Abstract
Lateral organ boundaries domain (LBD) proteins are plant-specific transcription factors that play a crucial role in growth and development, as well as metabolic processes. However, knowledge of the function of LBD proteins in Camellia sinensis is limited, and no systematic investigations of the LBD family have been reported. In this study, we identified 54 LBD genes in Camellia sinensis. The expression patterns of CsLBDs in different tissues and their transcription responses to exogenous hormones and abiotic stress were determined by RNA-seq, which showed that CsLBDs may have diverse functions. Analysis of the structural gene promoters revealed that the promoters of CsC4H, CsDFR and CsUGT84A, the structural genes involved in flavonoid biosynthesis, contained LBD recognition binding sites. The integrative analysis of CsLBD expression levels and metabolite accumulation also suggested that CsLBDs are involved in the regulation of flavonoid synthesis. Among them, CsLOB_3, CsLBD36_2 and CsLBD41_2, localized in the nucleus, were selected for functional characterization. Yeast two-hybrid assays revealed that CsLBD36_2 and CsLBD41_2 have self-activation activities, and CsLOB_3 and CsLBD36_2 can directly bind to the cis-element and significantly increase the activity of the CsC4H, CsDFR and CsUGT84A promoter. Our results present a comprehensive characterization of the 54 CsLBDs in Camellia sinensis and provide new insight into the important role that CsLBDs play in abiotic and flavonoid biosynthesis.
Subject terms: Plant stress responses, Plant stress responses, Secondary metabolism, Secondary metabolism
Introduction
Plants have evolved a variety of biochemical and physiological mechanisms to survive under temporary or continuous environmental challenges1,2. Transcription factor (TF) families play important roles in plant growth, development and environmental stress responses3,4. As plant specific TFs, LATERAL ORGAN BOUNDARIES (LBD) genes can be identified by a highly conserved LBD domain, which acts in the boundary of plant organs to regulate the development of leaves, inflorescences, roots and microspores5,6. LBD genes also play important roles in metabolic processes in higher plants, such as anthocyanin and nitrogen metabolism7.
As the complete reference genomes of more species are sequenced, the LBD gene family has been identified in several plants. In Arabidopsis and zea mays, 43 and 44 LBD members have been found, respectively, 35 have been identified in rice, 57 in poplar, 58 in malus domestica, 28 in brachypodium and 46 in tomato6,8–13. Generally, LBD proteins are defined by an N-terminal LBD domain. The characteristic LBD domain comprises a C-domain containing four conserved cysteines with spacing (CX2CX6CX3C) required for DNA binding activity. Moreover, the LBD domain contains a Gly-Ala-Ser (GAS) block and a leucine zipper-like coiled-coil motif (LX6LX3LX6L), which includes five hydrophobic amino acids separated by six variable amino acid residues responsible for protein dimerization14. According to characteristic sequence motifs, LBD genes are divided into two classes. The majority of LBD genes belong to Class I, as they contain a perfectly conserved CX2CX6CX3C zinc finger-like domain and an LX6LX3LX6L leucine zipper-like coiled-coil motif. Usually, members of the Class I group are involved in plant development and auxin signal transduction cascades14–16. In contrast, Class II LBD genes, which possess a conserved zinc finger-like domain cannot form coiled-coil structures8,14.
In Arabidopsis, several LBDs have been characterized. For example, AtASL4 (AtLOB) is predominantly expressed in the proximal base of lateral tissues and interacts with various transcription factors and proteins to participate in early leaf development5. The genes AtLBD17, AtLBD18 and AtLBD29 can regulate the development of lateral roots and callus formation, and can establish a molecular link between auxin signaling and the plant regeneration program17–19. In Class II, AtAS2 (AtLBD6) not only participates in leaf near-paraxial polarity, but also plays a role in floral development by synergistically regulating the differentiation of border cells in flower organs through AS1 and JAG20. AtLBD38 and AtLBD39 play a role in nitrogen metabolism and anthocyanin synthesis7,21. AtLBD20 is a root specific LBD gene that negatively regulates the responses to fungal infection22. The functions of LBD genes have also been studied in other species. For example, OsIG1 (homologous to AtAS2) can affect leaf lateral growth by regulating the division and differentiation of vesicular cells between vascular bundles23. OsLBD37 and OsLBD38 are involved in the regulation of rice plant heading date and crop yield24. ZmIG1 plays a key role in the regulation of female gamete development and leaf axial differentiation25. LBD1 and LBD4 in poplars act together on the secondary phloem, while PtaLBD15 and PtaLBD18 are specifically expressed in the secondary xylem, indicating that the LBD family is involved in secondary growth during xylem formation10.
Tea is the world’s most popular beverage and offers a wealth of health benefits. Previous studies have documented that flavonoids have strong antioxidant activity as well as many other medicinal properties that act against a variety of human diseases26. Catechins are a major component of flavonoids and are synthesized through the flavonoid pathway, which has been intensively investigated in several plant species27–30. Firstly, conversion of phenylalanine to chalcon, a common precursor in the flavonoid biosynthetic pathway, is catalyzed by phenylalanine ammonia-lyse (PAL), cinnamate 4-hydroxylase (C4H), 4-coumarate-CoA ligase (4CL) and chalcone synthase (CHS)31,32. Subsequently, chalcon is converted into to leucoanthocyanidin under the catalyzation of chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), flavonoid 3′5′-hydroxylase (F3′5′H), and dihydroflavonol 4-reductase (DFR)33,34. Then, leucoanthocyanidins are either catalyzed by leucoanthocyanidin 4-reductase (LAR) to produce Catechins (C) and gallocatechin (GC), or by the sequential action of anthocyanidin synthase (ANS) and anthocyanidin reductase (ANR) to form epicatechin (EC) and epigallocatechin (EGC), respectively35. Proanthocyanidins (PAs, also called condensed tannins) are oligomers and polymers of non-galloylated catechins (C, GC, EC and EGC), which can be catalyzed by galloyl-1-O-β-D-glucosyltransferase (UGGT) and epicatechin: 1-O-galloyl-β-D-glucose O-galloyltransferase (ECGT) to synthesis galloylated catechins (catechin gallate, epicatechin gallate, epigallocatechin gallate and gallocatechin gallate)36.
Catechin content is greatest in new tea shoots, and gradually decreases with the maturation of tissues and the growth of plants37. LBD genes play essential roles in plant growth and development and are involved in anthocyanin synthesis and nitrate metabolism7,21. However, the function of LBD genes in Camellia sinensis remains largely unexplored. Completion of the genome-sequencing project for Camellia sinensis has made it possible to identify LBD genes on a genome-wide scale38,39. We identified 54 LBD genes in the Camellia sinensis genome through database searches, and classified them according to their homology with LBD genes in Arabidopsis. We analyzed their sequence phylogeny, genomic structure, conserved domains and evolutionary mechanisms. We also investigated CsLBD gene expression patterns in different tissues and in response to MeJA and other different abiotic stresses. Furthermore, we characterized the function of selected LBD genes by subcellular localization, transactivation analysis, yeast one-hybrid assay and dual-luciferase assay to demonstrate that three LBD members have different effects on flavonoid synthesis. Our findings will serve as a foundation for further research into the roles of CsLBDs in flavonoid synthesis.
Results
Identification and annotation of LBD genes in tea plant
To identify the LBD gene family members in tea plant, 78 LBD protein sequences, 43 from Arabidopsis and 35 from O. sativa, were chosen to screen the tea genome database (Camellia sinensis var. sinensis). FGENESH (http://www.softberry.com/berry.phtml) was applied to confirm these predicated LBD sequences and the ExPASy proteomics server (http://www.expasy.ch/prosite/) was used to check the domains of the LBD sequences. A total of 54 LBD genes were identified in Camellia sinensis var. sinensis. These LBD genes were predicted to encode proteins 123–401 amino acids in length, with putative molecular weights (MWs) ranging from 13.39 to 45.04 and isoelectric points (pI) ranging from 4.7 to 9.37 (Supplementary Dataset 1). To avoid confusion, the 54 CsLBDs were named according to their homology with Arabidopsis LBDs. However, not every LBD member of Camellia sinensis corresponded to a gene in Arabidopsis. Genes with the same homology were further distinguished by an extra number.
Sequence alignment and phylogenetic analysis of LBD genes
Multiple sequence alignment of LBDs in Camellia sinensis revealed the presence of the CX2CX6CX3 zinc finger-like domain signature in the N terminus of all genes with the exception of CsLBD29. This domain, is required for DNA-binding activityGly-Ala-Ser (GAS) block and leucine zipper-like coiled-coil (LX6LX3LX6L) motifs were located at the C terminus, and are responsible for protein dimerization. In the GAS block, CsLBD23 and CsLBD24 had no corresponding sequences, whereas all CsLBD proteins contained conserved residues in the (D/N) PX2G motif. As in other species, the leucine zipper-like motif (LX6LX3LX6L) was only observed in CsLBD class I proteins, suggesting that the classes might have distinct functions (Supplementary Fig. S1).
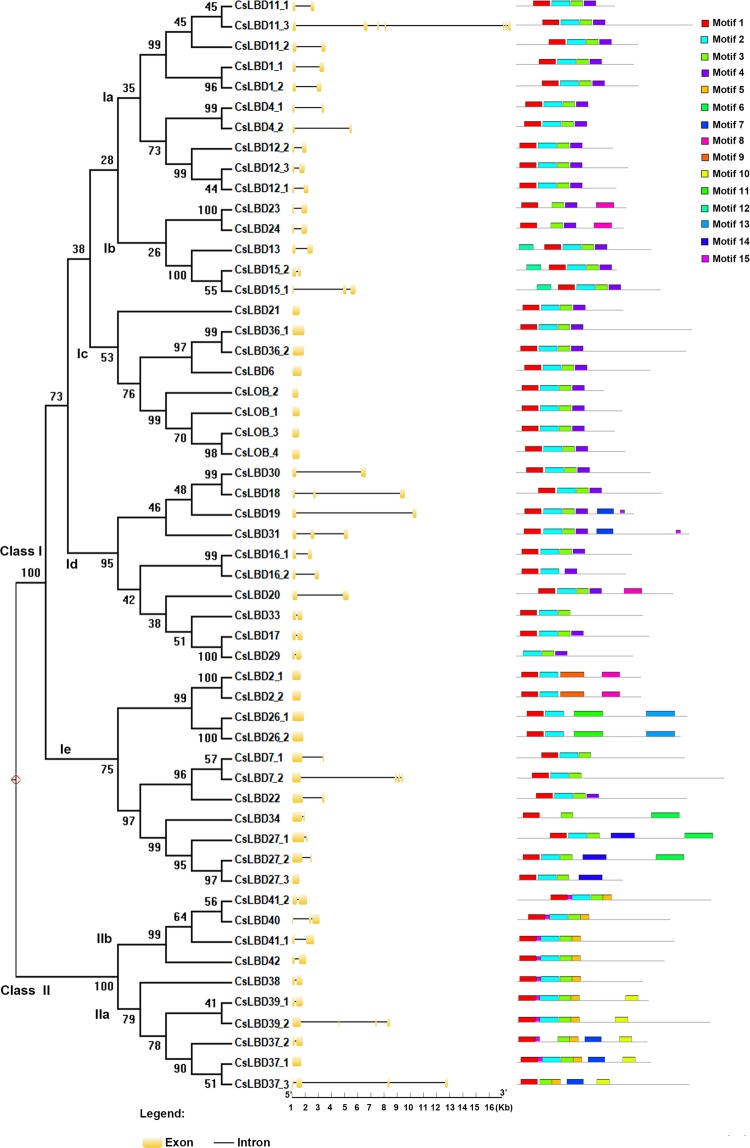
Exon/intron analysis showed that the number of CsLBD exons ranged from one to seven. The majority of the CsLBD genes contained two exons (31 genes), 17 had no introns, 4 had two introns, CsLBD37_3 and CsLBD7_2 had four introns, CsLBD39_2 had five and CsLBD11_3 had seven. The size of LBD gene loci ranged from 459 (CsLBD_2) to 16697 (CsLBD11_3) nucleotides (Fig. 1). Most of the LBD exon/intron structures were clustered together in the phylogenetic tree, indicating evolutionary conservation of the gene structure.
Figure 1.
Phylogenetic analysis, and identification of intron-exon and conserved motifs in 54 LBDs in Camellia sinensis. A phylogenetic tree of 54 CsLBDs was constructed using MEGA X by the NJ method with 1000 bootstrap replicates. Introns and exons are represented by a black line and orange box, respectively. Conserved motifs are indicated by a colored box numbered from 1 to 15.
MEME online software was used to predict the motifs in CsLBD protein sequences, and fifteen motifs were indicated in the LBD protein structure (Fig. 1). Nearly all members of the CsLBD family contained motifs 1, 2 and 3, suggesting that these motifs are essential for the functions of the LBD gene family. Motif 5 was only present in Class II. LBD proteins were clustered into subgroups based on their motif compositions and exon/intron structures.
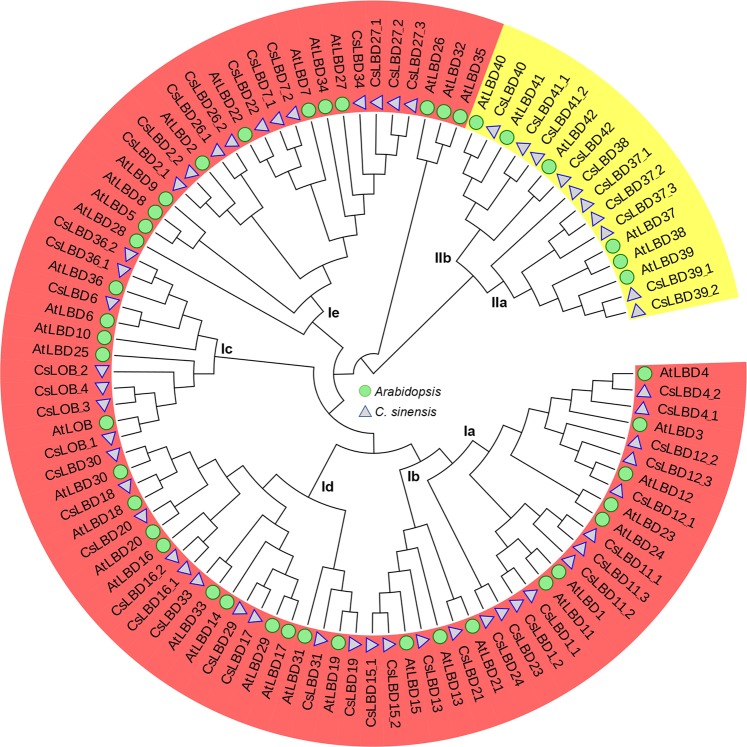
A phylogenetic tree was constructed with the full length protein sequences to examine the evolutionary patterns of 54 LBDs in Camellia sinensis and 43 in Arabidopsis using the MEGA X program. As shown in Fig. 2, all LBD proteins were divided into two classes, named Class I and Class II. These were further divided into five (Ia, Ib, Ic, Id and Ie) and two (IIa and IIb) subgroups, respectively. Class I comprised 44 CsLBDs and 36 AtLBDs, while Class II comprised 10 CsLBDs and 6 AtLBDs (Fig. 2).
Figure 2.
Phylogenetic tree of LBDs from Camellia sinensis and Arabidopsis. Amino acid sequences were aligned using Clustal W, and MEGA X software was used to construct the phylogenetic tree by the NJ method with 1000 bootstrap replicates. All LBD proteins were divided into Class I and Class II and then divided into five (Ia, Ib, Ic, Id and Ie) and two (IIa and IIb) subgroups, respectively.
CsLBD expression patterns of in Camellia sinensis
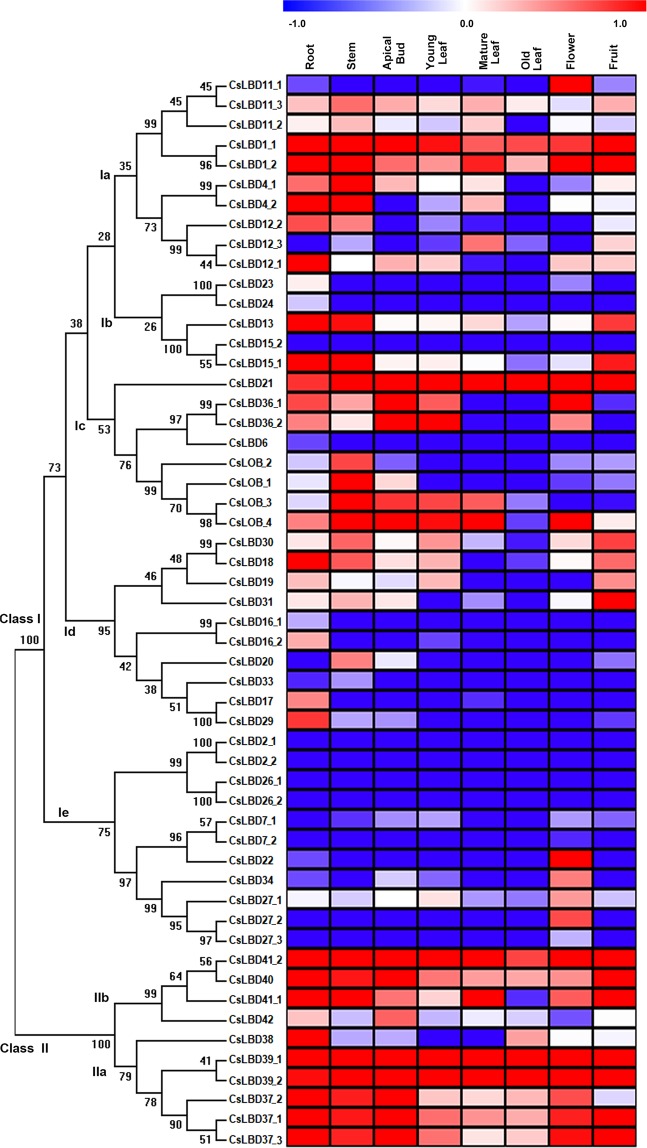
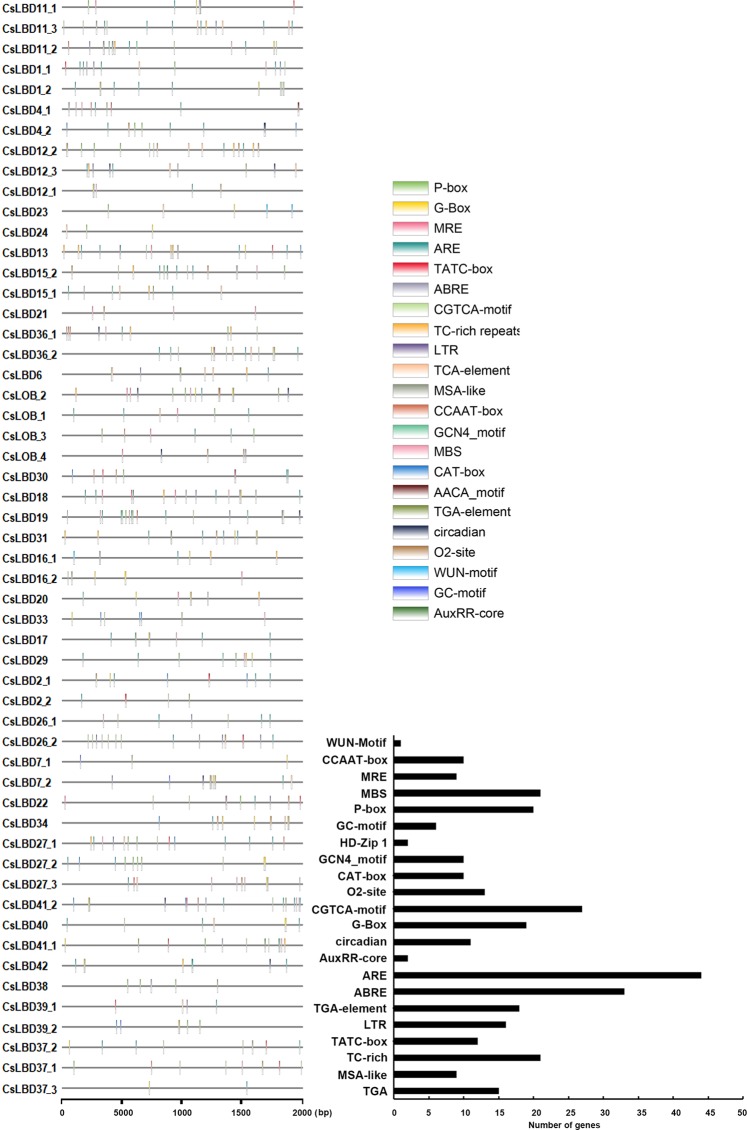
To explore the organ-specificity of LBD family members, we examined the abundance of 54 CsLBD transcripts in eight representative tissues of Camellia sinensis cv. Shuchazao, comprising apical bud, young leaf, mature leaf, flower, fruit, old leaf, stem and root tissue (Fig. 3). The expression of six members in subclass Ia was detected at least in one of the organs. CsLBD1_1 and CsLBD1_2 shared similar transcript profiles and were mainly expressed in fruit; CsLBD4_1 and CsLBD4_2 were mainly expressed in stems; and CsLBD11_1 and CsLBD12_1 were predominantly expressed in flower and root tissues, respectively. Only two members of subclass Ib (CsLBD13 and CsLBD15_1) showed high expression levels in root. All members of subclass Ic were detected in at least one organ, with the exception of CsLOB_2 and CsLBD6. Transcripts of CsLBD21 were detected in all organs; CsLBD36_1 was mainly expressed in apical bud and flower tissue, while CsLBD36_2 accumulated in apical bud and young leaf tissue; and CsLBD_1 and CsLBD_4 shared similar expression patterns and were predominantly expressed in root tissue. Members of subclasses Id and Ie showed very low or undetectable expression levels in all tested tissues, excepted CsLBD18, CsLBD31 and CsLBD22. CsLBD18 was mainly expressed in roots, CsLBD31 in fruit and CsLBD22 in flowers. All LBDs in Class II except CsLBD42 showed high expression in most organs. Among them, transcripts of CsLBD39_1, CsLBD39_2 and CsLBD41_2 were detected in the whole plant, whereas CsLBD37_1/2/3, CsLBD38, CsLBD40 and CsLBD41_1 were only highly expressed in roots. Furthermore, we examined the transcript abundance of 10 selected CsLBDs in different tissues of tea plant cultivar (Longjing 43) tissues using quantitative RT-PCR (qRT-PCR) (Supplementary Fig. S2). The expression patterns of nine genes were highly consistent with their transcriptomic profile from RNA-seq and their Pearson’s correlation coefficient R = 0.84. Both q-RT-PCR and RNA-seq data indicated that the no CsLBD26_2 transcripts were present levels in tested tissues. Previous studies have indicated that cis-acting elements present in gene promoter regions are closely related to their own expression pattern40,41. A number of common cis-elements were identified, most of which were involved in plant growth and development. CAT-box and GCN4_motifs were identified in ten CsLBD promoters, and are related to meristem and endosperm expression respectively. Both CsLOB_3 and CsLBD36_2 possessed with GCN4_motif and highly expressed in apical bud and young leaf. Six promoters contained MSA-Like elements, which are involved in cell cycle regulation (Fig. 4, Supplementary Dataset 2). High levels CsLBD21 and CsLBD41_2 transcripts were detected throughout all tissues, and their promoters contained growth-related cis-elements (circadian, CAT-box and MSA-like). This suggests that CsLBDs play important roles in biological processes, as well as regulating plant growth and development.
Figure 3.
Expression patterns of CsLBDs in different tissues in Camellia sinensis. The expression levels of CsLBD genes in eight tissues (Root, Stem, Old leaf: germinated in previous years, Mature leaf: geminate in the spring and are harvested in the autum, Young leaf: the first and second leaf follows the apical bud, Apical bud: unopened leaves on the top of activity growing shoots, Flower and Fruit) of tea plant were calculated using Log10(FPKM).
Figure 4.
Cis-element analysis of the CsLBD promoters. Plant CARE was used to identify the putative cis-acting element distribution in 2000 bp promoter sequences of 54 CsLBDs.
Expression profiles of CsLBDs in response to abiotic stress
Many CsLBD promoters included cis-elements responsible for stress responses, including 16 low-temperature responsive elements (LTR-element), 21 defense and stress response elements (TC-rich repeats), and nine MYB binding sites related to drought-inducibility (Fig. 4, Supplementary Dataset 2). To investigate the potential functions of CsLBD genes in response to MeJA and abiotic stresses, we searched the published literature for relevant microarray data. Expression patterns of the CsLBD genes in response to MeJA and abiotic stress (Cold stress, NaCl stress and drought stress) are shown in Fig. 5. Among the 54 predicted genes, 24 gene expression profiles were obtained in the Genevestigator analysis. It is possible that either the transcript abundance of the 30 genes was too low to be detected or there were no changes following treatments. Following MeJA treatment, the expression of most CsLBDs was unchanged or repressed; only CsLBD38 and CsLBD39_2 were induced significantly. Among 24 CsLBDs, 17 showed similar responses to three abiotic stresses: ten were induced, six LBDs were suppressed, and CsLOB_1 was unchanged. CsLBD13 was induced by drought and repressed by cold stress, CsLBD_4, CsLBD41_2, CsLBD38 and CsLBD39_2 were repressed by both NaCl and drought stress but induced by cold stress. CsLBD40 and CsLBD41_1 were induced by NaCl stress and suppressed by drought and cold stress.
Figure 5.
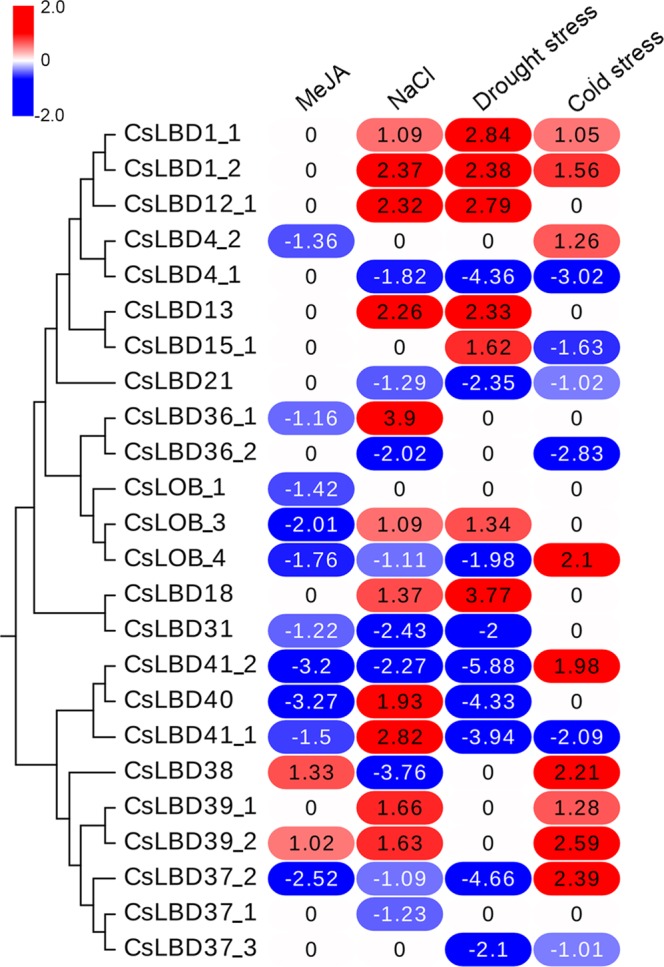
Expression pattern of CsLBDs in response to MeJA and abiotic stress. Genevestigator analysis of CsLBDs using the Log2(Experiment/Control) in response to MeJA, NaCl, drought and low temperature.
Integrative analysis of CsLBD expression levels and metabolite accumulation in tea tissues
Transcriptomic profiles and metabolite activities of eight representative tissues of tea plant was used to establish gene to metabolite networks. Among 54 CsLBDs, no expression of CsLBD2_1, CsLBD2_2, CsLBD26_1 and CsLBD26_2 was detected in any of the tested tissues. We preformed correlation analysis of 13 flavonoid metabolites and another 50 LBD transcripts and identified 47 positive correlations (R > 0.5) and 126 negative correlations (R < −0.5) (Fig. 6). There were 19 CsLBDs that had clear negative correlations with total catechins, of which four members belonged to subclass Ia, five to subclass Ib, four to subclass Id, three to subclass IIa and three to subclass IIb. Particularly interesting is the fact that only CsLBD36_2 and CsLBD42 were positively correlated with total catechins. Eight CsLBDs were found to have positive correlations with the other metabolites, while 19 had no correlations with the studied metabolites. These CsLBDs were distributed throughout all subclasses, except subclass Ib. CsLBD6 and CsLBD16_2 were negatively correlated with these metabolites.
Figure 6.
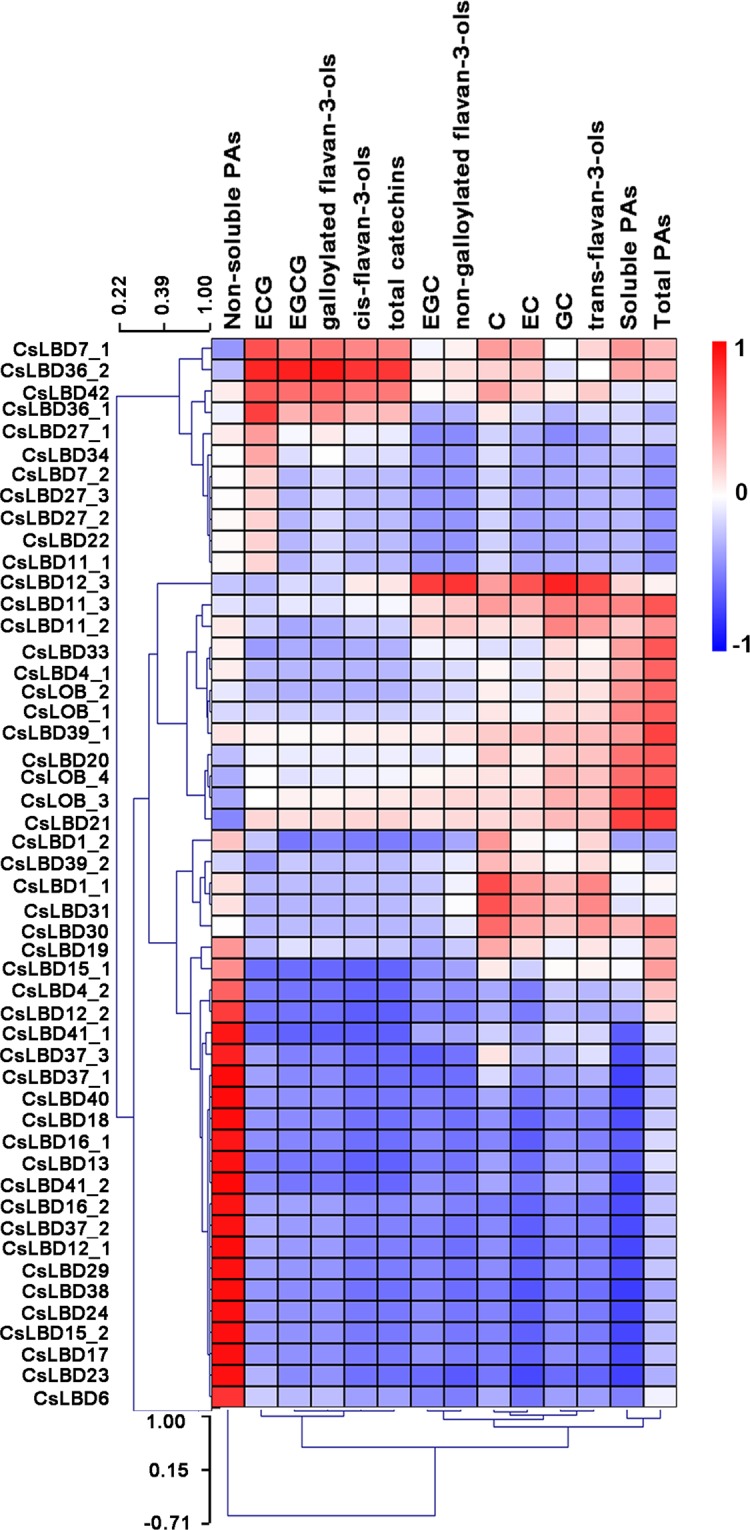
Integrative analysis of CsLBD expression levels and metabolite accumulation in tea tissues. Correlation analysis with 13 metabolites and 50 transcripts of CsLBD. R > 0.5: Positive correlations; R < −0.5: negative correlation.
Tea (Camellia sinensis) has a remarkable content, PAs are produced by the branched flavonoid pathway and can be classified as soluble or non-soluble. The relationship between CsLBDs and soluble PAs was opposite to that between CsLBDs and non-soluble PAs. Galloylated catechins (ECG, galloylated flavan-3-ols, EGCG cis-flavan-3-ols) and total catechins were correlated with CsLBD expression, and CsLBD36_2 and CsLBD42 expression were shown positive correlation with the accumulation of these metabolites. In addition, C, EC, trans-flavan-3-ols, GC, EGC and non-galloylated flavan-3-ols showed correlations with the expression of CsLOB_3, CsLBD21, CsLBD11_2, CsLBD11_3, CsLBD12_3. Detailed correlation analyses of CsLBD genes and metabolites is shown in Supplementary Dataset 3.
Subcellular localization and transactivation activity analysis of CsLBDs
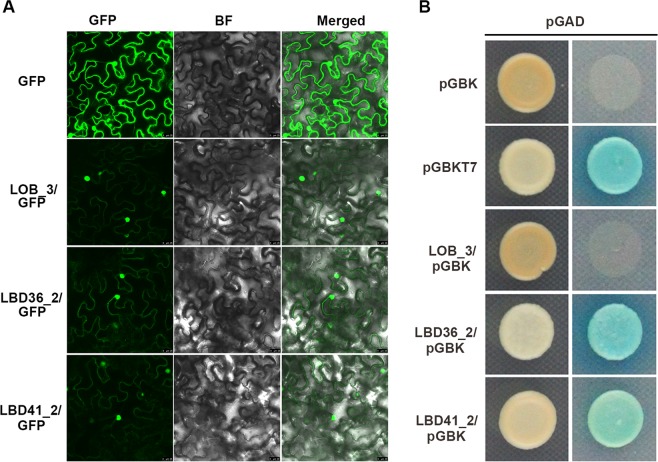
Correlation analysis between the transcriptomic profile and metabolites suggested that CsLOB_3 and CsLBD36_2 are positively correlated with total PAs and catechines, respectively, whereas CsLBD41_2 was negatively correlated with soluble PAs. CsLOB_3 and CsLBD36_2 were predominantly expressed in apical buds and young leaves, and CsLBD41_2 accumulated in tea flowers and roots. We were particularly interested in the detailed function of LBD genes, and cloned these genes from the tea plant cultivar ‘Longjing 43’. The open reading frames (ORFs) were inserted into the GFP reporter gene under the control of the CaMV 35S promoter. The GFP recombinant constructs and the CsLOB_3-GFP, CsLBD36_2-GFP and CsLBD41_2-GFP fusion proteins were introduced into tobacco. CsLOB_3-GFP, CsLBD36_2-GFP and CsLBD41_2-GFP were specifically localized in the nucleus (Fig. 7A), which is consistent with the predicted role of these genes as TFs. The GFP signal from the empty vector showed ubiquitous distribution throughout the cell.
Figure 7.
The potential function of LBDs in tea plant. (A) Subcellular localization of CsLOB_3, CsLBD36_2 and CsLBD41_2 and GFP as a control, which were transiently expressed in N. tabacum leaves. GFP: Green fluorescence image, BF: Bright-field microscopy image, Merge: Merged bright-field and green fluorescence images. (B) Transactivational analyses of CsLBDs in yeast. Positive control, negative control and the fusion constructs were transformed into the AH109 strain and successively incubated in SD/-Leu/-Trp media and SD/-Ade/-Leu/-Trp/-His plate supplemented with X-α-GAL.
To examine the transactivation activity of CsLBDs, we made the constructs containing CsLOB_3, CsLBD36_2 or CsLBD41_2 together with a DNA binding domain and each CsLBD-pGBK-pGAD pair was individually co-transformed into yeast cells AH109. Co-transformation of pGBKT7 and pGAD was used as a positive control, while pGBK and pGAD were the negative control. All of these transformants could readily grow on the SD/-Leu/-Trp medium and the resulting colonies were further selected on quadruple dropout medium supplemented with X-α-Gal. As shown in Fig. 7B, CsLOB_3 and the negative control did not grow, whereas CsLBD36_2, CsLBD41_2 and the positive control grew well and turned blue. This indicated that only CsLBD36_2 and CsLBD41_2 had transcriptional activity in these yeast strains.
Analysis of the promoter regions of the structural genes involved in the flavonoid pathway in tea plant
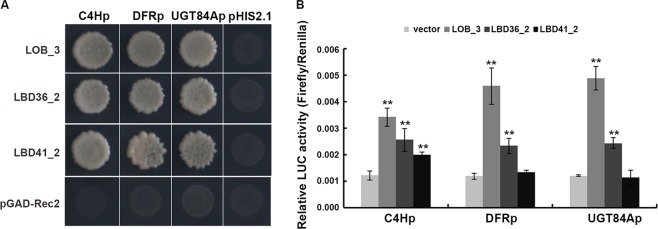
To explore the downstream molecular events behind the metabolic processes triggered by LBD TFs, we further analyzed the promoter regions of the structural genes involved in the flavonoid pathway. A total of 52 genes were analyzed and LBD dominant binding sites (G:HCGGCG or GCGGCW) were present in the promoter regions of three flavonoid biosynthesis related genes, TEA034002.1 (C4H), TEA010588.1 (DFR) and TEA026127.1 (UGT84A) (Supplementary Fig. S3). To identify whether CsLBD could bind to the promoter of these three structural genes, the CsC4H, CsDFR and CsUGT84A promoter regions were used as bait, and those yeast cells co-transformed with three CsLBD-pGADT7-Rec2 vectors, were tested on SD/-Trp/-Leu/-His + 30 mM 3-AT media, respectively (Fig. 8A). The result showed that all three CsLBDs bound to the cis-element in the promoter of CsC4H, CsDFR and CsUGT84A. We then cloned the promoter regions in a vector harboring the LUC reporter gene and analyzed the effect of CsLBDs on gene transcription. Compared to the background, the promoter activity of CsC4H was significantly elevated 2.8-fold, the CsDFR promoter was up-regulated by 3.8-fold and the CsUGT84A promoter was raised 4-fold when introduced to CsLOB_3. In addition, the LUC activity of three promoters was increased by different levels by CsLBD36_2, whereas CsLBD41_2 showed little impact on these promoter activities, only the CsC4Hp activity increased 1.6-fold (Fig. 8B). Taken together, our data suggest that CsLBD proteins are involved in flavonol biosynthesis.
Figure 8.
The transcriptional regulation of CsLBDs on the CsC4H, CsDFR and CsUGT84A promoter. (A) Yeast one-hybrid assays of the interactions between CsLBDs and CsC4H, CsDFR and CsUGT84A promoter fragments. The empty vectors of pGADT7-Rec were used as a negative control. The concentration of 3-amino-1,2,4-triazole was 30 mM. (B) Relative LUC/REN ratio from transient expression assays. The relative LUC activities were normalized to a 35S: REN internal control. Error bars indicate the SD of five biological replicates. **Significant difference at P < 0.01.
Discussion
Tea is the world’s most popular beverage and offers a wealth of health benefits38,39. Catechins, anthocyanidins and proanthocyanidins are important secondary metabolites that are synthesized via the flavonoid pathway37. In higher plants, flavonoid biosynthesis is not only regulated by structural genes but also involves a number of transcription factors. LBD transcription factors exist ubiquitously in plants and are involved in mediating plant-specific processes14,42. Following the completion of the tea plant genome sequence, 54 LBD genes from Camellia sinensis were identified. Comparative studies of other plant species show similar numbers of LBDs to that in Camellia sinensis, which indicates the evolutionary diversification of the LBD family through extensive expansion43.
Structural analysis is a powerful method of mining valuable information concerning protein function. The sequence alignment of 54 LBD proteins showed that all contained four Cys residues in the C block, except CsLBD29, which lacks the last three Cys residues. CsLBD23 and CsLBD24 lack the GAS-block, and the leucine zipper-like coiled-coil of CsLBD16_2, CsLBD2_1, CsLBD2_2, CsLBD7_1, CsLBD7_2, CsLBD26_1, CsLBD26_2, CsLBD27_1, CsLBD27_2 and CsLBD27_3 varied in amino acid content. Similar results were found in Soybean, L. japonicus9,44. However, LBDs in Arabidopsis, rice, apple and mulberry contain invariant residues in their conserved domains, indicating that LBD proteins in Camellia sinensis have more variation. Exon/intron structure and gene length analysis showed that most LBD genes in Camellia sinensis had similar gene structures within the same subgroup, a pattern also observed in Arabidopsis, rice and soybean6,44. We also demonstrated that CsLBD proteins generally possess similar protein motifs within each subgroup. Motif 1 contained the conserved CX2CX6CX3C zinc finger-like motif, motif 2 and motif 3 comprised the GAS-block and (D/N) PX2G motifs, respectively, and motif 4 contained the leucine zipper-like coiled-coil (LX6LX3LX6L) domain. Nearly all members of class I possessed motifs 1–4, whereas Class II contained motifs 1, 2, 3 and 5. This suggests that the CsLBDs in the same group or subgroup have similar functions to their homologs.
Recently, it has been reported that low temperature, NaCl, drought and other abiotic stressors promote the production of secondary metabolites in other species45–48. The identification of cis-elements showed that 31 CsLBDs have at least one of the HD-Zip1, G-box, GC-motif, MBS, MRE and WUN-motif elements, which are mainly involved in responses to abiotic stress and light. Eleven and seven CsLBDs were induced by NaCl and drought stress, respectively. The promoter of CsLBD1_1, CsLBD1_2, CsLBD12_1, CsLBD13 and CsLBD18 contained ABRE and MBS, which mainly participate in drought-inducibility, and all of them were accumulated after NaCl and drought treatments. Only six CsLBDs were downregulated after the low temperature treatment, suggesting that LBD may play diverse roles in response to different stresses. A total of 47 CsLBD promoters were found to have hormone responsiveness elements, including TATC-box and P-box elements related to gibberellin responsiveness, and ABRE and CGTCA elements, which are associated with abscisic acid and MeJA responsiveness, respectively. Recently, ABA has been reported to promote the biosynthesis of flavonols49,50. There were one to six ABRE elements distributed in the promoter of 27 CsLBDs, which indicates that these CsLBDs are regulated by ABA and flavonoid synthesis. It has also been reported that ABA and MeJA promote anthocyanin accumulation, also GA plays positive roles in the flavonoid pathway in apple and tea plant51–53. Expression analysis of CsLBDs after MeJA treatment, showed that only CsLBD38 and CsLBD39_2 were upregulated and ten CsLBDs were repressed by MeJA, five of which contained the gibberellin-responsive element in promoter region. Since expression pattern is closely related to the gene function47,51, these results suggested that these CsLBDs mainly repress flavonoid biosynthesis.
LBD proteins play central roles in a wide range of metabolic, physiological and developmental processes5,7,17,25. There may be a “bridge” between the transcription factor, the biosynthetic gene and the secondary metabolite, and integrated analyses of gene expression and metabolites to select key genes involved in metabolite processes have been reported54–56. We found that CsLBD36_2 was highly correlated with total catechins, with a correlation coefficient of 0.79. CsLOB_3 and CsLBD21 showed clear positive correlations with soluble PAs, with correlation coefficients of 0.69 and 0.74, respectively, and 18 CsLBDs showed negative correlations with soluble PAs, with the correlation coefficient values were between −0.76 to −0.5. Combined promoter, stress response and correlation analysis of transcription and metabolites suggested that LBDs are involved in plant secondary metabolism, and act as hormones to mediate plant development and defense responses, which is consistent with other species17,22. There are extensive studies of the function of LBDs in plant growth and development, whereas knowledge about the mechanisms by which LBD genes control secondary metabolism is limited. Thus far, only AtLBD37, AtLBD38 and AtLBD39 are known to act as repressors of anthocyanin synthesis and N availability signals7,21. OsLBD37 is also associated with nitrogen metabolism7,24. The functions of CsLOB_3, CsLBD36_2 and CsLBD41_2 in flavonoid synthesis were further researched, and were localized to the nucleus, while CsLBD36_2 and CsLBD 41_2 were shown to have self-activation activities. Previous studies showed that LBDs can dominantly recognize HCGGCG/GCGGCW to mediate the transcription of genes involved in plant growth, development and metabolic processes7,54. Here, we present that all three tested CsLBDs can directly bind to the cis-element in the promoter of CsC4H, CsDFR and CsUGT84A to regulate their transcriptomic level. Several class I members of the AtLBD family have been implicated in plant development, for example, AtLOB was shown to regulate early leaf development and AtLBD36 functions in regulating proximaldistal patterning of petals5,54. Our results show that CsLOB_3 and CsLBD36_2 positively regulate flavonoid synthesis, suggesting that CsLBDs play distinct roles in Camellia sinensis flavonoid regulation.
Conclusions
We identified and systematically analyzed 54 CsLBDs genes in the tea plant genome. Bioinformatics and expression pattern analysis of CsLBDs indicated that these genes have potential functions in growth, development and metabolic process in tea plant. Correlation analysis between the expression levels of CsLBDs and the content of secondary metabolites in the flavonoid pathway suggest that CsLBDs are involved in the flavonoid biosynthesis pathway. Moreover, candidate genes, CsLOB_3 and CsLBD36_2, were confirmed to be involved in the flavonoid biosynthesis pathway. Our report provides an important foundation for further functional studies on the CsLBDs and contributes to illuminating the regulatory mechanisms of the flavonoid synthesis pathway in tea plant.
Materials and Methods
Characterization of the CsLBD gene family
Forty-three annotated AtLBD genes and proteins were downloaded from the Arabidopsis information resource (http://www.Arabidopsis.org/), and were used in a multiple database search against the tea plant genome, which was downloaded from the tea plant information archive (http://tpia.teaplant.org/index.html). The programs INTERPROSCAN, SMART, MOTIF and PLANTSP were employed to examine the protein sequences that were derived from the candidate CsLBD genes. The ExPASy proteomics server (http://expasy.org/) was used to predict the isoelectric points and molecular weights of the CsLBD proteins. The Clustal X1.83 program was employed to alignment the protein sequences of CsLBD, MEGA X was used to construct a phylogenetic tree by the neighbor-joining method with bootstrap set to 100057. Gene structure display server2.0 was used to define the exon/intron structures of individual CsLBD genes by aligning the cDNA sequences to their corresponding genomic DNA sequences. MEME (http://meme-suite.org/) was used to characterize the conserved motifs of CsLBDs with the following parameters: the maximum number of motifs = 15.
Analysis of promoter regions and cis-acting elements
Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to determine the putative cis-acting element distribution in the 2000-bp promoter sequence of 54 CsLBDs and 51 structural genes in the flavonoid pathway.
Correlation of gene expression and metabolite accumulation analysis
Transcriptome data and metabolite data from tea cultivar Shuchazao were downloaded from the tea plant information archive (http://tpia.teaplant.org/index.html). Fragments per-kilobase of exon per million fragments (FPKM) was used to estimate the gene expression level in eight tea plant tissues (root, stem, old leaf, mature leaf, young leaf, apical bud, flower and fruit). The expression levels of CsLBD genes in each tissue were calculated using Log10(FPKM value). Mev4.9.0 (https://sourceforge.net/projects/mev-tm4/) was used to display the CsLBD expression patterns. The samples of eight tissues of tea cultivar Shuchazao in RNA-seq experiments were also used for detecting the metabolites by high-performance liquid chromatography (HPLC) analysis39. To screen LBDs associated with the main flavonoids, correlation analysis between CsLBD genes and 13 representative metabolites was performed using Pearson’s correlation coefficient. Correlations with the value of correlation coefficients |R| > 0.5 and a p-value < 0.05 were considered statistically significant.
Expression analysis
For total RNA extraction from tea plants a Trizol reagent (Invitrogen) was used according to the manufacturer’s instructions. Purified RNA (1 μg) was reverse transcribed using the transcription system (Vazyme). Beacon Designer 7.0 was used to design primers for the CsLBDs of interest, the primers are listed in Table S1. Real-time PCR was performed on an ABI7900HT Sequence Detection System (Applied Biosystems, CA, USA) using SYBR Green (Roche, Switzerland) in accordance with the manufacturer’s instructions, and the gene expression level was normalized to Glyceraldehyde-3-phosphate dehydrogenase (GAPDH, accession number: KA295375.1).
Subcellular localization of CsLBDs
Plant-mPloc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) was used to predict protein subcellular localization. The CsLBD sequences were amplified and cloned into the binary vector pCV-eGFP-N1 digested with Kpn I and BamH I (Supplementary Dataset 4). The recombinant binary constructs and pCV-eGFP-N1 (control) were individually introduced into Agrobacterium tumefaciens strain GV3101. Agrobacterium cultures carrying the recombinant vectors were grown overnight at 28 °C, cells were pelleted and resuspended in infiltration buffer (10 mM MgCl2, 10 mM MES and 150 µM acetosyringone) with OD600 = 1. The cell suspensions were incubated at room temperature and then infiltrated into 4–6 week old N. benthaminan leaves. Expression of fluorescent proteins was observed with a Leica TCS SP5 confocal laser scanning microscope system (Leica Microsystems, Bannockburn, IL, USA) at 48 h post-agroinfiltration. Fluorescence of GFP was observed at 495–545 nm.
Analyses of CsLBD transactivation activity
The CsLOB_3, CsLBD36_2 and CsLBD41_2 sequences were cloned into the pGBKT7 vector and pGBKT7-53 + pGADT7-T acted as a positive control, while pGBKT7-Lam+ pGADT7-T was a negative control. The transactivation assay was performed following instructions given in the yeast transformation system 2 user manual. BD-CsLBDs and pGBKT7-53 were individually transformed into the yeast strain AH109 and selected on SD/-Leu/-Trp plates. Colonies were further transferred to selective SD/-Ade/-Leu/-Trp/-His/X-α-Gal medium and incubated at 30 °C for 3–5 days.
Yeast one-hybrid assay
The PCR products of CsLOB_3, CsLBD36_2 and CsLBD41_2 were purified and inserted into a pGADT7-Rec vector, The truncated derivatives of the CsC4H, CsDFR and CsUGT84A promoters were inserted into a pHIS2.1 vector. CsLBD-pGADT7-Rec (pGADT7-Rec plasmid digested by Sma I) was co-transformed with bait vectors into yeast stain Y187 and plated on SD/-Leu/-Trp medium. Colonies were then transferred to SD/-Leu/-Trp/-His deficient medium with 30 mM 3-AT for 3 days.
Dual-luciferase assay for CsLBDs
Dual-luciferase assays were performed as described previously37. The CsLBDs sequences were cloned into the YUKKS vector. The 1.5 kb promoter regions of CsC4H, CsDFR and CsUGT84A were cloned into a pGreenII 0800-LUC vector. 35S::REN (Renilla luciferase) in the vector was used as an internal control. The activities of the CsC4H, CsDFR and CsUGT84A promoters with and without the effector transcription factors were measured using a Dual-Luciferase Reporter Assay System (Promega) following the manufacturer’s instructions.
Main conclusion
Our study presents a comprehensive characterization of the 54 CsLBDsin Camellia sinensis and demonstrates the involvement of CsLBDs in the regulation of flavonoid biosynthesis.
Supplementary information
Acknowledgements
This work was funded by the National Natural Science Foundation of China (31500242, 31670291, 31800249), Zhejiang Provincial Natural Science Foundation of China (LY16C020004), State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-products (2010DS700124-ZZ1901), and National Key Laboratory of Plant Molecular Genetics.
Author contributions
X.Y.Z. and Y.Q.H. designed the research and performed most of the experiments. W.D.H. and H.S. performed part of the experiments. P.X., G.J.H. and Y.F.W. were responsible for data analysis and writing of the manuscript. The authors agree with the content of the manuscript.
Data availability
All supporting data can be found within the manuscript and its additional files.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Xueying Zhang and Yuqing He.
Contributor Information
Gaojie Hong, Email: gjhong@126.com.
Ping Xu, Email: zdxp@zju.edu.cn.
Supplementary information
is available for this paper at 10.1038/s41598-019-52027-6.
References
- 1.Aida M, Tasaka M. Morphogenesis and patterning at the organ boundaries in the higher plant shoot apex. Plant Mol. Biol. 2006;60:915–928. doi: 10.1007/s11103-005-2760-7. [DOI] [PubMed] [Google Scholar]
- 2.Mittler R. Abiotic stress, the field environment and stress combination. Trends Plant Sci. 2006;11:0–19. doi: 10.1016/j.tplants.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Riechmann JL, Ratcliffe OJ. A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 2000;3:423–34. doi: 10.1016/S1369-5266(00)00107-2. [DOI] [PubMed] [Google Scholar]
- 4.Gao G, et al. DRTF: a database of rice transcription factors. Bioinformatics. 2006;22:1286–1287. doi: 10.1093/bioinformatics/btl107. [DOI] [PubMed] [Google Scholar]
- 5.Iwakawa H, et al. The ASYMMETRIC LEAVES2 Gene of Arabidopsis thaliana, Required for Formation of a Symmetric Flat Leaf Lamina, encodes a Member of a Novel Family of Proteins Characterized by Cysteine Repeats and a Leucine Zipper. Plant Cell Physiol. 2002;43:467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 6.Shuai B, Reynaga-Peña CG, Springer PS. The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant physiol. 2002;129:747–761. doi: 10.1104/pp.010926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD Family of Transcription Factors Repress Anthocyanin Synthesis and Affect Additional Nitrogen Responses in Arabidopsis. Plant Cell. 2009;21:3567–3584. doi: 10.1105/tpc.109.067041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang YM, Zhang SZ, Zheng CC. Genome-wide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J Genet. 2014;93:79–91. doi: 10.1007/s12041-014-0342-7. [DOI] [PubMed] [Google Scholar]
- 9.Yang Y, Yu X, Wu P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006;39:248–262. doi: 10.1016/j.ympev.2005.09.016. [DOI] [PubMed] [Google Scholar]
- 10.Yordanov YS, Regan S, Busov V. Members of the LATERAL ORGAN BOUNDARIES DOMAIN Transcription Factor Family Are Involved in the Regulation of Secondary Growth in Populus. Plant Cell. 2010;22:3662–3677. doi: 10.1105/tpc.110.078634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang X, Zhang S, Su L, Liu X, Hao YA. Genome-Wide Analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) Gene Family in Malus domestica with a Functional Characterization of MdLBD11. PLoS ONE. 2013;8:e57044. doi: 10.1371/journal.pone.0057044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gombos M, et al. Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep. 2017;36:61–79. doi: 10.1007/s00299-016-2057-0. [DOI] [PubMed] [Google Scholar]
- 13.Wang XF, et al. Identification, Evolution and Expression Analysis of the LBD Gene Family in Tomato. Scientia Agricultura Sinica. 2013;46:2501–2513. [Google Scholar]
- 14.Majer C, Hochholdinger F. Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci. 2011;16:47–52. doi: 10.1016/j.tplants.2010.09.009. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, et al. ARL1, a LOB-domain protein required for adventitious root formation in rice. Plant J. 2005;43:47–56. doi: 10.1111/j.1365-313X.2005.02434.x. [DOI] [PubMed] [Google Scholar]
- 16.Feng Z, Zhu J, Du X, Cui X. Effects of three auxin-inducible LBD members on lateral root formation in Arabidopsis thaliana. Planta. 2012;236:1227–1237. doi: 10.1007/s00425-012-1673-3. [DOI] [PubMed] [Google Scholar]
- 17.Fan M, Xu C, Xu K, Hu Y. LATERAL ORGAN BOUNDARIES DOMAIN transcription factors direct callus formation in Arabidopsis regeneration. Cell Res. 2012;22:1169–1180. doi: 10.1038/cr.2012.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee HW, Kim NY, Lee DJ, Kim J. LBD18/ASL20 Regulates Lateral Root Formation in Combination with LBD16/ASL18 Downstream of ARF7 and ARF19 in Arabidopsis. Plant Physiol. 2009;151:1377–1389. doi: 10.1104/pp.109.143685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee HW, Cho C, Kim J. Lateral Organ Boundaries Domain16 and 18 act downstream of the AUXIN1 and LIKE-AUXIN3 auxin influx carriers to control lateral root development in Arabidopsis. Plant Physiol. 2015;168:1792–1806. doi: 10.1104/pp.15.00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semiarti E, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development. 2001;128:1771–1783. doi: 10.1242/dev.128.10.1771. [DOI] [PubMed] [Google Scholar]
- 21.Albinsky D, et al. Metabolomic Screening Applied to Rice FOX Arabidopsis, Lines Leads to the Identification of a Gene-Changing Nitrogen Metabolism. Mol. Plant. 2010;3:125–142. doi: 10.1093/mp/ssp069. [DOI] [PubMed] [Google Scholar]
- 22.Thatcher LF, Powell JJ, Aitken EA, Kazan K, Manners JM. The lateral organ boundaries domain transcription factor LBD20 functions in Fusarium wilt Susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012;160:407–418. doi: 10.1104/pp.112.199067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, et al. Down-regulation of a LBD-like gene, OsIG1, leads to occurrence of unusual double ovules and developmental abnormalities of various floral organs and megagametophyte in rice. J. Exp. Bot. 2015;66:99–112. doi: 10.1093/jxb/eru396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li C, et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of, Ehd1, in rice. Biochem. Biophys. Res. Commun. 2017;486:720–725. doi: 10.1016/j.bbrc.2017.03.104. [DOI] [PubMed] [Google Scholar]
- 25.Evans MM. The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo sac and leaf development. Plant Cell. 2007;19:46–62. doi: 10.1105/tpc.106.047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman M, et al. Structure-activity relationships of tea compounds against human cancer cells. J. Agric. Food Chem. 2007;55:243–253. doi: 10.1021/jf062276h. [DOI] [PubMed] [Google Scholar]
- 27.Kubasek WL, et al. Regulation of flavonoid biosynthetic genes in germinating Arabidopsis seedlings. Plant Cell. 1992;4:1229–1236. doi: 10.2307/3869409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boss PK, Davies C, Robinson SP. Analysis of the expression of anthocyanin pathway genes in developing vitis vinifera L. cv Shiraz grape berries and the implications for pathway regulation. Plant Physiol. 1996;111:1059–1066. doi: 10.1104/pp.111.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takos AM, et al. Light‐induced expression of a MYB gene regulates anthocyanin biosynthesis in red apples. Plant Physiol. 2006;142:1216–1232. doi: 10.1104/pp.106.088104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer TC, et al. Flavonoid genes of pear (Pyrus communis) Trees. 2007;21:521–529. doi: 10.1007/s00468-007-0145-z. [DOI] [Google Scholar]
- 31.Winkel-Shirley B. Flavonoid biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dao TTH, Linthorst HJM, Verpoorte R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011;10:397–412. doi: 10.1007/s11101-011-9211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang X, et al. Characterisation of anthocyanidin reductase from Shuchazao green tea. J. Sci. Food. Agr. 2012;92:1533–1539. doi: 10.1002/jsfa.4739. [DOI] [PubMed] [Google Scholar]
- 34.Pang Y, et al. Functional Characterization of Proanthocyanidin Pathway Enzymes from Tea and Their Application for Metabolic Engineering. Plant Physiol. 2013;161:1103–1116. doi: 10.1104/pp.112.212050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang, P. Q. et al. Functional demonstration of plant flavonoid carbocations proposed to be involved in the biosynthesis of proanthocyanidins. Plant J. 10.1111/tpj.14515 (2019). [DOI] [PubMed]
- 36.Wang Y, et al. Novel insight into the role of withering process in characteristic flavor formation of teas using transcriptome analysis and metabolite profiling. Food Chem. 2019;272:313–322. doi: 10.1016/j.foodchem.2018.08.013. [DOI] [PubMed] [Google Scholar]
- 37.Zhang X, et al. AtHB2, a class II HD-ZIP protein, negatively regulates the expression of CsANS, which encodes a key enzyme in Camellia sinensis catechin biosynthesis. Physiol. Plant. 2019;166:936–945. doi: 10.1111/ppl.12851. [DOI] [PubMed] [Google Scholar]
- 38.Xia EH, et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant. 2017;10:866–877. doi: 10.1016/j.molp.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Wei C, et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. USA. 2018;115:E4151–E4158. doi: 10.1073/pnas.1719622115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi SK, Shinozaki K. Transcriptional regulatory networks in cellular response and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 41.Deokar AA, Bunyamin T. Genome-Wide Analysis of the Aquaporin Gene Family in Chickpea (Cicer arietinum L.) Front Plant Sci. 2016;7:1802. doi: 10.3389/fpls.2016.01802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu C, Luo F, Hochholdinger F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016;21:159–167. doi: 10.1016/j.tplants.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 43.Richardt S, Lang D, Reski R, Frank W, Rensing SA. PlanTAPDB, a Phylogeny-Based Resource of Plant Transcription-Associated Proteins. Plant Physiol. 2007;143:1452–1466. doi: 10.1104/pp.107.095760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang H, et al. Genome wide analysis of soybean LATERAL ORGAN BOUNDARIES Domain-Containing genes: A Functional Investigation of GmLBD12. Plant. Genome. 2017;10:1–19. doi: 10.3835/plantgenome2016.07.0058. [DOI] [PubMed] [Google Scholar]
- 45.Azuma A, Yakushiji H, Koshita Y, Kobayashi S. Flavonoid biosynthesis-related genes in grape skin are differentially regulated by temperature and light conditions. Planta. 2012;236:1067–1080. doi: 10.1007/s00425-012-1650-x. [DOI] [PubMed] [Google Scholar]
- 46.Petropoulos SA, et al. Salinity effect on nutritional value, chemical composition and bioactive compounds content of Cichorium spinosum L. Food Chem. 2017;214:129–136. doi: 10.1016/j.foodchem.2016.07.080. [DOI] [PubMed] [Google Scholar]
- 47.Sarker U, Oba S. Drought stress enhances nutritional and bioactive compounds, phenolic acids and antioxidant capacity of Amaranthus leafy vegetable. BMC Plant Biol. 2018;18:258. doi: 10.1186/s12870-018-1484-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gharibi S, et al. The effect of drought stress on polyphenolic compounds and expression of flavonoid biosynthesis related genes in Achillea pachycephala Rech.f. Phytochemistry. 2019;162:90–98. doi: 10.1016/j.phytochem.2019.03.004. [DOI] [PubMed] [Google Scholar]
- 49.Berli FJ, et al. Abscisic acid is involved in the response of grape (Vitis vinifera L.) cv. Malbec leaf tissues to ultraviolet‐B radiation by enhancing ultraviolet‐absorbing compounds, antioxidant enzymes and membrane sterols. Plant Cell Environ. 2010;33:1–10. doi: 10.1111/j.1365-3040.2009.02044.x. [DOI] [PubMed] [Google Scholar]
- 50.Berli FJ, Fanzone M, Piccoli P. Solar UV-B and ABA Are Involved in Phenol Metabolism of Vitis vinifera L Increasing Biosynthesis of Berry Skin Polyphenols. J. Agric. Food Chem. 2011;59:4874–4884. doi: 10.1021/jf200040z. [DOI] [PubMed] [Google Scholar]
- 51.Loreti E, et al. Gibberellins, jasmonate and abscisic acid modulate the sucrose-induced expression of anthocyanin biosynthetic genes in Arabidopsis. New Phytol. 2008;179:1004–1016. doi: 10.1111/j.1469-8137.2008.02511.x. [DOI] [PubMed] [Google Scholar]
- 52.Cheng H, et al. Gibberellin Acts through Jasmonate to Control the Expression of, MYB21, MYB24, and, MYB57, to Promote Stamen Filament Growth in Arabidopsis. PLoS Genet. 2009;5:e1000440. doi: 10.1371/journal.pgen.1000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun B, et al. Purple foliage coloration in tea (Camellia sinensis L.) arises from activation of the R2R3-MYB transcription factor CsAN1. Sci. Rep. 2016;6:32534. doi: 10.1038/srep32534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chalfun JA, et al. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximaldistal patterning in Arabidopsis petals. Plant Mol. Biol. 2005;57:559–575. doi: 10.1007/s11103-005-0698-4. [DOI] [PubMed] [Google Scholar]
- 55.Chen R, et al. Gene-to-metabolite network for biosynthesis of lignans in MeJA-elicited Isatis indigotica hairy root cultures. Front Plant Sci. 2015;6:952. doi: 10.3389/fpls.2015.00952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu H, Guo W, Yang D, Hou Z, Liang Z. Transcriptional Profiles of SmWRKY Family Genes and Their Putative Roles in the Biosynthesis of Tanshinone and Phenolic Acids in Salvia miltiorrhiza. Int. J. Mol. Sci. 2018;19:1593. doi: 10.3390/ijms19061593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supporting data can be found within the manuscript and its additional files.