Abstract
Recent anatomical evidence suggests a functionally significant back-projection pathway from the subiculum to CA1. Here we show that the afferent circuitry of CA1-projecting subicular neurons is biased by inputs from CA1 inhibitory neurons as well as visual cortex, but lacks input from entorhinal cortex. Efferents of the CA1-projecting subiculum neurons also target perirhinal cortex, an area strongly implicated in object-place learning. We identify a critical role for CA1-projecting subicular neurons in object-location learning and memory, and show that this projection modulates place-specific activity of CA1 neurons as well as their responses to displaced objects. Together, these experiments reveal a novel pathway by which cortical inputs, particularly those from visual cortex, reach hippocampal output region CA1, and our findings implicate this circuitry in the formation of complex spatial representations and learning of object-place associations.
The role of the hippocampus in spatial cognition and episodic memory has frequently been examined by reference to three prominent schemes by which it is anatomically organized: the tri-synaptic pathway, the transverse axis, and the longitudinal axis1–3. Historically, the greatest focus has been to examine function along the tri-synaptic pathway and to consider it as a feed-forward, unidirectional circuit. However, new viral genetic based mapping approaches for examining connectivity between structures are poised to identify novel circuits within and between hippocampus and cortex4,5. In principle, such circuits may serve to broaden the scope of neural functions carried out by interactions between hippocampus and cortex, complementing neural processing along the tri-synaptic pathway and across the transverse and longitudinal axes.
In examining how hippocampal output may impact cortical targets, emphasis is often placed on sub-region CA1 as the primary output. Yet, the subiculum (SUB) sub-region of the hippocampal formation can be considered as a continuation of the tri-synaptic or ‘canonical’ pathway through hippocampus. SUB receives dense input from CA1 as well as entorhinal cortex (EC) and sends efferents to areas such as retrosplenial cortex and perirhinal cortex6. These cortical targets are critically implicated in multiple forms of spatial cognition and memory6–8. SUB therefore stands as a second major output sub-region of hippocampus whose function remains elusive despite early work implicating SUB in spatial processing necessary for navigational performance9.
Recent studies identified a prominent return or ‘non-canonical’ projection from SUB to CA14,5. The projection is organized according to the transverse axis that splits sub-region CA1 into distal and proximal components5. The functional role of this pathway has been considered only with respect to theta-frequency oscillations that temporally organize nearly all hippocampal spiking dynamics10. These data challenge traditional views emphasizing a unidirectional projection between CA1 and SUB, and identify a feedback circuit by which SUB may directly modulate CA1 dynamics.
In the present work, we examine the synaptic circuit organization and function of the subiculum-CA1 back-projection pathway in the mouse. We apply modern anterograde and retrograde tracing techniques to show that visual cortex efferents can directly reach and impact both hippocampal sub-region CA1 and perirhinal cortex through a sub-population of SUB neurons. We use genetically targeted neuronal inactivation in conjunction with in vivo GCaMP6-based calcium imaging of CA1 in freely moving animals to show that CA1-projecting subicular neurons impact the magnitude of CA1 place cell activity. Furthermore, we examine the impact of this circuit on object-location learning within the environment. Activation and inactivation of this specific circuitry yields improvements and impairments, respectively, in encoding of object-location relationships, a key form of spatial cognition relevant to both human and animal navigation.
RESULTS
We first reveal the cortico-hippocampal circuitry associated with SUB projections to CA1. Monosynaptic rabies tracing identifies a significant SUB-CA1 back-projection pathway in mice (Fig. 1a–d), suggested previously in other mammalian species using less strict mapping methods11,12. We operationally define an input connection strength index (CSI) as the ratio of the number of presynaptic neurons in a brain region of interest (e.g., SUB) versus the number of postsynaptic (starter) neurons in CA1. Our quantitative analyses show that SUB inputs to CA1 are relatively strong with high circuit connection strengths. The CSI of subicular back-projections to distal CA1 excitatory neurons is quantitatively similar to that of medial EC inputs to proximal CA1 excitatory neurons (CSIs: SUB to CA1: 1.33 ± 0.27; EC to CA1: 1.51 ± 0.24; N = 6–7 cases). There is a weak-to-strong SUB input strength gradient along the proximal-distal axis of CA1 with an input bias to distal CA15.
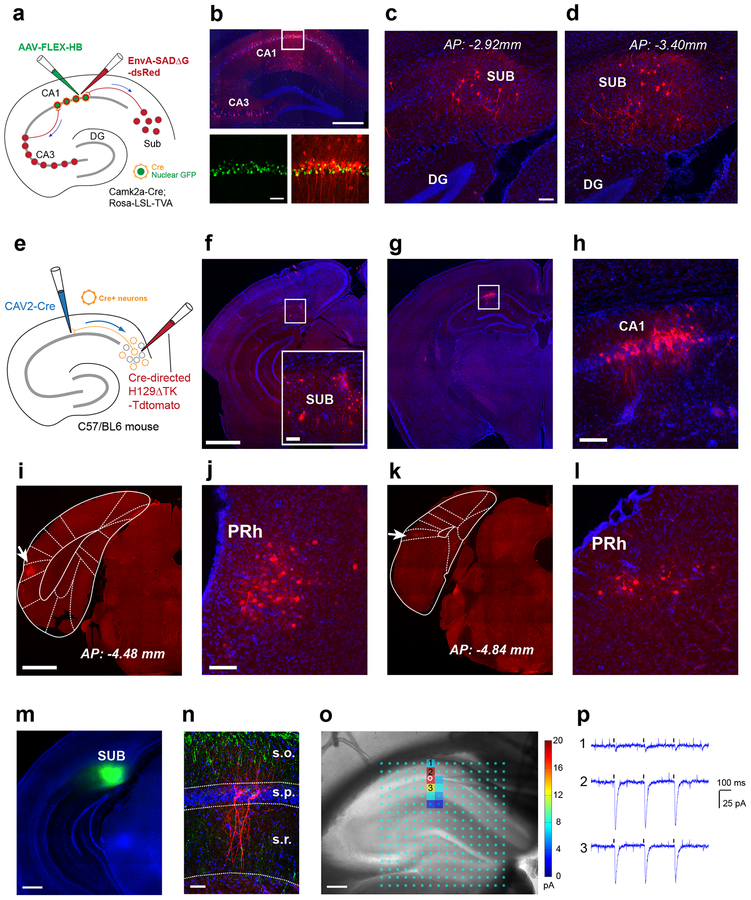
Fig. 1. The cortico-hippocampal circuitry involving a non-canonical subiculum - CA1 pathway is identified anatomically by retrograde monosynaptic rabies tracing and anterograde herpes simplex virus (H129) tracing, and verified functionally by channelrhodopsin-2 (ChR2) assisted circuit mapping.
a-d, Direct subiculum-CA1 backprojections are shown by monosynaptic retrograde rabies tracing. This experiment was independently repeated in 12 mice, each with similar results. a, The scheme for our Cre-dependent, monosynaptic rabies tracing approach (see details in the Methods). Using Camk2a-Cre; TVA mice, we map direct presynaptic input connections onto Camk2a-Cre expressing excitatory neurons in hippocampal CA1 in the intact brain. Starter neurons in dorsal hippocampal CA1 are shown (b, top panel), labeled by both EGFP and dsRed expression from both AAV and rabies infection (b, bottom panels). Their presynaptic partners (e.g., local interneurons and CA3 neurons) are labeled by the red fluorescent protein dsRed from rabies infection. The scale bars for the top and bottom panels are 500 μm and 50 μm. c-d, Retrogradely labeled subicular neurons presynaptic to CA1 excitatory neurons are seen in sections of dorsal subiculum at different anterior-posterior positions (c, AP: −2.92 mm; d, AP: −3.40 mm). Scale bar = 50 μm. e-h, Time-limited anterograde-directed HSV tracing supports subiculum-CA1 projections. This experiment was independently repeated in 5 mice, each with similar results. e, The scheme for anterograde tracing by combined use of CAV2-Cre injection in CA1 and the injection of Cre-dependent H129 (H129ΔTK-tdTomato) in SUB to map projections of CA1-projecting SUB excitatory neurons. f, H129 infected neurons at the injection site in the subiculum are shown in red; DAPI staining in blue. The scale bar on the left (1mm) applies to the lower magnification panels while the scale bar for the enlarged insert panel on the right is 200 μm. g-h, Postsynaptic neuronal labeling is robustly seen in hippocampal CA1 ipsilaterally at 48 hours post H129 viral injection. Scale bar = 200 μm in h. i-l, Besides CA1, postsynaptic neuronal labeling by H129 is seen in the perirhinal cortex (PRh) ipsilaterally. This experiment was independently repeated in 5 mice, each with similar results. i, An example of perirhinal labeling, with a white arrow pointing to the atlas aligned brain structure of PRh. The scale bar (1mm) applies to both i and k. j, An enlarged view of perirhinal neuronal labeling in i. The scale bar (200 μm) applies to both j and l. k-l, Perirhinal labeling from a different animal. m-p, Physiological mapping indicate that CA1 pyramidal neurons receive excitatory subicular inputs. This experiment was independently repeated in 8 cells from 5 mice, each with similar results. m, Spatially localized iontophoretic injection of AAV1-ChR2-Venus in the subiculum (green). n, Post-hoc verification of biocytin-filled recorded pyramidal neurons (red) along with the distribution of ChR2/Venus expressing subicular axons (green) counterstained with DAPI in a hippocampal slice. o-p, Direct subicular innervation of CA1 excitatory neurons is shown by postsynaptic current responses to local photoactivation of ChR2-expressing subicular axons in the presence of TTX and 4-AP, which block Na+ channels required for generating axonal action potentials and K+ channels critical for axonal membrane potential repolarization, respectively. o, Highly localized excitatory subicular inputs impinged onto the recorded excitatory pyramidal neuron (indicated by the white circle). The photoactivation sites (light cyan dots) are superimposed on the slice image, with the strength of evoked input sites scored with a color coded heat map for average integrated input strength within the analysis window (>10 ms to 160 ms post photostimulation), with the baseline spontaneous responses subtracted from the photostimulation response of the same site (for more details see Methods). p, Raw ChR2 photoactivation responses recorded from the pyramidal neuron in response to 3 repeated laser flashes (473 nm, 1 ms) at the oriens (1), pyramidale (2), and radiatum (3) layer of CA1, respectively.
We also used anterograde directed herpes simplex virus (H129) to identify SUB-CA1 back projections and to determine whether or not such neurons also send collateral projections to other brain regions (Fig. 1e–l). Selective labeling of CA1-projecting excitatory SUB neurons is achieved by canine adenovirus 2 (CAV2)-mediated retrograde Cre expression13. CAV2 expressing Cre (CAV2-Cre) is injected into CA1, yielding retrograde expression of Cre recombinase in CA1-projecting SUB neurons (Fig. 1e; Supplementary Fig. 1a,b). The Cre labeled CA1-projecting SUB neurons express tdTomato using the Ai9 reporter mouse (Supplementary Fig. 1c–e). Excitatory SUB neurons are predominantly labeled, as none of the CAV2-labeled cells in SUB are GABA positive and 90% of CAV2-labelled SUB neurons are immuno-positive for Ca2+/calmodulin-dependent protein kinase IIa (CaMKIIa) (Supplementary Fig. 1f). The output projections of CA1-projecting SUB neurons are mapped using Cre-dependent anterograde directed herpes simplex virus (H129 strain). Initial replication of this recombinant H129 strain14 is dependent upon Cre recombination of the expression cassette containing the codon of modified HSV thymidine kinase gene (TK) and the tdTomato reporter. After Cre recombination, the HSV viral genome permanently expresses TK and synaptically connected cells are labeled. We use the conservative time control of 48 hours post-injection to limit labeling to directly-connected postsynaptic neurons14. CA1 projections from CAV2-Cre labeled SUB neurons are confirmed by robust postsynaptic labeling of hippocampal CA1 neurons following 48 hours post H129 viral injection (Fig. 1f–h). Efferents of the CA1-projecting SUB neurons also target perirhinal cortex (Fig. 1i–l), an area strongly implicated in object-place learning15.
The neurophysiological impact of the SUB-CA1 projection is verified functionally by channelrhodopsin-2 assisted circuit mapping (CRACM)16 of SUB-CA1 projections (Fig. 1m–p). AAV expressing ChR2 is injected in SUB in a spatially restricted manner to express ChR2 in SUB neurons (Fig. 1m). Three weeks after the viral injection, the ChR2-Venus expressing axons can be seen across CA1 laminae (Fig. 1n). Using live hippocampal slices for CRACM experiments, whole-cell recordings from CA1 pyramidal neurons (n = 8 cells) show clear excitatory synaptic input responses, with the presence of TTX and 4-AP, to photoactivation of ChR2 expressing SUB axons locally in CA1 (Fig. 1o–p).
Complementing the unique efferent circuitry of SUB neurons projecting to CA1, we observe that they are also unique in the composition of their afferents (Fig. 2). Afferents to CA1-projecting SUB neurons were mapped and compared to the larger population of excitatory SUB neurons genetically defined by Camk2a-Cre expression17 (Fig. 2a,b). We employed Cre-dependent monosynaptic rabies tracing4 and CAV2-mediated retrograde Cre expression to selectively reveal sources of afferents to CA1-projecting SUB neurons in wild type mice (C57/B6). Both Camk2a-Cre defined and CA1-projecting SUB excitatory neurons receive the majority of their inputs from CA1, but differ quantitatively in this respect as assessed by the CSI and the percentage of total labeled neurons providing extrinsic inputs to SUB (Fig. 2c; Supplementary Table 1). CA1 excitatory neuronal input to SUB accounts for 86.7 ± 1.6% versus 60.3 ± 4.4% of the total inputs, respectively, to the excitatory SUB neurons in Camk2a-Cre mice (N = 5 cases) and CA1-projecting excitatory SUB neurons selectively labeled by CAV2-Cre (N = 6 cases) (p = 0.0006; Supplementary Table 1). The CSIs of CA1 excitatory inputs to Camk2a-Cre and CA1-projecting excitatory SUB neurons differ significantly at 47.72 ± 2.69 versus 14.9 ± 1.25 (p = 9.13 × 10−7; Supplementary Table 1). Although CA1-projecting SUB neurons receive less CA1 excitatory input than the Camk2a-Cre labelled population of excitatory SUB neurons, they receive a far denser CA1 inhibitory input, particularly from inhibitory neurons in stratum oriens (16.2 ± 1.6% versus 1.6 ± 0.4%; CSI: 4 ± 0.44 versus 0.9 ± 0.22; p = 0.0002). Many of these input mapped CA1 inhibitory neurons have cell body morphology resembling oriens-lacunosum-moleculare interneurons (Fig. 2a, third panel of the 2nd row; Supplementary Fig. 2a, last panel of the 2nd row); they also are immuno-positive for somatostatin (Supplementary Fig. 2a, last panel of the third row). Furthermore, dense axon plexuses in CA1 stratum lacunosum-moleculare appear to come from rabies-labeled CA1 stratum oriens neurons (Supplementary Fig. 2a, third row). Together this supports the idea that some of the rabies labeled CA1 inhibitory interneurons share local axonal projection features of the well-known oriens-lacunosum-moleculare interneurons18. It remains to be determined whether other CA1 stratum oriens neurons also contribute inhibitory inputs to CA1-projecting SUB neurons.
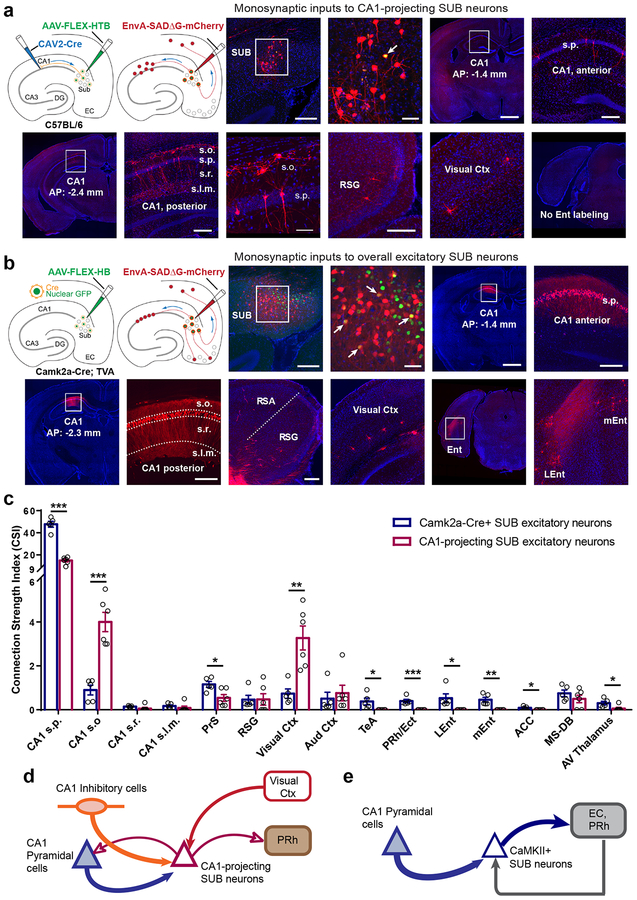
Fig. 2. CA1-projecting SUB excitatory neurons differ in circuit connections from larger populations of SUB excitatory neurons defined by Camk2a-Cre expression.
a, Global mapping of input connections to CA1-projecting SUB excitatory neurons using a combinatorial viral-genetic tracing method. The approach using CAV2-Cre and Cre-dependent monosynaptic rabies tracing is illustrated schematically (see details in Methods). The first two image panels show the injection site in the subiculum. Scale bars are 200 and 50 μm, respectively. DAPI staining is blue; rabies labeled neurons are red. The starter neuron (yellow) indicated by the white arrow appears to receive strong local SUB inputs, as it is surrounded by a cluster of other SUB neurons. Input mapped neurons in dorsal CA1, retrosplenial granular cortex (RSG), and visual cortex are shown in subsequent panels (also see Supplementary Fig. 2). CA1 projecting SUB excitatory neurons receive both excitatory and inhibitory CA1 inputs and do not receive direct input from entorhinal cortex. The scale bar (1mm) applies to low magnification image panels. The scale bar (50 μm) applies for the enlarged view of stratum oriens interneurons and pyramidal cells. All other scale bars = 200 μm. Note that all the labeled neurons shown in the figure are ipsilateral to the injection site, and very few contralateral labeled neurons were seen across all experimental cases. s.o., stratum oriens; s.p., stratum pyramidale; s.r., stratum radiatum; s.l.m., stratum lacunosum-moleculare. This experiment was independently repeated in 6 mice, each with similar results. b, Global mapping of input connections to the full population of SUB excitatory neurons using Camk2a-Cre; TVA mice. The Cre-dependent monosynaptic rabies tracing method is illustrated in the schematics on the left. The first two image panels show the injection site in the subiculum. The scale bars are 200 and 50 μm, respectively. DAPI staining is blue; rabies labeled neurons are red and starter neurons are yellow. Input mapped neurons in anterior (AP: −1.4 mm) and posterior (AP: −2.3 mm) dorsal CA1, retrosplenial granular cortex (RSG), visual cortex, and medial and lateral entorhinal cortex (mEnt and LEnt) are shown in the subsequent panels (see Supplementary Fig. 2 for more input mapped regions). The scale bar (1mm) applies to low magnification image panels. All other scale bars = 200 μm. Panels of RSG, visual cortex, and Lent/mEnt share the same scale. This experiment was independently repeated in 5 mice, each with similar results. c, Quantitative analysis of input connection strengths of CA1-projecting and Camk2a-Cre SUB excitatory neuron types, showing the connectivity strength index (CSI) for each input mapped brain structure. Data are measured from Camk2a-Cre expressing SUB excitatory neurons (N = 5 cases), and CA1-projecting SUB excitatory neurons (N = 6 cases), and are presented as mean ± SE. Two-tailed t-tests are used to test significance of differences for each input region. See detailed data in Supplementary Table 1b. *P < 0.05; **P < 0.01; ***P < 0.001. s.p., stratum pyramidale; s.o., stratum oriens; s.r., stratum radiatum; s.l.m., stratum lacunosum-moleculare. PrS: Presubiculum, RSG: Retrosplenial granular cortex, Vis Ctx: Visual cortex, Au Ctx: Auditory cortex, TeA: Temporal association cortex, PRh: Perirhinal cortex, Ect: ectorhinal cortex, Lent: Lateral entorhinal cortex; mEnt: Medial entorhinal cortex, ACC: anterior cingular cortex, MS-DB: Medial septum and diagonal band of Broca. d–e. Schematic highlights the major input connections and output projections of CA1-projecting SUB neurons versus CaMKII+ SUB excitatory neurons, based on our tracing data shown in Figs. 1 and 2 as well as incorporation of relevant literature on SUB efferent projections. Note that CA1-projecting SUB neurons are GABA immuno-negative and 90% of them are CaMKIIa positive (see Supplementary Fig. 1).
In addition to the differences in CA1 excitatory and inhibitory inputs, CA1-projecting SUB excitatory neurons receive much greater input from visual cortex and weaker input from presubiculum. These neurons also receive inputs from retrosplenial granular cortex (RSG). In further contrast to the Camk2a-Cre defined population of SUB excitatory neurons (Fig 2b, c), CA1-projecting SUB neurons receive no input from temporal association cortex, perirhinal cortex, ectorhinal or entorhinal cortex (Fig. 2a, c, Supplementary Table 1). The overall population of excitatory SUB neurons receives substantial input from these regions as reported in prior work6,7,19.
Thus, projection-specific anterograde and retrograde tracing techniques reveal a unique afferent/efferent circuitry associated with CA1-projecting SUB neurons (Fig. 2d). This SUB-CA1 projection can theoretically be considered as a direct feedback system for CA1 output. At the same time, the connectivity pattern shows previously undisclosed pathways by which cortical information, particularly from visual cortex, can reach and impact activity in CA1. Prior conceptions have most often considered EC to serve as the primary or even sole integrator of cortical input to the hippocampal system (Fig. 2e).
The direct path from visual cortex to CA1-projecting SUB neurons and their output to CA1 and perirhinal cortex strongly suggest that one role for this circuitry may be to provide critical information necessary for conjunctive object-place representations in hippocampus and perirhinal cortices. To test this, we used DREADDs (designer receptors exclusively activated by designer drugs) mediated inactivation of CA1-projecting excitatory SUB neurons to determine whether they are necessary for object-location learning behavior (Fig. 3; Supplementary Fig. 3). Inhibitory hM4D DREADDs20 were expressed selectively in CA1-projecting SUB neurons using dual CAV2-Cre injection in CA1 and AAV2-DIO-hM4D-mCherry injection in SUB of wild type mice (Fig. 3a,b). Sustained inactivation of CA1-projecting SUB neurons during training was achieved using the hM4D ligand (clozapine-N-oxide, CNO) delivered by systemic injection, which was validated by in vitro testing (Fig. 3c; Supplementary Fig. 4a–c).
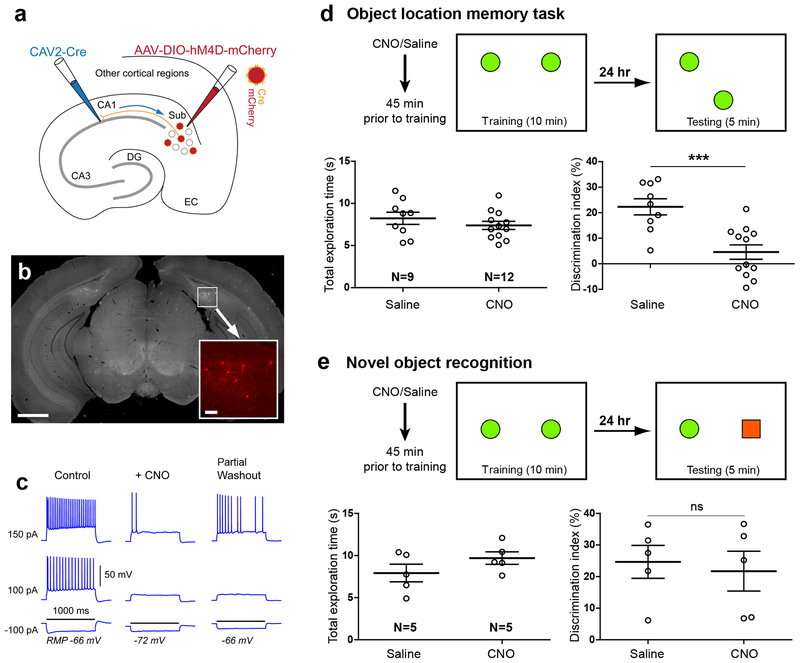
Fig. 3. Genetically targeted inactivation and activation of CA1-projecting excitatory SUB neurons modulates object-location memory.
a, Schematic illustration of genetic inactivation of CA1-projecting SUB excitatory neurons using dual CAV2-Cre injection in CA1 and AAV2-DIO-hM4D-mCherry injection in the SUB. b, Histological analysis in coronal brain sections verifies bilateral, spatially restricted hM4D-mCherry expression in SUB. Scale bar = 1 mm. The bottom right insert shows a higher magnification view of the white square region in b with hM4D-mCherry expressing, CA1-projecting SUB neurons in red. The insert scale bar = 100 μm. The viral injection experiment was independently repeated in 21 mice, each with similar results. c, Example electrophysiological data demonstrating in vitro validation of DREADDs-mediated SUB neuronal inactivation. The 5 μM CNO application hyperpolarized the cell’s resting membrane potential (RMP) and suppressed action potential firing by intrasomatic current injections. The black horizontal line indicates 1000 ms of the current injection duration with different strengths (i.e., −100 pA, 100 pA and 150 pA). The 5 μM concentration matched the CNO dosage of 1.5 mg/kg used for in vivo DREADDs experiments. Given hM4D is a G-protein coupled receptor, CNO effects can only be partially reversed with washout of 30–45 minutes, which is consistent with published results. The experiment was independently repeated in 10 cells from 4 mice, each with similar results. See more validation data in Supplementary Fig. 4a–c. d. Scheme for experimental design and results of location-dependent object recognition task following CNO-activated inhibition of CA1-projecting SUB excitatory neurons during the training phase. The box represents the open field arena, and the green filled circles indicate the training (left) and test (right) object locations. Before the experiment, mice were handled and habituated to the context in the absence of objects. Mice received a single dose i.p. injection of control saline or experimental CNO treatment (1.4mg/kg) 45 min prior to the training. Left graph: total exploration time of the animals during the testing session. Right graph: discrimination index for the testing session 24 hours after training. CNO treated mice show no preference for the moved object in contrast to saline treated controls. Data are presented as mean ± SE. *** p =0.0005 (two-tailed t-test). e, Test of novel object recognition. In training sessions, two identical objects (green filled circles) are placed in the arena, while for testing, distinct objects (green filled circle and red filled square) are placed. Mice with inhibition of CA1-projecting SUB neurons show equal preference for the novel object, similar to the saline treated controls. Data are presented as mean ± SE. n.s., not significant p = 0.73 (two-tailed t-test). For more data information, see Supplementary Table 2a.
Compared to mice with control saline treatment, mice with targeted inactivation of CA1-projecting SUB neurons during the training session displayed much lower object location discrimination (saline control: 22.31 ± 3.16 %; CNO: 4.60 ± 2.82%; p = 0.0005) during the test period when the location of one of the two objects was altered (Fig. 3d; Supplementary Table 2a). This DREADDs-mediated inactivation effect is specific to object location memory, as additional experiments showed that inactivating CA1-projecting SUB neurons had no effect on investigation of a novel object (Fig. 3e; Supplementary Table 2a).
We also asked whether stimulation of SUB inputs to CA1 and perirhinal cortex could enhance object-location memory. We applied a subthreshold training (3 min) design to test if optogenetic stimulation at a theta frequency enhances object-location memory (Fig. 4a,b). Slice electrophysiology experiments show that ChR2-expressing SUB neurons respond to blue laser stimulation (6 Hz, 50 ms) for 3 minutes, and remain healthy by the criteria of stable resting membrane potential and action potential height. The majority of the recorded cells faithfully spike in response to laser stimulation (Fig. 4c,d; Supplementary Fig. 4d–f), supporting effective optogenetic stimulation of SUB neurons in vivo under the same stimulation protocol. As expected, non-stimulation control mice fail to establish memory for object location during the brief training-period exposure, showing no bias to reinvestigation of a relocated object on the subsequent test day (Fig. 4e,f). The same result is true for control mice with viral expression of EGFP and with laser stimulation applied. In contrast, optogenetic stimulation of CA1-projecting SUB neurons during training leads to robust object-location memory formation; these mice spent more time investigating the relocated object on the subsequent test day (Fig. 4e,f; Supplementary Table 2b). Thus, there is bidirectional modulation of memory formation through inactivation and stimulation of CA1-projecting SUB neurons.
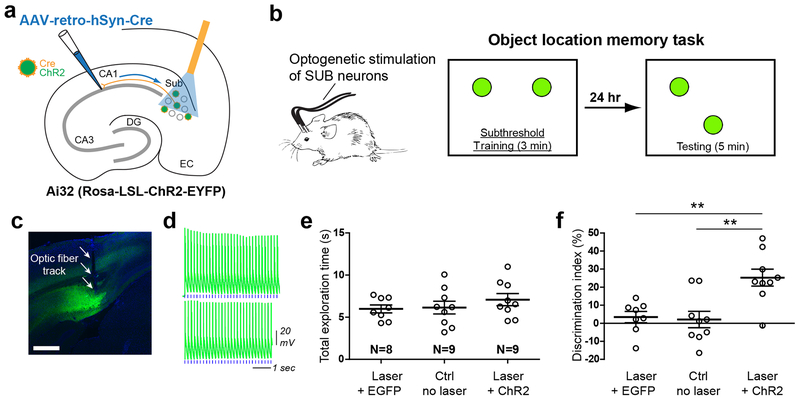
Fig. 4. Optogenetically activating CA1 projecting SUB neurons enhances object-location memory.
a, Scheme for optogenetic activation of CA1-projecting subicular neurons using AAV-retro-hSyn-Cre in Ai32 (Rosa-LSL-ChR2-EYFP) mice. b, Left, schematic depicting a mouse bilaterally connected with optic fibers for delivery of laser stimulation to the subiculum. Right, the experimental design of the object location task for testing the memory enhancement. The box represents the open field arena and the green filled circles indicate initial and shifted object locations. Object exploration was for only 3 min in training, which normally is sub-threshold for long-term memory formation; testing in response to a moved object location following subthreshold training is shown at the far right. c, A coronal brain section image shows the expression of ChR2-EYFP (green) in the subiculum and an optic fiber track. The viral injection experiment was independently repeated in 18 mice, each with similar results. d, Example responses of a ChR2-expressing SUB neuron to 473 nm blue laser stimulation (6 Hz, 50 ms) for 3 minutes with a matched light intensity of in vivo behavioral experiments. Each blue tick beneath the response trace indicates one stimulation. The experiment was independently repeated in 15 cells from 4 mice, each with similar results. See more relevant data in Supplementary Fig. 4d–f. e, The total exploration time with objects during the testing session is similar for each condition. f, The stable versus displaced object discrimination index during testing for each condition is shown. Mice that received subicular laser stimulation show strongly increased preference for the moved object in contrast to unstimulated controls or EGFP controls with laser stimulation. Data are presented as mean ± SE and a two-tailed t-test was used. ** p = 0. 0018 (top bar) and 0.0027 (bottom bar), respectively.
To further characterize behavioral functions of the SUB-CA1 projection, we examined how DREADDs-based inactivation of CA1-projecting SUB neurons impacts mouse performance on a dry-land version of the often-used Morris water maze test (Supplementary Fig. 5a–d)21. Temporary inactivation of SUB projections to CA1 does not impair learning and memory for a spatial location in the dry-land maze task (Supplementary Fig. 5e–j). Compared to controls, CNO treated animals exhibit comparable latencies and traveling distances to reach locations flagged by objects. Further, CNO in these same animals does not impact their ability to find a non-flagged spatial target on either the training day or, 24-hrs later, on the testing day.
The data above indicate that inhibition of CA1-projecting SUB neurons does not impair novel object recognition nor navigation to a remembered location defined by distal visual cues. Thus, the observed impairment in object-location memory produced by the same manipulation is more likely due to impairment in connecting objects to their locations in allocentric space. To discern how CA1-projecting SUB neurons might impact object-location learning, we first examined whether their inactivation impacts CA1 encoding of the animal’s environmental location.
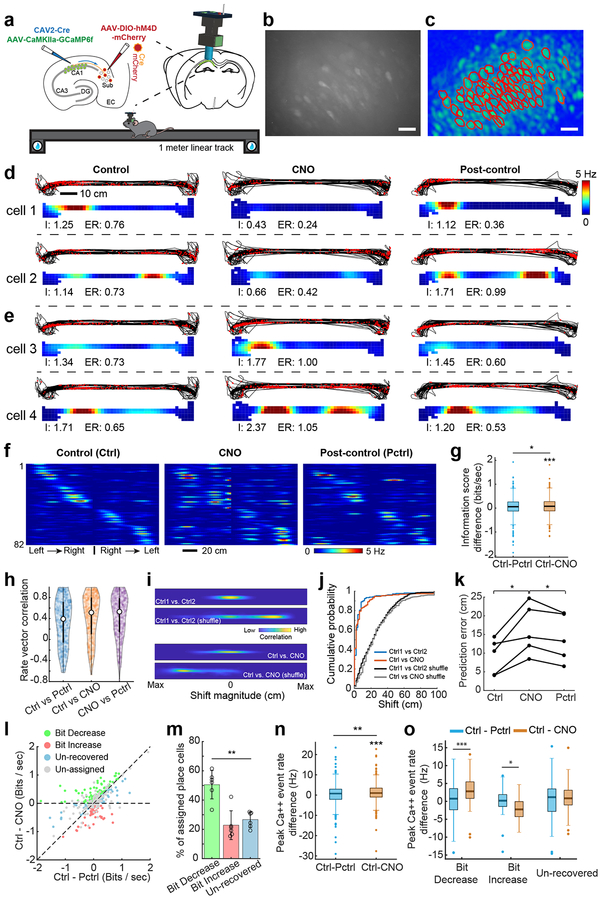
We examined the activity patterns of CA1 neuron populations using in vivo GCaMP6-based calcium imaging with miniature fluorescent microscopes22,23 (Fig. 5; Supplementary Figs. 6–8). GCaMP6f was expressed selectively in CA1 excitatory neurons using AAV1-Camk2a-GCaMP6f injection; dual CAV2-Cre in CA1 and AAV2-DIO-hM4D-mCherry injection in the SUB of wild type mice was used to express DREADDs in CA1-projecting SUB neurons (Fig. 5a). We tracked calcium transient activity as mice ran along a linear track for water reward at each end (Fig. 5a–c). Calcium transients in individual CA1 neurons are often localized to one or two locations along a track (Fig. 5d–f), consistent with the well-described ‘place-specific’ action potential firing of CA1 neurons. By identifying the same population of neurons across multiple days (Supplementary Fig. 7a), we were able to compare the location specificity and density of calcium transient activity before, during, and after inactivation of CA1-projecting SUB neurons. Three recording sessions, spaced 2 or 3 days apart, were obtained for each animal (N= 6 mice) with the middle recording sessions involving DREADDs-based inactivation of CA1-projecting SUB neurons and the first and last serving as pre- and post-DREADDs controls (Supplementary Fig. 7b). Additionally, we performed three recording sessions under control conditions (N = 3 mice, Supplementary Fig. 7c) and found stable hippocampal representations across days.
Fig. 5. CA1-projecting SUB neurons modulate place-specific activity of CA1 neurons in the linear track space.
Experiments described in the figure were independently repeated in 6 CNO-treated mice, each with similar results obtained. For control data, experiments were independently repeated in 3 mice, each with similar results obtained. a, Top left, a schematic illustration of AAV1 injection for targeted expression of GCaMP6f in CA1 excitatory neurons, and a second dual injection of CAV2-Cre in CA1 and AAV2-DIO-hM4D in SUB for targeted expression of DREADDs in the CA1-projecting SUB excitatory neurons. Top right, a schematic depicts a miniaturized fluorescent microscope (miniscope) used to image in vivo calcium signals in CA1 neurons in awake behaving mice. The implanted GRIN lens (shown in blue) and fixed miniscope baseplate allow reliable, repeated imaging of the same group of neurons over 2 weeks. The lower cartoon depicts a mouse running on a 1-meter linear track during in vivo calcium imaging. Water rewards are placed on both ends of the linear track. b, Representative maximum intensity projected image showing recorded CA1 neurons from three combined 15 min imaging sessions (control, CNO, post-control) across days. The calcium imaging videos were motion corrected and aligned across sessions, scale bar = 25 μm. c, A spatial footprint profile image shows extracted neurons (red contours) using the CNMF-E algorithm (see the Methods for details) based on the combined video in b, scale bar = 25 μm. d, Two sets of panels display tracking data (black lines) with superimposed red dots depicting sites where Ca++ events occurred (upper) and corresponding calcium activity rates (lower) for two example cells (cell 1 and cell 2) from the bit-decrease group (see classification below) on the linear track. Each bottom panel is a color-coded rate map showing the averaged spatial distribution of calcium event rates of the same CA1 neuron mapped to animal position on the linear track. Each line shows the event plot and rate maps of control (left), CNO (center), and post-control (right) sessions from the same CA1 place cell over about 2 weeks (2–3 days between sessions). “I” indicates the spatial information score (bits/second), “ER” indicates the mean calcium event rate. e, Same organization as d, these panels show the activities of two example cells (cell 3 and cell 4) from the bit-increase (see classification below) group. f, A color coded population event rate map organized by spatial position for left to right track traversals and then right to left traversals of all place cells from one representative mouse during control (Ctrl, left), CNO (center), and post-control (Pctrl, right) sessions. The depicted cell order is unchanged across the sessions of control, CNO and post-control as determined initially in the control session. Each line shows activity of one place cell. Color indicates event rate (scale bar). The overall rate map correlation values (Pearson’s) between control and post control sessions, and between control and CNO sessions are, respectively, 0.61 and 0.54. g, Comparison of the difference of spatial information scores (bits/second) between Ctrl and Pctrl (Ctrl –Pctrl, two-tailed t-test against zero: p = 0.15), and between Ctrl and CNO (Ctrl – CNO, two-tailed t-test against zero, p = 1.3 × 10−4) across all 347 place cells recorded from 6 mice. Differences between Ctrl – Pctrl and Ctrl – CNO are also observed (two-tailed, paired t-test, p = 0.03). h, A violin plot showing the distribution of individual place cell’s rate vector correlation coefficients (Pearson’s) for all session combinations (“control versus CNO”, “control versus post-control”, and “CNO versus post-control”). The median value for Ctrl vs Pctrl, Ctrl vs CNO, and CNO vs PCtrl are 0.40, 0.51, and 0.53 (n = 347 place cells from 6 mice). The white points indicate median values, and thin black lines extend to the most extreme values within 1.5 times of the interquartile range of the median. The filled color width represents a density estimate of the distribution of values along the y axis. i, Spatial cross-correlograms of example CA1 cells from the saline-treated experiment (Ctrl1 vs Ctrl2) and CNO-treated experiment (Ctrl vs CNO). Shuffled examples were obtained by randomly pairing rate maps across the same comparison sessions as for the example cells. j, The distributions of correlation-peak shift magnitudes for the place cells in the saline experiment (blue line) and the CNO experiment (red line) differ significantly from the corresponding shuffled distributions (p = 3.17 × 10−64, two-tailed, two-sample Kolmogorov–Smirnov test (KS), n = 174 cells from Ctrl1 vs Ctrl2 and 1000 shuffles from Ctrl1 vs Ctrl2; p = 1.48 × 10−101, two-sample KS, two-tailed, n = 347 cell from Ctrl vs CNO and 1000 shuffles from Ctrl vs CNO). Shuffled distributions were obtained by randomly pairing place maps 1,000 times across the indicated sessions. There is no significant difference between the distribution of Ctrl vs CNO (red line) and the Ctrl1 vs Ctrl2 (blue line) (p = 0.38, two-sample KS, two-tailed). k, Quantification of the prediction errors between predicted trajectories and actual trajectories for decoding accuracy using the trained model based on the first control session, which supports the observations in Supplementary Fig. 8a. Each line represents the prediction errors of Ctrl, CNO and Pctrl sessions from one mouse. Significantly higher prediction errors are observed in CNO sessions compared to those in Ctrl (p = 0.016, two-tailed, paired t-test) and Pctrl (p = 0.015, two-tailed, paired t-test) sessions. n = 5 mice. l, Recorded CA1 place cells can be classified into 3 non-overlapping groups termed bit-decrease, bit-increase, and un-recovered (see Methods for more information about the group classification), based on the statistical significance of differences in information scores (bit/sec) between CNO and Ctrl, and between CNO and Pctrl. Statistical testing employed a jackknife resampling method for each place cell with appropriate corrections for error terms. Un-assigned place cells did not pass the statistical test and were excluded from further categorization analysis. On the scatter plot, the x-axis is Ctrl – Pctrl (the difference of spatial information scores between Ctrl and Post-ctrl) and the y-axis is Ctrl – CNO (the difference of information scores between Ctrl and CNO). m, Of the 201 place cells that show significant differences (assigned place cells) from 6 mice, 50% show decreased information scores in CNO sessions compared to the control and post-control sessions (bit-decrease group, green bar). A smaller subset (~ 23%) show increased information scores in CNO compared to the control and post-control (bit-increase group, red bar). The remaining ones are the unrecovered group which accounts for ~ 27% of place cells. Comparing the mean percentages of each type seen in each mouse, a significant difference in the % of place cells among these three groups is observed (p = 0.002, repeated measures ANOVA, n = 6 mice). Data are presented as mean ± SE in the bar plot. n, Comparison of the difference of peak calcium event rates between Ctrl and Pctrl (Ctrl –Pctrl, two-tailed t-test against zero, p = 0.32), and between Ctrl and CNO (Ctrl –CNO, two-tailed t-test against zero, p = 1.6 × 10−5) across all 347 place cells recorded from 6 mice. Differences between Ctrl – Pctrl and Ctrl – CNO are also observed (two-tailed, paired t-test, p = 0.004). o, Comparisons of peak calcium event rates between Ctrl – Pctrl and Ctrl – CNO in bit decrease (two-tailed, paired t-test, p = 3 × 10−7, n = 97 cells), bit increase (two-tailed, paired t-test, p = 0.027, n = 48 cells) and un-recovered groups (two-tailed, paired t-test, p = 0.20, n = 56 cells), respectively. For the box plots throughout the figure, the three box lines from top to bottom represent the 25th, 50th (median), and 75th percentile of data values of the samples. The whiskers extend to the most extreme values within 1.5 times of the interquartile range of the median.
From CA1 neurons with place-specific activity tracked across all three sessions, we examined spatial information, an often-used metric for place-specific activity24. CNO-induced inactivation of CA1-projecting SUB neurons produced overall reductions in the spatial information metric bits/second (Fig. 5g). No such change in spatial information was seen across multiple control (CNO-free) days in 3 animals (Supplementary Fig. 8). Furthermore, CNO itself, at our dosage, does not directly affect spatial information or calcium event amplitudes in place cells in control animals25,26 (Supplementary Fig. 8). In contrast to the observed changes in bits/second, no CNO-induced changes were observed for the spatial information metric bits/spike (hereafter termed bits/event in keeping with the calcium transient “events” we imaged; Supplementary Fig. 9).
The contrast between the bits/second and bits/event metrics provides a clue to the specific impact of CA1-projecting SUB neurons on CA1 mapping of environmental location. A key difference between the two measures lies in their sensitivity to changes in the magnitude of activity within a place field (as modeled in Supplementary Fig. 9). For simulated firing fields, the bits/second metric is sensitive, in linear fashion, to a wide range of different in-field event rates while the bits/event metric is not. This suggests that inhibition of CA1-projecting SUB neurons may alter CA1 neuron in-field rates while not impacting event locations.
Consistent with the lack of effect of CNO treatment on the bits/event measure, the location-specific patterning of activity among neurons is largely unchanged as assessed by four different methodologies. First, mean correlations between positional rate vectors of individual neurons are similar for pairings of control and post-control days to values from either control day paired with testing under CNO (Fig. 5h). This result is inconsistent with either large-scale changes in firing field locations or dissolution of place-specific activity itself. Second, ensemble positional rate vector correlations for odd versus even trial runs and for the first versus second halves of recordings do not vary across control and CNO conditions (Supplementary Fig. 8j–m) indicating strong within-session reliability of location-specific activity. Third, direct measurement of field shifts across sessions reveals minimal variation in place field locations under treatment with CNO versus control sessions (Fig. 5i,j). Actual shifts are far less than one would expect if field locations shifted randomly in response to CNO treatment and were similar to the minimal field shifts seen between control sessions. Fourth, using an artificial neural network to decode the position of the animal in the second and third recording sessions (i.e., CNO and post-control sessions) based on a trained model from the first control session, we find only slightly reduced decoding accuracy in the CNO versus post-control session (Fig. 5k, Supplementary Fig. 8a). Thus, changes in the locations of place-specific activity are much smaller than one might expect if the place specific activity of CA1 neurons had undergone complete or ‘global’ remapping’27 wherein the firing field locations of individual neurons vary randomly across sessions. The results are also incompatible with a general dissolution of place-specific activity.
An alternative form of modulation in spatially-specific activity is termed ‘rate-remapping’, wherein in-field activity rates of CA1 place cells are altered despite no change in the locations of spatially specific activity27. Such positive and negative modulation of in-field place-specific firing allows hippocampal place cells to encode both locations and the context associated with being in those locations. Because the observed changes in the bits/second metric for CA1 place-specific activity is more compatible with this form of remapping, we next examined the possibility that inactivation of CA1-projecting SUB neurons impacts in-field firing.
To organize our analysis of rate remapping, we segregated neurons according to their individual positive or negative changes in cross-session spatial information scores (bits/second). For each neuron, we examined whether the spatial information score in the CNO session was significantly different from that in both the control and post-control sessions using a jackknife resampling method28. Each track-running session was divided into 10 equal-duration sub-sessions. We then applied the jackknife resampling to the full recording session, each time calculating bits/second values based on 9 of the 10 sub-sessions. The now extended sets of bits/second values produced by this resampling procedure were then subjected to statistical testing (see Methods for detail). A place cell is classified as ‘bit decrease’ only when its spatial information in the CNO session is significantly lower than in both the control and post-control sessions. A ‘bit increase’ neuron bears CNO session bits/second values that are higher than each control session. A place cell that passes the statistical test in the CNO versus control and the CNO versus post-control comparisons, but with opposite directions of change, is classified into the ‘un-recovered’ group. The rest of the place cells, which fail at least one of the statistical tests, are un-assigned and excluded from the category analysis.
Among the 201 place cells (from 6 mice) showing significant differences in CNO versus Ctrl and CNO versus Pctrl, we find 73% that exhibit either information decreases (‘bit decrease’ group – 50%) or information increases (‘bit increase’ group – 23%) relative to both control and post-control sessions (Fig. 5l, m). The remainder (27%), were characterized as ‘un-recovered’, showing non-uniform changes in spatial information in CNO versus control sessions. The number of neurons falling into the bit decrease category is equal to the bit increase and un-recovered categories combined, a bias consistent with the overall decrease in bits/second (Fig. 5g). Furthermore, this bias was reliable across animals (Fig. 5m). Finally, in 174 place cells recorded under three consecutive control sessions (Supplementary Fig. 7c), we observed no significant change in spatial information for the second session (Supplementary Fig. 8d). The percentage of assigned place cells characterized as ‘bit-decrease’ in the second control session relative to the first and third control sessions is not significantly greater than that of the ‘bit-increase’ or ‘un-recovered’ category and it is significantly lower than that in CNO treated animals (Supplementary Fig. 8c,f,g).
Thus, by several measures, inactivation of CA1-projecting SUB neurons minimally impacts the locations at which individual CA1 neurons are highly active. Yet, there is an overall significant bias toward reduction in the bits/second spatial information metric at both the population and single neuron levels of analysis. The former result indicates that the latter is likely not explained by increased scattering of calcium events across track space, but instead reflects changes in the peak in-field magnitude of location-specific activity. Consistent with this, we find that peak in-field calcium event rates exhibit a similar pattern with bits/second changes across the control, CNO and post-control sessions. Overall peak event rate differences are seen in comparing CNO to control, but not in comparing post-control to control (Fig. 5n). No peak event rate differences are observed in the three consecutive control sessions (Supplementary Fig. 8h). As expected, based on the sensitivity of the bits/second metric to event rates (Supplementary Fig. 9e–f), the neuron sub-population exhibiting significant decreases in bits/second values under CNO treatment is, overall, observed to have significantly reduced peak activity rates. The sub-population with spatial information increases exhibits rate increases (Fig 5o). Together, the findings evidence reliable alterations in the magnitude of location-specific activity in CA1 neurons subsequent to DREADDs-based inactivation of their SUB inputs. CA1-projecting SUB neurons can therefore be considered to contribute to the peak in-field event rates of CA1 neurons under control conditions. Their inhibition induces a form of ‘rate-remapping’ in CA1 with a bias toward reduced in-field activity.
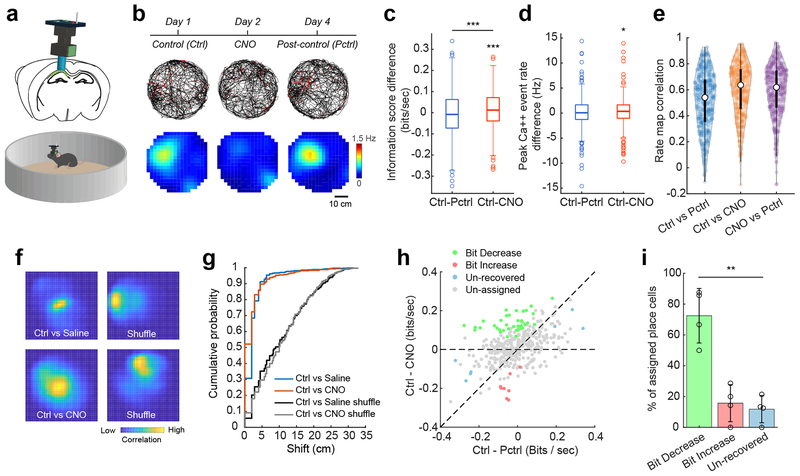
We further characterized the impact of inactivation of the SUB-CA1 back-projection on CA1 neural activity during open-field exploratory behavior using a separate cohort of mice (Fig. 6 and Supplementary Fig. 10). As on the linear track, DREADDs-based inactivation of CA1-projecting SUB neurons reduces the bits/second, but not bits/event spatial information scores among CA1 place cells (Fig. 6c, Supplemental Fig. 10a–d). Overall peak in-field firing is depressed under CNO compared to control, but differences in control versus post-control values are not observed (Fig. 6d). In close agreement to the data obtained during track-running, CNO does not significantly change the locations of CA1 place fields as assessed through cross-session correlation of event rate maps and direct measurement of field shifts (Fig. 6e–g; Supplementary Fig. 10a–d).
Fig. 6. Inactivation of CA1-projecting SUB neurons impacts CA1 neural activities in an open field.
A cohort of mice with bilateral hM4D expression in CA1-projecting SUB neurons were used for these experiments. Miniscope imaging of CA1 place cell activities were obtained while animals explored an open arena. The experiments were independently repeated in 4 mice of each treatment group, each with similar results. a, A schematic illustration of the CA1 Ca++ imaging experiment in a circular arena. b, Top: Example position sampling (grey lines) and locations of Ca++ events of a single neuron (red dots) for control, CNO, and post-control sessions. Bottom: Calcium event rate maps for the same example cell across sessions. The experimental time line is shown at the top. c, Comparisons of the difference of spatial information score(bits/second) between Ctrl and Pctrl (Ctrl – Pctrl, two-tailed t-test against zero: p = 0.26), and between Ctrl and CNO (Ctrl - CNO, two-tailed t-test against zero, p = 1.82 × 10−4) across 379 place cells from 4 mice. Significant differences between Ctrl – Pctrl and Ctrl – CNO are also observed (two-tailed, paired t-test, p = 2.73 × 10−5). For the box plots in the figure, the three box lines from top to bottom represent the 25th, 50th (median), and 75th percentile of data values of the samples. The whiskers extend to the most extreme values within 1.5 times of the interquartile range of the median. d, Comparisons of the difference of individual place cells’ peak calcium event rates between Ctrl and Pctrl (Ctrl – Pctrl, two-tailed t-test against zero, p = 0.32), and between Ctrl and CNO (Ctrl - CNO, two-tailed t-test against zero, p = 0.02). e, A violin plot showing the distribution of individual place cell’s rate map correlation coefficients (Pearson’s) for all session combinations. The median values for Ctrl vs Pctrl, Ctrl vs CNO, and CNO vs PCtrl are 0.51, 0.60, and 0.59 (n = 379 place cells from 4 mice). The overall high correlation indicates consistent and stable positional coding between different conditions. In the violin plot, the white points indicate median values, and thin black lines extend to the most extreme values within 1.5 times of the interquartile range of the median. The filled color width represents a density estimate of the distribution of values along the y axis. f, Spatial cross-correlograms of example CA1 cells from the saline-treatment experiment (Ctrl vs Saline) and CNO-treatment experiment (Ctrl vs CNO). Shuffled examples were obtained by randomly pairing place maps across the same comparison sessions as for the example cells. g, The distributions of correlation-peak shift magnitudes for the place cells in the saline experiment (blue line) and the CNO experiment (red line) differ significantly from the corresponding shuffled distributions (p = 1.94 × 10−107, two-sample Kolmogorov–Smirnov test (KS), two-tailed, n = 606 place cells from Ctrl vs Saline and 1000 shuffles from Ctrl vs Saline; p = 2.05 × 10−97, two-sample KS, two-tailed, n = 379 place cells from Ctrl vs CNO and 1000 shuffles from Ctrl vs CNO). Shuffled distributions were obtained by randomly pairing place maps 1,000 times across the indicated sessions. The distribution of Ctrl vs CNO (red line) is slightly left shifted than Ctrl vs Saline (blue line) (p = 1.69 × 10−10, two-sample KS, two-tailed). h, In the CNO inactivation experiment in the open field, recorded CA1 place cells can be classified into 3 non-overlapping groups termed bit-decrease, bit-increase, and un-recovered, based on the statistical significance of differences in information scores (bits/second) between CNO and Ctrl, and between CNO and Pctrl subject to a jackknife resampling test for each place cell. Un-assigned place cells did not pass the statistical test with jackknife resampling and were excluded from further categorization analysis. On the scatter plot, the x-axis is Ctrl – Pctrl (the difference of spatial information scores between Ctrl and Post-ctrl) and the y-axis is Ctrl – CNO (the difference of information scores between Ctrl and CNO). i, Of the 52 place cells that passed the statistical test from 4 mice, on average, 72% show decreased information scores in CNO sessions compared to the control and post-control sessions (bit-decrease group, green bar); a smaller subset (16%) show increased information scores in CNO compared to the control and post-control (bit-increase group, red bar), and the remaining 12% belong to the unrecovered group (blue bar). There is a significant difference across the group percentage values (p = 0.0033, repeated measures ANOVA, n = 4 mice), while the saline control experiment does not show this difference (Supplementary Fig. 10f). Data are presented as mean ± SE in the bar plots.
Further replication of the open field-based data is observed when information changes in individual neurons were assessed in category analysis using the jackknife resampling approach. Of the 52 assigned place cells, we consistently observe a bias toward larger numbers of neurons that exhibit significant information decreases (72%, Fig. 6h,i). Thus, more than three times as many place cells fall into the bit-decrease (lower bits/second) than the bit-increase (16%) or un-recovered (12%) category. This bias is not seen in the control animal group (Supplementary Fig. 10e–g).
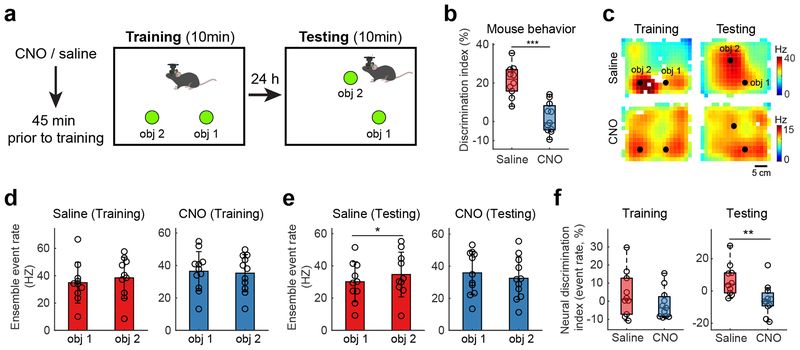
To more directly assess the contribution of SUB projections to CA1 on object-location memory, we also conducted miniscope imaging experiments in behaving mice during the object-location memory task itself (Fig. 7a). In this cohort of mice, we again observe impairment in object-location memory following inactivation of CA1-projecting SUB neurons (Fig. 7b). DREADDs-based inactivation of this sub-group of SUB neurons during the training session leads to significant impairment in long-term location-dependent memory as assessed on the subsequent testing day. Similar to the original cohort of animals undergoing this test (Fig. 3d), saline controls spend significantly more time in interaction with the newly displaced object than with the unmoved object, while CNO treated animals spend equal time in interaction with each object (Fig. 7a–b; Supplementary Table 3).
Fig. 7. Inactivation of CA1-projecting SUB neurons modulates CA1 neural activity correlated with impaired object-location memory performance.
a, Task schematic and timeline. b, CNO-treated mice (n = 11) exhibit significantly impaired object-location memory (OLM) performance, compared with saline-controls (n = 10), as reflected in the lower discrimination index (two-tailed Welch’s t-test, p = 2 × 10−5). c, Ensemble Ca++ event rate maps of all cells during training and testing phases from one saline-control mouse (343 cells) and one CNO-treated mouse (182 cells), respectively. The ensemble event rates are the summation of events from all recorded neurons, normalized by time spent at each location. d, No significant differences of ensemble Ca++ event rates between the two objects for both saline-control mice (n = 10, p = 0.274) and CNO-treated mice (n = 11, p = 0.576) are seen during the OLM training phase. P-values are from two-tailed paired t-tests. e, Ensemble Ca++ event rates during the OLM testing phase. We observe significantly higher Ca++ event rates around the displaced object (object 2) compared to that around the stable object (object 1) for saline-control mice (n = 10, p = 0.032). There is no difference of Ca++ event rates around object 2 versus object 1 for CNO-treated mice (n = 11, p = 0.14). P-values are from two-tailed paired t-tests. f, CNO-treated mice (n = 11) do not differ from saline-control mice (n = 10) in terms of the neural discrimination index calculated with ensemble Ca++ event rates during the training session (p = 0.244, two-tailed Welch’s t-test), but show decreased neural discrimination during the testing session (p = 0.0099, two-tailed Welch’s t-test). This neural discrimination index is based on relative event rates and expressed as (ERobject2 – ERobject1) / (ERobject2 + ERobject1) ×100%) wherein ERobject2 and ERobject1 are the ensemble event rates associated with the two objects, respectively. For the box plots throughout the figure, the three box lines from top to bottom represent the 25th, 50th (median), and 75th percentile of data values of the samples. The whiskers extend to the most extreme values within 1.5 times of the interquartile range of the median. Data are presented as mean ± SE in the bar plots.
To determine whether neural responses to object displacement parallel behavioral responses, we generated ensemble-wide Ca++ event rate maps of CA1 neurons for each recording (Fig. 7c). The ensemble event rates are the summation of events from all recorded neurons, normalized by time spent at each location. We observe no significant differences in the ensemble-wide rate of Ca++ events at the two object locations during training sessions for either saline-control mice or CNO-treated mice (Fig. 7d). However, for control mice, displacement of one object on the testing day produces a small but significant increase in CA1 activity at the site of that object as compared to the unmoved object (p = 0.032, Fig. 7e). In contrast, there is no such difference in Ca++ event rates in CNO-treated mice. The event rate values at the displaced and unmoved object positions are compared using the neural discrimination index presented in Fig. 7f. For each animal, the ensemble wide event rate discrimination index is calculated: (event rate at object 2 – event rate at object 1) / (event rate at object 2 + event rate at object 1) X100%. The discrimination index is positive for saline-treated animals, indicating enhanced neural responses to the displaced object, but slightly negative for CNO-treated animals indicating weak discrimination of the two objects. This effect was significant for up to a 3 cm space surrounding each object (Supplementary Fig. 10h). Thus, we find that DREADDs-based inactivation of CA1-projecting SUB neurons significantly reduces the discrimination in activity levels at the relocated versus stable object (Fig. 7f, p = 0.005). To further test specific contributions of place cells in the object-location experiment, for each animal, we determined the ratio of place cell fields (see Methods) in relation to each object and compared these ratios for the saline and CNO treated groups. Example neuron event rate maps are shown in Supplementary Fig. 10i–j. Both saline and CNO treated groups exhibit roughly the same number of fields for both objects during the training session (Supplementary Fig. 10k). We find a significant difference in these ratios evidencing higher numbers of place fields associated with the displaced object for the saline group and lower numbers of place fields related to the displaced object in the CNO group during the testing session (Supplementary Fig. 10l). Together, the results parallel the changes observed in behavioral discrimination. Targeted inactivation of the SUB-CA1 back-projection during OLM training impacts ensemble neural activity associated with long-term object-location dependent memory.
Discussion
In this work, we identify a distinct sub-population of SUB neurons forming a pathway from visual cortex to CA1 and perirhinal cortex, and demonstrate that this pathway plays a critical role in object-location learning and memory. We refer to this circuit as ‘non-canonical’ in that it runs opposite the feedforward pathway leading from CA1 to SUB to retrosplenial cortex. CA1-projecting SUB neurons are simultaneously poised to act in a feedback loop to CA1 and in a pathway for visual information to directly impact the two major output structures of hippocampus, CA1 and subiculum. By combining trans-synaptic viral tracing techniques, in vivo GCaMP6-based calcium imaging, projection-specific manipulations and behavioral testing, the present set of experiments reveal the presence of a cortico-hippocampal circuit that impacts specific learning and memory behaviors.
The unique afferent/efferent circuitry of CA1-projecting SUB neurons and their bidirectional modulation of CA1 in-field activity rates is consistent with the previously observed roles of CA1 neurons in formation of object-location memories. It is now established that a small population of CA1 neurons exhibit ‘landmark vector’ responses reflecting the animal’s location relative to objects29. Thus, one role for visual cortex inputs to CA1-projecting SUB neurons may be to provide a direct avenue by which information concerning the visual presence of objects is directly routed to CA1 for integration with neurons that encode current environmental location. Here, it is notable that some neurons of rat visual cortex exhibit reliable position-specific firing rates during track running, consistent with encoding of position-specific visual experience30.
Precisely how CA1-projecting SUB neurons mediate object-location memory at the level of synapses remains to be determined. Their inactivation does not affect navigation according to distal visual boundary cues in an object-free environment (see our dry-land experiments), a result consistent with the largely unchanged locations of place field activities under the same manipulation. The larger effect on CA1 dynamics is found to be on peak in-field calcium event rates. During inactivation of CA1-projecting SUB neurons, weaker responses for a majority of CA1 neurons are observed. Inactivation during initial exposure to two objects in an open field leads to a subsequent failure of CA1 populations to generate stronger responses to spatially displaced objects. This implies that object-location memory formation fails when CA1 neurons cannot integrate place-specific responses with the inputs of CA1-projecting SUB neurons.
The afferent circuitry of CA1-projecting SUB neurons includes the retrosplenial cortex, which encodes conjunctions among multiple types of spatial information31,32 and is required when visual cues are used flexibly to determine location33. Under conditions of associative learning of visual stimuli, retrosplenial cortex is a prominent contributor to activity patterns observed in visual cortex regions that project to SUB34. Therefore, retrosplenial cortex could influence SUB and CA1 through direct connections and indirectly through visual cortex.
As compared to the full population of excitatory SUB neurons, CA1-projecting SUB neurons receive especially dense input from CA1 inhibitory neurons. This projection is mainly from inhibitory neurons in stratum oriens. Many of these neurons also send efferents to CA1 stratum lacunosum-moleculare, similar to the projection pattern of some somatostatin-expressing stratum oriens neurons18,35. This aspect of the anatomical data is remarkable considering several parallel features of hippocampal and rhinal cortex anatomy and physiology. First, CA1-projecting SUB neurons also project to perirhinal cortex, a structure providing extensive input to lateral entorhinal cortex (LEC)36. Second, recent data indicate that both LEC and MEC neuron sub-populations exhibit distinct responses to objects37,38. Third, somatostatin-expressing CA1 stratum oriens interneuron firing is locked to similar phases of theta frequency oscillations as for CA1 pyramidal and entorhinal cortex neurons18,39. Fourth, some CA1 stratum oriens neurons innervate CA1 pyramidal neurons’ distal apical dendrites receiving LEC and MEC inputs as well as CA1-projecting SUB neurons. Finally, CA1 pyramidal cell inputs to stratum oriens neurons are subject to long term potentiation40. Therefore, CA1 stratum oriens neurons are well positioned to provide learning-dependent feedback regulation that can temporally organize processing of object-location information across a wide network including CA1-projecting SUB neurons, CA1, and perirhinal and entorhinal cortices.
These fine-scale features of circuitry and physiology connecting SUB, hippocampus, entorhinal cortex, and perirhinal cortex are more interesting when considering the results of lesion studies on memory formation and neurophysiological features of CA1, LEC, and perirhinal cortex41,42. Here we note the opposing and complementary topographic connectivity gradients revealed by quantitative analysis of canonical and non-canonical hippocampal CA1 inputs; non-canonical subiculum inputs co-track the weak-to-strong lateral entorhinal inputs along CA1’s transverse axis5. Recent work reveals extensive encoding of object locations relative to the animal in LEC38. It follows that, to some degree, such responses to objects are transmitted to CA1 neurons through terminals in stratum lacunosum moleculare where some somatostatin-expressing stratum oriens interneuron axons also terminate. Further, interactions between CA1 and perirhinal cortex are necessary for development of object-place memory41,43,44, which is now supported by our finding that CA1-projecting SUB neurons also project to perirhinal cortex.
In light of our circuit mapping results, it is clear that visual and retrosplenial cortical input can reach perirhinal cortex through direct projections from CA1-projecting SUB neurons. Like CA1, perirhinal cortex neurons respond to objects42,45 and lesions produce impairments in expression of object-place memory15,46. Beta-frequency and gamma-frequency paced stimulation of perirhinal cortex bi-directionally modulates recognition of objects as familiar versus novel47. Perirhinal cortex in turn densely innervates LEC, and LEC efferents more densely innervate distal CA1 where SUB-to-CA1 projections are most dense5. Notably, sub-populations of LEC neurons exhibit object responses persisting for days following object removal48, and inactivation of LEC reduces rate-remapping in CA1 neurons and alters their responses to visual objects49. The present findings indicate that CA1-projecting SUB neurons form a nexus point critical to object-place memory formation. These neurons are anatomically interconnected with several brain regions that are strongly implicated in encoding of object location relationships, and can modulate spatially-specific activity patterns of CA1 neurons in a manner that incorporates object place information into episodic memories formed by cortico-hippocampal circuits.
The circuitry associated with CA1-projecting SUB neurons and its role in object-location learning and place-specific activity of CA1 neurons bears some analogy to the influence of prefrontal cortex (via thalamic nucleus reuniens) on trajectory-specific modulation of CA1 place-specific activity50. Together, these findings highlight the need to consider the potential that multiple “top-down” cortical circuits influence CA1 output through intermediaries including SUB and nucleus reuniens. Such cortico-hippocampal circuits can be considered key avenues by which sensory input and planned action are bi-directionally integrated with cognitive maps in hippocampus and entorhinal cortex to yield intelligent behavior.
Methods
Animals
All experiments were conducted according to the National Institutes of Health guidelines for animal care and use and were approved by the Institutional Animal Care and Use Committee and the Institutional Biosafety Committee of the University of California, Irvine. Please see Supplementary Table 4 for the animal strains, viral injections and experiments performed. No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those reported in previous publications4,5,51–53. The data collection was randomized with experimental conditions / groups whenever possible. Experimenters were not blinded during data acquisition, but imaging data and behavioral data analyses were performed blind to treatment.
No animals or data points were excluded from the analyses unless specified in the Life Sciences Reporting Summary.
Viral injections
The general procedure has been previously described4,5,54. Mice were anaesthetized and placed in a rodent stereotax (Leica Angle Two™ for mouse) with continuous 1% isoflurane anesthesia with the head secured. A three-axis micromanipulator guided by a digital atlas was used to determine coordinates for the bregma and lambda. The injection coordinates targeting CA1 intermediate subfield are anteroposterior (AP) −1.94 mm, lateromedial (ML) −1.40 mm; dorsoventral (DV) −1.35 mm (all values given relative to the bregma). The coordinates targeting the subiculum are AP: −3.40; ML: −1.88; DV: −1.70. A small drill hole was made in the skull over the injection site, exposing the pia surface. A glass pipette (tip diameter, ~20–30 μm) was loaded with virus and then lowered into the brain at the appropriate coordinates. A Picospritzer (General Valve, Hollis, NH) was used to pulse virus into the brain at a rate of 20 – 30 nl/min with 10 ms pulse duration. To prevent backflow of virus, the pipette remained in the brain for 5 min after completion of the injection. Once the injection pipette was withdrawn, the mouse was removed from the stereotax, and the incision was closed with tissue adhesive (3M Vetbond, St. Paul, MN). Mice were taken back and recovered in their home cages.
In order to target larger populations of excitatory neurons in CA1 or subiculum for rabies tracing, we crossed the LSL-R26Tva-lacZ mouse line55 with Camk2a-Cre (T29) mouse line17. We term the double transgenic mice as Camk2a-Cre; TVA mice in which Cre-expressing cells also express TVA to restrict initial infection of EnvA-SADΔG rabies virus. Mice 8–12 weeks old (either sex) were used for experiments and had free access to food and water in their home-cages before and after surgeries. Genetically modified rabies viruses used for the experiments are deletion-mutant rabies and are based on a vaccine strain (SADB19). Rabies was locally made at UC Irvine with required cell lines and seeding viruses from Dr. Ed Callaway’s group at the Salk Institute for Biological Studies. For monosynaptic rabies tracing from CA1 or subicular excitatory neurons, a total of 0.1 μl of the helper virus (AAV8-EF1a-FLEX-HB, 2 × 1011 genome units per ml; H: histone EGFP; B: B19 rabies glycoprotein) (Addgene, Plasmid 37452) was injected into CA1 or the subiculum of Camk2a-Cre; TVA mice. After 3 weeks of the AAV injection which allowed for the infected neurons to express high contents of rabies glycoproteins (RG) and EGFP, the pseudotyped, RG-deleted rabies virus (EnvA-SADΔG-dsRed rabies, 0.2 μl, 2 × 109 infectious units per ml) was injected into the same location of the previous injection. The rabies virus was allowed to replicate and retrogradely spread from targeted Cre+ cell types to directly connected presynaptic cells for 9 days before the animals were perfused for tissue processing.
To map circuit input connections of CA1-projecting subicular neurons, 0.3 μl of CAV2-Cre virus (3 × 1012 infectious units per ml; purchased from E.J. Kremer’s Institute in France) was delivered into the CA1 region of the wild type C57BL/6 mouse in order to target CA1-projecting subicular neurons. CAV2-Cre is able to retrogradely transport into the subiculum and express Cre selectively in CA1-projecting subicular neurons. Then 0.1 μl of AAV8-EF1a-FLEX-HTB (2 × 1011 genome units per ml; H: histone GFP; T: TVA: B: rabies B19 glycoprotein) was delivered into the subiculum during the same surgery session. After three weeks, 0.2 μl of EnvA-SADΔG-mCherry rabies was injected into the same subiculum location. For control experiments to verify the retrograde efficiency and test viral tropism of CAV2-Cre, we injected the CAV2-Cre virus (0.4 μl) into CA1 of the Ai9 tdTomato reporter line56. The Ai9 animals are perfused for tissue processing 3 weeks after injection.
To map output projections of CA1-projecting subicular neurons, CAV2-Cre virus was first delivered into the CA1 region. Two weeks after the CAV2-Cre injections, 0.4 μl of Cre-dependent anterograde directed herpes simplex virus (H129ΔTK-TT14, kindly provided by Dr. Lynn Enquist at Princeton University) was injected into the subiculum. The H129 virus was allowed to replicate and anterogradely spread from targeted Cre+ cell types to directly connected postsynaptic cells within 48 hours before the animals were perfused for tissue processing.
For genetic inactivation of CA1-projecting subicular neurons with DREADDs, 0.3 μl of the CAV2-Cre virus was bilaterally delivered into the intermediate CA1 region (AP −1.94 mm, ML ±1.40 mm; DV −1.35 mm) in wild type C57BL/6J mice, followed by bilateral injection of 0.3 μl of AAV2-DIO-hM4D-mCherry (3.7 × 1012 genome units per ml; UNC Vector Core) in the subiculum bilaterally (AP −3.40 mm, ML ±1.96 mm, DV −1.67 mm) in the same surgery session. Mice were then allowed to recover in their home cages for three weeks before behavior experiments. For genetic activation of CA1-projecting subicular neurons, 0.2 μl of rAAV2-retro-hSyn-Cre virus57 (packaged by ViGene Biosciences, 1.9 × 1013 GC/ml) was bilaterally delivered into the intermediate CA1 region of the Ai32 mice58 that express an improved channelrhodopsin-2/EYFP fusion protein following exposure to Cre recombinase. rAAV2-retro in the CA1 injection site can retrogradely transport into the subiculum and express Cre to activate ChR2 expression specifically in CA1-projecting subicular neurons. Our histological examination indicates that different from CAV2-Cre, rAAV2-retro-hSyn-Cre does not infect local cell bodies in CA1, which prevents ChR2 expression in the axons of CA1 neurons. For control experiments with EGFP expression, rAAV2-retro-hSyn-EGFP virus was used instead.
To image in vivo calcium transients of CA1 neurons with inactivation of CA1-projecting subicular neurons on the linear track, 0.2 μl of AAV1-CaMKII-GCaMP6f-WPRE-SV40 (Penn Vector Core; 3.7 × 1013 GC/ml) was injected into CA1 (0.2 mm posterior and lateral to the injection site of the CAV2-Cre); 0.3 μl of CAV2-Cre virus was delivered into the right intermediate CA1 region of C57BL/6 mice and 0.3 μl of AAV2-DIO-hM4D-mCherry was delivered into the ipsilateral side of the subiculum during the same surgery session. We opted to use DREADDs-mediated inactivation over optogenetic silencing for imaging experiments because of the convenience of systemic ligand delivery and lack of light interference with miniature microscopic imaging. To image in vivo calcium transients of CA1 neurons with inactivation of CA1-projecting subicular neurons in the open field and during object location memory (OLM) tasks, similar viral injection strategy was used but CAV2-Cre and AAV2-DIO-hM4D-mCherry was delivered bilaterally in order to match the OLM behavioral experiments.
Histology, immunochemical staining, viral tracing data analysis
The mice were transcardially perfused; their brains were removed and left in 4% paraformaldehyde overnight, then transferred into 30% sucrose in PBS in the next day. The brain was sectioned coronally in 30 μm thickness on a freezing microtome (Leica SM2010R, Germany). Every third section was mounted for examination and quantification of starter cells and their presynaptic cells in different brain structures. For the cases of mapping input connections to CA1-projecting subicular neurons, selected sections were stained with a GFP antibody to amplify EGFP signal resulting from the helper AAV expression for robust identification of starter cells. A chicken anti-GFP primary antibody (Aves Labs, 1:500 dilution) followed with an Alexa Fluor (AF) 488-conjugated donkey anti-chicken secondary antibody (Jackson ImmunoResearch, 1:200 dilution) was applied to the sections. To immunochemically identify neuronal cells, NeuN immunostaining was used with a mouse anti-NeuN primary antibody (Millipore, 1:100) followed with an AF488 -conjugated donkey anti-mouse secondary antibody (Jackson ImmunoResearch, 1:200). To immunochemically identify GABAergic cells, GABA immunostaining was performed with a rabbit anti-GABA primary antibody (Sigma-Aldrich, 1:1000) followed with an AF488 or AF647-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch, 1:200 dilution). For the H129 tracing experiments, we performed immunostaining to amplify tdTomato signals with a rabbit anti-dsRed antibody (Clontech, 1:250 dilution) followed with a Cy3-conjugated donkey anti-rabbit secondary antibody (Jackson ImmunoResearch, 1:200 dilution). For CaMKII immunostaining, a rabbit polyclonal antibody (Santa Cruz Biotechnology, 1:500 dilution) was used followed by an AF488-conjugated donkey anti-rabbit secondary antibody. For somatostatin (SOM) immunostaining, a rat monoclonal primary antibody (EMD Millipore, 1:200 dilution) was used and followed by an AF488-conjugated donkey anti-rat secondary antibody. Sections were counter-stained with 10 μM DAPI, then mounted and cover-slipped. Please see our previous publications4,52,53,59–61 for detailed information regarding the relevant antibody characterization and specificity in these and other immunostaining experiments. Also see more information in the Life Sciences Reporting Summary.
Brain slice images were acquired by using an automated slide scanning and analysis software (Metamorph, Inc.) in a high-capacity computer coupled with a fluorescent BX61 Olympus microscope and a high-sensitivity Hamamatsu CCD camera. In addition, we imaged labeled cells in selected sections with a confocal microscope (LSM 700/780, Carl Zeiss Microscopy, Nussloch, Germany) coupled with z-stack and tile scanning features under a 20× objective lens. Quantitative examinations across the series of sections were conducted for complete and unbiased analyses of virally labeled neurons by using either Metamorph or Adobe Photoshop software (Adobe Systems, San Jose, CA).
Slice electrophysiology
To complement virus-based circuit mapping, we used our established functional mapping approach to perform ChR2-assisted circuit mapping experiment (CRACM) using the protocol of Petreanu et al. (2009)16. First, AAV1-CAG-ChR2-Venus was injected into the dorsal subiculum iontophoretically by using a Digital Midgard Precision Current Source (Stoelting Co. Wood Dale, IL). The virus was delivered by applying currents of positive 3 μA in an alternative mode with a cycle of 7s on and 7s off for 5 min. After recovery, mice were housed in the animal facility for 3 weeks before slice experiments.
Coronal sections of 400 μm were cut from dorsal hippocampus with a vibratome (VT1200S, Leica Biosystems, Buffalo Grove, IL) in sucrose containing ACSF (in mM: 85 NaCl, 75 sucrose, 2.5 KCl, 25 glucose, 1.25 NaH2PO4, 4 MgCl2, 0.5 CaCl2, and 24 NaHCO3). Slices were incubated for at least 30 minutes in normal ACSF (in mM: 126 NaCl, 2.5 KCl, 26 NaHCO3, 2 CaCl2, 2 MgCl2, 1.25 NaH2PO4, and 10 glucose) at 32 °C before transfer into slice recording chambers. Throughout the cutting, incubation and recording, the solutions were continuously supplied with 95% O2–5% CO2.
Whole cell recordings were performed under a DIC/fluorescent Olympus microscope (BX51WI, Olympus, Japan). To target whole cell recordings, cells were visualized at high magnification (60× objective, 0.9 NA; LUMPlanFl/IR, Olympus). Cell bodies of recorded neurons were at least 50 μm below the surface of the slice. Patch pipettes (4 – 6 MΩ resistance) made of borosilicate glass were filled with an internal solution containing (in mM) 126 K-gluconate, 4 KCl, 10 HEPES, 4 ATP-Mg, 0.3 GTP-Na, and 10 phosphocreatine (pH 7.2, 300 mOsm). Electrodes also contained 0.1% biocytin for post-hoc cell labeling and further morphological identification. Once stable whole-cell recordings were achieved with good access resistance (usually <30 MΩ), basic electrophysiological properties were examined through hyperpolarizing and depolarizing current injections.
During CRACM experiments, TTX (1μM) and 4-AP (100 μM) were added to bath solution to block Na+ channels and the K+ channels that are critical for repolarization of the axon so that spatially restricted activation of ChR2-expressing subicular axons with a 473 nm blue laser (~3 mW/5.5 mm2, 0.25ms; laser spot diameter, ~50 μm) only occurred in the vicinity of the laser beam (with action potentials blocked), allowing for functional assessments of direct and monosynaptic synaptic connectivity. We used sequential stimulation of the blue laser at 16 × 16 different sites arranged in a mapping grid covering most of the hippocampal slice in a nonraster, nonrandom sequence to avoid revisiting the vicinity of recently stimulated sites. Simultaneous whole-cell voltage-clamp recordings were made from CA1 pyramidal neurons to measure optogenetic stimulation-evoked excitatory postsynaptic current (EPSC) responses at the holding potential around −70mV. The EPSC response from each stimulation site was the measurement of the sum of total EPSCs within the analysis window (>10 ms to 160 ms post photostimulation), with the baseline spontaneous response subtracted from the photostimulation response of the same site. The value was normalized with the duration of the analysis window (i.e., 150 ms) and expressed as average integrated amplitudes in picoamperes (pA). For the color-coded map display, data were plotted as the average integrated EPSCs amplitude per pixel location (stimulation site), with the color scale coding input strength. After physiological assays had been completed, brain slices were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) overnight and transferred to 30% sucrose solution in PBS. The slices were stained against biocytin with 1:1,000 Cy3-conjugated streptavidin (Jackson ImmunoResearch) to show the morphology of the recorded cells. Neuron reconstructions were computer-assisted and based on stacks of optical sections acquired by a confocal microscope (LSM 700/780, Carl Zeiss).
Further, we provided direct evidence from in vitro recordings of SUB neurons bearing DREADDs to show that CNO potently inhibits these neurons. Camk2a-Cre mice were injected with AAV2-DIO-hM4D-mCherry injection in the subiculum. Three weeks following viral injection (time matched with the behavioral experimental time point), slice electrophysiology experiments were conducted. The hM4D-expressing neurons were targeted based upon their mCherry fluorescence. We also recorded from a few non-mCherry-expressing neurons for control experiments. To examine hM4D/CNO-mediated inactivation, 5 μM CNO in the recording solution was applied. The ligand application for 20 minutes was estimated to produce full hM4D/CNO effects, while the washout of 30–45 minutes was used to remove the CNO to assess post-control responses. Given hM4D is a G-protein coupled receptor, CNO effects can only be partially reversed with this washout, which is consistent with published results62–64. We focused on analyzing neuronal resting membrane potentials, spiking thresholds, and spike numbers at different steps of depolarizing current pulses at baseline, in the presence of CNO, and after its washout.
In addition, we performed in vitro electrophysiological recording of SUB neurons in response to blue laser stimulation in brain slices. Camk2a-Cre mice were injected with AAV1-DIO-ChR2-YFP injection in SUB. Three weeks following viral injection, slice electrophysiology experiments were conducted. ChR2-expressing SUB neurons were recorded, and examined with the ChR2 stimulation protocol (6 Hz, 50 ms for 3–15 minutes) applying a matched light intensity of in vivo behavioral experiments. ChR2-expressing neurons were modulated by blue laser stimulation and remained healthy. About half of the 15 recorded cells could faithfully spike following laser stimulation, while the other half of cells responded less reliably with spikes (but with clear modulation of subthreshold membrane potentials).
Mouse behavior experiments of object-location memory, novel object recognition and the dry land version of Morris water maze task
Prior to experiments, all mice were handled for 2 min/day for 6 days and were habituated to the experimental apparatus for 6 min/day for 3 consecutive days without objects. The experimental chamber is a rectangular open field arena with fixed local and distal cues (20 cm × 40 cm × 20 cm, manufactured by a local workshop at the University of California, Irvine). There were two identical objects presented to the animal in the box during the training phase of the object location memory (OLM) and novel object recognition experiments. Animals were allowed to freely explore the box and the objects for 10 min. During a 5 min test 24 hours after the training, animals were allowed to freely explore the box with one of the objects from training having been moved to a new location. For novel object recognition experiments, animals were allowed to freely explore the box with one of the objects from training replaced with another object with distinct geometry and appearance. Two different objects were used in a counterbalanced manner during object recognition experiments.
For the behavioral experiments with in vivo optogenetic stimulation, fiber optic ferrules were bilaterally implanted targeting dorsal subiculum at 2 weeks after the virus injection. During habituation and behavioral testing sessions for all mice, one end of the bifurcated optic fiber was connected to a rotary joint (Doric Lenses) on the ceiling of the behavioral box, the other end was connected to the bilateral implants. Laser delivery was at 6 Hz, 50 ms duration using a 450 nm laser during the entire 3-min training session for the optogenetic stimulation group and the control group with the EGFP control vector injected. The laser was off for the no-laser control group during training. Our decision to use theta-frequency patterned stimulation was based on prior work showing that theta-patterned stimulation in septal nucleus could improve performance on spatial tasks and that stronger theta-frequency signaling in hippocampus is correlated with stronger spatial performance65,66. It is also the case that both the hippocampus and SUB share strong co-modulation (coherence) of spatial firing according to theta-frequency oscillations10. For these reasons, we felt that theta-paced as opposed to randomly-paced stimulation might be better with respect to not disrupting theta rhythms. There was no laser delivered for all groups during the 5 min retention test. The laser power at the implant fiber tip was 2–3 mW /5.5 mm2 at a steady state, and it was measured 0.7–0.9 mW/5.5 mm2 with the delivery of 6 Hz, 50 ms duration. For behavioral experiments with DREADDs inactivation, mice were intraperitoneally injected with clozapine-N-oxide (CNO, 1.4 mg/kg) or saline 45 min before training. As the optimal use of CNO as a DREADDs ligand has been a topic of important discussion67,68, we examined the parameters that reflect animal’s behavioral states such as maximum running speeds and the number of running laps across control, CNO, and post-control session, and did not find any significant differences (Supplementary Fig. 7d–f). Further, the administration of CNO at our dosage to control mice that expressed no DREADDs was not shown to influence their object location memory performance69.
Experimental videos were recorded with ANY-MAZE software (Stoelting Co.) and analyzed offline by using customized software to score the retention times to each object with standardized criteria as previously described51,70,71. Interaction/exploration time of a mouse with the object is defined as the mouse’s nose being within 1 cm of the object and pointing directly at the object so that an elongated line from eyes to nose would hit the object. However, the following behaviors are excluded from exploration time. 1) Times for which the mouse is not approaching the object (e.g., if the mouse re-orientates itself and the nose accidentally comes close to the object). 2) The mouse is mounted on top of the object (even if it is looking down at the object). 3) The mouse rears on the object but is looking over the object (e.g. looking at the ceiling). 4) The mouse is engaged in a repetitive behavior (like digging close to the object or biting the object). The discrimination index (DI) is define as follows: (Tmoved – Tunmoved)/ (Tmoved + Tunmoved)×100% or (Tnovel − Tfamiliar)/ (Tnovel + Tfamiliar)×100%.
To enhance characterization of behavioral function of the SUB-CA1 projection, we implemented a dry-land version of the often-used Morris water maze test (an adaptation to21) to test how DREADD-based inactivation of CA1-projecting subiculum neurons impacts performance on a spatial navigation task (Supplementary Fig. 5). In the dry-land task, like the Morris water maze task, solving the task demands localization of a target in the frame of reference defined by distal visual cues (commonly known as allocentric space). It does not demand knowledge of the location of proximal cues and the possible use of proximal cues is avoided by careful attention to clearing the maze surface of odors between trials and by rotating the surface relative to the environment. Thus this behavioral task allows for a comparison of performance on the object-location task with that on a more purely spatial task related to localization in allocentric space. As illustrated in Supplemental Fig. 5c, a round board has a diameter of 60 cm with 37 wells each holding an interchangeable plastic cup (3 cm in diameter each). The wells are uniformly spread across it in a squared grid pattern. Following handling, water deprived mice were allowed to explore on the board and drink water from cups for 10 minutes of habituation for two days; all cups in the board except the 16 most peripheral ones were filled with 20 μl water each. During the first phase of pre-training (days 1–2), only one of the cups in a semi-randomly chosen quadrant of the arena had 60 μl water and it was flagged with a silver metal post close to the well (Supplementary Fig. 5c, left). The animal was released facing the edge of the board from the 3 other quadrants with 8 trials per day. The water cup location was changed to a new quadrant on each pre-training day. For each trial, the animal was allowed to explore, look for the target and drink water until a total of 2 minutes had expired. During the second phase of the pre-training period (days 3–4), the procedure was identical to the previous two days, except for the exploration time; we ended trials once the animal located and drink water from a target cup. On training day (day 5), the cohort of mice (10 mice) were either injected with CNO or saline (5 mice each) 45 minutes before trials began. The first 4 trials were run with the flag marker set close to the target cup location, and the last 4 trials were run without the flag at the same location (Supplementary Fig. 5c, right). The water cup location differed from those used during pre-training, but remained in the same location for the subsequent testing (day 6). Animals ran 8 trials with the target cup location unmarked on the testing day. Subsequently, animals remained in their home cages for two days, and then began a second pair of training and testing (days 9–10) using a new location with reversed CNO or saline treatment for different groups. A video camera was positioned directly above the round board, and all trials were videotaped and tracked.
Latency to find the targeted cup location and distance traveled to find the location were measured. Tracking in the video images was performed using a custom-written Matlab software. First, the external boundary of the dry-land maze platform was segmented from the background, and the area inside the boundary was regarded as the region of interest (ROI) for trajectory detection. The ears and tail of a mouse were segmented based on their grayscale values. Two manually selected thresholds defined the grayscale intervals where the ears and tail could be distinguished from the remaining elements inside the ROI. The ears were further distinguished from the tail by their ratios of the major axis length and minor axis length of the segmented elements. The element with a ratio less than 3 was regarded as the ear, while the other region was regarded as the tail. The trajectory was tracked with the ear positions in the video. Given that the time when the mouse was released on the platform was time 1 and the time when the mouse reached the targeted water well was time 2, the latency time of reaching the target was defined as the difference between time 2 and time 1. The corresponding distance was defined as the total travelling distance of the mouse between time 1 and time 2.
Miniscope imaging preparation, GRIN lens implantation, baseplate placement, and mouse linear track and open field mapping
At two weeks after AAV1-CaMKII-GCaMP6f injection, a gradient refractive index (GRIN) lens was implanted at the injection site in CA1. A 1.8 mm-diameter circular craniotomy was centered at the coordinates (AP −2.30 mm and ML +1.75 mm relative to bregma). ACSF was repeatedly applied to the exposed tissue; the cortex directly below the craniotomy was aspirated with a 27-gauge blunt syringe needle attached to a vacuum pump. The unilateral cortical aspiration might affect part of the anteromedial visual area determined using Allen Brain Atlas (www.brain-map.org/), but the procedure left the primary visual area intact. The GRIN lens (0.25 pitch, 0.55 NA, 1.8 mm diameter and 4.31 mm in length, Edmund Optics) was slowly lowered with a stereotaxic arm to CA1 with a depth of −1.60 mm relative to the bregma. Next, a skull screw was used to anchor the GRIN lens to the skull. Both the GRIN lens and skull screw were fixed with cyanoacrylate and dental cement. Kwik-Sil (World Precision Instruments) was used to cover the lens. Two weeks later, a small aluminum baseplate was cemented onto the animal’s head atop the existing dental cement. A miniscope was fitted into the baseplate and locked in a position so that the field of view was in focus to visualize GCaMP6f expressing neurons and visible landmarks, such as blood vessels.
In order to motivate animals to run on a linear track, access to water was regulated between every Sunday afternoon and every Friday afternoon. Mice were provided water starting at 1ml per day and with adjustments up or down until weights stabilized around 82–85% of their original body weight. In the meantime, mice were handled 5 min per day for 3 days. As illustrated in Supplementary Fig. 7, mice were trained to run on a 1-meter linear track with water rewards placed at both ends of the track for 60 laps per session for 5 days. Mice were then trained with a head-mounted miniscope (without recording) to run on the linear track with water reward for 60 laps per session for another 5 days. After all training was completed, in vivo GCaMP6f-based calcium imaging of population CA1 neurons in an awake behaving mouse was performed for the linear track experiments with miniscope recordings for 2 sessions of baseline control, 2 sessions of CNO inactivation, and 2 sessions of post-baseline control. In order to avoid GCaMP6f fluorescence bleaching, mice usually ran 1 session every other day, with 15 min recording. CNO was administered 45 min through intraperitoneal injection prior to imaging, saline was administered during control and post-control sessions in the same way. Intervals between the last CNO session and first post-control session were longer than 48 hours to ensure clearing of CNO.
For open field experiments, mice were handled for at least one week and habituated for i.p. injections and head-mounted miniscopes. Mice were then habituated for 3 days in an open field arena with miniscopes for 15 min on each day. The open field arena is a circular environment (36 cm in diameter) with fixed local and distal cues. Mice were randomly divided into two groups, with one group receiving CNO injection for DREADD-based inactivation of CA1-projecting subiculum neurons. On day 1, mice from both groups run in the open field arena for 15 min each while calcium signals from CA1 neurons are recorded. These recordings act as baseline controls. On day 2, mice from the saline control group received a single dose of saline injections 45min prior to the open field recording, while mice from the CNO treated group received a single dose of CNO injections (1.4 mg/kg) 45 min prior to the open field recording. Day 3 was off to ensure clearance of CNO. Experiments on day 4 were similar to day 1; mice from both groups explored in the open field arena for 15 min each with miniscope imaging. These recordings were used as post-controls.
For miniscope imaging during object location memory (OLM) tasks, mice were handled for at least one week and habituated for i.p. injections and head-mounted miniscopes. Mice were then habituated for 3 days in the OLM behavior box with miniscopes for 10 min on each day. For baseline recordings, animals were put in the OLM behavior box without objects and released for 10 min. At 45 min prior to training, mice were randomly divided into two groups, and one group received saline injections while the other group received CNO (1.4 mg/kg) injections. During recordings in training, two identical objects were presented to the animal during the 10 min training session. For the retention test, one of the two objects were moved to a new location and animals were allowed to explore for 10 min in order to get sufficient coverage for calcium imaging analysis. With respect to behavior, we found that the 10 min retention test gave rise to a more robust measurement with less variability in the discrimination index compared to the data analyzed based on the 1st 5 min of recording.
The linear track, open field, as well as the OLM experiments were placed in a dedicated animal behavior testing room. Two distinct shelving racks (195-cm height × 122-cm width versus 178-cm height × 152-cm width) with different objects and visual cue decorations were placed against the north and east room walls. Two identical fear-conditioning boxes (60 cm tall) were placed against the west room wall. The linear track was placed in parallel to the north rack with distinct visual cues at each end of the track. The open field enclosure was placed against the southwest edge of the east rack with cue cards inside to serve as local cues. The OLM testing arena was placed against the northwest edge of the east rack with 5 cm wide cue tape on the east wall of the enclosure. All the environments/cues remain constant during the entire series of the experiments.
Miniscope imaging data acquisition and analysis
Please refer to www.miniscope.org/22 for technical details of our custom-constructed miniscopes. The head-mounted scope has a mass of about 3 grams and uses a single, flexible coaxial cable to carry power, control signals, and imaging data to custom open source Data Acquisition (DAQ) hardware and software. Under our experimental conditions, the miniscope has a 700 μm × 450 μm field of view with a resolution of 752 pixels × 480 pixels (~1 μm per pixel). The electronics packaged the data to comply with the USB video class (UVC) protocol and then transmitted the data over Super Speed USB to a PC running custom DAQ software. The DAQ software was written in C++ and uses Open Computer Vision (OpenCV) libraries for image acquisition. Images were acquired at ~30 frames per second (fps) and recorded to uncompressed .avi files. The DAQ software simultaneously recorded animal’s behavior through a high definition webcam (Logitech) at ~30 fps, with time stamping both video streams for offline alignment.
Calcium imaging data processing.
Miniscope videos of individual sessions (i.e., control, CNO and post-control sessions) were first concatenated and down-sampled by a factor of 2 using NIH ImageJ software, then motion corrected using the NoRMCorre MATLAB package72. Next we visually inspected the maximum intensity projection images of individual video sessions, and manually performed minor linear translations to align videos of CNO and post-control sessions to the control session videos. Then we created a large combined data set by concatenating all aligned video images of control, CNO and post-control sessions, using NIH ImageJ. While this large data concatenation requires significant computer resources (i.e., 28 cores, 128GB dynamic random-access memory, 1TB solid state drive, 8TB hard disk drive), it greatly enhances our ability to obtain cross-session cell tracking from different days (Supplementary Fig. 7a), which makes unnecessary to perform individual map alignment or cell registration across sessions and facilitates hippocampal activity re-mapping experiments. Subsequent analysis was performed using custom MATLAB scripts. We adopted the newly developed method of extended constrained non-negative matrix factorization for endoscopic data (CNMF-E)73 to extract individual neuron’s calcium activity. The CNMF-E is based on the CNMF framework74, which enables simultaneously denoising, deconvolving and demixing of calcium imaging data. Its key features include modeling the large rapidly fluctuating background that has a low spatial-frequency structure and allows a good separation of single-neuron signals from this background (see Ref73 for details). After iteratively solving a constrained matrix factorization problem, CNMF-E extracts the spatial footprints of neurons and their associated temporal calcium activity. Specifically, the first step of estimating neuron’s temporal activity (a scaled version of dF/F, which is a metric used in most calcium imaging literature) is computing the weighted average of fluorescence intensities after subtracting other neurons’ temporal activity within the neuron’s region of interest. Then a deconvolution algorithm OASIS75 is applied to obtain the denoised neuron activity and deconvolved spiking activity, as illustrated in Supplementary Fig. 6b. Using the animal’s behavioral tracking video data, the position and speed of the animal was determined with a custom MATLAB script. We identified the time points at which the speed of the animal was lower than 3 cm/s and excluded them from further analysis. We then temporally aligned the position data to the calcium imaging data using linear interpolation.
Calculation of positional calcium event rates.
Once the deconvolved spiking activity of the neurons was extracted, we localized effective neuronal calcium events using a threshold (i.e., 10% to the maxima in linear track experiments) to deconvolved spiking activity across all the sessions. To characterize the neural activity, we calculated individual neuron’s positional event rate by dividing the total number of events of each neuron at each bin location by the total occupancy time at each location, where locations were defined by sorting the behavior tracking data into 2.7 cm × 2.7 cm bins after excluding 5% of the total length on either end of the linear track. Calcium event rate maps were smoothed using a 2D convolution with a Gaussian filter with a standard deviation of 2 cm. To find the in-field peak event rates for a place cell, the unsmoothed event rate maps were used, and the bin locations with a total occupancy time less than 0.2 sec were excluded. To determine the place field shift of the same place cell longitudinally, we calculated 2D cross-correlations of the smoothed event rate maps across different days. The cross-correlogram between event rate maps across two different sessions of matched neurons were computed, and then the Euclidian distance of the bin with the highest correlation was found from physical centers of the cross-correlogram was determined. This method76 circumvents the difficulty to match individual fields across days, which becomes inaccurate when multiple place fields are detected in at least one of the recording sessions.
Spatial information statistics.
To quantify the spatial location specificity of calcium events in terms of the information content of cell activity, spatial information scores in bits per second and bits per event were calculated for each neuron according to the following formula24,
where is the probability of the mouse occupying the i-th bin for the neuron, is the neuron’s event rate in the i-th bin, while is the mean rate of the neuron across the whole session. Bins with total occupancy time less than 0.2 sec are excluded from the calculation. To define place cells, event locations for each neuron were shuffled 1000 times and spatial information (bits / sec) was recalculated for each shuffle. This generated a distribution of shuffled information scores for each individual neuron. The value at 95% of each shuffled distribution was selected as the threshold for designation of the corresponding individual place cell. Neurons with less than ten calcium events for a given session were excluded.
Classification of place cells.
We categorized place cells into different groups according to their individual changes in cross-session spatial information scores (bits/second). For each place cell, we examined whether the spatial information score in the CNO session was significantly different from that in both the control and post-control sessions using a jackknife resampling technique28. To do so, each track-running session was divided into 10 equal-duration sub-sessions. We then applied the jackknife resampling to the full recording session, each time calculating bits/second values by omitting one sub-session. Therefore, for each place cell, the extended set of bits/second values, produced by this jackknife resampling procedure, include 10 values, which are denoted by xi (i = 1,…,10). The mean and variance of this sampling distribution is
After calculating and from the CNO session, we contrast it against the output from another session, e.g. and from the control session. Then we can construct a jackknife-based t-test, where the statistical test-value Q could be calculated as
A p-value was calculated for Q from a standard normal distribution, and a significance level of 0.05 was used to determine whether they are significantly different. Each place cell is assigned to an appropriate group if the spatial information in the CNO session is significantly different from that in both the control and post-control sessions. A place cell can be classified as ‘bit decrease’ only when its spatial information in the CNO session is significantly lower than both the control and post-control sessions, and vice versa for the ‘bit increase’. A place cell that passes the statistical test in the CNO vs. control and vs. post-control in the opposite directions is classified into ‘un-recovered’ groups. The rest of the place cells, which failed at least one of the statistical tests, are un-assigned and excluded for the category analysis.
Open field data analysis.
Most of the analysis conducted for open field experiments is similar to linear track experiments. Briefly, the time points at which the speed of the animal was lower than 2 cm/s were excluded from further analysis. Once the temporal activity of the neurons was extracted, we localized neuronal calcium events using a threshold of 3 times of the standard deviation of the calcium signals across all the sessions. To characterize the neural activity, we calculated individual neuron’s positional event rate by dividing the total number of events of each neuron at each bin location by the total occupancy time at each location (each 2 cm × 2 cm bin of behavior tracking data). In calculating spatial information scores of neural activity, bins with occupancy times less than 0.2 sec were excluded.
Miniscope imaging data analysis of OLM tasks.
We constructed calcium event rate maps in the same manner as the linear track experiment by sorting the behavior tracking data into bins of 1.5 cm × 1.5 cm. The bins with a total occupancy of less than 0.1s were regarded as unvisited and were not included in the rate map. For each mouse, the ensemble event rates are the summation of events from all neurons, normalized by time spent at each location. Of note, the individual neuron’s event rate associated with the object was quantified as the average event rate across bins within 3 cm surrounding the object. The total sum of individual neuron’s event rates of all CA1 neurons was defined as the ensemble event rate associated with the object. The relative event rate was expressed as a discrimination index (DI = (ER moved – ER unmoved) / (ER moved + ER unmoved)×100%) wherein ER moved and ER unmoved are the ensemble event rates associated with the moved and unmoved objects, respectively.
The determination of place cells is similar to the open field experiment. Place cell fields were determined using the approach in a previous study77, and were located by finding the centers of calcium activity from the binned event rate maps. The fields too close to each other (defined as less than 70% of the half distance between the center point of the spatial autocorrelation map and the closest local maximum) were removed. Place fields with in-filed event rates that did not reach 25% of the peak event rate of the rate map were also excluded. We measured individual place fields whose peaks were located within a 3-cm radius of either object.
Position decoding using a neural network model
We built a neural network model to predict the position of mice during track-running given the temporal activity of a specific set of CA1 neurons. Feedforward neural networks involve sequences of nonlinear transforms, composed with linear transformations (weights) of input data, forming layers of hidden units. Using the Tensorflow library, we created a fully connected feedforward neural network with 3 hidden layers, with rectified linear unit activations after each hidden layer as the nonlinearities. We included dropout (at 50%), L2 regularization, and batch normalization to regularize the model78,79, to decrease generalization error. The network was trained using the ADAM optimizer80. 10% of the linear track from both ends were removed, and the neural data were normalized using ‘z-scores’. As input to the neural network, we took 10–20 timesteps of neural activity data from each side of the temporal point at which we wished to predict position. The hyperparameters (i.e., the number of neurons per layer and the amount of neural data as input) of the model were determined by 5-fold cross-validation. We split the data from the first control session into a training set (80% of data) and a contiguous testing set (20% of data). After training, the model can be used to predict position on the testing set. We applied the trained model based on the first control session to other sessions (e.g., CNO and post-control), to investigate the effects of subicular inactivation on position decoding. Prediction (decoding) error was then computed as the mean absolute difference between the predicted and actual position on the horizontal track, which was formulated as where represents the actual position of the animal at the t-th timestep, is the position decoded by our model, and N is the number data points in the test set for any given session (i.e., 20% of the control data, and 100% of the CNO and post-control data).
Statistical Analysis
Data were presented as mean ± SE, mean ± SD or median ± interquartile range (IQR), as indicated. For statistical comparisons between groups, the data were checked for normality of distribution. If the criteria were met, a t-test was performed to compare two groups; otherwise, a Mann–Whitney U-test was used. For statistical comparisons across more than two groups, One-Way ANOVA and related multiple comparison tests were used for group comparisons. For paired statistical comparisons, a paired t-test was used. For data with repeated measurements for more than two groups, repeated measures ANOVA and related multiple comparison tests were used to compare between groups. In all experiments, the level of statistical significance was defined as p ≤ 0.05.
Code Accessibility
The custom code used for the analyses for the current study is available from the corresponding author upon reasonable request.
Data Availability
The datasets generated for the current study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgements
This work was supported by NIH grants including a BRAIN Initiative Grant (NS104897, X.X., D.A.N), other grants NS078434 (X.X.), MH105427 (X.X, P.G.), NS095355 (Q.N.), and a NSF grant DMS1763272 (Q.N.) and a Simons Foundation grant (594598, QN). TCH is supported by R35 GM127102. P.Z is supported by NIH NIBIB R01EB022913, NSF NeuroNex award DBI-1707398 and Gatsby Foundation. This work was also made possible, in part, through access to the confocal facility of the Optical Biology Shared Resource of the Cancer Center Support Grant (CA-62203) at UC, Irvine.
Footnotes
Competing Interests
The authors declare no competing interests.
References
- 1.Henriksen EJ et al. Spatial representation along the proximodistal axis of CA1. Neuron 68, 127–137, doi: 10.1016/j.neuron.2010.08.042 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kjelstrup KB et al. Finite scale of spatial representation in the hippocampus. Science 321, 140–143, doi: 10.1126/science.1157086 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Xu X, Sun Y, Holmes TC & Lopez AJ Noncanonical connections between the subiculum and hippocampal CA1. J Comp Neurol, doi: 10.1002/cne.24024 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sun Y et al. Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell Rep 7, 269–280, doi: 10.1016/j.celrep.2014.02.030 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Nitz DA, Holmes TC & Xu X Opposing and Complementary Topographic Connectivity Gradients Revealed by Quantitative Analysis of Canonical and Noncanonical Hippocampal CA1 Inputs. eNeuro 5, doi: 10.1523/ENEURO.0322-17.2018 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Witter MP Connections of the subiculum of the rat: topography in relation to columnar and laminar organization. Behav Brain Res 174, 251–264, doi: 10.1016/j.bbr.2006.06.022 (2006). [DOI] [PubMed] [Google Scholar]
- 7.Cembrowski MS et al. Dissociable Structural and Functional Hippocampal Outputs via Distinct Subiculum Cell Classes. Cell, doi: 10.1016/j.cell.2018.03.031 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Naber PA, Witter MP & Lopes Silva FH Networks of the hippocampal memory system of the rat. The pivotal role of the subiculum. Ann N Y Acad Sci 911, 392–403 (2000). [DOI] [PubMed] [Google Scholar]
- 9.Morris RG, Schenk F, Tweedie F & Jarrard LE Ibotenate Lesions of Hippocampus and/or Subiculum: Dissociating Components of Allocentric Spatial Learning. Eur J Neurosci 2, 1016–1028 (1990). [DOI] [PubMed] [Google Scholar]
- 10.Jackson J et al. Reversal of theta rhythm flow through intact hippocampal circuits. Nat Neurosci 17, 1362–1370, doi: 10.1038/nn.3803 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Berger TW, Swanson GW, Milner TA, Lynch GS & Thompson RF Reciprocal anatomical connections between hippocampus and subiculum in the rabbit evidence for subicular innervation of regio superior. Brain Res 183, 265–276 (1980). [DOI] [PubMed] [Google Scholar]
- 12.Shao LR & Dudek FE Electrophysiological evidence using focal flash photolysis of caged glutamate that CA1 pyramidal cells receive excitatory synaptic input from the subiculum. J Neurophysiol 93, 3007–3011, doi: 10.1152/jn.00877.2004 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Schwarz LA et al. Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92, doi: 10.1038/nature14600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo L & Anderson DJ A Cre-dependent, anterograde transsynaptic viral tracer for mapping output pathways of genetically marked neurons. Neuron 72, 938–950, doi:S0896–6273(11)01044–0 [pii]10.1016/j.neuron.2011.12.002 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heimer-McGinn VR, Poeta DL, Aghi K, Udawatta M & Burwell RD Disconnection of the Perirhinal and Postrhinal Cortices Impairs Recognition of Objects in Context But Not Contextual Fear Conditioning. J Neurosci 37, 4819–4829, doi: 10.1523/JNEUROSCI.0254-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petreanu L, Mao T, Sternson SM & Svoboda K The subcellular organization of neocortical excitatory connections. Nature 457, 1142–1145, doi:nature07709 [pii] 10.1038/nature07709 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsien JZ et al. Subregion- and cell type-restricted gene knockout in mouse brain. Cell 87, 1317–1326 (1996). [DOI] [PubMed] [Google Scholar]
- 18.Klausberger T & Somogyi P Neuronal diversity and temporal dynamics: the unity of hippocampal circuit operations. Science 321, 53–57, doi: 10.1126/science.1149381 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roy DS et al. Distinct Neural Circuits for the Formation and Retrieval of Episodic Memories. Cell 170, 1000–1012 e1019, doi: 10.1016/j.cell.2017.07.013 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roth BL DREADDs for Neuroscientists. Neuron 89, 683–694, doi: 10.1016/j.neuron.2016.01.040 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kesner RP, Farnsworth G & Kametani H Role of parietal cortex and hippocampus in representing spatial information. Cereb Cortex 1, 367–373 (1991). [DOI] [PubMed] [Google Scholar]
- 22.Cai DJ et al. A shared neural ensemble links distinct contextual memories encoded close in time. Nature 534, 115–118, doi: 10.1038/nature17955 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ziv Y et al. Long-term dynamics of CA1 hippocampal place codes. Nat Neurosci 16, 264–266, doi: 10.1038/nn.3329 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skaggs WE, McNaughton BL, Wilson MA & Barnes CA Theta phase precession in hippocampal neuronal populations and the compression of temporal sequences. Hippocampus 6, 149–172, doi: 10.1002/(SICI)1098-1063(1996)6:2<149::AID-HIPO6>3.0.CO;2-K (1996). [DOI] [PubMed] [Google Scholar]
- 25.Kanter BR et al. A Novel Mechanism for the Grid-to-Place Cell Transformation Revealed by Transgenic Depolarization of Medial Entorhinal Cortex Layer II. Neuron 93, 1480–1492 e1486, doi: 10.1016/j.neuron.2017.03.001 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Miao C et al. Hippocampal Remapping after Partial Inactivation of the Medial Entorhinal Cortex. Neuron 88, 590–603, doi: 10.1016/j.neuron.2015.09.051 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Leutgeb S et al. Independent codes for spatial and episodic memory in hippocampal neuronal ensembles. Science 309, 619–623, doi: 10.1126/science.1114037 (2005). [DOI] [PubMed] [Google Scholar]
- 28.Crowley PH Resampling Methods for Computation-Intensive Data-Analysis in Ecology and Evolution. Annu Rev Ecol Syst 23, 405–447, doi:DOI 10.1146/annurev.es.23.110192.002201 (1992). [DOI] [Google Scholar]
- 29.Deshmukh SS & Knierim JJ Influence of local objects on hippocampal representations: Landmark vectors and memory. Hippocampus 23, 253–267, doi: 10.1002/hipo.22101 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji D & Wilson MA Coordinated memory replay in the visual cortex and hippocampus during sleep. Nat Neurosci 10, 100–107, doi: 10.1038/nn1825 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Alexander AS & Nitz DA Retrosplenial cortex maps the conjunction of internal and external spaces. Nat Neurosci 18, 1143–1151, doi: 10.1038/nn.4058 (2015). [DOI] [PubMed] [Google Scholar]
- 32.Jacob PY et al. An independent, landmark-dominated head-direction signal in dysgranular retrosplenial cortex. Nat Neurosci 20, 173–175, doi: 10.1038/nn.4465 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Czajkowski R et al. Encoding and storage of spatial information in the retrosplenial cortex. Proc Natl Acad Sci U S A 111, 8661–8666, doi: 10.1073/pnas.1313222111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makino H & Komiyama T Learning enhances the relative impact of top-down processing in the visual cortex. Nat Neurosci 18, 1116–1122, doi: 10.1038/nn.4061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klausberger T GABAergic interneurons targeting dendrites of pyramidal cells in the CA1 area of the hippocampus. Eur J Neurosci 30, 947–957, doi: 10.1111/j.1460-9568.2009.06913.x (2009). [DOI] [PubMed] [Google Scholar]
- 36.Agster KL & Burwell RD Hippocampal and subicular efferents and afferents of the perirhinal, postrhinal, and entorhinal cortices of the rat. Behav Brain Res 254, 50–64, doi: 10.1016/j.bbr.2013.07.005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoydal OA, Skytoen ER, Andersson SO, Moser MB & Moser EI Object-vector coding in the medial entorhinal cortex. Nature 568, 400–404, doi: 10.1038/s41586-019-1077-7 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Wang C et al. Egocentric coding of external items in the lateral entorhinal cortex. Science 362, 945–949, doi: 10.1126/science.aau4940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klausberger T et al. Brain-state- and cell-type-specific firing of hippocampal interneurons in vivo. Nature 421, 844–848, doi: 10.1038/nature01374 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Maccaferri G & McBain CJ Long-term potentiation in distinct subtypes of hippocampal nonpyramidal neurons. J Neurosci 16, 5334–5343 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barker GR & Warburton EC When is the hippocampus involved in recognition memory? J Neurosci 31, 10721–10731, doi: 10.1523/JNEUROSCI.6413-10.2011 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deshmukh SS, Johnson JL & Knierim JJ Perirhinal cortex represents nonspatial, but not spatial, information in rats foraging in the presence of objects: comparison with lateral entorhinal cortex. Hippocampus 22, 2045–2058, doi: 10.1002/hipo.22046 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Barker GR et al. Separate elements of episodic memory subserved by distinct hippocampal-prefrontal connections. Nat Neurosci 20, 242–250, doi: 10.1038/nn.4472 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Bussey TJ, Duck J, Muir JL & Aggleton JP Distinct patterns of behavioural impairments resulting from fornix transection or neurotoxic lesions of the perirhinal and postrhinal cortices in the rat. Behav Brain Res 111, 187–202 (2000). [DOI] [PubMed] [Google Scholar]
- 45.Taylor KI, Moss HE, Stamatakis EA & Tyler LK Binding crossmodal object features in perirhinal cortex. Proc Natl Acad Sci U S A 103, 8239–8244, doi: 10.1073/pnas.0509704103 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu P & Bilkey DK The effect of excitotoxic lesions centered on the hippocampus or perirhinal cortex in object recognition and spatial memory tasks. Behav Neurosci 115, 94–111 (2001). [DOI] [PubMed] [Google Scholar]
- 47.Ho JW et al. Bidirectional Modulation of Recognition Memory. J Neurosci 35, 13323–13335, doi: 10.1523/JNEUROSCI.2278-15.2015 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tsao A, Moser MB & Moser EI Traces of experience in the lateral entorhinal cortex. Curr Biol 23, 399–405, doi: 10.1016/j.cub.2013.01.036 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Scaplen KM, Ramesh RN, Nadvar N, Ahmed OJ & Burwell RD Inactivation of the Lateral Entorhinal Area Increases the Influence of Visual Cues on Hippocampal Place Cell Activity. Front Syst Neurosci 11, 40, doi: 10.3389/fnsys.2017.00040 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ito HT, Zhang SJ, Witter MP, Moser EI & Moser MB A prefrontal-thalamo-hippocampal circuit for goal-directed spatial navigation. Nature 522, 50–55, doi: 10.1038/nature14396 (2015). [DOI] [PubMed] [Google Scholar]
Methods-only references
- 51.Haettig J, Sun Y, Wood MA & Xu X Cell-type specific inactivation of hippocampal CA1 disrupts location-dependent object recognition in the mouse. Learn Mem 20, 139–146, doi: 10.1101/lm.027847.112 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun Y et al. Neuregulin-1/ErbB4 Signaling Regulates Visual Cortical Plasticity. Neuron 92, 160–173, doi: 10.1016/j.neuron.2016.08.033 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuhlman SJ et al. A disinhibitory microcircuit initiates critical-period plasticity in the visual cortex. Nature 501, 543–546, doi:nature12485 [pii]10.1038/nature12485 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun Y, Grieco SF, Holmes TC & Xu X Local and Long-Range Circuit Connections to Hilar Mossy Cells in the Dentate Gyrus. eNeuro 4, doi: 10.1523/ENEURO.0097-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seidler B et al. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proc Natl Acad Sci U S A 105, 10137–10142, doi: 10.1073/pnas.0800487105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madisen L et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140, doi: 10.1038/nn.2467 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tervo DG et al. A Designer AAV Variant Permits Efficient Retrograde Access to Projection Neurons. Neuron 92, 372–382, doi: 10.1016/j.neuron.2016.09.021 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Madisen L et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci 15, 793–802, doi:nn.3078 [pii]10.1038/nn.3078 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen AQ, Dela Cruz JA, Sun Y, Holmes TC & Xu X Genetic cell targeting uncovers specific neuronal types and distinct subregions in the bed nucleus of the stria terminalis. J Comp Neurol, doi: 10.1002/cne.23954 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xu X, Roby KD & Callaway EM Mouse cortical inhibitory neuron type that coexpresses somatostatin and calretinin. J Comp Neurol 499, 144–160, doi: 10.1002/cne.21101 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Xu X, Roby KD & Callaway EM Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol 518, 389–404, doi: 10.1002/cne.22229 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ikrar T, Shi Y, Velasquez T, Goulding M & Xu X Cell-type specific regulation of cortical excitability through the allatostatin receptor system. Front Neural Circuits 6, 2, doi: 10.3389/fncir.2012.00002 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ferguson SM et al. Transient neuronal inhibition reveals opposing roles of indirect and direct pathways in sensitization. Nat Neurosci 14, 22–24, doi: 10.1038/nn.2703 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krashes MJ et al. Rapid, reversible activation of AgRP neurons drives feeding behavior in mice. J Clin Invest 121, 1424–1428, doi: 10.1172/JCI46229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Richard GR et al. Speed modulation of hippocampal theta frequency correlates with spatial memory performance. Hippocampus 23, 1269–1279, doi: 10.1002/hipo.22164 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buzsaki G Theta oscillations in the hippocampus. Neuron 33, 325–340 (2002). [DOI] [PubMed] [Google Scholar]
- 67.Gomez JL et al. Chemogenetics revealed: DREADD occupancy and activation via converted clozapine. Science 357, 503–507, doi: 10.1126/science.aan2475 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mahler SV & Aston-Jones G CNO Evil? Considerations for the Use of DREADDs in Behavioral Neuroscience. Neuropsychopharmacology 43, 934–936, doi: 10.1038/npp.2017.299 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lopez AJ et al. Promoter-Specific Effects of DREADD Modulation on Hippocampal Synaptic Plasticity and Memory Formation. J Neurosci 36, 3588–3599, doi: 10.1523/JNEUROSCI.3682-15.2016 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bui AD et al. Dentate gyrus mossy cells control spontaneous convulsive seizures and spatial memory. Science 359, 787–790, doi: 10.1126/science.aan4074 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vogel-Ciernia A & Wood MA Examining object location and object recognition memory in mice. Curr Protoc Neurosci 69, 8 31 31–17, doi: 10.1002/0471142301.ns0831s69 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pnevmatikakis EA & Giovannucci A NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J Neurosci Methods 291, 83–94, doi: 10.1016/j.jneumeth.2017.07.031 (2017). [DOI] [PubMed] [Google Scholar]
- 73.Zhou P et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife 7, doi: 10.7554/eLife.28728 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pnevmatikakis EA et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285–299, doi: 10.1016/j.neuron.2015.11.037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Friedrich J, Zhou P & Paninski L Fast online deconvolution of calcium imaging data. PLoS Comput Biol 13, e1005423, doi: 10.1371/journal.pcbi.1005423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mallory CS, Hardcastle K, Bant JS & Giocomo LM Grid scale drives the scale and long-term stability of place maps. Nat Neurosci 21, 270–282, doi: 10.1038/s41593-017-0055-3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ismakov R, Barak O, Jeffery K & Derdikman D Grid Cells Encode Local Positional Information. Curr Biol 27, 2337–2343 e2333, doi: 10.1016/j.cub.2017.06.034 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Srivastava N, Hinton G, Krizhevsky A, Sutskever I & Salakhutdinov R Dropout: A Simple Way to Prevent Neural Networks from Overfitting. J Mach Learn Res 15, 1929–1958 (2014). [Google Scholar]
- 79.Ioffe Sergey & Szegedy C Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift. arXiv:1502.03167 (2015). [Google Scholar]
- 80.Kingma Diederik P. & Ba J Adam: A Method for Stochastic Optimization. arXiv:1412.6980 (2017). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated for the current study are available from the corresponding author upon reasonable request.