Abstract
We examined the effects of dietary supplementation of a Saccharomyces cerevisiae-based direct-fed microbial (DFM) on the growth performance, whole-blood immune gene expression, serum biochemistry, and plasma metabolome of newly weaned beef steers during a 42 d receiving period. Forty newly weaned Angus crossbred steers (7 d post-weaning; 210 ± 12 kg of BW; 180 ± 17 d of age) from a single source were stratified by BW and randomly assigned to 1 of 2 treatments: basal diet with no additive (CON; n = 20) or a basal diet top-dressed with 19 g of the DFM (PROB; n = 20). Daily DMI and weekly body weights were measured to calculate average daily gain (ADG) and feed efficiency (FE). Expression of 84 immune-related genes was analyzed on blood samples collected on days 21 and 42. Serum biochemical parameters and plasma metabolome were analyzed on days 0, 21, and 42. On day 40, fecal grab samples were collected for pH measurement. Compared with CON, dietary supplementation of PROB increased final body weight (P = 0.01) and ADG (1.42 vs. 1.23 kg; P = 0.04) over the 42 d feeding trial. There was a tendency for improved FE with PROB supplementation (P = 0.10). No treatment effect (P = 0.24) on DMI was observed. Supplementation with PROB increased (P ≤ 0.05) the concentrations of serum calcium, total protein, and albumin. Compared with CON, dietary supplementation with PROB increased (P ≤ 0.05) the expression of some immune-related genes involved in detecting pathogen-associated molecular patterns (such as TLR1, TLR2, and TLR6), T-cell differentiation (such as STAT6, ICAM1, RORC, TBX21, and CXCR3) and others such as TNF and CASP1, on day 21 and/or day 42. Conversely, IL-8 was upregulated (P = 0.01) in beef steers fed CON diet on day 21. Plasma untargeted plasma metabolome analysis revealed an increase (P ≤ 0.05) in the concentration of metabolites, 5-methylcytosine and indoleacrylic acid involved in protecting the animals against inflammation in steers fed PROB diet. There was a tendency for lower fecal pH in steers fed PROB diet (P = 0.08), a possible indication of increased hindgut fermentation. This study demonstrated that supplementation of PROB diet improved the performance, nutritional status, and health of newly weaned beef steers during a 42 d receiving period.
Keywords: beef steer, direct-fed microbial, immune genes, indoleacrylic acid, performance, plasma metabolome
Introduction
The feedlot-receiving period is characterized by several stressors caused by separation from the mother, commingling, transportation, vaccination, exposure to pathogens, and changes in diet and environment (Arthington et al., 2013). These stressors cause reduced feed intake, poor calf performance, morbidity, and compromised immune response (Duff and Galyean, 2007). Feed intake of newly weaned calves is low during the first 2 wk after weaning (Galyean and Hubbert, 1995); this causes low nutrient intake which contributes to compromised immune function of the calves (Duff and Galyean, 2007).
Research studies have focused on evaluating several nutritional strategies to optimize animal performance and immunity during the receiving period (McAllister et al., 2011). One such strategy is the use of direct-fed microbials (DFMs) (McDonald et al., 2005; McAllister et al., 2011; Deters et al., 2018). DFMs improve the animal gut health by modulating ruminal fermentation, enhancing the establishment of beneficial microbial populations and/or enhancing post-ruminal digestion and absorption (Beauchemin et al., 2003; Raeth-Knight et al., 2007). DFM products have also been reported to improve immune-competence of livestock during stress periods (Duff and Galyean, 2007; McAllister et al., 2011). In order to ensure multi-factorial response to supplementation of DFMs, DFM products are formulated to contain several combinations of microorganisms including lactic acid-producing bacteria (such as Lactobacillus spp. Pediococcus spp., and Enterococcus spp.), yeast (such as Saccharomyces cerevisiae), fungi (such as Aspergillus spp.) and spore-forming Bacillus (McAllister et al., 2011). However, responses to DFMs are inconsistent across several studies possibly due to supplementation with different strains of organisms, inclusion level, diet, and animal factors (McAllister et al., 2011; Uyeno et al., 2015; Plaizier et al., 2018). For instance, Kenney et al. (2015) reported no effects of dietary supplementation of a mixture of Lactobacillus acidophilus, Enterococcus faecium, Pediococcus acidilacticii, Lactobacillus brevis, and Lactobacillus plantarum on the growth performance of receiving steers. In another study, Fink et al. (2014) reported no effects of dietary supplementation of S. cerevisiae-based DFMs on the performance of beef steers during a 56 d receiving period.
Some studies have also evaluated the use of fermentation products of microorganisms such as L. acidophilus, Lactobacillus casei, Aspergillus spp. and S. cerevisiae in the diets of beef cattle (Tricarico et al., 2007; Deters et al., 2018; Hall et al., 2018). Fermentation products are sources of nutritional metabolites such as B-vitamins, enzymes, amino acids, nucleotides, lipids, and organic acids that enhance the growth and activities of beneficial microbes in the gut (Rai et al., 2013). Like DFMs, responses of animals to supplementation of fermentation products are inconsistent (Deters et al., 2018; Hall et al., 2018). Hall et al. (2018) reported that dietary supplementation with a L. acidophilus fermentation production increased growth and intake of recently weaned beef cattle compared with a diet supplemented with monensin. In another study, no benefit of feeding a fermentation product of S. cerevisiae was observed in newly weaned beef steers (Deters et al., 2018).
To date, little emphasis has been placed on evaluating a blend of DFM containing multiple microorganisms and their fermentation products on the performance and health of recently weaned beef cattle. Moreover, inconsistent responses to feeding DFMs in ruminant production systems and continuing development of different strains of DFMs emphasize the need for more research studies to understand their underlying mechanisms. Therefore, we hypothesized that a blend of multiple strain-DFM and their fermentation products would improve the growth performance and health of newly weaned steers. The objective of this study was to evaluate the effects of dietary supplementation of a blend of DFMs and their fermentation products on performance, immunity, serum biochemistry, and plasma metabolome of newly weaned beef steers during a 42 d receiving period.
Materials and Methods
The Institutional Animal Care and Use Committees of Kentucky State University approved all research procedures.
Animals, Housing, and Feeding
Forty newly weaned Angus crossbred steers (7 d post-weaning; 210 ± 12 kg of BW; 180 ± 17 d of age) from a single source were stratified by BW into four weight blocks. Within each weight block, steers were randomly assigned to 1 of 2 treatments and housed in individual slatted floor pens (2.44 × 14.63 m2; 20 pens per treatment). The steers were assigned to receive a basal diet with no additive (CON; n = 20) or a basal diet supplemented with 19 g of Commence Feed Additive (PROB; n = 20) for a period of 42 d. Commence Feed Additive (PMI, Arden Hills, MN) is an optimized blend of 6.2 × 1011 cfu/g of S. cerevisiae, 3.5 × 1010 cfu/g of a mixture of Enterococcus lactis, Bacillus subtilis, Enterococcus faecium, and L. casei, and the fermentation products of these aforementioned microorganisms as well as those of Aspergillus oryzae and A. niger. The basal diet was fed ad libitum (to achieve approximately 10% ort) daily as a total mixed ration (TMR) at 0800 hours (Table 1). The additive was top-dressed daily on the TMR in the form of a premix using dried distillers grains with solubles for the PROB treatment while a similar premix with no additive was top-dressed for the CON treatment. The steers had free access to water. Adjacent pens shared a common water source, as such, adjacent pens were assigned to steers in the same treatment in order to reduce DFM exposure across treatments.
Table 1.
Ingredient and chemical composition of the basal diet1
| Ingredient | % of dietary DM |
|---|---|
| Corn silage | 79.7 |
| Dehydrated distillers grain | 9.06 |
| Soybean meal | 9.28 |
| Limestone | 0.42 |
| Deccox2 | 0.03 |
| Vitamin and mineral premix3 | 1.51 |
| Nutrient analysis4 | |
| DM, % | 44.5 |
| CP, % | 14.7 |
| aNDF, % | 38.6 |
| ADF, % | 21.5 |
| EE, % | 3.50 |
| Ca, % | 0.87 |
| P, % | 0.63 |
| TDN, % | 72.6 |
| NEm, Mcal/kg | 1.72 |
| NEg, Mcal/kg | 1.10 |
1Chemical composition of complete diets calculated from analysis and concentration of individual ingredients.
2Contains 6% decoquinate for the prevention of coccidiosis (Zoetis Inc.).
3Guaranteed analysis: 15% Ca; 7.5% P; 20% salt; 1% Mg; 1% K; 3,600 mg/kg Mn; 12 mg/kg Co; 1,200 mg/kg Cu; 3,600 mg/kg Zn; 27 mg/kg Se; 60 mg/kg I; 660,000 IU/kg vitamin A; 660 IU/kg vitamin E; and 66,000 IU/kg vitamin D.
4DM = dry matter; CP = crude protein; aNDF = neutral detergent fiber (amylase treated); ADF = acid detergent fiber; EE = ether extract; TDN = total digestible nutrients; NEm = net energy of maintenance; NEg = net energy of gain.
Sample Collection and Measurements
Dry matter intake
The quantity of feed offered to each steer was recorded daily. Diet refused (as fed) was also measured daily. Diet DM refused and offered were obtained by drying weekly samples of diets refused and offered in a forced-air oven at 56 °C for 48 h. Daily DMI was determined by subtracting the daily DM refused from the daily DM offered and then dividing by 7 d. Samples of feed ingredients and TMR collected weekly were dried for 48 h at 60 °C in a forced-air oven, ground to pass through a 1-mm screen (Wiley Mill; Arthur H. Thomas Co.), and sent to a commercial laboratory (Dairy One Forage Laboratory, Ithaca, NY) for analysis of the chemical composition, which included DM (method 930.15; AOAC International, 2000), N (method 990.03; AOAC International, 2000), ADF (method 989.03; AOAC International, 2000), NDF (Van Soest et al., 1991), ether extract using diethyl ether (method 2003.05; AOAC International, 2000), and Ca and P (Sirois et al., 1994).
Body weight
Body weights of steers were obtained before morning feeding on days 0, 21 and 42. Average daily gain (ADG) was determined by subtracting the initial weight on day 0 from the final weight on day 42 and then dividing by the duration of the experiment (42). ADGs from days 1 to 21 and 22 to 42 were also determined.
Blood and feces sample collection
15 mL of blood was taken before the morning feeding on days 0, 21, and 42. The blood samples were taken from the coccygeal vessels into tubes containing sodium heparin (Fisher Scientific Company) and a glass tube containing no anticoagulant (Fisher Scientific Company). Tubes containing sodium heparin were placed on ice immediately after collection, and a sub-sample of the whole blood (approximately 500 µL) was transferred into RNA-protect tubes (cat. no. 76554; Qiagen) which contain a reagent that lyses blood cells and stabilizes intracellular RNA. These samples were stored at −20 °C until RNA extraction and expression analysis of innate and adaptive immune-related genes. Thereafter, plasma samples were immediately prepared from the remaining samples by centrifugation at 2500 × g for 20 min at 4 °C, and stored at −80 °C until targeted and untargeted quantitative metabolomics were done.
Blood samples in the glass tubes with no anticoagulant were allowed to sit at room temperature for 120 min to coagulate. Serum was then separated by centrifugation at 2500 × g for 20 min for subsequent analysis of serum biochemical indices.
On day 40, fecal grab samples were collected from the steers at approximately 4 h after morning feeding for pH measurement.
Serum Biochemistry Indices
Serum biochemical parameters were analyzed using a VetScan Chemistry Analyzer (Abaxis, Inc., CA, United States) which provides an in vitro quantitative analysis of total protein concentration (TP), albumin concentration (ALB), urea nitrogen, creatine kinase, phosphorus, calcium, magnesium, alanine aminotransferase activity, gamma glutamyl transferase activity, and alkaline phosphatase activity (Marchal et al., 2012). Globulin concentration was calculated as the difference between TP and ALB.
Whole Blood Immune Gene Expression Analysis
RNA extraction and cDNA preparation
RNA was extracted using RNeasy Protect Animal Blood kit (cat. no. 73224; Qiagen). Briefly, the RNAprotect Animal Blood Tubes containing the blood samples collected on days 21 and 42 were centrifuged at 3200 × g for 10 min. The supernatant was discarded, and the pellet was resuspended in 240 µL buffer RSB, followed by digestion with 20 µL proteinase K. The mixture was thereafter homogenized by centrifugation through QIAshredder spin columns. After addition of 240 µL ethanol (96–100%), the homogenized mixture was centrifuged through RNeasy MinElute spin columns (where total RNA binds to the RNeasy MinElute silica membrane). The bound total RNA was subjected to DNase digestion to remove genomic DNA contamination, and then washed with 350 µL buffer RW1, followed by 500 µL buffer RPE. Pure RNA was thereafter eluted in 30 µL buffer REB. All samples had >100 ng/µL total RNA. The RNA was used to synthesize complementary DNA (cDNA) using the RT2 First Strand Kit (cat. no. 330401; Qiagen) following the manufacturer’s instructions.
RT2 profiler PCR array
Expression of 84 genes related to innate and adaptive immune responses was analyzed using the RT2 Profiler cow innate and adaptive immune responses PCR Array (PABT-052ZA; Qiagen) according to the manufacturer’s instructions. Briefly, RT2 SYBR Green ROX qPCR Mastermix (Cat No. 330523; Qiagen) and RNase-free water were mixed with cDNA synthesis reaction volume. Then, 25 µL of the PCR component mix was placed into each well of the 96-well plate PCR array. The array contained 84 adaptive and innate immune-related genes, five housekeeping genes (β-actin, glyceraldehyde-3-phosphate dehydrogenase, hypoxanthine phosphoribosyltransferase 1, TATA box binding protein, and tyrosine 3-monooxygenase), one genomic DNA control to detect gDNA contamination, three reverse transcription controls to control for impurities that may degrade RNA, and three positive PCR controls to verify efficiency of the PCR amplification of cDNA template. Real-time PCR was performed on a QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Foster City, CA) using the following cycling conditions: 95 °C for 10 min, 40 cycles of denaturation at 95 °C for 15 s and 60 °C 1 min. The expression levels of the genes (delta Ct-values) were calculated by subtracting the average cycle threshold (Ct) value of the housekeeping genes from the Ct value of the genes-of-interest.
LC-MS/MS-Based Targeted Metabolomics Analysis
Plasma samples (20 µL) collected on days 0, 21, and 42 were analyzed using a commercial kit that uses LC-MS/MS according to the procedure described by Zhang et al., (2017). The kit assay (AbsoluteIDQ p180) was used with an ABI 4000 Q-Trap mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). This kit allows quantification of several metabolites, including 21 amino acids, 20 biogenic amines, 40 acylcarnitines, 34 glycerophospholipids, 15 organic acids, and 8 other metabolites. The list of identified metabolites are provided in Supplementary Table S1.
CIL/LC-MS-Based Untargeted Metabolomics Analysis
In-depth untargeted metabolome profile of the plasma samples collected on day 42 was done using a chemical isotope labeling (CIL)/ liquid chromatography–mass spectrometry (LC-MS)-based technique. This technique uses 12C and 13C-isotope dansylation labeling to target the amine and phenol-containing submetabolome. The workflow of the differential 12C- and 13C-isotope dansylation LC-MS method for analyzing the amine/phenol-containing submetabolome of the plasma is shown in Supplementary Figure S1. Sample amount normalization was done using liquid chromatography–ultraviolet (LC-UV) quantification of the dansyl-labeled metabolites (Wu and Li, 2012) and relative quantification of the metabolites based on peak ratio values was performed on an Agilent 1100 LC system (Palo Alto, CA) connected to a Bruker Impact HD quadrupole time-of-flight (QTOF) MS (Billercia, MA). Detailed information of sample preparation methods, dansylation protocol, LC-UV and LC-MS setup, and concentration measurement have been previously reported (Mung and Li, 2017). One of the CON samples was not analyzed due to poor quality, therefore, a total number of 47 LC-MS data files were generated (4 blank group samples, 4 quality control samples, 20 PROB samples, and 19 CON samples).
Metabolite data processing
The 47 LC-MS data files (in profile mode) were converted to text file (in centroid mode) using Bruker DataAnalysis software 4.4. Raw data processing was performed using IsoMS Pro 1.0 according to procedures described by Mung and Li, 2017. Peak pairs whose mean (sample)/mean (blank) was ≤ 4.0 were filtered out. Peak pairs with no data present in at least 80% of the samples were filtered out. The final metabolite-intensity table was generated using IsoMS-Quant (Huan and Li, 2015).
Metabolite identification
Three-tier identification approach was used to perform metabolite identification. In tier 1, peak pairs were searched against a labeled metabolite library (CIL Library) based on accurate mass and retention time (Huan and Li, 2015). The CIL Library (amine/phenol channel) contains 712 experimental entries, including metabolites and dipeptides. In tier 2, linked identity library (LI Library) was used for identification of the remaining peak pairs. LI Library includes over 2,000 human endogenous metabolites from 68 metabolic pathways, providing high-confidence putative identification results based on accurate mass and predicted retention time matches (Li et al. 2013). In tier 3, the remaining peak pairs were searched, based on accurate mass match, against the MyCompoundID (MCID; www.MyCompoundID.org) library which is composed of 8,021 known human endogenous metabolites and their predicted metabolic products from one metabolic reaction (375,809 compounds) and two metabolic reactions (10,583,901 compounds) (Li et al. 2013). The mass accuracy tolerance window was set at 5 ppm for all searches, and the retention time tolerance window was set to 30 s for CIL and MCID libraries, and 192 s for LI library.
Fecal pH
A sub-sample of the feces (approximately 10 g) was mixed with de-ionized water at a ratio of 1:10 and then vortexed. Fecal pH was immediately measured with a pH meter.
Statistical Analysis
All variables, except immune gene expression and untargeted metabolomics data, were analyzed using the GLIMMIX procedure of SAS (SAS Institute Inc., Cary, NC), with treatment included as a fixed effect and block included as a random effect. Significant effects were declared at P ≤ 0.05 and tendencies for significance were declared at 0.05 < P ≤ 0.10. For the performance data, values of initial weight of the steers were included as a covariate for the final body weight. For the serum biochemistry and plasma targeted metabolome data (LC-MS/MS), values on day 0 were included in the model as a covariate for days 21 and 42.
For the CIL/LC-MS data, univariate (volcano plot) and multivariate statistical analysis (Partial least squares discriminant analysis [PLS-DA] scores plot) were generated using IsoMS Pro 1.0 (Mung and Li, 2017). The volcano plot was constructed by plotting the fold change (FC; PROB/CON) of each metabolite against P-value. Metabolites with FC ≥ 1.2 or ≤ 0.83 having P-value ≤ 0.05 were considered to be differentially increased or decreased relative to CON, respectively.
The GeneGlobe Data Analysis Center (Qiagen, Valencia, USA) was used to analyze the gene expression data using the delta-delta-Ct (ΔΔCt) method [(CTgene of interest – CThousekeeping genes)PROB – (CTgene of interest – CThousekeeping genes)CON] with normalization of the raw data using the arithmetic mean of the 5 housekeeping genes. Genes with FC ≥ 1.2 or ≤ 0.83 having P-value ≤ 0.05 were considered to be differentially upregulated or downregulated relative to CON, respectively.
Results and Discussion
The effects of PROB supplementation on the growth performance of the steers are shown in Table 2. Compared with CON, dietary supplementation of PROB increased final body weight (P = 0.01) and ADG (P = 0.04) over the 42 d feeding trial. There was no effect on DMI (P = 0.24); consequently, there was a tendency for improved FE with PROB supplementation (P = 0.10). In contrast to previous studies (Elam et al. 2003; Baah et al., 2009) who reported improved growth of beef cattle during the first few days of feeding DFM, the overall growth efficiency of the steers fed supplemental PROB in this study was more pronounced during the last 21 d of this experiment (P = 0.02, ADG: 1.50 vs. 1.23 and P = 0.05, FE = 0.230 vs. 0.196). No treatment effects on growth measures (P > 0.05) were found during the first 21 d (days 1–21), though there were numerical increases in ADG and DMI for steers fed PROB diet. The mechanism by which certain DFMs and their fermentation products enhance the performance of animals is multifaceted and their effect is inconsistent due to supplementation with different multiple strains of organisms, doses, diets, and animal factors such as health and stress status (McAllister et al., 2011). The additive used in this study contained a mixture of S. cerevisiae, Bacillus subtilis, lactic acid bacteria (E. faecium, E. lactis, and L. casei), the fermentation products of these aforementioned microorganisms as well as those of Aspergillus oryzae and A. niger. Saccharomyces cerevisiae favors the activities of lactic acid-utilizing bacteria, via supply of micronutrients (vitamins and amino acids) essential for their growth, and fiber-degrading bacteria by increasing rumen pH (Martin and Nisbet, 1992; Callaway and Martin, 1997). Lactic acid bacteria produce lactic acid in the rumen, which promotes the growth of lactic acid-utilizing bacteria that can metabolize lactate to propionate, the major precursor for glucose synthesis in ruminants (Nagaraja and Titgemeyer, 2007; McAllister et al., 2011). Therefore, it is reasonable to speculate that the possible mechanism of action of PROB includes improving the energy status of the animals by increasing ruminal production of propionate, a major precursor for glucose synthesis in ruminants, which probably explains increased feed efficiency (FE) of the steers fed diet supplemented with PROB. Moreover, the fermentation products of these microorganisms such as peptides, amino acids, organic acids, vitamins, and enzymes are thought to be sources of beneficial metabolites for the growth of rumen bacteria, feed digestion, and modulation of immune function. Generally, DFM products containing several microorganisms including S. cerevisiae and lactic acid bacteria are fed to optimize the gut health of animals by competing against undesirable microorganisms, stimulating the production of beneficial bacteria, and increasing intestinal nutrient uptake (Beauchemin et al., 2003; Raeth-Knight et al., 2007).
Table 2.
Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products on the performance of steers during a 42 d receiving period1
| Item | CON | PROB | SEM | P-value |
|---|---|---|---|---|
| Initial weight, kg | 209 | 210 | 8.08 | 0.95 |
| Final weight, kg | 260b | 270a | 2.67 | 0.01 |
| Days 1–42 | ||||
| ADG, kg/d | 1.23b | 1.42a | 0.06 | 0.04 |
| DMI, kg/d | 5.86 | 6.11 | 0.15 | 0.24 |
| Feed efficiency | 0.209y | 0.232x | 0.01 | 0.10 |
| Days 1–21 | ||||
| ADG, kg/d | 1.24 | 1.32 | 0.10 | 0.56 |
| DMI, kg/d | 5.43 | 5.72 | 0.18 | 0.25 |
| Feed efficiency | 0.226 | 0.232 | 0.01 | 0.82 |
| Days 22–42 | ||||
| ADG, kg/d | 1.23b | 1.50a | 0.08 | 0.02 |
| DMI, kg/d | 6.30 | 6.50 | 0.17 | 0.41 |
| Feed efficiency | 0.196b | 0.230a | 0.01 | 0.05 |
1CON = control; PROB = a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products fed at 19 g/steer/d (PMI, Arden Hills, MN); SEM = standard error of mean; ADG = average daily gain; DMI = dry matter intake.
a,bWithin a row, treatment means with different superscripts differ, P ≤ 0.05.
x,yWithin a row, treatment means with different superscripts tend to differ, 0.05 < P ≤ 0.10.
The result of this study is consistent with previous studies that reported improved gains and/or FE of beef and dairy cattle fed DFMs containing either one or several combinations of microorganisms such as S. cerevisiae, E. faecium, Bacillus subtilis, E. lactis, and L. casei (Baah et al., 2009; Qiao et al., 2010; Chiquette et al., 2015; Oh et al., 2019). Conversely, other studies have reported no effects of DFM supplementation. Kenney et al. (2015) revealed that supplementation of a multistrain DFM (L. acidophilus and E. faecium, Pediococcus acidilacticii, L. brevis, and L. plantarum) at a rate of 109 cfu per steer/d had no effects on growth performance and FE of receiving beef steers; however, the authors reported increased growth efficiency during the early receiving period when the diet’s degradable intake protein was greater than the animals’ requirement, confirming that diet types can affect the response of animals to DFM supplementation. In another study, supplementation with S. cerevisiae-based DFM at 1010 cfu/g did not affect the ADG and FE of beef steers during a 56 d receiving period (Fink et al., 2014). Similarly, Cole et al. (1992) reported no effects of supplementation with a blend of S. cerevisiae and its fermentation product on the growth performance of feeder calves and lambs.
No treatment effects were found for serum biochemical indices on day 21 (data not shown). The result of the effects of PROB supplementation on the serum biochemical indices of the beef steers on day 42 is shown in Table 3. Serum enzyme activities and metabolites are commonly used as a diagnostic tool to assess the health and nutritional status of animals (Prvulovic et al., 2012). Supplementation with PROB increased (P < 0.05) the concentrations of serum total protein and globulin (Table 3). Increased concentrations of serum total protein and globulin in animals indicate improved health status (Bobbo et al., 2017) that is possibly associated with greater feed intake and/or increased nutrient supply; however, this was probably caused by increased nutrient absorption because DMI was not affected. Increased serum globulin concentration suggests a better immune status because it provides an assessment of circulating immunoglobulins in the blood (Kaneko, 1997). This result is consistent with the improved performance and FE of the steers fed PROB diet in this study. This agrees with a recent study (Sallam et al., 2019), which reported increased serum total protein in growing lambs fed diet supplemented with S. cerevisiae-based DFM.
Table 3.
Effects of a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products on the serum biochemical indices of beef steers during a 42 d receiving period1
| Item | CON | PROB | SEM | P-value |
|---|---|---|---|---|
| Albumin, g/dL | 3.51 | 3.57 | 0.05 | 0.41 |
| Alkaline phosphatase, U/L | 158.0 | 155.0 | 8.68 | 0.80 |
| Aspartate aminotransferase, U/L | 89.8 | 84.3 | 7.13 | 0.60 |
| Calcium, mg/dL | 10.3b | 10.6a | 0.10 | 0.01 |
| γ-glutamyltransferase, U/L | 13.8 | 13.9 | 1.07 | 0.96 |
| Total protein, g/dL | 6.57b | 7.01a | 0.04 | 0.01 |
| Globulin concentration, g/dL | 3.15b | 3.43a | 0.06 | 0.01 |
| Urea N, mg/dL | 9.97 | 9.82 | 0.45 | 0.82 |
| Phosphorus, mg/dL | 9.86 | 9.94 | 0.18 | 0.75 |
| Magnesium, mg/dL | 2.21 | 2.25 | 0.03 | 0.43 |
1CON = control; PROB = a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products fed at 19 g/steer/d (PMI, Arden Hills, MN); SEM = standard error of mean.
a,bWithin a row, treatment means with different superscripts differ, P ≤ 0.05.
Serum calcium concentration was greater (P = 0.01) in steers fed PROB diet (Table 3). It is well known that low serum Ca levels (<8.5 mg/dL; Oetzel and Goff, 1998) are indicative of hypocalcemia which causes depressed rumen and muscle function (Santos et al., 2016). Moreover, blood calcium plays a vital role in optimizing the immune system of animals. However, the serum calcium level observed in this study is within the normal physiological range for clinically healthy ruminants (8.5–11 mg/dL; Oetzel and Goff, 1998), therefore, the increased serum calcium concentration (10.6 vs. 10.3 mg/dL) observed may not be of biological significance. The concentrations of liver enzymes (alkaline phosphatase, aspartase aminotransferase, and γ-glutamyltransferase) were unaffected (P > 0.05), indicating no effect on liver function.
The results of the effect of PROB supplementation on the expression of immune-related genes are shown in Table 4. The immune status of receiving beef cattle is critical for their performance and adaptability to the feedlot environment (Galyean et al., 1999; Carroll and Forsberg, 2007). Improving the immune status of livestock by feeding immunomodulatory supplements such as DFMs is a popular management strategy to reduce health challenges and production losses (Branson et al., 2016). There was no change detected in the expression of the immune genes on day 0 (1.2 > FC > 0.83; P > 0.05; Supplementary Table S2). Compared with CON, dietary supplementation with PROB increased the expression of Toll-like receptor 1 (TLR1) on day 42 (P = 0.01; FC = 2.01), TLR2 on day 21 and day 42 (P = 0.02, FC = 1.46 and P = 0.01, FC = 1.82, respectively), and TLR6 on day 21 and day 42 (P = 0.01, FC = 1.27 and P = 0.01, FC = 1.52, respectively). Most DFMs modulate immune function via the activation of the TLR (Yan and Polk, 2011). The TLR genes are involved in detecting pathogen-associated molecular patterns such as bacterial lipopolysaccharides and peptidoglycans (Mogensen, 2009). This suggests that PROB supplementation enhanced the ability of the immune cells to recognize the presence of pathogens.
Table 4.
Effects of a blend of S. cerevisiae-direct-fed microbial and fermentation products on blood immune gene expression in beef steers during a 42 d receiving period1
| Day 21 | Day 42 | ||||
|---|---|---|---|---|---|
| Gene Name | Gene symbol | Fold change2 | P-value | Fold change2 | P-value |
| Toll-like receptor 2 | TLR2 | 1.46 | 0.02 | 1.82 | 0.01 |
| Toll-like receptor 6 | TLR6 | 1.27 | 0.01 | 1.52 | 0.01 |
| Tumor Necrosis Factor | TNF | 1.22 | 0.05 | 1.53 | 0.02 |
| Signal transducer and activator of transcription 6 | STAT6 | 1.32 | 0.02 | 1.22 | 0.04 |
| Caspase 1 | CASP1 | 1.38 | 0.04 | 1.29 | 0.02 |
| Interleukin 8 | IL-8 | 0.43 | 0.01 | ||
| Intercellular adhesion molecule 1 | ICAM1 | 1.29 | 0.02 | ||
| Retinoic acid-related orphan receptor C | RORC | 1.25 | 0.03 | ||
| Transcription factor T-bet | TBX21 | 1.44 | 0.01 | ||
| Toll-like receptor 1 | TLR1 | 2.01 | 0.01 | ||
| C-X-C motif chemokine receptor 3 | CXCR3 | 1.20 | 0.02 | ||
1CON = control; PROB = a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products fed at 19 g/steer/d (PMI, Arden Hills, MN).
2Fold change (relative to control) = 2-ΔΔCt = [(CTgene of interest – CTreference genes)PROB – (CTgene of interest – CTreference genes)CON]. Only genes with both fold change ≥ 1.2 or ≤ 0.83, relative to Control and P ≤ 0.05 are shown.
Dietary supplementation with PROB increased expression of Signal transducer and activator of transcription 6 on days 21 and 42 (P = 0.02, FC = 1.32 and P = 0.04, FC = 1.22, respectively), and those of Intercellular adhesion molecule 1 (P = 0.02, FC = 1.29), Retinoic acid-related orphan receptor C (P = 0.03, FC = 1.25), Transcription factor T-bet (P = 0.01, FC = 1.44), and C-X-C motif chemokine receptor 3 (P = 0.02, FC = 1.20) on day 42 (Table 4). These genes are either directly or indirectly involved in T-cell differentiation (Mesri et al., 1994; Zhu and Paul, 2008). Signal transducer and activator of transcription 6 enhance T helper 2 (Th2) cell differentiation (Zhu and Paul, 2008). C-X-C motif chemokine receptor 3 gene induces migration of Th1 cells (Qin et al., 1998). Intercellular adhesion molecule 1 gene enhances activation of T-cells by binding to lymphocyte function-associated antigen-1 (Mesri et al., 1994). Retinoic acid-related orphan receptor gamma, an isoform of Retinoic acid-related orphan receptor C gene, is a regulator of T helper 17 (Th17) cell differentiation (Ivanov et al., 2007). Transcription factor T-bet drives T helper 1 (Th1) immune response by enhancing the expression of interferon-gamma gene during the immune challenge (Nakayamada et al., 2011). Naive T cells have to be activated to become Th1, Th2, and Th17 cells in order to eliminate pathogens and protect the host (Reiner et al., 2007). T helper 1 cells are involved in defending the host against intracellular pathogens (Nakayamada et al., 2011) whereas Th2 and Th17 cells defend the host against extracellular pathogens (Ivanov et al., 2007; Zhu and Paul, 2008).
Dietary supplementation of PROB increased the expression of Tumor necrosis factor (TNF; P = 0.05, FC = 1.22 and P = 0.02, FC = 1.53, respectively) and Caspase1 (P = 0.04, FC = 1.38 and P = 0.02, FC = 1.29, respectively) on both days 21 and 42 (Table 4). After pathogen recognition by pattern recognition receptors, TNF activates neutrophils for microbicidal activity against the pathogen (Futosi et al., 2013). Caspase1 plays a critical role in promoting the maturation of proinflammatory cytokines, such as IL-1β and IL-18 (Latz et al., 2013). Taken together, these results suggest that dietary supplementation of PROB promotes the animal’s immune status towards intracellular and extracellular pathogens. Similar studies that have attempted to evaluate the effects of multi-strain DFM products on the expression of immune-related genes in cattle could not be found; however, a previous study in chicken showed that dietary supplementation of a DFM product containing of L. casei, L. acidophilus, Bifidobacterium thermophilum, and E. faecium increased the expression of an inflammatory cytokine, Interleukin-6 (Chichlowski et al. 2007). Some studies have reported the immunomodulatory effects of DFMs containing only S. cerevisiae and its fermentation products. For instance, Branson et al. (2016) reported that dietary supplementation of an additive containing S. cerevisiae induced the expression of TLR1, TLR4, and TLR6 in the whole blood of rodent. Studies in pigs and broilers showed that supplementation with S. cerevisiae fermentation products induced expression of TLR2 and TLR4 in the ileal tissue (Weedman et al., 2011; Yitbarek et al., 2012). In addition, previous studies have shown that the spores of Bacillus subtilis, one of the constituents of the DFM used in this study, could serve as a microparticle adjuvant that can instruct and promote a balanced Th1 and Th2 immune response without stimulating an allergic reaction (Kosaka et al., 1998; Barnes et al., 2007; Sun et al., 2010).
Expression of Interleukin 8 (IL-8) was upregulated (P = 0.01; FC = 0.43) in beef steers fed CON diet relative to PROB on day 21, but its expression was unaffected on day 42 (P > 0.05). Interleukin 8 promotes migration of neutrophils, basophils, and T-cells to the site of infection (Zhang and An, 2007); however, IL-8 is almost never expressed in non-induced healthy cells but only expressed in response to inflammatory stress such as bacterial and viral ligands (Hoffman et al., 2002). Santos et al. (2002) reported elevated IL-8 expression in calves exposed to Salmonella infection. Inflammatory stress associated with separation from the mother and exposure to a new environment has been reported to be evident till 35 d after weaning (Lynch et al., 2011). Therefore, it is reasonable to speculate that elevated IL-8 expression is probably an indication of greater inflammatory stress in steers fed CON diet relative to PROB diet. It is important to note that this study reported immune gene expression in whole blood that does not entirely indicate functional changes. Therefore, these results should be interpreted with caution and further research study is needed to confirm the effects of PROB diet on the expression of genes related to innate and adaptive immunity using specific immune cells such as neutrophils, rather than whole blood.
Several studies have shown the usefulness of metabolomics to assess the health and nutritional status of livestock (Saleem et al., 2012; Ogunade et al., 2018; Yang et al., 2018). This study utilized LC-MS/MS-based targeted metabolomics to quantify 139 plasma metabolites. No treatment effects were detected (P > 0.10) for all the metabolites (data not reported), except plasma glucose concentration which was greater in steers fed PROB diet on day 42 (5.23 vs. 4.72 mM, SE = 0.13, P = 0.01). Increased plasma glucose confirms the improved energy status of the steers, which supports the possible mode of action of PROB.
In order to evaluate a greater number of metabolites, we utilized dansylation CIL/LC-MS-based untargeted metabolomics to assess thousands of amine and phenol-containing submetabolome in the plasma samples collected on day 42. Dansylation CIL/LC-MS-based untargeted metabolomics has a significant advantage over conventional LC-MS-based metabolomics method due to its ability to detect thousands of metabolites (Mung and Li, 2019). In addition, the use of isotope labeling in CIL/LC-MS allows for accurate relative quantification compared to semi-quantification analysis obtained by using conventional LC-MS (Mung and Li, 2019).
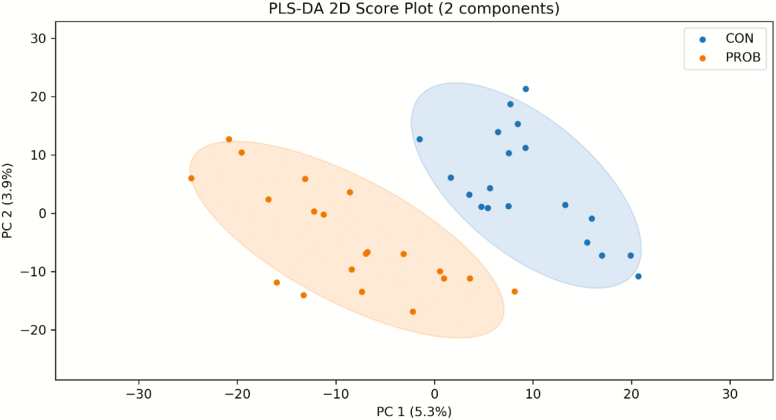
Using dansylation CIL/LC-MS analysis, a total number of 2,605 unique peak pairs (representing different compounds) were detected (Supplementary Table S3). Approximately 88.1% (2,296 peak pairs) of the 2,605 unique peak pairs were positively identified or putatively matched. Among them, 127 peak pairs were positively identified in tier 1 (CIL library; Supplementary Table S4), 102 peak pairs were putatively identified with high-confidence in tier 2 (LI library; Supplementary Table S5), and 2067 peak pairs were matched in tier 3 (MCID library). The PLS-DA scores plot shows a clear separation between the CON and PROB samples (Figure 1), indicating that PROB supplementation altered the plasma metabolome of the beef steers. The permutation test result of P = 0.02 confirms the validity of the PLS-DA model (Supplementary Figure S2).
Figure 1.
Partial least squares discriminant analysis (PLS-DA) scores plot of the two treatments. CON = control; PROB = a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products fed at 19 g/steer/d (PMI, Arden Hills, MN).
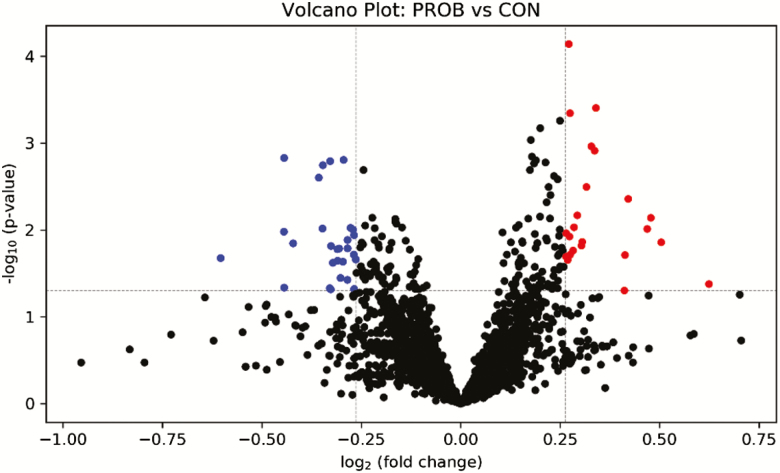
The results of the volcano plot analysis showed that 28 peak pairs (FC > 1.2, P ≤ 0.05) were increased by PROB supplementation, whereas 24 peak pairs (FC < 0.83, P ≤ 0.05) were reduced (Figure 2). Among those that were differentially altered, 2 peak pairs were positively identified in tier 1 using CIL Library, 5 peak pairs were putatively identified with high confidence in tier 2 using LI Library, and 37 peak pairs were matched using MCID library (Supplementary Table S6). The metabolites that were positively and putatively identified with high confidence are shown in Table 5. Two metabolites (5-methylcytosine and indoleacrylic acid) were differentially increased (FC > 1.2, P ≤ 0.05) in steers fed PROB diet, whereas five metabolites (5-aminopentanoic acid, 4-methylaminobutyrate, 3,4-dihydroxyphenylethylene glycol, trans-2,3-dihydroxycinnamate, and 2-hydroxy-3-(4-hydroxyphenyl)propenoate) were differentially reduced (FC < 0.83, P ≤ 0.05).
Figure 2.
Volcano plot showing the differential metabolites. FC > 1.2, P-value ≤ 0.05 (in red): significantly increased; FC < 0.83, P-value ≤ 0.05 (in blue): significantly reduced relative to CON. CON = control; PROB = a blend of Saccharomyces cerevisiae-based direct-fed microbial and fermentation products fed at 19 g/steer/d (PMI, Arden Hills, MN).
Table 5.
Identified peak pairs (tier 1 and tier 2) that were affected by dietary supplementation of a blend of S. cerevisiae-based direct-fed microbial and fermentation products1
| Item | Normalized RT | FC | P-value | Identification level |
|---|---|---|---|---|
| 5-Methylcytosine | 529 | 1.20 | 0.02 | Tier 2 |
| Indoleacrylic acid | 1,247 | 1.26 | 0.01 | Tier 1 |
| 5-Aminopentanoic acid | 524 | 0.72 | 0.01 | Tier 1 |
| 4-Methylaminobutyrate | 554 | 0.79 | 0.01 | Tier 2 |
| 3,4-Dihydroxyphenylethyleneglycol | 636 | 0.80 | 0.01 | Tier 2 |
| Trans-2,3-Dihydroxycinnamate | 842 | 0.81 | 0.01 | Tier 2 |
| 2-Hydroxy-3-(4-hydroxyphenyl)propenoate | 872 | 0.81 | 0.02 | Tier 2 |
1Normalized RT (retention time) shows the corrected retention time of the peak pair with Universal RT Calibrant data. FC: fold change relative to Control; P-value was calculated from student’s t-test; Tier 1 - Positive Identification (CIL Library); Tier 2 - High Confidence Putative Identification (LI Library). Only metabolites with both fold change ≥ 1.2 or ≤ 0.83, relative to Control and P ≤ 0.05 are shown.
5-methylcytosine is a product of DNA methylation that is essential for optimum function of several biological processes, such as regulation of gene expression in mammals (Moore et al., 2013). An oxidation product of 5-methylcytosine, 5-hydroxymethylcytosine, has been reported to promote cytokine production during Th cell differentiation (Ansel et al., 2006; Ichiyama et al., 2015). The gut microbiota influences host immunity by promoting the production of tryptophan metabolite, indoleacrylic acid, which has been reported to enhance intestinal epithelial barrier function and prevent inflammatory responses (Wikoff et al., 2009; Wlodarska et al., 2017), indicating that PROB diet promotes an immunity-promoting function of the gut microbiota (Larsson et al., 2011). The fact that the plasma concentrations of these two metabolites (5-methylcytosine and indoleacrylic acid) were differentially increased in steers fed PROB diet suggests a better immune status of the steers and is consistent with the results of the immune gene expression in this study.
The decreased concentrations of 5-aminopentanoic acid, a degradation product of bacterial catabolism of lysine (Rohles et al., 2016), 3,4-dihydroxyphenylethylene glycol and 2-hydroxy-3-(4-hydroxyphenyl)propenoic acid, products of tyrosine metabolism (Hartman et al., 1955), and trans-2,3-dihydroxycinnamate, a product of phenylalanine metabolism (Ziarrusta et al., 2018) suggest that PROB supplementation possibly altered the metabolisms of lysine, tyrosine, and phenylalanine. In fact, tyrosine and phenylalanine metabolic pathways are related because phenylalanine is hydroxylated into tyrosine in cells via phenylalanine hydroxylase (Li et al., 2011). Since the relative concentrations of these amino acids (phenylalanine, tyrosine, and lysine) were not affected by treatment, the biological significance of the changes in their metabolites is unknown. Further studies are needed to determine the roles of these metabolites as well as those that were putatively matched using MCID library on the health and performance of animals.
There was a tendency for lower fecal pH in steers fed PROB diet (P = 0.08; 6.46 vs. 6.62; SE = 0.06). An increased microbial fermentation in the large intestine causes greater production of organic acids, which consequently lowers fecal pH (Gressley et al., 2011). Lower pH indicates that PROB diet possibly increased post-ruminal microbial fermentation of carbohydrates that may have escaped ruminal fermentation and small intestinal digestion, thereby resulting to greater total-tract nutrient digestion. Certain DFMs containing Lactobacillus spp. and Bacillus subtilis have been shown to alter microbial fermentation in the hindgut because these microorganisms are able to survive low pH in the small intestine (Jenny et al., 1991; Abu-Tarbouch et al., 1996). For instance, an increased abundance of fecal lactobacilli was reported in dairy calves fed a DFM containing lactobacilli (Abu-Tarbouch et al., 1996). Future studies should examine the effect of PROB supplementation on hindgut fermentation and microbiome.
Conclusion
This study demonstrated that PROB diet improved the growth and FE of newly weaned beef steers during the receiving period. The increased growth and FE was supported by increased expression of genes responsible for promoting the animal’s immune response toward intracellular and extracellular pathogens. In addition, plasma untargeted metabolomic profiling of the steers fed PROB diet revealed an increase in the concentration of metabolites involved in protecting the animals against inflammation. Fecal pH of steers fed PROB diet tended to be lower relative to CON, suggesting an increased microbial fermentation in the hindgut. The results of this study showed that, under the condition of this experiment, a blend of multi-strain DFMs and their fermentation products can improve the performance and health of newly weaned beef cattle during the receiving period. Further studies are needed to evaluate the effects of PROB on rumen fermentation, microbiota, and metabolome to better understand its underlying mechanisms of action.
Supplementary Material
Footnotes
The study was funded by PMI. Additional funding support was provided by the National Institute of Food and Agriculture (Evans Allen project 1008985).
Literature Cited
- Abu-Tarboush H. M., Al-Saiady M. Y., and Keir El-Din A. H.. . 1996. Evaluation of diet containing lactobacilli on performance, fecal coliform, and lactobacilli of young dairy calves. Anim. Feed Sci. Tech. 57:39–49. doi: 10.1016/0377-8401(95)00850-0 [DOI] [Google Scholar]
- Ansel K. M., Djuretic I., Tanasa B., and Rao A.. . 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24:607–656. doi: 10.1146/annurev.immunol.23.021704.115821 [DOI] [PubMed] [Google Scholar]
- AOAC International 2000. Official methods of analysis. 17th ed Arlington (VA): AOAC International. [Google Scholar]
- Arthington J. D., Cooke R. F., Maddock T. D., Araujo D. B., Moriel P., Dilorenzo N., and Lamb G. C.. . 2013. Effects of vaccination on the acute-phase protein response and measures of performance in growing beef calves. J. Anim. Sci. 91:1831–1837. doi: 10.2527/jas.2012-5724 [DOI] [PubMed] [Google Scholar]
- Baah J., Wang Y., and McAllister T. A.. . 2009. Impact of a mixed culture of Lactobacillus casei and L. lactis on in vitro ruminal fermentation and the growth of feedlot steers fed barley-based diets. Can. J. Anim. Sci. 89:263–271. doi.org/10.4141/CJAS08117 [Google Scholar]
- Barnes A. G., Cerovic V., Hobson P. S., and Klavinskis L. S.. . 2007. Bacillus subtilis spores: a novel microparticle adjuvant which can instruct a balanced Th1 and Th2 immune response to specific antigen. Eur. J. Immunol. 37:1538–1547. doi: 10.1002/eji.200636875 [DOI] [PubMed] [Google Scholar]
- Beauchemin K. A., Yang W. Z., Morgavi D. P., Ghorbani G. R., Kautz W., and Leedle J. A.. . 2003. Effects of bacterial direct-fed microbials and yeast on site and extent of digestion, blood chemistry, and subclinical ruminal acidosis in feedlot cattle. J. Anim. Sci. 81:1628–1640. doi: 10.2527/2003.8161628x [DOI] [PubMed] [Google Scholar]
- Bobbo T., Fiore E., Gianesella M., Morgante M., Gallo L., Ruegg P. L., Bittante G., and Cecchinato A.. . 2017. Variation in blood serum proteins and association with somatic cell count in dairy cattle from multi-breed herds. Animal 11:2309–2319. doi: 10.1017/S1751731117001227 [DOI] [PubMed] [Google Scholar]
- Branson J. A., McLean D. J., Forsberg N. E., and Bobe G.. . 2016. Yeast-containing feed additive alters gene expression profiles associated with innate immunity in whole blood of a rodent model. Innate Immun. 22:249–256. doi: 10.1177/1753425916640326 [DOI] [PubMed] [Google Scholar]
- Callaway E. S., and Martin S. A.. . 1997. Effects of a Saccharomyces cerevisiae culture on ruminal bacteria that utilize lactate and digest cellulose. J. Dairy Sci. 80:2035–2044. doi: 10.3168/jds.S0022-0302(97)76148-4 [DOI] [PubMed] [Google Scholar]
- Carroll J. A., and Forsberg N. E.. . 2007. Influence of stress and nutrition on cattle immunity. Vet. Clin. North Am. Food Anim. Pract. 23:105–149. doi: 10.1016/j.cvfa.2007.01.003 [DOI] [PubMed] [Google Scholar]
- Chichlowski M., Croom J., McBride B. W., Daniel L., Davis G., and Koci M. D.. . 2007. Direct-fed microbial PrimaLac and salinomycin modulate whole-body and intestinal oxygen consumption and intestinal mucosal cytokine production in the broiler chick. Poult. Sci. 86:1100–1106. doi: 10.1093/ps/86.6.1100 [DOI] [PubMed] [Google Scholar]
- Chiquette J., Lagrost J., Girard C. L., Talbot G., Li S., Plaizier J. C., and Hindrichsen I. K.. . 2015. Efficacy of the direct-fed microbial Enterococcus faecium alone or in combination with Saccharomyces cerevisiae or Lactococcus lactis during induced subacute ruminal acidosis. J. Dairy Sci. 98:190–203. doi: 10.3168/jds.2014-8219 [DOI] [PubMed] [Google Scholar]
- Cole N. A., Purdy C. W., and Hutcheson D. P.. . 1992. Influence of yeast culture on feeder calves and lambs. J. Anim. Sci. 70:1682–1690. doi: 10.2527/1992.7061682x [DOI] [PubMed] [Google Scholar]
- Deters E. L., Stokes R. S., Genther-Schroeder O. N., and Hansen S. L.. . 2018. Effects of a Saccharomyces cerevisiae fermentation product in receiving diets of newly weaned beef steers. I. Growth performance and antioxidant defense. J. Animal Sci. 96:3897–905. doi.org/10.1093/jas/sky246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff G. C., and Galyean M. L.. . 2007. Board-invited review: recent advances in management of highly stressed, newly received feedlot cattle. J. Anim. Sci. 85:823–840. doi: 10.2527/jas.2006-501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elam N. A., Gleghorn J. F., Rivera J. D., Galyean M. L., Defoor P. J., Brashears M. M., and Younts-Dahl S. M.. . 2003. Effects of live cultures of Lactobacillus acidophilus (strains NP45 and NP51) and Propionibacterium freudenreichii on performance, carcass, and intestinal characteristics, and Escherichia coli strain O157 shedding of finishing beef steers. J. Anim. Sci. 81:2686–2698. doi: 10.2527/2003.81112686x [DOI] [PubMed] [Google Scholar]
- Fink D. N., Ribeiro F. R. B., Burdick N. C., Parr S. L., Carroll J. A., Young T. R., Bernhard B. C., Corley J. R., Estefan A. G., Rathmann R. J., and Johnson B. J.. . 2014. Yeast supplementation alters the performance and health status of receiving cattle. Prof. Anim. Sci. 30:333–341. doi.org/10.15232/S1080-7446(15)30125-X [Google Scholar]
- Futosi K., Fodor S., and Mócsai A.. . 2013. Reprint of Neutrophil cell surface receptors and their intracellular signal transduction pathways. Int. Immunopharmacol. 17:1185–1197. doi: 10.1016/j.intimp.2013.11.010 [DOI] [PubMed] [Google Scholar]
- Galyean M. L., and Hubbert M. E.. . 1995. Effects of season, health, and management on feed intake by beef cattle. In: Owens F. N., editor. Symposium: Intake by Feedlot Cattle Oklahoma Agric. Exp. Stn., Stillwater, OK: Oklahoma State University, p. 226–234. [Google Scholar]
- Galyean M. L., Perino L. J., and Duff G. C.. . 1999. Interaction of cattle health/immunity and nutrition. J. Anim. Sci. 77:1120–1134. doi: 10.2527/1999.7751120x [DOI] [PubMed] [Google Scholar]
- Gressley T. F., Hall M. B., and Armentano L. E.. . 2011. Ruminant Nutrition Symposium: productivity, digestion, and health responses to hindgut acidosis in ruminants. J. Anim. Sci. 89:1120–1130. doi: 10.2527/jas.2010-3460 [DOI] [PubMed] [Google Scholar]
- Hall J. B., Laarman A. H., Reynolds M. K., and Smith W. K.. . 2018. Performance of backgrounding steers fed diets containing monensin or a lactobacillus fermentation product. Transl. Anim. Sci. 2:130–133. doi: 10.1093/tas/txy035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman W. J., Akawie R. I., and Clark W. G.. . 1955. Competitive inhibition of 3, 4-dihydroxyphenylalanine (DOPA) decarboxylase in vitro. J. Biol. Chem. 216:507–529. [PubMed] [Google Scholar]
- Hoffmann E, Dittrich-Breiholz O., Holtmann H., and Kracht M.. 2002. Multiple control of interleukin-8 gene expression. J. Leukoc. Biol. 72:847–855. doi.org/10.1189/jlb.72.5.847 [PubMed] [Google Scholar]
- Huan T., and Liang Li. . 2015. Quantitative metabolome analysis based on chromatographic peak reconstruction in chemical isotope labeling liquid chromatography mass spectrometry. Anal. Chem. 87:7011–7016. doi.org/10.1021/acs.analchem.5b01434 [DOI] [PubMed] [Google Scholar]
- Ichiyama K., Chen T., Wang X., Yan X., Kim B. S., Tanaka S., Ndiaye-Lobry D., Deng Y., Zou Y., Zheng P., . et al. 2015. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 42:613–626. doi: 10.1016/j.immuni.2015.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I. I., Zhou L., and Littman D. R.. . 2007. Transcriptional regulation of Th17 cell differentiation. Semin. Immunol. 19:409–417. doi: 10.1016/j.smim.2007.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenny B. F., Vandijk H. J., and Collins J. A.. . 1991. Performance and fecal flora of calves fed a Bacillus subtilis concentrate. J. Dairy Sci. 74:1968–1973. doi: 10.3168/jds.S0022-0302(91)78364-1 [DOI] [PubMed] [Google Scholar]
- Kaneko J. J. 1997. Serum proteins and the dysproteinemias. In: Kaneko J. J., editor. Clinical biochemistry of domestic animals. 5th ed London (UK): Academic Presss; p. 117–138. doi: 10.1016/B978-012396305-5/50006-3 [DOI] [Google Scholar]
- Kenney N. M., Vanzant E. S., Harmon D. L., and McLeod K. R.. . 2015. Effect of direct-fed microbials on utilization of degradable intake protein in receiving steers. J. Anim. Sci. 95:93–102. doi: 10.4141/cjas-2014-021 [DOI] [PubMed] [Google Scholar]
- Kosaka T., Maeda T., Nakada Y., Yukawa M., and Tanaka S.. . 1998. Effect of Bacillus subtilis spore administration on activation of macrophages and natural killer cells in mice. Vet. Microbiol. 60:215–225. doi: 10.1016/s0378-1135(97)00102-8 [DOI] [PubMed] [Google Scholar]
- Larsson J. M., Karlsson H., Crespo J. G., Johansson M. E., Eklund L., Sjövall H., and Hansson G. C.. . 2011. Altered O-glycosylation profile of MUC2 mucin occurs in active ulcerative colitis and is associated with increased inflammation. Inflamm. Bowel Dis. 17:2299–2307. doi: 10.1002/ibd.21625 [DOI] [PubMed] [Google Scholar]
- Latz E., Xiao T. S., and Stutz A.. . 2013. Activation and regulation of the inflammasomes. Nat. Rev. Immunol. 13:397–411. doi: 10.1038/nri3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Ilangovan U., Daubner S. C., Hinck A. P., and Fitzpatrick P. F.. . 2011. Direct evidence for a phenylalanine site in the regulatory domain of phenylalanine hydroxylase. Arch. Biochem. Biophys. 505:250–255. doi: 10.1016/j.abb.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li R., Zhou J., Zuniga A., Stanislaus A. E., Wu Y., Huan T., Zheng J., Shi Y., Wishart D. S., . et al. 2013. MyCompoundID: using an evidence-based metabolome library for metabolite identification. Anal. Chem. 85:3401–3408. doi: 10.1021/ac400099b [DOI] [PubMed] [Google Scholar]
- Lynch E. M., McGee M., Doyle S., and Earley B.. . 2011. Effect of post-weaning management practices on physiological and immunological responses of weaned beef calves. Irish J. Agr. Food Res. 50:161–174. https://www.jstor.org/stable/41549249 [Google Scholar]
- Marchal J., Dorieux O., Haro L., Aujard F., and Perret M.. . 2012. Characterization of blood biochemical markers during aging in the Grey Mouse Lemur (Microcebus Murinus): impact of gender and season. BMC Vet. Res. 8:211. doi: 10.1186/1746-6148-8-211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. A., and Nisbet D. J.. . 1992. Effect of direct-fed microbials on rumen microbial fermentation. J. Dairy Sci. 75:1736–1744. doi: 10.3168/jds.S0022-0302(92)77932-6 [DOI] [PubMed] [Google Scholar]
- McAllister T., Beauchemin K. A., Alazzeh A., Baah J., Teather R., and Stanford K.. . 2011. Review: the use of direct fed microbials to mitigate pathogens and enhance production in cattle. Can. J. Anim. Sci. 91:193–211. doi: 10.4141/cjas10047 [DOI] [Google Scholar]
- McDonald A., Andersen P., Defoor P., and Botts R.. . 2005. Direct-fed microbials improve health, performance of cattle. Feedstuffs 77:12–13. [Google Scholar]
- Mesri M., Liversidge J., and Forrester J. V.. . 1994. ICAM-1/LFA-1 interactions in T-lymphocyte activation and adhesion to cells of the blood-retina barrier in the rat. Immunology 83:52–57. [PMC free article] [PubMed] [Google Scholar]
- Mogensen T. H. 2009. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 22:240–73, Table of Contents. doi: 10.1128/CMR.00046-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore L. D., Le T., and Fan G.. . 2013. DNA methylation and its basic function. Neuropsychopharmacology 38:23–38. doi: 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mung D., and Li L.. . 2017. Development of chemical isotope labeling LC-MS for milk metabolomics: comprehensive and quantitative profiling of the Amine/Phenol submetabolome. Anal. Chem. 89:4435–4443. doi: 10.1021/acs.analchem.6b03737 [DOI] [PubMed] [Google Scholar]
- Mung D., and Li L.. . 2019. Chemical isotope labeling liquid chromatography mass spectrometry for investigating acute dietary effects of cow milk consumption on human urine metabolome. J. Food Drug Anal. 27:565–574. doi: 10.1016/j.jfda.2018.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraja T. G., and Titgemeyer E. C.. . 2007. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J. Dairy Sci. 90(Suppl 1):E17–E38. doi: 10.3168/jds.2006-478 [DOI] [PubMed] [Google Scholar]
- Nakayamada S., Kanno Y., Takahashi H., Jankovic D., Lu K. T., Johnson T. A., Sun H. W., Vahedi G., Hakim O., Handon R., . et al. 2011. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35:919–931. doi: 10.1016/j.immuni.2011.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetzel G. R., and Goff J. P.. . 1998. Milk fever (parturient paresis) in cows, ewes, and doe goats. In: Current Veterinary Therapy 4: Food Animal Practice. Philadelphia (PA): WB Saunders Co, pp. 215–218. [Google Scholar]
- Ogunade I. M., Jiang Y., Adeyemi J., Oliveira A., Vyas D., and Adesogan A. T.. . 2018. Biomarker of aflatoxin ingestion: 1H NMR-based plasma metabolomics of dairy cows fed aflatoxin B1 with or without sequestering agents. Toxins 10:545. doi.org/10.3390/toxins10120545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh J., Harper M., Melgar A., Compart D. M. P., and Hristov A. N.. . 2019. Effects of Saccharomyces cerevisiae-based direct-fed microbial and exogenous enzyme products on enteric methane emission and productivity in lactating dairy cows. J. Dairy Sci. 102:6065–6075. doi: 10.3168/jds.2018-15753 [DOI] [PubMed] [Google Scholar]
- Plaizier J. C., Danesh Mesgaran M., Derakhshani H., Golder H., Khafipour E., Kleen J. L., Lean I., Loor J., Penner G., and Zebeli Q.. . 2018. Review: enhancing gastrointestinal health in dairy cows. Animal 12(s2):s399–s418. doi: 10.1017/S1751731118001921 [DOI] [PubMed] [Google Scholar]
- Prvulovic D., Kosarcic S., Popovic M., Dimitrijev D., and Gruborlaj G.. . 2012. The influence of hydrated aluminosilicate on biochemical and haematological blood parameters, growth performance and carcass traits of pigs. J. Anim. Vet. Adv. 11:134–140. doi.org/10.3923/javaa.2012.134.140 [Google Scholar]
- Qiao G. H., Shan A. S., Ma N., Ma Q. Q., and Sun Z. W.. . 2010. Effect of supplemental Bacillus cultures on rumen fermentation and milk yield in Chinese Holstein cows. J. Anim. Physiol. Anim. Nutr. (Berl). 94:429–436. doi: 10.1111/j.1439-0396.2009.00926.x [DOI] [PubMed] [Google Scholar]
- Qin S., Rottman J. B., Myers P., Kassam N., Weinblatt M., Loetscher M., Koch A. E., Moser B., and Mackay C. R.. . 1998. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Invest. 101:746–754. doi: 10.1172/JCI1422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raeth-Knight M. L., Linn J. G., and Jung H. G.. . 2007. Effect of direct-fed microbials on performance, diet digestibility, and rumen characteristics of Holstein dairy cows. J. Dairy Sci. 90:1802–1809. doi: 10.3168/jds.2006-643 [DOI] [PubMed] [Google Scholar]
- Rai V., Yadav B., and Lakhani G. P.. . 2013. Applications of probiotic and prebiotic in animal production: a review. Environ. Ecol. 31:873–876. [Google Scholar]
- Reiner S. L., Sallusto F., and Lanzavecchia A.. . 2007. Division of labor with a workforce of one: challenges in specifying effector and memory T cell fate. Science 317:622–625. doi: 10.1126/science.1143775 [DOI] [PubMed] [Google Scholar]
- Rohles C. M., Gießelmann G., Kohlstedt M., Wittmann C., and Becker J.. . 2016. Systems metabolic engineering of Corynebacterium glutamicum for the production of the carbon-5 platform chemicals 5-aminovalerate and glutarate. Microb. Cell Fact. 15:154. doi: 10.1186/s12934-016-0553-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem F., Ametaj B. N., Bouatra S., Mandal R., Zebeli Q., Dunn S. M., and Wishart D. S.. . 2012. A metabolomics approach to uncover the effects of grain diets on rumen health in dairy cows. J. Dairy Sci. 95:6606–6623. doi: 10.3168/jds.2012-5403 [DOI] [PubMed] [Google Scholar]
- Sallam M. A., Kholif A. E., Amin K. A., Adel N. M., ElDin N., Attia M. F. A., Matloup O. H., and Anele U. Y.. . 2019. Effects of microbial feed additives on feed utilization and growth performance in growing Barki lambs fed diet based on peanut hay. Anim. Biotech. 30:1–8. doi: 10.1080/10495398.2019.1616554 [DOI] [PubMed] [Google Scholar]
- Santos J. E. P., Martinez N., Vieira-Neto A., Lopera C., and Nelson C.. . 2016. Dietary manipulations and interventions to improve calcium metabolism. Florida Ruminant Nutrition Symposium 27th Annual Meeting, Gainesville, Florida, pp 140–149. [Google Scholar]
- Santos R. L., Zhang S., Tsolis R. M., Bäumler A. J., and Adams L. G.. . 2002. Morphologic and molecular characterization of Salmonella typhimurium infection in neonatal calves. Vet. Pathol. 39:200–215. doi: 10.1354/vp.39-2-200 [DOI] [PubMed] [Google Scholar]
- Sirois P. K., Reuter M. J., Laughlin C. M., and Lockwood P. J.. . 1994. A method for determining macro and micro elements in forages and feeds by inductively coupled plasma atomic emission spectrometry. Spectroscopist. 3:6–9. [Google Scholar]
- Sun P., Wang J. Q., and Zhang H. T.. . 2010. Effects of Bacillus subtilis Natto on performance and immune function of preweaning calves. J. Dairy Sci. 93:5851–5855. doi: 10.3168/jds.2010-3263 [DOI] [PubMed] [Google Scholar]
- Tricarico J. M., Abney M. D., Galyean M. L., Rivera J. D., Hanson K. C., McLeod K. R., and Harmon D. L.. . 2007. Effects of a dietary Aspergillus oryzae extract containing alpha-amylase activity on performance and carcass characteristics of finishing beef cattle. J. Anim. Sci. 85:802–811. doi: 10.2527/jas.2006-427 [DOI] [PubMed] [Google Scholar]
- Uyeno Y., Shigemori S., and Shimosato T.. . 2015. Effect of probiotics/prebiotics on cattle health and productivity. Microbes Environ. 30:126–132. doi: 10.1264/jsme2.ME14176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Soest P. J., Robertson J. B., and Lewis B. A.. . 1991. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 74:3583–3597. doi: 10.3168/jds.S0022-0302(91)78551-2 [DOI] [PubMed] [Google Scholar]
- Weedman S. M., Rostagno M. H., Patterson J. A., Yoon I., Fitzner G., and Eicher S. D.. . 2011. Yeast culture supplement during nursing and transport affects immunity and intestinal microbial ecology of weanling pigs. J. Anim. Sci. 89:1908–1921. doi: 10.2527/jas.2009-2539 [DOI] [PubMed] [Google Scholar]
- Wikoff W. R., Anfora A. T., Liu J., Schultz P. G., Lesley S. A., Peters E. C., and Siuzdak G.. . 2009. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. U. S. A. 106:3698–3703. doi: 10.1073/pnas.0812874106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wlodarska M., Luo C., Kolde R., d’Hennezel E., Annand J. W., Heim C. E., Krastel P., Schmitt E. K., Omar A. S., Creasey E. A., . et al. 2017. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 22:25–37.e6. doi: 10.1016/j.chom.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., and Li L.. . 2012. Determination of total concentration of chemically labeled metabolites as a means of metabolome sample normalization and sample loading optimization in mass spectrometry-based metabolomics. Anal. Chem. 84:10723–10731. doi: 10.1021/ac3025625 [DOI] [PubMed] [Google Scholar]
- Yan F., and Polk D. B.. . 2011. Probiotics and immune health. Curr. Opin. Gastroenterol. 27:496–501. doi: 10.1097/MOG.0b013e32834baa4d [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Dong G., Wang Z., Wang J., Zhang Z., and Liu J.. . 2018. Rumen and plasma metabolomics profiling by UHPLC-QTOF/MS revealed metabolic alterations associated with a high-corn diet in beef steers. PLoS ONE 13(11): e0208031. doi: 10.1371/journal.pone.0208031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yitbarek A., Echeverry H., Brady J., Hernandez-Doria J., Camelo-Jaimes G., Sharif S., Guenter W., House J. D., and Rodriguez-Lecompte J. C.. . 2012. Innate immune response to yeast-derived carbohydrates in broiler chickens fed organic diets and challenged with Clostridium perfringens. Poult. Sci. 91:1105–1112. doi: 10.3382/ps.2011-02109 [DOI] [PubMed] [Google Scholar]
- Zhang G., Dervishi E., Dunn S. M., Mandal R., Liu P., Han B., Wishart D. S., and Ametaj B. N.. . 2017. Metabotyping reveals distinct metabolic alterations in ketotic cows and identifies early predictive serum biomarkers for the risk of disease. Metabolomics 13:43. doi: 10.1007/s11306-017-1180-4 [DOI] [Google Scholar]
- Zhang J. M., and An J.. . 2007. Cytokines, inflammation, and pain. Int. Anesthesiol. Clin. 45:27–37. doi: 10.1097/AIA.0b013e318034194e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., and Paul W. E.. . 2008. CD4 T cells: fates, functions, and faults. Blood 112:1557–1569. doi: 10.1182/blood-2008-05-078154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziarrusta H., Mijangos L., Picart-Armada S., Irazola M., Perera-Lluna A., Usobiaga A., Prieto A., Etxebarria N., Olivares M., and Zuloaga O.. . 2018. Non-targeted metabolomics reveals alterations in liver and plasma of gilt-head bream exposed to oxybenzone. Chemosphere 211:624–631. doi: 10.1016/j.chemosphere.2018.08.013 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.