Abstract
Purpose:
While FGFR1 amplification has been described in breast cancer, the optimal treatment approach for FGFR1-amplified (FGFR1+) metastatic breast cancer (MBC) remains undefined.
Experimental Design:
We evaluated clinical response to endocrine and targeted therapies in a cohort of patients with HR+/HER2− MBC and validated the functional role of FGFR1-amplification in mediating response/resistance to hormone therapy in vitro.
Results:
In the clinical cohort (N=110), we identified that patients with FGFR1+ tumors were more likely to have PR-negative disease (47% vs 20%; p=0.005), co-existing TP53 mutations (41% vs. 21%; p = .05), and exhibited shorter time to progression with endocrine therapy alone and in combination with CDK4/6 inhibitor, but not with a TORC1 inhibitor (everolimus), adjusting for key prognostic variables in multivariate analysis. Furthermore, mTOR-based therapy resulted in a sustained radiological and molecular response with decrease in FGFR1 circulating tumor DNA (ctDNA) in an index case of FGFR1+ HR+/HER2− MBC. In pre-clinical models, ER+/FGFR1 amplified CAMA1 human breast cancer cells were only partially sensitive to fulvestrant, palbociclib, alpelisib, but highly sensitive to everolimus. In addition, transduction of a FGFR1 expression vector into ER+ T47D cells induced resistance to fulvestrant that could be overcome by added TORC1 inhibition, but not PI3K or CDK4/6 inhibition.
Conclusion:
Collectively, these findings suggest that while FGFR1 amplification confers broad resistance to ER, PI3K, and CDK4/6 inhibitors, TORC1 inhibitors might have a unique therapeutic role in the treatment of patients with ER+/FGFR1+ MBC.
Keywords: FGFR1 amplification, breast cancer, endocrine resistance, targeted therapy
INTRODUCTION
Approximately 70% of breast cancers are classified as hormone-receptor positive (HR+) and anti-estrogen therapy (also called endocrine therapy) is the mainstay of treatment.1 However, resistance to endocrine therapy is a well-documented challenge in the treatment of HR+ breast cancer.2,3,4 The molecular underpinnings of endocrine therapy resistance are still being elucidated, however several studies have implicated the activation of growth factor signaling pathways as one key-mediator of estrogen-independent cell growth and proliferation.4
Fibroblast Growth Factor Receptor 1 (FGFR1) is a transmembrane protein and member of the fibroblast growth factor receptor family.5 Molecular aberrations of the FGFR1 gene, located on chromosome 8p11–12, are widely implicated in cancer pathogenesis.6,7 In breast cancer, amplifications in FGFR1 have been identified in 7.5–17% of all cases, and as many as 16–27% of aggressive HR+ (luminal-B type) tumors.8,9,10 FGFR1 signaling via the MAPK and PI3K pathways appears to play a central role in proliferation, migration, and survival of mammary cells.11,12,13 Upregulation of FGFR1 in culture and mouse models results in increased cell dysplasia and invasion, and inhibition of FGFR1 via siRNAs or small molecules appears to reverse these effects.14,15 In previous studies, FGFR1 amplification has been implicated as a predictor of decreased overall survival in patients with HR+ breast cancer.16,17
While FGFR1 amplification has been described in breast cancer, the clinical impact of this alteration in modulating response to standard therapies is not well established. This is particularly relevant with the clinical development of multiple therapies for HR+ breast cancer including CDK4/6 inhibitors, PI3K inhibitors, and TORC inhibitors. However currently there is a lack of biomarker-driven strategies available to guide the selection of a matched therapy. Furthermore, while few clinical trials of multi-targeted FGFR tyrosine kinase inhibitors in breast cancer have been completed, the therapeutic efficacy has been limited, thus, targeted treatment for FGFR1 amplified HR+ breast cancer remains an unmet clinical need.17
In this study, we evaluated clinical response to endocrine and targeted therapies in a cohort of patients with metastatic HR+ breast cancer, and validated the functional role of FGFR1-amplification in mediating response/resistance to hormone therapy in vitro. We show herein that FGFR1 gene amplification in HR+ breast cancer drives resistance to endocrine therapy and multiple other targeted therapies including CDK4/6 inhibitors, but is not associated with resistance to inhibition of TORC1.
MATERIALS AND METHODS
Selection and Description of Study Participants
All patients in this study were seen at the Massachusetts General Hospital Cancer Center for treatment of metastatic HR+/HER2− breast cancer and were offered genetic tumor profiling as part of their routine clinical care before the study close date of March 31st 2018. This profiling included Fluorescence In-Situ Hybridization (FISH) analysis of the FGFR gene, as well as SNAPSHOT tumor genotyping. After written patient consent, Profiling was completed on either primary or metastatic tumor tissue depending on specimen availability. The cohort included both de-novo metastatic patients and those with recurrent disease. Start date and end date of treatment was evaluated for first line endocrine therapy with/without CDK 4/6 inhibitor, and first exposure for chemotherapy and endocrine therapy with mTOR inhibitor. Demographic and clinical data were obtained by researchers who had not participated in the clinical care of the included patients, after approval by Partners IRB (Institutional Review Board). The study was conducted according to International Ethical Guidelines for Biomedical Research Involving Human Subjects.
Clinical Biomarkers, FGFR1 FISH, Genotyping, and Circulating Tumor DNA
For tissue specimens, clinical markers of the estrogen receptor and progesterone receptor were measured and defined according to ASCO/CAP guidelines.
For each FGFR1 FISH specimen, 5-micron sections of formalin-fixed paraffin-embedded tumor material were prepared and an H&E section reviewed to select regions for hybridization that contain a majority of tumor cells. A dual-color FISH assay was performed using Bacterial Artificial Chromosome probe CTD-2288L6 (chromosome 8p, FGFR1 locus, home-brew) and a copy number probe (centromere 8; Abbott-Vysis 06J54–018 or 06J37–018). Signal quantitation was used to generate a FGFR1/centromere 8 ratio. FGFR1 to Centromere 8 ratios of ≥ 2 were considered amplified.
The SNAPSHOT assay utilizes “Anchored Multiplex PCR” for single nucleotide variant (SNV) and insertion/deletion (indel) detection in genomic DNA using next generation sequencing (NGS). Genomic DNA was isolated from a formalin-fixed paraffin embedded tumor specimen (after histological review for tumor enrichment), then sheared with the Covaris M220 instrument, followed by end-repair, adenylation, and ligation with an adapter. A sequencing library targeting hotspots and exons in 39 genes was generated using two hemi-nested PCR reactions. IlluminaMiSeq 2 × 151 base paired-end sequencing results were aligned to the hg19 human genome reference using BWA-MEM. MuTect and a laboratory-developed insertion/deletion analysis algorithm were used for SNV and indel variant detection, respectively.29 This assay has been validated to detect SNV and indel variants at 5% allelic frequency or higher in target regions with sufficient read coverage.
For ctDNA testing, we utilized a CLIA certified assay Guardant, a NGS based commercially available assay that detects ctDNA down to 0.1% allelic fraction with a clinical sensitivity of 85% (compared to 80.7% in tissue) and 99.8% specificity.30 Since use of ctDNA to detect copy number alterations is influenced by the number of copies in the tissue and the amount of DNA shedding into circulation, in order to reduce the number of biases, we normalized the results for aneuploidy to reduce any variations in tumor shedding that may occur over the course of a patient’s disease and therapy. In addition, since digital sequencing enables tracking and quantification of cfDNA fragments overlapping specific genomic sites, PCR duplicates and artifacts of amplification during library preparation can easily be removed and within-run (repeatability) and between-run (reproducibility) precision portion of the validation study has been shown to be > 90% concordant between different runs. For the index patient, ctDNA was measured prior to treatment, and 11 months into treatment.
Definitions of Clinical Treatments and Outcomes
Time to progression is defined as the time elapsing from the initiation of a given therapy, until that therapy was discontinued due to physician-judged disease progression. Endocrine therapy alone included any combination of selective estrogen receptor modulators, selective estrogen receptor degraders, aromatase inhibitors, or ovarian suppression, but excludes patients who received inhibitors of mTOR, CDK 4/6, or PI3K.
Statistical Methods
Patient and treatment characteristics were compared between groups using Wilcoxon rank-sum test for continuous variables and Fisher’s exact test or Pearson’s chi-squared test for categorical variables. Actuarial analysis of time to progression was performed using univariate and multivariable Cox regression. Figures of time to progression were calculated using Kaplan-Meier methodology. All tests were two-sided, and p value less than 0.05 was considered statistically significant.
Cell Culture Model and Lentivirus Transduction
T47D and CAMA-1 cells were obtained from ATCC and were cultured in DMEM supplemented with 10% fetal bovine serum (FBS). FGFR1− and GFP-expressing lentiviruses were generated by co-transfecting 4-mg proviral pLX302-FGFR1 or pLX302-GFP plasmids (Open BioSystems), 3 mg psPAX2 (plasmid encoding gag, pol, rev, and Tat genes), and 1 mg pMDG2 envelope plasmid (Sigma Aldrich) into 293FT cells using Lipofectamine 2000 (Thermo Fisher). 293FT cells were fed with 10% DMEM-FBS 24 h after transfection; virus-containing supernatants were harvested 48 and 72 h after transfection, diluted 1:4 and applied to target cells with 8 mg/mL polybrene (Sigma Aldrich). Target cells were selected with 1 mg/mL puromycin.
Immunodetection
Cells were plated in DMEM/10% FBS supplemented with 2 ng/mL FGF2 and treated for 6 h with inhibitors or vehicle. Lysates were generated as previously described28. For immunoblot analysis, the following antibodies were used: phospho-FRS2, phospho-p44/42 MAPK (Erk1/ 2), total p44/p42 MAPK (Erk½), and actin (Cell Signaling Technologies); FGFR1 antibody was obtained from Abcam; ERa antibody was obtained from SantaCruz. Immunoreactive proteins were visualized by enhanced chemiluminescence (Pierce).
Clonogenic assays
CAMA-1, T47DGFP and T47DFGFR1 cells, seeded into 6-well plates in DMEM/10% FBS/2 ng/mL FGF2, were treated with vehicle, fulvestrant 1 μM, alpelisib 1 μM, lucitanib 1 μM, selumetinib 1 μM, everolimus 25 nM, paclitaxel 10 nM, each alone or in combination. Media and inhibitors were replenished every 3 days, and cells were grown for 12 weeks till the untreated wells achieved 80% confluence. Cells were fixed and monolayers stained in 20% methanol with 0.5% crystal violet and washed with water. The intensity of the staining was quantitated by spectrophotometric detection as previously described28. Drugs were obtained from Selleck Chemicals.
Gene expression analysis
RNA was purified from cells using Maxwell RSC simplyRNA Cells Kit (Promega Corporation, Madison, WI, USA) and cDNA was generated using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA). qPCR was performed with a cDNA equivalent of 50 ng RNA, 1 µM each of the forward and reverse primers and SYBR Green PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA), using a QuantStudio 3 Real-Time PCR System machine (Applied Biosystems), and primers were used against the following targets: GAPDH (QIAGEN – PPH00150F), PGR (QIAGEN – PPH01007F). CT (threshold cycle) values were determined in triplicate samples by subtracting the target gene CT from the GAPDH CT; 2ΔCT was used to determine the expression of PGR relative to GAPDH.
RESULTS
Patient Characteristics and Genomic Landscape
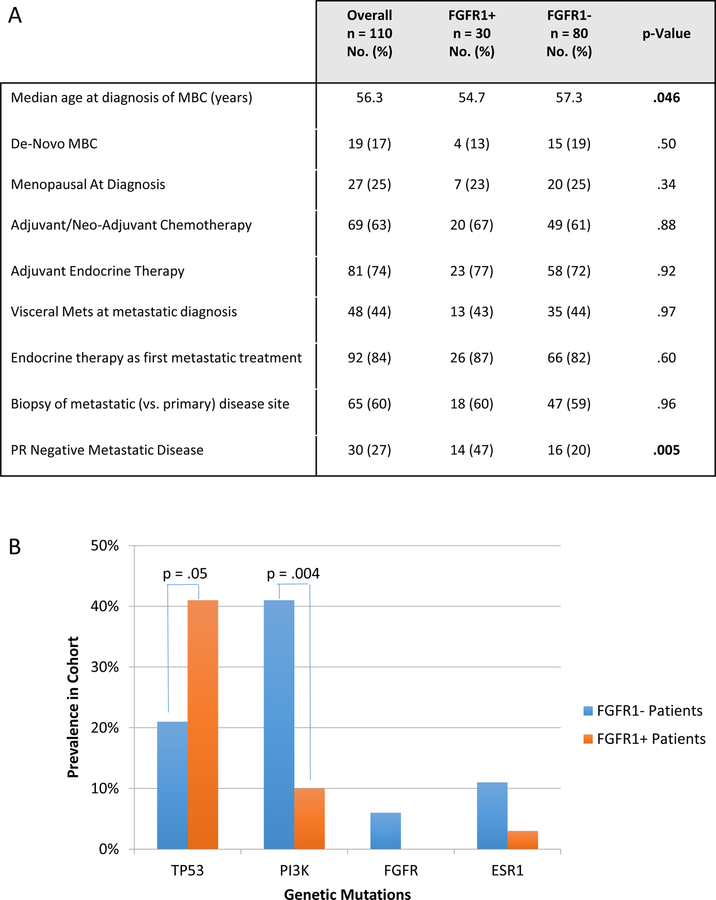
We first evaluated the clinical and genomic characteristics associated with FGFR1 amplification, based on a clinical cohort of patients with HR+/HER2‒ metastatic breast cancer (N=110) who had undergone molecular profiling utilizing FGFR1 FISH. Sixty-five (59%) of the FGFR1 FISH tests were performed on metastatic specimens, and the remainder were performed on primary specimens. Patients with HR+/HER2‒ breast cancer that also harbor FGFR1 amplification (FGFR+) (27.3%), as compared to FGFR1 non-amplified (FGFR1‒) patients tended to be slightly younger (median age at diagnosis of MBC: 54.7 vs. 57.3 years; p=0.046), and more likely to have PR-negative disease (47% vs 20%; p=0.005). Otherwise, FGFR1+ patients did not differ from patients with FGFR1‒ tumors in the likelihood of presentation with de-novo metastatic breast cancer, menopausal status at diagnosis, prior exposure to adjuvant endocrine or chemotherapy, presence of visceral metastases, biopsy site, or first-line treatment approach (Fig. 1A).
Figure 1. Clinical and genomic characteristics associated with FGFR1 amplification in patients with HR+/HER2− Metastatic Breast Cancer (MBC).
1A) Clinical characteristics of patients with metastatic HR+/HER2− breast cancer, harboring FGFR1 amplification (FGFR1+) versus non-amplified patient (FGFR1-). PR = Progesterone Receptor.
1B) Prevalence of tumor genomic alterations in FGFR+ vs. FGFR1− patients based on SNAPSHOT profiling.
Among those patients who had additional sequencing of their tumor specimens (N = 95), 26 (27.3%) patients had mutations in TP53, 30 (31.6%) had mutations in PIK3CA, 8 (8.4%) had mutations in ESR1, and 4 (4.2%) had mutations in other genes in the FGFR pathway. FGFR1+ tumors were more likely to have co-existing TP53 mutations (41% vs. 21%; p = .05), and less likely to have PIK3CA mutations (10% vs. 41%; p= .004) than FGFR1‒ tumors (Fig. 1B; Supplementary Fig. 1A–B). Eight patients were found to have mutations in ESR1, only one of whom had a concurrent FGFR1 amplification; there was no correlation observed between ESR1 mutations and median time to progression on endocrine therapy or chemotherapy in our cohort.
Clinical Outcomes
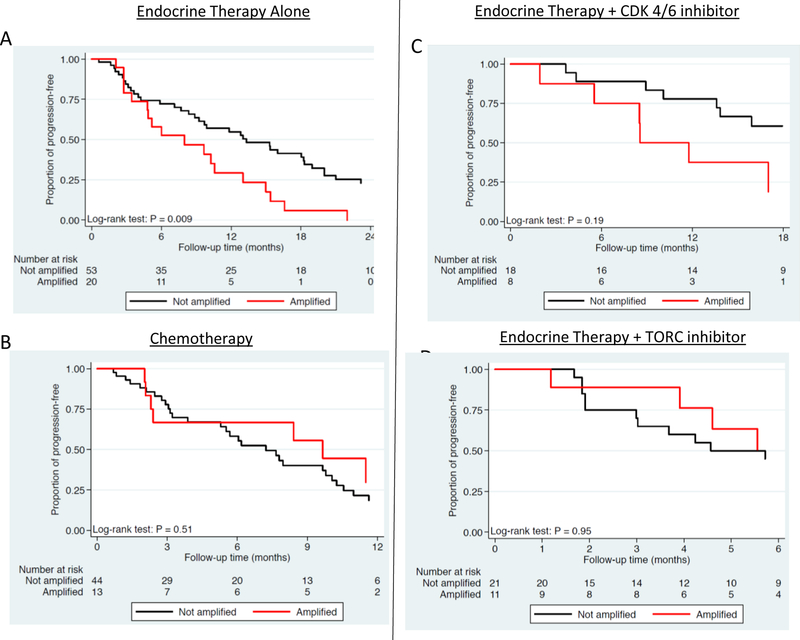
We then evaluated the association between FGFR1 amplification and time to progression (TTP) on anti-estrogen therapy alone and in combination with CDK4/6 inhibitors and with mTOR inhibitors, as well as chemotherapy, among patients with HR+/HER2− MBC (N=110). Patients with FGFR1+ tumors exhibited a shorter TTP than patients with FGFR‒ tumors on first line endocrine therapy (8.0 vs. 13.3 months; p=0.009, Fig 2A), but not first-line chemotherapy (8.4 vs. 10.9 months; p=0.99, Fig. 2B). A shorter TTP was also seen in patients with FGFR1+ tumors who received first line endocrine therapy in combination with a CDK4/6 inhibitor (8.5 vs. 25.3 months; p=0.19; Fig. 2C), though this was statistically underpowered due to small sample size (only 8 patients with FGFR1 amplified tumors). Interestingly, patients with FGFR1+ tumors who received endocrine therapy with mTOR inhibitor (2nd line and beyond) did not have worse TTP as compared to patients with non-amplified FGFR1 (7.1 vs. 4.5 months, p=0.95; Fig. 2D), highlighting lack of clinical resistance to mTOR based therapy.
Figure 2. Time to Progression with Various Therapies Among Patients with Metastatic HR+/HER2− Breast cancer, Stratified by FGFR1 Amplification.
1A) Time to Progression (TTP) on first line Endocrine-based Therapy in patients with metastatic HR+/HER2− breast cancer, stratified by FGFR1 amplification status.
1B) TTP on first exposure to Chemotherapy among patients with metastatic HR+/HER2− breast cancer, stratified by FGFR1 amplification status.
1C) TTP on first line Endocrine Therapy in combination with CDK 4/6 inhibitor among patients with metastatic HR+/HER2− breast cancer, stratified by FGFR1 amplification status.
1D) TTP on first exposure to Endocrine Therapy in combination with mTOR inhibitor among patients with metastatic HR+/HER2− breast cancer, stratified by FGFR1 amplification status.
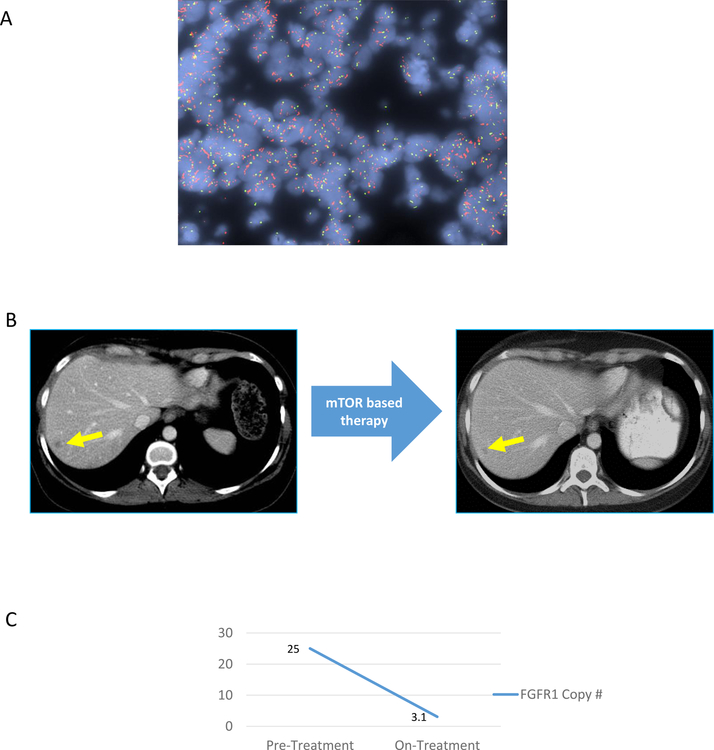
Furthermore, in an index case of a patient with FGFR1+/HR+/HER2− metastatic breast cancer, we observed a remarkable clinical and molecular response with mTOR based therapy. This patient had metastatic FGFR1-amplified HR+/HER2− breast cancer (FGFR1 copy number greater than 25; Fig. 3A), and prior disease progression after 11 months of first-line therapy with letrozole and a CDK 4/6 inhibitor (palbociclib). She then enrolled in a clinical trial of endocrine therapy with a CDK 4/6 inhibitor and mTOR inhibitor (everolimus). Despite prior CDK 4/6 exposure, the patient had a major clinical response with a 70.9% reduction in target lesions per RECIST after 13 cycles of mTOR based therapy (Fig. 3B), as well as a major molecular response (FGFR1 copy number in cfDNA decreased to 3.1; Fig 3B), highlighting the specific contribution of mTOR based therapy. The patient is currently doing well on the clinical trial (treatment ongoing at 14 months; data cut-off July 15th 2018).
Figure 3. Radiological and molecular response with mTOR based therapy in an index patient with FGFR1 amplified metastatic breast cancer.
2A) Restaging CT scan obtained before and during treatment with endocrine therapy in combination with CDK 4/6 inhibitor and mTOR inhibitor, in an index patient with FGFR1 amplified metastatic breast cancer. Target lesions in liver are indicated with yellow arrow.
2B) Changes in the FGFR1 copy number based on cfDNAbefore and during treatment with endocrine therapy in combination with CDK 4/6 inhibitor and mTOR inhibitor, in an index patient with FGFR1-amplified metastatic breast cancer.
2C) FGFR1 FISH of primary tumor tissue. The FGFR1 probe is in red, with control probe (centromere 8) in green.
Finally, we conducted a multivariate analysis to evaluate the clinical impact of FGFR1 amplification on therapeutic response, accounting for known prognostic variables, including age, de novo metastatic disease, presence of visceral metastases, and PR expression (Supplementary Fig. 2A). FGFR1 amplification emerged as the independent predictive variable for worse TTP with first line endocrine therapy in HR+ metastatic breast cancer (HR for progression 3.21; p = .006). However, FGFR1 amplification was not associated with worse TTP in patients receiving mTOR inhibitors, in a multivariate analysis (Supplementary Fig. 2B), supporting the clinical observations that both FGFR1+ and FGFR1− tumors are clinically sensitive to mTOR-based therapy.
Functional validation in pre-clinical models
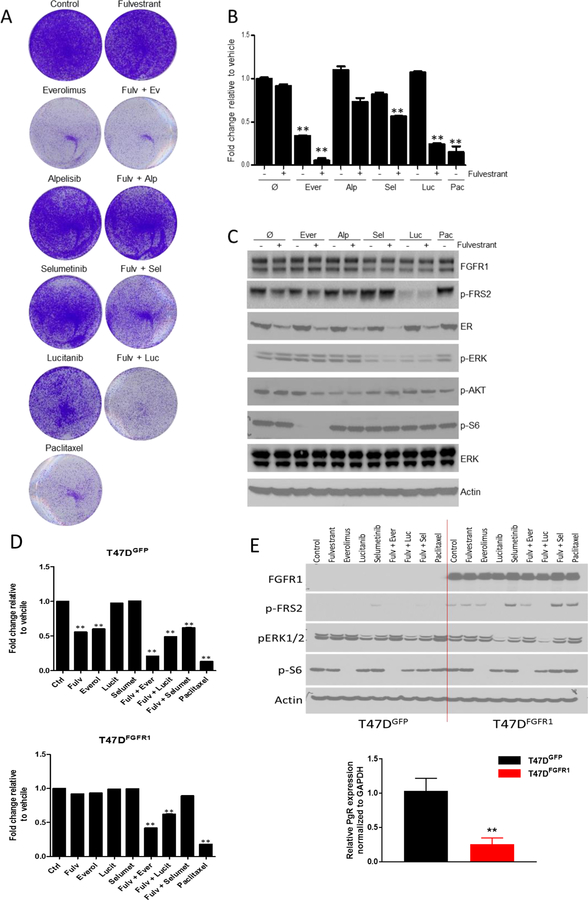
To provide functional validity to the clinical observations, we further explored the association of FGFR1 amplification with anti-estrogen resistance and sensitivity to the TORC1 inhibitor in ER+/FGFR1+ CAMA-1 breast cancer cells. CAMA-1 cells were relatively insensitive to single agent fulvestrant, the PI3Kα inhibitor alpelisib, the MEK½ inhibitor selumetinib, and the FGFR (TKI) lucitanib, but their growth was significantly inhibited by everolimus or paclitaxel (Fig. 4A–B, Supplementary Fig. 3).
Figure 4.
4A-B) Growth inhibition of ER+/FGFR1 amplified CAMA-1 breast cancer cells by various inhibitors. Representative images (A) and quantification (B) of integrated intensity are shown (**, P < 0.01 vs. control, t test).
4C) Pharmacodynamic impact on cell-signaling in ER+/FGFR1 amplified CAMA-1 breast cancer cells by various inhibitors. Immunoblot analysis with the indicated antibodies of lysates treated for 6 hours with vehicle, fulvestrant, everolimus, alpelisib, selumetinib, lucitanib, paclitaxel and the combination in 10% DMEM-FBS plus 2ng/mL FGF2.
4D) Growth inhibition of ER+ T47D cells with a FGFR1 expression vector by various inhibitors. Representative images (A) and quantification (B) of integrated intensity are shown (**, P < 0.01 vs. control, t test).
4E) Pharmacodynamic impact on cell-signaling in ER+ T47D cells with a FGFR1 expression vector by various inhibitors. Immunoblot analysis with the indicated antibodies of lysates treated for 6 hours with vehicle, fulvestrant, everolimus, alpelisib, selumetinib, lucitanib, paclitaxel and the combination in 10% DMEM-FBS plus 2ng/mL FGF2. PR mRNA levels from T47D cells with/without a FGFR1 expression vector were analyzed by qRT-PCR.
Mechanistically, treatment with lucitanib suppressed p-FRS2 and pERK consistent with impact on MAPK pathway, but surprisingly direct inhibition of MAPK pathway (by MEK inhibitor leading to suppressed pERK) paradoxically prompted upregulation of FGFR1 signaling activity, possibly via a compensatory feedback loop. Treatment with mTOR inhibitor (everolimus) suppressed both p-FRS2 and P-S6 consistent with impact on mTOR pathway (Fig. 4C).
To directly assess the contribution of FGFR1, we stably transduced ER+ T47D cells with an FGFR1 expression vector. T47D-FGFR1 cells exhibited higher levels of p-FRS2 and lower level of PR compared to T47DGFP control cells (Fig. 4D–E). As anticipated, FGFR1 expression was sufficient to confer resistance of T47D cells to fulvestrant and reducing sensitivity to everolimus. However, the combination of fulvestrant with either everolimus or with lucitanib overcame this resistance and attenuated growth of T47D-FGFR1 cells, as did treatment with paclitaxel (Fig. 4D–E).
Finally, we evaluated the impact of triplet combination therapy on ER+/FGFR1 amplified CAMA-1 breast cancer cells. The addition of everolimus had a marked impact on the arrest of the growth in response to fulvestrant plus palbociclib, but the addition of palbociclib had minimal effect on the arrest of the growth in response to fulvestrant plus everolimus (Supplementary Fig. 4A–C).
Discussion
We present a translational investigation of FGFR1 amplification in HR+ breast cancer. We report that in both clinical and pre-clinical investigations, breast malignancies harboring FGFR1 amplifications exhibit resistance to endocrine treatment alone or in combination with CDK 4/6 inhibitor, but remain relatively sensitive to endocrine therapy used in combination with a TORC inhibitor.
Our results support the previously reported association between FGFR1 amplification and luminal B type breast cancer, in that FGFR1+ patients were more likely to have aggressive disease that is PR negative, harbors p53 mutations, and lacks PIK3CA mutations.18 The connection between p53 mutation and FGFR1 amplification remains unclear, but has been demonstrated in other cancer types, including squamous cell carcinoma of the lung.19 We note that p53 mutations have been shown to correlate with a higher frequency of DNA copy number abnormalities, raising the possibility that mutations in p53 are permissive for gene amplifications such as FGFR1.20 Notably, p53 mutations did not have major prognostic value in our cohort—correlation between p53 mutations and time to progression on endocrine therapy did not reach significance in univariate analysis (13.1 vs. 9.7 months in TP53 mutant vs. WT patients respectively; Hazard Ratio .67, p= 0.2) despite having the same prevalence as FGFR1 amplifications. Because of this, p53 mutations were not included in multivariate analyses, and were not considered to be a true confounder in our cohort. Deeper investigation of the relationship between p53 mutations and FGFR1 amplifications is warranted.
With respect to PR expression, upregulation of growth factor pathways can cause downregulation of PR in vitro, and increased growth factor signaling is associated with decreased PR expression in vivo, supporting the hypothesis that PR negativity results from (and could be used as a surrogate marker for) the same growth factor signaling pathways that underlie resistance to endocrine therapy.21
Our preclinical data support the hypothesis that FGFR1 signals at least in part via an estrogen-receptor-independent pathway that involves MAPK/MEK/ERK signaling, as previously demonstrated by Turner et al. (Cancer Res 2010). However, we show that effective inhibition of either FGFR1 or its downstream targets (MEK½ in this case) is insufficient to inhibit growth of FGFR1 amplified cells in vitro. Notably, MEK inhibition prompted a compensatory upregulation of FGFR1 signaling activity in our model, a phenomenon that has been previously reported.22 These results resonate with several early clinical trials of monotherapy with FGFR inhibitors across multiple solid tumor subtypes, including breast, lung, bladder, head/neck, and cholangiocarcinoma, which demonstrated at best very modest antitumor activity when used as monotherapy.23
The addition of a TORC inhibitor to fulvestrant in vitro successfully inhibited cell growth more than any other modality besides chemotherapy. However, dampening proximal FGFR1 signaling does not seem to affect TORC activity, suggesting that TORC activation is not a simple downstream effect of FGFR1 signaling. It is not clear exactly why TORC inhibition is effective in inhibiting the growth of FGFR1 amplified cells, but other work has demonstrated the same phenomenon, raising the possibility that FGFR1 amplification itself underlies TORC pathway dependence that could be broadly targetable.24 While the mechanism behind this connection remains elusive, it appears that neither downstream suppression of the MAPK pathway nor upstream suppression of the PI3K/AKT pathway is sufficient to overcome FGFR1-associated cell growth and division. This insinuates the existence of compensatory cross-talk between the two known growth factor signaling pathways serving FGFR1, which is yet to be characterized and warrants additional evaluation.
Our clinical data provides evidence that FGFR1-amplified tumors have shorter time to progression with various endocrine-based therapies, including combination with CDK 4/6 inhibitors. This finding was also observed in retrospective biomarker analysis of the MONALEESA-2 trial where FGFR1+ patients who were treated with ribociclib (an inhibitor of CDK4/6) and letrozole had significantly shorter progression free survival (PFS) than FGFR1 WT patients (10.6 vs. 24.8 months; p= 0.075).25 In addition, our clinical data is consistent with the retrospective biomarker analysis of the landmark BOLERO-2 trial, which revealed that the addition of everolimus to endocrine therapy improved PFS in all patients, including those with FGFR1 amplification (PFS = 6.8 vs 2.8 months; Hazard Ratio = 0.39), highlighting that FGFR1 amplification does not result in clinical resistance to everolimus based therapy.26
Importantly, we do not claim that patients with FGFR1-amplified tumors are more (or less) sensitive to everolimus than patients with cancer without FGFR1 amplification. Instead, our findings imply that in FGFR1-amplified breast cancers, endocrine therapy in combination with TORC1 inhibition may be still be active and warrants further evaluation.
No study has prospectively compared endocrine therapy plus everolimus against endocrine therapy plus an inhibitor of CDK4/6 in breast cancer, let alone in FGFR1+ patients. Our preclinical and clinical data collectively suggest that FGFR1 amplification is associated broadly with resistance to various therapies, but still retains relative sensitivity to mTOR inhibition. Our translational results complement recent publications on role of FGFR1 amplification in mediating endocrine therapy resistance, including combination with CDK 4/6 inhibitors,26,27,28 and highlight that FGFR1 amplification could be used to guide rational genotype-based decision-making in HR+ MBC. Further prospective studies are needed to confirm these results and develop more effective therapeutic strategies for patients with FGFR1+ HR+/HER2− MBC.
Supplementary Material
Supplemental Figure 1.
Supp. 1A) Prevalence of mutations in FGFR1+ vs. FGFR1− patients by SNAPSHOT with associated p-values.
Supp. 1B) Co-existing mutations in FGFR1-Amplified patients as detected by targeted genotyping (SNAPSHOT). Mutations are denoted by red boxes.
Supplemental Figure 2.
Supp. 2A) Association between Time to Progression (TTP) on Endocrine Therapy alone with FGFR1 amplification in multivariate analysis adjusting for known prognostic variables in metastatic HR+/HER2− breast cancer.
Supp. 2B) Association between Time to Progression (TTP) on endocrine and mTOR inhibitor combination with FGFR1 amplification in multivariate analysis adjusting for known prognostic variables in metastatic HR+/HER2− breast cancer.
Supplemental Figure 3.
Average of the fold change in 2D growth with increasing combination doses of fulvestrant, selumetinib, alpelisib, lucitanib (in all cases 0 to 1000nM) and everolimus (0 to 500 nM) relative to untreated controls from three independent experiments.
Supplemental Figure 4.
Supp. 3A-B) Growth inhibition of ER+/FGFR1 amplified CAMA-1 breast cancer cells by triplet combinations of targeted therapies.
Supp. 3C) Pharmacodynamic impact on cell-signaling in ER+/FGFR1 amplified CAMA-1 breast cancer cells by triplet combinations of targeted therapies.
STATEMENT OF SIGNIFICANCE:
This work elucidates the clinical and functional role of FGFR1 amplification in hormone-receptor positive breast cancer, and provides early evidence that FGFR1 amplification mediates resistance to endocrine therapy, but not TORC inhibition.
Acknowledgements:
We thank all the patients who participated in this study. We are grateful to Massachusetts General Hospital (MGH) nurses for their help with the study. This work was supported by Susan G Komen grant CCR15224703 (A.B), K12 5K12CA087723 (A.B.), KL2 TR001100 (L.M.S) and ASCO Young Investigator Award (L.M.S).
Footnotes
Conflict of Interest disclosure:
DJ: Consultant/advisory board: Novartis, Genentech, Eisai, Ipsen, EMD Serono.
LF: Grant Support: Lilly.
LS: Consultant/advisory board: Novartis.
DS: Ownership interests and Intellectual property rights/inventor/patent holder: Biotheronostics.
SJI: Consultant/advisory board: Abbvie, PharaMar, Genentech/Roche, Myriad Genetics, Hengrui Therapuetics, Puma Biotech, Immunomedics.
BM: Consultant/advisory board: MOTUS GI (spouse).
AJI: Consultant/advisory board: Roche, Chugai, Constellation, Pfizer. Ownership interests and Intellectual property rights/inventor/patent holder: ArcherDx.
CLA: Stock options: Provista, Y-Trap; Advisory Roles: Symphogen, Daiichi Sankyo, Taiho Oncology, PUMA Biotechnology, Novartis, Merck, Lilly, Radius, Sanofi, H3Biomedicine, OrigiMed. Grant Support: Pfizer, Lilly, Radius.
AB: Consultant/advisory board: Novartis, Pfizer, Genentech/Roche, Radius Health, Merck, Spectrum pharma, Immunomedics, Taiho Oncology, Sanofi, Diiachi, Puma. Research Grant (self): Biothernostics. Research Grant (institution): Novartis, Pfizer, Genentech/Roche, Radius Health, Merck, Immunomedics, Sanofi.
References
- 1.Harvey Jennet M., et al. “Estrogen receptor status by immunohistochemistry is superior to the ligand-binding assay for predicting response to adjuvant endocrine therapy in breast cancer.” Journal of clinical oncology 175 (1999): 1474–1474. [DOI] [PubMed] [Google Scholar]
- 2.Clarke Robert, Tyson John J., and Dixon J. Michael. “Endocrine resistance in breast cancer–an overview and update.” Molecular and cellular Endocrinology 418 (2015): 220–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Osborne C. Kent, and Schiff Rachel. “Mechanisms of endocrine resistance in breast cancer.” Annual review of medicine 62 (2011): 233–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Musgrove Elizabeth A., and Sutherland Robert L.. “Biological determinants of endocrine resistance in breast cancer.” Nature reviews. Cancer 99 (2009): 631. [DOI] [PubMed] [Google Scholar]
- 5.Johnson Daniel E., and Williams Lewis T.. “Structural and functional diversity in the FGF receptor multigene family.” Advances in cancer research 60 (1992): 1–41. [DOI] [PubMed] [Google Scholar]
- 6.Chang Jinjia, et al. “Prognostic value of FGFR gene amplification in patients with different types of cancer: a systematic review and meta-analysis.” PloS one 98 (2014): e105524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babina Irina S., and Turner Nicholas C.. “Advances and challenges in targeting FGFR signaling in cancer.” Nature Reviews Cancer 175 (2017): 318–332. [DOI] [PubMed] [Google Scholar]
- 8.André Fabrice, and Cortés Javier. “Rationale for targeting fibroblast growth factor receptor signaling in breast cancer.” Breast cancer research and treatment 1501 (2015): 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacquemier Jocelyne, et al. “Expression of the FGFR1 gene in human breast‐carcinoma cells.” International journal of cancer 593 (1994): 373–378. [DOI] [PubMed] [Google Scholar]
- 10.Courjal Frank, et al. “Mapping of DNA amplifications at 15 chromosomal localizations in 1875 breast tumors: definition of phenotypic groups.” Cancer research 5719 (1997): 4360–4367. [PubMed] [Google Scholar]
- 11.Xian Wa, et al. “Pleiotropic effects of FGFR1 on cell proliferation, survival, and migration in a 3D mammary epithelial cell model.” The Journal of cell biology 1714 (2005): 663–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suyama Kimita, et al. “A signaling pathway leading to metastasis is controlled by N-cadherin and the FGF receptor.” Cancer cell 24 (2002): 301–314. [DOI] [PubMed] [Google Scholar]
- 13.Haugsten Ellen Margrethe, et al. “Roles of fibroblast growth factor receptors in carcinogenesis.” Molecular Cancer Research 811 (2010): 1439–1452. [DOI] [PubMed] [Google Scholar]
- 14.Wang Wei, et al. “A Versatile Tumor Gene Deletion System Reveals a Crucial Role for FGFR1 in Breast Cancer Metastasis.” Neoplasia 195 (2017): 421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reis-Filho Jorge Sergio, et al. “FGFR1 emerges as a potential therapeutic target for lobular breast carcinomas.” Clinical Cancer Research 1222 (2006): 6652–6662. [DOI] [PubMed] [Google Scholar]
- 16.Turner Nicholas, et al. “FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer.” Cancer research (2010): 0008–5472. [DOI] [PMC free article] [PubMed]
- 17.André F, et al. Targeting FGFR with dovitinib (TKI258): preclinical and clinical data in breast cancer. Clin Cancer Res 2013. July 1;19(13):3693–702. [DOI] [PubMed] [Google Scholar]
- 18.Ades Felipe, et al. “Luminal B breast cancer: molecular characterization, clinical management, and future perspectives.” Journal of Clinical Oncology 3225 (2014): 2794–2803. [DOI] [PubMed] [Google Scholar]
- 19.Heist Rebecca S., et al. “FGFR1 amplification in squamous cell carcinoma of the lung.” Journal of thoracic oncology 712 (2012): 1775–1780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jain Ajay N., et al. “Quantitative analysis of chromosomal CGH in human breast tumors associates copy number abnormalities with p53 status and patient survival.” Proceedings of the National Academy of Sciences 9814 (2001): 7952–7957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arpino Grazia, et al. “Estrogen receptor–positive, progesterone receptor–negative breast cancer: association with growth factor receptor expression and tamoxifen resistance.” Journal of the National Cancer Institute 9717 (2005): 1254–1261. [DOI] [PubMed] [Google Scholar]
- 22.Yang X, Webster JB, Kovalenko D, Nadeau RJ, Zubanova O, Chen PY, Friesel R. Sprouty genes are expressed in osteoblasts and inhibit fibroblast growth factor-mediated osteoblast responses. Calcif Tissue Int 2006;78(4):233–240. [DOI] [PubMed] [Google Scholar]
- 23.Schram Alison M., Voss Martin H., and Hyman David M.. “Genome-Driven Paradigm for the Development of Selective Fibroblast Growth Factor Receptor Inhibitors” (2016): 131–134. [DOI] [PubMed]
- 24.Singleton Katherine R., et al. “KinomeRNAi screens reveal synergistic targeting of MTOR and FGFR1 pathways for treatment of lung cancer and HNSCC.” Cancer research 7520 (2015): 4398–4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formisano Luigi, et al. “Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer.” Nature communications 101 (2019): 1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hortobagyi Gabriel N., et al. “Correlative analysis of genetic alterations and everolimus benefit in hormone receptor–positive, human epidermal growth factor receptor 2–negative advanced breast cancer: results from BOLERO-2.” Journal of Clinical Oncology 345 (2016): 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Razavi P, et al. The Genomic Landscape of Endocrine-Resistant Advanced Breast Cancers. Cancer Cell 2018. September 10;34(3):427–438.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Formisano L, et al. Nat Commun. Aberrant FGFR signaling mediates resistance to CDK4/6 inhibitors in ER+ breast cancer 2019. March 26;10(1):1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng Z, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med December;20(12):1479–84. [DOI] [PubMed] [Google Scholar]
- 30.Lanman RB, et al. Analytical and clinical validation of a digital sequencing panel for quantitative, highly accurate evaluation of cell-free circulating tumor DNA. PLoS ONE 2015;10:e0140712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1.
Supp. 1A) Prevalence of mutations in FGFR1+ vs. FGFR1− patients by SNAPSHOT with associated p-values.
Supp. 1B) Co-existing mutations in FGFR1-Amplified patients as detected by targeted genotyping (SNAPSHOT). Mutations are denoted by red boxes.
Supplemental Figure 2.
Supp. 2A) Association between Time to Progression (TTP) on Endocrine Therapy alone with FGFR1 amplification in multivariate analysis adjusting for known prognostic variables in metastatic HR+/HER2− breast cancer.
Supp. 2B) Association between Time to Progression (TTP) on endocrine and mTOR inhibitor combination with FGFR1 amplification in multivariate analysis adjusting for known prognostic variables in metastatic HR+/HER2− breast cancer.
Supplemental Figure 3.
Average of the fold change in 2D growth with increasing combination doses of fulvestrant, selumetinib, alpelisib, lucitanib (in all cases 0 to 1000nM) and everolimus (0 to 500 nM) relative to untreated controls from three independent experiments.
Supplemental Figure 4.
Supp. 3A-B) Growth inhibition of ER+/FGFR1 amplified CAMA-1 breast cancer cells by triplet combinations of targeted therapies.
Supp. 3C) Pharmacodynamic impact on cell-signaling in ER+/FGFR1 amplified CAMA-1 breast cancer cells by triplet combinations of targeted therapies.