ABSTRACT
Tissue fibrosis is a chronic disease driven by persistent fibroblast activation that has recently been linked to epigenetic modifications. Here, we screened a small library of epigenetic small-molecule modulators to identify compounds capable of inhibiting or reversing TGFβ-mediated fibroblast activation. We identified pracinostat, an HDAC inhibitor, as a potent attenuator of lung fibroblast activation and confirmed its efficacy in patient-derived fibroblasts isolated from fibrotic lung tissue. Mechanistically, we found that HDAC-dependent transcriptional repression was an early and essential event in TGFβ-mediated fibroblast activation. Treatment of lung fibroblasts with pracinostat broadly attenuated TGFβ-mediated epigenetic repression and promoted fibroblast quiescence. We confirmed a specific role for HDAC-dependent histone deacetylation in the promoter region of the anti-fibrotic gene PPARGC1A (PGC1α) in response to TGFβ stimulation. Finally, we identified HDAC7 as a key factor whose siRNA-mediated knockdown attenuates fibroblast activation without altering global histone acetylation. Together, these results provide novel mechanistic insight into the essential role HDACs play in TGFβ-mediated fibroblast activation via targeted gene repression.
KEY WORDS: Myofibroblasts, Fibrosis, Epigenetics, Chromatin
Summary: TGFβ, a master regulator of fibrosis, epigenetically represses anti-fibrotic gene expression via HDACs to promote fibroblast activation.
INTRODUCTION
During normal physiological wound healing, fibroblasts become transiently activated into myofibroblasts to aid in tissue repair. Upon successful repair and injury resolution, fibroblasts normally return to their quiescent state (Bainbridge, 2013; Darby et al., 2014). However, during fibrosis, myofibroblasts maintain an activated state resulting in sustained extracellular matrix deposition and remodeling (Gabbiani, 2003; Wynn, 2008). In diseases such as idiopathic pulmonary fibrosis (IPF), a progressive and deadly disease with no curative therapeutic available (Raghu et al., 2011), pathological progression is characterized by persistent deposition of extracellular matrix, ultimately impairing lung function resulting in death (Raghu et al., 2011). Fibroblasts cultured from IPF lung tissue exhibit altered proliferation and excess extracellular matrix (ECM) deposition that persists stably in cell culture (Moodley et al., 2003; Ramos et al., 2001). However, these cells are not clonal or genetically altered, suggesting that the persistent activated state of disease-derived fibroblasts may reflect alterations in their epigenome.
Variations in the epigenome have been shown to accompany fibrosis disease progression in multiple tissues (Coward et al., 2009; Page and Mann, 2015; Yang and Schwartz, 2015). Specifically, diseased lung fibroblasts have been shown to exhibit causative alterations in DNA methylation, histone acetylation and micro (mi)RNAs when compared to their non-diseased counterparts (Coward et al., 2018, 2014, 2009; Huang et al., 2014; Khalil et al., 2015; Lino Cardenas et al., 2013; Liu et al., 2010; Miao et al., 2018; Sanders et al., 2012, 2017; Tang et al., 2013; Xiao et al., 2016; Yang et al., 2014, 2013). Recently, we identified several epigenetic regulators in an siRNA screen of fibroblast activation (Oh et al., 2018). Taken together, these studies demonstrate that fibroblast activation is a complex phenomenon regulated by multiple epigenetic mechanisms. Understanding how epigenetic changes regulate persistent fibroblast activation may lead to the development of novel therapeutic interventions to halt disease progression.
Here, we screened a broad range of small-molecule epigenetic modifiers and evaluated their role in TGFβ-mediated fibroblast activation in vitro. From this screen, we identified a histone deacetylase (HDAC) inhibitor, pracinostat, as a potent attenuator of lung fibroblast activation. We then sought to better understand how TGFβ signaling utilizes HDAC function to disrupt fibroblast homeostasis and initiate and maintain myofibroblast activation. Specifically, we aimed to identify the molecular targets of HDAC inhibition during fibroblast activation and also to identify key HDACs which mediate fibroblast activation. Transcriptomic and chromatin immunoprecipitation (ChIP) analysis revealed that TGFβ signaling acts as a major repressor of gene expression, including repression of multiple known or potential inhibitors of fibroblast activation and fibrosis, and demonstrated that this repressive function of TGFβ is HDAC dependent. Investigation of individual HDACs revealed HDAC7 as a key enzyme facilitating TGFβ regulation of gene expression without globally perturbing histone acetylation. Taken together, these results demonstrate the important role of TGFβ-mediated epigenetic repression via HDACs, and provide important insight into new avenues for inhibiting or reversing fibroblast activation.
RESULTS
Identification of pracinostat as a potent attenuator of human fibroblast activation
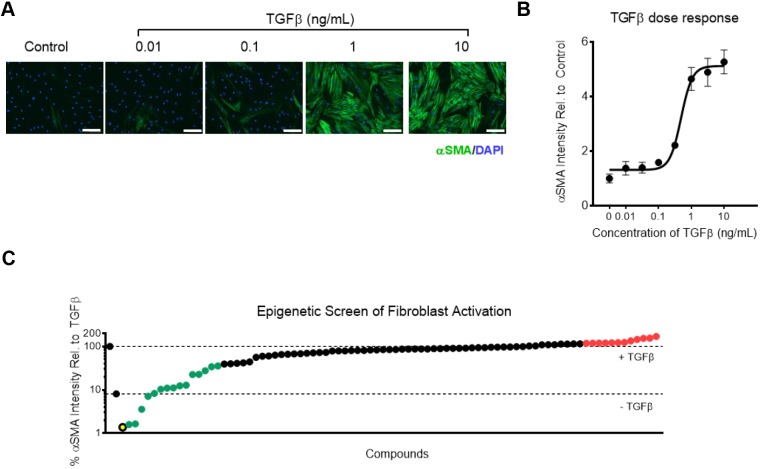
To identify epigenetic regulators of fibroblast activation, we developed an image-based screening assay using α-smooth muscle actin (αSMA) immunofluorescence intensity as a primary outcome. Cytoskeletal αSMA is critical for contractility, and has previously been shown to be an effective metric for evaluating fibroblast activation (Hinz et al., 2001; Oh et al., 2018; Tatler and Jenkins, 2012). Using IMR90 human fetal lung fibroblasts, we validated the range and sensitivity of this assay by quantifying αSMA intensity as a dose response to TGFβ stimulation (Fig. 1A,B). Based on this dose response, TGFβ stimulation at 2 ng/ml was used for the small-molecule screen (Fig. 1B). To evaluate the relative effectiveness of epigenetic small-molecule modulators in regulating fibroblast activation, we tested and rank-ordered 99 modulators of epigenetic regulating enzymes based on their effects on αSMA intensity at concentration of 2.5 μM and 500 nM (Fig. 1C; Tables S1–S3). We tested a broad range of molecules targeting the major classes of epigenetic regulating enzymes, including epigenetic writers, erasers, readers and chromatin remodelers. Screening at both concentrations identified a diverse array of small-molecule inhibitors and activators capable of either reducing or enhancing αSMA intensity (Tables S1–S3). Among the most effective compounds at reducing αSMA were inhibitors of lysine demethylase 1A (SP2509), protein arginine methyltransferase 3 (XY1), and lethal (3) malignant brain tumor-like protein 1 (UNC669). Lysine demethylase 1A acts as both a transcriptional repressor and activator through its ability to demethylate both lysine-4 and -9 of histone 3 (H3K4 and H3K9, respectively). Protein arginine methyltransferase 3 catalyzes mono- and asymmetric di-methylation of arginine residues on proteins and plays a role in ribosomal biosynthesis. Lethal (3) malignant brain tumor-like protein 1 is a member of the polycomb group and reads and binds to methylated lysine-20 on histone 4 (H4K20) resulting in gene repression. The wide variety of enzyme modulators found effective in this screen highlights the likely broad participation of multiple complex epigenetic mechanisms in TGFβ-mediated fibroblast activation. Among those compounds identified as hits in the screen were also several previously identified as being anti-fibrotic, including JQ1, a bromodomain inhibitor whose action attenuates lung fibroblast activation in vitro and experimental lung fibrosis in vivo (Tang et al., 2013), UNC1999, which attenuates liver fibrosis through inhibition of EZH2 (Martin-Mateos et al., 2019), and SIRT1720, which ameliorates experimental renal fibrosis by activating SIRT1 (Ren et al., 2017).
Fig. 1.
Identification of pracinostat as a novel attenuator of human lung fibroblast activation. (A) Immunofluorescence images (10× objective magnification) of IMR90 cells treated with indicated doses of TGFβ for 3 days. Scale bars: 100 µm. (B) αSMA intensity with increasing TGFβ dose in IMR90 cells (n=4 biological replicates). (C) Rank ordered plot of 99 small-molecule modulators of epigenetic regulating enzymes. The screen was performed at 2.5 μM in IMR90 cells in technical triplicates. Compounds shown in green are hits which attenuated fibroblast activation. Compounds shown in red are hits that accentuated fibroblast activation. The compound shown in yellow is pracinostat. Toxic compounds are not shown. Dots represent mean values and error bars represent s.e.m.
The most effective compound at the 2.5 μM dosing, and amongst the most effective at 0.5 μM dosing, was pracinostat, an HDAC inhibitor (panHDAC except HDAC6), which is known to attenuate experimental renal fibrosis (Kang et al., 2017). HDACs primarily function by catalyzing the de-acetylation of histones resulting in gene repression; thus, inhibition of HDACs should result in an increase in transcriptional activity. Interestingly, while there is widespread evidence that HDAC inhibitors are effective in both in vitro and in vivo fibrosis models (Barter et al., 2010; Bombardo et al., 2018; Conforti et al., 2017; Glenisson et al., 2007; Guo et al., 2009; Kang et al., 2017; Khalil et al., 2015; Kim et al., 2018), there is a lack of understanding of how HDAC inhibition reduces fibroblast activation and fibrosis. Moreover, while several HDAC inhibitors were highly effective at reducing αSMA in our screen, others were less so (entinostat and tacedinaline), while still others actually enhanced the level of αSMA (CS-055, RSC133 and TC-H 106), highlighting the fact that specific HDACs and HDAC inhibitors likely act via distinct targets and mechanisms to regulate fibroblast biology. Thus, we focused the remainder of this study on deciphering how HDAC inhibition through pracinostat functions to attenuate fibroblast activation and which specific HDACs are involved.
Pracinostat inhibits IPF-derived fibroblast contractility and ECM deposition
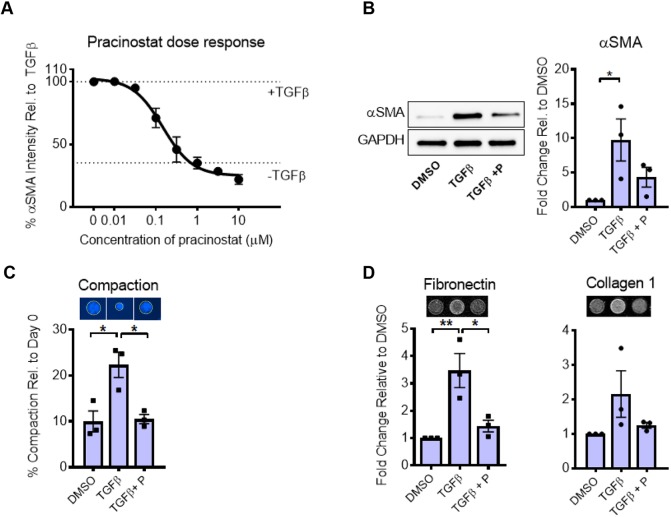
While our screen was performed in IMR90 fetal lung fibroblasts, our ultimate goal is to identify biological mechanisms and targets relevant to human disease. Therefore, to test whether pracinostat is effective in primary human diseased lung fibroblasts, we first performed a dose response study of pracinostat in primary patient-derived IPF fibroblasts stimulated with TGFβ as in our primary screen. These studies confirmed the strong dose-dependent inhibitory effect of pracinostat on αSMA in primary IPF fibroblasts, and revealed an optimally effective dose of pracinostat to be between 1–3 µM (Fig. 2A). Pracinostat also potently diminished the increase of αSMA protein after 24 h of stimulation with TGFβ (Fig. 2B) as determined by western blotting, confirming our immunofluorescence imaging results. While αSMA protein expression is a hallmark of myofibroblast differentiation and linked to cell contractility (Desmouliere et al., 1993; Hinz et al., 2001; Tatler and Jenkins, 2012), we sought to evaluate the broader functional effectiveness of pracinostat in inhibiting myofibroblast phenotypes associated with fibrotic tissue remodeling. To examine whether pracinostat alters fibroblast contractile function, we performed gel compaction assays using collagen gels, and found that pracinostat significantly blocked myofibroblast-mediated gel compaction in the presence of TGFβ (Fig. 2C). To test whether pracinostat also attenuates ECM deposition, we used an antibody-based method to probe for ECM protein deposition. In the presence of TGFβ plus pracinostat, deposition of both fibronectin and collagen I was strongly reduced compared to TGFβ treatment alone (Fig. 2D) demonstrating that ECM deposition by fibroblasts in response to TGFβ is dependent on HDAC function. Thus, pracinostat is broadly effective in inhibiting TGFβ-mediated fibroblast activation including contractile function and extracellular matrix deposition in primary IPF fibroblasts.
Fig. 2.
Pracinostat mitigates IPF fibroblast activation. (A) Dose–response curve of pracinostat inhibition of αSMA intensity in IPF fibroblasts (n=4 biological replicates) with TGFβ present. (B) Representative western blot showing the effect of pracinostat (+P) on TGFβ-mediated induction of αSMA, and quantification from independent replicate experiments (n=3). (C) Collagen gel compaction assay of IPF lung fibroblasts treated with TGFβ and pracinostat for 72 h. Data represent the percentage area compaction of the gel compared to day 0. Representative images are shown, as are quantitative results from independent replicate experiments (n=3). (D) ECM deposition of lung fibroblasts treated with TGFβ and pracinostat for 72 h. Representative images are shown, as are quantitative results from independent replicate experiments (n=3). All experimental replicates were performed with three separate IPF lung fibroblast donor lines. Error bars represent s.e.m. *P<0.05, **P<0.01 (one-way ANOVA with Tukey's multiple comparison test).
HDAC inhibition blocks gene expression changes associated with TGFβ-induced fibroblast activation
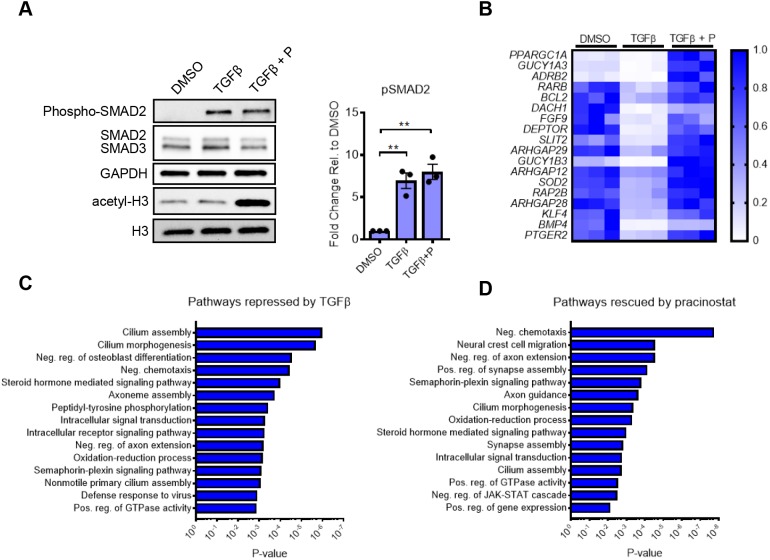
Because HDACs act on a multitude of substrates including non-histone proteins, before testing its effects on gene expression we first tested whether pracinostat directly alters canonical TGFβ signaling. We confirmed that pracinostat did not alter phosphorylated (phospho)-SMAD2 levels upon TGFβ stimulation (Fig. 3A). In contrast, pracinostat strongly increased global histone acetylation at the same time point (45 min), confirming histones as an early direct target of pracinostat (Fig. 3A). Based on these observations, we hypothesized that pracinostat acts primarily by preventing or reversing gene repression in response to TGFβ signaling. To test this concept, we performed RNA sequencing of IPF-derived fibroblasts treated with TGFβ alone or in the presence of pracinostat. We found that TGFβ repressed 2638 genes at 12 h by at least 2-fold [false discovery rate (FDR)<0.1], indicating that a major component of early TGFβ signaling is not only activation of pro-fibrotic genes, but also repression of genes, including potentially putative anti-fibrotic genes that prevent myofibroblast activation or maintain fibroblast quiescence. Remarkably, pracinostat treatment simultaneous with TGFβ prevented the repression of 1103 of these genes (42%), demonstrating that HDACs are critical enzymes mediating TGFβ-dependent gene repression. Examples of putative and confirmed fibrosis suppressing genes repressed by TGFβ and rescued by HDAC inhibition include PGC1α (also known as PPARGC1A), ARHGAP12, ARHGAP29, SOD2, DACH1 and FGF9 (Fig. 3B). PGC1α is a transcriptional co-activator responsible for regulating genes required for mitochondrial biogenesis and cell energy expenditure, and is protective in experimental lung fibrosis (Yu et al., 2018). ARHGAP12 and ARHGAP29 are small Rho-binding subfamily members whose function has recently been highlighted as being critical for YAP/TAZ inactivation (Meng et al., 2018), major regulators of fibroblast activation in fibrosis (Liu et al., 2015). SOD2 is a major intracellular antioxidant (Kwak et al., 2015), and oxidative stress is essential to TGFβ-mediated fibroblast activation (Cheresh et al., 2013). DACH1 has been shown to interfere with TGFβ signaling through binding of SMAD4 and NCOR1 (Wu et al., 2003). FGF9 inhibits TGFβ induced upregulation of ACTA2 and collagen 1 in human lung fibroblasts (Joannes et al., 2016). Collectively, these results highlight multiple genes whose function is potentially inhibitory to fibroblast activation, and whose expression is repressed by TGFβ in a HDAC-dependent fashion. More broadly, gene ontology (GO) analysis revealed pracinostat prevented TGFβ-mediated repression of genes involved in negative chemotaxis, oxidation-reduction process, cilium morphogenesis and positive regulation of GTPase activity (Fig. 3C,D). Several of these groups, such as cilium morphogenesis and oxidation–reduction process have been linked to fibroblast activation and tissue fibrosis (Cheresh et al., 2013; Villalobos et al., 2019). Together, these results suggest that TGFβ-mediated repression is an important contributor to lung fibroblast activation, and that HDACs play an essential role in this response.
Fig. 3.
TGFβ-mediated gene repression requires HDAC activity. (A) IPF fibroblasts were stimulated with TGFβ with and without pracinostat (+P) for 45 min. Representative blots and quantification of phospho (p)SMAD2 from independent replicates (n=3) are shown. Blots were quantified relative to total SMAD2. (B) RNA sequencing was performed in IPF lung fibroblasts from three separate donors. IPF lung fibroblasts were treated with TGFβ with and without pracinostat (1 µM) for 12 h. The heatmap shows the expression changes of putative and confirmed fibrosis suppressing mediators that are significantly repressed by TGFβ and reversed by pracinostat (according to color scale on the right). Each row is normalized to itself. Each column, per treatment condition, represents an individual IPF lung fibroblast donor (n=3). (C) Gene Ontology analysis via DAVID6.8 of the 2638 genes which TGFβ down regulates ≥2-fold at 12 h compared to DMSO control. (D) Gene Ontology analysis via DAVID6.8 of the 1103 genes which pracinostat rescued ≥2-fold when compared to the TGFβ treatment. Error bars represent s.e.m. **P<0.01 (one-way ANOVA with Tukey's multiple comparison test).
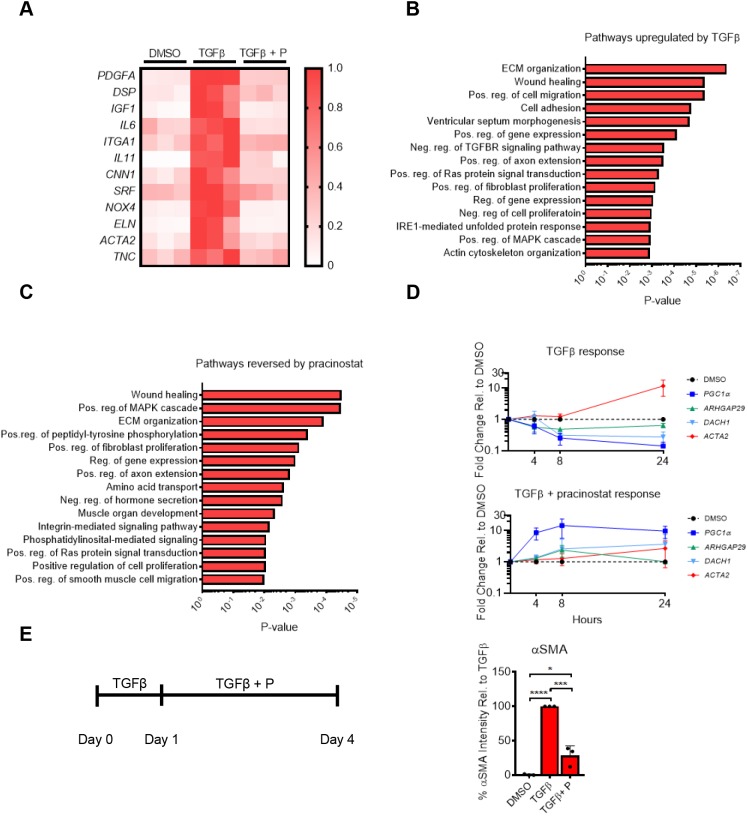
In addition to the broad repressive effects, we found that TGFβ upregulated 505 genes by at least 2-fold compared to control (FDR<0.1). Pracinostat prevented the upregulation of 287 of these genes (57%). Because HDACs function predominantly in gene repression, these effects of pracinostat are likely indirect, reflecting the combined effect of the above programs of genes whose repression by TGFβ are prevented by pracinostat. Prominent examples of well-characterized profibrotic genes whose expression was elevated by TGFβ and reduced by pracinostat include: ACTA2, TNC, IL6, IL11 and PDGFA (Fig. 4A). Many of these genes are strongly linked to myofibroblast activation and fibrotic tissue remodeling (Bhattacharyya et al., 2016; Estany et al., 2014; Henderson and Sheppard, 2013; Hung et al., 2013; Le et al., 2014; Oh et al., 2018; Schafer et al., 2017; Trojanowska, 2008; Tschumperlin et al., 2018), consistent with our prior functional assays demonstrating that pracinostat reduces pro-fibrotic cellular functions. GO analysis using DAVIDv6.8 revealed that pracinostat blocked the TGFβ induction of pro-fibrotic pathways, such as wound healing, positive regulation of MAPK cascade and ECM organization (Fig. 4B,C).
Fig. 4.
TGFβ-induced gene activation requires HDAC activity. RNA-seq was performed in IPF lung fibroblasts from three separate donors. IPF lung fibroblasts were treated with TGFβ with and without pracinostat (1 µM) for 12 h. (A) Heatmap showing expression changes of known pro-fibrotic mediators that are increased significantly by TGFβ and reversed by pracinostat (+P) (according to color scale on the right). Each row is normalized to itself. Each column, per treatment condition, represents an individual IPF lung fibroblast donor. (B) Gene Ontology analysis via DAVID6.8 of the 504 genes that TGFβ upregulates ≥2-fold at 12 h compared to DMSO control. (C) Gene Ontology analysis via DAVID6.8 of the 201 genes which pracinostat reduced ≥2-fold when compared to the TGFβ treatment. (D) Temporal PCR analysis of IPF fibroblasts treated with TGFβ alone or TGFβ and pracinostat (1 µM). Data are normalized to DMSO-treated control at each time point. (E) Immunofluourescence analysis of αSMA intensity in IPF fibroblasts pre-activated with TGFβ for 24 h, then treated with pracinostat (1 µM) and TGFβ for an additional 72 h. Data normalized to DMSO control. All cell culture experiments were performed with cells from three separate IPF lung fibroblast donors. Error bars represent s.e.m. ****P<0.0001, ***P<0.001, *P<0.05 (one-way ANOVA with Tukey's multiple comparison test).
To investigate whether TGFβ-mediated repression of anti-fibrotic genes precedes TGFβ-mediated upregulation of pro-fibrotic genes, we examined the effect of TGFβ on select genes identified in Figs 3B and 4A over time. For anti-fibrotic genes, we selected PGC1α, ARHGAP12 and DACH1 due to their reported anti-fibrotic effect in fibrosis and fibroblasts (Lynch et al., 2018; Peng et al., 2017; Sandner and Stasch, 2017; Yu et al., 2018) in addition to their robust upregulation in the presence of pracinostat (Fig. 3B). For pro-fibrotic genes, we focused on ACTA2. Temporal analysis of gene expression revealed that TGFβ signaling repressed anti-fibrotic genes (PGC1α, ARHGAP12 and DACH1) prior to upregulation of ACTA2 (Fig. 4D). Interestingly, simultaneous treatment with TGFβ and pracinostat resulted in increased expression of these anti-fibrotic genes that preceded the muted TGFβ-mediated upregulation of pro-fibrotic genes (Fig. 4D). These data are consistent with the concept that early TGFβ-mediated gene repression is an essential step necessary for later activation of pro-fibrotic genes and that the prevention of such repression via pracinostat could explain its anti-fibrotic properties. However, such results raise the question of whether targeting of HDAC-mediated repression could be effective when applied to cells that were already activated. We thus sought to investigate whether pracinostat could reverse activation of fibroblasts already stimulated with TGFβ. After 24 h of TGFβ treatment, addition of pracinostat (still in the presence of TGFβ) was able to attenuate activation of fibroblasts when compared to TGFβ treatment alone (Fig. 4E), indicating that HDAC-mediated repression is critical not only for initiation of TGFβ-mediated fibroblast activation, but also maintenance of this activated state.
TGFβ signaling represses PGC1α through histone deacetylation
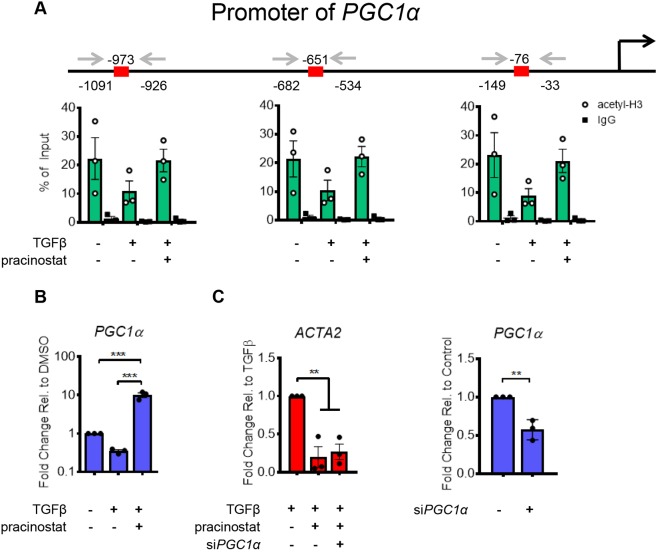
To confirm that TGFβ signaling directly regulates repression of an anti-fibrotic gene through histone alterations, we performed ChIP analyses on specific target histone–DNA interactions in the promoter regions of PGC1α. To identify differences in histone acetylation, we performed PCR analysis following immunoprecipitation of chromatin bound to acetylated histone 3 (acetyl-H3). We found that TGFβ stimulation strongly reduced acetyl-H3 levels along the proximal promoter of PGC1α (Fig. 5A) and pracinostat increased acetyl-H3 at these regions, consistent with the changes we observed in the transcript levels under the same conditions (Fig. 5B). Interestingly, the genomic regions amplified in Fig. 5B also contain putative SMAD2–SMAD3–SMAD4-binding sites, indicating TGFβ downstream signaling molecules could be directing HDAC enzymatic function to these genomic regions.
Fig. 5.
TGFβ mediates histone deacetylation on promoter of PGC1a. (A) ChIP analysis of the PGC1α promoter. Sequences in the promoter were amplified after chromatin immunoprecipitation to observe changes in histone 3 acetylation (acetyl-H3). Species-appropriate IgG was used as the control. Data are normalized, and are given as percentage of input. Red boxes on the genomic track represent putative SMAD2–SMAD3–SMAD4-binding sites according to EPD (Dreos et al., 2017, 2015). (B) PCR validation of HDAC-mediated repression of PGC1α (C) ACTA2 gene expression in IPF fibroblasts treated with TGFβ, pracinostat (1 µM) and siRNA for PGC1a as indicated. PGC1α knockdown levels are also shown. All experiments were performed with cells from three separate IPF lung fibroblast donors. Error bars represent s.e.m. ***P<0.001, **P<0.01 (one-way ANOVA with Tukey's multiple comparison test). For PGC1α knockdown analysis, an unpaired t-test was performed to test for significance.
The broad program of known and putative anti-fibrotic genes repressed by TGFβ and rescued by pracinostat suggests that there may be widespread redundancy in the pathways that maintain fibroblast in an inactive state. To test whether the inhibitory effects of pracinostat on fibroblast activation were specifically dependent on PGC1α, a strong regulator of fibroblast quiescence (Caporarello et al., 2019; Ligresti et al., 2019), we used siRNA to knockdown PGC1α and then measured anti-fibrotic efficacy of pracinostat on ACTA2 expression. Interestingly, pracinostat maintained its inhibitory effect on ACTA2 transcript levels (Fig. 5C) even in the presence of reduced PGC1α expression, demonstrating that pracinostat's effects are robust and resistant to single gene modulation. Together these findings highlight the concept that pracinostat likely acts on many gene targets that collectively and redundantly function to blunt fibroblast activation and protect against the aberrant and sustained pathological actions of these cells.
HDAC7 is required for TGFβ-induced fibroblast activation
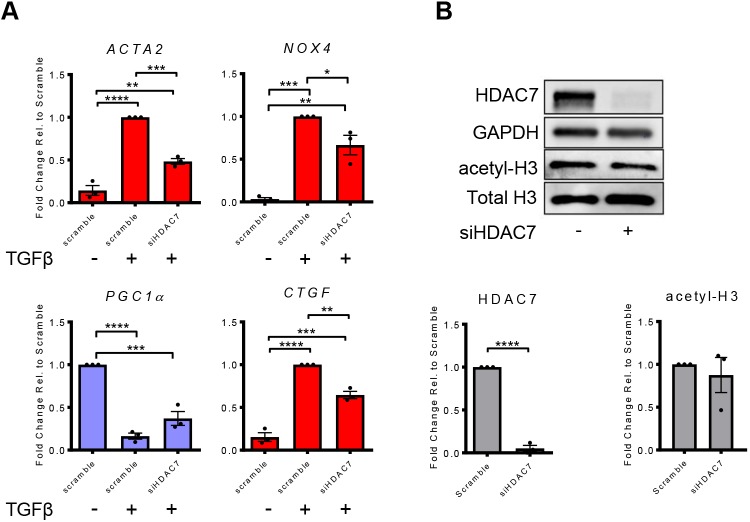
While HDAC inhibitors are powerful tools, their biological effects are widespread and potentially confound efforts to safely target them functionally in a class-wide fashion in the setting of chronic disease. As progress is being made in identifying more-selective inhibitors of specific groups or individual HDACs (Eckschlager et al., 2017), we sought to determine critical individual HDACs essential to TGFβ-mediated activation of lung fibroblasts. In the presence of TGFβ, we used siRNA to knockdown each individual HDAC that pracinostat has been reported to inhibit (Wang et al., 2011). Using the ACTA2 transcript level as a readout, HDAC7 siRNA was the most effective in reducing fibroblast activation when compared to TGFβ treatment alone (Table S2). To evaluate whether HDAC7 regulates transcription of other HDACs, we tested expression level of all other HDACs following HDAC7 knockdown under TGFβ stimulation. HDAC7 knockdown modestly but significantly reduced expression of HDAC2, HDAC6, HDAC8 and HDAC10, and increased expression of HDAC9 (Fig. S1C); these changes were small compared to the direct effect on HDAC7 protein (Fig. 6B). To test whether reducing HDAC7 has similar transcriptional effects to using pracinostat, we examined the effect of knocking down HDAC7 on select genes identified in Figs 3B and 4A. HDAC7 knockdown significantly attenuated TGFβ upregulation of the fibroblast activation genes NOX4, CTGF and ACTA2 (Fig. 6A; Fig. S1B). In addition, knocking down HDAC7 diminished the reduction of PGC1α induced by TGFβ treatment (Fig. 6A; Fig. S1B), although the magnitude of the effect was reduced relative to pracinostat, and did not reach statistical significance. These results suggest that TGFβ-mediated regulation of key pro- and anti-fibrotic genes requires the specific actions of HDAC7. Interestingly, while HDAC7 silencing resulted in strong reduction of protein levels of HDAC7, there was no significant change in total acetyl-H3 protein (Fig. 6B; Fig. S1A), suggesting that HDAC7 acts relatively specifically on a subset of genomic loci, rather than as a global modulator of histone acetylation.
Fig. 6.
HDAC7 is required for TGFβ-mediated fibroblast activation. (A) qPCR analysis of IPF lung fibroblasts treated with siRNA targeting HDAC7 (siHDAC7) for 72 h then treated with TGFβ for 24 h. Pro-fibrotic genes are shown in red. Putative and confirmed anti-fibrotic genes are shown in light blue. (B) Western blot confirmation of protein knockdown of HDAC7 and its relative effect on acetyl-H3 are shown from a representative blot and quantification from independent replicates. All experiments were performed with cells from three separate IPF lung fibroblast donors. Error bars represent s.e.m. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001 (one-way ANOVA with Tukey's multiple comparison test). For HDAC7 knockdown analysis, an unpaired t-test was performed to test for significance.
Motif discovery in TGFβ-repressed gene programs
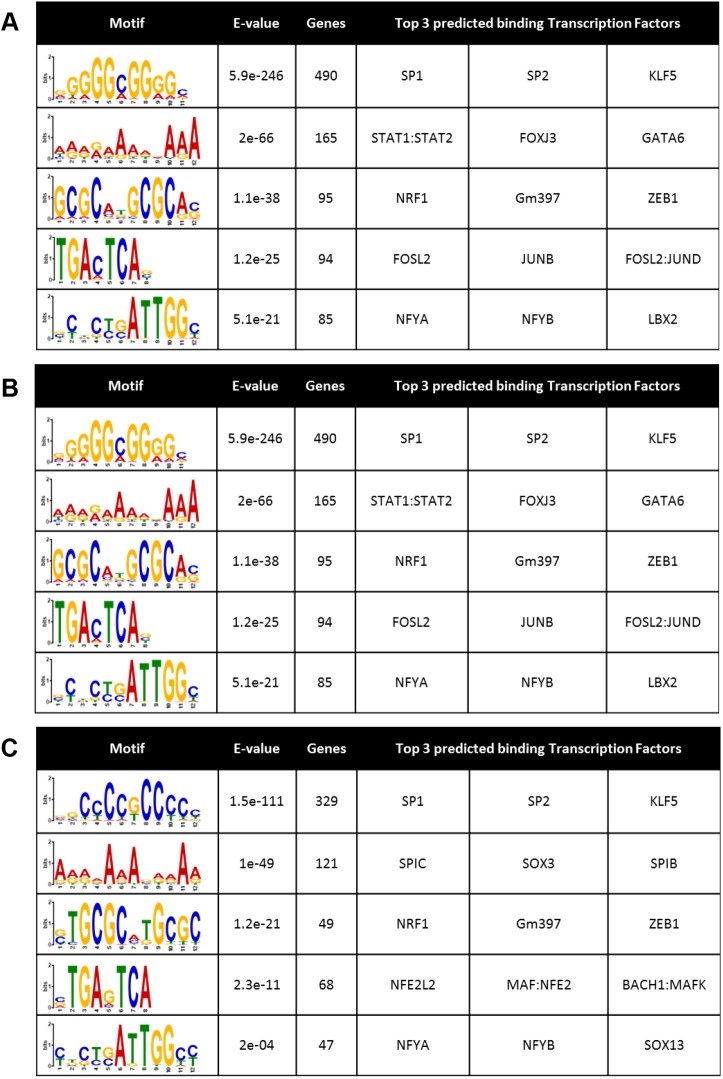
The transcriptional effects of TGFβ are known to be mediated by a complex network of canonical and non-canonical pathways and transcriptional complexes including SMAD (or non-canonical) transcriptional effectors that may link to HDACs to mediate repressive functions (Derynck and Budi, 2019). Thus, we sought to determine whether genes which were repressed by TGFβ and rescued by HDAC inhibition exhibited transcription factor (TF) motif enrichment in their promoters as a potential mechanism for this behavior. Scanning the promoters (±2 kb) of genes that were repressed by TGFβ, and also separately those that were and were not rescued by pracinostat, we cross-referenced these genomic areas with publically available DNA accessibility sites (ENCODE Project Consortium, 2012; Roadmap Epigenomics Consortium et al., 2015) to determine TF motif enrichments. SP1, a TF with widespread genomic interactions (ENCODE Project Consortium, 2012), was the top hit in all groups examined (Fig. 7A–C), implicating it in TGF-β-mediated repression, but not specifically in HDAC-mediated effects. Interestingly, the SMAD3 DNA-binding motif was identified as being highly enriched in the promoter region of genes whose transcription was suppressed by TGFβ and de-repressed by pracinostat (Fig. 7B), suggesting HDAC inhibition may disrupt SMAD-dependent co-repressive activity. This is consistent with our findings that TGFβ treatment leads to deacetylation of the PGC1α promoter in an HDAC-dependent manner around putative SMAD2–SMAD3–SMAD4-binding sites (Fig. 5A), and with prior work linking SMADs to HDAC-mediated gene repression (Kim and Lassar, 2003; Wotton et al., 1999). To further explore the concept of SMAD-dependent repressive activity, we analyzed publically available SMAD2/3 ChIP-seq data (Table S7) (Kim et al., 2011). Interestingly, of the 1103 genes whose expression pracinostat rescued from TGFβ repression, 332 of these are direct SMAD2/3 targets as identified by ChIP-seq (Table S7). Examples of these 332 genes include: PGC1α, DACH1, ARHGAP12, ARHGAP29, SOD2, BMP4 and PTGER2, broadly overlapping with the panel of putative and known fibrosis suppressing genes identified as repressed by TGFβ and rescued by pracinostat in Fig. 3. Together, these data support our central finding that TGFβ-mediated gene repression is essential to the initiation and maintenance of fibroblast activation, potentially via SMAD-directed repression of anti-fibrotic mediators.
Fig. 7.
TF motif enrichment analysis. (A) TF motifs enriched in the promoter region of genes repressed by TGFβ identified via RNA sequencing analysis represented by a sequence logo diagram where the size of the letter represent the frequency at which each nucleotide occurs. (B) TF motifs enriched in the promoter region of genes repressed by TGFβ and rescued by HDAC inhibition. (C) TF motifs enriched in the promoter region of genes repressed by TGFβ and not rescued by HDAC inhibition. The E-value is a statistical measure to describe the likelihood of finding a motif within random background genomic sequences.
An outstanding remaining question raised by these findings is how SMAD binding differentially results in repression versus transcription at specific genomic loci. Beyond SMAD3, several TFs, such as ZNF384 and members of the AP-1 complex, were also revealed to be enriched in the promoters of genes repressed by TGFβ and rescued by pracinostat in our study. These results set the stage for future work aimed at deciphering how TGFβ signaling results in repression of specific genes and activation of others through TF complex formation and epigenetic modifications, and how these integrated effects influence TGFβ-mediated fibroblast activation.
DISCUSSION
Fibroblast activation is central to wound healing, and persistent fibroblast activation underlies progressive fibrotic diseases, such as IPF (Raghu et al., 2011). Here, we showed, through a small-molecule epigenetic library screen, that fibroblast activation is altered by inhibiting a variety of epigenetic functions (i.e. gene activation/repression), highlighting the complexity of fibroblast biology and TGFβ-mediated epigenetic alterations. In particular, we show that HDAC-mediated gene repression is a critical event that precedes TGFβ-induced pro-fibrotic gene expression and identified the pan-HDAC inhibitor pracinostat as a powerful attenuator of TGFβ-mediated gene repression and fibroblast activation. Transcriptional analysis revealed that TGFβ requires HDAC function to promote the repression of a broad program of putative or known anti-fibrotic genes. Interestingly, pracinostat remains effective even when applied to already activated cells, implicating HDAC function in the maintenance of fibroblast activation. In addition, we show TGFβ signaling deacetylates histones on the promoter of PGC1α, a gene it represses, and that HDAC inhibition increases histone acetylation at these regions. We further demonstrate the specific importance of HDAC7 in TGFβ signaling in lung fibroblasts. Taken together, our findings showing that TGFβ-mediated repression of anti-fibrotic gene expression is critical to fibroblast activation should motivate further work to decode the repressive mechanisms and key targets of these effects, as they could serve as novel avenues for inhibiting or reversing fibroblast activation in progressive fibrotic diseases.
Epigenetic mechanisms are well known to play a key role in fibrosis (Helling and Yang, 2015; Page and Mann, 2015; Yang and Schwartz, 2015). Specifically in pulmonary fibrosis and fibroblast biology, changes in DNA methylation (Huang et al., 2014; Qu et al., 2018; Sanders et al., 2012; Yang et al., 2014), microRNAs (Khalil et al., 2015; Liu et al., 2010; Miao et al., 2018; Yang et al., 2013) and histone-modifying enzymes (Bai et al., 2019; Bombardo et al., 2018; Conforti et al., 2017; Coward et al., 2014, 2009; Glenisson et al., 2007; Hemmatazad et al., 2009; Khalil et al., 2015; Korfei et al., 2015; Ota et al., 2015; Sanders et al., 2014, 2017) have been shown to accompany and drive lung fibroblast activation and disease progression. Our screen interrogated the effects of a multitude of epigenetic-modifying small molecules, and identified effects of a variety of epigenetic writers, erasers and readers, demonstrating the complexity and importance of epigenetic regulators in controlling fibroblast biology. Supporting the utility of our screen, several enzymes already reported to be important in fibroblast biology and fibrosis, such as bromodomain-containing proteins (e.g. BRD4) (Tang et al., 2013) and EZH2 (Coward et al., 2018; Xiao et al., 2016) were identified. We chose to focus on pracinostat because it was most effective in our primary assay outcome, and because HDAC inhibition has proven effective in fibrosis models (Barter et al., 2010; Bombardo et al., 2018; Conforti et al., 2017; Glenisson et al., 2007; Guo et al., 2009; Kang et al., 2017; Khalil et al., 2015; Kim et al., 2018; Ota et al., 2015) but its anti-fibrotic effects remain poorly understood. However, we also note that inhibitors of several enzymes not previously linked to fibrosis were identified in our screen, such as L3MBTL3, a reader of methylated-lysine on histones (Min et al., 2007; Trojer et al., 2007). Further validation and investigation of these additional screen hits is needed to delineate their respective role in fibroblast biology.
While our epigenetic library screened a wide-variety of small-molecule enzyme modulators, a limitation of our screening approach was the use of αSMA as a single read-out of in vitro fibroblast activation. A recent study using single-cell RNA sequencing identified heterogeneous populations of activated fibroblasts in experimental lung fibrosis in mice, not all of which are αSMA positive (Peyser et al., 2019). Future work could incorporate additional markers of fibroblast activation (e.g. LTBP1) to more completely evaluate effective modulators of fibroblast phenotype. To increase confidence in the physiologic relevance of our primary screen hit, we did confirm significant effects of pracinostat on hydrogel compaction, collagen I and fibronectin deposition, important functional indicators of fibrogenic fibroblast activation.
While inhibitors of classes of epigenome-editing enzymes are used in clinically approved therapies, these drugs carry many global side-effects (Subramanian et al., 2010) complicating their use for long-term tissue-specific chronic diseases such as fibrosis. Understanding specific gene targets of pro-fibrotic signaling pathways and epigenetic enzymes could yield promising gene-specific therapies for fibrosis through the use of precise tools such as CRISPR-Cas9 epigenome editing (Hilton et al., 2015; Klann et al., 2017). Here, we describe the epigenetic silencing of specific anti-fibrotic genes by pro-fibrotic pathways, including PGC1α, DACH1 and ARHGAP12. Interestingly, the anti-fibrotic efficacy of pracinostat was maintained even when PGC1α was silenced (Fig. 4F), suggesting potentially broad redundancy in the anti-fibrotic targets rescued by pracinostat, consistent with previously published efforts that failed to identify monogenic mechanisms of inhibitors of epigenetic-regulating enzymes (Bhadury et al., 2014). Because pracinostat inhibits numerous HDACs, its broad effects are likely due to the concurrent inactivation of multiple HDAC family members. Thus, the identification of specific HDACs critical in sustaining the activation of diseased fibroblasts is required to improve the design of potential therapeutics for lung fibrosis.
To identify critical HDACs involved in TGFβ signaling, we knocked down individual HDACs in the presence of TGFβ and identified HDAC7 as an important player in TGFβ gene regulation (Fig. 6A). Previous studies have identified HDAC7 as being a potential key player in fibrosis owing to TGFβ signaling upregulating its gene expression and its upregulation in fibroblastic foci in IPF lung tissue (Hemmatazad et al., 2009; Kang et al., 2018; Korfei et al., 2015), although functional studies have been lacking. Interestingly, we found that knockdown of HDAC7 in the presence of TGFβ also modestly reduced expression of HDAC8 (Fig. S1C), which has previously been shown to drive fibroblast activation in vitro and in vivo (Saito et al., 2019), suggesting an additional target through which HDAC7 may regulate fibroblast activation. HDAC7 knockdown strongly attenuated TGFβ upregulation of fibrosis-associated genes (ACTA2, NOX4 and CTGF) (Fig. 6A; Fig. S1B). We found that HDAC7 knockdown had a modest but not statistically significant effect on PGC1α expression following TGFβ stimulation (Fig. 6A; Fig. S1B), suggesting that HDAC7 knockdown effects on fibroblast activation likely reflect coordinated regulation of multiple anti-fibrotic genes, as seen for pracinostat (Figs 3 and 5). While our work suggests a role of HDAC7 in fibroblast activation, further work is needed to characterize the mechanism through which TGFβ engages HDAC7 function, along with other HDACs, to exert its effects on gene transcription and fibroblast activation.
Most HDACs do not contain a DNA-binding motif (Seto and Yoshida, 2014). Thus, these enzymes depend on the formation of complexes with other DNA-binding proteins to direct their enzymatic function. Thus, we set out to identify candidate DNA-binding proteins that potentially mediate gene repression by TGFβ and HDACs using TF motif enrichment. Importantly, the DNA-binding motif for SMAD3 was significantly highly enriched in the promoters of genes whose expression was repressed by TGFβ and rescued by pracinostat (Fig. 7B) indicating SMAD–HDAC-mediated gene repression may be a key component of lung fibroblast activation. In addition, our analysis of publicly available SMAD2/3 ChIP-seq data confirms SMAD2/3 binding directly to many putative anti-fibrotic mediators (Table S6) (Kim et al., 2011). Previous work has shown that SMADs form repressive complexes with HDAC1 (Kim and Lassar, 2003; Wotton et al., 1999); however, no work has previously identified SMAD3 working together with HDAC7. SMAD3 is also known to mediate gene expression, thus the specific mechanisms that mediate repression and activation of genomic loci downstream of TGFβ will require further investigation. Notably, our motif analysis identified several additional TF motifs enriched in genes repressed by TGFβ in an HDAC-dependent fashion. Few of these have been implicated previously in fibrosis and fibroblast activation (e.g. MAFK and NFYA), although Jun has previously been associated with both fibrosis and SMAD signaling (Wernig et al., 2017; Wong et al., 1999). Further investigation will be required to understand the mechanisms by which TGFβ-mediated transcriptional and epigenetic mechanisms converge on gene repression and expression to drive fibroblast activation, with a particular emphasis on TGFβ-mediated repression of genes that function to limit or reverse fibroblast activation.
In summary, our work has identified a key role for HDACs in mediating TGFβ activation of lung fibroblasts through their function as regulators of gene repression. Our work also has initiated efforts to identify individual HDACs, HDAC gene targets and potential HDAC-interacting TFs important to fibroblast activation. These data further our understanding of how TGFβ signaling interacts with epigenetic mechanisms in the regulation of fibroblast biology, an essential step toward new and more-specific epigenetic-targeted therapeutics for fibrotic disease.
MATERIALS AND METHODS
Cell culture
IMR90 human lung fibroblasts were purchased from ATCC (Manassas, VA) and were maintained in EMEM (ATCC, Manassas, VA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA) and antibiotic-antimycotic (Thermo Fisher Scientific, Waltham, MA) unless otherwise noted. Primary human lung fibroblasts isolated by explant culture from the lungs of subjects diagnosed with IPF who underwent lung transplantation were kindly provided by Peter Bitterman and Craig Henke at the University of Minnesota under a protocol approved by the University of Minnesota Institutional Review Board and by Carol Feghali-Bostwick at the Medical University of South Carolina under a protocol approved by the University of Pittsburgh Institutional Review Board. Informed consent was obtained for all tissue donors and all investigation has been conducted according to the principles expressed in the Declaration of Helsinki. Experiments were done with IPF fibroblasts between passage 4–7. IPF-derived human lung fibroblasts were maintained in DMEM (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) and antibiotic-antimycotic (Thermo Fisher Scientific). IPF lung fibroblasts were treated with 2 ng/ml TGFβ (Thermo Fisher Scientific) and also the noted concentrations of pracinostat (Caymen Chemical). IMR-90 and primary cells were subject to routine mycoplasma testing.
Epigenetic small-molecule screen
Small-molecule screening of fibroblast activation was performed in two separate doses using a library of 99 epigenetic modifying compounds selected and annotated by Nanosyn, Inc. IMR-90 lung fibroblasts were plated in 96-well plates (10,000 cells/well) and allowed to attach in EMEM+10% FBS for 6 h. Medium was then exchanged for EMEM+0.1% FBS and cells were then treated with 2 ng/ml of TGFβ plus the compound of interest (500 nM or 2.5 μM) for 72 h. Cells were then fixed in 4% formalin for 10 min, permeabilized with 0.25% Triton X-100 (Sigma-Aldrich, St Louis, MO, USA), blocked with 1% BSA (Sigma-Aldrich), and incubated with primary antibodies (listed in Table S5) overnight at 4°C. Cells were then incubated with secondary antibodies (listed in Table S5) and DAPI (Biolegend, San Diego, CA) for 1 h at room temperature. Images were acquired using a Cytation5 microscope (BioTek, Winooski, VT, USA) with a 10× objective. The cell number for each condition was determined by identifying independent objects on the DAPI channel. αSMA intensity was used to assess the overall protein expression, and was reported relative to cell number. Background was defined as the total intensity observed in an identical well prepared without cells. Small-molecule modulators were classified as a ‘hit’ if the αSMA intensity was at least one standard deviation away from the mean αSMA intensity of all compounds measured. Small-molecular modulators were defined to be toxic and omitted from further analysis if the percentage of total cells relative to control (+TGFβ) was less than or equal to the mean minus one standard deviation. Full screen results can be found in Table S3 (2.5 μM) and Table S4 (500 nM).
Protein lysis and blotting
For protein analysis, cells were lysed with RIPA buffer (Thermo Fisher Scientific) supplemented with Halt™ protease and phosphatase inhibitor cocktail (Thermo Fisher Scientific). Total protein content was quantified with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). Protein samples were run on 4–15% gradient SDS-PAGE gels, transferred to PVDF membranes and incubated overnight at 4°C with primary antibodies (listed in Table S5). Blots were then incubated with appropriate HRP-conjugated IgG secondary antibody (listed in Table S5) for 1 h at room temperature. Protein bands were acquired using Super Signal West Pico Plus (Thermo Fisher Scientific) and a ChemiDoc Imaging System (Bio-Rad, Hercules, CA). Each antibody produced one clear band, or two bands in the case of SMAD 2/3, and HDAC7 antibody was validated through siRNA experiments (Fig. 6B; Fig. S1A).
Compaction assay
Cell-embedded collagen micro tissues were generated as previously described (Brett et al., 2016). Briefly, IPF fibroblasts (4×106 cells/ml) were diluted in rat-tail collagen type-I (6 mg/ml) (Corning, Corning, NY). Polystyrene beads (1:50) (Thermo Fisher Scientific) of 1 μm in diameter were added to the solution to visualize the droplet. Emulsions were formed by flow-focusing the collagen/cell solution and oil (fluorocarbon oil FC-40; Sigma-Aldrich) with 2% FluoroSurfactant (RAN Biotechnologies, Beverly, MA, USA) into a microfluidic PDMS device. The size of the droplet was controlled by fixing the inner aqueous phase flow rate to 250 μl/h and outer oil phase flow rate to 800 μl/h. Droplet formations were performed at 4°C while the droplets were recovered at 37°C. Oil was aspirated and the droplets were transferred to a pre-made microfluidic capture device. The droplets were incubated at 37°C in medium containing either DMSO or 2 ng/ml TGFβ in the absence or presence of pracinostat (1 µM) for 3 days. The area (μm2) of the droplet containing 1 μm Nile Blue beads was measured with a 2.5× objective (plane size 2881×2127 μm) with a Cytation 5 instrument.
ECM deposition assay
IPF-derived human lung fibroblasts were plated in 96-well plates (10,000 cells/well) and allowed to attach in EMEM+10% FBS for 6 h. Media was then exchanged for EMEM+0.1% FBS treated with 2 ng/ml TGFβ and pracinostat (3 µM) for 72 h. Cells were then fixed in 4% formalin and blocked in Odyssey blocking buffer for 1 h and then incubated with primary antibody (listed in Table S5) overnight at 4°C. Cells were then incubated with secondary goat anti-mouse IgG IRDye™ 800 antibody and goat anti-rabbit IgG IRDye™ 680 antibody (1:750 dilution, 45 min, room temperature). The 96-well plates were then scanned with the Odyssey CLx Infrared Imaging System (LI-COR Biosciences, Lincoln, NE), and integrated fluorescence intensities were acquired using the software provided with the imaging station (Odyssey Software Version 3.0, LI-COR Biosciences).
RNA isolation
mRNA was isolated using an RNeasy mini kit (Qiagen); the concentration of mRNA was measured using a Nanodrop spectrophotometer (Thermo Fisher Scientific). cDNA was synthesized using SuperScript VILO (Thermo Fisher Scientific, Waltham, MA); RT-PCR was performed using FastStart Essential DNA Green Master (Roche Diagnostics, Mannheim, Germany) and analyzed using a LightCycler 96 (Roche Diagnostics, Mannheim, Germany). Primers used for qRT-PCR are listed in Table S6.
RNA-sequencing and data analysis
RNA quality was determined with a Fragment Analyzer (Agilent, Santa Clara, CA) instrument. RNA samples that had RQN values ≥6 were approved for library preparation and sequencing. RNA libraries were prepared using 200 ng of total RNA according to the manufacturer's instructions for the TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA), employing poly(A) mRNA enrichment using oligo(dT) magnetic beads. The final adapter-modified cDNA fragments were enriched by 12 cycles of PCR using Illumina TruSeq PCR primers. The concentration and size distribution of the completed libraries were determined using a Fragment Analyzer (Agilent) and Qubit fluorometry (Invitrogen, Carlsbad, CA). Libraries were sequenced as 100×2 paired end reads on an Illumina HiSeq 4000, following Illumina's standard protocol with the Illumina cBot and HiSeq 3000/4000 PE Cluster Kit, which yielded ∼43×106–65×106 reads per sample. Base-calling was performed using Illumina's RTA version 2.7.3.
Data was analyzed using MAPRSeq version 3.0.0, which is an integrated RNA-seq analysis pipeline developed at the Mayo Clinic (Kalari et al., 2014). As part of the MAPRSeq pipeline, reads were aligned to the hg38 (GRCh38) build of the human reference genome using STAR with default parameter setting (Dobin et al., 2013). The aligned reads were then used for Ensembl (v78) gene and exon expression quantification using featureCounts from the Subread package to obtain raw counts as well as normalized [reads per kilobase of exon per million mapped reads (RPKM)] expression values (Liao et al., 2013). Extensive quality control analysis of the aligned RNA-Seq reads was performed using the RSeQC package to assess metrics such as insert size, gene body coverage, and junction saturation (Wang et al., 2012). For group comparisons, P-values were adjusted for multiple tests using the Benjamini and Hochberg method, herein termed the false-discovery rate (FDR). RNA-seq data generated in this study are available through the Gene Expression Omnibus under accession code GSE136534.
To identify differentially expressed genes regulated by TGFβ and HDACs, lists of genes whose expression was activated or suppressed by TGFβ were first generated using the following criteria: RPKMs >0 in all groups, RPKM (in DMSO control group) ≥0.01, a fold change of ≥2 (activated) or ≤0.5 (suppressed) when compared to DMSO control, and a false-discovery rate ≤0.1. These two gene lists were then compared to the TGFβ plus pracinostat (1 µM) results to identify the effect of pracinostat on TGFβ-mediated gene regulation. Genes in the activated gene list were classified as being dependent on HDACs if TGFβ/TGFβ+pracinostat ≥2 with an FDR≤0.1. Genes suppressed by TGFβ were classed as being dependent on HDAC function if TGFβ/TGFβ+pracinostat≤0.5 with an FDR≤0.1. Gene lists identified above were used in gene ontology analysis to identify enriched gene sets and pathways with David v6.8 (Huang et al., 2009a,b).
To identify enrichment of TF-binding motifs in the promoter regions of genes repressed by TGFβ and rescued, or not rescued by pracinostat, we downloaded the DNase I hypersensitive sites (DHSs) generated by the NIH Roadmap and Encyclopedia of DNA Elements (ENCODE) epigenomics projects in 53 cell and tissue types (ENCODE Project Consortium, 2012; Roadmap Epigenomics et al., 2015), which are available at https://egg2.wustl.edu/roadmap/data/byFileType/peaks/consolidated/narrowPeak. BEDTools (Quinlan and Hall, 2010) was used to intersect the above DHSs with the promoters (defined as the transcription start site ±2 kb) from the Ensembl gene annotation. If multiple DHSs overlapped the same promoter, then a single DHS with the highest confidence (i.e. lowest FDR) was retained. To identify TF-binding motifs (6–12 base pairs wide) within the DHSs, we used the MEME program (v4.11.1) (Bailey et al., 2009) to scan the 100-bp centered on each of the DHSs within promoters, at a P-value cutoff of 103. We included all motifs identified by MEME whose E-value, a statistical measure of the probability a motif would be identified in random genomic background, was less than 0.05. We used TOMTOM in the MEME Suite web server to identify the TFs with the highest likelihood of binding to those motifs. The top three TFs for each motif were then retained.
ChIP-PCR
ChIP-PCR was carried out using the Pierce Magnetic ChIP kit (Thermo Fisher Scientific) according to the manufacturer's protocol. Briefly, cells were fixed with 1% formaldehyde for 10 min and then neutralized with the supplied 10× glycine for 5 min. Cells were washed and then collected in ice-cold PBS with Halt™ protease and phosphatase inhibitor cocktail. Cells were centrifuged at 3000 g for 5 min. Nuclei were isolated, incubated with MNase, and lysed by sonication. Protein–DNA lysates were then incubated with primary antibodies (listed in Table S5) overnight at 4°C with mixing. Precipitated DNA fragments were then amplified according to the manufacturer's protocol with primers listed in Table S6. Primers were designed around putative SMAD2–SMAD3–SMAD4-binding sites in the promoter region of PGC1α using the Eukaryotic Promoter Database (EPD) (Dreos et al., 2017, 2015).
ChIP-seq analysis
Publically available SMAD2/3 ChIP-seq data (GSE29422) in human embryonic stem cells was analyzed to observe direct gene targets of SMAD2/3 (Kim et al., 2011). Paired-end reads were aligned to the hg19 build of the human genome with bowtie2 (Langmead and Salzberg, 2012) using default parameters. Peaks were called using MACS2 callpeaks (Zhang et al., 2008) using default settings with a q-value threshold of ≤0.05. Peak locations were annotated using Chipseek (Chen et al., 2014). Genes repressed by TGFβ and rescued, or not rescued, by pracinostat were cross-referenced with the SMAD2/3 ChIP-seq annotated data set to identify putative direct targets of SMAD2/3 repressive activity.
siRNA transfections
RNA interference was performed with SMARTpool: ON-TARGETplus siRNAs (Dharmacon, Lafayette, CO) specific for the genes listed in Table S2, PGC1α and non-targeting control, by using Lipofectamine RNAiMAX reagent (Thermo Fisher Scientific). An additional single ON-TARGETplus siRNA was used to confirm HDAC7 findings. Cells were transfected in DMEM with 10% FBS for 72 h, then placed in DMEM with 0.1% FBS overnight. The following morning, TGFβ was administered for 24 h and RNA was collected and analyzed as described above.
Statistical analysis
GraphPad Prism 7.0 (GraphPad Software, La Jolla, CA) was used for generating Figs 1–6 and statistical analyses. Data are represented as mean±s.e.m. Statistical analysis was performed using either an unpaired two-tailed t-test, when directly comparing two groups, or one-way ANOVA with Tukey multiple-comparison adjustment when comparing three or more groups. For RNA-seq, P-values were adjusted by the Benjamini and Hochberg method (Benjamini and Hochberg, 1995).
Supplementary Material
Acknowledgements
We would like to thank the members of the Y. S. Prakash laboratory for assistance with reagents and thoughtful discussions.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Author contributions
Conceptualization: D.L.J., A.J.H., N.C., X.V., T.O., G.L., D.J.T.; Methodology: D.L.J., A.J.H., N.C., K.M.C., G.L.; Formal analysis: D.L.J., A.J.H., N.C., K.M.C., Z.Y., H.Y., G.L.; Investigation: D.L.J., A.J.H., N.C., K.M.C., G.L.; Data curation: D.L.J., A.J.H., N.C., K.C., Z.Y., H.Y., D.J.T.; Writing - original draft: D.L.J., D.J.T.; Writing - review & editing: D.L.J., H.Y., X.V., T.O., G.L., D.J.T.; Funding acquisition: D.J.T.
Funding
Funding support provided by the National Institutes of Health (NIH) grants HL105355 (D.L.J.), HL142596 (G.L.), HL092961 (D.J.T.) and HL133320 (D.J.T.), and an American Lung Association Senior Research Training Fellowship (A.J.H.). Deposited in PMC for release after 12 months.
Data availability
RNA-seq data generated in this study are available through the Gene Expression Omnibus under accession code GSE136534.
Supplementary information
Supplementary information available online at http://jcs.biologists.org/lookup/doi/10.1242/jcs.233486.supplemental
References
- Bai L., Bernard K., Tang X., Hu M., Horowitz J. C., Thannickal V. J. and Sanders Y. Y. (2019). Glutaminolysis epigenetically regulates anti-apoptotic gene expression in IPF fibroblasts. Am. J. Respir. Cell Mol. Biol. 60, 49-57. 10.1165/rcmb.2018-0180OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W. and Noble W. S. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202-W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge P. (2013). Wound healing and the role of fibroblasts. J. Wound Care 22, 407-408, 410-412 10.12968/jowc.2013.22.8.407 [DOI] [PubMed] [Google Scholar]
- Barter M. J., Pybus L., Litherland G. J., Rowan A. D., Clark I. M., Edwards D. R., Cawston T. E. and Young D. A. (2010). HDAC-mediated control of ERK- and PI3K-dependent TGF-beta-induced extracellular matrix-regulating genes. Matrix Biol. 29, 602-612. 10.1016/j.matbio.2010.05.002 [DOI] [PubMed] [Google Scholar]
- Benjamini Y. and Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289-300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- Bhadury J., Nilsson L. M., Muralidharan S. V., Green L. C., Li Z., Gesner E. M., Hansen H. C., Keller U. B., McLure K. G. and Nilsson J. A. (2014). BET and HDAC inhibitors induce similar genes and biological effects and synergize to kill in Myc-induced murine lymphoma. Proc. Natl. Acad. Sci. USA 111, E2721-E2730. 10.1073/pnas.1406722111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya S., Wang W., Morales-Nebreda L., Feng G., Wu M., Zhou X., Lafyatis R., Lee J., Hinchcliff M., Feghali-Bostwick C. et al. (2016). Tenascin-C drives persistence of organ fibrosis. Nat. Commun. 7, 11703 10.1038/ncomms11703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bombardo M., Chen R., Malagola E., Saponara E., Hills A. P., Graf R. and Sonda S. (2018). Inhibition of class I histone deacetylases abrogates tumor growth factor beta expression and development of fibrosis during chronic pancreatitis. Mol. Pharmacol. 94, 793-801. 10.1124/mol.117.110924 [DOI] [PubMed] [Google Scholar]
- Brett M.-E., Crampton A. L. and Wood D. K. (2016). Rapid generation of collagen-based microtissues to study cell–matrix interactions. Technology 4, 80-87. 10.1142/S2339547816400094 [DOI] [Google Scholar]
- Caporarello N., Meridew J. A., Jones D. L., Tan Q., Haak A. J., Choi K. M., Manlove L. J., Prakash Y. S., Tschumperlin D. J. and Ligresti G. (2019). PGC1alpha repression in IPF fibroblasts drives a pathologic metabolic, secretory and fibrogenic state. Thorax 74, 749-760. 10.1136/thoraxjnl-2019-213064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.-W., Li H.-P., Lee C.-C., Gan R.-C., Huang P.-J., Wu T. H., Lee C.-Y., Chang Y.-F. and Tang P. (2014). ChIPseek, a web-based analysis tool for ChIP data. BMC Genomics 15, 539 10.1186/1471-2164-15-539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheresh P., Kim S.-J., Tulasiram S. and Kamp D. W. (2013). Oxidative stress and pulmonary fibrosis. Biochim. Biophys. Acta 1832, 1028-1040. 10.1016/j.bbadis.2012.11.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti F., Davies E. R., Calderwood C. J., Thatcher T. H., Jones M. G., Smart D. E., Mahajan S., Alzetani A., Havelock T., Maher T. M. et al. (2017). The histone deacetylase inhibitor, romidepsin, as a potential treatment for pulmonary fibrosis. Oncotarget 8, 48737-48754. 10.18632/oncotarget.17114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward W. R., Watts K., Feghali-Bostwick C. A., Knox A. and Pang L. (2009). Defective histone acetylation is responsible for the diminished expression of cyclooxygenase 2 in idiopathic pulmonary fibrosis. Mol. Cell Biol. 29, 4325-4339. 10.1128/MCB.01776-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward W. R., Feghali-Bostwick C. A., Jenkins G., Knox A. J. and Pang L. (2014). A central role for G9a and EZH2 in the epigenetic silencing of cyclooxygenase-2 in idiopathic pulmonary fibrosis. FASEB J. 28, 3183-3196. 10.1096/fj.13-241760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward W. R., Brand O. J., Pasini A., Jenkins G., Knox A. J. and Pang L. (2018). Interplay between EZH2 and G9a regulates CXCL10 gene repression in idiopathic pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 58, 449-460. 10.1165/rcmb.2017-0286OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby I. A., Laverdet B., Bonte F. and Desmouliere A. (2014). Fibroblasts and myofibroblasts in wound healing. Clin. Cosmet. Investig. Dermatol. 7, 301-311. 10.2147/CCID.S50046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derynck R. and Budi E. H. (2019). Specificity, versatility, and control of TGF-beta family signaling. Sci. Signal. 12, eaav5183 10.1126/scisignal.aav5183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmouliere A., Geinoz A., Gabbiani F. and Gabbiani G. (1993). Transforming growth factor-beta 1 induces alpha-smooth muscle actin expression in granulation tissue myofibroblasts and in quiescent and growing cultured fibroblasts. J. Cell Biol. 122, 103-111. 10.1083/jcb.122.1.103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M. and Gingeras T. R. (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15-21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R., Ambrosini G., Périer R. C. and Bucher P. (2015). The eukaryotic promoter database: expansion of EPDnew and new promoter analysis tools. Nucleic Acids Res. 43, D92-D96. 10.1093/nar/gku1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreos R., Ambrosini G., Groux R., Perier R. C. and Bucher P. (2017). The eukaryotic promoter database in its 30th year: focus on non-vertebrate organisms. Nucleic Acids Res. 45, D51-D55. 10.1093/nar/gkw1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckschlager T., Plch J., Stiborova M. and Hrabeta J. (2017). Histone deacetylase inhibitors as anticancer drugs. Int. J. Mol. Sci. 18, E1414 10.3390/ijms18071414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2012). An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57-74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estany S., Vicens-Zygmunt V., Llatjós R., Montes A., Penín R., Escobar I., Xaubet A., Santos S., Manresa F., Dorca J. et al. (2014). Lung fibrotic tenascin-C upregulation is associated with other extracellular matrix proteins and induced by TGFbeta1. BMC Pulm. Med. 14, 120 10.1186/1471-2466-14-120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabbiani G. (2003). The myofibroblast in wound healing and fibrocontractive diseases. J. Pathol. 200, 500-503. 10.1002/path.1427 [DOI] [PubMed] [Google Scholar]
- Glenisson W., Castronovo V. and Waltregny D. (2007). Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim. Biophys. Acta 1773, 1572-1582. 10.1016/j.bbamcr.2007.05.016 [DOI] [PubMed] [Google Scholar]
- Guo W., Shan B., Klingsberg R. C., Qin X. and Lasky J. A. (2009). Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am. J. Physiol. Lung Cell Mol. Physiol. 297, L864-L870. 10.1152/ajplung.00128.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling B. A. and Yang I. V. (2015). Epigenetics in lung fibrosis: from pathobiology to treatment perspective. Curr. Opin. Pulm. Med. 21, 454-462. 10.1097/MCP.0000000000000191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemmatazad H., Rodrigues H. M., Maurer B., Brentano F., Pileckyte M., Distler J. H. W., Gay R. E., Michel B. A., Gay S., Huber L. C. et al. (2009). Histone deacetylase 7, a potential target for the antifibrotic treatment of systemic sclerosis. Arthritis Rheum. 60, 1519-1529. 10.1002/art.24494 [DOI] [PubMed] [Google Scholar]
- Henderson N. C. and Sheppard D. (2013). Integrin-mediated regulation of TGFbeta in fibrosis. Biochim. Biophys. Acta 1832, 891-896. 10.1016/j.bbadis.2012.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilton I. B., D'Ippolito A. M., Vockley C. M., Thakore P. I., Crawford G. E., Reddy T. E. and Gersbach C. A. (2015). Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 33, 510-517. 10.1038/nbt.3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinz B., Celetta G., Tomasek J. J., Gabbiani G. and Chaponnier C. (2001). Alpha-smooth muscle actin expression upregulates fibroblast contractile activity. Mol. Biol. Cell 12, 2730-2741. 10.1091/mbc.12.9.2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1-13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T. and Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44-57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Huang S. K., Scruggs A. M., McEachin R. C., White E. S. and Peters-Golden M. (2014). Lung fibroblasts from patients with idiopathic pulmonary fibrosis exhibit genome-wide differences in DNA methylation compared to fibroblasts from nonfibrotic lung. PLoS ONE 9, e107055 10.1371/journal.pone.0107055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C. F., Rohani M. G., Lee S.-S., Chen P. and Schnapp L. M. (2013). Role of IGF-1 pathway in lung fibroblast activation. Respir. Res. 14, 102 10.1186/1465-9921-14-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joannes A., Brayer S., Besnard V., Marchal-Sommé J., Jaillet M., Mordant P., Mal H., Borie R., Crestani B. and Mailleux A. A. (2016). FGF9 and FGF18 in idiopathic pulmonary fibrosis promote survival and migration and inhibit myofibroblast differentiation of human lung fibroblasts in vitro. Am. J. Physiol. Lung Cell Mol. Physiol. 310, L615-L629. 10.1152/ajplung.00185.2015 [DOI] [PubMed] [Google Scholar]
- Kalari K. R., Nair A. A., Bhavsar J. D., O'Brien D. R., Davila J. I., Bockol M. A., Nie J., Tang X., Baheti S., Doughty J. B. et al. (2014). MAP-RSeq: mayo analysis pipeline for RNA sequencing. BMC Bioinformatics 15, 224 10.1186/1471-2105-15-224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S.-W., Lee S.-M., Kim J.-Y., Kim S.-Y., Kim Y.-H., Kim T.-H., Kang M.-S., Jang W.-H. and Seo S.-K. (2017). Therapeutic activity of the histone deacetylase inhibitor SB939 on renal fibrosis. Int. Immunopharmacol. 42, 25-31. 10.1016/j.intimp.2016.11.008 [DOI] [PubMed] [Google Scholar]
- Kang D. H., Yin G. N., Choi M.-J., Song K.-M., Ghatak K., Minh N. N., Kwon M.-H., Seong D.-H., Ryu J.-K. and Suh J.-K. (2018). Silencing histone deacetylase 7 alleviates transforming growth factor-beta1-induced profibrotic responses in fibroblasts derived from Peyronie's plaque. World J. Mens Health 36, 139-146. 10.5534/wjmh.170005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil W., Xia H., Bodempudi V., Kahm J., Hergert P., Smith K., Peterson M., Parker M., Herrera J., Bitterman P. B. et al. (2015). Pathologic regulation of collagen I by an aberrant protein phosphatase 2A/histone deacetylase C4/MicroRNA-29 signal axis in idiopathic pulmonary fibrosis fibroblasts. Am. J. Respir. Cell Mol. Biol. 53, 391-399. 10.1165/rcmb.2014-0150OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-W. and Lassar A. B. (2003). Smad-dependent recruitment of a histone deacetylase/Sin3A complex modulates the bone morphogenetic protein-dependent transcriptional repressor activity of Nkx3.2. Mol. Cell Biol. 23, 8704-8717. 10.1128/MCB.23.23.8704-8717.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. W., Yoon S.-J., Chuong E., Oyolu C., Wills A. E., Gupta R. and Baker J. (2011). Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 357, 492-504. 10.1016/j.ydbio.2011.06.009 [DOI] [PubMed] [Google Scholar]
- Kim D. J., Dunleavey J. M., Xiao L., Ollila D. W., Troester M. A., Otey C. A., Li W., Barker T. H. and Dudley A. C. (2018). Suppression of TGFbeta-mediated conversion of endothelial cells and fibroblasts into cancer associated (myo)fibroblasts via HDAC inhibition. Br. J. Cancer 118, 1359-1368. 10.1038/s41416-018-0072-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klann T. S., Black J. B., Chellappan M., Safi A., Song L., Hilton I. B., Crawford G. E., Reddy T. E. and Gersbach C. A. (2017). CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 35, 561-568. 10.1038/nbt.3853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korfei M., Skwarna S., Henneke I., MacKenzie B. A., Klymenko O., Saito S., Ruppert C., von der Beck D., Mahavadi P., Klepetko W. et al. (2015). Aberrant expression and activity of histone deacetylases in sporadic idiopathic pulmonary fibrosis. Thorax 70, 1022-1032. 10.1136/thoraxjnl-2014-206411 [DOI] [PubMed] [Google Scholar]
- Kwak H.-B., Lee Y., Kim J.-H., Van Remmen H., Richardson A. G. and Lawler J. M. (2015). MnSOD overexpression reduces fibrosis and pro-apoptotic signaling in the aging mouse heart. J. Gerontol. A Biol. Sci. Med. Sci. 70, 533-544. 10.1093/gerona/glu090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. and Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357-359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le T.-T. T., Karmouty-Quintana H., Melicoff E., Le T.-T. T., Weng T., Chen N.-Y., Pedroza M., Zhou Y., Davies J., Philip K. et al. (2014). Blockade of IL-6 Trans signaling attenuates pulmonary fibrosis. J. Immunol. 193, 3755-3768. 10.4049/jimmunol.1302470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G. K. and Shi W. (2013). The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 41, e108 10.1093/nar/gkt214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ligresti G., Caporarello N., Meridew J. A., Jones D. L., Tan Q., Choi K. M., Haak A. J., Aravamudhan A., Roden A. C., Prakash Y. S. et al. (2019). CBX5/G9a/H3K9me-mediated gene repression is essential to fibroblast activation during lung fibrosis. JCI Insight 5, 127111 10.1172/jci.insight.127111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lino Cardenas C. L., Henaoui I. S., Courcot E., Roderburg C., Cauffiez C., Aubert S., Copin M.-C., Wallaert B., Glowacki F., Dewaeles E. et al. (2013). miR-199a-5p Is upregulated during fibrogenic response to tissue injury and mediates TGFbeta-induced lung fibroblast activation by targeting caveolin-1. PLoS Genet. 9, e1003291 10.1371/journal.pgen.1003291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Friggeri A., Yang Y., Milosevic J., Ding Q., Thannickal V. J., Kaminski N. and Abraham E. (2010). miR-21 mediates fibrogenic activation of pulmonary fibroblasts and lung fibrosis. J. Exp. Med. 207, 1589-1597. 10.1084/jem.20100035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Lagares D., Choi K. M., Stopfer L., Marinković A., Vrbanac V., Probst C. K., Hiemer S. E., Sisson T. H., Horowitz J. C. et al. (2015). Mechanosignaling through YAP and TAZ drives fibroblast activation and fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 308, L344-L357. 10.1152/ajplung.00300.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. R., Tran M. T. and Parikh S. M. (2018). PGC1alpha in the kidney. Am. J. Physiol. Ren. Physiol. 314, F1-F8. 10.1152/ajprenal.00263.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Mateos R., De Assuncao T. M., Arab J. P., Jalan-Sakrikar N., Yaqoob U., Greuter T., Verma V. K., Mathison A. J., Cao S., Lomberk G. et al. (2019). Enhancer of Zeste homologue 2 inhibition attenuates TGF-beta dependent hepatic stellate cell activation and liver fibrosis. Cell Mol. Gastroenterol. Hepatol. 7, 197-209. 10.1016/j.jcmgh.2018.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Z., Qiu Y., Lin K. C., Kumar A., Placone J. K., Fang C., Wang K.-C., Lu S., Pan M., Hong A. W. et al. (2018). RAP2 mediates mechanoresponses of the Hippo pathway. Nature 560, 655-660. 10.1038/s41586-018-0444-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C., Xiong Y., Zhang G. and Chang J. (2018). MicroRNAs in idiopathic pulmonary fibrosis, new research progress and their pathophysiological implication. Exp. Lung Res. 44, 178-190. 10.1080/01902148.2018.1455927 [DOI] [PubMed] [Google Scholar]
- Min J., Allali-Hassani A., Nady N., Qi C., Ouyang H., Liu Y., MacKenzie F., Vedadi M. and Arrowsmith C. H. (2007). L3MBTL1 recognition of mono- and dimethylated histones. Nat. Struct. Mol. Biol. 14, 1229-1230. 10.1038/nsmb1340 [DOI] [PubMed] [Google Scholar]
- Moodley Y. P., Scaffidi A. K., Misso N. L., Keerthisingam C., McAnulty R. J., Laurent G. J., Mutsaers S. E., Thompson P. J. and Knight D. A. (2003). Fibroblasts isolated from normal lungs and those with idiopathic pulmonary fibrosis differ in interleukin-6/gp130-mediated cell signaling and proliferation. Am. J. Pathol. 163, 345-354. 10.1016/S0002-9440(10)63658-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh R. S., Haak A. J., Smith K. M. J., Ligresti G., Choi K. M., Xie T., Wang S., Walters P. R., Thompson M. A., Freeman M. R. et al. (2018). RNAi screening identifies a mechanosensitive ROCK-JAK2-STAT3 network central to myofibroblast activation. J. Cell Sci. 131, jcs209932 10.1242/jcs.209932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ota C., Yamada M., Fujino N., Motohashi H., Tando Y., Takei Y., Suzuki T., Takahashi T., Kamata S., Makiguchi T. et al. (2015). Histone deacetylase inhibitor restores surfactant protein-C expression in alveolar-epithelial type II cells and attenuates bleomycin-induced pulmonary fibrosis in vivo. Exp. Lung Res. 41, 422-434. 10.3109/01902148.2015.1060275 [DOI] [PubMed] [Google Scholar]
- Page A., and Mann D. A. (2015). Epigenetic regulation of liver fibrosis. Clin. Res. Hepatol. Gastroenterol. 39 Suppl. 1, S64-S68. 10.1016/j.clinre.2015.05.013 [DOI] [PubMed] [Google Scholar]
- Peng H., Wang Q., Lou T., Qin J., Jung S., Shetty V., Li F., Wang Y., Feng X. H., Mitch W. E. et al. (2017). Myokine mediated muscle-kidney crosstalk suppresses metabolic reprogramming and fibrosis in damaged kidneys. Nat. Commun. 8, 1493 10.1038/s41467-017-01646-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyser R., MacDonnell S., Gao Y., Cheng L., Kim Y., Kaplan T., Ruan Q., Wei Y., Ni M., Adler C. et al. (2019). Defining the activated fibroblast population in lung fibrosis using single-cell sequencing. Am. J. Respir. Cell Mol. Biol. 61, 74-85. 10.1165/rcmb.2018-0313OC [DOI] [PubMed] [Google Scholar]
- Qu J., Zhu L., Zhou Z., Chen P., Liu S., Locy M. L., Thannickal V. J. and Zhou Y. (2018). Reversing mechanoinductive DSP expression by CRISPR/dCas9-mediated epigenome editing. Am. J. Respir. Crit. Care Med. 198, 599-609. 10.1164/rccm.201711-2242OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan A. R. and Hall I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841-842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G., Collard H. R., Egan J. J., Martinez F. J., Behr J., Brown K. K., Colby T. V., Cordier J.-F., Flaherty K. R., Lasky J. A., et al. (2011). An official ATS/ERS/JRS/ALAT statement: idiopathic pulmonary fibrosis: evidence-based guidelines for diagnosis and management. Am. J. Respir. Crit. Care Med. 183, 788-824. 10.1164/rccm.2009-040GL [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos C., Montaño M., García-Alvarez J., Ruiz V., Uhal B. D., Selman M. and Pardo A. (2001). Fibroblasts from idiopathic pulmonary fibrosis and normal lungs differ in growth rate, apoptosis, and tissue inhibitor of metalloproteinases expression. Am. J. Respir. Cell Mol. Biol. 24, 591-598. 10.1165/ajrcmb.24.5.4333 [DOI] [PubMed] [Google Scholar]
- Ren Y., Du C., Shi Y., Wei J., Wu H. and Cui H. (2017). The Sirt1 activator, SRT1720, attenuates renal fibrosis by inhibiting CTGF and oxidative stress. Int. J. Mol. Med. 39, 1317-1324. 10.3892/ijmm.2017.2931 [DOI] [PubMed] [Google Scholar]
- Roadmap Epigenomics Consortium, Kundaje A., Meuleman W., Ernst J., Bilenky M., Yen A., Heravi-Moussavi A., Kheradpour P., Zhang Z., Wang J., Ziller M. J. et al. (2015). Integrative analysis of 111 reference human epigenomes. Nature 518, 317-330. 10.1038/nature14248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Zhuang Y., Suzuki T., Ota Y., Bateman M. E., Alkhatib A. L., Morris G. F. and Lasky J. A. (2019). HDAC8 inhibition ameliorates pulmonary fibrosis. Am. J. Physiol. Lung Cell Mol. Physiol. 316, L175-L186. 10.1152/ajplung.00551.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders Y. Y., Ambalavanan N., Halloran B., Zhang X., Liu H., Crossman D. K., Bray M., Zhang K., Thannickal V. J. and Hagood J. S. (2012). Altered DNA methylation profile in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 186, 525-535. 10.1164/rccm.201201-0077OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders Y. Y., Hagood J. S., Liu H., Zhang W., Ambalavanan N. and Thannickal V. J. (2014). Histone deacetylase inhibition promotes fibroblast apoptosis and ameliorates pulmonary fibrosis in mice. Eur. Respir. J. 43, 1448-1458. 10.1183/09031936.00095113 [DOI] [PubMed] [Google Scholar]
- Sanders Y. Y., Liu H., Scruggs A. M., Duncan S. R., Huang S. K. and Thannickal V. J. (2017). Epigenetic regulation of Caveolin-1 gene expression in lung fibroblasts. Am. J. Respir. Cell Mol. Biol. 56, 50-61. 10.1165/rcmb.2016-0034OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandner P. and Stasch J. P. (2017). Anti-fibrotic effects of soluble guanylate cyclase stimulators and activators: a review of the preclinical evidence. Respir. Med. 122 Suppl. 1, S1-S9. 10.1016/j.rmed.2016.08.022 [DOI] [PubMed] [Google Scholar]
- Schafer S., Viswanathan S., Widjaja A. A., Lim W.-W., Moreno-Moral A., DeLaughter D. M., Ng B., Patone G., Chow K., Khin E. et al. (2017). IL-11 is a crucial determinant of cardiovascular fibrosis. Nature 552, 110-115. 10.1038/nature24676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto E. and Yoshida M. (2014). Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 6, a018713 10.1101/cshperspect.a018713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S., Bates S. E., Wright J. J., Espinoza-Delgado I. and Piekarz R. L. (2010). Clinical toxicities of histone deacetylase inhibitors. Pharmaceuticals (Basel) 3, 2751-2767. 10.3390/ph3092751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Peng R., Phillips J. E., Deguzman J., Ren Y., Apparsundaram S., Luo Q., Bauer C. M., Fuentes M. E., DeMartino J. A. et al. (2013). Assessment of Brd4 inhibition in idiopathic pulmonary fibrosis lung fibroblasts and in vivo models of lung fibrosis. Am. J. Pathol. 183, 470-479. 10.1016/j.ajpath.2013.04.020 [DOI] [PubMed] [Google Scholar]
- Tatler A. L. and Jenkins G. (2012). TGF-beta activation and lung fibrosis. Proc. Am. Thorac Soc. 9, 130-136. 10.1513/pats.201201-003AW [DOI] [PubMed] [Google Scholar]
- Trojanowska M. (2008). Role of PDGF in fibrotic diseases and systemic sclerosis. Rheumatology (Oxf.) 47 Suppl. 5; v2-v4. 10.1093/rheumatology/ken265 [DOI] [PubMed] [Google Scholar]
- Trojer P., Li G., Sims R. J. III, Vaquero A., Kalakonda N., Boccuni P., Lee D., Erdjument-Bromage H., Tempst P., Nimer S. D et al. . et al. (2007). L3MBTL1, a histone-methylation-dependent chromatin lock. Cell 129, 915-928. 10.1016/j.cell.2007.03.048 [DOI] [PubMed] [Google Scholar]
- Tschumperlin D. J., Ligresti G., Hilscher M. B. and Shah V. H. (2018). Mechanosensing and fibrosis. J. Clin. Invest. 128, 74-84. 10.1172/JCI93561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos E., Criollo A., Schiattarella G. G., Altamirano F., French K. M., May H. I., Jiang N., Nguyen N. U. N., Romero D., Roa J. C. et al. (2019). Fibroblast primary cilia are required for cardiac fibrosis. Circulation 139, 2342-2357. 10.1161/CIRCULATIONAHA.117.028752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yu N., Chen D., Lee K. C. L., Lye P. L., Chang J. W. W., Deng W., Ng M. C. Y., Lu T., Khoo M. L. et al. (2011). Discovery of (2E)-3-{2-butyl-1-[2-(diethylamino)ethyl]-1H-benzimidazol-5-yl}-N-hydroxyacrylami de (SB939), an orally active histone deacetylase inhibitor with a superior preclinical profile. J. Med. Chem. 54, 4694-4720. 10.1021/jm2003552 [DOI] [PubMed] [Google Scholar]
- Wang L., Wang S. and Li W. (2012). RSeQC: quality control of RNA-seq experiments. Bioinformatics 28, 2184-2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Wernig G., Chen S.-Y., Cui L., Van Neste C., Tsai J. M., Kambham N., Vogel H., Natkunam Y., Gilliland D. G., Nolan G. et al. (2017). Unifying mechanism for different fibrotic diseases. Proc. Natl. Acad. Sci. USA 114, 4757-4762. 10.1073/pnas.1621375114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong C., Rougier-Chapman E. M., Frederick J. P., Datto M. B., Liberati N. T., Li J.-M. and Wang X.-F. (1999). Smad3-Smad4 and AP-1 complexes synergize in transcriptional activation of the c-Jun promoter by transforming growth factor beta. Mol. Cell Biol. 19, 1821-1830. 10.1128/MCB.19.3.1821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wotton D., Lo R. S., Lee S. and Massagué J. (1999). A Smad transcriptional corepressor. Cell 97, 29-39. 10.1016/S0092-8674(00)80712-6 [DOI] [PubMed] [Google Scholar]
- Wu K., Yang Y., Wang C., Davoli M. A., D'Amico M., Li A., Cveklova K., Kozmik Z., Lisanti M. P., Russell R. G. et al. (2003). DACH1 inhibits transforming growth factor-beta signaling through binding Smad4. J. Biol. Chem. 278, 51673-51684. 10.1074/jbc.M310021200 [DOI] [PubMed] [Google Scholar]
- Wynn T. A. (2008). Cellular and molecular mechanisms of fibrosis. J. Pathol. 214, 199-210. 10.1002/path.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Senavirathna L. K., Gou X., Huang C., Liang Y. and Liu L. (2016). EZH2 enhances the differentiation of fibroblasts into myofibroblasts in idiopathic pulmonary fibrosis. Physiol. Rep. 4, e12915 10.14814/phy2.12915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I. V. and Schwartz D. A. (2015). Epigenetics of idiopathic pulmonary fibrosis. Transl. Res. 165, 48-60. 10.1016/j.trsl.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Cui H., Xie N., Icyuz M., Banerjee S., Antony V. B., Abraham E., Thannickal V. J. and Liu G. (2013). miR-145 regulates myofibroblast differentiation and lung fibrosis. FASEB J. 27, 2382-2391. 10.1096/fj.12-219493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang I. V., Pedersen B. S., Rabinovich E., Hennessy C. E., Davidson E. J., Murphy E., Guardela B. J., Tedrow J. R., Zhang Y., Singh M. K. et al. (2014). Relationship of DNA methylation and gene expression in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 190, 1263-1272. 10.1164/rccm.201408-1452OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G., Tzouvelekis A., Wang R., Herazo-Maya J. D., Ibarra G. H., Srivastava A., de Castro J. P. W., DeIuliis G., Ahangari F., Woolard T. et al. (2018). Thyroid hormone inhibits lung fibrosis in mice by improving epithelial mitochondrial function. Nat. Med. 24, 39-49. 10.1038/nm.4447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C. A., Eeckhoute J., Johnson D. S., Bernstein B. E., Nussbaum C., Myers R. M., Brown M., Li W. et al. (2008). Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.