Abstract
Genome editing by the CRISPR/Cas9 system has recently been used to produce gene knockout lines in many plant species. We applied this system to analyze Japanese gentian plants that produce blue flowers because of the accumulation of a polyacylated anthocyanin, gentiodelphin. Mutant lines in which anthocyanin modification genes were knocked out were examined to assess the contribution of each gene to the blue pigmentation of flowers. The targeted genes encoded anthocyanin 5-O-glycosyltransferase (Gt5GT), anthocyanin 3′-O-glycosyltransferase (Gt3′GT), and anthocyanin 5/3′-aromatic acyltransferase (Gt5/3′AT). The Gt5GT knockout lines accumulated delphinidin 3G, whereas the Gt3′GT knockout lines accumulated delphinidin 3G-5CafG as the major flower pigment. Knocking out Gt5/3′AT resulted in the accumulation of delphinidin 3G-5G-3′G and delphinidin 3G-5G as the primary and secondary pigments, respectively. These results indicated the existence of two pathways mediating the modification of delphinidin 3G-5G in flowers, with one involving a glycosylation by 3′GT and the other involving an acylation by 5/3′AT. The Gt5GT, Gt3′GT, and Gt5/3′AT transformants produced pale red violet, dull pink, and pale mauve flowers, respectively, unlike the vivid blue flowers of wild-type plants. Thus, the glycosylation and subsequent acylation of the 3′-hydroxy group of the B-ring in delphinidin aglycone is essential for the development of blue gentian flowers.
Subject terms: Molecular engineering in plants, Secondary metabolism
Introduction
Flavonoids, carotenoids, betalains, and chlorophylls, which are the four major pigments in higher plants1, are distributed in almost all plant organs, including flowers, fruits, seeds, roots, stems, and leaves, wherein they play important roles in plant development, propagation and survival. Specifically, the coloration of flowers and fruits that helps attract pollinators and seed dispersers is crucial for plant reproduction under natural conditions. Among these pigments, anthocyanins, belonging to the large flavonoid family, produce diverse colors such as red, purple, and blue2. This diversity is mostly due to modifications of the anthocyanidin skeleton with sugars and aromatic acids as well as the number and position of hydroxyl and methoxyl groups attached to the basic anthocyanidin aglycone molecule. More than 100 kinds of polyacylated anthocyanins with several aromatic acids have been isolated3. The thorough characterization of the flavonoid biosynthetic pathway in higher plants has revealed that the genes responsible for the biosynthesis of anthocyanidin aglycone are highly conserved among plant species4. However, the genes responsible for the subsequent modifications after the initial 3-O-glycosylation are highly diverse among plant species and plant organs. The accumulated anthocyanins vary even within the same plant species, with flowers, in particular, producing various anthocyanin types.
Blue-flowered Japanese gentians accumulate a polyacylated anthocyanin, gentiodelphin [delphinidin 3-O-β-d-glucosyl-5-O-(6-O-caffeoyl-β-d-glucoside)-3′-O-(6-O-caffeoyl-β-d-glucoside)], as a major pigment5,6. All of the flavonoid biosynthetic structural genes that contribute to gentiodelphin biosynthesis have been identified7,8. Furthermore, several transcription factor genes, such as MYB and bHLH, regulating flavonoid biosynthesis have been isolated and characterized9–11. For example, a previous transient expression assay demonstrated that GtMYB3, which belongs to the R2R3MYB family, binds to the promoters of the gentian F3′5′H and 5/3′AT (5,3′-aromatic acyltransferase) genes to upregulate expression9. Additionally, an analysis of the expression of GtMYB3 with the SRDX motif, which is a dominant negative regulatory domain, confirmed that GtMYB3 is also a positive regulator of several downstream flavonoid biosynthetic genes in gentian flowers12.
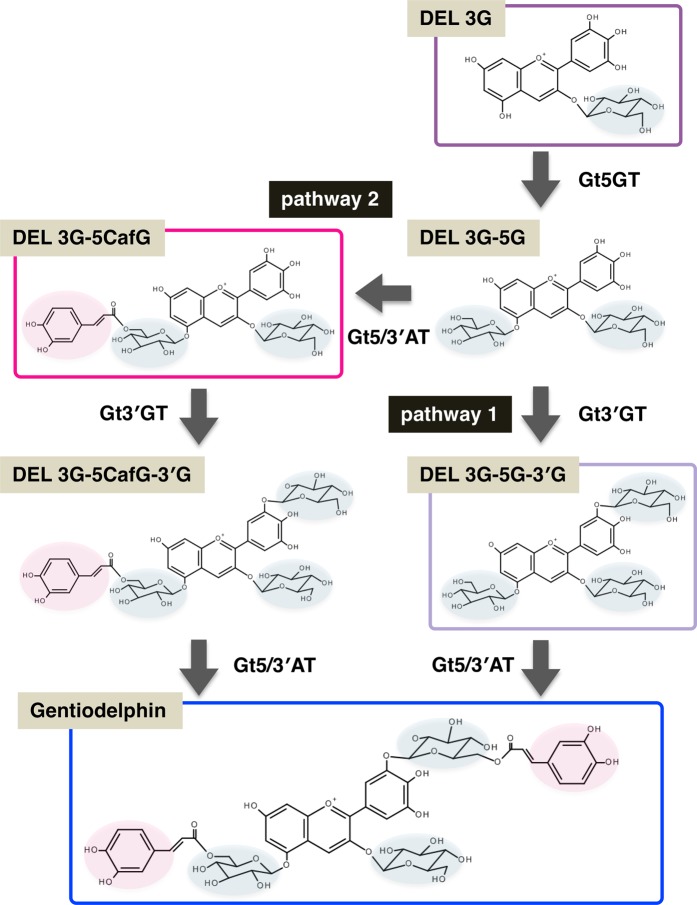
The conversion of delphinidin to gentiodelphin involves four modification steps (Fig. 1). Namely, delphinidin 3-O-glucoside is formed by the glycosylation of delphinidin via Gt3GT activity. Thereafter, additional sugar and acyl modifications are catalyzed by three key enzymes, including two glycosyltransferases, Gt5GT and Gt3′GT, and one acyl transferase, Gt5/3′AT, to produce gentiodelphin (Fig. 1). Regarding the polyacylated gentiodelphin structure, in vitro biochemical analyses suggested that the glycosylation and subsequent acylation of the 3′-hydroxy group of the B-ring of delphinidin aglycone are important for the stabilization and intramolecular sandwich-type stacking of two gentiodelphin caffeic acids, ultimately leading to a more intense blue coloration5,13–15. However, the functions of these modification enzymes in vivo have not been comprehensively characterized in gentian because of a lack of available lines in which these genes have been mutated. We previously generated mutant lines in which the expression levels of one (Gt5/3′AT) or two (Gt5/3′AT and GtF3′5′H) genes were knocked down by RNA interference (RNAi), and revealed that two transgenic lines exhibited abnormal flower coloration due to the decreased acylation of anthocyanins16. However, the effects were somewhat ambiguous because of the insufficient suppression of the target genes. The expression of the target genes was only downregulated rather than eliminated in the RNAi lines, and we were unable to analyze the effects of the complete inhibition of the expression of the anthocyanin modification genes. Furthermore, there are no available reports describing the suppression of Gt5GT and Gt3′GT expression.
Figure 1.
Gentiodelphin biosynthetic pathway. Gt5GT, UDP-glucose:anthocyanin 5-O-glucosyltransferase; Gt3′GT, UDP-glucose:anthocyanin 3′-O-glucosyltransferase; Gt5/3′AT, anthocyanin 5,3′-aromatic acyltransferase.
Genome editing by the clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) system has recently been used to target specific genes in various higher plants, including ornamental plants, such as DFR in Japanese morning glory (Ipomoea nil)17, F3H in torenia18, and PDS in gentian19. Therefore, it is now possible to produce and analyze gene knockout mutant lines targeting genes of interest even in horticultural plant species.
In this study, we used the CRISPR/Cas9 system to target three anthocyanin modification genes (Gt5GT, Gt3′GT, and Gt5/3′AT) and analyze the effects of the knockout of these genes on the pigmentation of gentian flowers. We generated genome-edited lines in which the three targeted anthocyanin modification genes were knocked out and subsequently evaluated the effects on flower color. We confirmed the genome was accurately edited by next-generation sequencing (NGS) before examining flower colors with a spectrophotometer and by a compositional analysis of anthocyanin pigments via HPLC. Our results verified the utility of CRISPR/Cas9-based genome editing and the importance of anthocyanin modification genes for the blue coloration of gentian flowers.
Results
Production of genome-edited gentian plants
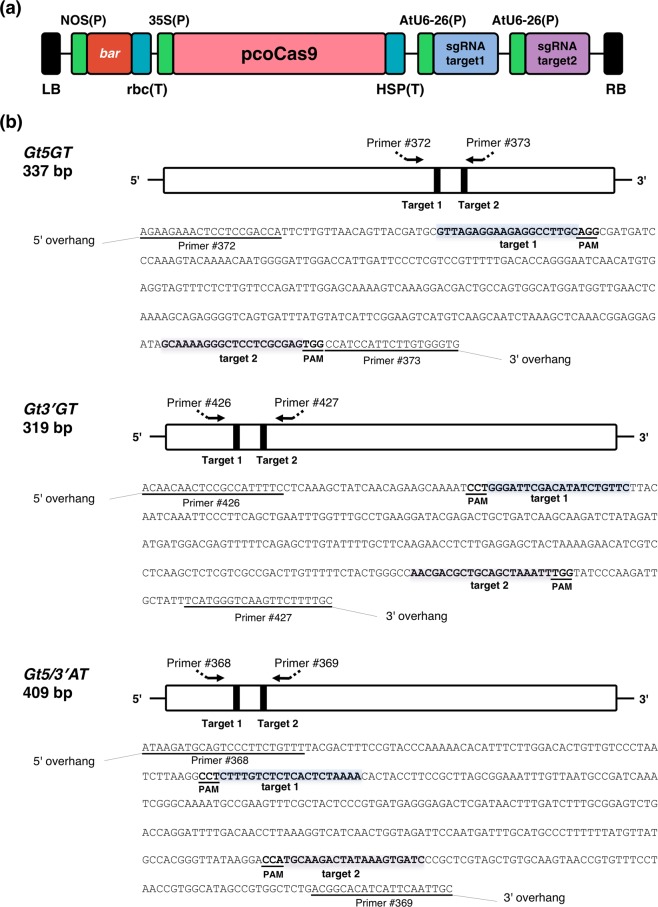
Adventitious shoots were selected on medium containing 0.5 or 0.75 mg l−1 bialaphos. A total of 35, 41, and 37 bialaphos-resistant putative transgenic lines were obtained for the transformation experiments involving the Gt5GT, Gt3′GT, and Gt5/3′AT constructs, respectively, of which 23, 36, and 31 lines, respectively, were confirmed to be transgenic by the PCR amplification of part of the pcoCas9 sequence using crude DNA samples. The genome-edited shoots were then screened by a direct Sanger sequence analysis of the PCR products amplified by the primer sets presented in Fig. 2b. Several clones of the PCR products from potential genome-edited shoots were subjected to further subcloning, and the sequences, including the target sites, were determined. Finally, we obtained two, two, and five lines that may have biallelic mutations at the Gt5GT, Gt3′GT, and Gt5/3′AT target sites, respectively. For each transformed construct, two genome-edited knockout lines were selected and grown in a closed greenhouse until flowering.
Figure 2.
Schematic diagram of the CRISPR/Cas9 construct and the target sites of three anthocyanin modification genes. (a) Schematic diagram of the T-DNA region of the CRISPR/Cas9 binary vector carrying pcoCas9, which is a codon-optimized Cas9 for plants. Two sgRNA expression cassettes are tandemly aligned. (b) Schematic diagrams of the Gt5GT, Gt3′GT, and Gt5/3′AT genes and their sequences, including the sequences of the amplified regions. Shaded boxes represent targets 1 and 2, whereas PAM refers to the protospacer adjacent motif (NGG). The primers used for the PCR amplification or direct sequencing are presented.
Next-generation sequencing analysis of genome-edited gentian plants
The results of the amplicon sequence analyses of all target sites in the transformants are summarized in Table 1. We excluded non-specific or low-copy fragments that were thought to be derived from PCR and/or sequence errors, and determined the number of major fragments of each allele. The number of the major Gt5GT, Gt3′GT, and Gt5/3′AT genomic DNA amplicons in the WT plants represented 68.8% (24,402/35,448), 72.7% (21,762/29,916), and 75.8% (33,661/44,435) of the total number of fragments, respectively. Analyses of SNP sequences revealed the gentian genome includes two alleles for Gt5GT and Gt5/3′AT, but four alleles for Gt3′GT. The counted read numbers among individual gene alleles of RNA were almost equal (less than 2.5-fold) in WT plants, except for Gt3′GT allele 4, which was counted approximately 5- to 10-fold higher than that of alleles 1–3. Amplicon sequence analyses also indicated that each transformant contained several characteristic mutations, such as deletions of 1–30 bases or more than 200 bases between the target 1 and target 2 sequences, depending on the transgenic lines and target genes (Table 1). All Gt5GT and Gt5/3′AT transformants were confirmed to have heterozygous biallelic mutations. Interestingly, the inversion of a 202-base sequence and a 27-base deletion were also confirmed in one allele of Gt5/3′AT #50. Additionally, we were unable to distinguish between alleles 2, 3, and 4 in Gt3′GT #11 because of a lack of SNPs due to a 221-bp deletion. Regarding Gt3′GT #35, some alleles were not detected, probably because of one or more large deletions or base changes to primer sites. However, for both Gt3′GT lines, the remaining amplified alleles were affected by mutations that caused frameshifts. Although the ratio of fragment counts for each allele differed between the genomic DNA and transcripts, we detected a consistent mutation pattern between the genomic DNA and transcripts in all transgenic lines. Therefore, these transgenic lines were confirmed as gene knockout lines, and they were analyzed further.
Table 1.
NGS analysis of PCR amplicons of DNA and RNA of genome-edited lines.
| Target gene | Line | Allele | No. of counts (DNA) | No. of counts (RNA) | Target 1 | Target 2 | ||
|---|---|---|---|---|---|---|---|---|
| Sequence | In/Del | Sequence | In/Del | |||||
| Gt5GT | WT | 1 | 12,79–8 | 50,836 | GTTAGAGGAAGAGGCCTTGC AGG | 0 | GCAAAAGGGCTCCTCGCGAG TGG | 0 |
| 2 | 11,604 | 26,024 | GTTAGAGGAAGAGGCCTTGC AGG | 0 | GCAAAAGGGCTCCTCGCGAG TGG | 0 | ||
| #47 | 1 | 9,611 | 6,443 | GTTAGAGGAAGAGGCCTTGC AGG | 0 | GCAAAAGGGCT-----CGAG TGG | −5 | |
| 2 | 9,605 | 36,768 | GTTAGAGGAAGAGGCCTTGC AGG | 0 | GCAAAAGGG---------AG TGG | −9 | ||
| #106 | 1 | 18,816 | 29,042 | CGTTAGAGGAAGAGGCC-TGC AGG | −1 | GCAAAAGGG-------CGAG TGG | −7 | |
| 2 | 30,010 | 261,382 | CGTTAGAGGAAGAGGC----------------||----------------GAG TGG | −256 | ||||
| Gt3′GT | WT | 1 | 5,666 | 4,852 | CCT GGGATTCGACATATCTGTTC | 0 | AACGACGCTGCAGCTAAATT TGG | 0 |
| 2 | 5,419 | 2,055 | CCT GGGATTCGACATATCTGTTC | 0 | AACGACGCTGCAGCTAAATT TGG | 0 | ||
| 3 | 5,825 | 2,018 | CCT GGGATTCGACATATCTGTTC | 0 | AACGACGCTGCAGCTAAATT TGG | 0 | ||
| 4 | 4,852 | 22,326 | CCT GGGATTCGACATATCTGTTC | 0 | AACGACGCTGCAGCTAAATT TGG | 0 | ||
| #11* | 1 | 20,194 | 35,543 | CCT GGG------CATATCTGTTC | −6 | AACGACGCTGCAGCT-AATT TGG | −1 | |
| 2 | 34,884 | 461,515 | CCT GGG---------------------||--------------------AATT TGG | −221 | ||||
| 3 | ||||||||
| 4 | ||||||||
| #35 | 1 | 7,186 | 3,004 | CCT -------GACATATCTGTTC | −7 | AACGACGCTGCAGCTAAATT TGG | 0 | |
| 2 | 7,223 | 33,855 | --||--ATTCGACATATCTGTTC | −10 | AACGACGCTGCAGCTAAATT TGG | 0 | ||
| 3 | — | — | not amplified | — | not amplified | — | ||
| 4 | — | — | not amplified | — | not amplified | — | ||
| Gt5/3′AT | WT | 1 | 15,403 | 12,453 | CCT CTTTGTCTCTCACTCTAAAA | 0 | CCA TGCAAGACTATAAAGTGATC | 0 |
| 2 | 18,258 | 20,721 | CCT CTTTGTCTCTCACTCTAAAA | 0 | CCA TGCAAGACTATAAAGTGATC | 0 | ||
| #50 | 1 | 16,376 | 15,213 | CCT CTTTGTCTCTCACTCTAAAA | 0 | ------||-------AAGTGATC | −30 | |
| 2 | 19,913 | 11,267 | CCT CTT-----||------ [202 base reversed] AGACTATAAAGTGATC | −27 | ||||
| #60 | 1 | 7,872 | 1,272 | CCT CTTTGTCTCTCACTCTAAAA | 0 | CCA TGC----CTATAAAGTGATC | −4 | |
| 2 | 6,874 | 22,102 | CCT C---GTCTCTCACTCTAAAA | −3 | CCA TGC–ACTATAAAGTGATC | −3 | ||
Double underlines show PAM sequences.
*Allele 2, 3 and 4 are indistinguishable from each other due to large deletion.
Petal phenotypes of the genome-edited gentian plants
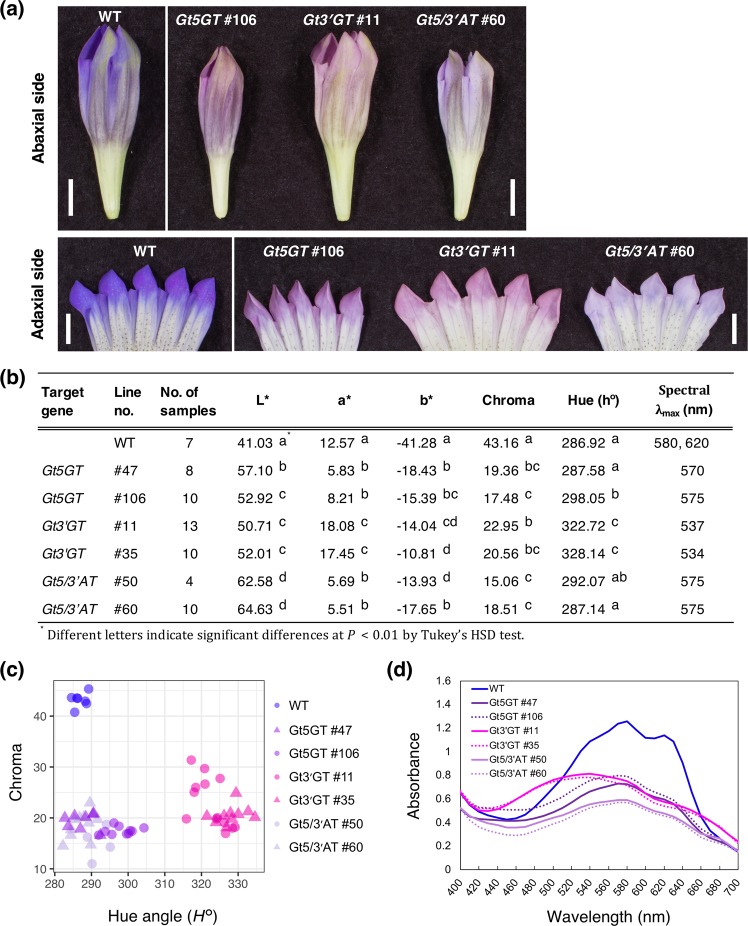
At approximately 4 months after plants were first acclimated, we detected changes to the flower color phenotypes in the transformants in a closed greenhouse. Figure 3a presents the abaxial and adaxial sides of flower petals of representative transformants. Specifically, the Gt5GT #106, Gt3′GT #11, and Gt5/3′AT #60 lines produced pale red violet, dull pink, and pale mauve flowers, respectively. The colorimetric values of the adaxial side of the flower limb area for the Gt5GT (#47 and #106), Gt3′GT (#11 and #35), and Gt5/3′AT (#50 and #60) transformants are summarized in Fig. 3b. The L*, a*, b*, and C* values for all transformants were significantly higher and/or lower than the corresponding values for the WT plants (Fig. 3b). The hue angles of Gt3′GT (#11 and #35) plants, which produced flowers with dull pink petals, exceeded 300°. The C* values and hue angles are presented in a scatter plot to visualize the color distribution (Fig. 3c). Although the two-dimensional coordinated positions of the Gt5GT and Gt5/3′AT lines overlapped (Fig. 3c), the L* values of the Gt5/3′AT lines were significantly higher than those of the Gt5GT lines (Fig. 3b). Moreover, the hue angles were significantly greater in the Gt3′GT lines than in the WT plants and in the other transformants. Furthermore, the C* value was significantly lower in all transformants than in the WT plants. Thus, the color distribution clearly indicated that all transformants differed from the WT plants. The absorption spectra of the transformants are provided in Fig. 3d. The absorbance values were high for the WT plants, with two peaks (at 580 and 620 nm). In contrast, all transformants had absorbance values less than 1.0, with a slight decrease at 600 nm. The absorbance of the Gt3′GT lines shifted more intensely at shorter wavelengths than the Gt5GT and Gt5/3′AT lines.
Figure 3.
Characteristics of flower colors in genome-edited gentian plants. (a) Abaxial and adaxial sides of the flower petals of the wild-type (WT) plants and the Gt5GT, Gt3′GT, and Gt5/3′AT lines. Bar = 1 cm. (b) The L* a* b* color values and visible spectral data measured spectrophotometrically of the surface of fresh petals are provided. Different letters indicate significant differences at P < 0.01 according to Tukey’s HDS test. (c) Color distribution visualized by a 2-dimensional scatter plot based on L* and chroma values. (d) Absorption spectra of the surface of fresh petals in WT and genome-edited knockout lines measured spectrophotometrically. The spectra are based on the mean data for the petals described in (b).
Analysis of anthocyanins in genome-edited gentian plants
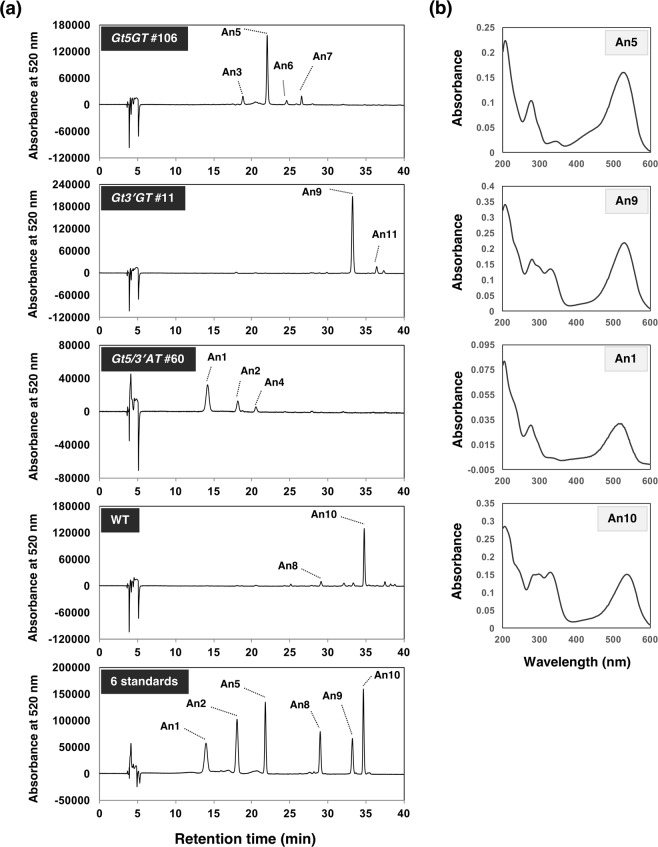
The petal extracts of the transformants and WT control were separated by HPLC (at 520 nm). The chromatograms and major peaks of Gt5GT #106, Gt3′GT #11, Gt5/3′AT #60, and WT plants (i.e., the same lines as in Fig. 3a) are presented in Fig. 4a,b. Eleven peaks were detected in the samples. Minor peaks (peak area <100,000) were excluded from analyses. We designated the peaks as An1 to An11 based on the retention times. A co-chromatography and analysis of absorbance spectra indicated peaks An1, An2, An5, An8, An9, and An10 corresponded to six standards. Anthocyanin compositions calculated based on the peak area at 520 nm are summarized in Table 2. In WT plants, one major peak (An10) corresponding to gentiodelphin accounted for 91% of the total peak area, whereas gentiodelphin was undetectable in all transformants. In the Gt5GT, Gt3′GT, and Gt5/3′AT lines, the major peaks were An5, An9, and An1, which corresponded to delphinidin 3-glucoside (61% or 74%), delphinidin 3-glucosyl-5-caffeoyl-glucoside (90% or 100%), and delphinidin 3,5,3′-triglucosides (57% or 75%), respectively. The presence of minor peaks implied a small amount of unidentified anthocyanins accumulated in the transformants (Fig. 4a and Table 2). Similar results were obtained for other genome-edited lines (Supplementary Fig. S1). The anthocyanin concentrations in flower petal extracts were calculated and expressed as delphinidin equivalents (Supplementary Fig. S2). Gt5GT-edited lines showed significant increase in anthocyanin concentrations compared with WT and other genome-edited lines.
Figure 4.
Anthocyanin profiles of the flower petals of the genome-edited gentian plants. (a) HPLC chromatograms (at 520 nm) of the anthocyanins extracted from the petals of the Gt5GT #106, Gt3′GT #11, and Gt5/3′AT #60 lines. An1, delphinidin 3,5,3′-triglucoside; An2, delphinidin 3,5-diglucoside; An5, delphinidin 3-glucoside; An8, delphinidin 3-glucosyl-5-caffeoyl-glucosyl-3′-glucoside; An9, delphinidin 3-glucosyl-5-caffeoyl-glucoside; An10, delphinidin 3-glucosyl-5-caffeoyl-glucosyl-3′-caffeoyl-glucoside (gentiodelphin); An3, 4, 6, 7, and 11 are unknown anthocyanins. (b) Spectra of the major peaks in Gt5GT#106 (An5), Gt3′GT#11 (An9), Gt5/3′AT #60 (An1), and WT (An10).
Table 2.
Anthocyanin-content ratio calculated from peak area of HPLC at 520 nm (%).
| plant | An1 | An2 | An3 | An4 | An5 | An6 | An7 | An8 | An9 | An10 | An11 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DEL 3G-5G-3′G | DEL 3G-5G | DEL 3 G | DEL 3G-5CafG-3′G | DEL 3G-5CafG | Gentiodelphin | |||||||
| WT | 1 | 8 | 91 | |||||||||
| Gt5GT | #47 | 29 | 61 | 10 | ||||||||
| #106 | 11 | 74 | 8 | 7 | ||||||||
| Gt3′GT | #11 | 90 | 10 | |||||||||
| #35 | 100 | |||||||||||
| Gt5/3′AT | #50 | 57 | 35 | 8 | ||||||||
| #60 | 75 | 25 | ||||||||||
Discussion
In this study, we used the CRISPR/Cas9 system to edit three anthocyanin modification genes (Gt5GT, Gt3′GT, and Gt5/3′AT) in the Japanese gentian genome. In preliminary experiments, we obtained genome-edited gentian plants by applying one sgRNA targeting one site. However, because the frequency of the genome editing greatly varied depending on the genes and target sites, we decided to use two sgRNAs to target two sites on a single gene (Fig. 2b). The use of multiple sgRNAs reportedly increases the efficiency of Cas9-mediated genome editing by increasing the mutation rate of the target genes20–22. It is also expected to introduce large fragment deletions between the two target sites. In the current study, we obtained one line (Gt3′GT #11) with a large sequence deletion (Table 1). Additionally, the biallelic mutation efficiencies based on a Sanger sequence analysis were 5.7% (2/35 lines), 4.9% (2/41 lines), and 8.6% (5/58 lines) for the Gt5GT, Gt3′GT, and Gt5/3′AT transformants, respectively (data not shown). These efficiencies are much lower than the reported efficiencies for other plants, such as torenia (80%)18, morning glory (75%)17, and rice (67.6%)23. The reason for this difference remains unclear, but one possibility is that the CaMV 35 S promoter-mediated expression of SpCas9 is considerably silenced in gentian24. Another possibility is that the high heterogeneity of the large gentian genome (approximately 5 Gb, 2n = 2 × = 26) contributes to the observed inefficiencies. Our NGS analysis of DNA and RNA revealed that the number of major allele fragments represented 66.9%–95.4% of the total number of fragments (Supplementary Table S2). The minor fragments contained SNPs and/or In/dels around the target sequence, but these variations were likely derived from PCR or NGS errors. Alleles 3 and 4 in Gt3′GT-edited line #35 were not detected in our NGS analysis, suggesting the mutations were located outside the sequenced region amplified with primers #426 and #427. An analysis of RNA amplicons indicated that the number of non-mutated WT allele fragments accounted for less than 0.05% of the total number of fragments. This result may have been due to technical errors during the NGS analysis or because of the extremely low chimerism of the remaining unedited cells. However, the expression of a few non-edited sequences in gentian flowers is unlikely to affect the absorbance of the petal surface evaluated with a spectrophotometer or the peak area values determined by HPLC. Thus, we concluded that Gt5GT-, Gt3′GT-, and Gt5/3′AT-edited lines can be used to evaluate flower coloration during an in vivo analysis. To increase the efficiency of gentian genome editing, we are now assessing the effects of several modifications to the CRISPR/Cas9 system, including to the promoters, the sgRNA scaffold structure, and the selection conditions during the A. tumefaciens-mediated transformation step.
Knocking out Gt3′GT induced the accumulation of delphinidin 3G-5CafG as the major anthocyanin in the petal tissue of lines #11 (90%) and #35 (100%) (Table 2). The Gt3′GT-edited knockout lines did not accumulate delphinidin 3G-5G in petals, suggesting the glucose moiety at the 5-position was efficiently acylated by Gt5/3′AT. In contrast, the Gt5/3′AT-edited knockout lines accumulated not only delphinidin 3G-5G-3′G, but also some delphinidin 3G-5G (Fig. 4, Table 2), likely because of the incomplete Gt3′GT-catalyzed modification of delphinidin 3G-5G. As presented in Fig. 1, the modification pathway, which had been biochemically verified in a previous study15, branches into two pathways so that delphinidin 3G-5G may be glucosylated by Gt3′GT (pathway 1) or acylated by Gt5/3′AT (pathway 2). Additionally, the residual delphinidin 3G-5G may be used as a substrate for pathway 2. Thus, our analyses of the Gt3′GT- and Gt5/3′AT-knockout gentian plants indicate that both pathways contribute to gentiodelphin biosynthesis in gentian petals. Unidentified anthocyanins also accumulated in Gt5GT-edited knockout lines. It is probable that any of An3, An6, and An7 corresponds to delphinidin 3,3′-diglucoside or delphinidin 3-glucosyl-3′-caffeoyl-glucoside, because the recombinant Gt3′GT protein has slight glucosylation activity against delphinidin 3-glucoside15. It is also notable that Gt5/3′AT lines accumulated no acylated anthocyanins and 5GT lines accumulated anthocyanins where there are no side groups at the 5 position as major pigments, suggesting that CRISPR/Cas9 system could produce knockout lines for the specific glucosyl and acyl transferase genes.
The absorption spectrum of the surface of the fresh petals of WT plants revealed the highest absorbance occurred at 580 nm, with an additional peak at 620 nm (Fig. 3d). The characteristic peaks detected for the WT gentian petals were also observed for the petals of other plant species with blue flowers, including Commelina communis (metal complex pigment), Platycodon grandiflorus (polyacylated anthocyanin), and Felicia amelloides (intermolecular co-pigmentation with flavone C-glycosides) (Supplementary Fig. S3). However, the absorption spectra of the Gt5GT-, Gt3′GT-, and Gt5/3′AT-edited knockout lines were lower than that of the WT control, with a slight decrease at 600 nm. The a*, b*, L*, and C* values of the genome-edited knockout lines accumulating intermediate delphinidin derivatives varied from the corresponding values of the WT plants (Fig. 3b). Specifically, the b* value for blue–yellow color components (blue in the negative direction) considerably increased in all transformants.
Flower color is determined by not only the anthocyanin kinds but also the anthocyanin amounts in petals. Our result indicated that Gt5GT-edited lines showed about 2-fold increase in anthocyanin concentrations compared with WT and other lines (Supplementary Fig. S2). This may also contribute to the observed change of flower color in Gt5GT lines. Although the reason remains unknown, anthocyanin transport activity and/or anthocyanin stability in vacuole may be different among anthocyanin molecules. Further analysis is needed to elucidate the effects of anthocyanin modifications on flower color pigmentation in gentian. However, our results clearly confirmed the actual gentiodelphin biosynthetic pathway and its contribution to blue color development previously only reported from in vitro chemical analysis.
In summary, we confirmed the importance of gentiodelphin for the blue coloration of gentian flowers by analyzing genome-edited lines in which specific anthocyanin modification genes were knocked out by the CRISPR/Cas9 system. Our data provide novel insights regarding the gentiodelphin biosynthetic pathway and verify the co-pigmentation bluing effects on flower petals. We are now developing new CRISPR/Cas9 vectors and optimizing the method for selecting genome-edited knockout lines. We are also generating various genome-edited knockout lines suitable for horticultural, physiological, and pathological studies. We believe the CRISPR/Cas9 system will accelerate future gentian studies.
Materials and Methods
Plant materials and transformation
The blue-flowered gentian cv. Albireo (Gentiana triflora × Gentiana scabra) plants that were transformed in this study were maintained in an in vitro culture. Half-strength Murashige and Skoog (MS) medium containing 3% (w/v) sucrose and 0.2% (w/v) gellan gum was used as the basal culture medium. The growth conditions in an incubation room were as follows: 22 °C under a 16-h light/8-h dark photoperiod, with light supplied by cool white fluorescent lamps. Plants were transformed according to a modified version of a previously described Agrobacterium tumefaciens-mediated procedure24. Briefly, leaf sections were excised and co-cultivated with A. tumefaciens cells harboring a CRISPR/Cas9 vector (Fig. 2a) on half-strength MS medium supplemented with 100 μM acetosyringone. After a 5-day co-cultivation, the leaf sections were transferred to selection medium containing 0.5 or 0.75 mg l−1 bialaphos (Meiji-Seika Co. Ltd., Tokyo, Japan), 500 mg l−1 cefotaxime (Claforan; Aventis Pharma, France), 10 mg l−1 thidiazuron [N-phenyl-N′-(1,2,3-thiadiazol-5-yl) urea], and 1 mg l−1 1-naphthaleneacetic acid. Adventitious shoots were induced from calli dedifferentiated from the edge of leaves on selection medium and transferred to half-strength MS medium solidified with 0.25% (w/v) gellan gum and supplemented with 3% (w/v) sucrose, 500 mg l−1 cefotaxime, and 0.75 mg l−1 bialaphos for rooting. The production of transgenic material was first confirmed by PCR, which was completed with crude extracts from shoots, MightyAmp DNA polymerase (TaKaRa, Shiga, Japan), and primers #61_Cas9U1898 and #62_Cas9L2260 specific for pcoCas9 (Supplementary Table S1). The regenerated plants were acclimated as previously described25.
Construction of binary vectors for editing the gentian genome
Binary CRISPR/Cas9 vectors (pSbar-pcoCas9_AtU6-26p-target-gRNA_AtU6-26p-target-gRNA) (Fig. 2) targeting gentian Gt5GT, Gt3′GT, and Gt5/3′AT genes were constructed for a subsequent plant transformation. A basic vector, pSbar-pcoCas9, was first constructed by replacing the uidA (gus) gene of pSKAN221-HT26 with the plant codon-optimized SpCas9 (pcoCas9)27. Two single-guide RNA (sgRNA) expression cassettes targeting the gentian Gt5GT, Gt3′GT, and Gt5/3′AT genes were introduced into pSbar-pcoCas9 in tandem via a restriction enzyme digestion and ligation. Target sites for each gene are presented in Fig. 2b. Finally, three binary vectors, pSbar-pcoCas9-AtU6-26p-Gt5GTtarget1gRNA-AtU6-26p-t5GTtarget2gRNA, pSbar-pcoCas9-AtU6-26p-Gt5/3′ATtarget1gRNA-AtU6-26p-Gt5/3′ATtarget2gRNA, and pSbar-pcoCas9-AtU6-26p-Gt3′GTtarget1gRNA-AtU6-26p-Gt3′GTtarget2gRNA, were inserted into A. tumefaciens strain EHA101 cells by electroporation.
Measurement of gentian petal colors
To evaluate the flower color phenotypes of the genome-edited knockout lines and the untransformed wild-type (WT) plants, the adaxial surface of freshly collected petals was analyzed spectrophotometrically. For each line, the colorimetric values (L*, a*, and b*) and the absorption spectra of the adaxial surface of the limb area of fresh petals were measured with the CM-3600A spectrophotometer (Konica Minolta, Tokyo, Japan). The chroma C* value and the hue angle were also calculated. Tukey’s HSD test was performed to test differences among sample means for significance.
Anthocyanin analysis
The anthocyanin compositions of petal extracts were analyzed by HPLC. Fresh petals were collected from greenhouse-grown plants and stored at −80 °C until used. The petals were soaked in 80% methanol containing 0.1% (v/v) trifluoroacetic acid to extract the anthocyanins, which were then separated with an HPLC–photodiode array detector system [PU-4180 PUMP, MD-4015 photodiode array detector, and ChromNAV (version 2.02) software; JASCO, Tokyo, Japan] equipped with a COSMOSIL PBr column (4.6 mm internal diameter × 250 mm; Nacalai Tesque, Kyoto, Japan). We used a linear gradient elution (2 ml min−1) of 10%–80% acetonitrile in 1% aqueous phosphoric acid over 50 min.
Total anthocyanin concentrations were estimated based on the molar absorptivity of delphinidin (ε mol = 27,940 at 520 nm, evaluated in 80% MeOH containing 0.1% TFA). Delphinidin chloride, delphinidin 3-O-glucoside and delphinidin 3,5-O-diglucoside were purchased from Funakoshi Co. (Tokyo, Japan). Additionally, delphinidin 3-O-β-d-glucoside 5-O-(6-O-caffeoyl-β-d-glucoside) 3′-O-β-d-glucoside, delphinidin 3-O-β-d-glucoside 5-O-(6-O-caffeoyl-β-d-glucoside), delphinidin 3,5,3′-O-triglucoside (kindly provided by Drs. Tanaka and Nakamura, Suntory Ltd.), and gentiodelphin [delphinidin 3-O-β-d-glucoside 5-O-(6-O-caffeoyl-β-d-glucoside) 3′-O-(6-O-caffeoyl-β-d-glucoside)] were used as authentic standards.
Sanger sequencing analysis
The leaves of transgenic plant lines were analyzed by PCR with MightyAmp DNA polymerase according to the manufacturer’s instructions. Details regarding the primers designed for the Gt5GT, Gt3′GT, and Gt5/3′AT genes are provided in Supplementary Table S1. The PCR products were purified and sequenced with the BigDye Terminator (version 1.1) Cycle Sequencing Kit and the ABI PRISM 3130xl or 3500 Genetic Analyzers (Applied Biosystems, Foster City, CA, USA).
Next-generation sequencing analysis
The genomic DNA and transcripts in the petals of the genome-edited lines and the WT control underwent an Illumina NGS analysis. Genomic DNA was isolated from the leaves of transgenic and WT gentian plants with the GeneElute Genome DNA Isolation system (Sigma-Aldrich, St Louis, MO, USA). Total RNA was isolated from the petals of transgenic gentian plants with the RNeasy Plant Mini kit (Qiagen, GmbH, Hilden, Germany) and treated with DNase I (TaKaRa) according to the manufacturer’s instructions. The extracted RNA samples were reverse transcribed with PrimeScript II (TaKaRa). The subsequent library preparation, PCR amplification, amplification check, and library quantification check were performed as previously described18. The primer positions and target sites are indicated in Fig. 2b. Additional details regarding the primers (e.g., index) are listed in Supplementary Table S1. A bioinformatics analysis was performed as previously described18. Briefly, the resulting raw sequence reads were pre-processed with the FASTX toolkit, and the read pairs were merged into a single contiguous sequence (fragment) with a fastq-join script28. The unique fragments were then counted.
Supplementary information
Acknowledgements
We thank Dr. Holger Puchta (Botanical Institute, Karlsruhe Institute of Technology, Germany) for providing pcoCAS9. We also thank Drs. Noriko Nakamura and Yoshikazu Tanaka (Suntory Ltd.) for providing anthocyanin standards. The authors are grateful to Mses. H. Harumi and A. Fujisaki, Iwate Biotechnology Research Center, for their technical support. Computational analyses were partially performed on the NIG supercomputer at the ROIS National Institute of Genetics. This work was financially supported by Iwate Prefecture and by Grants-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (16K18654). This work was also supported in part by “Strategic program on agricultural research and innovation”, Ministry of Agriculture, Forestry and Fisheries, Japan. We thank Edanz Group (www.edanzediting.com/ac) for editing a draft of this manuscript.
Author contributions
K.T., N.S. and M.N. designed research, K.T., A.H., A.W., N.S. and M.N. performed experiments, K.T., N.S. and M.N. analyzed data, and K.T. and M.N. wrote the paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-51808-3.
References
- 1.Delgado-Vargas F, Jimenez AR, Paredes-Lopez O. Natural pigments: carotenoids, anthocyanins, and betalains–characteristics, biosynthesis, processing, and stability. Crit Rev Food Sci Nutr. 2000;40:173–289. doi: 10.1080/10408690091189257. [DOI] [PubMed] [Google Scholar]
- 2.Tanaka Y, Sasaki N, Ohmiya A. Biosynthesis of plant pigments: anthocyanins, betalains and carotenoids. Plant J. 2008;54:733–749. doi: 10.1111/j.1365-313X.2008.03447.x. [DOI] [PubMed] [Google Scholar]
- 3.Honda T, Saito N. Recent progress in the chemistry of poly-acylated anthocyanins as flower color pigments. Heterocycles. 2002;56:633–692. doi: 10.3987/REV-01-SR(K)2. [DOI] [Google Scholar]
- 4.Grotewold, E. The Science of Flavonoids. (Springer-Verlag, 2006).
- 5.Goto T, et al. Structure of gentiodelphin, an acylated anthocyanin from Gentiana maikinoi, that is stable in dilute aqueous solution. Tetrahedron Lett. 1982;23:3695–3698. doi: 10.1016/S0040-4039(00)88660-8. [DOI] [Google Scholar]
- 6.Hosokawa K, et al. Seven acylated anthocyanins in blue flowers of Gentiana. Phytochemistry. 1997;45:167–171. doi: 10.1016/S0031-9422(96)00775-3. [DOI] [PubMed] [Google Scholar]
- 7.Nishihara, M., Nakatsuka, T., Mizutani-Fukuchi, M., Tanaka, Y. & Yamamura, S. In Floricultural and Ornamental Biotechnology V (ed Teixeira da Silva, J. A.) 57-67 (Global Science Books, 2008).
- 8.Nakatsuka T, Sato K, Takahashi H, Yamamura S, Nishihara M. Cloning and characterization of the UDP-glucose:anthocyanin 5-O-glucosyltransferase gene from blue-flowered gentian. J Exp Bot. 2008;59:1241–1252. doi: 10.1093/jxb/ern031. [DOI] [PubMed] [Google Scholar]
- 9.Nakatsuka T, et al. Identification and characterization of R2R3-MYB and bHLH transcription factors regulating anthocyanin biosynthesis in gentian flowers. Plant Cell Physiol. 2008;49:1818–1829. doi: 10.1093/pcp/pcn163. [DOI] [PubMed] [Google Scholar]
- 10.Nakatsuka T, et al. Isolation and characterization of GtMYBP3 and GtMYBP4, orthologues of R2R3-MYB transcription factors that regulate early flavonoid biosynthesis, in gentian flowers. J Exp Bot. 2012;63:6505–6517. doi: 10.1093/jxb/ers306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakatsuka T, Sasaki N, Nishihara M. Transcriptional regulators of flavonoid biosynthesis and their application to flower color modification in Japanese gentians. Plant Biotechnol. 2014;31:389–399. doi: 10.5511/plantbiotechnology.14.0731a. [DOI] [Google Scholar]
- 12.Tasaki K, Atsumi G, Nishihara M, Sekine KT. Development of a Broad bean wilt virus 2-based expression vector for gentian. Sci Hort. 2016;201:279–286. doi: 10.1016/j.scienta.2016.01.045. [DOI] [Google Scholar]
- 13.Yoshida K, Toyama Y, Kameda K, Kondo T. Contribution of each caffeoyl residue of the pigment molecule of gentiodelphin to blue color development. Phytochemistry. 2000;54:85–92. doi: 10.1016/S0031-9422(00)00049-2. [DOI] [PubMed] [Google Scholar]
- 14.Yoshida K, Mori M, Kondo T. Blue flower color development by anthocyanins: from chemical structure to cell physiology. Nat Prod Rep. 2009;26:884–915. doi: 10.1039/b800165k. [DOI] [PubMed] [Google Scholar]
- 15.Fukuchi-Mizutani M, et al. Biochemical and molecular characterization of a novel UDP-glucose:anthocyanin 3′-O-glucosyltransferase, a key enzyme for blue anthocyanin biosynthesis, from gentian. Plant Physiol. 2003;132:1652–1663. doi: 10.1104/pp.102.018242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakatsuka T, et al. Genetic engineering of novel flower colour by suppression of anthocyanin modification genes in gentian. J Plant Physiol. 2010;167:231–237. doi: 10.1016/j.jplph.2009.08.007. [DOI] [PubMed] [Google Scholar]
- 17.Watanabe K, et al. CRISPR/Cas9-mediated mutagenesis of the dihydroflavonol-4-reductase-B (DFR-B) locus in the Japanese morning glory Ipomoea (Pharbitis) nil. Sci Rep. 2017;7:10028. doi: 10.1038/s41598-017-10715-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishihara M, Higuchi A, Watanabe A, Tasaki K. Application of the CRISPR/Cas9 system for modification of flower color in Torenia fournieri. BMC Plant Biol. 2018;18:331. doi: 10.1186/s12870-018-1539-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishihara M, Tasaki K, Sasaki N, Takahashi H. Development of basic technologies for improvement of breeding and cultivation of Japanese gentian. Breed Sci. 2018;68:14–24. doi: 10.1270/jsbbs.17074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma X, et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Molecular Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 21.Yan W, Chen D, Kaufmann K. Efficient multiplex mutagenesis by RNA-guided Cas9 and its use in the characterization of regulatory elements in the AGAMOUS gene. Plant Methods. 2016;12:23. doi: 10.1186/s13007-016-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hashimoto R, Ueta R, Abe C, Osakabe Y, Osakabe K. Efficient multiplex genome editing induces precise, and self-ligated type mutations in tomato plants. Front Plant Sci. 2018;9:916. doi: 10.3389/fpls.2018.00916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mikami M, Toki S, Endo M. Comparison of CRISPR/Cas9 expression constructs for efficient targeted mutagenesis in rice. Plant Mol Biol. 2015;88:561–572. doi: 10.1007/s11103-015-0342-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mishiba K, et al. Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 2005;44:541–556. doi: 10.1111/j.1365-313X.2005.02556.x. [DOI] [PubMed] [Google Scholar]
- 25.Hosokawa K, Matsuki R, Oikawa Y, Yamamura S. Production of transgenic gentian plants by particle bombardment of suspension-culture cells. Plant Cell Rep. 2000;19:454–458. doi: 10.1007/s002990050755. [DOI] [PubMed] [Google Scholar]
- 26.Hamachi A, et al. Overexpression of geraniol synthase induces heat stress susceptibility in Nicotiana tabacum. Planta. 2019;249:235–249. doi: 10.1007/s00425-018-3054-z. [DOI] [PubMed] [Google Scholar]
- 27.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 28.Aronesty E. Comparison of sequencing utility programs. Open Bioinform J. 2013;7:1–8. doi: 10.2174/1875036201307010001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.