The composition of post-translational modifications attached to the crystallizable fragment (Fc) of IgG antibodies are widely known to be associated with disease and inflammation, however, much less is known about the composition of antibodies bound to circulating immune cells or the composition of the antibody-binding receptors themselves. This report characterizes the composition of asparagine-linked glycans from the predominant antibody-binding receptor expressed by natural killer cells, Fc γ receptor 3a/CD16a and it's bound IgG1 ligand.
Keywords: glycoproteomics, immunology, glycoprotein secretion, post-translational modifications, membranes, CD16, Fc gamma receptor, immunoglobulin G
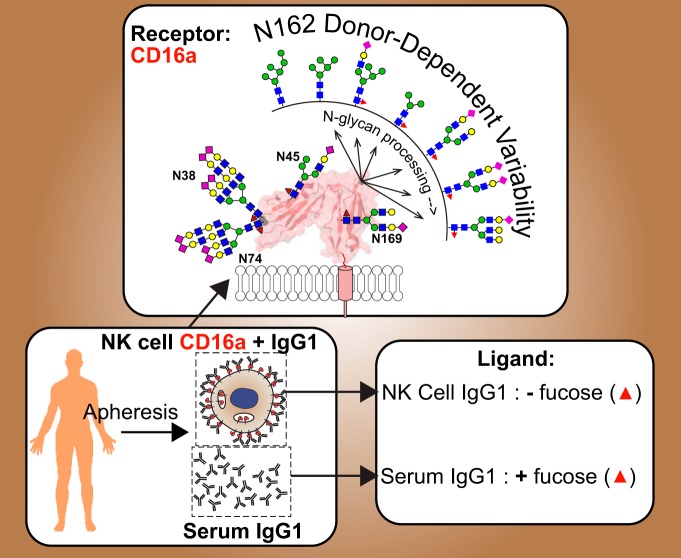
Graphical Abstract
Highlights
Enough CD16a from NK cells can be isolated from a single donor following apheresis.
NK cell CD16a shows variability between different donors.
NK cells preferentially bind IgG1 that lacks core fucosylation.
Abstract
Post-translational modification confers diverse functional properties to immune system proteins. The composition of serum proteins such as immunoglobulin G (IgG) strongly associates with disease including forms lacking a fucose modification of the crystallizable fragment (Fc) asparagine(N)-linked glycan that show increased effector function, however, virtually nothing is known about the composition of cell surface receptors or their bound ligands in situ because of low abundance in the circulating blood. We isolated primary NK cells from apheresis filters following plasma or platelet donation to characterize the compositional variability of Fc γ receptor IIIa/CD16a and its bound ligand, IgG1. CD16a N162-glycans showed the largest differences between donors; one donor displayed only oligomannose-type N-glycans at N162 that correlate with high affinity IgG1 Fc binding whereas the other donors displayed a high degree of compositional variability at this site. Hybrid-type N-glycans with intermediate processing dominated at N45 and highly modified, complex-type N-glycans decorated N38 and N74 from all donors. Analysis of the IgG1 ligand bound to NK cell CD16a revealed a sharp decrease in antibody fucosylation (43.2 ± 11.0%) versus serum from the same donors (89.7 ± 3.9%). Thus, NK cells express CD16a with unique modification patterns and preferentially bind IgG1 without the Fc fucose modification at the cell surface.
Molecules at the cell surface mediate intercellular interactions and reveal characteristics fundamental to the lineage and function of that cell. These antigens are often also critical to cell function. For example, Fc γ receptor IIIa (FcγRIIIa/CD16a) is a protein expressed on natural killer (NK)1 cells that binds immunoglobulin G (IgG), triggering a protective immune response (Fig. 1A). The levels of CD16a and CD56 reflect the developmental stage with mature NK cells exhibiting a CD56DIM/CD16+ phenotype (1). Similar approaches to define and isolate cell types overlook one known source of variability that contributes to function: post-translational modification. Every individual protein at the cell surface is potentially unique because of the compositional variability of post-translational modifications (PTMs) at multiple sites that are not directly encoded in the genome (2). Further, composition of the PTMs impacts protein function and possibly immune cell response including CD16a and CD56 on NK cells, CD64 (FcγRI) as well the T cell receptor on T cells among others not covered here (3–7). Aberrant PTMs associate with many diseases but the native composition of only a few endogenous proteins is defined (including, but not limited to ref (8–11)). Thus, a complete description must include a definition of PTMs to understand the potential plasticity of any cell response.
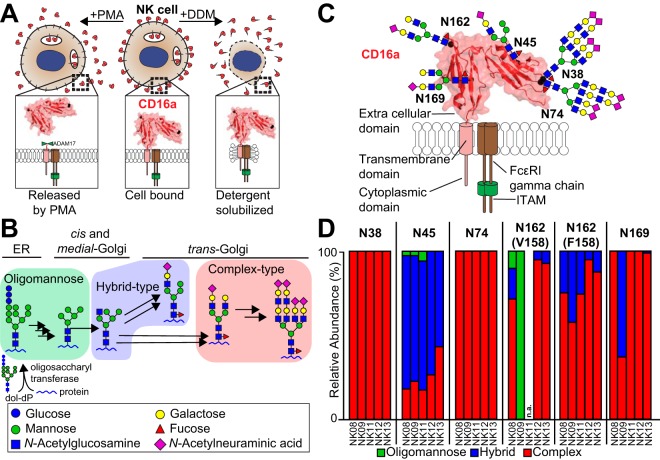
Fig. 1.
CD16a is the primary Fc γ receptor expressed on NK cells and is heavily modified with N-glycans. A, CD16a was isolated by either NK cell stimulation with PMA to release CD16a or by lysing the cell with detergent (DDM). B, An abbreviated representation of the N-glycan remodeling pathway in mammals is shown that generates a diverse repertoire of modifications. C, A cartoon of CD16a topology shows the extracellular, transmembrane and cytoplasmic domains as well as the most abundant N-glycans at the five sites, scaled to the size of the protein. D, The relative abundance of the three N-glycan types identified at each CD16a site from each of five donors (donor identifier is listed below the bar and N-glycosylation site above). Peptides containing N162 glycosylation also reveal donor allotype (V158 v. F158) and are reported separately. n.a., not applicable. See also supplemental Tables S1–S2 and supplemental Figs. S1 and S2.
PTM composition cannot be predicted from a DNA sequence. The predominant mammalian PTM, asparagine-linked (N-)glycosylation, is heavily altered following the initial protein modifying reaction by any number of hundreds of glycosyltransferases and glycosylhydrolases in the ER and Golgi, generating a large distribution that may include any of the more than 3,400 known human N-glycoforms at any single site (Fig. 1B) (12). This heterogeneity presents an obstacle to defining protein composition along with the tiny amount of any individual receptor in the human body. Further, many integral membrane proteins require detergents during purification, reducing the sensitivity of mass spectrometry (MS)-based approaches (13). Unfortunately, recombinant expression appears to provide no salvation as the expressing cell dramatically impacts processing (6).
Little is known about CD16a processing, though composition impacted the affinity for IgG1 Fc in vitro (6, 7, 14). Presenting minimally remodeled N-glycans at N162 increased affinity by up to 50 fold (14, 15). N45 glycosylation also contributes to affinity by stabilizing CD16a structure and roles for the three remaining N-glycans remain undefined (15). Though it is not clear how CD16a PTMs impact immune function, a wealth of evidence indicates that the 2–4-fold affinity increase because of a common polymorphism (V158) improved clinical outcome following treatment with therapeutic monoclonal antibodies (mAbs) when compared with patients expressing F158 (16–21). Thus, there is a possibility that CD16a PTM composition impacts antibody binding affinity in vivo.
IgG1 from serum is likely the predominant ligand for CD16a under normal conditions. IgG3 exhibits higher affinity but is unlikely the major ligand because the concentration in serum is much less than IgG1 (17, 22–24). IgG, like CD16a, is modified. N-glycosylation of the IgG crystallizable fragment (Fc) is required for receptor binding and changes in Fc N-glycan composition are associated with disease (including, but not limited to ref (8, 25–29).). Of the potential modifications to the Fc N-glycan, fucosylation of the core N-acetylglucosamine (GlcNAc) residue occurs at high levels in human serum (∼85–95%) though antibodies lacking this modification bind CD16a with higher affinity and elicit antibody-dependent cell-mediated cytotoxicity at lower antibody concentrations (30, 31). IgG1 is abundant in serum (∼10 mg/ml) and though changes in serum IgG and antigen-specific IgG composition are known (29, 32, 33), the distribution of IgG glycoforms that are bound to effector cells remain undefined.
We adopted a targeted approach to define the composition of CD16a and its bound IgG ligand from NK cells. NK cell CD16a is the primary receptor for mAb therapies though CD16a is also expressed by “non-classical” monocytes and macrophages (34). A preliminary study showed the unexpected presence of minimally remodeled N-glycans from NK cell CD16a, though it was unclear if these forms occupied N162 (6). We obtained mononuclear leukocytes from individual donors and isolated NK cells that express high levels of CD16a. NK cells constitute only a minor fraction of the total peripheral blood cell count (1–6% of leukocytes (35)). We then purified CD16a and IgG1 to analyze the composition of PTMs at each modified site to demonstrate that apheresis is a viable source of cell type specific cell surface receptors and their bound ligands.
EXPERIMENTAL PROCEDURES
Experimental Design
The healthy human donors were de-identified and not directly enrolled for the study but rather randomly selected based on scheduled donations at the blood centers. Additionally, one donation provides enough material for only one analysis thus, it was not possible to replicate the analysis with material from the same donor. The consistent rate of migration of CD16a in SDS-PAGE gel from over 20 donors in our laboratory and in other studies suggested relatively homogenous distribution of PTMs on different donors thus, biological replicates are represented by five donors.
Cell Isolation
Apheresis filters were obtained from the DeGowin Blood Center (Iowa City, IA) and LifeServe Blood Center (Ames, IA). Donors were provided a health screen including measurements of body temperature, blood pressure, pulse and blood count. A review exemption from the Institutional Review Board at Iowa State University was obtained prior to sample collection. An aliquot of cells (50 μl) was centrifuged at 3000 × g for 10 min, serum and cells were collected and stored separately at −80 °C. NK cells were isolated and validated as described (6).
NK Cell Activation
NK cells were washed with PBS, then incubated at a density of 5 × 106 cells/ml in RPMI 1640 medium (Thermo, Waltham, MA) with 0.1% FBS (pretreated to remove IgG) and 1 μg/ml phorbol 12-myristate 13-acetate (PMA; Millipore Sigma, Burlington, MA) for 60 min at 37 °C. Medium containing CD16a was centrifuged at 1000 × g for 10 min and stored at −80 °C.
CD16a Immunoprecipitation
NK cells were lysed with 100 mm Tris, 100 mm sodium chloride, 5 mm EDTA, 5 mm oxidized glutathione, 10 μm potassium ferricyanide, 1 mm 4-(2-aminoethyl)benzenesulfonyl fluoride, 10 mg/ml dodecyl maltoside (DDM), pH 8.0, 4 °C; 250 μl per 1 × 107 cells. The lysate was clarified by centrifugation at 10,000 × g for 10 min at 4 °C and the supernatant frozen at −80 °C. Protein G Sepharose resin (GE Healthcare, Chicago, IL) was added to thawed lysate (5 μl per 200 μl lysate) for 60 min at 4 °C with mixing and then removed by centrifugation at 500 × g for 5 min. The supernatant was sonicated in bath sonicator for 1 min. Anti-hCD16 (3G8)-coupled resin was prepared as described (6) but as an aglycosylated variant (N297Q) and coupled at 2.3–2.6 mg/ml, then incubated with lysate (10 μl per 200 μl lysate) overnight at 4 °C with mixing. Resin was washed 2 × 600 μl of lysis buffer by centrifugation (500 × g for 5 min) to pellet the resin. This was followed by 2 × 1 volume washes with 50 mm Tris, 100 mm sodium chloride, pH 8.0 and 2 × 1 volume with 50 mm ammonium carbonate, pH 8.0. CD16a was eluted with 200 μl of 54.9% acetonitrile (ACN), 45% water and 0.1% trifluoroacetic acid (TFA). Each elution fraction was neutralized by with 26 μl 1 m ammonium carbonate. CD16a was purified from the medium following PMA treatment in a similar manner. Except the medium was sequentially passed through columns containing 50–100 μl protein G Sepharose followed by 100 μl 3G8 agarose resin. CD16a yields were assessed by Western blotting as described (6).
IgG Immunoprecipitation from Serum and NK Cell Lysate
Serum IgG was isolated by diluting 2 μl serum into 500 μl 100 mm Tris, 100 mm sodium chloride, 10 mg/ml dodecyl maltoside, pH 8.0 and incubating with 50 μl Protein G Sepharose resin for 60 min at 4 °C with mixing. NK cell IgG was obtained from Protein G Sepharose resin incubated with the NK cell lysate as described above. Resin was washed as described above but IgG was eluted with 5 × 300 μl 100 mm Formic Acid, pH 2.5 and immediately neutralized with 150 μl 1 m ammonium carbonate, pH 8.0 (27). IgG yields were determined with western blots using 5% of the eluted fraction and a rabbit anti-hIgG antibody (Thermo, 0.24 μg/ml) with goat anti-rabbit IgG HRP antibody (Research and Diagnostic Systems, Minneapolis, MN, 1:1000 dilution).
CD16a and IgG Proteolysis
The CD16a ACN:water:TFA elutions were combined, lyophilized and resuspended in 120 μl 50 mm ammonium carbonate, 10% (v/v) methanol, pH 8.0. The samples were boiled at 95 °C for 5 min and cooled on ice. Sequencing grade chymotrypsin (Millipore Sigma) and GluC (Promega, Madison, WI) were reconstituted according to the manufacturer's guidelines and added to the CD16a sample at an enzyme:CD16a molar ratio of 1:30 and 1:20, respectively, and incubated at 25 °C for 18 h. Then, 75 ng of each enzyme was added followed by incubation at 37 °C for 4 h. Chymotrypsin cleaves C-terminal to phenylalanine, tyrosine, and tryptophan and less efficiently at leucine. Glu-C cleaves C-terminal to glutamic acid.
Elution fractions from IgG isolation were similarly combined, lyophilized, resuspended in 120 μl 50 mm ammonium carbonate, pH 8.0 and boiled. Sequencing grade Trypsin (Promega) was reconstituted and added at an enzyme:IgG molar ratio of 1:20 before incubation at 37 °C for 18 h. Trypsin cleaves C-terminal to the lysine and arginine residues. Next, dithiothreitol was added (5 mm final) to CD16a and IgG samples and incubated at 37 °C for 1 h, followed by iodoacetamide (15 mm final) and incubation at 25 °C for 1 h in the dark. The samples were centrifuged at 10,000 × g for 3 min and the supernatant was lyophilized.
Glycopeptide Enrichment and Detergent Removal
Samples were resuspended in 10 μl water and purified using a glycopeptide enrichment kit (Millipore Sigma) except that the elution was repeated three more times than suggested. The eluted glycopeptide fractions were combined and lyophilized. The supernatant from CD16a samples containing mostly unmodified peptides was washed with saturated ethyl acetate in water and lyophilized (36).
Mass Spectrometry
Samples were resuspended in 5 μl of 5% ACN, 0.1% TFA acid in water then separated on a 75 μm × 20 cm fused silica capillary (Agilent Technologies, Santa Clara, CA) column/emitter attached to an EASY nLC-1200 LC system with a Nanospray FlexIon source (Thermo). The tip of this column was fabricated using a laser puller (Sutter P-200, Novato, CA) and was packed with 5 μm Agilent Zorbax C18 medium using a pressure cell (Next Advance, Troy, NY). The remainder of the column was packed with either the same material or with 3 μm UChrom C18 medium (nanoLCMS Solutions, Oroville, CA). Samples were eluted with a 0% to 35% gradient of Buffer B (80% ACN and 0.1% formic acid in water) in Buffer A (0.1% formic acid in water) at 300 nl/min over 60 min. Buffer B was then increased to 70% over 10 min, increased to 100% for 10 min and held for 3 min. 20 μl of 100% Buffer A (0.1% formic acid in water) was then passed through the column at 400 bar to equilibrate the column. Each sample run was followed by a blank run with 5 μl of Buffer A injected instead of sample. The LC eluent was analyzed on a Q Exactive Hybrid Quadrupole-Orbitrap Mass Spectrometer with an HCD fragmentation cell (Thermo). MS spectra were collected in positive ion mode with a scan range of 600–2500 m/z (for glycopeptides) or 266–2500 m/z (for peptides). Data-dependent acquisition was obtained using an 80 ms MS1 window and up to 20 of the most abundant ions were selected for HCD fragmentation at a stepped normalized collision energy (NCE) of 17, 27, and 37 eV (for glycopeptides) or 27 eV only (for peptides). Ions visible at 600 m/z in the MS2 spectra are carryover from a calibration standard.
Data Analysis
The CD16a glycopeptides were initially identified using Byonic (Protein Metrics, Cupertino, CA, v. 2.12.0; search details given below) that identified two of the five N-glycopeptides (N45 and N162). The remaining glycopeptides were identified through manual evaluation of the spectra by matching expected and observed parent ion mass, presence of characteristic oxonium ion species and the presence of a fragment corresponding to peptide with one HexNAc in MS2 spectra. Once retention times for each glycopeptide were identified, we generated a comprehensive mass list for all possible CD16a glycopeptides based on species identified by manual assignment in one NK cell data set (NK13) and N-glycans identified in a previous NK cell CD16a glycomics study as well as any potential N-glycan variations (6). These mass lists were used to probe raw MS1 data to identify other species not originally found in the manual and Byonic searches. MS2 spectra were available for most species and annotated using Glycoworkbench (see the supplemental MS2 annotations) (37). For the minority of species that did not give rise to high quality MS2 spectra, identifications were made based on masses observed in the MS1 spectra (with errors < 0.05 Da), comparable retention times to species with the same peptide backbone, and predictable differences to identified peaks in the same data set (e.g. with a mass difference of 365.13 Da). The IgG1 glycopeptide mass list was generated from previously identified serum IgG1 glycoforms and annotated MS2 spectra for each species are likewise available as supplemental MS2 spectra (27). Tier 3 quantification of glycopeptides was performed for all data sets using these mass lists. MS1 isotope mass and intensities over a retention time range unique for each ion were extracted from the RAW spectrum file into a .cvs file using Xcalibur Software (Thermo Fisher Scientific). Glycopeptide intensities were measured using an R script (see the Supplement) to report intensities of first seven isotopologue peaks from the extracted raw MS1 data. Each species was validated for mass accuracy, isotopolog intensity distribution and MS2 spectra (where available). Intensities from different charge states were summed and the relative abundance was calculated using the summed intensity of all observed glycoforms on same peptide as a reference.
Peptides were identified from the glycopeptide enrichment flow-through fraction using Protein Metrics Byonic software (Version 2.12.0). The search parameters included Cleavage site (FYWEL), Cleavage side (C-terminal), Digestion specificity (Semi specific (slow)), Missed cleavages (2), Precursor mass tolerance (5 ppm), Fragment mass tolerance (0.05 Dalton), Recalibration (lock mass) (none), maximum precursor mass (5, 000), Precursor and mass charge assignments (Compute from MS1), Maximum # of precursors per scan (10), smoothing width (0.01 m/z) and the following modifications included (in addition to N- and O-glycan databases): Carbamidomethyl/+57.021464 @ C | fixed; Deamidated/+0.984016 @ N | common1; Acetyl/+42.010565 @ Protein N-term | rare1; Phospho/+79.966331 @ C, D, H, K, R, S, T,Y | common2; HexNAc/+203.079373 @ N,S,T common1. Each identified glycopeptide was manually validated. Monosaccharides were determined by mass, and isomers were not distinguished.
IgG N-glycan composition was quantified using the completely digested peptide (EEQYNSTYR), however, N-glycans on NK cell IgG from NK12 and NK13 were quantified using a partially digested peptide (TKPREEQYNSTYR) because of its higher abundance in these two data sets.
Donor Genotyping
Total RNA was isolated from frozen pellets using the TRIzol reagent (Thermo). cDNA was synthesized from 1 μg total RNA using the High-Capacity RNA-to-cDNA kit (Thermo) according to the manufacturer protocol. Full-length fcgr3a cDNA was amplified using forward (5′-CAGTGTGGCATCATGTGGCAG-3′) and reverse (5′-TTTGTCTTGAGGGTCCTTTCT-3′) primers and sequenced at the ISU DNA Facility. Genomic DNA was isolated from the interphase layer formed during RNA isolation with the TRIzol reagent. Genomic segments covering CD16a exon 4 (partial), intron 4–5 and exon 5 (partial) were amplified using forward (5′-CACAGCTGGAAGAACACTGC-3′) and reverse (5′-GAAAGTCCAGGCACACCACTC-3′) primers for the upstream section and forward (5′-TGAAGTAACCGAGGTGCAAA-3′) and reverse (5′-TTTGTCTTGAGGGTCCTTTCT-3′) primers for the downstream section that partially overlaps with the first fragment. The segments were cloned into pGEM-T Easy Vector (Promega) and sequenced.
RESULTS
NK Cell CD16a from Apheresis
We recovered 2.5–25 × 107 NK cells from each of five healthy donors following apheresis (Table I; supplemental Fig. S1) (38). CD16a from three donors was purified following detergent lysis with a yield of 1.7 μg, 0.7 μg and 2.0 μg from the NK08, NK09 and NK11 donors, respectively. The eluted protein migrated between 53 and 59 kDa in SDS-PAGE (supplemental Fig. S2). We also purified CD16a without detergent following PMA activation to release nearly the entire extracellular portion of the mature receptor that we expect is predominantly borne on the cell surface, yielding 0.3 μg and 0.5 μg of CD16a from the NK12 and NK13 donors, respectively (Fig. 1A and supplemental Fig. S2) (39–41).
Table I. NK cell and serum donor information.
| Donor | Gender | Age | ABO group | Rh factor | Date of collection | NK cell count (×106) | NK cell Viability (%) | CD16a genotype | CD16a form |
|---|---|---|---|---|---|---|---|---|---|
| NK00 | M | 40 | A | + | 1/25/2017 | 56.9 | 87 | n.d. | n.a. |
| NK01 | M | 64 | AB | + | 3/15/2018 | 74 | 73 | V/F | n.a. |
| NK02 | F | 33 | B | + | 3/15/2018 | 43.2 | 70 | V/F | n.a. |
| NK04 | M | 43 | B | + | 3/15/2018 | 80 | 88 | F/F | n.a. |
| NK06 | M | 23 | A | + | 5/22/2018 | 84.5 | 68 | V/V | n.a. |
| NK08 | F | 37 | O | + | 5/31/2018 | 91.7 | 75 | V/F | Detergent solubilized |
| NK09 | M | 59 | O | - | 6/7/2018 | 64.8 | 88 | V/F | Detergent solubilized |
| NK11 | M | 63 | A | + | 6/7/2018 | 252 | 68 | F/F | Detergent solubilized |
| NK12 | F | 28 | A | + | 8/28/2018 | 25.4 | 58 | V/F | Released by PMA |
| NK13 | M | 49 | O | + | 8/28/2018 | 157 | 82 | V/F | Released by PMA |
n.a. - not applicable.
n.d. - not determined.
LC-MS/MS Analysis of CD16a
We identified peptides corresponding to nearly the entire CD16a sequence except for the hydrophobic transmembrane region that was not identified (supplemental Fig. S3). The analysis of identifying peaks in MS2 spectra let to the definitive identification of peptides and PTM composition, and in some cases these spectra also provided clues to PTM configuration (Fig. 2 and supplemental Fig. S4). N-glycans were the only type of CD16a PTM observed. We identified 211 unique CD16a glycopeptides from five donors (Table II and supplemental Table S1). Though detergent extraction led to greater protein yields, PMA-released CD16a provided a greater number of ions. This is likely because of an inability to completely remove detergent following lysis; no detergent was used for purifying PMA-released CD16a. In addition to the identification of N-glycan modifications, two peptides showed partial deamidation of the asparagine residue, though it is not clear if these modifications occurred in situ or during the purification (LQNGKGRKY and FHHNSDFY; supplemental Fig. S3).
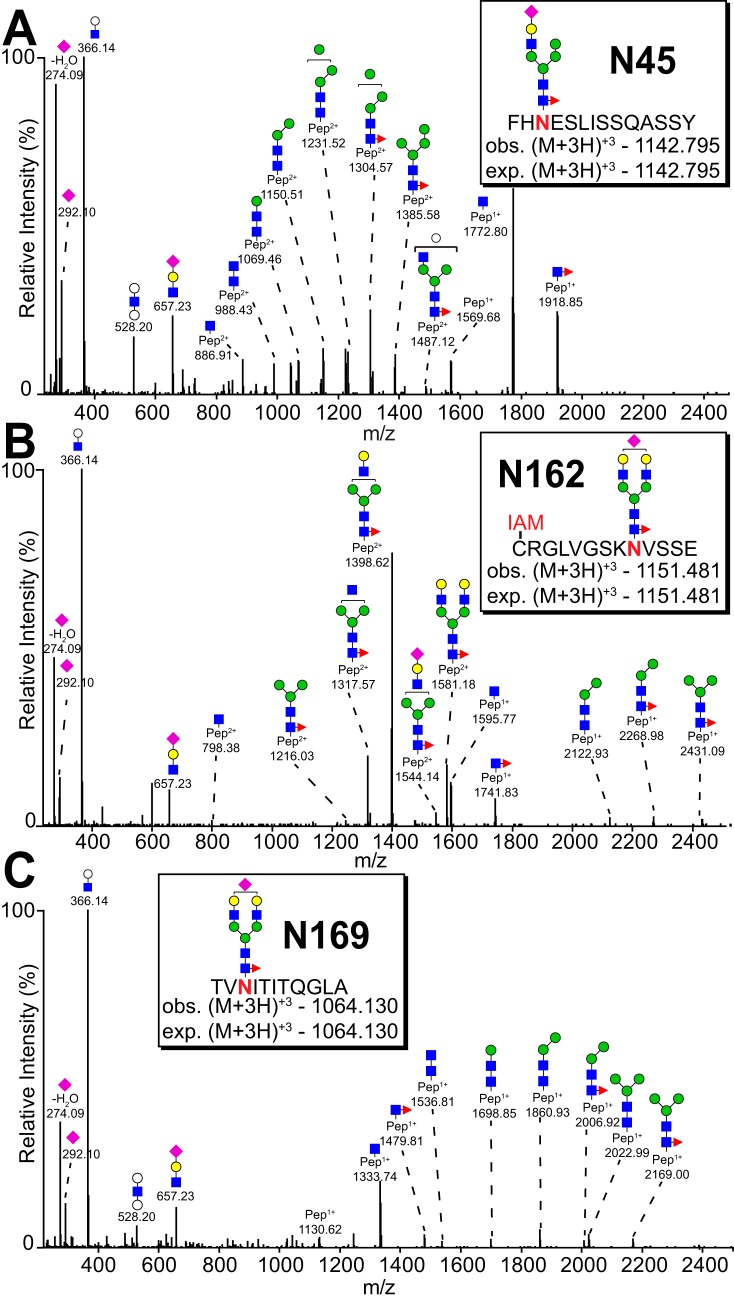
Fig. 2.
MS2 spectra for the most abundant N-glycan at N45, N162 and N169. A, N45 glycopeptide (from NK13). B, N162 glycopeptide (from NK08). A cysteine residue modified by iodoacetamide is indicated with a red “IAM.” C, N169 glycopeptide (from NK13). Inserts indicate the parent ion and charge as well as the observed and expected mass. Identification was based on mass of the parent ion, mass of fragmented ions and characteristic oligosaccharide oxonium ions. See also Table II and supplemental Fig. S4.
Table II. CD16a N-glycopeptides identified in this study. V/V, V/F or F/F indicate donor allotype.
| Site | # | Peptide | Detergent solubilized |
Released by PMA |
# of unique glycoforms | |||
|---|---|---|---|---|---|---|---|---|
| NK08 (V/F) | NK09 (V/F) | NK11 (F/F) | NK12 (V/F) | NK13 (V/F) | ||||
| N38 | 1 | KDSVTLKCQGAYSPEDNSTQW | 13 | 10 | n.d. | 4 | 6 | 16 |
| 2 | KCQGAYSPEDNSTQW | 9 | 4 | 4 | 4 | 17 | 18 | |
| N45 | 1 | FHNE | 16 | 8 | 27 | n.d. | 2 | 29 |
| 2 | FHNESLISSQASSY | 20 | 25 | 6 | 15 | 37 | 40 | |
| N74 | 1 | RCQTNLSTLSDPVQLE | 10 | 4 | 4 | 8 | 17 | 17 |
| 2 | RCQTNLSTL | 8 | 2 | 7 | n.d. | n.d. | 10 | |
| N162 (V158) | 1 | VGSKNVSSE | 2 | n.d. | n.a. | 5 | 5 | 6 |
| 2 | CRGLVGSKNVSSE | 16 | 4 | n.a. | 8 | 20 | 29 | |
| 3 | FCRGLVGSKNVSSE | 1 | n.d. | n.a. | 2 | 5 | 6 | |
| N162 (F158) | 4 | GSKNVSSE | 3 | 2 | 8 | 1 | 1 | 9 |
| 5 | FGSKNVSSE | n.d. | n.d. | n.d. | 2 | 3 | 3 | |
| 6 | CRGLFGSKNVSSE | n.d. | n.d. | n.d. | 2 | 2 | 3 | |
| N169 | 1 | TVNITITQGL | 1 | n.d. | 3 | n.d. | n.d. | 3 |
| 2 | TVNITITQGLA | n.a. | n.a. | n.a. | 7 | 16 | 17 | |
| 3 | TVNITITQG | n.a. | 5 | n.a. | n.a. | n.a. | 5 | |
n.a. - not applicable.
n.d. - not detected.
CD16a Site Impacts Processing
N-glycopeptides from the same site of different donors showed greater similarity than different N-glycopeptides from the same donor (Fig. 1D). N-glycans at N38, N74 and N169 showed minimal variability between donors and were almost exclusively complex types. N-glycans at N45 and N162 showed greater variability between donors with N45 displaying predominantly hybrid types with some complex-type glycans and N162 with mainly complex-type glycans showed the greatest variability between different donors.
The profiles of detergent-solubilized CD16a and CD16a released by PMA activation showed important differences. CD16a released from PMA-activated NK cells contained no oligomannose forms in contrast to detergent solubilized CD16a with 2–6% of N45 glycopeptides containing oligomannose forms. N45 is ideal for such comparisons because of superior ion yields from each donor (Table II). This result suggests that a small amount of folded but immature CD16a is retained in NK cells following PMA-induced activation.
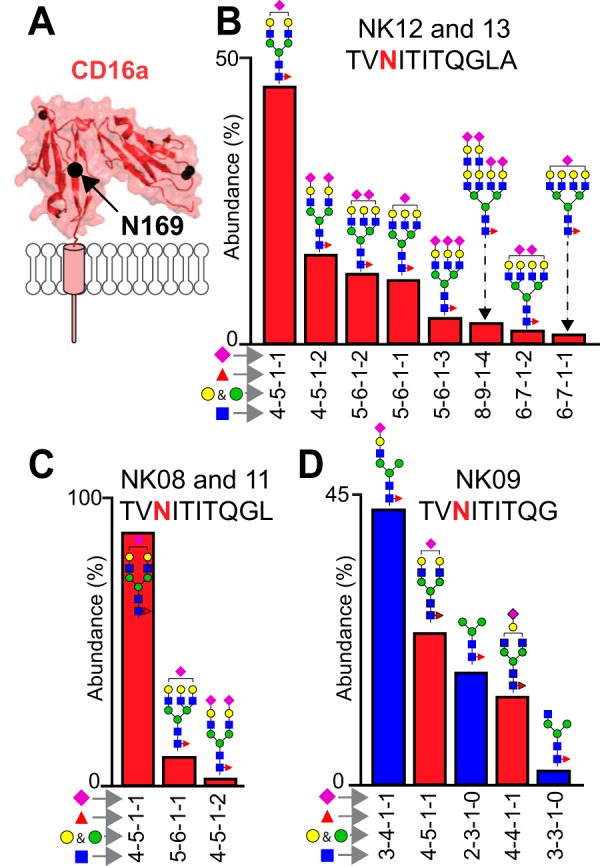
Donor-To-Donor Variability of the N162-glycan Composition
This proteolysis strategy links V/F158 and N162 on a peptide containing a single glycan (Table II). Donor allotype was confirmed for four of the five donors by sequencing CD16a cDNA; however, we observed peptides corresponding to both alleles from the NK09 donor but only found cDNA corresponding to F158 (supplemental Fig. S5) that may be explained by the greater abundance of the F158-containing transcripts in the NK09 NK cells biasing the cDNA sequencing results. If both alleles are present with equal copy numbers, no such variability is expected in the genomic DNA. Sequencing the genomic DNA segment including part of exon 4, intron 4–5 and exon 5 that covers the site encoding V158 and the protein C terminus from the NK09 donor confirmed the presence of both alleles along with multiple mutations in both coding and non-coding regions of the V158 allele when compared with the consensus CD16a sequence (supplemental Fig. S5).
CD16a glycopeptides with the F158 residue yielded less intensity and as a result a lower number of identified glycoforms (14 forms) when compared with V158 peptides in heterozygous donors (33 forms) suggesting either a reduced abundance of the F158 peptide or lower ionization efficiency of this glycopeptide (Table I and supplemental Table S1).
The predominant N162 glycopeptide contained a monosialylated, core fucosylated, biantennary complex-type N-glycan (Fig. 3 and supplemental Table S1), though a comparison of donors showed substantial variability in the abundance of under-processed forms. Donors contained a range from 5% to 42% of hybrid-type N162-glycans, except for NK09 N162(V158) that showed only oligomannose-type N-glycans, as confirmed by MS2 spectra (supplemental Fig. S4). One could argue that the NK09 donor expressed only under-processed CD16a V158 that was trapped in intracellular vesicles. If this were true, the other four sites should include roughly 50% of under-processed forms when in most cases none appeared (Fig. 1D). Thus, NK09 expressed CD16a with minimal processing at the N162(V158) and N169 (discussed below) sites but with processing comparable to the other four donors at the three other N-glycosylation sites. Oligomannose-type N162-glycans also appeared in spectra from the NK08 donor but at a lower level compared with NK09.
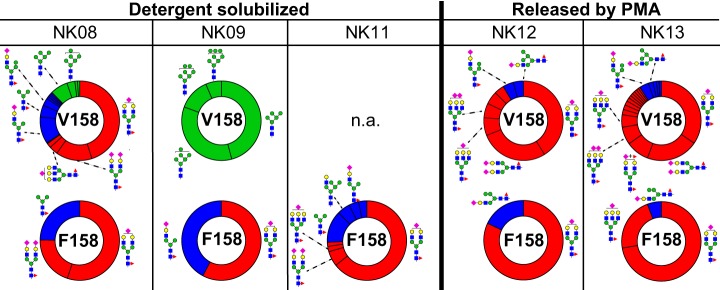
Fig. 3.
Donor-to-donor variability of the N162-glycan composition. Each wheel represents the relative abundance of individual N-glycans from five donors and from both CD16a allotypes (V158 v. F158). Glycans are grouped by oligomannose (green), hybrid (blue) or complex type (red). The abundance of N162 glycans from CD16a-V158 was determined using N162 glycopeptide 2. Glycan abundance for CD16a-F158 were summed from the intensities of N162 glycopeptide 4 (NK08, NK09 and NK11) and N162 glycopeptide 5 (NK12 and NK13). The N-glycan cartoons represent one possible configuration based on composition. n.a., not applicable. See also Table II and supplemental Fig. S4.
The appearance of minimally processed CD16a N-glycans in this analysis is consistent with our recent glycomics study that revealed a significant number of oligomannose forms in the pool of CD16a N-glycans from three older donors (67–79y (6)). The age comparison is notable because NK09 is one of the oldest in this pool of five donors at 59, however, the prevalence of oligomannose only at V158 protein might also be because of age independent factors such as unique point mutations in the sequence encoding the NK09 CD16a V158 protein (Table I and supplemental Fig. S5). The presence of oligomannose N-glycans at N162, as noted above, increases affinity for IgG1 Fc (6, 14). Thus, NK08 and NK09 may be expressing CD16a glycoforms that bind with greater affinity than NK11, NK12, and NK13 with higher proportions of complex-type N162-glycans. It is unclear how the hybrid type N162-glycans impact antibody binding affinity.
A paucimannose N162-glycan appeared in detergent solubilized samples with F158 glycopeptides showing higher levels than V158 (GlcNAc2Mannose3Fucose1; Fig. 3 and supplemental Fig. S4). We observed this glycoform in our recent glycomics analysis (6), though it is not generally expected on mature N-glycoproteins and likely results from partial degradation in the cell (2). Partially degraded paucimannose glycans also recently appeared with proteins localized to the neutrophil granule (42).
CD16a N45 Displays Predominantly Hybrid-type N-glycans
The N45-glycan is unique among the five CD16a modifications because it forms measurable intramolecular interactions between glycan and polypeptide residues that restrict mobility and likely limit glycan processing (15). As expected, N45 glycoforms showed the highest proportion of minimally processed forms from all five donors among 47 unique glycoforms identified (Table II, Figs. 1D and 4).
Fig. 4.
CD16a N45 displays predominantly hybrid-type N-glycans. Bars show the relative abundance of N45-glycan species accounting for 95.3% of the total observed glycopeptide intensity from five donors. The abundance of each species was determined by first measuring the relative abundance of each glycan within each dataset, then averaging the relative abundance from the five datasets. Glycopeptide 1 (NK08, NK09, NK11 and NK13) and glycopeptide 2 (all data sets) were used for these comparisons. The N-glycan cartoons represent one possible configuration based on composition. Glycan types are specified by color as in Fig. 3. See also Table II and supplemental Fig. S4.
Though the N45 glycan shows restricted processing, the termini received a high degree of sialylation. Monosialylated hybrid types accounted for the majority of the N45-glycans and 47–68% of the total forms at this site (Fig. 4; supplemental Table S1). Complex-type bi- and tri-antennary glycans comprised the second most abundant group (15–26%) (supplemental Fig. S4). The relative abundance of hybrid and complex glycoforms were highly comparable for four of the five donors; NK13 had higher abundance of complex-type N-glycans (43%) compared with an average of 21% for the other donors. NK13, unlike the other donors, also contained N45-glycans with highly processed N-acetyllactosamine (LacNAc) repeats (10% of the complex-type N-glycans at this site from this donor). Oligomannose forms were only observed in the detergent solubilized samples and produced the lowest ion intensities of any form, representing less than 6% of the N45-glycoforms.
We identified two features of the N45 glycopeptides that complicated data analysis. First, N45-glycan composition impacted proteolysis in the presence of residual detergent that remained following specific washes to remove lipids and detergents. Less fucose appeared in the smaller N45 glycopeptide #1 than the longer N45 glycopeptide #2 (Table II; supplemental Fig. S4). CD16a purified following PMA activation did not reveal a similar bias.
Second, we observed some N45 glycopeptide ions in samples from all donors with an additional mass of 79.96 Da (supplemental Fig. S4). Similar modifications were not observed at other sites or with recombinant CD16a. The species eluted later than unmodified N45 glycopeptides in detergent-solubilized CD16a. Analysis of MS2 spectra from relevant species did not reveal the modification location, or whether it is covalently attached to the carbohydrate or amino acid residues. N-glycans are known to contain phosphate or sulfate modifications that correspond to a mass change of 79.9663 or 79.9658 Da, respectively (10).
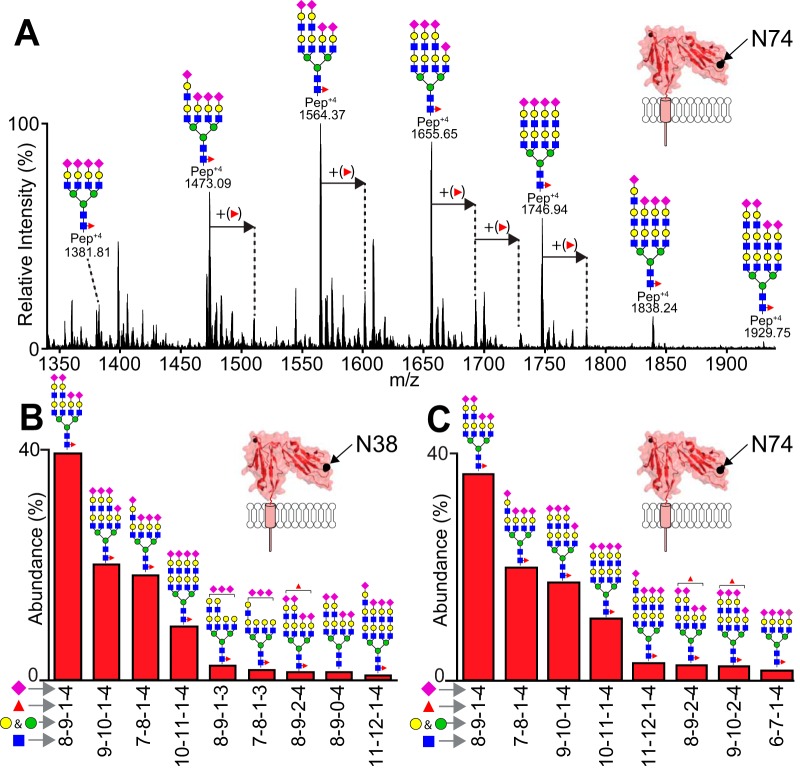
The predominant N38 and N74 glycans contain multiple N-acetyllactosamine repeats
As the most highly processed sites identified for NK cell CD16a, N38 and N74 displayed only highly modified, complex-type N-glycans (Fig. 5). One characteristic features is the presence of LacNAc repeats at both sites that is evident from the glycan mass as well as a characteristic oxonium ion in MS2 spectra consisting of a single N-acetylneuraminic acid, two hexoses and two N-acetylhexosamines (mass = 1022.36 Da; supplemental Fig. S4 and supplemental Table S1). Further, we believe the N38 and N74 glycans are predominantly tetraantennary because of the presence of four, and no more than four, sialic acids on 86 and 99% of the forms found at N38 and N74, respectively, as well as the complete absence of characteristic polysialic acid fragments in the MS2 spectra.
Fig. 5.
The predominant N38 and N74 glycans contain multiple N-acetyllactosamine motifs. A, An ESI-MS spectrum shows the distribution of N74 glycopeptides that eluted from 49.20–61.10 min (IAM-modified glycopeptide 1; NK13 dataset). B, The predominant N38 glycans account for 95.8% of the total observed N38 glycopeptide intensity (IAM-modified N38 glycopeptide 2). C, The predominant N74 glycans account for 96.1% of the total observed N74 glycopeptide intensity (IAM-modified N74 glycopeptide 1). The N-glycan cartoons represent one possible configuration based on composition. See also Table II and supplemental Fig. S4.
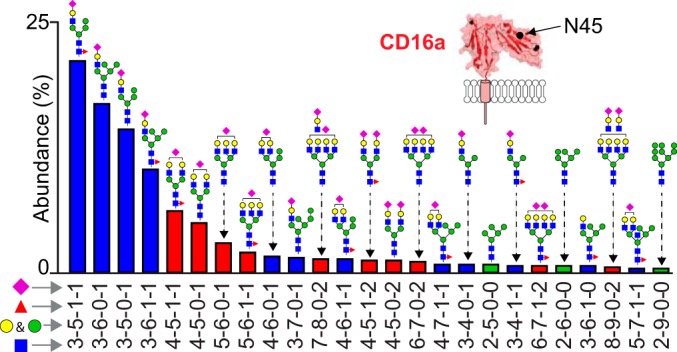
The N169 Glycan Is A Complex Type
CD16a purified following PMA activation provided greater intensity and depth of coverage than CD16a solubilized with detergent because of ADAM17-catalyzed cleavage that reduced the size of the PMA-activation peptide (40, 41). In both situations, however, a monosialylated, biantennary, core fucosylated complex-type glycan proved to be the predominant form, representing as much as 43% to 47% of the N169 total in PMA released samples. This position was preserved among all samples, except that from NK09 (Fig. 6). Apart from an atypical truncated form of the N169 peptide that is not consistent with ADAM17 cleavage or chymotrypsin/Glu-C cleavage, donor NK09 displayed predominantly hybrid type N169-glycans (63%) with fewer complex types (37%) (Table II; supplemental Fig. S4). This restricted processing of the NK09 N169 site is like the nearby N162 of the NK09 donor that showed predominantly under-processed forms (Fig. 6D). It is unclear if the mutations present in the NK09 CD16a V158 genomic sequence impact N162 and N169 N-glycan processing (supplemental Fig. S5).
Fig. 6.
The N169 glycan is a complex type. A, Ribbon diagram of CD16a highlighting N169. B, The two different CD16a isolation techniques generate two different glycopeptides that show similar processing. The predominant N169 glycans from CD16a released with PMA ionizes well. These forms account for 96.2% of the total observed N169 intensity from the NK12 and NK13 donors (N169 glycopeptide 2). C, N169 glycopeptides isolated from CD16a purified using detergent lysis of NK cells provided fewer observations from the NK08 and NK11 donors (N169 glycopeptide 1). D, CD16a glycopeptides from the NK09 donor showed a peptide lacking the L residue. N-glycan cartoons represent one possible configuration based on composition. See also Table II and supplemental Fig. S4.
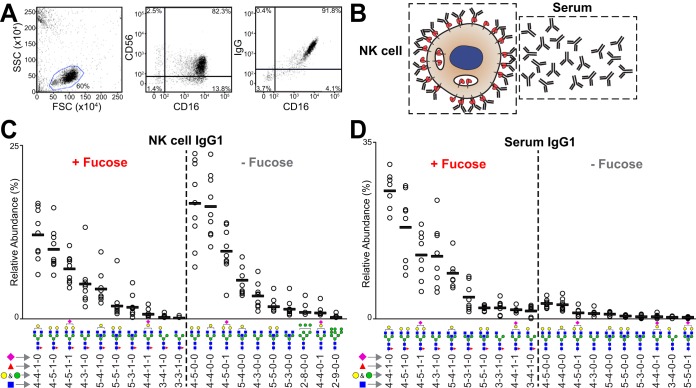
NK Cells Retain IgG1 Lacking Core Fucose
Freshly isolated NK cells exhibited IgG staining in a FACS experiment that correlate with amount of CD16a staining, indicating NK cells bound IgG primarily through CD16a (Fig. 7A). Data from the right-hand panel of Fig. 7A fit to a line with an R-squared value of 0.70 and a repeat experiment showed similar results with an R-squared of 0.75 (data not shown). A high average percentage of IgG purified from the NK cell lysates of nine donors (45% of the potential IgG, based on the mass of CD16a in the lysate; Table I). We also purified IgG from the serum of each donor for comparison (NK06 excepted; Table I). IgG1 purified from NK cells showed a striking lack of fucose (56.8 ± 11.0%) compared with serum (10.3 ± 3.9%) (Fig. 7). The rank order of the top fucosylated and afucosylated forms remained unchanged between the NK cell and serum fractions, revealing fucosylation level as the primary difference. Further, oligomannose Fc N-glycans lacking core fucosylation appeared on NK cells. We did not detect these minimally processed N-glycans in serum samples.
Fig. 7.
NK cells retain IgG1 lacking core fucose. A, Flow cytometry showing the collinearity of IgG and CD16a staining on primary human NK cells following cell isolation. B, Sources of IgG molecules (depicted as Y-shaped symbols). The top ten fucosylated and afucosylated IgG1 glycoforms isolated from primary NK cells (C) and serum (D). See also supplemental Tables S3 and S4.
DISCUSSION
Powerful connections between the post-translational modification of serum proteins and immune system function are emerging as new technologies provide resolution at the level of a single donor and at a single modified site (27, 43, 44). These prior studies likely represent a tiny sample of the larger group of molecules that display clinically relevant and variable modifications. Little is known about PTMs on endogenous human membrane proteins not because it is unimportant, but because of low abundance in a single donor. Here we characterized the antibody-binding receptor CD16a from primary human NK cells recovered from apheresis filters and discovered unexpected variability between donors. We believe this strategy is applicable to characterize PTMs of immune cell receptors and will lead to studies that will define how PTMs vary and impact immune system function.
Enrichment of IgG Lacking Fucose
The functional receptor-binding unit of IgG1 is the Fc, a dimer that binds asymmetrically to CD16a (45). This asymmetric mode will bind to one of the two Fc chains providing the greatest affinity. For example, Fc containing both a fucosylated N-glycan and a glycan lacking fucose will preferentially bind CD16a in a manner that places the Fc half lacking a fucose adjacent to the CD16a N162-glycan (46). Further, IgG1 N-glycans on the same molecule are not necessarily identical (47) but it is possible that a subset of the serum IgG is symmetrically afucosylated thus dominating the antibodies on the NK cells; high levels of afucosylation were observed for antigen-specific responses (29, 44, 48). We expect symmetrically afucosylated IgG to be a minority of the total serum IgG and apply a distributive model for analysis. Thus, at 10% serum fucosylation, we expect that 95% of the NK cell-bound IgGs contain one fucosylated N-glycan and one N-glycan lacking fucose with a maximum possible level of fucose of 47% given a distributive model. This calculated value is comparable to the measured value of 43.2 ± 11.0%. Thus, the majority of CD16a on NK cells is likely bound to IgG1 with at least one N-glycan lacking a core fucose residue.
The enrichment of IgG1 lacking fucose on the surface of NK cells may provide the body a mechanism to prioritize a specific response. It is known that antigen-specific antibodies can exhibit dramatically different glycan composition when compared with bulk serum IgG in the same body, including large differences in fucosylation (29, 32, 33). These substantial differences in antibody composition predict that an antigen specific antibody response lacking fucose would result in NK cells binding more of that antibody than other serum antibodies with higher levels of fucosylation, potentially priming NK cells. This prioritized response is expected to be specific to NK cells because CD16a is the only FcγR expressed on NK cells from most donors in contrast to monocytes, neutrophils or resident macrophages that express a mixture of FcγRs including CD32a and CD64 that do not exhibit a preference for IgG antibodies that lack fucose (49, 50).
Donor-specific Variability in CD16a Processing
Even though N-glycosylation is affected by a range of different factors, the similarities between the N-glycosylation profiles of NK cell CD16a from five relatively diverse donors suggests the presence of conserved biological features that serve specific irreplaceable functions. CD16a with the most abundant N-glycans identified at each site is shown in Fig. 1C. These modifications are roughly scaled to the size of the protein to demonstrate that the modification adds ∼13 kDa to the 20 kDa extracellular domain.
Modifications at N162 showed the greatest variability in processing after comparing the results from different donors. As described above, the composition of the N162-glycan affects affinity of CD16a for IgG1 (6, 14, 51, 52). Though some compositional variability was expected, the degree of donor specific variability in the abundance of minimally processed forms identified here was not anticipated (Fig. 3). This observation suggests that NK cells have the capability to tune IgG binding and potentially the threshold for activating ADCC by changing the N-glycan composition. Conditions in the body, including cytokine levels, are known to affect N-glycan composition (53–57). A demonstration of similar or greater variability in a much larger number of apheresis filters from healthy and inflamed individuals would support this hypothesis and supports the importance of similar studies with site-specific resolution of CD16a from a single donor.
Composition of the N45-glycan does not appear to impact affinity, but its presence is required for high affinity interactions (14, 15). The appearance of a high percentage of hybrid forms is unexpected. N45 is often the least processed CD16 N-glycan, and this restriction may be because of intramolecular interactions that reduce N45-glycan accessibility during processing (15). It was further unexpected that the NK09 donor showed even less processing at N162 than N45. This indicates that additional CD16a-specific factors might contribute to processing.
Though N-glycosylation at N38 and N74 does not impact affinity in vitro, these N-glycans may serve critical, likewise tunable roles in CD16a-mediated cell activation (14). For example, the observed LacNAc repeats have the potential to regulating receptor clustering at the NK cell surface. LacNAc repeats can serve as ligands for galectins, which were shown to modulate T-cell activation by preventing T-cell receptor clustering (4). LacNAc repeats are added following the MGAT5-catalyzed addition of a GlcNAc residue to the N-glycan, forming a new branch point. IL-10 dependent expression of MGAT5 in CD8+ T-cells is known to modulate the immune response (53). Similarly, MGAT5 is expressed in NK cells and may contribute to the NK cell response (58).
We noted substantial differences in a comparison of the primarily hybrid-type NK cell CD16a N45-glycans and the predominance of oligomannose forms at comparable sites of CD16b expressed by neutrophils (59, 60). Further, CD16b lacked complex-type N45-glycans that represented 25% of the NK cell CD16a forms (Fig. 1D). Other differences include CD16b N162 that showed higher branch fucosylation. CD16b revealed fewer hybrid N162 forms and the appearance of the fucosylated paucimannose N-glycan seen in CD16a. Even the highly processed sites N38 and N74 consistently showed less sialylation in CD16b compared with CD16a. These PTM differences likely represent important features that contribute to functional differences between CD16a and CD16b, in addition to the presence of a glycosylphosphatidylinositol anchor on CD16b and the G129 residue that provides high antibody binding affinity to CD16a (52). Considering the high structural and sequence homology of CD16a and CD16b, it will be informative to determine how conditions in the secretory pathway contribute to the observed N-glycan processing differences.
Two recent studies characterized N-glycans from the related receptor CD16b. These studies purified soluble CD16b from a large volume of pooled serum or only the CD16b N45 and N162 glycans from neutrophils of individual donors (59, 60). Neutrophils and CD16b are much more abundant (30–80% of leukocytes), requiring a much smaller amount of blood to isolate enough mass of receptor (∼20 ml). Our studies represent a substantial advance toward characterizing cell surface proteins from a less abundant cell type.
Implications of the Approach
This present study identified several important considerations for future studies of endogenous receptors. First, core fucosylation reduces cleavage at proximal protease sites, introducing potential bias in the analysis of peptides resulting from missed cleavages are not identified. For CD16a, the presence of a core fucose residue at N45 inhibited GluC activity at the adjacent amino acid residue (E46). Similar composition-dependent inhibition was identified in trypsinized IgA (61) and alpha-1-acid glycoprotein (62). Second, the parallel analysis of both the peptide and glycopeptide-enriched fractions prevents glycopeptide ion suppression and provides the opportunity to identify less hydrophilic modifications. Third, signal intensity is enhanced by reducing the levels of residual detergent. Our observation that avoiding detergent in all purification steps increased signal was unexpected considering that the methods included extensive washes to remove residual detergent. This situation was avoided through PMA induction of NK cells that shed CD16a into the medium. Finally, analyzing peptides provides broad protein coverage and can identify functionally relevant polymorphisms as shown for CD16a allotype that impacts mAb therapies and may be a risk factor for autoimmunity (17, 63–66).
In summary, these results demonstrate the feasibility of characterizing, in a donor, cell-type and site-specific manner, PTMs from receptors and ligands on the surface of low-abundance leukocytes in the peripheral blood, including NK cells, T and B lymphocytes, and monocytes.
DATA AVAILABILITY
Raw data available at MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the identifier MSV000083890 or via ftp://massive.ucsd.edu/MSV000083890/.
Supplementary Material
Acknowledgments
We thank donors and staff at the DeGowin Blood Center (Iowa City, IA) and LifeServe Blood Center (Ames and Des Moines, IA) for providing materials. We also thank Dr. Sean Rigby (ISU Flow Cytometry Facility) for performing the flow cytometry measurements, Jacob Roberts and Alison Warnell (ISU) for preparing the 3G8 N297Q construct and Jacob Roberts for assistance during NK cell isolation. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Institutes of Health.
Footnotes
* This material is based upon work supported by the National Institutes of Health under Award No. R01 GM115489 (NIGMS) and R21 AI142122 (NIAID). The authors declare no conflicts of interest.
 This article contains supplemental Figures, Supplementary PDF, and Tables.
This article contains supplemental Figures, Supplementary PDF, and Tables.
1 The abbreviations used are:
- NK
- natural killer
- ADCC
- antibody-dependent cell mediated cytotoxicity
- ER
- endoplasmic reticulum
- ESI-MS
- electrospray ionization-tandem mass spectrometry
- PTMs
- post-translational modifications
- GlcNAc
- N-acetylglucosamine
- PMA
- phorbol 12-myristate 13-acetate
- DDM
- dodecyl maltoside
- LacNAc
- N-acetyllactosamine.
REFERENCES
- 1. Nagler A., Lanier L. L., Cwirla S., and Phillips J. H. (1989) Comparative studies of human FcRIII-positive and negative natural killer cells. J. Immunol. 143, 3183–3191 [PubMed] [Google Scholar]
- 2. Moremen K. W., Tiemeyer M., and Nairn A. V. (2012) Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drake P. M., Nathan J. K., Stock C. M., Chang P. V., Muench M. O., Nakata D., Reader J. R., Gip P., Golden K. P., Weinhold B., Gerardy-Schahn R., Troy F. A. 2nd, and Bertozzi C. R. (2008) Polysialic acid, a glycan with highly restricted expression, is found on human and murine leukocytes and modulates immune responses. J. Immunol. 181, 6850–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Demetriou M., Granovsky M., Quaggin S., and Dennis J. W. (2001) Negative regulation of T-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature 409, 733–739 [DOI] [PubMed] [Google Scholar]
- 5. Kuball J., Hauptrock B., Malina V., Antunes E., Voss R. H., Wolfl M., Strong R., Theobald M., and Greenberg P. D. (2009) Increasing functional avidity of TCR-redirected T cells by removing defined N-glycosylation sites in the TCR constant domain. J. Exp. Med. 206, 463–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Patel K. R., Roberts J. T., Subedi G. P., and Barb A. W. (2018) Restricted processing of CD16a/Fc gamma receptor IIIa N-glycans from primary human NK cells impacts structure and function. J. Biol. Chem. 293, 3477–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes J. M., Frostell A., Karlsson R., Muller S., Martin S. M., Pauers M., Reuss F., Cosgrave E. F., Anneren C., Davey G. P., and Rudd P. M. (2017) Identification of Fc gamma receptor glycoforms that produce differential binding kinetics for rituximab. Mol. Cell. Proteomics 16, 1770–1788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Parekh R. B., Dwek R. A., Sutton B. J., Fernandes D. L., Leung A., Stanworth D., Rademacher T. W., Mizuochi T., Taniguchi T., Matsuta K., and et al. (1985) Association of rheumatoid arthritis and primary osteoarthritis with changes in the glycosylation pattern of total serum IgG. Nature 316, 452–457 [DOI] [PubMed] [Google Scholar]
- 9. Varshney S., and Stanley P. (2018) Multiple roles for O-glycans in Notch signalling. FEBS Lett. 592, 3819–3834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Varki A. (2017) Essentials of glycobiology, Third edition , Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- 11. Pang X., Li H., Guan F., and Li X. (2018) Multiple roles of glycans in hematological malignancies. Front. Oncol. 8, 364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tiemeyer M., Aoki K., Paulson J., Cummings R. D., York W. S., Karlsson N. G., Lisacek F., Packer N. H., Campbell M. P., Aoki N. P., Fujita A., Matsubara M., Shinmachi D., Tsuchiya S., Yamada I., Pierce M., Ranzinger R., Narimatsu H., and Aoki-Kinoshita K. F. (2017) GlyTouCan: an accessible glycan structure repository. Glycobiology 27, 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chandler K. B., and Costello C. E. (2016) Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis 37, 1407–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Subedi G. P., and Barb A. W. (2018) CD16a with oligomannose-type N-glycans is the only “low-affinity” Fc gamma receptor that binds the IgG crystallizable fragment with high affinity in vitro. J. Biol. Chem. 293, 16842–16850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Subedi G. P., Falconer D. J., and Barb A. W. (2017) Carbohydrate-polypeptide contacts in the antibody receptor CD16A identified through solution NMR spectroscopy. Biochemistry 56, 3174–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bibeau F., Lopez-Crapez E., Di Fiore F., Thezenas S., Ychou M., Blanchard F., Lamy A., Penault-Llorca F., Frebourg T., Michel P., Sabourin J. C., and Boissiere-Michot F. (2009) Impact of Fc{gamma}RIIa-Fc{gamma}RIIIa polymorphisms and KRAS mutations on the clinical outcome of patients with metastatic colorectal cancer treated with cetuximab plus irinotecan. J. Clin. Oncol. 27, 1122–1129 [DOI] [PubMed] [Google Scholar]
- 17. Bruhns P., Iannascoli B., England P., Mancardi D. A., Fernandez N., Jorieux S., and Daeron M. (2009) Specificity and affinity of human Fc gamma receptors and their polymorphic variants for human IgG subclasses. Blood 113, 3716–3725 [DOI] [PubMed] [Google Scholar]
- 18. Zhang W., Gordon M., Schultheis A. M., Yang D. Y., Nagashima F., Azuma M., Chang H. M., Borucka E., Lurje G., Sherrod A. E., Iqbal S., Groshen S., and Lenz H. J. (2007) FCGR2A and FCGR3A polymorphisms associated with clinical outcome of epidermal growth factor receptor expressing metastatic colorectal cancer patients treated with single-agent cetuximab. J. Clin. Oncol. 25, 3712–3718 [DOI] [PubMed] [Google Scholar]
- 19. Cartron G., Dacheux L., Salles G., Solal-Celigny P., Bardos P., Colombat P., and Watier H. (2002) Therapeutic activity of humanized anti-CD20 monoclonal antibody and polymorphism in IgG Fc receptor FcgammaRIIIa gene. Blood 99, 754–758 [DOI] [PubMed] [Google Scholar]
- 20. Weng W. K., and Levy R. (2003) Two immunoglobulin G fragment C receptor polymorphisms independently predict response to rituximab in patients with follicular lymphoma. J. Clin. Oncol. 21, 3940–3947 [DOI] [PubMed] [Google Scholar]
- 21. Musolino A., Naldi N., Bortesi B., Pezzuolo D., Capelletti M., Missale G., Laccabue D., Zerbini A., Camisa R., Bisagni G., Neri T. M., and Ardizzoni A. (2008) Immunoglobulin G fragment C receptor polymorphisms and clinical efficacy of trastuzumab-based therapy in patients with HER-2/neu-positive metastatic breast cancer. J. Clin. Oncol. 26, 1789–1796 [DOI] [PubMed] [Google Scholar]
- 22. Vance B. A., Huizinga T. W., Wardwell K., and Guyre P. M. (1993) Binding of monomeric human IgG defines an expression polymorphism of Fc gamma RIII on large granular lymphocyte/natural killer cells. J. Immunol. 151, 6429–6439 [PubMed] [Google Scholar]
- 23. Mota G., Moldovan I., Calugaru A., Hirt M., Kozma E., Galatiuc C., Brasoveanu L., and Boltz-Nitulescu G. (2004) Interaction of human immunoglobulin G with CD16 on natural killer cells: ligand clearance, Fc gamma RIIIA turnover and effects of metalloproteinases on Fc gamma RIIIA-mediated binding, signal transduction and killing. Scand. J. Immunol. 59, 278–284 [DOI] [PubMed] [Google Scholar]
- 24. Edberg J. C., and Kimberly R. P. (1997) Cell type-specific glycoforms of Fc gamma RIIIa (CD16): differential ligand binding. J. Immunol. 159, 3849–3857 [PubMed] [Google Scholar]
- 25. Nose M., and Wigzell H. (1983) Biological significance of carbohydrate chains on monoclonal antibodies. Proc. Natl. Acad. Sci. U.S.A. 80, 6632–6636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Subedi G. P., and Barb A. W. (2015) The structural role of antibody N-glycosylation in receptor interactions. Structure 23, 1573–1583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pucic M., Knezevic A., Vidic J., Adamczyk B., Novokmet M., Polasek O., Gornik O., Supraha-Goreta S., Wormald M. R., Redzic I., Campbell H., Wright A., Hastie N. D., Wilson J. F., Rudan I., Wuhrer M., Rudd P. M., Josic D., and Lauc G. (2011) High throughput isolation and glycosylation analysis of IgG-variability and heritability of the IgG glycome in three isolated human populations. Mol. Cell. Proteomics 10, M111.010090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lauc G., Huffman J. E., Pucic M., Zgaga L., Adamczyk B., Muzinic A., Novokmet M., Polasek O., Gornik O., Kristic J., Keser T., Vitart V., Scheijen B., Uh H. W., Molokhia M., Patrick A. L., McKeigue P., Kolcic I., Lukic I. K., Swann O., van Leeuwen F. N., Ruhaak L. R., Houwing-Duistermaat J. J., Slagboom P. E., Beekman M., de Craen A. J., Deelder A. M., Zeng Q., Wang W., Hastie N. D., Gyllensten U., Wilson J. F., Wuhrer M., Wright A. F., Rudd P. M., Hayward C., Aulchenko Y., Campbell H., and Rudan I. (2013) Loci associated with N-glycosylation of human immunoglobulin G show pleiotropy with autoimmune diseases and haematological cancers. PLoS Genet. 9, e1003225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapur R., Kustiawan I., Vestrheim A., Koeleman C. A., Visser R., Einarsdottir H. K., Porcelijn L., Jackson D., Kumpel B., Deelder A. M., Blank D., Skogen B., Killie M. K., Michaelsen T. E., de Haas M., Rispens T., van der Schoot C. E., Wuhrer M., and Vidarsson G. (2014) A prominent lack of IgG1-Fc fucosylation of platelet alloantibodies in pregnancy. Blood 123, 471–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shields R. L., Lai J., Keck R., O'Connell L. Y., Hong K., Meng Y. G., Weikert S. H., and Presta L. G. (2002) Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human Fcgamma RIII and antibody-dependent cellular toxicity. J. Biol. Chem. 277, 26733–26740 [DOI] [PubMed] [Google Scholar]
- 31. Shinkawa T., Nakamura K., Yamane N., Shoji-Hosaka E., Kanda Y., Sakurada M., Uchida K., Anazawa H., Satoh M., Yamasaki M., Hanai N., and Shitara K. (2003) The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 278, 3466–3473 [DOI] [PubMed] [Google Scholar]
- 32. Mahan A. E., Jennewein M. F., Suscovich T., Dionne K., Tedesco J., Chung A. W., Streeck H., Pau M., Schuitemaker H., Francis D., Fast P., Laufer D., Walker B. D., Baden L., Barouch D. H., and Alter G. (2016) Antigen-specific antibody glycosylation is regulated via vaccination. PLoS Pathog. 12, e1005456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lofano G., Gorman M. J., Yousif A. S., Yu W. H., Fox J. M., Dugast A. S., Ackerman M. E., Suscovich T. J., Weiner J., Barouch D., Streeck H., Little S., Smith D., Richman D., Lauffenburger D., Walker B. D., Diamond M. S., and Alter G. (2018) Antigen-specific antibody Fc glycosylation enhances humoral immunity via the recruitment of complement. Sci. Immunol. 3, pii: eaat7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grilo A. L., and Mantalaris A. (2019) The increasingly human and profitable monoclonal antibody market. Trends Biotechnol. 37, 9–16 [DOI] [PubMed] [Google Scholar]
- 35. Neron S., Thibault L., Dussault N., Cote G., Ducas E., Pineault N., and Roy A. (2007) Characterization of mononuclear cells remaining in the leukoreduction system chambers of apheresis instruments after routine platelet collection: a new source of viable human blood cells. Transfusion 47, 1042–1049 [DOI] [PubMed] [Google Scholar]
- 36. Yeung Y. G., and Stanley E. R. (2010) Rapid detergent removal from peptide samples with ethyl acetate for mass spectrometry analysis. Curr. Protoc. Protein Sci. Chapter 16, Unit 16 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ceroni A., Maass K., Geyer H., Geyer R., Dell A., and Haslam S. M. (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J. Proteome Res. 7, 1650–1659 [DOI] [PubMed] [Google Scholar]
- 38. Cooper M. A., Fehniger T. A., and Caligiuri M. A. (2001) The biology of human natural killer-cell subsets. Trends Immunol. 22, 633–640 [DOI] [PubMed] [Google Scholar]
- 39. Trinchieri G., O'Brien T., Shade M., and Perussia B. (1984) Phorbol esters enhance spontaneous cytotoxicity of human lymphocytes, abrogate Fc receptor expression, and inhibit antibody-dependent lymphocyte-mediated cytotoxicity. J. Immunol. 133, 1869–1877 [PubMed] [Google Scholar]
- 40. Lajoie L., Congy-Jolivet N., Bolzec A., Gouilleux-Gruart V., Sicard E., Sung H. C., Peiretti F., Moreau T., Vie H., Clemenceau B., and Thibault G. (2014) ADAM17-mediated shedding of FcgammaRIIIA on human NK cells: identification of the cleavage site and relationship with activation. J. Immunol. 192, 741–751 [DOI] [PubMed] [Google Scholar]
- 41. Jing Y., Ni Z., Wu J., Higgins L., Markowski T. W., Kaufman D. S., and Walcheck B. (2015) Identification of an ADAM17 cleavage region in human CD16 (FcgammaRIII) and the engineering of a non-cleavable version of the receptor in NK cells. PLoS ONE 10, e0121788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thaysen-Andersen M., Venkatakrishnan V., Loke I., Laurini C., Diestel S., Parker B. L., and Packer N. H. (2015) Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J. Biol. Chem. 290, 8789–8802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wuhrer M., Stam J. C., van de Geijn F. E., Koeleman C. A., Verrips C. T., Dolhain R. J., Hokke C. H., and Deelder A. M. (2007) Glycosylation profiling of immunoglobulin G (IgG) subclasses from human serum. Proteomics 7, 4070–4081 [DOI] [PubMed] [Google Scholar]
- 44. Ackerman M. E., Crispin M., Yu X., Baruah K., Boesch A. W., Harvey D. J., Dugast A. S., Heizen E. L., Ercan A., Choi I., Streeck H., Nigrovic P. A., Bailey-Kellogg C., Scanlan C., and Alter G. (2013) Natural variation in Fc glycosylation of HIV-specific antibodies impacts antiviral activity. J. Clin. Invest. 123, 2183–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sondermann P., Huber R., Oosthuizen V., and Jacob U. (2000) The 3.2-A crystal structure of the human IgG1 Fc fragment-Fc gammaRIII complex. Nature 406, 267–273 [DOI] [PubMed] [Google Scholar]
- 46. Shatz W., Chung S., Li B., Marshall B., Tejada M., Phung W., Sandoval W., Kelley R. F., and Scheer J. M. (2013) Knobs-into-holes antibody production in mammalian cell lines reveals that asymmetric afucosylation is sufficient for full antibody-dependent cellular cytotoxicity. MAbs 5, 872–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bondarenko P. V., Second T. P., Zabrouskov V., Makarov A. A., and Zhang Z. (2009) Mass measurement and top-down HPLC/MS analysis of intact monoclonal antibodies on a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom 20, 1415–1424 [DOI] [PubMed] [Google Scholar]
- 48. Wang T. T., Sewatanon J., Memoli M. J., Wrammert J., Bournazos S., Bhaumik S. K., Pinsky B. A., Chokephaibulkit K., Onlamoon N., Pattanapanyasat K., Taubenberger J. K., Ahmed R., and Ravetch J. V. (2017) IgG antibodies to dengue enhanced for Fc gamma RIIIA binding determine disease severity. Science 355, 395–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Subedi G. P., and Barb A. W. (2016) The immunoglobulin G1 N-glycan composition affects binding to each low affinity Fc gamma receptor. MAbs 8, 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dekkers G., Treffers L., Plomp R., Bentlage A. E. H., de Boer M., Koeleman C. A. M., Lissenberg-Thunnissen S. N., Visser R., Brouwer M., Mok J. Y., Matlung H., van den Berg T. K., van Esch W. J. E., Kuijpers T. W., Wouters D., Rispens T., Wuhrer M., and Vidarsson G. (2017) Decoding the human immunoglobulin G-glycan repertoire reveals a spectrum of Fc-receptor- and complement-mediated-effector activities. Front. Immunol. 8, 877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Drescher B., Witte T., and Schmidt R. E. (2003) Glycosylation of FcgammaRIII in N163 as mechanism of regulating receptor affinity. Immunology 110, 335–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roberts J. T., and Barb A. W. (2018) A single amino acid distorts the Fc gamma receptor IIIb/CD16b structure upon binding immunoglobulin G1 and reduces affinity relative to CD16a. J. Biol. Chem. 293, 19899–19908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Smith L. K., Boukhaled G. M., Condotta S. A., Mazouz S., Guthmiller J. J., Vijay R., Butler N. S., Bruneau J., Shoukry N. H., Krawczyk C. M., and Richer M. J. (2018) Interleukin-10 directly inhibits CD8(+) T cell function by enhancing N-glycan branching to decrease antigen sensitivity. Immunity 48, 299–312.e295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bassaganas S., Allende H., Cobler L., Ortiz M. R., Llop E., de Bolos C., and Peracaula R. (2015) Inflammatory cytokines regulate the expression of glycosyltransferases involved in the biosynthesis of tumor-associated sialylated glycans in pancreatic cancer cell lines. Cytokine 75, 197–206 [DOI] [PubMed] [Google Scholar]
- 55. Parker B. L., Thaysen-Andersen M., Fazakerley D. J., Holliday M., Packer N. H., and James D. E. (2016) Terminal galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol. Cell. Proteomics 15, 141–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Colomb F., Vidal O., Bobowski M., Krzewinski-Recchi M. A., Harduin-Lepers A., Mensier E., Jaillard S., Lafitte J. J., Delannoy P., and Groux-Degroote S. (2014) TNF induces the expression of the sialyltransferase ST3Gal IV in human bronchial mucosa via MSK1/2 protein kinases and increases FliD/sialyl-Lewis(x)-mediated adhesion of Pseudomonas aeruginosa. Biochem. J. 457, 79–87 [DOI] [PubMed] [Google Scholar]
- 57. Padro M., Mejias-Luque R., Cobler L., Garrido M., Perez-Garay M., Puig S., Peracaula R., and de Bolos C. (2011) Regulation of glycosyltransferases and Lewis antigens expression by IL-1beta and IL-6 in human gastric cancer cells. Glycoconj. J. 28, 99–110 [DOI] [PubMed] [Google Scholar]
- 58. Benson V., Grobarova V., Richter J., and Fiserova A. (2010) Glycosylation regulates NK cell-mediated effector function through PI3K pathway. Int. Immunol. 22, 167–177 [DOI] [PubMed] [Google Scholar]
- 59. Yagi H., Takakura D., Roumenina L. T., Fridman W. H., Sautes-Fridman C., Kawasaki N., and Kato K. (2018) Site-specific N-glycosylation analysis of soluble Fc gamma receptor IIIb in human serum. Sci. Rep. 8, 2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Washburn N., Meccariello R., Duffner J., Getchell K., Holte K., Prod'homme T., Srinivasan K., Prenovitz R., Lansing J., Capila I., Kaudinya G., Manning A. M., and Bosques C. J. (2018) Characterization of endogenous human FcgammaRIII by mass spectrometry reveals site, allele and sequence specific glycosylation. Mol. Cell. Proteomics [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Deshpande N., Jensen P. H., Packer N. H., and Kolarich D. (2010) GlycoSpectrumScan: fishing glycopeptides from MS spectra of protease digests of human colostrum sIgA. J. Proteome Res. 9, 1063–1075 [DOI] [PubMed] [Google Scholar]
- 62. Lee J. Y., Kim J. Y., Park G. W., Cheon M. H., Kwon K. H., Ahn Y. H., Moon M. H., Lee H. J., Paik Y. K., and Yoo J. S. (2011) Targeted mass spectrometric approach for biomarker discovery and validation with nonglycosylated tryptic peptides from N-linked glycoproteins in human plasma. Mol. Cell. Proteomics 10, M111.009290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Trotta A. M., Ottaiano A., Romano C., Nasti G., Nappi A., De Divitiis C., Napolitano M., Zanotta S., Casaretti R., D'Alterio C., Avallone A., Califano D., Iaffaioli R. V., and Scala S. (2016) Prospective evaluation of cetuximab-mediated antibody-dependent cell cytotoxicity in metastatic colorectal cancer patients predicts treatment efficacy. Cancer Immunol. Res. 4, 366–374 [DOI] [PubMed] [Google Scholar]
- 64. Wu J., Edberg J. C., Redecha P. B., Bansal V., Guyre P. M., Coleman K., Salmon J. E., and Kimberly R. P. (1997) A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. J. Clin. Invest. 100, 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Koene H. R., Kleijer M., Swaak A. J., Sullivan K. E., Bijl M., Petri M. A., Kallenberg C. G., Roos D., von dem Borne A. E., and de Haas M. (1998) The Fc gammaRIIIA-158F allele is a risk factor for systemic lupus erythematosus. Arthritis Rheum. 41, 1813–1818 [DOI] [PubMed] [Google Scholar]
- 66. Dong C., Ptacek T. S., Redden D. T., Zhang K., Brown E. E., Edberg J. C., McGwin G. Jr, Alarcon G. S., Ramsey-Goldman R., Reveille J. D., Vila L. M., Petri M., Qin A., Wu J., and Kimberly R. P. (2014) Fcgamma receptor IIIa single-nucleotide polymorphisms and haplotypes affect human IgG binding and are associated with lupus nephritis in African Americans. Arthritis Rheumatol. 66, 1291–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw data available at MassIVE (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp) with the identifier MSV000083890 or via ftp://massive.ucsd.edu/MSV000083890/.