Abstract
Chemical modification of nucleotide bases in DNA provides one mechanism for conveying added information to the genetic code. 5-methylcytosine (5mC), represents the most common chemically modified base in eukaryotic genomes. Sometimes referred to simply as DNA methylation, in eukaryotes, 5mC is most prevalent at CpG dinucleotides and is frequently associated with transcriptional repression of transposable elements. However, 5mC levels and distributions are variable across phylogenies, and emerging evidence suggests functions for DNA methylation may be more diverse and complex than previously appreciated. Here, we summarize our current understanding of DNA methylation profiles and functions in different eukaryotic lineages.
GLOSSARY: 5-methylcytosine, DNA methylation, DNA methyltransferases, heterochromatin, gene body DNA methylation, DNMT, genomic imprinting, genome integrity
Expanding roles of DNA methylation throughout eukaryotes:
DNA methylation has been the subject of intense investigation for decades. Interest in this modification was stimulated in the 1970s, when it was proposed that DNA methylation might be a mechanism for controlling multicellular development, though at the time there was no experimental evidence to support this idea [1, 2]. Interest in DNA methylation continued to grow following key findings that 5mC is required in certain plant and animal species for proper development, as well as transposon silencing in plants, animals, and some fungi [3, 4]. Additional work uncovered roles for 5mC in mammalian X-chromosome inactivation and mono-allelic expression of imprinted genes in mammals and plants [5–7]. Despite decades of work, however, many key questions about how 5mC is controlled and how this modification functions in eukaryotic genomes remain unanswered. Most early work on DNA methylation was restricted to a handful of model systems, but the emergence of new technologies has facilitated studies of 5mC in diverse organisms and provided new and surprising insights into the control of DNA methylation and its diverse functions in eukaryotes.
Mechanisms for establishment and maintenance of DNA methylation:
DNA methyltransferase enzymes are responsible for formation of 5mC through the transfer of a methyl group from the cofactor S-Adenosyl-L methionine (SAM) to the 5’-position of the cytosine ring in DNA [8]. At the amino acid level, DNA methyltransferases are identified by a series of highly conserved motifs associated with catalytic activity [9]. Most DNA methyltransferase enzymes can be categorized into one of two groups. The first group comprises de novo DNA methyltransferases, which are primarily responsible for establishing 5mC at previously unmethylated sites. The second group includes proteins that function primarily to maintain already established DNA methylation marks during DNA replication [3, 4].
The mechanisms responsible for targeting 5mC establishment to specific sequences are only partially understood. There is evidence to suggest that recruitment by select histone tail modifications, pairing of repetitive sequences, and small RNA pathways can all be involved in guiding the establishment of 5mC [10–12]. In addition, transcription factor binding provides a potent mechanism for shaping the global landscape of 5mC establishment through occlusion of potential DNA methyltransferase target sequences [13–16]. The relative importance of these targeting approaches appears to vary between species, and different mechanisms may be used to direct 5mC to different sequences within the same species.
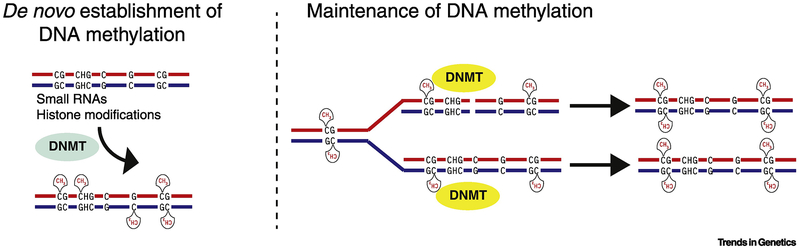
The process of DNA replication presents a challenge for the propagation of methylation states through mitosis. In some cases, this problem may be solved through the de novo reestablishment of 5mC at target sites during each round of cell division [4, 17]. More commonly, maintenance mechanisms promote preservation of 5mC patterns at symmetrical CpG sites following replication (Figure 1). Propagation of methylation at CpGs is achieved by recruitment of maintenance methyltransferases to hemimethylated CpG sites at replication forks [18–20]. This recruitment, in turn drives methylation of reciprocal unmethylated cytosines in the newly synthesized DNA. The high fidelity of this maintenance mechanism may explain why methylation of CpGs predominates over other dinucleotide contexts in most species with methylated genomes.
Figure 1. Schematic diagram of de novo and maintenance DNA methylation.
Methylation of cytosines de novo is independent of existing 5mC and can be targeted to cytosines in CpG and non-CpG contexts. Recruitment of de novo cytosine methyltransferases is regulated by small RNAs and/or specific chromatin modifications. Maintenance methylation involves recognition of hemi-methylated CpG sites generated during DNA replication. Maintenance methyltransferases target CpG sequences on the newly synthesized strand to generate a fully methylated CpG site.
Methods to detect DNA methylation:
Bisulfite sequencing represents the current gold standard method for detection of 5mC in DNA. In this approach, treatment with sodium bisulfite preferentially deaminates unmethylated cytosines in DNA [21, 22]. Deaminated cytosines are subsequently converted to uracil through desulfonation and replaced by thymines during PCR amplification. Methylated cytosines are protected from the bisulfite reaction, allowing their detection through sequencing of the converted, amplified DNA. In the past decade, the coupling of the bisulfite reaction to high-throughput sequencing has made it possible to map genome-wide cytosine methylation states at single-base resolution for any species that has a publicly available reference genome [23,24], and new innovations are extending the power of these approaches to species that lack reference genomes [25]. Although powerful, these types of Whole Genome Bisulfite Sequencing (WGBS) approaches have some limitations. For example, vertebrate genomes also harbor low levels of the modified base 5-hydroxymethylcytosine, which cannot be distinguished from 5mC using standard bisulfite sequencing approaches. It is also important to recognize that in genomes with very low levels of 5mC or extensive 5mC in non-CpG contexts, it may be difficult to distinguish background levels of bisulfite non-conversion from true methylation events. Controls including known methylated and unmethylated DNA standards can provide useful context in these cases.
Diversity of methylation profiles and functions across eukaryotic genomes:
Traditional views of 5mC distribution and function in eukaryotes have been heavily influenced by early analysis of a few species including humans, mouse, the flowering plant Arabidopsis thaliana, and the filamentous fungus Neurospora crassa. Today, WGBS data exists for more than 150 eukaryotic genomes. This wealth of new sequencing data has revealed more extensive taxonomic diversity among methylomes and methyltransferase enzymes than previously appreciated. Below we summarize our current understanding of 5mC distribution and function in different eukaryotic lineages and discuss similarities and differences across species.
Vertebrates:
Vertebrate genomes are extensively methylated, with 5mC detected at more than 70% of CpGs in somatic tissues [26]. Low levels of non-CpG methylation have also been reported in some cellular contexts, most prominently in neurons and embryonic stem cells [27–29]. At the sequence level, transposable elements and satellite repeats near centromeres and telomeres are commonly cited as being highly enriched in 5mC in vertebrate genomes. However, it may be more accurate to describe vertebrate genomes as being methylated at CpGs in all sequences, with two types of exceptions. The first exception is non-methylated islands (NMIs). Located near gene promoters, NMIs represent the only sequence class that consistently escapes DNA methylation in vertebrate species [30]. In mammals, NMIs are often referred to as CpG islands due to their high CpG density, whereas in other vertebrates, NMIs may exhibit significantly lower CpG densities [30, 31]. In general, there is a relatively small fraction of CpG dinucleotides in vertebrate genomes that exhibit dynamic changes in 5mC levels between different tissues and developmental stages [32]. These differentially methylated regions (DMRs) often include binding sites for transcription factors, and their hypomethylation correlates with active transcription of nearby genes.
Cytosine methylation in vertebrates is achieved through a combination of de novo and maintenance methyltransferases (Figure 2). Most vertebrate genomes encode for one maintenance DNA methyltransferase of the Dnmt1 family and a variable number of de novo DNA methyltransferases of the Dnmt3 family. In mammals, two Dnmt3 proteins, Dnmt3a and Dnmt3b, cooperate to establish the bulk of 5mC [33]. An additional Dnmt3 gene, Dnmt3c is specifically found in rodents, and is important for the methylation of young retrotransposons in the male germ line [34, 35]. In addition to these catalytically active de novo methyltransferases, mammalian genomes also encode for a stimulatory cofactor, DNMT3L, which shares homology to other Dnmt3 proteins but lacks key catalytic motifs. This cofactor is critical for de novo methylation in the germline [36, 37]. Other non-mammalian vertebrate lineages appear to lack Dnmt3L orthologs, but exhibit larger expansions of the Dnmt3 de novo methyltransferase family. For example, the zebrafish genome encodes for six Dnmt3 genes [38]. The reasons for this expansion remain unclear. The mechanisms that target vertebrate Dnmt3 enzymes to particular sequences are not completely understood. However, pathways associated with small RNAs called piRNAs have been implicated in establishing 5mC at transposons in germ cells [39]. Histone tail modifications have also been identified as potential modulators of 5mC deposition. For example, Dnmt3 proteins can directly bind H3K36me3 to facilitate de novo methylation[10, 40]. Recent evidence also suggests that methylation may actually be broadly targeted across vertebrate genomes, with protection by bound transcription factors serving as a dominant force driving hypomethylation at DMRs [13–16].
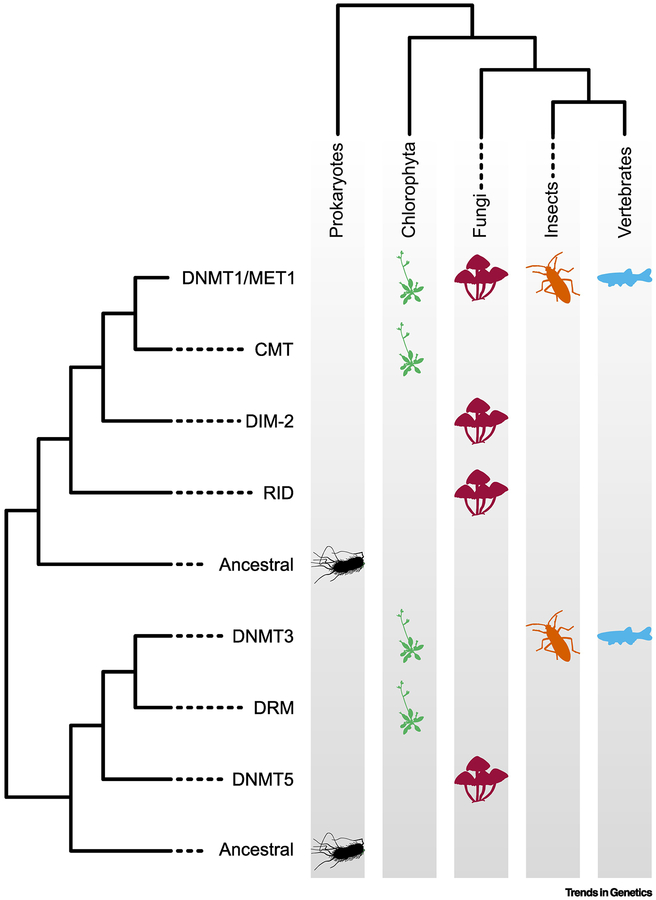
Figure 2: Evolutionary relationship of eukaryotic DNA methyltransferases.
DNMT1 homologs are found in essentially all eukaryotes that utilize 5mC, whereas lineage-specific losses and gains of DNA methyltransferases are found in specific taxa. This phylogeny is a representation and is not applicable to all species within each lineage due to the recurrent loss of DNA methylation machinery. Figure courtesy of Adam Bewick.
Mutation or inhibition of the maintenance DNA methyltransferase Dnmt1 leads to global loss of 5mC and embryonic lethality in all vertebrates tested to date [41–43]. Global loss of 5mC is associated with significant derepression of transposable elements, suggesting that a major function of the vertebrate DNA methyltransferase machinery is to suppress transcription from these parasitic elements [44]. In addition to roles in controlling transposon expression, 5mC has long-been hypothesized to be a key regulator of tissue-specific gene expression. However, while altered gene expression profiles at autosomal, biallelically expressed genes have been noted in vertebrate genomes following 5mC depletion, expression of most genes remains unaffected and not all observed expression changes can be easily attributed to methylation changes at corresponding DMRs [45–47]. Where DNA methylation changes impact transcription, these changes are likely mediated by sequence-specific DNA binding proteins that preferentially bind either methylated or unmethylated recognition sites [13]. In mammals, 5mC is also important for monoallelic expression from imprinted genes with high levels of 5mC typically accumulating on the silenced allele [5, 6]. Similarly, the silent X chromosome of mammalian females is associated with high levels of DNA methylation [7]. Global reprogramming of 5mC patterns is observed in the mammalian germ line and the early embryo. In contrast, the zebrafish methylome does not appear to undergo similar widespread global demethylation during embryogenesis [48, 49].
Insects:
To date, methylomes of more than 40 insect species have been reported, with representation from at least six different orders [26, 50–61]. 5mC is not detected in all insect genomes, and 5mC and the DNA methyltransferases that mediate this mark seem to have been independently lost multiple times in the insect lineage. Most notably, 5mC appears to be absent from the genomes of dipteran insects [50] including the popular laboratory model Drosophila melanogaster. When methylation is present in insects, it predominates in CpG contexts. However, the levels and localization of CpG methylation are quite distinct from vertebrates. Methylation levels in insects are typically much lower than in vertebrate genomes, as most insect genomes exhibit methylation at fewer than 15% of CpGs [50]. 5mC is consistently enriched in exon sequences of expressed insect genes, with genes that possess housekeeping functions most likely to be methylated [56, 57, 62]. Methylation of repetitive sequences is highly variable in the insect lineage. Transposons and other repetitive sequences are not primary targets of DNA methyltransferases in holometabolous insects such as the honey bee, Apis mellifera and the jewel wasp, Nasonia vitripennis, while similar repeats are often methylated in hemimetabolous insects such as the milkweed bug, Oncopeltus fasciatus and the termite, Zootermopsis nevadensis [51, 57]. The mechanistic basis for this distinction is unknown, but it likely involves how DNA methyltransferases are recruited to their targets.
Although 5mC distributions vary significantly between vertebrates and insects, the methyltransferase enzymes that mediate this modification are closely related in both phylogenies. Insect species with 5mC typically encode for one or more maintenance DNA methyltransferase of the Dnmt1 family and at least one de novo methyltransferase with high similarity to vertebrate Dnmt3 proteins (Figure 2) [50]. Duplication of Dnmt1 has occurred in certain families within the Hymenoptera and other lineage-specific duplications have occurred [50]. Curious exceptions have been noted in which only Dnmt1 or Dnmt3 methyltransferase genes can be detected in a given insect genome. This observation raises the possibility that in some insects, DNA methyltransferase enzymes of the Dnmt1 or Dnmt3 family may have developed the capacity to efficiently perform both de novo and maintenance functions. Alternatively, in some cases the loss of Dnmt1 or Dnmt3 orthologs may reflect an intermediate stage in the lineage specific decay or adaptation of the DNA methylation machinery.
To date, there have been few studies addressing the functional roles for DNA methylation and DNA methyltransferases in insects. The lack of tractable insect species for reverse genetics that also harbor 5mC has represented one challenge to such exploration. However, new studies have begun to fill this void. Knockdown of Dnmt1 in the milkweed bug, O. fasciatus, was recently used to successfully reduce genome wide 5mC levels in ovary tissues, providing an experimental framework for assessing function in an insect genome [51]. Affected females produced only limited numbers of poor-quality eggs, which developed abnormally when fertilized. Yet, loss of methylation within transposable elements or genes did not appear to significantly affect their expression [51]. Intriguingly, similar developmental arrest of progeny was also noted following maternal depletion of Dnmt1 in the red flour beetle Tribolium castaneum, even though the Tribolium genome appears to lack 5mC [63]. Together, these two experiments raise the possibility that Dnmt1 may have DNA methylation independent functions in at least some insect species. Future reverse genetic studies of 5mC pathway components will enable testing of hypothesized effects of DNA methylation on gene regulation in insect genomes.
Plants:
5mC has been found in all examined plant species. In plants, DNA methylation is primarily found at transposons and other repetitive sequences. In some angiosperm species, DNA methylation at CpG sites is also common in exons of genes that are moderately and broadly expressed [64–66]. This type of methylation is referred to as gene body methylation (gbM) and is not associated with silencing of gene expression [67, 68]. Although its function, if any, is unknown, angiosperm gene body methylation shares similarities with the exonic DNA methylation of highly conserved housekeeping genes noted in insects [69, 70]. Curiously, gbM has been lost from the genomes of some angiosperm species, further adding to the mysterious nature of this subclass of methylated loci [71].
The composition of DNA methyltransferases present in plants is somewhat similar to animals, in that there are maintenance and de novo enzymes (Figure 2). However, there are also features of the plant DNA methylation machinery that are unique. DNA methyltransferases of the MET 1 family, which are orthologous with Dnmt1 methyltransferases found in animals, are responsible for maintaining methylation at CpG sites in all plant species examined. De novo methyltransferase proteins with strong similarity to animal Dnmt3 enzymes can be detected in the genomes of some early land plants such as mosses [72], but clear Dnmt3 orthologs are absent from the majority of plant species studied to date. Instead, most plants encode for domains rearranged methyltransferases of the DRM2 family. Although related to Dnmt3 proteins, the motifs important for methyltransferase activity are rearranged in DRM2 proteins compared to Dnmt3 enzymes [73, 74]. In addition, unlike Dnmt3 proteins, DRM2 methyltransferases appear similarly effective in catalyzing de novo methylation at CpHpH sites (H = A, C or T) with increased activity at CpG, CpT and CpA sites [74]. In contrast to mammals, targeting of this unique family of de novo DNA methyltransferases appears to be mediated almost exclusively through small RNAs via the RNA directed DNA methylation pathway [4]. These small RNAs are typically 24 nucleotides long and are generated by the repetitive sequences that are ultimately methylated by these enzymes. Plant genomes also possess DNA methyltransferases called chromomethylases, which are mostly responsible for methylating CpHpG sites in repetitive DNA [75]. To date, these methyltransferases have not been found outside of the plant kingdom. Chromomethylases in angiosperm species are recruited to sequences by H3 histone tail methylation at lysine 9 (H3K9me2) [76, 77]. Following recruitment, CMT-dependent methylation is subsequently able to recruit the enzymes that mediate H3K9 methylation to these same sequences [78]. In this way, CMT methyltransferases participate in a feed forward loop that allows long-term propagation of both non-CpG and histone H3K9 methylation at target loci.
The majority of our knowledge regarding the function of DNA methylation in plants is derived from studies in two angiosperm species, Arabidopsis thaliana and Zea mays. Somewhat remarkably, viable A. thaliana plants with significant depletion of methylation in CpG or CpHpG contexts can be isolated, although they do not develop normally. In contrast, significant depletion of 5mC is lethal in Z. mays [79, 80]. In both species, decreases in 5mC levels are associated with increased expression of transposable elements and aberrant expression of some gene loci. However, there is currently only a limited understanding of how these changes drive the development abnormalities observed in 5mC depleted plants. DNA methylation is also involved in genome imprinting in the endosperm of flowering plants [81], though it should be noted that imprinting evolved independently in flowering plants and vertebrates despite mechanistic similarities in both groups.
Fungi:
DNA methylation is absent from genomes of several extensively studied fungi including Saccharomyces cerevisiae, Schizosaccharomyces pombe, and Aspergillus nidulans. However, a recent broad survey of DNA methyltransferases and DNA methylomes from representative fungal species indicates that 5mC is more widespread than previously appreciated in this kingdom, with highest levels of 5mC detected among basidiomycetes [82]. Although gene body methylation is a common feature of animal and plant genomes, it is notably absent in the vast majority of fungal genomes and instead methylation is localized primarily to repeat sequences [82]. In most fungal species where 5mC is observed, methylation of cytosines in all dinucleotide contexts can be detected. Four classes of fungal DNA methyltransferases have been observed: DNMT1, DNMT5, DIM-2 and RID (Figure 2). Fungal Dnmt1 orthologs share strong homology to Dnmt1/MET1 enzymes from plants and animals, suggesting extensive conservation of this maintenance machinery across eukaryotes. Phylogenetic evidence would suggest DNMT5 proteins are likely maintenance enzymes. There is evidence to suggest DIM-2 and RID can function as a de novo methyltransferases, though the methylation capacities of RID homologs are not well defined [83, 84]. As a general rule, ascomycete fungi encode homologs of DIM-2, whereas basidiomycetes encode DNMT1 and DNMT5 homologs, but all possible combinations of DNMTs are observed, presumably due to horizontal transfer of DNMT genes between fungi [82].
To date, most functional studies of DNA methylation in fungi have been performed using the ascomycete fungus N. crassa. Most 5mC in N. crassa is associated with repeat sequences that have been altered by a homology-based genome defense system called repeat-induced point mutation (RIP) [85]. RIP was first identified in N. crassa and involves DNA methylation and subsequent mutation of duplicated sequences in DNA [86, 87]. RIP has been reported to occur in other ascomycete fungi [88–93], and a RIP-like process called MIP (methylation induced premeiotically) methylates but does not mutate repeats during sexual development in the ascomycete Ascobolus immersus [84, 94, 95]. Interestingly, homologs of RID are required for sexual development in A. immersus and Aspergillus nidulans, but the precise role of these proteins in meiotic tissues is not understood [84, 96].
In N. crassa, 5mC in is not limited to symmetrical sites and a mechanism for maintenance methylation does not appear to exist, instead it is likely that 5mC is reestablished de novo at every round of cell division [97–102]. This de novo targeting is mediated primarily by H3K9 methylation of AT-rich targets [99, 101, 103], although antisense transcripts have been implicated in targeting 5mC to certain genes that are GC-rich and present as single-copies [104]. In N. crassa, loss of 5mC is not associated with significant gene expression changes, but 5mC is required to restrict the mobility of functional transposons [105]. 5mC was shown to inhibit transcriptional elongation in N. crassa, but the mechanism is not understood [106].
Concluding Remarks and Future Perspectives:
The ever-increasing availability of eukaryotic methylomes allows for a more holistic analysis of DNA methylation in eukaryotes than could be performed just a decade ago. Still, there are many lineages that remain under or unexplored. For example, analysis of 5mC in the parasitic nematode Trichinella spiralis has recently challenged the long-standing assumption that the nematode lineage lacks this modification [107]. In the ciliate, Oxytricha trifallx, DNA methylation is likely required for DNA elimination supporting function independent of gene regulation [108]. Many other protostome lineages have been similarly neglected and almost nothing is known regarding methylation in cnidaria, porifera and entire groups of protists. These omissions suggest that there is still much to learn regarding the diversity of methylomes and DNA methyltransferase systems in eukaryotes.
There are numerous outstanding questions in this field (Box 1). The question of why there is such outstanding natural diversity in eukaryotic DNA methyltransferases and 5mC distributions remains an open question. The loss of a subset of methylation pathways in addition to the complete loss of methylation in some lineages emphasizes that 5mC is not absolutely required for gene regulation, transposon control or survival in eukaryotes, suggesting that compensatory pathways can support these functions in some contexts. Still, the essential nature of 5mC in vertebrates and Z. mays argues that in other organisms such redundancies are lacking or are insufficient to compensate for newly evolved 5mC requirements. Specialist functions for 5mC such as imprinted gene regulation in mammals and in the endosperm of flowering plants, repeat induced point mutation in N. crassa and gene body methylation in insects and angiosperms suggest that the base functions of 5mC can readily be usurped to support unique requirements in different lineages. Indeed, the lack of 5mC in repetitive sequences of some insect species suggests that common functions for 5mC in transposon control may not always be retained as the predominant 5mC function in species with 5mC. There is clearly much more to learn about 5mC in eukaryotes. However, new methylome data underscores the risks of broadly extrapolating findings regarding DNA methylation and DNA methyltransferase function from one species to the next.
Outsanding Questions:
Is there a specialized role for DNA methylation during meiosis? In mammals, disruption of DNA methylation leads to failed meiosis. Similarly, knockdown of DNMT1 in the milkweed bug, Oncopeltus fasciatus, caused females to cease laying eggs, and eggs that were preduced failed to develop. In fungi, homologs of RID are required for sexual development in two ascomycetes, Ascobolus immersus and Aspergillus nidulans.
Is there a function of gene body DNA methylation in plants and insects? Gene body DNA methylation is found within moderately and constitutively expressed housekeeping genes in flowering plants and insects. It is often evolutionary conserved, yet a well-defined function has yet to be determined.
Why are DNA methyltransferases and DNA methylation lost in numerous independent insect taxa? Curiously some insect species have only lost Dnmt1 or Dnmt3, yet still retain DNA methylation. These findings suggest that additional undiscovered enzymatic activities of DNA methyltransferases might exist or that compensatory mechanisms have evolved to allow the loss of DNA methylation.
Does DNA methylation regulate gene expression beyond transponson silencing in fungi? In N. crassa, a small number of protein-coding genes are associated with DNA methylation directed by a class of small RNAs, and in several basidiomycetes, large methylated domains found on chromosome arms span multiple protein coding genes. These observations raise the possibility that 5mC could regulate gene expression during development or in response to changing environmental conditions.
Highlights:
Although DNA methyltransferase content is generally conserved across eukaryotes, there is exists extensive variation in how this modified base is used for a variety of cellular processes.
The manner in which DNA methyltransferases are recruited to target sequences leads to a diversity of genome-wide DNA methylation patterns between eukaryotes.
Continued exploration of DNA methylation patterns and DNA methyltransferase content in diverse eukaryotic lineages will lead to an expanded understanding of mechanism by which the modified base functions in genomes.
Acknowledgements.
This work was supported by the National Institutes of Health (R01GM110092) to MGG and by the National Science Foundation (MCB-1856143) and by the Technical University of Munich-Institute for Advanced Study funded by the German Excellent Initiative and the European Seventh Framework Programme under grant agreement no. 291763 to RJS. RJS is a Pew Scholar in the Biomedical Sciences, supported by The Pew Charitable Trusts.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References cited
- 1.Holliday R, and Pugh JE (1975). DNA modification mechanisms and gene activity during development. Science 187, 226–232. [PubMed] [Google Scholar]
- 2.Riggs AD (1975). X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet 14, 9–25. [DOI] [PubMed] [Google Scholar]
- 3.Goll MG, and Bestor TH (2005). Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74, 481–514. [DOI] [PubMed] [Google Scholar]
- 4.Law JA, and Jacobsen SE (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11, 204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Elhamamsy AR (2017). Role of DNA methylation in imprinting disorders: an updated review. J Assist Reprod Genet 34, 549–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Satyaki PR, and Gehring M (2017). DNA methylation and imprinting in plants: machinery and mechanisms. Crit Rev Biochem Mol Biol 52, 163–175. [DOI] [PubMed] [Google Scholar]
- 7.Disteche CM, and Berletch JB (2015). X-chromosome inactivation and escape. J Genet 94, 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen L, MacMillan AM, Chang W, Ezaz-Nikpay K, Lane WS, and Verdine GL (1991). Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30, 11018–11025. [DOI] [PubMed] [Google Scholar]
- 9.Posfai J, Bhagwat AS, Posfai G, and Roberts RJ (1989). Predictive motifs derived from cytosine methyltransferases. Nucleic Acids Res 17, 2421–2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dhayalan A, Rajavelu A, Rathert P, Tamas R, Jurkowska RZ, Ragozin S, and Jeltsch A (2010). The Dnmt3a PWWP domain reads histone 3 lysine 36 trimethylation and guides DNA methylation. J Biol Chem 285, 26114–26120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gladyshev E, and Kleckner N (2017). DNA sequence homology induces cytosine-tothymine mutation by a heterochromatin-related pathway in Neurospora. Nat Genet 49, 887–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matzke MA, and Mosher RA (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15, 394–408. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, Wang G, and Qian J (2016). Transcription factors as readers and effectors of DNA methylation. Nat Rev Genet 17, 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domcke S, Bardet AF, Adrian Ginno P, Hartl D, Burger L, and Schubeler D (2015). Competition between DNA methylation and transcription factors determines binding of NRF1. Nature 528, 575–579. [DOI] [PubMed] [Google Scholar]
- 15.Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Scholer A, van Nimwegen E, Wirbelauer C, Oakeley EJ, Gaidatzis D, et al. (2011). DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature 480, 490–495. [DOI] [PubMed] [Google Scholar]
- 16.Krebs AR, Dessus-Babus S, Burger L, and Schubeler D (2014). High-throughput engineering of a mammalian genome reveals building principles of methylation states at CG rich regions. Elife 3, e04094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, Ueda Y, Dodge JE, Wang Z, and Li E (2003). Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol 23, 5594–5605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bostick M, Kim JK, Esteve PO, Clark A, Pradhan S, and Jacobsen SE (2007). UHRF1 plays a role in maintaining DNA methylation in mammalian cells. Science 317, 1760–1764. [DOI] [PubMed] [Google Scholar]
- 19.Woo HR, Pontes O, Pikaard CS, and Richards EJ (2007). VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization. Genes Dev 21, 267–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, et al. (2007). The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature 450, 908–912. [DOI] [PubMed] [Google Scholar]
- 21.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, and Paul CL (1992). A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci U S A 89, 1827–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clark SJ, Harrison J, Paul CL, and Frommer M (1994). High sensitivity mapping of methylated cytosines. Nucleic Acids Res 22, 2990–2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD, Pradhan S, Nelson SF, Pellegrini M, and Jacobsen SE (2008). Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lister R, O’Malley RC, Tonti-Filippini J, Gregory BD, Berry CC, Millar AH, and Ecker JR (2008). Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133, 523–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bewick AJ, Hofmeister BT, Lee K, Zhang X, Hall DW, and Schmitz RJ (2015). FASTmC: A Suite of Predictive Models for Nonreference-Based Estimations of DNA Methylation. G3 6, 447–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng S, Cokus SJ, Zhang X, Chen PY, Bostick M, Goll MG, Hetzel J, Jain J, Strauss SH, Halpern ME, et al. (2010). Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A 107, 8689–8694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ziller MJ, Muller F, Liao J, Zhang Y, Gu H, Bock C, Boyle P, Epstein CB, Bernstein BE, Lengauer T, et al. (2011). Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 7, e1002389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo JU, Su Y, Shin JH, Shin J, Li H, Xie B, Zhong C, Hu S, Le T, Fan G, et al. (2014). Distribution, recognition and regulation of non-CpG methylation in the adult mammalian brain. Nat Neurosci 17, 215–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, and Ecker JR (2015). Non-CG Methylation in the Human Genome. Annu Rev Genomics Hum Genet 16, 55–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grutzner F, Odom DT, Patient R, Ponting CP, et al. (2013). Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2, e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bird A (1987). CpG islands as gene markers in the vertebrate nucleus. Trends Genet 3, 342–347. [Google Scholar]
- 32.Luo C, Hajkova P, and Ecker JR (2018). Dynamic DNA methylation: In the right place at the right time. Science 361, 1336–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okano M, Bell DW, Haber DA, and Li E (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99, 247–257. [DOI] [PubMed] [Google Scholar]
- 34.Jain D, Meydan C, Lange J, Claeys Bouuaert C, Lailler N, Mason CE, Anderson KV, and Keeney S (2017). rahu is a mutant allele of Dnmt3c, encoding a DNA methyltransferase homolog required for meiosis and transposon repression in the mouse male germline. PLoS Genet 13, e1006964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Herault Y, Guillou F, and Bourc’his D (2016). The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Science 354, 909–912. [DOI] [PubMed] [Google Scholar]
- 36.Bourc’his D, Xu GL, Lin CS, Bollman B, and Bestor TH (2001). Dnmt3L and the establishment of maternal genomic mprints. Science 294, 2536–2539. [DOI] [PubMed] [Google Scholar]
- 37.Bourc’his D, and Bestor TH (2004). Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431, 96–99. [DOI] [PubMed] [Google Scholar]
- 38.Goll MG, and Halpern ME (2011). DNA methylation in zebrafish. Prog Mol Biol Transl Sci 101, 193–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, and Hannon GJ (2008). A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31, 785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baubec T, Colombo DF, Wirbelauer C, Schmidt J, Burger L, Krebs AR, Akalin A, and Schubeler D (2015). Genomic profiling of DNA methyltransferases reveals a role for DNMT3B in genic methylation. Nature 520, 243–247. [DOI] [PubMed] [Google Scholar]
- 41.Anderson RM, Bosch JA, Goll MG, Hesselson D, Dong PD, Shin D, Chi NC,Shin CH, Schlegel A, Halpern M, et al. (2009). Loss of Dnmt1 catalytic activity reveals multiple roles for DNA methylation during pancreas development and regeneration. Dev Biol 334, 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stancheva I, and Meehan RR (2000). Transient depletion of xDnmt1 leads to premature gene activation in Xenopus embryos. Genes Dev 14, 313–327. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 43.Li E, Bestor TH, and Jaenisch R (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell 69, 915–926. [DOI] [PubMed] [Google Scholar]
- 44.Yoder JA, Walsh CP, and Bestor TH (1997). Cytosine methylation and the ecology of intragenomic parasites. Trends Genet 13, 335–340. [DOI] [PubMed] [Google Scholar]
- 45.Jackson-Grusby L, Beard C, Possemato R, Tudor M, Fambrough D, Csankovszki G, Dausman J, Lee P, Wilson C, Lander E, et al. (2001). Loss of genomic methylation causes p53-dependent apoptosis and epigenetic deregulation. Nat Genet 27, 31–39. [DOI] [PubMed] [Google Scholar]
- 46.Chernyavskaya Y, Mudbhary R, Zhang C, Tokarz D, Jacob V, Gopinath S, Sun X, Wang S, Magnani E, Madakashira BP, et al. (2017). Loss of DNA methylation in zebrafish embryos activates retrotransposons to trigger antiviral signaling. Development 144, 2925–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gutierrez-Arcelus M, Lappalainen T, Montgomery SB, Buil A, Ongen H, Yurovsky A, Bryois J, Giger T, Romano L, Planchon A, et al. (2013). Passive and active DNA methylation and the interplay with genetic variation in gene regulation. eLife 2, e00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Potok ME, Nix DA, Parnell TJ, and Cairns BR (2013). Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153, 759–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y, Liu S, Chen X, Cao Y, and Tao Y (2013). Genome-wide distribution of DNA methylation and DNA demethylation and related chromatin regulators in cancer. Biochim Biophys Acta 1835, 155–163. [DOI] [PubMed] [Google Scholar]
- 50.Bewick AJ, Vogel KJ, Moore AJ, and Schmitz RJ (2017). Evolution of DNA Methylation across Insects. Mol Biol Evol 34, 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bewick AJ, Sanchez Z, McKinney EC, Moore AJ, Moore PJ, and Schmitz RJ (2019). Dnmt1 is essential for egg production and embryo viability in the large milkweed bug, Oncopeltus fasciatus. Epigenetics & chromatin 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bonasio R, Li Q, Lian J, Mutti NS, Jin L, Zhao H, Zhang P, Wen P, Xiang H, Ding Y, et al. (2012). Genome-wide and caste-specific DNA methylomes of the ants Camponotus floridanus and Harpegnathos saltator. Curr Biol 22, 1755–1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Libbrecht R, Oxley PR, Keller L, and Kronauer DJ (2016). Robust DNA Methylation in the Clonal Raider Ant Brain. Curr Biol 26, 391–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Glastad KM, Arsenault SV, Vertacnik KL, Geib SM, Kay S, Danforth BN, Rehan SM, Linnen CR, Kocher SD, and Hunt BG (2017). Variation in DNA Methylation Is Not Consistently Reflected by Sociality in Hymenoptera. Genome Biol Evol 9, 1687–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Standage DS, Berens AJ, Glastad KM, Severin AJ, Brendel VP, and Toth AL (2016). Genome, transcriptome and methylome sequencing of a primitively eusocial wasp reveal a greatly reduced DNA methylation system in a social insect. Mol Ecol 25, 1769–1784. [DOI] [PubMed] [Google Scholar]
- 56.Rehan SM, Glastad KM, Lawson SP, and Hunt BG (2016). The Genome and Methylome of a Subsocial Small Carpenter Bee, Ceratina calcarata. Genome Biol Evol 8, 1401–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Glastad KM, Gokhale K, Liebig J, and Goodisman MA (2016). The caste - and sex-specific DNA methylome of the termite Zootermopsis nevadensis. Sci Rep 6, 37110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiang H, Zhu J, Chen Q, Dai F, Li X, Li M, Zhang H, Zhang G, Li D, Dong Y, et al. (2010). Single base-resolution methylome of the silkworm reveals a sparse epigenomic map. Nat Biotechnol 28, 516–520. [DOI] [PubMed] [Google Scholar]
- 59.Wang X, Wheeler D, Avery A, Rago A, Choi JH, Colbourne JK, Clark AG, and Werren JH (2013). Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet 9, e1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cunningham CB, Ji L, Wiberg RA, Shelton J, McKinney EC, Parker DJ, Meagher RB, Benowitz KM, Roy-Zokan EM, Ritchie MG, et al. (2015). The Genome and Methylome of a Beetle with Complex Social Behavior, Nicrophorus vespilloides (Coleoptera: Silphidae). Genome Biol Evol 7, 3383–3396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zemach A, McDaniel IE, Silva P, and Zilberman D (2010). Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science 328, 916–919. [DOI] [PubMed] [Google Scholar]
- 62.Hunt BG, Glastad KM, Yi SV, and Goodisman MA (2013). The function of intragenic DNA methylation: insights from insect epigenomes. Integr Comp Biol 53, 319–328. [DOI] [PubMed] [Google Scholar]
- 63.Schulz NKE, Wagner CI, Ebeling J, Raddatz G, Diddens-de Buhr MF, Lyko F, and Kurtz J (2018). Dnmt1 has an essential function despite the absence of CpG DNA methylation in the red flour beetle Tribolium castaneum. Sci Rep 8, 16462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Niederhuth CE, Bewick AJ, Ji L, Alabady MS, Kim KD, Li Q, Rohr NA, Rambani A, Burke JM, Udall JA, et al. (2016). Widespread natural variation of DNA methylation within angiosperms. Genome Biol 17, 194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bewick AJ, Niederhuth CE, Ji L, Rohr NA, Griffin PT, Leebens-Mack J, and Schmitz RJ (2017). The evolution of CHROMOMETHYLASES and gene body DNA methylation in plants. Genome Biol 18, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bewick AJ, and Schmitz RJ (2017). Gene body DNA methylation in plants. Curr Opin Plant Biol 36, 103–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tran RK, Henikoff JG, Zilberman D, Ditt RF, Jacobsen SE, and Henikoff S (2005). DNA methylation profiling identifies CG methylation clusters in Arabidopsis genes. Curr Biol 15, 154–159. [DOI] [PubMed] [Google Scholar]
- 68.Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. (2006). Genome-wide high-resolution mapping and functional analysis of DNA methylation in arabidopsis. Cell 126, 1189–1201. [DOI] [PubMed] [Google Scholar]
- 69.Takuno S, and Gaut BS (2012). Body-methylated genes in Arabidopsis thaliana are functionally important and evolve slowly. Mol Biol Evol 29, 219–227. [DOI] [PubMed] [Google Scholar]
- 70.Takuno S, and Gaut BS (2013). Gene body methylation is conserved between plant orthologs and is of evolutionary consequence. Proc Natl Acad Sci U S A 110, 1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bewick AJ, Ji L, Niederhuth CE, Willing EM, Hofmeister BT, Shi X, Wang L, Lu Z, Rohr NA, Hartwig B, et al. (2016). On the origin and evolutionary consequences of gene body DNA methylation. Proc Natl Acad Sci U S A 113, 9111–9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yaari R, Katz A, Domb K, Harris KD, Zemach A, and Ohad N (2019). RdDM-independent de novo and heterochromatin DNA methylation by plant CMT and DNMT3 orthologs. Nature communications 10, 1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao X, and Jacobsen SE (2002). Role of the arabidopsis DRM methyltransferases in de novo DNA methylation and gene silencing. Curr Biol 12, 1138–1144. [DOI] [PubMed] [Google Scholar]
- 74.Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, and Jacobsen SE (2014). Molecular mechanis m of action of plant DRM de novo DNA methyltransferases. Cell 157, 1050–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, and Jacobsen SE (2001). Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292, 2077–2080. [DOI] [PubMed] [Google Scholar]
- 76.Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, et al. (2004). Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23, 4286–4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Du J, Zhong X, Bernatavichute YV, Stroud H, Feng S, Caro E, Vashisht AA, Terragni J, Chin HG, Tu A, et al. (2012). Dual binding of chromomethylase domains to H3K9me2-containing nucleosomes directs DNA methylation in plants. Cell 151, 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jackson JP, Lindroth AM, Cao X, and Jacobsen SE (2002). Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature 416, 556–560. [DOI] [PubMed] [Google Scholar]
- 79.Li Q, Eichten SR, Hermanson PJ, Zaunbrecher VM, Song J, Wendt J, Rosenbaum H, Madzima TF, Sloan AE, Huang J, et al. (2014). Genetic perturbation of the maize methylome. Plant Cell 26, 4602–4616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fu FF, Dawe RK, and Gent JI (2018). Loss of RNA-Directed DNA Methylation in Maize Chromomethylase and DDM1-Type Nucleosome Remodeler Mutants. Plant Cell 30, 1617–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gehring M, and Satyaki PR (2017). Endosperm and Imprinting, Inextricably Linked. Plant Physiol 173, 143–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bewick AJ, Hofmeister BT, Powers RA, Mondo SJ, Grigoriev IV, James TY, Stajich JE, and Schmitz RJ (2019). Diversity of cytosine methylation across the fungal tree of life. Nat Ecol Evol 3, 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kouzminova EA, and Selker EU (2001). dim-2 encodes a DNA-methyltransferase responsible for all known cytosine methylation in Neurospora. EMBO Journal 20, 4309–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Malagnac F, Wendel B, Goyon C, Faugeron G, Zickler D, Rossignol JL, Noyer-Weidner M, Vollmayr P, Trautner TA, and Walter J (1997). A gene essential for de novo methylation and development in Ascobolus reveals a novel type of eukaryotic DNA methyltransferase structure. Cell 91, 281–290. [DOI] [PubMed] [Google Scholar]
- 85.Lewis ZA, Honda S, Khlafallah TK, Jeffress JK, Freitag M, Mohn F, Schubeler D, and Selker EU (2009). Relics of repeat-induced point mutation direct heterochromatin formation in Neurospora crassa. Genome research 19, 427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Aramayo R, and Selker EU (2013). Neurospora crassa, a model system for epigenetics research. Cold Spring Harb Perspect Biol 5, a017921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Selker EU, Cambareri EB, Jensen BC, and Haack KR (1987). Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell 51, 741–752. [DOI] [PubMed] [Google Scholar]
- 88.Goldfarb M, Santana MF, Salomao TM, Queiroz MV, and Barros EG (2016). Evidence of ectopic recombination and a repeat-induced point (RIP) mutation in the genome of Sclerotinia sclerotiorum, the agent responsible for white mold. Genet Mol Biol 39, 426–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Rouxel T, Grandaubert J, Hane JK, Hoede C, van de Wouw AP, Couloux A, Dominguez V, Anthouard V, Bally P, Bourras S, et al. (2011). Effector diversification within compartments of the Leptosphaeria maculans genome affected by Repeat-Induced Point mutations. Nat Commun 2, 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Clutterbuck AJ (2011). Genomic evidence of repeat-induced point mutation (RIP) in filamentous ascomycetes. Fungal Genet Biol 48, 306–326. [DOI] [PubMed] [Google Scholar]
- 91.Hamann A, Feller F, and Osiewacz HD (2000). The degenerate DNA transposon Pat and repeat-induced point mutation (RIP) in Podospora anserina. Mol Gen Genet 263, 1061–1069. [DOI] [PubMed] [Google Scholar]
- 92.Nielsen ML, Hermansen TD, and Aleksenko A (2001). A family of DNA repeats in Aspergillus nidulans has assimilated degenerated retrotransposons. Mol Genet Genomics 265, 883–887. [DOI] [PubMed] [Google Scholar]
- 93.Ikeda K, Nakayashiki H, Kataoka T, Tamba H, Hashimoto Y, Tosa Y, and Mayama S (2002). Repeat-induced point mutation (RIP) in Magnaporthe grisea: implications for its sexual cycle in the natural field context. Mol Microbiol 45, 1355–1364. [DOI] [PubMed] [Google Scholar]
- 94.Goyon C, and Faugeron G (1989). Targeted transformation of Ascobolus immersus and de novo methylation of the resulting duplicated DNA sequences. Mol Cell Biol 9, 2818–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rhounim L, Rossignol JL, and Faugeron G (1992). Epimutation of repeated genes in Ascobolus immersus. EMBO J 11, 4451–4457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lee DW, Freitag M, Selker EU, and Aramayo R (2008). A cytosine methyltransferase homologue is essential for sexual development in Aspergillus nidulans. PLoS One 3, e2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Selker EU, Jensen BC, and Richardson GA (1987). A portable signal causing faithful DNA methylation de novo in Neurospora crassa. Science 238, 48–53. [DOI] [PubMed] [Google Scholar]
- 98.Miao VP, Freitag M, and Selker EU (2000). Short TpA-rich segments of the zeta-eta region induce DNA methylation in Neurospora crassa. J Mol Biol 300, 249–273. [DOI] [PubMed] [Google Scholar]
- 99.Tamaru H, and Selker EU (2001). A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature 414, 277–283. [DOI] [PubMed] [Google Scholar]
- 100.Tamaru H, and Selker EU (2003). Synthesis of Signals for De Novo DNA Methylation in Neurospora crassa. Mol Cell Biol 23, 2379–2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Freitag M, Hickey PC, Khlafallah TK, Read ND, and Selker EU (2004). HP1 is essential for DNA methylation in neurospora. Mol Cell 13, 427–434. [DOI] [PubMed] [Google Scholar]
- 102.Honda S, and Selker EU (2008). Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol Cell Biol 28, 6044–6055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, and Selker EU (2003). Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet 34, 75–79. [DOI] [PubMed] [Google Scholar]
- 104.Dang Y, Li L, Guo W, Xue Z, and Liu Y (2013). Convergent transcription induces dynamic DNA methylation at disiRNA loci. PLoS Genet 9, e1003761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhou Y, Cambareri EB, and Kinsey JA (2001). DNA methylation inhibits expressionand transposition of the Neurospora Tad retrotransposon. Mol Genet Genomics 265, 748–754. [DOI] [PubMed] [Google Scholar]
- 106.Rountree MR, and Selker EU (1997). DNA methylation inhibits elongation but not initiation of transcription in Neurospora crassa. Genes & development 11, 2383–2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gao F, Liu X, Wu XP, Wang XL, Gong D, Lu H, Xia Y, Song Y, Wang J, Du J, et al. (2012). Differential DNA methylation in discrete developmental stages of the parasitic nematode Trichinella spiralis. Genome Biol 13, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Bracht JR, Perlman DH, and Landweber LF (2012). Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol 13, R99. [DOI] [PMC free article] [PubMed] [Google Scholar]