Abstract
Atrial fibrillation (AF), defined by disorganized atrial cardiac rhythm, is the most prevalent cardiac arrhythmia worldwide. Recent genetic studies have highlighted a major heritable component and identified numerous loci associated with AF risk, including the cardiogenic transcription factor genes TBX5, GATA4, and NKX2-5. We report that Tbx5 and Gata4 interact with opposite signs for atrial rhythm controls compared with cardiac development. Using mouse genetics, we found that AF pathophysiology caused by Tbx5 haploinsufficiency, including atrial arrhythmia susceptibility, prolonged action potential duration, and ectopic cardiomyocyte depolarizations, were all rescued by Gata4 haploinsufficiency. In contrast, Nkx2-5 haploinsufficiency showed no combinatorial effect. The molecular basis of the TBX5/GATA4 interaction included normalization of intra-cardiomyocyte calcium flux and expression of calcium channel genes Atp2a2 and Ryr2. Furthermore, GATA4 and TBX5 showed antagonistic interactions on an Ryr2 enhancer. Atrial rhythm instability caused by Tbx5 haploinsufficiency was rescued by a decreased dose of phospholamban, a sarco/endoplasmic reticulum Ca2+-ATPase inhibitor, consistent with a role for decreased sarcoplasmic reticulum calcium flux in Tbx5-dependent AF susceptibility. This work defines a link between Tbx5 dose, sarcoplasmic reticulum calcium flux, and AF propensity. The unexpected interactions between Tbx5 and Gata4 in atrial rhythm control suggest that evaluating specific interactions between genetic risk loci will be necessary for ascertaining personalized risk from genetic association data.
Keywords: Cardiology, Genetics
Keywords: Arrhythmias, Calcium, Transcription
Introduction
Atrial fibrillation (AF) is the most common sustained cardiac arrhythmia, affecting more than 7 million Americans and 33 million people worldwide (1). AF is characterized by an irregular pattern of atrial depolarization, resulting in rapid and disorganized atrial conduction and lack of effective atrial chamber contraction. The rhythm abnormality in patients with AF manifests with circulatory deficits and systemic thromboembolism that greatly increase morbidity and mortality. Because age is an independent risk factor for AF, its prevalence is expected to rise significantly as the population ages. AF has become a major clinical and economic burden, owing to the limitations and side effects associated with current AF therapies. Although AF most often manifests in the context of pre-existing cardiac pathologies, such as hypertension and cardiomyopathy, idiopathic or lone AF forms have indicated a heritable component (2). Genome-wide association studies (GWAS) have to date identified more than 100 AF-associated loci, including many transcription factor (TF) loci, suggesting that transcriptional control of atrial rhythm is an important mediator of AF risk (3, 4).
Recent work has illuminated a role for abnormal cardiomyocyte calcium (Ca2+) handling in the cellular pathophysiology of AF. The current paradigm of AF causation describes ectopic (triggered) atrial activity, mediated by early and delayed afterdepolarization (EAD and DAD) events and a fibrillogenic substrate that propagates the abnormal triggers causing arrhythmia. EADs and DADs have been associated with abnormal cardiomyocyte Ca2+ handling, including RYR2 dysfunction, reduced sarco/endoplasmic reticulum Ca2+-ATPase 2 (SERCA2) (encoded by ATP2A2) activity, and/or increased Na+/Ca2+ exchanger (NCX) activity (5–9). It remains to be defined how alterations of RYR2, ATP2A2, or NCX expression in AF models contribute to AF risk.
GWAS and familial inheritance have implicated transcription factor genes in AF pathogenesis, including the cardiogenic transcription factors (TFs) TBX5, GATA4, and NKX2-5 (2, 10). Autosomal dominant mutations in the T-box TF TBX5 cause Holt-Oram syndrome, characterized by upper limb malformations, congenital heart defects, cardiac conduction system abnormalities, and increased AF risk (11, 12). In addition, GWAS has identified common risk variants associated with PR interval and increased AF susceptibility in intergenic or intronic regions of TBX5 (13–16). Adult-specific Tbx5 deletion in mice leads to spontaneous and sustained AF, characterized by slowed conduction and decrements in the expression of ion channels linked to AF (17). TBX5 drives the atrial expression of Pitx2, and TBX5 and PITX2 comodulate the expression of cardiac rhythm effector genes, including Ryr2 and Atp2a2. These findings indicated that interactions between TBX5 and PITX2 provide tight control of an atrial rhythm gene regulatory network and that perturbation of this network triggered AF susceptibility (17). This example suggested that cardiac TFs implicated in AF by genetic association may coregulate a gene regulatory network for atrial rhythm homeostasis. GATA4 and NKX2-5 are particularly relevant candidates, given that they both interact physically and genetically with TBX5 during cardiac development. GATA4, a zinc finger transcription factor, plays critical roles in heart development and cardiomyocyte differentiation (18–20). Several GATA4 loss-of-function mutations have been reported to underlie AF susceptibility in humans (10, 21–23). GATA4 is highly expressed in adult cardiomyocytes; however, its specific role in atrial cardiac rhythm has not been investigated. NKX2-5, a homeodomain containing TF has been implicated in AF by GWAS and family studies (13, 24–26). Common variants associated with PR prolongation, a marker for increased AF risk, have also been identified close to NKX2-5 by GWAS (13). Heterozygous mutations in NKX2-5 are associated with a spectrum of congenital heart diseases (CHDs) in humans and mice (27–31). NKX2-5 also has been shown to regulate a number of target genes involved in the cardiomyocyte action potential, suggesting that it may play a direct role in AF predisposition (32).
Combinatorial interactions between TBX5, GATA4, and NKX2-5 are critical for heart development. TBX5, GATA4, and NKX2-5 physically interact, and CHD-causing (but not CHD-sparing) mutations in TBX5 abrogate these interactions (33). Mutations in GATA4 that disrupt transcriptional cooperativity with TBX5 result in CHD and impaired cardiac gene expression, leading to aberrant chromatin states and gene expression (33, 34). Tbx5/Gata4 double-heterozygous mice develop cardiac defects that are more severe than Tbx5 or Gata4 haploinsufficient mice alone, providing evidence for a cooperative interaction (35). TBX5, NKX2-5, and GATA4 synergistically activate multiple cardiac enhancers and promoters (33, 36–45). Cooperative interactions between TBX5, NKX2-5, and GATA4 on cardiac gene expression may rely on interdependent binding of these factors genome-wide, enabling co-regulation of the cardiac differentiation program (46). Removal of Tbx5 or Nkx2-5 in mouse embryonic stem cell cardiac differentiations or GATA4 in human induced pluripotent stem cardiac differentiations resulted in inappropriate distribution of the other TFs to lineage-inappropriate sites, inducing ectopic gene regulation (34, 46). This paradigm suggested that heterotypic interactions between these cardiogenic TFs may influence atrial rhythm control.
We investigated the combinatorial genetic interactions between the cardiogenic transcription factors Tbx5, Gata4, and/or Nkx2-5 in murine atrial rhythm control. We hypothesized that adult-specific combined Tbx5, Gata4, or Nkx2-5 haploinsufficiency may alter AF susceptibility and illuminate AF pathophysiology. Surprisingly, we found that Gata4 haploinsufficiency rescued the cardiac rhythm, cardiomyocyte electrophysiology, and molecular defects caused by Tbx5 haploinsufficiency. In contrast, introducing Nkx2-5 haploinsufficiency had no observed effect on the penetrance or severity of defects caused by Tbx5 haploinsufficiency alone. Gata4 haploinsufficiency rescued the calcium-handling defects and Ryr2 and Atp2a2 gene expression deficits caused by Tbx5 haploinsufficiency. We found that GATA4 negatively modulated TBX5 activation of a cis-regulatory element at Ryr2. This observation indicated that GATA4 could repress TBX5-dependent transcriptional activation in some contexts and provided a molecular model for the rescue of Tbx5 haploinsufficiency by Gata4 haploinsufficiency. We tested the hypothesis that rescue of Tbx5 haploinsufficiency was mediated by rescue of calcium homeostasis. Introduction of haploinsufficiency for phospholamban (Pln), a SERCA inhibitor, into Tbx5-haploinsufficient mice caused normalization of cardiac rhythm and rescue of SERCA activity. Rescue of the Tbx5-haploinsufficient phenotype by Gata4 deficiency illuminated a TF genetic interaction in atrial rhythm control and provided a molecular model for transcriptional control of cardiomyocyte calcium flux as a central component of atrial rhythm homeostasis.
Results
Reduced Gata4 rescues atrial arrhythmias caused by reduced Tbx5.
We previously demonstrated that adult-specific Tbx5 deletion or haploinsufficiency caused spontaneous AF or AF susceptibility, respectively, linking TBX5 dose with AF risk (17). GATA4 and NKX2-5 physically and genetically interact with TBX5 during cardiac development, and all 3 cardiogenic TFs are strongly expressed in the adult atrium and are genetically linked to AF risk in humans (Figure 1A and refs. 2, 33, 35, 41, 46–48). We therefore hypothesized that GATA4 and NKX2-5 would genetically interact with TBX5 in adult atrial rhythm control. We employed a conditional knockout strategy to establish haploinsufficiency of Gata4, Tbx5, and Nkx2-5 singly or in combination in the adult mouse, affording normal gene dosage throughout development to circumvent early lethality or structural heart defects observed with each mouse model (43, 49–51). We combined TF floxed alleles (Gata4fl/fl; or Tbx5fl/fl or Nkx2-5fl/fl) with a tamoxifen (TM)-inducible Cre recombinase allele at the Rosa26 locus (R26CreERT2) to generate adult compound haploinsufficient Gata4/Tbx5 (Gata4fl/+;Tbx5fl/+;R26CreERT2), Gata4/Nkx2-5 (Gata4fl/+;Nkx2-5fl/+;R26CreERT2), Tbx5/Nkx2-5 (Tbx5fl/+;Nkx2-5fl/+;R26CreERT2), and triple-haploinsufficient (Tbx5fl/+;Gata4fl/+;Nkx2-5fl/+;R26CreERT2) mice (28, 43, 52). All TF allelic combinations were generated and evaluated as littermates in a mixed genetic background. Mice were treated with TM at 6 weeks of age and loss of Gata4, Tbx5, and/or Nkx2-5 expression was confirmed in the left atrium by qPCR 2 weeks following TM treatment (Figure 1B and Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI124231DS1). Specifically, Gata4 decrements were not observed in Tbx5 or Nkx2-5 single heterozygotes, indicating that TBX5 or NKX2-5 alone does not regulate expression of Gata4 (Figure 1B and Supplemental Figure 1A). Similarly, Tbx5 (or Nkx2-5) expression is not regulated by GATA4 or NKX2-5 (or TBX5) alone. However, a greater reduction in Gata4 expression was observed in Tbx5/Nkx2-5 compound heterozygotes compared with their respective single-haploinsufficient controls (P = 0.195 vs Tbx5fl/+;R26CreERT2, P = 0.038 vs Nkx2-5fl/+;R26CreERT2, respectively), suggesting that TBX5 and NKX2-5 may cooperatively regulate GATA4.
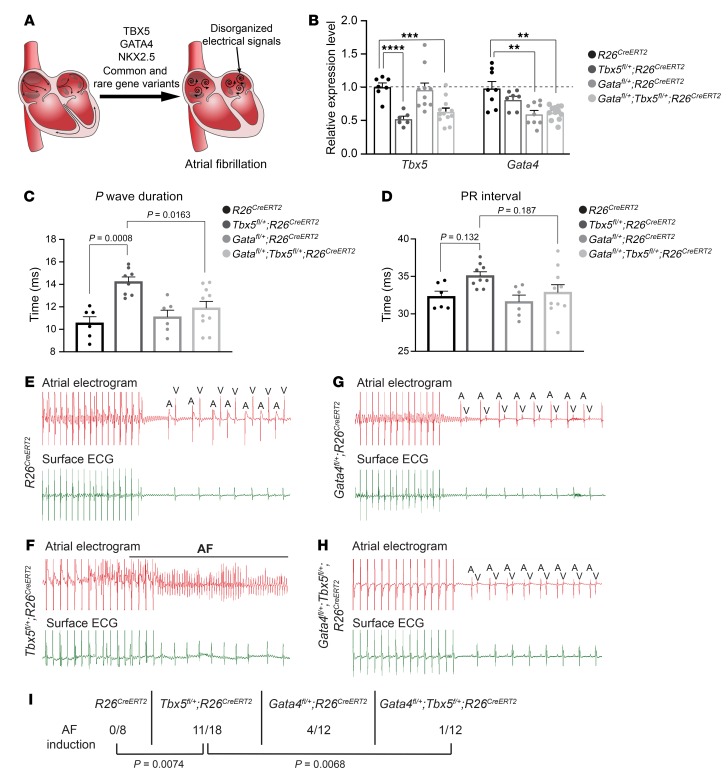
Figure 1. Gata4 haploinsufficiency rescues atrial arrhythmias caused by Tbx5 haploinsufficiency.
(A) Common and rare gene variants in the transcription factors TBX5, GATA4, and NKX2-5 have been linked to increased AF susceptibility. (B) Relative transcript expression by qPCR in the left atrium of Tbx5fl/+;R26CreERT2, Gata4fl/+;R26CreERT2, and Gata4/Tbx5 compound heterozygotes 2 weeks after TM treatment. Data are represented as means ± SEM normalized to GAPDH and relative to R26CreERT2 mice (set as 1) (n = 7–8 R26CreERT2, n = 6–7 Tbx5fl/+;R26CreERT2, n = 9–10 Gata4fl/+;R26CreERT2, n = 12 Gata4fl/+;Tbx5fl/+;R26CreERT2). Experiments were performed in technical duplicates. P values were determined by 1-way ANOVA followed by Tukey post-hoc test. **P = 0.01; ***P = 0.001; ****P = 0.0001. (C and D) P-wave duration and PR interval calculated from ambulatory telemetry ECG recordings from R26CreERT2 (n = 6), Tbx5fl/+;R26CreERT2 (n = 8–9), Gata4fl/+;R26CreERT2 (n = 6), and Gata4fl/+;Tbx5fl/+;R26CreERT2 (n = 10) mice. Tbx5fl/+;R26CreERT2 adult mice displayed significant increase in P-wave duration (C) and prolongation of the PR interval compared with R26CreERT2 littermate controls (D). P values were determined by 1-way ANOVA followed by post-hoc Tukey test. (E–H) Intracardiac atrial electrogram recordings and corresponding surface ECG of R26CreERT2 (n = 8), Tbx5fl/+;R26CreERT2 (n = 18), Gata4fl/+;R26CreERT2 (n = 12), and Gata4fl/+;Tbx5fl/+;R26CreERT2 (n = 12) mice. Tbx5 heterozygotes displayed an irregular atrial electrogram, consistent with lack of P wave on surface ECG, which is representative of AF (F). A, atrial electrical signal; V, far-field ventricular electrical signal. (I) Pacing induction by intra-atrial pacing of mice in D–G. AF was reproducibly induced in 11 of 18 Tbx5 heterozygotes (60%) in contrast to 1 of 12 Gata4/Tbx5 compound heterozygotes, indicating rescue of atrial arrhythmias. P values were determined by Fisher’s exact test.
We first confirmed that atrial rhythm was sensitive to Tbx5 dosage in these mixed-background crosses by examining adult Tbx5 heterozygotes (Tbx5fl/+;R26CreERT2). Significant prolongation of the P-wave duration representing atrial depolarization (AF-associated finding in human studies) was observed in Tbx5fl/+;R26CreERT2 mice compared with control littermates 2 weeks after TM treatment by conscious ambulatory telemetry ECG (P = 0.0008) (Figure 1C and Supplemental Table 1). The PR interval, representing the period between initiation of atrial and ventricular depolarization, was unchanged in Tbx5fl/+;R26CreERT2 compared with R26CreERT2 mice (P = 0.132) (Figure 1D and Supplemental Table 1), consistent with our previous publication (17). We further interrogated propensity to atrial arrhythmias by catheter-directed intracardiac pacing. Tbx5fl/+;R26CreERT2 adult mice were highly susceptible to AF induction, depicted by the irregular atrial electrogram, by intracardiac atrial pacing using either programmed single extra-stimulus or burst pacing. AF was reproducibly induced in 11 of 18 Tbx5fl/+;R26CreERT2 mice but in 0 of 8 R26CreERT2 mice following atrial burst pacing (P = 0.0074) (Figure 1, E, F, and I). Of these, 7 of 11 Tbx5fl/+;R26CreERT2 mice displayed atrial tachycardia (AT) and/or AF episodes lasting greater than 1,000 ms (P = 0.0128), with a mean duration of 15,963 ms. In addition, 4 of 11 Tbx5 heterozygotes (P = 0.103) displayed rapid atrial irregular rhythm for less than 1,000 ms, with a mean duration of 678 ms. Remarkably, 5 of 11 Tbx5fl/+;R26CreERT2 mice displayed spontaneous atrial arrhythmias in addition to induced irregular atrial rhythm (Supplemental Figure 2). Tbx5fl/+;R26CreERT2 mice showed normal cardiac function 2 weeks after TM treatment, with no difference in left ventricular ejection fraction compared with R26CreERT2 adult mice (Supplemental Figure 3). These findings establish that Tbx5 haploinsufficiency causes atrial conduction deficits and increased AF risk.
Atrial rhythm was not sensitive to adult-specific Gata4 or Nkx2-5 haploinsufficiency. Adult-specific Gata4 (Gata4fl/+;R26CreERT2) or Nkx2-5 (Nkx2-5fl/+;R26CreERT2) haploinsufficiency caused no abnormalities of P-wave duration (P = 0.928 R26CreERT2 vs Gata4fl/+;R26CreERT2 and P = 0.08 R26CreERT2 vs Nkx2-5fl/+;R26CreERT2) or PR interval (P = 0.956 R26CreERT2 vs Gata4fl/+;R26CreERT2 and P = 0.985 R26CreERT2 vs Nkx2-5fl/+;R26CreERT2) by conscious ambulatory telemetry ECG and showed no sign of beat-to-beat variability by Poincaré analysis in comparison with R26CreERT2 control littermates (P = 0.946 R26CreERT2 vs Gata4fl/+;R26CreERT2 and P = 0.992 R26CreERT2 vs Nkx2-5fl/+;R26CreERT2) (Figure 1, C and D, Supplemental Figure 1, and Supplemental Table 1). Unlike reduced Tbx5 dosage, Gata4 or Nkx2-5 haploinsufficient mice were not vulnerable to atrial arrhythmias by pacing induction (Figure 1, F, G, and I, and Supplemental Figure 4). Specifically, 4 of 12 Gata4fl/+;R26CreERT2 mice (P = 0.116) and 1 of 8 Nkx2-5fl/+;R26CreERT2 mice (P = 0.5) experienced AF compared with 0 of 9 R26CreERT2 mice and both showed statistically less AF inducibility than Tbx5 haploinsufficiency (P = 0.0074) (Figure 1, G and I, and Supplemental Figure 4B).
Remarkably, atrial arrhythmicity caused by reduced Tbx5 dose was rescued by reduced Gata4 dose. Specifically, the increased P-wave duration observed in Tbx5-haploinsufficient mice was rescued in combined Gata4/Tbx5-haploinsufficient mice (P = 0.0163 Gata4/Tbx5- compound heterozygote versus Tbx5fl/+;R26CreERT2) (Figure 1C). Gata4/Tbx5-compound heterozygotes were not susceptible to AF induction by intracardiac burst pacing. Only 1 of 12 Gata4fl/+;Tbx5fl/+;R26CreERT2 mice reproducibly paced into AF, demonstrating rescue of AF inducibility compared with 11 of 18 for Tbx5fl/+;R26CreERT2 mice (P = 0.0068) and no greater propensity for AF induction compared with R26CreERT2 controls (0 of 8 R26CreERT2, P = 0.999) (Figure 1, H and I).
Nkx2-5 haploinsufficiency had no discernable effect on atrial rhythm, by itself or in combination with Tbx5 or Gata4 haploinsufficiency. We analyzed adult-specific Tbx5/Nkx2-5 and Gata4/Nkx2-5 compound heterozygotes and Gata4/Tbx5/Nkx2-5 triple heterozygotes for atrial conduction deficits or arrhythmia inducibility. No significant P-wave duration or PR interval differences were detected in any of these genotypes compared with their respective single- or double- haploinsufficient controls (Supplemental Figure 1, B and C, and Supplemental Table 1). Furthermore, no significant differences in atrial arrhythmia inducibility were observed in Gata4/Nkx2-5 (P = 0.999 vs Gata4fl/+;R26CreERT2 and P = 0.999 vs Nkx2-5fl/+;R26CreERT2) or Tbx5/Nkx2-5 (P = 0.361 vs Tbx5fl/+;R26CreERT2 and P = 0.323 vs Nkx2-5fl/+;R26CreERT2) compound heterozygotes compared with their respective single- heterozygote controls or in Gata4/Tbx5/Nkx2-5 triple heterozygotes compared with Gata4/Tbx5 double-heterozygote controls (P = 0.999) (Supplemental Figure 4). Specifically, 1 of 12 Gata4fl/+;Nkx2-5fl/+;R26CreERT2 mice paced into AF compared with control littermates, suggesting that GATA4 and NKX2-5 do not interact together in the adult heart for control of atrial rhythm (Supplemental Figure 4, D and F). Tbx5fl/+;Nkx2-5fl/+;R26CreERT2 mice were susceptible to AF induction by intracardiac burst pacing, with prevalence identical to that of Tbx5 heterozygotes, indicating that TBX5 and NKX2-5 do not interact synergistically or antagonistically in the adult atrium (Supplemental Figure 4, C and F). Finally, mice lacking 1 copy of Gata4, Tbx5, and Nkx2-5 (Tbx5fl/+;Gata4fl/+;Nkx2-5fl/+;R26CreERT2) were not vulnerable to AF induction (Supplemental Figure 4, E and F). Thus, the AF susceptibility observed in Tbx5/Nkx2-5 compound heterozygotes was rescued by reducing Gata4 gene dosage, similar to Tbx5fl/+;Gata4fl/+;R26CreERT2 mice. In each genetic context, Nkx2-5 was dispensable, causing no worsening or improvement of atrial rhythm in adult mice compared with littermate controls.
Abnormal atrial electric activity observed in Tbx5 heterozygotes is normalized in Gata4/Tbx5 compound heterozygotes.
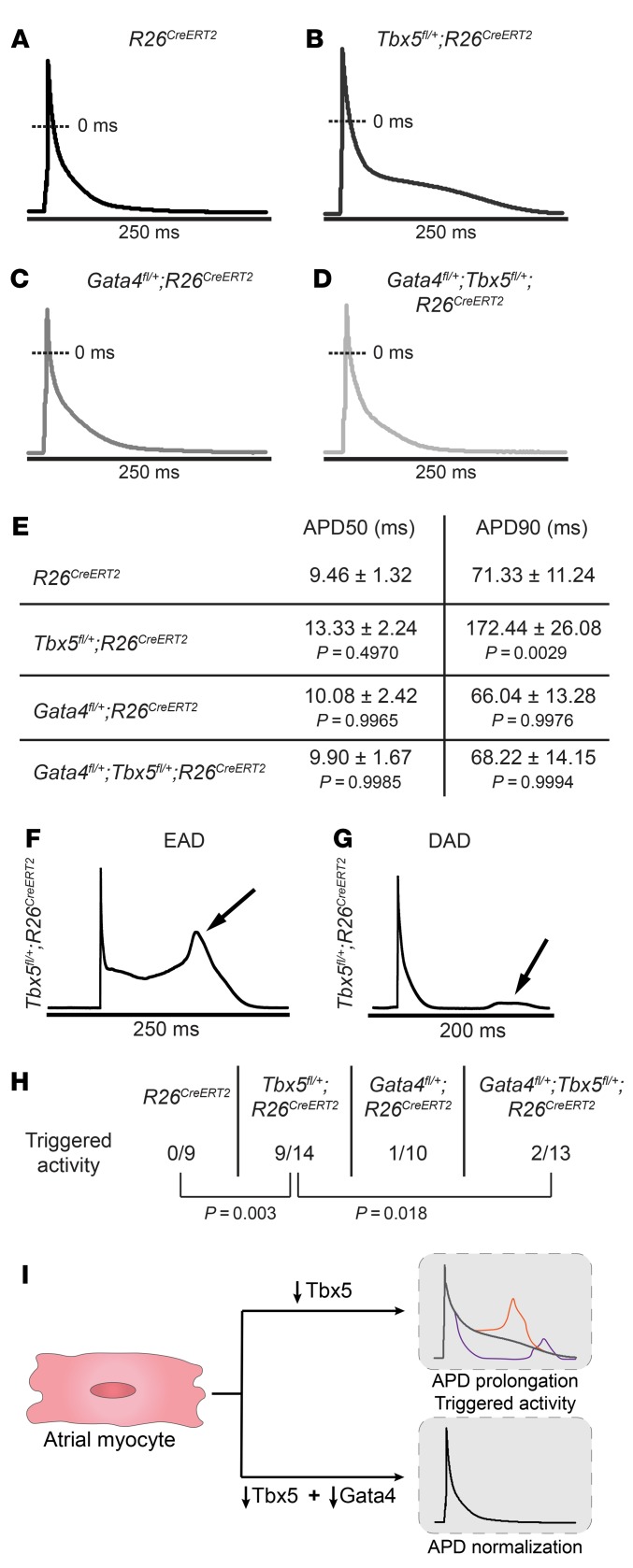
We sought to define the cellular basis by which reduced Gata4 dose rescued the atrial conduction deficits and arrhythmia propensity caused by reduced Tbx5 dose. We previously showed that adult-specific Tbx5 haploinsufficiency caused prolonged atrial cardiomyocyte (CM) action potentials (APs) and abnormal spontaneous depolarizations of atrial myocytes, electrophysiological deficits that can cause or contribute to AF (17). We hypothesized that Gata4 haploinsufficiency may rescue the cellular electrophysiology defects caused by Tbx5 haploinsufficiency. APs from atrial myocytes isolated from Tbx5fl/+;R26CreERT2 mice 2 weeks after TM treatment were significantly prolonged, specifically in phases 2 and 3 of the AP and time to 90% repolarization (APD90), compared with R26CreERT2 atrial myocytes (P = 0.0029) (Figure 2, A, B, and E, and Supplemental Table 2). EADs and DADs were frequently observed in Tbx5fl/+;R26CreERT2 atrial myocytes but never in control littermates (9 of 14 Tbx5fl/+;R26CreERT2 vs 0 of 9 R26CreERT2 atrial myocytes, P = 0.003), consistent with our previous study (Figure 2, F–H, and ref. 17). Adult-specific Gata4 haploinsufficiency caused no aberrations in AP measurements or inappropriate afterdepolarizations (Figure 2, C, E, and H, and Supplemental Table 2). Remarkably, decreased Gata4 dose rescued the cellular electrophysiology abnormalities observed in Tbx5fl/+;R26CreERT2 mice. Atrial AP duration was rescued in Gata4fl/+;Tbx5fl/+;R26CreERT2 myocytes (P = 0.0006) (Figure 2, B, D, and E). Additionally, propensity of EADs and DADs was rescued in Gata4fl/+;Tbx5fl/+;R26CreERT2 compared with Tbx5fl/+;R26CreERT2 atrial myocytes (P = 0.018), and Gata4fl/+;Tbx5fl/+;R26CreERT2 atrial myocytes showed no increased propensity for inappropriate depolarizations compared with R26CreERT2 control atrial myocytes (P = 0.493) (Figure 2H). We conclude that the cellular electrophysiology defects caused by decreased Tbx5 dose were rescued by reduced Gata4 dose (Figure 2I).
Figure 2. Abnormal atrial cardiomyocyte electrical activity caused by Tbx5 haploinsufficiency is rescued by Gata4 haploinsufficiency.
(A–D) Representative AP recordings from atrial myocytes isolated from R26CreERT2, Tbx5fl/+;R26CreERT2, Gata4fl/+;R26CreERT2, and Gata4fl/+;Tbx5fl/+;R26CreERT2 mice 2 weeks after receiving TM. Prolongation of phase 2 and 3 of the AP was observed exclusively in Tbx5fl/+;R26CreERT2 mice (B) but not in Gata4fl/+;Tbx5fl/+;R26CreERT2 littermates (D), suggesting rescue of AP abnormalities. (E) Corresponding properties of AP from mice in A–D. Tbx5 heterozygotes showed significant prolongation of APD90 in comparison with R26CreERT2 controls. These defects were completely rescued in Gata4/Tbx5 compound heterozygote mice (APD90, P = 0.006). APD50, APD at 50% repolarization; APD90, APD at 90% repolarization. Data represent mean ± SEM (n = 3–5 animals per genotype; n = 9 cardiomyocytes R26CreERT2, n = 14 Tbx5fl/+;R26CreERT2, n = 10 Gata4fl/+;R26CreERT2, and n = 13 Gata4fl/+;Tbx5fl/+;R26CreERT2). P values of APD50 and APD90 were determined by 1-way ANOVA followed by Tukey post-hoc test. (F and G) Representative tracings of EADs and DADs in Tbx5fl/+;R26CreERT2 atrial myocytes. (H) Representative abnormal depolarization events (EADs and DADs) observed in atrial myocytes. Reduced Gata4 gene dosage rescued abnormal triggers observed in Tbx5- haploinsufficient atrial myocytes. Total number of ectopic depolarization events were recorded in R26CreERT2 (n = 9), Tbx5fl/+;R26CreERT2 (n = 14), Gata4fl/+;R26CreERT2 (n = 10), and Gata4fl/+;Tbx5fl/+;R26CreERT2 (n = 13) atrial myocytes from n > 4 for each genotype. P values of triggers were determined by Fisher’s exact test. (I) The cellular conduction deficits caused by Tbx5 haploinsufficiency, including AP prolongation and triggered activity and normalization by reduced Gata4 dose.
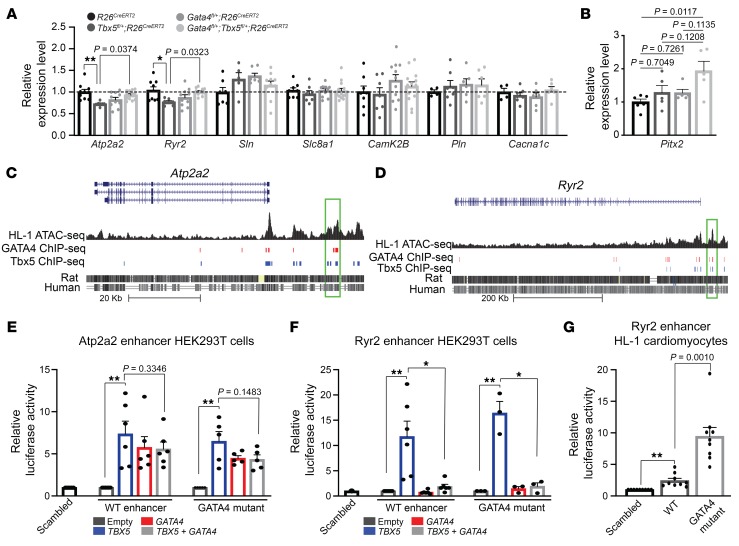
Atrial myocyte ectopic activity, implicated as a mechanism of AF induction, can result from abnormal calcium handling. We therefore investigated altered expression of genes controlling cardiomyocyte calcium flux as a potential molecular mechanism for paroxysmal AF induction in Tbx5 heterozygote mice. We focused on calcium-handling genes involved in phase 2 and 3 of the AP, given our observation of specific deficits during these AP phases in Tbx5 adult haploinsufficient mice (Figure 2). Adult-specific Tbx5 heterozygote mice showed significantly diminished left atrial expression of Atp2a2 and Ryr2 compared with R26CreERT2 mice (P = 0.0038 for Atp2a2; P = 0.0163 for Ryr2, respectively), whereas expression of other calcium-handling genes, including Sln, Pln, Slc8a1, and Cacna1c were not significantly altered (Figure 3A and Supplemental Table 3). Interestingly, Atp2a2 and Ryr2 expression was normalized in Gata4/Tbx5 compound heterozygotes (P = 0.0232 for Atp2a2; P = 0.0323 for Ryr2, respectively) (Figure 3A and Supplemental Table 3). In addition, we also assessed expression of potassium-handling genes previously linked to AF. RNA expression of Kcnj3 (P = 0.0443), Kcnj5 (P = 0.0039), and Kcnh2 (P = 0.0182) showed significant reduction in left atrial tissue of Tbx5fl/+;R26CreERT2 mice compared with control littermates (Supplemental Figure 5), whereas expression of Kcna5, Kcnd3, Kcnk2, Kcnn3, and Kcnq1 were unchanged. However, Kcnj3, Kcnj5, or Kcnh2 gene expression was not normalized in Gata4fl/+;Tbx5fl/+;R26CreERT2 mice (P = 0.7001 for Kcnj3, P = 0.4254 for Kcnj5 and P = 0.5286 for Kcnh2, respectively). Overall, these results suggested that atrial arrhythmogenesis in adult Tbx5 heterozygous mice may be mediated by altered sarcoplasmic reticulum (SR) calcium flux, mediated by reduced expression of Atp2a2 and Ryr2.
Figure 3. Antagonistic interactions between TBX5 and GATA4 on Atp2a2 and Ryr2 expression and on an Ryr2 enhancer.
(A) Relative gene expression by qPCR of known AF calcium genes and calcium-interacting proteins from left atrium of R26CreERT2 (n = 6–10) Tbx5fl/+;R26CreERT2 (n = 5–7), Gata4fl/+;R26CreERT2 (n = 7–10), and Gata4fl/+;Tbx5fl/+;R26CreERT2 (n = 9–13) mice 2 weeks after TM treatment. Tbx5 heterozygotes showed 20% decrease in Atp2a2 and Ryr2 gene expression, which was normalized in Gata4/Tbx5 compound heterozygotes. Data are normalized to GAPDH and relative to R26CreERT2. P values were determined by 1-way ANOVA followed by Tukey post-hoc test. (B) Relative transcript expression of Pitx2 by qPCR in the left atrium of Tbx5 heterozygotes, Gata4 heterozygotes, and Gata4/Tbx5 compound heterozygotes 2 weeks after TM treatment. Data are represented as means ± SEM normalized to GAPDH and relative to R26CreERT2 mice (set as 1) (n = 6 R26CreERT2, n = 5 Tbx5fl/+;R26CreERT2, n = 5 Gata4fl/+;R26CreERT2, and n = 5 Gata4fl/+;Tbx5fl/+;R26CreERT2). Experiments were performed in technical duplicates. P value was determined by 1-way ANOVA followed by post-hoc Tukey test. (C and D) Atp2a2 and Ryr2 genomic locus (Mm9) aligned with published ATAC-seq dataset from HL-1 cardiomyocytes and ChIP-seq dataset for TBX5 and GATA4. Green rectangle denotes the cis-regulatory regions with overlapping open chromatin as well as TBX5- and GATA4-binding motifs. (E–G) In vitro luciferase response assay of Atp2a2 and Ryr2 candidate enhancers in HEK293T cells cotransfected with TBX5 and/or GATA4 or HL-1 atrial cardiomyocytes and corresponding GATA mutant enhancer. Data are means ± SEM, normalized to scrambled vector. Experiments were performed in technical triplicates (n = 5 for Atp2a2 enhancer; n = 4 for Ryr2 in HEK293T cells and n = 5 for Ryr2 in HL-1 cardiomyocytes). P values were determined by 1-way ANOVA followed by Tukey post-hoc test. *P < 0.05; **P < 0.01.
We previously demonstrated that TBX5 drives atrial expression of Pitx2 and that TBX5 and PITX2 oppositely modulate the expression of cardiac rhythm effector genes, including Ryr2 and Atp2a2 (17). This finding suggested that the rescue of Tbx5 haploinsufficiency by Gata4 haploinsufficiency could occur through diminished Pitx2 levels. Pitx2 mRNA expression remained unchanged in Tbx5fl/+;R26CreERT2 and Gata4fl/+;R26CreERT2 mice 2 weeks after TM treatment, as previously described (P = 0.7049 Tbx5fl/+;R26CreERT2 compared with R26CreERT2 and P = 0.7261 Gata4fl/+;R26CreERT2 compared with R26CreERT2) (Figure 3B and ref. 17). Levels of Pitx2 mRNA were slightly higher in Gata4fl/+;Tbx5fl/+;R26CreERT2 mice compared with WT littermates (P = 0.0117); however, this increase was not significant compared with Tbx5fl/+;R26CreERT2 or Gata4fl/+;R26CreERT2 mice (P = 0.1208 vs Tbx5fl/+;R26CreERT2 and P = 0.1135 vs Gata4fl/+;R26CreERT2 mice, respectively). This observation suggested that the rescue of Tbx5 haploinsufficiency by Gata4 haploinsufficiency was not mediated by a reduction of Pitx2 expression.
We hypothesized that TBX5 and GATA4 directly coregulate expression of Atp2a2 and Ryr2. To test this hypothesis, we defined regions with overlapping chromatin occupancy for both TBX5 and GATA4 from published ChIP datasets (Figure 3, C and D, Supplemental Figures 6 and 7, and ref. 53). Candidate cis-regulatory elements (CREs) were refined by open chromatin regions from the HL-1 cardiomyocyte ATAC-seq dataset (54). We functionally interrogated these CREs for enhancer activity in the presence of TBX5 and/or GATA4. The Atp2a2 enhancer (mm9 Chr5: 122970476-122971591) demonstrated activation in response to TBX5 expression in human embryonic kidney (HEK) 293T cells, as previously described (Figure 3E and ref. 54). GATA4 similarly activated the enhancer and coexpression of TBX5, and GATA4 had no additive or synergistic effects by in vitro luciferase reporter assay in HEK293T cells (Figure 3E). In contrast, the Ryr2 enhancer was activated by TBX5 (P = 0.0013) but not GATA4 alone (P = 0.5923) in HEK293T cells (Figure 3F). Remarkably, co-expression of GATA4 suppressed TBX5-dependent transactivation of this Ryr2 enhancer (P = 0.0681 compared with TBX5 alone), suggesting antagonistic interactions of TBX5 and GATA4 on this CRE. We further tested the Ryr2 CRE in HL-1 cardiomyocytes, which possesses expression of cardiac transcription factors, including TBX5 and GATA4 (55). This enhancer previously demonstrated TBX5-dependent activity in HL-1 cells, and consistently, this enhancer alone showed activation in HL-1 cardiomyocytes (Figure 3G and ref. 54). Interestingly, mutation of the 4 GATA binding motifs within this enhancer caused an increase of enhancer activity in HL-1 cardiomyocytes (P = 0.0010 vs WT enhancer) (Figure 3G). These observations indicated that GATA4 represses TBX5-dependent activity of the Ryr2 enhancer.
Reduced SR load and SERCA function caused by Tbx5 haploinsufficiency is rescued by Gata4 haploinsufficiency.
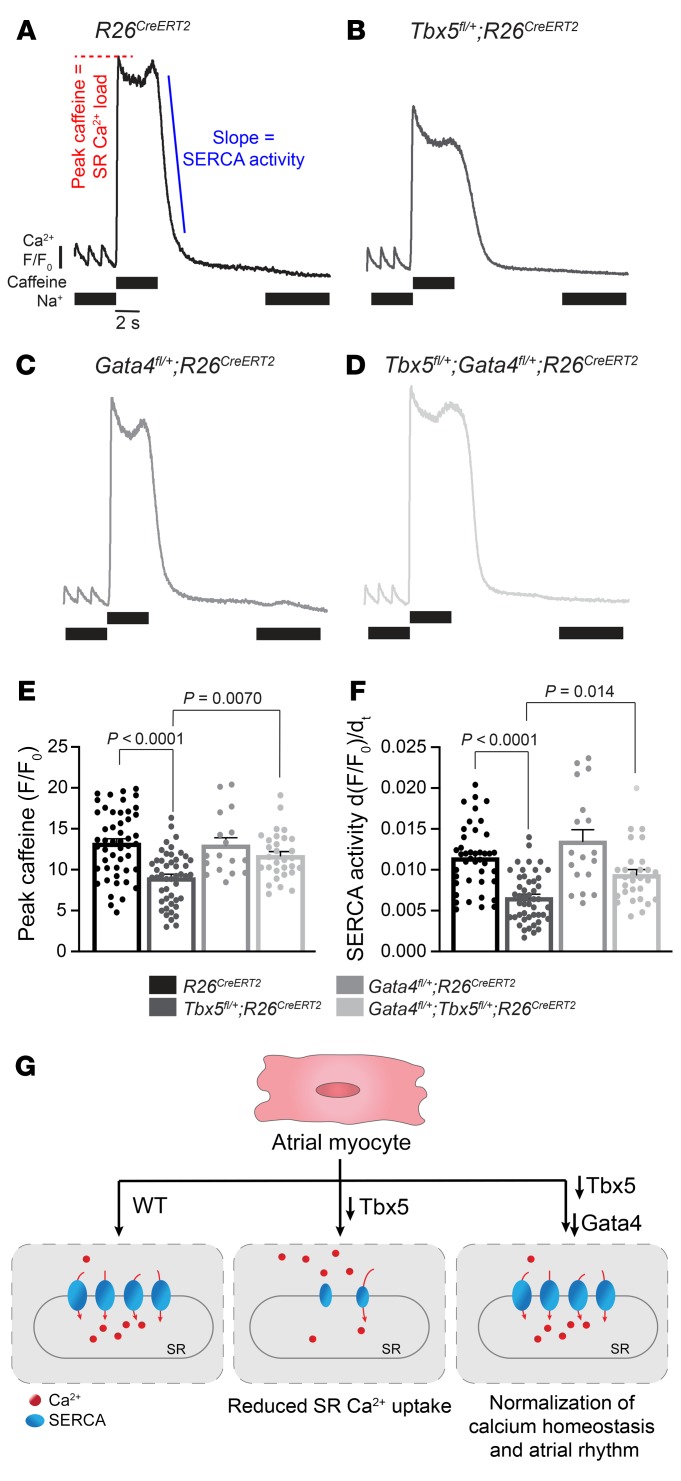
Abnormal CM membrane depolarizations, including DADs, can be induced by Ca2+-driven Na+/Ca2+ exchanger (NCX) activity in the setting of reduced SERCA-mediated SR Ca2+ uptake, causing membrane depolarization (56). We therefore hypothesized that the ectopic depolarizations caused by Tbx5 haploinsufficiency would be associated with depressed SERCA function and slowed SR Ca2+ uptake. To test this hypothesis, we measured SR load and SERCA activity in Tbx5 heterozygote atrial myocytes, 2 weeks after TM treatment. We used nonfluorescent acetoxymethyl ester (Fluo-4 AM) to study cytosolic calcium-handling kinetics and Fura-2 acetoxymethyl ester (Fura-2 AM) to study diastolic calcium levels. We defined SR Ca2+ uptake by loading myocytes with Fluo-4 AM and pacing with a train-of-field stimuli to achieve a steady-state load followed by application of caffeine to synchronize ryanodine receptor opening, resulting in a maximum release of SR calcium into the cytosol (Figure 4, A–D). We observed reduced caffeine-induced calcium-transient amplitudes in Tbx5fl/+;R26CreERT2 in comparison with R26CreERT2 mice, indicating reduced SR calcium levels (P < 0.0001) (Figure 4, A, B, and E). To assess SERCA activity, we measured [Ca]i decay rate after caffeine in the absence of external sodium (NCX inactive), allowing measurement of isolated SR uptake. Consistent with reduced left atrial expression of Atp2a2, we observed decreased SERCA activity in Tbx5-haploinsufficient mice (P < 0.0001) (Figure 4, A, B, and F, and Supplemental Figure 8). Furthermore, no significant differences were observed in resting cytosolic calcium between Tbx5fl/+;R26CreERT2 and R26CreERT2 cardiomyocytes (1.001 Tbx5fl/+;R26CreERT2 vs 0.992 R26CreERT2, P = 0.515) (Supplemental Figure 8). This finding suggests that the observed differences in SERCA function are not due to alterations in cytosolic calcium levels or calcium buffering capacity, but rather to TBX5-depedent regulation of SERCA. Overall, our findings indicate that Tbx5 haploinsufficiency causes reduced SERCA function, providing a molecular mechanism for ectopic CM depolarizations (Figure 4G).
Figure 4. Reduced SERCA function caused by Tbx5 haploinsufficiency is rescued by Gata4 haploinsufficiency in atrial myocytes.
(A–D) Representative SERCA2 traces after steady-state field stimulation at 1 Hz and application of caffeine in the absence of Nao provides a measurement of SR load. Removal of caffeine in the absence of external Nao provides a measure of SERCA2-mediated SR calcium uptake. (E) SR load, determined from peak caffeine transients was diminished in Tbx5fl/+;R26CreERT2 compared with R26CreERT2 mice and was completely rescued in Gata4fl/+;Tbx5fl/+;R26CreERT2 mice (n = 49 R26CreERT2, n = 47 Tbx5fl/+;R26CreERT2, n = 18 Gata4fl/+;R26CreERT2 and n = 30 Gata4fl/+;Tbx5fl/+;R26CreERT2 atrial myocytes from 3–5 mice for each genotype). P values were determined by 1-way ANOVA followed by post-hoc Tukey test. (F) SERCA activity determined from the maximal rate of calcium decay was diminished in Tbx5fl/+;R26CreERT2 compared with R26CreERT2 and normalized by Gata4 haploinsufficiency. P values were determined by 1-way ANOVA followed by post-hoc Tukey test. (G) Calcium homeostasis in control atrial myocytes is disrupted by Tbx5 haploinsufficiency and rescued by decreasing Gata4 gene dosage.
Decreased Gata4 dose rescued abnormal SERCA function observed in Tbx5-haploinsufficient mice. Specifically, SR calcium load in Gata4fl/+;Tbx5fl/+;R26CreERT2 myocytes was not different from R26CreERT2 control cardiomyocytes (P = 0.312), and Gata4fl/+;Tbx5fl/+;R26CreERT2 myocytes showed a significantly increased SR load compared with Tbx5fl/+;R26CreERT2 cardiomyocytes (P = 0.0070) (Figure 4, B, D, and E). Furthermore, SERCA activity was normalized in Gata4fl/+;Tbx5fl/+;R26CreERT2 myocytes as observed by a normal decay rate upon removal of caffeine, compared with Tbx5fl/+;R26CreERT2 cardiomyocytes (P = 0.0145) (Figure 4, B, D, and F). Additional analysis of the calcium dependence of SERCA activity showed decreased activity at all levels of cytosolic calcium in Tbx5fl/+;R26CreERT2 compared with R26CreERT2, and this was normalized in Gata4fl/+;Tbx5fl/+;R26CreERT2 mice (Supplemental Figure 8). We conclude that the atrial calcium homeostasis defects caused by reduced Tbx5 dose were rescued by reduced Gata4 dose, providing a molecular mechanism for the physiological rescue (Figure 4G).
Modulation of SERCA function eliminates TF-driven arrhythmogenic phenotype.
The observation that reduced Gata4 dose rescued the defects caused by a reduced Tbx5 dose at the level of cardiac rhythm control, cellular electrophysiology, gene expression, and reduced SR calcium flux suggested that the pathophysiology of reduced Tbx5 dose on atrial rhythm control may be entirely mediated by reduced SR calcium uptake.
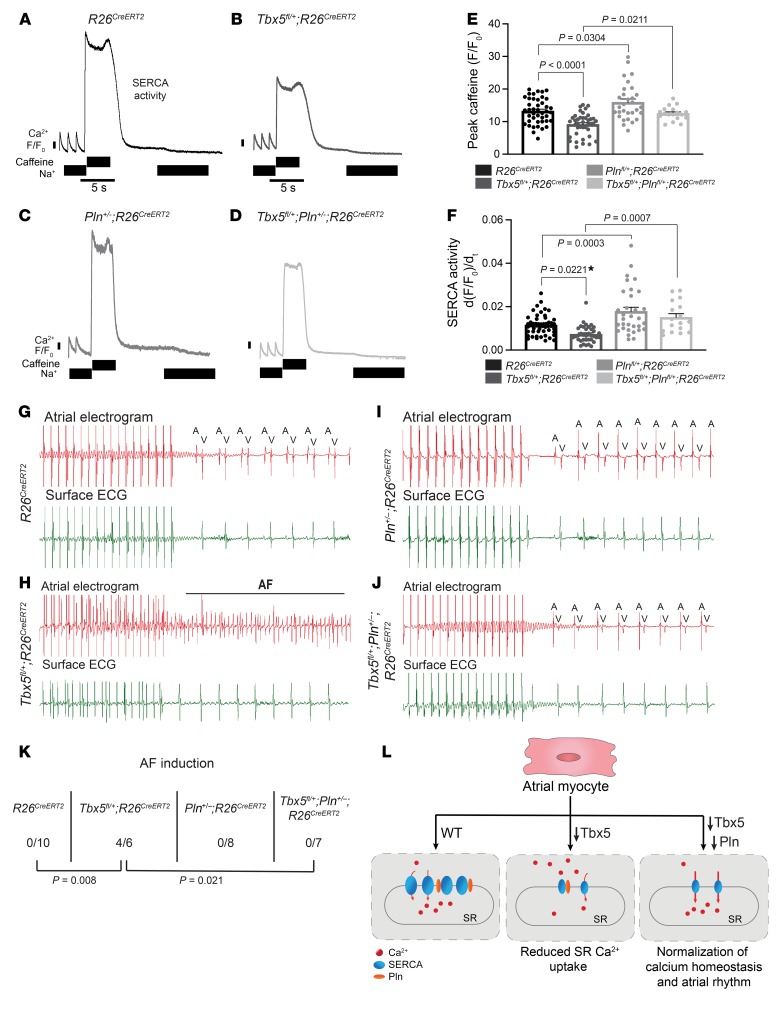
PLN inhibits SERCA activity in its nonphosphorylated state. Pln is essential for atrial calcium homeostasis, and human genetic variants at PLN associate with AF risk, suggesting that PLN activity contributes to AF susceptibility (3, 57). We hypothesized that reducing Pln gene levels may restore SERCA activity and rescue atrial calcium abnormalities caused by reduced Tbx5 dose. We compared SR uptake, SERCA function, and AF inducibility in adult-specific littermate Tbx5-haploinsufficient, Pln-haploinsufficient or Tbx5/Pln double-haploinsufficient mice 2 weeks after TM treatment. We observed depressed SR load (P < 0.0001) and SERCA activity (P = 0.0221) in atrial cardiomyocytes from Tbx5fl/+;R26CreERT2 mice compared with those from R26CreERT2 littermate controls, as expected (Figure 5, A, B, E, and F). Pln heterozygote atrial cardiomyocytes showed a significant increase in SR load (P = 0.0304) and SERCA function (P = 0.0003) compared with those from R26CreERT2 (Figure 5, A, C, E, and F). Both SR load (P = 0.0211) and SERCA activity (P = 0.0007) were normalized in Tbx5/Pln compound heterozygotes (Figure 5L).
Figure 5. Normalization of SERCA2 function eliminates AF susceptibility in Tbx5fl/+;R26CreERT2.
(A–D) Representative SERCA2 traces of R26CreERT2 (A), Tbx5fl/+;R26CreERT2 (B), Pln+/–;R26CreERT2 (C), and Tbx5fl/+;Pln+/–;R26CreERT2 (D) atrial myocytes. (E and F) Representative SR load and SERCA2 measurements from A–D. SR load (E) and SERCA2 function (F) was low in Tbx5fl/+;R26CreERT2 and normalized in Tbx5/Pln compound heterozygous mice (n = 43 R26CreERT2, n = 40 Tbx5fl/+;R26CreERT2, n = 30 Pln+/–;R26CreERT2, and n = 18 Tbx5fl/+;Pln+/–;R26CreERT2) For SR load and SERCA2 measurements, 3–4 mice per genotype were analyzed. (G–J) Intracardiac atrial electrogram recordings and corresponding surface ECG of R26CreERT2 (n = 9), Tbx5fl/+;R26CreERT2 (n = 6), Pln+/–;R26CreERT2 (n = 8), and Pln+/–;Tbx5fl/+;R26CreERT2 (n = 7) mice. Tbx5 heterozygotes displayed irregular atrial electrogram, representative of AF (H). A, atrial electrical signal; V, far-field ventricular electrical signal. (K) Pacing induction by intra-atrial pacing of mice in G–J. AF was reproducibly induced in 4 of 6 Tbx5 heterozygotes (60%) in contrast to 0 of 7 Pln/Tbx5 compound heterozygotes, indicating complete rescue of atrial arrhythmias. P values were determined by Fisher’s exact test; *P < 0.05. (L) Calcium homeostasis in control atrial myocytes is disrupted by Tbx5 haploinsufficiency and rescued by decreasing Pln gene dosage.
We hypothesized that normalization of SERCA function and SR load in Tbx5/Pln compound heterozygotes may rescue the AF predisposition caused by decreased Tbx5 dose. Tbx5 heterozygotes were highly susceptible to atrial arrhythmias, consistent with our previous observations (Figure 1, E and H). AF was induced in 4 of 6 Tbx5fl/+;R26CreERT2 mice compared with 0 of 10 for R26CreERT2 controls (P = 0.008) (Figure 5, H and K). Pln heterozygotes demonstrated no AF inducibility (0 of 8 mice; P = 0.999) (Figure 5, I and K). Remarkably, Tbx5fl/+;Pln+/–;R26CreERT2 adult mice were not susceptible to AF induction by intracardiac pacing (0 of 7 Tbx5fl/+;Pln+/–;R26CreERT2 mice; P = 0.021 vs Tbx5fl/+;R26CreERT2) (Figure 5, G–K). Overall, we conclude that decreased Pln gene dose normalizes SERCA activity and rescues AF susceptibility caused by Tbx5 haploinsufficiency, indicating that diminished SERCA activity is a primary deficit causing arrhythmogenesis in the setting of Tbx5 haploinsufficiency (Figure 5L).
Discussion
We report that TBX5 and GATA4, both implicated in human AF, genetically interact in mice for atrial rhythm control with unanticipated results. Surprisingly, reducing Gata4 gene dosage rescued defects caused by Tbx5 haploinsufficiency, including atrial arrhythmias, prolonged APs, abnormal ectopic cellular depolarizations, SR Ca2+ load and SERCA calcium flux. Rescue of both P-wave prolongation and ectopic cardiomyocyte depolarizations suggests that Gata4 haploinsufficiency may rescue both the vulnerable substrate and arrhythmia trigger propensity caused by Tbx5 haploinsufficiency. The identification of SERCA function as the nexus of the Tbx5/Gata4 genetic interaction suggested the more general hypothesis that calcium flux deficits caused by Tbx5 haploinsufficiency were the primary mechanism for the observed atrial rhythm disturbance. This hypothesis is supported by the ability of the reduced dose of pln, encoding a direct-binding SERCA inhibitor, to also rescue the Tbx5 heterozygote phenotype. By providing insights into the coregulation of atrial rhythm and calcium homeostasis by TBX5 and GATA4, this work illuminates complex genetic interactions that will undergird efforts toward personalized care in human rhythm control. The work also identifies SERCA calcium flux as the pathophysiologic basis of increased AF risk caused by decreased Tbx5 dose and therefore, a possible pathophysiologic mechanism germane to AF risk more generally.
We defined a calcium-handling network underlying cardiomyocyte prolonged action potentials and ectopy observed with decreased Tbx5 dose and rescued by reduced Gata4 dose. We hypothesized that normalization of SERCA expression caused normalization of AP duration and elimination of ectopic depolarization events. Consistently, reduction in Atp2a2 expression, SERCA activity, and SR Ca2+ load observed in Tbx5 haploinsufficient mice were all rescued by Gata4 haploinsufficiency (Figure 4). Recent studies investigating the cellular mechanisms of AF have suggested that abnormal atrial calcium handling contributes to AF pathogenesis, leading to trigger formation (56, 58–60). Although SR calcium content is affected by both SERCA uptake and RYR2-mediated calcium leak, our previous study suggested that Ryr2 binding was not affected in Tbx5 knockout mice, suggesting a primary role of SERCA in TBX5-dependent calcium dysfunction (61). Consistent with this model, increased SERCA activity by reduced PLN, an SERCA inhibitor, rescued the Tbx5- haploinsufficient phenotype (Figure 5). Human PLN mutations have been previously linked to AF and cardiomyopathy (62–64). Reductions in SERCA2 expression/activity or enhancement of the inhibitory effects of PLN are also hallmarks of heart failure (HF) (65, 66). Interestingly, PLN inhibition has been shown to alleviate cardiomyopathy in several animal models and improve cardiomyocyte contractility in patients with HF (67–69). Here, we show that decreased Pln dose can rescue the arrhythmogenic phenotype caused by Tbx5 haploinsufficiency, by restoring SERCA function and normalizing SR Ca2+ content (Figures 4 and 6). Zhu et al. observed rescue of heart failure and ventricular SERCA function following ablation of PLN in Tbx5vdel/+ mice (70). TBX5 therefore regulates SERCA in both the atria and ventricles for both cardiac function and rhythm. The observation that decreased Pln normalizes SR Ca2+ uptake and SERCA activity in a mouse model of AF provides a direct molecular link between calcium flux perturbations observed in HF and AF risk, which are strongly associated, regardless of genetic background. These observations suggest that PLN inhibition and more generally, the modulation of SR calcium flux, may be considered as possible therapeutic approaches for the treatment of AF in patients with HF.
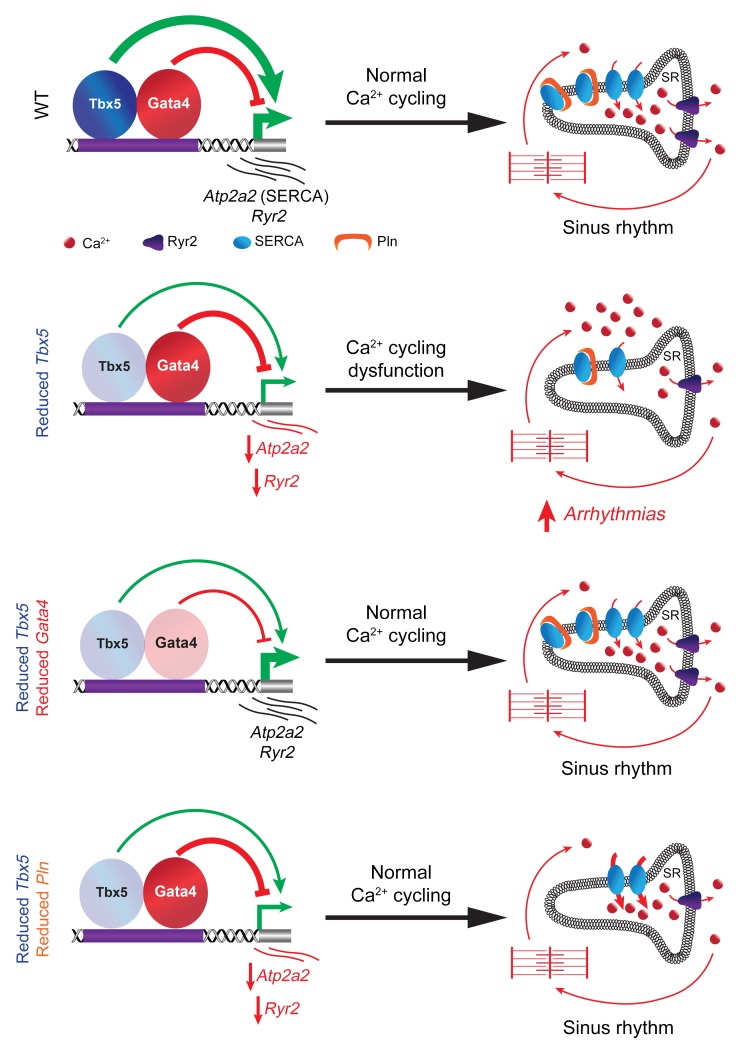
Figure 6. TBX5 and GATA4 are key regulators of atrial calcium homeostasis.
Proteins involved in Ca2+ handling of atrial myocytes. In the healthy atrium, calcium homeostasis is maintained through a tight balance between Ca2+ fluxes across the membrane. Reducing Tbx5 gene dosage in the adult heart results in depressed Atp2a2 expression and SERCA2 function, affecting SR Ca2+ influx. Abnormal Ca2+ handling leads to Ca2+ accumulation in the cytosol, contributing to ectopic atrial activity, prolonged APs, and atrial arrhythmias observed with adult-specific Tbx5 haploinsufficiency. Reducing Gata4 gene dosage rescues Atp2a2 expression, SERCA2 function and SR Ca2+ uptake, restoring sinus rhythm. SERCA2 and its inhibitory protein PLN play a fundamental role in Ca2+ handling within atrial myocytes. Reducing Pln gene dosage, as a means to modulate SERCA2 function, normalizes SERCA2 activity and SR Ca2+ uptake, resulting in restoration of sinus rhythm in combined Tbx5/Pln haploinsufficiency.
Understanding of the genetic basis of cardiac rhythm control has benefited greatly from highly powered genome-wide association studies, which have identified more than 100 loci contributing to the heritability of AF. A mechanistic and actionable understanding of how the identified loci affect cardiac rhythm control requires the transition from genetic implication to functional investigation (71). The implication of the cardiogenic TFs TBX5, GATA4, and NKX2-5 by GWAS is an exciting feature, suggesting a shared transcriptional kernel in cardiac development and adult cardiac rhythm control. In the context of cardiac development, Tbx5, Gata4, and Nkx2-5 interact genetically in a synergistic fashion, their encoded TFs physically associate, and they cooperatively drive embryonic cardiac gene expression (33–44). These detailed studies provided a clear paradigm for the cooperative interaction between these TFs in adult rhythm control. Remarkably, we find that these TFs interact very differently in the adult mouse: Tbx5 and Gata4 act oppositely, and Nkx2-5 has no appreciable impact.
Understanding how TBX5 and GATA4 levels are integrated at the molecular level to afford genetic rescue of the Tbx5 mutant phenotype by reduced Gata4 activity will be essential for AF risk prediction. We identified an enhancer at Ryr2 that molecularly integrates GATA4 and TBX5 with opposite activity. Previous work has characterized multiple promoters or enhancers coregulated by TBX5 and GATA4, and in each case, positive interactions were observed, either additive or synergistic (33, 36–41). In a few cases, however, GATA4 has been reported to possess repressive molecular activity on cardiac enhancers. For example, GATA4 recruits Hdac1/2 to deacetylate specific atrioventricular canal loci, thereby repressing AV canal identity during cardiac chamber development (37). GATA4 can also cooperate with -catenin on TCFL2-enhancers in the adult heart, to maintain normal homeostasis through repression of TCFL2-driven loci (72). Therefore, GATA4 harbors repressive potential in some contexts in the heart. Here, we identified a repressive role for GATA4 at an enhancer of Ryr2, on which GATA4 opposes TBX5-dependent activation (Figure 3). We observed that TBX5 activation and GATA4 antagonism of the Ryr2 enhancer was dependent on GATA-binding sites in HL-1 cells but not HEK293T cells. One possibility for these different results is the distinct physiological conditions of these cell lines. Because HEK cells lack endogenous expression of TBX5 and GATA4, TF overexpression may overcome the effect of the binding-site mutation because TBX5 and GATA4 physically interact (33). In contrast, HL-1 cells, possessing endogenous physiologic expression of the cardiogenic kernel of TFs, including TBX5 and GATA4, may be more sensitive to the necessity of the GATA binding sites. It remains to be elucidated how GATA4 antagonizes TBX5-dependent function in the adult atrium, perhaps by recruiting repressive chromatin-remodeling enzymes or cardiac corepressors. Nonetheless, our observations provide a model for the integration of TBX5 and GATA4 dose on gene expression by the opposite modulation of single- target gene enhancers. Understanding the molecular mechanisms underlying the opposite action of TBX5 and GATA4 on cardiac gene expression will improve our understanding of the molecular basis of adult cardiac rhythm control.
This work unveiled complex genetic interactions between genes individually implicated in AF by human genetic studies. The opposite sign with which decreased Gata4 dose affected AF risk in the context of decreased Tbx5 dose indicates the importance of unveiling specific genetic interactions between genetic risk loci for understanding the combined impact of genetic variants across genomes for disease risk prediction. Human genetic studies have been underpowered for the identification of multigenic interactions to date, highlighting the importance of gene-gene interaction studies in model systems such as those included in this study. This work suggests that interaction studies between genetic risk loci will be an essential component of understanding personalized risk from genetic association studies.
Methods
Experimental design.
This study was designed to investigate the oligogenic interaction of TFs that have been linked to AF by GWAS studies. We used murine models of conditional Tbx5 deletion, conditional Gata4 deletion, conditional Nkx2-5 deletion, and germline Pln heterozygosity for their similarities to human AF phenotypes. The number of mice per genotype depended on the experiments and is specified in figure legends. For animal studies, littermates were used as controls, and mice were grouped when appropriate. Endpoints for studies were selected according the phenotype of adult Tbx5-haploinsufficient mice. For single-cell electrophysiology and calcium flux measurements, 3–5 mice per genotype were analyzed, and a total of 10 cells was recorded for every mouse. All experiments, recordings, and analysis were performed in a blinded fashion. Outliers were excluded if samples/replicates were greater than 2 SDs from the population mean.
Transgenic mice.
Tbx5fl/fl, Gata4fl/fl, Nkx2-5fl/fl, Pln+/–, and Rosa26CreERT2 mice have all been previously described (28, 43, 52, 73, 74). Mice were maintained on a mixed genetic background, harboring 1 copy of the Cre recombinase. All experiments involving Tbx5, Gata4, NKx2-5, and Pln heterozygotes and Gata4/Tbx5, Tbx5/Nkx2-5, Gata4/Nkx2-5, and Tbx5/Pln compound heterozygotes and Tbx5/Gata4/Nkx2-5 triple heterozygotes were performed 2 weeks after TM treatment, and age-matched littermate controls (R26CreERT2) were used for comparison. A total of 200 μl of tamoxifen (20 mg/ml in corn oil) was administered by i.p. injection for 3 consecutive days at 6 to 8 weeks of age, as previously described (17).
Telemetry ECG recordings.
Ambulatory ECG studies were performed on 8- to 10-week-old mice. Mice were anesthetized using isoflurane, and telemetry transmitters (ETA-F10, Data Science International [DSI]) were implanted s.c. in the back with leads tunneled to the right upper and left lower thorax, as previously described (75). After the postimplant recovery period of 1 day, baseline recordings were collected by DSI telemetric physiological monitor system. P-wave duration, PR interval, and Poincare plots were calculated using Ponemah Physiology Platform (DSI) and custom Python script.
Intracardiac electrophysiology studies.
Animals underwent catheter-based intracardiac recordings 2 weeks after receiving TM, as previously described (17). Mice were anesthetized with isoflurane, and right atrial and ventricular electrograms as well as surface electrograms were recorded using a 1.1-F octapolar catheter (EPR-800, Millar Instruments) inserted via the right jugular vein. Programmed extra-stimulation protocols and burst pacing were used to induce AT and AF in animal subjects. Programmed right atrial extra stimulation was carried out using S1 drive trains of 80 ms followed by 5 extra stimulations at 50 ms each. Burst pacing was performed by applying a series of single extra stimulus delivered at a constant pacing rate of 15–20 ppm (900–1200 bpm). Inducibility was considered positive if 2 or more series of AT and/or fibrillation was observed following burst pacing in the same animal. Fibrillation was considered noninducible if 0 or 1 cycle or irregular atrial rhythm was observed in the animal.
Echocardiography.
Transthoracic echocardiography was performed 2 weeks after TM treatment under inhaled isoflurane, delivered via a nose cone, as previously described (76). Briefly, animals were imaged with a VisualSonics Vevo 770 machine (VisualSonics) using a 30-MHz high-frequency transducer. Body temperature was maintained using a heated imaging platform. Two-dimensional images were recorded in parasternal long- and short-axis projections, with guided M-mode recordings at the midventricular level in both views. M-mode echocardiographic images were obtained at the midpapillary muscle level in the parasternal short-axis view and measurement calculated at the same level from M- and/or B-mode images of long- or short-axis view. Left ventricular dimensions in both diastole and systole were measured from at least 3 different beats from each projection and averaged to calculate the left ventricular ejection fraction.
Calcium flux measurement.
Single-cell atrial cardiomyocytes were isolated by Langendorff perfusion with 2 mg/ml of Collagenase Type 2 at 5 ml/min. Atrial myocytes were plated on laminin-coated glass-bottom dishes and incubated at room temperature for 30 minutes prior to incubation with Fluo-4 AM (Molecular Probes/Invitrogen). Cells were then incubated with 10-μM Fluo-4 AM for 1 hour in normal Tyrode’s solution containing (in mM): 140 NaCl, 4 KCl, 10 glucose, 10 HEPES, and 1 MgCl2, 1 CaCl2, with pH 7.4 using NaOH, followed by a 10-minute perfusion wash with pre-warmed Tyrode’s solution. SERCA and NCX measurements were performed using an Olympus microscope with a x20 objective lens, a LAMBDA DG-4 power source with 488-nm excitation and 515-nm emission filters and a PMT (Microphotometer) to record whole-cell signal, with electrical field stimulation (Grass stimulator; Astro-Med) at 1 Hz. SERCA and NCX activities were measured as follows: 10-mM caffeine- containing solution was applied in the absence of extracellular sodium and the cells returned to normal Tyrode’s solution at the end of the recording. In the presence of sodium-free caffeine Tyrode’s solution, the intracellular calcium remains elevated, and the peak value is a measure of SR calcium load that was released into the cytosol.
Whole-cell electrophysiological recordings.
Action potentials were recorded using the whole-cell patch-clamp method (17). Whole-cell action potentials were recorded using an Axopatch-200B amplifier connected to a Digidata1550A acquisition system (Axon Instruments). Atrial myocytes were isolated by Langendorff perfusion, plated on laminin-coated glass- bottom dishes and incubated at room temperature for 30 minutes before recordings. Tyrode’s solution (140-mM NaCl, 4-mM KCl, 1-mM MgCl2, 1-mM CaCl2, 10-mM HEPES, and 10-mM glucose, pH 7.4 with NaOH) was used to perfuse atrial cardiomyocytes during recordings at 37°C. Internal pipette solution contained 20-mM KCl, 100-mM K-glutamate, 10-mM HEPES, 5-mM MgCl2, 10-mM NaCl, 5-mM Mg-ATP, and 0.3-mM Na-GTP. Action potentials were triggered using 0.5 nA × 2-ms current clamp pulses following liquid junction potential correction. All recordings were filtered at a frequency of 2 kHz using a built-in Bessel filter and sampled at 10 kHz. Results were analyzed using pCLAMP10 (Axon Instruments).
Diastolic calcium measurements.
Atrial myocytes were isolated by Langendorff perfusion, plated on laminin-coated glass-bottom dishes, and incubated at room temperature for 30 minutes before staining with Fura-2 AM. Cells were then incubated with 1-μM Fura-2 AM for 10 minutes in normal Tyrode’s solution. Diastolic calcium measurements were performed at a fluorescence emission of 510 nm and recorded using an Olympus IX81 Inverted Widefield Microscope. [Ca]i was calculated by the ratio of emissions following excitation at 340 nm and 380 nm.
Relative luciferase assays.
HEK293T and HL-1 cells were cotransfected as described previously (17). The Atp2a2 and Ryr2 cis-regulatory elements were amplified from C57/B6 mouse genomic DNA. The sequence was confirmed by sequencing and then cloned into the pGL4.23 enhancer luciferase response vector with a minimal promoter. Mutagenesis of the GATA4-binding motifs was performed by PCR cloning, confirmed by sequencing and then cloned into the pGL4.23 vector. Cells were co-transfected with the luciferase response vector and pRL control using lipofectamine 3000, then lysed and assayed following 48 hours using the Dual-Luciferase Reporter Assay system (Promega).
Quantitative real-time PCR.
Total RNA was extracted from the left atrial wall of 8- to 10-week-old adult mice (2 weeks after receiving TM) using Trizol (Invitrogen) combined with RNeasy mini-kit (Qiagen), according to the manufacturer’s instructions. First-strand cDNA was synthesized using the qScript cDNA synthesis kit (Quanta) according to the manufacturer’s protocol. Gene expression was assayed using the Power SYBR Green PCR Master Mix (Applied Biosystems) and run on an Applied Biosystems AB7500 machine in 96-well plates. Relative fold changes were calculated using the comparative threshold cycle method (2–ΔCt) using glyceraldehyde-3-phosphate dehydrogenase (Gapdh) as an internal control. Primer sequences used in this study are listed in Supplemental Table 3.
Statistics.
All data are represented as means ± SEM. For comparison of conscious ECG parameters, action potential duration (APD50 and APD90), SR load, and SERCA activity, a 1-way ANOVA followed by post-hoc Tukey analysis was used to test significance. For gene-expression studies and in vitro luciferase assays, a 1-way ANOVA followed by internal Student’s t test was used to test significance. AF inducibility and triggered activity (from AP measurements) counts were analyzed with a Fisher’s exact test (2-tailed).
Study approval.
All animal experiments were performed in accordance with national and institutional guidelines and were approved by the University of Chicago Institutional Animal Care and Use Committee.
Author contributions
BL and WD were involved in experimental design, execution and analysis, wrote the manuscript, and performed statistical analysis. BL performed and analyzed ambulatory ECG, whole-animal electrophysiology studies, and gene expression. WD performed and analyzed calcium flux measurements. LT performed and analyzed single-cell electrophysiology experiments. SL, KMS, and MG performed and analyzed luciferase assays. MTB performed and analyzed intracardiac electrophysiology studies. CRW performed and analyzed single-cell electrophysiology experiments and wrote the manuscript. IPM was involved in the design, execution, and analysis of experiments and wrote the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the NIH (R01 HL148719 and R01 HL147571 to IPM; K08HL129073 to MTB), the American Heart Association (7CSA33610126 to IPM) and the Leducq Foundation (to IPM). We thank Evangelia Kranias for the Phospholamban mice (NIH R01 HL26057). This research was supported in part by the NIH through resources provided by the Computation Institute and the Biological Sciences Division of the University of Chicago and Argonne National Laboratory, under grant 1S10OD018495-01.
Version 1. 10/14/2019
Electronic publication
Version 2. 11/01/2019
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2019, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2019;129(11):4937–4950.https://doi.org/10.1172/JCI124231.
Contributor Information
Brigitte Laforest, Email: laforeb27@gmail.com.
Wenli Dai, Email: wenli.dai10@gmail.com.
Leonid Tyan, Email: leo.zurzmansor@gmail.com.
Sonja Lazarevic, Email: dabizljevic@uchicago.edu.
Kaitlyn M. Shen, Email: kmshen@uchicago.edu.
Margaret Gadek, Email: mgggadek@gmail.com.
Christopher R. Weber, Email: christopher.weber@uchospitals.edu.
References
- 1.Nishida K, Datino T, Macle L, Nattel S. Atrial fibrillation ablation: translating basic mechanistic insights to the patient. J Am Coll Cardiol. 2014;64(8):823–831. doi: 10.1016/j.jacc.2014.06.1172. [DOI] [PubMed] [Google Scholar]
- 2.Tucker NR, Ellinor PT. Emerging directions in the genetics of atrial fibrillation. Circ Res. 2014;114(9):1469–1482. doi: 10.1161/CIRCRESAHA.114.302225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roselli C, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50(9):1225–1233. doi: 10.1038/s41588-018-0133-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nielsen JB, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50(9):1234–1239. doi: 10.1038/s41588-018-0171-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobrev D, Nattel S. New insights into the molecular basis of atrial fibrillation: mechanistic and therapeutic implications. Cardiovasc Res. 2011;89(4):689–691. doi: 10.1093/cvr/cvr021. [DOI] [PubMed] [Google Scholar]
- 6.El-Armouche A, et al. Molecular determinants of altered Ca2+ handling in human chronic atrial fibrillation. Circulation. 2006;114(7):670–680. doi: 10.1161/CIRCULATIONAHA.106.636845. [DOI] [PubMed] [Google Scholar]
- 7.Grandi E, et al. Human atrial action potential and Ca2+ model: sinus rhythm and chronic atrial fibrillation. Circ Res. 2011;109(9):1055–1066. doi: 10.1161/CIRCRESAHA.111.253955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neef S, et al. CaMKII-dependent diastolic SR Ca2+ leak and elevated diastolic Ca2+ levels in right atrial myocardium of patients with atrial fibrillation. Circ Res. 2010;106(6):1134–1144. doi: 10.1161/CIRCRESAHA.109.203836. [DOI] [PubMed] [Google Scholar]
- 9.Mauritz C, et al. Generation of functional murine cardiac myocytes from induced pluripotent stem cells. Circulation. 2008;118(5):507–517. doi: 10.1161/CIRCULATIONAHA.108.778795. [DOI] [PubMed] [Google Scholar]
- 10.Mahida S, Ellinor PT. New advances in the genetic basis of atrial fibrillation. J Cardiovasc Electrophysiol. 2012;23(12):1400–1406. doi: 10.1111/j.1540-8167.2012.02445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moskowitz IP, et al. The T-Box transcription factor Tbx5 is required for the patterning and maturation of the murine cardiac conduction system. Development. 2004;131(16):4107–4116. doi: 10.1242/dev.01265. [DOI] [PubMed] [Google Scholar]
- 12.Bruneau BG, et al. Chamber-specific cardiac expression of Tbx5 and heart defects in Holt-Oram syndrome. Dev Biol. 1999;211(1):100–108. doi: 10.1006/dbio.1999.9298. [DOI] [PubMed] [Google Scholar]
- 13.Pfeufer A, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42(2):153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holm H, et al. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42(2):117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 15.Tan N, et al. Weighted gene coexpression network analysis of human left atrial tissue identifies gene modules associated with atrial fibrillation. Circ Cardiovasc Genet. 2013;6(4):362–371. doi: 10.1161/CIRCGENETICS.113.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zang X, et al. SNP rs3825214 in TBX5 is associated with lone atrial fibrillation in Chinese Han population. PLoS One. 2013;8(5):e64966. doi: 10.1371/journal.pone.0064966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadadur RD, et al. Pitx2 modulates a Tbx5-dependent gene regulatory network to maintain atrial rhythm. Sci Transl Med. 2016;8(354):354ra115. doi: 10.1126/scitranslmed.aaf4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oka T, et al. Cardiac-specific deletion of Gata4 reveals its requirement for hypertrophy, compensation, and myocyte viability. Circ Res. 2006;98(6):837–845. doi: 10.1161/01.RES.0000215985.18538.c4. [DOI] [PubMed] [Google Scholar]
- 19.Bisping E, et al. Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proc Natl Acad Sci U S A. 2006;103(39):14471–14476. doi: 10.1073/pnas.0602543103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heineke J, et al. Cardiomyocyte GATA4 functions as a stress-responsive regulator of angiogenesis in the murine heart. J Clin Invest. 2007;117(11):3198–3210. doi: 10.1172/JCI32573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang YQ, et al. GATA4 loss-of-function mutations in familial atrial fibrillation. Clin Chim Acta. 2011;412(19–20):1825–1830. doi: 10.1016/j.cca.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 22.Jiang JQ, Shen FF, Fang WY, Liu X, Yang YQ. Novel GATA4 mutations in lone atrial fibrillation. Int J Mol Med. 2011;28(6):1025–1032. doi: 10.3892/ijmm.2011.783. [DOI] [PubMed] [Google Scholar]
- 23.Posch MG, et al. Mutations in the cardiac transcription factor GATA4 in patients with lone atrial fibrillation. Eur J Med Genet. 2010;53(4):201–203. doi: 10.1016/j.ejmg.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 24.Boldt LH, et al. Mutational analysis of the PITX2 and NKX2-5 genes in patients with idiopathic atrial fibrillation. Int J Cardiol. 2010;145(2):316–317. doi: 10.1016/j.ijcard.2009.11.023. [DOI] [PubMed] [Google Scholar]
- 25.Huang RT, Xue S, Xu YJ, Zhou M, Yang YQ. A novel NKX2.5 loss-of-function mutation responsible for familial atrial fibrillation. Int J Mol Med. 2013;31(5):1119–1126. doi: 10.3892/ijmm.2013.1316. [DOI] [PubMed] [Google Scholar]
- 26.Xie WH, et al. Prevalence and spectrum of Nkx2.5 mutations associated with idiopathic atrial fibrillation. Clinics (Sao Paulo) 2013;68(6):777–784. doi: 10.6061/clinics/2013(06)09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Briggs LE, et al. Perinatal loss of Nkx2-5 results in rapid conduction and contraction defects. Circ Res. 2008;103(6):580–590. doi: 10.1161/CIRCRESAHA.108.171835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pashmforoush M, et al. Nkx2-5 pathways and congenital heart disease; loss of ventricular myocyte lineage specification leads to progressive cardiomyopathy and complete heart block. Cell. 2004;117(3):373–386. doi: 10.1016/s0092-8674(04)00405-2. [DOI] [PubMed] [Google Scholar]
- 29.Chung IM, Rajakumar G. Genetics of congenital heart defects: the NKX2-5 gene, a key player. Genes (Basel) 2016;7(2):E6. doi: 10.3390/genes7020006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jay PY, et al. Nkx2-5 mutation causes anatomic hypoplasia of the cardiac conduction system. J Clin Invest. 2004;113(8):1130–1137. doi: 10.1172/JCI19846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jay PY, et al. Haploinsufficiency of the cardiac transcription factor Nkx2-5 variably affects the expression of putative target genes. FASEB J. 2005;19(11):1495–1497. doi: 10.1096/fj.04-3064fje. [DOI] [PubMed] [Google Scholar]
- 32.Furtado MB, et al. A novel conditional mouse model for Nkx2-5 reveals transcriptional regulation of cardiac ion channels. Differentiation. 2016;91(1–3):29–41. doi: 10.1016/j.diff.2015.12.003. [DOI] [PubMed] [Google Scholar]
- 33.Garg V, et al. GATA4 mutations cause human congenital heart defects and reveal an interaction with TBX5. Nature. 2003;424(6947):443–447. doi: 10.1038/nature01827. [DOI] [PubMed] [Google Scholar]
- 34.Ang YS, et al. Disease model of GATA4 mutation reveals transcription factor cooperativity in human cardiogenesis. Cell. 2016;167(7):1734–1749.e22. doi: 10.1016/j.cell.2016.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maitra M, et al. Interaction of Gata4 and Gata6 with Tbx5 is critical for normal cardiac development. Dev Biol. 2009;326(2):368–377. doi: 10.1016/j.ydbio.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nadeau M, et al. An endocardial pathway involving Tbx5, Gata4, and Nos3 required for atrial septum formation. Proc Natl Acad Sci U S A. 2010;107(45):19356–19361. doi: 10.1073/pnas.0914888107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stefanovic S, Barnett P, van Duijvenboden K, Weber D, Gessler M, Christoffels VM. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat Commun. 2014;5:3680. doi: 10.1038/ncomms4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takeuchi JK, et al. Tbx5 specifies the left/right ventricles and ventricular septum position during cardiogenesis. Development. 2003;130(24):5953–5964. doi: 10.1242/dev.00797. [DOI] [PubMed] [Google Scholar]
- 39.Ding B, et al. p204 is required for the differentiation of P19 murine embryonal carcinoma cells to beating cardiac myocytes: its expression is activated by the cardiac Gata4, Nkx2.5, and Tbx5 proteins. J Biol Chem. 2006;281(21):14882–14892. doi: 10.1074/jbc.M511747200. [DOI] [PubMed] [Google Scholar]
- 40.Linhares VL, et al. Transcriptional regulation of the murine Connexin40 promoter by cardiac factors Nkx2-5, GATA4 and Tbx5. Cardiovasc Res. 2004;64(3):402–411. doi: 10.1016/j.cardiores.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misra C, Chang SW, Basu M, Huang N, Garg V. Disruption of myocardial Gata4 and Tbx5 results in defects in cardiomyocyte proliferation and atrioventricular septation. Hum Mol Genet. 2014;23(19):5025–5035. doi: 10.1093/hmg/ddu215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kasahara H, Benson DW. Biochemical analyses of eight NKX2.5 homeodomain missense mutations causing atrioventricular block and cardiac anomalies. Cardiovasc Res. 2004;64(1):40–51. doi: 10.1016/j.cardiores.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 43.Bruneau BG, et al. A murine model of Holt-Oram syndrome defines roles of the T-box transcription factor Tbx5 in cardiogenesis and disease. Cell. 2001;106(6):709–721. doi: 10.1016/S0092-8674(01)00493-7. [DOI] [PubMed] [Google Scholar]
- 44.Moskowitz IP, et al. A molecular pathway including Id2, Tbx5, and Nkx2-5 required for cardiac conduction system development. Cell. 2007;129(7):1365–1376. doi: 10.1016/j.cell.2007.04.036. [DOI] [PubMed] [Google Scholar]
- 45.Postma AV, et al. A gain-of-function TBX5 mutation is associated with atypical Holt-Oram syndrome and paroxysmal atrial fibrillation. Circ Res. 2008;102(11):1433–1442. doi: 10.1161/CIRCRESAHA.107.168294. [DOI] [PubMed] [Google Scholar]
- 46.Luna-Zurita L, et al. Complex interdependence regulates heterotypic transcription factor distribution and coordinates cardiogenesis. Cell. 2016;164(5):999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahida S. Transcription factors and atrial fibrillation. Cardiovasc Res. 2014;101(2):194–202. doi: 10.1093/cvr/cvt261. [DOI] [PubMed] [Google Scholar]
- 48.Pradhan L, et al. Intermolecular interactions of cardiac transcription factors NKX2.5 and TBX5. Biochemistry. 2016;55(12):1702–1710. doi: 10.1021/acs.biochem.6b00171. [DOI] [PubMed] [Google Scholar]
- 49.Molkentin JD, Lin Q, Duncan SA, Olson EN. Requirement of the transcription factor GATA4 for heart tube formation and ventral morphogenesis. Genes Dev. 1997;11(8):1061–1072. doi: 10.1101/gad.11.8.1061. [DOI] [PubMed] [Google Scholar]
- 50.Kuo CT, et al. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11(8):1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- 51.Lyons I, et al. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2-5. Genes Dev. 1995;9(13):1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- 52.Watt AJ, Battle MA, Li J, Duncan SA. GATA4 is essential for formation of the proepicardium and regulates cardiogenesis. Proc Natl Acad Sci U S A. 2004;101(34):12573–12578. doi: 10.1073/pnas.0400752101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108(14):5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang XH, et al. Transcription-factor-dependent enhancer transcription defines a gene regulatory network for cardiac rhythm. Elife. 2017;6:e31683. doi: 10.7554/eLife.31683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.White SM, Constantin PE, Claycomb WC. Cardiac physiology at the cellular level: use of cultured HL-1 cardiomyocytes for studies of cardiac muscle cell structure and function. Am J Physiol Heart Circ Physiol. 2004;286(3):H823–H829. doi: 10.1152/ajpheart.00986.2003. [DOI] [PubMed] [Google Scholar]
- 56.Nattel S, Dobrev D. Electrophysiological and molecular mechanisms of paroxysmal atrial fibrillation. Nat Rev Cardiol. 2016;13(10):575–590. doi: 10.1038/nrcardio.2016.118. [DOI] [PubMed] [Google Scholar]
- 57.Christophersen IE, et al. Large-scale analyses of common and rare variants identify 12 new loci associated with atrial fibrillation. Nat Genet. 2017;49(6):946–952. doi: 10.1038/ng.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Voigt N, et al. Cellular and molecular mechanisms of atrial arrhythmogenesis in patients with paroxysmal atrial fibrillation. Circulation. 2014;129(2):145–156. doi: 10.1161/CIRCULATIONAHA.113.006641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Voigt N, et al. Enhanced sarcoplasmic reticulum Ca2+ leak and increased Na+-Ca2+ exchanger function underlie delayed afterdepolarizations in patients with chronic atrial fibrillation. Circulation. 2012;125(17):2059–2070. doi: 10.1161/CIRCULATIONAHA.111.067306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nattel S, Shiroshita-Takeshita A, Brundel BJ, Rivard L. Mechanisms of atrial fibrillation: lessons from animal models. Prog Cardiovasc Dis. 2005;48(1):9–28. doi: 10.1016/j.pcad.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 61.Dai W, et al. A calcium transport mechanism for atrial fibrillation in Tbx5-mutant mice. Elife. 2019;8:e41814. doi: 10.7554/eLife.41814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu GS, et al. A novel human R25C-phospholamban mutation is associated with super-inhibition of calcium cycling and ventricular arrhythmia. Cardiovasc Res. 2015;107(1):164–174. doi: 10.1093/cvr/cvv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt JP, et al. Dilated cardiomyopathy and heart failure caused by a mutation in phospholamban. Science. 2003;299(5611):1410–1413. doi: 10.1126/science.1081578. [DOI] [PubMed] [Google Scholar]
- 64.van der Zwaag PA, et al. Phospholamban R14del mutation in patients diagnosed with dilated cardiomyopathy or arrhythmogenic right ventricular cardiomyopathy: evidence supporting the concept of arrhythmogenic cardiomyopathy. Eur J Heart Fail. 2012;14(11):1199–1207. doi: 10.1093/eurjhf/hfs119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hayward C, Banner NR, Morley-Smith A, Lyon AR, Harding SE. The current and future landscape of SERCA gene therapy for heart failure: a clinical perspective. Hum Gene Ther. 2015;26(5):293–304. doi: 10.1089/hum.2015.018. [DOI] [PubMed] [Google Scholar]
- 66.Sikkel MB, Hayward C, MacLeod KT, Harding SE, Lyon AR. SERCA2a gene therapy in heart failure: an anti-arrhythmic positive inotrope. Br J Pharmacol. 2014;171(1):38–54. doi: 10.1111/bph.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morihara H, et al. Phospholamban inhibition by a single dose of locked nucleic acid antisense oligonucleotide improves cardiac contractility in pressure overload-induced systolic dysfunction in mice. J Cardiovasc Pharmacol Ther. 2017;22(3):273–282. doi: 10.1177/1074248416676392. [DOI] [PubMed] [Google Scholar]
- 68.Hoshijima M, et al. Chronic suppression of heart-failure progression by a pseudophosphorylated mutant of phospholamban via in vivo cardiac rAAV gene delivery. Nat Med. 2002;8(8):864–871. doi: 10.1038/nm739. [DOI] [PubMed] [Google Scholar]
- 69.Kaneko M, Hashikami K, Yamamoto S, Matsumoto H, Nishimoto T. Phospholamban ablation using CRISPR/Cas9 system improves mortality in a murine heart failure model. PLoS One. 2016;11(12):e0168486. doi: 10.1371/journal.pone.0168486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu Y, et al. Tbx5-dependent pathway regulating diastolic function in congenital heart disease. Proc Natl Acad Sci U S A. 2008;105(14):5519–5524. doi: 10.1073/pnas.0801779105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Roberts JD. Noncoding genetic variation and gene expression: deciphering the molecular drivers of genome-wide association study signals in atrial fibrillation. Circ Genom Precis Med. 2018;11(3):e002109. doi: 10.1161/CIRCGEN.118.002109. [DOI] [PubMed] [Google Scholar]
- 72.Iyer LM, et al. A context-specific cardiac β-catenin and GATA4 interaction influences TCF7L2 occupancy and remodels chromatin driving disease progression in the adult heart. Nucleic Acids Res. 2018;46(6):2850–2867. doi: 10.1093/nar/gky049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ventura A, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445(7128):661–665. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 74.Luo W, et al. Targeted ablation of the phospholamban gene is associated with markedly enhanced myocardial contractility and loss of beta-agonist stimulation. Circ Res. 1994;75(3):401–409. doi: 10.1161/01.RES.75.3.401. [DOI] [PubMed] [Google Scholar]
- 75.Wheeler MT, Allikian MJ, Heydemann A, Hadhazy M, Zarnegar S, McNally EM. Smooth muscle cell-extrinsic vascular spasm arises from cardiomyocyte degeneration in sarcoglycan-deficient cardiomyopathy. J Clin Invest. 2004;113(5):668–675. doi: 10.1172/JCI20410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arnolds DE, et al. TBX5 drives Scn5a expression to regulate cardiac conduction system function. J Clin Invest. 2012;122(7):2509–2518. doi: 10.1172/JCI62617. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.