Ethanol is a common product of microbial fermentation, and the Pseudomonas aeruginosa response to and utilization of ethanol are relevant to our understanding of its role in microbial communities. Here, we report that the putative alcohol dehydrogenase AdhA is responsible for ethanol catabolism and acetate accumulation under low-oxygen conditions and that it is regulated by Anr.
KEYWORDS: AdhA, ExaA, Pseudomonas aeruginosa, ethanol, lasR
ABSTRACT
Pseudomonas aeruginosa has a broad metabolic repertoire that facilitates its coexistence with different microbes. Many microbes secrete products that P. aeruginosa can then catabolize, including ethanol, a common fermentation product. Here, we show that under oxygen-limiting conditions P. aeruginosa utilizes AdhA, an NAD-linked alcohol dehydrogenase, as a previously undescribed means for ethanol catabolism. In a rich medium containing ethanol, AdhA, but not the previously described PQQ-linked alcohol dehydrogenase, ExaA, oxidizes ethanol and leads to the accumulation of acetate in culture supernatants. AdhA-dependent acetate accumulation and the accompanying decrease in pH promote P. aeruginosa survival in LB-grown stationary-phase cultures. The transcription of adhA is elevated by hypoxia and under anoxic conditions, and we show that it is regulated by the Anr transcription factor. We have shown that lasR mutants, which lack an important quorum sensing regulator, have higher levels of Anr-regulated transcripts under low-oxygen conditions than their wild-type counterparts. Here, we show that a lasR mutant, when grown with ethanol, has an even larger decrease in pH than the wild type (WT) that is dependent on both anr and adhA. The large increase in AdhA activity is similar to that of a strain expressing a hyperactive Anr-D149A variant. Ethanol catabolism in P. aeruginosa by AdhA supports growth on ethanol as a sole carbon source and electron donor in oxygen-limited settings and in cells growing by denitrification under anoxic conditions. This is the first demonstration of a physiological role for AdhA in ethanol oxidation in P. aeruginosa.
IMPORTANCE Ethanol is a common product of microbial fermentation, and the Pseudomonas aeruginosa response to and utilization of ethanol are relevant to our understanding of its role in microbial communities. Here, we report that the putative alcohol dehydrogenase AdhA is responsible for ethanol catabolism and acetate accumulation under low-oxygen conditions and that it is regulated by Anr.
INTRODUCTION
Pseudomonas aeruginosa has broad catabolic potential that contributes to its ability to thrive in diverse environments that often include other microbes. One environment in which P. aeruginosa metabolism has been well studied is in the mucus that accumulates during chronic infections in the lungs of individuals with cystic fibrosis. In this low-oxygen setting, P. aeruginosa encounters other pathogens, many of which are known to produce ethanol by fermentation (1). Ethanol has been detected in the volatiles expired from the lungs of cystic fibrosis (CF) patients infected with P. aeruginosa (2, 3), and several previous studies have shown that the addition of ethanol to the growth medium can alter P. aeruginosa metabolism, motility, and virulence factor production (4–7).
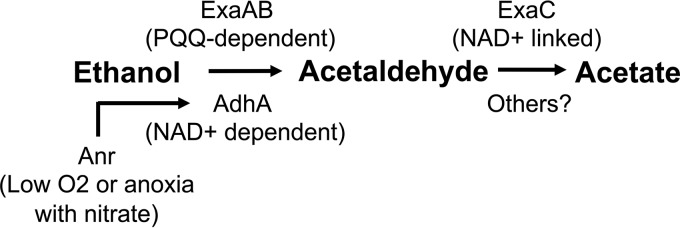
Ethanol metabolism in P. aeruginosa has mainly been studied with ethanol supplied as a sole carbon source under aerobic conditions, and that focus has been on the pyrroloquinoline quinone (PQQ)-dependent ethanol-oxidizing enzyme ExaA, which donates electrons directly to the cytochrome 550 protein, ExaB (8–11) (Fig. 1). Görisch and colleagues (8–10) described aerobic ethanol catabolism in detail, including a complex regulatory cascade initiated by ErbR/AgmR. The disruption of exaA, exaB, or genes involved in PQQ biosynthesis eliminates the ability to grow aerobically on ethanol as a sole carbon source on agar medium (11).
FIG 1.
Ethanol catabolism in P. aeruginosa. ExaA is a PQQ-linked enzyme that, with a soluble cytochrome c (ExaB), catalyzes the oxidation of ethanol to acetaldehyde. ExaC then oxidizes acetaldehyde to acetate. In this work, we show that AdhA is regulated by Anr in response to oxygen limitation and that AdhA can also catalyze ethanol oxidation as a growth substrate or for conversion to acetate.
The P. aeruginosa genome encodes putative alcohol dehydrogenases other than ExaA, but the roles of these others in the oxidation of ethanol have not been clearly established. The protein encoded by P. aeruginosa PAO1 gene PA5427 was named AdhA based on the high structural similarity of its crystal structure to those of diverse characterized alcohol dehydrogenase enzymes that use NAD+ as a cofactor (12). AdhA contains a catalytic domain (amino acids 1 to 157 and 292 to 342) with a Zn2+ active site and a coenzyme-binding domain (residues 158 to 291) with a Rossmann fold motif that is characteristic of NAD+ coenzyme-binding proteins (12, 13). The biological activity of this enzyme as an ethanol dehydrogenase in P. aeruginosa, however, was not described.
In P. aeruginosa, adhA transcripts are markedly elevated during hypoxic growth and under anoxic conditions in the presence of nitrate (14–17). Furthermore, adhA expression was greatly reduced upon deletion of anr, which encodes the oxygen-responsive transcription factor that controls the expression of genes involved in anaerobic and microoxic metabolism (14, 15). Eschbach and colleagues (18) examined P. aeruginosa AdhA for the potential to participate in the conversion of pyruvate to produce ethanol when oxygen is limiting. Although lactic acid and acetic acid were detected, no evidence for ethanol formation was found.
In this work, we show that in rich medium amended with ethanol, ethanol was oxidized with the accumulation of acetate under oxygen-limiting conditions and this was not dependent on ExaA but did require AdhA (see Fig. 1 for pathways). We also found that acetate accumulation and the concomitant reduction in pH increased survival during stationary phase. The ∆adhA mutant was also incapable of growth with ethanol as the sole carbon source and electron donor under microoxic or anoxic conditions with nitrate as an electron acceptor. We provide evidence that the transcription of adhA is regulated by Anr both in LB medium and during growth on ethanol as a sole carbon source in low-oxygen or anoxic environments. Together, these data suggest that AdhA is capable of ethanol oxidation for growth and for conversion to acetate and that it can work in conjunction with ExaAB to enable P. aeruginosa to catabolize ethanol in diverse environments.
RESULTS
AdhA is responsible for accumulation of acetate from ethanol in LB.
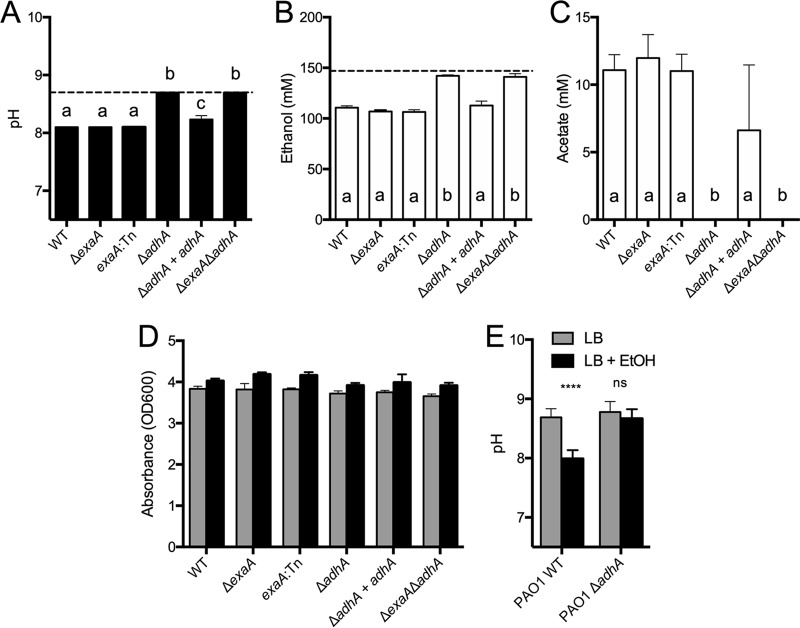
We previously reported that P. aeruginosa strain PA14 grown in LB medium with 1% ethanol yields a lower final pH in stationary-phase culture supernatants than cultures grown in LB alone (4). Due to the production of ammonia from amino acid catabolism, the culture pH from early stationary-phase P. aeruginosa strain PA14 cultures (12 h, optical density [OD] of ∼4) was 1.5 pH units higher than that in uninoculated medium (pH 8.7 versus pH 7.2 for uninoculated medium) (Fig. 2A, dashed line). When ethanol was present in the medium, the final culture pH was lower than when cultures were grown in LB alone (pH 8.1 versus pH 8.7) (Fig. 2A). High-pressure liquid chromatography (HPLC) analysis of P. aeruginosa wild-type (WT) culture supernatants at this time point (12 h) revealed that 36.3 mM ethanol had been consumed (Fig. 2B) and 11.1 mM acetate had accumulated in the medium (Fig. 2C). No acetate was detected in LB cultures without added ethanol, and no lactate, pyruvate, or succinate was detected in either the LB or LB plus ethanol culture supernatants by the HPLC method used (data not shown). The final ODs of cultures grown in LB plus 1% ethanol were similar for cultures grown with and without ethanol. All strains yielded a slightly higher OD at 600 nm (OD600) value in the presence of ethanol, but the difference did not reach statistical significance for the wild type (Fig. 2D).
FIG 2.
Ethanol catabolism by P. aeruginosa leads to lower medium pH through the accumulation of acetate. P. aeruginosa PA14 strains, including wild-type, ΔexaA, exaA::TnM, ΔadhA, ΔadhA adhA, and ΔexaA ΔadhA strains, were grown in LB for 12 h in tubes on a roller drum. (A) pH values of cultures, measured using pH indicator strips, in LB (dotted line) and in LB plus ethanol (bars). (B) Ethanol remaining in culture supernatants relative to that in uninoculated LB plus 1% ethanol (dotted line). (C) Acetate concentrations in supernatants in LB plus 1% ethanol in the same cultures analyzed in panels A and B. (D) Optical density of strains grown in LB (gray) and LB plus 1% ethanol (black). (E) Final culture pH values of P. aeruginosa PAO1 wild-type and ΔadhA strains after 16 h of growth in LB or in LB with 1% ethanol (EtOH). All cultures were grown at 37°C. In panels A to C, samples with different lowercase letters are significantly different, P < 0.05. In panel D, values for ethanol-grown cultures were slightly but significantly higher (P < 0.05) for each strain except WT and the ΔadhA mutant. ****, P < 0.001; ns, not significant.
To determine if ethanol catabolism in LB was due to activity of the characterized ethanol dehydrogenase ExaAB, we examined ethanol catabolism in a mutant with an in-frame deletion of exaA and a validated exaA::TnM insertion mutant (5, 19), both of which were generated in the strain PA14 background. Both mutants acidified the culture medium, consumed ethanol, and accumulated acetate in cultures with ethanol present, suggesting that ExaA was not required and that another ethanol catabolic enzyme was present (Fig. 2A to C). In light of this result, we examined whether AdhA, a putative NAD+-linked alcohol dehydrogenase encoded by PA14_71630 (PA5427 in strain PAO1) contributed to ethanol catabolism and acetate accumulation. We constructed and analyzed an in-frame mutant lacking adhA and found that it did not have a lower pH when grown in LB plus 1% ethanol than when grown in LB alone (Fig. 2A). The ΔadhA mutant showed only a minor decrease in the ethanol concentration of the medium (4.7 mM), and no acetate was detected (Fig. 2B and C). The defect in the adhA mutant was complemented by replacement of the adhA gene at the native locus (Fig. 2A to C), while a ΔadhA ΔexaA double mutant phenocopied the ΔadhA mutant. Together, these data indicate that ethanol was required for the accumulation of acetate in P. aeruginosa cultures grown in LB and that this phenomenon was dependent on adhA, but not exaA, in P. aeruginosa strain PA14. A similar requirement for adhA for acidification of LB medium in cultures with ethanol was observed in P. aeruginosa strain PAO1, a genetically distinct laboratory strain (Fig. 2E).
Consistent with our observation that AdhA activity was responsible for the lower culture pH in LB with ethanol at 12 h (Fig. 2), culture pH in LB with ethanol at 24 h showed even greater differences between the wild type and the adhA or adhA exaA mutants (pH 6.5 for wild type and 8.7 for mutants).
Ethanol catabolism through AdhA increases stationary-phase survival.
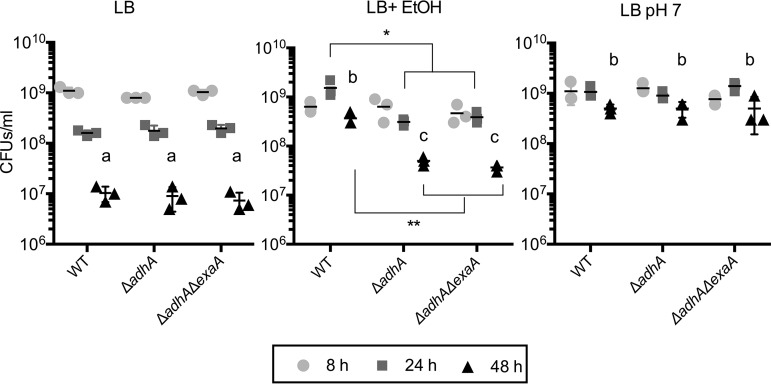
On the basis of our initial observations that cultures grown with ethanol remained turbid for longer in stationary phase, and in light of previous studies in Escherichia coli that show ethanol can promote stationary-phase survival in rich media such as LB (20), we tested the hypothesis that ethanol catabolism by AdhA promoted stationary-phase survival. In LB cultures with no ethanol, strain PA14 wild type, and ΔadhA and ΔadhA ΔexaA mutants all showed a significant decrease in viability over time (8 h, 24 h, and 48 h), with no significant differences between strains (Fig. 3, LB). In LB with 1% ethanol, the wild-type cultures showed the absence of or a reduction in viability loss compared to that of LB cultures without ethanol, indicating that ethanol was protective. The ΔadhA and ΔadhA ΔexaA mutants did not receive the same benefit from the presence of ethanol in the medium (Fig. 3, LB+EtOH). To determine if ethanol was protective in late stationary phase because of its effects on culture pH or because of other factors, the experiment also included cultures grown in LB that was buffered to pH 7. We found that maintaining a pH of 7 was sufficient to mitigate the loss of viability in late stationary phase relative to that in unbuffered medium in the wild-type and all mutant strains (Fig. 3, LB pH 7 versus LB), similarly to what has been observed for E. coli (21). We also performed a kinetic analysis of the contribution of AdhA to P. aeruginosa culture pH and growth in LB in the absence and presence of ethanol by measuring culture density and culture pH over the course of 30 h for both the WT and the adhA deletion mutant (see Fig. S1 in the supplemental material). We observed a reduction in pH in the presence of added ethanol only in the WT beginning at ∼8 h postinoculation. The decrease in the high stationary-phase OD exhibited by the WT without ethanol and the ΔadhA mutant with and without ethanol was not observed until 16 h, by which time a decrease in pH by almost ∼1 pH unit was observed only in the WT with ethanol (Fig. S1A and B). Analysis of the culture densities and pH values at 12 h and 26 h revealed some small level of protection by ethanol even in the ΔadhA mutant in the absence of a pH change, suggesting that the effects of ethanol are multifactorial (Fig. S1C and D). Together, these data suggest that the ability of AdhA to convert ethanol to acetate in the late stationary phase of growth even in rich media can mitigate the toxic alkaline pH condition and its effects on cells.
FIG 3.
Ethanol and pH effects on P. aeruginosa stationary-phase survival in LB. The CFU values from 8-h, 24-h, and 48-h cultures were measured for PA14 wild-type (WT), ΔadhA, and ΔexaA ΔadhA strains in LB, LB with 1% ethanol (EtOH), and LB buffered to pH 7 in a single experiment shown in three panels for ease of comparison. *, P < 0.05; **, P < 0.01. Lowercase letters indicate significant differences between 48-h cultures across the three conditions tested; a or b, P < 0.05. Data shown are from a single experiment with three independent replicates at each time point. The experiment was repeated with similar results two additional times.
Anr regulates AdhA-dependent ethanol oxidation in wild-type and ΔlasR strains.
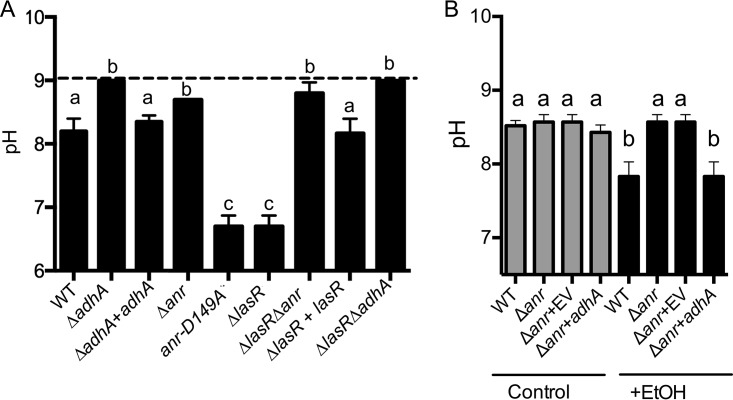
We and others showed previously that adhA transcript levels are as much as 40-fold lower in a Δanr mutant than in the wild type during growth under microoxic or anoxic conditions (14, 15). Thus, we speculated that Anr was regulating adhA in dense LB cultures when oxygen levels decreased due to cell respiration. Indeed, like the ΔadhA strain, the Δanr mutant culture had a higher pH after growth in LB plus 1% ethanol after 16 h than the wild type or the adhA-complemented strain, though the defect in acidification was not quite as striking as in the adhA strain (Fig. 4). We previously reported the creation of a strain in the PAO1 background in which native anr is replaced with an allele that encodes Anr-D149A, a variant that is active even in the presence of oxygen, and demonstrated that the activity is 2 to 4 times higher than that of native Anr in microoxic environments (22). For the present study, we constructed a PA14 strain in which anr was replaced with anr-D149A and saw greater acidification of LB plus 1% ethanol medium than with the otherwise isogenic wild-type PA14, indicating that increased Anr activity leads to even more ethanol catabolism and acetate accumulation in LB medium (Fig. 4A). To determine if AdhA was sufficient to restore ethanol conversion of acetate in the Δanr strain, we tested whether expression of adhA under the control of an arabinose-inducible promoter supported medium acidification in LB amended with ethanol. Indeed, this was the case. The Δanr mutant with adhA, but not with the empty vector, restored medium acidification in LB plus 1% ethanol (Fig. 4B).
FIG 4.
Culture pH after growth in LB plus 1% ethanol for 16 h. (A) The pH values for cultures of P. aeruginosa PA14 wild-type (WT), ΔadhA, ΔadhA adhA, Δanr, anr-D149A, ΔlasR, ΔlasR Δanr, ΔlasR ΔadhA, and ΔlasR lasR strains after growth in LB (dashed line) or LB with 1% ethanol (bars). Significance for comparisons of the LB plus ethanol cultures was performed using a one-way ANOVA and Tukey’s multiple-comparison test; samples with the same lowercase letter were not significantly different (P > 0.05). (B) The overexpression of adhA in the Δanr mutant was sufficient to restore medium acidification in the presence of ethanol. WT, Δanr, and Δanr strains with an empty vector (EV) or overexpressing adhA were grown without (Control) or with (+EtOH) ethanol for 16 h followed by analysis of the culture pH. The OD values of the strains bearing either vector were, on average, 1 OD600 unit lower than the WT or Δanr strains, but the EV and adhA-expressing culture densities were similar.
LasR is a global transcriptional regulator of the acyl-homoserine lactone quorum sensing system (23), and P. aeruginosa isolates with loss-of-function mutations in lasR are commonly recovered from both the clinic and in the environment (24–27). LasR loss-of-function mutants isolated from CF airways have been shown to confer growth advantages on certain carbon and nitrogen sources (25). Previous studies have shown that under microoxic conditions, Anr activity is higher in both engineered and naturally occurring lasR-defective strains than in their counterparts with functional LasR (14, 24). In the study by Hammond et al. (14), adhA was found to be 5-fold higher (P < 0.005) in lasR mutants than in their genetically similar counterparts with lasR intact, across four pairs of strains, including strains PA14 and PAO1. Thus, we tested whether ethanol-dependent acidification of the medium was greater in ΔlasR mutant cultures. Like the strain bearing the Anr-D149A variant, ΔlasR cultures showed greater acidification at 16 h than the wild type or the lasR-complemented strain (Fig. 4A). Deletion of either adhA or anr in the ΔlasR background eliminated the acidification phenotype (Fig. 4A). Together, these data strongly suggest that Anr induction of AdhA promotes ethanol oxidation to acetate, resulting in reduced medium pH, and that conditions or backgrounds that increase Anr activity will enhance the rate of ethanol conversion to acetate in LB.
Anr-regulated AdhA can support growth of P. aeruginosa strains PA14 and PAO1 on ethanol as a sole carbon source under low-oxygen and anoxic conditions with nitrate.
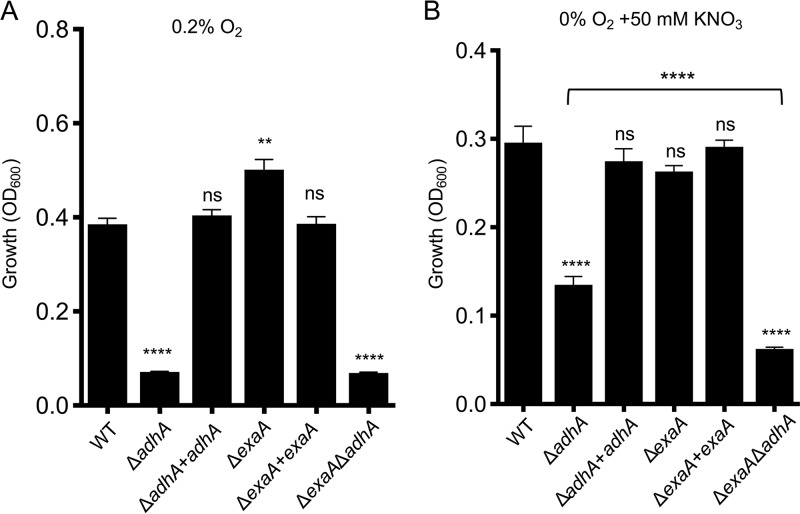
In light of adhA regulation by Anr (Fig. 4A) and its ability to oxidize ethanol to acetate in LB (Fig. 2B), we sought to determine if AdhA would be required to support growth on ethanol as a sole carbon source in low-oxygen environments. The ΔadhA mutant was grown under microoxic conditions (a 0.2% oxygen atmosphere) in M63 liquid medium with 1% ethanol as the sole carbon source. Under these low-oxygen conditions, the ΔadhA strain showed a marked defect in growth that was fully complemented by reconstitution of the mutant with the native adhA gene (Fig. 5A). The ΔexaA mutant did not have a defect in microoxic growth and in fact showed a slight significant increase in growth that was complemented by the reconstitution of the exaA gene at the native locus. The ΔexaA ΔadhA mutant was similar in growth to the adhA single mutant (Fig. 5A). All strains grew equally well in M63 medium with glucose as the sole carbon source in 0.2% oxygen, with an average OD600 of 0.71 ± 0.09 across strains. The low final OD600 of ∼0.07 in the adhA mutant strains was likely an inoculation remnant, since we added inoculum to an OD of 0.05 (Fig. 5 and 6).
FIG 5.
Growth of P. aeruginosa PA14 strains on ethanol as a sole carbon source. (A) Growth in M63 liquid medium with ethanol as the sole carbon source in 0.2% oxygen. (B) P. aeruginosa strains grown in M63 liquid medium with ethanol as a sole carbon source in anoxia with 50 mM KNO3. Data are representative of at least three independent experiments, each with three or more biological replicates. Statistics are based on one-way ANOVA and Tukey’s multiple-comparison test; **, P < 0.01; ****, P < 0.001; ns, not significant. Statistics presented indicate differences from the WT unless otherwise indicated.
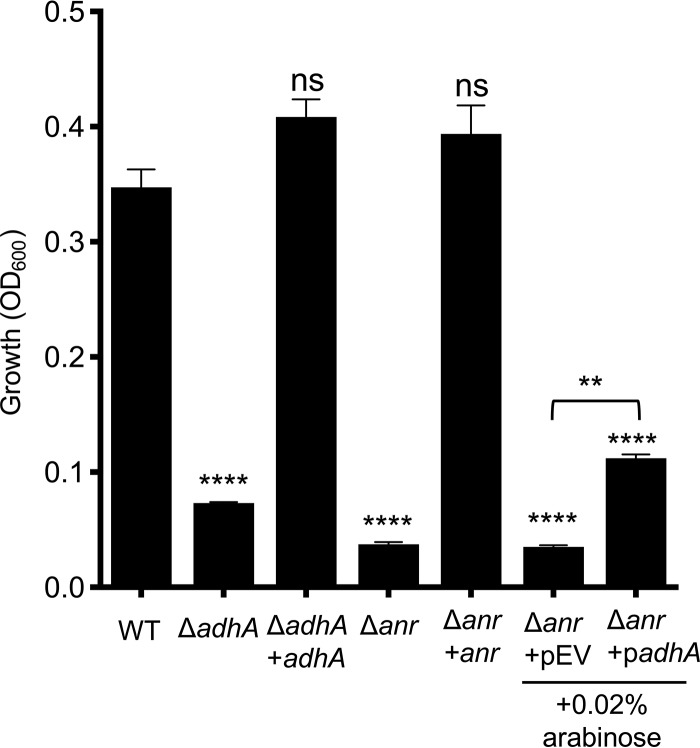
FIG 6.
Anr regulation of adhA expression. P. aeruginosa strains grown in M63 liquid medium with ethanol as the sole carbon source in a 0.2% oxygen atmosphere. The strains included wild-type strain PA14 (WT) and ΔadhA, ΔadhA adhA, Δanr, and Δanr anr mutants as well as a Δanr mutant containing an empty vector (pEV) or a plasmid with adhA under the control of an arabinose-inducible promoter (padhA). Statistics based on one-way ANOVA and Tukey’s multiple-comparison test; unless otherwise indicated, statistical comparisons are to the WT culture. ****, P < 0.0001; **, P < 0.01.
AdhA also supported ethanol utilization as the sole carbon source in P. aeruginosa when growing anoxically using nitrate respiration for energy generation (Fig. 5B). The wild type, the ΔadhA mutant, and the ΔadhA-complemented strain were grown in M63 liquid medium with 1% ethanol as the sole carbon source and 50 mM nitrate. This experiment showed that adhA significantly contributes to growth on ethanol under anoxic conditions. Although the exaA deletion mutant grew quite well, the slightly reduced growth of the ΔexaA strain relative to that of the wild type and the reduced growth of the ΔexaA ΔadhA double mutant relative to that of the ΔadhA single mutant suggested that exaA contributed to a small amount of the mostly AdhA-dependent ethanol utilization as the sole carbon source under anoxic denitrifying conditions (Fig. 5B). Further study would be needed to evaluate this interesting possibility. Under anoxic conditions in medium with glucose as the sole carbon source and with nitrate as the terminal electron acceptor, all strains grew similarly to the wild type, with an OD600 of 0.39 ± 0.05, except for the Δanr mutant which did not grow. Our finding that AdhA is needed for growth on ethanol as the sole carbon source under anoxic denitrifying conditions is consistent with observations that ethanol can be utilized as an electron donor under this same condition. Gliese et al. (28) discussed unpublished results showing that an NAD(P)-linked ethanol dehydrogenase was expressed under these denitrifying conditions, but no data or details about the exact nature of this ethanol dehydrogenase were reported in their paper.
Like the ΔadhA mutant, the Δanr mutant was unable to grow on ethanol as a sole carbon source at 0.2% oxygen, and that defect was complemented by restoring anr to the strain (Fig. 6). To determine if the defect in growth in the Δanr mutant on ethanol in low oxygen was due to the inability to induce adhA, the Δanr strain expressing adhA was again employed. In this background, overexpression of adhA was sufficient to partially rescue growth of a Δanr mutant relative to that of a Δanr strain bearing the empty vector (Fig. 6). The inability to completely restore growth to the Δanr mutant upon overexpression of adhA suggests that other Anr-regulated factors such as the high-affinity cytochrome c oxidases are required for fitness in low-oxygen environments during growth on ethanol.
To complement our analysis of the role of AdhA in ethanol catabolism in liquid M63 with ethanol as the sole carbon source under microoxic and anoxic conditions, we also analyzed the role of AdhA in growth on ethanol under normoxic culture conditions. Consistent with previously published results (5, 9, 10), exaA was necessary and sufficient for growth on ethanol on agar medium at 21% oxygen in both strains PA14 and PAO1 (see Fig. S2A). The adhA mutants in both PA14 and PAO1 showed no defect in aerobic growth on ethanol on an agar medium and the phenotype of the ΔadhA ΔexaA mutant was similar to that of the ΔexaA single mutant (Fig. S2A). When ethanol was replaced with 0.2% glucose as sole C source, all strains grew equally well on the M63 agar medium (Fig. S2A). However, in liquid medium (M63 with ethanol as the sole carbon source in 5-ml cultures in test tubes on a roller drum), both exaA and adhA were required for growth equal to that in the WT PA14 and WT PAO1 backgrounds (Fig. S2B and C, respectively). These data suggest that even under normoxic atmospheric conditions, metabolic activity in cells within liquid media in culture tubes created sufficient oxygen restriction to induce adhA and require its participation, along with exaA, to achieve maximal growth under these conditions of limited aeration. All strains grew equally well in M63 medium with glucose as the sole carbon source, with an average OD600 of 0.94 ± 0.2. Together, these data suggest that AdhA, under the control of Anr, can contribute to ethanol catabolism under diverse culture conditions and, in some cases, can work in parallel with the ExaAB ethanol catabolic complex.
DISCUSSION
Pseudomonas aeruginosa is remarkable in its ability to grow in diverse environments. Although, for years, only one pathway for ethanol oxidation and utilization in this organism has been known (10), it is unsurprising that P. aeruginosa possesses other enzymes that carry out the same metabolic process, regulated by entirely different signals. Through this work, we show that AdhA contributes to the catabolism of ethanol in a rich medium (LB plus 1% ethanol) and in a minimal medium with ethanol as the sole carbon source under oxygen-limiting conditions. In the absence of oxygen, ethanol also supported growth with the electron acceptor nitrate, a phenomenon also observed in the complex microbial communities of wastewater treatment systems (29). AdhA-dependent ethanol catabolism in LB was concomitant with acetate accumulation in culture supernatants, and the resulting acidification contributed to increased P. aeruginosa survival by counteracting the alkalinization that develops due to deamination of amino acids.
We suggest that AdhA-dependent accumulation of acetate from ethanol had not been previously observed in P. aeruginosa because previous studies largely investigated the aerobic oxidation and utilization of ethanol as a sole carbon source under oxic conditions. In the present study, we observed acetate accumulation when we added ethanol to LB, a rich medium, and incubated it with Pseudomonas in a test tube on a roller drum apparatus. We hypothesize that the rapid respiratory activity of cells after reaching late log phase had depleted much of the dissolved oxygen in the broth on the roller tube apparatus. This would be expected to increase the level of AdhA activity, which is markedly elevated by hypoxia as explained above. The obvious question that arises is why the acetate formed was not further metabolized under the LB medium conditions. We hypothesize that this is due to the presence of amino acids in the medium activating the previously characterized catabolite repression of acetate utilization in P. aeruginosa (9, 30).
It is interesting to consider why ethanol requires AdhA for catabolism under microoxic conditions. We and others found that microoxic growth of P. aeruginosa is sufficient to markedly elevate levels of transcripts that encode the NAD+-linked alcohol dehydrogenase adhA (PA5427) while not elevating the levels of transcripts associated with the PQQ-dependent ethanol oxidation pathway (14–17). This difference may explain why AdhA is active under these conditions but does not explain why the PQQ-dependent ExaA is not. We do not yet know if ExaA is expressed but not active or if it is not produced in cells grown under low oxygen. ExaA is localized to the periplasm and donates electrons to a soluble cytochrome c550, ExaB, which mediates electron transfer between ExaA and an unknown cytochrome c oxidase (10). One might speculate that this activity may not function under microoxic conditions. In contrast, AdhA is predicted to be cytosolic with only NAD+ as the electron acceptor. The reoxidation of NADH may be more favorable than the reoxidation of reduced factors in the ExaAB complex when oxygen is at very low levels or under denitrifying conditions.
Published findings suggest that an additional factor, catabolite repression, may repress elements of the Exa pathway, since the amino acids present in LB have been shown capable of exerting catabolite repression on several P. aeruginosa enzymatic activities (31). There is direct evidence for catabolite repression of some of the enzymes of the PQQ ethanol oxidation pathway (9, 28, 30). However, there is no evidence for the catabolite repression of AdhA, and there is indirect evidence suggesting that growth on amino acids does not cause repression of the marked elevation of adhA transcript levels induced by microoxic growth conditions (14, 16). Therefore, we suggest that future studies may find that many nutrient compounds exert catabolite repression on exaA and other genes encoding enzymes in the PQQ ethanol oxidation pathway but that the oxidation of ethanol to acetate by AdhA is not similarly subject to catabolite repression. Thus, in environments like the cystic fibrosis lung environment, where oxygen gradients are steep, nutrients are abundant, and ethanol-producing microbes are often present, Anr and AdhA may be particularly important factors to consider in determining if locally produced ethanol is catabolized by P. aeruginosa.
The oxygen-responsive transcription factor Anr can be active at high cell densities or in biofilms under normoxic conditions as well as under microoxic or anoxic conditions, and several published transcriptomics analyses of the Anr regulon and gene expression comparisons between oxic and microoxic or anoxic environments showed that Anr is a key regulator of adhA (14–16, 32). It is perhaps curious that an NAD+-linked ethanol oxidizing system is activated by Anr and hypoxia, since low-oxygen conditions limit the oxidation of NADH. We, like others (18), have considered the possibility that P. aeruginosa uses AdhA to ferment pyruvate to ethanol, regenerating NAD+, but we never observed ethanol in supernatants from LB-grown cultures, even when extracellular acetate was provided in the medium. We noted that the ΔadhA mutant, compared to the WT, overproduced phenazines in both the PA14 and PAO1 backgrounds (see Fig. S2A in the supplemental material). This observation may indicate an imbalance in the NAD+/NADH ratio upon deletion of adhA (given the role of phenazines in redox homeostasis) (33). Furthermore, as ethanol addition to strain PA14 causes overproduction of some phenazine pigments apparent on mature colony growth on agar plates (5, 6), we suggest that ethanol concentrations are higher in the strain lacking AdhA and that this leads to higher levels of pyocyanin and perhaps other phenazines than in the wild type.
The finding that P. aeruginosa accumulates significant amounts of acetic acid from added ethanol has not been reported previously. Acetate accumulations in other species such as an Acetobacter sp. and Gluconobacter spp., which are used for the commercial production of vinegar, generate large amounts of extracellular acetic acid from ethanol oxidation using an ExaAB-like system, including a PQQ-linked alcohol dehydrogenase (28, 34). A very early study reported that many Pseudomonas fluorescens strains accumulated acetic acid from ethanol in a peptone medium; but in that study, the P. aeruginosa strain did not, although it did use ethanol as a sole carbon source (35).
Previous studies in E. coli may help to understand our finding of the effect of ethanol to increase the survival of P. aeruginosa in LB. In E. coli, a similar viability loss in late stationary phase occurs, and this is prevented by ethanol addition to the LB. In these studies, the addition of ethanol to LB also caused an acidification and decrease in the highly alkaline pH that develops in LB. Further work led to the conclusion that in E. coli, ethanol protection against viability loss is due to more than just an indirect effect of ethanol on pH through the enzymatic conversion of ethanol to acetic acid (20, 36) and that very low concentrations of ethanol are protective. Ferraro and Finkel (37) suggest that very low concentrations of ethanol may also increase long-term viability in LB of some other genera, including Pseudomonas, but not the viability of all genera tested. Together, these data suggest that ethanol is at times an important resource as an inducer, a carbon source, a compound that can be used to modulate environmental pH, and an agent that increases long-term bacterial persistence.
MATERIALS AND METHODS
Strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table S1 in the supplemental material. Bacteria were maintained on 1.5% agar LB (lysogeny broth) plates (38). Where stated, ethanol (200 proof) was added to the medium (liquid or molten agar) to a final concentration of 1%. For LB medium buffered to pH 7, 100 mM HEPES acid was added to LB and the pH brought up to 7 by titration with NaOH. The exaA::TnM strain was maintained on LB with 60 μg/ml gentamicin (19).
Construction of in-frame deletions, complementation, and plasmids.
The construction of plasmids, including in-frame deletion and complementation constructs, was completed using yeast cloning techniques in Saccharomyces cerevisiae as previously described (39) unless otherwise stated. The primers used for plasmid construction are listed in Table S2. In-frame deletion and single-copy complementation constructs were made using the allelic replacement vector pMQ30 (39). Promoter fusion constructs were made using a modified pMQ30 vector with lacZ-GFP fusion integrating at the neutral att site on the chromosome. All plasmids were purified from yeast using Zymoprep Yeast Plasmid Miniprep II according to the manufacturer’s protocol and transformed into electrocompetent E. coli strain S17 by electroporation. Plasmids were introduced into P. aeruginosa by conjugation, and recombinants were obtained using sucrose counterselection and genotype screening by PCR. The primers are listed in Table S2. The exaA::TnM strain was confirmed with locus-specific primers as previously published (5).
Growth on ethanol as a sole carbon source.
For the assessment of growth on ethanol as a sole carbon source, M63 medium with 1% ethanol was used (40). Microoxic cultures were grown in 1 ml medium in a single well of a 12-well plastic culture plate in a chamber with a controlled atmosphere (Coy Products) at 37°C set to maintain a 0.2% O2 atmosphere. M63 minimal medium with either 0.2% glucose or 1% ethanol as the sole carbon source was used. Cultures were inoculated to an initial OD600 of 0.05 in fresh medium from overnight liquid cultures grown in LB. OD600 was measured 48 h postinoculation using a Genesys 6 spectrophotometer. Normoxic cultures grown in 5 ml of M63 minimal medium with either 0.2% glucose or 1% ethanol as the sole carbon source were inoculated at an initial OD600 of 0.05 from overnight liquid cultures grown in LB and incubated in 18-mm by 150-mm borosilicate culture tubes at 37°C on a roller drum. The OD600 was measured 24 h postinoculation using a Genesys 6 spectrophotometer. Anoxic cultures were grown in 2 ml of M63 minimal medium with either 0.2% glucose or 1% ethanol as the sole carbon source and supplemented with 50 mM KNO3. Cultures were inoculated at an initial OD600 of 0.05 into fresh medium from overnight liquid cultures grown in LB in 18-mm borosilicate culture tubes in 15-ml conical screw-cap tubes (Sarstedt) at 37°C on a roller drum. Tubes were placed in 2.5-liter AnaeroPack system rectangular jars (Mitsubishi Gas Chemical Co., Inc.) with one GasPak EZ sachet (BD). These cultures were grown for 5 days at 37°C prior to measuring OD600 using a Genesys 6 spectrophotometer.
Ethanol catabolism experiments in LB medium.
For experiments in which P. aeruginosa was grown in liquid LB, LB plus 1% ethanol, or LB buffered to pH 7, strains were first grown overnight in LB for 14 to 16 h, and ∼100 μl of the overnight culture was used to inoculate 5 ml of the test medium to an initial OD of 0.05. For pH measurements at the designated time points, culture supernatant was applied to Millipore pH indicator strips (pH 6.5 to 10.0); colors were compared to the reference and values were recorded immediately.
HPLC analysis of culture supernatants.
For HPLC, culture supernatants were collected after pelleting cells by centrifugation for 1 min at 13,000 rpm. Culture supernatant (400 μl) plus 40 μl of 10% sulfuric acid solution was added to Costar Spin-X 0.22-μm filter centrifuge tubes and spun for 5 min at 13,000 rpm. One hundred microliters of each sample was transferred to an HPLC autosampler vial (Chemglass CV-1042-1232), and one 40-μl injection of each sample was run for 30 min. Chemical peaks were detected by refractive index using an Aminex HPX-87H column (Bio-Rad, Hercules, CA) with a 2.5 mM sulfuric acid solution mobile phase. Chemical concentrations were determined using calibration curves based on three dilution standards of pyruvate, lactate, acetate, and ethanol.
Statistics.
Unless otherwise stated, data are based on three biological replicates with the means and standard deviations calculated. Data are representative of at least three independent experiments containing multiple replicates. Unless stated otherwise, means and standard deviations were calculated in GraphPad Prism 8, and analyses were completed using a one-way analysis of variance (ANOVA) and Tukey’s multiple-comparison test, with P values indicated in figure legends.
Supplementary Material
ACKNOWLEDGMENTS
Research reported in this publication was supported by National Institutes of Health (NIH) grant R01 GM108492 to D.A.H. and NIAID T32AI007519 to C.E.H. DNA Sequencing is supported in part by a Cancer Center Core Grant (P30CA023108) from the National Cancer Institute. Support for the project was also provided by the NIGMS P20GM113132 through the Molecular Interactions and Imaging Core (MIIC) and the CF RDP STANTO19R0.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
We thank Judy Jacobs for important insights, literature reviews, and comments on the manuscript. We also thank Daniel G. Olson for invaluable assistance with HPLC and Monique Porter for experimental support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00393-19.
REFERENCES
- 1.Phan J, Meinardi S, Barletta B, Blake DR, Whiteson K. 2017. Stable isotope profiles reveal active production of VOCs from human-associated microbes. J Breath Res 11:017101. doi: 10.1088/1752-7163/aa5833. [DOI] [PubMed] [Google Scholar]
- 2.Bos LD, Meinardi S, Blake D, Whiteson K. 2016. Bacteria in the airways of patients with cystic fibrosis are genetically capable of producing VOCs in breath. J Breath Res 10:047103. doi: 10.1088/1752-7163/10/4/047103. [DOI] [PubMed] [Google Scholar]
- 3.Montuschi P, Paris D, Melck D, Lucidi V, Ciabattoni G, Raia V, Calabrese C, Bush A, Barnes PJ, Motta A. 2012. NMR spectroscopy metabolomic profiling of exhaled breath condensate in patients with stable and unstable cystic fibrosis. Thorax 67:222–228. doi: 10.1136/thoraxjnl-2011-200072. [DOI] [PubMed] [Google Scholar]
- 4.Harty CE, Martins D, Doing G, Mould DL, Clay ME, Occhipinti P, Nguyen D, Hogan DA. 2019. Ethanol stimulates trehalose production through a SpoT-DksA-AlgU-dependent pathway in Pseudomonas aeruginosa. J Bacteriol 201:e00794-18. doi: 10.1128/JB.00794-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen AI, Dolben EF, Okegbe C, Harty CE, Golub Y, Thao S, Ha DG, Willger SD, O'Toole GA, Harwood CS, Dietrich LEP, Hogan DA. 2014. Candida albicans ethanol stimulates Pseudomonas aeruginosa WspR-controlled biofilm formation as part of a cyclic relationship involving phenazines. PLoS Pathog 10:e1004480. doi: 10.1371/journal.ppat.1004480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morales DK, Jacobs NJ, Rajamani S, Krishnamurthy M, Cubillos-Ruiz JR, Hogan DA. 2010. Antifungal mechanisms by which a novel Pseudomonas aeruginosa phenazine toxin kills Candida albicans in biofilms. Mol Microbiol 78:1379–1392. doi: 10.1111/j.1365-2958.2010.07414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lewis KA, Baker AE, Chen AI, Harty CE, Kuchma SL, O’Toole GA, Hogan DA. 2019. Ethanol decreases Pseudomonas aeruginosa flagellar motility through the regulation of flagellar stators. J Bacteriol 201:e00285-19. doi: 10.1128/JB.00285-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gliese N, Khodaverdi V, Schobert M, Gorisch H. 2004. AgmR controls transcription of a regulon with several operons essential for ethanol oxidation in Pseudomonas aeruginosa ATCC 17933. Microbiology 150:1851–1857. doi: 10.1099/mic.0.26882-0. [DOI] [PubMed] [Google Scholar]
- 9.Mern DS, Ha SW, Khodaverdi V, Gliese N, Gorisch H. 2010. A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: indication of four levels of sensor kinases and response regulators. Microbiology 156:1505–1516. doi: 10.1099/mic.0.032847-0. [DOI] [PubMed] [Google Scholar]
- 10.Gorisch H. 2003. The ethanol oxidation system and its regulation in Pseudomonas aeruginosa. Biochim Biophys Acta 1647:98–102. doi: 10.1016/s1570-9639(03)00066-9. [DOI] [PubMed] [Google Scholar]
- 11.Schobert M, Gorisch H. 1999. Cytochrome c550 is an essential component of the quinoprotein ethanol oxidation system in Pseudomonas aeruginosa: cloning and sequencing of the genes encoding cytochrome c550 and an adjacent acetaldehyde dehydrogenase. Microbiology 145:471–481. doi: 10.1099/13500872-145-2-471. [DOI] [PubMed] [Google Scholar]
- 12.Levin I, Meiri G, Peretz M, Burstein Y, Frolow F. 2004. The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol. Protein Sci 13:1547–1556. doi: 10.1110/ps.03531404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rossmann MG, Moras D, Olsen KW. 1974. Chemical and biological evolution of nucleotide-binding protein. Nature 250:194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- 14.Hammond JH, Dolben EF, Smith TJ, Bhuju S, Hogan DA. 2015. Links between Anr and quorum sensing in Pseudomonas aeruginosa biofilms. J Bacteriol 197:2810–2820. doi: 10.1128/JB.00182-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trunk K, Benkert B, Quack N, Munch R, Scheer M, Garbe J, Jansch L, Trost M, Wehland J, Buer J, Jahn M, Schobert M, Jahn D. 2010. Anaerobic adaptation in Pseudomonas aeruginosa: definition of the Anr and Dnr regulons. Environ Microbiol 12:1719–1733. doi: 10.1111/j.1462-2920.2010.02252.x. [DOI] [PubMed] [Google Scholar]
- 16.Alvarez-Ortega C, Harwood CS. 2007. Responses of Pseudomonas aeruginosa to low oxygen indicate that growth in the cystic fibrosis lung is by aerobic respiration. Mol Microbiol 65:153–165. doi: 10.1111/j.1365-2958.2007.05772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamath KS, Krisp C, Chick J, Pascovici D, Gygi SP, Molloy MP. 2017. Pseudomonas aeruginosa proteome under hypoxic stress conditions mimicking the cystic fibrosis lung. J Proteome Res 16:3917–3928. doi: 10.1021/acs.jproteome.7b00561. [DOI] [PubMed] [Google Scholar]
- 18.Eschbach M, Schreiber K, Trunk K, Buer J, Jahn D, Schobert M. 2004. Long-term anaerobic survival of the opportunistic pathogen Pseudomonas aeruginosa via pyruvate fermentation. J Bacteriol 186:4596–4604. doi: 10.1128/JB.186.14.4596-4604.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liberati NT, Urbach JM, Miyata S, Lee DG, Drenkard E, Wu G, Villanueva J, Wei T, Ausubel FM. 2006. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A 103:2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vulic M, Kolter R. 2002. Alcohol-induced delay of viability loss in stationary-phase cultures of Escherichia coli. J Bacteriol 184:2898–2905. doi: 10.1128/jb.184.11.2898-2905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Farrell MJ, Finkel SE. 2003. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol 185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jackson AA, Gross MJ, Daniels EF, Hampton TH, Hammond JH, Vallet-Gely I, Dove SL, Stanton BA, Hogan DA. 2013. Anr and its activation by PlcH activity in Pseudomonas aeruginosa host colonization and virulence. J Bacteriol 195:3093–3104. doi: 10.1128/JB.02169-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Whiteley M, Greenberg EP. 2001. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J Bacteriol 183:5529–5534. doi: 10.1128/JB.183.19.5529-5534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hammond JH, Hebert WP, Naimie A, Ray K, Van Gelder RD, DiGiandomenico A, Lalitha P, Srinivasan M, Acharya NR, Lietman T, Hogan DA, Zegans ME. 2016. Environmentally endemic Pseudomonas aeruginosa strains with mutations in lasR are associated with increased disease severity in corneal ulcers. mSphere 1:e00140-16. doi: 10.1128/mSphere.00140-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D’Argenio D, Wu M, Hoffman L, Kulasekara H, Déziel E, Smith E, Nguyen H, Ernst R, Larson Freeman T, Spencer D, Brittnacher M, Hayden H, Selgrade S, Klausen M, Goodlett D, Burns J, Ramsey B, Miller S. 2007. Growth phenotypes of Pseudomonas aeruginosa lasR mutants adapted to the airways of cystic fibrosis patients. Mol Microbiol 64:512–533. doi: 10.1111/j.1365-2958.2007.05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoffman LR, Kulasekara HD, Emerson J, Houston LS, Burns JL, Ramsey BW, Miller SI. 2009. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros 8:66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith EE, Buckley DG, Wu Z, Saenphimmachak C, Hoffman LR, D'Argenio DA, Miller SI, Ramsey BW, Speert DP, Moskowitz SM, Burns JL, Kaul R, Olson MV. 2006. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A 103:8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gliese N, Khodaverdi V, Gorisch H. 2010. The PQQ biosynthetic operons and their transcriptional regulation in Pseudomonas aeruginosa. Arch Microbiol 192:1–14. doi: 10.1007/s00203-009-0523-6. [DOI] [PubMed] [Google Scholar]
- 29.Shen Z, Zhou Y, Wang J. 2013. Comparison of denitrification performance and microbial diversity using starch/polylactic acid blends and ethanol as electron donor for nitrate removal. Bioresour Technol 131:33–39. doi: 10.1016/j.biortech.2012.12.169. [DOI] [PubMed] [Google Scholar]
- 30.Kretzschmar U, Khodaverdi V, Adrian L. 2010. Transcriptional regulation of the acetyl-CoA synthetase gene acsA in Pseudomonas aeruginosa. Arch Microbiol 192:685–690. doi: 10.1007/s00203-010-0593-5. [DOI] [PubMed] [Google Scholar]
- 31.La Rosa R, Behrends V, Williams HD, Bundy JG, Rojo F. 2016. Influence of the Crc regulator on the hierarchical use of carbon sources from a complete medium in Pseudomonas. Environ Microbiol 18:807–818. doi: 10.1111/1462-2920.13126. [DOI] [PubMed] [Google Scholar]
- 32.Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J, Hwang SH, Lau GW, Limbach PA, Rowe JJ, Lieberman MA, Barraud N, Webb J, Kjelleberg S, Hunt DF, Hassett DJ. 2008. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J Bacteriol 190:2739–2758. doi: 10.1128/JB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Glasser NR, Kern SE, Newman DK. 2014. Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Mol Microbiol 92:399–412. doi: 10.1111/mmi.12566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toyama H, Mathews FS, Adachi O, Matsushita K. 2004. Quinohemoprotein alcohol dehydrogenases: structure, function, and physiology. Arch Biochem Biophys 428:10–21. doi: 10.1016/j.abb.2004.03.037. [DOI] [PubMed] [Google Scholar]
- 35.Stanier RY. 1947. Oxidation of ethanol to acetic acid by fluorescent Pseudomonads. J Bacteriol 54:191–194. [PubMed] [Google Scholar]
- 36.Gonidakis S, Finkel SE, Longo VD. 2010. E. coli hypoxia-inducible factor ArcA mediates lifespan extension in a lipoic acid synthase mutant by suppressing acetyl-CoA synthetase. Biol Chem 391:1139–1147. doi: 10.1515/BC.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ferraro CM, Finkel SE. 2019. Physiological, genetic, and transcriptomic analysis of alcohol-induced delay of Escherichia coli death. Appl Environ Microbiol 85:e02113-18. doi: 10.1128/AEM.02113-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shanks RMQ, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. 2006. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol 72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neidhardt FC, Bloch PL, Smith DF. 1974. Culture medium for enterobacteria. J Bacteriol 119:736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.