Abstract
Insect odorant-binding proteins (OBPs) are a large, diverse group of low-molecular weight proteins secreted into the fluid bathing olfactory and gustatory neuron dendrites. The best-characterized OBP, LUSH (OBP76a) enhances pheromone sensitivity enabling detection of physiological levels of the male-specific pheromone, 11-cis vaccenyl acetate. The role of the other OBPs encoded in the Drosophila genome is largely unknown. Here, using clustered regularly interspaced short palindromic repeats/Cas9, we generated and characterized the loss-of-function phenotype for two genes encoding homologous OBPs, OS-E (OBP83b) and OS-F (OBP83a). Instead of activation defects, these extracellular proteins are required for normal deactivation of odorant responses to a subset of odorants. Remarkably, odorants detected by the same odorant receptor are differentially affected by the loss of the OBPs, revealing an odorant-specific role in deactivation kinetics. In stark contrast to lush mutants, the OS-E/F mutants have normal activation kinetics to the affected odorants, even at low stimulus concentrations, suggesting that these OBPs are not competing for these ligands with the odorant receptors. We also show that OS-E and OS-F are functionally redundant as either is sufficient to revert the mutant phenotype in transgenic rescue experiments. These findings expand our understanding of the roles of OBPs to include the deactivation of odorant responses.
Keywords: olfaction, olfactory, perireceptor
INSECT odorant-binding proteins (OBPs) are abundant proteins secreted into the sensillum lymph that bathes the chemosensory neuron dendrites (Vogt et al. 1991; Leal 2013). These low-molecular weight proteins are synthesized and secreted by nonneuronal chemosensory support cells [reviewed in Ha and Smith (2009), Ronderos and Smith (2009), and Smith (2012)]. The function of this family of proteins has been a subject of speculation since their discovery as extracellular proteins that interact directly with pheromone molecules in moths (Vogt and Riddiford 1981). Postulated roles include the transport of hydrophobic pheromone molecules to the odorant receptors (Kaissling et al. 1985; Vogt et al. 1985; Du and Prestwich 1995; Krieger and Breer 1999; Wojtasek and Leal 1999; Sandler et al. 2000; Horst et al. 2001; Vogt 2003), clearance of odorant from the sensillum lymph (Steinbrecht and Müller 1971; Vogt and Riddiford 1981; Ziegelberger 1995), protection of odorants from degrading enzymes in the lymph (Kaissling 1996, 1998, 2001), acting as filters or buffers (Pelosi and Maida 1990; Larter et al. 2016), and functioning as components of the ligand that actives the neuronal receptors (Pophof 2002; Xu et al. 2005; Laughlin et al. 2008). In Drosophila, 52 OBPs have been identified in the genome (Galindo and Smith 2001; Graham and Davies 2002; Hekmat-Scafe et al. 2002). These proteins are expressed in chemosensory sensilla in both olfactory and gustatory sensilla (Galindo and Smith 2001; Jeong et al. 2013). The function of most of the OBPs is unknown.
The best-studied Drosophila OBP is LUSH (OBP76a), a 14-kDa protein important for olfactory sensitivity to the male-specific pheromone, 11-cis vaccenyl acetate (cVA) (Xu et al. 2005; Laughlin et al. 2008). lush mutants are insensitive to physiological levels of cVA pheromone, revealing a role for this OBP in sensitizing neurons to pheromones (Xu et al. 2005; Laughlin et al. 2008). X-ray crystal structures of LUSH bound to cVA revealed that cVA is encapsulated by LUSH, consistent with its postulated role in the transport of the hydrophobic pheromone though the sensillum lymph to the receptors on the neuronal cilia (Laughlin et al. 2008). While transport of cVA pheromone molecules through the lymph is likely one important component of LUSH function, genetic analysis revealed that LUSH has a more intimate role in the activity of the pheromone-sensing neurons. lush mutants have a striking and unexplained 400-fold reduction in spontaneous activity (spikes in the absence of pheromone), specifically in the Or67d pheromone-sensing neurons. This defect is inconsistent with an exclusive role in pheromone transport, as this phenotype is manifested in the absence of pheromones (Xu et al. 2005). Introduction of recombinant LUSH into the lush mutant sensillum lymph restores the spontaneous rate and normal pheromone sensitivity of Or67d neurons (Xu et al. 2005). This suggests that LUSH may have a direct role in these events, perhaps as a coligand at Or67d receptors (Xu et al. 2005; Laughlin et al. 2008). Similar defects in pheromone sensitivity have recently been reported for OBP mutants in other insects (Dong et al. 2017; Ye et al. 2017; Zhang et al. 2017). However, the role for OBPs is not limited to sensitization and other functions have been reported, including antagonizing chemosensory receptor function (Jeong et al. 2013; Swarup et al. 2014).
OS-E and OS-F (OBP83b and OBP83a) were identified as abundant, antenna-enriched transcripts in subtraction hybridization experiments (McKenna et al. 1994; Pikielny et al. 1994). The genes encoding these OBPs are coexpressed in a subset of sensilla exclusively in the Drosophila antenna (Hekmat-Scafe et al. 1997; Shanbhag et al. 2001; Larter et al. 2016). However, the biological functions of OS-E and OS-F have not been previously elucidated. We set out to generate mutants in OS-E and OS-F to evaluate the effects that loss of these proteins might confer on the function of the neurons present in sensilla that express these OBPs. Our results reveal an unexpected phenotype associated with the loss of these OBPs that implicates them in the deactivation of odorant responses.
Materials and Methods
Drosophila stocks
Wild-type flies were an isogenized w1118 strain (BS3605; Bloomington Drosophila Stock Center). OS-E/F mutants were backcrossed for five generations to this wild-type stock to minimize genetic background effects. Nos>Cas9 flies were generated by Kondo and Ueda (2013). Hsp70>Cre flies were obtained from the Bloomington Drosophila Stock Center (BS34516) and were used to delete the Lox-3xP3>RFP-Lox marker from the deletion mutants. Or47b mutants are described in Wang et al. (2011), Or88a and Or65abc mutants are described in Pitts et al. (2016), and the Or83c mutants (Or83cMB11142) are described in Ronderos et al. (2014). The lush mutants (lush1) are described in Kim et al. (1998). Snmp1 mutants (Snmp1Z0429) are described in Jin et al. (2008). Flies of both sexes were used in these experiments.
Generation and validation of OS-E/F mutants
Clustered regularly interspaced short palindromic repeats (CRISPR) targets were identified upstream and downstream of the OS-E and OS-F genes using the CRISPR Optimal Target Finder (Gratz et al. 2014). Overlapping oligonucleotides were annealed for each target site and cloned into the pU6-Bbs1-chiRNA plasmid, as described in Gratz et al. (2014). Approximately 1 kb of sequence upstream and downstream of the cleavage targets were cloned using PCR, and inserted into pHD-DsRed-attP (Addgene). The DNAs were diluted to final concentrations of 20 ng/µl for the U6 DNAs and 250 µg/µl for the targeting DNA in injection buffer (1 mM NaPO4 pH 6.8 and 50 mM KCl), injected into Nos>Cas9 embryos (Kondo and Ueda 2013), and the resulting flies were crossed to Balancer Chromosome stocks (TM6b). The balanced progeny were screened for red fluorescent protein (RFP) expression in the eye. Independent mutant lines were established from three lines, and all were homozygous viable and fertile. The phenotypes reported here were confirmed in independent lines. OS-E/F mutant stocks in which the 3xP3>RFP was excised using Cre recombinase were also used in some experiments. No differences in phenotype were observed whether 3xP3>RFP was present or not.
Primers used for mutant generation and validation
CRISPR upstream target oligonucleotides:
5′ CTTCGGCCCTTTTATGAGATTACT 3′
5′ AAACAGTAATCTCATAAAAGGGCC 3′
CRISPR downstream target oligonucleotides:
5′ CTTCGTCAAGAGTTGTTTGCGCCG 3′
5′ AAACCGGCGCAAACAACTCTTGAC 3′
Upstream homology domain primers:
5′ GCATGCCTGGTGCAGTTGCTGTTGCATCGG 3′
5′ GCGGCCGCATTACTGGGGCTCCATTTC 3′
Downstream homology domain primers:
5′ ACTAGTGCGCCGTGGCAAAAACTTGTATAAAAAC 3′
5′ CTCGAGTAAATTTAAAAATCTTTGACTTTAATTCG 3′
Validation primers:
Primers to validate upstream integration:
5′ AATTATATTGCCCCATCCCC 3′
5′ CGATGAACTTCACCTTGTAG 3′
Primers to validate downstream integration:
5′ CGCGACTCTAGATCATAATC 3′
5′ CCTTCCAGGGAATAAAGTAC 3′
Primers specific to OS-E and OS-F genes:
Primers for OS-E:
5′ GGACAGATTTGGTAAGTAGC 3′
5′ GAGCCCCAGTAATCTCATAA 3′
Primers for OS-F:
5′ TGGCTTTGAATGGCTTTGG 3′
5′ ATTGTCGTCCACCACTTCG 3′
Quantitative RT-PCR
RNA was extracted from 5- to 10-day-old Drosophila antennae. A PicoPure RNA Isolation Kit (Applied Biosystems, Foster City, CA) was used for RNA extraction. From each genotype, 50 antennae were dissected and collected in 50 µl of extraction buffer. The antennae were homogenized using Bead Ruptor4 (Omni International) and precipitated in ethanol. DNA contamination was removed with RNase-free DNase (QIAGEN, Valencia, CA). First-strand complementary DNA synthesis was performed using First-Strand Synthesis SuperMix for qRT-PCR (QPCR) (Invitrogen, Carlsbad, CA). QPCR was performed in an Applied Biosystems 7500 Real-Time PCR Systems with Fast SYBR Green Master Mix (Applied Biosystems). Three replicates were performed together with a no-reverse transcriptase control and a no-template control. Melting curve analysis and primer efficiency tests were performed for all primer sets.
Primers for QPCR
OS-E:
5′ GCTCCCAAAACTGGCGTTAC 3′
5′ GAGAAGGTCTTGAACGCCATT 3′
OS-F:
5′ CTTTGGTCGGCGTGTCAG 3′
5′ CCAAGCCCTTCCACGACG 3′
5′ GCGTGGGTTTGTGATCAGTT 3′
5′ GATCTTCTCCTTGCCCATCC 3′
The EF1 primers are as described in Ponton et al. (2011).
Genomic rescue
A 10.6-kb DNA fragment (containing the OS-E and OS-F genes, and all noncoding DNA extending to the next identified locus) was isolated by high-fidelity PCR from wild-type DNA. No other known transcription units are encoded by this fragment and the correct coding sequences for these two genes were confirmed as lacking PCR errors by DNA sequencing. The fragment was cloned into pCasper4 (Pirrotta 1988) and used to generate transgenic flies as previously described (Spradling and Rubin 1982). For single-gene rescue, deletions were produced in the rescuing transgene using a Q5 site-directed mutagenesis kit (New England Biolabs, Beverly, MA).
Genomic rescue primers
5′ CTCGAGAAGCTGGCAACTGAATCCGA 3′
5′ GCGGCCGCTTCGAGTTCCAGTTGCAGTT 3′
Q5 OS-E-coding deletion oligonucleotides
5′ TTTGAAACTACAATGAATGG 3′
5′ AATTTATTTACATTTATATTAACATTTAATTG 3′
Q5 OS-F-coding deletion oligonucleotides
5′ TTTAATGTGGCTCTTTCCGTTTC 3′
5′ ACACCTGGGCCACCTTTC 3′
Myc-tagged OS-E
We added a BglII site into the genomic rescue construct one codon after the predicted signal cleavage site (Almagro Armenteros et al. 2019) in OS-E using the Q5 system (New England Biolabs). The 2× myc tag linker EQKLISEEDLEQKLISEEDL(GGS)8 was inserted in frame by annealing and ligating four overlapping oligonucleotides into the BglII site, and sequencing several clones to identify inserts in the proper orientation.
Q5 primers to introduce BglII site
5′ AGATCTCTGGGCAGCGGCACAGCC 3′
5′ GAACCAAGGCGCGATGGAGAGG 3′
2× Myc tag primers
5′ GATCTCAGGAACAAAAACTCATCTCAGAAGAGGATCTGGAACAAAAACTCATCTCA 3′
5′ AGATCCTCTTCTGAGATGAGTTTTTGTTCCAGATCCTCTTCTGAGATGAGTTTTTGTTCCTGA 3′
(GlyGlySer)8 linker
5′ GAAGAGGATCTGGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCGGCGGCAGCA 3′
5′ GATCTGCTGCCGCCGTGCCGCCGCTGCCGCCGTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCCGCTGCCGCCC 3′
Single-sensillum recordings
Single-sensillum recordings were performed on 3–6-day-old flies as previously described (Xu et al. 2005; Laughlin et al. 2008; Pitts et al. 2016). Briefly, a single fly (3–6-days old) was fixed under a humidified charcoal-filtered air stream. A reference electrode was placed in the eye and a recording electrode was placed into an individual sensillum. Odorant samples were diluted in water or paraffin oil, and 30 μl was spotted onto a 1 cm2 Wattman paper and inserted into a pipet over which the stimulus air pulse was passed in a constant air stream 1.0 cm from the fly using a computer-controlled trigger. Figures denoting dilution % represent the dilution on the paper, not the actual stimulus concentration at preparation, which was much less. Odorants were applied for 300 msec for all odorants, except for four ligands that were applied for 1 sec.
Spontaneous activity was calculated as the number of spikes per second occurring in a 10-sec period prior to odorant presentation divided by 10. Elicited activity (Δ spikes/s) was calculated as the number of spikes occurring in the 1 sec following odorant exposure, from which spontaneous activity for the 1 sec prior to odorant application was subtracted. In all cases, a given odorant was only tested once on a single fly, though multiple odorants were tested on single flies.
The deactivation time constant, τ, was calculated by binning individual traces into 50-msec intervals. The bin with the largest number of spikes was counted as time point 0 and subsequent bins were used to plot the exponential decay curve from which τ was derived. τ was calculated using the formula N(t)=Noe−t/τ, where N(t) is the quantity of spikes at time t and τ is the time at which the population of spikes is reduced to 1/e times the initial value. Latency to activation was determined by identifying the time point after odor presentation at which a cluster of spikes was observed, measured in milliseconds after the initiation of odor presentation. This latency represents the time from activation of the valve initiating the flow of odorant to the preparation to the arrival of the odorant molecules at the receptors, producing spikes. cVA was obtained from Pherobank (Wijk bij Duurstede, The Netherlands). The general odorants used in this study were obtained from Sigma ([Sigma Chemical], St. Louis, MO) and were of the highest purity available.
Immunocytochemistry
Immunocytochemistry was performed on 10 μm Drosophila frozen head tissue sections from male and female flies, as previously described (Jin et al. 2008). A Myc monoclonal antibody (Developmental Studies Hybridoma Bank, Iowa City, IA) was diluted to 1:1000 for immunofluorescence and was detected with goat anti-mouse Alexa 555 antibody (Molecular Probes, Eugene, OR). Confocal images were obtained using a Zeiss ([Carl Zeiss], Thornwood, NY) LSM 510 confocal microscope. A C-terminal Myc-tagged Acinus line was used as a positive control (Nandi et al. 2017).
Statistical analysis
Wild-type and mutant genotypes were compared using two-tailed Student’s t-tests. Multiple genotype values were compared using one-way ANOVA with Tukey’s post hoc test to correct for multiple comparisons. Analysis was performed using GraphPad Prism 7 and Origin 8.5 (OriginLab).
Data availability
Fly strains and plasmids are available upon request. Supplemental Figures include Supplemental Material, Figures S1–S4. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9773615.
Results
Generation of an OS-E/F null mutant
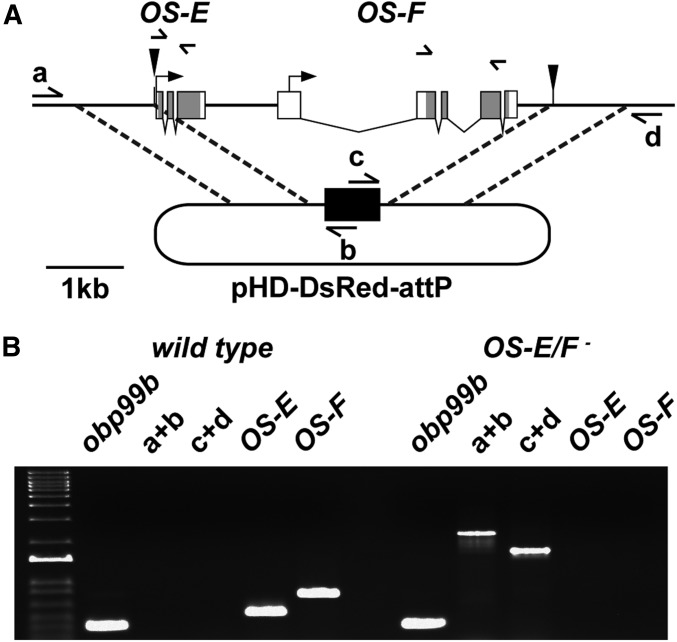
OS-E and OS-F are closely related, and located <1 kb apart on the third chromosome, suggesting that they arose from a gene duplication event (Hekmat-Scafe et al. 1997, 2000). Due to the amino acid sequence similarity and coexpression within the same sensilla (Shanbhag et al. 2001), OS-E and OS-F could have redundant functions. Therefore, to evaluate potential roles in olfaction, we decided to excise both OS-E and OS-F genes using CRISPR/Cas9-mediated homologous recombination to probe for phenotypes associated with the loss of these gene products (see Materials and Methods). Briefly, unique Cas9 cleavage sites were identified at either end of the OS-E/F coding region (Gratz et al. 2014), and used to replace OS-E and OS-F with an eye-specific promoter driving the RFP gene (3xP3>RFP) (Figure 1A). Correct integration of the RFP gene was confirmed using a set of PCR primers with one primer external to the region of homology and one primer within the RFP construct (Figure 1B). We further validated deletion of the OS-E and OS-F genes in the mutants using gene-specific primers (Figure 1, A and B), and by showing there are no transcripts using QPCR (Figure S1). The OS-E/F mutants were backcrossed for five generations to a control genetic background (w1118) to minimize differences in genetic background, resulting in the control (wild-type) and mutant (OS-E/F−) lines that were compared using odor-evoked electrophysiology.
Figure 1.
Generation of OS-E/F null mutants. (A) Map of the OS-E/F genomic region on the right arm of the third chromosome. CRISPR-mediated replacement of the OS-E and OS-F genes with 3xP3>dsRed (black rectangle) is depicted. Solid triangles indicate the position of CRISPR/Cas9 cleavage sites. The dashed lines denote the regions of homology upstream and downstream of the OBP genes that were cloned into the donor vector (see Materials and Methods for details). Labeled arrows (a–d) indicate the positions and orientations of primers for PCR reactions used to identify correct integration of the DsRed gene (black rectangle) into the OS-E/F locus. Unlabeled arrows indicate gene-specific primers used to determine the presence of the OS-E and OS-F genes. (B) Agarose gel image of PCR fragments generated with the primers depicted in (A). PCR fragment sizes from controls and OS-E/F mutants confirm correct integration of the DsRed gene, and loss of the OS-E and OS-F genes in the mutant. Markers in left lane are a 1-kb ladder (Thermo Fisher Scientific). CRISPR, clustered regularly interspaced short palindromic repeats; OBP, odorant-binding protein.
OS-E/F mutants are defective for spontaneous activity and odorant sensitivity to a subset of odorants
OS-E and OS-F proteins are highly expressed, and are secreted into the sensillum lymph of three classes of intermediate and two classes of trichoid sensillae (Shanbhag et al. 2001, 2005). Recent reports have indicated that these OBPs are also expressed in a subset of basiconic sensilla (Larter et al. 2016); however, we observed that the expression of these OBPs was relatively low at these sites (Figure S2), and we detected no differences in odorant-induced responses or spontaneous activity in these neurons in the absence of OS-E and OS-F (data not shown), so they were not considered further.
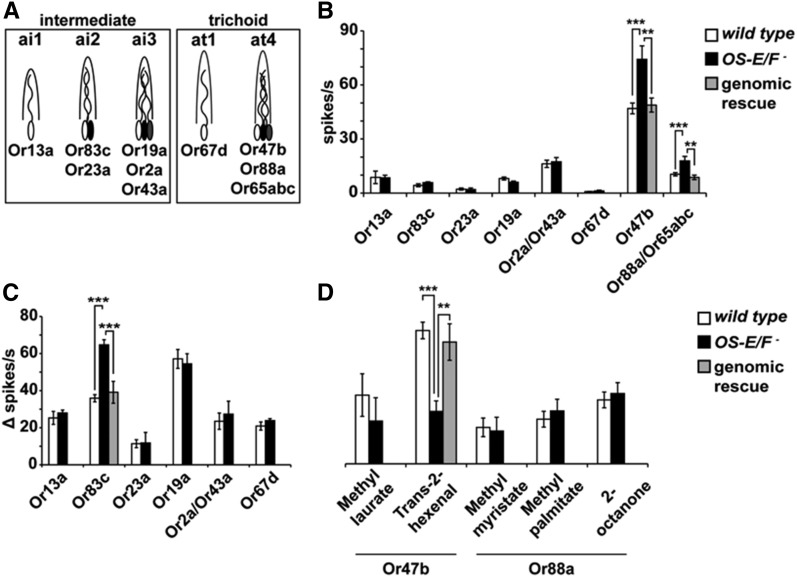
The five classes of trichoid and intermediate sensillae contain the dendrites of a total of 10 classes of olfactory neurons, based on odorant receptor expression (Couto et al. 2005) (Figure 2A). Because lush mutants have a 400-fold reduction in spontaneous activity (Xu et al. 2005), we first characterized the spontaneous activity rates for each of the 10 olfactory neuron classes from wild-type and OS-E/F mutant flies normally exposed to high levels of these OBPs. Figure 2B shows that we observed no reductions in spontaneous activity in the OS-E/F mutants; however, we did see a significant increase in spontaneous activity in both the large- and small-spiking neurons from four sensilla in the OS-E/F mutants. The increased spontaneous activity phenotypes were rescued by a genomic transgene containing both OS-E and OS-F genes, demonstrating that these defects result from loss of OS-E and/or OS-F (Figure 2B).
Figure 2.
Spontaneous and evoked activity from wild-type and OS-E/F mutants. (A) Cartoon of two trichoid and three intermediate sensilla classes normally expressing OS-E and OS-F, depicting the neurons expressing the characteristic odorant receptors defining each neuron class. (B) Average spontaneous activity of individual trichoid and intermediate neurons of the indicated genotypes in the absence of odorants. Or2a and Or43a, as well as Or88a and Or65abc, neuron responses were combined due to similarities in spike amplitudes. Or88a and Or65abc spontaneous rates were determined in the Or47b mutant (Wang et al. 2011). Or83c wild-type = 4.34 ± 0.8 spikes/s; Or83c OS-E/F mutant = 8.36 + 1.93 spikes/s (P = 1.4 × 10−7). Or88a/Or65abc wild-type = 10.46 ± 0.97 spikes/s; Or88a/Or65abc OS-E/F mutant = 20.31 ± 2.47 spikes/s (P = 0.0003). Or47b wild-type = 46.97 ± 3.00 spikes/s; Or47b OS-E/F mutant = 74.25 ± 7.42 spikes/s (P = 0.00029). n = 10–20 for each genotype. (C) Odor-induced responses of trichoid and intermediate neurons to the best-known activating ligands for each neuron type (n = 5–22) (Hallem et al. 2004; Galizia and Sachse 2010; Dweck et al. 2013; Ronderos et al. 2014; Pitts et al. 2016). Or83c wild-type Δ spikes/s = 35.9 ± 1.94; Or83c OS-E/F mutant Δ spikes/s = 64.65 ± 2.8 (P = 1.15 × 10−7). Or47b wild-type Δ spikes/s = 72.38 ± 4.48; Or47b OS-E/F mutant Δ spikes/s = 28.45 ± 5.73 (P = 3.21 × 10−6). Odorants and dilutions used to test each neuron were as follows: Or13a, 10% 1-octen-3-ol; Or83c, 1% farnesol; Or23a, 10% cyclohexanone; Or19a, 1% limonene, Or2a/Or43a, 10% benzaldehyde; Or67d, 1% cVA; and Or47b, 10% trans-2-hexenal. Error bars indicate SEM. All P-values determined by one-way ANOVA with post hoc Tukey’s test. (D) Responses of at4 neurons to known activating ligands (Dweck et al. 2015; Pitts et al. 2016). All odorants were used at a 10% dilution on the filter paper (see Materials and Methods for details). Or47b response to trans-2-hexenal: wild-type Δ spikes/s = 72.38 ± 4.48, OS-E/F mutant Δ spikes/s = 28.45 ± 5.73, and genomic rescue Δ spikes/s = 66.1 ± 9.91. ANOVA P-values: wild-type to OS-E/F mutant = 3.21 × 10−6, wild-type to genomic rescue = 0.51, and OS-E/F mutant to genomic rescue = 0.0033. n = 10–22. Error bars indicate SEM. All P-values determined by one-way ANOVA with post hoc Tukey’s test. cVA, 11-cis vaccenyl acetate.
We next examined odorant responses from these 10 neuron classes in wild-type and OS-E/F mutants using previously identified activating ligands for each neuron (Hallem et al. 2004; Galizia and Sachse 2010; Dweck et al. 2013; Ronderos et al. 2014; Pitts et al. 2016). While the odorant responses from most neurons were unaffected by the loss of OS-E and OS-F, we did detect a significant increase in spiking rate in the mutant Or83c neurons in response to farnesol (Figure 2C) and an apparent reduced response from Or47b olfactory sensory neurons in the mutants in response to trans-2-hexenal (Figure 2D). These defects were reverted by introducing a transgenic copy of a wild-type genomic transgene. While differences in OS-E/F mutant Or47b neuron responses to trans-2-hexenal were observed, there were no differences in the responses to methyl laurate, a previously reported pheromone component detected by this neuron (Dweck et al. 2015). We did test additional Or88a ligands including methyl palmitate and methyl myristate, but there were no significant differences observed in responses between wild-type and OS-E/F mutants (Figure 2D).
Examination of the traces from OS-E/F mutant Or83c and Or47b neurons revealed that the altered responses do not actually reflect altered sensitivity, but instead stem from a delay in neuronal deactivation that affects the Δ spikes per second measurement because of the 1-sec bin used for this calculation (Figure 3). Thus, while Δ spikes calculations were different, this defect actually reflects a striking defect in the deactivation kinetics.
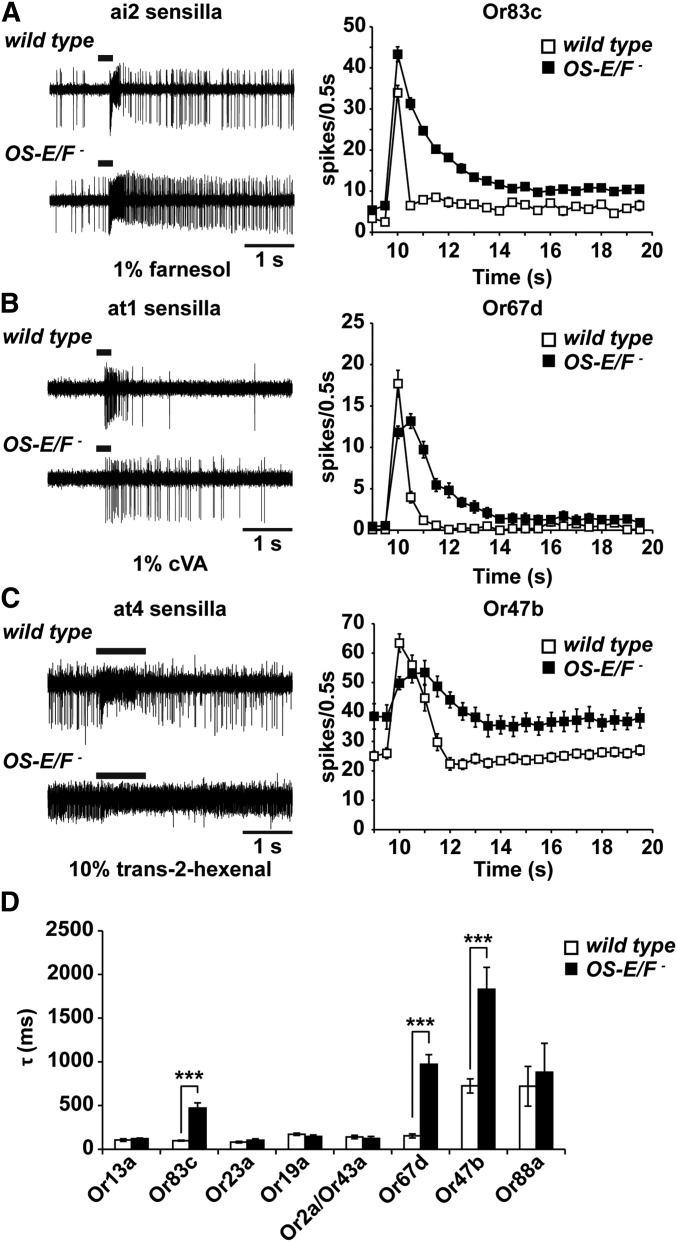
Figure 3.
Deactivation kinetics are abnormal in OS-E/F mutants to a subset of odorants. (A) Comparison mutant aI2a neuronal responses to farnesol. (B) Comparison of wild-type and OS-E/F mutant Or67d neurons to cVA. C) Comparison of wild-type and OS-E/F mutant at4 neuronal responses to trans-2-hexenal. In (A–C), representative traces are shown on the left, time courses of activation and deactivation as measured by binning spikes in 0.5-sec bins are shown on the right. Note delayed return to baseline activity in the mutants. (D) Time constant τ of deactivation of all tested olfactory neurons. Or83c wild-type = 99.2 ± 4.6 msec; Or83c OS-E/F mutant = 472.1 ± 58.2 msec (P = 5.14 × 10−6). Or67d wild-type = 153.7 ± 22.4 msec; Or67d OS-E/F mutant = 971.7 ± 112.5 msec (P = 1.69 × 10−6). Or47b wild-type = 725.69 ± 81 msec; Or47b OS-E/F mutant = 1830 ± 250.8 msec (P = 6.95 × 10−6). n = 10–20. Error bars indicate SEM. All P-values determined by two-tailed Student’s t-test. cVA, 11-cis vaccenyl acetate.
We reexamined the odor-induced electrophysiological responses from the 10 potentially affected olfactory neurons in wild-type and OS-E/F mutants for deactivation defects by calculating the deactivation time constant τ for the best-known activating ligand for each neuron (Figure 3D). This revealed that Or83c, Or47b, and Or67d neurons stimulated with farnesol, trans-2-hexenal, and cVA, respectively, had poststimulus spiking activity that persisted for much longer in the OS-E/F mutants than for controls (Figure 3, A–C). To quantify the deactivation kinetics, we binned individual traces into 50-msec intervals, and quantified the number of spikes in each bin and calculated the τ values. Wild-type Or83c neurons stimulated with 1% farnesol had a τ value of 99.2 ± 4.5 msec while the OS-E/F mutant had a τ of 472.1 ± 58 msec, almost five times slower (Figure 3). We also noted delayed deactivation kinetics in cVA responses in OS-E/F mutants stimulated with cVA compared to wild-type controls. The deactivation time constant for wild-type Or67d neurons was 160.7 ± 17.2 msec and for OS-E/F mutants the time constant was 1249.6 ± 246.5 msec, a sevenfold slowing in deactivation kinetics. Finally, Or47b neurons exposed to trans-2-hexenal had altered deactivation kinetics as well. For wild-type flies, τ was 725.7 ± 81 msec and for the OS-E/F mutants it was 1830 ± 102 msec, a 2.5-fold delay for the mutants (P = 6.95 × 10−7). The deactivation defects were reverted to wild-type upon introduction of a rescuing genomic transgene into the mutant background (Figure 2 and Figure 7). We did not observe abnormal deactivation kinetics in the other seven classes of neurons activated with their best-known ligands (Figure 3). This could reflect a lack of requirement for OS-E/F or perhaps different activating ligands that are currently unknown may require these OBPs for normal deactivation. We conclude that a subset of odorant responses depend upon OS-E and/or OS-F for normal deactivation kinetics.
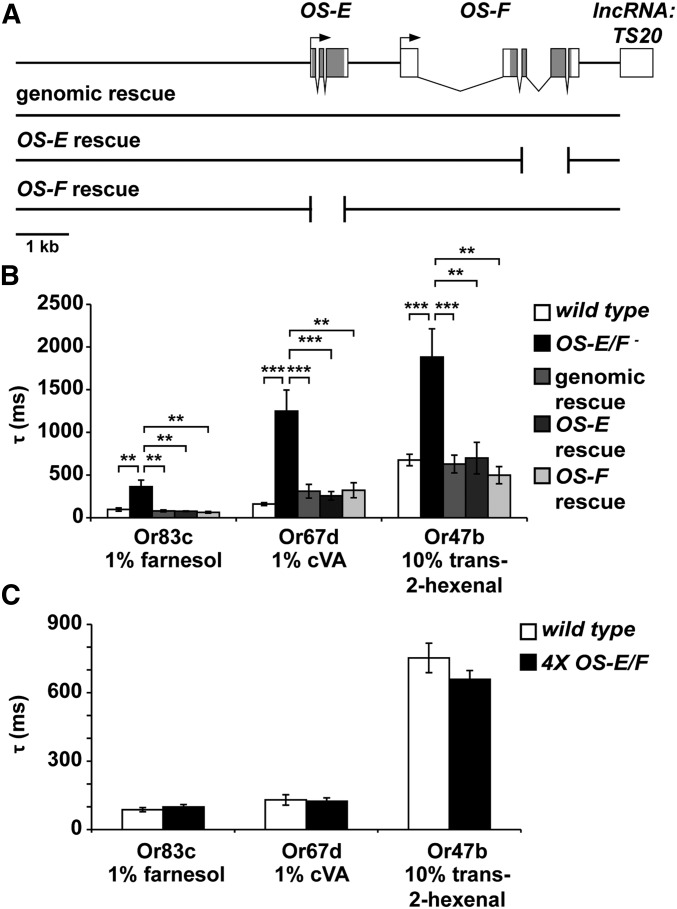
Figure 7.
OS-E and OS-F are functionally redundant for rapid neuronal deactivation. (A) Map of the OS-E/F gene region with black rectangles depicting coding regions. Arrows indicate transcription start sites. The lines beneath the map depict the DNA deletions that were used to evaluate the function of individual OBP genes. Breaks in the lines showing individual rescues show where the coding regions of either OS-E or OS-F were excised from the genomic rescue construct (see Materials and Methods for details). (B) Time constants of deactivation for Or83c, Or67d, and Or47b neurons with the genotypes indicated. Delayed deactivation present in the OS-E/F mutants is reversed by all three forms of the rescuing transgene. Deactivation time constants for Or83c neurons to farnesol (left side of graph): wild-type = 96.36 ± 19.24 msec, OS-E/F mutant = 375.98 ± 83.94 msec, genomic rescue = 75.34 ± 12.31 msec, OS-E rescue = 76.54 ± 4.15 msec, and OS-F rescue = 63.79 ± 9.48 msec. ANOVA P-values: wild-type to OS-E/F mutant = 0.0013, OS-E/F mutant to genomic rescue = 0.001, OS-E/F mutant to OS-E rescue = 0.0013, and OS-E/F mutant to OS-F rescue = 0.003, all nonsignificant P-values = 0.99. For Or67d responses to cVA (center part of graph): wild-type = 160.73 ± 17.23, OS-E/F mutant = 1249.58 ± 246.56, genomic rescue = 310.43 ± 80.34, OS-E rescue = 257.52 ± 50.11, and OS-F rescue = 322.24 ± 88.65. ANOVA P-values: wild-type to OS-E/F mutant = 9.8 × 10−5, OS-E/F mutant to genomic rescue = 0.0019, OS-E/F mutant to OS-E rescue = 0.0016; OS-E/F mutant to OS-F rescue = 0.0013, wild-type to genomic rescue = 0.90, wild-type to OS-E rescue = 0.98, wild-type to OS-F rescue = 0.94, genomic rescue to OS-E rescue = 0.99, genomic rescue to OS-F rescue = 0.99, and OS-E rescue to OS-F rescue = 0.99. For Or47b responses to trans-2-hexenal (right part of graph): wild-type = 676.42 ± 64.24 msec, OS-E/F mutant = 1882.5 ± 328.9 msec, genomic rescue = 629.12 ± 102.74 msec, OS-E rescue = 699.18 ± 184.7 msec, and OS-F rescue = 499.1 ± 99.4 msec. ANOVA P-values: wild-type to OS-E/F mutant = 2.5 × 10−4, OS-E/F mutant to genomic rescue = 3.8 × 10−4, OS-E/F mutant to OS-E rescue = 0.004, OS-E/F mutant to OS-F rescue = 0.001, wild-type to genomic rescue = 0.70, wild-type to OS-E rescue = 0.90, wild-type to OS-F rescue = 0.14, genomic rescue to OS-E rescue = 0.73, genomic rescue to OS-F rescue = 0.40, and OS-E rescue to OS-F rescue = 0.39. n = 5–10. Error bars indicate SEM. All P-values determined by one-way ANOVA with post hoc Tukey’s test. (C) Extra copies of OS-E and OS-F do not alter deactivation kinetics. Time constants of deactivation for Or83c, Or67d, and Or47b neurons with the genotypes indicated. Deactivation time constants for Or83c neurons to farnesol (left side of graph): wild-type = 87.13 ± 9.2 msec, 4X OS-E/F= 99.6 ± 9.9 msec, P = 0.43. For Or67d responses to cVA (center part of graph): wild-type = 130.1 ± 22.9 msec, 4X OS-E/F = 124.3 ± 14.2, P = 0.83. For Or47b responses to trans-2-hexenal (right part of graph): wild-type = 752.65 ± 64.6 msec, 4X OS-E/F = 658.7 ± 38.8 msec, P = 0.52. cVA, 11-cis vaccenyl acetate; lncRNA, long noncoding RNA; OBP, odorant-binding proteins.
Deactivation kinetic abnormalities are odor- and not receptor-specific
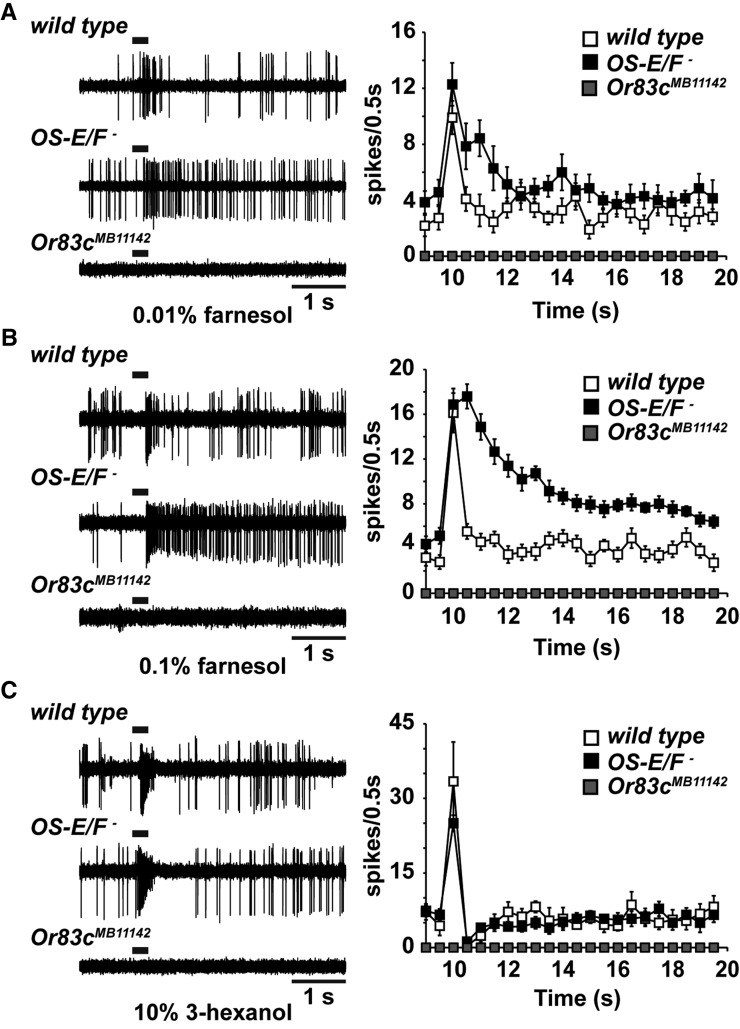
Only a subset of olfactory sensilla expressing OS-E and OS-F contain olfactory neurons with delayed deactivation kinetics to the activators we applied. We sought to establish whether this reflects a requirement for these binding proteins exclusively for those neurons, or if perhaps the OBPs are required for specific receptors or function with specific olfactory neurons. Or67d neurons are tuned to the cVA pheromone, and no other ligands have been observed to potently activate these neurons (Clyne et al. 1997; Xu et al. 2005). However, Or83c neurons are potently activated by farnesol, but they are also strongly activated by 3-hexanol (Ronderos et al. 2014). To establish whether the deactivation kinetic abnormalities we observed in the OS-E/F mutants could be specific to a receptor or neuron, we measured the deactivation kinetics of 3-hexanol in Or83c neurons.
Exposure of Or83c neurons to 3-hexanol produced robust responses in both wild-type and OS-E/F mutants (Figure 4C). Remarkably, while the deactivation kinetics to farnesol were much slower in the OS-E/F mutants compared to wild-type controls, the deactivation kinetics for 3-hexanol in Or83c neurons from OS-E/F mutants were not significantly different from wild-type controls (Figure 4). The τ values were 136.7 ± 21.6 msec for wild-type and 115.2 ± 14.6 msec for OS-E/F mutants (P = 0.88 by Student’s t-test). Therefore, the abnormal deactivation kinetics we observed for farnesol in OS-E/F mutant Or83c neurons do not reflect a requirement for OS-E or OS-F on overall Or83c neuron function.
Figure 4.
Odorant specificity of the Or83c deactivation defect. (A–C) representative traces are shown on the left, time courses of responses assayed by binning spikes in 500-msec bins on the right. (A) Responses of Or83c neurons from wild-type, and OS-E/F and Or83c mutant flies in response to 1% farnesol. Deactivation time constant (τ) = 92.3 ± 16.2 msec for wild-type and 404.0 ± 80 msec for OS-E/F mutants (P = 0.0015, n = 10). (B) Responses of wild-type, and OS-E/F and Or83c mutant flies to 0.1% farnesol. Despite lower peak activation, a prominent deactivation defect is still present in the mutants. τ = 109.4 ± 42.2 msec for wild-type and 870.5 ± 152 msec for OS-E/F mutants (P = 0.0014). n = 5. (C) Responses of wild-type, and OS-E/F and Or83c mutant flies to 10% 3-hexanol. No differences in deactivation are apparent between wild-type and OS-E/F mutants. Or83c mutants do not respond to 3-hexanol. τ = 115.2 ± 14.5 msec for wild-type, 109.6 ± 32.9 msec for OS-E/F mutants (P = 0.88). n = 5. Error bars represent SEM. All P-values determined by two-tailed Student’s t-test.
It is possible that OS-E or OS-F are required for the normal functioning of the Or83c receptor itself. The results above exclude a global deactivation effect for the OBPs on the Or83c neurons, but it is possible that there is a specific requirement for Or83c receptor function. For example, farnesol is known to be detected by Or83c receptors (Ronderos et al. 2014), but perhaps 3-hexanol is detected by a different, presently unknown receptor coexpressed in Or83c neurons that does not require OS-E/F function. We examined farnesol- and 3-hexanol-induced responses in Or83c receptor mutants (Ronderos et al. 2014). Deletion of the Or83c receptor gene abolished responses to both odorants (Figure 4, A–C). Therefore, 3-hexanol is also detected by the Or83c receptors. This finding eliminates the possibility that OS-E/F-binding proteins function in a obligate, receptor-specific manner. Finally, we were concerned that the deactivation defects for farnesol might reflect the potency of this activator on Or83c neurons. We examined the response of these neurons to a 10-fold higher dilution of farnesol. At this dilution, farnesol activates the Or83c neurons to action potential frequencies similar to those induced by 10% 3-hexanol. Remarkably, there was still a prominent deactivation defect to the diluted farnesol, but the response deactivation kinetics to 3-hexanol were not different from controls (Figure 4B). Finally, we tested additional odorants structurally related to farnesol on Or83c neurons. Farnesal, farnesene, and geraniol are all weak activators of Or83c neurons. Interestingly, farnesal and farnesene also displayed deactivation defects in the OS-E/OS-F mutants, while geraniol did not (Figure S4). Together, these data indicate that the deactivation kinetic abnormalities in OS-E/F mutants are odorant-specific, and do not stem from a role in regulating the intrinsic function of either the olfactory neurons or the odorant receptors.
Activation kinetics are not altered in OS-E/F mutants
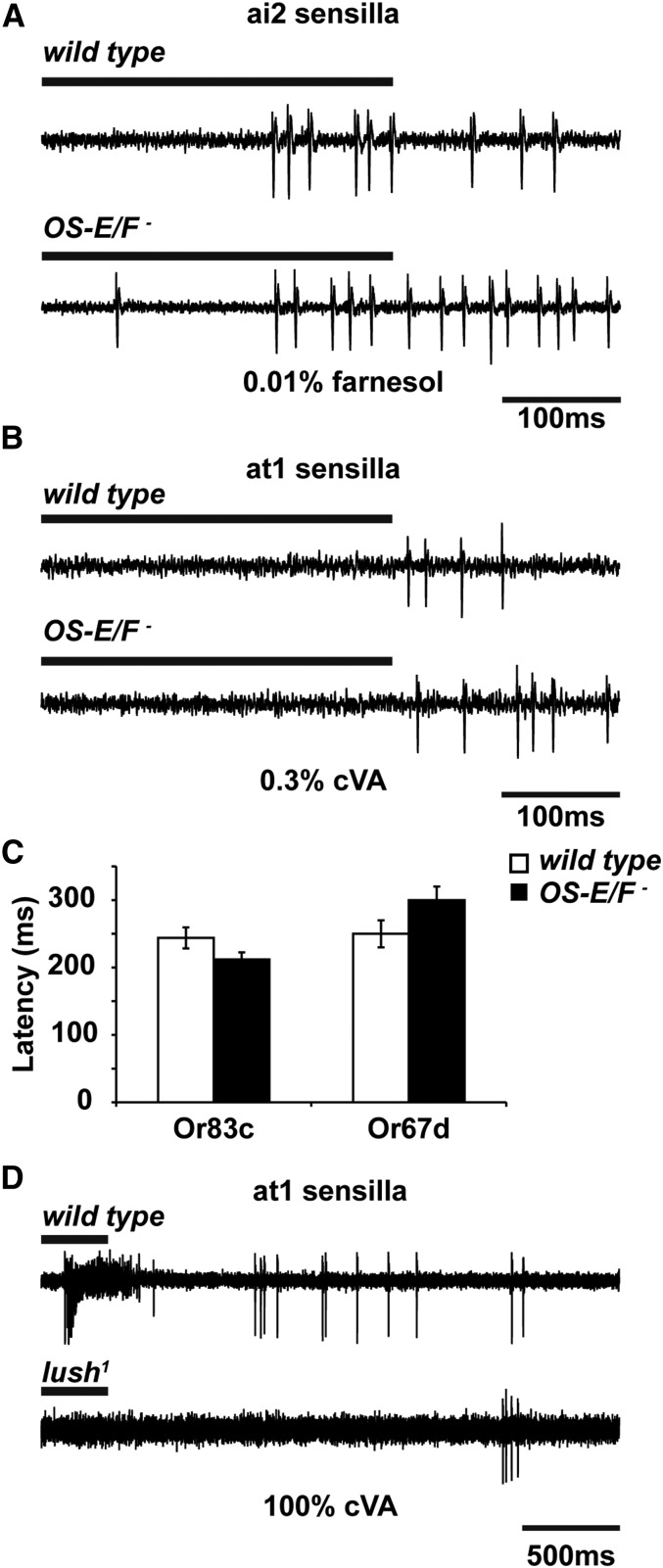
While it does not explain the defects in spontaneous activity, one explanation for the odorant-specific defects we observed in the OS-E/F mutants is that the efficient clearance of these odorants from the extracellular sensillum lymph requires these OBPs. Thus, in the absence of these OBPs, there is a delay in neuronal response termination stemming from persistence of the ligands in the lymph. If this model is true, then the OBPs and odorant receptors should be competing for odorant molecules in the sensillum lymph as odorants arrive at the antenna. In the absence of the abundant extracellular OBPs, odorant molecules might reach the odorant receptors faster and perhaps induce a more robust initial response in the OS-E/F mutants, especially at low stimulus concentrations. Indeed, an enhanced odorant response was recently reported for OBP28a mutants in ab8 sensilla exposed to 1-octanol (Larter et al. 2016).
We carefully analyzed the activation kinetics, and the peak responses, for wild-type and OS-E/F mutant flies responding to threshold-activating applications of cVA or farnesol. Figure 5 shows that the latency for activation of OS-E/F mutant Or83c neurons to 0.01% farnesol was not significantly different between wild-type and OS-E/F mutants. Figure 5C shows that wild-type activation occurred in 228.3 ± 2.1 msec and that of OS-E/F mutants in 225.9 ± 4.9 msec (P = 0.65). Similarly, the latency for activation of OS-E/F mutant Or67d neurons to 0.3% cVA was indistinguishable from control (Figure 5, B and C). Wild-type flies exposed to 0.3% cVA had a latency of 109.8 ± 2.2 msec and for OS-E/F mutants it was 109.7 ± 11.7 msec (P = 0.99). Responses to higher doses were also unaffected by loss of OS-E/OS-F (Figure S3). This suggests that odorant receptors are not competing with OBPs for odorant ligands in the lymph.
Figure 5.
Activation kinetics for farnesol and cVA are not affected by loss of OS-E and OS-F. (A) Representative 500-msec traces from wild-type and OS-E/F mutant-aI2 sensilla responses to 0.01% farnesol. (B) Representative 3-sec traces from wild-type, OS-E/F mutant, and lush1 Or67d neurons to 0.3% cVA. (C) Analysis of latency to activation for neurons and genotypes indicated. Or83c activation latency for wild-type is 228.3 ± 2.1 msec; for OS-E/F mutants, 225.8 ± 4.9 msec (P = 0.65). n = 10. Latency for Or67d activation is not significantly different in wild-type and OS-E/F mutants. Or67d activation latency in wild-type is 223 ± 34 msec and for OS-E/F mutants 299.8 ± 20 msec (P = 0.99). (D) Wild-type Or67d neurons exposed to 100% cVA have a latency for activation of 109.8 ± 2.2 msec. For lush1 mutants, the latency for activation is 1941.7 ± 414.5 msec. lush1 mutants are significantly different from wild-type (P = 4.25 × 10−5), n = 5–10 for each genotype. Error bars indicate SEM. All P-values are determined by two-tailed Student’s t-test. cVA, 11-cis vaccenyl acetate.
By contrast, we examined the activation latency for lush mutants and wild-type controls to 100% cVA (Figure 5D). lush mutants do not respond to physiologic concentrations of cVA; therefore, 100% cVA was used to produce reliable spiking activity in the lush mutants (van der Goes van Naters and Carlson 2007; Laughlin et al. 2008; Gomez-Diaz et al. 2013). For wild-type Or67d neurons exposed to 100% cVA, the latency for activation was 109.8 ± 2.2 msec. For the lush mutants exposed to the identical stimulus, the latency was 1941 ± 414 msec, almost 18-fold slower (significantly different by Student’s t-test, P = 4.25 × 10−5). This activation latency defect is consistent with a role for LUSH in sensitizing these neurons to the cVA pheromone and is strikingly different from the defects observed for the OS-E/F mutants. Together, these findings reveal that OS-E/F proteins are required for normal deactivation kinetics for a subset of odorant responses, but are not involved in the activation of the olfactory neurons.
No genetic interactions between OS-E/F and Snmp1
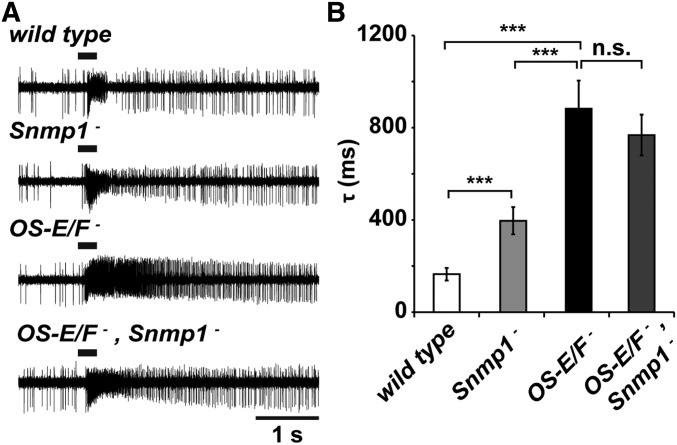
The phenotype of the OS-E/F mutants is reminiscent of the deactivation defects observed in Snmp1 mutants (Benton et al. 2007; Jin et al. 2008; Li et al. 2014). SNMP1 (Sensory neuron membrane protein 1) is a CD36 homolog expressed in the plasma membrane of a small subset of Drosophila olfactory neurons, which includes both farnesol- and cVA-detecting neurons (Benton et al. 2007; Jin et al. 2008; Ronderos et al. 2014). We wondered if there is a functional link between these OBPs and SNMP.
In the absence of SNMP, activation of Or67d neurons to cVA was extremely slow (minutes to hours) and deactivation never occurred (Li et al. 2014). Farnesol responses were less affected by a loss of SNMP1, but did show a striking deactivation defect (Ronderos et al. 2014). Antibodies to SNMP1 infused into the sensillum lymph of Or83c neurons also resulted in abnormal deactivation consistent with SNMP1 functioning on the cilia surface exposed to the sensillum lymph (Ronderos et al. 2014). We made OS-E/F,Snmp1 triple mutants to test whether the triple mutants had slower deactivation responses to farnesol than Snmp1 or OS-E/F mutants alone. If these components act in parallel, a more severe deactivation defect might be apparent in the triple mutants. Figure 6 shows that the Snmp1 mutants have a clear deactivation defect compared to wild-type controls, but that the deactivation defects in the OS-E/F mutants even more severe. The triple mutant is not significantly different from the OS-E/F mutant phenotype (Figure 6B).
Figure 6.
No genetic interactions between Snmp1 and OS-E/F mutants. (A) Representative 5-sec traces from wild-type, Snmp1 mutants, OS-E/F double mutants, or Snmp1/OS-E/F triple mutants in response to 100% farnesol. (B) Time constants of deactivation with the genotypes indicated: wild-type = 165.16 ± 27.1 msec, Snmp1 mutant = 396.7 ± 58.9 msec, OS-E/F mutant = 882.31 ± 121.92 msec, and Snmp1/OS-E/F mutant = 768.31 ± 88.7 msec. ANOVA P-values: wild-type to Snmp1 mutant = 0.0031; wild-type to OS-E/F mutant = 9.27 × 10−5; wild-type to Snmp1/OS-E/F triple mutant = 2.91 × 10−5; Snmp1 mutant to OS-E/F mutant = 0.006; Snmp1 mutant to Snmp1/OS-E/F mutant = 0.0064; and OS-E/F mutant to Snmp1/OS-E/F mutant = 0.46. n = 6–7 for each genotype. Error bars indicate SEM. All P-values were determined by one-way ANOVA with post hoc Tukey’s test.
OS-E and OS-F are functionally redundant
OS-E and OS-F have 79% amino acid similarity (McKenna et al. 1994). We were curious whether one or both OBP genes were required for rescue of the defective response kinetics in the OS-E/F mutants. Therefore, we modified the genomic rescue construct to excise large portions of the coding regions of either OS-E or OS-F (Figure 7), and generated transgenic flies with these modified rescuing transgenes in the OS-E/F mutant background. Figure 7B shows that expression of either OS-E or OS-F alone was sufficient to restore deactivation kinetics to normal (Figure 7B). These data reveal that OS-E and OS-F are functionally redundant. Finally, we tested whether extra copies of OS-E and OS-F (4× flies) would have faster deactivation kinetics compared to 2×, wild-type animals. Flies with the wild-type endogenous OS-E and OS-F genes, and homozygous for the OS-E/OS-F rescuing transgene, had deactivation kinetics that were not statistically different from controls (Figure 7).
Discussion
We identified a redundant role for OS-E and OS-F in accelerating the deactivation kinetics for three odorants detected by three different classes of olfactory neurons. This phenotype is different from any previously demonstrated defect for a member of the OBP family. Mutants defective for OS-E and OS-F expression have delayed deactivation kinetics to farnesol, trans-2-hexenal, and to cVA, three relatively large, hydrophobic odorants. Or83c, a receptor that detects both farnesol and 3-hexanol, is defective for deactivation to farnesol, but not 3-hexanol. This illustrates the odorant-specific nature of the OBP requirement. The deactivation defects in OS-E/F mutants are in stark contrast to the lush OBP mutants that have no apparent defect in deactivation, but instead have a striking delay in activation kinetics and impaired sensitivity to the cVA pheromone (Figure 5, Xu et al. 2005).
How do OS-E and OS-F affect deactivation kinetics? One possibility is that they clear specific odorants—including farnesol, trans-2-hexenal, and cVA—from the sensillum lymph bathing the olfactory neuron dendrites in these sensillae. This model is consistent with the receptor-independent, odorant-specific phenotypes we observe. However, it does not explain the defects in spontaneous activity that occur in the absence of odorants. Alternatively, OS-E/F might have a more intimate role, for example, by interacting directly with receptors, and acting to strip odorants off of the receptors following receptor binding and activation. The fact that activation kinetics are unaffected in OS-E/F mutants by farnesol, trans-2-hexenal, and cVA suggests that the OBPs are not competing with the odorant receptors for odorants in the sensillum lymph. OBPs are highly expressed in the sensillum lymph [estimated at ≤10 mM (Klein 1987)]. If these OBPs function exclusively to clear ligands from the sensillum lymph, they should compete for odorants in the lymph with the odorant receptors, slowing or reducing the magnitude of activation. This would be reflected in an enhanced or more rapid odorant response in the OBP mutants. Surprisingly, we see no change in activation latency or sensitivity to support this role. These data are more consistent with a receptor-stripping model whereby the OBPs function to displace odorants only after ligands have interacted with the receptor. If the OBPs associate with the receptors, even in the absence of ligands, this could alter the spontaneous activity of the receptors. Future studies will be required to explore the biochemical mechanisms of how these OBPs affect the kinetics of specific odorants.
An odorant-specific role was first postulated for OBPs upon their initial discovery and direct binding of OBPs to odorant ligands is well established (Vogt and Riddiford 1981; Pelosi 1994; Du and Prestwich 1995; Prestwich and Du 1995; Laughlin et al. 2008). Mutants in lush provided the first genetically defined function for an insect OBP and revealed that it is an essential sensitization factor (Xu et al. 2005). Indeed, misexpression of Or67d receptors in the Or47b neurons confers sensitivity to physiologically relevant cVA exposures, but only when LUSH is present (Ha and Smith 2006). Pheromone sensitivity phenotypes similar to those observed in lush mutants have been observed for other insect OBPs. Mutants in the striped stem borer Chilo suppressalis PBP1 and the cotton bollworm Helicoverpa armigera PBP1 have impaired electrophysiological responses to pheromones (Dong et al. 2017; Ye et al. 2017), and RNA interference (RNAi) knockdown of OBP3 and OBP7 in the aphid Acyrthosiphon pisum impairs detection and behavioral responses to alarm pheromones (Zhang et al. 2017). Thus, in the case of pheromone signaling, OBPs appear to function primarily as sensitizing factors. However, other OBPs have been implicated in altering the responses of chemosensory neurons in other ways.
OBP28a mutants have olfactory neuron responses that are initially more robust than those of wild-type flies, consistent with a role in the clearance of odorant from the sensillum lymph. However, these mutants also unexpectedly deactivate faster than controls (Larter et al. 2016). The phenotypes of OS-E/F mutants are distinct from that of the OBP28a mutants, and have little effect on the amplitude of the initial response, but maintain higher firing rates compared to controls following the termination of the stimulus.
Another Drosophila OBP, OBP59a, is required for normal humidity detection (Sun et al. 2018). How this OBP mediates sensitivity to humidity is unclear, but perhaps osmolarity changes are detected through the OBP. In Drosophila taste neurons, OBP49a is required to suppress activation of sweet-detecting gustatory neurons in the presence of bitter compounds (Jeong et al. 2013). In this case, the OBP–ligand complex appears to antagonize sweet receptors (Jeong et al. 2013; Swarup et al. 2014). RNAi knock down of several OBPs was shown to alter chemosensory behavior, often reducing behavioral responses to subsets of odorants (Swarup et al. 2011). Thus, OBPs appear to function in diverse ways to shape chemosensory responses. Many of these phenotypes are only explainable if there are direct OBP–receptor interactions.
The studies reported here are the first to demonstrate a role for OBPs in the deactivation kinetics of chemosensory responses. It is worth noting that even small changes in olfactory neuron firing rates can have striking effects on behavior (Bhandawat et al. 2007). Future experiments will explore the behavioral consequences of these deactivation defects. The findings reported here expand our understanding of the roles of invertebrate OBPs to include the deactivation of responses to subsets of odorants and highlight another phenotype to be explored as additional members of this diverse gene family are functionally dissected.
Acknowledgments
We thank Kishor Kunwar for assistance with the QPCR analysis. This work was supported by National Institutes of Health grants R01 DC-015230 to D.P.S. and 5T32 GM-008203 to E.A.S. The authors declare no competing financial interests.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9773615.
Communicating editor: D. Andrew
Literature Cited
- Almagro Armenteros J. J., Tsirigos K. D., Sonderby C. K., Petersen T. N., Winther O. et al. , 2019. SignalP 5.0 improves signal peptide predictions using deep neural networks. Nat. Biotechnol. 37: 420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., and Vosshall L. B., 2007. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450: 289–293. 10.1038/nature06328 [DOI] [PubMed] [Google Scholar]
- Bhandawat V., Olsen S. R., Gouwens N. W., Schlief M. L., and Wilson R. I., 2007. Sensory processing in the Drosophila antennal lobe increases reliability and separability of ensemble odor representations. Nat. Neurosci. 10: 1474–1482. 10.1038/nn1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne P., Grant A., O’Connell R., and Carlson J. R., 1997. Odorant response of individual sensilla on the Drosophila antenna. Invert. Neurosci. 3: 127–135. 10.1007/BF02480367 [DOI] [PubMed] [Google Scholar]
- Couto A., Alenius M., and Dickson B. J., 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15: 1535–1547. 10.1016/j.cub.2005.07.034 [DOI] [PubMed] [Google Scholar]
- Dong X. T., Liao H., Zhu G. H., Khuhro S. A., Ye Z. F. et al. , 2017. CRISPR/Cas9-mediated PBP1 and PBP3 mutagenesis induced significant reduction in electrophysiological response to sex pheromones in male Chilo suppressalis. Insect Sci. 26: 388–399. 10.1111/1744-7917.12544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du G., and Prestwich G. D., 1995. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry 34: 8726–8732. 10.1021/bi00027a023 [DOI] [PubMed] [Google Scholar]
- Dweck H. K., Ebrahim S. A., Kromann S., Bown D., Hillbur Y. et al. , 2013. Olfactory preference for egg laying on citrus substrates in Drosophila. Curr. Biol. 23: 2472–2480. 10.1016/j.cub.2013.10.047 [DOI] [PubMed] [Google Scholar]
- Dweck H. K., Ebrahim S. A., Thoma M., Mohamed A. A., Keesey I. W. et al. , 2015. Pheromones mediating copulation and attraction in Drosophila. Proc. Natl. Acad. Sci. USA 112: E2829–E2835. 10.1073/pnas.1504527112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo K., and Smith D. P., 2001. A large family of divergent odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics 159: 1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galizia C. G., and Sachse S., 2010. Odor coding in insects, pp. 35–70 in Neurobiology of Olfaction, edited by Menini A. CRC Press, Boca Raton, FL. [PubMed] [Google Scholar]
- Gomez-Diaz C., Reina J. H., Cambillau C., and Benton R., 2013. Ligands for pheromone-sensing neurons are not conformationally activated odorant binding proteins. PLoS Biol. 11: e1001546 10.1371/journal.pbio.1001546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham L. A., and Davies P. L., 2002. The odorant-binding proteins of Drosophila melanogaster: annotation and characterization of a divergent family. Gene 292: 43–55. 10.1016/S0378-1119(02)00672-8 [DOI] [PubMed] [Google Scholar]
- Gratz S. J., Ukken F. P., Rubinstein C. D., Thiede G., Donohue L. K. et al. , 2014. Highly specific and efficient CRISPR/Cas9-catalyzed homology-directed repair in Drosophila. Genetics 196: 961–971. 10.1534/genetics.113.160713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. S., and Smith D. P., 2006. A pheromone receptor mediates 11-cis-vaccenyl acetate-induced responses in Drosophila. J. Neurosci. 26: 8727–8733. 10.1523/JNEUROSCI.0876-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha T. S., and Smith D. P., 2009. Odorant and pheromone receptors in insects. Front. Cell. Neurosci. 3: 10 10.3389/neuro.03.010.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A., Ho M. G., and Carlson J. R., 2004. The molecular basis of odor coding in the Drosophila antenna. Cell 117: 965–979. 10.1016/j.cell.2004.05.012 [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe D.S., Steinbrecht R. A., and Carlson J. R., 1997. Coexpression of two odorant binding homologs in Drosophila: implications for olfactory coding. J. Neurosci. 17: 1616–1624. 10.1523/JNEUROSCI.17-05-01616.1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe D. S., Dorit R. L., and Carlson J. R., 2000. Molecular evolution of odorant-binding protein genes OS-E and OS-F in Drosophila. Genetics 155: 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hekmat-Scafe D. S., Scafe C. R., McKinney A., and Tanouye M. A., 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster. Genome Res. 12: 1357–1369. 10.1101/gr.239402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horst R., Damburger F., Peng G., Lunigbuhl P., Guntert P. et al. , 2001. NMR structure reveals intramolecular regulation mechanism for pheromone binding and release. Proc. Natl. Acad. Sci. USA 98: 14374–14379. 10.1073/pnas.251532998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong Y. T., Shim J., Oh S. R., Yoon H. I., Kim C. H. et al. , 2013. An odorant-binding protein required for suppression of sweet taste by bitter chemicals. Neuron 79: 725–737. 10.1016/j.neuron.2013.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Ha T. S., and Smith D. P., 2008. SNMP is a signaling component required for pheromone sensitivity in Drosophila. Proc. Natl. Acad. Sci. USA 105: 10996–11001. 10.1073/pnas.0803309105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaissling K. E., 1996. Peripheral mechanisms of pheromone reception in moths. Chem. Senses 21: 257–268. 10.1093/chemse/21.2.257 [DOI] [PubMed] [Google Scholar]
- Kaissling K.-E., 1998. Olfactory transduction in moths: I. Generation of receptor potentials and nerve impulses, II. Extracellular transport, deactivation and degradation of stimulus molecules, pp. 98–112 in From Structure to Information in Sensory Systems, edited by Taddei-Ferretti C. and Musio C.. World Scientific, Singapore. [Google Scholar]
- Kaissling K.-E., 2001. Olfactory perireceptor events in moths: a kinetic model. Chem. Senses 26: 125–150. 10.1093/chemse/26.2.125 [DOI] [PubMed] [Google Scholar]
- Kaissling K. E., Klein U., deKramer J. J., Keil T. A., Kanaujia S. et al. , 1985. Insect olfactory cells: electrophysiological and biochemical studies, pp. 173–183 in Proceedings of the International Symposium in Memmory of D. Nachmansohn, edited by Changeux J. P., Hucho F., Maelicke A., and Neumann E.. Walter de Gruyter, Berlin: 10.1515/9783110855630-017 [DOI] [Google Scholar]
- Kim M. S., Repp A., and Smith D. P., 1998. LUSH odorant-binding protein mediates chemosensory responses to alcohols in Drosophila melanogaster. Genetics 150: 711–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., 1987. Sensillum-lymph proteins from antennal olfactory hairs of the moth Antherea polyphemus (Saturniidae). Insect Biochem. 17: 1193–1204. 10.1016/0020-1790(87)90093-X [DOI] [Google Scholar]
- Kondo S., and Ueda R., 2013. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics 195: 715–721. 10.1534/genetics.113.156737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger J., and Breer H., 1999. Olfactory reception in invertebrates. Science 286: 720–723. 10.1126/science.286.5440.720 [DOI] [PubMed] [Google Scholar]
- Larter N. K., Sun J. S., and Carlson J. R., 2016. Organization and function of Drosophila odorant binding proteins. Elife 5: e20242. 10.7554/eLife.20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laughlin J. D., Ha T. S., Jones D. N., and Smith D. P., 2008. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone-binding protein. Cell 133: 1255–1265. 10.1016/j.cell.2008.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal W. S., 2013. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58: 373–391. 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- Li Z., Ni J. D., Huang J., and Montell C., 2014. Requirement for Drosophila SNMP1 for rapid activation and termination of pheromone-induced activity. PLoS Genet. 10: e1004600 10.1371/journal.pgen.1004600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M. P., Hekmat-Scafe D. S., Gaines P., and Carlson J. R., 1994. Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 269: 16340–16347. [PubMed] [Google Scholar]
- Nandi N., Tyra L. K., Stenesen D., and Krämer H., 2017. Stress-induced Cdk5 activity enhances cytoprotective basal autophagy in Drosophila melanogaster by phosphorylating acinus at serine437. Elife 6: e30760. 10.7554/eLife.30760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi P., 1994. Odorant-binding proteins. Crit. Rev. Biochem. Mol. Biol. 29: 199–228. 10.3109/10409239409086801 [DOI] [PubMed] [Google Scholar]
- Pelosi P., and Maida R., 1990. Odorant-binding proteins in vertebrates and insects: similarities and possible common function. Chem. Senses 15: 205–215. 10.1093/chemse/15.2.205 [DOI] [Google Scholar]
- Pikielny C. W., Hasan G., Rouyer F., and Rosbash M., 1994. Members of a family of Drosophila putative odorant-binding proteins are expressed in different subsets of olfactory hairs. Neuron 12: 35–49. 10.1016/0896-6273(94)90150-3 [DOI] [PubMed] [Google Scholar]
- Pirrotta V., 1988. Vectors for P-mediated transformation in Drosophila, pp. 437–456 in Vectors: A Survey of Molecular Cloning Vectors and Their Uses, edited by Rodriguez R. L. and Denhart D. T.. Butterworth, London: 10.1016/B978-0-409-90042-2.50028-3 [DOI] [PubMed] [Google Scholar]
- Pitts S., Pelser E., Meeks J., and Smith D., 2016. Odorant responses and courtship behaviors influenced by at4 neurons in Drosophila. PLoS One 11: e0162761 [corrigenda: PLoS One 13: e0190805 (2018)]. 10.1371/journal.pone.0162761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponton F., Chapuis M. P., Pernice M., Sword G. A., and Simpson S. J., 2011. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J. Insect Physiol. 57: 840–850. 10.1016/j.jinsphys.2011.03.014 [DOI] [PubMed] [Google Scholar]
- Pophof B., 2002. Moth Pheromone binding proteins contribute to the excitation of olfactory receptor cells. Naturwissenschaften 89: 515–518. 10.1007/s00114-002-0364-5 [DOI] [PubMed] [Google Scholar]
- Prestwich G. D., and Du G., 1995. Pheromone binding proteins, pheromone recognition, and signal transduction in moth olfaction in Insect Pheromone Research: New Directions, edited by Carde R. T. and Minks A. K.. Chapman & Hall, New York. [Google Scholar]
- Ronderos D. S., and Smith D. P., 2009. Diverse signaling mechanisms mediate volatile odorant detection in Drosophila. Fly (Austin) 3: 290–297. 10.4161/fly.9801 [DOI] [PubMed] [Google Scholar]
- Ronderos D. S., Lin C. C., Potter C. J., and Smith D. P., 2014. Farnesol-detecting olfactory neurons in Drosophila. J. Neurosci. 34: 3959–3968. 10.1523/JNEUROSCI.4582-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler B. H., Nikonova L., Leal W. S., and Clardy J., 2000. Sexual attraction in the silkworm moth: structure of the pheromone-binding-protein-bombykol complex. Chem. Biol. 7: 143–151. 10.1016/S1074-5521(00)00078-8 [DOI] [PubMed] [Google Scholar]
- Shanbhag S. R., Hekmat-Scafe D., Kim M. S., Park S. K., Carlson J. R. et al. , 2001. Expression mosaic of odorant-binding proteins in Drosophila olfactory organs. Microsc. Res. Tech. 55: 297–306. 10.1002/jemt.1179 [DOI] [PubMed] [Google Scholar]
- Shanbhag S. R., Smith D. P., and Steinbrecht R. A., 2005. Three odorant-binding proteins are co-expressed in sensilla trichodea of Drosophila melanogaster. Arthropod Struct. Dev. 34: 153–165. 10.1016/j.asd.2005.01.003 [DOI] [Google Scholar]
- Smith D. P., 2012. Volatile pheromone signalling in Drosophila. Physiol. Entomol. 37 DOI: 10.1111/j.1365-3032.2011.00813.x. 10.1111/j.1365-3032.2011.00813.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling A. C., and Rubin G. M., 1982. Transposition of cloned P elements into Drosophila germline chromosomes. Science 218: 341–347. 10.1126/science.6289435 [DOI] [PubMed] [Google Scholar]
- Steinbrecht R. A., and Müller B., 1971. On the stimulus conducting structures in insect olfactory receptors. Z. Zellforsch. Mikrosk. Anat. 117: 570–575. 10.1007/BF00330716 [DOI] [PubMed] [Google Scholar]
- Sun J. S., Larter N. K., Chahda J. S., Rioux D., Gumaste A. et al. , 2018. Humidity response depends on the small soluble protein Obp59a in Drosophila. Elife 7: e39249. 10.7554/eLife.39249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Williams T. I., and Anholt R. R., 2011. Functional dissection of Odorant binding protein genes in Drosophila melanogaster. Genes Brain Behav. 10: 648–657. 10.1111/j.1601-183X.2011.00704.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S., Morozova T. V., Sridhar S., Nokes M., and Anholt R. R., 2014. Modulation of feeding behavior by odorant-binding proteins in Drosophila melanogaster. Chem. Senses 39: 125–132. 10.1093/chemse/bjt061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Goes van Naters W., and Carlson J. R., 2007. Receptors and neurons for fly odors in Drosophila. Curr. Biol. 17: 606–612. 10.1016/j.cub.2007.02.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt R. G., 2003. Insect Pheromone Biochemistry and Moleuclar Biology, the Biosynthesis and Detection of Pheromones and Plant Volatiles. Elsevier, New York. [Google Scholar]
- Vogt R. G., and Riddiford L. M., 1981. Pheromone binding and inactivation by moth antennae. Nature 293: 161–163. 10.1038/293161a0 [DOI] [PubMed] [Google Scholar]
- Vogt R. G., Riddiford L. M., and Prestwich G. D., 1985. Kinetic properties of a pheromone degrading enzyme: the sensillar esterase of Antherea polyphemus. Proc. Natl. Acad. Sci. USA 82: 8827–8831. 10.1073/pnas.82.24.8827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt R. G., Prestwich G. D., and Lerner M. R., 1991. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 22: 74–84. 10.1002/neu.480220108 [DOI] [PubMed] [Google Scholar]
- Wang L., Han X., Mehren J., Hiroi M., Billeter J. C. et al. , 2011. Hierarchical chemosensory regulation of male-male social interactions in Drosophila. Nat. Neurosci. 14: 757–762. 10.1038/nn.2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojtasek H., and Leal W. S., 1999. Conformational change in the pheromone-binding protein from Bombyx mori induced by pH and interaction with membranes. J. Biol. Chem. 274: 30950–30956. 10.1074/jbc.274.43.30950 [DOI] [PubMed] [Google Scholar]
- Xu P., Atkinson R., Jones D. N., and Smith D. P., 2005. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45: 193–200. 10.1016/j.neuron.2004.12.031 [DOI] [PubMed] [Google Scholar]
- Ye Z. F., Liu X. L., Han Q., Liao H., Dong X. T. et al. , 2017. Functional characterization of PBP1 gene in Helicoverpa armigera (Lepidoptera: Noctuidae) by using the CRISPR/Cas9 system. Sci. Rep. 7: 8470 10.1038/s41598-017-08769-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Wang B., Grossi G., Falabella P., Liu Y. et al. , 2017. Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 27: 55–61. 10.1016/j.cub.2016.10.013 [DOI] [PubMed] [Google Scholar]
- Ziegelberger G., 1995. Redox-shift of the pheromone binding protein in the silkworm Antherea polyphemus. Eur. J. Biochem. 232: 706–711. 10.1111/j.1432-1033.1995.tb20864.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Fly strains and plasmids are available upon request. Supplemental Figures include Supplemental Material, Figures S1–S4. Supplemental material available at FigShare: https://doi.org/10.25386/genetics.9773615.