Abstract
How food production first entered eastern Africa ~5000 years ago and the extent to which people moved with livestock is unclear. We present genome-wide data from 41 individuals associated with Later Stone Age, Pastoral Neolithic (PN), and Iron Age contexts in what are now Kenya and Tanzania to examine the genetic impacts of the spreads of herding and farming. Our results support a multi-phase model in which admixture between northeastern African-related peoples and eastern African foragers formed multiple pastoralist groups, including a genetically homogeneous PN cluster. Additional admixture with northeastern and western African-related groups occurred by the Iron Age. These findings support several movements of food producers, while rejecting models of minimal admixture with foragers and of genetic differentiation between makers of distinct PN artifacts.
One sentence summary:
DNA from 41 ancient eastern Africans documents demic diffusion and admixture with local groups during the spread of food production into sub-Saharan Africa.
Introduction
Domestic sheep, goats, and cattle of southwest Asian origin were first introduced to northeastern Africa ~8000 years before present (BP), and spread into eastern Africa beginning ~5000 BP, ultimately reaching southernmost Africa by ~2000 BP (1, 2). How pastoralism – a way of life centered on herding animals – spread into eastern Africa is unclear. Livestock appear in northern Ethiopia and Djibouti relatively late, ~4500–4000 BP (3), and are poorly documented elsewhere in the Horn of Africa and in South Sudan. Instead, the earliest known domesticated animals in sub-Saharan Africa are found in Kenya at the beginning of the Pastoral Neolithic (PN; ~5000–1200 BP) era near Lake Turkana, where archaeological evidence documents groups that pursued fishing and herding and constructed elaborate monumental cemeteries (4–6). Although livestock spread quickly through the Turkana Basin, herding practices were not transmitted farther south for many hundreds of years. Sheep, goats, and pottery typical of Turkana began to trickle into Kenya’s south-central Rift Valley ~4200 BP (7, 8), but it was not until ~3300 BP that specialized pastoralism spread across Kenya and northern Tanzania, transforming the economic, social, and physical landscapes of the region (9–11).
The core PN era (~3300–1200 BP) in Kenya and Tanzania witnessed the development of diverse herder societies, some heavily reliant on livestock (2). However, pastoralism did not fully replace Later Stone Age (LSA) economies present in the region since ~50,000 BP, creating a mosaic of herding and foraging communities on the landscape. Two contemporaneous pastoralist traditions have been identified: Elmenteitan and Savanna Pastoral Neolithic (SPN) (12, 13). Elmenteitan sites are found between the central Rift Valley and the western Lake Victoria basin of Kenya. Occupants used a particular obsidian source, and left behind distinctive lithic and ceramic traditions, and primarily cremation burials. By contrast, SPN sites are found across a wider part of Kenya and Tanzania. Occupants used different obsidian sources, had greater diversity in material culture, and mainly buried their dead in cairns. The heterogeneous SPN category likely encompasses multiple groups. Some distinctions between SPN and Elmenteitan traditions, such as mortuary practices, are variable (6), and relationships between PN groups – both cultural and genetic – remain uncertain. In addition, little is known about herder interactions with LSA foragers, or about relationships among later PN herders and the first iron-using herders after ~1200 BP. By this time, farming is also documented in the region (14, 15).
Archaeologists have debated the cultural and genetic affinities of the first pastoralists in eastern Africa and the role that movement of people played in the spread of herding to the region. Because the oldest instances of livestock remains and associated pottery and stone tool traditions have been found near Lake Turkana, it has been hypothesized that pastoralism was introduced by migrants from Sudan and/or Ethiopia, potentially in a series of small movements, and that their descendants gave rise to PN traditions farther south (12, 13, 15, 16). However, there are no unambiguous cultural connections between Kenya’s earliest herders and northern groups, and archaeological evidence supports the local adoption of herding to some degree (8, 16, 17). Other archaeological and linguistic evidence has been jointly used to hypothesize two expansions into eastern Africa: an initial expansion of herders speaking Afro-Asiatic (specifically proto-Southern Cushitic) languages from the Horn of Africa linked with the SPN, and a second expansion of herders speaking Nilo-Saharan (specifically Nilotic) languages linked with the Elmenteitan.
People of the latter expansion have also been hypothesized to be ancestral to some Iron Age groups (18, 19). One subset of Rift Valley sites is designated Pastoral Iron Age (PIA; ~1200 BP to recent) on the basis of material culture and evidence for herding, whereas other sites appear connected to farming and are classified into early, middle, and later Iron Age (IA; ~2500 BP to recent) variants (2, 14). Iron-working first entered eastern Africa via the Lake Victoria Basin ~2500 BP and spread toward the coast by 2000 BP (14). This may have brought early IA farmers – thought to have spoken Bantu languages originating in equatorial western Africa – into contact with PN herders, although iron-working is not widely attested among herders until ~1200 BP at PIA sites (2, 15). Alternatively, PIA sites may reflect other iron-working traditions entering from the north, potentially associated with movements of Nilotic-speaking pastoralists into and within the Rift (2). This complex mosaic of foragers, herders, and farmers gave rise to much of the present day ethno-linguistic landscape of eastern Africa (20).
Rigorous testing of models for how herding spread has been inhibited by several factors. A spatial and chronological gap exists between the earliest evidence of pastoralism in the Turkana Basin and the later PN expansion, with few material culture similarities between them. Additionally, relationships among PN and diverse Iron Age groups remain poorly understood. Human skeletal material from relevant contexts tends to be fragmentary, limiting bioarchaeological analysis, and reliable radiocarbon dates are rare. Finally, the persistence of foraging groups raises questions about interaction networks during this period (12, 15), and whether food production spread primarily via demic expansion or via local adoption of novel practices and livestock.
To address these debates, we generated genome-wide ancient DNA data from individuals buried at sites associated with LSA (n=3), early pastoral and PN (n=31), IA (n=1), and PIA (n=6) archaeological traditions in what are now Kenya and Tanzania (Fig. 1, Tables 1, S1). We extracted DNA from a combination of tooth and bone samples and enriched for a targeted set of ~1.2 million single nucleotide polymorphisms (SNPs; (21)). Surprisingly, given the tropical climate and variable curatorial conditions, we obtained excellent data quality, with a median of approximately 0.51× coverage, or 440,000 SNPs covered by at least one sequence, for the 41 newly reported individuals (from a total of 67 sequencing libraries; Table S2). The data scored well in standard ancient DNA authenticity metrics for all but two individuals (I12391 and I13970, whom we excluded from genome-wide analyses but for whom we obtained Y chromosome and mitochondrial DNA [mtDNA] haplogroups; (21)). We also generated direct radiocarbon dates for 35 individuals (Tables S3–S4, Fig. S1). We analyze these data jointly with sequences from published ancient African individuals (22–27), as well as from people living in eastern Africa today (28–31).
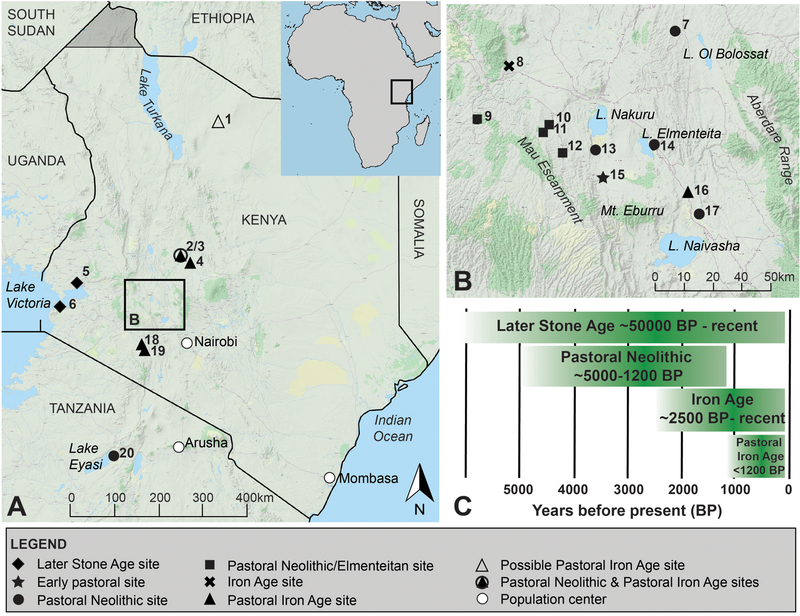
Figure 1. Map of the study area and regional chronology.
Locations of sampled archaeological sites in Kenya and Tanzania (A), with detail of the south-central Rift Valley (B). A timeline for eastern African archaeological traditions (C) highlights their degree of overlap and diffuse endpoints. Numbers in (A) and (B) correspond to sites listed in Table 1. Location of (7) is approximate. Terrain basemaps © www.thunderforest.com, data © www.osm.org/copyright, adapted under CC-BY-SA 2.0.
Table 1.
Ancient individuals reported in this study, ordered by start of calibrated radiocarbon date range
| Lab ID | Site1 | Map # | Lat | Long | Archaeological association2 | Genetic cluster | Sex | mtDNA haplogroup | Y chromosome haplogroup | Cover-age on target | Uncalibrated years before present (BP) (lab number) | Calibrated years before present (cal BP), 2σ3 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| I12533 | Prettejohn’s Gully (GsJi11) | 15 | −0.545 | 36.106 | Early pastoral? | PN outlier | M | K1a | E2(xE2b); E-M75 | 0.83 | 3670±20 (PSUAMS-4982) | 4080–3890 |
| I12534 | Prettejohn’s Gully (GsJi11) | 15 | −0.545 | 36.106 | Early pastoral? | PN outlier | F | L3f1b | 0.69 | 3640±20 (PSUAMS-4983) | 4060–3860 | |
| I8874 | Cole’s Burial (GrJj5a) | 14 | −0.442 | 36.267 | PN | PN cluster | M | L3i2 | E1b1b1a1a1b1; E-CTS3282 | 3.90 | 3070±20 (PSUAMS-4723) | 3350–3180 |
| I8809 | Kisima Farm, A5/Porcupine Cave | 2 | 0.458 | 36.709 | PN | PN cluster | M | M1a1 | E1b1b1b2b2a1; E-M293 | 3.48 | 2855±20 (PSUAMS-4510) | 3030–2860 |
| I8820 | Kisima Farm, A5/Porcupine Cave | 2 | 0.458 | 36.709 | PN | PN cluster | F | M1a1f | 0.07 | 2675±20 (PSUAMS-4717) | 2840–2740 | |
| I12398/94 | Rigo Cave (GrJh3) | 12 | −0.464 | 35.971 | PN/ELM | PN cluster | M | L3f | E1b1b1b2b2a1; E-M293 | 0.89 | 2480±20 (PSUAMS-4945); 2570±15 (PSUAMS-4946) | 2710–2380; 2750–2510 |
| I8759 | Naishi Rockshelter | 13 | −0.458 | 36.081 | PN | PN outlier | M | L3×1a | E1b1b1b2b; E-V1515 (prob. E-M293) | 0.07 | 2550±15 (PSUAMS-4715) | 2750–2500 |
| I13980 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN cluster | M | HV1b1 | E1b1b1a1b2; E-V22 | 2.72 | 2530±20 (PSUAMS-5655) | 2740–2490 |
| I13981 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN cluster | F | L0a | 0.36 | 2510±20 (PSUAMS-5656) | 2730–2460 | |
| I8758 | Naishi Rockshelter | 13 | −0.458 | 36.081 | PN | PN cluster | M | L0a2d | A1b(xA1b1b2a); A-P108 | 0.29 | 2470±15 (PSUAMS-4624) | 2700–2370 |
| I8804 | Keringet Cave?5 | 9 | −0.358 | 35.699 | PN | PN cluster | M | L4b2a1 | A1b1b2; A-L427 | 0.50 | 2465±20 (PSUAMS-4716) | 2700–2360 |
| I8923 | Rigo Cave (GrJh3) | 12 | −0.464 | 35.971 | PN/ELM | PN cluster | M | M1a1b (likely) | E1b1b1b2b2; E-V1486 (prob. E-M293) | 0.15 | 2440±20 (PSUAMS-4512) | 2690–2350 |
| I13979 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN cluster | F | L3×1 | 2.56 | 2410±20 (PSUAMS-5654) | 2490–2350 | |
| I8922 | Rigo Cave (GrJh3) | 12 | −0.464 | 35.971 | PN/ELM | PN cluster | M | L4b2a2c | E1b1b1b2b2a1; E-M293 | 2.79 | 2400±15 (PSUAMS-4725)6 | c. 2460–2350 |
| I8814 | Naivasha Burial Site | 17 | −0.663 | 36.410 | PN | PN cluster | F | L4b2a2b | 2.53 | 2400±20 (PSUAMS-4784) | 2480–2340 | |
| I13978 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN outlier | F | L4b2a1 | 0.56 | 2355±20 (PSUAMS-5653) | 2400–2310 | |
| I8830 | Naivasha Burial Site | 17 | −0.663 | 36.410 | PN | PN cluster | M | M1a1b | xBT (prob. A) | 0.10 | 2320±20 (PSUAMS-4720) | 2360–2210 |
| I8920 | Naivasha Burial Site | 17 | −0.663 | 36.410 | PN | PN cluster | M | L3h1a1 | E1b1b1b2b2a1; E-M293 | 1.68 | 2310±15 (PSUAMS-4724) | 2350–2210 |
| I8919 | Naivasha Burial Site | 17 | −0.663 | 36.410 | PN | PN cluster | M | L4a1 | A1b1b2b; A-M13 | 1.84 | 2255±20 (PSUAMS-4789) | 2340–2160 |
| I8918 | Naivasha Burial Site | 17 | −0.663 | 36.410 | PN | PN cluster | M | L3×1a | E1b1b1b2b2a1; E-M293 | 2.45 | 2235±20 (PSUAMS-4744) | 2320–2150 |
| I13762 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN cluster | M | L3i2 | E1b1b1b2b2a1; E-M293 | 1.81 | 2140±15 (PSUAMS-5458) | 2150–2020 |
| I10719 | Njoro River Cave II | 11 | −0.389 | 35.917 | PN/ELM | PN cluster | F | L3h1a2a1 | 1.11 | 2070±15 (PSUAMS-4758) | 2110–1930 | |
| I13970 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | N/A | F | L3h1a2a1 | 0.03 | 2030±20 (PSUAMS-5650) | 2000–1900 | |
| I13977 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN cluster | M | L0f2a1 | E1b1b1b2b2; E-V1486 (prob. E-M293) | 0.30 | 2005±20 (PSUAMS-5652) | 2000–1890 |
| I8808 | Jawuoyo Rockshelter | 5 | −0.067 | 34.667 | LSA | Forager cline | M | L4b2a2c | E1b1b1a1b2; E-V22 | 1.37 | 1895±15 (PSUAMS-4783) | 1880–1750 |
| I8805 | Egerton Cave (GrJh10) | 10 | −0.375 | 35.933 | PN/ELM | PN cluster | F | L0a1d | 3.79 | 1880±15 (PSUAMS-4741) | 1870–1740 | |
| I12384 | Ol Kalou | 7 | −0.3007 | 36.4007 | PN | PN cluster | M | L3d1d | E1b1b1b2b2a1; E-M293 | 0.51 | 1800±20 (PSUAMS-4940) | 1810–1620 |
| I13972 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | PN outlier | M | T2+150 | E1b1b1b2b2; E-V1486 (prob. E-M293) | 0.09 | 1780±25 (PSUAMS-5651) | 1740–1580 |
| I12394 | Keringet Cave (GrJg4) | 9 | −0.358 | 35.699 | PN/ELM | PN cluster | F | K1a | 0.42 | 1585±15 (PSUAMS-4943) | 1530–1400 | |
| I8892 | Ilkek Mounds | 16 | −0.603 | 36.374 | PIA | PIA cluster | M | L0f2a | E2(xE2b); E-M75 | 0.10 | 1170±15 (PSUAMS-4788) | 1170–980 |
| I8802 | Deloraine Farm (GqJh6) | 8 | −0.183 | 35.809 | IA | IA other | M | L5b1 | E1b1a1a1a1a; E-M58 | 2.65 | 1160±15 (PSUAMS-4625) | 1170–970 |
| I8901 | Kisima Farm, C4 | 3 | 0.458 | 36.709 | PIA | PIA cluster | M | L3h1a1 | E2(xE2b); E-M75 | 0.02 | 1110±15 (PSUAMS-4743) | 1060–940 |
| I12391 | Kasiole 2 (GvJh54) | 18 | −1.326 | 35.939 | PIA | N/A | M | L3h1a2a1 | E1b1b1b2b; E-V1515 (prob. E-M293) | 0.02 | 1110±15 (PSUAMS-4942) | 1060–940 |
| I12381 | Laikipia District Burial (GoJl45) | 4 | 0.380 | 36.893 | PIA | PIA cluster | F | L0a1c1 | 0.92 | 635±15 (PSUAMS-4939) | 650–560 | |
| I12379 | Emurua Ole Polos (GvJh122) | 19 | −1.396 | 35.983 | PIA/recent | PIA cluster | M | L3h1a2a1 | E1b1b1b2b2a1; E-M293 | 3.38 | 270±15 (PSUAMS-4938) | 420–160 |
| I13763 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | Forager cline | F | .. | 0.01 | insufficient collagen | N/A | |
| I13982 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | Forager cline | F | .. | 0.02 | insufficient collagen | N/A | |
| I13983 | Gishimangeda Cave | 20 | −3.476 | 35.348 | PN | Forager cline | M | .. | BT (low cov.; prob. B) | 0.02 | insufficient collagen | N/A |
| I8904 | Kokurmatakore | 1 | 3.132 | 37.433 | PIA?8 | PN outlier? | M | L3a2a | E1b1b1; E-M35 (not E-M293) | 0.09 | insufficient collagen | N/A |
| I8930 | White Rock Point (GrJb2) | 6 | −0.450 | 34.321 | LSA | Forager cline | M | L2a4 | BT(xCT) (low cov.; prob. B) | 0.03 | insufficient collagen | N/A |
| I8931 | White Rock Point (GrJb2) | 6 | −0.450 | 34.321 | LSA | Forager cline | F | L0a2 (likely) | 0.03 | insufficient collagen | N/A |
Site codes, where available, follow a standardized system for Africa (49).
PN, Pastoral Neolithic; ELM, Elmenteitan; IA, Iron Age; PIA, Pastoral Iron Age; LSA, Later Stone Age.
Calibrated in OxCal v.4.3.2 (50), modeling for an unspecified mixture of IntCal13 (51) and SHCal13 (52) curves, and rounding to the nearest decade.
Samples are from the same individual, but provided slightly different radiocarbon dates.
The context of this individual is uncertain; see (21) for detail.
Indirect date on a bone that may be from a different individual (see Table S1). All other dates are direct on the individual for whom DNA data is reported.
Approximate location.
New attempts to date this individual failed, but a published date on bone apatite from this individual suggests a PIA association, despite genetic clustering with PN individuals; see (21) for detail.
Overview of genetic affinities of ancient eastern Africans
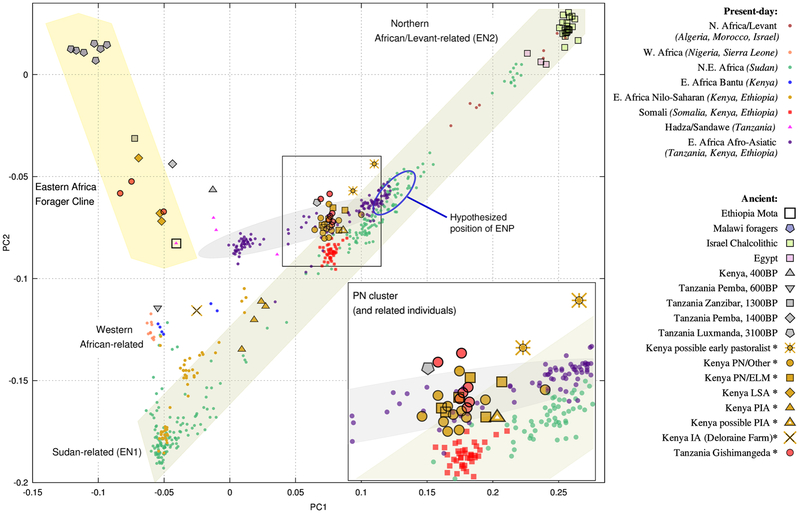
We visualized the genetic structure of the ancient individuals using principal component analysis (PCA) of the genome-wide data (Fig. 2; Table S5). We defined PCs using a small set of present-day groups (southern Africans, northeastern Africans, and non-Africans; (21)) and projected a large number of diverse individuals onto these axes. An alternative analysis with western Africans additionally used to compute the axes yielded almost identical results (Fig. S2). Present-day groups from Sudan mostly lie along a cline extending from Copts (upper right, near individuals from northern Africa and the Levant) to Nilotic speakers such as Dinka and Nuer (bottom left). Afro-Asiatic speakers (mostly from Ethiopia) form a second cline, with the right end near Sudanese Beja and Nubians, and the left end extending toward eastern African foragers (who themselves form a south-to-north gradient; cf. ref. (22)). Present-day Kenyans largely fall in the space between Sudanese, Ethiopians, and western Africans, with their language family affiliations tending to predict their broad-scale genetic affinities.
Figure 2. Principal component analysis.
Shaded regions are drawn to highlight notable clines of ancestry. Ancient individuals from this study are indicated in the legend with asterisks. Dates for published ancient individuals outside of Kenya and Tanzania are ~4500 BP for Mota, ~8100–2500 BP for Malawi foragers, ~6500–5800 BP for Israel Chalcolithic, and ~3600–2000 BP for Egypt. ELM, Elmenteitan; LSA, Later Stone Age; PN, Pastoral Neolithic; IA, Iron Age (I8802); PIA, Pastoral Iron Age; ENP, early northeastern pastoralists. See (21) for more details and Table S5 for the full list of individuals shown.
The positions of ancient eastern Africans on the PCA strongly correlate with archaeological associations. The three individuals from LSA cultural contexts all cluster with previously reported ancient foragers, falling intermediate between those from southern Ethiopia (Mota) and coastal Tanzania (Zanzibar and Pemba Islands), consistent with their geographic position (22, 24). Individuals from pastoralist contexts (including one from Luxmanda in Tanzania (22)) are highly differentiated from foragers, with the exception of three individuals uncertainly assigned a pastoral context at Gishimangeda Cave in Tanzania, who cluster with foragers. PN individuals, including Elmenteitan and those within the heterogenous SPN category (whom we refer to as “other PN”), mostly form a tight cluster near present-day Afro-Asiatic speakers, with a small number of modest outliers, including the two individuals buried at Prettejohn’s Gully, whose earlier date (~4000 BP) coincides with the initial limited spread of herding into the area. Finally, five Iron Age individuals are shifted to the left in the PCA: four PIA individuals toward Nilotic speakers, and an IA child from Deloraine Farm (I8802) – the earliest agricultural site in Kenya’s Rift Valley (32) – toward western Africans and Bantu speakers.
We also examined the uniparentally inherited loci (mtDNA and Y chromosomes) of the sampled individuals. The most striking pattern is the high frequency among the PN individuals (7–12 out of 17 males; Table S6) of the E-M293 haplogroup (E1b1b1b2b2a1), a Y chromosome lineage that has been hypothesized to be associated with the spread of pastoralism in the Horn of Africa, Kenya, and Tanzania and from there to southern Africa, on the basis of its present-day distribution and diversity (33, 34). Other males also carried haplogroups most frequently found in present-day eastern Africa, with the exception of E-M58 (E1b1a1a1a1a; predominantly western African) in the IA individual I8802, consistent with his position in PCA. The observed mtDNA lineages form more of a mosaic pattern, including types most closely associated with eastern and northeastern Africans, eastern African foragers, and northern Africans and western Eurasians (Table S7).
Formal modeling of admixture
To obtain quantitative inferences about the genetic relationships among the ancient and present-day individuals, we used qpAdm (35, 36), which provides a flexible framework for testing admixture models and estimating mixture proportions. Guided by the PCA, we began by using three groups of individuals—present-day Dinka (28), ancient Chalcolithic-period individuals from Israel (25), and the ~4500 BP forager from Mota, southern Ethiopia (24)—to represent distinct components of ancestry plausibly found in ancient and present-day eastern Africans, with present-day western Africans among the outgroups (21). We note that the use of these proxy groups in qpAdm modeling does not imply an assumption that they are directly ancestral to the true sources contributing to the individuals we analyzed. Instead, for a model to be properly formulated, the reference groups only need to be more closely related to the true sources than are the outgroups, without substantially different admixture (35). Thus, for example, ancestry related to the Chalcolithic Israel reference individuals could plausibly have originated anywhere in northeastern Africa or the Levant, and could have been present in northeastern Africa for many thousands of years. We use the Chalcolithic individuals in this study because we lack genetic data from a phylogenetically adjacent reference group from Egypt, Sudan/South Sudan, or the Horn. Additionally, qpAdm does not require any assumptions regarding the internal phylogeny relating the references and outgroups, and it provides both standard errors for mixture proportion estimates and a p-value for overall model fit quality (35).
Our qpAdm modeling reveals that the PN individuals had substantial proportions of all three ancestry components (~40% each for those represented by Dinka and by the Chalcolithic Israel individuals, and ~20% related to Mota; Fig. 3, Tables S8–S9), with no evidence of western African-related ancestry. The individuals from Prettejohn’s Gully can also be well modeled using the same three components, but in a modestly different ratio. The Iron Age group as a whole (including the more recent ~300 BP individual from Emurua Ole Polos, but excluding the possible PIA individual from Kokurmatakore) does not fit well under a three-way model, but the fit improves markedly when we exclude the Deloraine Farm individual I8802 (p = 0.009 versus p = 0.0003). The remaining four individuals (who are confidently assigned to PIA contexts) are inferred to have substantially more Sudan (Dinka)-related ancestry (~60%) than is seen in the PN. We also observe similar patterns for present-day groups falling near the ancient individuals in PCA (using data from ref. (31)), whereby the three-way model fits better for Afro-Asiatic- and Nilo-Saharan-speaking groups than for Bantu-speaking groups (Table S8). Consistent with the PCA results, Afro-Asiatic speakers are inferred (as in PN) to have relatively even proportions of the components represented by Dinka and by Chalcolithic Israel (but with varying proportions of Mota-related ancestry), while Nilo-Saharan speakers are inferred to have more Sudan-related ancestry. Alternative model formulations in which we either use ancient individuals from Taforalt in Morocco (27) in place of the Chalcolithic Israel group, or present-day Lemande from Cameroon (28) in place of Dinka (with Dinka moved to the outgroup set), fit significantly worse for most test groups (Table S8).
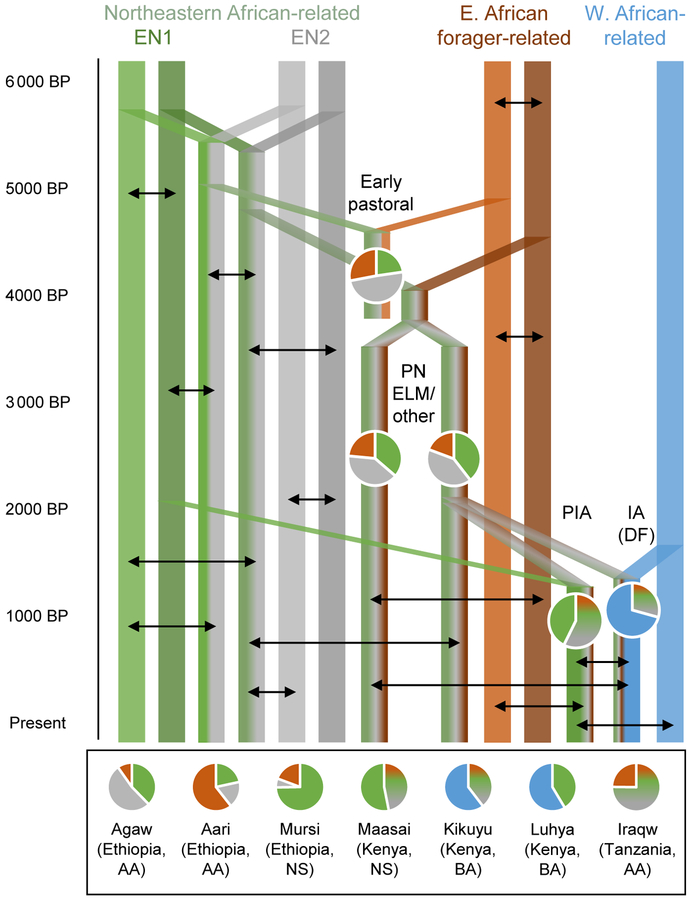
Figure 3. Proposed model of admixture for ancestry in eastern Africans.
Solid-color bars represent lineages of northeastern African (EN1/Sudan-related in green, EN2 in gray), eastern African forager-related (orange), and western African-related (blue) ancestry, and mixed-color bars represent admixed populations (hypothesized early northeastern pastoralists, or ENP, as green plus gray). Pie charts show ancestry proportions for sampled ancient (embedded in figure, at approximate date points) and present-day (bottom) populations inferred from qpAdm (PN-related ancestry as mixed-color sections). Black arrows represent likely ongoing interactions and not specific admixture events inferred from the data. EN1/EN2, early northeastern pastoralist source populations; PN, Pastoral Neolithic; ELM, Elmenteitan; PIA, Pastoral Iron Age; IA/DF, Iron Age (Deloraine Farm); AA, Afro-Asiatic; NS, Nilo-Saharan; BA, Bantu.
From these results, we formulated a four-part hypothesis to explain the origins of the ancestries in the sampled eastern African groups. First, admixture in northeastern Africa, likely associated with the spread of pastoralism, created groups (as yet unsampled with ancient DNA) with approximately equal proportions of ancestry related to (1) present-day Nilotic speakers such as Dinka and Nuer, and (2) sampled ancient and present-day groups from northern Africa and the Levant. We refer to this combination as early northeastern African pastoralist-associated ancestry (henceforth “early northeastern pastoralist,” or ENP), and the two sub-components as EN1 and EN2. Second, descendants of these groups mixed with local foragers in eastern Africa, leading to the ~20% Mota-related ancestry in the PN individuals. Third, an additional period of Sudan-related gene flow occurred before the Iron Age and contributed to PIA groups. Fourth, close to the same time, western African-related ancestry related to present-day Bantu speakers (also seen in an individual buried on Pemba Island ~600 BP (22)) appeared in the Rift Valley (notably at Deloraine Farm), in association with the spread of farming.
To test these hypotheses and gain further insight into changes in ancestry over time, we carried out a second round of analysis in qpAdm using pairs of reference groups linked more closely with each historical phase. For the initial spread of pastoralism, we used Hadendowa (Sudanese Beja (29)) plus Mota. It is likely that the genetic landscape of northeastern Africa has changed substantially since the time of the events we are modeling, so we do not propose that Hadendowa are descended directly from ENP; rather, we chose them to serve as a proxy for the (approximate) mixture of ancestries hypothesized to be present in the true ENP-related source (on the basis of the PCA and qpAdm results above). This two-way model yields a good fit for the PN individuals (p = 0.45), but not for Iron Age individuals (either the PIA cluster or the IA individual from Deloraine Farm), nor for present-day Nilotic- and Bantu-speaking groups (all p < 1e-6; Table S8). We also attempted to fit PN as a mixture of possible early Kenyan pastoralist (represented by Prettejohn’s Gully) and forager-related ancestry, but this combination was rejected (p < 1e-6 using either Mota or the three Kenyan LSA individuals to represent the forager component), suggesting that the two ancient pastoralist groups are not simply differentiated by their proportions of forager-related ancestry.
Finally, to study later transformations, we built models using PN as one proxy source and either Dinka (Sudan-related), Mota (forager-related), or Lemande (western African-related) as the other. We obtain improved fits for the Iron Age individuals and for present-day Kenyan Nilotic- and Bantu-speaking groups in this framework: the PIA cluster can be fitted as a mixture of ~57% PN-related and ~43% Sudan-related ancestry, while the Deloraine Farm individual can be modeled as a mixture of ~29% PN-related and ~71% western African-related ancestry (Fig. 3). Similar models also yield good fits for present-day Maasai (~47% PN-related and ~53% Sudan-related) and Kikuyu (~40% PN-related and ~60% western African-related), while Luhya can be fit as a mixture of Sudan-related (~41%) and western African-related (~59%) ancestry (Fig. 3).
We also investigated the fine-scale genetic structure of the PN cluster and related individuals using direct tests of asymmetry in allele frequencies (Table S10). First, in agreement with their co-localization in PCA, we do not detect any significant differences in allele-sharing between Elmenteitan and other PN individuals relative to a set of 27 comparison groups, including the Iron Age and possible early pastoralist groups from this study (max nominal Z = 2.1; see also Fig. 4). There are hints of differentiation (max Z = 2.6) between the main PN cluster and individual I8904 from Kokurmatakore (previously dated to the PIA (37)), but this individual’s ancestry is much more similar to other PN individuals than to PIA (Fig. 2, Table S10). We also find only minor asymmetry between the primary Kenyan and Tanzanian PN clusters (max Z = 2.5). However, four PN-period individuals who appear as outliers on PCA do have statistically significant ancestry differences as compared to the PN cluster. In particular, two individuals from Gishimangeda Cave (I13972 and I13978) and the previously reported ~3100 BP pastoralist individual from Luxmanda in Tanzania (22) have evidence of more or different forager-related ancestry relative to Sudan-related ancestry (e.g., f4(Ancient South African foragers, Dinka; X, PN) > 0, Z = 3.2, 5.1, and 6.8, respectively; Fig. 4, Table S8, Table S10), while individual I8759 (an early PN individual buried at Naishi Rockshelter in Kenya) has evidence of less forager-related ancestry (e.g., f4(Ancient South African foragers, Europeans; I8759, PN) < 0, Z = −4.4). We also confirm the differences in ancestry between the PN cluster and the two possible early pastoralist individuals from Prettejohn’s Gully (both Z > 5). While the individuals from Prettejohn’s Gully fall relatively far apart on PCA (Fig. 2), their ancestry is only weakly differentiated via f-statistics (max Z = 2.2; Table S10).
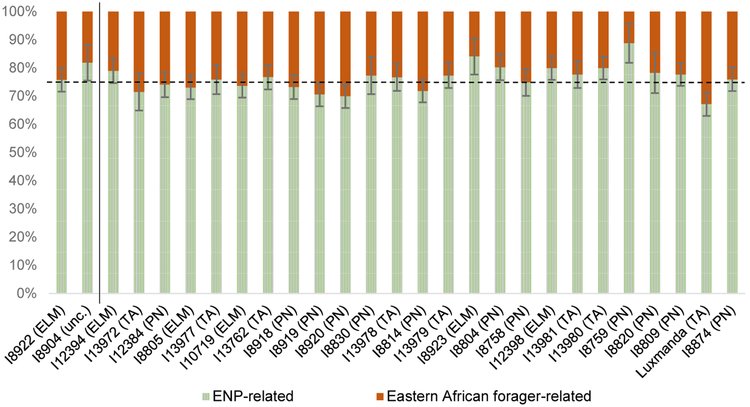
Figure 4. Mixture proportions for PN individuals.
Results are from a two-component qpAdm model using Sudanese Beja (green-and-gray striped) and the ancient individual from Mota, Ethiopia (orange) as proxy sources (for early northeastern pastoralist, or ENP, and eastern African forager-related ancestry, respectively). Radiocarbon-dated individuals (to the right of the solid line) are ordered from most ancient on the right (I8874, 3350–3180 BP) to most recent on the left (I12394, 1530–1400 BP). Bars show two standard errors in each direction. The dotted line represents the Kenya PN group-level estimate (74.7 ± 1.0% ENP-related ancestry). We note that the linear regression coefficient for forager-related ancestry as a function of date is not significantly nonzero (R2 = 0.03, p = 0.39), nor as a function of latitude (R2 = 0.03, p = 0.37). ELM, Elmenteitan; PN, other Kenyan Pastoral Neolithic; TA, Tanzanian PN (including the Luxmanda individual from ref. (22)); unc., uncertain.
Dates of admixture
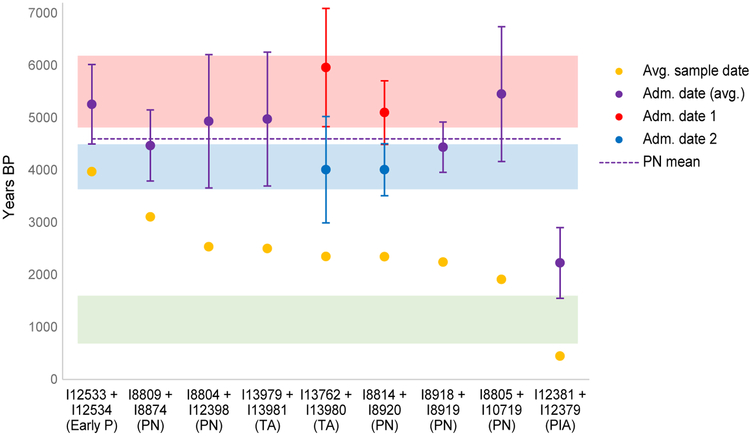
The fact that we observe tight clustering of PN individuals via PCA and qpAdm, with little if any spatial or temporal structure as revealed by direct dating (Fig. 4, Table S9), suggests that the admixture responsible for forager-related ancestry in the PN had largely ceased before the lifetimes of our sampled individuals. To test this hypothesis, we used MALDER (38, 39) to estimate dates of admixture for pairs of high-coverage individuals with similar direct radiocarbon dates and locations (21). All pairs have inferred dates that point to quite distant average times of admixture (mean ~4600 BP for PN and ~5300 BP for Prettejohn’s Gully; Fig. 5, Table S10), with the concordance among the PN estimates providing an independent line of evidence for a lack of substantial ongoing mixture. We infer a more recent average date (~2200 BP) for two late PIA individuals, likely associated with additional Sudan-related ancestry (Table S11). Our power to detect multiple waves of admixture is limited with ancient data, but for one pair of PN individuals from Naivasha Burial Site, we are confidently able to identify two separate events, the first at ~5100 BP and the second at ~4000 BP. We also infer two waves for a pair of individuals from Gishimangeda Cave, dating to ~6000 BP and ~4000 BP. In light of our qpAdm results, and given the associated MALDER amplitudes (Table S11), these multiple dates plausibly represent estimates of the times of (1) the formation of admixed ENP ancestry, and (2) admixture in eastern Africa between local foragers and descendants of the first mixture, leading to the three-component ancestry of PN individuals. In this context, the single (and intermediate) estimated dates for other PN pairs can be interpreted as averages of these two processes (Fig. 5).
Figure 5. Dates of admixture inferred for pairs of ancient individuals.
Bars show two standard errors in each direction. The shaded areas represent implied periods of admixture: from top to bottom, ENP (early northeastern pastoralist; red), forager-related (blue), and additional Sudan-related (green). Early P, early pastoralists; PN, Pastoral Neolithic; PIA, Pastoral Iron Age; TA, Tanzanian PN. See also Table S11.
Incorporating genetic evidence into models for the spread of food production
The four-phase model emerging from our genetic and radiocarbon dating results builds upon archaeological reconstructions for the spread of herding into eastern Africa, supporting some theories while rejecting others that until now have been considered plausible. Under a proposed ‘moving frontier’ model, herders entering new environments would interact in diverse ways with indigenous foragers, resulting in varying cultural responses and blurred archaeological boundaries as groups adopted some of each other’s cultural practices. In the ensuing ‘static frontier,’ more intensive herding and/or competition would transform initial relationships into more stable, long-term patterns (40). Archaeologists interpret the construction of cemeteries by the first herders in the Turkana Basin and the apparent trickle of people with similar material culture into the south-central Rift Valley as part of a moving frontier, whereas the explosion of pastoralist cultures in the PN reflects more established, static herder-forager relationships (6, 15).
On the basis of genetic data from a wide sampling of PN individuals, we infer two phases of admixture associated with the spread of pastoralism: the first likely ~6000–5000 BP in northeastern Africa, and the second ~4000 BP between this admixed ENP group and eastern African foragers. Archaeological data show that the Nile Valley was important for herders seeking reliable water sources toward the end of the African Humid Period (~7000–6000 BP) (1, 41). Herders plausibly traced the White Nile southward, following unknown trajectories through South Sudan and/or southern Ethiopia to arrive in the Turkana Basin ~5000 BP (5, 6). Alternatively, they may have moved via the Horn of Africa, but current evidence for herding in that region postdates that of Turkana (3). Our results support archaeological hypotheses that no matter the routes they took, early herders interacted with local foragers as they spread (16, 42). In eastern Africa, extensive forager-herder interactions have been proposed both in the Turkana Basin and during the initial trickle of herding into the south-central Rift Valley (6–10, 16, 17). Either area, or another unsampled region, could have witnessed the admixture we document between descendants of the (already admixed) ENP group and local foragers, giving rise to the groups who then developed the PN cultural traditions of southern Kenya and northern Tanzania.
Our attempts to extract DNA from early herders in Turkana were unsuccessful (21), so the genetic ancestr(ies) of the first eastern African pastoralist groups remain uncertain. However, some lineages may be reflected in a man and a woman buried at Prettejohn’s Gully ~4000 BP. There are few associated artifacts, but the individuals’ genetic profiles suggest they may represent an initial limited dispersal of herders into the south-central Rift Valley that did not leave large numbers of descendants. Previously, evidence of herding prior to ~3300 BP was limited to Turkana-related Nderit pottery found sporadically and usually undated in the area (8), and to sheep or goat remains associated with a date of ~4200 BP at a site 33 km south of Prettejohn’s Gully (7). Genetically, the two individuals are most similar to those from PN sites, but they fall outside the range of sampled PN (and present-day) variation and cannot be modeled as directly related to PN. They also have an older date of admixture, and the male individual (I12533) carries a Y chromosome haplogroup (E2; E-M75) not found in any of our sampled PN individuals. Our results thus imply at least two chronologically distinct movements of herders through eastern Africa, consistent with archaeological evidence of complex spreads (2), while at the same time adding new information by showing that one group (the one that gave rise to the PN cluster) was eventually much more demographically successful than the others.
While Prettejohn’s Gully may represent a limited trickle of herders into the south-central Rift Valley, numerous PN sites after ~3300 BP attest to successful specialized pastoralism. Archaeologists attribute this florescence to herder innovations that allowed them to overcome environmental and disease barriers, likely facilitated by strong social networks reflected in widespread material cultural traditions (8–11). The dense cluster of PN individuals in our PCA – including burials >450 km apart – suggests these networks formed among people with shared ancestry, with the close relatedness perhaps reinforced by ongoing mobility and gene flow. Moreover, it is striking that individuals in our sample buried with distinctive Elmenteitan material culture display minimal genetic differentiation from those of other PN burials. Strong Elmenteitan material cultural traditions may reflect maintenance of social boundaries, but our results do not support the view that these people were genetically distinct (12, 18). In comparison to present-day groups, all PN individuals (associated with both the SPN and Elmenteitan material cultures) show the greatest genetic affinity to Afro-Asiatic speakers, supporting the hypothesis that the initial large-scale expansion of pastoralism in eastern Africa was linked to the spread of Afro-Asiatic languages (18, 19).
With regard to the ‘moving frontier’ model, we find that while sampled PN individuals carry ~20% admixture from local forager groups, this gene flow almost all occurred well before the core PN era, as herders entered new environments. By contrast, the rapid spread of pastoralists into Kenya and Tanzania after ~3300 BP involved minimal gene flow between herders and foragers, plausibly due to the formation of a static frontier along which social barriers prevented large-scale gene flow, despite possible social and economic interaction (8, 15). Alternatively, population densities of foragers may have been so low that gene flow between the groups had little demographic impact on the more numerous pastoralists (12). Static frontiers were not absolute, however, in agreement with ethnographic and ethnohistoric records that testify to some intermarriage between foragers and food producers (e.g., 43, 44)). Today, for example, the Eyasi Basin is an important interaction zone for diverse foraging and food producing groups (44), and is home to speakers of each of the four main African language phyla. In our data, the ancestries of the individual buried at Luxmanda (22), the southernmost known PN site (11), and of two newly reported individuals from Gishimangeda Cave in the Eyasi Basin, all attest to additional admixture with foragers beyond the events contributing to the possible early pastoralists from Prettejohn’s Gully and to the main PN cluster. Furthermore, at Gishimangeda Cave, we observe three individuals clustering genetically with foragers, which may reflect social ties between people with different ancestry and/or ways of life. However, given that the three forager-related individuals produced insufficient collagen for radiocarbon dating and substantially lower-coverage genetic data than the pastoralist-related individuals, we speculate that the observation of distinct ancestry types is more probably a consequence of multiple burial phases at the site (i.e., greater antiquity for the likely forager individuals).
Low frequency of genetic adaptation to milk consumption
To test whether the success of PN groups in eastern Africa was aided by genetic adaptations linked to diet, we also evaluated the sequenced individuals for presence or absence of genetic variants associated with adult lactase persistence (LP; Table S12). While our coverage is limited for some individuals and some SNPs, we only observe one instance of an LP-conferring mutation, in individual I13762, from Gishimangeda Cave in Tanzania. This individual, who falls within the main PN genetic cluster and lived during the later PN (2150–2020 cal BP), carried the derived allele at the rs145946881 (G/C-14010) SNP, which is the most common LP mutation found among eastern African groups today. The other ancient individuals could possibly have carried different variants conferring the same phenotype, but the assayed SNPs are found at high frequencies in some present-day eastern African groups and thus are likely to have been important historically (45). This suggests that eastern African pastoralists were mostly lactose intolerant as recently as 3000–1000 years ago and that the LP alleles only recently rose in frequency, although our results also demonstrate that the G/C-14010 mutation was present and could have been a target for natural selection by the PN period. Direct evidence for dairying is currently lacking in the region, despite the specialized pastoralist lifestyle inferred from faunal remains at PN sites (8). However, culinary innovations such as fermentation could have enabled dairy consumption even in the absence of LP.
Increasing complexity in the Iron Age
The eastern African Iron Age can be summarized archaeologically as a mosaic in which foragers, herders, and early farmers with distinct traditions and ways of life overlapped in space and time (2, 14, 15, 19). This complexity is reflected in the ancient individuals we analyzed. The young boy buried at Deloraine Farm – the site with the earliest direct evidence of farming in the Rift Valley (32) – shows affinity to western Africans and speakers of Bantu languages (both genome-wide and on the Y chromosome). This is the earliest documentation of western African-related ancestry in eastern Africa, in a region where today such ancestry is widespread and the majority of people speak Bantu languages (46).
The Deloraine Farm child’s genetic distinctiveness as compared to the PN cluster is notable in light of similarities in artifacts between Elmenteitan sites and Deloraine Farm, viewed as evidence of continuity from the Elmenteitan to the Iron Age (32, 47). By contrast, four PIA individuals spanning an ~800-year period show greater affinity to present-day Nilotic speakers and are associated with an influx of Sudan (Dinka)-related ancestry. Similarities between archaeologically and ethnographically documented material culture suggest that PIA sites may be associated with ancestors of present-day Kenyan Nilotic speakers such as the Kalenjin or Maasai (32, 47). Both the PIA individuals and present-day Maasai retain substantial components of PN-related ancestry, showing that the ancestry composition of PIA and more recent pastoralists reflects mixture with previously established herder groups in eastern Africa. For other groups, such as Luhya (who speak a Bantu language), there is little evidence of PN-related ancestry, suggesting that their ancestors spread into Kenya without mixing substantially with local herders. Boundaries between foragers and food producers may have been maintained during the Iron Age, as we do not observe a significant increase in forager ancestry in the PIA or IA individuals, but we cannot rule out a small proportion of additional forager-associated admixture. Overall, we caution that these interpretations are limited by small sample sizes and may not reflect the more nuanced regional dynamics within this mosaic.
Genetic diversity of eastern African foragers
Archaeological evidence of foragers across Holocene eastern Africa encompasses an array of material culture and subsistence traditions (48). This study adds to our understanding of LSA genetic variation by reporting ancient DNA from additional foragers without pastoralist-related admixture, including from fisher-foragers near Lake Victoria who may have been living contemporaneously with PN herders in the broader region. These individuals fall in an intermediate position between Ethiopian and Tanzanian foragers on a genetic cline that is well correlated (among sampled ancient individuals) with geographical location (22). Broadly, however, the similarity of foragers buried in the Victoria and Eyasi Basins to individuals living on the Kenya coast and in Ethiopia and coastal Tanzania (22, 24) suggests that shared forager ancestry extended widely across the region, as also attested by present-day genetic data (20).
Conclusions
Genome-wide data from 41 ancient eastern Africans show that archaeological complexity during the spreads of herding and farming is also reflected in genetic patterns, which indicate multiple movements of and gene flow among ancestrally distinct groups of people. We identify three components of ancestry harbored by ancient pastoralists and propose a sequence of admixture events to explain our observations; future archaeological and ancient DNA research in the Turkana Basin, the Horn of Africa, and other parts of northeastern Africa will be necessary to confirm the earliest stages of the spread of herding into the region. At the other end of our timeframe, we show that multiple admixture events impacted Iron Age groups associated with heterogeneous economic, cultural, and linguistic patterns. This complexity can be further explored through additional comparisons of genetic and archaeological diversity. Ancient DNA offers a new source of information about eastern African Holocene prehistory, and an important next direction is to integrate this information rigorously with insights provided by the longer-established disciplines of archaeology and linguistics.
Materials and Methods
Human skeletal remains from East African archaeological sites, including the petrous portion of the skull, teeth, and other bones, were sampled from the National Museums of Kenya and Tanzania and the Livingstone Museum in Zambia, following protocols to minimize both destruction and contamination. Bioarchaeological data on the analyzed individuals, along with detailed information about archaeological context, are provided in the full Materials and Methods (21). DNA was extracted from bone powder in dedicated clean rooms at Harvard Medical School using protocols optimized for ancient DNA. Illumina sequencing libraries were prepared with uracil-DNA-glycosylase (UDG) to reduce deamination-induced errors. Before sequencing, libraries were enriched for molecules overlapping approximately 1.2 million genome-wide SNPs. Of the 77 samples processed for this study, 43 (from 41 distinct individuals) provided genome-wide ancient DNA data. Direct radiocarbon dates were generated at the Pennsylvania State University (PSU) Radiocarbon Laboratory via accelerator mass spectrometry (AMS).
Raw sequencing data were filtered and aligned to the human reference genome. One sequence per individual was chosen randomly from those overlapping each targeted SNP to represent that individual at that position. On the basis of authenticity metrics, two individuals were excluded from genome-wide analyses. The other 39 individuals were analyzed in conjunction with published genetic data from ancient individuals and present-day groups, using a variety of statistical approaches. Multiple population genetics methods were applied to investigate proportions, sources, and dates of admixture, with a particular emphasis on testing of proposed admixture models through the qpAdm software.
Supplementary Material
Acknowledgments
We thank S. Mallick for consultation on data analysis and M. Meyer for sharing optimized oligonucleotide sequences for single-stranded library preparation. We thank R. Pinhasi for advising on techniques for minimally invasive sample preparation.
Funding: M.E.P. started this research as a fellow of the Radcliffe Institute for Advanced Study at Harvard University. D.R. was supported by U.S. National Institutes of Health grant GM100233, by an Allen Discovery Center grant, and by a grant from the John Templeton Foundation, and is an investigator of the Howard Hughes Medical Institute. Radiocarbon work was supported by the NSF Archaeometry program (grant BCS-1460369) to D.J.K. and B.J.C.
Footnotes
Competing interests: Authors declare no competing interests.
Data and materials availability: The aligned sequences are available via the European Nucleotide Archive under accession number PRJEB31373. The genotype datasets used for analyses are included as supplementary materials (Data S1). Skeletal samples were exported and repatriated under materials transfer agreements with the curating institutions; see Materials and Methods for details on research permissions.
Publisher's Disclaimer: This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS
References and Notes
- 1.Marshall F, Hildebrand E, Cattle before crops: the beginnings of food production in Africa. Journal of World Prehistory 16, 99–143 (2002). [Google Scholar]
- 2.Lane P, in Pastoralism in Africa: Past, present and future, Bollig M, Schnegg M, Wotzka H-P, Eds. (Berghahn, Oxford, 2013), pp. 105–143. [Google Scholar]
- 3.Lesur J, Hildebrand EA, Abawa G, Gutherz X, The advent of herding in the Horn of Africa: New data from Ethiopia, Djibouti and Somaliland. Quaternary International 343, 148–158 (2014). [Google Scholar]
- 4.Barthelme J, Fisher-Hunters and Neolithic Pastoralists in East Turkana, Kenya. (B.A.R. International Series, Oxford, 1985), vol. 254. [Google Scholar]
- 5.Hildebrand EA et al. , A monumental cemetery built by eastern Africa’s first herders near Lake Turkana, Kenya. Proceedings of the National Academy of Sciences 115, 8942–8947 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sawchuk EA, Goldstein ST, Grillo KM, Hildebrand EA, Cemeteries on a moving frontier: mortuary practices and the spread of pastoralism from the Sahara into eastern Africa. Journal of Anthropological Archaeology 51, 187–205 (2018). [Google Scholar]
- 7.Ambrose SH, Chronology of the Later Stone Age and Food Production in East Africa. Journal of Archaeological Science 25, 377–392 (1998). [Google Scholar]
- 8.Gifford-Gonzalez D, Early pastoralists in East Africa: ecological and social dimensions. Journal of Anthropological Archaeology 17, 166–200 (1998). [Google Scholar]
- 9.Gifford-Gonzalez D, “Animal disease challenges” fifteen years later: the hypothesis in light of new data. Quaternary International 436, 283–293 (2017). [Google Scholar]
- 10.Marshall F, Grillo KM, Arco L, in Sustainable Lifeways: Cultural Persistence in an Ever-Changing Environment,, Miller N, Moore, Ryan, Eds. (University of Pennsylvania Museum of Archaeology and Anthropology, Philadelphia, 2011), pp. 38–73. [Google Scholar]
- 11.Grillo KM et al. , Pastoral Neolithic settlement at Luxmanda, Tanzania. Journal of Field Archaeology 43, 102–120 (2018). [Google Scholar]
- 12.Ambrose SH, in From Hunters to Farmers: The Causes and Consequences of Food Production in Africa, Clark JD, Brandt SA, Eds. (University of California Press, Berkeley, CA, 1984), pp. 212–239. [Google Scholar]
- 13.Ambrose SH, in Encyclopedia of Prehistory. Volume 1: Africa, Peregrine PN, Ember M, Eds. (Kluwer Academic Publishers, New York, 2001), pp. 97–109. [Google Scholar]
- 14.Crowther A, Prendergast ME, Fuller DQ, Boivin N, Subsistence mosaics, forager-farmer interactions, and the transition to food production in eastern Africa. Quaternary International 489, 101–120 (2018). [Google Scholar]
- 15.Lane P, The ‘moving frontier’ and the transition to food production in Kenya. Azania 39, 243–264 (2004). [Google Scholar]
- 16.Bower J, The Pastoral Neolithic of East Africa. Journal of World Prehistory 5, 49–82 (1991). [Google Scholar]
- 17.Marean C, Hunter to herder: Large mammal remains from the hunter-gatherer occupation at Enkapune ya Muto rockshelter. African Archaeological Review 10, 65–127 (1992). [Google Scholar]
- 18.Ehret C, in From Hunters to Farmers, Clark JD, Brandt SA, Eds. (University of California Press, Berkeley, 1984), pp. 26–35. [Google Scholar]
- 19.Ambrose SH, in The Archaeological and Linguistic Reconstruction of African History, Ehret C, Posnansky M, Eds. (University of California Press, Berkeley, 1982), pp. 104–157. [Google Scholar]
- 20.Scheinfeldt LB et al. , Genomic evidence for shared common ancestry of East African hunting-gathering populations and insights into local adaptation. Proc Natl Acad Sci U S A, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Materials and methods are available as supplementary materials.
- 22.Skoglund P et al. , Reconstructing prehistoric African population structure. Cell 171, 59–71.e21 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schlebusch CM et al. , Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358, 652–655 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Gallego Llorente M et al. , Ancient Ethiopian genome reveals extensive Eurasian admixture in Eastern Africa. Science 350, 820 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Harney É et al. , Ancient DNA from Chalcolithic Israel reveals the role of population mixture in cultural transformation. Nature Communications 9, 3336 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuenemann VJ et al. , Ancient Egyptian mummy genomes suggest an increase of Sub-Saharan African ancestry in post-Roman periods. Nat Commun 8, 15694 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Loosdrecht M et al. , Pleistocene North African genomes link Near Eastern and sub-Saharan African human populations. Science 360, 548–552 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Mallick S et al. , The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538, 201 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hollfelder N et al. , Northeast African genomic variation shaped by the continuity of indigenous groups and Eurasian migrations. PLOS Genetics 13, e1006976 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pagani L et al. , Ethiopian genetic diversity reveals linguistic stratification and complex influences on the Ethiopian gene pool. The American Journal of Human Genetics 91, 83–96 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fan S et al. , African evolutionary history inferred from whole genome sequence data of 44 indigenous African populations. Genome Biology 20, 82 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambrose SH, Collett D, Marshall F, Excavations at Deloraine, Rongai, 1978. Azania 19, 79–104 (1984). [Google Scholar]
- 33.Trombetta B et al. , Phylogeographic refinement and large scale genotyping of human Y chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent. Genome Biology and Evolution 7, 1940–1950 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Henn BM et al. , Y-chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. Proceedings of the National Academy of Sciences 105, 10693 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haak W et al. , Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522, 207–211 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazaridis I et al. , Genetic origins of the Minoans and Mycenaeans. Nature 548, 214 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stiles D, Munro-Hay SC, Stone cairn burials at Kokurmatakore, northern Kenya. Azania 16, 151–166 (1981). [Google Scholar]
- 38.Loh P-R et al. , Inferring admixture histories of human populations using linkage disequilibrium. Genetics 193, 1233 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pickrell JK et al. , Ancient west Eurasian ancestry in southern and eastern Africa. Proceedings of the National Academy of Sciences 111, 2632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alexander J, in Early Frontiers in Southern Africa, Hall M, Avery, Avery DM, Williams ML, Humphreys AJB, Eds. (B.A.R. International Series, Oxford, 1984), vol. 207, pp. 12–23. [Google Scholar]
- 41.Haaland R, Haaland G, in Oxford Handbook of African Archaeology, Mitchell P, Lane P, Eds. (Oxford University Press, Oxford, 2013), pp. 541–553. [Google Scholar]
- 42.Hassan F, in The Origins and Development of African Livestock: Archaeology, Genetics, Linguistics, Ethnography, Blench R, MacDonald K, Eds. (UCL Press, London, 2000), pp. 61–86. [Google Scholar]
- 43.Cronk L, From Mukogodo to Maasai: Ethnicity and Cultural Change In Kenya. (Westview, Boulder, 2004). [Google Scholar]
- 44.Marlowe F, The Hadza : Hunter-Gatherers of Tanzania. (Berkeley, 2010). [Google Scholar]
- 45.Ranciaro A et al. , Genetic Origins of Lactase Persistence and the Spread of Pastoralism in Africa. The American Journal of Human Genetics 94, 496–510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tishkoff SA et al. , The genetic structure and history of Africans and African Americans. Science 324, 1035–1044 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sutton JEG, Deloraine: further excavations and the Iron Age sequence of the central Rift. Azania 17, 103–125 (1993). [Google Scholar]
- 48.Kusimba SB, in The Oxford Handbook of African Archaeology, Mitchell P, Lane P, Eds. (Oxford University Press, Oxford, 2013). [Google Scholar]
- 49.Nelson CM, A standardized site enumeration system for the continent of Africa. Nyame Akuma 40, 62–67 (1993). [Google Scholar]
- 50.Bronk Ramsey C, Bayesian analysis of radiocarbon dates. Radiocarbon 51, 337–360 (2009). [Google Scholar]
- 51.Reimer PJ et al. , INTCAL13 and MARINE13 radiocarbon age calibration curves 0–50,000 years cal BP. Radiocarbon 55, 1869–1887 (2013). [Google Scholar]
- 52.Hogg A et al. , SHCal13 Southern Hemisphere calibration, 0–50,000 years cal BP. Radiocarbon 55, 1889–1903 (2013). [Google Scholar]
- 53.Prendergast ME, Sawchuk E, Boots on the ground in Africa’s ancient DNA ‘revolution’: archaeological perspectives on ethics and best practices. Antiquity 92, 1–13 (2018). [Google Scholar]
- 54.Marsh EJ et al. , IntCal, SHCal, or a mixed curve? Choosing a 14C calibration curve for archaeological and paleoenvironmental records from tropical South America. Radiocarbon 60, 925–940 (2018). [Google Scholar]
- 55.Schepartz L, From Hunters to Herders: Subsistence Pattern and Morphological Change in Eastern Africa, PhD, University of Michigan, (1987). [Google Scholar]
- 56.Ambrose SH, DeNiro MJ, Reconstruction of African human diet using bone collagen carbon and nitrogen isotope ratios. Nature 319, 321–324 (1986). [Google Scholar]
- 57.Ambrose SH, Stable carbon and nitrogen isotope analysis of human and animal diet in Africa. Journal of Human Evolution 15, 707–731 (1986). [Google Scholar]
- 58.Cohen M, Deloraine Farm: a new type of pottery. Azania 7, 161–167 (1972). [Google Scholar]
- 59.Chittick N, Excavations at Deloraine, Rongai. Azania 14, 162–163 (1979). [Google Scholar]
- 60.Collett D, Robertshaw P, Pottery traditions of early pastoral communities in Kenya. Azania 18, 107–125 (1983). [Google Scholar]
- 61.Leakey MD, Leakey LSB, Excavations at the Njoro River Cave Stone Age Cremated Burials in Kenya Colony. Leakey LSB, Ed., (Oxford, 1950). [Google Scholar]
- 62.Faugust PM, Sutton JEG, The Egerton Cave on the Njoro River. Azania 1, 149–153 (1966). [Google Scholar]
- 63.Cohen M, A Reassessment of the stone bowl cultures of the Rift Valley, Kenya. Azania: Archaeological Research in Africa 5, 27–38 (1970). [Google Scholar]
- 64.Ikeda J, Hayama S , The Hadza and the Iraqw in northern Tanzania: dermatographical, anthropometrical, odontometrical and osteological approaches. African Study Monographs 2, 1–26 (1982). [Google Scholar]
- 65.Mehlman MJ, Later Quaternary Archaeological Sequences in Northern Tanzania, PhD, University of Illinois at Urbana-Champaign, Urbana-Champaign, IL: (1989). [Google Scholar]
- 66.Sawchuk E, Social Change and Human Population Movements – Dental Morphology in Holocene Eastern Africa, PhD, University of Toronto, (2017). [Google Scholar]
- 67.Brown J, The excavation of a group of burial mounds at Ilkek near Gilgil, Kenya. Azania 1, 59–77 (1966). [Google Scholar]
- 68.Bower J, Nelson CM, Waibel AF, Wandibba S, The University of Massachusetts’ Later Stone Age/Pastoral Neolithic comparative study in Central Kenya: an overview. Azania 12, 119–146 (1977). [Google Scholar]
- 69.Bower J, Nelson CM, Early pottery and pastoral cultures of the Central Rift Valley, Kenya. Man 13, 554–566 (1978). [Google Scholar]
- 70.Leakey MD, Leakey LSB, Game PM, Goodwin AJH. Report on the Excavations at Hyrax Hill, Nakuru, Kenya Colony, 1937–38. Transactions of the Royal Society of South Africa 30, 271–409 (1943). [Google Scholar]
- 71.Gabel C, Six rockshelters on the Northern Kavirondo shore of Lake Victoria. African Historical Studies II, 205–254 (1969). [Google Scholar]
- 72.Frahm E, Goldstein ST, Tryon CA, Late Holocene forager-fisher and pastoralist interactions along the Lake Victoria shores, Kenya: perspectives from portable XRF of obsidian artifacts. Journal of Archaeological Science: Reports 11, 717–742 (2017). [Google Scholar]
- 73.Lane P, Ashley C, Oteyo G, New dates for Kansyore and Urewe Wares from Northern Nyanza, Kenya. Azania 41, 123–138 (2006). [Google Scholar]
- 74.Prendergast ME, Lane PJ, Middle Holocene fishing strategies in East Africa: zooarchaeological analysis of Pundo, a Kansyore shell midden in northern Nyanza, Kenya. International Journal of Osteoarchaeology 20, 88–112 (2010). [Google Scholar]
- 75.Merrick HV, An annotated bibliography of Kenyan prehistoric archaeology, 1896–1981. African Archaeological Review 1, 143–177 (1983). [Google Scholar]
- 76.Siiriäinen A, Later Stone Age investigation in the Laikipia Highlands, Kenya: a preliminary report. Azania 12, 162–186 (1977). [Google Scholar]
- 77.Goldstein ST, Shaffer CM, Experimental and archaeological investigations of backed microlith function among Mid-to-Late Holocene herders in southwestern Kenya. Archaeological and Anthropological Sciences, 1–22 (2016). [Google Scholar]
- 78.Collett D, Robertshaw P, Problems in the interpretation of radiocarbon dates: the pastoral Neolithic of East Africa. African Archaeological Review 1, 57–74 (1983). [Google Scholar]
- 79.Ryan K, Tracking East African cattle herders from prehistory to the present. Expedition 51, http://www.penn.museum/sites/expedition/?p=11700 (2009). [Google Scholar]
- 80.Lane P, in Conserving Wildlife in African Landscapes: Kenya’s Ewaso Ecosystem, Georgiadis N, Moss J, Malleret-King D, Eds. (Smithsonian Institution Scholarly Press, Washington, 2011). [Google Scholar]
- 81.Kitson E, A study of the Negro skull with special reference to the crania from Kenya Colony. Biometrika 23, 271–314 (1931). [Google Scholar]
- 82.Leakey LSB, The Stone Age Races of Kenya. (Oxford University Press, London, 1935). [Google Scholar]
- 83.Clark JD, “An Archaeologist at Work in African Prehistory and Early Human Studies,” an oral history conducted in 2000–2001 by Suzanne Riess, Regional Oral History Office, The Bancroft Library,” (University of California, Berkeley, Berkeley, 2002). [Google Scholar]
- 84.Merrick HV, Monaghan MC, The date of the cremated burials in Njoro River Cave. Azania 19, 7–11 (1984). [Google Scholar]
- 85.Barnes J, The Ghosts of Happy Valley: Searching for the Lost World of Africa’s Infamous Aristocrats (Aurum Press, London, 2014). [Google Scholar]
- 86.McCall GJH, “Geology of the Nakuru-Thomson’s Falls-Lake Hannington area. Report No. 78.,” (Geological Survey of Kenya, 1967). [Google Scholar]
- 87.Leakey LSB, The Stone Age Cultures of Kenya Colony. (Cambridge University Press, London, 1931). [Google Scholar]
- 88.Butzer KW, Isaac GL, Richardson JL, Washbourn-Kamau C, Radiocarbon dating of East African lake levels. Science 175, 1069–1076 (1972). [DOI] [PubMed] [Google Scholar]
- 89.Isaac G, Merrick H, Nelson CM, in Palaeoecology of Africa, The Surrounding Islands and Antartica, Volume VI , van Zinderen Bakker EM, Ed. (Balkema, Cape Town, 1972), pp. 225–232. [Google Scholar]
- 90.Washbourn-Kamau C, Late Quaternary lakes in the Nakuru-Elmenteita Basin, Kenya. The Geographical Journal 137, 522–535 (1971). [Google Scholar]
- 91.Wandibba S, Excavations at Rigo Cave in the Central Rift Valley, Kenya. Azania 18, 81–92 (1983). [Google Scholar]
- 92.Robertshaw P, Collett D, Gifford D, Mbae NB, Shell middens on the Shores of Lake Victoria. Azania 18, 1–43 (1983). [Google Scholar]
- 93.Dale D, Ashley CZ, Holocene hunter-fisher-gatherer communities: new perspectives on Kansyore using communities of western Kenya. Azania: Archaeological Research in Africa 45, 24–48 (2010). [Google Scholar]
- 94.Prendergast M, Kansyore fisher-foragers and transitions to food production in East Africa: the view from Wadh Lang’o, Nyanza Province. Azania: Archaeological Research in Africa 45, 83–111 (2010). [Google Scholar]
- 95.Kennett DJ, Archaeogenomic evidence reveals prehistoric matrilineal dynasty. Nature Communications 8, 14115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.McClure SB, Puchol OG, Culleton BJ, AMS dating of human bone from Cova De La Pastora: New evidence of ritual continuity in the prehistory of eastern Spain. Radiocarbon 52, 25–32 (2010). [Google Scholar]
- 97.Lohse JC, Culleton BJ, Black SL, Kennett DJ, A precise chronology of middle to late Holocene bison exploitation in the far southern Great Plains. Journal of Texas Archeology and History 1, 94–126 (2014). [Google Scholar]
- 98.van Klinken GJ, Bone collagen quality indicators for palaeodietary and eadiocarbon measurements. Journal of Archaeological Science 26, 687–695 (1999). [Google Scholar]
- 99.Santos GM, Southon JR, Druffel-Rodriguez KC, Griffin S, Mazon M, Magnesium Perchlorate as an Alternative Water Trap in AMS Graphite Sample Preparation: A Report On Sample Preparation at Kccams at the University of California, Irvine. Radiocarbon 46, 165–173 (2004). [Google Scholar]
- 100.Stuiver M, Polach HA, Discussion reporting of 14C Data. Radiocarbon 19, 355–363 (1977). [Google Scholar]
- 101.Wright DK, Accuracy vs. precision: understanding potential errors from radiocarbon dating on African landscapes. African Archaeological Review 34, 303–319 (2017). [Google Scholar]
- 102.Harding L, History of modern man unravels as German scholar is exposed as fraud. The Guardian, 19 February (2005). [Google Scholar]
- 103.Protsch R, The chronological position of Gamble’s Cave II and Bromhead’s Site (Elmenteita) of the Rift Valley, Kenya. Journal of Human Evolution 7, 101–109 (1978). [Google Scholar]
- 104.Protsch R, The Naivasha hominid and its confirmed late Upper Pleistocene age. Anthropologischer Anzeiger 35, 97–102 (1976). [PubMed] [Google Scholar]
- 105.Protsch R, The absolute dating of Upper Pleistocene subSaharan fossil hominids and their place in human evolution. Journal of Human Evolution 4, 297–322 (1975). [Google Scholar]
- 106.Sirak KA et al. , A minimally-invasive method for sampling human petrous bones from the cranial base for ancient DNA analysis. BioTechniques 62, 283–289 (2017). [DOI] [PubMed] [Google Scholar]
- 107.Dabney J et al. , Complete mitochondrial genome sequence of a Middle Pleistocene cave bear reconstructed from ultrashort DNA fragment. Proceedings of the National Academy of Sciences of the United States of America 110, 15758–15763 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Korlevic P et al. , Reducing microbial and human contamination in DNA extractions from ancient bones and teeth. BioTechniques 59, 87–93 (2015). [DOI] [PubMed] [Google Scholar]
- 109.Rohland N, Glocke I, Aximu-Petri A, Meyer M, Extraction of highly degraded DNA from ancient bones, teeth and sediments for high-throughput sequencing. Nature protocols 13, 2447–2461 (2018). [DOI] [PubMed] [Google Scholar]
- 110.Rohland N, Harney E, Mallick S, Nordenfelt S, Reich D, Partial uracil-DNA-glycosylase treatment for screening of ancient DNA. Philos Trans R Soc Lond B Biol Sci 370, 20130624 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.DeAngelis MM, Wang DG, Hawkins TL, Solid-phase reversible immobilization for the isolation of PCR products. Nucleic Acids Res 23, 4742–4743 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rohland N, Reich D, Cost-effective, high-throughput DNA sequencing libraries for multiplexed target capture. Genome research 22, 939–946 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gansauge MT et al. , Single-stranded DNA library preparation from highly degraded DNA using T4 DNA ligase. Nucleic Acids Res 45, e79 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Maricic T, Whitten M, Paabo S, Multiplexed DNA sequence capture of mitochondrial genomes using PCR products. PLoS One 5, e14004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fu Q et al. , DNA analysis of an early modern human from Tianyuan Cave, China. Proceedings of the National Academy of Sciences 110, 2223–2227 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Fu Q et al. , An early modern human from Romania with a recent Neanderthal ancestor. Nature 524, 216–219 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mathieson I et al. , Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528, 499–503 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Li H, Durbin R, Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Behar DM et al. , A “Copernican” reassessment of the human mitochondrial DNA tree from its root. American journal of human genetics 90, 675–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Skoglund P et al. , Separating endogenous ancient DNA from modern day contamination in a Siberian Neandertal. Proceedings of the National Academy of Sciences of the United States of America 111, 2229–2234 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fu Q et al. , A revised timescale for human evolution based on ancient mitochondrial genomes. Current biology : CB 23, 553–559 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Korneliussen TS, Albrechtsen A, Nielsen R, ANGSD: Analysis of next generation sequencing data. BMC bioinformatics 15, 356–356 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Li H, A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics (Oxford, England) 27, 2987–2993 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Weissensteiner H et al. , HaploGrep 2: mitochondrial haplogroup classification in the era of high-throughput sequencing. Nucleic acids research 44, W58–W63 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.van Oven M, Kayser M, Updated comprehensive phylogenetic tree of global human mitochondrial DNA variation. Human mutation 30, E386–394 (2009). [DOI] [PubMed] [Google Scholar]
- 126.Skoglund P, Storå J, Götherström A, Jakobsson M, Accurate sex identification of ancient human remains using DNA shotgun sequencing. Journal of Archaeological Science 40, 4477–4482 (2013). [Google Scholar]
- 127.Patterson N et al. , Ancient admixture in human history. Genetics 192, 1065 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patterson N, Price AL, Reich D, Population structure and eigenanalysis. PLoS Genet 2, e190 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Liu LT, Dobriban E, Singer A, ePCA: High dimensional exponential family PCA. arXiv:1611.05550, (2016). [Google Scholar]
- 130.Lazaridis I et al. , Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513, 409–413 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fu Q et al. , Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514, 445–449 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lipson M et al. , Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551, 368 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.