Abstract
AUTS2 was originally discovered as the gene disrupted by a translocation in human twins with Autism spectrum disorder, intellectual disability, and epilepsy. Since that initial finding, AUTS2-linked mutations and variants have been associated with a very broad array of neuropsychiatric disorders, sugg esting that AUTS2 is required for fundamental steps of neurodevelopment. However, genotype-phenotype correlations in this region are complicated, because most mutations could also involve neighboring genes. Of particular interest is the nearest downstream neighbor of AUTS2, GALNT17, which encodes a brain-expressed N-acetylgalactosaminyltransferase of unknown brain function. Here we describe a mouse (Mus musculus) mutation, T(5G2;8A1)GSO (abbreviated 16Gso), a reciprocal translocation that breaks between Auts2 and Galnt17 and dysregulates both genes. Despite this complex regulatory effect, 16Gso homozygotes model certain human AUTS2-linked phenotypes very well. In addition to abnormalities in growth, craniofacial structure, learning and memory, and behavior, 16Gso homozygotes display distinct pathologies of the cerebellum and hippocampus that are similar to those associated with autism and other types of AUTS2-linked neurological disease. Analyzing mutant cerebellar and hippocampal transcriptomes to explain this pathology, we identified disturbances in pathways related to neuron and synapse maturation, neurotransmitter signaling, and cellular stress, suggesting possible cellular mechanisms. These pathways, coupled with the translocation’s selective effects on Auts2 isoforms and coordinated dysregulation of Galnt17, suggest novel hypotheses regarding the etiology of the human “AUTS2 syndrome” and the wide array of neurodevelopmental disorders linked to variance in this genomic region.
Keywords: Neurological development, Autism, Translocation, Gene regulation, Syndromic phenotypes
Autism Susceptibility Candidate 2 (AUTS2) was originally identified as the gene disrupted by reciprocal translocation in human twins with Autism spectrum disorder (ASD), intellectual disability (ID), and epilepsy (Sultana et al. 2002). Since that first report, hundreds of mutations or variants in the AUTS2 genomic region have been identified, most of which correspond to translocations, duplications, deletions or other types of genomic rearrangements. Detailed analyses of genotype-phenotype associations have revealed ID and developmental delay as core features of the “AUTS2 syndrome”. However, other phenotypes, including the stereotypic behaviors and abnormal social development characteristic of ASD, feeding difficulties, reduced postnatal growth, epilepsy, microcephaly and characteristic craniofacial abnormalities are also observed in substantial subsets of patients (Beunders et al. 2013; Beunders et al. 2016). In addition to the syndromic phenotypes associated with genome rearrangements, AUTS2-linked single nucleotide polymorphisms (SNPs) have been associated with Attention Deficit Hyperactive Disorder (Elia et al. 2010), dyslexia (Girirajan et al. 2011), schizophrenia (Zhang et al. 2014a), bipolar disorder (Hamshere et al. 2009), depression (Myung et al. 2015), language disorders (Chen et al. 2017; Amarillo et al. 2014) and addiction (Kapoor et al. 2013; Chen et al. 2016; Chen et al. 2013). Association with this very wide range of psychiatric disorders suggests that mutations in this region disrupt the functions of genes with central roles in neurodevelopment.
Most attention has focused on the AUTS2 gene itself, a complex locus spanning more than 1.5 million base pairs (Mbp) that generates distinct protein isoforms from several alternative promoters. Patients carrying two rare exonic deletions in the longest AUTS2 isoform have been studied in depth, and display central AUTS2 syndrome features including ID and developmental delay as well as ASD, feeding difficulties, growth deficits, microcephaly, and craniofacial abnormalities, supporting the importance of AUTS2 in these phenotypes (Beunders et al. 2015). Studies in Drosophila (Schumann et al. 2011), zebrafish (Oksenberg et al. 2013) and mice (Hori et al. 2015; Hori et al. 2014; Gao et al. 2014) have further demonstrated important roles for Auts2 in neurogenesis and neuron differentiation. In particular, the mouse studies have revealed that cytoplasmic AUTS2 protein isoforms control the formation of lamellipodia and filopodia, thus impacting neuronal migration and neurite formation (Hori et al. 2014), whereas nuclear AUTS2 serves as a co-factor in a novel PRC1gene regulatory complex (Gao et al. 2014). Global Auts2 loss-of-function (LOF) is lethal in homozygous form, associated with grossly disrupted cortical layering and neurite extension deficits (Hori et al. 2014). On the other hand, heterozygotes are viable with decreased exploratory activity, reduced anxiety-related behaviors, developmental delay and memory impairments (Hori et al. 2015). Furthermore, mice homozygous for a conditional knockout affecting only the longest Auts2 isoform are viable but small, with abnormal vocalization and delayed development of righting reflexes (Gao et al. 2014).
Despite these important clues and an intensive focus on human patients over the past several years, many questions regarding genotype-phenotype relationships in this genomic region remain unanswered. The complexity added by the existence of multiple AUTS2 isoforms is clearly a significant part of this puzzle. Additionally, most mutations correspond to genomic rearrangements that could possibly affect the expression of neighboring genes. In this regard, the immediate downstream neighbor of AUTS2, GALNT17 (previously named WBSCR17), is of particular interest. GALNT17 encodes a brain-expressed N-acetylgalactosaminyl transferase (GalNAcT) that, like AUTS2, regulates the formation of lamellipodia, with impact on cell adhesion and motility (Nakayama et al. 2012). However, Galnt17 mutants have not been examined and virtually nothing is known about the expression or functions of this GalNacT protein in neurons or the brain.
Here we present the characterization of a novel laboratory mouse mutation identified in a genetic screen, T(5G2;8A1)GSO (or 16Gso), a reciprocal translocation breaking between Auts2 and Galnt17 that, presumably through the disruption of regulatory interactions, leads to the mis-expression of both genes. We investigated the behavioral and neuropathological phenotypes of mutant animals and integrated these data with gene expression analysis to develop new hypotheses regarding the functions of both genes, and new clues to the neurodevelopmental correlates of AUTS2 syndrome phenotypes.
Material and Methods
Animals and Tissue Collection
The generation of 16Gso mice has been described previously (Elso et al. 2008; Weisner 2015). The genotypes studied here were in mice maintained on a C57BL/6 X C3H/He F1(B6C3) genetic background. In order to efficiently collect animals of each genotype for testing, 16Gso homozygotes (simplified as 16GsoT/+) were generated by breeding homozygous parents; heterozygotes were collected separately in crosses involving 16Gso homozygotes (abbreviated as 16GsoT/T) and wild type (WT) mice. Animals were housed under standard conditions (12h light/dark cycle, group-housed). This study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health. The protocol was approved by the Institutional Animal Care and Use Committee of the University of Illinois (Animal Assurance Number: A3118-01; approved IACUC protocol number 18240).
Mapping and sequencing the 16Gso translocation breakpoint
Metaphase chromosome FISH was performed as previously described (Elso et al. 2004). BACs from the RP23 and RP24 BAC libraries were used as template for FISH probes. The breakpoint region was narrowed by determining BACs that were above, below or split by the breakpoint region by co-hybridization with a control BAC used to mark proximal regions of chr5 and chr8. The analysis identified two overlapping chr5 BAC clones (RP23-7a8 and RP23-283H6) and two chr8 clones (RP24-234k3 and RP24-234a5) that spanned breakpoint sites. We chose ∼500 kb intervals surrounding the regions of BAC overlap for hybrid selection and deep sequencing. Both hybrid selection and sequencing were performed by Otogenetics Inc. (Atlanta), and approximately 30X depth of paired-end Illumina sequence reads from two 16GsoT/T and two WT animals for each region was returned for our analysis. We mapped reads to the mm9 genome build using Bowtie software (Langmead et al. 2009), and identified sets of paired ends that mapped to different chromosomes. Of those reads, we found clusters of end-pairs with one end mapping to a region of chr5 and the other end mapping to chr8; these were the only endpairs to map repeatedly to any genomic region. Finally, we developed primers from regions of chr5 and 8 covered by the clustered clones, and used PCR to generate and sequence chr5;8 and chr8;5 breakpoint spanning regions. New primer sets were designed around the breakpoint site for reliable PCR genotyping.
Mouse Phenotypic tests
WT, 16GsoT/+ and 16GsoT/T pups were tested for developmental delay at postnatal day 5 (P5), P6, P7, P8 and P9, in the form of the righting reflex; the same animals were tested for freezing, repetitive behavior and novel object recognition tasks at 6-8 weeks old. The weight for each animal was also measured at P5, P7, P9 and P35.
Righting reflex:
righting reflex test was modified from a previous study (Castelhano-Carlos et al. 2010). The pups were flipped and the time for them to turn back to their original position was recorded. Different degrees of performance on turning back were assigned to specific score for each day of the testing, score 0 = failed to turn back within 15 sec; score 1 = pups turned back between 10 to 15 sec; score 2 = pups successfully made right turn within 10 sec for the first time; score 3 = pups successfully made right turn within 10 sec for the second time.
Freezing behavior:
the freezing time was measured when the mouse was placed in a novel environment, a 35 cm × 30 cm paper box, for 10 min.
Repetitive behavior (grooming):
The average grooming time per episode in 10 min interval was measured, when mice were placed in an empty box as used in freezing tests.
Novel object recognition (NOR):
NOR tasks were followed as previously described (Botton et al. 2010). Tasks included three sessions: habituation, training and retention. In the habituation session, the mouse was placed in an empty 35 cm × 30 cm paper box for 10 min, and returned to its home cage. The training session was held 24 hr after the habituation session. In the training session, two novel objects (Legos were used in this experiment) were placed in the box, and the exploratory behavior was monitored for 10 min. In the retention session, which was carried out 24 hr after the training session, one of the familiar Legos from training session was exchanged for a new Lego, and the mouse was allowed to explore both objects for 5 min. Exploratory behavior was defined as sniffing or touching the objects. The exploratory index was calculated as exploration time on novel object to total exploration time, as a percentage.
Home cage activity:
Home cage activity was also monitored for adult animals between three and seven months of age. Activity was monitored continually for 7 days, during which animals were singly housed, using overhead cameras under white or red light and Topscan software to track animal movement.
Skull Measurements:
mouse skulls were cleaned of skin and viscera, and the snout was measured from the distal tip to the anterior notch of the frontal process, lateral to the infraorbital fissure. Measurements were also taken from the distal tip of the snout to the bregma, the midline intersection of the frontal and parietal bones, and to the intersection of the interparietal bones and occipital bones at the base of the skull.
Immunohistochemistry
Isolated tissues were fixed in fresh 4% paraformaldehyde, embedded in paraffin, and cut into 5-6 µm sections using a Leica RM2155 microtome and Super Plus charged slides. Cerebellum (CB) slices were sectioned into sagittal sections, and hippocampus (HC) sections were in coronal orientation. For IHC, the primary antibody was used at optimal ratio in Antibody Diluent Reagent Solution (Life Technologies) and incubated overnight at 4°. Slides were then washed and incubated with secondary antibody (1:200) in antibody diluent for 1 hr at room temperature. Slides were further washed and stained with Hoecsht 3342 and scanned under Zeiss LSM 710 Confocal Microscope for further analysis. Primary antibodies: MAP2 (Abcam ab5392, 1:300), AUTS2 (Sigma HPA000390, 1:200), WBSCR17 (LSBio LS-B14501, 1:250), Calbindin D-28K (Sigma C9848, 1:300), Calretinin (Santa Cruz sc-365989, 1:300), CNPase (Millipore MAB326, 1:250), DRD2 (Millipore AB5084P, 1:200), GFAP (Invitrogen 180063, 1:300), GAD 65/67 (Santa Cruz sc-365180, 1:200), Tbr2 (Abcam ab23345, 1:100), cFos (Abcam ab190298, 1:200); Secondary antibodies (Thermo-Fisher Scientific): Goat anti-Mouse IgG (Alexa Fluor 488, A11001), Goat anti-Rabbit IgG (Alexa Fluor 594, A11012), Donkey anti-Rat IgG (Alexa Fluor 594, A21209).
Histopathology
Paraffin-embedded slides were stained with hematoxylin and eosin (H&E). For CB, sections were taken from the midline, spaced at 600 micrometer intervals in adults, 300 micrometer intervals in P14 animals, and 200 micrometer intervals in P7 animals. Purkinje cells (PCs) were counted by two independent researchers who were blinded to the genotypes of the animals (n = 3 of each genotype and age), and who were trained to identify cells based on size and morphology. For the hippocampal cell density experiment, the cells were counted in randomly assigned 90 µm × 90 µm squares in dentate gyrus, CA3, and CA1 on 20X magnification, from slides taken from dorsal regions of the HC (n = 3 mice of each genotype and age). The thickness of the cell layer was measured through NanoZoomer Digital Pathology software.
RNA-seq analysis
CB and HC tissues were collected from P14 and P35 WT and 16GsoT/T, and RNA was purified using Trizol followed by 45 min of RNase-free DNase I treatment (NEB) and RNA Clean & Concentrator Kit (Zymo Research). The quality of RNA was tested by Agilent BioAnalyzer, and Illumina libraries were generated using the KAPA Stranded mRNA-Seq kit with MRNA Capture Beads (Kapa Biosystems). Sequencing was performed on Illumina Hi-Seq 2000 sequencers. Reads were mapped using Tophat2 software (Kim et al. 2013), transcripts quantified using HT-seq count (Anders et al. 2015), and differential expression determined using the edgeR package (Robinson et al. 2010) as we have previously described (Saul et al. 2017). Differentially expressed genes determined with fdr ≤ 0.05 were analyzed for functional enrichment using DAVID (Huang da et al. 2009).
Quantitative RT-PCR (qRT-PCR)
RNA was prepared from snap frozen tissues and purified using Trizol (Invitrogen) extraction followed by 45 min of RNase-free DNase I treatment (NEB) and RNA Clean & Concentrator Kit (Zymo Research). cDNA was then prepared using a total of 2 ug RNA in a 20 µl reaction with Random Primer Mix (NEB) and M-MuLV Reverse Transcriptase (NEB). qRT-PCR was then carried out using custom-designed primers sets specific for Auts2 isoform transcripts (Table S2) with Power SYBR Green PCR master mix (Applied Biosystems) in a final volume of 10 μl. Reactions were run on an ABI 7900 thermocycler for 40 cycles with an annealing temperature of 60°. Expression values were normalized relative to the Pgk1 control in each sample and compared using standard methods (Livak and Schmittgen 2001).
Data availability
All data and resources reported in this manuscript are publicly available. Sequencing data reported in this paper has been deposited into the GEO database under accession number GSE132684. The results of sequence analysis are presented in Table S2 and full listing of enriched functional categories from DAVID analysis in Table S3. Oligonucleotide primers used for qRT-PCR are list in Table S4. Data from behavioral tests and represented as graphs in this manuscript are presented in Table S1, and H&E stains of 16Gso T/T and WT cortex are presented in Fig. S1. Sequence confirming the expression of novel Auts2 isoform T6, which is identical to that in an ENSEMBL model and mouse ESTs as reported, is also provided in Supplemental Text. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9887540.
Results
Genetic and molecular analysis of the 16Gso mutation
The 16Gso mutation arose in a genetic screen of the offspring of males exposed to bis-acrylamide, a treatment that efficiently generates reciprocal translocations in late stage spermatocytes (Stubbs et al. 1997). Karyotype analysis confirmed that the animals carried a reciprocal translocation affecting chromosome 5 (chr5) band G2 and chromosome 8 (chr8) band A1 (Stubbs et al. 1997). Heterozygotes for the 16Gso translocation (hereafter abbreviated as 16GsoT/+ for simplicity) were overtly normal, and although most 16Gso homozygotes (16GsoT/T) survived to adulthood and were fertile, they were generally smaller than wild type (WT) littermates, and were otherwise frankly abnormal in appearance and behavior.
To map the 16Gso breakpoint more precisely, we used methods described previously (Elso et al. 2004; Robinson et al. 2005; Elso et al. 2013; Bolt et al. 2014), selecting bacterial artificial chromosome (BAC) clones from chr5 and chr8 to use as probes in fluorescent in situ hybridization (FISH). This narrowed the breakpoint to BAC overlap regions of approximately 130 kb on chr5 (chr5:131,812,581-131,945,233 in NCBI build 37) and ∼193kb on chr8 (chr8:12,244,884-12,437,596). Next, we enriched DNA spanning approximately 500 kb around both breakpoint regions with hybrid-selection based-methods and focused on paired ends mapping to different chromosomes - with one end on chr5 and the other on chr8 – to pinpoint the location of the breakpoint. Finally, we used PCR to confirm, isolate and sequence trans-breakpoint DNA fragments to reveal the structure of the translocation breakpoints.
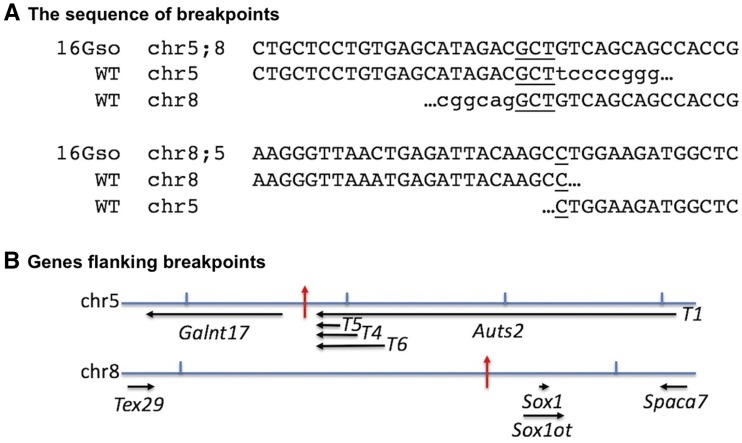
The sequence of these breakpoint junction fragments confirmed the fusion of chr5 and chr8 sequences in 16Gso DNA (Figure 1A). The 16Gso breakpoint is located about 70kb upstream of Sox1 on chr8 (mm9 coordinates chr8:12325249); on chr5 it is situated between Galnt17 and Auts2, ∼72 kb upstream and 60 kb downstream of those genes, respectively (chr5:131854998) (Figure 1B, red arrows). Like other germline translocations we have sequenced (Elso et al. 2008; Elso et al. 2013; Bolt et al. 2014; Elso et al. 2004; Robinson et al. 2005), the T(8;5) and T(5;8) 16Gso breakpoints are simple in structure, with small deletions (6 and 8 bp on chr 5 and chr8, respectively) and short (1-3 nucleotide) duplications at the breakpoint fusion sites, the classic signature of non-homologous end-joining repair. Aside from these short deletions and duplications at the breakpoint site, we saw no evidence of further rearrangement in the DNA sequence or in extensive FISH and genomic PCR across both breakpoint regions.
Figure 1.
Sequence and genomic map location of the 16Gso translocation breakpoints. (A) Sequences obtained by PCR across the 16Gso breakpoint aligned with the WT sequence at the same genomic sites. Sequences that align between WT chr5 and chr8 and the mutant DNA sequences are shown in caps, while lower case letters represent WT sequences beyond the breakpoint site. The alignment revealed the deletions on both translocated chromosomes (underlined), involving one copy of a homologous triplet (GCT) shared by the two WT chromosomes in the translocated chr5;8, and deletion of a single C at the chr8;5 breakpoint site. (B) The breakpoint-flanking sequences mapped between Galnt17 and Auts2 genes on chr5, and upstream of Sox1 on chr8 (red arrows on each chromosome map). Genes are drawn to scale within each map; arrows indicate direction of transcription for each gene. For Auts2, transcripts T1, T4, T5, and T6 are labeled at the locations of their promoters. Above both maps, vertical lines demark a genomic distance of 500 kb.
Initial phenotypic characterization of 16Gso mutants
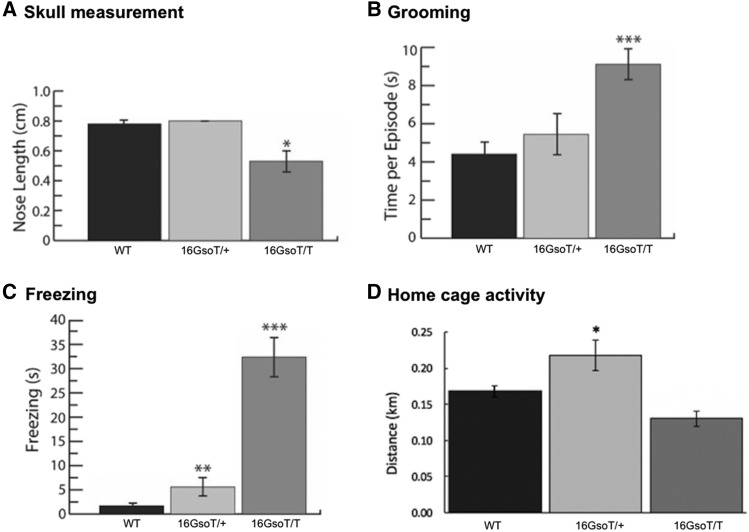
16GsoT/T mice were selected for study because they displayed several obvious physical and behavioral phenotypes, including a relatively shortened skull due specifically to reduced length of the snout (Figure 2A; Table 1; Table S1). In addition, 16GsoT/T adults displayed stereotypic behaviors, such as repetitive self-grooming (Figure 2B). Mature 16GsoT/T animals froze in place for extended periods in response to even minor stressors such as handling or after placement in novel environments, suggesting increased levels of anxiety (Figure 2C). Furthermore, some older 16GsoT/T adults (>6 months old) displayed obvious seizures under stressful (e.g., testing) conditions. In contrast, younger 16GsoT/T animals never displayed this seizure phenotype, and none of the many 16GsoT/+ mice we have observed have exhibited seizures at any age.
Figure 2.
Phenotypic features in 16GsoT/T mutants. (A) Skull measurement: the length from base to tip of the nose was significantly decreased in 16GsoT/T (n = 3) compared to WT (n = 4) and 16GsoT/+ mice (n = 3). (B) Repetitive grooming behavior: the average grooming time for each episode within a 10 min experimental period was significantly increased in 16GsoT/T (n = 9) compared to WT (n = 11) and 16GsoT/+ (n = 12). (C) Freezing behavior: 16GsoT/T (n = 14) froze more in the novel environment compared to WT (n = 10) or 16GsoT/+ (n = 14) in 10 min experimental period. (D) Home cage activity: the average distance traveled (quantified through Topscan software) was increased in 16GsoT/+ (n = 10), but there was no significant difference between WT (n = 9) and 16GsoT/T (n = 5) animals. Skull measurement: ANOVA, *P < 0.05. Grooming and freezing: t- Test, **P < 0.01; ***P < 0.001. Home cage activity: Tukey’s test, *P < 0.05.
Table 1. Phenotypes detected in 16GsoT/T mice compared with those identified in published KO mutants, AUTS2 syndrome, and ASD human patients.
| Phenotype | 16Gso T/T | Sox1 −/−a,b | Auts2−/−c | Auts2-/+d | Auts2-T1−/−e | Related phenotypes in AUTS2 or ASD patientsf |
|---|---|---|---|---|---|---|
| Small size, low postnatal growth | + | — | nr | + | + | Low birth weight, short stature |
| Delayed righting | + | nr | . | + | nr | Developmental delay |
| Learning and memory deficits | + | — | . | + | nr | Intellectual disability |
| Cortical layering defects | — | — | + | - | nr | Intellectual disability |
| Abnormal skull/nose length | + | — | nr | nr | nr | Dysmorphic features |
| Freezing behavior, anxiety | + | + | . | nr | nr | Adverse response to novelty, anxiety |
| Reduced exploratory activity | + | nr | . | + | nr | Intellectual disability |
| Abnormal nesting | + | nr | . | nr | nr | Abnormal social behavior |
| Repetitive grooming behaviors | + | — | . | nr | nr | Stereotypic behavior, obsessive compulsive behavior |
| Seizures | + | + | . | nr | nr | Epilepsyf,g |
| Abnormal Purkinje Cell differentiation | + | — | . | nr | nr | ASDh |
| Abnormal Purkinje Cell dendrites | + | — | . | nr | nr | ASDh |
| Purkinje Cell loss | + | — | . | nr | nr | ASDh |
| Abnormal numbers and packing of hippocampal neurons | + | — | . | nr | nr | ASDh |
in each column denotes that the phenotype has been observed; - denotes that the phenotype was investigated but not detected; nr = not reported; . = not applicable (animals die around the time of birth).
Data taken from aMalas et al., 2003
Hori et al., 2016
Summarized from (Beunders et al. 2013) unless otherwise noted
In contrast to these abnormal behaviors under stress, 16GsoT/T mice displayed mostly normal behaviors when left undisturbed in their home cage, with no differences from WT animals in overall levels of activity throughout the day (Figure 2D). In their home cages, the mutants can be observed grooming cagemates and sleeping in pairs in a manner not significantly different than WT mice. For the above-mentioned phenotypes and others described below, we tested males and females separately and noted some quantitative and age-dependent differences, but males and females followed very similar trends. The single exception was repetitive self-grooming, which male but not female 16GsoT/T animals displayed (Table S1).
16GsoT/+ mice were normal for most of the 16GsoT/T phenotypes we documented, supporting a primarily recessive inheritance. However, the fact that 16GsoT/+ male (but not female) animals display a slight increase in freezing behavior (Figure 2C) and home cage activity (Figure 2D) indicated the possibility of subtle semi-dominant effects.
Phenotypic comparisons to mutations in specific breakpoint-spanning genes
We next sought to test 16Gso mutants for phenotypes already associated with the breakpoint-flanking genes. Galnt17 mutants have not been characterized to date in any species, and phenotypes related to Galnt17 mutations could not be considered here. However, knockout (KO) mutations of both Sox1 and Auts2 have been reported. Sox1 KO homozygotes display seizures beginning at late juvenile stages (age 4-6 weeks), and show evidence of freezing as well as other anxiety-related behaviors – all suggestive of phenotypes already noted for 16GsoT/T mice (Table 1). However, the behavior of Sox1 KO animals is otherwise not different from wild type animals (Malas et al. 2003).
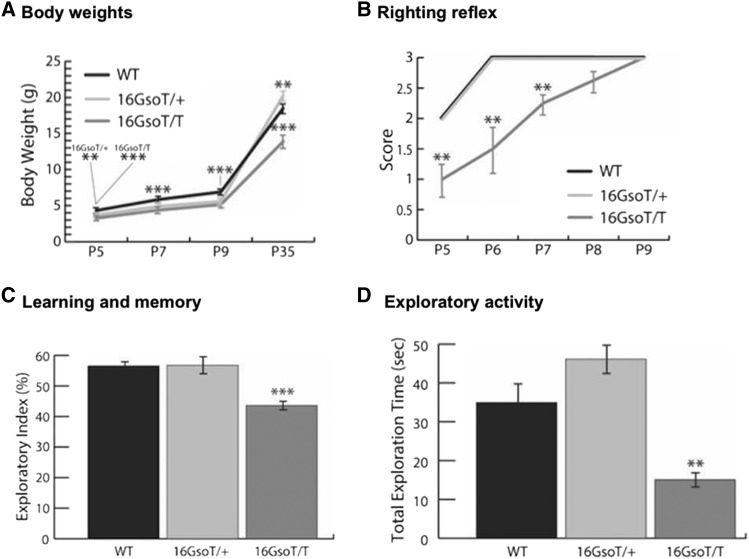
Additionally, Auts2 KO mice – including one affecting all transcript isoforms (Hori et al. 2014) and another specifically targeting the longest Auts2 isoform (referred to here as T1, Figure 1; (Gao et al. 2014)) have been characterized in some detail. These KO animals display growth retardation, delayed righting reflex, and learning deficits, all of which are relevant to human AUTS2 syndrome phenotypes (Table 1). To quantitate these phenotypes in 16Gso mutants, we completed three additional tests: (1) body weight and length as a function of age, (2) developmental timing of the righting reflex, and (3) novel object recognition (NOR) test, which utilizes the natural exploratory behavior of mice to quantify learning and object recognition. The results confirmed that 16GsoT/T animals were significantly smaller than WT controls from juvenile stages through adulthood as measured by both weight (Figure 3A) and overall body length (Table S1). Furthermore, we found a significant delay in development of the righting reflex in 16GsoT/T pups (Figure 3B). In the NOR test, we found that 16GsoT/T mice showed significant deficits in learning and memory (Figure 3C), and lower levels of object exploration during the training phase compared to WT controls (Figure 3D). Except in the case of body weight at juvenile stages, 16GsoT/+ animals were not significantly different than WT confirming predominantly recessive inheritance (Figure 3A-D). Thus, 16GsoT/T mice are similar to Auts2 KO heterozygotes in displaying small body size, delayed righting, lower levels of exploratory activity, and deficits in the NOR test, suggesting a possible role for Auts2 loss-of-function in these phenotypes.
Figure 3.
16GsoT/T mutants show lighter weight, developmental delay, impaired learning and memory, and reduced exploratory activity. In each case, 16GsoT/T and 16GsoT/+ were compared to WT age-matched controls. (A) Body weight was measured at P5, P7, P9 and P35 for each genotype (WT n = 9-17; 16GsoT/T n = 9-19; 16GsoT/+ n = 11-21 depending on age). Both 16GsoT/+ and 16GsoT/T weighed significantly less than WT at P5, P7, and P9. At P35, 16GsoT/T was lighter than WT, but 16GsoT/+ animals weighed slightly more than WT. (B) Developmental delay: The 16GsoT/T mice (n = 14) showed delayed development of the righting reflex, while there was no difference between WT (n = 14) and 16GsoT/+ (n = 14) males. (C) Novel object recognition test revealed that 16GsoT/T males (n = 8) showed impaired learning and memory, while WT (n = 8) and 16Gs T/+ animals (n = 9) have normal learning and memory. (D) Exploratory activity was determined in the training session of the NOR test, and showed significantly lower levels of object exploration by 16GsoT/T mice. The number of animals tested in this experiment are identical to that for (C). P-values determined with the t-Test, **P < 0.01; ***P < 0.001.
Normal cortical layering, but subtle hippocampal abnormalities and clear cerebellar pathology in 16Gso adults
To ask whether these 16GsoT/T behavioral phenotypes might be associated with obvious brain pathologies, we examined hematoxylin and eosin (H&E)-stained sectioned brains of adult 16GsoT/T and age-matched WT animals. We found marked differences in the structure of hippocampus (HC) and the cerebellum (CB) of the 16GsoT/T mice, two brain regions closely tied to ASD, ID, and neuropsychiatric pathology. In contrast to Auts2 KO animals (Hori et al. 2014), we observed no obvious pathologies in layering or structure of the cortex (Fig. S1), and no obvious malformations elsewhere in mutant brains. Details of the cerebellar and hippocampal pathologies are detailed further as follows.
Abnormal development of cerebellar Purkinje cells and Bergmann Glia, and age-dependent Purkinje cell loss:
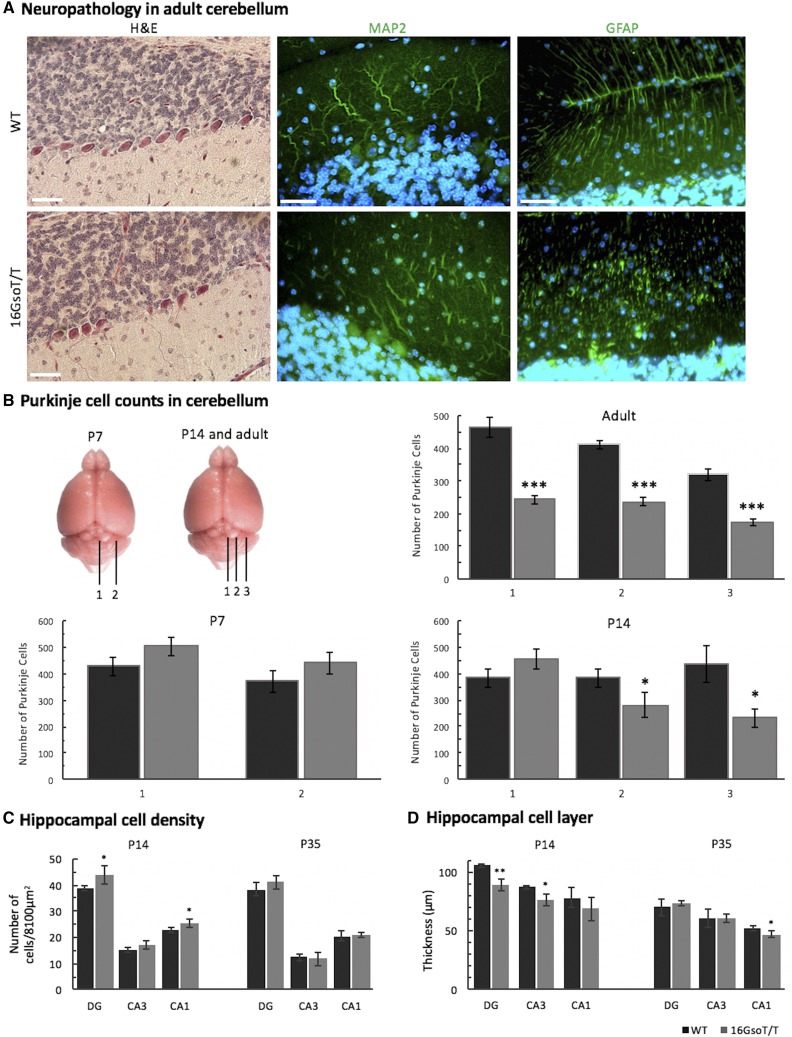
First and most obvious, as revealed by H&E-stained sections, mature 16GsoT/T adults had fewer cerebellar Purkinje cells (PCs), ranging from 52–58% of the number found in WT mice; the cells were also irregularly spaced throughout the PC layer (Figure 4A, B). The adjacent granule cell layer was identical in thickness and cell morphology to WT animals (data not shown). To determine the developmental time course of this cerebellar pathology, we then examined H&E stained sections at postnatal days 7 (P7), 14 (P14) and adult. We saw no difference in PC numbers or structure at P7. However, small but significant reductions in PC number were observed in lateral sections of the 16GsoT/T CB at P14, and became severe in adult (Figure 4B).
Figure 4.
Cerebellar and hippocampal pathology in 16GsoT/T mutants. (A) H&E staining showed that PCs are fewer in number and irregularly spaced in adult 16GsoT/T CB as compared to WT; MAP2 staining reveals that the 16GsoT/T PCs have sparse, irregularly branching dendritic trees, and GFAP staining highlights structural abnormalities of the Bergmann glia. (B) The number of PCs was quantified at different positions throughout the CB at different stages of development (P7, P14 and adult) in WT (black) and 16GsoT/T mutants (gray). Position 1 is at the midline, 1 and 2 each are 200 microns apart at P7, and 1, 2 and 3 each are 300 microns apart at P14, and 600 microns apart in adult. (C) Cell density in DG, CA3 and CA1 was quantified in WT (black) and 16GsoT/T (gray) at P14 and P35. 16GsoT/T has significantly increased cell number in DG and CA1 at P14. (D) Thickness of cell layer was measured in DG, CA3 and CA1 in WT (black) and 16GsoT/T (gray) at P14 and P35. n, WT = 16GsoT/T = 3 males at P7 and P14; WT = 16GsoT/T = 4-5 males in adult. ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001. Scale bar = 50 μm.
Immunohistochemistry (IHC) with the MAP2 antibody showed PCs in WT adults to be aligned neatly along the interface between granule cell and molecular layers, extending elaborate dendritic trees with thick primary branches. In contrast, 16GsoT/T PCs were not aligned, and lacked the thick primary branches, with more disorganized dendritic trees (Figure 4A). In contrast to WT animals, PCs in 16GsoT/T mice showed fragmented, disrupted MAP2 staining, indicating abnormal dendritic morphology. Additionally, the GFAP antibody showed radial glial processes of Bergmann glia, which form a scaffold for PC dendrites, also appeared abnormal in 16GsoT/T CB, lacking the organized, linear processes seen in WT sections (Figure 4A). This pathology was first observed at P14 (not shown), but also documented at later stages (Figure 4A). No pathology was documented in 16GsoT/+ mice at any developmental stage (not shown).
Together these data indicated that in 16GsoT/T mice, PCs are generated in normal numbers and migrate to their final location, but do not develop normally and are lost as the animals age. This pathology is coupled to abnormalities of the Bergmann glia, the development of which is intimately linked to that of PCs (Chrobak and Soltys 2017; Yamada and Watanabe 2002).
As mentioned, other than microcephaly, brain pathology has not been reported for human AUTS2 patients. However, PC loss is the most common brain pathology observed in postmortem analysis of ASD patients (Amaral et al. 2008; Pierce and Courchesne 2001; Varghese et al. 2017); PC loss is also associated with behavioral rigidity (Dickson et al. 2010) and repetitive behaviors in mice (Martin et al. 2010). Therefore, the 16GsoT/T PC phenotype is highly relevant to the clinical manifestations of ASD. However, this pathology has not been reported to be associated with any of the breakpoint-linked genes. Specifically, CB has been examined and is normal in Sox1 KO homozygotes (Sottile et al. 2006) and cerebellar abnormalities have not been reported for Auts2 KO animals of either type (Table 1). Focused studies with these animals or with Galnt17 mutants could potentially resolve this discrepancy.
Abnormal cell numbers and densities in the hippocampal dentate gyrus and Ammon’s horn:
In the P14 16GsoT/T HC, we identified a second set of cellular pathologies with excellent potential to explain certain 16GsoT/T behavioral traits. These included abnormalities in cell layer size, cell numbers, and neuron cell density across the dentate gyrus (DG) and Ammon’s Horn region cornu ammonis 1 and 3 (CA1, CA3). Each region displayed distinct structural phenotypes and different trajectories over the postnatal time course (Figure 4C, D).
Specifically, in P14 16GsoT/T mutants, DG granule cells (GCs) and CA1 pyramidal cells (PyCs) were unusually densely packed; in contrast, cell density was normal in the mutant CA3. Furthermore, although the DG and CA3 layers were both thinner in P14 mutants compared to WT, the CA1 cell layer was of normal size. Putting these data together, the mutant DG had normal numbers of abnormally packed GCs, while mutant CA1 had significantly increased numbers of abnormally packed cells, and mutant CA3 contained significantly decreased numbers of normally spaced PyCs than WT mice.
This hippocampal pathology shifted as the animals matured. At P35, the 16GsoT/T DG and CA3 regions were not distinguishable from WT controls. However, while cell density had normalized in the mutant CA1, the PyC layer was significantly thinner than that of WT adult animals. Therefore, although other HC regions had normalized over time, in 16GsoT/T mutants there was a clear shift from increased numbers of CA1 PyC neurons at juvenile stages, to significantly decreased numbers of CA1 PyC in adults (Figure 4C, D).
These features are reminiscent of human neuropathology documented in association with ID, ASD, epilepsy, and other AUTS2-linked disorders (Varghese et al. 2017; Goodman et al. 2014; Otten and Meeter 2015; McKinnon et al. 2009). In the case of ASD, dense packing of hippocampal neurons has been linked to aberrant timing of neuronal maturation (Varghese et al. 2017; Amaral et al. 2008). Like the cerebellar abnormalities, these pathologies may be unique to 16Gso mutants since Sox1 KO homozygotes display normal hippocampal structure (Malas et al. 2003), and hippocampal defects have not been reported for either Auts2 KO mice or human patients to date.
Global gene expression reveals region- and time-selective down-regulation of the Auts2 and Galnt17 genes
With the goal of gaining a clearer picture of genetic factors involved in these neuropathologies, we carried out global analysis of gene expression (RNA-seq) in CB and HC of 16GsoT/T mice and age-matched WT controls. For both regions, we examined gene expression at P14, when pathology is first observed in both brain regions, and at P35 to examine longer-term effects. Because male and female phenotypes were documented to be very similar (see above; Table S1) we focused all expression analyses on males (Table S2). In both brain regions, we saw differential expression of Galnt17 and Auts2 at one or both time points. In contrast, mis-expression of Sox1 was not detected in either experiment. Results from each brain region are detailed below.
Cerebellum - dysregulation of extracellular matrix and signals of cellular stress:
In the P14 CB of 16GsoT/T animals, we saw significant down-regulation of Galnt17 (-2.07 fold change) with no detectable effects on Auts2 or Sox1 (Table S2). Focusing more globally on the most highly differentially expressed genes (DEGs) in P14 CB (fdr < 0.05, ≥ 1.5 absolute value of FC), up-regulated DEGs were enriched in gene ontology (GO) functional categories related to corticotropin response and transcriptional activation, mostly related to immediate early genes (IEGs), such as Fos, that mark activated neurons (Table 2; Table S3). P14 down-regulated DEGs were not enriched in any functional categories, but included genes associated with AUTS2-linked neurological disorders such as Hif3a (Zhang et al. 2014b), Prodh, Hbegf and Fkbp5 (Yang et al. 2015; de Koning et al. 2015; Sasaki et al. 2015; Valdés-Moreno et al. 2017; Shao and Vawter 2008; Fani et al. 2013; Menke et al. 2013).
Table 2. DAVID Gene Ontology and Pathway analysis of DEGs (fdr0.05, ≥ 1.5FC) in HC and CB at P14 and P35.
| Brain region | Age | Functional Category |
|---|---|---|
| Upregulated | ||
| cerebellum | P14 | cellular response to corticotropin-releasing hormone stimulus |
| positive regulation of transcription from RNA polymerase II promoter | ||
| cerebellum | P35 | myelin sheath |
| cellular response to interleukin-4 | ||
| growth cone | ||
| neuron development neuron projection | ||
| synaptic vesicle cycle | ||
| unfolded protein binding | ||
| calcium ion-regulated exocytosis of neurotransmitter | ||
| axon guidance | ||
| nervous system development | ||
| inclusion body | ||
| glucocorticoid receptor binding regulation of synaptic plasticity response to heat | ||
| hippocampus | P14 | structural constituent of myelin sheath |
| glycosaminoglycan biosynthesis - heparan sulfate / heparin | ||
| cilium movement | ||
| cilium organization | ||
| hippocampus | P35 | response to cAMP |
| cellular response to calcium ion | ||
| amphetamine addiction Downregulated | ||
| cerebellum | P14 | no enriched categories |
| cerebellum | P35 | cell adhesion |
| proteinaceous extracellular matrix | ||
| ECM-receptor interaction | ||
| growth factor binding | ||
| cilium morphogenesis | ||
| negative regulation of cell migration | ||
| hippocampus | P14 | response to hypoxia |
| response to amphetamine | ||
| synaptic transmission, dopaminergic | ||
| hippocampus | P35 | postsynaptic density |
| glutamatergic synapse | ||
| long-term potentiation response to cAMP | ||
| cellular response to calcium ion | ||
Representative categories are listed in order of their enrichment false discovery rate; for a full list of all enriched categories and statistics, see Table S4.
By P35, CB gene expression was much more dramatically dysregulated. Galnt17 down-regulation was still observed but at a much lower level (-1.24 fold change); a slight up-regulation of Auts2 (1.38 fold change) was also detected in CB at this later time point. More globally, and consistent with P14, P35 up-regulated DEGs were enriched in the GO category “cellular response to corticotropin-releasing hormone stimulus” as well as other signals of cellular stress, including “cellular response to interleukin-4”, “unfolded protein binding”, “response to heat” and “inclusion body”. Up-regulated P35 genes were also enriched in functions related to myelination, neurotransmitter exocytosis, axon guidance, synaptic plasticity and, neuron development (Table 2; Table S3).
P35 down-regulated CB DEGs were very strongly enriched in functions related to extracellular matrix but also categories such as cilium morphogenesis and motility, suggesting involvement of the ciliated ependymal cells (Table 2). Notably given that Galnt17 functions as a GalNAcT, both CB DEGs included genes encoding enzymes central to protein glycosylation of various types (Gcnt1, Extl1, Fut10, Ogt, Gcnt2, Man2c1, B3galnt2, B3galt2, B3gnt2, B4galtn4, B4galt1, Chpf2, Eogt) and genes involved in downstream sulfation or sialylation of glycosylated sidechains (Ndst3, St3gal6, St6galnac2, St8sia4). The data suggested a broad dysregulation of glycosylation-related functions and a persistent signal of cellular stress. The significant up-regulation of genes related to neurodevelopment, growth cone, and synapses at P35 further suggested that in the mutant CB, neuron maturation occurs, but is significantly delayed.
Hippocampus - dysregulated neurotransmitter signaling and further signs of neuronal stress:
Looking next at HC datasets, we also observed significant Galnt17 down-regulation at both time points in this region (-1.38 and -1.54 fold change at P14 and P35, respectively). In addition, we observed the down-regulation of Auts2 transcripts, exclusively at P14 (measured collectively at -1.38 fold change). As in CB, Sox1 was not among the DEGs at either time point. Globally, we identified certain DEGs and functions consistent with those identified in CB, but also clear evidence of unique HC effects. For example, genes including disease-related Hif3a, Prodh, Fkbp5, and Hbegf, were commonly down-regulated in P14 CB and HC, while others - including Fos - were consistently up-regulated (Table S2). Although at different ages (P14 in HC, P35 in CB), genes involved in oligodendrocyte development and myelination were also up-regulated in both brain regions. Cilia-related functions were also enriched in both brain regions although in opposite directions, being down-regulated in P35 CB but up-regulated in P14 HC. However, most enriched gene functions were brain region-specific. Most notably, genes involved in dopamine signaling - including the gene encoding dopamine receptor DRD2 - were significantly down-regulated in the HC of 16GsoT/T mice at the P14 time point (Table 2).
By P35, genes involved in neuron functions - including post-synaptic densities and long-term potentiation - were down-regulated in the 16GsoT/T HC. Genes related to glutamatergic signaling were also robustly and significantly down-regulated. Together with this signal of neurotransmitter imbalance, Fos and other IEGs were very highly up-regulated (Table S2). This expression pattern - echoing the upregulation of IEGs in mutant CB - strongly suggests a response to chronic neurological stress (Schreiber et al. 1991; Jett et al. 2017).
Brain region- and time-specific dysregulation of specific Auts2 isoforms:
Considering that the translocation could possibly dysregulate expression of Auts2 isoforms differentially, we designed and calibrated primers to measure expression of the transcripts individually. In the present data set and in a previous project (Saul et al. 2017), we had noticed evidence for a novel Auts2 promoter also included in a transcript model (ENSMUSG00000029673) and confirmed expression from this novel promoter by sequencing an qRT-PCR product from both CB and HC (Supplemental Text). We refer to this novel transcript as T6 (Figure 1B), and we also designed primers to detect its expression (Table S4).
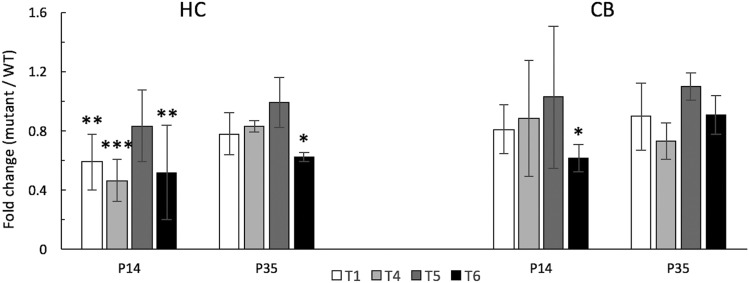
Differential expression of Auts2 isoforms was detected by qRT-PCR in both brain regions, although in transcript-, region- and time-specific patterns. Transcripts T1 and T4 were both significantly down-regulated in P14 HC, whereas novel isoform T6 was down-regulated in both brain regions, with significant differences in HC at both P14 and P35 (Figure 5). In contrast, isoform T5 was not differentially expressed at either time point or brain region. We did not detect evidence of Auts2 isoform up-regulation at P35 as suggested by RNA-seq data, possibly related to the relatively low levels of Auts2 expression at the later time point and the small amounts of expression-level change. Nevertheless, the data confirmed that several Auts2 transcripts are down-regulated by the 16Gso mutation during early postnatal development, with peak effects around P14.
Figure 5.
Cerebellar and hippocampal expression of Auts2 transcripts in 16GsoT/T compared to WT controls at P14 and P35. Auts2 isoforms (T1, T4, T5 and T6) expression in 16GsoT/T HC and CB at P14 and P35 was normalized to WT levels, which were set at 1. The expression of Auts2 isoforms T1, T4 and T6 was decreased in P14 16GsoT/T HC, while in CB, only Auts2 isoform T6 was significantly reduced. At P35, Auts2 isoform T6 was significantly decreased in HC, and no Auts2 isoform is dysregulated in CB. n = 3 for each genotype and sample. ANOVA, *P < 0.05; **P < 0.01; ***P < 0.001. CB = cerebellum; HC = hippocampus.
Auts2 and Galnt17 are co-expressed in a broad array of brain cell types
These data suggested primary roles for Auts2 and Galnt17 in the postnatal development of 16GsoT/T brain pathologies. To further assess these potential roles, it was important to document the cell types in which AUTS2 and GALNT17 proteins are expressed. Auts2 expression has been documented in detail: the gene is highly expressed in both cerebellar PCs and hippocampal granule and pyramidal cells in adults, with a highly dynamic pattern across the brain during development (Bedogni et al. 2010). However, brain expression or function has never been investigated for Galnt17.
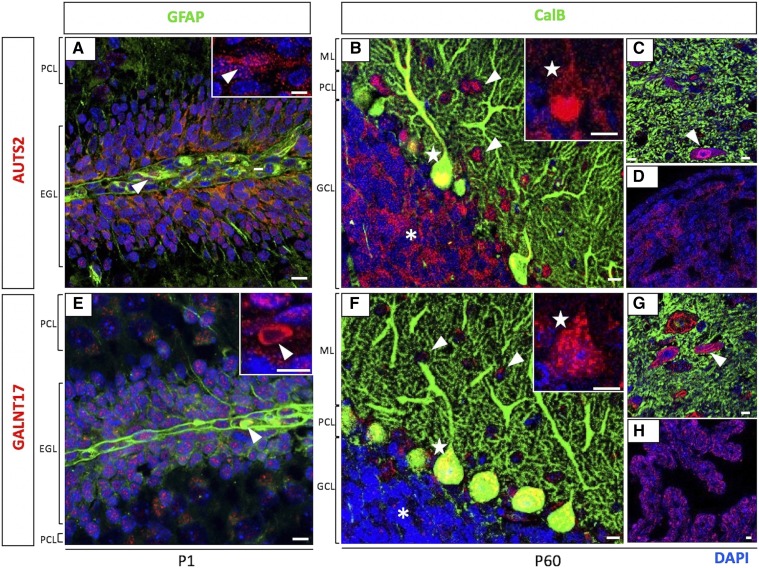
To examine protein expression, we used IHC with antibodies to AUTS2 and GALNT17 in CB and HC sections from WT animals. As revealed by these experiments, AUTS2 and GALNT17 proteins are both highly expressed in PCs, co-localizing with Calbindin (CalB) in juvenile (not shown) and adult brain sections (stars in Figure 6B, F). We also saw co-expression in the smaller CalB-negative basket and stellate cells nestled among the PC dendrites, with AUTS2 expressed in the nucleus and GALNT17 expressed in the cytoplasm, respectively (arrowheads in Figure 6B, F). AUTS2 and GALNT17 were also detected in the cytoplasm of the cerebellar granule cells (asterisks in Figure 6B, F). We noted three other co-expression sites in the cerebellar region. First, as reported previously (Bedogni et al. 2010), AUTS2 was detected in selected neurons within the deep cerebellar nuclei; GALNT17 was also detected at high levels in these cells (arrowheads in Figure 6C, G). Also, consistent with previous data on AUTS2, both proteins were detected in the choroid plexus and ependymal cells (Figure 6D, H). Particularly relevant to the 16Gso cerebellar pathology but not previously reported, both AUTS2 and GALNT17 were also detected in Bergmann glia, as confirmed by co-localization with marker GFAP in Bergmann glia cell bodies at P1 (arrowheads in Figure 6A, E). Notably, every cell type that we found to express AUTS2 also expressed GALNT17.
Figure 6.
Co-expression of AUTS2 and GALNT17 in different cell types in CB. Expression of AUTS2 (red; top) and GALNT17 (red; bottom) in various cell types was determined by co-staining with antibodies to different markers, and by cell shape and location in CB. Both proteins are expressed in Bergmann glia (A and E; arrowheads) as marked by GFAP in green at P1, and in PCs (B and F; stars) as marked by Calbindin (CalB) in green at P60. Both proteins were also co-expressed in cerebellar GCs (B and F; asterisks), stellate cells (B and F; arrowheads), large neurons in the deep cerebellar nuclei (C and G; arrowheads), and choroid plexus (D and H). scale bar = 10 μm. PCL = Purkinje cell layer; EGL = external granule layer; ML = molecular layer; GCL = granule cell layer.
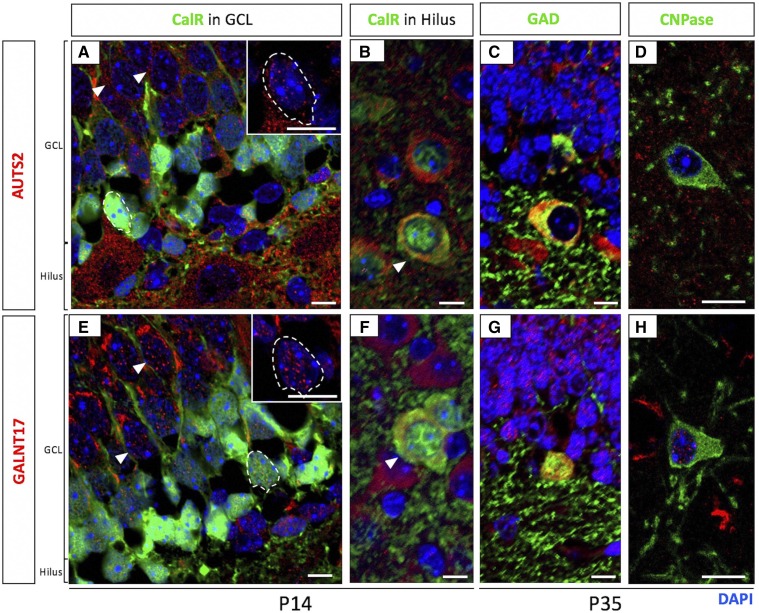
In postnatal HC, AUTS2 protein has been reported to co-localize with markers of Calretinin (CalR)-expressing immature granule neurons in the subgranular zone (SGZ) and granule cell layer (GCL) of the dentate gyrus (DG) as well as mature GCs in the GCL (Bedogni et al. 2010). We confirmed AUTS2 and demonstrated GALNT17 protein expression in both cell populations at P14 as well (Figure 7A, E). In mature GC, the GALNT17 protein was concentrated into bright punctate foci (Figure 7E, arrows), consistent with previous reports of the protein’s concentration within the Golgi apparatus (Nakayama et al. 2012). However, in immature cells in the SGZ, the GALNT17 signal was more evenly distributed including staining in the nucleus (Figure 7E, dashed circles). These cells were also CalR+, confirming that GALNT17 and AUTS2 are co-expressed GC from a very early stage in their differentiation (dashed circles in Figure 7A, E). AUTS2 and GALNT17 proteins were also found together in PyC of the CA1 and CA3 regions of the HC (not shown). Additionally, we saw expression of both proteins in GABA-ergic (GAD 65/67 positive, or GAD+) cells, including the pyramidal basket cells located at the edge of the DG (Figure 7C, G) and scattered cells inside the DG hilus. Inside the ventral HC hilus, we also detected expression of both proteins in the large, scattered CalR+ mossy cells (Figure 7B, F, arrowheads). We further detected expression of both AUTS2 and GALNT17 proteins colocalizing with the marker CNPase (Figure 7D, H), indicating expression in oligodendrocytes, which has not been reported for AUTS2 previously. Therefore, many of the cell types thought to be abnormal based on RNA-seq experiments – including neurons as well as oligodendrocytes and ependymal cells – co-express AUTS2 and GALNT17.
Figure 7.
Co-expression of AUTS2 and GALNT17 in different cell types in HC. Expression of AUTS2 (red; top) and GALNT17 (red; bottom) in various cell types was determined by co-staining with antibodies to different markers, by cell shape, and by location in HC. Both proteins are expressed in immature GCs (A and E; dashed circles) marked by Calretinin (CalR) and in mature GCs (A and E; arrowheads), as well as mossy cells (B and F; arrowheads) marked by CalR, GABAergic pyramidal basket cells (C and G) marked by GAD 65/67, and oligodendrocytes (D and H) marked by CNPase. Scale bar = 10 μm. GCL = granule cell layer.
Leveraging clues from transcriptomic data for new mechanistic insights
Based on RNA-seq data, we hypothesized that Galnt17 LOF and global dysregulation of glycosylation is a major contributor to the unfolded protein response and cellular stress, the delayed differentiation of PCs, and failed development of Bergmann glia in mutant mice. We conjectured that the 16GsoT/T hippocampal pathology could also reflect abnormal developmental timing based on several bits of evidence. For example, RNA-seq data suggested delays in differentiation of neuronal intermediate progenitor cells (IPCs), based on down-regulation of markers of later differentiation stages - including Neurod1 and Prox1 (later intermediate progenitors), a post-mitotic marker for immature GC, Calb2 (encoding CalR), and mature GC marker Calb1 (CalB) (Table S3). Furthermore, as mentioned above, the abnormally dense packing of GCs and PyCs we observed is reminiscent of pathologies observed in the HC of ASD patients where it is thought to reflect abnormal timing of neuron maturation, dendrite formation, and synaptic connectivity (Amaral et al. 2008; Varghese et al. 2017). Importantly, mis-timed maturation of hippocampal neurons could potentially explain key 16GsoT/T behavioral phenotypes -including impaired learning and memory as well as seizures – as well as related human AUTS2 syndrome phenotypes (Beunders et al. 2013).
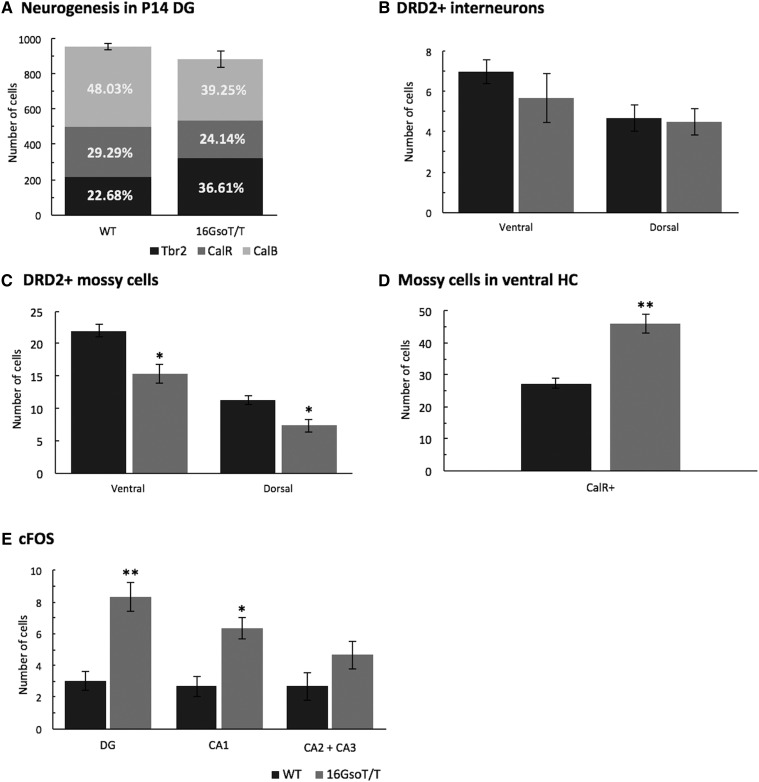
To address this possibility, we examined the developmental trajectory of DG GCs, which are marked by location and well-established markers (reviewed by (Khalaf-Nazzal and Francis 2013; Nicola et al. 2015)). We used antibodies to TBR2, CalR, and CalB to identify and count the numbers of cells of each type in the DG of P14 mutants and controls. Whereas the DG of P14 16GsoT/T and WT mice contain similar numbers of neurons overall (Figure 8A), P14 mutants contain higher relative numbers of TBR2+ IPCs and fewer CalR+ and CalB+ cells than controls (Figure 8A). By P35, the numbers of TBR2+, CalR+ and CalB+ neurons in mutant DG had normalized (not shown) along with cell density (Figure 4C, D). This finding is consistent with the idea that in 16GsoT/T mutants, the dense packing of GC at P14 is a signature of the cells’ immaturity, and that hippocampal GC differentiation is significantly delayed.
Figure 8.
Molecular confirmation of 16GsoT/T hippocampal pathologies. (A) Percentage of IPCs (Tbr2+), immature GC (CalR+), and mature GC (CalB+) were measured in the DG at P14, with 16GsoT/T (gray bars) compared to age-matched WT controls (black bars). 16GsoT/T animals showed increased numbers of Tbr2+ IPCs (36.61% of the total numbers of neurons), but decreased numbers of immature (24.14%) and mature (39.25%) GC compared with WT (black). However, there was no difference in total number of cells in WT and 16GsoT/T hippocampal regions examined. (B) Number of DRD2+, GAD+ interneurons in ventral and dorsal HC. We detected no significant difference in numbers of interneurons between WT (black bars) and 16GsoT/T (gray bars) P14 animals. (C) Number of DRD2+ mossy cells in ventral and dorsal HC. Our counts revealed that 16GsoT/T animals have significantly fewer DRD2+ mossy cells in both dorsal and ventral HC than WT controls. (D) Number of overall mossy cells are significantly more in 16GsoT/T than in WT in ventral HC. (E) Number of cFOS+ cells in DG, CA1, and CA2 and CA3 hippocampal regions at P35. 16GsoT/T P35 HC contained more cFOS+ cells in DG and CA1 regions than in WT controls, but similar numbers in CA2/3. n = 3 for both genotypes. DG = dentate gyrus; HC = hippocampus. ANOVA, *P < 0.05; **P < 0.01.
A second transcriptomic signal we sought to investigate was the reduced dopaminergic signaling detected in the P14 16GsoT/T animals – centered around the dopamine receptor type 2 gene, Drd2. In the HC, Drd2 is expressed primarily in DG hilar mossy cells and interneurons, both of which interact intimately with and regulate hippocampal GC and PyC. To identify the cell types responsible for reduced DRD2 expression, we stained P14 16GsoT/T and WT hippocampal sections with a DRD2 antibody together with markers for interneurons (GAD+) to clearly distinguish them from mossy cells. The results revealed that DRD2+, GAD+ interneurons were present in normal numbers in 16GsoT/T mutant mice (Figure 8B). In contrast, DRD2+, GAD- mossy cells were present in significantly lower numbers in 16GsoT/T mutant dorsal and ventral HC compared to WT controls (Figure 8C). However, when we used CalR to identify hilar mossy cells independently of the DRD2 signal, we saw that the mossy cells were actually present in higher numbers in 16GsoT/T than in WT animals in ventral HC (Figure 8D). Therefore, mossy cells are generated in higher numbers, and migrate properly to the hilus in 16GsoT/T HC, DRD2 protein is expressed at significantly reduced levels in that population of cells.
Finally, we used an antibody to cFOS to identify the sources of the increased IEG expression in 16GsoT/T HC and CB. In P35 HC, increased cFOS signal was detected clearly in the DG GCL (Figure 8E) and in CA1 PyCs. However, we found no significant difference in cFOS signals between 16GsoT/T mutants and WT in CA2 or CA3 regions. Together with the histopathological findings, these data suggest that 16GsoT/T hippocampal pathology is primarily focused in the DG and CA1 regions, and that despite the normalized cell numbers in adult DG, cellular pathology persists in both regions into adulthood.
Discussion
Here we provide a detailed molecular, behavioral, cellular, and neuropathological analysis of mice inheriting T(5G2;8A1)GSO (16Gso), a novel reciprocal translocation involving chr5 and chr8. On chr5, the breakpoint falls in the intergenic region between Galnt17 and Auts2; the translocation affects expression of Galnt17 as well as specific Auts2 alternative promoters in a conditional manner, with distinct spatial and temporal effects on each. Importantly, this dysregulation was detected in brain regions that show distinct neuropathologies in 16GsoT/T mice and at the developmental time points during which these pathologies first appear. Although Galnt17 mutations have not been investigated, several phenotypes detected in 16GsoT/T mice overlap obviously with those of Auts2 KO mutants, and share features with human AUTS2 syndrome patients as well (Table 1).
On chr8, the 16Gso translocation breakpoint falls upstream of the Sox1 gene; mis-expression of Sox1 was not detected in the brain regions or postnatal time points we examined, but Sox1 KO homozygotes display seizures, freezing, and anxiety-related behaviors also expressed by 16GsoT/T mice (Table 1). Although we cannot rule out a subtle role for this gene in the 16Gso behavioral phenotypes, several lines of evidence argue against Sox1 as a central driver. In particular, neither Sox1 KO homozygotes (Malas et al. 2003; Sottile et al. 2006) nor 16Gso/+, Sox1/+ double heterozygotes (Weisner 2015) display the distinctive brain pathologies that we detected in 16GsoT/T mice. Moreover, genetic complementation tests between the recessive 16Gso and Sox1 KO alleles have been completed, and neither brain pathologies, seizures nor exaggerated freezing behaviors were observed in 16Gso/+, Sox1-/+ compound heterozygotes (Weisner 2015).
Here we should note that epilepsy is one of the phenotypes shared by 16Gso mutants and some AUTS2 syndrome patients (Table 1), suggesting that even this phenotype could be related to chr5 breakpoint-spanning genes. However, the specific hippocampal and cerebellar pathologies we identified in 16GsoT/T mice have not been previously associated with any Auts2 mouse mutations and other than generalized microcephaly, specific brain pathologies have not been described for human AUTS2 patients to date. On the other hand, severe cortical abnormalities that have been documented for Auts2 KO homozygotes (Hori et al. 2014) are not seen in 16GsoT/T mice. We hypothesize that these differences in behavior and pathology are likely due to the translocation’s partial, isoform-specific, and spatially- and temporally-limited effects on Auts2 expression, with a possible contribution from the coordinated dysregulation of Galnt17.
Testing these conjectures will require further investigation with conditional mutants, including a first look at the brains and behaviors of Galnt17 mice. Nevertheless, the unique 16GsoT/T cerebellar and hippocampal pathologies - including the mistiming of both PC and hippocampal neuron differentiation and PC loss – hold significant potential for human clinical relevance. These 16GsoT/T phenotypes bear some very striking similarities to those observed in ASD patients (Amaral et al. 2008; Varghese et al. 2017; Fatemi et al. 2002; Courchesne and Pierce 2005; Courchesne et al. 2005), and related cellular phenotypes have been tied to temporal lobe epilepsy, bipolar disorder, depression, schizophrenia, and ADHD as well (Chambers 2013; Goodman et al. 2014; McKinnon et al. 2009; Otten and Meeter 2015; Hester and Danzer 2014; Finch et al. 2002; Maloku et al. 2010; Strata 2015). The similarities suggest a broad relevance to AUTS2-linked disease.
Importantly, 16Gso neuropathologies involve cell populations that express AUTS2 and GALNT17 during the critical postnatal period when the pathologies first appear. Indeed, we show that the two proteins are highly co-expressed in brain; in fact, we could not identify a single cell type in which one was expressed and the other was not. One possible explanation for this tight co-expression could be that Galnt17 is a target of AUTS2 nuclear isoforms; however, arguing against this possibility, Galnt17 binding has not been indicated by AUTS2 chromatin precipitation to date (Oksenberg et al. 2014), and we found Galnt17 to be highly down-regulated in P14 CB in the absence of effects on Auts2. Rather, we posit, the high level of co-expression suggests that the two genes are controlled within a common three-dimensional regulatory structure, perhaps involving interactions with shared local enhancers. This possibility is supported by the fact that three-dimensional interactions between Auts2 and Caln1 (located directly downstream of Galnt17) are regulated by cocaine in striatal spiny neurons (Engmann et al. 2017). Furthermore, the cellular functions attributed to GALNT17 – ECM interactions, cell adhesion, cell motility, formation of lamellipodia and filopodia, and endocytosis (Nakayama et al. 2012) - dovetail well with those identified for AUTS2 (Hori et al. 2014); these data suggest that the proteins could be coordinated to act in the same pathways in developing neuronal cells.
The current study provides novel mechanistic clues to these pathways and possible roles of each gene. For example, RNA-seq data implicate Galnt17 LOF as the major contributor to the delayed maturation and age-related loss of PCs, whereas reduced expression of Auts2, Galnt17, or both genes in concert could contribute to the 16GsoT/T hippocampal pathology. Interestingly, common themes of delayed neuronal maturation and cellular stress were detected in both brain regions. Connecting brain pathology to behaviors, we surmise that the abnormal development of hippocampal neurons is linked to 16GsoT/T learning deficits, and suggest that similar pathologies may be relevant to ID in AUTS2 syndrome patients as well. Furthermore, the failure of hippocampal connectivity, PC loss, or a combination of the two could contribute to developmental delay, anxious and repetitive behaviors, and seizures. The present data can be used to design experiments involving strategic combinations of conditional mutants together with new tools, such as single-cell sequencing, to address these hypotheses.
As mentioned, genotype-phenotype relationships in the AUTS2 region are complicated, as has been studied and summarized in great depth by expert groups (Beunders et al. 2016; Beunders et al. 2013; Oksenberg and Ahituv 2013). Our data suggest that different combinations of AUTS2 isoform mis-expression, perhaps combined with effects on co-regulated GALNT17, could explain some of the phenotypic heterogeneity. Interestingly, in addition to a multigenic and Auts2 intragenic deletions and duplications, human duplications contained completely within the intergenic region between AUTS2 and GALNT17 have been reported in public databases, and are associated with generalized developmental delay (for example, in the ClinVar database (Landrum et al. 2016)). Data from the 16Gso translocation would predict that these and perhaps other noncoding variants in the region could impact expression of both genes. However, gene expression has not been investigated in conjunction with any AUTS2 syndrome mutations; fortunately, questions raised by this study can be addressed by further studies in mouse and other model organisms.
It is now well-established that even small, intragenic mutations can disrupt enhancers and impact expression of neighboring genes (Dixon et al. 2015; Dixon et al. 2012; Griffin et al. 2002; Kleinjan et al. 2006; Kleinjan and Coutinho 2009), and we propose that a clear picture of genotype-phenotype associations in this region will emerge with incorporation of a detailed regulatory map. An elegant study in zebrafish has identified several conserved Auts2-region enhancers (Oksenberg and Ahituv 2013), but additional regulatory elements active in specific brain cell types and at additional developmental time points are certain to emerge. Fortunately, tools for investigating genome-wide regulatory relationships are becoming increasingly accessible, and can possibly provide key missing pieces to this genetic puzzle.
Since Galnt17 expression and in vivo functions have not previously been characterized, our data, revealing the protein’s broad co-expression with AUTS2 and its potential role in regionally-associated neuropsychiatric disorders, are both novel and significant. This study builds groundwork for understanding the neurodevelopmental underpinnings of AUTS2 syndrome behavioral phenotypes and of other types of regionally-associated neurological disease, and suggests roles for each gene in these phenotypes. More generally, this study raises the possibility that other clusters of topologically-linked, coregulated genes could explain natural variance in expression many types of neurodevelopmental disorders, including those associated with mutations within the boundaries of well-studied genes.
Acknowledgments
The authors are grateful to Chris Seward and Yunshu Song for critical comments on the manuscript, to Jon Cerqua and Diana Fan for excellent research assistance, and to Justin Rhodes and Paula Bucko for invaluable help with and training in activity monitoring experiments. This research was funded by the U.S. National Institute of Mental Health under grant numbers MH107200 and MH114600 (awarded to L.S).
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9887540.
Communicating editor: A. McCallion
Literature Cited
- Amaral D. G., Schumann C. M., and Nordahl C. W., 2008. Neuroanatomy of autism. Trends Neurosci. 31: 137–145. 10.1016/j.tins.2007.12.005 [DOI] [PubMed] [Google Scholar]
- Amarillo I. E., Li W. L., Li X., Vilain E., and Kantarci S., 2014. De novo single exon deletion of AUTS2 in a patient with speech and language disorder: a review of disrupted AUTS2 and further evidence for its role in neurodevelopmental disorders. Am. J. Med. Genet. A. 164A: 958–965. 10.1002/ajmg.a.36393 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedogni F., Hodge R. D., Nelson B. R., Frederick E. A., Shiba N. et al. , 2010. Autism susceptibility candidate 2 (Auts2) encodes a nuclear protein expressed in developing brain regions implicated in autism neuropathology. Gene Expr. Patterns 10: 9–15. 10.1016/j.gep.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G., de Munnik S. A., Van der Aa N., Ceulemans B., Voorhoeve E. et al. , 2015. Two male adults with pathogenic AUTS2 variants, including a two-base pair deletion, further delineate the AUTS2 syndrome. Eur. J. Hum. Genet. 23: 803–807. 10.1038/ejhg.2014.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beunders G., van de Kamp J., Vasudevan P., Morton J., Smets K. et al. , 2016. A detailed clinical analysis of 13 patients with AUTS2 syndrome further delineates the phenotypic spectrum and underscores the behavioural phenotype. J. Med. Genet. 53: 523–532. 10.1136/jmedgenet-2015-103601 [DOI] [PubMed] [Google Scholar]
- Beunders G., Voorhoeve E., Golzio C., Pardo L. M., Rosenfeld J. A. et al. , 2013. Exonic deletions in AUTS2 cause a syndromic form of intellectual disability and suggest a critical role for the C terminus. Am. J. Hum. Genet. 92: 210–220. 10.1016/j.ajhg.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolt C. C., Elso C. M., Lu X., Pan F., Kispert A. et al. , 2014. A distant downstream enhancer directs essential expression of Tbx18 in urogenital tissues. Dev. Biol. 392: 483–493. 10.1016/j.ydbio.2014.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botton P. H., Costa M. S., Ardais A. P., Mioranzza S., Souza D. O. et al. , 2010. Caffeine prevents disruption of memory consolidation in the inhibitory avoidance and novel object recognition tasks by scopolamine in adult mice. Behav. Brain Res. 214: 254–259. 10.1016/j.bbr.2010.05.034 [DOI] [PubMed] [Google Scholar]
- Castelhano-Carlos M. J., Sousa N., Ohl F., and Baumans V., 2010. Identification methods in newborn C57BL/6 mice: a developmental and behavioural evaluation. Lab. Anim. 44: 88–103. 10.1258/la.2009.009044 [DOI] [PubMed] [Google Scholar]
- Chambers R. A., 2013. Adult hippocampal neurogenesis in the pathogenesis of addiction and dual diagnosis disorders. Drug Alcohol Depend. 130: 1–12. 10.1016/j.drugalcdep.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S. J., Liao D. L., Shen T. W., Yang H. C., Chen K. C. et al. , 2016. Genetic signatures of heroin addiction. Medicine (Baltimore) 95: e4473 10.1097/MD.0000000000004473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. S., Reader R. H., Hoischen A., Veltman J. A., Simpson N. H. et al. , 2017. Next-generation DNA sequencing identifies novel gene variants and pathways involved in specific language impairment. Sci. Rep. 7: 46105 10.1038/srep46105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y. H., Liao D. L., Lai C. H., and Chen C. H., 2013. Genetic analysis of AUTS2 as a susceptibility gene of heroin dependence. Drug Alcohol Depend. 128: 238–242. 10.1016/j.drugalcdep.2012.08.029 [DOI] [PubMed] [Google Scholar]
- Chrobak A. A., and Soltys Z., 2017. Bergmann Glia, Long-Term Depression, and Autism Spectrum Disorder. Mol. Neurobiol. 54: 1156–1166. 10.1007/s12035-016-9719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E., and Pierce K., 2005. Brain overgrowth in autism during a critical time in development: implications for frontal pyramidal neuron and interneuron development and connectivity. Int. J. Dev. Neurosci. 23: 153–170. 10.1016/j.ijdevneu.2005.01.003 [DOI] [PubMed] [Google Scholar]
- Courchesne E., Redcay E., Morgan J. T., and Kennedy D. P., 2005. Autism at the beginning: microstructural and growth abnormalities underlying the cognitive and behavioral phenotype of autism. Dev. Psychopathol. 17: 577–597. 10.1017/S0954579405050285 [DOI] [PubMed] [Google Scholar]
- de Koning M. B., van Duin E. D., Boot E., Bloemen O. J., Bakker J. A. et al. , 2015. PRODH rs450046 and proline x COMT Val158Met interaction effects on intelligence and startle in adults with 22q11 deletion syndrome. Psychopharmacology (Berl.) 232: 3111–3122. 10.1007/s00213-015-3971-5 [DOI] [PubMed] [Google Scholar]
- Dickson P. E., Rogers T. D., Del Mar N., Martin L. A., Heck D. et al. , 2010. Behavioral flexibility in a mouse model of developmental cerebellar Purkinje cell loss. Neurobiol. Learn. Mem. 94: 220–228. 10.1016/j.nlm.2010.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Jung I., Selvaraj S., Shen Y., Antosiewicz-Bourget J. E. et al. , 2015. Chromatin architecture reorganization during stem cell differentiation. Nature 518: 331–336. 10.1038/nature14222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y. et al. , 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J., Gai X., Xie H. M., Perin J. C., Geiger E. et al. , 2010. Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Mol. Psychiatry 15: 637–646. 10.1038/mp.2009.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elso C., Lu X., Morrison S., Tarver A., Thompson H. et al. , 2008. Germline translocations in mice: unique tools for analyzing gene function and long-distance regulatory mechanisms. J. Natl. Cancer Inst. Monogr. 2008: 91–95. 10.1093/jncimonographs/lgn008 [DOI] [PubMed] [Google Scholar]
- Elso C., Lu X., Weisner P. A., Thompson H. L., Skinner A. et al. , 2013. A reciprocal translocation dissects roles of Pax6 alternative promoters and upstream regulatory elements in the development of pancreas, brain, and eye. Genesis 51: 630–646. [DOI] [PubMed] [Google Scholar]
- Elso C. M., Lu X., Culiat C. T., Rutledge J. C., Cacheiro N. L. et al. , 2004. Heightened susceptibility to chronic gastritis, hyperplasia and metaplasia in Kcnq1 mutant mice. Hum. Mol. Genet. 13: 2813–2821. 10.1093/hmg/ddh307 [DOI] [PubMed] [Google Scholar]
- Engmann O., Labonte B., Mitchell A., Bashtrykov P., Calipari E. S. et al. , 2017. Cocaine-Induced Chromatin Modifications Associate With Increased Expression and Three-Dimensional Looping of Auts2. Biol. Psychiatry 82: 794–805. 10.1016/j.biopsych.2017.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fani N., Gutman D., Tone E. B., Almli L., Mercer K. B. et al. , 2013. FKBP5 and attention bias for threat: associations with hippocampal function and shape. JAMA Psychiatry 70: 392–400. 10.1001/2013.jamapsychiatry.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi S. H., Halt A. R., Realmuto G., Earle J., Kist D. A. et al. , 2002. Purkinje cell size is reduced in cerebellum of patients with autism. Cell. Mol. Neurobiol. 22: 171–175. 10.1023/A:1019861721160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch A. J., Nicolson R. I., and Fawcett A. J., 2002. Evidence for a neuroanatomical difference within the olivo-cerebellar pathway of adults with dyslexia. Cortex 38: 529–539. 10.1016/S0010-9452(08)70021-2 [DOI] [PubMed] [Google Scholar]
- Gao Z., Lee P., Stafford J. M., von Schimmelmann M., Schaefer A. et al. , 2014. An AUTS2-Polycomb complex activates gene expression in the CNS. Nature 516: 349–354. 10.1038/nature13921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girirajan S., Brkanac Z., Coe B. P., Baker C., Vives L. et al. , 2011. Relative burden of large CNVs on a range of neurodevelopmental phenotypes. PLoS Genet. 7: e1002334 10.1371/journal.pgen.1002334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman J., Marsh R., Peterson B. S., and Packard M. G., 2014. Annual research review: The neurobehavioral development of multiple memory systems–implications for childhood and adolescent psychiatric disorders. J. Child Psychol. Psychiatry 55: 582–610. 10.1111/jcpp.12169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin C., Kleinjan D. A., Doe B., and van Heyningen V., 2002. New 3′ elements control Pax6 expression in the developing pretectum, neural retina and olfactory region. Mech. Dev. 112: 89–100. 10.1016/S0925-4773(01)00646-3 [DOI] [PubMed] [Google Scholar]
- Hamshere M. L., Green E. K., Jones I. R., Jones L., Moskvina V. et al. , 2009. Genetic utility of broadly defined bipolar schizoaffective disorder as a diagnostic concept. Br. J. Psychiatry 195: 23–29. 10.1192/bjp.bp.108.061424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hester M. S., and Danzer S. C., 2014. Hippocampal granule cell pathology in epilepsy - a possible structural basis for comorbidities of epilepsy? Epilepsy Behav. 38: 105–116. 10.1016/j.yebeh.2013.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Nagai T., Shan W., Sakamoto A., Abe M. et al. , 2015. Heterozygous Disruption of Autism susceptibility candidate 2 Causes Impaired Emotional Control and Cognitive Memory. PLoS One 10: e0145979 10.1371/journal.pone.0145979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hori K., Nagai T., Shan W., Sakamoto A., Taya S. et al. , 2014. Cytoskeletal regulation by AUTS2 in neuronal migration and neuritogenesis. Cell Reports 9: 2166–2179. 10.1016/j.celrep.2014.11.045 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., and Lempicki R. A.. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jett J. D., Bulin S. E., Hatherall L. C., McCartney C. M., and Morilak D. A., 2017. Deficits in cognitive flexibility induced by chronic unpredictable stress are associated with impaired glutamate neurotransmission in the rat medial prefrontal cortex. Neuroscience 346: 284–297. 10.1016/j.neuroscience.2017.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor M., Wang J. C., Wetherill L., Le N., Bertelsen S. et al. , 2013. A meta-analysis of two genome-wide association studies to identify novel loci for maximum number of alcoholic drinks. Hum. Genet. 132: 1141–1151. 10.1007/s00439-013-1318-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalaf-Nazzal R., and Francis F., 2013. Hippocampal development - old and new findings. Neuroscience 248: 225–242. 10.1016/j.neuroscience.2013.05.061 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R. et al. , 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan D. A., Seawright A., Mella S., Carr C. B., Tyas D. A. et al. , 2006. Long-range downstream enhancers are essential for Pax6 expression. Dev. Biol. 299: 563–581. 10.1016/j.ydbio.2006.08.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinjan D. J., and Coutinho P., 2009. Cis-ruption mechanisms: disruption of cis-regulatory control as a cause of human genetic disease. Brief. Funct. Genomics Proteomics 8: 317–332. 10.1093/bfgp/elp022 [DOI] [PubMed] [Google Scholar]
- Landrum M. J., Lee J. M., Benson M., Brown G., Chao C. et al. , 2016. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 44: D862–D868. 10.1093/nar/gkv1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., and Salzberg S. L., 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Malas S., Postlethwaite M., Ekonomou A., Whalley B., Nishiguchi S. et al. , 2003. Sox1-deficient mice suffer from epilepsy associated with abnormal ventral forebrain development and olfactory cortex hyperexcitability. Neuroscience 119: 421–432. 10.1016/S0306-4522(03)00158-1 [DOI] [PubMed] [Google Scholar]
- Maloku E., Covelo I. R., Hanbauer I., Guidotti A., Kadriu B. et al. , 2010. Lower number of cerebellar Purkinje neurons in psychosis is associated with reduced reelin expression. Proc. Natl. Acad. Sci. USA 107: 4407–4411. 10.1073/pnas.0914483107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin L. A., Goldowitz D., and Mittleman G., 2010. Repetitive behavior and increased activity in mice with Purkinje cell loss: a model for understanding the role of cerebellar pathology in autism. Eur. J. Neurosci. 31: 544–555. 10.1111/j.1460-9568.2009.07073.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon M. C., Yucel K., Nazarov A., and MacQueen G. M., 2009. A meta-analysis examining clinical predictors of hippocampal volume in patients with major depressive disorder. J. Psychiatry Neurosci. 34: 41–54. [PMC free article] [PubMed] [Google Scholar]
- Menke A., Klengel T., Rubel J., Bruckl T., Pfister H. et al. , 2013. Genetic variation in FKBP5 associated with the extent of stress hormone dysregulation in major depression. Genes Brain Behav. 12: 289–296. 10.1111/gbb.12026 [DOI] [PubMed] [Google Scholar]
- Myung W., Kim J., Lim S. W., Shim S., Won H. H. et al. , 2015. A genome-wide association study of antidepressant response in Koreans. Transl. Psychiatry 5: e633 10.1038/tp.2015.127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama Y., Nakamura N., Oki S., Wakabayashi M., Ishihama Y. et al. , 2012. A putative polypeptide N-acetylgalactosaminyltransferase/Williams-Beuren syndrome chromosome region 17 (WBSCR17) regulates lamellipodium formation and macropinocytosis. J. Biol. Chem. 287: 32222–32235. 10.1074/jbc.M112.370932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicola Z., Fabel K., and Kempermann G., 2015. Development of the adult neurogenic niche in the hippocampus of mice. Front. Neuroanat. 9: 53 10.3389/fnana.2015.00053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N., and Ahituv N., 2013. The role of AUTS2 in neurodevelopment and human evolution. Trends Genet. 29: 600–608. 10.1016/j.tig.2013.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N., Haliburton G. D., Eckalbar W. L., Oren I., Nishizaki S. et al. , 2014. Genome-wide distribution of Auts2 binding localizes with active neurodevelopmental genes. Transl. Psychiatry 4: e431 10.1038/tp.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksenberg N., Stevison L., Wall J. D., and Ahituv N., 2013. Function and regulation of AUTS2, a gene implicated in autism and human evolution. PLoS Genet. 9: e1003221 10.1371/journal.pgen.1003221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten M., and Meeter M., 2015. Hippocampal structure and function in individuals with bipolar disorder: a systematic review. J. Affect. Disord. 174: 113–125. 10.1016/j.jad.2014.11.001 [DOI] [PubMed] [Google Scholar]
- Pierce K., and Courchesne E., 2001. Evidence for a cerebellar role in reduced exploration and stereotyped behavior in autism. Biol. Psychiatry 49: 655–664. 10.1016/S0006-3223(00)01008-8 [DOI] [PubMed] [Google Scholar]
- Robinson K. O., Petersen A. M., Morrison S. N., Elso C. M., and Stubbs L., 2005. Two reciprocal translocations provide new clues to the high mutability of the Grid2 locus. Mamm. Genome 16: 32–40. 10.1007/s00335-004-2423-z [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Omotuyi O. I., Ueda M., Shinohara K., and Ueda H., 2015. NMDA receptor agonists reverse impaired psychomotor and cognitive functions associated with hippocampal Hbegf-deficiency in mice. Mol. Brain 8: 83 10.1186/s13041-015-0176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul M. C., Seward C. H., Troy J. M., Zhang H., Sloofman L. G. et al. , 2017. Transcriptional regulatory dynamics drive coordinated metabolic and neural response to social challenge in mice. Genome Res. 27: 959–972. 10.1101/gr.214221.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber S. S., Tocco G., Shors T. J., and Thompson R. F., 1991. Activation of immediate early genes after acute stress. Neuroreport 2: 17–20. 10.1097/00001756-199101000-00004 [DOI] [PubMed] [Google Scholar]
- Schumann G., Coin L. J., Lourdusamy A., Charoen P., Berger K. H. et al. , 2011. Genome-wide association and genetic functional studies identify autism susceptibility candidate 2 gene (AUTS2) in the regulation of alcohol consumption. Proc. Natl. Acad. Sci. USA 108: 7119–7124. 10.1073/pnas.1017288108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L., and Vawter M. P., 2008. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol. Psychiatry 64: 89–97. 10.1016/j.biopsych.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottile V., Li M., and Scotting P. J., 2006. Stem cell marker expression in the Bergmann glia population of the adult mouse brain. Brain Res. 1099: 8–17. 10.1016/j.brainres.2006.04.127 [DOI] [PubMed] [Google Scholar]
- Strata P., 2015. The emotional cerebellum. Cerebellum 14: 570–577. 10.1007/s12311-015-0649-9 [DOI] [PubMed] [Google Scholar]
- Stubbs L., Carver E. A., Cacheiro N. L., Shelby M., and Generoso W., 1997. Generation and characterization of heritable reciprocal translocations in mice. Methods 13: 397–408. 10.1006/meth.1997.0546 [DOI] [PubMed] [Google Scholar]
- Sultana R., Yu C. E., Yu J., Munson J., Chen D. et al. , 2002. Identification of a novel gene on chromosome 7q11.2 interrupted by a translocation breakpoint in a pair of autistic twins. Genomics 80: 129–134. 10.1006/geno.2002.6810 [DOI] [PubMed] [Google Scholar]
- Valdés-Moreno M. I., Alcantara-Alonso V., Estrada-Camarena E., Mengod G., Amaya M. I. et al. , 2017. Phosphodiesterase-7 inhibition affects accumbal and hypothalamic thyrotropin-releasing hormone expression, feeding and anxiety behavior of rats. Behav. Brain Res. 319: 165–173. 10.1016/j.bbr.2016.11.027 [DOI] [PubMed] [Google Scholar]
- Varghese M., Keshav N., Jacot-Descombes S., Warda T., Wicinski B. et al. , 2017. Autism spectrum disorder: neuropathology and animal models. Acta Neuropathol. 134: 537–566. 10.1007/s00401-017-1736-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisner P. A., 2015. The role of AUTS2 in neurodevelopment and neurological disease. Ph.D. Disertation, University of Illinois at Urbana-Champaign, Champaign, IL. [Google Scholar]
- Yamada K., and Watanabe M., 2002. Cytodifferentiation of Bergmann glia and its relationship with Purkinje cells. Anat. Sci. Int. 77: 94–108. 10.1046/j.0022-7722.2002.00021.x [DOI] [PubMed] [Google Scholar]
- Yang S. L., Wu C., Xiong Z. F., and Fang X., 2015. Progress on hypoxia-inducible factor-3: Its structure, gene regulation and biological function. (Review) Mol. Med. Rep. 12: 2411–2416. 10.3892/mmr.2015.3689 [DOI] [PubMed] [Google Scholar]
- Zhang B., Xu Y. H., Wei S. G., Zhang H. B., Fu D. K. et al. , 2014a Association study identifying a new susceptibility gene (AUTS2) for schizophrenia. Int. J. Mol. Sci. 15: 19406–19416. 10.3390/ijms151119406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Yao Q., Lu L., Li Y., Chen P. J. et al. , 2014b Hypoxia-inducible factor 3 is an oxygen-dependent transcription activator and regulates a distinct transcriptional response to hypoxia. Cell Reports 6: 1110–1121. 10.1016/j.celrep.2014.02.011 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data and resources reported in this manuscript are publicly available. Sequencing data reported in this paper has been deposited into the GEO database under accession number GSE132684. The results of sequence analysis are presented in Table S2 and full listing of enriched functional categories from DAVID analysis in Table S3. Oligonucleotide primers used for qRT-PCR are list in Table S4. Data from behavioral tests and represented as graphs in this manuscript are presented in Table S1, and H&E stains of 16Gso T/T and WT cortex are presented in Fig. S1. Sequence confirming the expression of novel Auts2 isoform T6, which is identical to that in an ENSEMBL model and mouse ESTs as reported, is also provided in Supplemental Text. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9887540.