Abstract
Repellent odors are widely used to prevent insect-borne diseases, making it imperative to identify the conserved molecular underpinnings of their olfactory systems. Currently, little is known about the molecules supporting odor signaling beyond the odor receptors themselves. Most known molecules function in one of two classes of olfactory sensilla, single-walled or double-walled, which have differing morphology and odor response profiles. Here, we took two approaches to discover novel genes that contribute to insect olfaction in the periphery. We transcriptionally profiled Drosophila melanogaster amos mutants that lack trichoid and basiconic sensilla, the single-walled sensilla in this species. This revealed 187 genes whose expression is enriched in these sensilla, including pickpocket ion channels and neuromodulator GPCRs that could mediate signaling pathways unique to single-walled sensilla. For our second approach, we computationally identified 141 antennal-enriched (AE) genes that are more than ten times as abundant in D. melanogaster antennae as in other tissues or whole-body extracts, and are thus likely to play a role in olfaction. We identified unambiguous orthologs of AE genes in the genomes of four distantly related insect species, and most identified orthologs were expressed in the antenna of these species. Further analysis revealed that nearly half of the 141 AE genes are localized specifically to either single or double-walled sensilla. Functional annotation suggests the AE genes include signaling molecules and enzymes that could be involved in odorant degradation. Together, these two resources provide a foundation for future studies investigating conserved mechanisms of odor signaling.
Keywords: Gene Expression, Drosophila, Insect, Olfaction, Antenna
Vector borne diseases sicken hundreds of millions of people worldwide each year. Most infections are contracted through the bites of blood-feeding arthropods (World Health Organization 2017). The principal cues that disease vectors use to locate humans are kairomones, odors released from human sweat, skin, and breath (Takken and Knols 2010; Ray 2015). Many of the most effective means to reduce disease burden prevent vectors from contacting hosts through manipulation of their olfactory driven behaviors (Takken and Knols 2010).
The general anatomy of insect olfactory systems is conserved. Odors are detected by odor receptors located on the dendrites of olfactory receptor neurons (ORNs). A small number of ORNs and non-neuronal auxiliary cells are housed within sensory hairs known as sensilla, and hundreds of these sensilla densely cover the primary insect olfactory organ, the antenna (Shanbhag et al. 1999). There are two major morphological classes of sensilla, single-walled and double-walled (Keil 1999). In most organisms, pheromones and food and plant odors are detected by single-walled sensilla (Steinbrecht 1997). Many microbial products, such as acids and amines, are detected by double-walled sensilla (Steinbrecht 1997). In the widely studied olfactory system of Drosophila melanogaster, basiconics and trichoids are types of single-walled sensilla and coeloconics are double-walled sensilla (Shanbhag et al. 1999).
Insects utilize a large number of receptors to detect and discriminate the immense number and variety of odors that exist in the environment. Research over the past two decades has characterized two large odor receptor families: Ionotropic Receptors (IRs) and Odorant Receptors (ORs) (Joseph and Carlson 2015; Robertson 2019). In addition to odor receptor co-receptors, most ORNs express only one or a few members of one receptor family and these largely determine the ORN’s odor response profile (Couto et al. 2005; Fishilevich and Vosshall 2005; Benton et al. 2009; Joseph and Carlson 2015). In Drosophila melanogaster, OR receptors are expressed by ORNs in single-walled sensilla, and IR are receptors expressed in double-walled sensilla (Joseph and Carlson 2015). Recent studies suggest this segregation also holds in other insect species (Pitts et al. 2004; Yang et al. 2012; Guo et al. 2014).
Numerous studies have examined the contribution of individual odor receptors to the detection of particular odorants. Such studies have been primarily carried out in Drosophila due to the number of available genetic tools that support molecular manipulations of specific receptors and neurons (Hallem and Carlson 2006). More recently, genomic studies and antennal transcriptome analysis have led to identification of odor receptors in non-model organisms, including mosquitoes and other vectors of disease (Grosse-Wilde et al. 2011; Liu et al. 2012; Rinker et al. 2013; Hansen et al. 2014; Matthews et al. 2016). With the exception of odor receptor co-receptors, most odor receptors are poorly conserved across species (Croset et al. 2010; Hansson and Stensmyr 2011; Andersson et al. 2015), making them poor targets for broad-spectrum insect repellents. This lack of conservation is consistent with the consensus that odor receptors evolve rapidly to reflect the ecology of particular organisms (Carey et al. 2010; Robertson 2019).
Currently there is a surprising lack of knowledge regarding specific molecules other than odor receptors that play critical roles in mediating the first steps of odor detection in the antenna. Such molecules could underlie signal amplification, adaptation, odor degradation, signal termination, the transepithelial electrical potential, or other signaling processes (Stengl et al. 1999; Leal 2013; Guo and Smith 2017). Importantly, such genes may show broader conservation across species than the odor receptors themselves.
Genes with a role in insect olfaction have been sought previously using behavioral screens to identify smell impaired mutants (McKenna et al. 1989; Anholt et al. 1996; Arya et al. 2015). These screens could not distinguish genes that play a role in olfactory circuits in the brain from those that contribute to odor responses in the antenna. More recent work has characterized gene expression in the antenna using RNA-Seq transcriptional profiling in Drosophila and other species (Grosse-Wilde et al. 2011; Liu et al. 2012; Rinker et al. 2013; Hansen et al. 2014; Menuz et al. 2014; Younus et al. 2014; Matthews et al. 2016). The challenge lies in identifying those genes that play a specific role in olfaction rather than mediating functions that generally support neuronal or antennal physiology. Genes that play critical roles in olfaction are not necessarily those with the highest antennal expression; for example, most odor receptors are expressed at relatively low levels (<50 FPKM) (Shiao et al. 2013; Menuz et al. 2014). One successful approach to overcoming this challenge is transcriptional profiling of genetic mutants. By comparing antennal gene expression in wild-type flies and atonal mutants that lack double-walled sensilla, we previously identified an uncharacterized transporter with an unexpectedly critical role in maintaining ORN activity (Menuz et al. 2014).
Here, we took two approaches to identify genes that are likely to play a role in antennal olfactory signaling. We first identified genes that are enriched in single-walled sensilla by generating an antennal transcriptome dataset for amos flies, which selectively fail to develop these sensilla (zur Lage et al. 2003). This dataset revealed 187 amos-depleted genes, including 35 expected members of the OR family. We then took a computational approach to identify 141 genes whose expression is greatly enriched in the Drosophila antenna. Orthologs of the antennal-enriched genes were identified in four insect species separated by >300 million years of evolution, and most orthologs were expressed in antennal transcriptomes of their respective species. Half of the AE genes could be mapped to either single or double walled sensilla, and functional annotation revealed new candidates for mediating signal amplification, adaptation, and odor degradation. Together, our two approaches have identified dozens of novel candidate olfaction genes that will be a boon for future studies of olfactory signaling in the periphery.
Materials and Methods
Drosophila melanogaster stocks
Mutant amos1 pr1/Cyo flies (zur Lage et al. 2003) were obtained from Richard Benton, and the line was outcrossed for ten generations to Canton-S wild-type flies to remove a lethal mutation. These outcrossed homozygous amos mutant flies and Canton-S wild-type flies were used for RNA-Seq studies. Canton-S flies were used for qRT-PCR experiments.
Total RNA extraction from fly tissues
Canton-S and amos flies (3-5 days old) were frozen in liquid nitrogen, and tissues were dissected into 1.5 ml Eppendorf tubes kept in liquid nitrogen. For RNA sequencing, each sample consisted of 300-400 antennae from approximately equal numbers of males and females. Analysis of the expression of known auditory organ genes found in the second antennal segment indicates that our samples predominantly contained antennal third (olfactory) segments as intended. Many of the most highly expressed genes in the second (auditory) segment of the antennae are not expressed in our CS dataset (File S1, “auditory gene expression”) (Senthilan et al., 2012).
For quantitative reverse transcriptase PCR (qRT-PCR), tissue samples consisted of legs (∼50), antennae (∼200), entire bodies without heads and legs (∼10), and heads (∼10). Tissues were ground using disposable RNAse-free pestles and a QIAshredder column (Qiagen). Total RNA was extracted using RNeasy Micro Kits (Qiagen) for qRT-PCR and an RNeasy Mini Kit (Qiagen) for RNA-Seq. For both, the manufacturer’s RNeasy Mini Kit protocol was followed as it gave better yield and purity. To increase the yield, the RNeasy spin column was rolled horizontally during the RW1 and RPE steps to ensure no sample remained on the wall and cap of the RNeasy spin column, and RLT was gently warmed before use.
Next generation sequencing
Three independent antennal RNA samples were collected from Canton-S and amos flies and then cleared of genomic DNA using the DNAse in an iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad). The six cleared RNA samples (∼0.5 µg each) were sent to the Center for Genome Innovation at the University of Connecticut for quality control, library preparation, and NextGen sequencing. Libraries were prepared using the Illumina mRNA sample prep kit for non-stranded RNA, and sequencing was carried out on an Illumina NextSeq 500 with 150 cycles to produce paired-end reads.
RNA-Seq quantification and differential expression analysis
Raw reads were downloaded from BaseSpace Sequence Hub (Illumina). Quality control on the reads was performed using Sickle with the default parameters to produce trimmed reads (Joshi 2011). The reference Drosophila melanogaster genome and gene annotation file were downloaded from NCBI (dm6, 2014, Release 6 plus ISO1 MT) (dos Santos et al. 2014). STAR was used with the default parameters to align trimmed reads to the reference genome with mitochondrial genes removed (Dobin et al. 2013). Programs were executed using the University of Connecticut High Performance Computing cluster. All mapped reads generated by STAR have been submitted to the NCBI SRA and are available under the BioProject accession number PRJNA532415.
Two methods were used to call gene expression. For differential expression analysis comparing CS and amos flies, HTSeq was used to obtain raw counts of reads mapping uniquely to each gene (Anders et al. 2015). HTSeq-count was used with the “intersection-nonempty” and “nonunique-none” modes to handle reads mapping to more than one feature (File S1, “HTSeq”). Raw counts were then analyzed using EdgeR v3.8 package in R studio v3.4.2 to identify differential gene expression (Robinson et al. 2010) (File S1, “amos EdgeR analysis”); this was the same method as used previously for atonal analysis (Menuz et al. 2014). Genes with a FDR <0.01 and more than fourfold change in expression between amos and CS flies were considered differentially expressed. Of these 341 genes, 187 were reduced in amos flies.
Using analogous criteria, we also identified 154 genes that were upregulated in amos mutants (File S1, “amos EdgeR analysis”). Closer analysis revealed that for 68 of these genes, all wt samples and two amos samples had very low expression (average RPM 1.8), whereas there was some increased expression in amos sample 3 likely due to slight contamination by an additional tissue. Therefore, we considered the remaining 86 genes to be those that were truly upregulated in amos mutants (File S1, “amos upregulated-genes”). To be certain that this variability in amos sample 3 did not influence our identification of amos-depleted genes, we verified that the expression values of each of the 187 amos-depleted genes were similar in all three samples of a particular genotype.
We used Cufflinks to compare gene expression across tissues because this was similar to the data processing used for the ModEncode datasets (Graveley et al. 2011; Trapnell et al. 2012; Brown et al. 2014) and because Cufflinks could provide values for gene expression as fragments per kilobase per million-mapped reads (FPKM). Cufflinks handles ambiguous reads by estimating transcript abundances, considering biases such as non-uniform distribution along the gene length. Library type was selected as “fr-firststrand” and the maximum number of fragments per locus was set to 1 billion in order to include genes with very high number of reads mapped to them, such as some OBPs. All other parameters were set to default. Per sample, 84–92% of total reads were aligned, for a total of ∼4.3 to 7.6 million aligned reads per sample (File S1, “Cufflinks”).
Identifying antennal enriched genes in Drosophila
There were 4,129 genes in CS wild-type antennae that are expressed >1 FPKM in each of the three samples and with an average expression of at least 10 FPKM. Expression levels of these genes in other tissues were obtained from datasets generated by the modENCODE consortium and downloaded from FlyBase (Roy et al. 2010; Graveley et al. 2011; Brown et al. 2014; Thurmond et al. 2019). These included transcriptomes of different Drosophila melanogaster tissues (larval digestive system, 4 days old adult digestive system, larval fat body, pupal fat body, larval salivary gland, pupal salivary gland, larval central nervous system, pupal central nervous system, 4 days old adult male head, and 4 days old adult female head) and whole bodies (embryo 22-24 hr, larva L1, larva L3, pupa P5, pupa P15, 5 days old adult male and 5 days old adult female). Custom scripts written in Spyder3.2.3, a scientific Python development environment from the Anaconda navigator, were used to create two tables: 1) the 4,129 antennal genes, their antennal expression, and their expression in other tissues, and 2) the 4,129 antennal genes, their antennal expression, and their expression in whole bodies over development. Genes with >10-fold higher expression in antennae compared to the ten tissue datasets and genes with >10-fold higher expression in antennae compared to seven whole body developmental time points were identified. The two gene datasets were then merged to identify 177 genes enriched in both. Automatic annotation using FlyBase determined that 141 of these genes encoded proteins, and these were considered the set of coding antennal-enriched (AE) genes for further analysis (File S1, “antennal-enriched genes”).
Quantitative real-time PCR validation of AE genes
Seven of the AE genes that had not been studied previously were selected for validation with qRT-PCR experiments. Equal amounts of RNA extracted from dissected fly tissues (200 ng per sample) were used to generate cDNA using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad). The cDNA was used as a template for qRT-PCR with the SsoFast EvaGreen Supermix (Bio-Rad), similar to (Menuz et al. 2014). The housekeeping gene eIF1A was used to normalize total cDNA levels between tissues. A CFX96 thermocycler (Bio-Rad) was used for the qRT-PCR assays. Gene expression in each tissue was measured relative to the expression of eIF1A in that sample. Each reaction was run on three to five independent tissue replicates. Differences between samples were analyzed with ANOVA followed by the Dunnett’s post hoc test using GraphPad Prism 7.
The following primer pairs were used:
eIF1A: ATCAGCTCCGAGGATGACGC and GCCGAGACAGACGTTCCAGA
CG10357: GCAGTCATGTTCTACGCTG and CCTTAGGCAGTCCTCTACA
CG14142: GCACTTAACATGCCTAAAAGT and CGAAAGTATTCACCGCCGA
CG34309: CCATTTGCGGATTTGTACTTC and ATTCGTTCTTCAGTCAGCGG
CG42284: CCAGAGTCAGAGAACGATAACG and CCATTGCTGATGTGAGTGAC
CG14661: CCTGTTGATCTGTTTTGTAGC and CAGTTCCTTAATGCCCTTGG
CG9541: CCAGTCGCCGATTATTTCG and GTATCCCTCTGCCCATACC
CG14445: GTGCGAAAAGAACGAGTTCC and CGTAGGATGGGGTCAATAGG
Analysis of ortholog expression in the antennae of other insect species
We searched for orthologs in other insect species for 96 of the AE genes. Known chemoreceptors (IRs/ORs/Grs) and OBPs were excluded because their conservation has been extensively researched in prior studies. The UniProt ID for each of the 96 genes was obtained using the Flybase gene symbol as a query in UniProtKB (UniProt Consortium 2018). Orthologs of these proteins were identified using InParanoid 8 using the Drosophila AE gene UniProt ID as the query (Sonnhammer and Östlund 2014). Four insect species were selected: Harpegnathos saltator (ants), Tribolium castaneum (beetles), Anopheles gambiae (mosquitos), and Apis mellifera (bees). These were selected because they represent a wide distribution along the insect phylogenetic tree, and because antennal transcriptomes for these species could be found in the NCBI SRA (Leinonen et al. 2010; Missbach et al. 2014). Conservative criteria were applied to identify unambiguous orthologs and to exclude inparalogs. Orthologs were identified when the bootstrap value for the match was >90% and the identified ortholog mapped back to the original Drosophila gene when used in a BLAST search of Drosophila melanogaster proteins.
The NCBI SRA was utilized to determine if the AE gene orthologs are expressed in the antennae of their respective insect species. To do this, the UniProt gene IDs of the orthologs were first used to obtain the corresponding RefSeq mRNA sequences. These mRNA sequences were used to query the antennal transcriptomes of the respective species using NCBI SRA-BLAST (Harpegnathos saltator, SRX1164271; Tribolium castaneum, SRX757109; Apis mellifera, SRX518058; Anopheles gambiae, SRX765675) (Zhou et al. 2012; Dippel et al. 2014; Hodges et al. 2014; Jasper et al. 2014). We later interrogated two additional transcriptomes for Figure S1 using a second Tribolium castaneum dataset, SRX757107, and one from Bombyx mori, SRX3181880 (Dippel et al. 2014; Qiu et al. 2018). Perfect matches (32-100 bp depending on the deposited read lengths) were used to identify expression of each ortholog. The number of reads mapping to a gene was used to estimate its antennal expression in FPKM based on the mRNA length and total number of reads in the sample. Genes were considered expressed in the antenna if they had expression >3 FPKM. To analyze expression of random genes in the antennal transcriptomes, we randomly selected genes from Drosophila, identified orthologs in the other insect genomes, and determined the percentage of orthologs expressed in the antennal transcriptomes, similar to the analysis of the AE gene orthologs.
We note that after completing the study, we learned that the original paper for the Anopheles transcriptome was retracted due to contamination of samples from the maxillary palp, but not those from the antenna (Hodges et al. 2015). Therefore, our analysis of the antennal dataset should not be affected.
Functional annotation of amos-depleted and AE genes
FlyBase, NCBI BLAST, and DAVID were used to manually curate protein sequences (Huang et al. 2009; Thurmond et al. 2019). FlyBase integrates information from different databases including GO terms, UniProt protein families, and InterPro protein domains. SignalP 4.1 and TMHMM v.2.0 were used to identify signal peptide sequences and transmembrane domains and DeepLoc to infer cellular localization (Möller et al. 2001; Almagro Armenteros et al. 2017; Nielsen 2017). Significant enrichment of GO terms in the amos-depleted dataset was determined using PANTHER 14.0 in AmiGO 2 with GO database release 2018-12-01 (Ashburner et al. 2000; Carbon et al. 2009; Mi et al. 2016; Gene Ontology Consortium 2019).
Data availability
All raw read data are available at BioProject accession number PRJNA532415. Analyzed data reporting gene expression are available in File S1. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9820682.
Results
Identification of genes highly expressed in single-walled sensilla
Given the morphological and molecular differences of the single and double-walled sensilla, it is likely that they have unique molecular signaling pathways. The function of single-walled sensilla has been of particular interest due to their importance in the detection of pheromones by many insects and host-derived odors by insect vectors of disease (Kaissling et al. 1989; Degennaro et al. 2013; Mcmeniman et al. 2014; Ghaninia et al. 2018; Chahda et al. 2019). Previously we successfully used transcriptional profiling of atonal antennae, which lack double-walled sensilla, to identify a novel gene essential for the response of ORNs to ammonia (Menuz et al. 2014). We therefore decided to take a similar genetic approach to identify genes enriched in Drosophila single-walled sensilla.
To do this, we used RNA-Seq to compare antennal expression of genes in wild-type and amos antennae, which lack single-walled sensilla including basiconic and trichoid sensilla, but have normal numbers of double-walled sensilla (Figure 1A) (zur Lage et al. 2003). These flies entirely lack single-walled sensilla cells, including ORNs and support cells that derive from sensory organ precursors (SOPs) in development (zur Lage et al. 2003). We first outcrossed the amos line for ten generations to wild-type (wt) Canton-S flies to remove a nearby lethal mutation. We then carried out next generation transcriptional profiling on three sets of antennae from amos and wt flies (File S1, “HtSeq”). As expected, most genes are expressed at similar levels in the two genotypes (Figure 1B). This is consistent with the idea that many genes have a ubiquitous role across the sensillar populations. However, the expression of 187 genes is statistically reduced at least fourfold in amos mutants (File S1, “amos edgeR analysis”). We considered these genes amos-depleted.
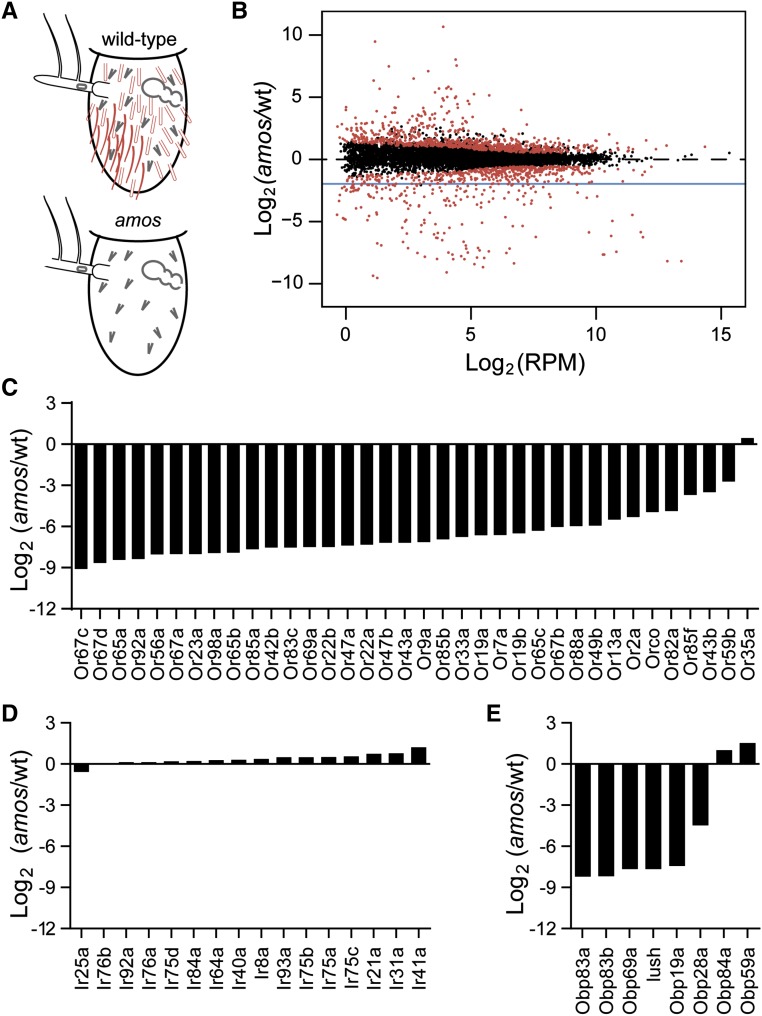
Figure 1.
Identification of genes selectively expressed in single-walled sensilla. (A) amos flies fail to develop single-walled sensilla, including trichoid (red filled) and basiconic (red outline) subtypes in contrast to wild-type flies, whereas coeloconic sensilla are present in normal numbers (gray). Schematic based on findings from zur Lage et al., 2003 and adapted from Menuz et al., 2014. (B) Scatterplot showing the Drosophila genes (dots) plotted based on their average expression level in RPM and the expression ratio between amos and wt flies. Genes were considered amos-depleted if they were differentially expressed with FDR <0.01 (red) and were at least fourfold reduced in amos flies (below blue line). (C) Expression of antennal OR genes is greatly reduced in amos mutants, but (D) IR expression is not. (E) Six OBPs found in single-walled sensilla (Obp83a, Obp83b, Obp69a, lush, Obp19a, and Obp28a) have greatly reduced expression in amos antennae, but two Obps found in double-walled sensilla (Obp84a and Obp59a) do not.
We then examined the expression of genes known to be localized to single-walled sensilla to validate the specificity of the amos mutation and our approach. Our wt dataset contained 36 members of the OR receptor family that were expressed in the wild-type antenna, and all but one gene was amos-depleted (Figure 1C). The exception, Or35a, is the only member of the OR family expressed in double-walled sensilla (Couto et al. 2005). Conversely, none of the 16 IR family members were identified as amos-depleted (Figure 1D). We also examined the predicted localization of eight antennal Odorant Binding Proteins (OBPs) that were detected by in situ hybridization in either single or double-walled sensilla (Larter et al. 2016). The six OBPs detected in single-walled sensilla were amos-depleted, whereas the two OBPs found in double-walled sensilla were not (Figure 1E). These data confirm that single-walled sensilla are indeed selectively lost in amos antennae and that our approach identifies genes enriched in single-walled sensilla. We note that although most of these 187 genes are likely enriched in single-walled sensilla, there could be some remaining genetic background differences between amos and wt flies after outcrossing, and this could contribute to some of the observed differences in expression for some genes.
Many of the single-wall sensilla enriched genes are transmembrane proteins
Having validated the use of amos mutants, we next looked more closely at the 187 genes identified as amos-depleted (File S1, “amos edgeR analysis”). The ten genes reduced with the greatest statistical significance included five genes previously shown to be localized to single-walled sensilla: three members of the OR family, Obp28a, and an enzyme Jhedup (Figure 2A) (Couto et al. 2005; Larter et al. 2016; Steiner et al. 2017). The localization of the other four genes had not been studied in the antenna previously.
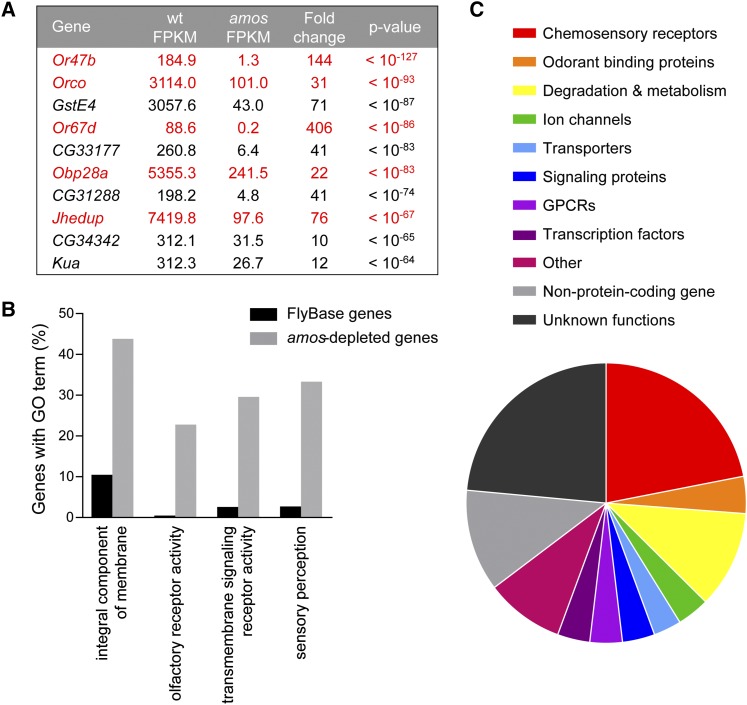
Figure 2.
amos-depleted genes fulfill a variety of molecular functions. (A) The top ten differentially expressed genes based on p-value include five genes with known localization to single-walled sensilla (red). (B) GO term enrichment analysis of the 187 amos-depleted genes compared to all Drosophila genes listed in FlyBase reveals several enriched GO terms. (C) The chart depicts the frequency of amos-depleted genes falling into different functional categories.
We then turned to Gene Ontology annotation to determine if particular categories of genes were enriched among the amos-depleted genes. Using AmiGO 2, we found that 162 of 187 amos-depleted genes had been previously annotated with Gene Ontology (GO) terms (Ashburner et al. 2000; Carbon et al. 2009; Gene Ontology Consortium 2019). The frequency of GO terms in our dataset was statistically compared to their frequency among all FlyBase genes using PANTHER version 14 (Figure 2B) (Mi et al. 2016). This revealed that the amos-depleted genes are particularly enriched in transmembrane proteins; the GO term “integral component of membrane” was associated with 43.8% of amos-depleted genes vs. 10.5% of FlyBase genes (P < 10−23). This is in part due to the enrichment of genes labeled with the molecular functions “olfactory receptor activity” (22.8% in amos-depleted vs. 0.5% of FlyBase genes, P < 10−41) and “transmembrane signaling receptor activity” (29.6% in amos-depleted vs. 2.6% of FlyBase genes, P < 10−31). Genes associated with the GO biological process term “sensory perception” were also highly enriched among the amos-depleted genes (33.3%) compared to 2.8% of FlyBase genes (P < 10−37).
Functional annotation of the amos-depleted genes was accomplished with BLAST searches and FlyBase tools (Figure 2C, Table 1) (Thurmond et al. 2019). Putative functions could be assigned to ∼75% of the genes. In addition to the large number of OR receptors, our dataset included six members of the GR gustatory receptor family (Gr21a, Gr43a, Gr63a, Gr64a, Gr64f, and Gr93a). Together OR and GR chemoreceptors represent ∼20% of the amos-depleted genes.
Table 1. 187 genes depleted in amos antennae.
| FBgn ID | Symbol | Function |
|---|---|---|
| CHEMOSENSORY RECEPTORS | ||
| FBgn0030715 | Or13a | OR chemosensory receptor |
| FBgn0041626 | Or19a | OR chemosensory receptor |
| FBgn0062565 | Or19b | OR chemosensory receptor |
| FBgn0026398 | Or22a | OR chemosensory receptor |
| FBgn0026397 | Or22b | OR chemosensory receptor |
| FBgn0026395 | Or23a | OR chemosensory receptor |
| FBgn0023523 | Or2a | OR chemosensory receptor |
| FBgn0026392 | Or33a | OR chemosensory receptor |
| FBgn0033043 | Or42b | OR chemosensory receptor |
| FBgn0026389 | Or43a | OR chemosensory receptor |
| FBgn0026393 | Or43b | OR chemosensory receptor |
| FBgn0026386 | Or47a | OR chemosensory receptor |
| FBgn0026385 | Or47b | OR chemosensory receptor |
| FBgn0028963 | Or49b | OR chemosensory receptor |
| FBgn0034473 | Or56a | OR chemosensory receptor |
| FBgn0034865 | Or59b | OR chemosensory receptor |
| FBgn0041625 | Or65a | OR chemosensory receptor |
| FBgn0041624 | Or65b | OR chemosensory receptor |
| FBgn0041623 | Or65c | OR chemosensory receptor |
| FBgn0036009 | Or67a | OR chemosensory receptor |
| FBgn0036019 | Or67b | OR chemosensory receptor |
| FBgn0036078 | Or67c | OR chemosensory receptor |
| FBgn0036080 | Or67d | OR chemosensory receptor |
| FBgn0041622 | Or69a | OR chemosensory receptor |
| FBgn0030016 | Or7a | OR chemosensory receptor |
| FBgn0041621 | Or82a | OR chemosensory receptor |
| FBgn0037399 | Or83c | OR chemosensory receptor |
| FBgn0037576 | Or85a | OR chemosensory receptor |
| FBgn0037590 | Or85b | OR chemosensory receptor |
| FBgn0037685 | Or85f | OR chemosensory receptor |
| FBgn0038203 | Or88a | OR chemosensory receptor |
| FBgn0038798 | Or92a | OR chemosensory receptor |
| FBgn0039551 | Or98a | OR chemosensory receptor |
| FBgn0030204 | Or9a | OR chemosensory receptor |
| FBgn0037324 | Orco | OR chemosensory receptor |
| FBgn0041250 | Gr21a | GR chemosensory receptor |
| FBgn0041243 | Gr43a | GR chemosensory receptor |
| FBgn0035468 | Gr63a | GR chemosensory receptor |
| FBgn0045479 | Gr64a | GR chemosensory receptor |
| FBgn0052255 | Gr64f | GR chemosensory receptor |
| FBgn0041229 | Gr93a | GR chemosensory receptor |
| ODORANT BINDING PROTEINS | ||
| FBgn0020277 | lush | Odorant Binding Protein |
| FBgn0010403 | Obp83b | Odorant Binding Protein |
| FBgn0011283 | Obp28a | Odorant Binding Protein |
| FBgn0046878 | Obp83cd | Odorant Binding Protein |
| FBgn0011281 | Obp83a | Odorant Binding Protein |
| FBgn0031109 | Obp19a | Odorant Binding Protein |
| FBgn0046876 | Obp83ef | Odorant Binding Protein |
| FBgn0011279 | Obp69a | Odorant Binding Protein |
| TRANSPORTERS | ||
| FBgn0025709 | CNT2 | SLC28 nucleoside transporter |
| FBgn0033685 | OSCP1 | Organic solute carrier partner 1 (OSCP1) |
| FBgn0032879 | CarT | SLC22 carcinine transporter |
| FBgn0039915 | Gat | SLC6 GABA transporter |
| FBgn0038716 | CG7342 | SLC22 transporter |
| FBgn0031998 | SLC5A11 | SLC5 sodium/sugar transporter |
| ION CHANNELS | ||
| FBgn0003861 | trp | transient receptor potential channel |
| FBgn0031803 | ppk14 | sodium channel |
| FBgn0039679 | ppk19 | sodium channel |
| FBgn0053289 | ppk5 | sodium channel |
| FBgn0034489 | ppk6 | sodium channel |
| FBgn0053349 | ppk25 | sodium channel |
| FBgn0036235 | CG6938 | anoctamin |
| G-PROTEIN COUPLED RECEPTORS | ||
| FBgn0087012 | 5-HT2A | serotonin receptor |
| FBgn0261929 | 5-HT2B | serotonin receptor |
| FBgn0050106 | CCHa1-R | CCHamide neuropeptide receptor |
| FBgn0052447 | CG32447 | class C GPCR |
| FBgn0030437 | hec | GPCR involved in mating |
| FBgn0016650 | Lgr1 | hormone receptor |
| FBgn0037546 | mAChR-B | acetylcholine receptor |
| SIGNALING MOLECULES | ||
| FBgn0010223 | Galphaf | G protein |
| FBgn0050274 | CG30274 | kinase |
| FBgn0032083 | CG9541 | kinase |
| FBgn0037167 | CG11425 | lipid phosphatase |
| FBgn0011676 | Nos | nitric oxide synthase |
| FBgn0250862 | CG42237 | phospholipase A2 |
| FBgn0002937 | ninaB | retinal isomerase |
| DEGRADATION & METABOLISM | ||
| FBgn0031426 | CG18641 | carboxylic ester hydrolase |
| FBgn0033395 | Cyp4p2 | cyp450 |
| FBgn0000473 | Cyp6a2 | cyp450 |
| FBgn0038029 | GstD11 | glutathione S transferase |
| FBgn0010044 | GstD8 | glutathione S transferase |
| FBgn0063496 | GstE4 | glutathione S transferase |
| FBgn0034076 | Jhedup | carboxylesterase |
| FBgn0000075 | amd | carboxylase |
| FBgn0263830 | CG40486 | estradiol 17-beta-dehydrogenase |
| FBgn0069973 | CG40485 | estradiol 17-beta-dehydrogenase |
| FBgn0039131 | CG12268 | fatty acyl coA reductase |
| FBgn0085371 | CG34342 | fatty acyl coA reductase |
| FBgn0000078 | Amy-d | amylase |
| FBgn0000079 | Amy-p | amylase |
| FBgn0050502 | fa2h | oxidoreductase |
| FBgn0035743 | Acbp6 | acyl-CoA-binding protein (ACBP) |
| FBgn0265268 | CG18234 | peptidyl-proline dioxygenase |
| FBgn0033521 | CG12896 | peroxiredoxins |
| FBgn0051115 | CG31115 | S-methyl-5-thioadenosine phosphorylase |
| FBgn0053177 | CG33177 | glutathione peroxidase |
| FBgn0051644 | CG31644 | mitochondrial cytochrome-c oxidase |
| TRANSCRIPTION FACTORS | ||
| FBgn0002633 | E(spl)m7-HLH | basic helix-loop-helix transcription factor |
| FBgn0034096 | CG7786 | basic leucine zipper transcription factor |
| FBgn0041105 | nerfin-2 | C2H2 zinc finger transcription factor |
| FBgn0019650 | toy | homeobox transcription factor |
| FBgn0015561 | unpg | homeobox transcription factor |
| FBgn0020378 | Sp1 | Sp1/Klf family transcription factor |
| FBgn0000233 | btd | Sp1/Klf family transcription factor |
| OTHER FUNCTIONS | ||
| FBgn0010385 | Def | antibacterial peptide |
| FBgn0004781 | Ccp84Ac | cuticle component |
| FBgn0033597 | Cpr47Ea | cuticle component |
| FBgn0039347 | CG5071 | cyclophilin family peptidylprolyl isomerase |
| FBgn0001174 | halo | kinesin-1 co-factor |
| FBgn0260004 | Snmp1 | pheromone detection/lipoprotein receptor |
| FBgn0004511 | dy | zona pellucida domain protein family |
| FBgn0037433 | CG17919 | phosphatidylethanolamine-binding protein |
| FBgn0029977 | hdm | DNA binding protein/repair |
| FBgn0035608 | blanks | RNA binding protein |
| FBgn0035626 | lin-28 | RNA binding protein |
| FBgn0029843 | Nep1 | metallopeptidase |
| FBgn0031560 | CG16713 | serine endopeptidase inhibitor |
| FBgn0262721 | CG43165 | serine endopeptidase inhibitor |
| FBgn0013433 | beat-Ia | axon guidance |
| FBgn0038498 | beat-IIa | axon guidance |
| FBgn0259210 | prom | photoreceptor positioning |
| UNKNOWN FUNCTIONS | ||
| FBgn0050488 | antr | cystein rich secretory protein |
| FBgn0014000 | Hf | helical cytokine/innate immunity |
| FBgn0032850 | Kua | transmembrane protein 189 |
| FBgn0261534 | l(2)34Fc | insect defense protein |
| FBgn0033855 | link | secreted protein, localized near midline |
| FBgn0042129 | OS9 | olfactory specific, small secreted protein |
| FBgn0037427 | Osi17 | insect Osiris transmembrane protein |
| FBgn0037414 | Osi7 | insect Osiris transmembrane protein |
| FBgn0037415 | Osi8 | insect Osiris transmembrane protein |
| FBgn0033042 | Tsp42A | tetraspannin four transmembrane protein |
| FBgn0033127 | Tsp42Ef | tetraspannin four transmembrane protein |
| FBgn0033135 | Tsp42En | tetraspannin four transmembrane protein |
| FBgn0038028 | CG10035 | |
| FBgn0032843 | CG10730 | |
| FBgn0039297 | CG11852 | |
| FBgn0040688 | CG12483 | |
| FBgn0030886 | CG12672 | |
| FBgn0033501 | CG12911 | |
| FBgn0037013 | CG13250 | |
| FBgn0039319 | CG13659 | |
| FBgn0031219 | CG13694 | |
| FBgn0030277 | CG1394 | |
| FBgn0032734 | CG15169 | |
| FBgn0032733 | CG15170 | |
| FBgn0039723 | CG15522 | |
| FBgn0032719 | CG17321 | |
| FBgn0036923 | CG17732 | |
| FBgn0050339 | CG30339 | |
| FBgn0050356 | CG30356 | |
| FBgn0051097 | CG31097 | |
| FBgn0051288 | CG31288 | |
| FBgn0036459 | CG3349 | |
| FBgn0085195 | CG34166 | |
| FBgn0259831 | CG34309 | |
| FBgn0260657 | CG42540 | |
| FBgn0261834 | CG42766 | |
| FBgn0262858 | CG43222 | |
| FBgn0263656 | CG43647 | |
| FBgn0264443 | CG43861 | |
| FBgn0034128 | CG4409 | |
| FBgn0039346 | CG5079 | |
| FBgn0030913 | CG6123 | |
| FBgn0039728 | CG7896 | |
| FBgn0032085 | CG9555 | |
| NON-PROTEIN CODING GENES | ||
| FBgn0263453 | asRNA:CR43476 | antisense RNA CG43476 |
| FBgn0264874 | asRNA:CR44065 | antisense RNACG10874 |
| FBgn0266144 | asRNA:CR44850 | antisense RNA l(2)34Fc |
| FBgn0266275 | asRNA:CR44960 | antisense RNA CG33178 |
| FBgn0266402 | asRNA:CR45042 | antisense RNA Gr64d, Gr64e |
| FBgn0263444 | asRNA:CR43467 | antisense RNA Or46a |
| FBgn0265168 | asRNA:CR44237 | antisense RNA Or88a |
| FBgn0050009 | lncRNA:CR30009 | long non-coding RNA |
| FBgn0263509 | lncRNA:CR43498 | long non-coding RNA |
| FBgn0264382 | lncRNA:CR43834 | long non-coding RNA |
| FBgn0264438 | lncRNA:CR43856 | long non-coding RNA |
| FBgn0264835 | lncRNA:CR44043 | long non-coding RNA |
| FBgn0265312 | lncRNA:CR44285 | long non-coding RNA |
| FBgn0265376 | lncRNA:CR44317 | long non-coding RNA |
| FBgn0265718 | lncRNA:CR44525 | long non-coding RNA |
| FBgn0265734 | lncRNA:CR44541 | long non-coding RNA |
| FBgn0266828 | lncRNA:CR45290 | long non-coding RNA |
| FBgn0266859 | lncRNA:CR45320 | long non-coding RNA |
| FBgn0267058 | lncRNA:CR45502 | long non-coding RNA |
| FBgn0267938 | lncRNA:CR46218 | long non-coding RNA |
| FBgn0267965 | lncRNA:CR46245 | long non-coding RNA |
| FBgn0263472 | snoRNA:2R:9445205 | small nucleolar RNA |
Interestingly, many amos-depleted genes have potential roles in neuronal signaling (Table 1). We identified 13 transporters and ion channels, including five members of the pickpocket family of degenerin/epithelial Na+ channels (ENaCs). There are also seven GPCRs, including serotonin and acetylcholine receptors that are known to regulate presynaptic terminals in other neuronal systems (Hoyer et al. 1994; Caulfield and Birdsall 1998). Additionally, there are also several signaling molecules such as kinases that are amos-depleted.
Similar to genes that were enriched in double-walled coeloconic sensilla, many genes enriched in single-walled sensilla are likely to function in odor degradation and metabolism such as cytochrome p450s and reductases (Table 1) (Leal 2013; Menuz et al. 2014). Such genes comprise more than 10% of the amos-depleted genes. Most have not been studied previously, although a few such as the carboxylesterase Jhedup and the glutathione S transferases (GSTs) GstD11 and GstD8 were shown to be enriched in the antenna compared to other tissues (Younus et al. 2014; Steiner et al. 2017).
Single-walled sensilla also selectively express proteins with a variety of other functions. Some genes could play a developmental role such as axon guidance molecules from the beat family and seven transcription factors, including CG7786 and Sp1. The amos-depleted genes also include more than 20 non-protein coding genes, the majority of which are long non-coding RNAs. Interestingly, three are antisense RNAs which target OR and GR receptors.
We also looked at the putative functions of the 86 amos-upregulated genes (File S1, “amos-upregulated genes”). This set of genes included 12 non-coding RNAs, 17 biotransformation enzymes, four GPCRs, and five chitin-related genes in addition to others with diverse predicted functions. There were also four genes implicated in immune system activity and nine genes related to serine proteases and their inhibitors, which can also be involved in defense responses. This could indicate that the amos mutants are infected by microbes, consistent with their reduced fitness compared to wild-type flies. Other upregulated genes may be expressed in the ectopic mechanosensory bristles found on amos antennae (zur Lage et al. 2003). Some genes may be upregulated in the remaining coeloconic sensilla to compensate for the loss of olfactory sensing mediated by single-walled sensilla, which contain more than 3/4 of all ORNs (Shanbhag et al. 1999). For example, such compensation could explain the upregulation of the Odorant Binding Protein Obp57e and the chemosensory receptor Gr98b.
Identification of antennal-enriched genes in Drosophila
We then sought a different approach to identify genes that are involved specifically in insect olfaction, particularly those that play a conserved role across insect species. We reasoned that many of these molecules are likely to be highly enriched in the antenna compared to other tissues in the same insect. Recently, there has been an explosion of RNA-Seq datasets quantifying the transcriptomes of various tissues (Grosse-Wilde et al. 2011; Liu et al. 2012; Rinker et al. 2013; Hansen et al. 2014; Menuz et al. 2014; Younus et al. 2014; Matthews et al. 2016). The RNA-Seq technique provides a quantitative measure (“FPKM”) of gene expression that reflects the portion of transcripts in a given transcriptome from that particular gene. A key advantage of RNA-Seq compared to earlier microarray technology is its quantitative nature, allowing expression of a given gene to be compared across tissues and datasets (Wang et al. 2009).
In order to identify conserved antennal-enriched genes, we first focused on Drosophila melanogaster because we could leverage the high-quality transcriptome datasets generated by the ModEncode consortium for this species (Graveley et al. 2011; Brown et al. 2014). The ModEncode consortium has systematically characterized gene expression in Drosophila at a high sequencing depth across several tissues and whole-body extracts at multiple developmental time points. Such depth is critical for accurate quantification of gene expression in tissues with low expression.
In order to compare ModEncode data with our antennal data, we re-quantified gene expression in our wild-type samples using a similar computational pipeline as the ModEncode datasets (File S1, “Cufflinks”) (Graveley et al. 2011; Brown et al. 2014). We then compared the mean expression of each gene expressed in the antenna with its expression in ten ModEncode tissue datasets including heads, digestive systems, fat bodies, etc. (Figure 3) (Brown et al. 2014). We specifically chose several tissues enriched with neurons to rule out genes involved in neuronal function rather than specific roles in olfaction. By comparing expression across datasets, we identified genes whose expression was at least 10-fold higher in antennae compared to all other tissues examined. Given that the ModEncode data has integer values (0, 1, 2 FPKM, etc.), we conservatively required that any identified antennal-enriched genes should also have greater than 10 FPKM expression in the antenna to ensure it was truly 10-fold enriched. With these strict criteria, we identified 280 genes more than 10 times as abundant in the antennae compared to other tissues (Figure 3).
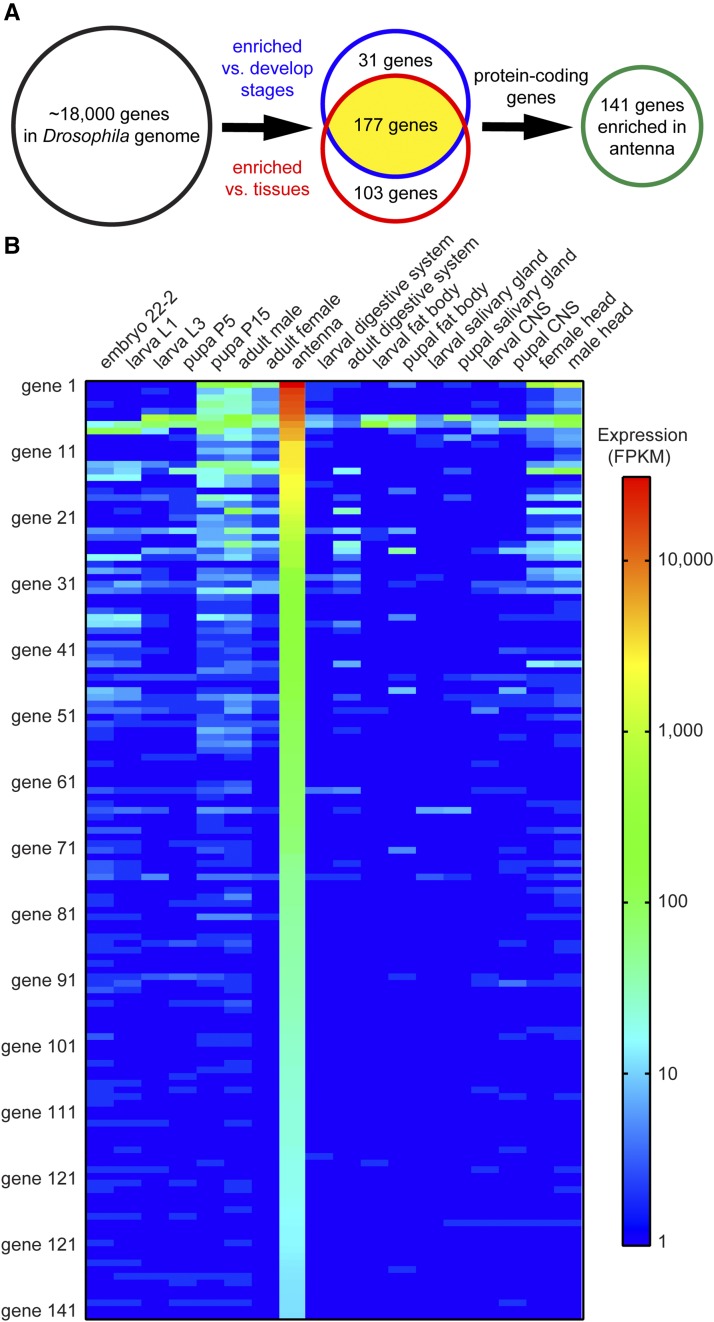
Figure 3.
Computational identification of 141 antennal-enriched genes in Drosophila. (A) Summary of bioinformatic pipeline to identify antennal-enriched (AE) genes in Drosophila. Of >17,000 Drosophila genes, expression of 280 genes was >10-fold higher in antennae compared to any of ten tissue samples, and expression of 208 genes was >10-fold higher than seven samples of whole-bodies from different developmental stages. A subset of 177 genes was found in both groups, and among these were 141 protein-encoding genes. (B) Heat map of expression in FPKM of the 141 AE genes in the tissues and whole-body samples examined.
The ModEncode consortium has also profiled gene expression across whole body extracts from multiple developmental stages in Drosophila (Graveley et al. 2011). Genes with specific olfactory functions should also be more than 10-fold enriched in antennae compared to expression across the body at any one developmental stage. Using the same methods as above, we compared antennal expression with expression in six developmental stages ranging from embryos to adults. We included both male and female adults, in case there is sexual dimorphism in gene expression. We identified 208 genes whose expression was 10-fold enriched in antennae compared to these developmental stages (Figure 3).
Finally, we merged the two gene lists to identify 177 genes that are enriched in the antenna compared to both the tissue and developmental datasets. Computational characterization of the genes using FlyBase indicated that 36 genes are non-protein coding. Further analysis focused on the remaining 141 “antennal-enriched” (AE) protein-coding genes (Figure 3B, Table 2, File S1, “antennal enriched genes”).
Table 2. Function and localization of 141 antennal-enriched (AE) genes.
| FBgn ID | Symbol | FPKM | amos-depleted | ato-depleted | Function |
|---|---|---|---|---|---|
| CHEMOSENSORY RECEPTORS | |||||
| FBgn0031634 | Ir25a | 49 | IR chemosensory receptor | ||
| FBgn0051718 | Ir31a | 11 | a | IR chemosensory receptor | |
| FBgn0035604 | Ir64a | 17 | X | IR chemosensory receptor | |
| FBgn0036757 | Ir75a | 44 | X | IR chemosensory receptor | |
| FBgn0036829 | Ir75d | 30 | X | IR chemosensory receptor | |
| FBgn0036937 | Ir76b | 70 | X | IR chemosensory receptor | |
| FBgn0052704 | Ir8a | 25 | X | IR chemosensory receptor | |
| FBgn0030715 | Or13a | 13 | X | OR chemosensory receptor | |
| FBgn0062565 | Or19b | 16 | X | OR chemosensory receptor | |
| FBgn0026398 | Or22a | 36 | X | OR chemosensory receptor | |
| FBgn0026397 | Or22b | 25 | X | OR chemosensory receptor | |
| FBgn0026395 | Or23a | 11 | X | OR chemosensory receptor | |
| FBgn0028946 | Or35a | 18 | X | OR chemosensory receptor | |
| FBgn0033043 | Or42b | 76 | X | OR chemosensory receptor | |
| FBgn0026389 | Or43a | 21 | X | OR chemosensory receptor | |
| FBgn0026386 | Or47a | 45 | X | OR chemosensory receptor | |
| FBgn0026385 | Or47b | 86 | X | OR chemosensory receptor | |
| FBgn0034473 | Or56a | 21 | X | OR chemosensory receptor | |
| FBgn0034865 | Or59b | 82 | X | OR chemosensory receptor | |
| FBgn0041625 | Or65a | 28 | X | OR chemosensory receptor | |
| FBgn0041624 | Or65b | 32 | X | OR chemosensory receptor | |
| FBgn0041623 | Or65c | 10 | X | OR chemosensory receptor | |
| FBgn0036009 | Or67a | 14 | X | OR chemosensory receptor | |
| FBgn0036078 | Or67c | 13 | X | OR chemosensory receptor | |
| FBgn0036080 | Or67d | 41 | X | OR chemosensory receptor | |
| FBgn0041622 | Or69a | 14 | X | OR chemosensory receptor | |
| FBgn0030016 | Or7a | 23 | X | OR chemosensory receptor | |
| FBgn0037576 | Or85a | 17 | X | X, b | OR chemosensory receptor |
| FBgn0037590 | Or85b | 21 | X | OR chemosensory receptor | |
| FBgn0038798 | Or92a | 85 | X | OR chemosensory receptor | |
| FBgn0039551 | Or98a | 39 | X | OR chemosensory receptor | |
| FBgn0030204 | Or9a | 35 | X | OR chemosensory receptor | |
| FBgn0037324 | Orco | 894 | X | OR chemosensory receptor | |
| FBgn0045502 | Gr10a | 23 | c | GR chemosensory receptor | |
| FBgn0041250 | Gr21a | 11 | X | GR chemosensory receptor | |
| FBgn0052255 | Gr64f | 31 | X | GR chemosensory receptor | |
| ODORANT BINDING PROTEINS | |||||
| FBgn0020277 | lush | 1,718 | X | Odorant Binding Protein | |
| FBgn0031109 | Obp19a | 2,997 | X | Odorant Binding Protein | |
| FBgn0011280 | Obp19d | 30,847 | Odorant Binding Protein | ||
| FBgn0011283 | Obp28a | 5,444 | X | Odorant Binding Protein | |
| FBgn0034766 | Obp59a | 1,261 | X | Odorant Binding Protein | |
| FBgn0011279 | Obp69a | 3,026 | X | Odorant Binding Protein | |
| FBgn0011281 | Obp83a | 15,113 | X | Odorant Binding Protein | |
| FBgn0010403 | Obp83b | 17,591 | X | Odorant Binding Protein | |
| FBgn0011282 | Obp84a | 2,156 | X | Odorant Binding Protein | |
| SMALL SECRETED PROTEINS, unknown functions | |||||
| FBgn0010401 | Os-C | 12,694 | X | ||
| FBgn0011293 | a10 | 13,648 | X | ||
| FBgn0259831 | CG34309 | 190 | X | ||
| FBgn0259098 | CG42246 | 46 | |||
| FBgn0040688 | CG12483 | 96 | X | X | |
| FBgn0261501 | CG42649 | 39 | |||
| FBgn0262540 | CG43094 | 140 | |||
| FBgn0262541 | CG43095 | 11 | |||
| FBgn0085209 | CG34180 | 40 | |||
| FBgn0263654 | CG43645 | 88 | |||
| FBgn0034486 | CG13869 | 15 | |||
| FBgn0262856 | CG43220 | 604 | |||
| JUVENILE HORMONE BINDING PROTEIN | |||||
| FBgn0037288 | CG14661 | 2,725 | haemolymph juvenile hormone binding protein | ||
| FBgn0039298 | to | 1,554 | haemolymph juvenile hormone binding protein | ||
| FBgn0038850 | CG17279 | 145 | X | haemolymph juvenile hormone binding protein | |
| CILIA-RELATED PROTEINS | |||||
| FBgn0034446 | Arl6 | 67 | BBSome | ||
| FBgn0033578 | BBS4 | 44 | BBSome | ||
| FBgn0037280 | BBS5 | 39 | BBSome | ||
| FBgn0034622 | BBS9 | 37 | BBSome | ||
| FBgn0032119 | CG3769 | 14 | dynein | ||
| FBgn0033140 | CG12836 | 19 | X | dynein | |
| FBgn0034481 | CG11041 | 32 | X | dynein regulatory complex | |
| FBgn0050259 | CG30259 | 67 | X | dynein regulatory complex | |
| FBgn0263076 | Klp54D | 18 | kinesin | ||
| FBgn0050441 | IFT20 | 35 | intraflagellar transport | ||
| FBgn0032692 | IFT46 | 20 | intraflagellar transport | ||
| FBgn0031829 | IFT52 | 31 | intraflagellar transport | ||
| FBgn0038358 | Ttc26 | 21 | intraflagellar transport | ||
| FBgn0038814 | CG15923 | 11 | meckelin | ||
| FBgn0038170 | CG14367 | 117 | cilium assembly | ||
| FBgn0033685 | OSCP1 | 97 | X | cilium assembly | |
| DEGRADATION & METABOLISM | |||||
| FBgn0026268 | antdh | 2,954 | oxidoreductase | ||
| FBgn0034076 | Jhedup | 2,263 | X | carboxylesterase | |
| FBgn0051809 | CG31809 | 79 | oxidoreductase | ||
| FBgn0085371 | CG34342 | 76 | X | oxidoreductase | |
| FBgn0263830 | CG40486 | 994 | X | oxidoreductase | |
| FBgn0030949 | Cyp308a1 | 178 | cyp450 | ||
| FBgn0000473 | Cyp6a2 | 608 | X | cyp450 | |
| FBgn0033696 | Cyp6g2 | 115 | X | cyp450 | |
| FBgn0033697 | Cyp6t3 | 89 | cyp450 | ||
| FBgn0038029 | GstD11 | 179 | X | glutathione S transferase | |
| FBgn0010044 | GstD8 | 185 | X | glutathione S transferase | |
| FBgn0063496 | GstE4 | 2,287 | X | glutathione S transferase | |
| FBgn0053177 | CG33177 | 176 | X | glutathione S transferase | |
| FBgn0053178 | CG33178 | 339 | glutathione S transferase | ||
| FBgn0026314 | Ugt35b | 674 | UDP-glucoronosyl transferase | ||
| FBgn0034605 | CG15661 | 147 | UDP-glucoronosyl transferase | ||
| FBgn0038350 | AOX4 | 52 | X | aldehyde oxidase | |
| FBgn0038732 | CG11391 | 1,072 | acyl-coenzyme A (CoA) synthetase | ||
| FBgn0051216 | Naam | 134 | X | nicotinamide amidase | |
| FBgn0035453 | CG10357 | 204 | X | lipase | |
| KINASES | |||||
| FBgn0032083 | CG9541 | 96 | X | kinase | |
| FBgn0050274 | CG30274 | 47 | X | kinase | |
| FBgn0031730 | CG7236 | 66 | kinase | ||
| FBgn0259712 | CG42366 | 12 | kinase | ||
| FBgn0031800 | CG9497 | 2,031 | kinase | ||
| TRANSCRIPTION FACTORS | |||||
| FBgn0034520 | lms | 12 | transcription factor | ||
| FBgn0003513 | ss | 35 | transcription factor | ||
| FBgn0052006 | CG32006 | 38 | transcription factor | ||
| PEPTIDASE-RELATED PROTEINS | |||||
| FBgn0052271 | CG32271 | 94 | peptidase | ||
| FBgn0053159 | CG33159 | 79 | X | peptidase | |
| FBgn0040532 | CG8369 | 6,966 | proteinase inhibitor | ||
| PROTEINS WITH OTHER FUNCTIONS | |||||
| FBgn0259231 | CCKLR-17D1 | 33 | neuropeptide GPCR | ||
| FBgn0260004 | Snmp1 | 294 | X | CD36 lipoprotein receptor | |
| FBgn0033110 | CG9447 | 18 | acyl transferase | ||
| FBgn0031254 | CG13692 | 21 | ARF/SAR small GTPase | ||
| FBgn0032181 | CG13133 | 188 | X | chaperone | |
| FBgn0028658 | Adat1 | 143 | adenosine deaminase | ||
| FBgn0039864 | CG11550 | 657 | lipophilic ligand binding protein | ||
| FBgn0020660 | eIF4B | 14 | translation initiation factor | ||
| FBgn0039386 | CG5948 | 108 | superoxide dismutase | ||
| FBgn0035434 | Drsl5 | 10,249 | anti-fungal peptide | ||
| FBgn0038309 | Amt | 209 | X | ammonium transporter | |
| FBgn0053349 | ppk25 | 28 | X | X | ppk sodium channel |
| FBgn0036235 | CG6938 | 23 | X | anoctomin family | |
| PROTEINS WITH UNKNOWN FUNCTIONS | |||||
| FBgn0259179 | CG42284 | 151 | ankyrin repeats | ||
| FBgn0052668 | CG32668 | 17 | Arm repeat | ||
| FBgn0031289 | CG13950 | 77 | X | galectin domain | |
| FBgn0037836 | CG14692 | 20 | IQ motif | ||
| FBgn0038397 | CG10185 | 44 | X | WD40 repeat | |
| FBgn0039673 | CG7568 | 25 | X | WD40 repeat | |
| FBgn0042129 | OS9 | 5,104 | X | ||
| FBgn0011294 | a5 | 2,995 | X | ||
| FBgn0033851 | CG13332 | 15 | |||
| FBgn0035000 | CG13578 | 152 | |||
| FBgn0036143 | CG14142 | 92 | X | ||
| FBgn0038579 | CG14313 | 26 | |||
| FBgn0263656 | CG43647 | 23 | X | ||
| FBgn0039523 | CG12885 | 22 | |||
| FBgn0032850 | Kua | 80 | X | ||
| FBgn0033629 | Tsp47F | 138 | |||
| FBgn0263655 | CG43646 | 17 | |||
| FBgn0050285 | CG30285 | 234 | |||
| FBgn0037014 | CG13251 | 12 | |||
| FBgn0028740 | CG6362 | 26 | |||
| FBgn0037492 | CG10050 | 278 | X | ||
IR31a is known to be expressed in ato-dependent ac1 sensilla (Benton et al. 2009; Menuz et al. 2014). It nearly met the threshold for ato-depletion (3.9-fold reduced vs. 4-fold reduced).
Or85a is known to be expressed in single-walled ab2 sensilla (Couto et al. 2005; Fishilevich and Vosshall 2005). Its reduction in ato flies is likely due to its location under the Df(3R)p13 deficiency used with ato1 (Menuz et al. 2014).
Gr10a is known to be expressed in amos-sensitive ab1 sensilla (Fishilevich and Vosshall 2005). Due to overlapping annotation with Or10a, it was not detected by HTSeq.
Validation of the bioinformatic identification approach
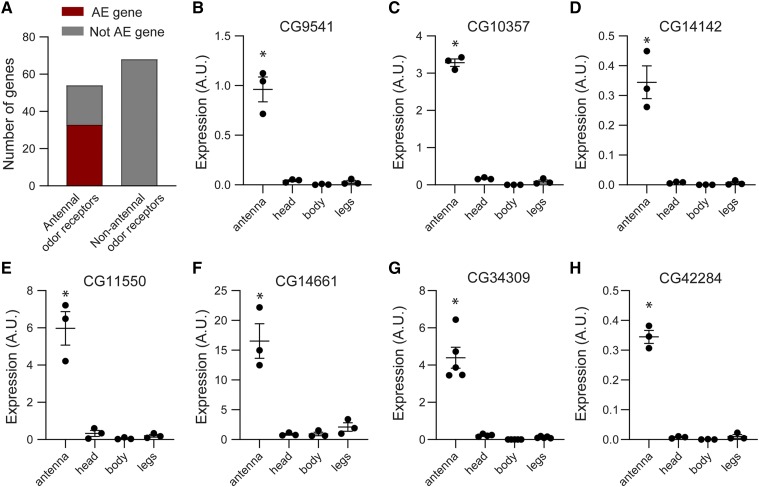
Our strategy relies on the ability to compare gene expression levels in our dataset to the ones generated by ModEncode previously. We therefore used two methods to validate our list of AE genes. First, we considered members of the OR and IR receptor families that are known to be expressed in the antenna (Figure 4A) (Couto et al. 2005; Fishilevich and Vosshall 2005; Silbering et al. 2011). Of these, ∼2/3 are contained within the set of AE genes. The other known antennal odor receptors generally had very low expression in the wild-type antennae, consistent with previous studies (Shiao et al. 2013; Menuz et al. 2014), and were therefore automatically excluded from the list of AE genes. We also looked at other OR and IR family members that are not expressed in the adult antenna, but rather in gustatory tissues or larvae (Couto et al. 2005; Sánchez-Alcañiz et al. 2018). As expected, none of these genes was identified as antennal-enriched.
Figure 4.
Validation of newly identified AE genes. (A) Graph of IR and OR family odor receptors, some of which are known to be expressed in the antenna and others that are not. The AE gene list includes ∼2/3 of known antennal odor receptors, whereas no non-antennal odor receptor is included. (B-H) Expression of seven AE genes was compared in antennae, heads, bodies and legs with qRT-PCR. For each, antennal expression is significantly higher than in the other tissues (n = 3-5, P < 0.001, one-way ANOVA with Dunnett’s post hoc test).
As a second validation method, we used quantitative real-time PCR (qRT-PCR) to quantify the expression levels of antennal-enriched genes in tissues from wild-type flies. We investigated seven AE genes whose expression and function have never been directly examined. Each of these seven genes was detected at a significantly higher level in antennae compared to heads, legs, and bodies (P < 0.001, n = 3-5) (Figure 4B-H). Gene expression in each tissue for each gene was more than 10-fold lower than in antennae, with the exception of CG14661 expression in legs. CG14661 was detected at only ∼8 fold lower levels in legs than in antennae, perhaps due to the presence of leg gustatory sensilla which share many similarities to olfactory sensilla.
Orthologs of AE genes have conserved antennal expression
We next sought to determine if these 141 AE genes found in Drosophila are also expressed in the antennae of distantly related insect species, which would be consistent with a conserved olfactory function. We chose to focus on the 96 genes that were not known odor receptors or OBPs, as the antennal expression of these gene families has been extensively studied in multiple insect species. We identified existing antennal RNA-Seq datasets on the NCBI Sequence Read Archive (SRA) from four insect species: mosquitoes (Anopheles gambiae, Diptera), bees (Apis mellifera, Hymenoptera), ants (Harpegnathos saltator, Hymenoptera), and beetles (Tribolium castaneum, Coleoptera). These are a diverse set of insects with a nearest common ancestor over 300 million years ago (Missbach et al. 2014).
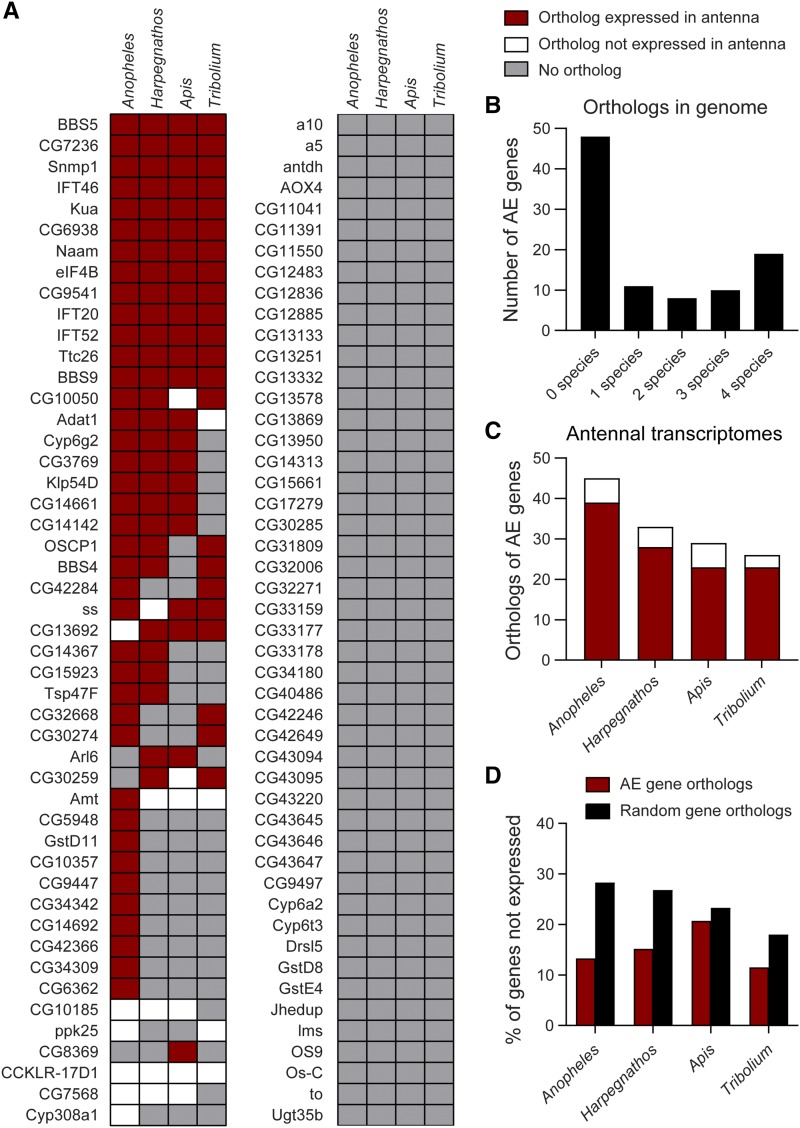
We used the InParanoid 8 program to identify unambiguous orthologs of the 96 AE genes in the proteome of each of the four target insect species (Sonnhammer and Östlund 2014). With this program, we found an ortholog in at least one of the four insect species for half of the 96 AE genes, with 18 genes having orthologs in all four species (Figure 5A, B). Other genes either had no orthologs or had gene duplication events (paralogs), making it unclear which would be the functional ortholog of the Drosophila AE gene. The species with the most identified orthologs was the Anopheles mosquito, the species most closely related to Drosophila flies (Figure 5C).
Figure 5.
Orthologs of AE genes are expressed in the antennae of other insect species. (A) At least one of the four other insect genomes examined (Anopheles, Harpegnathos, Apis, Tribolium) contained an unambiguous ortholog for 48 of the 96 AE genes examined. The box shading indicates if a genomic ortholog of the AE gene was identified (white or red) or not (gray) in a particular species. Red signifies that the ortholog is expressed in the antenna of that species, whereas white indicates the ortholog is not expressed. (B) The graph depicts the number of AE genes with genomic orthologs found in 0, 1, 2, 3 or 4 of the examined species. (C) The graph shows the number of genomic orthologs of AE genes found in each insect species and the proportion of those orthologs that were detected in the antennal transcriptome of that species. Similar coloring as in A. (D) The graph shows the percentage of the AE gene orthologs and orthologs of randomly selected genes not expressed in each species.
We then used the coding sequences of these orthologous genes to interrogate the antennal transcriptomes of the relevant species using NCBI SRA-BLAST. In each of the four species, 79–89% of the orthologs were antennal-expressed (Figure 5A, C). Although this fraction is higher than the fraction of genes expressed in a typical tissue (60–70%) (Ramsköld et al. 2009), it is possible that highly conserved genes are particularly likely to be expressed. To test this possibility, we applied the same analysis to randomly selected Drosophila genes (Figure S1A). In each transcriptome analyzed, a greater fraction of the orthologs of random genes were not expressed than the fraction of AE genes (Figure 5D). If we raised the threshold for defining antennal expression to become more stringent, the difference between AE and random orthologs grew even larger (Figure S1B).
Surprisingly few random genes were not expressed in Tribolium (18%), and we wondered if this might reflect contamination of the sample, and thereby bias our interpretation of the AE gene expression. We therefore identified a second Tribolium antennal dataset with a smaller fraction of randomly selected genes expressed. Again, the AE genes were more often expressed than the randomly selected genes (Figure S1C). We note that the fraction of AE orthologs expressed in the Apis transcriptome was only slightly more than the fraction of expressed random orthologs, unlike the other three species. We therefore expanded our analysis to a fifth species, Bombyx mori. We found that the fraction of expressed AE orthologs was much greater than that of the randomly selected genes, similar to Anopheles, Harpegnathos, and Tribolium (Figure S1D). Together, our data support the possibility that more AE gene orthologs are expressed in the antennal transcriptomes than would be expected by chance alone.
Differential localization of antennal-enriched genes
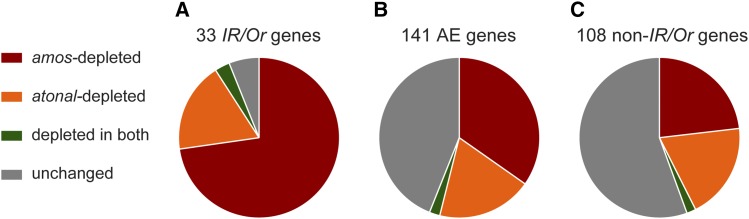
Having identified 141 AE genes in Drosophila, we wondered whether some of the genes might function within signaling pathways specific to single or double-walled sensilla, similar to the odor receptor families themselves. Our first dataset revealed genes that are depleted in amos mutants and are therefore likely to be enriched in single-walled sensilla. We had previously generated a similar dataset for atonal flies, which selectively lack double-walled sensilla (Jarman et al. 1994; Menuz et al. 2014). We therefore inferred the localization of the 141 AE genes by cross-referencing the amos-depleted and atonal-depleted gene sets. The accuracy of this localization was examined by considering the 33 odor receptor AE genes. Nearly all of the OR and IR genes were depleted in either amos or atonal antennae in a manner consistent with their known localization (Figure 6A).
Figure 6.
Localization of AE genes to morphological subtypes of sensilla. (A) The majority of IR and OR odor receptors could be localized to either single or double-walled sensilla based on their depletion in either amos or atonal antennae. (B) Over half of the 141 AE genes are depleted in either amos or atonal flies. A few genes were depleted in both genotypes (green) and half were unchanged in both mutants (gray). (C) Nearly half of the 108 non-chemoreceptor AE genes could be localized to a sensilla class.
Including the odor receptors, we found that 49 of the AE genes were amos-depleted and 27 were atonal-depleted (Figure 6B, Table 2). Considered alone, nearly half of the 108 non-odor receptor AE genes could be localized to one of the two morphological sensilla classes, with a quarter of the genes depleted in either amos or atonal antennae (Figure 6B, Table 2). Further confirming our genetic localization strategy, we correctly mapped seven amos-depleted OBPs that had been previously detected in single-walled sensilla and two atonal-depleted OBPs that were previously found in double-walled sensilla (Table 2) (Larter et al. 2016). Obp19a, the only OBP AE gene not depleted in amos or atonal antennae, was previously shown to be expressed in epithelial cells that are not specific to either sensilla class (Larter et al. 2016). We expect that some other “non-localized” AE genes also play a role in cells outside of sensilla; another possibility is that some contribute to the function of both single and double-walled sensilla.
Molecular functions of antennal-enriched genes
We next examined the putative molecular functions and predicted subcellular localization of the 141 AE genes (Table 2). We did this using web-based tools including DAVID, FlyBase, and DeepLoc (Huang et al. 2009; Almagro Armenteros et al. 2017; Thurmond et al. 2019). For some, we also ran BLAST searches to identify similar genes with known functions in other species.
The 141 AE genes include many known antennal chemosensory receptors, which represent 25% of the AE genes. Interestingly, 17% of the AE genes are small, secreted proteins with poorly characterized functions. This includes 9 OBPs, 3 members of the haemolymph juvenile hormone-binding protein family, and 12 additional proteins of unknown function. Of these, the OBPs have been best studied. Their exact function is unclear, but they have been hypothesized to be involved in solubilizing hydrophobic odorants, protecting odorants from degradation, odorant buffering, or delivery of odorants to receptors (Leal 2013; Larter et al. 2016). It is possible that the newly identified small, secreted proteins have similar roles. One difference is that nearly all OBPs were enriched in either single- or double-walled sensilla, whereas nearly all of the other small, secreted proteins distribute more broadly.
Insect antennae are densely covered with olfactory sensilla, and these sensilla have internal structures resembling cilia (Keil 1999; Keil 2012). It is therefore unsurprising that 16 (11%) of the AE genes are related to ciliary function, such as components of the BBSome and intraflagellar transport complexes (Table 2). Three genes related to the motor dynein are atonal-depleted genes, indicating that cytoskeletal transport may differ in the two sensilla classes.
Genes that may be related to odorant degradation comprise another 14% of the AE genes. A few of these genes, including antdh, Jhedup, and Cyp308a1, have been previously identified as antennal enriched (Wang et al. 1999; Younus et al. 2014; Steiner et al. 2017). More than half of these genes are localized to either single- or double-walled sensilla.
Smaller numbers of AE genes have more specialized functions. For example, three are transcription factors and three are related to peptidases. Other predicted functions include kinases, ion channels, transporters, and receptors. There are also a large number (15%) of AE genes with no predicted function.
Discussion
We have generated two resources for studying the molecular basis of insect olfaction beyond the function of the odor receptors themselves. We identified 187 genes enriched in single-walled sensilla that could contribute to the detection of food odors, pheromones, and attractive human-derived odors. Most of these genes are previously unstudied in the antenna, and their predicted functions of many suggest they could have roles in regulating olfactory neuron signaling. We also describe 141 antennal-enriched genes, of which some including lush, Snmp1, Amt, spineless and OR/IR odor receptors have established olfactory roles (Dong et al. 2002; Xu et al. 2005; Benton et al. 2007; Menuz et al. 2014). Thus, we expect many of the other AE genes will also contribute to olfaction in Drosophila. Excitingly, most of the identified orthologs of AE genes were expressed in the antennae of distantly related insect species, suggesting that many may have evolutionarily conserved roles.
Our study of amos-depleted genes unexpectedly identified a number of non-protein coding RNAs that are differentially expressed in single and double-walled sensilla. Several are antisense RNAs that target chemosensory receptors (Or46a, Or88a, Gr64d, and Gr64e). In other systems, antisense RNAs are increasingly reported to regulate gene expression (Kashi et al. 2016), but to our knowledge a role for antisense RNAs in olfactory physiology has not been studied. Such a regulatory system could underlie changes in odor receptor activity in response to circadian rhythms and prolonged odor exposure (Krishnan et al. 1999; von der Weid et al. 2015).
Recent reports indicate that several members of the gustatory receptor family are unexpectedly found in the antenna (Menuz et al. 2014; Fujii et al. 2015). The localization and function of most antennal GRs is unknown. Our data from amos flies indicate that many GRs are found within single-walled sensilla. It will be interesting to determine if they are co-expressed with OR receptors within ORNs, such as Or10a and Gr10a (Fishilevich and Vosshall 2005), or if there are unidentified ORNs that exclusively use these receptors such as those utilizing Gr21a/Gr63a (Jones et al. 2007). There is at least one antennal lobe glomerulus (VA7m) that is not mapped to a chemoreceptor (Grabe et al. 2016); perhaps a GR-expressing ORN projects to this glomerulus.
We observed that many of the genes enriched in single-walled sensilla could comprise distinct molecular signaling pathways that underlie their unique olfactory functions. These include seven GPCRs that could serve to differentially regulate the activity of ORNs in response to neuromodulators or hormones (Wilson 2013; Gadenne et al. 2016). The discovery of metabotropic serotonin and acetylcholine receptors enriched in single-walled sensilla suggests that these neuromodulators could act on ORN terminals in the antennal lobe similar to the previously described metabotropic GABAB receptors (Olsen and Wilson 2008). Other amos-depleted GPCRs such as hec, implicated in mating behavior, and Lgr1, responsive to hormones, could affect the known activation of some single-walled sensilla ORNs to mating pheromones (Hauser et al. 1997; Li et al. 2011). Our dataset also identified signaling molecules, such as the G-protein Gαf, the kinases CG30274 and CG9541, and the phospholipase A2 CG42237, which could be components of intracellular signal transduction cascades. Five members of the pickpocket family of ligand-gated cation channels are also amos-depleted. Although the functions of most family members are poorly understood, some are gated by neuropeptides and other small molecules and could act as neuromodulators (Zelle et al. 2013). Based on the enrichment of particular receptors and signaling molecules in single-walled sensilla, we speculate that neuromodulators differentially regulate the activity of ORNs in different morphological classes of sensilla, an area ripe for future investigations.
We found that a large number of AE genes encode enzymes with potential roles in odor degradation and that these are often depleted in either atonal or amos antennae. Additionally, there are a large number of such enzymes that are either amos-depleted or atonal-depleted, even if they are not antennal enriched. Together this indicates that different sensilla classes may utilize different degradation enzymes. One reason this might occur is sensilla may selectively express enzymes needed to metabolize odors their ORNs detect, with the segregation of enzymes for alcohols, esters, and long-chain hydrocarbon pheromones to single-walled sensilla and enzymes for amines and acids to double-walled sensilla (Steinbrecht 1997; Hallem and Carlson 2006; Silbering et al. 2011). An alternative possibility is that odors only accumulate in the peri-neuronal sensillar lymph in particular sensillar classes, perhaps due to restricted access by pores on the sensillar shaft (Steinbrecht 1997). If so, then all sensilla with similar pores, such as single- or double-walled sensilla, might need similar enzymes to prevent odor accumulation.
The structure of insect ORN outer dendrites is thought to resemble a modified cilium (Keil 1999; Keil 2012), but only limited work has characterized the effect of ciliary mutants on olfactory function. We identified 16 AE genes involved in ciliary function, including four members of the basal body complex (BBSome) and four involved in intraflagellar transport that both contribute to the assembly and maintenance of cilia (Avidor-Reiss et al. 2004; Jana et al. 2016). Only a few ciliary genes are differentially expressed in amos and atonal mutants, suggesting similar ciliary structures are found in both single- and double-walled sensilla. Further, antennal expression of ciliary gene orthologs is highly conserved in the four insect species examined.
Although our approach sought to identify genes with exclusive roles in olfaction, it is clear that ciliary genes also contribute to the function of other ciliated cells and biological processes. Ciliary genes were most likely detected as antennal-enriched due to the dense tiling of olfactory sensilla on the antennal surface (Shanbhag et al. 1999), making the density of ciliated cells in this tissue far greater than in other tissues examined. More broadly, this also indicates that some AE genes may play a role in both the antennae and in other tissues.
Our study identified a large number of known olfactory genes and many more novel genes with potential roles in olfaction, but it is likely that some genes with relevance for olfaction were overlooked. By design, our computational approach discarded many genes that are important for olfaction and contribute to the physiology of other tissues. This may be the reason why we did not detect genes that are likely to maintain the transepithelial potential, an electrical potential that drives odor receptor transduction and likely relies on potassium ion movement (Kuppers and Thurm 1979; Stengl et al. 1999). Our approach also discarded genes with low abundance in the antenna to ensure that genes were truly antennal enriched. This conservative approached prevented some known antennal odor receptors from being included among the AE genes. Our strategy pooled approximately equal numbers of male and female antennae, preventing additional consideration of sexual dimorphism in gene expression, such as has been observed previously (Zhou et al. 2009). It will be interesting to determine if any of the identified genes are highly enriched in either males or females.
A key advantage of our approach is the identification of genes with potentially conserved roles in olfactory signaling. We discovered many AE genes with orthologs expressed in the antennae of all insects we examined, making them prime targets for future mechanistic investigations. When combined with information from our amos-depleted and atonal-depleted datasets, our identification of AE genes reveals new avenues for examining the mechanistic underpinnings of poorly understood processes that support faithful odor detection in the antenna.
Acknowledgments
We thank Andrew Jarman and Richard Benton for the amos1 pr1/Cyo fly line. We thank Bo Reese at the Center for Genome Innovation at the University of Connecticut for sample preparation and sequencing services. We thank Anastasios Tzingounis for providing insightful comments on the manuscript.
Footnotes
Supplemental material available at FigShare: https://doi.org/10.25387/g3.9820682.
Communicating editor: T. Mackay
Literature Cited
- Almagro Armenteros J. J., Sønderby C. K., Sønderby S. K., Nielsen H., and Winther O., 2017. DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics 33: 3387–3395. 10.1093/bioinformatics/btx431 [DOI] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., and Huber W., 2015. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M. N., Löfstedt C., and Newcomb R. D., 2015. Insect olfaction and the evolution of receptor tuning. Front. Ecol. Evol. 3: 53 10.3389/fevo.2015.00053 [DOI] [Google Scholar]
- Anholt R., Lyman R., and Mackay T., 1996. Effects of Single P-Element Insertions on Olfactory Behavior in Drosophila melanogaster. Genetics 143: 293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya G. H., Magwire M. M., Huang W., Serrano-Negron Y. L., Mackay T. F. et al. , 2015. The genetic basis for variation in olfactory behavior in Drosophila melanogaster. Chem. Senses 40: 233–243. 10.1093/chemse/bjv001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H. et al. , 2000. Gene ontology: tool for the unification of biology. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T. et al. , 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117: 527–539. 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., Gomez-Diaz C., and Vosshall L. B., 2009. Variant Ionotropic Glutamate Receptors as Chemosensory Receptors in Drosophila. Cell 136: 149–162. 10.1016/j.cell.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benton R., Vannice K. S., and Vosshall L. B., 2007. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature 450: 289–293. 10.1038/nature06328 [DOI] [PubMed] [Google Scholar]
- Brown J. B., Boley N., Eisman R., May G. E., Stoiber M. H. et al. , 2014. Diversity and dynamics of the Drosophila transcriptome. Nature 512: 393–399. 10.1038/nature12962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S., Ireland A., Mungall C. J., Shu S., Marshall B. et al. , 2009. AmiGO: online access to ontology and annotation data. Bioinformatics 25: 288–289. 10.1093/bioinformatics/btn615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey A. F., Wang G., Su C. Y., Zwiebel L. J., and Carlson J. R., 2010. Odorant reception in the malaria mosquito Anopheles gambiae. Nature 464: 66–71. 10.1038/nature08834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caulfield M. P., and Birdsall N. J., 1998. International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 50: 279–290. [PubMed] [Google Scholar]
- Chahda J. S., Soni N., Sun J. S., Ebrahim S. A., Weiss B. L. et al. , 2019. The molecular and cellular basis of olfactory response to tsetse fly attractants. PLoS Genet. 15: e1008005 10.1371/journal.pgen.1008005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto A., Alenius M., and Dickson B. J., 2005. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr. Biol. 15: 1535–1547. 10.1016/j.cub.2005.07.034 [DOI] [PubMed] [Google Scholar]
- Croset V., Rytz R., Cummins S. F., Budd A., Brawand D. et al. , 2010. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLoS Genet. 6: e1001064 10.1371/journal.pgen.1001064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGennaro M., McBride C. S., Seeholzer L., Nakagawa T., Dennis E. J. et al. , 2013. orco mutant mosquitoes lose strong preference for humans and are not repelled by volatile DEET. Nature 498: 487–491. 10.1038/nature12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippel S., Oberhofer G., Kahnt J., Gerischer L., Opitz L. et al. , 2014. Tissue-specific transcriptomics, chromosomal localization, and phylogeny of chemosensory and odorant binding proteins from the red flour beetle Tribolium castaneum reveal subgroup specificities for olfaction or more general functions. BMC Genomics 15: 1141 10.1186/1471-2164-15-1141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C. et al. , 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong P. S., Dicks J. S., and Panganiban G., 2002. Distal-less and homothorax regulate multiple targets to pattern the Drosophila antenna. Development 129: 1967–1974. [DOI] [PubMed] [Google Scholar]
- dos Santos G., Schroeder A. J., Goodman J. L., Strelets V. B., Crosby M. A. et al. , 2014. FlyBase: introduction of the Drosophila melanogaster Release 6 reference genome assembly and large-scale migration of genome annotations. Nucleic Acids Res. 43: D690–D697. 10.1093/nar/gku1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishilevich E., and Vosshall L. B., 2005. Genetic and functional subdivision of the Drosophila antennal lobe. Curr. Biol. 15: 1548–1553. 10.1016/j.cub.2005.07.066 [DOI] [PubMed] [Google Scholar]
- Fujii S., Yavuz A., Slone J., Jagge C., Song X. et al. , 2015. Drosophila sugar receptors in sweet taste perception, olfaction, and internal nutrient sensing. Curr. Biol. 25: 621–627. 10.1016/j.cub.2014.12.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadenne C., Barrozo R. B., and Anton S., 2016. Plasticity in insect olfaction: to smell or not to smell? Annu. Rev. Entomol. 61: 317–333. 10.1146/annurev-ento-010715-023523 [DOI] [PubMed] [Google Scholar]
- Gene Ontology Consortium , 2019. The Gene Ontology resource: 20 years and still GOing strong. Nucleic Acids Res. 47: D330–D338. 10.1093/nar/gky1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaninia M., Berger S. L., Reinberg D., Zwiebel L. J., Ray A. et al. , 2018. Antennal Olfactory Physiology and Behavior of Males of the Ponerine Ant Harpegnathos saltator. J. Chem. Ecol. 44: 999–1007. 10.1007/s10886-018-1013-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabe V., Baschwitz A., Dweck H. K., Lavista-Llanos S., Hansson B. S. et al. , 2016. Elucidating the neuronal architecture of olfactory glomeruli in the Drosophila antennal lobe. Cell Reports 16: 3401–3413. 10.1016/j.celrep.2016.08.063 [DOI] [PubMed] [Google Scholar]
- Graveley B. R., Brooks A. N., Carlson J. W., Duff M. O., Landolin J. M. et al. , 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471: 473–479. 10.1038/nature09715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse-Wilde E., Kuebler L. S., Bucks S., Vogel H., Wicher D. et al. , 2011. Antennal transcriptome of Manduca sexta. Proc. Natl. Acad. Sci. USA 108: 7449–7454. 10.1073/pnas.1017963108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., and Smith D. P., 2017. Odorant Receptor Desensitization in Insects. J. Exp. Neurosci. 11: 1–5. 10.1177/1179069517748600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Krieger J., Große-Wilde E., Mißbach C., Zhang L. et al. , 2014. Variant ionotropic receptors are expressed in olfactory sensory neurons of coeloconic sensilla on the antenna of the desert locust (Schistocerca gregaria). Int. J. Biol. Sci. 10: 1–14. 10.7150/ijbs.7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallem E. A., and Carlson J. R., 2006. Coding of odors by a receptor repertoire. Cell 125: 143–160. 10.1016/j.cell.2006.01.050 [DOI] [PubMed] [Google Scholar]
- Hansen I. A., Rodriguez S. D., Drake L. L., Price D. P., Blakely B. N. et al. , 2014. The odorant receptor co-receptor from the bed bug, Cimex lectularius L. PLoS One 9: e113692 10.1371/journal.pone.0113692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson B. S., and Stensmyr M. C., 2011. Evolution of insect olfaction. Neuron 72: 698–711. 10.1016/j.neuron.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Hauser F., Nothacker H.-P., and Grimmelikhuijzen C. J., 1997. Molecular cloning, genomic organization, and developmental regulation of a novel receptor from Drosophila melanogaster structurally related to members of the thyroid-stimulating hormone, follicle-stimulating hormone, luteinizing hormone/choriogonadotropin receptor family from mammals. J. Biol. Chem. 272: 1002–1010. 10.1074/jbc.272.2.1002 [DOI] [PubMed] [Google Scholar]
- Hodges T. K., Cosme L. V., Athrey G., Pathikonda S., Takken W. et al. , 2014. Species-specific chemosensory gene expression in the olfactory organs of the malaria vector Anopheles gambiae. BMC Genomics 15: 1089 10.1186/1471-2164-15-1089 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hoyer D., Clarke D. E., Fozard J. R., Hartig P. R., Martin G. R. et al. , 1994. International Union of Pharmacology classification of receptors for 5-hydroxytryptamine (Serotonin). Pharmacol. Rev. 46: 157–203. [PubMed] [Google Scholar]
- Huang D. W., Sherman B. T., and Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jana S. C., Bettencourt-Dias M., Durand B., and Megraw T. L., 2016. Drosophila melanogaster as a model for basal body research. Cilia 5: 22 10.1186/s13630-016-0041-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarman A. P., Grell E. H., Ackerman L., Jan L. Y., and Jan Y. N., 1994. Atonal is the proneural gene for Drosophila photoreceptors. Nature 369: 398–400. 10.1038/369398a0 [DOI] [PubMed] [Google Scholar]
- Jasper W. C., Linksvayer T. A., Atallah J., Friedman D., Chiu J. C. et al. , 2014. Large- scale coding sequence change underlies the evolution of postdevelopmental novelty in honey bees. Mol. Biol. Evol. 32: 334–346. 10.1093/molbev/msu292 [DOI] [PubMed] [Google Scholar]
- Jones W. D., Cayirlioglu P., Kadow I. G., and Vosshall L. B., 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445: 86–90. 10.1038/nature05466 [DOI] [PubMed] [Google Scholar]
- Joseph R. M., and Carlson J. R., 2015. Drosophila Chemoreceptors: A Molecular Interface Between the Chemical World and the Brain. Trends Genet. 31: 683–695. 10.1016/j.tig.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N. A., and J. N. Fass, 2011 Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files, Version 1.33.
- Kaissling K. E., Hildebrand J. G., and Tumlinson J. H., 1989. Pheromone receptor cells in the male moth Manduca sexta. Arch. Insect Biochem. Physiol. 10: 273–279. 10.1002/arch.940100403 [DOI] [Google Scholar]
- Kashi K., Henderson L., Bonetti A., and Carninci P., 2016. Discovery and functional analysis of lncRNAs: methodologies to investigate an uncharacterized transcriptome. Biochim. Biophys. Acta 1859: 3–15. 10.1016/j.bbagrm.2015.10.010 [DOI] [PubMed] [Google Scholar]
- Keil T. A., 1999. Morphology and development of the peripheral olfactory organs, pp. 5–47 in Insect olfaction, edited by Hansson B. S. Springer, New York: 10.1007/978-3-662-07911-9_2 [DOI] [Google Scholar]
- Keil T. A., 2012. Sensory cilia in arthropods. Arthropod Struct. Dev. 41: 515–534. 10.1016/j.asd.2012.07.001 [DOI] [PubMed] [Google Scholar]
- Krishnan B., Dryer S. E., and Hardin P. E., 1999. Circadian rhythms in olfactory responses of Drosophila melanogaster. Nature 400: 375–378. 10.1038/22566 [DOI] [PubMed] [Google Scholar]
- Kuppers J., and Thurm U., 1979. Active ion transport by a sensory epithelium. I. Transepithelial short circuit current, potential difference, and their dependence on metabolism. J. Comp. Physiol. 134: 131–136. [Google Scholar]
- Larter N. K., Sun J. S., and Carlson J. R., 2016. Organization and function of Drosophila odorant binding proteins. eLife 5: e20242 10.7554/eLife.20242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal W. S., 2013. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 58: 373–391. 10.1146/annurev-ento-120811-153635 [DOI] [PubMed] [Google Scholar]
- Leinonen R., Sugawara H., Shumway M., and Collaboration I. N. S. D., 2010. The sequence read archive. Nucleic Acids Res. 39: D19–D21. 10.1093/nar/gkq1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hoxha V., Lama C., Dinh B. H., Vo C. N. et al. , 2011. The hector G-protein coupled receptor is required in a subset of fruitless neurons for male courtship behavior. PLoS One 6: e28269 10.1371/journal.pone.0028269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Gu S., Zhang Y., Guo Y., and Wang G., 2012. Candidate olfaction genes identified within the Helicoverpa armigera Antennal Transcriptome. PLoS One 7: e48260 10.1371/journal.pone.0048260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews B. J., McBride C. S., DeGennaro M., Despo O., and Vosshall L. B., 2016. The neurotranscriptome of the Aedes aegypti mosquito. BMC Genomics 17: 32 10.1186/s12864-015-2239-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna M., Monte P., Helfand S. L., Woodard C., and Carlson J., 1989. A simple chemosensory response in Drosophila and the isolation of acj mutants in which it is affected. Proc. Natl. Acad. Sci. USA 86: 8118–8122. 10.1073/pnas.86.20.8118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMeniman C. J., Corfas R. A., Matthews B. J., Ritchie S. A., and Vosshall L. B., 2014. Multimodal integration of carbon dioxide and other sensory cues drives mosquito attraction to humans. Cell 156: 1060–1071. 10.1016/j.cell.2013.12.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menuz K., Larter N. K., Park J., and Carlson J. R., 2014. An RNA-seq screen of the Drosophila antenna identifies a transporter necessary for ammonia detection. PLoS Genet. 10: e1004810 10.1371/journal.pgen.1004810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H., Huang X., Muruganujan A., Tang H., Mills C. et al. , 2016. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 45: D183–D189. 10.1093/nar/gkw1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missbach C., Dweck H. K., Vogel H., Vilcinskas A., Stensmyr M. C. et al. , 2014. Evolution of insect olfactory receptors. eLife 3: e02115 10.7554/eLife.02115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N. et al. , 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797. 10.1126/science.1198374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Möller S., Croning M. D., and Apweiler R., 2001. Evaluation of methods for the prediction of membrane spanning regions. Bioinformatics 17: 646–653. 10.1093/bioinformatics/17.7.646 [DOI] [PubMed] [Google Scholar]
- Nielsen H., 2017. Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611: 59–73. 10.1007/978-1-4939-7015-5_6 [DOI] [PubMed] [Google Scholar]
- Olsen S. R., and Wilson R. I., 2008. Lateral presynaptic inhibition mediates gain control in an olfactory circuit. Nature 452: 956–960. 10.1038/nature06864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitts R. J., Fox A. N., and Zwiebel L. J., 2004. A highly conserved candidate chemoreceptor expressed in both olfactory and gustatory tissues in the malaria vector Anopheles gambiae. Proc. Natl. Acad. Sci. USA 101: 5058–5063. 10.1073/pnas.0308146101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu C. Z., Zhou Q. Z., Liu T. T., Fang S. M., Wang Y. W. et al. , 2018. Evidence of peripheral olfactory impairment in the domestic silkworms: insight from the comparative transcriptome and population genetics. BMC Genomics 19: 788 10.1186/s12864-018-5172-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsköld D., Wang E. T., Burge C. B., and Sandberg R., 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLOS Comput. Biol. 5: e1000598 10.1371/journal.pcbi.1000598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A., 2015. Reception of odors and repellents in mosquitoes. Curr. Opin. Neurobiol. 34: 158–164. 10.1016/j.conb.2015.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinker D. C., Zhou X., Pitts R. J., Rokas A., and Zwiebel L. J., 2013. Antennal transcriptome profiles of anopheline mosquitoes reveal human host olfactory specialization in Anopheles gambiae. BMC Genomics 14: 749 10.1186/1471-2164-14-749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. M., 2019. Molecular evolution of the major arthropod chemoreceptor gene families. Annu. Rev. Entomol. 64: 227–242. 10.1146/annurev-ento-020117-043322 [DOI] [PubMed] [Google Scholar]
- Robinson M. D., McCarthy D. J., and Smyth G. K., 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. 10.1093/bioinformatics/btp616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Alcañiz J. A., Silbering A. F., Croset V., Zappia G., Sivasubramaniam A. K. et al. , 2018. An expression atlas of variant ionotropic glutamate receptors identifies a molecular basis of carbonation sensing. Nat. Commun. 9: 4252 10.1038/s41467-018-06453-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senthilan P. R., Piepenbrock D., Ovezmyradov G., Nadrowski B., Bechstedt S. et al. , 2012. Drosophila auditory organ genes and genetic hearing defects. Cell 150: 1042–1054. [DOI] [PubMed] [Google Scholar]
- Shanbhag S., Muller B., and Steinbrecht A., 1999. Atlas of olfactory organs of Drosophila melanogaster. 1. Types, external organization, innervation and distribution of olfactory sensilla. Int. J. Insect Morphol. Embryol. 28: 377–397. 10.1016/S0020-7322(99)00039-2 [DOI] [Google Scholar]
- Shiao M.-S., Fan W.-L., Fang S., Lu M.-Y. J., Kondo R. et al. , 2013. Transcriptional profiling of adult Drosophila antennae by high-throughput sequencing. Zool. Stud. 52: 42 10.1186/1810-522X-52-42 [DOI] [Google Scholar]
- Silbering A. F., Rytz R., Grosjean Y., Abuin L., Ramdya P. et al. , 2011. Complementary function and integrated wiring of the evolutionarily distinct Drosophila olfactory subsystems. J. Neurosci. 31: 13357–13375. 10.1523/JNEUROSCI.2360-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnhammer E. L., and Östlund G., 2014. InParanoid 8: orthology analysis between 273 proteomes, mostly eukaryotic. Nucleic Acids Res. 43: D234–D239. 10.1093/nar/gku1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbrecht R. A., 1997. Pore structures in insect olfactory sensilla: A review of data and concepts. Int. J. Insect Morphol. Embryol. 26: 229–245. 10.1016/S0020-7322(97)00024-X [DOI] [Google Scholar]
- Steiner C., Bozzolan F., Montagné N., Maïbèche M., and Chertemps T., 2017. Neofunctionalization of “Juvenile Hormone Esterase Duplication” in Drosophila as an odorant-degrading enzyme towards food odorants. Sci. Rep. 7: 12629 10.1038/s41598-017-13015-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stengl M., Ziegelberger G., Boekhoff I., and Krieger J., 1999. Perireceptor events and transduction mechanisms in insect olfaction, pp. 49–66 in Insect olfaction, edited by Hansson B. S. Springer, New York: 10.1007/978-3-662-07911-9_3 [DOI] [Google Scholar]
- Takken W., and Knols B. G. J., 2010. Olfaction in vector-host interactions, Wageningen Academic Publishers, Wageningen: 10.3920/978-90-8686-698-4 [DOI] [Google Scholar]
- Thurmond J., Goodman J. L., Strelets V. B., Attrill H., Gramates L. S. et al. , 2019. FlyBase 2.0: the next generation. Nucleic Acids Res. 47: D759–D765. 10.1093/nar/gky1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D. et al. , 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium , 2018. UniProt: the universal protein knowledgebase. Nucleic Acids Res. 46: 2699 10.1093/nar/gky092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Weid B., Rossier D., Lindup M., Tuberosa J., Widmer A. et al. , 2015. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci. 18: 1455–1463. 10.1038/nn.4100 [DOI] [PubMed] [Google Scholar]
- Wang Q., Hasan G., and Pikielny C. W., 1999. Preferential expression of biotransformation enzymes in the olfactory organs of Drosophila melanogaster, the antennae. J. Biol. Chem. 274: 10309–10315. 10.1074/jbc.274.15.10309 [DOI] [PubMed] [Google Scholar]
- Wang Z., Gerstein M., and Snyder M., 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10: 57–63. 10.1038/nrg2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R. I., 2013. Early olfactory processing in Drosophila: mechanisms and principles. Annu. Rev. Neurosci. 36: 217–241. 10.1146/annurev-neuro-062111-150533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization, October 31, 2017 Vector-borne diseases, https://www.who.int/news-room/fact-sheets/detail/vector-borne-diseases.
- Xu P., Atkinson R., Jones D. N., and Smith D. P., 2005. Drosophila OBP LUSH is required for activity of pheromone-sensitive neurons. Neuron 45: 193–200. 10.1016/j.neuron.2004.12.031 [DOI] [PubMed] [Google Scholar]
- Yang Y., Krieger J., Zhang L., and Breer H., 2012. The olfactory co-receptor Orco from the migratory locust (Locusta migratoria) and the desert locust (Schistocerca gregaria): identification and expression pattern. Int. J. Biol. Sci. 8: 159–170. 10.7150/ijbs.8.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younus F., Chertemps T., Pearce S. L., Pandey G., Bozzolan F. et al. , 2014. Identification of candidate odorant degrading gene/enzyme systems in the antennal transcriptome of Drosophila melanogaster. Insect Biochem. Mol. Biol. 53: 30–43. 10.1016/j.ibmb.2014.07.003 [DOI] [PubMed] [Google Scholar]
- Zelle K. M., Lu B., Pyfrom S. C., and Ben-Shahar Y., 2013. The genetic architecture of degenerin/epithelial sodium channels in Drosophila. G3 (Bethesda) 3: 441–450. 10.1534/g3.112.005272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou S., Stone E. A., Mackay T. F., and Anholt R. R., 2009. Plasticity of the chemoreceptor repertoire in Drosophila melanogaster. PLoS Genet. 5: e1000681 10.1371/journal.pgen.1000681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Slone J. D., Rokas A., Berger S. L., Liebig J. et al. , 2012. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLoS Genet. 8: e1002930 10.1371/journal.pgen.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- zur Lage P. I., Prentice D. R., Holohan E. E., and Jarman A. P., 2003. The Drosophila proneural gene amos promotes olfactory sensillum formation and suppresses bristle formation. Development 130: 4683–4693. 10.1242/dev.00680 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All raw read data are available at BioProject accession number PRJNA532415. Analyzed data reporting gene expression are available in File S1. Supplemental material available at FigShare: https://doi.org/10.25387/g3.9820682.