Abstract
Single nucleotide polymorphisms (SNPs) are the most common genetic variation in mammalian cells with prognostic potential. Anillin-actin binding protein (ANLN) has been identified as being involved in PI3K/PTEN signaling, which is critical in cell life/death control, and kinase insert domain receptor (KDR) encodes a key receptor mediating the cancer angiogenesis/metastasis switch. Knowledge of the intrinsic connections between PI3K/PTEN and KDR signaling, which represent two critical transitions in carcinogenesis, led the present study to investigate the effects of the potential synergy between ANLN and KDR on breast cancer outcome and identify relevant SNPs driving such a synergy at the genetic level. The survival associations of SNPs from KDR and ANLN were assessed through pairwise interaction survival analysis, quantitative trait loci analysis, pathway enrichment analysis and network construction, and the interactions between ANLN and KDR were validated in vitro. It was found that both rare homozygotes in the ANLN:rs12535394 and KDR:rs11133360 SNP pair are prognostic of favorable breast cancer survival and underpin the prominent roles of the immune response in cancer state control. This study contributes to breast cancer prognosis and therapeutic design by providing genetic evidence of interactions between ANLN and KDR, and suggesting the prominent role of the immune response in driving the synergies between the cancer cell life/death and angiogenesis/metastasis transitions during carcinogenesis.
Keywords: ANLN, KDR, single nucleotide polymorphisms, survival, breast cancer
Introduction
Breast cancer is the leading cause of cancer-related mortality amongst women worldwide (1), with a mortality rate of ~627,000 annually estimated in 2018 (2). Uncontrolled proliferative growth and angiogenesis are two basic cancer hallmarks governing the critical transitions towards malignancy during carcinogenesis (3). PI3K/PTEN signaling, frequently altered in breast carcinoma (4), confers a survival advantage to tumor cells (5). Anillin, encoded by anillin actin-binding protein (ANLN), is an actin-binding protein, which has been identified as being involved in the PI3K/PTEN pathway (6,7). It is an F-actin binding protein, which maintains podocyte cytoskeletal dynamics, cell motility and signaling through its interaction with CD2-associated protein, which stimulates the phosphorylation of AKT at serine 473 (6,8). The inhibition of PI3K/AKT activity in non-small cell lung cancer cells decreased ANLN stability and reduced nuclear levels, suggesting the critical involvement of ANLN in PI3K/AKT signaling (7). ANLN also serves a significant role in pulmonary carcinogenesis through PI3K/AKT pathway-dependent nuclear function (7). The nuclear expression of ANLN in tumor cells is independently prognostic of a poor outcome in patients with breast cancer (9,10), and ANLN mutations are suggestive of estrogen receptor-positive breast cancer tumorigenesis and endocrine therapy resistance (11) due to the hyperactivation of PI3K/PTEN (6). VEGFR-2, also known as kinase insert domain receptor (KDR), promotes angiogenesis (12,13). PI3K/PTEN activation enhances VEGF signaling, forming a positive feedback loop leading to uncontrolled progressive signaling in tumor cells (4,5). This body of evidence is indicative of the potential synergy between ANLN and KDR influencing breast cancer prognosis.
The majority of breast malignancies are caused by acquired and uncorrected genetic adjustments in somatic genes due to inherited gene shuffling (14). Single nucleotide polymorphisms (SNPs) represent a predominant genetic variation in the human genome (15), a large number of which are associated with various types of cancer (16). ANLN undergoes genetic changes, including amplification, deletion and SNP mutations, in patients with several types of cancer; mutation rates vary between 0.2% in clear cell renal cell carcinoma and 19.6% in prostate cancer (17,18). A total of 27 mutations, including 12 amplifications, two deletions and 13 SNPs, were identified in lung adenocarcinoma (19). The potential functional genetic variant rs10013228 in KDR is a prognostic marker of resected colorectal cancer (20) and renal cell carcinoma (21). SNPs rs10020464, rs11941492 and rs12498529 of KDR are associated with KRAS2-mutated tumors, which are also microsatellite instable and CpG island methylator phenotype-positive (22), and rs11941492 is significantly associated with the early onset of esophageal adenocarcinoma (23). The genetic variant rs1870377 of KDR is associated with sunitinib-mediated overall survival (OS) rate (24). However, no SNPs of KDR with clinical implications have been reported for breast cancer (25). In the present study, the potential synergy between ANLN and KDR and its effect on breast cancer outcome were investigated, and relevant SNPs driving this synergy at the genetic level were identified.
Materials and methods
Datasets
A total of 14,481 SNPs for ANLN and 11,704 SNPs for KDR were retrieved from the dbSNP NCBI database (26). Among these, 20 SNPs of ANLN and 13 SNPs of KDR were mapped to the Affymetrix SNP6.0 Array, which was used in The Cancer Genome Atlas (TCGA; http://cancergenome.nih.gov). Genotype data of the 33 SNPs covering 501 samples were retrieved from the TCGA. The gene expression and clinical data were retrieved from the TCGA bioportal (http://www.cbioportal.org/), containing 20,440 genes and 1,102 samples.
Pairwise SNP survival analysis
Breast cancer OS analysis was conducted on interactions between SNPs of ANLN and KDR using the Cox proportional hazard model. The recessive and dominant models were tested in the pairwise SNP association analysis. In the recessive model, the heterozygote is combined with the common homozygote, assuming that the disease-associated phenotype is caused by the concomitant presence of both rare alleles; the dominant model combines the heterozygote with the rare homozygote, assuming that the disease-associating phenotype is caused by the presence of the rare allele. A 10-year breast cancer OS analysis was performed utilizing the ‘survival’ package (27) in R software (28) and a log-rank test was used to assess the statistical significance of the association between SNPs and the clinical outcome. An SNP pair was considered interactive if the P-values of the Cos regression model and the interaction term were both <0.05 and the number of iterations was <10.
Expression quantitative trait loci analysis (eQTL) and function predictions
To identify those genes for which expression was significantly affected by the identified disease-associating SNPs, eQTL analysis was performed using a linear model in R to identify genes associated with complex phenotypes (29). Whether the allele status (rare homozygote, heterozygote or common homozygote) of a given SNP was linearly associated with the expression of a given gene was assessed. The top 25 percentile of the SNPs with a P-value in the linear model at P<0.05 were considered eQTLs of a gene.
The combined impact of ANLN and KDR on genes identified from the eQTL analysis was investigated by stratifying the expression of the genes of interest by the combined expression of ANLN and KDR. One-way ANOVA and a Least Significant Difference (LSD) post hoc test were applied using R (version 3.5.2) to assess the statistical significance, with P<0.05 used as the threshold suggestive of a significant trilateral correlation.
The PredictSNP version 2.1 interface (30) was used to predict the functional interaction network of the SNPs of interest, which uses a series of tools and databases for SNP functional prediction. PredictSNP2 provides easy access to binary predictions and uniform confidence values for the five best-performing prediction tools CADD, DANN, FATHMM, FunSeq2 and GWAVA, and the results obtained from these tools are combined into a consensus score (31). CADD (32) estimates the relative pathogenicity of human genetic variants, DANN (33) uses a deep learning approach for annotating the pathogenicity of genetic variants, GWAVA (34) is designed for the analysis of regulatory variants, and the FunSeq2 (31) framework annotates and prioritizes non-coding regulatory variants in cancer.
Pathway analysis and network construction
In order to investigate the biologically functional consequences introduced by SNPs, pathway enrichment analysis was performed using genes affected by SNPs with statistical significance. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway (http://www. genome.jp/kegg/) (35) and Gene Ontology (GO) term enrichment analyses were performed using the packages clusterProfiler (36) and org.Hs.eg.db (37) in R, and Chi-square and Fisher's exact tests were used for statistical assessment.
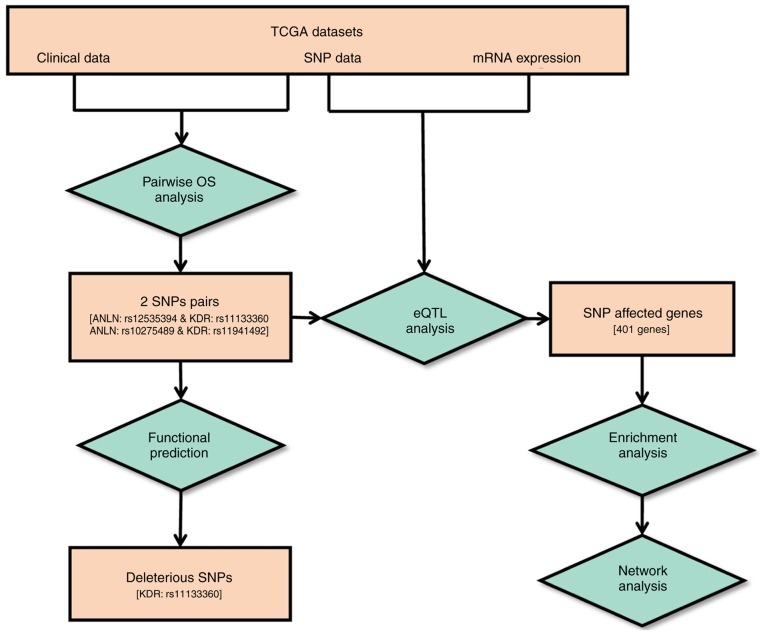
The genes identified from the enriched pathways were collected for gene regulatory network construction using GeneMANIA (http://www.genemania.org) (38), which uses the label propagation algorithm to predict gene-gene interactions at seven levels (co-expression, co-localization, genetic interaction, physical interaction, shared protein domain, pathway and predicted). The interactions at the co-localization, genetic interaction, physical interaction, shared protein domain and pathway levels were used for network construction. The output comprises a regulatory network that uses the user-defined gene list based on databases and publications from multiple resources (38,39). The ‘max resultant genes’ was set as five, the ‘max resultant attributes’ was set as 10, and the GO weighting system was used, which uses biological process (BP)-based, molecular function (MF)-based and cellular component (CC)-based approaches. The complete workflow is illustrated in Fig. 1.
Figure 1.
Workflow of the study. The square boxes represent data or results and the diamond boxes show the analytical processes. Values shown within square brackets represent the quantity of the results obtained. SNPs, single nucleotide polymorphisms; TCGA, The Cancer Genome Atlas; OS, overall survival; eQTL, expression quantitative trait loci analysis.
Experimental validation
Cell culture
One human normal mammary epithelial cell line (MCF10A) and one breast cancer cell line (SUM149PT) were purchased from ATCC and used in the present study. The MCF10A cells were cultured in DMEM/F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 5% charcoal-stripped horse serum (Gibco; Thermo Fisher Scientific, Inc.), 10 µg/ml insulin (PeproTech, Inc.), 20 ng/ml epithelial growth factor (PeproTech, Inc.) and 1.4×10−6 mol/l hydrocortisone (PeproTech, Inc.). The SUM149PT cells were cultured in F12 (Gibco; Thermo Fisher Scientific, Inc.) supplemented with 5% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 20 µg/ml insulin (PeproTech, Inc.), 1% HEPES (PeproTech, Inc. and 2.8×10−6 mol/l hydrocortisone (PeproTech, Inc.). Assay-ready cells were prepared by culturing the cells in a large batch and aliquoting them into ampules that were kept in liquid nitrogen in solution containing 90% FBS and 10% DMSO. Immediately prior to transfection, the cells were thawed and washed with culture medium and the cell number was counted using a hemocytometer (Thermo Fisher Scientific, Inc.).
SgRNA preparation
pLenti-U6-sgRNA-PGK-Neo (cat. no. K019), piLenti-EF1a-dCas9-SAM (cat. no. K015) and piLenti-EF1a-dCas9-KRAB (cat. no. K203) were purchased from Applied Biological Materials, Inc. The sgRNAs were designed using the publicly available software CHOPCHOP (version 3, http://chopchop.cbu.uib.no/) (40) (Table SI), supplemented with the BbsI restriction site, and synthesized from Applied Biological Materials (ABM), Inc. Each sgRNA was added with a sequence complementary to the sticky ends of BbsI, following ligation with the pLenti-U6-sgRNA-PGK-Neo vector digested using BbsI, and the recombinant plasmid was amplified in DH5 Escherichia coli (Sigma-Aldrich; Merck KGaA). The plasmids containing sgRNAs were validated using PCR, enzyme digestion and sequencing, and transfected together with piLenti-EF1a-dCas9-SAM or piLenti-EF1a-dCas9-KRAB into cells. The controls were designed as cells concomitantly transfected with all sgRNAs modulating the target gene alone, without piLenti-EF1a-dCas9-SAM or piLenti-EF1a-dCas9-KRAB.
Cell transfection
A total of 1×106 cells per well were added in 2 ml of culture medium and transferred to 6-well plates (Nalgene, cat. no. 167018). The cells were incubated overnight and were at 70–80% confluence prior to transfection. The medium was replaced with 2 ml serum-free medium prior to transfection. Subsequently, 100 µl Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.) containing 1 µg sgRNA (ABM, Inc.) and 1 µg CRISPR/dCas9 (ABM, Inc.) plasmids were added to 100 µl Opti-MEM containing 6 µl lipo2000 transfection reagent per well and mixed for 15–20 min prior to transfection. The mixture was transferred to 6-well plate and incubated at 37°C for 5–8 h in the presence of 5% CO2 (HERA Cell 150i, Thermo Fisher Scientific, Inc.). The serum-free medium was replaced with 2 ml medium containing 10% serum. The cells were incubated at 37°C for 24 h, followed by the addition of G418 and puromycin and incubation for 48 h.
Reverse transcription-quantitative PCR (RT-qPCR) assay
Following transfection, the cells were collected and extracted for total RNA using TRIzol reagent (Tiangen Biotech Co., Ltd.) 3 days after transfection. The cDNA was synthesized using PrimeScript RT reverse transcriptase as per the manufacturer's protocol (Takara Bio, Inc.). The primers used for RT-qPCR analysis are listed in Table SII. The qPCR sample consisted of 5 µl 2X SYBR premix ex Taq, 0.4 µl, 10 µm forward and reverse primers, 0.2 µl ROX reference dye, 2 µl cDNA and 2 µl H2O. The detailed procedure for RT-qPCR was as follows: Initial denaturation at 95°C for 5 min, 45 cycles of denaturation at 95°C for 5 sec, annealing at 57°C for 30 sec, extension at 72°C for 15 sec. The absorbance value was recorded at the extension stage. The relative expression level was calculated using the 2−∆∆Cq method (41). All RT-qPCR experiments were performed using the ABI StepOnePlus Real-Time PCR system (ABI; Thermo Fisher Scientific, Inc.).
Statistical analysis
One-way ANOVA coupled with Scheffe's post hoc test were conducted using R software (version 3.5.2) to evaluate the significance of changes in the expression level of ANLN or KDR following genetic modulations compared with each corresponding control where the P-value threshold was set as 0.05.
Results
SNPs of ANLN and KDR synergistically affect breast cancer survival rates
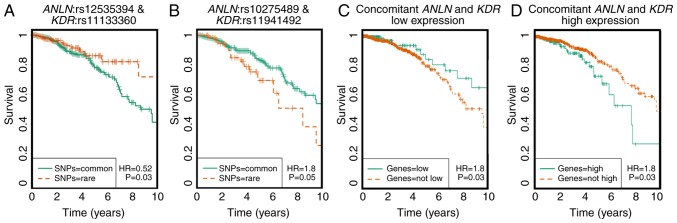
Pairwise SNP survival analysis was performed using multivariate Cox regression models and statistical significance was assessed using the likelihood ratio test. Of all 260 SNP pairs, two pairs were identified with significant synergistic effects on breast cancer survival rate from the dominant model (Fig. 2A-D). The two pairs were ANLN:rs12535394 and KDR:rs11133360 and ANLN:rs10275489 and KDR:rs11941492. The concurrent presence of both rare homozygotes in the ANLN:rs12535394 and KDR:rs11133360 SNP pair is associated with favorable clinical outcomes (HR=0.52, P=0.03), whereas the ANLN:rs10275489 and KDR:rs11941492 SNP pair was associated with poor breast cancer prognosis (HR=1.8, P=0.05). These four SNPs are non-linkage disequilibrium (LD) linked and were defined as disease-associated SNPs. Of these four disease-associated SNPs, KDR:rs11133360 was identified as intronic (chr4:55116585) and its rare allele was predicted as deleterious by the FunSeq2 and GWAVA tools (Table I).
Figure 2.
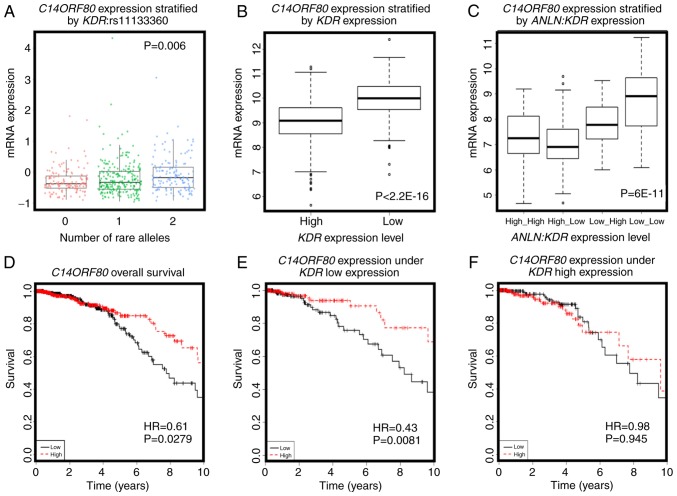
Kaplan-Meier plots showing the prognostic value of the identified disease-associated SNP pair and the corresponding gene pair. (A) Overall breast cancer survival for the SNP pair ANLN:rs12535394 and KDR:rs11133360. (B) Overall breast cancer survival for the SNP pair ANLN:rs10275489 and KDR:rs11941492. ‘SNPs=rare’ refers to rare homozygote, ‘SNPs=common’ represents either a common homozygote or heterozygote. (C) Overall breast cancer survival with concomitantly low expression of ANLN and KDR. ‘Genes=low’ refers to concomitant low expression of both genes, ‘Genes=not low’ includes cases where expression of at least one gene is not low. (D) Overall breast cancer survival with concomitantly high expression of ANLN and KDR. ‘Genes=high’ refers to concomitant high expression of both genes, ‘Genes=not high’ includes cases where expression of at least one gene is not high. SNP, single nucleotide polymorphism; ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor.
Table I.
Effects of the identified disease-associated single nucleotide polymorphisms predicted using PredictSNP2.
| KDR: rs11133360 (position: chr4:55116585) | KDR: rs11941492 (position: chr4:55112043) | |||||
|---|---|---|---|---|---|---|
| Function predictor | Prediction | Score | Expected accuracy | Prediction | Score | Expected accuracy |
| PredictSNP2 | Neutral | −1 | 0.88 | Neutral | −1 | 0.88 |
| CADD | Neutral | 8.242 | 0.76 | Neutral | 3.493 | 0.82 |
| DANN | Neutral | 0.3796 | 0.8 | Neutral | 0.5371 | 0.82 |
| FATHMM | Neutral | 0.1461 | 0.91 | Neutral | 0.0614 | 0.95 |
| FunSeq2 | Deleterious | 2.3903 | 0.67 | – | 0.7762 | 0.45 |
| GWAVA | Deleterious | 0.49 | 0.64 | Neutral | 0.15 | 0.79 |
For KDR: rs11133360 and KDR: rs11941492, the common allele was C, the rare allele was T and the region was intronic. PredictSNP2 provides easy access to other tools, including CADD, DANN, FATHMM, FunSeq2 and GWAVA, with binary predictions and uniform confidence values as the outputs.
Genes affected by disease-associated SNPs
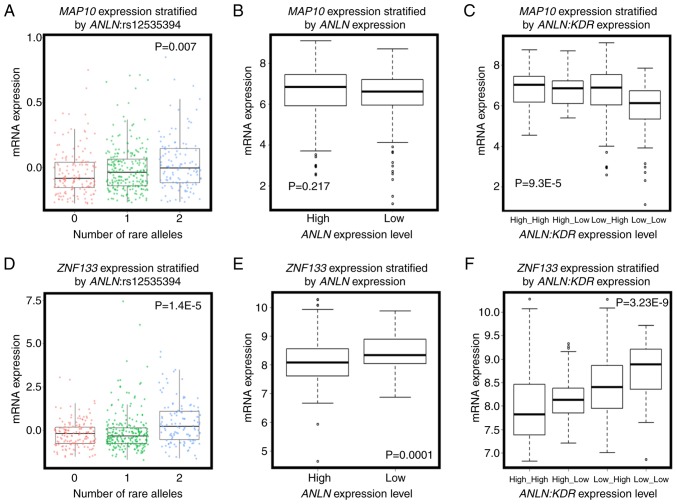
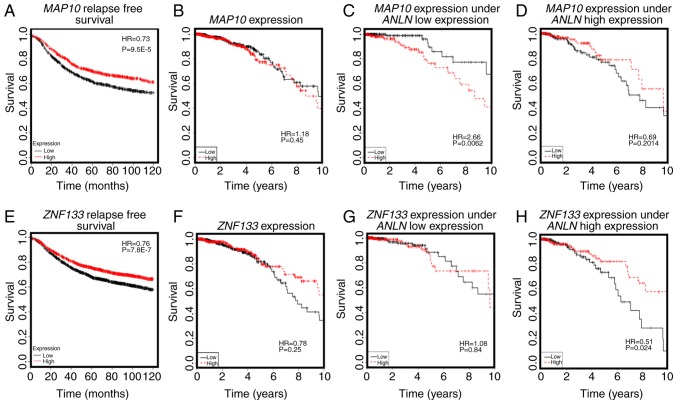
The eQTL analysis of the four disease-associated SNPs resulted in 401 genes with KEGG annotations (Tables SIII and SIV; Figs. 3 and 4). Amongst these genes, the expression of microtubule-associated protein 10 [MAP10; correlation (cor)=0.554, P=0.001] was significantly positively correlated with the number of rare alleles of ANLN:rs12535394 (P=0.007, Fig. 3A), and interacted with ANLN at the transcriptional level to predict breast cancer OS (Fig. 4C and D). Furthermore, the overexpression of MAP10 conveyed favorable clinical outcomes (HR=0.73, P=9.5E-5, Fig. 4A), but was associated with poor breast cancer OS when the expression of ANLN was concomitantly low (HR=2.66, P=0.0062, Fig. 4C). The expression of MAP10 was not directly associated with that of ANLN (cor=0.04, P=0.198, Fig. 3B), but was expressed at a low level under the concomitant low expression of both ANLN and KDR (P=9.3E-5, Fig. 3C), further suggesting the involvement of MAP10 in the synergy created between ANLN and KDR at the transcriptional level. Zinc finger protein 133 (ZNF133) was the top gene whose expression was significantly associated with the allele status of ANLN:rs12535394. The SNP rare allele was associated with a high and statistically significant expression of ZNF133 (P=1.4E-5). The rare allele status of this SNP was positively correlated with the expression of ZNF133 (cor=0.14, P=0.3.8E-6, Fig. 3D) and negatively correlated with the expression of ANLN. The expression of ZNF133 was stratified into distinct expression levels by the expression of ANLN (P=0.0001); the expression of ZNF133 and expression of ANLN were negatively correlated (cor=−0.22, P=1.6E-12, Fig. 3E), suggesting a negative correlation between the rare allele of ANLN:rs12535394 and the expression of ANLN. Such a negative association was exemplified by the low expression of KDR, i.e., the concomitant underexpression of ANLN and KDR was associated with the overexpression of ZNF133 (P=3.23E-9, Fig. 3F). Given the favorable clinical outcome associated with the high expression of ZNF133 (HR=0.76, P=7.8E-7, Fig. 4E), it was reasoned that the ANLN rare allele is associated with desirable breast cancer relapse-free survival. Furthermore, ZNF133 interacted with ANLN to influence breast cancer OS at the transcriptional level (Fig. 4G and H), providing further evidence of the association between the allele status of ANLN:rs12535394 and the expression and prognostic value of ANLN. Similar to ZNF133, C14ORF80 was the top gene whose expression was significantly positively correlated with the rare allele expression of KDR:rs11133360 (cor=0.2, P=0.001). The SNP rare allele was significantly associated with a high expression of C14ORF80 (P=0.006, Fig. 5A) and negatively correlated with the expression of KDR (cor=−0.4, P<2.2E-16). The expression of C14ORF80 was stratified into distinct expression levels by the expression of KDR (P<2.2E-16, Fig. 5B), indicating a negative correlation between the rare allele of KDR:rs11133360 and expression of KDR. In addition, the concomitant high expression of ANLN and KDR was associated with a high expression of C14ORF80 (P=6E-11, Fig. 5C), which was prognostic of favorable breast cancer OS (HR=0.61, P=0.0279, Fig. 5D), suggesting the favorable prognostic value of KDR:rs11133360. Furthermore, C14ORF80 was shown to interact with KDR to influence breast cancer OS at the transcriptional level (Fig. 5E and F), supporting the association between the allele status of KDR:rs11133360 and the expression and prognostic value of KDR. The rare alleles of ANLN:rs12535394 and KDR:rs11133360 have been implicated with favorable clinical outcomes, which is consistent with the observations at the SNP level in the present study (HR=0.52, P=0.0313, Fig. 2A), and have been associated with a low expression of ANLN and KDR at the transcriptional level, which was in accordance with what was observed at the transcriptional level in the present study (Fig. 2C).
Figure 3.
Evidence of associations between ANLN:rs12535394 and the expression of ANLN. (A) Association between the allele status of ANLN:rs12535394 and the gene expression of MAP10. (B) Expression of MAP10 stratified by that of ANLN. (C) Expression of MAP10 stratified by the joint expression of ANLN and KDR. (D) Association between the allele status of ANLN:rs12535394 and the gene expression of ZNF133. (E) Expression of ZNF133 stratified by that of ANLN. (F) Expression of ZNF133 stratified by the joint expression of ANLN and KDR. 0, 1 and 2 represent the numbers of rare alleles in ANLN:rs12535394. The Cancer Genome Atlas data was used for producing all panels. ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor; MAP10, microtubule-associated protein 10; ZNF133, zinc finger protein 133.
Figure 4.
Prognostic value of key genes associated with ANLN:rs12535394 and their impact on the interactions between ANLN and KDR on breast cancer survival. (A) Prognostic value of MAP10 on breast cancer relapse-free survival, drawn using Kaplan Meier Plotter. (B) Prognostic value of MAP10 on breast cancer overall survival drawn using TCGA data. (C) Expression of MAP10 on breast cancer survival under ANLN low expression, drawn using TCGA data. (D) Expression of MAP10 on breast cancer survival under high expression of ANLN, drawn using TCGA data. (E) Prognostic value of ZNF133 on breast cancer relapse-free survival, drawn using Kaplan Meier Plotter. (F) Prognostic value of ZNF133 on breast cancer overall survival, drawn using TCGA data. (G) Expression of ZNF133 on breast cancer survival under low expression of ANLN, drawn using TCGA data. (H) Expression of ZNF133 on breast cancer survival under high expression of ANLN, drawn using TCGA data. ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor; MAP10, microtubule-associated protein 10; ZNF133, zinc finger protein 133; TCGA, The Cancer Genome Atlas.
Figure 5.
Evidence of associations between KDR:rs11133360 and the expression of KDR and the prognostic value of a key gene associated with ANLN:rs12535394 and its impact on interactions between ANLN and KDR on breast cancer survival. (A) Association between the allele status of KDR:rs11133360 and the gene expression of C14ORF80. 0, 1 and 2 represents the number of rare alleles in KDR:rs11133360. (B) Expression of C14ORF80 stratified by that of KDR. (C) Expression of C14ORF80 stratified by the joint expression of ANLN and KDR. (D) Prognostic value of C14ORF80 on breast cancer overall survival, drawn using TCGA data. (E) Expression of C14ORF80 and breast cancer survival under low expression of KDR. (F) Expression of C14ORF80 and breast cancer survival under high expression of KDR. TCGA data was used for drawing all panels. The prognostic value of C14ORF80 on breast cancer overall survival drawn using Kaplan Meier Plotter is not present as this gene was not available in that database. ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor; TCGA, The Cancer Genome Atlas.
No genes were found to be significantly associated with the ANLN:rs10275489 allele status and expression of ANLN, or with the KDR:rs11941492 allele status and expression of KDR.
Pathway and network construction using eQTL genes influenced by disease-associated SNPs
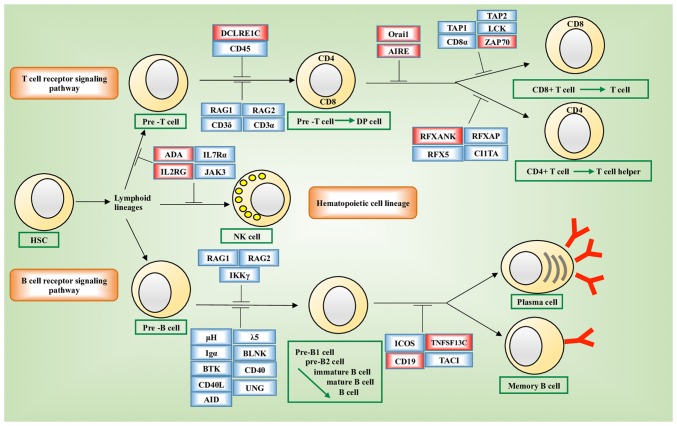
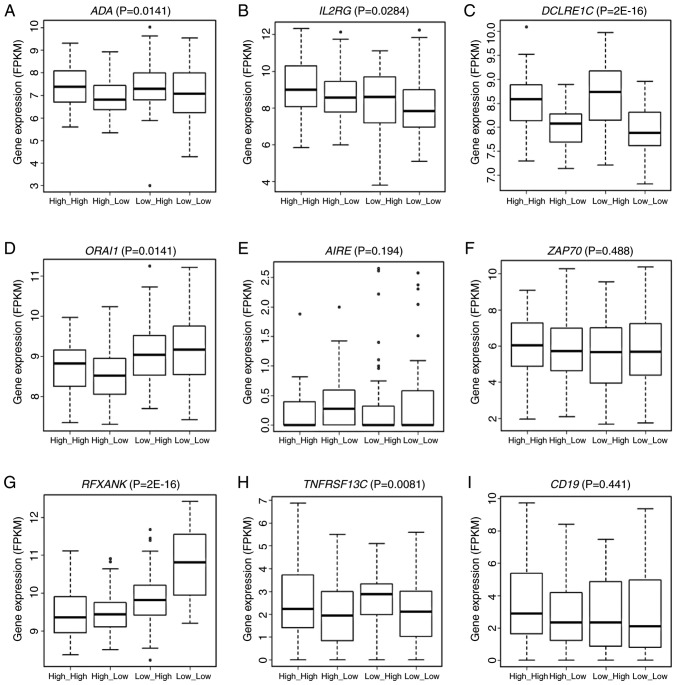
Pathway enrichment analysis showed that these genes were significantly enriched in the primary immunodeficiency disorder (PID) pathways (hsa05340, P=10−4; Fig. 6). Genes enriched in hsa05340 included ORAI1, DCLRE1C, IL2RG, RFXANK, ADA, AIRE, CD19, TNFRSF13C and ZAP70. The expression of six of these nine genes, i.e., ADA (P=0.0141), IL2RG (P=0.0284), DCLRE1C (P=2E-16), ORAI1 (P=0.0141), RFXANK (P=2E-16) and TNFRSF13C (P=0.0081) varied significantly across the groups stratified by the joint assessment of ANLN and KDR expression (Fig. 7A-I). Significant pairs were ‘high_high vs. low_high’, ‘high_low vs. low_high’, ‘high_low vs. low_low’ in ADA, ‘high_high vs. low_high’ and ‘high_high vs. low_low’ in IL2RG, ‘high_high vs. low_high’, ‘high_low vs. low_high’, ‘high_high vs. low_low’, ‘high_low vs. low_low’ in DCLRE1C, ‘high_high vs. high_low’, ‘high_high vs. low_high’, ‘high_high vs. low_low’, ‘high_low vs. low_high’, ‘low_high vs. low_low’ in ORAI1, ‘high_high vs. high_low’, ‘high_low vs. low_high’, ‘high_high vs. low_low’, ‘high_low vs. low_low’, ‘low_high vs. low_low’ in RFXANK, and ‘high_low vs. low_high’ and ‘high_low vs. low_low’ in TNFRSF13C. All statistical P-values from ANOVA and the LSD test for pairwise comparisons are listed in Table SV.
Figure 6.
Pathway enrichment analysis of genes with eQTL associations with identified disease-associated SNPs. (A) Enriched pathways of genes with eQTL associations with identified disease-associated SNPs. The ‘primary immunodeficiency pathway (hsa05340)’ was enriched from the Kyoto Encyclopedia of Genes and Genomes (36) and is illustrated. Genes whose expression were quantitatively associated with the identified disease-associating SNPs are highlighted in red. Cell developmental stages are annotated using green boxes. HSC, hematopoietic stem cell; DP, CD4+CD8+; NK, natural killer; eQTL, expression quantitative trait loci.
Figure 7.
Expression of PID pathway genes (A-I) with expression quantitative trait loci associations stratified by the expression of ANLN and KDR. ANOVA and the LSD post hoc test were used to assess the statistical significance of variations in the expression of one PID pathway gene stratified by the joint expression of ANLN and KDR. The expression of 9 genes in total was presented in boxplot where data were stratified by high/low expression of ANLN and KDR. PID, primary immunodeficiency disorder; ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor.
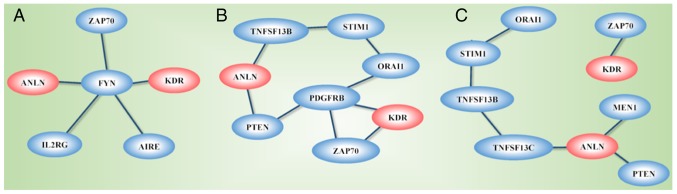
The network constructed from GeneMANIA using KDR, ANLN and the genes affected by the disease-associated SNPs as the initial input exhibited different topological structures when constructed using different GO-weighted approaches (Fig. 8A-C). In addition to ANLN and KDR, the network included PTEN, PDGFRB, ZAP70, ORAI1, STIM1, TNFSF13B and TNFRSF13C when BP was used as the weighting approach, FYN, ZAP70, IL2RG and AIRE when the weighting criteria was based on MF, and PTEN, MEN1, TNFRSF13C, TNFSF13B, STIM1, ORAI1 and ZAP70 when CC was used in weighting. The network was segregated into two areas when CC was used as the weighting method.
Figure 8.
Networks of ANLN and KDR. Networks were constructed using GeneMania (39) with (A) molecular function, (B) biological process, (C) cellular component as the weighting strategies. ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor.
Experimental validation of interactions between ANLN and KDR
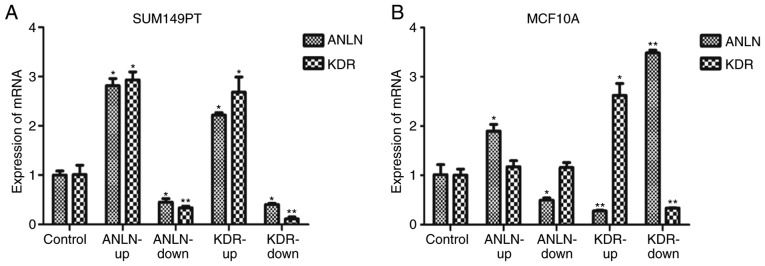
In the in vitro experiments, ANLN did not affect the expression of KDR in the MCF10A normal breast epithelial cells (P=0.27 when ANLN was upregulated, P=0.09 when ANLN was downregulated, Fig. 9B), but positively regulated the expression of KDR in the SUM149PT breast cancer cells (P=2.74E-6 when ANLN was upregulated, P=1.47E-2 when ANLN was downregulated, Fig. 9A). KDR had an opposing effect on the expression of ANLN in the MCF10A cells (P=2.82E-4 when KDR was upregulated, P=3.36E-9 when KDR was downregulated, Fig. 9B), but positively influenced the expression of ANLN in SUM149PT cells (P=1.82E-7 when KDR was upregulated, P=1.25E-4 when KDR was downregulated, Fig. 9A). All statistical P-values from ANOVA with Scheffe's test for all pairwise comparisons are listed in Table SVI.
Figure 9.
Interactions between ANLN and KDR as validated using SUM149PT and MCF10A cells. (A) Expression of KDR on modulating the expression of ANLN and the expression of ANLN after modulating KDR in SUM149PT breast cancer cells. (B) Expression of KDR after modulating the expression of ANLN and the expression of ANLN after modulating KDR in MCF10A normal breast epithelial cells. Bars represent the mean ± SD from at least three independent experiments (*0.01<P<0.05 and **P<0.01 by ANOVA and Scheffe's post hoc test). Gene expression in each experimental group was normalized by the corresponding control. ANLN-up, dCas9-SAM and sgRNA upregulating ANLN were co-transfected into SUM149PT or MCF10A cells; ANLN-down, dCas9-KRAB and sgRNA downregulating ANLN co-transfected into cells; KDR-up, dCas9-SAM and sgRNA upregulating KDR co-transfected into cells; KDR-down, dCas9-KRAB and sgRNA downregulating KDR co-transfected into cells; Control, sgRNAs up- and downregulating ANLN and KDR co-transfected into cells; ANLN, anillin actin-binding protein; KDR, kinase insert domain receptor.
Discussion
Through pairwise interactive OS analyses of the SNPs of ANLN and KDR, the present study identified four disease-associated SNPs (ANLN:rs12535394, KDR:rs11133360, ANLN:rs10275489 and KDR:rs11941492), where ANLN:rs12535394 paired with KDR:rs11133360, and ANLN:rs10275489 paired with KDR:rs11941492 to synergistically influence the clinical outcome of breast cancer. Of the two SNP pairs, the allele status of ANLN:rs12535394 and KDR:rs11133360 was associated with the expression of ANLN and KDR, respectively, via ZNF133 and C14ORF80, and were synergistically prognostic of a favorable clinical outcome in breast cancer.
The quantity of the rare allele of ANLN:rs12535394 was positively associated with the expression of ZNF133 and negatively correlated with the expression of ANLN with statistical significance, and the overexpression of ZNF133 was prognostic for a favorable clinical outcome. These results indicate that the rare allele of ANLN:rs12535394 is protective and associated with a low expression of ANLN via ZNF133. Therefore, ZNF133 interacts with ANLN to affect breast cancer survival, in which the protective effect of the overexpression of ZNF133 is amplified under the high expression of ANLN (Fig. 4F and H). Few ZNF133 functionalities have been reported in cancer, but the transcriptionally suppressive activity of zinc finger protein has been reported for its overamplification in neuroblastoma cells (42) and overexpression in chronic myeloid leukemia (43) in two independent high-throughput studies. These findings collectively suggest the tissue-specific pathological functionalities of ZNF133, i.e., ZNF133 is tumor suppressive in breast cancer, particularly in situations under a high expression of ANLN, which warrants experimental validation.
Furthermore, the quantity of the rare allele of ANLN:rs12535394 was positively associated with the expression of MAP10. MAP family proteins regulate microtubule properties (44) and serve an important role in an array of cellular processes, including cell division, cell motility, intracellular trafficking, microtubule stability and cell morphology maintenance (45). The overexpression and post-translational modifications of MAPs contribute to the dysregulation of microtubule dynamics and the development of serious diseases including human breast cancer (46–50). In the present study, the overexpression of MAP10 was not pathologically relevant but conveyed a poor clinical outcome when the expression of ANLN was low (Fig. 4C), suggesting the conditional prognostic value of MAP10 in breast cancer, i.e., protective when the expression of ANLN is high and risky when the expression of ANLN is low.
Similarly, the quantity of the rare allele of KDR:rs11133360 was positively associated with the expression of C14ORF80, which was significantly negatively correlated with the expression of KDR, and the overexpression of C14ORF80 was prognostic for favorable breast cancer OS (Fig. 5); these results indicate that the rare allele of KDR:rs11133360 is negatively associated with the expression of KDR via the overexpression of C14ORF80 and conveys desirable prognostic value on the clinical outcome of breast cancer. The prognostic value of C14ORF80 was increased under a low expression of KDR, suggesting interactions between these genes. C14ORF80, also termed TEDC1, has not previously been annotated nor shown to be associated with cancer. There are currently no publications available on this gene from the Web of Science. The present study highlights the potential prognostic role of C14ORF80 in predicting breast cancer survival rates and its involvement in KDR-mediated tumor angiogenesis.
The synergy created amongst SNPs affecting breast cancer survival may involve more than two SNPs. However, it is plausible to include fewer indicators in the diagnostic panel for the sake of clinical convenience. Therefore, pairwise interaction models were used initially in the present study to examine the potential synergy; establishing multivariate models was a consideration if no specific results were obtained from the pairwise interaction analysis.
A total of 401 genes were obtained for which the expression profiles were significantly affected by the disease-associated SNPs and these were annotated in the KEGG. These genes were enriched in the PID pathway. PID is a diverse group of illnesses characterized by defects in the function of one or more components of the immune system, which predisposes affected individuals to an increased incidence of infections, autoimmunity and malignancies (51). Patients with PID are at increased risk of developing malignancies compared with healthy individuals in the population (52), and the overall risk for developing cancer in patients with PID was estimated to be up to 25% (53). The majority of these identified genes have immune-related functionalities. For example, ADA encodes an enzyme that increases the rate of hydrolyzation of adenine to inosine and serves a potential role in the development of the immune system and maturation of mammalian cells (54). IL2RG encodes a protein that is an important signaling component of numerous interleukin receptors, including those of interleukin-2, −4, −7 and −21. Importantly, of the nine genes involved in the PID pathway and transcriptionally associated with the rare allele status of these disease-associated SNPs, six are significantly affected by interactions between ANLN and KDR at the transcriptional level. Together, these results suggest the prominent role of the immune response in the synergies created between ANLN and KDR on their prognostic value in breast cancer survival, which may be the driving force behind the life/death control of cells and angiogenesis/metastasis transition during carcinogenesis.
The network constructed using ANLN, KDR and genes affected by the identified disease-associated SNPs as the input revealed differential networks depending on the weighting approaches used. ANLN and KDR were connected through PTEN and PDGFRB when BP weighting was used, were linked through FYN when MF weighting was used, and did not connect when CC was used as the weighting approach. The differential topological structures obtained through the use of different GO weighting approaches suggest that ANLN and KDR interact and create synergies in BPs and MFs, but do not share the use or functionalities of CCs. PDGFRB encodes a typical receptor tyrosine kinase, PDGFRβ, which physically interacts with PTEN according to an in situ proximity ligation assay (55). PTEN is a representative molecule in PI3K/PTEN signaling that shares the same biological pathway with ANLN (7). FYN physically interacts with ANLN according to the human interactome generated from quantitative proteomics (56), and FYN shares similar oncological roles with ANLN, i.e., the overexpression of FYN promotes cell proliferation, migration and invasion in breast cancer cells (57,58).
Experimentally the present study showed that ANLN and KDR interact in normal breast epithelial cells and breast cancer cells. ANLN alterations did not affect the expression of KDR, however, modulating the expression of KDR led to the inverse regulation of ANLN in normal breast epithelial cells, suggesting that KDR is an upstream regulator of ANLN under normal conditions. When the cells were attracted in the malignant state, altering either ANLN or KDR led to regulation of the other gene in the same direction, suggesting the formation of a feed-forward loop that may lead to oncological signal amplification. This is clinically plausible as the concomitant low expression of ANLN and KDR, which is associated with favorable breast cancer survival, was easily achieved by targeting ANLN or KDR alone, with ANLN being a more plausible therapeutic target than KDR as it is a downstream effector of KDR in non-malignant cells. By contrast, a change in the interaction mode of the two genes in normal and cancer cells implicates the importance of the ANLN-KDR interaction in the transition of cells between normal and cancerous states and on breast cancer clinical outcomes. Whether ANLN-KDR interactions constitute to or are the consequence of carcinogenesis remain to be elucidated.
The present study used a triple-negative breast cancer cell line to experimentally validate interactions between ANLN and KDR, as the main effect of the synergy is driven by that in triple-negative breast cancer (Fig. S1). In addition, the use of SNP/gene pairs with synergistic prognostic values for triple-negative breast cancer carriers is more plausible clinically than other subtypes, as the triple-negative subtype is highly malignant and lacks targeted therapies (59,60). However, investigating the association between ANLN-KDR synergy and breast cancer subtyping is worthwhile and remains the subject of future investigations.
In conclusion, the concurrent presence of both rare homozygotes in ANLN:rs12535394 and KDR:rs11133360 was identified as prognostic for favorable survival in breast cancer. The quantity of the rare allele of ANLN:rs12535394 was negatively associated with the expression of ANLN, and that of KDR:rs11133360 was negatively associated with the expression of KDR, both of which are protective. Novel roles of genes that bridge the gap between SNPs and corresponding genes were revealed and merit in-depth investigation, including the potential tissue-specific tumor suppressive roles of ZNF133 in breast cancer, the conditional effects of MAP10 on breast cancer survival rates and the possible suppressive role of C14ORF80 (a gene not being annotated) during tumor angiogenesis. Pathways controlling cell proliferation/apoptosis and angiogenesis/migration genetically interact and ultimately influence immune responses and patient clinical outcomes, suggesting the intrinsic connection amongst cancer hallmarks and the prominent role for immunotherapy in cancer state transition. Experimental validations confirmed the roles of the ANLN-KDR interaction in the transition of cells between normal and cancerous states and in breast cancer prognosis, and implicate the therapeutic potential of ANLN.
Supplementary Material
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural Science Foundation of China (grant no. 81972789), National Science and Technology Major Project of China (grant. no. 2018ZX10302205-004-002), the Natural Science Foundation of Jiangsu Province (grant. no. BK20161130), the Six Talent Peaks Project in Jiangsu Province (grant. no. SWYY-128), the Technology Development Funding of Wuxi (grant. no. WX18IVJN017), the Postgraduate Education Reform Project of Jiangsu Province, and Research Funds for the Medical School of Jiangnan University ESI Special Cultivation Project (grant. no. 1286010241170320).
Availability of data and materials
The datasets generated and/or analyzed in the present study were retrieved from TCGA (http://cancergenome.nih.gov) and (http://www.cbioportal.org/).
Authors' contributions
XD designed, supervised and financed the project, analyzed the results and drafted the manuscript. XC and OH conducted the computational analysis. YM performed the experiments. XD, XC and YM prepared the figures and tables. All authors have read and approved the content of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Torre LA, Islami F, Siegel RL, Ward EM, Jemal A. Global cancer in women: Burden and Trends. Cancer Epidemiol Biomarkers Prev. 2017;26:444–457. doi: 10.1158/1055-9965.EPI-16-0858. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA. Hallmarks of cancer: The next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Li X, Zhang R, Liu Z, Li S, Xu H. The genetic variants in the PTEN/PI3K/AKT pathway predict susceptibility and CE(A)F chemotherapy response to breast cancer and clinical outcomes. Oncotarget. 2017;8:20252–20265. doi: 10.18632/oncotarget.15690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang BH, Liu LZ. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2009;102:19–65. doi: 10.1016/S0065-230X(09)02002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hall G, Lane BM, Khan K, Pediaditakis I, Xiao J, Wu G, Wang L, Kovalik ME, Chryst-Stangl M, Davis EE, et al. The Human FSGS-causing ANLN R431C mutation induces dysregulated PI3K/AKT/mTOR/Rac1 signaling in podocytes. J Am Soc Nephrol. 2018;29:2110–2122. doi: 10.1681/ASN.2017121338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki C, Daigo Y, Ishikawa N, Kato T, Hayama S, Ito T, Tsuchiya E, Nakamura Y. ANLN plays a critical role in human lung carcinogenesis through the activation of RHOA and by involvement in the phosphoinositide 3-kinase/AKT pathway. Cancer Res. 2005;65:11314–11325. doi: 10.1158/0008-5472.CAN-05-1507. [DOI] [PubMed] [Google Scholar]
- 8.Huber TB, Hartleben B, Kim J, Schmidts M, Schermer B, Keil A, Egger L, Lecha RL, Borner C, Pavenstädt H, et al. Nephrin and CD2AP associate with phosphoinositide 3-OH kinase and stimulate AKT-dependent signaling. Mol Cell Biol. 2003;23:4917–4928. doi: 10.1128/MCB.23.14.4917-4928.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou W, Wang Z, Shen N, Pi W, Jiang W, Huang J, Hu Y, Li X, Sun L. Knockdown of ANLN by lentivirus inhibits cell growth and migration in human breast cancer. Mol Cell Biochem. 2015;398:11–19. doi: 10.1007/s11010-014-2200-6. [DOI] [PubMed] [Google Scholar]
- 10.Magnusson K, Gremel G, Rydén L, Pontén V, Uhlén M, Dimberg A, Jirström K, Pontén F. ANLN is a prognostic biomarker independent of Ki-67 and essential for cell cycle progression in primary breast cancer. BMC Cancer. 2016;16:904. doi: 10.1186/s12885-016-2923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciruelos Gil EM. Targeting the PI3K/AKT/mTOR pathway in estrogen receptor-positive breast cancer. Cancer Treat Rev. 2014;40:862–871. doi: 10.1016/j.ctrv.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 12.Guo S, Colbert LS, Fuller M, Zhang Y, Gonzalez-Perez RR. Vascular endothelial growth factor receptor-2 in breast cancer. Biochim Biophys Acta. 2010;1806:108–121. doi: 10.1016/j.bbcan.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi Y, Kitadai Y, Bucana CD, Cleary KR, Ellis LM. Expression of vascular endothelial growth factor and its receptor, KDR, correlates with vascularity, metastasis, and proliferation of human colon cancer. Cancer Res. 1995;55:3964–3968. [PubMed] [Google Scholar]
- 14.Mahdi KM, Nassiri MR, Nasiri K. Hereditary genes and SNPs associated with breast cancer. Asian Pac J Cancer Prev. 2013;14:3403–3409. doi: 10.7314/APJCP.2013.14.6.3403. [DOI] [PubMed] [Google Scholar]
- 15.Nelson MR, Marnellos G, Kammerer S, Hoyal CR, Shi MM, Cantor CR, Braun A. Large-scale validation of single nucleotide polymorphisms in gene regions. Genome Res. 2004;14:1664–1668. doi: 10.1101/gr.2421604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deng N, Zhou H, Fan H, Yuan Y. Single nucleotide polymorphisms and cancer susceptibility. Oncotarget. 2017;8:110635–110649. doi: 10.18632/oncotarget.22372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beltran H, Prandi D, Mosquera JM, Benelli M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV, Varambally S, et al. Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat Med. 2016;22:298–305. doi: 10.1038/nm.4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cancer Genome Atlas Research, Network, corp-author. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Long X, Zhou W, Wang Y, Liu S. Prognostic significance of ANLN in lung adenocarcinoma. Oncol Lett. 2018;16:1835–1840. doi: 10.3892/ol.2018.8858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong G, Guo X, Fu X, Wan S, Zhou F, Myers RE, Bao G, Burkart A, Yang H, Xing J. Potentially functional genetic variants in KDR gene as prognostic markers in patients with resected colorectal cancer. Cancer Sci. 2012;103:561–568. doi: 10.1111/j.1349-7006.2011.02194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garrigós C, Espinosa M, Salinas A, Osman I, Medina R, Taron M, Molina-Pinelo S, Duran I. Single nucleotide polymorphisms as prognostic and predictive biomarkers in renal cell carcinoma. Oncotarget. 2017;8:106551–106564. doi: 10.18632/oncotarget.22533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Slattery ML, Lundgreen A, Wolff RK. VEGFA, FLT1, KDR and colorectal cancer: Assessment of disease risk, tumor molecular phenotype, and survival. Mol Carcinog. 2013;53(Suppl 1):E140–E150. doi: 10.1002/mc.22058. [DOI] [PubMed] [Google Scholar]
- 23.Wu IC, Zhao Y, Zhai R, Liu G, Ter-Minassian M, Asomaning K, Su L, Liu CY, Chen F, Kulke MH, et al. Association between polymorphisms in cancer-related genes and early onset of esophageal adenocarcinoma. Neoplasia. 2011;13:386–392. doi: 10.1593/neo.101722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao C, Cao J, Wang Y, Liu B, Wang Z. Effects of VEGF and VEGFR polymorphisms on the outcome of patients with metastatic renal cell carcinoma treated with sunitinib: A systematic review and meta-analysis. Oncotarget. 2017;8:68854–68862. doi: 10.18632/oncotarget.19924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beeghly-Fadiel A, Shu XO, Lu W, Long J, Cai Q, Xiang YB, Zheng Y, Zhao Z, Gu K, Gao YT, Zheng W. Genetic variation in VEGF family genes and breast cancer risk: A report from the Shanghai Breast Cancer Genetics Study. Cancer Epidemiol Biomarkers Prev. 2011;20:33–41. doi: 10.1158/1055-9965.EPI-10-0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Therneau T. A package for survival analysis in S. version 2.38. https://CRAN.Rproject.org/package=survival. 2015
- 28.R Development Core Team, corp-author. R: A language and environment for statistical computing. R Foundation for Statistical Computing. https://www.gbif.org/tool/81287/r-a-language-and-environment-for-statistical-computing 2009
- 29.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. Trait-associated SNPs are more likely to be eQTLs: Annotation to enhance discovery from GWAS. PLoS Genet. 2010;6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bendl J, Musil M, Štourač J, Zendulka J, Damborský J, Brezovský J. PredictSNP2: A unified platform for accurately evaluating SNP effects by exploiting the different characteristics of variants in distinct genomic regions. PLoS Comput Biol. 2016;12:e1004962. doi: 10.1371/journal.pcbi.1004962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fu Y, Liu Z, Lou S, Bedford J, Mu XJ, Yip KY, Khurana E, Gerstein M. FunSeq2: A framework for prioritizing noncoding regulatory variants in cancer. Genome Biol. 2014;15:480. doi: 10.1186/s13059-014-0480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Quang D, Chen Y, Xie X. DANN: A deep learning approach for annotating the pathogenicity of genetic variants. Bioinformatics. 2015;31:761–763. doi: 10.1093/bioinformatics/btu703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ritchie GR, Dunham I, Zeggini E, Flicek P. Functional annotation of noncoding sequence variants. Nat Methods. 2014;11:294–296. doi: 10.1038/nmeth.2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, Yamanishi Y. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36 (Database Issue) 2008:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G, Wang LG, Han Y, He QY. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carlson M. org.Hs.eg.db: Genome wide annotation for Human. 2019 [Google Scholar]
- 38.Franz M, Rodriguez H, Lopes C, Zuberi K, Montojo J, Bader GD, Morris Q. GeneMANIA update 2018. Nucleic Acids Res. 2018;46:W60–W64. doi: 10.1093/nar/gky311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Labun K, Montague TG, Krause M, Torres Cleuren YN, Tjeldnes H, Valen E. CHOPCHOP v3: Expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 2019;47:W171–W174. doi: 10.1093/nar/gkz365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Heiskanen MA, Bittner ML, Chen Y, Khan J, Adler KE, Trent JM, Meltzer PS. Detection of gene amplification by genomic hybridization to cDNA microarrays. Cancer Res. 2000;60:799–802. [PubMed] [Google Scholar]
- 43.Li H, Jie S, Zou P, Zou G. CDNA microarray analysis of chronic myeloid leukemia. Int J Hematol. 2002;75:388–393. doi: 10.1007/BF02982130. [DOI] [PubMed] [Google Scholar]
- 44.Brouhard GJ, Rice LM. Microtubule dynamics: An interplay of biochemistry and mechanics. Nat Rev Mol Cell Biol. 2018;19:451–463. doi: 10.1038/s41580-018-0009-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dogra N, Kumar A, Mukhopadhyay T. Fenbendazole acts as a moderate microtubule destabilizing agent and causes cancer cell death by modulating multiple cellular pathways. Sci Rep. 2018;8:11926. doi: 10.1038/s41598-018-30158-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Parker AL, Kavallaris M, McCarroll JA. Microtubules and their role in cellular stress in cancer. Front Oncol. 2014;4:153. doi: 10.3389/fonc.2014.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226:352–364. doi: 10.1002/path.3022. [DOI] [PubMed] [Google Scholar]
- 48.Visochek L, Castiel A, Mittelman L, Elkin M, Atias D, Golan T, Izraeli S, Peretz T, Cohen-Armon M. Exclusive destruction of mitotic spindles in human cancer cells. Oncotarget. 2017;8:20813–20824. doi: 10.18632/oncotarget.15343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dong X, Liu F, Sun L, Liu M, Li D, Su D, Zhu Z, Dong JT, Fu L, Zhou J. Oncogenic function of microtubule end-binding protein 1 in breast cancer. J Pathol. 2010;220:361–369. doi: 10.1002/path.2662. [DOI] [PubMed] [Google Scholar]
- 50.Liu M, Wang X, Yang Y, Li D, Ren H, Zhu Q, Chen Q, Han S, Hao J, Zhou J. Ectopic expression of the microtubule-dependent motor protein Eg5 promotes pancreatic tumourigenesis. J Pathol. 2010;221:221–228. doi: 10.1002/path.2706. [DOI] [PubMed] [Google Scholar]
- 51.McCusker C, Upton J, Warrington R. Primary immunodeficiency. Allergy Asthma Clin Immunol. 2018;14(Suppl 2):S61. doi: 10.1186/s13223-018-0290-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shapiro RS. Malignancies in the setting of primary immunodeficiency: Implications for hematologists/oncologists. Am J Hematol. 2011;86:48–55. doi: 10.1002/ajh.21903. [DOI] [PubMed] [Google Scholar]
- 53.Filipovich AH, Mathur A, Kamat D, Shapiro RS. Primary immunodeficiencies: Genetic risk factors for lymphoma. Cancer Res 52 (19 Suppl) 1992:5465S–5467S. [PubMed] [Google Scholar]
- 54.Moriwaki Y, Yamamoto T, Higashino K. Enzymes involved in purine metabolism-a review of histochemical localization and functional implications. Histol Histopathol. 1999;14:1321–1340. doi: 10.14670/HH-14.1321. [DOI] [PubMed] [Google Scholar]
- 55.Chen TC, Lin KT, Chen CH, Lee SA, Lee PY, Liu YW, Kuo YL, Wang FS, Lai JM, Huang CY. Using an in situ proximity ligation assay to systematically profile endogenous protein-protein interactions in a pathway network. J Proteome Res. 2014;13:5339–5346. doi: 10.1021/pr5002737. [DOI] [PubMed] [Google Scholar]
- 56.Hein MY, Hubner NC, Poser I, Cox J, Nagaraj N, Toyoda Y, Gak IA, Weisswange I, Mansfeld J, Buchholz F, et al. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 57.Xie YG, Yu Y, Hou LK, Wang X, Zhang B, Cao XC. FYN promotes breast cancer progression through epithelial-mesenchymal transition. Oncol Rep. 2016;36:1000–1006. doi: 10.3892/or.2016.4894. [DOI] [PubMed] [Google Scholar]
- 58.Lee GH, Yoo KC, An Y, Lee HJ, Lee M, Uddin N, Kim MJ, Kim IG, Suh Y, Lee SJ. FYN promotes mesenchymal phenotypes of basal type breast cancer cells through STAT5/NOTCH2 signaling node. Oncogene. 2018;37:1857–1868. doi: 10.1038/s41388-017-0114-y. [DOI] [PubMed] [Google Scholar]
- 59.Dai X, Li T, Bai Z, Yang Y, Liu X, Zhan J, Shi B. Breast cancer intrinsic subtype classification, clinical use and future trends. Am J Cancer Res. 2015;5:2929–2943. [PMC free article] [PubMed] [Google Scholar]
- 60.Dai X, Xiang L, Li T, Bai Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer. 2016;7:1281–1294. doi: 10.7150/jca.13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed in the present study were retrieved from TCGA (http://cancergenome.nih.gov) and (http://www.cbioportal.org/).