Abstract
Herpesviruses can rewire cellular signaling in host cells by expressing viral G protein–coupled receptors (GPCRs). These viral receptors exhibit homology to human chemokine receptors, but some display constitutive activity and promiscuous G protein coupling. Human cytomegalovirus (HCMV) has been detected in multiple cancers, including glioblastoma, and its genome encodes four GPCRs. One of these receptors, US28, is expressed in glioblastoma and possesses constitutive activity and oncomodulatory properties. UL33, another HCMV-encoded GPCR, also displays constitutive signaling via Gαq, Gαi, and Gαs proteins. However, little is known about the nature and functional effects of UL33-driven signaling. Here, we assessed UL33's signaling repertoire and oncomodulatory potential. UL33 activated multiple proliferative, angiogenic, and inflammatory signaling pathways in HEK293T and U251 glioblastoma cells. Notably, upon infection, UL33 contributed to HCMV-mediated STAT3 activation. Moreover, UL33 increased spheroid growth in vitro and accelerated tumor growth in different in vivo tumor models, including an orthotopic glioblastoma xenograft model. UL33-mediated signaling was similar to that stimulated by US28; however, UL33-induced tumor growth was delayed. Additionally, the spatiotemporal expression of the two receptors only partially overlapped in HCMV-infected glioblastoma cells. In conclusion, our results unveil that UL33 has broad signaling capacity and provide mechanistic insight into its functional effects. UL33, like US28, exhibits oncomodulatory properties, elicited via constitutive activation of multiple signaling pathways. UL33 and US28 might contribute to HCMV's oncomodulatory effects through complementing and converging cellular signaling, and hence UL33 may represent a promising drug target in HCMV-associated malignancies.
Keywords: G protein-coupled receptor (GPCR), cell signaling, oncogene, herpesvirus, glioblastoma, constitutive signaling, human cytomegalovirus (HCMV), UL33
Introduction
Human cytomegalovirus (HCMV),2 Epstein–Barr virus (EBV), and Kaposi's sarcoma–associated herpesvirus (KSHV) are members of Herpesviridae linked to oncogenesis. EBV and KSHV are oncogenic viruses (1, 2), whereas HCMV is considered an oncomodulatory virus that aggravates rather than initiates tumorigenesis (3). Each of the viruses contain genes encoding for one or more G protein–coupled receptors (GPCRs) showing homology to human chemokine receptors (4). Several viral GPCRs, i.e. HCMV-encoded US28 (5, 6), EBV-encoded BILF1 (7), and KSHV-encoded ORF74 (8), possess oncogenic or oncomodulatory properties. These viral receptors are constitutively active and show G protein promiscuity, whereas human chemokine receptors are only activated upon agonist stimulation and predominantly couple to Gαi proteins. By means of this constitutive GPCR signaling, herpesviruses have devised mechanisms to rewire cellular signaling of host cells to facilitate virus biology and consequent pathogenesis.
HCMV is a ubiquitous DNA virus with a seroprevalence of more than 50% in adults (9). Once acquired, this β-herpesvirus establishes a life-long latent infection, which is usually asymptomatic. However, under conditions in which the immune system is compromised (e.g. in AIDS patients or organ transplantation recipients or upon tumor-associated inflammation), reactivation of HCMV may result in severe pathologies (10). HCMV DNA and proteins have been detected in tumor samples of multiple human cancers (11–17) in which the virus exerts oncomodulatory properties (3, 15, 18–20). HCMV encodes four GPCRs: UL33, UL78, US27, and US28 (21). These receptors are present on the virion (22–26), and transcripts of each have been detected in peripheral blood mononuclear cells obtained from asymptomatic latently infected individuals (27). Ligands for UL33, UL78, and US27 have not been identified to date. US28, in contrast, binds to and internalizes a large number of human chemokines (e.g. CX3CL1 and CCL2–5, -7, -11, -13, -26, and -29), contributing to HCMV-mediated immune evasion (4, 28). At present, only UL33 and US28 have been shown to couple to G proteins and display G protein–dependent signaling (4, 29, 30).
US28 promiscuously couples to Gαs, Gαi, Gαq, and Gα12/13 proteins and constitutively activates downstream signaling, including proinflammatory, proangiogenic, and proliferative pathways (5, 31, 32). Additionally, US28 can modulate migration of macrophages and smooth-muscle cells in a ligand-dependent manner (33) and is essential for establishment of latent infection in hematopoietic progenitor cells and monocytes (34, 35). In vitro and in vivo studies have also demonstrated an oncomodulatory role for US28. For example, tumor formation was induced upon expression of US28 in xenograft models and transgenic mice (5, 6, 36), and US28 expression in glioblastoma cells accelerated tumor growth in an orthotopic mice model (20). Importantly, US28 expression is detected in brain tumor specimens obtained from HCMV-positive glioblastoma patients (16, 20, 31).
UL33 also signals in a constitutive manner. Expression of this viral GPCR results in production of inositol phosphate (InsP) and activation of CREB via coupling to Gαq/Gαi and Gαs proteins, respectively (29, 30). Furthermore, significant contribution of UL33 to HCMV-mediated CREB activation was demonstrated in HCMV-infected glioblastoma cells (30). Experiments using R33 and M33, UL33 homologs expressed by rodent-specific cytomegaloviruses, have illustrated pathological relevance of UL33 gene family members. Deletion of R33 from rat cytomegalovirus (RCMV) highlighted the contribution of R33 to virus replication in vivo. Moreover, lower mortality rates were observed in rats infected with R33-knockout virus (37). Constitutive G protein–dependent signaling by murine cytomegalovirus (MCMV) M33 was shown to promote migration of infected lung dendritic cells, which is required for entrance to the blood circulation and systemic spread of the virus (38). Accordingly, deletion of M33 from MCMV hampered virus replication in the salivary gland and spleen (39, 40). Establishing, maintaining, or reactivation from latency in the spleen and lungs was strongly dependent on constitutive signaling by M33 (40, 41). Interestingly, complementation with either UL33 or US28 partially rescued the attenuation of viral replication and establishment of latent infection upon deletion of M33 from MCMV, indicating conserved functionality between UL33 and M33 as well as functional overlap between UL33 and US28 (39, 40).
Although UL33 displays promiscuous G protein coupling and constitutive activity, limited information is available on the signaling properties and functional consequences of UL33. In this study, we explored the signaling repertoire of UL33 as well as its contribution to oncomodulation. Our data demonstrate that UL33 is capable of enhancing tumor growth via constitutive activation of proinflammatory, proliferative, and proangiogenic signal transduction pathways. Hence, UL33 could play an important role in the pathogenesis of HCMV-associated malignancies.
Results
UL33 constitutively activates oncogenic signaling pathways
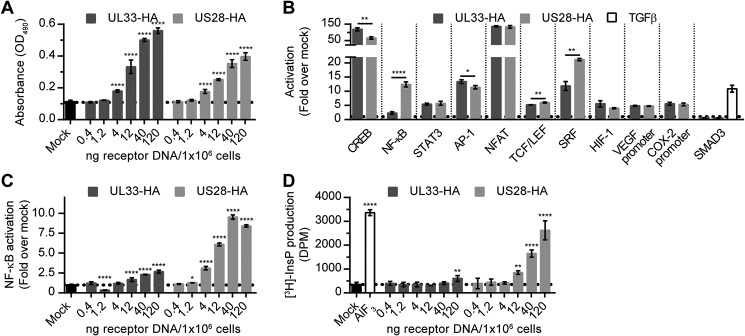
UL33 promiscuously couples to G proteins in a constitutive manner (30). We evaluated the ability of UL33 to activate inflammatory and oncogenic signaling pathways using luciferase-based reporter genes and compared it with US28-mediated signaling. Upon transfection of UL33 or US28 DNA in HEK293T cells, dose-dependent receptor expression was observed (Fig. 1A). To compare the intrinsic signaling capacity of UL33 and US28, signaling was evaluated at equal total protein levels of the receptors (Fig. S1A). UL33 constitutively induced activation of STAT3, AP-1, NFAT, TCF/LEF, HIF-1, the VEGF promoter, and the COX-2 promoter to a degree comparable with that of US28 (Fig. 1B). The SMAD3 reporter was refractory to input by both UL33 and US28, whereas TGFβ stimulation, serving as a positive control, resulted in a pronounced activation of this transcriptional readout. Interestingly, CREB, NF-κB, and SRF were differentially activated by the two GPCRs (Fig. 1B). UL33 enhanced CREB activity ∼120-fold, compared with 65-fold for US28. In contrast, activation of NF-κB and SRF was less pronounced for UL33 compared with US28 (2- versus 12-fold for NF-κB and 12- versus 21-fold for SRF). At higher expression levels, UL33-mediated NF-κB activation reached a maximum at 2.7-fold, compared with 10-fold for US28 (Fig. 1C). For comparison, a similar range of UL33 expression resulted in a dose-dependent UL33-driven COX-2 promoter activation (Fig. S1B). Previously, UL33 was reported to increase InsP production in COS-7 cells via coupling to Gαq/11 and Gαi proteins (29, 30). In HEK293T cells, UL33 stimulated InsP production only at the highest DNA dose used (1.7-fold), whereas US28 enhanced InsP levels in a dose-dependent fashion (up to 8-fold) (Fig. 1D). Together, these data indicate that UL33 constitutively activates multiple proliferative, angiogenic, and proinflammatory signaling pathways, which partially overlap with US28-mediated signaling despite some distinct differences.
Figure 1.
UL33 and US28 mediate constitutive activation of proliferative and proangiogenic signal transduction pathways in HEK293T cells. A, dose-dependent receptor expression in HEK293T cells transiently transfected with increasing amounts of DNA encoding UL33-HA or US28-HA (C-terminally epitope-tagged receptors), as assessed through anti-HA ELISA. B, HEK293T cells were transfected with 12 ng of UL33-HA or 40 ng of US28-HA/106 cells in combination with the corresponding luciferase reporter genes. At similar expression levels, the constitutive activation of multiple signaling pathways was assessed. Mock cells (transfected with reporter gene plasmid) were stimulated with 8 pm TGFβ for 24 h as a positive control for SMAD3 activation. Constitutive NF-κB activation (C) or InsP production (D) was assessed in HEK293T cells transfected with a series of DNA encoding UL33-HA or US28-HA. AlF3 was included as positive control in the InsP production assay. Graphs are representatives of at least three individual experiments performed in triplicate, and data are presented as mean ± S.D. (error bars). *, p < 0.05; **, p < 0.01; and ****, p < 0.0001.
UL33 stimulates transformation of NIH-3T3 cells
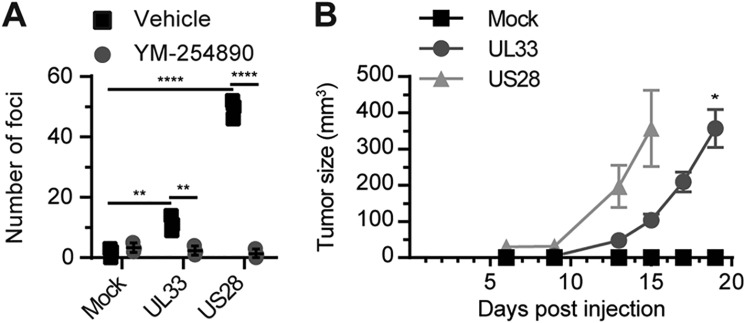
To evaluate oncomodulatory properties of UL33, a stable NIH-3T3 cell line expressing UL33-eGFP was generated (Fig. S2). Like expression of US28, but to a lesser extent, UL33 expression resulted in loss of contact inhibition in NIH-3T3 cells as evidenced by the formation of foci (Fig. 2A). Inhibition of Gαq-mediated signaling using YM-254890 abolished foci formation induced by both receptors. Because UL33 activated proliferative and proangiogenic signaling pathways and induced a transformed phenotype in vitro, UL33's potential to induce tumor formation in vivo was assessed. Tumors were observed 13 days after subcutaneous inoculation of UL33- or US28-expressing NIH-3T3 cells in both flanks of athymic nude mice (Fig. 2B). The mock-treated group did not develop tumors up to 75 days postinjection. Although the oncostimulatory potential of US28 was more pronounced, UL33 clearly induced tumor formation in vivo. These findings demonstrate that UL33, like US28, induces cellular transformation.
Figure 2.
UL33 induces an oncogenic phenotype in NIH-3T3 cells. A, the formation of foci by NIH-3T3 cells stably transfected with empty vector (Mock), UL33-eGFP, or US28 treated with vehicle (0.03% DMSO) or 300 nm YM-254890. B, tumor formation in nude mice flank-inoculated with NIH-3T3 cells stably expressing UL33-eGFP or US28 (six mice per cell line, inoculated in both flanks). Data are depicted as mean ± S.E. (error bars). *, p < 0.05; **, p < 0.01; and ****, p < 0.0001.
UL33 signaling in glioblastoma cells
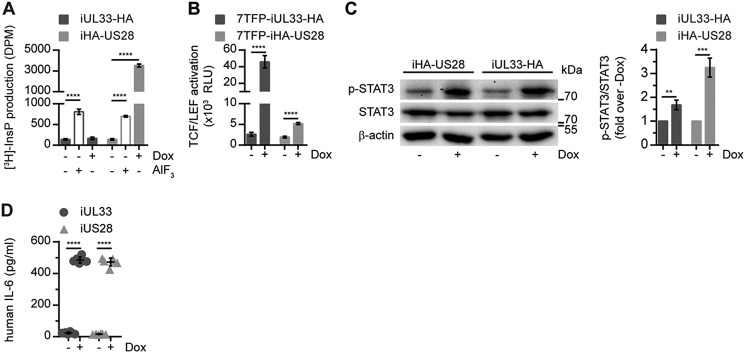
To probe the properties of the viral GPCRs in a disease-relevant cellular background, we generated U251 glioblastoma cell lines with inducible receptor expression (U251 iUL33, U251 iUL33-HA, U251 iUS28, and U251 iHA-US28). Upon induction of receptor expression using doxycycline, UL33 and US28 were expressed at similar levels (Fig. S3). Subsequently, UL33-mediated signaling was assessed in these glioblastoma cells. InsP levels were not increased by UL33 expression, whereas US28-expressing cells exhibited a 25-fold increase in InsP production (Fig. 3A). Both receptors induced activation of TCF/LEF, albeit to a different extent (17.5-fold for UL33 versus 2.7-fold for US28) (Fig. 3B). Additionally, expression of either UL33 or US28 resulted in a pronounced increase in STAT3 Tyr705 phosphorylation and IL-6 secretion (Fig. 3, C and D). US28 expression resulted in more STAT3 activation compared with UL33, whereas the receptors induced the secretion of IL-6 by a similar grade. Thus, UL33 also activates oncomodulatory signaling in U251 glioblastoma cells.
Figure 3.
UL33 signaling in U251 malignant glioma cells. The effects of UL33 and US28 expression on InsP production (A), TCF/LEF activation (B), STAT3 Tyr705 phosphorylation (C), and IL-6 secretion (D) were determined in U251 glioblastoma cell lines. Receptor expression was induced upon doxycycline (Dox) stimulation of the cells. AlF3 was included as a positive control for InsP production. Graphs are representatives of at least three individual experiments performed in triplicate (A and B) or pooled data of three individual experiments performed in singlicate (C) or duplicate (D). Data are presented as mean ± S.D. (error bars). **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001.
UL33 contributes to HCMV-mediated STAT3 activation in glioblastoma cells
Next, we set out to study UL33 and US28 in the context of HCMV-infected U251 cells. To facilitate our studies, we genetically engineered the bacterial artificial chromosome (BAC) of the clinically relevant HCMV Merlin strain (pAL1502 (42)). Introduction of an HA epitope at the C terminus of UL33 allowed for simultaneous detection of UL33 and US28 protein expression. UL33- or US28-knockout viruses were generated to evaluate the contribution of the respective receptors to HCMV-mediated effects. Integrity of the BACs was confirmed via endonuclease restriction profile analysis as well as sequencing of the UL33 and US28 gene regions, respectively (Fig. S4A).
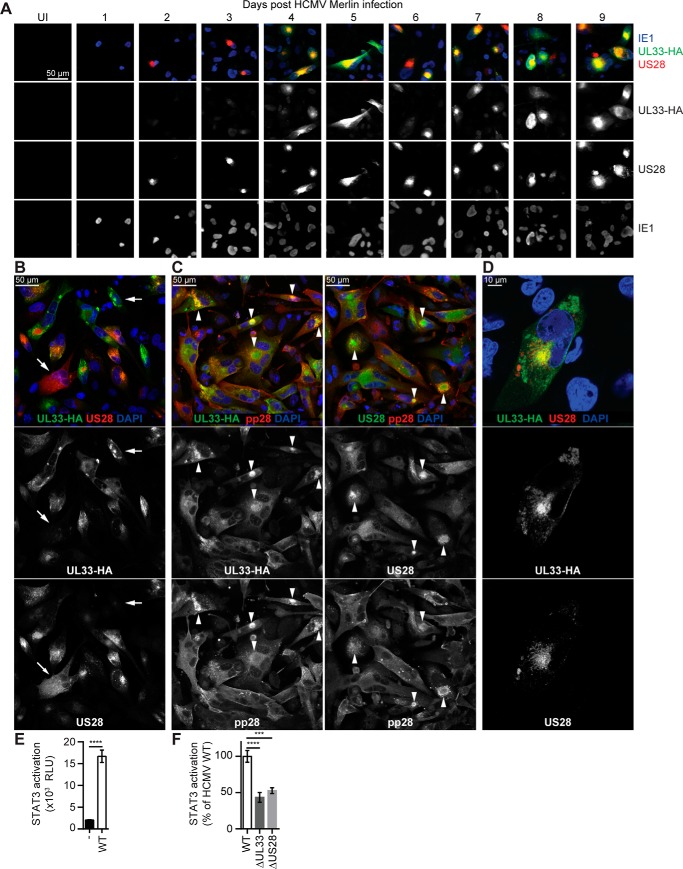
Upon infection with Merlin UL33-HA virus, UL33-HA and US28 protein expression was monitored each day postinfection (dpi) for a period of 9 consecutive days (Fig. 4A). HCMV immediate-early 1 (IE1) protein was detected in the nucleus of infected cells throughout the time frame of analysis. UL33 expression was apparent at 4–9 dpi, whereas low levels of US28 were already observed at 2 dpi, and intense US28 staining was evident from 3 to 9 dpi.
Figure 4.
UL33 is expressed and induces STAT3 activation in HCMV-infected U251 cells. U251 cells were infected with HCMV Merlin UL33-HA virus. A, every 24 h, over a time frame of 9 days, the expression of IE1 (blue), UL33 (anti-HA staining; green), and US28 (polyclonal anti-US28 antibodies; red) was monitored. B and D, 6 dpi the localization of UL33 (anti-HA staining) was compared with US28 (polyclonal anti-US28 antibodies). Arrows indicate examples of cells with high UL33 and low US28 abundance or vice versa. C, costaining of UL33 (anti-HA antibody) and US28 (polyclonal anti-US28 antibodies) with HCMV pp28 at 6 dpi. Arrows indicate examples of cells with a perinuclear VAC. E and F, STAT3 activity in U251–3SFP cells upon infection with WT (UL33-HA), UL33-deficient (ΔUL33), or US28-deficient (UL33-HA, ΔUS28) Merlin virus as determined 5 dpi. Graphs are representatives of two (F) and three (E) individual experiments performed in triplicate. Data are depicted as mean ± S.D. (error bars). ***, p < 0.001; and ****, p < 0.0001. UI, uninfected; RLU, relative light units.
The expression pattern of UL33 and US28 was variable between infected cells with some cells expressing high levels of both receptors, whereas in other cells the staining was less profound (Fig. 4B). Despite synchronized infection, part of the cells expressed one receptor more than the other, indicating that UL33 and US28 expression might be controlled by distinct regulatory mechanisms of the host cell. Previous research by Fraile-Ramos et al. (43, 44) revealed the presence of UL33 and US28 in the virion assembly compartment (VAC) of HCMV-infected fibroblasts. Although only a subset of infected U251 cells display a typical circular and perinuclear VAC, as identified by the viral tegument protein pp28 (45), both receptors colocalized in the VAC (Fig. 4C). Compared with the apparent strict perinuclear localization of US28 in cells where a VAC was present, UL33 protein was also more dispersed throughout the cell (Fig. 4D). In conclusion, US28 and UL33 are expressed upon HCMV infection of U251 glioblastoma cells but with distinct expression kinetics and subcellular localization.
Employing U251 cells stably expressing a reporter gene (U251–3SFP), STAT3-mediated signaling of UL33 and US28 in HCMV-infected glioblastoma cells was examined. HCMV Merlin infection strongly enhanced STAT3 activity 5 dpi when both receptors were expressed in U251 cells (9-fold) (Fig. 4E). At a similar infection rate (Fig. S4B), STAT3 activation was significantly impaired in cells infected with either UL33- or US28-knockout virus (57 and 47% reduction, respectively) (Fig. 4F). Parallel infection of U251–3SFP and U251 cells constitutively expressing firefly luciferase/mCherry (U251-FM) showed that the rise in STAT3 activity was not due to a general increase in transcriptional activity in HCMV-infected U251 cells (Fig. S4C). These results indicate that UL33, like US28, activates proliferative signaling in HCMV-infected U251 cells.
UL33 aggravates glioblastoma tumor growth
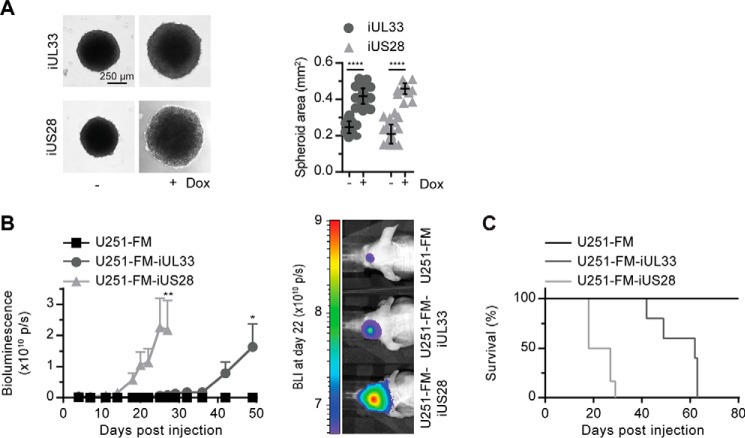
Finally, we determined the effect of UL33 on proliferation of glioblastoma cells. Expression of UL33 or US28 in U251 cells enhanced 3D growth, resulting in a 1.7- and 2.2-fold increase in spheroid size, respectively (Fig. 5A). An orthotopic glioblastoma mouse model was established to study the contribution of UL33 in glioblastoma progression in situ. To this end, U251, U251 iUL33, and U251 iUS28 cells constitutively expressing firefly luciferase/mCherry (FM) were constructed to quantify tumor size using bioluminescence imaging (20, 46). Following inoculation of these cells in the striatum of nude athymic mice, expression of UL33 or US28 induced tumor growth (Fig. 5B). No tumor development was observed in mice inoculated with U251-FM control cells within the time frame of the experiment (50 days). The onset of UL33-mediated tumor growth was delayed compared with tumor formation by US28-expressing cells (32 versus 14 days postinjection). A similar pattern was apparent for the mortality of the mice (Fig. 5C). This delayed onset of tumor growth for UL33-expressing cell lines could be attributed to lower receptor expression levels. Evaluation of receptor mRNA levels in the cells used for inoculation of the mice showed higher abundance of UL33 than US28 mRNA (Fig. S5), suggesting that differential signaling properties, not receptor expression levels, account for the delayed onset of UL33-mediated tumor growth. Taken together, these observations illustrate that UL33 can aggravate glioblastoma tumor growth, albeit to a lesser extent than US28.
Figure 5.
UL33 promotes tumorigenesis in U251 malignant glioma cells. A, U251 iUL33 and U251 iUS28 cells were cultured as 3D spheroids, and the effect of receptor expression was assessed. The graph depicts pooled data of three individual experiments performed in octuplicate, presented as mean ± S.D. (error bars). B, orthotopic glioblastoma model in which U251-FM iUL33, U251-FM iUS28, or U251-FM cells were injected in the striatum of mice. Mice were fed doxycycline in their drinking water to induce UL33 and US28 expression. Tumor size was measured via bioluminescence imaging (BLI) in six mice per cell line. Data are depicted as mean ± S.E. (error bars). C, survival of mice in the in vivo study. *, p < 0.05; **, p < 0.01; and ****, p < 0.0001.
Discussion
A growing body of evidence has substantiated the concept of oncomodulation by HCMV in the context of glioblastoma (18). Despite ongoing debates (47–49), multiple HCMV-encoded proteins and genes, including US28 (16, 20, 31), have been detected in glioblastoma tissue samples (18). Moreover, high expression of HCMV IE1 antigen in glioblastoma is associated with poor prognosis (19), and adjuvant antiviral therapy (50, 51) or HCMV-specific immunotherapy (52, 53) showed promising improvements in glioblastoma patient survival rates. In the current study, expression of another constitutively active HCMV-encoded GPCR, UL33, promoted a proliferative phenotype in U251 glioblastoma cells, as observed both in 3D cell culture and an orthotopic glioblastoma mouse model.
The transcription fingerprint of UL33 in HEK293T cells accentuated the oncomodulatory signaling properties of this receptor. Activation of transcription factors STAT3, AP-1, NFAT, TCF/LEF, SRF, HIF-1, and the VEGF and COX-2 promoters by UL33 is reported here for the first time and further substantiates UL33's potential to constitutively drive proliferative, proangiogenic, and proinflammatory signaling pathways. The activation of CREB by UL33 is in line with previous reports in COS-7 (29, 30) and HCMV-infected glioblastoma cells (30). However, in contrast to our observations in HEK293T cells, NF-κB and SRF were not activated by UL33 in COS-7 cells (29, 54). These findings imply different consequences of UL33 expression in different cell types and potentially in different tumor types.
In U251 cells, UL33 enhanced TCF/LEF- and STAT3-driven transcription as well as production of IL-6. Activation of these pathways is of pathological relevance given their roles in the biology of glioblastoma and HCMV. In glioma, cytoplasmic and nuclear expression of β-catenin, a transactivator of TCF/LEF, is positively correlated with tumor grade and negatively correlated with patient survival (55, 56). Knockdown of β-catenin inhibits proliferation of U251 and U87 glioblastoma cells in vitro as well as U251 tumor growth in a xenograft mouse model (55, 57). Moreover, activation of β-catenin and TCF/LEF is enhanced upon HCMV Titan 2B infection of U373 glioblastoma cells (58). Previously, a link among HCMV infection, IL-6 production, STAT3 Tyr705 phosphorylation, and prognosis was reported for glioblastoma patients, including an important role for US28 (16, 31). Similarly, UL33-mediated activation of the IL-6/STAT3 axis could be instrumental in the aggravation of HCMV-associated malignancy.
Based on our data, both UL33 and US28 constitutively activate oncomodulatory signaling pathways. However, the effect of UL33 on in vitro and in vivo tumor growth was not as pronounced as for US28. This is likely attributed to differential signaling by the receptors.
When comparing the signaling repertoire of UL33 with that of US28, we observed that the employed signaling networks partly overlap. Both receptors activated STAT3, AP-1, NFAT, TCF/LEF, HIF-1, and the VEGF and the COX-2 promoters in HEK293T cells and the IL-6/STAT3 axis in U251 cells. The latter was also confirmed in a viral setting. In addition, important differences in signaling output were observed. Although constitutively coupling to Gαq proteins is essential for the foci-forming capacity of both receptors in NIH-3T3 cells, InsP production, a common consequence of Gαq activity, was only stimulated by US28 in HEK293T and U251 cells. This suggests differences upstream in the signaling pathways activated by the receptors at the level of Gαq proteins or effector enzymes. In addition, the magnitude of TCF/LEF and STAT3 activation in U251 cells and activation of CREB and SRF in HEK293T cells differed between UL33 and US28. The dissimilitude in activation of STAT3 given similar excretion of IL-6 suggest that additional factors, besides IL-6, contribute to US28-mediated STAT3 activation in U251 cells. Also, NF-κB activation was only marginally enhanced in UL33-expressing cells where NF-κB is a key hub in US28-mediated signaling (4, 31). Remarkably, factors known to be regulated by NF-κB in the context of US28 signaling such as COX-2, IL-6, and STAT3 (5, 31) were up-regulated with similar efficacy by UL33 and US28. It would be of interest to study the difference in activation mechanism for these factors in further detail.
Upon infection with HCMV, we observed spatiotemporal differences in the expression of UL33 and US28 in glioblastoma cells. US28 is expressed early after infection, whereas both receptors were detected at later stages of HCMV infection. Coexpression of UL33 and US28 in infected cells might affect their reciprocal signaling characteristics by formation of UL33–US28 heteromers or via cross-talk at the level of signaling (59). In agreement with previous observations, both receptors localized mainly in the virion assembly compartment (43, 44). Nevertheless, direct comparison of UL33 and US28 in infected U251 cells showed differences in localization with UL33 less strictly localized to the perinuclear VAC. This difference in subcellular localization suggests that the receptors might engage different signaling pools. Additionally, the marked differences observed between UL33 and US28 expression levels in individual cells imply regulation of viral gene expression by host cell factors. Further studies are required to identify which cellular factors and/or differentiation state of tumor cells dictates the expression of UL33 and US28.
Although it would be interesting to study UL33 in viral context in vivo, such experiments are complicated by HCMV's strict species tropism. Nonetheless, mouse and rat CMVs are well characterized in vivo and encode homologs of UL33 (37–41). Furthermore, recombinant MCMV has been used as an in vivo model to study HCMV-encoded GPCR function (39, 40, 60). When translating findings obtained in these model systems to HCMV pathology, it is important to understand the functional similarities and differences between UL33 and its CMV homologs. With respect to the activation of CREB, NFAT, and NF-κB, UL33 signaling overlapped with that of MCMV M33 (29, 39). UL33 signaling also corresponded to NF-κB and SRF activation by RCMV R33 (61). UL33, like M33 (29) and R33 (61), was previously shown to induce the accumulation of InsP in COS-7 cells (29, 30). At best, we observed minimal elevation of InsP levels by UL33 in HEK293T or U251 cells, highlighting the significance of the cellular background. Besides, functional differences between UL33 homologs also exist. For example, CREB is activated by UL33 and inhibited by R33 (61). Taken together, UL33 signaling displays similarities, but also clear differences, with that of M33 and R33. This might be of particular importance when using MCMV and RCMV as model systems to study the function of UL33 in a viral context.
Besides US28 and UL33, also EBV- and KSHV-encoded GPCRs BILF1 (7) and ORF74 (8) play important roles in the pathogenesis of herpesvirus-associated malignancies. Hence, based on our data, UL33 appears to be the fourth herpesvirus-encoded GPCR with intrinsic oncogenic properties. These receptors share the capacity to activate proangiogenic, proliferative, and proinflammatory signaling pathways in a ligand-independent manner and thereby stimulate tumorigenicity. Thus, rewiring of cellular signaling through the expression of constitutively active GPCRs might be a common mechanism by which Herpesviridae members promote oncogenesis. This corresponds well with the transforming potential of constitutively active G protein mutants and the high occurrence of activating mutations in GPCRs and G proteins in human tumors (62).
In conclusion, the HCMV-encoded GPCR UL33 is able to exert oncomodulatory activity via the induction of proliferative, proinflammatory, and proangiogenic pathways. The differences in expression and functional characteristics of UL33 and US28 might suggest that HCMV has devised distinct means to control (host) cellular functions through the expression of different viral GPCRs. These insights improve our understanding of HCMV's ability to aggravate tumor progression in HCMV-associated malignancies.
Experimental procedures
Cell lines and cell culture
NIH-3T3 cells (ATCC, Manassas, VA) stably transfected with UL33-eGFP (AD169 strain) were generated and cultured as described previously for mock NIH-3T3 and NIH-3T3-US28 (VHL/E strain) cells (6). The U251 cell line was authenticated by short tandem repeat profiling (Baseclear B.V., Leiden, The Netherlands). U251 cell lines with inducible expression of (HA-) US28 (VHL/E strain) or UL33(-HA) (AD169 strain) and/or constitutive FM expression were generated by lentiviral transduction (20, 32). Linear dsDNA encoding STAT3 transcription recognition elements flanked by AscI and PstI cloning sites was obtained by single-cycle PCR using oligonucleotides A and B (Table 1). Lentiviral STAT3 firefly luciferase reporter gene plasmids (3SFP) were constructed by subcloning three STAT3 transcription recognition elements in the backbone of the lentiviral 7TFP reporter plasmid (63) using the above-mentioned cloning sites. To introduce the 7TFP and 3SFP reporters in U251-iHA-US28, U251-iUL33-HA, or U251 cells, lentiviruses were produced upon transfection of HEK293T cells with packaging vectors pMD2.G and psPAX2 (a gift from Didier Trono (Addgene plasmid 12260)) and 7TFP or 3SFP plasmids. The U251 cell lines were cultured as described previously (32), and receptor expression was induced using 1 μg/ml doxycycline (D9891, Sigma-Aldrich). HEK293T and HFFF TR cells (kindly provided by Dr. Richard J. Stanton) were cultured as described previously (20, 42).
Table 1.
Oligonucleotides used to generate the 3SFP plasmid
| Code | Sequence |
|---|---|
| A | 5′-ATACTGCAGGTCGACATTTCCCGTAAATCGTCGAGTCGACATTTCCCGTAAATCGTCGAGTCGACATTTCCCGTAAATCGTCGAGGCGCGCCGCCCCTCCCCCGTGCCTTCC-3′ |
| B | 5′-GGAAGGCACGGGGGAGGGGC-3′ |
HCMV Merlin BAC recombineering
SW102 Escherichia coli carrying the HCMV Merlin BAC pAL1502, a variant of BAC pAL1498 (42) lacking the eGFP tag, were acquired from Dr. Richard J. Stanton. Recombineering was performed using galK-positive/negative selection as described previously by Warming et al. (64). Adaptations to the protocol used to generate the HCMV Merlin BAC recombinants are specified below. The galK expression cassette was amplified from pgalK (Fredrick National Laboratory for Cancer Research) by PCR using oligonucleotide primers with homology arms targeting specific regions in the Merlin BACs (Table 2). Primers 1–4 were used to replace US28 and UL33 with galK, whereas introduction of galK in front of the stop codon of UL33 using primers 5 and 6 allowed epitope tagging of UL33. To remove galK and generate ΔUS28, ΔUL33, and UL33-HA BACs, dsDNA was generated by PCR using Pfu polymerase and oligonucleotides 7–12 (Table 2). The following PCR conditions were used: 94 °C for 15 s, 30 s at 57 °C for ΔUS28/54 °C for ΔUL33/60 °C for UL33-HA, 68 °C for 60 s, two cycles.
Table 2.
Primers used for BAC recombineering and sequencing
Primers 1–6 were used to introduce galK (homology arm is italicized and sequence recognizing galK is underlined), and primers 7–12 were used to remove galK (homology arm is italicized, complementary sequence is underlined, and glycine + HA tag is bold). The deletion of galK, US28, or UL33 was confirmed using primers 13–18. Primers 16 and 18–22 were employed for sequencing of the recombineering products. The oligonucleotides were purchased from Eurofins Genomics.
| Code | Name | Sequence |
|---|---|---|
| 1 | US28->galK F | 5′-GTGCGTGGACCAGACGGCGTCCATGCACCGAGGGCAGAACTGGTGCTATCCCTGTTGACAATTAATCATCGGCA-3′ |
| 2 | US28->galK R | 5′-ATCCATAACTTCGTATAATGTATGCTATACGAAGTTATAGCGCTTTTTTATCAGCACTGTCCTGCTCCTT-3′ |
| 3 | Ul33->galK F | 5′-CGGAAGCGTCGTCGCCCCGGACTGCGCCCGCGGTCTGCTATTCGTCCACGCCTGTTGACAATTAATCATCGGCA-3′ |
| 4 | Ul33->galK R | 5′-GGGAAATGGCGACGGGTTCTGGTGCTTTCTGAATAAAGTAACAGGAAAGCTCAGCACTGTCCTGCTCCTT-3′ |
| 5 | UL33galK F | 5′-CAAAAATCCCCCATCGACTCTCACAATCGCATCATAACCTCAGCGGGGTACCTGTTGACAATTAATCATCGGCA-3′ |
| 6 | UL33galK R | 5′-AAATGGCGACGGGTTCTGGTGCTTTCTGAATAAAGTAACAGGAAAGCTCATCAGCACTGTCCTGCTCCTT-3′ |
| 7 | galK->ΔUS28 F | 5′-CAGTCTCTCGGTGCGTGGACCAGACGGCGTCCATGCACCGAGGGCAGAACTGGTGCTATCTAAAAAAGCG-3′ |
| 8 | galK->ΔUS28 R | 5′-TTAATTAAGGATCCATAACTTCGTATAATGTATGCTATACGAAGTTATAGCGCTTTTTTAGATAGCACCA-3′ |
| 9 | galK->ΔUl33 F | 5′-CTCCAGAACCCGGAAGCGTCGTCGCCCCGGACTGCGCCCGCGGTCTGCTATTCGTCCACGGCTTTCCTGT-3′ |
| 10 | galK->ΔUl33 R | 5′-CGTATATGAGGGGAAATGGCGACGGGTTCTGGTGCTTTCTGAATAAAGTAACAGGAAAGCCGTGGACGAA-3′ |
| 11 | UL33galK->UL33HA F | 5′-CAAAAATCCCCCATCGACTCTCACAATCGCATCATAACCTCAGCGGGGTAGGCTACCCGTACGACGTCCCAGACTACGCC-3′ |
| 12 | UL33galK->UL33HA R | 5′-AAATGGCGACGGGTTCTGGTGCTTTCTGAATAAAGTAACAGGAAAGCTCAGGCGTAGTCTGGGACGTCGTACGGGTAGCC-3′ |
| 13 | galK internal F | 5′-CTCTGTTTGCCAACGCATTTGG-3′ |
| 14 | galK internal R | 5′-CGAAATCATCGCGCATAGAGG-3′ |
| 15 | US28 internal F | 5′-CGCATTTCCAGAATCGTTGC-3′ |
| 16 | Downstream US28 R | 5′-CCCTGAACATGTCCATCAGG-3′ |
| 17 | UL33 internal F | 5′-GCGTGTTACTCATCTCCTTCG-3′ |
| 18 | Downstream UL33 R | 5′-GGACGTCGTTTTTATCGTACCG-3′ |
| 19 | Upstream US28 F | 5′-GTAATTCGATCCTCTCTCACGC-3′ |
| 20 | Downstream US28 R | 5′-CTTTGCATACTTCTGCCTGC-3′ |
| 21 | Upstream UL33 F | 5′-CGCCGCATTTTTTCAGGATCTTGG-3′ |
| 22 | Downstream UL33 R | 5′-CGGTACGATAAAAACGACGTCC-3′ |
BAC isolation and verification
BAC DNA was isolated using a NucleoBond Xtra BAC kit (Machery Nagel, Düren, Germany). Recombinants were compared with pAL1502 after BamHI and HindIII digestion. The US28 and UL33 gene regions were sequenced (Eurofins Genomics, Ebersberg, Germany) upon PCR amplification using primers 13–22 (Table 2) and the following conditions: 94 °C for 30 s, 55 °C for 30 s, 72 °C for 3 min, 30 cycles.
Virus production
HFFF TR cells were transfected with BAC DNA (2 μg/2 × 106 cells) using the Amaxa basic fibroblast Nucleofector kit (Lonza, Basel, Switzerland) and an Amaxa Nucleofector (Lonza). Subsequent virus productions were initiated by infection of HFFF TR cells at m.o.i. 0.02. Of note, expression of RL13 and the UL128 locus was repressed during virus production in HFFF TR cells. IE1 staining in HFFF TR cells 3 dpi was used to determine virus titers.
Transfection
Transient transfections of HEK293T cells were performed using the polyethylenimine method (65). One day after plating, HEK293T cells were transfected with increasing amounts (0.4–120 ng) of pcDEF3-UL33-HA (AD169 strain) or pcDEF3-US28-HA (VHL/E strain) supplemented with empty pcDEF3 vector to a total of 2 μg of DNA/1 × 106 cells. For reporter gene assays, 1 μg of reporter gene plasmid was cotransfected.
Receptor ELISA
24 h post-transfection of HEK293T cells and 48 h post-receptor induction in the U251 cell lines, the cells were fixed and permeabilized. Expression of HA-tagged receptors was determined using anti-HA antibody (11867423001, Sigma-Aldrich) and anti-rat horseradish peroxidase–conjugated antibody (31470, Thermo Fisher Scientific, Waltham, MA). Absorbance was measured at 490 nm using a PowerWave X340 (BioTek, Winooski, VT).
Immunofluorescence microscopy
Stably transfected NIH-3T3 cells and U251 cell lines were fixed 1 day postseeding and -receptor induction. After permeabilization, UL33 and US28 expression was visualized using rabbit anti-UL33 (22) or rabbit anti-US28 antibodies (generated by Covance, Princeton, NJ (36)) and Alexa Fluor® 546–conjugated anti-rabbit (A11010, Thermo Fisher Scientific) or Alexa Fluor 488–conjugated anti-rabbit antibodies (A11008, Thermo Fisher Scientific), respectively. U251 cells were infected with HCMV Merlin at m.o.i. 3, and cells were fixed for immunofluorescence analysis every 24 h over a time course of 9 days. In Merlin-infected U251 cells, UL33 was stained using rat anti-HA and Alexa Fluor 488–linked anti-rat antibodies (A11006, Thermo Fisher Scientific). Visualization of US28 was performed as in NIH-3T3 cells. Mouse anti-immediate early antigen (MAB810R, Merck Millipore, Billerica, MA) and Alexa Fluor 350–conjugated anti-mouse (A11045, Thermo Fisher Scientific) antibodies were used to determine IE1 expression in infected U251 and HFFF TR cells. Epifluorescence imaging was performed using a Nikon TE200 microscope (Nikon, Tokyo, Japan).
For imaging at a higher resolution, confocal microscopy was performed on Merlin-infected U251 cells fixed at 5 dpi (m.o.i. 6). UL33 and US28 were stained using rat anti-HA and rabbit anti-US28 antibodies, respectively, as described above. pp28 was detected using mouse anti-pp28 (sc-69749, Santa Cruz Biotechnology). Cell nuclei were stained using DAPI (D9542, Sigma-Aldrich). Confocal laser-scanning microscopy was performed at room temperature on a Nikon A1R+ microscope (Nikon) equipped with a 60 × 1.4 oil-immersion objective. Samples were irradiated using the 405-, 488-, and 561-nm laser lines with 2, 0.5, and 1%, respectively. The 488 and 561 channels were detected with GaAsP photomultiplier tubes. The 405 channel (DAPI) was detected with a regular photomultiplier tube. The samples were scanned with a Nikon Galvano scanner at 2048 × 2048 pixels, corresponding to 51.5-nm pixel size. NIS-Elements AR 4.60.00 software (Nikon) was used for image acquisition. Fiji software was used for image analysis (66).
Western blotting
U251 cells were grown in serum-free conditions before lysis in native lysis buffer (32) 48 h post-receptor expression. A standard procedure was used for Western blot analysis. Membranes were probed with rabbit anti-STAT3 (4904, Cell Signaling Technology), rabbit anti-phospho-STAT3 (Tyr705) (9145, Cell Signaling Technology), and mouse anti-β-actin (A5316, Sigma-Aldrich) antibodies followed by horseradish peroxidase–conjugated secondary antibodies (170-6515 or 170-6516, Bio-Rad). Proteins were visualized using ECL substrate (NEL104001EA, PerkinElmer Life Sciences) and imaged using a ChemiDoc imager (Bio-Rad). Protein abundance was quantified using Image Studio software (LI-COR Biosciences, Lincoln, NE) and normalized to β-actin loading control.
Foci and spheroid formation
The formation of foci in NIH-3T3 cells and spheroid growth in U251 cells were evaluated as described before (20) with exception of Gαq inhibition. Treatment with Gαq inhibitor YM-254890 (300 nm) or DMSO vehicle was initiated 6 h after cell seeding, and treatment medium was refreshed every 3 days. Foci were quantified using Fiji software.
Reporter gene assays
Luciferase activity was measured 48 h after induction of receptor expression in U251 cell lines, under serum-free conditions, or 24 h post-transfection of HEK293T cells. U251–3SFP cells were infected with HCMV Merlin WT (UL33-HA), UL33-knockout (ΔUL33), or US28-knockout (UL33-HA, ΔUS28) virus at similar infection rates. Five dpi, after 2 days of serum starvation, luciferase activity was quantified. Similarly, luciferase activity was evaluated in U251–3SFP and U251-FM cells upon parallel infection at m.o.i. 6, which functioned as an external control for the STAT3 activation in infected U251 cells. Bioluminescence was measured using a Victor3 plate reader (PerkinElmer Life Sciences).
Inositol phosphate accumulation
HEK293T and U251 cells were labeled overnight in inositol-free medium supplemented with 1 μCi/ml myo[2-3H]inositol (PerkinElmer Life Sciences). One day post-transfection or -receptor induction, [3H]inositol phosphates were quantified (31).
IL-6 ELISA
Conditioned medium, accumulated from 24 to 48 h post-receptor induction, was collected from serum-starved U251 cell lines. IL-6 concentrations were measured using a human IL-6 Quantikine kit (R&D Systems, Minneapolis, MN).
Quantitative real-time PCR
U251, U251-FM-iUL33, and U251-FM-iUS28 cells were synchronized in serum-free growth medium supplemented with doxycycline. Culture medium was refreshed after 1 day, and another 24 h later cellular RNA was extracted using TRIzol reagent. mRNA was converted into cDNA through reverse transcription using the iScript cDNA Synthesis kit (Bio-Rad) according to the manufacturer's instructions. PCRs were performed using LC SYBR Green Master I reagent (Roche Applied Science) and the LightCycler 480 PCR detection system (Roche Applied Science) at 95 °C for 10 min followed by 45 cycles of 95 °C for 10 s, 60 °C for 15 s, and 70 °C for 15 s. UL33 and US28 transcripts were quantified using primers targeting the identical 3′-UTR of their mRNA: F, CCGCGGTTCGAAGGTAAG, R, CGATCATTACTAACCGGTACGC. ACTB was used as reference gene: F, TCCACCTTCCAGCAGATGTG; R, GCATTTGCGGTGGACGAT. Receptor mRNA abundance was calculated using the 2−ΔΔCt method and ACTB as an internal control.
Animal studies
For the xenograft model (6), stably transfected NIH-3T3 cells were injected subcutaneously into the flank of 8–10-week-old female athymic nude mice (Harlan/Envigo, Horst, The Netherlands). For the orthotopic glioblastoma model, U251 cells were stereotactically injected in the striatum of 6-week-old female athymic nude mice (Harlan/Envigo) as described previously (20).
Ethics statement
Animal experiments were conducted in compliance with Dutch law and European Community Council Directive 2010/63/EU for laboratory animal care and approved by the VU Medical Center animal experimentation commission (license number NCH13-03).
Data analysis
Student's t test (two-tailed, significance level of α = 0.05, using Holm–Šídák method) or analysis of variance with multiple comparisons test (Dunnett correction) was performed using Prism software (GraphPad, San Diego, CA).
Author contributions
J. R. v. S., R. L., T. W., M. S., and M. J. S. conceptualization; J. R. v. S. and M. P. B. data curation; J. R. v. S. and M. P. B. formal analysis; J. R. v. S. validation; J. R. v. S., M. P. B., T. S. F., R. H., N. D. B., P. v. G., E. V. L., E. S., and D. M. investigation; J. R. v. S., M. P. B., T. S. F., T. L., A. R., M. S.-v. W., R. J. P. M., G. A. M. S. v. D., C. S.-N., and T. W. methodology; J. R. v. S. writing-original draft; J. R. v. S., R. H., A. R., C. S.-N., M. S., and M. J. S. writing-review and editing; R. J. P. M. resources; G. A. M. S. v. D., C. S.-N., and M. S. supervision; M. S. and M. J. S. funding acquisition; M. J. S. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Richard Stanton for sharing HCMV Merlin BACs and excellent advice regarding virus work. Dr. Renée van Amerongen is acknowledged for providing the 7TFP plasmid, and Laura Smits-de Vries is acknowledged for assistance. We thank the AO|2 M microscopy core platform of VU University Medical Center Amsterdam for imaging support.
This work was supported by the Netherlands Organization for Scientific Research (NWO) Grants Vici 016.140.657 and Vidi 700.54.425 (to M. J. S.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S5.
- HCMV
- human cytomegalovirus
- GPCR
- G protein–coupled receptor
- EBV
- Epstein–Barr virus
- KSHV
- Kaposi's sarcoma–associated herpesvirus
- InsP
- inositol phosphate
- CREB
- cAMP-response element–binding protein
- RCMV
- rat cytomegalovirus
- MCMV
- murine cytomegalovirus
- NFAT
- nuclear factor of activated T cells
- Tcf
- T cell factor
- Lef
- lymphoid enhancer-binding factor
- VEGF
- vascular endothelial growth factor
- TGFβ
- transforming growth factor β
- SRF
- serum response factor
- BAC
- bacterial artificial chromosome
- HA
- hemagglutinin
- dpi
- day(s) postinfection
- IE1
- immediate-early 1
- VAC
- virion assembly compartment
- 3D
- three-dimensional
- FM
- firefly luciferase/mCherry
- CMV
- cytomegalovirus
- HFFF
- human fetal foreskin fibroblast
- eGFP
- enhanced GFP
- m.o.i.
- multiplicity of infection
- DAPI
- 4′,6-diamidino-2-phenylindole
- F
- forward
- R
- reverse
- TR
- Tet repressor.
References
- 1. Thompson M. P., and Kurzrock R. (2004) Epstein-Barr virus and cancer. Clin Cancer Res. 10, 803–821 10.1158/1078-0432.CCR-0670-3 [DOI] [PubMed] [Google Scholar]
- 2. Martin J. N. (2007) The epidemiology of KSHV and its association with malignant disease, in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) Chapter 54, Cambridge University Press, Cambridge, UK: [PubMed] [Google Scholar]
- 3. Michaelis M., Doerr H. W., and Cinatl J. (2009) The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia 11, 1–9 10.1593/neo.81178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Vischer H. F., Siderius M., Leurs R., and Smit M. J. (2014) Herpesvirus-encoded GPCRs: neglected players in inflammatory and proliferative diseases? Nat. Rev. Drug Discov. 13, 123–139 10.1038/nrd4189 [DOI] [PubMed] [Google Scholar]
- 5. Maussang D., Langemeijer E., Fitzsimons C. P., Stigter-van Walsum M., Dijkman R., Borg M. K., Slinger E., Schreiber A., Michel D., Tensen C. P., van Dongen G. A., Leurs R., and Smit M. J. (2009) The human cytomegalovirus-encoded chemokine receptor US28 promotes angiogenesis and tumor formation via cyclooxygenase-2. Cancer Res. 69, 2861–2869 10.1158/0008-5472.CAN-08-2487 [DOI] [PubMed] [Google Scholar]
- 6. Maussang D., Verzijl D., van Walsum M., Leurs R., Holl J., Pleskoff O., Michel D., van Dongen G. A., and Smit M. J. (2006) Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 13068–13073 10.1073/pnas.0604433103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lyngaa R., Nørregaard K., Kristensen M., Kubale V., Rosenkilde M. M., and Kledal T. N. (2010) Cell transformation mediated by the Epstein-Barr virus G protein-coupled receptor BILF1 is dependent on constitutive signaling. Oncogene 29, 4388–4398 10.1038/onc.2010.173 [DOI] [PubMed] [Google Scholar]
- 8. Bais C., Santomasso B., Coso O., Arvanitakis L., Raaka E. G., Gutkind J. S., Asch A. S., Cesarman E., Gershengorn M. C., and Mesri E. A. (1998) G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature 391, 86–89 10.1038/34193 [DOI] [PubMed] [Google Scholar]
- 9. Bate S. L., Dollard S. C., and Cannon M. J. (2010) Cytomegalovirus seroprevalence in the United States: the national health and nutrition examination surveys, 1988–2004. Clin. Infect. Dis. 50, 1439–1447 10.1086/652438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Griffiths P., Baraniak I., and Reeves M. (2015) The pathogenesis of human cytomegalovirus. J. Pathol. 235, 288–297 10.1002/path.4437 [DOI] [PubMed] [Google Scholar]
- 11. Harkins L., Volk A. L., Samanta M., Mikolaenko I., Britt W. J., Bland K. I., and Cobbs C. S. (2002) Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet 360, 1557–1563 10.1016/S0140-6736(02)11524-8 [DOI] [PubMed] [Google Scholar]
- 12. Samanta M., Harkins L., Klemm K., Britt W. J., and Cobbs C. S. (2003) High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J. Urol. 170, 998–1002 10.1097/01.ju.0000080263.46164.97 [DOI] [PubMed] [Google Scholar]
- 13. Brouchet L., Valmary S., Dahan M., Didier A., Galateau-Salle F., Brousset P., and Degano B. (2005) Detection of oncogenic virus genomes and gene products in lung carcinoma. Br. J. Cancer 92, 743–746 10.1038/sj.bjc.6602409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cox B., Richardson A., Graham P., Gislefoss R. E., Jellum E., and Rollag H. (2010) Breast cancer, cytomegalovirus and Epstein-Barr virus: a nested case-control study. Br. J. Cancer 102, 1665–1669 10.1038/sj.bjc.6605675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Baryawno N., Rahbar A., Wolmer-Solberg N., Taher C., Odeberg J., Darabi A., Khan Z., Sveinbjörnsson B., FuskevÅg O. M., Segerström L., Nordenskjöld M., Siesjö P., Kogner P., Johnsen J. I., and Söderberg-Nauclér C. (2011) Detection of human cytomegalovirus in medulloblastomas reveals a potential therapeutic target. J. Clin. Investig. 121, 4043–4055 10.1172/JCI57147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Soroceanu L., Matlaf L., Bezrookove V., Harkins L., Martinez R., Greene M., Soteropoulos P., and Cobbs C. S. (2011) Human cytomegalovirus US28 found in glioblastoma promotes an invasive and angiogenic phenotype. Cancer Res. 71, 6643–6653 10.1158/0008-5472.CAN-11-0744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wakefield A., Pignata A., Ghazi A., Ashoori A., Hegde M., Landi D., Gray T., Scheurer M. E., Chintagumpala M., Adesina A., Gottschalk S., Hicks J., Powell S. Z., and Ahmed N. (2015) Is CMV a target in pediatric glioblastoma? Expression of CMV proteins, pp65 and IE1–72 and CMV nucleic acids in a cohort of pediatric glioblastoma patients. J. Neurooncol. 125, 307–315 10.1007/s11060-015-1905-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dziurzynski K., Chang S. M., Heimberger A. B., Kalejta R. F., McGregor Dallas S. R., Smit M., Soroceanu L., Cobbs C. S., and HCMV and Gliomas Symposium (2012) Consensus on the role of human cytomegalovirus in glioblastoma. Neuro Oncol. 14, 246–255 10.1093/neuonc/nor227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fornara O., Bartek J. Jr, Rahbar A., Odeberg J., Khan Z., Peredo I., Hamerlik P., Bartek J., Stragliotto G., Landazuri N., and Söderberg-Nauclér C. (2016) Cytomegalovirus infection induces a stem cell phenotype in human primary glioblastoma cells: prognostic significance and biological impact. Cell Death Differ. 23, 261–269 10.1038/cdd.2015.91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heukers R., Fan T. S., de Wit R. H., van Senten J. R., De Groof T. W. M., Bebelman M. P., Lagerweij T., Vieira J., de Munnik S. M., Smits-de Vries L., van Offenbeek J., Rahbar A., van Hoorick D., Söderberg-Naucler C., Würdinger T., et al. (2018) The constitutive activity of the virally encoded chemokine receptor US28 accelerates glioblastoma growth. Oncogene 37, 4110–4121 10.1038/s41388-018-0255-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chee M. S., Satchwell S. C., Preddie E., Weston K. M., and Barrell B. G. (1990) Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature 344, 774–777 10.1038/344774a0 [DOI] [PubMed] [Google Scholar]
- 22. Margulies B. J., Browne H., and Gibson W. (1996) Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology 225, 111–125 10.1006/viro.1996.0579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Penfold M. E., Schmidt T. L., Dairaghi D. J., Barry P. A., and Schall T. J. (2003) Characterization of the rhesus cytomegalovirus US28 locus. J. Virol. 77, 10404–10413 10.1128/JVI.77.19.10404-10413.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Varnum S. M., Streblow D. N., Monroe M. E., Smith P., Auberry K. J., Pasa-Tolic L., Wang D., Camp D. G. 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R. D., and Nelson J. A. (2004) Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78, 10960–10966 10.1128/JVI.78.20.10960-10966.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Margulies B. J., and Gibson W. (2007) The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles. Virus Res. 123, 57–71 10.1016/j.virusres.2006.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. O'Connor C. M., and Shenk T. (2012) Human cytomegalovirus pUL78 G protein-coupled receptor homologue is required for timely cell entry in epithelial cells but not fibroblasts. J. Virol. 86, 11425–11433 10.1128/JVI.05900-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cheng S., Caviness K., Buehler J., Smithey M., Nikolich-Žugich J., and Goodrum F. (2017) Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc. Natl. Acad. Sci. U.S.A. 114, E10586–E10595 10.1073/pnas.1710522114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Randolph-Habecker J. R., Rahill B., Torok-Storb B., Vieira J., Kolattukudy P. E., Rovin B. H., and Sedmak D. D. (2002) The expression of the cytomegalovirus chemokine receptor homolog US28 sequesters biologically active CC chemokines and alters IL-8 production. Cytokine 19, 37–46 10.1006/cyto.2002.0874 [DOI] [PubMed] [Google Scholar]
- 29. Waldhoer M., Kledal T. N., Farrell H., and Schwartz T. W. (2002) Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 76, 8161–8168 10.1128/JVI.76.16.8161-8168.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Casarosa P., Gruijthuijsen Y. K., Michel D., Beisser P. S., Holl J., Fitzsimons C. P., Verzijl D., Bruggeman C. A., Mertens T., Leurs R., Vink C., and Smit M. J. (2003) Constitutive signaling of the human cytomegalovirus-encoded receptor UL33 differs from that of its rat cytomegalovirus homolog R33 by promiscuous activation of G proteins of the Gq, Gi, and Gs classes. J. Biol. Chem. 278, 50010–50023 10.1074/jbc.M306530200 [DOI] [PubMed] [Google Scholar]
- 31. Slinger E., Maussang D., Schreiber A., Siderius M., Rahbar A., Fraile-Ramos A., Lira S. A., Söderberg-Nauclér C., and Smit M. J. (2010) HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci. Signal 3, ra58 10.1126/scisignal.2001180 [DOI] [PubMed] [Google Scholar]
- 32. de Wit R. H., Mujić-Delić A., van Senten J. R., Fraile-Ramos A., Siderius M., and Smit M. J. (2016) Human cytomegalovirus encoded chemokine receptor US28 activates the HIF-1α/PKM2 axis in glioblastoma cells. Oncotarget 7, 67966–67985 10.18632/oncotarget.11817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vomaske J., Melnychuk R. M., Smith P. P., Powell J., Hall L., DeFilippis V., Früh K., Smit M., Schlaepfer D. D., Nelson J. A., and Streblow D. N. (2009) Differential ligand binding to a human cytomegalovirus chemokine receptor determines cell type-specific motility. PLoS Pathog. 5, e1000304 10.1371/journal.ppat.1000304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Humby M. S., and O'Connor C. M. (2015) Human cytomegalovirus US28 is important for latent infection of hematopoietic progenitor cells. J. Virol. 90, 2959–2970 10.1128/JVI.02507-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Krishna B. A., Poole E. L., Jackson S. E., Smit M. J., Wills M. R., and Sinclair J. H. (2017) Latency-associated expression of human cytomegalovirus US28 attenuates cell signaling pathways to maintain latent infection. MBio 8, e01754–17 10.1128/mBio.01754-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bongers G., Maussang D., Muniz L. R., Noriega V. M., Fraile-Ramos A., Barker N., Marchesi F., Thirunarayanan N., Vischer H. F., Qin L., Mayer L., Harpaz N., Leurs R., Furtado G. C., Clevers H., et al. (2010) The cytomegalovirus-encoded chemokine receptor US28 promotes intestinal neoplasia in transgenic mice. J. Clin. Investig. 120, 3969–3978 10.1172/JCI42563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beisser P. S., Vink C., Van Dam J. G., Grauls G., Vanherle S. J., and Bruggeman C. A. (1998) The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection. J. Virol. 72, 2352–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Farrell H. E., Bruce K., Lawler C., Oliveira M., Cardin R., Davis-Poynter N., and Stevenson P. G. (2017) Murine cytomegalovirus spreads by dendritic cell recirculation. MBio 8, e01264–17 10.1128/mBio.01264-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Case R., Sharp E., Benned-Jensen T., Rosenkilde M. M., Davis-Poynter N., and Farrell H. E. (2008) Functional analysis of the murine cytomegalovirus chemokine receptor homologue M33: ablation of constitutive signaling is associated with an attenuated phenotype in vivo. J. Virol. 82, 1884–1898 10.1128/JVI.02550-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Farrell H. E., Abraham A. M., Cardin R. D., Sparre-Ulrich A. H., Rosenkilde M. M., Spiess K., Jensen T. H., Kledal T. N., and Davis-Poynter N. (2011) Partial functional complementation between human and mouse cytomegalovirus chemokine receptor homologues. J. Virol. 85, 6091–6095 10.1128/JVI.02113-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Cardin R. D., Schaefer G. C., Allen J. R., Davis-Poynter N. J., and Farrell H. E. (2009) The M33 chemokine receptor homolog of murine cytomegalovirus exhibits a differential tissue-specific role during in vivo replication and latency. J. Virol. 83, 7590–7601 10.1128/JVI.00386-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stanton R. J., Baluchova K., Dargan D. J., Cunningham C., Sheehy O., Seirafian S., McSharry B. P., Neale M. L., Davies J. A., Tomasec P., Davison A. J., and Wilkinson G. W. (2010) Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J. Clin. Investig. 120, 3191–3208 10.1172/JCI42955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fraile-Ramos A., Kledal T. N., Pelchen-Matthews A., Bowers K., Schwartz T. W., and Marsh M. (2001) The human cytomegalovirus US28 protein is located in endocytic vesicles and undergoes constitutive endocytosis and recycling. Mol. Biol. Cell 12, 1737–1749 10.1091/mbc.12.6.1737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fraile-Ramos A., Pelchen-Matthews A., Kledal T. N., Browne H., Schwartz T. W., and Marsh M. (2002) Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3, 218–232 10.1034/j.1600-0854.2002.030307.x [DOI] [PubMed] [Google Scholar]
- 45. Seo J. Y., and Britt W. J. (2006) Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80, 5611–5626 10.1128/JVI.02630-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Rijn S., Nilsson J., Noske D. P., Vandertop W. P., Tannous B. A., and Würdinger T. (2013) Functional multiplex reporter assay using tagged Gaussia luciferase. Sci. Rep. 3, 1046 10.1038/srep01046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cobbs C. S., Harkins L., Samanta M., Gillespie G. Y., Bharara S., King P. H., Nabors L. B., Cobbs C. G., and Britt W. J. (2002) Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 62, 3347–3350 [PubMed] [Google Scholar]
- 48. Cobbs C. (2014) Response to “Human cytomegalovirus infection in tumor cells of the nervous system is not detectable with standardized pathologico-virological diagnostics.” Neuro Oncol. 16, 1435–1436 10.1093/neuonc/nou295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Taha M. S., Abdalhamid B. A., El-Badawy S. A., Sorour Y. M., Almsned F. M., and Al-Abbadi M. A. (2016) Expression of cytomegalovirus in glioblastoma multiforme: myth or reality? Br. J. Neurosurg. 30, 307–312 10.3109/02688697.2015.1119241 [DOI] [PubMed] [Google Scholar]
- 50. Stragliotto G., Rahbar A., Solberg N. W., Lilja A., Taher C., Orrego A., Bjurman B., Tammik C., Skarman P., Peredo I., and Söderberg-Nauclér C. (2013) Effects of valganciclovir as an add-on therapy in patients with cytomegalovirus-positive glioblastoma: a randomized, double-blind, hypothesis-generating study. Int. J. Cancer 133, 1204–1213 10.1002/ijc.28111 [DOI] [PubMed] [Google Scholar]
- 51. Peng C., Wang J., Tanksley J. P., Mobley B. C., Ayers G. D., Moots P. L., and Clark S. W. (2016) Valganciclovir and bevacizumab for recurrent glioblastoma: a single-institution experience. Mol. Clin. Oncol. 4, 154–158 10.3892/mco.2015.692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Batich K. A., Reap E. A., Archer G. E., Sanchez-Perez L., Nair S. K., Schmittling R. J., Norberg P., Xie W., Herndon J. E. 2nd, Healy P., McLendon R. E., Friedman A. H., Friedman H. S., Bigner D., Vlahovic G., et al. (2017) Long-term survival in glioblastoma with cytomegalovirus pp65-targeted vaccination. Clin. Cancer Res. 23, 1898–1909 10.1158/1078-0432.CCR-16-2057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Mitchell D. A., Batich K. A., Gunn M. D., Huang M. N., Sanchez-Perez L., Nair S. K., Congdon K. L., Reap E. A., Archer G. E., Desjardins A., Friedman A. H., Friedman H. S., Herndon J. E. 2nd, Coan A., McLendon R. E., et al. (2015) Tetanus toxoid and CCL3 improve dendritic cell vaccines in mice and glioblastoma patients. Nature 519, 366–369 10.1038/nature14320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Moepps B., Tulone C., Kern C., Minisini R., Michels G., Vatter P., Wieland T., and Gierschik P. (2008) Constitutive serum response factor activation by the viral chemokine receptor homologue pUS28 is differentially regulated by Gαq/11 and Gα16. Cell. Signal. 20, 1528–1537 10.1016/j.cellsig.2008.04.010 [DOI] [PubMed] [Google Scholar]
- 55. Pu P., Zhang Z., Kang C., Jiang R., Jia Z., Wang G., and Jiang H. (2009) Downregulation of Wnt2 and β-catenin by siRNA suppresses malignant glioma cell growth. Cancer Gene Ther. 16, 351–361 10.1038/cgt.2008.78 [DOI] [PubMed] [Google Scholar]
- 56. Liu C., Tu Y., Sun X., Jiang J., Jin X., Bo X., Li Z., Bian A., Wang X., Liu D., Wang Z., and Ding L. (2011) Wnt/β-catenin pathway in human glioma: expression pattern and clinical/prognostic correlations. Clin. Exp. Med 11, 105–112 10.1007/s10238-010-0110-9 [DOI] [PubMed] [Google Scholar]
- 57. Pulvirenti T., Van Der Heijden M., Droms L. A., Huse J. T., Tabar V., and Hall A. (2011) Dishevelled 2 signaling promotes self-renewal and tumorigenicity in human gliomas. Cancer Res. 71, 7280–7290 10.1158/0008-5472.CAN-11-1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Langemeijer E. V., Slinger E., de Munnik S., Schreiber A., Maussang D., Vischer H., Verkaar F., Leurs R., Siderius M., and Smit M. J. (2012) Constitutive β-catenin signaling by the viral chemokine receptor US28. PLoS One 7, e48935 10.1371/journal.pone.0048935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tschische P., Tadagaki K., Kamal M., Jockers R., and Waldhoer M. (2011) Heteromerization of human cytomegalovirus encoded chemokine receptors. Biochem. Pharmacol. 82, 610–619 10.1016/j.bcp.2011.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Farrell H. E., Abraham A. M., Cardin R. D., Mølleskov-Jensen A. S., Rosenkilde M. M., and Davis-Poynter N. (2013) Identification of common mechanisms by which human and mouse cytomegalovirus seven-transmembrane receptor homologues contribute to in vivo phenotypes in a mouse model. J. Virol. 87, 4112–4117 10.1128/JVI.03406-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Gruijthuijsen Y. K., Casarosa P., Kaptein S. J., Broers J. L., Leurs R., Bruggeman C. A., Smit M. J., and Vink C. (2002) The rat cytomegalovirus R33-encoded G protein-coupled receptor signals in a constitutive fashion. J. Virol. 76, 1328–1338 10.1128/JVI.76.3.1328-1338.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. O'Hayre M., Vázquez-Prado J., Kufareva I., Stawiski E. W., Handel T. M., Seshagiri S., and Gutkind J. S. (2013) The emerging mutational landscape of G proteins and G-protein-coupled receptors in cancer. Nat. Rev. Cancer 13, 412–424 10.1038/nrc3521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fuerer C., and Nusse R. (2010) Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One 5, e9370 10.1371/journal.pone.0009370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Warming S., Costantino N., Court D. L., Jenkins N. A., and Copeland N. G. (2005) Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 10.1093/nar/gni035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schlaeger E. J., and Christensen K. (1999) Transient gene expression in mammalian cells grown in serum-free suspension culture. Cytotechnology 30, 71–83 10.1023/A:1008000327766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.