Abstract
Rhesus cytomegalovirus (RhCMV)-based vaccines maintain effector-memory T cell responses (TEM) that protect ~50% of rhesus monkeys (RMs) challenged with simian immunodeficiency virus (SIV). Because human CMV (HCMV) causes disease in immune-deficient subjects, clinical translation will depend upon attenuation strategies that reduce pathogenic potential without sacrificing CMV’s unique immunological properties. We demonstrate that “intrinsic” immunity can be used to attenuate strain 68–1 RhCMV vectors without impairment of immunogenicity. The tegument proteins pp71 and UL35 encoded by UL82 and UL35 of HCMV counteract cell-intrinsic restriction via degradation of host transcriptional repressors. When the corresponding RhCMV genes, Rh110 and Rh59, were deleted from 68–1 RhCMV (ΔRh110 and ΔRh59), we observed only a modest growth defect in vitro, but in vivo, these modified vectors manifested little to no amplification at the injection site and dissemination to distant sites, in contrast to parental 68–1 RhCMV. ΔRh110 was not shed at any time post-infection and was not transmitted to naïve hosts by either close contact (mother to infant) or by leukocyte transfusion. In contrast, ΔRh59 was both shed and transmitted by leukocyte transfusion, indicating less effective attenuation than pp71 deletion. Importantly, the T cell immunogenicity of ΔRh110 was essentially identical to 68–1 RhCMV with respect to magnitude, TEM phenotype, epitope targeting, and durability. Thus, pp71 deletion preserves CMV vector immunogenicity while stringently limiting vector spread, making pp71 deletion an attractive attenuation strategy for HCMV vectors.
One sentence summary:
Live-attenuated, spread-deficient rhesus CMV-vectors retain T cell immunogenicity.
INTRODUCTION
The beta-herpesviruses HCMV and RhCMV are ubiquitous in human and RM populations, respectively (1,2). Although these viruses are specifically adapted to their respective host species, they share orthologous genomes and a remarkably similar biology, reflecting millions of years of host-viral co-evolution (3,4). Infection usually occurs in the first year of life and is asymptomatic in the vast majority of subjects, with viral replication and spread controlled by host immunity, particularly virus-specific T cell responses (5,6). At the same time, CMV uses complex immune evasion strategies to prevent host immunity from clearing the infection or inhibiting its ability to spread to new hosts, including super-infection of CMV+ hosts, resulting in permanent infections with one or more viral strains (7–9). CMV and their primate hosts thus establish a stable equilibrium characterized by 1) a low viral burden in infected individuals with mostly latent infection (albeit with occasional reactivation and sufficient shedding to maintain efficient host-to-host transmission) resulting in persistent infection, and 2) an extra-ordinarily high frequency of CD4+ and CD8+ CMV-specific T cells with a unique TEM -biased phenotype and function that contains the infection and thus tissue damage and disease (6,10,11).
Although other viruses can establish persistent infection, the lifelong, high frequency, tissue-based, TEM-biased T cell responses established by CMV are unique, which raised the possibility of exploiting CMV as a vaccine vector for other infectious diseases (12–14). In particular, we hypothesized that recombinant CMV expressing heterologous inserts would pre-position potent, effector-differentiated responses to a different pathogen in the portal of entry and sites of early spread of that pathogen. This early interception (prior to implementation of its immune evasive programs) would potentially have superior efficacy for immune evasive pathogens than typical vaccine-elicited memory T cell responses that require anamnestic expansion over many days, if not weeks, to mount peak effector responses in tissues (14). Indeed, we demonstrated that the cellular immune responses elicited by strain 68–1 RhCMV vectors expressing SIV inserts intercept and completely control SIV replication in the first week post-challenge in ~50% of vaccinated RMs, without anamnestic expansion of SIV-specific immune responses (15–17). Most remarkably, the residual infection in protected RMs was progressively cleared over weeks to months, ultimately leaving these RMs indistinguishable by virologic and immunologic criteria from vaccinated RMs that were never challenged (17). RhCMV vectors with Mycobacterium tuberculosis (Mtb) inserts elicit analogous CD4+ and CD8+ TEM responses to Mtb, and RMs vaccinated with these vectors show a nearly 70% reduction in Mtb disease after Erdman strain challenge, with 40% of vaccinated animals showing no detectable granulomatous disease at all (18).
These preclinical data support consideration of the possible development of CMV-vectored HIV and Mtb vaccines for use in people, creating an imperative to develop and test HCMV-based vectors in the clinic (19,20). However, clinical development of HCMV-based vectors for prophylactic use in healthy individuals will require a high standard of safety. Although HCMV does not cause overt disease in the vast majority of infections, the potential for symptomatic primary infection in HCMV-naïve individuals, and for serious disease in immunocompromised subjects or in mother-to-fetus transmission (21), is sufficiently high to contraindicate the use of unmodified HCMV vectors in a prophylactic vaccine setting. We therefore sought to develop an attenuation strategy that would reduce or, if possible, eliminate the pathogenic potential of HCMV, yet retain the unique, persistent immunogenicity, as well as the ability to super-infect CMV+ individuals. Furthermore, it is preferable for clinical vectors to not be transmissible by close contact or even transfusion, so as to prevent unintentional infection of individuals for which vaccination might pose a safety risk.
Encouragingly, mouse CMV (MCMV) rendered unable to spread from initially infected cells nonetheless maintained strong cell-mediated immunity, suggesting that viral dissemination may not be required for immunogenicity (22–24). However, it remains unclear whether highly debilitated RhCMV or HCMV vectors can establish and maintain persistent secondary infections in monkeys or humans. Indeed, the HCMV strain Towne, attenuated by serial in vitro passaging, seems to have lost its ability for persistent immune induction as indicated by steadily declining T cell responses (25). In addition, chimeras of Towne and a fibroblast-adapted derivative of the primary isolate Toledo were unable to establish secondary infections in HCMV+ individuals (26), and the T cell responses elicited by these chimeras in HCMV− individuals did not display the CMV-typical TEM phenotype and declined over time (27,28). These data suggest that traditional attenuation strategies may yield suboptimal HCMV vectors that lose the ability to maintain persistent immune stimulation.
Here, we examine a new strategy for CMV attenuation based on exploiting host proteins that provide “intrinsic” immunity. This term describes cellular intrinsic defense mechanisms involving host proteins such as death domain-associated protein (DAXX), alpha-thalassemia X-linked mental retardation protein (ATRX), BCL-associated factor 1 (BclAF1) and promyelocytic leukemia protein (PML) that combine to form nuclear ND10 bodies (29). ND10 proteins repress transcription of viral immediate early (IE) genes, which are critical for early (E) and late (L) gene expression and genome replication (30, 31). To counteract intrinsic immunity, herpesviruses encode proteins that eliminate or disperse ND10 components (32). HCMV evades intrinsic defenses through viral tegument proteins released during viral entry, thus facilitating IE expression (33). The tegument protein pp71, encoded by UL82, mediates the degradation of DAXX (34–36) and, together with UL35, BclAF1 (37) in the infected cell nucleus. Intrinsic immune repression of IE expression might enable the establishment and maintenance of latency by silencing viral genes needed for lytic replication (38–40). Thus, by eliminating viral counter-mechanisms targeting intrinsic immunity, it might be possible for CMV vectors to maintain persistent infection and long-term immune stimulation, while severely hampering lytic replication, cell-to-cell spread, and distant dissemination. To examine this possibility, we characterized the in vivo replication, spread, dissemination, and immunogenicity of RhCMV vectors lacking Rh110 and Rh59, the RhCMV orthologs of HCMV UL82 and UL35.
RESULTS
RhCMV pp71 mediates DAXX degradation and supports RhCMV growth in vitro.
HCMV pp71 has two DAXX interaction domains that are required for localization of pp71 to ND10 bodies (41) where pp71 dislocates ATRX and mediates the degradation of DAXX by the proteasome (38). Although Rh110, the predicted RhCMV pp71 ortholog, shares 41% identity with HCMV pp71 (42), these DAXX interaction domains are not conserved. Nevertheless, when RhCMV pp71 was expressed in telomerized rhesus fibroblasts (TRFs) under doxycycline (DOX) control, steady-state DAXX was reduced upon removal of DOX (Fig. 1A). When we examined the localization of rhesus DAXX, ATRX, and PML by immunofluorescence, we observed punctate nuclear co-staining of these proteins in the absence of pp71. Upon pp71 expression, both ATRX and DAXX no longer localized in punctate dots, whereas pp71 partially co-localized to PML bodies (Fig. S1). Thus, despite lacking canonical DAXX-interaction domains, RhCMV pp71 degrades and displaces DAXX from PML bodies similar to HCMV pp71 (32).
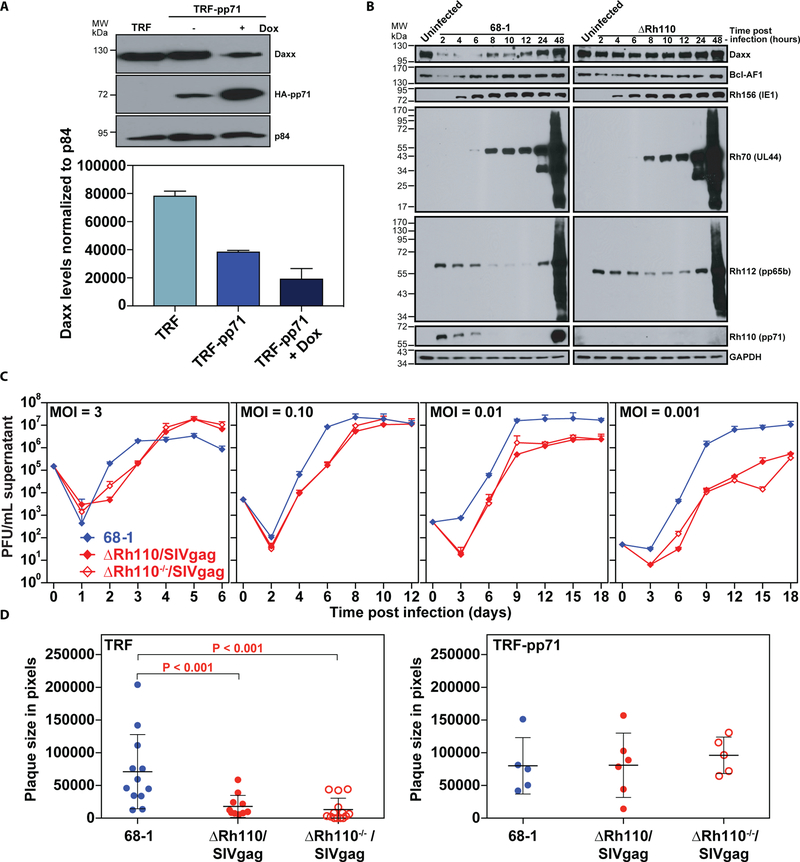
Figure 1: RhCMV pp71 degrades DAXX and absence of pp71 results in in vitro growth deficiency.
(A) TRFs or TRF-pp71 were untreated (−) or treated with 10 µg/ml doxycycline (+) to induce RhCMV pp71 expression for 24 hrs. Nuclear lysates were harvested and analyzed by immunoblot using anti-hemagglutinin (HA) antibodies to detect epitope-tagged pp71. The cellular nuclear matrix protein p84 was analyzed as a loading control. The graph shows mean (+ SD) with n = 2. (B) TRFs were infected with 68–1 or ΔRh110 (MOI=3) or left uninfected and cell lysates were harvested at the indicated time points, electrophoretically separated, and subjected to immunoblots with antibodies to the indicated host and viral proteins. Each time point represents an independent infection. (C) TRFs were infected with 68–1 or either complemented or uncomplemented (−/−) ΔRh110/SIVgag at the indicated MOI. Supernatants were harvested at the indicated times and titered by TCID50 (median tissue culture infectious dose) on TRF-pp71. Average titers from two experimental and two technical replicates (+ SD) are shown. (D) TRFs or TRF-pp71 were infected with 68–1, complemented ΔRh110/SIVgag, or uncomplemented ΔRh110 (ΔRh110−/−) at MOI = 0.001. Plaques were analyzed at 7 dpi using phase microscopy and plaque size was measured using Adobe Photoshop. Individual plaque sizes, as well as average (+/− SD) from one of two experiments are shown. The Kruskal-Wallis (KW) test was used to determine the significance of differences between the 3 groups [P = 0.0002, left; P = not significant (NS), right] with the Wilcoxon rank sum test used to perform pair-wise analysis if KW P-values were ≤0.05; brackets indicate pair-wise comparisons with two-sided Wilcoxon P-values ≤0.05. P-values ≤0.05 are considered statistically significant.
As reported for HCMV (31,35,36,43,44), DAXX and BclAF1 were reduced during early times of infection with parental strain 68–1 RhCMV (68–1) compared to uninfected cells (Fig. 1B) [unless otherwise noted, all RhCMV constructs are based on bacterial artificial chromosome (BAC)-derived strain 68–1 (45)]. This degradation was mediated by pp71, since DAXX abundance, and to some extent, BclAF1 abundance were restored in fibroblasts infected with 68–1 RhCMV lacking pp71 (ΔRh110) (Fig. 1B, Fig. S2A,B). At the high multiplicity of infection (MOI) used in this experiment, deletion of pp71 did not affect the kinetics or expression of IE, E, or L proteins, similar to previous reports showing that DAXX repression of HCMV gene expression is overcome at high MOI (36,43). This growth defect could be reversed by knockdown of DAXX expression (Fig. S2C), suggesting increased anti-viral activity by DAXX being the main reason for the observed growth defect and consistent with increased IE expression reported for HCMV upon DAXX siRNA treatment (36).
Interestingly, pp71-deleted RhCMV could be recovered upon BAC transfection of fibroblasts without the need for complementation, and high titer stocks could be generated in the absence of pp71 complementation. In contrast, UL82-deleted HCMV could only be grown in pp71-complementing cells (46) and guinea pig CMV lacking the UL82 homolog could not be recovered unless complemented (47). To determine the impact of pp71 deletion and complementation on viral growth in vitro, we compared 68–1 virus production upon infection of fibroblasts with RhCMV in which Rh110 had been replaced with SIVgag (ΔRh110/SIVgag) so that transcription was regulated by the native Rh110 promoter. ΔRh110/SIVgag was generated on complementing cells, which results in pp71 incorporation into the virus particle (Fig. S2D), or without complementation (−/−). There was little or no growth impairment of complemented or uncomplemented ΔRh110/SIVgag at high MOI (1 and 0.1), whereas a 10- to 100-fold reduction of viral yield was observed at low MOI (0.01 and 0.001) (Fig. 1C). In contrast, a 10- to 100-fold reduction in viral growth was reported for complemented or uncomplemented HCMV at high MOI and ~5-log reduction in viral growth at low MOI (46) suggesting that abrogation of pp71 expression impacts growth of RhCMV less severely than that of HCMV. Nevertheless, pp71 deletion clearly reduced spreading of virus from infected cells to neighboring cells since the average plaque sizes of complemented and uncomplemented ΔRh110/SIVgag at day 7 post-infection at low MOI (0.001) were strongly reduced compared to 68–1 in TRF, but not in TRF-pp71 (Fig. 1D). Thus, although pp71 is not absolutely required for RhCMV growth in vitro, it substantially enhances the efficiency of viral spreading in fibroblast cultures.
ΔRh110 RhCMV is spread-deficient in vivo but remains immunogenic for T cell responses.
To examine whether deletion of pp71 would affect viral dissemination in vivo, we subcutaneously inoculated RhCMV-naïve RMs with 107 plaque-forming units (PFU) of 68–1 expressing SIVgag in the left arm and complemented ΔRh110-expressing SIVrev/tat/nef/int (rtni) in the right arm (Fig. S2A). The viral constructs used in each of the in vivo experiments in this report are schematically depicted in Fig. S3. RMs were necropsied at 14, 21, or 28 days post-infection (dpi) and viral copy numbers were determined by ultra-sensitive, nested qPCR specific for SIVgag or SIVrtni sequences in multiple tissue samples (17, 49). SIVgag-containing genomes were detectable in most tissues in the RMs necropsied at 14 dpi, with particularly high genome copy numbers observed at the site of inoculation and associated draining lymph nodes, the contralateral injection site, and salivary glands (Table 1A), whereas genome copy numbers were progressively lower in RMs necropsied at 21 and 28 dpi, in keeping with progressive immune control of infection, but were still detectable in most tissues. In notable contrast, genomes for ΔRh110/SIVrtni were barely detectable in the same tissues at 14 dpi and below detection limits in almost all tissues at 21 and 28 dpi (Table 1A). Of note, similar frequencies of SIVgag- and SIVrtn-specific CD4+ and CD8+ T cell responses were measured in lymphoid and non-lymphoid tissues of all 3 RMs, with the possible exception of some tissues at 14 dpi where SIVgag-specific T cell responses were modestly higher (Fig. S4A). Thus, deletion of Rh110 markedly limited the genome replication and dissemination of RhCMV during primary infection, but had little to no effect on the initial development of vector-elicited, SIV-specific T cells.
Table 1: Rh110 (pp71) deletion reduces dissemination of RhCMV vectors.
(A) Three RhCMV-naïve RMs (T1A-1, T1A-2, T1A-3) were co-inoculated with 107 PFU each of 68–1/SIVgag (left arm) and ΔRh110/SIVrtni (right arm). One RM each was necropsied at 14, 21, or 28 dpi and viral genome copy numbers per 107 cell equivalents were determined in the indicated tissues using ultra-sensitive nested qPCR specific for SIVgag (68–1) or SIVrtni (ΔRh110). (B) Three naturally RhCMV-infected RMs (T1B-1, T1B-2, T1B-3) were co-inoculated with 107 PFU each of 68–1/SIVrtni (left arm) and ΔRh110/SIVgag (right arm). One RM each was necropsied at 14, 21, or 28 dpi and viral genome copy numbers per 107 cell equivalents in the indicated tissues were determined using ultra-sensitive nested qPCR specific for SIVgag (ΔRh110) or SIVrtni (68–1). (C) Two naturally RhCMV-infected RMs (T1C-1 and T1C-2) were co-inoculated with 107 PFU each of complemented ΔRh110 and uncomplemented ΔRh110 in different arms, with the SIVgag and SIVrtni inserts used to mark the complemented and uncomplemented vectors, respectively, in RM1 and the reverse in RM2. Both RMs were necropsied at 14 dpi and viral genome copy numbers were determined using ultra-sensitive nested qPCR specific for SIVgag vs. SIVrtni. Normalized to 1×107 cell equivalents. WT, wild type; GI, gastrointestinal; LN, lymph node; PLN, parietal lymph node.
| Table 1A | Primary infection: 68–1 vs. ∆Rh110 | |||||
|---|---|---|---|---|---|---|
| RM T1A-1 | RM T1A-2 | RM T1A-3 | ||||
| Tissue type | 14 dpi | 21 dpi | 28 dpi | |||
| 68–1/SIVgag | ∆Rh110/SIVrtni | 681/SIVgag | ∆Rh110/SIVrtni | 68–1/SIVgag | ∆Rh110/SIVrtni | |
| Skin Inj site-right (∆Rh110) | 19,975,462 | <1 | 13,227 | 2 | <1 | <1 |
| Skin Inj site-left (WT) | 1,013,192,441 | 3 | 14,453 | <1 | <1 | <1 |
| Axillary LN-right (∆Rh110 draining) | 4,547 | <1 | <1 | <1 | 7 | <1 |
| Axillary LN-left (WT draining) | 17,687,789 | <1 | 29,484 | <1 | 5,771 | <1 |
| PLN (except draining LN) | 265,298 | 49 | 3 | <1 | 40 | <1 |
| Mesenteric LN | <1 | 1 | 5 | <1 | 5 | <1 |
| GI Tract | 34 | 5 | 8 | <1 | 64 | <1 |
| Liver/Gallbladder | 11 | <1 | <1 | <1 | 41 | <1 |
| Heart/Lung/Kidney/BAL | 66 | 4 | 77 | <1 | <1 | <1 |
| BM/Spleen/Tonsil | 4 | 3 | 9,736 | <1 | <1 | <1 |
| Neuro/Endocrine | 14 | <1 | 8 | <1 | <1 | <1 |
| Genitourinary tract | 125 | 14 | 14,075 | <1 | 17 | <1 |
| Salivary Glands | 13,262 | <1 | 1,242 | <1 | 11 | <1 |
| PBMCs | <1 | 3 | <1 | <1 | <1 | <1 |
| Table 1B | Superinfection: 68–1 vs. ∆Rh110 | |||||
|---|---|---|---|---|---|---|
| RM T1B-1 | RM T1B-2 | RM T1B-3 | ||||
| Tissue type | 14 dpi | 21 dpi | 28 dpi | |||
| 68–1/SIVrtni | ∆Rh110/SIVgag | 68–1/SIVrtni | ∆Rh110/SIVgag | 68–1/SIVrtni | ∆Rh110/SIVgag | |
| Skin Inj site-right (WT) | 47 | 5 | 16,502 | <1 | 376 | <1 |
| Skin Inj site-left (∆Rh110) | 11 | 354 | 7 | <1 | 355 | <1 |
| Axillary LN-right (WT draining) | 117 | <1 | 7 | <1 | 509 | <1 |
| Axillary LN-left (∆Rh110 draining) | <1 | <1 | <1 | <1 | <1 | <1 |
| Peripheral LN | 1,434 | <1 | 41 | <1 | 2 | <1 |
| Mesenteric LN | <1 | 1 | 1 | <1 | <1 | <1 |
| GI tract | 3 | <1 | <1 | <1 | 12 | <1 |
| Liver/Gallbladder | 1 | <1 | 9 | <1 | 9 | <1 |
| Heart/Lung/Kidney/BAL | 5 | <1 | <1 | <1 | 8 | <1 |
| BM/Spleen/Tonsil | 15 | 5 | <1 | <1 | <1 | <1 |
| Neuro/Endocrine | <1 | <1 | 3 | <1 | 3 | <1 |
| Genitourinary tract | 3 | <1 | 4 | <1 | 1,047 | <1 |
| Salivary gland | 23 | 1 | <1 | <1 | <1 | <1 |
| Table 1C | ∆Rh110: Complemented vs. Uncomplemented | ||||
|---|---|---|---|---|---|
| RM T1C-1 | RM T1C-2 | ||||
| Tissue type | Uncomplemented
∆Rh110/SIVrtni |
Complemented
∆Rh110/SIVgag |
Tissue type | Uncomplemented
∆Rh110/SIVgag |
Complemented
∆Rh110/SIVrtni |
| Skin Inj site-right (compl.) | <1 | <1 | Skin Inj site-right (uncompl.) | <1 | <1 |
| Skin Inj site-left (uncompl.) | 13 | <1 | Skin Inj site-left (compl.) | <1 | <1 |
| Axillary LN-right (compl. draining) | <1 | <1 | Axillary LN-right (uncompl. draining) | <1 | <1 |
| Axillary LN-left (uncompl. draining) | 17 | <1 | Axillary LN-left (compl. draining) | <1 | <1 |
| PLN (except draining LN) | <1 | <1 | PLN (except draining LN) | <1 | <1 |
| Mesenteric LN | <1 | <1 | Mesenteric LN | <1 | <1 |
| GI Tract | <1 | <1 | GI Tract | <1 | <1 |
| Liver/Gallbladder | <1 | <1 | Liver/Gallbladder | <1 | <1 |
| Heart/Lung/Kidney/BAL | <1 | <1 | Heart/Lung/Kidney/BAL | <1 | <1 |
| BM/Spleen/Tonsil | <1 | <1 | BM/Spleen/Tonsil | <1 | <1 |
| Neuro/Endocrine | <1 | <1 | Neuro/Endocrine | <1 | <1 |
| Genitourinary tract | 5 | <1 | Genitourinary tract | <1 | 2 |
| Salivary Glands | <1 | <1 | Salivary Glands | <1 | <1 |
| PBMCs | <1 | <1 | PBMCs | <1 | <1 |
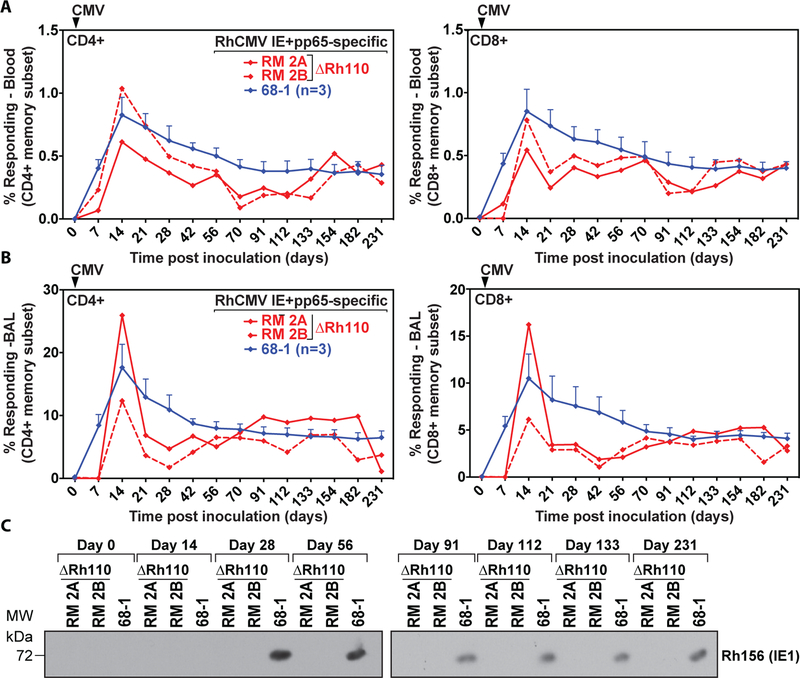
The inability to detect ΔRh110/SIVrtni by ultra-sensitive PCR at four weeks post-primary infection could indicate that RhCMV lacking pp71 was unable to persist, and thus might be unable to maintain immune stimulation. However, in two RhCMV-naïve RMs given 107 PFU of ΔRh110, the magnitude and kinetics of T cell responses to RhCMV IE1 and pp65a in peripheral blood mononuclear cells (PBMCs) and bronchoalveolar lavage (BAL) cell preparations were comparable to that of 4 RhCMV-naïve RMs inoculated with 107 PFU of 68–1 through 231 dpi (Fig. 2A,B). In contrast to 68–1, ΔRh110 was not shed in the urine of these monkeys at any time point (Fig. 2C), confirming the in vivo attenuation of ΔRh110. Thus, despite marked inhibition of viral spread in vivo, lack of pp71 did not affect viral immunogenicity in the first 7 months after primary infection.
Figure 2: Rh110 (pp71)-deleted RhCMV retains T cell immunogenicity but is no longer shed in urine.
(A) Frequencies of RhCMV-specific CD4+ and CD8+ T cell responses in PBMCs were determined at the indicated time points in two RM inoculated with 107 PFU of ΔRh110 and three RMs given the same dose of 68–1 RhCMV. RhCMV IE1- and pp65a-specific T cells were determined by flow cytometric ICS after stimulation with mixes of consecutive, overlapping peptides comprising the RhCMV IE1 and pp65a proteins using intracellular expression of CD69 and either or both of TNF-α and IFN-γ to define Ag-specific T cells. Shown are response frequencies to IE1 and pp65a within the memory subset after background subtraction for each of the two ΔRh110-inoculated RMs (RM 2A and RM 2B) and the mean (+ SEM) of these response frequencies for the three RMs given 68–1 RhCMV. (B) Frequencies of RhCMV-specific T cell responses in bronchoalveolar lavages (BAL) were determined as in panel A in the same animals at the indicated time points. (C) Urine was isolated at the indicated days post-infection from RM 2A and RM 2B or one RM inoculated with 68–1. The presence of virus in co-cultures was determined by immunoblot for RhCMV IE1 (see Materials and Methods).
A key feature of CMV vectors is their ability to overcome pre-existing anti-CMV immunity, permitting the use of CMV vectors regardless of prior CMV infection (7), which must be preserved in any attenuation strategy. To determine whether ΔRh110 retained this capability, we administered RhCMV-seropositive (RhCMV+) RMs with 107 PFU of 68–1/SIVrtni (subcutaneously, right arm) and the same dose of complemented ΔRh110/SIVgag (subcutaneously, left arm), and monitored their dissemination by qPCR in tissues of RMs necropsied at 14, 21 or 28 dpi. As shown in Table 1B, pre-existing immunity substantially reduced genome copy numbers of 68–1/SIVrtni in all tissues when compared to nonimmune RMs (Table 1A), consistent with anti-CMV immunity profoundly limiting replication and dissemination of Rh110-intact 68–1 RhCMV. However, similar to seronegative RMs, ΔRh110 was not detectable or barely detectable in any tissues at 14 dpi, except for the inoculation site, and in the RMs necropsied at 21 and 28 dpi, this vector was below the limit of detection in all tissues. Again, SIVgag- and SIVrtn-specific CD4+ and CD8+ T cell responses were detectable in most tissues of all 3 RMs and were of comparable magnitude (Fig. S4B), indicating that even in the setting of pre-existing anti-CMV immunity, the development of RhCMV vector-elicited, insert-specific T cell responses was not compromised by the Rh110 deletion. We also compared the spread of uncomplemented ΔRh110 with pp71-complementation, which would provide the incoming vector with pp71 in its tegument and increase the efficiency of the first round of vector replication. At 14 dpi, both complemented ΔRh110/SIVgag and uncomplemented ΔRh110/SIVrtni were largely undetectable in tissues in two RhCMV+ RMs (Table 1C), confirming the profound spread deficiency of pp71-deleted vectors.
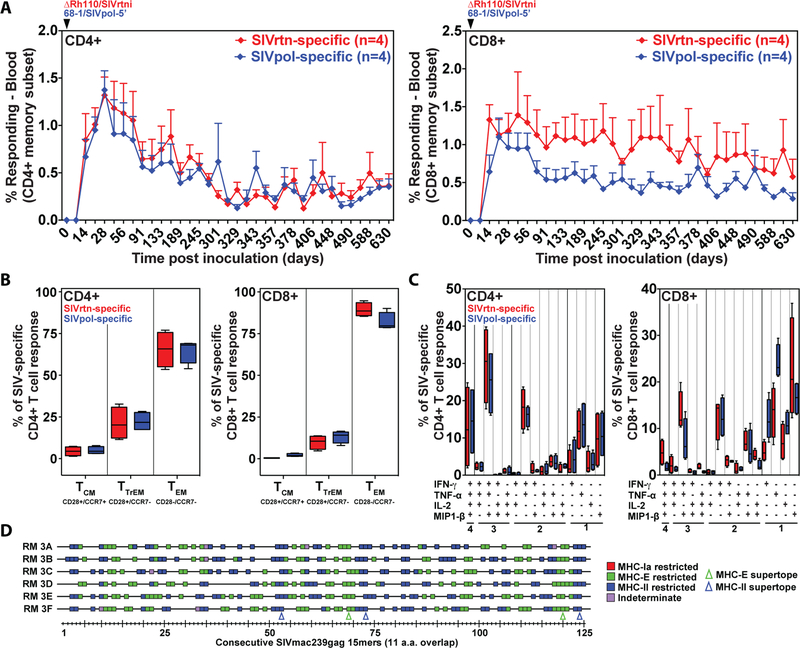
Persistent CMV infections, including infections with 68–1 RhCMV vectors, elicit and indefinitely maintain high frequency TEM, likely reflecting continuous or frequently recurring antigen exposure (13,15,18,48,50,51). To determine whether, despite limited spreading, pp71-deficient RhCMV maintained TEM with the same phenotype and function as 68–1 in CMV-immune RMs, we co-inoculated four RhCMV+ RMs with complemented ΔRh110/SIVrtni or 68–1/SIVpol-5’ so as to directly compare their immunogenicity in the same RM. 68–1/SIVpol-5’ was shed into urine at 28 dpi in all 4 RMs, and found in all urine samples thereafter, as expected (7), whereas ΔRh110/SIVrtni was not detected in urine at any time point through 490 dpi (Fig. S5). Despite this notable difference, the average peak and plateau phase frequencies of SIVrtn-specific CD4+ and CD8+ T cells elicited by ΔRh110/SIVrtni in blood and BAL were similar, if not higher, than the corresponding SIVpol-specific T cell responses elicited by 68–1/SIVpol-5’ (Fig. 3A; Fig. S6). Moreover, the circulating SIVrtn-specific CD4+ and CD8+ T cell responses elicited by ΔRh110 displayed the same highly TEM-biased phenotype (CCR7-, CD28-; Fig. 3B) as 68–1 vector-elicited responses and overlapping patterns of cytokine production that were commensurate with the TEM phenotype (high TNF-α, IFN-β, MIP-1β; low IL-2; Fig. 3C). We also previously reported that 68–1 RhCMV vectors elicit CD8+ T cells that very broadly target highly unconventional epitopes restricted by either MHC-II or MHC-E (52, 53). This broad, unconventional epitope targeting is not affected by pp71 deletion (Fig. 3D). Thus, the magnitude, durability, phenotype, cytokine synthesis function, and CD8+ T cell epitope targeting of ΔRh110-elicited SIV-specific T cell responses were essentially indistinguishable from that elicited by parental 68–1 RhCMV. We therefore conclude that, despite our inability to detect ΔRh110 in tissues 4 weeks following inoculation, pp71-deficient RhCMV persists in tissues at levels that support the maintenance of abundant highly TEM-biased T cells.
Figure 3: Durability, functional phenotype, and epitope targeting of SIV insert-specific T cell responses elicited by Rh110 (pp71)-deleted RhCMV vectors in naturally RhCMV-infected RMs.
(A) Four naturally RhCMV-infected RMs were co-inoculated with 107 PFU of 68–1/SIVpol-5’ and the same dose of ΔRh110/SIVrtni and flow cytometric ICS was used to follow the magnitude of the CD4+ and CD8+ T cell responses in peripheral blood to SIVpol and SIVrtn peptide mixes as described in Fig. 2. The mean + SEM of SIVpol- and SIVrtn-specific response frequencies within the memory CD4+ (left panel) and CD8+ (right panel) T cell populations are shown. (B) Boxplots compare the memory differentiation of the RhCMV-elicited CD4+ and CD8+ memory T cells in PBMCs (of the same RM shown in panel A) responding to SIVpol or SIVrtn with TNF-α and/or IFN-γ production at 630 dpi. Memory differentiation state was based on CD28 and CCR7 expression, delineating central memory (TCM), transitional effector memory (TTrEM), and effector memory (TEM), as designated. The Wilcoxon rank sum test was used to pairwise compare differences between the fraction of SIVpol- and SIVrtn-specific CD4+ and CD8+ T cells within each memory subset, with P = NS for all comparisons. (C) Boxplots compare the frequency of RhCMV-elicited CD4+ and CD8+ memory T cells in PBMCs of the same RM shown in panel A responding to SIVpol or SIVrtn peptides with TNF-α, IFN-γ, IL-2, and MIP1-β production, alone and in all combinations at 630 dpi. The Wilcoxon rank sum test was used to pairwise compare differences between the fraction of SIVpol- and SIVrtn-specific CD4+ and CD8+ T cells expressing 1, 2, 3 or 4 cytokines, with P = NS for all comparisons. (D) SIVgag-specific CD8+ T cells in the peripheral blood of six ΔRh110/SIVgag-inoculated RMs were epitope-mapped using a flow cytometric ICS assay (CD69, TNF-α, IFN-γ readout, as described above) to detect recognition of each consecutive, overlapping 15-mer peptide comprising the SIVgag protein. Peptides resulting in specific CD8+ T cell responses are indicated by a box, with the color of the box designating MHC restriction as determined by blocking with the anti-pan-MHC-I mAb W6/32, the MHC-E blocking peptide VL9 and the MHC-II blocking peptide CLIP, as previously described (52,53). The blue and green arrowheads indicate the positions of previously identified MHC-II- and MHC-E-restricted SIVgag supertopes, respectively.
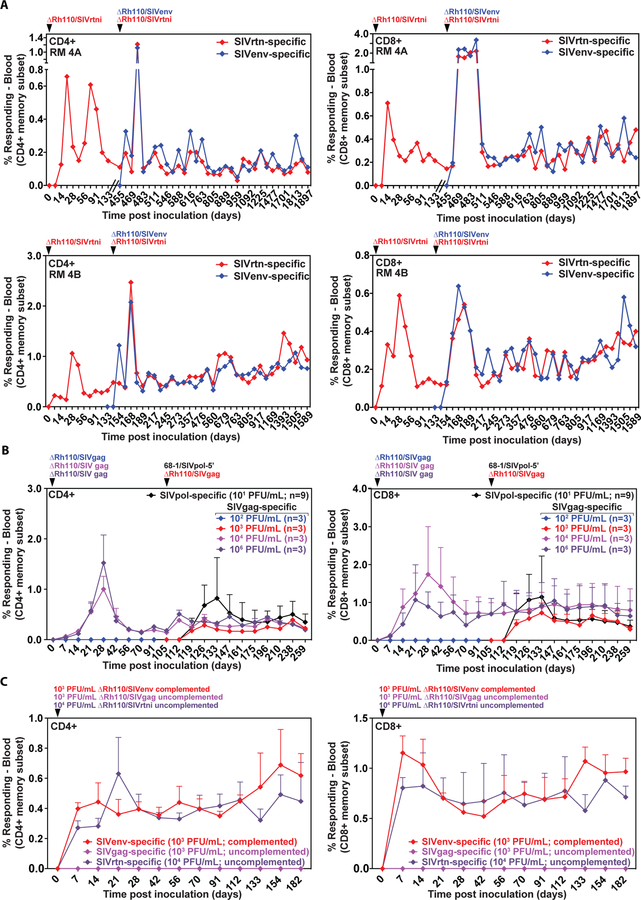
To confirm that ΔRh110 RhCMV vectors could be used repeatedly in the same RM, either to boost pre-existing T cell responses or elicit new T cell responses to different inserts, we inoculated 2 RMs that had previously been vaccinated with ΔRh110/SIVrtni with both a second dose of ΔRh110/SIVrtni and with a different pp71-deleted vector expressing an SIVenv insert. As shown in Fig. 4A, this repeat, dual vaccination resulted in both a transient boosting of the SIVrtn-specific CD4+ and CD8+ T cell responses, and the de novo induction of an SIVenv-specific CD4+ and CD8+ T cell response of similar magnitude. This outcome is essentially identical to the previously reported boosting and repeated de novo infection behavior of pp71-intact 68–1 RhCMV (15). Moreover, the magnitude of both the SIVrtn- and SIVenv-specific CD4+ and CD8+ T cell responses remained stable for at least 4 years (Fig. 4A), at which point immune monitoring was discontinued. Thus, pp71-deleted RhCMV vectors retain the unique ability of this vaccine platform to sequentially induce and maintain T cell responses to different antigens in the same host.
Figure 4: Potency of Rh110 (pp71)-deleted RhCMV vectors in superinfection.
(A) Two naturally RhCMV-infected RMs were inoculated with 107 PFU of ΔRh110/SIVrtni on day 0 and again on day 455 (for RM 4A) or day 133 (for RM 4B), the latter inoculation in combination with 107 PFU of ΔRh110/SIVenv. The panels show longitudinal analysis of the SIVrtn- and SIVenv-specific CD4+ and CD8+ T cell response frequencies among PBMCs determined by flow cytometric ICS (CD69, TNF-α, IFN-γ readout) for each animal. (B) At time point 0, three groups (n=3 per group) of naturally RhCMV-infected RMs were subcutaneously inoculated with the indicated dose (102, 104, and 106 PFU) of ΔRh110/SIVgag, complemented for pp71 by growing in TRFs-pp71. The three RMs given 102 PFU did not manifest a detectable SIVgag-specific T cell response through 112 days of observation and were re-inoculated with 103 PFU dose of the same vector. At the same time, all 9 RMs were inoculated with 101 PFU of 68–1/SIVpol-5’. The figures show longitudinal analysis of the mean + SEM of SIVgag- and SIVpol-specific CD4+ and CD8+ T cell response frequencies among PBMCs, determined as described in Panel A. (C) Three naturally RhCMV-infected RMs were subcutaneously inoculated with 103 PFU of pp71-complemented ΔRh110/SIVenv, 103 PFU of ΔRh110/SIVgag, and 104 PFU ΔRh110/SIVrtni, with the latter two vectors grown in TRFs and thus not complemented for pp71. SIVenv-, SIVgag-, and SIVrtn-specific CD4+ and CD8+ T cell responses within the peripheral blood memory compartment were followed by flow cytometric ICS as described above (mean + SEM shown at each time point).
CMV vectors are T cell-targeted vaccines, and RhCMV vectors elicit few, if any, insert-specific antibodies (Abs) in RMs (15–18). However, these vectors have the potential to elicit/boost Ab responses to RhCMV itself, raising the question of whether ΔRh110 vectors differ from parental 68–1 vectors in this activity. To address this, we compared RhCMV-specific Ab responses in cohorts of RhCMV+ RMs that were vaccinated with either a 68–1 (n=16) or ΔRh110 (n=14) RhCMV/SIV vector set composed of 5 vectors, each expressing one SIVmac239 insert (gag, env, rtn, pol-5’ and pol-3’) and subcutaneously administered at a dose 5×106 PFU per vector. Abs to whole RhCMV viral lysates were measured by ELISA prior to, and 4, 6, and 12 weeks post-vaccination. As shown in Fig. S7, both cohorts showed a significant (p<0.001), but transient, boost in antibody titers, the magnitude of which was not different between the monkeys vaccinated with ΔRh110 vs. 68–1 vectors. Thus, when administered in a high-dose vaccination regimen, pp71-deleted RhCMV vectors were able to boost antibody responses to a similar degree as the parental 68–1 vectors.
To determine the extent to which pp71 deletion affects the minimal dose required for T cell immunogenicity, we first inoculated 3 cohorts of seropositive RMs (n=3 each) with 102, 104, or 106 PFU of pp71-complemented ΔRh110/SIVgag and monitored the development of CD4+ and CD8+ T cell responses to SIVgag. As shown in Fig. 4B, ΔRh110/SIVgag induced de novo SIVgag-specific T cell responses at 104 and 106 PFU, but not at 102 PFU. To further refine the dose required for immunogenicity, we re-inoculated the 3 RMs that had failed to respond to 102 PFU with 103 PFU ΔRh110/SIVgag, and for comparison, we inoculated all 9 RMs with 101 PFU of 68–1/SIVpol-5’ (Fig. 4B). Remarkably, despite the extremely low dose of 68–1 administered, all 9 RMs manifested SIVpol-specific CD4+ and CD8+ T cell responses, and at 103 PFU, ΔRh110/SIVgag also induced both CD4+ and CD8+ SIVgag-specific T cells. Since this experiment was conducted with ΔRh110 vectors grown on complementing cells, we also determined the minimal dose required for uncomplemented ΔRh110 while, at the same time, confirming the minimal effective dose for complemented ΔRh110. We directly compared the immunogenicity of three different ΔRh110 constructs in three RhCMV+ RMs: 1) pp71-complemented ΔRh110/SIVenv at 103 PFU, 2) uncomplemented ΔRh110/SIVgag at 103 PFU, and 3) uncomplemented ΔRh110/SIVrtni at 104 PFU. As shown in Fig. 4C, the 103 PFU dose was immunogenic (elicited SIVenv-specific T cells) for the pp71-complemented ΔRh110/SIVenv vector, but not for the uncomplemented ΔRh110/SIVgag vector. However, at the 104 PFU dose, the uncomplemented ΔRh110/SIVrtni vector was immunogenic, indicating that pp71 complementation provides a 1-log dose sparing.
No evidence for recombination of ΔRh110 RhCMV/SIV vectors with endogenous RhCMV.
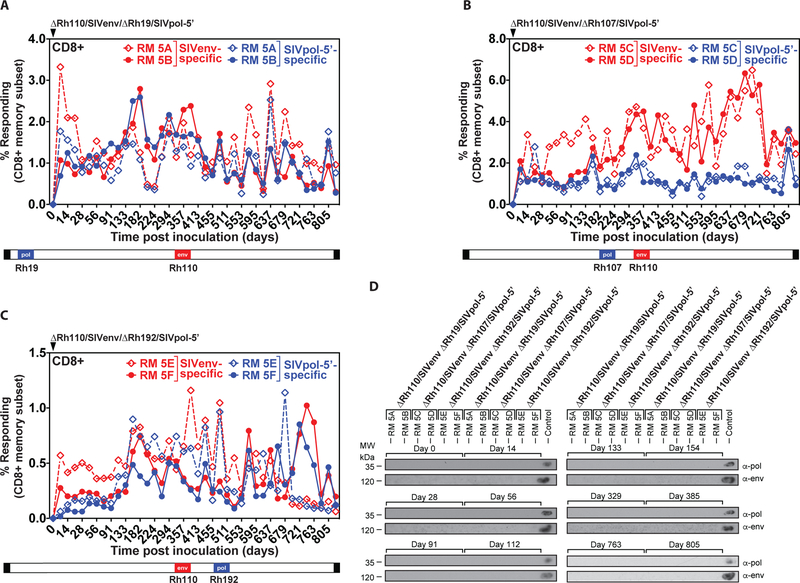
One concern with using CMV vectors in CMV+ hosts is the possibility of recombination between the vector and the endogenous virus that would revert the attenuation. Such in vivo recombination has been reported for alpha-herpesvirus vaccines and naturally circulating varicella zoster strains (54,55), and deep sequence analysis of primary HCMV isolates shows signs of past recombination events (56). To investigate propensity for recombination with endogenous virus, we designed three vectors each carrying two SIV antigens as immunogenic markers in different parts of the RhCMV genome. In all three vectors, SIVenv replaced Rh110, whereas SIVpol-5’ replaced the coding sequence of three different RhCMV genes – Rh19, Rh107, and Rh192 (corresponding, respectively, to the HCMV RL11 gene family, UL78 and US12) – in different parts of the genome. Each gene locus was shown to support insert expression and to be dispensable for vector replication in vitro (Fig. S8). Any repair of ΔRh110 by homologous recombination would result in a virus that lacks SIVenv, but retains SIVpol, and repair of Rh110 by other types of recombination would yield a pp71-intact vector with one or both of the SIV inserts included, any of which would likely be dissemination-competent and therefore expected to appear in the urine over time. Upon inoculation of two RhCMV+ RMs with each of the three recombinants, we observed de novo induction and long-term maintenance of CD8+ T cell (Fig. 5A–C) and CD4+ T cell (Fig. S9A–C) responses to both SIVenv and SIVpol indicating “take” and persistence of all three vectors, and functional expression of both inserts in each vector. In general, the magnitude of the SIVpol- and SIVenv-specific CD4+ and CD8+ T cell responses over >800 days of follow-up was similar in each RMs (with the possible exception of modest reduction in the CD8+ T cell response to Rh107-regulated pol), suggesting that T cell immunogenicity is not highly dependent on promoter type or location in the RhCMV genome. Most importantly, immunoblots of co-cultures of viruses isolated from urine of all 6 RMs were consistently negative for SIV antigens over the >800 days of observation (Fig. 5D), suggesting that the Rh110 deletion was not repaired by recombination with endogenous virus in any of these RMs over a >2-year period.
Figure 5: Genetic stability of Rh110 (pp71)-deleted RhCMV vectors in the setting of superinfection.
Two naturally RhCMV-infected RMs were each inoculated with 107 PFU of each dual insert-expressing ΔRh110/SIVenv/ΔRh19/SIVpol-5’ (A), ΔRh110/SIVenv/ΔRh107/SIVpol-5’ (B), or ΔRh110/SIVenv ΔRh192/SIVpol-5’ (C) and flow cytometric ICS was used to follow the SIVenv- and SIVpol-specific CD8+ T cell responses in peripheral blood, as described in Fig. 2. Dashed and solid lines each delineate the individual RM among the RM pairs given each vector. The relative position of the SIV antigens replacing endogenous genes in the viral genome is shown schematically below the graphs. CD4+ T cell responses from the same RM are shown in fig. S9. (D) Urine samples from the indicated time points post-vector inoculation were analyzed for vector shedding by viral co-culture followed by western blot analysis of SIVpol-5’ (top panels) or SIVenv (bottom panels) expression. Urine from RMs that previously received 68–1/SIVpol-5’ and 68–1/SIVenv vectors was included as a positive control.
Lack of ΔRh110 RhCMV/SIV vector transmission with close contact.
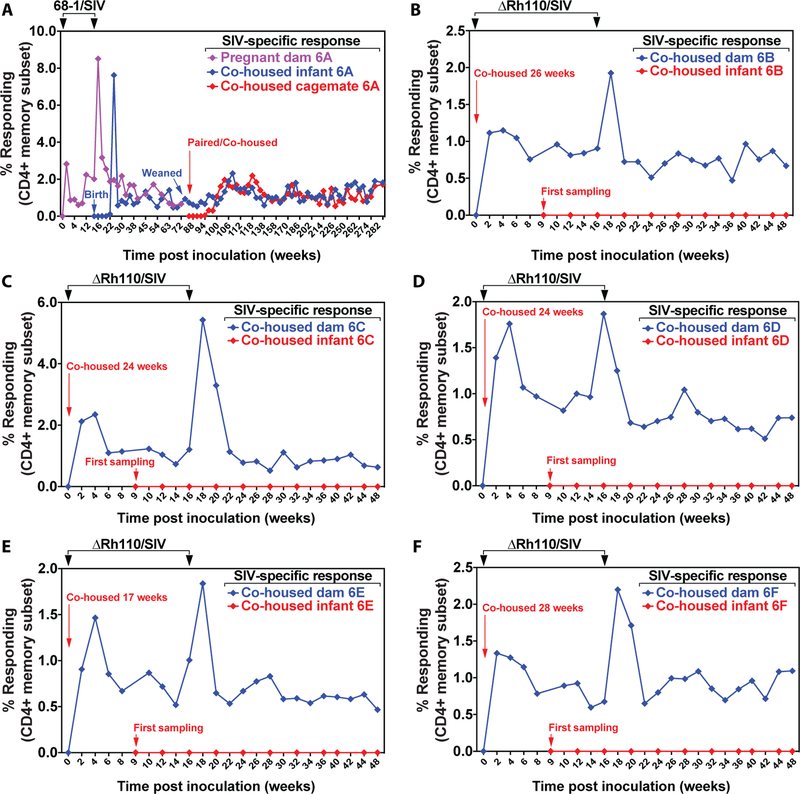
Transmission of RhCMV is highly efficient with essentially 100% of conventionally raised RMs becoming RhCMV+ in the first year of life (2). In keeping with this, we observed serial transmission of SIV-specific cellular immunity (using de novo SIV-specific T cell responses as a surrogate for vector transmission) from a 68–1/SIV vector-vaccinated dam to her newborn infant, and then, after weaning, from the infant to a cohoused cage mate (Fig. 6A, Fig. S10A). In contrast, the nursing infants of 5 naturally RhCMV-infected dams inoculated with ΔRh110 vectors (106 PFU each of 5 different vectors expressing SIVenv, pol-5’, pol-3’, gag and rtn), did not acquire SIV-specific CD4+ or CD8+ T cell responses to any insert, although in all 5 instances, de novo responses to RhCMV were observed indicating that endogenous RhCMV was transmitted from mother to infant (Fig. 6B–F, Fig. S10B–F, G). Thus, pp71-deleted RhCMV vectors are either not shed at all in urine, saliva, or breast milk or, if shedding occurs, the amount is insufficient to mediate transmission to even a highly susceptible naïve host over prolonged periods of close contact.
Figure 6: Lack of maternal-infant transmission of Rh110 (pp71)-deleted RhCMV vectors.
(A) A naturally RhCMV+ pregnant dam was inoculated twice, as shown, with a panel of five 68–1 vectors (5×106 PFU each) expressing SIVenv, SIVgag, SIVrtni, SIVpol-3’, or SIVpol-5′. The dam gave birth to a healthy infant at 16 weeks post first inoculation. The infant was co-housed with, and nursed from, the inoculated dam for 88 weeks, at which time the infant was weaned and co-housed with another, already naturally RhCMV+ juvenile RM. The mother, infant, and co-housed cagemate were followed for total SIV-specific CD4+ T cell responses (SIVgag + pol + rtn + env) in peripheral blood by flow cytometric ICS with the response frequencies in the memory compartment shown. (B to F) Five naturally RhCMV-infected female RMs were inoculated with a panel of five ΔRh110 vectors (5×106 PFU each) expressing SIVenv, SIVgag, SIVrtni, SIVpol-3’, or SIVpol-5’ while nursing 17–28-week-old infants and were re-inoculated 16 weeks with later with the same vectors at the same dose. Vaccinated mothers and nursing infants were co-housed for a total of 48 weeks. Both mothers and infants were longitudinally followed for total SIV-specific CD4+ T cell responses by flow cytometric ICS. For A-F, CD8+ T cell responses are shown in fig. S10.
Attenuation analysis of Rh59 (UL35)-deleted RhCMV/SIV vectors.
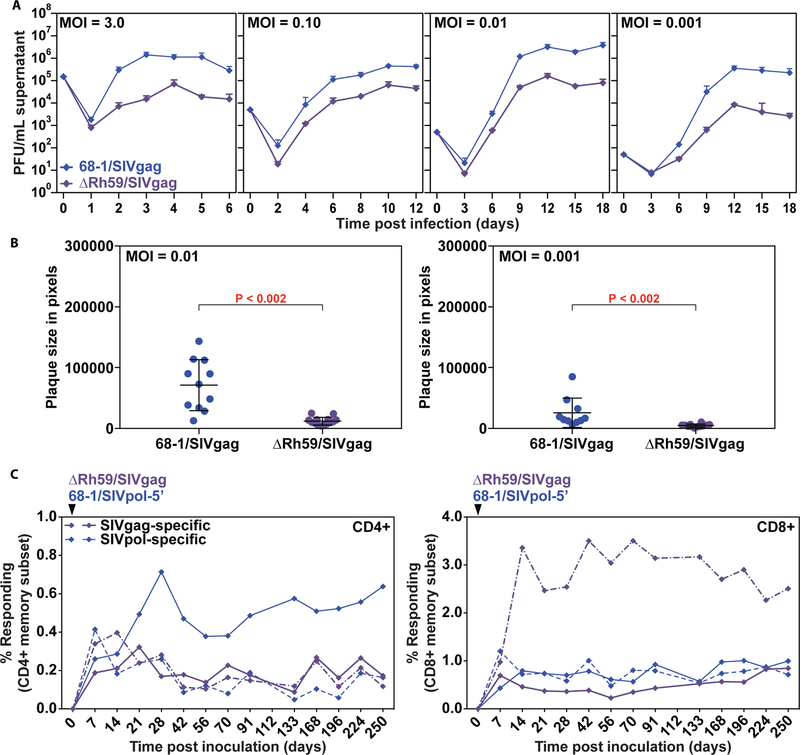
To determine whether the lack of viral dissemination, viral shedding and host-to-host transmission of pp71-deleted vectors was unique to this deletion or extended to other viral proteins that counteract intrinsic immunity, we evaluated the attenuation resulting from deletion of Rh59, the homolog of HCMV UL35, which cooperates with pp71 to counteract the cellular restriction factor BclAF1 (37). Similar to deletion of UL82, deletion of UL35 results in a modest growth defect at low MOI of HCMV (57). Rh59 was either deleted from 68–1/SIVrtni or the Rh59 open reading fame (ORF) was replaced with SIVgag (Fig. S11). Also similar to ΔRh110, we were able to recover both ΔRh59/SIVrtni and ΔRh59/SIVgag without the need for complementation. Interestingly, however, RhCMV lacking Rh59 showed a 10- to 100-fold reduction of viral yield at all MOIs tested (Fig. 7A) suggesting that deletion of Rh59 affects viral replication even at high MOI. This growth defect was also reflected by reduced plaque size measured at 7 dpi (Fig. 7B).
Figure 7: Deletion of the UL35 ortholog Rh59 from RhCMV results in growth deficiency in vitro while maintaining immunogenicity in vivo.
(A) TRFs were infected with 68–1/SIVgag or ΔRh59/SIVgag at the indicated MOI. The supernatant was harvested at the indicated dpi and titered by TCID50 on TRFs. Average titers from two experimental and two technical replicates (+ SD) are shown. (B) TRFs were infected with 68–1/SIVgag or ΔRh59/SIVgag at MOI = 0.01 or 0.001. Plaques were analyzed at 7 dpi using phase microscopy and plaque size was measured using Adobe Photoshop. Individual plaque sizes, as well as average (+/− SD) from one of three experiments are shown. Statistical significance was determined by the Wilcoxon rank-sum test with P ≤ 0.05 considered significant. (C) Two RMs were co-inoculated with 107 PFU of 68–1/SIVpol-5’ and ΔRh59/SIVgag, and the T cell responses to peptide mixes comprising SIVpol and SIVgag were longitudinally monitored in peripheral blood by flow cytometric ICS (CD69, TNF-α, IFN-γ readout) with the response frequencies in the memory compartment shown for each RM (one designated by solid lines, the other by dashed lines).
To evaluate ΔRh59 vector spread in vivo, we performed necropsies on 3 RhCMV+ RMs at 14, 21, and 28 days after subcutaneous inoculation with 107 PFU of ΔRh59/SIVgag (right arm) and the same amount of 68–1/SIVrtni in the left arm. As shown in Table 2, the genome copy numbers of ΔRh59/SIVgag were at or below detection limits in all tissues tested at all 3 time points, whereas 68–1/SIVrtni was detected at all time points in multiple tissues. As demonstrated for ΔRh110, SIVgag-specific CD4+ and CD8+ T cell responses in tissues at necropsy elicited by ΔRh59 were similar to the SIVrtn-specific responses elicited by 68–1 in the same RM (Fig. S12A). Longitudinal analysis of ΔRh59 vs. 68–1-elicited SIV-specific CD4+ and CD8+ T cells in blood (Fig. 7C), and BAL (Fig. S12B) further showed similar response magnitude and durability in two RMs over 250 days, suggesting that deletion of Rh59 did not affect vector immunogenicity despite substantially decreased dissemination in vivo.
Table 2: Rh59 (UL35) deletion reduces dissemination of 68–1 RhCMV vectors.
Three naturally RhCMV-infected RMs (T2–1, T2–2, T2–3) were co-inoculated with 107 PFU each of 68–1/SIVrtni (left arm) and ΔRh59/SIVgag (right arm). One RM was necropsied at 14, 21, or 28 dpi and viral genome copy numbers per 107 cell equivalents in the indicated tissues were determined using ultra-sensitive nested qPCR specific for SIVrtni (68–1) or SIVgag (ΔRh59).
| Table 2 | Super infection: 68–1 vs. ∆Rh59 | |||||
|---|---|---|---|---|---|---|
| RM T2–1 | RM T2–2 | RM T2–3 | ||||
| Tissue type | 14 dpi | 21 dpi | 28 dpi | |||
| 68–1/SIVrtni | ∆Rh59/SIVgag | 68–1/SIVrtni | ∆Rh59/SIVgag | 68–1/SIVrtni | ∆Rh59/SIVgag | |
| Skin Inj site-right (∆Rh59) | 12 | <1 | 193 | <1 | 36,110 | 7 |
| Skin Inj site-left (68–1) | 541 | <1 | 23,793 | <1 | 4,480,601 | <1 |
| Axillary LN | 3 | <1 | 13 | <1 | 2,515 | 7 |
| Peripheral LN | <1 | <1 | <1 | <1 | 18 | <1 |
| Liver/Gallbladder | <1 | <1 | <1 | <1 | 2 | <1 |
| Heart/Lung/Kidney/BAL | 2,933 | <1 | 3 | <1 | 6 | <1 |
| Bone marrow/Spleen/Tonsil | 6 | <1 | <1 | <1 | 8 | <1 |
| Neuro/Endocrine | 3 | <1 | <1 | <1 | 4 | 1 |
| Genitourinary tract | <1 | <1 | 3 | <1 | 9 | <1 |
| Salivary gland | 2 | <1 | 4 | <1 | 3 | <1 |
Normalized to 1×107 cell equivalents
ΔRh59, but not ΔRh110, RhCMV/SIV vectors are shed and transmitted upon leukocyte transfer.
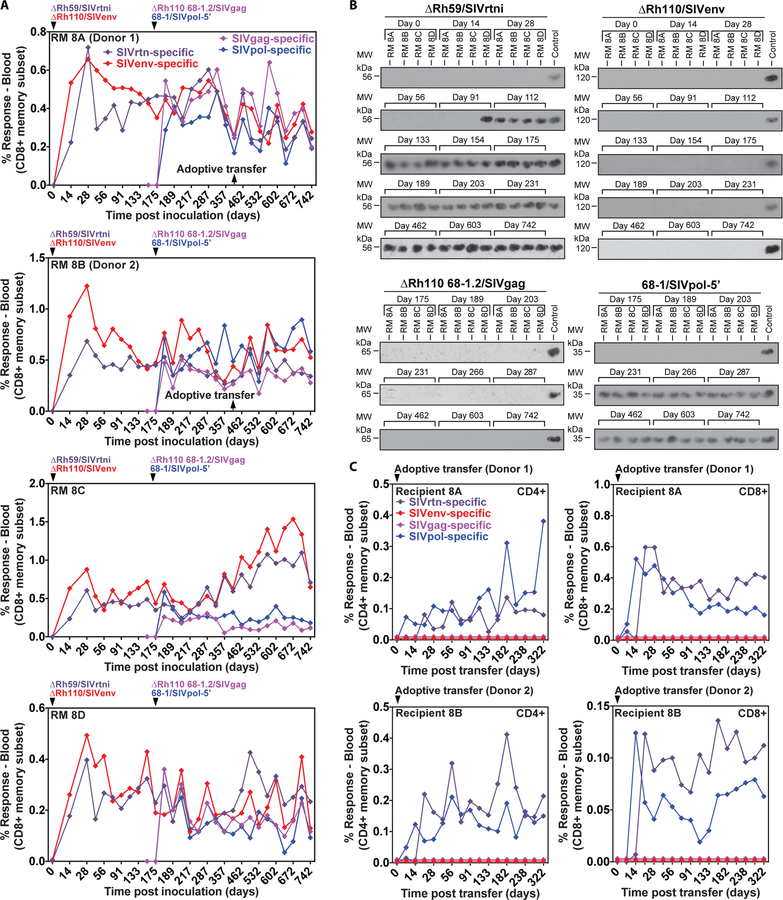
To further compare the relative attenuation of ΔRh110 and ΔRh59 in vivo, we first inoculated RhCMV+ RMs with 5×106 PFU of ΔRh59/SIVrtni and ΔRh110/SIVenv, and then 175 days later, inoculated these RMs with 68–1/SIVpol-5’ and ΔRh110 68–1.2/SIVgag. Compared to 68–1 RhCMV, 68–1.2 RhCMV displays broader cell tropism due to repair of Rh157.5 and Rh157.4 (homologous to the pentameric receptor complex subunits UL128 and UL130 in HCMV), as well as the anti-apoptotic gene Rh61/60 (UL36 in HCMV) (58). ΔRh110 68–1.2/SIVgag was thus generated to determine whether increased anti-apoptosis activity and the ability to more efficiently infect non-fibroblast cells due to an intact pentamer would counter the attenuation resulting from pp71 deletion. All 4 vectors induced de novo, persistent CD8+ (Fig. 8A) and CD4+ (Fig. S13A) T cell responses to their respective SIV inserts in all 4 RMs that were similar in magnitude in all tissues (Fig. S13B). In keeping with previous results, 68–1/SIVpol-5’ appeared in urine by 56 dpi (day 231 of the experiment) and at every subsequent time point, whereas ΔRh110/SIVenv was not detected in the urine at any time point through 742 dpi (Fig. 8B). Similarly, ΔRh110 68–1.2 /SIVgag was not found in urine through 567 dpi (day 742 of the experiment), indicating that repair of the pentameric complex and anti-apoptotic genes did not reverse the attenuation of pp71 deletion. However, in sharp contrast, ΔRh59/SIVrtni appeared in urine in one RM at 91 dpi, and in all RMs by 112 dpi, and was found in all subsequent urine samples (Fig. 8B). Thus, despite the in vitro growth deficiency and strongly reduced in vivo dissemination, Rh59-deficient RhCMV was shed from infected RMs, albeit with a modest delay compared to 68–1 RhCMV.
Figure 8: Comparison of the shedding and transmission upon leukocyte transfer of Rh110 (pp71)-deleted, Rh59 (UL35)-deleted and 68–1 RhCMV vectors.
(A) Four RMs were co-inoculated with 106 to 107 PFU of ΔRh59/SIVrtni, pentameric complex-repaired ΔRh110 68–1.2/SIVgag, ΔRh110/SIVenv, and 68–1/SIVpol-5’ at the designated time points and the CD8+ T cell responses to peptide mixes comprising each of the SIV antigens were longitudinally monitored in peripheral blood by flow cytometric ICS (CD69, TNFα, IFN-γ readout), with the response frequencies in the memory compartment shown (see also fig. S13B). (B) Immunoblots of viral co-cultures from urine samples obtained at the indicated days. Each of the SIV inserts carries a different epitope tag, allowing specific identification of each vector using tag-specific mAbs (see Materials and Methods). (C) Bone marrow and blood leukocytes from two of the RMs shown in A (1.9 × 107 bone marrow and 3.0 × 107 blood cells from RM 8A (donor 1); 0.8 × 107 bone marrow and 3.0 × 107 blood cells from RM 8B (donor 2); obtained at the indicated time point) were transferred to two naturally RhCMV+ (but vector-naïve) RMs to test the ability of leukocyte transfer to transmit each vector to a new host. Vector infection of the new host was determined by longitudinal assessment of CD4+ and CD8+ T cell responses to each of the four different SIV inserts, as described in (A).
CMV can readily spread from infected individuals to naïve recipients by blood transfusion or bone marrow transplantation, likely from latent viral reactivation in myeloid lineage cells, particularly monocytes (59–61). To more stringently compare the in vivo spread deficiency of ΔRh59 and ΔRh110, we adoptively transferred by intravenous infusion 8 or 19 × 106 bone marrow cells plus 30 × 106 peripheral blood leukocytes from 2 RMs inoculated with ΔRh59/SIVrtni, ΔRh110/SIVenv, 68–1/SIVpol-5’, and ΔRh110 68–1.2/SIVgag to two naturally RhCMV-infected, but vector-naïve, RMs, and then monitored these recipient RMs for induction of SIV-specific T cell responses indicating vector transmission (Fig. 8C). Strikingly, SIVpol- and SIVrtn-specific T cell responses rapidly appeared in both RMs. The SIVpol-specific responses arising from transmission of the 68–1 vector peaked a week earlier than the SIVrtn-specific responses arising from transmission of the ΔRh59 vector, whereas SIVenv and SIVgag responses were never detected, consistent with a lack of transmission of both 68–1- and 68–1.2-derived ΔRh110 vectors. These data indicate that ΔRh110 vectors were either not present in the transferred cells or that these attenuated viruses were unable to reactivate from latency and disseminate despite being transferred in myeloid cells. These data indicate that the long-term in vivo attenuation afforded by deletion of Rh110 (pp71) is greater than that afforded by deletion of Rh59 (UL35).
DISCUSSION
The goal of this study was to identify a strategy that would enhance the safety of CMV-based vectors while maintaining the unique immunologic features that characterize the CMV vector platform, in particular the establishment and maintenance of high frequency TEM in both CMV-naïve and naturally CMV-infected individuals. Since it is very likely that the indefinite maintenance of TEM requires long-term Ag exposure provided by persistent viral infection (62), attenuation strategies that eliminate viral persistence would not be suitable for this purpose. In this regard, the replication-deficient HCMV with in vivo-abrogated expression of IE1/2 and UL51 currently being developed as an HCMV vaccine (63) would not be expected to maintain high frequency TEM with appropriate long-term functionality. Moreover, since it is very likely that the unique phenotype and function of CMV vector-elicited responses reflect the composite interaction of the virus’ many immunomodulatory programs with the host immune system (64,65), attenuation strategies that interfere with these programs also run the risk of adversely affecting the quality of the CMV vector-elicited immune responses. For instance, we previously demonstrated that deletion of the gene region encoding RhCMV homologs of the MHC-I evasins US2–11 resulted in vectors that lost the ability to super-infect (7), whereas retention of natural killer cell-activating ligands by RhCMV ORF Rh159 (UL148) was required for primary infection (66). Therefore, in planning our CMV vector attenuation strategy, we sought an attenuation approach that would down-modulate, but not eliminate, CMV vector infectivity by markedly reducing its capacity to spread from cell-to-cell upon lytic replication or reactivation, but that would otherwise preserve its immunomodulatory capabilities and its ability to persist. Our goal was a stably attenuated vector that would be able to exceed the threshold required for persistent immunogenicity, but would be sufficiently spread restricted such that the infection would be kept well below the level required to cause disease, and even more stringently, below the level necessary to transmit vector from one individual to another. We also sought to accomplish this goal with discrete, difficult-to-reverse genetic modifications based on well-understood mechanisms of action at the molecular and cellular levels so as to avoid, as far as possible, unexpected off-target impacts on vector biology and function.
In view of these immunobiological and virological considerations, genetic deletion of viral tegument proteins such as pp71 that counter host intrinsic immunity was an attractive candidate attenuation strategy. First, attenuation based on deletion is much more difficult to reverse in vivo than attenuation based on point mutations or on adding attenuating genetic elements, especially with our strategy of replacing the pp71 coding sequence with the heterologous Ag. This strategy not only results in a more naturally regulated expression of the transgene than use of heterologous promoters, but also eliminates the possibility of reversing vector attenuation by homologous recombination with endogenous virus.
Second, the host intrinsic immune mechanisms under consideration are highly conserved in mammals and the relevant components, such as DAXX, are essential genes that are expressed in every cell type (29,67). These features increase the likelihood of successful translation from RMs to humans, and reduce concerns that genetic heterogeneity in the human population would reduce the effectiveness of this strategy in subpopulations, potentially placing subgroups of individuals at higher risk for adverse events after vaccination. Indeed, the universality of these mechanisms had led to them being proposed as a general approach to attenuate herpes viral vaccines and vectors (68).
Third, the pattern of in vitro attenuation reported for Δpp71 HCMV – reduction in viral growth at low MOI, but not high MOI (46) – also appeared to be advantageous for a TEM-targeted vaccine, as it suggested a relatively specific and early defect in viral infectivity that greatly reduces, but does not eliminate, the capacity for lytic, productive infection. Pp71 deletion does not appear to irrevocably inactivate CMV or change its fundamental immunobiology, but rather, appears to place a barrier to the triggering of its lytic genetic program that limits viral production and subsequent cell-to-cell spread during initial infection or upon subsequent reactivation from latency. Some Δpp71 CMV-infected cells (perhaps those exposed to high MOI) might bypass this restriction and progress to lytic infection, but pp71-deleted progeny of those few cells that produce virus would be severely handicapped in their ability to productively infect new cells, potentially resulting in a smoldering, low-level infection that persists, but cannot efficiently expand or disseminate. Moreover, Δpp71 CMV may go directly into latency (38,39), potentially maintaining the capacity for subsequent insert expression either during latency or in response to reactivation stimuli, and therefore providing for persistent or periodic Ag production and presentation from all infected cells, whether or not they are able to engage in productive infection (64).
These promising in vitro characteristics do not, however, indicate whether the degree to which a Δpp71 CMV vector would manifest the necessary balance between the infectivity and persistence (needed for immunogenicity) and the spread-restriction (needed for safety) would be appropriate for optimal vector function. The level of spread of Δpp71 in vivo might be too high or too low, resulting in insufficient attenuation or insufficient immunogenicity, respectively. In this regard, the relatively modest in vitro attenuation of ΔRh110 vectors relative to their HCMV counterparts (46), even at very low MOI, raised concern that the Δpp71 RhCMV vector design might be insufficiently attenuated in the RM model. However, this turned out not to be the case, as pp71-deleted 68–1 vectors showed a remarkable degree of in vivo attenuation relative to pp71-intact RhCMV based on strain 68–1. Whereas 68–1 RhCMV showed high level replication at the site of inoculation (>109 genome copies/107 cell equivalents at day 14) and robust dissemination in primary infection, co-injected ΔRh110 was barely detectable with a high-sensitivity assay at 14 dpi, and was essentially undetectable in tissues taken later. Indeed, the numbers of ΔRh110 genome copies were so low in both primary and super-infection (with no difference between primary and super-infection, in contrast to 68–1), that it remains unclear whether there was any spread of ΔRh110 in vivo; the genome copies detected at day 14 might simply reflect the ΔRh110 genomes from the injected (high dose) inoculum and/or the virus produced within the cells infected by this inoculum. The robust attenuation of ΔRh110 was also supported by our findings that whereas 68–1 is readily shed in urine and efficiently transferred from RM to RM with close (mother-to-infant) contact or blood cell transfusion, ΔRh110 was never found in urine and was not transferred from RM to RM with either close contact or cell transfer. These data may indicate that pp71 is essential for in vivo RhCMV infectivity, such that ΔRh110 would be too attenuated for use as an effective vector. However, this concern is allayed by comparable 68–1 and ΔRh110 CD4+ and CD8+ T cell immunogenicity, even in the setting of super-infection. Moreover, as shown in the companion article, we demonstrate that ΔRh110 vectors show efficacy against viral challenge that is as good or better than 68–1 vectors (69).
The only clear immunologic difference between ΔRh110 and parental RhCMV vectors was the dose required to establish immunogenicity. Whereas 68–1 can establish fully immunogenic super-infection with 10 injected PFU, the (uncomplemented) ΔRh110 vector required 104 PFU to establish immunogenicity in RhCMV+ RMs. This observation is consistent with the hypothesis that the primary defect of ΔRh110 is a reduction in its ability to spread from cell-to-cell. The lack of any shedding or transmission of ΔRh110 suggests that this vector never effectively seeds shedding sites such as the salivary glands and kidneys, or long-term latency sites such as hematopoietic stem cells and their myeloid lineage progeny in the bone marrow and in the circulation (60, 61). This, in turn, implies that robust CMV vector immunogenicity can be achieved and maintained with a local infection (sites of inoculation and draining lymph nodes) initiated by 104 PFU, and therefore possibly involving only thousands of infected cells or fewer.
The magnitude of the SIV-specific T cell responses elicited by the different effective doses of ΔRh110, with or without complementation, and by the pp71-intact 68–1 RhCMV were similar, suggesting that a threshold level of vector infection and associated insert expression is required to trigger immunogenicity, but once this threshold is achieved, the magnitude of resultant immune response is dose independent. Since complemented ΔRh110 has pp71 in its tegument to facilitate the first round of replication, the comparison between 68–1 and uncomplemented ΔRh110 better reflects the effect of pp71 deletion on in vivo spread, indicating that pp71 deletion compromises vector spread by at least 1000-fold in vivo (10,000 PFU vs. 10 PFU), a 10- to 100-fold higher deficit than the reduction of viral growth observed with low MOI in vitro.
It is also noteworthy that the Rh59 (UL35)-deleted RhCMV vector, which showed an MOI-independent 1–2-log growth defect in vitro and a similar spread deficiency after in vivo inoculation as ΔRh110, did not show the same overall attenuation as ΔRh110, as ΔRh59 was both shed in urine and readily transferred to naïve RMs by leukocyte transfer. Thus, even though UL35 is involved in countering the same intrinsic immune mechanism as pp71, the contribution of this protein to inactivating the host PML repressor complex would therefore appear to be less than pp71, at least for RhCMV. Alternatively, pp71 could support in vivo spread by countering additional host defense mechanisms. For instance, it was recently reported that pp71 of HCMV counters the innate immune signaling adaptor STING (70). From a vaccine development standpoint, the biologic phenotype of ΔRh59 (ΔUL35) RhCMV, though adequately immunogenic, is not satisfactorily attenuated for a primary CMV attenuation strategy.
Most of the ΔRh110 constructs analyzed in this study were based on BAC-cloned strain 68–1 (45) which shows CMV-typical signs of fibroblast adaptation including deletion of UL128 and UL130 homologous subunits of the tropism-determining pentameric receptor complex (Rh157.5 and Rh157.4), and a defect in the anti-apoptotic UL36 homolog Rh60/61 (4,71,72). These mutations are associated with reduced viremia, shedding and transmission of 68–1 compared to low passage RhCMV isolates (73), and thus potentially contribute to the attenuation of ΔRh110. However, repair of the homologs of UL128, UL130 and UL36 in strain 68–1.2 (58), did not enable Rh110-deleted RhCMV to be shed or transmitted by transfusion. This observation suggests that increased surveillance by intrinsic immunity resulting from Rh110 deletion substantially increases the attenuation afforded by the other deletions (particularly pentameric complex ablation) that spontaneously occurred in 68–1 during tissue culture adaptation.
The low level of in vivo infection manifested by ΔRh110 reduces the likelihood of co-infecting the same cell as endogenous RhCMV and potential repair of the Rh110 deletion by recombination. Homologous recombination of a ΔRh110 vector in which the Ag insert replaces the Rh110 coding sequence with an endogenous RhCMV would be expected to yield a pp71-intact, Ag-less vector and a pp71-deleted, Ag-containing version of the endogenous RhCMV strain, not an insert-expressing wildtype vector with the potential to spread. Thus, such a recombination event would have no consequence for either the vaccine recipient or unvaccinated close contacts. It is, however, theoretically possible that pp71 expression might be restored in Rh110-deleted RhCMV by non-homologous recombination, leaving a spread-competent CMV that also expresses the vaccine insert. While this would not subject the vaccine recipient to additional risk, since this individual already harbors wildtype CMV, it could lead to shedding and consequent transmission to unvaccinated close contacts of an insert-containing CMV vector. However, in experiments designed to detect Rh110 repair by either homologous or non-homologous gene exchange, we saw no evidence of such attenuation-reversal in 6 superinfected RMs followed over 800 days. These data suggest that if such Rh110-repairing recombination occurs at all, it is likely to be a very infrequent event. We would also note that the most likely outcome of Δpp71 CMV vector co-infection with a wildtype CMV is complementation of the Δpp71 CMV virions, as reported for MCMV lacking essential genes (74), which would increase their infectivity. However, since the vast majority of available target cells would not be infected, this enhancement, like complementation during in vitro production, would be lost in subsequent rounds of infection and therefore would not be expected to meaningfully reduce Δpp71 CMV vector attenuation.
Taken together, our data support the general conclusion that the indefinite maintenance of T cell immunity by CMV vectors can be uncoupled from viral spread within individual hosts and viral dissemination among hosts. A limitation of our study is that we only evaluated attenuated vectors in immunocompetent animals where RhCMV is non-pathogenic. Nevertheless, the results strongly suggest that the markedly reduced capacity to spread will also limit the ability of attenuated vectors to cause disease while preserving full CMV vector immunogenicity. Our findings in RhCMV are reminiscent of the finding in the mouse model where it was demonstrated that single-cycle MCMV viruses (i.e., viruses that replicate their genomes but are unable to generate infectious progeny) generated the MCMV-typical TEM inflation over time and protected against MCMV challenge (23,24), and are in contrast with findings in most other replication-defective viral vector systems in development (75). Although, so far, this finding is limited to animal models, the fact that similar observations were made in both murine and rhesus models, bodes well for the development of safe, HCMV-based vaccines for the human population. Moreover, the “pp71 deletion by insert replacement” design is a viable vector design strategy for initial assessment of attenuated HCMV vectors in humans. Given that UL82 (pp71)-deleted HCMV is more growth restricted in vitro than RhCMVΔRh110 (5-log vs. 2-log reduction relative to 68–1 at low MOI) (46), it is likely that a HCMVΔUL82 vector will be, if anything, more attenuated in humans than ΔRh110 in RMs, offering an extra margin of safety at the possible expense of requiring a higher dose for immunogenicity.
MATERIALS AND METHODS
Study Design.
The objective of this study was to evaluate the impact of viral attenuation by deletion of RhCMV genes that, when present, counteract host immune responses. In vitro studies were performed with life-extended primary rhesus fibroblasts to determine the impact of viral gene deletions on host cell protein expression and viral growth. The number of independent experiments and the number of replicates per experiment are indicated in the figure legends. Animal studies were approved by the Institutional Animal Care and Use Committee. To minimize the number of animals used in these experiments, most were designed with the goal to generate qualitative rather than quantitative comparisons with non-attenuated vectors. Because we observed highly consistent results among the 2–4 animals per group, these low numbers were sufficient to determine whether attenuated viral vectors lack viral shedding, viral transmission, and recombination with endogenous virus, while maintaining the ability to re-infect and elicit as well as maintain unconventionally restricted T cell responses. Furthermore, most key observations, e.g., lack of shedding, and all immunological parameters, were independently observed in multiple experiments whereas others, such as lack of spontaneous transmission, were independently confirmed by distinct experimental designs. To obtain quantitative comparisons with non-attenuated vectors, such as measurements of genome copy numbers or the determination of the minimal immunogenic dose, animal numbers were minimized by including internal controls, i.e., animals were co-inoculated with non-attenuated vectors. Primary data are reported in data file S1.
Statistical Analysis.
We used non-parametric testing to determine differences between groups. In comparisons of two groups only, we used two-sided Wilcoxon rank-sum tests with α=0.05 to detect differences between the groups. In comparisons of more than two groups, we used the Kruskal-Wallis rank-sum test to detect any differences between groups with α=0.05. If any differences were detected, we performed post-hoc two-sided Wilcoxon rank-sum tests with α=0.05 on the pairwise differences between groups. No multiplicity adjustments were performed for post-hoc analyses. All statistical analyses were performed using R (v. 3.2.2).
Supplementary Materials
Data File S1. Primary data.
Supplemental Materials and Methods
Table S1. Raw data used to generate graphs.
Fig. S1. Redistribution of corepressors by RhCMV pp71.
Fig. S2. Description and characterization of Rh110 (pp71)-deleted RhCMV vectors.
Fig. S3. Summary of recombinant RhCMV vectors used in vivo.
Fig. S4. Tissue distribution of SIV-insert-specific T cell responses elicited by ΔRh110 (Δpp71) vs. 68–1 vectors.
Fig. S5. Urine shedding of ΔRh110 (Δpp71) vs. 68–1 RhCMV vectors.
Fig. S6. T cell responses in lung airspace (bronchoalveolar lavage cells) upon super-infection with ΔRh110 (Δpp71) vs. 68–1 RhCMV vectors.
Fig. S7. Boosting of RhCMV antibody responses by ΔRh110 (Δpp71) RhCMV vectors.
Fig. S8. Description and characterization of dual insert-expressing ΔRh110 (Δpp71) RhCMV vectors.
Fig. S9. CD4+ T cell responses to dual antigen insert-expressing ΔRh110 (Δpp71) RhCMV vectors.
Fig. S10. Lack of maternal-infant transmission of ΔRh110 (Δpp71) RhCMV vectors.
Fig. S11. Description and characterization of Rh59 (UL35)-deleted RhCMV vectors.
Fig. S12. SIV-specific CD4+ and CD8+ T cell responses elicited by ΔRh59 (ΔUL35) vs. 68–1 RhCMV vectors in tissue sites.
Fig. S13. T cell responses to SIV antigens expressed by ΔRh110 (Δpp71) and ΔRh59 (ΔUL35) RhCMV vectors.
ACKNOWLEDGMENTS:
We thank T. Shenk (Princeton University) for reagents, and J. Shao (Fred Hutchinson Cancer Research Center) for help with statistical analysis. We also thank the OHSU molecular virology support core and monoclonal antibody core for technical service, P. Smith for help with the ELISA for RhCMV antibodies, A. Townsend for figure preparation, and A. Sylwester, S. Hagen, N. Whizin, K. Randall, A. Selseth, and L. Boshears for other technical or administrative assistance.
FUNDING: This work was supported by the Bill & Melinda Gates Foundation-supported Collaboration for AIDS Vaccine Discovery (OPP1033121, LJP); the National Institute of Allergy and Infectious Diseases (NIAID) (R01 AI095113, P01 AI094417, and R37 AI054292 to LJP; R01 AI059457 to KF; U42 OD023038 to MKA); and the NIH Office of the Director (U42OD010426 and P51OD011092).
Footnotes
COMPETING INTERESTS: OHSU, LJP, EEM SGH and KF have a substantial financial interest in Vir Biotechnology, Inc., a company that may have a commercial interest in the results of this research and technology. LJP, SGH and KF are co-inventors of patent PCT/US2011/036657 “Recombinant RhCMV and HCMV vectors and uses thereof” licensed to Vir. LJP, SGH and KF have received compensation for consulting for Vir. The potential individual and institutional conflicts of interest have been reviewed and managed by OHSU.
DATA AND MATERIALS AVAILABILITY: All data associated with this study are present in the paper or Supplementary Materials. Vector primer sequences have been deposited in Genbank (see Supplementary Materials for accession numbers). The computer code used to perform statistical analysis is available at https://doi.org/10.5281/zenodo.3242804. RhCMV/SIV vectors can be obtained through a materials transfer agreement.
REFERENCES AND NOTES:
- 1.Cannon MJ, Schmid DS, Hyde TB, Review of cytomegalovirus seroprevalence and demographic characteristics associated with infection. Rev Med Virol 20, 202–213 (2010). [DOI] [PubMed] [Google Scholar]
- 2.Yue Y, Barry PA, Rhesus cytomegalovirus a nonhuman primate model for the study of human cytomegalovirus. Adv Virus Res 72, 207–226 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Lockridge KM, Sequar G, Zhou SS, Yue Y, Mandell CP, Barry PA, Pathogenesis of experimental rhesus cytomegalovirus infection. J Virol 73, 9576–9583 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malouli D, Nakayasu ES, Viswanathan K, Camp DG 2nd, Chang WL, Barry PA, Smith RD, Früh K, Reevaluation of the Coding Potential and Proteomic Analysis of the BAC-Derived Rhesus Cytomegalovirus Strain 68–1. J Virol 86, 8959–8973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zanghellini F, Boppana SB, Emery VC, Griffiths PD, Pass RF, Asymptomatic primary cytomegalovirus infection: virologic and immunologic features. J Infect Dis 180, 702–707 (1999). [DOI] [PubMed] [Google Scholar]
- 6.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, Nelson JA, Picker LJ, Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J Exp Med 202, 673–685 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen SG, Powers CJ, Richards R, Ventura AB, Ford JC, Siess D, Axthelm MK, Nelson JA, Jarvis MA, Picker LJ, Früh K, Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328, 102–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ross SA, Arora N, Novak Z, Fowler KB, Britt WJ, Boppana SB, Cytomegalovirus reinfections in healthy seroimmune women. J Infect Dis 201, 386–389 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pokalyuk C, Renzette N, Irwin KK, Pfeifer SP, Gibson L, Britt WJ, Yamamoto AY, Mussi-Pinhata MM, Kowalik TF, Jensen JD, Characterizing human cytomegalovirus reinfection in congenitally infected infants: an evolutionary perspective. Mol Ecol 26, 1980–1990 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Kern F, Khatamzas E, Surel I, Frommel C, Reinke P, Waldrop SL, Picker LJ, Volk HD, Distribution of human CMV-specific memory T cells among the CD8pos. subsets defined by CD57, CD27, and CD45 isoforms. Eur J Immunol 29, 2908–2915 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Cicin-Sain L, Sylwester AW, Hagen SI, Siess DC, Currier N, Legasse AW, Fischer MB, Koudelka CW, Axthelm MK, Nikolich-Zugich J, Picker LJ, Cytomegalovirus-specific T cell immunity is maintained in immunosenescent rhesus macaques. J Immunol 187, 1722–1732 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jarvis MA, Hansen SG, Nelson JA, Picker LJ, Früh K, in Cytomegaloviruses: From Molecular Pathogenesis to Intervention Reddehase MJ, Ed. (Caister Academic Press, 2013), vol. 2, chap. 21. [Google Scholar]
- 13.Masopust D, Picker LJ, Hidden memories: frontline memory T cells and early pathogen interception. J Immunol 188, 5811–5817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picker LJ, Hansen SG, Lifson JD, New Paradigms for HIV/AIDS Vaccine Development. Annu Rev Med, 63, 95–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr., Lifson JD, Nelson JA, Jarvis MA, Picker LJ, Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 15, 293–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hansen SG, Ford JC, Lewis MS, Ventura AB, Hughes CM, Coyne-Johnson L, Whizin N, Oswald K, Shoemaker R, Swanson T, Legasse AW, Chiuchiolo MJ, Parks CL, Axthelm MK, Nelson JA, Jarvis MA, Piatak M Jr., Lifson JD, Picker LJ, Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen SG, Piatak M Jr., Ventura AB, Hughes CM, Gilbride RM, Ford JC, Oswald K, Shoemaker R, Li Y, Lewis MS, Gilliam AN, Xu G, Whizin N, Burwitz BJ, Planer SL, Turner JM, Legasse AW, Axthelm MK, Nelson JA, Früh K, Sacha JB, Estes JD, Keele BF, Edlefsen PT, Lifson JD, Picker LJ, Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen SG, Zak DE, Xu G, Ford JC, Marshall EE, Malouli D, Gilbride RM, Hughes CM, Ventura AB, Ainslie E, Randall KT, Selseth AN, Rundstrom P, Herlache L, Lewis MS, Park H, Planer SL, Turner JM, Fischer M, Armstrong C, Zweig RC, Valvo J, Braun JM, Shankar S, Lu L, Sylwester AW, Legasse AW, Messerle M, Jarvis MA, Amon LM, Aderem A, Alter G, Laddy DJ, Stone M, Bonavia A, Evans TG, Axthelm MK, Früh K, Edlefsen PT, Picker LJ, Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat Med 24, 130–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koff WC, Burton DR, Johnson PR, Walker BD, King CR, Nabel GJ, Ahmed R, Bhan MK, Plotkin SA, Accelerating next-generation vaccine development for global disease prevention. Science 340, 1232910 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hawn TR, Day TA, Scriba TJ, Hatherill M, Hanekom WA, Evans TG, Churchyard GJ, Kublin JG, Bekker LG, Self SG, Tuberculosis vaccines and prevention of infection. Microbiol Mol Biol Rev 78, 650–671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boppana SB, Britt WJ, in Cytomegaloviruses, Reddehase MJ, Ed. (Caister Academic Press, Norfolk, UK, 2013), vol. 2, pp. 1–25. [Google Scholar]
- 22.Loo CP, Snyder CM, Hill AB, Blocking Virus Replication during Acute Murine Cytomegalovirus Infection Paradoxically Prolongs Antigen Presentation and Increases the CD8+ T Cell Response by Preventing Type I IFN–Dependent Depletion of Dendritic Cells. J Immunol 98, 383–393 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snyder CM, Cho KS, Bonnett EL, Allan JE, Hill AB, Sustained CD8+ T cell memory inflation after infection with a single-cycle cytomegalovirus. PLoS Pathog 7, e1002295 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohr CA, Arapovic J, Muhlbach H, Panzer M, Weyn A, Dolken L, Krmpotic A, Voehringer D, Ruzsics Z, Koszinowski U, Sacher T, A spread-deficient cytomegalovirus for assessment of first-target cells in vaccination. J Virol 84, 7730–7742 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobson MA, Sinclair E, Bredt B, Agrillo L, Black D, Epling CL, Carvidi A, Ho T, Bains R, Adler SP Antigen-specific T cell responses induced by Towne cytomegalovirus (CMV) vaccine in CMV-seronegative vaccine recipients, J Clin Virol 35, 332–337 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Heineman TC, Schleiss M, Bernstein DI, Spaete RR, Yan L, Duke G, Prichard M, Wang Z, Yan Q, Sharp MA, Klein N, Arvin AM, Kemble G, A phase 1 study of 4 live, recombinant human cytomegalovirus Towne/Toledo chimeric vaccines. J Infect Dis 193, 1350–1360 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Adler SP, Manganello AM, Lee R, McVoy MA, Nixon DE, Plotkin S, Mocarski E, Cox JH, Fast PE, Nesterenko PA, Murray SE, Hill AB, Kemble G, A Phase 1 Study of Four Live, Recombinant Human Cytomegalovirus Towne/Toledo Chimera Vaccines in CMV Seronegative Men. J Infect Dis 214, 1341–1348 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Murray SE, Nesterenko PA, Vanarsdall AL, Munks MW, Smart SM, Veziroglu EM, Sagario LC, Lee R, Claas FHJ, Doxiadis IIN, McVoy MA, Adler SP, Hill AB, Fibroblast-adapted human CMV vaccines elicit predominantly conventional CD8 T cell responses in humans. J Exp Med, 214, 1889–1899 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindsay CR, Morozov VM, Ishov AM, PML NBs (ND10) and Daxx: from nuclear structure to protein function. Front Biosci 13, 7132–7142 (2008). [DOI] [PubMed] [Google Scholar]
- 30.Tang Q, Maul GG, Mouse cytomegalovirus immediate-early protein 1 binds with host cell repressors to relieve suppressive effects on viral transcription and replication during lytic infection. J Virol 77, 1357–1367 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A, PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol 80, 7995–8005 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Everett RD, Bell AJ, Lu Y, Orr A, The replication defect of ICP0-null mutant herpes simplex virus 1 can be largely complemented by the combined activities of human cytomegalovirus proteins IE1 and pp71. J Virol 87, 978–990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalejta RF, Functions of human cytomegalovirus tegument proteins prior to immediate early gene expression. Curr Top Microbiol Immunol 325, 101–115 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Tavalai N, Papior P, Rechter S, Leis M, Stamminger T, Evidence for a role of the cellular ND10 protein PML in mediating intrinsic immunity against human cytomegalovirus infections. J Virol 80, 8006–8018 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saffert RT, Kalejta RF, Inactivating a cellular intrinsic immune defense mediated by Daxx is the mechanism through which the human cytomegalovirus pp71 protein stimulates viral immediate-early gene expression. J Virol 80, 3863–3871 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preston CM, Nicholl MJ, Role of the cellular protein hDaxx in human cytomegalovirus immediate-early gene expression. J Gen Virol 87, 1113–1121 (2006). [DOI] [PubMed] [Google Scholar]
- 37.Lee SH, Kalejta RF, Kerry J, Semmes OJ, O’Connor CM, Khan Z, Garcia BA, Shenk T, Murphy E, BclAF1 restriction factor is neutralized by prteasomal degradation and microRNA repression during human cytomegalovirus infection. Proc Natl Acad Sci USA 109, 9575–9580 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Penkert RR, Kalejta RF, Tale of a tegument transactivator: the past, present and future of human CMV pp71. Future Virol 7, 855–869 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsai K, Thikmyanova N, Wojcechowskyj JA, Delecluse HJ, Lieberman PM, EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog 7, e1002376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qin Q, Penkert RR, Kalejta RF, Heterologous viral promoters incorporated into the human cytomegalovirus genome are silenced during experimental latency. J Virol 87, 9886–9894 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hofmann H, Sindre H, Stamminger T, Functional interaction between the pp71 protein of human cytomegalovirus and the PML-interacting protein human Daxx. J Virol 76, 5769–5783 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hansen SG, Strelow LI, Franchi DC, Anders DG, Wong SW, Complete sequence and genomic analysis of rhesus cytomegalovirus. J Virol 77, 6620–6636 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cantrell SR, Bresnahan WA, Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J Virol 80, 6188–6191 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cantrell SR, Bresnahan WA, Interaction between the human cytomegalovirus UL82 gene product (pp71) and hDaxx regulates immediate-early gene expression and viral replication. J Virol 79, 7792–7802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang WL, Barry PA, Cloning of the full-length rhesus cytomegalovirus genome as an infectious and self-excisable bacterial artificial chromosome for analysis of viral pathogenesis. J Virol 77, 5073–5083 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bresnahan WA, Shenk TE, UL82 virion protein activates expression of immediate early viral genes in human cytomegalovirus-infected cells. Proc Natl Acad Sci USA 97, 14506–14511 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McGregor A, Liu F, Schleiss MR, Molecular, biological, and in vivo characterization of the guinea pig cytomegalovirus (CMV) homologs of the human CMV matrix proteins pp71 (UL82) and pp65 (UL83). J Virol 78, 9872–9889 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen SG, Vieville C, Whizin N, Coyne-Johnson L, Siess DC, Drummond DD, Legasse AW, Axthelm MK, Oswald K, Trubey CM, Piatak M Jr, Lifson JD, Nelson JA, Jarvis MA, Picker LJ, Addendum: Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat Med 17, 1692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malouli D, Hansen SG, Nakayasu ES, Marshall EE, Hughes CM, Ventura AB, Gilbride RM, Lewis MS, Xu G, Kreklywich C, Whizin N, Fischer M, Legasse AW, Viswanathan K, Siess D, Camp DG 2nd, Axthelm MK, Kahl C, Defilippis VR, Smith RD, Streblow DN, Picker LJ, Früh K, Cytomegalovirus pp65 limits dissemination but is dispensable for persistence. J Clin Invest, 124 1928–1944 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klenerman P, Hill A, T cells and viral persistence: lessons from diverse infections. Nat Immunol 6, 873–879 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Lanzavecchia A, Sallusto F, Understanding the generation and function of memory T cell subsets. Curr Opin Immunol 17, 326–332 (2005). [DOI] [PubMed] [Google Scholar]
- 52.Hansen SG, Sacha JB, Hughes CM, Ford JC, Burwitz BJ, Scholz I, Gilbride RM, Lewis MS, Gilliam AN, Ventura AB, Malouli D, Xu G, Richards R, Whizin N, Reed JS, Hammond KB, Fischer M, Turner JM, Legasse AW, Axthelm MK, Edlefsen PT, Nelson JA, Lifson JD, Früh K, Picker LJ, Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hansen SG, Wu HL, Burwitz BJ, Hughes CM, Hammond KB, Ventura AB, Reed JS, Gilbride RM, Ainslie E, Morrow DW, Ford JC, Selseth AN, Pathak R, Malouli D, Legasse AW, Axthelm MK, Nelson JA, Gillespie GM, Walters LC, Brackenridge S, Sharpe HR, Lopez CA, Früh K, Korber BT, McMichael AJ, Gnanakaran S, Sacha JB, Picker LJ, Broadly targeted CD8(+) T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Norberg P, Depledge DP, Kundu S, Atkinson C, Brown J, Haque T, Hussaini Y, MacMahon E, Molyneaux P, Papaevangelou V, Sengupta N, Koay ES, Tang JW, Underhill GS, Grahn A, Studahl M, Breuer J, Bergstrom T, Recombination of Globally Circulating Varicella-Zoster Virus. J Virol 89, 7133–7146 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee SW, Markham PF, Coppo MJ, Legione AR, Markham JF, Noormohammadi AH, Browning GF, Ficorilli N, Hartley CA, Devlin JM, Attenuated vaccines can recombine to form virulent field viruses. Science 337, 188 (2012). [DOI] [PubMed] [Google Scholar]
- 56.Renzette N, Gibson L, Jensen JD, Kowalik TF, Human cytomegalovirus intrahost evolution-a new avenue for understanding and controlling herpesvirus infections. Curr Opin Virol 8, 109–115 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schierling K, Buser C, Mertens T, Winkler M, Human cytomegalovirus tegument protein ppUL35 is important for viral replication and particle formation. J Virol 79, 3084–3096 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lilja AE, Shenk T, Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc Natl Acad Sci USA 105, 19950–19955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hillyer CD, Emmens RK, Zago-Novaretti M, Berkman EM, Methods for the reduction of transfusion-transmitted cytomegalovirus infection: filtration versus the use of seronegative donor units. Transfusion 34, 929–934 (1994). [DOI] [PubMed] [Google Scholar]
- 60.Söderberg-Naucler C, Nelson JY, Human cytomegalovirus latency and reactivation - a delicate balance between the virus and its host’s immune system. Intervirol 42, 314–321 (1999). [DOI] [PubMed] [Google Scholar]
- 61.Sinclair J, Sissons P, Latency and reactivation of human cytomegalovirus. J Gen Virol 87, 1763–1779 (2006). [DOI] [PubMed] [Google Scholar]
- 62.Shin H, Blackburn SD, Blattman JN, Wherry EJ, Viral antigen and extensive division maintain virus-specific CD8 T cells during chronic infection. J Exp Med 204, 941–949 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang D, Freed DC, He X, Li F, Tang A, Cox KS, Dubey SA, Cole S, Medi MB, Liu Y, Xu J, Zhang ZQ, Finnefrock AC, Song L, Espeseth AS, Shiver JW, Casimiro DR, Fu TM, A replication-defective human cytomegalovirus vaccine for prevention of congenital infection. Sci Transl Med 8, 362ra145 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Poole E, Sinclair J, Sleepless latency of human cytomegalovirus. Med Microbiol Immunol 204, 421–429 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Powers C, DeFilippis V, Malouli D, Früh K, Cytomegalovirus immune evasion. Cur Top Microbiol Immunol 325, 333–359 (2008). [DOI] [PubMed] [Google Scholar]
- 66.Sturgill ER, Malouli D, Hansen SG, Burwitz BJ, Seo S, Schneider CL, Womack JL, Verweij MC, Ventura AB, Bhusari A, Jeffries KM, Legasse AW, Axthelm MK, Hudson AW, Sacha JB, Picker LJ, Früh K, Natural Killer Cell Evasion Is Essential for Infection by Rhesus Cytomegalovirus. PLoS Pathog 12, e1005868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michaelson JS, Bader D, Kuo F, Kozak C, Leder P, Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev 13, 1918–1923 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schreiner S, Wodrich H, Virion factors that target Daxx to overcome intrinsic immunity. J Virol 87, 10412–10422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hansen SG, Marshall E, Malouli D, Ventura AB, Hughes CM, Ainslie E, Ford JC, Morrow D, Gilbride RM, Bae JY, Legasse AW, Oswald K, Shoemaker R, Berkemeier B, Bosche WJ, Hull M, Womack J, Shao J, Edlefsen PT, Reed JS, Burwitz BJ, Sacha JB, Axthelm MK, Früh K, Lifson JL, Picker LJ, A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci Transl Med 11, eaaw2607 (2019). (companion paper) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fu YZ, Su S, Gao YQ, Wang PP, Huang ZF, Hu MM, Luo WW, Li S, Luo MH, Wang YY, Shu HB, Human Cytomegalovirus Tegument Protein UL82 Inhibits STING-Mediated Signaling to Evade Antiviral Immunity. Cell Host Microbe 21, 231–243 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Oxford KL, Eberhardt MK, Yang KW, Strelow L, Kelly S, Zhou SS, Barry PA, Protein coding content of the UL)b’ region of wild-type rhesus cytomegalovirus. Virol 373, 181–188 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCormick AL, Skaletskaya A, Barry PA, Mocarski ES, Goldmacher VS, Differential function and expression of the viral inhibitor of caspase 8-induced apoptosis (vICA) and the viral mitochondria-localized inhibitor of apoptosis (vMIA) cell death suppressors conserved in primate and rodent cytomegaloviruses. Virol 316, 221–233 (2003). [DOI] [PubMed] [Google Scholar]
- 73.Oxford KL, Strelow L, Yue Y, Chang WL, Schmidt KA, Diamond DJ, Barry PA, Open reading frames carried on UL/b’ are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys. J Virol 85, 5105–5114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cicin-Sain L, Podlech J, Messerle M, Reddehase MJ, Koszinowski UH, Frequent coinfection of cells explains functional in vivo complementation between cytomegalovirus variants in the multiply infected host. J Virol 79, 9492–9502 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dudek T, Knipe DM, Replication-defective viruses as vaccines and vaccine vectors. Virol 344, 230–239 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Borst EM, Benkartek C, Messerle M, Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr Protoc Immunol, Chapter 10, Unit 10 32 (2007). [DOI] [PubMed] [Google Scholar]
- 77.Hel Z, Johnson JM, Tryniszewska E, Tsai WP, Harrod R, Fullen J, Tartaglia J, Franchini G, A novel chimeric Rev, Tat, and Nef (Retanef) antigen as a component of an SIV/HIV vaccine. Vaccine 20, 3171–3186 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Lilja AE, Chang WL, Barry PA, Becerra SP, Shenk TE, Functional genetic analysis of rhesus cytomegalovirus: Rh01 is an epithelial cell tropism factor. J Virol 82, 2170–2181 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Burwitz BJ, Malouli D, Bimber BN, Reed JS, Ventura AB, Hancock MH, Uebelhoer LS, Bhusari A, Hammond KB, Espinosa Trethewy RG, Klug A, Legasse AW, Axthelm MK, Nelson JA, Park BS, Streblow DN, Hansen SG, Picker LJ, Früh K, Sacha JB, Cross-Species Rhesus Cytomegalovirus Infection of Cynomolgus Macaques. PLoS Pathog 12, e1006014 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cline AN, Bess JW, Piatak M Jr., Lifson JD, Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol 34, 303–312 (2005). [DOI] [PubMed] [Google Scholar]
- 81.Pitcher CJ, Hagen SI, Walker JM, Lum R, Mitchell BL, Maino VC, Axthelm MK, Picker LJ, Development and homeostasis of T cell memory in rhesus macaque. J Immunol 168, 29–43 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data File S1. Primary data.
Supplemental Materials and Methods
Table S1. Raw data used to generate graphs.
Fig. S1. Redistribution of corepressors by RhCMV pp71.
Fig. S2. Description and characterization of Rh110 (pp71)-deleted RhCMV vectors.
Fig. S3. Summary of recombinant RhCMV vectors used in vivo.
Fig. S4. Tissue distribution of SIV-insert-specific T cell responses elicited by ΔRh110 (Δpp71) vs. 68–1 vectors.
Fig. S5. Urine shedding of ΔRh110 (Δpp71) vs. 68–1 RhCMV vectors.
Fig. S6. T cell responses in lung airspace (bronchoalveolar lavage cells) upon super-infection with ΔRh110 (Δpp71) vs. 68–1 RhCMV vectors.
Fig. S7. Boosting of RhCMV antibody responses by ΔRh110 (Δpp71) RhCMV vectors.
Fig. S8. Description and characterization of dual insert-expressing ΔRh110 (Δpp71) RhCMV vectors.
Fig. S9. CD4+ T cell responses to dual antigen insert-expressing ΔRh110 (Δpp71) RhCMV vectors.
Fig. S10. Lack of maternal-infant transmission of ΔRh110 (Δpp71) RhCMV vectors.
Fig. S11. Description and characterization of Rh59 (UL35)-deleted RhCMV vectors.
Fig. S12. SIV-specific CD4+ and CD8+ T cell responses elicited by ΔRh59 (ΔUL35) vs. 68–1 RhCMV vectors in tissue sites.
Fig. S13. T cell responses to SIV antigens expressed by ΔRh110 (Δpp71) and ΔRh59 (ΔUL35) RhCMV vectors.