Abstract Abstract
Many species of Ganoderma exhibit a high phenotypic plasticity. Hence, particularly among them, the morphological species concept remains difficult to apply, resulting in a currently confused taxonomy; as a consequence, the geographical distribution range of many species also remains very uncertain. One of the areas with a strong uncertainty, as far as morphological species concept is concerned, is the Neotropics. It is common that names of species described from other regions, mainly from northern temperate areas, have been applied to Neotropical species. The aim of the present study was to determine which species might lay behind the G. weberianum complex in the Neotropics, using morphological studies and phylogenetic inferences based on both single (ITS) and multilocus (ITS, rpb2, and tef1-α) sequences. The results indicated that G. weberianumsensu Steyaert, which is the usually accepted concept for this taxon, was absent from the Neotropics. In this area, G. weberianumsensu Steyaert encompassed at least two phylogenetic species, which are tentatively, for the time being, identified as belonging to G. mexicanum and G. parvulum. These two species could be distinguished morphologically, notably by the ornamentation or its absence on their chlamydospores. The results also showed that additional species from the Neotropics might still exist, including, e.g., G. perzonatum, but their circumscription remains uncertain until now because of the paucity of material available. Furthermore, it was found that the current concept of G. resinaceum embraced a complex of species.
Keywords: Caribbean, Chlamydospores, Fomes weberianus, Ganodermataceae , Paleotropics, South America
Introduction
Ganoderma P. Karst. has always been considered as an extremely difficult group with many poorly circumscribed species, forming species complexes (Moncalvo and Ryvarden 1997). Early in the 20th century, Lloyd (1905) already emphasized the excessively confused taxonomy of Ganoderma stating “these fungi have been described and named over and over again, until the literature has become an almost unfathomable maze of meaningless and conflicting names”.
A century later, one can deduce that the situation has improved very little, if at all. Ryvarden (1991), for instance, still concluded that the taxonomic issue of the genus worldwide was very “chaotic”. Hitherto, there is no comprehensive Ganoderma study and the absence of a world monograph contributed to “problems with species circumscriptions and identification”, fideMoncalvo (2000).
Nowadays, about 220 species have been described in Ganoderma, over 400 taxa if one includes varieties, of which 167 apply to the so-called laccate species (Ryvarden 1991, Moncalvo and Ryvarden 1997, Index Fungorum http://www.indexfungorum.org/names/names.asp). Nonetheless, estimations based on the identification of terminal clades shown by phylogenetic analysis of a large ITS sequence data set gave a range of 60–80 terminal clades or phylogenetic species within the “laccate” Ganoderma spp. and 10–30 within the “non-laccate” Ganoderma spp. (Moncalvo 2000). Over the past two decades, phylogenetic studies have tried to elucidate the status of certain species and to better circumscribe their geographic distribution (e.g., Moncalvo 2000, Wang et al. 2009, Yao et al. 2013, de Lima-Junior et al. 2014, Zhou et al. 2014, Hapuarachchi et al. 2015, Loyd et al. 2018). However, the real species number and their distribution range remain largely unknown (Moncalvo and Ryvarden 1997, Moncalvo 2000).
Alliances of taxa, taxonomically informal but morphologically homogeneous and phylogenetically (variably) supported, also have been evidenced within Ganoderma (e.g., Moncalvo 2000, Hong and Jung 2004). Moncalvo (2000), for instance, as a result of phylogenetic analyses based on, so far, the most comprehensive ITS DNA sequences data set, and morphological characters, identified three core groups (1–3) and a bunch of residual species of uncertain affinities. The three core groups were furthermore divided into several subgroups. The core group 1 included most of the laccate species, and was divided into G. curtisii, G. lucidum, G. resinaceum, and G. tropicum lineages (Moncalvo 2000).
The G. resinaceum lineage (subgroup 1.2, Moncalvo 2000) comprised species having laccate pileus, basidiospores with “extremely fine ornament” (Pegler and Young 1973), and chlamydospores formed in their basidiomes and in pure cultures on artificial media. In this lineage, Moncalvo (2000) mentioned “genetically isolated populations”, from North and South America and the Old-World that could be equated to as many species or species complexes. Moncalvo (2000) also suggested that the G. weberianum complex would represent the tropical Asian “counterpart” of the northern temperate G. resinaceum complex.
Ganoderma weberianum (Bres. & Henn. ex Sacc.) Steyaert was established by Steyaert (1972) based on Fomes weberianus Bres. & Henn. ex Sacc. (Saccardo 1891). Steyaert (1972) built the description of this species on a presumed type specimen held in B, the type specimen of G. rivulosum Pat. & Har. (Patouillard and Hariot 1906), a name that he considered as a synonym, and numerous specimens from Africa and Southeast Asia. In his description, Steyaert (1972) emphasized the importance of chlamydospores or “gasterospores”, both the morphology and abundance of which were variable between specimens, being mainly double-walled, smooth or ornamented with “cristae” or “columns”, and scarce to extremely abundant. Steyaert (1972) then informally recognized two morphotypes within his concept of G. weberianum, characterized by singular combinations of cuticular cells length and abundance of chlamydospores.
Since then, the distribution range of G. weberianumsensu Steyaert remained uncertain as exposed by the following authors. Steyaert (1972) reported the species from Africa and Southeast Asia and suggested that it was “probably extant in tropical America”. Subsequently, the species was reported from all tropical areas (e.g., Corner 1983, Quanten 1997, Pan and Dai 2001, Wang et al. 2005, Mohanty et al. 2011, Kinge et al. 2012), including the Neotropics (Torres-Torres et al. 2012, 2015, Manzano et al. 2013, López-Peña et al. 2016), up to South-eastern USA (Loyd et al. 2017, 2018). Nevertheless, Moncalvo (2000) suggested that Steyaert’s concept should be narrowed to include specimens originating only from Southeast Asia, in addition to the type locality in Samoa (G. weberianumsensu Moncalvo). Smith and Sivasithamparam (2000, 2003) corroborated this distribution range, including also Australia. With regards to the Neotropics, Moncalvo (2000) found an isolated branch within the G. weberianum complex, which he tentatively identified as G. subamboinense Bazzalo & J.E. Wright ex Moncalvo & Ryvarden. Welti and Courtecuisse (2010) also reported G. subamboinense from the Lesser Antilles and suggested reassessing G. subamboinense var. laevisporum Bazzalo & J.E. Wright.
In the present study, we analyzed the status of G. weberianumsensu Steyaert in the Neotropics and, in particular, the statuses of G. subamboinense and G. subamboinense var. laevisporum. We also investigated through multilocus phylogenetic analysis, their phylogenetic relationships with specimens or species of the G. weberianum complex from other biogeographic zones.
Materials and methods
Studied materials
For this study, specimens from B, BAFC, BPI, ENCB, FH, IBUG, INBIO, MUCL, NY, S, and XAL herbaria (abbreviations follow Thiers, continuously updated), including the type specimens of Fomes weberianus, Ganoderma argillaceum Murrill, G. mexicanum Pat., G. microsporum R.S. Hseu, G. parvulum Murrill, G. perturbatum (Lloyd) Torrend, G. perzonatum Murrill, G. pulverulentum Murrill, G. rivulosum, G. sessiliforme Murrill, G. stipitatum (Murrill) Murrill, G. subamboinense var. subamboinense, G. subamboinense var. laevisporum, G. subincrustatum Murrill, and G. vivianimercedianum M. Torres were re-examined. Strains examined during this study were deposited at CBS, CIRM-CF, and BCCM/MUCL. The formation of chlamydospores was examined after growing the strains on malt extract agar medium at 25 °C over four weeks according to previous results of Bazzalo and Wright (1982).
The microscopic observations procedure followed Decock et al. (2007). Specimen sections were mounted in 5% KOH solution. Melzer’s reagent and cotton blue were used to test the amyloidity or dextrinoidity and cyanophyly of the microscopic structures, respectively. Microscopic characters were observed under a light microscope Axioscope 40 Carl Zeiss. Images were captured using Axio Vision 4 software on the same microscope. At least 30 structures of each mature specimen were measured. Basidiospores were measured without taking in account the apical umbo when not shrunk. Cuticular cells were measured from the middle part of the basidiome except in the case of some type materials, where only a fragment was received as loan. The 5% extremes of all microscopic measurements from each size range were given in parentheses and the arithmetic mean was provided in brackets. Color terms follow Kornerup and Wanscher (1963), and terms in descriptions are defined in Torres-Torres and Guzmán-Dávalos (2012).
DNA sequencing
Genomic DNA from herbarium specimens was extracted using three protocols: (I) CTab method with 1% PVP (Palomera et al. 2008), (II) salt-extraction method with 1% PVP (Aljanabi and Martinez 1997), and (III) Wizard Genomic DNA Purification Kit (Promega) with 1% PVP. Modifications of the protocols, following Doyle and Doyle (1987) or Palomera et al. (2008), were occasionally made. Genomic DNA was extracted from living cultures according to Amalfi et al. (2010, 2012).
The ITS region (ITS1, 5.8S, and ITS2) was amplified from dried specimens using the primer pairs G-ITS-F1/ITS4B (Cao et al. 2012) or ITS1F/ITS4B (Gardes and Bruns 1993), and from living cultures using the primer pairs ITS1F/ITS4 (White et al. 1990). The primers bRPB2–6F/bRPB2–7.1R (Matheny 2005) and CF2–EF983F/CR2–EF2218R (Rehner and Buckley 2005, Matheny et al. 2007) were used to amplify the rpb2 and tef1–α regions, respectively.
Polymerase chain reaction (PCR) to amplify the ITS from dried specimens followed Guzmán-Dávalos et al. (2003) with some modifications. Each 52 μl reaction solution contained 50 μl of PCR mix [35 μl of MilliQ water, 6 μl of 10 X Taq reaction buffer without MgCl2, 3 μl of 50 mM MgCl2, 3 μl of 5 mM dNTP, 3 μl of 2 μg/μl Bovine Serum Albumine (BSA)], 0.5 μl of each 10 μM primer, 0.15 μl of Taq DNA polymerase (5U/μl), and 1 μl of DNA template (1:5 dilution). PCR amplifications were performed in an ESCO Swift MaxPro thermocycler as described by Guzmán-Dávalos et al. (2003), except that the annealing temperature was following Cao et al. (2012). PCR products were purified with the GFXTM PCR DNA Purification Kit (GE Healthcare). Purified products (GFX) were sent to the Sequencing Department, University of Arizona and LaniVeg (CUCBA, University of Guadalajara). Polymerase chain reactions and amplification protocol of the ITS regions (including 5.8S), partial tef1–α gene, and the region of rpb2 from cultures were described in Amalfi et al. (2010, 2012) and Decock et al. (2013). Sequences were assembled and edited with SequencherTM 4.8 software (Gene Codes Corp., Ann Arbor, Michigan).
Phylogeny
Two DNA sequence data sets were compiled: an ITS data set and a concatenated ITS, rpb2, and tef1–α data set. The combined data set comprised DNA sequences from 71 specimens/cultures from Africa, Europe, Meso– and South America, and Southeast Asia (Table 1). It included sequences from the types of G. microsporum, G. subamboinense, and G. subamboinense var. laevisporum. Sequences of 35 ITS, nine rpb2, and 13 tef1–α were downloaded from GenBank (www.ncbi.nlm.nih.gov/genbank/). It was subdivided into 11 partitions: ITS1, 5.8S, ITS2; rpb2 and tef1–α introns, and –1st, –2nd, and –3rd codon positions of rpb2 and tef1–α. Ganoderma curtisii (Berk.) Murrill and G. lucidum (Curtis) P. Karst. were selected as the outgroup according to results shown by Moncalvo (2000).
Table 1.
DNA sequences of Ganoderma weberianumresinaceum complex and outgroup used in this study, with their voucher materials and geographic origin.
| Species name | Voucher/strain | Locality | GenBank accession numbers | Reference | ||
|---|---|---|---|---|---|---|
| ITS (ITS1/ITS2) | rpb2 | tef1-α | ||||
| G. austroafricanum | CBS 138724 | South Africa | KM507324 | MK611970 | Coetzee et al. 2015 | |
| G. curtisii | CBS 100132 | USA | JQ781848 | KJ143967 | KJ143927 | Cao et al. 2012 |
| G. hoehnelianum | Cui 13982 | China | MG279178 | MG367515 | MG367570 | Xing et al. 2018 |
| Dai 11995 | China | KU219988 | Xing et al. 2018 | |||
| Yuan 6337 | China | MG279160 | MG367498 | MG367551 | Xing et al. 2018 | |
| G. lucidum | K 175217 | UK | KJ143911 | KJ143971 | KJ143929 | Zhou et al. 2014 |
| MUCL 31549 | France | MK554777 | MK554765 | MK554730 | This study | |
| MUCL 35119 | France | MK554779 | MK554752 | MK554719 | This study | |
| G. mexicanum | D. Jarvio 143 | Mexico | MK531823 | This study | ||
| MUCL 49453 SW17 | Martinique | MK531811 | MK531836 | MK531825 | This study | |
| MUCL 55832 | Martinique | MK531815 | MK531839 | MK531829 | This study | |
| MUCL 57308/ BRFM1548 | Martinique | MK531818 | MK531842 | MK531831 | This study | |
| MUCL 57309/ BRFM1830 | Martinique | MK531819 | MK531843 | MK531833 | This study | |
| MUCL 57310/ BRFM1851 | Martinique | MK531820 | MK531844 | MK531832 | This study | |
| G. microsporum | RSH0821 (TYPE) | Taiwan | X78751/ X78772 | Moncalvo et al. 1995 | ||
| G. parvulum | E. Fletes 7619 | Costa Rica | MK531821 | This study | ||
| MUCL 43863 | Cuba | MK554769 | MK554745 | MK554739 | This study | |
| MUCL 44148 | Cuba | MK531132 | MK531845 | MK531834 | This study | |
| MUCL 46029 | Cuba | MK554767 | MK554749 | MK554725 | This study | |
| MUCL 47074 | Cuba | MK554782 | MK554759 | MK554729 | This study | |
| MUCL 47096 | Cuba | MK554783 | MK554742 | MK554721 | This study | |
| MUCL 52655 | French Guiana | MK554770 | MK554755 | MK554717 | This study | |
| MUCL 53123 | French Guiana | MK531814 | MK531837 | MK531827 | This study | |
| MUCL 53712 | French Guiana | MK531813 | MK531838 | MK531828 | This study | |
| MUCL 57307/ BRFM1043 | French Guiana | MK531817 | MK531841 | MK531830 | This study | |
| G. platense | BAFC384 | Argentina | AH008109 | Gottlieb et al. 2000 | ||
| G. polychromum | 330OR | USA | MG654196 | MG754742 | Loyd et al. 2018 | |
| MS343OR | USA | MG654197 | MG754743 | Loyd et al. 2018 | ||
| G. resinaceum | CBS 194.76 | Netherlands | KJ143916 | KJ143934 | Zhou et al. 2014 | |
| HMAS 86599 | UK | AY884177 | JF915435 | Wang et al. 2012 | ||
| MUCL 38956 | Netherlands | MK554772 | MK554747 | MK554723 | This study | |
| MUCL 40604 | Belgium | MK554766 | MK554743 | MK554722 | This study | |
| MUCL 51491 | Belgium | MK554775 | MK554741 | MK554733 | This study | |
| MUCL 52253 | France | MK554786 | MK554764 | MK554737 | This study | |
| Rivoire 4150 | France | KJ143915 | Zhou et al. 2014 | |||
| G. sessile | 111TX | USA | MG654306 | MG754866 | MG754747 | Loyd et al. 2018 |
| 117TX | USA | MG654309 | MG754868 | MG754749 | Loyd et al. 2018 | |
| BAFC2373 | Argentina | AH008111 | Gottlieb et al. 2000 | |||
| CBS 220.36 | USA | JQ520201 | Park et al. 2012 | |||
| JV 1209/27 | USA | KF605630 | KJ143976 | KJ143937 | Zhou et al. 2014 | |
| JV 1209/9 | USA | KF605629 | KJ143936 | Zhou et al. 2014 | ||
| MUCL 38061 | USA | MK554778 | MK554754 | MK554736 | This study | |
| NY 00985711 | USA | KJ143918 | Zhou et al. 2014 | |||
| G. stipitatum | THC 16 | Colombia | KC884264 | Submission to GenBank | ||
| G. subamboinense | Ule.2748/F 15183 (TYPE) | Brazil | MK531824/MK531822 | This study | ||
| G. subamboinense var. laevisporum | BAFC 745/ ATCC 52420 | Argentina | JQ520205 | Park et al. 2012 | ||
| BAFC 25225/ ATCC 52419/ BAFC 247/ (TYPE) | Argentina | X78736/ X78757 | Moncalvo et al. 1995 | |||
| FLASF59210 | USA | MG654371 | Loyd et al. 2018 | |||
| UMNFL100 | USA | MG654373 | MG754762 | Loyd et al. 2018 | ||
| UMNFL32 | USA | MG654372 | MG754761 | Loyd et al. 2018 | ||
| G. weberianum | 15–1048 | USA | KU214242 | Submission to GenBank | ||
| CBS 128581 | Taiwan | MK603805 | MK611971 | MK636693 | This study | |
| CBS 219.36 | Philippines | MK603804 | MK611972 | MK611974 | This study | |
| CCRC 37081 | Taiwan | Z37064/ Z37086 | Smith and Sivasithamparam 2000 | |||
| DFP8401 | Australia | EU239393 | Smith and Sivasithamparam 2000 | |||
| GanoTK16 | Cameroon | JN105704 | Kinge et al. 2012 | |||
| Guzmán–Dávalos 9569 | Mexico | MK554771 | This study | |||
| GW–10 | India | GU726934 | Mohanty et al. 2011 | |||
| GW–11 | India | GU726935 | Mohanty et al. 2011 | |||
| HMAS97365 | China | JF915411 | JF915434 | Wang et al. 2012 | ||
| SUT H2 | Australia | AY569451 | Submission to GenBank | |||
| B–18 | Cuba | JN637827 | Manzano et al. 2013 | |||
| Ganoderma sp. | MUCL 43285 | Cameroon | MK554773 | MK554762 | MK554731 | This study |
| MUCL 43522 | Cuba | MK554792 | MK554760 | MK554732 | This study | |
| MUCL 46912 | China | MK554791 | MK554758 | MK554734 | This study | |
| MUCL 47495 | Gabon | MK554785 | MK554753 | MK611976 | This study | |
| MUCL 47536 | Gabon | MK554768 | MK554746 | MK554724 | This study | |
| MUCL 47542 | Gabon | MK554780 | MK554757 | MK554716 | This study | |
| MUCL 47543 | Gabon | MK554774 | MK554763 | MK554718 | This study | |
| MUCL 47828 | China | MK554788 | MK554740 | MK554728 | This study | |
| MUCL 47835 | China | MK554781 | MK554756 | MK554727 | This study | |
| MUCL 49266 | Cameroon | MK554784 | MK554750 | MK554738 | This study | |
| MUCL 49272 | Cameroon | MK603806 | MK611973 | MK611975 | This study | |
| MUCL 49277 | Cameroon | MK554776 | MK554744 | MK554720 | This study | |
| MUCL 49980 | Congo DRC | MK554789 | MK554748 | MK554735 | This study | |
| MUCL 49981 | Congo DRC | MK554787 | MK554761 | MK554726 | This study | |
| MUCL 51856 | Taiwan | MK554790 | MK554751 | This study | ||
| MUCL 52843 | Gabon | MK531812 | MK531835 | MK531826 | This study | |
| MUCL 57035 | Kenya | MK531816 | MK531840 | This study | ||
| UH–L | Cuba | LT726730 | Torres-Farradá et al. 2017 | |||
| UH–M | Cuba | LT726731 | Torres-Farradá et al. 2017 | |||
Bold names= newly generated sequences for this study.
The ITS data set was composed by 30 specimens/cultures, of which 29 originated from the Neotropics (Table 1). It was subdivided into three partitions: ITS1, 5.8S, ITS2. In this case, G. austroafricanum M.P.A. Coetzee, M.J. Wingf., Marinc. & Blanchette was selected as outgroup according to the results obtained by Coetzee et al. (2015).
All sequences were automatically aligned with MUSCLE (Robert 2004) and manually adjusted using PhyDe (Müller et al. 2010). PartitionFinder (Lanfear et al. 2012) was used to determine the best evolutionary model for each gene using the corrected Akaike information criterion (AICc). Maximum Likelihood (ML) analyses were conducted using RAxML 7.0.4 (Stamatakis 2006) and Bayesian Inference (BI) analyses with MrBayes v.3.2.2 (Ronquist and Huelsenbeck 2003). In the ML analysis, the default priors were used, including individual parameters for each partition, performing 1000 replicates under the GTRGAMMA model. BI analyses were run on CIPRES Science Gateway (Miller et al. 2010). Two independent runs, with 4,000,000 generations each, were carried out with a sampling frequency every 1000 generations and a burn-in of 25%. A 50% majority rule consensus tree with posterior probabilities (PP) was obtained. Convergence of the Markov chains to a stationary distribution was assessed by visual examination of the log likelihood values in the program Tracer v1.7.1 (Rambaut et al. 2018). Nodes were considered supported when bootstrap values (BS) were ≥ 75% and the PP was ≥ 0.85. The final alignments were deposited in TreeBASE (www.treebase.org), under accession ID: 24140 (http://purl.org/phylo/treebase/phylows/study/TB2:S24140).
Results
Molecular phylogeny
The combined dataset contained 172 DNA sequences: 71 ITS, 50 rpb2, and 51 tef1–α. The final alignment comprised 526 bp in the ITS, 776 in the rpb2, and 1123 in the tef1–α. The concatenated data set (ITS + rpb2 + tef1–α) was 2425 bp long. From it, 23 ambiguous sites (12 from ITS1 and ITS2, 11 from tef1–α introns) were removed. The evolutionary models that best fit the individual dataset according to the AICc criterion were ITS1 = GTR+I+G, 5.8S = K80, ITS2 = GTR+I+G, rpb2 1st = GTR+I, 2nd = HKY+G, 3rd codon positions = HKY+G, rpb2 intron= K80, tef1–α 1st = GTR+I, 2nd = HKY+G, 3rd codon positions = GTR+G, and tef1–α intron = GTR+I. In BI analyses, the average standard deviation of split frequencies was 0.008100 in the concatenated data set and 0.008875 in the ITS data set. As far as our specimens from the Neotropics are concerned, the phylogenetic trees obtained from Bayesian (not shown) and Maximum likelihood inferences using the concatenated (Fig. 1) and the ITS (Fig. 2) data sets showed overall the same two clades, except for the unsupported branch of the specimen MUCL 43522 present in the concatenated ML and BI analyses, which collapsed in the ITS tree, and the placement of the specimen UMNFL100, G. subamboinense var. laevisporum, from Florida (Figs 1–2).
Figure 1.
Phylogeny of the Ganoderma weberianum-resinaceum complex based on concatenated ITS, rpb2, and tef1-α sequence data obtained by Maximum Likelihood (ML). Bayesian posterior probability (PP) above 0.85 and bootstrap values (BS) from ML above 75 % are shown (PP/ BS).
Figure 2.
Phylogeny of the Ganoderma weberianum complex from the Neotropics based on rDNA ITS sequence data obtained by Maximum Likelihood (ML). Bayesian posterior probability (PP) above 0.85 and bootstrap values (BS) from ML above 75 % are shown (PP/BS).
The Ganoderma weberianum-resinaceum lineage was resolved with strong support (PP 1, BS 100%). It was divided into two major clades, I and II (Fig. 1). Clade I (PP 1, BS 98%) corresponded to the G. resinaceum clade as defined by Moncalvo (2000), with G. austroafricanum, from South Africa, located in a basal position. This was further subdivided in an unsupported clade A (PP 0.66, BS 58) and moderately supported clade B (PP 0.89, BS 82). Clade A included specimens all originated from the temperate area of the Northern Hemisphere and a specimen from the highlands of central Kenya (MUCL 57035). Clade A was structured into two subclades, A1 (PP 1, BS 100), with the Kenyan specimen (MUCL 57035) in basal position, and A2 (PP 1, BS 89), with a specimen from China (MUCL 46912) in basal position. Clade B brought together several specimens originated from both North and South America, distributed into three well-supported subclades that corresponded to G. sessile Murrill and G. polychromum (Copel.) Murrill (PP 1, BS 89; PP 0.99, BS 100), as defined by Loyd et al. (2018). Two specimens from Argentina, tentatively identified as G. sessile and G. platense Speg., formed together a third distant well-supported subclade (PP 1, BS 99).
Clade II (PP 1/BS 99) was subdivided into two major clades: clade C (PP 1, BS 95) and clade D (PP 1, BS 100). Clade C corresponded to G. weberianumsensu Steyaert. It was further structured into two well-supported subclades, C1 (PP 1, BS 91) and C2 (PP 1, BS 74), with a geographic dichotomic pattern opposing the New World / Neotropics (C1) to the Old World / Paleotropics (C2).
New World / Neotropical clade (C1) had two low- to moderately supported terminal clades, C1.1 (PP 0.89, BS 86) and C1.2 (PP 0.80, BS 62). C1.1 included the ex-type strain of G. subamboinense var. laevisporum (BAFC 247, Bazzalo and Wright 1982) from Argentina and five specimens from Martinique, of which one was identified previously as G. subamboinense (Welti and Courtecuisse 2010) and three as G. weberianum (CIRM-CF, on-line catalog). Its sister clade, C1.2, comprised the type specimen of G. subamboinense (S F15183!) and ten specimens from Cuba and French Guiana, of which one was identified as G. weberianum (CIRM-CF, on-line catalog). It also comprised two specimens from Florida, which were both, alternatively identified as G. subamboinense var. laevisporum at GenBank, or as G. cf. weberianum in Loyd et al. (2018).
The Old World / Paleotropical clade (C2) contained specimens from both Africa and Asia. This clade was further subdivided into three well-supported subclades, viz. C2.1 (PP 0.99, BS 95), with two specimens from India, C2.2 (PP 1, BS 97) gathering specimens from Central Africa (Cameroon and Gabon), and C2.3 (PP 1, BS 91) with specimens from Australia and Southeast Asia (China, Philippines, and Taiwan). A specimen from Australia (DFP 8401) in clade C2.3 was identified as G. rivulosum by Steyaert (fideSmith and Sivasithamparam 2000) and reidentified as G. weberianum by Smith and Sivasithamparam (2000). The C2.3 also included the ex-type strain of G. microsporum (RSH0821, BPI) from Taiwan.
Finally, clade D (PP 1, BS 100) comprised specimens of Ganoderma sp. from Central Africa (DR Congo and Gabon) and from China, these latter tentatively identified as G. hoehnelianum Bres. at GenBank (Fig. 1).
The ITS data set comprised sequences from thirty specimens (Table 1) and the final alignment had 536 positions. Five positions, the alignments of which were judged to be ambiguous, were removed from the analyses. Clades C1.1 (PP 0.92, BS 100) and C1.2 (PP 0.78, BS 97) were confirmed by the ITS data set (Fig. 2). A specimen from Mexico (Guzmán-Dávalos 9569, IBUG), previously identified as G. weberianum by Torres-Torres et al. (2015), formed an additional branch, basal and sister to C1.1 and C1.2 (Fig. 2).
Taking in account both single and multilocus phylogenetic analyses, we considered that our Neotropical specimens formed two related but distinct, well-supported terminal clades, C1.1 and C1.2 (Figs 1–2) that could be equated each to a phylogenetic species. The additional branch formed by the single specimen from Mexico also might be equated to a distinct phylogenetic species.
Morphological studies
From a morphological perspective, specimens in the phylogenetic species C1.1 and C1.2 were very similar, characterized by an overall reddish brown to violet brown pileal surface, light, cork-colored context, occasionally paler in the upper zone —described as not fully homogenous by Torres-Torres et al. (2012)—, with none to several (up to 4) dense, brown stripes or continuous lines of resinous deposits extending from the base of the context toward the margin. The cuticular cells were mainly cylindrical to clavate, apically rounded, regular, amyloid, and the basidiospores ovoid to broadly ovoid, with free to subfree pillars, and chlamydospores (“gasterospores” in Bazzalo and Wright 1982) in their context (Figs 3–4).
Figure 3.
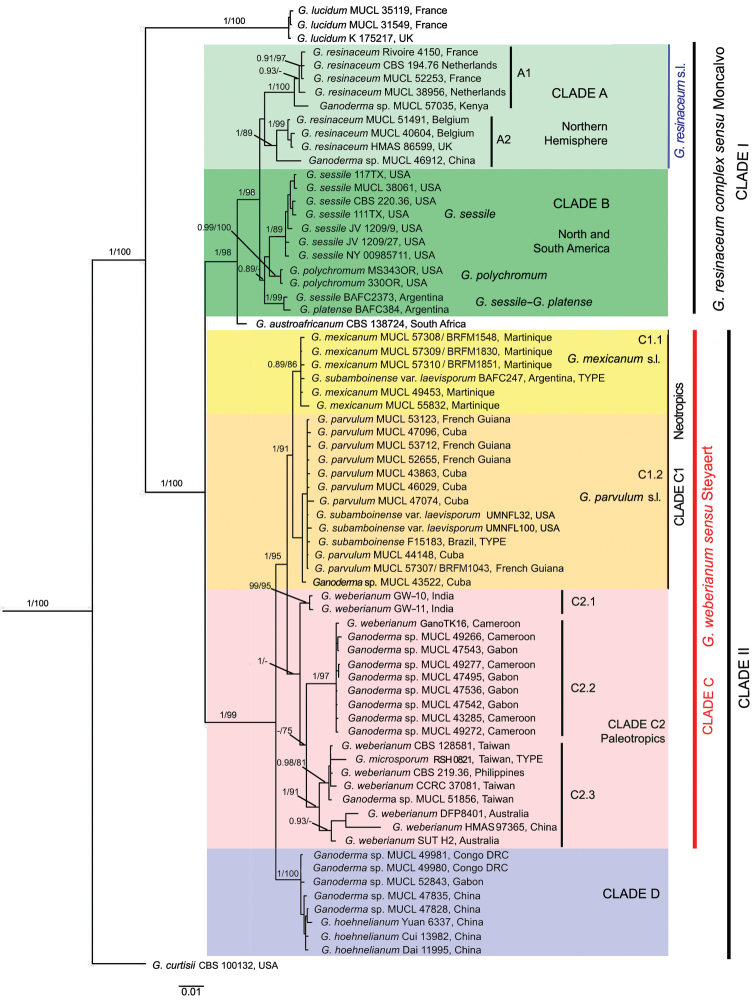
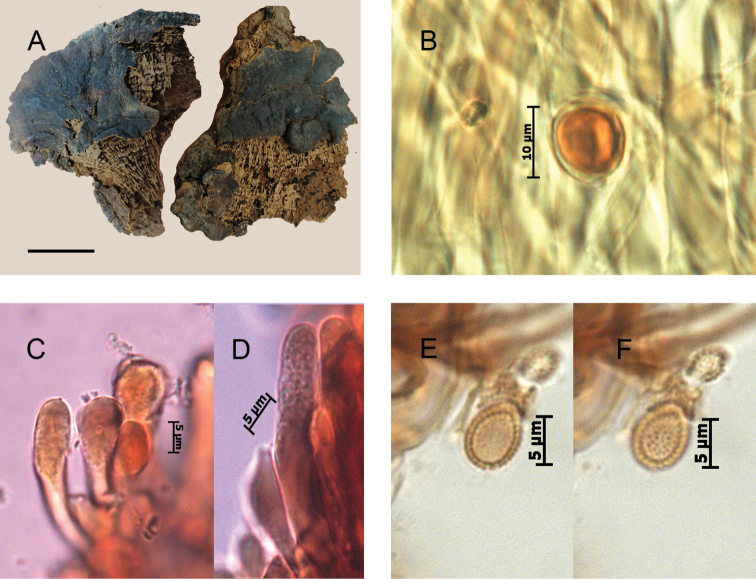
Morphological features and microscopic structures of Ganoderma mexicanumA–F J.P. Fiard SW 17 (as G. subamboinense in Welti and Courtecuisse 2010) A pileus and context not fully homogeneous with discrete bodies of the resin-like deposits (arrow) B cuticular cells C basidiospores with free to subfree pillars D–F smooth-walled chlamydospores D from context E from culture, in cotton blue F from culture, in Melzer reagent G–I BAFC 25525 (as G. subamboinense var. laevisporum, holotype) G cuticular cells H basidiospores with free to subfree pillars I smooth-walled chlamydospore from context.
Figure 4.
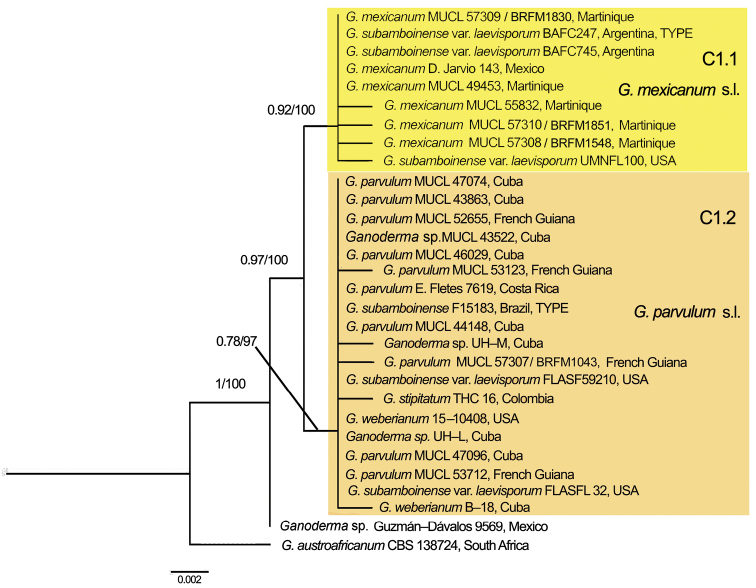
Morphological features and microscopic structures of Ganoderma parvulumA–H MUCL 53123 A pilear surface B cuticular cells C–D basidiospores with free to subfree pillars E–H chlamydospores ornamented with free to partially anastomosed ridges E–F from context G–H from culture I–K E. Ule 2748 (as G. subamboinense, holotype) I upper surface of basidiomata, copyright: Naturhistoriska riksmuseet, Stockholm J cuticular cells K chlamydospores ornamented with partially anastomosed ridges, from context, in KOH. Scale bars: 1 cm (A); 5 μm (B).
These chlamydospores were mostly subglobose in the context of the basidiomes and more variably shaped in pure culture on artificial media, thick-walled, hyaline to yellowish, and with dextrinoid content. In the specimens of C1.1, the chlamydospores were constantly, permanently smooth-walled (Fig. 3D–F, I) whereas in specimens of C1.2, they were smooth becoming roughened on aging, with free to partially anastomosed fine ridges with a meridian orientation (Fig. 4E–H, K). The Mexican specimen (Guzmán-Dávalos 9569, IBUG!) also presented chlamydospores in the context but they were punctuated, ornamented with thick pillars (Fig. 5).
Figure 5.
Ganoderma sp., Mexican specimen Guzmán-Dávalos 9569 (as G. weberianum in Torres-Torres et al. 2015) A stipitate basidioma B pilear surface C–D Detail of chlamydospore ornamented with pillars. Scale bar: 1 cm (A–B).
Basidiospores were slightly wider in specimens from C1.1 compared to those of C1.2, mainly 8–9 × 6–7 µm (averaging 8.5 × 6.5 µm, Fig. 3C, H) vs. 8–9.5 × 5.5–6.5 µm (averaging 8.9 × 6.0 µm, Fig. 4C–D). Cuticular cells were moderately longer in specimens from C1.2 (up to 100 µm long, Fig. 4B, J) than specimens in C1.1 (up to 65 µm long, Fig. 3B, G).
In general, the morphology allowed the distinction of three morphotypes, which could be considered as three morphospecies. Each of these also corresponded to a phylogenetic species.
Taxonomic conclusions
The present study, using single and multilocus phylogenetic inferences combined with morphological and in vitro culture studies, concordantly revealed two species of Ganoderma in the G. weberianumsensu Steyaert lineage, spanning over the Neotropics. Furthermore, a specimen from Mexico, represented only by the ITS sequence (Guzmán-Dávalos 9569, Figs 2 & 5), also could be equated to a morphological and phylogenetic species, pending confirmation when additional material is available. However, none of these three species could be equated to G. weberianumsensu Moncalvo (Moncalvo 2000), which is restricted to tropical Asia.
Clade C1.1 contained the ex-type strain of G. subamboinense var. laevisporum; hence, it could correspond to this taxon. The sister clade C1.2 contained the ex-type strain of G. subamboinense var. subamboinense; thus, it could correspond to the typical variety. Furthermore, both the molecular and morphological data would warrant recognition of both varieties at species level.
Ganoderma subamboinense was originally described by Hennings (1904) as Fomes subamboinense Henn. The type specimen originated from Brazil. Bazzalo and Wright (1982) accepted the species and distinguished a var. laevisporum from the typical variety. Both varieties were characterized by the presence of chlamydospores in their context, which were rough-walled with “veins anastomosing to form a sort of reticulum” in the typical variety, and smooth-walled in var. laevisporum, what was later confirmed by Gottlieb and Wright (1999). Ryvarden (2000), based on presumed morphological resemblance, reduced G. subamboinense s.l. (both varieties) as a synonym of G. multiplicatum (Mont.) Pat. Similarities between G. subamboinense var. laevisporum and G. multiplicatum var. vitalii Steyaert were previously reported (Bazzalo and Wright 1982). Inversely, Torres-Torres et al. (2012) recognized G. multiplicatum as an independent species that could be differentiated from G. subamboinense s.l., e.g., in having apically irregular cuticular cells, with many protuberances. Steyaert (1980) had already characterized G. multiplicatum var. vitalii with “mostly irregular” cuticular cells.
The revision of a fragment of the holotype of G. subamboinense var. subamboinense (S F15183!) (Fig. 4I–K) and of the holotype of G. subamboinense var. laevisporum (BAFC 25525!) (Fig. 3G–I) confirmed the previous observations (Bazzalo and Wright 1982, Gottlieb and Wright 1999). Both varieties are mainly characterized by a pale context with resinous incrustations or thin resinous brown bands, stretching from base towards the margin, cylindrical to clavate, apically regular, amyloid cuticular cells, ovoid to broadly ovoid basidiospores with free to subfree pillars, and chlamydospores in their context. The chlamydospores were striated in the typical variety and smooth-walled in var. laevisporum.
The study of the holotype of G. multiplicatum (K 123639!), originating from French Guiana, confirmed irregular cuticular cells with both lateral and apical protuberances, distinct from those of both varieties of G. subamboinense. Phylogenetic analyses inferred from an ITS data set (Bolaños et al. 2016) or a combined ITS–LSU data set (de Lima-Junior et al. 2014) also showed that G. subamboinense var. laevisporum (ITS sequence from the ex-type culture ATCC 52419) and G. multiplicatum (that should be considered as sensu auctores) formed two distinct clades, in two distant lineages. Hence, the synonymy of G. subamboinense and G. multiplicatum, as suggested by Ryvarden (2000), here also is rejected.
The macro- and microscopic features of both varieties of G. subamboinense, overall, corresponded to those of our specimens from C1.1 and C1.2 clades, as described above. Therefore, G. subamboinense var. subamboinense (C1.2) and G. subamboinense var. laevisporum (C1.1) could be applied to the taxa shown by these clades. However, for nomenclatural reasons, the varietal epithet laevisporum cannot be used, at any rank. The nomenclatural status of the epithet laevisporum was questioned; as previously highlighted, it was invalidly published (Moncalvo and Ryvarden 1997, Welti and Courtecuisse 2010). Moncalvo and Ryvarden (1997) noted that Bazzalo and Wright (1982) did not formally propose the combination G. subamboinense, making it invalid; consequently, the varietal epithet also was invalid. Moncalvo and Ryvarden (1997) validated the combination G. subamboinense but this did not automatically validate the varietal epithet, which, therefore, cannot be used. A name, therefore, should be found for the taxon shown by the C1.1.
Furthermore, several other names whose types originate from the Neotropics but currently of uncertain status or in the limbo of the G. weberianum-resinaceum complex (Moncalvo and Ryvarden 1997, Ryvarden 1985, 2000, 2004) also could be reconsidered for species represented by both clades. Taking into account the main features of our specimens as described above, such as a light-colored context, G. argillaceum, G. mexicanum, G. parvulum, G. perturbatum, G. perzonatum, G. praelongum Murrill, G. pulverulentum, G. sessiliforme, G. stipitatum, G. subincrustatum, and G. vivianimercedianum (Patouillard 1898, Murrill 1902, 1903, 1908, 1912, Steyaert 1962, 1980, Torres-Torres et al. 2008) were worth revisiting.
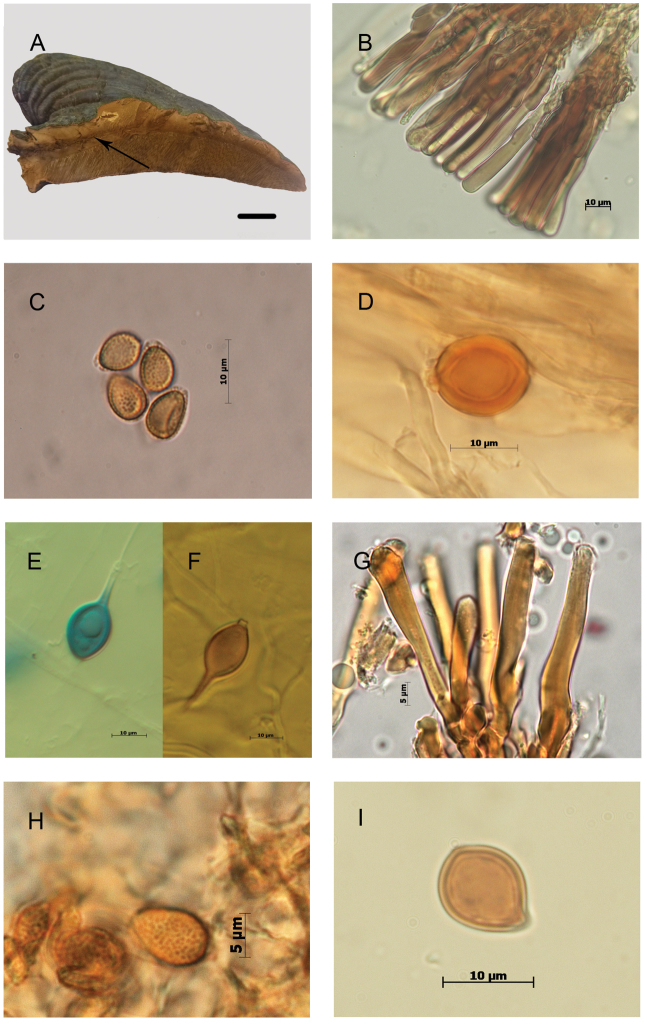
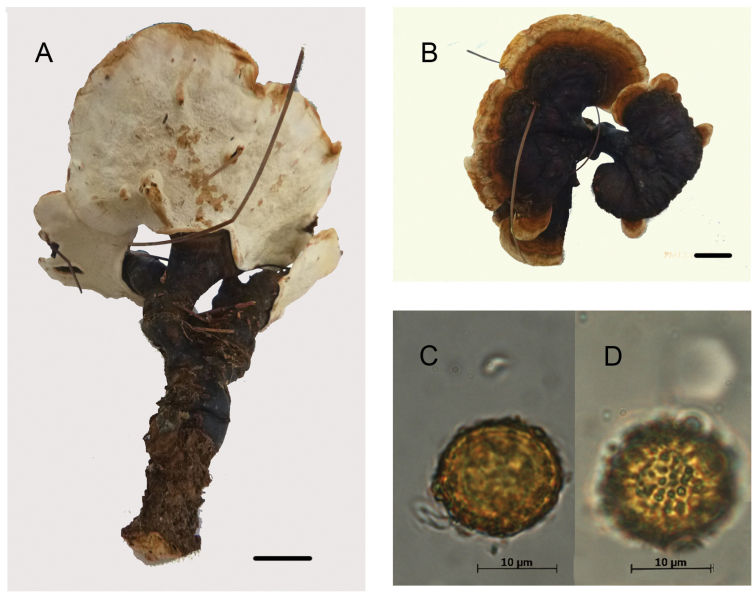
Ganoderma mexicanum (holotype: FH 458184!) is the earliest name potentially available. It should be treated together with G. sessiliforme (holotype: NY 98713!); both type specimens are originated from neighboring localities in Morelos State, Mexico, viz. Tepalcingo, D. de Jonacatepec (Patouillard 1898) and Cuernacava (Murrill 1912), respectively. Torres-Torres et al. (2012) emphasized the poorly conserved type of G. mexicanum (Fig. 6) and reported an additional specimen from Brazil. On the basis of these two specimens, Torres-Torres et al. (2012) described clavate to narrowly clavate, 35.2–72.4 × 6.8–10.5 μm cuticular cells with very thick wall, without apical granulations, and ellipsoid basidiospores, 9.3–10.6 × 6.2–7.4 μm, with subfree pillars. Our study of the type specimen of G. mexicanum showed smaller, clavate to narrowly clavate cuticular cells, 25–37 × 5–7.5 μm, averaging 28.5 µm long, with occasional apical granulations. Basidiospores were ovoid with free to subfree pillars, (6.5–) 7.8–9.4 × 5.2–6.5 (–7) μm, and smooth, dextrinoid chlamydospores, 10.3–15 μm, were observed in the context (Fig. 6).
Figure 6.
Morphological features and microscopic structures of the type specimen of Ganoderma mexicanum (P.J.B. Maury 4823, FH 458184), photographs by the authors, images courtesy of the Farlow Herbarium of Harvard University, Cambridge, Massachusetts, USA, A pilear surface B smooth chlamydospore C–D cuticular cells with incrustations C clavate D cylindrical E–F basidiospore with free to subfree pillars. Scale bars: 1 cm (A).
The study of the type specimen of G. sessiliforme (Fig. 7A–F) and of a second specimen collected in an area also neighboring the type locality (Guzmán 2078, ENCB, Fig. 7G–K, cf. Torres-Torres et al. 2015) revealed a pale context with a few resinous incrustations, scattered, smooth-walled, dextrinoid chlamydospores, (8–) 10–12 (–13.5) x 7–11 μm, clavate to narrowly clavate, smooth to sometimes faintly apically granulated cuticular cells, 25–38 × 5–9 µm, averaging 32 µm long, and ovoid basidiospores, 8–9.3 (–10.7) × 6–7.7 (–8) µm, with free to subfree pillars.
Figure 7.
Morphological features and microscopic structures of Ganoderma sessiliformeA–F NY 98713 (holotype) A pilear surface B context not fully homogeneous with discrete bodies of the resin-like deposits (arrow) C–D basidiospores with free to subfree pillars E–F cuticular cells in KOH E cylindrical F clavate, with narrow lumen (arrow) G–K Guzmán 2078, ENCB G pilear surface H–I basidiospores with free to subfree pillars J cuticular cells in Melzer reagent K smooth-walled chlamydospore from context in Melzer reagent.
Based on these observations and inversely to the previous conclusions of Torres-Torres et al. (2012, 2015), we did not found any consistent morphological difference between G. mexicanum and G. sessiliforme. Furthermore, the type specimen of both names originated from neighboring localities and, probably, related ecosystems. Therefore, G. sessiliforme and G. mexicanum are here considered as synonyms, the latter epithet (Patouillard 1898) having priority. However, the status and affinities of G. mexicanum are uncertain.
Patouillard (1898) suggested that G. mexicanum was a sessile form of G. lucidum but with smooth (“lisses”), ovoid basidiospores. There is also a typewritten, undated note from R. Singer in the type specimen folder emphasizing “This is merely Ganoderma sessile Murr.”, and then pencil corrected “same as [G. sessile]”. Previously, Murrill (1902) also pointed out similarities between G. sessiliforme and G. sessile. Loyd et al. (2018) also suggested that G. sessiliforme might represent a synonym of G. sessile.
Nevertheless, G. sessile has distinctly larger basidiospores, 11.2–14.4 (–16.4) × 7.2–8.8 µm (fideTorres-Torres et al. 2015) and a duplex and spongy context (Torres-Torres et al. 2012, 2015), both features which would justify distinguishing these species.
Gottlieb and Wright (1999) and Gottlieb et al. (2000) previously compared G. sessiliforme with G. subamboinense var. laevisporum. They argued that G. sessiliforme differed from G. subamboinense var. laevisporum by the size (up to 11 mm long) and ornamentation (“semirugose” under the SEM) of its basidiospores, and the lack of chlamydospores, both features that do not stand (cf. above). The characters of G. mexicanum, especially the light-colored context with resinous incrustations, the basidiospores size, and the presence of smooth chlamydospores, overall remind much those of G. subamboinense var. laevisporum and of our specimens from the clade C.1.1 but for the cuticular cells. The cuticular cells are more clavate and shorter, 25–38 μm in G. mexicanum compared to those of G. subamboinense var. laevisporum and specimens from C1.1, 30–50 μm (Figs 3G & 6C–D).
Ganoderma parvulum (type NY 985699!) is the second earliest name potentially available. It should be treated together with G. stipitatum; the types of both epithets were collected in Nicaragua by C.L. Smith (Murrill 1902, 1903), probably in neighboring localities. Steyaert (1980), based on type studies, although accepting these two taxa, considered they formed a species complex together with G. bibadiostriatum Steyaert, whose type was collected in Brazil (Steyaert 1962). Ryvarden, in a handwritten note dated from 1983, joint to the holotype of G. stipitatum (NY 985678!), concluded that G. parvulum and G. stipitatum, likely were synonyms (“the identity with G. parvulum Murr. is almost certain”), what he formalized later on, accepting a single species under G. stipitatum, with G. bibadiostriatum and G. parvulum as synonyms (Ryvarden 2000, 2004).
The examination of the type specimens of G. parvulum (NY 985699!) and G. stipitatum (NY 985678!) (Fig. 8A–E) revealed little developed, likely immature basidiomes. Murrill (1902) already suggested this, stating “It is possible that the specimens [of G. parvulum] I have are not quite mature”. The type of G. parvulum was characterized by a pale context (pale ochraceous, fideMurrill 1902; light “ochraceous buff”, fideSteyaert 1980), with resinaceous streaks, dark horny (fideMurrill 1902) or carob brown (fideSteyaert 1980). These resinaceous streaks were also present in the type of G. stipitatum (Murrill 1903, Steyaert 1980, Ryvarden 2004, pers. obs.). Cuticular cells were cylindrical to slightly clavate, average 50 µm long, and very few basidiospores were observed in holotype specimens. Ryvarden, in a handwritten note dated from 1983, emphasized the absence of basidiospores in the type of G. stipitatum (“spores are not present”, NY 985678!). Nonetheless, Steyaert (1980) reported basidiospores, although without commenting on their abundance, 7.5–8.5 × 5.5–6.5 µm, averaging 8.1 × 5.9 µm in G. parvulum and 7.0–10.5 × 4.5–6.5 µm, averaging 7.8 × 5.5 µm in G. stipitatum. The few basidiospores we observed were 7–9 × 5–6.5 µm in G. parvulum and 7–10 × 5–6.5 µm in G. stipitatum, with free to subfree and very thin pillars in both.
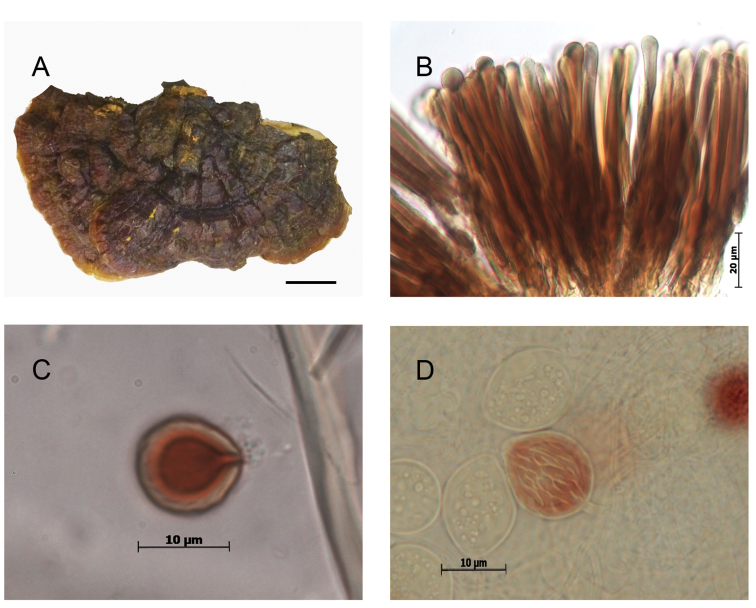
Figure 8.
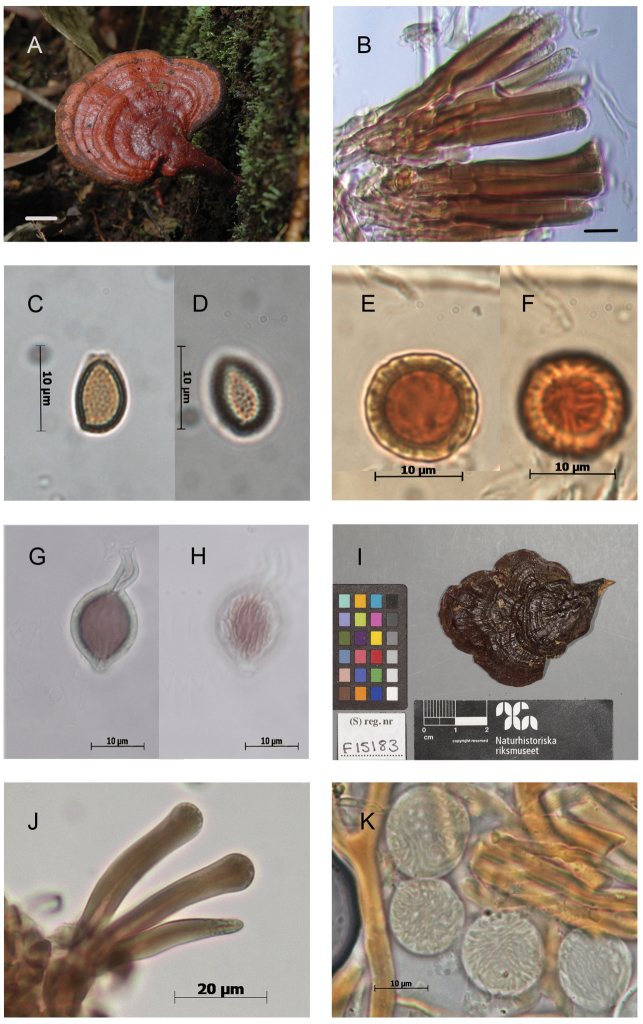
Morphological features and microscopic structures of type specimens of GanodermaA–EGanoderma stipitatum (NY 985678, holotype) A basidiomata B–C context B brown stripes of resinous deposits C numerous bodies of the resin-like deposits D–E chlamydospore, ornamented with partially anastomosed ridges, from context F–IGanoderma perzonatum (NY 985702, holotype), F basidiomata, copyright: NY Botanical Garden G cuticular cells, cylindrical, with incrustations H–I chlamydospore with fine longitudinal ridges, from context. Scale bar: 1 cm (A–C).
Chlamydospores were not reported in the literature for G. parvulum nor for G. stipitatum (Steyaert 1972, 1980, Gottlieb and Wright 1999, Ryvarden 2000, 2004, Welti and Courtecuisse 2010), with the sole exception of Torres-Torres et al. (2012). Torres-Torres et al. (2012) reported and illustrated double-walled chlamydospores with “inter-walled, very thick pillars” presumably from the type of G. parvulum (cf. Torres-Torres et al. 2012, fig. 22d). However, their fig. 8, which is captioned as type of G. parvulum, actually corresponds to the type of G. stipitatum (NY 985678!). The voucher specimen from which these punctuated chlamydospores were observed remained uncertain. Nonetheless, our study of the type of G. parvulum and G. stipitatum revealed scattered chlamydospores in the context of both. These chlamydospores were smooth-walled or also ornamented with anastomosed ridges (Fig. 8D–E).
The likely immaturity of the type of G. parvulum and G. stipitatum, to a certain extent, could prevent definitive taxonomic interpretations. Notwithstanding, we would follow Ryvarden (2000, 2004) in considering that these two epithets represent a single species. Furthermore, the main macro- and microscopic characteristics of G. stipitatum and to a lesser extent, of G. parvulum, as described above, overall, also correspond to those of G. subamboinense var. subamboinense. Therefore, G. parvulum, G. stipitatum, and G. subamboinense could be considered synonymous. In this case, the epithet parvulum (Murrill 1902) has priority over subamboinense (Hennings 1904) and stipitatum (basionym: Fomes stipitatusMurrill 1903), contrary to the conclusion of Ryvarden (2004).
Steyaert (1962, 1980), Bazzalo and Wright (1982), and Gottlieb and Wright (1999) recognized G. bibadiostriatum as a distinct species. Ganoderma bibadiostriatum was characterized by a distinctly brown (fideBazzalo and Wright 1982, or cinnamon, fideSteyaert 1962, 1980) context and basidiospores 7.0–11.0 × 5.5–7.5 µm, averaging 9.3 × 6.5 µm (fideSteyaert 1980), or 9–11 × 6–8 µm (fideGottlieb and Wright 1999). These features differed from those of G. parvulum. Through phylogenetic inferences based on ITS and LSU, de Lima-Junior et al. (2014) showed that Brazilian specimens identified as either G. parvulum or G. stipitatum (identifications that should be considered as sensu auctores) were gathered into a single clade, which nested within the G. tropicum clade sensu Moncalvo (2000), and were unrelated to G. subamboinense var. laevisporum, hence unrelated to the G. weberianum-resinaceum lineage. Therefore, contrary to Ryvarden (2000, 2004) but following Gottlieb and Wright (1999), we also rejected the synonymy of G. bibadiostriatum with G. parvulum and G. stipitatum. We suggest that the identity of the G. parvulum–G. stipitatum clade shown by de Lima-Junior et al. (2014) should be re-evaluated, and that it might well represent G. bibadiostriatum.
The status and affinities of G. perzonatum have been debated and still are uncertain. Ganoderma perzonatum was described from Cuba (Murrill 1908). Moncalvo and Ryvarden (1997) first related it to G. parvulum. Previously, Steyaert in 1962 and Wright in 1967, in two notes joint to the type specimen of G. stipitatum (NY 985678!) and to a second specimen annotated “probable type” [of G. stipitatum] (NY 985716!) also informally suggested that G. perzonatum and G. parvulum were synonymous. However, later on, Ryvarden (2004) retained the species that he associated to the G. resinaceum complex. The revision of the type specimen (NY 985702!) (Fig. 8F–I) confirmed the main features (Ryvarden 2004): a sessile, dimidiate habit with superposed pilei, a pale corky context with dark, resinous streaks, cuticular cells up to 100 µm long, and basidiospores 8.5–9.5 (–10) × 6–7 µm. Furthermore, chlamydospores with smooth or then ornamented with fine longitudinal ridges (Fig. 8H–I), also were observed, a feature previously unnoticed. These characteristics brought it back to G. parvulum, as first suggested by Moncalvo and Ryvarden (1997).
However, G. perzonatum would differ from G. parvulum in having larger, sessile, dimidiate basidiomes and markedly cylindrical, longer cuticular cells. A specimen originating from the type locality of G. perzonatum (MUCL 43522, La Havana, Cuba, Fig. 9) shared these characters. It also produced striated chlamydospores, both in the context of the basidiome and in pure culture, similar to those of G. parvulum. However, this specimen formed a short, isolated branch, basal to the C1.2 clade, in phylogenetic inferences of the combined data set (Fig. 1). Ganoderma perzonatum remains of uncertain interpretation. It could be included, for the time being, in the concept of G. parvulum (hence G. parvulum s.l.).
Figure 9.
Morphological features and microscopic structures of the Cuban specimen of Ganoderma MUCL 43522 A upper surface of basidiomata B cuticular cells C–D chlamydospore with longitudinal ridges C from context D from culture. Scale bar: 1 cm (A).
The taxonomic statuses of G. argillaceum, G. perturbatum, G. praelongum, G. pulverulentum, and G. subincrustatum also were widely debated. Ganoderma argillaceum and G. praelongum were considered as synonyms of G. resinaceum by Steyaert (1980). Bazzalo and Wright (1982) also included G. pulverulentum and G. subincrustatum to the list of the G. resinaceum presumed synonyms, to which Ryvarden (2000) added G. perturbatum and G. sessiliforme. Inversely, at the other extreme, Torres-Torres and Guzmán-Dávalos (2012) recognized under these epithets as many independent species. Gottlieb and Wright (1999) accepted G. praelongum with G. pulverulentum (Murrill 1908) as a synonym, whereas Welti and Courtecuisse (2010) recognized G. pulverulentum as an independent species.
Ganoderma argillaceum, G. praelongum, and G. pulverulentum differed from G. parvulum, G. mexicanum, and our Neotropical specimens in having larger basidiospores (respectively 9–11 × 6–8 µm, fideGottlieb and Wright 1999; 9–11 × 6–8.5 µm, fideGottlieb and Wright 1999; 9.6–12.8 × 6.2–8 µm, fideTorres-Torres et al. 2012). Ganoderma argillaceum (holotype NY, 01293316!) had basidiospores with abundant and thin pillars and lacked chlamydospores (Gottlieb and Wright 1999), whereas G. praelongum and G. pulverulentum had basidiospores with partially anastomosed pillars (Gottlieb and Wright 1999, Torres-Torres et al. 2012). Ganoderma vivianimercedianum also differed from our specimens in having larger basidiospores, 8.8–11.2 (–12) × 6.5–8 µm, and absence of chlamydospores (Torres-Torres et al. 2008). These names remained of uncertain status and affinities. They are most likely not synonyms of G. resinaceum s.s. from Europe (Bazzalo and Wright 1982) but affinities with North American species of the G. resinaceum clade, viz. G. sessile and G. polychromum (Loyd et al. 2018), should not be excluded.
Ganoderma perturbatum (BPI!) differs from our specimens from both clades (C1.1 and C1.2) in having larger basidiospores 10–12.8 × 8–9.4 µm with subacute apex and partially anastomosed pillars or short crest-like ornamentations. Ganoderma subincrustatum has cuticular cells generally with short and thick protuberances and basidiospores with partially anastomosed pillars (Torres-Torres and Guzmán-Dávalos 2012, Torres-Torres et al. 2015, pers. obs.). Their status and affinities also remained uncertain.
In conclusion, we are of the opinion that G. mexicanum could be selected as the earliest name available for the specimens of the clade C1.1. It is morphologically very similar if not identical to G. subamboinense var. laevisporum. We therefore suggest, for the time being, pending new material and DNA sequences data, to apply G. mexicanum to the clade C1.1. We also concluded that the name G. parvulum could be retained as the earliest name available for the taxa represented by the clade C1.2, previously reported in the literature as G. subamboinense var. subamboinense. Ganoderma perzonatum could represent another closely related taxon in the vicinity of G. mexicanum / G. parvulum.
Taxonomy
Ganoderma mexicanum
Pat., Bull. Soc. Mycol. Fr. 14: 54 (1898)
51387CD7-093B-578F-8C3A-12F25C8606D4
469325
≡ Fomes mexicanus (Pat.) Sacc., Syll. Fung. 14: 184 (1899) [MB166450]
= Ganoderma sessiliforme Murrill, Bull. New York Bot. Gard. 8: 149 (1912) [MB469342]
= Ganoderma subamboinense var. laevisporum Bazzalo & J.E. Wright, Mycotaxon 16(1): 302 (1982) [MB117102], invalid.
Description.
Basidiome annual, sessile, occasionally stipitate, solitary, light in weight, consistency corky-woody; pileus projecting 4–8 cm, 6–14 cm wide, up to 1.5–1.8 cm thick at the base, 0.3–0.4 cm at the margin, dimidiate, flabelliform to conchate in pole view, applanate or slightly convex in section; stipe absent or 2 × 0.5–3 cm, horizontal, short and thick, slightly swollen at the base, laccate, smooth, reddish brown (8F6) to violet brown (11F7); pileal surface laccate, smooth, radially zonate, with dark lines or with concentric variably deep sulcations, reddish brown (8F6) to dark brown (8F5), lighter towards the margin; margin likely white when young, entire to slightly lobulated, sometimes incurved; pore surface yellowish white to greyish yellow (4C7), yellowish brown (5E7), or brownish orange (5C3), bruising dark brown (6F5), sometimes marked with spots of same aspect as pilear surface (laccate, reddish brown, 8F6); pores round, 4–6 (–7) per mm; context 0.2–1 cm thick, fibrous, homogeneous to slightly heterogeneous (not fully homogeneous fideTorres-Torres et al. 2012), almost overall white to light yellow (4A4) or light yellow (4A4) to greyish orange (5B4) toward the crust, yellowish brown (5D6) to light brown (7D3) in a narrow zone above the tubes, with few to several resinous incrustations or thin resinous dark bands stretching from the basis to the margin; tubes 0.1–0.8 cm long, unstratified, concolorous with lower part of the context.
Hyphal system dimitic; generative hyphae 1–3 µm diam., septate, thin-walled with clamp connections, little branched, hyaline to yellowish; somatic hyphae as arboriform skeleto-binding hyphae, golden yellow, composed of a basal stalk arising from a clamp, unbranched, thick-walled but with a visible lumen, with several secondary processes, branches gradually tapering from 6 µm wide in the primary processes to 1.5–2 µm wide at the thin-walled apices, thick-walled to solid. Pileipellis a crustohymeniderm; cuticular cells clamped at the basal septum, shortly to moderately pedicelated then cylindrical a clavate, occasionally slightly apically capitate, rarely with 1–2 lateral branches, with rounded apices, thick-walled, smooth or with a fine apical granulation, amyloid, 25- [~ 40] -50 (-65) × 5-7 um. Hymenium: basidia not seen; basidiospores ovoid to broadly ovoid, the apex shrunken, appearing truncate, exosporium with thick, free to subfree pillars, (7.5–) 8– [8.5] –9 (–10.5) × (4.2–) 6– [6.5] –7 (–8) µm, Q = 1.33– [1.30] –1.28; spore print light brown (6E5) (estimated from spore deposit on the pileus). Chlamydospores in the basidiomata absent, rare, to variably abundant, only in the context, subspherical, ellipsoid, or citriform, terminal or intercalated; with smooth thick-wall; sometimes guttulate, dextrinoid, 9.5–13 (–16) × 8–10 µm. Chlamydospores always abundant in pure culture on malt agar, spherical to more often ellipsoid, terminal or intercalary, when terminal with the apex occasionally papillated; with smooth, hyaline to pale golden brown, single or double wall; sometimes with densely guttulate contents, often dextrinoid, 11–16 × 9.5–12 µm.
Holotype.
MEXICO. Estado de México: D. de Jonacatepec, Tepalcingo, 22 Oct 1890, P.J.B. Maury 4823 (FH 458184!).
Known distribution.
Argentina, Brazil, Martinique, Mexico.
Specimens examined.
ARGENTINA. Buenos Aires: Tigre, on Platanus sp., 15 May 1980, Connon (as holotype of G. subamboinense var. laevisporum, BAFC 25525, culture ex. type BAFC n° 247 = ATCC 52419). MARTINIQUE. Prêcheur: Anse Couleuvre, sentier de la cascade de la rivière Couleuvre, on Artocarpus altilis, in mature, secondary mesophylic forest, 13 Aug 2007, J.P. Fiard SW 17 (LIP, culture ex. MUCL 49453). Rivière–Pilote: Morne Aca, on a lying trunk, in meso-xerophylic forest, 14 Aug 2007, S. Welti, SW 19 (LIP). La Caravelle, xerophylic forest, on dead fallen trunk, 12 Aug 2015, C. Decock, MA/15–45 (MUCL 55832, culture ex. MUCL 55832). MEXICO. Morelos: Municipality of Cuernavaca, on dead wood, 24–27 December 1909, W.A. & E.L. Murrill 392 (as holotype of G. sessiliforme, NY 985713). Mpio. of Tepoztlán, Tepoztlán, Estación del Ferrocarril El Parque, w/o date, G. Guzmán 2078 (ENCB). Veracruz: San Andrés Tlalnelhuayocan, alrededores de San Antonio Hidalgo, bosque mesófilo de montaña, 1400 m, D. Jarvio 143 (XAL).
Ganoderma parvulum
Murrill, Bull. Torrey Bot. Club. 29: 605 (1902)
5E793C67-2277-5C89-B1DF-1E1E85EDF9AE
241944
≡ Fomes parvulus (Murrill) Sacc. & D. Sacc, Syll. Fung. (Abellini). 17: 123 (1905) [MB241944]
= Fomes stipitatus Murrill, Bull. Torrey Bot. Club. 30(4): 229 (1903) [MB241804]
≡ Ganoderma stipitatum (Murrill) Murrill, N. Amer. Fl. (New York) 9(2): 122 (1908) [MB451185]
= Fomes subamboinensis Henn., Hedwigia 43(3): 175 (1904) [MB148868]
≡ Ganoderma subamboinense (Henn.) Bazzalo & J.E. Wright ex Moncalvo & Ryvarden, Synopsis Fungorum 11: 82 (1997) [MB249603]
≡ Ganoderma subamboinense var. subamboinense Bazzalo & J.E. Wright Mycotaxon 16(1): 302 (1982) [MB417363] (invalid)
Description.
Basidiome annual, sessile or stipitate, solitary or sometimes concrescent or forming several (up to 3) pileus, light in weight, consistency corky-woody; pileus projecting 4.5–8 cm, 6.5–15 cm wide, up to 0.8–3 cm thick at the base, 0.5–0.7 cm at the margin; dimidiate, flabelliform to conchate in pole view, applanate or convex in section; stipe absent or 1.5–4.5 (–8) × 0.5–3 cm, horizontal or dorsally lateral, short and thick or long and tortuous, slightly swollen at the base, laccate, reddish brown (9F6) to dark brown (9F4) or violet brown (10F8) to almost black, stumpy or cylindrical, sometimes with laterals branches; pileal surface smooth, laccate, radially rugose or with concentric deep sulcations or occasionally, slightly zonate with dark lines, fully reddish brown (9F8) to violet brown (10F8), or gradually lighter towards the margin with a yellowish orange (5A7) band; margin white to pale yellow (4A3) or greyish yellow (4C7) to yellowish orange (5A7), entire to slightly lobulated, sometimes incurved; pore surface white, yellowish white (3A2), dull yellow (3B3), or sun yellow (2A5) when fresh and actively growing, greyish yellow (4C7), yellowish brown (5E7), or brownish orange (5C3) on drying, bruising dark brown (6F5), sometimes marked with spots of same aspect than pilear surface (laccate, reddish brown, 9F8); pores 4–5 per mm, round to mainly angular; context 0.3–2.4 cm thick, fibrous, homogeneous to slightly heterogeneous, sometimes zonate, greyish yellow (4B5) to greyish orange (5B3) toward the crust, and brownish orange (6C4) to light brown (6D4) in a narrow zone above the tubes, changing to yellow when cut in fresh specimens, with none to few to several (up to 4) resinous incrustations or occasionally resinous bands (up to 4), sometimes with yellow (3B8) spots throughout the context, with a yellow (3B8) to yellowish orange (4A7) thin line just below the crust; tubes 0.2–0.6 cm long, unstratified, concolorous with lower part of the context.
Hyphal system dimitic; generative hyphae 1.6–3.2 µm diam., septate, thin-walled with clamp connections, non-branched, hyaline to yellowish; somatic hyphae as arboriform skeleto–binding hyphae, golden yellow, composed of a basal stalk arising from a clamp, with several secondary processes, branches gradually tapering from 6 µm wide in the primary processes to 1.5–2 µm wide at the thin-walled apices, thick-walled to solid. Pileipellis a crustohymeniderm; cuticular cells clamped at the basal septum, pedicelated, mainly cylindrical to clavate, occasionally slightly apically capitate, rarely with 1–2 lateral protuberances, with regular, rounded end, thick-walled to almost solid, amyloid, the apex occasionally with a radial fine granulation, 40- [~ 60] -75 (-100) x 5-10 um. Hymenium: basidia not seen; basidiospores ovoid to broadly ovoid, the apex shrunken, appearing truncate, exosporium with thick, free to subfree pillars, (6–) 8– [8.9] –9.5 × (4.8–) 5.5– [6.0] –6.5 (–7) µm, Q = 1.45– [1.48] –1.46, ovoid; spore print (6E5), light brown (estimated from spore deposit on the pileus). Chlamydospores in the basidiomata absent, rare, to variably abundant, only in the context, subspherical, ellipsoid, or citriform, sometimes spindle-shaped, terminal or intercalated; smooth-walled to roughened with fine, isolated to partially anastomosed ridges, having a meridian orientation, variably stretching between the two extremities, totally dextrinoid or with dextrinoid content and golden wall, 7–13 × 6–12 µm. Chlamydospores always abundant in pure culture on malt agar, spherical to ellipsoid, sometimes spindle-shaped, often truncated at both ends; terminal or intercalary; when terminal with the apex occasionally papillated; single or golden double walled; with several large guttulae, with dextrinoid contents; smooth first then variably roughened, ornamented with fine partial or continuous ridges, isolated to partially anastomosed, 11–16 (–17.5) × 9–14.5 (–16) µm.
Holotype.
NICARAGUA. C.L. Smith s.n. (NY 985699!).
Known distribution.
Brazil, Colombia, Costa Rica, Cuba, French Guiana, Mexico, Nicaragua, South–eastern USA (Florida).
Specimens examined.
BRAZIL. St. Clara: Río Juruá, Oct 1900, E. Ule 2748, (under Fomes subamboinensis as type of G. subamboinense, F15183 (S). COSTA RICA. Puntarenas: Isla del Coco, orilla del río Genio, represa hidroeléctrica, 0–100 m s.n.m., 5 Jun 2005, E. Fletes–7619, Lote: 84813 (INB 3976555); Osa, P.N. Corcovado, Estación Sirena, Sendero Guanacaste, bosque primario, 10 m s.n.m., E. Fletes–266, Lote: 53967 (INB 1546586), río Madrigal, quebrada Ceniza, 200 a 300 m s.n.m., 19 Mar 2003, E. Fletes–4943, Lote: 73208 (INB 3700175). CUBA. Province La Habana: Municipality Boyeros, Zoológico Nacional de Cuba, on base of a living trunk, 22 Aug 2001, C. Decock and S. Oliva, MUCL 43522 (culture ex. MUCL 43522); Finca La Chata, on base of a living trunk of Casuarina equisetifolia, 27 May 2002, C. Decock w/o #, MUCL 43863 (culture ex. MUCL 43863); Province Villa Clara: Falcon, Carretera Central, dead stump of Casuarina equisetifolia, Aug 2002, C. Decock, CU–02/14, MUCL 44148 (culture ex 44148 = CRGF 715); Province Sancti Spiritus: Topes de Collantes, on the way to the Caburni, dead trunk, unidentified angiosperm, Sep 2004, C. Decock, CU–04/12, MUCL 46029 (culture ex 46029 = CRGF 202); Sep 2005, C. Decock, CU–05/196 (MUCL 47074, culture ex 47074 = CRGF 719); Province Pinar del Río: La Palma, near the Motel La Ciguaraya, decaying stump, unidentified angiosperm, Oct 2005, C. Decock, CU–05/246, MUCL 47096 (culture ex 47096 = CRGF 722). FRENCH GUIANA. Nouragues National Reserve, Inselberg CNRS research station, dead fallen trunk, unidentified angiosperm, Aug 2010, C. Decock, FG/10-283, MUCL 53123 (culture ex. 53123); 2011, C. Decock, FG/11–481, MUCL 53712 (culture ex. 53712). MEXICO. State of Veracruz: Zentla, camino Huatusco–Maromilla, a la altura de Puentecilla, bosque mesófilo de montaña, alt. 860 m s.n.m. (as G. lucidum), A. Sampieri 84 (XAL). NICARAGUA. C.L. Smith s.n., as F. stipitatus (holotype of G. stipitatum, NY 985678); C.L. Smith s.n., as F. stipitatus (“TYPE” of G. stipitatum, NY 985679); without data, C.L. Smith s.n., as F. stipitatus (“Probable TYPE” of F. stipitatus, det. as G. parvulum by Steyaert 1961, NY 985716).
Additional species examined.
BRAZIL. Rio Grande do Sul: Lageado, without date, R. Rick s.n. (holotype of G. perturbatum) (BPI). CUBA. Province La Habana: Municipality Santiago de Las Vegas, on mango log, 1904, F.S. Earle 309 (holotype of G. perzonatum, NY 985702); on dead mango, 5 Jul 1904, F.S. Earle 658 (holotype of G. argillaceum, NY 01293316). GRENADA. Without data, on dry manchinell, 14 Sep 1905, W.E. Broadway s.n. (holotype of G. pulverulentum, NY 00985705). INDONESIA. Java: without data, P. Serre s.n. (type of G. rivulosum, F181158, S). MEXICO. Estado de México: valle del Tepeite, 10 km NE of Santa María, 10 Aug 1986, E. Bastidas-Varela s.n. (holotype of G. vivianimercedianum, ENCB). SAMOA ISLAND. Without data, Weber s.n. (as “TYPUS” of Fomes weberianus F15098, S); without data, Weber s.n., as Fomes weberianus (B 700021870), “Fomes weberi”), without data, Weber s.n., as Fomes weberianus (B 700007410, “TYPE” of G. weberianum). TAIWAN. Taipei: on Salix babylonica Linn. (Salicaceae), 21 Aug 1983, R.-S. Hseu (isotype of G. microsporum, HMAS 57945, frag. in BR!).
Remarks: Ganoderma mexicanum and G. parvulum have sessile to stipitate basidiomes, more frequently stipitate in the latter, with a basal and horizontal stipe. The type specimens of G. subamboinense (Fig. 4I–K) and of G. stipitatum (Fig. 8A–E), overall, have the same basidiome habit. The two specimens of G. parvulum from the rainforest of French Guiana also were morphologically very homogeneous, stipitate, with a basal and horizontal stipe. In the Greater Antilles (Cuba), G. parvulum was found mostly in anthropic or urban environment and had sessile, dimidiate basidiomes.
The context of both G. mexicanum and G. parvulum was light-colored, usually very pale toward the crust and darker just above the tubes, with none to several brown resinous incrustations or resinous bands variably stretching through the context from the base to the margin. The context in G. parvulum sometimes showed yellow, scattered spots and a thin yellow line just below the crust. Both species have chlamydospores in their context and in pure culture on artificial media. There are not many morphological characters to differentiate them except for the ornamentation of their chlamydospores. However, chlamydospores are sometimes very scarce and difficult to observe in the basidiome. Nonetheless, they are always present, and frequent, in pure culture on artificial media.
The basidiospores were, on average, marginally wider in G. mexicanum in comparison to those of G. parvulum, viz. on average 8.6 × 6.4 µm or 9.0 × 6.0 µm, respectively. The cuticular cells were cylindrical to claviform, occasionally with 1–2 short lateral branches, strongly amyloid, usually smooth or with a fine apical granulation, which was more consistently present in G. parvulum. The cuticular cells also were marginally longer in G. parvulum (up to 100 µm long) compared to those of G. mexicanum (up to 65 µm long).
The distribution ranges and ecologies of both species are still little known. Ganoderma parvulum, as here interpreted, had been observed from the Brazilian Amazon, Colombia, and French Guiana in South America, Costa Rica and Nicaragua in Mesoamerica, and up to Cuba in the Caribbean. Loyd et al. (2017, 2018) reported G. cf. weberianum from the subtropical southern Florida (USA) on the basis of two specimens (UMNFL 32 and UMNFL 100), which DNA sequences, nevertheless, were deposited in GenBank under G. subamboinense var. laevisporum. Loyd et al. (2018) described striated chlamydospores in the context of these specimens, which points toward G. parvulum. Our multilocus phylogenetic inferences showed that these Florida specimens nested within the G. parvulum clade (Fig. 1). However, there was incongruence between the topology resulting from the multilocus-based phylogenies and the ITS-based inferences (Fig. 2) regarding the position of UMNFL 100. The ITS sequences of this showed a change in three nucleotide positions, that could represent a misreading of the sequencer. Notwithstanding, these reports extend the distribution range of G. parvulum northerly to the subtropical, south–eastern USA. This ample distribution would imply a broad ecological range, but also could encompass a hidden diversity.
In French Guiana, G. parvulum has been observed at the Nouragues Nature Reserve (~4°04′18″N, 52°43′57″W), a spot of primary, very humid (3000 mm of rain / year), tropical rainforest characteristic of the Guianas shield, which belongs to the larger Amazonian rain forest phytochorion. Locally, this species was uncommon; three basidiomes only were observed during six, two- to three-weeks long surveys of polypores. These three specimens were found emerging from dead, fallen trunks. In French Guiana, it has been observed also once in an anthropic, semi-urban environment (culture BRFM 1043, voucher specimen and data on the substrate and host unavailable). The type specimen of G. parvulum, originating from Brazil, was also, most likely, collected in the same phytochorion. In Cuba, Greater Antilles, the species has been observed mostly in anthropic, urban or semi-urban environments (cf. list of specimens examined).
Ganoderma mexicanum, as here interpreted, has been observed from Argentina, Brazil, Martinique (Lesser Antilles), and Mexico. In Mexico, the species is known from a rather restricted area of the Morelos State, which is the type locality of G. mexicanum and G. sessiliforme, and of a third additional specimen collected in secondary tropical forest with Quercus sp. (Torres-Torres et al. 2015). Raymundo et al. (2013) and López-Peña et al. (2016) also reported G. sessiliforme from xerophylic vegetation with Quercus sp. in Sonora, but the voucher specimens were not available for confirmation. In Martinique, the species was found in mesophylic to distinctly xerophylic forests, which could represent, locally, its preferential habitat. Several collections came from The Caravelle Peninsula, which is characterized by a seasonally dry season.
Discussion
The current morphological concept of Ganoderma weberianum dated back from Steyaert (1972) and was based on Fomes weberianus. However, on one hand, the very identification of F. weberianus remained questioned. As raised by Yombiyeni and Decock (2017), there was confusion around the modern interpretation of this taxon and its generic placement was debated; the species was either considered in Ganoderma, following Steyaert (1972), or in Phylloporia, following Ryvarden (1972). On another hand, the circumscription of G. weberianumsensu Steyaert remained questioned and, consequently, its distribution range remained uncertain.
Fomes weberianus, originating from Samoa (“in insula Samoa”), was first described by Saccardo (1891). This author did not specify a type or any reference specimens, mentioning only “Exempl. in Museo berolin” (nowadays B). The current concept of G. weberianum was developed based on a specimen held in B (#700007410) stamped as type; this specimen represents indeed a species of Ganoderma, hence G. weberianumsensu stricto (Steyaert 1972). However, the same year, Ryvarden (1972) recombined F. weberianus into Phylloporia, although without citing any reference specimen.
Nonetheless, in addition to the type cited by Steyaert (1972), two other specimens annotated as “Fomes weberianus, Weber, Samoa” exist, of which one also is stamped as type. One of these two specimens is located at B [Samoa Island, Weber “Fomes Weberi” det. P. Henn., Fomes weberianus (#700021870!)], and the second in the Bresadola herbarium in S [Samoa Island, Weber, det. P. Henn. and Bresadola as Fomes weberianus “n. sp.” “Typus!” F15098!]. These two latter specimens do not represent a species of Ganoderma but a species of Phylloporia; their morphological features agree very well with the modern, morphological concept of this genus (e.g., Wagner and Ryvarden 2002). Moreover, and essentially, the morphological characters of these two specimens are in complete agreement with the original diagnosis of F. weberianus (Saccardo 1891).
This diagnosis was, partly, a copy of a handwritten description, contemporary to, or previous to Saccardo (1891), and which is still present within the folder #700007410 in B. It emphasized a duplex context (“strato duplice”) made of an upper tomentose to floccose layer (“superiori tomentoso–floccoso”) and a lower, corky layer (“inferiori suberoso–lignoso”), separated one from the other by a thin black line (“a superiore linea nigra limitato”). Saccardo (1891), following the above-cited note, related F. weberianus to Polyporus circinatus (Fr.) Fr. and P. tomentosus Fr., two Hymenochaetaceae nowadays accepted in Onnia (Ryvarden 1990). Subsequent early interpretations of F. weberianus (e.g., Bresadola 1914, 1916, 1925, Lloyd 1915, Cunningham 1950) also associated this species to taxa that are, mainly, akin to species of Phylloporia as currently accepted. As far as we had been able to ascertain, there was no pre-Steyaert (1972) interpretation of this taxon as a species of Ganoderma.
This casted doubts on the interpretation of F. weberianus; considering the original diagnosis and both specimens from B and Bresadola herbarium in S, Phylloporia weberianasensuRyvarden (1972), most likely, is the correct interpretation. Hence, a lectotype should be designated. This will be discussed in more detail later on.
Our results, following Moncalvo (2000), confirmed that G. weberianumsensu Steyaert was polyphyletic and encompassed several species. Multilocus phylogenetic inferences had shown distinct, well-supported clades and an overall phylogenetic structure corresponding to a geographical pattern (Fig. 1). The G. weberianumsensu Steyaert lineage was divided into two main sublineages, viz. a Neotropical and Paleotropical sublineages.
As far the Neotropics are concerned, at least two species were confirmed, G. mexicanum (previously variably reported as G. sessiliforme and G. subamboinense var. laevisporum) and G. parvulum (previously known as G. subamboinense). Furthermore, the specimen MUCL 43522 from Cuba could represent G. perzonatum. Although we are of the opinion that G. perzonatum may well represent a species on its own, more material, ideally from various localities, and DNA sequences, is necessary to draw a definitive conclusion. On the other hand, the specimen Guzmán-Dávalos 9569 (IBUG!) from Mexico, basal to the G. parvulum s.l. / G. mexicanum s.l. clade in the ITS-based phylogenetic inferences (Fig. 2) and with chlamydospores ornamented with isolated pillars, also could represent a distinct taxon. This demonstrates a likely higher than known phylogenetic and morphological diversity and, ahead, taxonomic diversity. Several additional names also remain of uncertain status and, if any, unknown affinities; it includes G. argillaceum and G. praelongum, or still G. multiplicatum and G. vivianimercedianum. Collections from their type localities and DNA sequences data are highly needed.
The Paleotropical sublineage was further divided into three clades, including an African, an Indian, and a tropical Asian / Australasian clade, representing, at the least, as many phylogenetic species or species complexes. As regard to the situation in Central Africa, at least one species could be segregated from the G. weberianumsensu Steyaert. Ganoderma carocalcareum Douanla-Meli (Douanla-Meli and Langer 2009) could apply for this taxon but three previous priority names might apply too, which will require a revision of their type specimens.
On the basis of our phylogenetic inferences, we conclude that G. weberianum in Southeast Asia and Australia (which would correspond to G. weberianumsensuMoncalvo 2000) also is a complex of species. It would include, at least, G. rivulosum (S F181158!) and G. microsporum (isotype BPI!). Moncalvo et al. (1995) suggested that there are few differences between G. microsporum and G. weberianum, and later on, Moncalvo (2000) considered both names as synonyms (Moncalvo 2000), which was also the opinion of Smith and Sivasithamparam (2003) and Wang et al. (2005). However, this synonymy needs to be ascertained.
The sister clade of G. weberianumsensu Steyaert lineage is, in our phylogenetic analyses, hitherto, the clade D, which comprised specimens from Central Africa and China, the latter referenced at GenBank as G. hoehnelianum. Ganoderma hoehnelianum was described by Bresadola (1912) from Java (Indonesia) having basidiomes with a “crusta, tenui, opaca”. This “crust” was observed in the type specimen (S F181067!) and is different from the laccate pileal surface of the G. weberianum complex, made of cuticular cells organized in a dense palisade. Therefore, the identity of this clade also should be ascertained.
This study also confirmed that G. resinaceum sensu auctores from China, East Africa, Europa, and both North and South America represented a species complex. Loyd et al. (2018) showed that G. resinaceumsensu American auctores encompassed at least two distinct species, viz. G. polychromum and G. sessile. These results agreed with those of Moncalvo (2000), who distinguished European and North American “populations” of G. resinaceum, on the basis of which it was proposed that these “disjunct and genetically isolated [populations]” “may warrant recognition at the species level”. Our study showed that the G. resinaceumsensu European auctores also represented a species complex, with two well-supported phylogenetic species (Fig. 1); thus G. resinaceum in Europe also could hide a larger than expected diversity.
As emphasized by Moncalvo (2000) and Richter et al. (2015), the identification of species in Ganoderma was commonly based on the microanatomy of the pileus surface, the basidiospores morphology and size, and in some cases, the host relationships. The occurrence of chlamydospores in the basidiomes or in in vitro cultures also was highlighted as a valuable feature (Moncalvo 2000, Hong and Jung 2004, Richter et al. 2015, Loyd et al. 2019), although this character, as suggested by Steyaert (1980), also could be environment dependent. Our results concerning the Neotropical species of the G. weberianum complex confirmed the presence of chlamydospores, in basidiomes and in in vitro cultures, and their ornamentation, as pertinent taxonomic features for the systematic of this group. Three ornamentation types, smooth, with pillars, or with ridges have been observed in the Neotropical species. A similar situation could occur in G. weberianumsensu Moncalvo in Southeast Asia. Steyaert (1972) described G. weberianum with both smooth and ornamented chlamydospores with pillars, “columns”, or “partitions”. Smith and Sivasithamparam (2003) confirmed these observations based on examination of the type specimen (cited by Steyaert 1972) and specimens from Australia and the south Pacific regions.
Supplementary Material
Acknowledgments
The authors thank the curators of the following herbaria for the loan of types and other materials, B, BAFC, BPI, ENCB, FH, IBUG, INBIO, MUCL, NY, S, and XAL. Milay Cabarroi Hernández gratefully acknowledges financial support received from the Rufford Foundation, UK (project 17535–2), from the Université Catholique de Louvain, Belgium (Scholarship from the Conseil de l’Action Internationale (CAI): Coopération au développement), the University of Guadalajara, and the CONACYT, Mexico (National Scholarship). Cony Decock gratefully acknowledges the financial support received from the Belgian State – Belgian Federal Science Policy through the BCCM research program, the FNRS / FRFC (convention FRFC 2.4544.10) and the Nouragues Travel Grant “MYcorrhizal COmmon network of Trees on INselbergs” (program 2013) that allowed fungal diversity studies in French Guiana, and the CIUF/CUD through a PIC program (project CIUF–CUD–MUCL–Cuba) that allowed fungal diversity studies in Cuba. Cony Decock also thanks Dr. Anne Corval and Dr. Annaïg Le Guen, respectively previous and present Director of the “CNRS Guiana”, for granting authorization and facilities for field research at the CNRS Nouragues “Inselberg” and “Pararé” forest plots, and the CNRS staff members in Cayenne and at both camps, viz. Dorothée Deslignes, Laeticia Proux, Philippe Gaucher, Patrick Châtelet, Gilles Peroz, Wemo Betian, and Stéphane Ricatte. Cony Decock also thanks Prof. Régis Courtecuisse for his invitation to participate to the Mycoflora of Martinique project, and Dr. Anne Favel for providing strains from CIRM-CF Marseille. Laura Guzmán-Dávalos recognizes the support of the University of Guadalajara, Mexico. Our gratitude also is extended to Dr. Pilar Zamora Tavares from LaNiVeg at University of Guadalajara who provided Sanger Sequencing. Thanks are extended also to Stéphanie Huret for her help in the sequencing program and Philipe Charue for his help in the laboratory work, as well as Dr. Eduardo Ruiz and Dr. Gerardo Hernández Vera for their advice about the phylogenetic work.
Citation
Cabarroi-Hernández M, Villalobos-Arámbula AR, Torres-Torres MG, Decock C, Guzmán-Dávalos L (2019) The Ganoderma weberianum-resinaceum lineage: multilocus phylogenetic analysis and morphology confirm G. mexicanum and G. parvulum in the Neotropics. MycoKeys 59: 95–131. https://doi.org/10.3897/mycokeys.59.33182
Contributor Information
Cony Decock, Email: cortezzuma1511@yahoo.fr.
Laura Guzmán-Dávalos, Email: lguzman@cucba.udg.mx.
References
- Aljanabi SM, Martinez I. (1997) Universal and rapid salt extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Research 25: 4692–4693. 10.1093/nar/25.22.4692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amalfi M, Yombiyeni P, Decock C. (2010) Fomitiporia in sub-Saharan Africa: morphology and multigene phylogenetic analysis support three new species from the Guineo-Congolian rainforest. Mycologia 102: 1303–1317. 10.3852/09-083 [DOI] [PubMed] [Google Scholar]
- Amalfi M, Raymundo T, Valenzuela R, Decock C. (2012) Fomitiporia cupressicola sp.nov., a parasite on Cupressus arizonica, and additional unnamed clades in the southern USA and northern Mexico, determined by multilocus phylogenetic analyses. Mycologia 104: 880–893. 10.3852/11-196 [DOI] [PubMed] [Google Scholar]
- Bazzalo ME, Wright JE. (1982) Survey of the Argentine species of the Ganoderma lucidum complex. Mycotaxon 16: 293–325. [Google Scholar]
- Bolaños AC, Bononi VLR, Gugliotta AM, Muñoz JE. (2016) New records of Ganoderma multiplicatum (Mont.) Pat. (Polyporales, Basidiomycota) from Colombia and its geographic distribution in South America. Check List 12(4): 1948. 10.15560/12.4.1948 [DOI] [Google Scholar]
- Bresadola G. (1912) Polyporaceae Javanicae. Annales Mycologici 10: 492–508. [Google Scholar]
- Bresadola G. (1914) Fungi nonnulli exotici ex Museo Berolinensi. Annales Mycologici 12: 539–544. [Google Scholar]
- Bresadola G. (1916) Synonymia et adnotanda mycologia. Annales Mycologici 14: 221–242. [Google Scholar]
- Bresadola G. (1925) New species of fungi. Mycologia 17: 68–77. 10.1080/00275514.1925.12020457 [DOI] [Google Scholar]
- Cao Y, Wu S, Dai Y. (2012) Species clarification of the prize medicinal Ganoderma mushroom “Lingzhi”. Fungal Diversity 56: 49–62. 10.1007/s13225-012-0178-5 [DOI] [Google Scholar]
- Coetzee MPA, Marincowitz S, Muthelo VG, Wingfield MJ. (2015) Ganoderma species, including new taxa associated with root rot of the iconic Jacaranda mimosifolia in Pretoria, South Africa. IMA Fungus 6: 249–256. 10.5598/imafungus.2015.06.01.16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corner EJH. (1983) Ad Polyporaceas I, Amauroderma and Ganoderma. Beihefte zur Nova Hedwigia 75: 1–182. [Google Scholar]
- Cunningham GH. (1950) Australian Polyporaceae in herbaria of Royal Botanic Gardens, Kew, and British Museum of Natural History. Proceedings of the Linnean Society of New South Wales 75: 214–249.
- Decock C, Herrera-Figueroa S, Robledo G, Castillo G. (2007) Fomitiporia punctata (Basidiomycota, Hymenochaetales) and its presumed taxonomic synonyms in America: taxonomy and phylogeny of some species from tropical/subtropical area. Mycologia 99: 733–752. 10.1080/15572536.2007.11832537 [DOI] [PubMed] [Google Scholar]
- Decock C, Amalfi M, Robledo G, Castillo G. (2013) Phylloporia nouraguensis (Hymenochaetales, Basidiomycota), an undescribed species from the Neotropics. Cryptogamie, Mycologie 34: 15–27. 10.7872/crym.v34.iss1.2013.15 [DOI] [Google Scholar]
- Douanla-Meli C, Langer E. (2009) Ganoderma carocalcareus sp. nov., with crumbly-friable context parasite to saprobe on Anthocleista nobilis and its phylogenetic relationship in G. resinaceum group. Mycological Progress 8: 145–155. 10.1007/s11557-009-0586-4 [DOI] [Google Scholar]
- Doyle JJ, Doyle JL. (1987) A rapid isolate procedure from small quantities of fresh leaf tissue. Phytochemical Bulletin 19: 11–15. [Google Scholar]
- Gardes M, Bruns TD. (1993) ITS primers with enhanced specificity for basidiomycetes – application to the identification of mycorrhizae and rusts. Molecular Ecology 2: 113–118. 10.1111/j.1365-294X.1993.tb00005.x [DOI] [PubMed] [Google Scholar]
- Gottlieb AM, Wright JE. (1999) Taxonomy of Ganoderma from South America: subgenus Ganoderma. Mycological Research 103: 661–673. 10.1017/S0953756298007941 [DOI] [Google Scholar]
- Gottlieb AM, Ferrer E, Wright JE. (2000) rDNA analyses as an aid to the taxonomy of species of Ganoderma. Mycological Research 104: 1033–1045. 10.1017/S095375620000304X [DOI] [Google Scholar]
- Guzmán-Dávalos L, Mueller GM, Cifuentes J, Miller AN, Santerre A. (2003) Traditional infrageneric classification of Gymnopilus is not supported by ribosomal DNA sequence data. Mycologia 95: 1204–1214. 10.1080/15572536.2004.11833028 [DOI] [PubMed] [Google Scholar]
- Hapuarachchi KK, Wen TC, Deng CY, Kang JC, Hyde KD. (2015) Mycosphere essays 1: taxonomic confusion in the Ganoderma lucidum species complex. Mycosphere 6: 542–559. 10.5943/mycosphere/6/5/4 [DOI] [Google Scholar]
- Hennings P. (1904) Fungi amazonici I. a cl. Ernesto Ule collection. Hedwigia 43: 154–186. [Google Scholar]
- Hong SG, Jung HS. (2004) Phylogenetic analysis of Ganoderma based on nearly complete mitochondrial small-subunit ribosomal DNA sequences. Mycologia 96: 742–755. 10.1080/15572536.2005.11832922 [DOI] [PubMed] [Google Scholar]
- Kinge TR, Mih AM, Coetzee MPA. (2012) Phylogenetic relationships among species of Ganoderma (Ganodermataceae, Basidiomycota) from Cameroon. Australian Journal of Botany 60: 526–538. 10.1071/BT12011 [DOI] [Google Scholar]
- Kornerup A, Wanscher JH. (1981) Methuen handbook of colour. 3rd ed. Methuen, London, 252 pp. [Google Scholar]
- Lanfear R, Calcott B, Ho S Y, Guindon S. (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Molecular Biology and Evolution, 29: 1695–1701. 10.1093/molbev/mss020 [DOI] [PubMed] [Google Scholar]
- de Lima-Junior N, Baptista G, Malosso E. (2014) Delimitation of some Neotropical laccate Ganoderma (Ganodermataceae): molecular phylogeny and morphology. Revista de Biología Tropical 62: 1197–1208. 10.15517/rbt.v62i3.12380 [DOI] [PubMed] [Google Scholar]
- Lloyd CG. (1905) Synopsis of the genus Ganoderma. Mycological Writings 1.
- Lloyd CG. (1915) Synopsis of the genus Fomes. Mycological Writings 4: 209–288. [Google Scholar]
- López-Peña D, Gutiérrez A, Hernández-Navarro E, Valenzuela R, Esqueda M. (2016) Diversidad y distribución de Ganoderma (Polyporales: Ganodermataceae) en Sonora, México. Botanical Sciences 94: 431–439. 10.17129/botsci.463 [DOI] [Google Scholar]
- Loyd AL, Smith JA, Richter BS, Blanchette RA, Smith ME. (2017) The laccate Ganoderma of the Southeastern United States: a cosmopolitan and important genus of wood decay fungi. UF’s EDIS. http://edis.ifas.ufl.edu.
- Loyd AL, Held BW, Barnes CW, Schink MJ, Smith ME, Smith JA, Blanchette RA. (2018) Elucidating ‘lucidum’: distinguishing the diverse laccate Ganoderma species of the United States. PLoS One 13(7): e0199738. 10.1371/journal.pone.0199738 [DOI] [PMC free article] [PubMed]
- Loyd AL, Linder ER, Smith ME, Blanchette RA, Smith JA. (2019) Cultural characterization and chlamydospore function of the Ganodermataceae present in the eastern United States. Mycologia 111: 1–12. 10.1080/00275514.2018.1543509 [DOI] [PubMed] [Google Scholar]
- Manzano AM, Torres G, González A, Banguela A, Ramos-González PL, Valiente PA, Sánchez MI, Sánchez-Lamar Á, Rochefort D, Mclean MD, Ramos-Leal M, Guerra G. (2013) Role of laccase isozymes in textile dye decolorization and diversity of laccase genes from Ganoderma weberianum B–18. Journal of Applied Sciences in Environmental Sanitation 8: 237–242. [Google Scholar]
- Matheny PB. (2005) Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe, Agaricales). Molecular Phylogenetics and Evolution 35: 1–20. 10.1016/j.ympev.2004.11.014 [DOI] [PubMed] [Google Scholar]
- Matheny PB, Wang Z, Binder M, Curtis JM, Lim YW, Nilsson RH, Hughes KW, Petersen RH, Hofstetter V, Ammirati JF, Schoch C, Langer GE, Mclaughlin DJ, Wilson AW, Crane PE, Frøslev T, Ge ZW, Kerrigan RW, Slot JC, Vellinga EC, Liang ZL, Aime MC, Baroni TJ, Fischer M, Hosaka K, Matsuura K, Seidl MT, Vaura J, Hibbett DS. (2007) Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution 43: 430–451. 10.1016/j.ympev.2006.08.024 [DOI] [PubMed] [Google Scholar]
- Miller MA, Pfeiffer W, Schwartz T. (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Proceedings of the Gateway Computing Environments Workshop (GCE), New Orleans, pp. 1–8. 10.1109/GCE.2010.5676129 [DOI]
- Mohanty PS, Harsh NSK, Pandey A. (2011) First report of Ganoderma resinaceum and G. weberianum from north India based on ITS sequence analysis and micromorphology. Mycosphere 2: 469–474. [Google Scholar]
- Moncalvo JM. (2000) Systematics of Ganoderma. In: Flood P, Bridge D, Holderness M. (Eds) Ganoderma diseases of perennial crops.CABI, Wallingford, 23–45. 10.1079/9780851993881.0023 [DOI]
- Moncalvo JM, Ryvarden L. (1997) A nomenclatural study of the Ganodermataceae Donk. Synopsis Fungorum 11: 1–114. [Google Scholar]
- Moncalvo JM, Wang HH, Hseu RS. (1995) Phylogenetic relationships in Ganoderma inferred from the internal transcribed spacers and 25S ribosomal DNA sequences. Mycologia 87: 223–238. 10.1080/00275514.1995.12026524 [DOI] [Google Scholar]
- Müller J, Müller K, Neinhuis C, Quandt D. (2010) PhyDe® Phylogenetic Data Editor. http://www.phyde.de
- Murrill WA. (1902) The Polyporaceae of North America, part I. The genus Ganoderma. Bulletin of the Torrey Botanical Club 29: 599–608. 10.2307/2478682 [DOI] [Google Scholar]
- Murrill WA. (1903) The Polyporaceae of North America. III. The genus Fomes. Bulletin of the Torrey Botanical Club 30: 225–232. 10.2307/2478780 [DOI] [Google Scholar]
- Murrill WA. (1908) Additional Philippine Polyporaceae. Bulletin of the Torrey Botanical Club 35: 391–416. 10.2307/2479285 [DOI] [Google Scholar]
- Murrill WA. (1912) Polyporaceae of Mexico. Bulletin of New York Botanical Garden 8: 137–153. [Google Scholar]
- Palomera V, Castro-Félix P, Villalobos-Arámbula AR. (2008) High yield of high quality DNA from vegetative and sexual tissues of Mexican white pine (Pinus ayacahuite). African Journal of Biotechnology 7: 51–54. [Google Scholar]
- Pan HY, Dai YC. (2001) Ganoderma weberianum newly recorded from mainland of China. Fungal Science 16: 31–34. [Google Scholar]
- Park YJ1, Kwon OC, Son ES, Yoon DE, Han W, Yoo YB, Lee CS. (2012) Taxonomy of Ganoderma lucidum from Korea Based on rDNA and Partial β-Tubulin Gene Sequence Analysis. Mycobiology 40(1): 71–5. 10.5941/MYCO.2012.40.1.071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patouillard N. (1898) Quelques champignons nouveaux récoltés au Mexique par Paul Maury. Bulletin de la Société mycologique de France 14: 54.
- Patouillard N, Hariot P. (1906) Fungorum novorum. Decas secunda. Bulletin de la Société mycologique de France 22: 118–119. [Google Scholar]
- Pegler DN, Young TWK. (1973) Basidiospore form in the British species of Ganoderma Karst. Kew Bulletin 28: 351–364. 10.2307/4108879 [DOI] [Google Scholar]
- Quanten E. (1997) The polypores (Polyporaceae s.l.) of Papua New Guinea. Opera Botanica Belgica 11: 1–352. [Google Scholar]
- Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. (2018) Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Systematic Biology. 10.1093/sysbio/syy032 [DOI] [PMC free article] [PubMed]
- Raymundo T, Valenzuela R, Gutiérrez A, Coronado ML, Esqueda M. (2013) Agaricomycetes xilófagos de la planicie central del desierto Sonorense. Revista Mexicana de Biodiversidad 84: 417–424. 10.7550/rmb.30828 [DOI] [Google Scholar]
- Rehner SA, Buckley E. (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1–a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97: 84–98. 10.3852/mycologia.97.1.84 [DOI] [PubMed] [Google Scholar]
- Richter C, Wittstein K, Kirk P, Stadler M. (2015) An assessment of the taxonomy and chemotaxonomy of Ganoderma. Fungal Diversity 71: 1–15. 10.1007/s13225-014-0313-6 [DOI] [Google Scholar]
- Robert EC. (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research 32: 1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. 10.1093/bioinformatics/btg180 [DOI] [PubMed] [Google Scholar]
- Ryvarden L. (1972) A critical checklist of the Polyporaceae in tropical East Africa. Norwegian Journal of Botany 19: 229–238. [Google Scholar]
- Ryvarden L. (1985) Type studies in the Polyporaceae. 17. Species described by W.A. Murrill. Mycotaxon 23: 169–198. [Google Scholar]
- Ryvarden L. (1990) Type studies in the Polyporaceae. 22. Species described by C.G. Lloyd in Polyporus. Mycotaxon 38: 83–102. [Google Scholar]
- Ryvarden L. (1991) Genera of polypores. Nomenclature and taxonomy. Synopsis Fungorum 5: 1–363. [Google Scholar]
- Ryvarden L. (2000) Studies in Neotropical polypores. 2. A preliminary key to Neotropical species of Ganoderma with a laccate pileus. Mycologia 92: 180–191. 10.1080/00275514.2000.12061142 [DOI] [Google Scholar]
- Ryvarden L. (2004) Neotropical polypores. Part 1. Introduction, Ganodermataceae and Hymenochaetaceae. Synopsis Fungorum 19: 1–229. [Google Scholar]
- Saccardo PA. (1891) Sylloge Fungorum IX. – [Hymenomyceteae, Polyporeae, Fomes]. Padua, 174.
- Smith BJ, Sivasithamparam K. (2000) Internal transcribed spacer ribosomal DNA sequence of five species of Ganoderma from Australia. Mycological Research 104: 943–951. 10.1017/S0953756200002458 [DOI] [Google Scholar]
- Smith BJ, Sivasithamparam K. (2003) Morphological studies of Ganoderma (Ganodermataceae) from the Australasian and Pacific regions. Australian Systematic Botany 16: 487–503. 10.1071/SB02001 [DOI] [Google Scholar]
- Stamatakis A. (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Steyaert RL. (1962) Genus Ganoderma (Polyporaceae). Taxa nova 2. Bulletin du Jardin Botanique de l’État à Bruxelles 32: 89–104. 10.2307/3667315 [DOI] [Google Scholar]
- Steyaert RL. (1972) Species of Ganoderma and related genera mainly of the Bogor and Leiden Herbaria. Persoonia 7: 55–118. [Google Scholar]
- Steyaert RL. (1980) Study of some Ganoderma species. Bulletin du Jardin Botanique National de Belgique 50: 135–186. 10.2307/3667780 [DOI] [Google Scholar]
- Thiers B. (continuously updated) Index Herbariorum: a global directory of public herbaria and associated staff. New York Botanical Garden’s Virtual Herbarium. http://sweetgum.nybg.org/science/ih/. Accessed 2018.
- Torres-Farradá G, Manzano León AM, Rineau F, Ledo Alonso LL, Sánchez-López MI, Thijs S, Colpaert J, Ramos-Leal M, Guerra G, Vangronsveld J. (2017) Diversity of ligninolytic enzymes and their genes in strains of the genus Ganoderma: Applicable for biodegradation of xenobiotic compounds. Frontiers in Microbiology 8: 898. 10.3389/fmicb.2017.00898 [DOI] [PMC free article] [PubMed]
- Torres-Torres MG, Guzmán-Dávalos L. (2012) The morphology of Ganoderma species with a laccate surface. Mycotaxon 119: 201–216. 10.5248/119.201 [DOI] [Google Scholar]
- Torres-Torres MG, Guzmán-Dávalos L, Guggliota AM. (2008) Ganoderma vivianimercedianum sp. nov. and the related species, G. perzonatum. Mycotaxon 105: 447–454. [Google Scholar]
- Torres-Torres MG, Guzmán-Dávalos L, Guggliota AM. (2012) Ganoderma in Brazil: known species and new records. Mycotaxon 121: 93–132. 10.5248/121.93 [DOI] [Google Scholar]
- Torres-Torres MG, Guzmán-Dávalos L, Ryvarden L. (2015) Ganoderma subgénero Ganoderma en México. Revista Mexicana de Micología 41: 27–45. [Google Scholar]
- Wagner T, Ryvarden L. (2002) Phylogeny and taxonomy of the genus Phylloporia (Hymenochaetales). Mycological Progress 1: 105–116. 10.1007/s11557-006-0009-8 [DOI] [Google Scholar]
- Wang DM, Wu SH, Su CH, Peng JT, Shih YH. (2009) Ganoderma multipileum, the correct name for “G. lucidum” in tropical Asia. Botanical Studies 50: 451–458. [Google Scholar]
- Wang DM, Zhang XQ, Yao YJ. (2005) Type studies of some Ganoderma species from China. Mycotaxon 93: 61–70. [Google Scholar]
- Wang XC, Xi RJ, Li Y, Wang DM, Yao YJ. (2012) The species identity of the widely cultivated Ganoderma, ‘G. lucidum’ (Ling-zhi), in China. PLoS ONE 7(7): e40857. 10.1371/journal.pone.0040857 [DOI] [PMC free article] [PubMed]
- Welti S, Courtecuisse R. (2010) The Ganodermataceae in the French West Indies (Guadeloupe and Martinique). Fungal Diversity 43: 103–126. 10.1007/s13225-010-0036-2 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, New York, 315–322. 10.1016/B978-0-12-372180-8.50042-1 [DOI]
- Xing JH, Sun YF, Han YL, Cui BK, Dai YC. (2018) Morphological and molecular identification of two new Ganoderma species on Casuarina equisetifolia from China. MycoKeys 34: 93–108. 10.3897/mycokeys.34.22593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao YJ, Wang XC, Ang B. (2013) Epitypification of Ganoderma sichuanense J.D. Zhao & X.Q. Zhang (Ganodermataceae). Taxon 62: 1025–1031. 10.12705/625.10 [DOI] [Google Scholar]
- Yombiyeni P, Decock C. (2017) Hymenochaetaceae (Hymenochaetales) from the Guineo-Congolian phytochorion: Phylloporia littoralis sp. nov. from coastal vegetation in Gabon, with an identification key to the local species. Plant Ecology and Evolution 150: 160–172. 10.5091/plecevo.2017.1289 [DOI] [Google Scholar]
- Zhou LW, Cao Y, Wu SH, Vlasák J, Li DW, Li MJ, Dai YC. (2014) Global diversity of the Ganoderma lucidum complex (Ganodermataceae, Polyporales) inferred from morphology and multilocus phylogeny. Phytochemistry 114: 7–15. 10.1016/j.phytochem.2014.09.023 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.