Abstract
Legionella pneumophila (Lp) is a flagellated, intracellular bacterium that can cause Legionnaires’ disease (LD). Lp activates multiple innate immune receptors, and TOLLIP dampens MyD88-dependent signaling and may influence susceptibility to LD. We evaluated the effect of TOLLIP on innate immunity, pneumonia severity, and LD susceptibility in mouse lungs and human populations. To accomplish this, we evaluated the effect of TOLLIP on lung-specific Lp control and immune response and associated a common functional TOLLIP variant with Lp-induced innate immune responses and LD susceptibility in humans. After aerosol Lp infection, Tollip−/− mice demonstrated significantly fewer bacterial CFU and increased cytokine responses from BAL fluid. Tollip−/− macrophages also suppressed intracellular Lp replication in a flagellin-independent manner. The presence of a previously characterized, functionally active SNP associated with decreased TOLLIP mRNA transcript in monocytes was associated with increased TNF and IL-6 secretion after Lp stimulation of PBMC ex vivo. This genotype was separately associated with decreased LD susceptibility (309 controls, 88 cases, p = 0.008, OR 0.36, 95% CI 0.16 – 0.76) in a candidate gene association study. These results suggest that TOLLIP decreases lung-specific TLR responses to increase LD susceptibility in human populations. Better understanding of TOLLIP may lead to novel immunomodulatory therapies.
Introduction
An outbreak of severe pneumonia at a Philadelphia American Legion convention in 1976 led to identification of Legionella spp. as a cause of pneumonia.1 Legionella pneumonia ranges from a mild illness to a severe multifocal pneumonia termed Legionnaires’ disease (LD) with significant mortality.2 In mammals, Lp infects alveolar macrophages after inhalation into the deep airways.3 Lp is unique from other pulmonary pathogens; it contains flagellin, displays LPS that avoids detection by TLR4, and secretes multiple immunoregulatory proteins into the cytosol of infected macrophages to impair host immune responses.4–6 To survive intracellularly, Lp alters the phagolysosome within the macrophage, creating the Legionella-containing vacuole (LCV).6 Lp recruits proteins to the LCV that inhibit macrophage proinflammatory responses and permit its intracellular survival.7,8 IL-1β and TNF produced by infected and bystander macrophages induce antibacterial defense by activating and recruiting macrophages to the infected site 9, 10 Macrophages recognize Lp via multiple pathogen recognition receptors, including TLR2, TLR5, NLRC4, and NAIP5 in mice.11–15 Understanding the factors that influence signaling activity of these pathways may provide insight into unique mechanisms of host resistance and susceptibility to infection.
Toll-Interacting Protein (TOLLIP) regulates many innate immune processes that influence Lp immunity, including TLR2 and IL-1R, autophagy, and endosomal transport.16–19 In humans, common genetic variation in the TOLLIP gene is associated with susceptibility to multiple intracellular infections.20–22 How TOLLIP influences infectious disease host defense within the lung is not well understood. We hypothesized that TOLLIP, acting as a ubiquitin-binding autophagy receptor, negatively regulates the innate immune response after Lp infection, which results in increased susceptibility to LD.
Host genetic variation influences innate immune responses and susceptibility to many infections.23 Functional polymorphisms in TLR5 and STING are associated with altered cytokine responses and susceptibility to LD, but assessing genetic associations are challenging given the episodic nature of pneumonia outbreaks.24, 25 In prior studies, we identified a functionally active SNP at rs5743854 that is associated with decreased TOLLIP mRNA expression in monocytes and increased TNF after TLR2 stimulation.20, 26 In this paper, we characterize effect of this variant on Lp-specific macrophage immune responses in a cohort of healthy volunteers and evaluate the genetic association of this variant on LD susceptibility in a second well-characterized cohort from the Netherlands. Overall, using a mouse model, cellular studies, human cellular immunology, and a case-control population study of highly exposed individuals, we herein evaluate the role of TOLLIP on Lp pathogenesis and critical host immune mechanisms of control.
Methods
Human Subjects
Approval for human study protocols was obtained from the human subjects’ review boards at the University of Amsterdam Medical Center and the University of Washington Medical Center. All participants gave written informed consent. Genomic DNA was purified form peripheral blood leukocytes from 10ml of blood. Enrollment of the cases and controls from a Legionnaires’ disease (LD) outbreak at a flower show in the Netherlands was described previously.25, 27 Of the 188 cases identified in the original investigation of the flower show outbreak, 141 consented for the study. 18 individuals died and no DNA was available for genotyping. 88 cases were available with both DNA and epidemiologic data for TOLLIP genotyping. Control subjects (N=309) were drawn from the exhibitioners who worked at the same flower show and were at high risk for exposure to Legionella pneumophila. Studies in Seattle included 62 healthy volunteers who donated peripheral blood for immunogenetic analysis. The ethnic composition of these subjects was 73% Caucasian (45/62) and 27% East Asian (17/62).
Genotyping
Genomic DNA was isolated from whole blood and genotyping of selected SNPs was performed using a Fluidigm Biomark 96 × 96 chip (Fluidigm, Inc.). Cluster plots were visually inspected to ensure accurate genotyping calls. SNPs were manually assessed for data quality and only high-quality calls were accepted. 88 cases and 309 controls had high-quality genotyping data available for analysis. Genotypes were assessed for HWE with a Chi-square test comparing observed and expected frequencies in the control population. No SNPs violated HWE (P<0.001).
Genetic Analysis
We assessed the genetic association between the TOLLIP rs5743854 G/G genotype and Legionnaires’ disease with a recessive genetic model using Stata 13 (Stata Corp, College Station, TX) and the user-written package “genass.”28
Reagents
RPMI Medium 1640 and DMEM was purchased from Invitrogen (Carlsbad, CA.). Ultrapure LPS was purified from Salmonella minnesota R595 (List Biological Laboratories). Flagellin C (FliC) was purified from Salmonella typhimurium strain TH4778, which is flijB/fliC+. 29 Flagellin A (FlaA) was purified from L. pneumophila Corby strain by the same method with bacteria grown as described below on buffered charcoal yeast extract (BCYE) agar plates. Bone marrow was harvested from mice and grown in DMEM supplemented with 10% heat inactivated FCS (Atlas Bio, Fort Collins, CO) and 20% LCL-conditioned medium. Bone marrow-derived macrophages (BMDM) were used after 4–10 days of culture. Bronchoalveolar fluid was obtained by intratracheal lavage with cold PBS (10ml). Cells were then plated in RPMI 1640 supplemented with 10% heat-inactivated FCS. After a 4hr adherence step, the cells were washed six times with PBS before stimulation immediately or the next day.
Microbiology
Legionella pneumophila (Lp) was cultured in buffered charcoal yeast extract (BCYE) medium was prepared with α-ketoglutarate.30 Bacteria were inoculated from a frozen stock onto BCYE agar for four days at 35 C. Bacterial cultures used for cellular stimulation or replication assays were placed in RPMI culture medium supplemented with 10% FCS and thymidine. Aerosol infection experiments with WT Lp used Legionella pneumophila Philadelphia-01 strain (ATCC 33152). Macrophage experiments with mutant Lp used the Legionella pneumophila serogroup 1 JR32-derived (rpsLK88R and hsdR) strain lacking either flaA (ΔflaA Lp) or dot/icm locus (ΔdotA Lp).31, 32
Knockout Mouse Generation and Screening
Embryos from B6.Cg-Tolliptm1Kbns/Cnrm (Tollip−/−) mice were obtained from the European Mutant Mouse Archive (www.infrafrontier.eu), which were generated by Didierlaurent et al. 33 Mice were backcrossed 8 times on C57BL/6J background and were confirmed to be >95% C57BL/6J genetically by screening 150 SNP ancestry informative markers (Jax Genome Scanning Services, Jax, Inc). Genotyping was performed using DNA primers for neomycin (Forward sequence: AGG ATC TCC TGT CAT CTC ACC TTG CTC CTG; Reverse sequence AAG AAC TCG TCA AGA AGG CGA TAG AAG GCG) and the first exon of TOLLIP (Forward sequence: AGC TAC TGG GAG GCC ATA CA; Reverse sequence: CGT GTA CGG GAG ACC CAT TT). TOLLIP protein expression was confirmed in both knockout and control mice by Western blot (Supplemental Figure 1). TOLLIP antibodies for Western blotting were polyclonal (#4748, Cell Signaling). All wild type control mice were age-matched littermates of Tollip−/− mice to ensure a common genetic background.
Model of pneumonic legionellosis
The University of Washington Institutional Animal Care and Use Committee approved all animal protocols. Buffered charcoal yeast extract (BCYE) medium was prepared with alpha-ketoglutarate as described elsewhere.25 Bacteria were inoculated from a frozen stock onto BCYE agar for four days at 35C, harvested by rinsing plates with PBS, pelleted by centrifugation, and resuspended in PBS to a concentration of 1010 cfu/ml (estimated by optical density at 540nm and confirmed by quantitative culture).25 The mice were exposed to aerosolized bacteria in a whole animal exposure chamber (Biaera Technologies, Hagerstown, MD) with aerosols generated by a mini-Heart Hi-Flo jet nebulizers (Westmed, Tucson, AZ). 34 Bacterial deposition in the lungs was determined from quantitative culture of homogenized lung tissue harvested from mice euthanized with pentobarbital and exsanguinated immediately after aerosol exposure. At subsequent time points, mice were euthanized and exsanguinated for tissue harvests. The trachea was cannulated and the right lung was lavaged with 0.85% saline/0.6mml/L EDTA to determine cell counts and differentials. The left lung was homogenized in PBS and serially diluted in Mueller-Hinton broth for quantitative culture on BCYE agar. The remaining lung homogenate was mixed 1:1 with lysis buffer containing 2x protease inhibitor mixture, incubated for 30min on ice, clarified by centrifugation at 2500rpm, and then the supernatant was saved at −80C.
Histology
To prepare organs for histology, the lung was inflated to 15cm pressure with 4% paraformaldehyde, fixed in the same solution, embedded in paraffin, and then 4μm sections were generated. Sections stained with hematoxylin and eosin were examined by a pathologist blinded to mouse genotype.
Cytokine analysis
Multiplex cytokine analysis was performed as follows: samples of WT and Tollip−/− mouse bronchoalveolar lavage fluid were thawed on ice and centrifuged at 10,000g for 5–10 minutes to remove particulates. 50μL of each sample was added in triplicate to a 96-well plate. Samples were then added to 50μL of magnetic Simplex beads and incubated with shaking for 1hr at room temperature. Beads were washed 3X and stained with detection antibody cocktail (IFNγ; IL-12p70; IL-13; IL-1β; IL-2; IL-4; IL-5; IL-6; TNF; GM-CSF; IL-18; IL-10; IL-17A; IL-22; IL-23; IL-27; IL-9; GROα; IP-10; MCP-1; MCP-3; MIP-1α; MIP-1β; MIP-2; RANTES; Eotaxin; ThermoFisher Scientific) and incubated with shaking for 30min at room temperature. Beads were washed 3X and incubated with streptavidin-HRP with shaking for 30min at RT. Beads with sample cytokines and detection antibody were resuspended in 120μL of Reading Buffer and cytokine data was acquired immediately on Luminex™ 100/200. Analytes were captured by Ab-coated, fluorochrome-embedded microspheres and detected by biotin-streptavidin-PE using reagents purchased from R&D or Luminex. For single cytokine analysis, concentrations were determined with ELISA (R&D Systems) according to the manufacturer’s instructions.
Western blotting
Cell extracts were prepared from BMDM by lysis with 100μl Cell Extraction Buffer (Life Technologies) along with protease inhibitors (Thermo Scientific). Lysates were stored at −20° C. Western blotting of lysates was performed on 4–15% polyacrylamide gels and transferred onto PVDF membrane. Antibodies included anti-TOLLIP antibody (Pierce PA5–30747) at 1:1000 dilution, anti-GAPDH (Sigma, catalog A2066) at 1:2000 dilution, and anti-rabbit IgG-HRP secondary antibody (Sigma, catalog A0545) at 1:2000 dilution.
Statistics
For in vivo CFU count analysis, comparisons between groups were made by the Mann-Whitney test without presupposition of normality of the data. For mouse experiments, groups of 5 mice were compared with one another unless otherwise indicated. Cytokine measurements and cell counts were compared using a two-sided Students’ t-test. A value of p < 0.05 was considered a statistically significant result. Statistics were calculated using Prism version 7.1 (GraphPad, Inc.).
Results
Aerosolized lung infection of mice with Lp
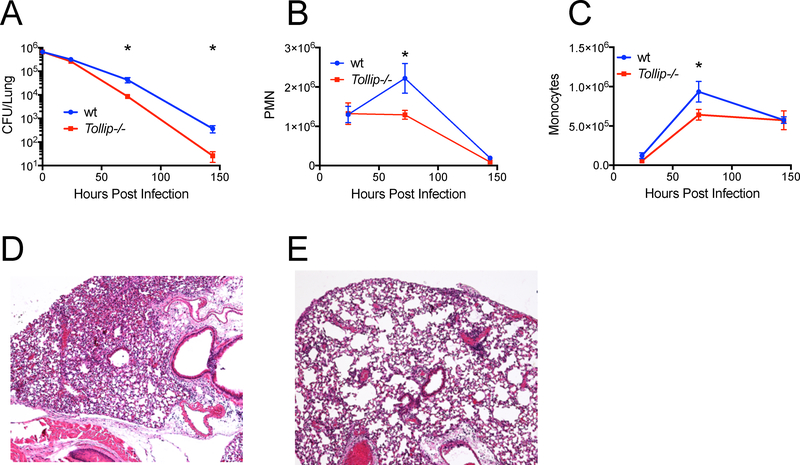
To understand the role of TOLLIP in Lp pathogenesis, we re-derived C57Bl/6 mice lacking the Tollip gene from embryos and confirmed lack of TOLLIP expression by Western blot (Supplemental Figure 1). We infected groups of 5 WT and 5 Tollip−/− mice with Lp Philadelphia-1 strain and measured lung CFU at 0, 24, 72, and 144 hours after infection. Tollip−/− mice demonstrated significantly fewer CFU in the lung compared with WT littermates at 72 and 144hr after infection (Figure 1A, mean WT 4.3×105 CFU, mean Tollip−/− 8.5×104 CFU at 72hr, p = 0.014). We also examined the recruitment of neutrophils and mononuclear cells to the lungs after infection in these mice. We collected bronchoalveolar lavage (BAL) fluid and measured cell counts at 24, 72, and 144 hours after infection. We did not detect significant differences in cell counts in Tollip−/− mice at 24 hours after infection, but by 72h after infection significantly fewer PMN were present in BAL samples harvested from Tollip−/− lungs. (Figure 1B, WT mean 2.2×106 cells/ml, mean Tollip−/− 1.3×106 cells/ml, p = 0.046). Similarly, monocyte counts were increased in WT mice compared with Tollip−/− mice after 72hr of infection (Figure 1C, mean WT 9.4×105 cells/ml, mean Tollip−/− 6.4×105 cells/ml, p = 0.018). After 72h, Tollip−/− mice demonstrated reduced airspace filling and leukocyte infiltration (Figure 1D) compared with controls (Figure 1E) by histopathology observations. In summary, Tollip−/− mice demonstrated accelerated bacterial clearance and decreased pulmonary inflammation after inhalation of Lp compared to WT controls.
Figure 1. TOLLIP-deficient mice demonstrate enhanced clearance of L. pneumophila (Lp) after aerosol infection.
Mice were infected with approximately 106 CFU WT Lp Philadelphia-1 strain and at 24, 72, and 144 hours post infection, lungs were harvested. A) Bacterial counts were measured by CFU on BCYE agar plates. B) Total polymorphonuclear cell counts from bronchoalveolar fluid. C) Total mononuclear cells from bronchoalveolar fluid samples. D) Hematoxylin and eosin staining of representative sections of D) WT and E) Tollip−/− mouse lung 72hr after Lp aerosol infection. * p< 0.05, Student’s t-test, two sided. Error bars represent +/− SEM. N = 5 mice in each group at each time point. Data are representative of three independent experiments.
In Vivo Cytokine Responses After Lp Infection
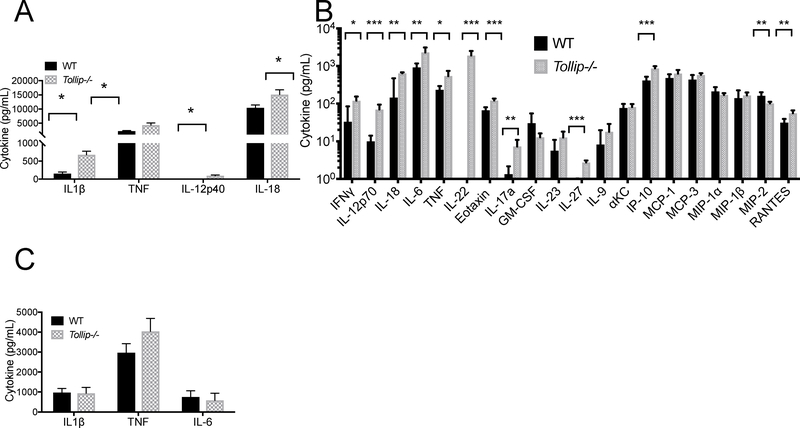
Given TOLLIP’s known role regulating innate immune responses, we next evaluated whether TOLLIP influenced lung cytokine concentrations after Lp infection. Twenty-four hours after aerosol infection with 106 CFU Lp, we euthanized mice (n = 5 per group) and measured cytokine responses from BAL fluid and serum. Tollip−/− mice demonstrated significantly increased levels of multiple cytokines critical for Lp control in BAL fluid, including IL-1β (Figure 2A; p = 0.001), TNF (p = 0.025), IL-12p40 (p = 0.001), and IL-18 (p = 0.04) by ELISA. We also evaluated a panel of 26 cytokines from BAL fluid in WT and Tollip−/− mice after Lp infection by multiplex array. We found that Tollip−/− mice produce significantly more IFNγ, IL-12p70, IL-18, IL-6, TNF, IL-22, IL-17A, Eotaxin, IL-27, and IP-10 (p<0.05 with false discovery rate < 1%, Figure 2B). We detected significantly decreased MIP-2 in Tollip−/− mice, but no differences in GM-CSF, IL-23, IL-9, αKC, MCP-1, MCP-3, MIP-1α, and MIP-1β. IL-13, IL-2, IL-4, IL-5, and IL-10 cytokine concentrations were below the limit of detection (data not shown). We did not detect statistically significant differences between WT and Tollip−/− cytokine concentrations in serum (Figure 2C). In summary, Tollip−/− mice demonstrated broadly increased cytokine responses from BAL fluid but not in serum after Lp infection.
Figure 2. Cytokine Responses from WT and Tollip−/− mice after L. pneumophila (Lp) aerosol infection.
Mice were infected with 106 CFU WT Lp Philadelphia-1 strain and after 24 hours, cytokine concentrations were measured from bronchoalveolar (BAL) fluid and serum. A) BAL cytokine concentrations measured by ELISA from WT and Tollip−/− mice. * p < 0.05, Student’s two-sided t-test. B) BAL cytokines, measured by Luminex cytokine assay 24 hours after Lp aerosol infection. * p < 0.05, ** p < 0.01, *** p < 0.001, at 1% false discovery rate. Error bars represent +/− SEM. C) Serum cytokine concentrations measured by ELISA. * p < 0.05, Student’s two-sided t-test. Error bars represent +/− SEM. Data are representative of three independent experiments with N = 5 in each group.
Pattern-recognition Receptor (PRR) Responses and TOLLIP
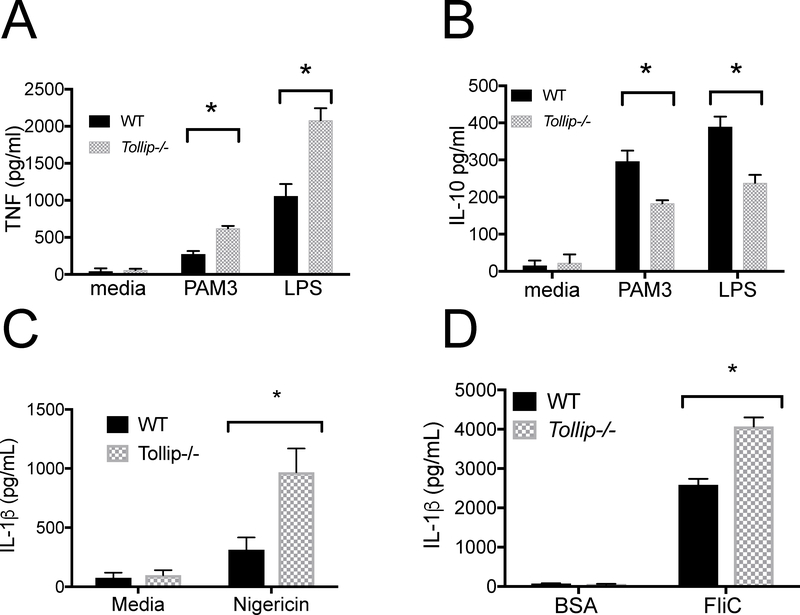
Macrophages are essential reservoirs for Lp in vivo35 TOLLIP downregulates PRR signaling in macrophages, however, and which specific PRR signaling complexes explain the improved control of Lp infection is not known. We hypothesized that multiple PRRs that are necessary for Lp control, including TLR2, TLR4, NLRC4, and NLRP3, are influenced by TOLLIP in macrophages. To evaluate the role of TOLLIP on TLR signaling, we stimulated WT and Tollip−/− peritoneal macrophages (PEM) with PAM3 (TLR2/1 agonist) or LPS (TLR4 agonist) for 24 hours, then measured TNF and IL-10 cytokine concentrations from cell culture supernatants by ELISA. Tollip−/− macrophages secreted increased concentrations of TNF after PAM3 (Figure 3A, p = 0.002) or LPS stimulation (Figure 3A, p = 0.01) when compared with WT macrophages. Conversely, Tollip−/− PEM secreted significantly less IL-10 than WT PEM after 24 hours’ stimulation with either PAM3 (p = 0.019) or LPS (p = 0.012, Figure 3B).
Figure 3. WT and Tollip−/− Macrophage Cytokine Responses after TLR and NLR Stimulation.
Bone marrow-derived macrophages (BMDM) were isolated from WT and Tollip−/− mice and stimulated with media, PAM3 (250ng/ml), a TLR 2/1 agonist, or LPS (10ng/ml), TLR4 agonist, for 24hr, and concentrations of A) TNF and B) IL-10 were measured from cellular supernatants. C) We stimulated mouse BMDM with LPS 100pg/ml for 2 hours, then stimulated overnight with C) media control or the NLRP3 ligand nigericin (100μg/ml) and D) NLRC4 ligand Salmonella flagellin (FliC, 100ng/ml) or bovine serum albumin (100ng/ml), coincubated with Lipofectamine 2000 to permit cytosolic translocation of each protein. IL-1β concentrations from cellular supernatants were measured by ELISA. Error bars are means ± SEM; statistical analyses were performed by paired t-tests using Prism 7.0e. * p < 0.05, Student’s two-sided t-test. The experiment shown is representative of three independent experiments, each conducted in triplicate.
We also evaluated the effect of TOLLIP deficiency on NLRP3- and NLRC4-induced cytokine responses. We stimulated WT and Tollip−/− BMDM with 100pg/ml LPS, followed 2 hours later by nigericin (NLRP3 ligand; 10μg/mL). We found that BMDM isolated from Tollip−/− mice demonstrated significantly increased IL-1β release when compared to BMDM from WT mice (Figure 3C, p = 0.026). Next, we compared IL-1β secretion from BMDM transfected with flagellin (NLRC4 ligand) via coincubation with cationic lipids as compared to bovine serum albumin-transfected BMDM controls. As with nigericin, we found that Tollip−/− BMDM secreted significantly more IL-1β after overnight incubation (Figure 3D, p = 0.005). No BMDM, from either WT or Tollip−/− mice, produced detectable TNF or IL-6 after flagellin stimulation without transfection (50 ng/ml, data not shown), which is consistent with prior data demonstrating that murine BMDM do not express the flagellin receptor TLR5 on the cell surface.36 These data demonstrate that TOLLIP-deficient macrophages induce increased TNF and IL-1β after TLR and NLR stimulation, responses that may contribute to the in vivo phenotype observed in Figure 1.
Cytokine Responses to Live Lp Macrophage Infection
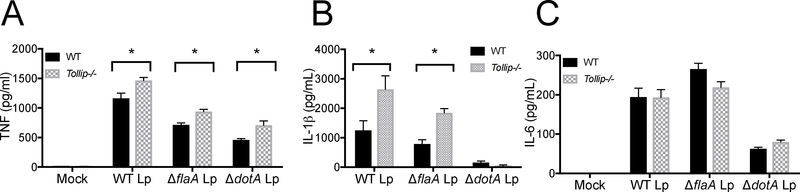
Lp influences immune activation in macrophages, so we tested the hypothesis that cytokine responses in TOLLIP-deficient macrophages were increased after live Lp infection in vitro. We plated 105 BMDM from WT and Tollip−/− mice and infected them at MOI 10 with wild-type Lp, ΔflaA Lp, or ΔdotA Lp that lacks the Type IV secretion system, making them incapable of intracellular replication, and collected supernatants after 24 hours’ infection. As compared to WT BMDM controls, BMDM lacking TOLLIP secreted significantly more TNF after infection with WT (Figure 4A, p = 0.025), ΔflaA Lp (Figure 4A, p = 0.004), and ΔdotA Lp (Figure 4A, p = 0.018). Further, IL-1β was increased in Tollip−/− BMDM after WT Lp (Figure 4B, p = 0.047) and ΔflaA Lp (Figure 4B, p = 0.002) infection, but no IL-1β was detected after ΔDotA Lp incubation. IL-6 secretion was not significantly different between WT and Tollip−/− BMDM after infection with any of the Lp straines tested (Figure 4C). These data show that TOLLIP deletion induces more TNF and IL-1β after infection with WT or ΔflaA Lp, suggesting that flagellin is not necessary for altered Lp cytokine responses by TOLLIP.
Figure 4. WT and Tollip−/− Macrophage Cytokine Responses after L. pneumophila (Lp) Infection in vitro.
Bone marrow-derived macrophages were isolated and infected with either negative control (mock), WT L. pneumophila (WT Lp), flagellin-deficient L. pneumophila (ΔflaA Lp), or dot-icm-locus-deficient L. pneumophila (ΔdotA Lp) on the JR32 background at MOI 10 overnight and A) TNF, B) IL-1β, and C) IL-6 concentrations were measured from cellular supernatants. * p < 0.05, Student’s two-sided t-test. Experiment shown is representative of three independent experiments, each performed in triplicate. Error bars represent +/− SEM of mean.
Intracellular Replication in Murine Macrophages
Although TOLLIP influences the innate immune response to Lp, many of these responses may be induced by bystander macrophages. Further, Lp induces an immune signature by inhibiting host protein translation.37 We assessed overall control of Lp within macrophages by measuring Lp intracellular replication in macrophages. We infected WT and Tollip−/− BMDM with ΔflaA Lp (MOI 0.02), which is replication-competent in macrophages.38 Tollip−/− macrophages markedly suppressed replication ΔflaA Lp in comparison with WT BMDM (Figure 5A, p < 0.001). These data demonstrate that Lp replication is suppressed within Tollip−/− macrophages.
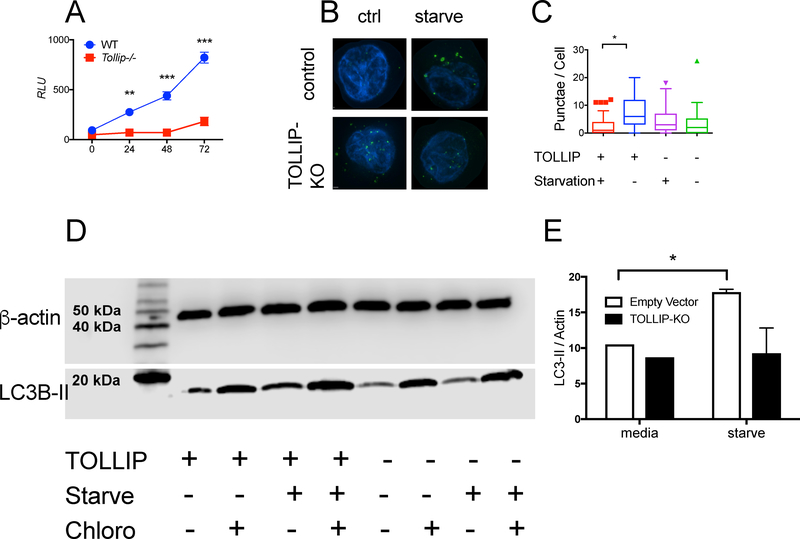
Figure 5. Intracellular L. pneumophila (Lp) Replication and Nonspecific Autophagy in WT and Tollip−/− Macrophages.
A) Bone marrow-derived macrophages (BMDM) from WT and Tollip−/− mice were infected with flagellin-deficient L. pneumophila (JR32 strain) expressing the luminescent lux gene. Relative luminescence was measured over time. B) Representative images depicting LC3+ organelles (green) and DAPI-stained nuclei (blue) from control (EV) or THP-1 cells with TOLLIP deleted via CRISPR-Cas9 gene editing (TOLLIP-KO) in RPMI supplemented with 10% FCS (ctrl) or EBSS (starve) for 4 hours. C) Tukey plot of the number of autophagosomes counted from 100 cells in EV and TOLLIP-KO cells in nutrient-enriched or starvation medium after 4 hours. Box demonstrates median and interquartile range of values. * p < 0.05, Mann-Whitney U-test, data is representative of three independent experiments. D) Western blot of THP-1 cells comparing LC3-II expression with β-actin as a control in TOLLIP-KO and control cell lines under nutrient-rich and starvation conditions, with or without chloroquine. E) Quantitative analysis of LC3-II expression, normalized to β-actin, in TOLLIP-deficient cells in control and TOLLIP-KO cells. Data are expressed as mean +/− SD of three separate experiments grouped together. * p < 0.05 by Student’s t test.
Nonselective Autophagy in Macrophages
The mechanism by which TOLLIP influences Lp-specific cytokine responses and controls intracellular replication is not clear. TOLLIP is an autophagy receptor and influences clearance of lipids from atherosclerotic plaques and insoluble protein aggregates in brain tissue.18, 39 Macrophages use autophagy for direct antimicrobial killing, but also to dampen immune responses.40 However, the role of TOLLIP on nonspecific autophagy in macrophages has not been established. We cultured THP-1 cells with TOLLIP deleted (TOLLIP-KO, Supplemental Figure 2) using CRISPR-Cas9 gene editing, along with empty vector control (EV) cells, for four hours in a nutrient rich (RPMI supplemented with 10% FCS) or starvation (EBSS) medium. Under typical conditions, starvation inhibits mTOR, a constitutive inhibitor of autophagic flux, leading to transient induction and organization of LC3-II, which forms the structural backbone of the autophagosome and attracts autophagy receptors to coordinate fusion with the lysosome.41 Starvation of EV cells for four hours induced increased numbers of LC3+ autophagosomes (Figure 5B and 5C, p < 0.001) compared to starvation of TOLLIP-KO cells. There were no significant differences in the number of autophagosomes present in EV and TOLLIP-KO cells in nutrient-rich conditions. Furthermore, EV cells, but not TOLLIP-KO cells incubated in starvation medium (EBSS) for four hours demonstrated significantly more LC3-II protein expression than EV cells incubated with nutrient-rich media (RPMI; Figure 5D and 5E, p < 0.05). Addition of chloroquine, a known inhibitor of lysosomal fusion and LC3 degradation, led to increased LC3-II levels in both EV and TOLLIP-KO cells (Figure 5D and 5E). We did not detect differences in ubiquitin staining between EV and TOLLIP-KO cells (Supplemental Figure 3). This data suggests that TOLLIP deficiency leads to reduced autophagic flux in monocytes and offers insight into the potential role of TOLLIP on specific autophagy functions.
TOLLIP SNP rs5743854 is Associated with Increased Cytokine Responses to Live Lp
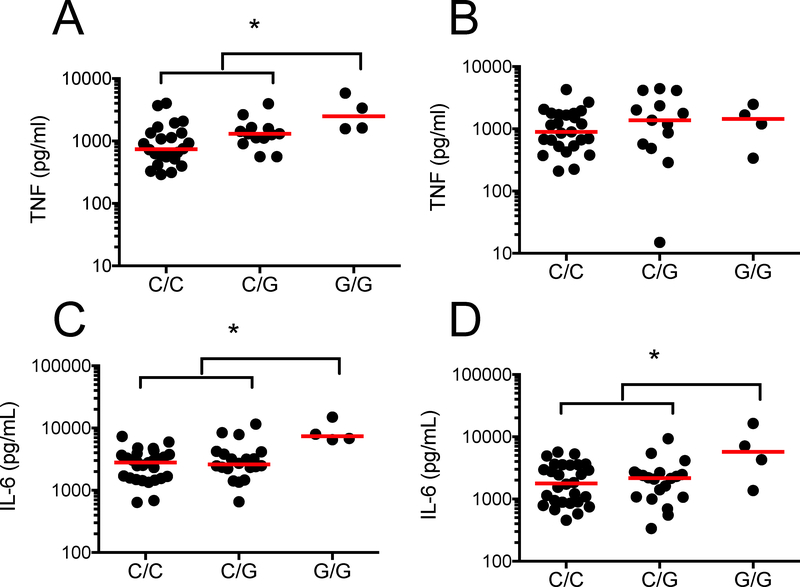
In prior studies, we characterized a functionally active promoter-region SNP in the TOLLIP gene region, rs5743854, that is associated with decreased TOLLIP expression in monocytes.42 We hypothesized that macrophages from individuals with this SNP would be associated with increased TNF responses after WT Lp and ΔflaA Lp infection. We isolated peripheral blood mononuclear cells from 62 healthy volunteers in the Seattle area and stimulated them with live WT and ΔflaA Lp (MOI 1) overnight, then measured IL-6 and TNF concentrations in cellular supernatants. Individuals with rs5743854 G/G genotype demonstrated increased TNF responses to WT Lp (Figure 6A p = 0.013; C/C = 34, G/C = 24, G/G = 4) but not ΔflaA Lp (Figure 6B, p = 0.82) when compared to those with either C/C or G/C genotype. Similarly, the same individuals demonstrated increased IL-6 responses to WT Lp (Figure 6C, p = 0.0007) and ΔflaA Lp (Figure 6D, p = 0.037). Thus, the presence of a functionally active common genetic variant that is associated with decreased TOLLIP mRNA expression in monocytes is also associated with increased TNF secretion after Lp infection.
Figure 6. rs5743854 G/G allele is associated with increased proinflammatory cytokine secretion after L. pneumophila (Lp) infection.
Peripheral blood mononuclear cells were isolated from 62 healthy volunteers in Seattle and stimulated with media, WT Lp (MOI 1) or ΔflaA Lp (MOI 1) for 24 hours. Secreted TNF and IL-6 levels were measured in supernatants via ELISA. TNF responses after A) WT Lp or B) ΔflaA Lp infection, stratified by rs5743854 genotype. Number of individuals tested per genotype -- C/C = 34, G/C = 24, G/G = 4. IL-6 responses after C) WT Lp or D) ΔflaA Lp infection, stratified by rs5743854 genotype. Dots represent cytokine values for individual volunteers. Median value represented by red lines. * p < 0.05 by Mann-Whitney U-test in a recessive genetic model (GG vs (CC + CG)).
TOLLIP SNP rs5743854 is Associated with Decreased Risk for LD
We further hypothesized that TOLLIP-deficiency (genotype G/G) is associated with susceptibility to LD in humans.16, 18 We performed a candidate gene case-control study on a cohort of individuals (N=88 LD cases and 309 controls) were exposed to Lp during a flower show.43 The TOLLIP SNP rs5743854 G/G genotype was associated with decreased LD susceptibility in a recessive model (p = 0.032; OR 0.44, 95% confidence interval (CI) 0.22 – 0.96). After adjustment for smoking, age, gender, and comorbidities as covariates, the significance of this association increased (Table 1, p = 0.008, OR 0.36, 95% CI 0.16 – 0.76). We evaluated the this association in relation to other LD susceptibility SNPs in TLR525 and in STING.24 We conducted a logistic regression evaluating the association between rs5743854 G/G genotype and LD with presence of the TLR5 stop codon allele or the HAQ allele of STING as covariates. In the presence of inactivated TLR5, the association of rs5743854 G/G genotype remains robust (p = 0.009, 95% CI 0.16 – 0.77, but the presence of STING deficiency abolishes the association (p = 0.273, 95% CI 0.17 – 1.65). Overall, these data demonstrate that the presence of the TOLLIP deficiency SNP is associated with increased cytokine responses after Lp infection and decreased risk for LD after exposure in a well-characterized case-control study.
Table 1.
Distribution of TOLLIP rs5743854 G allele in Netherlands cohort.
| Control | Cases | P value | Odds Ratio (95% CI) | |
|---|---|---|---|---|
| N | 309 | 88 | ||
| Male, f* (n) | 0.51 (159) | 0.52 (44) | 0.128 | |
| Age (median, IQR) | 49.6 (35.2 – 56.1) | 64.7 (54.2 – 71.5) | < 0.001 | |
| Smoker, f*(n) | 0.31 | 0.49 | 0.022 | |
| COPD, f (n) | 0.10 (24) | 0.07 (6) | ||
| Diabetes Mellitus, f (n) | 0.04 (3) | 0.09 (8) | ||
| Cancer, f(n) | 0.02 (2) | 0.04 (4) | ||
| Hx. Transplant, f(n) | 0.02 (2) | 0 (0) | ||
| Autoimmune Dz, f(n) | 0 (0) | 0.02 (2) | ||
| Alcohol Use, f(n) | 0.73 (61) | 0.35 (32) | ||
| Rs5743854 genotype | 0.008 | 0.35 (0.16 – 0.76)† | ||
| C/C f(n) | 0.32 (99) | 0.33 (29) | ||
| G/C | 0.41 (127) | 0.52 (46) | ||
| G/G | 0.27 (83) | 0.14 (12) |
f=frequency
recessive genetic model adjusted for gender, age, and comorbidities.
Discussion
Deletion of the TOLLIP gene in mice leads to faster clearance of Lp after aerosol infection and increased proinflammatory cytokine responses within the lungs of Lp-infected mice. Importantly, TOLLIP-deficient macrophages are resistant to Lp intracellular replication independently of flagellin, suggesting that TOLLIP acts independently of TLR5 and NLRC4 to influence immune responses. We link this finding to human immune responses and susceptibility to LD by demonstrating that a common, functionally active variant associated with TOLLIP deficiency (rs5743854 G/G genotype) is associated with increased cytokine responses to Lp and decreased risk for developing LD in a genetic case-control cohort from the Netherlands. Thus, we found that TOLLIP acts broadly to diminish innate immune responses and influence Lp pathogenesis.
We hypothesize that TOLLIP influences TLR2 signaling primarily to influence Lp disease. Lp is recognized by TLR2 (via Lp peptidoglycan-associated lipoprotein), TLR5, NLRP3, NLRC4, and AIM2. TNF is increased and IL-10 is decreased in TOLLIP-deficient macrophages after TLR2 stimulation, which both suppress intracellular Lp replication. Infection of Tollip−/− macrophages with ΔflaA Lp induces increased TNF and IL-1β and diminished intracellular replication, further supporting the critical effects of TOLLIP on TLR2 activity as sufficient for TOLLIP’s effect on Lp pathogenesis. Several questions remain, however. We demonstrate that TOLLIP acts upon TLR signaling and also influences IL-1β induction, even after infection with ΔflaA Lp. IL-1β is induced by the inflammasome, and inflammasome activation requires two signals: a priming signal, often MyD88-dependent, that upregulates expression of inflammasome components and pro-IL-1β, followed by an activation signal induced by ATP or nigericin for NLRP3, or cytosolic flagellin for NLRC4.44 TOLLIP-deficient macrophages demonstrate hyperactive induction of the first signal. TOLLIP may diminish the second signal of inflammasome activation as well. TOLLIP contains the CUE ubiquitin-binding domain, which has multiple binding partners and may thus influence signaling across multiple innate immune pathways. Other autophagy proteins, such as Atg16L, diminish inflammasome activation.45 Further, linear ubiquitin binding accessory protein SHARPIN suppresses both TLR and inflammasome activation directly.46 Detailed study of TOLLIP’s activity after intracellular infection may provide insight into how ubiquitin signaling may influence innate immunity.
Lp avoids antimicrobial killing via multiple mechanisms, including by impairing autophagy.47 However, we found that TOLLIP deficient macrophages demonstrated impaired starvation-induced autophagic flux, while simultaneously inhibiting Lp replication. To reconcile these seemingly contradictory data, we hypothesize that TOLLIP deficiency impairs the capacity for autophagy-induced immune inhibition. Depletion of critical autophagy proteins LC3 or Beclin leads to increased IL-1β production via two mechanisms: 1) decreased degradation of pro-IL-1β and 2) impaired degradation of inflammasome components. 48 If TOLLIP similarly influences immune responses, then different autophagy receptors may impact specific functions. One of the challenges to using small molecule autophagy inhibitors in vivo are side effects from off target functions. Therefore, this insight may identify novel drug targets that influence autophagy innate immune responses with minimal non-immune activity.
Intriguingly, we identified that TLR2 signaling in the absence of TOLLIP induces increased TNF and pro-inflammatory cytokines but diminished anti-inflammatory IL-10. The mechanism of this discrepancy is unclear. First, TOLLIP may selectively suppresses proinflammatory cytokine production in the TLR signaling cascade via novel, non-IRAK binding partners. Selective TRIF signaling, which occurs via nonclassical TLR4 activation induces Type I interferon activation followed by IL-10 induction.49 Alternately, induction of ER stress from infection in the setting TOLLIP deficiency may alter the cytokine balance in macrophages. ER stress occurs during the unfolded protein response (UPR), and a characteristic of ER stress is the selective induction of proinflammatory cytokines and inflammation.50 TOLLIP is a critical receptor for recycling insoluble protein aggregates, suggesting its importance in the UPR.18 Further understanding of how TOLLIP selectively influences proinflammatory cytokine induction will provide novel insights into immune regulation.
TOLLIP SNP rs5743854 was associated with increased cytokine responses to Lp and decreased susceptibility to LD. This finding is consistent with prior observations that the TOLLIP rs5743854 G allele was functionally active, associated with decreased TOLLIP mRNA expression in monocytes, and altered the innate and adaptive immune responses to mycobacteria. 26 Currently, genetic variation in TOLLIP is also associated with susceptibility to tuberculosis, leprosy, leishmaniasis, and malaria. However, presence of the G/G allele is associated with increased risk for tuberculosis but decreased risk for LD and increased proinflammatory cytokine responses after infection with both pathogens. Given this well-characterized phenotype, we believe that the differences in susceptibility demonstrate the differential survival strategies of these two pathogens. Lp induces decreased inflammation to survive but M. tuberculosis requires inflammation in later stages of disease. Further comparative studies across diseases will improve our understanding of the role of inflammation in multiple infections.
Limitations of our study included the possible confounding of the genetic findings by heterogeneity of the population substructure.51 We performed logistic regression by self-identified ethnicity and comorbidity and report these results in this paper. Another possible limitation could be misclassification of cases and controls. However, study participants were followed for a year to determine case status. rs5743854 G/G genotype is a functionally active SNP associated with TOLLIP deficiency in monocytes26 and is associated with increased Lp-specific PBMC cytokine responses, suggesting that this data represents effects of TOLLIP deficiency. We used different Lp strains for in vivo infection experiments and macrophage assays – Philadelphia-1 and JR32 for ΔflaA and ΔdotA. These strains demonstrate some differences in the host response, apart from the presence of flagellin, including the presence of the lvh locus.52−54 We confirmed TOLLIP’s effect using specific ligands and live bacteria, and we controlled each infection with a comparison with WT littermates. Despite this, TOLLIP diminished TLR2-dependent activity and impaired the immune response to Lp in vivo and in human populations.
In this paper, we demonstrate that TOLLIP deficiency increases Lp clearance in macrophages and in vivo by influencing TLR2 activity. We also found that presence of a known TOLLIP deficiency genotype is associated with decreased susceptibility to LD. Further studies targeting TOLLIP as a target for therapeutics may lead to novel treatments for intracellular infections and inflammatory disorders.
Supplementary Material
Acknowledgements
We would like to thank the families who participated in the study. We thank Kevin Hybiske, Meghan Zuck, and Stephen Hawn for valuable intellectual contributions and assay development. We would also acknowledge the support of the Cell Analysis Flow Cytometry and Imaging Core in the Department of Immunology at the University of Washington. This work was supported by R01 AI136971, K08 AI102971, and VA R&D to JAS, R01 AI093646 to SJS, K24 AI137310 to TRH, K08 HL127075 to CFH. Lp strains were a gift of Klaus Heuner and Russell Vance.
Footnotes
Conflict of Interest: The authors declare no financial or personal conflicts of interest to report.
References
- 1.Phin N, Parry-Ford F, Harrison T, Stagg HR, Zhang N, Kumar K et al. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect Dis 2014; 14(10): 1011–1021. [DOI] [PubMed] [Google Scholar]
- 2.Dominguez A, Alvarez J, Sabria M, Carmona G, Torner N, Oviedo M et al. Factors influencing the case-fatality rate of Legionnaires’ disease. Int J Tuberc Lung Dis 2009; 13(3): 407–412. [PubMed] [Google Scholar]
- 3.Horwitz MA. Phagocytosis of the Legionnaires’ disease bacterium (Legionella pneumophila) occurs by a novel mechanism: engulfment within a pseudopod coil. Cell 1984; 36(1): 27–33. [DOI] [PubMed] [Google Scholar]
- 4.Bornstein N, Marmet D, Dumaine MH, Surgot M, Fleurette J. Detection of flagella in 278 Legionella strains by latex reagent sensitized with antiflagellum immunoglobulins. J Clin Microbiol 1991; 29(5): 953–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Girard R, Pedron T, Uematsu S, Balloy V, Chignard M, Akira S et al. Lipopolysaccharides from Legionella and Rhizobium stimulate mouse bone marrow granulocytes via Toll-like receptor 2. J Cell Sci 2003; 116(Pt 2): 293–302. [DOI] [PubMed] [Google Scholar]
- 6.Sherwood RK, Roy CR. Autophagy Evasion and Endoplasmic Reticulum Subversion: The Yin and Yang of Legionella Intracellular Infection. Annu Rev Microbiol 2016; 70: 413–433. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Moreira E, Helbig JH, Swanson MS. Membrane vesicles shed by Legionella pneumophila inhibit fusion of phagosomes with lysosomes. Infect Immun 2006; 74(6): 3285–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hempstead AD, Isberg RR. Inhibition of host cell translation elongation by Legionella pneumophila blocks the host cell unfolded protein response. Proceedings of the National Academy of Sciences of the United States of America 2015; 112(49): E6790–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziltener P, Reinheckel T, Oxenius A. Neutrophil and Alveolar Macrophage-Mediated Innate Immune Control of Legionella pneumophila Lung Infection via TNF and ROS. PLoS Pathog 2016; 12(4): e1005591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kawamoto Y, Morinaga Y, Kimura Y, Kaku N, Kosai K, Uno N et al. TNF-alpha inhibits the growth of Legionella pneumophila in airway epithelial cells by inducing apoptosis. J Infect Chemother 2017; 23(1): 51–55. [DOI] [PubMed] [Google Scholar]
- 11.Akamine M, Higa F, Arakaki N, Kawakami K, Takeda K, Akira S et al. Differential roles of Toll-like receptors 2 and 4 in in vitro responses of macrophages to Legionella pneumophila. Infect Immun 2005; 73(1): 352–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Molofsky AB, Byrne BG, Whitfield NN, Madigan CA, Fuse ET, Tateda K et al. Cytosolic recognition of flagellin by mouse macrophages restricts Legionella pneumophila infection. J Exp Med 2006; 203(4): 1093–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zamboni DS, Kobayashi KS, Kohlsdorf T, Ogura Y, Long EM, Vance RE et al. The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol 2006; 7(3): 318–325. [DOI] [PubMed] [Google Scholar]
- 14.Hawn TR, Berrington WR, Smith IA, Uematsu S, Akira S, Aderem A et al. Altered inflammatory responses in TLR5-deficient mice infected with Legionella pneumophila. Journal of immunology (Baltimore, Md: 1950) 2007; 179(10): 6981–6987. [DOI] [PubMed] [Google Scholar]
- 15.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature 2011; 477(7366): 596–600. [DOI] [PubMed] [Google Scholar]
- 16.Burns K, Clatworthy J, Martin L, Martinon F, Plumpton C, Maschera B et al. Tollip, a new component of the IL-1RI pathway, links IRAK to the IL-1 receptor. Nature cell biology 2000; 2(6): 346–351. [DOI] [PubMed] [Google Scholar]
- 17.Zhang G, Ghosh S. Negative Regulation of Toll-like Receptor-mediated Signaling by Tollip. Journal of Biological Chemistry 2002; 277(9): 7059–7065. [DOI] [PubMed] [Google Scholar]
- 18.Lu K, Psakhye I, Jentsch S. Autophagic clearance of polyQ proteins mediated by ubiquitin-Atg8 adaptors of the conserved CUET protein family. Cell 2014; 158(3): 549–563. [DOI] [PubMed] [Google Scholar]
- 19.Jongsma ML, Berlin I, Wijdeven RH, Janssen L, Janssen GM, Garstka MA et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell 2016; 166(1): 152–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shah JA, Vary JC, Chau TT, Bang ND, Yen NT, Farrar JJ et al. Human TOLLIP Regulates TLR2 and TLR4 Signaling and Its Polymorphisms Are Associated with Susceptibility to Tuberculosis. J Immunol 2012; 189(4): 1737–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araujo FJ, Silva LD, Mesquita TG, Pinheiro SK, Vital Wde S, Chrusciak-Talhari A et al. Polymorphisms in the TOLLIP Gene Influence Susceptibility to Cutaneous Leishmaniasis Caused by Leishmania guyanensis in the Amazonas State of Brazil. PLoS Negl Trop Dis 2015; 9(6): e0003875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shah JA, Berrington WR, Vary JC Jr., Wells RD, Peterson GJ, Kunwar CB et al. Genetic Variation in Toll-Interacting Protein Is Associated With Leprosy Susceptibility and Cutaneous Expression of Interleukin 1 Receptor Antagonist. J Infect Dis 2016; 213(7): 1189–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casanova JL, Abel L. The genetic theory of infectious diseases: a brief history and selected illustrations. Annu Rev Genomics Hum Genet 2013; 14: 215–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruiz-Moreno JS, Hamann L, Shah JA, Verbon A, Mockenhaupt FP, Puzianowska-Kuznicka M et al. The common HAQ STING variant impairs cGAS-dependent antibacterial responses and is associated with susceptibility to Legionnaires’ disease in humans. PLoS Pathog 2018; 14(1): e1006829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. JExp Med 2003; 198(10): 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah JA, Musvosvi M, Shey M, Horne DJ, Wells RD, Peterson GJ et al. A Functional TOLLIP Variant is Associated with BCG-Specific Immune Responses and Tuberculosis. Am J Respir Crit Care Med 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Den Boer JW, Yzerman EP, Schellekens J, Lettinga KD, Boshuizen HC, Van Steenbergen JE et al. A large outbreak of Legionnaires’ disease at a flower show, the Netherlands, 1999. EmergInfectDis 2002; 8(1): 37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nahid P, Horne DJ, Jarlsberg LG, Reiner AP, Osmond D, Hopewell PC et al. Racial differences in tuberculosis infection in United States communities: the coronary artery risk development in young adults study. Clin Infect Dis 2011; 53(3): 291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ibrahim GF, Fleet GH, Lyons MJ, Walker RA. Method for the isolation of highly purified Salmonella flagellins. J Clin Microbiol 1985; 22(6): 1040–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edelstein PH. [Biological diagnosis of Legionnaire’s disease]. Rev Mal Respir 1984; 1(1): 11–14. [PubMed] [Google Scholar]
- 31.Didierlaurent A, Brissoni B, Velin D, Aebi N, Tardivel A, Kaslin E et al. Tollip regulates proinflammatory responses to interleukin-1 and lipopolysaccharide. Mol Cell Biol 2006; 26(3): 735–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walters KA, Olsufka R, Kuestner RE, Wu X, Wang K, Skerrett SJ et al. Prior infection with Type A Francisella tularensis antagonizes the pulmonary transcriptional response to an aerosolized Toll-like receptor 4 agonist. BMC Genomics 2015; 16: 874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Copenhaver AM, Casson CN, Nguyen HT, Fung TC, Duda MM, Roy CR et al. Alveolar macrophages and neutrophils are the primary reservoirs for Legionella pneumophila and mediate cytosolic surveillance of type IV secretion. Infect Immun 2014; 82(10): 4325–4336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uematsu S, Jang MH, Chevrier N, Guo Z, Kumagai Y, Yamamoto M et al. Detection of pathogenic intestinal bacteria by Toll-like receptor 5 on intestinal CD11c+ lamina propria cells. Nat Immunol 2006; 7(8): 868–874. [DOI] [PubMed] [Google Scholar]
- 35.Vinzing M, Eitel J, Lippmann J, Hocke AC, Zahlten J, Slevogt H et al. NAIP and Ipaf control Legionella pneumophila replication in human cells. J Immunol 2008; 180(10): 6808–6815. [DOI] [PubMed] [Google Scholar]
- 36.Chen K, Yuan R, Zhang Y, Geng S, Li L. Tollip Deficiency Alters Atherosclerosis and Steatosis by Disrupting Lipophagy. J Am Heart Assoc 2017; 6(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deretic V, Saitoh T, Akira S. Autophagy in infection, inflammation and immunity. Nat Rev Immunol 2013; 13(10): 722–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunologic manifestations of autophagy. J Clin Invest 2015; 125(1): 75–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shah JA, Musvosvi M, Shey M, Horne DJ, Wells RD, Peterson GJ et al. A Functional Toll-Interacting Protein Variant Is Associated with Bacillus Calmette-Guerin-Specific Immune Responses and Tuberculosis. Am J Respir Crit Care Med 2017; 196(4): 502–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to Legionnaires’ Disease. J Exp Med 2003; 198: 1563–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fontana MF, Vance RE. Two signal models in innate immunity. Immunol Rev 2011; 243(1): 26–39. [DOI] [PubMed] [Google Scholar]
- 42.Saitoh T, Fujita N, Jang MH, Uematsu S, Yang BG, Satoh T et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008; 456(7219): 264–268. [DOI] [PubMed] [Google Scholar]
- 43.Nastase MV, Zeng-Brouwers J, Frey H, Hsieh LT, Poluzzi C, Beckmann J et al. An Essential Role for SHARPIN in the Regulation of Caspase 1 Activity in Sepsis. Am J Pathol 2016; 186(5): 1206–1220. [DOI] [PubMed] [Google Scholar]
- 44.Rolando M, Escoll P, Buchrieser C. Legionella pneumophila restrains autophagy by modulating the host’s sphingolipid metabolism. Autophagy 2016; 12(6): 1053–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol 2011; 12(3): 222–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A et al. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun 2003; 306(4): 860–866. [DOI] [PubMed] [Google Scholar]
- 47.Zhang K, Kaufman RJ. From endoplasmic-reticulum stress to the inflammatory response. Nature 2008; 454(7203): 455–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cordell HJ, Clayton DG. Genetic association studies. Lancet 2005; 366(9491): 1121–1131. [DOI] [PubMed] [Google Scholar]
- 49.Samrakandi MM, Cirillo SL, Ridenour DA, Bermudez LE, Cirillo JD. Genetic and phenotypic differences between Legionella pneumophila strains. J Clin Microbiol 2002; 40(4): 1352–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shames SR, Liu L, Havey JC, Schofield WB, Goodman AL, Roy CR. Multiple Legionella pneumophila effector virulence phenotypes revealed through high-throughput analysis of targeted mutant libraries. Proc Natl Acad Sci U S A 2017; 114(48): E10446–E10454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burstein D, Amaro F, Zusman T, Lifshitz Z, Cohen O, Gilbert JA et al. Genomic analysis of 38 Legionella species identifies large and diverse effector repertoires. Nature genetics 2016; 48(2): 167–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.