Abstract
Understanding gene regulation networks in multicellular organisms is crucial to decipher many complex physiological processes ranging from development to aging. One technique to characterize gene expression with tissue-specificity in whole organisms is single-molecule fluorescence in situ hybridization (smFISH). However, this protocol requires lengthy incubation times, and it is challenging to achieve multiplexed smFISH in a whole organism. Multiplexing techniques can yield transcriptome-level information, but they require sequential probing of different genes. The inefficient macromolecule exchange through diffusion-dominant transport across dense tissues is the major bottleneck. In this work, we address this challenge by developing a microfluidic/electrokinetic hybrid platform to enable multicycle smFISH in an intact model organism, Caenorhabditis elegans. We integrate an ion concentration polarization based ion pump with a microfluidic array to rapidly deliver and remove gene-specific probes and stripping reagents on demand in individual animals. Using our platform, we can achieve rapid smFISH, an order of magnitude faster than traditional smFISH protocols. We also demonstrate the capability to perform multicycle smFISH on the same individual samples, which is impossible to do off-chip. Our method hence provides a powerful tool to study individual-specific, spatially resolvable, and large-scale gene expression in whole organisms.
I. INTRODUCTION
Regulation of gene expression plays a critical role in many physiological processes. From embryogenesis to development to aging, gene expressions reflect unique profiles at each stage.1–3 From simple yeast and bacterial cells to complex mammalian systems, gene expression remains to be a key element that governs higher-level processes. Understanding gene regulation in multicellular organisms carries significant implications as these gene networks are often conserved across species.4
Investigations using model organisms, such as C. elegans, Drosophila, and zebrafish, have shown that gene expression is cell-/tissue-, age-, and individual-dependent.5–8 One promising technique to capture all these pieces of information is single-molecule fluorescence in situ hybridization (smFISH). Fluorescently labeled short nucleic acids act as probes; they are delivered into a fixed and permeabilized sample. These probes bind to the target mRNA. Imaging the sample can then reveal individual, fluorescent puncta, each corresponding to an individual mRNA molecule.9 To prevent false-positive signals from nonspecific binding, the probes are designed to be sequence-specific. Each probe set consists of 20–40 fluorescently labeled short nucleic acids, and a punctum only becomes resolvable once 20–30 probes bind to the same mRNA molecule. This local molecular crowding effect ensures specificity.9 This enables us to quantify and localize the expression at a cellular to subcellular level. In mounted cells and thin tissue slices, some have advanced smFISH technology with multiplexing and barcoding techniques to quantify the entire transcriptome.10,11 This approach requires multiple rounds of hybridizing the smFISH probes, stripping the probes, and rehybridizing them in the same sample. Although robustly achieved in these simple systems, adapting multiplexing techniques to whole organisms presents significant transport challenges to achieve a sufficient probe concentration in the organism. Even single rounds usually require long incubation times (e.g., days) for probe hybridization due to the combinatorial probing nature of smFISH strategy.10 Therefore, when using smFISH in whole organisms, the number of genes studied is usually limited by the number of fluorophores the imaging setup can resolve.
Intact whole organisms have dense tissues. Even after fixation and permeabilization, transporting reagents in and out of the sample can be difficult. In the traditional approach, multiple organisms are typically incubated together in a single small tube with smFISH probes, and the reagent delivery is diffusion-dependent and individual identities are lost. smFISH probes are expensive and maintaining individuality (i.e., using a separate tube for each animal) is extremely costly, time-consuming, and labor intensive. Although the traditional approach is sufficient for a single round of smFISH, multiplexing requires multiple reagents to be delivered in and out of the sample over multiple steps while tracking the same individual: (1) the original probes need to be delivered, (2) the enzymatic stripping reagents need to be delivered, (3) the dehybridized probes and stripping reagents need to be removed, and (4) the new probes need to be redelivered. With the ability to perform multicycle smFISH, we can potentially examine complete gene networks and expression dynamics with tissue- and cellular-specificity within individual whole organisms; however, the major limitation to adapting these higher-level strategies is the precise reagent delivery and removal needed in different steps.
To perform multicycle smFISH on intact multicellular organisms, macromolecular probes need to be transported efficiently and on demand through different tissues, while having precise control as each step requires different reagents and incubation times. The short nucleic acid probes are expected to be concentrated only in the organism during the probe hybridization step, while completely removed after each probing cycle. Recently, enabling electrokinetic technologies for such localized molecular concentration and extraction process have been reported. These technologies are based on a unique spatially inhomogeneous ion concentration polarization (ICP) phenomenon of ion-selective media12,13 and find a wide range of applications in precision medicine.14 For instance, programmable isotachophoresis and free-flow isoelectric focusing were made possible by a highly localized concentration gradient in microfluidic chips, integrated with perm-selective membranes or polyelectrolytes for concentration, isolation, and identification of proteins or nucleic acid biomarkers in cell lysates or plasma samples.15–20
Here, we report a platform that combines the advantages of microfluidics and electrokinetics to enable rapid and multicycle smFISH in whole organisms, using C. elegans as a model. Our device consists of two components: a microfluidic trap array21 and an ICP-based ion pump using a nanoporous cation-exchange membrane (CEM). A group of individual fixed C. elegans can be isolated in trackable locations in the microfluidic trap array. We show that the ICP-based ion pump can rapidly deliver the short nucleic acid probes to and remove from the whole worms by selectively inducing ion concentration polarization in the traps. By efficiently transporting and enriching probes across tissue barriers in the whole worm, we shorten the smFISH process by an order of magnitude. By reprobing a neuronally expressed gene in two separate rounds in adult C. elegans, we demonstrate that our method can reliably profile gene expression repeatedly in the same individuals. Our platform, therefore, lays the foundation of large-scale, multiplexed smFISH to study individual-specific, system-wide, and spatially resolvable gene expression in multicellular organisms.
II. EXPERIMENTAL METHODS AND MATERIALS
A. C. elegans strains, culture, and sample preparation
The worms used in this work were N2 strain (QLN2). All worms were grown on standard NGM agar plates with Escherichia coli (E. coli) OP50 lawns and maintained at 20 °C. Hermaphrodite worms were synchronized to Day-2 adults before being washed off plate for experiments using S-Basal. The population of worms was then sacrificed and fixed in 3.7% formaldehyde for 45 min; this process cross-links the biomolecules including native mRNAs with the surrounding tissues. After fixation, they were washed and incubated in 70% ethanol overnight at 4 °C for permeabilization of the tissues.
B. Device fabrication and materials
The master mold of the microfluidic device was fabricated with a negative photoresist, SU-8 2050 (Microchem) by UV photolithography. The microfluidic device was fabricated in polydimethylsiloxane (PDMS, Dow Corning Sylgard 184) by soft lithography. Briefly, we spin-cast the microchannel layer PDMS (10:1 ratio between the elastomer and the curing agent) on the master mold at 750 rpm for 30 s. This microchannel layer was partially cured at 75 °C for 15 min. Cut pipette tips with a filter paper at the bottom as buffer reservoirs and a 1.5 × 25 mm CEM strip (Mega a.s., Czech Republic) were placed at the designated positions on the partially cured PDMS layer, fixed by Kapton tape. More PDMS (10:1) was poured onto the microchannel layer to embed the cation-exchange membrane and buffer reservoirs. The entire device was then cured at 75 °C for 90 min. After peeling off the device from the SU-8 master, through-holes for electrical connection were cut underneath the CEM and the buffer reservoirs, and the through-holes for fluidic connection were punched by 19-gauge needles (McMaster-Carr). Finally, the device was bonded onto a precleaned coverslip using corona discharge followed by thermal bonding at 75 °C for 24 h. After the device was completed, 200 μl 2% agarose gel was placed in the input reservoir to prevent fluid leaking from the channel into the reservoir. All devices were filled with 1× S-basal buffer for 48 h to let the CEM swell properly prior to the test.
C. Electrical and imaging setup for characterizing probe delivery and removal
The external electric voltage signal was generated by a Keithley 2450A SourceMeter Instrument (Tektronix) and transmitted into the microfluidic device using platinum electrodes. The electrodes were placed in buffer reservoirs separated from the main microfluidic channel by 2% agarose gel plugs to prevent electrochemical byproducts, such as bubbles and pH changes, leaking into the microfluidic channel.
We used fluorescein isothiocyanate (FITC)-Dextran (1 mg/ml) in either the hybridization buffer or the wash buffer for smFISH to characterize the probe delivery and removal to the worms. FITC-Dextran was chosen as an approximation of smFISH probes in this characterization because it had a much cheaper reagent cost and was more easily acquired commercially. In addition, this fluorescent reagent has a similar molecular weight and charge to smFISH probes. The probe transport dynamic by fluorescence was recorded with whole field illumination on a fluorescence dissecting scope (Leica, MZ16F) with a frame rate of 5 fps and a 100 ms exposure time. To quantify the dynamics and intensities, we used a custom MATLAB code. The plots and statistics were created using GraphPad Prism.
D. Single-molecule fluorescence in situ hybridization in C. elegans
Probe design and the protocol to perform smFISH in C. elegans have been previously described.9 In our study, we used custom Stellaris smFISH probes for specific targeting gpa-3 labeled with Cal Fluor Red 610. The fixed and permeabilized samples were incubated in S-Basal for 5 min. Next, they were washed with wash buffer (10% formamide, 2× SSC, in nuclease-free water) for 5 min. For off-chip experiments, the worms were incubated in custom probes (1.25 μM) in hybridization buffer [1 g dextran sulfate, 40 μl RNase-free bovine serum albumin (BSA) (50 mg/ml), 10% formamide, 9 ml nuclease-free water] at 30 °C. For on-chip experiments, worms were first manually loaded into the device. To transport the probes into the worms, we flowed the Stellaris smFISH probes into the device using a syringe pump at 15 μl/min for 3 min, applied −150 V from the input reservoir for 5 min, and continued flow for 2 min. The device was then incubated at 30 °C while under slow flow (10 μl/h) to prevent evaporation. Prior to imaging, the worms were incubated in wash buffer for 30 min, following the previous literature.9 This short washing step was to remove free-floating probes in the microchannels which might interfere with the fluorescence imaging.
To strip the probes from the sample, we prepared a DNase I stripping reagent (0.5 U/μl). The reagent was flowed into the device at 15 μl/min for 3 min, then 10 μl/h at 30 °C for 4 h. To remove the stripped probes and DNase I, we applied +150 V from the input reservoir for 5 min. Prior to imaging, we washed the worms in wash buffer for 30 min at 15 μl/min. To rehybridize the worms, we followed the same protocol as above.
E. smFISH imaging and quantification
Imaging was performed on a spinning disk confocal microscope (PerkinElmer UltraVIEW VoX) using a Hamamatsu FLASH 4 sCMOS camera. Images were analyzed and puncta were identified using a standard software, FISH-quant.22 To quantify the signal-to-noise ratio (SNR), we measured the maximum intensity of each puncta (signal) and divided by the standard deviation of the intensities of the head of the worm, anterior to the gut (noise), using ImageJ. To quantify the signal-to-background ratio, we measured the maximum intensity of each puncta (signal) and divided by the average intensity of the head of the worm, anterior to the gut (background), using ImageJ. The plots and statistics were created using GraphPad Prism.
III. RESULTS AND DISCUSSION
A. Device design and demonstration of the operating principle
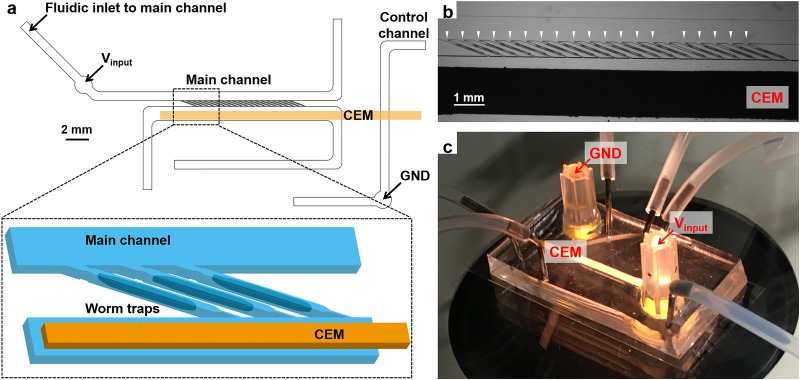
To incorporate smFISH multiplexing techniques of whole organisms, it requires the preservation of individual identities of the animals throughout the entire process. In our platform, this is guaranteed by isolating individual C. elegans into an array of trackable worm traps [Fig. 1(a)]. A group of age-synchronized adult worms can be loaded into the traps within a minute by allowing fluidic flow across the trap array [Fig. 1(b)]. As shown in Fig. 1(a), the ion pump is built by placing a strip of CEM on top of the main channel, closely adjacent to the end of the traps and bridging the main channel to a separated control channel. To induce the ion concentration polarization and control the probe transport, DC electric fields are introduced to the worm traps through the CEM by inserting one electrode in the input reservoir on the main channel and another electrode in the ground reservoir on the control channel [Figs. 1(a) and 1(c)].
FIG. 1.
(a) Schematic of the microfluidic/electrokinetic hybrid device. (b) Micrograph of individually loaded worms in worm traps. White arrowheads indicate individually loaded adult worms. (c) Image of the device setup.
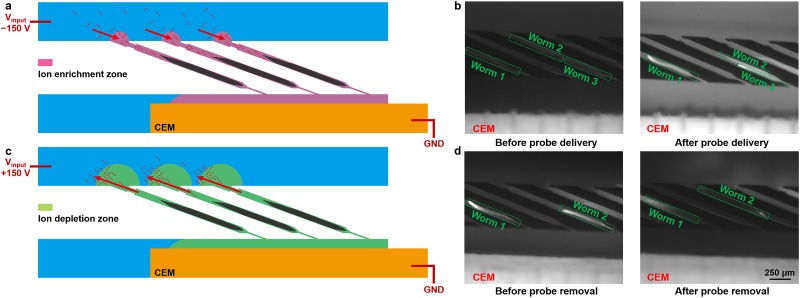
Because of the charge selectivity of the cation-exchange membrane, ion concentration polarization can be established under DC electric fields in the vicinity of the CEM,12,13,23 which is, in our case, in the worm traps. When imposing a negative voltage at the input electrode, negatively charged nucleic acid probes are continuously transported by electrophoresis into the worm traps, where an ion enrichment zone is established [Fig. 2(a)]. The electrophoretic force efficiently drives the probes through the cuticle and dense tissues of the fixed and permeabilized worms, and the negative gradient of the electric field from the main channel to the traps ensures the probes concentrated in the worms. To characterize the probe enrichment process, we used fluorescein isothiocyanate (FITC)-Dextran as a model for the smFISH probes because they have a similar molecular weight and charge. Figure 2(b) shows that after 5 min of enrichment by the ICP-based ion pump, FITC-Dextran molecules are predominately concentrated in the loaded worms, characterized by the elevated fluorescence intensity in the worm (Fig. S1 in the supplementary material).
FIG. 2.
(a) Operating principle of probe delivery. (b) Fluorescent images of FITC-Dextran (model of smFISH probes) transport into individual worms before and after the probe delivery process. The input reservoir received −150 V for 5 min. (c) Operating principle of probe removal. (d) Fluorescent images of FITC-Dextran transport out of individual worms before and after the probe removal process. The input reservoir received +150 V for 7 min. The image settings (brightness and contrast) are consistent for all fluorescent images in (b) and (d).
This electrokinetic probe enrichment strategy can drastically reduce the time required for smFISH in two stages, probe delivery and probe hybridization. In the first stage, the electrophoretically driven transport accelerates the probe delivery to target mRNA molecules in the worm, compared with the traditional diffusion-dominated method. We can define the “acceleration” of probe transport by the ratio of characteristic diffusion-based probe delivery time, , to the characteristic electrokinetically-enhanced time, . Based on the geometrical design of our microfluidic chip, the “acceleration” is scaled with (see the supplementary material for details). Here, and are the diameter and length of the worm, respectively, and is the applied electrical potential. Considering the aspect ratio of the worm and our ∼150 V applied voltage, the probe delivery can be enhanced by at least 20 folds.
In the second stage, after probes are delivered into the worm, they are enriched; this higher probe concentration results in a rapid hybridization between probes and the target mRNA molecules. The kinetics of such hybridization can be approximated as a second-order reaction.24,25 The characteristic time for 90% target mRNA molecules hybridized with at least 20 probes, , is given by , where is the enriched initial concentration of probes and is the hybridization rate constant (see the supplementary material for details). Based on the numerical simulation of ion concentration profile near an ion-selective media26,27 and the ion current profile during our experiments (data not shown), we estimate that the ion and probe concentrations in the worm are enriched by 3–5 folds during the nonequilibrium probe delivery step. In contrast, using the traditional diffusion-based protocol, the maximum probe concentration in worms can only reach as high as the equilibrium concentration in the buffer. The elevated ion concentration does not affect the probe hybridization in the hybridization buffer we used28 nor the penetration of the probes into the worms.29 Thus, the subsequent incubation time required for sufficient probe hybridization onto the target mRNAs can be reduced; this time is scaled inversely proportional to the enriched probe concentration. Traditional diffusion-based protocols require overnight incubation (12–24 h) in order to complete the probe hybridization. In contrast, combining the enhancement by probe delivery and probe hybridization with our nonequilibrium method, we can shorten the probing step during each round of smFISH to a few hours. Hence, in later smFISH experiments, we chose to perform probe delivery with our ICP-based ion pump for 5 min and a subsequent 4-h incubation for probe hybridization.
After the target mRNAs are probed and imaged, the probes can be stripped from the in situ mRNA with DNase I; this then allows for profiling of other genes. Once stripped, (1) the DNase I must be removed to allow for future probe hybridization and (2) the dehybridized fluorescent probes need to be removed from the animals; otherwise, the stripped probes are likely to elevate the background fluorescence level, which may overwhelm the true FISH signals. To accomplish this, we reverse the polarity of the input voltage to positive to develop an ion depletion zone in the traps for the probe removal. Within the ion depletion zone, the ionic strength can be reduced by 2–3 orders of magnitude.27 As a result, the high electric field in the depletion zone can focus the charged probes at the depletion front isotachophoretically,17,20,30,31 illustrated in Fig. 2(c). As the depletion zone grows and the depletion front passes through the loaded worms, charged unbound macromolecules can be removed from the animals almost completely, demonstrated by FITC-Dextran in Fig. 2(d). We designed the CEM surface to be parallel to the bottom wall of the microchannel with only 50 μm separation. Therefore, the electroconvection vortices are effectively suppressed by the large hydrodynamic resistance to avoid arresting the depletion front.
B. Characterization of macromolecular transport through intact tissues
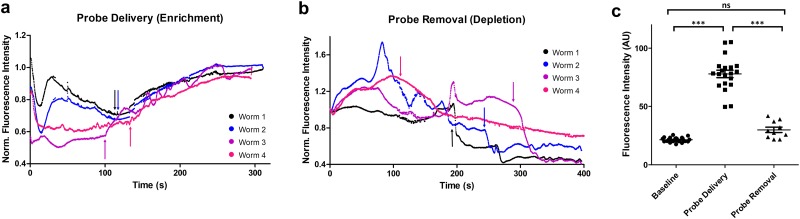
Using FITC-Dextran as a model, we further characterized the dynamics and robustness of the probe delivery [Fig. 3(a)] and removal process [Fig. 3(b)]. We measured the fluorescence intensity in the whole worm over time. During the probe delivery step and before the onset of the ion enrichment, FITC-Dextran molecules that were preloaded in the traps but not in the worms were driven out of the traps, evident by the initial decrease of the fluorescence intensity [Fig. 3(a)]. However, when the ion enrichment zone was established after about 2 min, the samples started to become brighter consistently due to the accumulation of FITC-Dextran molecules in the tissues [Fig. 3(a)]. After 5 min of electrokinetic-assisted probe delivery, the samples in the device showed a significantly higher fluorescence intensity [Fig. 3(c)]. Similarly, during the probe removal step, we observed a sudden drop of fluorescence intensity from the worms in Fig. 3(b), demonstrating the removal of these accumulated FITC-Dextran molecules by the depletion front. While we noticed that FITC-Dextran molecules were removed at different rates in the samples, we found that after 7 min of electrokinetic-assisted probe removal, the fluorescence intensity of all the worms returned to the baseline level [Fig. 3(c)]. The heterogeneity was expected as tissue compositions varied from worm-to-worm, and depletion fronts developed at different time points across the trap array. The fast dynamics enabled by the ICP-based ion pump allows us to rapidly send charged molecular probes into the whole worms repeatedly and robustly, regardless of the tissue barrier of the multicellular organisms.
FIG. 3.
(a) Quantification of fluorescence intensity dynamics in individual worms during the probe delivery process. −150 V was applied at the input reservoir. Arrows indicate when the ion enrichment zone started to establish for each worm. (b) Quantification of fluorescence intensity dynamics in individual worms during the probe removal process. +150 V was applied at the input reservoir. Arrows indicate when the ion depletion front passed through each worm. (c) Quantification of fluorescence intensity of individual worms at baseline and after probe delivery and removal. A one-way ANOVA and Tukey's multiple comparison tests were performed. (***P < 0.001; ns, not significant).
C. Rapid multicycle smFISH
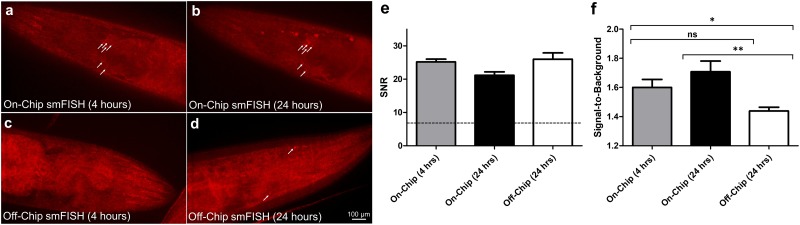
To demonstrate the utility of this device for profiling gene expression, we performed smFISH for a neuronal-expressed gene, gpa-3, and compared its performance to the traditional off-chip approach (Fig. 4). gpa-3's expression is age-related and its function is involved in food-sensing, suggesting that it plays an important role in many biological processes such as learning and development.32,33 Using our device, we improved the probe delivery and increased its local concentration into the worms, shortening the time required for smFISH hybridization step to only 4 h [Fig. 4(a)] (see Sec. II for puncta identification). After 24 h of incubation on-chip, we found that the quality of smFISH did not change [Fig. 4(b)], confirming that our rapid on-chip hybridization step was sufficient to visualize the hybridized probes. In contrast, the diffusion-dominated off-chip approach was unable to produce any obvious puncta after 4 h of incubation [Fig. 4(c)]. Instead, for the off-chip condition, we were only able to resolve puncta after the 24 h incubation period, which is the typical overnight protocol [Fig. 4(d)]. In all the samples, we found the signal-to-noise ratio (SNR) to be much higher than 7 [Fig. 4(e)], the minimal acceptable SNR previously reported,22,34 allowing us to confidently identify each punctum. Since unbound probes may contribute to the background fluorescence observed, we quantified the signal-to-background ratio at each time point to validate that the background fluorescence does not become too high to overwhelm the true signal [Fig. 4(f)]. We observed that the signal-to-background ratio was significantly higher even with only 4 h on-chip when compared to the 24 h off-chip condition [Fig. 4(f)], indicating that smFISH on our platform achieves higher quality signal with less time required. Thus, by increasing the local concentration of probes and improving the probe delivery into the animals, we were able to shorten the time required for smFISH without compromising the quality.
FIG. 4.
Fluorescent images of individual puncta, each corresponding to a gpa-3 mRNA molecule in the neuronal cells. (a) and (b) On-chip probe delivery by the ICP-based ion pump in the same worm after (a) 4 h and (b) 24 h. (c) and (d) Off-chip diffusion-dominated delivery. (c) Individual puncta were not resolved after 4 h of off-chip incubation. (d) Puncta are visible only after 24 h of off-chip incubation. White arrows are added to aid puncta visualization. (e) Signal-to-noise ratio (SNR) of the puncta at each condition. The dotted line corresponds to the minimum value acceptable (SNR = 7). (f) Signal-to-background ratio of each punctum at each condition. An unpaired, two-tailed T test was performed (**P < 0.01, *P < 0.05; ns, not significant).
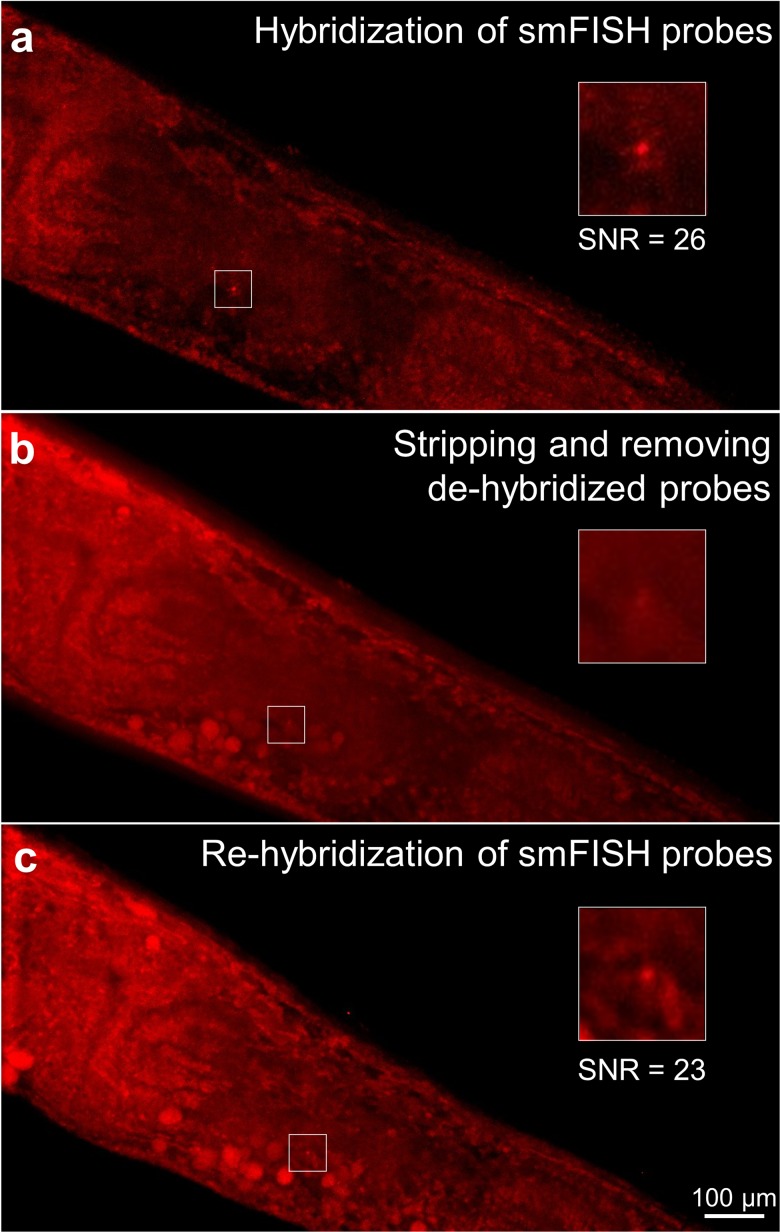
Next, we aimed to engineer our platform to enable multicycle smFISH. We probed for gpa-3, removed the probe using a DNase I stripping reagent, and reprobed gpa-3 while tracking single animals. We showed that this complex level of transport could not be achieved using traditional smFISH off-chip strategies (Fig. S2 in the supplementary material). In contrast, we achieved multicycle smFISH on-chip; first, we probed for gpa-3 using the ICP-enhanced delivery, and we were able to visualize puncta as expected, shown by a representative sample [Fig. 5(a)]. Next, we delivered DNase I to strip the hybridized probes for 4 h. Using depletion-assisted isotachophoresis, we removed both the DNase I and stripped probes [Fig. 5(b)]. We then reprobed for gpa-3 and found its expression location and SNR are similar to the initial hybridization [Fig. 5(c); Fig. S3 in the supplementary material]. The quality of the signal (SNR) was not compromised during the multicycle smFISH assay across all samples we tested (Fig. S3 in the supplementary material). If the stripping reagent was still present in the system, the readded probes would not be able to hybridize to the target mRNA molecules. Thus, this experiment demonstrated that we can efficiently deliver and remove both the probes and stripping reagents to perform multicycle smFISH.
FIG. 5.
Multicycle smFISH of gpa-3 within the same worm using ICP-enhanced smFISH, and puncta were identified using FISH-quant. (a) Individual puncta were resolved after initial hybridization. (b) After DNase I treatment for 4 h, probes were dehybridized and removed by the ICP-based ion pump. (c) The same puncta were resolved again after rehybridization enhanced by ICP. Insets show the magnified area at the vicinity of the target mRNA molecule.
IV. CONCLUSIONS
In this work, we engineered a microfluidic/electrokinetic hybrid platform to fluidically isolate whole organisms and efficiently manipulate macromolecule delivery in and out of the animals with controlled spatial and temporal resolution using an ICP-based ion pump. This unique approach has broad applications for many biological investigations. In this paper, we demonstrate its ability to enhance smFISH and enable smFISH multiplexing strategies. Using this device with an optimized protocol, we are able to perform smFISH in whole C. elegans on a much faster time scale at only 4 h. With the capability of removing charged molecules on demand, we also demonstrate that our platform is suitable to perform multiplexing strategies, which is impossible to achieve with traditional smFISH methods. This platform provides a tool for us to study large-scale gene expression within individual animals. We envision that the level of information we can collect from this method will enable us to understand complex physiological processes ranging from learning to memory to development.
SUPPLEMENTARY MATERIAL
See the supplementary material for (1) detailed mechanism explanations to estimate the enhancement of the probe delivery and probe hybridization in ICP-enhanced smFISH in whole organisms, (2) raw fluorescence intensity profile of images (shown in Fig. 2) characterizing the probe delivery into and removal from worms, (3) inefficient DNase I treatment for probe removal using the traditional off-chip smFISH method, and (4) signal-to-noise ratio (SNR) quantification of individual puncta in the multicycle smFISH assay.
ACKNOWLEDGMENTS
We acknowledge U.S. National Science Foundation (No. 1707401) and National Institutes of Health (NIH) (Nos. R01AG056436 and R01GM088333) for funding support. J.W. would like to acknowledge his support from the American Federation for Aging Research and the National Science Foundation Graduate Research Fellowships Program (No. DGE-1650044). We thank Dr. Guillaume Aubry for helping in the fabrication of the device master. G.S. would also like to thank Professor Satyajyoti Senapati and Professor Hsueh-Chia Chang at University of Notre Dame for providing us the ion exchange membrane.
Note: This article is part of the special topic, Festschrift for Professor Hsueh-Chia Chang.
REFERENCES
- 1.Schmid M., Davison T. S., Henz S. R., Pape U. J., Demar M., Vingron M., Scholkopf B., Weigel D., and Lohmann J. U., Nat. Genet. 37(5), 501–506 (2005). 10.1038/ng1543 [DOI] [PubMed] [Google Scholar]
- 2.Tomancak P., Beaton A., Weiszmann R., Kwan E., Shu S., Lewis S. E., Richards S., Ashburner M., Hartenstein V., Celniker S. E., and Rubin G. M., Genome Biol. 3(12), research0088 (2002). 10.1186/gb-2002-3-12-research0088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lund J., Tedesco P., Duke K., Wang J., Kim S. K., and Johnson T. E., Curr. Biol. 12(18), 1566–1573 (2002). 10.1016/S0960-9822(02)01146-6 [DOI] [PubMed] [Google Scholar]
- 4.Guarente L. and Kenyon C., Nature 408(6809), 255–262 (2000). 10.1038/35041700 [DOI] [PubMed] [Google Scholar]
- 5.Elowitz M. B., Levine A. J., Siggia E. D., and Swain P. S., Science 297(5584), 1183–1186 (2002). 10.1126/science.1070919 [DOI] [PubMed] [Google Scholar]
- 6.Raj A. and van Oudenaarden A., Cell 135(2), 216–226 (2008). 10.1016/j.cell.2008.09.050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rea S. L., Wu D., Cypser J. R., Vaupel J. W., and Johnson T. E., Nat. Genet. 37(8), 894–898 (2005). 10.1038/ng1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wernet M. F., Mazzoni E. O., Celik A., Duncan D. M., Duncan I., and Desplan C., Nature 440(7081), 174–180 (2006). 10.1038/nature04615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj A., van den Bogaard P., Rifkin S. A., van Oudenaarden A., and Tyagi S., Nat. Methods 5, 877 (2008). 10.1038/nmeth.1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eng C. L., Lawson M., Zhu Q., Dries R., Koulena N., Takei Y., Yun J., Cronin C., Karp C., Yuan G. C., and Cai L., Nature 568(7751), 235–239 (2019). 10.1038/s41586-019-1049-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen K. H., Boettiger A. N., Moffitt J. R., Wang S., and Zhuang X., Science 348(6233), aaa6090 (2015). 10.1126/science.aaa6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Slouka Z., Senapati S., and Chang H.-C., Annu. Rev. Anal. Chem. 7(1), 317–335 (2014). 10.1146/annurev-anchem-071213-020155 [DOI] [PubMed] [Google Scholar]
- 13.Kim S. J., Wang Y.-C., Lee J. H., Jang H., and Han J., Phys. Rev. Lett. 99(4), 044501 (2007). 10.1103/PhysRevLett.99.044501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slouka Z., Senapati S., Shah S., Lawler R., Shi Z., Stack M. S., and Chang H.-C., Talanta 145, 35–42 (2015). 10.1016/j.talanta.2015.04.083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun G., Pan Z., Senapati S., and Chang H.-C., Phys. Rev. Appl. 7(6), 064024 (2017). 10.1103/PhysRevApplied.7.064024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun G., Senapati S., and Chang H.-C., Lab Chip 16(7), 1171–1177 (2016). 10.1039/C6LC00026F [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marczak S., Senapati S., Slouka Z., and Chang H.-C., Biosens. Bioelectron. 86, 840–848 (2016). 10.1016/j.bios.2016.07.093 [DOI] [PubMed] [Google Scholar]
- 18.Cheng L.-J. and Chang H.-C., Biomicrofluidics 5(4), 46502–465028 (2011). 10.1063/1.3657928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng L.-J. and Chang H.-C., Lab Chip 14(5), 979–987 (2014). 10.1039/c3lc51023a [DOI] [PubMed] [Google Scholar]
- 20.Qu Y., Marshall L. A., and Santiago J. G., Anal. Chem. 86(15), 7264–7268 (2014). 10.1021/ac501299a [DOI] [PubMed] [Google Scholar]
- 21.Lee H., Kim S. A., Coakley S., Mugno P., Hammarlund M., Hilliard M. A., and Lu H., Lab Chip 14(23), 4513–4522 (2014). 10.1039/C4LC00789A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., and Zimmer C., Nat. Methods 10(4), 277–278 (2013). 10.1038/nmeth.2406 [DOI] [PubMed] [Google Scholar]
- 23.Rubinstein I., Electro-Diffusion of Ions (SIAM, Philadelphia, 1990). [Google Scholar]
- 24.Bercovici M., Han C. M., Liao J. C., and Santiago J. G., Proc. Natl. Acad. Sci. U.S.A. 109(28), 11127–11132 (2012). 10.1073/pnas.1205004109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsourkas A., Behlke M. A., Rose S. D., and Bao G., Nucleic Acids Res. 31(4), 1319–1330 (2003). 10.1093/nar/gkg212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yossifon G. and Chang H.-C., Phys. Rev. Lett. 101(25), 254501 (2008). 10.1103/PhysRevLett.101.254501 [DOI] [PubMed] [Google Scholar]
- 27.Yossifon G., Mushenheim P., Chang Y. C., and Chang H. C., Phys. Rev. E 79(4), 046305 (2009). 10.1103/PhysRevE.79.046305 [DOI] [PubMed] [Google Scholar]
- 28.Tsuruoka M., Yano K., Ikebukuro K., Nakayama H., Masuda Y., and Karube I., J. Biotechnol. 48(3), 201–208 (1996). 10.1016/0168-1656(96)01510-6 [DOI] [PubMed] [Google Scholar]
- 29.Fujimoto B. S., Miller J. M., Ribeiro N. S., and Schurr J. M., Biophys. J. 67(1), 304–308 (1994). 10.1016/S0006-3495(94)80481-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quist J., Janssen K. G. H., Vulto P., Hankemeier T., and van der Linden H. J., Anal. Chem. 83(20), 7910–7915 (2011). 10.1021/ac2018348 [DOI] [PubMed] [Google Scholar]
- 31.Quist J., Vulto P., van der Linden H., and Hankemeier T., Anal. Chem. 84(21), 9065–9071 (2012). 10.1021/ac301612n [DOI] [PubMed] [Google Scholar]
- 32.Hahm J.-H., Kim S., and Paik Y.-K., Aging Cell 8(4), 473–483 (2009). 10.1111/j.1474-9726.2009.00495.x [DOI] [PubMed] [Google Scholar]
- 33.Meisel J. D., Panda O., Mahanti P., Schroeder F. C., and Kim D. H., Cell 159(2), 267–280 (2014). 10.1016/j.cell.2014.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bharadwaj R., Santiago J. G., and Mohammadi B., Electrophoresis 23(16), 2729–2744 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See the supplementary material for (1) detailed mechanism explanations to estimate the enhancement of the probe delivery and probe hybridization in ICP-enhanced smFISH in whole organisms, (2) raw fluorescence intensity profile of images (shown in Fig. 2) characterizing the probe delivery into and removal from worms, (3) inefficient DNase I treatment for probe removal using the traditional off-chip smFISH method, and (4) signal-to-noise ratio (SNR) quantification of individual puncta in the multicycle smFISH assay.