Introductory
Ammonia-oxidizing archaea of the phylum Thaumarchaeota are among the most abundant marine microorganisms1. These organisms thrive in the oceans despite ammonium being present at low nanomolar concentrations2,3. Some Thaumarchaeota isolates have been shown to utilize urea and cyanate as energy and N-sources through intracellular conversion to ammonium4–6. Yet, it is unclear whether patterns observed in culture extend to marine Thaumarchaeota, and whether Thaumarchaeota in the ocean directly utilize urea and cyanate or rely on co-occurring microorganisms to break these substrates down to ammonium. Urea utilization has been reported for marine ammonia-oxidizing communities7–10, but no evidence of cyanate utilization exists for marine ammonia oxidizers. Here, we demonstrate that in the Gulf of Mexico, Thaumarchaeota use urea and cyanate both directly and indirectly as energy and N-sources. We observed substantial and linear rates of nitrite production from urea and cyanate additions, which often persisted even when ammonium was added to micromolar concentrations. Furthermore, single cell analysis revealed that the Thaumarchaeota incorporated ammonium-, urea- and cyanate-derived N at significantly higher rates than most other microorganisms. Yet, no cyanases were detected in thaumarchaeal genomic data from the Gulf of Mexico. Therefore, we tested cyanate utilization in Nitrosopumilus maritimus, which also lacks a canonical cyanase, and showed that cyanate was oxidized to nitrite. Our findings demonstrate that marine Thaumarchaeota can use urea and cyanate as both an energy and N-source. Based on these results we hypothesize that urea and cyanate are substrates for ammonia-oxidizing Thaumarchaeota throughout the ocean.
Keywords: Ammonia oxidation, nitrification, metabolic versatility, dissolved organic nitrogen, single cell
Nitrification, the stepwise oxidation of ammonia to nitrate, plays a key role linking the most reduced and oxidized species of the nitrogen (N) cycle. In marine systems, the first step of nitrification, ammonia oxidation, is predominantly carried out by ammonia-oxidizing archaea (AOA) belonging to the phylum Thaumarchaeota1,11. Marine Thaumarchaeota have generally been considered to be metabolically restricted organisms that use ammonia as a substrate for energy generation. However, dissolved organic N (DON, here defined as N-compounds containing at least one C atom) can provide additional substrates for ammonia oxidizers via intracellular conversion of DON to ammonium. The simple DON-compounds urea and cyanate are present ubiquitously in marine systems9,12–15. From the limited set of measurements available, urea concentrations appear to be of the same order of magnitude as ammonium concentrations, while cyanate concentrations are generally less than 30 % of ammonium (Supplementary Fig. 1)12–16. Urea is an intracellular metabolite and component of nitrogenous waste from both prokaryotes and eukaryotes and is released during remineralization of organic matter12,16, while sources of cyanate include urea, cyanide, and thiocyanate decomposition and photoproduction13,17. Some marine Thaumarchaeota cultures have been shown to encode a urease (e.g. 4,5), and there is evidence that marine thaumarchaeal communities use urea as an alternative energy source7–10. To date however, only one Thaumarchaeon, the terrestrial Nitrososphaera gargensis, has been shown to encode a cyanase, which seems to have been acquired via lateral gene transfer6. Cyanate utilization by nitrifiers has not been investigated so far in the marine environment, even though ammonia oxidation has been hypothesized to be a main factor shaping cyanate concentration profiles13,14.
The continental shelves are regions with high ammonia oxidation rates, which sustain the nutrient turnover that drives disproportionately high primary production in these regions, which despite their small surface area, account for 20 to 30 % of total marine primary productivity18. We aimed to assess whether Thaumarchaeota supplement their ammonia requirement in the continental shelf waters of the Gulf of Mexico by utilizing urea and cyanate. Furthermore, we investigated whether the Thaumarchaeota were directly utilizing urea and cyanate, or whether they were relying on co-occurring microorganisms to break down these substrates to ammonium and therefore utilizing them indirectly. During a cruise in 2016 to the GoM, bottom waters were hypoxic (< 63 μmol kg-1 dissolved oxygen) due to summertime eutrophic conditions, which recur yearly19 (Fig. 1a and Supplementary Fig. 2). Ammonium, urea, and cyanate were present at variable concentrations in the water column along the entire east-west sampling transect and were generally highest in the hypoxic bottom waters. The median ammonium, urea and cyanate concentrations were 320 nM, 69 nM and 11.5 nM, respectively. The ratios of these three N-compounds fell in the range observed across other shelf regions (Fig. 1b and Supplementary Fig. 1, 3)9,12–16. The median concentration of total DON was 12,100 nM (Supplementary Fig. 3), similar to previous measurements in the GoM and other shelf regions12. These compounds could therefore all potentially serve as energy and N-sources for microorganisms in the GoM.
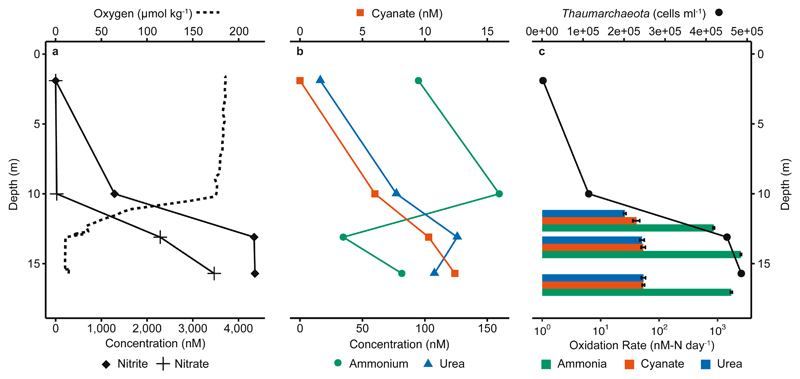
Fig. 1. Depth distribution of nutrient and oxygen concentrations, Thaumarchaeota cell counts and oxidation rates from Station 2.
a) Nitrite, nitrate and oxygen concentrations. b) Ammonium, urea and cyanate concentrations. c) Thaumarchaeota depth distribution based on CARD-FISH counts and measured ammonia and urea- and cyanate-derived oxidation rates (without added 14N-ammonium), calculated from slopes across all time points and triplicate incubations. Oxidation rates are depicted on a log-axis. Rate experiments were carried out at 12 m, 14 m and 16.5 m. Error bars for rates represent standard errors of slopes calculated across all biological triplicates and all timepoints. All rates were significant (see Supplementary Table 1). Thaumarchaeota abundance was determined from CARD-FISH counts (n for DAPI-stained / Thaumarchaeota cells = 9,247 / 7; 13,296 / 389; 17,253 / 1,541; 16,770 / 1,660 from 1.7 m, 10.1 m, 13.0 m and 15.7 m depth, respectively). Data from an additional 2 stations are shown in Fig. 2 d-f and Supplementary Fig. 3, 4 and 8.
Thaumarchaeota have previously been identified as the dominant ammonia oxidizers in the GoM20,21; this was also the case in summer 2016, when Thaumarchaeota cell counts (determined by CARD-FISH) were up to 4.9 × 105 cells ml-1, approximately 10 % of total cell counts (Fig. 1c, Supplementary Fig. 4, Supplementary Discussion). 16S rRNA gene amplicon sequencing confirmed that Thaumarchaeota were the only detectable ammonia oxidizers in the GoM, with reads clustering primarily into one operational taxonomic unit (OTU, 97% sequence similarity cluster). This OTU was closely related to Nitrosopumilus sp., which previous 16S-based approaches have revealed to be a dominant ammonia oxidizer in continental shelf waters (22–24, Supplementary Fig. 5). Using metagenomic sequencing, we generated six Thaumarchaeota metagenome-assembled genomes (MAGs). Four of these were more than 90% complete, with the most abundant MAGs branching confidently with Nitrosopumilus sp. and Nitrosomarinus sp. in a phylogenetic analysis based on 34 single-copy marker genes (Supplementary Fig. 6, Supplementary Table 2). Furthermore, five of the MAGs contained amoA, the gene encoding the structural subunit of ammonia monooxygenase (see Supplementary Table 2). We also investigated amoA transcription; all amoA transcripts retrieved from metatranscriptomes were phylogenetically affiliated with Thaumarchaeota and also clustered with the obtained MAGs (Supplementary Fig. 7).
The use of ammonia, urea and cyanate as energy sources was investigated using 15N13C-tracer incubations at three depths and three stations. Upon addition of 15N-ammonium, we observed linear production of 15N-nitrite over time in the dark under in-situ oxygen and temperature conditions. Ammonia oxidation rates ranged between 80 – 2,500 nM-N d-1 (Fig. 1c, Fig. 2, Supplementary Fig. 8), comparable to rates previously measured in the region21,25 and in other shelf and oxygen-depleted systems9,26,27. Although all of the measured rates are potential rates due to the addition of 15N-tracers, the short length of the incubations (< 24h) and the linearity of the rates from the beginning indicate that the ammonia oxidizers were active in-situ. Moreover, rates showed a strong positive correlation with in-situ nitrite concentrations (Fig. 2d), indicating that ammonia oxidation is a major determinant of nitrite concentration in the GoM.
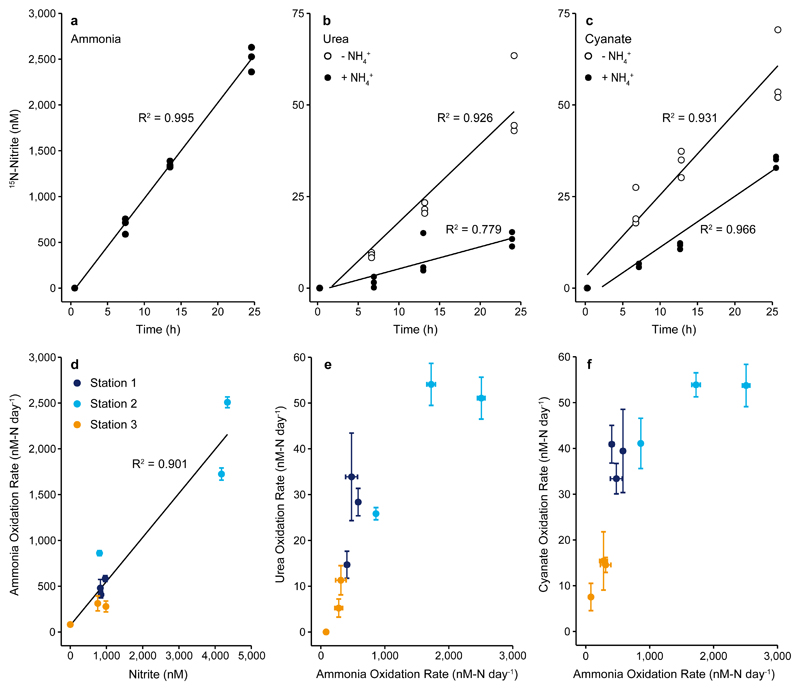
Fig. 2. Production of 15N-nitrite over time and correlations between rates obtained from various substrate additions.
Upper panels: 15N-nitrite concentration over time after addition of a) 15N-ammonium, b) 15N-urea, and c) 15N-cyanate without (open circles) and with (filled circles) added 14N-ammonium (5 μM) at Station 2, 14 m depth. Data points represent biological triplicates. Lower panels: Correlations between d) ammonia oxidation rate and in-situ nitrite concentration, e) ammonia oxidation rate and urea-derived oxidation rate, and f) ammonia oxidation rate and cyanate-derived oxidation rate (from incubations without added 14N-ammonium). Black lines in panels a) – d) are linear regressions, R2 was calculated based on Pearson Correlations, and was significant in all analyses (a-c), one-sided t-test; a) t = 42.66, DF = 10, p = 1.20×10-13, b) t = 11.184, DF = 10, p = 5.65×10-7 and t = 5.931, DF = 10, p = 1.45×10-4 for urea without and with added ammonium, respectively, c) t = 11.634, DF = 10, p = 3.91×10-7 and t = 16.935, DF = 10, p = 1.08×10-8 for cyanate without and with added ammonium, respectively, d) two-sided t-test; t = 8.002, DF = 7, p = 9.10×10-5. For panels e) and f), Spearman rank correlations were calculated and were significant for both e) S = 10, ρ = 0.917, p = 0.001 and f) S = 10, ρ = 0.917, p = 0.001. Error bars in panels (d-f) represent standard errors of slopes calculated across all biological triplicates and all timepoints.
Significant and linear production of 15N-nitrite was also observed after addition of 15N13C-urea or 15N13C-cyanate. The maximum rates measured were similar for both compounds (up to 54 nM-N d-1), although rates varied between stations and depths (Fig. 1c and Fig. 2, Supplementary Fig. 8). The measured urea-derived oxidation rates are in the range of those previously reported from the marine environment (see Supplementary Discussion for per cell rates)9,10. Until now there were no nitrifier-associated cyanate-derived oxidation rates from the marine environment. Urea- and cyanate-derived oxidation rates constituted up to 7% and 10%, respectively, of the measured ammonia oxidation rates (Fig. 1c, and Fig. 2, Supplementary Fig. 8) and showed a strong positive correlation with ammonia oxidation rates (Fig. 2e, 2f).
In principle, the production of 15N-nitrite from additions of 15N13C-urea or 15N13C-cyanate could indicate both direct and indirect utilization of these substrates by Thaumarchaeota. Indirect utilization could result from either abiotic or biotic breakdown of urea and cyanate. In water, abiotic urea and cyanate breakdown to ammonium and carbon dioxide can occur through a temperature and pH dependent process (6,17; see Supplementary Discussion) and the resulting ammonium can subsequently be used by microorganisms. The measured abiotic breakdown rates of urea to ammonium were insignificant in GoM seawater, and cyanate breakdown was minor (Supplementary Table 3 and Supplementary Discussion). Alternatively, other microorganisms might intracellularly break down urea or cyanate to carbon dioxide and ammonium (biotic breakdown), which is subsequently released to the environment and used by Thaumarchaeota. Such cross-feeding has been demonstrated in co-culture experiments with urea- and cyanate-degrading nitrite oxidizers and ammonia-oxidizing bacteria6,28. In our experiments, breakdown of urea and cyanate would progressively increase the amount of 15N in the extracellular ammonium pool, which would lead to exponential production of 15N-nitrite from ammonia oxidation over time in the case of indirect utilization.
We quantified how much of the observed nitrite production could be assigned to direct substrate utilization by Thaumarchaeota or to the breakdown of urea and cyanate into the extracellular ammonium pool by biotic or abiotic breakdown (indirect utilization). We ran parallel incubations which were identical except for the addition of a large 14N-ammonium pool (ammonium pool incubations). These were intended to reduce the likelihood that 15N-ammonium formed from biotic or abiotic breakdown of urea or cyanate, would be oxidized to 15N-nitrite by Thaumarchaeota. In the ammonium pool incubations, the rates of 15N-nitrite production were still linear, although they were lower than those in the incubations without added 14N-ammonium (Fig. 2b, 2c, Supplementary Fig. 9). We could observe the production of 15N-ammonium in the ammonium pool incubations, some of which was still oxidized to 15N-nitrite, due to the high ammonia oxidation rates (indirect utilization). However, by quantifying the amount of 14N and 15N-ammonium at each time point and combining this with the known ammonia oxidation rates, we were able to calculate the proportion of 15N-nitrite production that could have stemmed from indirect utilization of 15N-urea or 15N-cyanate. Thereby, we were able to quantify the direct utilization rates and can confidently show that there were significant rates of 15N-nitrite production as a result of direct utilization (up to 9.9 nM d-1, Supplementary Table 3, Supplementary Fig. 10). These rates are likely to be underestimations, as it is possible that the Thaumarchaeota utilized less urea and cyanate in response to the large ammonium addition in the ammonium pool incubations.
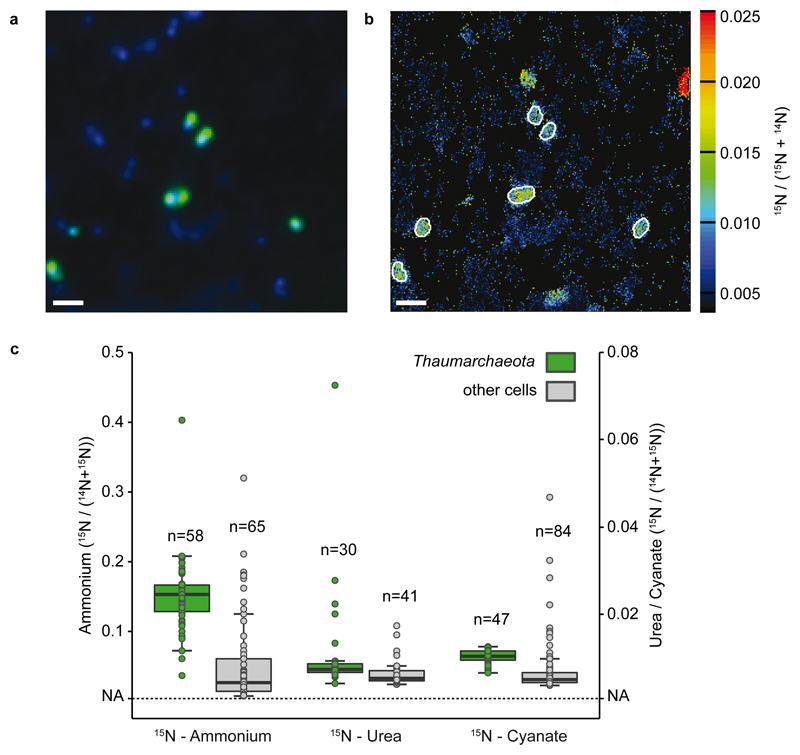
We used nanoSIMS to determine the incorporation of ammonium, urea and cyanate into Thaumarchaeota cells in the GoM, which enabled us to gain insights into metabolic heterogeneity within the Thaumarchaeota community at a single cell level. All measured Thaumarchaeota cells (n=58) incorporated 15N from ammonium (average 14.4 amol-N cell-1 d-1) and 13C from bicarbonate (average 19.8 amol-C cell-1 d-1) and were significantly more enriched than the surrounding microorganisms (Fig. 3, Mann-Whitney U test, p<0.01). Thaumarchaeota also assimilated 15N from urea (29 of 30 measured cells) and cyanate (all measured cells, n=47). Rates of N assimilation from urea and cyanate were up to two orders of magnitude lower (average 0.72 amol-N cell-1 d-1 urea and 0.64 amol-N cell-1 d-1 cyanate) than rates of N-assimilation from ammonium, but significantly higher than those of surrounding cells (Fig. 3, Mann-Whitney U test, p<0.01). Average bulk rates of assimilation by Thaumarchaeota in the GoM were 6.0 nM-N d-1 for ammonium, 0.3 nM-N d-1 for urea and 0.3 nM-N d-1 for cyanate. Thaumarchaeota assimilation of 15N from urea was more heterogeneous than from ammonium and cyanate, with some cells showing a distinctly higher enrichment of 15N from urea compared to others (Fig. 3, Supplementary Fig. 11). We could not detect any 13C-incorporation from additions of 15N13C-urea or 15N13C-cyanate. This is likely due to a combination of the small amount of 13C-CO2 produced from urea and cyanate degradation and strong dilution by the 2 mM ambient dissolved inorganic carbon (DIC). Although this nanoSIMS analysis cannot distinguish between direct and indirect assimilation, the data show that most Thaumarchaeota cells were active and metabolically versatile, using urea and cyanate as additional N-sources.
Fig. 3. Thaumarchaeota single cell ammonium, urea and cyanate uptake determined by nanoSIMS at Station 2, 14m depth.
a) Representative CARD-FISH image of Thaumarchaeota (green; counterstained by DAPI, blue) with a specific probe (Thaum726). b) corresponding nanoSIMS image of 15N/(14N+15N) enrichment after addition of 15N-cyanate. Thaumarchaeota are marked by white outlines. Scale bar is 1 μm. In total, 9, 6 and 8 fields of view were analyzed by nanoSIMS for the 15N-cyanate, 15N-ammonium and 15N-urea treatment. c) 15N/(14N+15N) enrichment of Thaumarchaeota (green) and non-targeted cells (grey) after incubation with 15N-ammonium (left), 15N-urea (middle, without added 14N-ammonium) or 15N-cyanate (right, without added 14N-ammonium). Note the different scales for 15N-ammonium and 15N-urea and 15N-cyanate, respectively. Number of cells analyzed per category is indicated above each boxplot. Boxplots depict the 25 – 75 % quantile range, with the center line depicting the median (50% quantile); whiskers encompass data points within 1.5 × the interquartile range. NA is the natural abundance 15N/(14N+15N) value (0.0037). Four non-Thaumarchaeota cell values in the 15N-urea treatment are not depicted and have 15N/(14N+15N) values of 0.326, 0.095, 0.118 and 0.139, these cells were included in all calculations. More ammonium was assimilated than urea and cyanate by the Thaumarchaeota, and the Thaumarchaeota assimilated significantly more 15N compared to surrounding cells in all treatments (one-sided Mann-Whitney U Test, U = 3348.5, p = 6.19×10-14; U = 873, p = 0.001; U = 3409, p = 2.91×10-12 for ammonium, urea and cyanate, respectively).
Using the nanoSIMS results, we calculated single cell N-based growth rates for Thaumarchaeota of 0.23 ± 0.012 (SE) d-1 for ammonium, similar to previous measurements in marine systems (0.21 – 0.47 d-1, 7,29) and in enrichments or pure cultures4,11. This ammonium growth was supplemented by urea- and cyanate-based growth (0.011 ± 0.0035 (SE) d-1 and 0.009 ± 0.0003 (SE) d-1, respectively). Interestingly, 13DIC-based growth rates were about 6-fold lower than ammonium-based growth rates (0.04 ± 0.005 (SE) d-1). This could be an artifact due to the small size of the Thaumarchaeota (see methods) or could indicate that Thaumarchaeota in the GoM did not meet all of their C-demand from autotrophic C-fixation. However, more work is required to resolve this. The single cell uptake and growth rates provide further evidence that Thaumarchaeota in the GoM have the capability to use N from urea and cyanate, directly or indirectly, in addition to ammonium, however, they seem to do so at lower rates compared to when using ammonium.
To examine how Thaumarchaeota in the GoM might be utilizing urea and cyanate, we screened both the metagenome assemblies and the Thaumarchaeota MAGs for ureases (ureC) and cyanases (cynS), the enzymes responsible for intracellular breakdown of urea and cyanate to ammonia, respectively. Detected ureC sequences were very diverse (Supplementary Fig. 12), with 10.2 % associated with Thaumarchaeota. Based on the recovery of thaumarchaeal ureC versus thaumarchaeal 16S rRNA genes and amoA genes, we estimated that approximately 10-15% of Thaumarchaeota cells contain a urease, which is similar to the ratio reported previously for coastal Georgia9. Of the metagenomic ureC identified, 1.1% could be assigned to GoM MAG1 (putatively assigned to the genus Nitrosopelagicus (Fig. 4a and Supplementary Fig. 6)) while others were related to the genus Nitrosopumilus (Supplementary Fig. 12) but did not bin into one of the six almost complete MAGs. These ureases were similar to those identified in coastal Georgia9, many of which were related to Nitrosopumilus sediminis strain AR2. The transcribed ureases were also diverse, with 5.9 % of ureC transcripts associated with Thaumarchaeota and clustering either with GoM MAG1 or Nitrosopumilus related ureC sequences (Supplementary File 1-6). Metagenomic analyses therefore indicated that only a sub-population of Thaumarchaeota in the GoM have known ureases, consistent with the single-cell observation that some Thaumarchaeota cells assimilated significantly more N from urea than others (Fig. 3, Supplementary Fig. 11).
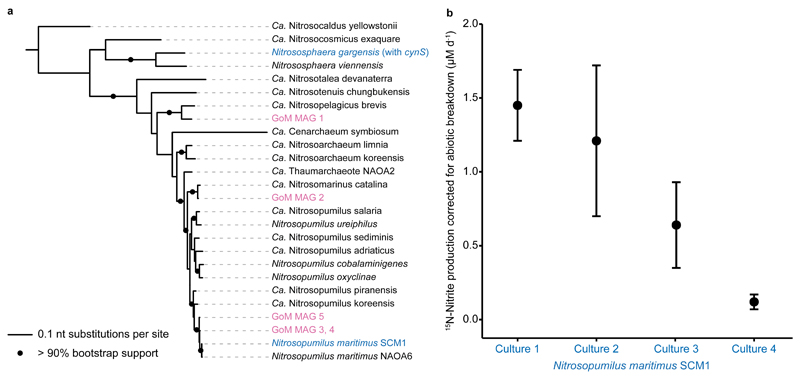
Fig. 4. amoA based phylogeny of Thaumarchaeota MAGs recovered in this study and cyanate utilization by the marine Thaumarchaeon Nitrosopumilus maritimus.
a) Phylogenetic placement of the amoA sequences from Gulf of Mexico metagenome assembled genomes (GoM MAGs, magenta) and Thaumarchaeota cultures that are able to utilize cyanate (blue). N. gargensis is the only Thaumarchaeon that encodes a known cyanase. Reference Thaumarchaeota amoA sequences are shown in black. Tree was constructed using IQ-TREE60 with automated model selection from near full-length amoA sequences and confidence was assessed with ultrafast bootstrapping (1,000 iterations). The scale bar represents nt substitutions per site, bootstrap support values >90% are depicted. b) 15N-nitrite production rate by the marine Thaumarchaeon N. maritimus SCM1 incubated with 15N-cyanate and a 14N-ammonium pool, corrected for abiotic breakdown of cyanate to ammonium in the culture medium. Error bars are the standard errors of the slope across all time points of one biological replicate. Rates were calculated based on linear regressions (one-sided t-test, t = 6.13, DF = 2, p = 0.012; t = 2.38, DF = 2, p = 0.070; t = 2.22, DF = 2, p = 0.078; t = 3.72, DF = 2, p = 0.033, for culture 1, 2, 3 and 4, respectively). When data were fitted with an exponential regression, p was <0.001 for all four cultures. Differences in rates between biological replicates correlate with the different starting biomass in each culture.
We could not assign any meta-omics cynS sequences to Thaumarchaeota, and no sequences related to cyanases were detected in the Thaumarchaeota MAGs (Supplementary Fig. 13). To reconcile this result with the indications that cyanate was utilized directly in the GoM, we examined cyanate utilization in four cultures of Nitrosopumilus maritimus SCM1. Identifiable cyanases are absent from the genome of Nitrosopumilus maritimus SCM1, and from the genomes available for other marine Thaumarchaeota (Fig. 4a). However, when we incubated Nitrosopumilus maritimus with 15N13C-cyanate, we observed production of 15N-nitrite. This production occurred at much higher rates than could be accounted for by abiotic breakdown of cyanate to ammonium (Fig. 4b). Interestingly, we observed linear 15N-ammonium production in all four cultures, which was far above the abiotic breakdown rate. When this production rate was taken into account in a modelling approach similar to that used above, it appears that almost all of the cyanate utilized by the Thaumarchaeota cultures could have entered the extracellular ammonium pool prior to oxidation (Supplementary Fig. 14). This could suggest that cyanate breakdown by Thaumarchaeota occurs extracellularly, or that there was equilibration between intra- and extracellular ammonium pools (see Supplementary Discussion). These results indicate that Nitrosopumilus maritimus is capable of utilizing cyanate, even though it does not have a canonical cyanase. Currently, the biochemical pathway involved in cyanate utilization is unclear.
Until recently, marine Thaumarchaeota were considered to be metabolically restricted organisms that only use ammonia as a substrate for energy conservation9,10. Here we show that in the GoM, Thaumarchaeota can use urea and cyanate to supplement their N- and energy requirements. The presence and transcription of urease in a sub-population of the Thaumarchaeota, combined with the single cell uptake data and rate determinations, suggest that part of the Thaumarchaeota community directly utilize urea as a substrate. Similar evidence was obtained for cyanate utilization; however, we could not detect Thaumarchaeota cyanases. In fact, no known marine Thaumarchaeota have an identifiable cyanase. Yet, we show that Nitrosopumilus maritimus, which is closely related to GoM Thaumarchaeota, can oxidize cyanate to nitrite. This indicates that cyanate can be utilized by marine Thaumarchaeota from distinct geographical regions, even when known cyanases cannot be detected. Considering that the GoM has ratios of cyanate, urea and ammonium typical of shelf regions (Supplementary Fig. 1, 9,12–15) and a thaumarchaeal community representative of continental shelves22–24, the use of urea and cyanate to supplement N-requirements could be a widespread trait. In the oligotrophic gyres where ratios of DON to ammonium are higher relative to the shelf seas16, we hypothesize that urea and cyanate are also important substrates for ammonia oxidizers.
Materials and Methods
Sampling
Sampling took place on the Louisiana Shelf in the northern Gulf of Mexico aboard the R/V Pelican, cruise PE17-02, from July 23rd to August 1st, 2016, spanning a west-east transect from 92°48'4'' W to 90°18'7'' W. Seawater was collected in 20 L Niskin bottles on a rosette equipped with a CTD and SBE 43 oxygen sensor. Nutrient profiles spanning the water column (surface water to water-sediment interface at max. 19 m) were determined at nine stations. Process rate measurements, molecular and FISH analyses were carried out at three of the nine stations (Supplementary Fig. 2).
Nutrient analyses
Ammonium concentrations were measured in unfiltered seawater samples to avoid sample contamination by the filtration process (Supplementary Discussion and Supplementary Fig. 15). For all other nutrient measurements, seawater was prefiltered using 0.22 μm PES syringe filters (Millex, Millipore, Supplementary Discussion and Supplementary Fig. 15). Ammonium concentrations were measured fluorometrically by the orthophthaldialdehyde method (limit of detection (LOD) 10 nM in a 1 cm cuvette)30. Nitrite (LOD 50 nM in a 1 cm cuvette) and urea (LOD 30 nM in a 10 cm cuvette) concentrations were measured photometrically onboard using the Griess and diacetylmonoxime methods respectively31,32. Samples for cyanate concentration measurements (LOD 1.5 nM) were derivatized onboard and stored at -20°C until return to the laboratory, where samples were stored at -80°C until analysis using high performance liquid chromatography (Dionex, ICS-3000 system coupled to fluorescence detector, Thermo Scientific, Dionex Ultimate 3000)33. Samples for nitrate measurements (LOD 50 nM) were stored at -20°C and concentrations were determined upon return with a chemiluminescence NO/NOx analyzer after reduction to NO with acidic vanadium (II) chloride34. Samples for total dissolved nitrogen (TDN) concentrations were filtered through pre-combusted GF/F filters (Whatman) in HCl-cleaned filter holders by gravity filtration from Niskin bottles, acidified with HCl and subsequently stored at 4°C in the dark until measurement by chemiluminescence (Shimadzu TOC-VCPH)35. Dissolved organic nitrogen (DON) was calculated by subtraction of measured ammonium, nitrite and nitrate concentrations.
Process rate experiments
Process rate measurements were carried out as described in Bristow et al.21 and were determined at three stations at three depths in and below the oxycline (Supplementary Fig. 2, 8). Water was sampled directly from the Niskin bottle into 250 ml serum bottles, which were sealed bubble-free with deoxygenated butyl rubber stoppers36. Bottles were stored at in-situ temperature (28°C) in the dark until the start of the experiments (< 7 h after sampling). Exposure to natural light during sampling was minimized and all further handling took place under red light to prevent assimilation by phytoplankton.
For each amendment and depth, tracer was added to triplicate serum bottles. Amendments were designed to test for ammonia (15NH4+), urea (15N13C-urea)- and cyanate (15N13C-cyanate)-derived oxidation rates (Supplementary Table 5) and were made as 5 μM additions. Additionally, in ammonia oxidation experiments, 200 μM 13C-bicarbonate (DIC) was added. Abiotic breakdown of urea and cyanate to ammonium was determined upon return, using sterile filtered bottom water from Station 1, and its potential contribution to the observed oxidation rates was calculated (see Supplementary Discussion). To further test the contribution of biotic and abiotic breakdown of urea and cyanate and subsequent use of the resulting extracellular 15N-ammonium by ammonia oxidizers, additional ammonium pool incubations with a 14N-ammonium amendment (5 μM) were performed to dilute any extracellular 15N-ammonium formed (see Supplementary Discussion).
15N13C-tracer solutions and 14N-pools (Supplementary Table 5) were added using gas tight syringes (Hamilton). Pre-weighed aliquots of 15N13C-tracers were dissolved in sterile filtered seawater just before the start of every experiment to minimize abiotic breakdown. After tracer amendments and subsequent gentle shaking for approximately 10 seconds, a volume of 40 ml was removed and replaced with helium (He). The headspaces were then flushed with He twice, before adding pure oxygen according to Garcia and Gordon37 to match in-situ oxygen concentrations. Triplicate serum bottles per depth contained optode spots (Firesting, Pyroscience), enabling us to monitor oxygen concentrations, which remained within 20% of in-situ concentrations throughout the experiment. The removed 40 ml were sterile filtered (0.22 μm, PES, Q-Max, Frisenette ApS) and stored at -20°C as time zero samples. The amended 15N-tracer concentrations were determined by concentration measurements and subtraction of in-situ values. For determination of 13C-DIC labeling percentage, unfiltered samples were filled bubble-free into exetainers (Labco, UK) and preserved with saturated mercury(II) chloride solution (50 μl per 6 ml sample). After sampling, serum bottles were incubated in a recirculated water bath at in-situ temperature (28°C), in the dark. After 6 h, 12 h and 24h, 20 ml of seawater was sampled and replaced with He. Samples were sterile filtered and frozen. Serum bottle headspaces were flushed with He twice and oxygen was added to match in-situ concentrations as before. After 24 h, the remaining seawater of triplicates was combined, and 20 ml were fixed and filtered onto 0.22 μm GTTP filters for FISH and 0.22 μm gold sputtered GTTP filters for nanoSIMS, respectively.
15N-rate measurements and determination of 13C-DIC labeling percentage
Ammonia, urea- and cyanate-derived oxidation rates were determined from 15N-nitrite increase over time. Nitrite was converted to N2 with sulfamic acid38 and 29N2 was measured by gas-chromatography isotope ratio mass spectrometry (GC-IRMS) on a customized TraceGas coupled to a multicollector IsoPrime100 (Manchester, UK). Abiotic breakdown rates of 15N13C-urea and 15N13C-cyanate to 15N-ammonium were measured according to Zhang et al.39, combining hypobromite oxidation of ammonium to nitrite and subsequent neutralization by HCl before reduction to N2 by sulfamic acid (see section below). All rates were inferred from the slopes of linear regressions across all time points and replicates and were corrected for initial 15N-labeling percentage. Only slopes that were significantly different from zero are reported (p < 0.05, one-sided Student t-test). Non-significant regressions are reported as not detected rates. Initial 13C-DIC labeling percentages were determined after acidification40 by 13C-CO2/12C-CO2 measurements using cavity ring-down spectroscopy (G2201-i coupled to a Liaison A0301, Picarro Inc., Santa Clara, USA, connected to an AutoMate Prep Device, Bushnell, USA).
Hypobromite conversions for 15N-ammonium measurement in cyanate and urea samples
15N-ammonium concentrations in samples containing 15N13C-urea and 15N13C-cyanate were determined following a modified protocol from Zhang et al.39 where ammonium was oxidized to nitrite by hypobromite, and subsequently converted to N2 by sulfamic acid38. This method minimizes concurrent conversion of urea and cyanate (see below). After hypobromite conversion and sodium arsenite addition, the sample pH must be set to pH 8-9 by addition of 6 N HCl. This step is crucial because a too acidic pH results in spontaneous oxidation of nitrite to nitrate and a too basic pH interferes with the subsequent reduction of nitrite to N2 by sulfamic acid41. After sulfamic acid conversion of nitrite to N2, 29N2 was measured on a customized TraceGas Isotope Ratio Mass Spectrometer (TraceGas IRMS) coupled to a multicollector IsoPrime100 (Manchester, UK). Detection limits were estimated from the median of the standard error of the slope, multiplied by the t value for p = 0.05.
We did not remove the 15N-nitrite prior to hypobromite conversion, and therefore measured combined 15N-ammonium + 15N-nitrite. Prior nitrite removal was omitted because this requires a sulfamic acid conversion, which is carried out at low pH. Low pH leads to increased abiotic decay of cyanate to ammonium6, which impedes subsequent accurate 15N-ammonium measurement. To obtain 15N-ammonium values, we measured 15N-nitrite (following the method of Füssel et al.38) in a separate sample aliquot and subtracted the obtained 15N-nitrite values from the combined 15N-ammonium + 15N-nitrite values.
For all hypobromite conversions, freshly prepared 15N13C-urea and 15N13C-cyanate standards were converted concurrently. Across conversions, only minor proportions of 15N13C-urea-N (0.42% ± 0.06%) and 15N13C-cyanate-N (0.14% ± 0.03%) were converted to 29N2. This 29N2 can either stem from a minor 15N-ammonium contamination of the stocks, or from direct conversion of 15N13C-urea and 15N13C-cyanate during the hypobromite protocol. While we did not detect ammonium based on concentration measurements in 15N13C-urea stocks, we did detect a minor ammonium contamination in 15N13C-cyanate stocks, corresponding to the 29N2 measured in hypobromite conversions of cyanate standards. Therefore, we assume that the 29N2 detected in hypobromite conversions of 15N13C-urea does not represent actual ammonium contamination but rather direct conversion of 15N13C-urea. As the labeling percentage within our experiments did not vary across time points, the direct conversion of 15N13C-urea was simply accounted for by using the slope of change in 29N2 concentration with time to calculate the rate. However, if the labeling percentage were to change across time points, each time point would need to be corrected individually for this direct conversion, before a rate could be calculated. In contrast, 29N2 detected in hypobromite conversions of 15N13C-cyanate is likely due to a small 15N-ammonium contamination of the 15N13C-potassium cyanate salt.
Additionally, we found that some 15N13C-urea was also converted to 30N2 during hypobromite conversions. To test if hypobromite combines the amide groups of one and the same or two different urea molecules, hypobromite conversions of equimolar (5 μM) concentrations of 15N13C-urea and 14N12C-urea were performed. We detected no increase in 29N2 in these samples, indicating that hypobromite combines the two amide groups of a single urea molecule. Therefore, we recommend that if analysis of 30N2 after hypobromite conversions on samples containing 15N-urea is required, the sample can be stripped of 30N2 before sulfamic acid conversion by prolonged bubbling of the sample liquid with He.
DNA and RNA analyses
Nucleic acid extraction
Seawater for molecular analyses was collected from the same casts and depths as seawater for process rate measurements. For each sample, a peristaltic pump was used to directly filter 1L of seawater onto 0.22 μm cartridge filters (Sterivex™, Millipore). An upstream prefilter was not used, thereby avoiding potential bias in taxon representation due to prefilter clogging42. Replicate cartridges for DNA analysis (16S rRNA gene sequencing and metagenomics) were filled with lysis buffer (50 mM Tris-HCl, 40 mM EDTA, 0.73 M sucrose) and stored at -20°C. Replicates for RNA analysis (metatranscriptomics) were filled with RNA stabilizing buffer (25 mM sodium citrate, 10 mM EDTA, 5.3 M ammonium sulfate, pH 5.2), flash frozen in liquid nitrogen, and stored at -80°C.
DNA was extracted from Sterivex cartridges using a phenol:chloroform protocol, as described previously43. Cells were lysed by adding lysozyme (2 mg in 50 μl of lysis buffer per filter) directly to the cartridges, sealing the cartridges, and incubating for 45 min at 37°C. Proteinase K (1 mg in 100 μL lysis buffer, 100 μl 20% SDS) was added, and the cartridges were resealed and incubated for 2 hours at 55°C. The lysate was removed, and DNA was extracted once with phenol:chloroform:isoamyl alcohol (25:24:1) and once with chloroform:isoamyl alcohol (24:1) and then concentrated by spin dialysis using Ultra-4 (100 kDa, Amicon) centrifugal filters.
RNA was extracted from cartridges using a modification of the mirVana™ miRNA Isolation kit (Ambion)44. Cartridges were thawed on ice, RNA stabilizing buffer was then expelled and discarded, and cells were lysed by adding Lysis buffer and miRNA Homogenate Additive (Ambion) directly to the cartridges. Following vortexing and incubation on ice (10 min), lysates were transferred to RNAase-free tubes and processed through an acid-phenol:chloroform extraction according to the kit protocol. The TURBO DNA-free™ kit (Ambion) was used to remove DNA, and the extract was purified using the RNeasy MinElute Cleanup Kit (Qiagen).
16S rRNA gene sequencing and analysis
The proportional abundances of microbial taxa were assessed at all experimental depths and stations using 16S rRNA gene amplicon sequencing, following an established pipeline (e.g. 42,43). Briefly, amplicons were generated by PCR using equal amounts of DNA template (1 ng), Platinum® PCR SuperMix (Life Technologies), and primers F515 and R806 encompassing the V4 region of the 16S rRNA gene45. Despite a mismatch in the 515F primer to most Thaumarchaeota, the 16S rRNA gene of this group is recovered in PCR assays using this primer46. This is in line with in-silico coverage tests (test-prime, arb-silva) allowing for 1 mismatch overall, but 0 mismatches for at least 5 bases at the 3’ end of the primer, suggesting an estimated recovery of 95% of known marine Thaumarchaeota (Nitrosopumilales and Marine benthic group1). This indicates that the Thaumarchaeota in the Gulf of Mexico were well covered despite a mismatch in the forward primer (see Supplementary Discussion). Both forward and reverse primers were barcoded and appended with Illumina-specific adapters. Thermal cycling involved: denaturation at 94°C (3 min), followed by 30 cycles of denaturation at 94°C (45 sec), primer annealing at 55°C (45 sec) and primer extension at 72°C (90 sec), followed by extension at 72°C for 10 min. Amplicons were analyzed by gel electrophoresis to verify size (~400 bp) and purified using Diffinity RapidTip2 pipette tips (Diffinity Genomics, NY). Amplicons from different samples were pooled at equal concentrations and sequenced on an Illumina MiSeq using a 500-cycle Nano kit.
Barcoded sequences were de-multiplexed, trimmed (length cutoff 100 nt), and filtered to remove low quality reads (average Phred score < 25) using Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Paired-end reads were merged using FLASH47, with a minimum average length of 250 nt for each read, minimum average length of 300 nt for paired read fragments, and maximum allowable fragment standard deviation of 30 nt. The number of trimmed and merged reads per sample ranged from 11,842 – 21,970. Chimeric sequences were detected by reference-based searches using USEARCH48 and removed from the sequence pools. Operational Taxonomic Units (OTUs) were defined by clustering at 97% sequence identity using open-reference picking with the UCLUST algorithm48 in QIIME149. The average number of sequences assigned per OTU was 836 (range 646 – 1,138). Taxonomy was assigned to OTUs using the Greengenes database50. Singleton sequences and sequences affiliated with mitochondria and chloroplasts were removed from any further analysis. Proportional abundances of orders constituting >0.5% of the community were calculated after rarefaction based on the sample with the lowest number of reads (11,842 reads).
OTUs identified as Thaumarchaeota were analyzed further to assess the diversity and abundance of the ammonia-oxidizing community. The Thaumarchaeota sequences, which constituted the only known ammonia oxidizers in the dataset, were analyzed by placement into a reference phylogeny composed of near full-length reference Thaumarchaeota 16S rRNA sequences compiled from the ribosomal database project (RDP). To identify additional sequences for inclusion in the reference phylogeny, all Thaumarchaeota OTU sequences were queried against the NCBI non-redundant database via BLASTN. Top matching sequences and 16S rRNA sequences of cultured representatives were sorted by size and near-full sequences (≥1,300 nt), including both RDP and BLASTN matches (n=32), were aligned with MUSCLE51 and then used to generate a phylogenetic tree (Supplementary Fig. 5) in RAxML52 via maximum likelihood estimation with the “GTRGAMMA” model and rapid bootstrapping (1,000 iterations). Short sequences consisting of both the Thaumarchaeota OTU amplicon sequences and non-full length top matches (<1,300 nt, identified via BLASTN) were placed into the phylogenetic tree using the Evolutionary Placement Algorithm (EPA)53. The resulting tree was visualized in FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/).
Metagenome sequencing, assembly and binning
Metagenomic libraries were constructed using NEBNext® UltraTM II FS DNA Library Prep Kit for Illumina, creating average fragment sizes of 550 bp. Samples were sequenced on one lane of an Illumina HiSeq using 2x250 bp cycle kit at Georgia Tech’s High-Throughput DNA Sequencing core facility.
Paired-end Illumina reads were pre-processed with bbduk (BBMap - Bushnell B. - sourceforge.net/projects/bbmap/) to remove adapters and residual phiX sequences. Reads were further quality-filtered with bbduk (ktrim=r k=21 mink=11 hdist=2 minlen=149 qtrim=r trimq=15). Quality-filtered reads were assembled with Metaspades (-k 21,33,55,77,99,127)54. BBMap (BBMap - Bushnell B. - sourceforge.net/projects/bbmap/) was used to map each individual read set to each assembly to assist in differential-coverage genome binning. Large (>2 kb) scaffolds were clustered into Metagenome-Assembled Genomes (MAGs) by oligonucleotide frequency (k=4) and read coverage using Maxbin255 and Metabat256. Redundant bins were subsequently dereplicated and evaluated using dRep57 with a completeness cutoff of 40%, contamination cutoff of 10% and minimum genome size of 200kb. Thaumarchaeal genomes were identified using a phylogenetic tree calculated using FastTree258 based on an automated alignment generated by CheckM59 and containing sequences from the dereplicated MAGs and known Thaumarchaeota. Metagenome sequencing statistics and information on dereplicated thaumarchaeal MAGs are listed in Supplementary Table 6 and 2, respectively.
MAG phylogenetic reconstruction
A concatenated alignment of 34 universal single-copy marker genes was generated using CheckM for published Thaumarchaeota and thaumarchaeal MAGs as well as representative Bathyarchaeota and Aigarchaeota (as outgroups). Phylogenetic reconstruction was carried out using IQ-TREE60 with automated model selection and confidence was assessed with ultrafast bootstrapping (1,000 iterations).
Metatranscriptome sequencing
Community RNA (metatranscriptome) from Station 2, where the measured oxidation rates were highest, was analyzed for evidence of ammonium, urea, and cyanate utilization. To enrich for mRNA, ribosomal RNA (rRNA) was depleted from total RNA using the Ribo-Zero™ rRNA Removal Kit for bacteria (Epicentre). mRNA-enriched total RNA was converted to cDNA and prepared for sequencing using the ScriptSeq™ v2 RNA-Seq Library preparation kit (Epicentre) and sequenced on an Illumina MiSeq using a 600 cycle kit. Metatranscriptomes were separated into ribosomal and non-ribosomal partitions using SortMeRNA61. Metatranscriptome sequencing statistics are listed in Supplementary Table 7.
Single-gene phylogenetic reconstruction
Small subunit rRNA sequences from metagenomes were identified in metagenomic assemblies using nhmmer against rfam databases for small subunit rRNAs (RFAM: RF00177, RF01959, RF01960), requiring at least 300 nucleotides to match the model. Sequences were classified using the RDPclassifer62 as implemented in Mothur.
Prodigal63 was used to generate gene predictions from each metagenomic assembly, using the metagenome option (-p meta). Assemblies were screened for marker genes of ammonium, urea, and cyanate utilization: ammonia monooxygenase subunit alpha (amoA), urease subunit alpha (ureC), and cyanate lyase/hydratase (cynS), respectively. hmmsearch64, which identifies protein sequences based on pfam hmm models, was used to identify genes of interest (Archaeal amoA (PF12942.2); cynS (PF02560.9) and ureC (PF00449.15)), with the requirement that the protein sequence and hmm model align over at least 70% of the length of the model and that the reverse search of the identified protein sequence against the pfam database returned the target model as the best hit. Metagenome encoded genes of interest were used as queries against the NCBI non-redundant (nr) protein database (as of March, 2018) using default settings and the hits were filtered to remove sequences with less than 50% sequence coverage of the query gene. Hits were then clustered at 90% identity using Usearch48 and added to custom amoA nucleotide, UreC amino acid, and CynS amino acid sequence databases. The CynS database was previously compiled6. The amoA and UreC databases were compiled from Pfam entries65. Metagenomic-encoded amoA, UreC and CynS sequences were added to the custom databases and aligned with mafft-linsi66 and trimmed using trimal -automated167. Phylogenetic reconstruction was calculated with IQ-TREE60 with automated model selection and confidence assessed with ultrafast bootstrapping (1,000 iterations). Resulting trees were visualized using ITOL68.
Metagenomic and metatranscriptomic reads were used to quantify amoA, ureC and cynS in these datasets. mRNA reads were screened by BLASTX against the datasets assembled for phylogenetic analysis (see above). Positive BLASTX matches were defined by a bit score ≥ 50 and amino acid identity ≥ 40%. Reads were added to alignments used for calculating phylogeny of each gene of interest using the --add-fragments option in mafft and placed into single gene trees using the evolutionary placement algorithm53. Fragments per kilobase per million reads (FPKM) values were calculated based on the number of read pairs for which one or both reads placed into a specified location in the tree, divided by the average gene length in the reference alignment (in kb) divided by the number of total metagenomic read pairs or ribosomal-RNA free metatranscriptomic read pairs (in millions). Gene lengths for the target genes are as follows: amoA (593 nt), ureC (1,477 nt), cynS (462 nt).
The percentage of ureC-containing Thaumarchaeota was estimated for each metagenomic dataset using a method similar to that of Tolar et al. and Santoro et al.9,10, which involved comparing the FPKM for urease genes (FPKMureC) classified as thaumarchaeal ureC and the FPKM for thaumarchaeal amoA (FPKMamoA) and SSU (16S rRNA genes, FPKMSSU) genes, under the assumption that amoA and SSU were universally present in all Thaumarchaeota as single copy genes. The percentage of ureC Thaumarchaeota was then calculated as FPKMureC / FPKMamoA and/or as FPKMureC / FPKMSSU.
Thaumarchaeota quantification by CARD-FISH
For Thaumarchaeota quantification, seawater samples were fixed with 1% paraformaldehyde (PFA, without methanol, EMS) for 12 to 24 h at 4°C before filtration (< 400 mbar) onto 0.22 μm GTTP filters (Millipore) and washing with sterile filtered seawater. Filters were stored frozen at -20°C. Thaumarchaeota abundances were determined by CARD-FISH following Pernthaler et al. 200469 using the horseradish peroxidase (HRP) labeled probe Thaum726 (GCTTTCATCCCTCACCGTC, for probe specifications see below and Supplementary Table 8) and unlabeled competitor probes (Thaum726_compA: GCTTTCGTCCCTCACCGTC, Thaum726_compB: GCTTTCATCCCTCACTGTC)70,71. For CARD-FISH, cells were immobilized on the filters by embedding in 0.2% low gelling agarose and endogenous peroxidases were inactivated by incubation in 0.01 M HCl for 10 min. Cells were permeabilized by HCl (0.1 M HCl for 1 min) and lysozyme (10 mg ml-1 in 50 mM EDTA and 100 mM Tris-HCl at 37°C for 1 h). Filter pieces were hybridized with HRP probes and the respective competitor probes at 25% formamide concentration at 46°C for up to 3.5 h. After a 5 min washing step at 48°C and HRP probe equilibration in 1x PBS for 15 min, signal amplification was done with OregonGreen488 labeled tyramides at 48°C for 15 to 30 min. Before enumeration on an epifluorescence microscope (Axioplan 2, Zeiss), cells were counterstained with DAPI (10 μg ml-1, 5 min at room temperature). For each CARD-FISH experiment, positive controls using probes EUB338 I-III72,73 and negative controls with the probe NonEUB74 on separate filter pieces were included to exclude non-specific binding of oligonucleotides or insufficient inactivation of endogenous peroxidases.
The probe used for CARD-FISH (Thaum726) targeted all recovered GoM Thaumarchaeota OTUs except for three, which together made up only 0.07% of all Thaumarchaeota 16S rRNA gene reads. Thaum726 also targeted all Thaumarchaeota 16S rRNA gene fragments obtained by metagenomics that had sequence information at the probe binding site. Furthermore, Thaum726 targets 94.6% of Nitrosopumilales Thaumarchaeota (Marine Group 1, sequences included in ARB Silva database SSURef_NR99_128_SILVA_07_09_16, Supplementary Table 8). To ensure specificity to Thaumarchaeota for our samples, Thaum726 was also screened for non-target matches against the entire retrieved GoM 16S rRNA gene amplicon dataset. We found that it targeted only one non-target Crenarchaeota OTU present only at Station 1, 16m depth with a relative abundance of 0.01%. It is therefore unlikely that we under- or overestimated Thaumarchaeota present in the GoM due to mismatches in the probe binding site in Thaumarchaeota, or probe binding to non-target organisms.
nanoSIMS Analyses and Calculation of Single Cell Growth and N-uptake Rates
For nanoSIMS analyses, unfixed seawater samples were gently filtered (< 100 mbar) onto gold sputtered 0.22 μm GTTP filters (Millipore), and subsequently fixed in 3% PFA (in sterile filtered seawater) for 30 min at room temperature, washed twice in sterile filtered seawater and then stored at -20°C until CARD-FISH and nanoSIMS. CARD-FISH targeting Thaumarchaeota was done without embedding filters in agarose. After counterstaining with DAPI, regions of interest were marked on a laser microdissection microscope (6000 B, Leica) and images of CARD-FISH signals were acquired on an epifluorescence microscope (Axioplan 2, Zeiss).
Single cell 15N- and 13C-uptake from 15N-ammonium and 13C-bicarbonate, 15N13C-urea and 15N13C-cyanate were determined for Station 2, 14m depth, using a nanoSIMS 50L (CAMECA), as previously described75. Instrument precision for detection of 15N/14N and 13C/12C isotope ratios was monitored daily on Graphite Planchet and regularly on caffeine standards. Due to the small size of Thaumarchaeota (<1 μm), samples were only briefly (10 s) pre-sputtered with a Cs+ beam (~300 pA) before measurement. Measurements were done on a field size of 10 × 10 μm or 15 × 15 μm, with a dwelling time of 2 ms per pixel and 256 × 256 pixel resolution over 40 planes. Analysis of the acquired data was performed using the Look@NanoSIMS software package76 as previously described75. Ratios of 15N/(15N+14N) and 13C/(13C+12C) of Thaumarchaeota and non-Thaumarchaeota cells were used for calculation of growth rates only when the overall enrichment Poisson error across all planes of a given cell was <5%. The variability in 15N/(15N+14N) ratios across measured Thaumarchaeota and non-Thaumarchaeota cells was calculated following Svedén et al. 77 (see section below).
Single cell growth rates were calculated as previously described75, where cell 15N- and 13C-atom% excess was calculated by subtracting natural abundance 15N/(15N+14N) and 13C/(13C+12C) values (0.37% and 1.11%, respectively). To be conservative in our calculations, we did not take the isotopic dilution of 15N/(15N+14N) and 13C/(13C+12C) ratios due to CARD-FISH into account78,79 (see section below). For calculation of per-cell N-uptake rates, an average carbon content of 9 fg-C per Thaumarchaeota cell80 and a C:N ratio following Redfield (C:N = 6.625:1) were assumed, resulting in an average N-content of 1.36 fg-N per Thaumarchaeota cell. N-uptake rates were calculated by:
| (1) |
| (2) |
where 15Nat%excesscell and 15Nat%excesslabel are 15N-atom% of a given measured cell and of the 15N-enriched seawater during the incubation after subtraction of natural abundance 15N-atom% (0.37%), fgNcell is the assumed N-content per cell, and time is the incubation time in days81.
Analysis of 15N/(14N+15N) ratio variability and isotopic dilution due to CARD-FISH
To test if sufficient cells have been measured by nanoSIMS, we calculated the mean and standard error for randomly subsampled ROIs according to Svedén et al.77, who propose that the error of randomly subsampled ROIs of one population should be <10%. Our analysis showed that the standard error for Thaumarchaeota 15N/(14N+15N) ratios was < 10% after random subsampling of 5 and 3 cells respectively, for ammonium and cyanate incubations (Supplementary Fig. 11). This indicates a highly homogenous 15N-uptake by the measured Thaumarchaeota population. However, the spread in 15N/(14N+15N) ratios was much larger for Thaumarchaeota in the urea incubation (20%, Supplementary Fig. 11). This could be either due to too few measured cells, or, alternatively, due to the presence of several Thaumarchaeota subpopulations, which have different activities on urea. The latter is in line with the presence of urease in only approximately 10-15% of the Thaumarchaeota based on metagenomics (see main text), indicating that the spread in 15N/(14N+15N) ratios in the urea treatment is an inherent feature of the Thaumarchaeota community. The spread in 15N/(14N+15N) ratios was higher in all treatments for non-Thaumarchaeota than for Thaumarchaeota, likely due to the diversity of microorganisms and their physiology in this class.
CARD-FISH has previously been shown to introduce isotopic dilution of both target and, to a lesser extent, non-target cells 78,79. The extent of isotopic dilution is affected by growth stage and hypothesized to also depend on washing steps and the CARD-FISH protocol used. We have not accounted for isotopic dilution of Thaumarchaeota cells due to CARD-FISH in our experiments to be conservative, as the isotopic dilution effect for complex environmental samples is still not well constrained. Thaumarchaeota are therefore likely to be even higher enriched in 15N and 13C compared to other cells than reported, and in-situ growth rates may have been underestimated.
Cyanate oxidation by cultures of Nitrosopumilus maritimus
Experiments were carried out to assess cyanate use by four cultures of the marine Thaumarchaeon N. maritimus SCM182. All cultures were inoculated in 100 ml synthetic Crenarchaeota medium2 containing 1 mM NH4Cl. Cultures were incubated in the dark at 28°C without shaking. After consumption of >0.7 mM NH4+, cultures were amended with freshly prepared, sterile filtered (0.2 μm) 200 μM 14N-NH4+ and 40 μM 13C15N-cyanate. At the time of tracer addition, cell numbers were 1.11×108, 1.35×108, 7.89 ×107 and 9.42 ×107 cells ml-1 in culture 1, 2, 3 and 4, respectively. At each time point (0, 3, 6 and 12 h), 20 ml samples were taken under sterile conditions, sterile filtered and stored at -80°C until concentration analysis (cyanate, ammonium, nitrite), and GC-IRMS (15N-ammonium and 15N-nitrite) measurements. The purity of cultures was confirmed at the end of the experiment using CARD-FISH (probe Thaum726 and DAPI staining), as described above. Abiotic breakdown of cyanate was assessed in sterile filtered media and a further experiment assessed the breakdown of cyanate in the culture supernatant of N. maritimus SCM1 after growth on 1 mM ammonium (supernatant controls, see Supplementary Discussion). The contribution of abiotic cyanate breakdown to the observed cyanate-derived oxidation rates in the N. maritimus cultures was calculated analogously to field experiments (Supplementary Discussion).
Supplementary Material
Supplementary information is available for this paper.
Acknowledgements
The authors thank the captain and crew of the R/V Pelican PE17-02 cruise. The authors are grateful to G. Klockgether, D. Tienken, I. Ulber, L. Seidl, W. Neweshy, N. Alrubeay, M. Philippi and D.J. Parris for technical support; G. Lavik, J. Milucka, W. Mohr, N. Lehnen and S. Ahmerkamp for fruitful discussions. This research was funded by the Max-Planck-Society, the European Research Council Advanced Grant project NITRICARE 294343 (to M.W) and the National Science Foundation grants 1558916 and 1564559 (to F.J.S.)
Footnotes
Data Availability
All sequence data and thaumarchaeal MAGs generated in this study are deposited in NCBI under BioProject number: PRJNA397176. Metatranscriptomes are deposited under BioSample numbers SAMN07461123-SAMN07461125; 16S amplicon sequencing under SAMN07461114-SAMN07461122; metagenomes under SAMN10227777-SAMN10227781, and MAGs under SAMN10233969 – SAMN10233974. Accession numbers of sequences used for tree calculations (16S rRNA gene, amoA, UreC, CynS, and genome sequences) are given in Supplementary Table 9. CTD data, measured nutrient concentrations, process rates, Thaumarchaeota relative abundance based on 16S rRNA gene amplicon sequencing and Thaumarchaeota specific CARD-FISH counts are given in Supplementary Table 10.
Author Contributions
L.A.B., K.K., H.K.M., M.M.M.K. and M.W. designed the study. K.K., L.A.B. and H.K.M. performed experiments, S.L. and A.K. ran nanoSIMS analyses. K.K., L.A.B., H.K.M. and P.H. analyzed samples and data. C.C.P. sampled for and performed molecular analyses with contribution from C.W.H. and F.J.S. Cyanate concentrations were measured by M.M. and A.R., TDN was analyzed by J.N. Cultures were provided by S.P. and M.K. The manuscript was written by K.K., L.A.B. and H.K.M., with contributions from all co-authors.
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Francis CA, Roberts KJ, Beman JM, Santoro AE, Oakley BB. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc Natl Acad Sci U S A. 2005;102:14683–14688. doi: 10.1073/pnas.0506625102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 3.Horak REA, et al. Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J. 2013;7:2023–2033. doi: 10.1038/ismej.2013.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin W, et al. Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci U S A. 2014;111:12504–12509. doi: 10.1073/pnas.1324115111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayer B, et al. Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J. 2016;10:1051–1063. doi: 10.1038/ismej.2015.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palatinszky M, et al. Cyanate as an energy source for nitrifiers. Nature. 2015;524:105–108. doi: 10.1038/nature14856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alonso-Saez L, et al. Role for urea in nitrification by polar marine Archaea. Proc Natl Acad Sci U S A. 2012;109:17989–17994. doi: 10.1073/pnas.1201914109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Connelly TL, Baer SE, Cooper JT, Bronk DA, Wawrik B. Urea Uptake and Carbon Fixation by Marine Pelagic Bacteria and Archaea During the Arctic Summer and Winter Seasons. Appl Environ Microbiol. 2014;80:6013–6022. doi: 10.1128/AEM.01431-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tolar BB, Wallsgrove NJ, Popp BN, Hollibaugh JT. Oxidation of urea-derived nitrogen by thaumarchaeota-dominated marine nitrifying communities. Environ Microbiol. 2017;19:4838–4850. doi: 10.1111/1462-2920.13457. [DOI] [PubMed] [Google Scholar]
- 10.Santoro AE, et al. Thaumarchaeal ecotype distributions across the equatorial Pacific Ocean and their potential roles in nitrification and sinking flux attenuation. Limnol Oceanogr. 2017;62:1984–2003. [Google Scholar]
- 11.Wuchter C, et al. Archaeal nitrification in the ocean. Proc Natl Acad Sci U S A. 2006;103:12317–12322. doi: 10.1073/pnas.0600756103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sipler RE, Bronk DA. Dynamics of Dissolved Organic Nitrogen. Biogeochemistry of Marine Dissolved Organic Matter: Second Edition. Elsevier Inc; 2014. [DOI] [Google Scholar]
- 13.Widner B, Mulholland MR, Mopper K. Distribution, Sources, and Sinks of Cyanate in the Coastal North Atlantic Ocean. Environ Sci Technol Lett. 2016;3:297–302. [Google Scholar]
- 14.Widner B, Mulholland MR. Cyanate distribution and uptake in North Atlantic coastal waters. Limnol Oceanogr. 2017;62:2538–2549. [Google Scholar]
- 15.Widner B, Mordy CW, Mulholland MR. Cyanate distribution and uptake above and within the Eastern Tropical South Pacific oxygen deficient zone. Limnol Oceanogr. 2018;63:S177–S192. [Google Scholar]
- 16.Antia NJ, Harrison PJ, Oliveira L. The role of dissolved organic nitrogen in phytoplankton nuitrition, cell biology and ecology. Phycologia. 1991;30:1–89. [Google Scholar]
- 17.Dirnhuber P, Schütz F. The isomeric transformation of urea into ammonium cyanate in aqueous solutions. Biochem J. 1948;42:628–632. [PubMed] [Google Scholar]
- 18.Liu K-K, Atkinson L, Quiñones R, Talaue-McManus L. Carbon and Nutrient Fluxes in Continental Margins - A Global Synthesis. Carbon and nutrient fluxes in continental margins. Springer; 2010. [DOI] [Google Scholar]
- 19.Rabalais NN, Turner RE, Wiseman WJ. Hypoxia in the Gulf of Mexico. J Environ Qual. 2001;30:320–329. doi: 10.2134/jeq2001.302320x. [DOI] [PubMed] [Google Scholar]
- 20.Tolar BB, King GM, Hollibaugh JT. An analysis of Thaumarchaeota populations from the northern Gulf of Mexico. Front Microbiol. 2013;4:1–72. doi: 10.3389/fmicb.2013.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bristow LA, et al. Biogeochemical and metagenomic analysis of nitrite accumulation in the Gulf of Mexico hypoxic zone. Limnol Oceanogr. 2015;60:1733–1750. [Google Scholar]
- 22.Woebken D, Fuchs BM, Kuypers MMM, Amann R. Potential interactions of particle-associated anammox bacteria with bacterial and archaeal partners in the Namibian upwelling system. Appl Environ Microbiol. 2007;73:4648–4657. doi: 10.1128/AEM.02774-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Galand PE, Gutiérrez-Provecho C, Massana R, Gasol JM, Casamayor EO. Inter-annual recurrence of archaeal assemblages in the coastal NW Mediterranean Sea (Blanes Bay Microbial Observatory) Limnol Oceanogr. 2010;55:2117–2125. [Google Scholar]
- 24.Liu Q, et al. Light and temperature control the seasonal distribution of thaumarchaeota in the South Atlantic bight. ISME J. 2018;12:1473–1485. doi: 10.1038/s41396-018-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carini SA, McCarthy MJ, Gardner WS. An isotope dilution method to measure nitrification rates in the northern Gulf of Mexico and other eutrophic waters. Cont Shelf Res. 2010;30:1795–1801. [Google Scholar]
- 26.Ward BB. In: Nitrogen in the Marine Environment. Capone DG, Bronk DA, Mulholland MR, Carpenter EJ, editors. Elsevier Inc; 2008. pp. 199–261. [DOI] [Google Scholar]
- 27.Lam P, et al. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc Natl Acad Sci U S A. 2009;106:4752–4757. doi: 10.1073/pnas.0812444106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koch H, et al. Expanded metabolic versatility of ubiquitous nitrite-oxidizing bacteria from the genus Nitrospira. Proc Natl Acad Sci U S A. 2015;112:11371–11376. doi: 10.1073/pnas.1506533112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Herndl GJ, et al. Contribution of Archaea to Total Prokaryotic Production in the Deep Atlantic Ocean. Appl Environ Microbiol. 2005;71:2303–2309. doi: 10.1128/AEM.71.5.2303-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holmes RM, Aminot A, Kérouel R, Hooker BA, Peterson BJ. A simple and precise method for measuring ammonium in marine and freshwater ecosystems. Can J Fish Aquat Sci. 1999;56:1801–1808. [Google Scholar]
- 31.Grasshoff K, Kremling K, Ehrhardt M. Methods of seawater analysis. Vol. 7 Wiley-VCH; 1999. [Google Scholar]
- 32.Mulvenna PF, Savidge G. A modified manual method for the determination of urea in seawater using diacetylmonoxime reagent. Estuar Coast Shelf Sci. 1992;34:429–438. [Google Scholar]
- 33.Widner B, Mulholland MR, Mopper K. Chromatographic determination of nanomolar cyanate concentrations in estuarine and sea waters by precolumn fluorescence derivatization. Anal Chem. 2013;85:6661–6666. doi: 10.1021/ac400351c. [DOI] [PubMed] [Google Scholar]
- 34.Braman RS, Hendrix SA. Nanogram Nitrite and Nitrate Determination in Environmental and Biological Materials by Vanadium(III) Reduction with Chemiluminescence Detection. Anal Chem. 1989;61:2715–2718. doi: 10.1021/ac00199a007. [DOI] [PubMed] [Google Scholar]
- 35.Seidel M, et al. Composition and Transformation of Dissolved Organic Matter in the Baltic Sea. Front Earth Sci. 2017;5:1–20. [Google Scholar]
- 36.De Brabandere L, L, T B, Revsbech NP, Foadi R. A critical assessment of the occurrence and extend of oxygen contamination during anaerobic incubations utilizing commercially available vials. J Microbiol Methods. 2012;88:147–154. doi: 10.1016/j.mimet.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Garcia HE, Gordon LI. Oxygen Solubility in Seawater: Better Fitting Equations. Limnol Oceanogr. 1992;37:1307–1312. [Google Scholar]
- 38.Füssel J, et al. Nitrite oxidation in the Namibian oxygen minimum zone. ISME J. 2012;6:1200–1209. doi: 10.1038/ismej.2011.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Altabet MA, Wu T, Hadas O. Sensitive Measurement of NH4+ 15N/14N (δ15NH4+) at Natural Abundance Levels in Fresh and Saltwaters. Anal Chem. 2007;79:5589–5595. doi: 10.1021/ac070106d. [DOI] [PubMed] [Google Scholar]
- 40.Torres ME, Mix AC, Rugh WD. Precise δ13C analysis of dissolved inorganic carbon in natural waters using automated headspace sampling and continuous-flow mass spectrometry. Limnol Oceanogr Methods. 2005;3:349–360. [Google Scholar]
- 41.Granger J, Sigman DM. Removal of nitrite with sulfamic acid for nitrate N and O isotope analysis with the denitrifier method. Rapid Commun Mass Spectrom. 2009;23:3753–3762. doi: 10.1002/rcm.4307. [DOI] [PubMed] [Google Scholar]
- 42.Padilla CC, et al. Standard filtration practices may significantly distort planktonic microbial diversity estimates. Front Microbiol. 2015;6:1–10. doi: 10.3389/fmicb.2015.00547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Padilla CC, et al. NC10 bacteria in marine oxygen minimum zones. ISME J. 2016;10:2067–2071. doi: 10.1038/ismej.2015.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Frias-Lopez J, et al. Microbial community gene expression in ocean surface waters. Proc Natl Acad Sci. 2008;105:3805–3810. doi: 10.1073/pnas.0708897105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caporaso JG, et al. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci. 2011;108:4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parada AE, Needham DM, Fuhrman JA. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol. 2016;18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 47.Magoč T, Salzberg SL. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 49.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 53.Berger SA, Krompass D, Stamatakis A. Performance, accuracy, and web server for evolutionary placement of short sequence reads under maximum likelihood. Syst Biol. 2011;60:291–302. doi: 10.1093/sysbio/syr010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017;27:824–834. doi: 10.1101/gr.213959.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu YW, Simmons BA, Singer SW. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics. 2015;32:605–607. doi: 10.1093/bioinformatics/btv638. [DOI] [PubMed] [Google Scholar]
- 56.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Olm MR, Brown CT, Brooks B, Banfield JF. DRep: A tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 2017;11:2864–2868. doi: 10.1038/ismej.2017.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS, Arkin AP. FastTree 2 - Approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kopylova E, Noé L, Touzet H. SortMeRNA : Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics. 2012;28:3211–3217. doi: 10.1093/bioinformatics/bts611. [DOI] [PubMed] [Google Scholar]
- 62.Wang Q, Garrity GM, Tiedje JM, Cole JR. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finn RD, et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pernthaler A, Pernthaler J, Amann R. In: Molecular Microbial Ecology Manual. Kowalchuk G, de Bruijn FJ, Head IM, Akkermans ADL, van Elsas JD, editors. Kluwer Academic Publishers; 2004. pp. 711–726. [DOI] [Google Scholar]
- 70.Beam JP. Geobiological interactions of archaeal populations in acidic geothermal springs of Yellowstone National Park, WY, USA. Montana State University; 2015. [Google Scholar]
- 71.Sauder LA, et al. Cultivation and characterization of Candidatus Nitrosocosmicus exaquare, an ammonia-oxidizing archaeon from a municipal wastewater treatment system. ISME J. 2017;11:1142–1157. doi: 10.1038/ismej.2016.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Amann RI, et al. Combination of 16S rRNA-Targeted Oligonucleotide Probes with Flow Cytometry for Analyzing Mixed Microbial Populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daims H, Brühl A, Amann R, Schleifer K-H, Wagner M. The Domain-specific Probe EUB338 is Insufficient for the Detection of all Bacteria: Development and Evaluation of a more Comprehensive Probe Set. Syst Appl Microbiol. 1999;22:434–444. doi: 10.1016/S0723-2020(99)80053-8. [DOI] [PubMed] [Google Scholar]
- 74.Wallner G, Amann R, Beisker W. Optimizing fluorescent in situ hybridization with rRNA-targeted oligonucleotide probes for flow cytometric identification of microorganisms. Cytometry. 1993;14:136–143. doi: 10.1002/cyto.990140205. [DOI] [PubMed] [Google Scholar]
- 75.Martínez-Pérez C, et al. The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.163. 16163. [DOI] [PubMed] [Google Scholar]
- 76.Polerecky L, et al. Look@NanoSIMS - a tool for the analysis of nanoSIMS data in environmental microbiology. Environ Microbiol. 2012;14:1009–1023. doi: 10.1111/j.1462-2920.2011.02681.x. [DOI] [PubMed] [Google Scholar]
- 77.Svedén JB, et al. High cell-specific rates of nitrogen and carbon fixation by the cyanobacterium Aphanizomenon sp. at low temperatures in the Baltic Sea. FEMS Microbiol Ecol. 2015;91:1–10. doi: 10.1093/femsec/fiv131. [DOI] [PubMed] [Google Scholar]
- 78.Musat N, Foster R, Vagner T, Adam B, Kuypers MMM. Detecting metabolic activities in single cells, with emphasis on nanoSIMS. FEMS Microbiol Rev. 2012;36:486–511. doi: 10.1111/j.1574-6976.2011.00303.x. [DOI] [PubMed] [Google Scholar]
- 79.Woebken D, et al. Revisiting N2 fixation in Guerrero Negro intertidal microbial mats with a functional single-cell approach. ISME J. 2015;9:485–496. doi: 10.1038/ismej.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Berg C, Listmann L, Vandieken V, Vogts A, Jürgens K. Chemoautotrophic growth of ammonia-oxidizing Thaumarchaeota enriched from a pelagic redox gradient in the Baltic Sea. Front Microbiol. 2015;5:1–10. doi: 10.3389/fmicb.2014.00786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Krupke A, et al. The effect of nutrients on carbon and nitrogen fixation by the UCYN-A – haptophyte symbiosis. ISME J. 2015;9:1635–1647. doi: 10.1038/ismej.2014.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.