Abstract
Injured tubular epithelium exhibits cellular plasticity in that it can dedifferentiate, re-enter the cell cycle and subsequently either redifferentiate or adopt a chronically injured phenotype. Although some nephrogenic genes are re-expressed during injury and repair, developmental pathways are only partially recapitulated and the process is more accurately viewed as an entirely new program intrinsic to the regenerative response to injury. Recent advances in our understanding of the molecular circuitry underpinning epithelial plasticity have come from bulk, cell-specific and single cell transcriptomic analyses. These results have begun to define the signaling pathways and gene regulatory networks governing the epithelial injury response. In this review, we highlight recent transcriptomic analyses in kidney injury, repair and fibrosis and outline the ways that these studies are improving our understanding of kidney regeneration.
Introduction
Acute kidney injury (AKI) is highly prevalent and occurs in one in five adults and one in three children hospitalized with acute illness (1). An episode of AKI can predict not only the future development of CKD, but also mortality (2). It is increasingly recognized that incomplete or maladaptive repair, characterized by ongoing localized tubule injury, might explain the epidemiologic link between AKI and future risk of CKD (3–5). Technological advances in transcriptomics, such as RNA-sequencing (RNA-seq), ribosome profiling and single cell RNA-seq (scRNA-seq) are now providing much more comprehensive information about the molecular events occurring during AKI and repair than previously possible (6). These advances should ultimately enable development of therapeutic strategies to harness repair mechanisms to promote successful recovery from AKI while suppressing maladaptive responses that can lead to failed repair and fibrosis (7, 8).
Bulk RNA-seq studies in AKI
Two recent studies from the McMahon Laboratory have used bulk RNA-seq to characterize the full course of human AKI in great detail. This approach entails extraction of all mRNA from a kidney sample followed by cDNA library generation and sequencing. Its advantages include simplicity, ability to perform on cryopreserved samples and the ability to sequence very deeply and detect nearly all transcripts present. The primary disadvantage is that this approach is unable to identify the response of an individual kidney cell type because gene expression signatures are integrated from all the cells present in the sample. In these two studies, they analyzed the transcriptomes from 42 kidney transplant recipients at 4 separate time points: before and after implantation and 3 and 12 months after transplantation.
In one study, Cippà et al. applied an elegant computational approach based on single cell RNA-seq analysis to identify and characterize shared trajectories of disease progression among these 42 disparate datasets (9). The primary finding was that distinct gene signatures differentiate successful repair after reperfusion (the transplant itself was appropriately categorized as an AKI) from failed repair characterized by progression to chronic kidney disease (CKD). In particular, genes related to mitochondrial homeostasis, senescence and inflammation predominated in biopsies from patients that progressed to CKD.
A second study analyzed this same dataset in more detail, focusing on B cell-associated transcripts that segregated with patients that developed CKD compared with those that underwent successful repair (10). They sought to answer whether a B cell alloimmune response was an intrinsic component of CKD progression, or alternatively that the host immune response actively causes allograft damage, sustaining CKD transition. They garnered several independent lines of evidence supporting the latter interpretation, ie that the late B cell immune response is pathogenic, related to ongoing tissue injury and driving fibrosis. These lines of evidence included (a) a lack of histologic evidence of B cell-mediated immunity, (b) the presence of ectopic lymphoid tissue composed predominantly of B cells in late stages after AKI, (c) evidence for clonal expansion of B cell subsets, based on B cell receptor repertoire sequencing and (d) the presence of autoantibodies in a long term mouse model or AKI – despite the fact that this was an autologous, non–transplant model. This important result shows the power of transcriptomic analyses to reveal novel (and in this case somewhat unexpected) biologic insight.
Proximal tubule-specific RNA-seq in AKI
Recently we have applied the Translating Ribosome Affinity Purification (TRAP) method to transcriptionally profile injured proximal tubule during AKI. This method involves the affinity purification of mRNA bound to ribosomes by expression of a tagged ribosomal fusion protein expressed specifically in proximal tubule but not other kidney cell types. We have previously validated this approach in both podocytes and myofibroblasts (11, 12). Using a Kim-1-CreERt2 knockin mouse line that we generated, we could specifically profile injured proximal tubule over the course of acute injury and repair, since Kim-1 (gene name Havcr-1) is only expressed in injured proximal tubule (13).
We validated our approach by showing that acutely injured and genetically labeled proximal tubule co-expressed Kim-1, Vimentin, Sox9 and Ki67, indicating a dedifferentiated and proliferative state. Translational profiling during injury and repair revealed signatures of both successful and unsuccessful maladaptive repair. It is also possible that some of these transcriptomic changes represent neither successful nor maladaptive repair, but rather adaptive responses of the cell to the injury environment. We were particularly interested in the gene regulatory network governing repair, and so we specifically analyzed upregulated transcription factors. This identified forkhead box protein M1 (FoxM1) as one of the most highly upregulated transcription factors in injured proximal tubule. FoxM1 in some cellular contexts regulates cell cycle progression. We could also identify known downstream targets of FoxM1 Ccnb1, Plk1, Aurkb, which are also all cell cycle related proteins. To test whether FoxM1 regulates epithelial proliferation, we performed siRNA knockdown of Foxm1 in human proximal tubular epithelial cells which indeed suppressed proliferation in vitro. These results implicate FoxM1 as a novel proximal tubule-specific regulator of injury-induced cell proliferation.
scRNA-seq
Finally, recent technological advances now allow massively parallel scRNA-seq, and this is beginning to be applied to better understand mammalian kidney injury and repair. An important limitation has been the generation of a single cell suspension that contains all kidney cell types and that preserves RNA integrity. Unfortunately, generating single cell suspensions has turned out to be very difficult in adult kidney, and even more challenging in postischemic kidney which contains inflammatory cells and can be fibrotic. We have recently shown that generating nuclear preparations circumvents these limitations, because the dissociation uses NP40 detergent and dounce homogenization – which frees even cells embedded in fibrotic matrix – while preventing RNA degradation (14).
We used nuclear preparations to perform scRNA-seq on mouse kidney subjected to unilateral ureteral obstruction. We could identify many proximal tubule epithelia in this dataset, including two novel proximal tubule subclusters. We annotated one of these subclusters as ‘proliferating proximal tubule,’ because it expressed a proliferation cell signature. Reassuringly, this cluster exclusively expresses FoxM1, validating our prior identification of this transcription factor by TRAP of injured proximal tubule (Figure 1).
Figure 1. Cell-type expression pattern of FoxM1 by single nucleus RNA-seq of day 14 unilateral ureteral obstruction (UUO) mouse kidney.
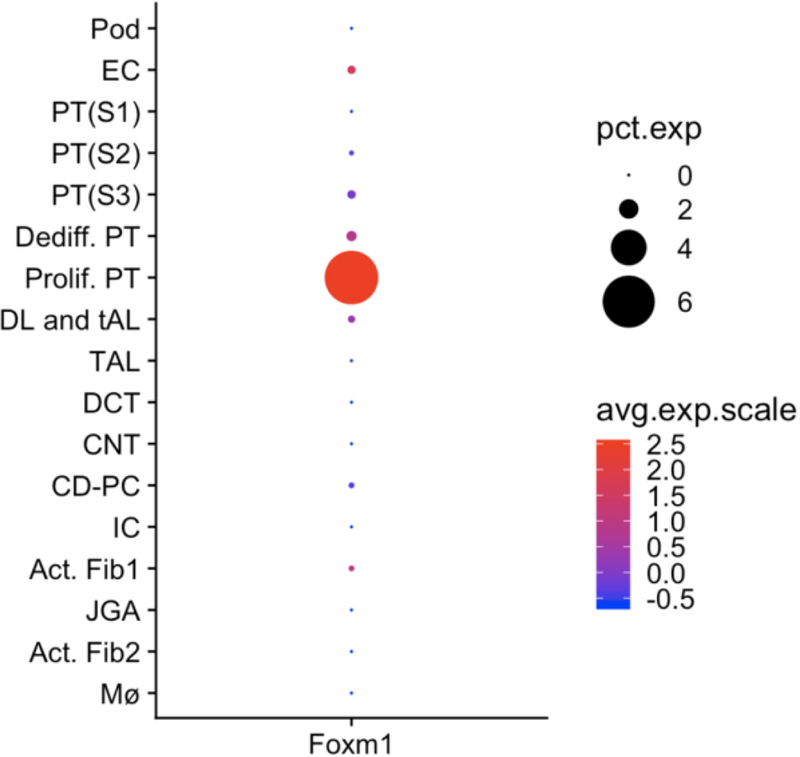
The only kidney cell type that expresses appreciable FoxM1 is the ‘proliferating proximal tubule’ cell cluster, suggesting a role for FoxM1 in regulating proximal tubule proliferation after injury. Data is from the Kidney Interactive Transcriptomics website (http://humphreyslab.com/SingleCell/). Pct. Exp., percent expression; avg. exp. Scale, average expression scale; Pod, podocyte; EC, endothelial cell; PT, proximal tubule; DL, descending loop; tAL, thin ascending loop; TAL, thick ascending loop; DCT, distal convoluted tubule; CNT, connecting segment; CD-PC, collecting duct – principal cell; IC, intercalated cell; Act. Fib., activated fibroblasts; JGA, juxtaglomerular apparatus; Mϕ, macrophage.
Future questions
We expect that the first transcriptional atlases of kidney injury and repair at the single cell level will become available in the near term. These are sure to lead to greater insight concerning the molecular pathways that underlie successful and failed epithelial repair. But transcriptomic data may not answer the more fundamental question of whether epithelial cell plasticity during repair reflects different transient cell states, or whether injury can induce a fundamentally different epithelial cell type. For example, for the failed proximal tubule gene signature that we have recently identified, does this represent a transient injury state that persists because of ongoing injury, or is it an entirely new, stable cell type that promotes fibrosis?
One way to conceptualize the difference between a transient cell state and a distinct cell type is the spectrum of genes that are available to be expressed by that cell at any moment. Chromatin accessibility is one determinant of whether a gene can be expressed. For example, inactive genes maintained in an open chromatin configuration are immediately available to be expressed by that cell, but genes sequestered in closed chromatin are not. So one way to ask whether a proximal tubule cell that has failed to repair is a fundamentally new cell type is whether its epigenetic profile – its global chromatin accessibility profile – is different from either an acutely injured or a healthy proximal tubule cell. A variety of new technologies are now available to scan the epigenome – including at the single cell level. We predict that these techniques represent a potentially powerful new approach to understand the fundamental cell biology of kidney injury and repair. Since drugs targeting the epigenome (15) are in development, these kinds of studies hold promise for novel therapeutic approaches in AKI.
Acknowledgements
Work in the Humphreys Lab is supported by NIH/NIDDK grants DK103740 and DK107374 and by the NIDDK Diabetic Complications Consortium (DiaComp, www.diacomp.org) grants DK076169 and DK115255 (all to B.D.H).
References
- 1.Susantitaphong P, Cruz DN, Cerda J, Abulfaraj M, Alqahtani F, Koulouridis I, et al. World incidence of AKI: a meta-analysis. Clin J Am Soc Nephrol 2013;8(9):1482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coca SG, Yusuf B, Shlipak MG, Garg AX, Parikh CR. Long-term risk of mortality and other adverse outcomes after acute kidney injury: a systematic review and meta-analysis. Am J Kidney Dis 2009;53(6):961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amdur RL, Chawla LS, Amodeo S, Kimmel PL, Palant CE. Outcomes following diagnosis of acute renal failure in U.S. veterans: focus on acute tubular necrosis. Kidney Int 2009;76(10):1089–97. [DOI] [PubMed] [Google Scholar]
- 4.Hsu RK, Hsu CY. The Role of Acute Kidney Injury in Chronic Kidney Disease. Semin Nephrol 2016;36(4):283–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basile DP, Bonventre JV, Mehta R, Nangaku M, Unwin R, Rosner MH, et al. Progression after AKI: Understanding Maladaptive Repair Processes to Predict and Identify Therapeutic Treatments. J Am Soc Nephrol 2016;27(3):687–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu H, Humphreys BD. The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int 2017;92(6):1334–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang-Panesso M, Humphreys BD. Cellular plasticity in kidney injury and repair. Nature reviews Nephrology 2017;13(1):39–46. [DOI] [PubMed] [Google Scholar]
- 8.Humphreys BD. Mechanisms of Renal Fibrosis. Annual review of physiology 2018;80:309–26. [DOI] [PubMed] [Google Scholar]
- 9.Cippa PE, Sun B, Liu J, Chen L, Naesens M, McMahon AP. Transcriptional trajectories of human kidney injury progression. JCI Insight 2018;3(22). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cippa PE, Liu J, Sun B, Kumar S, Naesens M, McMahon AP. A late B lymphocyte action in dysfunctional tissue repair following kidney injury and transplantation. Nature communications 2019;10(1):1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grgic I, Genovese G, Pollak MR, Humphreys BD, editors. Translational Profiling of Podocytes in Focal Segmental Glomerulosclerosis. American Society of Nephrology; 2011; Philadelphia, Pennsylvania. [Google Scholar]
- 12.Grgic I, Krautzberger AM, Hofmeister A, Lalli M, Dirocco DP, Fleig SV, et al. Translational Profiles of Medullary Myofibroblasts during Kidney Fibrosis. J Am Soc Nephrol 2014. [DOI] [PMC free article] [PubMed]
- 13.Chang-Panesso M, Kadyrov FF, Lalli M, Wu H, Ikeda S, Kobayashi A, et al. FoxM1 drives proximal tubule proliferation during repair from acute kidney injury. bioRxiv 2018:436576. [DOI] [PMC free article] [PubMed]
- 14.Wu H, Kirita Y, Donnelly EL, Humphreys BD. Advantages of Single-Nucleus over Single-Cell RNA Sequencing of Adult Kidney: Rare Cell Types and Novel Cell States Revealed in Fibrosis. J Am Soc Nephrol 2019;30(1):23–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heerboth S, Lapinska K, Snyder N, Leary M, Rollinson S, Sarkar S. Use of epigenetic drugs in disease: an overview. Genet Epigenet 2014;6:9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]


