Abstract
The identification of novel herbicides is of crucial importance to modern agriculture. We developed an efficient in vivo assay based on oxygen evolution measurements using suspensions of chlorenchyma cells isolated from the single-cell C4 plant Bienertia sinuspersici to identify and characterize inhibitors of C4 photosynthesis. This novel approach fills the gap between conventional in vitro assays for inhibitors targeting C4 key enzymes and in vivo experiments on whole plants. The assay addresses inhibition of the target enzymes in a plant context thereby taking care of any reduced target inhibition due to metabolization or inadequate uptake of small molecule inhibitors across plant cell walls and membranes. Known small molecule inhibitors targeting C4 photosynthesis were used to validate the approach. To this end, we tested pyruvate phosphate dikinase inhibitor bisindolylmaleimide IV and phosphoenolpyruvate carboxylase inhibitor okanin. Both inhibitors show inhibition of plant photosynthesis at half-maximal inhibitory concentrations in the sub-mM range and confirm their potential to act as a new class of C4 selective inhibitors.
Keywords: C4 photosynthesis, Inhibitor, phosphoenolpyruvate carboxylase, pyruvate phosphate dikinase, Bienertia sinuspersici, cell suspension, screening
Introduction
A number of different types of photosynthesis — C3, C4, and crassulacean acid metabolism (CAM) — are known for terrestrial plants. Each of them is specialized to accommodate different environmental conditions with C3 photosynthesis being the most common one. CAM and C4 plants adapted to hot and dry climate by establishing CO2 concentrating mechanisms that circumvent the oxygenase reactivity of ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) which is the key enzyme of photosynthetic carbon fixation within the Calvin-Benson-Bassham (CBB) cycle. In C4 plants, this CO2 concentrating mechanism is implemented by a spatial separation of CO2 uptake from the CBB cycle. Normally, this separation is established across two cell types: bundle sheath (BS) cells which are surrounded by a layer of leaf mesophyll (LM) cells forming the so-called Kranz anatomy. Primary CO2 fixation takes place in the LM cells, where atmospheric CO2 is fixed after conversion to HCO3− by phosphoenolpyruvate carboxylase (PEPC) yielding the four-carbon organic acid oxaloacetate that is subsequently converted species-specifically into malate or aspartate. These diffuse to the BS cells where CO2 is released by decarboxylation and hereby enriched within the BS cells. The local increase in CO2 concentration greatly disfavors the oxygenase activity of RuBisCO which then uses the released CO2 during the CBB cycle to form triose phosphates. Pyruvate formed in the process of decarboxylation diffuses back into the LM cells and is regenerated to phosphoenolpyruvate (PEP) by the pyruvate phosphate dikinase (PPDK). The regenerated PEP then may be reused to fix another molecule of CO2. With the discovery of plants capable of performing single-cell C4 photosynthesis (SCC4), it has been shown however that Kranz anatomy is not a mandatory requirement for C4 photosynthesis (Sage, 2002; Voznesenskaya et al., 2002).
Many important crop plants like soybean, sugar beet or wheat are C3 plants, while some of the most abundant weeds such Amaranthus retroflexus or Echinochloa crus-galli utilize C4 photosynthesis. With the rise of known resistances of weeds to commercially available herbicides over the past decades the effects of resistant weeds on modern agriculture are becoming an increasingly pressing issue (Heap, 2014; Busi et al., 2018; Heap, 2019). In addition to herbicide resistances, the predicted increase in average temperatures caused by global warming and rising levels of atmospheric CO2 are thought to shift the odds in favor of C4 plants which will eventually render C4 weeds even more competitive in the context of C3 crops (Fernando et al., 2016; Carboni et al., 2017; Waryszak et al., 2018). To combat this development, new herbicides that are specifically designed to target C4 weeds are needed.
The identification and characterization of novel inhibitors interfering with mandatory biochemical pathways in plants are key requirements for the development of new herbicides. This process usually includes in vitro assays which assess the efficiency and efficacy toward a specific target enzyme followed by extensive in vivo studies using whole plants. However, compounds that prove to be effective in vitro may not be effective at all on whole plants due to reduced bioavailability which might be caused by several issues such as slow uptake into the plant tissue, solubility problems or detoxification of the active compound within the cells (Shimabukuro, 1985). Furthermore, in vivo studies with whole plants are cumbersome — particularly in terms of set-up and repetitions, human resources, and technical facilities required for a controlled environment. Consequently, an alternative in vivo method for the preliminary validation of biological effectiveness of compounds is needed for pre-screening purposes.
In recent years, a number of inhibitory compounds targeting C4 photosynthesis have been identified (Haines et al., 2005; Nguyen et al., 2016; Dick et al., 2017; Minges and Groth, 2017). These compounds inhibit PPDK or PEPC activity, which are key enzymes in Kranz C4 and SCC4 pathways. Effects of C4 inhibitors have been previously studied in vitro using purified enzymes in spectrophotometric assays (Doyle et al., 2005; Motti et al., 2007; Nguyen et al., 2016; Minges and Groth, 2017). While in vitro experiments are usually fast and convenient to perform and scale-up well (Feng et al., 2005; Bailey et al., 2018), they represent a dramatic simplification of the larger biological context in which the studied biochemical reaction takes place. Here, plant related metabolization or uptake limitations of the compound due to the plant cuticula, cell walls or cell membrane are largely ignored. These restrictions are avoided when leaf cuttings or whole plants are used and compounds are tested at in vivo conditions (Haines et al., 2005; Motti et al., 2007). Compared to whole plant toxicity assays, oxygen evolution measurements on leaf cuttings are relatively easy and straightforward to perform using Clark-type electrodes with buffer-filled reaction chambers (Clark, 1956; Delieu and Walker, 1972). In this set-up the cuticular barrier is bypassed as water-soluble compounds are able to freely diffuse into the cells and within the symplast by the plasmodesmata exposed on the cut surfaces. Nevertheless, plant related metabolization of the compound may be still observable in these studies as most of the plant tissue is still largely intact. However, ensuring homogeneous illumination of all leaf cuttings for all repetitions and across different studies still remains challenging in this set-up. Stirring of the buffer solution naturally leads to unpredictable and random movement of the leaf slices, leaving no possibility to ensure that individual slices do not shade each other from the light source. Aside from these purely mechanical issues, earlier studies questioned the biological availability of dissolved CO2 (Jones and Osmond, 1973). The apparent Km determined for CO2 was larger than observed in gas phase experiments (Jones and Slatyer, 1972) or on isolated chloroplasts (Jensen and Bassham, 1966). Jones and Slatyer (1972) pointed out that this effect is largely dependent on the experimental procedure and plant species used to prepare the leaf slices and is likely to be caused by diffusion problems of CO2 from the buffer to the photosynthetically active cells. This dependence on experimental setup and biological system used for the experiments severely hampers the interpretation of comparative studies (Jones and Osmond, 1973).
On the other hand, whole plant toxicity assessments are rather time-consuming — ranging from days to weeks for each replicate — to set up and to perform (Lein et al., 2004; Burgos, 2015). The outcome strongly depends on general growth conditions which can be difficult to control and/or other parameters that are not immediately discernible to the experimenter. In whole plant assays, inhibitor uptake across the cuticular barrier into the apoplast is usually the success-limiting factor (Tice, 2001). Hence, whole plant experiments are commonly considered in later stages of the development of new herbicides. Today, a simple, scalable assay albeit as close as possible to in planta conditions is still missing. While leaf cuttings provide a more complex biological system with intact photosynthetic apparatus, technical (e.g. uniform illumination of the sample) as well as biological (free diffusion of compounds) limits confine their application. Here, we present a novel in vivo test system using intact plant cell suspensions that builds up on previous studies using leaf cuttings (Haines et al., 2005; Motti et al., 2007; Minges and Groth, 2017).
Toxicity tests using heterotrophic plant cell cultures are routinely used (Olofsdotter et al., 1994) and do not suffer from the same disadvantages as whole plant or leaf cutting assays. However they are not applicable to the analysis of photosynthesis inhibitors. Similarly, isolated chloroplasts have been used to study photosynthesis and photosynthetic oxygen evolution (POE) (Jensen and Bassham, 1966). However, cellular environment such as membranes and detoxification mechanisms is missing. In the context of C4 photosynthesis, its complex apparatus of CO2 concentration mechanisms is absent and as such isolated chloroplasts are of limited use for the identification of C4-specific inhibitors of photosynthesis.
Bienertia sinuspersici is a succulent that belongs to the Suaedoideae subfamily of Amaranthaceae and features SCC4 within dimorphic chloroplasts (Freitag and Stichler, 2002; Akhani et al., 2005). Furthermore, B. sinuspersici chlorenchyma cells occur in loosely packed layers which simplifies the preparation of a functional cell suspension (Akhani et al., 2005; Offermann et al., 2011). Previous studies suggest that the difference between SCC4 and Kranz-type C4 is mainly of morphological, but not of biochemical nature (Lara et al., 2006; Lara et al., 2008; Park et al., 2010; Offermann et al., 2011; Offermann et al., 2015). Hence, most findings regarding biochemical mechanisms and key enzymes of C4 photosynthesis should generally be transferable between SCC4 and Kranz-type C4 plants.
Materials and Methods
Plant Material
B. sinuspersici plants were grown in growth chambers (CLF Plant Climatics, DE) as previously described (Leisner et al., 2010; Offermann et al., 2011) with day/night cycle (30°C/18°C, 14 h/10 h) and a photosynthetic flux density of 500 µmol quanta m−2 s−1. Plants were watered twice a week. Liquid fertilizer (0.2 % (v/v), NPK 12-4-6, Wuxal Top N, Manna, DE), and 100 mM NaCl were provided once per week while watering the plants. Zea mays was grown in the greenhouse under natural light conditions.
Preparation of Chlorenchyma Cell Suspension
Medium-sized and well-illuminated mature leaves were harvested from the upper third of young B. sinuspersici plants. Leaves were gently squeezed with mortar and pestle in cold extraction buffer (750 mM betaine, 2.5 mM MgCl2, 2.5 mM NaH2PO4, 0.5 mM MnCl2, 25 mM HEPES, pH 7.5) for 5 to 10 min. A total volume of 5 ml buffer was used for the extraction of 50 leaves per preparation. The obtained cell suspension was centrifuged twice (100 × g, 4°C, 5 min). The supernatant of each step was discarded to segregate intact cells from cell debris and free chloroplasts. The green-colored sediment was re-suspended in the same volume of degassed extraction buffer used for the initial cell extraction step (Offermann et al., 2011). Chlorenchyma cell suspensions were kept in the dark on ice until usage in the O2 evolution assay. If necessary, cells were further diluted to yield an O2 evolution rate of approx. 10 nmol ml−1 min−1.
Quality of Cell Suspensions
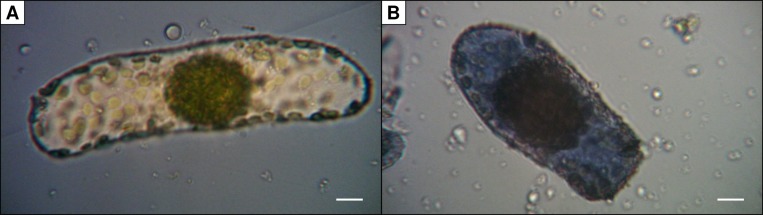
Vital staining of B. sinuspersici cells was performed by mixing 45 µl of cell suspensions with 5 µl of 2% (w/v) trypan blue staining solution (Tennant, 1964). A fifth of the stained cell suspension was transferred to a Neubauer counting chamber. A light microscope (M11, Wild, Heerbrugg, CH) with attached digital camera (MikrOkular, Bresser, DE) was used for analysis and documentation ( Figure 1 ). The ratio of intact (unstained) and damaged cells (stained) was calculated. In preparations used for measurements of oxygen evolution rate, 50% to 80% of cells were intact judged by trypan blue staining with a cell concentration about 100,000 to 450,000 cells ml−1.
Figure 1.
Trypan blue staining of isolated Bienertia sinuspersici chlorenchyma cells. Damaged cells (B) are stained due to diffusion of the dye into the cell whereas intact cells (A) remain unstained (scale bars = 10 µm).
Measurements of Oxygen Evolution Rates
Measurements were performed using a Clark-type oxygen electrode (Chlorolab 2+ with light source LED1, Hansatech Instruments, Norfolk, UK) (Delieu and Walker, 1972). Pyruvate (1 mM) and NaHCO3 (1 mM) were added to 1 ml B. sinuspersici chlorenchyma cell suspension in extraction buffer to prevent substrate limitation. The sample was then incubated at 30°C for 5 min and added to the pre-heated (30°C) electrode chamber. Oxygen evolution in the dark was monitored until stability (dark rate). Two factors contribute to the dark rate: oxygen depletion by the electrode (electrode drift) and mitochondrial respiration. Subsequently the electrode chamber was illuminated at 1,000 µmol m−2 s−1 and light-dependent oxygen evolution (light rate) was recorded after signal stabilization. The POE which is indicative for the photosynthetic activity was calculated as the difference between light and dark rate.
Inhibition experiments were performed in the presence of known inhibitors of PEPC or PPDK. Here we used the chalcone okanin (Carbosynth, Compton, UK) — an allosteric inhibitor of PEPC activity — and bisindolylmaleimide IV (BIM4) (Cayman Chemicals, Ann Arbor, US) — an active-site inhibitor competing with ATP for the nucleotide binding site of PPDK.
Inhibitors were added to the electrode chamber after the system reached equilibrium. The rate was recorded (inhibited light rate), once a stable POE was reached again after addition of the compound. The relative inhibition was then calculated according to equation 1.
| (1) |
For okanin a substantial light-independent oxygen consumption was observed immediately after addition of the compound. However, this oxygen scavenging effect is well-known for other hydroxylated chalcones (Rathmell and Bendall, 1972; Haraguchi et al., 1998). To correct for this effect, the rate of oxygen consumption in the dark after addition of okanin was determined at every titration step and subtracted from the inhibited light rates.
Half-maximal (50%) inhibitory concentrations (IC50) were calculated by measuring the activity in the presence of different concentrations of each compound. Nine inhibitor concentrations in the range of 0 to 350 µM (BIM4) or 0 to 3,000 µM (okanin) were measured in triplicates as described above. A log-logistic dose response curve with three parameters (LL3) was globally fitted to the individual replicates using the R software collection v3.5.0 (R Core Team, 2018) and the R package drc v3.0-1 (Ritz et al., 2015).
Comparison of Cell Suspension and Leaf Cutting Assays
Samples of B. sinuspersici cell suspensions and Z. mays leaves were taken on different days over the course of 6 h resulting in three measurement series for each type of sample. B. sinuspersici cell suspensions were prepared once for each measurement series as described previously and kept on ice. Samples of Z. mays originating from the same leaf and sized approx. 2 cm2 were harvested from the plant immediately before each measurement. The midrib was removed before samples were further processed into slices of approx. 1 mm thickness and placed in the measurement chamber filled with 1 ml of buffer containing 0.33 M sorbitol, 2.5 mM MgCl2, 2.5 mM NaH2PO4, 25 mM HEPES/KOH (pH 7.5), 50 μM MnCl2, and 2.5 mM dithiothreitol (Burnell and Hatch, 1988; Haines et al., 2005). Sodium bicarbonate and sodium pyruvate were added at 4 mM to initiate C4-driven photosynthesis. POE rates were monitored as stated before Leaf slices were recovered from the chamber and transferred to 1 ml of 80 % acetone (v/v) for chlorophyll extraction. Samples in acetone were kept in the dark at 4°C for 48 h. Absorption was measured using a DU800 spectrophotometer (Beckman Coulter, USA) at 645 and 663 nm and the total chlorophyll amount was calculated as stated in equation 2 (Arnon, 1949). Chlorophyll content was subsequently used for normalization of the POE rates expressed in nmol min−1 μg chl−1.
| (2) |
Each measurement of a series was further normalized to the respective mean of the series. Data were processed with the R software collection v3.5.0 (R Core Team, 2018). Data were verified to be normally distributed using a Shapiro-Wilk’s test in combination with visual inspection of density and quantile-quantile plots. Variances of both sample types were statistically analyzed using an F-test with a significance level of α = 0.05.
Multiple Sequence Alignment
Protein sequences of PEPC and PPDK from several C4 plants were selected on the basis of sequence quality and availability of PPDK and PEPC sequences for each species. They were aligned using MAFFT v7.407 in L-INS-i mode with default parameters (Katoh and Standley, 2013). Sequences used for the alignments are (UniProt identifiers if not stated otherwise): Bs (B. sinuspersici, PEPC: GenBank ABG20459.1, PPDK: GenBank GCEP01054245.1/GCEP01053558.1), Ft (Flaveria trinervia, PEPC: P30694, PPDK: P22221), So (Saccharum officinarum, PEPC: Q9FS96, PPDK: Q9SNY6), Sb (Sorghum bicolor, PEPC: P15804, PPDK: Q84N32), Zm (Z. mays, PEPC: P04711, PPDK: P11155), Ah (Amaranthus hypochondriacus, PEPC: E2JE39, PPDK: F8UU33), Ec (E. crus-galli, PEPC: Q52NW0, PPDK: Q4JIY1), and Si (Setaria italica, PEPC: Q8S2Z8, PPDK: A0A290Y0Z5). Figures of multiple sequence alignment (MSA) were prepared using TeXshade (Beitz, 2000).
Homology Modeling and Ligand Docking
Ten homology models each of B. sinuspersici PEPC and PPDK were generated using MODELLER v9.21 (Webb and Sali, 2016). The models were ranked based on their normalized DOPE score (zDOPE) (Shen and Sali, 2006; Pieper et al., 2010) and the top-ranked model was used after visual inspection for docking of inhibitors into the proposed binding sites. Templates used for modeling of BsPEPC include PDB IDs 3ZGE, 3ZGB, 4BXH, 4BXC, 1JQO, 1QB4, and 5VYJ. Template selection for modeling of BsPPDK was limited to PDB ID 5JVL due to a high conformational diversity between the deposited structures. Ligands bound to the inhibitor and/or substrate binding sites were left in place during modeling to preserve side chain conformations and interactions within the binding sites.
Okanin and BIM4 were docked into the malate/aspartate binding site of BsPEPC or the nucleotide binding pocket of BsPPDK respectively. Charges and protonation state of the C4 enzymes were added by tools from the UCSF Chimera software suite (Pettersen et al., 2004). A total of 200 initial conformers of each compound, okanin and BIM4, were generated using RDKit (RDKit, 2019). Conformers with RMSDs below 0.1 Å were removed and the remaining set was used for docking using the AutoDock vina-derived software smina and the vinardo scoring function (Trott and Olson,2009; Koes et al., 2013). Side chains within a distance of 3 Å of the compound present in the original models (aspartate for BsPEPC and 2′-Br-dAppNHp for BsPPDK) were treated as being flexible during docking. The bounding box was sized 30 Å in each dimension and centered at the Cα atom of bound aspartate (BsPEPC) or the 4′-carbon atom of 2′-Br-dAppNHp (BsPPDK), respectively. Energy minimized docking poses were subsequently ranked according to their predicted binding affinities. Figures of molecular models were prepared using PyMOL (Schrödinger, 2017).
Results and Discussion
Photosynthetic Oxygen Production in SCC4 Cells Responds to Known C4 Inhibitors
The photosynthetic activity of B. sinuspersici chlorenchyma cell suspensions was determined by a Clark type oxygen electrode as described above. The yields of typical preparations were in the range of 1,000 to 7,200 intact cells per leaf judged by trypan blue staining. This results in a count of 50,000 to 360,000 intact cells used per measurement. During the initial incubation phase in the dark, a clear decrease in oxygen concentration due to electrode drift and mitochondrial respiration was observed. Upon illumination oxygen concentration in the electrode chamber increased due to photosynthetic activity of the cell suspension, allowing the analysis of C4 photosynthesis of B. sinuspersici cells in vivo. Optimal ranges of common control parameters such as pH, osmolarity, and temperature were identified allowing measurements of cell suspensions with stable oxygen rates for several hours. Thereby effects of known inhibitors of key enzymes of the C4 photosynthetic cycle can be tested at cellular conditions.
The two inhibitors analyzed in this study target different key enzymes of C4 photosynthesis. While the non-competitive inhibitor okanin binds to an allosteric regulatory site in PEPC, the competitive inhibitor BIM4 targets the nucleotide binding site of PPDK. Sequence alignments and phylogenetic analysis of both target enzymes across different C4 species including B. sinuspersici suggest that key motifs such as regulatory and active side residues are highly conserved. Hence, results obtained on B. sinuspersici cell suspension system presented here, may be transferred and generalized to other C4 species including typical weed species. Notwithstanding the observed conservation on sequence level, subtle differences across species — in particular for the allosteric feedback inhibition site of PEPC — have to be kept in mind.
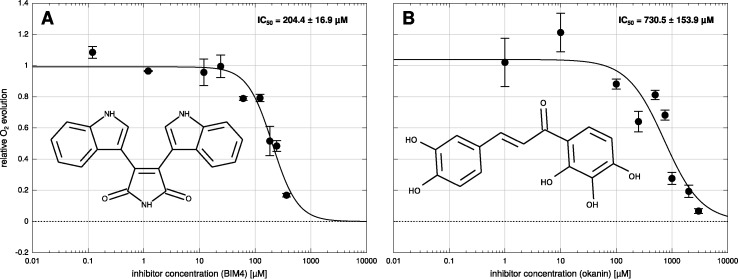
Measurements on isolated B. sinuspersici chlorenchyma cell suspensions show that both inhibitors significantly decrease photosynthetic oxygen production. For both compounds, inhibition is concentration dependent. From the corresponding activity curves ( Figure 2 ) IC50 of 204 ± 17 µM were obtained for BIM4 and 731 ± 154 µM for okanin. In contrast, IC50 concentrations in the upper nanomolar range have been reported for both inhibitors for the purified enzymes — precisely 0.8 ± 0.2 µM for BIM4 and 0.6 ± 0.1 µM for okanin (Nguyen et al., 2016; Minges et al., 2017). The IC50 concentrations observed under in vitro and in vivo conditions differ by almost three orders of magnitude which is probably owed to uptake limitations across the cell membrane and/or plant related metabolization of the active compounds. For okanin the higher IC50 observed in the in vivo studies on plant cell suspension might also issue from sequence variation in a key residue of the allosteric binding site. In B. sinuspersici this residue at position 884 (Flaveria numbering) corresponds to arginine, whereas the Flaveria C4 model used in the in vitro studies (Paulus et al., 2013a; Paulus et al., 2014; Nguyen et al., 2016) carries a glycine at this position allowing unrestricted access to the allosteric binding site.
Figure 2.
Dose response curve of Bienertia sinuspersici cell suspension. Bisindolylmaleimide IV (BIM4) (A) or okanin (B) were added in nine different concentrations to the cell suspension and photosynthetic oxygen evolution was measured. IC50 values were calculated by a globally fitting a three parameter log-logistic dose response curve to the individual data points. Errors shown are standard errors of the mean (SEM).
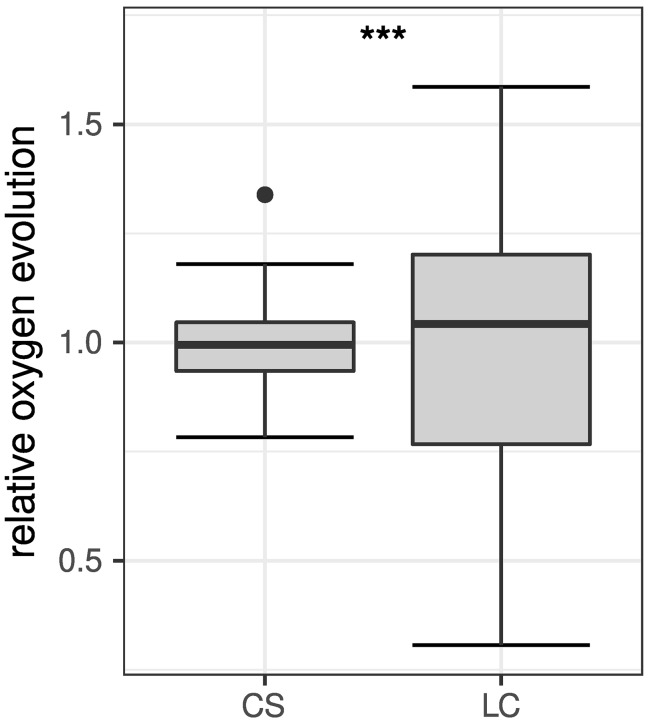
To explore whether the cell suspension assay is able to yield more accurate data compared to leaf cutting assays, measurements of POE were repeated on different days over the course of 6 h as described in as described in Material and Methods For better comparability, POE rates obtained from both methods were normalized to the respective mean of each measurement series ( Figure 3 ). Analysis of variances for both methods clearly shows a narrower distribution of data for the cell suspension assay [standard deviation (sd) = 0.12] than for the leaf slice assay (sd = 0.33). Statistical analysis using an F-test suggest that the observed difference in variances between both methods is highly significant (F = 0.12332, p = 6.998 · 10−7).
Figure 3.
Comparison of relative POE obtained from cell suspension (CS, n = 30) and leaf cutting (LC, n = 21) assays. Data was normalized to the respective mean of each measurement series for better comparability and are shown as boxplots (Tukey 1977). The box represents the interquartile range (IQR) while the median is depicted by a solid line. Whiskers include data within 1.5 × IQR. Data points beyond this range are shown individually depicted by a filled circle (•). The difference between variances of both datasets is statistically highly significant (***p ≤ 0.001) according to F-test results (F = 0.12332, p = 6.988 · 10−7).
Structural Comparison of PEPC and PPDK Inhibitor Binding Sites Among Different C4 Species
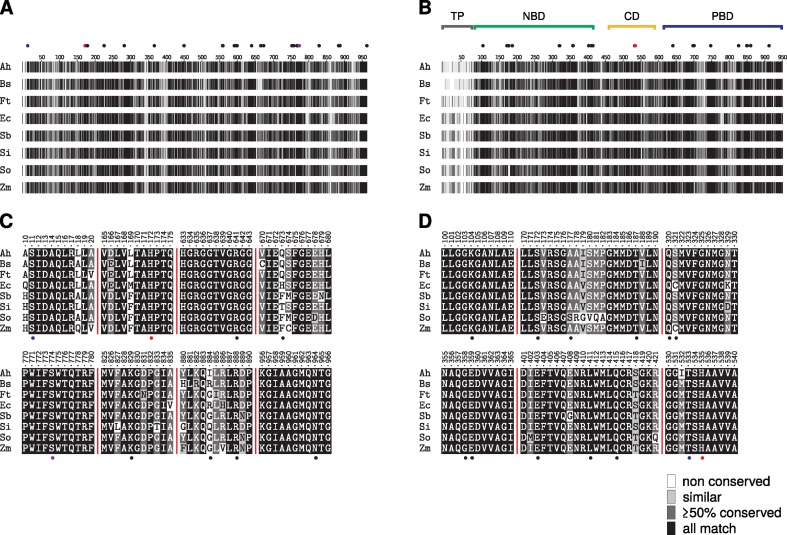
A critical prerequisite to generalize and transfer findings obtained with the B. sinuspersici cell suspension assay presented in this study is that key enzymes and mechanisms in the SCC4 plant function in a similar manner as in conventional Kranz-type C4 plants. To identify possibly meaningful differences on sequence level, a MSA of the two key enzymes of C4 photosynthesis — PEPC and PPDK — was computed for a range of C4 species ( Figure 4 ). The obtained MSA (see Supplementary Material) confirms that primary sequences of both C4 enzymes are highly conserved between the analyzed C4 plants (Figures 4A, B) in particular their substrate and effector binding sites or their catalytic core ( Figures 4C, D). For PPDK, substrate binding can be allocated to two distinct domains: the N-terminal nucleotide-binding domain (NBD) and the C-terminal PEP/pyruvate-binding domain ( Figure 4B ). Both are separated by a flexible central domain which houses a regulatory threonine and a catalytic histidine residue (Carroll et al., 1994; Herzberg et al.,1996). Previous studies indicated that the PPDK inhibitor BIM4 binds to the NBD as an ATP competitive inhibitor (Minges and Groth, 2017). Consequently, the analysis of the PPDK MSA was focused on regions and residues directly involved in nucleotide binding ( Figure 4D ).
Figure 4.
Multiple sequence alignment of phosphoenolpyruvate carboxylase (PEPC) (A, C) and pyruvate phosphate dikinase (PPDK) (B, D) sequences of representative C4 plants. The complete alignment is visualized as a condensed fingerprint in (A and B). Sequence similarity according to BLOSUM62 substitution matrix is depicted as grayscale gradient, ranging from black (full match) to white (not conserved). Sequence numbering corresponds to Flaveria trinervia (Ft) PEPC and PPDK. The substrate binding domains of PPDK (nucleotide: NBD, PEP/pyruvate: PBD), its catalytic central domain (CD). and the non-conserved chloroplastic transit peptide (TP) are marked as bars above the fingerprint (B). Cut-outs of regions important for inhibitor binding or catalytic activity are shown in panels C (PEPC) and D (PPDK) respectively. Residues involved in substrate or effector binding are depicted as solid black dots while catalytic residues are colored in red. Residues marked with blue dots are of regulatory function. S774 of PEPC (A, C) depicted in purple has been described as a specific marker for C4-type PEPCs (Svensson et al., 2003). All highlighted positions are located in highly conserved regions with the exception of positions 673 and 884 in PEPC (C). Full-length sequence alignment is available from the Supplementary Material .
While residues are fully conserved at the PPDK nucleotide binding site and the PEPC C-terminal tetrapeptide needed for optimal catalytic activity (Dong et al., 1999; Kai et al., 2003) the MSA indicates obvious variance at positions 673 and 884 (F. trinervia numbering) for PEPC. This region is part of the PEPC allosteric aspartate/malate inhibitor site and is likewise involved in binding the inhibitor okanin (Nguyen et al., 2016). Although, previous studies disclosed that residues at position 673 contribute to the binding of PEPC feedback inhibitors aspartate and malate (Paulus et al., 2013b), no particular role of those residues for C4-typical kinetics has been demonstrated yet. Notably position 884 has been demonstrated in previous in vitro studies of critical importance to modulate the binding affinity of C4 acids malate and aspartate (Paulus et al., 2013a; Paulus et al., 2013b). C3-type PEPCs have a conserved R884 at this position that further stabilizes aspartate/malate binding at the inhibitor site. Crystallographic studies in combination with mutational analysis and activity studies have shown that substitution of R884 by glycine significantly decreased the binding affinity of PEPC for aspartate and malate and thereby the sensitivity of PEPC toward feedback inhibition (Paulus et al., 2013a; Paulus et al., 2013b). Many C4-type PEPCs carry glycine at position 884, but also serine, glutamine, and glutamate are found for C4-type PEPCs at this position which also provide decreased feedback inhibition (Paulus et al., 2013a). Previous studies suggest that a bulky amino acid such as arginine at position 884 interferes with the binding of the C4-specific inhibitor okanin (Paulus et al., 2014; Nguyen et al., 2016). Remarkably enough, the C3-typical arginine is also found at position 884 in a range of C4 PEPCs particularly in the C4 grass lineages as indicated by the MSA ( Figure 4C ). Though, it is not clear yet whether other mutations at the allosteric inhibitor site counteract the higher binding affinity for C4 acids aspartate/malate caused by R884 for these enzymes. Moreover it has been hypothesized that C4 characteristics may rather not be determined by the presence of C4-typical amino acids, but by the absence of non-C4 ones at certain positions at the active or allosteric sites (Christin et al., 2007). Along the same lines, in recent studies, Rosnow et al. (2014; 2015) compared PEPC sequences in the C4 Suaedoideae clade and did not notice any convergence in amino acid substitutions at the allosteric feedback inhibitor binding site. However, they observed a convergence of kinetic parameters such as a lower PEP affinity of C4 PEPCs compared to their C3 isoforms which presumably allows for the accumulation of higher PEP concentrations in the C4 Suaedoideae (Bläsing et al., 2000).
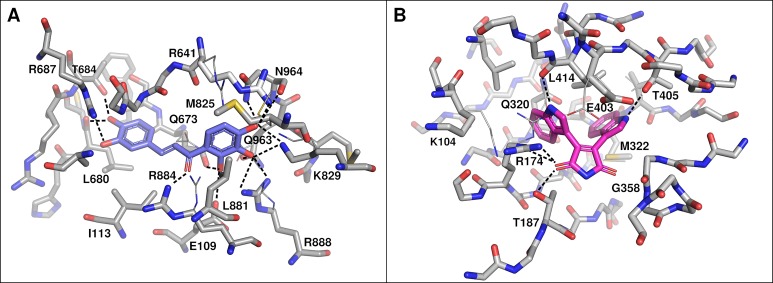
To verify the possibility of okanin binding to BsPEPC despite containing an arginine at position 884 and to verify the proposed binding mode of BIM4 within the nucleotide binding site of BsPPDK, homology models of both enzymes were calculated. Combined with virtual docking of okanin and BIM4, these models substantiated the structural similarity of the two key enzymes of C4 photosynthesis in B. sinuspersici compared to those of other C4 plants. In its best-scored docking pose, okanin forms polar contacts to the side chains of E109, R641, T684, R687, K829, R884, and N964 (Figure 5A ). In this context, the involvement of R884 is unexpected as it is thought to be the key switch in mediating sensitivity of C4 PEPCs toward their feedback inhibitors aspartate and malate as well as for the selective binding of okanin to C4 PEPCs (Paulus et al., 2013a; Paulus et al., 2013b; Nguyen et al., 2016). In its orientation resolved in the crystal structures of model C4 PEPC from F. trinervia, R884 indeed provides substantial steric hindrance and thus may shield the site from okanin binding (Paulus et al., 2013b; Schlieper et al., 2014). Interestingly though, our recent docking studies on the BsPEPC homology model suggest that the R884 side chain is displaced upon okanin binding. The observed side chain flexibility might account for the inhibitory effects of okanin on PEPC observed in the in vitro (Nguyen et al., 2016) and in vivo experiments presented in this study.
Figure 5.
Docked binding poses of okanin (A) and bisindolylmaleimide IV (BIM4) (B). Residues within 3 Å of the compound are labeled (Flaveria trinervia nomenclature) and polar interactions are shown as dashed lines. Side chain conformations after docking are shown as sticks. For side chains that have been displaced during flexible docking, the original conformation is shown in line representation. In case of okanin and BsPEPC (A), residues M825, R641, and R884 assumed a substantially different side chain conformation. However, only the original orientation of R884 would have been of steric hindrance. In case of BIM4 and BsPPDK, residues K104, R174, and E403 have been displaced with the original side chain orientation of the latter one being of steric hindrance to BIM4 binding.
For BsPPDK, docking results of BIM4 are largely congruent with those previously obtained for F. trinervia PPDK (Minges and Groth, 2017) with predicted binding affinities up to −50.208 kJ mol−1 (−12.0 kcal mol−1). The docking suggests that BIM4 is bound via polar side chain contacts to R174, T187, and T405 ( Figure 5B ). Similar to the situation observed for okanin, individual side chains (R174 and E403) are shifted upon inhibitor binding in comparison to their initial conformation in the homology models.
Conclusion
Suspensions of intact B. sinuspersici chlorenchyma cells provide an easy to use, but yet powerful system to study the selective effect of small molecule inhibitors on C4 photosynthesis in a plant context. Compared to measurements on leaf cuttings the Bienertia system is easier in handling, higher in performance and has better reproducibility. The homogeneous Bienertia cell suspensions used in this set-up ensure an uniform distribution of compounds and constant illumination across the sample. Moreover, inhibitor uptake from the apoplast, but also the cell membrane are still valid in this system as isolated cells have intact cell membranes. The cell suspension assay itself is easily scalable. The assay volume per measurement is limited only by the type of electrode chamber. Setups with smaller or larger volumes are possible as well as parallel measurements using multiple Clark-type electrodes or fluorescence-based assays in a 96 well scale (Deshpande et al., 2004; Arain et al., 2005). Altogether, the in vivo approach presented in this study expands our methodical repertoire to further study the molecular processes of C4 photosynthesis and to foster the development of C4-specific inhibitors.
Data Availability Statement
The datasets generated and analyzed for this study can be found in the Zenodo repository doi: 10.5281/zenodo.2635517.
Author Contributions
GG designed research. SO and AM contributed to experimental design. DJ performed cell preparation and oxygen measurements. DJ and AM performed data analysis. AM performed bioinformatical analysis. AM, DJ and GG wrote the manuscript. All authors read and edited the manuscript prior to publication.
Funding
Funded in part by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany´s Excellence Strategy — EXC 2048/1 — Project ID: 390686111. Work in the S.O. lab was funded by the “Cooperative Research Program for Agriculture Science & Technology Development (Project No. 0109532016)”. Rural Development Administration, Republic of Korea.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Computational support and infrastructure was provided by the Centre for Information and Media Technology†(ZIM) at the University of Düsseldorf (Germany).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.01350/full#supplementary-material
References
- Akhani H., Barroca J., Koteeva N., Voznesenskaya E., Franceschi V., Edwards G., et al. (2005). Bienertia sinuspersici (chenopodiaceae): a new species from southwest asia and discovery of a third terrestrial c plant without kranz anatomy. Syst. Bot. 30, 290–301. 10.1600/0363644054223684 [DOI] [Google Scholar]
- Arain S., Weiss S., Heinzle E., John G. T., Krause C., Klimant I. (2005). Gas sensing in microplates with optodes: Influence of oxygen exchange between sample, air, and plate material. Biotechnol. Bioeng. 90, 271–280. 10.1002/bit.20348 [DOI] [PubMed] [Google Scholar]
- Arnon D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15. 10.1104/pp.24.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey D. C., Buckley B. P., Chernov M. V., Gulick A. M. (2018). Development of a high-throughput biochemical assay to screen for inhibitors of aerobactin synthetase IucA. Advancing Life Sci. R.D. 23, 1070–1082. 10.1177/2472555218787140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beitz E. (2000). TeXshade: shading and labeling of multiple sequence alignments using LaTeX2e. Bioinf. 16, 135–139. 10.1093/bioinformatics/16.2.135 [DOI] [PubMed] [Google Scholar]
- Bläsing O. E., Westhoff P., Svensson P. (2000). Evolution of C4 phosphoenolpyruvate carboxylase in Flaveria, a conserved serine residue in the carboxyterminal part of the enzyme is a major determinant for C4-specific characteristics. J. Biol. Chem. 275, 27917–27923. 10.1074/jbc.M909832199 [DOI] [PubMed] [Google Scholar]
- Burgos N. R. (2015). Whole-plant and seed bioassays for resistance confirmation. Weed Sci. 63, 152–165. 10.1614/WS-D-14-00019.1 [DOI] [Google Scholar]
- Burnell J., Hatch M. (1988). Photosynthesis in phosphoenolpyruvate carboxykinase-type C4 plants: Photosynthetic activities of isolated bundle sheath cells from Urochloa panicoides. Arch Biochem. Biophys. 260, 177–186. 10.1016/0003-9861(88)90439-0 [DOI] [PubMed] [Google Scholar]
- Busi R., Goggin D. E., Heap I. M., Horak M. J., Jugulam M., Masters R. A., et al. (2018). Weed resistance to synthetic auxin herbicides. Pest. Manage.Sci. 74, 2265–2276. 10.1002/ps.4823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carboni M., Guéguen M., Barros C., Georges D., Boulangeat I., Douzet R., et al. (2017). Simulating plant invasion dynamics in mountain ecosystems under global change scenarios. Global Change Biol. 24, e289–e302. 10.1111/gcb.13879 [DOI] [PubMed] [Google Scholar]
- Carroll L. J., Xu Y., Thrall S. H., Martin B. M., Dunaway-Mariano D. (1994). Substrate binding domains in pyruvate phosphate dikinase. Biochem. 33, 1134–1142. 10.1021/bi00171a012 [DOI] [PubMed] [Google Scholar]
- Christin P.-A., Salamin N., Savolainen V., Duvall M. R., Besnard G. (2007). C4 Photosynthesis Evolved in Grasses via Parallel Adaptive Genetic Changes. Curr. Biol. 17, 1241–1247. 10.1016/j.cub.2007.06.036 [DOI] [PubMed] [Google Scholar]
- Clark L. C. (1956). Monitor and control of blood and tissue oxygen tension. Trans.Am. Soc. Artif. Intern Organs 2, 41–48. [Google Scholar]
- Delieu T., Walker D. A. (1972). An improved cathode for the measurement of photosynthetic oxygen evolution by isolated chloroplasts. New Phytol 71, 201–225. 10.1111/j.1469-8137.1972.tb04068.x [DOI] [Google Scholar]
- Deshpande R. R., Wittmann C., Heinzle E. (2004). Microplates with integrated oxygen sensing for medium optimization in animal cell culture. Cytotechnology 46, 1–8. 10.1007/s10616-004-6401-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick M., Erlenkamp G., Nguyen G. T., Förster K., Groth G., Gohlke H. (2017). Pyrazolidine-3, 5-dione-based inhibitors of phosphoenolpyruvate carboxylase as a new class of potential C4plant herbicides. FEBS Lett. 591, 3369–3377. 10.1002/1873-3468.12842 [DOI] [PubMed] [Google Scholar]
- Dong L., Patil S., Condon S. A., Haas E. J., Chollet R. (1999). The Conserved C-Terminal Tetrapeptide of Sorghum C4 Phosphoenolpyruvate Carboxylase Is Indispensable for Maximal Catalytic Activity, but Not for Homotetramer Formation. Arch Biochem. Biophys. 371, 124–128. 10.1006/abbi.1999.1433 [DOI] [PubMed] [Google Scholar]
- Doyle J. R., Burnell J. N., Haines D. S., Llewellyn L. E., Motti C. A., Tapiolas D. M. (2005). A rapid screening method to detect specific inhibitors of pyruvate orthophosphate dikinase as leads for C4 plant-selective herbicides. J. Biomol. Screening 10, 67–75. 10.1177/1087057104269978 [DOI] [PubMed] [Google Scholar]
- Feng B. Y., Shelat A., Doman T. N., Guy R. K., Shoichet B. K. (2005). High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148. 10.1038/nchembio718 [DOI] [PubMed] [Google Scholar]
- Fernando N., Manalil S., Florentine S. K., Chauhan B. S., Seneweera S. (2016). Glyphosate Resistance of C3 and C4 Weeds under Rising Atmospheric CO2. Front Plant Sci. 7, 910. 10.3389/fpls.2016.00910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag H., Stichler W. (2002). Bienertia cycloptera Bunge ex Boiss., chenopodiaceae, another C4 Plant without Kranz Tissues. Plant Biol. 4, 121–132. 10.1055/s-2002-20444 [DOI] [Google Scholar]
- Haines D. S., Burnell J. N., Doyle J. R., Llewellyn L. E., Motti C. A., Tapiolas D. M. (2005). Translation of in Vitro Inhibition by Marine Natural Products of the C4 Acid Cycle Enzyme Pyruvate P(i) Dikinase to in vivo C4 Plant Tissue Death. J. Agric. Food Chem. 53, 3856–3862. 10.1021/jf048010x [DOI] [PubMed] [Google Scholar]
- Haraguchi H., Ishikawa H., Mizutani K., Tamura Y., Kinoshita T. (1998). Antioxidative and superoxide scavenging activities of retrochalcones in Glycyrrhiza inflata. Bioorg. Med. Chem. 6, 339–347. 10.1016/S0968-0896(97)10034-7 [DOI] [PubMed] [Google Scholar]
- Heap I. (2014). Herbicide Resistant Weeds. Dordrecht: Springer Neth. 3, 281–301. 10.1007/978-94-007-7796-5 [DOI] [Google Scholar]
- Heap I. (2019). The international survey of herbicide resistant weeds. Online. Internet. http://www.weedscience.org, Online; accessed.
- Herzberg O., Chen C. C., Kapadia G., McGuire M., Carroll L. J., Noh S. J., et al. (1996). Swiveling-domain mechanism for enzymatic phosphotransfer between remote reaction sites. Proc. NatAcad Sci 93, 2652–2657. 10.1073/pnas.93.7.2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen R. G., Bassham J. A. (1966). Photosynthesis by isolated chloroplasts. Proc. NatAcad Sci 56, 1095–1101. 10.1073/pnas.56.4.1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H., Osmond C. (1973). Photosynthesis by thin leaf slices in solution i. properties of leaf slices and comparison with whole leaves. Aust. J. Biol. Sci. 26, 15. 10.1071/BI9730015 [DOI] [Google Scholar]
- Jones H. G., Slatyer R. O. (1972). Estimation of the transport and carboxylation components of the intracellular limitation to leaf photosynthesis. Plant Physiol. 50, 283–288. 10.1104/pp.50.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kai Y., Matsumura H., Izui K. (2003). Phosphoenolpyruvate carboxylase: three-dimensional structure and molecular mechanisms. Arch Biochem. Biophys. 414, 170–179. 10.1016/S0003-9861(03)00170-X [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 30, 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes D. R., Baumgartner M. P., Camacho C. J. (2013). Lessons Learned in Empirical Scoring with smina from the CSAR 2011 Benchmarking Exercise. J. Chem. Inf. Model. 53, 1893–1904. 10.1021/ci300604z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara M. V., Chuong S. D., Akhani H., Andreo C. S., Edwards G. E. (2006). Species having C4 single-cell-type photosynthesis in the chenopodiaceae family evolved a photosynthetic phosphoenolpyruvate carboxylase like that of kranz-type C4 species. Plant Physiol. 142, 673–684. 10.1104/pp.106.085829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lara M. V., Offermann S., Smith M., Okita T. W., Andreo C. S., Edwards G. E. (2008). Leaf development in the single-cell C4 system in bienertia sinuspersici: expression of genes and peptide levels for C4 metabolism in relation to chlorenchyma structure under different light conditions. Plant Physiol. 148, 593–610. 10.1104/pp.108.124008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein W., Börnke F., Reindl A., Ehrhardt T., Stitt M., Sonnewald U. (2004). Target-based discovery of novel herbicides. Curr Opin. Plant Biol. 7, 219–225. 10.1016/j.pbi.2004.01.001 [DOI] [PubMed] [Google Scholar]
- Leisner C. P., Cousins A. B., Offermann S., Okita T. W., Edwards G. E. (2010). The effects of salinity on photosynthesis and growth of the single-cell C4 species Bienertia sinuspersici (Chenopodiaceae). Photosynth. Res. 106 (3), 201–214. 10.1007/s11120-010-9595-z [DOI] [PubMed] [Google Scholar]
- Minges A., Ciupka D., Winkler C., Höppner A., Gohlke H., Groth G. (2017). Structural intermediates and directionality of the swiveling motion of pyruvate phosphate dikinase. Sci. Rep. 7, 45389. 10.1038/srep45389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minges A., Groth G. (2017). Small-molecule inhibition of pyruvate phosphate dikinase targeting the nucleotide binding site. Plos One 12, e0181139. 10.1371/journal.pone.0181139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motti C. A., Bourne D. G., Burnell J. N., Doyle J. R., Haines D. S., Liptrot C. H., et al. (2007). Screening marine fungi for inhibitors of the C4 plant enzyme pyruvate phosphate dikinase: unguinol as a potential novel herbicide candidate. Appl. Environ. Microbiol. 73, 1921–1927. 10.1128/AEM.02479-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen G. T. T., Erlenkamp G., Jäck O., Küberl A., Bott M., Fiorani F., et al. (2016). Chalcone-based Selective Inhibitors of a C4 Plant Key Enzyme as Novel Potential Herbicides. Sci. Rep. 6, 27333. 10.1038/srep27333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Offermann S., Friso G., Doroshenk K. A., Sun Q., Sharpe R. M., Okita T. W., et al. (2015). Developmental and Subcellular Organization of Single-Cell C4 Photosynthesis in Bienertia sinuspersici Determined by Large-Scale Proteomics and cDNA Assembly from 454 DNA Sequencing. J. Proteome Res. 14, 2090–2108. 10.1021/pr5011907 [DOI] [PubMed] [Google Scholar]
- Offermann S., Okita T. W., Edwards G. E. (2011). Resolving the Compartmentation and Function of C4 Photosynthesis in the Single-Cell C4 Species Bienertia sinuspersici. Plant Physiol. 155, 1612–1628. 10.1104/pp.110.170381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olofsdotter M., Olesen A., Andersen S. B., Streibig J. C. (1994). A comparison of herbicide bioassays in cell cultures and whole plants. Weed Res. 34, 387–394. 10.1111/j.13653180.1994.tb02034.x [DOI] [Google Scholar]
- Park J., Okita T. W., Edwards G. E. (2010). Expression profiling and proteomic analysis of isolated photosynthetic cells of the non-Kranz C4 species Bienertia sinuspersici. Funct. Plant Biol. 37, 1. 10.1071/FP09074 [DOI] [Google Scholar]
- Paulus J. K., Förster K., Groth G. (2014). Direct and selective small-molecule inhibition of photosynthetic PEP carboxylase: New approach to combat C4 weeds in arable crops. FEBS Lett. 588, 2101–2106. 10.1016/j.febslet.2014.04.043 [DOI] [PubMed] [Google Scholar]
- Paulus J. K., Niehus C., Groth G. (2013. a). Evolution of C4 phosphoenolpyruvate carboxylase: enhanced feedback inhibitor tolerance is determined by a single residue. Mol. Plant 6, 1996–1999. 10.1093/mp/sst078 [DOI] [PubMed] [Google Scholar]
- Paulus J. K., Schlieper D., Groth G. (2013. b). Greater efficiency of photosynthetic carbon fixation due to single amino-acid substitution. Nat. Commun. 4, 1518. 10.1038/ncomms2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., et al. (2004). UCSF chimera - a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Pieper U., Webb B. M., Barkan D. T., Schneidman-Duhovny D., Schlessinger A., Braberg H., et al. (2010). ModBase, a database of annotated comparative protein structure models, and associated resources. Nucleic Acids Res. 39, D465–D474. 10.1093/nar/gkq1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team (2018). R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- Rathmell W. G., Bendall D. S. (1972). The peroxidase-catalysed oxidation of a chalcone and its possible physiological significance. Biochem. J. 127, 125–132. 10.1042/bj1270125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RDKit (2019). RDKit: Open-source cheminformatics. [Online; accessed].
- Ritz C., Baty F., Streibig J. C., Gerhard D. (2015). Dose-response analysis using R. Plos One 10, e01460210146021. 10.1371/journal.pone.0146021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow J. J., Edwards G. E., Roalson E. H. (2014). Positive selection of kranz and non-kranz c4 phosphoenolpyruvate carboxylase amino acids in suaedoideae (chenopodiaceae). J. Exp. Bot. 65, 3595–3607. 10.1093/jxb/eru053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosnow J. J., Evans M. A., Kapralov M. V., Cousins A. B., Edwards G. E., Roalson E. H. (2015). Kranz and single-cell forms of C4 plants in the subfamily Suaedoideae show kinetic C4 convergence for PEPC and Rubisco with divergent amino acid substitutions. J. Exp. Bot. 66, 7347–7358. 10.1093/jxb/erv431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage R. F. (2002). C4 photosynthesis in terrestrial plants does not require Kranz anatomy. Trends Plant Sci. 7, 283–285. 10.1016/S1360-1385(02)02293-8 [DOI] [PubMed] [Google Scholar]
- Schlieper D., Förster K., Paulus J. K., Groth G. (2014). Resolving the activation site of positive regulators in plant phosphoenolpyruvate carboxylase. Mol. Plant 7, 437–440. 10.1093/mp/sst130 [DOI] [PubMed] [Google Scholar]
- Schrödinger L. L. C. (2017). The PyMOL molecular graphics system, version 2.0 PyMOL.
- Shen M., Sali A. (2006). Statistical potential for assessment and prediction of protein structures. Protein Sci 15, 2507–2524. 10.1110/ps.062416606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimabukuro R. H. (1985). “Detoxication of herbicides,” in Weed Physiology, vol. 2 (S. O. Duke: CRC Press; ), 215–240. Herbicide Physiology. 10.1201/9781351077736-9 [DOI] [Google Scholar]
- Svensson P., Bläsing O. E., Westhoff P. (2003). Evolution of C4 phosphoenolpyruvate carboxylase. Arch Biochem. Biophys. 414, 180–188. 10.1016/S0003-9861(03)00165-6 [DOI] [PubMed] [Google Scholar]
- Tennant J. R. (1964). Evaluation of the trypan blue technique for determination of cell viability. Transplantation 2, 685–694. 10.1097/00007890-196411000-00001 [DOI] [PubMed] [Google Scholar]
- Tice C. M. (2001). Selecting the right compounds for screening: does lipinski’s rule of 5 for pharmaceuticals apply to agrochemicals? Pest Manage.Sci. 57, 3–16. [DOI] [PubMed] [Google Scholar]
- Trott O., Olson A. J. (2009). AutoDock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. NA-NA. 1st edn 31, 455–461. 10.1002/jcc.21334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey J. W. (1977). Exploratory Data Analysis (Reading, MA: Addison-Wesley; ), [Google Scholar]
- Voznesenskaya E. V., Franceschi V. R., Kiirats O., Artyusheva E. G., Freitag H., Edwards G. E. (2002). Proof of C4 photosynthesis without Kranz anatomy in Bienertia cycloptera (Chenopodiaceae). Plant J. 31, 649–662. 10.1046/j.1365-313X.2002.01385.x [DOI] [PubMed] [Google Scholar]
- Voznesenskaya E. V., Franceschi V. R., Kiirats O., Freitag H., Edwards G. E. (2001). Kranz anatomy is not essential for terrestrial C4 plant photosynthesis. Nat. 414, 543–546. 10.1038/35107073 [DOI] [PubMed] [Google Scholar]
- Waryszak P., Lenz T. I., Downey P. O. (2018). Herbicide effectiveness in controlling invasive plants under elevated CO2: sufficient evidence to rethink weeds management. J. Environ. Manage. 226, 400–407. 10.1016/j.jenvman.2018.08.050 [DOI] [PubMed] [Google Scholar]
- Webb B., Sali A. (2016). Comparative protein structure modeling using modeller. Curr. Protoc. Bioinf. 54, 5.6.1–5.6.37. 10.1002/cpbi.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and analyzed for this study can be found in the Zenodo repository doi: 10.5281/zenodo.2635517.