Abstract
Purpose
To study the impact of advanced paternal age on embryo aneuploidy.
Methods
This is a multicenter international retrospective case series of couples undergoing assisted reproduction via in vitro fertilization using donor eggs to control for maternal factors and preimplantation genetic testing for aneuploidy via next-generation sequencing at Igenomix reproductive testing centers. The main outcome measure was the prevalence of embryo aneuploidy in egg donor cycles. Semen analysis data was retrieved for a small subset of the male patients.
Results
Data from 1202 IVF/ICSI egg donor cycles using ejaculated sperm (total 6934 embryos) evaluated using PGT-A between January 2016 and April 2018 in a global population across all Igenomix centers were included. No significant association was identified between advancing paternal age and the prevalence of embryo aneuploidy overall and when analyzing for each chromosome. There was also no significant association between advancing paternal age and specific aneuploid conditions (monosomy, trisomy, partial deletion/duplication) for all chromosomes in the genome.
Conclusions
This is the largest study of its kind in an international patient population to evaluate the impact of advancing paternal age on embryo aneuploidy. We conclude there is no specific effect of paternal age on the prevalence of embryo aneuploidy in the context of embryo biopsies from egg donor cycles.
Electronic supplementary material
The online version of this article (10.1007/s10815-019-01549-z) contains supplementary material, which is available to authorized users.
Keywords: Advanced paternal age, Advanced maternal age, Aneuploidy, In vitro fertilization, Egg donor
Introduction
In recent years, there has been growing interest in the study of advanced paternal age (APA) and its impact on male fertility potential, reproductive success, and the health of offspring. This is of particular relevance as the age at which men and women choose to start families has become increasingly later than their parents due to a variety of sociocultural factors including increasing life expectancy, changing roles for women, advanced age at time of marriage, and improving access to assisted reproductive technologies (ART). Recent epidemiologic data in the USA published by Khandwala et al. demonstrated that the mean paternal age increased from 27.4 to 30.9 years on a review of more than 168 million live births between 1972 and 2016 [1]. While the impact of advanced maternal age on reproductive success is well understood, only more recently has the consequence of advanced paternal age become more tangible. McPherson et al. published retrospective data in 2018 on 4057 in vitro fertilization (IVF) cycles confirming that there is an additive negative effect of advanced age in both parents on pregnancy and live birth rate [2]. With regard to male reproductive health specifically, advanced age has demonstrated negative consequences for testicular function [3], the male reproductive endocrine axis [4], semen parameters [5–8], and the integrity of the sperm genome and epigenome [9–12]. Associations between APA and morbidity in offspring have been documented since the 1980s, including increased risk of fetal demise [13]; congenital anomalies [14]; single genetic disorders [15–19]; malignancies of the breast, central nervous system, and hematopoietic system [20–22]; and neurocognitive disorders such as autism, bipolar affective disorder, and schizophrenia [23–28].
Single-gene mutations appear to be the mode of disease inheritance most strongly associated with APA with parent-offspring trio studies demonstrating an average de novo mutation rate of + 2 point mutations per year of advancing paternal age and a doubling of paternally inherited mutations every 16.5 years [29]; however, paternally linked aneuploid conditions have also been reported, but the evidence remains controversial. Aneuploidy, a condition of abnormal chromosome number, is a result of non-disjunction during meiosis with trisomy being the most common class of aneuploidy. Studies on human sperm, specifically, have shown that all chromosomes are susceptible to non-disjunction with chromosomes 21, 22, and the sex chromosomes having an increased frequency of aneuploidy [30]. While most embryo aneuploidies are a result of maternally inherited aberrations, with prevalence increasing from 30% in women in their early 30s to nearly 90% for women > 44 years of age [31], there is some data to suggest an age-related paternal contribution. For example, there is a known paternal contribution to trisomy 21 (Down syndrome) in 10% of cases and work by Zaragoza et al. reported an additive effect of APA on the increased prevalence of trisomy 21 when maternal age is also advanced > 35 years [32]. Conversely, others have found no association between APA and the prevalence of trisomy 21 [33]. As most autosomal aneuploidies are non-viable, sex chromosome aneuploidies are more commonly seen in live births. Approximately 55% of sex chromosome aneuploidies are of paternal origin, including 80% of Turner syndrome 45,X0, 6% of 47,XXX, 100% of 47,XYY, and 50% of Klinefelter syndrome 47,XXY [34]. Despite the obvious importance of paternal contribution to sex chromosome aneuploidy, there has been no demonstrated effect of APA on the prevalence of these conditions in offspring [33, 35, 36]. A study of 7549 blastocysts during 2826 cycles with preimplantation genetic testing specifically examined the prevalence of 47,XXY karyotypes and found a significant association with advancing maternal age but not advancing paternal age. The authors concluded that a meiotic missegregation specifically in oogenesis was implicated [37].
To best approach the question of how paternal age affects the prevalence of aneuploidy, it is essential to control for maternal factors. Oocyte donation for use in IVF represents an ideal model for the study of the impact of paternal age as maternal age, and health factors are optimized through the use of donated eggs from young women. This practice is shown to result in high implantation rates, high pregnancy rates, and good obstetric outcomes [38–40]. We hypothesized advancing paternal age would be associated with increased embryo aneuploidy, and the aim of this study was to investigate the impact of advanced paternal age on the prevalence of embryo aneuploidy in IVF cycles using egg donors as a control for maternal factors by means of preimplantation genetic testing for aneuploidy (PGT-A) and next-generation sequencing (NGS).
Materials and methods
This is a retrospective non-randomized study on data obtained from sperm injection (ICSI) cycles with donated oocytes in conjunction with preimplantation genetic testing for aneuploidy (PGT-A) using next-generation sequencing (NGS) across Igenomix testing centers in Spain, USA (Florida, California, and New York), Canada, Brazil, Mexico, India, and the United Arab Emirates between January 2016 and April 2018. This study received an Institutional Review Board waiver from the University of Miami Health System as all patient data was de-identified.
Semen analysis and semen collection, donor oocyte selection and harvest, and intracytoplasmic sperm injection were performed at each respective referring IVF center according to individual protocol. All donated oocytes were from females aged 35 years or younger with proven fertility or otherwise normal physical and gynecologic examinations. Trophectoderm biopsy was performed at days 5/6 blastocyst and a standardized protocol was employed for processing of biopsy specimens and transport to Igenomix testing centers. At the time of biopsy, samples were successively washed in washing buffer and ultimately placed into sterile 200-μl PCR tubes aliquoted with 2.5 μl of buffer prior to transport at room temperature.
The NGS platform used in the current study was IonChef™ and S5 sequencer from ThermoFisher Scientific. Ion Reproseq™ PGS Kit was used to lyse the samples, amplify DNA, and generate DNA fragment libraries. The template and loading of the libraries was carried out automatically by the IonChef™ equipment. We used Ion 520 and Ion 530 chips with capacity for 24 and 96 embryo samples, respectively. Once a run was sequenced, the quality parameters of the sequencing were examined, and if quality measures were met, they were used for the following analyses. Those samples that did not meet minimum quality values were discarded from the study. Once sequencing analysis was performed, raw data was transferred to Torrent Suite Software v5.4 and Ion Reporter Software for translation of raw sequence data into the embryo ploidy output.
Only cycles using ejaculated sperm and egg donors were included in the study, and only centers using PGT-A for all cycles were included. The majority of cycles utilized fresh ejaculated sperm compared to frozen/thawed specimens. Paternal age was stratified into six subgroups [18–60, and 60+ years] based on clinically appropriate categorical thresholds [41, 42]. The Mantel-Maenszel chi-square test for trends was used to assess for significance in trends when comparing increasing paternal age and percent aneuploid embryos. Analysis of variance (ANOVA) was used to assess differences in mean sperm concentration, data for which was available only for a subset of patients. R version 3.4.1 (The R Foundation, Vienna, Austria) was used for statistical analyses. P < 0.05 was considered statistically significant.
Results
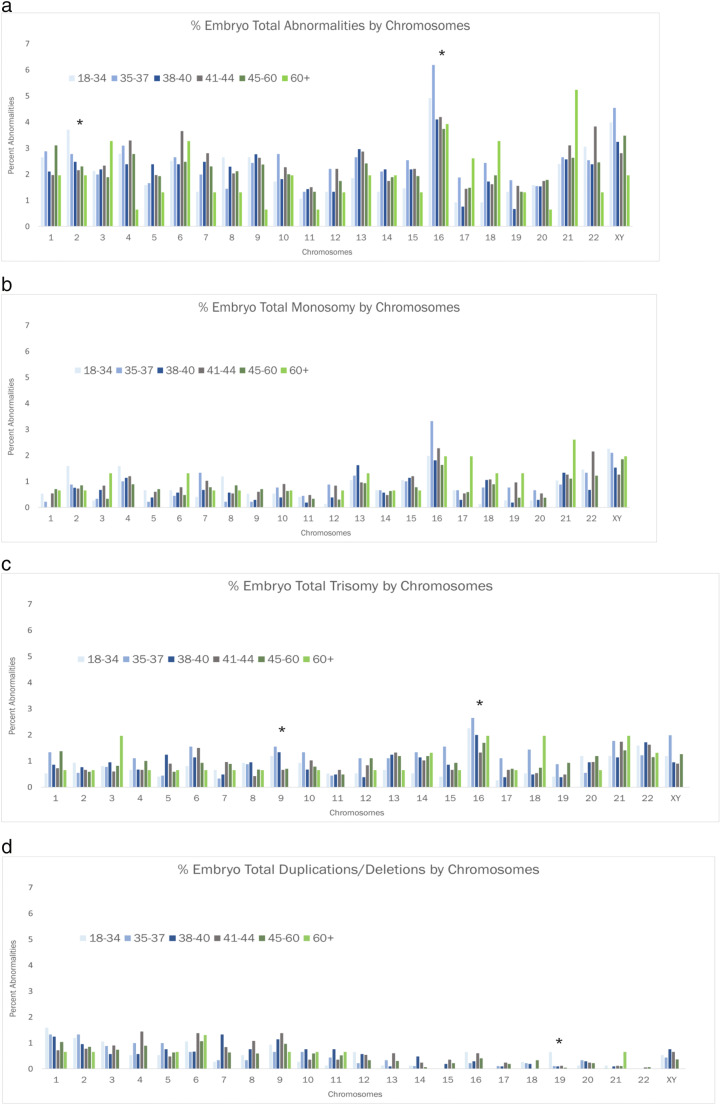
Results for percent aneuploidy are tabulated in Fig. 1 for 6934 embryos obtained from 1202 ICSI cycles using donated oocytes in conjunction with PGT-A. These represent data from over 150 referring IVF clinics. There was no significant trend in the prevalence of aneuploidy across the various paternal age subgroups (p = 0.67). This was the case regardless of paternal age subgroup stratification. We also analyzed the prevalence of overall aneuploidy and specific aneuploidy conditions (monosomy, trisomy, and partial deletion or duplication) by each chromosome across the individual paternal age subgroups (Fig. 2a–d, Supplemental Table 1). Again, we found no significant relationship between advancing paternal age and increased prevalence of aneuploid conditions for specific chromosomes. Notably, the prevalence of aneuploidy for chromosome 21 did appear to be enriched in the paternal age subgroup for men aged 60+ years; however, when considering the specific embryo aneuploidy types, this did not appear to be represented by trisomy 21 (Down syndrome). Additionally, sperm concentration was only available for a subset of male patients (n = 137) representing individuals from each age subgroup. While there was no significant trend in sperm concentration with increasing age (p = 0.81), these data are underpowered to draw a clinically meaningful conclusion regarding age and sperm concentration, and the relationship between sperm concentration and aneuploidy rates in egg donor cycles. Other semen analysis parameters were not available for analysis. While aneuploidy rates could not be tabulated for each of the > 150 referring IVF clinics, there was consistency in aneuploidy rates across the nine global participating Igenomix testing centers (data not shown).
Fig. 1.
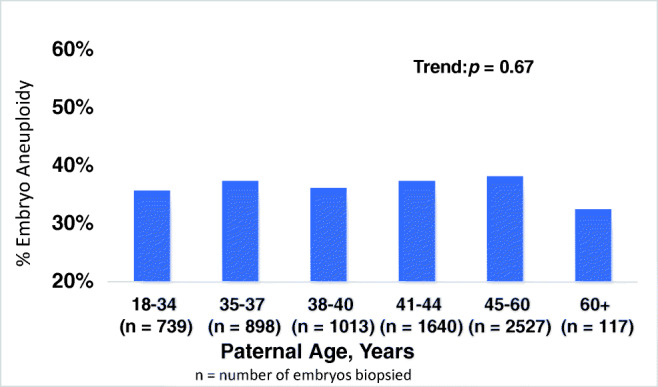
Overall percent embryo aneuploidy stratified by paternal age
Fig. 2.
Prevalence of specific aneuploid condition for each chromosome by paternal age subgroup. a Percent total aneuploidy. b Percent monosomy. c Percent trisomy. d Percent partial deletion or duplication. *Statistically significant trend. For individual p values, see Supplemental Data.
Discussion
In this study, we investigated the impact of APA on the prevalence of embryo aneuploidy in IVF cycles using donor eggs as a means of controlling for maternal factors. Our cohort is the largest and most diverse multi-national patient population studied in this manner to date. The data we obtained demonstrates no effect of advancing paternal age on the prevalence of embryo aneuploidy when controlling for maternal factors across the entire genome, which stands in contrast to early data indicating increased risk for certain aneuploid conditions such as trisomy 21. Our dataset is also the first to utilize the high-fidelity next-generation sequencing in the study of the paternal-age effect on aneuploidy across each individual human chromosome.
The paternal contribution to aneuploid conditions in human offspring has long been established as an observable phenomenon through cytogenetic studies [43, 44], but the specific effect of paternal age remains a subject of controversy. As early as the 1960s, studies have demonstrated at best a modest paternal-age effect [45–47]. Major weaknesses in these studies is the inability to control for maternal age through careful patient selection, small cohort size, observational study design in the era prior to cytogenetic testing, and reliance on limited or subjective methods such as fluorescence in situ hybridization (FISH) [36, 48]. One recent attempt to control for maternal-age effect through an original case-control dataset using maternal age matching to within 6 months found no statistically significant association between Down syndrome and paternal age [49], similar to other findings when proper controls are used [50, 51]. In the last 2 years, investigators have shifted to using egg donor ICSI cycles with preimplantation genetic testing (PGT) and new approaches such as next-generation sequencing (NGS) to mitigate these challenges. In proceedings from the 2017 Scientific Congress of the American Society of Reproductive Medicine, Tapia et al. presented a cohort of 243 egg donor ICSI cycles (2171 embryos) coupled with PGT-A and NGS. This was the first study to employ such methodology to study the effect of paternal age, and they found no paternal-age effect on embryo aneuploidy [41]. A recent study by Capelouto et al. in 2018 similarly examined the impact of various paternal factors including age on IVF outcomes in egg donor cycles. They found no relationship between advancing paternal age (> 35 years) and implantation rate, clinical pregnancy rate, live birth rate, infant birth weight, and preterm deliveries [42]. In contrast, two studies were published recently by one group of investigators using FISH for preimplantation genetic diagnosis in egg donor cycles. Their 2015 study involved a very small cohort and claimed that paternal age > 50 years was associated with a statistically significant increase in overall aneuploidy [52]. A 2018 study by the same group, utilizing the same methods and cohort from a single center, reported a significant increase in the prevalence of trisomy 13, trisomy 18, and trisomy 21 when the paternal age is > 50 years [53]. The conclusion that APA is associated with an increased prevalence of aneuploidy in these two studies is limited by the small nature of these cohorts and the method of genetic diagnosis.
While advancing paternal age has been correlated with declining semen parameters as well as sperm DNA fragmentation index (DFI), neither of these paternal characteristics have been clearly linked to increased aneuploidy in offspring [54, 55]. A recent large-scale observational cohort study involving 1219 consecutive ICSI cycles and a multiplex DNA amplification process for PGT-A examined the aneuploidy rate in relation to various male semen factors including normozoospermia, moderate oligospermia (sperm concentration between 5 × 106 and 15 × 106/mL), severe oligospermia (sperm concentration < 5 × 106/mL), obstructive azoospermia, and non-obstructive azoospermia. Cycles with severe female factor or female genetic conditions were excluded from analysis. There was no significant difference in the percent aneuploidy across the five study groups, and the investigators hypothesized that either these abnormal semen parameters do not contribute to abnormal embryo ploidy or that there are corrective factors in the egg leading to early demise of affected aneuploid embryos prior to the blastocyst stage [54]. Another recent retrospective study investigated the relationship between elevated sperm DNA fragmentation and embryo aneuploidy in 177 IVF-ICSI cycles. While not a controlled study, there were no significant differences in the prevalence of female factor across three study groups: DFI > 30%, DFI 15–30%, and DFI < 15%. The authors identified a significant association between advancing age, and increasing DFI and patients with higher DFI also had significantly worse sperm concentration and motility, and tended to obese. On performance of PGT-A with a high-throughput array-comparative genomic hybridization method for all chromosomes, however, they found no significant difference in embryo aneuploidy rate across the three DFI subgroups [55].
The potential importance of sperm aneuploidy itself and its contribution to embryo aneuploidy as it relates to paternal age is less well studied. It is generally accepted that sex chromosome aneuploidy is more common than autosome aneuploidy, and that sperm disomy for the implicated chromosome is increased in fathers of children with Down syndrome and Klinefelter syndrome when the condition is known to be inherited paternally [56]. A small (n = 10) 2016 study of sperm aneuploidy rates in men < 40 years and men aged > 60 years found no evidence that sperm aneuploidy rates increase with advancing age using a whole-genome dual-probe FISH analysis to increase the accuracy of detection [57]. Conversely, another group found sperm aneuploidy to be elevated above the age of 40 years in a group of 83 men using only five chromosome FISH hybridization probes. They did not assess embryo aneuploidy specifically following ICSI [48]. Certainly, larger studies utilizing higher fidelity methods and with an attention to reproductive outcomes are needed.
With regard to methodology, FISH represents the earliest means by which aneuploidy could be assessed as part of preimplantation genetic diagnosis beginning in the early 1990s [58]. The first applications of FISH involved assessment of chromosome copy number for chromosomes 13, 16, 18, 21, 22, X, and Y, which are most commonly associated with live birth defects and spontaneous abortion, but overall, the number of chromosomes that could be assessed was limited by spectral resolution of filter sets and the number of available fluorochromes (red, yellow, aqua, blue, and green). Disadvantages to FISH include the inability to screen the entire genomic complement of chromosomes (thus an inability to capture 60–80% of all aneuploid embryos) [59], as well as the subjectivity inherent in the method that may lead to increased false-positive and false-negative results. It is thought that these limitations of FISH technology may be the principal reason that randomized controlled trials failed to show a benefit of preimplantation genetic testing for improving live birth rates [60, 61]. Next-generation sequencing (NGS) is a recently introduced alternative to older methods like FISH as a means of testing for embryo aneuploidy and mosaicism. After whole-genomic amplification, embryo-specific sequences are identified in a barcoding step and broken down into smaller fragments which are then massively sequenced in parallel [62]. Aneuploidy is assessed by the number of reads per chromosome which is proportional to the copy number of each chromosome. The method is low-cost and high throughput and allows for simultaneous evaluation of aneuploidy, single-gene mutations, mosaicism, and translocations. NGS has an accuracy for identifying aneuploidy of 98%, but its inherently increased sensitivity may theoretically decrease its specificity, and recent studies indicate potential for high false-positive results approaching 33% [63]. Taking this into consideration, our results demonstrated no effect of advancing paternal age despite the tendency for NGS to potentially overcall the prevalence of aneuploidy.
Limitations of the current study include the inability to capture and standardize individual center methods of semen analysis, donor oocyte selection, and ICSI. Embryology data such as egg number from each donation cycle, number of fertilized eggs per cycle, and number of fertilized eggs reaching blastocyst stage were not transmitted by the > 150 referring IVF clinics; thus, we cannot rule out a potential natural culture selection process acting on aneuploid embryos prior to the blastocyst stage. Regardless, data of that sort is beyond the scope of this study to ascertain as routine biopsies are not performed on embryos that fail to reach blastocyst stage. We also were limited by our inability to obtain data regarding original indications for PTG-A in these couples, assisted reproduction outcomes, or genetic diagnosis for point mutations, sperm aneuploidy, and epigenetic changes that may be associated with advancing paternal age. Additionally, we lack semen analysis data on the majority of patients and thus variation in semen parameters between age groups may be a confounding factor. The strengths of this study involve an unprecedented large and diverse cohort involving multiple international fertility centers and the use of a centralized laboratory with standardized, high-fidelity genome-wide sequencing method to provide accurate diagnosis across each chromosome for a variety of aneuploid states.
Conclusions
In summary, we present the largest reported cohort of egg donor ICSI cycles with preimplantation genetic testing to investigate the effect of APA on embryo aneuploidy in biopsied blastocysts. Although the body of evidence suggests APA is associated with a host of other genetic and epigenetic disease states in offspring, the paternal contribution to abnormal chromosome number does not appear to be a matter of age.
Electronic supplementary material
P values for trends by chromosome and aneuploid condition (PDF 53 kb)
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Khandwala YS, Zhang CA, Lu Y, Eisenberg ML. The age of fathers in the USA is rising: an analysis of 168 867 480 births from 1972 to 2015. Hum Reprod. 2017;32(10):2110–2116. doi: 10.1093/humrep/dex267. [DOI] [PubMed] [Google Scholar]
- 2.McPherson NO, Zander-Fox D, Vincent AD, Lane M. Combined advanced parental age has an additive negative effect on live birth rates-data from 4057 first IVF/ICSI cycles. J Assist Reprod Genet. 2018;35(2):279–287. doi: 10.1007/s10815-017-1054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Handelsman DJ, Staraj S. Testicular size: the effects of aging, malnutrition, and illness. J Androl. 1985;6(3):144–151. doi: 10.1002/j.1939-4640.1985.tb00830.x. [DOI] [PubMed] [Google Scholar]
- 4.Feldman HA, Longcope C, Derby CA, Johannes CB, Araujo AB, Coviello AD, Bremner WJ, McKinlay JB. Age trends in the level of serum testosterone and other hormones in middle-aged men: longitudinal results from the Massachusetts male aging study. J Clin Endocrinol Metab. 2002;87(2):589–598. doi: 10.1210/jcem.87.2.8201. [DOI] [PubMed] [Google Scholar]
- 5.Kidd SA, Eskenazi B, Wyrobek AJ. Effects of male age on semen quality and fertility: a review of the literature. Fertil Steril. 2001;75(2):237–248. doi: 10.1016/s0015-0282(00)01679-4. [DOI] [PubMed] [Google Scholar]
- 6.Eskenazi B, Wyrobek AJ, Sloter E, Kidd SA, Moore L, Young S, et al. The association of age and semen quality in healthy men. Hum Reprod. 2003;18(2):447–454. doi: 10.1093/humrep/deg107. [DOI] [PubMed] [Google Scholar]
- 7.Brahem S, Mehdi M, Elghezal H, Saad A. The effects of male aging on semen quality, sperm DNA fragmentation and chromosomal abnormalities in an infertile population. J Assist Reprod Genet. 2011;28(5):425–432. doi: 10.1007/s10815-011-9537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johnson SL, Dunleavy J, Gemmell NJ, Nakagawa S. Consistent age-dependent declines in human semen quality: a systematic review and meta-analysis. Ageing Res Rev. 2015;19:22–33. doi: 10.1016/j.arr.2014.10.007. [DOI] [PubMed] [Google Scholar]
- 9.Moskovtsev SI, Willis J, Mullen JB. Age-related decline in sperm deoxyribonucleic acid integrity in patients evaluated for male infertility. Fertil Steril. 2006;85(2):496–499. doi: 10.1016/j.fertnstert.2005.05.075. [DOI] [PubMed] [Google Scholar]
- 10.Broer L, Codd V, Nyholt DR, Deelen J, Mangino M, Willemsen G, Albrecht E, Amin N, Beekman M, de Geus EJC, Henders A, Nelson CP, Steves CJ, Wright MJ, de Craen AJM, Isaacs A, Matthews M, Moayyeri A, Montgomery GW, Oostra BA, Vink JM, Spector TD, Slagboom PE, Martin NG, Samani NJ, van Duijn CM, Boomsma DI. Meta-analysis of telomere length in 19,713 subjects reveals high heritability, stronger maternal inheritance and a paternal age effect. Eur J Hum Genet. 2013;21(10):1163–1168. doi: 10.1038/ejhg.2012.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichman NE, Teitler JO. Paternal age as a risk factor for low birthweight. Am J Public Health. 2006;96(5):862–866. doi: 10.2105/AJPH.2005.066324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Curley JP, Mashoodh R, Champagne FA. Epigenetics and the origins of paternal effects. Horm Behav. 2011;59(3):306–314. doi: 10.1016/j.yhbeh.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alio AP, Salihu HM, McIntosh C, August EM, Weldeselasse H, Sanchez E, Mbah AK. The effect of paternal age on fetal birth outcomes. Am J Mens Health. 2012;6(5):427–435. doi: 10.1177/1557988312440718. [DOI] [PubMed] [Google Scholar]
- 14.Lian ZH, Zack MM, Erickson JD. Paternal age and the occurrence of birth defects. Am J Hum Genet. 1986;39(5):648–660. [PMC free article] [PubMed] [Google Scholar]
- 15.Orioli IM, Castilla EE, Scarano G, Mastroiacovo P. Effect of paternal age in achondroplasia, thanatophoric dysplasia, and osteogenesis imperfecta. Am J Med Genet. 1995;59(2):209–217. doi: 10.1002/ajmg.1320590218. [DOI] [PubMed] [Google Scholar]
- 16.Jones KL, Smith DW, Harvey MA, Hall BD, Quan L. Older paternal age and fresh gene mutation: data on additional disorders. J Pediatr. 1975;86(1):84–88. doi: 10.1016/s0022-3476(75)80709-8. [DOI] [PubMed] [Google Scholar]
- 17.Wynn J, King TM, Gambello MJ, Waller DK, Hecht JT. Mortality in achondroplasia study: a 42-year follow-up. Am J Med Genet A. 2007;143A(21):2502–2511. doi: 10.1002/ajmg.a.31919. [DOI] [PubMed] [Google Scholar]
- 18.Wilkie AO, Slaney SF, Oldridge M, Poole MD, Ashworth GJ, Hockley AD, et al. Apert syndrome results from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9(2):165–172. doi: 10.1038/ng0295-165. [DOI] [PubMed] [Google Scholar]
- 19.Toriello HV, Meck JM, PPaG C. Statement on guidance for genetic counseling in advanced paternal age. Genet Med. 2008;10(6):457–460. doi: 10.1097/GIM.0b013e318176fabb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemminki K, Kyyrönen P. Parental age and risk of sporadic and familial cancer in offspring: implications for germ cell mutagenesis. Epidemiology. 1999;10(6):747–751. [PubMed] [Google Scholar]
- 21.Yip BH, Pawitan Y, Czene K. Parental age and risk of childhood cancers: a population-based cohort study from Sweden. Int J Epidemiol. 2006;35(6):1495–1503. doi: 10.1093/ije/dyl177. [DOI] [PubMed] [Google Scholar]
- 22.Murray L, McCarron P, Bailie K, Middleton R, Davey Smith G, Dempsey S, McCarthy A, Gavin A. Association of early life factors and acute lymphoblastic leukaemia in childhood: historical cohort study. Br J Cancer. 2002;86(3):356–361. doi: 10.1038/sj.bjc.6600012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.D'Onofrio BM, Rickert ME, Frans E, Kuja-Halkola R, Almqvist C, Sjölander A, et al. Paternal age at childbearing and offspring psychiatric and academic morbidity. JAMA Psychiatry. 2014;71(4):432–438. doi: 10.1001/jamapsychiatry.2013.4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichenberg A, Gross R, Weiser M, Bresnahan M, Silverman J, Harlap S, Rabinowitz J, Shulman C, Malaspina D, Lubin G, Knobler HY, Davidson M, Susser E. Advancing paternal age and autism. Arch Gen Psychiatry. 2006;63(9):1026–1032. doi: 10.1001/archpsyc.63.9.1026. [DOI] [PubMed] [Google Scholar]
- 25.Buizer-Voskamp JE, Laan W, Staal WG, Hennekam EA, Aukes MF, Termorshuizen F, et al. Paternal age and psychiatric disorders: findings from a Dutch population registry. Schizophr Res. 2011;129(2–3):128–132. doi: 10.1016/j.schres.2011.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hare EH, Moran PA. Raised parental age in psychiatric patients: evidence for the constitutional hypothesis. Br J Psychiatry. 1979;134:169–177. doi: 10.1192/bjp.134.2.169. [DOI] [PubMed] [Google Scholar]
- 27.Malaspina D, Harlap S, Fennig S, Heiman D, Nahon D, Feldman D, Susser ES. Advancing paternal age and the risk of schizophrenia. Arch Gen Psychiatry. 2001;58(4):361–367. doi: 10.1001/archpsyc.58.4.361. [DOI] [PubMed] [Google Scholar]
- 28.Frans EM, Sandin S, Reichenberg A, Lichtenstein P, Långström N, Hultman CM. Advancing paternal age and bipolar disorder. Arch Gen Psychiatry. 2008;65(9):1034–1040. doi: 10.1001/archpsyc.65.9.1034. [DOI] [PubMed] [Google Scholar]
- 29.Kong A, Frigge ML, Masson G, Besenbacher S, Sulem P, Magnusson G, Gudjonsson SA, Sigurdsson A, Jonasdottir A, Jonasdottir A, Wong WSW, Sigurdsson G, Walters GB, Steinberg S, Helgason H, Thorleifsson G, Gudbjartsson DF, Helgason A, Magnusson OT, Thorsteinsdottir U, Stefansson K. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488(7412):471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin RH. Meiotic chromosome abnormalities in human spermatogenesis. Reprod Toxicol. 2006;22(2):142–147. doi: 10.1016/j.reprotox.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 31.Franasiak JM, Forman EJ, Hong KH, Werner MD, Upham KM, Treff NR, Scott RT., Jr The nature of aneuploidy with increasing age of the female partner: a review of 15,169 consecutive trophectoderm biopsies evaluated with comprehensive chromosomal screening. Fertil Steril. 2014;101(3):656–63.e1. doi: 10.1016/j.fertnstert.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 32.Zaragoza MV, Jacobs PA, James RS, Rogan P, Sherman S, Hassold T. Nondisjunction of human acrocentric chromosomes: studies of 432 trisomic fetuses and liveborns. Hum Genet. 1994;94(4):411–417. doi: 10.1007/BF00201603. [DOI] [PubMed] [Google Scholar]
- 33.Sloter E, Nath J, Eskenazi B, Wyrobek AJ. Effects of male age on the frequencies of germinal and heritable chromosomal abnormalities in humans and rodents. Fertil Steril. 2004;81(4):925–943. doi: 10.1016/j.fertnstert.2003.07.043. [DOI] [PubMed] [Google Scholar]
- 34.Tempest HG. Meiotic recombination errors, the origin of sperm aneuploidy and clinical recommendations. Syst Biol Reprod Med. 2011;57(1–2):93–101. doi: 10.3109/19396368.2010.504879. [DOI] [PubMed] [Google Scholar]
- 35.Lanfranco F, Kamischke A, Zitzmann M, Nieschlag E. Klinefelter’s syndrome. Lancet. 2004;364(9430):273–283. doi: 10.1016/S0140-6736(04)16678-6. [DOI] [PubMed] [Google Scholar]
- 36.Fonseka KG, Griffin DK. Is there a paternal age effect for aneuploidy? Cytogenet Genome Res. 2011;133(2–4):280–291. doi: 10.1159/000322816. [DOI] [PubMed] [Google Scholar]
- 37.Mazzilli R, Cimadomo D, Rienzi L, Capalbo A, Levi Setti PE, Livi C, Vizziello D, Foresta C, Ferlin A, Ubaldi FM. Prevalence of XXY karyotypes in human blastocysts: multicentre data from 7549 trophectoderm biopsies obtained during preimplantation genetic testing cycles in IVF. Hum Reprod. 2018;33(7):1355–1363. doi: 10.1093/humrep/dey110. [DOI] [PubMed] [Google Scholar]
- 38.Wong IL, Legro RS, Lindheim SR, Paulson RJ, Sauer MV. Efficacy of oocytes donated by older women in an oocyte donation programme. Hum Reprod. 1996;11(4):820–823. doi: 10.1093/oxfordjournals.humrep.a019260. [DOI] [PubMed] [Google Scholar]
- 39.Sauer MV, Kavic SM. Oocyte and embryo donation 2006: reviewing two decades of innovation and controversy. Reprod BioMed Online. 2006;12(2):153–162. doi: 10.1016/s1472-6483(10)60855-3. [DOI] [PubMed] [Google Scholar]
- 40.Budak E, Garrido N, Soares SR, Melo MA, Meseguer M, Pellicer A, et al. Improvements achieved in an oocyte donation program over a 10-year period: sequential increase in implantation and pregnancy rates and decrease in high-order multiple pregnancies. Fertil Steril. 2007;88(2):342–349. doi: 10.1016/j.fertnstert.2006.11.118. [DOI] [PubMed] [Google Scholar]
- 41.Tapia LG, Rubino P, Ruiz de Assin R, Thiel A, Li X, Kolb B, et al. Advanced paternal age does not affect embryo aneuploidy rate in egg donor cycles [ASRM abstract P-49] Fertil Steril. 2017;108(3, Supp):e126. [Google Scholar]
- 42.Capelouto SM, Nagy ZP, Shapiro DB, Archer SR, Ellis DP, Smith AK, Spencer JB, Hipp HS. Impact of male partner characteristics and semen parameters on in vitro fertilization and obstetric outcomes in a frozen oocyte donor model. Fertil Steril. 2018;110(5):859–869. doi: 10.1016/j.fertnstert.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 43.Wagenbichler P, Killian W, Rett A, Schnedl W. Origin of the extra chromosome no. 21 in Down's syndrome. Hum Genet. 1976;32(1):13–16. doi: 10.1007/BF00569971. [DOI] [PubMed] [Google Scholar]
- 44.Griffin DK. The incidence, origin, and etiology of aneuploidy. Int Rev Cytol. 1996;167:263–296. doi: 10.1016/s0074-7696(08)61349-2. [DOI] [PubMed] [Google Scholar]
- 45.Mantel N, Stark CR. Paternal age in Down’s syndrome. Am J Ment Defic. 1967;71(6):1025–1027. [PubMed] [Google Scholar]
- 46.Erickson JD. Paternal age and Down syndrome. Am J Hum Genet. 1979;31(4):489–497. [PMC free article] [PubMed] [Google Scholar]
- 47.Lowe X, Eskenazi B, Nelson DO, Kidd S, Alme A, Wyrobek AJ. Frequency of XY sperm increases with age in fathers of boys with Klinefelter syndrome. Am J Hum Genet. 2001;69(5):1046–1054. doi: 10.1086/323763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaarouch I, Bouamoud N, Madkour A, Louanjli N, Saadani B, Assou S, Aboulmaouahib S, Amzazi S, Copin H, Benkhalifa M, Sefrioui O. Paternal age: negative impact on sperm genome decays and IVF outcomes after 40 years. Mol Reprod Dev. 2018;85(3):271–280. doi: 10.1002/mrd.22963. [DOI] [PubMed] [Google Scholar]
- 49.De Souza E, Alberman E, Morris JK. Down syndrome and paternal age, a new analysis of case-control data collected in the 1960s. Am J Med Genet A. 2009;149A(6):1205–1208. doi: 10.1002/ajmg.a.32850. [DOI] [PubMed] [Google Scholar]
- 50.Hatch M, Kline J, Levin B, Hutzler M, Warburton D. Paternal age and trisomy among spontaneous abortions. Hum Genet. 1990;85(3):355–361. doi: 10.1007/BF00206761. [DOI] [PubMed] [Google Scholar]
- 51.Hook EB, Regal RR. A search for a paternal-age effect upon cases of 47, +21 in which the extra chromosome is of paternal origin. Am J Hum Genet. 1984;36(2):413–421. [PMC free article] [PubMed] [Google Scholar]
- 52.García-Ferreyra J, Luna D, Villegas L, Romero R, Zavala P, Hilario R, et al. High aneuploidy rates observed in embryos derived from donated oocytes are related to male aging and high percentages of sperm DNA fragmentation. Clin Med Insights Reprod Health. 2015;9:21–27. doi: 10.4137/CMRH.S32769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.García-Ferreyra J, Hilario R, Dueñas J. High percentages of embryos with 21, 18 or 13 trisomy are related to advanced paternal age in donor egg cycles. JBRA Assist Reprod. 2018;22(1):26–34. doi: 10.5935/1518-0557.20180004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mazzilli R, Cimadomo D, Vaiarelli A, Capalbo A, Dovere L, Alviggi E, Dusi L, Foresta C, Lombardo F, Lenzi A, Tournaye H, Alviggi C, Rienzi L, Ubaldi FM. Effect of the male factor on the clinical outcome of intracytoplasmic sperm injection combined with preimplantation aneuploidy testing: observational longitudinal cohort study of 1,219 consecutive cycles. Fertil Steril. 2017;108(6):961–72.e3. doi: 10.1016/j.fertnstert.2017.08.033. [DOI] [PubMed] [Google Scholar]
- 55.Gat I, Tang K, Quach K, Kuznyetsov V, Antes R, Filice M, Zohni K, Librach C. Sperm DNA fragmentation index does not correlate with blastocyst aneuploidy or morphological grading. PLoS One. 2017;12(6):e0179002. doi: 10.1371/journal.pone.0179002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Templado C, Vidal F, Estop A. Aneuploidy in human spermatozoa. Cytogenet Genome Res. 2011;133(2–4):91–99. doi: 10.1159/000323795. [DOI] [PubMed] [Google Scholar]
- 57.Donate A, Estop AM, Giraldo J, Templado C. Paternal age and numerical chromosome abnormalities in human spermatozoa. Cytogenet Genome Res. 2016;148(4):241–248. doi: 10.1159/000446724. [DOI] [PubMed] [Google Scholar]
- 58.Munné S, Lee A, Rosenwaks Z, Grifo J, Cohen J. Diagnosis of major chromosome aneuploidies in human preimplantation embryos. Hum Reprod. 1993;8(12):2185–2191. doi: 10.1093/oxfordjournals.humrep.a138001. [DOI] [PubMed] [Google Scholar]
- 59.Munné S, Fragouli E, Colls P, Katz-Jaffe M, Schoolcraft W, Wells D. Improved detection of aneuploid blastocysts using a new 12-chromosome FISH test. Reprod BioMed Online. 2010;20(1):92–97. doi: 10.1016/j.rbmo.2009.10.015. [DOI] [PubMed] [Google Scholar]
- 60.Mastenbroek S, Twisk M, van Echten-Arends J, Sikkema-Raddatz B, Korevaar JC, Verhoeve HR, Vogel NEA, Arts EGJM, de Vries JWA, Bossuyt PM, Buys CHCM, Heineman MJ, Repping S, van der Veen F. In vitro fertilization with preimplantation genetic screening. N Engl J Med. 2007;357(1):9–17. doi: 10.1056/NEJMoa067744. [DOI] [PubMed] [Google Scholar]
- 61.Twisk M, Mastenbroek S, Hoek A, Heineman MJ, van der Veen F, Bossuyt PM, Repping S, Korevaar JC. No beneficial effect of preimplantation genetic screening in women of advanced maternal age with a high risk for embryonic aneuploidy. Hum Reprod. 2008;23(12):2813–2817. doi: 10.1093/humrep/den231. [DOI] [PubMed] [Google Scholar]
- 62.Knapp M, Stiller M, Meyer M. Generating barcoded libraries for multiplex high-throughput sequencing. Methods Mol Biol. 2012;840:155–170. doi: 10.1007/978-1-61779-516-9_19. [DOI] [PubMed] [Google Scholar]
- 63.Goodrich D, Tao X, Bohrer C, Lonczak A, Xing T, Zimmerman R, Zhan Y, Scott Jr RT, Treff NR. A randomized and blinded comparison of qPCR and NGS-based detection of aneuploidy in a cell line mixture model of blastocyst biopsy mosaicism. J Assist Reprod Genet. 2016;33(11):1473–1480. doi: 10.1007/s10815-016-0784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
P values for trends by chromosome and aneuploid condition (PDF 53 kb)