Abstract
Methods that increase the total emission per fluorophore would provide increased sensitivity and a wider dynamic range for chemical analysis, medical diagnostics, and in vivo molecular imaging. The use of fluorophore-metal interactions has the potential to dramatically increase the detectability of single fluorophores for bioanalytical monitoring. The fabrication and single-molecule analysis of fluorophore-labeled DNA molecules tethered to silver island films are described in this article. The single-molecule spectroscopic method reveals some insightful information on the behaviors of single molecules, rather than an ensemble of molecules. Analysis of fluorescence images, intensity profiles, total emitted photons, and lifetime distributions reveals some of sample heterogeneities. Investigations of time-dependent emission characteristics of single molecules indicate that the total number of emitted photons on the silvered surface is more than 10 times greater than on free labeled DNA molecules on a glass substrate. In addition, time-correlated single-photon counting results reveal the reduced lifetimes of single molecules tethered to silver island films.
The importance of DNA-based assays for sensitive detection of biohazards, diseases, and genome sequencing is progressively growing.1 Currently, most microarray imaging and analysis are primarily based on the detection of fluorescence from organic fluorophores. Fluorophores can be easily attached to a broad range of target molecules, including DNA, antibodies, proteins, and other biomacromolecules.2,3 Because the amount of genetic materials available for analysis is extremely limited and the reaction volumes used are in the microliter or nanoliter level, it is important to develop highly sensitive methods for microarray-based analysis.
Single-molecule screening provides the ultimate level of sensitivity, which may help to establish molecular profiles for species that are present at low concentration levels or small quantities.4–7 As single fluorescent molecules are sensitive to their surrounding environments, profiling individual molecules and probing single molecules including fluorescence intensity time traces, spectra, and fluorescence lifetimes can provide unique information about the chemical and physical characteristics of the surrounding nanoenvironment.8–11 In addition, single-molecule detection is becoming increasingly important in in vivo cell imaging as well as providing essential links to the fundamental physics. Recent advances have allowed direct imaging and dynamic studies of single biomacromolecules (e.g., DNA, RNA, and proteins) as well as multicomponent molecular assemblies.12–14
However, single-molecule methodologies have achieved only limited success for applications in chemical analysis and medical diagnostics.6,7 The use of organic fluorophores for routine single-molecule studies suffers from photobleaching, low signal intensities, and random “on/off” blinking.15 In contrast to photobleaching, blinking is characterized by a reversible transition between an emission state and a dark or dim state. Blinking is most often light induced15 and can be a result of conformational changes,11 molecular rotation,16 or reversible metastable species.17 However, these complex photophysical or photochemical behaviors make the use of organic fluorophores as single-molecule probes challenging. It is important to achieve strong and stable fluorescence signals for microarray-based bioanalysis. Methods that increase the total emission per labeled biomolecule would provide improved signal-to-noise ratio and offer less problems with the unwanted background from the nonspecific molecules, matrixes, or optical components.
The use of fluorescence detection schemes in combination with the resonant excitation of surface plasmon has been shown to increase the sensitivity for bioanalyte monitoring in ensemble experiments. The obtainable enormous enhancement of the optical field at and near the metal/dielectric interface can be subsequently used to improve the S/N ratio for the analysis of biorecognition and interfacial binding events in sensor formats.18,19 These reported works exploited the so-called plasmonic enhancement effect,20–23 whereby large fluorescence enhancements can be observed from fluorophores that are in the vicinity of a metallic surface. When considering the fluorescence detection of molecules near thin metallic films, several factors need to be considered. The emission can be quenched due to radiation energy transfer to the metal as molecules adsorbed directly on the surface. Another consideration is the enhanced fluorescence due to increased electromagnetic field of surface plasmon, enhanced quantum yield, or both.24 By placing a fluorophore near a metallic particle, the spectral properties of the fluorophore can be altered, increasing the sensitivity through an increase of quantum yield and photostability of the fluorophore. This effect is due to the surface plasmon resonance associated with the metal particle, which modifies the photonic mode density around the fluorophore and thereby increases the detected fluorescence signal.25
In our experiment, we show the applicability of single-molecule imaging for detection of small amounts of oligonucleotides immobilized on silvered surfaces. A configuration used in standard DNA assays is adopted.26 A labeled double-stranded DNA (ds-DNA) molecule is formed by hybridization with the complementary sequences, being tagged with a single Cy5 probe, and tethered to silver island films (SIFs) deposited on amino-coated glass slides. Silver island films consist of a layer of subwavelength-size silver particles and make excellent substrates for this study, as they can be simply and reproducibly fabricated, and their intrinsic optical properties are easy to be measured and well studied.27,28 These films display the plasmon resonance typical for metal colloids, have a blue-green color, but are not reflective.21 In this report, we make a comparison of the signal from the adsorbates in an electromagnetically inert environment (bare glass surfaces) to that from an equivalent coverage on a silver island film. Our experiment shows that fluorescence is greatly enhanced near a silvered surface on a molecule-by-molecule basis. The majority of identified fluorophores bound to DNA molecules contributes to somewhat stronger emission upon the proximity to the silver particles. One effect that makes a substantial contribution to the enhancement, in most instances, is the through-space electromagnetic interactions among the optical fields, the nearby molecules, and the electronic plasma resonances localized on the roughness features of the metal surface. We anticipate that a wide variety of biomolecule binding assays can be developed using the phenomenon of silver particle-enhanced fluorescence.
EXPERIMENTAL SECTION
Sample Preparation.
All oligonucleotides were obtained from the Biopolymer Core Facility at the University of Maryland School of Medicine. Nanopure water, purified using Millipore Milli-Q gradient system, was used for all experiments. All other compounds were purchased from Sigma-Aldrich and used as received. The labeled oligomers with Cy5 on the 5′ ends were obtained from Biopolymer Core facility of University of Maryland Baltimore. The coverslips used in the experiments were first soaked in a 10:1 (v/v) mixture of concentrated H2SO4 and 30% H2O2 overnight, extensively rinsed with water, sonicated in absolute ethanol for 2 min, and dried with air stream. The purity was checked by fluorescence measurements at single-molecule levels.
The sample configuration is sketched in Scheme 1. First, the glass slides were rigorously cleaned and coated with amino groups by dipping the slides in 2% aqueous solution of (3-aminopropyl)-triethoxysilane (APS) for 30 min at room temperature. The slides were thoroughly rinsed with water and air-dried prior to silver deposition. Only one side of each slide was coated with silver islands. To prevent nonspecific binding of DNA to the glass surface, the free amino groups remaining on the quartz surface were blocked with succinic anhydride by dipping in a freshly prepared solution of 0.111 g of succinic anhydride in 7 mL of 1-methyl-2-pyrrolidone and 0.77 mL of 0.2 M sodium borate buffer (pH 8). After a 15-min incubation at room temperature, slides were washed thoroughly with water.
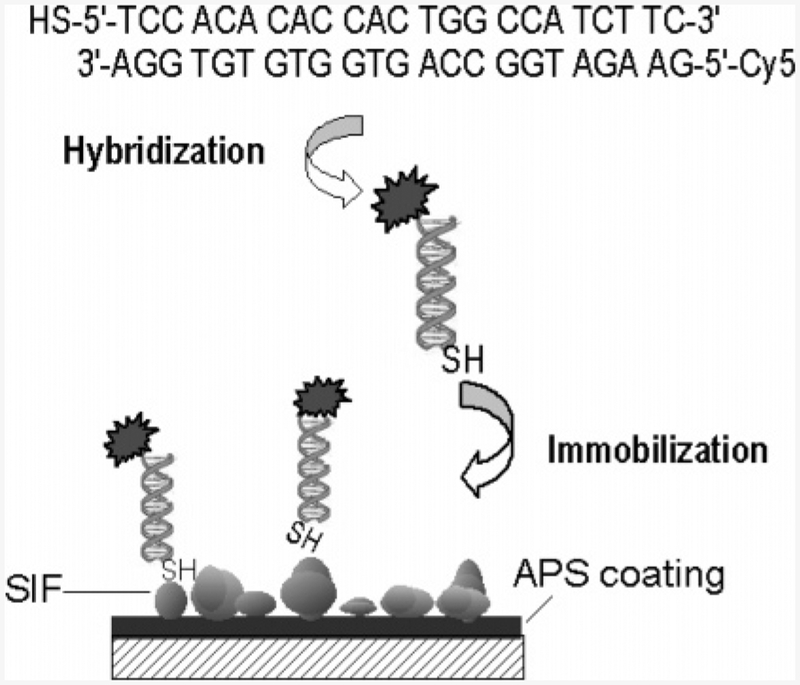
Scheme 1.
Schematic Illustration of DNA Immobilization Process in Our Experimentsa
a The Cy5-labeled oligonucleotide is hybridized to the complementary thiolated oligonucleotide. After incubation, labeled DNA molecules are tethered to silver island nanostructures on a APS-treated glass substrate.
Solutions of dsDNA samples were prepared by mixing complementary oligonucleotides in 5 mM Hepes (pH 7.5), 0.1 M KCl, and 0.25 mM EDTA buffer to a final concentration of 5 nM and cooling very slowly after incubation at 70 °C for 2 min. Immobilization of DNA samples on silver islands was accomplished by the method reported.29 Each slide was placed in a solution of thio-derivatized dsDNA for 48 h at 5 °C. A final washing step largely removed the unbound DNA probes from the substrate.
Bulk Fluorescence Measurements.
Emission spectra from DNA-SIF layers and DNA layers were collected in front-face geometry on a SLM 8000 spectrofluorometer with 635-nm excitation from a xenon lamp. Immobilization of DNA samples on both SIF and glass substrates was accomplished by the same scheme as described above. The final concentration of the DNA sample in the incubation solution was 75 nM.
Single-Molecule Imaging and Lifetime Studies.
All single-molecule studies were performed with a time-resolved confocal microscope (MicroTime 200, PicoQuant). A single-mode pulsed laser diode (635 nm, 100 ps, 40 MHz) (PDL800, PicoQuant) was used as the excitation light. The collimated laser beam was spectrally filtered by an excitation filter (D637/10, Chroma) before directing into an inverted microscope (Olympus, IX 71). An oil immersion objective (Olympus, 100×, 1.3 NA) was used both for focusing laser light onto sample and for collecting fluorescence emission from the sample. The fluorescence that passed a dichroic mirror (Q655LP, Chroma) was focused onto a 75-μm pinhole for spatial filtering to reject out-of-focus signals. To further isolate single-molecule emission and reduce background, the desired spectral detection range was selected by placing a long-pass filter (HQ685/70, Chroma) in front of the single-photon avalanche diode (SPAD). Images were recorded by raster scanning (in a bidirectional fashion) the sample over the focused spot of the incident laser with a pixel integration of 0.6 ms. The excitation power into the microscope was maintained less than 2 μW. Time-dependent fluorescence data were collected with a dwell time of 50 ms. The fluorescence lifetime of single molecules was measured by time-correlated single-photon counting (TCSPC) with the TimeHarp 200 PCI-board (PicoQuant). The data were stored in the time-tagged time-resolved mode, which allows recording every detected photon with its individual timing information. Total instrument response function widths of ~300 ps fwhm can be obtained in combination with a pulsed diode laser, which permits recording of subnanosecond fluorescence lifetimes extendable to less than 100 ps with reconvolution. Lifetimes were estimated by fitting to a χ2 value of less than 1.2 and with a residuals trace that was fully symmetrical about the zero axis. All measurements were performed in a dark compartment at room temperature.
RESULTS AND DISCUSSION
Fluorescence Spectra for Cy5-DNA on Glass and Cy5-DNA Tethered to SIF.
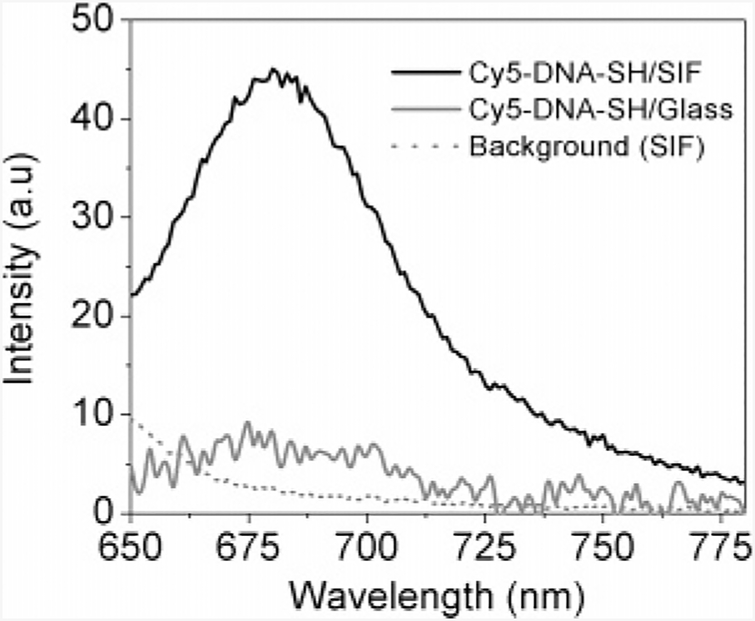
Figure 1 show the bulk emission spectra obtained for DNA samples immobilized both on glass and on SIF after complete hybridization, as well as a spectrum for a silver island film. The incubation concentrations for DNA samples used herein were ~25 times higher for those used in single-molecule studies. Each DNA sample emits most strongly near 680 nm. This spectral range was adopted for single-molecule studies to further isolate single-molecule emission and reduce background. The initial ensemble result indicates that the Cy5 dyes deposited on glass exhibit similar fluorescence characteristics compared to those tethered to SIF. Numerous studies have reported theoretically30 and empirically31 that high SERS activity can only be observed when illuminating the substrates with wavelengths close to the surface plasmon frequency. As clearly depicted in Figure 1, the presence of silver particle did not distort the Cy5 spectrum at a red excitation far away from its resonance wavelength. Furthermore, some recent studies have revealed that a high silver surface coverage can result in an increase in the SERS activity.32 However, our SEM data (Supporting Information) and previous AFM results20,23,26,29 indicate that SIFs consist of a thin layer of subwavelength-size silver particles and do not form a continuous silver coating on glass substrate. Thus, we assume that the contribution of SERS is insignificant in our case. In fact, more intense fluorescence emission is clearly observed in the case of DNA tethered to SIF. The comparison of the spectra shows that the emission from dye-labeled DNA sample is ~6-fold brighter on the SIF compared to that on glass.
Figure 1.
Fluorescence emission spectra of Cy5-labeled dsDNA immobilized on glass (gray line) and SIF (black line). The dot line represents the emission background from SIF. Excitation wavelength, 635 nm.
Fluorescence Images and Time Traces.
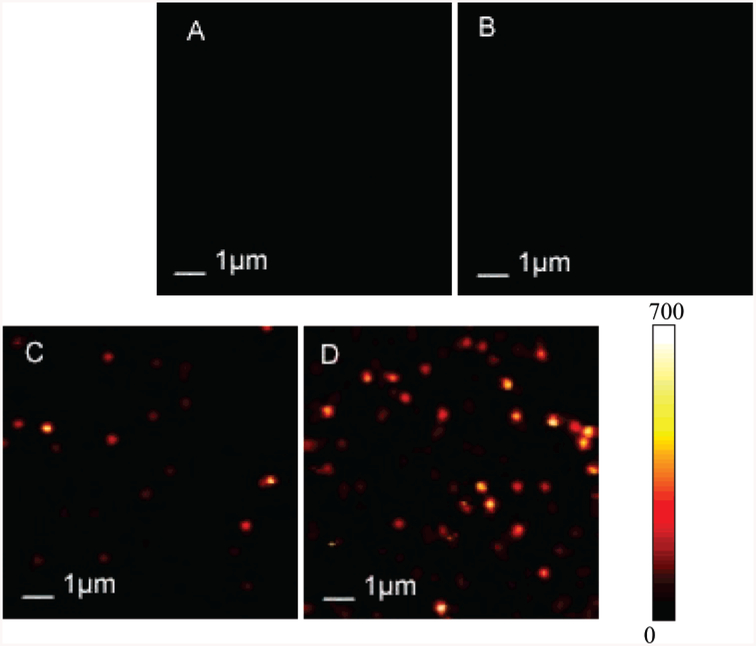
Figure 2A presents a typical 10 × 10 μm fluorescence image of Cy5-labeled dsDNA molecules, which were spin-dispersed (6000 rpm) on a clean glass coverslip, and Figure 2B shows an image acquired from a glass coverslip deposited with SIF. Each pixel has a 0.6-ms dwell time, and the total number of photons counted in that time is displayed in a colorized scale, ranging from dark (fewer integrated photons) to light. The apparent emission intensities from free labeled DNA molecules (Figure 2A) and silver nanostructure (Figure 2B) are generally less then 100 arbitrary units (au) per pixel, which are hardly observable under the marked contrast scale as shown in Figure 2. With application of DNA molecules tethered to the silver island nanostructure, the brightness of round spots greatly increases as shown in Figure 2C,D. In a fluorescence image, a bright spot will be considered as the fluorescence emission from a single fluorophore if it meets several criteria:33 (1) The spot size is in the same order as the diffraction-limited size of the laser focus in the confocal setup, which is ~300 nm in this experiment. (2) The signal intensity is consistent with that expected for single-molecule emission considering the properties of the molecule, the excitation intensity, and the efficiency of optical filters. (3) The density of the spots is proportional to the concentration of dye in the solution. (4) The photobleaching process should occur in a single step with an abrupt drop to the background level. We were aware of SIF background in the experiment. Silver and gold nanopartilcles are known to be photoactivated with light and emit characteristic “blinking” behaviors.34 To be certain that the observed bright spots arise from single dye molecules and not from silver nanoparticles scattering or other optical process, we have investigated time transients for these round bright spots. Continuous illuminating silver nanoparticles always results in strong intensity fluctuations35 and “nondestructive” blinking34 as shown in Figure 3, which can easily identify themselves from those observed from single dye molecules. The light brightness from SIF, although somewhat stronger than those from the free probe molecules, is significantly lower than the bright signal of Cy5 molecules tethered to SIF as clearly depicted in Figure 2. Thus, single dye-labeled DNA molecules are easily recognized from silvered surfaces. In addition, both the density of spots observed on unsilvered surface (not shown) and silvered surface increases with higher incubation dye concentrations also confirm the observation of single molecules. Images in Figure 2C,D clearly depict two samples incubated with two different dye concentrations. As dye concentration in the incubation solution increases, an increase in the density of bright spots is observed until a spotting saturation of the image is reached at a relatively high concentration. Intensities of round bright spots presented in Figure 2C and D vary dramatically from less than a hundred to hundreds of arbitrary units per pixel. This can be explained by an inhomogeneous immobilization process, proximity of probes to metal surface, and possibilities of photobleaching before the microscopic study on the substrate. Nevertheless, we propose that most of the observed bright spots in Figure 2C and D should be attributed to single probe molecules tethered to silver nanostructures.
Figure 2.
Typical 10 × 10 μm fluorescence images. These two-dimensional images are 150 × 150 pixels and were acquired in less than 1 min. Each pixel has a 0.6-ms dwell time, and the fluorescence intensity is displayed in a colorized scale, ranging from dark to light. (A) Cy5-labeled dsDNA molecules spin-dispersed on a glass coverslip; (B) silver island film deposited on a glass coverslip; (C) Cy5-labeled dsDNA-SH molecules tethered to silver island film deposited on a glass coverslip with an incubation concentration of 0.5 nM; (D) Cy5-labeled dsDNA-SH molecules tethered to silver island film deposited on a glass coverslip with incubation concentration of 1.5 nM. Illumination intensity was maintained at 1 μW.
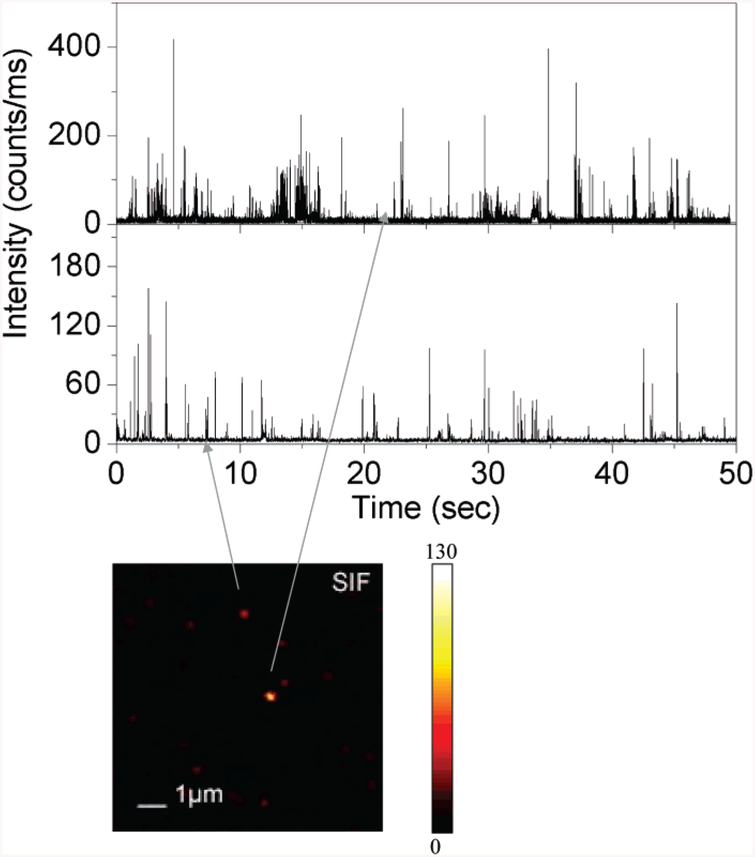
Figure 3.
Typical time traces of blinking behaviors from silver island films. Illumination intensity, 1 μW. The SIF image was rescaled from Figure 2B.
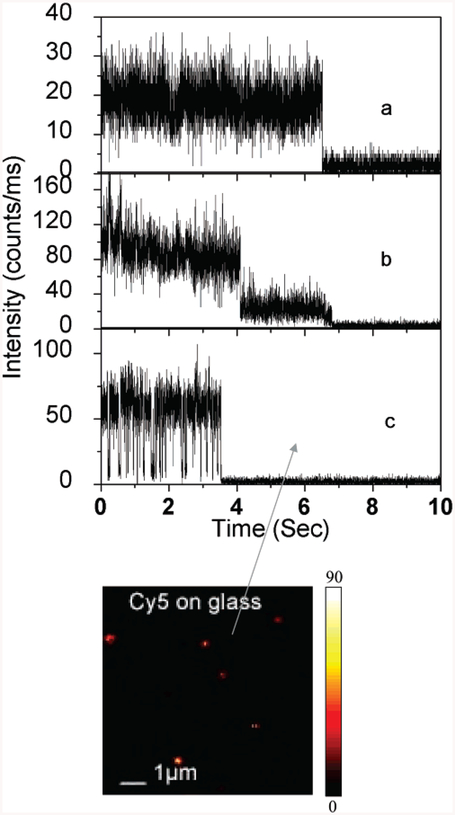
The time traces were collected using nearly the same excitation power during the experiment. The time profiles illustrate the overall trend observed from more than 100 single molecules in each environment. As a control experiment, Cy5-labeled dsDNA molecules were spin-cast on glass substrate. Among 85% of time traces collected for these investigated molecules show clearly one-step photobleaching, corresponding to the typical behavior expected for a single Cy5 molecule as shown in Figure 4a. The intensity is fairly constant until it drops abruptly to the background level in a single step. Most of the Cy5 molecules on glass substrate were bleached away within several seconds. Figure 4b presents a rarely observed two-step photobleaching example. During the first several seconds, dye molecule emits constant photons with relatively high count rate and then drops to a stable lower intensity level until an irreversible photobleaching, showing a possibility that two or more molecules were present under the focusing volume. The other example shown in Figure 4c is also a classical situation encountered in our single-molecule experiment. The emission switches between a high-intensity count level and a lower or background intensity level. This well-known behavior corresponds to “blinking”. The vast majority of causes is related to environmental and dye molecule dynamics. Included are triplet blinking36 and time-dependent variation in the fluorescence quantum yield;13 physical processes such as translational and rotational molecular motions may also contribute.37,38
Figure 4.
Representative time traces of single Cy5-labeled dsDNA molecules immobilized on a glass coverslip. Illumination intensity, 1 μW. The image was rescaled from Figure 2A.
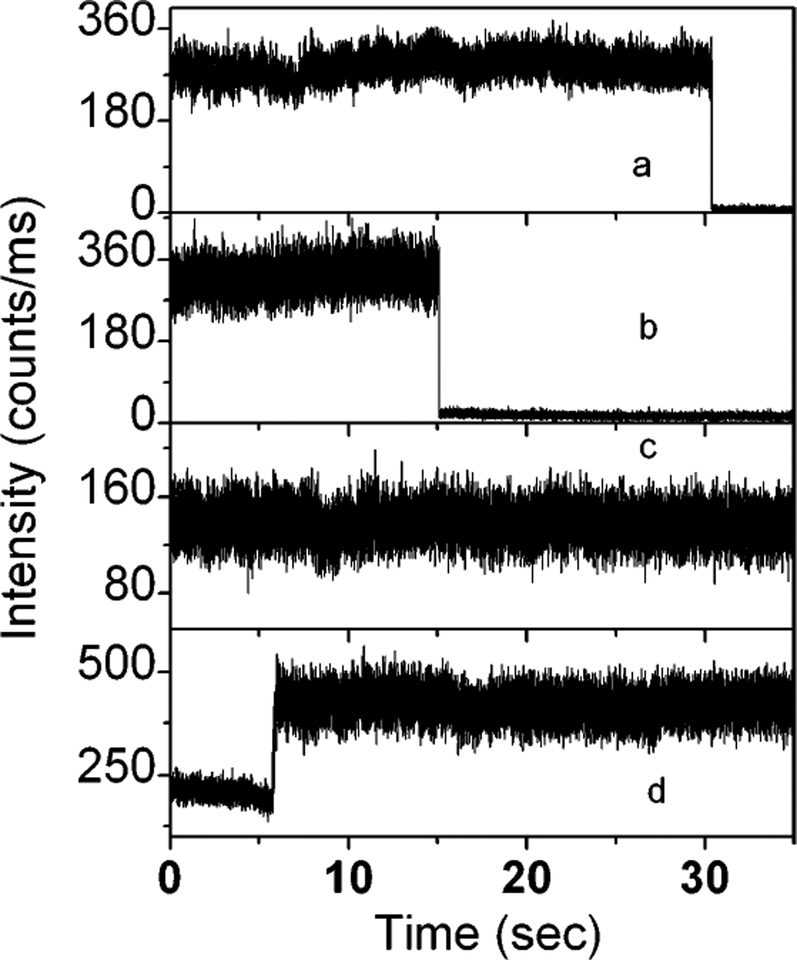
Typical time traces acquired from tethered samples are presented in Figure 5. The various time-dependent behaviors indicate the heterogeneity of the surface properties. These molecules demonstrate impressive photostability on SIF. About 72% of observed time traces recorded from DNA molecules tethered to the SIF show one-step photobleaching behaviors similar to those observed the on glass surface (Figure 5a and b). However, much higher and fairly constant emission rates are observed from these time profiles, which are generally more than 10-fold from those observed in the absence of SIF. The molecule shown in Figure 5a photobleached after ~30 s of continuous illumination. In fact, a few ultrabright spots can be monitored indefinitely at the single-molecule level without photobleaching as shown in Figure 5c. No photobleaching occurred during a 60-s period observed in 18% of the investigated single molecules. In fact, different sites on the silvered surface in which the probe molecules were tethered are moderately varied. In a rare case presented in Figure 5d, the molecule may have undergone a change in its quantum yield after 5-s continuous illumination, emitting higher and steady photons in the subsequent observed period. The attached fluorophore is flexible after tethering to SIF and is at a certain distance from the metallic particle; both the incident field and radiated fields may be varied during a period. We made an assumption that when the molecule is excited, part of its energy can be converted into rotation or into translation and the molecule can extend to a position or rotate to a location where enhancement is favored.
Figure 5.
Representative time traces of single Cy5-labeled dsDNA molecules tethered to silver island films deposited on a glass coverslip. Illumination intensity, 1 μW.
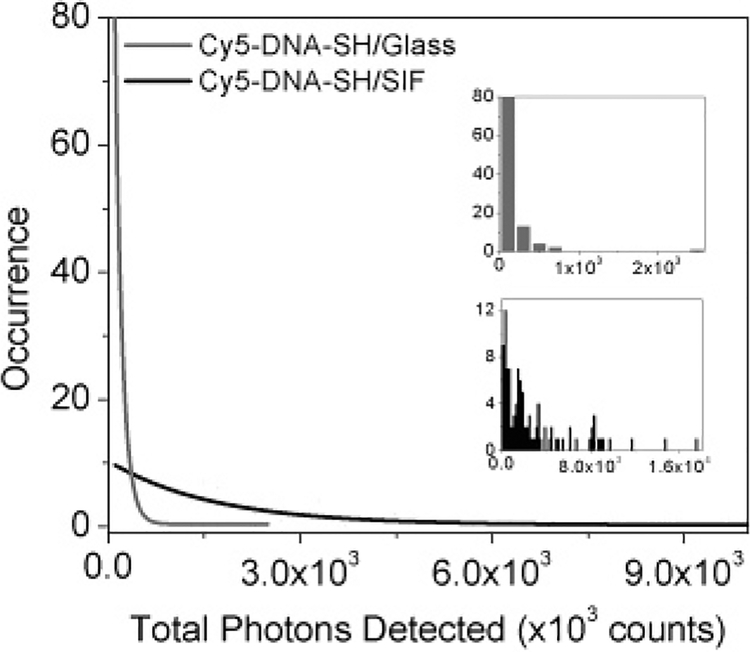
We have derived histograms of the total number of photons emitted by each molecule before photobleaching. The total number of detected photons was determined by integrating of individual single-molecule time transients as shown in Figure 4a and Figure 5a, b. This was done by taking as many 10 × 10 μm2 fluorescence images as necessary. These measured single dye molecules were all photobleached within the illumination period of 60 s. First characteristics of the underlying properties can be derived from a qualitative comparison of the histograms. It is clear from these data that dye molecules tethered to SIF emit significantly more total photons than do those adsorbed directly on glass before photobleaching. Exponential fits of histograms are presented in Figure 6 and indicate that the average photon in the tethered sample and free dyes on glass are approximately 1.6 × 106 (dark line) and 1.1 × 105 (gray line), respectively. The values reflect only the total numbers that were detected prior to photobleaching. There are in fact lower count rates observed on bare glass slides; most of molecules emit photons at a rate less than 30 counts/ms as compared to a few hundred counts per millisecond on the tethered samples. Additionally, the majority of Cy5-dsDNA molecules deposited on glass survived less than roughly 10 s. The increase in total emitted photons on the silvered surface indicates an increase in the fluorescence quantum yield of the tethered dye molecules, and also an increase in photostability is observed as frequently manifested by the longer survival time.
Figure 6.
Exponential fits of total number of photons detected before photobleaching. Insets: histograms of free Cy5-labeled dsDNA molecules (top, gray bars) and Cy5-labeled dsDNA molecules tethered to silver island films (bottom, black bars).
Fluorescence Lifetime.
A fluorophore typically emits a photon within nanoseconds after absorption of the excitation photon. The decay law is often monoexponential, characterized by a lifetime
| (1) |
where Г and knr are the radiative and nonradiative decay rates, respectively. The lifetime is a sensitive probe of the local environment of the molecule. The changes in knr are typically due to changes in a fluorophore’s environment, quenching or FRET.25 The radiative decay rate Г is constant, and any changes are primarily due to changes in refractive index. Proximity of flurophores to metals can result in an increase in the total radiative decay rate. The lifetime near the metal then becomes18
| (2) |
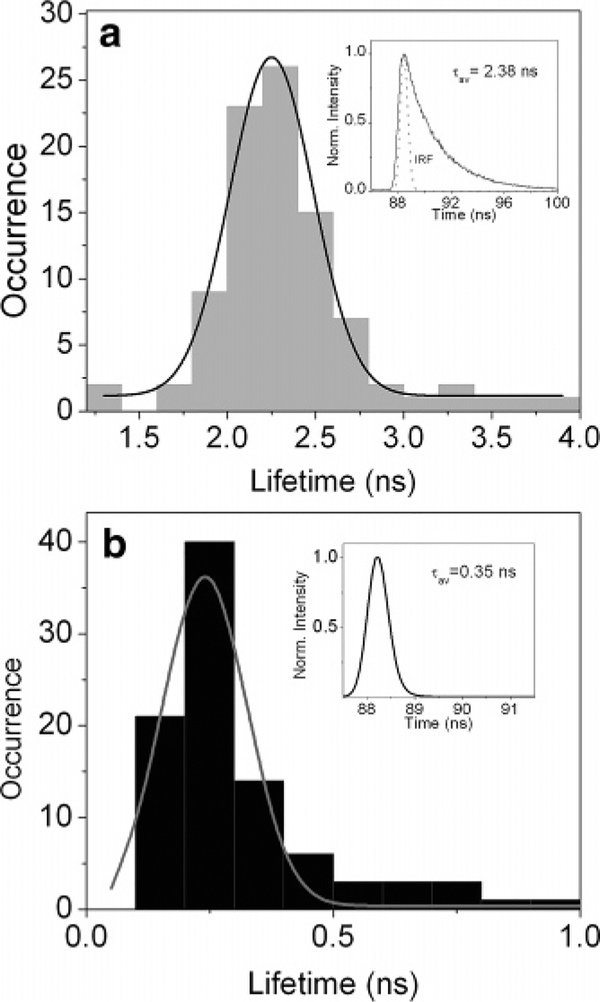
The radiative rate is given by Г + Гm, when Гm is the part due to the metal. The change in radiative rate results in remarkable effects such as the increase in quantum yield and decrease in lifetime.25 We implemented the TCSPC measurement on a single fluorescent spot. The insets in Figure 7 show typical fluorescence decay curves of two probe molecules immobilized on glass (Figure 7a) and tethered to SIF (Figure 7b). The curves decay exponentially with averaged time constants of τav) = 2.38 ns for the free labeled DNA molecule and τav) 0.35 ns for the DNA molecule tethered to SIFs, respectively (fitting parameters are given in Supporting Information). The nearly single-exponential decay of fluorescence observed for different molecules suggests that most of the molecules adsorbed on glass are in a relatively homogeneous environment. In the case of SIF (insert in Figure 7b), double-exponential decay analysis yields two short decay components of about 0.14 and 0.77 ns. However, the intensity amplitudes differ considerably. Multiexponential single-molecule fluorescence decays have been investigated previously and were related to fluctuating lifetimes during the measurement.39 We observed a predominant intensity contribution (>85%) from a short-lifetime component (<0.5 ns) for the investigated single molecules in the presence of SIF. It seems reasonable therefore to assume that the longer lifetime component arises from double-stranded DNA molecules exposed to the glass substrate and its contribution decreases where the surface becomes occupied by a metallic nanostructure. The multicomponent decay suggests variations in the photonic mode density around the probe as a result of the local density fluctuations since tethered fluorophores were relatively flexible than those adsorbed directly on glass surface.
Figure 7.
Histograms of lifetimes. (a) Free Cy5-labeled dsDNA molecules (inset: a typical TCSPC decay curve with an averaged lifetime of 2.38 ns); (b) Cy5-labeled dsDNA molecules tethered to silver island films (inset: a typical TCSPC decay curve with an averaged lifetime of 0.35 ns). The fitting parameters are provided in Supporting Information.
Figure 7 also illustrates histograms of fluorescence lifetimes acquired from ~100 single molecules on the silvered surface and unsilvered surface, respectively. To aid interpretation, lifetime distributions were fit to Gaussian functions. A symmetric distribution with a center value equal to 2.2 ns is obtained for dye molecules immobilized on glass coverslips, in contrast to 0.25 ns in the presence of silver island films. The symmetric distribution of a lifetime histogram as depicted in Figure 7a indicates the sample on glass substrate is relatively homogeneous. Lifetimes of tethered dyes are shifted well to the lower values as shown in Figure 7b. In contrast to the symmetric distribution shown in Figure 7a, the fluorescence lifetime distribution in Figure 7b is slightly off-center. The lifetime ranges from hundreds of picoseconds to 1 ns. The histogram shows a long tail and can only be fit approximately. The skewed distribution of lifetimes is in line with a large heterogeneity of the sample and random orientation of the probes. This involved situation is due to the likely existence of two or more different subpopulations of single-molecule species arising from diverse electromagnetic interactions. It can also be assumed that the spread of different tethered sites accounts for lifetime and intensity variation from molecule to molecule, which is in good agreement with the observed multiexponential fluorescence decays.
CONCLUSION
In summary, we demonstrate the capability to detect metal-enhanced fluorescence at a single-molecule level. Large fluorescence enhancements can be obtained from fluorophores that are in the vicinity of metallic nanoparticles. The excited fluorophore induces a charge distribution on the metal surface. If wavevector matching allows the plasmon to radiate, then the fluorophore emission is observed as metal-enhanced fluorescence. Increased quantum yield and reduced lifetime all increase the brightness. Our experiment results demonstrate that both emission rates and lifetimes are greatly affected by silvered surfaces on molecular scales. Single dye molecules tethered to SIFs are brighter than their free counterparts as observed. Time profiles also indicate higher photostability in these tethered samples. Most of the photons detected on bare glass coverslips range from 104 to 105 photons. In contrast, measured counts for single molecules on silvered surface are in the range of a few 106, and these values depict that dye molecules tethered to SIF are very efficient emitters. The broad distribution of photon counts is in agreement with a larger heterogeneity of the geometric shapes of a SIF nanostructure, various locations of the molecule relative to the particle, and the random orientation of the probe molecules. The lifetime data on silvered surfaces are skewed toward lower values as expected. The proximity to a metallic nanostructure markedly modifies the fluorescence lifetime via perturbations of the intramolecular transition of the molecule. Fluorophores can become more photostable, less prone to optical saturation, and have higher emission rates, which are consistent with time transients observed. In addition, the shorter lifetime of the fluorophore result in less time for photochemistry while in the excited state and thus more excitation-emission cycles prior to photobleaching. These studies have provided a better understanding of types of microenvironments present in silver nanostructures. In the future, such studies would benefit many applications to research, such as clinical sensing, proteomics and genomics, high-sensitivity detection, and single-molecule counting.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the National Center for Research Resources, RR-08119 and National Human Genome Research Institute, HG-002655. The authors thank Dr. Chandran R. Sabanayagam for SEM technique support and critical discussion of the manuscript.
Footnotes
SUPPORTING INFORMATION AVAILABLE
SEM image of SIF and lifetime fitting parameters. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Brown PO; Bostein D Nat. Genet. Suppl 1999, 21, 33–37. [DOI] [PubMed] [Google Scholar]
- (2).Deyholos MK; Galbrainth DW Cytometry 2001, 43, 229–238. [PubMed] [Google Scholar]
- (3).Schena M; Heller RA; Theriault TP; Konrad K; Lachenmerier E; Davis RW Trends Biotechnol 1998, 16, 301–306. [DOI] [PubMed] [Google Scholar]
- (4).Trabesinger W; Schutz GJ; Gruber HJ; Schindler H; Schmidt T Anal. Chem 1999, 71, 279–283. [DOI] [PubMed] [Google Scholar]
- (5).Osborne MA; Furey WS; Klenerman D; Balasubramanian S Anal. Chem 2000, 72. [DOI] [PubMed] [Google Scholar]
- (6).Osborne MA; Barnes CL; Balasubramanian S; Klenerman DJ Phys. Chem. B 2001, 105, 3120–3126. [Google Scholar]
- (7).Ma Y; Shortreed MR; Li H; Huang W; Yueng ES Electrophoresis 2001, 22, 421–426. [DOI] [PubMed] [Google Scholar]
- (8).Xie XS; Trautman JK Annu. Rev. Phys. Chem 1998, 49, 441–480. [DOI] [PubMed] [Google Scholar]
- (9).Wennmalm S; Rigler RJ Phys. Chem. B 1999, 103, 2516–2519. [Google Scholar]
- (10).Julien C; Debarre A; Nutarelli D; Richard A; Tchenio PJ Phys. Chem. B 2006. [DOI] [PubMed] [Google Scholar]
- (11).Huang Z; Ji D; Xia A Colloids Surf., A 2005, 257–258, 203–209. [Google Scholar]
- (12).Weiss S Science 1999, 283, 1676–1683. [DOI] [PubMed] [Google Scholar]
- (13).Tinnefeld P; Sauer M Angew. Chem., Int. Ed 2005, 44, 2642–2671. [DOI] [PubMed] [Google Scholar]
- (14).Peterman EJG; Brasselet S; Moerner WE J. Phy. Chem. A 1999, 103, 10553–10560. [Google Scholar]
- (15).Dickson RM; Cubitt AB; Tsien RY; Moerner WE Nature 1997, 388, 355–358. [DOI] [PubMed] [Google Scholar]
- (16).Ha T; Glass J; Enderle T; Chemla DS; Weiss S Phys. Rev. Lett 1998, 103, 6839. [Google Scholar]
- (17).Zondervan R; Kulzer F; Orlinksii SB; Oritt M Phys. Chem. A 2003, 107, 6770–6776. [Google Scholar]
- (18).Libebermann T; Knoll W Langmuir 2003, 19, 1657–1572. [Google Scholar]
- (19).Stranik O; McEvoy HM; McDonagh C; MacCraith BD Sens. Actuators, B 2005, 107, 148–152. [Google Scholar]
- (20).Lakowicz JR Anal. Biochem 2001, 298, 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Sokolov K; Chumanov G; Cotton TM Anal. Chem 1998, 70, 3898–3905. [DOI] [PubMed] [Google Scholar]
- (22).Liebermann T; Knoll W Colloids Surf 2000, 171, 115–130. [Google Scholar]
- (23).Malicka J; Gryczynski I; Gryczynski Z; Lakowicz JR Anal. Biochem 2003, 315, 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Stefani FD; Vasilev K; Bocchio N; Stoyanova N; and Kreiter M Phys. Rev. Lett 2005, 94, 1–4. [DOI] [PubMed] [Google Scholar]
- (25).Lakowicz JR; Malicka J; Gryczynski I; Gryczynski Z; Geddes CJ Phys. D: Appl. Phys 2003, 36, R240–R249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mirkin CA; Letsinger RL; Mucic RC; Storhoff JJ Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- (27).Lakowicz JRMJ; Gryczynski I BioTechniques 2003, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Tarcha PJ; DeSaja-Gonzalez J; Rodriguez-LIorente S; Aroca R Appl. Spectrosc 1999, 53, 43–48. [Google Scholar]
- (29).Lukomska J; Malicka J; Gryczynski I; Leonenko Z; Lakowicz JR Biopolymers 2005, 77, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kerker M; Wang D-S; Chew H; Siiman O; Bumm LA Suface Enhanced Raman Scattering; Plenum Press: New York, 1993. [Google Scholar]
- (31).Bergman JG; Chemla DS; Liao PF; Glass AM; Pinczuk A; Hart RM; Olson DH Opt. Lett 1981, 6, 33. [DOI] [PubMed] [Google Scholar]
- (32).Stockle RM; Deckert V; Fokas C; Zenobi R Appl. Spectrosc 2000, 54, 1577–1583. [Google Scholar]
- (33).Mei E; Bardo AM; Collinson MM; Higgins DA J. Phys. Chem. B 2000, 104, 9973–9980. [Google Scholar]
- (34).Peyser LA; Vinson AE; Bartko AP; Dickson RM Science 2001, 291, 103–106. [DOI] [PubMed] [Google Scholar]
- (35).Geddes CD; Parfenov A; Gryczynski I; Lakowicz JR J. Phys. Chem. B 2003, 107, 9980–9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Huang Z; Ji D; Wang S; Xia A; Koberling F; Pattings M; Erdmann RJ Phys. Chem. A 2006, 110, 45–45. [DOI] [PubMed] [Google Scholar]
- (37).Higgins DA; Collinson MM; Saroja G; Bardo AM Chem. Mater 2002, 14, 3734–3744. [Google Scholar]
- (38).Wirth MJ; Swinton DJ J. Phys. Chem. B 2001, 105, 1472–1477. [Google Scholar]
- (39).Loos D; Cotlet M; Schryver FD; Habuchi S; Hofkens J Biophys. J 2004, 87, 2598–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.