. Neurobiol Aging 2014;35:1510.e19–e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Liu Y, Yu JT, Wang HF, Hao XK, Yang YF, Jiang T, et al. Association between NME8 locus polymorphism and cognitive decline, cerebrospinal fluid and neuroimaging biomarkers in Alzheimer’s disease. PLoS One 2014;9:e114777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Louwersheimer E, Ramirez A, Cruchaga C, Becker T, Kornhuber J, Peters O, et al. The influence of genetic variants in SORL1 gene on the manifestation of Alzheimer’s disease. Neurobiol Aging 2015; 36:1605.e13–e20. [DOI] [PubMed] [Google Scholar]
- [187].Zhang DF, Li J, Wu H, Cui Y, Bi R, Zhou HJ, et al. CFH Variants Affect Structural and Functional Brain Changes and Genetic Risk of Alzheimer’s Disease. Neuropsychopharmacology 2016; 41:1034–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Ebbert MT, Boehme KL, Wadsworth ME, Staley LA, Mukherjee S, Crane PK, et al. Interaction between variants in CLU and MS4A4E modulates Alzheimer’s disease risk. Alzheimers Dement 2016; 12:121–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Ge T, Nichols TE, Ghosh D, Mormino EC, Smoller JW, Sabuncu MR. A kernel machine method for detecting effects of interaction between multidimensional variable sets: An imaging genetics application. Neuroimage 2015;109:505–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Wang C, Wang HF, Tan MS, Liu Y, Jiang T, Zhang DQ, et al. Impact of Common Variations in PLD3 on Neuroimaging Phenotypes in Non-demented Elders. Mol Neurobiol 2016;53:4343–51. [DOI] [PubMed] [Google Scholar]
- [191].Yan J, Kim S, Nho K, Chen R, Risacher SL, Moore JH, et al. Hippocampal transcriptome-guided genetic analysis of correlated episodic memory phenotypes in Alzheimer’s disease. Front Genet 2015;6:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Warstadt NM, Dennis EL, Jahanshad N, Kohannim O, Nir TM, McMahon KL, et al. Serum cholesterol and variant in cholesterolrelated gene CETP predict white matter microstructure. Neurobiol Aging 2014;35:2504–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Leduc V, De Beaumont L, Theroux L, Dea D, Aisen P, Petersen RC, et al. HMGCR is a genetic modifier for risk, age of onset and MCI conversion to Alzheimer’s disease in a three cohorts study. Mol Psychiatry 2015;20:867–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Roussotte FF, Gutman BA, Hibar DP, Madsen SK, Narr KL, Thompson PM. Carriers of a common variant in the dopamine transporter gene have greater dementia risk, cognitive decline, and faster ventricular expansion. Alzheimers Dement 2015;11:1153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Hibar DP, Stein JL, Jahanshad N, Kohannim O, Hua X, Toga AW, et al. Genome-wide interaction analysis reveals replicated epistatic effects on brain structure. Neurobiol Aging 2015;36:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [196].Biffi A, Sabuncu MR, Desikan RS, Schmansky N, Salat DH, Rosand J, et al. Genetic variation of oxidative phosphorylation genes in stroke and Alzheimer’s disease. Neurobiol Aging 2014; 35:1956.e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Koran ME, Hohman TJ, Thornton-Wells TA. Genetic interactions found between calcium channel genes modulate amyloid load measured by positron emission tomography. Hum Genet 2014; 133:85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Benedet AL, Labbe A, Lemay P, Zimmer ER, Pascoal TA, Leuzy A, et al. Epistasis analysis links immune cascades and cerebral amyloidosis. J Neuroinflammation 2015;12:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Hohman TJ, Koran ME, Thornton-Wells TA. Interactions between GSK3beta and amyloid genes explain variance in amyloid burden. Neurobiol Aging 2014;35:460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Escott-Price V, Bellenguez C, Wang LS, Choi SH, Harold D, Jones L, et al. Gene-wide analysis detects two new susceptibility genes for Alzheimer’s disease. PLoS One 2014;9:e94661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Perez-Palma E, Bustos BI, Villaman CF, Alarcon MA, Avila ME, Ugarte GD, et al. Overrepresentation of glutamate signaling in Alzheimer’s disease: network-based pathway enrichment using meta-analysis of genome-wide association studies. PLoS One 2014; 9:e95413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Wang W, Mandel J, Bouaziz J, Commenges D, Nabirotchkine S, Chumakov I, et al. A Multi-Marker Genetic Association Test Based on the Rasch Model Applied to Alzheimer’s Disease. PLoS One 2015;10:e0138223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Jiang S, Yang W, Qiu Y, Chen HZ. Identification of novel quantitative traits-associated susceptibility loci for APOE epsilon 4 non-carriers of Alzheimer’s disease. Curr Alzheimer Res 2015;12:218–27. [DOI] [PubMed] [Google Scholar]
- [204].Kim JH, Song P, Lim H, Lee JH, Lee JH, Park SA. Gene-based rare allele analysis identified a risk gene of Alzheimer’s disease. PLoS One 2014;9:e107983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Zieselman AL, Fisher JM, Hu T, Andrews PC, Greene CS, Shen L, et al. Computational genetics analysis of grey matter density in Alzheimer’s disease. BioData Min 2014;7:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Yankner BA. REST and Alzheimer disease. Ann Neurol 2015; 78:499. [DOI] [PubMed] [Google Scholar]
- [207].Dols-Icardo O, Vilaplana E, Sampedro F, Alcolea D, Belbin O, Camacho V, et al. Effect of REST on brain metabolism in the Alzheimer disease continuum. Ann Neurol 2015;78:661–2. [DOI] [PubMed] [Google Scholar]
- [208].Li K, Jiang Q, Xu A, Liu G. REST rs3796529 variant does not confer susceptibility to Alzheimer’s disease. Ann Neurol 2015;78:835–6. [DOI] [PubMed] [Google Scholar]
- [209].Jiang Q, Liu G. REST rs3796529 variant does not influence human subcortical brain structures. Ann Neurol 2015;79:334–5. [DOI] [PubMed] [Google Scholar]
- [210].Moon SW, Dinov ID, Zamanyan A, Shi R, Genco A, Hobel S, et al. Gene interactions and structural brain change in early-onset Alzheimer’s disease subjects using the pipeline environment. Psychiatry Investig 2015;12:125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Moon SW, Dinov ID, Hobel S, Zamanyan A, Choi YC, Shi R, et al. Structural Brain Changes in Early-Onset Alzheimer’s Disease Subjects Using the LONI Pipeline Environment. J Neuroimaging 2015; 25:728–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Khondoker M, Newhouse S, Westman E, Muehlboeck JS, Mecocci P, Vellas B, et al. Linking Genetics of Brain Changes to Alzheimer’s Disease: Sparse Whole Genome Association Scan of Regional MRI Volumes in the ADNI and AddNeuroMed Cohorts. J Alzheimers Dis 2015;45:851–64. [DOI] [PubMed] [Google Scholar]
- [213].Printy BP, Verma N, Cowperthwaite MC, Markey MK. Effects of genetic variation on the dynamics of neurodegeneration in Alzheimer’s disease. Conf Proc IEEE Eng Med Biol Soc 2014;2014:2464–7. [DOI] [PubMed] [Google Scholar]
- [214].Moon SW, Dinov ID, Kim J, Zamanyan A, Hobel S, Thompson PM, et al. Structural Neuroimaging Genetics Interactions in Alzheimer’s Disease. J Alzheimers Dis 2015;48:1051–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Xu Z, Shen X, Pan W. Longitudinal analysis is more powerful than cross-sectional analysis in detecting genetic association with neuroimaging phenotypes. PLoS One 2014;9:e102312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Koran ME, Hohman TJ, Meda SA, Thornton-Wells TA. Genetic interactions within inositol-related pathways are associated with longitudinal changes in ventricle size. J Alzheimers Dis 2014;38:145–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Nho K, Kim S, Risacher SL, Shen L, Corneveaux JJ, Swaminathan S, et al. Protective variant for hippocampal atrophy identified by whole exome sequencing. Ann Neurol 2015;77:547–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, et al. APOE and BCHE as modulators of cerebral amyloid deposition: a florbetapir PET genome-wide association study. Mol Psychiatry 2014;19:351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Souza MB, Araujo GS, Costa IG, Oliveira JR. Combined Genome-Wide CSF Abeta-42’s Associations and Simple Network Properties Highlight New Risk Factors for Alzheimer’s Disease. J Mol Neurosci 2016;58:120–8. [DOI] [PubMed] [Google Scholar]
- [220].Chouraki V, De Bruijn RF, Chapuis J, Bis JC, Reitz C, Schraen S, et al. A genome-wide association meta-analysis of plasma Abeta peptides concentrations in the elderly. Mol Psychiatry 2014;19:1326–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Ramanan VK, Risacher SL, Nho K, Kim S, Shen L, McDonald BC, et al. GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain 2015;138:3076–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Ramirez A, van der Flier WM, Herold C, Ramonet D, Heilmann S, Lewczuk P, et al. SUCLG2 identified as both a determinator of CSF Abeta1–42 levels and an attenuator of cognitive decline in Alzheimer’s disease. Hum Mol Genet 2014;23:6644–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Li QS, Parrado AR, Samtani MN, Narayan VA Alzheimer’s Disease Neuroimaging Initiative. Variations in the FRA10AC1 Fragile Site and 15q21 Are Associated with Cerebrospinal Fluid Abeta1–42 Level. PLoS One 2015;10:e0134000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Hohman TJ, Koran ME, Thornton-Wells TA. Genetic modification of the relationship between phosphorylated tau and neurodegeneration. Alzheimers Dement 2014;10:637–645.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Kauwe JS, Bailey MH, Ridge PG, Perry R, Wadsworth ME, Hoyt KL, et al. Genome-wide association study of CSF levels of 59 alzheimer’s disease candidate proteins: significant associations with proteins involved in amyloid processing and inflammation. PLoS Genet 2014;10:e1004758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Sherva R, Tripodis Y, Bennett DA, Chibnik LB, Crane PK, de Jager PL, et al. Genome-wide association study of the rate of cognitive decline in Alzheimer’s disease. Alzheimers Dement 2014; 10:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Mukherjee S, Kim S, Ramanan VK, Gibbons LE, Nho K, Glymour MM, et al. Gene-based GWAS and biological pathway analysis of the resilience of executive functioning. Brain Imaging Behav 2014;8:110–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Hibar DP, Stein JL, Renteria ME, Arias-Vasquez A, Desrivieres S, Jahanshad N, et al. Common genetic variants influence human subcortical brain structures. Nature 2015;520:224–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Chen CH, Peng Q, Schork AJ, Lo MT, Fan CC, Wang Y, et al. Large-scale genomics unveil polygenic architecture of human cortical surface area. Nat Commun 2015;6:7549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Roussotte FF, Jahanshad N, Hibar DP, Thompson PM. Altered regional brain volumes in elderly carriers of a risk variant for drug abuse in the dopamine D2 receptor gene (DRD2). Brain Imaging Behav 2015;9:213–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Roussotte FF, Jahanshad N, Hibar DP, Sowell ER, Kohannim O, Barysheva M, et al. A commonly carried genetic variant in the delta opioid receptor gene, OPRD1, is associated with smaller regional brain volumes: replication in elderly and young populations. Hum Brain Mapp 2014;35:1226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Lampert EJ, Roy Choudhury K, Hostage CA, Rathakrishnan B, Weiner M, Petrella JR, et al. Brain atrophy rates in first degree relatives at risk for Alzheimer’s. Neuroimage Clin 2014;6:340–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, et al. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet 2011;43:436–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Shin S, Kim JH, Cho JH, Kim GS, Choi SA, Lee JH. Mild cognitive impairment due to alzheimer disease is less likely under the age of 65. Alzheimer Dis Assoc Disord 2015;29:26–31. [DOI] [PubMed] [Google Scholar]
- [235].Campion D, Dumanchin C, Hannequin D, Dubois B, Belliard S, Puel M, et al. Early-onset autosomal dominant Alzheimer disease: prevalence, genetic heterogeneity, and mutation spectrum. Am J Hum Genet 1999;65:664–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Mukherjee S, Kim S, Gibbons LE, Nho K, Risacher SL, Glymour MM, et al. Genetic architecture of resilience of executive functioning. Brain Imaging Behav 2012;6:621–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Kim J, Pan W. A cautionary note on using secondary phenotypes in neuroimaging genetic studies. Neuroimage 2015;121:136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Radmanesh F, Devan WJ, Anderson CD, Rosand J, Falcone GJ. Accuracy of imputation to infer unobserved APOE epsilon alleles in genome-wide genotyping data. Eur J Hum Genet 2014;22:1239–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Lin JA, Zhu H, Mihye A, Sun W, Ibrahim JG. Functional-mixed effects models for candidate genetic mapping in imaging genetic studies. Genet Epidemiol 2014;38:680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Huang M, Nichols T, Huang C, Yu Y, Lu Z, Knickmeyer RC, et al. FVGWAS: Fast voxelwise genome wide association analysis of large-scale imaging genetic data. Neuroimage 2015;118:613–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Hua WY, Nichols TE, Ghosh D. Multiple comparison procedures for neuroimaging genomewide association studies. Biostatistics 2015; 16:17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Beaton D, Dunlop J, Abdi H. Partial Least Squares Correspondence Analysis: A Framework to Simultaneously Analyze Behavioral and Genetic Data. Psychol Methods 2015;21:21. [DOI] [PubMed] [Google Scholar]
- [243].Hao X, Yu J, Zhang D. Identifying genetic associations with MRI-derived measures via tree-guided sparse learning. Med Image Comput Comput Assist Interv 2014;17:757–64. [DOI] [PubMed] [Google Scholar]
- [244].Zhang Y, Xu Z, Shen X, Pan W. Testing for association with multiple traits in generalized estimation equations, with application to neuroimaging data. Neuroimage 2014;96:309–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Sun Q, Zhu H, Liu Y, Ibrahim JG. SPReM: Sparse Projection Regression Model For High-dimensional Linear Regression. J Am Stat Assoc 2015;110:289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Sheng J, Kim S, Yan J, Moore J, Saykin A, Shen L. Data synthesis and method evaluation for brain imaging genetics. Proc IEEE Int Symp Biomed Imaging 2014;2014:1202–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Sherif FF, Zayed N, Fakhr M. Discovering Alzheimer Genetic Biomarkers Using Bayesian Networks. Adv Bioinformatics 2015; 2015:639367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Xu Z, Pan W Alzheimer’s Disease Neuroimaging Initiative. Approximate score-based testing with application to multivariate trait association analysis. Genet Epidemiol 2015;39:469–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Nho K, West JD, Li H, Henschel R, Bharthur A, Tavares MC, et al. Comparison of Multi-Sample Variant Calling Methods for Whole Genome Sequencing. IEEE Int Conf Systems Biol 2014;2014:59–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Hua WY, Ghosh D. Equivalence of kernel machine regression and kernel distance covariance for multidimensional phenotype association studies. Biometrics 2015;71:812–20. [DOI] [PubMed] [Google Scholar]
- [251].Zhu H, Khondker Z, Lu Z, Ibrahim JG. Bayesian Generalized Low Rank Regression Models for Neuroimaging Phenotypes and Genetic Markers. J Am Stat Assoc 2014;109:977–90. [PMC free article] [PubMed] [Google Scholar]
- [252].Cao Y, Wei P, Bailey M, Kauwe JS, Maxwell TJ. A versatile omnibus test for detecting mean and variance heterogeneity. Genet Epidemiol 2014;38:51–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Weiss TL, Zieselman A, Hill DP, Diamond SG, Shen L, Saykin AJ, et al. The role of visualization and 3-D printing in biological data mining. BioData Min 2015;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [254].Zhang Z, Huang H, Shen D. Integrative analysis of multi-dimensional imaging genomics data for Alzheimer’s disease prediction. Front Aging Neurosci 2014;6:260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [255].Kong D, Giovanello KS, Wang Y, Lin W, Lee E, Fan Y, et al. Predicting Alzheimer’s Disease Using Combined Imaging-Whole Genome SNP Data. J Alzheimers Dis 2015;46:695–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [256].Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 2002; 297:353–6. [DOI] [PubMed] [Google Scholar]
- [257].Altmann A, Ng B, Landau SM, Jagust WJ, Greicius MD. Regional brain hypometabolism is unrelated to regional amyloid plaque burden. Brain 2015;138:3734–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].Jack CR Jr, Knopman DS, Jagust WJ, Petersen RC, Weiner MW, Aisen PS, et al. Tracking pathophysiological processes in Alzheimer’s disease: an updated hypothetical model of dynamic biomarkers. Lancet Neurol 2013;12:207–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Beckett LA, Donohue MC, Wang C, Aisen P, Harvey DJ, Saito N. The Alzheimer’s Disease Neuroimaging Initiative phase 2: Increasing the length, breadth, and depth of our understanding. Alzheimers Dement 2015;11:823–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Franklin EE, Perrin RJ, Vincent B, Baxter M, Morris JC, Cairns NJ. Brain collection, standardized neuropathologic assessment, and comorbidity in Alzheimer’s Disease Neuroimaging Initiative 2 participants. Alzheimers Dement 2015;11:815–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [261].Susanto TA, Pua EP, Zhou J. Cognition, brain atrophy, and cerebrospinal fluid biomarkers changes from preclinical to dementia stage of Alzheimer’s disease and the influence of apolipoprotein e. J Alzheimers Dis 2015;45:253–68. [DOI] [PubMed] [Google Scholar]
- [262].Young AL, Oxtoby NP, Daga P, Cash DM, Fox NC, Ourselin S, et al. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain 2014;137:2564–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [263].Toledo JB, Xie SX, Trojanowski JQ, Shaw LM. Longitudinal change in CSF Tau and Abeta biomarkers for up to 48 months in ADNI. Acta Neuropathol 2013;126:659–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Bertens D, Knol DL, Scheltens P, Visser PJ. Temporal evolution of biomarkers and cognitive markers in the asymptomatic, MCI, and dementia stage of Alzheimer’s disease. Alzheimers Dement 2015; 11:511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [265].Araque Caballero MA, Brendel M, Delker A, Ren J, Rominger A, Bartenstein P, et al. Mapping 3-year changes in gray matter and metabolism in Abeta-positive nondemented subjects. Neurobiol Aging 2015;36:2913–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [266].Insel PS, Mattsson N, Donohue MC, Mackin RS, Aisen PS, Jack CR Jr, et al. The transitional association between beta-amyloid pathology and regional brain atrophy. Alzheimers Dement 2015;11:1171–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [267].Kerbler GM, Fripp J, Rowe CC, Villemagne VL, Salvado O, Rose S, et al. Basal forebrain atrophy correlates with amyloid beta burden in Alzheimer’s disease. Neuroimage Clin 2015;7:105–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Grothe MJ, Ewers M, Krause B, Heinsen H, Teipel SJ. Basal forebrain atrophy and cortical amyloid deposition in nondemented elderly subjects. Alzheimers Dement 2014;10:S344–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Teipel S, Heinsen H, Amaro E Jr, Grinberg LT, Krause B, Grothe M. Cholinergic basal forebrain atrophy predicts amyloid burden in Alzheimer’s disease. Neurobiol Aging 2014;35:482–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Dowling NM, Johnson SC, Gleason CE, Jagust WJ. The mediational effects of FDG hypometabolism on the association between cerebrospinal fluid biomarkers and neurocognitive function. Neuroimage 2015;105:357–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Mattsson N, Insel PS, Aisen PS, Jagust W, Mackin S, Weiner M. Brain structure and function as mediators of the effects of amyloid on memory. Neurology 2015;84:1136–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [272].Byun MS, Kim SE, Park J, Yi D, Choe YM, Sohn BK, et al. Heterogeneity of Regional Brain Atrophy Patterns Associated with Distinct Progression Rates in Alzheimer’s Disease. PLoS One 2015; 10:e0142756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Mattsson N, Insel PS, Nosheny R, Tosun D, Trojanowski JQ, Shaw LM, et al. Emerging beta-amyloid pathology and accelerated cortical atrophy. JAMA Neurol 2014;71:725–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Fortea J, Vilaplana E, Alcolea D, Carmona-Iragui M, Sanchez-Saudinos MB, Sala I, et al. Cerebrospinal fluid beta-amyloid and phospho-tau biomarker interactions affecting brain structure in preclinical Alzheimer disease. Ann Neurol 2014;76:223–30. [DOI] [PubMed] [Google Scholar]
- [275].Song Z, Insel PS, Buckley S, Yohannes S, Mezher A, Simonson A, et al. Brain amyloid-beta burden is associated with disruption of intrinsic functional connectivity within the medial temporal lobe in cognitively normal elderly. J Neurosci 2015;35:3240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Donohue MC, Jacqmin-Gadda H, Le Goff M, Thomas RG, Raman R, Gamst AC, et al. Estimating long-term multivariate progression from short-term data. Alzheimers Dement 2014;10:S400–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Mattsson N, Tosun D, Insel PS, Simonson A, Jack CR Jr, Beckett LA, et al. Association of brain amyloid-beta with cerebral perfusion and structure in Alzheimer’s disease and mild cognitive impairment. Brain 2014;137:1550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Toledo JB, Weiner MW, Wolk DA, Da X, Chen K, Arnold SE, et al. Neuronal injury biomarkers and prognosis in ADNI subjects with normal cognition. Acta Neuropathol Commun 2014;2:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Toledo JB, Bjerke M, Chen K, Rozycki M, Jack CR Jr, Weiner MW, et al. Memory, executive, and multidomain subtle cognitive impairment: clinical and biomarker findings. Neurology 2015;85:144–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Bondi MW, Edmonds EC, Jak AJ, Clark LR, Delano-Wood L, McDonald CR, et al. Neuropsychological criteria for mild cognitive impairment improves diagnostic precision, biomarker associations, and progression rates. J Alzheimers Dis 2014;42:275–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Edmonds EC, Delano-Wood L, Clark LR, Jak AJ, Nation DA, McDonald CR, et al. Susceptibility of the conventional criteria for mild cognitive impairment to false-positive diagnostic errors. Alzheimers Dement 2015;11:415–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Edmonds EC, Delano-Wood L, Galasko DR, Salmon DP, Bondi MW. Subjective cognitive complaints contribute to misdiagnosis of mild cognitive impairment. J Int Neuropsychol Soc 2014;20:836–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [283].Peter J, Abdulkadir A, Kaller C, Kummerer D, Hull M, Vach W, et al. Subgroups of Alzheimer’s disease: stability of empirical clusters over time. J Alzheimers Dis 2014;42:651–61. [DOI] [PubMed] [Google Scholar]
- [284].Park MH, Han C. Is there an MCI reversion to cognitively normal? Analysis of Alzheimer’s disease biomarkers profiles. Int Psychogeriatr 2015;27:429–37. [DOI] [PubMed] [Google Scholar]
- [285].Pravata E, Tavernier J, Parker R, Vavro H, Mintzer JE, Spampinato MV. The neural correlates of anomia in the conversion from mild cognitive impairment to Alzheimer’s disease. Neuroradiology 2016;58:59–67. [DOI] [PubMed] [Google Scholar]
- [286].Nettiksimmons J, DeCarli C, Landau S, Beckett L. Biological heterogeneity in ADNI amnestic mild cognitive impairment. Alzheimers Dement 2014;10:511–521.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Madsen SK, Gutman BA, Joshi SH, Toga AW, Jack CR Jr, Weiner MW, et al. Mapping ventricular expansion onto cortical gray matter in older adults. Neurobiol Aging 2015;36:S32–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [288].Albert MS, Dekosky ST, Dickson D, Dubois B, Feldman HH, Fox NC, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement 2011;7:270–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [289].Gifford KA, Liu D, Damon SM, Chapman WG, Romano Iii RR, Samuels LR, et al. Subjective memory complaint only relates to verbal episodic memory performance in mild cognitive impairment. J Alzheimers Dis 2015;44:309–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Jack CR Jr, Knopman DS, Chetelat G, Dickson D, Fagan AM, Frisoni GB, et al. Suspected non-Alzheimer disease pathophysiology -concept and controversy. Nat Rev Neurol 2016;12:117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Caroli A, Prestia A, Galluzzi S, Ferrari C, van der Flier WM, Ossenkoppele R, et al. Mild cognitive impairment with suspected nonamyloid pathology (SNAP): Prediction of progression. Neurology 2015;84:508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Shaw LM, Vanderstichele H, Knapik-Czajka M, Clark CM, Aisen PS, Petersen RC, et al. Cerebrospinal fluid biomarker signature in Alzheimer’s disease neuroimaging initiative subjects. Ann Neurol 2009;65:403–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [293].Landau SM, Lu M, Joshi AD, Pontecorvo M, Mintun MA, Trojanowski JQ, et al. Comparing PET imaging and CSF measurements of Ass. Ann Neurol 2013;74:826–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Debette S, Markus HS. The clinical importance of white matter hyperintensities on brain magnetic resonance imaging: systematic review and meta-analysis. BMJ 2010;341:c3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Ramirez J, McNeely AA, Scott CJ, Masellis M, Black SE. White matter hyperintensity burden in elderly cohort studies: The Sunnybrook Dementia Study, Alzheimer’s Disease Neuroimaging Initiative, and Three-City Study. Alzheimers Dement 2016;12:203–10. [DOI] [PubMed] [Google Scholar]
- [296].Lorius N, Locascio JJ, Rentz DM, Johnson KA, Sperling RA, Viswanathan A, et al. Vascular disease and risk factors are associated with cognitive decline in the alzheimer disease spectrum. Alzheimer Dis Assoc Disord 2015;29:18–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Tosto G, Zimmerman ME, Carmichael OT, Brickman AM. Predicting aggressive decline in mild cognitive impairment: the importance of white matter hyperintensities. JAMA Neurol 2014;71:872–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Lin F, Lo RY, Cole D, Ducharme S, Chen DG, Mapstone M, et al. Longitudinal effects of metabolic syndrome on Alzheimer and vascular related brain pathology. Dement Geriatr Cogn Dis Extra 2014;4:184–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Morris JK, Vidoni ED, Honea RA, Burns JM. Impaired glycemia increases disease progression in mild cognitive impairment. Neurobiol Aging 2014;35:585–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Scott JA, Braskie MN, Tosun D, Thompson PM, Weiner M, DeCarli C, et al. Cerebral Amyloid and Hypertension are Independently Associated with White Matter Lesions in Elderly. Front Aging Neurosci 2015;7:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Durazzo TC, Mattsson N, Weiner MW, Korecka M, Trojanowski JQ, Shaw LM. History of cigarette smoking in cognitively-normal elders is associated with elevated cerebrospinal fluid biomarkers of oxidative stress. Drug Alcohol Depend 2014;142:262–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Durazzo TC, Mattsson N, Weiner MW. Interaction of Cigarette Smoking History With APOE Genotype and Age on Amyloid Level, Glucose Metabolism, and Neurocognition in Cognitively Normal Elders. Nicotine Tob Res 2016;18:204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [303].Makedonov I, Chen JJ, Masellis M, MacIntosh BJ. Physiological fluctuations in white matter are increased in Alzheimer’s disease and correlate with neuroimaging and cognitive biomarkers. Neurobiol Aging 2016;37:12–8. [DOI] [PubMed] [Google Scholar]
- [304].Willette AA, Calhoun VD, Egan JM, Kapogiannis D. Prognostic classification of mild cognitive impairment and Alzheimer’s disease: MRI independent component analysis. Psychiatry Res 2014; 224:81–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Nettiksimmons J, Beckett L, Schwarz C, Carmichael O, Fletcher E, Decarli C. Subgroup of ADNI normal controls characterized by atrophy and cognitive decline associated with vascular damage. Psychol Aging 2013;28:191–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Hohman TJ, Samuels LR, Liu D, Gifford KA, Mukherjee S, Benson EM, et al. Stroke risk interacts with Alzheimer’s disease biomarkers on brain aging outcomes. Neurobiol Aging 2015; 36:2501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Moran C, Beare R, Phan TG, Bruce DG, Callisaya ML, Srikanth V. Type 2 diabetes mellitus and biomarkers of neurodegeneration. Neurology 2015;85:1123–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Coutu JP, Goldblatt A, Rosas HD, Salat DHAlzheimer’s Disease Neuroimaging Initiative. White Matter Changes are Associated with Ventricular Expansion in Aging, Mild Cognitive Impairment, and Alzheimer’s Disease. J Alzheimers Dis 2015;49:329–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Lindemer ER, Salat DH, Smith EE, Nguyen K, Fischl B, Greve DN. White matter signal abnormality quality differentiates mild cognitive impairment that converts to Alzheimer’s disease from nonconverters. Neurobiol Aging 2015;36:2447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Wolf D, Fischer FU, Scheurich A, Fellgiebel A. Non-Linear Association between Cerebral Amyloid Deposition and White Matter Microstructure in Cognitively Healthy Older Adults. J Alzheimers Dis 2015;47:117–27. [DOI] [PubMed] [Google Scholar]
- [311].Zhou Y, Yu F, Duong TQ. White matter lesion load is associated with resting state functional MRI activity and amyloid PET but not FDG in mild cognitive impairment and early Alzheimer’s disease patients. J Magn Reson Imaging 2015;41:102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Chiang GC, Cruz Hernandez JC, Kantarci K, Jack CR Jr, Weiner MW. Cerebral Microbleeds, CSF p-Tau, and Cognitive Decline: Significance of Anatomic Distribution. AJNR Am J Neuroradiol 2015;36:1635–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Tosto G, Zimmerman ME, Hamilton JL, Carmichael OT, Brickman AMAlzheimer’s Disease Neuroimaging Initiative. The effect of white matter hyperintensities on neurodegeneration in mild cognitive impairment. Alzheimers Dement 2015;11:1510–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [314].Delbeuck X, Van der Linden M, Collette F. Alzheimer’s disease as a disconnection syndrome? Neuropsychol Rev 2003;13:79–92. [DOI] [PubMed] [Google Scholar]
- [315].Reid AT, Evans AC. Structural networks in Alzheimer’s disease. Eur Neuropsychopharmacol 2013;23:63–77. [DOI] [PubMed] [Google Scholar]
- [316].Henkins KM, Sokolow S, Miller CA, Vinters HV, Poon WW, Cornwell LB, et al. Extensive p-tau pathology and SDS-stable p-tau oligomers in Alzheimer’s cortical synapses. Brain Pathol 2012;22:826–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Takahashi RH, Capetillo-Zarate E, Lin MT, Milner TA, Gouras GK. Co-occurrence of Alzheimer’s disease ss-amyloid and tau pathologies at synapses. Neurobiol Aging 2010;31:1145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Toga AW, Thompson PM. Connectomics sheds new light on Alzheimer’s disease. Biol Psychiatry 2013;73:390–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Bullmore E, Sporns O. Complex brain networks: graph theoretical analysis of structural and functional systems. Nat Rev Neurosci 2009;10:186–98. [DOI] [PubMed] [Google Scholar]
- [320].van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci 2013;17:683–96. [DOI] [PubMed] [Google Scholar]
- [321].Lee SH, Coutu JP, Wilkens P, Yendiki A, Rosas HD, Salat DH. Tract-based analysis of white matter degeneration in Alzheimer’s disease. Neuroscience 2015;301:79–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [322].Sun X, Salat D, Upchurch K, Deason R, Kowall N, Budson A. Destruction of white matter integrity in patients with mild cognitive impairment and Alzheimer disease. J Investig Med 2014; 62:927–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [323].Ouyang X, Chen K, Yao L, Hu B, Wu X, Ye Q, et al. Simultaneous changes in gray matter volume and white matter fractional anisotropy in Alzheimer’s disease revealed by multimodal CCA and joint ICA. Neuroscience 2015;301:553–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [324].Patil RB, Ramakrishnan S. Analysis of sub-anatomic diffusion tensor imaging indices in white matter regions of Alzheimer with MMSE score. Comput Methods Programs Biomed 2014;117:13–9. [DOI] [PubMed] [Google Scholar]
- [325].Nishioka C, Poh C, Sun SW. Diffusion tensor imaging reveals visual pathway damage in patients with mild cognitive impairment and Alzheimer’s disease. J Alzheimers Dis 2015;45:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [326].Lee SH, Seo J, Lee JM, Park H. Differences in early and late mild cognitive impairment tractography using a diffusion tensor MRI. Neuroreport 2014;25:1393–8. [DOI] [PubMed] [Google Scholar]
- [327].Prescott JW, Guidon A, Doraiswamy PM, Roy Choudhury K, Liu C, Petrella JR. The Alzheimer structural connectome: changes in cortical network topology with increased amyloid plaque burden. Radiology 2014;273:175–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [328].Daianu M, Jahanshad N, Nir TM, Jack CR Jr, Weiner MW, Bernstein MA, et al. Rich club analysis in the Alzheimer’s disease connectome reveals a relatively undisturbed structural core network. Hum Brain Mapp 2015;36:3087–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [329].Nir TM, Jahanshad N, Toga AW, Bernstein MA, Jack CR Jr, Weiner MW, et al. Connectivity network measures predict volumetric atrophy in mild cognitive impairment. Neurobiol Aging 2015; 36:S113–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [330].Fischer FU, Wolf D, Scheurich A, Fellgiebel A. Altered whole-brain white matter networks in preclinical Alzheimer’s disease. Neuroimage Clin 2015;8:660–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [331].Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer’s disease from healthy aging: evidence from functional MRI. Proc Natl Acad Sci U S A 2004; 101:4637–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [332].Liang P, Xiang J, Liang H, Qi Z, Li K. Alzheimer’s Disease Neuro-Imaging I. Altered amplitude of low-frequency fluctuations in early and late mild cognitive impairment and Alzheimer’s disease. Curr Alzheimer Res 2014;11:389–98. [DOI] [PubMed] [Google Scholar]
- [333].Cai S, Huang L, Zou J, Jing L, Zhai B, Ji G, et al. Changes in thalamic connectivity in the early and late stages of amnestic mild cognitive impairment: a resting-state functional magnetic resonance study from ADNI. PLoS One 2015;10:e0115573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [334].McKenna F, Koo BB, Killiany RAlzheimer’s Disease Neuroimaging Initiative. Comparison of ApoE-related brain connectivity differences in early MCI and normal aging populations: an fMRI study. Brain Imaging Behav 2016;10:970–83. [DOI] [PubMed] [Google Scholar]
- [335].Grothe MJ, Teipel SJ. Alzheimer’s Disease Neuroimaging I. Spatial patterns of atrophy, hypometabolism, and amyloid deposition in Alzheimer’s disease correspond to dissociable functional brain networks. Hum Brain Mapp 2016;37:35–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [336].Jones DT, Knopman DS, Gunter JL, Graff-Radford J, Vemuri P, Boeve BF, et al. Cascading network failure across the Alzheimer’s disease spectrum. Brain 2016;139:547–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [337].Myers N, Pasquini L, Gottler J, Grimmer T, Koch K, Ortner M, et al. Within-patient correspondence of amyloid-beta and intrinsic network connectivity in Alzheimer’s disease. Brain 2014;137:2052–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [338].Montembeault M, Rouleau I, Provost JS, Brambati SM. Altered Gray Matter Structural Covariance Networks in Early Stages of Alzheimer’s Disease. Cereb Cortex 2016;26:2650–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [339].Yao Z, Hu B, Zheng J, Zheng W, Chen X, Gao X, et al. A FDG-PET Study of Metabolic Networks in Apolipoprotein E epsilon4 Allele Carriers. PLoS One 2015;10:e0132300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [340].Son SJ, Kim J, Seo J, Lee JM, Park H. Connectivity analysis of normal and mild cognitive impairment patients based on FDG and PiB-PET images. Neurosci Res 2015;98:50–8. [DOI] [PubMed] [Google Scholar]
- [341].Teipel S, Grothe MJ Alzheimer’s Disease Neuroimaging Initiative. Does posterior cingulate hypometabolism result from disconnection or local pathology across preclinical and clinical stages of Alzheimer’s disease? Eur J Nucl Med Mol Imaging 2016;43:526–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [342].Palop JJ, Mucke L. Amyloid-beta-induced neuronal dysfunction in Alzheimer’s disease: from synapses toward neural networks. Nat Neurosci 2010;13:812–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [343].Frost B, Jacks RL, Diamond MI. Propagation of tau misfolding from the outside to the inside of a cell. J Biol Chem 2009;284:12845–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [344].Raj A, LoCastro E, Kuceyeski A, Tosun D, Relkin N, Weiner M. Network Diffusion Model of Progression Predicts Longitudinal Patterns of Atrophy and Metabolism in Alzheimer’s Disease. Cell Rep 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [345].Raj A, Kuceyeski A, Weiner M. A network diffusion model of disease progression in dementia. Neuron 2012;73:1204–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [346].Iturria-Medina Y, Sotero RC, Toussaint PJ, Evans AC. Epidemic spreading model to characterize misfolded proteins propagation in aging and associated neurodegenerative disorders. PLoS Comput Biol 2014;10:e1003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [347].Villemagne VL, Furumoto S, Fodero-Tavoletti MT, Mulligan RS, Hodges J, Harada R, et al. In vivo evaluation of a novel tau imaging tracer for Alzheimer’s disease. Eur J Nucl Med Mol Imaging 2014; 41:816–26. [DOI] [PubMed] [Google Scholar]
- [348].Qi Z, Goryawala M, Cabrerizo M, Barker W, Loewenstein D, Duara R, et al. Multivariate Analysis of structural MRI and PET (FDG and 18F-AV-45) for Alzheimer’s disease and its prodromal stages. Conf Proc IEEE Eng Med Biol Soc 2014;2014:1051–4. [DOI] [PubMed] [Google Scholar]
- [349].Apostolova LG, Hwang KS, Kohannim O, Avila D, Elashoff D, Jack CR Jr, et al. ApoE4 effects on automated diagnostic classifiers for mild cognitive impairment and Alzheimer’s disease. Neuroimage Clin 2014;4:461–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [350].Steenland K, Zhao L, Goldstein F, Cellar J, Lah J. Biomarkers for predicting cognitive decline in those with normal cognition. J Alzheimers Dis 2014;40:587–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [351].Alexopoulos P, Kriett L, Haller B, Klupp E, Gray K, Grimmer T, et al. Limited agreement between biomarkers of neuronal injury at different stages of Alzheimer’s disease. Alzheimers Dement 2014; 10:684–9. [DOI] [PubMed] [Google Scholar]
- [352].Teipel SJ, Kurth J, Krause B, Grothe MJ. The relative importance of imaging markers for the prediction of Alzheimer’s disease dementia in mild cognitive impairment - Beyond classical regression. Neuroimage Clin 2015;8:583–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [353].Trzepacz PT, Yu P, Sun J, Schuh K, Case M, Witte MM, et al. Comparison of neuroimaging modalities for the prediction of conversion from mild cognitive impairment to Alzheimer’s dementia. Neurobiol Aging 2014;35:143–51. [DOI] [PubMed] [Google Scholar]
- [354].Gomar JJ, Conejero-Goldberg C, Davies P, Goldberg TE. Extension and refinement of the predictive value of different classes of markers in ADNI: four-year follow-up data. Alzheimers Dement 2014; 10:704–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [355].Muñoz-Ruiz MA, Hall A, Mattila J, Koikkalainen J, Herukka SK, Vanninen R, et al. Comparing Predictors of Conversion to Alzheimer’s Disease Using the Disease State Index. Neurodegener Dis 2014;13:200–2. [DOI] [PubMed] [Google Scholar]
- [356].Barnes DE, Cenzer IS, Yaffe K, Ritchie CS, Lee SJ. A point-based tool to predict conversion from mild cognitive impairment to probable Alzheimer’s disease. Alzheimers Dement 2014;10:646–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [357].Mattila J, Koikkalainen J, Virkki A, Simonsen A, van Gils M, Waldemar G, et al. A disease state fingerprint for evaluation of Alzheimer’s disease. J Alzheimers Dis 2011;27:163–76. [DOI] [PubMed] [Google Scholar]
- [358].Hall A, Munoz-Ruiz M, Mattila J, Koikkalainen J, Tsolaki M, Mecocci P, et al. Generalizability of the disease state index prediction model for identifying patients progressing from mild cognitive impairment to Alzheimer’s disease. J Alzheimers Dis 2015;44:79–92. [DOI] [PubMed] [Google Scholar]
- [359].Dukart J, Sambataro F, Bertolino A. Accurate Prediction of Conversion to Alzheimer’s Disease using Imaging, Genetic, and Neuropsychological Biomarkers. J Alzheimers Dis 2015;49:1143–59. [DOI] [PubMed] [Google Scholar]
- [360].Lee SJ, Ritchie CS, Yaffe K, Stijacic Cenzer I, Barnes DE. A clinical index to predict progression from mild cognitive impairment to dementia due to Alzheimer’s disease. PLoS One 2014;9:e113535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [361].Marshall GA, Zoller AS, Kelly KE, Amariglio RE, Locascio JJ, Johnson KA, et al. Everyday cognition scale items that best discriminate between and predict progression from clinically normal to mild cognitive impairment. Curr Alzheimer Res 2014;11:853–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [362].Chincarini A, Bosco P, Gemme G, Esposito M, Rei L, Squarcia S, et al. Automatic temporal lobe atrophy assessment in prodromal AD: Data from the DESCRIPA study. Alzheimers Dement 2014; 10:456–67. [DOI] [PubMed] [Google Scholar]
- [363].Varon D, Barker W, Loewenstein D, Greig M, Bohorquez A, Santos I, et al. Visual rating and volumetric measurement of medial temporal atrophy in the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort: baseline diagnosis and the prediction of MCI outcome. Int J Geriatr Psychiatry 2015;30:192–200. [DOI] [PubMed] [Google Scholar]
- [364].Ferreira D, Cavallin L, Larsson EM, Muehlboeck JS, Mecocci P, Vellas B, et al. Practical cut-offs for visual rating scales of medial temporal, frontal and posterior atrophy in Alzheimer’s disease and mild cognitive impairment. J Intern Med 2015;278:277–90. [DOI] [PubMed] [Google Scholar]
- [365].Ferreira D, Cavallin L, Granberg T, Lindberg O, Aguilar C, Mecocci P, et al. Quantitative validation of a visual rating scale for frontal atrophy: associations with clinical status, APOE e4, CSF biomarkers and cognition. Eur Radiol 2016;26:2597–610. [DOI] [PubMed] [Google Scholar]
- [366].Coupe P, Fonov VS, Bernard C, Zandifar A, Eskildsen SF, Helmer C, et al. Detection of Alzheimer’s disease signature in MR images seven years before conversion to dementia: Toward an early individual prognosis. Hum Brain Mapp 2015;36:4758–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [367].Khan W, Westman E, Jones N, Wahlund LO, Mecocci P, Vellas B, et al. Automated Hippocampal Subfield Measures as Predictors of Conversion from Mild Cognitive Impairment to Alzheimer’s Disease in Two Independent Cohorts. Brain Topogr 2015;28:746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [368].Elahi S, Bachman AH, Lee SH, Sidtis JJ, Ardekani BA. Corpus callosum atrophy rate in mild cognitive impairment and prodromal Alzheimer’s disease. J Alzheimers Dis 2015;45:921–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [369].Jefferson AL, Gifford KA, Damon S, Chapman GW, Liu D, Sparling J, et al. Gray & white matter tissue contrast differentiates Mild Cognitive Impairment converters from non-converters. Brain Imaging Behav 2015;9:141–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [370].Martinez-Torteya A, Rodriguez-Rojas J, Celaya-Padilla JM, Galvan-Tejada JI, Trevino V, Tamez-Pena J. Magnetization-prepared rapid acquisition with gradient echo magnetic resonance imaging signal and texture features for the prediction of mild cognitive impairment to Alzheimer’s disease progression. J Med Imaging (Bellingham) 2014;1:031005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [371].Retico A, Bosco P, Cerello P, Fiorina E, Chincarini A, Fantacci ME. Predictive Models Based on Support Vector Machines: Whole-Brain versus Regional Analysis of Structural MRI in the Alzheimer’s Disease. J Neuroimaging 2015;25:552–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [372].Min R, Wu G, Cheng J, Wang Q, Shen D. Multi-atlas based representations for Alzheimer’s disease diagnosis. Hum Brain Mapp 2014; 35:5052–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [373].Min R, Cheng J, Price T, Wu G, Shen D. Maximum-margin based representation learning from multiple atlases for Alzheimer’s disease classification. Med Image Comput Comput Assist Interv 2014; 17:212–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [374].Liu M, Zhang D, Adeli E, Shen D. Inherent Structure-Based Multiview Learning With Multitemplate Feature Representation for Alzheimer’s Disease Diagnosis. IEEE Trans Biomed Eng 2016; 63:1473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [375].Khedher L, Ramirez J, Gorriz JM, Brahim A. Automatic classification of segmented MRI data combining Independent Component Analysis and Support Vector Machines. Stud Health Technol Inform 2014;207:271–9. [PubMed] [Google Scholar]
- [376].Guo R, Ahn M, Zhu H. Spatially Weighted Principal Component Analysis for Imaging Classification. J Comput Graph Stat 2015; 24:274–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [377].Ben Ahmed O, Mizotin M, Benois-Pineau J, Allard M, Catheline G, Ben Amar C. Alzheimer’s disease diagnosis on structural MR images using circular harmonic functions descriptors on hippocampus and posterior cingulate cortex. Comput Med Imaging Graph 2015; 44:13–25. [DOI] [PubMed] [Google Scholar]
- [378].Ziegler G, Ridgway GR, Dahnke R, Gaser C. Individualized Gaussian process-based prediction and detection of local and global gray matter abnormalities in elderly subjects. Neuroimage 2014; 97:333–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [379].Guerrero R, Wolz R, Rao AW, Rueckert D. Manifold population modeling as a neuro-imaging biomarker: application to ADNI and ADNI-GO. Neuroimage 2014;94:275–86. [DOI] [PubMed] [Google Scholar]
- [380].Ortiz A, Gorriz JM, Ramirez J, Martinez-Murcia FJ. Automatic ROI selection in structural brain MRI using SOM 3D projection. PLoS One 2014;9:e93851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [381].Li M, Oishi K, He X, Qin Y, Gao F, Mori S. An efficient approach for differentiating Alzheimer’s disease from normal elderly based on multicenter MRI using gray-level invariant features. PLoS One 2014;9:e105563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [382].Moradi E, Tohka J, Gaser C. Semi-supervised learning in MCI-to-ad conversion prediction—When is unlabeled data useful? Pattern Recognition in Neuroimaging, 2014 International Workshop on: IEEE; 2014. p. 1–4. [Google Scholar]
- [383].Doyle OM, Westman E, Marquand AF, Mecocci P, Vellas B, Tsolaki M, et al. Predicting progression of Alzheimer’s disease using ordinal regression. PLoS One 2014;9:e105542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [384].Ganz M, Greve DN, Fischl B, Konukoglu E. Relevant feature set estimation with a knock-out strategy and random forests. Neuroimage 2015;122:131–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [385].Liu M, Zhang D, Shen D. Identifying informative imaging biomarkers via tree structured sparse learning for AD diagnosis. Neuroinformatics 2014;12:381–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [386].Andrade de Oliveira A, Carthery-Goulart MT, Oliveira Junior PP, Carrettiero DC, Sato JR. Defining multivariate normative rules for healthy aging using neuroimaging and machine learning: an application to Alzheimer’s disease. J Alzheimers Dis 2015; 43:201–12. [DOI] [PubMed] [Google Scholar]
- [387].Bron EE, Smits M, Niessen WJ, Klein S. Feature Selection Based on the SVM Weight Vector for Classification of Dementia. IEEE J Biomed Health Inform 2015;19:1617–26. [DOI] [PubMed] [Google Scholar]
- [388].Hidalgo-Munoz AR, Ramirez J, Gorriz JM, Padilla P. Regions of interest computed by SVM wrapped method for Alzheimer’s disease examination from segmented MRI. Front Aging Neurosci 2014;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [389].Beheshti I, Demirel H. Probability distribution function-based classification of structural MRI for the detection of Alzheimer’s disease. Comput Biol Med 2015;64:208–16. [DOI] [PubMed] [Google Scholar]
- [390].Liu M, Zhang D, Shen D. Hierarchical fusion of features and classifier decisions for Alzheimer’s disease diagnosis. Hum Brain Mapp 2014;35:1305–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [391].Bibo S, Zhewei W, Jundong L. Distance-informed metric learning for Alzheimer’s disease staging. Conf Proc IEEE Eng Med Biol Soc 2014;2014:934–7. [DOI] [PubMed] [Google Scholar]
- [392].Tong T, Wolz R, Gao Q, Guerrero R, Hajnal JV, Rueckert D. Multiple instance learning for classification of dementia in brain MRI. Med Image Anal 2014;18:808–18. [DOI] [PubMed] [Google Scholar]
- [393].Zhang X, Hu B, Ma X, Moore P, Chen J. Ontology driven decision support for the diagnosis of mild cognitive impairment. Comput Methods Programs Biomed 2014;113:781–91. [DOI] [PubMed] [Google Scholar]
- [394].Gorji HT, Haddadnia J. A novel method for early diagnosis of Alzheimer’s disease based on pseudo Zernike moment from structural MRI. Neuroscience 2015;305:361–71. [DOI] [PubMed] [Google Scholar]
- [395].Zhang J, Zhou L, Wang L, Li W. Functional Brain Network Classification With Compact Representation of SICE Matrices. IEEE Trans Biomed Eng 2015;62:1623–34. [DOI] [PubMed] [Google Scholar]
- [396].Jiang X, Zhang X, Zhu D. Intrinsic functional component analysis via sparse representation on Alzheimer’s disease neuroimaging initiative database. Brain Connect 2014;4:575–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [397].Shen D, Zhu H. Spatially Weighted Principal Component Regression for High-Dimensional Prediction. Inf Process Med Imaging 2015; 24:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [398].Gray KR, Wolz R, Heckemann RA, Aljabar P, Hammers A, Rueckert D. Multi-region analysis of longitudinal FDG-PET for the classification of Alzheimer’s disease. Neuroimage 2012;60:221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [399].Rodrigues F, Silveira M. Longitudinal FDG-PET features for the classification of Alzheimer’s disease. Conf Proc IEEE Eng Med Biol Soc 2014;2014:1941–4. [DOI] [PubMed] [Google Scholar]
- [400].Liu S, Cai W, Wen L, Feng DD, Pujol S, Kikinis R, et al. Multi-Channel neurodegenerative pattern analysis and its application in Alzheimer’s disease characterization. Comput Med Imaging Graph 2014;38:436–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [401].Li R, Perneczky R, Yakushev I, Forster S, Kurz A, Drzezga A, et al. Gaussian Mixture Models and Model Selection for [18F] Fluorodeoxyglucose Positron Emission Tomography Classification in Alzheimer’s Disease. PLoS One 2014;10:e0122731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [402].Lange C, Suppa P, Frings L, Brenner W, Spies L, Buchert R. Optimization of Statistical Single Subject Analysis of Brain FDG PET for the Prognosis of Mild Cognitive Impairment-to-Alzheimer’s Disease Conversion. J Alzheimers Dis 2015;49:945–59. [DOI] [PubMed] [Google Scholar]
- [403].Lebedev AV, Westman E, Van Westen GJ, Kramberger MG, Lundervold A, Aarsland D, et al. Random Forest ensembles for detection and prediction of Alzheimer’s disease with a good between-cohort robustness. Neuroimage Clin 2014;6:115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [404].Moradi E, Pepe A, Gaser C, Huttunen H, Tohka J. Machine learning framework for early MRI-based Alzheimer’s conversion prediction in MCI subjects. Neuroimage 2015;104:398–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [405].Zhu X, Suk HI, Shen D. A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage 2014;100:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [406].Fiot JB, Raguet H, Risser L, Cohen LD, Fripp J, Vialard FX. Longitudinal deformation models, spatial regularizations and learning strategies to quantify Alzheimer’s disease progression. Neuroimage Clin 2014;4:718–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [407].Li H, Liu Y, Gong P, Zhang C, Ye J. Hierarchical interactions model for predicting Mild Cognitive Impairment (MCI) to Alzheimer’s Disease (AD) conversion. PLoS One 2014;9:e82450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [408].Suk HI, Shen D. Clustering-induced multi-task learning for AD/MCI classification. Med Image Comput Comput Assist Interv 2014; 17:393–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [409].Suk HI, Lee SW, Shen D. Subclass-based multi-task learning for Alzheimer’s disease diagnosis. Front Aging Neurosci 2014;6:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [410].Zhu X, Suk HI, Wang L, Lee SW, Shen D. Alzheimer’s Disease Neuroimaging I. A novel relational regularization feature selection method for joint regression and classification in AD diagnosis. Med Image Anal 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [411].Zu C, Jie B, Liu M, Chen S, Shen D, Zhang D, et al. Label-aligned multi-task feature learning for multimodal classification of Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav 2016;10:1148–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [412].Yu G, Liu Y, Shen D. Graph-guided joint prediction of class label and clinical scores for the Alzheimer’s disease. Brain Struct Funct 2016; 221:3787–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [413].Chen T, Zeng D, Wang Y. Multiple kernel learning with random effects for predicting longitudinal outcomes and data integration. Bio-metrics 2015;71:918–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [414].Cheng B, Liu M, Zhang D, Munsell BC, Shen D. Domain Transfer Learning for MCI Conversion Prediction. IEEE Trans Biomed Eng 2015;62:1805–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [415].Cheng B, Liu M, Suk HI, Shen D, Zhang D. Multimodal manifold-regularized transfer learning for MCI conversion prediction. Brain Imaging Behav 2015;9:913–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [416].Pillai PS, Leong TY. Fusing Heterogeneous Data for Alzheimer’s Disease Classification. Stud Health Technol Inform 2015; 216:731–5. [PubMed] [Google Scholar]
- [417].Suk HI, Lee SW, Shen D. Alzheimer’s Disease Neuroimaging I. Deep sparse multi-task learning for feature selection in Alzheimer’s disease diagnosis. Brain Struct Funct 2016;221:2569–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [418].Suk HI, Lee SW, Shen D. Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 2014;101:569–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [419].Li F, Tran L, Thung KH, Ji S, Shen D, Li J. A Robust Deep Model for Improved Classification of AD/MCI Patients. IEEE J Biomed Health Inform 2015;19:1610–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [420].Liu S, Liu S, Cai W, Che H, Pujol S, Kikinis R, et al. Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease. IEEE Trans Biomed Eng 2015;62:1132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [421].Sotolongo-Grau O, Pesini P, Valero S, Lafuente A, Buendia M, Perez-Grijalba V, et al. Association between cell-bound blood amyloid-beta(1–40) levels and hippocampus volume. Alzheimers Res Ther 2014;6:56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [422].Burnham SC, Faux NG, Wilson W, Laws SM, Ames D, Bedo J, et al. A blood-based predictor for neocortical Abeta burden in Alzheimer’s disease: results from the AIBL study. Mol Psychiatry 2014; 19:519–26. [DOI] [PubMed] [Google Scholar]
- [423].Nazeri A, Ganjgahi H, Roostaei T, Nichols T, Zarei M. Imaging proteomics for diagnosis, monitoring and prediction of Alzheimer’s disease. Neuroimage 2014;102 Pt 2:657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [424].McIntyre JA, Ramsey CJ, Gitter BD, Saykin AJ, Wagenknecht DR, Hyslop PA. Antiphospholipid autoantibodies as blood biomarkers for detection of early stage Alzheimer’s disease. Autoimmunity 2015;48:344–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [425].Lehallier B, Essioux L, Gayan J, Alexandridis R, Nikolcheva T, Wyss-Coray T, et al. Combined Plasma and Cerebrospinal Fluid Signature for the Prediction of Midterm Progression From Mild Cognitive Impairment to Alzheimer Disease. JAMA Neurol 2015;:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [426].Bron EE, Steketee RM, Houston GC, Oliver RA, Achterberg HC, Loog M, et al. Diagnostic classification of arterial spin labeling and structural MRI in presenile early stage dementia. Hum Brain Mapp 2014;35:4916–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [427].Prasad G, Joshi SH, Thompson PM. Optimizing brain connectivity networks for disease classification using epic. Proceedings/IEEE International Symposium on Biomedical Imaging: from nano to macro IEEE International Symposium on Biomedical Imaging. 2014;2014:834–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [428].Zhan L, Liu Y, Wang Y, Zhou J, Jahanshad N, Ye J, et al. Boosting brain connectome classification accuracy in Alzheimer’s disease using higher-order singular value decomposition. Front Neurosci 2015; 9:257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [429].Ranjan P, Ramakrishnan S. Proposal of a new tractographic feature for analysis of white matter in Alzheimer diffusion mr images. Biomed Sci Instrum 2014;50:322–7. [PubMed] [Google Scholar]
- [430].Prasad G, Joshi SH, Nir TM, Toga AW, Thompson PM. Brain connectivity and novel network measures for Alzheimer’s disease classification. Neurobiol Aging 2015;36:S121–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [431].Nir TM, Villalon-Reina JE, Prasad G, Jahanshad N, Joshi SH, Toga AW, et al. Diffusion weighted imaging-based maximum density path analysis and classification of Alzheimer’s disease. Neurobiol Aging 2015;36:S132–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [432].Wee CY, Yang S, Yap PT, Shen D. Alzheimer’s Disease Neuroimaging I. Sparse temporally dynamic resting-state functional connectivity networks for early MCI identification. Brain Imaging Behav 2016; 10:342–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [433].Zippo AG, Castiglioni I, Borsa VM, Biella GE. The Compression Flow as a Measure to Estimate the Cognitive Impairment Severity in Resting State fMRI and 18FDG-PETAlzheimer’s Disease Connectomes. Front Comput Neurosci 2015;9:148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [434].Ni H, Zhou L, Ning X, Wang L. Exploring multifractal-based features for mild Alzheimer’s disease classification. Magn Reson Med 2016; 76:259–69. [DOI] [PubMed] [Google Scholar]
- [435].Lillemark L, Sorensen L, Pai A, Dam EB, Nielsen M. Alzheimer’s Disease Neuroimaging I. Brain region’s relative proximity as marker for Alzheimer’s disease based on structural MRI. BMC Med Imaging 2014;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [436].Friedman EJ, Young K, Asif D, Jutla I, Liang M, Wilson S, et al. Directed progression brain networks in Alzheimer’s disease: properties and classification. Brain Connect 2014;4:384–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [437].Ortiz A, Munilla J, Alvarez-Illan I, Gorriz JM, Ramirez J. Exploratory graphical models of functional and structural connectivity patterns for Alzheimer’s Disease diagnosis. Front Comput Neurosci 2015;9:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [438].Phillips DJ, McGlaughlin A, Ruth D, Jager LR, Soldan A. Graph theoretic analysis of structural connectivity across the spectrum of Alzheimer’s disease: The importance of graph creation methods. Nat Commun 2015;7:377–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [439].Schwarz CG, Reid RI, Gunter JL, Senjem ML, Przybelski SA, Zuk SM, et al. Improved DTI registration allows voxel-based analysis that outperforms tract-based spatial statistics. Neuroimage 2014; 94:65–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [440].Yan J, Li T, Wang H, Huang H, Wan J, Nho K, et al. Cortical surface biomarkers for predicting cognitive outcomes using group l2,1 norm. Neurobiol Aging 2015;36:S185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [441].Zhu X, Suk HI, Shen D. Multi-modality canonical feature selection for Alzheimer’s disease diagnosis. Med Image Comput Comput Assist Interv 2014;17:162–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [442].Zhu X, Suk HI, Lee SW, Shen D. Canonical feature selection for joint regression and multi-class identification in Alzheimer’s disease diagnosis. Brain Imaging Behav 2016;10:818–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [443].Wan J, Zhang Z, Rao BD, Fang S, Yan J, Saykin AJ, et al. Identifying the neuroanatomical basis of cognitive impairment in Alzheimer’s disease by correlation- and nonlinearity-aware sparse Bayesian learning. IEEE Trans Med Imaging 2014;33:1475–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [444].Tsao S, Gajawelli N, Zhou J, Shi J, Ye J, Wang Y, et al. Evaluating the Predictive Power of Multivariate Tensor-based Morphometry in Alzheimers Disease Progression via Convex Fused Sparse Group Lasso. Proc SPIE Int Soc Opt Eng 2014;9034:90342L. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [445].Zhu X, Suk HI, Lee SW, Shen D. Subspace Regularized Sparse Multitask Learning for Multiclass Neurodegenerative Disease Identification. IEEE Trans Biomed Eng 2016;63:607–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [446].Mendelson AF, Zuluaga MA, Thurfjell L, Hutton BF, Ourselin S. The empirical variance estimator for computer aided diagnosis: lessons for algorithm validation. Med Image Comput Comput Assist Interv 2014;17:236–43. [DOI] [PubMed] [Google Scholar]
- [447].Dong T, Tian L. Confidence Interval Estimation for Sensitivity to the Early Diseased Stage Based on Empirical Likelihood. J Biopharm Stat 2015;25:1215–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [448].Dong T, Attwood K, Hutson A, Liu S, Tian L. A new diagnostic accuracy measure and cut-point selection criterion. Stat Methods Med Res 2015. [DOI] [PubMed] [Google Scholar]
- [449].Falahati F, Ferreira D, Soininen H, Mecocci P, Vellas B, Tsolaki M, et al. The Effect of Age Correction on Multivariate Classification in Alzheimer’s Disease, with a Focus on the Characteristics of Incorrectly and Correctly Classified Subjects. Brain Topogr 2016; 29:296–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [450].Coart E, Barrado LG, Duits FH, Scheltens P, van der Flier WM, Teunissen CE, et al. Correcting for the Absence of a Gold Standard Improves Diagnostic Accuracy of Biomarkers in Alzheimer’s Disease. J Alzheimers Dis 2015;46:889–99. [DOI] [PubMed] [Google Scholar]
- [451].Garcia Barrado L, Coart E, Burzykowski T. Development of a diagnostic test based on multiple continuous biomarkers with an imperfect reference test. Stat Med 2016;35:595–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [452].Li S, Ning Y. Estimation of covariate-specific time-dependent ROC curves in the presence of missing biomarkers. Biometrics 2015; 71:666–76. [DOI] [PubMed] [Google Scholar]
- [453].Dubey R, Zhou J, Wang Y, Thompson PM, Ye J. Analysis of sampling techniques for imbalanced data: An n = 648 ADNI study. Neuroimage 2014;87:220–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [454].Xiang S, Yuan L, Fan W, Wang Y, Thompson PM, Ye J, et al. Bi-level multi-source learning for heterogeneous block-wise missing data. Neuroimage 2014;102 Pt 1:192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [455].Thung KH, Wee CY, Yap PT, Shen D. Neurodegenerative disease diagnosis using incomplete multi-modality data via matrix shrinkage and completion. Neuroimage 2014;91:386–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [456].Yu G, Liu Y, Thung KH, Shen D. Multi-task linear programming discriminant analysis for the identification of progressive MCI individuals. PLoS One 2014;9:e96458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [457].Li R, Zhang W, Suk HI, Wang L, Li J, Shen D, et al. Deep learning based imaging data completion for improved brain disease diagnosis. Med Image Comput Comput Assist Interv 2014;17:305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [458].Shu H, Nan B, Koeppe R. Multiple testing for neuroimaging via hidden Markov random field. Biometrics 2015;71:741–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [459].Samtani MN, Raghavan N, Novak G, Nandy P, Narayan VA. Disease progression model for Clinical Dementia Rating-Sum of Boxes in mild cognitive impairment and Alzheimer’s subjects from the Alzheimer’s Disease Neuroimaging Initiative. Neuropsychiatr Dis Treat 2014;10:929–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [460].Ito K, Hutmacher MM. Predicting the time to clinically worsening in mild cognitive impairment patients and its utility in clinical trial design by modeling a longitudinal clinical dementia rating sum of boxes from the ADNI database. J Alzheimers Dis 2014;40:967–79. [DOI] [PubMed] [Google Scholar]
- [461].Dodge HH, Zhu J, Harvey D, Saito N, Silbert LC, Kaye JA, et al. Biomarker progressions explain higher variability in stage-specific cognitive decline than baseline values in Alzheimer disease. Alzheimers Dement 2014;10:690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [462].Sperling RA, Rentz DM, Johnson KA, Karlawish J, Donohue M, Salmon DP, et al. The A4 study: stopping AD before symptoms begin? Sci Transl Med 2014;6:228fs13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [463].Schmidt ME, Chiao P, Klein G, Matthews D, Thurfjell L, Cole PE, et al. The influence of biological and technical factors on quantitative analysis of amyloid PET: Points to consider and recommendations for controlling variability in longitudinal data. Alzheimers Dement 2015;11:1050–68. [DOI] [PubMed] [Google Scholar]
- [464].Carbonell F, Zijdenbos AP, Charil A, Grand’Maison M, Bedell BJ. Optimal Target Region for Subject Classification on the Basis of Amyloid PET Images. J Nucl Med 2015;56:1351–8. [DOI] [PubMed] [Google Scholar]
- [465].Schreiber S, Landau SM, Fero A, Schreiber F, Jagust WJ. Comparison of Visual and Quantitative Florbetapir F 18 Positron Emission Tomography Analysis in Predicting Mild Cognitive Impairment Outcomes. JAMA Neurol 2015;72:1183–90. [DOI] [PubMed] [Google Scholar]
- [466].Hutton C, Declerck J, Mintun MA, Pontecorvo MJ, Devous MD Sr, Joshi AD. Quantification of 18F-florbetapir PET: comparison of two analysis methods. Eur J Nucl Med Mol Imaging 2015; 42:725–32. [DOI] [PubMed] [Google Scholar]
- [467].Shokouhi S, Claassen D, Kang H, Ding Z, Rogers B, Mishra A, et al. Longitudinal progression of cognitive decline correlates with changes in the spatial pattern of brain 18F-FDG PET. J Nucl Med 2013;54:1564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [468].Shokouhi S, Rogers BP, Kang H, Ding Z, Claassen DO, McKay JW, et al. Modeling clustered activity increase in, amyloid-beta positron emission tomographic images with statistical descriptors. Clin Interv Aging 2015;10:759–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [469].Hill DL, Schwarz AJ, Isaac M, Pani L, Vamvakas S, Hemmings R, et al. Coalition Against Major Diseases/European Medicines Agency biomarker qualification of hippocampal volume for enrichment of clinical trials in predementia stages of Alzheimer’s disease. Alzheimers Dement 2014;10:421–429.e3. [DOI] [PubMed] [Google Scholar]
- [470].Yu P, Sun J, Wolz R, Stephenson D, Brewer J, Fox NC, et al. Operationalizing hippocampal volume as an enrichment biomarker for amnestic mild cognitive impairment trials: effect of algorithm, test-retest variability, and cut point on trial cost, duration, and sample size. Neurobiol Aging 2014;35:808–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [471].Ithapu VK, Singh V, Okonkwo OC, Chappell RJ, Dowling NM, Johnson SC. Imaging-based enrichment criteria using deep learning algorithms for efficient clinical trials in mild cognitive impairment. Alzheimers Dement 2015;11:1489–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [472].Huang Y, Ito K, Billing CB Jr, Anziano RJ. Development of a straightforward and sensitive scale for MCI and early AD clinical trials. Alzheimers Dement 2015;11:404–14. [DOI] [PubMed] [Google Scholar]
- [473].Donohue MC, Sperling RA, Salmon DP, Rentz DM, Raman R, Thomas RG, et al. The preclinical Alzheimer cognitive composite: measuring amyloid-related decline. JAMA Neurol 2014;71:961–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [474].Caroli A, Prestia A, Wade S, Chen K, Ayutyanont N, Landau SM, et al. Alzheimer Disease Biomarkers as Outcome Measures for Clinical Trials in MCI. Alzheimer Dis Assoc Disord 2015;29:101–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [475].Leung KK, Clarkson MJ, Bartlett JW, Clegg S, Jack CR Jr, Weiner MW, et al. Robust atrophy rate measurement in Alzheimer’s disease using multi-site serial MRI: tissue-specific intensity normalization and parameter selection. Neuroimage 2010;50:516–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [476].Gutman BA, Wang Y, Yanovsky I, Hua X, Toga AW, Jack CR Jr, et al. Empowering imaging biomarkers of Alzheimer’s disease. Neurobiol Aging 2015;36:S69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [477].Gutman BA, Hua X, Rajagopalan P, Chou YY, Wang Y, Yanovsky I, et al. Maximizing power to track Alzheimer’s disease and MCI progression by LDA-based weighting of longitudinal ventricular surface features. Neuroimage 2013;70:386–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [478].Insel PS, Mattsson N, Mackin RS, Kornak J, Nosheny R, Tosun-Turgut D, et al. Biomarkers and cognitive endpoints to optimize trials in Alzheimer’s disease. Ann Clin Transl Neurol 2015;2:534–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [479].Hua X, Ching CR, Mezher A, Gutman BA, Hibar DP, Bhatt P, et al. MRI-based brain atrophy rates in ADNI phase 2: acceleration and enrichment considerations for clinical trials. Neurobiol Aging 2016;37:26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [480].Hua X, Hibar DP, Ching CR, Boyle CP, Rajagopalan P, Gutman BA, et al. Unbiased tensor-based morphometry: improved robustness and sample size estimates for Alzheimer’s disease clinical trials. Neuroimage 2013;66:648–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [481].Berres M, Kukull WA, Miserez AR, Monsch AU, Monsell SE, Spiegel R. A Novel Study Paradigm for Long-term Prevention Trials in Alzheimer Disease: The Placebo Group Simulation Approach (PGSA): Application to MCI data from the NACC database. J Prev Alzheimers Dis 2014;1:99–109. [PMC free article] [PubMed] [Google Scholar]
- [482].Jack CR Jr, Knopman DS, Jagust WJ, Shaw LM, Aisen PS, Weiner MW, et al. Hypothetical model of dynamic biomarkers of the Alzheimer’s pathological cascade. Lancet Neurol 2010; 9:119–28. [DOI] [PMC free article] [PubMed] [Google Scholar]