Significance
Food is the only source of the essential vitamin B1 for humans, but many microorganisms such as yeast and bacteria can synthetize it themselves. Here we report on a group of yeasts that have lost part of the vitamin B1 biosynthetic pathway in the past but have managed to rebuild it by capturing multiple genes from bacteria through horizontal gene transfer (HGT). We show a mosaic pathway composed of yeast and bacterial genes working coordinately to accomplish the synthesis of an essential nutrient. This involved adaptation of the bacterial genes to the very different expression rules in their new environment using several different mechanisms. Our results endorse HGT as an important mechanism for evolutionary adaptation in eukaryotes.
Keywords: horizontal gene transfer, horizontal operon transfer, yeast metabolism, gene fusion, thiamine
Abstract
Horizontal acquisition of bacterial genes is presently recognized as an important contribution to the adaptation and evolution of eukaryotic genomes. However, the mechanisms underlying expression and consequent selection and fixation of the prokaryotic genes in the new eukaryotic setting are largely unknown. Here we show that genes composing the pathway for the synthesis of the essential vitamin B1 (thiamine) were lost in an ancestor of a yeast lineage, the Wickerhamiella/Starmerella (W/S) clade, known to harbor an unusually large number of genes of alien origin. The thiamine pathway was subsequently reassembled, at least twice, by multiple HGT events from different bacterial donors involving both single genes and entire operons. In the W/S-clade species Starmerella bombicola we obtained direct genetic evidence that all bacterial genes of the thiamine pathway are functional. The reconstructed pathway is composed by yeast and bacterial genes operating coordinately to scavenge thiamine derivatives from the environment. The adaptation of the newly acquired operons to the eukaryotic setting involved a repertoire of mechanisms until now only sparsely documented, namely longer intergenic regions, post-horizontal gene transfer (HGT) gene fusions fostering coordinated expression, gene relocation, and possibly recombination generating mosaic genes. The results provide additional evidence that HGT occurred recurrently in this yeast lineage and was crucial for the reestablishment of lost functions and that similar mechanisms are used across a broad range of eukaryotic microbes to promote adaptation of prokaryotic genes to their new environment.
Horizontal transfer of genes (HGT) from bacteria to various eukaryotic settings is well documented over the entire scope of eukaryotes (1–4), including eukaryotic microbes comprising fungi, protists, and algae (5–9). Notwithstanding this, the events leading to the selection and fixation of xenologous genes in their new environment are still relatively poorly understood. One important hurdle that needs to be overcome for protein coding genes is the fact that while expression is a prerequisite for selection, mechanisms of gene expression are widely distinct between eukaryotes and prokaryotes. This is particularly relevant for horizontally acquired traits that require the action of more than one gene product, because this means that all genes involved must be functional in order for the selectable phenotype to be expressed.
Functionally related genes are often located in genomic proximity. In bacteria, they are organized in operons and are transcribed as polycistronic mRNAs, while in eukaryotes they may be found in clusters of independently transcribed genes (10–12). This is thought to facilitate horizontal transfer of function related genes between similar organisms in a single event (13–21). Remarkably, this also seems to apply to horizontal transfers of operons from bacteria to eukaryotes according to previous reports (6, 22–25). Bacterial operons transferred to eukaryotic organisms have been proposed to fulfill important roles such as changes in the nutritional status of the host (22) or adaptation to extreme environments (23, 25). Different mechanisms presumed to have facilitated a transition from bacterial operon transcription to eukaryotic-style gene expression were proposed, such as gene fusion giving rise to multifunctional proteins (6, 23, 24), increase in intergenic distances between genes to generate room for eukaryotic promoters, and independent transcription producing mRNAs with poly(A) tails have been demonstrated (22). In the best documented study, which concerns a bacterial siderophore biosynthesis operon acquired by yeasts belonging to the Wickerhamiella/Starmerella (W/S) clade, the bacterial genes acquired as an operon were shown to be functional (22).
Thiamine, commonly known as vitamin B1, is essential for all living organisms because its active form, thiamine pyrophosphate (TPP), is an indispensable cofactor of enzymes participating in amino acid and carbohydrate metabolism (26–30). Some organisms that are unable to synthesize thiamine de novo are nevertheless capable of using a salvage pathway to rescue the pyrimidine (hydroxymethylpyrimidine or HMP) and thiazole (hydroxyethylthiazole or HET) precursors and similar compounds that result from natural thiamine degradation in the surrounding environment (31, 32).
In the present study we describe a composite thiamine salvage pathway made up of yeast and bacterial genes found in several species of the yeast W/S clade. Our recent work (5) revealed that the W/S-yeast clade harbors an unusual large number of HGT events in yeasts, mostly as single genes, a finding independently confirmed in a large study in which ∼300 yeast genomes were examined (9). Here we show that in W/S-clade species most of the genes in the thiamine salvage pathway were originally acquired from bacteria as part of an operon and we use genetic dissection to link each of the transferred genes with an observable phenotype in the yeast setting. Moreover, we present evidence for the occurrence of 2 independent horizontal operon transfer (HOT) events in distinct subclades within the W/S lineage that, complemented by the independent horizontal acquisition of single functionally related genes, endow the yeast host with the ability to salvage a wide range of thiamine precursors.
Results
Thiamine Genes in the W/S-Yeast Clade.
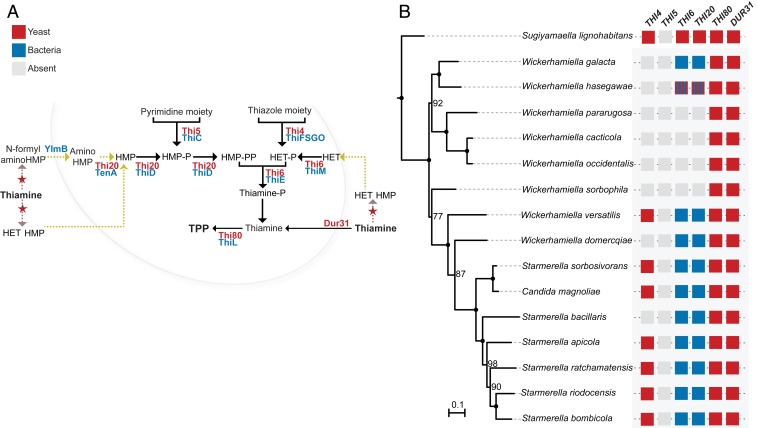
Many organisms lack the genes encoding enzymes of the 2 upper branches of the pathway used to generate the pyrimidine and thiazole precursors for de novo biosynthesis of thiamine, while maintaining only the genes composing the salvage pathway for thiamine or its precursors (31–33). Thi5 and Thi4 are the most important enzymes in each of the 2 pathway branches required for de novo thiamine synthesis (34) (Fig. 1A). THI5 is absent in all W/S clade species examined and THI4 is missing in about half of the species in the clade. They maintain however the indispensable genes encoding a thiamine transporter, Dur31 (35), and the kinase Thi80, responsible for TPP synthesis (Fig. 1 A and B). Species that lack THI5, or both THI5 and THI4, often maintain the salvage pathway genes THI6 and THI20 that enable them to scavenge thiamine derivatives from the environment (36, 37). In W/S-clade species, significant tBLASTx hits (e-value < e−10) were retrieved from available genomes for the THI6 and THI20 genes (Fig. 1 A and B). These 2 genes are contiguous in the genome of all W/S-clade species, but not in the genomes of most Saccharomycotina yeasts (SI Appendix, Fig. S1). Thi20 seems to be encoded by 2 separate genes, while for Thi6 a single gene was predicted in most species. A subsequent BLASTp search using the 3 predicted proteins from St. bombicola (2 corresponding to different domains of THI20 and 1 corresponding to THI6) retrieved bacterial proteins as top hits (Fig. 1B and Dataset S1) and no yeast proteins among the first 1,000 hits. The same result was obtained for all W/S-clade THI6 or THI20 homologs, suggesting that these genes, when present, always have bacterial origin in the W/S clade. The proteins exhibiting the Thi6 and Thi20 enzymatic activities are both encoded by 2 genes each in most bacteria (thiE and thiM, and tenA and thiD, respectively), as depicted in Fig. 1A. It was not possible to ascertain the origin of THI6 and THI20 in Wickerhamiella hasegawae, because preliminary BLASTx searches using the C-terminal and N-terminal domains of the Thi6 and Thi20 from this species showed a mosaic-like pattern for both genes, where part of the protein exhibited high sequence similarity with bacterial proteins while the other domain presented homology with fungal proteins (Fig. 1B and Dataset S2).
Fig. 1.
(A) Thiamine biosynthetic pathway in bacteria and yeast. Genes involved in thiamine biosynthesis in B. subtilis are represented in blue while their counterparts in S. cerevisiae are represented in red. Yellow arrows represent the salvage pathways that can be present in both yeast and bacteria. The red stars represent thiamine degradation. (B) Presence and absence of the main genes involved in the de novo and salvage thiamine biosynthesis in the W/S clade. Gray squares denote missing genes; blue squares, genes of bacterial origin; and red squares, genes of fungal origin. The origin of the W. hasegawae THI6 and THI20 genes was unclear. Phylogenetic relationships between species are depicted based on a ML phylogeny constructed as described in Materials and Methods. Branches with 100% support are indicated by black dots while bootstrap values >75% are shown next to the respective branches.
Independent Acquisitions of Bacterial Thiamine Operons by W/S-Clade Species.
The fact that THI genes are found adjacent to each other in the genomes of W/S-clade yeasts led us to examine the possibility that they were acquired in a single event as an operon.
The thiamine operon is organized differently in distinct bacteria (38), which is thought to result from reshuffling during evolution and is observed in many other instances (39–41). Hence, gene order and content, as phylogenetic analysis, can be indicative of the plausibility of HOT occurrences as opposed to single gene acquisitions, as well as of the bacterial lineage that originated a putative HOT event.
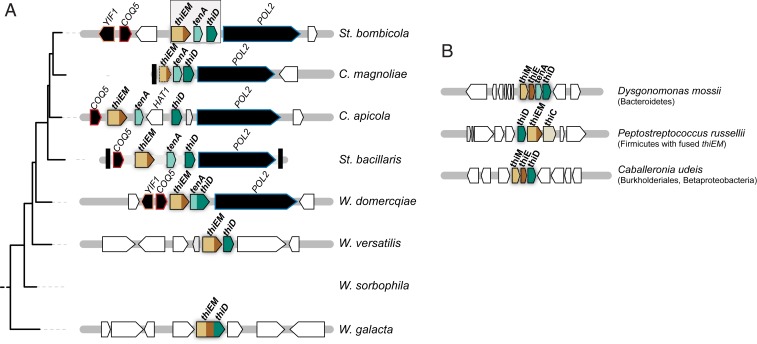
Since the results of BLASTp searches suggested that the possible bacterial donors of THI genes might belong to the Bacteroidetes or the Firmicutes for most W/S-clade species or the Burkholderiales (Betaproteobacteria) for Wickerhamiella galacta (Dataset S1), gene content and order of the thiamine clusters identified in the W/S clade were compared to operons found in extant representatives of these 3 bacterial lineages. For most W/S-clade species, the organization of the thiamine cluster resembled that of the thiamine operon in the Bacteroidetes (Fig. 2 A and B). The exceptions were W. galacta and Wickerhamiella versatilis in which tenA was found at a different genomic location (Fig. 2A). Hence, cluster organization in these 2 species resembled instead the Burkholderiales (Betaproteobacteria; Fig. 2B).
Fig. 2.
Thiamine operon organization. (A) Organization of THI genes in the genomes of representative W/S-clade species. Thiamine metabolism-related genes are represented by different colors. Syntenic genes between species are represented in black while nonsyntenic genes are represented in white. Black vertical bars represent ends of scaffolds. (B) Thiamine operon organization in putative donor lineages. A representative species belonging to each order/phylum is shown. Genes and intergenic regions are drawn to scale. Arrows denote direction of transcription.
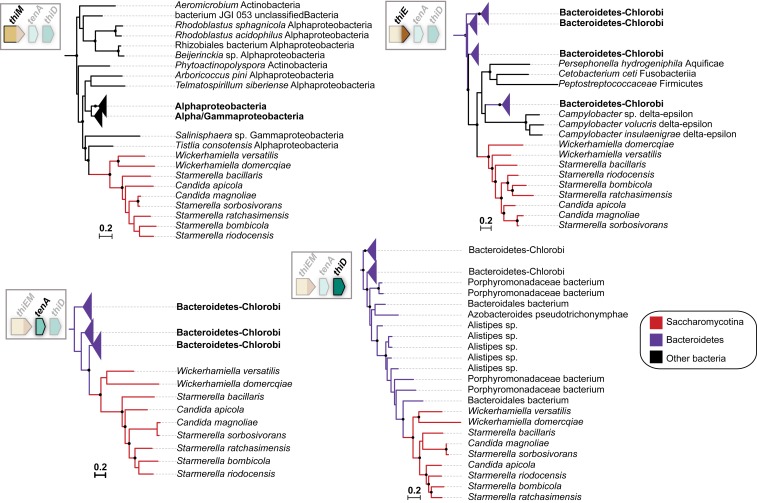
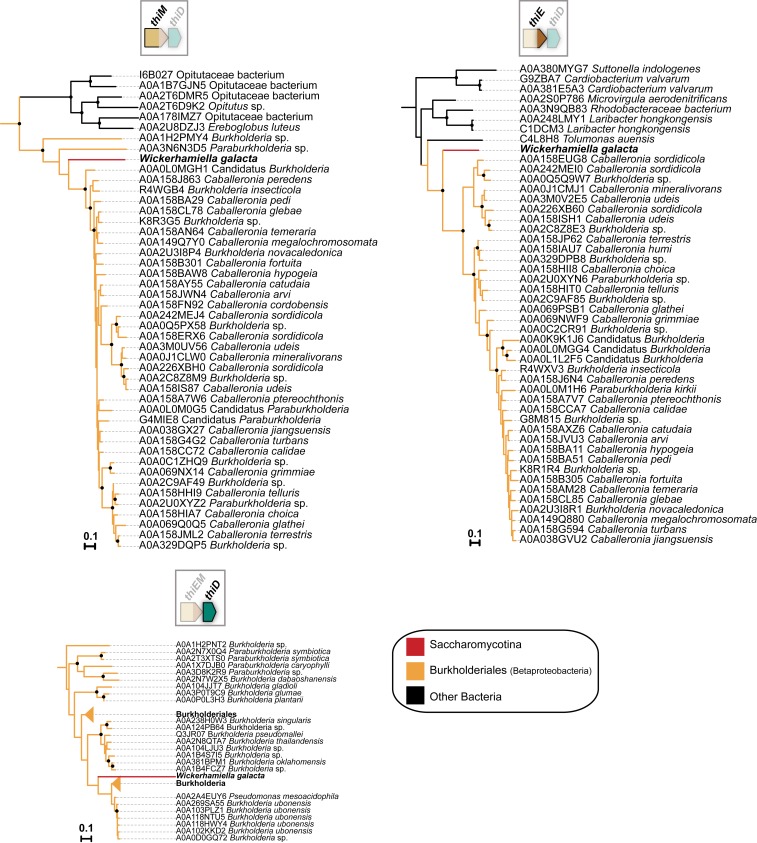
To investigate further the origin of the thiamine cluster found in the W/S clade, a detailed phylogenetic analysis was subsequently performed. The THI6 homolog (henceforth named thiEM) encompasses regions that are homologous to bacterial thiE (upstream portion of THI6) and thiM (downstream portion of THI6). Fusions between these 2 genes also occur in bacteria but are apparently uncommon (42). In our BLASTp searches using the fused version of ThiEM from St. bombicola, we could identify similar fusion events in Actinobacteria and in the Firmicutes (Dataset S1). However, preliminary phylogenetic analysis showed that the thiEM gene fusion found in the W/S clade is unrelated to the bacterial fusions in the 2 former bacterial phyla (SI Appendix, Fig. S2). Hence, independent phylogenies were constructed for the ThiE and ThiM portions of the ThiEM protein (Fig. 3). The phylogenetic signal for the 2 protein moieties in ThiEM was not consistent, either when protein moieties are analyzed separately (Fig. 3) or as a fusion protein (SI Appendix, Fig. S2). Nevertheless, the closest bacterial relatives suggested by the independent phylogenies (Fig. 3) are species in the Proteobacteria for ThiM and in the Bacteroidetes for ThiE. The latter is in line with the congruent phylogenies obtained for TenA and ThiD that also suggest a donor in the Bacteroidetes and is also consistent with the findings concerning gene order and content (Fig. 2). Given that the phylogenetic position of TenA from W. versatilis strongly supports a common origin with the remaining species in the St. bombicola subclade (Fig. 3 and SI Appendix, Fig. S2), the distinct genomic location of the tenA gene is likely the result of a postacquisition rearrangement. In Wickerhamiella domercqiae, a fusion between the tenA and thiD genes was observed (Fig. 2A). In independent phylogenies the TenA and ThiD moieties cluster with the cognate proteins in other species of the St. bombicola subclade, in positions consistent with the phylogenetic relationships between the species (Figs. 3 and 1B), strongly suggesting that the fusion occurred postacquisition.
Fig. 3.
Maximum likelihood phylogenies of Thi proteins in the W/S clade. Pruned phylogenies showing the closest relatives to W/S-clade proteins are shown. Branches with support higher than 95% (ultrafast bootstrap) are indicated by black dots.
In W. galacta, the thiD gene is fused instead to thiE and thiM, while tenA is in another genomic location. Again, independent phylogenies were constructed with the ThiD, ThiE, and ThiM homologous portions separately. The 3 protein moieties cluster with homologs in the Burkholderiales (Betaproteobacteria, see Fig. 4 and SI Appendix, Fig. S2), albeit with weak support for ThiD (Fig. 4), while the donor lineage in the case of tenA seems to be distinct, probably belonging to the Actinobacteria (SI Appendix, Figs. S3 and S2). Acquisition of a tenA-lacking operon from the Burkholderiales (Betaproteobacteria) by W. galacta, with the tenA gene originating from a different donor is also consistent with the gene order and content information shown in Fig. 2. The phylogenetic analyses including all putative bacterial donor lineages and transferred genes (SI Appendix, Fig. S2) also support the independent origin of the bacterial operons found in the St. bombicola and W. galacta subclades and of tenA in W. galacta.
Fig. 4.
Maximum likelihood phylogenies of Thi proteins from W. galacta. Pruned phylogenies showing the closest relatives to W. galacta Thi proteins encoded in the operon are shown. Branches with support higher than 95% (ultrafast bootstrap) are indicated by black dots. The phylogenetic tree for TenA is shown in SI Appendix, Fig. S3.
Hence, taken together, our data support the identification in the W/S clade of 2 independent HOT events, as well as the independent acquisition of tenA by W. galacta.
Horizontally Acquired THI Genes Participate in a Thiamine Salvage Pathway.
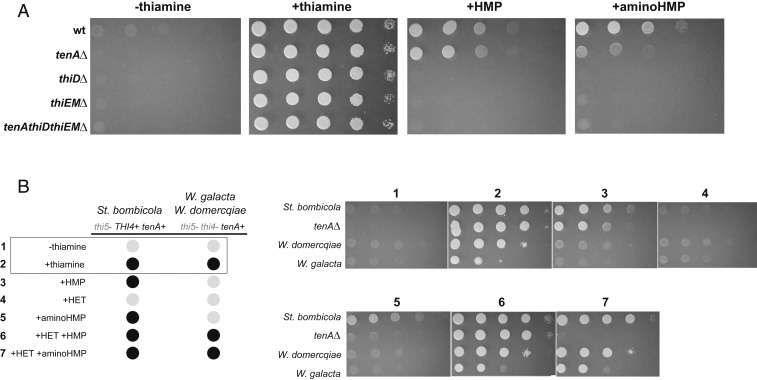
All W/S-clade species examined are predicted to be auxotrophic for thiamine due to the absence of THI5 (Fig. 1B). However, they may be able to rescue 4-amino-2-methyl-5-(phosphooxymethyl)pyrimidine (HMP) from the environment if their salvage pathway of bacterial origin composed of thiEM and thiD (Fig. 1A) is functional. In accordance with this, wild-type (WT) St. bombicola was shown to grow on medium lacking thiamine but supplemented with HMP (Fig. 5A). Next, St. bombicola deletion mutants (tenA∆, thiD∆, thiEM∆, and tenA∆thiD∆thiEM∆) were constructed and their ability to grow was assessed in medium supplemented with different thiamine precursors.
Fig. 5.
Assimilation of thiamine derivatives by St. bombicola WT and deletion mutants. (A) Growth assays for St. bombicola strains (WT and mutants) cultivated on YNB without thiamine and supplemented with 0.2 µM of thiamine, 0.2 µM of HMP or 0.02 µM of aminoHMP, after 5 d of incubation at 25 °C. (B) Growth assays for other W/S-clade species in the presence of 0.2 µM of HET, 0.2 µM of HMP, or 0.02 µM of aminoHMP and different combinations of these compounds.
As expected from their predicted role in the putative salvage pathway, deletion of thiD and/or thiEM rendered the strains unable to use HMP (Fig. 5A). Deletion of tenA did not have an effect on growth on HMP-supplemented medium, in line with this protein having an amino-hydrolase activity capable of producing HMP from 4-amino-5-aminomethyl-2-methylpyrimidine (aminoHMP) (43, 44), and therefore acting upstream of HMP synthesis (Fig. 1A). To assess this possibility, growth in the presence of aminoHMP was also evaluated. As shown in Fig. 5A, the tenAΔ mutant grows notably worse than the WT strain in medium containing aminoHMP as a source of thiamine (Fig. 5 A and B), although some residual growth of the mutant was observed (Fig. 5A). Growth on aminoHMP as sole source of thiamine was also evaluated in liquid media (SI Appendix, Fig. S4), where the WT attained much higher cell densities than the tenAΔ mutant. As expected, no differences in growth were observed in thiamine- or HMP-supplemented media in this mutant (SI Appendix, Fig. S4). Together, these results show that aminoHMP is a substrate for TenA, but that an additional source of aminohydrolase activity, possibly unspecific, seems to be present, supporting residual growth of the tenAΔ mutant in aminoHMP particularly in solid medium.
Assimilation tests were also used to evaluate the functionality of the thiamine salvage pathway in 2 additional W/S-clade species, W. domercqiae and W. galacta. Both species lack THI4, so that to test assimilation of HMP or aminoHMP, it was also necessary to supplement the medium with 5-(2-hydroxyethyl)-4-methylthiazole (HET) (Figs 1A and 5 B, Left). As shown in Fig. 5 B, Right, when either HET, HMP, or aminoHMP were provided separately, no growth was observed for these species, as expected in the absence of THI4. Also as predicted, a combination of HMP and HET restores growth in both species. Notably, W. galacta was also able to grow when aminoHMP and HET were added to the growth medium showing that although tenA is not part of the thiamine cluster, and was likely acquired from a different bacterial lineage, it is operating as part of the salvage pathway by supplying the pyrimidine moiety of thiamine. Similar results were obtained for W. domercqiae, which could also grow when HET and aminoHMP were simultaneously supplied (Fig. 5 B, Right).
In Bacillus subtilis, in addition to TenA that is responsible for the conversion of aminoHMP to HMP, the thiamine salvage pathway involves another protein, YlmB, which is essential for the deformylation of N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine (N-formyl-aminoHMP), a compound commonly resulting from thiamine degradation (43, 44). YlmB converts this compound in aminoHMP, thus acting upstream of TenA (Fig. 1A) (43). Interestingly, we found evidence for the presence of a putative YlmB (annotated as an acetylornithine deacetylase)-encoding gene in most W/S-clade species (SI Appendix, Fig. S5). This gene was also horizontally acquired from bacteria, but from a donor belonging to the Acetobacteraceae family (Alphaproteobacteria; SI Appendix, Fig. S5).
Expression of Thiamine Cluster Genes.
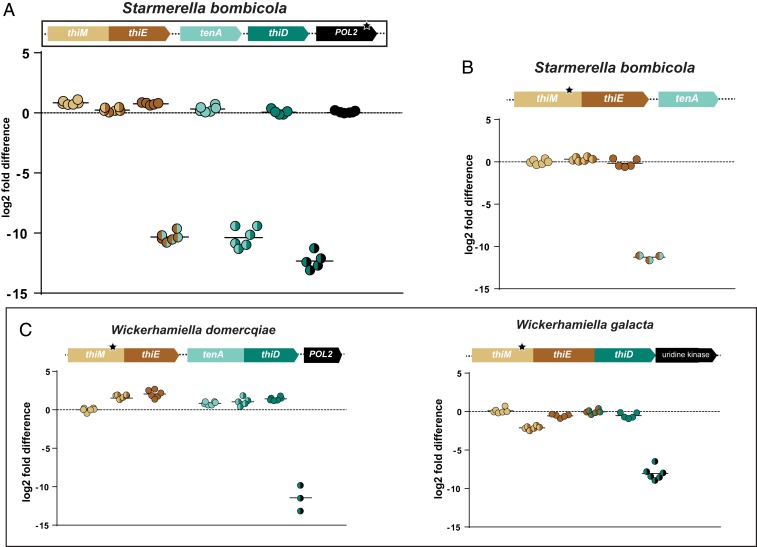
If the salvage pathway operating in extant W/S-clade species is derived from a bacterial operon, its expression can be presumed to be currently adapted to eukaryotic canonical transcription. Polycistronic mRNAs (and operons) are rare in eukaryotes (45–47), the few known cases involving processing of extended polycistronic pre-mRNAs into monocistronic mRNAs by 3′ end formation and polyadenylation and subsequent transsplicing by a small nuclear ribonucleoprotein (48–50). The siderophore biosynthesis gene cluster described also in W/S-clade yeasts is the best studied case of adaptation of a bacterial operon to the eukaryotic setting (22). In this case, intergenic regions were shown to be lengthier than observed in the putative bacterial donors, which was also observed for THI genes in W/S-clade species (Fig. 2A). Expression of the THI genes was first investigated in St. bombicola using reverse transcription qPCR and gene-specific primers (Fig. 6A). Similar levels of expression were observed within gene amplicons while expression of fragments spanning any 2 independent genes was several orders of magnitude lower (Fig. 6A). Nevertheless, the fragment spanning the fusion point between the thiM and thiE moieties in the thiEM gene was slightly less expressed (∼4-to 5-fold), which could result from the coexistence of a mRNA spanning the 2 fused genes with shorter mRNAs encompassing only 1 of the genes. To find out to which extent these transcripts conformed with a canonical eukaryotic structure, we repeated the experiment for St. bombicola but using an oligo(dT)20 primer to synthesize the first cDNA strand, followed by qPCR with gene-specific primers, which will reveal only mRNAs possessing poly(A) tails. Results were very similar, emphasizing that most transcripts have poly(A) tails (Fig. 6B). However, this time expression of the fragment encompassing the thiM/thiE fusion was similar to the expression within gene amplicons, implying that some of the transcripts spanning only the thiM or the thiE moieties may lack poly(A) tails. Next, we examined poly(A)-tailed transcripts along the fused THI genes in 2 additional species, W. dormercqiae and W. galacta (Fig. 6C). Unlike the observations for St. bombicola, the results indicate that in some cases, in addition to transcripts potentially encompassing the complete fused genes encoding multidomain proteins, shorter poly(A)-tailed mRNA molecules are also produced. For example, in W. galacta, transcripts containing the fusion site between the thiM and thiE moieties are evidently less abundant than transcripts containing the other regions probed within the triple fusion between the thiM, thiE, and thiD genes, suggesting that poly(A) transcripts comprising only the thiM gene on the one hand and the thiE/thiD fusion gene on the other hand, are also generated in significant amounts.
Fig. 6.
Expression profiles of THI genes in W/S-clade species. (A) Expression of THI genes (depicted as the log2 fold difference) in St. bombicola relative to the internal reference POL2 (black star) using only gene-specific primers. Expression levels of intergenic regions are also depicted. (B) Expression of the thiEM fusion gene in St. bombicola after preselection of poly(A)-tailed mRNAs. (C) Expression of THI genes in W. domercqiae and W. galacta after preselection of poly(A)-tailed mRNAs. Mean values are represented by the black horizontal lines. Raw data can be accessed in Dataset S3. The black stars indicate the gene that served as reference in each case.
Discussion
De novo synthesis of thiamine is impaired in W/S-clade species because they lack the THI5 and in some cases also the THI4 genes, responsible respectively for synthesis of the pyrimidine (HMP) and thiazole (HET) precursors of thiamine. In addition, yeast orthologs of downstream components of the pathway, THI6 and THI20, are also missing in this lineage. However, we showed here unequivocally for one W/S-clade species using genetic evidence and for other species by examining their ability to salvage various thiamine precursors, that this does not render these organisms dependent solely on import of thiamine from the outside. Instead, clusters of THI genes originating from bacterial operons, complemented by other individual genes, form functional salvage pathways capable of rescuing thiamine degradation products from the environment thereby providing cells with sufficient TPP synthesis capacity to support growth.
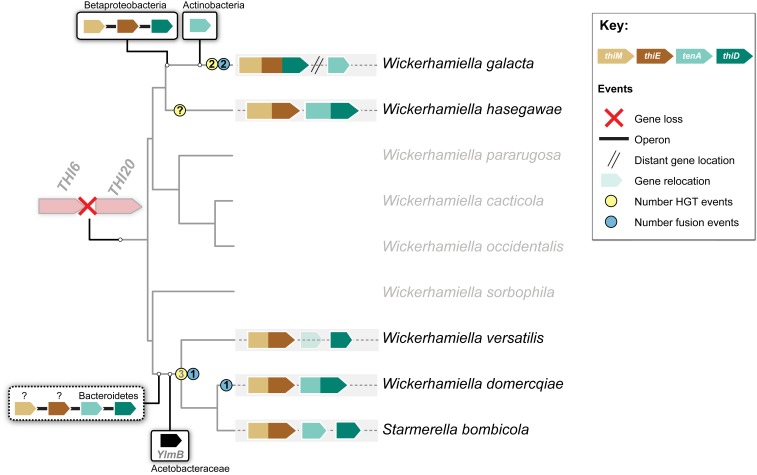
Our findings suggest an evolutionary model (Fig. 7) in which the THI6 and THI20 genes, that are rarely found clustered in yeast genomes, were lost in an ancestor of the W/S clade, in addition to THI5, resulting initially in obligate reliance on external sources of thiamine. THI4 was also lost in several species, which could be related to its mode of action as a suicide enzyme (51). We found within the W/S clade several different subsequent outcomes of the loss of thiamine prototrophy. Three of the species examined belonging to 1 of the 2 sister lineages within the W/S clade represented in this study (Wickerhamiella cacticola, Wickerhamiella pararugosa, and Wickerhamiella occidentalis) still lack any gene resembling THI6 and THI20, while the 2 remaining species (W. hasegawae and W. galacta) examined in this subclade present evidence of horizontal acquisition of xenologs, in the case of W. galacta from the Burkholderiales (Betaproteobacteria). In 9 of the 10 species forming the second subclade, THI6 and THI20 homologs also seem to be of bacterial origin, but from a different phylum, the Bacteroidetes. According to our evidence, this transfer event took place in the common ancestor of the 9 species, which is consubstantiated by the fact that the phylogenies of THI genes recapitulate the species phylogeny.
Fig. 7.
Main events in the evolution of THI genes in the W/S clade. Schematic phylogenetic relationships between W/S species are depicted based on the ML phylogeny presented in Fig. 1B. Loss of native THI6 and THI20 genes in the most recent common ancestor (MRCA) of the W/S clade is indicated by a red cross. Putative horizontal gene transfer events are represented by boxes above/below which the putative donor lineage is indicated. The number of putative HGT events and fusion events are indicated in yellow and blue circles, respectively; numbers in gray represent cases where the number of HGT events could not be asserted with certainty. Operons are represented by a straight black line linking the genes, while the uncertainty surrounding the origin of thiE and thiM in the St. bombicola subclade is denoted by question marks. Horizontal acquisition of the putative N-formyl-4-amino-5-aminomethyl-2-methylpyrimidine deformylase (YlmB) is also shown. The origin of the W. hasegawae genes is elusive.
The activities of the THI20 and THI6 gene products are encoded by 4 genes of which at least 3 are organized in an operon in both putative donor lineages, the Bacteroidetes and the Burkholderiales (Betaproteobacteria). The identity of the donor lineages suggested by the phylogeny is further supported by the fact that gene content and gene order in W/S-clade THI clusters is identical to the operons found in extant representatives of the 2 donor lineages. This in turn strongly suggests that an entire operon was transferred in a single event in both cases. However, inference of the donor lineage originating the THI cluster in St. bombicola and neighboring species was based on phylogenies obtained for the TenA and ThiD proteins only, because it was not possible to obtain a reliable phylogenetic signal for either the ThiE/M fused proteins or the separate ThiE and ThiM moieties, leaving the possibility open that these genes were transferred in an independent event from the tenA and thiD genes and possibly from a different donor. It seems more likely, however, that such a putative second event did not occur entirely independently of the first since sequence similarity may have promoted recombination involving the thiE/thiM genes acquired initially from the Bacteroidetes as part of the operon transfer and homologous genes from a distinct unidentified bacterial lineage, as observed before (52–55). Alternatively, recombination events in the bacterial donor lineage prior to the acquisition by yeasts might also have resulted in the conflicting phylogenetic signal observed for ThiEM. Even more elusive is the origin of the THI6 and THI20 homologs from W. hasegawae, which seem to be mosaics of uncertain origin exhibiting regions of homology with both bacterial and fungal proteins (Dataset S2). Interestingly, no evidence for mosaicism was reported for the siderophore biosynthesis gene cluster in W/S-clade yeasts (22).
In addition to HOTs related to thiamine metabolism we also detected complementary single acquisitions of functionally related genes from bacteria that further extended the range of thiamine precursors that could be salvaged by W/S-clade species (Fig. 7). The best example of this in our data is W. galacta that acquired a tenA-lacking operon from the Burkholderiales (Betaproteobacteria), but nevertheless seems to have been able to acquire a tenA gene from the Actinobacteria. Other W/S-clade species also seem to have acquired a putative ylmB gene from the Acetobacteraceae (Alphaproteobacteria) in addition to a tenA-containing operon from the Bacteroidetes. These 2 genes encode enzymes that mediate the utilization of aminoHMP (tenA) and N-formyl-aminoHMP (ylmB) as sources of HMP. These compounds are originated by thiamine degradation which can occur naturally (43) or by the action of microorganisms (56). The formation of HMP as a degradation product of thiamine has been reported to occur also under different pH and temperature conditions in laboratory experiments (57, 58). Consequently, the products of thiamine degradation (such as HMP, HET, aminoHMP, etc.) may be more abundant than thiamine itself in the environment (56). Moreover, some thiamine breakdown products, such as N-formyl-aminoHMP or aminoHMP, can be toxic (59, 60) so that TenA and YlmB may also have a detoxification role.
Horizontally transferred genes are probably quickly lost if they are not selected for, and for protein coding genes this means that they have to be expressed. Eukaryotic canonical gene expression imposes very different prerequisites from those found in bacterial operons, namely monocystronic mRNAs with appropriate 3′ and 5′ modifications and markedly different promoters. These adaptations were recently studied for the first time for the siderophore biosynthesis gene cluster described in W/S-clade yeasts (22). The occurrence of multiple HOT events in this clade probably reflects the previously reported large number of HGT events in general (5, 9, 22, 61). Common to both events in the W/S clade is the observed increase in intergenic spacing possibly to accommodate de novo evolved promoters. However, adaptation of the W/S-clade siderophore biosynthesis operon did not involve postacquisition gene fusions, of which at least 3 independent examples were observed in the W/S-thiamine cluster (Fig. 7). We interpreted this as an effective means to achieve coordinated expression of the bacterial genes. Gene fusions were also observed in HOT events in protists (6, 23, 24) implying that this may be a general and frequent mechanism employed to transpose coordinated prokaryotic expression to the eukaryotic context. The absence of gene fusions in the long siderophore gene cluster may be due to restrictions imposed by proper functioning of the enzymes which may be incompatible with fusion.
Our data may suggest that individual expression of the genes, possibly driven by spurious promoters, preceded the fusions, because in the pool of poly(A)-tailed mRNAs, shorter transcripts seem to coexist with the mRNAs spanning the complete fusion genes. Overlapping mRNAs were also detected for the siderophore biosynthesis gene cluster from W. versatilis (22).
Taken together, our observations revealed examples of mechanisms facilitating expression of xenologous genes observed so far sporadically in HOT occurrences in various eukaryotic microbes, namely multiple instances of post-HGT gene fusions, increased spacing between genes, dispersion of genes to different genomic locations, and likely instances of recombination leading to the formation of mosaic genes. These observations support the idea that the repertoire of mechanisms for adaptation of gene expression are consistently and frequently used across the eukaryotic domain to functionalize genes horizontally acquired from bacteria. Moreover, they show that successive layers of horizontal transfer events were involved in fine tuning a previously acquired metabolic pathway.
Materials and Methods
Strains.
Yeast strains were obtained from the Portuguese Yeast Culture Collection, Caparica, Portugal (PYCC) except for W. versatilis JCM 5958, which was kindly provided by the Japan Collection of Microorganisms, Tsukuba, Japan (JCM), and W. galacta NRRL Y-17645, which was obtained from Agricultural Research Service Culture Collection, Peoria, IL (ARS-NRRL). All strains were maintained in YMA medium [1% (wt/vol) glucose, 0.3% (wt/vol) malt extract (wt/vol), 0.3% yeast extract, 0.5% (wt/vol) peptone and 2% (wt/vol) agar].
Identification of Genes Involved in TPP Biosynthesis.
Genes related with TPP de novo biosynthesis (THI4, THI5, THI6, THI7, THI20, and THI80) were searched in the genomes of the W/S clade using Saccharomyces cerevisiae S288C homologs as queries (NC_001139.9, NC_001138.5, NC_001148.4, NC_001144.5, NC_001147.6, and NC_001147.6, respectively). The respective best hits were subsequently blasted against the National Center for Biotechnology Information (NCBI) nonredundant (nr) database. Whenever the best hit in NCBI corresponded to the identity of the query gene, it was assumed that the gene was present.
Phylogenetic Analyses.
The species phylogeny represented in Fig. 1B was constructed using the same dataset as in Gonçalves et al. (5) (see SI Appendix, Table S1 for the complete list of taxa used in the phylogeny) based on a previously described methodology (62) with the addition of other W/S-clade species whose genomes were recently published in the context of the Y1000+ Project (9, 22). Briefly, Rpa1, Rpa2, Rpb1, Rpb2, Rpc1, and Rpc2 protein sequences for each species were used to construct the maximum likelihood (ML) tree with RAxML (63) v7.2.8 using the PROTGAMMAILG model of amino acid substitution and 1,000 rapid bootstraps. Branch support values (>75%) are displayed. This tree is in agreement with the recently published phylogeny from Kominek et al. (22).
Independent phylogenies were constructed for the N-terminal (ThiM) and C-terminal (ThiE) domains of the ThiEM protein. For the putative proteins from bacterial origin TenA, ThiD, ThiE, and ThiM a preliminary BLASTp search against the nr NCBI database was performed and it was confirmed that the top 1,000 hits only included bacterial proteins except for the ThiE portion for which some nonbacterial (fungi, plants) sequences were also recovered (Dataset S1). Preliminary phylogenies using the top 5,000 hits were constructed (SI Appendix, Fig. S2) and confirmed that all Thi proteins clustered with bacterial homologs. A more detailed phylogeny was subsequently performed using only the top 750 bacterial hits found in UniProtKB (UNIPROTKB_BACTERIA) using St. bombicola proteins as queries (TenA, ThiD, ThiM, and ThiE portions). The protein sequences for other W/S-clade species were obtained by tBLASTx in the local genome databases. Sequences with more than 98% similarity were removed with CD-HIT v4.6.7 (64) and the remaining sequences were aligned with MAFFT v7.222 (65) using an iterative refinement method (L-INS-i). Poorly aligned sequences were removed with trimAl v1.2 (66) using its “gappyout” option. Phylogenies were constructed with IQ-TREE v1.6.6 (67) using the LG+I+G4 model of substitution (found as the best fitting model for all of the 4 alignments) and ultrafast bootstrap (-bb 1,000) (68) for branch support determination.
Because both the BLASTp results and preliminary phylogenies for Thi proteins from W. galacta showed that they were probably acquired from a lineage belonging to the Burkholderiales (Betaproteobacteria), independent phylogenies were also constructed with their closest relatives. For that, the top 750 hits found in UniProtKB (UNIPROTKB_BACTERIA) to W. galacta Thi proteins were used. Alignments were trimmed and phylogenies were constructed as aforementioned.
The overall organization of THI genes was investigated for 2 species belonging to the respective putative donor bacterial lineages: Caballeronia udeis and Burkholderia novazelandica (Burkholderiales, putative donor to the THI operon in W. galacta), Dysgonomonas mossi and Mangrovibacterium marinum (Bacteroidetes, putative donor to the THI operon in other W/-clade species), and Peptostreptococcus russelli and Veillonella dispar (Firmicutes encoding thiEM fusion genes). See SI Appendix, Table S2 for information on the genome assemblies that were used.
Complete phylogenies, alignment files, and trimmed files can be accessed in Figshare (69).
Investigation of Operon-Like Features and Gene Fusions in W/S-Clade Species.
St. bombicola PYCC 5882, W. domercqiae PYCC 3067, and W. galacta NRRL Y-17645 cells were grown in YPD [1% (wt/vol) yeast extract, 2% (wt/vol) peptone, 2% (wt/vol) glucose] for 24 h. Total RNA was extracted using the standard TRIzol method followed by purification using the RNA Clean & Concentrator-5 kit from Zymoresearch (Irvine, CA) which included a DNase treatment for genomic DNA removal. Absence of residual gDNA in RNA samples was confirmed by PCR with all of the primer pairs used for qPCR reactions. A total of 1 µg of total RNA was used for first-strand cDNA synthesis using the Maxima H Minus Reverse Transcriptase (Thermo Fisher Scientific) following the manufacturer’s guidelines and an oligo(dT)20 primer. Real-time quantitative RT-PCR (RT-qPCR) assays were performed using the SensiFAST SYBR No-Rox Kit from Bioline (London), with 20-µL reactions with 70 nM of each primer (SI Appendix, Table S3) and 50 ng of cDNA. Primers used are listed in SI Appendix, Table S3. The reaction consisted of a first step at 95 °C for 2 min followed by 40 cycles of 5 s at 95 °C, 10 s at 60 °C, and 10 s at 72 °C. Total RNA was also used in a 1-step amplification using the SensiFAST SYBR No-Rox One-Step Kit from Bioline. The reaction consisted of a first step at 45 °C for 10 min, followed by 1 step at 95 °C for 2 min. These steps were followed by 40 cycles of 5 s at 95 °C, 10 s at 60 °C, and 10 s at 72 °C. Two independent RNA and cDNA samples were obtained and 3 replicates per qPCR reaction were used. The efficiency of each primer pair was calculated using 5 10-fold DNA dilutions and are presented in SI Appendix, Table S3. Relative expression was calculated using the 2−∆Ct method where ∆Ct = (Cttest − Ctref) and Cttest is the Ct of the expression of the fragment under study and Ctref is the Ct of POL2 (Fig. 6A) or thiM (Fig. 6 B and C). Raw data are shown in Dataset S3.
Construction of St. bombicola Deletion Mutants.
Standard molecular biology techniques were performed essentially as described in ref. 70 using Escherichia coli DH5ɑ as host. St. bombicola PYCC 5882 was used in all procedures involving this species. Knockout cassetes (SI Appendix, Table S4) were constructed as described in Gonçalves et al. (5) using hygromycin as the selective marker. Transformation of St. bombicola was performed as described in ref. 5. After transformation, mutants were selected on YPD plates containing 650 µg/mL of hygromycin (InvivoGen). Correct integration of the disruption cassettes was verified by appropriate PCR reactions and by sequencing. Two different transformants from 2 independent gene disruption transformations were used for the phenotypic assays.
Growth Assays on Plates.
St. bombicola WT and mutants and other W/S-clade species were tested for growth in the presence of HMP, aminoHMP, and HET. Strains were first grown for 24 h on YMA [1% (wt/vol) glucose, 0.3% (wt/vol) malt extract (wt/vol), 0.3% yeast extract, 0.5% (wt/vol) peptone, and 2% (wt/vol) agar] medium and transferred to YNB without amino acids and without thiamine (Formedium, Norfolk, UK) plates to exhaust intracellular thiamine pools. Cells were washed with sterile water and resuspended in water to a final OD640n of 0.5. Cell suspensions were subsequently serially diluted 10-fold, spotted onto YNB plates without amino acids and without thiamine (Formedium), and supplemented with CSM (complete supplement mixture, MP Biomedicals), 0.2 µM of thiamine, 0.2 µM of HMP, 0.02 µM of aminoHMP, and 0.2 µM of HET combined with 0.2 µM of HMP or with 0.02 µM of aminoHMP (for strains that lack both THI4 and THI5). Plates were incubated at 25 °C for 5 d.
Supplementary Material
Acknowledgments
We thank PYCC for providing the strains used in this work; undergraduate students João Sousa, Ana Sousa, and Andreia Barro for performing some of the preliminary growth assays; and members of the YeastGenomicsLab, in particular Patrícia Brito, for fruitful discussions. We would also like to acknowledge the Y1000+ Project which was the source of genome data used in this work concerning 8 W/S-clade species. This work was supported by the UCIBIO-Unidade de Ciências Biomoleculares Aplicadas, which is financed by national funds from Fundação para a Ciência e Tecnologia, Ministério da Ciência, Tecnologia e Ensino Superior (FCT/MCTES; https://www.fct.pt/) UID/Multi/04378/2019 and grants FructYEAST - LISBOA-01-0145-FEDER-029529/PTDC/BIA-MIC/29529/2017 (to P.G.) and SFRH/BD/89489/2012 (to C.G.), both from FCT/MCTES.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All the alignment files and complete phylogenies and can be accessed in figshare (DOI: 10.6084/m9.figshare.9800636).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909844116/-/DCSupplemental.
References
- 1.Gladyshev E. A., Meselson M., Arkhipova I. R., Massive horizontal gene transfer in bdelloid rotifers. Science 320, 1210–1213 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Husnik F., McCutcheon J. P., Functional horizontal gene transfer from bacteria to eukaryotes. Nat. Rev. Microbiol. 16, 67–79 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Keeling P. J., Palmer J. D., Horizontal gene transfer in eukaryotic evolution. Nat. Rev. Genet. 9, 605–618 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Savory F., Leonard G., Richards T. A., The role of horizontal gene transfer in the evolution of the oomycetes. PLoS Pathog. 11, e1004805 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gonçalves C., et al. , Evidence for loss and reacquisition of alcoholic fermentation in a fructophilic yeast lineage. eLife 7, e33034 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stairs C. W., Roger A. J., Hampl V., Eukaryotic pyruvate formate lyase and its activating enzyme were acquired laterally from a Firmicute. Mol. Biol. Evol. 28, 2087–2099 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Schönknecht G., Weber A. P., Lercher M. J., Horizontal gene acquisitions by eukaryotes as drivers of adaptive evolution. BioEssays 36, 9–20 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Marcet-Houben M., Gabaldón T., Acquisition of prokaryotic genes by fungal genomes. Trends Genet. 26, 5–8 (2010). [DOI] [PubMed] [Google Scholar]
- 9.Shen X. X., et al. , Tempo and mode of genome evolution in the budding yeast subphylum. Cell 175, 1533–1545.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hurst L. D., Pál C., Lercher M. J., The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 5, 299–310 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Keller N. P., Turner G., Bennett J. W., Fungal secondary metabolism–From biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Boycheva S., Daviet L., Wolfender J. L., Fitzpatrick T. B., The rise of operon-like gene clusters in plants. Trends Plant Sci. 19, 447–459 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Wisecaver J. H., Rokas A., Fungal metabolic gene clusters-caravans traveling across genomes and environments. Front. Microbiol. 6, 161 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khaldi N., Collemare J., Lebrun M.-H., Wolfe K. H., Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 9, R18 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richards T. A., Genome evolution: Horizontal movements in the fungi. Curr. Biol. 21, R166–R168 (2011). [DOI] [PubMed] [Google Scholar]
- 16.Reynolds H. T., et al. , Horizontal gene cluster transfer increased hallucinogenic mushroom diversity. Evolution lett. 2, 88–101 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lawrence J., Selfish operons: The evolutionary impact of gene clustering in prokaryotes and eukaryotes. Curr. Opin. Genet. Dev. 9, 642–648 (1999). [DOI] [PubMed] [Google Scholar]
- 18.Lawrence J. G., Roth J. R., Selfish operons: Horizontal transfer may drive the evolution of gene clusters. Genetics 143, 1843–1860 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lawrence J. G., Selfish operons and speciation by gene transfer. Trends Microbiol. 5, 355–359 (1997). [DOI] [PubMed] [Google Scholar]
- 20.Slot J. C., Rokas A., Multiple GAL pathway gene clusters evolved independently and by different mechanisms in fungi. Proc. Natl. Acad. Sci. U.S.A. 107, 10136–10141 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marcet-Houben M., Gabaldón T., Horizontal acquisition of toxic alkaloid synthesis in a clade of plant associated fungi. Fungal Genet. Biol. 86, 71–80 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kominek J., et al. , Eukaryotic acquisition of a bacterial operon. Cell 176, 1356–1366.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsaousis A. D., et al. , Evolution of Fe/S cluster biogenesis in the anaerobic parasite Blastocystis. Proc. Natl. Acad. Sci. U.S.A. 109, 10426–10431 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersson J. O., Roger A. J., Evolutionary analyses of the small subunit of glutamate synthase: Gene order conservation, gene fusions, and prokaryote-to-eukaryote lateral gene transfers. Eukaryot. Cell 1, 304–310 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Posewitz M. C., et al. , Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J. Biol. Chem. 279, 25711–25720 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Friedrich W., “Thiamin, Vitamin B1, Aneurin” in Vitamins (de Gruyter, New York, 1988), pp. 353–359. [Google Scholar]
- 27.Butterworth R. F., Thiamin deficiency and brain disorders. Nutr. Res. Rev. 16, 277–284 (2003). [DOI] [PubMed] [Google Scholar]
- 28.Jordan F., Current mechanistic understanding of thiamin diphosphate-dependent enzymatic reactions. Nat. Prod. Rep. 20, 184–201 (2003). [DOI] [PubMed] [Google Scholar]
- 29.Trevelyan W. E., Harrison J. S., Studies on yeast metabolism. IV. The effect of thiamine on yeast fermentation. Biochem. J. 57, 561–566 (1954). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jurgenson C. T., Begley T. P., Ealick S. E., The structural and biochemical foundations of thiamin biosynthesis. Annu. Rev. Biochem. 78, 569–603 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karunakaran R., et al. , Thiamine is synthesized by a salvage pathway in Rhizobium leguminosarum bv. viciae strain 3841. J. Bacteriol. 188, 6661–6668 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gutowska M. A., et al. , Globally important haptophyte algae use exogenous pyrimidine compounds more efficiently than thiamin. MBio 8, e01459-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wightman R., Meacock P. A., The THI5 gene family of Saccharomyces cerevisiae: Distribution of homologues among the hemiascomycetes and functional redundancy in the aerobic biosynthesis of thiamin from pyridoxine. Microbiology 149, 1447–1460 (2003). [DOI] [PubMed] [Google Scholar]
- 34.Nosaka K., Recent progress in understanding thiamin biosynthesis and its genetic regulation in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 72, 30–40 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Donovan P. D., et al. , TPP riboswitch-dependent regulation of an ancient thiamin transporter in Candida. PLoS Genet. 14, e1007429 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Onozuka M., Konno H., Kawasaki Y., Akaji K., Nosaka K., Involvement of thiaminase II encoded by the THI20 gene in thiamin salvage of Saccharomyces cerevisiae. FEMS Yeast Res. 8, 266–275 (2008). [DOI] [PubMed] [Google Scholar]
- 37.Begley T. P., Ealick S. E., McLafferty F. W., Thiamin biosynthesis: Still yielding fascinating biological chemistry. Biochem. Soc. Trans. 40, 555–560 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taboada B., Ciria R., Martinez-Guerrero C. E., Merino E., ProOpDB: Prokaryotic operon dataBase. Nucleic Acids Res. 40, D627–D631 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Price M. N., Huang K. H., Alm E. J., Arkin A. P., A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33, 880–892 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Itoh T., Takemoto K., Mori H., Gojobori T., Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 16, 332–346 (1999). [DOI] [PubMed] [Google Scholar]
- 41.Belda E., Moya A., Bentley S., Silva F. J., Mobile genetic element proliferation and gene inactivation impact over the genome structure and metabolic capabilities of Sodalis glossinidius, the secondary endosymbiont of tsetse flies. BMC Genomics 11, 449 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Henry C. S., et al. , Systematic identification and analysis of frequent gene fusion events in metabolic pathways. BMC Genomics 17, 473 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jenkins A. H., Schyns G., Potot S., Sun G., Begley T. P., A new thiamin salvage pathway. Nat. Chem. Biol. 3, 492–497 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Bettendorff L., At the crossroad of thiamine degradation and biosynthesis. Nat. Chem. Biol. 3, 454–455 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Blumenthal T., Operons in eukaryotes. Brief. Funct. Genomics Proteomics 3, 199–211 (2004). [DOI] [PubMed] [Google Scholar]
- 46.Gordon S. P., et al. , Widespread polycistronic transcripts in fungi revealed by single-molecule mRNA sequencing. PLoS One 10, e0132628 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yue Q., et al. , Functional operons in secondary metabolic gene clusters in Glarea lozoyensis (Fungi, Ascomycota, Leotiomycetes). MBio 6, e00703 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blumenthal T., Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 11, 132–136 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Blumenthal T., Trans-splicing and operons in C. elegans. WormBook 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pouchkina-Stantcheva N. N., Tunnacliffe A., Spliced leader RNA-mediated trans-splicing in phylum Rotifera. Mol. Biol. Evol. 22, 1482–1489 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Chatterjee A., et al. , Saccharomyces cerevisiae THI4p is a suicide thiamine thiazole synthase. Nature 478, 542–546 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hao W., Richardson A. O., Zheng Y., Palmer J. D., Gorgeous mosaic of mitochondrial genes created by horizontal transfer and gene conversion. Proc. Natl. Acad. Sci. U.S.A. 107, 21576–21581 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Husnik F., McCutcheon J. P., Repeated replacement of an intrabacterial symbiont in the tripartite nested mealybug symbiosis. Proc. Natl. Acad. Sci. U.S.A. 113, E5416–E5424 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu B., Hao W., Horizontal transfer and gene conversion as an important driving force in shaping the landscape of mitochondrial introns. G3 (Bethesda) 4, 605–612 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Omelchenko M. V., Makarova K. S., Wolf Y. I., Rogozin I. B., Koonin E. V., Evolution of mosaic operons by horizontal gene transfer and gene displacement in situ. Genome Biol. 4, R55 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dwivedi B. K., Arnold R. G., Chemistry of thiamine degradation in food products and model systems: A review. J. Agric. Food Chem. 21, 54–60 (1973). [DOI] [PubMed] [Google Scholar]
- 57.Windheuser J. J., Higuchi T., Kinetics of thiamine hydrolysis. J. Pharm. Sci. 51, 354–364 (1962). [DOI] [PubMed] [Google Scholar]
- 58.Boissier J. R., Tillement J. P., Study of the stabilization by histidine of the thiamine in aqueous solution at 37 degrees C. Ann. Pharm. Fr. 27, 743–748 (1969). [PubMed] [Google Scholar]
- 59.Iwashima A., Kawasaki Y., Kimura Y., Transport of 2-methyl-4-amino-5-hydroxymethylpyrimidine in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1022, 211–214 (1990). [DOI] [PubMed] [Google Scholar]
- 60.Haughton B. G., King H. K., Toxo-pyrimidine phosphate as an inhibitor of bacterial enzyme systems that require pyridoxal phosphate. Biochem. J. 70, 660–665 (1958). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gonçalves C., Coelho M. A., Salema-Oom M., Gonçalves P., Stepwise functional evolution in a fungal sugar transporter family. Mol. Biol. Evol. 33, 352–366 (2016). [DOI] [PubMed] [Google Scholar]
- 62.Parrent J. L., James T. Y., Vasaitis R., Taylor A. F., Friend or foe? Evolutionary history of glycoside hydrolase family 32 genes encoding for sucrolytic activity in fungi and its implications for plant-fungal symbioses. BMC Evol. Biol. 9, 148 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 64.Li W., Godzik A., Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22, 1658–1659 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Katoh K., Standley D. M., MAFFT: Iterative refinement and additional methods. Methods Mol. Biol. 1079, 131–146 (2014). [DOI] [PubMed] [Google Scholar]
- 66.Capella-Gutiérrez S., Silla-Martínez J. M., Gabaldón T., trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S., UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gonçalves C., Gonçalves P., “Complete phylogenies and alignment files.” Figshare. https://figshare.com/articles/Phylogenies_Files/9800636. Accessed 11 September 2019.
- 70.Sambrook J., Molecular cloning: A laboratory manual (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, ed. 3, 2001). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.