This study advances our understanding of BQ inhibition behavior and the mechanism of microbial tolerance to this inhibitor and identifies the key genes responsible for BQ detoxification. The insights here into BQ toxicity and tolerance provide the basis for future synthetic biology to engineer industrial fermentation strains with enhanced BQ tolerance.
KEYWORDS: p-benzoquinone, lignocellulose, tolerance mechanism, metabolic engineering, hydroquinone
ABSTRACT
p-Benzoquinone (BQ) is a lignin-derived inhibitor of biorefinery fermentation strains produced during pretreatment of lignocellulose. Unlike the well-studied inhibitors furan aldehydes, weak acids, and phenolics, the inhibitory properties of BQ, the microbial tolerance mechanism, and the detoxification strategy for this inhibitor have not been clearly elucidated. Here, BQ was identified as a by-product generated during acid pretreatment of various lignocellulose feedstocks, including corn stover, wheat straw, rice straw, tobacco stem, sunflower stem, and corncob residue. BQ at 20 to 200 mg/liter severely inhibited the cell growth and fermentability of various bacteria and yeast strains used in biorefinery fermentations. The BQ tolerance of the strains was found to be closely related to their capacity to convert BQ to nontoxic hydroquinone (HQ). To identify the key genes responsible for BQ tolerance, transcription levels of 20 genes potentially involved in the degradation of BQ in Zymomonas mobilis were investigated using real-time quantitative PCR in BQ-treated cells. One oxidoreductase gene, one hydroxylase gene, three reductase genes, and three dehydrogenase genes were found to be responsible for the conversion of BQ to HQ. Overexpression of the five key genes in Z. mobilis (ZMO1696, ZMO1949, ZMO1576, ZMO1984, and ZMO1399) accelerated its cell growth and cellulosic ethanol production in BQ-containing medium and lignocellulose hydrolysates.
IMPORTANCE This study advances our understanding of BQ inhibition behavior and the mechanism of microbial tolerance to this inhibitor and identifies the key genes responsible for BQ detoxification. The insights here into BQ toxicity and tolerance provide the basis for future synthetic biology to engineer industrial fermentation strains with enhanced BQ tolerance.
INTRODUCTION
Inhibitory compounds derived from pretreatment of lignocellulose biomass are a major challenge to the efficient bioconversion of lignocellulose biomass to biofuels and biochemicals (1–3). Such inhibitors include furan aldehydes (furfural and 5-hydromethylfurfural [HMF]), weak organic acids (acetic acid, formic acid, levulinic acid), and phenolic acids and aldehydes (4-hydroxybenzaldehyde, vanillin, syringaldehyde) (4, 5). However, the list of inhibitors is still expanding (3, 6). Lignin-derived quinone and its derivatives have been identified in various lignocellulose hydrolysates as follows: p-benzoquinone (BQ) and 2,6-dimethoxybenzoquinone (DMBQ) in the hydrolysates of acid-pretreated agricultural residues, softwood, and hardwood (7); methoxyhydroquinone in the hemicellulose hydrolysate of steam-pretreated birch (8); and hydroquinone (HQ) in the hydrolysates of acid-hydrolyzed Norway spruce, grasses, agricultural residues, and agave (9, 10).
These quinones are generated by the oxidation of phenolic compounds. For example, BQ comes from the oxidation of p-hydroxyphenyl compounds of 4-hydroxybenzoic acid, 4-hydroxybenzyl aldehyde, and 4-hydroxybenzyl alcohol (11); methoxybenzoquinone (MBQ) is produced from guaiacyl compounds of vanillic acid, vanillin, and vanillyl alcohol (12); and 2,6-dimethoxyhydroquinone (DMHQ) is produced from syringic acid, syringaldehyde, and syringyl alcohol (12). Quinones were found to strongly inhibit Saccharomyces cerevisiae. Just 0.02 g/liter BQ and 0.2 g/liter DMBQ completely inhibit the cell growth of S. cerevisiae (7, 13). BQ shows high toxicity to animal cells by increasing its reactive oxygen species (ROS) level, oxidizing DNA, breaking DNA double strands, and decreasing Ogg1 transcript levels (14–16). However, compared with the extensive studies on furan aldehydes, weak organic acids, and phenolic aldehydes, only a few studies have reported on microbial inhibition by quinones (7, 13, 17). Metabolic pathways for the conversion of quinones have not been clearly characterized, and strategies to lessen quinone inhibition in biorefinery fermentation strains are not well developed.
In this study, the presence of the toxic (inhibitory) quinone BQ was identified in various lignocellulose feedstocks after acid pretreatment, and the inhibitory effect on five typical biorefinery fermentation strains was examined. The tolerance mechanism of the strains to BQ was established based on metabolism and transcriptional analysis. Enhancement of BQ tolerance and ethanol production were evaluated by overexpressing key genes responsible for BQ conversion in an engineered Zymomonas mobilis strain. This study provides a complete understanding of BQ inhibition behavior and the mechanism of microbial tolerance and identifies key genes responsible for BQ detoxification. This information is an important basis for development of highly BQ-tolerant fermentative strains for lignocellulose biorefining.
RESULTS AND DISCUSSION
Identification and detoxification of p-benzoquinone in pretreated lignocellulose feedstocks.
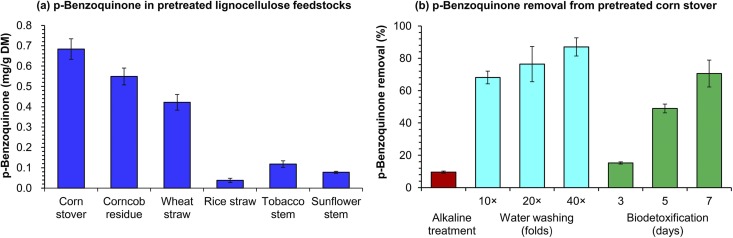
The presence of BQ in six typical lignocellulose feedstocks including corn stover, wheat straw, rice straw, tobacco stem, sunflower stem, and corncob residue after dry acid pretreatment was shown by gas chromatography-mass spectrometry (GC-MS) (retention time, 5.64 min; m/z 108) (Fig. S1). The BQ content was measured by reverse-phase high-performance liquid chromatography (HPLC) (Fig. 1a) and found to be in the range of 0.04 to 0.68 mg/g dry feedstock, approximately equivalent to 12 to 205 mg/liter when a high solids loading condition (30%, wt/wt) is applied for enzymatic hydrolysis and fermentation of the pretreated lignocellulose feedstock.
FIG 1.
Identification and removal of p-benzoquinone (BQ) inhibitor in the pretreated lignocellulose feedstocks. (a) BQ content in the six different lignocellulose feedstocks after dry acid pretreatment, including the pretreated corn stover, corncob residue, wheat straw, rice straw, tobacco stem, and sunflower stem; (b) BQ removal from the pretreated corn stover by physical water washing, chemical alkali treatment, and biological detoxification. The alkali treatment was performed by elevating pH to 9.0 using NH4OH for 1 h at 30°C and then reducing to 5.5 using H2SO4. The biodetoxification was performed by A. resinae ZN1 at 28°C, pH 5.5, for 3 to 7 days. Error bars represent two standard deviations.
Removal of BQ from pretreated lignocellulose feedstock (corn stover) was attempted using three detoxification methods, including alkaline treatment, water washing, and biodetoxification (Fig. 1b). Alkaline treatment by ammonium hydroxide was the least effective method for BQ removal. Water washing removed approximately 65% of the BQ (when using 10-fold fresh water compared with the amount of solids) and 87% of the BQ (40-fold), but the BQ removal was, therefore, accompanied by the cost of huge amounts of fresh water and wastewater generation, as well as considerable sugar loss (18). Unlike the fast and complete removal of furan aldehydes and weak organic acids, solid-state biodetoxification by Amorphotheca resinae ZN1 only resulted in limited BQ removal (15% after 3 days and 70% after 7 days). The prolonged biodetoxification led to reduced process efficiency and the accelerated consumption of fermentable sugars.
These results reveal that BQ is an inevitable by-product of lignocellulose pretreatment, at least acid-based pretreatment, and the current detoxification methods are not efficient for removal of BQ from the pretreated lignocellulose feedstocks at a reasonable cost. Noticeably, the biodetoxification approach, which has high efficiency for detoxifying most of the inhibitory furan and phenolic aldehyde compounds, did not work well for BQ.
Tolerance of various fermentation strains to BQ inhibitor.
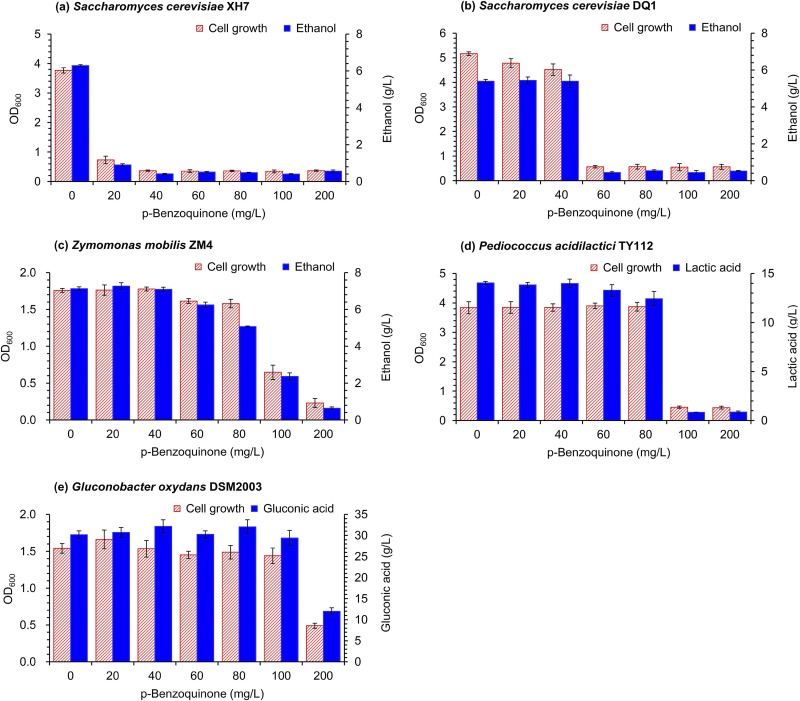
Inhibition of biorefinery fermentation strains by BQ was examined in detail in five representative strains used for production of cellulosic ethanol and organic acids (Fig. 2). The BQ concentration in the culture medium ranged from 20 to 200 mg/liter, based on the BQ content in the acid-pretreated lignocellulose hydrolysate (Fig. 1a). The selected biorefinery fermentation strains included two yeast strains (thermal-tolerant S. cerevisiae DQ1 and xylose-utilizing S. cerevisiae XH7) and three bacterial strains (Z. mobilis ZM4 for ethanol fermentation, Pediococcus acidilactici TY112 for l-lactic acid fermentation, and Gluconobacter oxydans DSM2003 for gluconic acid fermentation). The two yeast strains showed only weak tolerance to BQ; the cell growth of and ethanol fermentability by S. cerevisiae XH7 were reduced by 81% and 86%, respectively, at 20 mg/liter BQ compared with that in the absence of BQ (Fig. 2a), while S. cerevisiae DQ1 started showing strong inhibition by 60 mg/liter BQ (Fig. 2b). However, Z. mobilis ZM4 showed high tolerance to BQ; obvious inhibition of cell growth and ethanol generation appeared only when BQ was present at 100 to 200 mg/liter (Fig. 2c). P. acidilactici TY112 also showed relatively high BQ tolerance (80 mg/liter) (Fig. 2d). G. oxydans DSM2003 showed the highest tolerance to BQ; even at 200 mg/liter BQ, cell growth and gluconic acid fermentation were still maintained at approximately one-third of the level in the absence of BQ (Fig. 2e).
FIG 2.
Inhibition of p-benzoquinone (BQ) on cell growth and product fermentation of the five different yeasts and bacterial strains. (a) Engineered S. cerevisiae XH7 with glucose and xylose cofermentation into ethanol. (b) Thermotolerant strain S. cerevisiae DQ1 for ethanol fermentation. (c) Wild Z. mobilis ZM4 for ethanol fermentation. (d) Engineered P. acidilactici TY112 for l-lactic acid fermentation. (e) Wild G. oxydans DSM2003 for gluconic acid fermentation. The cell growth and product generation were measured at 12 h for all of the five strains. Conditions were as follows: 10% (vol/vol) inoculum size, 30°C, and 150 rpm for S. cerevisiae XH7 and S. cerevisiae DQ1; 30°C in static-state culture for Z. mobilis ZM4; 42°C and 150 rpm for P. acidilactici TY112; and 30°C and 220 rpm for G. oxydans DSM2003. Error bars represent two standard deviations. OD600, optical density at 600 nm.
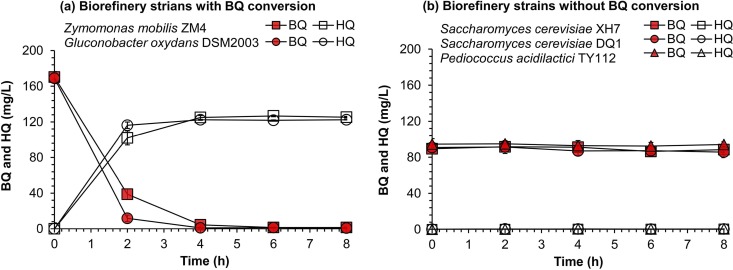
Microbial tolerance is generally understood in terms of capacity to convert inhibitors into less toxic substances (19, 20). The BQ conversion capacity of the five selected strains was examined in phosphate-buffered saline (pH 7.0) containing 20 g/liter glucose and 200 or 100 mg/liter BQ (Fig. 3). The two bacteria with the highest BQ tolerance, Z. mobilis ZM4 and G. oxydans DSM2003, showed quick conversion of 200 mg/liter BQ into HQ (Fig. 3a). Compounds with retention times of 5.13 and 9.17 min in gas chromatography-mass spectrometry (GC-MS) were identified as BQ (m/z 108) and HQ (m/z 110), respectively (Fig. S2). The yeasts S. cerevisiae XH7 and S. cerevisiae DQ1 and the lactic acid bacterium P. acidilactici TY112 failed to show any BQ conversion (Fig. 3b). For further degradation of HQ, only some special strains, such as Burkholderia sp. strain AK-5, can further metabolize HQ through the 2-hydroxy-1,4-benzoquinone and 1,2,4-trihydroxybenzene pathway to produce water and carbon dioxide (21).
FIG 3.
Conversion of p-benzoquinone (BQ) into hydroquinone (HQ) by the five different fermentation strains. (a) Z. mobilis ZM4 and G. oxydans DSM2003 with BQ conversion capacity. (b) S. cerevisiae XH7, S. cerevisiae DQ1, and P. acidilactici TY112 without BQ conversion capacity. The strains were cultured to the exponential growth phase and then harvested, washed by phosphate-buffered saline (PBS) (pH 7.0), and resuspended in PBS before inoculated into the fresh PBS containing 20 g/liter of glucose and 200 mg/liter of BQ for Z. mobilis ZM4 and G. oxydans DSM2003 or 100 mg/liter of BQ for S. cerevisiae XH7, S. cerevisiae DQ1, and P. acidilactici TY112. Conditions were as follows: 10% (vol/vol) inoculum size; 30°C and 150 rpm for S. cerevisiae XH7 and S. cerevisiae DQ1; 30°C in static-state culture for Z. mobilis ZM4; 42°C and 150 rpm for P. acidilactici TY112; and 30°C and 220 rpm for G. oxydans DSM2003. Error bars represent two standard deviations.
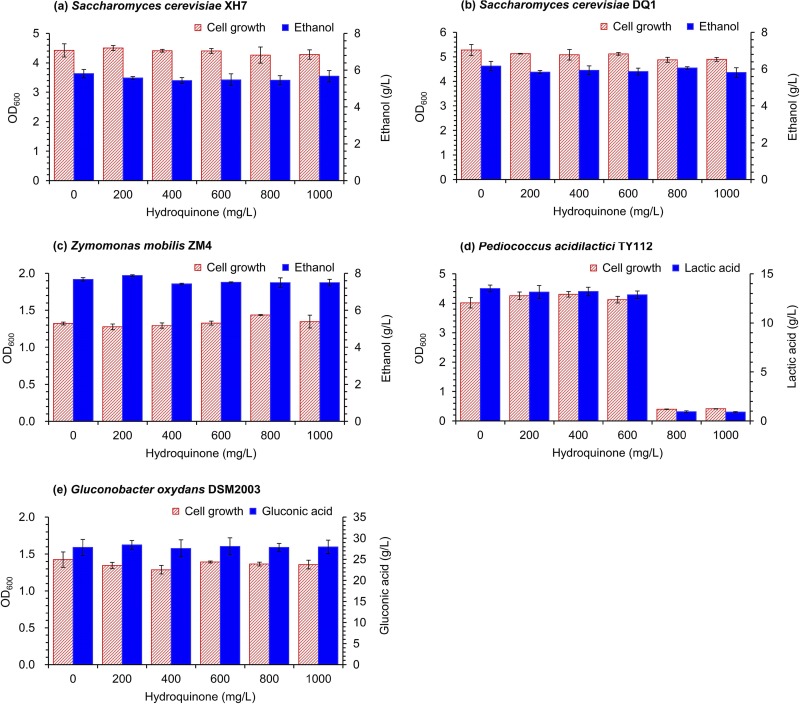
The inhibitory behavior of HQ toward the five fermentation strains was examined. No inhibition was observed at up to 1,000 mg/liter HQ, except for slight inhibition of P. acidilactici TY112 at high BQ concentration (Fig. 4). These results suggest that the high BQ tolerance of the biorefinery strains Z. mobilis ZM4 and G. oxydans DSM2003 came from their capacity for BQ conversion to nontoxic HQ.
FIG 4.
Inhibitory effect of hydroquinone (HQ) on cell growth and product formation of the five different fermentation strains, S. cerevisiae XH7 (a), S. cerevisiae DQ1 (b), Z. mobilis ZM4 (c), P. acidilactici TY112 (d), and G. oxydans DSM2003 (e). The cell growth and product production were measured at 12 h for all of the five strains. Conditions were as follows: 10% (vol/vol) inoculum size, 30°C, and 150 rpm for S. cerevisiae XH7 and S. cerevisiae DQ1; 30°C in static-state culture for Z. mobilis ZM4; 42°C and 150 rpm for P. acidilactici TY112; and 30°C and 220 rpm for G. oxydans DSM2003. Error bars represent two standard deviations.
Inhibitor conversion by fermentation microorganisms provides a practical option for complete and ultimate removal of the residual lignin-derived phenolic compounds. These phenolic inhibitors, such as BQ, are toxic but generally at low concentration and low water solubility. Therefore, BQ generally gives less stress on cell growth and metabolism of fermentation strains. When high conversion ratio and high product titer are required, however, the fermenting strains should demonstrate all potentials, and the residual and minimum BQ clearly plays negatively. Conversely, conventional washing is unable to wash out the BQ inhibitors due to its low water solubility and the huge amount of water that is required. Biodetoxification by specific fungi is effective but very slow, and this slow process consumes considerable xylose sugar and reduces the conversion ratio of the target product, ethanol.
Mining the genes responsible for BQ tolerance in Z. mobilis.
The following two important results were obtained from the above experiments: the toxic inhibitor BQ is not easily removed from pretreated lignocellulose feedstock by the available detoxification methods, and the BQ tolerance of biorefinery strains comes from the conversion of BQ to HQ. To effectively release the inhibition of BQ toward fermenting strains, we tried the strategy of converting BQ into nontoxic HQ by the fermentation strains during the fermentation process, instead of direct removal of BQ from the feedstock. We selected the highly BQ-tolerant strain Z. mobilis as the target strain, and mined the key genes responsible for conversion of BQ to HQ based on genome annotation analysis and a gene library involved in conversion of phenolic aldehydes into corresponding alcohols (22). The genes encoding the oxidoreductases could be identified by mining the well-sequenced and clearly annotated genome information of Z. mobilis (23, 24). Under such data support, the real-time quantitative PCR (qRT-PCR) method is sufficient to identify the key genes with strong responses to the conversion of BQ to HQ.
Twenty genes from the engineered strain Z. mobilis 8b putatively responsible for the conversion of BQ to HQ were selected, including three oxidoreductase genes, six reductase genes, six dehydrogenase genes, one hydroxylase gene, and four hypothetical protein genes predicted to be responsible for reduction of phenolic aldehydes into the corresponding alcohols (Table 1).
TABLE 1.
Genes and primers used in the qRT-PCR assay
| Gene | Annotation | Primer sequence (5′–3′) |
|
|---|---|---|---|
| Forward | Reverse | ||
| Primer for the oxidoreductase gene | |||
| ZMO1116 | Oxidoreductase | TGTGGTTTGGGCCATCCG | TGTCGGTGCGTCCTGTTTGT |
| ZMO1772 | NAD(P)H quinone oxidoreductase | GGTCGCCTTGTCATTGTCG | GGCTGTTCTGGCACGCAT |
| ZMO1885 | NADH flavin oxidoreductase | TGGAGTGATGCCCAAGTAGAAG | CACTGACATTAGACGGCACCATA |
| Primer for the reductase gene | |||
| ZMO1993 | NADPH quinone reductase | GCGGTGTCGGTAGCTTGTT | GCCTTCGCCGTGATTCTG |
| ZMO0833 | UDP-N-acetylenolpyruvoylglucosamine reductase | ATCGCCTGCGTTGTGGTG | GCATTCATGCGGATCATACCA |
| ZMO1222 | 3-Oxoacyl-(acyl-carrier-protein) reductase | TTAGCCGTGCCGTCATCAGA | CGATCATGCCTGCCTTTGC |
| ZMO1303 | Pyrroline-5-carboxylate reductase | ACAACCCTGATTTCTATTCTTGCC | AACAACGCCCTTCCCTAACG |
| ZMO1984 | Aldo-keto reductase | TCGCCATTTGTCAGCCTATC | CAGCAAAGTTACTGCTACCCAC |
| ZMO1254 | Redoxin domain-containing protein | CGCTATGGTAATCCCTATCAGGC | GCTCATAAGCTGTGGCAAATCC |
| Primer for the dehydrogenase gene | |||
| ZMO1335 | NAD(P)H dehydrogenase (quinone) | CCCGATTTGGGCGTTTG | AGACCGCTCCGACCTTTCC |
| ZMO1949 | NAD(P)H dehydrogenase (quinone) | TGGAGGCAAGCGGTTCTAC | GCAACAGCGGTTTGAGCAT |
| ZMO0157 | d-Isomer-specific 2-hydroxyacid dehydrogenase | TATGGCTGTCGGGTTATTCC | TTCGGCGATGGCTTGG |
| ZMO0788 | d-Sorbitol dehydrogenase | CAGTTGTCACGGTCGCAATG | GCCGCAATCTGACTATCGTTTA |
| ZMO1576 | Short-chain dehydrogenase/reductase | TCTATCGTCGCTAAGGGTGGTC | GCTGCAATCCCATAAAGAACG |
| ZMO1696 | Zinc-binding alcohol dehydrogenase | TCCGAAGACAGGCGAAGAATG | CCTCGTCACCGACCTTAAATAGC |
| Primer for other genes | |||
| ZMO1399 | Fatty acid hydroxylase | GAAACGGATTCACTATGACCAC | CCGACAGCATAACCGATACA |
| ZMO0020 | Hypothetical protein | CCGATCTAGTAAGCCAATTCACC | TTTCAAATCTGTTGGTTGGGTGT |
| ZMO0021 | Hypothetical protein | CCACTTCATATCGCTTCTGTCG | GCGTAATCGGTGATCCCAAA |
| ZMO0074 | Hypothetical protein | CGATAAAGACCGCCCGACC | TCGCCAAGCAGGCATTCG |
| ZMO1821 | Hypothetical protein | AGAAAGCCGCCGCCATC | CACCATCATACCAACTGTCAACG |
| ZMOr003a | 16S rRNA gene | TTAAGTTGGGCACTTTAGAGGAAC | TGTCACCGCCATTGTAGCAC |
The 16S rRNA gene ZMOr003 was used as the internal control to normalize the difference of the data.
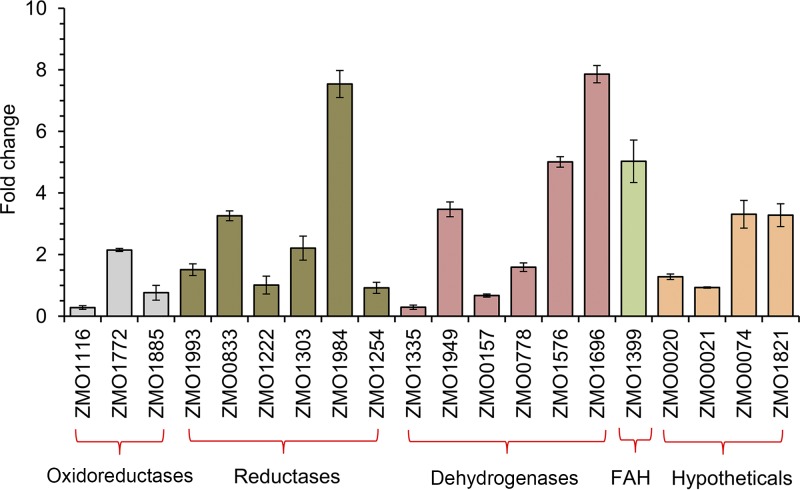
The transcription levels of these 20 genes during the conversion of BQ to HQ were analyzed using real-time quantitative PCR (qRT-PCR) (Fig. 5). Twelve genes among the 20 showed a strong response to BQ, among which 10 genes were significantly upregulated (i.e., by more than 2-fold) and two were downregulated.
FIG 5.
Transcription level analyses of the twenty selected relevant genes in Z. mobilis 8b in response to 200 mg/liter of p-benzoquinone (BQ) when cultured in RM. FAH, fatty acid hydroxylase. Gene functions and the primers used for the specific set of genes are shown in Table 1. Cell mass of Z. mobilis 8b was collected after treatment for 1 h by BQ. The cultures without BQ treatment served as the controls, and the fold change was obtained based on the normalized gene expression level between the BQ treatment and control samples. Three biological and technical replicates were run for the qRT-PCR analysis.
The significantly upregulated genes included the following: one oxidoreductase gene, ZMO1772, encoding an NAD(P)H quinone oxidoreductase; three reductase genes (ZMO0833, encoding a UDP-N-acetylenolpyruvoylglucosamine reductase; ZMO1303, encoding a pyrroline-5-carboxylate reductase; and ZMO1984, encoding an aldo-keto reductase); three dehydrogenase genes [ZMO1949, encoding an NAD(P)H dehydrogenase; ZMO1696, encoding a zinc-binding alcohol dehydrogenase; and ZMO1576, encoding a short-chain dehydrogenase/reductase]; one fatty acid hydroxylase gene, ZMO1399; and two hypothetical protein genes, ZMO0074 and ZMO1821. The two downregulated genes were oxidoreductase-encoding gene ZMO1116 and NAD(P)H dehydrogenase gene ZMO1335.
Enhancement of BQ tolerance and cellulosic ethanol fermentation by Z. mobilis.
Z. mobilis ZM4 is a wild-type strain and only utilizes glucose, fructose, and sucrose as the substrates. BQ is present only when lignocellulose biomass is pretreated, in which xylose is generated by partial hydrolysis of hemicellulose, at least in acid-based pretreatment operations, such as dilute acid, dry acid, steam explosion, hot water, SO2, etc. Therefore, a xylose-utilizing strain is a more reasonable choice for metabolic engineering of a practical useful engineered strain. Z. mobilis 8b was selected for its advantage of multiple sugar utilization on glucose, xylose, and arabinose. Enhancement of BQ tolerance of Z. mobilis 8b was achieved by overexpressing genes significantly upregulated in the presence of BQ, including ZMO1772, ZMO0833, ZMO1303, ZMO1984, ZMO1949, ZMO1696, ZMO1576, and ZMO1399. These genes were individually inserted into the expression vector pHW20a and introduced into Z. mobilis 8b to generate eight recombinant strains according to the protocols described by Dong et al. (25) (Fig. S4). The fermentability of the obtained recombinants was evaluated in BQ-containing synthetic medium and practical lignocellulose systems (Fig. 6). Among these lignocellulose biomasses, corn stover is relatively sensitive to pretreatment severity to release the monosaccharide sugars by cellulase enzymes and to generate more overdegradation products, such as phenolic aldehydes or acids, than wheat straw and rice straw (26). Figure 1a experimentally confirmed that the acid-pretreated corn stover generated the maximum amount of lignin-derived BQ compounds. To demonstrate the most significant performance of the gene candidates by constructing Z. mobilis recombinants, we selected corn stover as the testing biomass and used corn stover hydrolysate for testing BQ tolerance.
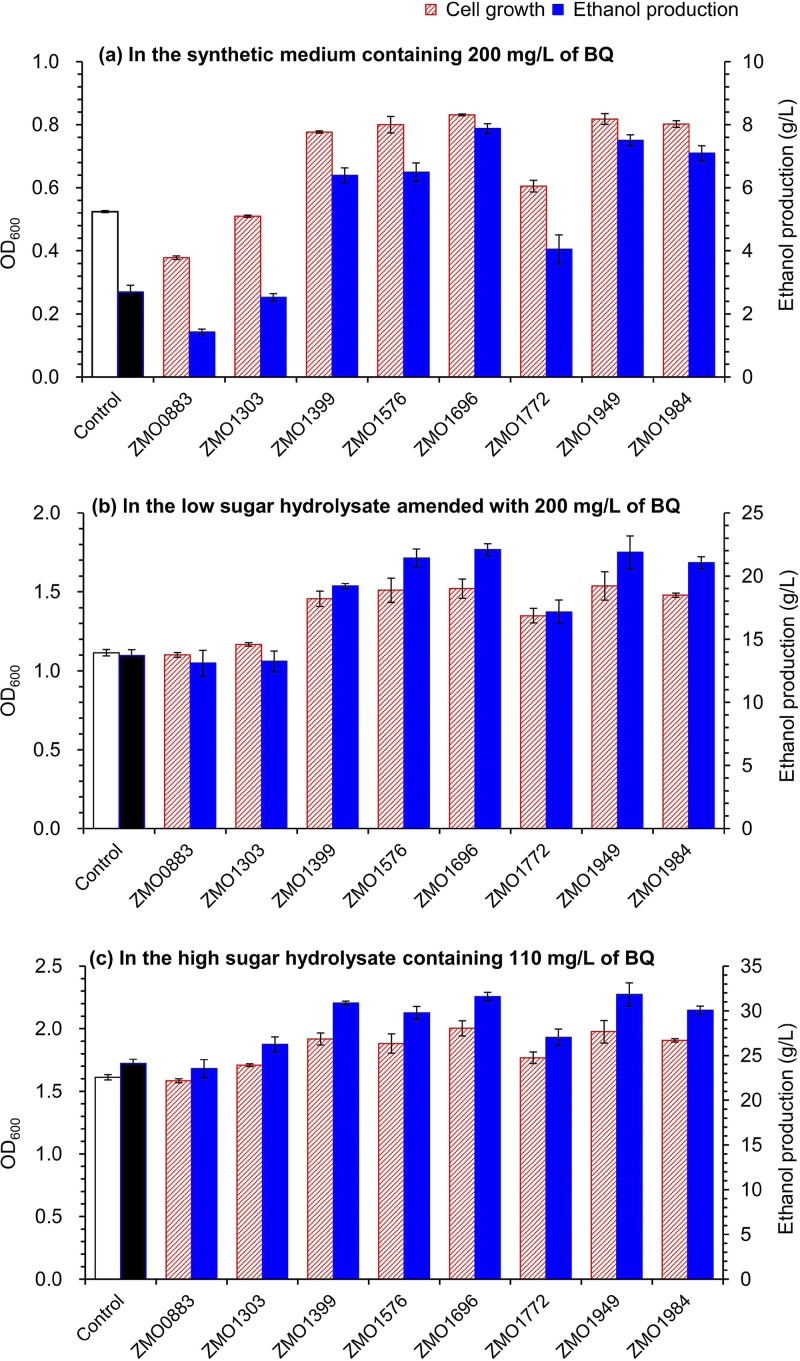
FIG 6.
Cell growth and ethanol fermentation of Z. mobilis recombinant strains in various culture systems containing different levels of p-benzoquinone (BQ). (a) In RM synthetic medium containing 200 mg/liter of BQ. (b) In low-sugar corn stover hydrolysate amended with 200 mg/liter of BQ. (c) In high-sugar corn stover hydrolysate containing 110 mg/liter of BQ without BQ addition. The recombinant strain harboring the empty pHW20a served as the control strain. The cell growth and ethanol production were measured at 16 h for RM medium fermentation, 24 h for low-sugar hydrolysate fermentation, and 72 h for high-sugar hydrolysate fermentation. Conditions were as follows: 10% (vol/vol) inoculum size and 30°C in static-state culture. Error bars represent two standard deviations.
The first test was conducted in synthetic medium containing 200 mg/liter BQ (Fig. 6a). The overexpression of zinc-binding alcohol dehydrogenase gene ZMO1696 significantly increased cell growth and ethanol production by 58% and 192%, respectively, compared with those of the control strain harboring empty pHW20a. Overexpression of the two dehydrogenase genes, the NAD(P)H dehydrogenase gene ZMO1949 and the short-chain dehydrogenase/reductase gene ZMO1576, led to increases of cell growth by 56% and 53% and ethanol generation by 179% and 141%, respectively. Overexpression of aldo-keto reductase gene ZMO1984 increased the cell growth and ethanol generation by 53% and 163%, respectively. However, overexpression of the other two reductase genes, ZMO0833 and ZMO1303, did not lead to an observable increase in cell growth or ethanol generation. Overexpression of the fatty acid hydroxylase gene ZMO1399 increased the cell growth and ethanol generation by 48% and 137%, respectively. Overexpression of the NAD(P)H quinone oxidoreductase gene ZMO1772 slightly improved the cell growth and ethanol generation by 15% and 50%, respectively.
The second test was carried out in low-sugar-containing corn stover hydrolysate supplemented with 200 mg/liter BQ (Fig. 6b). Similar results were obtained to those above. Overexpression of the dehydrogenase genes ZMO1696, ZMO1949, and ZMO1576 increased the cell growth by 36%, 38%, and 35% and the ethanol production by 61%, 60%, and 56%, respectively. Overexpression of the reductase gene ZMO1984, the hydroxylase gene ZMO1399, and the oxidoreductase gene ZMO1772 increased the cell growth by 33%, 31%, and 21% and the ethanol production by 54%, 40%, and 25%, respectively. Overexpression of the other two genes (ZMO0833 and ZMO1303) did not result in observable improvement in ethanol fermentation.
The third test was in high-sugar-containing hydrolysate without additional BQ supplementation (Fig. 6c). Because of the relatively low original BQ content (110 mg/liter, about half of that in the two previous tests), the cell growth and ethanol generation of the recombinants did not show significant increases when overexpressing the genes responsible for BQ conversion as they did in the high-BQ-containing medium or hydrolysate (each containing 200 mg/liter BQ). However, we still observed obvious increases in cell growth and ethanol fermentability. Overexpression of ZMO1696, ZMO1949, ZMO1576, ZMO1984, and ZMO1399 increased the cell growth by 17% to 24% and ethanol generation by 24% to 32%. Overexpression of ZMO1772 slightly increased the cell growth and ethanol generation by around 12%. The recombinants overexpressing ZMO0833 and ZMO1303 performed similarly to the control strain.
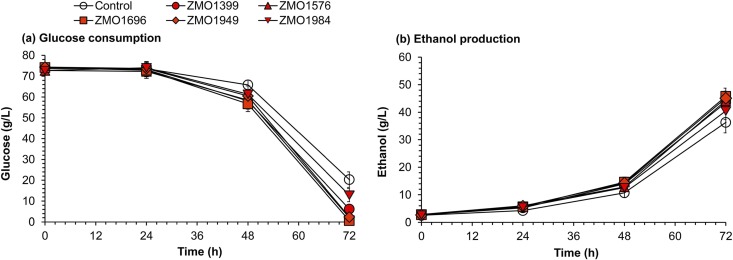
The final evaluation was conducted for practical simultaneous saccharification and cofermentation (SSCF) under high-dry-acid-pretreated and biodetoxified corn stover solid loading (Fig. 7). The five Z. mobilis recombinants harboring the genes (ZMO1696, ZMO1949, ZMO1576, ZMO1984, and ZMO1399) with the most obvious function in converting BQ were used. In the SSCF, all five recombinant strains showed obviously accelerated glucose consumption, and the ethanol generation increased by 26%, 24%, 22%, 20%, and 12% by overexpression of ZMO1696, ZMO1949, ZMO1576, ZMO1399, and ZMO1984, respectively, compared with that of the control strain. The maximum ethanol titer after SSCF for 72 h reached 46 g/liter for the Z. mobilis recombinant harboring ZMO1696 compared to 36 g/liter of ethanol for the control strain. These results confirmed the function of the mined genes in relieving BQ stress in the practical production of cellulosic ethanol from lignocellulose feedstock.
FIG 7.
Simultaneous saccharification and cofermentation (SSCF) of cellulosic ethanol by Z. mobilis recombinants harboring the ZMO1399, ZMO1576, ZMO1696, ZMO1949, and ZMO1984 genes. (a) Glucose consumption in SSCF; (b) ethanol production in SSCF. The feedstock used was 30% (wt/wt) solids loading of dry-acid-pretreated and biodetoxified corn stover. Corn stover feedstock was prehydrolyzed for 12 h at 4 mg total protein/g DM at 50°C with a pH of 4.8. Then, the seed broth was inoculated at 10% of the inoculation ratio to start the SSCF at 30°C and pH 5.0. Error bars represent two standard deviations.
The current study used real-time quantitative PCR to mine key genes responsible for BQ conversion in Z. mobilis. To demonstrate the most significant performance of the gene candidates by constructing Z. mobilis recombinants, we selected corn stover as the testing biomass and used corn stover hydrolysate for testing BQ tolerance. Finally, we found that some genes, such as ZMO1116 and ZMO1399, worked well for BQ tolerance (Fig. S3). These genes could be used for metabolic engineering to improve the BQ tolerance of other strains. This study further advances our understanding of BQ inhibition behavior and the mechanism of microbial tolerance to this inhibitor and identifies the key genes responsible for BQ detoxification. The insights here into BQ toxicity and tolerance provide the basis for future synthetic biology to engineer industrial fermentation strains with enhanced BQ tolerance.
Conclusions.
BQ was identified to be a by-product in acid-pretreated lignocellulose biomass and was toxic to various biorefining strains and, particularly, S. cerevisiae. Conversion of BQ to nontoxic HQ was an important mechanism of BQ tolerance in biorefinery strains. Oxidoreductase-, reductase-, and dehydrogenase-encoding genes were identified as key genes responsible for BQ conversion in Z. mobilis, and overexpression of these genes significantly improved BQ tolerance and accelerated cellulosic ethanol fermentation from dry-acid-pretreated and biodetoxified corn stover feedstock by Z. mobilis. The insights into BQ toxicity and tolerance in this study provide an important basis for development of high-BQ-tolerant fermentative strains for lignocellulose biorefining.
MATERIALS AND METHODS
Strains, plasmids, media, and culture conditions.
A. resinae ZN1 used for biodetoxification of pretreated corn stover feedstock was isolated in our previous work (27) and stored in the China General Microbiological Culture Collection Center (CGMCC; Beijing, China) with registration number 7452. A. resinae ZN1 was grown at 28°C on potato dextrose agar (PDA) slants containing 200 g/liter potato extract juice, 20 g/liter glucose, and 15 g/liter agar.
S. cerevisiae DQ1 used for ethanol fermentation (without xylose utilization) was developed in our previous study (28) and stored in the CGMCC with registration number 2528. S. cerevisiae XH7 used for ethanol fermentation from glucose and xylose was derived from the wild-type diploid S. cerevisiae strain BSIF through rationally designed genetic modifications combined with alternating adaptive evolutions (29). The S. cerevisiae strains were grown at 30°C, 150 rpm, in synthetic medium containing 2 g/liter KH2PO4, 1 g/liter (NH4)2SO4, 1 g/liter MgSO4·7H2O, 20 g/liter glucose, and 1 g/liter yeast extract.
Z. mobilis ZM4 (ATCC 31821) used for ethanol fermentation was purchased from the American Type Culture Collection (ATCC, Manassas, VA, USA). Z. mobilis 8b, used for ethanol fermentation from glucose and xylose (30), was also purchased from ATCC. The two Z. mobilis strains and derived recombinants were inoculated into rich medium (RM) containing 2 g/liter KH2PO4, 20 g/liter glucose, and 10 g/liter yeast extract and incubated at 30°C without shaking; 20 μg/ml tetracycline and 30 μg/ml nalidixic acid were added for culture of Z. mobilis recombinants.
G. oxydans DSM2003 used for gluconic acid fermentation was purchased from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). G. oxydans DSM2003 was incubated at 30°C, 220 rpm, in the synthetic medium containing 1.5 g/liter KH2PO4, 1.5 g/liter (NH4)2SO4, 0.5 g/liter MgSO4·7H2O, 20 g/liter yeast extract, and 80 g/liter sorbitol for seed culture or 80 g/liter glucose for fermentation.
P. acidilactici TY112 used for l-lactic acid fermentation was developed in our previous study (31) and stored in the CGMCC with registration number 8664. P. acidilactici TY112 was cultured at 42°C, 150 rpm, in de Man-Rogosa-Sharpe medium containing 2 g/liter KH2PO4, 0.58 g/liter MgSO4·7H2O, 0.25 g/liter MnSO4, 5 g/liter sodium acetate, 2 g/liter ammonium citrate dibasic, 20 g/liter glucose, 10 g/liter peptone, and 10 g/liter yeast extract.
Escherichia coli S17-1 λπ used for plasmid transformation (32) was cultured at 37°C, 220 rpm, in Luria-Bertani medium containing 10 g/liter NaCl, 5 g/liter yeast extract, and 10 g/liter tryptone; 20 μg/ml tetracycline was added to screen for positive E. coli recombinants.
Reagents and enzymes.
BQ and HQ were purchased from Kamai Shu Biotechnology (Shanghai, China). Tryptone and yeast extract were purchased from Oxoid (Hampshire, UK). Tetracycline and nalidixic acid were from Sigma-Aldrich (St. Louis, MO, USA). Glucose, xylose, and other chemical reagents were of analytical reagent grade and purchased from Lingfeng Chemical Reagent Co. (Shanghai, China).
RNAiso Plus and PrimeStar HS DNA polymerase were purchased from TaKaRa (Otsu, Japan). ReverTra Ace qPCR RT master mix and SYBR green real-time PCR master mix kits were from Toyobo (Osaka, Japan). A TIANamp bacterial DNA kit was purchased from Tiangen (Beijing, China). Restriction endonucleases and T4 DNA ligase were from Thermo Scientific (Wilmington, DE, USA).
Commercial cellulase Cellic CTec 2.0 was purchased from Novozymes (Beijing, China). The filter paper activity was 203.2 filter paper units (FPU)/ml determined using the NREL protocol LAP-006 (33), the cellobiase activity was 4,900 cellobiase activity units (CBU)/ml determined using the method of Ghose (34), and the total protein concentration was 87.3 mg total protein/ml cellulase solution determined by the Bradford method (35) using bovine serum albumin as a standard.
Raw materials and biorefinery processing.
Corn stover and sunflower stem were harvested from Bayan Nur, Inner Mongolia, China, in autumn 2015. Wheat straw was harvested from Dancheng, Henan, China, in summer 2013. Rice straw was harvested from Chang Zhou, Jiangsu, China, in autumn 2014. Tobacco stem was harvested from Luo Yang, Henan, China, in summer 2015. Corncob residue was the solid waste generated from the xylitol production line of Longlive Biotech (Yucheng, Shandong, China). After collection, the raw feedstocks were coarsely chopped; water washed to remove field dirt, stones, and metals; air dried to constant weight; and then milled to pass through a 10-mm-diameter mesh. The five lignocellulose feedstocks were pretreated by the dry acid pretreatment method (26, 36). Briefly, 1,200 g of dry feedstock and 600 g of 5% (wt/wt) dilute sulfuric acid solution were concurrently fed into a 20-liter pretreatment reactor under mild helical agitation and treated at 175°C for 5 min.
BQ detoxification from pretreated lignocellulose feedstock was performed as follows. For water washing detoxification, different amounts of fresh water were used to soak pretreated corn stover, which was stirred for 30 min at room temperature. Then, the slurry was filtered, air dried, and used to determine the BQ content. For alkaline treatment detoxification, dry-acid-pretreated corn stover was elevated to pH 9.0 using NH4OH for 1 h at 30°C and then adjusted to pH 5.5 using H2SO4. For biodetoxification by A. resinae ZN1, pretreated corn stover was adjusted to pH 5.5 using 20% Ca(OH)2. A. resinae ZN1 spores were cultured on PDA slants at 28°C for 4 days, and then the spores were washed with sterile deionized water and 20 ml of spore suspension (4 × 106 to 5 × 106 spores/ml) was inoculated onto 200 g pretreated corn stover (pH 5.5) for 6 days at 28°C to produce seed culture, which was then inoculated onto freshly pretreated feedstock (pH 5.5) at 10% (wt/wt) inoculum for 3 to 7 days at 28°C.
To prepare corn stover hydrolysate, corn stover feedstock was biodetoxified by A. resinae ZN1 for 4 days (as described above) to remove furfural, 5-hydromethylfurfural, acetic acid, and partial phenolic inhibitors. The residual BQ content was 0.35 mg/g dry solid matter (DM) after the biodetoxification. Hydrolysate was then prepared in a 5-liter bioreactor with a cellulase dosage of 4 mg total protein per gram of dry solid matter at 50°C, pH 4.8, with mild agitation for 48 h. The hydrolysate slurry was centrifuged at 9,300 × g for 10 min, autoclaved at 115°C for 20 min, and then filtered to yield the hydrolysate. Specific hydrolysates were prepared by hydrolyzing 15% (wt/wt) corn stover solids to yield low-sugar-containing hydrolysate and by hydrolyzing 30% (wt/wt) corn stover solids to yield high-sugar-containing hydrolysate. The low-sugar-containing hydrolysate contained 60 g/liter glucose, 15 g/liter xylose, 0.9 g/liter acetic acid, and 0.06 g/liter BQ. The high-sugar-containing hydrolysate contained 106 g/liter glucose, 27 g/liter xylose, 2.1 g/liter acetic acid, and 0.11 g/liter BQ.
SSCF was conducted in a 5-liter bioreactor equipped with a helical ribbon impeller (37). The feedstock used was 30% (wt/wt) solids loading of dry-acid-pretreated and biodetoxified corn stover. After prehydrolysis at 50°C, 150 rpm, pH 4.8, for 12 h using cellulase Cellic CTec 2.0 at a cellulase dosage of 4 mg total protein/g DM, a nutrient solution including 2 g/liter KH2PO4 and 10 g/liter yeast extract was added, and seed cultures were inoculated at 10% (vol/vol) inoculum. The SSCF lasted for 72 h at 30°C, 100 rpm, pH 5.0.
Sample collection, RNA extraction, and qRT-PCR assay.
Z. mobilis 8b was cultured in RM with or without 200 mg/liter BQ addition for 1 h, and then the cells were collected by centrifugation at 13,400 × g at 4°C for 10 min, washed three times with sterile deionized water, and quickly frozen in liquid nitrogen before being stored at −80°C. The total RNA was extracted according to the protocol for TRIzol reagent (RNAiso Plus; TaKaRa, Otsu, Japan), and the RNA quality was checked using a DU 800 spectrophotometer (Beckman Coulter, Fullerton, CA, USA). cDNA was synthesized according to the protocol of the ReverTra Ace qPCR RT master mix kit (Toyobo, Osaka, Japan). qRT-PCR amplification was performed using a SYBR green Realtime PCR master mix kit (Toyobo, Osaka, Japan) and a Bio-Rad CFX 96 system (Bio-Rad, Hercules, CA, USA) with the following procedure: 95°C for 1 min; 40 cycles of 95°C for 15 s, 55°C for 15 s, and 72°C for 30 s; and a final melting curve step heating from 65 to 95°C at 0.1°C per second. The genes and primers used for qRT-PCR assay are shown in Table 1. The 16S rRNA gene ZMOr003 was selected as an internal control for normalization of the data. The 2−ΔΔCT method was used to quantify the expression levels of the tested genes (38). Fold change (FC) of the tested genes was obtained based on difference in the normalized gene expression level between BQ treatment and control samples. Significantly up- or downregulated genes were respectively defined by the following criteria: FC ≥ 2.0 or ≤ 0.5. Three biological replicates were run for the qRT-PCR analysis.
Construction of Z. mobilis recombinants and fermentation evaluation.
Genomic DNA of Z. mobilis was extracted using a TIANamp bacterial DNA kit (Tiangen Biotech, Beijing, China). We used the genomic DNA to amplify the genes (ZMO1772, ZMO0833, ZMO1303, ZMO1984, ZMO1949, ZMO1696, ZMO1576, ZMO1399, ZMO0074, ZMO1821, ZMO1116, and ZMO1335) for overexpression. These key genes used for overexpression were individually inserted into the pHW20a plasmid (constructed by Dong et al. [25]), at HindIII and KpnI sites, and then the plasmid was introduced into Z. mobilis 8b (22, 25, 39–41). A single colony of the recombinant strain was inoculated into 5 ml RM and precultured at 30°C for 20 h, and then the whole culture broth was transferred into 50 ml of fresh RM and incubated overnight. Ethanol fermentation evaluations were conducted in 250-ml Erlenmeyer flasks containing 50 ml of synthetic medium or corn stover hydrolysate at 30°C and pH 5.0 without shaking.
HPLC and GC-MS analyses.
Glucose, xylose, acetic acid, furfural, 5-hydromethylfurfural, ethanol, and lactic acid were determined by HPLC (LC-20AD; Shimadzu, Kyoto, Japan) equipped with an RID-10A refractive index detector (Shimadzu, Kyoto, Japan) and a Bio-Rad Aminex HPX-87H column (Bio-Rad, Hercules, CA, USA). The column temperature was 65°C, and 5 mM H2SO4 solution was used as the mobile phase with a flow rate of 0.6 ml/min.
Gluconic acid measurement followed the method of Zhang et al. (42) using HPLC (LC-20AT; Shimadzu, Kyoto, Japan) with an SPD-20A UV-Vis detector (Shimadzu, Kyoto, Japan) and a Bio-Rad Aminex HPX-87H column at 55°C, using 5 mM H2SO4 as the mobile phase with a flow rate of 0.4 ml/min and detection at 210 nm.
BQ, HQ, vanillin, syringaldehyde, and 4-hydroxybenzaldehyde (HBA) were analyzed by HPLC (LC-20AT; Shimadzu, Kyoto, Japan) fitted with an SPD-20A detector and a YMC-Pack ODS-A column (YMC, Tokyo, Japan). For BQ and HQ, the mobile phases used were 0.1% (vol/vol) formic acid in water (eluent A) and 0.1% (vol/vol) formic acid in a mixture of acetonitrile and 2-propanol (3:1, vol/vol) (eluent B). Elution started at 30% eluent B, increased from 30% to 40% in 9 min, further increased from 40% to 50% in 8 min, and then decreased from 50% to 30% in 0.01 min before holding at 30% for 33 min. The flow rate, column temperature, and detector wavelength were 0.25 ml/min, 30°C, and 245 nm, respectively. For vanillin, syringaldehyde, and HBA, 0.1% (vol/vol) formic acid (eluent A) and acetonitrile (eluent B) served as the mobile phases. The elution started at 10% eluent B, increased from 10% to 35% in 4 min, and was maintained at 35% for 11 min, and then the eluent B concentration was decreased from 35% to 10% in 5 min and held at 10% for 10 min. The flow rate, column temperature, and detector wavelength were 1.0 ml/min, 35°C, and 270 nm, respectively.
BQ in pretreated lignocellulose feedstocks and HQ generated during BQ degradation were identified on an Agilent 6890-5973N GC-MS fitted with an HP-5 MS capillary column (30 m by 0.25 mm; film thickness, 0.25 μm) (Agilent, Santa Clara, CA, USA). The injector temperature was 280°C, and a 1-μl sample was injected in splitless mode. The temperature program was from 80°C (held for 4 min) to 280°C at a rate of 8°C/min. For the extract of pretreated corn stover, the compound with a retention time of 5.64 min was identified as BQ (m/z 108), while for the synthetic medium used for BQ conversion, the compounds with retention times of 5.13 and 9.17 min were identified as BQ (m/z 108) and HQ (m/z 110), respectively.
Supplementary Material
ACKNOWLEDGMENTS
This research was supported by the National Natural Science Foundation of China (21978083 and 31961133006).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.01443-19.
REFERENCES
- 1.Bellido C, Bolado S, Coca M, Lucas S, González-Benito G, García-Cubero MT. 2011. Effect of inhibitors formed during wheat straw pretreatment on ethanol fermentation by Pichia stipitis. Bioresour Technol 102:10868–10874. doi: 10.1016/j.biortech.2011.08.128. [DOI] [PubMed] [Google Scholar]
- 2.Franden MA, Pilath HM, Mohagheghi A, Pienkos PT, Zhang M. 2013. Inhibition of growth of Zymomonas mobilis by model compounds found in lignocellulosic hydrolysates. Biotechnol Biofuels 6:99. doi: 10.1186/1754-6834-6-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klinke HB, Thomsen AB, Ahring BK. 2004. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biotechnol 66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- 4.Jonsson LJ, Alriksson B, Nilvebrant NO. 2013. Bioconversion of lignocellulose: inhibitors and detoxification. Biotechnol Biofuels 6:16. doi: 10.1186/1754-6834-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palmqvist E, Hahn-Hägerdal B. 2000. Fermentation of lignocellulosic hydrolysates. II: inhibitors and mechanisms of inhibition. Bioresour Technol 74:25–33. doi: 10.1016/S0960-8524(99)00161-3. [DOI] [Google Scholar]
- 6.Jonsson LJ, Martin C. 2016. Pretreatment of lignocellulose: formation of inhibitory by-products and strategies for minimizing their effects. Bioresour Technol 199:103–112. doi: 10.1016/j.biortech.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Stagge S, Cavka A, Jönsson LJ. 2015. Identification of benzoquinones in pretreated lignocellulosic feedstocks and inhibitory effects on yeast. AMB Express 5:62. doi: 10.1186/s13568-015-0149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchert J, Niemela K, Puls J, Poutanen K. 1990. Improvement in the fermentability of steamed hemicellulose hydrolysate by ion exclusion. Process Biochem 25:176–180. [Google Scholar]
- 9.Larsson S, Reimann A, Nilvebrant NO, Jonsson LJ. 1999. Comparison of different methods for the detoxification of lignocellulose hydrolyzates of spruce. ABAB 77:91–103. doi: 10.1385/ABAB:77:1-3:91. [DOI] [Google Scholar]
- 10.Mitchell VD, Taylor CM, Bauer S. 2014. Comprehensive analysis of monomeric phenolics in dilute acid plant hydrolysates. Bioenerg Res 7:654–669. doi: 10.1007/s12155-013-9392-6. [DOI] [Google Scholar]
- 11.Saa JM, Morey J, Rubido C. 1986. An oxidative degradation approach to p-quinones. J Org Chem 51:4471–4473. doi: 10.1021/jo00373a025. [DOI] [Google Scholar]
- 12.Wozniak JC, Dimmel DR, Malcolm EW. 1989. The generation of quinones from lignin and lignin-related compounds. J Wood Chem Technol 9:491–511. doi: 10.1080/02773818908050312. [DOI] [Google Scholar]
- 13.Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant N-O, Jönsson LJ. 2000. Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. ABAB 84:617–632. doi: 10.1385/ABAB:84-86:1-9:617. [DOI] [PubMed] [Google Scholar]
- 14.Philbrook NA, Winn LM. 2015. Benzoquinone toxicity is not prevented by sulforaphane in CD-1 mouse fetal liver cells. J Appl Toxicol 36:1015–1024. doi: 10.1002/jat.3251. [DOI] [PubMed] [Google Scholar]
- 15.Lee S-K, Chung S-M, Lee M-Y, Lee J-Y, Bae O-N, Chung J-H. 2002. The roles of ATP and calcium in morphological changes and cytotoxicity induced by 1,4-benzoquinone in platelets. Biochim Biophys Acta 1569:159–166. doi: 10.1016/s0304-4165(01)00252-5. [DOI] [PubMed] [Google Scholar]
- 16.Irons RD. 1985. Quinones as toxic metabolites of benzene. J Toxicol Environ Health 16:673–678. doi: 10.1080/15287398509530777. [DOI] [PubMed] [Google Scholar]
- 17.Raj A, Nachiappan V. 2016. Exposure to benzene metabolites causes oxidative damage in Saccharomyces cerevisiae. Antonie Van Leeuwenhoek 109:841–854. doi: 10.1007/s10482-016-0684-7. [DOI] [PubMed] [Google Scholar]
- 18.Dong H, Bao J. 2010. Metabolism: biofuel via biodetoxification. Nat Chem Biol 6:316–318. doi: 10.1038/nchembio.355. [DOI] [PubMed] [Google Scholar]
- 19.Liu ZL, Ma M, Song M. 2009. Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282:233–244. doi: 10.1007/s00438-009-0461-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu ZL, Moon J. 2009. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 446:1–10. doi: 10.1016/j.gene.2009.06.018. [DOI] [PubMed] [Google Scholar]
- 21.Takenaka S, Koshiya J, Okugawa S, Takata A, Murakami S, Aoki K. 2011. Fe-superoxide dismutase and 2-hydroxy-1,4-benzoquinone reductase preclude the auto-oxidation step in 4-aminophenol metabolism by Burkholderia sp. strain AK-5. Biodegradation 22:1–11. doi: 10.1007/s10532-010-9369-5. [DOI] [PubMed] [Google Scholar]
- 22.Yi X, Gu H, Gao Q, Liu ZL, Bao J. 2015. Transcriptome analysis of Zymomonas mobilis ZM4 reveals mechanisms of tolerance and detoxification of phenolic aldehyde inhibitors from lignocellulose pretreatment. Biotechnol Biofuels 8:153. doi: 10.1186/s13068-015-0333-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seo JS, Chong H, Park HS, Yoon KO, Jung C, Kim JJ, Hong JH, Kim H, Kim JH, Kil JI, Park CJ, Oh HM, Lee JS, Jin SJ, Um HW, Lee HJ, Oh SJ, Kim JY, Kang HL, Lee SY, Lee KJ, Kang HS. 2005. The genome sequence of the ethanologenic bacterium Zymomonas mobilis ZM4. Nat Biotechnol 23:63–68. doi: 10.1038/nbt1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang S, Pappas KM, Hauser LJ, Miriam LL, Chen GL, Hurst GB, Pan C, Kouvelis VN, Typas MA, Pelletier DA, Klingeman DM, Chang YJ, Samatova NF, Brown SD. 2009. Improved genome annotation for Zymomonas mobilis. Nat Biotechnol 27:893–894. doi: 10.1038/nbt1009-893. [DOI] [PubMed] [Google Scholar]
- 25.Dong HW, Bao J, Ryu DDY, Zhong JJ. 2011. Design and construction of improved new vectors for Zymomonas mobilis recombinants. Biotechnol Bioeng 108:1616–1627. doi: 10.1002/bit.23106. [DOI] [PubMed] [Google Scholar]
- 26.Zhang J, Wang X, Chu D, He Y, Bao J. 2011. Dry pretreatment of lignocellulose with extremely low steam and water usage for bioethanol production. Bioresour Technol 102:4480–4488. doi: 10.1016/j.biortech.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 27.Zhang J, Zhu Z, Wang X, Wang N, Wang W, Bao J. 2010. Biodetoxification of toxins generated from lignocellulose pretreatment using a newly isolated fungus, Amorphotheca resinae ZN1, and the consequent ethanol fermentation. Biotechnol Biofuels 3:26. doi: 10.1186/1754-6834-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chu D, Zhang J, Bao J. 2012. Simultaneous saccharification and ethanol fermentation of corn stover at high temperature and high solids loading by a thermotolerant strain Saccharomyces cerevisiae DQ1. Bioenerg Res 5:1020–1026. doi: 10.1007/s12155-012-9219-x. [DOI] [Google Scholar]
- 29.Li H, Shen Y, Wu M, Hou J, Jiao C, Li Z, Liu X, Bao X. 2016. Engineering a wild-type diploid Saccharomyces cerevisiae strain for second-generation bioethanol production. Bioresour Bioprocess 3:51. doi: 10.1186/s40643-016-0126-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M, Eddy C, Deanda K, Finkelstein M, Picataggio S. 1995. Metabolic engineering of a pentose metabolism pathway in ethanologenic Zymomonas mobilis. Science 267:240–243. doi: 10.1126/science.267.5195.240. [DOI] [PubMed] [Google Scholar]
- 31.Yi X, Zhang P, Sun J, Tu Y, Gao Q, Zhang J, Bao J. 2016. Engineering wild-type robust Pediococcus acidilactici strain for high titer l- and d-lactic acid production from corn stover feedstock. J Biotechnol 217:112–121. doi: 10.1016/j.jbiotec.2015.11.014. [DOI] [PubMed] [Google Scholar]
- 32.Simon R, Priefer U, Puhler A. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat Biotechnol 1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 33.Adney B, Baker J. 1996. Measurement of cellulase activities. NREL/TP-510–42628 laboratory analytical procedure (LAP). National Renewable Energy Laboratory, Golden, CO. [Google Scholar]
- 34.Ghose TK. 1987. Measurement of cellulase activities. Pure Appl Chem 59:257–268. doi: 10.1351/pac198759020257. [DOI] [Google Scholar]
- 35.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 36.He Y, Zhang J, Bao J. 2014. Dry dilute acid pretreatment by co-currently feeding of corn stover feedstock and dilute acid solution without impregnation. Bioresour Technol 158:360–364. doi: 10.1016/j.biortech.2014.02.074. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Chu D, Huang J, Yu Z, Dai G, Bao J. 2010. Simultaneous saccharification and ethanol fermentation at high corn stover solids loading in a helical stirring bioreactor. Biotechnol Bioeng 105:718–728. doi: 10.1002/bit.22593. [DOI] [PubMed] [Google Scholar]
- 38.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Dong HW, Fan LQ, Luo Z, Zhong JJ, Ryu DD, Bao J. 2013. Improvement of ethanol productivity and energy efficiency by degradation of inhibitors using recombinant Zymomonas mobilis (pHW20a-fdh). Biotechnol Bioeng 110:2395–2404. doi: 10.1002/bit.24897. [DOI] [PubMed] [Google Scholar]
- 40.Wang X, Gao Q, Bao J. 2017. Enhancement of furan aldehydes conversion in Zymomonas mobilis by elevating dehydrogenase activity and cofactor regeneration. Biotechnol Biofuels 10:24. doi: 10.1186/s13068-017-0714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia J, Liu CG, Zhao XQ, Xiao Y, Xia XX, Bai FW. 2018. Contribution of cellulose synthesis, formation of fibrils and their entanglement to the self-flocculation of Zymomonas mobilis. Biotechnol Bioeng 115:2714–2725. doi: 10.1002/bit.26806. [DOI] [PubMed] [Google Scholar]
- 42.Zhang H, Liu G, Zhang J, Bao J. 2016. Fermentative production of high titer gluconic and xylonic acids from corn stover feedstock by Gluconobacter oxydans and techno-economic analysis. Bioresour Technol 219:123–131. doi: 10.1016/j.biortech.2016.07.068. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.