Abstract
Major advances in our understanding of cancer pathogenesis and therapy have come from efforts to catalog genomic alterations in cancer. A growing number of large-scale genomic studies have uncovered mutations that drive cancer by perturbing co- and post-transcriptional regulation of gene expression. These include alterations that impact each phase of RNA processing, including splicing, transport, editing, and decay of messenger RNA. The discovery of these events illuminates a number of novel therapeutic vulnerabilities generated by aberrant RNA processing in cancer, several of which have progressed to clinical development.
Keywords: Polyadenylation, PRMT1, PRMT5, RBM10, RBM39, RNA, Splicing, SF3B1, SRSF2, Sulfonamides, U2AF1, ZRSR2
INTRODUCTION
The advent of high-throughput transcriptome sequencing (RNA sequencing; RNA-seq) has provided a wealth of information on RNA splicing on a genome-wide scale. It is now understood from RNA-seq that >95% of human genes are subject to alternative splicing (AS), the enzymatic process by which a single gene has the potential to produce multiple, potentially functionally distinct pre-mRNA and protein isoforms. Splicing is considered to be a major mediator of proteome diversity through its ability to generate multiple transcripts with differing amino acid sequences from a single gene. Moreover, due to the link between splicing and nonsense-mediated mRNA decay (NMD), splicing also provides a means to regulate gene expression.
Systematic application of RNA-seq to an ever-expanding number of human tumors and matched normal tissues has now identified numerous means by which RNA processing is dysregulated in cancer. In this review we focus on how RNA splicing and polyadenylation are altered in cancer and functionally drive cancer initiation and maintenance. This is notably only a subset of the means by which RNA processing goes awry in cancer. Cancer-associated changes in RNA editing, RNA modifications, and expression of non-coding RNA species including micro-RNAs and long non-coding RNAs are covered in recent excellent publications.(1–4) Finally, motivated by recent data illustrating individual splicing alterations as well as mutations in RNA splicing factors as therapeutic vulnerabilities in cancer, we discuss ongoing and future efforts to target RNA splicing for cancer therapy.
BASIC MECHANISMS OF RNA SPLICING CATALYSIS
RNA splicing is a nuclear enzymatic process accomplished by a macromolecular machine composed of a large constellation of RNA binding proteins (RBPs) and additional splicing proteins combined with five small nuclear RNAs (snRNAs) in complexes known as small nuclear ribonucleoproteins (snRNPs) (reviewed recently (5–7)). snRNAs base pair with sequences within pre-mRNA that are critical in delineating exons from introns. These include the dinucleotides at the first two and last two positions of an intron (known as the 5’ and 3’ splice sites (ss), respectively) and a poorly conserved sequence within the intron known as the branchpoint (Figure 1A). The branchpoint is typically close to the 3’ ss and is usually an adenosine nucleotide. A stretch of pyrimidine nucleotides (the polypyrimidine tract) is often adjacent to the branchpoint and promotes the spliceosome’s recognition of the branchpoint. Deletion or mutation of these intronic sequences typically greatly abrogates splicing usage at these sites. In addition, sequences throughout the exon and intron, referred to as splicing enhancers and silencers, are bound by RBPs that recruit or repel spliceosome assembly to promote or inhibit splicing, respectively.
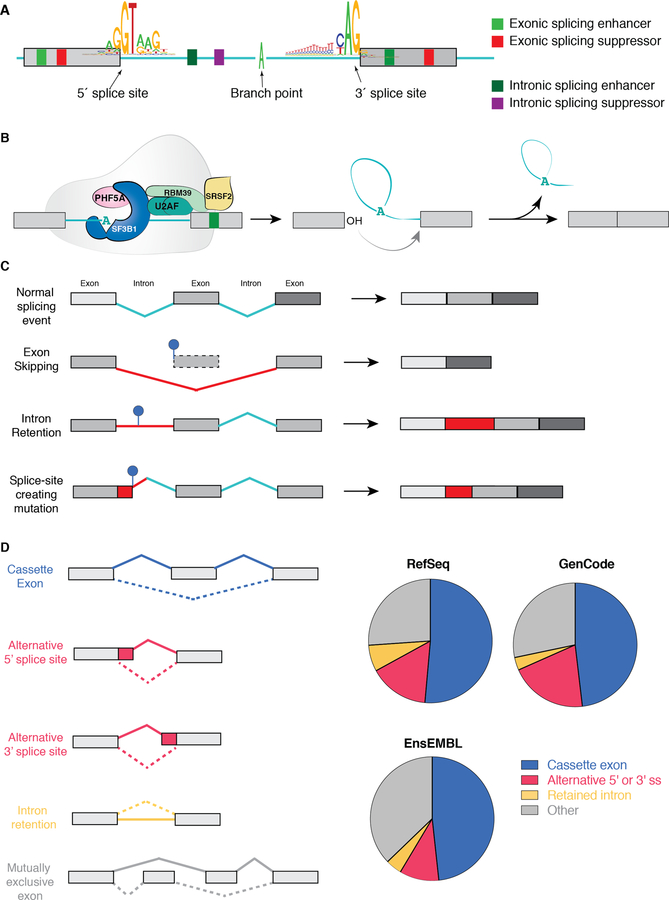
Figure 1. Fundamentals of RNA splicing and how mutations within genes alter splicing in cis.
(A) Diagram of an intron and two flanking exons with consensus sequences defining the 5’ splice site (ss), branchpoint, and 3’ ss. Colored boxes depict sequences within exons and introns that increase or decrease the likelihood of splice site recognition by RNA binding proteins (splicing enhancers or repressors, respectively). (B) Diagram of the SF3b complex of the spliceosome (which contains SF3B1), associated RNA binding proteins (the U2AF heterodimer and an accessory splicing factor RBM39) and the sequential reactions involved in removal of an intron (intron shown in teal and exons in grey). As shown, the SF3b complex is involved in recognizing the branchpoint (shown here as an adenosine nucleotide (“A”)) and is recruited to this site by the U2AF complex, which recognizes sequences at the 3’ ss. During splicing catalysis, the branchpoint A carries out a nucleophilic attack at the 5’ ss, forming a lariat, and then the 3’OH of the released 5’ exon performs a second nucleophilic attack at the last nucleotide of the intron at the 3’ ss, joining the exons and releasing the intron lariat. (C) Diagram of how single nucleotide variants near splice sites, throughout an exon, and deep within introns may disrupt splicing or generate novel aberrant splice sites in the mRNA of a gene in cis. (D) Pie charts depicting distribution of each category of splicing event shown on the left based on annotations of the human genome from RefSeq, GenCode, and EnsEMBL.(130) “Other” represents complex splicing events (>1 of the five categories found simultaneously) as well as the small proportion of splicing events represented by mutually exclusive exons.
The enzymatic process of splicing consists of two sequential transesterification reactions (Figure 1B). In the first step, the 2’-hydroxyl group of the branch point performs a nucleophilic attack on the phosphorous atom of the 5’ ss to generate a linear left exon and a right intron-exon branched sequence known as a lariat. In the second step, the 2’-hydroxyl group of the linear left exon attacks the phosphorous atom of the 3’ ss thereby concatenating two exons. An excised intron is simultaneously released within the lariat and later degraded by RNA debranching enzymes.
Over the last four years, there have been major advances in the mechanistic understanding of spliceosome function with the publication of at least 18 independent structures of the spliceosome at a resolution of 3.3–9.9 Å (reviewed recently (8–10)). While these initial efforts were mostly carried out using cryoelectron microscopy (cryo-EM) of yeast spliceosomes, initial structures of the human spliceosome followed in 2017. It is almost certain that high-resolution structures of the human spliceosome in each step of the splicing reaction will be elucidated in the very near future.
ALTERATIONS in RNA SPLICING in CANCER
Splicing changes in cancer relative to normal tissues
Prior to the advent of RNA-seq, evaluation of expressed sequence tag (EST) libraries from human cancer cells and other tissue types revealed that cancer cells have an elevated rate of stop codons relative to non-cancer cells.(11) These data suggested increased mis-splicing in cancer transcriptomes relative to normal tissues; the basis for which was not entirely clear. Over the last 10 years, however, a wealth of DNA and RNA-sequencing data from tumors and paired normal tissues as part of The Cancer Genome Atlas (TCGA) effort as well as distinct normal human tissues from the GTEx (Genotype-Tissue Expression) Program, have enabled systematic interrogation of splicing across cancers and evaluation for a potential genetic basis for splicing alterations in cancer. The largest of such studies, recently published by Kahles et al., reanalyzed whole exome sequencing (WES) and RNA-seq data across 32 TCGA cancer types from 8,705 patients.(12) This effort showed that tumor samples, on average, harbor ~20% more AS events than normal tissues. As such, many splicing changes identified in cancer cells can be linked to single-nucleotide variants (SNVs) within that gene that disrupt mRNA sequences required for normal splicing (Figure 1C). Heterozygous cis-acting splicing alterations act in an allele-specific manner such that the mutant allele shows abnormal splicing whereas the wild-type allele supports normal splicing. Interestingly, prior work from Supek et al. and Jung et al. found that SNVs resulting in frameshifts in mRNA sequence due to intron retention (IR) occur most commonly in tumor suppressors while sparing oncogenes.(13, 14) In contrast, in-frame exonic SNVs disrupting exon usage are most commonly enriched in oncogenes.
While most studies evaluating the effects of SNVs on splicing have focused on mutations abolishing splice sites, recent reanalysis of 8,000 tumor samples from 33 TCGA cancer types by Jayasinghe et al. identified many examples of mutations creating novel splice sites (Figure 1C).(15) These splice site-creating mutations impact many known cancer genes. Intriguingly, tumors bearing these mutations also appeared to be associated with increased expression of T-cell associated genes and mRNAs encoding the immune checkpoint blockade molecules PD-1 and PD-L1. These data suggest the potential for splicing to generate novel, immunogenic peptides (a topic discussed later in this review).
It is important to note that the most studies evaluating the impact of cis-acting mutations on splicing have utilized WES data and focused on mutations that occur at 5’ or 3’ exon-intron boundaries. However, in vitro screening of sequence variants across every nucleotide within model exons has shown that >50% of nucleotide substitutions in exons can induce splicing changes with similar effects expected from coding and synonymous (or “silent”) mutations.(16, 17) Consistent with this, analysis of >3,000 cancer exomes and >300 cancer genomes by Supek et al. identified recurrent selection for synonymous mutations in cancer, the majority of which impacted RNA splicing and occurred in oncogenes.(14) Notably, many regulatory elements required for ss recognition occur deep within introns (at sites including the branchpoint, polypyrimidine tract, and intronic splicing enhancers and silencers) and longer introns are more prone to splicing errors.(18) Thus, increased use of whole-genome sequencing paired with RNA-seq of the same tumor samples will be important in improving our knowledge of how mutations in these noncoding regions impact expression of protein-coding isoforms through splicing.
There have been a variety of efforts to quantify the distribution of splicing changes in cancer cells (Figure 1D). At least one report found no biases in recognition of cassette exons or 5’ or 3’ ss globally in cancer but almost all cancer types exhibit widespread increased IR.(19) While these findings are intriguing, they have not been uniformly replicated using alternative methods of splicing analyses.(20) It is important to note that the detection and quantification of splicing changes from RNA-seq data remains a formidable challenge for several reasons. First, while numerous informatics tools have been described to identify and quantitate splicing (reviewed recently (20–22)), no single bioinformatics method is uniformly used and these tools vary widely in their requirement for prior transcriptome annotation. Clearly, reliance on preannotated libraries of known transcripts limits detection of novel, unannotated splicing events. In addition, existing analysis tools for detecting splicing changes vary in their false positive rate for detection of certain splicing events and ability to detect complicated splicing events (consisting of multiple simultaneous events from those shown in Figure 1C).(20, 22) For example, there are many examples where use of alternative exons residing within an intron can be mistaken for IR.(20) Discrepancies in the description of global RNA splicing changes in cancer highlight the challenges of inferring splicing changes from short-read RNA-seq data. Future efforts applying long-read RNA-seq methodologies to capture data from entire mRNA transcript isoforms and methods of RNA-seq analyses which do not rely on pre-annotation of splicing isoforms will be crucial in further illuminating our understanding of altered splicing in cancer.
Mutations in RNA splicing factors in cancer
While the above data cataloging how mutations within genes impact their own splicing in cis elucidated important roles for splicing in cancer, the discovery of recurrent mutations in components of the RNA splicing machinery in 2011 further highlighted the importance of aberrant splicing in cancer.(23) Currently, hotspot, heterozygous mutations in the RNA splicing factors SF3B1, SRSF2, and U2AF1 are known to be enriched throughout myeloid leukemias, chronic lymphocytic leukemia (CLL), uveal (UVM) and mucosal melanomas (Figure 2A–B).(24–30) A recent study surveying mutations in 404 genes encoding splicing factors from 33 cancer types in the TCGA from investigators at H3 Biomedicine and the TCGA identified putative driver mutations in 119 splicing factor genes.(31) These include a number of additional splicing factors affected by recurrent hotspot mutations (PHF5A, PCBP1, and HNRNPCL1) as well as a larger number of splicing factors impacted by loss-of-function mutations (RBM10, FUBP1). While the overall mutational frequency in RNA splicing factors is low across unselected solid cancer types, these mutations recur at rates higher than expected by chance in the melanoma subtypes noted above in addition to lung adenocarcinoma (LUAD) and bladder cancer (BLCA).
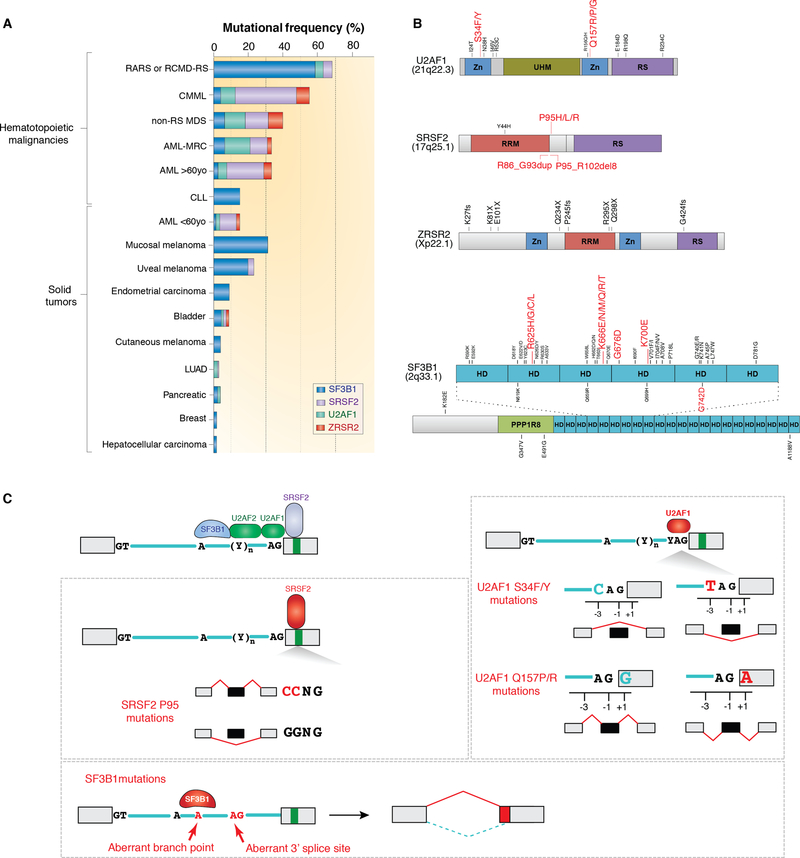
Figure 2. Frequency, location, and global impact on RNA splicing of recurrently mutated splicing factors in cancer.
(A) Histogram depicting frequency of mutations in SF3B1, SRSF2, U2AF, and ZRSR2 in hematopoietic malignancies and solid tumors. In addition to these four genes (which are the most frequently mutated in hematopoietic malignancies), a host of additional splicing factors affected by hotspot as well as presumed loss-of-function mutations are also mutated in cancer and not shown here. Abbreviations: RARS (refractory anemia with ring sideroblasts), RCMD-RS (refractory cytopenias with multilineage dysplasia and ring sideroblasts), MDS (myelodysplastic syndromes), CMML (chronic myelomonocytic leukemia), AML-MRC (acute myeloid leukemia with myelodysplasia related changes), CLL (chronic lymphocytic leukemia), LUAD (lung adenocarcinoma). (B) Protein diagrams of the four mutated splicing factors shown in (A) with location of mutant residues. Hotspot mutations are shown in red. Abbreviations: HD (HEAT repeat domain), Zn (Zinc finger), RRM (RNA recognition motif), RS (Serine/Arginine rich domain), UHM (U2AF homology motif). (C) Diagram of an intron, two flanking exons, and locations in RNA where the four factors from (A) bind (top left). Mutations in SRSF2 skew the binding avidity of SRSF2 such that mutants bind C-rich sequences more avidly to promote exon splicing while reducing binding affinity for G rich sequences (middle left). SF3B1 is responsible for recognition of the branchpoint. Mutations in SF3B1 cause recognition of an aberrant branchpoint leading to intron proximal alternative 3’ ss selection (bottom left). Finally, U2AF1 is responsible for recognition of the 3’ yAG|r dinucleotide (where “y” represents the C- or T- pyrimidine nucleotide immediately intronic to the AG and r represents the first nucleotide in the downstream exon (at the +1 position)). As shown on the right, U2AF1 S34F/Y mutations favor inclusion of cassette exons bearing a 3’ ss containing a C-nucleotide at the −3 position while Q157 mutants promote splicing of exons with G nucleotides at the +1 position. Panels A and B adapted from Dvinge et al., RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016 Jul;16(7):413–30 and used with permission.
The most commonly mutated splicing factor across all cancers is SF3B1. The initial discovery of mutations in SF3B1 was surprising and unexpected for a variety of reasons. First, this was the first example of mutations in a splicing factor positively selected for in clonal disorders. Second, mutations in SF3B1 have an exquisite and intriguing enrichment in otherwise unrelated cancer types including myelodysplastic syndromes (MDS), CLL, and UVM. Finally, within each of these disorders, SF3B1 had immediate relevance for disease diagnosis and/or prognostication. For example, SF3B1 mutations in MDS are present in >90% of patients with a subtype of MDS known as refractory anemia with ring sideroblasts (RARS) characterized by anemia, the finding of iron-laden mitochondria surrounding the nuclei of erythroid precursors (“ring sideroblasts”) jn the bone marrow, and an overall favorable prognosis.(23, 24) This form of MDS has been morphologically recognized for decades but a genetic basis was unknown until the discovery of SF3B1 mutations. In fact, SF3B1 mutations are now part of the diagnostic criteria for RARS, as SF3B1 mutations have >97% positive predictive value for patients suspected to have this form of MDS.(32)
Within MDS and other myeloid malignancies, SF3B1 mutations are present in the predominant clone and are selected early in the disease process. In contrast, SF3B1 mutations in CLL are most commonly subclonal and enriched in patients with more advanced and aggressive disease.(26, 33) SF3B1 mutations are present in 10–20% of patients refractory to chemotherapeutic agents such as fludarabine in contrast to <5% of patients with untreated CLL or the CLL precursor, monoclonal B-lymphocytosis (MBL, reviewed previously(34)). SF3B1 mutations are also more common in CLL patients with an unmated immunoglobulin heavy-chain variable region (IGHV) gene (a well-established adverse prognostic factor in this disease), and are enriched in CLL patients requiring therapeutic intervention over those not requiring therapy. Although SF3B1 mutations have been associated with adverse response to chemoimmunotherapy regimens in CLL, how SF3B1 mutations impact response to the many recently approved therapies in CLL such as ibrutinib, venetoclax, and phosphoinositide 3-kinase inhibitors is not clear.
In the setting of UVM, SF3B1 mutations occur in 10–21% of patients and are associated with specific favorable prognostic features including disomy 3 and lower age at diagnosis.(27–29) The impact of SF3B1 mutations in this specific subset of UVM patients has been conflicting, with some reports suggesting SF3B1 mutations are associated with an increased risk of metastasis amongst disomy 3 patients.
SF3B1 is a member of the U2 snRNP complex, where it physically associates with p14, PHF5A, SF3B3, and the U2 sRNA (Figure 1B). The U2 snRNP complex is important in recognizing the branchpoint within the intron. Consonant with this, global surveys of bulk RNA-seq data from cancer cells and mouse models bearing mutations in SF3B1 at the most commonly mutated residues in SF3B1, including the SF3B1 K700E and K666N mutations, reveal that SF3B1-mutant cells exhibit use of an aberrant intron-proximal 3’ ss (Figure 2C).(35–37) Further work by Darman et al., Tang et al., and Carrocci et al. have shown that this change in 3’ ss occurs due to reduced fidelity of branchpoint selection in cells expressing mutant SF3B1.(38–40) Each of the mutations in SF3B1 occurs within the 4th–7th HEAT repeat domains (Figure 2B). Recent mapping of cancer-associated mutations in SF3B1 onto the crystal structure of human SF3B1 by Cretu et al. suggest that SF3B1 mutations alter the conformation of the HEAT repeat domains and/or interactions with U2AF2 or the SF3b complex protein p14.(41) Ultimately these changes would modify how the U2 complex interacts with the branchpoint. As noted above, SF3B1 is affected by a number of distinct hotspot mutations, many of which are associated with specific lineages of cancer. The most common individual hospot mutation in SF3B1 is the SF3B1 K700E mutation, which is present across myeloid malignancies, CLL, and many solid tumors including breast cancer, pancreatic ductal adenocarcinoma and others. In addition, there are several mutated residues in SF3B1 that appear to be cancer-specific. These include SF3B1 R625 mutations, enriched in melanomas, SF3B1 E902 mutations, exclusive to bladder cancer, and SF3B1 G742 mutations, most enriched in CLL. The functional basis for the lineage specificity of individual hotspot mutations in SF3B1 is currently unknown. It is possible that each mutation is associated with distinct mis-splicing events that may be important in transforming specific tissue types, giving rise to histologically distinct cancers. For example, the bladder-cancer specific E902 hotspot mutation is not associated with the same intron-proximal 3’ ss usage as other SF3B1 mutational hotspots.(31) However, the mechanistic basis for this observation or how it relates to bladder cancer pathogenesis is unclear. Finally, it is important to note that lineage-specificity of mutations could also occur due to the potential for tissue-specific nucleotide mutability, gene mutation rates, and/or expression of interacting proteins.
Although SF3B1 is the most frequently mutated splicing factor across cancers, more has been learned about how hotspot mutations in the splicing factors U2AF1 and SRSF2 impact splicing and disease development. U2AF1 is an RBP and part of a heterodimeric U2AF complex with its partner, U2AF2, that serves to recruit the U2 snRNP to the branchpoint (Figure 2C). U2AF1 binds the AG dinucleotide at the 3’ ss while U2AF2 binds the polypyrimidine tract (Figure 2C). U2AF1 is affected by hotspot mutations at S34 and Q157 residues, each of which occur in one of its two zinc fingers (Figure 2B). Similar to the allele-specificity of mutations in SF3B1, each mutant residue of U2AF1 is associated with specific subtypes of cancer. For example, S34 mutations are recurrent in LUAD whereas Q157 mutations are not present in this disease.(42) Interestingly, work by Ilagan et al. and others showed that aberrant splicing driven by mutant U2AF1 occurs in an allele-specific manner at sequences surrounding the AG dinucleotide at the 3’ ss.(43) U2AF1 mutants skew normal splicing activity of U2AF such that U2AF1 S34 mutants promote exon inclusion at the 3’ ss when a C-nucleotide is located at the −3 position, while Q157 mutants promote exon inclusion when a G-nucleotide is in the +1 position (Figure 2C).
Unlike SF3B1 and U2AF1, which are core components of the spliceosome, SRSF2 is an auxiliary splicing factor, that is a member of the serine/arginine-rich (SR) family of RBPs that binds splicing enhancers to recruit the core spliceosome to promote exon splicing. In its wild-type state, SRSF2 physically binds CCNG and GGNG sequences within RNA equally well to promote splicing.(44) However, mutations in SRSF2, which occur as point mutations or in-frame deletions at Proline 95 (Figure 2B), skew this binding preference such that the mutant proteins promotes RNA binding and splicing at C-rich sequences while having reduced affinity for G-rich sequences (Figure 2C).(45, 46)
Despite the consistency of these findings that sequence-specific aberrant splicing changes are associated with point mutations in SF3B1, U2AF1, and SRSF2, to date there have been very few examples linking splicing alterations induced by mutant splicing factor mutations to functional consequences in a tumor model (see EZH2 aberrant splicing described below). These challenges likely relate to the lack of uniformity in methods for the detection of splicing alterations by RNA-seq as well as inherent limitations of certain splicing analysis tools noted earlier. Even once the full spectrum of reliable splicing changes induced by spliceosome gene mutations are identified, requisite functional studies to systematically understand the functional impact of individual splicing changes on protein expression and function, tumorigenesis, and cell differentiation are needed. In addition to evaluating the effect of aberrant splicing on the expression of individual proteins, it will also be critically important to systematically explore the effects of cancer-associated changes in splicing on the proteome. Such efforts would help elucidate the extent to which alterations in splicing result in downregulation of canonical protein abundance due either to the expression of nonsense mediated decay (NMD)-inducing transcripts or aberrant protein isoforms from novel splicing events.
In addition to SF3B1, SRSF2, and U2AF1, presumed loss-of-function mutations in a fourth RNA splicing factor, ZRSR2, also occur in myeloid malignancies (Figure 2B). Interestingly, ZRSR2 is the only protein of the four frequently mutated splicing factors in leukemias that primarily functions in the minor spliceosome. While most introns are spliced by the major spliceosome (so called “U2-type introns”), a small subset (<1%) of introns have distinct 5’ and 3’ splice sites which are recognized by a separate splicing complex known as the minor spliceosome. (47, 48) Mutations in ZRSR2, encoded on the X chromosome have a male predominance in MDS. ZRSR2 mutations occur throughout its open reading frame suggesting that mutations in ZRSR2 confer loss-of-function. Consequently, ZRSR2 deficient leukemia cells have been shown to have global increases in minor (or “U12-type”) intron retention. However, how these splicing changes in ZRSR2-mutant cells relate to those in cells bearing mutations in the other RNA splicing factors is currently unclear.(49) Likewise, which ZRSR2-regulated splicing events are important for myeloid leukemia pathogenesis, if any, are unknown.
How do mutations in splicing factors confer a cellular advantage?
The fact that the splicing factors mutated in cancer play essential roles in splicing catalysis and the mutations are clearly associated with global alterations in RNA splicing highlight the potential that aberrant splicing of specific transcripts is responsible for disease development. However, it is still unclear whether mis-splicing of hundreds of distinct mRNAs drives clonal selection as opposed to one or handfuls of individual mis-splicing events. Interestingly several recent studies from subjects with age-related clonal hematopoiesis, a condition associated with a risk of developing MDS and AML, have shown that mutations in the RNA splicing factors SRSF2 and U2AF1 are highly predictive for the eventual development of overt myeloid malignancies.(50, 51) These data, in combination with selection for specific mutant residues in these factors, further underscores the concept that mutant splicing factors confer clonal advantage to cells. Despite the strength of these human genetic data, currently there is very little evidence from experimental models that spliceosomal gene mutations confer a cellular benefit or are required for disease maintenance. For example, several studies in human cancer cell lines bearing naturally occurring mutations in RNA splicing factors have demonstrated that eradication of mutant U2AF1 or SF3B1 has no impact on cell growth.(52, 53) These data suggest the possibility that RNA splicing factor mutations may be required for cancer initiation while being dispensable for tumor maintenance. At the same time, expression of mutations in RNA splicing factors in conditional knockin mice has very consistently been associated with impaired self-renewal.(37, 45, 54) However, these data do not exclude the possibility that expression of mutant splicing factors may confer an advantage in specific genetic contexts or in response to certain non-cell autonomous stressors or microenvironmental stimuli that are not yet appreciated. To this end, it is important to note that mutations in RNA splicing factors have non-random mutational co-occurrences that may be important in understanding the role of mutant splicing factors in disease pathogenesis. There is also the distinct possibility that non-human experimental models of splicing factor mutations may not accurately capture the biological impact of these mutations owing to species-specific changes in splicing.
Currently, very few mRNAs mis-spliced by mutant splicing factors have been rigorously functionally linked to disease development or dissected in molecular detail. One notable exception is mis-splicing of EZH2 driven by mutant SRSF2. Mutations in SRSF2 promote expression of an unannotated isoform of EZH2 due to the presence of C-rich exon splicing enhancers within an exon that is typically skipped. This so-called “poison exon” contains premature termination codon, thus mutant SRSF2 promotes expression of a form of EZH2 mRNA that undergoes NMD. (45) Thus, these findings explain the paradoxical observations that loss-of-function mutations in EZH2 are enriched in the same constellation of myeloid neoplasms as mutations in SRSF2, however EZH2 and SRSF2 mutations are significantly mutually exclusive with one another.(55) Moreover, EZH2 loss functionally promotes myeloid malignancy development in vivo while restoration of EZH2 expression rescues the impaired hematopoiesis characteristic of mutant SRSF2. Despite this exquisite link between EZH2 aberrant splicing and mutant SRSF2, there are still hundreds of additional mRNAs mis-spliced by mutant SRSF2, and the relative importance of these additional events to the pathogenesis of SRSF2-mutant cancers is not clear.
In addition to altered splicing, mutant RNA splicing factors may play a role in disease development through effects on gene expression or pathways not strictly related to splicing. For example, there is increasing evidence that transcriptional alterations unrelated to splicing of individual mRNAs are perturbed in spliceosomal-mutant cells. Transcription and RNA splicing are intimately coupled in vivo. In fact, RNA splicing occurs co-transcriptionally with recruitment of splicing factors to splicing signals in nascent RNA following RNA polymerase II (RNAPII) elongation. Interestingly, it is known that wild-type SRSF2 is important in RNAPII transcriptional elongation and recent work has demonstrated that mutant SRSF2 is associated with impaired RNAPII pause release.(56, 57) As a result, SRSF2 mutant cells actually have increased DNA/RNA hybrids (so-called R-loops) at gene promoters and associated activation of the ATR DNA damage response signaling pathway.
Recent work by Chen et al. and Nguyen et al. demonstrated that augmented R loop formation and ATR activation is also seen in U2AF1-mutant cells.(57, 58) However, it is not precisely clear how mutant U2AF1 alters RNAPII pause release. R loop accumulation is also seen in the presence of compounds that impede splicing catalysis, suggesting a more general contribution of splicing fidelity to RNAPII elongation.(59) While R loops have been associated with increased genomic instability, myeloid neoplasms bearing mutations in splicing factors are not known to harbor high mutation burdens or aneuploidy. Although the exact contribution of R loops to leukemia pathogenesis is not yet clear, ATR activation by R loops may have therapeutic relevance in spliceosomal-mutant cells. Recent studies have demonstrated that U2AF1 mutant cells are preferentially sensitive to ATR inhibition.(59) This may have implications for ongoing clinical trials of ATR inhibitors for cancer patients.
It is also important to note that the earliest reports of RNA splicing factor mutations suggested a link between aberrant RNA splicing and NMD. Overexpression of mutant U2AF1 in cell lines was associated with increased expression of mRNAs encoding NMD components.(23) Subsequent evaluation of SF3B1-mutant cancer cell lines identified an 8 to 10 fold decrease in canonical protein expression of genes with transcripts mis-spliced by mutant SF3B1 and predicted to undergo NMD.(35) However, it is has yet to be demonstrated whether the actual enzymatic process of NMD is altered in any way in spliceosomal mutant cells or if these cells are preferentially reliant on NMD to process aberrant transcripts. Finally, there have been recent intriguing reports of roles for wild-type and mutant U2AF1 on translation of cytosolic mRNAs and alternative polyadenylation (APA).(60, 61) How these potential roles of U2AF1 relate to its well-established function in nuclear RNA splicing are not yet clear, nor is it clear if other recurrently mutated splicing factors can impact these processes as well.
In addition to mutations in genes encoding RNA splicing factors, there are numerous examples of alterations in expression of splicing factors that play a causative role in tumorigenesis (reviewed recently (62)). For example, several studies have identified that overexpression of SRSF1, which is upregulated in cancers with amplification of chromosome 17q23, can transform a variety of cell types.(63, 64) Although SRSF1 is involved in NMD, RNA export, and translation in addition to splicing, it has previously been identified that SRSF1’s oncogenic activity depends on its regulation of splice isoforms of critical regulators of apoptosis and cell survival. In addition to this well-studied example, there is an interesting recent description of a specific form of kidney cancer marked by fusions involving splicing factors. This condition, known as translocation renal cell carcinoma, contains fusions of the transcription factor MITF to a variety of partners, many of which are splicing regulatory factors including SFPQ, LUC7L3, KHSRP, and KHDRBS2.(65) Further efforts to determine whether and how RNA splicing is altered in this condition may be very informative.
ALTERATIONS IN mRNA PROCESSING BEYOND SPLICING: ALTERED POLYDENYLATION IN CANCER
In addition to RNA splicing, processing of 3’ ends of mRNA is also critical in the regulation of gene expression and function. As nascent mRNA is generated by RNAPII, the 3’ end of the RNA must undergo endonucleolytic cleavage followed by synthesis of a poly(A) tail. These two coupled reactions are referred to as cleavage and polyadenylation (or simply polyadenylation). It has been known for several decades that through APA, a single gene can give rise to multiple transcripts which differ only in the sequence at their 3’ termini through cleavage at distinct polyadenylation site (PASs). It is currently thought that at least 70% of mammalian mRNAs express APA isoforms (reviewed recently(66)). APA has many parallels to alternative splicing in that it is an enzymatic process which (i) exhibits tissue specificity(67), (ii) is regulated by cis elements embedded in mRNA sequences and trans acting proteins recognizing these sequences, (iii) can result in alternative mRNA isoforms, and (iv) occurs mostly co-transcriptionally. Pre-mRNA is cleaved 10–30 nucleotides downstream of a polyadenylation (pA) signal, a hexameric sequence located 10–35 nucleotides upstream of the PAS. The canonical pA signal is AAUAAA but this sequence can adopt variants more than 10-times weaker. Importantly, the splicing machinery is also critically important in promoting the use of PASs at the 3’ ends of genes. It is now understood that, in addition to helping to identify the 5’ ss, the U1 snRNA is also critically important in suppressing premature 3’ end cleavage and polyadenylation from cryptic PASs within introns (68). As such, blocking the binding of the U1 snRNA to the 5’ ss induces premature cleavage of transcripts within introns. This role of U1 in suppressing transcriptional termination increases the likelihood of the spliceosome identifying a downstream 3’ ss and is termed “telescripting.”
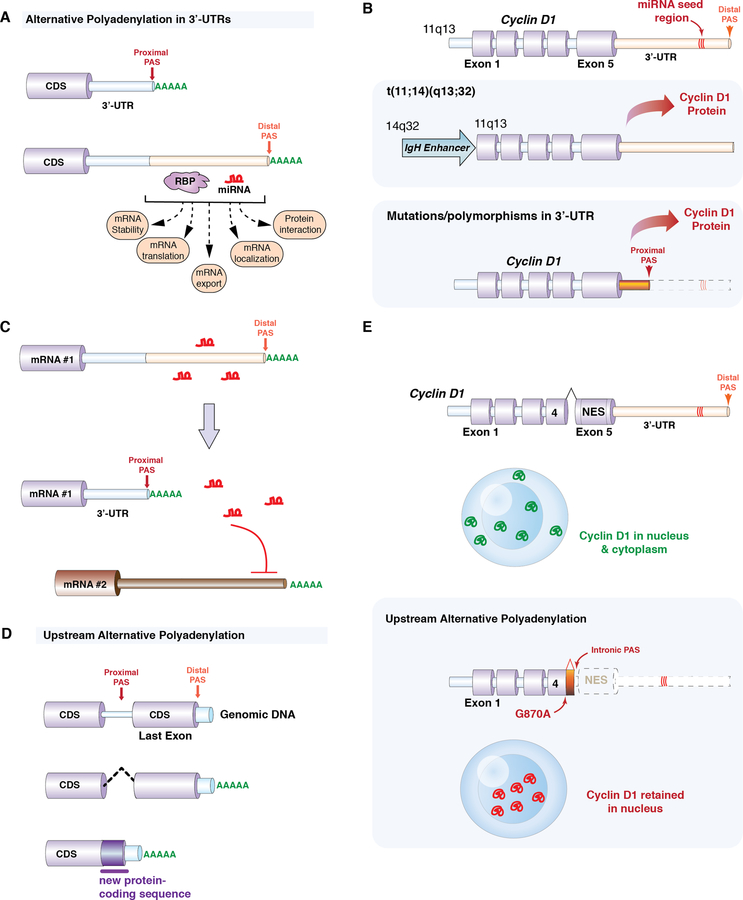
It is currently thought that most APA sites are located in 3’ untranslated regions (3’-UTRs).(67, 69) In such cases, the choice of one APA site over another does not change the protein-coding mRNA sequence, but results in mRNA species that differ in the sequences within the 3’-UTR (Figure 3A). Given that the 3’-UTR contains cis elements involved in mRNA stability, translation, nuclear export, and mRNA localization, altering 3’-UTR content can have strong effects on mRNA expression and function. For example, more than half of microRNA (miRNA) target sites are located in alternative UTRs.(70) Shortening of 3’-UTRs would therefore remove miRNA-mediated repression. There are several well-characterized examples of oncogenes upregulated by removal of miRNA repression sites through APA. For example, cyclin D1 is a well-known proto-oncogene in many cancers and undergoes ectopic expression in mantle cell lymphoma. In most cases, a chromosomal translocation brings the coding region of Cyclin D1 under the control of the IGHV promoter (the t(11;14)(q13;q32) translocation). However, in some mantle cell lymphoma patients that lack Cyclin D1 translocations, Cyclin D1 overexpression is due to truncation of its 3’-UTR with loss of miRNA repression sites (Figure 3B)(71). In some cases, 3’-UTR shortening is due to mutations creating a novel pA signal. Moreover, mutation of the miRNA seed regions in a normal length UTR revealed the same effects on upregulation of Cyclin D1 expression. Similar examples of upregulation of cancer-specific isoforms through loss of 3’-UTR regulatory regions have been described for Cyclin D2 (72) and CDC6 (73). In addition to shortening of 3’-UTRs to release mRNAs from miRNA-mediated repression, recent data also suggest that shortening of the 3’-UTR of an mRNA may allow release of miRNAs to suppress the expression of other mRNAs in trans (Figure 3C).(74) This idea is based on the finding that cancer cells with shortened 3’-UTRs are enriched in mRNAs proposed to act as competing endogenous RNAs (ceRNAs).
Figure 3. Altered gene regulation in cancer through alternative cleavage and polyadenylation of mRNAs.
(A) Schematic of how alternative polyadenylation (APA) of mRNAs in the 3’-untranslated region (3’-UTR) of mRNAs results in two distinct isoforms which differ only in their 3’-UTRs. The 3’-UTR can contain multiple potential polyadenylation sites (PASs) and additional sequences that may be recognized by RNA binding proteins (RBPs) and/or micro-RNAs (miRNAs). Altering 3’-UTR length may impact miRNA-mediated gene repression, protein-protein interactions, mRNA stability, translation, export, and localization. (B) Cyclin D1 upregulation is a well-studied example of how altering 3’-UTR length results in proto-oncogene activation. In a proportion of mantle cell lymphoma patients, Cyclin D1 is upregulated through polymorphisms and mutations in the 3’-UTR that result in the use of a proximal PAS and a shortened 3’-UTR that lacks a miRNA seed region. (C) Schematic illustrating how shortening of the 3’-UTR of an mRNA may allow release of miRNAs to suppress the expression of other mRNAs in trans. (D) APA may also utilize PAS upstream of the normal stop codon and thereby alter the coding sequence of mRNA. In this example, a proximal PAS site is located within an intron and use of this PAS site results in production of a shorter protein with a novel 3’ amino acid sequence. (E) In addition to alterations in 3’-UTR length, cyclin D1 is also subject to use of upstream PASs. Polymorphisms at the end of exon 4 (for example, the G870A polymorphism) may promote use of an intronic polyadenylation signal within intron 4. This cleaves both miRNA binding sites as well as sequences encoding the normal nuclear export signal (NES) from cyclin D1. As a result, this cyclin D1 protein isoform is restricted to the nucleus.
In addition to these specific examples of alternative 3’-UTRs regulating mRNA expression, several recent studies have provided surprising evidence that 3’-UTRs also mediate mRNA-protein interactions and protein localization. Given that RBPs interact with 3’-UTRs, Berkovits and Mayr hypothesized that altering 3’-UTR length could affect a “scaffold” function of 3’-UTRs and change nascent protein interactions with proteins that affect cellular trafficking. In one such example, APA of the 3’-UTR of the mRNA encoding the transmembrane protein CD47 resulted in short or long isoforms of CD47 mRNA that were translated in association with different protein complexes, leading to CD47 localization at either the plasma membrane (long 3’UTR) or the endoplasmic reticulum (short 3’UTR) (75).
Despite individual examples of altered APA events in cancer-associated mRNAs, there have been only a handful of surveys of global APA usage in cancer. Global APA evaluation has been limited by the fact that standard RNA-seq does not effectively capture 3’ ends of RNA and few informatic methods identify APA sites and polyA tail lengths from standard RNA-seq. One key study utilized a novel bioinformatics algorithm for de novo identification of APAs from standard RNA-seq and applied this to 358 TCGA tumor/normal pairs from seven cancer types.(76) This identified many tumor specific APA events in cancer cells, most of which utilize short 3’-UTRs and are generally associated with increased gene expression, likely due to escape from miRNA-mediated gene repression. At the same time, this study also revealed that APA appears to be highly tumor specific. For example, lung, uterus, breast, and bladder cancers have significantly more APAs than head/neck or kidney cancers. APA has also been shown to be tissue-specific in normal tissues.(77) These observations beg the question of what regulates APA. Recurrent genetic alterations in genes encoding the cleavage and polyadenylation machinery have not been identified and recurrent mutations impacting pA sites in cis have not been described in cancer, outside of the few examples noted above. Thus, it is currently believed that APA is regulated by changes in expression of the machinery responsible for APA. The core polyadenylation trans-factors include four multi-subunit protein complexes (CPSF, CstF, CFI, and CFII). CPFS recognizes the pA signal and CstF binds downstream U/GU-rich elements (mostly through the CstF64 subunit) which help mark the pA signal. Interestingly, Xia et al. found increased expression of CstF64 mRNA in most tumors, which is hypothesized to promote usage of proximal, weaker pA sites and prevent usage of stronger, distal pA sites.(76)
Although most APA sites occur in 3’-UTRs, there is increasing recognition that PASs may be located upstream of the last exon, usually within introns. Use of such upstream APA sites (also referred to as intronic polyadenylation (IpA) sites when occurring within introns), would be expected to alter the coding region of a gene, similar to how alternative splicing would alter the protein-coding mRNA sequence (Figure 3D). There is increasing recognition that cancer cells may utilize aberrant IpA sites more frequently than normal cells and that IpA may result in truncated, aberrant mRNAs with important functional impact.(78) It is important to note that IpA also occurs during normal physiologic regulation of gene expression. For example, as B-cells terminally differentiate into plasma cells, the mRNA encoding immunoglobulin M heavy chain (IgHM) undergoes an IpA event to skip exons encoding the transmembrane domains of IgHM.(79) This results in plasma cells expressing a form of IgHM that is soluble and excreted. Several recent studies have applied 3’ RNA-seq to normal immune cells in addition to multiple myeloma(77) and CLL.(78) While CLL was found to have increased IpA compared to normal B-cell subsets, myeloma cells had reduced IpA usage. In either instance, aberrant IpA events could be speculated to promote tumorigenesis. In addition to undergoing 3’ UTR shortening, cyclin D1 also is subject to IpA in a manner that promotes tumorigenesis (Figure 3E).(80, 81) A protein isoform of cyclin D1 (known as cyclin D1b) is produced when cyclin D1 mRNA is cleaved at an APA site within an intron. This removes both the 3’ UTR miRNA repression sites as well as sequences encoding the protein’s normal nuclear export signal. Generation of cyclin D1b may occur due to polymorphisms at the end of exon 4, which promote use of a polyA signal within an intron just downstream of exon 4.
As mentioned above, polyadenylation of mRNAs is coupled with mRNA cleavage. Polyadenylation, the template-independent addition of adenosine homopolymers to the 3’ end of mRNAs, is critical in regulating mRNA stability. The initial addition of the poly(A) tail occurs in the nucleus and is a fairly well-understood process. However, further additions to poly(A) tail length may occur in the cytoplasm by cytoplasmic poly(A) polymerases. This process and the RNA species regulated by it are less well understood. Interestingly, recent data suggest that one of the most commonly mutated genes in multiple myeloma, FAM46C, encodes a cytoplasmic poly(A) polymerase.(82) FAM46C, encoded at the 1p12 locus, is affected by deletions as well as homozygous and hemizygous mutations in approximately10% of myeloma patients. Work from two groups suggests that FAM46C loss regulates growth of myeloma cells across a variety of myeloma cell lines. (82, 83) However, the RNA substrates of FAM46C and why it is so uniquely mutated in myeloma, as opposed to other cancers, are not understood.
THERAPEUTIC TARGETING OF RNA PROCESSING IN CANCER
Rationale for Targeting Splicing for Cancer Therapy
As described earlier, it has been documented that cancer cells harbor widespread changes in RNA splicing compared to normal cells, including increased expression of known pro-oncogenic and anti-apoptotic isoforms of genes such as MDM2, Bcl(x), and VEGF. (84) These findings suggested that modulating RNA splicing of specific transcripts, or even globally, might have therapeutic benefit. The discovery of heterozygous hotspot mutations in spliceosome components further highlighted RNA splicing as a potential therapeutic vulnerability for cells bearing these mutations. Initial clues to this possibility came from human genetic data. For example, while hotspot mutations in BRAF, RAS, PIK3CA, and other signaling proteins, commonly undergo allelic imbalance in a manner that increases the dosage of the mutant allele, this conspicuously does not occur with hotspot mutations in SRSF2, SF3B1, or U2AF1.(85) Instead, cells bearing mutations in these genes consistently retain expression of the wild-type allele and never undergo loss-of-heterozygosity or become hemizygous. These data indicate haplo-essentiality of mutations in SF3B1, SRSF2, and U2AF1, and suggest that increased expression of the mutant allele is negatively selected in cells.
Consistent with this genetic requirement for the wild-type allele in splicing factor mutant cells, spliceosomal gene mutations are also highly mutually exclusive with one another.(23, 86) In MDS where mutations in the three most commonly mutated splicing factors, SF3B1, SRSF2, or U2AF1 are present in >50% of patients; less than1% of patients harbor a mutation in >1 of these genes simultaneously. These data again argue for the strong requirement of a certain level of normal splicing catalysis in splicing factor-mutant cells.
Each of the above genetic observations has now been evaluated in functional studies of the cellular and phenotypic effects of changing allelic ratios of splicing factor mutations. For example, expression of Srsf2P95H in a homozygous (Srsf2P95H/P95H) or hemizygous state (Srsf2P95H/null) led to immediate elimination of Srsf2 mutant hematopoietic cells.(87) Very similar findings have been seen in the setting of U2AF1S34F mutant lung cancer cell lines and SF3B1 mutant breast cancer cells.(52, 53) Moreover, induced simultaneous expression of Sf3b1K700E/WT and Srsf2P95H/WT mutations in hematopoietic cells in vivo resulted in synthetic lethality.(86) The precise mechanistic basis for why mutations in SRSF2, U2AF1, or SF3B1 are intolerable when expressed without the wild-type allele or combined in the same cell is still not fully understood.
In addition to a requirement for wild-type spliceosome function in splicing factor-mutant cells, several other genetic subtypes of cancer have been suggested as preferentially sensitive to alterations in splicing. These include several reports of a requirement for normal splicing in MYC amplified cancers, including glioblastoma, breast cancer, and lymphomas (88–91). Studies have shown that MYC upregulates transcription of the splicing regulatory proteins PTB and hnRNPA1/2, which in turn promote use of the pro-oncogenic isoform of pyruvate kinase (known as PKM2) while suppressing expression of the alternative, mutually exclusive isoform PKM1 (88, 91). More recently, it has been appreciated that MYC’s effects on splicing extend beyond this individual splicing event in PKM1/2 to create global alterations in splicing of many mRNAs.(89, 90) Consistent with this, expression of MYC has been identified as conferring sensitivity to genetic or pharmacologic inhibition of splicing.(92)
In addition to splicing factor-mutant cells and MYC driven cancers, given that RNA splicing is essential for cell survival, there have been several studies identifying splicing as a therapeutic vulnerability for cells with “bystander” genetic deletions of splicing factors. For example, breast cancer cell lines containing partial deletion of SF3B1 are highly sensitive to further down-regulation of SF3B1, while SF3B1 copy neutral cells are more tolerant to down-regulation of the SF3B1.(93) Similarly, cells with deletion of the exon-junction complex core member MAGOH, which commonly occurs in cancer cells containing chromosome 1p deletion, are sensitive to loss of the MAGOH paralog MAGOHB.(94)
Discovery of Chemical Modulators of Splicing: SF3b Binding Compounds
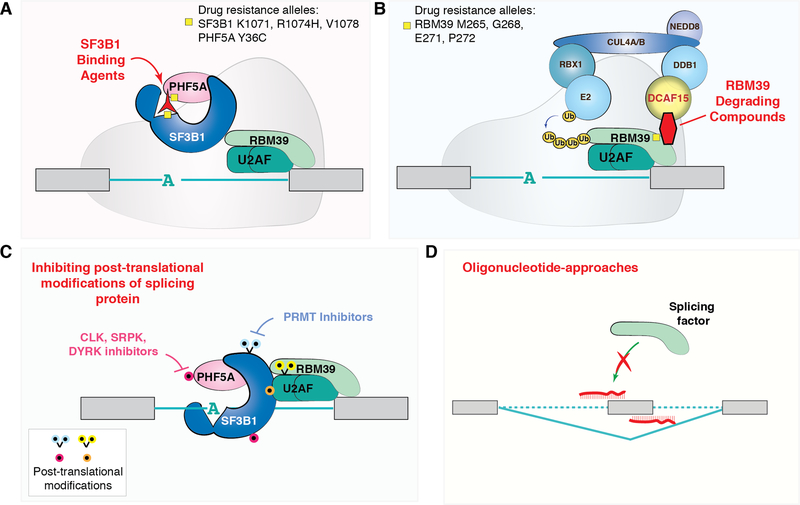
In parallel to the above data presenting a rationale for inhibition of splicing in cancer, a series of chemical biology studies uncovered diverse compounds that target the spliceosome. The initial discovery of compounds which selectively inhibit splicing came in 2007 when investigators from RIKEN and Eisai Co. demonstrated that the natural products FR901464 and pladienolides, and their derivatives including spliceostatin A and E7107, respectively, physically bind to the SF3b complex and inhibit pre-mRNA splicing at an early step in spliceosome assembly (Figure 4A).(95, 96) These compounds were originally isolated from bacteria (Pseudomonas sp. and Streptomyces sp.) and were known to exhibit potent cytotoxic effects against various solid tumor cell lines in vitro and in xenograft models. More recently, forward genetic experiments performed in established resistant cell lines identified specific residues in SF3B1 and PHF5A, both components of the SF3b complex that were associated with drug resistance.(97, 98) These findings affirmed the on-target specificity of SF3b-binding agents for the spliceosome and provided further clues to the exact structural basis for their mechanism of action. This culminated in publication of the crystal structure of SF3b in complex with pladienolide B and the cryo-EM structure of Sf3b bound to E7107.(99, 100) These structures definitively show that SF3b inhibitors act by binding to the SF3b complex and interfering with the ability of the complex to recognize branchpoint nucleotides within introns. Extensive RNA-seq studies of cells treated with these compounds further revealed widespread inhibition of splicing efficiency, with dose-dependent increases in cassette exon skipping and IR.(87, 101, 102)
Figure 4. Methods for therapeutic modulation of RNA splicing.
Pharmacologic means to perturb splicing include (A) drugs that physically bind the SF3b complex and disrupt its ability to recognize the branchpoint region of the intron. (B) More recently, anticancer sulfonamide compounds were discovered to cause the degradation of RBM39. These compounds physically link RBM39 to the DCAF15-CUL4 ubiquitin ligase, resulting in ubiquitinylation of RBM39 and its subsequent proteasomal degradation (of note, it is currently unknown whether degradation of RBM39 occurs while bound to U2AF and/or assembled on a 3’ splice site region). Specific mutations in SF3B1, PHF5A, and RBM39 that confer drug resistance to these molecules are shown. (C) The function, cellular localization, and assembly of a variety of splicing proteins depend on post-translational modifications and inhibitors of the enzymes placing these marks have been developed. These include protein arginine methyltransferase (PRMT) inhibitors as well as inhibitors of CLK, SRPK, and DYRK kinases. (D) Finally, oligonucleotides that modify splicing of specific transcripts by blocking the RNA–RNA base-pairing or protein–RNA binding interactions that occur between the splicing machinery and the pre-mRNA may be used to target individual aberrant splicing events in cancer.
Consistent with the studies above, which identified recurrent genetic subtypes of solid tumors that are particularly vulnerable to inhibition of splicing, hematopoietic cells bearing hotspot mutations in SRSF2, SF3B1, and U2AF1 have been shown to be preferentially sensitive to the SF3b-inhibitory compounds, E7107, H3B-8800 (an orally bioavailable analogue of E7107), and sudemycins (analogues of the natural compound FR901464).(37, 87, 101, 103) This has been demonstrated across numerous model systems including in vivo studies in splicing factor-mutant conditional knockin mice, acute and chronic myeloid leukemia patient-derived xenografts, and a variety of human splicing factor-mutant cancer cell lines. Owing to the large number of individual splicing events impacted by chemical inhibition of SF3b, in addition to the aberrant splicing caused by each individual mutation, the precise mechanistic basis for preferential sensitivity of splicing factor-mutant cells to these compounds is not entirely clear. However, these agents induce aberrant splicing of mRNAs encoding numerous RNA splicing factors.. Given the preferential dependence of splicing factor mutant cells on otherwise normal splicing catalysis, aberrant splicing of mRNAs encoding RNA splicing factor proteins by splicing modulatory drugs may partially explain the preferential effects of these compounds on splicing factor-mutant cells.(101) In addition, a number of studies have identified that the mRNAs encoding the expression of BCL2 family proteins are altered by SF3b inhibition. For example, SF3b inhibition promotes expression of pro-apoptotic isoforms of MCL1 as well as induction of non-functional alternatively spliced transcripts of BCL2 and BCL2A1.(104) As such, E7107 has been shown to induce apoptosis in cell lines normally dependent on these proteins for survival and has synergistic effects with BCL2 inhibition in some settings.(105)
While SF3b-inhibitory molecules provide a useful tool to investigate spliceosome assembly and function, the SF3b complex is essential in all cells and the therapeutic index of these compounds has been a major question. The first SF3b binding agent to be tested in patients was E7107, where 66 patients with refractory metastatic solid tumors were treated in two prior phase I clinical trials (106, 107). While some promising clinical responses were observed, clinical development of E7107 was halted due to the development of ocular toxicity in two patients, the cause of which was unclear. It is important to note that these trials were performed before the discovery that components of the spliceosome are recurrently mutated in various cancers, and there are no clinical reports to date of the effects of SF3b-inhibition on the cancer types most enriched in these mutations. To this end, a phase I clinical trial of H3B-8800 is now ongoing in patients with AML, MDS, and CMML that is relapsed/refractory to conventional therapy (clinicaltrials.gov identifier ). This trial will present an important opportunity to evaluate the safety of H3B-8800 in patients and to determine the degree of splicing modulation achievable in patients.
Splicing Inhibitor Sulfonamides: A New Class of Targeted Protein Degraders
Until recently, few chemical means to perturb splicing were known outside of SF3b inhibitors. In 2017, Han et al. and investigators from Eisai discovered that a series of sulfonamide-containing compounds induce proteasomal degradation of the accessory RNA splicing factor, RBM39.(108, 109) These compounds, which include the molecules indisulam, E7820, and chloroquinoxaline sulfonamide, had been known to have anticancer properties in vitro for decades, but the cellular mechanism of action was not fully understood. However, a chemical genetic approach to identify indisulam-resistant cells (by Han et al) and an expression proteomics effort (led by Uehara et al.) identified RBM39 as the cellular target of these compounds. This led to the discovery that the anticancer sulfonamides bind a substrate receptor of the CRL4 E3 ubiquitin ligase complex known as DCAF15 and direct ubiquitin-mediated degradation of RBM39 (Figure 4B). As such, deletion of DCAF15 renders cells resistant to these compounds, as does introduction of RBM39-drug resistant mutations.(108, 109)
The discovery of molecules that direct the CRL4-DCAF15 E3 ubiquitin ligase complex to degrade RBM39 was surprising for several reasons. Before this discovery, the only compounds known to induce targeted protein degradation were the IMiD molecules including lenalidomide.(110–112) While IMiDs normally engage an E3 ubiquitin ligase to target the substrates CRBN, IKZF1, and IKZF3 for proteasomal degradation, there are intense ongoing efforts to restructure these molecules in order to target additional proteins (so-called “PROteolysis-TArgeting Chimeras (PROTACs) strategies). Thus, the identification of a novel set of chemically diverse compounds that act via an analogous mechanism opens up new possibilities to design small molecules that can target “undruggable” disease-causing proteins for proteasomal degradation.
In addition to the broad implications of hijacking anticancer sulfonamides to degrade diverse substrates, this discovery has also raised interest in the known substrate of these compounds, RBM39. RBM39 (also referred to as CAPERα) is an RBP that contains classic RNA recognition motifs (RRMs) in addition to a unique class of protein recognition motifs known as U2AF homology motifs (UHMs). These domains are also shared by the paralogous proteins RBM23 (CAPERβ), PUF60, SPF45, and the well characterized splicing factors, U2AF1 and U2AF2. Although prior studies have suggested RBM39 physically interacts with the estrogen receptor, AP1, and other transcription factors, the only rigorous structural evidence regarding RBM39 interactors supports an interaction of RBM39 with SF3B1 and U2AF2.(113, 114)
Consistent with the structural data above, RBM39 degradation in vitro results in clear inhibition of splicing catalysis, analogous to that seen with direct SF3b inhibition. As such, splicing factor mutant leukemias have recently been shown to be as sensitive to RBM39 degradation as they are to SF3b inhibition.(102) However, unlike SF3b inhibitors, whose clinical safety and potential are unclear, numerous anticancer sulfonamides have already completed phase I and II clinical trials. Indisulam, for example, has been used in phase II clinical trials in patients with melanoma, non-small cell lung cancer, and AML with favorable safety profiles (115–117). At the same time, each of these studies was carried out before the mechanism of action of these compounds was known. Thus, it is not known whether the doses of drug utilized in the clinical studies actually caused RBM39 degradation. Likewise, there was no effort in prior studies to stratify patient enrollment based on predictors of sensitivity to these compounds, which include DCAF15 expression levels and the presence of hotspot mutations in splicing factors.(102, 108, 109) It will therefore be critical to revisit the clinical utility of these compounds with appropriate pharmacodynamic monitoring for RBM39 degradation and splicing inhibition, as well as stratification based on the aforementioned predictive biomarkers.
Targeting Post-translational Modifications of Splicing Proteins
In addition to inhibiting the interaction of the spliceosome with mRNA and degrading splicing proteins, there has been considerable interest in modulating splicing by modifying post-translational modifications (PTMs) required for spliceosome function. It is known that PTMs of a variety of splicing factors regulate spliceosome formation and splicing catalysis. For example, phosphorylation of SR proteins regulates the shuttling of SR proteins in and out of the nucleus to modulate splicing.(118–120) Phosphorylation of SR proteins is required for spliceosome complex formation but dephosphorylation of SR proteins allows splicing catalysis to occur and initiates nuclear export of SR proteins. In another example, symmetric arginine dimethylation (SDMA), catalyzed by PRMT5, is required for small nuclear ribonucleoprotein (snRNP) assembly.(121, 122) Consequently, a variety of chemical inhibitors of kinases and methyltransferases which act on splicing proteins, such as CLKs (cdc-like kinases), SRPKs (SR protein kinases), and PRMTs (protein arginine methyltransferases), amongst others, are being evaluated (Figure 4C).
Of the above approaches, the class of compounds furthest in clinical development and most heavily studied is the PRMT inhibitors. Genetic ablation or chemical inhibition of PRMT5 results in splicing inhibition and anticancer effects across a number of cancer types.(90, 123) Given these results, it will be important to determine whether malignancies with splicing factor mutations are preferentially sensitive to PRMT5 inhibition, and whether inhibition of Type I PRMT enzymes (which catalyze asymmetric arginine dimethylation (ADMA)) cause splicing changes similar to PRMT5 inhibition. Of note, consistent with the numerous cellular substrates for PRMT enzymes, many different mechanisms have been proposed to underlie the sensitivity of cancer cells to PRMT inhibitors beyond splicing. Moreover, given the number of splicing proteins and individual protein residues that undergo arginine methylation, discerning which individual arginine methylation events, if any, are responsible for the cellular effects of PRMT inhibitors has been a challenge.
The above mechanistic questions notwithstanding, at least three PRMT5 inhibitors are now in phase I clinical trials for patients with relapsed/refractory solid tumors. These include the compounds GSK3326595 (clinical trials.gov for solid tumors and B cell non-Hodgkin lymphomas; clinical trials.gov for MDS/AML), PF 06939999 (clinical trials.gov ), and JNJ-64619178 (clinical trials.gov ). In addition, a first-in-human trial of the type I PRMT inhibitor GSK3368715 has been initiated for patients with relapsed/refractory B cell non-Hodgkin lymphomas (clinical trials.gov ). These studies will provide an important opportunity to evaluate the safety of PRMT inhibition in vivo at levels which impact SDMA/ADMA levels.
Other Means to Target Splicing in Cancer
Beyond the therapeutic approaches noted above, inhibition of other enzymatic steps in RNA processing is being pursued. For instance, the observation that N6-adenonisone methylation of RNA is required for the survival of certain cancer types but dispensable for normal counterpart cells has led to great interest in identifying chemical inhibitors of the N6-adenonisone methyltransferase METTL3 (reviewed recently (3)). In addition, a recent functional genomic screen by Yamauchi et al. showed that the decapping enzyme scavenger DCPS, which catalyzes the final step of 3’ to 5’ mRNA decay, is required for the survival of AML cells but dispensable for normal hematopoietic cells.(124) Evidence for this preferential dependency was bolstered by the description of families with germline DCPS bi-allelic loss-of-function mutations who have normal hematopoiesis. Interestingly, several inhibitors of DCPS have been described. One such compound, RG3039,(125) has already completed a phase I clinical trial for spinal muscular atrophy (SMA) and is known to be safe in patients. Given these results, evaluating the clinical potential of DCPS inhibition in cancer patients will be an important next step.
In addition to chemical modulators of splicing, it is important to note the potential for oligonucleotide-based approaches to modify individual splicing events (Figure 4D). These approaches consist of modified nucleic acids that base pair with pre-mRNA and modify splicing by blocking RNA–RNA base-pairing or splicing factor–RNA binding interactions. Antisense oligonucleotide (ASO)-based approaches have met clinical success resulting in FDA-approval of the ASOs Nusinersen and Eteplirsen for SMA and Duchenne muscular dystrophy, respectively. However, whether there will be therapeutic benefit in targeting individual splicing events in disorders such as cancer, which are marked by many simultaneous genetic and splicing alterations, remains to be determined.
The Potential for Splicing Derived Neo-Epitopes
Checkpoint inhibitor-based immunotherapies have transformed clinical care for a number of malignancies. However, the majority of patients receiving checkpoint inhibitors do not derive benefit. This observation has spurred intense efforts to identify biomarkers of response to checkpoint blockade as well as pharmacologic approaches to increase response to these therapies. In this regard, a number of studies have shown that the burden of mutations occurring at the level of DNA increases the chances of generating immunogenic peptides that can be presented on MHC class I. Interestingly, a number of studies have also suggested that cancer-specific changes to RNA splicing may be an additional source of neo-epitopes. For example, the recent pan-cancer TCGA splicing analysis by Kahles et al. suggested that predicted neo-epitopes generated by tumor-specific alternative splicing events are far more abundant than those generated by somatic SNVs.(12)
While this is an intriguing idea, to date there is little functional evidence that cancer-associated aberrant splicing actually elicits tumor immunogenicity. There has been little experimental validation of the immunogenicity of neoantigens derived from altered splicing. Evaluation of the immunogenicity of tumors based on RNA expression of immune markers have differed in their conclusions about whether cancer-associated splicing changes are associated with reduced or increased immune cell infiltration (15, 31). Moreover, one recent analysis focused on neo-epitopes generated from IR failed to identify any association between retained intron neoepitope load and clinical benefit from checkpoint inhibitors in melanoma patients.(126) One important to point to note in studies of intron-retained mRNAs is that the fate of mRNAs with IR may not be immediately clear from RNA-seq alone. First, if the IR mRNA is exported to the cytoplasm and contains a premature termination codon it may undergo NMD instead of being translated into a novel protein isoform. Alternatively, it has recently been recognized that some RNAs with retained introns may remain in the nucleus and be subjected to degradation by the nuclear exosome or undergo splicing post-transcriptionally and later exported to the cytoplasm for translation.(127) mRNAs subject to splicing following transcription are referred to as “detained introns.” The mechanisms that detain incompletely spliced transcripts in the nucleus versus allowing their export to the cytoplasm are not entirely clear.
Given these results, further efforts to determine whether pharmacologic means to perturb splicing can induce the generation of neo-epitopes and boost the immunogenicity of tumors would be innovative. Moreover, it is still unclear whether introduction of mutations in RNA splicing factors might increase neo-epitope load and/or responsiveness to immune-checkpoint blockade.
Conclusions
Despite the major advances in our understanding of the genomics, molecular biology, and therapeutic implications of altered RNA processing in cancer, the full contribution of aberrant RNA splicing and polyadenylation to cancer pathogenesis has not been fully elucidated. Further efforts to comprehensively catalog genetic alterations in the vast noncoding regions within genes (both within introns and at 3’ UTRs) that have a regulatory impact on RNA splicing and PAS choice are still needed.
Moreover, greater systematic evaluation of the potential functional roles of distinct RNA isoforms, particularly aberrant, unannotated RNA isoforms produced in cancer cells, will be greatly informative for disease biology. Understanding the role of these isoforms could also be incredibly important for therapy. For example, it is currently recognized that acquired pathologic splice variants can confer therapeutic resistance to therapies as diverse as RAF inhibitors(128) and anti-CD19 chimeric antigen receptor T-cells.(129) These findings underscore the need to understand the dynamic role of RNA processing in untreated cancers as well as serially with therapy.
The identification of functionally important pathologic RNA isoforms opens up the possibility of oligonucleotide-based therapies for splicing alterations in cancer. In parallel, it is hopeful that the ongoing efforts to modulate splicing clinically may address the question of whether globally modifying RNA splicing is safe and therapeutically efficacious in patients. These include ongoing clinical trials to target the SF3B complex, degrade RBM39, and prevent protein arginine methylation of splicing proteins. Finally, the future application of novel methods of assessing splicing, including the use of splicing analysis within single cells and single-molecule RNA sequencing technologies, will hopefully provide new insights into splicing dysregulation in cancer.
STATEMENT OF SIGNIFICANCE.
There is increased recognition that genetic alterations impacting RNA splicing and polyadenylation are common in cancer and may generate novel therapeutic opportunities. Such mutations may occur within an individual gene or in RNA processing factors themselves, thereby influencing splicing of many downstream target genes. This review discusses the biological impact of these mutations on tumorigenesis and the therapeutic approaches targeting cells bearing these mutations.
ACKNOWLEDGEMNETS
We thank members of the Obeng and Abdel-Wahab for their reading and input on this manuscript.
Financial support: E.A.O. is supported by grants from the EvansMDS Foundation, the Leukemia Research Foundation, and the American Society of Hematology. O.A.-W. is supported by grants from the Leukemia and Lymphoma Society, the EvansMDS Foundation, the Henry & Marilyn Taub Foundation, grants from NIH/NHLBI (R01 HL128239) and NIH/NCI (1 R01CA201247-01A1), the Dept. of Defense Bone Marrow Failure Research Program (BM150092 and W81XWH-12-1-0041), an MSK Steven Greenberg Lymphoma Research Award, the Geoffrey Beene Research Center of MSK, and the Pershing Square Sohn Cancer Research Alliance.
Conflict of interest disclosure: O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc, Merck, and Janssen and is on the scientific advisory board of Envisgenics Inc.; O.A.-W. has received prior research funding from H3B Biomedicine unrelated to the current manuscript.
References
- 1.Han L, Diao L, Yu S, Xu X, Li J, Zhang R, et al. The Genomic Landscape and Clinical Relevance of A-to-I RNA Editing in Human Cancers. Cancer cell 2015;28:515–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell 2016;29:452–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vu LP, Cheng Y, Kharas MG. The Biology of m(6)A RNA Methylation in Normal and Malignant Hematopoiesis. Cancer discovery 2019;9:25–33. [DOI] [PubMed] [Google Scholar]
- 4.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther 2016;1:15004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y, Rio DC. Mechanisms and Regulation of Alternative Pre-mRNA Splicing. Annu Rev Biochem 2015;84:291–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollander D, Naftelberg S, Lev-Maor G, Kornblihtt AR, Ast G. How Are Short Exons Flanked by Long Introns Defined and Committed to Splicing? Trends Genet 2016;32:596–606. [DOI] [PubMed] [Google Scholar]
- 7.Matera AG, Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol 2014;15:108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan C, Wan R, Shi Y. Molecular Mechanisms of pre-mRNA Splicing through Structural Biology of the Spliceosome. Cold Spring Harb Perspect Biol 2019;11. [DOI] [PMC free article] [PubMed]
- 9.Plaschka C, Newman AJ, Nagai K. Structural Basis of Nuclear pre-mRNA Splicing: Lessons from Yeast. Cold Spring Harb Perspect Biol 2019;11. [DOI] [PMC free article] [PubMed]
- 10.Kastner B, Will CL, Stark H, Luhrmann R. Structural Insights into Nuclear pre-mRNA Splicing in Higher Eukaryotes. Cold Spring Harb Perspect Biol 2019. [DOI] [PMC free article] [PubMed]
- 11.Chen L, Tovar-Corona JM, Urrutia AO. Increased levels of noisy splicing in cancers, but not for oncogene-derived transcripts. Hum Mol Genet 2011;20:4422–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kahles A, Lehmann KV, Toussaint NC, Huser M, Stark SG, Sachsenberg T, et al. Comprehensive Analysis of Alternative Splicing Across Tumors from 8,705 Patients. Cancer cell 2018;34:211–24 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung H, Lee D, Lee J, Park D, Kim YJ, Park WY, et al. Intron retention is a widespread mechanism of tumor-suppressor inactivation. Nat Genet 2015;47:1242–8. [DOI] [PubMed] [Google Scholar]
- 14.Supek F, Miñana B, Valcárcel J, Gabaldón T, Lehner B. Synonymous mutations frequently act as driver mutations in human cancers. Cell 2014;156:1324–35. [DOI] [PubMed] [Google Scholar]
- 15.Jayasinghe RG, Cao S, Gao Q, Wendl MC, Vo NS, Reynolds SM, et al. Systematic Analysis of Splice-Site-Creating Mutations in Cancer. Cell reports 2018;23:270–81 e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ke S, Shang S, Kalachikov SM, Morozova I, Yu L, Russo JJ, et al. Quantitative evaluation of all hexamers as exonic splicing elements. Genome research 2011;21:1360–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Julien P, Minana B, Baeza-Centurion P, Valcarcel J, Lehner B. The complete local genotype-phenotype landscape for the alternative splicing of a human exon. Nat Commun 2016;7:11558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickrell JK, Pai AA, Gilad Y, Pritchard JK. Noisy splicing drives mRNA isoform diversity in human cells. PLoS Genet 2010;6:e1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dvinge H, Bradley RK. Widespread intron retention diversifies most cancer transcriptomes. Genome Med 2015;7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Q, Rio DC. JUM is a computational method for comprehensive annotation-free analysis of alternative pre-mRNA splicing patterns. Proc Natl Acad Sci U S A 2018;115:E8181–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ding L, Rath E, Bai Y. Comparison of Alternative Splicing Junction Detection Tools Using RNA-Seq Data. Curr Genomics 2017;18:268–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin KT, Krainer AR. PSI-Sigma: a comprehensive splicing-detection method for short-read and long-read RNA-seq analysis. Bioinformatics 2019. [DOI] [PMC free article] [PubMed]
- 23.Yoshida K, Sanada M, Shiraishi Y, Nowak D, Nagata Y, Yamamoto R, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature 2011;478:64–9. [DOI] [PubMed] [Google Scholar]
- 24.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 mutation in myelodysplasia with ring sideroblasts. N Engl J Med 2011;365:1384–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Graubert TA, Shen D, Ding L, Okeyo-Owuor T, Lunn CL, Shao J, et al. Recurrent mutations in the U2AF1 splicing factor in myelodysplastic syndromes. Nat Genet 2012;44:53–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L, Lawrence M, Wan Y, Stojanov P, Sougnez C, Stevenson K, et al. SF3B1 and other novel cancer genes in chronic lymphocytic leukemia. The New England journal of medicine 2011;365:2497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furney SJ, Pedersen M, Gentien D, Dumont AG, Rapinat A, Desjardins L, et al. SF3B1 mutations are associated with alternative splicing in uveal melanoma. Cancer discovery 2013;3:1122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harbour JW, Roberson ED, Anbunathan H, Onken MD, Worley LA, Bowcock AM. Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nature genetics 2013;45:133–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin M, Masshofer L, Temming P, Rahmann S, Metz C, Bornfeld N, et al. Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nature genetics 2013;45:933–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hintzsche JD, Gorden NT, Amato CM, Kim J, Wuensch KE, Robinson SE, et al. Whole-exome sequencing identifies recurrent SF3B1 R625 mutation and comutation of NF1 and KIT in mucosal melanoma. Melanoma Res 2017;27:189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seiler M, Peng S, Agrawal AA, Palacino J, Teng T, Zhu P, et al. Somatic Mutational Landscape of Splicing Factor Genes and Their Functional Consequences across 33 Cancer Types. Cell reports 2018;23:282–96 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malcovati L, Karimi M, Papaemmanuil E, Ambaglio I, Jadersten M, Jansson M, et al. SF3B1 mutation identifies a distinct subset of myelodysplastic syndrome with ring sideroblasts. Blood 2015;126:233–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quesada V, Conde L, Villamor N, Ordóñez G, Jares P, Bassaganyas L, et al. Exome sequencing identifies recurrent mutations of the splicing factor SF3B1 gene in chronic lymphocytic leukemia. Nature genetics 2012;44:47–52. [DOI] [PubMed] [Google Scholar]
- 34.Wan Y, Wu CJ. SF3B1 mutations in chronic lymphocytic leukemia. Blood 2013;121:4627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3’ Splice Site Selection through Use of a Different Branch Point. Cell reports 2015;13:1033–45. [DOI] [PubMed] [Google Scholar]
- 36.DeBoever C, Ghia EM, Shepard PJ, Rassenti L, Barrett CL, Jepsen K, et al. Transcriptome sequencing reveals potential mechanism of cryptic 3’ splice site selection in SF3B1-mutated cancers. PLoS computational biology 2015;11:e1004105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obeng EA, Chappell RJ, Seiler M, Chen MC, Campagna DR, Schmidt PJ, et al. Physiologic Expression of Sf3b1(K700E) Causes Impaired Erythropoiesis, Aberrant Splicing, and Sensitivity to Therapeutic Spliceosome Modulation. Cancer cell 2016;30:404–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Darman RB, Seiler M, Agrawal AA, Lim KH, Peng S, Aird D, et al. Cancer-Associated SF3B1 Hotspot Mutations Induce Cryptic 3? Splice Site Selection through Use of a Different Branch Point. Cell reports 2015;13:1033–45. [DOI] [PubMed] [Google Scholar]
- 39.Tang Q, Rodriguez-Santiago S, Wang J, Pu J, Yuste A, Gupta V, et al. SF3B1/Hsh155 HEAT motif mutations affect interaction with the spliceosomal ATPase Prp5, resulting in altered branch site selectivity in pre-mRNA splicing. Genes Dev 2016;30:2710–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carrocci TJ, Zoerner DM, Paulson JC, Hoskins AA. SF3b1 mutations associated with myelodysplastic syndromes alter the fidelity of branchsite selection in yeast. Nucleic acids research 2017;45:4837–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cretu C, Schmitzova J, Ponce-Salvatierra A, Dybkov O, De Laurentiis EI, Sharma K, et al. Molecular Architecture of SF3b and Structural Consequences of Its Cancer-Related Mutations. Molecular cell 2016;64:307–19. [DOI] [PubMed] [Google Scholar]
- 42.Brooks AN, Choi PS, de Waal L, Sharifnia T, Imielinski M, Saksena G, et al. A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events. PLoS One 2014;9:e87361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ilagan JO, Ramakrishnan A, Hayes B, Murphy ME, Zebari AS, Bradley P, et al. U2AF1 mutations alter splice site recognition in hematological malignancies. Genome research 2015;25:14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Daubner GM, Clery A, Jayne S, Stevenin J, Allain FH. A syn-anti conformational difference allows SRSF2 to recognize guanines and cytosines equally well. EMBO J 2012;31:162–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim E, Ilagan JO, Liang Y, Daubner GM, Lee SC, Ramakrishnan A, et al. SRSF2 Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer cell 2015;27:617–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang J, Lieu YK, Ali AM, Penson A, Reggio KS, Rabadan R, et al. Disease-associated mutation in SRSF2 misregulates splicing by altering RNA-binding affinities. Proc Natl Acad Sci U S A 2015;112:E4726–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tarn WY, Steitz JA. A novel spliceosome containing U11, U12, and U5 snRNPs excises a minor class (AT-AC) intron in vitro. Cell 1996;84:801–11. [DOI] [PubMed] [Google Scholar]
- 48.Hall SL, Padgett RA. Conserved sequences in a class of rare eukaryotic nuclear introns with non-consensus splice sites. J Mol Biol 1994;239:357–65. [DOI] [PubMed] [Google Scholar]
- 49.Madan V, Kanojia D, Li J, Okamoto R, Sato-Otsubo A, Kohlmann A, et al. Aberrant splicing of U12-type introns is the hallmark of ZRSR2 mutant myelodysplastic syndrome. Nat Commun 2015;6:6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Abelson S, Collord G, Ng SWK, Weissbrod O, Mendelson Cohen N, Niemeyer E, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature 2018;559:400–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Desai P, Mencia-Trinchant N, Savenkov O, Simon MS, Cheang G, Lee S, et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nature medicine 2018;24:1015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou Q, Derti A, Ruddy D, Rakiec D, Kao I, Lira M, et al. A chemical genetics approach for the functional assessment of novel cancer genes. Cancer research 2015;75:1949–58. [DOI] [PubMed] [Google Scholar]
- 53.Fei DL, Motowski H, Chatrikhi R, Prasad S, Yu J, Gao S, et al. Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and Is Required for Cell Survival. PLoS Genet 2016;12:e1006384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shirai CL, Ley JN, White BS, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer cell 2015;27:631–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Papaemmanuil E, Gerstung M, Malcovati L, Tauro S, Gundem G, Van Loo P, et al. Clinical and biological implications of driver mutations in myelodysplastic syndromes. Blood 2013;122:3616–27; quiz 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji X, Zhou Y, Pandit S, Huang J, Li H, Lin CY, et al. SR proteins collaborate with 7SK and promoter-associated nascent RNA to release paused polymerase. Cell 2013;153:855–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen L, Chen JY, Huang YJ, Gu Y, Qiu J, Qian H, et al. The Augmented R-Loop Is a Unifying Mechanism for Myelodysplastic Syndromes Induced by High-Risk Splicing Factor Mutations. Molecular cell 2018;69:412–25 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nguyen HD, Yadav T, Giri S, Saez B, Graubert TA, Zou L. Functions of Replication Protein A as a Sensor of R Loops and a Regulator of RNaseH1. Molecular cell 2017;65:832–47 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nguyen HD, Leong WY, Li W, Reddy PNG, Sullivan JD, Walter MJ, et al. Spliceosome Mutations Induce R Loop-Associated Sensitivity to ATR Inhibition in Myelodysplastic Syndromes. Cancer research 2018;78:5363–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park SM, Ou J, Chamberlain L, Simone TM, Yang H, Virbasius CM, et al. U2AF35(S34F) Promotes Transformation by Directing Aberrant ATG7 Pre-mRNA 3’ End Formation. Molecular cell 2016;62:479–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palangat M, Anastasakis DG, Fei DL, Lindblad KE, Bradley R, Hourigan CS, et al. The splicing factor U2AF1 contributes to cancer progression through a noncanonical role in translation regulation. Genes Dev 2019;33:482–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Urbanski LM, Leclair N, Anczukow O. Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip Rev RNA 2018;9:e1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Anczuków O, Rosenberg AZ, Akerman M, Das S, Zhan L, Karni R, et al. The splicing factor SRSF1 regulates apoptosis and proliferation to promote mammary epithelial cell transformation. Nature structural & molecular biology 2012;19:220–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Karni R, de Stanchina E, Lowe S, Sinha R, Mu D, Krainer A. The gene encoding the splicing factor SF2/ASF is a proto-oncogene. Nature structural & molecular biology 2007;14:185–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Malouf GG, Su X, Yao H, Gao J, Xiong L, He Q, et al. Next-generation sequencing of translocation renal cell carcinoma reveals novel RNA splicing partners and frequent mutations of chromatin-remodeling genes. Clin Cancer Res 2014;20:4129–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian B, Manley JL. Alternative polyadenylation of mRNA precursors. Nat Rev Mol Cell Biol 2017;18:18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lianoglou S, Garg V, Yang JL, Leslie CS, Mayr C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev 2013;27:2380–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaida D, Berg MG, Younis I, Kasim M, Singh LN, Wan L, et al. U1 snRNP protects pre-mRNAs from premature cleavage and polyadenylation. Nature 2010;468:664–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Derti A, Garrett-Engele P, Macisaac KD, Stevens RC, Sriram S, Chen R, et al. A quantitative atlas of polyadenylation in five mammals. Genome research 2012;22:1173–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sandberg R, Neilson JR, Sarma A, Sharp PA, Burge CB. Proliferating cells express mRNAs with shortened 3’ untranslated regions and fewer microRNA target sites. Science 2008;320:1643–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wiestner A, Tehrani M, Chiorazzi M, Wright G, Gibellini F, Nakayama K, et al. Point mutations and genomic deletions in CCND1 create stable truncated cyclin D1 mRNAs that are associated with increased proliferation rate and shorter survival. Blood 2007;109:4599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mayr C, Bartel DP. Widespread shortening of 3’UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 2009;138:673–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Akman BH, Can T, Erson-Bensan AE. Estrogen-induced upregulation and 3’-UTR shortening of CDC6. Nucleic acids research 2012;40:10679–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Park HJ, Ji P, Kim S, Xia Z, Rodriguez B, Li L, et al. 3’ UTR shortening represses tumor-suppressor genes in trans by disrupting ceRNA crosstalk. Nat Genet 2018;50:783–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Berkovits BD, Mayr C. Alternative 3’ UTRs act as scaffolds to regulate membrane protein localization. Nature 2015;522:363–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Xia Z, Donehower LA, Cooper TA, Neilson JR, Wheeler DA, Wagner EJ, et al. Dynamic analyses of alternative polyadenylation from RNA-seq reveal a 3’-UTR landscape across seven tumour types. Nat Commun 2014;5:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Singh I, Lee SH, Sperling AS, Samur MK, Tai YT, Fulciniti M, et al. Widespread intronic polyadenylation diversifies immune cell transcriptomes. Nat Commun 2018;9:1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lee SH, Singh I, Tisdale S, Abdel-Wahab O, Leslie CS, Mayr C. Widespread intronic polyadenylation inactivates tumour suppressor genes in leukaemia. Nature 2018;561:127–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Takagaki Y, Seipelt RL, Peterson ML, Manley JL. The polyadenylation factor CstF-64 regulates alternative processing of IgM heavy chain pre-mRNA during B cell differentiation. Cell 1996;87:941–52. [DOI] [PubMed] [Google Scholar]
- 80.Lu F, Gladden AB, Diehl JA. An alternatively spliced cyclin D1 isoform, cyclin D1b, is a nuclear oncogene. Cancer research 2003;63:7056–61. [PubMed] [Google Scholar]
- 81.Solomon DA, Wang Y, Fox SR, Lambeck TC, Giesting S, Lan Z, et al. Cyclin D1 splice variants. Differential effects on localization, RB phosphorylation, and cellular transformation. The Journal of biological chemistry 2003;278:30339–47. [DOI] [PubMed] [Google Scholar]
- 82.Mroczek S, Chlebowska J, Kulinski TM, Gewartowska O, Gruchota J, Cysewski D, et al. The non-canonical poly(A) polymerase FAM46C acts as an onco-suppressor in multiple myeloma. Nat Commun 2017;8:619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhu YX, Shi CX, Bruins LA, Jedlowski P, Wang X, Kortum KM, et al. Loss of FAM46C Promotes Cell Survival in Myeloma. Cancer Res 2017;77:4317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang J, Manley JL. Misregulation of pre-mRNA alternative splicing in cancer. Cancer discovery 2013;3:1228–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bielski CM, Donoghue MTA, Gadiya M, Hanrahan AJ, Won HH, Chang MT, et al. Widespread Selection for Oncogenic Mutant Allele Imbalance in Cancer. Cancer cell 2018;34:852–62 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lee SC, North K, Kim E, Jang E, Obeng E, Lu SX, et al. Synthetic Lethal and Convergent Biological Effects of Cancer-Associated Spliceosomal Gene Mutations. Cancer cell 2018;34:225–41 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lee SC, Dvinge H, Kim E, Cho H, Micol JB, Chung YR, et al. Modulation of splicing catalysis for therapeutic targeting of leukemia with mutations in genes encoding spliceosomal proteins. Nat Med 2016;22:672–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010;463:364–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015;525:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Koh CM, Bezzi M, Low DH, Ang WX, Teo SX, Gay FP, et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015;523:96–100. [DOI] [PubMed] [Google Scholar]
- 91.Clower CV, Chatterjee D, Wang Z, Cantley LC, Vander Heiden MG, Krainer AR. The alternative splicing repressors hnRNP A1/A2 and PTB influence pyruvate kinase isoform expression and cell metabolism. Proc Natl Acad Sci U S A 2010;107:1894–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hsu TY, Simon LM, Neill NJ, Marcotte R, Sayad A, Bland CS, et al. The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 2015;525:384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Paolella BR, Gibson WJ, Urbanski LM, Alberta JA, Zack TI, Bandopadhayay P, et al. Copy-number and gene dependency analysis reveals partial copy loss of wild-type SF3B1 as a novel cancer vulnerability. eLife 2017;6. [DOI] [PMC free article] [PubMed]
- 94.Viswanathan SR, Nogueira MF, Buss CG, Krill-Burger JM, Wawer MJ, Malolepsza E, et al. Genome-scale analysis identifies paralog lethality as a vulnerability of chromosome 1p loss in cancer. Nat Genet 2018;50:937–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kaida D, Motoyoshi H, Tashiro E, Nojima T, Hagiwara M, Ishigami K, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol 2007;3:576–83. [DOI] [PubMed] [Google Scholar]
- 96.Kotake Y, Sagane K, Owa T, Mimori-Kiyosue Y. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nature chemical biology 2007;3:570–5. [DOI] [PubMed] [Google Scholar]
- 97.Yokoi A, Kotake Y, Takahashi K, Kadowaki T, Matsumoto Y, Minoshima Y, et al. Biological validation that SF3b is a target of the antitumor macrolide pladienolide. FEBS J 2011;278:4870–80. [DOI] [PubMed] [Google Scholar]
- 98.Teng T, Tsai JH, Puyang X, Seiler M, Peng S, Prajapati S, et al. Splicing modulators act at the branch point adenosine binding pocket defined by the PHF5A-SF3b complex. Nat Commun 2017;8:15522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Finci LI, Zhang X, Huang X, Zhou Q, Tsai J, Teng T, et al. The cryo-EM structure of the SF3b spliceosome complex bound to a splicing modulator reveals a pre-mRNA substrate competitive mechanism of action. Genes Dev 2018;32:309–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cretu C, Agrawal AA, Cook A, Will CL, Fekkes P, Smith PG, et al. Structural Basis of Splicing Modulation by Antitumor Macrolide Compounds. Molecular cell 2018;70:265–73 e8. [DOI] [PubMed] [Google Scholar]
- 101.Seiler M, Yoshimi A, Darman R, Chan B, Keaney G, Thomas M, et al. H3B-8800, an orally available small-molecule splicing modulator, induces lethality in spliceosome-mutant cancers. Nature medicine 2018;24:497–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang E, Lu SX, Pastore A, Chen X, Imig J, Chun-Wei Lee S, et al. Targeting an RNA-Binding Protein Network in Acute Myeloid Leukemia. Cancer cell 2019. p. 369–84 e7. [DOI] [PMC free article] [PubMed]
- 103.Shirai CL, White BS, Tripathi M, Tapia R, Ley JN, Ndonwi M, et al. Mutant U2AF1-expressing cells are sensitive to pharmacological modulation of the spliceosome. Nat Commun 2017;8:14060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Aird D, Teng T, Huang CL, Pazolli E, Banka D, Cheung-Ong K, et al. Sensitivity to splicing modulation of BCL2 family genes defines cancer therapeutic strategies for splicing modulators. Nat Commun 2019;10:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ten Hacken E, Valentin R, Regis FFD, Sun J, Yin S, Werner L, et al. Splicing modulation sensitizes chronic lymphocytic leukemia cells to venetoclax by remodeling mitochondrial apoptotic dependencies. JCI Insight 2018;3. [DOI] [PMC free article] [PubMed]
- 106.Eskens FA, Ramos FJ, Burger H, O’Brien JP, Piera A, de Jonge MJ, et al. Phase I pharmacokinetic and pharmacodynamic study of the first-in-class spliceosome inhibitor E7107 in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 2013;19:6296–304. [DOI] [PubMed] [Google Scholar]
- 107.Hong DS, Kurzrock R, Naing A, Wheler JJ, Falchook GS, Schiffman JS, et al. A phase I, open-label, single-arm, dose-escalation study of E7107, a precursor messenger ribonucleic acid (pre-mRNA) splicesome inhibitor administered intravenously on days 1 and 8 every 21 days to patients with solid tumors. Invest New Drugs 2014;32:436–44. [DOI] [PubMed] [Google Scholar]
- 108.Han T, Goralski M, Gaskill N, Capota E, Kim J, Ting TC, et al. Anticancer sulfonamides target splicing by inducing RBM39 degradation via recruitment to DCAF15. Science 2017;356. [DOI] [PubMed]
- 109.Uehara T, Minoshima Y, Sagane K, Sugi NH, Mitsuhashi KO, Yamamoto N, et al. Selective degradation of splicing factor CAPERalpha by anticancer sulfonamides. Nat Chem Biol 2017;13:675–80. [DOI] [PubMed] [Google Scholar]
- 110.Lu G, Middleton RE, Sun H, Naniong M, Ott CJ, Mitsiades CS, et al. The myeloma drug lenalidomide promotes the cereblon-dependent destruction of Ikaros proteins. Science 2014;343:305–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Kronke J, Udeshi ND, Narla A, Grauman P, Hurst SN, McConkey M, et al. Lenalidomide causes selective degradation of IKZF1 and IKZF3 in multiple myeloma cells. Science 2014;343:301–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kronke J, Fink EC, Hollenbach PW, MacBeth KJ, Hurst SN, Udeshi ND, et al. Lenalidomide induces ubiquitination and degradation of CK1alpha in del(5q) MDS. Nature 2015;523:183–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Stepanyuk GA, Serrano P, Peralta E, Farr CL, Axelrod HL, Geralt M, et al. UHM-ULM interactions in the RBM39-U2AF65 splicing-factor complex. Acta Crystallogr D Struct Biol 2016;72:497–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Loerch S, Maucuer A, Manceau V, Green MR, Kielkopf CL. Cancer-relevant splicing factor CAPERalpha engages the essential splicing factor SF3b155 in a specific ternary complex. The Journal of biological chemistry 2014;289:17325–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Assi R, Kantarjian HM, Kadia TM, Pemmaraju N, Jabbour E, Jain N, et al. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 2018;124:2758–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Talbot DC, von Pawel J, Cattell E, Yule SM, Johnston C, Zandvliet AS, et al. A randomized phase II pharmacokinetic and pharmacodynamic study of indisulam as second-line therapy in patients with advanced non-small cell lung cancer. Clin Cancer Res 2007;13:1816–22. [DOI] [PubMed] [Google Scholar]
- 117.Smyth JF, Aamdal S, Awada A, Dittrich C, Caponigro F, Schoffski P, et al. Phase II study of E7070 in patients with metastatic melanoma. Ann Oncol 2005;16:158–61. [DOI] [PubMed] [Google Scholar]
- 118.Tacke R, Chen Y, Manley JL. Sequence-specific RNA binding by an SR protein requires RS domain phosphorylation: creation of an SRp40-specific splicing enhancer. Proc Natl Acad Sci U S A 1997;94:1148–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xiao SH, Manley JL. Phosphorylation-dephosphorylation differentially affects activities of splicing factor ASF/SF2. EMBO J 1998;17:6359–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Cho S, Hoang A, Sinha R, Zhong XY, Fu XD, Krainer AR, et al. Interaction between the RNA binding domains of Ser-Arg splicing factor 1 and U1–70K snRNP protein determines early spliceosome assembly. Proc Natl Acad Sci U S A 2011;108:8233–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Meister G, Eggert C, Buhler D, Brahms H, Kambach C, Fischer U. Methylation of Sm proteins by a complex containing PRMT5 and the putative U snRNP assembly factor pICln. Curr Biol 2001;11:1990–4. [DOI] [PubMed] [Google Scholar]
- 122.Brahms H, Meheus L, de Brabandere V, Fischer U, Luhrmann R. Symmetrical dimethylation of arginine residues in spliceosomal Sm protein B/B’ and the Sm-like protein LSm4, and their interaction with the SMN protein. RNA 2001;7:1531–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Banasavadi-Siddegowda YK, Welker AM, An M, Yang X, Zhou W, Shi G, et al. PRMT5 as a druggable target for glioblastoma therapy. Neuro Oncol 2018;20:753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yamauchi T, Masuda T, Canver MC, Seiler M, Semba Y, Shboul M, et al. Genome-wide CRISPR-Cas9 Screen Identifies Leukemia-Specific Dependence on a Pre-mRNA Metabolic Pathway Regulated by DCPS. Cancer cell 2018;33:386–400 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gogliotti RG, Cardona H, Singh J, Bail S, Emery C, Kuntz N, et al. The DcpS inhibitor RG3039 improves survival, function and motor unit pathologies in two SMA mouse models. Hum Mol Genet 2013;22:4084–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Smart AC, Margolis CA, Pimentel H, He MX, Miao D, Adeegbe D, et al. Intron retention is a source of neoepitopes in cancer. Nature biotechnology 2018;36:1056–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boutz PL, Bhutkar A, Sharp PA. Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 2015;29:63–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Poulikakos PI, Persaud Y, Janakiraman M, Kong X, Ng C, Moriceau G, et al. RAF inhibitor resistance is mediated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011;480:387–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Sotillo E, Barrett DM, Black KL, Bagashev A, Oldridge D, Wu G, et al. Convergence of Acquired Mutations and Alternative Splicing of CD19 Enables Resistance to CART-19 Immunotherapy. Cancer discovery 2015;5:1282–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Sammeth M, Foissac S, Guigo R. A general definition and nomenclature for alternative splicing events. PLoS computational biology 2008;4:e1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]